ABSTRACT
Here we describe protein-protein interactions between signaling components in the conserved self-incompatibility pathway from Brassica spp. and Arabidopsis lyrata. Previously, we had demonstrated that ARC1 is necessary in A. lyrata for the rejection of self-pollen by the self-incompatibility pathway. The results described here demonstrate that A. lyrata ARC1 interacts with A. lyrata S Receptor Kinase (SRK1) in the yeast 2-hybrid system. A. lyrata ARC1 also interacted with B. napus SRK910 illustrating that interactions in this pathway are conserved across species. Finally, we discuss how the more widely occurring interactions between SRK and ARC1-related family members may be modulated in vivo by expression and subcellular localization patterns resulting in a particular response.
KEYWORDS: Brassicaceae, plant U-box proteins (PUBs), protein-protein interactions, S-Domain 1 Receptor like Kinases (SD1-RLKs), self-incompatibility, yeast two-hybrid
Flowering plants employ a wide variety of strategies to achieve reproductive success and prevent self-pollination by selecting for desirable pollen grains. One way to encourage outcrossing is the development of a self-incompatibility system that allows for the recognition and rejection of self-pollen. Self-incompatibility systems have been studied in several major families of flowering plants including the Brassicaceae, Papaveraceae and Solanaceae and with recent genomics advances there has been a great deal of progress into the understanding of the molecular mechanisms that underlie self-incompatibility in these families.1,2 In the mustard family of plants (Brassicaceae), the majority of research into the self-incompatibility signaling pathway has been performed in Brassica species (B. napus, B. oleracea, B. rapa) but less so in other genera in comparison to Brassica spp. This previous research included the elucidation and establishment of the initial signaling events that recognize self- versus non-self-pollen.2 In the Brassicaceae the stigmas are a dry type, where the water for pollen grain hydration must be supplied from the female stigmatic papillae to the male pollen grain.3 When a self-incompatible pollen grain lands on the stigma surface, the pollen is rejected by the preventing both pollen grain hydration and pollen tube entry through the stigmatic papillae; thus, blocking fertilization. In Brassica spp., in the event of a self-incompatible pollination, the S-locus Receptor like Kinase (SRK) in the stigmatic papillae4,5 is able to recognize the pollen ligand, displayed on the pollen coat, known as S-locus Cysteine Rich/S-locus Protein 11 (SCR/SP11).6-9 The recognition of SCR by SRK leads to the phosphorylation and activation of SRK.10-12 Once phosphorylated and activated, SRK is able to phosphorylate M-locus Protein Kinase (MLPK), a cytoplasmic receptor like kinase.13,14 SRK and MLPK then are proposed to phosphorylate and activate an E3 ubiquitin ligase, Arm Repeat Containing 1 (ARC1).11,12,15,16 ARC1 is able to ubiquitinate Exo70A1, a component of the exocyst complex, which mediates secretory vesicle delivery to the point of compatible pollen contact. As ARC1 is able to ubiquitinate Exo70A1, it is likely either redirecting the localization of Exo70A1 or sending Exo70A1 to the 26S proteasome for degradation.17 Regardless of the mechanism by which Exo70A1 is removed from the point of pollen contact, the end result is that secretory vesicle delivery is blocked and therefore, self-compatible pollen grain hydration is prevented.18
Although all of this research was conducted in Brassica spp., the self-incompatibility system is found in many other species of the Brassicaceae. As a result of genome sequencing it has become possible to study these signaling components in species that are less closely related to Brassica spp such as Arabidopsis lyrata and Capsella grandiflora.19-21 When examining self-incompatibility in species such as A. lyrata there were several outstanding questions. Are all of the components in this pathway conserved across all of the Brassicaceae? Specifically, do the signaling pathways work the same way between A. lyrata and Brassica spp? This is in the context of the fact that Brassica and Arabidopsis diverged from each other approximately 20–40 MYA.22 Research in Brassica (Lineage II of the Brassicaceae) has clearly demonstrated that the SCR-SRK-ARC1 signaling pathway is required for self-pollen rejection while the requirement of the SCR-SRK-ARC1 signaling pathway for self-pollen rejection was not clear in a species in Lineage I such as A. lyrata.12,16,21,23
To determine if the SCR-SRK-ARC1 signaling pathway is conserved in Lineage I of the Brassicaceae,22 we selected self-incompatible A. lyrata to examine the role of SCR-SRK-ARC1. A. lyrata is closely related to A. thaliana which is self-compatible and is missing the self-incompatibility signaling components; SCR, SRK and ARC1.22-26 We found that ARC1 was necessary for self-pollen rejection, as when the expression of ARC1 was knocked down through RNAi in self-incompatible A. lyrata, the transgenic plants were able to accept self-pollen that it should have rejected.24 As well, through a comparative genomics analysis of several Brassicaceae genomes including A. lyrata, A. thaliana, A. arabacium, B. rapa, C. rubella, L. alabamica, S. irio, T. halophila, T. parvula, we discovered that the ARC1 gene was frequently deleted in compatible species.24 Thus, based on these findings ARC1 was proposed to play a major role in self-incompatibility systems and its presence in the genome was correlated with the presence of a self-incompatibility system in the Brassicaceae.27
Further experiments focused on the conservation of the SCR-SRK-ARC1 signaling pathway by using self-compatible A. thaliana as a heterologous system. The majority of A. thaliana ecotypes lack a functional ortholog to SCR, SRK and ARC1.24-26 When SCR-SRK-ARC1 were expressed in A. thaliana, this led to the reconstruction of a functional self-incompatibility signaling pathway where self-pollen was rejected and resulted in a significant decrease in seed set.27-29 These transgenic studies as well as others have been very through in examining the phenotype of self-pollinations when self-incompatibility signaling components are all expressed in A. thaliana. But the previous results from studies into self-incompatibility from Brassica species were underpinned by the use of yeast 2-hybrid assays.11,15,30-34 These yeast 2-hybrid screens lead to the initial identification of several signaling components including ARC1, as it was identified from a screen for interactors with the cytoplasmic domain of SRK.15,30 Therefore, following the recent work on transgenic A. lyrata RNAi plants and the self-incompatible transgenic SCR-SRK-ARC1 A. thaliana, an outstanding question is whether A. lyrata SRK and ARC1 interact similarly to the previously observed results for Brassica SRK and ARC1 in the yeast 2-hybrid system.15,31-34
To examine interactions between A. lyrata SRK and ARC1, a pairwise yeast 2-hybrid interaction screen was performed. ARC1 and 2 other Plant U-box (PUB) proteins, PUB17 and PUB29,11 were included in this experiment (Fig 1A). Bn-ARC1, Al-ARC1 and Al-PUB17 have the same domain organization with a N-terminal UND domain, U-box and a C-terminal ARM repeat domain (Fig 1A) and Al-ARC1 and Al-PUB17 are 66% similar based on protein identity.35,36 The UND domain likely contributes to PUB protein target specificity, the U-box domain is known to interact with the E2 conjugating enzyme, and the ARM repeat domain is also an interaction domain that can bind to cytosolic kinase domains from S Domain-1 (SD1) Receptor-Like Kinases (RLKs).11,15 A. thaliana At-PUB29 belongs to a smaller class of PUB proteins that lack a UND domain but still contain the conserved U-box and ARM repeat domain (Fig. 1A). At-PUB29 was previously found to have very little or no interactions with kinase domains from SD1-RLKs.11 The original interaction between Bn-SRK910 and Bn-ARC1 was demonstrated using the cytosolic kinase domain from Bn-SRK910 and the C-terminal ARM repeat domain from Bn-ARC1.15 Thus, ARM-repeat domains from ARC1, PUB17 and PUB29 were tested in this experiment (Fig. 1A). The cytosolic kinase domains used in this analysis were from A. lyrata SRK1, B. napus SRK910, and the A. thaliana LRR Receptor kinase, HAESA,37 which was previously used a negative control for SRK-ARC1 interactions11,15,32 (Fig. 1A). As in previous studies,11,15,32 the interactions of these proteins in the yeast nucleus were dependent on using constructs without the extracellular and transmembrane domains, but included the entire cytosolic domains starting right after the transmembrane domains as shown in Fig. 1A. Both the Al-SRK1 and Bn-SRK910 constructs started immediately after the tryptophan as we found that the ability of SRK to interact with the ARM repeat domains was drastically reduced when the residues of the juxtamembrane domain, between the transmembrane domain and the start of the kinase catalytic domain were excluded (data not shown).
Figure 1.
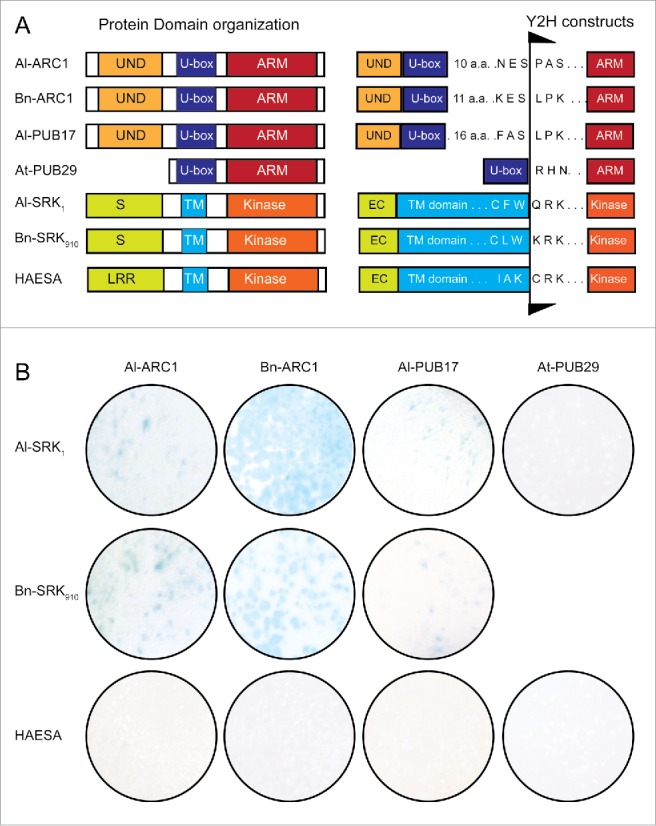
Yeast 2-hybrid interactions between A. lyrata SRK and ARC1. (A) Protein domains and yeast 2-hybrid constructs. For the PUB proteins, only the ARM domains were cloned into pVP16. For the receptor kinases, A. lyrata SRK1 and B. napus SRK910 both have an extracellular domains belonging to the SD1 subfamily while A. thaliana HAESA contains a LRR extracellular domain. All three transmembrane receptor kinases contain a cytosolic kinase domain at the C-terminus, and this region was cloned into pBTM116. (B) β-galactosidase activity from the yeast 2-hybrid filter lift assays. All proteins were analyzed in a pair-wise fashion of each kinase domain with each ARM domain. A. lyrata SRK1 interacts with A. lyrata ARC1, B. napus ARC1 and A. lyrata PUB17 but not A. thaliana PUB29. B. napus SRK910 interacts with A. lyrata ARC1, B. napus ARC1 and A. lyrata PUB17. B. napus SRK910 was previously shown not to interact with A. thaliana PUB29.11 The HAESA kinase domain does not interact with any of the PUB proteins.
Pairwise interactions were tested between the kinase domains and the ARM repeat domains (Fig. 1B), and as a direct comparison and control we included the previously characterized BnARC1 and BnSRK910. Interactions were assessed based on the activation of lacZ reporter gene (a blue color generated by the β-galactosidase activity, Fig. 1B). As predicted from previous studies, the yeast 2-hybrid results confirmed that the interaction of ARC1 with SRK is conserved within species and show cross-species interactions as well. Al-ARC1 was found to interact with both Al-SRK1 and Bn-SRK910 even though this latter SRK was from a different Brassicaceae species. Consistent with previous experiments, Al-ARC1 did not interact with kinase domain from HAESA (which is not involved in self-incompatibility37). Similarly, Bn-ARC1 interacted with both Al-SRK1 and Bn-SRK910 but not with HAESA (Fig. 1B). In addition to ARC1, we examined the interaction of the next most closely related PUB protein from A. lyrata, PUB17.24 Al-PUB17 showed a similar pattern of interactions with positive results for Al-SRK1 and Bn-SRK910, but not with HAESA. Lastly as expected, the more divergent At-PUB29 was not able to interact with any of the kinase domains (Fig. 1B).11 We then performed a yeast 2-hybrid dilution series with the same pairwise combinations on plates (Fig. S1). lacking histidine to assess for the activation of the second reporter gene, HIS3 (Fig. S1). Visible growth at the 1:100 dilution was seen for pairwise combinations between Bn-SRK910 and Bn-ARC1, Al-ARC1 or Al-PUB17, and for pairwise combinations between Al-SRK1 and Bn-ARC1, Al-ARC1 or Al-PUB17. Colonies were not seen at the 1:100 dilution for pairwise combinations between Bn-SRK910 or Al-SRK1 and At-PUB29 (Fig. S1). Thus, these results matched the interaction patterns detected from the lacZ reporter gene as shown in Fig. 1B.
In conclusion, by testing pairwise yeast 2-hybrid interactions, we were able to determine that Al-ARC1 could interact with Al-SRK1 as predicted from prior 2-hybrid interactions studies between Brassica ARC1s and SRKs.15,31,33,34 These results reinforce our functional transgenic studies examining the role of Al-ARC1 in self-incompatible A. lyrata with the Al-SRK1 haplotype. When Al-ARC1 was knocked down via RNAi in these A. lyrata plants, the transgenic pistils were able to accept self-pollen, instead of rejecting it.24 The 2-hybrid results presented here reinforce our hypothesis that the SRK-ARC1 pathway is conserved in the Brassicaceae through cross-species interactions observed between Al-ARC1 and Bn-SRK910, and Bn-ARC1 and Al-SRK1.24,27,28 This conservation was also see in our recent study where both Al-ARC1 and Bn-ARC1 were functional in producing self-incompatible A. thaliana plants when co-transformed with the Al-SRKb and Al-SCRb transgenes.28,29 Thus, these experiments demonstrate that despite millions of years of divergence between the Brassica and Arabidopsis species, the functional interaction between SRK and ARC1 is conserved.
One aspect of these results that may appear counterintuitive at first, was the interaction that was observed for Al-PUB17 with Al-SRK1 and Bn-SRK910, as Al-PUB17 does not appear to be involved in self-incompatibility.24 Previously, we have observed that 2 other closely related PUB proteins with the UND/U-box/ARM domain organization, PUB13 and 14, can interact with cytosolic domains from Bn-SRK910 and several Arabidopsis SD1-RLKs in a pairwise yeast 2-hybrid interaction panel.11 Thus, these closely related ARM domains likely represent binding modules to kinases from the SD1-RLK class, and perhaps a combination of expression patterns with other regulatory events confer greater specificity in planta. A recent study on PUB17 in potato and Nicotiana benthamiana plant immunity has shown that PUB17 is acting in the nucleus; thus illustrating that subcellular localization is an important parameter in defining PUB protein function.38 In this context, perhaps other proteins present in the stigmatic papillae help to facilitate a specific interaction between ARC1 and SRK. For example, recently a Brassica oleracea J domain protein, Bo-JDP1, was found to specifically interact with Bo-ARC1 and Bn-ARC1, but not with At-PUB14 and At-PUB17.39 Interestingly, the transient expression of a JDP1 subdomain (JDP169–344) with Bo-ARC1 in Arabidopsis protoplasts resulted in both proteins co-localizing to the plasma membrane.39 Thus, perhaps a protein such as JDP1, may play a role in mediating interactions between ARC1 and SRK at the stigmatic papillar plasma membrane, and proteins with a similar function to that of JDP1 may add greater specificity to interactions between SD1-RLKs and the PUB proteins in vivo.
Materials and methods
The LexA-VP16 system40 was used for this yeast 2-hybrid study as previously described.12,15,32 Specific yeast 2-hybrid constructs were generated with the respective domains shown in Fig. 1A (starting amino acid sequences for each protein domain is shown). All PCR products were amplified with Phusion Polymerase (Invitrogen), and first subcloned into pGEMTeasy (Promega) for verification by sequencing at the Genome Quebec facility. The Al-ARC1 and Al-PUB17 ARM domains were PCR amplified using primers with BamHI and NotI restriction sites for cloning into the pVP16.40 The forward and reverse primers used for Al-ARC1 were 5′-TGGATCCCTGCTTCGGTTCTTCAAACAAGA-3′ and 5′-GCGCGGCCGCTCACAAAACAGATACAGGTATAG-3′; and for Al-PUB17 were 5′-TGGATCCCCTTTGCTTCGGCTCTTCCGACG-3′ and 5′-GCGCGGCCGCTCACAACACAGGTACGGAGATTG-3′. The Al-SRK1 cDNA was cloned using 3′ RACE on A. lyrata stigma cDNA using the Clonetech RACE kit. The Al-SRK1 cytosolic domain was then PCR amplified using primers with BamHI and SalI restriction sites for directional cloning into pBTM116.40 Al-SRK1 forward and reverse primers were 5′-GGATCCCCCAAAGGAAACTGAAGCGAACAGGAGCAGC-3′ and 5′-GTCGACTTACCGAGCGTTGATGACTGAGG-3′. GenBank Accession numbers for Al-SRK1 is KF418159 and for Al-ARC1 is KF418158. The yeast 2-hybrid plasmids for At-PUB29 (in pVP16) and HAESA (in pBTM116) were from Samuel et al.11
For the yeast transformations, the Saccharomyces cerevisiae L40 strain was transformed using a single step lithium acetate protocol from Gietz and Woods.41 Transformants with both the pair-wise combination plasmids were selected for on synthetic complete drop out medium lacking leucine and tryptophan (SC-Leu,-Trp) and the plates with supplemented adenine hemisulfate (100 mg/L). Transformed yeast were grown at 30°C for 2 d and then examined using the yeast colony filter-lift assay for β-galactosidase activity (Clontech Yeast Protocols Handbook; PT3024-1, July 2009). Positive interactions were defined as turning blue before the known negative controls that should lack activation of the lacZ reporter gene. All photos were taken immediately upon a clear blue color being detected in the experimental samples, and the negative controls were photographed concurrently, in this system approximately 2 hrs after the start of the β-galactosidase activity assay. The yeast transformations and filter-lift assays for β-galactosidase activity were repeated several times to validate results, and representative images are shown in Fig. 1B. The yeast pairwise serial dilution assay tested for activation of the HIS3 reporter gene by plating transformed yeast on SC-Leu,-Trp,-His media as according to.42 The media was also supplemented with 5 mM 3-Amino-1,2,4-triazole (3-AT) to reduce background activation of the HIS3 reporter gene. Single colonies from a 2 day old plate were re-suspended to an OD600 = 1 (1 × 107 cells). The yeast were then diluted for a 1:10 and 1:100 dilution of the stock. Five uL of each series of dilutions were plated out, grown for 2 d at 30°C, and then photographed. This experiment was repeated 3 times with similar results each time.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by university start-up funding to E.I., and a grant from the Natural Sciences and Engineering Research Council of Canada to D.R.G.
References
- 1.Charlesworth D. Plant sex chromosomes. Gen Dynam 2008; 4:83-94; PMID:19056969; http://dx.doi.org/21968124 10.1159/000126008 [DOI] [PubMed] [Google Scholar]
- 2.Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol 2012; 15:78-83; PMID:21968124; http://dx.doi.org/ 10.1016/j.pbi.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 3.Heslop-Harrison J. An interpretation of the hydrodynamics of pollen. Amer J Bot 1979; 66:737-43; http://dx.doi.org/ 10.2307/2442418 [DOI] [Google Scholar]
- 4.Goring DR, Banks P, Fallis L, Baszczynski CL, Beversdorf WD, Rothstein SJ. Identification of an S-locus glycoprotein allele introgressed from B. napus ssp. rapifera to B. napus ssp. oleifera. Plant J 1992; 2:983-9; PMID:1302644; http;//dx.doi.org/ 10.1046/j.1365-313X.1992.t01-9-00999.x [DOI] [PubMed] [Google Scholar]
- 5.Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci U S A 1991; 88:8816-20; PMID:1681543; http://dx.doi.org/ 10.1073/pnas.88.19.8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 2001; 293:1824-6; PMID:11546871; http://dx.doi.org/ 10.1126/science.1062509 [DOI] [PubMed] [Google Scholar]
- 7.Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science 1999; 286:1697-700; PMID:10576728; http://dx.doi.org/ 10.1126/science.286.5445.1697 [DOI] [PubMed] [Google Scholar]
- 8.Takayama S, Shiba H, Iwano M, Asano K, Hara M, Che FS, Watanabe M, Hinata K, Isogai A. Isolation and characterization of pollen coat proteins of Brassica campestris that interact with S locus-related glycoprotein 1 involved in pollen-stigma adhesion. Proc Natl Acad Sci U S A 2000; 97:3765-70; PMID:10716697; http://dx.doi.org/ 10.1073/pnas.97.7.3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takayama S, Shimosato H, Shiba H, Funato M, Che FS, Watanabe M, Iwano M, Isogai A. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 2001; 413:534-8; PMID:11586363; http://dx.doi.org/ 10.1038/35097104 [DOI] [PubMed] [Google Scholar]
- 10.Giranton JL, Dumas C, Cock JM, Gaude T. The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc Natl Acad Sci U S A 2000; 97:3759-64; PMID:10725390; http://dx.doi.org/ 10.1073/pnas.97.7.3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol 2008; 147:2084-95; PMID:18552232; http://dx.doi.org/ 10.1104/pp.108.123380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone S, Anderson E, Mullen R, Goring D. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant cell 2003; 15:885-98; PMID:12671085; http://dx.doi.org/ 10.1105/tpc.009845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakita M, Shimosato H, Murase K, Isogai A, Takayama S. Direct interaction between the S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biotechnol 2007; 24:185-90; http://dx.doi.org/ 10.5511/plantbiotechnology.24.185 [DOI] [Google Scholar]
- 14.Murase K, Shiba H, Iwano M, Che FS, Watanabe M, Isogai A, Takayama S. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 2004; 303:1516-9; PMID:15001779; http://dx.doi.org/ 10.1126/science.1093586 [DOI] [PubMed] [Google Scholar]
- 15.Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci U S A 1998; 95:382-7; PMID:941938410576738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone SL, Arnoldo M, Goring DR. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 1999; 286:1729-31; PMID:10576738; http://dx.doi.org/ 10.1126/science.286.5445.1729 [DOI] [PubMed] [Google Scholar]
- 17.Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. The Plant cell 2009; 21:2655-71; PMID:19789280; http://dx.doi.org/ 10.1105/tpc.109.069740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safavian D, Goring DR. Secretory activity is rapidly induced in stigmatic papillae by compatible pollen, but inhibited for self-incompatible pollen in the brassicaceae. PloS One 2013; 8:e84286; PMID:24386363; http://dx.doi.org/ 10.1371/journal.pone.0084286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foxe JP, Slotte T, Stahl EA, Neuffer B, Hurka H, Wright SI. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci U S A 2009; 106:5241-5; PMID:19228944; http://dx.doi.org/0.1073/pnas.080767910619307580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo YL, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup MH. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci U S A 2009; 106:5246-51; PMID:19307580; http://dx.doi.org/ 10.1073/pnas.0808012106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al.. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 2011; 43:476-81; PMID:21478890; http://dx.doi.org/ 10.1038/ng.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci 2011; 16:108-16; PMID:21177137; http://dx.doi.org/ 10.1016/j.tplants.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 23.Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 2001; 13:627-43; PMID:11251101; http://dx.doi.org/ 10.1105/tpc.13.3.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Indriolo E, Tharmapalan P, Wright SI, Goring DR. The ARC1 E3 ligase gene is frequently deleted in self-compatible Brassicaceae species and has a conserved role in Arabidopsis lyrata self-pollen rejection. Plant cell 2012; 24:4607-20; PMID:23204404; http://dx.doi.org/ 10.1105/tpc.112.104943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitashiba H, Liu P, Nishio T, Nasrallah JB, Nasrallah ME. Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2011; 108:18173-8; PMID:22025723; http://dx.doi.org/ 10.1073/pnas.1115283108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, Stadler T, Suzuki G, Takayama S, Watanabe M, Shimizu KK. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 2010; 464:1342-6; PMID:20400945; http://dx.doi.org/ 10.1038/nature08927 [DOI] [PubMed] [Google Scholar]
- 27.Indriolo E, Goring DR. A conserved role for the ARC1 E3 ligase in Brassicaceae self-incompatibility. Front Plant Sci 2014; 5:181; PMID:24847339; http://dx.doi.org/ 10.3389/fpls.2014.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Indriolo E, Safavian D, Goring DR. The ARC1 E3 ligase promotes two different self-pollen avoidance traits in arabidopsis. Plant Cell 2014; 26:1525-43; PMID:24748043; http://dx.doi.org/ 10.1105/tpc.114.122879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goring DR, Indriolo E, Samuel MA. The ARC1 E3 ligase promotes a strong and stable self-incompatibility response in Arabidopsis species: response to the Nasrallah and Nasrallah commentary. Plant Cell 2014; 26:3842-6; PMID:25336510; http://dx.doi.org/ 10.1105/tpc.114.131243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR. Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 1996; 8:1641-50; PMID:8837514; http://dx.doi.org/http://dx.doi.org/ 10.1105/tpc.8.9.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang HC, Yang K, Zhu LQ, Yang YJ, Xue LY, Yang H, Chang DL, Gao QG, Ren XS, Li CQ, et al.. The interactions between the truncated fragments of ARM repeat conaining (ARC1) from Brassica oleracea var. acephala L and kinase domain of S-receptor kinase (SRK) from B. oleracea var. capitata L tested by a yeast two-hybrid system. J Ag Biotech 2011; 19:988-95 [Google Scholar]
- 32.Mazzurco M, Sulaman W, Elina H, Cock JM, Goring DR. Further analysis of the interactions between the Brassica S receptor kinase and three interacting proteins (ARC1, THL1 and THL2) in the yeast two-hybrid system. Plant Mol Biol 2001; 45:365-76; PMID:11292081; http://dx.doi.org/ 10.1023/A:1006412329934 [DOI] [PubMed] [Google Scholar]
- 33.Vanoosthuyse V, Tichtinsky G, Dumas C, Gaude T, Cock JM. Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol 2003; 133:919-29; PMID:14555783; http://dx.doi.org/ 10.1104/pp.103.023846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan XG, Yang J, Zhao X, Li YH. Isolation, Expression of ARC1 from Ornamental Kale and Interaction Analysis Between ARC1 and SRK. Acta Horticulturae Sinica 2011; 38:2342-8 [Google Scholar]
- 35.Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends Plant Sci 2001; 6:354-8; PMID:11495788; http://dx.doi.org/ 10.1016/S1360-1385(01)01960-4 [DOI] [PubMed] [Google Scholar]
- 36.Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 2004; 134:59-66; PMID:14657406; http://dx.doi.org/ 10.1104/pp.103.029553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Gen Dev 2000; 14:108-117; PMID:10640280; http://dx.doi.org/ 10.1101/gad.14.1.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Q, McLellan H, Boevink PC, Sadanandom A, Xie C, Birch PR, Tian Z. U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J Exp Bot 2015; PMID:25873665; http://dx.doi.org/25666275 10.1093/jxb/erv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan X, Yang J, Cao M, Wang Y, Kawabata S, Li Y. Isolation and characterization of a J domain protein that interacts with ARC1 from ornamental kale (Brassica oleracea var. acephala). Plant Cell Rep 2015; 34:817-29; PMID:25666275; http://dx.doi.org/ 10.1007/s00299-015-1744-6 [DOI] [PubMed] [Google Scholar]
- 40.Vojtek AB, Hollenberg SM. Ras-Raf interaction: two-hybrid analysis. Method Enzymol 1995; 255:331-42; PMID:8524119; http://dx.doi.org/ 10.1016/S0076-6879(95)55036-4 [DOI] [PubMed] [Google Scholar]
- 41.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Method Enzymol 2002; 350:87-96; PMID:12073338; http://dx.doi.org/ 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- 42.Van Criekinge W, Beyaert R. Yeast Two-Hybrid: State of the Art. Biol Proced Online 1999; 2:1-38; PMID:12734586; http://dx.doi.org/ 10.1251/bpo16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


