Abstract
Spinal cord injury (SCI) represents a major debilitating health issue with a direct socioeconomic burden on the public and private sectors worldwide. Although several studies have been conducted to identify the molecular progression of injury sequel due from the lesion site, still the exact underlying mechanisms and pathways of injury development have not been fully elucidated. In this work, based on OMICs, 3D matrix-assisted laser desorption ionization (MALDI) imaging, cytokines arrays, confocal imaging we established for the first time that molecular and cellular processes occurring after SCI are altered between the lesion proximity, i.e. rostral and caudal segments nearby the lesion (R1-C1) whereas segments distant from R1-C1, i.e. R2-C2 and R3-C3 levels coexpressed factors implicated in neurogenesis. Delay in T regulators recruitment between R1 and C1 favor discrepancies between the two segments. This is also reinforced by presence of neurites outgrowth inhibitors in C1, absent in R1. Moreover, the presence of immunoglobulins (IgGs) in neurons at the lesion site at 3 days, validated by mass spectrometry, may present additional factor that contributes to limited regeneration. Treatment in vivo with anti-CD20 one hour after SCI did not improve locomotor function and decrease IgG expression. These results open the door of a novel view of the SCI treatment by considering the C1 as the therapeutic target.
Spinal cord injury (SCI)1 belongs to the serious, currently incurable disorders of the central nervous system (CNS), that are often accompanied by a permanent disability (1). Most SCI are related to traumatic spinal cord damages induced by road trauma, falls, or sport injuries (diving). Among the hallmark features of SCI is the axonal disruption in the spinal cord, which is often caused by fractured intervertebral disc or vertebrate. This primary event is followed by a progressive cascade of secondary deleterious reactions spreading to the adjacent spared tissue leading to a worsening of the neurological status (2, 3). Although axonal regeneration is initiated, it is hampered by a combination of local factors that include severe inflammation, lack of trophic support and development of an inhibitory scar-forming environment. In fact, the regenerative capacity of the central nervous system is particularly challenged in SCI as multiple cues converge to act as a chemical and physical barrier for the repair process (4, 5). It is now acknowledged that inflammation is one of the major key player that determines abortive axonal repair in SCI. Thus, although the immune response is recognized as primordial to preserve tissue homeostasis, the spatio-temporal course of inflammation in SCI is not favorable to axonal regeneration.
Acute inflammation develops hours to days following initial spinal cord trauma and is triggered by the axonal damage and neuronal cell death at the lesion site. This is followed by a cellular and molecular inflammatory cascade that initiates the activation of resident glial cells (microglia, astrocytes), infiltration of blood-borne immune cells (lymphocytes, monocytes), and a massive release of chemokines/cytokines by microglia, macrophages and neuronal cells. During the acute inflammatory response, macrophages/microglia phagocyte cell debris and also release neurotoxic factors and stimulate the formation a glial scar that, on the long-term, prevents axonal regrowth. At this stage, a resolution of acute inflammation and a switch toward a neuroprotective cytokines/chemokines profile would favor tissue repair and limit glial scar extension. Instead, a chronic inflammatory process usually develops weeks after trauma and leads to both an aberrant tissue remodeling and a defective tissue repair. Interestingly, although there is currently no efficient therapy for SCI, one of the approved clinical treatments in the early phase of SCI is the administration of methylprednisolone treatment in order to prevent edema and to modulate inflammation (6). However, high-dose methylprednisolone is often associated with severe immunosuppression and side effects, such as pulmonary or urinary tract infections. Thus, there is an urgent need for identifying and testing novel neuro-therapeutic strategies that could prevent inflammation, limit scar extent, and stimulate tissue repair process.
In this context, a large array of molecules and therapies has been tested experimentally with the goal of targeting the healthy tissue adjacent to spinal cord lesion. Such a strategy is aimed at not only protecting this spared tissue from secondary lesion but also stimulating its regenerative potential in order to promote neuronal networks connectivity and axonal outgrowth. Among these proposed therapeutic strategies, cellular therapy belongs to the promising candidate approaches. Ideally, cell therapy strategies may allow to: (1) bridge spinal cord segments over any cavities or cysts formed at the lesion site, (2) replace lost neurons, oligodendroglia, and (3) create a favorable environment for axonal regeneration (7). Different cell therapy approaches include embryonic stem cells, induced pluripotent stem cells (iPS) and different categories of adult stem cells and progenitors such as olfactory unsheathing stem cells, neural progenitor cells (NPC) and mesenchymal stem cells (MSCs). In addition, graft of activated macrophages and transplantation of peripheral or central nervous tissue have been also proposed as an alternative to these stem cells based treatments (8). Comparative to cell therapy, other approaches including the use of exogenously-delivered neuroprotective molecules that would protect neurons from deleterious secondary processes, promote axonal growth and/or enhance nerve conduction in the preserved or regenerating axons. Different classes of molecules were shown to afford variable levels of clinical recovery in animal models of SCI. These comprise anti-inflammatory compounds such as minocycline, neurotrophic factors (BDNF, GDNF, NGF, erythropoietin) and molecules that alleviate regenerating axons from the inhibitory effects of extracellular matrix molecules (9, 10). In particular, chondroitinase ABC eliminates chondroitin sulfate proteoglycans (CSPG) that interact with the major membranous component NG2 and inhibit the regeneration of damaged axons (11). Also, Nogo-A is one of several neurite growth inhibitory receptors expressed by axons (12). Thereby Nogo neutralizing antibodies or blockers of the post-receptors components of RhoA are used to improve long-distance axon regeneration and sprouting (13). Of note, Rho pathway is important to control the neuronal response after CNS injury and the RhoA inhibitor cethrin is actually in phase I/II a clinical trial (14). However, although numerous therapies exhibit potentials to foster neuroprotection, stimulate neurite outgrowth and reduce inflammation, the translation to clinical side is still not crowned by success. Reasons for such a failure are multiple and reside notably in our relatively poor knowledge on the spatiotemporal kinetics of secondary molecular events that characterize the post-trauma phase. This holds particularly important with regard to inflammatory mechanisms that may greatly vary depending on the time point and spinal cord segment considered. Defining a time- and segment-specific window for effective treatment is a key knowledge for an appropriate neuro-therapeutic intervention.
In this work, we present the first exhaustive spatio-temporal proteomic and biochemical analysis performed along the entire spinal cord axis in rat model of SCI. We combined this global proteomic analysis with 3D molecular mass spectrometry imaging study, time course analysis of immune cells infiltration and cytokine microarrays quantification. The whole spectrum of the data allowed us to depict a complete scheme along the spinal cord axis of the cellular and molecular sequel of events occurring through the time course of inflammatory process and abortive regeneration. We identified specific markers for each segment at different time points (3, 7, and 10 days) of the biochemical-pathophysiological processes and observed that, surprisingly, segments caudal to the lesion site host a robust inflammatory process accompanied by the local synthesis of neuroprotective and regenerative molecules. In particular, we demonstrated that the caudal segment immediately adjacent to the lesion site possesses, at least temporarily, all the intrinsic components/features that may allow axonal regeneration to occur. Such a caudal-to-rostral altered regenerative potential is likely hampered by inhibitory signals such as glycans that are abundantly detected or even secreted at the lesion site. Finally, we provided evidence that immunoglobulins (IgGs) are present at the lesion site only 3 days after injury and that in vivo treatment of anti-CD20 did not diminished the presence of these antibodies and did not ameliorate the BBB score of the treated animals.
EXPERIMENTAL PROCEDURES
Reagents
Dulbecco's modified Eagle's medium (DMEM) media, phosphate buffer saline (PBS), fetal calf serum (FCS) were purchased from Invitrogen Life Technologies (Milan, Italy). Rat Cytokine Array Panel A was from R&D Systems (Minneapolis, MN). All chemicals were of the highest purity obtainable. Water, formic acid (FA), trifluoroacetic acid (TFA), acetonitrile (AcN) were purchased from Biosolve B.V. (Valkenswaard, The Netherlands). dl-dithiothreitol (DTT), Thiourea and iodoacetamide (IAA) were purchased from SIGMA (Saint-Quentin Fallavier, France). Trypsin/Lys-C Mix, Mass Spec Grade was purchased from Promega (Charbonnieres, France) and Dynabeads® Protein A from Novex (Life Technologies, France). Fluorescence conjugated secondary antibodies and DAPI were purchased from MolecularProbes (Eugene, OR).
Animals
The study was performed with approval and in accordance to the guidelines of the Institutional Animal Care and Use Committee of the Slovak Academy of Sciences and with the European Communities Council Directive (2010/63/EU) regarding the use of animals in Research, Slovak Law for Animal Protection No. 377/2012 and 436/2012.
Experimental Design and Statistical Rational
For the collection of the conditioned media n = 3 rats control (no balloon inflation, 0 day) were sacrificed, n = 3 rats after 3 days SCI, n = 3 rats after 7 days post injury and n = 3 rats after 10 days post SCI. For the control group all segments are in triplicate. For the group 3 days after SCI, R3 and C3 segments include 2 replicates and n = 3 for the other segments. For the group 7 days after SCI R3 and C3 segments include 2 replicates and n = 3 for the other segments. For the group 10 days post SCI all segments are in triplicate. For the cytokines arrays experiments n = 1 rat were sacrificed per condition. The experiments were performed in experimental replicates. For the IgG purification n = 3 rats per condition (control, 3, 7, and 10 days) were sacrificed. The experiments were performed in biological replicates. For the immunohistochemistry experiments n = 3 rats per condition were sacrificed. The analyses were performed in biological triplicates. For MALDI imaging experiment n = 1 rat was sacrificed 3 days post injury. Twenty-five sections for the complete 3D MALDI imaging experiments have been performed from R1 to C1.
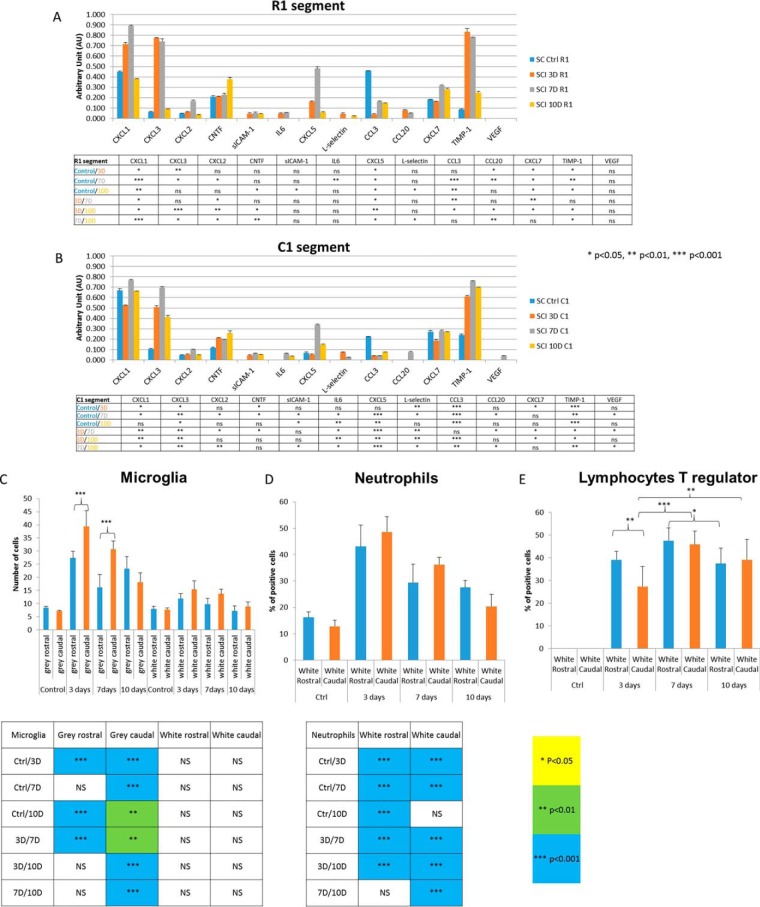
Statistical analysis: For the proteomic statistical analysis of conditioned media, only proteins presenting as significant by the ANOVA test were used with FDR 5%. Normalization was achieved using a Z-score with a matrix access by rows. The immunohistochemistry statistical analyses were based on one-way ANOVA followed by Tukey Kramer test, significant values were marked * p < 0.05, ** p < 0.01, *** p < 0.001. Quantification analyses of immunofluorescence staining for Iba1, FoxP3 and neutrophil elastase were performed on six sections from rostral and caudal/per condition (n = 3 each). Error bars represent the S.E. BBB score analysis was based on one-way ANOVA test. Values of p < 0.05 were considered statistically significant. For the cytokines array panel, the statistical analyses were performed using Student's t test *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent the S.E.
Experimental Spinal Cord Injury Procedure
The SCI was induced using the modified balloon compression technique in adult male Wistar rats, according to our previously published study (15). Manual bladder expression, 2 times a day, was required for 3–10 days after the injury. In the control group a 2-French Fogarty catheter was inserted at the same level of spinal cord, but the balloon was not inflated and no lesion was performed.
Collection of Conditioned Media (CM) from Control and Lesioned Spinal Cord Segments
Experimental SCI rats at 3, 7, and 10 days and sham-operated-control rats were sacrificed by isoflurane anesthesia followed by decapitation. The spinal cord was pressure expressed by injecting sterile saline (10 ml) throughout the vertebrate canal, along the caudo-rostral axis. Each spinal cord was macroscopically observed to check that lesion was well centered at the Th8-Th9 level on the longitudinal axis. Entire spinal cord was divided into transversally sectioned slides (∼1.0 cm thick each) obtained from the lesion site (Th7-Th11) and from the segments rostral (C1-Th6) and caudal (Th12-L6) to the site of injury. Slides were then chopped into 0.5 cm thick sections (2 sections per segment) and deposited into a 12-well culture plate containing 1 ml DMEM without FCS. After 24 h incubation in a humidified atmosphere with 5% CO2 at 37 °C, 1 ml of SCI-derived conditioned media CM (CM-SCI) were collected (rostral, lesion, caudal segments) and centrifuged 30 min at 15,000 rpm at 4 °C. The same procedure was performed for obtaining CM from control spinal cord tissue. Samples were stored at −80 °C. To address the degree of cell-to cell integrity and viability, additional cryostat sections were cut from incubated segments for 24 h and immersed into tissue-tek®. Afterward cryostat sections were processed to standard IHC with NeuN and GFAP antibodies. The data confirmed the cyto- architecture of neurons and glial cells (supplemental Fig. S1) and confirmed the well preserved neuro-glial integrity within cultured spinal cord segments 24 h in vitro. This confirms that the collected molecules are products of vital cells processes.
Conditioned Media Digestion
A volume of 150 μl of tissue supernatants were denatured with 2 m urea in 10 mm HEPES pH 8 by ultrasonication on ice. Protein reduction is realized by incubation with 10 mm DTT for 40 min at 56 °C followed by alkylation with 55 mm IAA for 40 min in the dark. IAA was quenched with 100 mm thiourea. Protein digestion was performed with 30 μg/ml LysC/Trypsin mix, overnight, at 37 °C. Digestion was stopped with 0.5% TFA. The solution was dried in a SpeedVac to reduce the volume. Peptides were desalted using C18 ziptips (Millipore). Elution peptides were dried in a SpeedVac and resuspended in 0.1% FA before injecting into nanoLC.
LC MS/MS Analysis of Conditioned Media
Samples were separated by online reversed-phase chromatography using a Thermo Scientific Proxeon Easy-nLC1000 system equipped with a Proxeon trap column (100 μm ID × 2 cm, Thermo Scientific) and a C18 packed-tip column (Acclaim PepMap, 75 μm ID × 15 cm, Thermo Scientific). Peptides were separated using an increasing amount of acetonitrile (5–35% over 120 min) at a flow rate of 300 nL/min. The LC eluent was electrosprayed directly from the analytical column and a voltage of 1.7 kV was applied via the liquid junction of the nanospray source. The chromatography system was coupled to a Thermo Scientific Q-exactive mass spectrometer programmed to acquire in a data-dependent mode Top 10 most intense ion method. The survey scans were done at a resolving power of 70,000 FWHM (m/z 400), in positive mode and using an AGC target of 3e6. Default charge state was set at 2, unassigned and +1 charge states were rejected and dynamic exclusion was enabled for 25 s. The scan range was set to 300–1600 m/z. For ddMS2, the scan range was between 200–2000 m/z, 1 microscan was acquired at 17,500 FWHM and an isolation window of 4.0 m/z was used.
MS Data Analysis
All the MS data were processed with MaxQuant (17) (version 1.5.1.2) using the Andromeda (18) search engine. Proteins were identified by searching MS and MS/MS data against Decoy version of the complete proteome for Rattus norvegicus of the UniProt database (19) (Release June 2014, 33,675 entries) combined with 262 commonly detected contaminants. Trypsin specificity was used for the digestion mode with N-terminal acetylation and methionine oxidation selected as the variable. Carbarmidomethylation of cysteines was set as a fixed modification, with up to two missed cleavages. For MS spectra, an initial mass accuracy of 6 ppm was selected, and the MS/MS tolerance was set to 20 ppm for HCD data. For identification, the FDR at the peptide spectrum matches (PSMs) and protein level was set to 1%. Relative, label-free quantification of proteins was performed using the MaxLFQ algorithm (20) integrated into MaxQuant with the default parameters. The data sets and the Perseus result files used for analysis were deposited at the ProteomeXchange Consortium (21) (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (22) with the data set identifier PXD003375. Analysis of the proteins identified was performed using Perseus software (http://www.perseus-framework.org/) (version 1.5.0.31). The file containing the information from identification was used with hits to the reverse database, and proteins only identified with modified peptides and potential contaminants were removed. Then, the LFQ intensity was logarithmized (log2[x]). Categorical annotation of rows was used to defined different groups after grouping replicates: (1) Temps (Control, 3, 7 and 10 days), (2) Segments (R3, R2, R1, L, C1, C2, and C3). Multiple-samples tests were performed using ANOVA test with a FDR of 5% and preserving grouping in randomization. Normalization was achieved using a Z-score with a matrix access by rows.
For the statistical analysis, only proteins presenting as significant by the ANOVA test were used for statistical analysis. Hierarchical clustering depending on time or segment was first performed using the Euclidean parameter for distance calculation and average option for linkage in row and column trees using a maximum of 300 clusters. For visualization of the variation of proteins expression depending to the condition, the profile plot tool was used with a reference profile and an automatic selection of the 10 or 15 correlated profiles. To quantify fold changes of proteins across samples, we used MaxLFQ. To visualize these fold changes in the context of individual protein abundances in the proteome, we projected them onto the summed peptide intensities normalized by the number of theoretically observable peptides. Specifically, to compare relative protein abundances between and within samples, protein lengths normalized to log 2 protein intensities (termed “iBAQ” value in MaxQuant) were added to the MaxLFQ differences. Functional annotation and characterization of identified proteins were obtained using PANTHER software (version 9.0, http://www.pantherdb.org) and STRING (version 9.1, http://string-db.org). Using the GeneMANIA Cytoscape plugin (23). 4 coexpression networks were generated from the proteomic values obtained by the analysis of control samples and caudal, rostral and lesion segments respectively. Each segment-specific coexpression network was calculated from data obtained at all-time points following SCI. Such an approach allowed then to perform a supervised clustering in order to identify functionally-relevant subnetworks that would constitute a segment-specific and time-independent molecular signature. In particular, to identify “Inflammation” subnetworks, Fibrinogen alpha (FgA) was chosen as a reference protein and, for each group of samples, networks were built that gathered the 100 proteins whose values were the most closely correlated with those of FgA. Lists of proteins and their encoding genes were then submitted to an enrichment analysis based on gene ontology (GO) annotations, using the open source tool EnrichR (24). Finally, coexpressed proteins whose encoding genes were annotated with the same GO term of interest (for instance “complement activation”), were extracted and subnetworks visualized on cytoscape. The same method was applied to identify “axon guidance” and “neuron differentiation regulation” subnetworks from the networks of proteins coexpressed with neurofilament proteins Nfl, Nfm and Nfh in each group of samples. For presentation purposes, nodes were assigned equal weights and subnetworks were slightly distorted to avoid node superimposition.
Subnetwork Enrichment Pathway Analyses and Statistical Testing
The Elsevier's Pathway Studio version 9.0 (Ariadne Genomics/Elsevier) was used to deduce relationships among differentially expressed proteomics protein candidates using the Ariadne ResNetdatabase (25, 26). “Subnetwork Enrichment Analysis” (SNEA) algorithm was selected to extract statistically significant altered biological and functional pathways pertaining to each identified set of protein hits (C1, C2, C3, R1, R2, and R3 sets). SNEA utilizes Fisher's statistical test used to determine if there are non-randomized associations between two categorical variables organized by specific relationship. SNEA starts by creating a central “seed” from all relevant entities in the database, and retrieving associated entities based on their relationship with the “seed” (i.e. binding partners, expression targets, protein modification targets, regulation). The algorithm compares the subnetwork distribution to the background distribution using one-sided Mann-Whitney U-Test, and calculates a p value indicating the statistical significance of difference between two distributions. In our analysis, “GenBank” ID and gene symbols from each set were imported to the software to form an experimental data set. For the reconstruction of networks of pathways, biological processes and molecular function were evaluated for each single protein hit and its associated targets (networks and pathways) (27, 28). Integrated Venn diagram analysis was performed using “the InteractiVenn”; a web-based tool for the analysis of complex data sets.
Cytokines Profile of Conditioned Medium
Cytokines and chemokines expression of CM from control, 3, 7, and 10 days for the segments R1 and C1 was performed by using a Rat Cytokine Array Panel A according to the manufacturer's instructions. Briefly, the array membranes were first incubated in the blocking buffer for 1 h. In the meantime, 200 μl of CM were mixed with the Detection Antibody Mixture and incubated for 1 h at room temperature. Then, after removing the block buffer, the sample/antibody mixture were added to array membranes and incubated overnight at 4 °C. After incubation, the membranes were washed 3 times with the wash buffer and then incubated with the Streptavidin-HRP solution for 30 min at room temperature. The membranes were finally washed with wash buffer for 3 times and the bound antibodies were detected by chemoluminescence using the chemireagent mix. The membranes were quantified by densitometry using ImageJ software. Background staining and spot size were analyzed as recommended by the manufacturer. Normalization was done with control expression.
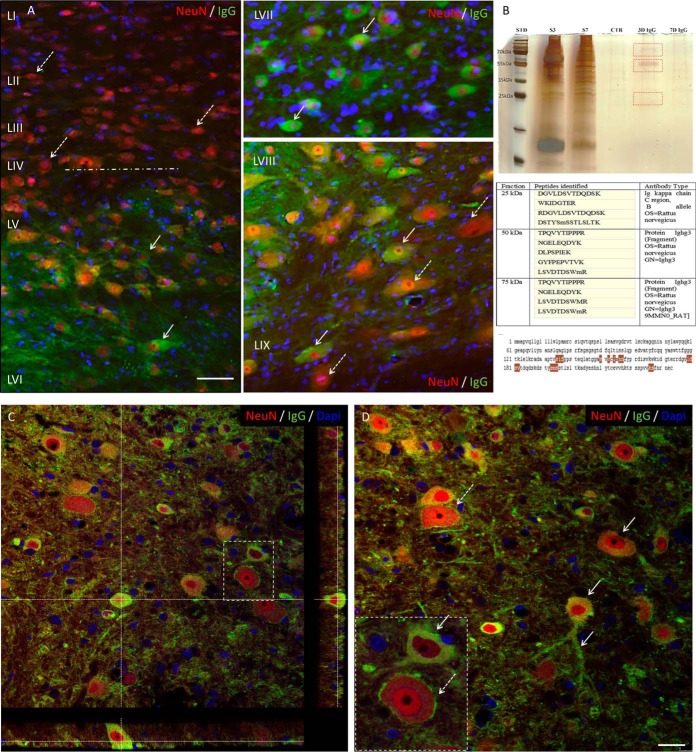
IgG Purification with Dynabeads Protein A
CM (150 μl) obtained from lesion site(∼1.5 mg of protein in 2 ml) after SCI3, 7 or10 days after SCI and from the corresponding segment of control spinal cord were bound with 50 μl of Dynabeads Protein-A 200 μl of PBS-Tween 20 and incubated 10 min with rotation at room temperature. The beads coupled with IgGs were harvested using a magnet (1 min) and the recovered supernatant solution was removed. The remaining beads-IgG complex was washed with PBS-Tween 20. Afterward, IgGs bound to microbeads were eluted by adding 30 μl of 0.1 m glycine (pH 3), incubated 10 min at 70 °C temperature to dissociate the beads-IgG complex and then neutralized by adding 30 μl of 1 m Tris-HCl (pH 8.5). Afterward, a magnet was used to remove the beads from the eluate containing IgGs. The eluate was then separated on SDS-PAGE (12% polyacrylamide gel slabs) followed by silver staining to check the bands corresponding to IgGs of the Mw of 25 kDa, 50 kDa and 75 kDa. Shot-gun mass spectrometry analysis was performed to identify the band corresponding to the appropriate Mw. Silver destaining was performed using 30 mm of potassium ferricyanide and 100 mm sodium thiosulfate (1:1) 200 μl per band for 20 min in the dark. Bands were washed 2 times for 15 min with Milli-Q water followed by an in-gel digestion as previously described (29). The samples were reconstituted with 10 μl of 5% AcN/0.1% FA and injected on a UPLC MS instrument. Separation was performed using an EASY-nLC 1000 UPLC (Thermo Scientific) equipped with a 75 μm × 2 cm Acclaim PepMap 100 pre-column with nanoViper fittings and a 50 μm ID x 150 mm Acclaim PepMap RSLC analytical column (Thermo Scientific). The peptides were eluted using a gradient of AcN starting from 5% to 30% over 1 h at a flow rate of 300 nl/min. The Q-Exactive instrument was set to acquire top 10 MS2. The survey scans were taken at 70,000 FWHM (at m/z 400) resolving power in positive mode and using a target of 1E6 and default charge state of +2. Unassigned and +1 charge states were rejected, and dynamic exclusion was enabled for 30 s. The scan range was set to m/z 300–1600 m/z. For the MS2, 1 microscan was obtained at 17,500 FWHM and isolation window of 3.0 m/z, using a scan range between m/z 200–2000 m/z. Tandem mass spectra were processed with Thermo Scientific Proteome Discoverer software version 1.3. Spectra were searched against UniprotKB/Swiss-Prot (version March 2014) filtered with Rattus norvegicus (25678 sequences) taxonomy using the SEQUEST HT algorithm (version 1.3.0.339). The search was performed choosing trypsin as the enzyme with one missed cleavage allowed. Precursor mass tolerance was 10 ppm, and fragment mass tolerance was 0.1 Da. N-terminal acetylation; and cysteine carbamidomethylation; methionine oxidation was set as variable modifications. Peptide validation was performed with the Percolator algorithm by filtering based on a q-value below 0.01, which corresponds to a false discovery rate (FDR) of 1%. The data sets used for analysis were deposited at the ProteomeXchange Consortium (21) (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (22) with the data set identifier PXD004247.
Tissue Processing and Immunohistochemical Analysis of Immune Cells
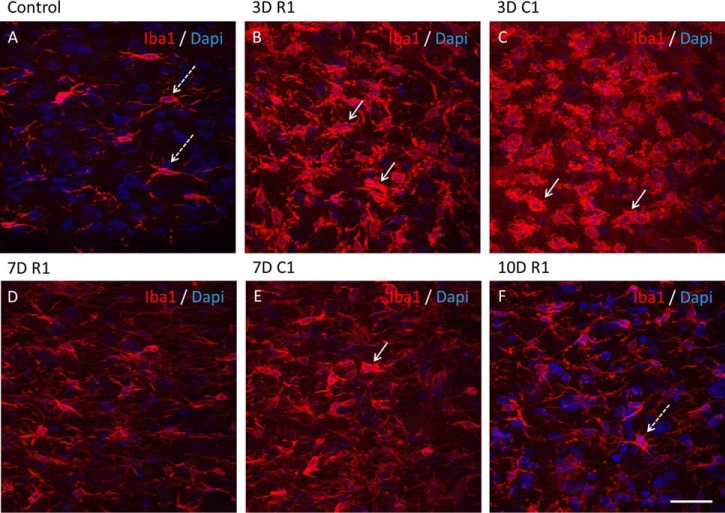
Rats following SCI at 3, 7 and 10 days and sham surgery were deeply anesthetized and perfused transcardially with saline followed by 4% paraformaldehyde (PFA) in 0.1 m phosphate buffer saline. Spinal cords were removed post-fixed in 4% PFA, embedded in gelatin-egg albumin protein matrix (10% ovalbumin, 0.75% gelatin and glutaraldehyde) and soaked overnight in 30% sucrose. Each spinal cord was dissected into 1 cm block (rostral and caudal segment) and 40 μm thick longitudinal cryostat (Leica Instruments, Heidelberg, Germany) sections (sagittal plane) were cut serially and standard immunohistochemistry technique was performed. Before incubation with primary antibodies for FoxP3 and neutrophil elastase, to reduce autofluorescence background, tissue sections were incubated 15 min at room temperature in Sudan Black B and then washed in PBS. Tissue sections were incubated in the following primary antibodies: rabbit anti-Iba1, a marker for microglia/macrophages (1:800; Wako Pure Chemical Industries, Osaka, Japan), rabbit anti-FoxP3, a marker for lymphocyte T regulator (1:200; Abcam, Cambridge, UK), rabbit anti-neutrophil elastase, a marker for neutrophil (1:200; Abcam). Tissues sections were washed in PBS and then incubated in the secondary fluorescent antibody goat anti-rabbit IgG or goat anti-mouse conjugated with AlexaFluor 594, goat anti-mouse IgG or goat anti- rabbit IgG conjugated with AlexaFluor488. Omission of the primary antibody served as the negative control. For nuclear staining, we used 4–6-diaminidino-2-phenylindol (DAPI) (1:10000). Finally, sections were washed in 0.1 m PBS, mounted, and coverslipped with Vectashield mounting medium (VectorLaboratories, Inc., Burlingame,CA) and observed under a fluorescence microscope (NikonEclipseTi, Japan) and confocal laser scanning microscope (Leica TCSSP5 AOBS, Leica Microsystems, Mannheim, Germany).
Quantification analyses of immunofluorescence staining for Iba1, FoxP3 and neutrophil elastase were performed on 6 sections from rostral and caudal/per condition (n = 3 each). Captured fluorescence digital images at 40x magnification were analyzed by ImageJ software. Iba1 quantification was determined by randomization counting cells manually with Image J software in gray and white matters of rostral and caudal segments. FoxP3 and neutrophil elastase quantification was determined by the percentage of positive cells compared with DAPI staining by counting cell manually with image J software.
Immunohistochemical Analysis of IgG Deposition
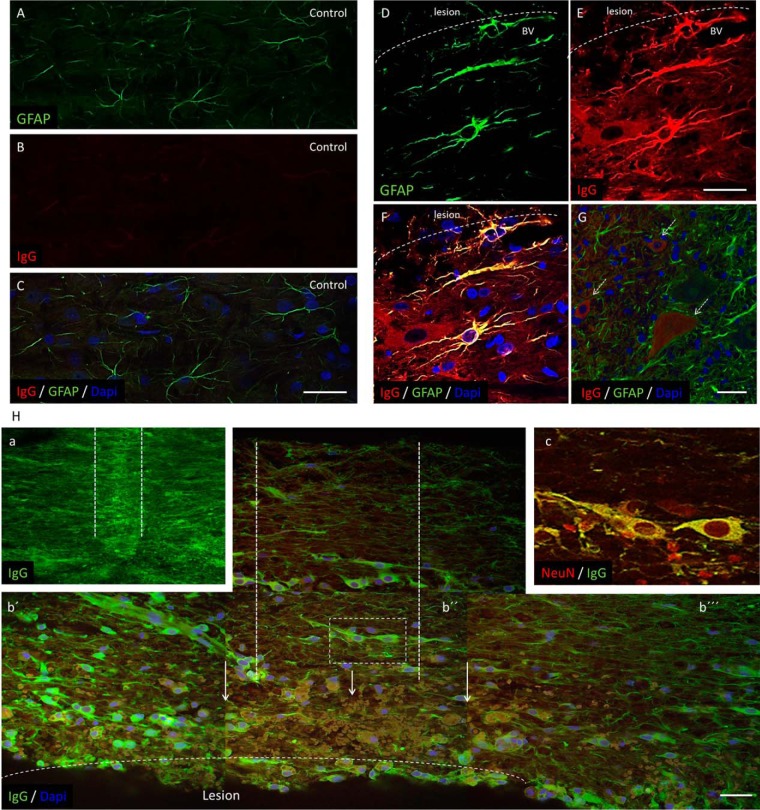
Tissue sections of 40 μm thickness from rostral and caudal segment adjacent to the lesion at the 3, 7 and 10 days post-injury and corresponding cuts for controls (embedded in gelatin-egg albumin protein matrix, same procedure as for IHC for immune response) were cut. One set of tissue sections were incubated with primary goat anti-rat IgG2a antibody (1:500, Bethyl Laboratories, Inc., Montgomery, TX) overnight at 4 °C. After 3 washing steps in PBS, sections were incubated with secondary fluorescent antibodies: rabbit anti-goat IgG conjugated with AlexaFlour594 or AlexaFlour 488 for one hour at room temperature. For double labeling, we have used the following primary antibodies: mouse anti-GFAP, (1/500; Upstate Biotech Millipore), rabbit anti-Iba1 (1:800), mouse anti-NeuN, marker for neurons, (1/1000; Upstate Biotech Millipore) overnight at 4 °C, followed with corresponding secondary antibodies. Another set of sections were labeled with secondary fluorescent antibody anti-rat IgG conjugated with (AlexaFlour 488), followed by incubation with primary antibodies for NeuN, GFAP and Iba1. Omission of the primary antibody served as the negative control. For nuclear staining, we used DAPI (1:10000). Finally, sections were washed with 0.1 M PBS, mounted, and coverslipped with Vectashield mounting medium and observed under a fluorescence microscope and confocal laser scanning microscope. Fluorescence or confocal images, including orthogonal views, were processed by using 0.5 μm optical sections at 40x or 60x magnification.
Intraspinal Delivery of CD-20 Antibody
One hour after SCI, in anesthetized rats with 1.5- 2% isoflurane we have microinjected intraspinally primary rabbit anti-rat CD-20 antibody (Biorbyt Ltd., Cambridge, United Kingdom), for local depletion of B- lymphocytes IgG production. Intraspina injections of (1) saline (n = 3) and (2) rabbit anti-rat CD-20 antibody, 0.5 μg/μl (n = 3) were performed using a 50-μl Hamilton syringe (30G needle, Cole Parmer, Anjou, Quebec) connected to UltraMicroPump III with Micro4 Controller, 4-Channel (World Precision Instruments, Inc., Sarasota, FL) and stereotactic device, with a delivery rate of 0.5 μl/min. 3 bilateral intraspinal injections of 1 μl per injection were performed at the lesion site, whereas one injection of 1 μl per injection was done at rostral and caudal segments. Each delivery was positioned 1 mm from the spinal cord midline and injected at a depth of 1.8–2 mm from the pial surface of the spinal cord. The distance between injections was 1 mm, avoiding vessels. Intraspinal injections were followed by procedure published in our study (16). After injecting the dose of saline or CD-20 antibody, the needle was maintained in the tissue for an additional 30 s. No antibiotic treatment was performed during animal's survival. All rats survived for 3 days.
Behavioral Testing After CD-20 Antibody Intraspinal Delivery
Animals were evaluated using Basso, Beattie, and Bresnahan (BBB) open-field test to assess motor function after SCI at day 0, 3, 7, 10, 14 and 28. Each rat was tested for 5 min by two blinded examiners. BBB test measures locomotor outcome (hind limb activity, body position, trunk stability, tail position and walking paw placement) of rats utilizing the rating scale ranges from 0 (no observable hind limbs movements) to a maximum of 21 (plantar stepping, coordination and trunk stability similar to control rats). All data are reported as mean ± S.E. Differences in mean BBB scores between the sham-controls and SCI groups at each survival interval were assessed using one-way ANOVA.
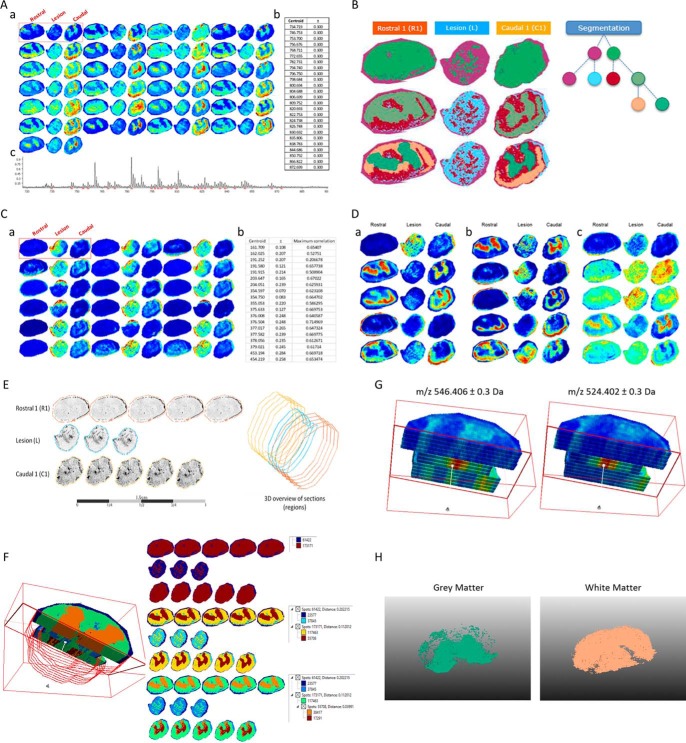
MALDI Imaging Data Analyses
Twelve micrometer tissue sections from the R1, lesion and C1 segments were obtained using a cryostat (Leica Microsystems, Nanterre, France). These were mounted on indium tin oxide (ITO)-coated slides and placed under vacuum in a dessicator for 15 min. 2,5-dihydroxybenzoic acid was used as matrix, and was prepared at a concentration of 20 mg/ml in 70:30 methanol/0.1% TFA in H2O. The matrix was deposited manually using a sprayer developed in-house at a flow rate of 300 μl/h for 5 min. Lipid imaging was performed on an UltraFlex II instrument (Bruker Daltonics, Bremen, Germany) equipped with a micro-channel plate (MCP) detector. The instrument was equipped with a Smartbeam™ laser capable of operating up to 200Hz and was controlled using FlexControl 3.3 (Build 108) software (Bruker Daltonics). The data sets were recorded in positive reflector mode and 500 laser shots were accumulated for each raster point. The laser focus was set to medium, and deflection of masses was deactivated. Spectra were acquired by oversampling at a lateral resolution of 20 μm. External calibration was performed using the PepMix standard (Bruker Daltonics). Spectra were acquired between m/z 60–1000.
The 2D MALDI Imaging data of rostral (R1) caudal (C1) and lesion (L) sections after 3 days of SCI (n = 1) were analyzed with SCiLS Lab 2015a (30). The baseline was removed by iterative convolution and the data were normalized based on the total ion count (TIC) method (31). Subsequently, the orthogonal matching pursuit was used to detect peaks. These peaks were aligned to the mean spectrum and the data were smoothed with a strong (lambda 0.5) Chambolle filter method. Automatic spatial segmentation was performed by using the bisecting k-means algorithm (32). Colocalized m/z values with the lesion region were elucidated by using Pearson's correlation analysis. With component analysis fundamental components of given spectra and m/z images can be discovered unsupervised. Here, the principal component analysis as well as the probabilistic latent semantic analysis were used (33). The PLSA was performed with random initialization with a threshold of 29.0588, maximum interval processing mode, lower m/z range threshold mode and individual spectrum mode. The PCA was performed with no scaling, a threshold of 29.0588, maximum interval processing mode, lower m/z range threshold mode and individual spectrum mode (30).
RESULTS
Workflow
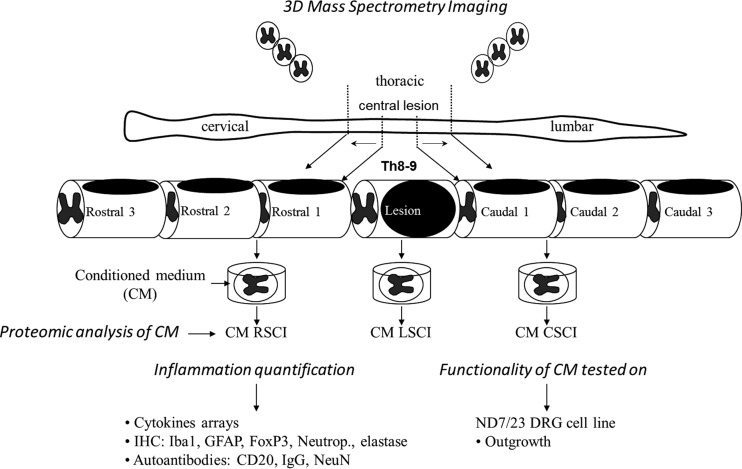
Our main goal was to establish a fine mapping of the molecular events that occur in a time- and segment-specific manner during sub-acute SCI. We chose to focus our analysis on the identification of soluble factors that may support intra- or intersegmental communications between distinct cellular compartments. For this purpose, we performed proteomic shot-gun triplicate analyses of the conditioned medium (CM) derived from seven spinal cord segments localized at the lesion site (L) or either caudal to the lesion (C1, C2, C3) or rostral (R1, R2, R3) to the lesion. Each of these analyses were performed on spinal cord segments obtained on days 3, 7, or 10 post-SCI giving rise to a set of 80 proteomic data covering an average of 1500 proteins identified par sample with at least 2 peptides per protein recognized and percentage of false positive (FDR) < 1% (supplemental Data S1). Proteomic data were then analyzed using complementary bioinformatics approaches that allowed unsupervised or supervised clustering of quantified proteins and the identification of differentially expressed molecules. Thus, both a comprehensive strategy without a priori and a focused one were applied to analyze proteomics data. In parallel, cytokine array measurements were performed and the whole of the proteomics data obtained from supernatant analyses was then correlated with immunohistological analyses and functional studies (Fig. 1).
Fig. 1.
Schematic design of the experimental procedure perform in this study.
Global Proteomic Study Along the Spinal Cord Axis at Key Time Points of SCI
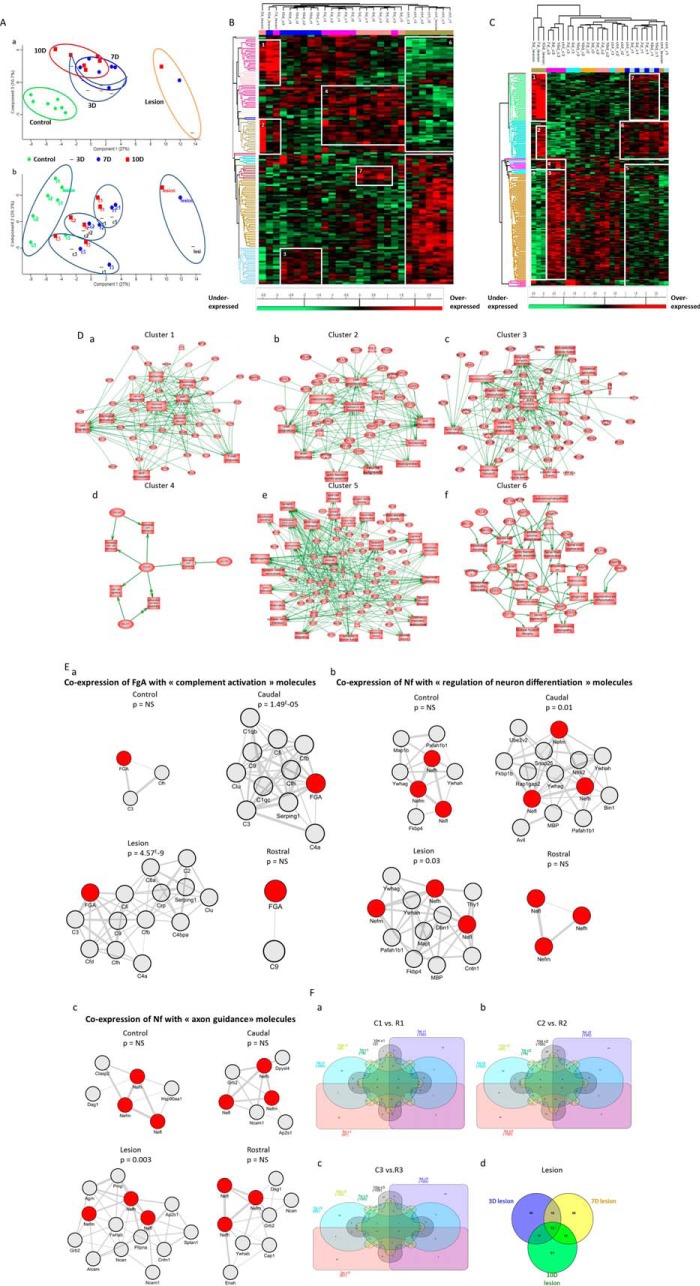
Principal component analysis of conditioned media proteomics data clearly demonstrated that profiles in SCI samples can be clustered depending on the segment localization, irrespective of the time point considered. In addition, control samples harbor a molecular profile that can be sharply distinguished from any of SCI profiles, including those of segments localized at distance from the lesion site (i.e. the R3 and C3 profiles) (Fig. 2A). Time course analyses show that control samples, in one hand, and lesion samples, on the other hand, are clearly distant from all other samples (Fig. 2Aa upper panel). The remaining profiles display a partial overlap, indicating that time point may not be the main parameter allowing discriminating the proteomic profiles of spinal cord segments in this model. In contrast, a clear clustering of profiles was obtained when analyzing samples on the basis of their spatial localization (Fig. 2Ab lower panel). Thus, independently from the time point considered, profiles from each group of segments analyzed (i.e. rostral versus caudal versus lesion) displayed intragroup similarities. This finding provides evidence that in the SCI model, proteomic profiles in spinal cord segments may be discriminated on the basis of spatial localization. This result also demonstrates that with regard to the secretome, spinal cord segments tend to coevolve in function of their distance to the lesion site rather than their caudal versus rostral localization. Indeed, in terms of proteins patterns, R3 and C3 samples cosegregated and were clearly distinguished from the R2 and C2 patterns, the R1 and C1 patterns and the lesion patterns.
Fig. 2.
Spatio-temporal organization of proteins. A, Principal component analysis (PCA) of conditioned media shows the representation of the different samples grouping regarding (a) time after injury and (b) spatial organization of the spinal cord. One point corresponds to all proteins identified according to their label free quantification for each segments and each time point. Heat map of proteins with different secretion profiles in conditioned media from different time after SCI (3D, 7D, 10D, and control) and from different segments (R3, R2, R1, L, C1, C2, C3). B, Hierarcherical clustering regarding time point. C, Hierarcherical clustering regarding spatial organization. Distinct clusters are highlighted. D, System biology analysis for network identification in each cluster issued from heat map of proteins with different secretion profiles in conditioned media from different time after SCI (3D, 7D, 10D, and control) and from different segment (R3, R2, R1, L, C1, C2, C3) regarding the spatial organization. E, Analysis of coexpression networks identify in inflammation, neuro-axonal and axonal guidance in conditioned media. The 100 genes which encoded molecules were the most tightly coregulated with fibrinogen- α (FgA), neurofilament L, M and H (Nf) respectively in conditioned media control, rostral, lesion and caudal were identified and assessed for gene set enrichment. Panel Ea showns subnetworks of genes annotated by the GO terms “inflammation”, caudal and lesion segments have significant p value unlike control and rostral segments. Panel Eb presents subnetworks of genes annotated by the GO terms “neuro-axonal,” caudal and lesion have significant p value unlike control and rostral segments. Panel Ec shows subnetworks of genes annotated by the GO terms “axonal guidance”, only lesion has significant p value unlike caudal, lesion and rostral segments. F, Differential distribution of unique and common/intersected biological and functional pathways among the three spinal cord rostral and caudal regions (C1, C2, C3 versus R1, R2, and R3) factored by time of SCI (3, 7, and 10 days). Each pair of spinal cord region (C1 versus R1) was analyzed across the three time points represented utilizing a comprehensive Venn analysis representation extracted from Subnetwork Enrichment Analysis.
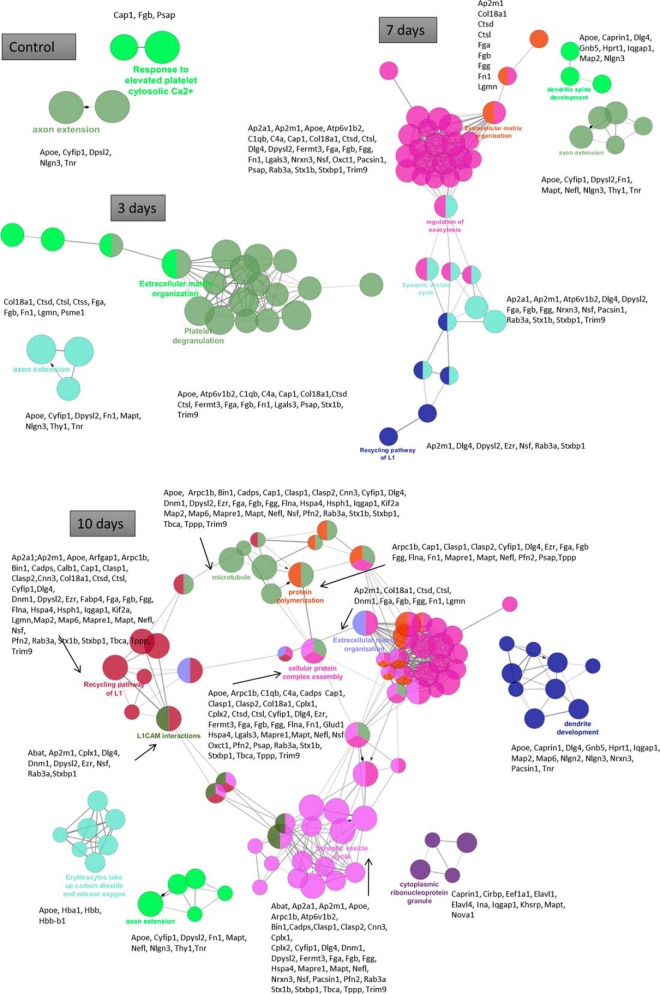
Hierarchical clustering depending on the time course post-injury (Fig. 2B, Table I) or the spatial localization along the spinal cord (Fig. 2C, Table II, supplemental Data S2) were first performed using the Euclidean parameter for distance calculation and average option for linkage in row and column trees using a maximum of 300 clusters. As a criterion of significance, we applied an ANOVA significance threshold of p < 0.05, and heat maps were created. A total of 147 proteins for time-dependent clustering and 193 proteins for segment-dependent clustering were considered reliable based on statistical analysis. In the time-dependent clustering (Fig. 2B, Table I), two major branches of the heat map separated the 3 day lesion segment in one branch from the others in the second branch. This latter was subdivided in two sub-branches: the first one gathering all controls whatever the segments and the second one all the other ones. This branch was itself subdivided in 4 branches, i.e. one for the lesion segments and the three other ones regroup all segments per time. Overall, this approach allowed a total of 6 clusters to be retrieved (Fig. 2B, Table I). The proteins in each cluster were then analyzed using PANTHER software (http://www.pantherdb.org) to determine the biological functions based on the protein classes. Control group were found to be characterized by two different clusters, i.e. cluster 5 signing an overexpression of tubulins network, enzymatic metabolism network and vesicule-trafficking network and cluster 6 corresponding to an under-expression of heat shock proteins network, neurofilaments and filamins networks and immune response factors (Table I). Lesion segments showed proteins overexpressed involved in neurogenesis, notably adducing and plectin, and in immune response (e.g. galectin 1 and 3, coronin) (cluster 1) (Table I). Cluster 2 corresponds to a clear increased expression of proteins involved in inflammation in the lesion segment after 3 days and a weak overexpression of these proteins at 7 days in the lesion segment as well as in other segments at 3 and 7 days (Table I). By comparison, inflammatory molecules were down-regulated in all segments at 10 days after SCI and in control samples. Cluster 4 corresponds to an overexpression of proteins in rostral and caudal segments on day 3 and day 7 post-SCI. Cluster 4 shows an overexpression of proteins involved in neurite outgrowth like SPARC, neurofilament, lamin and the ones involved in regeneration processes, but are underexpressed in control (cluster 6). Some proteins involved in these functions are found at 10 days overexpressed as CRMP-3 and transgelin 3, but these proteins are also present in control (Table I).
Table I. List of proteins identified by cluster after perseus analyses taking into account the time course.
| Time clustering | ||
|---|---|---|
| Accession number | Protein name | Gene name |
| Cluster 1 | ||
| O08557 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | Ddah1 |
| P02650 | Apolipoprotein E | Apoe |
| P06761 | 78 kDa glucose-regulated protein | Hspa5 |
| P08699 | Galectin-3 | Lgals3 |
| P09495 | Tropomyosin alpha-4 chain | Tpm4 |
| P10960 | Sulfated glycoprotein 1 | Psap |
| P11598 | Protein disulfide-isomerase A3 | Pdia3 |
| P11762 | Galectin-1 | Lgals1 |
| P30427 | Plectin | Plec |
| P31000 | Vimentin | Vim |
| P31232 | Transgelin | Tagln |
| G3V913 | Heat shock 27kDa protein 1 | Hspb1 |
| P55051 | Fatty acid-binding protein, brain | Fabp7 |
| P85972 | Vinculin | Vcl |
| Q5RKI0 | WD repeat-containing protein 1 | Wdr1 |
| Q5XFX0 | Transgelin-2 | Tagln2 |
| Q63610–2 | Tropomyosin alpha-3 chain | Tpm3 |
| Q6JE36 | Protein NDRG1 | Ndrg1 |
| Q9WVR7 | Protein phosphatase 1F | Ppm1f |
| B0K010 | Protein Txndc17 | Txndc17 |
| C0JPT7 | Filamin alpha | Flna |
| D3ZHA0 | Protein Flnc | Flnc |
| D3ZQP6 | Protein Sema7a | Sema7a |
| G3V852 | Protein Tln1 | Tln1 |
| G3V8L3 | Lamin A, isoform CRA_b | Lmna |
| G3V940 | Coronin | Coro1b |
| Q5XI38 | Lymphocyte cytosolic protein 1 | Lcp1 |
| Cluster 2 | ||
| B0BNN3 | Carbonic anhydrase 1 | Ca1 |
| P01048 | T-kininogen 1 | Map1 |
| P02091 | Hemoglobin subunit beta-1 | Hbb |
| P02680–2 | Fibrinogen gamma chain | Fgg |
| P04639 | Apolipoprotein A-I | Apoa1 |
| P06238 | Alpha-2-macroglobulin | A2m |
| P06866 | Haptoglobin | Hp |
| P08932 | T-kininogen 2 | |
| P14480 | Fibrinogen beta chain | Fgb |
| P17475 | Alpha-1-antiproteinase | Serpina1 |
| P20059 | Hemopexin | Hpx |
| Q63041 | Alpha-1-macroglobulin | A1m |
| Q9QX79 | Fetuin-B | Fetub |
| G3V7K3 | Ceruloplasmin | Cp |
| M0RBF1 | Complement C3 | C3 |
| O54854 | Kallikrein 6, isoform CRA_a | Klk6 |
| Q5EBC0 | Inter alpha-trypsin inhibitor, heavy chain 4 | Itih4 |
| Q5M7T5 | Protein Serpinc1 | Serpinc1 |
| Q62669 | Protein Hbb-b1 | Hbb-b1 |
| Q68FY4 | Group specific component | Gc |
| Q7TQ70 | Ac1873 | Fga |
| Cluster 3 | ||
| B0BNM1 | NAD(P)H-hydrate epimerase | Apoa1bp |
| B2RYG6 | Ubiquitin thioesterase OTUB1 | Otub1 |
| P05544 | Serine protease inhibitor A3L | Serpina3l |
| P10860 | Glutamate dehydrogenase 1, mitochondrial | Glud1 |
| P11030 | Acyl-CoA-binding protein | Dbi |
| P37805 | Transgelin-3 | Tagln3 |
| P50398 | Rab GDP dissociation inhibitor alpha | Gdi1 |
| P62161 | Calmodulin | Calm1 |
| P63086 | Mitogen-activated protein kinase 1 | Mapk1 |
| Q4KM74 | Vesicle-trafficking protein SEC22b | Sec22b |
| Q4KMA2 | UV excision repair protein RAD23 homolog B | Rad23b |
| Q62952–2 | Dihydropyrimidinase-related protein 3 | Dpysl3 |
| Q6AY84 | Secernin-1 | Scrn1 |
| Q7TP98 | Interleukin enhancer-binding factor 2 | Ilf2 |
| D4A5X8 | Protein Ahcyl1 | Ahcyl1 |
| F1M978 | Inositol monophosphatase 1 | Impa1 |
| F1M9V7 | Protein Npepps | Npepps |
| F1MAQ5 | Microtubule-associated protein | Map2 |
| G3V8G4 | Brevican, isoform CRA_a | Bcan |
| M0R686 | Protein Irgq | Irgq |
| M0RE01 | Uncharacterized protein | LOC100911107 |
| Cluster 4 | ||
| B0BNN3 | Carbonic anhydrase 1 | Ca1 |
| P01048 | T-kininogen 1 | Map1 |
| P02091 | Hemoglobin subunit beta-1 | Hbb |
| P02680–2 | Fibrinogen gamma chain | Fgg |
| P04639 | Apolipoprotein A-I | Apoa1 |
| P04692–5 | Tropomyosin alpha-1 chain | Tpm1 |
| P06238 | Alpha-2-macroglobulin | A2m |
| P06866 | Haptoglobin | Hp |
| P08592–2 | Amyloid beta A4 protein | App |
| P08932 | T-kininogen 2 | |
| P09606 | Glutamine synthetase | Glul |
| P11348 | Dihydropteridine reductase | Qdpr |
| P12839 | Neurofilament medium polypeptide | Nefm |
| P14480 | Fibrinogen beta chain | Fgb |
| P16975 | SPARC | Sparc |
| P17475 | Alpha-1-antiproteinase | Serpina1 |
| P19527 | Neurofilament light polypeptide | Nefl |
| P20059 | Hemopexin | Hpx |
| P21807 | Peripherin | Prph |
| P34058 | Heat shock protein HSP 90-beta | Hsp90ab1 |
| P50503 | Hsc70-interacting protein | St13 |
| P62703 | 40S ribosomal protein S4, X isoform | Rps4x |
| P82995 | Heat shock protein HSP 90-alpha | Hsp90aa1 |
| Q63041 | Alpha-1-macroglobulin | A1m |
| Q66HA8 | Heat shock protein 105 kDa | Hsph1 |
| Q9QX79 | Fetuin-B | Fetub |
| Q9QZA2 | Programmed cell death 6-interacting protein | Pdcd6ip |
| F1LM84 | Nidogen-1 | Nid1 |
| F1LRZ7 | Neurofilament heavy polypeptide | Nefh |
| G3V7K3 | Ceruloplasmin | Cp |
| G3V7U4 | Lamin-B1 | Lmnb1 |
| Q6PDW1 | 40S ribosomal protein S12 | Rps12 |
| M0RBF1 | Complement C3 | C3 |
| O54854 | Kallikrein 6, isoform CRA_a | Klk6 |
| Q5EBC0 | Inter alpha-trypsin inhibitor, heavy chain 4 | Itih4 |
| Q5M7T5 | Protein Serpinc1 | Serpinc1 |
| Q62669 | Protein Hbb-b1 | Hbb-b1 |
| Q68FY4 | Group specific component | Gc |
| Q6MGC4 | H2-K region expressed gene 2, rat orthologue | Pfdn6 |
| Q7TQ70 | Ac1873 | Fga |
| Cluster 5 | ||
| B0BNM1 | NAD(P)H-hydrate epimerase | Apoa1bp |
| B2RYG6 | Ubiquitin thioesterase OTUB1 | Otub1 |
| O08651 | d-3-phosphoglycerate dehydrogenase | Phgdh |
| O35077 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | Gpd1 |
| O35760 | Isopentenyl-diphosphate Delta-isomerase 1 | Idi1 |
| O88350 | Putative hydrolase RBBP9 | Rbbp9 |
| M0R590 | Protein LOC685186 | LOC685186 |
| P04905 | Glutathione S-transferase Mu 1 | Gstm1 |
| F1LND7 | Farnesyl pyrophosphate synthase | Fdps |
| P05544 | Serine protease inhibitor A3L | Serpina3l |
| P05708 | Hexokinase-1 | Hk1 |
| P08009 | Glutathione S-transferase Yb-3 | Gstm3 |
| P10860 | Glutamate dehydrogenase 1, mitochondrial | Glud1 |
| P10959 | Carboxylesterase 1C | Ces1c |
| P11030 | Acyl-CoA-binding protein | Dbi |
| P11980 | Pyruvate kinase PKM | Pkm |
| P12346 | Serotransferrin | Tf |
| P12785 | Fatty acid synthase | Fasn |
| P13233 | 2′,3′-cyclic-nucleotide 3′-phosphodiesterase | Cnp |
| P13668 | Stathmin | Stmn1 |
| G3V9G4 | ATP citrate lyase, isoform CRA_b | Acly |
| P17425 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | Hmgcs1 |
| P24155 | Thimet oligopeptidase | Thop1 |
| P24329 | Thiosulfate sulfurtransferase | Tst |
| P28037 | Cytosolic 10-formyltetrahydrofolate dehydrogenase | Aldh1l1 |
| P30904 | Macrophage migration inhibitory factor | Mif |
| P32232–2 | Cystathionine beta-synthase | Cbs |
| P37361 | Metallothionein-3 | Mt3 |
| P37805 | Transgelin-3 | Tagln3 |
| P50398 | Rab GDP dissociation inhibitor alpha | Gdi1 |
| P50408 | V-type proton ATPase subunit F | Atp6v1f |
| P51635 | Alcohol dehydrogenase [NADP(+)] | Akr1a1 |
| P84079 | ADP-ribosylation factor 1 | Arf1 |
| P62161 | Calmodulin | Calm1 |
| P62828 | GTP-binding nuclear protein Ran | Ran |
| P63086 | Mitogen-activated protein kinase 1 | Mapk1 |
| Q3KRE8 | Tubulin beta-2B chain | Tubb2b |
| Q497B0 | Omega-amidase NIT2 | Nit2 |
| Q4KM74 | Vesicle-trafficking protein SEC22b | Sec22b |
| Q4KMA2 | UV excision repair protein RAD23 homolog B | Rad23b |
| Q4V8C3–2 | Echinoderm microtubule-associated protein-like 1 | Eml1 |
| Q5BJP9 | Phytanoyl-CoA dioxygenase domain-containing protein 1 | Phyhd1 |
| Q5PQN7 | Protein LZIC | Lzic |
| Q5XI22 | Acetyl-CoA acetyltransferase, cytosolic | Acat2 |
| Q62952–2 | Dihydropyrimidinase-related protein 3 | Dpysl3 |
| Q66HG4 | Aldose 1-epimerase | Galm |
| Q6AY84 | Secernin-1 | Scrn1 |
| Q6AYS7 | Aminoacylase-1A | Acy1a |
| Q6P502 | T-complex protein 1 subunit gamma | Cct3 |
| Q6P9V9 | Tubulin alpha-1B chain | Tuba1b |
| Q7TP98 | Interleukin enhancer-binding factor 2 | Ilf2 |
| Q91XU1 | Protein quaking | Qki |
| Q9JHU0 | Dihydropyrimidinase-related protein 5 | Dpysl5 |
| Q9QXU9 | ProSAAS | Pcsk1n |
| Q9Z1B2 | Glutathione S-transferase Mu 5 | Gstm5 |
| B2GUZ9 | Fam49b protein | Fam49b |
| B4F7C2 | Protein Tubb4a | Tubb4a |
| B5DFN4 | Prefoldin 5 (Predicted), isoform CRA_a | Pfdn5 |
| D3ZFU9 | Protein Mylk | Mylk |
| D4AD67 | Protein Ktn1 | Ktn1 |
| D4A5X8 | Protein Ahcyl1 | Ahcyl1 |
| D4ACB8 | Chaperonin subunit 8 (Theta) (Predicted), isoform CRA_a | Cct8 |
| D4AEH9 | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (Glycogen debranching enzyme, glycogen storage disease type III) | Agl |
| F1LM42 | Protein Ank2 | Ank2 |
| F1M978 | Inositol monophosphatase 1 | Impa1 |
| F1M9V7 | Protein Npepps | Npepps |
| F1MAQ5 | Microtubule-associated protein | Map2 |
| F7F7H4 | Uncharacterized protein | Dock2 |
| G3V721 | WW domain binding protein 2, isoform CRA_b | Wbp2 |
| G3V7C6 | RCG45400 | Tubb4b |
| G3V8G4 | Brevican, isoform CRA_a | Bcan |
| G3V8V3 | Phosphorylase | Pygm |
| G3V9U0 | Acyl-CoA synthetase short-chain family member 2 (Predicted) | Acss2 |
| M0R686 | Protein Irgq | Irgq |
| M0RE01 | Uncharacterized protein | LOC100911107 |
| Q4KM55 | Protein Vta1 | Vta1 |
| Q642E5 | Diphosphomevalonate decarboxylase | Mvd |
| Cluster 6 | ||
| B0BNN3 | Carbonic anhydrase 1 | Ca1 |
| O08557 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | Ddah1 |
| P01048 | T-kininogen 1 | Map1 |
| P02091 | Hemoglobin subunit beta-1 | Hbb |
| P02650 | Apolipoprotein E | Apoe |
| P02680–2 | Fibrinogen gamma chain | Fgg |
| P04639 | Apolipoprotein A-I | Apoa1 |
| P04692–5 | Tropomyosin alpha-1 chain | Tpm1 |
| P06238 | Alpha-2-macroglobulin | A2m |
| P06761 | 78 kDa glucose-regulated protein | Hspa5 |
| P06866 | Haptoglobin | Hp |
| P08592–2 | Amyloid beta A4 protein | App |
| P08699 | Galectin-3 | Lgals3 |
| P08932 | T-kininogen 2 | |
| P09495 | Tropomyosin alpha-4 chain | Tpm4 |
| P09606 | Glutamine synthetase | Glul |
| P10960 | Sulfated glycoprotein 1 | Psap |
| P11348 | Dihydropteridine reductase | Qdpr |
| P11598 | Protein disulfide-isomerase A3 | Pdia3 |
| P11762 | Galectin-1 | Lgals1 |
| P12839 | Neurofilament medium polypeptide | Nefm |
| P14480 | Fibrinogen beta chain | Fgb |
| P16975 | SPARC | Sparc |
| P17475 | Alpha-1-antiproteinase | Serpina1 |
| P19527 | Neurofilament light polypeptide | Nefl |
| P20059 | Hemopexin | Hpx |
| P21807 | Peripherin | Prph |
| P30427 | Plectin | Plec |
| P31000 | Vimentin | Vim |
| P31232 | Transgelin | Tagln |
| P34058 | Heat shock protein HSP 90-beta | Hsp90ab1 |
| G3V913 | Heat shock 27kDa protein 1 | Hspb1 |
| P50503 | Hsc70-interacting protein | St13 |
| P55051 | Fatty acid-binding protein, brain | Fabp7 |
| P62703 | 40S ribosomal protein S4, X isoform | Rps4x |
| P82995 | Heat shock protein HSP 90-alpha | Hsp90aa1 |
| P85972 | Vinculin | Vcl |
| Q5RKI0 | WD repeat-containing protein 1 | Wdr1 |
| Q5XFX0 | Transgelin-2 | Tagln2 |
| Q63028 | Alpha-adducin | Add1 |
| Q63041 | Alpha-1-macroglobulin | A1m |
| Q63610–2 | Tropomyosin alpha-3 chain | Tpm3 |
| Q66HA8 | Heat shock protein 105 kDa | Hsph1 |
| Q68FQ2 | Junctional adhesion molecule C | Jam3 |
| Q6JE36 | Protein NDRG1 | Ndrg1 |
| Q9QX79 | Fetuin-B | Fetub |
| Q9QZA2 | Programmed cell death 6-interacting protein | Pdcd6ip |
| Q9WVR7 | Protein phosphatase 1F | Ppm1f |
| B0K010 | Protein Txndc17 | Txndc17 |
| C0JPT7 | Filamin alpha | Flna |
| D3ZHA0 | Protein Flnc | Flnc |
| D3ZQP6 | Protein Sema7a | Sema7a |
| F1LM84 | Nidogen-1 | Nid1 |
| F1LRZ7 | Neurofilament heavy polypeptide | Nefh |
| G3V7K3 | Ceruloplasmin | Cp |
| G3V7U4 | Lamin-B1 | Lmnb1 |
| G3V852 | Protein Tln1 | Tln1 |
| G3V8L3 | Lamin A, isoform CRA_b | Lmna |
| G3V940 | Coronin | Coro1b |
| Q6PDW1 | 40S ribosomal protein S12 | Rps12 |
| M0RBF1 | Complement C3 | C3 |
| O54854 | Kallikrein 6, isoform CRA_a | Klk6 |
| Q5EBC0 | Inter alpha-trypsin inhibitor, heavy chain 4 | Itih4 |
| Q5M7T5 | Protein Serpinc1 | Serpinc1 |
| Q5XI38 | Lymphocyte cytosolic protein 1 | Lcp1 |
| Q62669 | Protein Hbb-b1 | Hbb-b1 |
| Q68FY4 | Group specific component | Gc |
| Q6MGC4 | H2-K region expressed gene 2, rat orthologue | Pfdn6 |
| Q7TQ70 | Ac1873 | Fga |
| Cluster 7 | ||
| P05708 | Hexokinase-1 | Hk1 |
| P12346 | Serotransferrin | Tf |
| P30904 | Macrophage migration inhibitory factor | Mif |
| Q6P502 | T-complex protein 1 subunit gamma | Cct3 |
| D4ACB8 | Chaperonin subunit 8 (Theta) (Predicted), isoform CRA_a | Cct8 |
| D4AEH9 | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (Glycogen debranching enzyme, glycogen storage disease type III) | Agl |
| F7F7H4 | Uncharacterized protein | Dock2 |
| G3V721 | WW domain binding protein 2, isoform CRA_b | Wbp2 |
Table II. List of proteins identified by cluster after perseus analyses taking into account spatial localization.
| Segment clustering | ||
|---|---|---|
| Accession number | Protein name | Gene name |
| Cluster 1 | ||
| P02650 | Apolipoprotein E | Apoe |
| P04041 | Glutathioneperoxidase 1 | Gpx1 |
| P04642 | l-lactate dehydrogenase A chain | Ldha |
| P04785 | Proteindisulfide-isomerase | P4hb |
| P05197 | Elongation factor 2 | Eef2 |
| P07151 | Beta-2-microglobulin | B2m |
| P08649 | Complement C4 | C4 |
| P08699 | Galectin-3 | Lgals3 |
| P10960 | Sulfatedglycoprotein 1 | Psap |
| P22734–2 | Catechol O-methyltransferase | Comt |
| P23928 | Alpha-crystallin B chain | Cryab |
| Q6P6T6 | Cathepsin D | Ctsd |
| P31232 | Transgelin | Tagln |
| P37397 | Calponin-3 | Cnn3 |
| P62963 | Profilin-1 | Pfn1 |
| P85968 | 6-phosphogluconate dehydrogenase, decarboxylating | Pgd |
| P85970 | Actin-relatedprotein 2/3 complexsubunit 2 | Arpc2 |
| P85973 | Purine nucleoside phosphorylase | Pnp |
| Q3MIE4 | Synapticvesicle membrane protein VAT-1 homolog | Vat1 |
| Q4V7C7 | Actin-relatedprotein 3 | Actr3 |
| Q5I0D7 | Xaa-Pro dipeptidase | Pepd |
| Q5M7U6 | Actin-relatedprotein 2 | Actr2 |
| Q5XFX0 | Transgelin-2 | Tagln2 |
| Q63716 | Peroxiredoxin-1 | Prdx1 |
| Q64119 | Myosin light polypeptide 6 | Myl6 |
| Q68FS4–2 | Cytosol aminopeptidase | Lap3 |
| Q6AYC4 | Macrophage-cappingprotein | Capg |
| Q6B345 | Protein S100-A11 | S100a11 |
| Q6MG61 | Chlorideintracellularchannelprotein 1 | Clic1 |
| Q6Q0N1 | Cytosolic non-specificdipeptidase | Cndp2 |
| D3ZHA0 | Protein Flnc | Flnc |
| D3ZQP6 | Protein Sema7a | Sema7a |
| F1LP60 | Moesin | Msn |
| F1M5X1 | Protein Rrbp1 | Rrbp1 |
| F1M983 | ProteinCfh | Cfh |
| G3V852 | Protein Tln1 | Tln1 |
| G3V8V1 | Granulin, isoformCRA_c | Grn |
| M0R4S2 | Apolipoprotein D | Apod |
| Q5XI38 | Lymphocyte cytosolicprotein 1 | Lcp1 |
| Q6IN22 | Cathepsin B | Ctsb |
| Q6P9V7 | Proteasome (Prosome, macropain) activatorsubunit 1 | Psme1 |
| Q63798 | Proteasomeactivatorcomplexsubunit 2 | Psme2 |
| Cluster 2 | ||
| O35077 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | Gpd1 |
| P02688 | Myelin basic protein | Mbp |
| P32232–2 | Cystathionine beta-synthase | Cbs |
| P47819 | Glial fibrillaryacidicprotein | Gfap |
| P85845 | Fascin | Fscn1 |
| B0BNA5 | Coactosin-likeprotein | Cotl1 |
| P05370 | Glucose-6-phosphate 1-dehydrogenase | G6pdx |
| P14841 | Cystatin-C | Cst3 |
| P47875 | Cysteine and glycine-richprotein 1 | Csrp1 |
| P55051 | Fattyacid-binding protein, brain | Fabp7 |
| P55053 | Fattyacid-binding protein, epidermal | Fabp5 |
| P55067 | Neurocancoreprotein | Ncan |
| P62630 | Elongation factor 1-alpha 1 | Eef1a1 |
| P63259 | Actin, cytoplasmic 2 | Actg1 |
| P97584 | Prostaglandinreductase 1 | Ptgr1 |
| Q08163 | Adenylylcyclase-associatedprotein 1 | Cap1 |
| Q4G075 | Leukocyteelastaseinhibitor A | Serpinb1a |
| Q5M7W5 | Microtubule-associatedprotein 4 | Map4 |
| Q5U318 | Astrocyticphosphoprotein PEA-15 | Pea15 |
| Q5XI73 | Rho GDP-dissociation inhibitor 1 | Arhgdia |
| Q63544 | Gamma-synuclein | Sncg |
| Q64303 | Serine/threonine-protein kinase PAK 2 | Pak2 |
| Q68FP1–2 | Gelsolin | Gsn |
| Q6JE36 | Protein NDRG1 | Ndrg1 |
| Q7TP52 | Carboxymethylenebutenolidasehomolog | Cmbl |
| Q9EQS0 | Transaldolase | Taldo1 |
| Q9WUH4 | Four and a half LIM domainsprotein 1 | Fhl1 |
| D4A8F2 | Protein Rsu1 | Rsu1 |
| E9PT65 | ProteinRdx | Rdx |
| G3V8C4 | Chlorideintracellularchannel 4, isoformCRA_b | Clic4 |
| M0R4H5 | PDZ and LIM domainprotein 4 | Pdlim4 |
| Cluster 3 | ||
| O08838 | Amphiphysin | Amph |
| O08839–2 | Myc box-dependent-interactingprotein 1 | Bin1 |
| O35095 | Neurochondrin | Ncdn |
| O35179 | Endophilin-A1 | Sh3gl2 |
| O35264 | Platelet-activating factor acetylhydrolase IB subunit beta | Pafah1b2 |
| O35814 | Stress-induced-phosphoprotein 1 | Stip1 |
| O88989 | Malatedehydrogenase, cytoplasmic | Mdh1 |
| P00507 | Aspartate aminotransferase, mitochondrial | Got2 |
| P01830 | Thy-1 membrane glycoprotein | Thy1 |
| P04636 | Malatedehydrogenase, mitochondrial | Mdh2 |
| P07323 | Gamma-enolase | Eno2 |
| P07722 | Myelin-associatedglycoprotein | Mag |
| P09951 | Synapsin-1 | Syn1 |
| P10860 | Glutamate dehydrogenase 1, mitochondrial | Glud1 |
| P11348 | Dihydropteridinereductase | Qdpr |
| P12839 | Neurofilament medium polypeptide | Nefm |
| F1LNY3 | Neural celladhesionmolecule 1 | Ncam1 |
| P14408–2 | Fumarate hydratase, mitochondrial | Fh |
| P16617 | Phosphoglycerate kinase 1 | Pgk1 |
| P19527 | Neurofilament light polypeptide | Nefl |
| P21575–2 | Dynamin-1 | Dnm1 |
| P22062 | Protein-l-isoaspartate(d-aspartate) O-methyltransferase | Pcmt1 |
| P23565 | Alpha-internexin | Ina |
| P25113 | Phosphoglyceratemutase 1 | Pgam1 |
| P27605 | Hypoxanthine-guanine phosphoribosyltransferase | Hprt1 |
| P31016 | Disks large homolog 4 | Dlg4 |
| P39069 | Adenylate kinase isoenzyme 1 | Ak1 |
| P42123 | l-lactate dehydrogenase B chain | Ldhb |
| P47709 | Rabphilin-3A | Rph3a |
| P47728 | Calretinin | Calb2 |
| P47860 | 6-phosphofructokinase type C | Pfkp |
| P50554 | 4-aminobutyrate aminotransferase, mitochondrial | Abat |
| P53042 | Serine/threonine-protein phosphatase 5 | Ppp5c |
| P54690 | Branched-chain-amino-acidaminotransferase, cytosolic | Bcat1 |
| P60522 | Gamma-aminobutyricacidreceptor-associatedprotein-like 2 | Gabarapl2 |
| P61265 | Syntaxin-1B | Stx1b |
| P61765–2 | Syntaxin-binding protein 1 | Stxbp1 |
| P61983 | 14–3-3 protein gamma | Ywhag |
| P62632 | Elongation factor 1-alpha 2 | Eef1a2 |
| P62762 | Visinin-likeprotein 1 | Vsnl1 |
| P62815 | V-type proton ATPase subunit B, brainisoform | Atp6v1b2 |
| P62959 | Histidine triadnucleotide-binding protein 1 | Hint1 |
| P62966 | Cellular retinoicacid-binding protein 1 | Crabp1 |
| P63018 | Heatshockcognate 71 kDa protein | Hspa8 |
| P63041 | Complexin-1 | Cplx1 |
| P63329–2 | Serine/threonine-protein phosphatase 2B catalyticsubunit alpha isoform | Ppp3ca |
| P86252 | Transcriptionalactivatorprotein Pur-alpha (Fragments) | Pura |
| Q05140–2 | Clathrincoatassemblyprotein AP180 | Snap91 |
| Q07266–2 | Drebrin | Dbn1 |
| Q07310–14 | Neurexin-3 | Nrxn3 |
| Q5GFD9 | Protein IMPACT | Impact |
| Q5HZA6–2 | Prolylendopeptidase-like | Prepl |
| Q5XIT1 | Microtubule-associatedprotein RP/EB familymember 3 | Mapre3 |
| Q62717 | Calcium-dependentsecretionactivator 1 | Cadps |
| Q62813 | Limbic system-associated membrane protein | Lsamp |
| Q62910–5 | Synaptojanin-1 | Synj1 |
| Q63198 | Contactin-1 | Cntn1 |
| Q63560 | Microtubule-associatedprotein 6 | Map6 |
| Q63622–3 | Disks large homolog 2 | Dlg2 |
| Q63754 | Beta-synuclein | Sncb |
| Q7M767 | Ubiquitin-conjugating enzyme E2 variant 2 | Ube2v2 |
| Q80WA4 | RNA-binding protein Nova-1 | Nova1 |
| B2GV79 | Pdxpprotein | Pdxp |
| Q9EPH8 | Polyadenylate-binding protein 1 | Pabpc1 |
| Q9ER34 | Aconitate hydratase, mitochondrial | Aco2 |
| Q9QUL6 | Vesicle-fusing ATPase | Nsf |
| Q9QX69 | LanC-likeprotein 1 | Lancl1 |
| Q9R063–2 | Peroxiredoxin-5, mitochondrial | Prdx5 |
| G3V7I0 | Peroxiredoxin 3 | Prdx3 |
| F1LPP3 | Protein kinase C and casein kinase substrate in neuronsprotein 1 | Pacsin1 |
| A1L1M0 | Protein kinase, cAMP-dependent, catalytic, alpha | Prkaca |
| B0BN63 | LOC681996 protein | Ahsa1 |
| B0BNL2 | Peptidylprolyl cis/transisomerase, NIMA-interacting 1 | LOC364561 |
| B4F772 | Heatshock 70 kDa protein 4L | Hspa4l |
| B4F7A3 | Galectin | Lgalsl |
| D3ZC55 | Heatshock 70kDa protein 12A (Predicted), isoformCRA_a | Hspa12a |
| D3ZCA0 | Proline synthetaseco-transcribed (Predicted) | Prosc |
| D3ZD09 | Cytochrome c oxidasesubunit 6B1 | Cox6b1 |
| D3ZNW5 | Neurofascin | Nfasc |
| D3ZUY8 | Adaptorproteincomplex AP-2, alpha 1 subunit (Predicted) | Ap2a1 |
| D4A0I5 | DnaJ (Hsp40) homolog, subfamily C, member 6 (Predicted) | Dnajc6 |
| D4A133 | Protein Atp6v1a | Atp6v1a |
| D4A6C9 | Protein Tom1l2 | Tom1l2 |
| D4A8U7 | Dynactin 1, isoformCRA_a | Dctn1 |
| F1LRL9 | Microtubule-associatedprotein 1B | Map1b |
| F1LRZ7 | Neurofilament heavy polypeptide | Nefh |
| F1MAQ5 | Microtubule-associatedprotein | Map2 |
| F7EYB9 | ProteinOmg | Omg |
| G3V758 | Contactin 2 | Cntn2 |
| G3V774 | F-box onlyprotein 2 | Fbxo2 |
| G3V7L8 | ATPase, H+ transporting, V1 subunit E isoform 1, isoformCRA_a | Atp6v1e1 |
| G3V936 | Citrate synthase | Cs |
| G3V964 | Neurotrimin | Ntm |
| G3V9N8 | AP-1 complexsubunit beta-1 | Ap1b1 |
| M0RC65 | Cofilin 2, muscle (Predicted), isoformCRA_b | Cfl2 |
| Q52KS1 | 6-phosphofructokinase | Pfkm |
| Q5BJT9 | Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1b |
| Q5PQK2 | Fusion, derivedfrom t(1216) malignantliposarcoma (Human) | Fus |
| Q6AY48 | Poly(RC) binding protein 3 | Pcbp3 |
| Q6AYU5 | Poly(RC) binding protein 2 | Pcbp2 |
| Cluster 4 | ||
| P51635 | Alcoholdehydrogenase [NADP(+)] | Akr1a1 |
| Q4V898 | RNA-binding motif protein, X chromosome | Rbmx |
| Q9EPF2–2 | Cell surface glycoprotein MUC18 | Mcam |
| Q9QXU9 | ProSAAS | Pcsk1n |
| G3V803 | Cadherin-2 | Cdh2 |
| B2GUZ9 | Fam49b protein | Fam49b |
| P07897 | Aggrecancoreprotein | Acan |
| F1M7H7 | Membrane-associatedguanylate kinase, WW and PDZ domain-containingprotein 1 | Magi1 |
| Cluster 5 | ||
| M0RDM4 | Histone H2A | LOC680322 |
| O08838 | Amphiphysin | Amph |
| O08839–2 | Myc box-dependent-interactingprotein 1 | Bin1 |
| O35095 | Neurochondrin | Ncdn |
| O35179 | Endophilin-A1 | Sh3gl2 |
| O35264 | Platelet-activating factor acetylhydrolase IB subunit beta | Pafah1b2 |
| O35814 | Stress-induced-phosphoprotein 1 | Stip1 |
| O88989 | Malatedehydrogenase, cytoplasmic | Mdh1 |
| P00507 | Aspartate aminotransferase, mitochondrial | Got2 |
| P01830 | Thy-1 membrane glycoprotein | Thy1 |
| P02680–2 | Fibrinogen gamma chain | Fgg |
| P04636 | Malatedehydrogenase, mitochondrial | Mdh2 |
| P07323 | Gamma-enolase | Eno2 |
| P07722 | Myelin-associatedglycoprotein | Mag |
| P07897 | Aggrecancoreprotein | Acan |
| P09951 | Synapsin-1 | Syn1 |
| P10860 | Glutamate dehydrogenase 1, mitochondrial | Glud1 |
| P11348 | Dihydropteridinereductase | Qdpr |
| P12839 | Neurofilament medium polypeptide | Nefm |
| F1LNY3 | Neural celladhesionmolecule 1 | Ncam1 |
| P14408–2 | Fumarate hydratase, mitochondrial | Fh |
| P14480 | Fibrinogen beta chain | Fgb |
| P16617 | Phosphoglycerate kinase 1 | Pgk1 |
| P19527 | Neurofilament light polypeptide | Nefl |
| P21575–2 | Dynamin-1 | Dnm1 |
| P22062 | Protein-l-isoaspartate(d-aspartate) O-methyltransferase | Pcmt1 |
| P23565 | Alpha-internexin | Ina |
| P25113 | Phosphoglyceratemutase 1 | Pgam1 |
| P27605 | Hypoxanthine-guanine phosphoribosyltransferase | Hprt1 |
| P31016 | Disks large homolog 4 | Dlg4 |
| P39069 | Adenylate kinase isoenzyme 1 | Ak1 |
| P42123 | l-lactate dehydrogenase B chain | Ldhb |
| P47709 | Rabphilin-3A | Rph3a |
| P47728 | Calretinin | Calb2 |
| P47860 | 6-phosphofructokinase type C | Pfkp |
| P50554 | 4-aminobutyrate aminotransferase, mitochondrial | Abat |
| P51635 | Alcoholdehydrogenase [NADP(+)] | Akr1a1 |
| P53042 | Serine/threonine-protein phosphatase 5 | Ppp5c |
| P54690 | Branched-chain-amino-acidaminotransferase, cytosolic | Bcat1 |
| P60522 | Gamma-aminobutyricacidreceptor-associatedprotein-like 2 | Gabarapl2 |
| P61265 | Syntaxin-1B | Stx1b |
| P61765–2 | Syntaxin-binding protein 1 | Stxbp1 |
| P61983 | 14–3-3 protein gamma | Ywhag |
| P62632 | Elongation factor 1-alpha 2 | Eef1a2 |
| P62762 | Visinin-likeprotein 1 | Vsnl1 |
| P62815 | V-type proton ATPase subunit B, brainisoform | Atp6v1b2 |
| P62959 | Histidine triadnucleotide-binding protein 1 | Hint1 |
| P62966 | Cellular retinoicacid-binding protein 1 | Crabp1 |
| P63018 | Heatshockcognate 71 kDa protein | Hspa8 |
| P63041 | Complexin-1 | Cplx1 |
| P63329–2 | Serine/threonine-protein phosphatase 2B catalyticsubunit alpha isoform | Ppp3ca |
| P86252 | Transcriptionalactivatorprotein Pur-alpha (Fragments) | Pura |
| Q05140–2 | Clathrincoatassemblyprotein AP180 | Snap91 |
| Q07266–2 | Drebrin | Dbn1 |
| Q07310–14 | Neurexin-3 | Nrxn3 |
| F1M7H7 | Membrane-associatedguanylate kinase, WW and PDZ domain-containingprotein 1 | Magi1 |
| Q4V898 | RNA-binding motif protein, X chromosome | Rbmx |
| Q5GFD9 | Protein IMPACT | Impact |
| Q5HZA6–2 | Prolylendopeptidase-like | Prepl |
| Q5XIT1 | Microtubule-associatedprotein RP/EB familymember 3 | Mapre3 |
| Q62717 | Calcium-dependentsecretionactivator 1 | Cadps |
| Q62813 | Limbic system-associated membrane protein | Lsamp |
| Q62910–5 | Synaptojanin-1 | Synj1 |
| Q63198 | Contactin-1 | Cntn1 |
| Q63560 | Microtubule-associatedprotein 6 | Map6 |
| Q63622–3 | Disks large homolog 2 | Dlg2 |
| Q63754 | Beta-synuclein | Sncb |
| Q6PCT3 | Tumorprotein D54 | Tpd52l2 |
| Q7M767 | Ubiquitin-conjugating enzyme E2 variant 2 | Ube2v2 |
| Q80WA4 | RNA-binding protein Nova-1 | Nova1 |
| B2GV79 | Pdxpprotein | Pdxp |
| Q9EPF2–2 | Cell surface glycoprotein MUC18 | Mcam |
| Q9EPH8 | Polyadenylate-binding protein 1 | Pabpc1 |
| Q9ER34 | Aconitate hydratase, mitochondrial | Aco2 |
| Q9QUL6 | Vesicle-fusing ATPase | Nsf |
| Q9QX69 | LanC-likeprotein 1 | Lancl1 |
| Q9QXU9 | ProSAAS | Pcsk1n |
| Q9R063–2 | Peroxiredoxin-5, mitochondrial | Prdx5 |
| G3V7I0 | Peroxiredoxin 3 | Prdx3 |
| F1LPP3 | Protein kinase C and casein kinase substrate in neuronsprotein 1 | Pacsin1 |
| G3V803 | Cadherin-2 | Cdh2 |
| A1L1M0 | Protein kinase, cAMP-dependent, catalytic, alpha | Prkaca |
| B0BN63 | LOC681996 protein | Ahsa1 |
| B0BNL2 | Peptidylprolyl cis/transisomerase, NIMA-interacting 1 | LOC364561 |
| B2GUZ9 | Fam49b protein | Fam49b |
| B4F772 | Heatshock 70 kDa protein 4L | Hspa4l |
| B4F7A3 | Galectin | Lgalsl |
| D3ZC55 | Heatshock 70kDa protein 12A (Predicted), isoformCRA_a | Hspa12a |
| D3ZCA0 | Proline synthetaseco-transcribed (Predicted) | Prosc |
| D3ZD09 | Cytochrome c oxidasesubunit 6B1 | Cox6b1 |
| D3ZFG5 | Protein Pmp2 | Pmp2 |
| G3V9C7 | Histone H2B | Hist1h2bk |
| D3ZNW5 | Neurofascin | Nfasc |
| D3ZUY8 | Adaptorproteincomplex AP-2, alpha 1 subunit (Predicted) | Ap2a1 |
| D4A0I5 | DnaJ (Hsp40) homolog, subfamily C, member 6 (Predicted) | Dnajc6 |
| D4A133 | Protein Atp6v1a | Atp6v1a |
| D4A6C9 | Protein Tom1l2 | Tom1l2 |
| D4A8U7 | Dynactin 1, isoformCRA_a | Dctn1 |
| F1LRL9 | Microtubule-associatedprotein 1B | Map1b |
| F1LRZ7 | Neurofilament heavy polypeptide | Nefh |
| F1MAQ5 | Microtubule-associatedprotein | Map2 |
| F7EYB9 | ProteinOmg | Omg |
| G3V6P7 | Myosin, heavy polypeptide 9, non-muscle | LOC100911597 |
| G3V758 | Contactin 2 | Cntn2 |
| G3V774 | F-box onlyprotein 2 | Fbxo2 |
| G3V7L8 | ATPase, H+ transporting, V1 subunit E isoform 1, isoformCRA_a | Atp6v1e1 |
| G3V936 | Citrate synthase | Cs |
| G3V964 | Neurotrimin | Ntm |
| G3V9N8 | AP-1 complexsubunit beta-1 | Ap1b1 |
| M0RC65 | Cofilin 2, muscle (Predicted), isoformCRA_b | Cfl2 |
| Q52KS1 | 6-phosphofructokinase | Pfkm |
| Q5BJT9 | Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1b |
| Q5PQK2 | Fusion, derivedfrom t(1216) malignantliposarcoma (Human) | Fus |
| Q6AY48 | Poly(RC) binding protein 3 | Pcbp3 |
| Q6AYU5 | Poly(RC) binding protein 2 | Pcbp2 |
| Q7TQ70 | Ac1873 | Fga |
| Cluster 6 | ||
| B0BNA5 | Coactosin-likeprotein | Cotl1 |
| O35077 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | Gpd1 |
| P02688 | Myelin basic protein | Mbp |
| P05370 | Glucose-6-phosphate 1-dehydrogenase | G6pdx |
| P14841 | Cystatin-C | Cst3 |
| P32232–2 | Cystathionine beta-synthase | Cbs |
| P47819 | Glial fibrillaryacidicprotein | Gfap |
| P47875 | Cysteine and glycine-richprotein 1 | Csrp1 |
| P55051 | Fattyacid-binding protein, brain | Fabp7 |
| P55053 | Fattyacid-binding protein, epidermal | Fabp5 |
| P55067 | Neurocancoreprotein | Ncan |
| P62630 | Elongation factor 1-alpha 1 | Eef1a1 |
| P63259 | Actin, cytoplasmic 2 | Actg1 |
| P85845 | Fascin | Fscn1 |
| P97584 | Prostaglandinreductase 1 | Ptgr1 |
| Q08163 | Adenylylcyclase-associatedprotein 1 | Cap1 |
| Q4G075 | Leukocyteelastaseinhibitor A | Serpinb1a |
| Q5M7W5 | Microtubule-associatedprotein 4 | Map4 |
| Q5U318 | Astrocyticphosphoprotein PEA-15 | Pea15 |
| Q5XI73 | Rho GDP-dissociation inhibitor 1 | Arhgdia |
| Q63544 | Gamma-synuclein | Sncg |
| Q64303 | Serine/threonine-protein kinase PAK 2 | Pak2 |
| Q68FP1–2 | Gelsolin | Gsn |
| Q6JE36 | Protein NDRG1 | Ndrg1 |
| Q7TP52 | Carboxymethylenebutenolidasehomolog | Cmbl |
| D4ABI6 | Protein RGD1561252 | RGD1561252 |
| Q920J4 | Thioredoxin-likeprotein 1 | Txnl1 |
| Q9EQS0 | Transaldolase | Taldo1 |
| Q9WUH4 | Four and a half LIM domainsprotein 1 | Fhl1 |
| B0BMX3 | Protein S100a16 | S100a16 |
| D4A8F2 | Protein Rsu1 | Rsu1 |
| E9PT65 | ProteinRdx | Rdx |
| G3V8C4 | Chlorideintracellularchannel 4, isoformCRA_b | Clic4 |
| M0R4H5 | PDZ and LIM domainprotein 4 | Pdlim4 |
For the second clustering analysis taking into account the spatial distribution of proteins (Fig. 2C, Table II, supplemental. Data S2), we observed again that the lesion segments were clustered apart from the other groups of samples. The second branch is subdivided in two branches, i.e. one separating C3 at each time point from the others which are then subdivided again in two branches, i.e. one regrouping R3 (3 and 7 days after SCI) and the other one including segments with 2 major groups (R1 and C1 versus R2 and C2). In the lesion segments, proteins overexpressed are related to the immune response (Table II). These proteins are also expressed in R1 and C1 segments at each time point after SCI and under-expressed in the other segments, showing the correlation between lesion and the rostral and caudal adjacent segments. A total of 7 clear clusters were identified. Clusters 2 and 6 correspond to proteins overexpressed in lesion and in R1, C1, respectively, and include proteins involved in neurite outgrowth but also inhibitors of neurite outgrowth such as neurocan (Table II). These proteins are expressed in lesion after 7 and 10 days and also in segments R1 and C1 for each time point. Some proteins involved in neurite outgrowth are present in C3 segment but some are inhibitory like aggrecan protein. Proteins involved in neurogenesis and synaptogenesis are overexpressed in C3 segments, with lower expression in segments R3, R2, and C2 and under-expressed in lesion, R1 and C1 segments (cluster 3 and cluster 5). However, some neurite inhibitors are also present (Table II). For a more detailed analysis, we performed a systems biology analysis for network identification in each cluster identified in the segment-dependent clustering analysis (Fig. 2D, Table II). Differential pathways were generated using the “direct interaction” algorithm to map the functional relationships linking the identified proteins in each cluster. Regarding clusters identified as overexpressed in the lesion segment, the protein network formed by cluster 1 is implicated in immune response (i.e. cell migration, microglia activation and T cell response) as well as nerve regeneration whereas cluster 2 forms a protein network involved in neurite outgrowth, nerve regeneration, cell migration and cytoskeleton remodeling (Figs. 2Da and 2Db). By contrast, cluster 3, which corresponds to proteins under-expressed in lesion segment, is functionally related to synaptic functions and axonogenesis and collateral sprouting (Fig. 2Dc). Cluster 4 corresponds to proteins overexpressed in C3 segment and functionally associated with cell-adhesion N-cadherin-related functions involving notably aggrecan, and CD146 (Fig. 2Dd). Clusters 5 and 6 are formed by overexpressed proteins in both R1 and C1 segments. Cluster 5 is related to synaptogenesis, axon extension, and neuronal plasticity (Fig. 2De) whereas cluster 6 is linked to demyelination, oligodendrocytes differentiation, astrocystosis, and nerve regeneration, (Fig. 2Df). Overall, these clustering analysis data confirmed that (1) the lesion site is the most divergent from the other segments in terms of nature and functions of protein networks and (2) secretome profiles of spinal cord segments tend to coevolve depending on their relative distance to the lesion site (i.e. R3 with C3, R2 with C2, R1 with C1).
However, besides similarities, differences could be also identified between segments located at equidistance from the lesion site. This held of particular interest when comparing R1 versus C1 segments. Indeed, as compared with the R1 profile, the C1 profile was characterized by the overexpression of inflammation markers such as fibrinogen alpha, proteins involved in demyelination and molecules either favoring axonal regeneration or inhibiting neurite outgrowth. Searching for functionally-grouped networks overrepresented in the proteins obtained from segments C3, C2, to C1, from control to 3 days, 7 days and 10 days after SCI, using ClueGO, identified leading terms mostly related to axon regeneration, synaptogenesis, extracellular matrix organization, processes in time course (Fig. 3), and thus established neuron outgrowth and regeneration processes occurring after lesion in time course. These data confirm that a neurogenesis process occurs from caudal 3 to caudal 1 after SCI in time course.
Fig. 3.
ClueGo terms involved in proteins overexpressed in C3 compared with C2 and C1.
To complement these unsupervised bioinformatics analyses, we then proceeded to supervised coexpression analyses in order to confirm the unique functional profile of the caudal segments. Data obtained from our time course experiments were classified in four groups (i.e. control, lesion, rostral and caudal) from which coexpression interactions were identified. Starting from these coexpression networks, a supervised clustering was then performed using the pro-inflammatory factor Fibrinogen alpha (FgA) as a reference molecule. Results showed that, in the lesion and caudal segments only, FgA coexpression networks were significantly enriched in molecules annotated with “complement activation” gene ontology term (Fig. 2Ea). When performing a similar supervised clustering using now the axonal Neurofilament (Nf) molecules Nfl, Nfm, and Nfh, we also found in the lesion and caudal segments only, a significant enrichment in molecules involved in the “regulation of neuron differentiation” function (Fig. 2Eb). However, a significant enrichment in molecules involved in the “axon guidance” function was observed in the lesion segments with the exception of all other segments (Fig. 2Ec). These data confirm that, although similarities can be demonstrated between segments centered by the lesion site, the caudal segments express an unexpected pro-inflammatory profile and pro-regenerative profile.
We then sought to identify specific molecules and pathways that would provide a segment and time-specific proteomic signature of SCI. To achieve this goal, we analyzed the differential distribution of unique versus common/intersected biological and functional pathways among the three spinal cord rostral and caudal regions (C1,C2, C3 versus R1, R2, R3) factored by time points post-SCI (3, 7, and 10 days). Each pair of spinal cord segments (C1 versus R1, C2 versus R2, C3 versus R3) was analyzed across the three time points and a comprehensive Venn analysis diagram extracted from Subnetwork Enrichment Analysis (SEA) was generated. From the C1 versus R1 comparisons, unique statistically significant segment- and time-specific pathways were identified that included 32 pathways for C1 on day 3, 78 pathways for R1 on day 3, 53 pathways for C1 on day 7, 78 pathways for R1 on day 7, 14 pathways for R1 on day 10 and a single pathway for C1 on day 10 (Fig. 2Fa); see supplemental Data S3 for the identity of each of the unique pathways. In Fig. 2Fb, C2 versus R2 differential pathways were analyzed across the three time points (3, 7 and 10 days). Unique statistical significant pathways were identified including 48 pathways (3days, C2), 55 pathways (3days, R2), 48 pathways (7days, C2), 52 pathways (7days, R2), 66 pathways (10 days, C2) and 36 pathways (10 days, R2); see supplemental Data S4 for the identity of each of the unique pathways. In Fig. 2Fc, C3 versus R3 differential pathways were analyzed across the three time points (3, 7 and 10 days). Unique statistical significant pathways were identified including 30 pathways (3days, C3), 69 pathways (3days, R3), 58 pathways (7days, C3); 63 pathways (7days, R3), 52 pathways (10 days, C3) and 63 pathways (10 days, R3); refer to supplemental Data S5 for the identity of each of the unique pathways. In Fig. 2Fd, Lesion-specific unique and common/intersected biological and functional pathways are analyzed across the three time points. 59, 58 and 61 unique pathways were identified corresponding to the 3 days, 7 days and 10 days' time points respectively (supplemental Data S6). Moreover, taken into account both the spatial regionalization along the spinal cord and the time course after SCI, we established that R1 versus C1 harbored no common pathways whereas R2 versus C2 shared neurites outgrowth, cell proliferation, cell differentiation, cell death and cytoskeleton organization pathways. In R3 versus C3 analyses, common pathways included cell proliferation and endocytosis pathways were found (supplemental Data S7–S9). In the same way, when taking into account the time course, 40 specific proteins unique of each segment could be retrieved, i.e. 18 specific proteins in lesion segment, 2 in R1, 2 in C1, 3 in R2, 5 in C2, 2 in R3 and 8 in C3 (Table III). Interestingly, the chemokine Cxcl2 was identified as specific to R1, whereas in C1, Neurofascin and mediator of cell motility-1 (MEMO1). MEMO1 is implicated in a MEMO1-RhoA-DIAPH1 signaling pathway that plays an important role in ErbB2-dependent (34). Neurofascin is related to L1 family immunoglobulin cell adhesion molecule with multiple IgCAM and fibronectin domains. Neurosfacin is implicated in neurite outgrowth, neurite fasciculation and organization of the axon initial segment. Importantly, Neurofascin is also targeted by autoantibodies in patients with multiple sclerosis (MS) and its altered expression has been proposed as a contributing factor to axonal pathology in MS (35). Finally, 18 lesion-specific proteins were detected among which: the follistatin-like 1 (Fstl1), a molecule that modulates the action of several growth factors on cell proliferation and differentiation and Niban-like protein 1 (Fam129b), an inhibitor of apoptosis.
Table III. Specific protein per segments in time course.
| Segments | Protein ID | Protein Name | Identified per Time |
|---|---|---|---|
| Lesion | G3V7H3 | Cfd | 3d/7d |
| P16391 | RT1 | 3d/7d | |
| P18331 | Inhba | 3d/10d | |
| Q5FVH2 | Pld3 | 3d/7d | |
| Q62632 | Fstl1 | 3d/10d | |
| Q6P7C7 | Gpnmb | 3d/10d | |
| B2GVB9 | Fermt3 | 7d/10d | |
| B4F7E8 | Fam129b | 7d/10d | |
| E9PSY8 | Eps15 | 7d/10d | |
| G3V8L1 | Pycard | 7d/10d | |
| G3V8Q0 | Rgs10 | 7d/10d | |
| P97608 | Oplah | 7d/10d | |
| Q5BJY6 | Amdhd2 | 7d/10d | |
| Q641X3 | Hexa | 7d/10d | |
| Q66H59 | Npl | 7d/10d | |
| Q66HG3 | Cndp1 | 7d/10d | |
| Q6AYF2 | Lmcd1 | 7d/10d | |
| Q8K3F3 | Ppp1r14b | 7d/10d | |
| Rostral 1 | P30348 | Cxcl2 | 3d/10d |
| Q63507 | Rpl14 | 3d/7d | |
| Caudal 1 | F1LNE5 | Memo1 | 3d/10d |
| R9PY05 | Nfasc | 3d/7d | |
| Rostral 2 | Q5U1Z2 | Trappc3 | 7d/10d |
| G3V8D6 | Trim3 | 3d/7d | |
| P62836 | Rap1a | 3d/7d | |
| Caudal 2 | Q68A21 | Purb | 3d/10d |
| F1LN98 | Ewsr1 | 3d/10d | |
| P0DJJ3 | Sgip1 | 3d/7d | |
| Q5XI81 | Fxr1 | 3d/7d | |
| G3V928 | Lrp1 | ctrl/3d | |
| Rostral 3 | D4ACV3 | Hist2h2ac | 3d/7d |
| F1LMU0 | Myh4 | 3d/7d | |
| Caudal 3 | B2GV38 | Ubl4a | 7d/10d |
| P45479 | Ppt1 | 7d/10d | |
| P63045 | Vamp2 | 3d/10d | |
| Q1M168 | Atcay | 3d/10d | |
| F1LSU5 | Defa10 | 3d/10d | |
| Q920J3 | Coro6 | 3d/10d | |
| O88902 | Ptpn23 | 3d/7d | |
| D3ZH36 | LOC100910536 | 3d/7d |
Thus, these global proteomic analyses clearly demonstrate that lesion site and the proximal segments (R1 and C1) are different from the distal areas. These results are corroborated by MALDI mass spectrometry imaging (MALDI-MSI) analysis performed on R1, C1, and lesion tissues sections (Fig. 4). In fact, molecular 2D-maps based on metabolites and lipids will allow seeing the molecular regionalization in regards to physiological processes. The molecular imaging studies after SCI (3 days) show a complete disruption of gray matter-specific lipid signals along the lesion site (Fig. 4A). Spatial segmentation of MALDI-MSI data, dividing the spinal cord sections into anatomical regions allowed discriminating between different regions in an unsupervised method. The cluster analyses of rostral, lesion and caudal sections after 3 days of SCI confirm a disrupted structure of lipid distribution along the lesion site whereas the segmentation of rostral and caudal sections is consistently aligned with the spinal cord anatomy (Fig. 4B). The gray matter is shown in red and the white matter in green (Fig. 4B). m/z values exclusively localized with the lesion region were automatically elucidated and in Fig. 4C, 18 m/z images are shown. With component analysis fundamental components of the spectra and m/z images within the SCI MALDI-MSI data set can be discovered unsupervised. By that, the data are decomposed into a pre-specified small number of score images showing the main spatial features and a small set of component spectra showing the main spectral features. Furthermore a Receiver Operating Characteristic (ROC) analysis was performed for detecting m/z-values which allow a discrimination of different regions (C1 and R1); see Table IV for discriminating m/z values and Fig. 4Cb for a visualization of the spatial distribution of selected m/z values. In Fig. 4D, the results of the component analyses for the probabilistic latent semantic analysis (Figs. 4Da and 4Db) with random initialization and for principal component analysis (Fig. 4Dc) with five components are indicated. For verification of the findings obtained above, a second set of MALDI-MSI data was obtained for 24 slices of the caudal region. A 3D-visualization of selected m/z values, which were chosen according to the values stated in Table IV, confirms the significance of these molecular weights (Fig. 4E). Clustering has been performed through R1, C1, and lesion segments (Fig. 4F) and established that lesion is highly impacted compared with R1. Specific m/z at respectively 546.406 and 524.402 which have been retrieved at the lesion site but not in R1 and C1 segments confirm our hypothesis (Fig. 4G). In the same way, clustering analyses also allow to state that C1 is divergent from R1 (Fig. 4G). 3D map performed at the C1 segment from 24 sections showed that gray matter is more impacted compared with the white matter (Fig. 4H) and expressed specific lipids as the ones fully characterized by lipidomic in C1 and in lesion (Table V). In fact, comparison between the lesion and C1 in lipid composition showed difference in the level of expression between the two segments. These 2D- and 3D-molecular maps reinforce the proteomic analyses and point out that C1 is divergent from R1 in term of the molecular contents at different time points.
Fig. 4.
A, Comparison of signals across different spinal cord segments. (a) 25 m/z image sets (left rostral, middle lesion, right caudal) are shown with ± 0.3 Da and edge-preserving image denoising as well as automatic hot spot removal. (b) The respective m/z values are indicated in the table. (c) The localization of the respective peaks is shown in the mean spectrum below. B, Hierarchical clustering: regions which are conjectured to be gray respectively white matter tissue is clearly structured in rostral and caudal regions, no structure visible in lesion region. C, (a) Colocalized m/z image sets (left rostral, middle lesion, right caudal) and (b) respective m/z values in the lesion region with the respective Pearson's correlation values. D, Component analyses for the probabilistic latent semantic analysis (a and b) with random initialization and principal component analysis (c) with five components. E, 3D projection of lipid expression across the length of spinal cord. Initial 3D alignment to follow lipid spatial distribution with imaging. F, Cluster analyses (segmentation) for two, four and five clusters with the respective amount of spots per cluster and the distance values to reconstruct spinal cord. G, 3D projection of specific signals m/z 524.402 and 546.406 detected at the lesion site. H, 3D map at the level of the gray and white matters of the caudal 1 segment based on 24 consecutive sections.
Table IV. Discriminating m/z-values for lesion vs. rostal/ caudal region obtained by ROC.
| Centroid | ± | Maximum AUC | m/z at maximum |
|---|---|---|---|
| 86.005 | 0.245 | 0.744975 | 86.237 |
| 86.377 | 0.127 | 0.761224 | 86.263 |
| 86.686 | 0.182 | 0.674034 | 86.517 |
| 104.076 | 0.277 | 0.734237 | 104.328 |
| 104.630 | 0.277 | 0.727656 | 104.353 |
| 169.759 | 0.188 | 0.397301 | 169.921 |
| 170.055 | 0.108 | 0.458626 | 170.150 |
| 170.271 | 0.108 | 0.759526 | 170.278 |
| 170.456 | 0.076 | 0.691658 | 170.379 |
| 170.720 | 0.188 | 0.557438 | 170.532 |
| 174.917 | 0.258 | 0.394923 | 175.163 |
| 175.398 | 0.223 | 0.739125 | 175.392 |
| 175.885 | 0.264 | 0.251686 | 176.130 |
| 184.068 | 0.300 | 0.750433 | 184.322 |
| 191.828 | 0.300 | 0.239038 | 192.108 |
| 192.352 | 0.163 | 0.737723 | 192.261 |
| 192.678 | 0.163 | 0.186462 | 192.821 |
| 213.801 | 0.264 | 0.299782 | 214.066 |
| 214.244 | 0.178 | 0.749835 | 214.244 |
| 214.636 | 0.214 | 0.50978 | 214.422 |
| 221.982 | 0.277 | 0.721014 | 222.259 |
| 222.536 | 0.277 | 0.743943 | 222.310 |
| 240.018 | 0.300 | 0.622719 | 240.298 |
| 240.629 | 0.300 | 0.755234 | 240.400 |
| 782.731 | 0.300 | 0.643802 | 782.756 |
| 783.698 | 0.300 | 0.64038 | 783.748 |
| 798.684 | 0.300 | 0.61345 | 798.735 |
| 799.702 | 0.300 | 0.616393 | 799.727 |
Table V. Retention time, m/z, and identification of lipids expressed in C1 or lesion segment after lipidomics analyses.
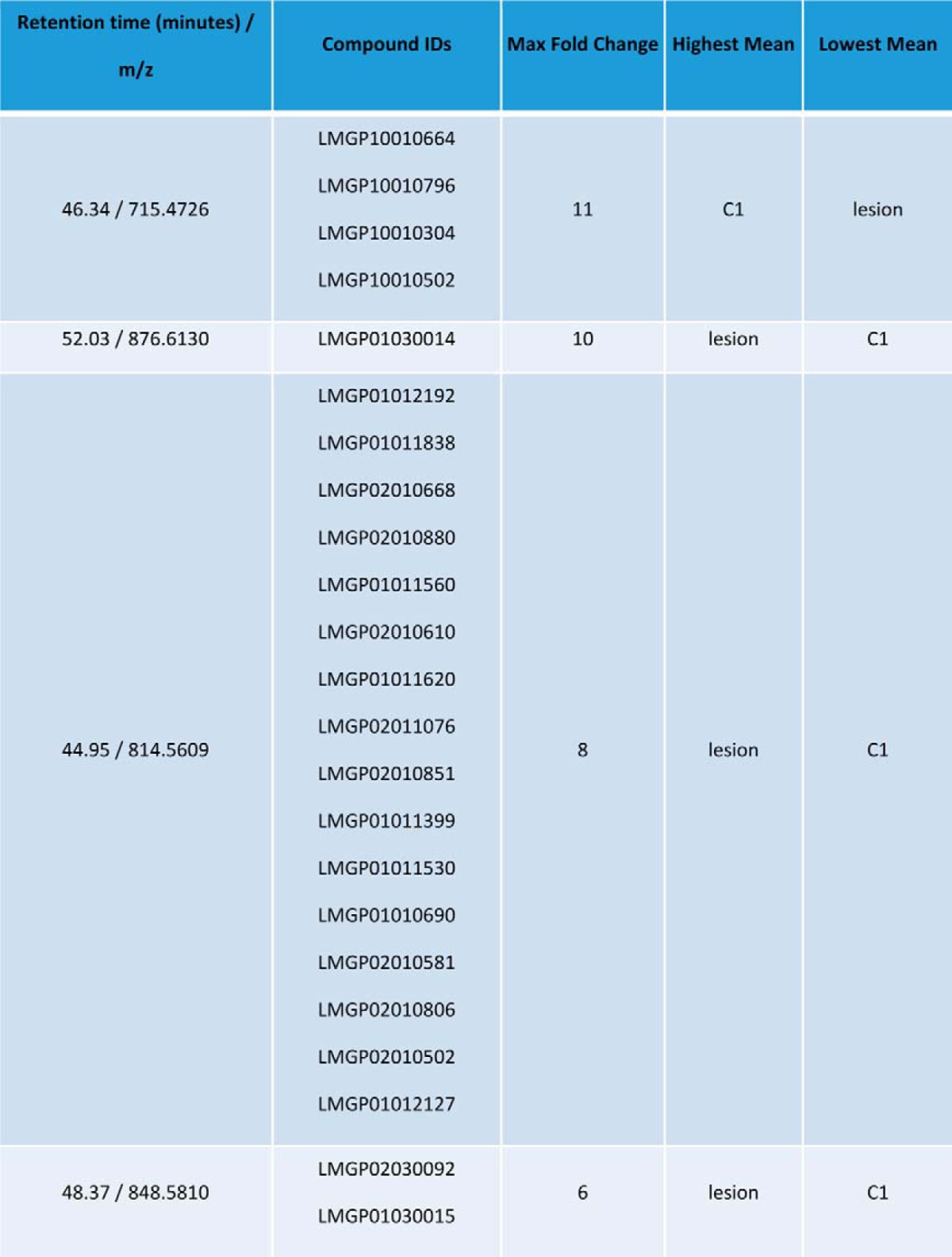
Time Course Analysis of Inflammatory and modify to regenerative Markers Along the Spinal Cord Axis
To complement the results obtained from our pan-proteomic approaches, we further focused our attention on immune and neurites outgrowth or axon guidance factors along the spinal cord in time course (Table VI). We established that, compared with control samples, immune-related proteins comprising galectins 1 and 3 (Lgals1 and 3), coronin 1b (Coro1b), macrophage migration inhibitory factor (MIF), granulin (Grn), lymphocyte cytosolic protein (Lcp1), complement system components (C3 and B2m) and cathepsins (Ctsb and Ctsd), are overexpressed in the lesion site 3 days after SCI, show further increased levels at 7 days and remain at high levels at 10 days. In contrast, the inflammation peak was observed at 3 days in R1 segments and at 7 days in C1 segments. For axon guidance and neuroprojection represented by microtubule-associated protein Tau (Mapt), Pak proteins (Pak 1 and 2), Ras-related C3 botulinum toxin substrate 1 (Rac1), neural cell adhesion molecule 1 (Ncam1), stathmins (Stmn 1 and 2), semaphorin 7a (Sema7a), dectin 1 (Dctn1), neurofilament light polypeptide (Nefl), profiling 1 (Pfn1), neurofascin (Nfasc), neurotrimin (Ntm), the level of these proteins was low at 3 days, increased at 7 days and stayed stable at 10 days in the lesion segment. At 3 days, the level is the highest in C2 and C3 segments compared with C1 and R1 segments. At 7 days, it increased in C1 to C3 segments. At 10 days, it decreased in lesion, C1 and C2 segments, stayed high in C3 and increased in R1 to R3 segments. For motoneurons degeneration represented by the super dismutase 1 (Sod1), the vesicle-associated membrane protein b (Vapb), dynactin-1 (Dctn1), the highest level is in C1 to C3 segments at 7 days and in R1 to R3 segments at 10 days. For neurites inhibition, i.e. RhoA, neurocan (Ncan), amphiphysin (Amph), the overexpression occurs in C1 segment in time course with a peak reaching at 7 days.
Table VI. Label free quantification (Intensity-based absolute quantification (IBAQ) value) of identified protein per segments and taking into account time after SCI. the. iBAQ values calculated by MaxQuant are proportional to the molar quantities of the proteins.
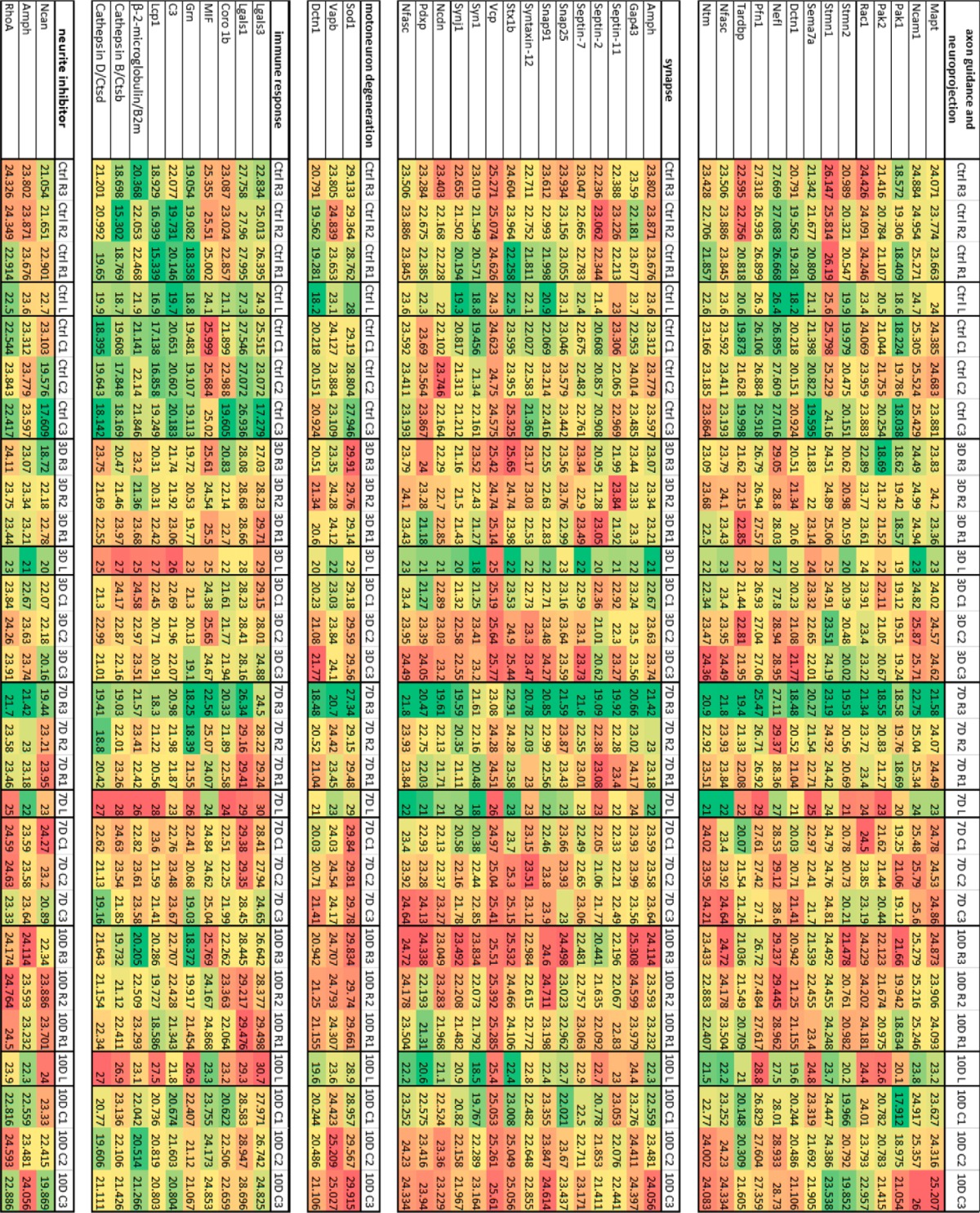
In order to extend such proteomic quantitative label free results, cytokines arrays (Figs. 5A–5B) and functional assays (Figs. 5C–5E) that compared the secretome of R1 versus C1 segments were performed. Cytokines arrays confirmed the time-dependent and localization-dependent synthesis of chemokines and cytokines in R1 and C1 segments. Compared with control, in R1 segment, CXCL1, CXCL3, CXCL5, CCL20, TIMP-1, and IL6 are overexpressed 3 days after SCI whereas CCL3 and CTNF decrease (Fig. 5A). At 7 days after SCI, some cytokines remain high (i.e. CXCL1, CXCL3, and TIMP1), increase (i.e. CXCL2, CXCL5, CXLC7, CCL3) or remain constant (i.e. IL6 and CCL20) (Fig. 5A). At 10 days, CXCL1, CXCL3, CXCL2, CXCL5, and TIMP 1 fall dramatically, IL6 and CCL20 disappear and CTNF and CXCL7 continue to increase (Fig. 5A). In C1, the evolution is different (Fig. 5B). At 3 days after SCI, level of CXCL3 and TIMP-1 increase but not as registered in R1, the level of CXCL7 and CXCL5 decrease, whereas IL6 and CCL20 are not detected (Fig. 5B). After 7 days, the cytokines pattern changed with an increase of CXCL1, CXCL2, CXCL3, CXCL5, and CXCL7 levels and re-apparition of IL6 together with CCL20. 10 days after SCI, CXCL1, CXCl3, CXCL5, TIMP 1 are even higher than in R1 segment and IL6 is always present in C1 but not in R1 segment. Taken together, these data showed that the cytokines pattern change in time course between R1 and C1. Specific chemokines (CXCL1, CXCL2, CXCL3, CXCL5, CXCL7, CCL3, CCL20, IL6) that are secreted by macrophages or epithelial cells after injury have the ability to attract neutrophils and lymphocytes, activate inflammation and stimulate extracellular matrix synthesis and tissue remodeling. This actually substantiates the hypothesis that the types of immune cells that are attracted along the spinal cord upon injury insult are quite different between rostral and caudal segments in time course, i.e. cells start to migrate toward R1 and then C1 segment as shown the proteomic data (Table VI). It has also to be noticed that IL6 and CCL20 are expressed firstly in R1 at 3 days after SCI and secondly appeared in C1 at 7 days. Moreover, CCL20 is also known to attract T regulator lymphocytes through CCR6 binding (36). In that context, it is necessary to correlate the cytokines expression with cellular presence in time course and along the spinal cord.
Fig. 5.
Chemokines and cytokines array after SCI in CM from R1 and C1 for control and 3, 7, and 10 days post injury. The rat cytokine array assay was performed in SC conditioned media. Blue bars show R1 CM, and orange indicates C1 CM. The bar diagrams represent the ratio of the spot mean pixel densities/reference point pixel densities. A, Comparison of cytokines and chemokines secretion in R1 segment at 3, 7 and 10 days after SCI and control. B, Comparison of cytokines and chemokines secretion in C1 segment at 3, 7, and 10 days after SCI and control. Significant differences were analyzed using Student's t test *p < 0.05, **p < 0.01, ***p < 0.001. Quantification of immune cells recruitment in control and 3, 7, and 10 days post-injury, comparison between R1 rostral and C1 caudal segments. C, Number of Iba1 positive microglia recruited in gray and white matters. D, Percentage of neutrophil elastase-positive neutrophils and E, FoxP3-positive Tregulators recruited in white matter. *p < 0.05, ** p < 0.001, *** p < 0.0001 One-way ANOVA.
Fluorescent immunohistochemistry and cell quantification were then performed in order to compare, in C1 versus R1, the post-SCI time course of microglial cells, neutrophils and FoxP3 positive T regulator lymphocytes (Tregs) infiltrations (Figs. 6 and 7). Quantification of microglia revealed that the highest density of Iba-1-positive cells could be observed in gray matter areas of caudal regions, at 3 days and 7 days post-SCI (Fig. 6C). No differences are observed between the R1 and C1 segments at 10 days (Fig. 6C). As compared with control samples, Iba-1-positive microglia, in all SCI samples harbor a rounded morphology indicative of an activation state (Fig. 6). Differences between the R1 and C1 segments, with regard to microglial cells density, reach significant levels only in gray matter areas, at 3 and 7 days post-SCI (Fig. 6B–6E). Irrespective of the segment considered, the density of microglial cells decreases at 7 days but still remains above the basal microglial density observed in control samples (Fig. 6C). Results from cytokine/chemokine arrays urged us to perform a similar immunohistochemical analysis for neutrophils and Tregs. Similar study was performed for neutrophils in regard to chemokines pattern found (Fig. 5D). Neutrophils were abundantly detected in both R1 and C1 segments with a peak reached at 3 days after SCI without any differences in term of amount between each segments. The level decreases in time course. However, immunohistochemistry based on anti-neutrophil elastase marker revealed that the shape of the cells is different in control and after SCI (Fig. 7). In controls, neutrophils are elongated and bipolar (Fig. 7A). Three days after SCI, cells start to make some ramifications and 7 days more cells have ramifications (Fig. 7A and 7C). Ten days later, the number of cells decreased, but the ones present are still ramified (data not shown). Analyses performed at the level of the blood vessel confirmed that lot of neutrophils are present 3 days (Fig. 7D) after SCI and less at 7 days. At higher magnification, we could discriminate sign of neutrophils extravasation, mimicking movement of cells out of blood vessels toward the injury (Fig. 7E). In comparison, Tregs are present 3 days after SCI, in rostral segment in higher amount than in caudal one. Their levels peak at 7 days for both segments and then decrease at 10 days (Fig. 7E). These results are in line with the cytokine phenotype expressed by the cells (IL6 and CCL20) (Fig. 5). At 3 days after SCI, Tregs presented ramifications (Fig. 7F) whereas at 7 and 10 days, they become round (data not shown). These data are clearly in line with the cytokines pattern observed in Fig. 5 where chemoattractant factors for neutrophils and microglia are detected. Neutrophils and microglia are recruited between 0 and 3 days after SCI in caudal segments and their levels decrease at 7 days. Similarly, Tregs are recruited between 3 and 7 days and peak at 7 days. These data are in line with the presence of CXCL1, CXCL3, CXCL5, CCL20, TIMP-1, and IL6 in R1, at 3 days, which are known to attract neutrophils and lymphocytes. In C1, a delay is observed for the recruitment of the Tregs, which are recruited 7 days after SCI which correlate with the detection of CCL20 in C1 only at 7 days, whereas neutrophils and microglial cells are already present at 3 days. Taken together the results showed that C1 is clearly different from R1 in term of cell types, molecular content in time course and revealed to be a target segment for therapy.
Fig. 6.
Representative longitudinal sections of spinal cord revealing Iba1 microglia positive cells rostrally (B, D, E) and caudally (C, F) to the lesion site after SCI (3D, 7D, 10D) in comparison to control (A). Note, activated microglia with hypertrophied cell body and retracted processes at 3 and 7 days at C1 segment (B, C, E arrows-full) in comparison to resting microglia in control (A, arrows-intermitted) or mild activation, re-appearing fine ramification at 10D (F). Scale bar = 25 μm.
Fig. 7.
Fluorescent immunohistochemistry performed on control spinal cord sections and after SCI (3 and 7 days) analyze for neutrophils with neutrophil elastase antibody and lymphocytes Tregulator with FoxP3 antibody. Infiltrated neutrophils showed either elongated (arrow- intermitted) or round shape morphology (arrow-full) at 3 days after SCI (B), whereas at 7 days cellular ramification was apparent in most neutrophils (C). Many blood vessels close to the injury site were filled with neutrophils at 3 and 7 days after SCI (D). At higher magnification, neutrophils extravasation, mimicking movement of cells out of blood vessels toward the injury can be detected (E). Scale bar A-F = 20 μm, D = 40 μm, E = 15 μm. (C3>C2>C1) for control, 3 days, 7 days, and 10 days.
Focus on C1 Biomarkers for a Targeted Treatment
Because of the presence of neurofascin and MEMO 1 as specific markers of C1 in time course, we investigated the role of these proteins in SCI. Neurosfacin could serve as autoantigen. Indeed, in patients sera suffering from inflammatory demyelinating polyneuropathy or multifocal motor neuropathy, specific autoantigens including neurofascin have been found6. Moreover, emerging data indicate that pathological sequelae that accompany central nervous system trauma, e.g. SCI, have characteristics of a self-directed immunological disease. Autoantibodies could exacerbate tissue damage impairing neurological recovery and amplify SCI injury. Moreover, the label free quantification (Intensity-based absolute quantification value) of identified protein per segments and taking into account time after SCI have revealed the presence of immunoglobulin (IgG2a) in lesion 3 days after SCI (Table VI). The fact that such cytotoxic immunoglobulin was detected so early after SCI is not in line with a classical adaptative response. In this context, we focused on tracking the occurrence of autoantibodies in time course after SCI. We performed double labeled immunofluorescence on frozen longitudinal sections after 3, 7, and 10 days post-SCI to establish the cell types (neurons, astrocytes and microglia) expressing IgGs. In spinal cord tissue, at 3 days postinjury, IgG-immunoreactivity using both antibodies was found in most neurons (labeled with anti-NeuN), with higher prevalence throughout the layers LV-LIX (Fig. 8A). However, in the case of motor neurons, we could see differences in the double staining expression (Fig. 8A). These IgGs have been characterized (Fig. 8B). In fact, the conditioned media collected at 3, 7, and 10 days post-SCI were incubated with protein A and afterward eluted proteins were separated on SDS-PAGE (Fig. 8B). After silver staining, clear bands are retrieved from the gel at 3 days, but not at 7 or 10 days after SCI. Each selected bands at 25 kDa, 55 kDa, and 75 kDa have been digested in gel before subjected to shot-gun analysis. The identification, performed by MS/MS, of the proteins confirms the presence of the IgGs kappa-chain VJC precursor of fragment (166–218) (Fig. 8B).
Fig. 8.
Specific expression of IgG positivity in neurons through the spinal cord laminae I-IX at 3D. A, Double immunofluorescence labeling revealed colocalization of NeuN-positive (red) and IgG-positive (green) cells that were restricted throughout laminae LV-LIX, whereas superficial laminae LI-LIV were IgG-negative. B, SDS-PAGE electrophoresis and silver staining confirmed the presence of autoantibodies at 3 days after injury by three bands localized at 70 kDa, 55 kDa and 25 kDa. After in-gel digestion, shot-gun proteomics analysis was performed to confirm the presence of IgGs, for each band (antibody type). BlastP has shown 62% of sequence alignment. Amino acid residues in brown consist of the polypeptide binding sites of the antibody, I.e., sites 135–137, 150, 152, 154, 156- 157, 179–182, 193–195, 226–227. Scale bar = 50 μm. Confocal images demonstrating the presence of IgGs in the spinal cord neurons. C, Orthogonal views confirmed that anti-IgG (green) and anti-NeuN (red) stainings overlap in some, but not all motor neurons. D, The distribution of IgGs was different across neuronal populations. Although in some neurons intense IgG positivity (green) was homogenously distributed with the cell body and processes (D, arrow-full), other neurons exhibited IgG expression limited to their membrane (arrow-intermitted). Scale bar = 25 μm.
Orthogonal views confirmed that anti-IgG (green) and anti-NeuN (red) staining overlap in some, but not all motor neurons (Fig. 8). Thus, some neurons diffusely stained with anti-NeuN were also heavily positive for IgG (Fig. 8C), whereas other neurons were lacking anti-IgG positivity completely, or exhibited labeling only on their surface membrane (Fig. 8D). In few cases we could distinguish also IgG-positive nerve processes. Similarly as in neurons, a double anti-GFAP and anti-IgG labeling confirmed positivity in astrocytes (Figs. 9D–9F). Many GFAP-positive astrocytes with hypertrophic appearance and thick processes indicating activated phenotype coexpressed IgG (yellow) primarily at the injury site. These reactive IgG-positive astrocytes were found around neuronal bodies and vessels (Figs. 9D–9F). Baseline expression of GFAP-positive astrocytes with the characteristic round small soma and slender, long processes, but lacking IgG-expression were seen in control spinal cord distributed throughout white and gray matters (Figs. 9A–9C) as well as in the regions out from injury site (Fig. 9G). Furthermore, no GFAP-positive astrocytes expressing IgG were detected in the spinal cord tissue from 7 days and 10 days after SCI. Interestingly, microglia did not express IgG in any of studied groups (data not shown). In addition, to exclude possible effect of B-cells mediated IgG production, we evaluated spinal cord tissue containing multiple in vivo injections of rabbit anti-rat CD-20 antibody. In this context, 1 h after SCI, 3 intraspinal injections per animal were applied bilaterally to the lesion site, at the level of the lesion cavity and at rostral and caudal segments. Basso, Beattie and Bresnahan studies at 0, 1, 3, 7, 14, 21 days until 28 days after SCI did not reveal any improvement compared with SCI without anti-CD20 treatment (Fig. 10). In sagittal sections double labeled with primary goat anti-rat IgG2a and mouse anti-NeuN, we did not observe significant attenuation of neurons expressing IgG. The injection tract was occupied with small amount of erythrocytes that were surrounded by neurons with intensely labeled IgGs as well as those farther from injection (Fig. 9H).
Fig. 9.
Representative images of GFAP-positive astrocytes expressing IgG. Control spinal cord sections contained typical astrocytes with star like thin processes (A), lacking IgG (red) expression (B), confirmed by double labeling (C). In contrast, reactive astrocytes with hypertrophic appearance and thick processes, coexpressed IgG (yellow) at the lesion site were found around neuronal bodies and vessels after 3days (D–F). In neighboring rostral and caudal segments, some neurons expressing IgG (red, arrows), but not GFAP-positive astrocytes (green) were detected (G). Scale bar = 25 μm. H) Intraspinal delivery of anti-CD-20 antibody in rats surviving 3days. (a) Identification of anti-CD-20 delivery tract within the spinal cord tissue labeled with goat-anti rat IgG (green). Note, there is no visible depletion of IgG labeling. (b, b′, b′′, b′′′) composite of four detailed figures showing the region adjacent to the lesion, where delivery of CD-20 was performed (outlined by two bars) and is accompanied with infiltrated erythrocytes (arrows) and neurons expressing IgG. At the injections site as well in rostro- caudal axis neurons and their processes expressing IgG were detected. (c) Detail of boxed area from b′′ shows double labeling of anti-IgG and anti NeuN confirmed neuronal expression of IgGs. Scale bar = 30 μm.
Fig. 10.
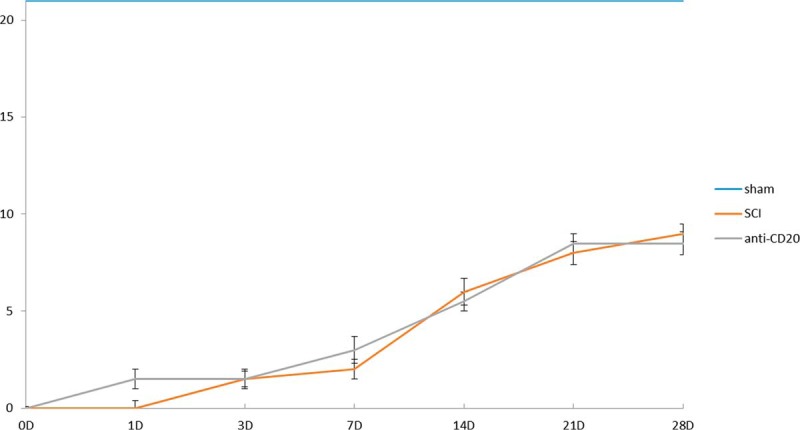
BBB studies at 0, 1, 3, 7, 10, 14, 21, and 28 days in following experimental groups: in sham, following SCI, and after SCI + anti-CD20 antibody injection. Note, no significant differences in motor function recovery between SCI and SCI+ anti-CD20 treatment during survival.
DISCUSSION
We have previously investigated the spectrum of released molecules in the conditioned media (CM) from the spinal cord central lesion and adjacent rostral and caudal segments at 3 days after spinal cord injury (SCI) in order to specify the molecular environment within the proximity of the injured tissue. Data suggested that regionalization in terms of inflammatory and neurotrophic responses may occur between rostral and caudal segments in acute SCI. Here, we extended our investigation both spatially and temporally using cellular, biochemical and proteomics techniques. We have shown that the lesion segment is the most divergent from the others ones and is mostly implicated in acute immune response through microglia and neutrophils activation, oxidative stress and lipid peroxidation. R1 and C1 segments present markers for inflammation, axonal regeneration, demyelination and even neurite inhibitors factors. By contrast, segments R3 and C3 and R2 and C2 expressed neurites outgrowth, cell proliferation, cell differentiation, endocytosis and cytoskeleton organization activated pathways. Moreover, these segments are very close in term of the molecular content in time course after lesion and are mostly divergent from R1 and C1 segments. A deeper analysis based on the differential distribution of the unique and common/intersected biological and functional pathways among the three spinal cord rostral and caudal regions (C1,C2, C3 versus R1, R2, R3) factored by time of SCI (3, 7, and 10 days) clearly demonstrate that R1 is different from C1. The Subnetwork Enrichment Analysis (SEA) showed that only 40 specific proteins unique can be retrieved from each segment taking into account the time course and the spatial localization nearby the lesion (Table III). In this context, the chemokine CXCL2 is considered as unique in R1 whereas in C1, the neurofascin and the mediator of cell motility 1 (MEMO 1), which control cell migration by relaying extracellular chemotactic signals to the microtubule cytoskeleton, are detected. The molecular 2D map obtained via MALDI-MSI analysis completed by a lipidomic study in both lesion and C1 (Table V) strengthens these results. The lipid profile detected in lesion is mostly different in term of level of expression to the one detected in C1. The 3D map of the C1 segment at the level of the gray and white matter confirms such differences.
The fine analysis of C1 segments showed, that in time course, C1 segments express neurite inhibitors like MEMO1, neurocan, as well as neurite stimulators such as neurofascin and amphiphysin proteins. MEMO1 is implicated in MEMO1-RHhoA-DIAPH1 signaling pathway which plays an important role in ErbB2-dependence (34). Neurocan is known to inhibit neurites outgrowth (37). After spinal cord injury, neurocan, brevican, and versican expression is increased within days in injured spinal cord parenchyma surrounding the lesion site and peaks at 2 weeks. Neurocan and versican are persistently elevated for 4 weeks post-injury, and brevican expression persisted for at least 2 months (37). On the contrary, neurofascin and amphiphysin, are two proteins known to be implicated in neurites outgrowth, are located at the level of the growth cone. However, these two proteins are also involved in autoimmunity as autoantigens (38–41).
With respect to the proteomic results our next step was to investigate the nature of cellular processes, and identify the cellular content responsible for production of detected molecules. It is well established that injury to the blood-brain barrier facilitates the extravasations of cellular components as well as immunoglobulins and complement proteins into the neural parenchyma (42, 43). The recruitment of blood-derived cellular components is dependent on inflammatory processes that are orchestrated by secondary damage at the lesion site. Thus, the cytokine environment in the central nervous system (CNS) may determine the phenotype of the inflammatory infiltrate. For example, intra-spinal injections of tumor necrosis factor (TNF)-α induced monocyte infiltration whereas interleukin (IL)-Iβ recruited mostly neutrophils (44, 45). Present data confirm that released cytokines and chemokines correlate with the activation of microglia and recruitment of neutrophils in segments anterior and posterior to the lesion site. These cells, along with the soluble mediators/proteins (e.g. cytokines, complement), interact to eliminate pathogenic elements in the affected site, while simultaneously priming the site for repair. Within the spinal cord, microglia are the resident tissue macrophages that primarily control the rate, magnitude and the ultimate fate (supporting regeneration or degeneration) of inflammatory processes at the injury site. However, in our study we have taken into account the cellular microenvironment in which microglia become activated (46). Therefore, we have analyzed response of microglia within the white and gray matters separately in rostro-caudal axis with time.
Present data confirmed striking differences in the number and morphology of microglia cells during injury time. The extent of microglia activation was significantly higher in gray than in white matter tracts in the caudal segments during the time period 3–7 days after injury, whereas at 10 days it dropped down. These discrepancies may be dependent on two factors: i) the metabolic changes in microglial function that are differentially affected at the gray matter necrotizing injury site and in degenerating white matter tracts, or ii) on the extent of blood-brain barrier injury, which varies between the gray and white matter (42, 47), and on the severity of injury. In this context, gray and white matters microglia express distinct morphologies and levels of cell surface antigens (48, 49), which most likely correspond with unique cell functions (50, 51). Indeed, the microenvironment in which microglia and/or macrophages are activated influences their neurotrophic or neurotoxic effector potential (52, 53) that correspond to M1 and M2 phenotypes. The predominance of microglia activation in caudal segments is in line with our previous and present studies confirming the occurrence of severe inflammation-associated tissue damage taking place in this segment and possibility to switch/or not from M1 to M2 phenotype (29).
In contrast to tissue resident microglia (54), neutrophils are the first inflammatory cells to arrive at the site of injury, with a peak at 24 h post injury. Recruitment of T regulators (Tregs) (Foxp3-positive) firstly occurs at 3 days and mainly at the level of the rostral segment and peaks at 7 days in both rostral and caudal sites. These results are in line with the presence of CCL20 in rostral segment and not in caudal at 3 days and then found at 7 days in both segments. In fact, CCL20 is known to recruit Tregs (55). Presence of recruitment of thymic-derived FoxP3-Tregs was recently suggested (56). Tregs can reduce inflammation and enhance CNS repair (57). Presence of such cells predominantly in rostral segment is in line with the cytokine medium which favor expression of macrophages expressing an M2 phenotype and production of neurotrophic factors (29). Moreover, the time difference of recruitment between rostral and caudal segments of these cells can also contribute to the less enhancement of neurites outgrowth in C1 compared with R1.
Thus, the spinal cord, as a part of CNS has generally been described as having immune privilege, but several facts motivated us to reexamine this principle. Particularly, in a search for presence of antibodies responding to neurofascin found in caudal segment, we have proven via immunohistochemistry that IgGs are expressed by interneurons and motor neurons, 3 days after spinal cord injury suggestive of autoimmunity. Furthermore, in CM from SCI, using biochemical and proteomic analysis, we showed by protein A, gel electrophoresis associated to mass spectrometry analyses, content of spinal IgGs at 3 days, but not at 7 or 10 days after injury. Here, we need to mention few important findings regarding neuronal IgG positivity and IgG sources. Firstly, spinal neurons revealed distinct expression of IgG throughout dorso-ventral axis. Although sensory neurons in laminae I-IV were negative, the neuronal populations corresponding to interneurons and motor neurons of laminae V-IX were most likely positive for IgG, with few discrepancies. The confocal orthogonal views showed that some neurons revealed very clear golgi-like staining of soma and processes, whereas in others the IgG expression was found solely in the neuronal soma, or was restricted to superficial membrane outlining the neuronal body. We suggest that divergence of neuronal population IgG response may be caused by their different vulnerability to injury or by the extent and severity of impairment in situ. Thus, it is more likely that we can see early response of most vulnerable interneurons and motor neurons expressing IgG that are probably behaving to secondary damage processes, similarly as are responding CA1 vulnerable neurons in hippocampus (58). Furthermore, also spinal cord ischemia leads to selective loss of highly vulnerable inhibitory GABAergic interneurons followed by motor neurons resulting to spasticity (59). There are other studies further confirming vulnerability of interneurons, and motor neurons to stress, ischemia, injury or various neurodegenerative diseases. Secondly, it is important to identify the source of IgGs. Because, the motor neurons are projecting outside the CNS, it is assumed that they can take up IgGs from peripheral tissues, by retrograde transport from nerve terminals (60, 61). However, this may not be the case for the interneurons that terminate only within the spinal cord. But, they may have the ability to take up IgGs from the cerebrospinal fluid (CSF) (62).
On other hand, there are studies suggesting productions of IgGs by neurons (63). Present results document specific transient neuronal expression of IgGs, but we do not have proof of their sources, which needs further transcriptomics studies. Another important issue that needs to be mentioned is the functionality of IgGs and their epitopes. There are two contradictory hypotheses that should be taken into account, where IgGs pose as neuroprotective (64) or neurodestructive (60). The first one suggests that neuron-derived IgGs has the ability to protect neurons from early apoptosis and cell death induced by complement. The second one refers to IgGs as detrimental factors of neurons, attacking the regeneration of axons after spinal cord injury (60)(B cells produce pathogenic antibodies and impair recovery after spinal cord injury in mice (65)). Our results are more likely in a favor of the first one, because we identify early, transient expression of IgGs at 3 days. Why we did not see IgGs at 7 or 10 days, we do not have reasonable explanation. Moreover, injection at 1 h after lesion of anti-CD20 did not attenuate the expression of IgGs detected at 3 days post-SCI and Basso, Beattie and Bresnahan motor score of the animals was similar to the one with preliminary treatment. These data tend to reveal that antibodies are not produced by active B cells. Nevertheless, to better understand this interesting finding; we need to follow up with further studies.
Taken together, we described for the first time the molecular and cellular processes occurring after SCI on the temporal and spatial levels. We established that differences in term of molecular pathways occurred between lesion, C1, R1, and the other segments. In fact, R2, C2, and R3, C3 coexpressed the molecules implicated in same physiological pathways. Only 40 specific proteins for each segment and taking into account time after injury have been characterized. Except the lesion, C1 is the most divergent. The presence of neurites outgrowth inhibitors in C1, with a delay in recruitment of Tregs, favors the lack of regeneration process in C1. Moreover, the presence of IgGs at the lesion site, at 3 days post-SCI, can also be one of the factors that contribute to limitation of the regeneration process. However, treatment of anti-CD20 did not showed any impact in vivo in IgGs presence and in enhancement of locomotor function. These results open the door to a novel view of the SCI treatment by considering the C1 as the therapeutic target in order to modulate inflammation and stimulate regeneration process.
Supplementary Material
Footnotes
Author contributions: D.C. and M.S. designed research; S.D., D.C., J.Q., J.F., S.N., L.P., L.H., P.M., J.H.K., F.K., L.S., J.B., V.C., I.F., and M.S. performed research; S.D., D.C., J.Q., J.F., S.N., L.P., L.H., P.M., J.H.K., F.K., C.M., M.W., L.S., J.B., V.C., I.F., and M.S. analyzed data; S.D. D.C. and M.S. wrote the paper; D.C., I.F., and M.S. received funding for this project.
* This research was supported by a collaboration between the Fundamental and Applied Biology Mass Spectrometry Laboratory (MS) and grants from Ministère de L'Education Nationale, L'Enseignement Supérieur et de la Recherche, INSERM, Région Nord-Pas de Calais (to S.D.), SIRIC ONCOLille Grant INCa-DGOS-Inserm 6041aa (IF) and Université de Lille 1 (S.D.), VEGA 2/0125/15 (DC), APVV SK-FR-2015–0018 (DC), Stefanic 2016 (MS) and APVV 0472–11 (DC).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SCI
- Spinal cord injury.
REFERENCES
- 1. Beattie M. S., Hermann G. E., Rogers R. C., and Bresnahan J. C. (2002) Cell death in models of spinal cord injury. Prog. Brain Res. 137, 37–47 [DOI] [PubMed] [Google Scholar]
- 2. Tator C. H. (1995) Update on the pathophysiology and pathology of acute spinal cord injury. Brain Pathol. 5, 407–413 [DOI] [PubMed] [Google Scholar]
- 3. Schwab M. E., and Bartholdi D. (1996) Degeneration and regeneration of axons in the lesioned spinal cord. Physiol. Rev. 76, 319–370 [DOI] [PubMed] [Google Scholar]
- 4. Schwab M. E., and Bartholdi D. (1996) Degeneration and regeneration of axons in the lesioned spinal cord. Physiol. Rev. 76, 319–370 [DOI] [PubMed] [Google Scholar]
- 5. Rossignol S., Schwab M., Schwartz M., and Fehlings M. G. (2007) Spinal cord injury: time to move? J. Neurosci. 27, 11782–11792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leypold B. G., Flanders A. E., Schwartz E. D., and Burns A. S. (2007) The impact of methylprednisolone on lesion severity following spinal cord injury. Spine 32, 373–378; discussion 379–381 [DOI] [PubMed] [Google Scholar]
- 7. Rowland J. W., Hawryluk G. W., Kwon B., and Fehlings M. G. (2008) Current status of acute spinal cord injury pathophysiology and emerging therapies: promise on the horizon. Neurosurg Focus 25, E2. [DOI] [PubMed] [Google Scholar]
- 8. Hawryluk G. W., Rowland J., Kwon B. K., and Fehlings M. G. (2008) Protection and repair of the injured spinal cord: a review of completed, ongoing, and planned clinical trials for acute spinal cord injury. Neurosurg Focus 25, E14. [DOI] [PubMed] [Google Scholar]
- 9. Wells J. E., Hurlbert R. J., Fehlings M. G., and Yong V. W. (2003) Neuroprotection by minocycline facilitates significant recovery from spinal cord injury in mice. Brain 126, 1628–1637 [DOI] [PubMed] [Google Scholar]
- 10. Huang H., Fan S., Ji X., Zhang Y., Bao F., and Zhang G. (2009) Recombinant human erythropoietin protects against experimental spinal cord trauma injury by regulating expression of the proteins MKP-1 and p-ERK. J. Int. Med. Res. 37, 511–519 [DOI] [PubMed] [Google Scholar]
- 11. Bradbury E. J., Moon L. D., Popat R. J., King V. R., Bennett G. S., Patel P. N., Fawcett J. W., and McMahon S. B. (2002) Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640 [DOI] [PubMed] [Google Scholar]
- 12. Schwab M. E. (2004) Nogo and axon regeneration. Curr. Opin. Neurobiol. 14, 118–124 [DOI] [PubMed] [Google Scholar]
- 13. Schwab M. E. (2002) Repairing the injured spinal cord. Science 295, 1029–1031 [DOI] [PubMed] [Google Scholar]
- 14. Fehlings M. G., and Vawda R. (2011) Cellular treatments for spinal cord injury: the time is right for clinical trials. Neurotherapeutics 8, 704–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanicky I., Urdzikova L., Saganova K., Cizkova D., and Galik J. (2001) A simple and reproducible model of spinal cord injury induced by epidural balloon inflation in the rat. J. Neurotrauma 18, 1399–1407 [DOI] [PubMed] [Google Scholar]
- 16. Grulova I., Slovinska L., Blasko J., Devaux S., Wisztorski M., Salzet M., Fournier I., Kryukov O., Cohen S., and Cizkova D. (2015) Delivery of Alginate Scaffold Releasing Two Trophic Factors for Spinal Cord Injury Repair. Sci. Reports 5, 13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 18. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 19. UniProt C. (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montojo J., Zuberi K., Rodriguez H., Bader G. D., and Morris Q. (2014) GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res 3, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., and Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yuryev A., Kotelnikova E., and Daraselia N. (2009) Ariadne's ChemEffect and Pathway Studio knowledge base. Expert Opin. Drug Discov. 4, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 26. Bonnet A., Lagarrigue S., Liaubet L., Robert-Granie C., Sancristobal M., and Tosser-Klopp G. (2009) Pathway results from the chicken data set using GOTM, Pathway Studio and Ingenuity softwares. BMC Proc. 3, S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pyatnitskiy M., Mazo I., Shkrob M., Schwartz E., and Kotelnikova E. (2014) Clustering gene expression regulators: new approach to disease subtyping. PLoS ONE 9, e84955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daraselia N., Wang Y., Budoff A., Lituev A., Potapova O., Vansant G., Monforte J., Mazo I., and Ossovskaya V. S. (2012) Molecular signature and pathway analysis of human primary squamous and adenocarcinoma lung cancers. Am. J. Cancer Res. 2, 93–103 [PMC free article] [PubMed] [Google Scholar]
- 29. Cizkova D., Le Marrec-Croq F., Franck J., Slovinska L., Grulova I., Devaux S., Lefebvre C., Fournier I., and Salzet M. (2014) Alterations of protein composition along the rostro-caudal axis after spinal cord injury: proteomic, in vitro and in vivo analyses. Front. Cell Neurosci. 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thiele H., Heldmann S., Trede D., Strehlow J., Wirtz S., Dreher W., Berger J., Oetjen J., Kobarg J. H., Fischer B., and Maass P. (2014) 2D and 3D MALDI-imaging: conceptual strategies for visualization and data mining. Biochim. Biophys. Acta 1844, 117–137 [DOI] [PubMed] [Google Scholar]
- 31. Bonnel D., Longuespee R., Franck J., Roudbaraki M., Gosset P., Day R., Salzet M., Fournier I. (2011) Multivariate analyses for biomarkers hunting and validation through on-tissue bottom-up or in-source decay in MALDI-MSI: application to prostate cancer. Anal Bioanal Chem. 401(1), 149–165 [DOI] [PubMed] [Google Scholar]
- 32. Chambolle A. (2004) An algorithm for total variation minimization and applications. J. Mathematical Imaging Vision 20, 89–97 [Google Scholar]
- 33. Zhang J., Li D., Hu W., Chen Z., and Yuan Y. (2014) Multilabel image annotation based on double-layer PLSA model. ScientificWorldJournal 2014, 494387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zaoui K., Honore S., Isnardon D., Braguer D., and Badache A. (2008) Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J. Cell Biol. 183, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mathey E. K., Derfuss T., Storch M. K., Williams K. R., Hales K., Woolley D. R., Al-Hayani A., Davies S. N., Rasband M. N., Olsson T., Moldenhauer A., Velhin S., Hohlfeld R., Meinl E., and Linington C. (2007) Neurofascin as a novel target for autoantibody-mediated axonal injury. J. Exp. Med. 204, 2363–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han G., Wu D., Yang Y., Li Z., Zhang J., and Li C. (2015) CrkL meditates CCL20/CCR6-induced EMT in gastric cancer. Cytokine 76, 163–169 [DOI] [PubMed] [Google Scholar]
- 37. Jones L. L., Margolis R. U., and Tuszynski M. H. (2003) The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp. Neurol. 182, 399–411 [DOI] [PubMed] [Google Scholar]
- 38. Neshige S., Hara N., Takeshima S., Iwaki H., Shimoe Y., Takamatsu K., and Kuriyama M. (2014) Anti-amphiphysin antibody-positive paraneoplastic neurological syndrome with a longitudinally extensive spinal cord lesion of the dorsal column. Clin. Neurol. 54, 572–576 [DOI] [PubMed] [Google Scholar]
- 39. Levin M. C., Lee S., Gardner L. A., Shin Y., Douglas J. N., and Cooper C. (2013) Autoantibodies to non-myelin antigens as contributors to the pathogenesis of multiple sclerosis. J. Clin. Cell. Immunol. 213, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huijbers M. G., Querol L. A., Niks E. H., Plomp J. J., van der Maarel S. M., Graus F., Dalmau J., Illa I., and Verschuuren J. J. (2015) The expanding field of IgG4-mediated neurological autoimmune disorders. Eur. J. Neurol. 22, 1151–1161 [DOI] [PubMed] [Google Scholar]
- 41. Derfuss T., Linington C., Hohlfeld R., and Meinl E. (2010) Axo-glial antigens as targets in multiple sclerosis: implications for axonal and grey matter injury. J. Mol. Med. 88, 753–761 [DOI] [PubMed] [Google Scholar]
- 42. Popovich P. G., Horner P. J., Mullin B. B., and Stokes B. T. (1996) A quantitative spatial analysis of the blood-spinal cord barrier. I. Permeability changes after experimental spinal contusion injury. Exp. Neurol. 142, 258–275 [DOI] [PubMed] [Google Scholar]
- 43. Noble L. J., and Wrathall J. R. (1989) Distribution and time course of protein extravasation in the rat spinal cord after contusive injury. Brain Res. 482, 57–66 [DOI] [PubMed] [Google Scholar]
- 44. Schnell L., Fearn S., Schwab M. E., Perry V. H., and Anthony D. C. (1999) Cytokine-induced acute inflammation in the brain and spinal cord. J. Neuropathol Exp. Neurol. 58, 245–254 [DOI] [PubMed] [Google Scholar]
- 45. Schnell L., Fearn S., Klassen H., Schwab M. E., and Perry V. H. (1999) Acute inflammatory responses to mechanical lesions in the CNS: differences between brain and spinal cord. Eur. J. Neurosci. 11, 3648–3658 [DOI] [PubMed] [Google Scholar]
- 46. Zhou X., He X., and Ren Y. (2014) Function of microglia and macrophages in secondary damage after spinal cord injury. Neural Regen Res. 9, 1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Horner P. J., Popovich P. G., Mullin B. B., and Stokes B. T. (1996) A quantitative spatial analysis of the blood-spinal cord barrier. II. Permeability after intraspinal fetal transplantation. Exp. Neurol. 142, 226–243 [DOI] [PubMed] [Google Scholar]
- 48. Perry M. J., and Lawson S. N. (1993) Neurofilaments in rat and cat spinal cord; a comparative immunocytochemical study of phosphorylated and non-phosphorylated subunits. Cell Tissue Res. 272, 249–256 [DOI] [PubMed] [Google Scholar]
- 49. Perry J. R., Deodhare S. S., Bilbao J. M., Murray D., and Muller P. (1993) The significance of spinal cord compression as the initial manifestation of lymphoma. Neurosurgery 32, 157–162 [DOI] [PubMed] [Google Scholar]
- 50. Popovich P. G., Wei P., and Stokes B. T. (1997) Cellular inflammatory response after spinal cord injury in Sprague-Dawley and Lewis rats. J. Comp. Neurol. 377, 443–464 [DOI] [PubMed] [Google Scholar]
- 51. Streit W. J., Semple-Rowland S. L., Hurley S. D., Miller R. C., Popovich P. G., and Stokes B. T. (1998) Cytokine mRNA profiles in contused spinal cord and axotomized facial nucleus suggest a beneficial role for inflammation and gliosis. Exp. Neurol. 152, 74–87 [DOI] [PubMed] [Google Scholar]
- 52. Zietlow R., Dunnett S. B., and Fawcett J. W. (1999) The effect of microglia on embryonic dopaminergic neuronal survival in vitro: diffusible signals from neurons and glia change microglia from neurotoxic to neuroprotective. Eur. J. Neurosci. 11, 1657–1667 [DOI] [PubMed] [Google Scholar]
- 53. Zhang S. C., and Fedoroff S. (1996) Neuron-microglia interactions in vitro. Acta. Neuropathol. 91, 385–395 [DOI] [PubMed] [Google Scholar]
- 54. Kurihara D., Ueno M., Tanaka T., and Yamashita T. (2010) Expression of galectin-1 in immune cells and glial cells after spinal cord injury. Neurosci. Res. 66, 265–270 [DOI] [PubMed] [Google Scholar]
- 55. Villares R., Cadenas V., Lozano M., Almonacid L., Zaballos A., Martinez A. C., and Varona R. (2009) CCR6 regulates EAE pathogenesis by controlling regulatory CD4+ T-cell recruitment to target tissues. Eur. J. Immunol. 39, 1671–1681 [DOI] [PubMed] [Google Scholar]
- 56. Raposo C., Graubardt N., Cohen M., Eitan C., London A., Berkutzki T., and Schwartz M. (2014) CNS repair requires both effector and regulatory T cells with distinct temporal and spatial profiles. J. Neurosci. 34, 10141–10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Austin P. J., Kim C. F., Perera C. J., and Moalem-Taylor G. (2012) Regulatory T cells attenuate neuropathic pain following peripheral nerve injury and experimental autoimmune neuritis. Pain 153, 1916–1931 [DOI] [PubMed] [Google Scholar]
- 58. Blanco-Suarez E., and Hanley J. G. (2014) Distinct subunit-specific alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor trafficking mechanisms in cultured cortical and hippocampal neurons in response to oxygen and glucose deprivation. J. Biol. Chem. 289, 4644–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cizkova D., Kakinohana O., Kucharova K., Marsala S., Johe K., Hazel T., Hefferan M. P., and Marsala M. (2007) Functional recovery in rats with ischemic paraplegia after spinal grafting of human spinal stem cells. Neuroscience 147, 546–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ankeny D. P., and Popovich P. G. (2010) B cells and autoantibodies: complex roles in CNS injury. Trends Immunol. 31, 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ankeny D. P., Guan Z., and Popovich P. G. (2009) B cells produce pathogenic antibodies and impair recovery after spinal cord injury in mice. J. Clin. Invest. 119, 2990–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gini B., Lovato L., Cianti R., Cecotti L., Marconi S., Anghileri E., Armini A., Moretto G., Bini L., Ferracci F., and Bonetti B. (2008) Novel autoantigens recognized by CSF IgG from Hashimoto's encephalitis revealed by a proteomic approach. J. Neuroimmunol. 196, 153–158 [DOI] [PubMed] [Google Scholar]
- 63. Huang J., Sun X., Mao Y., Zhu X., Zhang P., Zhang L., Du J., and Qiu X. (2008) Expression of immunoglobulin gene with classical V-(D)-J rearrangement in mouse brain neurons. Int. J. Biochem. Cell Biol. 40, 1604–1615 [DOI] [PubMed] [Google Scholar]
- 64. Fehlings M. G., and Nguyen D. H. (2010) Immunoglobulin G: a potential treatment to attenuate neuroinflammation following spinal cord injury. J. Clin. Immunol. 30 Supplemental 1, S109–S112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kobeissy F., and Moshourab R. A. (2015) Autoantibodies in CNS Trauma and Neuropsychiatric Disorders: A New Generation of Biomarkers. In: Kobeissy F. H., ed. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects, Boca Raton (FL) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.