Abstract
To investigate mechanisms for increased malignant properties in malignant melanomas by ganglioside GD3, enzyme-mediated activation of radical sources and subsequent mass spectrometry were performed using an anti-GD3 antibody and GD3-positive (GD3+) and GD3-negative (GD3-) melanoma cell lines. Neogenin, defined as a GD3-neighbored molecule, was largely localized in lipid/rafts in GD3+ cells. Silencing of neogenin resulted in the reduction of cell growth and invasion activity. Physical association between GD3 and neogenin was demonstrated by immunoblotting of the immunoprecipitates with anti-neogenin antibody from GD3+ cell lysates. The intracytoplasmic domain of neogenin (Ne-ICD) was detected in GD3+ cells at higher levels than in GD3− cells when cells were treated by a proteasome inhibitor but not when simultaneously treated with a γ-secretase inhibitor. Exogenous GD3 also induced increased Ne-ICD in GD3− cells. Overexpression of Ne-ICD in GD3− cells resulted in the increased cell growth and invasion activity, suggesting that Ne-ICD plays a role as a transcriptional factor to drive malignant properties of melanomas after cleavage with γ-secretase. γ-Secretase was found in lipid/rafts in GD3+ cells. Accordingly, immunocyto-staining revealed that GD3, neogenin, and γ-secretase were co-localized at the leading edge of GD3+ cells. All these results suggested that GD3 recruits γ-secretase to lipid/rafts, allowing efficient cleavage of neogenin. ChIP-sequencing was performed to identify candidates of target genes of Ne-ICD. Some of them actually showed increased expression after expression of Ne-ICD, probably exerting malignant phenotypes of melanomas under GD3 expression.
Keywords: γ-secretase, ganglioside, lipid raft, melanoma, signaling, transcription factor
Introduction
It has been long known that malignant transformation of cells brings about changes in the carbohydrate structures in the glycoproteins and glycolipids expressed on the cell surface (1). In particular, there have been a number of reports on the expression of unique glycosphingolipids in neuroectoderm-derived cancer cells (2), osteosarcomas (3, 4), small lung cancer cells (5, 6), and T-cell leukemia cells (7–9). Some of them have been utilized in the clinical field as tumor markers (10) or target molecules of antibody therapy (11).
Because ganglioside GD3 was reported to be neo-antigen in melanomas (12–14), it has attracted the interests of researchers in cancer immunology field, and its expression in normal and cancer tissues has been analyzed (15). The effects of anti-GD3 antibody therapy in melanomas (16) and various aspects of its roles in melanomas have been also reported (17–19). These results suggest that GD3 plays important roles in the enhancement of malignant properties of melanomas (20, 21) by regulating glycolipid-enriched microdomain (GEM)2/rafts (22). However, precise mechanisms for GD3 function in cancers have not yet been clarified.
To investigate how gangliosides are involved in the regulation of cell signaling, we have applied enzyme-mediated activation of radical sources (EMARS) method (23) to identify membrane molecules interacting with gangliosides in the vicinity of cell membrane. One of representative molecules associating with GD3 in melanoma cells was neogenin (24). Neogenin belongs to immunoglobulin (Ig) superfamily and has homology to deleted in colorectal cancer (DCC) (25). Although it has been well known as a receptor for axon repulsion in neuroscience research field (26), little is known about roles in cancers (27) despite its expression in wide range of cancer cells (28).
In this study we investigated functions of neogenin and its intracytoplasmic domain (Ne-ICD) in the phenotypes of GD3-positive melanoma cells and its association with GD3 in GEM/rafts. These results might be the first report on the concrete example of effectors to exert malignant properties of melanomas under melanoma-specific ganglioside, GD3.
Results
Expression of Neogenin in GD3+ Cells and Control Cells
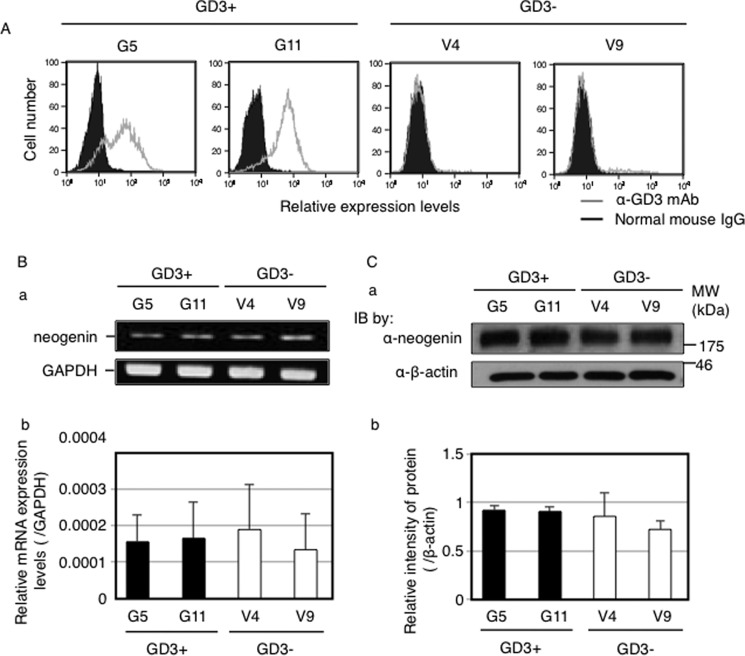
At first, we checked GD3 expression on GD3+ cells (G5, G11) and control cells (V4, V9) by flow cytometry. G5 and G11 cells expressed GD3 strongly, whereas GD3 was scarcely detected in V4 and V9 cells (Fig. 1A). Using these cells, we analyzed mRNA and protein expression levels of neogenin by RT-PCR (Fig. 1B) and Western blotting (Fig. 1C), respectively. Neogenin expression was almost equivalent among all GD3+ cells and control cells.
FIGURE 1.
Neogenin expression was equivalent among all GD3+ cells and control cells. A, GD3 expressions on GD3+ cells (G5, G11) and control cells (V4, V9). Cells (3 × 105) were incubated with anti-GD3 mAb (R24) for 1 h at 4 °C. After washing with PBS, cells were incubated with an FITC-labeled anti-mouse IgG antibody for 45 min at 4 °C. Then GD3 expression levels were analyzed using FACS CaliburTM. Gray lines indicate staining with an anti-GD3 antibody. Black lines with solid peaks indicate the staining with normal mouse IgG. B, mRNA expression mRNA levels of neogenin in GD3+ cells (G5, G11) and control cells (V4, V9) examined by RT-PCR. a, PCR products were observed by agarose gel electrophoresis. b, quantitative PCR analysis was performed. Expression mRNA levels of neogenin was normalized by the GAPDH gene (n = 3). C, protein expression levels of neogenin in GD3+ cells (G5, G11) and control cells (V4, V9) examined by Western blotting. a, total 12.5 μg of proteins were separated in SDS-PAGE. After blotting onto PVDF membranes, membranes were incubated with an anti-neogenin antibody (upper panel) or an anti-β-actin antibody (lower panel). IB, immunoblot. b, the intensities of bands in a were measured by Image JTM software and plotted. The intensities of bands of neogenin were normalized by β-actin (n = 3).
Neogenin Was Involved in Malignant Phenotypes in Melanoma Cells
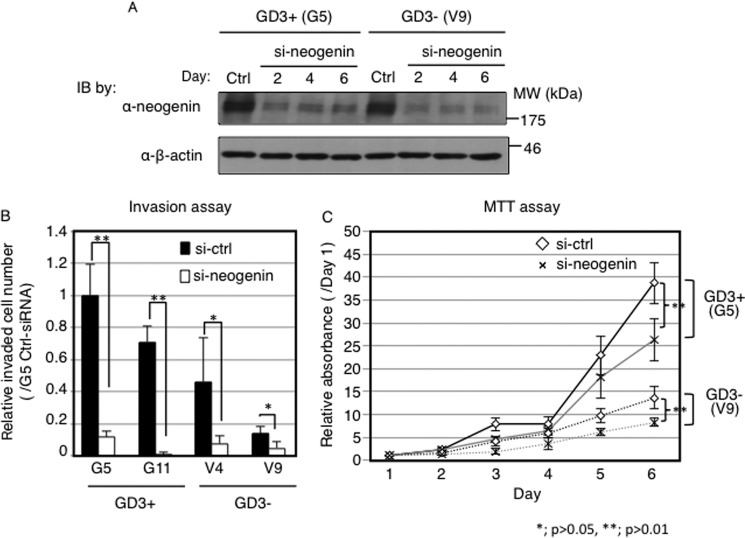
To clarify whether neogenin expression is involved in malignant phenotypes in melanoma cells, we suppressed neogenin expression by transfection of anti-neogenin siRNA. At 48 h after transfection, knockdown efficiency was examined by Western immunoblotting, showing strong suppression of neogenin expression (Fig. 2A). Using these siRNA-treated cells, invasion and cell proliferation assays were performed. As shown in Fig. 2B, numbers of invaded cells into the bottom side of chambers were dramatically reduced after knockdown of neogenin in all cell lines (G5, G11, V4, V9). The proliferation rates were also decreased in neogenin-silenced cells compared with control siRNA-transfected cells (Fig. 2C). These results suggested its novel function as a promoting molecule of cancer properties.
FIGURE 2.
Neogenin was involved in malignant phenotypes in melanoma cells. A, protein expression levels of neogenin after transfection with anti-neogenin siRNA. Neogenin siRNA and scrambled siRNA were transfected using Lipofectamine 2000 TM. Samples were blotted with an anti-neogenin antibody. IB, immunoblot. B and C, function of neogenin in melanoma cells. Phenotypes after neogenin knockdown were analyzed using an invasion assay (B) and MTT assay (C). B, 8 ×103 cells after siRNA transfection were plated in BD Falcon Cell Culture InsertsTM covered with 100 μg of Matrigel. After 24 h incubation, non-invaded cells were removed with a cotton bud, and invaded cells were stained with 1/40 Giemsa staining solution and counted. C, cells (0.5–1 × 104) were plated into multiwell plates (96-well), and MTT solution (5 mg/ml) was added and incubated for 4 h at days 1–6. Absorbance at 590/620 nm was measured, and relative absorbance (/day1) was plotted. Solid lines indicate GD3+ cells (G5), and dotted lines indicate control cells (V9). Gray lines indicate samples with transfection of neogenin siRNA, and black lines mean samples with transfection of scrambled siRNA.
A High Amount of Neogenin Was Localized in GEM/Rafts in GD3+ Cells in Contrast with GD3− Cells
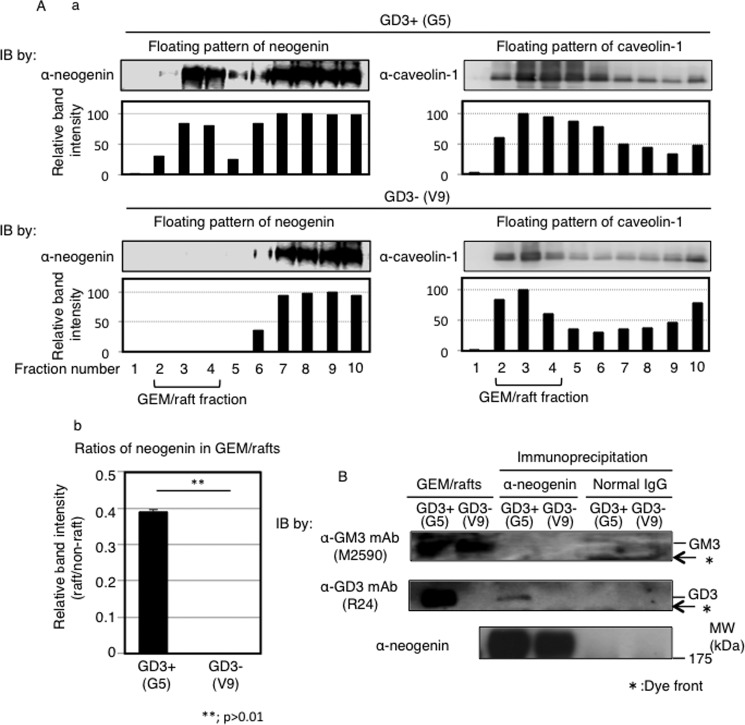
Next, we analyzed floating pattern of neogenin using fractions prepared from 1% Triton X-100 extracts by sucrose density gradient ultracentrifugation. As shown in Fig. 3A, there were approximately 50 times more neogenin in GEM/raft fractions in GD3+ cells (G5) than those in control cells (V9). These data suggested that neogenin shifted to GEM/rafts under GD3 expression.
FIGURE 3.
GEM/raft localization of neogenin in melanoma cells and association of GD3 with neogenin. A, a, total cell lysates (G5 and V9) with Triton X-100 were fractionated by sucrose-density gradient ultracentrifugation as described under “Experimental Procedures.” Fractions were analyzed by Western blotting (IB) with an anti-neogenin antibody and an anti-caveolin-1 antibody. Caveolin-1 was used as a GEM/rafts marker. The band intensities of neogenin and caveolin-1 were measured by Image JTM software and are presented. b, ratios of neogenin in GEM/rafts were determined by comparing band intensities in GEM/rafts and non-GEM/rafts fractions. B, association between neogenin and GM3 or GD3 was analyzed by immunoprecipitation and subsequent Western blotting. Total cell lysates (600 μg) were used for immunoprecipitation with an anti-neogenin antibody or normal goat IgG, and immunoprecipitates were analyzed by Western blotting with an anti-GM3 mAb (M2590) (upper panel), an anti-GD3 mAb (R24) (middle panel), or an anti-neogenin antibody (lower panel). GEM/raft fractions from G5 and V9 were also analyzed as shown at the left end.
GD3 Associates with Neogenin
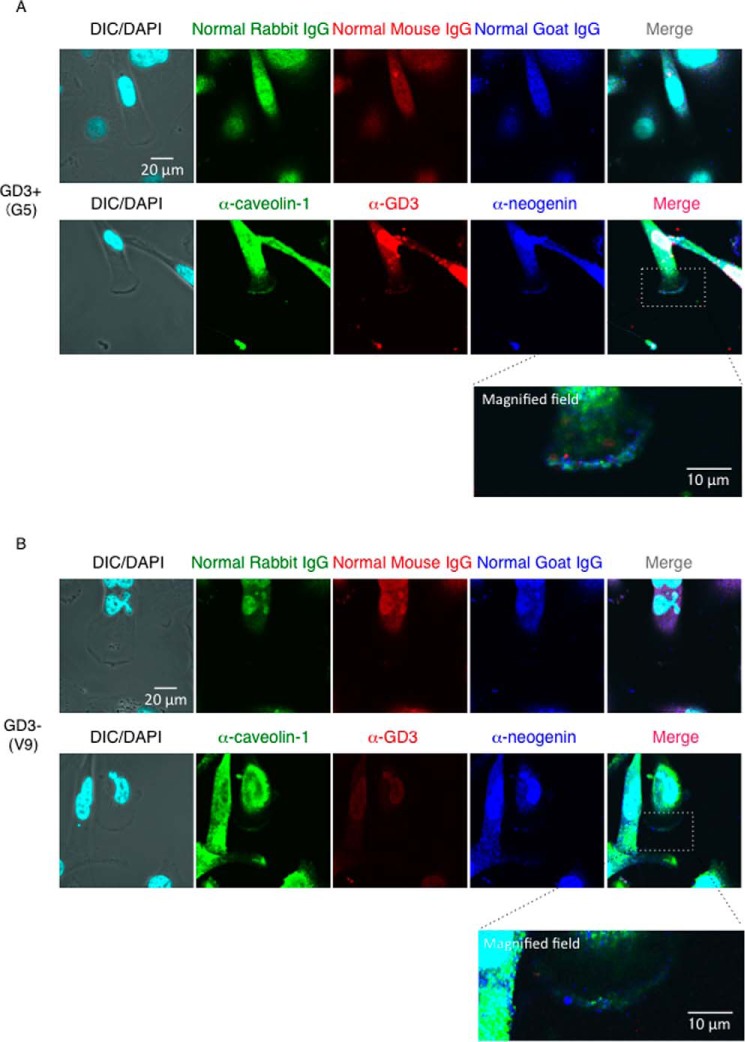
Next, we tried to clarify whether GD3 physically associates with neogenin. To address this issue, we performed immunoprecipitation with an anti-neogenin antibody and subsequent immunoblotting with an anti-GM3 monoclonal antibody (mAb) or an anti-GD3 mAb. GD3 was detected in immunoprecipitates only from GD3+ cells, but GM3 was not detected in either GD3+ cell-derived immunoprecipitates or GD3− cell-derived immunoprecipitates despite of abundant presence (Fig. 3B). To examine intracellular localization of caveolin-1, neogenin, and GD3, immunocytochemistry was performed using an anti-neogenin antibody, anti-caveolin antibody, and an anti-GD3 mAb (Fig. 4). Because nonspecific binding of normal IgG/2nd antibodies was very high, staining of nuclei was neglected here and in Fig. 7. Neogenin was co-localized with GD3 at ceveolin-1-positive leading edge of GD3+ cells (G5), whereas neogenin and caveolin-1 were not necessarily co-localized in GD3− cells (V9) (Fig. 4B). These results suggested that neogenin associates with GD3, and this association is important for GEM/raft localization, i.e. a switching of neogenin localization in the microdomain may occur based on the GD3 expression in melanoma cells. Neogenin was cleaved by γ-secretase in melanoma cells, and its intracytoplasmic domain (named Ne-ICD) was detected more in GD3+ cells than in control cells.
FIGURE 4.
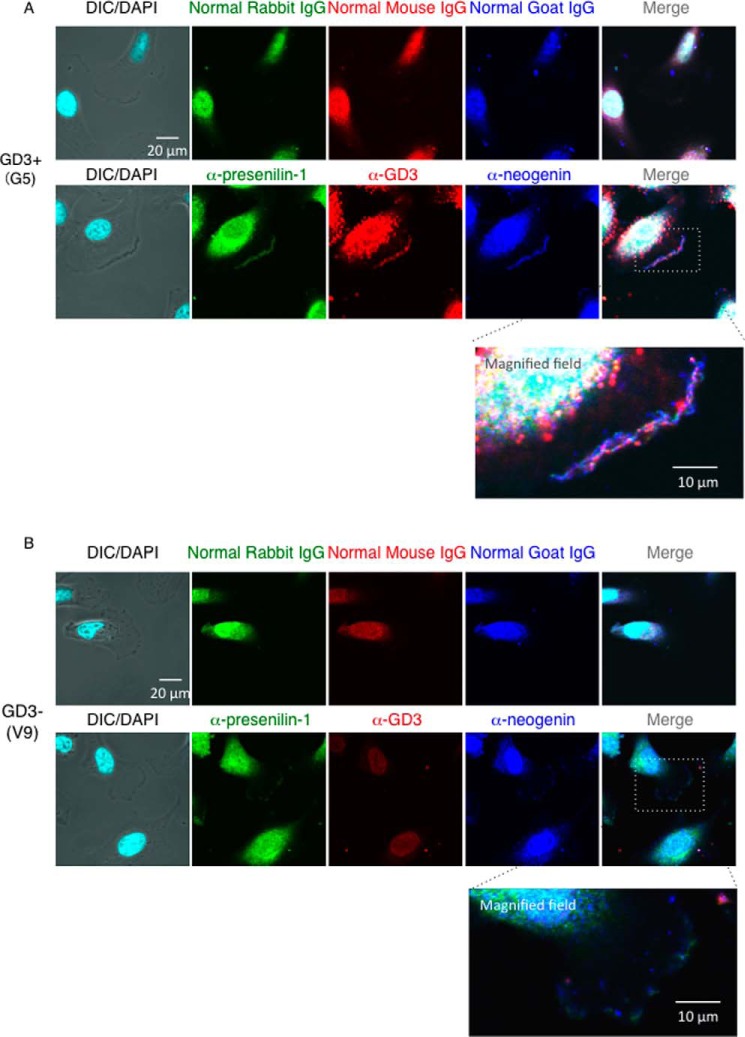
Neogenin was co-localized with GD3 at caveolin-1-positive leading edge of GD3+ melanoma cells. Immunocytochemical analysis of G5 (A) and V9 (B) cells with an anti-caveolin-1 antibody (green), an anti-neogenin antibody (blue), and an anti-GD3 mAb (R24) (red). Bar, 20 μm or 10 μm as indicated. High-magnified images of dashed square areas in merged image were shown at the bottom of each image. DIC, differential interference contrast.
FIGURE 7.
Neogenin was co-localized with GD3 and presenilin-1 in melanoma cells. Cells were fixed by 4% PFA and permeabilized with 0.1% Triton X-100. After blocking with 2.5% BSA, cells were incubated with a mouse anti-GD3 mAb (R24), a goat anti-neogenin antibody, or a rabbit anti-presenilin-1 (NTF) antibody overnight at 4 °C. Immunostaining of GD3+ cells (G5) (A) and control cells (V9) (B). Green, red, and blue indicate presenilin-1, GD3, and neogenin, respectively (lower panels). Immunostaining with normal IgG was also performed for negative control (upper panels). High-magnified fields of dashed square areas in merged image were shown at the bottom. Bar, 20 μm or 10 μm as indicated. DIC, differential interference contrast.
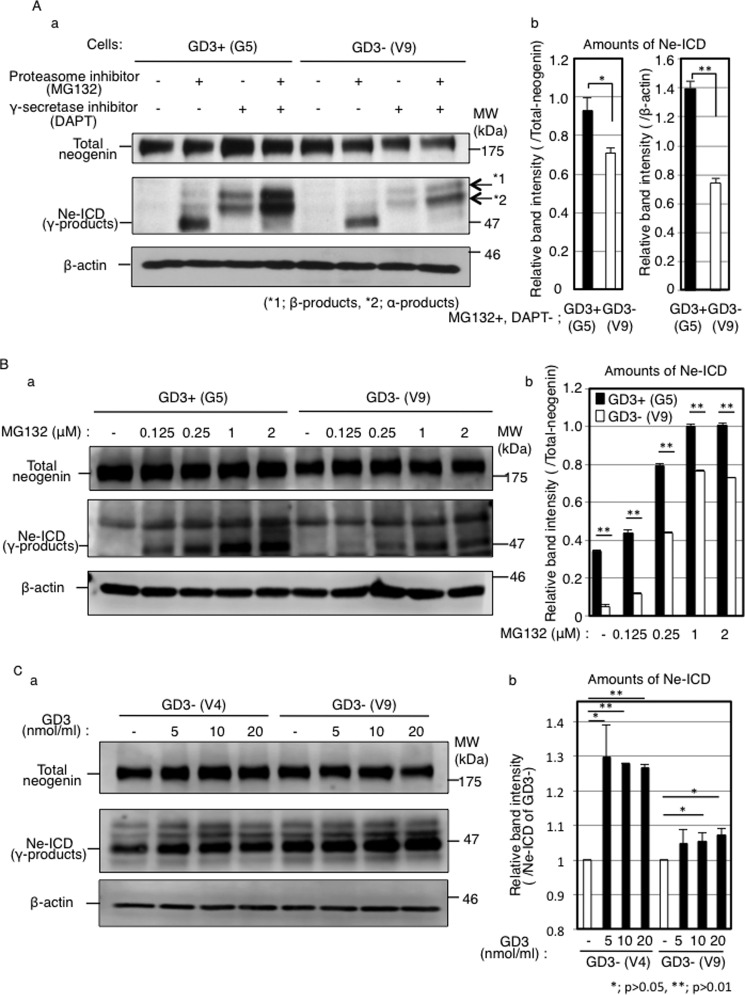
It is known that a part of γ-secretase is localized in GEM/rafts (30, 31) Goldschneider et al. (32) reported that neogenin was a substrate of γ-secretase and the cleaved product (Ne-ICD) functioned as a transcription factor in HEK293T cells. To analyze whether neogenin is cleaved by γ-secretase also in melanoma cells, we treated cells with proteasome inhibitor (MG132) and/or γ-secretase inhibitor (DAPT, N-[N-[(3,5-difluorophenyl)acetyl]-l-alanyl]-l-phenyl-glycine tert-butyl ester). In the absence of MG132, there was no band of the cleaved product (Ne-ICD) in both GD3+ cells (G5) and control cells (V9) (Fig. 5A, first and fifth lanes from the left end). However, in the presence of MG132, Ne-ICD was detected in GD3+ cells (G5) and control cells (V9) (Fig. 5A, second and sixth lanes from the left end). In the presence of DAPT, we could not detect Ne-ICD in any cell lines regardless of MG132 (Fig. 5A, third and fourth lanes and seventh and eighth lanes from the left end). These results indicated that neogenin was cleaved by γ-secretase. Interestingly, the amounts of Ne-ICD were more in the GD3+ cells (G5) than in control cells (V9) (Fig. 5Ab), although equivalent expression of total-neogenin and presenilin-1 was observed in GD3+ cells (G5) and control cells (V9) (Figs. 1, B and C, 5Aa (first and fifth lanes from the left end), and 6A). This difference in amounts was apparent especially at the low concentration (0.125–0.25 μm) treatment of MG132 (Fig. 5B). To study the effect of GD3 on the production of Ne-ICD, GD3 was directly added to GD3− cells. The amounts of Ne-ICD increased after the addition of GD3 in a dose-dependent manner in two GD3− cells (Fig. 5C). These results suggested that localization changes of neogenin to GEM/rafts by expression or exogenous addition of GD3 increased the efficiency of neogenin cleavage by γ-secretase.
FIGURE 5.
Neogenin was cleaved by γ-secretase, and the Ne-ICD levels were higher in GD3+ melanoma cells and increased in GD3− cells after exogenous addition of GD3. A, a, detection of Ne-ICD in GD3+ cells (G5) and control cells (V9) by Western blotting. Cells (2 × 105) were incubated with or without a proteasome inhibitor (MG132, 1 μm) and/or a γ-secretase inhibitor (DAPT, 1 μm). After incubation for 4 h, cells were lysed, and proteins (12.5 μg) were analyzed by Western blotting using an anti-neogenin antibody or an anti-β-actin antibody. b, band intensities of Ne-ICD were measured by Image JTM software and plotted. The intensities were normalized by those of β-actin (n = 3). B, a, levels of Ne-ICD after the treatment with various concentration of MG132. Cells (2 × 105) were incubated with various concentrations of a proteasome inhibitor, MG132 (0–2 μm). After incubation for 4 h, cells were lysed and analyzed by Western blotting as shown in A. b, band intensities were also measured and normalized by β-actin. C, a, amounts of Ne-ICD after the addition of GD3. Cells (2 × 105) were incubated with different concentration of exogenous GD3 for 48 h and used for analysis of Ne-ICD by Western blotting. b band intensities were measured and normalized by Ne-ICD of non-treated cells.
Presenilin-1 Was Localized in the GEM/Rafts of GD3+ Cells
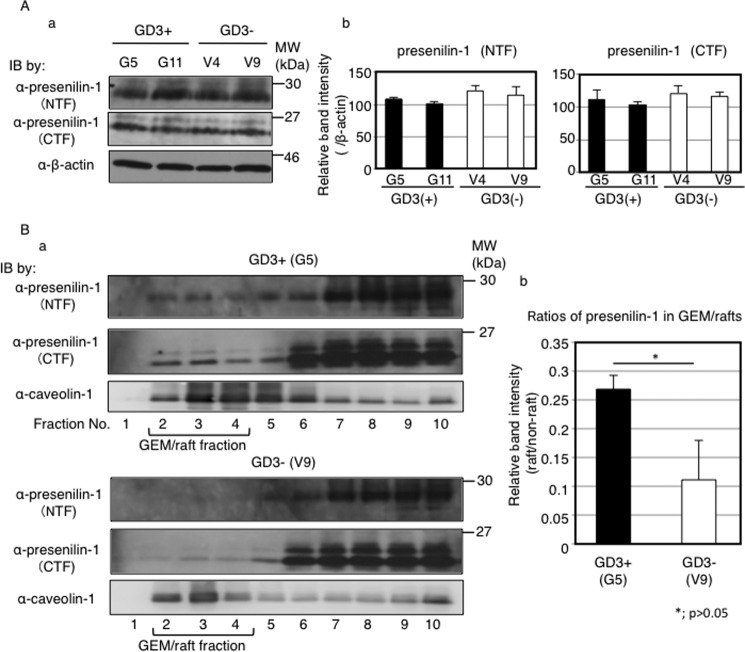
To analyze intracellular localization of presenilin-1 in melanoma cells, fractions prepared from 1% Triton X-100 extracts were analyzed by Western blotting with an anti-presenilin-1 antibody. Both C-terminal fragment and N-terminal fragment of presenilin-1 were definitely detected in the GEM/raft fraction of GD3+ cells compared with GD3− cells (Fig. 6B). These results suggested the possibility that cleavage of neogenin by γ-secretase occurs in the microdomains of GD3+ cells. Therefore, the physical association between GD3 and neogenin might be important for the cleavage of neogenin by γ-secretase.
FIGURE 6.
Presenilin-1 expression levels were equivalent among all GD3+ cells and control cells but localized in GEM/rafts of GD3+ melanoma cells. A, protein expression levels of presenilin-1 in GD3+ cells (G5, G11) and control cells (V4, V9) examined by Western blotting (IB). a, total cell lysates (12.5 μg) were blotted and analyzed using an anti-presenilin-1 C terminus (CTF) and an anti-presenilin-1 N terminus (NTF) antibodies. b, band intensities were measured by Image JTM software and plotted. The intensities were normalized by β-actin. B, a, floating patterns of presenilin-1 in GD3+ cells (G5) and control cells (V9). Total cell lysates were fractionated by sucrose-density gradient ultracentrifugation as described under “Experimental Procedures.” Fractions were analyzed by Western blotting with an anti-presenilin-1 antibody or an anti-caveolin-1 antibody. Caveolin-1 was used as a GEM/rafts marker. b, band intensities of presenilin-1 (CTF) were measured by Image J software and plotted. Ratios of presenilin-1 in GEM/rafts were determined by comparing its band intensities in GEM/rafts and in non-GEM/rafts fractions.
Presenilin-1, Neogenin, and GD3 Were Co-localized in GD3+ Melanoma Cells
To examine the intracellular localization of neogenin, GD3, and γ-secretase, we performed immunocytochemistry analysis. Neogenin, GD3, and presenilin-1 were co-stained at the leading edge of GD3+ cells (G5), suggesting possible association of these molecules. In the GD3− cells, presenilin-1 and neogenin were almost separately stained at the leading edge (Fig. 7).
Overexpression of Ne-ICD Enhanced Malignant Properties of Melanoma Cells
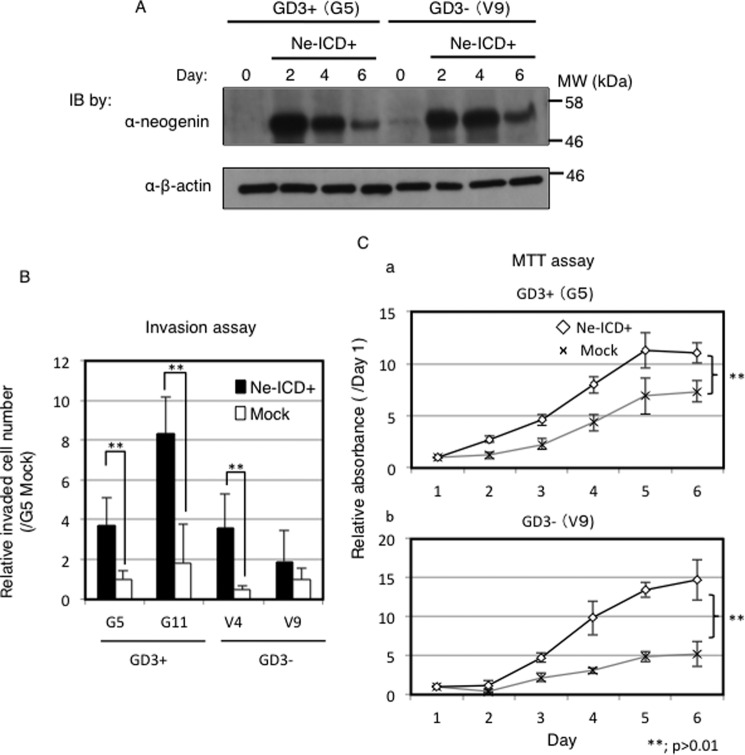
To clarify roles of Ne-ICD in melanoma cells, Ne-ICD cDNA was transfected into GD3+ cells and control cells. Expression levels of Ne-ICD after transfection were shown in Fig. 8A. Ne-ICD was detected even in the absence of MG132. And neogenin was detected at least for 6 days. Consequently, overexpression of Ne-ICD considerably increased the number of invaded cells and increased cell proliferation as shown in Fig. 8, B and C. Namely, Ne-ICD overexpression enhanced tumor phenotypes, such as invasion and cell proliferation in GD3+ cells (G5) and control cells (V9).
FIGURE 8.
Ne-ICD enhanced tumor phenotypes in melanoma cells. Function of Ne-ICD in melanoma cells. A, Ne-ICD expression after transfection of Ne-ICD cDNA. Cells (2 × 105) were transfected with Ne-ICD (L1455A) with Lipofectamine 2000TM. After incubation for 48∼144 h, cells were lysed and analyzed by Western blotting (IB) with an anti-neogenin antibody. Cancer phenotypes after overexpression of Ne-ICD were analyzed using an invasion assay (B) and MTT assay (C). B, at 48 h after transfection of the Ne-ICD expression vector, 8 ×103 cells were analyzed using invasion assay as described in Fig. 1E. Relative numbers of invaded cells are presented. C, cells (0.5–1 × 104) were analyzed by MTT assay as described in Fig. 1F. Black lines indicate cells with overexpression of Ne-ICD, and gray lines mean vector controls.
Target Genes of Ne-ICD Were Identified by Chromatin Immunoprecipitation (ChIP)
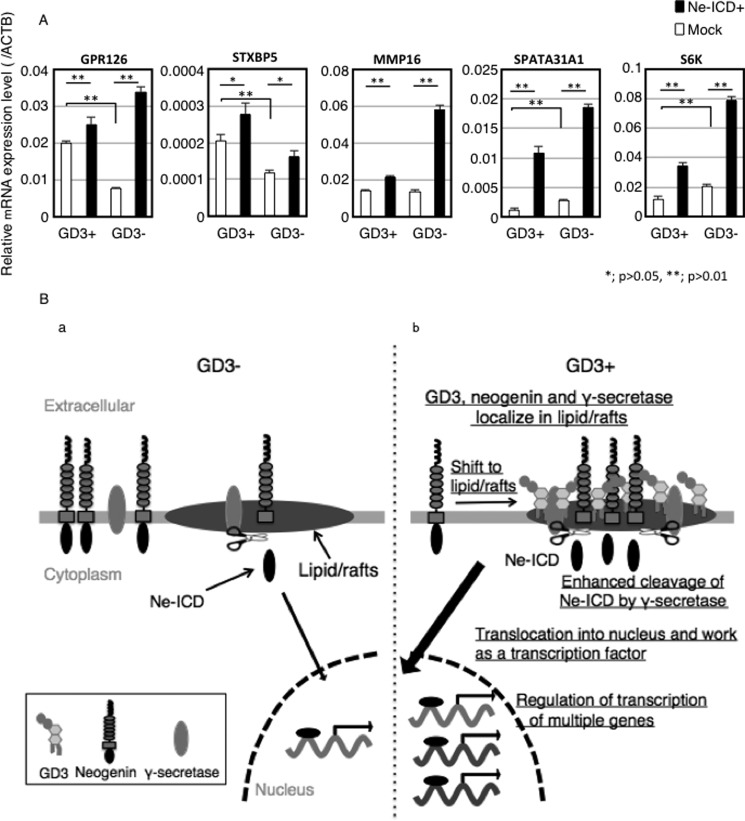
We tried to identify the target genes of Ne-ICD in melanoma cells by ChIP. We identified 17 genes that might be regulated by Ne-ICD in melanoma cells (Table 1). To examine the effects of Ne-ICD on the gene expression of these molecules, RT-quantitative PCR was performed using RNA from GD3+ and GD3− cells before and after transfection of a Ne-ICD expression vector. As shown in Fig. 9A, overexpression of Ne-ICD resulted in the up-regulation of the candidate genes such as GPR126, ST6BP5, MMP-16, SPATA31A1, and S6K etc. Therefore, these molecules seemed to be actual effectors for enhancement of the malignant properties induced under GD3 expression in melanomas.
TABLE 1.
A list of target genes of Ne-ICD identified by ChIP in GD3+ (G5) cells
Cells were transfected FLAG-tagged Ne-ICD. After incubation for 24 h, cells were fixed with DMA and formaldehyde. DNA was digested by micrococcal nuclease, then incubated with an anti-FLAG antibody. Collected DNA was ligated into pCR 2.1-TOPO and picked up colonies were applied for sequencing.
| Locus | Closest gene | Fragment |
|---|---|---|
| 8q21.3 | MMP16 (matrix metallopeptidase 16) | Intron 1 |
| 16p11.2 | MAPK3, ERK1 (mitogen-activated protein kinase 3) | Intron 4 |
| 6q24.1 | GPR126 (G protein-coupled receptor 126) | Intron 23 |
| 9p22.1 | NCKX2, SLC24A2 (sodium/potassium/calcium exchanger 2) | Intron 2 |
| 17p12 | COX10 (cytochrome c oxidase assembly homolog 10) | Intron 5 |
| 14q31-q32.1 | S6K (ribosomal protein S6 kinase) | Intron 7 |
| 14q24.3 | RGS6 (regulator of G-protein signaling 6) | 14 kb 3′ |
| 14q24.2 | DPF3, CERD4 (zinc and double PHD fingers, family 3) | 16 kb 3′ |
| 9q22.3 | PTCH1 (Patched-1) | 70 kb 5′ |
| 9q22.32 | RAD26L2,ERCC6L2 (excision repair cross- complementing rodent repair deficiency, complementation group 6-like 2) | 290 kb 5′ |
| 9p13.1 | ZFN658 (zinc finger protein 658) | 33 kb 5′ |
| 9q12 | ANKRD20A2 (ankyrin repeat domain-containing protein-20A2) | 1540 kb 3′ |
| 8q24.11 | Exostosin-1 (exostosin glycosyltransferase 1) | Intron 2 |
| 6q24.3 | ADGB (androglobin) | 50 kb 3′ |
| 6q24.3 | STXBP5 (syntaxin-binding protein 5, tomosyn) | 330 kb 5′ |
| 17p12 | CMT1A (peripheral myelin protein 22, CDRT1, PMP22) | Intron 12 |
| 9p13.1 | SPATA31A1 (spermatogenesis-associated protein- 31A) | 137 kb 3′ |
FIGURE 9.
Ne-ICD regulates gene expression. A, expression of target genes of Ne-ICD was examined before and after Ne-ICD overexpression. Ne-ICD cDNA was transfected into GD3+ cells (G5) and control cells (V9) using Lipofectamine 2000TM. After incubation for 48 h, total RNA was extracted, and RT-PCR was performed using each primer set of the target gene of Ne-ICD as listed in Table 2. B, a schema to summarize results. a, shows cells without GD3 expression. b, presents enhanced malignant properties in melanoma cells under GD3 expression via Ne-ICD generated in GEM/rafts with γ-secretase. Under GD3 expression, neogenin shifted to lipid/rafts and co-localized with GD3 and presenilin-1 in melanoma cells. The amounts of Ne-ICD were more in the GD3+ cells than in control cells. Exogenously added GD3 also increased the efficiency of neogenin cleavage by γ-secretase. Ne-ICD translocation to nucleus and works as a transcription factor and regulates transcription of multiple genes. Therefore, these molecules seemed to be actual effectors for enhancement of the malignant properties induced under GD3 expression in melanomas. ACTB, β-actin.
Discussion
Ganglioside GD3 has been considered as a melanoma-associated glycolipid, and its expression and roles in a wide variety of biological processes have been rigorously studied. In particular, roles in the enhancement of malignant properties of various cancers and mechanisms for such functions have been reported by our group and others. Because understanding of molecular mechanisms by which membrane glycolipids modulate signals transduced via various membrane receptors was considered to be essential for the application of these antigens for cancer treatment, we have studied mechanisms by which cancer-associated glycolipids enhance signaling pathways leading to the malignant phenotypes of cancer cells (33).
Using a GD3-deficient melanoma cell line N1, it was demonstrated that GD3+ cells undergo increased activation signals such as p130Cas, paxillin, Akt (20), and focal adhesion kinase (21). GD3 expression also resulted in the enhanced cell adhesion to various extracellular matrices (17). Involvement of increased phosphorylation of a Src kinase family, Yes, due to GD3, was also demonstrated in melanomas (18) and in gliomas (34).
Although a number of reports on the functions of cancer-associated glycolipids have been published to date, no mechanisms by which gangliosides directly modulate functions of associating molecules in the vicinity of cell membrane have been reported. To challenge this issue, we have tried to identify GD3-associating molecules using EMARS/MS methods (23). Neogenin was one of the identified candidates as GD3− associated molecules in melanoma cells (24). In the present study we have clarified concrete functions of neogenin in melanomas.
Neogenin is expressed in some normal tissues and plays in critical roles in the regulation of diverse developmental processes (35, 36) and embryogenesis (37). In particular, neogenin has been known as a repulsion receptor for axon guidance (38). As for roles of neogenin in cancer cells, there have been increasing reports on its expression in various cancers (28, 39–44) and on its functions in cancers (28, 44). However, implication of neogenin in cancer phenotypes is now controversial, i.e. neogenin was considered as a suppressor gene as deleted in colorectal cancer (DCC) in breast cancers (42) and gliomas (43), and it was also reported as a cancer-promoting gene in gastric cancers (45), medulloblastomas (40), and esophageal cancers (46).
In this study it was clearly demonstrated that neogenin, particularly its intra-cytoplasmic domain (Ne-ICD), plays as a driver to promote various cancer phenotypes as shown in the MTT (3-(4,5-dimethyl-thiazolyl-2)-2,5-diphenyltetrazolium bromide) assay and invasion assay using overexpressing and/or silenced cells. Furthermore, not only co-localization of neogenin with GD3 in GEM/rafts, but also physical association of neogenin with GD3, could be shown, suggesting a possibility that formation of molecular complex including GD3 and neogenin might be critical for its localization in GEM/rafts and for enhancement of cleavage of Ne-ICD via γ-secretase activation (Fig. 9B).
There were a number of reports to suggest that sphingolipids play critical roles in Alzheimer disease-associated proteins for lipid rafts (47). In deed, there have been some reports on the regulation of secretases by glycosphingolipids. Exogenous GM1 enhanced secretion of soluble Aβ from primary cultures of rat cortical neurons transiently transfected with human APP695 cDNA (48). Stimulation of proteolytic activity of BACE was demonstrated in the presence of cerebrosides, anionic glycerol-phospholipids and cholesterol (49). Furthermore, inhibition of glycosphingolipid synthesis reduced secretion of amyloid precursor protein (APP) and Aβ (50). As for γ-secretase, Holmes et al. (51) reported that gangliosides increased γ-secretase activity and elevated the Ab42:Ab40 ratio by reconstituting purified human γ-secretase into detergent-free proteoliposomes. All these results suggest that ganglioside profiles largely affect the activity of γ-secretase probably on the cell membrane. In the present study it was clearly demonstrated that increased γ-secretase activity was involved in the generation of Ne-ICD under GD3 expression. Co-localization of GD3, neogenin, and γ-secretase at the cell membrane suggested this ternary molecular association exerts generation of Ne-ICD, leading to increased expression of multiple genes and enhance malignant properties of cells (Fig. 9B) as described in HEK293T previously (32), whereas precise mechanisms for the activation of presenilin as a main subunit of γ-secretase in the ternary molecular association remain to be investigated.
Malignant melanomas have been one of the most refractory diseases despite of a number of clinical trials toward complete cure (52, 53). mAbs reactive with GD3 has been expected to open a new dimension in the melanoma therapy (54) and showed some promising results (16, 55) by modulating the molecular forms (56–58). However, it seems hard to achieve complete remission in many melanoma patients. Based on the results obtained in this study, neogenin might be also a target in melanoma therapy. Under expression of GD3, ∼30% of neogenin was localized in GEM/rafts, suggesting that it may associate and cooperate with GD3 to exert enhancement of malignant properties. Therefore, in addition to anti-GD3 mAb, combined use of anti-neogenin antibody might be able to disturb the ternary molecular association and suppress the generation of Ne-ICD, leading to the development of novel therapeutics of melanomas with less adverse effects.
As shown in Fig. 9A, many genes showed increased expression after transfection of a cDNA expression vector of Ne-ICD in both GD3+ and GD3− cells, confirming that these genes were driven by Ne-ICD under GD3 expression (Fig. 9B). GPR126 (G protein-coupled receptor 126) is involved in the angiogenesis by regulating proliferation and migration of endothelial cells (59). STXBP5 is syntaxin-binding protein 5, also named tomosyn. This protein is involved in the formation of SNARE complex, probably regulating secretion of vesicles by forming a complex with syntaxin-1, SNAP-25, and synaptotagmin (60). This function may modulate malignant properties of melanomas. MMP-16 is a member of metallopeptidase family and mediates a proteolytic switch to promote cell-cell adhesion, collagen alignment, and lymphatic invasion in melanoma (61). SPATA31A1 is spermatogenesis-associated protein-31A and was defined as one of resticular tumor-associated genes (62). Because testicular antigens are frequently linked to tumor-associated antigens, this gene may be involved in the natures of melanomas. S6K, ribosomal protein S6 kinase, is a well known downstream kinase of mTOR that regulates protein activation/translation leading to increased cell migration (63), glutamine metabolism (64), and many other physiological and pathological processes (65).
Consequently, we have identified molecules that might actually exert enhancing effects on various malignant properties of melanomas. Actual roles of these gene products in malignant melanomas and precise mechanisms by which Ne-ICD regulates transcription of these genes remain to be investigated.
Experimental Procedures
Antibodies and Reagents
The following antibodies were used in this study. Anti-GD3 mAb R24 was kindly provided by Dr. L. J. Old at Memorial Sloan-Kettering Cancer Center (New York). mAb R24 has been verified to react specifically with ganglioside GD3 containing NeuAc-NeuAc- and NeuAc-NeuGc-type disialyl structures (29). Other antibodies were purchased from commercial sources, i.e. mouse anti-GM3 mAb M2590 was from Cosmo Bio (Tokyo, Japan). Rabbit anti-caveolin-1, goat anti-neogenin, and rabbit anti-presenilin-1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), mouse anti-β-actin was from Sigma, rabbit anti-FLAG was from Cell Signaling Technology (Beverly, MA), sheep anti- mouse IgG antibody conjugated with HRP was from Amersham Biosciences, goat anti-rabbit IgG antibody conjugated with HRP was from Cell Signaling Technology (Beverly, MA), horse anti-goat IgG conjugated with HRP was from Vector Laboratories (Burlingame, CA), and Alexa 488-conjugated donkey anti-rabbit IgG (H+L) secondary antibody, Alexa 594-conjugated donkey anti-mouse IgG (H+L) secondary antibody, and Alexa647-conjugated donkey anti-goat IgG were from Invitrogen. Protein G-Sepharose and protein A-Sepharose beads were purchased from GE Healthcare. Proteasome inhibitor MG132 was from Sigma. γ-Secretase inhibitor (DAPT) was from Peptide Institute, Inc. (Osaka, Japan).
Cell Culture
GD3+ cells and control cells were established as described (20). Namely, SK-MEL-28-derived N1-cell lacking GD3 expression was transfected with cDNA expression plasmid (pMIKneo) with EffecteneTM (Qiagen, Valencia, CA) after single cell cloning with a limiting dilution technique. Consequently, after selection in the medium containing G418 (400 μg/ml), all clones were screened on GD3 expression using flow cytometry as described below. GD3-positive transfectant cells such as G5 and G11 and GD3-negative sublines such as V4 and V9 were established as derivatives from a common parent N1 clone. They were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 7.5% fetal calf serum (FCS) at 37 °C in a humidified atmosphere containing 5% CO2. Stable transfectants were maintained in the presence of 400 μg/ml G418 (Sigma). Expression of GD3 has been checked every a few months.
Flow Cytometry
Cell surface expression of GD3 was analyzed by flow cytometry (BD Biosciences). Cells were incubated with mAb R24 for 45 min on ice and then stained with FITC-labeled anti-mouse IgG antibody. Control samples were prepared by using non-relevant mAb with the same subclass.
RT-Quantitative PCR
RNAs were extracted using TRIzolTM reagent (Ambion by Life Technologies). Then, cDNA was generated using oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Invitrogen). quantitative PCR was performed using SsoAdvanced™ Universal SYBR Green SupermixTM (Bio-Rad) and CFX ConnectTM Real-Time System (Bio-Rad). Primers used in this study were listed in Table 2.
TABLE 2.
A list of primers used in this study
| Gene | Forward | Reverse |
|---|---|---|
| Neogenin | CTGGAAGGCGAGGAATGAGA, | TGTTTGAGACGAAGAGGCTG |
| GAPDH | ACTTCAACAGCGACACCCAC, | CAACTGTGAGGAGGGGAGAT |
| GPR126 | CAACCCAAGGGACCTCTCAC, | AAGCCAAGCTGGGTAATGCT |
| SPATA31A1 | TCAAAGAAAAAAAGCAAGCC, | ATTCTGCCGTGTTCTGAGTA |
| RGS6 | GCTTCTTCCCTGGCAGGATT, | CTTTTTGAGGCTGGTGGTGC |
| MMP16 | ATGTACGCAAGGGCCAAGA, | AACCCTCCCACAAGCAAACA |
| STXBP5 | ATCGGAGAGAACCCCGATCT, | TTCTGCTTTTGGTCTTCACCT |
| S6K | TGCAGCAGCCTACTGAGAAC, | TTGGTCAGGCAGTCAACACT |
| COX10 | CTGGATTTGCATTGGCTCCG, | CAAGTGGCAAAGGACACAGC |
| DPF3 | CTGAAAGCGCTCGGGGAC, | GTGCTCTCTGAGGTGAACCC |
| MAPK3 | TACCTACAGTCTCTGCCCTCC, | TGCTGTCTCCTGGAAGATGAG |
| ZNF658 | CCGTTTATTCAGGTGGGGCT, | ACACAGGACCCAAGTTTGCT |
| RAD26L2 | TGTCGGTGCCAATGTTGTTG, | AAGAAGTTCACACCTCCAGGAT |
| ANKD20A2 | GCTCAATGCCGAACTGTTGA, | GCTGTGTTTGGCTTAGGTCG |
| EXT1 | GAGGACGTGGGGTTTGACAT, | TTTGCCAGTCTTTGCCATGC |
| ADGB | TTAGGAAGCCCAGACTCCCA, | GCTCTTCCTGGCTCAGTTGT |
| CMT1 | TTTTCAGTGGTGTGTAGCAG, | GCAGTGTGATTGAGTTGAGT |
Construction of a cDNA Expression Vector
Human Ne-ICD cDNA was amplified using primers containing restriction sites for PstI and HindIII by KOD FX (TOYOBO, Tokyo, Japan). After gel purification, TA cloning was performed, and the product was ligated into pCR 2.1-TOPOTM vector (Invitrogen) Then, Ne-ICD was ligated into pCMV2B (Stratagene, La Jolla, CA) between PstI and HindIII sites. A mutated Ne-ICD (L1455A) was generated using a KOD-Plus-MutagenesisTM kit (TOYOBO).
Gene Transfection and Neogenin Knockdown
Cells (2 × 105) were plated in a 60-mm plastic plate (Greiner Bio-one, Frickenhausen, Germany) and incubated for 24 h. The Ne-ICD (L1455A) expression vector (4 μg) or anti-neogenin siRNAs were transfected into 1 × 106 cells using Lipofectamine 2000TM reagent (Invitrogen). The sequence of siRNAs for neogenin was 5′-CAUCAUGACUGAUACUCCATT-3′. This siRNA was purchased from Sigma. Effects of overexpression of Ne-ICD or down-regulation of neogenin were assessed at 48–144 h after the transfection by Western immunoblotting. For invasion assay and MTT assay, cells were collected at 48 h after the transfection.
Western Immunoblotting
Cells (2 × 105) were plated in a 60-mm plastic plate (Greiner Bio-one) and incubated for 24 h in DMEM supplemented with 7.5% FCS. After washing 3 times with PBS, cells were lysed by cell lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin) (Cell Signaling Technology Inc., Danvers, MA) supplemented with protease inhibitor mixture (Calbiochem) and I mm PMSF. Insoluble materials were removed by centrifugation at 10,000 × g for 10 min. Cell lysates (5–20 μg) were separated by SDS-PAGE using 6–15% gels. The separated proteins were transferred onto an Immobilon PTM membrane (Millipore, Billerica, MA). Blots were blocked with 5% BSA in PBS containing 0.05% Tween 20 for 1 h at room temperature. The membrane was probed with 1st antibody. After being washed with PBS containing 0.05% Tween 20, the blots were incubated with a goat anti-mouse IgG, a goat anti-rabbit IgG, or a rabbit anti-goat IgG conjugated with HRP for 45 min to 12 h. Conjugates were visualized with an Enhanced ChemiluminescenceTM detection system (PerkinElmer Life Sciences).
Immunoprecipitation
Cells were lysed with 500 μl of cell lysis buffer. Lysates were bounced 10 times with Digital HomogenizerTM (AsOne, Osaka, Japan). After removing insoluble materials by centrifugation at 10,000 × g for 10 min, lysates were incubated with an anti-neogenin antibody (2 μg) for 12 h at 4 °C with rotation. Then, protein G-Sepharose or protein A-Sepharose was added and rotated for 2 h at 4 °C. The beads were washed 3 times with immunoprecipitation washing buffer (50 mm Tris·HCl, pH 7.4, 150 mm NaCl, 0.5% Triton X-100, 1 mm Na3VO4). Finally, the precipitates were analyzed by Western immunoblotting.
Immunocytochemistry
Cells (5 × 103) were plated on a glass-bottom dish (Iwaki, Tokyo, Japan) and incubated for 24 h in DMEM supplemented with 7.5% FCS. After washing three times with PBS, cells were fixed with paraformaldehyde (4% in PBS) for 10 min at room temperature and then incubated with Triton X-100 (0.1% in PBS) for 10 min at room temperature. Nonspecific binding was blocked with BSA (2.5% in PBS) for 12 h at 4 °C. Cells were incubated with an anti-presenilin-1, an anti-caveolin-1, and anti-neogenin antibodies or mAb R24 in PBS containing 0.5% BSA for 12 h at 4 °C. After washing with PBS, cells were incubated with an Alexa FluorTM 488-conjugated donkey anti-rabbit IgG, an Alexa FluorTM 594-conjugated donkey anti-mouse IgG, or an Alexa FluorTM 647-conjugated donkey anti-goat IgG for 12 h at 4°C. Then cells were imaged using a confocal microscope (Fluoview FV10TM, Olympus, Tokyo, Japan).
Fractionation of GEM/Rafts
Cells (3 × 106) were washed with PBS 3 times and lysed with the lysis buffer. Lysates were Dounce-homogenized 10 times with a Digital HomogenizerTM (AS One, Osaka, Japan). After removing insoluble materials by centrifugation, lysates were mixed with an equal volume of 80% sucrose in TNE buffer (25 mm Tris·HCl, 1 mm EDTA, 150 mm NaCl, 1 mm Na3VO4). Then 30% sucrose and 5% sucrose in TNE buffer were overlaid sequentially. The gradient was formed by ultracentrifugation for 16 h at 4 °C at 100,000 × g using a MLS50 rotor (Beckman Coulter Inc., Brea, CA). After ultracentrifugation, 0.5 ml each of fraction was collected from the top of the gradient to yield 10 fractions and used for Western immunoblotting.
ChIP
Cells (4 × 106) were plated in a 15-cm dish (Greiner, Bio-one), and FLAG-Ne-ICD (L1455A) cDNA was transfected. Adherent cells were fixed by dimethyl adipimidate-2 HCl (2.5 mg/ml) in PBS for 5 min at room temperature. Formaldehyde (final 1%) was added and incubated for 10 min at room temperature. Glycine was added to the cells to stop fixation. After washing with PBS, fixed cells were scraped and pelleted by centrifugation at 1500 rpm. Cells were resuspended in ice-cold buffer A and pelleted by centrifugation at 3000 rpm. Cells were resuspended in 200 μl of ice-cold buffer B, and DNA was digested by micrococcal nuclease (0.5 μl/immunoprecipitation) for 20 min at 37 °C. EDTA was added to stop the digest, and cells were pelleted by centrifugation at 13,000 rpm. ChIP buffer was added to the pellet and incubated for 10 min at 4 °C and sonicated by Sonicator W-375 Cell DisruptorTM (Heat Systems-Ultrasonics, Inc., Plainview, NY).
Cell extracts were incubated overnight at 4 °C with anti-FLAG antibody or normal mouse IgG (5 μg) as a negative control and then precipitated by protein G-Sepharose. Beads were washed with low and high salt solution and eluted by ChIP elution buffer (Cell Signaling Technology). Cross-linking of DNA and proteins were reversed by thermomixer (Eppendorf, Tokyo, Japan) (1200 rpm) for 30 min at 65 °C. After digesting proteins by proteinase K for 2 h at 65 °C, DNA was purified by DNA purification spin column (Cell Signaling Technology) followed by cloning/sequencing.
Cloning and Sequencing of ChIPed-DNA
Taq-polymerase and dNTPs were added into purified DNA and incubated at 60 °C for 10 min to add adenine nucleotide. The DNA fragment was ligated to pCR 2.1-TOPO vector (Invitrogen) and transformed into DH5α competent cells (TOYOBO). Then lysates were incubated for 12 h on the X-Gal (5-bromo-4-chloro-3-indolyl- β-d-galactoside) containing LB plate. X-gal-positive colonies were picked up, and plasmids were purified then sequenced by M13 reverse primer (cag gaa aca gct atg ac).
Invasion Assay and MTT Assay
Invasion and MTT assays were performed as described previously (20).
Exogenous Addition of GD3 to GD3− Cells
Cells (2 × 105) were plated in a 60-mm plastic plate and incubated for 24 h in DMEM supplemented with 7.5% FCS. GD3 (≧98% purity) (Cayman Chemical, Ann Arbor, MI) was solubilized (1 mg/ml) in a mixture of methanol-chloroform (1:1) and dried in a grass tube by N2 gas stream. Then it was resuspended in DMEM supplemented with insulin, transferrin, selenium (BD Biosciences) by vortexing and sonication with a bath-type sonicator (BransonicTM, Branson M5800-J, Emerson-Japan, Kanagawa, Japan) for 10 min in the ice-cold water. This GD3 suspension medium (0, 5, 10, 20 nmol/ml) was added after removing old medium and washing twice with DMEM to cells. After incubation for 48 h, cells were used for Western immunoblotting.
Statistics
Mean values were compared using an unpaired Student's two-tailed t test; significance was set at p > 0.05 (*) and p > 0.01 (**).
Author Contributions
K. K., Y. Ohkawa, and Y. Ohmi performed the gene expression analysis, cell function analysis, immunoblotting, and immunoprecipitation analysis. Keiko Furukawa performed immunocytostaining. M. O. and T. O. performed ChIP-sequencing analysis to identify the targets of Ne-ICD. N. H., K. H., and N. K. performed the EMARS/MS analysis and fractionation of Triton X-100 extracts. Koichi Furukawa designed the experiments and supervised and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank T. Mizuno and Y. Nakayasu for technical assistance.
This work was supported by Grants-in-aid 15H04696, 15K15080, 25670141, and 24390078 from the Ministry of Education, Culture, Sports, and Technology of Japan (MEXT). This work was also supported by a scholarship from Nakamura-Sekizen-kai. The authors declare that they have no conflicts of interest with the contents of this article.
- GEM
- glycolipid-enriched microdomain
- EMARS
- enzyme-mediated activation of radical sources
- Ne-ICD
- neogenin intracytoplasmic domain
- DAPT
- N-[N-[(3,5-difluorophenyl)acetyl]-l-alanyl]-l-phenyl-glycine tert-butyl ester
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Hakomori S. I., and Murakami W. T. (1968) Glycolipids of hamster fibroblasts and derived malignant-transformed cell lines. Proc. Natl. Acad. Sci. U.S.A. 59, 254–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lloyd K. O., and Old L. J. (1989) Human monoclonal antibodies to glycolipids and other carbohydrate antigens: dissection of the humoral immune response in cancer patients. Cancer Res. 49, 3445–3451 [PubMed] [Google Scholar]
- 3. Chang H. R., Cordon-Cardo C., Houghton A. N., Cheung N. K., and Brennan M. F. (1992) Expression of disialogangliosides GD2 and GD3 on human soft tissue sarcomas. Cancer 70, 633–638 [DOI] [PubMed] [Google Scholar]
- 4. Shibuya H., Hamamura K., Hotta H., Matsumoto Y., Nishida Y., Hattori H., Furukawa K., and Ueda M. (2012) Enhancement of malignant properties of human osteosarcoma cells with disialyl gangliosides GD2/GD3. Cancer Sci. 103, 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheresh D. A., Rosenberg J., Mujoo K., Hirschowitz L., and Reisfeld R. A. (1986) Biosynthesis and expression of the disialoganglioside GD2, a relevant target antigen on small cell lung carcinoma for monoclonal antibody-mediated cytolysis. Cancer Res. 46, 5112–5118 [PubMed] [Google Scholar]
- 6. Yoshida S., Fukumoto S., Kawaguchi H., Sato S., Ueda R., and Furukawa K. (2001) Ganglioside G(D2) in small cell lung cancer cell lines: enhancement of cell proliferation and mediation of apoptosis. Cancer Res. 61, 4244–4252 [PubMed] [Google Scholar]
- 7. Siddiqui B., Buehler J., DeGregorio M. W., and Macher B. A. (1984) Differential expression of ganglioside GD3 by human leukocytes and leukemia cells. Cancer Res. 44, 5262–5265 [PubMed] [Google Scholar]
- 8. Merritt W. D., Casper J. T., Lauer S. J., and Reaman G. H. (1987) Expression of GD3 ganglioside in childhood T-cell lymphoblastic malignancies. Cancer Res. 47, 1724–1730 [PubMed] [Google Scholar]
- 9. Furukawa K., Akagi T., Nagata Y., Yamada Y., Shimotohno K., Cheung N. K., and Shiku H. (1993) GD2 ganglioside on human T-lymphotropic virus type I-infected T cells: possible activation of β-1,4-N-acetylgalactosaminyltransferase gene by p40tax. Proc. Natl. Acad. Sci. U.S.A. 90, 1972–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung N. K., Saarinen U. M., Neely J. E., Landmeier B., Donovan D., and Coccia P. F. (1985) Monoclonal antibodies to a glycolipid antigen on human neuroblastoma cells. Cancer Res. 45, 2642–2649 [PubMed] [Google Scholar]
- 11. Hakomori S. (1991) New directions in cancer therapy based on aberrant expression of glycosphingolipids: anti-adhesion and ortho-signaling therapy. Cancer Cells 3, 461–470 [PubMed] [Google Scholar]
- 12. Carubia J. M., Yu R. K., Macala L. J., Kirkwood J. M., and Varga J. M. (1984) Gangliosides of normal and neoplastic human melanocytes. Biochem. Biophys. Res. Commun. 120, 500–504 [DOI] [PubMed] [Google Scholar]
- 13. Portoukalian J., Zwingelstein G., Abdul-Malak N., and Doré J. F. (1978) Alteration of gangliosides in plasma and red cells of humans bearing melanoma tumors. Biochem. Biophys. Res. Commun. 85, 916–920 [DOI] [PubMed] [Google Scholar]
- 14. Pukel C. S., Lloyd K. O., Travassos L. R., Dippold W. G., Oettgen H. F., and Old L. J. (1982) GD3, a prominent ganglioside of human melanoma: detection and characterisation by mouse monoclonal antibody. J. Exp. Med. 155, 1133–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dippold W. G., Dienes H. P., Knuth A., and Meyer zum Büschenfelde K. H. (1985) Immunohistochemical localization of ganglioside GD3 in human malignant melanoma, epithelial tumors, and normal tissues. Cancer Res. 45, 3699–3705 [PubMed] [Google Scholar]
- 16. Houghton A. N., Mintzer D., Cordon-Cardo C., Welt S., Fliegel B., Vadhan S., Carswell E., Melamed M. R., Oettgen H. F., and Old L. J. (1985) Mouse monoclonal IgG3 antibody detecting GD3 ganglioside: a phase I trial in patients with malignant melanoma. Proc. Natl. Acad. Sci. U. S. A. 82, 1242–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohkawa Y., Miyazaki S., Hamamura K., Kambe M., Miyata M., Tajima O., Ohmi Y., Yamauchi Y., Furukawa K., and Furukawa K. (2010) Ganglioside GD3 enhances adhesion signals and augments malignant properties of melanoma cells by recruiting integrins to glycolipid-enriched microdomains. J. Biol. Chem. 285, 27213–27223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamamura K., Tsuji M., Hotta H., Ohkawa Y., Takahashi M., Shibuya H., Nakashima H., Yamauchi Y., Hashimoto N., Hattori H., Ueda M., Furukawa K., and Furukawa K. (2011) Functional activation of Src family kinase yes protein is essential for the enhanced malignant properties of human melanoma cells expressing ganglioside GD3. J. Biol. Chem. 286, 18526–18537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Furukawa K., Kambe M., Miyata M., Ohkawa Y., Tajima O., and Furukawa K. (2014) Ganglioside GD3 induces convergence and synergism of adhesion and hepatocyte growth factor/Met signals in melanomas. Cancer Sci. 105, 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hamamura K., Furukawa K., Hayashi T., Hattori T., Nakano J., Nakashima H., Okuda T., Mizutani H., Hattori H., Ueda M., Urano T., Lloyd K. O., and Furukawa K. (2005) Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proc. Natl. Acad. Sci. U. S. A. 102, 11041–11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamamura K., Tsuji M., Ohkawa Y., Nakashima H., Miyazaki S., Urano T., Yamamoto N., Ueda M., Furukawa K., and Furukawa K. (2008) Focal adhesion kinase as well as p130Cas and paxillin is crucially involved in the enhanced malignant properties under expression of ganglioside GD3 in melanoma cells. Biochim. Biophys. Acta 1780, 513–519 [DOI] [PubMed] [Google Scholar]
- 22. Hakomori S. I. (2000) Cell adhesion/recognition and signal transduction through glycosphingolipid microdomain. Glycoconj. J. 17, 143–151 [DOI] [PubMed] [Google Scholar]
- 23. Kotani N., Gu J., Isaji T., Udaka K., Taniguchi N., and Honke K. (2008) Biochemical visualization of cell surface molecular clustering in living cells. Proc. Natl. Acad. Sci. U. S. A. 105, 7405–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hashimoto N., Hamamura K., Kotani N., Furukawa K., Kaneko K., Honke K., and Furukawa K. (2012) Proteomic analysis of ganglioside-associated membrane molecules: substantial basis for molecular clustering. Proteomics 12, 3154–3163 [DOI] [PubMed] [Google Scholar]
- 25. Vielmetter J., Kayyem J. F., Roman J. M., and Dreyer W. J. (1994) Neogenin, an avian cell surface protein expressed during terminal neuronal differentiation, is closely related to the human tumor suppressor molecule deleted in colorectal cancer. J. Cell Biol. 127, 2009–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Monnier P. P., Sierra A., Macchi P., Deitinghoff L., Andersen J. S., Mann M., Flad M., Hornberger M. R., Stahl B., Bonhoeffer F., and Mueller B. K. (2002) RGM is a repulsive guidance molecule for retinal axons. Nature 419, 392–395 [DOI] [PubMed] [Google Scholar]
- 27. Nikolopoulos S. N., and Giancotti F. G. (2005) Netrin-integrin signaling in epithelial morphogenesis, axon guidance and vascular patterning. Cell Cycle 4, e131–e135 [PubMed] [Google Scholar]
- 28. Meyerhardt J. A., Look A. T., Bigner S. H., and Fearon E. R. (1997) Identification and characterization of neogenin, a DCC-related gene. Oncogene 14, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 29. Furukawa K., Yamaguchi H., Oettgen H.F., Old L.J., and Lloyd K. O. (1989) Human monoclonal antibodies reacting with the major gangliosides of human melanomas and comparison with corresponding mouse monoclonal antibodies. Cancer Res. 49, 191–196 [PubMed] [Google Scholar]
- 30. Vetrivel K. S., Cheng H., Lin W., Sakurai T., Li T., Nukina N., Wong P. C., Xu H., and Thinakaran G. (2004) Association of γ-secretase with lipid rafts in post-Golgi and endosome membranes. J. Biol. Chem. 279, 44945–44954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Urano Y., Hayashi I., Isoo N., Reid P. C., Shibasaki Y., Noguchi N., Tomita T., Iwatsubo T., Hamakubo T., and Kodama T. (2005) Association of active γ-secretase complex with lipid rafts. J. Lipid Res. 46, 904–912 [DOI] [PubMed] [Google Scholar]
- 32. Goldschneider D., Rama N., Guix C., and Mehlen P. (2008) The neogenin intracellular domain regulates gene transcription via nuclear translocation. Mol. Cell. Biol. 28, 4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Furukawa K., Hamamura K., Ohkawa Y., Ohmi Y., and Furukawa K. (2012) Disialyl gangliosides enhance tumor phenotypes with differential modalities. Glycoconj. J. 29, 579–584 [DOI] [PubMed] [Google Scholar]
- 34. Ohkawa Y., Momota H., Kato A., Hashimoto N., Tsuda Y., Kotani N., Honke K., Suzumura A., Furukawa K., Ohmi Y., Natsume A., Wakabayashi T., and Furukawa K. (2015) Ganglioside GD3 enhances invasiveness of gliomas by forming a complex with platelet-derived growth factor receptor α and Yes kinase. J. Biol. Chem. 290, 16043–16058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cole S. J., Bradford D., and Cooper H. M. (2007) Neogenin: a multi-functional receptor regulating diverse developmental processes. Int. J. Biochem. Cell Biol. 39, 1569–1575 [DOI] [PubMed] [Google Scholar]
- 36. Wilson N. H., and Key B. (2007) Neogenin: one receptor, many functions. Int. J. Biochem. Cell Biol. 39, 874–878 [DOI] [PubMed] [Google Scholar]
- 37. Gad J. M., Keeling S. L., Wilks A. F., Tan S. S., and Cooper H. M. (1997) The expression patterns of guidance receptors, DCC and Neogenin, are spatially and temporally distinct throughout mouse embryogenesis. Dev. Biol. 192, 258–273 [DOI] [PubMed] [Google Scholar]
- 38. Braisted J. E., Catalano S. M., Stimac R., Kennedy T. E., Tessier-Lavigne M., Shatz C. J., and O'Leary D. D. (2000) Netrin-1 promotes thalamic axon growth and is required for proper development of the thalamocortical projection. J. Neurosci. 20, 5792–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu X., Li Y., Wan X., Kayira T. M., Cao R., Ju X., Zhu X., and Zhao G. (2012) Down-regulation of neogenin accelerated glioma progression through promoter methylation and its overexpression in SHG-44 induced apoptosis. PLoS ONE 7, e38074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Milla L. A., Arros A., Espinoza N., Remke M., Kool M., Taylor M. D., Pfister S. M., Wainwright B. J., and Palma V. (2014) Neogenin1 is a Sonic Hedgehog target in medulloblastoma and is necessary for cell cycle progression. Int. J. Cancer 134, 21–31 [DOI] [PubMed] [Google Scholar]
- 41. Akino T., Han X., Nakayama H., McNeish B., Zurakowski D., Mammoto A., Klagsbrun M., and Smith E. (2014) Netrin-1 promotes medulloblastoma cell invasiveness and angiogenesis, and demonstrates elevated expression in tumor tissue and urine of patients with pediatric medulloblastoma. Cancer Res. 74, 3716–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Q., Liang F., Ke Y., Huo Y., Li M., Li Y., and Yue J. (2015) Overexpression of neogenin inhibits cell proliferation and induces apoptosis in human MDA-MB-231 breast carcinoma cells. Oncol. Rep. 34, 258–264 [DOI] [PubMed] [Google Scholar]
- 43. Xing W., Li Q., Cao R., and Xu Z. (2014) Neogenin expression is inversely associated with breast cancer grade in ex vivo. World J. Surg. Oncol. 12, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lai Wing Sun K., Correia J. P., and Kennedy T. E. (2011) Netrins: versatile extracellular cues with diverse functions. Development 138, 2153–2169 [DOI] [PubMed] [Google Scholar]
- 45. Kim S. J., Wang Y. G., Lee H. W., Kang H. G., La S. H., Choi I. J., Irimura T., Ro J. Y., Bresalier R. S., and Chun K. H. (2014) Up-regulation of neogenin-1 increases cell proliferation and motility in gastric cancer. Oncotarget 5, 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu Y. C., Lam K. Y., Law S., Wong J., and Srivastava G. (2001) Identification of differentially expressed genes in esophageal squamous cell carcinoma (ESCC) by cDNA expression array: overexpression of Fra-1, Neogenin, Id-1, and CDC25B genes in ESCC. Clin. Cancer Res. 7, 2213–2221 [PubMed] [Google Scholar]
- 47. van Echten-Deckert G., and Walter J. (2012) Sphingolipids: critical players in Alzheimer's disease. Prog. Lipid Res. 51, 378–393 [DOI] [PubMed] [Google Scholar]
- 48. Zha Q., Ruan Y., Hartmann T., Beyreuther K., and Zhang D. (2004) GM1 ganglioside regulates the proteolysis of amyloid precursor protein. Mol. Psychiatry 9, 946–952 [DOI] [PubMed] [Google Scholar]
- 49. Kalvodova L., Kahya N., Schwille P., Ehehalt R., Verkade P., Drechsel D., and Simons K. (2005) Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J. Biol. Chem. 280, 36815–36823 [DOI] [PubMed] [Google Scholar]
- 50. Tamboli I. Y., Prager K., Barth E., Heneka M., Sandhoff K., and Walter J. (2005) Inhibition of glycosphingolipid biosynthesis reduces secretion of the β-amyloid precursor protein and amyloid β-peptide. J. Biol. Chem. 280, 28110–28117 [DOI] [PubMed] [Google Scholar]
- 51. Holmes O., Paturi S., Ye W., Wolfe M. S., and Selkoe D. J. (2012) Effects of membrane lipids on the activity and processivity of purified γ-secretase. Biochemistry 51, 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wargo J. A., Reuben A., Cooper Z. A., Oh K. S., and Sullivan R. J. (2015) Immune effects of chemotherapy, radiation, and targeted therapy and opportunities for combination with immunotherapy. Semin Oncol. 42, 601–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Johnson D. B., and Sosman J. A. (2015) Therapeutic advances and treatment options in metastatic melanoma. JAMA Oncol. 1, 380–386 [DOI] [PubMed] [Google Scholar]
- 54. Dippold W. G., Knuth A., and Meyer zum Büschenfelde K. H. (1984) Inhibition of human melanoma cell growth in vitro by monoclonal anti-GD3-ganglioside antibody. Cancer Res. 44, 806–810 [PubMed] [Google Scholar]
- 55. Kirkwood J. M., Mascari R. A., Edington H. D., Rabkin M. S., Day R. S., Whiteside T. L., Vlock D. R., and Shipe-Spotloe J. M. (2000) Analysis of therapeutic and immunologic effects of R(24) anti-GD3 monoclonal antibody in 37 patients with metastatic melanoma. Cancer 88, 2693–2702 [PubMed] [Google Scholar]
- 56. Scott A. M., Lee F. T., Hopkins W., Cebon J. S., Wheatley J. M., Liu Z., Smyth F. E., Murone C., Sturrock S., MacGregor D., Hanai N., Inoue K., Yamasaki M., Brechbiel M. W., Davis I. D., et al. (2001) Specific targeting, biodistribution, and lack of immunogenicity of chimeric anti-GD3 monoclonal antibody KM871 in patients with metastatic melanoma: results of a phase I trial. J. Clin. Oncol. 19, 3976–3987 [DOI] [PubMed] [Google Scholar]
- 57. Forero A., Shah J., Carlisle R., Triozzi P. L., LoBuglio A. F., Wang W. Q., Fujimori M., and Conry R. M. (2006) A phase I study of an anti-GD3 monoclonal antibody, KW-2871, in patients with metastatic melanoma. Cancer Biother. Radiopharm. 21, 561–568 [DOI] [PubMed] [Google Scholar]
- 58. Lo A. S., Ma Q., Liu D. L., and Junghans R. P. (2010) Anti-GD3 chimeric sFv-CD28/T-cell receptor ζ designer T cells for treatment of metastatic melanoma and other neuroectodermal tumors. Clin. Cancer Res. 16, 2769–2780 [DOI] [PubMed] [Google Scholar]
- 59. Cui H., Wang Y., Huang H., Yu W., Bai M., Zhang L., Bryan B. A., Wang Y., Luo J., Li D., Ma Y., and Liu M. (2014) GPR126 protein regulates developmental and pathological angiogenesis through modulation of VEGFR2 receptor signaling. J. Biol. Chem. 289, 34871–34885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Katoh M., and Katoh M. (2004) Identification and characterization of human LLGL4 gene and mouse Llgl4 gene in silico. Int. J. Oncol. 24, 737–742 [PubMed] [Google Scholar]
- 61. Tatti O., Gucciardo E., Pekkonen P., Holopainen T., Louhimo R., Repo P., Maliniemi P., Lohi J., Rantanen V., Hautaniemi S., Alitalo K., Ranki A., Ojala P. M., Keski-Oja J., and Lehti K. (2015) MMP16 mediates a proteolytic switch to promote cell-cell adhesion, collagen alignment, and lymphatic invasion in melanoma. Cancer Res. 75, 2083–2094 [DOI] [PubMed] [Google Scholar]
- 62. Luk J. M., Lee N. P., Shum C. K., Lam B. Y., Siu A. F., Che C. M., Tam P. C., Cheung A. N., Yang Z. M., Lin Y. N., Matzuk M. M., Lee K. F., and Yeung W. S. (2006) Acrosome-specific gene AEP1: identification, characterization and roles in spermatogenesis. J. Cell. Physiol. 209, 755–766 [DOI] [PubMed] [Google Scholar]
- 63. Raimondi C., and Falasca M. (2011) Targeting PDK1 in cancer. Curr. Med. Chem. 18, 2763–2769 [DOI] [PubMed] [Google Scholar]
- 64. Csibi A., Lee G., Yoon S. O., Tong H., Ilter D., Elia I., Fendt S. M., Roberts T. M., and Blenis J. (2014) The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr. Biol. 24, 2274–2280 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65. Chung J., Grammer T. C., Lemon K. P., Kazlauskas A., and Blenis J. (1994) PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature 370, 71–75 [DOI] [PubMed] [Google Scholar]