Abstract
Background. The extent to which ectomycorrhizal fungi mediate primary production, carbon storage, and nutrient remineralization in terrestrial ecosystems depends upon fungal community composition. However, the factors that govern community composition at the root system scale are not well understood. Here, we explore a potential tradeoff between ectomycorrhizal fungal competitive ability and enzymatic function.
Methods. We grew Pinus muricata (Bishop Pine) seedlings in association with ectomycorrhizal fungi from three different genera in a fully factorial experimental design. We measured seedling growth responses, ectomycorrhizal abundance, and the root tip activity of five different extracellular enzymes involved in the mobilization of carbon and phosphorus.
Results. We found an inverse relationship between competitiveness, quantified based on relative colonization levels, and enzymatic activity. Specifically, Thelephora terrestris, the dominant fungus, had the lowest enzyme activity levels, while Suillus pungens, the least dominant fungus, had the highest.
Discussion. Our results identify a tradeoff between competition and function in ectomycorrhizal fungi, perhaps mediated by the competing energetic demands associated with competitive interactions and enzymatic production. These data suggest that mechanisms such as active partner maintenance by host trees may be important to maintaining “high-quality” ectomycorrhizal fungal partners in natural systems.
Keywords: Niche partitioning, Extracellular enzymes, Mycorrhizae, Tree-fungal mutualism, Pinus muricata, Mutualism
Introduction
Ectomycorrhizal fungi (EMF), the belowground mutualistic partners of most of the world’s temperate tree species, are key regulators of primary production and nutrient remineralization in terrestrial ecosystems. EMF mediate the transfer of water and nutrients from the soil to their host trees (Brownlee et al., 1983; Leake et al., 2004) and serve as a pathway for photosynthetically fixed carbon into the soil community (Talbot, Allison & Treseder, 2008). These fungi also affect the extent of carbon storage (Averill, Turner & Finzi, 2014; Clemmensen et al., 2015) and the rate of nutrient cycling (Leake et al., 2004; Koide, Fernandez & Malcolm, 2014) belowground.
The species composition of EMF communities affects these functions. For example, some taxa utilize distinct foraging strategies, such as the formation of rhizomorphs which allow the transport of water and nutrients over long distances (Brownlee et al., 1983; Agerer, 2001; Agerer, 2006), accumulate relatively high amounts of mycelial biomass belowground (Hobbie, 2006), and are linked with efficient nitrogen mobilization and low carbon sequestration (Clemmensen et al., 2015). EMF that form melanized hyphae may be more tolerant of water stress (Fernandez & Koide, 2013) and resistant to decomposition (Fernandez & Koide, 2014; Koide, Fernandez & Malcolm, 2014) than other taxa. EMF also differ in the extent to which they produce extracellular enzymes that break down soil organic matter and release nitrogen and phosphorus (Courty et al., 2006; Courty, Franc & Garbaye, 2010; Jones et al., 2010; Tedersoo et al., 2012). Because fungal traits are linked through their function to ecosystem processes (Koide, Fernandez & Malcolm, 2014; Treseder & Lennon, 2015), understanding the factors controlling ectomycorrhizal community composition on host tree root systems is of major importance.
While both abiotic (e.g., edaphic environment (Moeller, Peay & Fukami, 2014)) and biotic (e.g., plant community context (Bogar & Kennedy, 2013; Moeller et al., 2015)) conditions may play a primary role in filtering members of the ectomycorrhizal community, recent work has highlighted the importance of fungal species interactions to ectomycorrhizal community assembly (Kennedy, 2010). Competitive interactions, in particular, may locally structure fungal communities within a host plant’s root system, leading to competitive exclusion (Villeneuve, Le Tacon & Bouchard, 1991; Kennedy & Bruns, 2005; Kennedy, Peay & Bruns, 2009) or spatial segregation (Taylor & Bruns, 1999; Pickles et al., 2012). Over longer time scales, such competitive interactions may contribute to the successional patterns observed within ectomycorrhizal communities (Mason et al., 1983; Visser, 1995; Nara et al., 2003), likely in part through a tradeoff between colonization and competitive abilities (Lilleskov & Bruns, 2003; Kennedy et al., 2011).
Some functional traits such as foraging type (Peay, Kennedy & Bruns, 2011; Clemmensen et al., 2015) and propagule persistence (Baar et al., 1999; Taylor & Bruns, 1999) are associated with successional stage; however, it is not clear whether these differences in functionality result in differing competitive abilities. Here, we tested for a tradeoff between competitiveness and nutrient acquisition ability (measured as enzymatic activity) across three different ectomycorrhizal fungal genera. By holding host tree age, inoculum potential, and environmental conditions constant, we experimentally tested three hypotheses.
First, we hypothesized that a dominance hierarchy exists among the three fungal taxa used in our study, Rhizopogon occidentalis, Suillus pungens, and Thelephora terrestris. Based on prior greenhouse experimental work, we expected R. occidentalis to be competitively dominant to S. pungens (Kennedy et al., 2007; Kennedy et al., 2011), and we expected T. terrestris, an aggressive seedling colonizer (Mason et al., 1983; Velmala et al., 2013), to be competitively dominant to both these species. Second, we expected this dominance hierarchy to be inversely related to fungal enzymatic function. We based this hypothesis on the rationale that fungi experience an energetic tradeoff: they can either produce metabolically costly extracellular enzymes, or they can invest in chemical defenses against their competitors. Third, we hypothesized that this tradeoff would impact host tree seedling growth: seedlings would accrue the highest biomass when associating with the least dominant, most highly enzymatically functional fungi.
Materials and Methods
Experimental design
We worked with Pinus muricata (Bishop Pine) and three ectomycorrhizal fungi (Rhizopogon occidentalis, Suillus pungens, and Thelephora terrestris) known to associate with this tree in its native range (Peay et al., 2007). We selected these fungi because they are among the most common and abundant in early successional pine forests in Point Reyes National Seashore (PRNS), where we worked (Peay et al., 2007). We tested the competitive abilities, enzyme expression levels, and effects on seedling growth of these fungi grown in isolation and in competition.
P. muricata seeds were obtained from PRNS. Prior to the start of the experiment, seeds were surface sterilized and germinated in autoclave-sterilized perlite. Within 7 days, germinated seedlings were transplanted into conetainers filled with a 50:50 mix of autoclaved sand and soil from PRNS. Spores from each of the three EMF were obtained from sporocarps collected at PRNS. Sporocarps were incubated spore-side down overnight on foil at room temperature. Spores were collected by washing the foil with sterile (distilled, autoclaved) water and refrigerated at 4°C for three weeks until inoculation. Prior to inoculation, hemocytometer counts followed by serial dilutions in sterile water were used to obtain a concentration of 1,000 spores per mL for each of the three fungal species. We did not conduct spore viability stain assays; however, spore storage time was short (Bruns et al., 2009), and handling was consistent with other studies using spore inoculum from these species (Kennedy & Bruns, 2005; Kennedy & Peay, 2007; Kennedy, Peay & Bruns, 2009; Peay et al., 2012), so we expected a high proportion of viable spores in the inoculation slurries.
We inoculated the P. muricata seedlings with zero, one, two, or three fungal species in all possible combinations. We randomly assigned five seedlings to each treatment group (for a total of 5 seedlings × 7 treatments + 1 control = 40 seedlings). Each seedling received a total of 3-mL of inoculum. For control (non-mycorrhizal) seedlings, this consisted of 3-mL of sterile water. Seedlings inoculated with EMF received 1-mL of inoculum per fungal species (so that seedlings in the single-fungus treatments received 1-mL of spore inoculum and 2-mL of distilled water). Thus, seedlings in the three-species treatment received a total of 3,000 spores (1,000 per species).
Seedlings were maintained in a greenhouse at Stanford University for five months between inoculation and harvest. This experimental duration was chosen because it approximates the length of the main growth and fruiting season of EMF in PRNS, and because previous greenhouse studies of P. muricata seedlings and their EMF have been of similar duration (Kennedy & Bruns, 2005; Kennedy & Peay, 2007; Peay, Garbelotto & Bruns, 2009; Kennedy, Peay & Bruns, 2009). At harvest, each seedling’s root system was separated from the shoot at the root collar. The root system was washed clear of adhering soil using tap water. Roots were then cut into 3-cm segments, homogenized, and a subset was examined under a dissecting microscope. This entire subset was scored for mycorrhization using the grid-line intersection method: root segments were randomly arranged over a 1-cm grid, and every grid crossing was scored as mycorrhizal or non-mycorrhizal based on the presence or absence of fungal hyphae (Giovannetti & Mosse, 1980). Note that, because not all of the root system is comprised of fine root tips, total mycorrhization levels are always <100%. Following examination, eight mycorrhizal root tips from each seedling were randomly selected for fungal identification, and ten mycorrhizal tips were selected for enzyme assays. Enzyme assay tips were also collected from control seedlings to establish non-mycorrhizal baseline activity levels. Following root system processing, seedling root systems and shoots were dried at 65°C for 48 h and then weighed to determine biomass.
Fungal identification and enzyme assays
We used Sanger sequencing to assign mycorrhizal root tips to species. To extract DNA, tips were heated in 10 µL Extraction Solution (SKU E7526; Sigma-Aldrich Co. LLC, St. Louis, MO, USA) for 10 min at 65°C, then 10 min at 95°C, before addition of 10 µL of Neutralization Solution B (SKU N3910; Sigma-Aldrich Co. LLC). The internal transcribed spacer (ITS) region of the nuclear ribosomal RNA genes of each root tip was amplified using the ITS-1F (Gardes & Bruns, 1993) and ITS-4 primers (White et al., 1990) and sequenced by Beckman Coulter Genomics (Danvers, MA, USA). The resultant sequences were assigned to one of the three species using the Basic Local Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov).
We used fluorimetric assays to quantify the activity levels of five different enzymes: α-glucosidase (which hydrolyzes starch and glycogen), β-glucosidase (which hydrolyzes cellobiose), N-acetyl-glucosaminidase (which breaks down chitin), β-xylosidase (which breaks down xylose), and acid phosphatase (which releases phosphate). Each root tip was placed in an individual well of a 96-well-screen-bottom plate, and enzyme activities were sequentially measured according to the protocol outlined in Pritsch et al. (2011). Total per-tip activity was calculated based on a standard curve using 4-methylumbelliferone and normalized to surface area calculated using WinRHIZO. Following enzyme assays, DNA was also extracted from these tips, amplified, sequenced, and used either to assign root tips to a fungal taxon or to confirm lack of mycorrhization in the controls.
Data analysis
We used single-species treatments to verify the viability of our spore inoculum. In multi-species treatments, we determined the competitively dominant fungus as the one with the greatest relative colonization (i.e., greatest proportion of sequences in our randomly selected set of root tips). To determine differences in enzyme activity and seedling growth, we compared treatment means using Tukey’s Honestly Significant Difference tests to correct for multiple hypothesis testing. All statistical calculations were performed using R (R Core Team, 2014). To quantify variance in enzymatic activity, we used a principal components analysis to compress data from all five enzymatic assays into two dimensions for visualization (package bpca; Faria, Demetrio & Allaman, 2016). We then computed the Euclidean distance between pairs of tips from the same treatments (vegan, function betadisper; Oksanen et al., 2013) and performed a Tukey’s Honestly Significant Difference test to compare within-treatment variance.
Results
Fungal mycorrhization levels
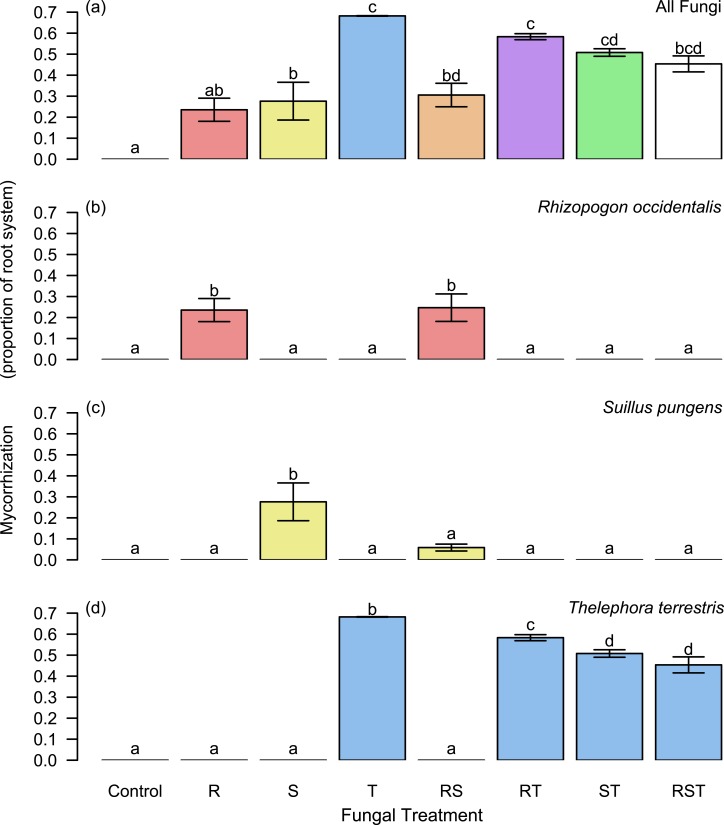
In single-species treatments, all fungi established associations with P. muricata seedlings. Mycorrhization levels differed by fungal taxon (Fig. 1). Thelephora terrestris had the highest mycorrhization level of 68.2 ± 0.00159% (mean ± standard deviation) root length colonized. Rhizopogon occidentalis (23.5 ±0.0952%) and Suillus pungens (27.6 ± 15.6%) were similar to one another in abundance. No ectomycorrhizal fungi were detected on the control seedlings.
Figure 1. Mycorrhization levels across experimental treatments.
(A) Total mycorrhization was highest in treatments that included Thelephora terrestris. No fungal contamination was present in the controls. (B–C) Fungi exhibited a dominance hierarchy, with T. terrestris as the only fungus present in multi-species treatments, and Rhizopogon occidentalis suppressing Suillus pungens growth in the two-species combination treatment. Bar heights indicate means across seedlings within a treatment group; whiskers give standard error. Letters indicate statistically significant differences in mean (Tukey’s HSD, P < 0.05). Colors represent species: Red, R. occidentalis, yellow, S. pungens, and blue, T. terrestris; color blends represent species combinations (e.g., green, S. pungens + T. terrestris in (A), where total mycorrhization is plotted).
Hypothesis 1: dominance hierarchy
Our data supported our hypothesized dominance hierarchy: T. terrestris competively excluded both R. occidentalis and S. pungens. R. occidentalis was competitively dominant (in terms of root mycorrhization) to, but did not completely exclude, S. pungens in the two-species treatment containing these fungi (Fig. 1).
Hypothesis 2: dominance-function tradeoff
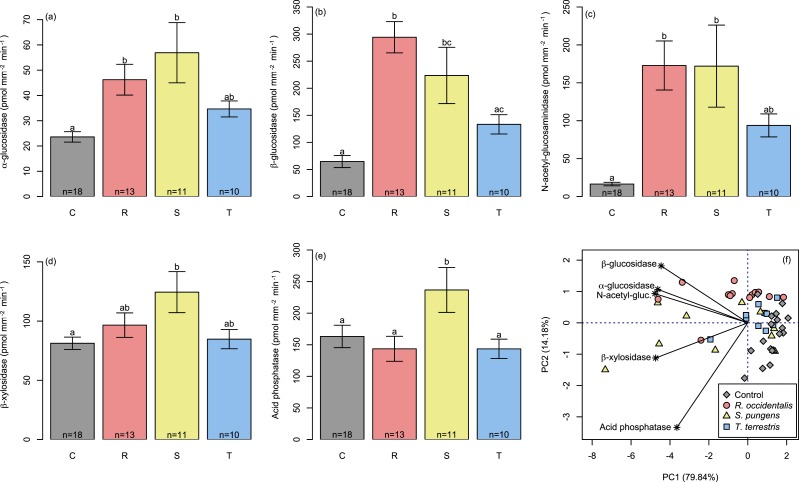
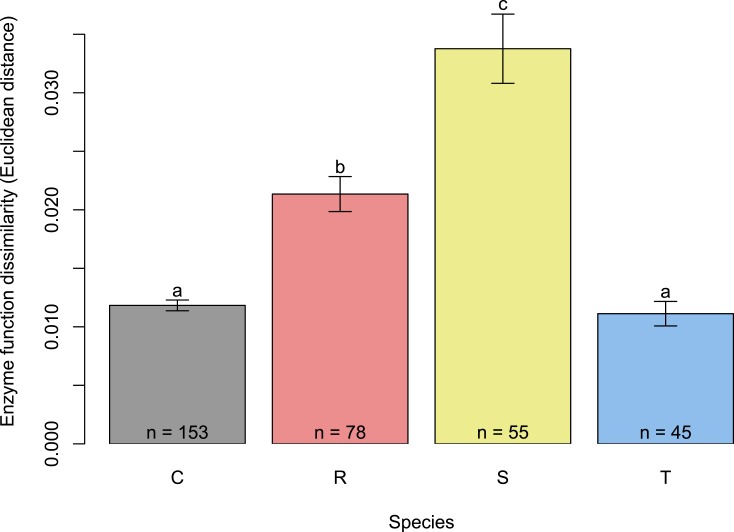
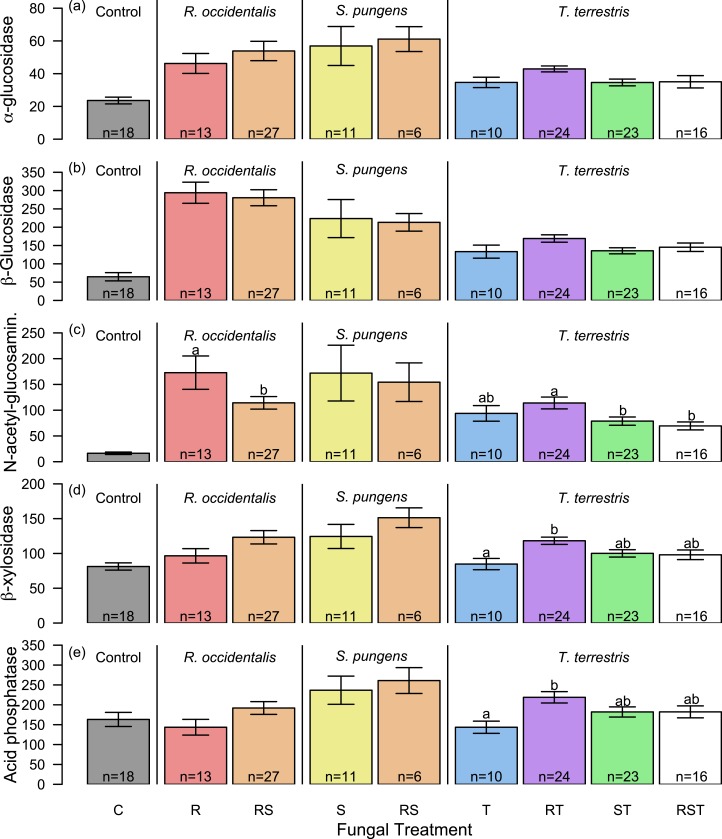
Enzyme profiles from single-species treatments supported our hypothesis that the least competitive fungus, S. pungens, would have the highest enzyme activity levels. S. pungens enzymatic activity was higher than non-mycorrhizal control root tips for all five extracellular enzymes assayed (Fig. 2, yellow bars). In contrast, T. terrestris, the competitively dominant fungus, had enzymatic activity levels indistinguishable from controls (Fig. 2, blue bars), and R. occidentalis, of intermediate competitive ability, had elevated enzymatic activity levels only for α- and β-glucosidase and N-acetyl-glucosaminidase (Fig. 2, red bars). Fungi varied significantly in their overall enzymatic profiles (Fig. 2F, Table 1; see Fig. S1 for additional PCA axes). Variance in enzymatic activity was greatest for S. pungens, intermediate for R. occidentalis, and lowest (equivalent to control tips) for T. terrestris (Fig. 3). In multi-species treatments, β-Xylosidase and Acid Phosphatase expression levels were elevated for T. terrestris-infected tips (Fig. 4).
Figure 2. Enzyme activities of root tips colonized by the three fungal taxa compared with control (non-mycorrhizal) root tips.
Data are from single-fungus inoculations (i.e., Treatment = R, S, or T). Across the five enzymes tested, only Suillus pungens-associated root tips showed consistently elevated activity relative to nonmycorrhizal tips (A–E Tukey’s HSD, P < 0.05). Association with Rhizopogon occidentalis elevated α- and β-glucosidase and N-acetyl-glucosaminidase enzyme activities relative to control tips. A principal component analysis (F) revealed that fungi differed in their enzymatic assays (PERMANOVA, P < 0.05).
Table 1. Principal components analysis of enzymatic activity for single-species treatments.
| Principal component axis | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Percent explained | 79.84 | 14.18 | 4.06 | 1.11 | 0.800 |
| Loadings | |||||
| α-glucosidase | −6.61 | 1.53 | −2.10 | 0.0609 | 0.775 |
| β-glucosidase | −6.36 | 2.60 | 1.67 | −0.981 | 0.113 |
| N-acetylglucosaminidase | −6.82 | 1.33 | 0.965 | 1.29 | −0.289 |
| β-xylosidase | −6.77 | −1.60 | −1.19 | −0.437 | −1.01 |
| Acid phosphatase | −5.21 | −4.77 | 0.91 | −0.00634 | 0.568 |
Figure 3. Variance in fungal enzymatic activity by species measured as Euclidean distance across all five enzymatic assays.
Data are from single-fungus inoculations (as in Fig. 2). S. pungens had the greatest variation in enzymatic function, R. occidentalis intermediate, and T. terrestris the least (Tukey’s HSD, P < 0.05).
Figure 4. Changes in enzyme activity by fungal associate and treatment.
N-acetyl-glucosaminidase activity was elevated for Rhizopogon occidentalis-associated root tips when in isolation relative to competition with Suillus pungens (C). β-xylosidase and acid phosphatase activities were elevated for Thelephora terrestris-associated tips when in competition with R. occidentalis (D–E). Bar heights indicate means across root tips within a treatment group (measured in pmol mm−2 min−1); whiskers give standard error. Letters indicate statistically significant differences in mean within species by treatment (Tukey’s HSD, P < 0.05). Colors represent treatments: red, R. occidentalis; yellow, S. pungens; and blue, T. terrestris; color blends represent species combinations (e.g., green, S. pungens + T. terrestris).
Hypothesis 3: seedling growth response
Seedling aboveground, belowground, and total biomass did not differ by treatment; thus our third hypothesis was not supported. Overall, we did observe a positive relationship between mycorrhization level and seedling biomass (Fig. 5).
Figure 5. Relationship between seedling growth and mycorrhization level.
Overall, total seedling biomass was positively correlated with mycorrhization, measured as the proportion of root length with hyphal structures (A, R2 = 0.433, P < 0.001). This was the result of both above- and belowground effects of mycorrhization on plant growth: both shoot (B, R2 = 0.213, P < 0.05) and root (C, R2 = 0.558, P < 0.001) dry weight increased with increasing mycorrhization. Points are color-coded by treatment as in Figs. 1–3.
Discussion
In this study, we present an experimental test for competition-function tradeoffs across three genera of ectomycorrhizal fungi (EMF). We observed a clear competitive dominance hierarchy among the EMF that was inversely related to their extracellular enzymatic activities. Specifically, the most competitively dominant EMF, Thelephora terrestris, had enzymatic activities indistinguishable from non-mycorrhized tree roots, whereas the competitively inferior Suillus pungens exhibited elevated enzymatic activity across all five enzymes assayed. A number of studies have previously documented differences in enzymatic activity profiles across (Courty et al., 2005; Courty et al., 2006; Buée et al., 2007; Courty, Franc & Garbaye, 2010; Kipfer et al., 2012; Velmala et al., 2013; Walker, Ward & Jones, 2016) and within (Jones et al., 2010) fungal genera. These differences, especially when observed in the field among members of assembled EMF communities, are suggestive of functional complementarity (Courty et al., 2005; Buée et al., 2007; Jones et al., 2010). In some cases, e.g., Rhizopogon species, enzymatic activities are disproportionately high relative to species abundance (Walker et al., 2014). However, to our knowledge, this study is the first to experimentally link these differences in enzymatic activity with competitive ability. Because our study, like others that quantify species-specific enzymatic activity, used root tips to obtain fungal tissues, our enzymatic data are representative only of root-tip associated exoenzyme activities, which may be different than expression levels in other parts of the soil (e.g., at the edges of hyphal extent furthest from the host tree where fungi are foraging for nutrients, or at local hotspots of resource availability in the heterogeneous soil environment) (Wright et al., 2005; Liao et al., 2014). Thus, much work remains to be done to determine the whole organism’s functional potential.
That T. terrestris was the most competitively dominant EMF in our study was not surprising given its proclivity for vigorous growth under greenhouse conditions (Mason et al., 1983; Velmala et al., 2013). However, S. pungens and R. occidentalis are key community members on Pinus muricata in the field, including at Point Reyes National Seashore where the seeds, soils, and fungal spores used in this study were collected (Peay et al., 2007). All three species are capable of colonizing seedlings through spore dispersal, with S. pungens and R. occidentalis more consistently observed on small tree islands than T. terrestris (Peay et al., 2007). R. occidentalis colonizes through generation of a long-lived spore bank (Bruns et al., 2009), while S. pungens and T. terrestris are the two most prolific aerial spore dispersers in the system (Peay et al., 2012). Our competitive dominance hierarchy, which is the inverse of this dispersal hierarchy, suggests a competition-colonization tradeoff among these taxa similar to that observed by Kennedy et al. (2011), at least in terms of root system abundance. While EMF may compete in other ways, such as by competing for nutrient and water resources in the soil or by competing for plant carbon resources (delivery of which may vary by root tip occupancy), in this case the observed complete exclusion of other EMF by T. terrestris suggests strong competitive dominance. However, in cases where exclusion is incomplete measurements of carbon and nitrogen acquisition may be necessary to determine competitive dominance. Although competitively excluded from seedling root systems by T. terrestris in our study, the persistence of S. pungens and R. occidentalis in the field (at least through the first decade of succession) could be the result of several mechanisms. First, less competitive fungi may be maintained through active partner maintenance. This could be the case if trees allocate carbon to their fungal partners in proportion to their partners’ provision of resources (Hoeksema & Kummel, 2003), which would likely be greater for highly enzymatically active EMF like S. pungens. Second, trees with larger root systems are likely to support a greater diversity of EMF than the seedlings in our study (Nara et al., 2003). Third, competition among EMF can be context dependent (Kennedy, Peay & Bruns, 2009), and while T. terrestris is dominant under greenhouse conditions, this is unlikely to be the case for all biotic and abiotic conditions.
Greenhouse growth conditions may also be responsible for the homogeneity of the seedling growth response. Although Kipfer et al. (2012) found that the most enzymatically active fungus in their study, Suillus granulatus, had a positive effect on seedling growth, we did not observe statistically significant differences in growth by treatment in our study. In part, this was likely due to the homogeneous, high-quality soil environment created by autoclaving the experimental soils, which can release substantial amounts of nutrients into plant-accessible pools, reducing the impact of EMF on seedling growth (Peay, Bruns & Garbelotto, 2010) and plant carbon allocation to EMF (Hobbie, 2006). Other studies have found no relationship between EMF competitiveness and seedling growth (Kennedy, Peay & Bruns, 2009). Perhaps, at least at the early seedling stage, growth effects are obscured by conflicting mechanisms. “High-quality” partners like S. pungens may be more expensive thanks to the energetic demands of producing extracellular enzymes; this additional carbon cost may negate any benefits to the host seedling, particularly in nutrient-rich soils.
Although T. terrestris was the only fungus observed by harvest time in all treatments that included it as a source of inoculum, we did observe reductions in its mycorrhization level and increases in some of its enzymatic activities in multi-species treatments relative to monoculture. These differences may be due to a time lag in competitive displacement of R. occidentalis similar to that observed by Lilleskov & Bruns (2003) when R. occidentalis was in competition with Tomentella sublilacina (like T. terrestris, a member of the Thelephoraceae), and/or shifts in T. terrestris enzymatic function induced by the presence of other EMF. Further experimental manipulation of community composition is likely to elucidate the roles of such mechanisms and clarify the role of fungal identity in mutualism function.
We also observed differences in absolute mycorrhization levels among the EMF in our single-species treatments. By our metric (percent of total root length colonized), T. terrestris had mycorrhization levels that were almost double those of the other EMF. In part, this may be due to different root growth forms associated with R. occidentalis and S. pungens, which tend to induce the production of tightly bunched clusters of root tips (HV Moeller & KG Peay, pers. obs., 2011; see also images in the DEtermination of EctoMYcorrhizae database, http://www.deemy.de/) whose prevalence would be underestimated by our grid-intersect sampling method. In contrast, T. terrestris exhibits greater spatial extent along the length of fine roots, and the formation of root tip clusters is not observed. While this difference in mycorrhization may also be due to interspecific differences in spore inoculum viability or rates of vegetative spread across root systems, prior studies have observed similar levels of mycorrhization for the three genera we studied (Browning & Whitney, 1993; Kennedy & Bruns, 2005). Indeed, Kennedy & Bruns (2005) observed ∼30% mycorrhization levels for Rhizopogon species within two months of inoculation. The evolutionary and ecological reasons for these differences in colonization strategy remain unclear, but a fuller understanding of EMF spatial extent and enzymatic function beyond the plant’s immediate root zone will likely shed light on these questions.
Supplemental Information
Axis loadings are given in Table 1.
Acknowledgments
The authors thank Jennifer Talbot for assistance in performing the enzymatic assays. We also thank Roger Koide and Peter Avis for thoughtful review of our manuscript.
Funding Statement
HVM received funding from the United States National Science Foundation through a Graduate Research Fellowship, a Doctoral Dissertation Improvement Grant, and a Postdoctoral Research Fellowship in Biology (DBI-1401332). KGP received funding from the National Science Foundation Dimensions of Biodiversity Program (DBI-1045658). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Holly V. Moeller and Kabir G. Peay conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Data Availability
The following information was supplied regarding data availability:
The raw data has been supplied as Supplemental Information.
References
- Agerer (2001).Agerer R. Exploration types of ectomycorrhizae. Mycorrhiza. 2001;11:107–114. doi: 10.1007/s005720100108. [DOI] [Google Scholar]
- Agerer (2006).Agerer R. Fungal relationships and structural identity of their ectomycorrhizae. Mycological Progress. 2006;5:67–107. doi: 10.1007/s11557-006-0505-x. [DOI] [Google Scholar]
- Averill, Turner & Finzi (2014).Averill C, Turner BL, Finzi AC. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature. 2014;505:543–545. doi: 10.1038/nature12901. [DOI] [PubMed] [Google Scholar]
- Baar et al. (1999).Baar J, Horton T, Kretzer AM, Bruns T. Mycorrhizal colonization of Pinus muricata from resistant propagules after a stand-replacing wildfire. New Phytologist. 1999;143:409–418. doi: 10.1046/j.1469-8137.1999.00452.x. [DOI] [Google Scholar]
- Bogar & Kennedy (2013).Bogar LM, Kennedy PG. New wrinkles in an old paradigm: neighborhood effects can modify the structure and specificity of Alnus-associated ectomycorrhizal fungal communities. FEMS Microbiology Ecology. 2013;83:767–777. doi: 10.1111/1574-6941.12032. [DOI] [PubMed] [Google Scholar]
- Browning & Whitney (1993).Browning M, Whitney RD. Infection of containerized jack pine and black spruce by Laccaria species and Thelephora terrestris and seedling survival and growth after outplanting. Canadian Journal of Forest. 1993;23:330–333. doi: 10.1139/x93-046. [DOI] [Google Scholar]
- Brownlee et al. (1983).Brownlee C, Duddridge JA, Malibari A, Read DJ. The structure and function of mycelial systems of ectomycorrhizal roots with special reference to their role in forming inter-plant connections and providing pathways for assimilate and water transport. Plant and Soil. 1983;71:433–443. doi: 10.1007/BF02182684. [DOI] [Google Scholar]
- Bruns et al. (2009).Bruns T, Peay K, Boynton P, Grubisha L. Inoculum potential of Rhizopogon spores increases with time over the first 4 yr of a 99-yr spore burial experiment. New Phytologist. 2009;181:463–470. doi: 10.1111/j.1469-8137.2008.02652.x. [DOI] [PubMed] [Google Scholar]
- Buée et al. (2007).Buée M, Courty P, Mignot D, Garbaye J. Soil niche effect on species diversity and catabolic activities in an ectomycorrhizal fungal community. Soil Biology and Biochemistry. 2007;39:1947–1955. doi: 10.1016/j.soilbio.2007.02.016. [DOI] [Google Scholar]
- Clemmensen et al. (2015).Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD. Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytologist. 2015;205:1525–1536. doi: 10.1111/nph.13208. [DOI] [PubMed] [Google Scholar]
- Courty, Franc & Garbaye (2010).Courty P-E, Franc A, Garbaye J. Temporal and functional pattern of secreted enzyme activities in an ectomycorrhizal community. Soil Biology and Biochemistry. 2010;42:2022–2025. doi: 10.1016/j.soilbio.2010.07.014. [DOI] [Google Scholar]
- Courty et al. (2006).Courty P-E, Pouysegur R, Buee M, Garbaye J. Laccase and phosphatase activities of the dominant ectomycorrhizal types in a lowland oak forest. Soil Biology and Biochemistry. 2006;38:1219–1222. doi: 10.1016/j.soilbio.2005.10.005. [DOI] [Google Scholar]
- Courty et al. (2005).Courty P-E, Pritsch K, Schloter M, Hartmann A, Garbaye J. Activity profiling of ectomycorrhiza communities in two forest soils using multiple enzymatic tests. New Phytologist. 2005;167:309–319. doi: 10.1111/j.1469-8137.2005.01401.x. [DOI] [PubMed] [Google Scholar]
- Faria, Demetrio & Allaman (2016).Faria JC, Demetrio CGB, Allaman IB. bpca: Biplot of multivariate data based on principal components analysis. Sāo Paulo: University of Sāo Paulo/ESALQ; 2016. [Google Scholar]
- Fernandez & Koide (2013).Fernandez CW, Koide RT. The function of melanin in the ectomycorrhizal fungus Cenococcum geophilum under water stress. Fungal Ecology. 2013;6:479–486. doi: 10.1016/j.funeco.2013.08.004. [DOI] [Google Scholar]
- Fernandez & Koide (2014).Fernandez CW, Koide RT. Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biology and Biochemistry. 2014;77:150–157. doi: 10.1016/j.soilbio.2014.06.026. [DOI] [Google Scholar]
- Gardes & Bruns (1993).Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Giovannetti & Mosse (1980).Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist. 1980;84:489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x. [DOI] [Google Scholar]
- Hobbie (2006).Hobbie EA. Carbon allocation to ectomycorrhizal fungi correlates with belowground allocation in culture studies. Ecology. 2006;87:563–569. doi: 10.1890/05-0755. [DOI] [PubMed] [Google Scholar]
- Hoeksema & Kummel (2003).Hoeksema JD, Kummel M. Ecological persistence of the plant-mycorrhizal mutualism: a hypothesis from species coexistence theory. The American Naturalist. 2003;162:S40–S50. doi: 10.1086/378644. [DOI] [PubMed] [Google Scholar]
- Jones et al. (2010).Jones MD, Twieg BD, Ward V, Barker J, Durall DM, Simard SW. Functional complementarity of Douglas-fir ectomycorrhizas for extracellular enzyme activity after wildfire or clearcut logging. Functional Ecology. 2010;24:1139–1151. doi: 10.1111/j.1365-2435.2010.01699.x. [DOI] [Google Scholar]
- Kennedy (2010).Kennedy P. Ectomycorrhizal fungi and interspecific competition: species interactions, community structure, coexistence mechanisms, and future research directions. New Phytologist. 2010;187:895–910. doi: 10.1111/j.1469-8137.2010.03399.x. [DOI] [PubMed] [Google Scholar]
- Kennedy & Bruns (2005).Kennedy PG, Bruns TD. Priority effects determine the outcome of ectomycorrhizal competition between two Rhizopogon species colonizing Pinus muricata seedlings. New Phytologist. 2005;166:631–638. doi: 10.1111/j.1469-8137.2005.01355.x. [DOI] [PubMed] [Google Scholar]
- Kennedy et al. (2011).Kennedy PG, Higgins LM, Rogers RH, Weber MG. Colonization-competition tradeoffs as a mechanism driving successional dynamics in ectomycorrhizal fungal communities. PLoS ONE. 2011;6:e2270. doi: 10.1371/journal.pone.0025126.t002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy et al. (2007).Kennedy PG, Hortal S, Bergemann SE, Bruns TD. Competitive interactions among three ectomycorrhizal fungi and their relation to host plant performance. Journal of Ecology. 2007;95:1338–1345. doi: 10.1111/j.1365-2745.2007.01306.x. [DOI] [Google Scholar]
- Kennedy & Peay (2007).Kennedy PG, Peay KG. Different soil moisture conditions change the outcome of the ectomycorrhizal symbiosis between Rhizopogon species and Pinus muricata. Plant and Soil. 2007;291:155–165. doi: 10.1007/s11104-006-9183-3. [DOI] [Google Scholar]
- Kennedy, Peay & Bruns (2009).Kennedy PG, Peay KG, Bruns TD. Root tip competition among ectomycorrhizal fungi: are priority effects a rule or an exception? Ecology. 2009;90:2098–2107. doi: 10.1890/08-1291.1. [DOI] [PubMed] [Google Scholar]
- Kipfer et al. (2012).Kipfer T, Wohlgemuth T, Van Der Heijden MGA, Ghazoul J, Egli S. Growth response of drought-stressed pinus sylvestris seedlings to single- and multi-species inoculation with ectomycorrhizal fungi. PLoS ONE. 2012;7:e2270. doi: 10.1371/journal.pone.0035275.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide, Fernandez & Malcolm (2014).Koide RT, Fernandez C, Malcolm G. Determining place and process: functional traits of ectomycorrhizal fungi that affect both community structure and ecosystem function. New Phytologist. 2014;201:433–439. doi: 10.1111/nph.12538. [DOI] [PubMed] [Google Scholar]
- Leake et al. (2004).Leake J, Johnson D, Donnelly D, Muckle G, Boddy L, Read D. Networks of power and influence: the role of mycorrhizal mycelium in controlling plant communities and agroecosystem functioning. Canadian Journal of Botany. 2004;82:1016–1045. doi: 10.1139/b04-060. [DOI] [Google Scholar]
- Liao et al. (2014).Liao HL, Chen Y, Bruns TD, Peay KG, Taylor JW, Branco S, Talbot JM, Vilgalys R. Metatranscriptomic analysis of ectomycorrhizal roots reveals genes associated with Piloderma-Pinussymbiosis: improved methodologies for assessing gene expression in situ. Environmental Microbiology. 2014;16:3730–3742. doi: 10.1111/1462-2920.12619. [DOI] [PubMed] [Google Scholar]
- Lilleskov & Bruns (2003).Lilleskov EA, Bruns TD. Root colonization dynamics of two ectomycorrhizal fungi of contrasting life history strategies are mediated by addition of organic nutrient patches. New Phytologist. 2003;159:141–151. doi: 10.1046/j.0028-646x.2003.00794.x. [DOI] [PubMed] [Google Scholar]
- Mason et al. (1983).Mason PA, Wilson J, Last FT, Walker C. The concept of succession in relation to the spread of sheathing mycorrhizal fungi on inoculated tree seedlings growing in unsterile soils. Plant and Soil. 1983;71:247–256. doi: 10.1007/BF02182659. [DOI] [Google Scholar]
- Moeller et al. (2015).Moeller HV, Dickie IA, Peltzer D, Fukami T. Mycorrhizal co-invasion and novel interactions depend on neighborhood context. Ecology. 2015;96:2336–2347. doi: 10.1890/14-2361.1. [DOI] [PubMed] [Google Scholar]
- Moeller, Peay & Fukami (2014).Moeller HV, Peay KG, Fukami T. Ectomycorrhizal fungal traits reflect environmental conditions along a coastal California edaphic gradient. FEMS Microbiology Ecology. 2014;87:797–806. doi: 10.1111/1574-6941.12265. [DOI] [PubMed] [Google Scholar]
- Nara et al. (2003).Nara K, Nakaya H, Wu B, Zhou Z, Hogetsu T. Underground primary succession of ectomycorrhizal fungi in a volcanic desert on Mount Fuji. New Phytologist. 2003;159:743–756. doi: 10.1046/j.1469-8137.2003.00844.x. [DOI] [PubMed] [Google Scholar]
- Oksanen et al. (2013).Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara B, Simpson GL, Solymos P, Stevens M, Wagner H. vegan: community ecology package. 2013. https://CRAN.R-project.org/package=vegan https://CRAN.R-project.org/package=vegan
- Peay, Bruns & Garbelotto (2010).Peay KG, Bruns TD, Garbelotto M. Testing the ecological stability of ectomycorrhizal symbiosis: effects of heat, ash and mycorrhizal colonization on Pinus muricata seedling performance. Plant and Soil. 2010;330:291–302. doi: 10.1007/s11104-009-0200-1. [DOI] [Google Scholar]
- Peay et al. (2007).Peay KG, Bruns TD, Kennedy PG, Bergemann SE, Garbelotto M. A strong species–area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecology Letters. 2007;10:470–480. doi: 10.1111/j.1461-0248.2007.01035.x. [DOI] [PubMed] [Google Scholar]
- Peay, Garbelotto & Bruns (2009).Peay KG, Garbelotto M, Bruns TD. Spore heat resistance plays an important role in disturbance-mediated assemblage shift of ectomycorrhizal fungi colonizing Pinus muricata seedlings. Journal of Ecology. 2009;97:537–547. doi: 10.1111/j.1365-2745.2009.01489.x. [DOI] [Google Scholar]
- Peay, Kennedy & Bruns (2011).Peay KG, Kennedy PG, Bruns TD. Rethinking ectomycorrhizal succession: are root density and hyphal exploration types drivers of spatial and temporal zonation? Fungal Ecology. 2011;4:233–240. doi: 10.1016/j.funeco.2010.09.010. [DOI] [Google Scholar]
- Peay et al. (2012).Peay KG, Schubert MG, Nguyen NH, Bruns TD. Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Molecular Ecology. 2012;21:4122–4136. doi: 10.1111/j.1365-294X.2012.05666.x. [DOI] [PubMed] [Google Scholar]
- Pickles et al. (2012).Pickles BJ, Genney DR, Anderson IC, Alexander IJ. Spatial analysis of ectomycorrhizal fungi reveals that root tip communities are structured by competitive interactions. Molecular Ecology. 2012;21:5110–5123. doi: 10.1111/j.1365-294X.2012.05739.x. [DOI] [PubMed] [Google Scholar]
- Pritsch et al. (2011).Pritsch K, Courty PE, Churin J-L, Cloutier-Hurteau B, Ali MA, Damon C, Duchemin M, Egli S, Ernst J, Fraissinet-Tachet L, Kuhar F, Legname E, Marmeisse R, Müller A, Nikolova P, Peter M, Plassard C, Richard F, Schloter M, Selosse M-A, Franc A, Garbaye J. Optimized assay and storage conditions for enzyme activity profiling of ectomycorrhizae. Mycorrhiza. 2011;21:589–600. doi: 10.1007/s00572-011-0364-4. [DOI] [PubMed] [Google Scholar]
- R Core Team (2014).R Core Team R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Talbot, Allison & Treseder (2008).Talbot JM, Allison SD, Treseder KK. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Functional Ecology. 2008;22:955–963. doi: 10.1111/j.1365-2435.2008.01402.x. [DOI] [Google Scholar]
- Taylor & Bruns (1999).Taylor D, Bruns T. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: minimal overlap between the mature forest and resistant propagule communities. Molecular Ecology. 1999;8:1837–1850. doi: 10.1046/j.1365-294x.1999.00773.x. [DOI] [PubMed] [Google Scholar]
- Tedersoo et al. (2012).Tedersoo L, Naadel T, Bahram M, Pritsch K, Buegger F, Leal M, Kõljalg U, Põldmaa K. Enzymatic activities and stable isotope patterns of ectomycorrhizal fungi in relation to phylogeny and exploration types in an afrotropical rain forest. New Phytologist. 2012;195:832–843. doi: 10.1111/j.1469-8137.2012.04217.x. [DOI] [PubMed] [Google Scholar]
- Treseder & Lennon (2015).Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiology and Molecular Biology Reviews. 2015;79:243–262. doi: 10.1128/MMBR.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmala et al. (2013).Velmala SM, Rajala T, Heinonsalo J, Taylor AFS, Pennanen T. Profiling functions of ectomycorrhizal diversity and root structuring in seedlings of Norway spruce (Picea abies) with fast- and slow-growing phenotypes. New Phytologist. 2013;201:610–622. doi: 10.1111/nph.12542. [DOI] [PubMed] [Google Scholar]
- Villeneuve, Le Tacon & Bouchard (1991).Villeneuve N, Le Tacon F, Bouchard D. Survival of inoculated Laccaria bicolor in competition with native ectomycorrhizal fungi and effects on the growth of outplanted Douglas-fir seedlings. Plant and Soil. 1991;135:95–107. doi: 10.1007/BF00014782. [DOI] [Google Scholar]
- Visser (1995).Visser S. Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytologist. 1995;129:389–401. doi: 10.1111/j.1469-8137.1995.tb04309.x. [DOI] [Google Scholar]
- Walker et al. (2014).Walker JKM, Cohen H, Higgins LM, Kennedy PG. Testing the link between community structure and function for ectomycorrhizal fungi involved in a global tripartite symbiosis. New Phytologist. 2014;202:287–296. doi: 10.1111/nph.12638. [DOI] [PubMed] [Google Scholar]
- Walker, Ward & Jones (2016).Walker JKM, Ward V, Jones MD. Ectomycorrhizal fungal exoenzyme activity differs on spruce seedlings planted in forests versus clearcuts. Trees: Structure and Function. 2016;30:497–508. doi: 10.1007/s00468-015-1239-7. [DOI] [Google Scholar]
- White et al. (1990).White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic Press, Inc.; New York: 1990. pp. 315–322. [Google Scholar]
- Wright et al. (2005).Wright DP, Johansson T, Le Quéré A, Söderström B, Tunlid A. Spatial patterns of gene expression in the extramatrical mycelium and mycorrhizal root tips formed by the ectomycorrhizal fungus Paxillus involutus in association with birch (Betula pendula) seedlings in soil microcosms. New Phytologist. 2005;167:579–596. doi: 10.1111/j.1469-8137.2005.01441.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Axis loadings are given in Table 1.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as Supplemental Information.