Abstract
The outer membrane (OM) of Gram-negative bacteria is a unique asymmetric lipid bilayer in which the outer leaflet is composed of lipopolysaccharide (LPS) and the inner leaflet is formed by glycerophospholipid (GPL). The OM plays a fundamental role in protecting Gram-negative bacteria from harsh environments and toxic compounds. The transport and assembly pathways for phospholipids of bacterial OM are unknown. Cardiolipin (CL) plays an important role in OM biogenesis and pathogenesis, and the inner membrane (IM) protein PbgA, containing five transmembrane domains and a globular domain in periplasm has been recently identified as a CL transporter from the IM to the OM with an unknown mechanism. Here we present the first two crystal structures of soluble periplasmic globular domain of PbgA from S. typhimurium and E. coli, which revealed that the globular domains of PbgA resemble the structures of the arylsulfatase protein family and contains a novel core hydrophobic pocket that may be responsible for binding and transporting CLs. Our structural and functional studies shed an important light on the mechanism of CL transport in Gram-negative bacteria from the IM to the OM, which offers great potential for the development of novel antibiotics against multi-drug resistant bacterial infections.
Gram-negative bacteria contain two membranes, the inner membrane (IM) and the outer membrane (OM) with the solvent filled periplasm and the peptidoglycan1,2. The peptidoglycan, which is much thinner than that found in Gram-positive bacteria, is bound to the OM by lipoproteins, but it remains strong and elastic to protect the bacteria against harsh environmental conditions2,3. The OM is an asymmetric lipid bilayer in which the inner leaflet is composed of glycerophospholipids (GPL) while the outer leaflet consists primarily of the glycolipid lipopolysaccharide (LPS), which functions as a barrier protecting bacteria from antibiotics and other harmful hydrophobic chemicals1,2,4,5. The OM also contains OM proteins and lipoproteins. The pathways of transport and assembly of LPS, lipoprotein and OMP from the IM to the OM have been identified and studied extensively; seven proteins LptA-G form a trans-envelope complex and transport LPS from the IM to the OM1,2,6,7,8,9,10,11,12,13. Five proteins LolA-E are responsible for sorting and transporting lipoproteins from the IM to the OM14,15,16,17, while five proteins BamA-E form a OMP insertion machine, which inserts unfolded OMP into the OM18,19,20,21,22. The proteins involved in phospholipid transport and assembly in the OM, however remain unknown.
In contrast, the IM is a symmetrical bilayer and is composed of phospholipids that maintain the barriers’ permeability for hydrophilic molecules and support the functions of the IM proteins3. The GPL molecules include phosphatidylethanolamine (PE), phosphatidylglycerols (PG) and cardiolipin (CL) in an approximately 75:20:5 ratio, respectively, in most enterobacterial membranes. CL is also known as diphosphatidylglycerol because it appears to contain two overlapping PG units (Supplementary Fig. S1). CL is also located in the OM of most Gram-negative bacteria, where it is essential for bacterial pathogenesis and survival in different environments under the regulatory control of the PhoPQ two-component system23,24. However, CL is synthesized at the IM, and so must be transported by lipid transfer proteins from the IM to the OM25,26.
Interestingly, CL is also presented in the IM of mitochondria, where it consists of about 20% of the total lipids, and is required for the mitochondrial function27,28. In yeast, the UPS1 and Mdm35 complex has been identified to transport phosphatidic acid (PA) from the mitochondria OM to the IM, and the structure of the UPS1 and Mdm35 complex has been recently solved29,30. In Gram-negative bacteria the Mla pathway, containing six proteins MlaA-F, has been suggested to be responsible for phospholipids transport from the OM to the IM31. In addition, the trigalactosyldiacylglycerol (TGD) pathway has been reported to deliver phosphatidic acid from the OM to the IM of chloroplasts32. However, there is no report about how the phospholipids are transported and assembled from the IM to the OM.
During infection, Salmonella typhimurium and Escherichia coli PhoPQ two-component regulatory system coordinates OM remodeling by activating genes encoding proteins and enzymes that increase OM-lipid hydrophobicity and decrease OM-lipid negative charge on the bacterial surface; this prevents binding and insertion of cationic antimicrobial peptides (CAMP) that kill the microbe23. S. typhimurium PbgA is an inner membrane protein containing five transmembrane helices and a globular periplasmic domain, which binds CL to promote PhoPQ-regulated trafficking of CL from the IM to the OM24.
Here we present the crystal structures of the globular domain of PbgA from S. typhimurium and E. coli, as well as functional assays. The structures and functional assays revealed the molecular basis of how the globular domain transports CL from the IM to the OM. To our knowledge, this is the first protein structure reported involved in phospholipid transport from the IM to the OM in bacteria.
Results
Two structures of PbgA
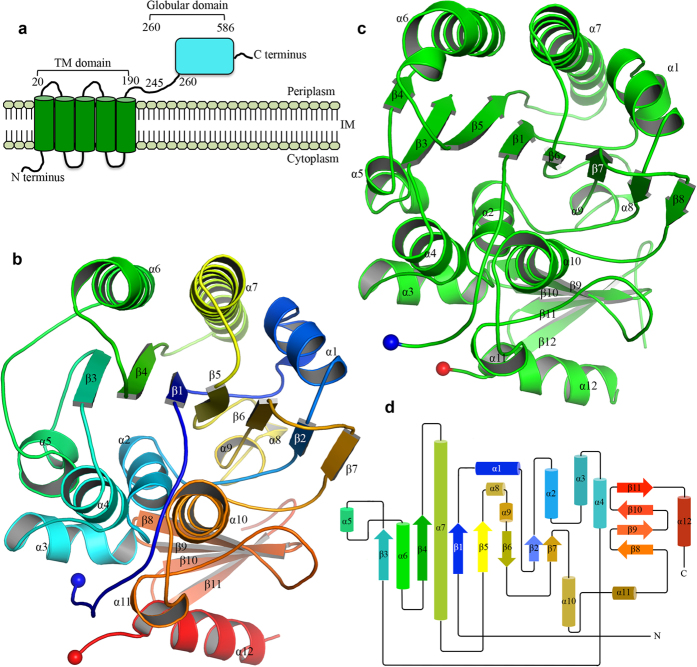
The PbgA (the original name YejM) protein was predicted to be an inner membrane protein containing five transmembrane domains (residues 1–190) and a C-terminal globular domain (residues 191–586) (Fig. 1a). To crystallize the PbgA proteins, the full length PbgA proteins and two N-terminal transmembrane domains truncation constructs (residues 191–586, and 245–586) named PbgA191-586 and PbgA245-586, from S. typhimurium Strain LT2 and E. coli K12 were cloned into pHISTEV plasmids. All the proteins were purified, and the PbgA245-586 from S. typhimurium Str. LT2 (StPbgA245-586) and E. coli (EcPbgA245-586) were crystallized, while the purified PbgA191-586 showed degradation and crystallized with in situ limited proteolysis using elastase (Supplementary Fig. S2a,b). The crystal of StPbgA254-586 belongs to space group P21 with cell dimensions a = 51.60 Å, b = 196.09 Å, c = 70.27 Å, α = γ = 90° and β = 95.70° (Table 1). The structure of StPbgA245-586 was determined to 1.64 Å resolution by single-wavelength anomalous dispersion (SAD) using selenomethionine (SeMet) incorporated protein. 28 selenium positions were identified in an asymmetric unit using SHELXD33. The electron density map was modified by SHELXE34,35. The initial model was built by Buccaneer36, and the full model was built manually using Coot37. Structure refinements were performed using REFMAC5 with the final model with the Rfactor of 0.19 and Rfree of 0.22 (Table 1).
Figure 1. Two crystal structures of cardiolipin transport protein PbgA.
The structures of PbgA resemble those of arylsulfatases and lipoteichoic acid synthases. (a) Scheme of PbgA of S. typhimurium strain LT2. (b) Cartoon representation of crystal structure of StPbgA245-586. The PbgA structure is colored in rainbow. (c) Cartoon representation of crystal structure of ECPbgA245-586 in green. (d) Scheme representation of structure of StPbgA245-586.
Table 1. Data collection and structure refinement statistics.
| Data collection | EcPbgA245 | StPbgA191 | StPbgA245 (SAD) |
|---|---|---|---|
| Resolution (Å) | 47.83–1.67 (1.71 −1.67) | 65.22–2.19 (2.25–2.19) | 65.85–1.64 (1.68 −1.64) |
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 |
| Space Group | P3121 | P21 | P21 |
| Completeness (%) | 99.6 (99.2)* | 99.4 (99.4) | 95.1 (89.2) |
| I/σ | 17.8 (1.1) | 5.8 (1.8) | 15.5 (1.1) |
| Multiplicity | 9.6 (7.6–9.9) | 6.1 (6.4–4.8) | 11.8 (13.1–6.1) |
| CC1/2 (%) | 100 (59.5) | 98.7 (54.5) | 99.9 (49.5) |
| Unique reflections | 44003 (4034) | 51747 (5044) | 66831 (6540) |
| Rmerge (%) | 14.3 (26.40) | 16.50 (27.80) | 15.2 (25.03) |
| Unit Cell (Å) | a = b = 57.42, c = 191.31 | a = 51.48 b = 195.65, c = 70.92 | a = 51.60 b = 196.09, c = 70.27 |
| α = β = 90°, γ = 120° | α = γ = 90°, β = 96.39° | α = γ = 90°, β = 95.79° | |
| Refinement | |||
| &Rfactor | 0.18 (0.21) | 0.20 (0.23) | 0.19 (0.20) |
| Rfree | 0.21 (0.25) | 0.24 (0.27) | 0.22 (0.24) |
| Number of atoms | |||
| Protein | 2604 | 10724 | 10745 |
| Waters | 498 | 961 | 1243 |
| R.M.S. deviation | |||
| Bond lengths (Å) | 0.014 | 0.02 | 0.017 |
| Bond angles (°) | 1.90 | 1.93 | 1.80 |
| Ramachandran statistics | |||
| Favoured (%) | 97 | 93 | 95 |
| Allowed (%) | 3 | 7 | 5 |
| PDB code | 5I5H | 5I5F | 5I5D |
*Values in parentheses are represents for the highest-resolution shell.
&Rfactor = Σ || Fobs|−| Fcal||/Σ |Fobs|, where Fobs and Fcal are observed all reflection measured and calculated currently model as structure factors, respectively. Rfree is calculated using 5% of total reflections, which is randomly selected not used in refinement.
There are four protomers of the StPbgA245-586 in the asymmetric unit. The structure of protomer A in the asymmetric unit is very similar to the other three protomers, B, C and D, with root-mean-square deviations (RMSD) of 0.285, 0.472 and 0.451 Å over 338 Cα atoms, respectively. The model of the StPbgA245-586 contains residues Q245-I563 and P568-A585, while the electron densities of the loop between I563-P568 are not visible, indicating that this loop is disordered (Fig. 1b,d).
The structure of StPbgA245-586 has the α/β fold, which looks like a burger, where helices α1, α6 and α7 are at the top, β-strands 1, 2, 3, 4, 5, 6, 7, 8, α8 and α9 form the second layer, α-helices 2, 4, 5 and 10 form the third layer, and β-strands 8, 9, 10, 11, α11 and α12 form the bottom layer (Fig. 1b).
The crystals of StPbgA191-586 belong to the space group P21 with cell dimensions a = 41.49 Å, b = 196.39 Å, c = 45.66 Å, α = γ = 90° and β = 107.53°. The StPbgA191-586 structure was determined by molecular replacement to 2.19 Å resolution; residues 191–245 has no visible electron density so were either disordered or cleaved. There are four protomers in the asymmetric unit, where the protomer A is superimposed well with protomers B, C and D with RMSD of 0.2642 Å, 0.4338 Å and 0.5627 Å over 340 Cα atoms, respectively. The StPbgA191-586 structure is very similar to StPbgA245-586 structure with RMSD of 0.5945 over 340 Cα atoms (Supplementary Fig. S3a,b).
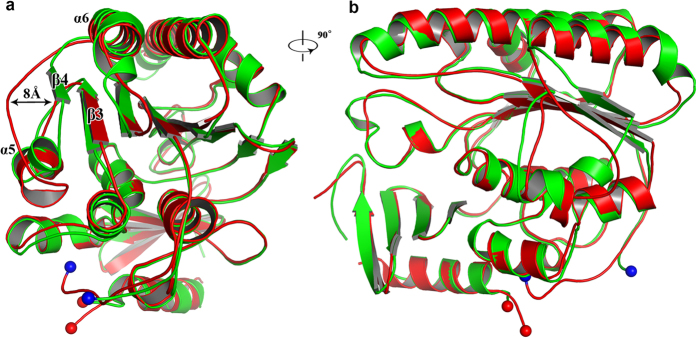
The crystals of EcPbgA245-586 belong to the space group of P3121 with cell dimensions a = b = 56.86 Å, c = 191.81 Å, α = β = 90° and γ = 120° (Table 1). The structure was determined by molecular replacement using the StPbgA245-586 structure as the search model to 1.67 Å resolution (Fig. 1c). There is one monomer in the asymmetric unit. The EcPbgA-245-586 structure resembles StPbgA254-586 structure with a RMSD of 0.9264 Å over 340 Cα atoms. In particular, the β4 of EcPbgA-245-586 moves away approximately 8 Å (Fig. 2a,b).
Figure 2. Comparison of structures of StPbgA and EcPbgA.
(a) StPbgA and EcPbgA are superimposed well. StPbgA colored in red, and EcPbgA colored in green. (b) Rotates 90° along y-axis relative to the left panel. Cα atoms of Loop S371-S360 moves away maximum of approximate 8 Å from of StPbgA to EcPbgA, strongly suggesting that this loop may involve in CL binding.
PbgA resembles arylsulfatase family proteins and lipoteichoic acid synthase
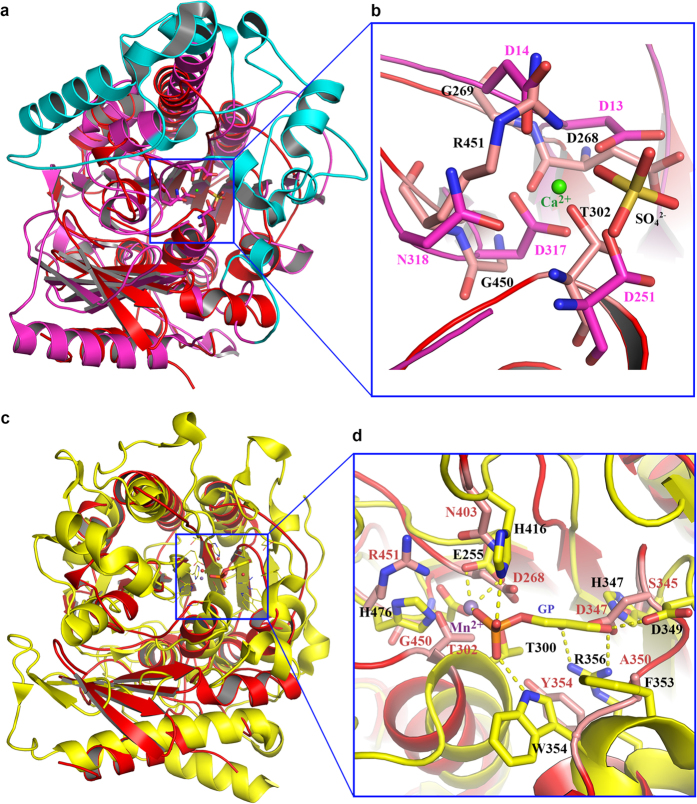
To find similar structures to PbgA, we performed searches in the Protein Data Bank using the Dali server using the StPbgA245-586 structure as the template. The PbgA structure is very similar to sulfatase family enzymes, where arylsulfatase (PDB code 1HDH)38 has the Dali-score of 29.9 with root-mean-square deviation (RMSD) of 2.9 Å over 296 Cα atoms to the StPbgA245-586 structure (Fig. 3a). The sulfatases hydrolyse a variety of sulphate esters utilising an active site containing Cα-formylglycine, a conserved and oxidized cysteine or serine residue, and metal binding residues D317, N318, D13 and D14 (PDB code 1HDH) (Fig. 3b). Although the overall structure is similar by superimposition of the two structures, StPbgA245-586 does not have the cysteine or serine at the position of the Cα-formyglycine of the sulfatase. We also did not observe any cysteine or serine residue of StPbgA245-586 around the position of the sulfatase’s active site. Instead, the StPbgA245-586 structure has residues T302, G450, R451, G269 and D268 at the corresponding positions in the sulfatase active site, respectively (Fig. 3a,b); this is consistent with the report that the PbgA does not have the sulfatase activity24. The major differences between the two structures are that the sulfatase has additional α-helices38 and loops that form a “gate” to close the active site (Fig. 3a).
Figure 3. PbgA structure is similar to arylsulfatase and lipoteichoic acid synthase, but do not contain the enzyme catalytic residues.
(a) StPbgA (red) is superimposed to arylsulfatase (matanga). The arylsulfatase has additional α-helices and loops (cyan), forming the substrate binding “gate”. (b) Superimposition of StPbgA and arylsulfatase at the active site. (c) StPbgA (red) is superimposed to lipoteichoic acid synthase (yellow). (d) Superimposition of StPbgA and lipoteichoic acid synthase at the active site.
The structure of lipoteichoic acid synthase of Staphylococcus aureus (LtaS, PDB code 2W5T)39, is also very similar to the StPbgA245-586 structure, with the Dali score of 28.4 and a RMSD of 3.0 Å over 284 Cα atoms. Despite only 17% sequence identity to StPbgA, LtaS is also a membrane protein with five transmembrane domains and a globular soluble domain with a structure strikingly similar to the StPbgA245-586 structure (Fig. 3c). The LtaS structure has revealed the active site pocket for binding a Mn2+ ion and Beta-Glycerol pho-sphate (sn-glycerol-2-phosphate), where residues T300, H476 and E255 bind Mn2+ ion, and H416, W354, F353, D349, H347 and R356 are involved in the Beta-Glycerol pho-sphate (sn-glycerol-2-phosphate) binding (Fig. 3c,d). As described above, the corresponding residues T302, G450, R451 and D268 of StPbgA are unlikely to form the metal binding site (Mn2+). The residues H347, D349, R356 and W354 of LtaS involving in Beta-Glycerol pho-sphate (sn-glycerol-2-phosphate) binding were confirmed to be important for the activity of LtaS39. The arrangement of the corresponding residues N403, S345, D347, A350 and Y354 of StPbgA does not correspond to the Beta-Glycerol pho-sphate (sn-glycerol-2-phosphate) binding site (Fig. 3d).
StPbgA245-586 does not bind metal ions and Beta-Glycerol pho-sphate (sn-glycerol-2-phosphate)
To confirm whether StPbgA245-586 binds heavy atoms and Beta-Glycerol pho-sphate (sn-glycerol-2-phosphate), we have performed co-crystallization the StPbgA245-586 in presence of the 20mM Mn2+ and 20 mg/ml sn-glycerol-2-phosphate. However, no extra electron density is visible in the solved structure, suggesting that PbgA may not bind metal or sn-glycerol-2-phosphate.
Potential CL binding site
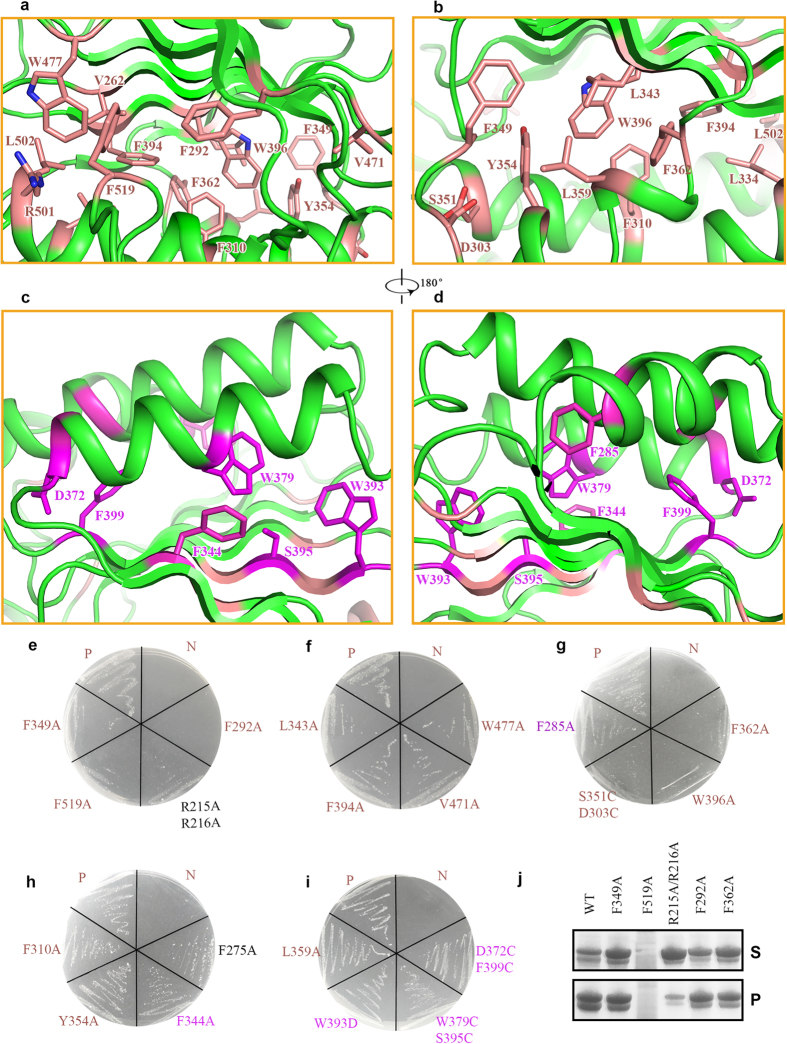
From the structures of the sulfatase and LtaS, their active sites are between the second layer and the third layer. In PbgA, the second layer consists of the β-sheet formed by strands 1–7 and the third layer is composed of of α-helices 2, 4, 5 and 10. This region is a highly hydrophobic core, which is formed by the hydrophobic residues L343, W396, F394, V262, W477, L473, V471, and F292 from the β-sheet of the second layer, and residues F349, Y354, L334, F310, L309, and F519 from the α-helices and loops of the third layer (Fig. 4a,b). PbgA transports CL from the IM to the OM24. We speculate that the hydrophobic core between the second and third layers might be the CL binding site. To explore this possibility, single alanine substitutions of residues L359, F292, F349, F519, L334, L343, F394, W396, V471 and W477, as well as a double alanine substitution R215/R216, were generated, and the PbgA global domain (191–586) inactivated strain NRD183 was used for the functional assay40.
Figure 4. PbgA potential CL binding site.
(a) the CL potential binding site between layers 2 and 3. The hydrophobic residues are colored in deep salmon. (b) Rotation 180° along y axis relative to the left panel. (c) Hydrophobic residues between layer 1 and 2 are colored in purple, which may do not involve in CL binding. (d) Rotation 180° along y axis relative to the left panel. (e–i) The functional assays of PbgA mutants and the drug resistance against 50 ug/ml vancomycin. (j) The CL binding assays were performed by co-sedimentation. All functional assays performed at least three times.
The deletion of PbgA periplasmic globular domain causes the outer membrane defect, which increases the OM permeability. The NRD183 strain was unable to grow in 50 μg/ml vancomycin, and the full length PbgA can rescue the NRD183 strain. The F292A mutant is lethal, and F349A, R215A/R216A, W396A, F362A and F275A severely impaired bacterial growth in 50 μg/ml vancomycin (Fig. 4e,g,h), while mutants L343A, F394A, V471A, W477A have minor effect on cell growth (Fig. 4f), and F310A, Y354A, L359A has no effect on cell growth (Fig. 4i), suggesting that residues F292, F349, R215/R216, W396, F362 and F275 are important for CL transport. The functional assay result of R215A/R216A mutant is consistent with the previous report that the residues R215/R21624 are involved in CL binding (Fig. 4e).
Residues F344, W379, W393 and F399 are located in the hydrophobic core between the α-helices 6 and 7 of the first layer and the β-strands 1 and 4 of the second layer of EcPbgA (Fig. 4c,d). In contrast, single mutant F344A, W393D, and double mutants F399C/D372C and W397C/S395C did not have any impact on cell growth, strongly suggesting that these residues are not involved in the CL binding and transfer (Fig. 4h,i).
CL binding residues were further confirmed using CL co-sedimentation assays. Mutant proteins (F349A, R215A/R216A, F292A and F362A) of EcPbgA 191–586 were over-expressed and purified. Mutants F349A, R215A/R216A, F292A, F362A have reduced CL co-sedimentation compared to the wild type (Fig. 4J); this is consistent with our functional assays, supporting that CL binding by these mutants is impaired. The F519A mutant forms an inclusion body so could not be assessed by CL co-sedimentation.
Potential CL-binding switch lid
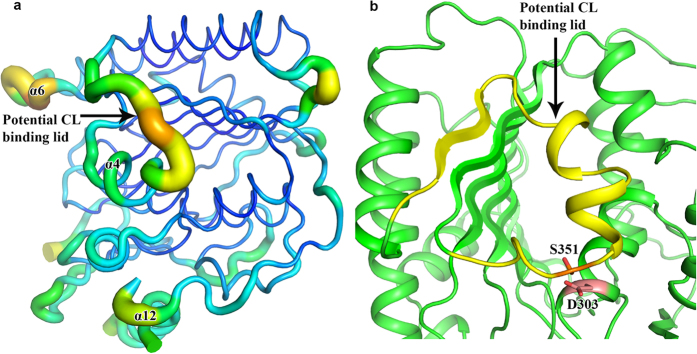
Comparison of the crystal structures of EcPbgA245-586 and StPbgA245-586 revealed a conformational change of the loop between helices α5 and α6 (Fig. 2a). In EcPbgA245-586 this loop closes the hydrophobic core between the second (β-strands 1–8, α8 and α9) and third layers (α-helices 2, 4, 5 and 10) and the residues P365-R368 form β-strand 4 to interact with β-strand 3, whereas the strand β4 is disordered and away from the hydrophobic core between the second and third layers in StPbgA245-586 (Figs 1b,c and 2a). Residue F349 is solvent exposed and F362 is located in the hydrophobic core in EcPbgA245-586, while the residue F349 rotates toward the hydrophobic core and the F362 is solvent exposed in StPbgA245-586 (Fig. 4a,b). Analysis of the StPbgA245-586 structure revealed that there is a loop formed by residues D347-Q370 that has high B-factors between helices α4 and α6, indicating that this loop is flexible and may act as a lid for the CL binding and transport (Fig. 5a). To explore this possibility, we generated a double cysteine mutant S251C/D303C, which would lock the switch and significantly influence the CL binding. Indeed the mutant S251C/D303C impairs the cell growth (Fig. 4g).
Figure 5. Potential CL binding lid.
(a) The PbgA loop consisting of residues D347-Q370 has higher B-factor comparing to the local residues, suggesting that the loop is flexible and may be a lid for the CL binding. (b) Cartoon representation of potential CL binding lid colored in yellow and the double cysteine mutation residues S351C/D303C may form a disulphide bond and prevent the lid to open for CL binding.
Discussion
The OM of Gram-negative bacteria is an asymmetric membrane composed of phospholipids and lipopolysaccharide. The lipids in the OM play important roles in the pathogenesis and drug resistance of Gram-negative bacteria. Despite the extensively studied LPS transport pathway in Gram-negative bacteria, the mechanisms for phospholipids transport and assembly in the OM remain to be characterized at the structural level.
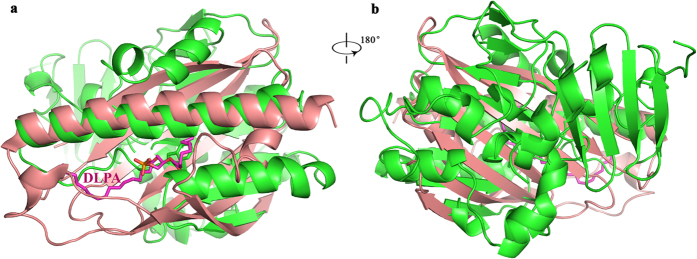
CL plays an essential role in OM remodelling in bacteria and is critical for mitochondrial function41. PbgA transports CL from the IM to the OM in Gram-negative bacteria24. Mitochondria evolved from bacteria and the UPS1/Mdm35 complex has been identified for phospholipid transfer from the OM to the IM in yeast. The recent structures of the UPS1 of yeast in complex with phospholipids have been determined29,30. The structure is quite different from those of PbgA with a RMSD of 3.7 Å over 62 Cα atoms (Fig. 6a,b). The UPS1 contains a concaved 7-antiparellel stranded β-sheet, which wraps PA molecules inside the cavity. Only the long helix α7 of PbgA is superimposed well with the helix α3 of UPS1; the conformation of the β-sheet at the second layer of PbgA is different from that of the UPS1, which does not fold to form a cavity (Fig. 6a), indicating that this area between the first and second layers of PbgA may not be the CL binding site. Indeed, the functional assays using alanine substitution of aromatic residues between the first and the second layer showed that all the mutants did not impair bacterial growth, which suggest that the PbgA-mediated transport of CL is different from UPS1-mediated transport of PA in mitochondria.
Figure 6. The PbgA structure is quite different from the UPS1 of mitochondria of yeast.
(a) Only the long helix α7 of PbgA is superimposed well with the helix α3 of UPS1. (b) Rotation 180° along y axis relative to the left panel. DLPA, 1,2-dilauroyl-sn-glycero-3-phosphate (12:0–12:0).
The structure of EcPbgA245-586/StPbgA245-586 is strikingly similar to arylsulfatases and lipoteichoic acid synthases, but it lacks their enzymatic activities39. Metal ions, such as Mg2+, Mn2+ or Ca2+ are required for their catalytic function. Superimposition of PbgA structures over those of arylsulfatase and lipoteichoic acid synthase suggested that PbgA is unlikely to form the required metal ion-binding site found at the active sites of arylsulfatase and lipoteichoic acid synthase.
The functional studies showed that single hydrophobic residue mutants F292A, F349A and F362A caused the cell death (Fig. 4e) or impaired cell growth in 50 μg/ml vancomycin, indicating that these residues are involved in CL transport. This was further confirmed by the co-sedimentation assays (Fig. 4J). All these residues are located in the hydrophobic core between the layers 2 and 3 of PbgA245-586, strongly suggesting that this hydrophobic core is the potential CL binding site. Additionally, single mutant F344A, W393D, and double mutants F399C/D372C and W397C/S395C between the first layer and the second layer did not have any influence on cell growth (Fig. 4h,i), which further confirms that the hydrophobic space between layer 2 and 3 may be responsible for the CL transport.
There is a significant conformational change of residues D347-Q370 between the EcPbgA245-586 and StPbgA245-586 (Fig. 2a), and these residues’ B-factors are higher than neighbouring residues (Fig. 5a), indicating that they are flexible. We speculated that this region may be the lid for the CL binding, which can open to allow the CL access the binding site between the second and third layers. The double cysteine mutant S351C/D303C may form a disulphide bond and prevent the lid to open for CL binding (Fig. 5b). This mutant is lethal, suggesting that the loop may be the lid for the CL binding (Fig. 4g). The B-factors of the last α-helix 12 are also higher compared to other local residues (Fig. 5a), indicating that this helix is flexible, which may suggest that it interacts with other proteins or OM components.
Phospholipid transport and assembly in the OM is a basic biological process. Our structural and functional studies have revealed that the hydrophobic core between the layer 2 and 3 may be the CL binding site and that the residues D347-Q370 may form a lid for the CL transport in the structures of PbgA245-586 of E. coli and S. typhinium, and. This is a first structure of a bacterial protein involved in phospholipid transport from the inner membrane to the OM. This work is important for understanding how bacteria pathogenesis and drug resistance is achieved by remodelling the OM, in addition to clarifying how Gram-negative bacteria assemble their OM.
Experimental Procedures
Cloning, expression, and purification of PbgA245–585
The pbgA gene (yejM) encoding PbgA245-586 and PbgA191-586 from S. typhimurium strain LT2 and E. coli strain K-12 were amplified and inserted into plasmid pEHISTEV with a NcoI restriction sites at 5′-end and EcoR1 sites at 3′-end respectively. This plasmid includes a hexahistidine tag (6× His) and a tobacco etch virus (TEV) protease cleavage site between the Histag and the N-terminus of the cloned genes. The recombinant plasmids were transformed into soluble BL21(DE3) strain (Novagen) for protein expressions. The transformed soluble BL21(DE3) cells were grown in Luria broth (LB) supplemented with antibiotic (Kanamycin 50 μg/ml) at 37 °C until the optical density of the culture reached 0.5–0.8 at a wavelength of 600 nm (OD600nm). The proteins were induced by addition of 0.1 mM isopropyl β-d-thiogalactopyranoside (IPTG) and incubated for 12 hours at 20 °C. For over-expression of selenomethionine (SeMet) labeled PbgA245-586, the protein was expressed in M9 medium supplemented with SeMet (Generon) to a final concentration of 100 μg/ml using the methionine inhibition method42. The cells were harvested by centrifugation at 5000 × g for 20 mins, resuspended in buffer containing 20 mM Tris-Cl, pH 7.8, 10% glycerol and 500 mM NaCl, supplemented with cOmplete (Roche), 1 mM DNase (Sigma-Aldrich) and 1 mM phenylmethylsulphonyl fluoride (PMSF, Sigma-Aldrich). The cells were broken using a cell disruption at 30,000 psi (Constant Systems Ltd). The Cell debris was removed by centrifugation at 120,000 × g for 25 mins at 4 °C. The supernatant was then being loaded onto a nickel-nitrilotriacetate affinity resin column (Ni-NTA, Qiagen) and the column was washed with 20 mM Tris-Cl, pH 7.8, 500 mM NaCl, 30 mM imidazole and 10% glycerol. The recombinant proteins were eluted with 20 mM Tris-Cl, pH 7.8, 500 mM NaCl, 300 mM imidazole and 10% glycerol. The protein buffer was changed to 20 mM Tris-Cl, pH 7.8, 500 mM NaCl, 10% glycerol and 10 mM imidazole using a desalting column (Hi-PrepTM 26/10, GE Healthcare) to prevent protein precipitation. The (6 × His) tag was removed by TEV protease, and the PbgA protein was obtained by applying the samples through a Ni-NTA column. The protein was further purified using size exclusion chromatography with a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) in a running buffer containing 20 mM Tris-Cl, pH 7.8 and 150 mM NaCl. Fractions with the highest purity of PbgA were pooled and concentrated to 10 mg/ml.
Crystallization and Data collection
Protein crystallization trails were performed using 1 μl of protein mixed with 1 μl of reservoir solution by the sitting-drop vapour diffusion technique at room temperature. The best crystals of SeMet incorporated StPbgA245-586 were obtained in 0.1 M Bis-tris pH 5.5 and 16% PEG3350. The best crystals of in EcPbgA245-586 were obtained in 0.2 M lithium sulphate, 0.1 M sodium acetate pH 4.3 and 48% PEG400. The best native crystals of StPbgA191-586 were obtained in 0.1 M Bis-tris pH 6.5 and 25% PEG3350. The crystals were harvested after 21 days and cryoprotected by supplementing the crystallization solution with 20% glycerol in the crystallization solutions, before being flash frozen in liquid nitrogen. Single-wavelength anomalous dispersion (SAD) data were collected at Diamond Light Source, UK at beam station I04. All data were processed using XDS43 and the StPbgA245-586 crystals that diffracted to 1.69 Angostram resolution belong to space group P21 with unit-cell dimensions: a = 52.6 Å, b = 196.09 Å, c = 70.27 Å and β = 95.79°. The native EcPbgA245-586 crystal data were collected to 1.65 Å resolution and the crystals belong to space group P3121 with unit-cell dimensions: a = 56.86 Å, b = 56.86 Å, c = 191.81 Å and γ = 120°.
Structure determination
The structure of StPbgA245-586 was solved by the SAD method using SHELX suite34. The initial model of the StPbgA245-586 was built automatically using the program Buccaneer36 with the phases obtained form the SHELX34. The native structure (Escherichia coli strain K-12) was determined by molecular replacement using structure from Salmonella as a search model using Phase44. The structures were manually built by using Coot37 and the structures were refined using REFMAC545. The structures were validated by Molprobity46. The statistics of the data collection and the structure refinement are summarized in Table 1.
Site-directed mutagenesis and functional assays
All single or double mutations were generated following the protocol47. The mutations were amplified by PCR using Q5® hot start high fidelity DNA polymerase (New England) and the pACYCDuet plasmid (Novagen), containing a C-terminal (6× His) tag of PbgA E. coli gene with an NcoI restriction site at the 5′-end and Hind III site at the 3′-end as the template for the pbgA mutagenesis.
The NRD183 strain still contains the transmembrane domain and only the periplasmic global domain is depleted. The NRD183 also contains a plasmid pNRD217, which carries the whole pbgA gene under pBAD control. As the NRD183 contains the N-terminal transmembrane domain, the strain is a conditional lethal strain. The strain is killed in presence of 50 ug/ml vancomycin without PbgA expression.
The mutations were confirmed by DNA sequencing. Once the full length of the PbgA was shown to rescue the NRD183 strain, these single or double mutants were transformed into E. coli NRD18340, respectively. The transformed E. coli cells were grown on LB agar plate supplemented with antibiotics (kanamycin 50 μg/ml, chloramphenicol 34 μg/ml, and ampicillin 50 μg/ml). Single colonies of each transformation was inoculated into 5 ml LB medium supplemented with above antibiotics and 0.2% L-arabinose. Subculture cells were used for functional assay. E. coli NRD183 with the empty plasmid pACYCDuet was used as a negative control. For functional assays, cells were harvested, washed one time and diluted in sterile LB medium to an absorbance (OD600 nm) of 0.5 and streaked onto LB agar plates supplemented with vancomycin 50 μg/ml, kanamycin 50 μg/ml and chloramphenicol 34 μg/ml and 5mM IPTG. Cell growths were recorded after overnight culture in 37 °C. All the assays have been repeated in triplicate.
Protein-Lipid co-sedimentation assays
Cardiolipins (CL) in chloroform (Sigma) was dried under nitrogen gas, and then re-suspended in 20 mM Tris (pH 7.8), and 150 mM NaCl. Subsequently, the CL was sonicated until the solution to be clear. The mixture (Lipid-protein) were incubated at 37 °C for 1 hour at a 10:1 molar ratio (20 mM: 2 mM), respectively following the reported method24. The reactions were centrifuged at 13.2 k rpm for 10 min. The supernatants were collected and the pellets were re-suspended in 20 μl of buffer in 20 mM Tris (pH 7.8), and 150 mM NaCl. The pellets (10 μl) and supernatants (10 μl) were boiled in the SDS-containing sample buffer and separated on 12% SDS Tris-HCl PAGE. The proteins were visualized by Coomassie staining.
Additional Information
Accession codes: The atomic coordinates and structure factors of EcPbgA245-585, StPbgA245-585 and StPbgA191-585 are deposited at the Protein Data Bank under access code 5I5H, 5I5D and 5I5F.
How to cite this article: Dong, H. et al. Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci. Rep. 6, 30815; doi: 10.1038/srep30815 (2016).
Supplementary Material
Acknowledgments
We thank Gareth Ashworth for the critical reading. We appreciate the staff at I24, I02 and I03 of Diamond Light Source UK for beamtime (proposal mx9475) and their assistance with data collection. CJD is a recipient of the Wellcome Trust investigator award (WT106121MA), and is supported by Medical Research Council (G1100110/1).
Footnotes
Author Contributions C.D. and H.D. conceived and designed the experiments. H.D., Z.Z., X.T., S.H. and H.L. expressed, purified and crystallized the EcPbgA and StPbgA. Z.Z., X.T. and H.D. did the mutagenesis and the functional assays. Z.Z., X.T. and B.P. performed the protein-Lipid co-sedimentation assays. H.D. undertook data collection and structure determination. H.D. generated the constructs for the protein expression. C.D. and H.D. wrote the manuscript.
References
- Raetz C. R. H. & Whitfield C. Lipopolysaccharide endotoxins. Annual review of biochemistry 71, 635–700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N., Kahne D. & Silhavy T. J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nature reviews. Microbiology 7, 677–683 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Kahne D. & Walker S. The bacterial cell envelope. Cold Spring Harbor perspectives in biology 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited Molecular Basis of Bacterial Outer Membrane Permeability Revisited. 67, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. & Trent M. S. Biosynthesis and Export of Bacterial Lipopolysaccharides. Annual review of biochemistry 83, 99–128 (2014). [DOI] [PubMed] [Google Scholar]
- Dong H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014). [DOI] [PubMed] [Google Scholar]
- Qiao S., Luo Q., Zhao Y., Zhang X. C. & Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014). [DOI] [PubMed] [Google Scholar]
- Wu T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005). [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiology and molecular biology reviews: MMBR 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N., Gronenberg L. S., Kahne D. & Silhavy T. J. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 105, 5537–5542 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R. et al. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. Journal of bacteriology 195, 1100–1108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freinkman E., Chng S. & Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms. (2010). doi:10.1073/pnas.1015617108/-/DCSupplemental. www.pnas.org/cgi/doi/10.1073/pnas.1015617108 [DOI] [PMC free article] [PubMed]
- Sperandeo P., Dehò G. & Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochimica et biophysica acta 1791, 594–602 (2009). [DOI] [PubMed] [Google Scholar]
- Ito Y., Kanamaru K., Taniguchi N., Shigehiko M. & Tokuda H. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Molecular Microbiology 62, 1064–1075 (2006). [DOI] [PubMed] [Google Scholar]
- Okuda S. & Tokuda H. Lipoprotein Sorting in Bacteria. Annual Review of Microbiology 65, 239–259 (2011). [DOI] [PubMed] [Google Scholar]
- Tokuda H. Biogenesis of outer membranes in Gram-negative bacteria. Bioscience, biotechnology, and biochemistry 73, 465–473 (2009). [DOI] [PubMed] [Google Scholar]
- Tokuda H. & Matsuyama S. I. Sorting of lipoproteins to the outer membrane in E. coli. Biochimica et Biophysica Acta-Molecular Cell Research 1693, 5–13 (2004). [DOI] [PubMed] [Google Scholar]
- Hagan C. L. & Kahne D. The reconstituted escherichia coli bam complex catalyzes multiple rounds of? -barrel assembly. Biochemistry 50, 7444–7446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci D. P. & Silhavy T. J. The Bam machine: a molecular cooper. Biochimica et biophysica acta 1818, 1067–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan C. L., Silhavy T. J. & Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annual review of biochemistry 80, 189–210 (2011). [DOI] [PubMed] [Google Scholar]
- Hagan C. L., Westwood D. B. & Kahne D. Bam lipoproteins assemble BamA in vitro. Biochemistry 52, 6108–6113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N., Rollauer S. E. & Buchanan S. K. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Current Opinion in Structural Biology 31, 35–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Matamouros S., Whittington D., Bishop R. E. & Miller S. I. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proceedings of the National Academy of Sciences of the United States of America 111, 1963–1968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D. et al. Delivery of Cardiolipins to the Salmonella Outer Membrane Is Necessary for Survival within Host Tissues and Virulence. Cell Host & Microbe 17, 441–451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nature reviews. Molecular cell biology 11, 739–750 (2010). [DOI] [PubMed] [Google Scholar]
- Schlattner U. et al. Mitochondrial cardiolipin/phospholipid trafficking: the role of membrane contact site complexes and lipid transfer proteins. Chemistry and physics of lipids 179, 32–41 (2014). [DOI] [PubMed] [Google Scholar]
- Osman C., Voelker D. R. & Langer T. Making heads or tails of phospholipids in mitochondria. Journal of Cell Biology 192, 7–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies G., Paradies V., De Benedictis V., Ruggiero F. M. & Petrosillo G. Functional role of cardiolipin in mitochondrial bioenergetics. Biochimica et Biophysica Acta-Bioenergetics 1837, 408–417 (2014). [DOI] [PubMed] [Google Scholar]
- Yu F. et al. Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups 1 -Mdm 35 complex. 16, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miliara X. et al. Structural insight into the TRIAP 1/PRELI-like domain family of mitochondrial phospholipid transfer complexes. EMBO reports 1–12 doi: 10.15252/embr.201540229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni J. C. & Silhavy T. J. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proceedings of the National Academy of Sciences of the United States of America 106, 8009–8014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus D. et al. Digalactosyldiacylglycerol synthesis in chloroplasts of the Arabidopsis dgd1 mutant. Plant physiology 128, 885–895 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T. R. & Sheldrick G. M. research papers Substructure solution with SHELXD. 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallographica Section D: Biological Crystallography 66, 479–485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. A short history of SHELX. Acta crystallographica. Section A, Foundations of crystallography 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallographica Section D: Biological Crystallography 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Boltes I. et al. 1.3 Å structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure 9, 483–491 (2001). [DOI] [PubMed] [Google Scholar]
- Lu D. et al. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proceedings of the National Academy of Sciences of the United States of America 106, 1584–1589 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N. R. & Cronan J. E. Genetic Interaction Between the Escherichia coli AcpT Phosphopantetheinyl Transferase and the YejM Inner Membrane Protein. Genetics 178, 1327–1337 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. T. et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature cell biology 15, 1197–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods in Enzymology 276, 523–530 (1997). [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. Journal of applied crystallography 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica Section D: Biological Crystallography 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I. W. et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Research 35, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. & Naismith J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC biotechnology 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.