Abstract
Signal transduction by the IL-36 receptor (IL-36R) is linked to several human diseases. However, the structure and function of the IL-36R is not well understood. A molecular model of the IL-36R complex was generated and a cell-based reporter assay was established to assess the signal transduction of recombinant subunits of the IL-36R. Mutational analyses and functional assays have identified residues of the receptor subunit IL-1Rrp2 needed for cytokine recognition, stable protein expression, disulfide bond formation and glycosylation that are critical for signal transduction. We also observed that, overexpression of ectodomain (ECD) of Il-1Rrp2 or IL-1RAcP exhibited dominant-negative effect on IL-36R signaling. The presence of IL-36 cytokine significantly increased the interaction of IL-1Rrp2 ECD with the co-receptor IL-1RAcP. Finally, we found that single nucleotide polymorphism A471T in the Toll-interleukin 1 receptor domain (TIR) of the IL-1Rrp2 that is present in ∼2% of the human population, down-regulated IL-36R signaling by a decrease of interaction with IL-1RAcP.
Keywords: cytokine, immunology, membrane protein, psoriasis, receptor structure-function, signal transduction
Introduction
Signaling by the IL-36 receptor (IL-36R)2 has been linked to inflammation-associated diseases, such as psoriasis (1–6). However, the structure and function of IL-36R are poorly understood. We seek to contribute to understanding of the features of IL-36R needed for signal transduction.
IL-36R is a member of the IL-1 receptor family that contains six receptor proteins that form four signaling complexes: IL-1RI, IL-18R, IL-33R, and IL-36R, and two decoy receptors and two negative regulators (7). IL-36R is a heterodimer that consists of a receptor subunit named IL-1Rrp2 (also known as IL-1RL2) and a co-receptor subunit Interleukin-1 receptor accessory protein, IL-1RAcP (7, 8). The receptor can recognize three different agonists, IL-36α, IL-36β, and IL-36γ (also known as IL-1F6, IL-1F8, and IL-1F9), to induce the expression of inflammatory cytokines (8–10). There are also two receptor antagonists, IL-36Ra and IL-38, which bind to IL-36 receptor and decrease the expression of inflammatory cytokines (9, 11).
IL-1Rrp2 contains a signaling peptide, an extracellular domain (ECD), a transmembrane helix and an intracellular Toll/IL-1 receptor (TIR) domain. The protein is localized to the plasma membrane as a single-pass transmembrane protein with the ECD on the cell surface and the TIR domain in the cytoplasm.
The presumed mechanism of activation is that agonist cytokines will interact with the ECD of IL-1Rrp2 to trigger the recruitment of the co-receptor IL-1RAcP (7, 8). Stable formation of the IL-1Rrp2 and IL-1RAcP complex allows the interaction of the TIR domains of the two proteins. Additional association with the TIR domain of the adaptor protein MyD88 as well as the TIR domains of adaptors IRAK1 and IRAK2 forms a signaling platform, leading to the activation of the stress-associated transcription factor NF-κΒ that will traffic to the nucleus and alter transcription of numerous genes, including those that encode proinflammatory cytokines.
The crystal structures of ECDs of IL-1R, IL-18R, and IL-33R ECD have been elucidated (12–15). The ECDs of IL-1R, IL-18R, and IL-33R and their co-receptors IL-1RAcP all contain three immunoglobulin (Ig)-like domains. The crystal structure of IL-36γ has been resolved (16), however, the structure of IL-1Rrp2 or IL-1Rrp2/IL-1RAcP/ligand complex is not available.
Post-translational modifications of innate immune receptors play critical roles in signal transduction (17). The structures of IL-1R and IL-18R reveal that the disulfide bond formations in the ECDs are essential to maintain the Ig-like structure. The IL-36R also actively traffics and its endocytosis is linked to signal transduction (18), indicating that glycosylation could be required for IL-36R signaling. In addition, glycosylation of IL-18R has been proposed to contact the co-receptor IL-18RAcP and possibly the ligand (14).
In this study, we modeled the ECDs of the human IL-1Rrp2 in complex with IL-1RAcP, their association with the three IL-36 cytokines and examined the function of post-translational modification on IL-36R-mediated signaling using a cell-based reporter assay. We determined regions of the IL-1Rrp2 that function in cytokine recognition and IL-36R subunit interactions and found that N-linked glycosylation in IL-1Rrp2 could affect the NF-κΒ reporter activation through regulating IL-1Rrp2 cell surface expression. Finally, we examined single nucleotide polymorphisms of IL-1Rrp2 and identified that an A471T substitution in the TIR domain down-regulated IL-36R signaling by decreasing the interaction with IL-1RAcP.
Results and Discussion
Molecular Modeling of IL-36R ECD in Complex with Ligands
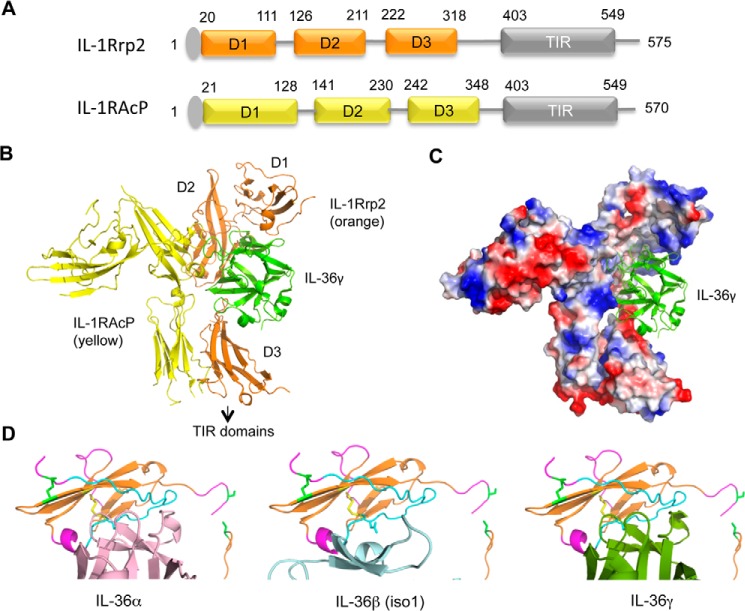
The sequences of IL-1Rrp2 and IL-1RAcP are predicted to have three immunoglobulin-like domains (D1-D3) joined to a cytoplasmic C-terminal TIR domain (Fig. 1, A and B). To model the structures of the IL-36R ECD in complex with ligand, we used the deposited crystal structures of the IL-1R/IL-1RAcP as templates (PDB code 4DEP) to generate homology models for the polypeptide backbones of IL-1Rrp2 and IL-1RAcP, followed by the addition of specific amino acids. The models of IL-1Rrp2 and IL-1RAcP both resembles an upside down “L” where D1 is present at the short part of the L and D2 and D3 together make up the longer portion of the L. Notably, the edge of IL-1RAcP D2 abuts the face of the IL-1Rrp2 D2 subdomain. The interface of this edge-to-face arrangement can potentially act as a pivot to allow IL-1RAcP to rotate with respect to the IL-1Rrp2.
FIGURE 1.
Molecular models of the IL-36 receptor and its interaction with IL-36 cytokines. A, schema of the domain organization for IL-1Rrp2 and co-receptor IL-1RAcP. The N-terminal signal peptides are colored gray. The three Ig-like domains and the Toll/interleukin-1 receptor domains are labeled as D1, D2, D3, and TIR, respectively. The numbers above the schematics denote the approximate residues demarcating the domains. B, ribbon structures of the model of IL-1Rrp2 ectodomain in complex with IL-1RAcP ectodomain and IL-36γ. The TIR domains are not shown. C, surface charge of the IL-1Rrp2 and IL-1RAcP complex bound to IL-36γ (green ribbon structure). D, molecular models of D1 of IL-1Rrp2 that contacts IL-36α, IL-36β, and IL-36γ.
The modeled IL-1Rrp2 ligand-binding pocket is comprised of segments from each of the three Ig-like domains. Specifically, part of D1 forms the top of the pocket, an edge of D2 contributes to the backside, and D3 forms the bottom of the pocket (Fig. 1B). A distinct distribution of acidic and basic patches lining the ligand pocket suggests that the binding affinity for IL-36 cytokines is likely driven by electrostatic attractions (Fig. 1C). There is noticeable space between the bottom of the ligand IL-36γ and D3 (Fig. 1B) that would be unfavorable for any electrostatic interactions at this interface. This space is also evident in models for the complex with other IL-36 cytokines (data not shown). These observations raise the possibility that a large rotation within the IL-36R ECD complex could close the gap by repositioning D3 for binding.
To further probe how different cytokines might interact with the IL-36R ECD complex, we modeled the interface between D1 of IL-1Rrp2 and IL-36 ligands. This was accomplished by performing rigid body rotation of ligands to find the best match between oppositely charged electrostatic surfaces at the ligand/pocket interface. We found that IL-36α and IL-36γ made similar contacts with elements in D1 (Fig. 1D). In contrast, the positioning of IL-36β (isoform 1) was distinctly different from the other two cytokines. This could mean that D1 has different associations with the IL-36 cytokines. Features of the model were thus tested below.
A Cell-based Reporter Assay to Examine IL-36R Signaling
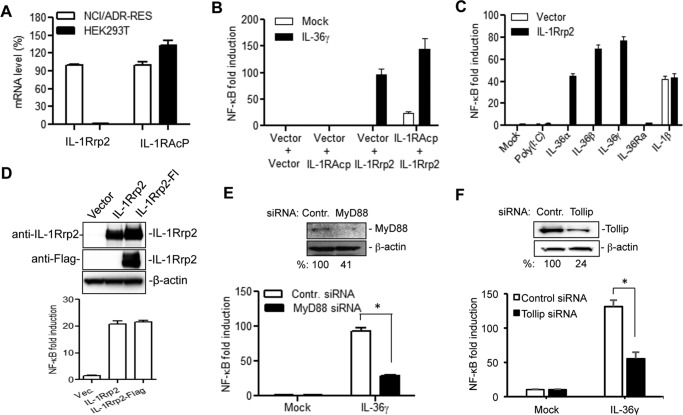
To examine the function of IL-36R and changes in IL-1Rrp2, we used HEK 293T cells that can be transfected with high efficiency. 293T cells lack expression of several innate immune receptors (19). The expression of endogenous IL-1Rrp2 and IL-1RAcP was examined using real-time RT-PCR. 293T cells expressed comparable levels of IL-1RAcP as did NCI/ADR-RES cells, but had undetectable levels of endogenous IL-1Rrp2 (Fig. 2A).
FIGURE 2.
A cell-based reporter assay to examine the function of IL-1Rrp2 in IL-36R signaling. A, HEK human embryonic kidney 293T cells express an undetectable level of IL-1Rrp2. The mRNA levels of IL-1Rrp2 and IL-1RAcP in 293T cells were quantified using real-time RT-PCR and adjusted to the levels from NCI/ADR-RES cells. The data were normalized to the GAPDH mRNA control from each sample. B, transfection of IL-1Rrp2 plasmid triggered NF-κΒ promoter activation upon IL-36 agonist addition. 293T cells were transfected to express IL-1Rrp2 and reporter constructs and then mock-treated or stimulated with 2 ng/ml of IL-36γ overnight. The luciferase activities are plotted as fold induction relative to cells transfected with the vector. The white and black bars denote the mock-treated and IL-36γ-stimulated activity. All results shown represent the means and standard deviations from at least three independent samples. C, effects of different ligands on the NF-κΒ activity in cells transfected to express IL-1Rrp2. D, effect of a FLAG-tagged IL-1Rrp2 on protein accumulation and signal transduction in 293T cells. Equal amounts of IL-1Rrp2 and FLAG-tagged IL-1Rrp2 plasmids were transfected into 293T cells, and the protein level was determined by Western blotting analysis probed with either goat anti-IL-1Rrp2 or rat anti-FLAG antibody. The NF-κΒ activation was determined as described above. E, siRNA knockdown of endogenous MyD88 in 293T cells significantly decreased IL-36γ mediated NF-κB activation. 293T cells were first transfected with 40 nm of either control siRNAs or MyD88 siRNAs for 48 h and then transfected to express IL-1Rrp2 for 24 h. The cells were mock-treated or treated with 2 ng/ml of IL-36γ. A significant reduction of MyD88 protein by the siRNA knockdown was confirmed by Western blotting analysis. The asterisk denotes that the two samples differed by a p value of < 0.05 in the Student's t test. F, effect of knockdown of endogenous Tollip on IL-36R signaling.
293T cells were transfected with a plasmid that can express IL-1Rrp2 along with plasmids that contain a firefly luciferase cDNA driven by a promoter containing NF-κΒ elements and a Renilla luciferase cDNA driven by constitutive thymidine kinase promoter, respectively. Expression of recombinant IL-1Rrp2 resulted in significant activation of NF-κΒ promoter when the cells were provided with exogenous IL-36γ (Fig. 2B). Co-transfection of plasmids encoding IL-1Rrp2 and IL-1RAcP further increased ligand-dependent NF-κΒ activation. The NF-κΒ signaling was activated by IL-36α, IL-36β, and IL-36γ ligand to similar extents. However, the IL-36Ra or the TLR3 ligand poly(I:C) had only minimal effects on signaling (Fig. 2C). NF-κB signaling was activated when the 293T cells were treated with IL-1β, indicating that the IL-1RI was expressed in 293T cells. However, IL-1β-induced IL-1RI signaling was not affected by the expression of IL-1Rrp2 (Fig. 2C).
To facilitate the detection of IL-1Rrp2 protein, a FLAG tag was engineered into the C-terminal of the IL-1Rrp2 construct. This construct expressed a comparable level of IL-1Rrp2 and exhibited similar activity to that of the un-tagged construct (Fig. 2D). Similar results were obtained with IL-1Rrp2 tagged with a Myc-epitope.
Adaptor proteins MyD88 and Tollip are required for IL-36R signaling in NCI/ADR-RES cells (18). SiRNA knockdown of MyD88 and Tollip in 293T cells significantly decreased IL-36γ/IL-1Rrp2 mediated NF-κB activation (Fig. 2, E and F), demonstrating that the reconstitution of IL-36R signaling in 293T cells could mimic requirements for IL-36R signal transduction.
Cysteine Residues in ECD Are Essential for IL-1Rrp2 Accumulation and Signaling
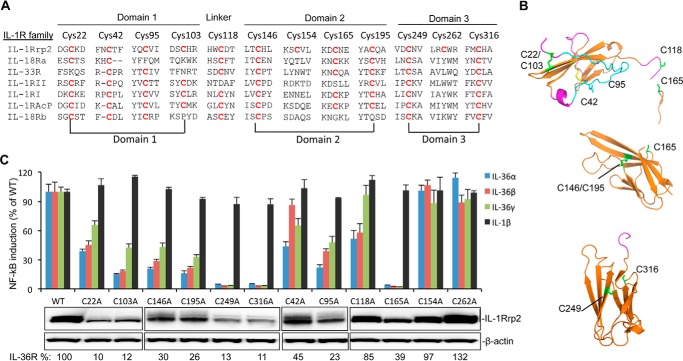
We seek to use the cell-based reporter assay to examine the features predicted in the model of the IL-1Rrp2. Disulfide bonds in the ECD of IL-1R family are essential for the formation of the Ig-like structure (13, 14). The ECD of IL-1Rrp2 contains 12 cysteine residues. Sequence alignment of members of the IL-1R family revealed that several cysteine residues are highly conserved, but that Cys-154 and Cys-262 are unique to IL-1Rrp2 (Fig. 3A). The molecular model places Cys-22 sufficiently close to Cys-103 to form a disulfide bond in D1, while Cys-146-Cys-195 form a disulfide bond in D2 and Cys-249-Cys-316 form a disulfide bond in D3 (Fig. 3B).
FIGURE 3.
Effects of cysteine to alanine substitutions in the IL-1Rrp2 ectodomain on protein accumulation and signaling. A, sequence alignment of cysteine residues in the IL-1R family. The IL-1Rrp2 ECD sequence is shown on the top, and residues in the IL-1R family that have conserved cysteine are marked in red. Cys pairs that are predicted to form disulfide bonds are connected by lines below the sequences. B, molecular model of potential disulfide bonds in three Ig-like domains of IL-1Rrp2. The cysteine residues, including the side chains, are in green. C, effects of cysteine mutations on IL-36R signaling and protein expression. Equal amounts of either WT IL-1Rrp2 or mutants were transfected into 293T cells along with the reporter plasmids as described. The cells were mock-treated or stimulated with 2 ng/ml of either IL-36α, IL-36β, IL-36γ, or IL-1β. The data were plotted relative to WT IL-1Rrp2 and expressed as a mean of three independent samples and the range for standard error. The levels of accumulated WT IL-1Rrp2 and cysteine mutants were determined by Western blotting analysis. β-Actin from each sample was detected as a loading control. The relative amount of protein was quantified by ChemiDoc software (Bio-Rad).
Should these cysteine residues form disulfide bonds, it would indicate that molecular model captured gross features of the ECD structure. We substituted these cysteine residues to alanine and examined the effects on protein expression. For all three pairs of cysteine, mutations resulted in similar decreases in protein accumulation, suggesting that the lack of proper disulfide bond formation affects the stability of the IL-1Rrp2 (Fig. 3C). The reporter assay showed that each pair of mutants also had fairly similar decreases in NF-κΒ activation (Fig. 3C).
We also assessed the functional roles of cysteine residues in the IL-1Rrp2 ECD that were not predicted to be involved in disulfide bond formation. Mutants C42A, C95A, C118A, and C165A resulted in varying decreases in both the accumulation of proteins and signaling (Fig. 3C). Notably, C42A was defective for signaling in response to IL-36α and IL-36γ, but did not significantly affect signaling in response to IL-36β. The differential effects will be examined below. Mutations in the cysteine residues Cys-154 and Cys-262 that are not conserved had only minimal effects on protein accumulation and signaling.
Glycosylation of IL-1Rrp2 Is Critical for Signaling
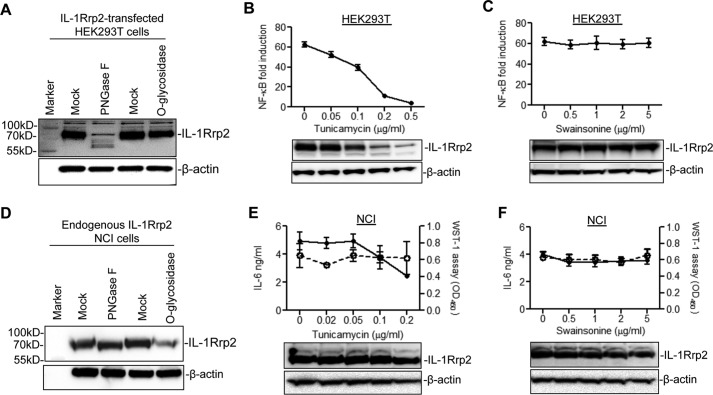
The molecular weight of IL-1Rrp2 is about 65 kDa, but it migrates to the position of ∼85 kDa in denaturing protein gels, suggesting that it is post-translationally modified (Fig. 4A). To examine whether IL-1Rrp2 is glycosylated, the lysate from IL-1Rrp2-transfected 293T cells was treated with PNGase F to remove N-linked glycans or with O-glycosidase to removes O-linked glycans. The results of the treatment were analyzed by Western blots probed to detect IL-1Rrp2. The PNGase F treatment significantly decreased the electrophoretic migration of IL-1Rrp2 while the treatment with O-glycosidase had no obvious effect on the protein mobility (Fig. 4A). The N-linked glycosylation inhibitor tunicamycin, which inhibits glycan attachment, the first step of N-linked glycosylation, suppressed NF-κΒ signaling by IL-36γ in a dose-dependent manner (Fig. 4B). However, the accumulation of IL-1Rrp2 was also decreased, presumably due to glycosylation being required for the stability of IL-1Rrp2. Swainsonine, which inhibits the processing of the oligosaccharide chains of N-linked glycosylation, did not affect either NF-κΒ activation or IL-1Rrp2 accumulation (Fig. 4C).
FIGURE 4.
N-linked glycosylation in IL-1Rrp2 is required for IL-36R signaling. A, IL-1Rrp2 contains N-linked glycans. 293T cells were transfected with IL-1Rrp2 for 24 h, and the cell lysate was mock-treated or treated with PNGase F or O-glycosidase. The lysate was separated using SDS-PAGE, and IL-1Rrp2 was detected by Western blotting analysis. B, effects of tunicamycin treatment on NF-κΒ activation. 293T cells were transfected with IL-1Rrp2 for 6 h, and treated with increasing concentrations of tunicamycin for an additional 18 h, followed by stimulation with IL-36γ. The data were plotted as NF-κB fold induction relative to mock-treated sample. The IL-1Rrp2 level in the cell lysate was determined by Western blotting analysis. C, effects of swainsonine on NF-κB activation. D, effects of PNGase F or O-glycosidase treatment on the migration of endogenous IL-1Rrp2 in NCI cells. The level of IL-1Rrp2 in the cell lysate was detected by Western blotting analysis. E, effects of tunicamycin on IL-36R signaling in NCI cells. NCI cells treated with increasing concentrations of tunicamycin for 24 h were stimulated with IL-36γ. The IL-6 level in the medium (solid line) was determined by ELISA. Cell proliferation (dashed line) was assessed by WST-1 assay according to the manufacturer's protocol. IL-1Rrp2 accumulation was detected by Western blotting analysis. F, effects of swainsonine on IL-36R signaling in NCI cells. The solid line and dashed line denoted IL-6 level and cell proliferation, respectively. The Western blotting analysis showed the effects of swainsonine on IL-1Rrp2 accumulation.
PNGase F treatment was used to examine the glycosylation of the endogenous IL-1Rrp2 in NCI cells, a cell-line that expresses functional IL-36R. The migration of endogenous IL-1Rrp2 from NCI cells was also changed upon PNGase F treatment, but the change in electrophoretic mobility was more modest when compared with that of recombinant IL-1Rrp2 in 293T cells (Fig. 4D). Tunicamycin did significantly suppress IL-6 secretion into the medium of NCI cells treated with IL-36γ (Fig. 4E, upper panel), and also reduced the electrophoretic migration of IL-1Rrp2 (Fig. 4E, lower panel). A similar inhibitory effect was observed for IL-8 secretion (data not shown). Addition of tunicamycin up to 0.2 μg/ml did not obviously affect the proliferation of NCI cells (Fig. 4E). Consistent with the observation in IL-1Rrp2 transfected 293T cells (Fig. 4C), swainsonine did not decrease the IL-6 and IL-8 secretion in NCI cells (Fig. 4F and data not shown). These results demonstrate that endogenous IL-1Rrp2 contains N-linked glycans and that glycosylation is crucial for the IL-36R signaling in transfected 293T cells and endogenously expressed NCI cells. Based on the effects of the PNGase F on the electrophoretic mobilities of the IL-1Rrp2 in the two cell lines, the degree of glycosylation likely differ between cell types.
Glycosylation Affects IL-1Rrp2 Cell Surface Expression
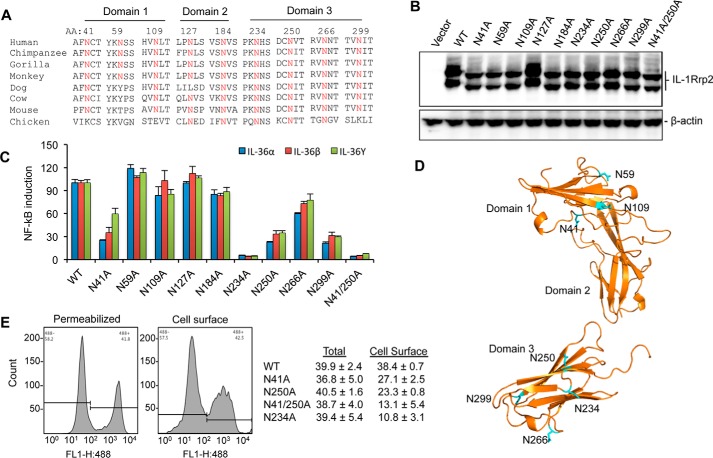
The extracellular domain of IL-1Rrp2 was predicted to contain nine N-linked glycosylation motifs (Fig. 5A). Most of asparagine residues are highly conserved in IL-1Rrp2 among different species (Fig. 5A). To identify the importance of the potential glycosylation sites to IL-1Rrp2 function, we made constructs wherein each of the predicted asparagine residues was substituted with alanine. None of the glycosylation mutants significantly decreased the accumulation of IL-1Rrp2 in the cell lysates (Fig. 5B). However, all the mutants migrated as lower molecular weight proteins when compared with that of WT except for mutant N127A. A double mutant N41A/N250A further reduced its electrophoretic migration. These results suggest that eight of the nine asparagine residues in IL-1Rrp2 ECD are indeed glycosylated in 293T cells.
FIGURE 5.
N-linked glycosylation in IL-1Rrp2 is critical for signaling and trafficking to the cell surface. A, sequence alignment of predicted N-linked glycosylation sites in IL-1Rrp2 and orthologs from other species. Residues in IL-1Rrp2 predicted to be glycosylated are shown above the IL-1Rrp2 sequence. B, effects of mutations in the predicted N-linked glycosylation residues on the electrophoretic migration of the resultant IL-1Rrp2 proteins. C, effects of substitution of predicted glycosylation sites in the IL-1Rrp2 on signal transduction by all three IL-36 cytokines. 293T cells were transfected with equal amounts of the plasmids expressing WT or mutant IL-1Rrp2. The cells were mock-treated or stimulated with 2 ng/ml of the IL-36 agonists. The data were plotted relative to WT IL-1Rrp2 from two independent experiments, each performed with three independent samples. D, model of the localization of glycosylation sites in the IL-1Rrp2 ECD. The asparagine residues are marked in blue. E, cellular location of WT or mutant IL-1Rrp2 was determined by flow cytometry. Total amount of IL-1Rrp2 was determined in detergent-permeabilized cells while IL-1Rrp2 present on the cell surface was detected in non-permeablilized cells. Flow cytometry was performed as described in “Experimental Procedures.” The data were presented as averages of four independent samples.
Next, we examined the effect of glycosylation on IL-36R signaling in 293T cells. Mutants N41A, N250A, N266A, and N299A decreased the signaling by 30–60% compared with that of WT, while N234A and N41/250A mutants decreased the NF-κΒ promoter activation to near background (Fig. 5C). We noted that mutations in D3 resulted in the most significant decrease in signaling. Mutants N109A and N184A only modestly decreased signaling while N127A, which does not appear to be glycosylated, and N59A did not affect IL-36R signaling in the cell-based assay (Fig. 6C).
FIGURE 6.
Residues in the IL-1Rrp2 ectodomain can differentially recognize three IL-36 cytokines. A, locations of the residues in IL-1Rrp2 that had differential response in signaling by the three IL-36 cytokines. The locations of the side chains are shown in blue in the ribbon diagram. The inset shows the overall electrostatic charges are shown as the envelope, with basic areas in blue and acidic ones in red. B, effects of increasing concentrations of the N41A mutant on IL-36R signaling. 293T cells were transfected with increasing concentrations of either WT or N41A along with constant amounts of reporter plasmids for 24 h and stimulated with 2 ng/ml of IL-36α, IL-36β, or IL-36γ. Signal transduction level was assessed using the NF-κΒ promoter that drives firefly luciferase expression. The transfection of 0.5 ng of plasmid of WT IL-1Rrp2 was plotted as 100%. The protein level was determined by Western blotting analysis. C, effects of increasing concentrations of the C42A mutant on IL-36R signaling. D, effects of the C118A mutant on IL-36R signaling. The level of C118A protein produced in the cells was shown in the Western blotting analysis.
Glycosylation could regulate the function of membrane-associated receptors by affecting protein folding, cell surface expression and/or ligand binding (20). Molecular modeling of IL-1Rrp2 ECD docked with the IL-36 cytokines revealed that most of the glycosylation sites were not involved in direct contact with three ligands (Fig. 5D). The defect in signaling for the mutant proteins prompted us to examine whether glycosylation could affect IL-1Rrp2 trafficking to cell surface. Cell surface localization of IL-1Rrp2 in 293T cells was quantified using flow cytometry. Permeabilized cells were used to quantify the overall level of IL-1Rrp2. All of the mutants expressed a similar level of IL-1Rrp2 in permeabilized cells when compared with cells expressing wild-type IL-1Rrp2. However, N41A and N250A decreased IL-1Rrp2 expression on the cell surface by 30–40% while N41/250A decreased the cell-surface receptor accumulation by 70% compared with that of WT IL-1Rrp2 (Fig. 5E). The N234A mutant, which significantly affected IL-36R signaling, decreased cell surface expression by ∼70% (Fig. 5E). These results suggest that glycosylation at residues Asn-41, Asn-234, and Asn-250 was critical for proper IL-1Rrp2 trafficking to the cell surface.
IL-1Rrp2 Residues That Contribute to Specific Recognition of Three IL-36 Ligands
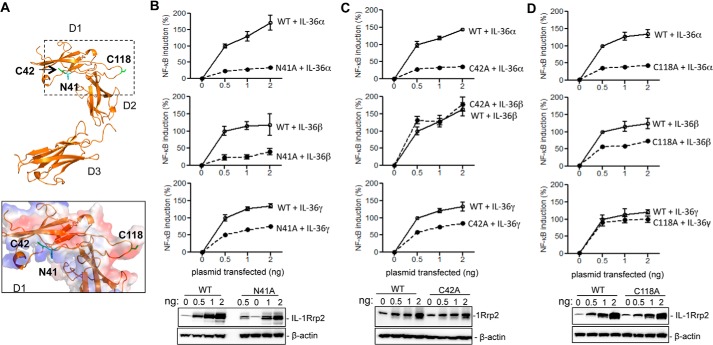
Substitutions in two adjacent residues, N41A and C42A in D1, exhibited distinct signaling in the presence of three IL-36 cytokines (Figs. 3C and 5C). Molecular modeling showed that D1 of IL-1Rrp2 could contact the three IL-36 cytokines in distinct ways (Fig. 6A). To extend these observations, we expressed increasing amounts of either the WT or mutants N41A, C42A in 293T cells and examined the responses to a constant level of each cytokine. N41A had lower NF-κΒ signaling in response to IL-36α and IL-36β when compared with that of IL-36γ (Fig. 6B). C42A had decreased signaling in response to IL-36α and IL-36γ, but no obvious effect on IL-36β signaling was observed (Fig. 6C).
The IL-1Rrp2 ECD forms a concave surface and residues Asn-41 and Cys-42 lie within the “roof” of the concave surface. This surface was modeled to find the residues that contribute to specific recognition of the cytokines (Fig. 6A). Notably, residue Asn-41 of IL-1Rrp2 is also glycosylated in 293T cells and contributes to cell surface localization. The reduced cell surface localization of the N41A mutant more specifically affected the recognition of IL-36α and IL-36β than that of IL-36γ. It is possible that IL-36γ may be recognized with higher affinity by N41A mutation, hence the level of signaling was not perturbed even with reduced cell surface abundance of IL-1Rrp2.
Cys-118 is located in the flexible linker between Domains 1 and 2 (Fig. 6A). The substitution of Cys-118 to an alanine appears to have differential signaling with the three IL-36 cytokines (Fig. 3C). Increased expression of the C118A mutant decreased signaling by IL-36α by 75% and IL-36β by 50%, but only modestly affected IL-36γ-mediated signaling (Fig. 6D). These results suggest that the linker sequence between D1 and D2 of IL-1Rrp2 contributes to differential recognition of the three IL-36 ligands (Fig. 6A).
The ECDs of IL-1Rrp2 and IL-1RAcP Exhibit Dominant-negative Effect on Signaling
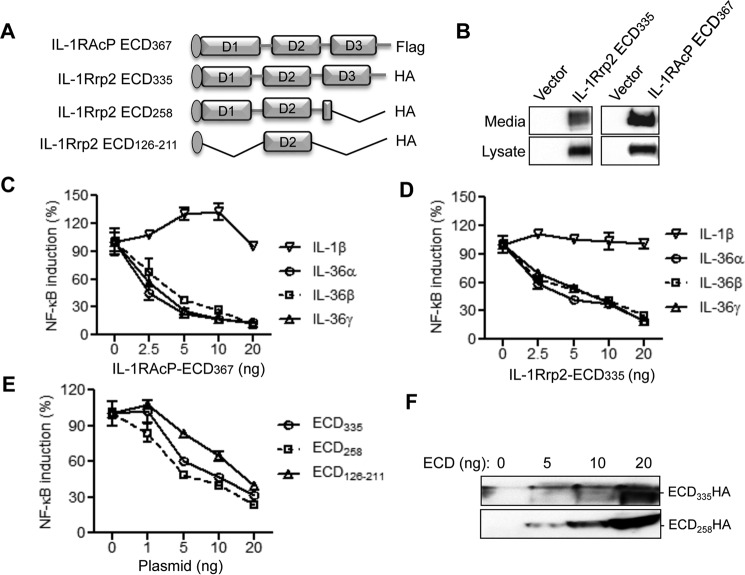
To examine whether expression of ECD from IL-1RAcP or IL-1Rrp2 affects IL-36R signaling, we constructed IL-1RAcP ECD367 and IL-1Rrp2 ECD335 that contain the signaling peptide and all three immunoglobulin-like domains of IL-1RAcP or IL-1Rrp2 (Fig. 7A). IL-1RAcP ECD367 and IL-1Rrp2 ECD335 expressed in 293T cells were found in both the cell lysate and in the culture medium, demonstrating that they could be transported out of the cells (Fig. 7B). Because of the lack of the TIR domain, both IL-1RAcP ECD367 and IL-1Rrp2 ECD335 are incapable of signaling (data not shown). However, expression of IL-1RAcP ECD367 or IL-1Rrp2 ECD335 in the presence of the WT IL-1Rrp2 decreased signal transduction by all three IL-36 cytokines in a dose-dependent manner (Fig. 7, C and D). The effect is specific to IL-1Rrp2 since IL-1RI signaling was only modestly affected by the overexpression of IL-1RAcP or IL-1Rrp2 ECD (Fig. 7, C and D). The dominant negative effect was also observed with overexpression of IL-1Rrp2 ECD258, which contains intact D1, D2 and partial D3 of IL-1Rrp2 (Fig. 7E). To determine whether D2 was sufficient to suppress IL-36R signaling, we made IL-1Rrp2 ECD126–211 construct, which contains the signaling peptide and D2 of IL-1Rrp2 ECD. Overexpress of ECD126–211 inhibited signaling by the WT IL-1Rrp2 (Fig. 7E). Increased expression of secreted IL-1Rrp2 ECD was detected in the cell culture medium by Western blotting analysis (Fig. 7F), suggesting that secreted ECDs from IL-1RAcP and/or IL-1Rrp2 negatively regulated IL-36 cytokines mediated signaling.
FIGURE 7.
Overexpression of ECDs of IL-1Rrp2 and IL-1RAcP exhibit dominant-negative effect on IL-36R signaling. A, schematic of ECDs of IL-1RAcP and IL-1Rrp2. The IL-1RAcP ECD367 contains the signal peptide at the N terminus and a FLAG tag at the C terminus, whereas the full-length and truncated IL-1Rrp2 ECDs contain the HA tags at the C terminus of the constructs. B, transient expression of IL-1RAcP ECD367 and IL-1Rrp2 ECD335. Western blotting analysis shows the abundances of ECD335 in the cell lysate and in the medium of cell culture. C, overexpression of IL-1RAcP ECD367 can inhibit IL-36R signaling induced by IL-36 cytokines. HEK293T cells were co-transfected with constant amount of IL-1Rrp2 plasmid along with increasing concentrations of IL-1RAcP ECD367. The cells were then treated with 2 ng/ml of either IL-1β, IL-36α, IL-36β, or IL-36γ and the data were plotted as percent of IL-1RAcP ECD367 mock-transfected control. D, overexpression of ECD335 inhibits IL-36R signaling induced by IL-36 cytokines. E, effects of expression of IL-1Rrp2 ECD258 and ECD126–211 on IL-36R signaling. F, levels of IL-1Rrp2 ECD335 and ECD258 in the culture medium. The secreted ECDs in medium were detected as input control.
Heterodimeric Interaction between the ECDs of IL-1RAcP and IL-1Rrp2
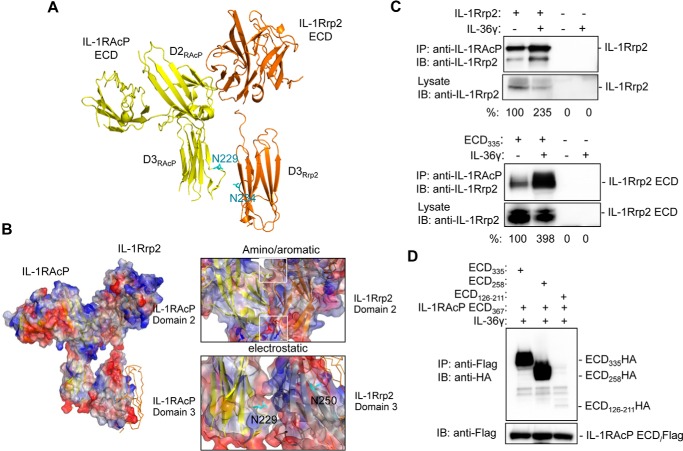
IL-1Rrp2 interacts with IL-1RAcP in the presence of ligand to trigger signal transduction (8). Molecular modeling suggests that the D2 of the IL-1Rrp2 ECD can interact with the D2 of IL-1RAcP ECD (Fig. 8A). The chemical basis for the heterotypic D2-D2 interaction is the stacking of aromatic residues in the two proteins as well as electrostatic interactions (Fig. 8B). Modeling of the IL-1Rrp2 interaction with IL-1RAcP suggests D3 of IL-1RAcP interacts with D3 of IL-1Rrp2 and cause the latter to undergo a conformational change to decrease the opening of the concave surface of IL-1Rrp2. This should have the effect on increasing the affinity of IL-36R with the cytokine.
FIGURE 8.
Interaction between IL-1Rrp2 and IL-1RAcP is increased in the presence of the IL-36 cytokine. A, molecular model of the interaction between the IL-1RAcP ECD and the IL-1Rrp2 ECD. B, chemical basis for the interaction between the ECDs of IL-1RAcP and IL-1Rrp2. The two molecules are shown with the positively charged residues in blue and the negatively charged residues in red. The blue arrow shows the position of the IL-1Rrp2 domain 3 in the absence of interaction with IL-1RAcP. C, IL-1RAcP forms a complex with the IL-1Rrp2. Upper panel: results of the co-IP assay with full-length IL-1Rrp2 and IL-1RAcP. Lower panel: results of a co-IP assay with IL-1Rrp2 ECD335 and IL-1RAcP. Where present, IL-36γ was added to cells at 20 ng/ml. D, interaction of IL-1RAcP ECD367 with IL-1Rrp2 ECD335. The cells were co-transfected with IL-1RAcP ECD367 along with either the full-length IL-1Rrp2 ECD335, truncated ECD258 or ECD126–211. After treated with IL-36γ, the complex in the culture medium was immunoprecipitated with anti-FLAG antibody and the interaction was detected by anti-HA antibody.
Whether IL-1Rrp2 can interact with IL-1RAcP was examined by co-immunoprecipitation assay using lysates of 293T cells that overexpressed the two proteins. In the absence of IL-36γ, the two proteins did co-immunoprecipitate. However, in the presence of IL-36γ, co-immunoprecipitation was increased by 2.4-fold (Fig. 8C, upper panel). Next, we tested whether IL-1RAcP could co-immunoprecipitate with IL-1Rrp2 ECD335. IL-1RAcP was able to interact with ECD335 in the absence of exogenous IL-36γ, while in the presence of IL-36γ, the amount of co-precipitated complex increased by 4-fold (Fig. 8C, lower panel). These results suggest that IL-1RAcP can interact with IL-1Rrp2 in the absence of ligand, but the presence of ligand will improve the strength of the interaction between IL-1Rrp2 and IL-1RAcP, leading to activation of signal transduction.
To determine the domain(s) in the IL-1Rrp2 ECD required to interact with IL-1RAcP, we tested the interaction of full-length and truncated IL-1Rrp2 ECDs with IL-1RAcP ECD367. 293T cells were co-transfected with IL-1RAcP ECD-FLAG along with either IL-1Rrp2 ECD335, truncated ECD258 or ECD126–211. The cells were treated with IL-36γ and the IL-1rAcP ECD367 in the culture medium was immunoprecipitated with anti-FLAG antibody. IL-1Rrp2 ECD335 and ECD258 could bind to IL-1RAcP ECD367 with the comparable level. However, D2 of the IL-1Rrp2 D2 did not efficiently interact with IL-1RAcP ECD367 (Fig. 8D), suggesting that although the C-terminal of D3 of IL-1Rrp2 ECD might not be critical for the heterodimer interaction, other portions of IL-1Rrp2 ECD contributes to the interaction with IL-1RAcP ECD as well as IL-36 ligands.
Single Nucleotide Polymorphism A471T Down-regulates IL-1Rrp2 Signal Transduction
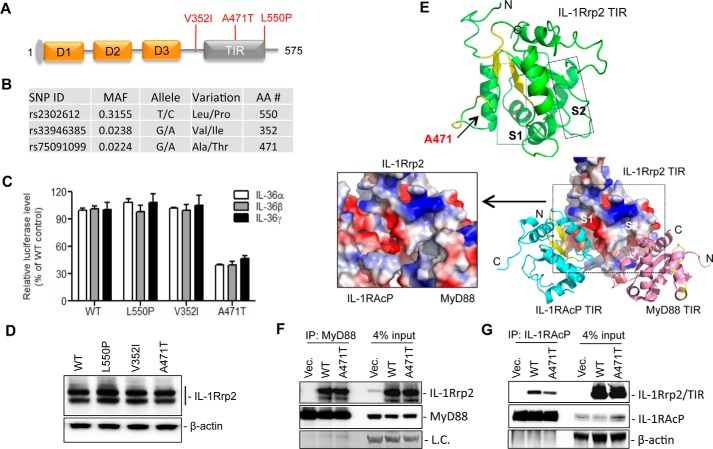
Single nucleotide polymorphisms (SNPs) in the genes encoding ST2 and IL-33 in the IL-1 and IL-1R family have been reported to be associated with asthma and inflammatory bowel disease (21, 22). The NCBI SNP database listed three major non-synonymous SNPs for IL-1Rrp2 in the human population: V352I, A471T, and L550P that are present, respectively, at frequencies of 31.5, 2.3, and 2.2% (Fig. 9, A and B). Val-352 is located in the transmembrane domain of IL-1Rrp2, and Leu-550 is within the flexible sequence near the C-terminal region. Ala-471, however, lies in the IL-1Rrp2 TIR domain that could potentially interact with other TIR domains.
FIGURE 9.
Effects of IL-1Rrp2 SNPs on signal transduction. A, locations of three non-synonymous SNPs in the IL-1Rrp2. B, properties of the IL-1Rrp2 SNPs. SNP data were derived from NCBI SNP database. C, comparisons of the effects of SNPs on IL-36R signaling in 293T cells. Cells were transfected with plasmids encoding WT IL-1Rrp2 or the SNPs. The cells were mock-treated or stimulated with three IL-36 ligands, respectively. The data were plotted relative to WT IL-1Rrp2. D, analysis of SNP expression in 293T cells. E, molecular modeling the TIR domain of IL-1Rrp2 and its interaction with TIR domain of MyD88 and IL-1RAcP. S1 and S2 denote the two grooves in the IL-1Rrp2 TIR domain that are modeled to interact, respectively, with the IL-1RAcP TIR and the MyD88 TIR. Ala-471 is located in the S1 groove. F, immunoprecipitation to examine the interaction between the FLAG-tagged WT or A471T IL-1Rrp2 TIR domain with MyD88. 293T cells were co-transfected with MyD88 along with either the WT IL-1Rrp2 or A471T for 24 h and then mock-treated or stimulated by IL-36γ for 5 h. The antibody used for IP was targeted against MyD88. MyD88 levels in the lysate and immunoprecipitated complex were detected as the control. G, immunoprecipitation assay to examine the interaction between the FLAG-tagged WT and A471T IL-1Rrp2 TIR domain with IL-1RAcP. Immunoprecipitation used the anti-IL-1RAcP antibody. The TIR domain of IL-1Rrp2 was detected with an anti-FLAG antibody. The data represent three independent experiments with consistent results.
To examine whether the three SNPs could affect the signaling transduction, plasmids encoding either WT IL-1Rrp2 or each of the SNPs were transfected into 293T cells, and NF-κΒ activation was determined using the reporter assay. SNPs V352I and L550P had only modest effects on signaling in the presence of all three IL-36 cytokines (Fig. 9C). In contrast, A471T reduced NF-κΒ promoter activation by ∼60% compared with that of WT. The reduction was comparable for all three cytokines (Fig. 9C). All three SNPs had similar levels of protein accumulation as detected by Western blotting analysis (Fig. 9D).
The TIR domain of IL-1RI interacts with TIR domains of IL-1RAcP and MyD88 to form a signaling complex (23, 24). Molecular modeling of the IL-1Rrp2 TIR domain revealed that it contains two grooves, S1 and S2, which could, respectively, dock with the TIR domains of IL-1RAcP and MyD88 (Fig. 9E). Ala-471 is located in the S1 groove, which has compatible charge and shape for interaction with the IL-1RAcP TIR (Fig. 9E). S2 of the IL-1Rrp2 TIR domain had a more compatible fit with the MyD88 TIR domain.
We made a construct of the TIR domain of IL-1Rrp2 and also one that contains the A471T substitution to examine for the interaction with the TIR domain of either MyD88 or IL-1RAcP. The WT and A471T TIR domain could co-immunoprecipitate with the TIR domain of MyD88 at similar level (Fig. 9F). However, the A471T TIR domain had a ∼50% decrease in interaction with IL-1RAcP TIR when compared with that of WT. This result is consistent in three independent experiments, suggesting that the reduction in signaling of SNP A471T was due to a decreased interaction with the IL-1RAcP TIR domain.
Summary
In this report, we modeled the IL-36R ECD in complex with IL-36 cytokines and examined the structure and function of IL-36R using a cell-based reporter assay. We found that disulfide bonds and N-linked glycosylation in the ECD of IL-1Rrp2 could affect signaling transduction by regulating protein accumulation and IL-36R trafficking to the cell surface, respectively. D1 in the ECD and the linker between D1 and D2 appear to contribute to specific cytokine recognition. Of particular interest is that residue Asn41 that plays a role in the recognition of specific ligands appears to be glycosylated. The modeling also suggests that there is space for a glycan to be present to contribute to IL-36β recognition.
The presence of IL-36γ significantly increased the interaction between IL-1Rrp2 ECD and the accessory protein IL-1RAcP. However, the secreted IL-1Rrp2 ECD or IL-1RAcP ECD in the cell culture medium exhibited dominant-negative effects on IL-36R signaling. The results together contribute to a model for IL-36R signaling wherein IL-36 cytokine binding increases the interaction between IL-1Rrp2 and IL-1RAcP leading to the signaling transduction. Upon activation, the secreted ECD of IL-1Rrp2 and/or IL-1RAcP could bind to the ECD of co-receptor as well as IL-36 cytokine, and then serve to feedback down-regulate the inflammation response.
Finally, we observed that SNP A471T in the TIR domain down-regulated the signaling via decreasing the interaction with IL-1RAcP. It will be of interest to determine whether individuals possessing this SNP will be affected in inflammation-associated diseases and/or infectious diseases.
Experimental Procedures
Reagents
Recombinant human IL-36α (Cat. 6995-IL/CF), IL-36β (Cat. 6834-IL/CF), IL-36γ (Cat. 6835-ILC/CF), IL-36Ra (1275-IL-025/CF), and IL-1β (Cat. 201-LB-005/CF) were purchased from R&D System. Glycosylation inhibitors tunicamycin and swainsonine were from Sigma. PNGase F and O-glycosidase were from New England Biolabs. Human MyD88, Tollip siRNA, and antibodies were from Santa Cruz Biotechnology. Anti-human IL-1Rrp2 antibody (Cat. AF872) was from R&D Systems and IL-1RAcP antibody (Cat PA5–19921) was from Pierce. Anti-FLAG and anti-HA antibodies were from Life Technology and Roche, respectively.
Cell Culture
HEK 293T cells were cultured in DMEM with 10% fetal bovine serum (FBS) and incubated at 37 °C and 5% CO2. NCI/ADR-RES cells (to be named NCI cells) were cultured in RPMI 1640 + l-glutamine and 10% FBS.
Plasmid Constructions
cDNA for human IL-1Rrp2, IL-1RAcP and a C-terminal FLAG epitope version of IL-1Rrp2 were cloned into HindIII/XhoI sites of pcDNA3.1 (Invitrogen). All the IL-1Rrp2 mutants, including cysteine mutants, asparagine mutants, and SNP mutants were constructed by site-directed mutagenesis. The full-length of IL-1Rrp2 ECD335, IL-1RAcP ECD367 with the C-terminal FLAG-tag and IL-1Rrp2 ECD335, truncated ECDs (ECD258 and ECD126–211) with the signaling peptides in the N-terminal and HA-tags at the C-terminal, respectively, were constructed in pcDNA vector. A construct to express the TIR domain of IL-1Rrp2 that contains a C-terminal HA-tag and one with a A471T mutation were also constructed in the HindIII/XhoI sites of pcDNA3.1 vector. The MyD88 plasmid is from Addgene (19). The sequences of all constructs were determined by DNA sequencing using the BigDye Kit (Invitrogen).
Real-time RT-PCR
The endogenous levels of IL-1Rrp2 and IL-1RAcP from HEK 293T and NCI cells were determined by quantitative real-time RT-PCR. Total RNAs were isolated from 293T and NCI cells using the TRIZOL reagent (Invitrogen) according to the manufacturer's protocol. Approximately 1 μg of total RNA was reverse-transcribed into cDNA using a M-MuLV (New England Biolab) and 4 μm of the 9-nt random primer mix. Real-time PCR was performed with IQTM SYBR Green Kit (Bio-Rad) and the IL-1Rrp2 and IL-1RAcP primers (Santa Cruz Biotechnology). The mRNA levels of IL-1Rrp2 and IL-1RAcP in 293T cells were normalized to the GAPDH level and compared with that of NCI cells using the 2−ΔΔCt method (26).
Cell-based Reporter Assays
293T were seeded for 24 h in CoStar White 96-well plates prior to transfection with a constant amount of the reporter plasmid NF-κΒ-firefly luciferase plasmids (15 ng), phRL-TK-Renilla luciferase (2 ng) and varied amounts of either WT IL-1Rrp2 or mutant plasmid. Transfection used Lipofectamine 2000 (Invitrogen). All transfections were amended with empty pcDNA3.1, as needed, to allow that equal amounts of plasmid were transfected. After a 24-h incubation, ligands were added into the medium at 2 ng/ml and incubated overnight. The luciferase activity was analyzed using the Dual-Glo Luciferase Reporter Assay (Promega) in the BioTek Synergy2 plate reader. The data were plotted either as fold induction of the firefly luciferase expressed from the NF-κΒ promoter relative to that of the cells expressing only the vector control, or percentage of decreased firefly luciferase activity relative to that of the WT IL-1Rrp2.
Western Blotting Analysis
Cells were lysed in the RIPA buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P40, and a mixture of proteinase inhibitors (Roche). The lysate was clarified by centrifugation at 16,000 × g for 20 min. About 30 μg of the soluble protein was separated by 4–12% SDS-PAGE and then transferred to PVDF membrane. After blocking with 5% skim milk in Tris-buffered saline (TBS), the blots were incubated with primary antibody overnight at 4 °C. After three washes with TBST (0.2% Tween-20), the membrane was probed with HRP-labeled secondary antibody, and the signal was detected with ECL prime detection reagent (GE Healthcare). β-actin acted as the sample loading control. The signal was quantified using software from the ChemiDoc System (Bio-Rad).
siRNA Knockdown
293T cells were seeded at T-25 flask overnight. The cells were transfected with 40 nm of either control siRNA or MyD88 or Tollip siRNA using RNAiMax (Life Technology) according to the manufacturer's protocol. After 48 h, the cells were trypsinized and seeded on a 96-well plate. The cells were then super-transfected with IL-1Rrp2 along with reporter plasmids as described above. The levels of endogenous MyD88 and Tollip in the 293T cells were examined by Western blotting analysis. The luciferase activity was analyzed and plotted as described above.
PNGase F and O-Glycosidase Treatment in Vitro
Approximately 40 μg of cell lysate was solubilized in the glycoprotein denaturation buffer for 10 min at 95 °C. The lysate was then treated with either PNGase F or O-glycosidase for overnight at 37 °C prior to gel electrophoresis using SDS-PAGE. The migration of treated IL-1Rrp2 was detected by Western blotting analysis.
Glycosylation Inhibitor Treatment and IL-6/IL-8 Cytokine Quantification in Cells
293T cells were first transfected with IL-1Rrp2 for 6 h and treated with increasing concentrations of tunicamycin or swainsonine for 18 h. The cells were then stimulated with 2 ng/ml of IL-36γ overnight, and luciferase activity was determined. For NCI cells, 2 × 104 cells were seeded in 96-well plates for 24 h and then treated with increasing concentrations of inhibitors for 24 h. The IL-36γ ligand was added at a final concentration of 2 ng/ml for 16 h. The supernatant was collected and spun for 5 min at 1000 rpm. IL-6 and IL-8 levels in the supernatant were quantified by ELISA using the Human OptEIATM kit according to the protocol by the manufacturer (BD Biosciences).
Cell Proliferation Assay
NCI and HEK 293T cells were treated in parallel with glycosylation inhibitors as described above. After ligand treatment, 10 μl of the WST-1 reagent (Clontech) was added into the 96-well plate and incubated for an additional 2 h at 37 °C. The plates were read at A450 and A630, and data were plotted relative to mock-treated controls.
Flow Cytometry
The cell surface expression of IL-1Rrp2 in 293T cells was quantified using flow cytometry. 293T cells were seeded into T-25 flasks and transfected with 1 μg of either WT pcDNA-IL-1Rrp2 or mutants for 24 h. The cells were detached by enzyme-free cell dissociation buffer (Gibco) and suspended in the flow cytometry fixation buffer (R&D System). After a wash with PBS, the cells were either mock-treated or permeabilized with flow cytometry permeabilization/wash buffer for 1 h. Then the cells were stained with goat anti-IL-1Rrp2 primary antibody (1:500) for 1 h, followed by donkey anti-goat IgG conjugated with AlexaFluor 488 (1:200; Life Technologies). After another wash with PBS, suspensions of 293T cells were analyzed by FACS Calibur. The data were presented as an average of two independent experiments with two replicates each. The data and figures were processed using Flowjo software (BD Biosciences).
Co-immunoprecipitation Assay
293T cells were seeded into 6-well plates at a density of 1.5 × 106/well. Cells were transfected with 0.5 μg of plasmid for 24 h. For the ligand treatment, IL-36γ was added at final concentration of 20 ng/ml for overnight. The cells were lysed in RIPA buffer, and the supernatant was incubated with anti-IL-1RAcP or the indicated antibody and magnetic beads (Thermo Scientific). To examine the secreted ECD-ECD interaction, immunoprecipitation was performed using the medium from cells co-transfected with FLAG- and HA-tagged IL-1Rrp2 ECD plasmids. After washing with TBST, the beads were suspended in 1× SDS running buffer and heated for 5 min at 95 °C. The solution was separated by SDS-PAGE followed by Western blotting analysis using the appropriate antibodies.
Single Nucleotide Polymorphism (SNP) Analysis
SNP data were derived from the NCBI SNP database. Only non-synonymous SNPs of IL-1Rrp2 were analyzed.
Molecular Modeling
Homology models were generated using the Phyre2 (Protein Homology/analogy Recognition Engine V 2.0 (27). The quality of protein models was assessed by a suite of programs run under Phyre2 Investigator. Query sequences for the human proteins were: Interleukin-1 receptor-like 2 precursor (IL-1Rrp2) NP_003845.2; Interleukin-1 receptor accessory protein (IL-1RAcP) UniProtKB/Swiss-Prot: Q9NPH3.2; Interleukin-36 β isoform 2 NP_775270.1; and interleukin-36 β isoform 1 NP_055253.2. Rigid and flexible regions were predicted by Flexpred (28). Model rendering, backbone superpositions, and electrostatic potential surface calculations were performed using PyMol (The PyMol Molecular Graphics System, Version 1.7.4 Schrodinger, LLC).
Homology models used to generate the IL-36R ECD complex were determined by a multi-stage process implemented in Phyre 2. Briefly, in stage 1, a large number of diverse true homologues are collected to produce an evolutionary profile that reflects residue preferences at each position in the primary sequence. In stage 2, the profile, together with a predicted secondary structure, is converted to a hidden Markov model (HMM). This HMM is scanned against a database of HMMs library of designated folds to generate crude backbone models that do not contain amino acid side chains. At this point, the models typically have some level of insertions and deletions (indels) that must be addressed. In stage 3, loops are modeled into indels using a library of fragments of known protein structures. Finally in stage 4, side chains are added to complete the homology model. This final step is estimated to be ∼80% accurate if the backbone is correctly determined (27).
The quality of each molecular model was evaluated by a suite of algorithms run in Phyre2 Investigator. ProQ2 (29) did not flag serious problems in the local or global quality of the homology models. Minor issues that were identified tended to occur in longer turns or loops. Any side chain clashes, the use of non-ideal rotamers, and the geometry of the backbone phi/psi angles were checked by using the Molprobity Progam (30) included in the Phyre2 Investigator.
TIR Domain Docking
PyDock was used to perform rigid body docking of TIR domain models. The IL-1Rrp2 TIR domain was designated the receptor protein and the TIR domain of IL-1RAcP or MyD88 was assigned the ligand protein in the PyDock setup. In the first stage of generating docking poses between receptor and ligand proteins, ftdock (fast Fourier transform method) was run. The resulting output was used to generate a rotation and translation matrix to build possible complex conformations. The dockser module in PyDock was implemented to score and rank all positions. In the case without restraints, the electrostatic, desolvation, and van Der Waals contributions are calculated for each of the 10,000 conformations, and these contributions are summed to find the total binding energy. The top 100 solutions are saved for inspection. A docking issue that must be addressed is how to get the rigid body search to find poses that orient the transmembrane helices of docked TIR domains in all the same direction. With this constraint in mind, we visually inspected our models and identified candidate binding surfaces on IL-1Rrp2 that could dock the TIR domains of IL-1RAcP and MyD88. From this different sets of trial restraints were produced to test in PyDock searches. As control, PyDock runs were performed without any restraints and in all cases the orientation of transmembrane helices between docking pairs were incorrect, indicating restraints were required to yield plausible docking poses. In PyDock, a restraint was deemed satisfied when the center of coordinates of the specified side chain was within 6 Å from any non-hydrogen atom of the partner molecule. The amount of energy specified restraints can additionally contribute to the total binding energy depends on whether the distance requirement is met. To avoid biasing the search, the number of restraints used was held to a minimum. For a given docking pair, restraints that yielded the highest calculated total binding energy, and therefore ranked number 1 out of 10,000 ranked poses, were favored. The top ranking poses are shown in Fig. 9.
Author Contributions
CC. C. K. and M. L. M. conceived and coordinated the study, analyzed all results and wrote the paper. G. Y. acquired the results in Figs. 1A, 2B–E, 3A, 3C, 4A–F, 5A–C, 5E, 6B–D, 7A–D, 7F, 8C, 8D, and 9A–D and 9E, 9F, analyzed the results, and wrote the paper. J. Y. acquired the models in Figs. 1, B–D, 3B, 5D, 6A, 8A–B, 9E and contributed to the writing. S. S. acquired the results in Figs. 2A, 2F, 7E and helped analyzed the results. G. C. and E. R. provided important reagents and analyzed the results. R. G. contributed to the writing of the paper and analysis of the results.
Acknowledgments
We thank Laura Kao for editing the manuscript and the members of the Kao laboratory and our colleagues at Boehringer-Ingelheim, especially Jay Fine, for support and helpful discussions throughout this work.
The authors declare that they have no conflicts of interest with the contents of this article.
- IL-36R
- IL-36 receptor
- ECD
- ectodomain
- TIR
- Toll-interleukin 1 receptor domain
- SNP
- single nucleotide polymorphism.
References
- 1. Towne J. E., and Sims J. E. (2012) IL-36 in psoriasis. Curr. Opin. Pharmacol. 12, 486–490 [DOI] [PubMed] [Google Scholar]
- 2. Gresnigt M. S., and van de Veerdonk F. L. (2013) Biology of IL-36 cytokines and their role in disease. Semin. Immunol. 25, 458–465 [DOI] [PubMed] [Google Scholar]
- 3. Blumberg H., Dinh H., Trueblood E. S., Pretorius J., Kugler D., Weng N., Kanaly S. T., Towne J. E., Willis C. R., Kuechle M. K., Sims J. E., and Peschon J. J. (2007) Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J. Exp. Med. 204, 2603–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gabay C., and Towne J. E. (2015) Regulation and function of interleukin-36 cytokines in homeostasis and pathological conditions. J. Leukoc. Biol. 97, 645–652 [DOI] [PubMed] [Google Scholar]
- 5. Medina-Contreras O., Harusato A., Nishio H., Flannigan K. L., Ngo V., Leoni G., Neumann P. A., Geem D., Lili L. N., Ramadas R. A., Chassaing B., Gewirtz A. T., Kohlmeier J. E., Parkos C. A., Towne J. E., Nusrat A., and Denning T. L. (2016) Cutting Edge: IL-36 Receptor Promotes Resolution of Intestinal Damage. J. Immunol. 196, 34–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Foster A. M., Baliwag J., Chen C. S., Guzman A. M., Stoll S. W., Gudjonsson J. E., Ward N. L., and Johnston A. (2014) IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J. Immunol. 192, 6053–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garlanda C., Dinarello C. A., and Mantovani A. (2013) The interleukin-1 family: back to the future. Immunity 39, 1003–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Towne J. E., Garka K. E., Renshaw B. R., Virca G. D., and Sims J. E. (2004) Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-κB and MAPKs. J. Biol. Chem. 279, 13677–13688 [DOI] [PubMed] [Google Scholar]
- 9. Towne J. E., Renshaw B. R., Douangpanya J., Lipsky B. P., Shen M., Gabel C. A., and Sims J. E. (2011) Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36α, IL-36β, and IL-36γ) or antagonist (IL-36Ra) activity. J. Biol. Chem. 286, 42594–42602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Debets R., Timans J. C., Homey B., Zurawski S., Sana T. R., Lo S., Wagner J., Edwards G., Clifford T., Menon S., Bazan J. F., and Kastelein R. A. (2001) Two novel IL-1 family members, IL-1 delta and IL-1 epsilon, function as an antagonist and agonist of NF-κB activation through the orphan IL-1 receptor-related protein 2. J. Immunol. 167, 1440–1446 [DOI] [PubMed] [Google Scholar]
- 11. van de Veerdonk F. L., Stoeckman A. K., Wu G., Boeckermann A. N., Azam T., Netea M. G., Joosten L. A., van der Meer J. W., Hao R., Kalabokis V., and Dinarello C. A. (2012) IL-38 binds to the IL-36 receptor and has biological effects on immune cells similar to IL-36 receptor antagonist. Proc. Natl. Acad. Sci. U.S.A. 109, 3001–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas C., Bazan J. F., and Garcia K. C. (2012) Structure of the activating IL-1 receptor signaling complex. Nat. Struct. Mol. Biol. 19, 455–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang D., Zhang S., Li L., Liu X., Mei K., and Wang X. (2010) Structural insights into the assembly and activation of IL-1β with its receptors. Nat. Immunol. 11, 905–911 [DOI] [PubMed] [Google Scholar]
- 14. Tsutsumi N., Kimura T., Arita K., Ariyoshi M., Ohnishi H., Yamamoto T., Zuo X., Maenaka K., Park E. Y., Kondo N., Shirakawa M., Tochio H., and Kato Z. (2014) The structural basis for receptor recognition of human interleukin-18. Nat. Commun. 5, 5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X., Hammel M., He Y., Tainer J. A., Jeng U. S., Zhang L., Wang S., and Wang X. (2013) Structural insights into the interaction of IL-33 with its receptors. Proc. Natl. Acad. Sci. U.S.A. 110, 14918–14923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Günther S., and Sundberg E. J. (2014) Molecular determinants of agonist and antagonist signaling through the IL-36 receptor. J. Immunol. 193, 921–930 [DOI] [PubMed] [Google Scholar]
- 17. Deribe Y. L., Pawson T., and Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 [DOI] [PubMed] [Google Scholar]
- 18. Saha S. S., Singh D., Raymond E. L., Ganesan R., Caviness G., Grimaldi C., Woska J. R. Jr., Mennerich D., Brown S. E., Mbow M. L., and Kao C. C. (2015) Signal transduction and intracellular trafficking by the interleukin 36 receptor. J. Biol. Chem. 290, 23997–24006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdörfer B., Giese T., Endres S., and Hartmann G. (2002) Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 168, 4531–4537 [DOI] [PubMed] [Google Scholar]
- 20. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moffatt M. F., Gut I. G., Demenais F., Strachan D. P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W. O., and Gabriel Consortium (2010) A large-scale, consortium-based genomewide association study of asthma. New Eng. J. Med. 363, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Latiano A., Palmieri O., Pastorelli L., Vecchi M., Pizarro T. T., Bossa F., Merla G., Augello B., Latiano T., Corritore G., Settesoldi A., Valvano M. R., D'Incà R., Stronati L., Annese V., and Andriulli A. (2013) Associations between genetic polymorphisms in IL-33, IL1R1 and risk for inflammatory bowel disease. PloS one 8, e62144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wesche H., Henzel W. J., Shillinglaw W., Li S., and Cao Z. (1997) MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 24. Dunne A., and O'Neill L. A. (2003) The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Science's STKE: Sig. Trans. re3. [DOI] [PubMed] [Google Scholar]
- 25. Jiang Z., Georgel P., Li C., Choe J., Crozat K., Rutschmann S., Du X., Bigby T., Mudd S., Sovath S., Wilson I. A., Olson A., and Beutler B. (2006) Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 10961–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuznetsov I. B., and McDuffie M. (2008) FlexPred: a web-server for predicting residue positions involved in conformational switches in proteins. Bioinformatics 3, 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ray A., Lindahl E., and Wallner B. (2012) Improved model quality assessment using ProQ2. BMC Bioinformatics 13, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]