Summary
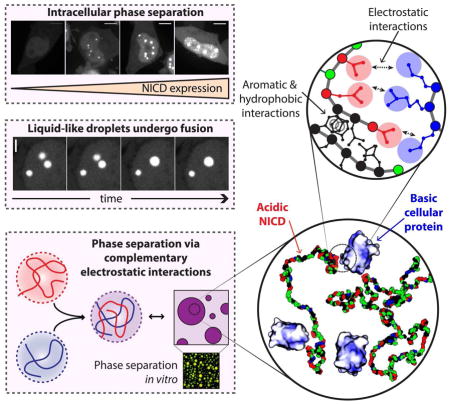
Liquid-liquid phase separation, driven by collective interactions among multivalent and intrinsically disordered proteins, is thought to mediate the formation of membrane-less organelles in cells. Using parallel cellular and in vitro assays we show that the Nephrin intracellular domain (NICD), a disordered protein, drives intracellular phase separation via complex coacervation, whereby the negatively charged NICD co-assembles with positively charged partners to form protein-rich dense liquid droplets. Mutagenesis reveals that the driving force for phase separation depends on the overall amino acid composition and not the precise sequence of NICD. Instead, phase separation is promoted by one or more regions of high negative charge density and aromatic/hydrophobic residues that are distributed across the protein. Many disordered proteins share similar sequence characteristics with NICD, suggesting that complex coacervation may be a widely used mechanism to promote intracellular phase separation.
Classification: Biological Sciences, Biochemistry
Graphical abstract
Introduction
Membrane-less organelles such as nucleoli, Cajal bodies, and stress granules are involved in diverse biological processes, from ribosome assembly, to gene regulation to signal transduction. These micron-sized organelles are found throughout the cell (Spector, 2006), and like their lipid-membrane-bound counterparts, provide distinct cellular compartments, concentrate select proteins and nucleic acids, and may localize specific biological reactions (Hyman et al., 2014).
Many membrane-less organelles have liquid-like physical properties. They are spherical, undergo fusion/fission, drip under shear stress, and exchange contents rapidly with the surrounding medium. Such structures include P granules in C. elegans embryos (Brangwynne et al., 2009; Elbaum-Garfinkle et al., 2015; Wang et al., 2014), nucleoli of Xenopus oocytes (Brangwynne et al., 2011) and C. elegans embryos (Berry et al., 2015), stress granules in mammalian cells (Kroschwald et al., 2015), and ribonucleoprotein granules in fungi (Zhang et al., 2015). Analogous liquid-like structures have also been generated by expressing various multivalent or disordered proteins (Li et al., 2012; Molliex et al., 2015; Nott et al., 2015; Patel et al., 2015). Their physical properties and condensation/dissolution behavior suggest that at least some of these structures assemble via a liquid-liquid phase transition.
Many membrane-less organelles are enriched in proteins containing large intrinsically disordered regions (IDRs) (Wright and Dyson, 2015). IDRs that lack charged residues phase separate due to a preference for homotypic self-associations over interactions with the solvent (Brangwynne et al., 2015). In polymer science, such phase separation is known as simple coacervation, and produces a dense phase enriched in a single polymer that is in equilibrium with a polymer-depleted phase (Veis, 2011). Most IDRs known to phase separate in vitro do so via simple coacervation. Tropoelastin, an extracellular matrix protein composed of imperfect repeats of VGVAPG, undergoes simple coacervation in vitro through interactions among its hydrophobic residues (Miao et al., 2003). IDRs that phase separate are also enriched in RNA binding proteins, where they often possess a limited set of amino acids—F, Y, S, G, Q, N (Couthouis et al., 2012; Han et al., 2012; Kato et al., 2012). Ddx4, an RNA binding protein localized to germ granules, contains IDRs that phase separate in vitro and in cells. In Ddx4, electrostatic interactions between clusters of opposing charge and cation-pi interactions between FG and RG motifs have been invoked as important driving forces for phase separation (Nott et al., 2015). The RNA binding proteins, FUS, hnRNPA1, hnRNPA2 and the mitotic spindle protein BugZ, also phase separate in vitro and/or in cells. For several of these, Tyr residues promote phase separation or recrutiment into phase separated structures (Burke et al., 2015; Jiang et al., 2015; Kato et al., 2012; Molliex et al., 2015; Patel et al., 2015). At high concentrations, many proteins containing low complexity IDRs form amyloid-like filaments consisting of repeated cross-beta strand elements (Han et al., 2012; Kato et al., 2012; Lin et al., 2015; Molliex et al., 2015). An intriguing postulate is that cross-beta elements, when occurring in small numbers between otherwise disordered chains, constitute the adhesive structures that promote liquid phase separation (Xiang et al., 2015). In proteins that contain modular binding domains and IDRs, module-ligand and IDR-IDR interactions act synergistically to promote phase separation (Lin et al., 2015).
When polymers are enriched in one type of charge, they repel one another, and this inhibits simple coacervation. However, when mixed with multivalent counterions, complexes of oppositely charged polymers drive phase separation via a process known as complex coacervation. The oppositely charged molecules phase separate together, to lower the net charge. In the resultant polymer-rich droplet phase both species are highly concentrated and this dense phase is in equilibrium with a polymer-depleted bulk phase (Veis, 2011). Here we have examined the intracellular phase separation of a negatively charged IDR, and find that it forms nuclear bodies via complex coacervation. Our investigations are based on the serendipitous observation that the disordered intracellular domain of the adhesion receptor Nephrin forms nuclear bodies when expressed as an autonomous entity in mammalian cells. Using quantitative microscopy, we show that nuclear bodies formed by the Nephrin intracellular domain (NICD) behave as phase separated liquid droplets, and are likely de novo nuclear structures. Cellular and biochemical data indicate that bodies/droplets form through non-specific associations of the negatively charged NICD with positively charged partners. Using deletion mutants and de novo sequence designs, we show that NICD phase separation is promoted by one or more blocks of high negative charge density and by aromatic/hydrophobic residues distributed along the sequence. The data demonstrate that the amino acid composition of NICD is more important than the precise sequence for phase separation. Proteome-wide analysis of sequence features suggests that complex coacervation of negatively charged IDRs may be a general phenomenon that contributes to the formation of membrane-less organelles.
Results
NICD forms nuclear bodies that are phase-separated liquids
The adaptor protein, Nck, and the actin nucleation-promoting factor, N-WASP, can phase separate when mixed together, due to multivalent interactions between SH3 domains on Nck and proline rich motifs on N-WASP. The Nck SH2 domain can also bind to phosphotyrosine sites on phosphorylated NICD. In vitro this interaction facilitates phase separation by scaffolding multiple Nck proteins, thereby increasing the effective valency of Nck SH3 domains (Banjade and Rosen, 2014; Li et al., 2012). We initially sought to recapitulate these phenomena in HeLa cells by overexpressing the NICD (residues 1077–1241) tagged at its C-terminus with a fluorescent protein (YPet) (Figure 1A, red box). We expressed either wild type NICD or, as an intended control, a non-phosphorylatable mutant (Y3F). Surprisingly, both proteins formed micron-scale nuclear bodies (Figures 1B, C; note that percent cells showing puncta is normalized to that of WT NICD throughout this report). Thus, these structures do not require the binding of Nck to phosphotyrosine motifs on Nephrin, and are unrelated to the previously described phase separation of the ternary system.
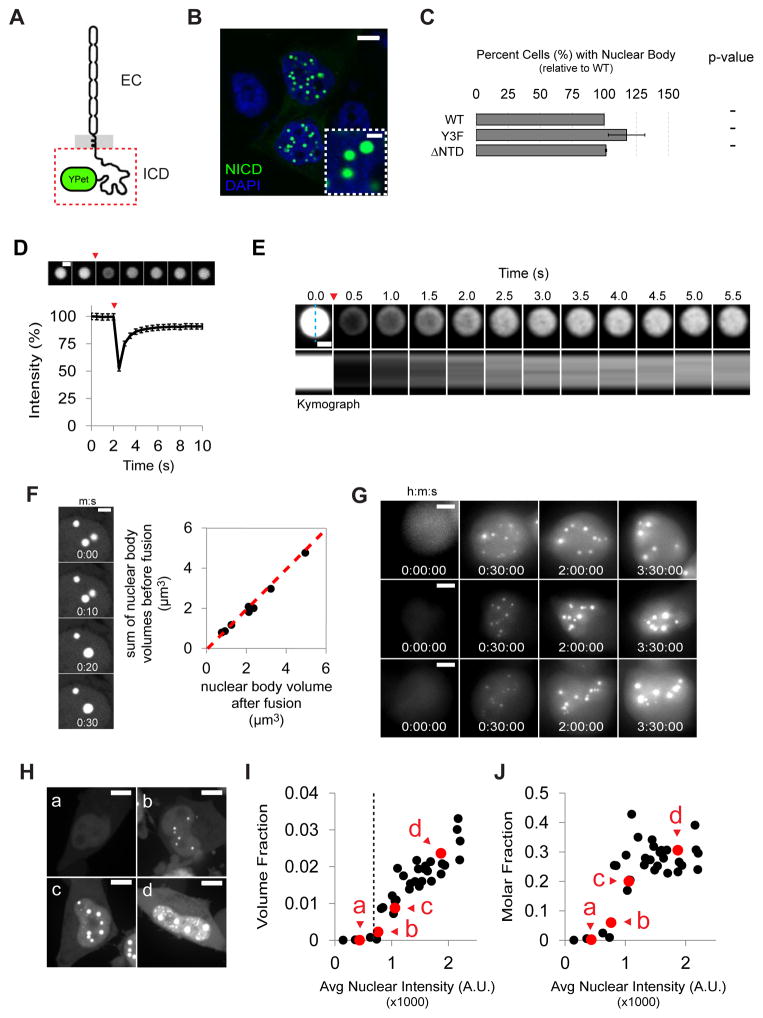
Figure 1. NICD nuclear bodies are phase separated liquids.
(A) Schematic representation of nephrin, including extracellular region (EC) and intracellular cytoplasmic domain (ICD). NICD (red box), expressed as a soluble protein, is C-terminally tagged with YPet. (B) Spherical micron-sized nuclear bodies (inset) form in the nuclei of HeLa cells expressing NICD (green). Nuclei were stained with DAPI (blue). Scale bars in main and inset panels are 5 and 1 μm, respectively. (C) Normalized (to WT) percent of HeLa cells containing nuclear bodies when expressing NICD mutants Y3F (Y100F, Y117F, and Y141F) or ΔNTD (residues 63–166). Data are represented as mean ± SEM. (p values for comparison to WT NICD: ‘-‘ indicates p > 0.05). (D) Confocal imaging and quantification of NICD fluorescence recovery after photobleaching (n=34 bodies). Red arrowhead marks photobleaching event. Data are represented as mean ± SEM. Scale bar = 1 μm. (E) Upper: recovery of fluorescence intensity in a larger nuclear body. Red arrowhead marks photobleaching event. Scale bar = 1 μm. Lower: kymographs of fluorescence recovery across the diameter of the nuclear body (cyan dashed line). Note that that total recovery at t = 5.5 sec is 73%, similar to that expected for complete recovery, based on photobleaching effects during the imaging time course (86%), but appears lower because of the contrast settings used to illustrate the radial recovery pattern. (F) Left: time lapse imaging showing fusion of two nuclear bodies. Scale bar = 2 μm. Right: volume is conserved during coalescence (n=8 bodies) (slope of red dashed line = 1). (G) Time lapse imaging of nuclear body assembly in transiently transfected HeLa cells expressing NICD. Scale bar = 5 μm. (H) Maximum projection images of HeLa cells expressing different levels of NICD (expression level increases from (a) to (d)). Scale bar = 5 μm. (I, J) Quantification of volume fraction (I) and molar fraction (J) of nuclear bodies in cells (n=36) expressing different levels of NICD. Each symbol represents a single cell. Volume and molar fraction of cells shown in (a) through (d) are indicated by red symbols. Saturation concentration at ~600–750 arbitrary units is indicated by the black vertical dashed line in I. See also Supplemental Figure S1.
NICD nuclear bodies varied in number across cells, varied in size within a given cell, and had liquid-like physical properties. They were spherical at the resolution of light microscopy (Figure 1B). NICD-YPet fluorescence in bodies recovered rapidly after photobleaching, with τ < 1 s for structures ~1 μm in diameter, consistent with a molecular diffusion constant of 0.5 μm2/s (Figure 1D) and rapid exchange with the surrounding nucleoplasm. NICD-YPet fluorescence returned to 90% of its initial value due to photobleaching of the cells during the recovery period (equivalent time lapse imaging decreased the intensity of unperturbed nuclear bodies by ~14%, not shown). In larger bodies monitored by confocal microscopy, fluorescence recovery was observed initially at the periphery, followed by radial spreading to the center (Figure 1E). Thus, these bodies are filled objects, and constituent NICD molecules exchange with the nucleoplasm faster than they diffuse within the bodies. Finally, NICD nuclear bodies also fuse rapidly (< 10 s, Figure 1F, left panel), and with conservation of volume (Figure 1F, right panel). Thus, NICD nuclear bodies behave as liquid droplets.
To examine the concentration dependence of body formation, we initially imaged transiently transfected cells over time as they began to express NICD. In most cells, numerous, small nuclear bodies formed abruptly ~6–8 h after transfection, when expression levels were relatively low (Figure 1G and S1A; the time point preceding the first instance of a nuclear body was set to t=0). Nuclear bodies increased in size and decreased in number over time (Figure 1G and S1B), likely due in part to merging (cf. Figure 1F). We also examined how the total summed volume of NICD nuclear bodies in a given cell varied with nuclear expression level. HeLa cells expressing different levels of NICD were imaged in 3D by confocal microscopy (Figure 1H, left panels (a–d)). For each cell, we determined the total fraction of the nucleus occupied by NICD bodies. Below an average nuclear fluorescence intensity of ~600–750 A.U., cells never contained observable nuclear bodies (Figure 1H, a). However, bodies appeared sharply above this value (Figures 1H, I, b), and total nuclear volume fraction increased steadily with increasing nuclear fluorescence (NICD concentration; (Figures 1H, I, c and d)). Nuclear bodies comprised 1–3.5% of the nuclear volume (Figure 1I), and retained up to ~25–40% of total nuclear NICD molecules (Figure 1J). These data demonstrate that NICD phase separates to generate liquid-like nuclear bodies.
We next asked if NICD nuclear bodies are formed de novo or if NICD is absorbed into an existing nuclear body. In HeLa cells, NICD bodies do not co-localize with markers of Cajal bodies, nucleoli, PML bodies or several other nuclear puncta (Figure S1A). However, they co-localize with nuclear paraspeckle markers NONO/p54nrb and SFPQ/PSF (Figure S1B). Nevertheless, NICD bodies form equally well in cells where NEAT1 has been deleted (Figure S1C), which lack paraspeckles (Figure S1D and (Clemson et al., 2009)), and in parental cells containing NEAT1. Thus, they are probably de novo structures that absorb NONO/p54nrb and SFPQ/PSF (and perhaps paraspeckles). We note that the sharp appearance of NICD nuclear bodies with expression level is also inconsistent with simple partitioning into pre-existing paraspeckles. Although NICD nuclear bodies co-localize with paraspeckle proteins, they are distinct from classical NEAT1-dependent paraspeckles.
NICD nuclear bodies form according to complex coacervation
In simple coacervation, when the concentration of a solute is increased to its solubility limit, the solution separates into two phases: a solute-rich phase (droplet phase) of smaller volume, and a solute-poor phase (bulk phase) of larger volume. As the total solute is increased beyond the solubility limit, the volume of the droplet phase increases at the expense of the bulk phase. Importantly, the concentrations of both phases remain constant, so that the partition coefficient of the solute ([solute]droplet/[solute]bulk) is invariant to the total amount of solute in the system (Dill and Bromberg, 2010). We examined the NICD-YPet fluorescence intensity within nuclear bodies and the surrounding nucleoplasm in a population of cells expressing different levels of NICD. We limited this analysis to nuclear bodies larger than the transverse point spread function of our microscope and took care to correct for the size-dependent decrease in apparent intensity due to the point spread function (Figure S2A, Supplemental Experimental Procedures). NICD body fluorescence intensity is relatively independent of size (Figure S2A), and essentially invariant with total nuclear fluorescence (Figure 2A). In contrast, fluorescence intensity in the surrounding nucleoplasm is proportional to nuclear NICD expression level (Figure 2B). Thus, the partition coefficient of NICD varies with expression level (Figure 2C), a behavior that is inconsistent with simple coacervation.
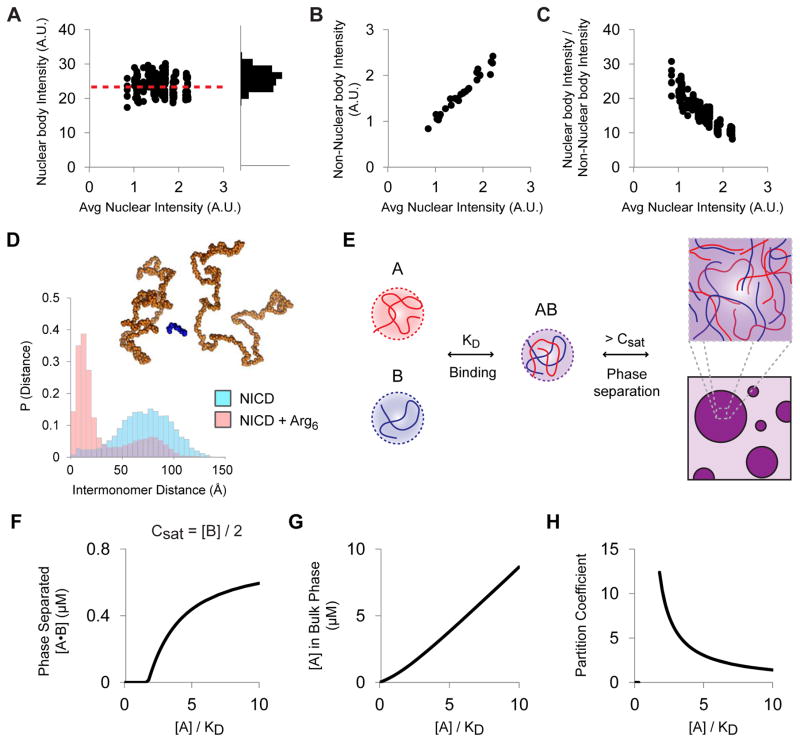
Figure 2. NICD nuclear bodies form according to complex coacervation.
(A) Left: nuclear body intensity in HeLa cells expressing different levels of NICD. Each symbol represents an individual nuclear body. Dashed red line indicates average for 239 nuclear bodies from 30 cells. Right: histogram of nuclear body intensities. (B) NICD intensity in the surrounding nucleoplasm (non-nuclear body intensity) for 30 cells, each indicated by a single symbol. (C) NICD partition coefficient (nuclear body intensity / non-nuclear body intensity) from cells expressing different levels of NICD (239 nuclear bodies from 30 cells). (D) Histograms of the distances of closest approach between NICD molecules with and without the inclusion of Arg6 peptides in atomistic Monte Carlo simulations. Purple bars indicate overlap between the two histograms. Inset: representative snapshot of the association of pairs of NICD molecules mediated by Arg6 peptide (see also Figure S2B). (E) Complex coacervation model of NICD nuclear body formation. Species A (red) binds to a partner B (blue) with an affinity (KD). The AB complex phase separates at concentrations greater than or equal to Csat, to form droplets enriched in A and B. (F) Modeling complex coacervation of AB. When KD = Csat = [B]/2, phase separated AB appears sharply above a threshold concentration of A. At higher concentrations of A, the amount of phase separated AB plateaus. (G) Except at low concentrations of total A, the concentration of unbound A, which remains in the bulk phase, rises nearly linearly with total amount of A added. (H) When the droplet phase is of constant concentration (cf. Figure 2A), the partition coefficient of A (as AB complex) decreases with total A. See also Supplemental Figure S2.
NICD is negatively charged, with an estimated isoelectric point (pI) of 4.3, net charge of −21 and net charge per residue of −0.13 at neutral pH, and fraction of charged residues (FCR) of 0.30. Initial atomistic simulations of NICD in the absence of multivalent counterions revealed the presence of intermolecular repulsions that were quantifiable in terms of the large distances between pairs of NICD molecules. We hypothesized that NICD phase separation might proceed via complex coacervation. We tested this postulate by performing simulations of NICD with positively charged oligo-lysine and oligo-arginine peptides of different valencies. We observed counterion-mediated self-association of NICD, demonstrating that NICD molecules can self-associate if electrostatic repulsions are diminished via counterion-mediated charge neutralization along NICD (Figures 2D and S2B).
We modeled complex coacervation phenomenologically by considering two species, A and B, that are soluble individually, but dimerize according to an equilibrium dissociation constant, KD, to generate a complex, AB, that phase separate beyond a saturation concentration, Csat (Figure 2E). As shown in Figure 2F, when KD is similar to Csat and B is limiting, the addition of A does not produce phase separated AB initially. However, when a threshold concentration of A is reached, AB phase separates to an extent that rises with additional A. At very high concentrations of A, the amount of phase separated AB plateaus as the concentration of B becomes limiting. Throughout this process, the concentration of unbound A rises nearly linearly with the total concentration of A (Figure 2G). Assuming the concentration of AB in the droplet phase is constant, this increase in the concentration of unbound A results in a steady decrease in the relative partitioning of A (as species AB) into the droplet phase (Figure 2H). These four behaviors are identical to our experimental observations in Figures 1I, 2B and 2C, respectively, and are observed qualitatively over a range of parameter values (Figure S2C). Taken together, these results suggest that NICD may form nuclear bodies via complex coacervation.
To test this hypothesis in vitro, we recombinantly expressed and purified human NICD. Ultraviolet circular dichroism, nuclear magnetic resonance spectroscopy, and gel filtration data indicate that the protein is disordered and likely monomeric in solution (Figures S3A–C). Additionally, atomistic simulations show that NICD adopts conformational ensembles that are expanded relative to globular, folded domains (Figure S3D). Isolated Alexa-568 labeled NICD remained uniformly dispersed in solution with no evidence of phase separation by fluorescence or light microscopy under a range of pH, salt and buffer conditions, both at room temperature and at 4°C (Figure 3, left panels; see also Figure S3E). However, solutions containing NICD plus a supercharged GFP mutant having net surface charge of +25 [scGFP(+25), (McNaughton et al., 2009)], as a generic positively-charged macroion, were opalescent, and contained spherical droplets enriched in both proteins (Figure 3, right panels; see also Figure S3E, rightmost panels). The inverse capillary velocity, which describes the ratio of effective viscosity to surface tension, was estimated to be 0.25 s/μm for phase separated droplets of wild type NICD and scGFP(+20) (Figure S3F), a value similar to that of phase separated droplets of the disordered protein, LAF-1 (Elbaum-Garfinkle et al., 2015). NICD also phase separated with positively charged oligoArg peptides (Figure S3G). Thus, in vitro, NICD phase separates via complex coacervation.
Figure 3. Complex coacervation of recombinant NICD in vitro with scGFP(+25).
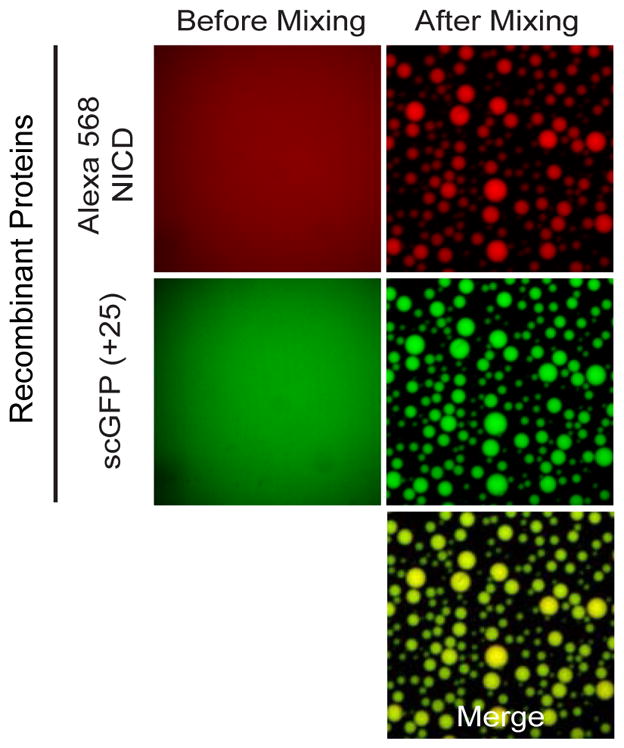
Alexa 568-NICD and scGFP(+25) form homogeneous solutions individually, but phase separate together into micron-sized spherical droplets when mixed. See also Supplemental Figure S3.
Phase separation of NICD is promoted by positive charge in partners
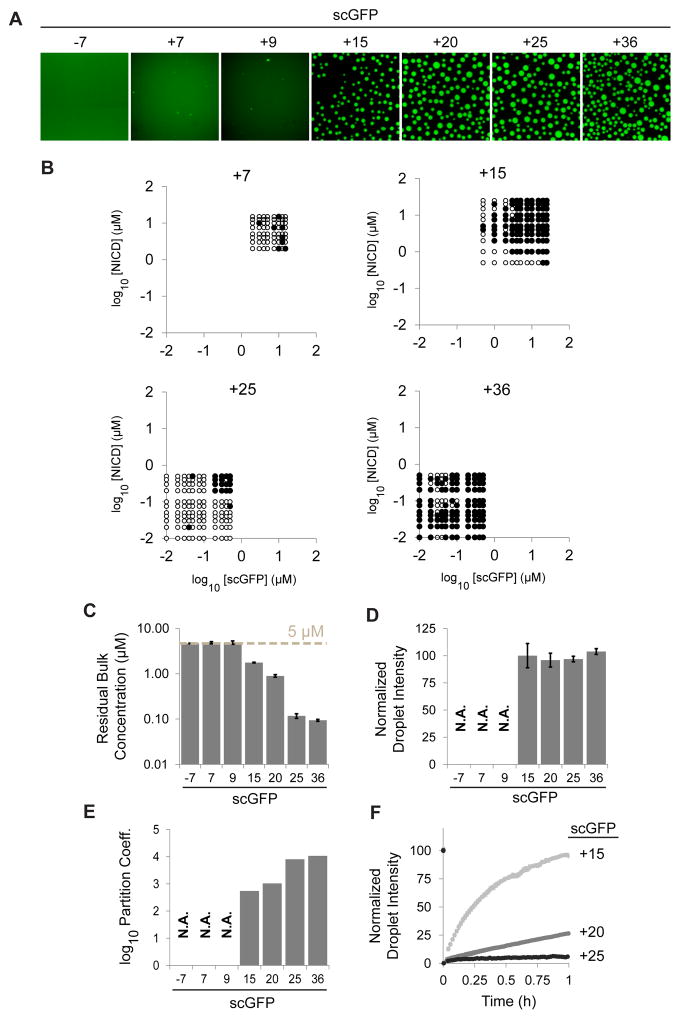
Theory and experiments with non-biological complex coacervation show that the driving force for phase separation and properties of the resulting concentrated phase depend on the charge density and valence of the complexing counterions (Veis, 2011). We explored this using a panel of supercharged GFPs: −7 (WT), +7, +9, +15, +20, +25, +36. We first examined the charge dependence of the saturation concentration for phase separation using two different measures – the appearance of phase separated droplets in fluorescence images across a grid of scGFP and NICD concentrations (Figures 4A, B), and the residual concentration of scGFP in bulk solution after phase separation of 5 μM each of NICD and scGFP (Figure 4C). The two measures showed good agreement. Both revealed that the saturation concentration decreases with increasing positive charge on scGFP, from ~5 μM for scGFP(+7) to 0.01–0.1 μM for scGFP(+36). WT GFP did not promote phase separation up to 10 μM concentrations. The concentration of NICD and (+)-scGFPs in droplets remained constant across the series (Figure 4D). Therefore, the partition coefficient also increased with scGFP charge (Figure 4E). In vitro Cin/Cout ratios (550–10,000) are significantly higher than the maximum in vivo Cin/Cout ratio (30), perhaps due to differences in solubility of the proteins in the cellular environment versus aqueous buffer or to a lower average positive charge on cellular ligands. Finally, FRAP analysis revealed that the movement of scGFP within droplets and its exchange between the droplet and bulk phase decreased with increasing positive charge (Figure 4F), suggesting increased strength of intermolecular interactions with higher charge. Thus, the charge of the added species affects the saturation concentration, degree of partitioning, and dynamics of complex coacervates formed by NICD.
Figure 4. Phase separation of NICD is promoted by positive charge in partners.
(A) Solutions containing 5 μM wild-type GFP (−7) or supercharged GFPs (scGFPs: +7, +9, +15, +20, +25, +36) and 5 μM NICD were imaged by fluorescence microscopy. (B) Different concentrations of NICD and scGFPs were mixed and scored for phase separation (black circle: phase separated; white circle: not phase separated). (C) Solutions containing 5 μM wild-type GFP (−7) or scGFPs (+7, +9, +15, +20, +25, +36) plus 5 μM NICD were clarified by centrifugation, and the concentration of supercharged GFP in the supernatant (residual bulk) was quantified. (D) Fluorescence intensities of phase separated droplets formed with different scGFPs. (E) Partition coefficients of scGFPs (droplet intensity / residual bulk concentration), 5 μM, in the presence of 5 μM NICD. (F) Fluorescence recovery after photobleaching analysis of scGFPs in phase separated droplets formed with NICD. In panels C, D, and F, data are represented as mean ± SEM.
Charge patterning of NICD affects nuclear body formation
We next examined how phase separation is affected by the linear patterning of charged residues in NICD. In the wild type protein, many negatively charged residues are grouped into a series of clusters across the sequence (Figure 5A). We highlight these clusters (Figure 5B), which we refer to as charge interacting elements (CIEs), by identifying regions of at least four consecutive residues with a net charge per residue < −0.35, averaging over a sliding window of five residues. This definition was based on previous work examining the conformational properties of highly charged IDRs (Das and Pappu, 2013) (see Supplemental Experimental Procedures). Although defining these elements represents a simplified approach for capturing the underlying electrostatic interactions, it allows us to quantify negatively charged clusters in a consistent manner, compare among NICD charge mutants, and identify NICD-like sequences based on charge features. There are four CIEs, with a combined length of 24 residues, in wild type NICD (Figures 5C, D). As initial cellular studies found that deletion of the amino-terminal 62 residues of NICD did not affect phase separation (Figure 1C), we focused on patterning in the remainder of the protein. By shuffling charged and polar uncharged residues with respect to one another, we designed NICD charge patterning mutants that increased (charge clustered mutant or CC) or decreased (charge scattered mutant or CS) the local charge density, which alter the CIE count and the total number of residues contained within CIEs (Figures 5A–D), while maintaining overall amino acid composition and hydrophobic/hydrophilic patterning.
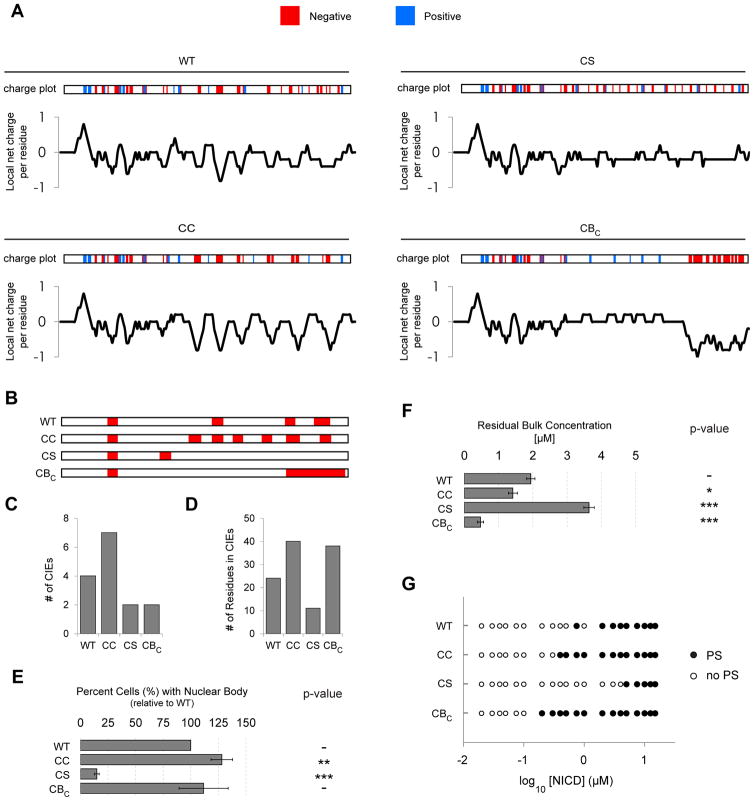
Figure 5. Charge patterning of NICD affects nuclear body formation.
(A) The positions of negatively (D, E; red) and positively (K, R; blue) charged residues (charge plots) and linear charge density scores are shown for wild-type (WT) NICD and charge mutants (CC, CS, CBC). (B) The positions and length of charge interaction elements (CIEs; red) are shown for WT NICD and charge mutants (CC, CS, CBC). (C) The number of CIEs in each sequence. (D) The number of residues in CIE regions for each sequence. (E) Normalized (to WT) percent of HeLa cells containing nuclear puncta when expressing wild type or mutant NICD. (F) The concentration of scGFP remaining in the supernatant (residual bulk) after clarification of solutions containing 5 μM NICD proteins plus 5 μM scGFP(+15). In E and F, data are represented as mean ± SEM and p-values for comparison to WT NICD represent: * < 0.05, ** < 0.01, *** < 0.001. (G) A range of equimolar concentrations of NICD and scGFP(+15) were mixed and scored for phase separation (black circle: phase separated; white circle: not phase separated). See also Supplemental Figures S4 and S5.
Mutant NICD proteins all expressed at similar levels in HeLa cells, and our qualitative conclusions are not dependent on expression level (Figures S4A–D). Wild type NICD formed nuclear bodies in ~70% of transfected cells (a value we normalized to 100% for comparison of different sequences). The CC mutant formed nuclear bodies in more cells than WT NICD (Figure 5E), whereas the CS mutant formed nuclear bodies in significantly fewer (Figure 5E). Similarly, in vitro measurements of the saturation concentration, with scGFPs between +15 and +36, found the CC and CS mutants phase separated at lower and higher concentrations than WT, respectively (Figures 5F, G; see also Figure S5C). Thus, in cells and in vitro, the phase separation of NICD correlates with sequence regions possessing high local charge density.
We also generated shuffled sequence mutants that clustered negatively charged residues into one region of NICD. These mutants are distinguished by the positions of the single, large charge block (CBN,I1,I2,C; Figure S5A). Three of the four CB mutants (CBI1, CBI2, CBC) efficiently formed nuclear bodies (Figures 5A and S5B; see also Discussion). We examined CBC in vitro, and found it to phase separate with equivalent or greater efficiency than WT NICD for a range of scGFPs (Figures 5F, G; see also Figure S5C). Thus, multiple clusters of negative charges distributed across the NICD sequence are not strictly necessary for complex coacervation in vitro or in cells. However, it appears that the total negative charge accumulated within CIEs reflects the determinants of complex coacervation and formation of nuclear bodies by NICD mutants. Specifically, more or denser clusters of negative charges appear to lead to stronger interactions with macro-cations, enabling the charge neutralization and non-covalent crosslinking needed for complex coacervation.
Specific residue types promote formation of nuclear bodies in a sequence-independent fashion
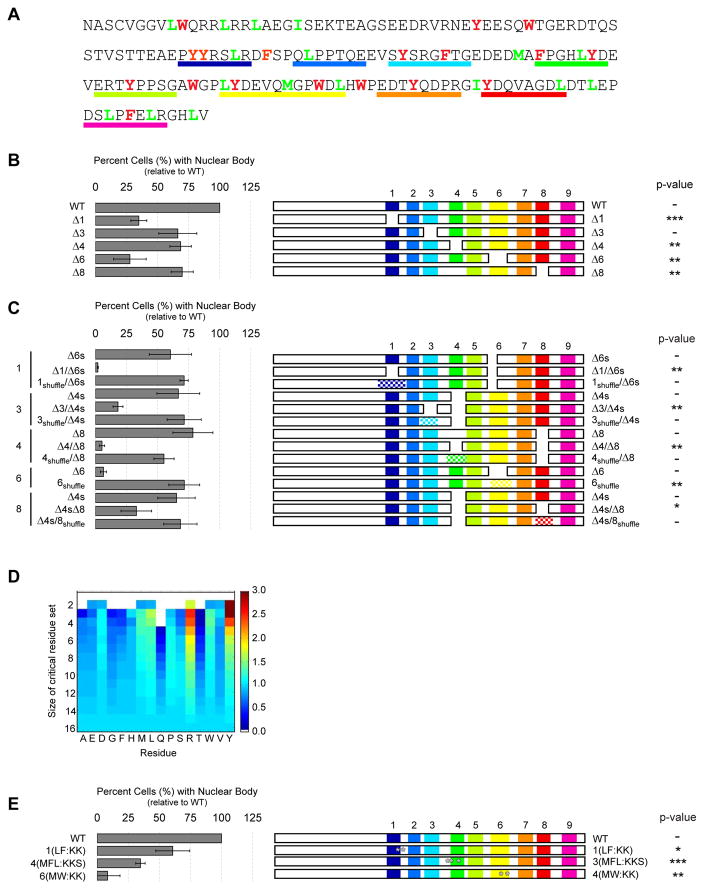
To identify other determinants of phase separation in NICD, we deleted a series of 6–12 amino acid segments (Figure 6A). Nearly all of these deletions reduced the formation of nuclear bodies (Figure 6B). We also generated mutants wherein different pairs of single deletions were combined (Figure S6A). With only a single exception (Δ1/Δ2), double deletions led to a greater reduction in nuclear body formation than the single deletions (Figure S6A). These data suggest that multiple elements distributed throughout the NICD sequence contribute to phase separation.
Figure 6. Identification of residue types that promote nuclear body formation.
(A) Amino acid sequence of NICD. Aromatic and hydrophobic residues are colored red and green, respectively. Regions deleted in panels B, C, and E are underlined in matching colors. (B) Normalized (to WT) percent of HeLa cells containing nuclear puncta (left) when expressing constructs deleted for individual sequence elements (schematically illustrated at right). (C) Quantification of nuclear body formation for locally shuffled sequences. The positions of shuffled sequences are checkered. (D) Heatmap of relative enrichment of specific residue types in deletions that correlated strongly with decreased cellular puncta. Warmer and cooler colors indicate enrichment and depletion, respectively, in critical residue sets. (E) Nuclear body formation by NICD point mutations. Positions of mutations are indicated by white asterisks. In panels B, C and E, data are represented as mean ± SEM and p-values for comparison to WT NICD, respective single deletion mutants and WT NICD, respectively are: * < 0.05, ** < 0.01, *** < 0.001). See also Supplemental Figure S6.
We next examined whether these elements contribute through specific sequences of amino acids, as is typical of most protein-protein interactions, or through more general physical properties that arise from amino acid content, as is more typical of polymer-polymer interactions. We shuffled the residues in individual functionally important regions, while maintaining overall amino acid composition. For each region, we generated multiple shuffled variants (Figure S6B), and compared their ability to form nuclear bodies to that of the native protein and the corresponding deletion mutant. We found that every shuffled variant formed nuclear bodies nearly as well as (and occasionally better than) the native NICD (Figure 6C; see also Figure S6B). Thus, regions of NICD that promote intracellular phase separation act in a sequence-independent fashion.
Previous studies showed that mutation of Tyr and Phe residues in IDRs from Ddx4 and BugZ inhibit their phase separation (Jiang et al., 2015; Nott et al., 2015). Additionally, mutation of Tyr residues in Fus and an IDR from hnRNPA2 prevented partitioning into RNA granules and phase separated droplets, respectively (Kato et al., 2012; Xiang et al., 2015). These studies focused specifically on aromatic residues because of their striking enrichment along with Ser, Gly and Gln residues in the IDRs of various RNA binding proteins (Han et al., 2012; Kato et al., 2012; Nott et al., 2015). NICD is not enriched in any one amino acid type, and has an amino acid compositionmore typical of “generic” IDRs (Figure S7). We thus asked whether any amino acid type contributed more significantly to phase separation, making no a priori assumptions regarding which residue may be of interest. As detailed in Supplemental Experimental Procedures, we addressed this issue through an unbiased statistical analysis of the residue types that are lost in each of the deletions and the effects of these losses on nuclear body formation. We first assessed all possible combination sets of residue types (NICD contains 16 unique residue types) for the correlation between the number of residues from a set lost upon each deletion and the fraction of cells with puncta associated with that deletion (Table S1). For the sets of residues that correlated most strongly (critical residue sets), we then determined the enrichment of each residue type within each set relative to a random prior in a set-size matched manner (Figure 6D and Table S2). Heatmap values greater than one indicate relative enrichment. Deletion of Tyr correlated most strongly with the loss of NICD bodies, followed by Arg, Leu, Met and Trp, and Asp (Figure 6D). We mutated aromatic or hydrophobic residues at several positions in the NICD sequence to test their impact on phase separation. In each case, the mutations reduced nuclear body formation (Figure 6E). Some point mutants had a greater effect than deletion of the corresponding residues and their neighbors (compare to deletions shown in Figures 6B, C), likely because of the introduction of positively charged Lys residues, which also decrease negative charge density (Figure S6C). These data suggest that aromatic and hydrophobic residues play important roles in increasing the driving force for phase separation.
Discussion
NICD forms nuclear bodies when expressed in mammalian cells. Our cellular and biochemical data indicate that NICD phase separates via complex coacervation, requiring interactions with positively charged partners. Charge neutralization, which is governed by the local linear charge density, and interactions involving aromatic and hydrophobic residues appear to be the main drivers of NICD phase separation; each driver correlates with the saturation concentration for phase separation and physical properties of droplets. Statistical analysis of deletions and mutagenesis indicate that aromatic (Tyr, Trp) and hydrophobic (Leu, Met) residues, which are distributed across the NICD sequence, contribute to phase separation. The overall amino acid composition coupled to charge patterning seems to suffice for driving phase separation, i.e., the precise sequence of NICD appears to matter little.
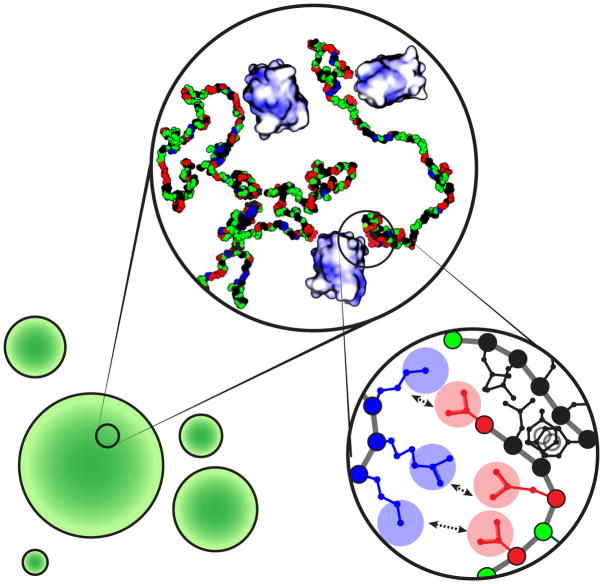
Our observations lead to a mechanistic model for NICD phase separation that is based on a hierarchy of interaction ranges and strengths (Figure 7). Long-range electrostatic repulsions among negatively charged interaction elements must be weakened in order to draw NICD into dense droplets. The dense phase is further stabilized by short-range interactions involving aromatic and hydrophobic residues. Electrostatic interactions with positively charged partners appear to neutralize the negative charge of NICD molecules and serve as non-covalent crosslinks (Figure 2D and S2B). In vitro we have observed NICD phase separation promoted by scGFP proteins and oligoArg peptides. In cells, it is likely that NICD has many partners, with varying degrees of charge and stoichiometry of interaction, that collectively promote phase separation in the nucleus. Notably, many RNA- and DNA-binding proteins, which are localized to the nucleus and are highly basic, could play this role. The strength of complementary electrostatic interactions and their contributions to phase separation increase with total positive charge on scGFP/oligoArg, and are modulated by the charge distribution along the NICD. Clustering Asp/Glu residues to produce one or more blocks with high local charge density promotes phase separation, whereas distributing these residues more evenly attenuates phase separation (see Figure 5 and S5). The exception is the sequence of CBN, which attenuates the driving force for phase separation. This warrants further investigation although atomistic simulations suggest that CBN is more collapsed than the other CB mutants. This collapse derives from intramolecular electrostatic contacts between the negative charge block and a region in the N-terminus. This might limit the accessibility of CIEs to multivalent counterions thus weakening the driving force for phase separation via complex coacervation.
Figure 7. Model depicting the hierarchies of interactions that drive NICD phase separation via complex coacervation.
Structures of phase separated droplets on three length scales, micrometer (bottom left), nanometer (top center), and atomic (bottom right). On the micrometer scale NICD forms liquid-like spherical droplets. Phase separation of NICD requires a multivalent counterion such as the positively charged supercharged GFP (scGFP). On the nanometer scale NICD is depicted as chains with a single bead per residue, which contact one another indirectly through scGFP counterions that likely bind the negatively charged clusters along the contour of NICD and directly through aromatic / hydrophobic interactions. In this representation, Asp / Glu residues are shown in red, Lys / Arg in blue, polar residues in green, and aromatic / hydrophobic residues in black. The scGFP(+36) molecules are shown in electrostatic surface representations. Dark blue patches indicate high positive surface charge. At the atomic scale, complementary electrostatic interactions as well as interactions involving aromatic (Tyr) and hydrophobic (Leu) residues physically crosslink NICD molecules. Note that the structural nature of these interactions remains unknown and should not be inferred from schematic image.
In addition to charge, Tyr, and to a lesser degree Leu, Met, and Trp residues also contribute to NICD phase separation. Interactions of hydrophobic residues are inherently short range, and mediate multivalent adhesions among NICD molecules. These contacts are weaker analogs of domain-ligand interactions that promote phase separation of modular signaling proteins (Banjade and Rosen, 2014; Banjade et al., 2015; Fromm et al., 2014; Li et al., 2012). The importance of these residue types in promoting phase separation and/or partitioning into the droplet phase is also suggested by mutagenesis of aromatic residues in the IDRs of Ddx4, FUS and BugZ (Jiang et al., 2015; Kato et al., 2012; Nott et al., 2015; Xiang et al., 2015). Rather than mutating specific residues, we approached the issue agnostically, making deletions across the sequence and then using statistical analyses to identify functionally important residue types. Moreover, unlike Ddx4 and FUS, which form cellular bodies as part of their normal functions, NICD has not evolved to form nuclear bodies (although see below). Nevertheless, we arrive at a similar conclusion, i.e., that Tyr is especially enriched in segments whose deletion is most deleterious to intracellular phase separation. This suggests that aromatic residues play an important role in promoting the phase separation of disordered proteins.
No single linear motif or specific amino acid sequence in NICD is crucial for driving phase separation. Instead the overall amino acid composition combined with the patterning of negatively charged residues and the distribution of aromatic residues along the linear sequence appears to be important. This indicates that adhesive structures form through non-specific interactions, perhaps through side-chain contacts alone, or through additional backbone contacts such as cross-beta strands. Thus, once drawn together by charge neutralization, NICDs appear to form labile complexes in which weak aromatic/hydrophobic and electrostatic interactions are made and broken rapidly, imparting liquid-like behavior to the phase separated state.
Recent studies of IDRs that phase separate have focused on two distinct archetypes. The first is enriched in polar residues such as Gly, Ser, Gln, and Asn, as in the IDRs of FUS and hnRNPA1. The second is enriched in charged residues, but the overall net charge per residue is low, and oppositely charged residues are segregated into blocks along the linear sequence, as in the IDRs of Ddx4 and LAF-1. Both archetypes phase separate via simple coacervation, and do not appear to require heterotypic interactions with a partner. NICD represents a third archetype. It is similar to Ddx4 and LAF-1 in that roughly a quarter of its residues are charged and the charges are clustered into blocks. However, NICD has a substantial net negative charge with more than twice as many negatively charged residues as positively charged ones. These properties lead to the requirement for positively charged multivalent ligands / counterions that drive phase separation via complex coacervation. The data to date indicate that three different strategies, based on the three archetypes of IDRs, can promote intracellular phase separation.
We analyzed the human proteome (UniProt: UP000005640_9606) using the consensus disorder prediction database MobiDB 2.0 (Potenza et al., 2015) to identify long (>100 residue) IDRs with sequence properties similar to NICD. We sought IDRs with a fraction of charged residues >0.25 and a two-fold excess of negatively charged residues over positively charged residues. We also required that at least twenty residues be encompassed by CIEs. Based on this analysis we identified 464 unique NICD-like IDRs from 443 unique proteins (Table S3). Further filtering of this list for sequences that had Tyr and/or Leu content >=6% of total residues in disordered regions identified 260 unique proteins. Performing Gene Ontology (GO) molecular function overrepresentation (experimental only) using PANTHER (Release 2015-08-06) (Mi et al., 2013) on these two sets identified significant enrichment in GO terms associated with nucleic acid binding (Table S4) and regulation of nucleic acid biosynthesis (Table S5). Furthermore, a number of these proteins are annotated as being localized in nuclear body structures, particularly the nucleolus (Tables S3, S4 and S5). It is possible that some of these proteins contribute to the formation of nuclear bodies through complex coacervation. In addition, since many positively charged proteins bind to RNA/DNA, it is possible that complex coacervation driven by NICD-like IDRs could compete with complex coacervation of RNA/DNA binding proteins and nucleic acids. This suggests the possibility that NICD-like IDRs might be effective at dissolving ribonucleoprotein bodies and/or sequestering molecules that otherwise partition into ribonucleoprotein bodies, as NICD appears to do with nuclear paraspeckles (Figure S1B).
We used NICD as a model system to understand the phase separation of an archetypal disordered protein. However, these findings also have potential implications for the biology of Nephrin. In mammals, Nephrin is a transmembrane adhesion receptor expressed primarily in podocyte cells of the kidney. Tyrosine phosphorylation of the intracellular domain of Nephrin, and consequent binding of Nck and assembly of cortical actin, are necessary for proper formation of the filtration barrier of the kidney (Jones et al., 2006). Interactions between phospho-Nephrin, Nck and N-WASP result in Nephrin phase separation into membrane attached puncta in vitro and in cells (Banjade and Rosen, 2014)(S. Kim and M.K. Rosen, unpublished). Assembly of these puncta has been attributed to multivalent SH2-phosphotyrosine and SH3-polyPro interactions. However, the SH2 domain, the first SH3 domain, and an adjacent linker region of Nck are basic (predicted pI ~9). N-WASP also has a poly-basic region. It is possible that these basic elements act analogously to scGFP or oligoArg peptides to promote Nephrin phase separation. Thus, non-specific charge-mediated interactions, along with specific modular domain interactions, could contribute to the formation of membrane puncta by the ternary Nephrin/Nck/N-WASP system. The disordered cytoplasmic regions of a number of transmembrane signaling proteins, including LAT, FAT1, and PDGFR, are also highly acidic, and interact with adaptor proteins that also have basic elements. Thus, it is possible that complex coacervation could contribute more broadly to the cytoplasmic clustering and intracellular phase separation of membrane-anchored proteins.
Experimental Procedures
Analysis of NICD nuclear bodies in cells
Human NICD was PCR amplified from a plasmid encoding CD16/7-Nephrin-GFP (a gift from Drs. Jones and Pawson) and inserted into a CMV promoter driven EGFP fusion vector. NICD mutants were produced using PCR with appropriate primers, and confirmed by sequencing (Table S6). HeLa cells were cultured in DMEM supplemented with 10% FBS, GlutaMAX and Pen/Strep mix. NEAT1 −/− and +/+ MEFs were cultured in a 1:1 mixture of DMEM and Ham’s F-12, with the same additives. For imaging experiments, cells were cultured in glass bottom dishes prior to transfection using Lipofectamine 2000 (Thermo Fisher). Live HeLa cells were imaged using a confocal or widefield microscope on a heated stage. For FRAP analysis, nuclear bodies were photobleached using a 488 nm laser, and custom ImageJ scripts were used to determine the area of and intensity within the photobleached region at each time point. NICD nuclear bodies in fixed HeLa cells were imaged in 3D at high-resolution using spinning disc confocal microscopy, and analyzed in a semi-automated fashion. All images were background-subtracted and flatfield-corrected, and cellular autofluorescence was determined to be negligible. The volumes of nuclear bodies (VNBs) larger than the transverse resolution limit were determined from their diameter and an assumption of spherical shape. Volumes of smaller nuclear bodies and intensities of all nuclear bodies (INBs) were calculated using a calibration that relates the effects of the point spread function on intensity. Molar fraction was calculated as [(Vtotal,NBs) * (Imean,NBs)] / [(Vtotal,nucleus) * (Imean,nucleoplasm)], and partition coefficients for each nuclear body were calculated as (Imean,NBs) / (Imean,nucleoplasm). Cell populations were scored for the presence of NICD nuclear bodies in transfected HeLa cells (which are fluorescent), when imaged with a Deltavision RT widefield microscope. Quantification of nuclear body formation was performed at unmatched and matched expression levels; data are shown as mean ± SEM. Detailed procedures are provided in Supplemental Experimental Procedures.
Atomistic simulations of NICD
Simulations used the CAMPARI Monte Carlo (MC) modeling suite (http://campari.sourceforge.net), with the ABSINTH implicit solvent model and force field paradigm (Vitalis and Pappu, 2009). Each simulation was initiated from a random, non-overlapping starting conformation. Single protein simulations used temperature replica exchange Monte Carlo (T-REMC), and all analysis was performed on conformations from the 310 K ensemble. Dimer simulations were run at 310 K. Detailed procedures for running simulations and their analysis are provided in Supplemental Experimental Procedures.
Sequence analysis
Sequence analysis for identifying CIEs was performed using localCIDER (Holehouse et al., 2015). The parameters used to define CIEs were based on previous work in polymer and polyampholyte physics. Pearson’s correlation coefficients between specific residue types and nuclear body formation scored how well the loss of unique combinations of residues affected the formation of nuclear bodies. All unique combinations of residues were considered. Variance in the experimental data was accounted for by calculating a distribution of correlation values using subsampled datasets. Significant differences were evaluated using unpaired Student’s t-tests, and residues which were consistently enriched relative to a normalized background were identified. Detailed procedures are provided in Supplemental Experimental Procedures.
Production of proteins and peptides
Recombinant NICD protein was expressed with two N-terminal tandem MBP domains and a C-terminal His8 tag, all Tev cleavable. Mutant NICD sequences (charge: CC, CS, CBC) were subcloned from the mammalian expression system, and expressed in the same fashion. NICD and scGFPs proteins were expressed in E. coli BL21(DE3)T1R and purified using a combination of affinity, ion exchange and gel filtration chromatography. NICD was labeled with Alexa Fluor 568 maleimide on the single cysteine. Two synthetic peptides, CR7 (sequence = CRRRRRRR), and CR20 (sequence = CRRRRRRRRRRRRRRRRRRRR), were synthesized and purified by GenScript (NJ, USA). Detailed procedures are provided in Supplemental Experimental Procedures.
Reconstitution and characterization of in vitro NICD phase transition
In vitro phase separation assays were mixed NICD and/or scGFP directly in 384-well plates at the indicated concentrations. After 24 hours at room temperature in the dark, mixtures were imaged by confocal microscopy. Saturation concentrations were determined by automated image scoring of a grid of NICD and scGFP concentrations for the presence of puncta. Alternatively, Alexa-568 NICD/scGFP solutions were clarified by centrifugation and the residual concentration of NICD/scGFP in the supernatant was determined by fluorescence using standards of known protein concentrations. Phase separated droplet sizes and normalized intensities were measured using custom ImageJ scripts, where droplet intensity values were normalized to those of WT NICD + scGFP(+15). Data are represented as mean ± SEM. FRAP analysis of phase separated droplets of NICD/scGFP used a 405 nm laser for photobleaching. Detailed procedures are provided in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Thomas Scheuermann for help with CD experiments and analysis; Kate Luby-Phelps and Abhijit Bugde in the UTSW Live Cell Imaging Facility for assistance with microscopy; Shinichi Nakagawa for sharing NEAT1 MEFs; Nina Jones and Tony Pawson for sharing Nephrin expression constructs; and Tyler Harmon for discussion. Research was supported by the Howard Hughes Medical Institute (M.K.R., D.R.L.), grants from the NIH (R01-GM56322, M.K.R., R01-NS056114, R.V.P., R01-GM095501, D.R.L. and F32-DK091074, C.W.P.), the Welch Foundation (I–1544, M.K.R.). The UTSW Live Cell Imaging Facility is a Shared Resource of the Harold C. Simmons Cancer Center and is supported in part by NCI Cancer Center Support Grant, 1P30 CA142543-01.
Footnotes
Author Contributions
Conceptualization, CWP, RVP, and MKR; Methodology, CWP, SBP, ASH, RVP, and MKR; Software, CWP, SBP, AM, and ASH; Formal Analysis, CWP and ASH; Investigation, CWP, MK, SBP, ASH, and RA; Resources, AY and DRL; Writing – Original Draft, CWP, RVP, and MKR; Writing – Review & Editing, CWP, SBP, ASH, RVP, and MKR; Visualization, CWP, SBP, ASH, RVP, and MKR; Funding Acquisition, RVP and MKR; Supervision, RVP and MKR.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014:3. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, Rosen MK. Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc Natl Acad Sci U S A. 2015;112:E6426–6435. doi: 10.1073/pnas.1508778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A. 2015;112:E5237–5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nature Physics. 2015;11:899–904. [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proceedings of the National Academy of Sciences. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K, Bromberg S. Molecular driving forces: statistical thermodynamics in biology, chemistry, physics, and nanoscience. Garland Science 2010 [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm SA, Kamenz J, Noldeke ER, Neu A, Zocher G, Sprangers R. In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew Chem Int Ed Engl. 2014;53:7354–7359. doi: 10.1002/anie.201402885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Holehouse AS, Ahad J, Das RK, Pappu RV. CIDER: Classification of Intrinsically Disordered Ensemble Regions. Biophysical Journal. 2015;108:228a. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, Zheng Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N, Blasutig IM, Eremina V, Ruston JM, Bladt F, Li H, Huang H, Larose L, Li SS, Takano T, et al. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature. 2006;440:818–823. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proc Natl Acad Sci U S A. 2009;106:6111–6116. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic acids research. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Bellingham CM, Stahl RJ, Sitarz EE, Lane CJ, Keeley FW. Sequence and structure determinants for the self-aggregation of recombinant polypeptides modeled after human elastin. J Biol Chem. 2003;278:48553–48562. doi: 10.1074/jbc.M308465200. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Potenza E, Di Domenico T, Walsh I, Tosatto SC. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic acids research. 2015;43:D315–D320. doi: 10.1093/nar/gku982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Veis A. A review of the early development of the thermodynamics of the complex coacervation phase separation. Advances in colloid and interface science. 2011;167:2–11. doi: 10.1016/j.cis.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A, Pappu RV. ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J Comput Chem. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.