The three CBF genes are essential for freezing tolerance and affect seedling development and salt tolerance.
Abstract
The three tandemly arranged CBF genes, CBF1, CBF2, and CBF3, are involved in cold acclimation. Due to the lack of stable loss-of-function Arabidopsis (Arabidopsis thaliana) mutants deficient in all three CBF genes, it is still unclear whether the CBF genes are essential for freezing tolerance and whether they may have other functions besides cold acclimation. In this study, we used the CRISPR/Cas9 system to generate cbf single, double, and triple mutants. Compared to the wild type, the cbf triple mutants are extremely sensitive to freezing after cold acclimation, demonstrating that the three CBF genes are essential for cold acclimation. Our results show that the three CBF genes also contribute to basal freezing tolerance. Unexpectedly, we found that the cbf triple mutants are defective in seedling development and salt stress tolerance. Transcript profiling revealed that the CBF genes regulate 414 cold-responsive (COR) genes, of which 346 are CBF-activated genes and 68 are CBF-repressed genes. The analysis suggested that CBF proteins are extensively involved in the regulation of carbohydrate and lipid metabolism, cell wall modification, and gene transcription. Interestingly, like the triple mutants, cbf2 cbf3 double mutants are more sensitive to freezing after cold acclimation compared to the wild type, but cbf1 cbf3 double mutants are more resistant, suggesting that CBF2 is more important than CBF1 and CBF3 in cold acclimation-dependent freezing tolerance. Our results not only demonstrate that the three CBF genes together are required for cold acclimation and freezing tolerance, but also reveal that they are important for salt tolerance and seedling development.
Low temperatures limit the geological distribution and growing season of plants. The freezing tolerance of Arabidopsis (Arabidopsis thaliana) and other plants from temperate regions can be increased by a prior exposure to low, nonfreezing temperatures, a process known as cold acclimation (Guy, 1990; Thomashow, 1999; Chinnusamy et al., 2003; Knight and Knight 2012). Cold acclimation is a complex process that involves extensive physiological, biochemical, and metabolic changes (Chinnusamy et al., 2007; Medina et al., 2011). These include changes in the stability of membranes and cytoskeleton (Nishida and Murata, 1996; Thalhammer and Hincha, 2014) and the production of cryoprotective proteins and of low Mr cryoprotectants, such as Suc, raffinose, and Pro (Thomashow, 1999; Cook et al., 2004; Guy et al., 2008). Most of these changes in response to low temperatures are the result of cold-responsive gene expression (Cook et al., 2004; Hannah et al., 2005; Maruyama et al., 2009).
The molecular basis of cold acclimation has been extensively studied over the last two decades. In Arabidopsis, three C-repeat binding factors (CBFs), including CBF1, CBF2, and CBF3 (also known as DREB1b, DREB1c, and DREB1a, respectively), play important roles in cold acclimation (Stockinger et al., 1997; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Medina et al., 1999; Gilmour et al., 2004). When Arabidopsis plants are exposed to low temperature (4°C), CBF genes are rapidly and transiently induced and generally reach their maximal level of expression after 1 to 2 h of cold treatment (Gilmour et al., 1998; Zarka et al., 2003; Novillo et al., 2004). CBF genes encode AP2/ERF (APETALA2/Ethylene-Responsive Factor)-type transcription factors that specifically bind to the C-repeat (CRT)/dehydration-responsive element (DRE; G/ACCGAC) and regulate the expression of downstream cold-responsive (COR) genes (Stockinger et al., 1997; Liu et al., 1998; Sakuma et al., 2002). An analysis of gene expression data sets derived from low temperature-treated plants detected 2,637 COR genes, of which 172 were induced or repressed by overexpression of at least one CBF gene (Park et al., 2015). Among these 172 COR genes, which were designated as CBF regulon genes, 133 are CBF-activated and 39 are CBF-repressed. The number of CBF regulon genes accounts for only 6.5% of the total number of COR genes, which suggests that CBF proteins regulate only a small portion of COR genes and that other transcription factors are also involved in the regulation of COR genes. Park et al. (2015) recently found that, in parallel with CBF genes, 27 other “first-wave” transcription factor genes were also highly up-regulated (>2-fold) at an early stage of cold treatment. Analysis of gene expression in transgenic plants overexpressing 11 of these first-wave transcription factors identified five proteins (HSFC1, ZAT12, ZF, ZAT10, and CZF1) that are also involved in the regulation of COR genes; more importantly, some COR genes were found to be coordinately regulated by two or more first-wave transcription factors (Vogel et al., 2005; Park et al., 2015; Zhao et al., 2015). These results suggest that COR genes are regulated by a complex low-temperature regulatory network.
Since the three CBF proteins show very high sequence similarity (86% identity), the extent of functional redundancy of CBF proteins is an intriguing question. An analysis of the CBF regulon genes controlled by each of the three CBF proteins showed that they regulate very similar gene sets (Park et al., 2015), suggesting that the three CBF proteins are redundant in the regulation of COR genes. However, distinct functions for the three CBF genes have also been implicated. First, the expression patterns of CBF1 and CBF3 differed from that of CBF2 during development and in response to low temperature (Novillo et al., 2007). At early stages of development, CBF1 and CBF3 genes were specifically expressed in roots, hypocotyls, and cotyledons, whereas CBF2 was expressed in hypocotyls, cotyledons, and in the first and second pairs of leaves, but not in roots. When plants were exposed to low temperatures, all three CBF genes were expressed in leaves, sepals, and siliques, but CBF2 was also expressed in stems (Novillo et al., 2007). Second, CBF1 and CBF3 reach their expression peak at around 1 h after low-temperature treatment. However, it takes approximately 2 h for CBF2 gene to reach its maximal expression level (Novillo et al., 2004). Moreover, a cbf2 T-DNA insertion mutant showed increased tolerance to freezing before and after cold acclimation, which is largely due to increased expression of CBF1 and CBF3 in the cbf2 mutant. These results suggested that CBF2 is a negative regulator of CBF1 and CBF3 expression (Novillo et al., 2004). How CBF2 may negatively regulate the expression of CBF1 and CBF3, however, is still unknown. In contrast, CBF1 or CBF3 RNAi lines and CBF1/CBF3 antisense lines were defective in cold acclimation. In these RNAi and antisense lines, the expression of CBF2 was not affected, suggesting that CBF1 and CBF3 are not involved in regulating the expression of other CBF genes (Novillo et al., 2007). Together, these results indicate that the functions of CBF1 and CBF3 in freezing tolerance are more similar to each other than to that of CBF2.
To date, researchers have only reported a T-DNA insertion line of cbf2 (T-DNA was inserted in the promoter region of CBF2), RNAi lines of CBF1 and CBF3, and antisense lines of CBF1/CBF3 (Novillo et al., 2004, 2007). The lack of additional, stable, loss-of-function cbf mutants, especially a cbf triple mutant, limits our ability to fully understand the roles of the three CBF genes in freezing tolerance and to assess the functional redundancy among the three CBF proteins. The three CBF genes are linked in tandem on chromosome 4 of Arabidopsis, which makes it almost impossible to obtain cbf double and triple mutants using traditional genetic methods. Obtaining these mutants is now possible, however, with the newly developed CRISPR/Cas9 technology. The CRISPR/Cas9 system can be used to efficiently edit target genes in a variety of organisms, including plants (Gaj et al., 2013; Mao et al., 2013). A multiplex CRISPR/Cas9 system was recently developed that allows the coexpression of multiple single-guide RNA (sgRNA) modules in one binary vector, which consequently enables the simultaneous edition of multiple genes (Zhang et al., 2015).
In this study, we used the CRISPR/Cas9 system to generate cbf single, double, and triple mutants. We analyzed the phenotypes of the cbf triple mutants in terms of development, freezing tolerance, chilling tolerance, and tolerances to other abiotic stresses. Using RNA-seq data derived from low temperature-treated wild-type plants and the cbf triple mutants, we analyzed the COR genes that are regulated by CBF proteins. Furthermore, by comparing cbf single, double, and triple mutants, we investigated the extent of redundancy of the three CBF proteins in freezing tolerance and regulation of COR genes.
RESULTS
Generation of cbf Triple Mutants
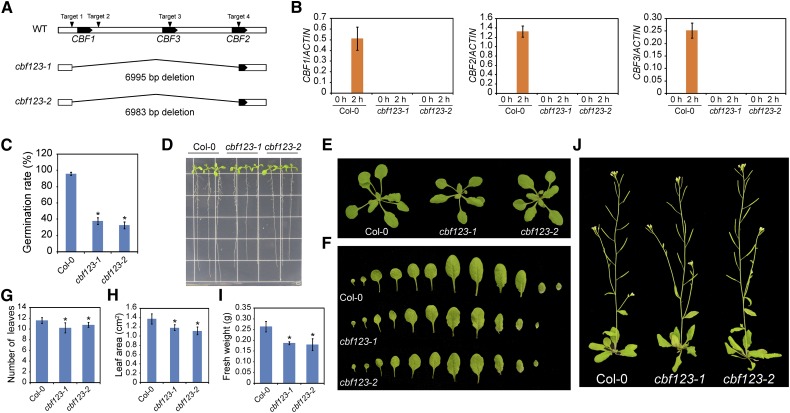
To generate cbf triple mutants, we explored the CRISPR/Cas9 system. Four sgRNAs targeting the promoter region and 3′ untranslated region (UTR) of CBF1 and the coding sequence (CDS) regions of CBF2 and CBF3 were designed and ligated in one binary vector, which was transformed into wild-type Col-0 to edit CBF genes (Fig. 1). In the T1 generation, approximately 40 hygromycin-resistant plants were individually collected. In the T2 generation of these plants, we screened for mutants that showed deletion of all three CBF genes by using PCR and Sanger sequencing. We identified two independent mutant lines, named cbf123-1 and cbf123-2, in which CBF1, CBF3, and partial CDS region of CBF2 were deleted. In cbf123-1, the sequence starting from the promoter region (216 bp upstream of ATG) of CBF1 to the 290 bp of the CDS region of CBF2 was deleted. For cbf123-2, the sequence starting from the promoter region (227 bp upstream of ATG) of CBF1 to the 290 bp of the CDS region of CBF2 was deleted (Fig. 1A). To confirm the deletion of the three CBF genes in the cbf triple mutants, we examined their transcript levels. After 2 h low-temperature (4°C) treatment, the expression of the three CBF genes was highly up-regulated in wild-type plants but was not induced in either of the cbf triple mutant lines (Fig. 1B), indicating that all three CBF genes were abolished in the cbf triple mutants.
Figure 1.
Growth phenotypes of the cbf triple mutants generated using the CRISPR/Cas9 system. A, Diagram showing that four sgRNAs targets were used in the CRISPR/Cas9 system to generate cbf triple mutants. Two independent cbf triple mutant lines with the deletion of large fragments were obtained. B, Accumulation of CBF1, CBF2, and CBF3 transcripts in plants that were treated with low temperature (4°C) for 2 h. Transcript accumulation was assessed by quantitative real-time RT-PCR, and ACTIN8 was used as the internal control. Error bars indicate the sd of three biological replicates. C, Germination rate of Col-0, cbf123-1, and cbf123-2. Seeds were sown on MS medium and placed in the incubator at 23°C, and germination was assessed after 2 d. D, Seedlings of Col-0, cbf123-1, and cbf123-2 grown on MS medium. Seeds were germinated on MS medium, and after 3 d, seedlings were transferred to new MS medium for further growth. Seedlings were photographed after 8 d of growth. E, Growth phenotype of Col-0, cbf123-1, and cbf123-2. Plants were grown in a greenhouse at 23°C and with a 15-h-light/9-h-dark regime. Plants were photographed after 4 weeks of growth. F, Morphological phenotype of the leaves of 4-week-old Col-0, cbf123-1, and cbf123-2 plants. G, Number of leaves produced per Col-0, cbf123-1, and cbf123-2 plant after the plants were grown in soil for 4 weeks. H, The area of the fifth rosette leaf of Col-0, cbf123-1, and cbf123-2 plants that were grown in soil for 4 weeks. Area was determined with ImageJ software. Error bars indicate the sd of three biological replicates. I, Fresh weight of Col-0, cbf123-1, and cbf123-2 plants that were grown in soil for 4 weeks. J, Flowering of Col-0, cbf123-1, and cbf123-2 plants that were grown in soil for 6 weeks. Asterisks indicate statistically significant differences (P < 0.05, one-way ANOVA).
CBF Genes Affect Seed Germination and Early Development
Overexpression of CBF genes retards plant growth and delays flowering (Gilmour et al., 2004; Park et al., 2015). To further investigate whether CBF genes contribute to plant development, we assessed the germination, growth, and flowering of the cbf triple mutants. For the germination assay, seeds were sown on Murashige and Skoog (MS) medium and stratified at 4°C for 3 d before they were transferred to a 23°C growth chamber; germination was assessed after 2 d. The germination rate of both cbf triple mutant lines was significantly lower than that of the wild type (Fig. 1C). To exclude the possibility that the decreased germination rate of the cbf triple mutants resulted from the stratification at 4°C for 3 d, we directly transferred the plates to 23°C without stratification before assessing germination. Similarly, the germination rate was significantly lower for the cbf mutants than for the wild type. These results suggest that CBF genes are required for optimal seed germination.
We observed the growth of seedlings on MS medium and found that the root growth was slightly slower for the cbf triple mutants than for the wild type (Fig. 1D). For seedlings in soil, growth was also slower for the cbf triple mutants than for the wild type (Fig. 1, E and F). Specifically, rosette leaf number and size were smaller and whole plant fresh weight was lower for the cbf triple mutants than for the wild type (Fig. 1, G–I). These results suggest that CBF genes contribute to seedling development and growth in Arabidopsis. However, flowering time was not significantly affected in the cbf triple mutants (Fig. 1J).
CBF Genes Play a Minor Role in Chilling Tolerance
Although CBF genes are known to be involved in freezing tolerance, whether they are also involved in chilling tolerance is unclear. We therefore investigated the chilling phenotype of the cbf triple mutants. After seedlings were grown on MS medium at 4°C for 2 months, root length of the cbf triple mutants was not shorter compared to that of the wild type (Supplemental Fig. S1, A and B). After seedlings were grown for 3 months on MS medium at 4°C, rosette leaf number was fewer but petioles were longer for the cbf triple mutants than for the wild type (Supplemental Fig. S1, C and D). The growth of plants in soil at 4°C was also observed, which showed that the growth rate and flowering time did not significantly differ between the cbf triple mutants and the wild type (Supplemental Fig. S1E). In addition, we examined the electrolyte leakage of 3-week-old plants after treatment with 4°C, and there was no difference between the cbf triple mutants and the wild type (Supplemental Fig. S1F). Together, these results suggest that the CBF genes do not have a major role in chilling tolerance.
The Three CBF Genes Are Critical for Freezing Tolerance
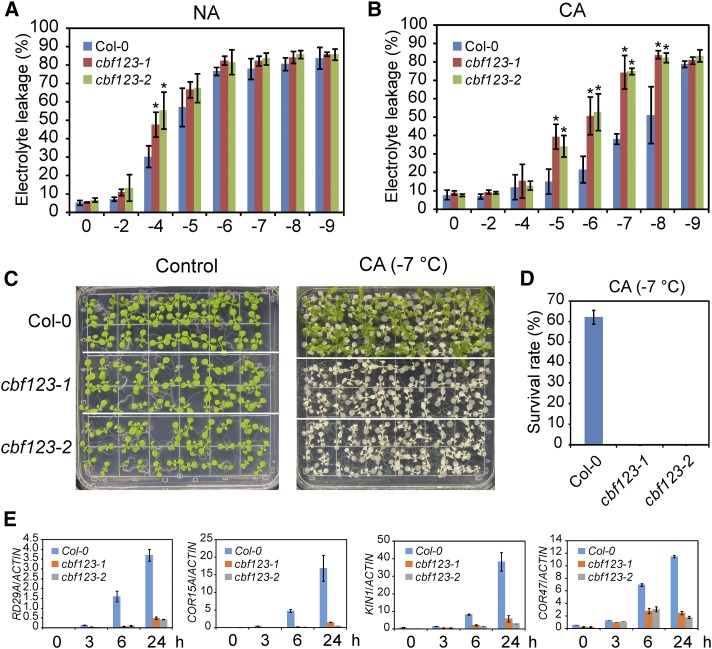
To examine the role of CBF proteins in freezing tolerance, we performed an electrolyte leakage assay for the wild type and cbf triple mutants before and after cold acclimation (4°C, 7 d). For plants without cold acclimation, the EL50 value (the temperature at which 50% of the cellular electrolytes were released due to freezing damage) was −4.8°C for the wild type and about −4°C for both of the cbf triple mutant lines (Fig. 2A). For plants after cold acclimation, the EL50 value was −8°C for the wild type and −6°C for the cbf triple mutants (Fig. 2B). These results suggest that the CBF genes are not only required for cold acclimation-dependent freezing tolerance but also affect basal freezing tolerance. Comparison of the EL50 values of nonacclimated wild-type plants with the cold-acclimated cbf triple mutants indicated that the cbf triple mutants did not completely lose the ability to undergo cold acclimation, suggesting that CBF-independent pathways are also involved in cold acclimation.
Figure 2.
cbf triple mutants are very sensitive to freezing. A, Electrolyte leakage assay conducted on nonacclimated (NA) Col-0, cbf123-1, and cbf123-2 plants grown at 23°C. B, Electrolyte leakage assay performed on Col-0, cbf123-1, and cbf123-2 plants after cold acclimation (CA; 4°C for 7 d). C and D, Freezing survival assay. Col-0, cbf123-1, and cbf123-2 seedlings (48 seedlings for each genotype) were grown on MS medium for 12 d and then transferred to 4°C for cold acclimation. The acclimated plants were subjected to −7°C for 1 h before they were transferred to 23°C for recovery. C shows the phenotype of seedlings before (left) and after (right) freezing treatment. D shows the survival rates of freezing-treated seedlings after 5 d of recovery at 23°C. The above experiments were performed at least three times with similar results, and results from a representative experiment are shown in A to C. Error bars indicate the sd (A and B, n = 3; D, n = 4). Asterisks indicate statistically significant differences (P < 0.05, one-way ANOVA). E, Eight-day-old seedlings of Col-0, cbf123-1, and cbf123-2 were treated at 4°C for 0, 3, 6, and 24 h. Expression of RD29A, COR15A, KIN1, and COR47 was assessed with quantitative real-time RT-PCR. ACTIN8 was used as the internal control. Error bars indicate the sd of three biological replicates.
To further confirm the roles of CBF genes in cold acclimation, we performed a freezing-survival assay. The cold-acclimated wild-type plants and the cbf triple mutants were treated at −7°C for 1 h, and the number of surviving plants was determined after 5 d of recovery at 23°C. The survival rate for the wild-type plants was about 60%, but for the cbf triple mutants, nearly all plants were dead after the freezing treatment (Fig. 2, C and D), confirming that CBF proteins are necessary for the freezing tolerance imparted by cold acclimation.
CBF Proteins Are Required for COR Gene Expression
To confirm that COR genes are regulated by CBF transcription factors, we tested the transcript level of four well-known COR genes: RD29A, COR15A, KIN1, and COR47. After treatment of 10-d-old seedlings of the wild type and cbf triple mutants with low-temperature (4°C) for 0, 3, 6, and 24 h, the expression of COR genes was assessed by real-time quantitative RT-PCR. The expression of the four COR genes was dramatically lower in the cbf triple mutants (Fig. 2E), which indicates that CBF proteins are essential for the regulation of the expression of these four COR genes.
We tested whether the expression of RD29A induced by abscisic acid (ABA), NaCl, and mannitol also depends on CBF proteins. We treated the wild type and cbf triple mutants with ABA for 0, 1, 3, and 6 h and then assessed the expression of RD29A. The result showed that ABA-induced expression of RD29A was partially reduced in the cbf triple mutants but that the reduction was less than cold-induced expression of RD29A (Supplemental Fig. S2A). Similarly, NaCl- and mannitol-induced expression of RD29A was also partially decreased in the cbf triple mutants (Supplemental Fig. S2, B and C). These results suggest that although CBF proteins mainly function in the cold induction of RD29A, they are also involved in ABA-, NaCl-, and mannitol-induced expression of RD29A.
Identification of CBF-Regulated Genes
As noted earlier, our findings indicated that CBF proteins are required for the regulation of expression of the four COR genes. To further determine the number of COR genes regulated by CBF proteins, we performed an RNA sequencing assay for both the wild type and the cbf123-1 triple mutant after low-temperature (4°C) treatment for 0, 3, and 24 h. Three biological replicates were performed for each sample at each time point. The RNA-seq data showed that 2,084 genes were up-regulated (q value < 0.05, fold change ≥ 2) and that 2,003 genes were down-regulated (q value < 0.05, fold change ≥ 2) in wild-type plants after low-temperature treatment (Supplemental Fig. S3A and Supplemental Table S1). These numbers were much greater than those identified by previous microarray assays, which identified 1297 up-regulated genes and 1427 down-regulated genes (Park et al., 2015). Comparing the COR genes identified by RNA-seq data and microarray data, 837 (64.5% of the genes identified in microarray data) up-regulated genes and 599 (42% of the genes identified in microarray data) down-regulated genes were identified in both studies (Supplemental Fig. S3B). We also compared the genes that were induced at an early stage (3 h) and at a late stage (24 h) of low-temperature treatment in wild-type plants. A total of 377 genes were up-regulated at both 3 h and 24 h, 121 genes were up-regulated only at 3 h, and 1,586 genes were up-regulated only at 24 h. A total of 182 genes were down-regulated at both 3 and 24 h, 96 genes were down-regulated only at 3 h, and 1725 genes were down-regulated only at 24 h (Supplemental Fig. S3C).
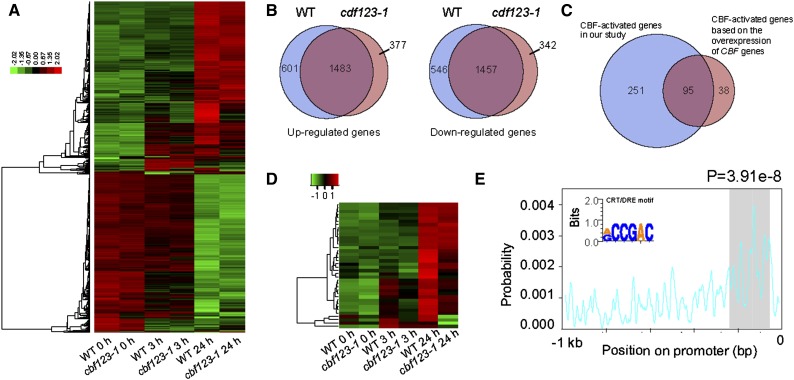
In the cbf123-1 triple mutant, 1,860 genes were up-regulated and 1,799 genes were down-regulated after low-temperature treatment (Supplemental Table S2). Comparison of the transcript profiles between the wild type and cbf123-1 showed that 1,483 (80% of the up-regulated genes in cbf123-1) up-regulated genes and 1,457 (81% of the down-regulated genes in cbf123-1) down-regulated genes were identified in both samples (Fig. 3, A and B), suggesting that the expression of a large number of COR genes was not affected in the cbf123-1 triple mutant. We further analyzed the genes that were regulated by CBF proteins. Two criteria were used to define CBF-regulated genes: (1) genes had to be up- or down-regulated (fold change of at least 2) after cold treatment for either 3 or 24 h in wild type plants; and (2) genes were either up- or down-regulated (fold change of at least 2) in the cbf123-1 triple mutants compared with wild-type plants. Based on these two criteria, 414 genes were identified as CBF-regulated genes. Among these 414 genes, 346 were considered as CBF-activated and 68 were considered as CBF-repressed genes (Supplemental Table S3). Based on the expression data from transgenic plants overexpressing CBF1, CBF2, or CBF3, 133 COR genes were previously identified as CBF-activated genes (Park et al., 2015), among which 95 were also identified as CBF-regulated genes in this study (Fig. 3C). For the remaining 38 CBF regulon genes that were not included in our list of CBF-regulated genes, we reanalyzed their gene expression pattern in the wild type and the cbf triple mutant using a hierarchical clustering approach; the results showed that most of these genes were also lower in the cbf triple mutant under low-temperature conditions (Fig. 3D). This was also supported by the finding that, when we relaxed the criteria for CBF-regulated genes (fold change of at least 1.5), 111 CBF regulon genes found in the previous study were identified in our study as CBF-regulated genes. Thus, the CBF-regulated genes identified by using cbf triple mutants nearly cover the CBF regulon genes identified by constitutive overexpression of CBF genes. Besides, the many more CBF-regulated genes identified in our study will broaden our view of the roles of CBF proteins in the regulation of COR genes. Gene Ontology (GO) enrichment analysis showed that CBF-regulated genes were significantly enriched in the categories of “response to cold”, “response to water deprivation,” and “response to abscisic acid stimulus” (Supplemental Fig. S4).
Figure 3.
Analysis of CBF-regulated genes. RNA-seq was conducted for wild-type plants and cbf123-1 triple mutants after 4°C treatment for 0, 3, and 24 h. Three biological replicates were performed for each sample at each time point. A, Hierarchical clustering of genes that were up-regulated or down-regulated after exposing wild-type and cbf triple mutants plants to 4°C for 3 h and 24 h. B, Venn diagrams indicating the number of genes that were up-regulated or down-regulated in both wild-type and cbf triple mutant plants. C, Venn diagram indicating the number and overlap of CBF-activated genes identified by RNA-seq data in this study and the number of CBF-activated genes identified by overexpression of CBF genes in a previous study (Park et al., 2015). D, Heat map showing the expression pattern of 38 CBF regulon genes that were not included in the list of CBF-regulated genes in this study. E, The distribution preference of the CRT/DRE motif in the promoter region of CBF-regulated genes.
It is well known that CBF proteins recognize and bind to the CRT/DRE (RCCGAC) motif and regulate downstream COR gene expression (Liu et al., 1998). We analyzed the CRT/DRE cis-element in the upstream regulatory regions (1,000 bp upstream of ATG start codon) of 346 CBF-activated genes and found that 212 (61.3%) contain the RCCGAC motif, and 71 of them have more than one motif in the promoter regions (Supplemental Table S3). An analysis of the location of the RCCGAC motif in the promoter regions showed that this cis-element is enriched in the region 150 bp upstream of start codon ATG (Fig. 3E). For the remaining 134 CBF-activated genes, no cis-elements significantly enriched in the promoter regions were identified, suggesting that these CBF-activated genes may not be the direct targets of CBF proteins. We also analyzed the CRT/DRE cis-element in the promoter regions of 68 CBF-repressed genes and found that only 10 genes (14.7%) contain the RCCGAC motif. Furthermore, an analysis of the locations of these CBF-regulated genes on the chromosomes of Arabidopsis showed that there was no significant distribution preference (Supplemental Fig. S5).
Putative Functions of CBF-Activated Genes
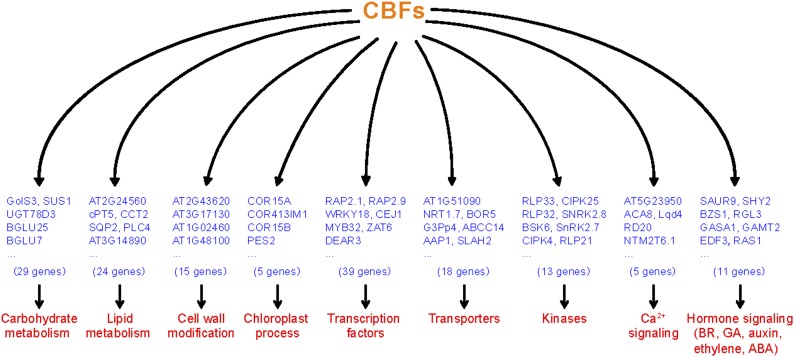
The galactinol synthase-encoding gene GOLS3, which has four CRT/DRE regulatory elements in the promoter region, was highly induced by overexpression of CBF1, CBF2, or CBF3 (Park et al., 2015). Here, we found that GOLS3 expression was decreased by about 55-fold in the cbf123-1 triple mutants, relative to its expression in the wild type. This result confirmed that cold-induced expression of GOLS3 is completely dependent on CBF proteins. In addition to GOLS3, 28 other genes involved in carbohydrate metabolism, including Suc synthase and β-glucosidases, were significantly down-regulated in the cbf123-1 triple mutant (Fig. 4; Supplemental Table S4). Another large group of genes regulated by CBF proteins are associated with lipid metabolism, and these include genes encoding GDSL-like lipases and phospholipases. Moreover, 39 transcription factors were identified that were significantly down-regulated in the cbf triple mutants, suggesting that multiple transcriptional cascades are involved in the cold regulatory network. Among these transcription factors, 11 (28.2%) belong to AP2/EREBP-type transcription factors. In addition, seven transcription factors were identified that are involved in hormone (ethylene, gibberellin [GA], auxin, brassinosteroid, and jasmonic acid)-mediated transcriptional regulation, which links CBF-mediated cold acclimation to hormonal responses. Significantly, genes involved in cell wall modification were also regulated by CBF proteins. These genes encode, for example, plant invertase/pectin methylesterase inhibitor, chitinase, and pectin lyase. This indicates that cell wall modification is involved in cold acclimation. Furthermore, many genes involved in chloroplast process, kinases, transporters, and Ca2+ signaling were also regulated by CBF proteins (Fig. 4). Previous studies showed that late embryogenesis abundant (LEA) proteins are involved in the response to freezing (Sasaki et al., 2014; Popova et al., 2015). Here, five LEA protein-encoding genes were significantly down-regulated in the cbf triple mutants.
Figure 4.
Functional categories of CBF-activated genes. CBF-activated genes were grouped according to their biological functions. Partial lists of genes of each category are shown.
Six First-Wave Transcription Factors Are Regulated by CBF Proteins
A previous study showed that, apart from CBF transcription factors, 27 first-wave transcription factors are also up-regulated at an early stage after cold treatment, and one of these transcription factors, ZF, also belongs to CBF regulon genes (Park et al., 2015). Using RNA-seq data, we reanalyzed the expression of these first-wave transcription factors and found that 26 of the genes were significantly up-regulated after 3 h of cold treatment (Supplemental Fig. S6). To determine whether CBF proteins are required for the expression of these first-wave transcription factor genes, we analyzed these genes in the cbf triple mutants; the results showed that the expression of six first-wave transcription factors (DEAR1, DREB, ZF, CZF2, ZAT10, and AZF2) was significantly down-regulated in the cbf123-1 triple mutant (Supplemental Table S3), suggesting that cold-induced expression of these genes depends at least partially on CBF proteins. However, for the first-wave transcription factors HSFC1, ZAT12, and CZF1, which are extensively involved in the regulation of COR genes (Vogel et al., 2005; Park et al., 2015), their expression was not affected in the cbf123 triple mutants.
Generation of cbf Single and Double Mutants
To understand the extent of functional redundancy of CBF proteins in the regulation of COR gene expression and freezing tolerance, we also generated cbf single and double mutants using CRISPR/Cas9. One cbf1 mutant line, in which the CBF1 gene was completely deleted, was obtained. Two independent mutant lines of cbf2, named cbf2-1 and cbf2-2, in which a single nucleotide was inserted or deleted in the CDS region, respectively, were obtained. One mutant line of cbf3, in which 15 nucleotides were deleted in the CDS region, was generated. For the cbf1 cbf3 double mutant, three independent mutant lines lacking the whole CDS of CBF1 and the partial CDS of CBF3 were obtained. For the cbf2 cbf3 double mutant, two mutant lines with different nucleotide insertion or deletion in the CDS regions of CBF2 and CBF3 were obtained. All of these point-mutated versions of CBFs lead to premature termination of translation. Detailed information about these cbf mutants is provided in Supplemental Figure S7. We tried several times to generate a cbf1 cbf2 double mutant using sgRNAs targeted to the CDS regions of CBF1 and CBF2, but we could not obtain mutant lines with point-mutated CBF1, which could be due to the resistance of the CDS region of CBF1 to CRISPR/Cas9-mediated gene editing.
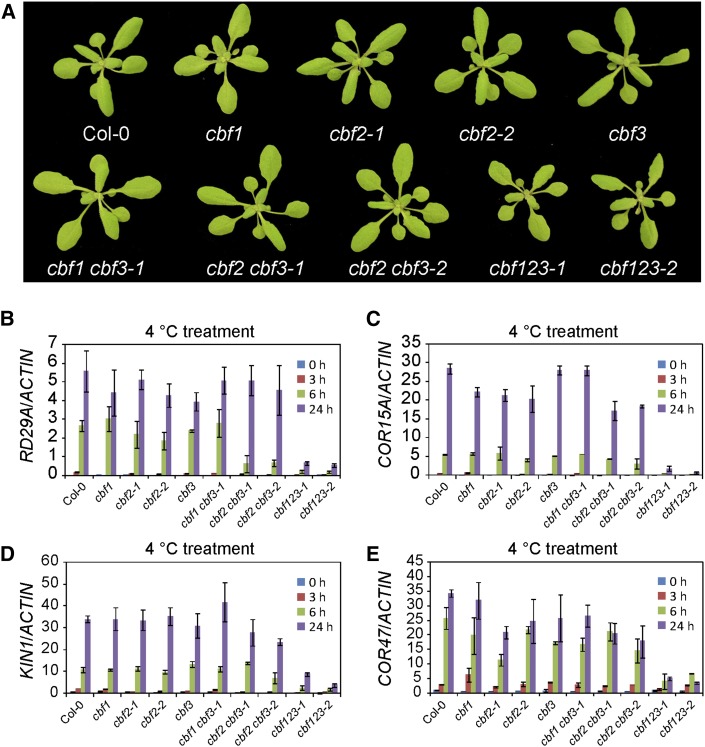
For all of the cbf mutants, we first determined the expression of the three CBF genes after low-temperature treatment for 3 h. For the cbf1 single mutants and cbf1 cbf3 double mutants, in which CBF1 and/or CBF3 were deleted, expression of CBF1 and/or CBF3 was not detectable or dramatically decreased (Supplemental Figs. S8 and S9A). In either cbf1 or cbf1 cbf3 double mutants, the expression of CBF2 was slightly increased at 3 h after cold treatment. For other point-mutated cbf single and double mutants, the expression of CBF genes was not significantly affected at 3 h after cold treatment (Supplemental Fig. S8). The morphologies of these single and double mutants did not significantly differ from that of the wild type (Fig. 5A; Supplemental Fig. S9B).
Figure 5.
CBF proteins are redundant in plant development and in the regulation of COR gene expression. A, Plants were photographed after they were grown in a greenhouse at 23°C and with a 15-h-light/9-h-dark regime for 4 weeks. B to E, Expression of RD29A, COR15A, KIN1, and COR47 genes in 8-d-old seedlings that were treated with 4°C for 0, 3, 6, and 24 h. Expression was assessed with quantitative real-time RT-PCR, and ACTIN8 was used as the internal control. Error bars indicate the sd of three biological replicates.
Redundancy of CBF Genes in the Regulation of COR Gene Expression
As we mentioned earlier, the cold-induced expression of the COR genes RD29A, COR15A, KIN1, and COR47 was dramatically down-regulated in the cbf triple mutants. To assess the degree of redundancy of the three CBF proteins in the regulation of these four COR genes, we treated all cbf mutants with low temperature for 0, 3, 6, and 24 h and tested the expression of these four COR genes using quantitative real-time RT-PCR. Our result showed that the expression of these four COR genes was not affected or was only slightly affected in either the cbf single or double mutants (Fig. 5, B–E), which suggests that the three CBF proteins are redundant in the regulation of these four COR genes.
CBF2 Is More Important Than CBF1 and CBF3 in Cold Acclimation
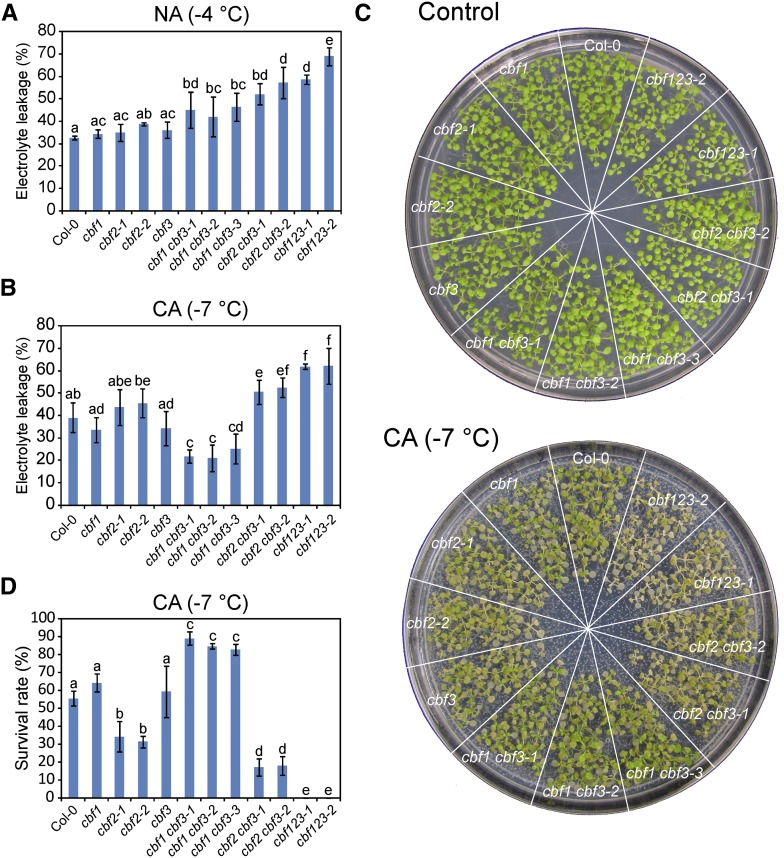
To investigate the redundancy of the three CBF proteins in freezing tolerance, we performed electrolyte leakage assays for all cbf mutants before and after cold acclimation (4°C, 7 d). For the plants without cold acclimation, cbf single mutants showed slightly increased ion leakage. cbf1 cbf3 and cbf2 cbf3 double mutants showed more ion leakage than wild-type plants and cbf single mutants, but less than cbf triple mutants (Fig. 6A). These results indicated that CBF proteins are involved in basal freezing tolerance and that CBF proteins are redundant in this process.
Figure 6.
CBF2 is more important than CBF1 or CBF3 in cold acclimation. A, Electrolyte leakage assay conducted on nonacclimated plants grown at 23°C. B, Electrolyte leakage assay conducted on cold-acclimated (4°C for 7 d) plants. C and D, Sensitivity of cold-acclimated plants to freezing. Plants were grown on MS medium for 12 d and were then transferred to 4°C for cold acclimation. The acclimated plants were subjected to freezing at −7°C for 1 h. The percentage of plants that survived was determined after 5 d of recovery at 23°C. C shows the phenotype of seedlings before (upper) and after (bottom) freezing treatment. D shows the survival rates of freezing-treated seedlings after 5 d of recovery at 23°C. All experiments were performed more than three times with similar results, and results from one representative experiment are shown. Error bars in A, B, and D indicate the sd (A and B, n = 3; D, n = 4). Different letters represent statistically significant differences (P < 0.05, one-way ANOVA).
We also investigated the freezing tolerance of plants after cold acclimation. Our results showed that cbf1 and cbf3 single mutants had slightly greater freezing tolerance than the wild type. In contrast, ion leakage was greater for both of the cbf2 mutant lines than for the wild type, suggesting that the mutation of CBF2 increases sensitivity to freezing. Unexpectedly, all three independent cbf1 cbf3 double mutant lines had increased freezing tolerance because less electrolytes were released after freezing treatment. In contrast, the two cbf2 cbf3 double mutant lines were hypersensitive to freezing (Fig. 6B). To further confirm the freezing phenotype of all of the cbf mutants after cold acclimation, we performed a freezing survival assay for seedlings, and the results of which were consistent with the results of the electrolyte leakage assay. Briefly, survival rate was slightly increased for the cbf1 and cbf3 single mutants, reduced for both of the cbf2 mutant lines, greatly increased for the cbf1 cbf3 double mutants, and substantially reduced for the cbf2 cbf3 double mutants. The survival rate of the cbf2 cbf3 double mutants under freezing was lower than that of the cbf2 single mutants but higher than that of the cbf triple mutants (Fig. 6, C and D). Together, these results indicate that all three CBF proteins are positive regulators in basal freezing tolerance and that they appear to play an equal role in this process. Importantly, our results suggest that the three CBF proteins have distinct functions in cold acclimation-dependent freezing tolerance. Both the electrolyte leakage assay and the freezing survival assay showed that CBF2 is more important than CBF1 and CBF3 in cold acclimation-dependent freezing tolerance.
The Expression of CBF2 Is Increased in the cbf1 cbf3 Double Mutant
To understand why cbf1 cbf3 double mutants showed increased freezing tolerance, we performed RNA-seq analysis for the wild type and cbf1 cbf3-1 double mutant after low-temperature treatment for 0, 3, and 24 h (Supplemental Table S5). We analyzed the expression of CBF1 and CBF3 and found that these two genes were not induced in cbf1 cbf3-1 double mutants after low-temperature treatment (Supplemental Table S6), which confirms that these two genes were deleted in the double mutant. The expression of CBF2 was slightly increased (1.2-fold) in the cbf1 cbf3-1 double mutant compared with the wild type after low-temperature treatment for 3 h. At 24 h of cold treatment, the expression of CBF2 in the cbf1 cbf3-1 double mutant was twice of that in the wild type (Supplemental Table S6), suggesting that the cold-induced expression of CBF2 lasts longer in the cbf1 cbf3-1 double mutant. To confirm this result, we determined the expression of CBF2 using real-time RT-PCR in the wild type and all three cbf1 cbf3 double mutant lines after low-temperature treatment for 0, 3, 6, and 24 h. The result showed that CBF2 expression was increased in all three cbf1 cbf3 double mutants, especially after cold treatment for 6 and 24 h (Supplemental Fig. S10).
To investigate how CBF-regulated genes may be affected in the cbf1 cbf3 double mutant, we analyzed the expression of all 346 CBF-activated genes. RNA-seq data showed that only eight of the genes were significantly down-regulated (fold change > 2), and only one of the genes was up-regulated (fold-change > 2) in the cbf1 cbf3-1 double mutant (Supplemental Table S7), which suggests that the three CBF proteins are redundant in regulating CBF-regulated genes and CBF2 protein alone is enough to induce the transcription of nearly all CBF-regulated genes. Out of the 346 CBF-activated genes, 200 were up-regulated in the cbf1 cbf3 double mutant compared with the wild type, although the increases were less than 2-fold for most of these 200 genes (Supplemental Table S7). These results suggest that the increased expression of CBF2 gene and some of the CBF-regulated genes in the cbf1 cbf3 double mutants contributes to the increased freezing tolerance of the double mutants.
cbf Triple Mutants Are Hypersensitive to Salt Stress
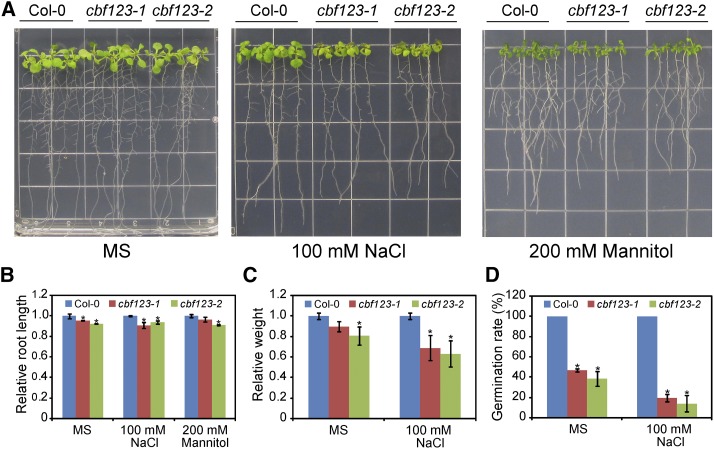
To examine whether CBF genes are also involved in the response to other abiotic stresses, we examined the phenotype of the cbf triple mutants under salt, osmotic, and drought stresses. Under salt stress, primary root elongation did not significantly differ between the wild type and the cbf triple mutants (Fig. 7, A and B). However, we found that cbf triple mutants had fewer lateral roots than the wild type under salt stress (Fig. 7A). After growing on NaCl medium, the leaves of the cbf triple mutants were yellower and smaller than those of the wild type (Fig. 7A). Quantitative measurement of the fresh weight of seedlings showed that the growth of cbf triple mutants was more inhibited under salt stress (Fig. 7C). In addition, the seed germination of cbf triple mutants was more inhibited under salt stress (Fig. 7D). These results suggest that the three CBF genes play a role in salt tolerance. Under osmotic stress induced by mannitol, however, the assessed phenotypes did not significantly differ between the cbf triple mutants and the wild type (Fig. 7, A and B). According to a water loss assay and a drought survival assay, drought resistance did not significantly differ between the cbf triple mutants and the wild type (Supplemental Fig. S11). Furthermore, we also observed the growth of cbf single and double mutants on salt medium and found that salt tolerance was unaffected in these mutants (Supplemental Figs. S9C and S12).
Figure 7.
The cbf triple mutants are sensitive to salt stress but not to osmotic stress. A, Growth of Col-0, cbf123-1, and cbf123-2 plants on MS, NaCl (100 mM), and mannitol (200 mM) media. The seedlings were first grown on MS medium for 3 d after germination before they were transferred to the treatment media. Seedlings were photographed after 10 d of growth on the treatment media. B, Relative root length of Col-0, cbf123-1, and cbf123-2 plants after the plants were grown on MS, NaCl (100 mM), and mannitol (200 mM) media. The root length of wild-type plants on each medium was set to 1. C, Relative fresh weight of Col-0, cbf123-1, and cbf123-2 plants after the plants were grown on MS and NaCl (100 mM) media. The fresh weight of wild-type plants on each medium was set to 1. D, Germination of Col-0, cbf123-1, and cbf123-2 on MS and NaCl (100 mM) media. Germination was assessed on each medium when the germination of wild type reached 100%. Asterisks indicate statistically significant differences (P < 0.05, one-way ANOVA).
DISCUSSION
Three CBF proteins play prominent roles in cold acclimation in Arabidopsis, which enables plants to increase their tolerance to freezing (Thomashow, 1999). Because of a lack of stable loss-of-function cbf mutants, the extent of contribution of CBF genes to freezing tolerance was not fully understood. In this study, we used the CRISPR/Cas9 technology to generate cbf single, double, and triple mutants, which made it possible to comprehensively study the roles of CBF proteins in development, freezing tolerance, chilling response, and regulation of COR gene expression. These mutants also enabled us to study the functional redundancy of the three CBF proteins in these processes.
The potential involvement of CBF proteins in plant development was indicated by the finding that overexpression of either CBF1, CBF2, or CBF3 retarded growth and delayed flowering (Gilmour et al., 2000, 2004; Park et al., 2015). These effects were due to the accumulation of DELLA proteins via the reduction of GA level by increased expression of GA-inactivating GA 2-oxidase genes (Achard et al., 2008). Our study showed that the cbf single and double mutants lacked significant morphological phenotypes but that the cbf triple mutants germinated and grew more slowly than the wild type, which suggests that CBF proteins affect seedling development and that the three CBF proteins are redundant in the regulation of development. However, the growth of the cbf triple mutants was less affected than that of transgenic plants that constitutively overexpress CBF genes. We suspect that constitutive overexpression of CBF genes to some extent mimics the cellular response to low temperatures, which induces the expression of downstream COR genes and inhibits plant growth. Apart from development, our study showed that the CBF genes are also involved in the response to salt stress, which suggests that CBF proteins have broad roles in abiotic stress responses (Supplemental Fig. S13).
For freezing-sensitive plants, the expression of CBF genes is increased after low-temperature treatment, but the up-regulation of CBF genes and downstream COR genes does little to increase freezing tolerance (Carvallo et al., 2011), which suggests that the increased expression of CBF genes in freezing-sensitive plants may play a role in chilling tolerance. In Arabidopsis, CBF genes have been well known to be involved in cold acclimation, but whether they may be important for chilling tolerance has been unclear. A previous study showed that disruption of the RNA helicase-encoding gene LOS4 leads to the down-regulation of CBF genes and downstream COR genes and increased sensitivity to chilling stress, which suggests that CBF proteins are also involved in chilling tolerance in plants (Gong et al., 2002). In rice (Oryza sativa), mutation of COLD1, which encodes a regulator of G-protein signaling, leads to reduced CBF gene expression and increased sensitivity to chilling (Ma et al., 2015). Two other studies showed that overexpression of Arabidopsis CBFs in chilling-sensitive plants, such as tomato (Solanum lycopersicum) and rice, can increase chilling tolerance (Hsieh et al., 2002; Oh et al., 2005). In this study, however, wild-type plants and cbf triple mutants did not differ in their sensitivity to chilling (4°C) in terms of electrolyte leakage or growth inhibition. A difference detected was that the petiole length was longer for the cbf triple mutants than for the wild-type plants after growing on MS medium under chilling conditions. Additionally, the roots of cbf triple mutant seedlings are shorter when grown under warm temperature but not shorter when grown under prolonged cold temperature, compared to the wild type. These observations could be explained by the possibility that some COR genes that have a growth suppressing role are induced under low temperature in wild-type plants but not in the cbf triple mutants. Because CBF proteins regulate only about 10% of COR genes, the sensitivity of los4 mutant plants to chilling could be due to the impairment of the expression of COR genes that are independent of CBF proteins. Alternatively, the chilling sensitivity of los4 mutant plants may involve a function of LOS4 that is independent of its role in regulating COR gene expression. CBF genes from Arabidopsis increase the chilling tolerance of tomato and rice; this may be because the chilling tolerance in the recipient species is more limited by CBF-dependent genes.
Constitutive overexpression of CBF1, CBF2, or CBF3 increases the freezing tolerance of nonacclimated plants (Jaglo-Ottosen et al., 1998; Gilmour et al., 2000). Furthermore, overexpression of a truncated version of CBF2, which imparts dominant-negative effects on the activity of the CBF-CRT/DRE regulatory module, reduces the freezing tolerance of cold-acclimated plants (Park et al., 2015). These results suggest that CBF proteins are positive regulators of cold acclimation. In this study, both the electrolyte leakage assay and the freezing survival assay showed that the cold-acclimated cbf triple mutants are much more sensitive to freezing than the cold-acclimated wild-type plants, adding strong support that CBF proteins are necessary for in cold acclimation. However, the cbf triple mutants do not completely lose their cold acclimation ability, indicating that CBF-independent pathways also contribute to cold acclimation. CBF-independent pathways may involve other first-wave transcription factors, such as HSFC1, ZAT12, and CZF1 (Vogel et al., 2005; Park et al., 2015).
The effect of CBF proteins on the freezing tolerance of nonacclimated plants remains poorly understood. One study showed that silencing of CBF1, CBF3, or CBF1 and CBF3 together did not affect the freezing tolerance of nonacclimated plants (Novillo et al., 2007). Another study showed that overexpression of a dominant-negative version of CBF2 had little effect on the freezing tolerance of nonacclimated plants (Park et al., 2015). These studies suggested that CBF proteins have a minor role in basal freezing tolerance. We found that nonacclimated cbf double and triple mutants were more sensitive to freezing than wild-type plants, suggesting that CBF proteins are also involved in basal freezing tolerance. Our data also showed that the three CBF proteins are redundant in the regulation of basal freezing tolerance. The roles of CBFs in basal freezing tolerance as well as in seedling development and salt tolerance are consistent with previous observations that CBF genes and some COR genes are expressed even without cold treatment and are regulated by circadian rhythm (Harmer et al., 2000).
Novillo et al. (2004) found that a cbf2 T-DNA insertion mutant displayed increased tolerance to freezing, which was due to the increased expression levels of CBF1 and CBF3, followed by the up-regulation of downstream COR genes, such as COR15A, COR47, and KIN1 (Novillo et al., 2004). The study by Kim et al. (2015) indicated that the T-DNA insertion mutation in CBF2 gene only increased the expression of CBF3, but not CBF1 gene. Chromatin immunoprecipitation assays indicated that CBF2 does not directly bind to the promoter region of CBF3. In addition, another study showed that the expression of COR15A gene was not affected in the cbf2 T-DNA insertion mutant after a cooling treatment (shifting from 28°C to 22°C; Wang and Hua, 2009). Our data indicate that the expression of CBF1 and CBF3 was not significantly affected in two independent cbf2 mutant lines generated by CRISPR/Cas9. These results suggest that the previously reported cbf2 T-DNA insertion mutant may be more complicated than it appeared. The study by Alonso-Blanco et al. (2005) showed that down-regulation of CBF2, caused by deletion of the promoter region of CBF2 in the Cape Verde Islands ecotype, reduced the expression of several CBF target genes and reduced freezing tolerance. A recent study indicated that an Arabidopsis ecotype collected from Italy (IT) had reduced freezing tolerance because the CBF2 gene in the IT plants encodes a nonfunctional protein (Gehan et al., 2015). Constitutive overexpression of CBF2 increased the freezing tolerance of nonacclimated plants (Gilmour et al., 2004). These studies suggest that the CBF2 protein is a positive regulator of freezing tolerance. In our study, two independent cbf2 mutant lines had significantly lower freezing tolerance than wild-type plants after cold acclimation, even though several tested COR genes, including COR15A, was not affected. Our results support that the CBF2 protein is a positive regulator of freezing tolerance.
Our study indicated that the CBF2 protein is more important than CBF1 and CBF3 in cold acclimation-dependent freezing tolerance. This conclusion is supported by the finding that the cbf2 single mutant, the cbf2 cbf3 double mutants, and the cbf123 triple mutants but not the cbf1 or cbf3 single mutants or the cbf1 cbf3 double mutants had increased sensitivity to cold acclimation-dependent freezing tolerance. The conclusion is also consistent with a previous study in which constitutive overexpression of CBF2 caused greater accumulation of Pro and sugar than constitutive overexpression of CBF1 and CBF3, and in which the freezing tolerance of nonacclimated plants was increased more in CBF2 overexpression plants than in CBF1 overexpression plants (Gilmour et al., 2004). An earlier study showed that cbf1/cbf3 antisense lines were sensitive to freezing after cold acclimation (Novillo et al., 2007). In our study, the three independent cbf1 cbf3 double mutants, in which CBF1 and partial CDS regions of CBF3 were deleted, all had significantly increased tolerance to freezing after cold acclimation. RNA-seq data and qRT-PCR analysis both showed that CBF2 gene expression is increased in cbf1 cbf3 double mutants, especially after cold treatment for 6 and 24 h. The result suggests that CBF1 and CBF3 may negatively affect the gene expression of CBF2. In addition, RNA-seq data showed that 200 CBF-activated genes are up-regulated in the cbf1 cbf3 double mutant compared to the wild type. In summary, our results showed that the three CBF proteins are required for acclimation-dependent freezing tolerance and also contribute to basal freezing tolerance. The three CBF proteins have similar roles in basal freezing tolerance, but CBF2 is more important than CBF1 or CBF3 in acclimation-dependent freezing tolerance (Supplemental Fig. S13).
COR genes have previously been identified by extensive transcriptome profiling based on microarray assays (Seki et al., 2001, 2002; Lee et al., 2005; Vergnolle et al., 2005; Park et al., 2015). In this study, we used RNA-seq to analyze cold-responsive genes and identified 4,087 COR genes, which is nearly twice the number identified from microarray data. By analyzing the gene expression in transgenic plants overexpressing CBF1, CBF2, or CBF3, Park et al. (2015) identified 172 CBF regulon genes. In this study, we compared the gene expression between wild-type plants and cbf triple mutants and found 414 genes that are putatively regulated by CBF proteins; this number is much higher than that identified previously. One probable reason is that the expression of some COR genes is not affected by the overexpression of CBF genes but is significantly affected in cbf triple mutants. Importantly, the CBF-regulated genes identified in this study include most of the CBF regulon genes identified previously, which indicated that CBF-regulated genes identified in our study are reliable. The additional CBF-regulated genes identified in this study will help us further understand CBF-regulated cellular processes that contribute to freezing tolerance.
An analysis of the functions of the CBF-regulated genes indicated that many are involved in sugar metabolism and lipid metabolism, which is consistent with the findings from metabolomic studies that low-temperature treatment or overexpression of CBF3 leads to the accumulation of Glc, inositol, galactinol, raffinose, and Suc (Gilmour et al., 2000; Taji et al., 2002; Cook et al., 2004). Remarkably, 39 transcription factors that are regulated by CBF proteins were identified, which suggests that transcriptional cascades are involved in cold acclimation and that many of the CBF-regulated genes are probably directly regulated by these transcription factors. Several CBF-regulated transcription factors are associated with hormone-mediated transcriptional regulation, which provides a link between cold stress and the regulation of hormonal responses and between cold tolerance and growth and development. Another group of CBF-regulated genes that have not previously been emphasized are those involved in the modification of cell walls. Our data showed that 15 cell wall-related genes were induced by low temperature but were significantly down-regulated in the cbf triple mutants. These genes encode plant invertase/pectin methylesterase inhibitor superfamily proteins, chitinase, and pectin lyase-like proteins, which are mainly involved in the degradation of cell wall components. This suggests that degradation of cell wall components could be beneficial for plants to survive freezing conditions. Other components that are regulated by CBF proteins include kinases and Ca2+ signaling related proteins, which are probably important for cellular signaling under cold conditions.
The three CBF proteins have been shown to be redundant in the regulation of COR genes (Park et al., 2015). Quantitative real-time RT-PCR demonstrated that four COR genes, RD29A, COR15A, KIN1, and COR47, are drastically down-regulated in the cbf triple mutants. RNA-seq data showed that 59 other genes were down-regulated by more than 5-fold in the cbf triple mutants, suggesting that the three CBF genes are essential for the regulation of these COR genes. Although RD29A, COR15A, KIN1, and COR47 were greatly down-regulated in the cbf triple mutants, their expression was not affected or was only slightly affected in the cbf single and double mutants. In generally, the expression level of these four COR genes is correlated with the freezing tolerance level. However, there are some potential complications. For example, disruption of a DEAD box RNA helicase led to increased expression of RD29A and COR15A but caused hypersensitivity to chilling and freezing (Guan et al., 2013). Our results showed that although these four COR genes are not greatly affected in the cbf2 single mutants and cbf2 cbf3 double mutants, these two mutants still had increased sensitivity to freezing after cold acclimation, which suggests that the down-regulation of other COR genes may cause the hypersensitivity of the cbf2 single and cbf2 cbf3 double mutants to freezing.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The plants used in this study are all in Col-0 background. cbf single, double, and triple mutants were generated by CRISPR/Cas9 technology. The mutation sites of cbf mutants were confirmed by PCR approach and Sanger sequencing. The mutation details of cbf mutants are described in Figure 1 and Supplemental Figure S7. Plants were grown at 23°C with long-day light cycle (16 h light/8 h dark).
Construction of Plasmids
The plasmids for CRISPR were designed according to the protocol described previously (Zhang et al., 2015). Briefly, two complementary oligonucleotides targeted to the 5′UTR and 3′UTR of CBF1, CDS regions of CBF2 and CBF3, were synthesized. These four oligo pairs were annealed to generate double-strand DNA, which was cloned into the BbsI site of either 18T-U6-sgR-Cas9 or 18T-U3b-sgR-Cas9 vector to generate 18T-U6-sgR-CBF1 5′UTR-Cas9, 18T-U6-sgR-CBF1 3′UTR-Cas9, 18T-U6-sgR-CBF2-Cas9, 18T-U3b-sgR-CBF2-Cas9, and 18T-U3b-sgR-CBF3-Cas9. For the construct used to edit the CBF1 gene, PCR was performed to amplify the U6-sgR-CBF1 3′UTR fragment using 18T-U6-sgR-CBF1 3′UTR-Cas9 vector as template. The PCR product and 18T-U6-sgR-CBF1 5′UTR vector were digested with KpnI and EcoRI, respectively. The digested products were ligated to generate 18T-U6-sgR-CBF1 5′UTR+U6-sgR-CBF1 3′UTR-Cas9, which was finally integrated into the binary vector pCambia1300 using HindIII and EcoRI restriction sites. For construct used to edit CBF2 or CBF3 gene, 18T-U3b-CBF2-Cas9 or 18T-U3b-CBF3-Cas9 was digested with HindIII and EcoRI and ligated with pCambia1300. For the construct used to simultaneously edit CBF1 and CBF3 genes, the U6-sgR-CBF1 5′UTR and U3b-sgR-CBF3 fragments were amplified from 18T-U6-sgR-CBF1 5′UTR-Cas9 and 18T-U3b-sgR-CBF3-Cas9 vectors, respectively. The U6-sgR-CBF1 5′UTR fragment was digested with HindIII and XhoI, and U3b-sgR-CBF3 fragment was digested with XhoI and XmaI. Both digested products were ligated with 18T-U6-sgRNA-Cas9 vector, which was digested with HindIII and XmaI, to generate the 18T-U6-sgRNA-CBF1 5′UTR+U3b-sgRNA-CBF3-Cas9 construct. The U6-sgRNA-CBF1 3′UTR fragment was then amplified from 18T-U6-sgRNA-CBF1 3′UTR vector and ligated with 18T-U6-sgRNA-CBF1 5′UTR+U3b-sgRNA-CBF3-Cas9 using KpnI and EcoRI restriction sites to generate 18T-U6-sgRNA-CBF1 5′UTR+U3b-sgRNA-CBF3+U6-sgRNA-CBF1 3′UTR-Cas9, which was integrated into pCambia1300 using HindIII and EcoRI restriction sites. For the construct used to simultaneously edit CBF2 and CBF3 genes, U6-sgR-CBF2 and U3b-sgR-CBF3 fragments were amplified and digested with HindIII/XhoI and XhoI/XmaI, respectively. Both digested products were ligated with 18T-U6-sgRNA-Cas9 (digested with HindIII and XmaI) to generate 18T-U6-sgRNA-CBF2+U3b-sgRNA-CBF3-Cas9, which was integrated into pCambia1300 using HindIII and EcoRI restriction sites. For the construct used to simultaneously edit CBF1, CBF2, and CBF3 genes, U6-sgRNA-CBF1 5′UTR and U3b-sgRNA-CBF2 fragments were amplified from 18T-U6-sgR-CBF1 5′UTR-Cas9 and 18T-U3b-sgR-CBF2-Cas9 vectors, respectively. The U6-sgRNA-CBF1 5′UTR fragment was digested with HindIII and XhoI, and U3b-sgRNA-CBF2 fragment was digested with XhoI and XmaI. Both digested products were ligated with 18T-U6-sgRNA-Cas9, which was digested with HindIII and XmaI, to generate 18T-U6-sgRNA-CBF1 5′UTR+U3b-sgRNA-CBF2-Cas9 construct. The fragments of U6-sgR-CBF1 3′UTR and U3b-sgR-CBF3 were amplified and digested with KpnI/XhoI and XhoI/EcoRI, respectively. Both digested products were ligated with 18T-U6-sgRNA-CBF1 5′UTR+U3b-sgRNA-CBF2-Cas9 (digested with KpnI and EcoRI) to generate 18T-U6-sgRNA-CBF1 5′UTR+U3b-sgRNA-CBF2+U6-sgR-CBF1 3′UTR+U3b-sgR-CBF3-Cas9, which was finally integrated into pCambia1300 using HindIII and EcoRI restriction sites. All assembled pCambia1300 constructs were transformed to wild-type Col-0 to edit CBF genes. The oligonucleotides used for CRISPR are listed in the Supplemental Table S8.
Germination and Plant Growth Assay
To test seed germination, around 120 seeds were surface-sterilized and placed on MS medium. The seeds were stratified for 3 d and then transferred to a chamber with 23°C. The seed germination was assessed after 2 d. To measure root length of seedlings, the seeds were germinated on MS medium. After 3 d, the seedlings were transferred to MS, NaCl, or mannitol medium for further growth. Fifteen seedlings were used to measure the root length. To evaluate plant growth, 4-week-old plants were used to analyze the number of leaves of each plant, the fresh weight, and leaf area. Statistical analysis was performed by using SPSS statistical software, and lsd was used to compare the mean separation significance among each genotype.
Quantitative Real-Time PCR Analysis
For low-temperature-induced gene expression analysis, 10-d-old seedlings grown on MS medium were treated with 4°C. Total RNA was extracted from whole seedlings using the TRIzol reagent (Invitrogen; Cat. No. 15596-018) according to the manufacturer’s instructions. The quantity and quality of RNA was evaluated by Nanodrop spectrophotometer (Thermo Scientific). cDNA was synthesized from 2 μg total RNA using a reverse transcription system (Promega). Synthesized cDNA was used as a template for quantitative real-time PCR using PerfeCTa SYBR Green Fastmix (Quanta Biosciences). Primers used for qRT-PCR are shown in Supplemental Table S8. The relative expression levels of CBF genes were determined by normalizing to the levels of ACTIN. First, the Ct values from both ACTIN and CBF genes were calculated by the formula POWER (2, -Ct) and then the obtained values of CBF genes were divided by the value of ACTIN to get the relative expression level.
Chilling Tolerance Assay
Three-day-old seedlings grown on MS medium were transferred to incubator with 4°C under constant light. The root length of plants was measured after 45 d treatment and the length of petioles were scored after 3 months. For plants grown in soil, 3-week-old plants were treated with low temperature (4°C) under constant light. The phenotype of plants was detected after 2 months. Electrolyte leakage assay was conducted as described with minor modification (Dong et al., 2006). Three-week-old plants grown under standard condition were transferred to a cold room with 4°C. Three rosette leaves were detached from each plant at the indicated time point and transferred to tubes with 15 mL of deionized water. Four biological replicates were performed. The conductivity of solution was measured after shaking overnight at room temperature. After measurement, the solution was autoclaved and shaken for additional 2 h, and the conductivity of solution was measured again. Finally, the percentage of electrolyte leakage was calculated by comparing the conductivity before and after autoclaving.
Freezing Tolerance Assay
Electrolyte leakage assay and survival assay were used to determine the freezing tolerance of plants in this study. The electrolyte leakage assay was performed according to the protocol described previously (Guo et al., 2002). Briefly, 3-week-old plants grown in soil were treated with or without cold acclimation (4°C, 7 d) before subjected to freezing treatment. A fully developed rosette leave was excised and placed in a small tube containing 100 μL deionized water. Three replicates were performed for each plant at each temperature point. A similar size of ice chip was added to each tube and the tube was immediately incubated in a freezing bath (model 1187; VWR Scientific) with temperature at 0°C. After placing all the tubes in the freezing bath, the program was run with 1°C decrements every 30 min until reaching to −9°C. The tubes were removed from the freezing bath when the experimental temperatures were reached and placed on ice to allow gradual thawing. The leaves and solutions were then transferred to large tubes with 25 mL deionized water. The tubes were gently shaken overnight, and the conductivity of solutions was measured. The tubes were then autoclaved at 121°C for 20 min. The autoclaved solutions were shaken for additional 3 h before the conductivity was measured again. Finally, the ratio of conductivity before and after autoclaving was calculated.
For freezing survival assay, 12-d-old seedlings grown on the MS medium were treated with cold acclimation for 7 d at 4°C before freezing treatment. The freezing treatment was performed in a freezing incubator with the following program: the temperature was set at 4°C and dropped to 0°C within 30 min and then the temperature was dropped 1°C every 1 h until reaching to the designated temperature. After treatment, the plates were transferred at 4°C for 12 h in the dark and then the seedlings were recovered at 23°C for 5 d, at which time the survival rates were counted.
Analysis of RNA-Seq Data
Reads of RNA-seq data were mapped to Arabidopsis reference genome (TAIR10) using TopHat (Trapnell et al., 2012). Differentially expressed genes were identified using cuffdiff in Cufflinks. Differentially expressed genes were defined based on the following criteria: q-value < 0.05 and at least 2-fold change of expression level compared with control samples. The criteria to identify CBF-regulated genes are as follows: (1) The genes were up- or down-regulated in either 3 or 24 h wild-type plants compared to 0 h wild-type plants; and (2) the genes were up- or down-regulated in either 0, 3, or 24 h in the cbf triple mutants compared with that in wild-type plants.
GO enrichment analysis was performed by AgriGO (Du et al., 2010). The histogram was generated using R (https://www.r-project.org/). AME (Analysis of Motif Enrichment) in the MEME Suite was used to investigate whether the CRT/DRE motif (RCCGAC) is enriched in the 1-kb promoters of the CBF-regulated genes (McLeay and Bailey, 2010). CentriMo (Local Motif Enrichment Analysis) was explored to identify the significant preference of the locations of CRT/DRE motif in the promoters of CBF-regulated genes (Bailey and Machanick 2012).
Accession Numbers
The data in this article have been deposited with links to BioProject accession number PRJNA316458 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chilling tolerance was tested for Col-0, cbf123-1, and cbf123-2.
Supplemental Figure S2. Gene expression of RD29A after ABA, NaCl, and mannitol treatment.
Supplemental Figure S3. Transcriptional profiling of wild-type plants after low-temperature treatment.
Supplemental Figure S4. GO enrichment analysis of CBF-activated genes.
Supplemental Figure S5. The distribution of CBF-regulated genes on the chromosomes of Arabidopsis.
Supplemental Figure S6. Gene expression of first-wave transcription factors.
Supplemental Figure S7. Generation of the cbf single and double mutants using CRISPR/Cas9 technology.
Supplemental Figure S8. Expression of CBF genes in the cbf mutants.
Supplemental Figure S9. Phenotype of the cbf 1 cbf3 double mutants.
Supplemental Figure S10. The expression of CBF2 gene in wild type and cbf1 cbf3 double mutants.
Supplemental Figure S11. CBF genes are not required for drought tolerance in Arabidopsis.
Supplemental Figure S12. Phenotype of the cbf single and double mutants under salt stress.
Supplemental Figure S13. A summary diagram showing the functions of three CBF genes.
Supplemental Table S1. Genes that are differentiated expressed in wild-type plants after low-temperature (4°C) treatment for 0, 3, and 24 h.
Supplemental Table S2. Genes that are differentiated expressed in cbf123-1 triple mutants after low-temperature (4°C) treatment for 0, 3, and 24 h.
Supplemental Table S3. Cold-responsive genes that are regulated by CBF proteins.
Supplemental Table S4. Functional categories of CBF-activated genes.
Supplemental Table S5. Genes that are expressed in wild type and cbf1 cbf3-1 double mutant after low-temperature (4°C) treatment for 0, 3, and 24 h.
Supplemental Table S6. Expression of CBF genes in wild type and cbf1 cbf3-1 double mutant after low-temperature (4°C) treatment for 0, 3, and 24 h.
Supplemental Table S7. Expression of CBF-regulated genes in wild type and cbf1 cbf3-1 double mutant after low-temperature (4°C) treatment for 0, 3, and 24 h.
Supplemental Table S8. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Rebecca Stevenson for technical assistance.
Glossary
- UTR
untranslated region
- CDS
coding sequence
- ABA
abscisic acid
- GO
Gene Ontology
- GA
gibberellin
- sgRNA
single-guide RNA
References
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martínez-Zapater JM (2005) Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 139: 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Machanick P (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvallo MA, Pino MT, Jeknic Z, Zou C, Doherty CJ, Shiu SH, Chen TH, Thomashow MF (2011) A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J Exp Bot 62: 3807–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12: 444–451 [DOI] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH, Zhu JK (2006) A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 26: 9533–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF III (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehan MA, Park S, Gilmour SJ, An C, Lee CM, Thomashow MF (2015) Natural variation in the C-repeat binding factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J 84: 682–693 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J (2013) A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 25: 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Ishitani M, Zhu JK (2002) An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc Natl Acad Sci USA 99: 7786–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK (2008) Metabolomics of temperature stress. Physiol Plant 132: 220–235 [DOI] [PubMed] [Google Scholar]
- Guy CL. (1990) Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41: 187–223 [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT (2002) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129: 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Kim YS, Lee M, Lee JH, Lee HJ, Park CM (2015) The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol Biol 89: 187–201 [DOI] [PubMed] [Google Scholar]
- Knight MR, Knight H (2012) Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol 195: 737–751 [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. (2015) COLD1 confers chilling tolerance in rice. Cell 160: 1209–1221 [DOI] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6: 2008–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeay RC, Bailey TL (2010) Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Pérez-Alonso M, Salinas J (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Catalá R, Salinas J (2011) The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci 180: 3–11 [DOI] [PubMed] [Google Scholar]
- Nishida I, Murata N (1996) Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu Rev Plant Physiol Plant Mol Biol 47: 541–568 [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Medina J, Salinas J (2007) Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci USA 104: 21002–21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF (2015) Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J 82: 193–207 [DOI] [PubMed] [Google Scholar]
- Popova AV, Rausch S, Hundertmark M, Gibon Y, Hincha DK (2015) The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim Biophys Acta 1854(10 Pt A): 1517–1525 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Christov NK, Tsuda S, Imai R (2014) Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol 55: 136–147 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Hincha DK (2014) A mechanistic model of COR15 protein function in plant freezing tolerance: integration of structural and functional characteristics. Plant Signal Behav 9: e977722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle C, Vaultier MN, Taconnat L, Renou JP, Kader JC, Zachowski A, Ruelland E (2005) The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol 139: 1217–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Wang Y, Hua J (2009) A moderate decrease in temperature induces COR15a expression through the CBF signaling cascade and enhances freezing tolerance. Plant J 60: 340–349 [DOI] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Mao Y, Ha S, Liu W, Botella JR, Zhu JK (2015) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep 35: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Lang Z, Zhu JK (2015) Cold responsive gene transcription becomes more complex. Trends Plant Sci 20: 466–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.