Abstract
Malignant peripheral nerve sheath tumors (MPNST) are rare soft tissue sarcomas that are a major source of mortality in neurofibromatosis type 1 (NF1) patients. To identify MPNST driver genes, we performed a lentiviral short hairpin (sh) RNA screen, targeting all 130 genes up-regulated in neurofibroma and MPNSTs versus normal human nerve Schwann cells. NF1 mutant cells show activation of RAS/MAPK signaling, so a counter-screen in RAS mutant carcinoma cells was performed to exclude common RAS-pathway driven genes. We identified 7 genes specific for survival of MPSNT cells, including MEIS1. MEIS1 was frequently amplified or hypomethylated in human MPSNTs, correlating with elevated MEIS1 gene expression. In MPNST cells and in a genetically engineered mouse model, MEIS1 expression in developing nerve glial cells was necessary for MPNST growth. Mechanistically, MEIS1 drives MPNST cell growth via the transcription factor ID1, thereby suppressing expression of the cell cycle inhibitor p27Kip and maintaining cell survival.
Keywords: MPNST, MEIS1, ID1, RNAi, p27Kip, G0/G1 arrest, Sarcoma
Highlights
-
•
Targeting over-expressed genes facilitates identification of sarcoma driver genes.
-
•
We identify MEIS1 as a MPNST oncogene.
-
•
MEIS1 suppresses p27Kip enabling MPNST survival.
We identify MEIS1 as a sarcoma oncogene, and identify an additional 7 genes specific for survival of malignant peripheral nerve sheath cells. MEIS1 was frequently amplified or hypomethylated in human tumors, correlating with elevated MEIS1 gene and protein expression. MEIS1 enables cell cycle progression in these tumor cells through downregulation of expression of a pro-cell death protein p27Kip. Thus, inhibitors targeting cell cycle checkpoints and/or upregulating p27Kip may have therapeutic value for these patients, and perhaps for other tumor types in which MEIS1 is an oncogene.
1. Introduction
Identification of the genes that drive carcinogenesis is essential to understand mechanisms that underlie tumor emergence and evolution, and to formulation of effective therapeutic strategies. This need is especially critical in sarcomas, because most are poorly characterized and lack effective therapies. Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive soft tissue sarcomas that occur sporadically or, in half of the cases occur in association with neurofibromatosis type 1 (NF1) (Ducatman et al., 1986, Widemann, 2009). This genetic link led to the understanding that MPNST in NF1 patients have biallelic NF1 mutations (Bottillo et al., 2009, Legius et al., 1993). This results in RAS pathway activation because the NF1 gene product, neurofibromin, binds to RAS proteins and accelerates their intrinsic GTPase activity (Bos et al., 2007). Further, sporadic MPNST also show NF1 loss or activating RAS or RAF mutations (Ratner and Miller, 2015). In spite of this understanding, the current treatment for MPNST patients remains surgical resection of the tumor, followed by non-specific high-dose chemotherapy (Gregorian et al., 2009, Katz et al., 2009). Unfortunately, without complete tumor resection MPNST are resistant to this therapeutic approach and fewer than 25% patients survive 5-years after diagnosis (Katz et al., 2009, Widemann, 2009).
MPNST cells, like many sarcomas, show complex hyperdiploid karyotypes, but most genetic changes that drive transformation are unknown. Importantly, when the MPNST precursor lesion called the atypical neurofibroma develops in an NF1 patient, the tumor suppressor gene CDKN2A is mutant or lost (Legius et al., 1993, Nielsen et al., 1999). An MPNST mouse model (GEM-PNST) mimics this combination of mutations. Consistent with a critical role for loss of the CDKN2A locus in driving MPSNT, 26% of Nf1+/−;Ink4a/Arf−/− mice form GEM-PNST (Joseph et al., 2008). In addition, mutations in epigenetic modifier genes of the PRC2 complex occur in many human MPNST. Functional data suggest that these mutations are under positive selection pressure (De Raedt et al., 2014, Lee et al., 2014). Human MPNST show inconsistent loss of function mutation in other tumor suppressor genes that inhibit the cell cycle, such as TP53 and RB (Legius et al., 1994). A more complete understanding of the molecular mechanisms that drive MPNST is necessary to facilitate development of new targeted therapies.
Lineage dependence, sometimes called lineage addiction, can identify genes that are important cancer targets (Garraway and Sellers, 2006). Gene expression profiling may thus demonstrate an enrichment of such genes, if overexpressed genes reflect the lineage of the tumor cells. Supporting this idea, Zuber et al. used shRNA screening combined with transcriptional analysis to define oncogenes to which leukemias are addicted (Zuber et al., 2011a, Zuber et al., 2011b). To identify MPNST drivers, we used available transcriptome data (Miller et al., 2009). Human neurofibroma and MPNST gene expression profiles were compared to those of normal human Schwann cells, because Schwann cells and/or their neural crest cell precursor like cells are believed to be the pathogenic drivers of MPNST (Buchstaller et al., 2012, Vogel et al., 1999). Analysis of this genome wide data set previously identified neural crest genes as abundantly expressed in human MPNSTs, and established that MPNST cells in vitro could be killed by inhibition of the SOX9 lineage gene (Miller et al., 2009). We hypothesized that other overexpressed genes could be identified among the overexpressed genes and necessary for MPNST cell growth and tumor maintenance.
We used RNA interference (RNAi), a common approach to inhibit gene expression, in which binding of sequence-specific small interfering (siRNA) targets transcript cleavage, resulting in transcript degradation (Rana, 2007). RNAi screening combines the power of genetic screens with phenotypic assays in vitro, making it possible to identify genes involved in a wide variety of biological processes, including identification of oncogenic drivers (Bernards et al., 2006, Zhuang and Hunter, 2012, Boutros and Ahringer, 2008, Zhuang and Hunter, 2012). Although RNAi technology has been used for more than a decade, large scale RNAi screens have not been reported in MPNST. We generated a lentiviral shRNA library to inhibit 130 genes identified as overexpressed in human neurofibroma and MPNSTs versus normal human Schwann cells. By screening this custom shRNA library, we identify the protein MEIS1 as being critically required for MPNST tumorogenesis.
MEIS1 encodes a homeobox protein belonging to the three amino acid loop extension (TALE) family of homeodomain-containing proteins. XMeis1 specifies neural crest identity in the frog embryo (Maeda et al., 2001), but MEIS1 has not been studied in mouse or human neural crest in detail. Mice lacking Meis1 do not show obvious neural crest defect, but rather show major defects in retina, lens morphogenesis, hematopoietic cells and vasculature, and die by embryonic day 14.5 (Hisa et al., 2004). During hematopoiesis, Meis1 expression levels are highest in self-renewing hematopoietic stem cells (HSCs) and decline with cell differentiation (Pineault et al., 2002, Chen et al., 2008). The Meis1 gene is a common target of retroviral insertion in a murine leukemia model and Meis1 overexpression in hematopoietic cells causes aggressive leukemia in mice, defining Meis1 as an oncogene (Nakamura et al., 1996, Thorsteinsdottir et al., 2001). MEIS1 is also frequently overexpressed and amplified in human neuroblastoma, a tumor of neural crest derived cells (Spieker et al., 2001). How Meis1 acts in any tumor type is unclear. Indeed, Meis1 regulated genes show little overlap in diverse systems, and target regulation is likely to be complex (Dardaei et al., 2015). Our results establish MEIS1 as a driver of MPNST cell growth, which acts on the cell cycle through ID1 and inhibition of p27Kip.
2. Experimental procedures
2.1. Tumor samples
Human MPNST were collected in accordance with institutional review board-approved protocols from discarded surgical specimens, and were received from the Cincinnati Children's Hospital Bio Bank as flash frozen or paraffin embedded samples.
2.2. Cell lines and reagents
MPNST cell lines STS26T (sporadic; NF1 wild type; RRID:CVCL_8917), and 8814 (RRID:CVCL_8916), 88-3, S462TY (RRID:CVCL_1Y70), T265 (RRID:CVCL_S805; all NF1 mutant), and immortalized human Schwann cells (iHSC) (Rahrmann et al., 2013), and normal human Schwann cells (NHSC) from autopsy specimens were obtained and maintained as described in (Miller et al., 2006, Watson et al., 2013, Declue et al., 1992).
2.3. Lentiviral transduction
MPNST cells were transduced with lentiviral particles at 50–60% confluence. Short hairpins (sh) RNAs targeting gene of interest and control non-targeting construct were from the Sigma Aldrich TRC library. The CCHMC Viral Vector Core produced virus using a 4-plasmid packaging system (http://www.cincinnatichildrens.org/research/div/exphematology/translational/vpf/vvc/default. html). Lentiviral particles were incubated with MPNST cells in the presence of polybrene (8 μg/mL; Sigma) for 24 h, followed by selection in 2 μg/mL puromycin, which killed uninfected cells within 3 days.
2.4. Lentiviral shRNA screening
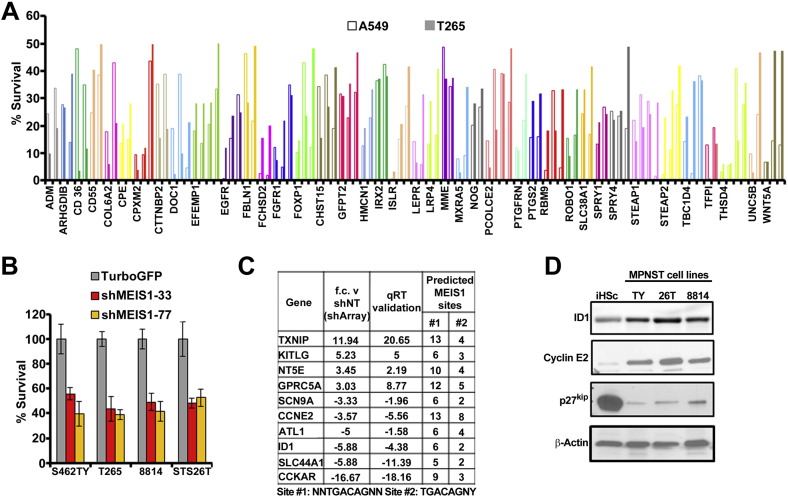
MPNST cells plated on day (− 1) at 1000 cell/well in 96 well plates. Once the cells successfully adhered to the plate (4 h post plating), cell were transduced with lentiviral shRNA. Puromycin is added on day (0) to select for cells successfully infected with the virus. Growth was measured on Day 5 using a metabolic assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega #G3582), measured as absorbance at 490 nm. Controls include - 1) lenti-virus encoding shSOX9 - positive control for death 2) non-targeting shRNA - negative control. shSOX9 treated MPNST cells show significant detrimental effect on survival by end point, thus this is used as a bench mark for an effect on cell growth. Screen I was conducted on only one MPNST cell line the T265 which are null for the NF1 gene. Screen II was conducted exactly as Screen I but with the addition of 8814, STS26T (sporadic MPNST cell line, wild type for NF1) and 88-3 MPNST cell lines. Screen III used 127shRNAs targeting genes of interest for analysis of growth for MPNST (T265) versus a lung adenocarcinoma cell line (A549) (dot plot, Fig. 1D).
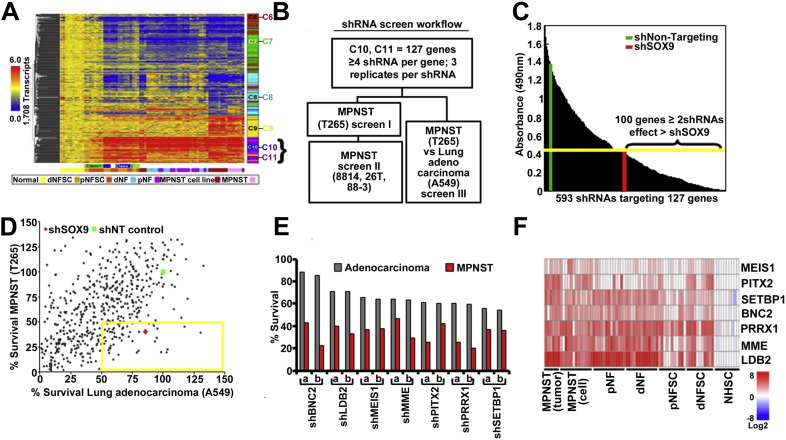
Fig. 1.
(A) K-means clustering of transcripts with increased expression in dermal neurofibroma Schwann cells (dNFSC), plexiform neurofibroma Schwann cell (pNFSC), dermal neurofibroma tumors (dNF), plexiform neurofibroma tumors (pNF), MPNST cell lines and MPNST solid tumors versus normal human Schwann cells (NHSC). The bar to the right of the heat map shows five k-means functional clusters (C6–C11). Genes overexpressed in neurofibroma and MPNST samples cluster in C10–C11. (B) A schematic representation of medium throughput shRNA screen targeting genes overexpressed > 3 fold in MPNSTs versus normal human Schwann cells. Each tested shRNA was used to infect cells in triplicate wells of a 96 well plate. Cell viability was measured for each well. Data is shown as the average of 3 wells. (C) Survival data from Screen I represented as a bar graph with cell growth measured in an MTS assay. Green vertical bar represents survival of MPNST with shNT; red vertical bar represents survival effects of shSOX9. Yellow horizontal bar represents cut off to select shRNAs showing detrimental effect on survival. (D) Screen II data shown in a dot plot. The percent survival of MPNSTs (T265) versus shNT is represented on the y axis, and the same statistic for lung adenocarcinoma (A549) is on the x axis. The yellow box encloses points representing shRNAs affecting MPNST cell survival > 50% but less effect on A549 cells. (E) Bar graph depicting survival for two shRNAs/gene (a,b) in the yellow box from D. (F) Expression data from panel A showing the 7 genes identified as specific to MPNST but not A549 cells.
2.5. Immunoblot
We washed cells with ice-cold PBS, lysed them in RIPA buffer, and boiled lysates for 5 min. Lysates were analyzed immediately or frozen at − 20 °C. Proteins were separated on 4–20% denaturing SDS-PAGE gradient gels (Bio-Rad #456-1094) and transferred to PVDF membranes (Millipore; Billerica, MA #IPFL00010). The membranes were blocked with 5% milk in 0.1 M Tris-buffered saline with 0.1% Tween (TBST), incubated with primary and secondary antibodies, and washed following manufacturer's instructions. Secondary antibodies were detected by chemi-luminescence (Millipore cat. #WBKLS0500). Primary antibodies were anti-MEIS1 (Abcam #ab19867, RRID: AB_776272), anti-cleaved PARP (Cell Signaling Technology Inc.) (CST; #9541, RRID: AB_331427), p27Kip (CST; #3686, RRID: AB_2077850), Phospho-Rb (Ser795) (CST #9301, RRID: AB_330013), Phospho-Rb (Ser807/811) (CST; 8516, RRID: AB_11178658) and ID1 (Abcam; #ab134163, RRID: AB_2572295). Anti-cyclin antibodies were from the cyclin sampler kit (CST; #9869, RRID: AB_1903944). Anti-rabbit IgG, HRP-linked (CST; #7074, RRID: AB_2099233) or anti-mouse IgG, HRP-linked (CST; #7076, RRID: AB_330924) were used appropriately depending on the host of the primary antibody. All membranes were stripped with stripping buffer for 5 min (Fisher #21059) to re-probe with HRP conjugated anti-β-actin (CST, #5125, RRID: AB_1903890) as a loading control.
2.6. Quantitative real time PCR (QRT)-PCR
Total cellular RNA was isolated with the RNeasy kit (Qiagen) and used as a template for cDNA synthesis (High-Capacity cDNA archive kit, Applied Biosystems) and QRT-PCR (ABI 7500 Sequence Detection System) as described (Patel et al., 2012). Primers were purchased as pre-validated qPCR sets from Integrated DNA Technologies. All qRT-PCR reactions were conducted multiple times with three or more replicates per experiment.
2.7. Immunohistochemistry
Tumors were dissected and fixed overnight in 10% formalin then cleared in 70% ethanol, dehydrated, and embedded in paraffin (http://www.cincinnatichildrens.org/research/cores/pathology-core/default/). Hematoxylin and eosin (H&E) and immunoperoxidase staining of 5 μm paraffin sections followed standard protocols. Antibodies used were rabbit anti-S100 (1:5000, DAKO; Z0311, RRID: AB_10013383) and rabbit anti-MEIS1 (1:1000 Abcam; #ab19867, RRID: AB_776272). For nuclear staining, sections were incubated in DAPI (SIGMA, 1:10,000) for 5 min, rinsed in 1X PBS thrice, and cover slipped in Flouromount G (EM Sciences, #17984-25, RRID: AB_2572296). Sections were photographed on a Nikon eclipse 80i bright field microscope.
2.8. Cell cycle and viability analysis
We transduced 2 × 105 MPNST cells with shMEIS1 or vector control. Cells were stained with propidium iodide and annexin V-alexa fluor 647 conjugate according to manufacturer's instructions (Life Technologies, Grand Island, NY, USA #A23204). Flow cytometry was performed on a FACSCanto (Becton Dickinson, Franklin Lakes, NJ, USA) and analyzed using FlowJo software.
2.9. shRNA growth assays
We plated MPNST cells on day (− 1) at 1000 cell/well in 96 well plates. Once cells adhered to the plate (5 h post plating) they were incubated with lentiviral shRNA for 24 h. Puromycin (2 μg/mL) was added on day (0) to select for cells infected with the virus. MTS reagent (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega #G3582) or Alamar blue (Life Technologies, #88951) was added at endpoint and growth measured as absorbance intensity per manufacturer instructions.
2.10. MPNST rescue experiment
Mouse Meis1 cDNA was (Addgene #21013) was cloned from MSCV-IRES-YFP into pCDH-RFP a lentivirus-compatible plasmid vector. The pCDH-Meis1-RFP was packaged into a lentivirus by the CCHMC viral vector core (https://research.cchmc.org/translationalcores/vector-production/pre-clinical-and-research-vector-products). MPNST cells were co-infected with shMEIS1 and pCDH-Meis1-RFP lentiviral particles at 60–70% confluence. Controls included co-infection of each shMEIS1 and pCDH-Meis1-RFP with shNon-targeting (shNT). For each condition, we split cells into three for cell survival in 96 well plates MTS assays and protein and RNA for further analysis.
2.11. Animal studies
Mice were housed in temperature and humidity controlled facilities with free access to food and water, on a 12-h dark–light cycle. The animal care and use committee of Cincinnati Children's Hospital Medical Center approved all animal use. Ink4a/Arf−/− mice were originally from the B6.129-Cdkn2atm1Rdp NCI; Mouse repository MMHCC strain #01XB1 (RRID:IMRS_NCIMR:01XB1). They were back crossed to pure C57BL/6J females for 5 generations before breeding for the compound mutant mice. Other mouse lines used are Nf1+/− (Brannan et al., 1994), and Meis1f/f (Unnisa et al., 2012), and Dhh-Cre (Jaegle et al., 2003). The Dhh-Cre allele was maintained on the male. This mouse line and Nf1+/− mice were maintained on the C57Bl/6 background for n > 8 generations before use. All mice were maintained on the C57BL/6J background.
2.12. Genotyping
Nf1 genotyping was performed using the oligonucleotides (all 5′ to 3′) GTATTG AATTGAAGCACCTTTGTT TG and CTGCCCAAGGCTCCCCCAG to detect the wildtype allele, and GCGTGTTCGAATTCG CCAATG to detect the targeted allele (neo) as described (Brannan et al., 1994). Ink4a/Arf mice (B6.129-Cdkn2atm1Rdp/NCI) were genotyped according to Mouse repository MMHCC strain #01XB1; Dhh-Cre Forward primer ACCCTGTTACGTATA GCCGA and 5′ CTCCGGTATTGA AACTCCAG; Meis1 Flox primers CCAAAGTAGCCACCAATA TCATGA and AGCGTCACTTGGAAAAGCAATGAT.
3. Results
3.1. MPNST cell survival is dependent on multiple overexpressed genes
We used a gene expression data set consisting of samples of primary neurofibroma Schwann cells, neurofibromas, MPNST cell lines and primary MPNST, compared to normal human Schwann cells (Miller et al., 2009). Expression levels were normalized to those of Schwann cells for each gene, because, of the tumor cell types in benign neurofibromas (Schwann cells, endothelial cells, fibroblasts, macrophages and mast cells), only neural crest-derived Schwann cells show biallelic mutations in the NF1 gene (Serra et al., 2000). We approached the identification of unique driver genes by focusing on genes in the C10/C11 K-means clusters; these clusters identify genes with increased expression in neurofibroma and also in MPNST (Fig. 1A). We hypothesized that some these 130 genes would be essential to MPNST cell growth.
A lentiviral shRNA library was used to downregulate each of the130 genes over-expressed > 3 fold. This library included 3–5 shRNA per gene (a total of 593 individual lentiviruses; Supplemental Table 1). ShSOX9, a known inhibitor of MPNST cell growth, was used as a positive control (Miller et al., 2009). A non-targeting shRNA (shNT) lentivirus was used as a negative control. A primary screen on a single NF1 mutant MPNST cell line (T265) identified 100 genes for which at least 2 shRNAs caused significant (> shSOX9) adverse effects on MPNST cell growth (Fig. 1B, C). Thus, Screen I showed that a remarkable number of shRNAs targeting overexpressed genes compromised MPNST cell growth. In Screen II, a screen of 42/127 randomly selected genes (shRNAs on 2 of 7 96-well plates containing virus) were tested in the additional MPNST cell lines 8814 (NF1−/−), S462TY (NF1−/−) and STS26T (sporadic MPNST wild-type for NF1; Supplemental Table 2); results for each shRNA were generally comparable among cell lines.
To exclude shRNAs that compromised MPNST cell growth due to off-target effects or through effects downstream of the well-studied RAS activation in MPNST cells, we infected an A549 lung adenocarcinoma cells with the same set of shRNAs (Screen III). A549 cells have an activating mutation in the K-Ras gene (Rodenhuis et al., 1987). They differ from MPNST cells in terms of lineage; MPNST cells are neural crest derived, while A549 cells are derived from ectoderm. We did this screen to exclude genes commonly induced by Ras activation, and to identify genes related to MPNST cell lineage.
Only 7 of the 100 genes affecting MPNST cell growth showed little effect on A549 cells (Fig. 1D; boxed yellow square). All 7 were targeted by at least 2 shRNAs, inhibited growth of MPNST cells by > 50%, but inhibited A549 cells by < 25% (Fig. 1E). A heatmap shows the expression of these 7 genes in benign neurofibromas and MPNSTs versus normal human Schwann cells (Fig. 1F). We decided to focus on MEIS1 because it acts as an oncogene in leukemia (Kumar et al., 2009, Kumar et al., 2010, Nakamura et al., 1996), yet is poorly studied in other settings.
3.2. MEIS1 is overexpressed in human tumors and MPNST cell lines
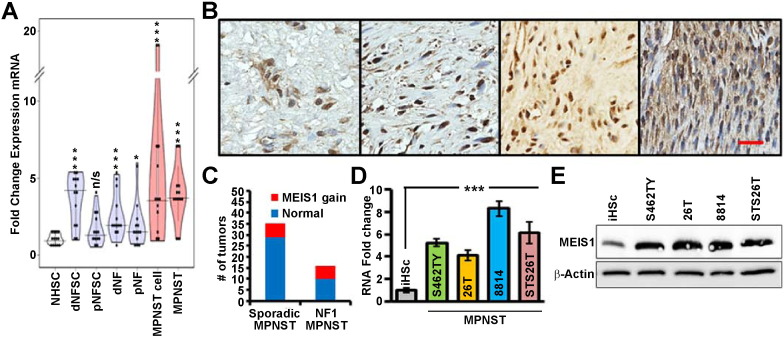
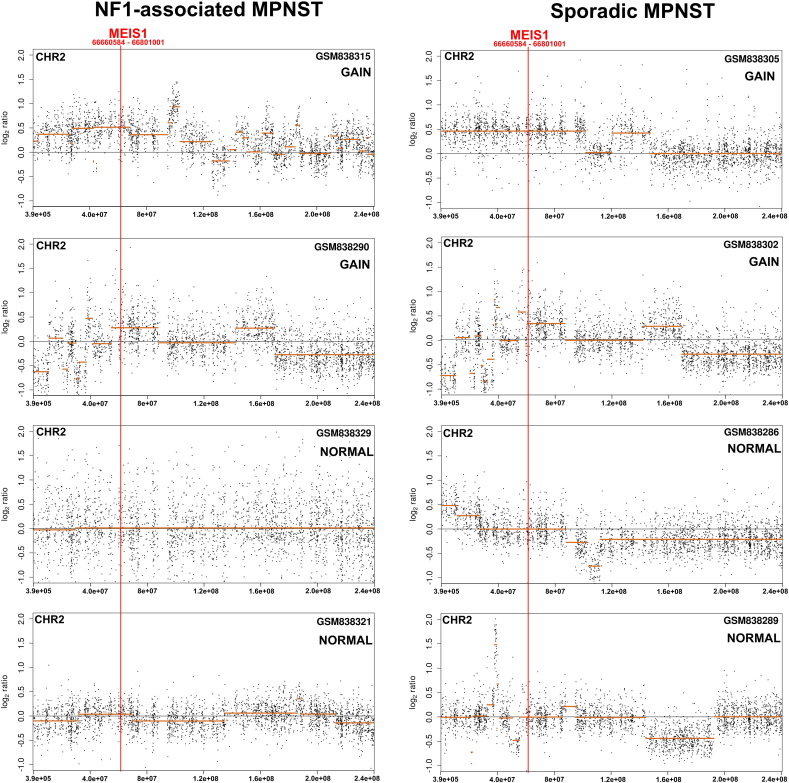
MEIS1 mRNA expression progressively increases in neurofibroma Schwann cells, neurofibroma, MPNST and MPNST cell lines. Its expression level varies among individual samples in each group (Fig. 2A). Using anti-MEIS1 antibody in immunohistochemistry we verified expression of MEIS1 protein in 9/9 MPNST solid tumors. Positive labelling for MEIS1 was of similar intensity in all labeled cells. However, the percent of tumor cells positive for MEIS1 varied (Fig. 2B). Analysis of single nucleotide polymorphisms (SNP) data revealed MEIS1 locus amplification in 6/35 (17.1%) of sporadic and in 6/16 (35.5%) of NF1-associated MPNSTs (Fig. 2C, Supplemental Table 3). Supplemental Fig. 2 shows SNP analysis of individual human MPNST, 4 of which show amplification and 4 of which do not. Overexpression of genes can also be linked to hypo-methylation. The MEIS1 locus on human chromosome 2 was identified as hypomethylated in MPNST (Table 1). In vitro, 4/4 human MPNST cell lines confirmed overexpression of MEIS1 at the mRNA and protein levels, versus iHSC (Fig. 2D, E).
Fig. 2.
MEIS1 expression is elevated in human MPNSTs. (A) Violin plot of MEIS1 gene expression data in each of seven sample types, normalized to expression in Schwann cells. Abbreviations = dermal neurofibroma Schwann cells (dNFSC), plexiform neurofibroma Schwann cell (pNFSC), dermal neurofibroma tumors (dNF), plexiform neurofibroma tumors (pNF), MPNST cell lines and MPNST solid tumors versus normal human Schwann cells (NHSC). (B) Representative anti-MEIS1 staining visualized with DAB (brown) counterstained with hematoxylin (blue); scale bar, 100 μm. (C) A bar graph displays gene amplification data of NF related MPNST and Sporadic MPNST. (D) Bar graph depicts MEIS1 mRNA normalized to iHSCs for MPNST cell lines (S462TY, T265, 8814, STS26T; p < 0.001 by ANOVA: single factor). (E) Immunoblot shows elevated MEIS1 protein is MPNST cell lines versus iHSC.
Fig. S2.
Copy-number change patterns across chromosome 2 (GRCh37 assembly) in MPNST patient's genome (GSM839315 in GSE33881 data series). The log2-ratio of probe intensities (black dots) of the query MPNST sample versus the combination of control/normal genomic DNAs and predicted segments (orange lines) are plotted using CGHcall package. The red arrowhead points the genomic location of MEIS1 gene.
Table 1.
Methylation analysis of the MEIS1 gene locus for 10 MPNST tumor samples from 9 patients as compared to normal human Schwann cells.
| UCSC ID | Symbol | CHR | Strand | Gene start (TSS) | The nearest CpG-IS from TSS | Distance between CpG-IS to TSS (bp) | Methylation status | ||
|---|---|---|---|---|---|---|---|---|---|
| uc002sdu.1 | MEIS1 | chr2 | + | 66516035 | 66653395 | 66515842 | 66517842 | 193 | Hypo |
| uc002sdv.1 | MEIS1 | chr2 | + | 66518311 | 66653395 | 66515842 | 66517842 | 469 | Hypo |
| uc002sdw.1 | MEIS1 | chr2 | + | 66520766 | 66649781 | 66515842 | 66517842 | 2942 | Hypo |
3.3. MPNST growth is sensitive to MEIS1 inhibition
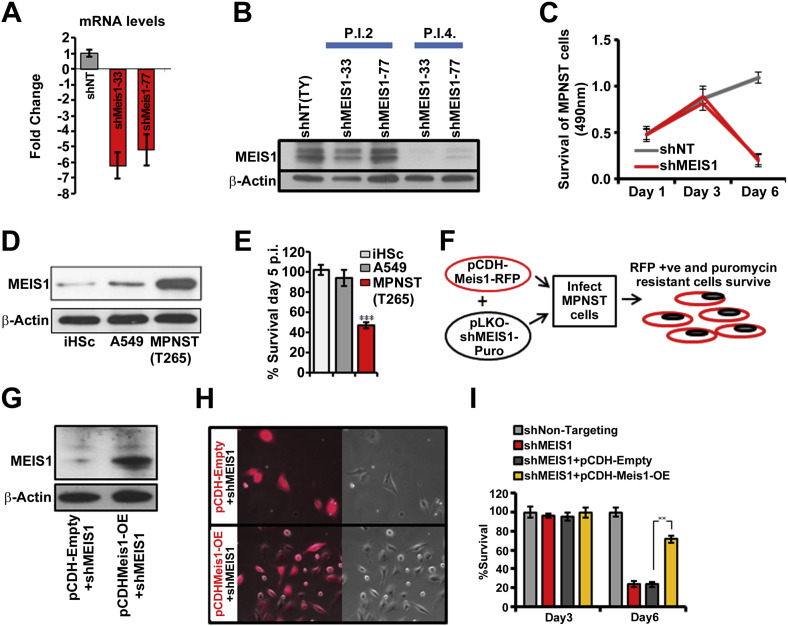
Screen I identified MEIS1 as necessary for survival of MPNSTs. We studied the effect of two MEIS1- shRNAs in detail. ShRNAs 33 and 77 decreased MEIS1 mRNA expression (5.1 and 6.3 fold, respectively; Fig. 3A). Inhibiting MEIS1 expression with either shRNA diminished expression of MEIS1 protein by day 4 after lentiviral infection (Fig. 3B). Both shRNAs reduced MPNST cell growth relative to shNT controls beginning after day 3 (Fig.3C); this effect was more pronounced by day 6. These growth effects were confirmed in 3 additional MPNST cell lines (Supplemental Fig. 1B). iHSC and A549 cells expressed MEIS1 at a significantly lower levels than MPNST cells, and down regulation of MEIS1 using lentiviral shRNA did not significantly affect iHSC or A549 cells (Fig. 3D, E). Thus, MEIS1knockdown inhibits MPNST cell survival while human Schwann cells are not significantly affected.
Fig. 3.
Reducing MEIS1 expression in MPNST cells (S462TY) inhibits cell growth. (A) Bar graph shows that shMEIS1 (shRNA#33 or #77) reduce MEIS1 mRNA in MPNST, by qRT-PCR. (B) Western blot shows that MEIS1 protein expression is reduced by day 4 post infection (p.i.4) with both shMEIS1 viruses. Anti-β-actin was used as a loading control. (C) Line graph depicts MPNST growth after transduction with shMEIS1 or shNT control at days 1, 3 and 6 in an MTS assay (p < 0.001). (D) An immunoblot shows that MEIS1 protein levels are higher in MPNST cells than in iHSC or A549 cells. (E) Bar graph depicts the increased effects of shMEIS1 on MPNST compared to iHSC or A549. (F) Schematic shows experiment designed to test the specificity of shMEIS1, using a mouse Meis1-RFP protein and a human-targeting shMEIS1. (G) Immunoblot shows Meis1 protein expression in MPNST cells infected with Meis1-RFP OE and shMEIS1 versus vector (Empty-RFP) with shMEIS1. (H) Fluorescent (red, left) images show rescue of cells expressing Meis1-RFP OE and shMEIS1 versus Empty-RFP and shMEIS1. (I) Bar graph quantifies cell survival in MTS assays conducted day 3 or 6 after infection. (p < 0.001 via ANOVA: single factor).
Fig. S1.
(A) Bar graph displays survival of MPNST (solid line) and A549 (clear bar) for 40 genes from screen data in Fig. 1D. (B) Bar graph depicts survival of 4 different MPNST cell lines shMEIS1 or shNT treatment for using an MTS assay. (p < 0.001; ANOVA: single factor) (C) Table representing genes differentially expressed in shMEIS1 treated samples. These genes represent genes that were validated for changes in expression via qRT-PCR and had putative MEIS1 binding sites in the promoter regions as shown in the table. (D) Immunoblot analysis shows endogenous expression levels of ID1, CCNE2 and p27Kip proteins in three different MPNST cell lines compared to iHSC.
In addition to effects on targeted genes, shRNAs can have off-target effects. To control for possible off-target effects, we generated a lentivirus encoding mouse Meis1 tagged with red fluorescent protein (pCDH-Meis1-RFP). MPNST cells infected with this virus expressed Meis1-RFP, which did not interfere with cell growth. Cells infected with Meis1-RFP alone did not survive puromycin selection, while puromycin-resistant cells infected with shMEIS1 alone die due to downregulation of MEIS1 expression (schematic in Fig. 3F). Simultaneous infection of MPNST cells with shMEIS1 and pCDH-Meis1-RFP rescued MPNST survival as assessed by visualization and quantified by MTS assay (Fig. 3G–I). Thus, shMEIS1 effects on MPNST cell survival are specific to knockdown of MEIS1. Overexpression of Meis1 in iHSC that are wildtype at the NF1 locus, or iHSC infected with shNF1 did not result in transformation of these cells as tested in a soft agar assay (not shown), suggesting that overexpression of Meis1 is not sufficient for transformation in this setting.
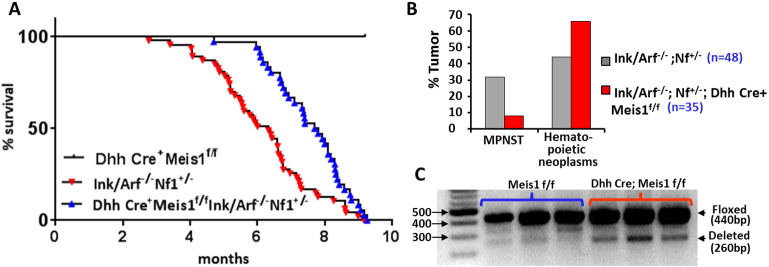
3.4. Meis1 downregulation attenuates tumor burden in an MPSNT mouse model
The shRNA screening described above tested if acute downregulation of MEIS1 is essential for cell survival in human cell lines. To test if loss of Meis1 affects MPNST growth in primary tumors with an intact microenvironment, we developed a genetically engineered mouse model. We used the Ink4a/Arf−/−;Nf1+/− GEM-PNST mouse model that recapitulates mutations in human MPNSTs. A quarter of these mice form GEM-PNST (Joseph et al., 2008). Since whole body Meis1 deletion is embryonically lethal, we bred Ink4a/Arf−/−;Nf1+/− with Meis1f/f mice, and with Dhh-Cre-mice, which express Cre-recombinase specifically in glia. Dhh-Cre is expressed beginning at day E12.5 in post-neural crest cells known as Schwann cell precursors (Wu et al., 2008). Nf1+/−;Ink4a/Arf−/−;Meis1f/f;DhhCre+ and littermate controls were followed for tumor formation. Nf1+/−;Ink4a/Arf−/− mice with Meis1f/f or Dhh-Cre alleles alone died by 10 months of age, as previously described for the Nf1+/−;Ink4a/Arf−/− model (Fig. 4A). GEM-PNSTs in these animals are grossly evident, typically at 4–6 months of age on the shoulders, ribs, or limbs of mice, in the vicinity of peripheral nerves. Importantly, significantly fewer GEM-PNSTs were observed in Nf1+/−;Ink4a/Arf−/−;Meis1f/f;DhhCre+ mice, where Meis1 deletion was restricted to the glial cells arising from Schwann cell precursors (Fig. 4B), as compared to Nf1+/−;Ink4a/Arf−/− mice. Hematopoietic neoplasms also develop in Nf1+/−;Ink4a/Arf−/− mice, as described by Joseph et al. Occurrence of hematopoietic neoplasms was not reduced with Meis1 loss in nerve glial cells. Both Nf1+/−;Ink4a/Arf−/− and Nf1+/−;Ink4a/Arf−/−;Meis1f/f;DhhCre+ mice succumbed to hematopoietic malignancies, with enlarged liver and/or spleen (Fig. 4B). The proportion of hematopoietic malignancies (44%) observed in Nf1+/−;Ink4a/Arf−/− mice was similar to that described in the previous report (Joseph et al., 2008). Hematopoietic malignancies typically occur in 6–8 month old Nf1+/−;Ink4a/Arf−/− mice. Nf1+/−;Ink4a/Arf−/−; Meis1f/f;DhhCre+ mice that failed to develop GEM-PNST developed hematopoietic disease, as read out by increase liver/spleen size (66%; Fig. 4B). Some mice with GEM-PNST might also have had sub-clinical hematopoietic disease that could be revealed by additional analysis.
Fig. 4.
Genetic deletion of Meis1 prevents formation of GEM-PNST. (A) Kaplan-Meier survival analysis shows that the lifespan of Nf1+/−;Ink4a/Arf−/−;Meis1f/f;DhhCre+ mice is significantly increased by Meis1 deficiency (p = 0.00024; log-rank Mantel-Cox test). (B) Bar graph shows the percent of mice in each cohort that succumbed to MPNSTs versus hematopoietic neoplasms. All mice were assessed at time of death for tumors and hematopoietic neoplasms represented by enlarged liver/spleen. Mouse found dead did not show GEM-PNST. (C) Agarose gel shows that mouse sciatic nerve from Meis1-flox mice with Dhh-Cre contain cells with Meis1 deletion. The 440 bp band is the floxed Meis1 allele; the 260 bp band is the deleted Meis1 allele.
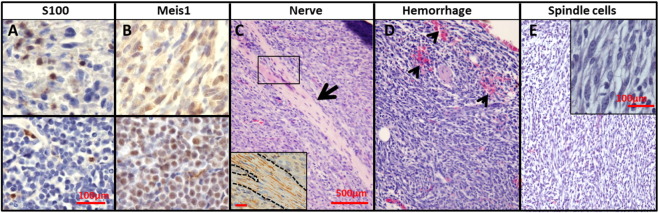
Histological analysis of MPNST-like tumors confirmed features typical of mouse GEM-PNST (Stemmer-Rachamimov et al., 2004), with scattered cells expressing the Schwann cell marker S100 (Supplemental Fig. 3A), nerve bundles (Supplemental Fig. 3C) positive for neurofilament staining (Supplemental Fig. 3C inset), and frequent regions of hemorrhage (Supplemental Fig. 3D). Cells often showed fascicular growth patterns with tightly packed spindle cells (Supplemental Fig. 3E, inset). Cells showed hyperchromatic nuclei and frequent mitoses (Fig. 4J). Two of 27 mice in the Nf1+/−;Ink4a/Arf−/−;Meis1f/f;DhhCre+ cohort developed GEM-PNST. While wild type mouse sciatic nerve did not show Meis1 expression (not shown), both these tumors stained positive for Meis1 (Supplemental Fig. 3), suggesting that they developed in nerve glial cells that had escaped inactivation of Meis1. Indeed, Dhh-Cre targets only about 50% of nerve glial cells (Wu et al., 2008). These results demonstrate an essential role for Meis1 in driving GEM-PNST, and confirm that Meis1 is an MPNST oncogene.
Fig. S3.
(A–E) Photomicrographs show features of mouse PNSTs, including cells positive for S100 (A), Meis1(B), and neurofilament (C). H&E staining reveals tumor integrated nerve bundles (large arrow, C), Hemorrhage (small arrow heads, D), and tumor areas with fascicular patterns of spindle cells (inset in E) are shown. Scale bars: A = 100 μm applies to A-B; C = 500 μm applies to C–E.
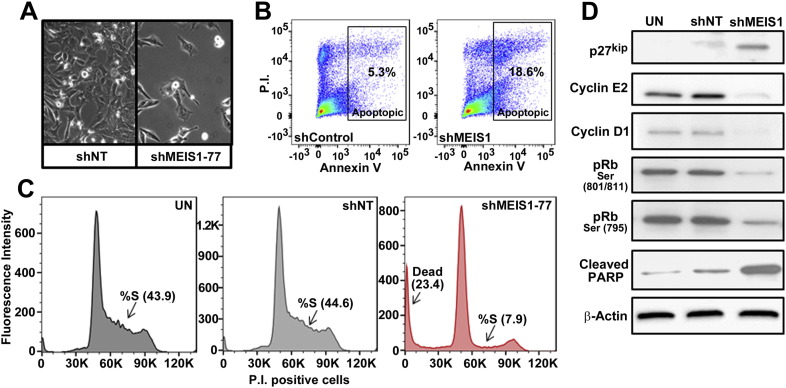
3.5. Downregulation of MEIS1 disrupts normal cycling of MPSNT cells in vitro
We studied cell cycle dynamics to understand the mechanisms of growth inhibition after MEIS1 down regulation (Fig. 5A). Human MPNST cells transduced with shMEIS1 were analyzed 4.5 days post infection by flow cytometry. Apoptotic cells (AnnexinV +) were also stained with propidium iodide, marking DNA. This analysis revealed a 3.7 fold increase in apoptotic cells in shMEIS1 samples (Fig. 5B). In addition, MEIS1 downregulation increased numbers of MPNST cells in the G1/G0 phase of the cell cycle (20% increase versus shNT), and a dramatic depletion of cells in S phase (70% decrease versus shNT) (Fig. 5C). These results suggest an arrest in proliferation upon downregulation of MEIS1 expression. This interpretation was confirmed by analysis of cycle regulator proteins. The decrease in S phase upon downregulation of MEIS1 expression correlated with increased levels of the cell cycle inhibitor p27Kip (Fig. 5D). Cyclin E2 and cyclin D1, markers of the late G1 phase of the cell cycle, were decreased. This correlated with hypo-phosphorylation of Rb, which is required for transition from G1 to S in normally cycling cells (Giacinti and Giordano, 2006). Increased cleaved PARP levels confirmed apoptotic cell death (Fig. 5D). Together, these data indicate that reduced proliferation is accompanied by increased cell death in MPNST upon MEIS1 downregulation.
Fig. 5.
Downregulation of MEIS1 causes MPNSTs cell cycle arrest in G1 and cell death. (A) Phase contrast images show fewer cells on plates infected with shMEIS1 versus shNT day 6 post infections. (B) Flow cytometry data for PI and AnnexinV stained S462-TY cells transduced with shControl or shMEIS1 4.5 days p.i. (C) Histogram displays data from flow cytometry analysis using the DNA dye propidium iodide (P.I.) showing the cell cycle phases including G1, S and G2. ShMEIS1 treated samples (red line) show fewer cells in S-phase and a significant increase in percent dead cells versus untreated (UN) or shNT infected cells all at post infection day 4.5. (D) Immunoblots show levels of cell cycle proteins p27Kip, phospho-Rb at designated phosphorylation sites, cyclinE2 and cyclinD1cell p.i day 5 in UN, shNT and shMEIS1 MPNST cells. Cleaved PARP marks cell death. Anti-β-actin was used as a loading control.
3.6. MEIS1 drives proliferation of MPNSTs via inhibition of p27Kip
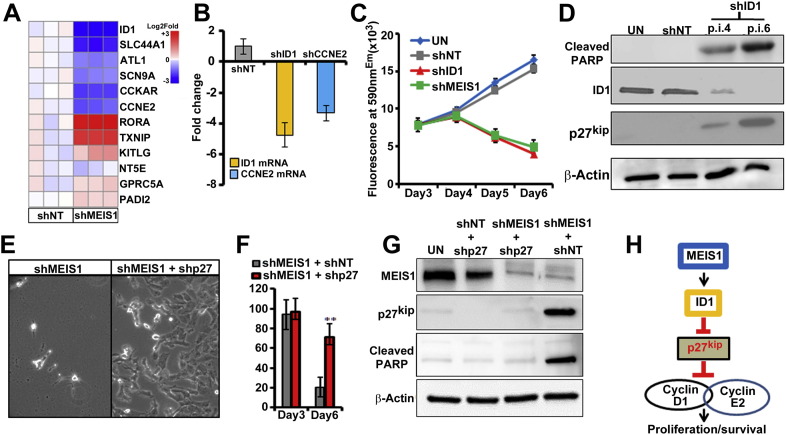
To define molecular pathways regulated by MEIS1 in MPNST cells, we compared the gene-expression profiles of shMEIS1 and shNT treated cells. We selected transduced MPNST cells in puromycin and analyzed RNA profiles by Affymetrix whole genome arrays at day 3.5 after infection, before cells had begun to die. Gene-set enrichment analysis revealed significant alterations in cell cycle genes, supporting our cell cycle analyses (Table 2). Interestingly, Inhibitor of DNA Binding 1 (ID1) and Cyclin E2 (CCNE2) were downregulated by shMEIS1 (Fig. 6A), and were in a subset of genes that we defined as being downregulated by shMEIS1 and containing predicted MEIS1 binding sites within 10 kb up- or down- stream of their transcriptional start sites. Differential expression of these two genes was validated using qRT-PCR (Supplemental Fig. 1C). ID1 can downregulate the cell cycle inhibitor p27Kip, which has roles in the G1/S transition (Ling et al., 2002, Manrique et al., 2015). Given the observed proliferation block after MEIS1 knockdown (Fig. 5C) and the concomitant ID1 and CCNE2 downregulation, we hypothesized that ID1 and/or CCNE2 might be critical mediators of the growth effects of MEIS1. We performed knockdown of ID1 and CCNE2 (Supplemental Fig. 1D) using lentiviral shRNAs to test if downregulation of either ID1 or CCNE2 causes MPNST growth arrest. Down regulation of CCNE2 resulted in minimal effects on MPNST growth (Fig. 6B, C). Importantly, however, downregulation of ID1 mimicked downregulation of MEIS1, and significantly inhibited growth of MPNST (Fig. 6C). In addition, like knockdown of MEIS1, knockdown of ID1 resulted in upregulation of p27kip and apoptosis, as shown by cleaved PARP (Fig. 6D).
Table 2.
Gene set enrichment analysis using genes with altered expression after shMEISl in MPNST cells. We applied an FDR < 0.05. The table lists non-redundant pathways identified at p < 0.002 that were identified using the sources listed.
| # | ID | Name | Source | p-Value | # Genes input/# genes in annotation | Enriched genes p < 0.002 |
|---|---|---|---|---|---|---|
| 2 | 920977 | RB in cancer | BioSystems: WikiPathways | 1.33E-04 | 8/87 | CCNE2 BARD1 KIF4A HMGB1 MCM3 CDC45 RBBP4 PCNA |
| 9 | 105683 | Activation of DNA fragmentation factor | BioSystems: REACTOME | 1.31E-03 | 3/13 | HMGB1 HIST1H1C HIST1H1D |
| 3 | 907939 | Tryptophan degradation via kynurenine | BioSystems: BIOCYC | 5.72E-04 | 3/10 | ID01 KYNII TD02 |
| 8 | 105682 | Apoptosis induced DNA fragmentation | BioSystems: REACTOME | 1.31E-03 | 3/13 | ACAT1 KYNU TD02 |
| 7 | SMP00016 | Propanoate metabolism | SMPDB | 1.02E-03 | 3/12 | ACAT1 ACSS3 PCCB |
| 10 | 106509 | Neurophilin interactions with VEGF/VEGFR | BioSystems: REACTOME | 1.77E-03 | 2/4 | NRP2 FLT1 |
| 13 | 833812 | ECM proteoglycans | BioSystems: REACTOME | 2.63E-03 | 5/55 | ITGA DCN ITGAV LAMA4 LRP4 |
| 14 | 83064 | TGF-beta signaling pathway | BioSystems: KEGG | 2.68E-03 | 6/80 | ID1 ID3 INHBA DCN FST RBX1 |
| 15 | 194384 | African trypanosomiasis | BioSystems: KEGG | 2.76E-03 | 4/34 | IL18 ID01 ICAM1 LAMA4 |
| 18 | 198828 | Hypertrophy model | BioSystems: WikiPathways | 4.78E-03 | 3/20 | IL18 NR4A3 IFRD1 |
Fig. 6.
(A) Heat map displays changes in gene after shMEIS1 in MPNST cells versus shNT. (B) Graph shows confirmation of reduced ID1 and CCNE2 mRNA in MPNST cells by qRT-PCR after exposure to respective shRNAs. (C) Survival p.i with shID1, shMEIS1, and shNT, analyzed using an Alamar blue fluorescence assay. (D) Immunoblots show ID1, cleaved PARP, p27Kip and β Actin in UN, shNT and shID1 treated MPNST cells (p.i. day 4, 6). (E) Phase contrast images show the rescue of cell survival in MPNST cells co-infected with shMEIS1 and shp27, versus shMEIS1/shNT. (F) Bar graph shows significant rescue in survival of MPNST in the presence of shp27. (G) Immunoblots confirm the downregulation of p27Kip expression in cells infected with shp27 concurrent with absence of cleaved PARP. (H) Schematic showing of how MEIS1 is driving survival of MPNST cells through downstream regulation of ID1 and p27Kip.
We therefore tested if p27Kip is necessary for the cell death observed on downregulation of MEIS1. Remarkably shCDKN1B (CDKN1B encodes the p27Kip protein) completely prevented shMEIS1 effects on MPNSTs cells (Fig. 6E, F). Immunoblots confirmed the successful downregulation of MEIS1 and p27Kip protein expression by the shRNAs (Fig. 6G). Knockdown of p27Kip was accompanied by a dramatic reduction in cleaved PARP. Thus, MEIS1 in MPNST cells inhibits p27Kip expression, enabling cell survival (Fig. 6H).
4. Discussion
Our screen for novel MPNST driver genes used data from neurofibroma and MPNST samples in comparison to Schwann cells (Miller et al., 2009), the most complete gene expression analysis data currently available for these tumors, and is a shRNA screen described in cells from this sarcoma type. We chose to down-regulate genes overexpressed in neurofibroma and MPNST. A counter screen in A549 cells resulted in identification of genes specific to MPNST, as well as a list of genes that are potentially relevant to other cells that depend on RAS signaling (Supplemental Fig. 1A). We identify MEIS1, a known leukemia oncogene, as an oncogenic driver of MPNST. Mechanistically, we found that MEIS1 downregulates the expression of p27Kip, which enables MPNST cell survival.
RNA interference is a powerful technique used for identification of driver genes in many common cancers. This methodology has pitfalls, including off-target effects of shRNAs and cellular cytotoxicity (Reviewed in Cullen, 2006). To minimize these drawbacks, we tested 3–5 shRNA per gene, predicting that they would not share off target effects. A gene was considered a positive hit only if multiple shRNA/gene had significant effects on cell survival. Supporting the specificity of shMEIS1, we reversed the effects by expression of a mouse Meis1 cDNA which was not targeted by the shRNA targeting human MEIS1. In addition, we recapitulated the effects of shMEIS1 in a loss-of-function GEM model. In contrast to the robust differential expression of MEIS1 in MPNST, MEIS3 is not differentially expressed in MPNST cells. MEIS2 is overexpressed in MPNSTs, and was part of our lentiviral survival screens. We required that at least two hairpins per gene effectively blocked growth of MPNST cells, and did not significantly alter A549 survival to define a hit. For shMEIS2, only one shRNA blocked MPNST cell growth, and this shMEIS2 was not specific to MPNST. Thus, we focused our efforts on MEIS1.
MEIS1 was the only known oncogene identified among 7 genes whose down-regulation diminished survival of MPNST cells. The other six genes necessary for MPNST but not A549 cell survival await further study. In a limited analysis, shPITX2 killed iHSC as well as MPNST cells (not shown). This result indicates that targeting some of the 6 genes, unlike targeting shMEIS1, causes effects on normal cells. Others of the 6 genes may act in the MEIS1 pathway, although none were identified in our shMEIS1 screen. All 5 cell lines tested, both NF1 and sporadic MPNST cells, were sensitive to downregulation of all 7 genes. This finding is consistent with the remarkable concordance in gene expression between NF1 patient and sporadic MPNST cell lines and tumors (Miller et al., 2006, Miller et al., 2009, Watson et al., 2004). Thus, some or all of the genes identified here as necessary for MPNST survival warrant further study.
MEIS1 was expressed in MPNST patient tumors and in MPNST cell lines. The MEIS1 locus was amplified in 23.5% of tested MPNST samples. MEIS1 is also amplified in neuroblastoma (Spieker et al., 2001), and upregulation of Meis1 is driven by retroviral insertion, causing leukemia (Nakamura et al., 1996). Methylation analysis of MPNST showed hypomethylation, indicating another possible mechanism underlying increased MEIS1 expression. Given the alterations in PRC complex genes in MPNST which alter methylation, we postulate that MEIS1 might be a target of PRC complex effects in some tumors (De Raedt et al., 2014, Lee et al., 2014). It remains to be determined if additional mechanisms contribute to MEIS1 over-expression in MPNSTs. In MLL leukemia, MEIS1 is a direct target of the MLL-fusion protein (Zeisig et al., 2004), which up-regulates gene-expression by multiple mechanisms, including H3K79 methylation and recruiting p-TEFb (Benedikt et al., 2011, Bernt et al., 2011). In leukemia, and in the developing hematopoietic system, Meis1 is a Hox co-factor (Penkov et al., 2013, Xiang et al., 2010). MEIS1 also acts by directly binding DNA. In development, there is evidence that MEIS1 has direct transcriptional ability (Zhang et al., 2006, Zhang et al., 2002). MEIS1 is also overexpressed in neuroblastoma, a tumor of neural crest-derived sympathetic cells, likely neuroblasts (Kiyonari and Kadomatsu, 2015, Geerts et al., 2003). The mechanisms underlying MEIS1 up-regulation in neuroblastoma are unknown.
MPNST are believed to be derived from Schwann cell precursors and/or their neural crest cell precursors, based on expression of neural crest markers SOX9 and TWIST1 and absence of markers of post-neural crest Schwann cell precursor cells, including SOX10 (Miller et al., 2009, Pekmezci et al., 2015, Vogel et al., 1999). We targeted Meis1 deletion in InkArf−/−;Nf1+/− mice with Dhh-Cre. This Cre driver is not activated in neural crest cells, but rather in more mature Schwann cell precursor cells. Therefore, Meis1 deletion in a more mature cell type attenuated MPNST tumor burden and increased mouse survival. MPNST may arise from more mature cells which take on characteristics of more undifferentiated neural crest cells. Alternatively, Meis1 overexpression may occur subsequent to tumor initiation driven by loss of the Nf1 and Ink/Arf tumor suppressors, and be necessary for tumor promotion. The latter interpretation is consistent with our finding that normal nerve cells do not express detectable Meis1, yet anti-Meis1 staining is robust in GEM-PNST.
MEIS1 knockdown kills MPNST cells concurrent with a G1 cell cycle arrest and upregulation of the cell cycle inhibitor p27Kip. p27Kip is essential for MPNST cell death, as its absence prevents shMEIS1-driven apoptosis (Fig. 6F). This result suggests that cell cycle perturbation due to deficient MEIS1 drives MPNST cell death. In MLL leukemia stem cells that depend on MEIS1, p27Kip represses cell proliferation, and enhances cell quiescence (Zhang et al., 2013). We were unable show that proliferation arrest precedes death, however, in spite of analyzing cells at 12 h intervals after lentiviral infection (unpublished). Rather, it appears that G1/S arrest and apoptotic cell death occur concurrently, at least in the unsynchronized population analyzed. The absence of putative MEIS1 binding sites on p27Kip suggests that MEIS1 regulates CDKN1B (p27Kip) indirectly, although new MEIS binding sites are still being identified (Dardaei et al., 2015). In contrast, however, we identified 6 NNTGACAGNN and 2 TGACAGNY putative Meis1 binding sites within 10 kb of the ID1 transcriptional start site, although we note that none were within 2 kb of the transcriptional start site. Which of these sites (if any) is directly regulated by MEIS1 remains to be determined, and it remains possible that MEIS1 regulates ID1 indirectly. Regardless, ID1 is likely involved in regulation p27Kip. ID1 can affect tumor growth, invasiveness, and angiogenesis (Ruzinova and Benezra, 2003, Yokota and Mori, 2002). As in MPNST cells, in leukemia, breast cancer and prostate cancer, down-regulation of ID1 leads to increased p27 Kip expression (Ling et al., 2002, Mern et al., 2010, Ouyang et al., 2002, Sharma et al., 2012, Tam et al., 2008). In support of a MEIS1, ID1, p27Kip survival pathway in MPNST, shMEIS1 downregulated ID1, and shID1, like shMEIS1, killed MPNST cells and increased levels of p27Kip (Fig. 6C, D). Thus, we postulate that MEIS1 regulates ID1 transcriptionally and that ID1, in turn, regulates expression of p27Kip.
We find that by perturbing the normal cycling of MPNST cells with shMEIS1, we render them highly sensitive to cell death, in comparison to iHSC which grow at a similar rate, or in comparison to A549 cells. Therefore, induction of proliferation arrest in MPNST cells may represent a possible therapeutic strategy. Our results identify MEIS1 as a transcription factor that drives MPNST cell growth, highlighting the need for the development of technology which enables successful targeting of the MEIS1 transcription factor for use in tumors including leukemia, neuroblastoma, and MPNST.
The following are the supplementary data related to this article.
Table showing the complete list of lentiviral shRNAs tested in Screens I and III.
Table showing that results of shRNAs are similar across MPNST cell lines. MTS reads are shown for 4 MPNST cell lines, for 42 genes.
Single nucleotide polymorphism (SNP) showing MPNST DNA with and without MEIS1 (MEIS1 italicized) locus amplification.
Supplementary material.
Funding sources
This work was supported by NIH grants R01NS28840 to NR and R01HL111192 to ARK. AVP was supported by Postdoctoral Fellowship W81XWH1110144 from the DAMD Program on Neurofibromatosis and a Pelotonia Postdoctoral Award.
Conflict of interest statement
DL is a co-founder, consultant, and co-owner of NeoClone Biotechnologies, Inc., Discovery Genomics, Inc., (recently acquired by Immusoft, Inc.), and B-MoGen Biotechnologies, Inc. Some of his laboratory work is funded by Genentech, Inc. He is a consultant to Surrogen, Inc., a company making porcine models of human diseases.
AVP, KEC, KM, AK and NR have no conflict of interest to disclose.
Author contributions
AVP designed and carried out most experiments, drafted the manuscript and figures, and contributed to manuscript revision. K.E.C. assisted with animal husbandry. K.C. carried out gene amplification and gene expression analyses. DL and AK provided scientific input and input on the manuscript. AK provided Meis1fl mice. NR conceived the project and contributed to experimental design and manuscript revision.
Acknowledgements
We thank Drs. Robert Hennigan and Paul Andreasson (Cincinnati Children's Hospital) for many helpful discussions. The Cincinnati Children's Hospital Research Foundation Flow Cytometry and Pathology Cores provided support for these studies (NIH P30 DK0909710551). iHSC were from M. Wallace; University of Florida, Gainesville.
Footnotes
The authors have no conflicts of interest to disclose.
References
- Benedikt A., Baltruschat S., Scholz B., Bursen A., Arrey T.N., Meyer B., Varagnolo L., Muller A.M., Karas M., Dingermann T., Marschalek R. The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia. 2011;25:135–144. doi: 10.1038/leu.2010.249. [DOI] [PubMed] [Google Scholar]
- Bernards R., Brummelkamp T.R., Beijersbergen R.L. shRNA libraries and their use in cancer genetics. Nat. Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- Bernt K.M., Zhu N., Sinha A.U., Vempati S., Faber J., Krivtsov A.V., Feng Z., Punt N., Daigle A., Bullinger L., Pollock R.M., Richon V.M., Kung A.L., Armstrong S.A. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Bottillo I., Ahlquist T., Brekke H., Danielsen S.A., Van Den Berg E., Mertens F., Lothe R.A., Dallapiccola B. Germline and somatic NF1 mutations in sporadic and NF1-associated malignant peripheral nerve sheath tumours. J. Pathol. 2009;217:693–701. doi: 10.1002/path.2494. [DOI] [PubMed] [Google Scholar]
- Boutros M., Ahringer J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Brannan C.I., Perkins A.S., Vogel K.S., Ratner N., Nordlund M.L., Reid S.W., Buchberg A.M., Jenkins N.A., Parada L.F., Copeland N.G. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Buchstaller J., Mckeever P.E., Morrison S.J. Tumorigenic cells are common in mouse MPNSTs but their frequency depends upon tumor genotype and assay conditions. Cancer Cell. 2012;21:240–252. doi: 10.1016/j.ccr.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Kumar A.R., Hudson W.A., Li Q., Wu B., Staggs R.A., Lund E.A., Sam T.N., Kersey J.H. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. Enhancing and confirming the specificity of RNAi experiments. Nat. Methods. 2006;3:677–681. doi: 10.1038/nmeth913. [DOI] [PubMed] [Google Scholar]
- Dardaei L., Penkov D., Mathiasen L., Bora P., Morelli M.J., Blasi F. Tumorigenesis by Meis1 overexpression is accompanied by a change of DNA target-sequence specificity which allows binding to the AP-1 element. Oncotarget. 2015;6:25175–25187. doi: 10.18632/oncotarget.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T., Beert E., Pasmant E., Luscan A., Brems H., Ortonne N., Helin K., Hornick J.L., Mautner V., Kehrer-Sawatzki H., Clapp W., Bradner J., Vidaud M., Upadhyaya M., Legius E., Cichowski K. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- Declue J.E., Papageorge A.G., Fletcher J.A., Diehl S.R., Ratner N., Vass W.C., Lowy D.R. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- Ducatman B.S., Scheithauer B.W., Piepgras D.G., Reiman H.M., Ilstrup D.M. Malignant peripheral nerve sheath tumors. A clinicopathologic study of 120 cases. Cancer. 1986;57:2006–2021. doi: 10.1002/1097-0142(19860515)57:10<2006::aid-cncr2820571022>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Garraway L.A., Sellers W.R. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Geerts D., Schilderink N., Jorritsma G., Versteeg R. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 2003;197:87–92. doi: 10.1016/s0304-3835(03)00087-9. [DOI] [PubMed] [Google Scholar]
- Giacinti C., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Gregorian C., Nakashima J., Dry S.M., Nghiemphu P.L., Smith K.B., Ao Y., Dang J., Lawson G., Mellinghoff I.K., Mischel P.S., Phelps M., Parada L.F., Liu X., Sofroniew M.V., Eilber F.C., Wu H. PTEN dosage is essential for neurofibroma development and malignant transformation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19479–19484. doi: 10.1073/pnas.0910398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisa T., Spence S.E., Rachel R.A., Fujita M., Nakamura T., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Jenkins N.A., Copeland N.G. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaegle M., Ghazvini M., Mandemakers W., Piirsoo M., Driegen S., Levavasseur F., Raghoenath S., Grosveld F., Meijer D. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N.M., Mosher J.T., Buchstaller J., Snider P., Mckeever P.E., Lim M., Conway S.J., Parada L.F., Zhu Y., Morrison S.J. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell. 2008;13:129–140. doi: 10.1016/j.ccr.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D., Lazar A., Lev D. Malignant peripheral nerve sheath tumour (MPNST): the clinical implications of cellular signalling pathways. Expert Rev. Mol. Med. 2009;11:e30. doi: 10.1017/S1462399409001227. [DOI] [PubMed] [Google Scholar]
- Kiyonari S., Kadomatsu K. Neuroblastoma models for insights into tumorigenesis and new therapies. Expert Opin. Drug Discov. 2015;10:53–62. doi: 10.1517/17460441.2015.974544. [DOI] [PubMed] [Google Scholar]
- Kumar A.R., Li Q., Hudson W.A., Chen W., Sam T., Yao Q., Lund E.A., Wu B., Kowal B.J., Kersey J.H. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.R., Sarver A.L., Wu B., Kersey J.H. Meis1 maintains stemness signature in MLL-AF9 leukemia. Blood. 2010;115:3642–3643. doi: 10.1182/blood-2010-01-264564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Teckie S., Wiesner T., Ran L., Prieto Granada C.N., Lin M., Zhu S., Cao Z., Liang Y., Sboner A., Tap W.D., Fletcher J.A., Huberman K.H., Qin L.X., Viale A., Singer S., Zheng D., Berger M.F., Chen Y., Antonescu C.R., Chi P. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legius E., Dierick H., Wu R., Hall B.K., Marynen P., Cassiman J.J., Glover T.W. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosom. Cancer. 1994;10:250–255. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- Legius E., Marchuk D.A., Collins F.S., Glover T.W. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat. Genet. 1993;3:122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- Ling M.T., Wang X., Tsao S.W., Wong Y.C. Down-regulation of Id-1 expression is associated with TGF beta 1-induced growth arrest in prostate epithelial cells. Biochim. Biophys. Acta. 2002;1570:145–152. doi: 10.1016/s0304-4165(02)00189-7. [DOI] [PubMed] [Google Scholar]
- Maeda R., Mood K., Jones T.L., Aruga J., Buchberg A.M., Daar I.O. Xmeis1, a protooncogene involved in specifying neural crest cell fate in Xenopus embryos. Oncogene. 2001;20:1329–1342. doi: 10.1038/sj.onc.1204250. [DOI] [PubMed] [Google Scholar]
- Manrique I., Nguewa P., Bleau A.M., Nistal-Villan E., Lopez I., Villalba M., Gil-Bazo I., Calvo A. The inhibitor of differentiation isoform Id1b, generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. Cancer Lett. 2015;356:899–909. doi: 10.1016/j.canlet.2014.10.035. [DOI] [PubMed] [Google Scholar]
- Mern D.S., Hoppe-Seyler K., Hoppe-Seyler F., Hasskarl J., Burwinkel B. Targeting Id1 and Id3 by a specific peptide aptamer induces E-box promoter activity, cell cycle arrest, and apoptosis in breast cancer cells. Breast Cancer Res. Treat. 2010;124:623–633. doi: 10.1007/s10549-010-0810-6. [DOI] [PubMed] [Google Scholar]
- Miller S.J., Jessen W.J., Mehta T., Hardiman A., Sites E., Kaiser S., Jegga A.G., Li H., Upadhyaya M., Giovannini M., Muir D., Wallace M.R., Lopez E., Serra E., Nielsen G.P., Lazaro C., Stemmer-Rachamimov A., Page G., Aronow B.J., Ratner N. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol. Med. 2009;1:236–248. doi: 10.1002/emmm.200900027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.J., Rangwala F., Williams J., Ackerman P., Kong S., Jegga A.G., Kaiser S., Aronow B.J., Frahm S., Kluwe L., Mautner V., Upadhyaya M., Muir D., Wallace M., Hagen J., Quelle D.E., Watson M.A., Perry A., Gutmann D.H., Ratner N. Large-scale molecular comparison of human Schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 2006;66:2584–2591. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Largaespada D.A., Shaughnessy J.D., JR., Jenkins N.A., Copeland N.G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat. Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- Nielsen G.P., Stemmer-Rachamimov A.O., Ino Y., Moller M.B., Rosenberg A.E., Louis D.N. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am. J. Pathol. 1999;155:1879–1884. doi: 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X.S., Wang X., Ling M.T., Wong H.L., Tsao S.W., Wong Y.C. Id-1 stimulates serum independent prostate cancer cell proliferation through inactivation of p16(INK4a)/pRB pathway. Carcinogenesis. 2002;23:721–725. doi: 10.1093/carcin/23.5.721. [DOI] [PubMed] [Google Scholar]
- Patel A.V., Eaves D., Jessen W.J., Rizvi T.A., Ecsedy J.A., Qian M.G., Aronow B.J., Perentesis J.P., Serra E., Cripe T.P., Miller S.J., Ratner N. Ras-driven transcriptome analysis identifies aurora kinase A as a potential malignant peripheral nerve sheath tumor therapeutic target. Clin. Cancer Res. 2012;18:5020–5030. doi: 10.1158/1078-0432.CCR-12-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekmezci M., Reuss D.E., Hirbe A.C., Dahiya S., Gutmann D.H., Von Deimling A., Horvai A.E., Perry A. Morphologic and immunohistochemical features of malignant peripheral nerve sheath tumors and cellular schwannomas. Mod. Pathol. 2015;28:187–200. doi: 10.1038/modpathol.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkov D., Mateos San Martin D., Fernandez-Diaz L.C., Rossello C.A., Torroja C., Sanchez-Cabo F., Warnatz H.J., Sultan M., Yaspo M.L., Gabrieli A., Tkachuk V., Brendolan A., Blasi F., Torres M. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 2013;3:1321–1333. doi: 10.1016/j.celrep.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Pineault N., Helgason C.D., Lawrence H.J., Humphries R.K. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- Rahrmann E.P., Watson A.L., Keng V.W., Choi K., Moriarity B.S., Beckmann D.A., Wolf N.K., Sarver A., Collins M.H., Moertel C.L., Wallace M.R., Gel B., Serra E., Ratner N., Largaespada D.A. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat. Genet. 2013;45:756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T.M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Ratner N., Miller S.J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhuis S., Van De Wetering M.L., Mooi W.J., Evers S.G., Van Zandwijk N., Bos J.L. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N. Engl. J. Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- Ruzinova M.B., Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Serra E., Rosenbaum T., Winner U., Aledo R., Ars E., Estivill X., Lenard H.G., Lazaro C. Schwann cells harbor the somatic NF1 mutation in neurofibromas: evidence of two different Schwann cell subpopulations. Hum. Mol. Genet. 2000;9:3055–3064. doi: 10.1093/hmg/9.20.3055. [DOI] [PubMed] [Google Scholar]
- Sharma P., Patel D., Chaudhary J. Id1 and Id3 expression is associated with increasing grade of prostate cancer: Id3 preferentially regulates CDKN1B. Cancer Med. 2012;1:187–197. doi: 10.1002/cam4.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieker N., Van Sluis P., Beitsma M., Boon K., Van Schaik B.D., Van Kampen A.H., Caron H., Versteeg R. The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics. 2001;71:214–221. doi: 10.1006/geno.2000.6408. [DOI] [PubMed] [Google Scholar]
- Stemmer-Rachamimov A.O., Louis D.N., Nielsen G.P., Antonescu C.R., Borowsky A.D., Bronson R.T., Burns D.K., Cervera P., Mclaughlin M.E., Reifenberger G., Schmale M.C., Maccollin M., Chao R.C., Cichowski K., Kalamarides M., Messerli S.M., Mcclatchey A.I., Niwa-Kawakita M., Ratner N., Reilly K.M., Zhu Y., Giovannini M. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res. 2004;64:3718–3724. doi: 10.1158/0008-5472.CAN-03-4079. [DOI] [PubMed] [Google Scholar]
- Tam W.F., Gu T.L., Chen J., Lee B.H., Bullinger L., Frohling S., Wang A., Monti S., Golub T.R., Gilliland D.G. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood. 2008;112:1981–1992. doi: 10.1182/blood-2007-07-103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Kroon E., Jerome L., Blasi F., Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnisa Z., Clark J.P., Roychoudhury J., Thomas E., Tessarollo L., Copeland N.G., Jenkins N.A., Grimes H.L., Kumar A.R. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood. 2012;120:4973–4981. doi: 10.1182/blood-2012-06-435800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K.S., Klesse L.J., Velasco-Miguel S., Meyers K., Rushing E.J., Parada L.F. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.A., Perry A., Tihan T., Prayson R.A., Guha A., Bridge J., Ferner R., Gutmann D.H. Gene expression profiling reveals unique molecular subtypes of neurofibromatosis type I-associated and sporadic malignant peripheral nerve sheath tumors. Brain Pathol. 2004;14:297–303. doi: 10.1111/j.1750-3639.2004.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A.L., Rahrmann E.P., Moriarity B.S., Choi K., Conboy C.B., Greeley A.D., Halfond A.L., Anderson L.K., Wahl B.R., Keng V.W., Rizzardi A.E., Forster C.L., Collins M.H., Sarver A.L., Wallace M.R., Schmechel S.C., Ratner N., Largaespada D.A. Canonical Wnt/beta-catenin signaling drives human Schwann cell transformation, progression, and tumor maintenance. Cancer Discov. 2013;3:674–689. doi: 10.1158/2159-8290.CD-13-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widemann B.C. Current status of sporadic and neurofibromatosis type 1-associated malignant peripheral nerve sheath tumors. Curr. Oncol. Rep. 2009;11:322–328. doi: 10.1007/s11912-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Williams J.P., Rizvi T.A., Kordich J.J., Witte D., Meijer D., Stemmer-Rachamimov A.O., Cancelas J.A., Ratner N. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008;13:105–116. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang P., Lo C., Argiropoulos B., Lai C.B., Rouhi A., Imren S., Jiang X., Mager D., Humphries R.K. Identification of E74-like factor 1 (ELF1) as a transcriptional regulator of the Hox cofactor MEIS1. Exp. Hematol. 2010;38:798. doi: 10.1016/j.exphem.2010.06.006. (808 e1-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Mori S. Role of Id family proteins in growth control. J. Cell. Physiol. 2002;190:21–28. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- Zeisig B.B., Milne T., Garcia-Cuellar M.P., Schreiner S., Martin M.E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Hess J.L., Slany R.K. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell. Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Friedman A., Heaney S., Purcell P., Maas R.L. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Rowan S., Yue Y., Heaney S., Pan Y., Brendolan A., Selleri L., Maas R.L. Pax6 is regulated by Meis and Pbx homeoproteins during pancreatic development. Dev. Biol. 2006;300:748–757. doi: 10.1016/j.ydbio.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Zhang J., Seet C.S., Sun C., Li J., You D., Volk A., Breslin P., Li X., Wei W., Qian Z., Zeleznik-Le N.J., Zhang Z., Zhang J. p27kip1 maintains a subset of leukemia stem cells in the quiescent state in murine MLL-leukemia. Mol. Oncol. 2013;7:1069–1082. doi: 10.1016/j.molonc.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J.J., Hunter C.P. RNA interference in Caenorhabditis elegans: uptake, mechanism, and regulation. Parasitology. 2012;139:560–573. doi: 10.1017/S0031182011001788. [DOI] [PubMed] [Google Scholar]
- Zuber J., Mcjunkin K., Fellmann C., Dow L.E., Taylor M.J., Hannon G.J., Lowe S.W. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Rappaport A.R., Luo W., Wang E., Chen C., Vaseva A.V., Shi J., Weissmueller S., Fellmann C., Taylor M.J., Weissenboeck M., Graeber T.G., Kogan S.C., Vakoc C.R., Lowe S.W. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table showing the complete list of lentiviral shRNAs tested in Screens I and III.
Table showing that results of shRNAs are similar across MPNST cell lines. MTS reads are shown for 4 MPNST cell lines, for 42 genes.
Single nucleotide polymorphism (SNP) showing MPNST DNA with and without MEIS1 (MEIS1 italicized) locus amplification.
Supplementary material.