Abstract
Human breast cancer susceptibility gene, BRCA2, encodes a 3418-amino acid protein that is essential for maintaining genomic integrity. Among the proteins that physically interact with BRCA2, Partner and Localizer of BRCA2 (PALB2), which binds to the N-terminal region of BRCA2, is vital for its function by facilitating its subnuclear localization. A functional redundancy has been reported between this N-terminal PALB2-binding domain and the C-terminal DNA-binding domain of BRCA2, which undermines the relevance of the interaction between these two proteins. Here, we describe a genetic approach to examine the functional significance of the interaction between BRCA2 and PALB2 by generating a knock-in mouse model of Brca2 carrying a single amino acid change (Gly25Arg, Brca2G25R) that disrupts this interaction. In addition, we have combined Brca2G25R homozygosity as well as hemizygosity with Palb2 and Trp53 heterozygosity to generate an array of genotypically and phenotypically distinct mouse models. Our findings reveal defects in body size, fertility, meiotic progression, and genome stability, as well as increased tumor susceptibility in these mice. The severity of the phenotype increased with a decrease in the interaction between BRCA2 and PALB2, highlighting the significance of this interaction. In addition, our findings also demonstrate that hypomorphic mutations such as Brca2G25R have the potential to be more detrimental than the functionally null alleles by increasing genomic instability to a level that induces tumorigenesis, rather than apoptosis.
Author Summary
Inheritance of a deleterious mutation in Breast Cancer2 (BRCA2) is a well-established factor associated with increased risk of hereditary breast and ovarian cancers. BRCA2 has numerous roles in maintaining the genome to prevent accumulation of mutations that can lead to cancer formation. Here, we describe the generation of a novel mouse model that has a single amino acid alteration in the BRCA2 protein that affects its interaction with another hereditary breast cancer protein, PALB2. We use these mice to examine the relevance of the interaction between BRCA2 and PALB2. We also examine the effect of disrupting one copy of Palb2 gene in the mutant mice. Our findings show that the loss of interaction between the two proteins increases cancer formation in mice. We conclude that not only are BRCA2 and PALB2 required for tumor suppression, their physical interaction is equally important.
Introduction
Error free DNA repair and replication along with proper cell cycle checkpoints are essential for maintaining the stability of the genome. The breast cancer susceptibility gene, BRCA2, is a major player involved in these functions. BRCA2 recruits the DNA recombinase, RAD51, to the site of double strand breaks (DSBs), which mediates repair by homologous recombination (HR) [1–3]. During replication stress, BRCA2 protects the nascent DNA strand at the stalled replication forks to prevent the forks from collapsing allowing their recovery [4–6]. BRCA2 is involved in the intra-S-phase as well as the G2/M checkpoints that prevent premature mitotic entry of cells with damaged DNA [7–9]. These critical functions of BRCA2 are key to its ability to maintain the genomic integrity and contribute to its role as a tumor suppressor. Although BRCA2 mutation carriers predominantly have an increased risk of hereditary breast and ovarian cancer (HBOC), they are also at risk of other cancers such as pancreatic, gastric, laryngeal and prostate cancers [10]. Biallelic mutations in BRCA2 are associated with Fanconi anemia (FA), a rare autosomal recessive disorder characterized by sensitivity to DNA crosslinking agents, bone marrow failure, developmental abnormalities, and increased tumorigenesis [11].
BRCA2 executes its basic functions through its interaction with a number of different proteins, of which bind to the various functional domains of BRCA2 (Fig 1A) [10, 12]. One of these proteins is Partner and Localizer of BRCA2 (PALB2), which binds to the N-terminal region of BRCA2 and is necessary for proper sub-nuclear localization of BRCA2 [13]. In vitro studies have suggested PALB2 to be essential for many functions of BRCA2, such as G2/M checkpoint, replication fork protections, as well as repair of double strand breaks by HR [7, 14–16].
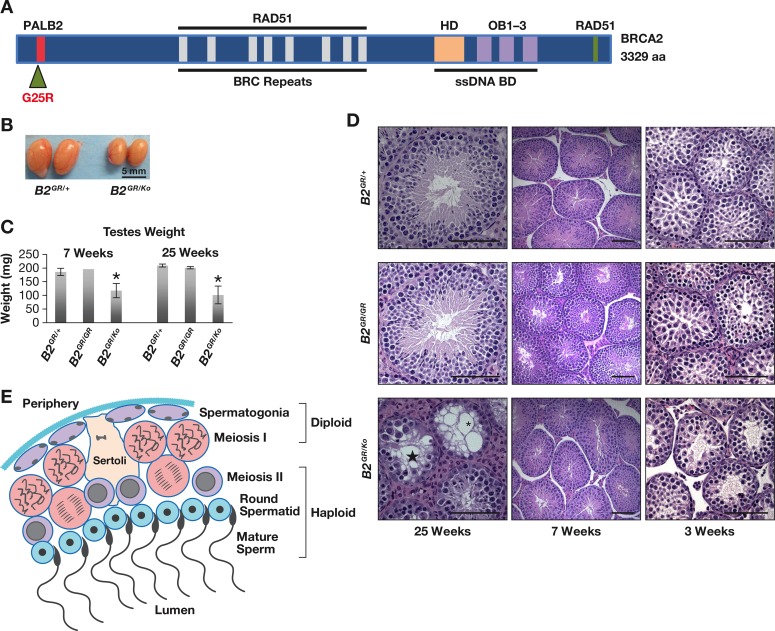
Fig 1. Brca2G25R/Ko mutant males have decreased fertility.
A. Structural and functional domains of murine BRCA2 consisting of 3329 amino acids with the N-terminal PALB2 binding domain (red), BRC repeats (gray) in the middle, helical domain (HD, orange), oligonucleotide binding (OB) regions 1–3 (purple), and a C-terminal RAD51 binding domain (green). Single-strand DNA binding domain (ssDNA, line). The dark green triangle marks the location of the G25R mutation. B. Representative images of mouse testes of indicated genotype (B2GR/+, left and B2GR/Ko, right) at 25 weeks of age. C. Average paired testes weight (mg) of the listed genotypes at 7 weeks and 25 weeks, error bars indicate SD. * p<0.05. D. Representative H&E stained histology of testes of the indicated genotypes (B2GR/+, upper, B2GR/GR, middle, and B2GR/Ko, lower) at 25 weeks (tubules lacking germ cells (*), and tubules arrest at meiosis (star), 7 weeks with normal spermatogenesis, and 3 weeks, with reduced haploid cells in B2GR/Ko testis sections. E. Cartoon of a portion of mouse adult seminiferous tubule cross-section showing spermatozoa at various developmental stages from periphery (top) to lumen (bottom). Scale Bar = 100 μM. Controls are represented by either: Brca2+/+, Brca2Ko/+, or Brca2G25R/+. Abbreviations: Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko.
Mutations in PALB2 are associated with increased risk of breast cancer, similar to that of BRCA2, making it an important hereditary breast cancer susceptibility gene [17]. Like BRCA2, PALB2 is also classified as an FA gene (FANCN). With so many similar and overlapping functions, it is not surprising that the Palb2 null mice exhibit an early embryonic lethal phenotype similar to Brca2 null embryos. K14-Cre-mediated deletion of Palb2 loss results in mammary tumors albeit with long latency, which is significantly reduced when combined with Trp53 loss. Interestingly, the genomic profiles of Palb2;Trp53-loss associated tumors are different from those associated with Brca2;Trp53-deficient tumors in mice [18–20].
The domains of BRCA2 and PALB2 interaction have been well characterized. In BRCA2, the highly conserved 10–40 residues near the N-terminus were shown to be necessary and sufficient for binding to PALB2. Within this region, three cancer-associated mutations affecting two amino acids, namely Gly25 (changed to Arg, G25R) and Trp31 (changed to Cys or Arg, W31C and W31R, respectively) were found to disrupt the interaction with PALB2. Furthermore, while W31C and W31R completely abolished the interaction with PALB2, G25R significantly reduced but did not completely abolish the interaction [13]. A mouse embryonic stem (mES) cell-based functional assay was used to examine the effect of these three mutations on BRCA2 function and results were consistent with biochemical studies [21]. The W31C and W31R mutations affected viability of mES cells. In contrast, the G25R mutation resulted in viable mES cells that exhibited sensitivity to DNA damaging agents and a significant reduction in homologous recombination. These results suggested that although G25R disrupted the interaction of BRCA2 with PALB2, the residual interaction was sufficient for cell viability, but not for DNA repair function [21].
Functional characterization of cancer-causing mutations in PALB2 suggested that the C-terminal region containing four WD40 motifs binds to BRCA2 [22]. It was subsequently shown that WD40 repeats 3 and 4 are required for this binding. X-ray crystallography of BRCA2 peptide (residues 21–39) and PALB2 (C-terminal region, residues 836–1186) co-crystals revealed that the core of the interaction is provided by Trp 31, Phe 32 and Leu 35 residues of BRCA2 [22, 23]. This explained the severity of the phenotype observed in mES cells expressing W31C and W31R mutant BRCA2. Interestingly, the N-terminal region of PALB2 binds to BRCA1, a breast cancer associated protein involved in DNA damage response and repair, and this interaction is impaired in G1 through the ubiquitination of PALB2 [16, 24]. PALB2 is believed to be a molecular adaptor that functionally link BRCA1 and BRCA2, which is apparently essential for BRCA2-mediated RAD51 recruitment and HR [16]. PALB2 also binds RAD51 in the same region as it binds BRCA2 [23, 25, 26].
While the binding of PALB2 to BRCA2 is considered vital for BRCA2 function, recent studies have suggested that there may exist a functional redundancy between the PALB2-binding domain and the C-terminal DNA-binding domain of BRCA2. In chicken DT40 cells, when the PALB2-binding N-terminal region of BRCA2 was deleted, it resulted in a mild reduction in HR and a mild sensitivity to IR, camptothecin, cisplatin as well as olaparib, a PARP1 inhibitor [14]. These cells proliferated with nearly normal kinetics and had a mild increase in the number of spontaneous chromosomal aberrations. In contrast, the loss of the BRCA2 C-terminal DBD, resulted in a moderate sensitivity to these DNA damaging agents as well as an increase in chromosomal aberrations and reduction in HR, suggesting that the N-terminal PALB2 binding region is dispensable but the DBD is necessary for DNA repair. Interestingly, when both regions were deleted, the effects were compounded resulting in a phenotype very similar to Brca2-null DT40 cells. These findings suggest that the two regions may be playing significantly overlapping roles [14]. Another study using BRCA2 polypeptides of various lengths revealed that having either the PALB2 binding region or the DBD is sufficient for BRCA2–mediated homologous recombination [15]. Taken together, these observations have raised concerns over the significance of the interaction between BRCA2 and PALB2, especially on the tumor suppressor function of BRCA2.
To examine the functional consequences of disrupting the interaction between BRCA2 and PALB2 and ascertain the requirement of this interaction for BRCA2 function in vivo, we have generated a knock-in mouse model, with a point mutation (c.73G>A) in exon 3 of Brca2 that changes the highly conserved glycine at position 25 to arginine (S1 Fig and Fig 1A). Although mice carrying the Brca2G25R mutant allele in a heterozygous or homozygous state exhibited no overt phenotype, we observed reduced fertility and a significant increase in tumor predisposition in hemizygous mutant mice. These phenotypes are significantly exacerbated when combined with Palb2 heterozygosity. Functional characterization of these mice unequivocally demonstrates the importance of BRCA2-PALB2 interaction on BRCA2-mediated genome stability as well as tumor suppression.
Results
Brca2G25R/G25R mice are viable
To examine the effect of disrupted interaction between BRCA2 and PALB2, we generated Brca2G25R/+ mice by introducing a G>A substitution in the first base of codon 25 of Brca2 in exon 3 in mouse embryonic stem (ES) cells by gene targeting (S1 Fig, Fig 1A). These mice are viable and fertile. When Brca2G25R/+ mice were intercrossed, homozygous mutants were obtained at expected Mendelian ratios (Table 1, p = 0.97). The Brca2G25R/G25R mice have a normal body size, both males and females are fertile, and have no observable phenotype.
Table 1. Observed and expected birth ratio and Chi Square analysis of offspring of various genotypes from Brca2GR/+ intercross.
| Brca2+/+ | Brca2GR/+ | Brca2GR/GR | |
|---|---|---|---|
| Observed | 54 | 106 | 55 |
| Expected | 54 | 108 | 54 |
χ2 p-value 0.97
We next examined the phenotype associated with the Brca2G25R allele in a hemizygous state, which mimics the physiological state of cells undergoing loss of heterozygosity in BRCA2 mutation carriers. We crossed Brca2G25R/G25R with mice heterozygous (Brca2Ko/+) for a Brca2 null allele, to obtain Brca2G25R/Ko mice. These mice are also viable and born at expected Mendelian ratios (Table 2, p = 0.90). Female mice are fertile with no histological or developmental defect, yet the male mice exhibit a decline in their fertility by 20–25 weeks of age. To determine the cause of the decline in fertility, we examined the testes of Brca2G25R/Ko males. At 25 and 7 weeks of age, we found the testes to be significantly reduced in size relative to the control littermates (48% and 60% respectively of control (wildtype and Brca2Ko/+ mice), Fig 1B and 1C, p = 0.01 and p<0.01). In addition, there was a 40% reduction in the sperm count. Histological analysis of the testis of Brca2G25R/Ko males at 25 weeks showed the presence of some seminiferous tubules that were filled with spermatocytes and appeared similar to the tubules present in Brca2G25R/+ and Brca2G25R/G25R males (Fig 1D, left panel). However, they also had some seminiferous tubules that were either completely devoid of germ cells (Fig 1D, asterisks) or with spermatocytes in meiotic arrest (Fig 1D, star, and Fig 1E). Histologically, the testis at 7 weeks appeared normal and indistinguishable from the testis of Brca2G25R/+ and Brca2G25R/G25R littermates (Fig 1D center panel), with the seminiferous tubules filled with spermatocytes at different developmental stages (Fig 1E).
Table 2. Observed and expected birth ratio and Chi Square analysis of offspring of various genotypes from Brca2GR/GR X Brca2Ko/+ cross.
| Brca2GR/+ | Brca2GR/Ko | |
|---|---|---|
| Observed | 30 | 29 |
| Expected | 30 | 30 |
χ2 p-value 0.90
To examine the testis morphology when the first wave of meiosis starts, we examined 3 week-old males (prior to completion of spermatogenesis). Histological analysis of Brca2G25R/Ko testis revealed the presence of more of the larger meiotic cells and less of the smaller haploid cells in the lumen when compared to their control littermates, suggesting a delay or defect in the cells to progress through meiosis (Fig 1D right panels, and Fig 1E).
Delayed meiotic progression due to persistent DNA damage
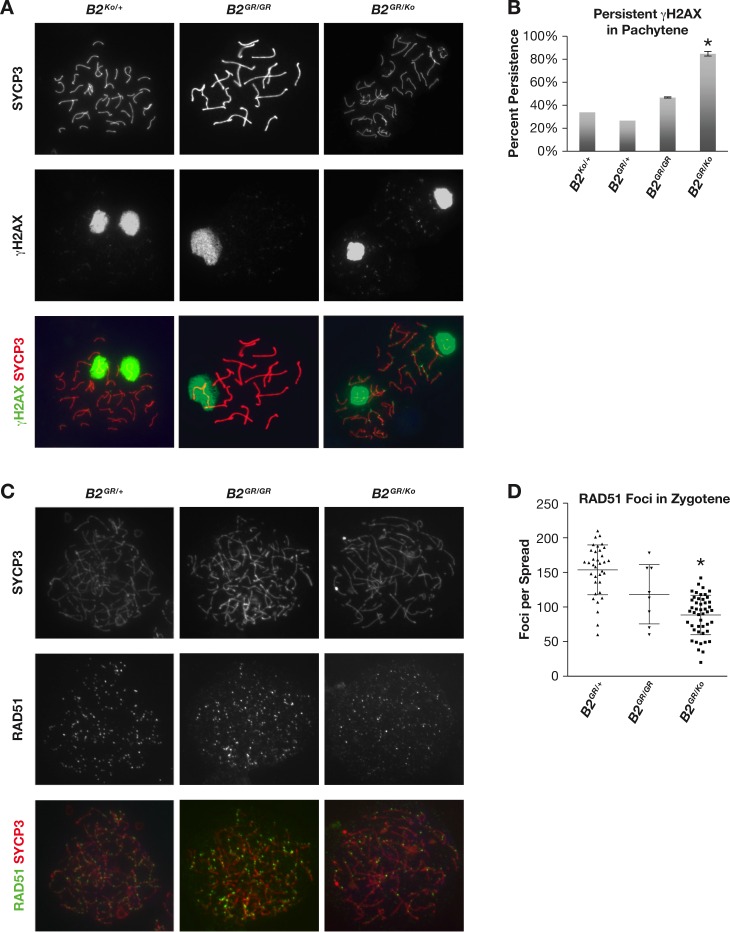
To determine the precise defect in meiotic progression, we examined meiocytes in pachynema stage of prophase I by staining with SYCP3, a marker of the synaptonemal complex, and γH2AX, a mark of unrepaired DNA double strand breaks (DSBs) [27]. As seen in Brca2Ko/+ and Brca2G25R/G25R chromosome spreads (Fig 2A), by this meiotic stage most DSBs are repaired and γH2AX staining is restricted to the unsynapsed sex chromosomes. In contrast, Brca2G25R/Ko meiocytes revealed a persistent staining of γH2AX (66% of spreads, p = 0.04) along the synapsed chromosomes suggesting the presence of unrepaired DSB breaks (Fig 2A and 2B). We also examined the presence of RAD51 foci at pachynema, which is restricted to 0–2 visible foci per pair of synapsed chromosomes and multiple RAD51 foci or a cloud along the sex chromosomes. In the Brca2G25R/Ko meiocytes the number of foci along the sex chromosomes is significantly reduced when compared to littermates (S2 Fig (arrows), p<0.001). Similarly, at zygonema, an earlier stage of meiotic prophase I, we observed an average of only 88 RAD51 foci per Brca2G25R/Ko meiocytes, which is significantly reduced compared to the controls at 154 foci (Fig 2C and 2D, p<0.001). These observations show that a decrease in BRCA2-PALB2 interaction affects RAD51 recruitment to the sites of DSBs during meiosis.
Fig 2. Brca2G25R/Ko decreased testis size due to persistent DSBs and decreased localization of RAD51 in meiocytes.
A. Representative images of pachytene meiocytes of (B2Ko/+, left, B2GR/GR, center, and B2GR/Ko, right) stained with SYCP3 (top, red), γH2AX (middle, green), and merged (bottom). B. Quantification of cells containing persistent γH2AX staining in pachynema. C. Representative images of zygotene meiocytes of (B2GR/+, left, B2GR/GR, center, and B2GR/Ko, right) stained with SYCP3 (top, red), RAD51 (middle, green), and merged (bottom). D. Quantification of RAD51 foci along SYCP3 stained meiotic cores at zygonema. Error bars: SD, * p<0.05. Controls are represented by either: Brca2+/+, Brca2Ko/+, or Brca2G25R/+. Abbreviations: Brca2Ko/+ = B2Ko/+, Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko.
Reduced Brca2G25R/Ko testis size due to increased apoptosis
With the delay of progression through meiosis and the persistence of DNA damage in meiotic spreads, we examined if this leads to increased apoptosis, which would account for the overall decrease in testis size. We performed TUNEL staining on 3-week old mouse testis sections but observed no significant difference in the number of TUNEL positive cells per tubule between different genotypes (S3A and S3C Fig). We then examined the testis of adult animals to see if the delay in meiotic progression may result in cells with unrepaired DNA were then cleared at a later time-point. Indeed, in 7-week old mice, we detected a significant increase in the number of TUNEL positive cells in the Brca2G25R/KO males (S3B and S3D Fig, p = 0.01) compared to littermate controls. These findings suggest that the declining fertility may be due to persistent DNA damage that leads to apoptosis of the spermatogonia, causing a gradual change in the composition of the seminiferous tubule.
Brca2G25R animals have increased tumor susceptibility
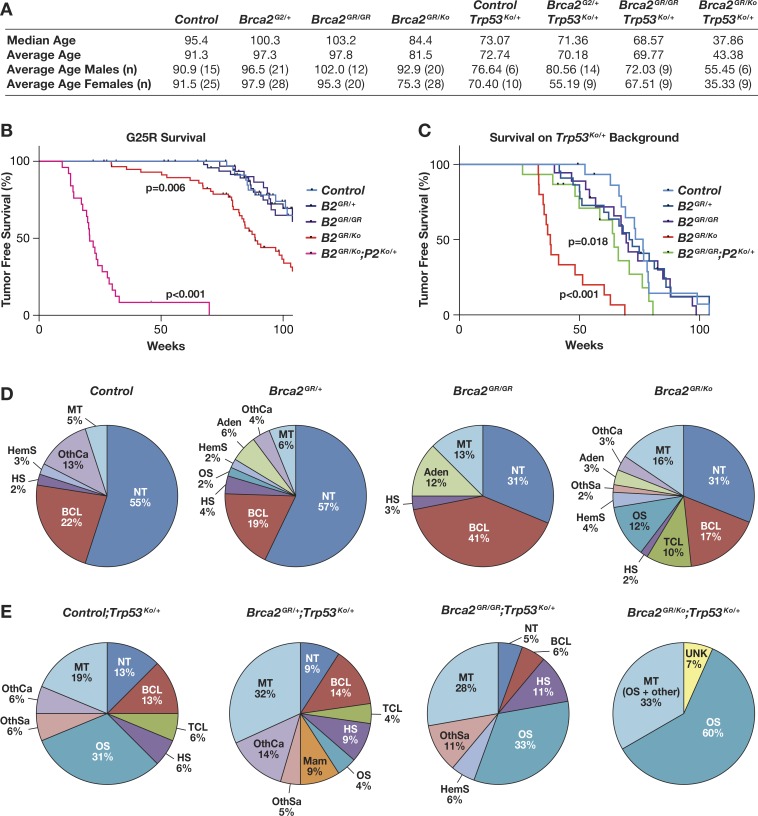
To examine the relevance of physical interaction between BRCA2 and PALB2 on the tumor suppressor function of BRCA2, we generated a cohort of control (wild-type and Brca2Ko/+), Brca2G25R/+, Brca2G25R/G25R, and Brca2G25R/Ko animals and monitored their tumor free survival for 2 yrs. The mean survival of the wild-type and Brca2Ko/+ mice (91 weeks) as well as Brca2G25R/+ mice (97 weeks) was not significantly different from the mean survival of Brca2G25R/G25R and Brca2G25R/Ko animals (98, 81 weeks, respectively, Fig 3A). Interestingly there is a difference in tumor latency of the Brca2G25R/Ko when broken down by gender. The mean survival of females is 75 weeks, whereas the mean survival of males is 93 weeks (Fig 3A). Based on a Kaplan-Meier curve, the tumor-free survival of Brca2G25R/Ko was significantly different from the control animals (Fig 3B, red line, Gehan-Breslow-Wilcoxcon p = 0.006). In the wild-type and Brca2Ko/+ mice, 55% of the mice did not exhibit any signs of neoplasia, however 22% of mice died due to B-cell lymphomas, with the remaining mice exhibiting various other tumor types, and 5% of these had multiple tumors (Fig 3D, S4 Fig). The cause of death and tumor spectrum of Brca2G25R/+ mice was similar to the controls (Fig 3D, S4 Fig) In contrast, 41% of the Brca2G25R/G25R animals died due to B-cell lymphomas, another 28% had various other tumor types, with only 31% of the mice not exhibiting any signs of neoplasia (Fig 3D, S4 Fig). The tumor spectrum of Brca2G25R/Ko mice was remarkably different from the other groups. While 17% of the mice developed B-cell lymphomas, an additional 10% of mice developed T-cell lymphomas, 42% with other tumor types and 16% of these had multiple tumors. However, 31% of these animals did not exhibit any signs of neoplasias (Fig 3D, S4 Fig,). This increased tumor predisposition of Brca2G25R/KO mice suggests that the physical interaction of BRCA2 with PALB2 is critical for their tumor suppressor function.
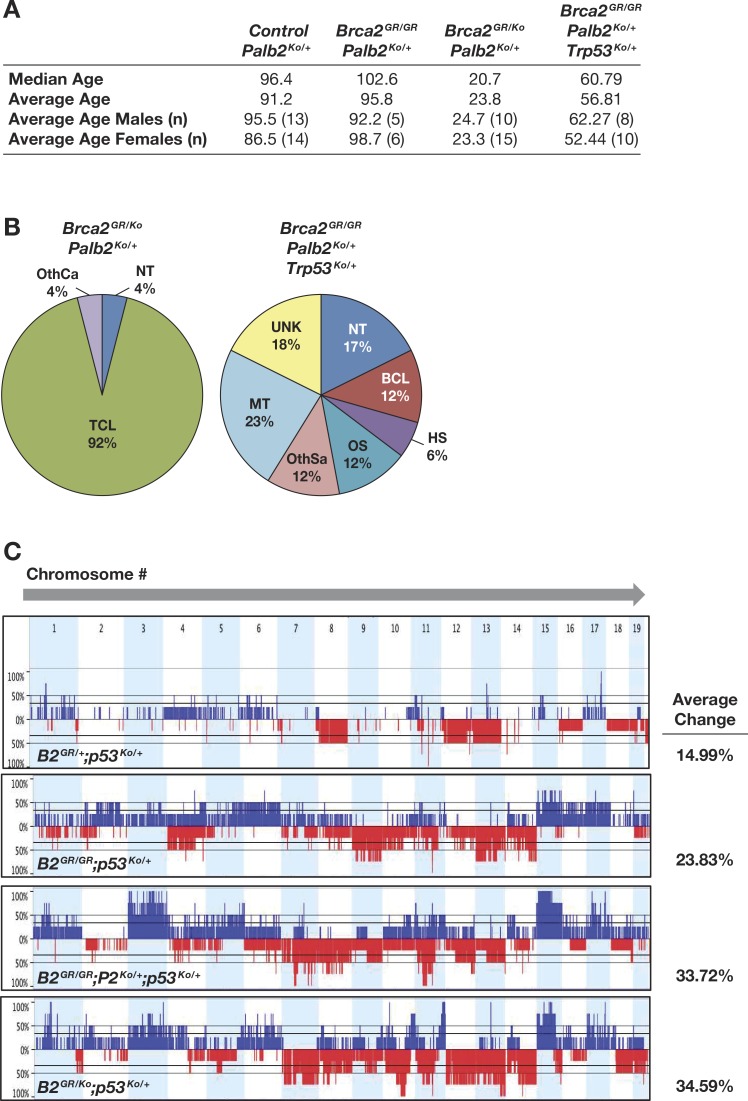
Fig 3. Increased tumor predisposition in Brca2G25R hemizygous mice.
A. Median and average age of death, including number of mice per genotype analyzed (n) and shown by gender. B. Kaplan-Meier tumor-free survival curves of Brca2 mutant animals (104 week study). C. Kaplan-Meier tumor-free survival curves of Brca2 mutant mice on a Trp53Ko/+ genetic background (104 week study). D. and E. Distribution of tumor types found in mice of indicated genotypes. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors, UNK = cause of death unknown. * UNK are included in the tumor-free survival curves as potential neoplasias. Controls are represented by: Brca2+/+ and Brca2Ko/+. Abbreviations: Brca2Ko/+ = B2Ko/+, Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko.
Brca2G25R hemizygous mice have a propensity towards osteosarcomas on a Trp53 heterozygous genetic background
It has been previously reported that the tumor susceptibility of Brca2 mutant mice is enhanced on a Trp53 mutant genetic background [28]. Therefore, we generated Brca2G25R mutant mice of various genotypes on a Trp53Ko/+ genetic background. The mean survival of Trp53Ko/+ mice was 73 weeks (Fig 3A). Osteosarcoma was detected in 31% of the mice, 19% developed lymphomas (B- and T-cell), 19% of the animals had multiple tumors, with only 13% showing no signs of neoplasias (Fig 3C, Fig 3E, S5 Fig). In this cohort, we observed a significant reduction in the mean survival (p<0.001) of Brca2G25R/Ko;Trp53Ko/+ mice to 43 weeks and a high incidence of tumor formation compared to mice of other genotypic classes (Fig 3C, Fig 3E, S5 Fig). Almost all Brca2G25R/Ko;Trp53Ko/+ mice succumbed to osteosarcomas (93% of the animals). Of these, a third also had additional tumors types (Fig 3E, S5 Fig). The Brca2G25R/+;Trp53Ko/+ mice had a mean survival of 70 weeks. 18% of the mice developed lymphomas (B- and T-cell), 37% had other carcinomas, 32% had multiple tumors, and only 4% developed osteosarcomas (Fig 3E, S5 Fig). While the Brca2G25R/G25R;Trp53Ko/+ mice had a comparable mean survival (70 weeks), 33% developed osteosarcomas. In addition, 17% had other carcinomas, with 22% of the animals having multiple tumors and 11% developed B-cell lymphomas (Fig 3E, S5 Fig). The difference in tumor latency based on gender is also evident in these mice; Brca2G25R/+;Trp53Ko/+ females exhibited an average latency of 55 weeks, yet it was 81 weeks for males. Similarly, Brca2G25R/Ko;Trp53Ko/+ males exhibited an average tumor latency of 55 weeks but the average female tumor latency was 35 weeks. Taken together, these data show a significant reduction in the tumor latency and a significant change in the tumor spectrum of Brca2G25R hemizygous mice on a Trp53 heterozygous genetic background that is also influenced by gender.
Synergistic interaction between Brca2G25R and Palb2 heterozygosity
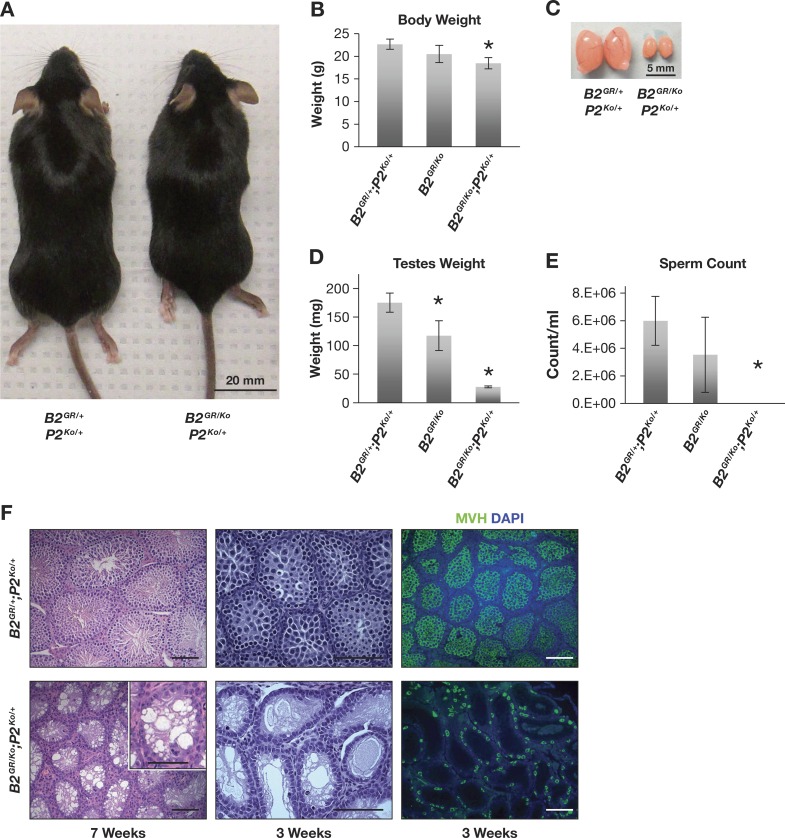
To fully appreciate whether the phenotypes observed in Brca2G25R/G25R as well as Brca2G25R/Ko mice are due to decreased interaction of BRCA2 with PALB2, we examined the effect of the loss of one functional allele of Palb2. We crossed Brca2G25R/Ko mice to Palb2 heterozygous mice carrying a gene trap allele known to render the gene non-functional. Palb2Ko/+ mice are viable and fertile with no known deleterious phenotype including tumor susceptibility. Palb2Ko/Ko mice are not viable and die during early embryogenesis [18–20]. Both Brca2G25R/G25R;Palb2Ko/+ (p = 0.24, Table 3) as well as Brca2G25R/Ko;Palb2Ko/+ mice are viable and born at expected Mendelian ratios (p = 0.48, Table 4). The Brca2G25R/G25R;Palb2Ko/+ mice are phenotypically indistinguishable from their control littermates. However, Brca2G25R/Ko;Palb2Ko/+ mice have significantly reduced body size compared to their normal littermates (19% reduction, p = 0.003, Fig 4A and 4B). In addition, the Brca2G25R/Ko;Palb2Ko/+ males are infertile with a significant reduction in testis size compared to the control at 6–8 weeks (14% of control, Fig 4C and 4D). No sperm was detected in these animals, suggesting a severe defect in spermatogenesis (Fig 4E).
Table 3. Observed and expected birth ratio and Chi Square analysis of offspring of various genotypes from Brca2GR/+;Palb2ko/+ X Brca2GR/+ cross.
| Brca2+/+ | Brca2GR/+ | Brca2GR/GR | ||||
|---|---|---|---|---|---|---|
| Palb2+/+ | Palb2Ko/+ | Palb2+/+ | Palb2Ko/+ | Palb2+/+ | Palb2Ko/+ | |
| Observed | 12 | 9 | 27 | 15 | 13 | 17 |
| Expected | 12 | 12 | 24 | 24 | 12 | 12 |
χ2 p-value = 0.24
Table 4. Observed and expected birth ratio and Chi Square analysis of offspring of various genotypes from Brca2GR/+;Palb2ko/+ X Brca2Ko/+ cross.
| Brca2+/+ | Brca2Ko/+ | Brca2GR/+ | Brca2GR/Ko | |||||
|---|---|---|---|---|---|---|---|---|
| Palb2+/+ | Palb2Ko/+ | Palb2+/+ | Palb2Ko/+ | Palb2+/+ | Palb2Ko/+ | Palb2+/+ | Palb2Ko/+ | |
| Observed | 21 | 19 | 18 | 18 | 14 | 13 | 16 | 9 |
| Expected | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
χ2 p-value = 0.48
Fig 4. Synergistic interaction between Brca2G25R mutation and Palb2 heterozygosity.
A. Image of decreased body size in B2GR/Ko;P2Ko/+ animals (right) compared to littermate controls (left). B. Average body weight of mice of indicated genotypes at 7 weeks. C. Seven-week old testes of B2GR/Ko;P2Ko/+ animals (right) are smaller compared to littermate controls (left). D. Average weight of testes of indicated genotypes at 7 weeks. E. Average sperm count of mice of indicated genotypes at 7 weeks. F. Representative images of testes sections from B2GR/+;P2Ko/+ (upper) and B2GR/Ko;P2Ko/+ (lower) animals. Left panel: 7 week H&E images, notice loss of germ cells in seminiferous tubules. Middle Panel: 3 week H&E stained images. Right panel: 3 week testis sections stained with MVH (green) and DAPI (blue) to visualize the germ cells. Controls are represented by either: Brca2+/+, Brca2Ko/+, or Brca2G25R/+ (with or without Palb2Ko/+). Abbreviations: Brca2G25R/+ = B2GR/+, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+.
Histological analysis of the testes of Brca2G25R/Ko;Palb2Ko/+ mice at 6–8 weeks of age revealed that the seminiferous tubules only had sertoli cells and were completely devoid of germ cells (Fig 4F, left panel), while the Brca2G25R/+;Palb2Ko/+, Brca2Ko/+;Palb2Ko/+, and Brca2GR/GR;Palb2Ko/+ males proceeded through spermatogenesis normally (Fig 4F and S6A Fig). The lack of any defect in Brca2Ko/+;Palb2Ko/+ mice suggests that the mutant phenotype observed in Brca2G25R/Ko;Palb2Ko/+ mice is likely due to the Brca2G25R mutation and not due to the reduced levels of both BRCA2 and PALB2 proteins. To determine whether there was a complete failure of germ cell formation, we examined the testes of mice at 3 weeks of age during initiation of meiosis. This was the age when we first observed defects in the Brca2G25R/Ko mice. In the testis section of 3-week old Brca2G25R/Ko;Palb2Ko/+ mice, in addition to sertoli cells along the basal lamina, we observed cells that appeared to be germ cells (Fig 4F, center panel, S6A Fig, Fig 1E). To confirm the identity of these cells, we stained the testis sections with a germ cell marker, mouse Vasa homologue (MVH) [29]. We observed several MVH-positive cells, which shows that the Brca2G25R/Ko;Palb2Ko/+ males are not devoid of germ cells (Fig 4F, right panels). Taken together, these results indicate that while the Brca2G25R/Ko;Palb2Ko/+ males are not lacking germ cells, there is an apparent failure of germ cell expansion. Overall, the significant enhancement of the mutant phenotype of Brca2G25R/Ko males on a Palb2Ko/+ background strongly suggests that Palb2 heterozygosity has a synergistic effect on the Brca2G25R mutation.
Decreased tumor latency in Brca2G25R and Palb2Ko mice
We examined the effect of Palb2 heterozygosity on the tumor predisposition of Brca2G25R/Ko animals that had a mean survival time of 80 weeks (Fig 3A). Consistent with the significant effect on the fertility of these animals, the Brca2G25R/Ko;Palb2Ko/+ animals (n = 25, p<0.001) exhibited a mean survival time of 24 weeks with the majority of the animals (92%) succumbing to T-cell lymphoblastic lymphomas (Fig 3B, Fig 5A, Fig 5B, S6D Fig). Although the effect of Palb2 heterozygosity on the tumor predisposition of Brca2G25R/G25R animals is not any different from the control animals (mean survival: 96 weeks, n = 11, S6B and S6C Fig), on a Trp53Ko/+ genetic background the Brca2G25R/G25R;Palb2Ko/+ mice showed a significant decrease in survival compared to the control (57 week vs. 70 weeks, p = 0.018, Fig 3C). These mice developed diverse neoplasias; 12% had lymphomas (B-cell), 12% osteosarcomas, 18% other carcinomas, with 23% of the animals having multiple tumors (Fig 5A, Fig 5B, S6D Fig). The decrease in survival and increase in tumor predisposition of Brca2G25R mutant mice on Palb2Ko/+ genetic background provides strong evidence to suggest that normal levels of interaction between BRCA2 and PALB2 is essential for tumor suppression.
Fig 5. Increased tumor formation in Brca2G25R mice in a Palb2Ko/+ genetic background.
A. Median and average age of mice, including the number of animals for each genotype. B. Distribution of tumors found in the of B2GR/Ko;P2Ko/+ and B2GR/GR;P2Ko/+ mutant mice. Tumor abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors, UNK = cause of death unknown. C. Array CGH analysis of tumors from mice of indicated genotypes. These show the percentage of any known copy number variation within all tumors of the given genotype. Numbers at the top indicate the chromosome numbers. Blue lines represent gains and red lines represent losses. Controls are represented by either: Brca2+/+, Brca2Ko/+, or Brca2G25R/+ (with or without Palb2Ko/+ and/or Trp53Ko/+). Abbreviations: Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+, Trp53Ko/+ = p53Ko/+.
Degree of genomic instability in Brca2G25R/Ko;Trp53Ko/+ tumors is similar to BrcaG25R/G25R;Palb2Ko/+;Trp53Ko/+ tumors
The difference in the tumor latency observed in mice of various genotypes, especially the significantly accelerated tumor formation in Brca2G25R/Ko;Trp53Ko/+ mice was correlated with the degree of genomic instability in the tumors. We examined the genomic profile of tumors by array comparative genomic hybridization (aCGH) analysis using an Agilent SurePrint G3 Mouse CGH Array (4X180K) chip. We examined tumors from Brca2G25R/+, Brca2G25R/G25R, Brca2G25R/Ko mice, all on Trp53Ko/+ genetic background. Based on the total region affected by copy number variation (CNV), we observed a mean genomic change of 14.99% in Brca2G25R/+;Trp53Ko/+ tumors (CNV average, 121, Fig 5C, S7 Fig, S8 Fig). The Brca2G25R/G25R;Trp53Ko/+ tumors had an average genomic alterations of 23.83% (p = 0.23) with an average CNV of 443 (p = 0.054) (Fig 5C, S7 Fig, S8 Fig). The tumors from Brca2G25R/Ko;Trp53Ko/+ mice revealed the presence of even greater genomic alterations (34.6% p = 0.052) and also had a number of regions with gains and losses (CNV = 413, p = 0.068) (Fig 5C, S7 Fig, S8 Fig). These observations suggest that the mutation that affects the binding of BRCA2 with PALB2 is associated with increased genomic alterations in tumors. To further demonstrate that this genomic instability is due to disruption of the interaction between BRCA2 and PALB2, we examined the tumors from Brca2G25R/G25R;Palb2Ko/+;Trp53Ko/+ mice. These mice exhibited a marked reduction in the mean tumor latency compared to Brca2G25R/G25R;Trp53Ko/+ mice. When we examined tumors from these mice by aCGH analysis, we observed an increase in genomic instability from 23.8% to 33.7% (p = 0.07). These tumors had gains and losses similar to those seen in Brca2G25R/Ko;Trp53Ko/+ tumors and the total CNVs observed were 424(p = 0.036) (Fig 5C, S7 Fig, S8 Fig). Interestingly, one of the common gains observed in both the Brca2G25R/Ko;Trp53Ko/+ and Brca2G25R/G25R;Palb2Ko/+;Trp53Ko/+ tumors was on mouse chromosome 15 centered around the Myc locus (S8A Fig). Amplification of the region surrounding Myc is commonly seen in BRCA2 tumors[30–33]. Taken together, these results demonstrate that the tumors that are genetically predicted to have the least interaction between BRCA2 and PALB2 have the highest levels of genomic instability.
Brca2G25R mouse embryonic fibroblasts (MEFs) under stress exhibit delayed cell cycle progression and decreased proliferation
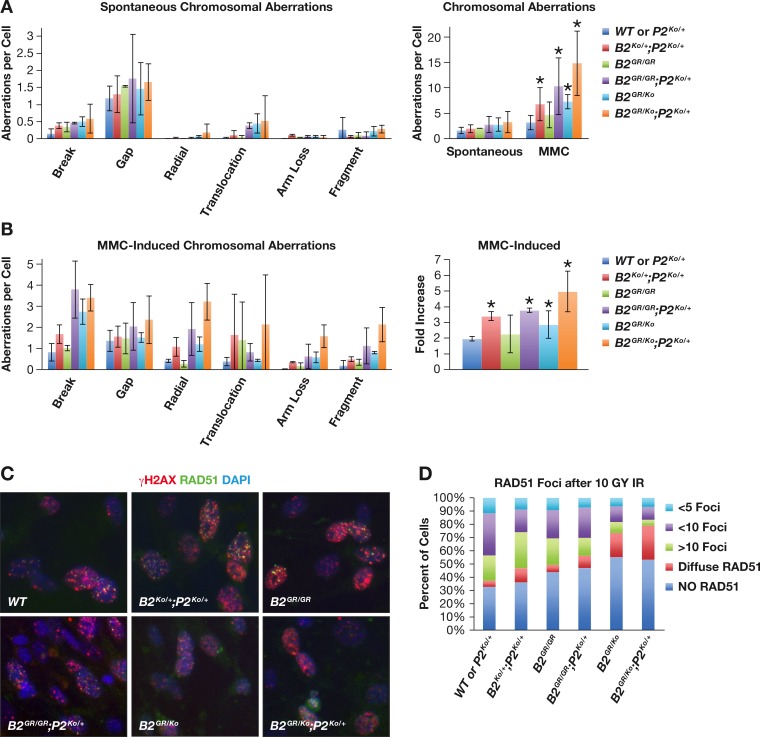
To evaluate the effect of disruption of BRCA2-PALB2 interaction at the cellular level, we generated mouse embryonic fibroblasts from embryos of various genotypes. We evaluated the growth of these cells (using at least 2 independent lines from each genotype) from early passages. We found a significant reduction in the growth of mutant MEFs of various genotypes relative to the control MEFs (WT and Palb2Ko/+). The 2 lines that grew the slowest were Brca2G25R/Ko and Brca2G25R/Ko;Palb2Ko/+ (S9A Fig, p<0.001 by day 5). To further evaluate the cause of this poor growth we examined the cell cycle profile of untreated and 100nM mitomycin C (MMC) treated MEFs. MMC is a DNA cross-linker that induces replication stress and leads to DSBs. There was no significant difference in the cell cycle profile of untreated MEF lines. Upon MMC treatment, we observed a modest increase in the number of cells at G2/M in Brca2Ko/+;Palb2Ko/+ (p = 0.028), Brca2G25R/Ko (p = 0.022), Brca2G25R/G25R;Palb2Ko/+ (p = 0.016), and Brca2G25R/Ko;Palb2Ko/+ (p = 0.17) MEFs compared to the controls (S9C Fig). We evaluated the proliferative ability of these cells by BrdU incorporation and found no difference in any of the untreated MEFs. In response to 100nM MMC treatment, there was a significant decrease in BrdU incorporation in Brca2G25R/Ko (p = 0.012) and Brca2G25R/Ko;Palb2Ko/+ MEFs (p = 0.009) (S9D and S9E Fig). Other lines were not statistically different from the controls. (S9D and S9E Fig) These results suggest that while there is persistent DNA damage in these cells upon MMC treatment, only the cells predicted to have the least interaction between BRCA2 and PALB2 exhibit a reduction in cell proliferation.
Brca2G25R MEFS exhibit genomic instability after MMC treatment
With the Brca2G25R mutant MEFs exhibiting delayed G2/M progression and reduced cell proliferation, we examined its effect on the genomic integrity of the cells. We examined metaphase spreads of untreated and MMC treated cells. We found very few chromosomal aberrations in untreated control MEFs (1.6–2.06 aberrations (ab)/cell). We observed a slight increase in breaks and translocations in Brca2G25R/G25R;Palb2Ko/+(2.76 ab/cell), Brca2G25R/Ko(2.74ab/cell), and Brca2G25R/Ko;Palb2Ko/+ (3.26ab/cell) (p = 0.0098, 0.0007, and 0.0001 respectively) (Fig 6A). After 100nM MMC treatment, Brca2G25R/G25R;Palb2Ko/+ (10.32 ab/cell), Brca2G25R/Ko,(7.28 ab/cell) and Brca2G25R/Ko;Palb2Ko/+ (14.84 ab/cell) MEFs showed a marked increase (3.8 fold, 2.9 fold, and 5.1 fold, respectively) in aberrations (breaks, radials, fragments and translocations) over controls MEFs (3.18–4.68 ab/cell) (Fig 6B, p<0.0001 for these three lines). Surprisingly, we also observed a 3.4 fold increase in breaks, radials, and translocations in Brca2Ko/+;Palb2Ko/+ MEFs (6.78 ab/cell) (Fig 6B, p = 0.0003). Taken together, these results show that BRCA2-PALB2 interaction is essential for maintenance of the genomic integrity.
Fig 6. Brca2G25R contributes to genomic instability and affects RAD51 foci formation.
A. Spontaneous chromosomal aberrations seen in MEFs of various genotypes (left) and total chromosomal aberrations seen in untreated and 100nM MMC treated MEFs (right). B. Chromosomal aberrations seen with in MEFS of various genotypes treated with 100nM MMC (left) and fold increase in aberrations seen after 100nM MMC treatment (right). Error Bars: SD, * p<0.05 C. Representative images of RAD51 and γH2Ax foci formation after irradiation in primary MEFs of various genotypes. D. Quantification of RAD51 foci seen in cells that also have γH2AX foci. Controls are represented by: Brca2+/+ (with or without Palb2Ko/+). Abbreviations: Brca2+/+ = WT, Brca2Ko/+ = B2Ko/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+.
Decrease in RAD51 foci formation in Brca2G25R MEFs
To examine the effect of disruption of BRCA2-PALB2 interaction on RAD51 recruitment in mitotic cells, we analyzed RAD51 foci formation in response to radiation induced DSBs. We irradiated cells with 1000 rads and immunostained after 3 hrs with γH2AX and RAD51. The percentage of cells that were γH2AX positive were very similar between the genotypes. Yet, there was a significant reduction in the number of cells lacking RAD51 foci in Brca2G25R/Ko (55% of cells) and Brca2G25R/Ko;Palb2Ko/+ (53% of cells) (both p<0.00001, Fig 6C and 6D) when compared to the controls (33% of the cells). Brca2G25R/G25R cells also had an increase in the number of cells lacking foci, (44% of cells, p = 0.0483) along with Brca2G25R/G25R;Palb2Ko/+ (47% of cells, Fig 6C and 6D, p = 0.0015). Interestingly, although Brca2Ko/+;Palb2Ko/+ cells exhibited a defect in cell growth and a moderate increase in chromosomal aberrations, they had normal RAD51 foci formation (Fig 6C and 6D). There were several Brca2G25R/Ko and Brca2G25R/Ko;Palb2Ko/+ cells that also showed diffused RAD51 staining (18% and 26% of cells respectively, Fig 6C and 6D, p<0.0001). It is possible that this represents the RAD51 that is relocalized into the nucleus, but is unable to form foci due to weakened BRCA2-PALB2 interaction. We also observed a modest increase in this diffuse staining in Brca2G25R/G25R;Palb2Ko/+ and Brca2Ko/+;Palb2Ko/+ MEFs (9% and 11%, Fig 6C and 6D, p = 0.034 and p = 0.0204). Consistent with these observations, we found a reduction in RAD51 foci in PCNA positive S-phase cells (S10A Fig). These results suggest that the mutation in the PALB2 binding domain decreases the ability of RAD51 recruitment.
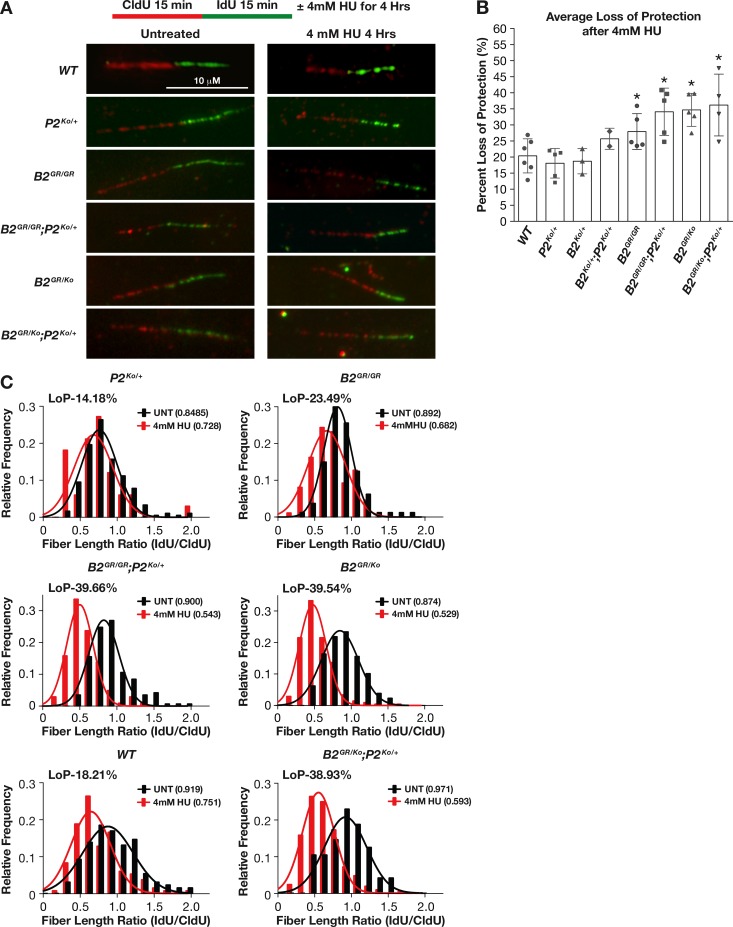
BRCA2-PALB2 interaction is essential for replication fork protection
BRCA2 and PALB2 are essential for the protection of the nascent DNA strands at stalled replication forks [5, 34]. To examine if a physical interaction between these two proteins is essential for fork protection as in the case of DSB repair, we examined the integrity of the nascent DNA strands under replicative stress in the primary MEFs. We labeled the replicating strand of DNA first with thymidine analog, CldU (red), and then with thymidine analog, IdU (green), for 15 minutes each followed by hydroxyurea (HU) treatment (which induces replication stress by depleting the dNTP pool) for 4 hours (Fig 7A). We measured the length of green and red tracks and estimated the loss of fork protection based on the ratios of IdU/CldU in the presence and absence of HU treatment. Loss of protection represents the change in average fiber length in response to replicative stress compared to when there is no fork degradation. It was calculated as the percent reduction in the ratio of DNA fiber length after HU treatment and before HU treatment compared to when the fork is fully protected and the ratio is expected to be 1. In the control cells, wildtype (n = 6), Brca2Ko/+(n = 3), and Palb2Ko/+ (n = 5), we observed a mean decrease in protection of 19% (3 independent experiments) (Fig 7). The loss of fork protection in Brca2Ko/+;Palb2Ko/+ MEFs was increased to 26% (n = 2). The Brca2G25R/G25R cells showed a 28% loss in fork protections (n = 5, p = 0.003), which increased significantly on a Palb2Ko/+ background (34%, n = 4, p<0.0001). Interestingly, the Brca2G25R/Ko MEFs exhibited 35% loss of protection (n = 5, p<0.0001) however, did not show any further loss when one allele of Palb2 was deleted (36%, n = 4) (Fig 7). While there is an apparent loss of fork protection with the disruption of BRCA2-PALB2 interaction, there seems to be a maximum loss that can be tolerated by the cells or it reflects the sensitivity of the DNA fiber assay. We also evaluated the ability of the DNA replication forks to restart after replication stress. We examined this in the MEFs that had exhibited the most severe cellular phenotype (Brca2G25R/KO;Palb2Ko/+). Control and mutant cells were first labeled with CldU, replication was then stalled with 0.5mM HU for 1.5hrs and subsequently resumed in the presence of IdU. These cells were then analyzed for the length of the CldU and IdU labeled DNA fibers to determine whether the stalled forks had restarted or there was a new origin of replication. Overall, here was no significant difference between control and Brca2G25R/KO;Palb2Ko/+ cells suggesting that fork restart was not affected in the mutant cells (S10B Fig). We conclude from these observations that similar to the role in HR-mediated DSB repair, BRCA2-PALB2 interaction is a major contributor to replication stress induced fork protection.
Fig 7. Brca2G25R mutation affects protection of stalled replication forks.
A. Representative images of the fork protection assay in primary MEFs of indicated genotypes in both untreated and 4mM HU treated conditions. The x-axis represents the fiber length ratio (IdU/CldU) and y-axis represents the frequency of the fibers that have the corresponding IdU/CldU ratio. B. Average loss of protection (LoP) for the various genotypes across multiple experiments. C. Representative frequency of IdU/CldU ratios in untreated (black) and HU treated (red) MEFs indicating the average ratio for untreated and HU treated (number in parenthesis) as well as percent of LoP. LoP = (1-a/b)X100, where a is the IdU/CldU ratio after HU treatment and b is the IdU/CldU ratio without HU treatment. Error bars: SD, * p<0.05. Controls are represented by: Brca2+/+, Brca2Ko/+, and Palb2Ko/+. Abbreviations: Brca2+/+ = WT, Brca2Ko/+ = B2Ko/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+.
Discussion
BRCA2 performs its function as a care-taker of the genome because of its ability to bind to a number of key proteins. One such binding partner is PALB2. Functional studies using knockout or knockdown of PALB2 have demonstrated that it is essential for BRCA2-mediated recruitment of RAD51 as well as for protection of stalled replication forks (review [35]). While these studies show the importance of PALB2, they do not shed light on the relevance of a direct interaction between these two proteins. Although three mutations in the PALB2-binding domain (G25R, W31C and W31R) that disrupt the interaction between the two proteins are linked to breast cancer development, their precise disease association remains uncertain due to limited family-linkage data [21].
To address the significance of the interaction between the two proteins and examine the effect of disruption of this interaction on tumor development, we generated a novel Brca2 knock-in mouse model carrying the G25R mutation, which results in DNA repair defects without affecting cell viability. Therefore, it provides a unique opportunity to dissect the relevance of the interaction between PALB2 and BRCA2 in various biological processes including tumorigenesis. We have generated mice of various genotypes, carrying one or two alleles of Brca2G25R. Furthermore, we have combined these genotypes with Palb2 heterozygosity to reduce PALB2 levels to gradually and systematically reduce the interaction without affecting viability. These mice exhibit mutant phenotypes that become increasingly severe as the interaction between BRCA2 and PALB2 is gradually reduced.
RAD51 recruitment and replication fork protection are impaired due to disruption of BRCA2-PALB2 interaction
Overall, Brca2G25R mutation did not affect viability or fertility of Brca2G25R/ G25R mice. However, Brca2G25R/Ko males exhibited a delay in meiotic progression due to a defect in RAD51 recruitment in spermatocytes. This resulted in persistence of unrepaired DNA damage and eventual cell death causing loss of germ cells. Interestingly, on a Palb2Ko/+ background, while the Brca2G25R/G25R mice did not exhibit any phenotypic defect, the Brca2G25R/Ko mice were relatively smaller in size and the males were infertile. The testes of Brca2G25R/Ko;Palb2Ko/+ males revealed the presence of germ cells at 3 weeks of age. However, germ cells were lost by 7 weeks of age leading to a germ cell-less phenotype. A lack of male germ cells has been previously reported in mice expressing a hypomorphic allele of Brca2 encoding a truncated protein but the timing or the mechanism of germ cell loss has not been described [36, 37].
Similar to the spermatocytes, a defect in PALB2-BRCA2 interaction also affects RAD51 foci formation in response to IR-induced DSBs in somatic cells. Brca2G25R/Ko and Brca2G25R/Ko;Palb2Ko/+ MEFs exhibited a significant reduction in RAD51 foci suggesting that the interaction between BRCA2 and PALB2 is essential for proper recruitment of RAD51 to the site of DSB.
Recent studies have suggested that heterozygosity of BRCA2 as well as PALB2 results in some loss of DNA replication fork protection and a defect in DNA damage response, which contributes to genomic instability [38, 39]. However, in the present study, single heterozygous mice and MEFs (Brca2Ko/+, Brca2G25R/+, and Palb2Ko/+) did not reveal any overt defects. We observed a small loss of fork protection in Brca2G25R/G25R and Brca2Ko/+;Palb2Ko/+ MEFs but Brca2G25R/G25R;Palb2Ko/+, Brca2G25R/Ko and Brca2G25R/Ko;Palb2Ko/+ MEFs exhibited a significant fork degradation. Our results show that although both BRCA2 and PALB2 have their own DNA binding motifs, as well as independent RAD51 binding motifs, their interaction is essential for proper recruitment of RAD51 to the sites of DNA damage and protection of stalled replication forks.
Genomic instability and tumor susceptibility due to disruption of BRCA2-PALB2 interaction
As observed in the MEFs derived from mice of various genotypes with mutations in Brca2 and Palb2, a defect in RAD51 recruitment as well as a loss of protection of the replication fork leads to genomic instability. Based on the tumor predisposition of these mice, it is evident that the genomic instability contributes to tumorigenesis. Brca2G25R/Ko mice had a significantly reduced tumor free survival compared to Brca2G25R/+ as well as Brca2G25R/G25R mice, which is further enhanced on a Trp53Ko/+ genetic background. Their tumor predisposition is reminiscent of the Brca2 mutant mice that lack exon 27 (Brca2Δ27/Δ27), in terms of latency, frequency of tumor formation and enhancement on a Trp53Ko/+ genetic background [28, 40, 41]. However, the Brca2G25R/Ko animals predominately developed osteosarcomas on the Trp53Ko/+ background, unlike the Brca2Δ27/Δ27;Trp53Ko/+ mice that developed a variety of tumor types. A recent sequence analysis of the exome of osteosarcomas reported increased incidence of BRCA2 mutations as well as signatures associated with BRCA2 loss suggesting the involvement of BRCA2 in the development of osteosarcomas [42–44]. The Brca2G25R/Ko;Palb2Ko/+ animals that result in the least interaction between BRCA2 and PALB2 and exhibited the shortest tumor latency, predominantly developed lymphoblastic T-cell lymphomas, similar to the mice carrying a truncating mutation in exon 11 of Brca2 [36, 37].
Overall, the tumor incidence and tumor latency in mice for the most part correspond with the degree of genomic instability observed in the MEFs. However, there were some exceptions. For example, the Brca2G25R/G25R MEFs exhibited a mild defect in RAD51 recruitment and fork protection but the survival of mice was not affected except they had a mild increase in tumor formation at 24 months of age. The tumor free survival of these mice was also not altered on a Trp53Ko/+ genetic background.
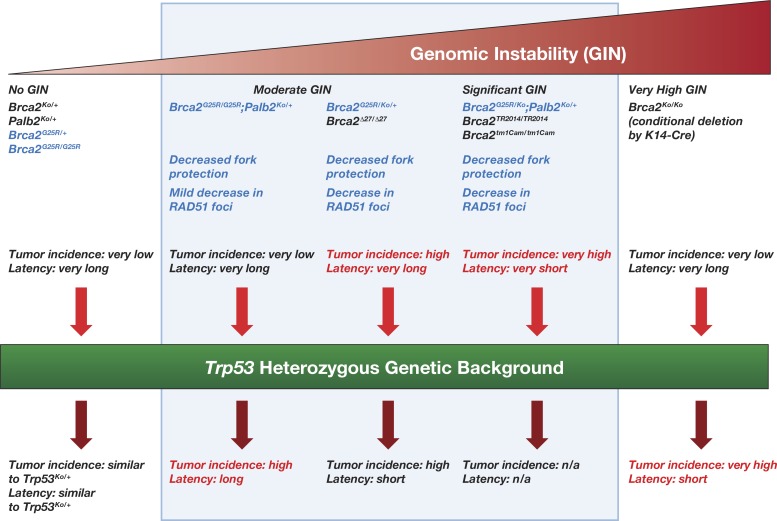
While an increase in genomic instability correlates well with an increase in tumor susceptibility, having a significantly high genomic instability may not necessarily enhance tumorigenesis (Fig 8). When Brca2 is rendered functionally null by CRE-mediated recombination in the epithelial cells using cytokeratin K14 promoter (which is expressed during embryogenesis), tumor incidence is low and tumor latency is very long [45]. Although the effect of germline inheritance of the mutant alleles in all cell lineages (as in the present study) versus loss in specific cell lineage cannot be ruled, the longer latency is believed to be due to the strong apoptotic response induced by the high genomic instability in cells lacking BRCA2 [46]. This is supported by the observation that the tumor incidence is increased and the tumor latency is significantly reduced when apoptosis is suppressed on a Trp53 heterozygous background [45]. The most striking phenotype in this present study is the high tumor incidence in Brca2G25R/Ko as well as Brca2G25R/Ko;Palb2Ko/+ mice with wild-type Trp53. Sixty-nine percent of Brca2G25R/Ko and 100% of Brca2G25R/Ko;Palb2Ko/+ mice developed tumors, with an average latency of 82 and 24 weeks, respectively. Considering these genotypes have no effect on the viability of mice, the genomic instability induced in cells must not be high enough to induce a strong apoptotic response but it must be significant enough to result in cellular transformation and tumorigenesis. Similar high tumor incidence has been reported in other mice carrying Brca2 hypomorphic alleles that result in viable mice, such as Brca2tm1Cam, Brca2Tr2014 [36, 37]. Our findings suggest that while complete loss of BRCA2 is clearly associated with increased risk of breast, ovarian and other cancers, hypomorphic alleles that disrupt key BRCA2 functions, are likely to be equally pathogenic. It is possible that such mutations may cause cancer in carriers at a younger age because they are not dependent on an additional mutation in TRP53 or other genes that have an anti-apoptotic response.
Fig 8. Relationship between genomic instability and tumorigenesis.
Mutations in Brca2 that result in mild genomic instability (GIN) do not contribute to tumorigenesis. Mutations that result in moderate GIN lead to tumor formation in mutant mice. However, mutations (e.g. functionally null allele, Brca2Ko) that cause severe GIN result in increased cell death and decreased tumor formation. Genotypes in blue indicate the mice described in the present study.
In conclusion, our results provide strong evidence to support the importance of a physical interaction between BRCA2 and PALB2. In spite of their ability to independently bind to RAD51, their heterodimerization via the N-terminus of BRCA2 and the C-terminus of PALB2 is essential for their function. Disruption of this interaction not only affects the DSB repair but also the ability to protect the stalled replication forks. These unique mouse models will help distinguish the importance of HR and fork protection in BRCA2’s and PALB2’s ability to protect genomic integrity. In addition, the unique mouse models described here will be useful for preclinical studies because of their high tumor incidence and low tumor latency.
Materials and Methods
Ethics statement
Mice were treated in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (The National Academies Press; Washington DC, 8th edition). The protocol was approved by the Animal Care and Usage Committee of NCI-Frederick (NIH) (Animal Study Proposals: 12–471 and 15–471).
Generation of G25R knock-in targeting vector
A single base substitution (G>A) was generated in the 6th nucleotide of exon 3 to change codon 25 (GGA> AGA, Gly25Arg) of Brca2 clone in BAC RPCI22-421-23A by recombineering in SW102 cells using the galK-based selection-counterselection method as described previously[21, 47]. The G>A substitution generated a MseI restriction enzyme recognition site. Next, a loxP-PGK-Tn5-neomycin-bp(A)-loxP cassette along with BanII and EcoRI restriction sites were inserted into intron 3 by recombineering. Finally, a 10.7 kb genomic region of Brca2 was retrieved by gap repair from the BAC into pBSK(+) plasmid containing the Thymidine Kinase (TK) gene under the control of the MC1 promoter. The resultant plasmid was the G25R knock-in targeting vector (S1 Fig).
Targeting Brca2 in ES cells and generation of Brca2G25R-Neo/+ mice
G25R knock-in targeting vector was linearized with NotI and electroporated into V6.4 (129 X C57BL/6J) ES cell line as described previously [48]. G418R, FIAUR ES cell clones were screened by Southern analysis of BanII and EcoRI restriction enzyme digested genomic DNA using 5’ and 3’ probes (S1 Fig). A few correctly targeted ES cell clones were injected into C57BL/6 blastocysts to generate chimeras. One of these chimeras transmitted the targeted allele in the germ line and Brca2G25R-Neo/+ pups were obtained.
Mice
To obtain Brca2G25R/+ mice, Brca2G25R-Neo/+ mice were crossed to β-actin-Cre-deleter strain [49]. To generate Palb2Ko/+ mice, CG0691 ES cells obtained from SIGTR were injected to into C57BL/6 blastocysts. Chimeras were crossed with C57BL/6 females to Palb2Ko/+ pups. Brca2G25R/Ko were obtained by breeding Brca2G25R/G25R or Brca2G25R/+ mice with Brca2ko/+ mice [50]. Brca2G25R/Ko mice were crossed with Palb2Ko/+ and Trp53Ko/+ mice to obtain mice of various genotypes used in the study. Mouse colony was maintained on a mixed C57BL/6 and 129/SvEv genetic background. All animal studies were performed as per the protocols outlined in the Guide for the Care and Use of Laboratory Animals and approved by the NCI-Frederick Animal Care and Use Committee.
Genotyping
For genotyping purposes, genomic DNA was extracted from tail biopsies, fresh or frozen tumor tissues following standard procedures. Genotyping was performed by PCR. Briefly, approximately 50 ng of DNA was amplified with the Taq DNA polymerase using primers listed in Supplementary Information (S1A and S1D Fig, S1 Table).
Histology and immunohistochemistry
For histology, tissues were fixed in 10% formalin overnight, paraffin-embedded, sectioned and stained with H&E (hematoxylin and eosin). Statistical differences in tumor types were assessed via Fisher's exact test, and survival curves were analyzed utilizing Prism Graphpad. For germ-cell visualization 3-week old testis sections were immunostained with germ cell marker (MVH) as previously described [51]. Antibodies: Rabbit anti-DDX4/MVH (Abcam ab13840; 1:400); goat anti-rabbit Alexa 488 conjugate (Molecular Probes A11008; 1:1,000).
Mouse embryonic fibroblasts
MEFs were generated from E13.5 embryos and cultured in 10% FBS at 5% O2. Cell growth analyses were done by seeding 10cm plates with 5 X 105 cells and counting the cells every day. Metaphases were collected after a 24 hour treatment with 100nM MMC and spread as previously described [52]. Immunofluorescence of RAD51 foci was done by seeding cells on coverslips, irradiating with 1000 rads the following day. Fixed cells were collected after 3 hours and cells were stained with Mouse anti-γH2AX (1:500, Millipore JBW301) and rabbit anti-RAD51 (1:250, Millipore PC130).
TUNEL staining
Paraffin sections of testes were TUNEL stained using the In Situ Cell Death Detection Kit (Roche 11684817910) following the directions included with the kit.
Meiotic chromosome analysis
Meiotic spreads were performed as previously described[53]. Primary antibodies used in this study: rabbit anti-SYCP3 (1:500, Abcam ab13840); mouse anti-γH2AX (1:500, Millipore JBW301); mouse anti-SYCP3 (1:500 Abcam ab20244); rabbit anti-RAD51 (1:250, Millipore PC130).
CGH array
A comparative genomic hybridization array was used for 4 tumors from the following genotypes, Brca2G25R/+;Trp53Ko/+ (2 Lymphomas, 2 Osteosarcoma), Brca2G25R/G25R;Trp53Ko/+, (2 Lymphomas, 2 Osteosarcoma) Brca2G25R/Ko;Trp53Ko/+ (4 Osteosarcomas) and Brca2G25R/G25R;Palb2Ko/+;Trp53Ko/+ (2 Sarcomas, 2 Osteosarcoma). The DNA was isolated using the Qiagen DNeasy Blood and Tissue Kit, wild-type mouse tail DNA was used as the control. The DNA was hybridized to an Agilent SurePrint G3 Mouse CGH Array 4X180K chip. The results from the array were analyzed using Nexus Copy Number from BioDiscovery.
DNA fibers
DNA fiber analysis was performed as previously described with minor modifications [54, 55]. Briefly, cells were seeded at 50% on the previous day. CldU was added to media for 15 min, cells were washed 3X with pre-warmed 1X PBS. Then IdU was added in pre-warmed media for 15 min. Cells were then treated with 4mM HU for 4 hrs. Cells were washed 2X with pre-warmed 1X PBS, trypsinized and placed in cold PBS at a concentration of 2.5 X 105 cells/ml. On pre-cleaned slides 7.5 μL of cell lysis solution (200mM Tris-HCl pH 7.4, 50mM EDTA, 0.5%SDS) was placed near the top of the slide. Then 2.5 μL of cells were added to this drop, mixed and then left at RT for 8 minutes. The slide was then tilted to allow spreading of fibers at approximately 45° angle to allow the drop to slowly run down the slide. The slides were then air-dried. Next, the slides were fixed in methanol/acetic acid 3:1 at 4° O/N. Slides were then re-hydrated with 1X PBS, denatured in 2.5M HCL for 1 hr, washed 5X with PBS. The slides were then blocked (2% BSA, 0.1% Tween 20, 1X PBS) for 40 minutes. The slides were then labeled with mouse anti-BrdU/IdU 1:100 (Becton Dickinson, #347580) and rat anti-BrdU/CldU (1:500) for 2.5hrs at RT or 4° O/N. Slides were then washed 5X with 0.2% PBS-Tween 20. Secondary antibodies, Alexa-fluors anti-rat 594, and anti-mouse 488 were used at 1:300 for 1 hr in dark at RT. Slides were washed again with 0.2% PBST, air-dried and mounted with anti-fade. Fibers were then visualized and imaged at 63X. Measurements of fibers were performed utilizing ImageJ software.
Supporting Information
A: Schematic representation of the gene targeting strategy to generate the Brca2G25R knock-in allele showing the wild type locus (Brca2WT), targeting vector, targeted allele (Brca2G25R-Neo) showing the presence of loxP-Neo-loxP cassette and the Brca2G25R knock-in allele with the point mutation and a single loxP site. The first four exons of Brca2 exons are indicated as boxes with corresponding numbers. Location of restriction sites used for Southern-based genotyping are indicated by B (BanII) and E (EcoRI). G25RGenoF and G25RGenoR designate location and direction of primers (in blue) used for PCR-based genotyping. Asterisk marks the location of the G>A mutation in the first base of codon 25 of Brca2 in exon 3. Location of probes used to detect 5′ and 3′ end targeting are shown with a solid box below the Brca2WT locus. B. Southern blot analysis showing correct gene targeting. D. Sequence read confirming G>A substitution in codon 25 of Brca2. D. Schematic representation of the Brca2 null allele (Brca2Ko/+) showing deletion of a portion of exon 11 which is replaced with human HPRT1 minigene. E. PCR-based genotyping of Brca2G25R allele utilizing the primers show in blue arrows in A.
(PDF)
A. Representative meiocytes at pachynema of (B2Ko/+, left and B2GR/Ko, right) stained with SYCP3 (top, red), and RAD51 (middle, green) and merged (Bottom), arrows indicate the XY Body. B. Quantification of RAD51 foci along the XY body. Error bars: SD, * p<0.05. Controls are represented by either: Brca2Ko/+ or Brca2G25R/+. Abbreviations: Brca2Ko/+ = B2Ko/+, Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko.
(PDF)
Representative images of testes with TUNEL staining (green) and counterstained with DAPI (blue) to show the cells that are undergoing apoptosis at 3 weeks (A.) and 7 weeks (B.). Quantification of TUNEL positive cells at 3 weeks (C.) and 7 weeks (D.). Error Bars: SD, *p<0.05.
(PDF)
Distribution of tumor types found in mice by gender of indicated genotypes. A. Females and B. Males. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors.
(PDF)
Distribution of tumor types found in mice of indicated genotypes. A. Females and B. Males. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors, UNK = cause of death unknown.
(PDF)
A. Testis cross section of Brca2Ko/+;Palb2Ko/+ mice showing normal spermatogenesis at 3 weeks and 7 weeks of age. Far left panel: Testis cross section of Brca2GR/GR;Palb2Ko/+ mouse at 25 weeks showing normal spermatogenesis. B. Kaplan-Meier tumor-free survival curves of Control, Control;Palb2Ko/+ and Brca2GR/GR;Palb2Ko/+ mutant animals (104 week study). C. Total distribution of tumors found in indicated genotypes. D. Tumor distribution broken down by gender of indicated genotypes. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors, UNK = cause of death unknown. Controls are represented by: Brca2+/+ and Brca2Ko/+. Abbreviations: Brca2G25R/G25R = B2GR/+, Palb2Ko/+ = P2Ko/+.
(PDF)
Individual copy number variation (CNV) graphs for each of the heterogeneous tumors across the genome. Chromosome number is at the bottom and amount of gain of loss is indicated at the Y-axis. (+1 or -1 indicates a 2 copy gain or 2 copy loss) Abbreviations: Ly = Lymphoma, OS = Osteosarcoma, HS = Histiocytic Sarcoma, Sarc = Sarcoma.
(PDF)
A. Image of amplified region around the myc locus in Brca2G25R/G25R Palb2Ko/+ and Brca2G25R/Ko tumors on a Trp53Ko/+ genetic background. B. Percent genome change, and C. total number of copy number changes in the individual tumors. Controls are represented by: Brca2G25R/+; Trp53Ko/+. Abbreviations: Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+, Trp53Ko/+ = p53Ko/+.
(PDF)
A. Fold change of cell growth over the course of 5 days of MEFs of various genotypes. B. Untreated and 100nM MMC treated cell cycle profiles of MEFs of the various genotypes. C. BrdU incorporation of untreated and 100nM MMC treated MEFs of the various genotypes. Error Bars: SD, *p<0.05. Controls are represented by: Brca2+/+ (with or without Palb2Ko/+). Abbreviations: Brca2+/+ = WT, Brca2Ko/+ = B2Ko/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+.
(PDF)
A. RAD51 foci quantification in cells that are PCNA positive in indicated genotypes. B. Fork restart assay: The cells were pulsed with CldU for 15 minutes, then 0.5mM HU was added for 1.5 hrs to stall replication and then released into media containing IdU for 15 minutes. CldU and IdU fibers lengths were measured and plotted for the indicated genotypes. The fibers were evaluated for continuing forks (CldU followed by IdU), stalled forks (CldU alone), or new forks (IdU alone) and graphed as percentage of all fibers evaluated. Controls are represented by: Brca2+/+, Brca2Ko/+.
(PDF)
(DOCX)
Acknowledgments
We thank members of our laboratory for helpful discussions and suggestions. We thank Drs. Jairaj Acharya, Ira Daar, Kajal Biswas for their comments and critical review of the manuscript. We also thank Drs. David Sun and Xiaolin Wu (NCI-Frederick Laboratory of Molecular Technology) for help with array CGH. Thanks to Dr. Arnab Ray Chaudhuri (NCI, Bethesda) for help with DNA fiber analysis and to Dr. Bing Xia (CINJ, Rutgers University) for reagents, Misun Kwon and Dr. Hyunsook Lee (Seoul National University) for help with BRCA2 expression studies. Thanks to Allen Kane (Leidos Biomedical Research, Inc., Scientific Publications, Graphics & Media Department) for help with the illustrations. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Data Availability
The raw array comparative genomic hybridization (aCGH) data generated in our study have been deposited in NCBI's GEO database and are publicly available (Accession: GSE84854).
Funding Statement
This research was sponsored by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, US National Institutes of Health under Contract No. HHSN261200800001E. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–45. Epub 2003/06/05. 10.1038/nrm1127 . [DOI] [PubMed] [Google Scholar]
- 2.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14(6):468–74. Epub 2007/05/23. 10.1038/nsmb1245 . [DOI] [PubMed] [Google Scholar]
- 3.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14(6):475–83. Epub 2007/05/23. 10.1038/nsmb1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle. 2015;14(3):342–53. Epub 2015/02/07. 10.4161/15384101.2014.987614 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–42. Epub 2011/05/14. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17(24):3017–22. Epub 2003/12/19. 10.1101/gad.279003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzel T, Nahse-Kumpf V, Kousholt AN, Klein DK, Lund-Andersen C, Lees M, et al. A genetic screen identifies BRCA2 and PALB2 as key regulators of G2 checkpoint maintenance. EMBO Rep. 2011;12(7):705–12. Epub 2011/06/04. 10.1038/embor.2011.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol Cell Biol. 2002;22(2):669–79. Epub 2002/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24(13):5850–62. Epub 2004/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12(1):68–78. Epub 2011/12/24. 10.1038/nrc3181 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, et al. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14(4):525–35. Epub 2006/07/25. 10.1016/j.ymthe.2006.05.018 . [DOI] [PubMed] [Google Scholar]
- 12.Martinez JS, Baldeyron C, Carreira A. Molding BRCA2 function through its interacting partners. Cell Cycle. 2015;14(21):3389–95. 10.1080/15384101.2015.1093702 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–29. Epub 2006/06/24. 10.1016/j.molcel.2006.05.022 . [DOI] [PubMed] [Google Scholar]
- 14.Al Abo M, Dejsuphong D, Hirota K, Yonetani Y, Yamazoe M, Kurumizaka H, et al. Compensatory functions and interdependency of the DNA-binding domain of BRCA2 with the BRCA1-PALB2-BRCA2 complex. Cancer Res. 2014;74(3):797–807. Epub 2013/11/29. 10.1158/0008-5472.CAN-13-1443 . [DOI] [PubMed] [Google Scholar]
- 15.Siaud N, Barbera MA, Egashira A, Lam I, Christ N, Schlacher K, et al. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet. 2011;7(12):e1002409 Epub 2011/12/24. 10.1371/journal.pgen.1002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–9. Epub 2009/03/10. 10.1016/j.cub.2009.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(6):497–506. Epub 2014/08/08. 10.1056/NEJMoa1400382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman-Colin C, Xia B, Bunting S, Klijn C, Drost R, Bouwman P, et al. Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proc Natl Acad Sci U S A. 2013;110(21):8632–7. Epub 2013/05/10. 10.1073/pnas.1305362110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouwman P, Drost R, Klijn C, Pieterse M, van der Gulden H, Song JY, et al. Loss of p53 partially rescues embryonic development of Palb2 knockout mice but does not foster haploinsufficiency of Palb2 in tumour suppression. J Pathol. 2011;224(1):10–21. Epub 2011/03/16. 10.1002/path.2861 . [DOI] [PubMed] [Google Scholar]
- 20.Rantakari P, Nikkila J, Jokela H, Ola R, Pylkas K, Lagerbohm H, et al. Inactivation of Palb2 gene leads to mesoderm differentiation defect and early embryonic lethality in mice. Hum Mol Genet. 2010;19(15):3021–9. Epub 2010/05/21. 10.1093/hmg/ddq207 . [DOI] [PubMed] [Google Scholar]
- 21.Biswas K, Das R, Eggington JM, Qiao H, North SL, Stauffer S, et al. Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum Mol Genet. 2012;21(18):3993–4006. Epub 2012/06/09. 10.1093/hmg/dds222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10(9):990–6. 10.1038/embor.2009.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JY, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, et al. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014;33(40):4803–12. 10.1038/onc.2013.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528(7582):422–6. 10.1038/nature16142 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17(10):1247–54. Epub 2010/09/28. 10.1038/nsmb.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17(10):1255–9. Epub 2010/09/28. 10.1038/nsmb.1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet. 2001;27(3):271–6. 10.1038/85830 . [DOI] [PubMed] [Google Scholar]
- 28.McAllister KA, Houle CD, Malphurs J, Ward T, Collins NK, Gersch W, et al. Spontaneous and irradiation-induced tumor susceptibility in BRCA2 germline mutant mice and cooperative effects with a p53 germline mutation. Toxicol Pathol. 2006;34(2):187–98. Epub 2006/03/21. 10.1080/01926230600611794 . [DOI] [PubMed] [Google Scholar]
- 29.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mechanisms of development. 2000;93(1–2):139–49. . [DOI] [PubMed] [Google Scholar]
- 30.Adem C, Soderberg CL, Hafner K, Reynolds C, Slezak JM, Sinclair CS, et al. ERBB2, TBX2, RPS6KB1, and MYC alterations in breast tissues of BRCA1 and BRCA2 mutation carriers. Genes Chromosomes Cancer. 2004;41(1):1–11. Epub 2004/07/06. 10.1002/gcc.20057 . [DOI] [PubMed] [Google Scholar]
- 31.Castro E, Jugurnauth-Little S, Karlsson Q, Al-Shahrour F, Pineiro-Yanez E, Van de Poll F, et al. High burden of copy number alterations and c-MYC amplification in prostate cancer from BRCA2 germline mutation carriers. Ann Oncol. 2015;26(11):2293–300. Epub 2015/09/09. 10.1093/annonc/mdv356 . [DOI] [PubMed] [Google Scholar]
- 32.Holstege H, van Beers E, Velds A, Liu X, Joosse SA, Klarenbeek S, et al. Cross-species comparison of aCGH data from mouse and human BRCA1- and BRCA2-mutated breast cancers. BMC Cancer. 2010;10:455 Epub 2010/08/26. 10.1186/1471-2407-10-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios J, Honrado E, Cigudosa JC, Benitez J. ERBB2 and MYC alterations in BRCA1- and BRCA2-associated cancers. Genes Chromosomes Cancer. 2005;42(2):204–5; author reply 6. Epub 2004/11/16. 10.1002/gcc.20125 . [DOI] [PubMed] [Google Scholar]
- 34.Nikkila J, Parplys AC, Pylkas K, Bose M, Huo Y, Borgmann K, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4:2578 10.1038/ncomms3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochimica et biophysica acta. 2014;1846(1):263–75. 10.1016/j.bbcan.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman LS, Thistlethwaite FC, Patel KJ, Yu VP, Lee H, Venkitaraman AR, et al. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 1998;58(7):1338–43. Epub 1998/04/16. . [PubMed] [Google Scholar]
- 37.Connor F, Bertwistle D, Mee PJ, Ross GM, Swift S, Grigorieva E, et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17(4):423–30. Epub 1997/12/17. 10.1038/ng1297-423 . [DOI] [PubMed] [Google Scholar]
- 38.Nikkila J, Parplys AC, Pylkas K, Bose M, Huo Y, Borgmann K, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4:2578 Epub 2013/10/25. 10.1038/ncomms3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wark L, Novak D, Sabbaghian N, Amrein L, Jangamreddy JR, Cheang M, et al. Heterozygous mutations in the PALB2 hereditary breast cancer predisposition gene impact on the three-dimensional nuclear organization of patient-derived cell lines. Genes Chromosomes Cancer. 2013;52(5):480–94. Epub 2013/01/24. 10.1002/gcc.22045 . [DOI] [PubMed] [Google Scholar]
- 40.Donoho G, Brenneman MA, Cui TX, Donoviel D, Vogel H, Goodwin EH, et al. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer. 2003;36(4):317–31. Epub 2003/03/06. 10.1002/gcc.10148 . [DOI] [PubMed] [Google Scholar]
- 41.McAllister KA, Bennett LM, Houle CD, Ward T, Malphurs J, Collins NK, et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 2002;62(4):990–4. Epub 2002/02/28. . [PubMed] [Google Scholar]
- 42.Guijarro MV, Ghivizzani SC, Gibbs CP. Animal models in osteosarcoma. Front Oncol. 2014;4:189 Epub 2014/08/08. 10.3389/fonc.2014.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng AJ, Mutsaers AJ, Baker EK, Walkley CR. Genetically engineered mouse models and human osteosarcoma. Clin Sarcoma Res. 2012;2(1):19 Epub 2012/10/06. 10.1186/2045-3329-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovac M, Blattmann C, Ribi S, Smida J, Mueller NS, Engert F, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun. 2015;6:8940 10.1038/ncomms9940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–25. 10.1038/ng747 . [DOI] [PubMed] [Google Scholar]
- 46.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11(10):1226–41. Epub 1997/05/15. . [DOI] [PubMed] [Google Scholar]
- 47.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic acids research. 2005;33(4):e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tessarollo L. Manipulating mouse embryonic stem cells. Methods Mol Biol. 2001;158:47–63. . [DOI] [PubMed] [Google Scholar]
- 49.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18(2):136–41. . [DOI] [PubMed] [Google Scholar]
- 50.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386(6627):804–10. Epub 1997/04/24. 10.1038/386804a0 . [DOI] [PubMed] [Google Scholar]
- 51.Reinholdt LG, Munroe RJ, Kamdar S, Schimenti JC. The mouse gcd2 mutation causes primordial germ cell depletion. Mechanisms of development. 2006;123(7):559–69. 10.1016/j.mod.2006.05.003 . [DOI] [PubMed] [Google Scholar]
- 52.Kuznetsov SG, Liu P, Sharan SK. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nature medicine. 2008;14(8):875–81. 10.1038/nm.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinholdt L, Ashley T, Schimenti J, Shima N. Forward genetic screens for meiotic and mitotic recombination-defective mutants in mice. Methods Mol Biol. 2004;262:87–107. 10.1385/1-59259-761-0:087 . [DOI] [PubMed] [Google Scholar]
- 54.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19(4):417–23. 10.1038/nsmb.2258. . [DOI] [PubMed] [Google Scholar]
- 55.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998;140(6):1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Schematic representation of the gene targeting strategy to generate the Brca2G25R knock-in allele showing the wild type locus (Brca2WT), targeting vector, targeted allele (Brca2G25R-Neo) showing the presence of loxP-Neo-loxP cassette and the Brca2G25R knock-in allele with the point mutation and a single loxP site. The first four exons of Brca2 exons are indicated as boxes with corresponding numbers. Location of restriction sites used for Southern-based genotyping are indicated by B (BanII) and E (EcoRI). G25RGenoF and G25RGenoR designate location and direction of primers (in blue) used for PCR-based genotyping. Asterisk marks the location of the G>A mutation in the first base of codon 25 of Brca2 in exon 3. Location of probes used to detect 5′ and 3′ end targeting are shown with a solid box below the Brca2WT locus. B. Southern blot analysis showing correct gene targeting. D. Sequence read confirming G>A substitution in codon 25 of Brca2. D. Schematic representation of the Brca2 null allele (Brca2Ko/+) showing deletion of a portion of exon 11 which is replaced with human HPRT1 minigene. E. PCR-based genotyping of Brca2G25R allele utilizing the primers show in blue arrows in A.
(PDF)
A. Representative meiocytes at pachynema of (B2Ko/+, left and B2GR/Ko, right) stained with SYCP3 (top, red), and RAD51 (middle, green) and merged (Bottom), arrows indicate the XY Body. B. Quantification of RAD51 foci along the XY body. Error bars: SD, * p<0.05. Controls are represented by either: Brca2Ko/+ or Brca2G25R/+. Abbreviations: Brca2Ko/+ = B2Ko/+, Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko.
(PDF)
Representative images of testes with TUNEL staining (green) and counterstained with DAPI (blue) to show the cells that are undergoing apoptosis at 3 weeks (A.) and 7 weeks (B.). Quantification of TUNEL positive cells at 3 weeks (C.) and 7 weeks (D.). Error Bars: SD, *p<0.05.
(PDF)
Distribution of tumor types found in mice by gender of indicated genotypes. A. Females and B. Males. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors.
(PDF)
Distribution of tumor types found in mice of indicated genotypes. A. Females and B. Males. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors, UNK = cause of death unknown.
(PDF)
A. Testis cross section of Brca2Ko/+;Palb2Ko/+ mice showing normal spermatogenesis at 3 weeks and 7 weeks of age. Far left panel: Testis cross section of Brca2GR/GR;Palb2Ko/+ mouse at 25 weeks showing normal spermatogenesis. B. Kaplan-Meier tumor-free survival curves of Control, Control;Palb2Ko/+ and Brca2GR/GR;Palb2Ko/+ mutant animals (104 week study). C. Total distribution of tumors found in indicated genotypes. D. Tumor distribution broken down by gender of indicated genotypes. Abbreviations: NT = No tumor observed, BCL = B-cell lymphoma, TCL = T-cell lymphoma, HS = Histiocytic sarcoma, OS = Osteosarcoma, HemS = Hemangiosarcoma, OthSa = Other sarcomas, AD = Adenomas, OthCa = Other carcinomas, MT = Multiple tumors, UNK = cause of death unknown. Controls are represented by: Brca2+/+ and Brca2Ko/+. Abbreviations: Brca2G25R/G25R = B2GR/+, Palb2Ko/+ = P2Ko/+.
(PDF)
Individual copy number variation (CNV) graphs for each of the heterogeneous tumors across the genome. Chromosome number is at the bottom and amount of gain of loss is indicated at the Y-axis. (+1 or -1 indicates a 2 copy gain or 2 copy loss) Abbreviations: Ly = Lymphoma, OS = Osteosarcoma, HS = Histiocytic Sarcoma, Sarc = Sarcoma.
(PDF)
A. Image of amplified region around the myc locus in Brca2G25R/G25R Palb2Ko/+ and Brca2G25R/Ko tumors on a Trp53Ko/+ genetic background. B. Percent genome change, and C. total number of copy number changes in the individual tumors. Controls are represented by: Brca2G25R/+; Trp53Ko/+. Abbreviations: Brca2G25R/+ = B2GR/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+, Trp53Ko/+ = p53Ko/+.
(PDF)
A. Fold change of cell growth over the course of 5 days of MEFs of various genotypes. B. Untreated and 100nM MMC treated cell cycle profiles of MEFs of the various genotypes. C. BrdU incorporation of untreated and 100nM MMC treated MEFs of the various genotypes. Error Bars: SD, *p<0.05. Controls are represented by: Brca2+/+ (with or without Palb2Ko/+). Abbreviations: Brca2+/+ = WT, Brca2Ko/+ = B2Ko/+, Brca2G25R/G25R = B2GR/GR, Brca2G25R/Ko = B2GR/Ko, Palb2Ko/+ = P2Ko/+.
(PDF)
A. RAD51 foci quantification in cells that are PCNA positive in indicated genotypes. B. Fork restart assay: The cells were pulsed with CldU for 15 minutes, then 0.5mM HU was added for 1.5 hrs to stall replication and then released into media containing IdU for 15 minutes. CldU and IdU fibers lengths were measured and plotted for the indicated genotypes. The fibers were evaluated for continuing forks (CldU followed by IdU), stalled forks (CldU alone), or new forks (IdU alone) and graphed as percentage of all fibers evaluated. Controls are represented by: Brca2+/+, Brca2Ko/+.
(PDF)
(DOCX)
Data Availability Statement
The raw array comparative genomic hybridization (aCGH) data generated in our study have been deposited in NCBI's GEO database and are publicly available (Accession: GSE84854).