Abstract
A validated animal model would assist with research on the immunological consequences of the chronic expression of stress keratins KRT6, KRT16, and KRT17, as observed in human pre-malignant hyperproliferative epithelium. Here we examine keratin gene expression profile in skin from mice expressing the E7 oncoprotein of HPV16 (K14E7) demonstrating persistently hyperproliferative epithelium, in nontransgenic mouse skin, and in hyperproliferative actinic keratosis lesions from human skin. We demonstrate that K14E7 mouse skin overexpresses stress keratins in a similar manner to human actinic keratoses, that overexpression is a consequence of epithelial hyperproliferation induced by E7, and that overexpression further increases in response to injury. As stress keratins modify local immunity and epithelial cell function and differentiation, the K14E7 mouse model should permit study of how continued overexpression of stress keratins impacts on epithelial tumor development and on local innate and adaptive immunity.
Keywords: Keratins, HPV16, E7, Hyperplasia
Highlights
-
•
Persistence of HPV infection in epithelial premalignancy is not fully understood.
-
•
HPV16 E7 expression in skin induces expression of “stress” keratin.
-
•
“Stress” keratin induction is also observed in human actinic keratosis.
-
•
Induction of “stress” keratins persists after skin grafting of E7-expressing skin.
-
•
Disruption of E7-Retinoblastoma binding abrogates keratin misregulation.
“Stress” keratins modify local immunity and epithelial cell function and differentiation. Thus, identification and validation of a suitable mouse model to study the influence of stress keratin expression normally and during tumor development is important. Mice that transgenically express high-risk HPV16 E7 oncoprotein in the skin (K14E7) presents with epithelial hyperplasia and is used for modeling HPV-associated pre-cancer. We show that “stress” keratins are induced in the K14E7 epithelium, similar to skin from patients with actinic keratosis. Thus the K14E7 mouse should permit study of how continued overexpression of stress keratins impacts on epithelial tumor development and on local immunity.
Non-Standard Abbreviations
| AK | Actinic keratosis |
| B-H | Benjamini and Hochberg |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
| dpg | Days-post-grafting |
| GO | Gene Ontology |
| GSEA | Gene set enrichment analysis |
| LFC | Log2-fold-change |
| ME | Module eigengene |
| WGCNA | Weighted Gene-coexpression Network Analysis |
1. Introduction
It is increasingly evident that prolonged misregulation of keratin expression contributes to the pathogenesis of the non-malignant hyperproliferative precursor condition, actinic keratosis (AK), as well as the eventual development of cutaneous squamous cell carcinoma (SCC) (Abbas and Bhawan, 2011, Abbas et al., 2011, Choi et al., 2010, Hameetman et al., 2013). Keratinocytes account for 95% of the cells in squamous epithelia (Quinn, 2004, Wickett and Visscher, 2006). Their cytoskeleton is composed of three types of filaments: microfilaments (actin filaments), microtubules and intermediate filaments. Intermediate filaments are encoded by about 70 genes in human, mouse and other mammalian genomes. Of all the intermediate filaments, keratins are the most diverse. There are 54 functional keratin genes in humans (Moll et al., 2008), 28 coding for type I acidic keratins (KRT9-10, 12-28 epithelial; KRT25-28, 31-40 hair/nail) and 26 coding for type II basic keratins (KRT1-8 epithelial; KRT71-86 hair/nail). All type I keratins except KRT8 are encoded on chromosome 17q21 and all type II keratins on chromosome 12q13 (Bragulla and Homberger, 2009). In mice, the orthologous genes are similarly clustered on chromosomes 11D and 15F (Hesse et al., 2004).
“High risk” human papillomaviruses (HPVs), including genotypes 16 and 18, infect anogenital and oral mucosal epithelial cells and promote development of cervical and oropharyngeal squamous cancers. Differentiation of HPV infected keratinocytes is delayed by the expression of viral non-structural proteins E6 and E7, allowing further replication of the HPV genome (McLaughlin-Drubin et al., 2012, zur Hausen, 1991). An established transgenic mouse model expressing HPV16 E7 oncoprotein in skin under the Keratin 14 promoter (K14E7) has been used as a model of epithelial premalignancy and chronic HPV infection. The HPV16 E7 oncogene is specifically expressed within keratinocytes during infection and inhibits binding of retinoblastoma (Rb) tumor suppressor protein to cell cycle regulatory proteins (Balsitis et al., 2005, McLaughlin-Drubin and Munger, 2009). Consequently, K14E7 transgenic animals are characterized by persistent epidermal hyperplasia (Herber et al., 1996, Moody and Laimins, 2010).
The impact of the HPV E7 protein as a transgene on cytoskeletal differentiation is phenotypically evident but the molecular basis of this phenotype and its impact on cell transformation and on local innate and adaptive immunity is uncertain. In this study, we define the keratin transcriptome of K14E7 and C57BL/6 mice, and compare this with the transcriptome in K14E7 mice without epithelial hyperproliferation, and with skin from patients with actinic keratosis (AK). We show that the pattern of stress keratin expression is similar in hyperproliferative K14E7 transgenic skin to that in human actinic keratosis. Further, we show that the altered keratin expression and the impact of injury on keratin expression is dependent on hyperproliferation rather than on expression of E7, validating the K14E7 mouse as a model of chronic hyperproliferative epithelium, allowing studies of the changes in local immune response associated with persisting expression of stress keratins.
2. Materials and Methods
2.1. Mice
C57BL/6 and K14E7 (C57BL/6 background) mice of different ages (2, 8 and 20 weeks of age; n = 3–4 age-match mice pairs per age group) were maintained at the Princess Alexandra Hospital Biological Resources Facility (BRF, Brisbane). C57BL/6 and K14E7 mice for grafting were obtained from the Animal Resources Centre (ARC, Perth). Heterozygous RbΔLXCXE/+ (RbΔL/+) transgenic mice were obtained from Dr. Fred Dick (Isaac et al., 2006), and were backcrossed to C57BL/6 for 10 generations. Mice homozygous for the transgene (RbΔL/ΔL) were bred and crossed to K14E7 mice (for 3 generations) to produce mice double transgenic for K14E7 and RbΔL/ΔL (Balsitis et al., 2005). Mice for grafting were between 10 and 15 weeks of age (n = 3 age-match mice pairs per grafting procedure). All mice were maintained under pathogen-free conditions and sex-matched (all female). All procedures were approved by the University of Queensland Animal Ethics Committee (AEC 290-10 and UQDI/367/13/NHMRC).
2.2. Grafting
Recipient mice were grafted as per procedures described previously (Choyce et al., 2013). Briefly, donor ear skin was split into dorsal and ventral halves, cartilage was removed, and the ear skin was then placed onto the thoracic flank (prepared matching the size of the graft) of anesthetized recipient mice. Grafts were secured using micropore tape, wrapped and bandaged. Bandages were removed after one week and the mice were monitored.
2.3. Sample Collection
Whole ear samples and grafts were collected from mice euthanized with 5% CO2. Samples were collected into Eppendorf tubes and immediately snap frozen on dry ice. Samples were kept in the − 80 °C freezer until used for RNA extraction.
2.4. Total RNA Extraction and Quality Assessment
All RNA extractions were performed using Qiagen RNeasy mini kit (catalogue #74106, Qiagen, Clifton Hill, VIC, Australia) following manufacturer's protocol and additional modifications adapted in our lab. Briefly, snap frozen ear was homogenized in 1 mL of Trizol on ice using a T10 basic ULTRA-TURRAX homogenizer (IKA, Germany). Lysate was transferred into a fresh Eppendorf tube and incubated at room temperature for 5 min. 0.2 mL of chloroform was added to the lysate, mixed by vigorously inverting for 15 s and incubated for 2–3 min at room temperature. Lysate mixture was centrifuged at 12,000 × g for 15 min at 4 °C. The upper aqueous phase containing RNA was transferred into a clean tube and mixed with one volume of freshly prepared 80% ethanol. The mixture was applied onto an RNeasy spin column and centrifuged at 8000 × g for 15 s to bind the RNA onto the membrane (all subsequent centrifugations were done at 8000 × g for 15 s unless stated otherwise). The column was washed with 700 μL of RW1 buffer and incubated with RNase-free DNase (0.5 μL DNA-se I in 35 μL of RDD buffer from Qiagen cat no. 79254) at room temperature for 15 min. Column was washed with another 350 μL of RW1 and twice with 500 μL of RPE buffer. RNA was eluted in 40 μL of RNase-free water. Quantification of purified total RNA samples was performed using nanodrop spectrophotometer. Based on the nanodrop concentration, RNA was diluted to about 200 ng/μL with RNase-free water and assessed for RNA integrity using the Agilent 2100 Bioanalyzer and RNA 6000 Nano LabChip kit according to manufacturer's protocol.
2.5. cDNA Library Preparation
cDNA library preparation from RNA samples was performed using Low Throughput Sample Protocol (suitable for 48 samples or less) from Illumina TruSeq RNA Sample Preparation Kit v2 (catalogue #15027387, #15025063 and #15027084 obtained from Illumina, San Diego, CA, USA). Briefly, mRNA was purified from total RNA using Ampure magnetic beads with attached poly-T oligos. Purified mRNA was fragmented with divalent cations and high temperature. Fragmented mRNA was mixed with random primers and reverse transcriptase and amplified through one PCR cycle (lid at 100 °C, 25 °C for 10 min, 42 °C for 50 min, 70 °C for 15 min, hold at 4 °C) to create first strand cDNA. First strand cDNA fragments were then incubated with DNA polymerase I at 16 °C for 1 h to synthesize second strand cDNA. Resultant double-stranded cDNA fragments were end repaired (3′ overhang was cleaved and 5′ overhang was filled with A to create blunt ends), and ligated to adapters (kit contains 24 different adapters). The products were purified and amplified by PCR (lid at 100 °C, 98 °C at 30 s, 10 cycles of: 98 °C for 10 s, 60 °C for 10 s, 72 °C for 10 s; 72 °C for 5 min, hold at 10 °C) to obtain the final cDNA library. Quality and concentration of cDNA libraries were checked using Agilent 2100 Bioanalyzer and DNA 1000 LabChip kit according to manufacturer's protocol.
2.6. Sequencing and Bioinformatics
Libraries were sequenced on Illumina HiSeq2000 machine with a PE 2 × 100 tag format. Raw reads of paired-end libraries were provided to QFAB in Fastq files for quality control, mapping and differential expression analysis. Raw read counts from sequenced libraries were quality checked with the FastQC tool. All libraries showed good quality profile with average quality (Phred) score of 36 per read slightly dropping at the end of the reads. Reads were trimmed using the Trimmomatic tool (Lohse et al., 2012). Read pairs were mapped to the mouse reference genome (mm10) using the Rsubread tool in R (Liao et al., 2013). Reads were also mapped to HPV16 E7 gene sequence to obtain expression level of E7 oncogene. Raw read counts per gene were normalized using the Limma-voom tool in R, which applies linear modeling to voom-transformed read counts (Law et al., 2014). Benjamini and Hochberg method was used for adjusting for multiple testing (Benjamini and Hochberg, 1995).
2.7. Database for Annotation, Visualization and Integrated Discovery (DAVID) Analysis
The entire gene list of significantly regulated genes identified from the RNA-seq analysis was uploaded onto the web documentation of DAVID. The DAVID documentation calculates the probability of ‘over-representation’ of most relevant biological terms using a modified Fisher exact probability to determine gene set-enrichment where P < 0.05 was considered significant (Huang da et al., 2009). It then organizes and condenses the large gene lists into biologically meaningful modules.
2.8. Gene Set Enrichment Analysis
GSEA was performed following a standard procedure (Subramanian et al., 2005). The gene signatures of AK (n = 4) and SCC (n = 4) samples were curated from a publicaly available microarray data set (GSE2503) (Nindl et al., 2006). The top ~ 300 genes that are up-regulated in each condition (versus normal, n = 6) were identified by differential gene expression using GEOquery and Limma R packages from the Bioconductor project.
2.9. Weighted Gene Co-expression Network Analysis (WGCNA)
Voom-transformed expression data from the RNA-seq experiments were used as input and a weighted gene network was generated by following the standard procedure of the WGCNA package (Zhang and Horvath, 2005). WGCNA provides a mean of identifying correlation patterns of genes across microarray or RNA-seq samples and subsequently finding modules of highly correlated genes/nodes via a hierarchical clustering approach coupled with topology overlap matrix-based dissimilarity measure. WGCNA constructs networks using a scale-free topology criterion where connectivity of genes/nodes follows a power-law distribution, highlighting strongly connected gene-gene pairs while penalizing weakly connected pairs. This was found to be useful for the identification of intrinsic biologically meaningful modules of coordinately expressed genes (Ravasz et al., 2002). Briefly, the similarity (concordance) between gene expression profiles across the experiments (samples) was measured using Pearson correlations. The co-expression similarities are converted into connection strengths and used to define the distance (dissimilarity) between genes (or nodes). Genes with coherent expression profiles were grouped into modules using average linkage hierarchical clustering coupled with the topological overlap dissimilarity measure. The gene modules correspond to the branches of the resulting dendrogram. To identify modules that are significantly associated with external traits, the summary profile (eigengene) of each module was correlated with predefined external traits and looked for the most significant associations. According to the WGCNA package, the module eigengene is the representative value for a module and is defined as the first principal component of a module; the normalized linear combination of genes with the maximum possible variability in a module. To facilitate biological interpretation, Gene Ontology functional enrichment analysis was performed for the genes contained in the various modules using the Bioconductor packages within R.
2.10. Immunohistochemistry-fluorescence Staining and Confocal Microscopy
Whole ears isolated from 8-week-old C57BL/6, K14E7 and K14E7/RbΔL/ΔL mice were frozen, embedded in OCT (Tissue-Tek) and cryo-sectioned at 10 μm thickness (n = 4 pairs of sex- and age-matched mice). Sections were fixed in acetone at − 20 °C for 20 min prior to staining. Blocking buffers and antibody diluents contained 10% normal goat serum, 1% bovine serum albumin and 0.025% Triton X-100 in PBS. Blocking was performed for 30 min, followed by overnight incubation at 4 °C with primary antibodies. Sections were then washed with PBS and incubated with secondary antibodies at room temperature. After washing with PBS, sections were then mounted in ProLong Gold anti-fade mounting reagent (ThermoFisher Scientific). Primary antibodies used for staining include: anti-keratin 5 (Poly19055, Biolegend), anti-cytokeratin 10 (DE-K10, Biolegend) and anti-cytokeratin 6 (ab24646, Abcam). Alexa Fluor 488- and 594-conjugated goat secondary antibodies were purchased from Jackson Immunoresearch. Images were acquired using an inverted confocal microscope (LSM510, Carl Zeiss, Inc) with a 63 × n.a. 1.4 oil objective using the associated ZEN software. Images were not cropped and Adobe Photoshop and Illustrator CC 2014 (Adobe Systems Incorporated, San Jose, USA) were used for preparation of figures.
3. Results
3.1. K14E7 Skin and Actinic Keratoses Shares Transcriptional Features
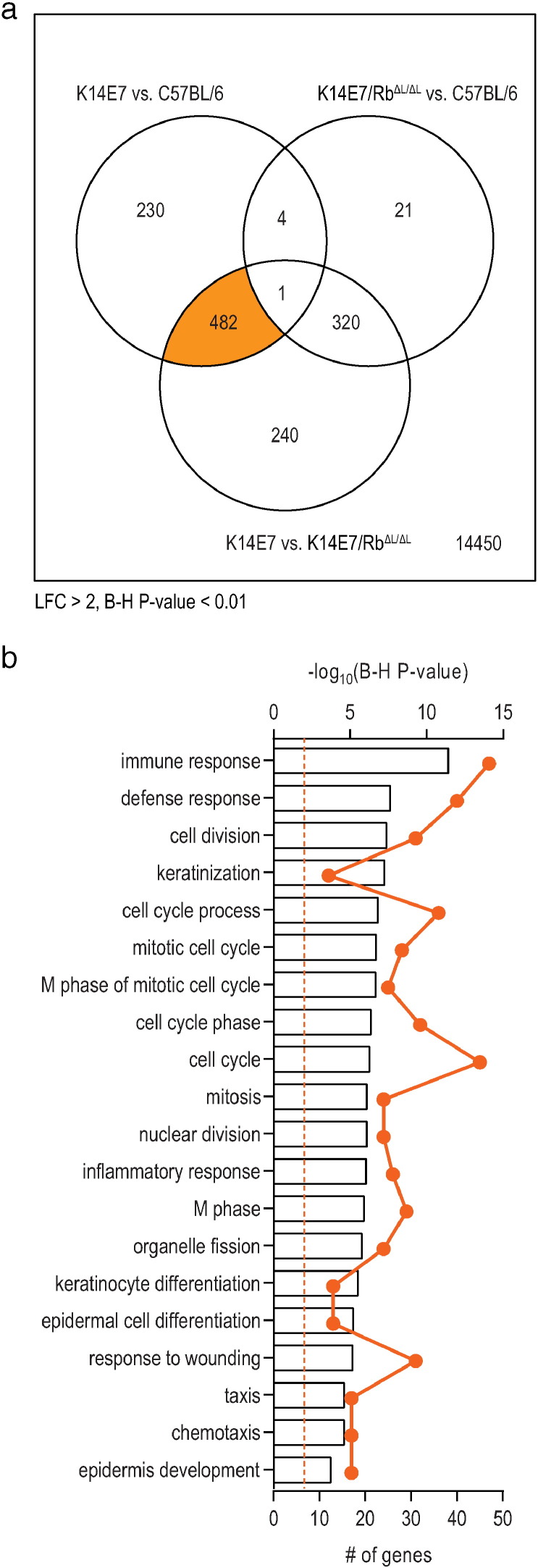
Mice heterozygous for the K14E7 transgene driven from a keratin 14 promoter have hyperproliferative epithelium, whereas K14E7 mice with a homozygous mutation in the Rb protein (RbΔLXCXE/ΔLXCXE) lack this phenotype (henceforth referred to as K14E7/RbΔL/ΔL mice) (Balsitis et al., 2005). To analyze the effects of E7 expression, epithelial proliferation, epithelial maturation, and wound healing on the skin transcriptome, we performed RNA-seq deep-sequencing analysis on ear skin from C57BL/6, K14E7 and K14E7/RbΔL/ΔL mice. We focused our analysis on the genes whose expression is altered when comparisons are made between K14E7 versus C57BL/6 and also between K14E7 versus K14E7/RbΔL/ΔL mice (Fig. 1a) as these should be genes whose expression are altered by hyperproliferation, as opposed to other effects of E7 expression, or effects of Rb mutation. We found that 482 genes in K14E7 mice were differentially expressed when compared to C57BL/6 and also when compared to K14E7/RbΔL/ΔL mice, using a conservative threshold of significance of Log2-fold-change of > 2 and a Benjamini and Hochberg (B–H) adjusted P-value of < 0.01 (Fig. 1a). We next utilized the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resource tool to discover enrichment of biological processes/functions defined by this subset of genes (Huang da et al., 2009). The top 20 significantly enriched Gene Ontology (GO) biological processes were identified (Fig. 1b), and included cell cycle and proliferation regulation, immune function, skin development and keratinocyte differentiation. These findings are consistent with the phenotype of the K14E7 mouse.
Fig. 1.
Differential gene expression and pathway analysis of RNA-seq data on ear skin from 8-week-old C57BL/6, K14E7 and K14E7/RbΔL/ΔL mice. (a) Differential gene expression was detected using Empirical Bayesian statistics embedded within the Limma-voom R package where genes that pass the cut-off of Log2-fold-change (LFC) > 2 and adjusted P < 0.01 were considered statistically significant. (b) Top 20 GO biological processes enriched in the orange intersect in K14E7 skin identified from DAVID analysis. Pathways are considered significant if the P-value (after B–H correction) is < 0.01 (orange dotted line).
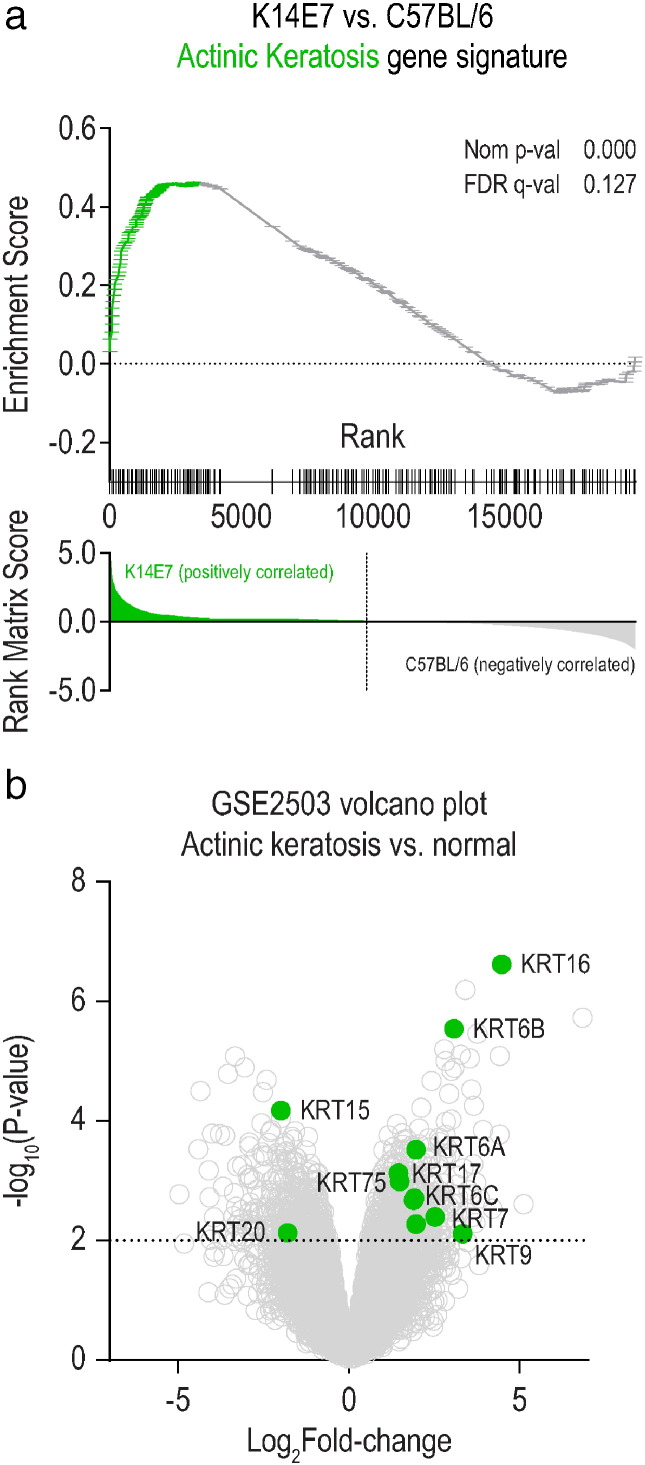
We further interrogated the RNA-seq expression data using the gene set enrichment analysis (GSEA) software package (Subramanian et al., 2005), to test whether the skin transcriptome of K14E7 mice would share transcriptional characteristics with hyperproliferative human squamous precancer actinic keratosis (AK) lesions and human squamous cell carcinomas (SCC). Transcriptional signatures of human AK and SCC lesions were curated from a publically available microarray data set (GSE2503) (Nindl et al., 2006), using the top ~ 300 genes up-regulated in each condition in comparison to normal skin. GSEA demonstrated that the AK signature (Fig. 2a), but not the SCC signature (data not shown), was significantly enriched in K14E7 skin when compared with normal mouse skin. Inspection of the “leading-edge” subset of genes in the AK gene set established that genes that encode for keratins, chemokines and chemokine receptors contribute the most to the enrichment (Subramanian et al., 2005). Thus, we examined the expression profile of keratin genes from the human AK/SCC microarray data set with a specific focus on AK data. Differential gene expression analysis revealed that keratin genes associated with epidermal growth, differentiation and stress response (KRT6A, KRT6B, KRT9, KRT16, KRT17) were significantly up-regulated in AK versus normal (Fig. 2b) as observed in our mouse model of epithelial premalignancy.
Fig. 2.
(a) Gene set enrichment analysis of a signature of ~ 300 up-regulated genes in human actinic keratosis patients in the ranked list of genes differentially expressed by K14E7 skin vs C57BL/6 skin. Genes in the leading edge are highlighted in green. (b) Volcano plot of all genes (grey) or keratin genes in the leading edge (green).
3.2. Effect of Epithelial Maturation on Keratin Expression in K14E7 and C57BL/6 Mice
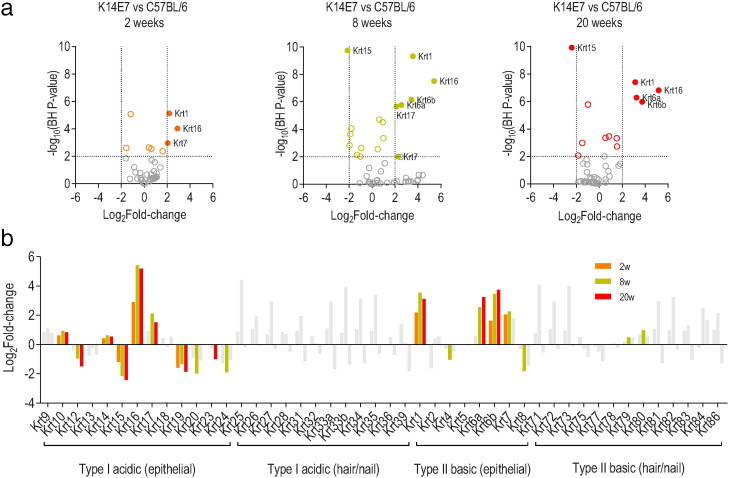
Keratin expression changes with maturation of epithelial cells. However, little is known about how chronic hyperproliferation affects expression of keratins across the epithelium over time. Therefore, we expanded the RNA-seq analysis to examine the expression of keratin genes in C57BL/6 mice and K14E7 mice of different ages. We observed significant differential expression of 8 genes that encode for keratin proteins at 2 weeks of age, 17 at 8 weeks and 12 at 20 weeks (adjusted P-value < 0.01) (Fig. 3a). The keratin genes that were significantly differentially expressed were largely restricted to the epithelial subgroups of both type I and type II keratins families (Fig. 3b). Notably, there were significant alterations with age in the transcription of the basal keratin Krt14, but not of the basal keratin Krt5, and of the keratins associated with cell differentiation (Krt1 and Krt10) (Fig. 3b). Consistent with the findings in Fig. 2b, keratin genes associated with epidermal injury (Krt6a, Krt6b, Krt16 and Krt17) were also significantly altered in the K14E7 mouse model over time (Fig. 3b).
Fig. 3.
Effect of HPV16 E7 on expression of keratin genes at 2 weeks, 8 weeks and 20 weeks of age in transgenic mouse whole ear skin. (a) Volcano plot of differential expression of keratin genes between K14E7 and C57BL/6 mice. Significant genes above the threshold of log-ratio ≥± 2 are as labeled as colored rings while those that also attain statistical significance (adjusted p-value < 0.01) are labeled as solid orange (2 weeks), gold (8 weeks) or red (20 weeks) circles. (b) Differential expression of keratin genes categorized into subfamilies. Genes that are differentially expressed in a significant manner (adjusted p-value < 0.01) are plotted in orange (2 weeks), gold (8 weeks) or red (20 weeks) while genes that did not attain statistical significance are plotted in gray.
Overall, these changes are consistent with the effect of E7 protein on promoting proliferation and delaying differentiation of basal keratinocytes in K14E7 mice. The results also suggests that presence of hyperproliferation mimics or induces epidermal injury-type responses in keratinocytes.
3.3. Effect of Grafting on Keratin Expression in Mouse Skin
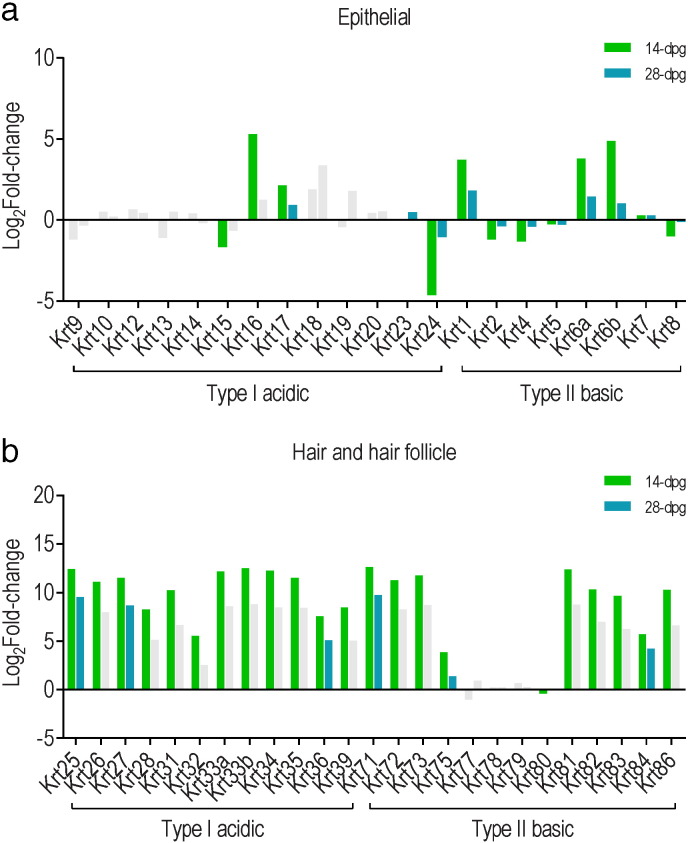
We have previously emulated localized chronic HPV infection by grafting skin of K14E7 mice onto syngeneic immune competent mice (Dunn et al., 1997, Frazer et al., 2001). This method of epithelial wounding allowed investigation into the effect of E7 on the regulation of expression of keratins associated with epidermal injury and repair. We analyzed the keratin transcriptome of skin grafted from C57BL/6 non-transgenic (C57) and E7-expressing transgenic (E7) animals to non-transgenic C57 recipients. C57BL/6 ear skin grafted onto C57BL/6 mice was analyzed for differential gene expression at day 14 post-grafting (14-dpg) and day 28 post-grafting (28-dpg). Transcription of keratin genes associated with injury and wound healing (Krt6a, Krt6b, Krt16 and Krt17) and epithelial differentiation (Krt1) were significantly induced in grafted skin at 14-dpg and to a lesser extent at 28-dpg (Fig. 4a) and significant up-regulation of multiple keratin genes associated with hair and hair follicle development was also observed, more evidently in 14-dpg skin than 28-dpg skin (Fig. 4b).
Fig. 4.
Differential expression of (a) epithelial, (b) hair and hair follicle specific or root sheath keratin genes between grafted and non-grafted skin. 14-dpg C57onC57 and 28-dpg C57onC57 samples were compared to 8 week old C57BL/6 sample (B–H adjusted P-value < 0.01).
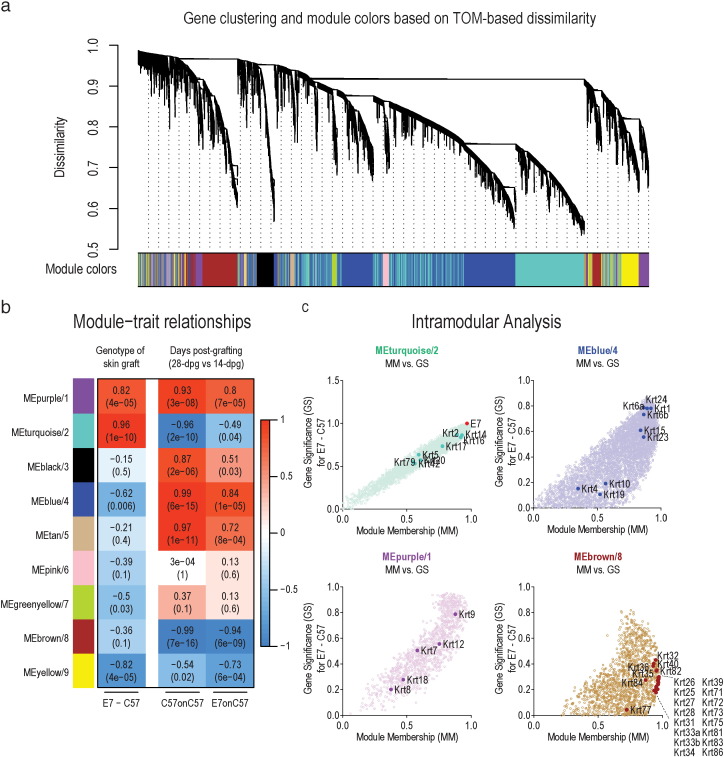
To discover gene expression changes within biologically significant gene networks in response to grafting, graft healing, and E7 expression, we performed weighted gene co-expression network analysis (WGCNA) (Zhang and Horvath, 2005) as this approach significantly alleviates the biases from multiple hypothesis testing. The analysis identified 9 modules (differentially colored) containing ~ 100–5000 genes per module (Fig. 5a). The module eigengene (ME) of each module was calculated and correlated with our predefined traits. Several of the modules displayed significant correlation or anti-correlation with the genotype of the skin graft or with days post-grafting (28-dpg versus 14-dpg) (P < 0.01) (Fig. 5b).
Fig. 5.
WGCNA analysis. (a) Clustering dendrogram of genes, with gene dissimilarity based on topological overlap, together with assigned module colors. (b) Module-trait relationship. Each row responds to a colored module eigengene (ME) detected by WGCNA analysis and each column to a trait. C57 and E7 skin grafts were distinguished as binary traits, with C57 set as 0 and E7 set as 1. The corresponding correlation (top) and p-value (bottom) is displayed in each cell. The table is color-coded by correlation according to the color key. (c) Scatterplot of the intramodular analysis (module membership versus gene significance) of genes found in turquoise, blue, purple and brown modules. E7 and keratin genes are highlighted as solid circles.
Specifically, purple and turquoise modules displayed significant correlation with E7 skin graft procedures while blue and yellow modules displayed significant anti-correlation (Fig. 5b). Similarly, modules corresponding to purple, black, blue and tan displayed significant correlation with days post-grafting while turquoise, brown and yellow displayed significant anti-correlation (Fig. 5b). We therefore performed GO functional enrichment analysis to broadly understand the biological significance of the identified modules. The top 10 significantly enriched GO functions for modules that displayed significant correlation or anti-correlation as highlighted above are shown in Table S1. By integrating the module-trait association analysis and functional enrichment analysis (Bonferroni adjusted P-value < 0.01 was considered significant), it was determined that E7 skin grafting was significantly correlated with antigen processing and major histocompatibility complex (MHC) regulation (MEpurple/1), and cell cycle, proliferation and DNA repair (MEturquoise/2). Conversely, C57 skin grafting was significantly correlated with, and E7 skin grafts are anti-correlated with, RNA processing, protein translation and cellular metabolic processes (MEblue/4), and cell migration and extracellular matrix regulation (MEyellow/9).
The modules that correlated or anti-correlated with time after grafting were generally common between C57 and E7 skin grafts, and displayed significant correlation with antigen processing (MEpurple/1), muscle development (MEblack/3), and RNA processing, protein translation and cellular metabolic processes (MEblue/4), and anti-correlation with cytoskeletal, hair and keratin filament regulation (MEbrown/8) and extracellular matrix regulation (MEyellow/9). Interestingly, WGCNA could detect that the turquoise module was significantly anti-correlated with the timing of graft healing in C57 grafts whereas this effect was slightly dampened in E7 grafts (Fig. 5b).
The keratin genes of interest were mostly associated within the modules with the most correlation or anti-correlation with the traits of interest (Fig. 5b and c). Keratin genes enriched in the brown module were hair and hair follicle associated type I and II keratins (Fig. 5c), likely as a result of hair regrowth after grafting of normal skin, and specific enrichment of this subset of keratin genes was not seen for E7 grafts. Epithelial keratin genes associated with epidermal injury (Krt6a, Krt6b, Krt16 and Krt17) and keratinocyte proliferation and differentiation (Krt1, Krt5, Krt9, Krt14 and Krt15) fell within turquoise, blue and purple modules (Fig. 5c) and showed high module membership (MM) (high intramodular connectivity) within their respective modules, and also high gene significance (GS) (biological significance), indicating that these genes are key genes associated with the effects of grafting E7 skin (Zhang and Horvath, 2005).
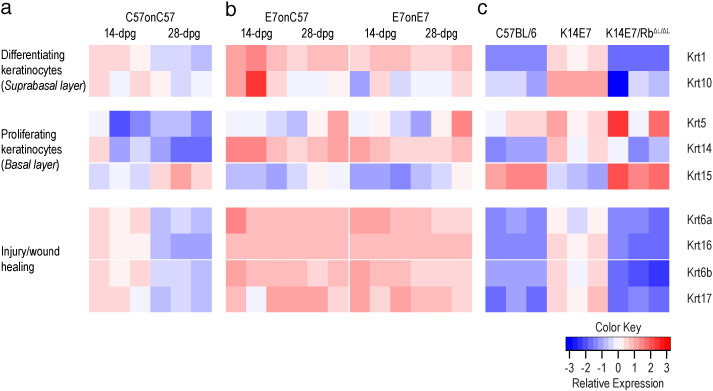
3.4. K14E7 Skin and Skin Graft is Characterized by Aberrant Regulation of Wound Healing-related Keratins
Profound differences in keratin gene expression patterns were noted during the course of graft healing. For C57 grafts on C57 recipients, induction of keratins associated with the differentiating layer of the epidermis, but not basal keratins, was observed 14 days after grafting (Fig. 6a). Keratins associated with “activated” keratinocytes, typically involved with injury and wound healing, were also induced at day 14 (Fig. 6a), and these effects were reduced by 28 days post grafting. Krt15, seen in mature/stable basal cells with lower rates of division in the stratified squamous epithelium (Porter et al., 2000), was in contrast more evident 28 days post grafting (Fig. 6a). In contrast, for E7 grafts to C57 or E7 recipients, marked up-regulation of the major keratins of basal/proliferating, differentiating and activated keratinocytes was observed at day 14 (Fig. 6b) and persisted at day 28, showing that E7 expression is associated with an exaggerated wound healing response (Fig. 6b).
Fig. 6.
Gene expression heatmap of keratins markers for keratinocyte differentiation, proliferation and injury/wound healing in (a) C57 grafts on C57 recipients, (b) E7 grafts on C57 recipients and E7 grafts on E7 recipients, and (c) 8-week-old C57BL/6, K14E7 and K14E7/RbΔL/ΔL mice. The intensity of the heatmap is scaled within each gene.
To distinguish the effects of E7 expression and E7-induced epithelial hyperplasia on regulation of keratin gene expression, K14E7/RbΔL/ΔL mice which express a mutant form of Rb that cannot be bound by E7, allowing for normal cell cycle regulation by Rb (Balsitis et al., 2005), were examined. The keratin expression profile in the ear skin of the K14E7/RbΔL/ΔL mouse closely resembled that of C57BL/6 mice (Fig. 6c), indicating that the alteration in keratin gene expression in K14E7 mice is a consequence of hyperplasia rather than other effects of E7. In summary, hyperplastic K14E7 skin demonstrates keratin expression reminiscent of a persistent wound healing response, and this effect is further enhanced in grafted E7 skin.
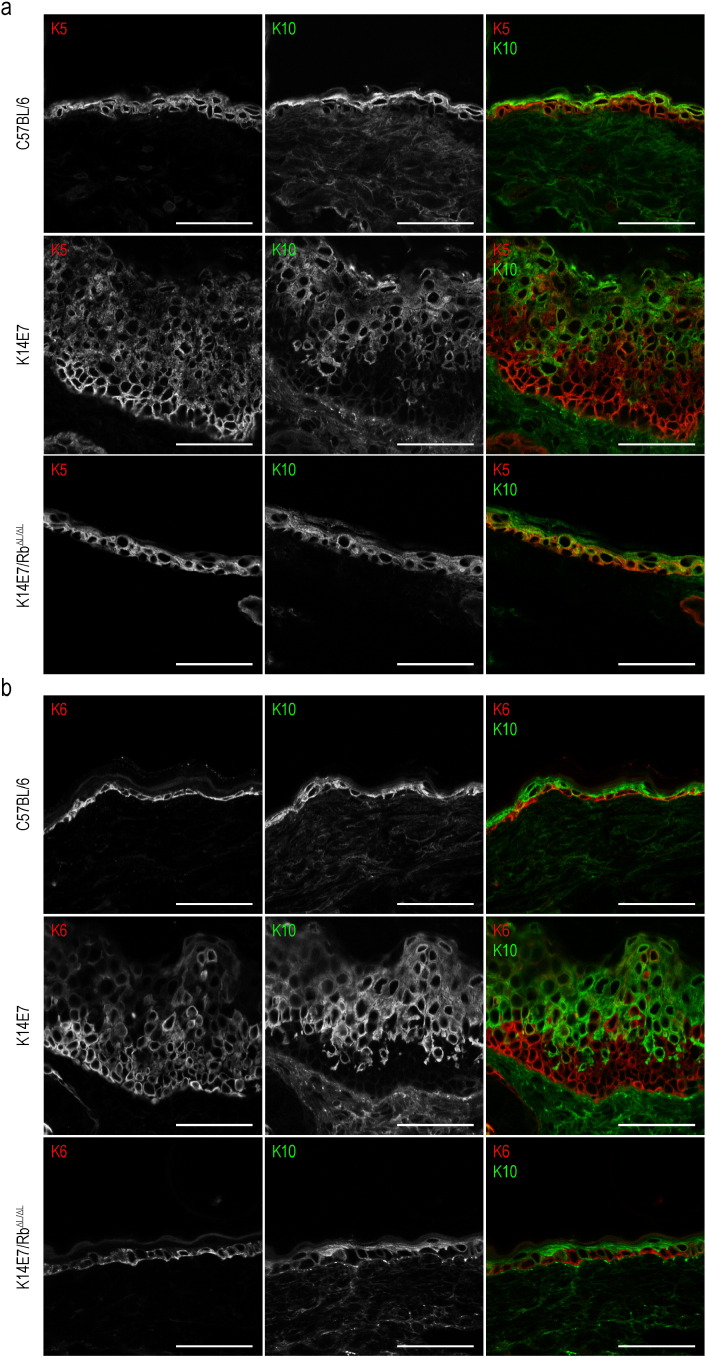
3.5. Disrupted Keratin Localization in K14E7 Ear Skin
Confocal microscopy was used to validate the transcriptomics findings, by determining the site of expression of keratin proteins. In C57BL/6 ear skin, Keratins 5 and 10 respectively defined the basal and suprabasal layers of the epidermis (Fig. 7a). In contrast, the localization of these keratin markers in K14E7 skin is more disorganized, coinciding with markedly expanded basal and spinous layers (Fig. 7a). In K14E7 epidermis, but not in C57BL/6 or K14E7/RbΔL/ΔL epidermis, Keratin 6, associated with injury and wound healing, was extensively expressed (Fig. 7b).
Fig. 7.
Localization of keratin proteins in C57BL/6, K14E7 and K14E7/RbΔL/ΔL ear skin. OCT-embedded sections were fixed with acetone prior to staining. Images were acquired using confocal microscopy whereby primary antibodies that target the keratin proteins are differentially labelled with fluorescently-conjugated secondary antibodies. Red denotes (a) keratin 5 or (b) keratin 6, while green denotes keratin 10. Scale bars = 50 μm.
In summary, HPV16 E7-induced epithelial premalignancy is associated with chronically disorganized keratin expression and localization resembling that seen acutely with epidermal injury or wound healing.
4. Discussion
In this study, we demonstrate that persisting epithelial hyperplasia induced by the HPV16 E7 protein induces a pattern of “stress” keratin expression in mouse epithelial cells resembling that seen in premalignant lesions of human epithelium (Nindl et al., 2006). This change is associated with epithelial proliferation induced by E7, and not directly by the E7 protein itself or by other biological effects associated with that protein. Further, the induced hyperplasia is associated with significant augmentation of expression of wound keratins in response to injury, when compared with normal skin. Epithelial cells, and transgenic mice, engineered to lack expression of particular keratins have been helpful to map out the biological function of “stress” keratins (Vijayaraj et al., 2007) including KRT6 (Loschke et al., 2016), KRT16 (Lessard and Coulombe, 2012) and KRT17 (Depianto et al., 2010). Animal models in which “stress” keratins are continuously overexpressed should similarly prove useful in defining the chronic consequences of their expression.
Chronic injury and tissue repair have long been recognized as predisposing to the development of epithelial cancers. Induction and maintenance of an injury/wound healing-like keratin response is a common characteristic in the pathogenesis of hyperproliferative disorders such as AK and SCCs, as well as HPV infection and associated premalignancy. Immunohistological analyses and gene expression profiling using microarray technology have independently identified that hyperproliferative disorders such as AK and SCCs consistently express the injury/wound healing related keratins KRT6, KRT16 and KRT17 (Chu and Weiss, 2002, Hameetman et al., 2013, Nindl et al., 2006).
It is recognized that wound healing keratins have multiple biological functions beyond their contribution to cell structure. KRT6 destabilizes desmosome mediated cell-cell adhesion by a mechanism involving sequestration of Src protein and inhibition of Src kinases (Rotty and Coulombe, 2012), enabling keratinocyte migration for wound healing. KRT16 regulates local induction of potentially tissue damaging innate immune responses (Lessard et al., 2013), and KRT17 polarizes adaptive immune responses away from potentially tissue damaging Th1 and Th17 type to antibody inducing Th2 type responses (Depianto et al., 2010). KRT17 also enables expression of chemokine ligands CXCL9, 10 and 11 (Chung et al., 2015) that through interaction with CXCR3, enhance epithelial growth and migration but also promote tumor development. These responses are, in the short term, beneficial to the injured skin of the host, preventing further cytotoxic damage. However, when maintained chronically, they can subvert potentially useful immune responses to persisting viral infections and tumors. We and others have defined some of the immunological consequences of such expression associated with chronically hyperplastic skin through use of K14E7 transgenic animals (Frazer et al., 2011). Skin grafts expressing HPV16 E7 protein placed onto syngeneic, non-transgenic mice to mimic HPV associated premalignancy and malignancy (Dunn et al., 1997, Frazer et al., 2001) are not rejected from immune competent hosts, despite expressing a non-self viral antigen. In contrast, transgenic skin grafts expressing another non-self antigen, chicken ovalbumin, are rejected, suggesting that E7 protein expression or the epithelial hyperplasia consequent on E7 expression induces immune evasion mechanisms to prevent rejection of E7 expressing cells (Zhou et al., 2011). Indeed, a general state of increased immune cell infiltrate accompanied with intense immunosuppression has been proposed as a dominant mechanism underscoring the tolerance towards K14E7 skin grafts (Bergot et al., 2014, Choyce et al., 2013, Frazer et al., 2001, Gosmann et al., 2014a, Gosmann et al., 2014b, Mattarollo et al., 2010a, Mattarollo et al., 2010b).
Using an unbiased deep-sequencing approach, the transcriptomics data from this study suggests that perhaps the state of perpetual epithelial wounding and healing is, via the mechanisms for subversion of cell mediated immune responses described above, another determinant of the persistence of E7 expressing skin grafts, providing a further model for analysis of the interaction of “stress” keratins with the host immune system.
The following are the supplementary data related to this article.
Gene ontologies of the genes in the modules identified in WGCNA and associated functional categories.
Funding Sources
This work is funded from public competitive grant awarding bodies with no involvement in the conduct of the research, or production of the manuscript. Specifically, this work was supported by research project grants from the National Health and Medical Research Council (NHMRC) and the Merchant Charitable Foundation. No payment was received from any pharmaceutical company or other agency to write this manuscript.
Author Contributions
Samal Zhussupbekova: Conceived the ideas, did the first data analysis, wrote the first draft of
the manuscript.
Rohit Sinha: Made the libraries, reviewed the manuscript.
Paula Kuo: Conceived the ideas, did further data analysis, reviewed the manuscript.
Paul F Lambert: Developed the mice, contributed to the data interpretation and discussion,
reviewed the manuscript.
Ian H Frazer: Conceived the ideas, contributed to the data interpretation, wrote the discussion,
reviewed the manuscript.
Zewen Kelvin Tuong: Did most of the data analysis, contributed to the data interpretation, revised and reviewed the manuscript.
Conflict of Interest
The authors state no conflict of interest.
Acknowledgments
We thank Rehan Villani and Kiarash Khosrotehrani for generously providing antibodies for anti-cytokeratin 6 and reagents for immunohistological experiments. We also thank the staff of the Biological Research Facility (Brisbane, QLD, Australia), Translational Research Institute microscopy core facility (Brisbane, QLD, Australia), QFAB (Brisbane, QLD, Australia) for their excellent technical assistance and animal care. This work was supported by the Australian Cancer Research Foundation (ACRF) for the Comprehensive Cancer Genomics Facility at The University of Queensland Diamantina Institute.
References
- Abbas O., Bhawan J. Expression of stem cell markers nestin and cytokeratin 15 and 19 in cutaneous malignancies. J. Eur. Acad. Dermatol. Venereol. 2011;25(3):311–316. doi: 10.1111/j.1468-3083.2010.03791.x. [DOI] [PubMed] [Google Scholar]
- Abbas O., Richards J.E., Yaar R., Mahalingam M. Stem cell markers (cytokeratin 15, cytokeratin 19 and p63) in in situ and invasive cutaneous epithelial lesions. Mod. Pathol. 2011;24(1):90–97. doi: 10.1038/modpathol.2010.180. [DOI] [PubMed] [Google Scholar]
- Balsitis S., Dick F., Lee D., Farrell L., Hyde R.K., Griep A.E., Dyson N., Lambert P.F. Examination of the pRb-dependent and pRb-independent functions of E7 in vivo. J. Virol. 2005;79(17):11392–11402. doi: 10.1128/JVI.79.17.11392-11402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate — a practical and powerful approach to multiple testing. J. R. Stat. Soc. B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Bergot A.S., Ford N., Leggatt G.R., Wells J.W., Frazer I.H., Grimbaldeston M.A. HPV16-E7 expression in squamous epithelium creates a local immune suppressive environment via CCL2- and CCL5-mediated recruitment of mast cells. PLoS Pathog. 2014;10(10):e1004466. doi: 10.1371/journal.ppat.1004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragulla H.H., Homberger D.G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009;214(4):516–559. doi: 10.1111/j.1469-7580.2009.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.H., Kim G.M., Kim S.Y. The keratin-14 expression in actinic keratosis and squamous cell carcinoma: is this a prognostic factor for tumor progression? Cancer Res. Treat. 2010;42(2):107–114. doi: 10.4143/crt.2010.42.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choyce A., Yong M., Narayan S., Mattarollo S.R., Liem A., Lambert P.F., Frazer I.H., Leggatt G.R. Expression of a single, viral oncoprotein in skin epithelium is sufficient to recruit lymphocytes. PLoS One. 2013;8(2):e57798. doi: 10.1371/journal.pone.0057798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P.G., Weiss L.M. Keratin expression in human tissues and neoplasms. Histopathology. 2002;40(5):403–439. doi: 10.1046/j.1365-2559.2002.01387.x. [DOI] [PubMed] [Google Scholar]
- Chung B.M., Arutyunov A., Ilagan E., Yao N., Wills-Karp M., Coulombe P.A. Regulation of C-X-C chemokine gene expression by keratin 17 and hnRNP K in skin tumor keratinocytes. J. Cell Biol. 2015;208(5):613–627. doi: 10.1083/jcb.201408026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depianto D., Kerns M.L., Dlugosz A.A., Coulombe P.A. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 2010;42(10):910–914. doi: 10.1038/ng.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn L.A., Evander M., Tindle R.W., Bulloch A.L., de Kluyver R.L., Fernando G.J., Lambert P.F., Frazer I.H. Presentation of the HPV16E7 protein by skin grafts is insufficient to allow graft rejection in an E7-primed animal. Virology. 1997;235(1):94–103. doi: 10.1006/viro.1997.8650. [DOI] [PubMed] [Google Scholar]
- Frazer I.H., De Kluyver R., Leggatt G.R., Guo H.Y., Dunn L., White O., Harris C., Liem A., Lambert P. Tolerance or immunity to a tumor antigen expressed in somatic cells can be determined by systemic proinflammatory signals at the time of first antigen exposure. J. Immunol. 2001;167(11):6180–6187. doi: 10.4049/jimmunol.167.11.6180. [DOI] [PubMed] [Google Scholar]
- Frazer I.H., Leggatt G.R., Mattarollo S.R. Prevention and treatment of papillomavirus-related cancers through immunization. Annu. Rev. Immunol. 2011;29:111–138. doi: 10.1146/annurev-immunol-031210-101308. [DOI] [PubMed] [Google Scholar]
- Gosmann C., Frazer I.H., Mattarollo S.R., Blumenthal A. IL-18, but not IL-12, induces production of IFN-gamma in the immunosuppressive environment of HPV16 E7 transgenic hyperplastic skin. J. Investig. Dermatol. 2014;134(10):2562–2569. doi: 10.1038/jid.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmann C., Mattarollo S.R., Bridge J.A., Frazer I.H., Blumenthal A. IL-17 suppresses immune effector functions in human papillomavirus-associated epithelial hyperplasia. J. Immunol. 2014;193(5):2248–2257. doi: 10.4049/jimmunol.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameetman L., Commandeur S., Bavinck J.N., Wisgerhof H.C., de Gruijl F.R., Willemze R., Mullenders L., Tensen C.P., Vrieling H. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer. 2013;13:58. doi: 10.1186/1471-2407-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber R., Liem A., Pitot H., Lambert P.F. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol. 1996;70(3):1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse M., Zimek A., Weber K., Magin T.M. Comprehensive analysis of keratin gene clusters in humans and rodents. Eur. J. Cell Biol. 2004;83(1):19–26. doi: 10.1078/0171-9335-00354. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Isaac C.E., Francis S.M., Martens A.L., Julian L.M., Seifried L.A., Erdmann N., Binne U.K., Harrington L., Sicinski P., Berube N.G., Dyson N.J., Dick F.A. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 2006;26(9):3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., Smyth G.K. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J.C., Coulombe P.A. Keratin 16-null mice develop palmoplantar keratoderma, a hallmark feature of pachyonychia congenita and related disorders. J. Investig. Dermatol. 2012;132(5):1384–1391. doi: 10.1038/jid.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J.C., Pina-Paz S., Rotty J.D., Hickerson R.P., Kaspar R.L., Balmain A., Coulombe P.A. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc. Natl. Acad. Sci. U. S. A. 2013;110(48):19537–19542. doi: 10.1073/pnas.1309576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Bolger A.M., Nagel A., Fernie A.R., Lunn J.E., Stitt M., Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40(Web Server issue):W622-7. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschke F., Homberg M., Magin T.M. Keratin isotypes control desmosome stability and dynamics through PKCalpha. J. Investig. Dermatol. 2016;136(1):202–213. doi: 10.1038/JID.2015.403. [DOI] [PubMed] [Google Scholar]
- Mattarollo S.R., Rahimpour A., Choyce A., Godfrey D.I., Leggatt G.R., Frazer I.H. Invariant NKT cells in hyperplastic skin induce a local immune suppressive environment by IFN-gamma production. J. Immunol. 2010;184(3):1242–1250. doi: 10.4049/jimmunol.0902191. [DOI] [PubMed] [Google Scholar]
- Mattarollo S.R., Yong M., Tan L., Frazer I.H., Leggatt G.R. Secretion of IFN-gamma but not IL-17 by CD1d-restricted NKT cells enhances rejection of skin grafts expressing epithelial cell-derived antigen. J. Immunol. 2010;184(10):5663–5669. doi: 10.4049/jimmunol.0903730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin M.E., Munger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384(2):335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin M.E., Meyers J., Munger K. Cancer associated human papillomaviruses. Curr. Opin. Virol. 2012;2(4):459–466. doi: 10.1016/j.coviro.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll R., Divo M., Langbein L. The human keratins: biology and pathology. Histochem. Cell Biol. 2008;129(6):705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10(8):550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Nindl I., Dang C., Forschner T., Kuban R.J., Meyer T., Sterry W., Stockfleth E. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol. Cancer. 2006;5:30. doi: 10.1186/1476-4598-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R.M., Lunny D.P., Ogden P.H., Morley S.M., McLean W.H., Evans A., Harrison D.L., Rugg E.L., Lane E.B. K15 expression implies lateral differentiation within stratified epithelial basal cells. Lab. Investig. 2000;80(11):1701–1710. doi: 10.1038/labinvest.3780180. [DOI] [PubMed] [Google Scholar]
- Quinn A.G. Biology of the skin and dermatological disease. Medicine. 2004;32(12):1–3. [Google Scholar]
- Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabasi A.L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297(5586):1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Rotty J.D., Coulombe P.A. A wound-induced keratin inhibits Src activity during keratinocyte migration and tissue repair. J. Cell Biol. 2012;197(3):381–389. doi: 10.1083/jcb.201107078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraj P., Sohl G., Magin T.M. Keratin transgenic and knockout mice: functional analysis and validation of disease-causing mutations. Methods Mol. Biol. 2007;360:203–251. doi: 10.1385/1-59745-165-7:203. [DOI] [PubMed] [Google Scholar]
- Wickett R.R., Visscher M.O. Structure and function of the epidermal barrier. Am. J. Infect. Control. 2006;34(10):S98–S110. [Google Scholar]
- Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. (p. Article17) [DOI] [PubMed] [Google Scholar]
- Zhou F., Leggatt G.R., Frazer I.H. Human papillomavirus 16 E7 protein inhibits interferon-gamma-mediated enhancement of keratinocyte antigen processing and T-cell lysis. FEBS J. 2011;278(6):955–963. doi: 10.1111/j.1742-4658.2011.08011.x. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184(1):9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene ontologies of the genes in the modules identified in WGCNA and associated functional categories.