The THO/TREX complex negatively regulates phosphate starvation-induced root-associated acid phosphatase activity by suppressing ethylene signaling in Arabidopsis.
Abstract
Induction and secretion of acid phosphatases (APases) is an adaptive response that plants use to cope with P (Pi) deficiency in their environment. The molecular mechanism that regulates this response, however, is poorly understood. In this work, we identified an Arabidopsis (Arabidopsis thaliana) mutant, hps8, which exhibits enhanced APase activity on its root surface (also called root-associated APase activity). Our molecular and genetic analyses indicate that this altered Pi response results from a mutation in the AtTHO1 gene that encodes a subunit of the THO/TREX protein complex. The mutation in another subunit of this complex, AtTHO3, also enhances root-associated APase activity under Pi starvation. In Arabidopsis, the THO/TREX complex functions in mRNA export and miRNA biogenesis. When treated with Ag+, an inhibitor of ethylene perception, the enhanced root-associated APase activity in hps8 is largely reversed. hpr1-5 is another mutant allele of AtTHO1 and shows similar phenotypes as hps8. ein2 is completely insensitive to ethylene. In the hpr1-5ein2 double mutant, the enhanced root-associated APase activity is also greatly suppressed. These results indicate that the THO/TREX complex in Arabidopsis negatively regulates root-associated APase activity induced by Pi starvation by inhibiting ethylene signaling. In addition, we found that the miRNA399-PHO2 pathway is also involved in the regulation of root-associated APase activity induced by Pi starvation. These results provide insight into the molecular mechanism underlying the adaptive response of plants to Pi starvation.
Although phosphorus (P) is abundant in most soils, the amount of inorganic P (Pi), the major form of P that plants assimilate, is limited (Raghothama, 2000). This is because the majority of P in these soils exists as organophosphates or is fixed with metals, which makes Pi highly immobile. To cope with Pi deficiency, plants activate a set of adaptive responses to reprioritize internal Pi use and to enhance external Pi acquisition (Yuan and Liu, 2008). Induction and secretion of acid phosphatases (APase) is a universal response of plants to Pi starvation and is regarded as an important way by which plants scavenge Pi from external organophosphates (Tran et al., 2010a). Indeed, knockout or overexpression of some APase genes significantly affects plant utilization of external organic P (Hurley et al., 2010; Robinson et al., 2012; Wang et al., 2009, 2011, 2014).
Pi starvation-induced (PSI) APases have been biochemically characterized in several plant species (Tran et al., 2010a) and have been extensively studied in Arabidopsis (Arabidopsis thaliana; del Pozo et al., 1999; Zhu et al., 2005; Veljanovski et al., 2006; Tran et al., 2010b; Wang et al., 2011, 2014; Robinson et al., 2012; Del Vecchio et al., 2014). Although there are 44 annotated APase genes in Arabidopsis (Li et al., 2002), three APases (AtPAP10, AtPAP12, and AtPAP26) of the Arabidopsis purple acid phosphatase (AtPAP) family together account for most of the PSI APase activity on the root surface (also called root-associated APase activity) (Wang et al., 2011, 2014; Robinson et al., 2012). Among these three APases, AtPAP12 and AtPAP26 are also major intracellular APases (Veljanovski et al., 2006; Tran et al., 2010b; Robinson et al., 2012; Wang et al., 2014). In contrast, AtPAP10 is mainly a secreted APase and is predominantly associated with the root surface after secretion (Wang et al., 2011). The triple knockout atpap10/12/26 mutant retains only about 20% of the root-associated APase activity of the wild type (Wang et al., 2014). Suc and ethylene have been identified as two positive regulators of PSI root-associated APase activity (Lei et al., 2011a, 2011b; Yu et al., 2012; Wang et al., 2012). Suc seems to act as a systemic signal that moves from shoots to roots to regulate transcription of AtPAP10 mRNA while ethylene serves as a local signal that mainly affects the secretion of AtPAP10 proteins or AtPAP10 enzymatic activity on the root surface (Zhang et al., 2014). In bacteria and yeast, the signaling pathway involved in this Pi scavenging system, the so-called PHO regulon, has been well established (Lenburg and O’Shea, 1996; Wanner, 1996). Although a plant PHO regulon had also been proposed (Goldstein et al., 1988), a different and more complex regulatory network is believed to control PSI root-associated APase activity in higher plants. Ethylene, for example, plays an important role in the regulation of root-associated APase in Arabidopsis but is not found in bacteria and yeast (Lei et al., 2011b; Wang et al., 2012; Yu et al., 2012).
MicroRNAs (miRNAs), which have 21 to 24 nucleotides, are a class of small regulatory RNAs. Biogenesis of miRNAs requires the activity of RNA polymerase II, which produces complementary, long, dsRNA precursors; these precursors are cleaved by DICER-LIKE (DCL) proteins and processed into mature miRNAs (Margis et al., 2006). The cleaved double-stranded miRNAs then become associated with ARGONAUTE proteins to inhibit the expression of their target genes through either degradation of target mRNAs or blockage of protein translation (Mallory and Vaucheret, 2006). These small RNAs participate in a variety of biological processes, including plant responses to external stresses. Several miRNAs have been determined to be involved in plant responses to nutrient deficiency. The level of miRNA395, for example, is up-regulated during plant responses to sulfur deprivation (Liang et al., 2010). This miRNA targets two families of genes, ATP sulfurylases and sulfate transporter 2;1, both of which are involved in sulfate metabolism. miRNA827 targets the mRNA of NITROGEN LIMITATION ADAPTATION, which encodes a ubiquitin E3 ligase (Kant et al., 2011). Interestingly, miRNA827 transcription is specifically induced by Pi deprivation. Subsequent research demonstrated that miRNA827 modulates Pi transporter activity by affecting the NITROGEN LIMITATION ADAPTATION-mediated degradation of Pi transporters (Lin et al., 2013). The miRNA whose function in response to nutrient deficiency has been best characterized is miRNA399 (Kuo and Chiou, 2011). miRNA399 is rapidly induced by Pi deficiency. It targets PHO2 mRNA, which encodes a ubiquitin E2 conjugase. When plants are exposed to Pi deficiency, increased miRNA399 enhances plant Pi uptake capacity by reducing the level of PHO2 proteins, thus stabilizing the Pi transporters on the root surface (Huang et al., 2013). Under Pi sufficiency, overexpression of miRNA399 or knockout of PHO2 leads to an overaccumulation of Pi in shoots. Although the role of the miRNA399-PHO2 pathway in Pi signaling has been well established, to date, its specific function in the control of PSI root-associated APase activity remains unknown.
In this work, we identified an Arabidopsis mutant, hps8, with enhanced PSI root-associated APase activity. The mutant phenotypes are caused by a mutation in AtTHO1, which encodes a component of the THO/TREX complex; this complex is involved in the export of mRNAs from the nucleus and in the biogenesis of miRNAs and siRNAs in Arabidopsis (Furumizu et al., 2010; Jauvion et al., 2010; Yelina et al., 2010). Our research indicates that the THO/TREX complex negatively regulates PSI root-associated APase activity by suppressing ethylene signaling. The analysis of miRNA expression profiles in hps8 also led us to discover that the miRNA399-PHO2 module is another important regulator of PSI root-associated APase activity.
RESULTS
Characterization of the Arabidopsis Mutant hps8
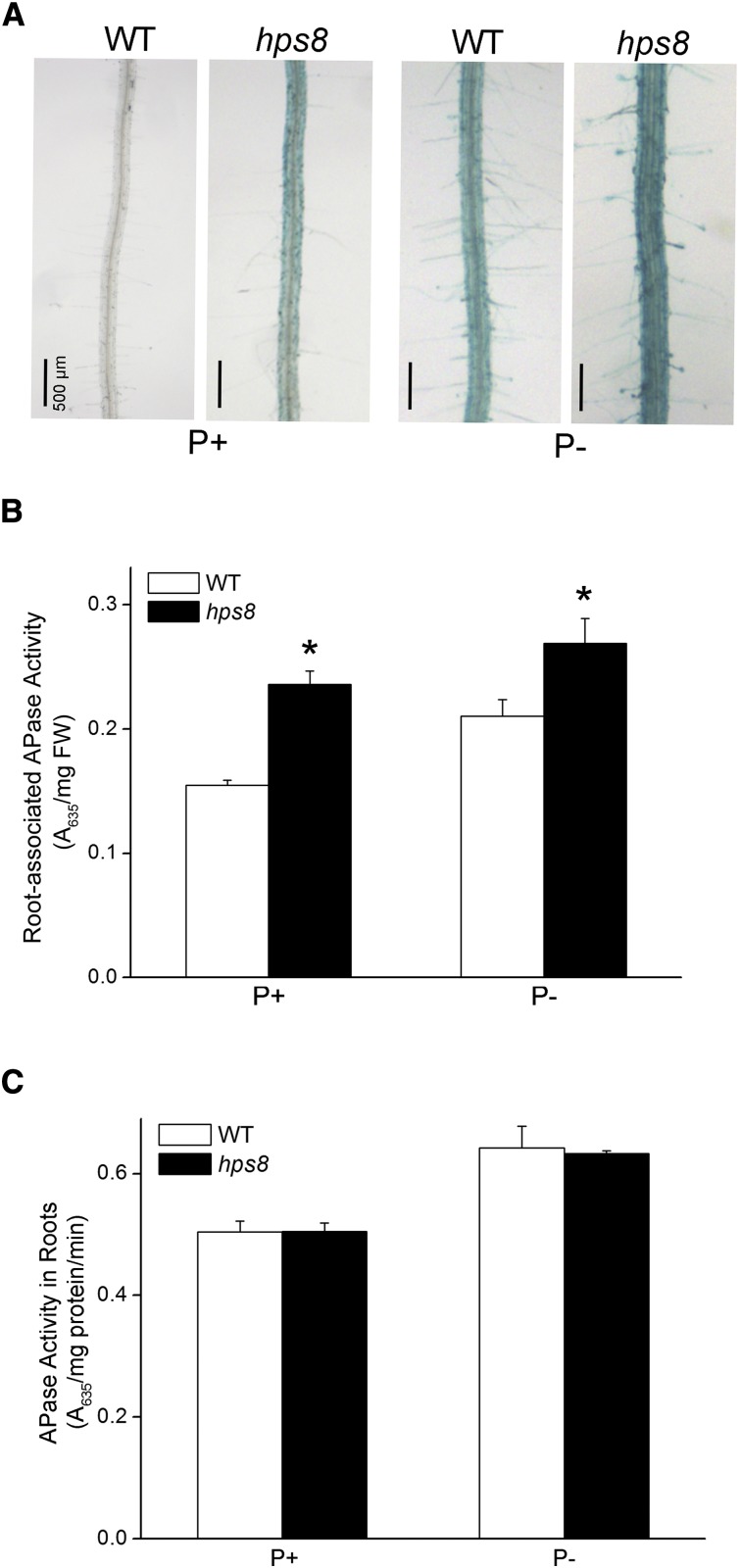
To identify novel molecular components involved in the control of PSI root-associated APase activity, we performed a genetic screen for Arabidopsis mutants with altered APase activity on the root surface. Arabidopsis seeds harvested from EMS-mutagenized plants were sown on Pi-deficient (P−) medium. Eight days after seed germination, an agar solution containing an APase substrate, 5-bromo-4-chloro-3-indolyl-P (BCIP), was overlaid on the roots of the seedlings. The cleavage of the substrate by the APases produces a blue precipitate. On the root surface of wild-type seedlings grown on Pi-sufficient (P+) medium, no blue staining was detected (Fig. 1A). In contrast, blue staining occurred on the root surface of wild-type seedlings grown on P− medium. Using this method, we identified an Arabidopsis mutant that showed enhanced BCIP staining on its root surface compared to that of the wild type under Pi deficiency (Fig. 1A). The mutant was designated hypersensitive to Pi starvation8 (hps8). hps8 also displayed light-blue staining when grown on P+ medium (Fig. 1A). The enhanced root-associated APase activity in hps8 was confirmed by quantitative analysis using BCIP as a substrate (Fig. 1B). To understand the cause of the enhanced root-associated APase activity in hps8, we analyzed the transcription of three major APase genes: AtPAP10, AtPAP12, and AtPAP26. The results showed that the transcription of these three APase genes in hps8 did not significantly differ from that of the wild type under P+ condition; under P− condition, however, the transcription of AtPAP10 and AtPAP12 was reduced 30% and 50%, respectively, in hps8, whereas transcription of AtPAP26 was unchanged (Supplemental Fig. S1). Interestingly, the total root intracellular APase activity of the wild type did not significantly differ from that of hps8 (Fig. 1C). These results suggested that the enhanced root-associated APase activity in hps8 might be due to altered posttranslational processes, such as increased secretion of APase proteins or increased APase enzymatic activity on the root surface.
Figure 1.
Analysis of Pi starvation-induced root APase activity in 8-d-old Arabidopsis seedlings of the wild type and hps8. A, Histochemical staining of APase activity on the root surface of the seedlings using BCIP as a substrate. B, Root surface-associated APase activity as determined with BCIP as the substrate. C, Root intracellular APase activity as determined with BCIP as the substrate. Values in B and C represent means with se of three replicates. Asterisks indicate significant difference (P < 0.05) from the wild type according to Student’s t test.
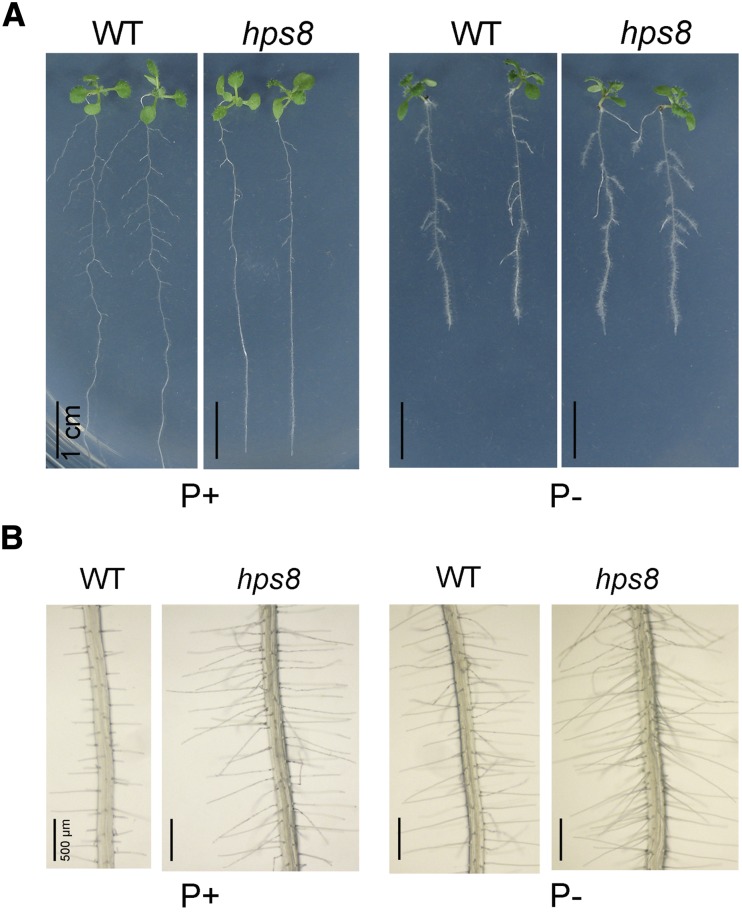
Pi deficiency reduces primary root growth and enhances the production of lateral roots and root hairs (Williamson et al., 2001; Linkohr et al., 2002). The growth of the primary root of the wild type and hps8 was similar on agar plates regardless of Pi conditions (Fig. 2A). The length of root cells and the size of the root apical meristem were also similar in the wild type and hps8 (Supplemental Fig. S2). On P+ medium, however, the hps8 seedlings had fewer and shorter lateral roots than the wild type. This difference in lateral roots was not evident when plants were grown on P− medium. hps8 also produced more and longer root hairs than the wild type under both P+ and P− conditions (Fig. 2B). When grown in soil, hps8 plants were smaller than the wild type and had serrated leaves (Fig. 3D).
Figure 2.
Root morphological phenotypes of 8-d-old Arabidopsis seedlings of the wild type and hps8 grown under Pi sufficiency (P+) and deficiency (P−) conditions. A, The whole seedlings showing root growth characteristics. B, Patterns of root hair formation.
Figure 3.
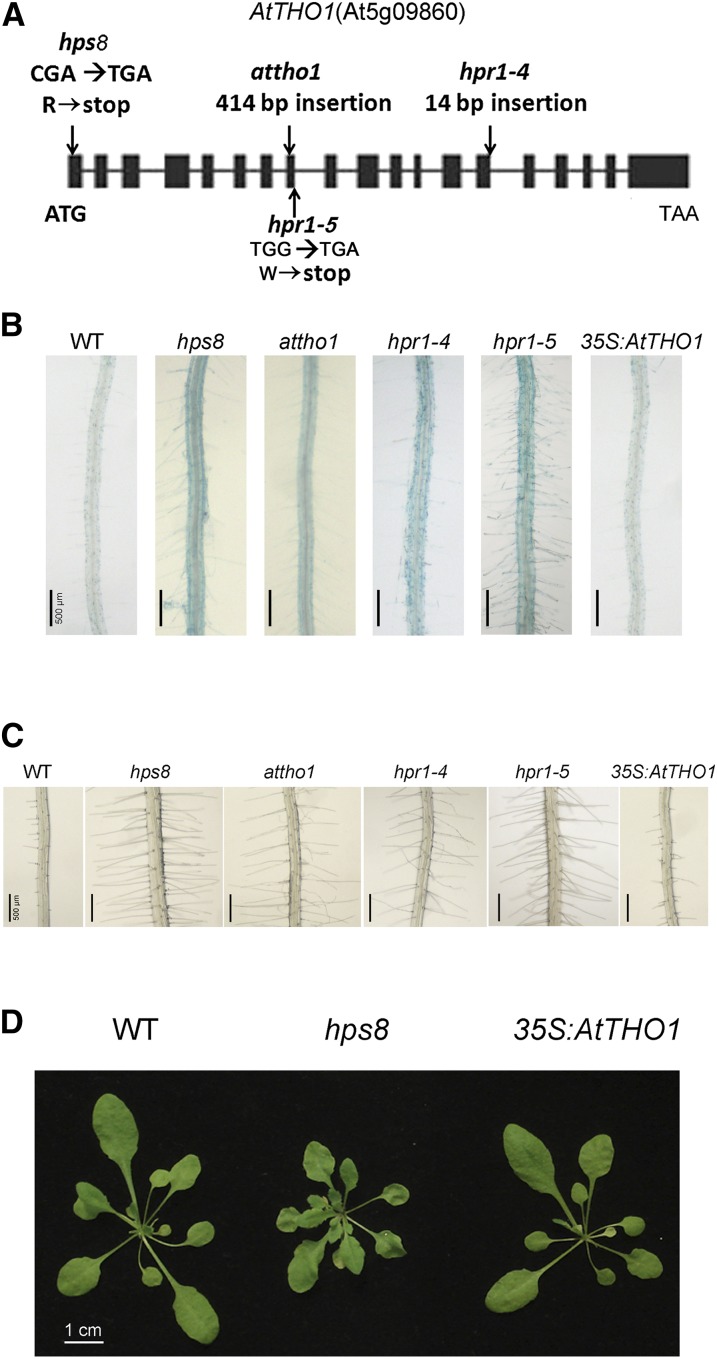
Molecular and genetic analyses of hps8. A, A diagram showing the structure of the AtTHO1 gene and the positions of the point mutation in the hps8 and hpr1-5 mutants and the insertions in the attho1 and hpr1-4 mutants. The black box and the horizontal line represent the exons and introns, respectively. The positions of start (ATG) and stop (TGA) codons are shown. B, Histochemical staining of APase activity on the root surface of 8-d-old seedlings of the wild type, hps8, attho1, hpr1-4, hpr1-5, and 35S:AtTHO1 grown on P+ medium. C, Root hair patterns of the seedlings corresponding to those shown in B. D, Morphologies of 3-week-old plants of the wild type, hps8, and 35S:AtTHO1 grown in soil.
The contents of total P and cellular Pi in shoots and roots did not significantly differ between hps8 and the wild type (Supplemental Fig. S3).
HPS8 Encodes a Subunit of the THO/TREX Complex
Analyses of F1 and F2 progeny derived from a cross between hp8 and a wild-type plant indicated that the mutant phenotypes of dark-blue BCIP-staining, enhanced root hair production, and small plant size were closely linked and could be attributed to a single recessive mutation. A map-based cloning approach was used to identify the molecular lesion in hps8. Using a set of molecular markers (Supplemental Table S1), we mapped the hps8 mutation to a 74-kb region on chromosome 5 (Supplemental Fig. S4). After all 20 annotated genes in this interval were sequenced, a point mutation was found in the first exon of the AtTHO1 gene (At5G09860; (Fig. 3A). This mutation caused a single nucleotide change of C to T, resulting in a conversion of an Arg to a stop codon. The introduced premature stop codon was located at the position of the 10th amino acid, which terminated the translation of almost the entire AtTHO1 protein. Hereafter, we refer to HPS8 as AtTHO1.
AtTHO1 shares homology with HPR1, a subunit of the THO/TREX protein complex that is conserved in yeast, animals, and humans. This protein complex functions in transcription elongation and mRNA export from the nucleus (Strässer et al., 2002; Rehwinkel et al., 2004; Katahira, 2012). The THO/TREX complex consists of seven subunits, and the counterparts of all seven subunits have been found in Arabidopsis. Five independently identified mutant alleles of AtTHO1 (attho1, hpr1-1, emu, hpr1-4, and hpr1-5) were previously reported (Yelina et al., 2010; Jauvion et al., 2010; Furumizu et al., 2010; Pan et al., 2012; Xu et al., 2015). Like hps8, the mutants attho1, hpr1-4, and hpr1-5 exhibited enhanced BCIP staining on their root surfaces and produced more root hairs than the wild type under both P+ and P− conditions (Fig. 3, B and C; data not shown). These results suggested that hps8 was another mutant allele of AtTHO1.
To further confirm that the mutant phenotypes of hps8 were caused by the mutation in the AtTHO1 gene, we introduced the wild-type genomic sequence of AtTHO1 into hps8 plants under the control of the Cauliflower Mosaic Virus 35S (CaMV 35S) promoter. The expression of the wild-type AtTHO1 gene fully restored the mutant phenotypes of hps8 with respect to BCIP staining, root hair production, and plant size (Fig. 3, B–D). Taken together, our results demonstrated that the mutation in the AtTHO1 gene was responsible for all of the hps8 phenotypes.
AtTHO1 Is Ubiquitously Expressed and Is Localized in the Nucleus
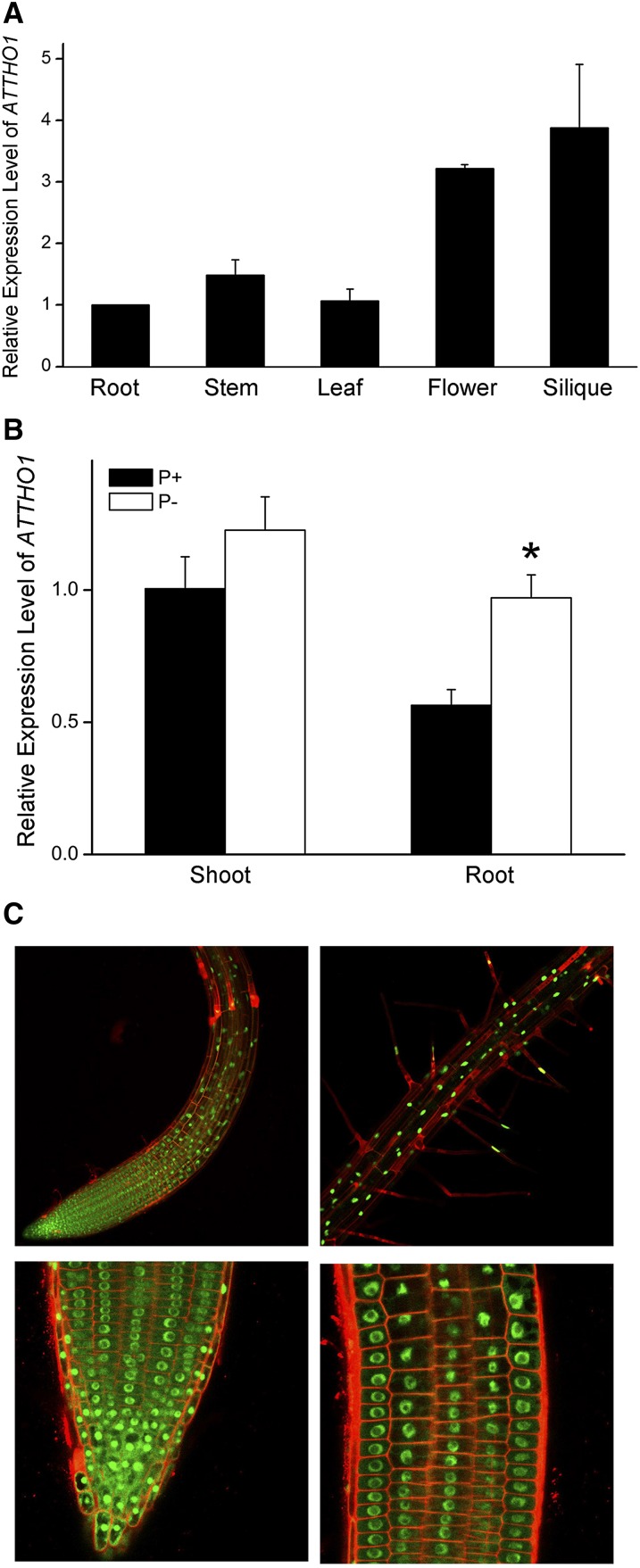
To analyze the expression patterns of AtTHO1, we performed qPCR using RNAs extracted from different tissues. The results showed that AtTHO1 expression was higher in flowers and siliques than in roots, stems, and leaves and was similar among the latter three organs (Fig. 4A). The expression of AtTHO1 mRNA was slightly increased by Pi starvation in roots but not in shoots (Fig. 4B).
Figure 4.
Expression pattern and subcellular localization of AtTHO1. A, Relative expression levels of AtTHO1 mRNA in roots, stems, leaves, flowers, and siliques. B, Relative expression levels of AtTHO1 mRNA in shoots and roots of 8-d-old wild-type seedlings grown under P+ and P− conditions. Asterisks indicate values that are significantly different from that of the wild type under the same growing conditions (Student’s t test, P < 0.05). C, Subcellular localization of GFP-AtTHO1 in roots of transgenic plants. Top, Root meristem and elongation zones (left) and maturation zone (right). Bottom, a magnified view of the meristematic region at the root tip (left) and near the elongation zone (right).
To determine the subcellular localization of AtTHO1 protein, we fused a GFP gene to the N terminus of AtTHO1 and transformed this fusion gene into wild-type plants under the control of the CaMV 35S promoter. In the root cells of the transgenic plants, a strong green fluorescence signal was observed in the nucleus (Fig. 4C), indicating that AtTHO1 is a nuclear-localized protein.
The THO/TREX Complex Negatively Regulates PSI Root-Associated APase Activity
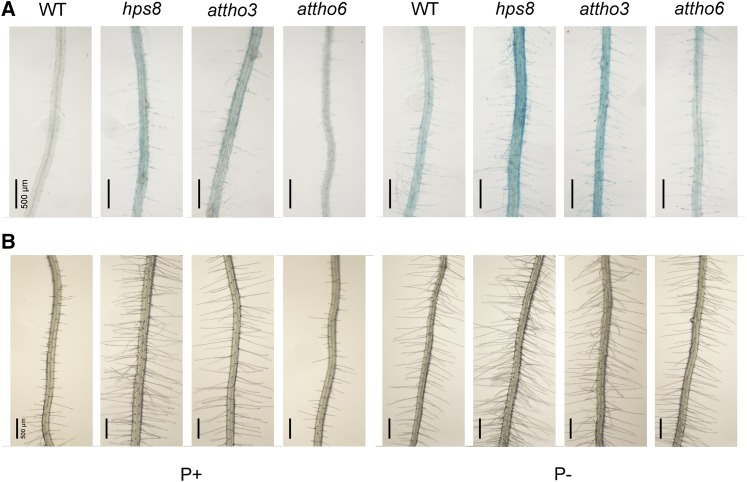
In Arabidopsis, four of the seven subunits of the THO/TREX complex (AtTHO1, AtTHO2, AtTHO3, and AtTHO6) are encoded by a single gene. The homozygote mutant of AtTHO2 (At1g24706) is embryo-lethal (Furumizu et al., 2010). To investigate whether the other two subunits of THO/TREX complex are involved in the regulation of PSI root-associated APase activity, we analyzed the effects of mutations of AtTHO3 (At5g56130) and AtTHO6 (At2g19430) genes. attho3 behaved like hps8 in exhibiting enhanced BCIP staining and overproduction of root hairs, but attho6 looked like the wild type (Fig. 5). These results indicated that the Arabidopsis THO/TREX complex negatively regulates PSI root-associated APase activity and that different components of this complex contribute differently to plant responses to Pi starvation.
Figure 5.
Root surface-associated APase activity and root hair patterns in various mutants. A, BCIP staining of APase activities on the root surface of 8-d-old seedlings of the wild type, hps8, attho3, and attho6 grown on P+ and P− media. B, Patterns of root hairs in the seedlings corresponding to those in A.
The hps8 Phenotypes Are Largely Reversed by the Inhibition of Ethylene Signaling
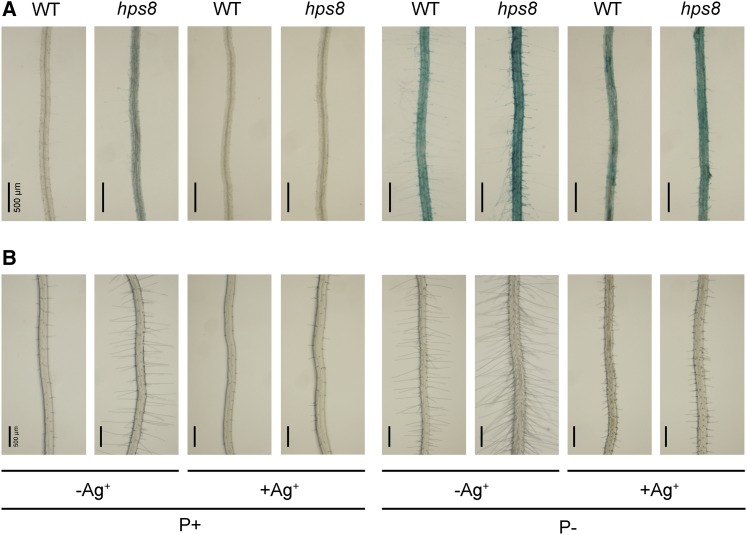
We previously showed that ethylene is a positive regulator of PSI root-associated APase activity (Lei et al., 2011b; Wang et al., 2012; Yu et al., 2012). Because the hpr1 mutants (hpr1-4 and hpr1-5) exhibit enhanced ethylene signaling (Pan et al., 2012., Xu et al., 2015), we wondered whether the hps8 mutant phenotypes were caused by enhanced ethylene signaling. Therefore, we first examined the effects of Ag+, an inhibitor of ethylene perception, on the PSI root-associated APase activity and root hair production in hps8. The seeds of the wild type and hps8 were directly sown on P+ and P− media with or without 10 μm Ag+, and phenotypes were analyzed 8 d after germination. Addition of Ag+ largely blocked the production of root hairs in the wild type under both P+ and P− conditions and reduced the BCIP staining on the wild-type root surface (Fig. 6A). For hps8, addition of Ag+ completely suppressed BCIP staining on the root surface on P+ medium and largely reduced such staining on P− medium. On P− medium with Ag+, however, the BCIP staining of hps8 was still stronger than that of the wild type. We also found that Ag+ could substantially reverse the enhanced production of root hairs by hps8 under both P+ and P− conditions, although the inhibition was not as complete as that observed for the wild type (Fig. 6B).
Figure 6.
Effects of the ethylene perception inhibitor Ag+ on the root-associated APase activity and root hair production in the wild type and hps8. A, BCIP staining of root-associated APase activity of 8-d-old seedlings of the wild type and hps8 grown on P+ and P− medium with or without addition of 10 μM Ag+. B, Root hair patterns of the seedlings corresponding to those shown in A.
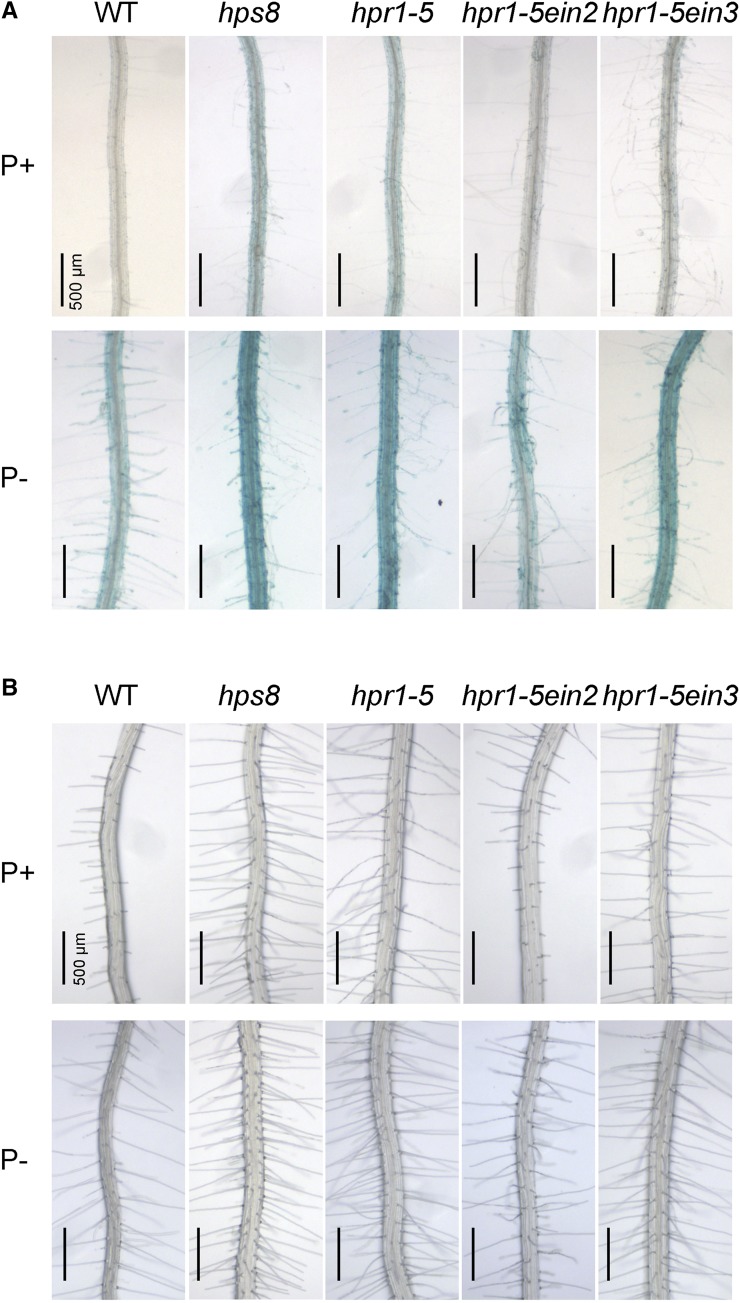
To obtain additional evidence that the enhanced APase activity and root hair production in hps8 was due to the increased ethylene signaling, we examined the root-associated APase activity and root hair phenotypes in an hpr1-5 single mutant and in hpr1-5ein2 and hpr1-5ein3 double mutants. EIN2 and EIN3 are two positive regulators of ethylene signaling (Zhao and Guo, 2011). ein2 is completely insensitive to ethylene while ein3 is only partially insensitive to ethylene. hpr1-5 exhibited phenotypes similar to those of hps8 in terms of enhanced root-associated APase activity and root hair production (Fig. 7). In hpr1-5ein2, the enhanced root hair production and root-associated APase activity were greatly suppressed under both P+ and P− conditions. In hpr1-5ein3, however, the enhanced root-associated APase activity and root hair production were only slightly suppressed under Pi sufficiency. This was probably because loss of EIN3 function does not completely block ethylene signaling.
Figure 7.
The root-associated APase activity and root hair production of the wild type and various mutants grown under P+ and P− conditions. A, Histochemical staining using BCIP as a substrate of APase activity on the root surface of 8-d-old seedlings. B, Root hair patterns of 8-d-old seedlings.
Taken together, these results indicated that increased ethylene signaling is largely responsible for the enhanced PSI root-associated APase activity and root hair production in hps8.
Regulated miRNA Biogenesis Is Important for the Control of PSI Root-Associated APase Activity
Because the altered ethylene signaling was not completely responsible for enhanced root-associated APase activity in hps8, we inferred that other factors must contribute to this mutant phenotype. Previous study showed that mutation in AtTHO1 reduced the accumulation of several miRNAs (Furumizu et al., 2010), suggesting that regulated biogenesis of miRNAs is important for the proper control of PSI root-associated APase activity. To test this hypothesis, we first examined root-associated APase activity in five mutants with defects in miRNA biogenesis. In Arabidopsis, the genes ABH1, HASTY, SE, DCL1, and HYL1 are involved in the processing of pri-miRNA to mature miRNA (Yang et al., 2006; Laubinger et al., 2008; Dong et al., 2008). The mutants for these five genes all displayed reduced root-associated APase activity under Pi deficiency as shown by BCIP staining (Supplemental Fig. S5), indicating that regulated miRNA biogenesis is important for the control of PSI root-associated APase activity (although the phenotypes of these mutants were opposite to those of hps8).
The Expression Profiles of miRNAs Are Altered in hps8
Next, we investigated how the mutation of AtTHO1 affects the accumulation of miRNAs at the genomic level. The abundance of mature miRNAs in the wild type and hps8 was compared using the miRNA-seq technique. Total RNAs were extracted from the roots of 8-d-old seedlings grown under P+ and P− conditions. The RNA samples were fractionated by electrophoresis, and the RNAs with 20 to 50 nucleotides were isolated and subjected to RNA deep sequencing. For four miRNA samples, we generated a total of 83,350,440 short reads (Supplemental Table S2). After the low quality data were filtered, more than four million reads per sample were used to map miRNAs in the Arabidopsis genome (TAIR 9.0). Among the reads for each sample, 79% to 85% could be aligned with the reference genome with perfect match (Supplemental Table S2). Two-hundred-thirty-one annotated miRNAs were detected in the sequenced samples, indicating a robust coverage of miRNAs. We first compared the miRNA expression profiles of the wild type grown under P+ and P− conditions. The differences in expression level between the two kinds of samples were represented with log2-transformed ratio. After normalization, only the miRNAs with more than 1 read/per million of mapped reads were used for comparison, and the log2 value of at least 1 was used as the cutoff to select miRNAs that differentially accumulated in P+ and P− wild-type seedlings (P value ≤ 0.01). Based on these criteria, 43 miRNAs were up-regulated and 23 miRNAs were down-regulated (Supplemental Table S3) in the roots of Pi-starved wild-type plants. The up-regulation of miRNA156, miRNA157, miRNA167, miRNA172, miRNA399, miRNA827, and miRNA2111, and the down-regulation of miRNA169 and miRNA395 were consistent with previous reports (Pant et al., 2009; Hsieh et al., 2009), indicating that the experimental condition we used for RNA-seq analysis was proper. We then compared the miRNA levels between the wild type and hps8 under normal growth conditions. Both enhanced and reduced accumulation of miRNAs were observed in hps8, indicating that the THO/TREX complex can both positively and negatively regulate the biogenesis of miRNAs. Finally, we examined the levels of the up- and down-regulated miRNAs in hps8 grown under P- condition. As shown in Supplemental Table S3, the accumulation of miRNA397, miRNA398, miRNA399, and miRNA408 was higher in P- hps8 than in P− wild type. Among the down-regulated miRNAs in P− wild type, the abundance of miRNA169 and miRNA395 was further down-regulated in P− hps8.
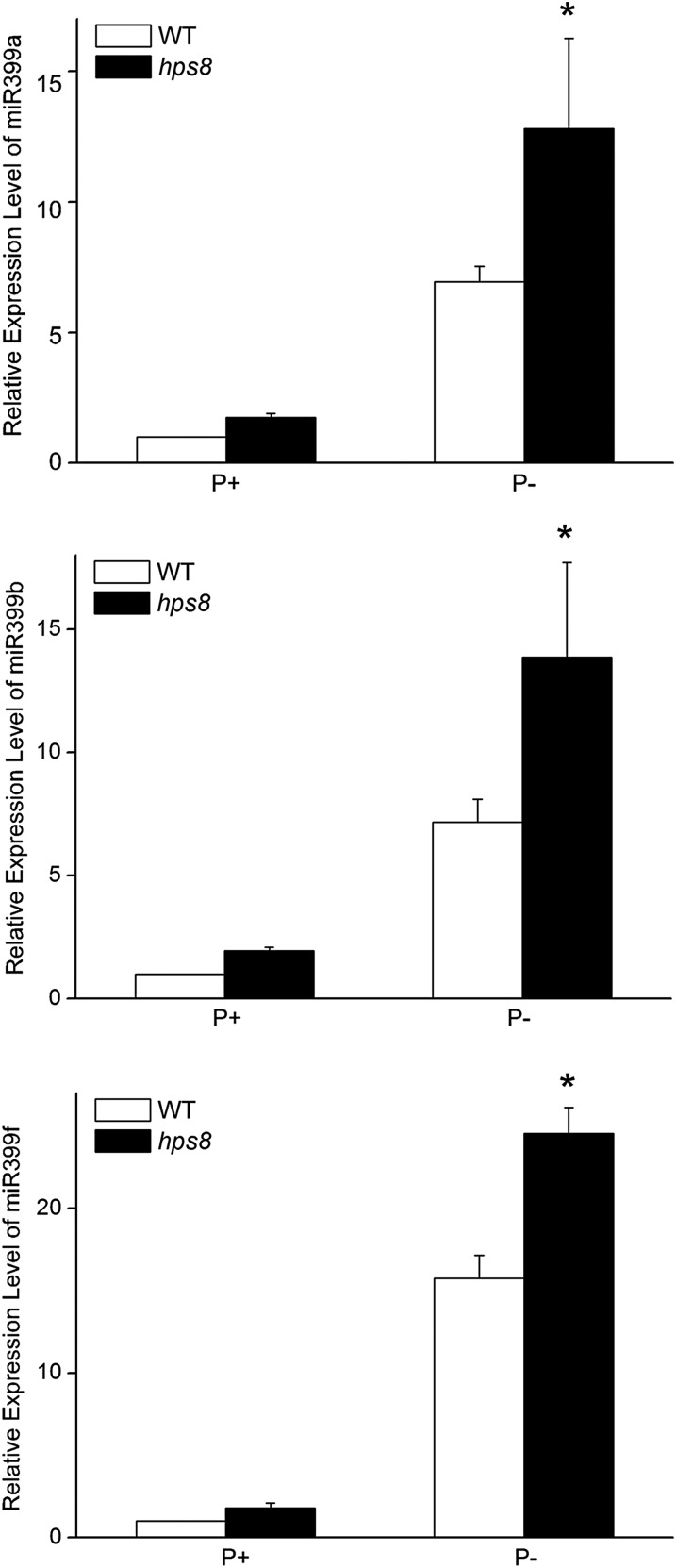
Among these differentially expressed miRNAs, miRNA399 plays a critical role in regulating Pi homeostasis in plants (Pant et al., 2008). In our RNA-seq analyses, we also found that the different members of the miRNA399 family were differentially induced by Pi starvation, i.e. miRNA399b, 399c, and 399f were the major contributors to the total amount of induced miRNA399s. Compared to their levels in P+ wild type, the levels of miRNA399b, 399c, and 399f were enhanced 30-, 31-, and 155- fold, respectively, in P− wild type. In P− hps8, however, the induction of these three miRNAs was increased by 39-, 38-, and 211-fold, respectively (Supplemental Table S3). The enhanced accumulation of these three mature miRNA399s in hps8 was confirmed by qPCR (Fig. 8).
Figure 8.
qPCR analysis of relative levels of three mature miRNA399s in the roots of 8-d-old seedlings of the wild type and hps8 grown under P+ and P− conditions. Values are means ± se with three replicates. Asterisks indicate that the mean is significantly different from that of the wild type (t-test, P < 0.05).
The miRNA399-PHO2 Pathway Is Involved in the Regulation of PSI Root-Associated APase Activity
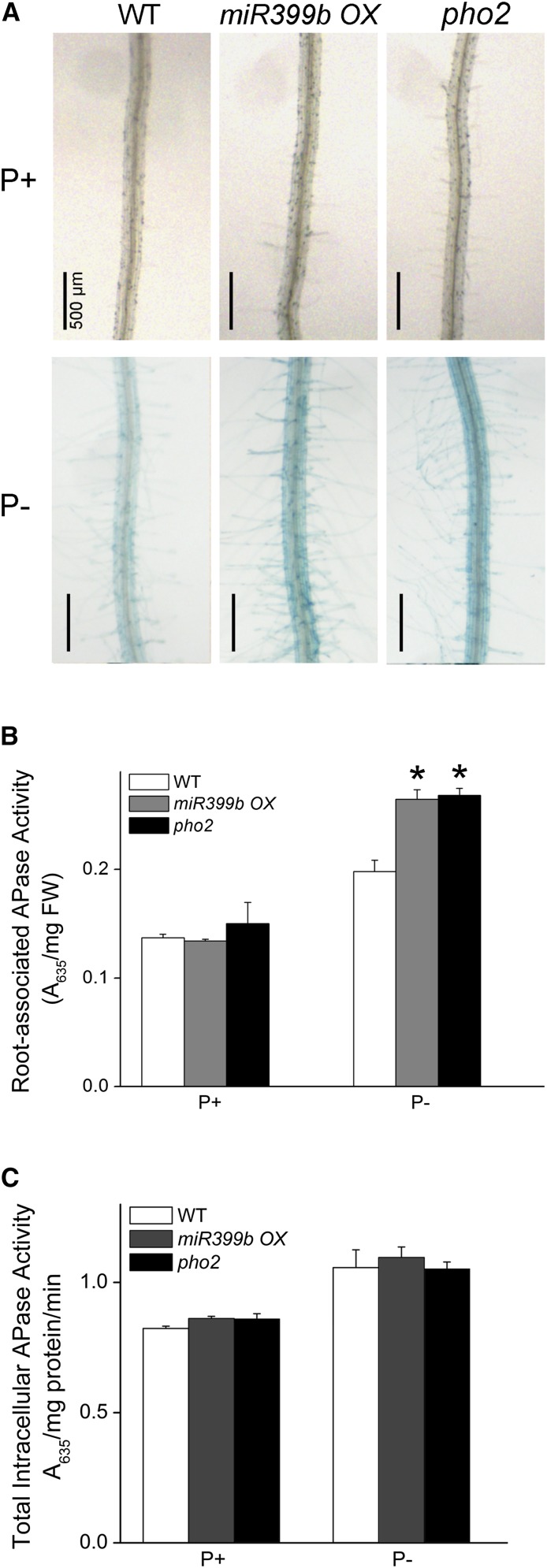
Our comparative miRNA-seq analyses suggested that the alteration of miRNA levels might affect root-associated APase activity. We then examined the root-associated APase activity in a miRNA399b-overexpressing line (hereafter referred as miRNA399 OX). There was no blue staining on the root surface of miRNA399b OX on P+ medium (Fig. 9A); under Pi starvation, however, BCIP staining was much stronger on the root surface of miRNA399 OX than of the wild type. PHO2 mRNA encodes a ubiquitin E2 conjugase and is the direct target of miRNA399. Thus, the phenotype of the pho2 mutant would be expected to be similar to that of miRNA399 OX. Indeed, pho2-1 (hereafter referred as pho2), which contains a point mutation in the sixth exon of the PHO2 gene (Aung et al., 2006), also showed enhanced BCIP staining on its root surface under Pi deficiency (Fig. 9A). The enhanced root-associated APase activity in miRNA399 OX and pho2 was confirmed by quantitative analysis of APase activity using BCIP as the substrate (Fig. 9B). To determine the cause of the enhanced root-associated APase activity in miRNA399 OX and pho2, we analyzed the transcription of three major APase genes: AtPAP10, AtPAP12, and AtPAP26. The results showed that the transcription of these three APase genes did not differ from that of the wild type under Pi sufficiency (Supplemental Fig. S6); under Pi deficiency, however, the transcription of AtPAP10 and AtPAP12 was slightly but significantly increased in miRNA399 OX and pho2 while that of AtPAP26 remained unchanged. As was the case with hps8, total root intracellular APase activity of miRNA399 OX and pho2 did not significantly differ from that of the wild type under both P+ and P− conditions (Fig. 9C). These results suggested that the miRNA399-PHO2 pathway positively regulates the secretion of APase proteins or their enzymatic activities on the root surface. Unlike hps8 and other ethylene-related mutants, miRNA399 OX and pho2 did not overproduce root hairs under normal growth conditions (Supplemental Fig. S7).
Figure 9.
Analysis of Pi starvation-induced root APase activity in 8-d-old Arabidopsis seedlings of the wild type, miRNA399 OX, and pho2 grown under P+ and P− conditions. A, Histochemical staining of APase activity on the root surface of the seedlings using BCIP as a substrate. B, Root surface-associated APase activity as determined with BCIP as the substrate. C, Root intracellular APase activity as determined with BCIP as the substrate. Values in B and C represent means with se of three replicates. Asterisks indicate a significant difference (P < 0.05) from the wild type according to Student’s t-test.
DISCUSSION
Induction and secretion of APases is an important strategy that plants use to cope with Pi deficiency in their environment. The regulatory mechanism underlying this adaptive response, however, has been largely unknown. To identify the molecular components involved in the regulation of PSI root-associated APase activity, we performed a genetic screen for Arabidopsis mutants with altered root-associated APase activity. From this screen, we obtained a series of hps (hypersensitive to Pi starvation) mutants with enhanced root-associated APase activity. hps2 is a new allele of CTR1, which is a key regulator of ethylene signaling (Lei et al., 2011a, 2011b). Knockout of the CTR1 gene causes plants to display constitutive ethylene responses (Kieber et al., 1993). HPS3 encodes ETHYLENE OVERPRODUCTION1 (Wang et al., 2004, 2012), which, when mutated, results in the overproduction of ethylene in Arabidopsis seedlings. hps4 contains a mutation in the SABRE gene. The SABRE protein was found to antagonistically interact with ethylene signaling to regulate Pi responses (Yu et al., 2012). We also obtained mutants with enhanced root-associated APase activity that did not result from altered ethylene biosynthesis or signaling. These mutants included hps7, hps9, and hps10. HPS7 is a tyrosylprotein sulfotransferase that suppresses the expression of many photosynthetic genes in roots (Kang et al., 2014). For hps9 and hps10, the mutated genes responsible for enhanced root-associated APase activity encode proteins localized in the Golgi apparatus and tonoplasts, respectively (unpublished data). These results indicated that the induction of root-associated APase activity by Pi starvation is regulated by multiple pathways.
In this work, we identified an Arabidopsis mutant, hps8, based on the alteration of root-associated APase activity. hps8 has enhanced root-associated APase activity under both Pi sufficiency and deficiency (Fig. 1). Although the transcription of two major APase genes, AtPAP10 and AtPAP12, was slightly changed, the total intracellular APase activity in hps8 is similar to that of the wild type. This suggested that the enhanced PSI root-associated APase activity in hps8 is due to the increased secretion of APase proteins or to the increased enzymatic activity of APases on the root surface. Compared to the wild type, this mutant also produces more root hairs and has smaller plant stature (Figs. 2 and 3). Our molecular and genetic analyses demonstrated that these mutant phenotypes result from a point mutation in the AtTHO1 gene (Fig. 3). AtTHO1 shares homology with HPR1, a subunit of the THO/TREX protein complex that is highly conserved in yeast, animals, and humans (Strässer et al., 2002; Rehwinkel et al., 2004; Katahira, 2012). In these organisms, the THO/TREX complex functions in transcription elongation and mRNA export from the nucleus. Previous research provided evidence that this complex also exists in Arabidopsis. First, the genes encoding all seven subunits of the THO/TREX complex have been identified in the Arabidopsis genome (Yelina et al., 2010). Second, the proteins corresponding to the six subunits of this complex have been found in the nuclei of Arabidopsis cells (Yelina et al., 2010). Our results also confirmed that AtTHO1 is a nucleus-localized protein (Fig. 4). Third, mutation in two subunits of this complex, AtTHO1 and AtTHO3, results in similar developmental defects and impaired biogenesis of tasiRNAs (Yelina et al., 2010; Jauvion et al., 2010). We also found that the mutants attho1 and attho3 both overproduce roots hairs and have enhanced root-associated APase activity under P+ and P− conditions (Fig. 5), further indicating that AtTHO1 and AtTHO3 exist as subunits of the same protein complex in Arabidopsis. In addition to functioning in mRNA export in Arabidopsis (Pan et al., 2012; Xu et al., 2015), this THO/TREX complex is involved in the biogenesis of miRNA (Yelina et al., 2010; Jauvion et al., 2010; Furumizu et al., 2010). As observed in the other mutant alleles of the AtTHO1 gene, the abundance of several miRNAs is altered in hps8 (Supplemental Table S3). Interestingly, the defects in the THO/TREX complex can either enhance or reduce the accumulation of mature miRNAs. This indicates that the roles of the THO/TREX complex in regulating miRNA biogenesis could differ depending on the species of miRNA.
As noted earlier, we previously demonstrated that ethylene is a positive regulator of root-associated APase activity induced by Pi starvation by analyzing the three hps mutants: hps2, hps3, and hps4 (Lei et al., 2011a, 2011b; Wang et al., 2012; Yu et al., 2012). All of these mutants show enhanced root-associated APase activity under Pi starvation. They also exhibit light-blue BCIP staining on their root surfaces, and they overproduce root hairs under normal growth conditions. Moreover, the total root intracellular APase activities of these three mutants do not significantly differ from that of the wild type under Pi deficiency, indicating that ethylene mainly affects the secretion of APases or the enzymatic activity of APases on the root surface. The APase phenotype of hps8 is very similar to the APase phenotypes of these three hps mutants, suggesting that the enhanced root-associated APase activity in hps8 might also result from enhanced ethylene signaling due to a mutation in the AtTHO1 gene. Consistent with this inference, hpr1-4 (a mutant allele of AtTHO1) has enhanced sensitivity to ethylene-induced senescence (Pan et al., 2012). Recently, Xu et al. (2015) reported that hpr1-5, another mutant allele of AtTHO1, has enhanced ethylene signaling. They further showed that AtTHO1 regulates the expression of REVERSION-TO-ETHYLENE1, which represses ethylene responses by promoting the activity of the ethylene receptor ETR1. When AtTHO1 is mutated, it reduces the expression of REVERSION-TO-ETHYLENE1, which derepresses ethylene responses by suppressing the activity of ETR1. Our experiments using the ethylene-perception inhibitor Ag+ suggest that the enhanced root-associated APase activity in hps8 is mainly caused by increased ethylene signaling (Fig. 6). Furthermore, we found that the enhanced root-associated APase activity in hpr1-5 is reversed in the hpr1-5ein2 and hpr1-5ein3 double mutants (Fig. 7). ein2 and ein3 are two ethylene-insensitive mutants that have defects in ethylene signaling (Zhao and Guo et al., 2011). ein2 is completely insensitive to ethylene whereas ein3 is only partially insensitive to ethylene. The levels of suppression of enhanced root-associated APase activity and root hair production in these two double mutants are correlated with the degree of their insensitivity to ethylene (hpr1-5ein2 > hpr1-5ein3). This experiment provided additional evidence that the enhanced root-associated APase activity in hps8 results from the increased ethylene signaling. Although increased root hair production may also contribute to the total root-associated APase activity in hps8, the enhanced BCIP staining in nonroot hair cells further indicated that the enhanced root-associated APase activity in hps8 was mainly due to the direct regulation by ethylene signaling. Based on these results, we conclude that the THO/TREX complex negatively regulates PSI root-associated APase activity by suppressing ethylene signaling.
Enhanced ethylene biosynthesis or signaling has previously been shown to inhibit primary root growth (Růzicka et al., 2007). Interestingly, this inhibition is not evident in hps8. We speculate that the enhancement of ethylene signaling in hps8 is too weak to inhibit primary root growth but is strong enough to enhance root hair production. In other words, production of root hairs might be more sensitive than primary root growth to ethylene.
Although enhanced ethylene signaling is largely responsible for the hps8 mutant phenotypes, the application of Ag+ did not completely reverse enhanced root-associated APase activity in hps8. Similarly, in hpr5-1ein2, the enhanced root-associated APase activity is not completely suppressed. These results suggest that other factors must also contribute to hps8 mutant phenotypes. Because the THO/TREX complex is involved in the biogenesis of miRNAs, we wondered whether miRNA homeostasis is important in regulating PSI root-associated APase activity. Indeed, we found that the five Arabidopsis mutants with defects in miRNA biogenesis had reduced root-associated APase activity (Supplemental Fig. S5). We then investigated whether the altered expression of miRNAs could affect the root-associated APase activity. Our miRNA-seq analyses revealed that the levels of 22 miRNA species, including several members of miRNA399 family, were elevated in hps8 under both P+ and P− conditions (Supplemental Table S3). miRNA399 has been shown to play a critical role in the maintenance of Pi homeostasis in plants (Liu et al., 2012; Huang et al., 2013). This prompted us to examine the phenotypes of the miRNA399 OX and the mutant of the PHO2 gene whose mRNA is the direct target of miRNA399. Our results showed that the miRNA399 OX and pho2 mutants both display enhanced root-associated APase phenotypes under Pi starvation. These results also demonstrate that the enhancement of root-associated APase activity caused by overexpression of miRNA399 is likely to be mediated by the degradation of PHO2 mRNA. Zhang et al. (2011) also reported that root-associated APase activity is enhanced in the pho2 mutant of rice, providing another line of evidence for the involvement of the miRNA399-PHO2 pathway in the control of PSI root-associated APase activity. However, miRNA399 OX and pho2 did not show light-blue BCIP staining under normal growth conditions (Fig. 9). In addition, ethylene does not affect the transcription of AtPAP10 under P− condition (Zhang et al., 2014), but the transcription of AtPAP10 is increased in miRNA399 OX and pho2 (Supplemental Fig. S6). These results suggest that the molecular mechanism by which the miRNA399-PHO2 pathway regulates the induction of root-associated APase activity by Pi starvation may be independent of ethylene production.
Finally, although the analysis of hps8 led us to discover the role of miRNA399-PHO2 pathway in the regulation of PSI root-associated APase activity, the results do not seem to indicate a direct link between the increased expression of miRNA399 and enhanced root-associated APase activity in hps8. These results include: (1) Pi overaccumulates in miRNA399 OX and pho2 shoots but not in hps8 shoots (Supplemental Fig. S3); and (2) In hps8, the level of PHO2 mRNA, which is the direct target of miRNA399, is not significantly reduced compared to that of the wild type (data not shown). Of course, we cannot exclude the possibility that a mild increase in the expression of miRNA399 is insufficient to perturb the Pi homeostasis in shoots but is sufficient to enhance PSI root-associated APase activity. Therefore, further investigation is needed to find components in addition to enhanced ethylene signaling that contribute to the increased root-associated APase activity in hps8.
In summary, we identified an Arabidopsis mutant, hps8, with enhanced PSI root-associated APase activity. The hps8 mutant carries a mutation in the THO/TREX complex, a complex that is involved in mRNA export and miRNA biogenesis in Arabidopsis. Subsequent analyses of hps8 indicated that the THO/TREX complex negatively regulates root-associated APase activity by suppressing ethylene signaling. In hps8, the expression of several miRNAs is altered, including the enhanced expression of miRNA399. The characterization of hps8 also led us to discover that the miRNA399-PHO2 signaling pathway is involved in the regulation of PSI root-associated APase activity. The next challenge will be to elucidate how the ethylene signaling and miRNA399-PHO2 pathways regulate this important adaptive response of plants to Pi starvation.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants of the Columbia ecotype were used as the wild type in this study. Half-strength MS medium with 1% (w/v) Suc and 1.2% (w/v) agar (Cat. no. A1296; Sigma-Aldrich) was used as the Pi-sufficient (P+) medium. The Pi-deficient medium (P−) was made by replacing the 0.625 mm KH2PO4 with 0.625 mm K2SO4 in the P+ medium. Seeds were surface-sterilized with 20% (v/v) bleach for 15 min. After four washes in sterile-distilled water, seeds were sown on petri plates containing P+ or P− medium. After the seeds were stratified at 4°C for 2 d, the agar plates were placed vertically in a growth room at 22 to 24°C and with a photoperiod of 16 h of light and 8 h of darkness. The light intensity was 100 μmol m−2 s−1.
Mutant Isolation
Approximately 100,000 EMS-mutagenized M2 seeds representing 6000 M1 plant lines were used for mutant screening. After the seedlings had grown on P− medium for 8 d, the roots were overlaid with a 0.5% agar solution containing 0.01% BCIP for 24 h at 23°C (Wang et al., 2011).The seedlings with enhanced BCIP staining relative to the wild type were identified as putative mutants and were transferred to soil. The plants were self-pollinated to produce progeny, and the mutant phenotypes were retested in the next generation. The mutants were back-crossed to the wild-type plants two times before they were further characterized.
Analysis of Root APase Activity
Histochemical and quantitative analyses of root intracellular and root-associated APase activities were performed as described by Wang et al. (2011).
Molecular Cloning of the HPS8 Gene
The mutant hps8 was crossed to a plant of the Landsberg erecta ecotype to generate a mapping population. The F2 progeny that displayed dark-blue BCIP staining on their root surfaces were selected, and DNAs from these seedlings were extracted for molecular mapping. A set of SSLP and CAPS markers was used to map the HPS8 gene. The sequences and chromosomal positions of the molecular markers are listed in Supplemental Table S1.
Vector Construction and Plant Transformation
For genetic complementation of the hps8 mutant, the wild-type genomic sequence of the AtTHO1 gene was amplified from the Arabidopsis genomic DNA using the primers 5′-GTCGACCCACTCTTCTTCAAGTTTGGG-3′ and 5′-ACTAGTACAAACTGAAAGCGTATTCA-3′. During amplification, the restriction enzyme sites of SalI and SpeI were added to the 5′ and 3′ ends, respectively, of the PCR product. The PCR product was ligated to the pEASY vector and verified for accuracy by sequencing. The AtTHO1 gene was then excised from the pEASY vector with SalI and SpeI enzymes and was used to replace the luciferase gene located between the 35S CaMV promoter and NOS terminator on the plant transformation vector pZhou. The resulting construct was named 35S CaMV:AtTHO1.
For subcellular localization of AtTHO1 protein, the protein coding sequence (CDS) of the AtTHO1 gene was PCR-amplified from Arabidopsis cDNAs using the primers 5′-CCCGGGATGGATGCATTTAGAGATGCTA-3′ and 5′-AAGCTTTCATGAGACGGGCATAGGA-3′. The XbaI and HindIII restriction enzyme sites were added to the 5′ and 3′ ends of the amplified CDS of the AtTHO1 gene. The amplified CDS was digested with XbaI and HindIII enzymes and cloned into the pEGAD vector to create the construct 35S CaMV:GFP-AtTHO1.
Both plant transformation vectors were mobilized into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998).
Quantitative Real-Time PCR
Total RNAs were extracted from 8-d-old seedlings, and quantitative real-time PCR was performed as described by Wang et al. (2011). The primers used for detection of the mRNA expression for HPS8 are as follows: 5′-TCCAGTGCCTCATTATGTTTGA-3′ (forward) and 5′-TGACTTAAGTTCTTCCTTCATGGTT-3 (reverse). The ACTIN gene was used as an internal control.
miRNA-Seq Analysis
Total RNAs were isolated from the roots of 8-d-old seedlings using the RNeasy Plant Mini Kit (Qiagen). The RNA samples were fractionated by electrophoresis, and the RNAs with 20 to 50 nucleotides were isolated and subjected to miRNA deep sequencing using a HiSEquation 2000 system (Illumina) with standard settings. The raw data of Illumina reads has been deposited into the NCBI Sequence Read Archive under accession no. SRP073352. The raw data were filtered to remove reads shorter than 18 nucleotides or longer than 28 nucleotides, as well as reads with poor quality. The remaining reads were mapped to the Arabidopsis reference genome (TAIR9) with exact match using bowtie. The expression counts of each miRNA were calculated and were normalized to reads per million (RPMs) mapped reads. Only the miRNAs with the expression level higher than 1.0 RPM were used for further analyses. The cutoff value for differential expression was a >2-fold change in expression with a P value ≤ 0.01.
qPCR Analysis for the Abundance of Mature miRNAs
Total RNAs were extracted with TRIzol reagent (Invitrogen) from 8-d-old seedlings grown on P+ and P− media. After the extracted RNAs were treated with RNase-free DNase I (Promega) to remove DNA contamination, 150 ng of total RNA was polyadenylated and reverse-transcribed using the Mir-X miRNA First Strand Synthesis Kit (Cat. no. 638315; Takara). miRNAs were detected using the Mir-X miRNA qRT-PCR SYBR Kit (Cat. no. 638314; Takara). Primers used for PCR analyses of each miRNA are listed in Supplemental Table S4.
Microscopy
The root hairs of the 8-d-old seedlings were examined with a stereomicroscope (model no. SZ61; Olympus).
For subcellular localization of the AtTHO1 protein, 8-d-old 35S::GFP-AtTHO1 seedlings were used for confocal microscopic analysis. The roots were excised from the seedlings and stained with 10 μM propidium iodide (PI) for 5 s and rinsed with distilled water. The specimens were examined with a confocal laser scanning microscope (710META; Zeiss). Excitation wavelengths of 546 and 488 nm were used to visualize the signals of PI staining and GFP, respectively. The emission wavelengths for PI and GFP were 573 and 507 nm.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database under the following accession numbers: AtTHO1 (At5g09860), AtTHO2 (At1g24706), AtTHO3 (At5g56130), AtTHO6 (At2g19430), EIN2 (At5g03280), and EIN3 (At5g03280).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative expression of three major APase genes in the roots of wild-type and hps8 seedlings.
Supplemental Figure S2. The root cell length and root meristem size of wild-type and hps8 seedlings grown under normal growth conditions.
Supplemental Figure S3. The contents of total P and cellular Pi in shoots and roots of the wild type and hps8.
Supplemental Figure S4. The mapping strategy for the HPS8 gene. The molecular markers used in the fine mapping are shown above the horizontal lines. The numbers below each horizontal line are the AGI coordinates on the chromosome. Distance unit: Mb.
Supplemental Figure S5. PSI root-associated APase activity of the wild type and five mutants that are defective in miRNA biogenesis.
Supplemental Figure S6. Relative expression of three major APase genes in roots of seedlings of the wild type, miRNA399 OX, and pho2.
Supplemental Figure S7. Patterns of root hairs of the wild type, miRNA399 OX, and pho2.
Supplemental Table S1. Molecular markers used for map-based cloning of the HPS8 gene.
Supplemental Table S2. Summary of the miRNA reads generated from miRNA-seq analysis.
Supplemental Table S3. Up- and down-regulated miRNAs in Pi-starved wild-type plants and their expression in hps8.
Supplemental Table S4. Sequences for the primers for qPCR analysis of mature miR399s.
Supplementary Material
Acknowledgments
We thank Dr. Chi-Kuang Wen for providing hpr1-5, hpr1-5ein2, and hpr1-5ein3; Dr. David Baulcombe for providing attho1 and attho6; Dr. Dingzhong Tang for providing hpr1-4; and Dr. Tzyy-Jen Chiou for providing miRNA399b OX and pho2-1.
Glossary
- PSI
Pi starvation-induced
Footnotes
Articles can be viewed without a subscription.
References
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Del Vecchio HA, Ying S, Park J, Knowles VL, Kanno S, Tanoi K, She YM, Plaxton WC (2014) The cell wall-targeted purple acid phosphatase AtPAP25 is critical for acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation. Plant J 80: 569–581 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumizu C, Tsukaya H, Komeda Y (2010) Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 16: 1809–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AH, Baertlein DA, McDaniel RG (1988) Phosphate starvation inducible metabolism in Lycopersicon esculentum: 1. Excretion of acid-phosphatase by tomato plants and suspension-cultured cells. Plant Physiol 87: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY, et al. (2013) Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley BA, Tran HT, Marty NJ, Park J, Snedden WA, Mullen RT, Plaxton WC (2010) The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol 153: 1112–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauvion V, Elmayan T, Vaucheret H (2010) The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 22: 2697–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Yu H, Tian C, Zhou W, Li C, Jiao Y, Liu D (2014) Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiol 165: 1156–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J. (2012) mRNA export and the TREX complex. Biochim Biophys Acta 1819: 507–513 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kuo HF, Chiou TJ (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D (2011a) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zhu C, Liu Y, Karthikeyan AS, Bressan RA, Raghothama KG, Liu D (2011b) Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol 189: 1084–1095 [DOI] [PubMed] [Google Scholar]
- Lenburg ME, O’Shea EK (1996) Signaling phosphate starvation. Trends Biochem Sci 21: 383–387 [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002) Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277: 27772–27781 [DOI] [PubMed] [Google Scholar]
- Liang G, Yang F, Yu D (2010) MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J 62: 1046–1057 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet (Suppl) 38: S31–S36 [DOI] [PubMed] [Google Scholar]
- Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PM (2006) The evolution and diversification of Dicers in plants. FEBS Lett 580: 2442–2450 [DOI] [PubMed] [Google Scholar]
- Pan H, Liu S, Tang D (2012) HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J 69: 831–843 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Köcher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E (2004) Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Robinson WD, Park J, Tran HT, Del Vecchio HA, Ying S, Zins JL, Patel K, McKnight TD, Plaxton WC (2012) The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J Exp Bot 63: 6531–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, Hurt E (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC (2010a) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14–27 [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC (2010b) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ 33: 1789–1803 [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, Knowles VL, Snedden WA, Plaxton WC (2006) Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol 142: 1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wang L, Dong J, Gao Z, Liu D (2012) The Arabidopsis gene hypersensitive to phosphate starvation3 encodes ethylene overproduction 1. Plant Cell Physiol 53: 1093–1105 [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu JW, Wang D, Liu D (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157: 1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D (2014) Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56: 299–314 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H (2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL. (1996) Phosphorus assimilation and control of the phosphate regulon. In FC Neidhardt , R Curtiss III, JL Ingraham, ECC Lin, KB Low, B Magasanik, WS Reznikoff, M Riley, M Schaechter, HE Umbrager, eds, Escherichia coli and Salmonella: Cellular and Molecular Biology, American Society for Microbiology, Washington, DC, pp 1357–1381 [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhou X, Wen CK (2015) HYPER RECOMBINATION1 of the THO/TREX complex plays a role in controlling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis. PLoS Genet 11: e1004956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yelina NE, Smith LM, Jones AME, Patel K, Kelly KA, Baulcombe DC (2010) Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci USA 107: 13948–13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Luo N, Sun L, Liu D (2012) HPS4/SABRE regulates plant responses to phosphate starvation through antagonistic interaction with ethylene signalling. J Exp Bot 63: 4527–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu D (2008) Signaling components involved in plant responses to phosphate starvation. J Integr Plant Biol 50: 849–859 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang C, Tian J, Li K, Shou H (2011) Identification of rice purple acid phosphatases related to phosphate starvation signalling. Plant Biol (Stuttg) 13: 7–15 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Lu S, Liu D (2014) A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J Exp Bot 65: 6577–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Guo HW (2011) Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol Plant 4: 626–634 [DOI] [PubMed] [Google Scholar]
- Zhu H, Qian W, Lu X, Li D, Liu X, Liu K, Wang D (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol Biol 59: 581–594 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.