Abstract
Desensitization is an important mechanism that curtails the activity of ligand-gated ion-channels (LGICs). Although the structural basis of desensitization is not fully resolved, it is thought to be governed by the physicochemical properties of the bound ligand. Here, we show the importance of an allosteric cation binding pocket in controlling transitions between activated and desensitized states of rat kainate-type (KAR) ionotropic glutamate receptors (iGluRs). Tethering a positive charge to this pocket sustains KAR activation, preventing desensitization, whereas mutations that disrupt cation binding eliminate channel gating. These different outcomes explain the structural distinction between deactivation and desensitization. Deactivation occurs when the ligand unbinds before the cation, whereas desensitization proceeds if a ligand is bound without cation pocket occupancy. This sequence of events is absent from AMPA-type iGluRs, identifying cations as gatekeepers of KAR gating, a role unique among even closely-related LGICs.
Structural and functional biologists have long sought to understand the mechanisms by which ligand-gated ion-channels (LGICs) respond to small chemical ligands and modulators. Seminal work established the general principle that LGICs are not only activated by biologically-derived molecules, such as the neurotransmitter acetylcholine 1, but they are also inactivated by prolonged exposure to these molecules through a process universally known as desensitization 2. Since this work, almost all LGICs have been shown to desensitize. For example, desensitization is thought to shape signaling within the vertebrate central nervous system (CNS) by impacting fast chemical transmission mediated by ionotropic glutamate receptors (iGluRs), along with GABAA and glycine receptors 3. From all of this work, it has been concluded that the conformational events that lead to the occurrence of deactivation and the onset of desensitization are governed by the physicochemical properties of the bound ligand 4. In support of this, pioneering work on native AMPA-type iGluRs (AMPARs) has shown that even modest changes to the ligand structure have profound effects on the rates and degree of desensitization 5.
During the last decade, structural and functional analyses of LGICs have revealed that the molecular basis of channel gating may be quite distinct for different ion-channel families 6–8. For the iGluR family, numerous mechanistic details of activation and desensitization have been identified and extensively commented upon 9–11. Following the elucidation of the ligand-binding domain (LBD) structure 12, a mechanism of iGluR desensitization was proposed, involving the separation of subunits that are assembled as dimers at the LBD 13. This mechanism has been supported by additional crystal structures, which captured AMPARs in different functional states 14. Accordingly, efforts to engineer iGluR receptors that lack desensitization have focussed on constraining movement at the LBD dimer interface. From this, covalent crosslinking of the dimer interface has been shown to generate AMPAR and kainate-type iGluRs (KARs) that yield non-decaying currents upon sustained agonist application 15, 16. Similar experiments on NMDA-type iGluRs have offered a more nuanced explanation of LBD function by studying the structural 17 and single-channel effects 18 of dimer crosslinking. Specifically, they propose that constricting the dimer interface primarily affects open-channel probability and not desensitization18. This observation suggests that a more in depth single-channel analysis of the mechanism of AMPAR and KAR desensitization is warranted.
Here, we set out to study the molecular basis of KAR desensitization by evaluating mutants that are proposed to block it 15,19. In both cases, the mutations are located in the GluK2 KAR LBD dimer interface, which not only is implicated in receptor desensitization, but also harbors binding pockets for both sodium and chloride ions 20, 21. Prior work from our lab shows that external ions are an absolute requirement for GluK2 receptor activation 22 yet their precise role in desensitization is unresolved 21, 23. Our present data identifies that desensitization of KARs only proceeds if a ligand is bound without cation pocket occupancy, whereas deactivation occurs when the ligand unbinds before the cation. This sequence of events identifies external cations as pivotal in directing KARs into active states or long-lived desensitized states.
RESULTS
KARs desensitize with or without prior channel activation
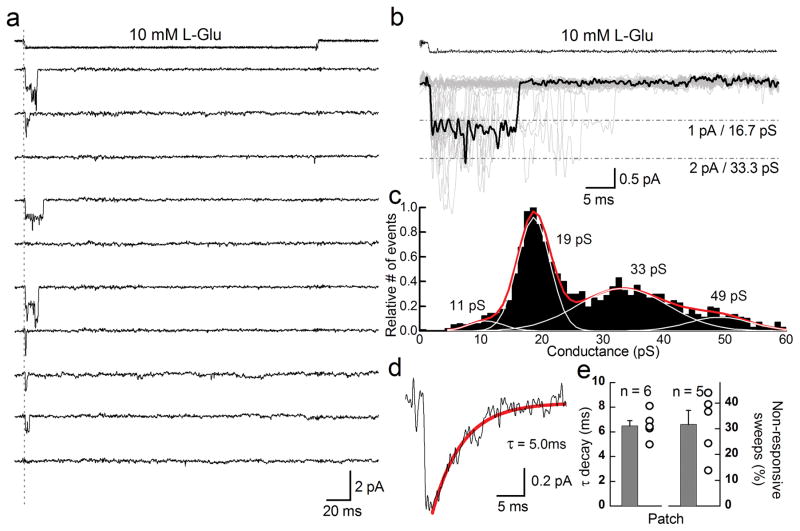
To observe the microscopic behavior of KAR desensitization, we excised outside-out patches from transfected mammalian cells expressing homomeric GluK2 receptors (see Methods). Using an ultrafast agonist perfusion system, we recorded single-channel events and then selected, for analysis, recordings where most responses corresponded to the conductance expected of a single channel 24. Although the actual number of active receptors per patch is not known, these single-channel recordings nevertheless reveal the different routes taken by KARs before entering into desensitization. In most cases, rapid application of saturating glutamate (10 mM L-Glu) activated GluK2 receptors, which open to one of several conductance levels (Fig. 1a–c). Once in the open state, KAR channels typically closed within tens of milliseconds, and did not re-open for any measurable duration of time afterwards indicating that the receptor desensitized. Since desensitization is not thought to occur directly from the open state, it presumably proceeded shortly after channel closure. In agreement with this latter point, ensemble averages of single-channel sweeps exhibited decay times constants (6.49 ± 0.41 ms, n = 6, Fig. 1d, e) which were statistically indistinguishable from decay rates of macroscopic responses (6.28 ± 0.43 ms, n = 9, p = 0.74), re-affirming that the onset of KAR desensitization is approximated by the duration of channel activity.
Figure 1.
Kainate receptor desensitization occurs with or without channel activation. (a) Typical GluK2 receptor unitary current events elicited by 10 mM L-Glu (250 ms pulse duration) in an outside-out patch recording (Patch # 12212p1, −60 mV). (b) Overlay of forty-five individual current records from the same patch shown in panel a. A typical opening elicited by L-Glu is shown in bold. (c) GluK2 conductance distributions plotted following time course fitting. (d) Averaging individual current records from the patch shown in panels a and b generated an ensemble response with a decay that could be fit by a single exponential function. (e, left) Decay time constants of ensemble responses from several patches and (right) the fraction of L-Glu applications that did not elicit a measureable response from receptors. Error bars, s.e.m. from five or six independent patch experiments.
In some cases, 10 mM L-Glu failed to elicit a measurable response during the entire 250 ms application (Fig. 1a) corresponding to about 31.7 ± 5.5 % of the 525 total sweeps from five patches (Fig. 1e). The apparent failure to respond to the agonist may reflect an intrinsic inability of L-Glu to reliably convert its energy of binding to activation. If this was the case, however, channel opening would eventually be observed, as the continued presence of L-Glu would ensure that the energy threshold for activation would be overcome. Consequently, the inability of L-Glu to activate GluK2 receptors must represent the onset of desensitization without prior passage through the open state(s).
The discrete molecular events that bring about desensitization are currently unresolved. Several studies, however, identify the ligand-binding domain (LBD) dimer interface 15 and the cation binding site 19, 25 in the conformational events that initiate KAR macroscopic desensitization. Whether one site or the other has a more direct effect on desensitization has yet to be directly studied. As discussed below, we examined this by studying the single-channel properties of two apparently non-desensitizing GluK2 receptors, namely the mutants D776K and Y521C L783C.
The D776K mutation abolishes GluK2 receptor desensitization
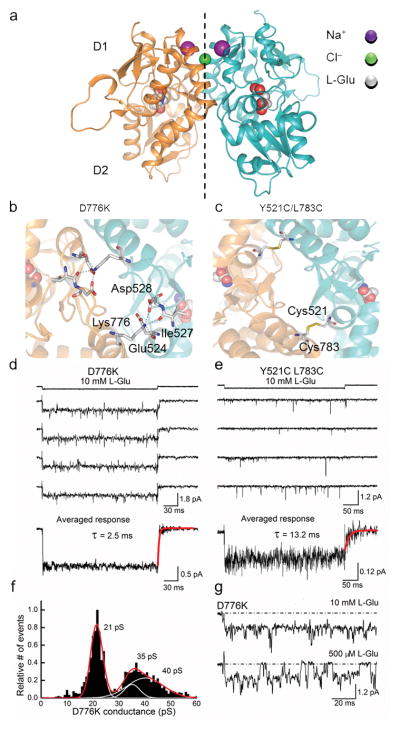
The LBD dimer interface of wildtype GluK2 receptors contains binding sites for two sodium ions (purple) and a single chloride ion (green) (Fig. 2a) 20, 21. Both GluK2 receptor mutations (D776K and Y521C L783C) are also located at the LBD dimer interface (Fig. 2b, c) where they are proposed to eliminate desensitization by constraining subunit movement. The positively-charged lysine of D776K establishes new inter-protomer contacts by tethering to the cation binding pocket (Fig. 2b) 25, whereas the cysteine residues of Y521C L783C are thought to achieve this through the formation of covalent disulfide bridges between subunits (Fig. 2c) 15. Since both mutant receptors are expected to affect the functional properties of KARs similarly, we were surprised to observe that their single-channel behavior was quite different.
Figure 2.
Mutation of Asp 776 to a Lys residue eliminates GluK2 receptor desensitization. (a) Crystal structure of the wildtype GluK2 LBD dimer (PDB 3G3F 42). (b) Top view of the GluK2 D776K LBD dimer interface showing electrostatic interactions between Lys 776 and the adjacent subunit (PDB 2XXX 25). (c) Top view of the GluK2 Y521C L783C LBD dimer interface showing covalent crosslinking between subunits (PDB 2I0C 15). (d) Typical current responses elicited by L-Glu acting on a single D776K channel (Patch # 12127p2, −60 mV). (e) Unitary current events elicited by L-Glu acting on Y521C L783C channels (Patch # 12322p3, −100 mV). In panels d and e, averaged ensemble responses were taken from 20 or 95 individual current records, respectively. Time constants of deactivation were obtained by fitting agonist-off current responses with a single exponential function. (f) GluK2 D776K conductance distributions plotted following time course fitting. (g) Individual current responses of a single GluK2 D776K receptor to 10 mM and 500 μM L-Glu (Patch # 12124p1).
Like wildtype receptors, 10 mM L-Glu rapidly activated single D776K channels. However, instead of opening only briefly prior to desensitization, agonist binding led to sustained activation of the 21–22 pS main open state (i.e. most-frequented) (Fig. 2d). In support of this, repetitive applications of 10 mM L-Glu to patches containing a single D776K receptor elicited activity in every case, demonstrating that this mutant GluK2 receptor displays close to the maximum probability of opening. Averaged ensemble responses were non-decaying in nature with rapid off-kinetics of about 2–3 ms due to L-Glu removal (Fig. 2d). These persistent openings were nevertheless interrupted by transient closures too brief to represent long-lived desensitized states and, consequently, must represent sojourns to lower conductance levels, or closed or unliganded states.
Unlike the D776K receptor, the double cysteine mutant did not yield persistent channel activity in saturating L-Glu. Instead, recordings were dominated by sub-millisecond openings that were separated by longer apparent closures (Fig. 2e) 26. Given the infrequent nature of gating, we concluded that responses observed in the excised patches were likely to originate from multiple channels. Despite the transient openings, averaging sweeps from many agonist applications generated a non-decaying ensemble response. The decay kinetics of the ensemble average current of Y521C L783C receptors were nevertheless at least five times slower (14.8 ± 2.9 ms, n = 4) than those of D776K receptors (Fig. 2e).
For GluK2 D776K, its consistent gating behavior allowed us to make additional inferences. Time-course fitting of resolvable single-channel events estimated conductance levels of 21, 35, and 40 pS which were calculated using a measured reversal potential of 0 mV (Fig. 2f). The open level most frequently visited was 21–22 pS, closely matching the predominant 19 pS conductance level of wildtype receptors, with the two largest conductance levels corresponding to brief sojourns from this state (i.e. 35 and 40 pS). Fitting Gaussian functions to an all-points histogram of D776K data further shows that more than 90 % of the analyzed records corresponded to the main open state (Supplementary Fig. 1). These conductance levels are likely to originate from single channels, rather than several channels opening simultaneously, as lowering the concentration of L-Glu interrupted openings to the 21–22 pS state with clear closures to baseline (Fig. 2g).
In summary, our single-channel data reveal that GluK2 D776K exhibits all the hallmarks expected of a non-desensitizing KAR: sustained activation, high unitary conductance, and an absence of long duration closures. GluK2 Y521C L783C responds quite differently and therefore, we could conclude that the structural basis of its functional behavior must be different. Since the Lys 776 residue is proposed to act as a tethered cation 25 we reasoned that occupancy of the ion binding pocket may be the key structural event that prevents the onset of desensitization. If true, cation interactions at the Y521C L783C receptor might therefore be unstable which would account for differences observed at the single-channel level. As explained below, we tested this hypothesis using molecular dynamic (MD) simulations to estimate the residency time of sodium bound to the cation binding pockets of both D776K and Y521C L783C receptors.
Lys 776 substitutes for sodium at the cation binding pocket
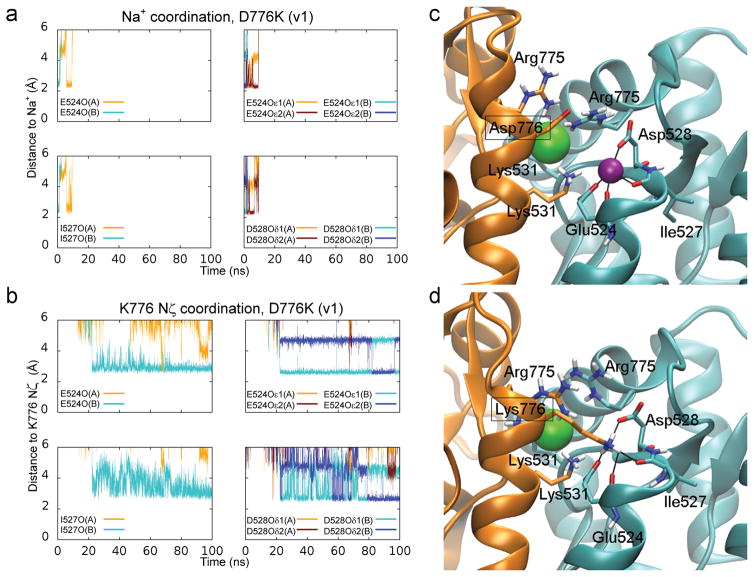
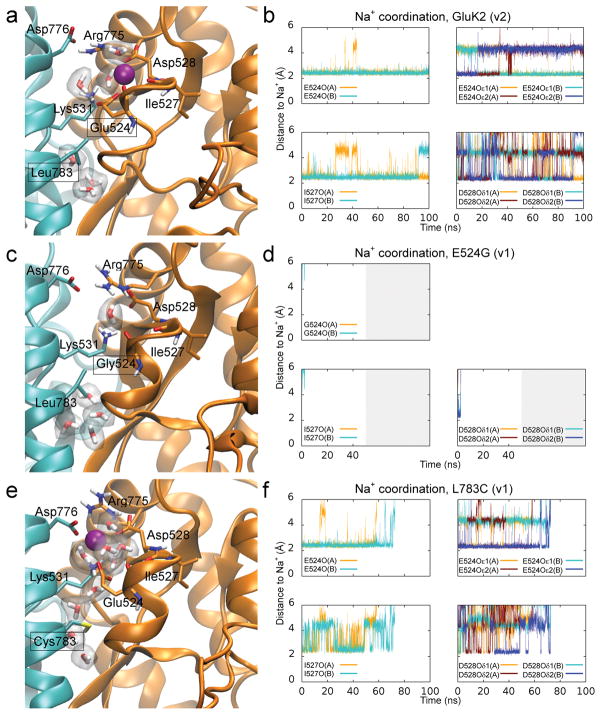
MD simulations were employed to explore how electrostatic interactions affect occupancy of the cation binding pocket, which cannot be achieved using X-ray crystal structures or electrophysiology. Over the course of each of two 100 ns simulations, the cation pockets of the D776K receptor first released both sodium ions and then formed new contact points with the amino groups of Lys 776 (Fig. 3a–d, Supplementary Movie 1). Consequently, the cation binding pocket was nearly continuously occupied by a positive charge during the entire simulation period, which is consistent with previous structural data 25. In contrast, simulations of the Y521C L783C receptor predict that these mutations destabilize sodium and chloride ion binding, facilitating rapid ion release in both simulations performed (Supplementary Fig. 2a, b, Supplementary Movie 2). There was also a tendency for water molecules to more readily occupy the cation pockets of Y521C L783C, which may explain the instability in sodium and chloride ion binding. Measurements of the surface area accessible to solvent indicated a much higher propensity for water molecules to interact with residues lining the cation pocket in the double cysteine mutant compared to wildtype GluK2 receptors (Supplementary Fig. 2c, d). If these simulations reflect the physiological behavior of kainate receptors, then activation could depend on occupancy of the cation pocket, while cation unbinding would promote channel closure and/or desensitization.
Figure 3.
Lys 776 can act as a tethered ion at the GluK2 cation binding pocket. (a) Coordination distances between sodium ions (bound to chains A and B) and several oxygen atoms found on residues lining the cation binding pocket (E524, I527, D528) during a 100 ns MD simulation of the D776K mutant. (b) Coordination distances for the positively charged Nζ of Lys 776. Distances were measured from oxygen atoms normally involved in sodium ion coordination. (c) Sodium ion coordination in the crystal structure of the wildtype GluK2 LBD. (d) Snapshot after 100 ns of MD simulation of the D776K mutant. Chain A and its residues are shown in orange, while chain B and its residues are shown in cyan. The sodium ion is shown in purple and the chloride ion in green. Coordination distances are indicated with black lines for the sodium ion (c) and the Lys 776 amine (d). Water molecules and non-polar hydrogen atoms are omitted.
GluK2 D776K receptors activate without external cations
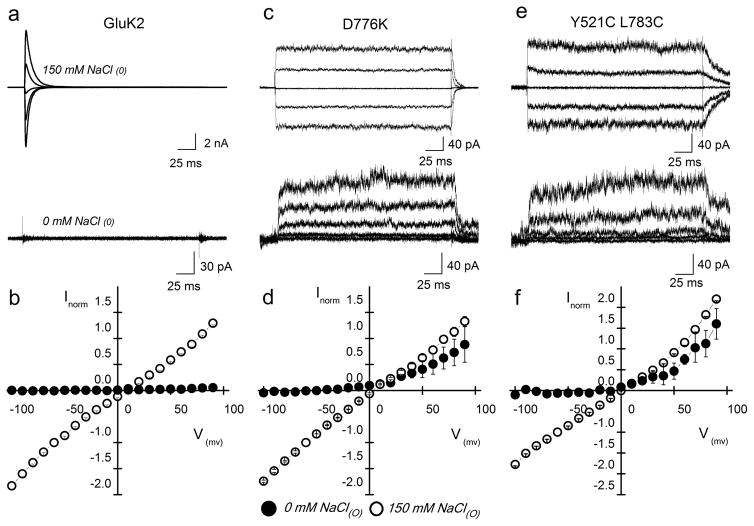
If occupancy of the cation binding pocket is a prerequisite for wildtype KAR activation, removal of all external ions should result in the absence of any detectable current. Although such recordings have already been shown to abolish wildtype KAR activity 22, this original finding has been disputed by more recent work claiming residual channel activity in ion-free conditions 21. To re-examine this issue, we repeated experiments comparing GluK2 receptors in the presence and absence of external ions. If Lys 776 acts as a tethered cation, as suggested by MD simulations (Fig. 3) and structural data 25, we reasoned that the GluK2 D776K would gate in the absence of external cations. In contrast, the instability of cation binding to GluK2 Y521C L783C suggests that this mutant would fail to gate in the absence of ions unless crosslinking the LBD dimer interface permits activation via a different mechanism. Consistent with the above predictions, wildtype GluK2 receptor activity was completely abolished by the removal of external monovalent ions (Fig. 4a, b) whereas the D776K receptor continued to gate (Fig. 4c, d) demonstrating that the wildtype GluK2 receptor gating mechanism has an absolute requirement for external cations. These data also further support the idea that the Lys 776 residue acts as a tethered cation, accounting for the ability of the D776K receptor to gate in the absence of external ions.
Figure 4.
GluK2 D776K receptors gate in the absence of external ions. (a, c, e) Membrane currents evoked by L-Glu acting on wildtype GluK2 (a), D776K (c), and Y521C L783C (e) receptors, in either 150 mM NaCl (top) or in nominal ion-free (bottom) external solution (Vm = −60, −30, 0, 30, and 60 mV). For wildtype GluK2, the same patch was recorded in both ionic conditions to show the complete abolition of current (Patch # 121106p2), whereas mutant responses were taken from different patches (D776K ion, Patch # 11510 p1; ion-free, Patch # 12925 p5; Y521C L783C ion, Patch # 121002 p2; ion-free, Patch # 121023 p2). (b, d, f) Averaged current-voltage plots in 0 mM (filled circles) and 150 mM (open circles) NaCl for wildtype GluK2 (b), D776K (d), and Y521C L783C (f) receptors. Currents were normalized to responses at −60 mV in 150 mM NaCl. Error bars, s.e.m. from three independent experiments for each receptor.
Interestingly, the Y521C L783C receptor was also able to gate in the absence of external cations (Fig. 4e, f). This finding is in agreement with a prior study 21 but inconsistent with the lack of responsiveness of wildtype GuK2 receptors in ion-free conditions (Fig. 4a, b), suggesting the need for an alternative explanation. With this in mind, we considered the possibility that crosslinking the dimer interface of the GluK2 receptor may eliminate the requirement of external cations for activation. We tested this possibility by identifying mutations in the LBD dimer interface that would disrupt cation binding without forming inter-protomer crosslinks.
Destabilizing cation binding impairs GluK2 activation
We studied disruption of the cation binding pocket by examining two mutant receptors, namely GluK2 E524G and L783C, which MD simulations suggest destabilize sodium binding to the cation binding pocket. Importantly, these mutations do not affect receptor surface expression (see Supplementary Fig. 3a, b). For E524G, which has a less electronegative cation pocket, two simulations of sodium coordination both estimated that sodium is released within 5 ns, unlike the wildtype receptor, which retained sodium for the duration of two, 100 ns simulations (Fig. 5a–d, Supplementary Movies 3 and 4). In this respect, E524G mimics the Y521C L783C receptor; however, it differs in that 10 mM L-Glu fails to elicit a measurable response in most excised patches (Supplementary Fig. 3c). We did observe responses in 3 out of the 18 patches tested but they were small (> −10 pA) in amplitude and thus consistent with the E524G mutation acting to destabilize cation binding.
Figure 5.
Occupancy of the GluK2 cation binding pocket is predicted to be disrupted by targeted mutation of the dimer interface. (a, c, e) Snapshots of sodium coordination in the wildtype GluK2 receptor (a), as well as mutants E524G (c) and L783C (e), all taken approximately 15 ns after the start of the MD simulation. (b, d, f) Sodium coordination plotted from MD simulations of the LBD dimer in the wildtype GluK2 receptor (b), and mutants E524G (d) and L783C (f).
Interestingly, when only one of the crosslinking residues (i.e. L783C) was mutated, 10 mM L-Glu failed to elicit a response in all cases whether we examined whole-cell recordings (unpublished, BAD & DB) or excised patches (n = 15) (Supplementary Fig. 3c). MD simulations suggested that the L783C mutant has a less pronounced effect than E524G on sodium stability, yet the ions managed to dissociate from their binding pockets within 100 ns in one of two simulations (Fig. 5e, f). One potential explanation for the sodium dissociation is that the L783C mutant permits access of additional water molecules into the cation binding pocket (Supplementary Movie 5), as observed in simulations of Y521C L783C. In comparison to the wildtype GluK2 receptor, the sodium ions in L783C interacted more frequently with water molecules, and less frequently with residues of the cation pocket (unpublished, MM & PCB). In both mutants, our data point to the lack of responsiveness of E524G and L783C arising from their disruptive effects on the cation binding pocket, a condition that may be similar to desensitization in a wildtype receptor. Because mutant receptors that disrupt L-Glu binding are retained within mammalian cells 27, we do not think that an inability to bind agonists can account for the phenotypes of E524G and L783C. As a result, an explanation is required to account for an additional cysteine (Y521C) restoring channel gating when introduced atop the L783C mutation. We conclude that the cation-independent activation of GluK2 Y521C L783C is due to its covalent crosslinking of the dimer interface, which circumvents normal gating requirements of the wildtype receptor (see also 26).
KAR desensitization proceeds after cation unbinding
MD simulations and single-channel data suggest that GluK2 D776K receptors are non-desensitizing because Lys 776 becomes tethered to the cation binding pocket. We therefore conclude that cation binding primes KARs for activation by the agonist. We also conclude that cation-unbound states are not primed for activation and thus, agonist-binding promotes entry into desensitized states as observed with the L783C and E524G mutant receptors. These different outcomes are important because they will determine the degree to which desensitization, and by implication cation unbinding, contributes to the wildtype KAR response. For example, during long agonist applications routinely used to measure desensitization rates, most receptors should desensitize because cations will eventually unbind with the agonist still bound. In contrast, with brief applications of L-Glu used to measure deactivation rates, fewer GluK2 receptors should desensitize because the agonist will unbind before the cation. Importantly, this sequence of events can be tested experimentally. Specifically, we predict that deactivation rates estimated with a brief agonist application should be minimally affected by the presence or absence of desensitization because decay from the peak response corresponds to agonist unbinding from the cation-bound state(s).
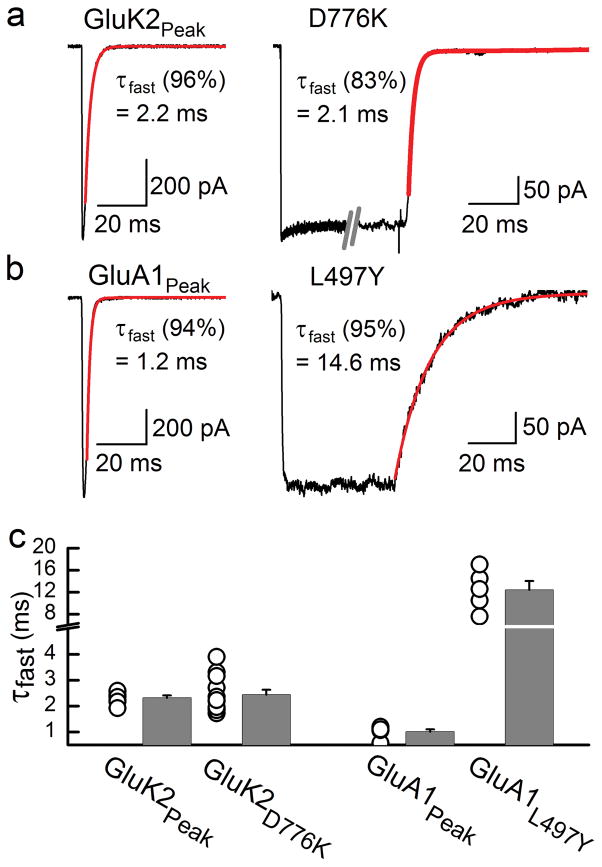
To examine the impact of desensitization on deactivation rates, we compared the relaxation kinetics observed following a brief application (i.e. 1 ms) of 10 mM L-Glu onto wildtype and non-desensitizing D776K KARs (Fig. 6a). For comparison, we also performed a similar analysis of wildtype and a mutant GluA1 AMPA receptor (i.e. L497Y) where single-channel desensitization is strongly inhibited 28 (Fig. 6b). Wildtype GluK2 receptors exhibited a fast exponential time constant of deactivation of 2.3 ± 0.1 ms (n = 7) (Fig. 6a) which was statistically indistinguishable from the off-kinetics of D776K receptors regardless of whether 1 ms (2.0 ± 0.2 ms, n=9) (p = 0.63) or 250 ms agonist pulses (2.4 ± 0.2 ms, n = 12) (p = 0.82) were applied (Fig. 6a, c). These observations support our assertion that KAR desensitization proceeds after cation unbinding. Accordingly, deactivation and desensitization can therefore be viewed as being structurally-distinct and separable processes. In contrast, the decay time constant observed following a 1 ms application of 10 mM L-Glu to GluA1 AMPARs had a fast exponential time constant of 1.0 ± 0.1 ms (n = 6) (Fig. 6b), which was about 10 times faster than the off-kinetics of the non-desensitizing L497Y mutant (12.4 ± 1.6 ms, n=5, Fig. 6b, c). This finding is consistent with the effect of the allosteric modulator, cyclothiazide, which also attenuates AMPAR desensitization 29.
Figure 6.
Desensitization and deactivation are uncoupled in GluK2 KARs. (a) Typical current decay observed following removal of 10 mM L-Glu from wildtype GluK2 (1 ms application, Patch # 00327p3) and GluK2 D776K (250 ms application, Patch # 11506p1) receptors. (b) Typical current decay observed following removal of 10 mM L-Glu from wildtype GluA1 (1 ms application, Patch # 00404p1, −55mV) and GluA1 L497Y (50 ms application, Patch # 99608p1, −55mV) receptors. For panels a–b, decay kinetics from saturating L-Glu were fit with a second-order exponential function (red) with representative values of the fast, dominant component displayed. (c) Distribution of off-kinetic rates show that the τfast values for the GluK2 peak response and D776K were statistically indistinguishable (P = 0.68), whereas the values for the GluA1 peak response and L497Y were statistically different (P < 0.001). Two-tailed Student’s t test performed (a = 0.05). Error bars, s.e.m. from five to twelve independent experiments.
To further test the impact of desensitization on the activation process, we compared the dose-response relationships of GluK2 D776K and wildtype receptors. We reasoned that because the absence of desensitization had little to no effect on GluK2 deactivation kinetics, rates of L-Glu unbinding should be high relative to rates of cation unbinding, which equate with desensitization. Under such circumstances, receptors would tend to enter desensitized states only during sustained L-Glu application. As such, the dose-response relationship of the peak response, occurring less than 1 ms after L-Glu exposure, should exhibit little change in the absence of desensitization.
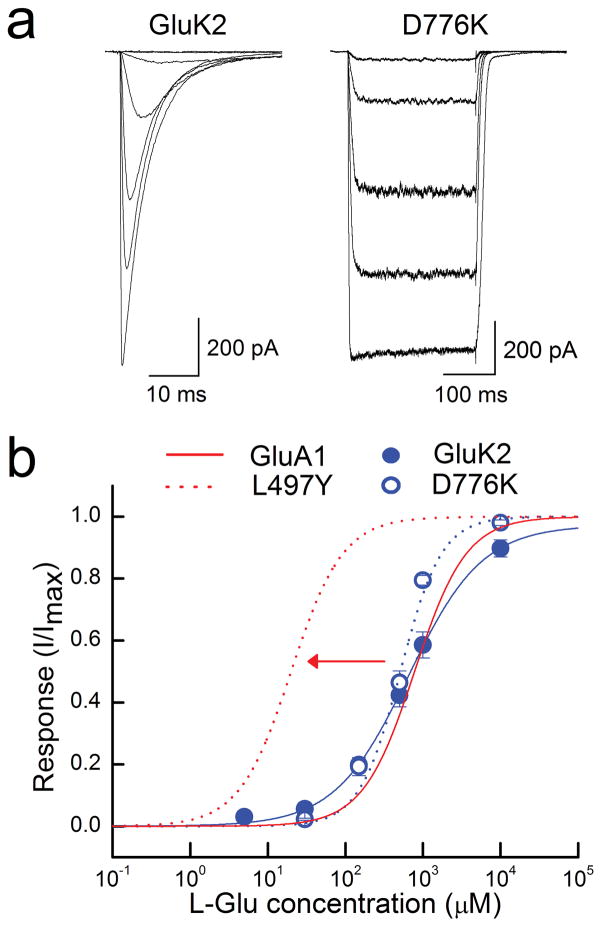
In agreement with our predictions, the EC50 (and nH) estimated from peak dose-response curves to L-Glu acting on wildtype GluK2 receptors were 652 ± 47 μM (nH = 0.87, n = 7), which closely matched that of D776K receptors, where the EC50 value was estimated to be 520 ± 91 μM (nH = 1.6, n = 8) (Fig 7a, b). These data differ from past work on AMPARs which has shown that mutations and allosteric modulators that reduce or eliminate desensitization cause progressive leftward shifts in the wildtype dose-response curve 28, 29. For example, one study noted a leftward shift of over an order of magnitude in the wildtype EC50 when studying GluA1 L497Y AMPARs 29 (Fig 7b). Our observations comparing wildtype and D776K receptors support the idea that desensitization has little impact on the time GluK2 receptors remain activated. This is, of course, to be expected if desensitization can only proceed after cation unbinding. Indeed, MD simulations reported here suggest that LBD dimer separation, a structural correlate of desensitization, is promoted for wildtype receptors in the absence of bound sodium ions (Supplementary Fig. 4). Our findings also suggest that desensitization impacts the time course of AMPAR activation which explains the effect of desensitization on both deactivation kinetics and agonist potency.
Figure 7.
Desensitization does not substantially shift peak agonist potency of GluK2 KARs. (a) Typical current responses elicited by L-Glu (10 μM – 10 mM) acting on wildtype GluK2 (Patch # 091204p2) and GluK2 D776K (Patch # 11610p1) receptors. (b) L-Glu dose-response relationships for KARs, normalized to the maximal current (Imax) of each patch, as well as simulated dose-response curves of wildtype and GluA1 L497Y receptors taken from previously reported values 29. Error bars, s.e.m. from seven and eight independent experiments.
DISCUSSION
The present study advances our understanding of iGluR gating in several substantial ways. First, we show that cation occupancy is the central requirement in keeping agonist-bound KARs in the activated state and out of desensitization. Second, we propose a structural model for the sequence of events that give rise to deactivation and desensitization. Deactivation is observed when the ligand unbinds from cation-bound states, whereas desensitization proceeds when the ligand is bound to cation-unbound states. Third and finally, closely-related AMPARs do not share this reliance on cation-dependent gating, as a result; desensitization appears able to curtail AMPAR channel activation. As discussed below, this unique property of KARs may provide clues to how subunit composition and/or auxiliary proteins affect native receptors at glutamatergic synapses.
The KAR dimer interface is a multi-faceted structure
It is remarkable that subunit crosslinking at two neighboring sites (residues 776 and 783) along the GluK2 LBD dimer interface produces very different functional consequences. The Y521C L783C mutation bridges opposing subunits, yet the crystal structure of its LBD suggests a separation of the upper D1 segment of the dimer interface 15. Although separation of the dimer interface is thought to underlie both KAR and AMPAR desensitization 13, it is not clear how much separation would be tolerable before channel activation could no longer be maintained. Given microscopic recordings showing that Y521C L783C channels cannot stably access the main open state of wildtype GluK2 26, we propose that this mutant is a mostly desensitized receptor typified by an open interface and/or a poorly activating receptor by virtue of its sporadic channel openings.
Targeted slightly higher along the LBD interface, the mutant residue Lys 776 occupies the GluK2 cation binding pocket and has two related consequences on receptor function; it increases open-channel probability to such an extent that no failures are observed and it sustains activation for the duration of agonist application. The latter effect supports the idea that the molecular events leading to desensitization are triggered at the apex of the interface, rather than being coordinated through the interface as a whole. Whether these interactions are further complicated by an emerging idea that KAR subunits desensitize with a tetrameric symmetry and not as a dimer of dimers 30, 31 awaits future study.
The cation binding pocket and its relation to gating events
Although structural rearrangements of the LBD accompany iGluR desensitization 13, it is presently unknown how such conformational changes are initiated. The matter is further complicated in KARs, where bound ions have been proposed to stabilize the LBD dimer interface 20. Here, we establish a framework to specify when KARs activate and desensitize by identifying the cation binding pocket as the molecular switch between these processes. In short, cation pocket occupancy maintains KAR activation, and by implication desensitization cannot occur until cations unbind. The link between cation binding and activation is based on several key observations reported above: the sustained single-channel activation observed in the GluK2 D776K mutation (Fig. 2), where the cation binding pocket is thought to be continuously occupied, the inability of GluK2 to activate in the absence of external ions (Fig. 4), and the gating deficiencies amongst mutants designed to disrupt cation binding (Fig. 5, Supplementary Fig. 3). Furthermore, the assertion that cation unbinding precedes desensitization can be deduced from other observations we reported. Specifically, we showed that deactivation kinetics of wildtype KARs were unaffected by desensitization confirming our assertion that the decay of the KAR peak response corresponds to agonist unbinding from the cation-bound state(s) (see Fig. 6a, c). This conclusion is consistent with previous work showing that GluK2 deactivation kinetics are made faster by lowering the external cation concentration or by replacing sodium with another cation 32. With long agonist applications (i.e. 250 ms), we propose that the decline in KAR activity is due cation unbinding since, besides the presence of the agonist, the only other known requirement of KARs to activate is allosteric ions 22. Given this, we concluded that their departure was the most plausible explanation to trigger the onset of desensitization. In accordance with this notion, MD simulations reported here (Supplementary Fig. 4) predict that removal of cations from the LBD dimer interface can induce structural changes associated with the desensitized state(s).
An alternative explanation for the observations above is that KAR desensitization is triggered by intrinsic rearrangements to the LBD structure, which are countered through the occupancy of bound cations. From this perspective, the relation between bound cations and decay kinetics is attributable to a direct modulation of the intrinsic rate of desensitization (by stabilizing LBD dimers) as has been suggested previously 21. This interpretation, however, is difficult to reconcile with several observations. To begin with, if desensitization is merely opposed, but not blocked by the presence of bound cations, some residual activation should be detected in solutions lacking external ions; which is not the case. Furthermore, from this perspective, the effect of cation species on deactivation kinetics would have to be explained by desensitization rates overlapping with those of deactivation. Experiments reported in this manuscript show that deactivation kinetics are unaffected by desensitization (i.e. comparing D776K to wildtype GluK2 receptors) (Fig. 6), which must therefore occur on a slower time scale. Thus, the two processes do not overlap, meaning activation must be directly regulated by cations.
Ion channels employ different strategies to desensitize
Desensitization of LGICs has been classically thought to arise from agonist molecules converting receptor complexes into non-reactive forms 33 in much the same way that even earlier work linked changes in membrane potential to voltage-gated ion-channel inactivation 34. Since then, structural explanations have emerged to account for how the processes of inactivation and desensitization occur at the amino acid level. Some of the first insights came from work on voltage-gated sodium and potassium channels, which were shown to possess intracellular inactivation gates 35, 36, whereas work on Cys-loop LGICs hinted at a broader re-arrangement of quaternary structure 37. Pioneering studies also identified coupling between activation and inactivation of voltage-gated channels 38, which has been more difficult to establish at LGICs. Such coupling might be expected to occur at iGluRs since closure in the agonist-binding domain initiated by ligand binding is thought to bring about both activation and then desensitization, as the agonist becomes entrapped in a stable, yet inactive conformation 12, 39. In keeping with this, data presented in this study suggest a tight coupling between these structural events in AMPARs. Interestingly, this is not the case for KARs, which uncouple the process of activation from desensitization through cation-dependent gating. This unique aspect of KAR gating provides an ideal target by which native receptor responses could be modulated at central synapses. For example, alterations in cation-affinity through protein-protein interactions could explain how heteromeric subunits 40 and/or auxiliary proteins 24 regulate the duration of synaptic KAR activity 41. Clearly, much still remains to be examined in future studies and how this allosteric cation binding pocket might be exploited to regulate KAR signaling within the vertebrate CNS.
ONLINE METHODS
Cell culture and transfection
HEK293T cells were transiently co-transfected with cDNA encoding wildtype or mutant GluK2(Q) KAR or GluA1(Q) AMPAR subunits and enhanced green fluorescent protein (eGFPS65T) as previously described32, or transfected with iGluR subunit cDNA on plasmids also encoding eGFP behind an internal ribosomal entry site. The cDNA for the mutant receptors was generated in two steps from wildtype plasmid using Quickchange II XL site-directed mutagenesis (Stratagene, LaJolla, CA). After transfection for 4 – 8 hrs using the calcium phosphate precipitation method, cells were washed twice with divalent-containing PBS and maintained in fresh medium (MEM containing Glutamax and 10% FBS). Electrophysiological recordings were performed 24 – 48 hrs later.
GluK2 receptor surface expression
To test for possible trafficking defects in mutants used in this study, we measured the fluorescence emitted by an ecliptic pHGFP genetically fused to the extracellular N-terminal of mutant or wildtype GluK2 receptors (Supplementary Fig. 3a, b). Unlike eGFP, the fluorescence emission of pHGFP is almost entirely quenched at pH 5.4542, which we used to evaluate the cellular location of the fluorophores43. Using this approach, a substantial but reversible attenuation in the fluorescence signal emitted by wildtype pHGFP-GluK2 was observed (n = 17 cells) following acidification of the external milieu (Supplementary Fig. 3a, b) demonstrating that most of the fluorescence signal was emitted by tagged GluK2 receptors on the plasma membrane. In contrast, acidification of the external solution had little effect on the weak fluorescence emitted by pHGFP-GluK2 R523A receptors (n = 6 cells) (Supplementary Fig. 3a, b), consistent with previous work showing that this mutant has poor surface expression27. Fluorescence emitted by pHGFP-GluK2 E524G and L783C receptors (n = 10 and 6 cells respectively) was robust, much like wildtype GluK2, and was reversibly attenuated by acidification (Supplementary Fig. 3a, b) suggesting that trafficking to the plasma membrane is not substantially perturbed for either mutant.
Electrophysiological solutions & recordings
External recording solutions typically contained (in mM): 150 NaCl, 5 HEPES, 0.1 CaCl2, 0.1 MgCl2, 2% phenol red. The internal recording solution contained (mM): 115 NaCl, 10 NaF, 5 HEPES, 5 Na4BAPTA, 0.5 CaCl2, 1 MgCl2, and 10 Na2ATP to chelate endogenous polyamines. The osmotic pressure was set to 295–300 mOsm using sucrose and the pH adjusted to 7.35 with 5 N NaOH. Agonist solutions were prepared by dissolving the agonist in external solution and adjusting the pH appropriately. In the case of recordings conducted in nominal external ions, the solution contained 100 μM of CaCl2 and MgCl2 to improve patch stability, sucrose to maintain the osmotic pressure at 295–300 mOsm, and 5 mM Tris to buffer pH. The pH was adjusted to 7.3–7.4 using 10 N HCl. To optimize recording stability in solutions of nominal ions, quartz electrodes were used to excise some outside-out patches. The outward current conveyed by receptors in such conditions was due to the efflux of sodium ions from the patch pipette. The lack of inward current in response to L-Glu confirmed that all cations were removed from the external milieu of the membrane patch.
All experiments were performed on excised membrane patches in the outside-out configuration. We used thin-walled borosilicate glass pipettes (3–5 MΩ, King Precision Glass, Inc.) coated with dental wax for macroscopic experiments. To obtain low noise or single-channel recordings, we used quartz glass (3–15 MΩ, King Precision Glass, Inc.) coated with Sylgard (Dow Corning). Agonist solutions were rapidly applied to outside-out patches for 250 ms at −60 mV (unless otherwise stated) using a piezo-stack driven perfusion system. Sufficient time between applications of L-Glu was allowed for complete recovery from macroscopic desensitization. Solution exchange time was determined routinely at the end of each experiment by measuring the liquid junction current (10–90 % rise-time = 100–400 μs). Series resistances (3–15 MΩ) were routinely compensated by 95%. For microscopic recordings, the headstage was set to the capacitive feedback recording mode. All recordings were performed at room temperature using an Axopatch 200B amplifier (Axon Instruments Inc., Foster City, CA, USA). Current records were filtered at 5 kHz for macroscopic responses and digitized at 25–50 kHz. Single-channel currents were all acquired at 50–100 kHz, low-pass filtered by an 8-pole Bessel filter at 10 kHz and digitally filtered offline at 1–3 kHz. The reference electrode was connected to the bath via an agar bridge of 3M KCl. Data were acquired using pClamp9 software (Axon Instruments Inc., Foster City, CA, USA), and illustrated using Origin 7 (OriginLab Corp., Northampton, MA, USA).
Macroscopic Response Analysis
Data were analyzed using Clampfit 9.0 and tabulated using Microsoft Excel. Curve fittings for determining the off-kinetic rates were performed using 1st or 2nd order exponential functions: y = Ai *exp(−x/ti). Dose-response data to L-Glu were normalized, pooled across patches, and fit with the logistic equation of the following form: I = Imax/(1+(EC50/[Glu])nH), where I is the normalized current at any agonist concentration, Imax is the interpolated maximal response, EC50 is the concentration of L-Glu that elicits the half-maximal response, and nH is the slope or Hill coefficient.
Single-Channel Analysis
For wildtype GluK2 receptors, analysis was conducted on patches (n = 5) from which fifty or more agonist applications were made at 15 s intervals. For GluK2 D776K, which displayed uniform current responses, analysis was limited to 58 agonist applications, which were divided among four patches. Single-channel data were subjected to digital low-pass filtering at 3 kHz (or 1kHz for presentation in figures), which resulted in root mean square baseline noise values that averaged 0.22 ± 0.024 pA (n = 5) and 0.22 ± 0.043 pA (n = 4) for wildtype and D776K receptors, respectively. These noise values corresponded to less than fifty percent of the smallest difference between adjacent conductance levels in the wildtype receptor. The 3kHz frequency was chosen on account of our data containing many rapid transitions between conductance levels, as described previously for AMPARs44. Accordingly, a resolution of two filter rise times (2 × 111 μs) was imposed to detect and account for brief events, while maintaining resolution of small conductances. Digitally-filtered data were exported to Signal 5.0 (Cambridge Electronic Design) to perform time-course fitting analysis with the program SCAN45. The idealized records were then used to provide information on response amplitudes, which could be fit with Gaussian functions, whose peaks reflect discrete conductance levels: y = Σ i=1…n (Ai/wi*sqrt(π/2)))*exp(−2*((x−xci)/wi)2 where A = area, xc = center of the peak, w = error associated with xc. From this analysis, the distribution and amplitude of single-channel events observed in patches containing a few channels (Fig. 2f) were similar to events measured at equilibrium in multichannel patches (Supplementary Fig. 5).
Molecular dynamics simulations
All crystal structures used in this manuscript were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) protein data bank. Two protein structures were used for building models for the MD simulations; an L-Glu-bound GluK2 LBD dimer (pdb-code: 3G3F (resolution 1.38 Å (ref. 46)) and an L-Glu-bound GluK2 Y521C L783C LBD dimer (pdb-code: 2I0C (resolution 2.25 Å (ref. 15)), respectively, which was used only for simulations concerning the double-cysteine mutant. Together with the crystallographically resolved water molecules, L-Glu ligands and ions were retained in the simulation setup, whereas two bound isopropyl alcohol molecules were deleted. In simulations of GluK2 without bound sodium ions (Supplementary Fig. 4), these were removed before system setup. The protein was solvated in water in a (90 Å) (ref. 43) box using the TIP3P water model47, whereafter the system was neutralized and 150 mM NaCl was added. Mutations, except for Y521C L783C, were imposed manually prior to simulation setup, either by editing/deleting atoms in the pdb-file or by using the mutate function of PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC) and adjusting the side chain rotamer. For the double cysteine mutant, the GluK2 double-cysteine (Y521C L783C) mutant structure was employed. This structure had no ions bound, so the interface-bound ions from the wildtype structure were added and rotamers for side chains surrounding the ion sites were optimised in PyMOL before solvation, neutralization and ionization as described above.
The MD simulations were performed in Gromacs 4.5 (ref. 48) with the OPLS all-atom force field49,50. The systems were first energy minimized until the maximum force on an atom was less than 100 kJ/mol/nm. Following energy minimization, a 200 ns restrained simulation with position restraints on protein heavy-atoms and on bound ions with a force constant of 1000 kJ mol−1 nm−2 was performed in the NVT ensemble with a temperature of 300 K maintained by a Berendsen thermostat51. Periodic boundary conditions were utilized and van der Waals interactions were cut off at 10 Å. Long-range electrostatics were accounted for by the Particle-Mesh Ewald method52. All bonds were treated as constraints using the LINCS algorithm, allowing a time step of 2 fs. Subsequently, 100 ns of production run were performed (only 30–50 ns for E524G). The NPT ensemble was employed with the temperature retained at 300 K and the pressure at 1 bar by using the Berendsen thermostat and barostat, respectively51. Two repeats for each mutational variant were produced. Analyses were performed using VMD53 and analysis tools of Gromacs48.
Statistical methods
Results are expressed as mean ± s.e.m. Statistical analyses of sample means were performed using two-tailed Student’s t tests. P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR) FRN-82804 (D.B.) and by the Leverhulme Trust RPG-059 (P.C.B.). G.B.D. and E.D.A. were supported by Natural Sciences and Engineering Research Council of Canada (NSERC) graduate fellowships, B.A.D. by a Chemical Biology CIHR postdoctoral award and M.R.P.A. by a CIHR Best & Banting doctoral award. D.B. is the recipient of a Canada Research Chair award. We thank the Oxford Supercomputing Centre for computer time. Finally, we wish to thank Dr. Jon Johnson for comments on the manuscript and Dr. Mark Fleck for discussions on AMPA receptors.
Footnotes
Author Contributions: G.B.D. designed and performed experiments, analyzed data, and wrote the paper; M.M., B.A.D. and M.R.P.A. designed and performed experiments, and analyzed data; E.D.A. analyzed data; P.C.B. designed experiments; and D.B. designed and performed experiments, analyzed data and wrote the paper.
Reference List
- 1.KATZ B, THESLEFF S. A study of the desensitization produced by acetylcholine at the motor end-plate. J Physiol. 1957;138:63–80. doi: 10.1113/jphysiol.1957.sp005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shelley C, Cull-Candy SG. Desensitization and models of receptor-channel activation. J Physiol. 2010;588:1395–1397. doi: 10.1113/jphysiol.2010.188664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones MV, Westbrook GL. The impact of receptor desensitization on fast synaptic transmission. Trends Neurosci. 1996;19:96–101. doi: 10.1016/s0166-2236(96)80037-3. [DOI] [PubMed] [Google Scholar]
- 4.Hille B. Ion channels of excitable membranes. Sinauer Associates Inc; Massachusetts, USA: 2001. Ligand-gated channels of fast chemical synapses; pp. 169–200. [Google Scholar]
- 5.Patneau DK, Mayer ML, Jane DE, Watkins JC. Activation and desensitization of AMPA/kainate receptors by novel derivatives of willardiine. J Neurosci. 1992;12:595–606. doi: 10.1523/JNEUROSCI.12-02-00595.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corringer PJ, et al. Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure. 2012;20:941–956. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Flynn GE, Johnson JP, Jr, Zagotta WN. Cyclic nucleotide-gated channels: shedding light on the opening of a channel pore. Nat Rev Neurosci. 2001;2:643–651. doi: 10.1038/35090015. [DOI] [PubMed] [Google Scholar]
- 9.Hansen KB, Yuan H, Traynelis SF. Structural aspects of AMPA receptor activation, desensitization and deactivation. Curr Opin Neurobiol. 2007;17:281–288. doi: 10.1016/j.conb.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Wollmuth LP, Sobolevsky AI. Structure and gating of the glutamate receptor ion channel. Trends Neurosci. 2004;27:321–328. doi: 10.1016/j.tins.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Madden DR. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci. 2002;3:91–101. doi: 10.1038/nrn725. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 15.Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- 16.Priel A, Selak S, Lerma J, Stern-Bach Y. Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron. 2006;52:1037–1046. doi: 10.1016/j.neuron.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Gielen M, et al. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borschel WF, Murthy SE, Kasperek EM, Popescu GK. NMDA receptor activation requires remodelling of intersubunit contacts within ligand-binding heterodimers. Nat Commun. 2011;2:498. doi: 10.1038/ncomms1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayeem N, Zhang Y, Schweppe DK, Madden DR, Green T. A nondesensitizing kainate receptor point mutant. Mol Pharmacol. 2009;76:534–542. doi: 10.1124/mol.109.056598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 21.Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong AY, Fay AM, Bowie D. External ions are coactivators of kainate receptors. J Neurosci. 2006;26:5750–5755. doi: 10.1523/JNEUROSCI.0301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowie D. Ion-dependent gating of kainate receptors. J Physiol. 2010;588:67–81. doi: 10.1113/jphysiol.2009.178863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, et al. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61:385–396. doi: 10.1016/j.neuron.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayeem N, Mayans O, Green T. Conformational flexibility of the ligand-binding domain dimer in kainate receptor gating and desensitization. J Neurosci. 2011;31:2916–2924. doi: 10.1523/JNEUROSCI.4771-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniels BA, Andrews ED, Aurousseau MR, Accardi MV, Bowie D. Crosslinking the ligand-binding domain dimer interface locks kainate receptors out of the main open state. J Physiol. 2013 doi: 10.1113/jphysiol.2013.253666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mah SJ, Cornell E, Mitchell NA, Fleck MW. Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci. 2005;25:2215–2225. doi: 10.1523/JNEUROSCI.4573-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern-Bach Y, Russo S, Neuman M, Rosenmund C. A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron. 1998;21:907–918. doi: 10.1016/s0896-6273(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell NA, Fleck MW. Targeting AMPA receptor gating processes with allosteric modulators and mutations. Biophys J. 2007;92:2392–2402. doi: 10.1529/biophysj.106.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowie D, Lange GD. Functional stoichiometry of glutamate receptor desensitization. J Neurosci. 2002;22:3392–3403. doi: 10.1523/JNEUROSCI.22-09-03392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schauder DM, et al. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proc Natl Acad Sci U S A. 2013;110:5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowie D. External anions and cations distinguish between AMPA and kainate receptor gating mechanisms. J Physiol. 2002;539:725–733. doi: 10.1113/jphysiol.2001.013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DEL CJ, KATZ B. Interaction at end-plate receptors between different choline derivatives. Proc R Soc Lond B Biol Sci. 1957;146:369–381. doi: 10.1098/rspb.1957.0018. [DOI] [PubMed] [Google Scholar]
- 34.HODGKIN AL, HUXLEY AF. The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J Physiol. 1952;116:497–506. doi: 10.1113/jphysiol.1952.sp004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 36.Stuhmer W, et al. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 37.Unwin N, Toyoshima C, Kubalek E. Arrangement of the acetylcholine receptor subunits in the resting and desensitized states, determined by cryoelectron microscopy of crystallized Torpedo postsynaptic membranes. J Cell Biol. 1988;107:1123–1138. doi: 10.1083/jcb.107.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armstrong CM, Bezanilla F. Inactivation of the sodium channel. II. Gating current experiments. J Gen Physiol. 1977;70:567–590. doi: 10.1085/jgp.70.5.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mano I, Lamed Y, Teichberg VI. A venus flytrap mechanism for activation and desensitization of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors. J Biol Chem. 1996;271:15299–15302. doi: 10.1074/jbc.271.26.15299. [DOI] [PubMed] [Google Scholar]
- 40.Barberis A, Sachidhanandam S, Mulle C. GluR6/KA2 kainate receptors mediate slow-deactivating currents. J Neurosci. 2008;28:6402–6406. doi: 10.1523/JNEUROSCI.1204-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copits BA, Swanson GT. Dancing partners at the synapse: auxiliary subunits that shape kainate receptor function. Nat Rev Neurosci. 2012;13:675–686. doi: 10.1038/nrn3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 43.Khiroug SS, et al. Dynamic visualization of membrane-inserted fraction of pHluorin-tagged channels using repetitive acidification technique. BMC Neurosci. 2009;10:141. doi: 10.1186/1471-2202-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Cho Y, Lolis E, Howe JR. Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J Neurosci. 2008;28:932–943. doi: 10.1523/JNEUROSCI.3309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colquhoun D, Sigworth FJ. Fitting and Statistical Analysis of Single Channel Records. In: Sakmann B, Neher E, editors. Single Channel Recording. Plenum Press; New York: 1995. pp. 483–587. [Google Scholar]
- 46.Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer ML. Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization. EMBO J. 2009;28:1518–1530. doi: 10.1038/emboj.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 48.Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society. 1996;118:11225–11236. [Google Scholar]
- 50.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. Journal of Physical Chemistry B. 2001;105:6474–6487. [Google Scholar]
- 51.Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics. 1984;81:3684–3690. [Google Scholar]
- 52.Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 53.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.