Abstract
Primary microcephaly is a neurodevelopmental disorder that is caused by a reduction in brain size as a result of defects in the proliferation of neural progenitor cells during development. Mutations in genes encoding proteins that localize to the mitotic spindle and centrosomes have been implicated in the pathogenicity of primary microcephaly. In contrast, the contractile ring and midbody required for cytokinesis, the final stage of mitosis, have not previously been implicated by human genetics in the molecular mechanisms of this phenotype. Citron kinase (CIT) is a multi-domain protein that localizes to the cleavage furrow and midbody of mitotic cells, where it is required for the completion of cytokinesis. Rodent models of Cit deficiency highlighted the role of this gene in neurogenesis and microcephaly over a decade ago. Here, we identify recessively inherited pathogenic variants in CIT as the genetic basis of severe microcephaly and neonatal death. We present postmortem data showing that CIT is critical to building a normally sized human brain. Consistent with cytokinesis defects attributed to CIT, multinucleated neurons were observed throughout the cerebral cortex and cerebellum of an affected proband, expanding our understanding of mechanisms attributed to primary microcephaly.
Keywords: cytokinesis, neurogenesis, primary microcephaly, lissencephaly, autosomal recessive, citron kinase, splicing mutation
Main Text
Primary microcephaly is a genetically heterogeneous neurodevelopmental disorder characterized by a severe reduction in brain growth.1 This decreased brain volume often stems from a primary defect in neurogenesis. The cerebral cortex is composed of neurons born from neural progenitor cells (NPCs) that reside adjacent to the lateral ventricle during early neurodevelopment.2 The mitotic machinery of NPCs is critical for rapid expansion of the progenitor pool underlying normal brain growth and maintenance of neural progenitor multipotency. Largely on the basis of human genetic findings, primary microcephaly has been attributed to defects in the fidelity of mitotic-spindle placement and centrosome stability.3, 4, 5, 6, 7, 8 Surprisingly, other steps in mitosis have not been implicated by neurogenetics in the pathophysiology of microcephaly, despite a series of rodent models showing that cleavage-furrow placement, constriction of the contractile ring, and abscission by the midbody are critical for neurogenesis.9, 10, 11
Citron kinase (CIT) is a multi-domain protein that localizes to the cleavage furrow and midbody, where it functions in cytokinesis, the final step of mitosis (Figure 1A).12, 13 CIT has an N-terminal kinase domain and a number of C-terminal domains that facilitate interactions between components of the contractile ring (anillin, actin, myosin, and RhoA). Although the endogenous substrates of CIT have yet to be confirmed, experimental data from multiple organisms have shown that the kinase activity and scaffolding functions of this protein are both important for successful completion of cytokinesis.12, 14, 15 Conversely, neurons with two or more fully formed nuclei can result from incomplete cytokinesis. The impact of cytokinesis defects on brain development and size are evident in the Cit knockout mouse and the Flathead rat, which has a spontaneous nonsense mutation in Cit exon 1. In rodents, CIT is critical for the proliferation of NPCs and male germ cell precursors. Cit-null animals are characterized by microcephaly, testicular hypoplasia, ataxia, growth deficiencies, and lethal seizures.10, 11 These phenotypes have been linked to cytokinesis defects and the presence of multinucleated cells throughout the cortex and cerebellum. Premature differentiation of early NPCs and widespread apoptosis accumulate across development, resulting in a cerebral cortex and cerebellum less than 50% of the size of a normal brain. Cortical hypoplasia and layer disorganization are predicted to account for the recurrent spontaneous seizures and premature death in these mice.16 Roles for CIT (MIM: 605629) in human brain development and primary microcephaly have been predicted but not yet reported.
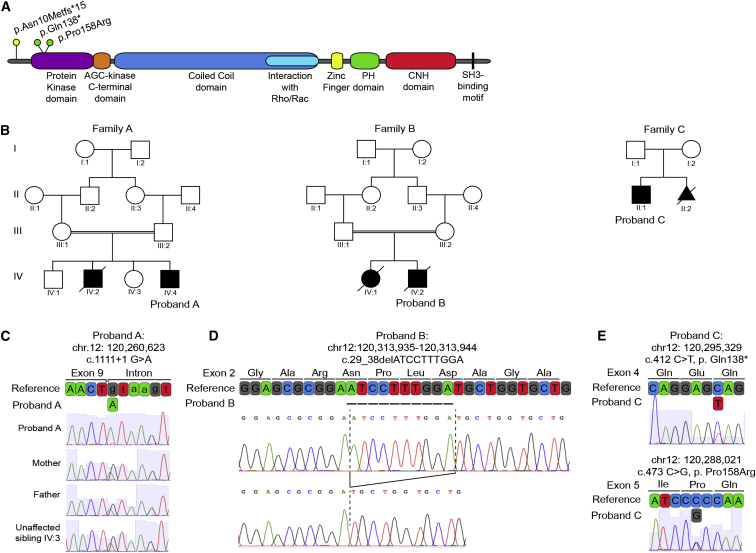
Figure 1.
Recessive CIT Variants in Microlissencephaly
(A) Domain organization of CIT shows pathogenic coding variants in relation to protein domains.
(B) Pedigrees of the families investigated; probands are noted.
(C) Chromatographs and schematic of the boundary between exon 9 and intron 9 of WT reference CIT, the homozygous splice donor variant from proband A (c.1111+1G>A), and the cryptic splice donor site four bases downstream. Parents and unaffected sibling IV:3 are heterozygous carriers.
(D) Chromatographs defining the homozygous 10 bp CIT exon 2 deletion (c.29_38delATCCTTTGGA; hg19 chr12: 120,313,935–120,313,944) amplified from proband B. This deletion creates a frameshift leading to a premature stop 15 codons downstream (not shown).
(E) Chromatographs and schematic of reference CIT exons 4 and 5 and the corresponding nonsense (c.412C>T [p.Gln138∗]) and missense (c.473C>G [p.Pro158Arg]) variants identified in proband C.
Here, we describe three independent families (A, B, and C) each with multiple affected children who present with severe microlissencephaly associated with neonatal mortality (Figure 1B). The clinical features of the probands are provided in Table 1. Egyptian family A is consanguineous with two microcephalic male siblings. The first affected child died in the neonatal period (Figure 1B). Proband A was delivered at term and weighed 2,600 g (2 SDs below the age and sex mean), consistent with the fifth percentile for growth. Head circumference, not measured until 3.5 months of age, was 27 cm (11–12 SDs below the age and sex mean). His length at 3.5 months was 53 cm (4–5 SDs below the age and sex mean), and he continued to experience failure to thrive thereafter. Dysmorphisms included hypotelorism, a sloping forehead, and relatively large ears (Figure 2A). He exhibited axial hypotonia, hypertonia of the upper and lower extremities, increased deep tendon reflexes, and a lack of head support. Microcephaly was confirmed by MRI, which revealed lissencephaly, enlarged ventricles, agenesis of the corpus callosum, cerebellar hypoplasia, and brainstem hypoplasia (Figure 2B). T2 hyperintensity was also noted throughout the white matter, consistent with spastic tetraplegia.
Table 1.
Genetic and Major Clinical Features
| Proband A | Proband B | Proband C | Subject II:2 of Family C | |
|---|---|---|---|---|
| CIT variant (hg19, GenBank: NM_007174.2) | chr12: 120,260,623 | chr12: 120,313,935–120,313,944 | chr12: 120,295,329 and chr12: 120,288,021 | chr12: 120,295,329 and chr12: 120,288,021 |
| c.1111+1G>A | c.29_38delATCCTTTGGA | c.412C>T and c.473C>G | c.412C>T and c.473C>G | |
| – | p.Asn10Metfs∗15 | p.Gln138∗ and p.Pro158Arg | p.Gln138∗ and p.Pro158Arg | |
| GERP score | 5.900 | NA | 4.660 and 5.280 | 4.660 and 5.280 |
| CADD score | 31.000 | NA | 38.000 and 27.800 | 38.000 and 27.800 |
| Gender | male | male | male | male |
| Gestational length | full term | full term | full term | pregnancy terminated at GW 29 + 2 |
| Birth weight | 2.6 kg (−2 SDs) | 1.730 kg (−4 SDs) | 2.92 kg (−1 SD) | NA |
| Birth length | unknown (home birth) | 27.2 cm crown to rump (−2.5 SD) | 46 cm (−2 SDs) | NA |
| Birth head circumference | unknown (home birth) | 24 cm (−8 SDs) | 30 cm (−3.5 SDs) | NA |
| Head circumference at most recent evaluation | 27 cm (−11 to −12 SDs) measured at 3.5 months | NA | 43 cm (−6.5 SDs) measured at 10 years | NA |
| Brain abnormalities | MRI revealed microlissencephaly, agenesis of corpus callosum, cerebellar and brainstem hypoplasia, and T2 hyperintensity of the whole white matter | autopsy revealed microlissencephaly, absent corpus callosum, hindbrain and cerebellum hypoplasia, cerebral cortex hypoplasia, and comparatively large ventricles; a detailed neuropathology is included in the main text | MRI revealed microcephaly, a simplified gyral pattern, and cerebellar and brainstem hypoplasia | fetal MRI revealed microcephaly, a biparietal diameter equivalent to that of a fetus of 23 GWs, and gyration equivalent to a that of a fetus of 26 GWs |
| Neurological findings | hypertonia of upper and lower extremities, axial hypotonia, increased DTRs, and spastic tetraplegia | NA | hypertonia of upper and lower extremities and brisk DTRs in the lower extremities | NA |
| Seizures | no | NA | no | NA |
| Intellectual disability | not assessed | NA | moderate to severe ID with mild autistic features | NA |
| Development | delayed | NA | delayed | NA |
| Dysmorphisms | hypotelorism, sloping forehead, and relatively large ears | prominent occiput, absent fontanelle, large ears, wide nasal bridge, prominent upper lip, high-arched palate, cloudy corneas, right-sided single palmar crease, and small nails | sloping forehead, prominent nose, and relatively large ears | NA |
| Status | unknown | died 1 day after birth | alive | terminated pregnancy |
World Health Organization Child Growth Standards were applied for weight, height, and head circumference. Abbreviations are as follows: DTR, deep tendon reflex; GW, gestational week; and NA, not available.
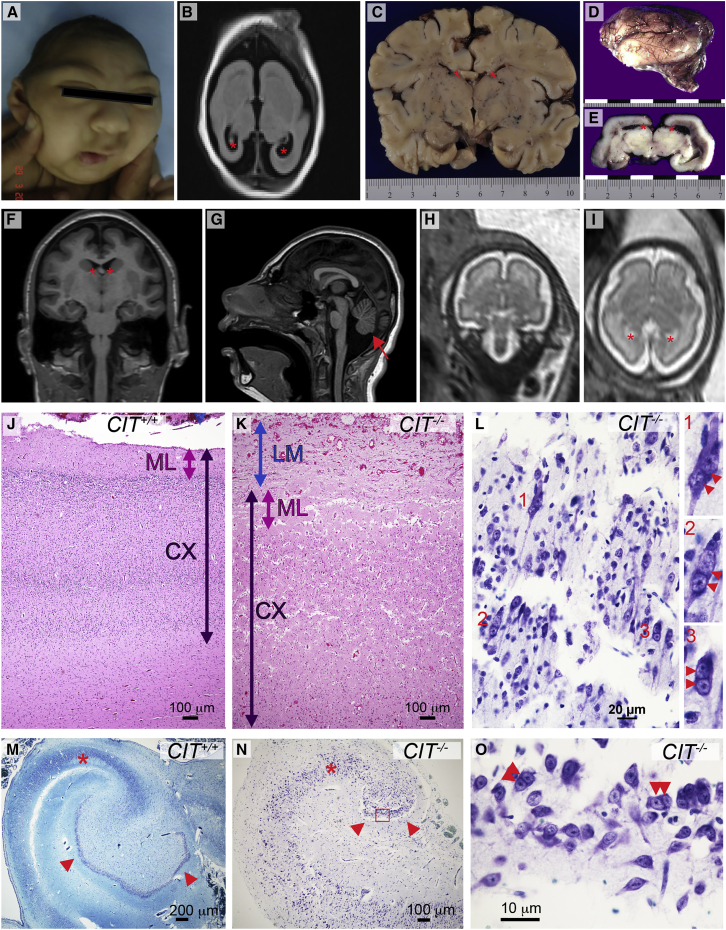
Figure 2.
Structural and Cellular Neocortical Phenotypes
(A) Proband A displays a microcephalic cranium, a sloping forehead, a wide nasal bridge, and hypotelorism.
(B) T2-weighted axial MRI of proband A at 3.5 months shows markedly reduced cerebral cortical size, simplified gyral folding, and enlarged ventricles (red asterisks).
(C) Coronal section of newborn control brain. Width is in centimeters, and lateral ventricles are marked by red asterisks.
(D) Lateral view of the lissencephalic newborn brain from proband B (scale in centimeters).
(E) Coronal section through the mid-thalamus of the brain of proband B shows enlarged lateral ventricles (red asterisks).
(F) Coronal brain MRI of proband C at 10 years of age shows microcephaly, a simplified gyral pattern, and enlarged ventricles (red asterisks).
(G) Mid-sagittal MRI of proband C shows a sloping forehead and a hypoplastic brainstem and cerebellum (red arrow).
(H and I) Coronal (H) and axial (I) fetal brain MRI of affected subject C:II:2 at 29 GWs shows reduced brain volume, reduced gyrification, and enlarged ventricles (red asterisks).
(J and K) Control (J) and proband B (K) cerebral cortex stained with H&E. Compared to the control, leptomeninges (LM; blue arrow) in the proband are excessively thick and contiguous with the molecular layer (ML; pink arrow). Compared to the control, the proband’s cortex (CX; dark purple arrow) is thickened, and the cortical layering is obscure.
(L) High magnification of proband B cortex stained with Kluver-Barrera. Disorganized parenchyma includes many multinucleated neurons: inserts show detail of multinucleated (red double arrowheads) cells labeled 1, 2, and 3.
(M and N) Kluver-Barrera-stained sections of control (M) and proband B (N) temporal lobe. The proband hippocampus is very small and has a hypoplastic pyramidal layer (red asterisk) and greatly reduced dentate gyrus (arrowheads). The normal control hippocampus was imaged at half the magnification to allow for a comparison of the overall hippocampal structure.
(O) Detail of the dentate gyrus from proband B (red box in N) shows binucleated granule cells (double arrowheads).
Family B has two affected children born to consanguineous parents from the United Arab Emirates. Male proband B was noted to have microcephaly, intrauterine growth retardation, and oligohydramnios by ultrasound at 30 gestational weeks (GWs). Echocardiography indicated cardiomegaly, biventricular dilatation, and tricuspid regurgitation. He was delivered by spontaneous vaginal delivery at 39 GWs with a head circumference of 24 cm (8 SDs below the age and sex mean) and a body length of 27.2 cm from crown to rump (2.5 SDs below the mean), consistent with 36 GWs. Despite Apgar scores of 8 at 1 min and 9 at 5 min, he died the following day.
At birth, the brain weight of proband B was 40 g (10% of the average newborn brain weight of 390 g). The cerebral hemispheres were lissencephalic apart from shallow Sylvian fissures. Large ventricles, small well-delineated basal ganglia and thalami, and agenesis of the corpus callosum were noted (Figures 2C–2E). The brain stem and cerebellum were hypoplastic with small cerebellar folia (Figure 4A). Dysmorphic facial features included a prominent occiput, an absent fontanelle, large ears, a wide nasal bridge, a prominent upper lip, a highly arched palate, cloudy corneas, a right-sided single palmar crease, and hypoplastic nails. Renal aplasia was also noted. These features were similar to those observed during the pregnancy and birth of an affected female sibling who presented with microcephaly and renal agenesis and survived only 4 hr after birth (Figure 1B).
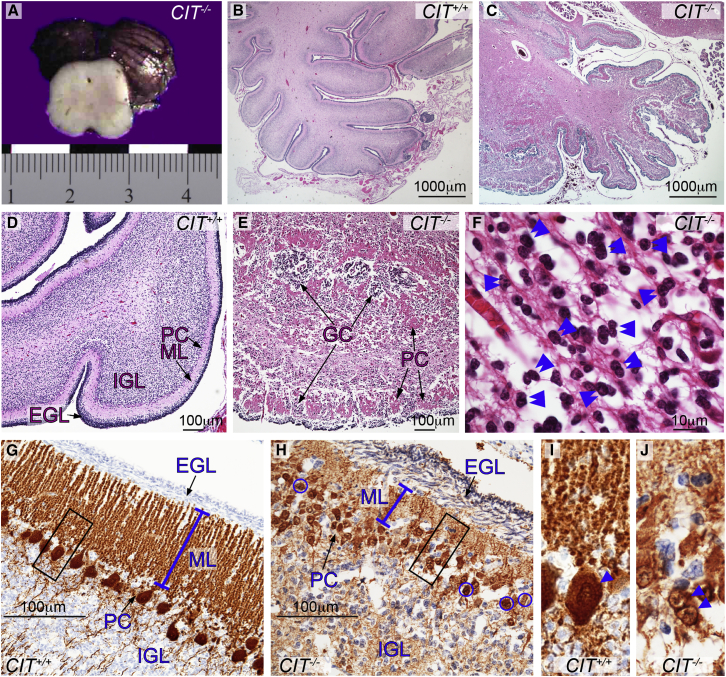
Figure 4.
Structural and Cellular Cerebellar CIT Phenotypes
(A) Section through the proband B midbrain shows an external view of the hypoplastic cerebellum. The scale is measured in centimeters.
(B–F) Histologic cerebellar sections stained with H&E. In proband B, the cerebellar folia are poorly developed and the cortex is disorganized (C, E, and H). Compared to control cerebellum (B and D), cerebellar lamination in proband B is disrupted by clustered Purkinje cells (PCs) interspersed with granule cells (GCs) within fused folia (C and E). Many GCs appear binucleated (F; double blue arrowheads).
(G–J) Cerebellum immunostained for calbindin shows that compared to the control (G), proband B has an abnormally thick EGL, a reduced ML (blue line), and multilayered collections of small PCs (H, arrows). The black boxes is (G) and (H) are enlarged to show that compared to control cells (I), PCs from proband B (J, blue arrowheads) are binucleated within a small soma. Abbreviations are as follows: EGL, external granule layer; IGL, internal granule layer; and ML, molecular layer.
Proband C is the first-born son of unrelated French parents and was delivered at full term. His birth weight was 2,900 g, his body length was 46 cm, and his head circumference was 30 cm (1, 2, and 3.5 SDs below the age and sex mean, respectively), consistent with a diagnosis of microcephaly. Proband C presented with a sloping forehead, a prominent nose, and relatively large ears. His neurological examination was remarkable for hypertonia of the upper and lower limbs and brisk tendon reflexes in the lower limbs. He walked independently at 18 months of age. He has developmental delay with moderate to severe ID and mild autistic features. No metabolic, electroretinographic, or electroencephalographic abnormalities were detected. By the age of 10 years, his head circumference was 6.5 SDs below the age and sex mean. MRI performed at this age showed a simplified gyral pattern and a hypoplastic cerebellum (Figures 2F and 2G). A second child in this family (II:2) was also confirmed to have microcephaly by fetal MRI at 29 GWs (Figures 2H and 2I). The biparietal diameter measured from the fetal MRI at that time was consistent with 23 GWs.
Although the microcephalic phenotype displayed by these probands aligns with a diagnosis of autosomal-recessive primary microcephaly (MCPH [MIM: 251200]), the additional syndromic features and neonatal mortality suggest that this is not classic MCPH. In order to identify a genetic diagnosis for these three probands, we employed whole-exome sequencing (WES) and candidate gene screening. Parents provided written informed consent that was approved by institutional review boards at the respective institutions. Quad WES (mother, father, proband, and unaffected sibling) was performed for family A to identify pathogenic variants. DNA was isolated from peripheral blood according to standard procedures. WES was performed at the Beijing Genomics Institute (BGI) according to standard protocols for Agilent SureSelect All Exon Kit V4 library preparation and Illumina HiSeq 2000 sequencing to a mean sequence depth of 98× for 99% exome coverage, and the sequence was analyzed with the SOAPaligner.17, 18 Variants were filtered against dbSNP, 1000 Genomes, the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (ESP6500), and the BGI SNP database. An average of 65,457 high-quality single-nucleotide variants and indels were identified per individual. Common variants (with a minor allele frequency [MAF] > 0.005) were removed, and rare variants were analyzed in relation to homozygous recessive, compound-heterozygous, and dominant modes of inheritance. Targeted analysis did not identify a causal mutation in any gene present in the OMIM database and was therefore followed by a complete analysis of all coding regions. A homozygous recessive G>A transition (c.1111+1G>A [GenBank: NM_007174.2]; hg19 chr12: 120,260,623) disrupting the donor splice site (5′ ss) of exon 9 in CIT was identified, consistent with consanguinity (Figure 1C). Segregation of this variant in the family was confirmed by Sanger sequencing, and both parents and the unaffected sibling were found to be heterozygous carriers.
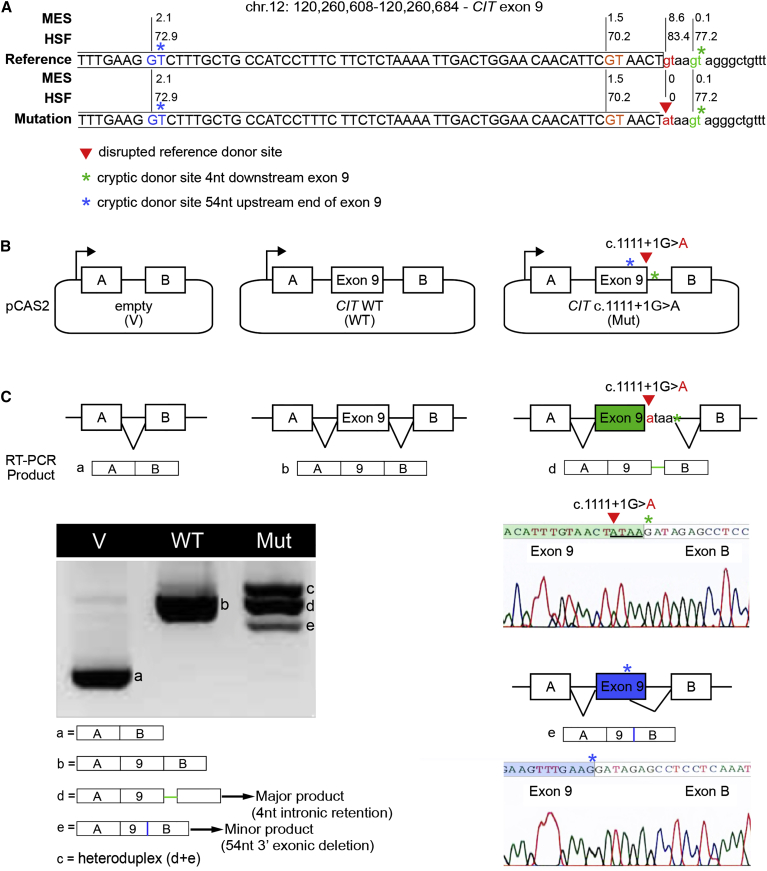
The c.1111+1G>A variant’s potential impact on RNA splicing was first evaluated by in silico approaches (Human Splicing Finder and MaxEntScan). This analysis predicted the destruction of the reference 5′ ss and revealed the presence of cryptic donor sites both upstream and downstream (Figure 3A). We evaluated the impact on splicing experimentally by performing a minigene reporter assay as previously described.19 We amplified the genomic region of interest (CIT exon 9 and at least 150 bp of flanking intronic sequences) from control and proband A (CIT c.1111+1G>A) genomic DNA and cloned it into the intron of the pCAS2 reporter plasmid to generate three-exon wild-type (WT) and mutant minigenes, respectively. Plasmids were transfected into HeLa cells, RNA was extracted 24 hr after transfection, and the splicing pattern of the minigenes was assessed by RT-PCR and sequencing of the gel-purified RT-PCR products.20 Our results confirmed that the c.1111+1G>A variant results in the production of aberrantly spliced transcripts as a result of the concomitant destruction of the reference 5′ ss and activation of two cryptic splice sites located nearby. The major aberrant transcripts induced by c.1111+1G>A result from activation of a cryptic splice site located downstream of the reference 5′ ss, leading to the aberrant retention of four nucleotides from intron 9 in the minigene’s transcripts (Figures 3B–3C and Figure S1). This alteration is expected to introduce a premature stop codon in exon 10 of CIT, creating a transcript that is predicted to undergo nonsense-mediated decay. The minor aberrant transcripts induced by c.1111+1G>A result from the activation of a cryptic splice site located upstream of the reference 5′ ss, leading to the aberrant deletion of 54 nucleotides from exon 9 in the minigene’s transcripts. This alteration is expected to produce an in-frame deletion of 18 amino acids in the kinase domain of CIT (CIT−18aa). The stability of mutant CIT−18aa has not been evaluated, but it is possible that the deletion disrupts kinase activity.
Figure 3.
Functional Analysis of CIT c.1111+1G>A Confirms Its Negative Impact on RNA Splicing
(A) Schematic representation of CIT sequence across the boundary between exon 9 and intron 9. The reference donor site (WT, red nucleotides) and cryptic donor sites (blue, orange, and green nucleotides) were delineated and scored with MaxEntScan (MES) and Human Splicing Finder (HSF) as shown in Figure S1. Only splice sites predicted by both algorithms in the WT sequence are shown. Exonic and intronic sequences are indicated by upper and lower case, respectively. The CIT c.1111+1G>A mutation from proband A (red arrowhead) disrupts the WT donor site. Asterisks indicate cryptic splice sites activated in the presence of c.1111+1G>A.
(B) A minigene splicing assay was performed as previously described. The structures of the CIT exon 9 minigenes were used in the splicing reporter assay. Arrows represent the CMV promoter. Boxes represent exons, and lines in between indicate introns.
(C) Analysis of WT and mutant (Mut) minigene splicing patterns. Transcripts were analyzed by RT-PCR after expression in HeLa cells with the use of primers specific to the minigene’s exons A and B (Table S1). RT-PCR products were separated on an agarose gel and sequenced. Aberrantly spliced products, accompanied by chromatographs, are illustrated on the right.
Resequencing of CIT coding exons and splice junctions in 35 additional probands with primary microcephaly identified two additional individuals, probands B and C, with recessive pathogenic variants. Proband B DNA was extracted from formalin-fixed, paraffin-embedded brain tissue with the QIAGEN QIAamp DNA FFPE Tissue Kit (catalog no. 56404) per the manufacturer’s instructions. The 46 coding exons of CIT on chromosome 12 were screened by dideoxy sequence analysis on an ABI 3730 sequencer (Applied Biosystems, Life Technologies), and sequence traces were analyzed with Sequencher 5.1 (Gene Codes Corporation). A homozygous pathogenic mutation consistent with consanguinity was identified in proband B. This 10 bp deletion in exon 2 of CIT creates a premature stop codon 15 codons downstream (c.29_38delATCCTTTGGA [p.Asn10Metfs∗15] [GenBank: NM_007174.2]; hg19 chr12: 120,313,935–120,313,944) and is predicted to function as a null allele (Figures 1A and 1D).
DNA from proband C and subject C:II:2 was isolated from peripheral blood according to standard procedures at the Robert Debré Hospital in Paris. CIT was included on a microcephaly gene panel for cohort screening by RainDance microdroplet PCR and 2 × 150 bp sequencing with an Illumina MiSeq system. Deep (142×) sequencing reads were mapped to the UCSC Genome Browser (hg19) with MiSeq analysis software, the Burrows-Wheeler Aligner, and the Genome Analysis Toolkit, and panel exons were 99% covered. Variants were filtered against MAF (>0.005), dbSNP, 1000 Genomes, the NHLBI ESP Exome Variant Server, and an in-house dataset. Rare variants were annotated for functional features of coding nucleotides with publically available databases outlined for WES. Both proband C and subject C:II:2 were found to carry compound-heterozygous CIT variants—one generating a nonsense variant in exon 4 (c.412C>T [p.Gln138∗]; hg19 chr12: 120,295,329) and the other generating a missense variant in exon 5 (c.473C>G [p.Pro158Arg]; hg19 chr12: 120,288,021), which encodes the kinase domain (Figures 1A and 1E). The CIT variants identified in proband B, proband C, and subject C:II:2 (c.29_38delATCCTTTGGA, c.412C>T, and c.473C>G, respectively) did not alter splicing (Figure S1). The p.Pro158Arg (c.473C>G) variant is predicted to disrupt kinase function, but not CIT accumulation. Compared with the homozygous null phenotypes in probands A and B, the severity of the microcephaly exhibited by proband C might be ameliorated by the accumulation of the p.Pro158Arg variant. The parents and unaffected relatives in families B and C were not available for carrier testing.
The CIT mutations presented here were absent from online genomic variant databases, including 1000 Genomes, the NHLBI ESP Exome Variant Server, dbSNP 141, and the Exome Aggregation Consortium (ExAC) Browser.21, 22, 23 Bioinformatic variant annotation by SeattleSeq Variation Annotation revealed that the point mutations in our probands affect highly conserved bases with high GERP and CADD scores that are predicted to be pathogenic (Table 1).24, 25 Further analysis of the genetic variation in CIT showed that loss-of-function alleles are infrequent. Homozygous nonsense or frameshift variants were not present in the ExAC Browser. However, 15 rare heterozygous truncating variants were detected (MAF < 0.001), suggesting that there is high evolutionary pressure against these alleles and supporting the deleteriousness of the recessive CIT variants described here. These findings correspond to a Residual Variation Intolerance Score (RVIS) of −2.35, indicating that CIT is in the top 1.14% of genes intolerant to common functional genetic variation.26
The cytokinesis functions of CIT have been conserved across evolution. Deficiencies of rodent CIT have been shown to disrupt NPC proliferation, implicating CIT in primary microcephaly. Although severe reductions in brain size are often accompanied by a simplified gyral pattern, gene mutations that disrupt neuronal migration are generally attributed to classical lissencephaly or smooth-brain disorders.1, 27, 28, 29 This combination of extreme reduction in brain volume and lissencephaly observed in probands A and B could provide evidence for a reduction in cortical surface area, which does not support gyri formation in the human brain. This idea is supported by the genotypic-phenotypic spectrum of microcephaly to microlissencephaly on the basis of recessive inheritance of CIT pathogenic variants, described here and in an accompanying study.30 Homozygous missense variants in the kinase domain encoded by CIT are associated with primary microcephaly with a simplified gyral pattern and few additional syndromic features. Conversely, homozygous null variants in CIT represent the severe end of the spectrum associated with extremely small lissencephalic brains and neonatal mortality.
Given the well-established role of CIT in cytokinesis, our three affected individuals provide evidence that this is an important pathogenic mechanism for microcephaly, a disorder previously linked primarily to gene products that localize to the centrosome or mitotic spindle.31 This cell biology was originally implicated by studies of ASPM, which localizes to the midbody, where it can be co-immunoprecipitated with CIT.32, 33, 34 Severe microcephaly with widespread multinucleated neurons is characteristic of the structural and cellular pathology observed in the Cit knockout mouse and the Flathead rat (Table S2).10, 11 In accordance with approval by institutional research boards and ethics committees, post-mortem analysis of proband B allowed the human cell and molecular neuropathology associated with pathogenic variants in CIT to be analyzed. All non-neural tissue examined had normal cytology, but multinucleated neurons were observed throughout the neuraxis of proband B, a hallmark of cytokinesis defects (Table S3). This neural-specific phenotype is phenocopied in the rodent models, yet the molecular details that underlie the tissue specificity of this feature have not been determined.
To analyze the cytoarchitechture, we stained brain sections from proband B with H&E and stained selected sections with Kluver-Barrera or calbindin. Microscopically, the profoundly microcephalic cerebral cortex showed both cytological and organizational abnormalities in many areas. The neocortex was excessively thick, and the six cortical layers were replaced by a molecular layer and two broad layers composed of loosely and irregularly scattered neurons (Figures 2J and 2K). The underlying white matter was unmyelinated and contained scattered ectopic neurons. The overlying leptomeninges were greatly thickened with reticulin and collagen fibers, were prominently vascularized, and contained heterotopic astrocytes and neurons, some of which were multinucleated (Figure 2L). The hippocampi were dysplastic, small, and under-rotated and showed hypoplastic dentate gyri (Figures 2M–2O). These cortical and hippocampal phenotypes are consistent with those observed in rodent models that have been attributed to massive apoptosis during neurodevelopment.11, 16, 35 Alternatively, although migration defects have not been described in association with Cit mutations in rodents, defective radial migration of human binucleated immature neurons is a novel, as yet untested explanation for this cortical disorganization.
Cortical hypoplasia and layer disorganization account for the recurrent spontaneous seizures associated with premature death in rodent models.16, 35 Extrapolating from these rodent models, the cytoarchitectural abnormalities observed in proband B are consistent with cytokinesis defects that contribute to reduced neuronal precursor proliferation, multinucleation of neurons, and increased cell death. Despite the cytoarchitectural similarities, pathogenic CIT variants have not been detected in epilepsy cohorts, including the Epi4K exome sequence collection of 264 epileptic encephalopathy trios.36
Neuropathology studies also revealed that the cerebellar cortex of proband B was hypoplastic and dysplastic (Figure 4). Laminar disorganization was more evident in the hemispheres than the vermis, where folia were fused and Purkinje cells were observed in multilayered islands interspersed with abnormal granule cell domains (Figures 4B–4H). At birth, the external granule cell layer (EGL) is usually compact with four to six rows of bipolar cells abutting the molecular layer (ML) (Figures 4B, 4D, and 4G). In proband B, the EGL was wider, the ML was narrower, and both were merged with the Purkinje cell layer (Figures 4G and 4H). In this case, the EGL was composed of two or three compact rows of cells overlying a looser band of horizontally oriented, thin, elongated cell bodies, some of which were clearly binucleated (Figures 4E, 4F, and 4H). Compared to control cells, Purkinje cells had short simplified dendritic arborization, and many were multinucleated (Figures 4G–4J). Likewise, the internal granular layer was severely hypo-cellular (Figures 4G and 4H).
In summary, we report the identification of pathogenic variants in CIT as a genetic basis for primary microcephaly, microlissencephaly, and cerebellar hypoplasia, whose phenotypic features are remarkably similar to those of Cit−/− rodent models. These findings highlight the evolutionarily conserved function of CIT in neurodevelopment and the disproportionate sensitivity of the neuraxis to pathogenic variants in this gene. Alternatively, the differences between non-neural phenotypes observed in proband B and the Cit−/− rodent models might represent the spectrum of species-specific tissues sensitive to homozygous truncating mutations in CIT (Table S2). Multinucleated neurons are rarely observed in the nervous system, apart from their occurrence in ganglion cell tumors. These genetic findings implicate novel mechanisms in the pathogenesis of primary microcephaly and allow us to describe the human presentation of this striking multinucleated neuronal phenotype.
Acknowledgments
We thank the families for their participation and contribution to this research. We appreciate Ritesh K.C. and Brian McGrath for stimulating discussion during manuscript preparation and Dr. Joseph Loturco, Dr. Joseph Gleeson, Dr. Ozgur Rosti, and Dr. Hongda Li for collaborations on CIT genetics and molecular biology. This work was supported by the NIH (R00HD069624 to S.L.B), the French Institut National du Cancer and Direction Générale de l’Offre de Soins (AAP/CFB/CI to A.M.), the French Ministry of Education (PhD fellowship to O.S.), the French Association Nationale de la Recherche et de la Technologie in the context of public-private partnership between INSERM and Interactive Biosoftware (CIFRE PhD fellowship to H.T.), and the Programme Hospitalizer de Recherché Clinique (grant no. P100128/IDRCB: 2010-A01481-38) and ERA-NET E-Rare-2 2013 (grant no. ANR-13-RARE-0007-01 2013) to S.D., A.V., V.E.G., P.G., and S.P. The sponsors of the study had no role in study design, data collection, analysis and interpretation, or writing of the manuscript.
Published: July 21, 2016
Footnotes
Supplemental Data include two figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.003.
Accession Numbers
The accession number for the c.29_38delATCCTTTGGA (chr12: 120,313,935–120,313,944) sequence reported in this paper is ClinVar: SCV000257393. The accession numbers for the CIT variants reported in this paper are ClinVar: SCV000257393, SCV000292379, SCV000292380, and SCV000292381.
Web Resources
1000 Genomes Project, http://browser.1000genomes.org
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
dbSNP 141, http://www.ncbi.nlm.nih.gov/projects/SNP/
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Residual Variation Intolerance Score (RVIS), http://genic-intolerance.org
SeattleSeq Variation Annotation, http://snp.gs.washington.edu/SeattleSeqAnnotation138/
UCSC Genome Browser, http://genome.ucsc.edu
World Health Organization Child Growth Standards, http://www.who.int/childgrowth/standards/en/
Supplemental Data
References
- 1.Gilmore E.C., Walsh C.A. Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:461–478. doi: 10.1002/wdev.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paridaen J.T., Huttner W.B. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014;15:351–364. doi: 10.1002/embr.201438447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond J., Roberts E., Mochida G.H., Hampshire D.J., Scott S., Askham J.M., Springell K., Mahadevan M., Crow Y.J., Markham A.F. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 4.Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A., Girimaji S.C., Duvvari M.R., Blanton S.H. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilgüvar K., Oztürk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A.O., Gökben S. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guernsey D.L., Jiang H., Hussin J., Arnold M., Bouyakdan K., Perry S., Babineau-Sturk T., Beis J., Dumas N., Evans S.C. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am. J. Hum. Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholas A.K., Khurshid M., Désir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier-Fisher A., Lin D.C., Greeve M., Kaplan D.R., Rottapel R., Miller F.D. Lfc and Tctex-1 regulate the genesis of neurons from cortical precursor cells. Nat. Neurosci. 2009;12:735–744. doi: 10.1038/nn.2339. [DOI] [PubMed] [Google Scholar]
- 10.Sarkisian M.R., Li W., Di Cunto F., D’Mello S.R., LoTurco J.J. Citron-kinase, a protein essential to cytokinesis in neuronal progenitors, is deleted in the flathead mutant rat. J. Neurosci. 2002;22:RC217. doi: 10.1523/JNEUROSCI.22-08-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Cunto F., Imarisio S., Hirsch E., Broccoli V., Bulfone A., Migheli A., Atzori C., Turco E., Triolo R., Dotto G.P. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/s0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 12.El Amine N., Kechad A., Jananji S., Hickson G.R. Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J. Cell Biol. 2013;203:487–504. doi: 10.1083/jcb.201305053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green R.A., Paluch E., Oegema K. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 14.Gai M., Camera P., Dema A., Bianchi F., Berto G., Scarpa E., Germena G., Di Cunto F. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell. 2011;22:3768–3778. doi: 10.1091/mbc.E10-12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassi Z.I., Verbrugghe K.J., Capalbo L., Gregory S., Montembault E., Glover D.M., D’Avino P.P. Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J. Cell Biol. 2011;195:595–603. doi: 10.1083/jcb.201105136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackman J.B., Ramos R.L., Sarkisian M.R., Loturco J.J. Citron kinase is required for postnatal neurogenesis in the hippocampus. Dev. Neurosci. 2007;29:113–123. doi: 10.1159/000096216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R., Li Y., Fang X., Yang H., Wang J., Kristiansen K., Wang J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 19.Soukarieh O., Gaildrat P., Hamieh M., Drouet A., Baert-Desurmont S., Frébourg T., Tosi M., Martins A. Exonic Splicing Mutations Are More Prevalent than Currently Estimated and Can Be Predicted by Using In Silico Tools. PLoS Genet. 2016;12:e1005756. doi: 10.1371/journal.pgen.1005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaildrat P., Killian A., Martins A., Tournier I., Frébourg T., Tosi M. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol. Biol. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- 21.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psaty B.M., O’Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C., Boerwinkle E., CHARGE Consortium Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M., Karczewski K., Minikel E., Samocha K., Banks E., Fennell T., O’Donnell-Luria A., Ware J., Hill A., Cummings B. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gleeson J.G., Allen K.M., Fox J.W., Lamperti E.D., Berkovic S., Scheffer I., Cooper E.C., Dobyns W.B., Minnerath S.R., Ross M.E., Walsh C.A. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 28.Fry A.E., Cushion T.D., Pilz D.T. The genetics of lissencephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:198–210. doi: 10.1002/ajmg.c.31402. [DOI] [PubMed] [Google Scholar]
- 29.Moon H.M., Wynshaw-Boris A. Cytoskeleton in action: lissencephaly, a neuronal migration disorder. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:229–245. doi: 10.1002/wdev.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Bielas S.L., Zaki M.S., Ismail S., Farfara D., Um K., Rosti R.O., Scott E.C., Tu S., Chi N.C. Biallelic Mutations in Citron Kinase Link Mitotic Cytokinesis to Human Primary Microcephaly. Am. J. Hum. Genet. 2016;99:501–510. doi: 10.1016/j.ajhg.2016.07.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris-Rosendahl D.J., Kaindl A.M. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH) Mol. Cell. Probes. 2015;29:271–281. doi: 10.1016/j.mcp.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Higgins J., Midgley C., Bergh A.M., Bell S.M., Askham J.M., Roberts E., Binns R.K., Sharif S.M., Bennett C., Glover D.M. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulvers J.N., Bryk J., Fish J.L., Wilsch-Bräuninger M., Arai Y., Schreier D., Naumann R., Helppi J., Habermann B., Vogt J. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc. Natl. Acad. Sci. USA. 2010;107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paramasivam M., Chang Y.J., LoTurco J.J. ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle. 2007;6:1605–1612. doi: 10.4161/cc.6.13.4356. [DOI] [PubMed] [Google Scholar]
- 35.Roberts M.R., Bittman K., Li W.W., French R., Mitchell B., LoTurco J.J., D’Mello S.R. The flathead mutation causes CNS-specific developmental abnormalities and apoptosis. J. Neurosci. 2000;20:2295–2306. doi: 10.1523/JNEUROSCI.20-06-02295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epi4K Consortium Epi4K: gene discovery in 4,000 genomes. Epilepsia. 2012;53:1457–1467. doi: 10.1111/j.1528-1167.2012.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.