DELAY OF GERMINATION18 QTL encodes a pseudophosphatase, involved in seed dormancy regulation.
Abstract
Seed dormancy controls the timing of germination, which regulates the adaptation of plants to their environment and influences agricultural production. The time of germination is under strong natural selection and shows variation within species due to local adaptation. The identification of genes underlying dormancy quantitative trait loci is a major scientific challenge, which is relevant for agricultural and ecological goals. In this study, we describe the identification of the DELAY OF GERMINATION18 (DOG18) quantitative trait locus, which was identified as a factor in natural variation for seed dormancy in Arabidopsis (Arabidopsis thaliana). DOG18 encodes a member of the clade A of the type 2C protein phosphatases family, which we previously identified as the REDUCED DORMANCY5 (RDO5) gene. DOG18/RDO5 shows a relatively high frequency of loss-of-function alleles in natural accessions restricted to northwestern Europe. The loss of dormancy in these loss-of-function alleles can be compensated for by genetic factors like DOG1 and DOG6, and by environmental factors such as low temperature. RDO5 does not have detectable phosphatase activity. Analysis of the phosphoproteome in dry and imbibed seeds revealed a general decrease in protein phosphorylation during seed imbibition that is enhanced in the rdo5 mutant. We conclude that RDO5 acts as a pseudophosphatase that inhibits dephosphorylation during seed imbibition.
Survival of plants depends on their control of seed germination timing because this indirectly determines the conditions during all subsequent phases of the life cycle. Seed dormancy prevents germination under (temporary) favorable conditions and can delay seedling establishment until the onset of the growth season. This makes it an important factor in the adaptation of plants to their local environment. The level of seed dormancy is influenced by environmental conditions experienced by the mother plant and by the seeds during storage in the seed bank, in particular temperature, humidity, light, and nitrate. In Arabidopsis (Arabidopsis thaliana), low temperatures experienced by the mother plant during seed maturation are known to enhance seed dormancy. In contrast, low temperatures during seed imbibition promote germination (Footitt et al., 2011; Kendall et al., 2011; Graeber et al., 2012; He et al., 2014). Changes in dormancy are coupled with an altered balance between the levels of the plant hormones abscisic acid (ABA) and gibberellin (GA), which are also reflected by the transcript levels of genes controlling hormone synthesis and breakdown. ABA is required for the induction of dormancy, whereas GA is necessary for germination. The biosynthesis and signaling pathways of both hormones negatively influence each other (Nambara et al., 2010; Graeber et al., 2012). Members belonging to clade A of the type 2C protein phosphatases (PP2Cs) have important roles in ABA-dependent dormancy by inhibiting the activity of SNF1-related kinases via dephosphorylation (Park et al., 2009).
Arabidopsis grows in large areas of the northern hemisphere and is adapted to different environments. This is reflected in seed dormancy variation between accessions and its underlying genetic variation (Springthorpe and Penfield, 2015). Therefore, it is not surprising that analyses of natural variation in seed dormancy between Arabidopsis accessions yielded several dormancy quantitative trait loci (QTLs; Alonso-Blanco et al., 2003; Bentsink et al., 2010). A major QTL, DELAY OF GERMINATION1 (DOG1), was identified in the progeny of a cross between the low dormant accession Landsberg erecta (Ler) and the highly dormant accession Cape Verde Islands (Cvi). DOG1 has been cloned and turned out to be a conserved key regulator of seed dormancy (Bentsink et al., 2006). The amount of DOG1 protein in freshly harvested seeds showed a strong positive correlation with dormancy levels. Interestingly, DOG1 transcript and protein levels are increased by low temperatures during seed maturation, corresponding with enhanced dormancy levels (Kendall et al., 2011; Nakabayashi et al., 2012).
Here, we report the cloning and analysis of a gene underlying the seed dormancy QTL DOG18, which was identified in crosses of Ler with three other accessions (Bentsink et al., 2010). DOG18 encodes a family member of the PP2C phosphatases named REDUCED DORMANCY5 (RDO5). We had previously identified this gene based on the low dormancy level of its mutant (Xiang et al., 2014). We now show that RDO5 functions as a pseudophosphatase that influences the seed phosphoproteome. DOG18 has extensive sequence variation among accessions and a high frequency of potential loss-of-function alleles with a geographic distribution limited to northwestern Europe. The influence of DOG18 on seed dormancy can be modified by other dormancy QTLs like DOG1 and DOG6, and by the temperature experienced by the mother plant during seed maturation.
RESULTS
Fine-Mapping of DOG18
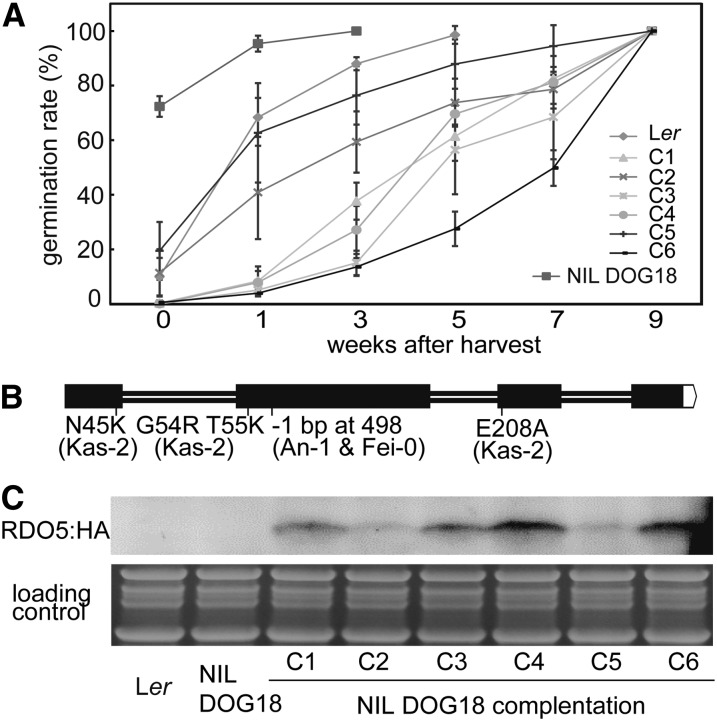
The seed dormancy QTL DOG18 has been identified in recombinant inbred line populations derived from crosses between Ler and the accessions Antwerpen-1 (An-1), Santa Maria da Feira-0 (Fei-0), and Kashmir-2 (Kas-2; Bentsink et al., 2010). DOG18 is a relatively weak QTL in comparison with DOG1 and its Ler allele enhances dormancy in all three populations. DOG18 has been located in a 2-cm region on chromosome 4 (Supplemental Fig. S1A; Bentsink et al., 2010). The Near-Isogenic Line DOG18-Fei-0 (NIL DOG18) contains an introgression of Fei-0 encompassing the DOG18 QTL in the Ler background and shows reduced seed dormancy compared to Ler (Fig. 1A). Fine-mapping of DOG18 using progeny from a cross between NIL DOG18 and Ler narrowed down its location to a 700-Kb region (Supplemental Fig. S1B). A recently cloned dormancy gene, RDO5, is located in this region making it a strong candidate for the gene underlying the QTL (Xiang et al., 2014). A comparison of the RDO5 genomic sequence among the four accessions showed that An-1 and Fei-0 contain an identical 1-bp deletion in the second exon, leading to a frame shift and early STOP codon. The Kas-2 allele of RDO5 still encodes a full-length protein, but has four amino-acid changes in comparison to the Ler allele (Fig. 1B; Supplemental Fig. S1C). Recently, the same gene was also identified to underlie the germination QTL IBO, which was detected in a cross between the accessions Eilenburg-0 and Loch Ness-0 (Lc-0). The Lc-0 allele enhances dormancy and has a, thus far unique, single-base pair change causing an A to E substitution in a highly conserved PP2C motif (Amiguet-Vercher et al., 2015).
Figure 1.
Complementation of DOG18 by RDO5. A, Germination after different periods of dry storage of seeds from Ler, NIL DOG18, and six independent transgenic NIL DOG18 lines containing the RDO5 gene from Ler. Shown are means ± se of six to eight independent batches of seeds for each genotype. B, Gene structure of RDO5 and natural polymorphisms identified in the An-1, Fei-0, and Kas-2 accessions compared to Ler. Exons are shown as black boxes and introns as lines. C, Immunoblot analysis of RDO5:HA protein accumulation in seeds of the six NIL DOG18 complementation transgenic lines from A. Coomassie Brilliant Blue staining was used as a loading control.
To obtain additional evidence that RDO5 is the gene underlying DOG18, complementation experiments were carried out. We transformed NIL DOG18 with a construct containing the RDO5 promoter and gene from Ler, fused with a C-terminal HA tag. Six independent transformants with a single introgression event were selected. All of these showed enhanced dormancy compared to NIL DOG18 (Fig. 1A). Moreover, the amount of RDO5:HA protein in the transformants correlated with their seed dormancy level confirming previous observations (Xiang et al., 2014). Lines2 and 5 showed dormancy levels similar to the Ler accession and contained less RDO5:HA protein in their seeds than the other four transformants. Line6 with a high RDO5:HA abundance showed a high dormancy level (Fig. 1, A and C). Additionally, the Fei-0 allele of RDO5 was introduced into the rdo5-1 mutant, which has very low dormancy levels (Xiang et al., 2014). Three independent homozygous single insertion transformants all showed a complete lack of complementation, indicating that the RDO5 Fei-0 allele is not functional (Supplemental Fig. S1D).
Loss-of-Function RDO5 Alleles Are Frequently Found in Northwestern European Accessions
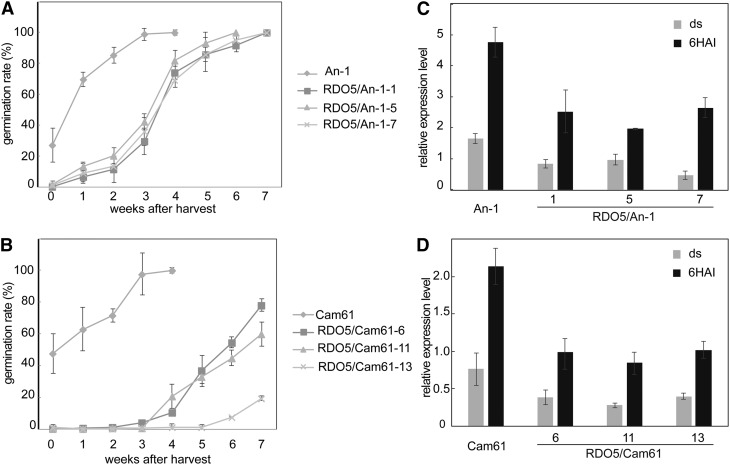
We were interested to find out whether the RDO5 Fei-0/An-1 specific allele is more widespread among Arabidopsis accessions. Therefore, a dCAPs marker was designed for this 1-bp deletion. A population of approximately 360 diverse accessions from the haplotype map collections (Li et al., 2010) was screened and the deletion was found in two additional accessions, Cam61 and Lac-3 (Supplemental Fig. S2). Fei-0 and Lac-3 are moderately dormant, but An-1 and Cam61 have low dormancy levels. We investigated whether the seed dormancy level of An-1 and Cam61 could be enhanced by introducing a genomic fragment containing the functional RDO5 allele from Ler driven by its native promoter. Three independent homozygous single insertion transformants with enhanced RDO5 transcript levels in seeds were obtained for both accessions(Supplemental Fig. 3, A and B). All transformants showed increased dormancy levels compared to the wild-type An-1 and Cam61 accessions (Fig. 2, A and B). We have previously shown that the reduced dormancy phenotype of the rdo5 mutant requires enhanced transcript levels of the Arabidopsis PUMILIO9 (APUM9) gene, which encodes a protein belonging to the conserved PUF family of RNA binding proteins (Xiang et al., 2014). Interestingly, introduction of the wild-type Ler RDO5 gene in the An-1 and Cam61 accessions caused a significant decrease in APUM9 transcript levels in both dry seeds and those imbibed 6 h after inoculation (6HAI; Fig. 2, C and D). This suggests that a reduction in APUM9 transcript levels by introduction of a functional RDO5 gene contributes to the enhanced dormancy levels in these two accessions.
Figure 2.
Complementation of An-1 and Cam61 by RDO5. A and B, Germination after different periods of dry storage of seeds from transgenic lines containing the RDO5 Ler allele in An-1 (A) or Cam61 (B) background. Shown are averages ± se of six to eight independent batches of seeds for each genotype. C and D, qRT-PCR analysis of APUM9 transcript levels in dry and 6HAI seeds in An-1 (C), Cam61 (D), and their transgenic lines. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
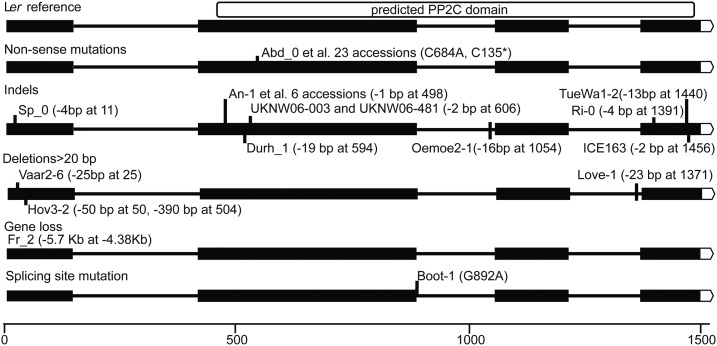
To identify additional alleles of RDO5, we analyzed sequences from nearly 870 accessions of the 1001 Genome Project with a world-wide distribution (http://1001genomes.org). Forty-two of these accessions contained RDO5 alleles that were predicted to have lost their function because of a single-base pair change leading to a STOP codon in the second exon, small deletions causing frame shifts and early STOP codons, large deletions, or a splice site mutation causing a frame shift (Fig. 3; Supplemental Table S1). Most of the accessions with non-sense mutations and indels are predicted to encode truncated proteins with a similar length to the protein encoded by the Fei-0 allele, for which we showed that it has lost its ability to induce dormancy (Supplemental Fig. S1D). Interestingly, nearly all of the 42 accessions were located in a relatively small region of Europe consisting of the United Kingdom, northern France, and southwestern Germany, as well as in coastal regions of Sweden (Fig. 4A). These regions have a mild oceanic climate with evenly dispersed rainfall and relatively small temperature changes during the year. Ri-0 and POG-0 originated from Canada and LP3413.41 originated from the United States. The accession Fei-0 originated from Portugal, but was collected in a very humid environment that is atypical for that region (personal communication, C. Alonso-Blanco). The genetic relationships among the potential loss-of-function RDO5 accessions were determined using a structure analysis, which indicated that the loss-of-function alleles of RDO5 had three different origins (Fig. 4B) and can be divided in three main haplotypes representative for Sweden, western Europe, and the United Kingdom (Supplemental Fig. S4). Accessions Ri-0 and POG-0 clustered with the haplotypes from western Europe and LP3413.41 with the UK, suggesting that these accessions originated from these regions (Fig. 4B). A haplotype network analysis of 132 different accessions, including the 32 predicted loss-of-function RDO5 accessions, revealed the existence of 14 independent haplotypes including one cluster of 22 identical alleles with a single-base pair change causing a STOP codon and a second cluster of three alleles with a 1-bp deletion including Fei-0 and An-1 alleles. In addition, multiple independent deletions causing frame shifts occurred (Fig. 4C). Therefore, independent RDO5 loss-of-function alleles seem to have arisen from different parental alleles.
Figure 3.
Natural loss-of-function mutations in the DOG18 gene. Natural mutations causing predicted DOG18 loss-of-function alleles are divided into five groups that are shown in separate rows. Location of mutations for the individual accessions are indicated. Exons (black boxes) are connected with horizontal lines representing intronic regions of DOG18. The location of the predicted PP2C domain is indicated at the top.
Figure 4.
Geographic distribution, population structure, and haplotype network of accessions containing RDO5 loss-of-function alleles. A, Geographical distribution of 42 accessions harboring RDO5 predicted loss-of-function alleles. B, Population structure of 95 accessions containing functional or predicted loss-of-function RDO5 alleles originating from Sweden, the UK, and western Europe at K = 3. Black stars indicate accessions carrying RDO5 loss-of-function alleles. C, RDO5 haplotype network. Haplotypes are represented by circles with sizes proportional to the number of populations containing that haplotype. Each node represents a single mutation. Black stars indicate accessions or groups carrying RDO5 loss-of-function alleles.
Factors Enhancing Seed Dormancy in Accessions Containing RDO5 Loss-of-Function Alleles
We analyzed seed dormancy in 11 accessions with potential RDO5 loss-of-function alleles and found considerable variation. Several accessions showed high dormancy levels (Fig. 5A), which was not expected considering the very low seed dormancy of the loss-of-function rdo5-1 mutant (Xiang et al., 2014). In addition, we observed extensive variation in dormancy within F2 progenies of crosses between the rdo5-2 mutant (in Col background) and the Fei-0 and Lac-3 accessions that contain RDO5 loss-of-function alleles (Supplemental Fig. S5). These observations indicated that a loss of RDO5 can be compensated by other genetic factors. Besides DOG18, 10 additional QTLs for seed dormancy have been found that could be candidates for these factors; among them, the two strongest are DOG1 and DOG6 (Bentsink et al., 2010).
Figure 5.
Genetic modifiers of the RDO5 phenotype. A, Dormancy level of 11 accessions with predicted loss-of-function RDO5 alleles. Col and Cvi are used as low- and high-dormancy controls. POG-0, HR-10, NFA-8, Vind-1, and NFA-10 belong to the nonsense mutation group; An-1, Fei-0, Lac-3, Cam61, and Durh-1 belong to the Indels group; and Fr-2 belongs to the gene-loss group. Shown are means ± se of six to eight independent batches of seeds for each genotype. DSDS50, days of seed dry storage required to reach 50% germination. B, Immunoblot analysis of DOG1 protein accumulation using a DOG1 antibody in the 11 accessions with predicted loss-of-function RDO5 alleles. A band from Coomassie Brilliant Blue staining was used as a loading control. The bottom panel indicates the DOG1 haplotype of the accessions, where ECCY is a strong allele and DSY is a weak allele (Nakabayashi et al., 2015). C, The correlation of dormancy level and DOG1 protein abundance (quantified from B and normalized by the loading control). D, A strong DOG6 allele from the Shahdara accession enhances dormancy of the rdo5-1 mutant (in Ler background). Germination percentages were determined in freshly harvested seeds. Shown are means ± se of six to eight independent batches of seeds for each genotype. **P < 0.01. E, qRT-PCR analysis of APUM9 transcript levels in Ler, rdo5-1 (Ler background), NIL DOG6, and rdo5-1 NIL DOG6 dry and 6HAI seeds. Inset, APUM9 expression in dry seeds from the different genotypes at a magnified scale. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
To study the role of DOG1 on dormancy in the absence of functional RDO5, DOG1 protein levels were analyzed in a set of RDO5 loss-of-function accessions with varying amounts of seed dormancy. In general, low dormant accessions showed a low abundance of DOG1 protein, whereas highly dormant accessions showed high levels of DOG1. A few exceptions, like Cam61 and Fr-2, showed relatively high DOG1 protein levels with low dormancy levels, whereas HR-10 showed relatively low DOG1 protein level and a high dormancy level (Fig. 5B). A comparison of the DOG1 alleles showed that two of the accessions (An-1 and Fr-2) contained the DSY haplotype, which has a strongly reduced function compared to the ECCY DOG1 haplotype that was present in the other eight accessions (Nakabayashi et al., 2015; Fig. 5B). When we considered the functionality of the DSY DOG1 allele to be one-tenth that of the ECCY DOG1 allele, a positive correlation was found between DOG1 protein level and dormancy level (R2 = 0.53, P = 0.01; Fig. 5C). Therefore, high DOG1 protein accumulation is one possible way for seeds to hide the phenotypic effect of RDO5 loss-of-function alleles.
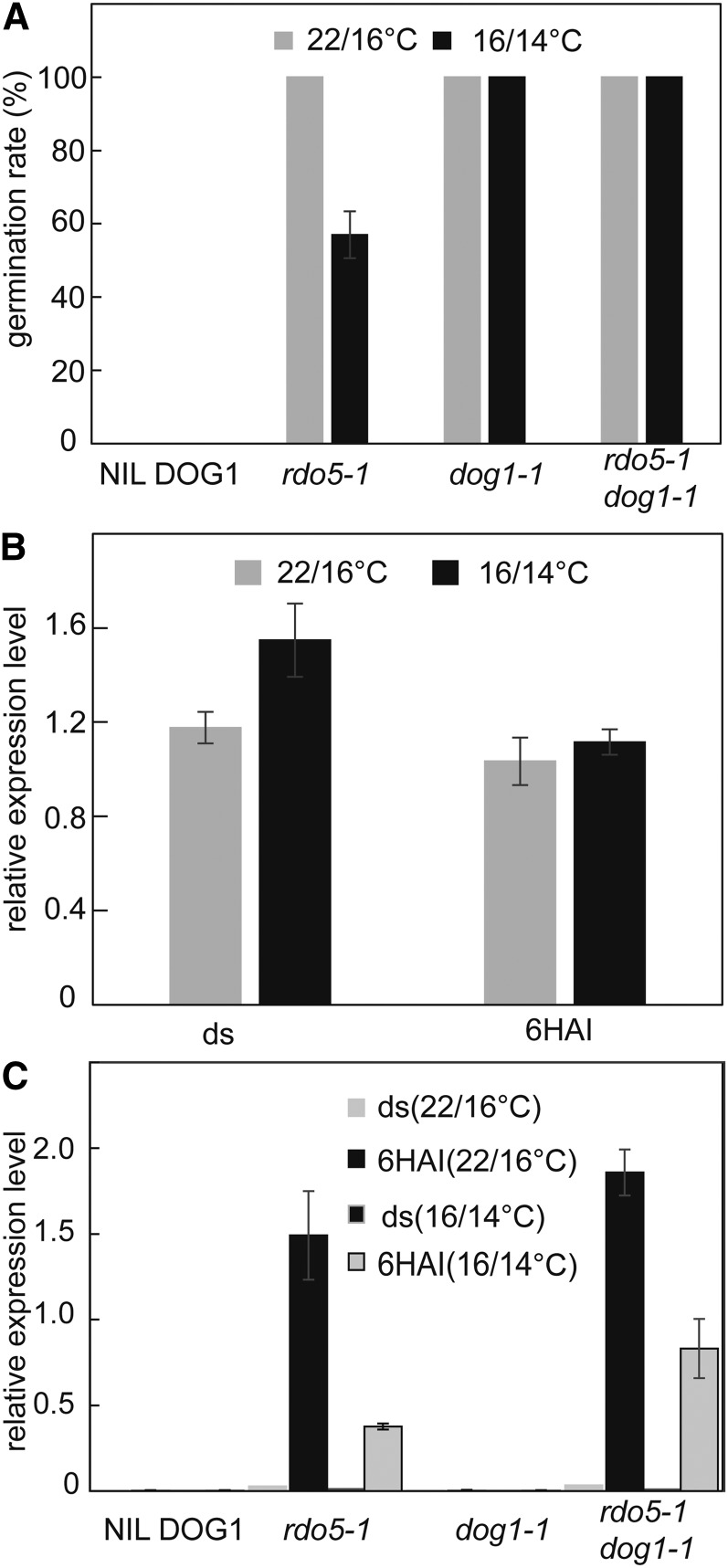
The restriction of natural loss-of-function RDO5 alleles to a relatively small region of northwestern Europe could be related to temperature. We tested germination of the rdo5-1 mutant and wild-type seeds maturing at higher (22/16°C) and lower (16/14°C) temperatures. The rdo5-1 mutant completely lacked dormancy at higher temperatures, but showed increased dormancy at the lower seed maturation temperature (Fig. 6A). This might be caused by enhanced accumulation of DOG1 induced by low temperatures (Kendall et al., 2011; Nakabayashi et al., 2012). Indeed, the enhanced dormancy level of rdo5-1 mutant seeds matured at low temperatures was lost in the dog1-1 mutant background. This indicated that DOG1 is required to enhance seed dormancy at low temperatures in the rdo5 mutant (Fig. 6A). Interestingly, in contrast with DOG1, the transcript levels of RDO5 were not affected by low temperatures (Fig. 6B).
Figure 6.
Maternal temperature affects the RDO5 dormancy phenotype. A, Germination of NIL DOG1, rdo5-1, dog1-1, and rdo5-1 dog1-1 freshly harvested seeds that matured under a d/n regime of 22/16°C or 16/14°C. Shown are means ± se of six to eight independent batches of seeds for each genotype. B, qRT-PCR analysis of RDO5 transcript levels in NIL DOG1 dry and 6HAI seeds that matured under a d/n regime of 22/16°C or 16/14°C. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se. C, qRT-PCR analysis of APUM9 transcript levels in NIL DOG1, rdo5-1, dog1-1, and rdo5-1 dog1-1 dry and 6HAI seeds that matured under a d/n regime of 22/16°C or 16/14°C. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
Because the RDO5 function is associated with decreased APUM9 transcript levels (Xiang et al., 2014), we analyzed APUM9 mRNA levels at both temperature regimes. As previously demonstrated (Xiang et al., 2014), APUM9 expression level is highly increased in the rdo5-1 background, especially in 6HAI seeds. The higher dormancy levels of seeds matured at lower temperatures was associated with reduced APUM9 transcript levels (Fig. 6C). However, the relation between reduced dormancy and enhanced APUM9 transcription was not observed in the dog1-1 mutant background, both in the presence and absence of rdo5-1. This suggests that DOG1 influences dormancy independently of APUM9.
The relationship of RDO5/DOG18 with DOG6 was assessed by combining the rdo5-1 mutant (in a Ler background) with an introgression from the Shahdara accession containing a strong DOG6 allele (Fig. 5D). Analysis of germination of freshly harvested seeds showed that DOG6 can enhance the dormancy level of rdo5-1, indicating an additive relation between DOG6 and RDO5/DOG18. We also analyzed the APUM9 transcript levels in the NIL DOG6 lines. Interestingly, we found that the enhancement in seed dormancy caused by the NIL DOG6 introgression was associated with strongly reduced APUM9 transcript levels, both in the wild type and the rdo5-1 mutant background (Fig. 5E). This indicated that APUM9 is not specific for the RDO5 pathway, but has a more general role in seed dormancy.
RDO5 Functions as a Pseudophosphatase
RDO5 is a member of the PP2C phosphatase family and enhances seed dormancy (Xiang et al., 2014). Although RDO5 has strong homology with PP2Cs, it does not show complete sequence conservation in amino acids essential for phosphatase activity. Amiguet-Vercher et al. (2005) described that, in particular, RDO5 lacks conserved residues for metal coordination and phosphate binding. Interestingly, the D111 residue in RDO5 is a conserved G in active PP2Cs. The replacement of this G to D in the clade A PP2C phosphatases HAB1, ABI1, ABI2 and AHG3 caused loss of activity (Rodriguez et al., 1998; Gosti et al., 1999; Robert et al., 2006; Yoshida et al., 2006). RDO5 also lacks about 40 amino acids in front of the MAPK docking site, which is important for protein interaction (Supplemental Figs. 6 and 7, A and B). Gosti et al. (1999) identified five amino-acid substitutions in this region in ABI1 that led to loss of activity, indicating that this domain is also important for activity. In addition, RDO5 lacks a Trp residue that is involved in the interaction with ABA in most ABA-responsive PP2Cs. This may explain why RDO5 does not affect ABA sensitivity (Xiang et al., 2014).
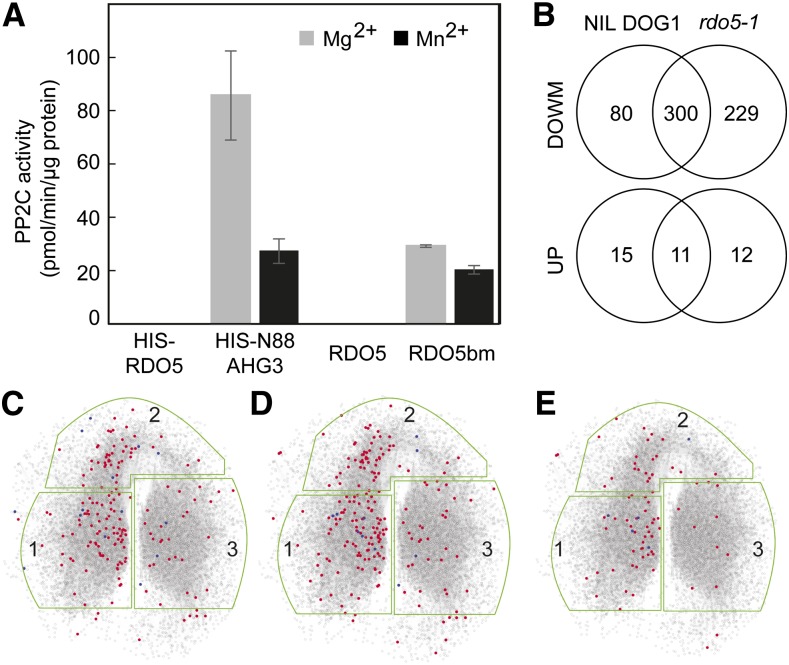
These observations suggest that RDO5 is not a catalytically active protein phosphatase. In accordance, Amiguet-Vercher et al. (2015) demonstrated very low phosphatase activity for RDO5 using in vitro phosphatase assays. We further confirmed these observations and did not detect any phosphatase activity for RDO5 using the synthetic phosphopeptide RRA(phosphoT)VA as substrate in the presence of either Mn2+ or Mg2+ as metal cofactor (Fig. 7A). We were interested whether the phosphatase activity of RDO5 could be restored by introducing back-mutations to recover all the missing residues known to affect PP2C activity (RDO5bm). Introduction of these back-mutations into the RDO5 sequence indeed restored the phosphatase activity of RDO5 (Fig. 7A; Supplemental Fig. S6). RDO5bm displayed a specific activity of 29.1 pmol.min−1.mg−1 and 20.2 pmol.min−1.mg−1 with Mg2+ or Mn2+ as co-factors, respectively. This activity is in a similar range to that of AHG3. These results suggest that RDO5 probably evolved from a functional PP2C phosphatase but has lost its phosphatase activity. Proteins homologous to phosphatases lacking essential amino acids for activity have previously been described as pseudophosphatases or antiphosphatase proteins that might prevent specific residues from becoming dephosphorylated or phosphorylated by protecting them from real phosphatases or kinases (Reiterer et al., 2014). Thus we studied whether RDO5 could be a pseudophosphatase indirectly implied in the control of protein phosphorylation levels by comparing the phosphoproteome of the rdo5-1 mutant with its wild-type NIL DOG1 in dry and 6HAI seeds. A label-free quantitative mass spectrometry analysis on three biological replicates for each genotype and treatment identified 1527 phosphorylation sites in 875 protein groups. Imbibition of seeds caused a general decrease in phosphorylation, which was enhanced in the rdo5 mutant (Fig. 7B).
Figure 7.
RDO5 functions as a pseudophosphatase. A, Phosphatase activity is restored in RDO5 after back-mutations. Phosphatase activity of RDO5 and RDO5 back-mutation (RDO5bm) proteins was measured in vitro using the RRA(phosphoT)VA peptide as a substrate. ΔC-terN88AHG3 was used as a positive control. Data are averages ± se from three replicates. B, Venn diagram analyses showing common and differential distribution of phosphorylated sites identified in NIL DOG1 and rdo5-1 after 6 h imbibition compared with dry seeds. C to E, Localization in the SeedNet network of differentially phosphorylated proteins from wild-type NIL DOG1 (C), rdo5-1 (D), and differentially phosphorylated proteins present in rdo5-1 but not in NIL DOG1 (E) after 6 h imbibition compared with dry seeds. The regions outlined in green correspond to clusters associated with dormancy (region 1) or germination (regions 2 and 3). The red dots represent proteins with decreased phosphorylation levels and the blue dots represent proteins with increased phosphorylation levels.
In the wild-type NIL DOG1 background, 380 phosphorylation sites in 238 proteins were significantly decreased after imbibition for 6 h, whereas 26 phosphorylation sites in 20 proteins increased in phosphorylation status compared to dry seed. In the rdo5-1 mutant, 529 phosphorylation sites in 289 proteins were significantly decreased and 23 phosphorylation sites in 17 proteins increased in phosphorylation status compared with dry seed (Supplemental Tables S2 and S3). In total, we found 229 phosphorylation sites with decreased phosphorylation levels and 12 phosphorylation sites with increased phosphorylation levels in rdo5-1 compared to the wild type (Fig. 7B). These results indicated a general decrease in protein phosphorylation during seed imbibition that is enhanced in the rdo5 mutant. We used SeedNet, a topological model of transcriptional interactions controlling dormancy and germination in Arabidopsis (Bassel et al., 2011), to study the relationship between the transcriptome and protein phosphorylation in imbibed seeds. The majority of differentially phosphorylated proteins 6HAI are located within region 1, which is associated with dormancy, and in a section of region 2 that does not contain germination-associated genes. (Fig. 7, C and D; Bassel et al., 2011). Proteins with altered phosphorylation levels specific in the rdo5 mutant showed the same pattern (Fig. 7E). These results suggest that the regulation of protein phosphorylation plays a role in the release of seed dormancy during early imbibition of seeds. Proteins directly involved in ABA signaling were not found among those with significant changes in phosphorylation level in rdo5-1 seeds compared to the wild-type. This confirmed our previous observations that RDO5 is probably not part of the ABA pathway (Xiang et al., 2014).
RDO5 controls protein phosphorylation levels despite its lack of phosphatase activity. This control might occur by protecting proteins from dephosphorylation during imbibition because we identified more proteins with decreased phosphorylation levels in the rdo5 mutant than in the wild type. Thus, our biochemical and proteomic results suggest that RDO5 acts as a pseudophosphatase.
In contrast to the changes observed in phosphoprotein abundance, from 5017 quantified protein groups only 106 (41 up-regulated and 65 down-regulated with a log2-fold change > 1) were significantly altered in abundance in wild type after 6 h imbibition in comparison to dry seeds. In the rdo5 mutant, 132 proteins groups (48 up-regulated and 84 down-regulated with a log2-fold change >1) were significantly altered upon imbibition (Supplemental Data Set 1). Interestingly, only 29 of these regulated proteins overlapped between both genotypes. A SeedNet analysis showed that differentially expressed proteins are evenly located in the network (Supplemental Fig. S8). Therefore, regulation of protein modifications seems to be more prominent during early seed imbibition compared to regulation of protein levels.
DISCUSSION
Seed dormancy has a high adaptive value for plants. Hence, it is not surprising that extensive natural variation exists for this trait. Exploring and identifying the underlying genes of this variation is important to understand plant adaptation and helpful to study the molecular mechanisms of dormancy. In Arabidopsis, several seed dormancy QTLs have been found. A major dormancy QTL locus, DOG1, was cloned nearly 10 years ago and has been shown to be conserved within the plant kingdom (Ashikawa et al., 2010; Graeber et al., 2010). Here, we report the identification of the underlying gene of a second dormancy QTL, DOG18. DOG18 encodes a member of the type 2C protein phosphatase family that we previously identified as RDO5, which is a positive regulator of seed dormancy (Xiang et al., 2014). Natural variation at this gene has recently been described by the identification of a specific allele, IBO, which has a unique mutation in the Loch Ness accession, probably causing enhanced activity (Amiguet-Vercher et al., 2015).
We identified predicted loss-of-function RDO5 alleles at a relatively high frequency in Arabidopsis accessions from northwestern Europe but could not detect any from other parts of the world (Fig. 4A). Absence of RDO5 function could be an adaptation to the northwestern European climate of mild temperatures with low fluctuations and evenly dispersed rainfall. Interestingly, the non-dormant phenotype of the rdo5-1 loss-of-function allele can be partially rescued when plants are grown at low temperatures. In addition, RDO5 enhances dormancy but is not essential for it, because accessions with loss-of-function RDO5 alleles vary considerably in their dormancy levels (Fig. 5A). This is consistent with the relatively weak effect on seed dormancy of the DOG18 QTL compared with DOG1 and DOG6 (Bentsink et al., 2010). We assume that RDO5 might have a neutral role or could even be under negative selection under specific environmental conditions prevalent in northwestern Europe. This could have led to the occurrence of natural loss-of-function alleles that subsequently spread through the population. Interestingly, a QTL analysis of dormancy in two accessions from north-central Sweden and central Italy identified DOG18 under greenhouse conditions but not in populations grown in the field. This is likely due to the lower temperatures in the field compared to the greenhouse, which mask the role of DOG18 in dormancy (Postma and Ågren, 2015).
We have identified two dormancy QTLs, DOG1 and DOG6, that can partially rescue the rdo5 mutant phenotype. Therefore, these QTLs contribute to explain differences in dormancy level between RDO5 loss-of-function accessions. These results also confirm that different QTLs have additive roles in seed dormancy, as previously suggested by Bentsink et al. (2010). Interestingly, in contrast to DOG18, naturally occurring loss-of-function alleles have never been identified for DOG1. This is consistent with the inability to induce seed dormancy in dog1 mutant plants grown at low temperatures during seed maturation and the absence of loci that enhance dormancy in dog1 loss-of-function alleles.
Despite its similarity to PP2C phosphatases, RDO5 is unlikely to be catalytically active as a phosphatase because it lacks several conserved residues. In accordance, we could not detect any phosphatase activity for RDO5. Amiguet-Vercher et al. (2015) also reported a very low phosphatase activity for IBO/RDO. It is likely that RDO5 has evolved from a functional phosphatase because we could recover its phosphatase activity by introducing amino-acid changes to restore critical residues needed for activity (Fig. 7A; Supplemental Fig. S6). These observations led us to propose that RDO5 could function as a pseudophosphatase. Pseudophosphatases have been described in humans and nematodes. For instance, human TAK-binding protein 1 is a member of the PPM family of protein phosphatases, which does not show phosphatase activity. TAK-binding protein1 possibly functions by binding to and controlling the accessibility of phosphorylated residues on a kinase protein or its downstream substrates, thereby regulating pro-inflammatory signaling pathways (Conner et al., 2006). Pseudophosphatases, as well as pseudokinases, are increasingly seen as regulators of signaling pathways that can act as modulators, competitors, or anchors of kinases and phosphatases (Reiterer et al., 2014).
Our comparison of the phosphoproteomes between wild-type NIL DOG1 and the rdo5 mutant showed that RDO5 influences the phosphorylation status of hundreds of proteins, despite its lack of phosphatase activity. This is in agreement with its function as a pseudophosphatase. We suggest that RDO5 may bind to its target proteins to prevent them from dephosphorylation by other active protein phosphatases. Such a role for RDO5 would suggest an opposite function compared to other phosphatases. This hypothesis is supported by the positive role of RDO5 in seed dormancy (Xiang et al., 2014), which is in contrast with the negative roles of clade A PP2Cs like ABI1, ABI2, AHG1, and HAI2 (Rodriguez et al., 1998; Gosti et al., 1999; Nishimura et al., 2007; Kim et al., 2013). Amiguet-Vercher et al. (2015) recently proposed a role for a unique natural allele of RDO5/IBO (from the Lc-0 accession) in ABA signaling because of its inhibitory influence on ABI phosphatase activity. However, our phosphoproteome analysis did not give support for a general role of RDO5 in the ABA pathway because proteins involved in ABA signaling were not found among those with significant changes in phosphorylation level in rdo5-1 mutant seeds. In addition, mutations in RDO5 neither affected ABA levels in dry and imbibed seeds nor ABA sensitivity during germination (Xiang et al., 2014). We observed a massive dephosphorylation during seed imbibition, which was enhanced in the rdo5 mutant. Therefore, RDO5 probably controls seed dormancy by preventing dephosphorylation during seed imbibition. Interestingly, changes in protein levels were relatively minor during imbibition compared to changes in phosphorylation levels.
We have previously identified the mRNA binding protein APUM9 to act downstream of RDO5 (Xiang et al., 2014). In this work, we identified a general role for APUM9 in seed dormancy that is not restricted to the RDO5 pathway. Factors that enhance dormancy like low seed maturation temperatures and the DOG6 QTL reduce APUM9 transcript levels (Figs. 6C and 5E). Therefore APUM9 might be a common downstream factor of these dormancy pathways. However, APUM9 is probably not part of the DOG1 dormancy pathway because its transcript levels are not affected by DOG1 (Fig. 6C). This again confirms regulation of dormancy by independent mechanisms.
The first two identified dormancy QTLs, DOG1 and RDO5, are both only expressed in seeds and their mutants exclusively show dormancy and germination defects without pleiotropic phenotypes (Bentsink et al., 2006; Xiang et al., 2014). It is probably no coincidence that these two modifiers of natural variation for seed dormancy are seed-specific. Sequence polymorphisms leading to changes in the expression levels of these genes or their protein function represent an excellent way for plants to adapt their dormancy levels to local environments without simultaneously altering other traits. The properties of DOG1 and RDO5 make them highly suitable target genes to manipulate and control seed dormancy levels in crop plants.
CONCLUSION
Germination timing is an important adaptive trait for plants and is controlled by seed dormancy. Arabidopsis accessions collected in nature show a high variation for dormancy, making it a typical quantitative trait. Finding the genes underlying dormancy quantitative trait loci is a major scientific challenge, which is relevant for agricultural and ecological goals. Here we identified the gene RDO5 to underlie the dormancy QTL DOG18. We show that the RDO5 protein can function as a pseudophosphatase. We found a relatively high number of predicted natural loss-of-function alleles for RDO5 that were restricted to northwestern Europe. The loss of RDO5 function could be compensated for by low temperatures and by strong alleles of the dormancy QTLs DOG1 and DOG6.
MATERIALS AND METHODS
Plant Materials
NIL DOG6 and NIL DOG18 were a gift from Leónie Bentsink (Wageningen University, Wageningen, The Netherlands). RDO5 loss-of-function accessions were obtained from the collection of Maarten Koornneef (Max Planck Institute for Planting Breeding Research, Cologne, Germany) and originated from the Nottingham Arabidopsis Stock Centre (http://www.arabidopsis.info/). The rdo5-1 mutant was described by Xiang et al. (2014) and has been obtained in a NIL DOG1 background consisting of the Ler accession with an introgression on chromosome 5, containing the DOG1 gene from Cvi (Alonso-Blanco et al., 2003). The rdo5-1 mutant allele in Ler background was obtained by crossing rdo5-1 with Ler and selection against the Cvi introgression fragment in the progeny. Germination tests were performed as described previously (Xiang et al., 2014). Information of primers used in this study can be found in Supplemental Table S4.
DOG18 Map-Based Cloning and Complementation Analysis
DOG18 mapping was performed by back-crossing NIL DOG18-Fei-0 with Ler. Genotyping F2 plants with cleaved-amplified polymorphic sequence markers narrowed the DOG18 interval into a 700-kb region on chromosome 4, which included the RDO5 gene. For NIL DOG18-Fei-0 complementation, a genomic fragment, including a 2.7-kb promoter sequence and the RDO5 coding sequence fused with the HA tag, was amplified from Ler using gene-specific primers (the HA tag was designed in the reverse primer), cloned into the pDONR207 vector, and transferred into pFASTR01 (Shimada et al., 2010). The binary construct was introduced by electroporation into Agrobacterium tumefaciens strain GV3101, which was subsequently used for transformation by floral dipping (Clough and Bent, 1998). T3 homozygous lines containing single insertion events were used for expression level detection and phenotyping.
Analysis of DOG18 Loss-of-Function Natural Variants
The RDO5 sequence from 855 available accessions was downloaded from Salk Arabidopsis thaliana 1001 Genomes. The different types of loss-of-function RDO5 alleles were confirmed by Sanger sequencing including non-sense mutations (POG-0, HR-10, NFA-8, NFA-10, and Vind-1), Indels (Cam61, Lac-3, An-1, Fei-0, and Durh-1), deletions (Vaar2-6), gene loss (Fr-2), and splicing site mutations (Boot-1). The whole RDO5 cDNA was sequenced in the accessions of the last four types to confirm the absence of mutations that might restore RDO5 function. For the structure analysis, population structure was inferred using model-based clustering algorithms implemented in the software Structure (http://pritchardlab.stanford.edu/structure_software/release_versions/v2.3.4/html/install.html), based on 149 genome-wide SNP markers (Lewandowska-Sabat et al., 2010; Platt et al., 2010), using the haploid setting and running 20 replicates with 50,000 and 20,000 Markov chain Monte Carlo iterations of burn-in and after-burning length, respectively (Hubisz et al., 2009). To determine the K number of significantly different genetic clusters, we applied the ΔK method in combination with the absolute value of ln P (X|K; Evanno et al., 2005) implanted in Structure Harvester (Earl and vonHoldt, 2012). For haplotype network analysis, the RDO5 nucleic acid sequence was translated into an amino acid sequence according to the standard genetic codes basing on the TAIR 10 ORF. The haplotype network of RDO5 was constructed using a Network 4.6 approach that implements a reduced median method (Bandelt et al., 1995).
RNA extraction, RT-PCR, protein extraction, and western blotting were performed as described in Xiang et al. (2014).
Three-Dimensional Protein Modeling
A three-dimensional model of full-length RDO5 sequence (UniProtKB: Q9T010 295 amino acids) was constructed using Swiss-Model software (http://swissmodel.expasy.org/; Arnold et al., 2006). The crystal structure of probable protein phosphatases from rice (UniProtKB: Q0JLP9/PDB: 4OIC chain B) was used as template. The model was superimposed on the crystal structure of the complex HAB1/SnRk2,6 (PDB: 3UJG chain B; Soon et al., 2012) using PDBeFold (http://www.ebi.ac.uk/msd-srv/ssm/).
Protein Purification and PP2C Activity Assay
The coding sequences of RDO5 from Ler and Fei-0 were amplified from cDNA using gene-specific primers. The full length of AHG3 was poorly soluble in Escherichia coli; therefore, an N-terminal-deleted AHG3 (ΔNter-N88AHG3) protein was cloned using gene-specific primers. N-terminal-deleted PP2Cs are reported to be active (Dupeux et al., 2011). RDO5 Ler and ΔNter-N88AHG3 coding sequences were introduced in a pDEST17 expression vector to generate an expression clone with proteins fused with a 6×His tag in the N terminus. For back-mutation (RDO5bm), mutated RDO5 sequences were synthesized by Life Technologies. To improve solubility of the back-mutated RDO5 protein, we generated a construct where RDO5bm is fused with HIS:MBP:TEV in the N-terminal. For this purpose, the synthesized sequence was elongated by PCR using gene-specific primers to introduce an N-terminal fusion TEV cleavage site. The PCR product was recombined in pDONR207 and subsequently in pDEST-HIS-MBP (Addgene), then constructs were introduced into a BL21-plyss strain. Fusion proteins were induced by addition of 0.3 mm IPTG in the medium. After overnight culture at 23°C, cells were harvested by centrifugation and pellets were stored at −80°C until purification. Pellets were resuspended in 50 mM HEPES (pH 7.5), 500 mm NaCl, 10% (v/v) glycerol, 5 mm DTT, 1 mm PMSF, and 25 mm imidazole, and cells were disrupted by sonication. The soluble fraction was obtained after centrifugation and applied on a 1-mL HisTrap column (GE Healthcare). The purification process was performed and monitored using ÄKTAprime and Primeview software (GE Healthcare).
Elution was performed with the resuspension buffer supplemented with 500 mm imidazole. Imidazole was removed from purified protein by gel filtration on PD-10 columns (GE Healthcare) and the protein fraction was concentrated to approximately 2 mg⋅mL−1 and stored at −80°C. Purified His:MBP:TEV-tagged recombinant protein was incubated with HIS:TEV protease and subsequently applied on Ni-NTA agarose (Qiagen) to remove the majority of the His:MBP:TEV tag and HIS:TEV protease. The purity of purified proteins was assessed by SDS-PAGE (Supplemental Fig. S7C). Phosphatase activity was measured using the RRA(phosphoT)VA peptide as a substrate. The Ser/Thr Phosphatase Assay system (Promega) was used for the phosphopeptide assay. Phosphatase assays were performed in 50-μL volumes in half-area, flat-bottom 96-well plates containing 50 mm HEPES buffer (pH 7.5), 10 mm MnCl2 or MgCl2, 1 mm DTT, 0.5 μm purified recombinant PP2C proteins, and 100 μm peptide substrate. Reactions were performed at 30°C for 30 min and stopped by addition of 50 µL of molybdate dye solution. Absorbance was read at 630 mm on a Multiscan Spektrum 96-well plate reader (Thermo Fisher Scientific).
Proteolytic Digestion and Desalting
Total protein was extracted in buffer containing 100 mm Tris-HCl (pH 7.5), 3% SDS, 100 mm DTT, and 1× protease inhibitor cocktail (Phosphatase Inhibitor Cocktail 2 and 3; Sigma-Aldrich). Seed protein extract (2 mg) was processed using the FASP method (Wiśniewski et al., 2009) as described in detail by Hartl et al. (2015). Cysteines were alkylated by incubation with 55 mm chloroacetamide in UA buffer (8 m urea and 0.1 m Tris/HCl, pH 8.5) for 30 min at room temperature in the dark followed by three washing steps with UA buffer. Incubation with LysC in UA buffer (1:50 enzyme/protein) was performed overnight at room temperature. The sample was diluted with ABC buffer (50 mm ammonium bicarbonate) to a final urea concentration of 2 m urea. Trypsin digestion (1:100 enzyme/protein) was performed for 4 h at room temperature. The sample was passed through the filter by centrifugation at 14,000g for 10 min and residual peptides eluted with 50 μL ABC. Formic acid was added to a final concentration of 0.5% and the sample was desalted using Sep-Pak SPE 1 cc/100 mg (Waters). Solvent was passed through by gravity flow. Columns were conditioned by successive addition of methanol and buffer B (80% ACN [acetonitrile], 0.5% FA [formic acid], and buffer A [0.5% FA]). The sample was loaded and washed three times with buffer A and eluted twice with buffer B. Eluates were concentrated in a centrifugal evaporator to remove acetonitrile. One-tenth of the eluted peptides were processed for whole proteome analysis, and the rest was subjected to titanium dioxide enrichment. For total proteome analysis, peptides were desalted and prefractionated before liquid chromatography-tandem mass spectrometry into three fractions using Empore Styrenedivenylbenzene Reversed Phase Sulfonate material (SDB-RPS; 3M), as described in detail by Kulak et al. (2014).
Phosphopeptide Enrichment
Phosphopeptides were enriched with a modified version of the protocol of Melo-Braga et al. (2015). Samples were diluted to 900 μL with 50% ACN, 1% TFA, and then a 100-μL loading buffer of 200 mg/mL DHB, in 50% ACN, 1% TFA, was added. Incubation steps were performed under gentle agitation at room temperature. Six milligrams of TiO2 beads (5 mm; GL Sciences) were suspended in 40-μL loading buffer (20 mg/mL DHB, 50% ACN, and 1% TFA) and incubated for 15 min. The first half of the suspension was added to the sample and incubated for 15 min. For removal of liquid, beads were pelleted by centrifugation at 3,000 rpm for 1 min. The supernatant was transferred to the second half of the beads suspension and incubated for 15 min. TiO2 beads from both incubations with bound phosphopeptides were resuspended with loading buffer and combined on C8 StageTips (Empore; 3M). Liquid was passed though the tips by centrifugation at 800g. The beads were washed twice with 80% ACN and 1% TFA and twice with 10% ACN and 0.1% TFA. Phosphopeptides were eluted three times with 5% ammonia and once with 30% ACN by centrifugation at 300g. Eluates were combined and concentrated in a centrifugal evaporator to remove ammonia. Samples were acidified by adding 1:10 volume of 20% ACN and 10% TFA and desalted with StageTips (Empore C18; 3M) as described by Rappsilber et al. (2007).
LC-MS/MS Data Acquisition
Dried peptides were redissolved in 2% ACN, 0.1% TFA for analysis and adjusted to a final concentration of 0.18 μg/μL. Samples were analyzed using an EASY-nLC 1000 system (Thermo Fisher Scientific) coupled to a Q Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were separated on 16-cm frit-less silica emitters (0.75 μm i.d.; New Objective), packed in house with reversed-phase ReproSil-Pur C18 AQ 3-μm resin (Dr. Maisch). Peptides (1 μg) were loaded on the column and eluted for 120 min using a segmented linear gradient of 0 to 95% solvent B (solvent A 5% ACN, 0.5% FA; solvent B 100% ACN, 0.5% FA) at a flow rate of 250 nL/min. Mass spectra were acquired in data-dependent acquisition mode with a TOP15 method. MS spectra were acquired in the Orbitrap analyzer (Thermo Fisher Scientific)) with a mass range of 300 to 1750 m/z at a resolution of 70,000 FWHM and a target value of 3 × 106 ions. Precursors were selected with an isolation window of 1.3 m/z. Higher energy collisional dissociation fragmentation was performed at a normalized collision energy of 25. MS/MS spectra were acquired with a target value of 105 ions at a resolution of 17,500 FWHM and a fixed first mass of m/z 100. Peptides with a charge of +1, greater than 6, or with an unassigned charge state were excluded from fragmentation for MS2; dynamic exclusion for 30 s prevented repeated selection of precursors.
MS Data Analysis
Raw data were processed using MaxQuant software (v. 1.5.3.12; http://www.maxquant.org/) with label-free quantification enabled (Cox et al., 2014). MS/MS spectra were searched by the Andromeda search engine against the Arabidopsis TAIR10_pep_20101214 database (ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/). Sequences of 248 common contaminant proteins and decoy sequences were automatically added during the search. Trypsin specificity was required and a maximum of two missed cleavages allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of Cys residues was set as fixed, and oxidation of Met and protein N-terminal acetylation as variable modifications. Phosphorylation of Ser, Thr, and Tyr were added as variable posttranscriptional modifications only for the TiO2-enriched samples. Peptide-spectrum matches and proteins were retained if they were below a false discovery rate of 1%. Subsequent quantitative statistical analyses were performed in Perseus (v. 1.5.2.6; MaxQuant). Identified proteins and phosphorylation sites were processed separately as follows: Hits were only retained if they were quantified in at least two of three replicates in any of the four conditions. Label-free quantification intensities (proteins) and peptide intensities (phosphorylation sites) were log2-transformed. Two-sample t tests were performed with a P value of 1% as cutoff. Log2 ratios were calculated by replacing missing intensity values with zero.
Distribution of Proteins in the SeedNet Network
Differently phosphorylated or expressed proteins were submitted with the software Cytoscape (http://www.cytoscape.org/) to load SeedNet network data files, which are publicly available at http://www.vseed.nottingham.ac.uk.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RDO5 (At4G11040), DOG1 (AT5G45830), APUM9 (At1G35730), and ACT8 (At1G49240).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Map-based cloning of DOG18.
Supplemental Figure S2. DOG18 Fei-0 allele screening.
Supplemental Figure S3. RDO5 transcript level detection in transgenic lines.
Supplemental Figure S4. Structure estimation of populations for K ranging from 1 to 14 by ΔK-values.
Supplemental Figure S5. Germination of rdo5 segregating populations.
Supplemental Figure S6. RDO5 sequence analysis.
Supplemental Figure S7. RDO5 topology structure analysis and SDS-PAGE gel of purified proteins.
Supplemental Figure S8. Localization of differently expressed proteins in the SeedNet network after 6-h imbibition in wild-type NIL DOG1-, rdo5-1-, and potential RDO5-specific targets.
Supplemental Table 1. Information regarding 42 loss-of-function DOG18 accessions including mutation type, accession name, ID, country, origin, habitat, and latitude and longitude of collection site.
Supplemental Table 2. Overview of quantitative mass spectrometry results.
Supplemental Table 3. Overview of regulated phosphosites and phosphorylated protein groups.
Supplemental Table 4. Primers used in this study.
Supplemental Data Set 1. Mass spectrometry results from the phosphoproteome analysis of rdo5-1 and NIL-DOG1.
Supplementary Material
Acknowledgments
We thank Leónie Bentsink and Maarten Koornneef for providing seed material and Christina Philipp and Anne Harzen for technical assistance. We also thank Carlos Alonso Blanco, William Hughes, and Maarten Koornneef for critical reading of the manuscript.
Glossary
- QTL
quantitative trait locus
Footnotes
Articles can be viewed without a subscription.
References
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiguet-Vercher A, Santuari L, Gonzalez-Guzman M, Depuydt S, Rodriguez PL, Hardtke CS (2015) The IBO germination quantitative trait locus encodes a phosphatase 2C-related variant with a nonsynonymous amino acid change that interferes with abscisic acid signaling. New Phytol 205: 1076–1082 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Ashikawa I, Abe F, Nakamura S (2010) Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci 179: 536–542 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conner SH, Kular G, Peggie M, Shepherd S, Schüttelkopf AW, Cohen P, Van Aalten DMF (2006) TAK1-binding protein 1 is a pseudophosphatase. Biochem J 399: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Antoni R, Betz K, Santiago J, Gonzalez-Guzman M, Rodriguez L, Rubio S, Park SY, Cutler SR, Rodriguez PL, Márquez JA (2011) Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol 156: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361 [Google Scholar]
- Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE (2011) Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA 108: 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Müller K, Wunchova A, Rott A, Leubner-Metzger G (2010) Cross-species approaches to seed dormancy and germination: conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol Biol 73: 67–87 [DOI] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35: 1769–1786 [DOI] [PubMed] [Google Scholar]
- Hartl M, König A-C, Finkemeier I (2015) Identification of lysine-acetylated mitochondrial proteins and their acetylation sites. Methods Mol Biol 1305: 107–121 [DOI] [PubMed] [Google Scholar]
- He H, de Souza Vidigal D, Snoek LB, Schnabel S, Nijveen H, Hilhorst H, Bentsink L (2014) Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J Exp Bot 65: 6603–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall SL, Hellwege A, Marriot P, Whalley C, Graham IA, Penfield S (2011) Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23: 2568–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lee Y, Park J, Lee N, Choi G (2013) HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol 54: 555–572 [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324 [DOI] [PubMed] [Google Scholar]
- Lewandowska-Sabat AM, Fjellheim S, Rognli OA (2010) Extremely low genetic variability and highly structured local populations of Arabidopsis thaliana at higher latitudes. Mol Ecol 19: 4753–4764 [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Braga MN, Ibáñez-Vea M, Larsen MR, Kulej K (2015) Comprehensive protocol to simultaneously study protein phosphorylation, acetylation, and n-linked sialylated glycosylation. Methods Mol Biol 1295: 275–292 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Bartsch M, Ding J, Soppe WJJ (2015) Seed dormancy in Arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet 11: e1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R, Seo M, Soppe WJJ (2012) The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24: 2826–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20: 55–67 [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. (2010) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma FM, Ågren J (2015) Maternal environment affects the genetic basis of seed dormancy in Arabidopsis thaliana. Mol Ecol 24: 785–797 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Reiterer V, Eyers PA, Farhan H (2014) Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol 24: 489–505 [DOI] [PubMed] [Google Scholar]
- Robert N, Merlot S, N’guyen V, Boisson-Dernier A, Schroeder JI (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett 580: 4691–4696 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springthorpe V, Penfield S (2015) Flowering time and seed dormancy control use external coincidence to generate life history strategy. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Mann M (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res 8: 5674–5678 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ (2014) Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26: 4362–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.