Abstract
KatA is the major catalase required for hydrogen peroxide (H2O2) resistance and acute virulence in Pseudomonas aeruginosa PA14, whose transcription is driven from the promoter (katAp1) located at 155 nucleotide (nt) upstream of the start codon. Here, we identified another promoter (katAp2), the +1 of which was mapped at the 51 nt upstream of the start codon, which was responsible for the basal transcription during the planktonic culture and down-regulated upon H2O2 treatment under the control by the master regulator of anaerobiosis, Anr. To dissect the roles of the dual promoters in conditions involving KatA, we created the promoter mutants for each -10 box (p1m, p2m, and p1p2m) and found that katAp1 is required for the function of KatA in the logarithmic growth phase during the planktonic culture as well as in acute virulence, whereas katAp2 is required for the function of KatA in the stationary phase as well as in the prolonged biofilm culture. This dismantling of the dual promoters of katA sheds light on the roles of KatA in stress resistance in both proliferative and growth-restrictive conditions and thus provides an insight into the regulatory impacts of the major catalase on the survival strategies of P. aeruginosa.
Pseudomonas aeruginosa is a non-fermentative bacterial pathogen that respires on oxygen as well as nitrogen oxides, and its mode of respiration is likely associated with the infection strategies: P. aeruginosa is capable of rapid colonization primarily through aerobic respiration during acute infections, which involve various virulence factors1; in contrast, P. aeruginosa survives persistently as a result of chronic infections promoted by biofilm mode of growth, which involve microaerobic or anaerobic respiration on nitrate (NO3−) under the hypoxic condition within the microbial population2. Thus, the characteristics of both infection strategies that P. aeruginosa exploits are quite distinct and known to be associated with a subset of virulence or persistence factors that should be orchestrated to support the survival of P. aeruginosa during each mode of interaction with the host environments3. For example, invasive functions such as secreted toxins and adhesion factors are required for acute virulence, but detrimental for chronic infection or persistence, depending on their cost-benefits4. This is demonstrated by accumulation of the mutations in the invasive functions over time, once infection is established5.
We previously reported the role of the major catalase, KatA in peroxide resistance, osmoprotection, and acute virulence of a P. aeruginosa strain6. Unlike other major bacterial catalases, KatA is highly stable and found in the extracellular milieu, which ensures the survival of P. aeruginosa cells in their biofilm7. The basal expression of KatA is relatively high, but its expression is further increased upon stationary growth phase and in response to H2O2 under the control of OxyR, the master regulator in response to H2O2. OxyR normally binds to the upstream region of the katA promoter as a functional repressor and switches to the activator upon H2O2 challenge8,9, where C199 has a critical role in both functions and in acute virulence10. Recently, KatA was shown to play a critical role in nitric oxide buffering under anaerobic respiration conditions, upon which KatA expression was increased by Anr, the master regulator of anaerobiosis required for dissimilatory NO3− reduction11. It has been suggested by Trunk et al.12 that KatA may belong to the Anr regulon, with a presumable Anr box located upstream of the KatA initiation codon. Although much has been being unveiled about the physiological roles of KatA and the regulatory mechanisms that involve OxyR and its cis-element of the katA promoter, more needs to be clarified to account for the involvement of Anr and the elevated expression of KatA upon anaerobic respiration for its roles to ensure the distinct infection strategies of P. aeruginosa.
In the present study, we have explored the regulatory module underlying the anaerobic regulation of KatA, by newly identifying a second promoter (katAp2) of the katA gene, which is responsible for the basal expression of KatA during the planktonic growth of P. aeruginosa strain PA14. This second promoter is regulated primarily by Anr. More importantly, the mutation analyses of each promoter revealed the separate roles of the dual promoters to cope with reactive oxygen and/or nitrogen species (ROS and RNS), which is dependent on the growth states.
Results
Identification of a second promoter (katAp2) of the katA gene
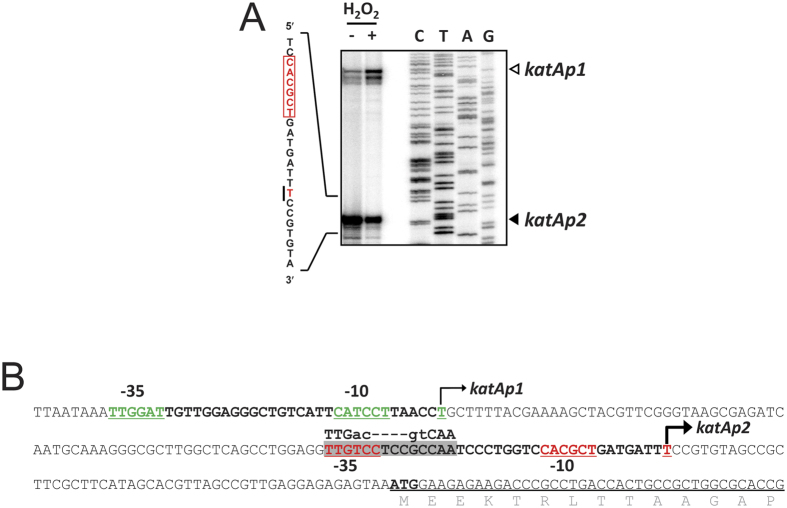
A transcription start site of the katA gene was identified at the 155 nucleotide (nt) upstream (T-155) of the katA initiation codon, whose transcription is induced upon H2O2 treatment. Despite over 5-fold induction of katAp1 transcription upon H2O2 exposure, the katA-lacZ fusion displayed a minor (~2-fold) induction8, which may be attributed to another level of regulation. Based on this speculation, we have identified another promoter of the katA gene, katAp2: from the high-resolution S1 nuclease mapping data, a second protective band was observed at the 51 nt upstream (T-51) of the translation initiation codon (Fig. 1A). The upstream of the position of T-51 could be a potential promoter by its context. It is of marked interest that the putative Anr box consensus (TTGac-N4-gtCAA) was identified, which overlaps with the potential −35 box of katAp2 (Fig. 1B), since KatA expression is deemed reduced in the anr mutant as previously assessed by the transcriptome and proteome analyses12. Identification of the katAp2 promoter and the potential Anr box suggests that the anr-dependent regulation of KatA might be most likely attributed to this second promoter. Moreover, the katAp2 transcript was slightly reduced upon H2O2 treatment (Fig. 1A; see below), which is in a good agreement with the fact that the Fe-S center of Anr is vulnerable to ROS and RNS with its transactivation activity compromised in the presence of O2−, H2O2, and NO13,14,15.
Figure 1. Promoter region of the katA gene.
(A) High-resolution S1 nuclease mapping. The 5′ end of the katA mRNA transcript was determined by high-resolution S1 nuclease mapping. Total RNA (50 μg) that had been prepared from the cells with (+) or without (−) 1 mM H2O2 treatment for 10 min at OD600 of 1.0 in LB were subjected to S1 nuclease mapping. The sequencing reactions (lanes C, T, A and G) were carried out as described in Methods. The two transcriptional start sites of the katA gene are indicated by open (katAp1) and closed (katAp2) arrows. The detailed transcriptional start site of katAp2 promoter is shown on the left by solid line, and its putative −10 box is enclosed in box. (B) Promoter elements of the katA gene. The sequence of the katA promoter region of the katA gene is shown, with the two 5′ ends of katAp1 and katAp2 promoters indicated by the bent arrows. The Anr binding consensus (TTGac-N4-gtCAA) is designated, whose center is located at −29 position from the katAp2 promoter. The putative −10 and −35 boxes are underlined. The N-terminal amino acid sequences of the katA gene are designated.
Regulation of katAp2 by Anr during aerobic NO3 − respiration
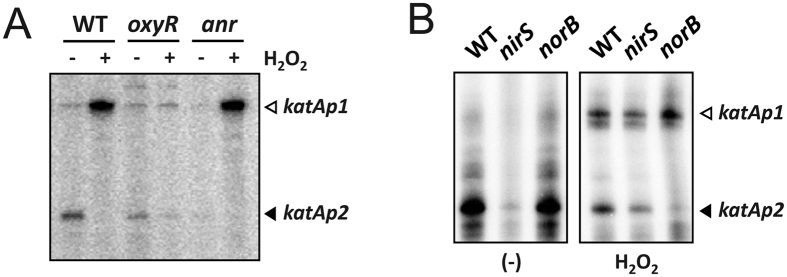
To verify the involvement of Anr in the regulation of katAp2, we examined the katA transcription in the anr null mutant, by using low-resolution S1 nuclease protection assay (Fig. 2A). The katAp1 transcription was highly elevated, whereas the katAp2 transcription was slightly reduced upon H2O2 treatment in the wild type bacteria. In contrast, however, the katAp2 transcription in the anr mutant was significantly reduced even in the absence of H2O2 treatment and the katAp1 transcription was not affected in the anr mutant. Furthermore, the H2O2-mediated decrease in katAp2-driven transcription is not associated with OxyR, in that the katAp2 transcription pattern in the oxyR null mutant did not significantly differ from that in the wild type bacteria. This result suggests that the dual promoters of katA were regulated separately by two global regulators, OxyR and Anr.
Figure 2. Transcription profiles of katA promoters in mutants.
The transcription patterns were assessed by low-resolution S1 nuclease protection assay with H2O2 treatments in oxyR and anr mutants grown in LB (A) and in nirS and norB mutants grown in LB amended with 15 mM KNO3 (B). Total RNA (50 μg) that had been prepared from the wild type (WT) and the mutant (oxyR, anr, nirS, and norB) cells with (+) or without (−) 1 mM H2O2 treatment for 10 min at OD600 of 0.5 were subjected to S1 nuclease assay. The two transcriptional start sites of the katA gene are indicated by open (katAp1) and closed (katAp2) arrows.
Since the katAp2 transcription is abolished in the anr mutant, accounting for the basal expression of KatA during the planktonic growth, we have tested if the Anr-dependent NO3− respiration during the planktonic growth may be associated with the katAp2 transcription as well. We have exploited the mutants for the genes encoding another downstream regulator, Dnr and the four nitrogen oxide reductases (Nar, Nir, Nor, and Nos) involved in dissimilatory denitrification16: NO3− is reduced to nitrite (NO2−) by Nar; NO2− is reduced to NO by Nir; NO is reduced to N2O by Nor; N2O is reduced to N2 by Nos. Denitrification is vital for growth and survival under microaerobic and anaerobic conditions as found in biofilms and microcolonies.
We found that only the nirS mutant displayed reduced katAp2 transcription (Figs 2B and S1). This is in good agreement with the previous observation by Su et al.11 that NO and Anr might be required for the increased KatA activity under anaerobic conditions.
Creation of the katA promoter mutants
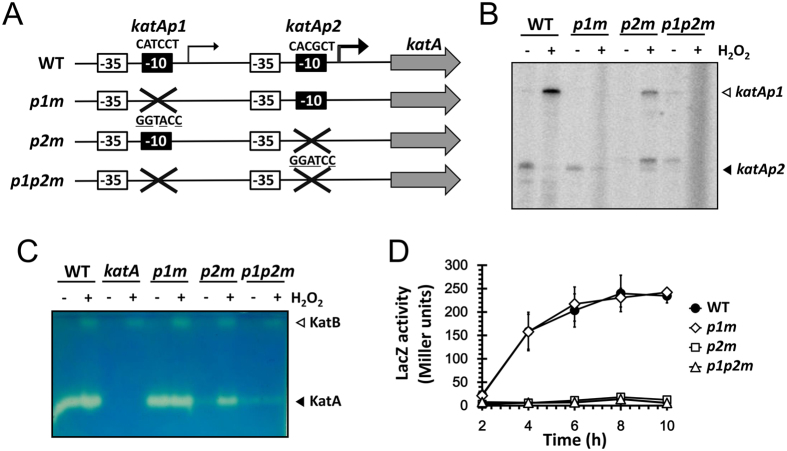
To explore the roles of the dual promoters in the expression of KatA that is required for several phenotypes such as stress resistance and virulence in P. aeruginosa6, we created the promoter mutants for each presumable −10 box, as described in Fig. 3A. The substitution mutations were introduced into the chromosome by pEX18T-based allelic exchange, which was confirmed by PCR followed by restriction enzyme digestions. The functional verification of the created promoter mutants (p1m, p2m and p1p2m) was performed based on the katA transcription and catalase activity profiles in the mutants (Fig. 3B,C). The katAp1 and katAp2 transcripts from the promoter mutants were analyzed by S1 nuclease protection assay, which verified that the introduced substitutions at the proposed −10 box resulted in the specific loss of the corresponding transcripts. A slower migrating band that was newly observed by the p2m mutation did not affect the catalase profiles in the mutants (Fig. 3C). As a result, the basal expression of KatA during the planktonic growth was abolished by the p2m mutation throughout the entire growth phases (Fig. 3D), whereas the p1m mutation contributed only to the induced expression upon H2O2 treatment, which was more evident in the p2m mutant (Figs. 3C and S2). These results indicate that the katAp2 promoter is involved in the basal as well as increased expression during the planktonic growth, whereas katAp1 is responsible for the H2O2-inducible expression of KatA. Because the katAp2 requires the transactivator, Anr for its transcription, we have postulated that Anr is somehow active even in the aerobic planktonic culture in complex media such as PIA that contains ~63 μM NO3− used for NO3− respiration17 (Fig. 2). However, as shown in Fig. S3, the stationary phase-induced transcription of katAp2 was not completely abolished in the double mutant for anr and rpoS, indicating that it involves more complicated regulatory networks such as quorum sensing and so on18.
Figure 3. Creation of the katA promoter mutants.
(A) Schematic representation. The katA gene and its potential promoter elements (−35 and −10 boxes) are designated. The sequences of each −10 box are indicated above the boxes. The katA promoter mutations were constructed by substituting the −10 box with the KpnI site (GGTACC) for the katAp1 mutant (p1m) or the BamHI site (GGATCC) for the katAp2 mutant (p2m) as indicated with the mutated nucleotides underlined. A double mutant for both promoters (p1p2m) was generated as well. (B) Transcription upon H2O2 induction. Total RNA (50 μg) isolated from the wild type (WT) and the katA promoter mutant (p1m, p2m, and p1p2m) cells with (+) or without (−) 1 mM H2O2 treatment for 10 min at OD600 of 0.5 in LB were subjected to low-resolution S1 nuclease assay. The two transcriptional start sites of the katA gene are indicated by open (katAp1) and closed (katAp2) arrows. (C) Catalase activity staining upon H2O2 induction. Catalase activities in cell extracts of the wild type (WT) and the katA null and promoter mutant (katA, p1m, p2m, and p1p2m) cells with (+) or without (−) 1 mM H2O2 treatment for 10 min as in (B) were visualized using 50 μg of proteins in each cell extract. The two catalase bands are indicated by open (KatB) and closed (KatA) arrows. (D) Promoter activity during aerobic planktonic growth. The wild type cells harboring one of the promoter transcription fusions (•, WT; ◇, p1m; □, p2m; ∆, p1p2m) were grown in LB amended with 15 mM KNO3. Culture aliquots were taken at every 2 h from 2 to 10 h post-inoculation and then subjected to β-galactosidase (LacZ) assay. The data represent the average of the means of three independent experiments (two cultures per experiment), with the error bars representing the standard deviations.
katAp1 is critical in the well-growing state, whereas katAp2 is critical in the growth-restricting state.
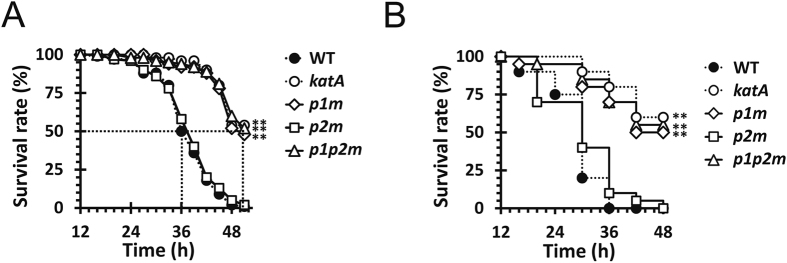
We first investigated the several stress-related phenotypes of the promoter mutants in comparison with the katA null mutant, which is hypersusceptible to H2O2 or acidified nitrite (aNO2)6,11,19. As shown in Fig. 4A, when the cells were tested during the logarithmic growth phase, the p1m mutant and the p1p2m mutant were hypersusceptible to H2O2 or aNO2 treatment, whereas the p2m mutant was no more susceptible than the wild type. In contrast, however, when cells were taken during the stationary growth phase or from the biofilm culture, the outcome did clearly differ: the p2m mutant as well as the p1p2m mutant was hypersusceptible to H2O2 or aNO2 treatment, whereas the p1m mutant was no more susceptible than the wild type (Fig. 4B,C). These results may be associated with the inducible expression of KatA by katAp1 when the basal expression of KatA by katAp2 might be insufficient, and also with the higher basal expression of KatA by katAp2 upon the stationary growth phase when the stress-induced expression of KatA through katAp1 is likely compromised in this general stress condition20.
Figure 4. Stress resistance of the katA promoter mutants.
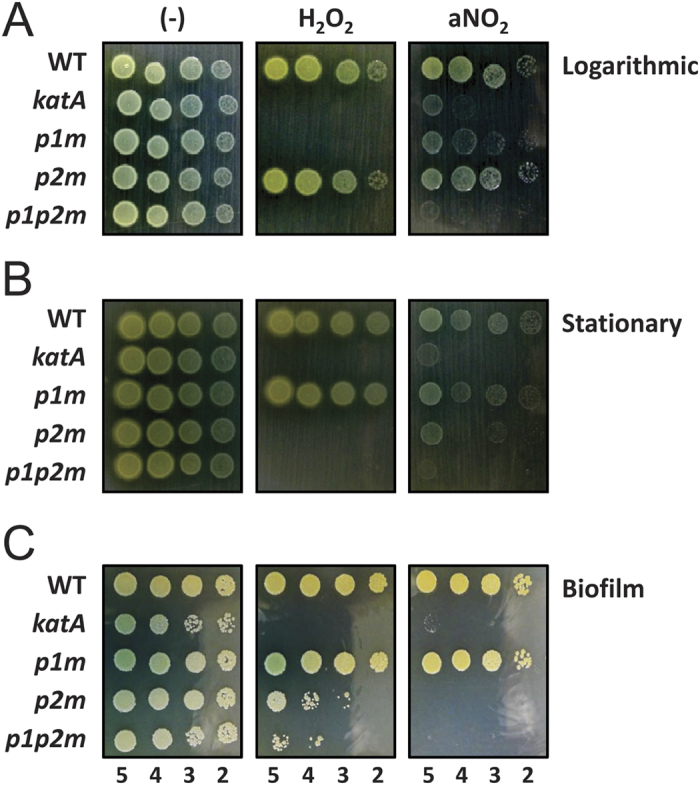
(A,B) Stress resistance in planktonic culture. Susceptibility to H2O2 and acidified NaNO2 (aNO2) were assessed for the wild type (WT) and the katA null and promoter mutant (katA, p1m, p2m, and p1p2m) cells that had been cultured for 3 h or for 6 h in LB broth (pH 6.5), and treated for 24 h with either 15 mM H2O2 and 200 μM NaNO2 or in LB broth (pH 6.5) (−). Ten-fold serial cell dilutions from the 3-h (A; logarithmic) and 6-h (B; stationary) cultures were spotted on LB agar plate. The numbers indicate the log (cfu) of the applied bacterial spots. (C) Stress resistance in static culture. The wild type (WT) and the katA null and promoter mutant (katA, p1m, p2m, and p1p2m) cells were statically and anaerobically cultured for 7 days in LB broth (pH 6.5) treated for 1 h with either 100 mM H2O2 and 1.2 M NaNO2 or in LB broth (pH 6.5) (−). Ten-fold serial cell dilutions from the cultures were spotted on LB agar plate. The numbers indicate the log(cfu) of the applied bacterial spots.
katAp1, but not katAp2, contributes to acute virulence.
We next measured the virulence potential of the promoter mutants using Drosophila and murine acute infection models, since the katA null mutant of PA14 strain was virulence-attenuated in the same infection models6. It is of marked interest that p1m and p1p2m, but not p2m displayed virulence attenuation in both infection models (Fig. 5), suggesting that the inducible KatA expression driven by katAp1, is critical in the host environments. Although we do not have any direct evidence on whether the katAp2 transcription would occur during the infection conditions, it is clear that the katAp2-mediated KatA expression is likely dispensable in the virulence pathways at least under our experimental conditions.
Figure 5. Acute virulence of the katA promoter mutants.
Virulence in Drosophila (A) and mouse peritoneal (B) infection models. The wild type (WT) and the mutant (katA, p1m, p2m, and p1p2m) cells were prepared and the survival of infected animals was determined as described in Methods. The values are the averages from five replicate experiments for Drosophila (A) and three for mouse (B) infections. Statistical significance based on the log-rank test is indicated (**p < 0.005).
katAp2 contributes to lowering endogenously generated RNS.
We hypothesized that KatA might have some surveillance function for homeostatic maintenance of cellular states regarding ROS and RNS by lowering the levels of endogenously generated ROS and RNS. We speculated that ROS and/or RNS could be accumulated in the p2m mutant, because katAp2 is required for the basal expression of KatA throughout the entire growth phases (Fig. 3D). We were able to measure the steady-state level of NO2− accumulated during the planktonic growth in LB broth supplemented with 15 mM NO3−, which can be used as an alternative electron acceptor through dissimilatory NO3− reduction involving three consecutive intermediates (NO2−, NO, and N2O) to the end product (N2)16. Among the intermediates in the denitrification process, NO is highly unstable and vulnerable to spontaneous oxidation to NO2− 21. Therefore, the amount of NO2− (either periplasmic or extracellular) may reflect the degree of NO3− respiration and the concomitant level of periplasmic RNS including NO22. Figure 6 shows that the katA null mutant as well as the katAp2 mutants (p2m and p1p2m) exhibited significantly higher levels (56.6~64.3 μM) of NO2− than the wild type and the p1m mutant (5.7~6.0 μM) during the planktonic growth. This result indicates the role of KatA as well as its katAp2-driven expression to lower the endogenous generation of RNS, under assumption that the NO3− respiration activity of the katA mutant does not significantly differ from that of the wild type bacteria, which requires more extensive investigation.
Figure 6. Accumulation of NO2− in the katA promoter mutants.
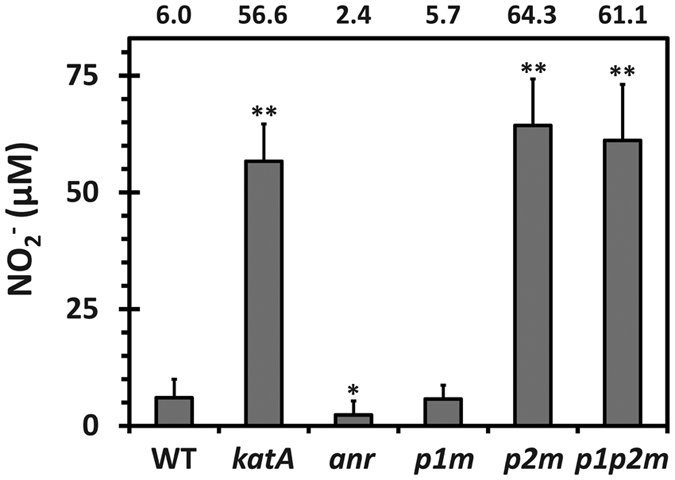
Steady-state level of extracellular NO2− was measured in the wild type (WT) and the katA null and promoter mutant (katA, p1m, p2m, and p1p2m) cells as well as anr mutant cells that had been grown to OD600 of 0.7. The amount (μM) of NO2− is calculated using the standard curve (r2 = 0.999) and the average values measured from the three independent experiments, with the error bars representing the one positive value of the standard deviations. Statistical significance based on the Student’s t-test is indicated (*p < 0.01; **p < 0.005).
Discussion
KatA is the major catalase of P. aeruginosa, whose “adaptive” function is to dismutate H2O2, an ROS either sporadically generated during the normal respiration on O2 or externally provided by redox-cycling agents or host factors. However, KatA exhibited higher activity under anaerobic conditions than aerobic conditions23, where no ROS could be potentially generated. This “enigmatic” expression profile of KatA under anaerobic conditions has been granted with the evidence that the KatA expression is lower in the anr mutant than in the wild type, with the potential Anr box located upstream of the katA coding region12. However, it had not been fully substantiated until the study about KatA function in anaerobiosis was reported11. They found that KatA contributes to the resistance to aNO2 that generates RNS like NO. They also proposed that KatA binds to NO to sequester and prevent it from its harmful effect, a new role of KatA in buffering free NO to ensure P. aeruginosa anaerobiosis, based on the investigation of direct NO-KatA interaction under anaerobic condition, as first demonstrated in bovine liver catalase able to complex with NO at the heme24. Although the anaerobic KatA expression could be justified by its proposed function, the exact regulatory mechanisms have remained elusive. In this study, we first connect the dots between the previous results regarding the expression and the function of KatA under anaerobic conditions, by identifying the second promoter, katAp2. This promoter is positively regulated by Anr and required to lower the endogenous generation of RNS during planktonic culture, which we now call the “surveillance” function of KatA.
In our previous study, P. aeruginosa KatA exhibits unusual properties ascribed to its metastability, high specific activity, and/or extracellular presence, unlike the other clade 3 monofunctional bacterial catalases such as Streptomyces coelicolor CatA and Bacillus subtilis KatA (BsKatA)7. At least one of those properties has been proposed to be implicated in biofilm life style of P. aeruginosa. P. aeruginosa respires on NO2− and/or NO3− under anaerobic conditions, which is in marked contrast to the two bacterial species: S. coelicolor is unable to grow under anaerobic conditions; B. subtilis is capable of fermentation under anaerobic conditions. Interestingly, BsKatA is subjected to activity modulation called “instant adaptation” by NO in response to ROS11,25. Although the careful comparison between KatA and the related catalases needs to be furthered in regards to the physiological traits of the cognate bacteria, the functions of KatA in the microaerobic and anaerobic life style of P. aeruginosa may be attributed to the unusual biochemical properties of KatA, most likely the extracellular presence, as previously suggested7. This aspect will open a new venue for an insight into molecular evolution of a clade 3 bacterial catalase to fulfill the new needs that the bacterium is faced with, by delving into the roles of the particular amino acid residues of KatA associated with the extracellular and/or periplasmic presence and the protective function to buffer the NO, because NO is normally generated in the periplasm by Nir during during microaerobic and/or biofilm growth of P. aeruginosa16.
Another important contribution of this work is to provide a new picture to associate the protective functions of KatA with its regulation governed by OxyR and Anr. It should be noted that OxyR and Anr work apparently under the opposite conditions: for example, OxyR is activated by H2O2 or NO26, while Anr is inactivated by H2O2 or NO, because its Fe-S cluster is vulnerable to H2O2 and NO13,14,15. Nevertheless, the dual promoters ensure the katA transcription in both balanced (i.e. surveillance function-requiring) and perturbed (i.e. adaptive function-requiring) conditions of ROS and/or RNS homeostasis. It is also likely that OxyR and Anr may cooperate for proper KatA expression during infection, due to the complicated respiration mode for P. aeruginosa growth that would be microaerophilic along the continuum between aerobic and anaerobic conditions in the human airways27,28.
Finally, the present study clearly demonstrates the discrete roles of the dual promoters in the KatA-associated phenotypes. The creation of the chromosomal mutations for the presumable −10 boxes worked well to disrupt each promoter activity and the subsequent KatA expression specifically from the corresponding promoters. Use of these promoter mutants helped clarify the roles of each promoter in the functions of KatA in P. aeruginosa physiology. Based on these properties regarding regulation and function, the pivotal roles of KatA during the growth and survival of P. aeruginosa warrant further verification. This can be done by using the promoter mutants created in the present study. Moreover, the complex involvement of multiple regulatory systems will be comprehensively explored at various regulatory levels, especially in response to stationary phase, quorum-sensing, and/or iron availability etc. All these aspects that will be more directly elucidated may provide a new insight into the therapeutic targets of this and the related bacteria associated with the stress responses toward ROS and RNS.
Methods
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli and P. aeruginosa strains were grown at 37 °C using Luria-Bertani (LB) broth or on 1.5% Bacto-agar solidified LB plates. For anaerobic growth, bacteria were grown in LB medium supplemented with 15 mM KNO3 in an anaerobic jar with AnaeroPack (MGC). Overnight-grown cultures were used as inoculum (1.6 × 107 cfu/ml) into fresh LB broth and grown at 37 °C to the OD600 as indicated and then used for the experiments described herein.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics or purposea | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PA14 | wild type laboratory strain; RifR | Lab collection |
| katA | PA14 with in-frame deletion of katA; RifR | Lee et al.6 |
| oxyR | PA14 with in-frame deletion of oxyR; RifR | Choi et al.32 |
| anr | PA14 with in-frame deletion of anr; RifR | This study |
| rpoS | PA14 with in-frame deletion of rpoS; RifR | Park et al.33 |
| anrrpoS | anr with in-frame deletion of rpoS; RifR | This study |
| narG | PA14 with MAR2 × T7 insertion at narG; RifR, GmR | Liberati et al.34 |
| nirS | PA14 with MAR2 × T7 insertion at nirS; RifR, GmR | Liberati et al.34 |
| norB | PA14 with MAR2 × T7 insertion at norB; RifR, GmR | Liberati et al.34 |
| nosZ | PA14 with MAR2 × T7 insertion at nosZ; RifR, GmR | Liberati et al.34 |
| dnr | PA14 with MAR2 × T7 insertion at dnr; RifR, GmR | Liberati et al.34 |
| p1m | PA14 with the chromosomal mutation (CATCCT to GGTACC)b at the −10 box of katAp1; RifR | This study |
| p2m | PA14 with the chromosomal mutation (CACGCT to GGATCC)b at the −10 box of katAp2; RifR | This study |
| p1p2m | p1m with the chromosomal mutation (CACGCT to GGATCC)b at the −10 box of katAp2; RifR | This study |
| E. coli | ||
| DH5α | multi-purpose cloning | Lab collection |
| S17-1 | conjugal transfer of mobilizable plasmid; TpR; SmR | Lab collection |
| Plasmids | ||
| pEX18T | Positive selection suicide vector for allelic exchange; CbR | Lab collection |
| pEX18T-∆anr | pEX18T with the in-frame deletion in the anr gene; CbR | This study |
| pEX18T-katAp1m | pUCP18 with the −10 box mutation (CATCCT to GGTACC)b of katAp1; CbR | This study |
| pEX18T-katAp2m | pUCP18 with the −10 box mutation (CACGCT to GGATCC)b of katAp2; CbR | This study |
| pQF50 | lacZ transcriptional fusion; CbR | Farinha and Kropinski35 |
| pQF50-katAp | pQF50 with the katA promoter; CbR | This study |
| pQF50-katAp1m | pQF50 with the −10 box mutation (CATCCT to GGTACC)b of katAp1; CbR | This study |
| pQF50-katAp2m | pQF50 with the −10 box mutation (CACGCT to GGATCC)b of katAp2; CbR | This study |
| pQF50-katAp1p2m | pQF50 with the −10 box mutation (CATCCT to GGTACC)b of katAp1 and (CACGCT to GGATCC)b of katAp2; CbR | This study |
aRif R, rifampicin-resistant; GmR, gentamicin-resistant; CbR, carbenicillin- and ampicillin-resistant; TpR, trimethoprim-resistant; SmR, stremptomycin-resistant.
bunderlines; mutated nucleotides at the presumable −10 boxes.
DNA oligonucleotide primers
The information on DNA oligonucleotide primers used for gene deletion, expression, and detection in this study are listed in Table S1.
RNA isolation and S1 nuclease protection analysis
P. aeruginosa strains were grown in LB or LB containing 15 mM KNO3 media and then the half of the culture was treated with 1 mM H2O2 for 10 min, with the remaining half used as the untreated control. Total RNA was isolated from 109 cells by using RNeasy kit (Qiagen) and 50 μg of RNA samples were used for S1 nuclease protection experiment as described elsewhere8. Briefly, the PCR-generated probe using the oligonucleotide primer pairs (katA-N10: 5′ end at −133 and katA-S1C1: 5′ end at +264) was labeled with [γ-32P] ATP by T4 polynucleotide kinase. For hybridization, the mixtures of RNA samples and labeled probe were incubated at 90 °C for 10 min for denaturation, and then slowly cooled down to 55 °C for hybridization. After overnight hybridization, S1 nuclease digestion was performed at 30 °C for 30 min by adding 8 units of S1 nuclease for each sample. The reaction was stopped and precipitated with 100% ethanol. The samples were dissolved and denatured at 90 °C for 5 min in formamide-dye solution, and then analyzed by 6% PAGE containing 7 M urea. For high resolution S1 mapping, the unlabeled katA-S1C1 was used to generate the nucleotide sequence ladder using Sequenase Version 2.0 DNA Sequencing Kit (USB) with [α-32P] dATP and pUCP-katA as the template7.
Creation of the katA promoter mutants
The katA promoter mutant allele for the katAp1 (−10 box; CATCCT to GGTACC, the KpnI site) or for the katAp2 (−10 box; CACGCT to GGATCC, the BamHI site) were generated by SOEing (splicing by overlap extension) PCR using 4 oligonucleotide primers. The resulting PCR products were cloned into pEX18T. These katA promoter mutant alleles were introduced into PA14 chromosome as described elsewhere29.
Creation of the lacZ fusions and β-galactosidase assay
All the lacZ fusions of the katA promoter regions were created by SOEing PCR using 4 oligonucleotide primers (Table S1). The primers were designed to generate transcriptional fusions of the katA promoter regions without its own ribosome-binding site (RBS) at the upstream of the lacZ RBS. Briefly, the katA promoter regions were prepared by using the primers pairs (katA-N3 and katA-lacZ-UC) and each of the chromosomes from the WT and the promoter mutant (p1m, p2m and p1p2m) bacteria as the templates. The lacZ coding region was generated by using the primer pairs (katA-lacZ-DN and pQF50-lacZ-C1) and the pQF50 plasmid as the template. The PCR products were fused by SOEing PCR using katA-N3 and pQF50-lacZ-C1 primers and the amplified fragments were cloned into pQF50. These promoter fusion constructs were introduced by electroporation and LacZ (β-galactosidase) activity was determined using the bacterial culture aliquots taken at the indicated time points as previously described30.
Catalase activity staining
Catalase activity staining was performed as described previously7. Briefly, cell extracts (40 μg) were applied to a 7% native polyacrylamide gel and electrophoresed. The gel was washed in distilled water and then treated with 1 mM H2O2 for 10 min. After treatment, the gel was rinsed and transferred to 1% (w/v) ferric chloride and 1% (w/v) potassium ferricyanide solution. The reaction was stopped by washing in distilled water.
Stress susceptibility measurement
Stress susceptibility was measured based on the survival of the P. aeruginosa cells upon 24 h treatment to 15 mM H2O2 or 200 μM NaNO2 for prolonged exposure. For pulse-treatment, cells were exposed to 100 mM H2O2 or 1.2 M NaNO2 in acidified LB (pH 6.5). After exposure, 10-fold serial dilutions (3 μl) of the cells were spotted onto an LB agar plate to enumerate the survivor bacteria.
Virulence measurement
Determination of mortality from Drosophila systemic infections and mouse peritonitis-sepsis caused by P. aeruginosa cells that had been grown to the OD600 of 3.0 was performed as previously described30. For Drosophila systemic infections, 3- to 6-day-old adult female flies (Oregon R) were infected by pricking at the dorsal thorax with a 10 mm needle (Ernest F. Fullam, Inc.). The needle dipped into bacterial suspensions diluted in 10 mM MgSO4 containing 107 cfu. Fly mortality was monitored for 54 h postinfection. For murine peritonitis-sepsis, bacterial cells were harvested, washed twice with phosphate buffered saline (PBS) (2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.0) and diluted to 2 × 106 cfu in 100 ml of PBS containing 1% mucin as an adjuvant. Anesthetized 4-week-old mice (ICR) were intraperitoneally infected according to the institutional protocol approved by the Institutional Animal Care and Use Committee at CHA University. Kaplan-Meier analysis and log-rank tests were used to compare the virulence difference between the groups30,31. A p value of less than 0.01 was considered to be significant.
NO2 − measurement
The steady-state level of NO2− that had been endogenously generated and transported during the normal growth in LB amended with 15 mM KNO3 was measured based on Griess reaction. Culture aliquots (300 μl) of the cells that had been grown to OD600 of 0.7 were mixed with the equal volume of Griess reagent (Sigma, USA) and incubated at 37 °C for 10 min. The NO2− level in the supernatant was measured by the absorbance at 550 nm. Statistical significance between the groups is indicated, based on a p value of less than 0.01 by the Student’s t test.
Additional Information
How to cite this article: Chung, I.-Y. et al. Dual promoters of the major catalase (KatA) govern distinct survival strategies of Pseudomonas aeruginosa. Sci. Rep. 6, 31185; doi: 10.1038/srep31185 (2016).
Supplementary Material
Acknowledgments
We are grateful to the former members for their technical assistance. This work was supported by the National Research Foundation of Korea (NRF) Grant (NRF-2015R1A2A1A15053007).
Footnotes
Author Contributions Y.-H.C. conceived and designed the research. I.-Y.C., B.-o.K. and H.-J.J. designed and performed the experiments, and collected and analyzed the experimental data. Y.-H.C. and I.-Y.C. wrote the manuscript. All authors reviewed the manuscript.
References
- Arai H. Regulation and Function of Versatile Aerobic and Anaerobic Respiratory Metabolism in Pseudomonas aeruginosa. Front Microbiol 2, 103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr T. L. & Greenberg E. P. The genetic basis for the commitment to chronic versus acute infection in Pseudomonas aeruginosa. Mol Cell 16, 497–498 (2004). [DOI] [PubMed] [Google Scholar]
- Furukawa S., Kuchma S. L. & O’Toole G. A. Keeping their options open: acute versus persistent infections. J Bacteriol 188, 1211–1217 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggan K. A. & Wolfgang M. C. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 14, 47–70 (2012). [PubMed] [Google Scholar]
- Smith E. E. et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 103, 8487–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-S., Heo Y.-J., Lee J.-K. & Cho Y.-H. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect Immun 73, 4399–4403 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. H., Choi Y. S. & Cho Y. H. Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA14 are associated with its biofilm peroxide resistance. J Bacteriol 190, 2663–2670 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo Y. J. et al. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J Bacteriol 192, 381–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo I. et al. Structural details of OxyR peroxide-sensing mechanism. Proc Natl Acad Sci USA 112, 6443–6448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H. W. & Cho Y. H. Mutational analysis of Pseudomonas aeruginosa OxyR to define the regions required for peroxide resistance and acute virulence. Res Microbiol 163, 55–63 (2012) [DOI] [PubMed] [Google Scholar]
- Su S. et al. Catalase (KatA) plays a role in protection against anaerobic nitric oxide in Pseudomonas aeruginosa. PLoS ONE 9, e91813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunk K. et al. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12, 1719–1733 (2010). [DOI] [PubMed] [Google Scholar]
- Galimand M., Gamper M., Zimmermann A. & Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol 173, 1598–1606 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribelli P. M., Nikel P. I., Oppezzo O. J. & Lopez N. I. Anr, the anaerobic global regulator, modulates the redox state and oxidative stress resistance in Pseudomonas extremaustralis. Microbiology 159, 259–268 (2013). [DOI] [PubMed] [Google Scholar]
- Yoon S. S. et al. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J 26, 3662–3672 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61, 533–616 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlitzsch D. et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109, 317–325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett D. J. et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol 34, 1082–1093 (1999). [DOI] [PubMed] [Google Scholar]
- Yoon S. S. et al. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest 116, 436–446 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A., Majdalani N. & Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65, 189–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignarro L. J., Fukuto J. M., Griscavage J. M., Rogers N. E. & Byrns R. E. Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from L-arginine. Proc Natl Acad Sci USA 90, 8103–8107 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhang X., Broderick M. & Fein H. Measurement of nitric oxide production in biological systems by using Griess reaction assay. Sensors 3, 276–284 (2003). [Google Scholar]
- Panmanee W. et al. The peptidoglycan-associated lipoprotein OprL helps protect a Pseudomonas aeruginosa mutant devoid of the transactivator OxyR from hydrogen peroxide-mediated killing during planktonic and biofilm culture. J Bacteriol 190, 3658–3669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwar N., McGarry J. M., Kostera J., Pacheco A. A. & Schmidt M. Interaction of nitric oxide with catalase: structural and kinetic analysis. Biochemistry 50, 4491–4503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I. & Nudler E. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci USA 102, 13855–13860 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen A., Privalle C. T., Keng T., DeAngelo J. & Stamler J. S. Nitrosative stress: activation of the transcription factor OxyR. Cell 86, 719–729 (1996). [DOI] [PubMed] [Google Scholar]
- Alvarez-Ortega C. & Harwood C. S. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65, 153–165 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra W., Kim E. J. & Zeng A. P. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology 148, 3195–3202 (2002). [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J. & Schweizer H. P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998). [DOI] [PubMed] [Google Scholar]
- Kim S.-H., Park S.-Y., Heo Y.-J. & Cho Y.-H. Drosophila melanogaster-based screening for multihost virulence factors of Pseudomonas aeruginosa PA14 and identification of a virulence-attenuating factor, HudA. Infect Immun 76, 4152–4162 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland J. M. & Altman D. G. The logrank test. BMJ 328, 1073 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.-S. et al. Identification of Pseudomonas aeruginosa genes crucial for hydrogen peroxide resistance. J Microbiol Biotechnol 17, 1344–1352 (2007). [PubMed] [Google Scholar]
- Park S.-Y., Heo Y.-J., Choi Y.-S., Deziel E. & Cho Y.-H. Conserved virulence factors of Pseudomonas aeruginosa are required for killing Bacillus subtilis. J Microbiol 43, 443–450 (2005). [PubMed] [Google Scholar]
- Liberati N. T. et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA 103, 2833–2838 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha M. A. & Kropinski A. M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol 172, 3496–3499 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.