Marina V. Rodnina is the winner of the 2015 Hans Neurath Award.Abstract
The cellular proteome is shaped by the combined activities of the gene expression and quality control machineries. While transcription plays an undoubtedly important role, in recent years also translation emerged as a key step that defines the composition and quality of the proteome and the functional activity of proteins in the cell. Among the different post‐transcriptional control mechanisms, translation initiation and elongation provide multiple checkpoints that can affect translational efficiency. A multitude of specific signals in mRNAs can determine the frequency of translation initiation, choice of the open reading frame, global and local elongation velocities, and the folding of the emerging protein. In addition to specific signatures in the mRNAs, also variations in the global pools of translation components, including ribosomes, tRNAs, mRNAs, and translation factors can alter translational efficiencies. The cellular outcomes of phenomena such as mRNA codon bias are sometimes difficult to understand due to the staggering complexity of covariates that affect codon usage, translation, and protein folding. Here we summarize the experimental evidence on how the ribosome—together with the other components of the translational machinery—can alter translational efficiencies of mRNA at the initiation and elongation stages and how translation velocity affects protein folding. We seek to explain these findings in the context of mechanistic work on the ribosome. The results argue in favour of a new understanding of translation control as a hub that links mRNA homeostasis to production and quality control of proteins in the cell.
Keywords: ribosome, tRNA, protein synthesis and folding, mRNA rare codons, translational pausing
Introduction
Protein synthesis is a fundamental step in gene expression, which largely determines the composition and quality of the cellular proteome. Transcription efficiency and mRNA stability shape the pool of cellular mRNAs. Translation of mRNAs into proteins is carried out by the ribosome with the help of translation factors, using aminoacyl‐tRNAs (aa‐tRNA) and GTP as substrates. The interplay between mRNA, ribosomes, translation factors, and aa‐tRNA pools defines how often a given mRNA is translated and ensures that proteins are produced in required amounts and assume their functionally active folds. The amount and quality of protein production are controlled at essentially all phases of translation, which includes translation initiation, elongation, termination, and ribosome recycling. Proteins emerging from the ribosome start to fold co‐translationally, while still attached to the ribosome. In many cases their correct folding requires the help of chaperones. The quality control machinery of the cell then controls the outcome of the protein production.
Translation initiation is a step at which the ribosome selects an mRNA and finds the start of the open reading frame (ORF) for translation. It is usually assumed that initiation is slow compared to elongation, although the rate of initiation may vary considerably for different mRNAs. Estimations for the duration of initiation range from 10–60 s in yeast1, 2 to 0.3–250 s in Escherichia coli.3, 4 Initiation is tightly regulated in eukaryotes by a variety of mechanisms, ranging from the control of mRNA recruitment to the modulation of the activity of initiation factors.5, 6, 7, 8 In bacteria the efficiency of initiation can be regulated as well, for example by protein‐, RNA‐, metabolite‐, and temperature‐regulated secondary structures of the mRNA that control access to the ribosome binding site (RBS) at the beginning of the ORF.9, 10 During translation elongation, the ribosome translates the ORF by selecting aa‐tRNAs corresponding to the sequence of the codons in the mRNA and synthesizes polypeptides in the peptidyl transferase center (PTC). The average rate of a single elongation cycle is between 1 and 40 amino acids/s. Thus, for an average protein in E. coli (ca., 300 amino acids), the synthesis time is about 20 s. However, the process of elongation is not uniform, with periods of rapid synthesis interrupted by pauses. Global and local translation velocities affect the amount and quality of proteins synthesized; however, the origin of translation pauses and the rules that define the local speed and accuracy of translation in different regions of a given mRNA are not fully understood. Finally, translation termination and ribosome recycling, which both are slow relative to the cycles of translation elongation, release the newly synthesized polypeptide and disassemble the ribosome into the small and large subunits, which are fed into the free ribosome pool for another round of initiation.
Recent advances in quantitative methods to study translation on a global level in vivo, combined with bioinformatic approaches and biochemical, biophysical, and genetic studies on the mechanisms of translation in vitro, suggest that translation is an important checkpoint for protein production in the cell and provide new insights into mechanisms of translation regulation. In this review, we discuss how the ribosome together with other components of translational machinery may alter translational efficiencies of mRNAs at the initiation and elongation steps and how translation velocity affects protein folding. We focus on recent findings; in‐depth overviews of the earlier literature can be found in previous reviews.11, 12, 13, 14, 15, 16
Translational Efficiency
The steady‐state levels of cellular proteins are controlled at multiple stages of gene expression, including transcription, mRNA processing and turnover, mRNA translation, and protein turnover (Fig. 1). In E. coli, copy numbers of individual proteins range from 10−1 to 104 per cell (i.e., from one protein molecules for 10 cells to 10,000 copies per cell), compared to mRNA copy numbers which range from 10−3 to 101 per cell.17 In baker's yeast, cells contain 103 to 106 copies of individual proteins that are synthesized by translating 10−1 to 102 copies of the respective mRNA.18 In mammalian cells, the copy numbers of protein and mRNA range from 101 to 108 and 1 to 104, respectively.19, 20 The differences in the abundance of proteins and mRNAs imply that there must be an amplification step that controls protein production posttranscriptionally. In part, different copy numbers of mRNAs and respective proteins can be explained by differences in the lifetimes of mRNAs and proteins. For example, in E. coli typical mRNA and protein half‐lives are 5 and 180 min, respectively, indicating that mRNA is degraded within minutes, whereas most proteins have lifetimes longer than the doubling time of the cells. However, this difference in the lifetimes between mRNAs and proteins (36‐fold on average) is still too small to explain the observed variations in numbers of proteins per mRNA in the range of 500 (E. coli) to >5000 (mammalian cells).18, 19 Thus, the observed amplification must arise at the translation level, resulting from repetitive rounds of translation of the same mRNA by several ribosomes. On the level of cell populations, a translation efficiency of >70% of yeast and about 50% of E. coli genes is mainly determined by transcription and mRNA stability,18, 21 whereas translation‐related effects account for regulation of only 13% (E. coli)21 or up to 40% (differentiating mammalian cells)22 of genes. Surprisingly, on single‐cell level, there appears to be little correlation between mRNA and protein levels.17, 19, 20, 23 This suggests that the cellular abundance of proteins either is predominantly controlled at the level of translation19, 20, 22 or results from stochastic switching of transcription in combination with mRNA degradation and the so‐called extrinsic noise in translation,24, 25 caused by for example availability of the ribosomes, translation factors, or mRNAs.
Figure 1.
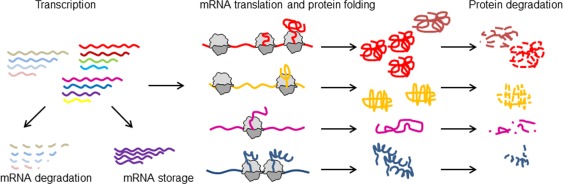
Homeostasis of mRNA and protein. The gene‐expression pathway comprises multiple regulatory levels. Following transcription, mRNA can be degraded, stored or used for translation. Translational efficiencies of individual mRNAs (different color) depend on their sequence signatures and the pools of translational components. Protein folding frequently starts co‐translationally. When translation is completed, proteins are released from the ribosome and fulfil their functions until they are degraded by cellular proteases. Each of these steps enables the selective regulation of expression of individual mRNAs. Translational efficiency can be regulated at initiation or elongation, resulting in different pattern of ribosome distribution on the mRNA. The pace of translation can regulate protein folding (bright red and dark red protein folds).
The translational efficiency of a given mRNA depends on a number of factors, such as concentration in the cell and sequence (mRNA signatures), as well as on the available cellular resources, including pools of free ribosomes, aa‐tRNAs and availability of translation factors (pools signatures). During the translation initiation phase, the ribosome recruits mRNA according to their concentrations and the properties of the RBS. Translational efficiency is further modulated at the elongation phase due to codon usage, mRNA secondary structures, and ribosome stalling signals. Both translation initiation and elongation contribute to the frequency of ribosome loading in polysomes, which can further change translational efficiency. In the following, we summarize the potential regulatory signatures in mRNAs and different mechanism by which the ribosome together with other components of the translational machinery may alter the translational efficiency of an mRNA.
Translation Initiation
Although intuitively initiation should play a dominant role in translation regulation, and there are numerous examples of translational control through initiation, the exact contribution of the initiation step to translation efficiency of mRNA is still under debate. A predictive model for the variation of protein abundance in bacteria suggests that the efficiency of initiation accounts for only a small fraction (1‐5%) of the variation.21, 26 In contrast, ribosome profiling experiments suggest that initiation may be rate‐limiting for protein production, as seen from the accumulation of ribosomes at start codons of mRNAs.2, 4, 27 We note that the degree of accumulation at the start codon—and hence the waiting times for initiation—may be overestimated due to cycloheximide treatment used in the ribosome profiling workflows to stop translation.28 Initiation process in bacteria is relatively simple and well‐understood.29 In comparison, initiation in eukaryotes is much more complex, requires many more factors whose function is not always entirely clear, involves complex interaction networks between factors, and is a target of multiple translation control mechanisms.5, 6, 7, 8, 30 Because bacterial systems are much better studied, in the following we focus on the mechanism of the RBS selection in bacteria.
The experience from protein expression in E. coli suggests that the RBS plays a very critical role. In fact, the available online tools to estimate translational efficiency based on the thermodynamic properties of the RBS yield reasonably good predictions.31 For example, synthetic RBSs can control translational efficiency over at least a 100,000‐fold range.32 Quantitation of global effects of RBS on the translation rate and on mRNA stability using >12,000 synthetic combinations of common promoters and RBSs suggests that 30% of variation can be explained by the properties of the RBS.33 Although the effects of the RBS on the initiation efficiency and mRNA stability may be coupled, the difference between the weakest and strongest RBS sequences (an 90‐fold increase in translation efficiency) corresponds to only an fourfold increase in mRNA stability.33 Thus, the contribution of translation initiation to the control of translational efficiency is substantial, and it is important to understand which parameters govern efficient initiation.
In bacteria, the RBS spans nucleotides −20 to +15 around the translation start codon.34 The features of the RBS that affect the initiation efficiency are: (1) the nature of the start codon; (2) the Shine‐Dalgarno (SD) sequence which is located upstream of the start codon and pairs with a complementary sequence near the 3′ end of 16S rRNA (anti‐Shine‐Dalgarno sequence, aSD); (3) the mRNA secondary structure near the start site; and (4) A/U‐rich elements in the mRNA which are recognized by protein bS1 of the small ribosomal subunit (30S). Although AUG is a most common initiation codon, GUG and UUG are also used rather frequently. On the other hand, the frequency of erroneous initiation at internal AUG, GUG, or UUG codons is very low, suggesting that the ribosome controls the selection of the initiation start site. The presence and the strength of the SD or the mRNA secondary structures are highly variable among the mRNAs. Although the strength of the SD–aSD interaction and the lack of the mRNA secondary structures in the RBS appear to be good predictors for high translation efficiency, each of these elements alone appears to have a limited effect or to control the efficiency of initiation only in a certain context. Usually a combination of these elements contributes to the recruitment of an mRNA to the ribosome, and the overall structure of the RBS can be thought as being an essential element determining the efficiency with which a given mRNA is recruited.35 The details of how this occurs mechanistically and what the contribution of each potential regulatory element in the RBS is not entirely clear, but may be explained in general terms by the multistep nature of the initiation process. This is described in the following.
Translation initiation in bacteria proceeds in three steps. In the first step, the 30S ribosomal subunit recruits the three translation initiation factors IF1, IF2, and IF3, initiator tRNA (fMet‐tRNAfMet) and mRNA to form a 30S preinitiation complex (30S PIC). Although the factors and mRNA bind to the 30S subunit in a random order, there is a kinetically favored predominant route for 30S PIC assembly. IF3 is the first factor to be recruited, with an arrival time of <1 ms, followed by IF2 at 1 ms and IF1 after 30 ms. (All values measured in vitro at conditions of efficient translation at 20°C and extrapolated to in vivo concentrations of the factors36). Initiator fMet‐tRNAfMet is the last to arrive after 100 ms. Binding of mRNA is independent of the other components in the 30S PIC. It is not necessary that the ribosome binds directly to the initiation start codon; a single‐stranded region of the mRNA in the vicinity of the start site is sufficient to recruit the ribosome, which then promotes unwinding of secondary structures of the mRNA around the RBS and aligns the start codon mRNA in the 30S subunit P site.37 The process of mRNA alignment, which may be similar to scanning the mRNA during translation initiation in eukaryotes,30 ceases when the start codon is recognized by the anticodon of fMet‐tRNAfMet, resulting in an affinity switch and the formation of a stable 30S initiation complex (30S IC).36 Next, the large ribosomal subunit (50S) joins the 30S IC. The docking of the 50S subunit on the 30S IC is a stepwise process that relies on shape recognition. IF2 promotes 50S subunit docking, whereas IF3 slows down the reaction. All three factors and the mRNA define the exact conformation of the 30S subunit and the position of fMet‐tRNAfMet in the complex, which modulates the rate of the 50S subunit recruitment38 and the stability of the 70S initiation complex (70S IC).39 50S subunit docking also depends on the RBS in the mRNA. An “optimal” RBS allows very rapid 50S subunit docking, with arrival times in the millisecond range, whereas a “nonoptimal” RBS may delay 70S IC formation by as long as 5 s.38 After the dissociation of the initiation factors (which takes about 300 ms40 and involves GTP hydrolysis by IF2), the mRNA in the 70S IC is committed for translation.
The complex pathway of translation initiation provides several kinetic checkpoints that control the efficiency of 70S IC formation (Fig. 2). The recruitment of mRNA to the 30S PIC depends on (i) the mRNA concentration in the cell (defined by transcription and mRNA stability); (ii) its rate constant of binding to the ribosome, and (iii) the kinetic stability of the complex. The association and dissociation rate constants vary in a relatively broad range, depending on the mRNA.36, 37 Therefore, the initial recruitment of the mRNA may already affect the translational efficiency (checkpoint 1). Binding of structured mRNAs to the stand‐by site at the platform of the 30S subunit41, 42 is followed by the unfolding of secondary structure elements and accommodation of the mRNA in the mRNA channel of the 30S subunit, which may be facilitated by protein bS1 of the 30S subunit.43, 44 The fate of the mRNA at this point depends on kinetic partitioning between mRNA dissociation (k−1 in Fig. 2) and unfolding (k2; checkpoint 2). Thus, an mRNA with strong secondary structure elements which unfold slowly is more likely to dissociate from the 30S PIC and be replaced by another mRNA than to proceed to the 30S IC. Notably, protein bS1 binds mRNA through AU‐rich sequences.45 This might explain why protein expression strongly depends on the presence of AU‐rich sequences in the ORF 5′ region.46 Next, the start codon selection operates the affinity switch that leads to locking of the mRNA and fMet‐tRNAfMet on the 30S subunit.36 This comprises the next checkpoint for mRNA selection which favors those mRNAs that have a canonical start codon (AUG) that is efficiently recognized by the anticodon of fMet‐tRNAfMet (high value of k3, Fig. 2) and disfavors mRNAs with noncanonical initiation codons (low k3 value) by delaying the transition to the active 70S IC (checkpoint 3). Finally, the transition from the 30S IC to the 70S IC provides the last checkpoint for the mRNA and tRNA selection which monitors the details of the SD‐aSD interaction and the presence of the cognate codon–anticodon pair (checkpoint 4).38, 47 Thus, the selection of mRNA and tRNA comprises several consecutive kinetic partitioning checkpoints that favor the forward steps for good substrates and disfavor or delay the entry of poor substrates further into initiation.
Figure 2.
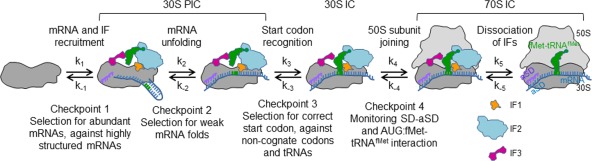
Kinetic partitioning mechanism of mRNA selection. Checkpoint 1, initial docking complex with mRNA bound to the platform of the 30S subunit.41 Checkpoint 2, formation of the mature 30S PIC. The step indicated as mRNA unfolding may entail a number of further intermediates, for example, the formation of a stable SD‐aSD interaction, rearrangements of the 30S subunit, and possibly sampling of the initial start codon. As the latter checkpoint, mainly the secondary structure of the RBS is monitored. Recruitment of fMet‐tRNAfMet to the 30S PIC, which is not shown as a separate step, constitutes a control checkpoint for the selection of fMet‐tRNAfMet against all other aa‐tRNAs due to specific interactions with IF2. Checkpoint 3, 30S IC formation. Codon recognition is a composite step that triggers the stabilization of fMet‐tRNAfMet binding and the destabilization of IF3 binding and promotes further conformational changes in the 30S IC; checkpoint 3 selects against mismatches in the codon‐anticodon complex. Checkpoint 4, the early 70S IC. Here, the properties of the RBS are sensed.38 After GTP hydrolysis by IF2, the factors dissociate, resulting in further tightening of the 30S‐50S interactions and the formation of a mature 70S IC ready for translation elongation.40 Modified from Ref. 35.
The kinetic partitioning mechanism for mRNA discrimination is radically different from models that take into account only the thermodynamic stability of the secondary structure at the RBS or the SD‐aSD interactions.31 The stability of the secondary structure is neither necessary nor sufficient to predict translation efficiency.33 Several groups reported a selection for structures with weak folding at the beginning of genes.46, 48, 49 The kinetic mechanism of mRNA selection predicts that, rather than the thermodynamic stability, the kinetic partitioning between mRNA unwinding and dissociation from the 30S subunit determines the efficiency of mRNA recruitment to the ribosome. Similarly, while in some simplified cases thermodynamic models can provide good predictions for the translation efficiencies, in particular when synthetic RBS elements are studied,32 many cellular mRNAs that have weak SD sequences are translated very efficiently. The kinetic partitioning model would predict that a strong SD–aSD interaction may have a stabilizing role at checkpoint 3 (Fig. 2), but may also affect 50S subunit joining; thus, the net effect of the strong SD may be difficult to predict. The overall duration of initiation, and thus the translation efficiency of a given mRNA, will depend on the speed and efficiency by which every checkpoint is passed, providing a large dynamic range of gradual responses to various RBS types.
Finally, the speed of translation elongation may depend on polysome formation. For example, studies of translation initiation in a cell‐free system suggests that the first initiation on an mRNA is slower than subsequent initiations,50 which may be due to changes in the mRNA structure upon translation by the first ribosome in polysomes or an interaction between ribosomes in polysomes.51 Taking into account the complex mechanism of mRNA selection may help to understand the contribution of each element in the RBS to the regulation of overall translation efficiency, allowing for a more accurate assessment for the translatability of mRNAs in vivo.
The processivity of elongation
Codon usage along the mRNA and amino acid composition contribute 12–26% of the total variation of mRNA–protein correlation in bacteria21, 26 and 30% in human cells.52, 53 For a given protein sequence, multiple degrees of freedom still remain that may allow evolution to tune the efficiency and fidelity of gene expression under various conditions.54 The elongation phase of protein synthesis can affect the overall translational efficiency in three ways. First, an increase of the speed of translation may allow for a faster ribosome turnover and more efficient loading of the following ribosome onto the mRNA, which would increase the density of ribosomes on a given mRNA. A rapid turnover of ribosomes appears to be important, as protein production, for instance in yeast cells, is typically limited by the availability of free ribosomes.55 The ribosome density on an mRNA correlates extremely well with protein abundance and provides a reliable indicator for the prediction of absolute translation efficiencies.56 Ribosome stalling, when occuring at the beginning of the mRNA, may result in ribosome queuing and inhibition of translation initiation.57
Second, the coding region of the mRNA may entail regulatory signals for translation bursts and pauses, which together define the synthesis time and lead to ribosome pile‐up at certain codons. The bursts and pauses may stem from many factors, such as codon‐specific rates of cognate aa‐tRNA delivery to the ribosome, the abundance of aa‐tRNAs, or codon context.58 Pauses can be caused by secondary structure elements in the mRNA.29, 48, 59 Peptide and aminoacyl moieties attached to the tRNAs in the P and A site, respectively, can attenuate the rates of peptide bond formation, thereby affecting the ribosome processivity.60, 61, 62 Finally, collisions between individual ribosomes in polysomes and the cooperation between translating ribosomes and the transcription machinery may also play a role.63 As a result of this complex interplay, some codons are translated faster or slower than the average, that is, the rate of translation is not uniform along an mRNA. In fact, ribosome profiling suggested that the ribosome density along the mRNA varies dramatically. However, the peaks of high occupancy do not correlate with particular codons,27, 64, 65 and it is unclear which features of the mRNA cause the peaks and troughs of ribosome occupancy.
Third, changes in local translational velocity may alter the fidelity of translation and affect the quality of the protein product, which can result in incorrect or misfolded proteins that have to be removed by the cellular quality control machinery. Particularly for the elongation phase, it is difficult to assign the observed effects specifically to individual mRNA signatures, because most of the effects can be caused by parallel variations in pool signatures. In the following, we will discuss how the elements of mRNA sequence and amino acid composition of the nascent protein can modulate the translation elongation rates.
Translation elongation entails three steps, decoding, peptide bond formation, and tRNA–mRNA translocation (Fig. 3). During the decoding step, aa‐tRNA in a ternary complex with EF‐Tu and GTP binds to the ribosome according to the mRNA codon exposed in the A site. Decoding is a multistep process that entails two major stages, initial selection of ternary complexes before GTP hydrolysis by EF‐Tu and aa‐tRNA accommodation after GTP hydrolysis and release from EF‐Tu–GDP. During the initial selection stage, many different ternary complexes compete for binding to the ribosome, allowing the ribosome to select the aa‐tRNA that is cognate to the mRNA codon. Codon‐anticodon interaction activates GTP hydrolysis by EF‐Tu, resulting in a conformational change in the complex that sets aa‐tRNA free from EF‐Tu. While the factor dissociates from the ribosome, the 3′ end of aa‐tRNA moves through the ribosome into the A site of the PTC on the 50S subunit making it ready to react with the peptidyl‐tRNA in the P site. Finally, during the translocation step the resulting deacylated tRNA and the new peptidyl‐tRNA extended by one amino acid move from the A and P to the P and E sites, respectively, with the help of EF‐G–GTP. Each of the three steps can affect the local rates of translation as described below.
Figure 3.
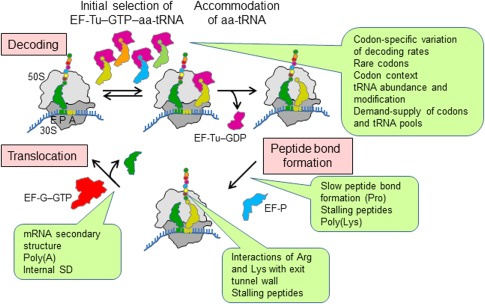
Links between translation elongation and non‐uniform translation. Elongation entails three steps, decoding, peptide bond formation and translocation. During decoding, EF‐Tu in bacteria (or eEF1α in eukaryotes) delivers aa‐tRNA to the A site of the ribosome. These factors are GTPases that in their GTP‐bound conformation form a high‐affinity ternary complex with aa‐tRNA and GTP which, in turn, binds to the ribosome and, after GTP hydrolysis, releases aa‐tRNA to accommodate in the PTC. The ribosome selects an aa‐tRNA that is cognate to the codon in the A site (yellow) among other aa‐tRNAs. These can be almost‐cognate (orange), near‐cognate (green) or non‐cognate (blue). Peptide bond formation between aa‐tRNA in the A site and pept‐tRNA in the P site is catalyzed by the ribosome and usually does not require auxiliary proteins. The peptidyl transfer reaction between two Pro residues requires EF‐P (eIF5A in eukaryotes). Translocation is catalyzed by EF‐G (eEF2 in eukaryotes) at the cost of GTP hydrolysis. Ribosomes, aa‐tRNA and factors are all active players, and the exact interplay between them determines the actual speed and fidelity of translation. Callouts summarize potential sources of translational non‐uniformity at each step of the elongation cycle.
Decoding, tRNA Pools, and Rare Codons
In each round of elongation, the aa‐tRNA that is cognate to the given codon constitutes only a small fraction of the total aa‐tRNA pool (Fig. 3). Near‐ and noncognate ternary complexes can compete with the cognate ones for the initial binding to the ribosome until the tRNA recognizes the codon in the A site.66 Cognate codon–anticodon complexes are stabilized by interactions with the ribosome.67 In contrast, if there is a mismatch in the codon–anticodon complex, the ribosome in most cases does not recognize the complex as cognate, and the ternary complex rapidly dissociates from the ribosome without GTP hydrolysis. The binding of different cognate ternary complexes to the ribosome has a largely uniform kinetics that is independent of the tRNA and amino acid68 or of the GC content of the codon–anticodon complex.69, 70, 71 Furthermore, the rates of the initial decoding steps, which include the initial binding of the ternary complex to the ribosome and codon reading, are similar for cognate, near‐cognate, and noncognate complexes,70 as differences are sensed only after the ribosome has checked the geometry of the codon–anticodon complex in the decoding site. Therefore, at the initial selection step, the effective rate of decoding depends on the concentrations of the cognate aa‐tRNA compared to all other aa‐tRNAs in the ternary complex pool. Furthermore, because there is a difference between the dissociation rates of the near and non‐cognate ternary complexes, it also matters how many ternary complexes with near‐cognate (as compared to non‐cognate) aa‐tRNAs can read a particular codon and what their concentrations are. The redundancy of the genetic code allows the choice between alternative codons for the same amino acid, or between isoacceptor aa‐tRNA. The rate differences in reading different cognate codons by a given tRNA isoacceptor are not large, usually within a factor of two, if the first two codon positions entail Watson‐Crick base pairs and the 3rd position Watson–Crick or wobble base pairs. In some cases, isoacceptor tRNAs can fairly efficiently read non‐Watson‐Crick and non‐wobble bases in the third codon position of the same codon family. For example, , which usually reads GCU, GCA, and GCG codons, can also read the GCC codon (normally read by ) as an almost‐cognate, with rates that are intermediate between cognate and near‐cognate ternary complexes.69 The effects of binding kinetics and concentration alone can result in non‐uniform decoding along the mRNA. The decoding process is further modulated by tRNA modifications and the ability of some pairs to undergo tautomerization, such as G‐U base pairs, which can assume conformations that the ribosome recognizes as cognate.72 Notably, because at the conditions of active protein synthesis the concentrations of free aa‐tRNAs are not high relative to ribosomes, the availability of a particular tRNA may depend on the overall profile of the transcriptome and may change with growth phase and conditions of stress. This may be particularly true for eukaryotic cells, where tRNA transcription (and probably tRNA modification73) is regulated. Thus, the composition of the tRNA pool may contribute to the translation efficiency by modulating the decoding rates.
One important source of non‐uniform translation is the presence of rare codons in mRNAs, or codons for which the cognate tRNA is present in low amount. In fact, frequencies of synonymous codons and the concentrations of the respective tRNAs may differ by more than an order of magnitude (for reviews, see Ref. 11). The presence of rare codons in mRNAs has well‐documented effects on mRNA stability,74, 75, 76 accuracy of amino acid incorporation, and protein folding. Rare codons are often found at the boundaries between protein domains, where ribosome pausing is thought to attenuate the vectorial folding of the nascent protein domains, and as a result determine how much correctly folded, soluble protein is present in the cell.12, 77 The effect can be direct, by slowing down translation at rare codons, because they are read by low‐abundance tRNAs, or indirect, due to co‐variates, such as altered mRNA structure and stability, changes in translation fidelity or appearance of internal SD‐like sequences as a result of a synonymous replacement.64 Direct measurements of translation rates on different rare codons suggest that they are in fact translated slower than the abundant codons, with the observed rate variation spanning a range between 3‐ and 25‐fold.78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88 In contrast, initial ribosome profiling experiments did not find any significant correlation between ribosome occupancy and rare codons.64, 65, 89 This may be because the effects are not very strong,11 or because synonymous codons do not slow down translation and the observed effects on protein production and solubility are due covariates. More recent reanalysis of ribosome profiling data suggested that the ribosomes indeed queue at rare codons, indicating a slower translation; however, the effect on ribosome occupancy is not large, twofold at most.90, 91 Thus, although there is a clear effect of rare codons on translation, its interpretation in terms of specific mRNA signatures remains open.
One potential explanation for the rare‐codon effect is that it acts through the tRNA pools. Surprisingly, the decoding times appear not to correlate with aa‐tRNA levels, but rather with the total tRNA concentrations91; the reasons and implications of this observation are not clear. The effect of codon usage may also depend on the balance of demand and supply in the cell. At conditions of normal growth, codon usage and tRNA abundance are sufficiently balanced with respect to the codon demand‐tRNA supply relationship, so that decoding of rare codons is not significantly slower than that of the abundant ones.4, 14, 65 When the flux through the ORFs is altered by stress, for example upon protein overexpression, rapid growth, nutrient starvation, or adaptation to a change of the environment, the concentrations of free ribosomes and tRNAs may change as well, causing shifts in the availability of single aa‐tRNAs and in the composition of the free tRNA pool. This may lead to the breakdown of the steady‐state level of demand‐supply and change the occupancy of the mRNA with ribosomes.4 At such conditions, translation rates of rare (and abundant codons) may change in a way that is difficult to predict and lead to the observed effects on protein production, solubility, and function. Also at optimal growth conditions, the demand–supply balance may be “non‐perfect,” because pausing may be needed to regulate protein folding.91 Thus, mRNA sequences and tRNA pools provide an additional, rich informational framework that may define the proteome of the cell beyond the rules of the genetic code.
The distribution of codons along the mRNA may be important as well. Clusters of identical codons may act as ‘tRNA sponges’ that deplete the pool of the respective tRNA and slow down translation of the respective downstream codons. Alternatively, it has been suggested that such clusters could lead to increased translation in eukaryotes due to local re‐use of tRNAs92; the details of such a mechanism and whether it operates in vivo remains unclear. In all kingdoms of life, rare codons seem to be overrepresented in the first 90–150 nucleotides of ORFs.49, 53 A “low‐efficiency ramp” consisting of rare codons at the beginning of the mRNA may control the rate of translation flow of the ribosomes by forming an early traffic bottleneck.93 However, recent ribosome profiling data do not support the translational ramp hypothesis and provide technical explanations for the appearance of the presumed ramps in ribosome profiling studies.94
Alternatively, slowing elongation may facilitate the recruitment of chaperones and nascent chain modification enzymes to the nascent peptide emerging from the ribosome.95, 96, 97 In fact, actively translating ribosomes attain a higher affinity to the signal recognition particle (SRP) than vacant ribosomes even before the signal anchor sequence in the nascent peptide, which is recognized the SRP as a signal for membrane targeting, emerges from the polypeptide exit tunnel,98 but the effects on other nascent‐peptide binding proteins is unknown. Clusters of rare codons 35–40 codons downstream of the SRP‐binding site, which is the length of the nascent peptide that spans the exit tunnel of the ribosome, may result in a local translation slowdown and kinetically enhance recognition by ribosome‐associated factors.95, 99
Probably the most likely explanation for the rare codons at the beginning of genes is provided by an expression analysis of >14,000 combinations of promoters, RBS sequences, and N‐terminal codons.29 The analysis suggests that the translation efficiency correlates most with a reduced propensity of an mRNA to form secondary structure elements.48, 100 Furthermore, analysis of complete genomes of 340 species including bacteria, archaea, fungi, plants, insects, fishes, birds, and mammals suggests that nearly all species show evidence for reduced mRNA secondary structure near the start codon.101 Thus, the particular distribution of rare codons at the beginning of ORFs may improve the efficiency of translation initiation in all organisms, rather than regulate ribosome traffic during elongation. The strategy to optimize the translation rate by avoiding mRNA secondary structure at the cost of maintaining rare codons may be an important constraint on codon choice.102
Peptide‐mediated Ribosome Stalling
Another cause of translational stalling is inefficient peptidyl transfer (Fig. 3). Because of differences in the physico‐chemical properties of amino acids, the intrinsic rates of peptide bond formation depend on the identity of the peptidyl‐tRNA and aa‐tRNA in the P‐ and A‐sites, respectively.62 However, for most combinations, the intrinsic rate of peptide bond formation is high, and the effective rate of amino acid incorporation is determined by the accommodation of the aa‐tRNA in the A site.66 Therefore, the chemistry step usually has little effect on the overall rate of translation. However, there are several notable exceptions. The amino acid proline (Pro) is a poor A‐site acceptor of peptidyl moiety during peptide bond formation103, 104 and makes a poor peptidyl donor in the P site.60, 61, 62 Indeed, there are many examples showing that that the presence of stretches of Pro codons in an mRNA can have a dramatic influence on translation. Poly(Pro) sequences cause stalling during translation in bacteria and eukaryotes and require specialized translation factors, EF‐P and eIF5A in bacteria and eukaryotes, respectively, to alleviate stalling.60, 105, 106 The minimum motif that elicits Pro‐dependent stalling contains two Pro residues within a special context, such as P/D/A‐P‐P or P‐P‐P/G/W/N/D.57, 107, 108 The reasons for the poor reactivity of Pro are not entirely clear. Most likely, the steric properties of Pro in combination with restrictions imposed by the peptidyl transferase center of the ribosome make it so exceptionally slow. In comparison, the electronic properties or the ability to undergo cis‐trans isomerization are less important.109 In some cases, Pro‐induced ribosome stalling leads to tmRNA‐mediated peptide tagging and degradation by the trans‐translation mechanism.110, 111, 112 Pro is present in many peptide sequences that are known to induce programmed regulatory stalling, such as the bacterial SecM and TnaC sequences, as well as the human cytomegalovirus (CMV) uORF2 of gp24.113 Such sequences can elicit ribosome stalling in response to certain metabolic conditions or the presence of antibiotics.114 Except for the direct effect of Pro, these stalling sequences specifically engage in the interactions with residues in the ribosomal peptide exit tunnel to elicit stalling.115 There are also stalling sequences that do not depend on Pro stalling, such as ErmCL and ErmBL.116 The common feature for all stalling sequences is that they inhibit the peptidyl transferase reaction by inducing a misalignment of reactive groups at the peptidyl transferase center of the ribosome.115 The interactions of the nascent peptide with the exit tunnel wall, in particular with the critical region at the constriction formed by the ribosomal proteins uL22/uL4, and the relay system that transmits these recognition events from the tunnel to the PTC are important features of the arrest signals.114, 115, 116
Pausing at Glycine Codons and Internal SD Sequences
Another amino acid that can cause slow peptide bond formation is glycine (Gly). In fact, ribosome profiling experiments revealed that Gly‐rich tripeptide motifs have the highest pause scores in E. coli.117 However, because Gly codons in the mRNA can act as internal SD‐like sequences (e.g., GGAGGA), these pauses are difficult to distinguish from those caused by presumably inefficient tRNA‐mRNA translocation induced by SD–aSD interaction. Single‐molecule experiments suggested that tRNA translocation is indeed slowed down by SD–aSD interactions.118, 119 Based on the positions of higher ribosome occupancy relative to Gly codons it appeared more likely that internal SD‐like sequences (rather than slow decoding of Gly codons or inefficient peptidyl transfer reaction with Gly) are the major determinant of translational pausing in bacteria.89 In contrast, a recent refined ribosome profiling study as well as biochemical assays suggests that internal SD motifs have little (if any) effect on elongation rates.120 The latter finding is in agreement with biochemical data suggesting that internal SD sequences do not have a significant effect on the translocation velocity at conditions of rapid translation.59 Furthermore, pauses in ribosome profiling datasets occurred when Gly codons reside in the E site, rather than at the position of the presumed SD‐aSD interaction. Moreover, stalling at Gly codons may be related to the action of chloramphenicol, an antibiotic that is commonly used to stop translation in ribosome profiling workflows.120
However, other data argue in favor of SD sequences being an important determinant for ribosome stalling. A Gly codon pair GGA‐GGU, which is a perfect SD‐aSD match, reduces translation speed when placed about 9 nucleotides upstream of the translational stall.87 Production of a bacterial metabolic enzyme negatively correlates with the frequency of motifs with high affinity for the aSD sequence.121 An internal SD sequence also modulates ribosome stalling at the frame shifting site of the bacterial dnaX.122 Overall, while Gly codons or internal SD sequences normally do not cause translational pausing, it seems that in certain contexts they might modulate the rate of translation, possibly in combination with some other, yet unknown factors.
Translation Pausing Mediated by Arginine and Lysine Codons
Translation is often downregulated by codons for arginine (Arg) and lysine (Lys), but because of a large amount of co‐variates it is often difficult to understand the mechanism. Poly‐basic amino acid stretches contribute to slowing of the ribosome and a peak of ribosome footprints on mRNAs.64 One of the Arg codons (AGG) is decoded by a tRNA which is very rare in E. coli; thus, the effect can result from slow decoding.81 Tandem Arg codons AGG and AGA can appear as an internal SD sequence, which may account for the translation pause in an engineered reported system in vivo.87 Furthermore, translation in rabbit reticulocyte lysate is inhibited by poly(Lys) and poly(Arg) sequences.125 Notably, peptide bond formation with single Lys and Arg residues is rapid62; thus, the effects at the peptidyl transferase center cannot account for the observed Arg/Lys‐mediated translation arrest. Positively charged peptide segments cause pausing when they are 10‐20 Å from the PTC (Fig. 4). Thus, the most likely explanation is that patches of positively charged residues in the nascent peptide interact with the negatively charged exit tunnel walls and these electrostatic interactions modulate translation rates.123, 124 This is supported by ribosome profiling data which indicate that regions with the highest ribosomal occupancy along the mRNA are often due to positive charges in the nascent peptide, and that this slowing effect cannot be accounted for by mRNA structure or by codon usage bias.64
Figure 4.
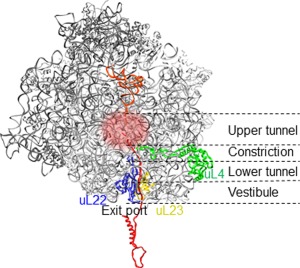
Schematic representation of a cross‐section of the ribosome with the nascent peptide. The peptide exit tunnel (overall length 100 Å) can be separated into three folding zones, as indicated.154 Ribosomal proteins uL4 and uL22 form the constriction. Protein uL23 and residues of 23S rRNA in the upper tunnel region (indicated by glow) can signal events in the tunnel to the PTC and the ribosome surface in the vicinity of the exit port.115
Stretches of consecutive AAA codons, one of the two codons encoding Lys, have yet another effect on translation. Insertion of consecutive AAA codons into reporters has a stronger negative impact on protein expression than insertion of an equivalent number of synonymous AAG codons in both eukaryotes and bacteria.125, 126 Peptide bond formation between two consecutive Lys residues is surprisingly slow, resulting in ribosome stalling.125 Ribosome stalling at Lys codons triggers ribosome sliding on successive AAA codons. When ribosomes resume translation, they may shift in an incorrect reading frame within the ORF. The ribosomes translating in the −1 or +1 frame usually quickly encounter out‐of‐frame stop codons that result in termination. In eukaryotes, such premature termination events target the mRNA for nonsense‐mediated decay.125 Although such sequences are underrepresented in genes from many eukaryotic organisms, the expression of about 2% of genes in the human genome may be subject to this form of regulation.126
Ribosome Pausing at mRNA Secondary Structure Elements
Translation pauses caused by mRNA secondary structure elements arguably are the least studied examples for non‐uniform translation. As described above, secondary structure at the RBS and the beginning of the ORF is an important indicator of translation efficiency.29, 48, 127 However, the role of secondary structures in the middle of ORFs is less clear. Chemical probing of the structure of yeast mRNAs suggested that out of >20,000 mRNA regions examined (representing ca. 2,000 transcripts) only about 4% were structured in vivo, compared to 24% in vitro.128 The remaining potential structures have to be disrupted during elongation to allow movement of the ribosome along the mRNA. Optical‐tweezer experiments suggested that translocation and mRNA unwinding are strictly coupled functions of the ribosome118; thus, the overall rate of translation may depend on the secondary structure of the mRNA at conditions when decoding is rapid. Thermodynamically stable elements in the mRNA secondary structure, such as pseudoknots and stem‐loops, are known to slow down ribosome movement along the mRNA, for example at sequences that cause programmed ribosome frameshifting.129, 130, 131 Kinetic experiments show that the pseudoknot present in the infectious bronchitis virus 1a/1b mRNA impedes ribosome dynamics by slowing down (by 15‐fold) the swiveling movement of the 30S head in the direction of the 3′ end of the mRNA.129 This movement is essential for tRNA‐mRNA movement in translocation.132 However, mRNA elements that stimulate frameshifting may be very special and may have evolved to stall the ribosome. In normal translation, ribosomes can unwind downstream helices of considerable stability, including unwinding of a perfect 27 base‐pair helix of predicted T m = 70°C.133 The helicase is formed by ribosomal proteins uS3, uS4, and uS5 at the mRNA entrance of the small ribosomal subunit and is expected to act on nucleotide +11 of the mRNA (counting from the first nucleotide of the P‐site codon).133 Although the existence of the helicase activity has been experimentally shown for bacterial ribosomes only, the mechanism is very likely to work in both pro‐ and eukaryotes, because the three proteins are universal and the residues in uS3 and uS5 that are directly involved in the helicase activity are evolutionary conserved.129 The ribosome helicase does not require GTP or ATP as an energy source.133 Optical‐tweezer experiments suggest that the translation rate is greatly influenced by the GC content of folded structures at the mRNA entry site.134 Unlike other helicases, the ribosome uses two distinct active mechanisms to unwind the mRNA. It destabilizes helix junctions at the mRNA entry site by biasing thermal fluctuations towards the open state, increasing the probability of translocation; additionally it mechanically pulls apart mRNA single strands of the closed junction during conformational changes that accompany translocation.134 In the cell, the mRNA structure also depends on the interactions with RNA‐binding proteins, with and without specific helicase activity, which may stabilize or disrupt secondary structure elements.
Elongation Processivity and Protein Folding
Variable translation rates may affect folding of newly synthesized proteins. For many proteins, folding begins co‐translationally, with the emerging nascent peptide attached to the translating ribosome. The ribosome can affect folding in a number of ways (Fig. 5). First, in contrast to post‐translation folding, co‐translational folding of nascent chains on the ribosome is vectorial, that is, it starts from the N terminus and involves elements that emerge successively from the N to the C terminus of the nascent protein. Because translation is slow relative to local folding events, vectorial folding in the confined space of, and interactions with, the exit tunnel may define the landscape of protein folding.135, 136, 137, 138 Second, the ribosome may stabilize folding intermediates that are short‐lived (or not existing) in solution.139, 140 Third, because emerging peptides can interact with the ribosome surface,139, 141, 142, 143, 144 the ribosome may have a chaperoning effect protecting the nascent chain from misfolding and aggregation until the protein is fully synthesized and extruded from the peptide exit tunnel.145 Fourth, the spatial proximity of ribosomes that synthesize proteins encoded in different ORFs within one operon may ensure their efficient co‐translational assembly in vivo, as shown for bacterial luciferase subunits LuxA and LuxB.146 The ribosome‐associated chaperone trigger factor delays the onset of co‐translational interactions until the LuxB dimer interface is fully exposed. These results show that coupling of protein assembly to translation can be crucial for the effective assembly of protein complexes.146 Finally, the ribosome provides a platform for protein biogenesis factors that interact with the emerging nascent peptide and effect its maturation and proper cellular localization.16, 147, 148, 149
Figure 5.
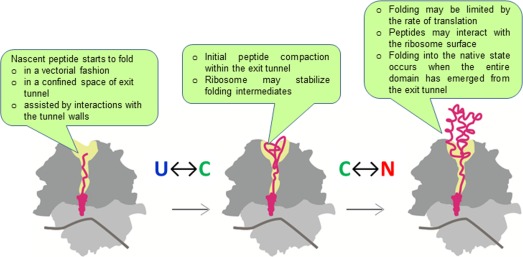
Effect of the ribosome on protein folding. U, unfolded protein; C, compact transient state or folding intermediate; N, native fold. Callouts summarize the potential effects at each step.
The nascent peptide can form small secondary structure elements, such as α‐helices or small α‐helical domains within the exit tunnel.150, 151, 152, 153 Small tertiary structure elements, such as tertiary hairpins of transmembrane helices, can form near the exit port in the vestibule of the tunnel.154, 155 Native‐like folds of larger domains can form when the whole domain has emerged from the ribosome.140, 144, 156 Tertiary structures fold in a hierarchical way, with tertiary subdomains folding sequentially, but not independently.154 For example, the first nucleotide‐binding domain of the cystic fibrosis transmembrane conductance regulator folds cotranslationally via the sequential compaction of N‐terminal, α‐helical, and α/β‐core subdomains, and the timing of these events is critical.157
Changes in local translation kinetics can influence the conformation of newly synthesized proteins. Most examples available so far link protein folding to the use of rare codons or to tRNA abundance. Naturally occurring synonymous single‐nucleotide polymorphisms (sSNPs) and synonymous mutations can affect the activity and post‐translational modification of a protein, altering its interactions with drugs and inhibitors, sensitivity to proteases, and aggregation propensity.77, 84, 88, 157, 158, 159, 160, 161 In some cases, these changes are associated with diseases.162, 163 Synonymous mutations that change elongation rates can even switch folding of some protein domains from post‐translational to cotranslational.164 Alterations in codon context caused by synonymous mutations may also induce the mis‐incorporation of amino acids, leading to protein misfolding.165
The analysis in vivo and in vitro of the structure and folding of gamma‐B crystallin showed that synonymous mutations can change the fraction of soluble protein in the cell, the sensitivity of the protein to protease digestion, and alter the conformational ensemble of the mature protein156 (Fig. 6). The presence of rare codons in the mRNA reduced the rate of translation, which resulted in a delayed emergence of the N‐terminal domain from the exit tunnel. This alone can, in principle, change protein folding, provided the pauses introduced by rare codons appear at positions where the nascent chain enters folding states that are far from equilibrium.166, 167 However, time‐resolved FRET experiments also showed a slower folding of the full‐length domain after it has emerged from the exit tunnel.156 It is possible that slow translation allows for partitioning of elemental folding events into alternative pathways.168 On the other hand, slower folding may be due to a lower rate of C‐terminal domain synthesis. In this case, the folding of N‐terminal domain may be retarded by interactions with the exit tunnel area until the domain moves away from the ribosome surface. These observations suggest that codon bias can alter local and global translation rates and can result in the formation of alternative conformations of the nascent protein on the ribosome and in solution after the release of the completed protein. Thus, the rate of translation—encoded by the use of synonymous codons—provides a code‐within‐the‐code that shapes the quality of the cellular proteome.
Figure 6.
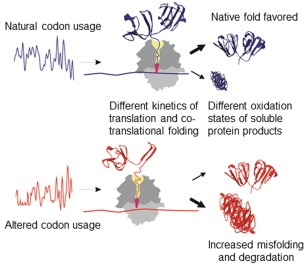
Synonymous codon usage directs co‐translational folding toward different protein conformations Reproduced from Ref. 156 with permission.
Perspectives
The data summarized in this review show that translational control entails a far broader spectrum of phenomena than previously thought. In addition to the regulation of mRNA stability and localization, as well as of translation initiation, which are traditionally associated with the term translation control, global, and local elongation rates turn out to define the composition and the quality of the cellular proteome. The staggering complexity of the mRNA signatures and dynamic changes of the pools of translational components make it difficult to dissect the networks into functional units and distinguish between covariates. Ribosome profiling provides an unprecedented view into the global translation in different cell types. While there is an on‐going discussion about the best workflows, it is clear that more ribosome profiling data at different conditions and a more detailed analysis of the data by bioinformatic tools can provide a wealth of new data and testable hypotheses as to the mechanisms behind the regulatory events. These datasets should be complemented with the results of other omics approaches, such as transcriptomics, nascentomics and tRNAomics, to obtain precise numbers as to pools of actively translated mRNAs and tRNAs. The understanding of the new translational control mechanisms is lagging behind, including many phenomena known to affect translation efficiency. For example, effects related to termination and ribosome recycling, interaction between ribosomes in polysomes, the interaction between the transcription and translation machineries (in bacteria) or cotranslational protein folding are insufficiently understood. Other important unresolved questions concern the accuracy of protein production and its effects on cellular fitness. Finally, the interplay between the translation and quality control machineries is a question of fundamental importance. Because proteome imbalance causes many human diseases, it is essential to understand the processes shaping the proteome. Deciphering the link between translation, proteome and disease will remain a challenging, but crucial task for the field.
Acknowledgment
The authors thanks Anton Komar and Wolfgang Wintermeyer for critical reading and Albena Draycheva for preparing Figure 4. The author declares no conflict of interests.
References
- 1. Ciandrini L, Stansfield I, Romano MC (2013) Ribosome traffic on mRNAs maps to gene ontology: genome‐wide quantification of translation initiation rates and polysome size regulation. PLoS Comput Biol 9:e1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siwiak M, Zielenkiewicz P (2013) Transimulation—protein biosynthesis web service. PLoS One 8:e73943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitarai N, Sneppen K, Pedersen S (2008) Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J Mol Biol 382:236–245. [DOI] [PubMed] [Google Scholar]
- 4. Subramaniam AR, Zid BM, O'Shea EK (2014) An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159:1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Richter JD, Sonenberg N (2005) Regulation of cap‐dependent translation by eIF4E inhibitory proteins. Nature 433:477–480. [DOI] [PubMed] [Google Scholar]
- 6. Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siddiqui N, Sonenberg N (2015) Signalling to eIF4E in cancer. Biochem Soc Trans 43:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serganov A, Nudler E (2013) A decade of riboswitches. Cell 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han Y, Liu L, Fang N, Yang R, Zhou D (2013) Regulation of pathogenicity by noncoding RNAs in bacteria. Future Microbiol 8:579–591. [DOI] [PubMed] [Google Scholar]
- 11. Chaney JL, Clark PL (2015) Roles for synonymous codon usage in protein biogenesis. Annu Rev Biophys 44:143–166. [DOI] [PubMed] [Google Scholar]
- 12. Komar AA (2009) A pause for thought along the co‐translational folding pathway. Trends Biochem Sci 34:16–24. [DOI] [PubMed] [Google Scholar]
- 13. Brar GA, Weissman JS (2015) Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16:651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gorgoni B, Marshall E, McFarland MR, Romano MC, Stansfield I (2014) Controlling translation elongation efficiency: tRNA regulation of ribosome flux on the mRNA. Biochem Soc Trans 42:160–165. [DOI] [PubMed] [Google Scholar]
- 15. Novoa EM, Ribas de Pouplana L (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28:574–581. [DOI] [PubMed] [Google Scholar]
- 16. Pechmann S, Willmund F, Frydman J (2013) The ribosome as a hub for protein quality control. Mol Cell 49:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS (2010) Quantifying E. coli proteome and transcriptome with single‐molecule sensitivity in single cells. Science 329:533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25:117–124. [DOI] [PubMed] [Google Scholar]
- 19. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2013) Corrigendum: global quantification of mammalian gene expression control. Nature 495:126–127. [DOI] [PubMed] [Google Scholar]
- 20. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. [DOI] [PubMed] [Google Scholar]
- 21. Guimaraes JC, Rocha M, Arkin AP (2014) Transcript level and sequence determinants of protein abundance and noise in Escherichia coli . Nucleic Acids Res 42:4791–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kristensen AR, Gsponer J, Foster LJ (2013) Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol Syst Biol 9:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, Westermark B, Landegren U (2016) Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep 14:380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albayrak C, Jordi CA, Zechner C, Lin J, Bichsel CA, Khammash M, Tay S (2016) Digital quantification of proteins and mRNA in single mammalian cells. Mol Cell 61:914–924. [DOI] [PubMed] [Google Scholar]
- 25. Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single‐cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441:840–846. [DOI] [PubMed] [Google Scholar]
- 26. Nie L, Wu G, Zhang W (2006) Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics 174:2229–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP (2016) Improved ribosome‐footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep 14:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goodman DB, Church GM, Kosuri S (2013) Causes and effects of N‐terminal codon bias in bacterial genes. Science 342:475–479. [DOI] [PubMed] [Google Scholar]
- 30. Hinnebusch AG (2014) The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812. [DOI] [PubMed] [Google Scholar]
- 31. Reeve B, Hargest T, Gilbert C, Ellis T (2014) Predicting translation initiation rates for designing synthetic biology. Front Bioeng Biotechnol 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM (2013) Composability of regulatory sequences controlling transcription and translation in Escherichia coli . Proc Natl Acad Sci USA 110:14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dreyfus M (1988) What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol 204:79–94. [DOI] [PubMed] [Google Scholar]
- 35. Milon P, Rodnina MV (2012) Kinetic control of translation initiation in bacteria. Crit Rev Biochem Mol Biol 47:334–348. [DOI] [PubMed] [Google Scholar]
- 36. Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV (2012) Real‐time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol 19:609–615. [DOI] [PubMed] [Google Scholar]
- 37. Studer SM, Joseph S (2006) Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell 22:105–115. [DOI] [PubMed] [Google Scholar]
- 38. Milon P, Konevega AL, Gualerzi CO, Rodnina MV (2008) Kinetic checkpoint at a late step in translation initiation. Mol Cell 30:712–720. [DOI] [PubMed] [Google Scholar]
- 39. MacDougall DD, Gonzalez RL Jr. (2015) Translation initiation factor 3 regulates switching between different modes of ribosomal subunit joining. J Mol Biol 427:1801–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goyal A, Belardinelli R, Maracci C, Milon P, Rodnina MV (2015) Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res 43:10700–10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP (2007) Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell 130:1019–1031. [DOI] [PubMed] [Google Scholar]
- 42. Yusupova G, Jenner L, Rees B, Moras D, Yusupov M (2006) Structural basis for messenger RNA movement on the ribosome. Nature 444:391–394. [DOI] [PubMed] [Google Scholar]
- 43. Qu X, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr. (2012) Ribosomal protein S1 unwinds double‐stranded RNA in multiple steps. Proc Natl Acad Sci USA 109:14458–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, Fabbretti A, Choulier L, Micura R, Klaholz BP, Romby P, Springer M, Marzi S (2013) Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 11:e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hajnsdorf E, Boni IV (2012) Multiple activities of RNA‐binding proteins S1 and Hfq. Biochimie 94:1544–1553. [DOI] [PubMed] [Google Scholar]
- 46. Allert M, Cox JC, Hellinga HW (2010) Multifactorial determinants of protein expression in prokaryotic open reading frames. J Mol Biol 402:905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Antoun A, Pavlov MY, Lovmar M, Ehrenberg M (2006) How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell 23:183–193. [DOI] [PubMed] [Google Scholar]
- 48. Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding‐sequence determinants of gene expression in Escherichia coli . Science 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tuller T, Waldman YY, Kupiec M, Ruppin E (2010) Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA 107:3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Underwood KA, Swartz JR, Puglisi JD (2005) Quantitative polysome analysis identifies limitations in bacterial cell‐free protein synthesis. Biotechnol Bioeng 91:425–435. [DOI] [PubMed] [Google Scholar]
- 51. Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W (2009) The native 3D organization of bacterial polysomes. Cell 136:261–271. [DOI] [PubMed] [Google Scholar]
- 52. Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two‐thirds of protein abundance variation in a human cell line. Mol Syst Biol 6:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brockmann R, Beyer A, Heinisch JJ, Wilhelm T (2007) Posttranscriptional expression regulation: what determines translation rates? PLoS Comput Biol 3:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gingold H, Pilpel Y (2011) Determinants of translation efficiency and accuracy. Mol Syst Biol 7:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB (2013) Rate‐limiting steps in yeast protein translation. Cell 153:1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li GW, Burkhardt D, Gross C, Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR (2015) High‐precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep 11:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH (2015) Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11:e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Borg A, Ehrenberg M (2015) Determinants of the rate of mRNA translocation in bacterial protein synthesis. J Mol Biol 427:1835–1847. [DOI] [PubMed] [Google Scholar]
- 60. Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV (2013) EF‐P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88. [DOI] [PubMed] [Google Scholar]
- 61. Muto H, Ito K (2008) Peptidyl‐prolyl‐tRNA at the ribosomal P‐site reacts poorly with puromycin. Biochem Biophys Res Commun 366:1043–1047. [DOI] [PubMed] [Google Scholar]
- 62. Wohlgemuth I, Brenner S, Beringer M, Rodnina MV (2008) Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem 283:32229–32235. [DOI] [PubMed] [Google Scholar]
- 63. Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS (2014) A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Charneski CA, Hurst LD (2013) Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol 11:e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Qian W, Yang JR, Pearson NM, Maclean C, Zhang J (2012) Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet 8:e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wohlgemuth I, Pohl C, Rodnina MV (2010) Optimization of speed and accuracy of decoding in translation. Embo J 29:3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902. [DOI] [PubMed] [Google Scholar]
- 68. Ledoux S, Uhlenbeck OC (2008) Different aa‐tRNAs are selected uniformly on the ribosome. Mol Cell 31:114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kothe U, Rodnina MV (2007) Codon reading by tRNAAla with modified uridine in the wobble position. Mol Cell 25:167–174. [DOI] [PubMed] [Google Scholar]
- 70. Gromadski KB, Daviter T, Rodnina MV (2006) A uniform response to mismatches in codon‐anticodon complexes ensures ribosomal fidelity. Mol Cell 21:369–377. [DOI] [PubMed] [Google Scholar]
- 71. Gromadski KB, Rodnina MV (2004) Kinetic determinants of high‐fidelity tRNA discrimination on the ribosome. Mol Cell 13:191–200. [DOI] [PubMed] [Google Scholar]
- 72. Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G (2015) Structural insights into the translational infidelity mechanism. Nat Commun 6:7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kirchner S, Ignatova Z (2015) Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16:98–112. [DOI] [PubMed] [Google Scholar]
- 74. Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J (2015) Codon optimality is a major determinant of mRNA stability. Cell 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boel G, Letso R, Neely H, Price WN, Wong KH, Su M, Luff JD, Valecha M, Everett JK, Acton TB, Xiao R, Montelione GT, Aalberts DP, Hunt JF (2016) Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 529:358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mishima Y, Tomari Y (2016) Codon usage and 3' UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61:874–885. [DOI] [PubMed] [Google Scholar]
- 77. Zhang G, Hubalewska M, Ignatova Z (2009) Transient ribosomal attenuation coordinates protein synthesis and co‐translational folding. Nat Struct Mol Biol 16:274–280. [DOI] [PubMed] [Google Scholar]
- 78. Pedersen S (1984) Escherichia coli ribosomes translate in vivo with variable rate. Embo J 3:2895–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sorensen MA, Kurland CG, Pedersen S (1989) Codon usage determines translation rate in Escherichia coli . J Mol Biol 207:365–377. [DOI] [PubMed] [Google Scholar]
- 80. Irwin B, Heck JD, Hatfield GW (1995) Codon pair utilization biases influence translational elongation step times. J Biol Chem 270:22801–22806. [DOI] [PubMed] [Google Scholar]
- 81. Bonekamp F, Jensen KF (1988) The AGG codon is translated slowly in E. coli even at very low expression levels. Nucleic Acids Res 16:3013–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Varenne S, Lazdunski C (1986) Effect of distribution of unfavourable codons on the maximum rate of gene expression by an heterologous organism. J Theor Biol 120:99–110. [DOI] [PubMed] [Google Scholar]
- 83. Varenne S, Buc J, Lloubes R, Lazdunski C (1984) Translation is a non‐uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol 180:549–576. [DOI] [PubMed] [Google Scholar]
- 84. Komar AA, Lesnik T, Reiss C (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462:387–391. [DOI] [PubMed] [Google Scholar]
- 85. Letzring DP, Dean KM, Grayhack EJ (2010) Control of translation efficiency in yeast by codon‐anticodon interactions. Rna 16:2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Curran JF, Yarus M (1989) Rates of aminoacyl‐tRNA selection at 29 sense codons in vivo. J Mol Biol 209:65–77. [DOI] [PubMed] [Google Scholar]
- 87. Chevance FF, Le Guyon S, Hughes KT (2014) The effects of codon context on in vivo translation speed. PLoS Genet 10:e1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sander IM, Chaney JL, Clark PL (2014) Expanding Anfinsen's principle: contributions of synonymous codon selection to rational protein design. J Am Chem Soc 136:858–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li GW, Oh E, Weissman JS (2012) The anti‐Shine‐Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B (2014) Measurement of average decoding rates of the 61 sense codons in vivo. Elife 3:e03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dana A, Tuller T (2014) The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res 42:9171–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y (2010) A role for codon order in translation dynamics. Cell 141:355–367. [DOI] [PubMed] [Google Scholar]
- 93. Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141:344–354. [DOI] [PubMed] [Google Scholar]
- 94. Heyer EE, Moore MJ (2016) Redefining the translational status of 80S monosomes. Cell 164:757–769. [DOI] [PubMed] [Google Scholar]
- 95. Pechmann S, Chartron JW, Frydman J (2014) Local slowdown of translation by nonoptimal codons promotes nascent‐chain recognition by SRP in vivo. Nat Struct Mol Biol 21:1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zalucki YM, Beacham IR, Jennings MP (2011) Coupling between codon usage, translation and protein export in Escherichia coli . Biotechnol J 6:660–667. [DOI] [PubMed] [Google Scholar]
- 97. Zalucki YM, Beacham IR, Jennings MP (2009) Biased codon usage in signal peptides: a role in protein export. Trends Microbiol 17:146–150. [DOI] [PubMed] [Google Scholar]
- 98. Bornemann T, Jockel J, Rodnina MV, Wintermeyer W (2008) Signal sequence‐independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol 15:494–499. [DOI] [PubMed] [Google Scholar]
- 99. Pechmann S, Frydman J (2013) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bentele K, Saffert P, Rauscher R, Ignatova Z, Bluthgen N (2013) Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol 9:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gu W, Zhou T, Wilke CO (2010) A universal trend of reduced mRNA stability near the translation‐initiation site in prokaryotes and eukaryotes. PLoS Comput Biol 6:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gorochowski TE, Ignatova Z, Bovenberg RA, Roubos JA (2015) Trade‐offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res 43:3022–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC (2009) Slow peptide bond formation by proline and other N‐alkylamino acids in translation. Proc Natl Acad Sci USA 106:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Johansson M, Ieong KW, Trobro S, Strazewski P, Aqvist J, Pavlov MY, Ehrenberg M (2011) pH‐sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A‐site aminoacyl‐tRNA. Proc Natl Acad Sci USA 108:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K (2013) Translation elongation factor EF‐P alleviates ribosome stalling at polyproline stretches. Science 339:82–85. [DOI] [PubMed] [Google Scholar]
- 106. Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE (2013) eIF5A promotes translation of polyproline motifs. Mol Cell 51:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Peil L, Starosta AL, Lassak J, Atkinson GC, Virumae K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN (2013) Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF‐P. Proc Natl Acad Sci USA 110:15265–15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Starosta AL, Lassak J, Peil L, Atkinson GC, Virumae K, Tenson T, Remme J, Jung K, Wilson DN (2014) Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res 42:10711–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Doerfel LK, Wohlgemuth I, Kubyshkin V, Starosta AL, Wilson DN, Budisa N, Rodnina MV (2015) Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J Am Chem Soc 137:12997–13006. [DOI] [PubMed] [Google Scholar]
- 110. Hayes CS, Bose B, Sauer RT (2002) Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem 277:33825–33832. [DOI] [PubMed] [Google Scholar]
- 111. Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR (2009) Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem 284:34809–34818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR (2013) Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA 110:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ito K, Chiba S (2013) Arrest peptides: cis‐acting modulators of translation. Annu Rev Biochem 82:171–202. [DOI] [PubMed] [Google Scholar]
- 114. Ito K, Chiba S, Pogliano K (2010) Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun 393:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wilson DN, Arenz S, Beckmann R (2016) Translation regulation via nascent polypeptide‐mediated ribosome stalling. Curr Opin Struct Biol 37:123–133. [DOI] [PubMed] [Google Scholar]
- 116. Gupta P, Liu B, Klepacki D, Gupta V, Schulten K, Mankin AS, Vazquez‐Laslop N (2016) Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol 12:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR (2013) Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA 110:E878–E887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I (2008) Following translation by single ribosomes one codon at a time. Nature 452:598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S (2007) Peptide bond formation destabilizes Shine‐Dalgarno interaction on the ribosome. Nature 446:454–457. [DOI] [PubMed] [Google Scholar]
- 120. Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR (2016) Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep 14:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Agashe D, Martinez‐Gomez NC, Drummond DA, Marx CJ (2013) Good codons, bad transcript: large reductions in gene expression and fitness arising from synonymous mutations in a key enzyme. Mol Biol E 30:549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Larsen B, Wills NM, Gesteland RF, Atkins JF (1994) rRNA‐mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol 176:6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lu J, Deutsch C (2008) Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lu J, Kobertz WR, Deutsch C (2007) Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol 371:1378–1391. [DOI] [PubMed] [Google Scholar]
- 125. Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R (2015) Ribosomes slide on lysine‐encoding homopolymeric A stretches. Elife 4:e05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Arthur L, Pavlovic‐Djuranovic S, Smith‐Koutmou K, Green R, Szczesny P, Djuranovic S (2015) Translational control by lysine‐encoding A‐rich sequences. Sci Adv 1:e1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D (2014) Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol 10:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS (2014) Genome‐wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV (2014) Programmed −1 frame shifting by kinetic partitioning during impeded translocation. Cell 157:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim HK, Liu F, Fei J, Bustamante C, Gonzalez RL Jr, Tinoco I Jr (2014) A frame shifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci USA 111:5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chen J, Petrov A, Johansson M, Tsai A, O'Leary SE, Puglisi JD (2014) Dynamic pathways of −1 translational frame shifting. Nature 512:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, Rodnina MV (2016) Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol 23:342–348. [DOI] [PubMed] [Google Scholar]
- 133. Takyar S, Hickerson RP, Noller HF (2005) mRNA helicase activity of the ribosome. Cell 120:49–58. [DOI] [PubMed] [Google Scholar]
- 134. Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr. (2011) The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. O'Brien EP, Vendruscolo M, Dobson CM (2012) Prediction of variable translation rate effects on cotranslational protein folding. Nat Commun 3:868. [DOI] [PubMed] [Google Scholar]
- 136. Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O'Brien EP (2013) In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc Natl Acad Sci USA 110:E132–E140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ziv G, Haran G, Thirumalai D (2005) Ribosome exit tunnel can entropically stabilize alpha‐helices. Proc Natl Acad Sci USA 102:18956–18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Clark PL, King J (2001) A newly synthesized, ribosome‐bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates. J Biol Chem 276:25411–25420. [DOI] [PubMed] [Google Scholar]
- 139. Kaiser CM, Goldman DH, Chodera JD, Tinoco I Jr, Bustamante C (2011) The ribosome modulates nascent protein folding. Science 334:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Holtkamp W, Kokic G, Jager M, Mittelstaet J, Komar AA, Rodnina MV (2015) Cotranslational protein folding on the ribosome monitored in real time. Science 350:1104–1107. [DOI] [PubMed] [Google Scholar]
- 141. Deckert A, Waudby CA, Wlodarski T, Wentink AS, Wang X, Kirkpatrick JP, Paton JF, Camilloni C, Kukic P, Dobson CM, Vendruscolo M, Cabrita LD, Christodoulou J (2016) Structural characterization of the interaction of alpha‐synuclein nascent chains with the ribosomal surface and trigger factor. Proc Natl Acad Sci USA. 113:5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Eichmann C, Preissler S, Riek R, Deuerling E (2010) Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc Natl Acad Sci USA 107:9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hsu ST, Fucini P, Cabrita LD, Launay H, Dobson CM, Christodoulou J (2007) Structure and dynamics of a ribosome‐bound nascent chain by NMR spectroscopy. Proc Natl Acad Sci USA 104:16516–16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Cabrita LD, Cassaignau AM, Launay HM, Waudby CA, Wlodarski T, Camilloni C, Karyadi ME, Robertson AL, Wang X, Wentink AS, Goodsell LS, Woolhead CA, Vendruscolo M, Dobson CM, Christodoulou J (2016) A structural ensemble of a ribosome‐nascent chain complex during cotranslational protein folding. Nat Struct Mol Biol 23:278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Evans MS, Sander IM, Clark PL (2008) Cotranslational folding promotes beta‐helix formation and avoids aggregation in vivo. J Mol Biol 383:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B (2015) Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350:678–680. [DOI] [PubMed] [Google Scholar]
- 147. Kim YE, Hipp MS, Bracher A, Hayer‐Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355. [DOI] [PubMed] [Google Scholar]
- 148. Gloge F, Becker AH, Kramer G, Bukau B (2014) Co‐translational mechanisms of protein maturation. Curr Opin Struct Biol 24:24–33. [DOI] [PubMed] [Google Scholar]
- 149. Ismail N, Hedman R, Linden M, von Heijne G (2015) Charge‐driven dynamics of nascent‐chain movement through the SecYEG translocon. Nat Struct Mol Biol 22:145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Woolhead CA, McCormick PJ, Johnson AE (2004) Nascent membrane and secretory proteins differ in FRET‐detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116:725–736. [DOI] [PubMed] [Google Scholar]
- 151. Lu J, Deutsch C (2005) Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol 12:1123–1129. [DOI] [PubMed] [Google Scholar]
- 152. Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R (2010) alpha‐Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol 17:313–317. [DOI] [PubMed] [Google Scholar]
- 153. Nilsson OB, Hedman R, Marino J, Wickles S, Bischoff L, Johansson M, Muller‐Lucks A, Trovato F, Puglisi JD, O'Brien EP, Beckmann R, von Heijne G (2015) Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep 12:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kosolapov A, Deutsch C (2009) Tertiary interactions within the ribosomal exit tunnel. Nat Struct Mol Biol 16:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. O'Brien EP, Hsu ST, Christodoulou J, Vendruscolo M, Dobson CM (2010) Transient tertiary structure formation within the ribosome exit port. J Am Chem Soc 132:16928–16937. [DOI] [PubMed] [Google Scholar]
- 156. Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA (2016) Synonymous codons direct cotranslational folding toward different protein conformations. Mol Cell 61:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, Skach WR (2015) Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348:444–448. [DOI] [PubMed] [Google Scholar]
- 158. Kimchi‐Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–528. [DOI] [PubMed] [Google Scholar]
- 159. Hu S, Wang M, Cai G, He M (2013) Genetic code‐guided protein synthesis and folding in Escherichia coli . J Biol Chem 288:30855–30861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y (2013) Non‐optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495:111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y (2015) Codon usage influences the local rate of translation elongation to regulate co‐translational protein folding. Mol Cell 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sauna ZE, Kimchi‐Sarfaty C (2011) Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12:683–691. [DOI] [PubMed] [Google Scholar]
- 163. Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi‐Sarfaty C (2014) Exposing synonymous mutations. Trends Genet 30:308–321. [DOI] [PubMed] [Google Scholar]
- 164. Nissley DA, Sharma AK, Ahmed N, Friedrich UA, Kramer G, Bukau B, O'Brien EP (2016) Accurate prediction of cellular co‐translational folding indicates proteins can switch from post‐ to co‐translational folding. Nat Commun 7:10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Drummond DA, Wilke CO (2008) Mistranslation‐induced protein misfolding as a dominant constraint on coding‐sequence evolution. Cell 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sharma AK, Bukau B, O'Brien EP (2016) Physical origins of codon positions that strongly influence cotranslational folding: a framework for controlling nascent‐protein folding. J Am Chem Soc 138:1180–1195. [DOI] [PubMed] [Google Scholar]
- 167. O'Brien EP, Vendruscolo M, Dobson CM (2014) Kinetic modelling indicates that fast‐translating codons can coordinate cotranslational protein folding by avoiding misfolded intermediates. Nat Commun 5:2988. [DOI] [PubMed] [Google Scholar]
- 168. O'Brien EP, Ciryam P, Vendruscolo M, Dobson CM (2014) Understanding the influence of codon translation rates on cotranslational protein folding. Acc Chem Res 47:1536–1544. [DOI] [PubMed] [Google Scholar]


