Abstract
Two nuclear receptors, the pregnane X receptor (PXR) and the constitutive androstane receptor (CAR), participate in the xenobiotic detoxification system by regulating the expression of drug-metabolizing enzymes and transporters in order to degrade and excrete foreign chemicals or endogenous metabolites. This review aims to expand the perceived relevance of PXR and CAR beyond their established role as master xenosensors to disease-oriented areas, emphasizing their modulation by small molecules. Structural studies of these receptors have provided much-needed insight into the nature of their binding promiscuity and the important elements that lead to ligand binding. Reports of species- and isoform-selective activation highlight the need for further scrutiny when extrapolating from animal data to humans, as animal models are at the forefront of early drug discovery.
Keywords: PXR, CAR, small molecule, xenobiotic, detoxification, binding promiscuity
1. Introduction
Organisms are constantly exposed to environmental chemicals that can profoundly affect their wellbeing, and they have developed mechanisms to metabolize and excrete these xenobiotics through the concerted transcriptional activation of an arsenal of genes [1]. The xenobiotic detoxification strategy involves nuclear receptors (NRs), such as pregnane X receptor (PXR) and constitutive androstane receptor (CAR), which serve as xenosensors and coordinate the expression of metabolizing enzymes and transporters [2]. Although these chemicals can be eliminated unchanged, the majority first undergo biotransformation through a series of processes that include oxidation and conjugation reactions.
Some of the metabolites that result from the degradation of exogenous substances have themselves the potential to cause inadvertent harmful effects, including toxicity [3]. In addition, the activation of this detoxification and disposition system can reduce the efficacy of concurrently administered drugs or exacerbate their side effects [2]. Importantly, these xenosensors act on endogenous chemicals to maintain appropriate physiologic levels. Among the members of the large NR superfamily, PXR and CAR are predominant in their ability to bind a vast array of chemicals, thereby leading to the upregulation of genes involved in biotransformation and homeostasis [4].
The molecules that can modulate PXR and CAR are diverse and pervasive, ranging from small synthetic organic compounds to large natural products [1]. Because the binding of these molecules to these NRs can result in important biologic effects, this review aims to summarize the relevant findings regarding the modulation of PXR and CAR by small molecules. Emphasis is placed on cross-species differences in PXR and CAR activation, the structural understanding of ligand binding, and the potential clinical impact of these modulators.
2. PXR
PXR (NR1I2), a member of the NR superfamily, is well characterized as a xenobiotic sensor because it binds to structurally diverse chemicals, including numerous clinical drugs and exogenous and endogenous substances [1]. PXR transcriptional activation regulates the expression of its target genes, which encode proteins involved in xenobiotic detoxification and endobiotic metabolism, such as drug-metabolizing enzymes and transporters [5]. PXR transcriptional activation is regulated through either direct binding by a ligand or indirect post-translational modification at key amino acid residues, for example, by phosphorylation [6–10]. Four major structural domains of PXR are involved in its function: a sequence-specific DNA-binding domain (DBD), a flexible hinge, a ligand-binding domain (LBD), and an activation function 2 (AF-2) domain located in the LBD [11]. Upon binding a ligand, the PXR LBD changes its conformation, allowing the AF-2 domain to dissociate from the co-repressor. The subsequent hPXR-co-activator interaction facilitates the binding of the DBD to site-specific DNA sequences, leading to the initiation of transcription of target genes. The heterodimerization of PXR with retinoid X receptor α (RXRα) is also required for ligand-induced PXR transcriptional activation [12].
One of the main physiologic functions of PXR is to regulate the expression of genes encoding drug-metabolizing enzymes and transporters in response to xenobiotic exposure [4, 5]. Therefore, PXR is known as a master regulator of the xenobiotic detoxification system. However, the unwanted activation of PXR by xenobiotics during pharmacotherapy may lead to deleterious reactions, such as adverse drug-drug interactions (DDIs), cancer drug resistance, and liver toxicity [13–15]. Therefore, researchers have long been interested in identifying and investigating the small molecule modulators that can control PXR transcriptional activation.
2.1 Species-selective PXR modulation
Ligand-induced PXR activation displays marked species selectivity. The LBDs of human and mouse PXR differ markedly in their amino acid sequences [16]. Therefore, ligands can selectively bind to the PXR of different species. For example, rifampicin and SR12813 can strongly bind to human PXR (hPXR) but not to mouse PXR (mPXR), whereas 5-pregnen-3β-ol-20-one-16α-carbonitrile (PCN) is a poor ligand for hPXR but a strong agonist for mPXR [17]. The species selectivity of PXR complicates the relevance of using mouse models to evaluate the PXR activation of a given hPXR ligand in vivo and to investigate the in vivo function of hPXR. To overcome the species-related differences in ligand recognition and to enable functional studies of hPXR to be conducted in mice, several humanized mouse models of PXR have been developed for investigating the in vivo activation of hPXR by its ligand. The first-generation hPXR mouse model was developed by randomly inserting the hPXR gene into a mouse genome from which the mPXR gene had been deleted. In this transgenic mouse model, the expression of the hPXR gene was under the control of either the liver-specific albumin promoter [18] or the rat fatty acid–binding protein promoter [19]. Similarly, the second-generation hPXR mouse model was also developed using a transgenic approach. A genomic fragment containing the entire hPXR gene and its promoter was cloned and randomly integrated into a mouse genome with a Pxr-null background [20]. To further improve this model, a double transgenic mouse model expressing hPXR and cytochrome P450 3A4 (CYP3A4) was generated by using bacterial artificial chromosome transgenesis in Pxr-null mice. In this model, rifampicin treatment robustly induces CYP3A4, mimicking the human response to rifampicin [21]. To precisely control the copy number and locus of hPXR genes inserted into the mouse genome, the third-generation hPXR mouse model was developed through knock-in strategies by simultaneously replacing the mPXR gene with the hPXR gene under the control of the endogenous mouse promoter [22]. The same knock-in strategy was used to develop the latest humanized PXR mouse model, in which the genomic sequence of the LBD of hPXR was homologously knocked into the mPXR gene, resulting in the expression of a chimeric PXR (mDBD-hLBD) [23]. This chimeric PXR mouse model was intended to negate the effects of potential differences between hPXRs and mPXRs in terms of the DNA binding affinity of their DBDs. These humanized PXR models have broad application in basic biology research for demonstrating the functions of PXR and in translational research for evaluating the in vivo toxicity and pharmacokinetics of PXR ligands during drug development.
2.2 Potential clinical use of PXR functional modulators
PXR agonists have been extensively investigated and are well documented; they include clinical drugs, phytochemicals, dietary constituents, and endogenous substances. In the clinic, people are cautioned about using PXR agonists because they may cause adverse drug-drug or diet-drug interactions during drug therapy. However, recent clinical and preclinical evidence suggests that some PXR agonists can be used to treat certain diseases, such as inflammatory bowel disease (IBD), through the activation of PXR transcriptional function [24]. In this section, we will highlight the beneficial effects of PXR agonists in treating IBD.
IBD is a group of chronic or recurring conditions characterized by an immune response and inflammation of the gastrointestinal tract. Early association studies of the pathogenesis of IBD revealed that the expression and activity of PXR and the expression of PXR transcriptional target genes were substantially reduced in the intestines of patients with IBD [25]. Moreover, genetic variation in the gene encoding PXR was associated with altered activity of PXR and was strongly associated with susceptibility to adult IBD [26]. By using a Pxr-null mouse model, Cheng and colleagues clearly showed that a PXR-dependent mechanism was involved in dextran sulfate sodium (DSS)–induced IBD and that treatment with PCN, an mPXR agonist, can protect mice from DSS-induced colitis (Cheng et al. 2010). Using a humanized PXR mouse model, the same group further demonstrated that rifaximin, a selective hPXR agonist, exhibited preventive and therapeutic effects on IBD induced by DSS and 2,4,6-trinitrobenzene sulfonic acid (TNBS) in mouse models [27]. Further clinical evidence indicated that rifaximin alone or in combination with other antibiotics was effective at alleviating active IBD, supporting the therapeutic value of PXR agonists in treating IBD [28]. The underlying mechanism by which PXR agonists treat IBD mainly involves reciprocal crosstalk between PXR and NF-κB. Upon PXR activation, the NF-κB signaling cascade can be inhibited, resulting in the suppression of the NF- κB–mediated proinflammatory response [29].
In pursuing the PXR activation approach to treating IBD, several PXR agonists derived from natural products have been identified, and their potential use in IBD therapy has been assessed. Solomonsterol A, isolated from the marine sponge Theonella swinhoei, is a potent PXR agonist and effectively protects humanized PXR mice from developing the clinical signs and symptoms of colitis through PXR-dependent inhibition of NF-κB activation [30]. Artemisinin, a natural compound used to treat malaria, was identified as a potential PXR agonist [31], and a recent report indicated that this drug can prevent and reduce IBD development in a mouse model of DSS-induced IBD by a mechanism associated with the transcriptional activation of PXR [32]. Chrysin, a flavonoid compound that can be extracted from many plants, can activate hPXR and mPXR in reporter gene assays and upregulate xenobiotic detoxification genes in the colon but not in the liver. Chrysin can ameliorate chemical-induced colitis in a mouse model through a PXR/NF-κB–dependent mechanism [33]. Isorhamnetin, an activator of the human PXR, has also been reported to attenuate TNBS-induced colitis in mice [34]. Because flavonoids are important components in many plants, foods, herbal medicines, and health supplements, a recent study systemically examined the effect of 3-hydroxyflavone and its structurally related analogs on PXR activation [35]. By using structure-activity relationship analysis, this study found that flavonols activated PXR in a species-dependent manner and identified 3-hydroxyflavone, galangin, quercetin, isorhamnetin, and tamarixetin as hPXR agonists; this scaffold may serve as a starting point for developing nutraceuticals to treat PXR-associated intestinal diseases, such as IBD.
Studies using genetically modified PXR-related animal models have established PXR as a therapeutic target for IBD. However, rifampicin, a well-known hPXR agonist, has no effect on experimentally induced IBD [27]. One possible explanation for this is that rifampicin is highly bioavailable, and its distribution in the liver can markedly reduce hepatic expression of stearoyl-CoA desaturase-1, leading to lower levels of anti-inflammatory unsaturated fatty acids in the plasma, which counteract the anti-inflammatory function of PXR in the gut [27]. Unlike rifampicin, rifaximin is a gut-specific agonist of hPXR [36]. It is concentrated in the intestine, and less than 0.2% of rifaximin is transported into the liver and kidneys. This suggests that the discovery and development of gut-specific hPXR agonists is needed to treat IBD. In evaluating the therapeutic efficacy of PXR agonists against IBD, both humanized PXR and Pxr-null mouse models should be used to ensure the on-target effect of the agonists.
PXR activation has been linked to several clinical phenomena, such as adverse reactions caused by DDI, drug-induced liver injury (DILI), chemoresistance in cancer patients, and increased risk of Type-2 diabetes [37]. Therefore, there is ongoing interest in developing modulators that can counteract PXR activation. In this review, we define this group of PXR modulators as PXR antagonists. It is anticipated these antagonists will be able to prevent the onset of PXR activation–related clinical symptoms and/or enable their progression to be managed.
Ecteinascidin 743 (ET 743), a natural product derived from the Caribbean marine tunicate Ecteinascidia turbinata, was the first compound reported to have a repressive effect on PXR agonist–mediated activation of PXR target genes, such as CYP3A4 and multidrug resistance protein 1 (MDR1) [38]. However, ET 743 was reported to bind to the minor groove of DNA [39] and can, therefore, interfere with DNA repair pathways, leading to perturbation of the cell cycle and microtubule disorganization [40]. Thus, it remains unclear whether ET 743 negatively modulates PXR activation through direct interaction with the receptor or through another, indirect mechanism(s). The clinical use of ET 743 as a PXR inhibitor is also limited because of its potent cytotoxic properties.
Phytochemicals in the daily diet or herbal medicines are thought to have low or no cytotoxicity and to be relatively safe for potential clinical use. Therefore, great efforts have been made to identify PXR antagonists among such compounds. Sulforaphane (SFN), a major component of certain cruciferous vegetables, was the first naturally occurring PXR antagonist to be reported. In cell-based assays, this compound has IC50 values of 12 to 14 μM against ligand-induced PXR transcriptional activation. SFN has been inferred to bind to the PXR LBD on the basis of scintillation proximity assays, yielding a Ki of 16 μM, and this is further evidenced by its disruption of co-activator recruitment when evaluated using the mammalian two-hybrid assay [41]. Computational results suggest that this molecule may also interact with the outer surface of PXR at the AF-2 domain [42]. Thus, the molecular mechanism and ligand-receptor binding model of SFN action on PXR are still exclusive. Moreover, a recent human clinical study suggested that this compound was not an effective antagonist of the hPXR in vivo [43]. Considering the antagonistic effect on PXR of SFN at relative high concentrations in vitro and its other pharmacologic activities, such as histone deacetylase inhibition [44], achieved at relatively lower concentrations, further investigation of the on-target effect of SFN against PXR is warranted, especially at physiologically relevant concentrations.
Coumestrol, a phytoestrogen prevalent in legumes and soy beans, is another naturally occurring chemical that affects PXR transcriptional activation [45]. Very similar to SFN, this compound can elicit an antagonistic effect on PXR activation at relative high concentrations, with an IC50 value of 12 μM in a PXR promoter reporter assay and a Ki value of 13 μM in competitive ligand binding assays of the PXR LBD [45]. Further mutagenesis studies have shown that the compound binds to the outer surface of the PXR LBD. Collectively, this evidence suggests that the binding of coumestrol to the PXR ligand binding pocket is weak [42, 45]. Other phytochemicals, including sesamin (a lignan found in sesame seeds) and camptothecin (a quinoline alkaloid isolated from the plant Camptotheca acuminata), have also been reported to functionally block PXR activation–mediated gene expression [46, 47]. However, there is still no clear evidence of their direct engagement with PXR. Therefore, this class of compounds can be regarded as functional blockers of PXR, as opposed to PXR antagonists that can directly affect the binding of an agonist to this target.
The most investigated synthetic inhibitors of PXR transactivation are the azole class of chemicals, including ketoconazole and its derivatives [48]. A pioneer study found that ketoconazole, an antifungal drug, was active in repressing ligand-induced PXR activation, and it can also decrease the interaction of PXR with the co-activator SRC-1 and co-repressor SMRT in a mammalian two-hybrid assay in HepG2 cells [49]. One group used biochemical and cell-based assays and molecular docking computations to gain further molecular and structural insights into the action of ketoconazole in regulating the transcriptional activity of PXR. They concluded that ketoconazole can bind at the AF-2 surface of PXR, rather than in the ligand-binding pocket, resulting in PXR transcriptional regulation [50–52]. By using a yeast two-hybrid screening strategy together with computational docking analysis, the same group recently identified a putative binding region on the AF-2 domain of PXR that allows the binding of ketoconazole to this receptor. The binding region includes Gln-272, Phe-264, and Ser-208 of PXR, which could be critical for disrupting the PXR–SRC-1 interaction due to the binding of ketoconazole to PXR [53]. Because ketoconazole is a well-documented inhibitor of the enzymatic activity of CYP3A4 and can somehow cause hepatic toxicity, a group of its analogs were synthesized in order to develop a safer PXR activation blocker with fewer off-target effects [54]. FLB-12, a ketoconazole analog, was discovered as a result of a structure-activity relationship study. FLB-12 exhibited relative selectivity against ligand-induced PXR activation in vitro and in vivo, no inhibition of CYP3A4 activity, and less toxicity than ketoconazole. In pharmacologic studies, FLB-12 abrogated the PXR-mediated resistance to 7-ethyl-10-hydroxycamptothecin (SN-38) of colon cancer cells in vitro and attenuated PXR-mediated acetaminophen hepatotoxicity in vivo [55]. In light of these studies, developing selective PXR antagonists appears to be a feasible approach for managing PXR-related adverse DDIs and cancer drug resistance.
PXR antagonists could, theoretically, act more selectively by directly competing with the binding of agonists to the ligand-binding pocket of PXR. However, no such compound has yet been reported, at least in part because of the promiscuous nature of this receptor for ligand binding. Because the PXR ligand-binding cavity is comparatively large, this receptor can bind compounds with diverse chemical structures. Therefore, it could be challenging to discover a compound that specifically and directly competes with the binding of structurally diverse PXR agonists to the ligand-binding pocket of PXR. However, our recent experience leads us to believe that large-scale high-throughput screening, using a large collection of structurally diverse compounds, might be an effective approach to discover this novel class of PXR antagonists characterized by 1) direct binding at the PXR binding pocket, 2) effective antagonism against variable well-characterized PXR agonists, 3) on-target efficacy in a humanized PXR mouse model, and 4) no obvious toxicity in vitro or in vivo.
3. CAR
Originally identified as MB67 [56], CAR belongs to the Type I NR family and is denoted as NR1I3. The structural domains of CAR are arranged much like those of other NRs, and the function of CAR is determined by its domains; the DBD, the dimerization domain, and the LBD. CAR is a transcription factor that binds to specific response elements in the promoter regions of CAR target genes. Activated CAR uses RXRs as heterodimeric partners for DNA binding [56, 57]. The best described CAR response element is the phenobarbital-responsive enhancer module (PBREM), which is present in the promoter regions of CAR target genes. For example, PBREM is found in the promoter region of the CYP2B6 gene, approximately 1.8 kb upstream of the transcriptional start site, and harbors two direct repeat separated by 4 nucleotides (DR4) motifs [58]. CAR also binds DNA at the xenobiotic responsive enhancer module (XREM), which is also found in the CYP2B6 distal regulatory regions and contains one DR4 motif surrounded by half sites [59]. CAR shares these binding sites with PXR [18, 60]. This commonality of DNA response elements, among other factors, allows CAR and PXR to co-regulate a subset of their target genes, though they also separately regulate other genes [61].
CAR is referred to as a xenobiotic sensor owing to its broad specificity for both endogenous and exogenous ligands with varying chemical structures. Solving the structure of the CAR LBD has enabled researchers to understand this phenomenon and has suggested that the large hydrophobic binding region of the CAR LBD sanctions binding to diverse chemical structures. The AF-2 region in the CAR LBD appears to be rigidly fixed in an active conformation, even in the absence of ligands, and ligand docking further supports the idea that conformational changes affect the transactivation activity of CAR [62].
3.1 CAR functions as a transcription factor
CAR expression is modulated by other NRs, such as hepatocyte nuclear factor 4 alpha (HNF4α), peroxisome proliferator-activated receptor alpha (PPARα), glucocorticoid receptor (GR), and possibly others [63–65]. It is well documented that glucocorticoids (such as dexamethasone) increase the amount of CAR mRNA in the liver [65]. CAR expression is increased by a variety of xenobiotics; BMS-665351 is an inhibitor of insulin-like growth factor 1 receptor (IGF-1R) that selectively induces CAR expression without activating GR, PPARα, or HNF4α, implying the involvement of as yet unknown mechanisms in the transcriptional regulation of CAR [66].
Activated CAR regulates the expression of genes involved in the metabolism and efflux of xenobiotics and endobiotics. These target genes are often phase I, II enzymes and transporters that regulate the metabolism and secretion of endogenous and exogenous signaling molecules, including many members of the CYP family, SULTs, UGTs, GST, membrane transporters, and others. Among the phase I enzymes, CYP2B6 is considered a primary target gene for CAR activation, as evidenced by the well-established CAR-binding promoter regions. CYP2B6 is also co-regulated by PXR. Additionally, even though CYP3A4 is thought to be mainly regulated by PXR, activated CAR can increase CYP3A4 levels [67, 68]. Studies have revealed that, together, CAR and PXR play key roles in xenobiotic and endobiotic metabolism, especially in liver and intestinal tissues, and in maintaining cellular and systemic homeostasis. Bilirubin breakdown and subsequent heme reabsorption is regulated by CAR through UGTs and membrane transporters that promote bilirubin excretion [69–71].
CAR activation also appears to change the metabolic environment in the liver in response to drugs. Phenobarbital (PB) activation of CAR causes a repression of hepatic gluconeogenic enzymes, such as phosphoenoylpyruvate carboxykinase 1 (PEPCK1) and glucose-6-phosphatase (G6Pase) [72], suggesting that CAR plays a critical role in regulating hepatic energy metabolism in response to drug exposure. CAR helps maintain hormone homeostasis by responding to androgens, estrogens, and other hormones that are metabolized by the CAR target genes SULTs and UGTs [73, 74]. CAR also directly regulates the synthesis of the thyroid hormone [75]. Such crosstalk between CAR and other NRs may explain the endocrine disruptions that result from exposure to certain xenobiotics. Thus, as a xenosensor, CAR can regulate several important physiologic processes that maintain systemic homeostasis.
3.2 Species selectivity of CAR
CAR exhibits remarkable species selectivity in its ligand binding and activation profiles. Human and rodent CARs share several common characteristics, such as nuclear translocation on direct or indirect activation and DNA response element sequences that recruit CAR. On the other hand, in contrast to most mammalian CARs, the activation of the Xenopus CAR seems to be translocation independent, as these receptors accumulate in the nucleus, even in the absence of activators [76]. CAR is also absent in most fish species [76]. Compared to the more than 20 functional hCAR splice variants identified, far fewer CAR variants have been detected in other species, and only approximately five in the rat [77] and pig [78], most of which generate truncated protein products. Comparative genomic analyses show that mice, rats, and marmosets do not have a conserved CAR2 site and are thus incapable of generating a protein analogous to hCAR2. CAR2 in human tissue has been shown to bind di(2-ethylhexyl) phthalate (DEHP) and acts as the main human DEHP receptor [79]. This means that rodent models of DEHP toxicity will not accurately profile potential human toxicity because of their inability to generate a CAR2-like transcript. Porcine CAR (pgCAR) might be more homologous to human CAR than is rodent CAR, as is evidenced by the responses of pgCAR and hCAR to ligands being more similar than are those of hCAR and mCAR [78].
All things considered, drug metabolism data generated from rodent models may not reflect human CAR functions accurately, and caution is necessary when directly extrapolating from mouse/rat data to humans. In a classic example of species selectivity, 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), an mCAR agonist, activates mouse but not human CAR, and pharmacologic concentrations of androstanol repress mouse but not human CAR [80, 81]. CITCO, an hCAR agonist, binds and activates hCAR but has no effect on mCAR. There is also an important species-related difference in hepatic energy metabolism between hCAR and mCAR. Whereas hCAR activation selectively inhibits gluconeogenesis without suppressing fatty acid synthesis, mCAR activation results in the downregulation of genes associated with gluconeogenesis (G6Pase and PEPCK1), fatty acid synthesis (Fas and Acc-α), and lipogenesis [sterol regulatory element-binding protein 1c (SREBP-1c) and Scd-1] [82].
The apparent species-specific differences between human CAR and CAR of other animals are particularly important when choosing animal models for analyzing the pharmacologic effect of new drugs on CAR induction and the subsequent activation of downstream CYPs. Rodent models are commonly used to evaluate the potential of clinical drugs and environmental agents for being tumor initiators or tumor promoters in humans. In the case of CAR, particular attention is needed when evaluating the carcinogenic potential of chemicals in animal models and humans, considering the reduced interspecies homology of CAR LBDs. For example, PB and other CYP2B inducers can act as nongenotoxic carcinogens and tumor promoters in mice and rats but appear to have a low likelihood of producing liver tumors in humans. In fact, Lake and colleagues postulated that human CAR activators do not increase replicative DNA synthesis in human hepatocytes, whereas the activation of rodent CAR results in liver hypertrophy, increased replicative DNA synthesis, altered hepatic foci, and liver tumors [83, 84].
3.3 Isoform selectivity of CAR modulation
The liver expresses high levels of CAR, and a lower level of CAR expression has been detected in brain, intestinal, heart, adrenal, testis, prostate, and kidney tissues [56, 85, 86]. CAR exists in many alternatively spliced variants [57, 87, 88] with diverse tissue expression profiles [85, 89–92], heterodimerization properties [89, 93], and ligand specificities [79, 94–98]. The organ-specific expression of CAR splice variants requires further exploration in order to understand the variations in drug metabolism and DDIs. The best studied human CAR variant is constitutively active when expressed in immortalized cells and is denoted as hCAR1 or simply CAR1 (348aa; NM_005122) by most researchers. Table 1 lists all annotated CAR isoforms identified so far.
Table 1.
List of annotated human CAR variants from the NCBI and Ensembl databases
| CAR splice variant |
Transcript variant |
NCBI ID | Transcript ID on Ensembl* |
Transcript length (nt) |
Protein length (aa) |
Annotated coding sequence |
Common name |
|
|---|---|---|---|---|---|---|---|---|
| 1 | NR1I3-036; NR1I3-201 | 1 | NM_001077482.2 | ENST00000367980 | 1074 | 357 | CCDS41429.1 | SV25 |
| 2 | NR1I3-002 | 2 | NM_001077480.2 | ENST00000367982 | 1059 | 352 | CCDS41430.1 | hCAR2; SV23 |
| 3 | NR1I3-001 | 3 | NM_005122.4 | ENST00000367983 | 1047 | 348 | CCDS1228.1 | hCAR1; WT; REF; SV0 |
| 4 | NR1I3-012 | 4 | NM_001077481.2 | ENST00000367985 | 945 | 314 | CCDS41428 | |
| 5 | NR1I3-011 | 5 | NM_001077471.2 | ENST00000367984 | 930 | 309 | CCDS44262 | |
| 6 | NR1I3-031 | 6 | NM_001077469.2 | ENST00000428574 | 1023 | 340 | CCDS44261.1 | |
| 7 | NR1I3-029 | 7 | NM_001077478.2 | ENST00000442691 | 1020 | 339 | CCDS 44260.1 | |
| 8 | NR1I3-032 | 8 | NM_001077474.2 | ENST00000505005 | 985 | 296 | CCDS53405 | |
| 9 | NR1I3-019 | 9 | NM_001077472.2 | ENST00000367981 | 975 | 324 | CCDS41427.1 | |
| 10 | NR1I3-015 | 10 | NM_001077479.2 | ENST00000511676 | 960 | 319 | CCDS53409 | |
| 11 | NR1I3-014 | 11 | NM_001077470.2 | ENST00000504010 | 944 | 280 | CCDS53407 | |
| 12 | NR1I3-030 | 12 | NM_001077473.2 | ENST00000412844 | 948 | 315 | CCDS53411 | |
| 13 | NR1I3-033 | 13 | NM_001077476.2 | ENST00000508740 | 977 | 311 | CCDS53410 | |
| 14 | NR1I3-025 | 14 | NM_001077477.2 | ENST00000437437 | 973 | 306 | CCDS53408.1 | |
| 15 | NR1I3-024 | 15 | NM_001077475.2 | ENST00000512372 | 861 | 267 | CCDS53406 | |
| 16 | NR1I3-X01 | X1 | XM_005245693.3 | -- | 1290 | 429 | CDS from 236..1525 | |
| 17 | NR1I3-X02 | X2 | XM_005245694.3 | -- | 1275 | 424 | CDS from 235..1509 | |
| 18 | NR1I3-X03 | X3 | XM_011510237.1 | -- | 1161 | 386 | CDS from 234..1394 | |
| 19 | NR1I3-X04 | X4 | XM_005245697.3 | -- | 1062 | 353 | CDS from 87..1148 | hCAR3; SV24 |
| 20 | NR1I3-008 | -- | -- | ENST00000515452 | 1093 | 238 | Protein coding | |
| 21 | NR1I3-009 | -- | -- | ENST00000506209 | 1441 | 297 | Protein coding | |
| 22 | NR1I3-013 | -- | -- | ENST00000508387 | 634 | 118 | Protein coding | |
| 23 | NR1I3-016 | -- | -- | ENST00000502985 | 774 | 147 | Protein coding | |
| 24 | NR1I3-021 | -- | -- | ENST00000515621 | 1130 | 273 | Protein coding | |
| 25 | NR1I3-026 | -- | -- | ENST00000511944 | 620 | 147 | Protein coding | |
| 26 | NR1I3-027 | -- | -- | ENST00000511748; ENST00000628566 | 357 | 118 | Protein coding | |
| 27 | NR1I3-010 | -- | -- | ENST00000506018 | 1380 | 274 | Nonsense- mediated decay | |
| 28 | NR1I3-017 | -- | -- | ENST00000512340 | 891 | 90 | Nonsense- mediated decay | |
| 29 | NR1I3-018 | -- | -- | ENST00000510951 | 774 | 90 | Nonsense- mediated decay | |
| 30 | NR1I3-020 | -- | -- | ENST00000502848 | 914 | 119 | Nonsense- mediated decay | |
| 31 | NR1I3-023 | -- | -- | ENST00000507215 | 620 | 90 | Nonsense- mediated decay | |
| 32 | NR1I3-004 | -- | -- | ENST00000488651 | 580 | No protein | Processed transcript | |
| 33 | NR1I3-005 | -- | -- | ENST00000464422 | 480 | No protein | Processed transcript | |
| 34 | NR1I3-006 | -- | -- | ENST00000479324 | 764 | No protein | Processed transcript | |
| 35 | NR1I3-035 | -- | -- | ENST00000503547 | 535 | No protein | Processed transcript | |
| 36 | NR1I3-003 | -- | -- | ENST00000491193 | 1435 | No protein | Retained intron | |
| 37 | NR1I3-034 | -- | -- | ENST00000505944 | 865 | No protein | Retained intron |
The investigation of hCAR activation by flavonoids revealed interesting information regarding ligand specificities for some hCAR splice variants. 3-hydroxyflavone indirectly activated the hCAR2 (denoted SV23; NM_001077480.2) and hCAR3 (denoted SV24; XM_005245697.3), but without transactivation through LBD binding or recruitment of steroid receptor co-activators (SRCs). 3-hydroxyflavone can also activate wild-type (WT) hCAR by directly binding to its LBD, along with other flavonols, such as galangin, quercetin, and tamarixetin, but has no effect on full-length hCAR (denoted SV1; NM_001077482.2) [98]. The DEHP is a highly potent and selective agonist of hCAR2 [79, 99]. The antimalarials artemisinin, artemether, and arteether activate hCAR2 (SV23) in the absence of fetal bovine serum, but the direction of their effect can be reversed if serum-free conditions are used during cell culture [95]. The antibacterial agent triclosan (2,4,4'-trichloro-2'-hydroxydiphenylether) is an inverse agonist of hCAR1 but a weak agonist of hCAR3 [100]. The antiretroviral drugs rilpivirine and etravirine activate hCAR in an isoform-dependent manner. Both drugs activate the constitutively expressed WT hCAR without direct binding to the LBD or SRC recruitment [97]. Most probably, this phenomenon is a result of indirect CAR activation mechanisms, as in the case of PB and phenytoin. Interestingly, however, neither drug has any effect on the inducible variants hCAR2 and hCAR3. In addition to splice variants, various clinical variants resulting from single-nucleotide polymorphisms in the CAR gene have also been identified [101], and these might confer differential functional effects, such as changes in bone mineral density or in the pharmacokinetics of the anti-HIV drug efavirenz [102, 103].
In one study of the splice-variant profile of CAR in the liver, researchers observed interethnic and interindividual variations among human liver samples from Koreans and Caucasians, including four novel splice variants in Korean livers [104]. The most predominant splice variant in the 30 Korean livers analyzed was, surprisingly, not the WT hCAR. These results suggest that there is a range of expressional variation of hCAR splice variants among different ethnic groups. Variations in CAR activity that result from genetic polymorphisms or from alternative splicing may influence the expression of CAR target genes and contribute to the overall interindividual variability in drug metabolism.
3.4 Indirect activation of CAR
Indirect activation of CAR with PB or phenytoin has been well documented [105, 106] and is now understood to be a common mechanism of CAR activation, with an increasing number of indirect activators having been identified [e.g., flavonoids [107], triclosan [108], or acetaminophen [109]]. These activators do not bind directly to CAR; instead, they activate CAR by stimulating its nuclear translocation in a ligand-independent manner. In rodent hepatocytes, a PB-mediated increase in CYP2B mRNA had been documented before the discovery of CAR, but the latter shed light on the mechanism of action underlying this phenomenon [58]. In the native hepatocyte environment, CAR is sequestered in the cytoplasm as part of a multiprotein complex that includes the heat-shock protein 90 [110]. Upon stimulation by indirect activators or direct ligand-binding, CAR disassociates from the cytoplasm-localized protein complex and moves into the nucleus, subsequent to dephosphorylation at Thr-38. The pathway of indirect CAR activation by PB involves the EGF receptor (EGFR) signaling pathway, in which PB binding to EGFR antagonizes EGF activation of EGFR, which eventually leads to the dephosphorylation of CAR at Thr-38 [111, 112]. Dephosphorylation of CAR activates nuclear translocation, and transcriptional activity ensues [112], even in the absence of direct ligands. Similarly, small molecule modulators of various kinase pathways (growth hormones and cytokines) that converge in changing the phosphorylation status of CAR (via ERK, MAPK, PP2A, etc.) are all capable of regulating CAR nuclear translocation and, ultimately, CAR function [113–116]. It should be noted that PB can indirectly activate NRs other than CAR, PXR [63], and PPARα [64], and hence the indirect method is not a specific mechanism of receptor activation.
The localization status of CAR in its —non-active state might be ambiguous. True to its name, CAR is indeed constitutively active when exogenously expressed in immortalized cell lines. Many research groups have shown that CAR is predominantly cytoplasmic in liver tissue of animals in the absence of ligands [66, 113, 117, 118]. While CAR spontaneously accumulates in the nucleus in immortalized cells, Lynch et al. have illustrated by using adenovirus expressing fluorescent-hCAR in human primary hepatocytes, that CAR is predominantly retained in the cytoplasm prior to chemical stimulation. However a small percentage (~7%) of total CAR protein was detected in the nucleus [119]. It is difficult to assess the transcriptional function of this low level of nuclear CAR, especially when using the expression of CAR target genes to monitor the activity of CAR, as these genes may be regulated by multiple transcription factors (e.g., CYP2B6 can be regulated by CAR, PXR, HNF4α and others). Moreover, CAR has been shown to be sensitive to the activation states of various signaling pathways including EGF, IGF , AMPK, and ERK-MAPK [120]. These pathways regulate the phosphorylation status of CAR, and dephosphorylation of a single CAR residue at Thr38 induces the nuclear translocation of CAR and stimulates its transcriptional activity [111]. This, as discussed earlier, has been shown to be the main mechanism by which indirect activators, such as PB, stimulate CAR function even in the absence of direct binding ligands. Translocation of CAR to the nucleus drives its transcriptional activation. It is conceivable that the ratio of CAR distributed between the cytoplasmic and nuclear compartments in the absence of ligands, is dependent on a fine balance of signaling molecules and hence needs to be taken into consideration when discussing the CAR localization.
3.5 Potential clinical use of CAR modulators
3.5.1 CAR in metabolic syndromes
Several reports have delineated the antilipogenic properties of CAR. CAR activation suppresses lipid metabolism and lowers serum triglyceride by inducing Insig-1, which in turn reduces the levels of the lipogenic transcription factor SREBP-1 [65]. CAR activation in mice is associated with an improved serum lipid profile and increased insulin sensitivity. The mouse CAR activator TCPOBOP markedly suppresses adipose deposition and weight gain induced by a high-fat diet in mice [121–123]. Previously, in human studies, it has been noted that PB decreases plasma glucose levels and improves insulin sensitivity in diabetic patients [124]. The anti-epileptic drug phenytoin, which is also an indirect activator of CAR, increases the levels of high-density lipoprotein (HDL) cholesterol [125, 126]. CAR activation in mice decreased HDL and plasma apolipoprotein A-I, suggesting a protective role for CAR in atherosclerosis, cardiovascular ailments, and obesity [60]. Because CAR plays a role in reducing insulin resistance, Masuyama and Hiramatsu considered its role in the pathogenesis of preeclampsia (a complication of pregnancy that is characterized by high blood pressure and proteinuria) in individuals with insulin resistance and adipocyte dysfunction. In pregnant mice in which obesity had been induced by a high-fat diet, CAR activation with TCPOBOP resulted in improved glucose tolerance, with significant changes in the expression of gluconeogenic, lipogenic, and adipocytokine genes and reduced hypertension and proteinuria [127]. This suggests that CAR is a potential therapeutic target for obese patients with preeclampsia and insulin resistance, though no parallel studies in humans have yet been conducted.
Much effort has been exerted to understand the role of CAR activation in bile acid homeostasis in mice. Increased bile acid metabolism and detoxification, together with reduced bilirubin and bile acid serum levels, have been observed in healthy mice upon CAR activation [128, 129]. Hepatic necrosis, increased alanine aminotransferase levels, and a corresponding increase in serum concentrations of bilirubin have been observed in CAR KO mice, suggesting an important role for CAR in hepatic maintenance [129]. CAR agonists could potentially be used to treat cholestasis in patients with bile acid dysregulation.
3.5.2 CAR in cancer
The function of CAR in tumor maintenance and the development of chemoresistance is less clear than are its roles in other processes. In rodent models, it is accepted that CAR activators may act as nongenotoxic carcinogens and tumor promoters. PB activation of CAR induces the Gadd45b gene in mouse liver. GADD45B represses apoptosis, and CAR interacts with GADD45B to repress TNFα-induced JNK1 phosphorylation and cell death, thus promoting the development of hepatocellular carcinoma in mice [130]. However, studies of human CAR have attributed no carcinogenic properties to CAR activators [131]; in fact, some reports suggest that chemotherapeutic efficacy may be increased upon CAR activation. For example, the chemotherapeutic agent cyclophosphamide is converted from the prodrug to the active form by the enzyme CYP2B6, which in turn is a target of CAR activation, thus making it advantageous to use CAR agonists to improve chemotherapeutic efficacy [132]. Similarly, CAR agonists modestly increased paclitaxel-induced tumor depletion in five human lung cancer cell lines [133]. The growth and expansion of brain tumor stem cells was inhibited by the human CAR agonist CITCO in isolated human gliospheres and in xenograft mouse studies by inducing cell cycle arrest and apoptosis [134]. Conversely, CAR is known to regulate the expression of MDR1, which functions as a transmembrane efflux pump for eliminating anticancer agents and plays a major role in the development of chemoresistance. In four ovarian cell lines expressing CAR, treatment with the CAR activator CITCO and an anticancer agent caused an upregulation of MDR1 and UGT1A1, decreased the efficacy of the anticancer agent, and reduced the levels of apoptosis to a greater extent than did the anticancer drug alone [135]. Similar observations were made for multiple anticancer agents. CAR inhibition reversed these effects and increased both chemotherapeutic efficacy and apoptosis.
In summary, the data from these investigations has advanced our understanding of the role of CAR: once merely a well-known xenobiotic sensor and endobiotic modulator, CAR is now seen as a promising drug target for therapy of metabolic disorders and cancer.
4. Structural features of the PXR and CAR LBDs
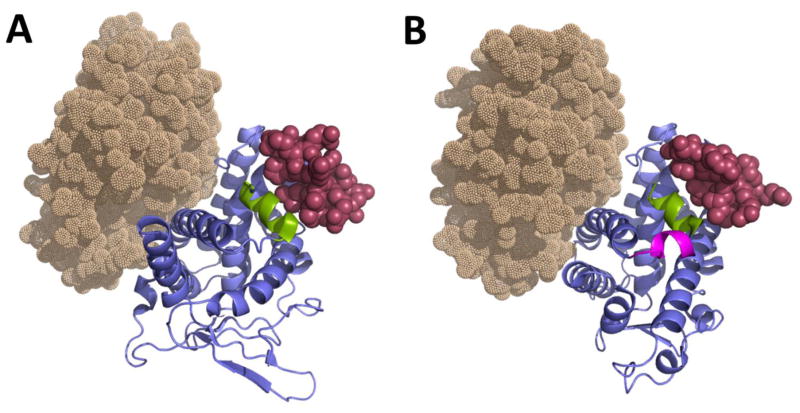
The PXR and CAR LBDs represent a compact α-helix sandwich fold (Figure 1), with the PXR LBD consisting of three sets of α-helices: α1/α3, α4/α5/α8/α9, and α7/α10 [136]. The CAR LBD consists of 11 α-helices [62, 137], with the two 310 helices α2 and α2′ being believed to contribute to the hydrophobic character of the binding pocket while possibly forming part of the ligand access gate [138]. In contrast to the three β-strands present in CAR, PXR contains a layer of five-stranded antiparallel β-sheets, which include the two novel β-strands (β1 and β1′) that are not observed in other NRs. It encompasses an insert of approximately 60 amino acids between the α1 and α3 helices, which cover β1, β1′, and the novel helix α2. There is evidence that PXR homodimerizes through the terminal β1′ strands from each monomer by interlocking the corresponding Trp-223 and Tyr-225 residues [139]. Trp-223Ala and Tyr-225Ala double mutants prevent homodimerization, with an associated reduction in CYP3A4 induction.
Figure 1.
Crystal structures of hPXR LBD (PDB code 4J5X) and hCAR LBD (PDB code 1XV9). The LBDs of PXR (A) and CAR (B) are depicted in blue (cartoon representation), showing their respective AF-2 helices in green and the co-activator SRC-1 peptide as maroon spheres. The —helix x that is notable in the CAR structure is shown in pink. The RXRα LBD is shown as light brown dots. Images of all the protein structures have been generated using the program Pymol.
Co-activators and co-repressors, such as SRC-1 and SMRT, bind the AF-2 surface of the LBD through the Leu-Xxx-Xxx-Leu-Leu (for co-activators) and Ile/Leu-Xxx-Xxx-Ile/Val-Ile (for co-repressors) motifs [140]. The interaction of the agonist with the LBD leads to the exposure of a hydrophobic surface for co-activator binding (Figure 1). Crystal structures show that the SRC-1 peptide is buried in a groove on the surface of the PXR LBD that is composed of the AF-2 helix, α3, and α4, with all the Leu residues of the peptide being in contact with the surface of the protein [141]. Polar interactions involving Leu-259 and Glu-427 of hPXR contribute to the formation of a —charge clamp that is believed to stabilize contacts with the co-activator peptide. The co-activator recruitment appears to play a vital role in fixing the ligand in the correct arrangement. The structure of PXR when binding the cholesterol-lowering drug SR12813 in the absence of SRC-1 reveals that the ligand occupies the binding cavity at three distinct positions [136], which corroborates the flexible nature of the LBD. However, in structures consisting of PXR-SR12813 in complex with an SRC-1 peptide, the agonist is constrained to a single binding orientation [141]. The idea that SRC-1 is necessary in order to restrict the bound ligand to a single active conformation led to the generation of binding proteins constructed using the fibronectin type III domain (Adnectins) that attach in the same location as do SRC-1 peptides, in the belief that these biologics would provide additional advantages over the SRC-1 peptides as chaperones for crystallographic studies [142]. It was anticipated that problems arising from the lack of electron density resolution observed in some structures when using small peptides would be ameliorated by the use of the larger Adnectins, and their higher affinity for PXR would increase the chances of trapping the ligand in a single binding mode.
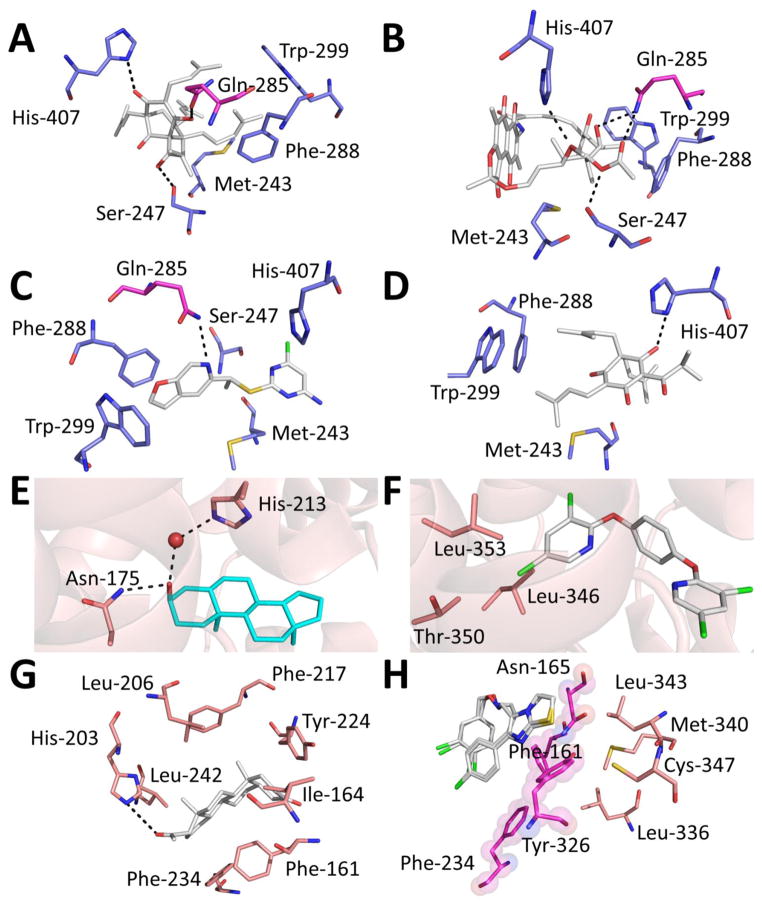
Even though agonist binding to the LBD forces the AF-2 helix to be positioned in the active conformation for co-activator recruitment, a review of the large set of available hPXR and CAR structures revealed that not all agonists interact directly with residues in the AF-2 helix. In the case of PXR, TO901370 and compound 1 interact with Met-425 [142, 143], and PNU-142721 and estradiol interact with Met-425 and Phe-429 [144, 145]. In addition to Met-425 and Phe-429, SR12813 makes contact with Phe-420, which is adjacent to the AF-2 helix [141]. Colupulone also interacts with Phe-420 in addition to Met-425 [146]. However, no interaction has been observed with hyperforin [147]. When in complex with hCAR, 5β-pregnanedione and CITCO did not form direct contacts with the AF-2 helix [62], which was blocked from the binding cavity by a barrier composed of the residues Phe-161, Asn-165, Phe-234, and Tyr-326 [62]. Interestingly, the mCAR superagonist TCPOBOP penetrated this barrier, allowing it to interact with the AF-2 residue Leu-353 and the linker helix residues Leu-346 and Thr-350 [137].
Important features of the constitutive nature of CAR include the short and rigid AF-2 helix with a missing C-terminal extension, as is found in other NRs [62, 137, 138], which results in further stabilization of the active AF-2 conformation due to an interaction between the free carboxylate and Lys-195 (hCAR). A short —helix x composed of only four to six residues replaces the extended loop linking α10 and the AF-2 (Figure 1). Because only a single amino acid separates helix x from the AF-2 helix, the latter is restricted to favoring the active orientation. This hypothesis requires validation, as other NRs exhibit a comparable helix with no basal activity [148]. The structure of mCAR when in complex with the inverse agonist androstenol provides additional insights about its constitutive activity and its repression by this inhibitor [138]. In the active conformation, CAR exhibits α10 and α11 as a single continuous helix, but androstenol causes the formation of two distinct helices separated by a —kink that ultimately leads to the destabilization of the AF-2 helix [138].
RXR heterodimerizes with a number of NRs, including PXR and CAR. Association with RXR was shown to increase the affinity of CAR for ligands and the co-activator [137]. It has been suggested that the large heterodimerization surface contributes to the constitutive nature of CAR by stabilizing the active conformation [149]. The affinity of the heterodimeric PXR and RXR duplex for the co-activator peptide is greater than that of either individual receptor, but this cooperativity does not apparently involve significant structural changes during heterodimerization [150]. Ligand binding appeared to stabilize the complex and enhance co-activator recruitment. Therefore, NR activation involves more than just a sequence of events starting with ligand binding; instead, there is dynamic interplay among all the participating elements.
Unlike other NRs that have evolved to bind very selective ligands, PXR and CAR do not discriminate between molecules on the basis of size or chemical composition. This is largely due to the highly flexible and fluid LBD. The binding cavity of PXR is considerably larger than that of other NRs. The volume of the binding cavity of the apo-PXR is approximately 1150 Å3 [136]. Ligand binding changes the shape of the pocket to fit the bound molecule, which is generally accompanied by an overall volume expansion. The size of the binding cavity increases to 1544 Å3 with hyperforin [147] and to 1344 Å3 with SR12813 [141]. However, the particular area that is in direct contact with SR12813 shrinks when the SRC-1 peptide is bound to PXR, thus constraining the ligand to being positioned in a single orientation, in contrast to the multiple modes observed in the absence of the co-activator. The change in volume upon ligand docking follows an internal rearrangement of certain structural features, such as helix α2. The observation that CAR is less promiscuous than PXR could be explained, to a large extent, by its smaller ligand pocket size, which ranges between 525 Å3 and 675 Å3 [62, 137, 138].
The ligand pockets of PXR and CAR are lined by mostly hydrophobic residues, but there are a few polar amino acids present that can form significant interactions with ligands. The cavity of PXR encompasses 28 amino acids, of which eight have polar or charged side chains [136]. Similarly, the 27 residues of the CAR pocket create a highly hydrophobic environment, with only a quarter of them being polar [138]. In both receptors, amino acid charge effects are further neutralized in the interior or vicinity of the ligand site by salt bridges, such as those between Glu-321 and Arg-410, between Asp-205 and Arg-413 in hPXR [136], or between the pairs Asp-238/Arg-156 and Glu-225/Lys-235 in mCAR [138].
Because the binding pocket is composed mainly of nonpolar residues, it is no surprise that hydrophobic and van der Waals forces play crucial roles in the interactions of the ligand with PXR and CAR protein residues. In the case of PXR, other important pairwise contacts have been observed in many of the structures elucidated thus far, as illustrated in Figures 2 and 3 [141–143, 145–147, 151]. In several cases, such as with hyperforin, SR12813, and rifampicin, Ser-247 and His-407 in hPXR participate jointly in hydrogen bonding. Gln-285 forms direct hydrogen bonds with hyperforin, TO901370, rifampicin, compound 1, and via a water molecule with colupulone. Far less common involvement was noticed for Arg-410 (interacting with 17β-estradiol) and His-327 (coupling to TO901370). Aromatic side chains have been implicated in forming additional attractive forces: Trp-299 forms π-stacking contact with SR12813, TO901370, and compound 1, with Phe-288 and Tyr-306 participating with less frequency. In sharp contrast to PXR, hydrogen bonding between ligands and residues lining the cavity of CAR is not that common: CITCO [62] and TCPOBOP form no hydrogen bonds [137], 5β-pregnanedione participates in a single hydrogen bond with His-203 in hCAR [62], and androstenol forms a direct bond with Asn-175 and a water-mediated interaction with His-213 in mCAR [138]. Hence, most of the CAR ligands co-crystallized thus far depend heavily on hydrophobic contacts for binding.
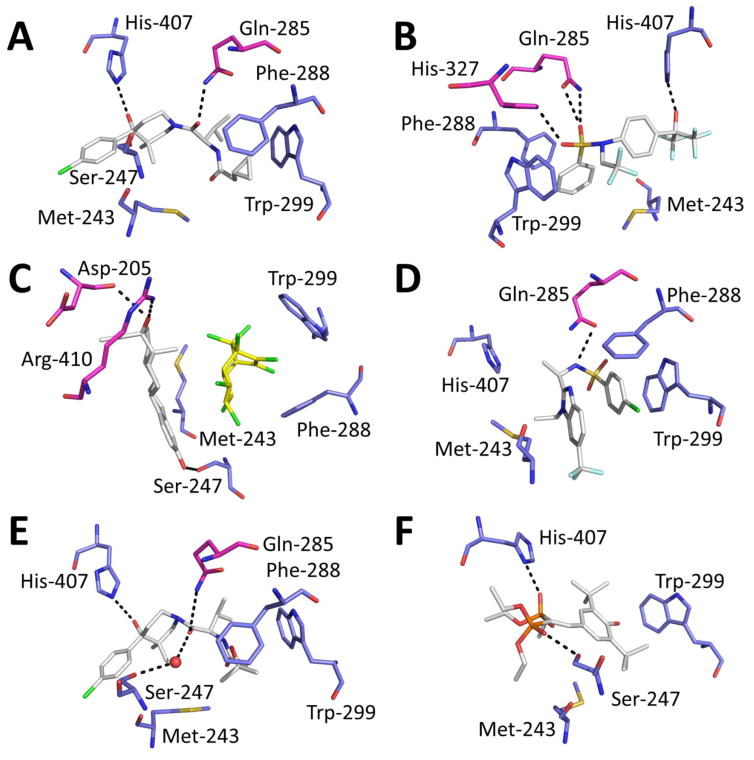
Figure 2.
Crystal structure of agonists in complex with hPXR. (A) Compound 1, PDB code 4XHD; (B) TO901317, PDB code 2O9I; (C) 17α-ethinylestradiol (white) and trans-nonachlor (yellow), PDB code 4X1G; (D) a sphingosine 1-phosphate receptor antagonist, PDB code 5A86; (E) BMS817399, PDB code 4NY9; and (F) hPXR-SR12813, PDB code 1NRL. Color code: white: ligand carbon; red: oxygen; blue: nitrogen; green: chlorine; pale blue: fluorine; and yellow: sulfur. The protein residues represented in blue (carbon) are commonly found in most reported crystal structures. The residues shown in pink are less frequently observed in other structures but participate in hydrogen bonding (indicated with a dashed line).
Figure 3.
(A–D) hPXR in complex with agonists (continued from Figure 2): (A) hPXR-hyperforin, PDB code 1M13; (B) hPXR-rifampicin, PDB code 1SKX; (C) hPXR- PNU142721, PDB code 3R8D; and (D) hPXR-colupulone, PDB code 2QNV. (E) mCAR complexed with the inverse agonist androstenol (PDB code 1XNX), showing direct and water-mediated hydrogen bonding. (F) mCAR complexed with the superagonist TCPOBOP (PDB code 1XLS) interacting with residues at the AF-2 helix. (G) hCAR complexed with the agonist 5β-pregnanedione (PDB code 1XV9) surrounded by interacting protein residues. (H) hCAR complexed with the agonist CITCO (PDB code 1XVP), showing the barrier residues (pink) that block access to residues in the AF-2 helix (orange). CITCO is depicted in the two modeled conformations.
The initial PXR structure provides clues to the importance of certain residues in species-selective PXR activation. SR12813 is an efficient agonist of hPXR but not of the mouse ortholog. On the other hand, PCN preferentially activates mPXR over hPXR. Based on the interactions of hPXR with SR12813, four amino acids were mutated in the mPXR expression system to the corresponding hPXR residues (Leu-206, Ser-208, His-407, and Arg-410), resulting in a reversed response profile: SR12813 became an agonist, whereas PCN had no noticeable effect [136]. This would explain the species-selective responsiveness to xenobiotics and the interesting fact that few residues are critically involved in this process.
The synergistic activation of PXR has been observed for the pesticide trans-nonachlor (TNC) and the active ingredient of contraceptive pills 17α-ethinylestradiol (EE2) [152]. Although each of these chemicals displays weak potency by itself, their simultaneous use results in a much stronger biological response, which correlates with cooperativity, as opposed to merely additive effects. Structural comparisons of PXR-EE2, PXR-TNC, and PXR-EE2-TNC revealed unambiguous binding of EE2 in both the binary and ternary complexes, but TNC could be observed only when bound in combination with EE2. Even after EE2 binding, a substantial portion of the ligand-binding site remains unoccupied, and this space is large enough to accommodate ligands of the size of TNC. In fact, the space filled by the EE2-TNC pair appears to overlap with that occupied by SR12813 or rifampicin. In addition to nonpolar contacts between TNC and PXR, TNC is further stabilized by van der Waals interactions with EE2. The notion of functional enhancement as a result of interligand interactions within the pocket leads the authors to propose the concept of a —supramolecular ligand. Because of the large size of the PXR ligand pocket, it is plausible that more of these multiligand assemblies will be discovered, particularly as other reports of crystal structures have shown that large portions of the pocket are left unoccupied by the bound ligand. For instance, 17β-estradiol and TO901317 leave vacant volumes of 981 Å3 and 442 Å3, respectively [143, 145].
5. PXR and CAR inhibitors
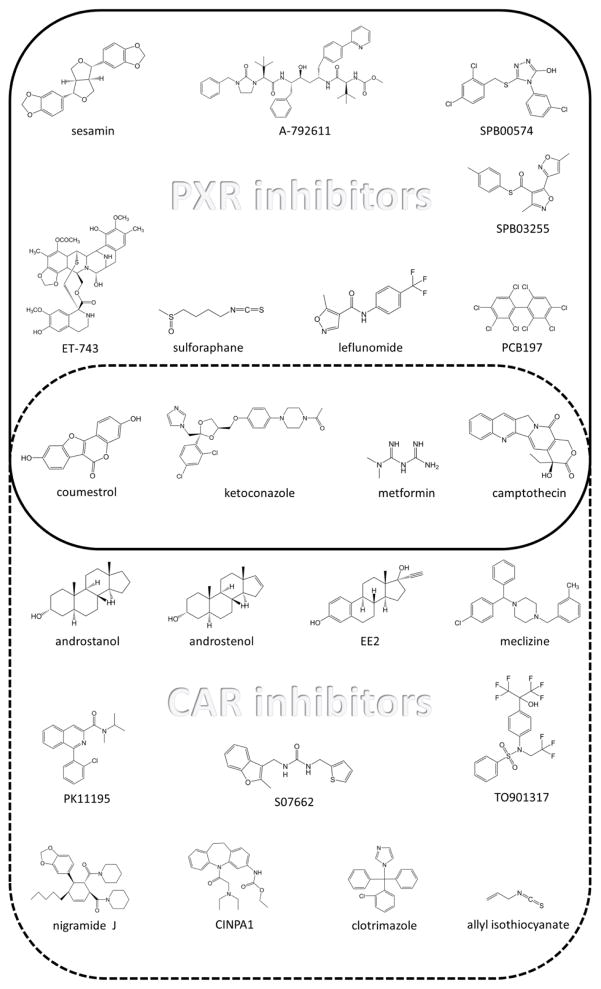
There is great impetus to develop PXR and CAR inhibitors in order to prevent the reduction of drug efficacy and prevent drug-induced toxicity. Several classes of chemicals have been found to mitigate the activity of PXR and CAR (Figure 4), but they all have major drawbacks, including cytotoxicity, poor pharmacologic properties, lack of selectivity, or poor potency.
Figure 4.
Reported PXR (top) and CAR (bottom) inhibitors. Compounds described as inhibiting both NRs are enclosed in the center.
NR activity can be repressed by a number of mechanisms. In addition to displacing an agonist by directly positioning itself at the ligand pocket, an inhibitor can attach to the outer surface of the protein. This directly or allosterically blocks the recruitment of partner proteins, such as co-activators or RXR, or it strengthens the interactions with a co-repressor. It is also foreseeable that upstream events can be modulated by, for example, posttranslational modifications that mitigate the activity of the NR.
Given the large and flexible pockets of PXR and CAR and the extensive collection of agonists discovered so far, it is not surprising that a relatively large number of inhibitors have also been reported. However, based on biochemical assays that measure displaced radiolabeled ligands for PXR, it is believed that only a few of these inhibitors, such as SFN [41], polychlorinated biphenyls [153], and coumestrol [45], bind at the ligand pocket. The antifungal drug clotrimazole was reported to attenuate the activity of hCAR by directly competing with the agonist [17]. Coumestrol not only competes with a PXR agonist for binding to the ligand-binding cavity, but also likely binds to a surface outside the ligand binding pocket [45]. Computational studies cannot definitely confirm the binding of the inhibitor to the ligand cavity, but they can provide a plausible scenario in which this event can occur. This approach was applied to the hCAR inverse agonist S07662 by using molecular dynamics simulations [154].
Inhibitors that probably interact with the outer surface of PXR ligand binding pocket include ketoconazole [48], SPB03255, and SPB00574 [42], which are thought to reside at the AF-2 surface, preventing the recruitment of co-activators. Most of the remaining reported inhibitors cannot be assigned a particular inhibition modality, because only cell-based data is available, with no confirmation of direct PXR binding. Crystal structures would provide definite validation of the mechanism behind the antagonism, but the only structure obtained so far for an inhibitor in complex with either NR is for the mCAR inverse agonist androstenol; this provides insight into the structural features that lead to decreased basal activity [138].
A number of PXR and CAR inhibitors are of natural origin. Some of these compounds, such as sesamin [46], are present in a regular diet and, therefore, are not thought to be highly toxic. However, others exhibit potent cytotoxicity, such as ET-743 [40]. These natural products range widely in size and chemical structure complexity, as is evident when the structure of ET-743 is compared with that of SFN (Figure 4).
Some of the reported PXR inhibitors were previously described as possessing other biological activity. Leflunomide was portrayed as the first FDA-approved compound to be repurposed for PXR inhibition [42], which is now approved for treating rheumatoid arthritis and psoriatic arthritis. Metformin is a marketed biguanide compound used as an antihyperglycemic agent to treat diabetes [155]. A-792611 is an HIV protease inhibitor, which not only suppresses PXR activity but is also metabolized by and inhibits CYP3A4 [156]. These types of novel PXR inhibitors, which have proved potent in preclinical studies for other indications, would have the added benefits of their established safety and desirable PK/PD profiles. However, their non-PXR related activities indicate that they are not specific for PXR.
Small-molecule modulators of PXR can most likely affect the activity of CAR, and vise versa. The difficulty in obtaining modulators specific to one of the xenobiotic receptors is by and large due to their ligand promiscuity. For instance, most of the reported CAR inhibitors have been shown to be medium to potent activators of PXR. Hence, these inhibitors would be bound to be ineffective in cases where both receptors are present, as the same sets of genes to be silenced by one of the receptors would be activated by the counterpart. Recently, the chemical CINPA1 and its analogs were identified as potent CAR selective inhibitors that do not activate PXR [157, 158]. This novel class of compounds enables receptor-selective inhibition to address the biology of a particular receptor without interference from other NRs.
6. Conclusion
PXR and its close relative CAR are recognized as master xenobiotic sensors and are notorious for their characteristic ligand promiscuity. Indeed, their reported agonists differ greatly in molecular size, structure, and physiochemical properties. The ability of PXR and CAR to regulate the expression of a number of overlapping or distinctive sets of genes that are responsible for xenobiotic detoxification can be seen as a double-edged sword. Foreign chemicals that can harm the organism are degraded and excreted out of the system, and endogenous substances are maintained at appropriate levels. However, PXR and CAR activation can also lead to undesirable DDIs, and xenobiotic metabolites (including therapeutics) can result in the generation of toxic by-products.
These ligands have displayed species and—mainly for CAR—isoform preference. Investigating cross-species preference is vital in order to be able to extrapolate in vivo findings of DDIs from animal models to humans, and organ-specific expression of the variants would enhance our understanding of variations in drug metabolism. The discovery of novel agonists and antagonists with species and isoform selectivity would provide important tools to greatly broaden our knowledge of these issues and would serve as a basis for developing therapeutic agents to counter adverse reactions in drug coadministration.
Crystal structures provide insights into the molecular basis of promiscuity for PXR and CAR, and the constitutive character of CAR, laying the groundwork for developing antagonists to be used as cotherapeutics. An increase in the number of known structures with different classes of ligands would greatly improve the prediction of potential PXR or CAR activators, enabling the identification of compounds that may cause adverse drug effects. All of the structures reported so far involve only the LBD, but studies encompassing full-length proteins would provide an essential understanding of the interplay among ligands, target DNA, and partner proteins. Structural characterization on the other isoforms is lacking, particularly when the activity of CAR vary across isoforms.
Although there is convincing evidence to support the use of PXR and CAR as therapeutic targets, their roles beyond being xenobiotic receptors need to be expanded to include their effects on health and diseases such as inflammation, diabetes and cancer.
HIGHLIGHTS.
PXR and CAR are the major xenobiotic sensors that initiate detoxification.
PXR and CAR bind diverse chemical classes and exhibit species and isoform selectivity.
Crystal structures reveal the molecular basis of binding promiscuity.
Structure analysis enables prediction of modulators of PXR and CAR.
PXR and CAR are important therapeutic targets in a number of diseases.
Acknowledgments
We thank other members of the Chen research laboratory for valuable discussions, and Dr. Keith A. Laycock (St. Jude Department of Scientific Editing) for editing the manuscript. This work was supported by ALSAC, St. Jude Children’s Research Hospital, and the National Institutes of Health [Grants R01GM086415, R01GM110034, and P30-CA21765]. The funding sources had no involvement in the writing or the decision to submit the manuscript for publication.
ABBREVIATIONS
- AF-2
activation function 2
- CAR
constitutive androstane receptor
- CYP
cytochrome P450
- DBD
DNA-binding domain
- DDI
drug-drug interactions
- DEHP
di(2-ethylhexyl) phthalate
- DILI
drug-induced liver injury
- DR4
direct repeat separated by 4 nucleotides
- DSS
dextran sulfate sodium
- EE2
17α-ethinylestradiol
- EGFR
EGF receptor
- G6Pase
glucose-6-phosphatase
- GR
glucocorticoid receptor
- GST
glutathione S-transferase
- HDL
high-density lipoprotein
- HNF4α
hepatocyte nuclear factor 4 alpha
- IBD
inflammatory bowel disease
- IGF-1R
insulin-like growth factor-1 receptor
- LBD
ligand-binding domain
- MDR1
multidrug resistance protein 1
- NR
nuclear receptor
- PB
Phenobarbital
- PBREM
phenobarbital-responsive enhancer module
- PCN
5-pregnen-3β-ol-20-one-16α-carbonitrile
- PEPCK1
phosphoenoylpyruvate carboxykinase 1
- PPARα
peroxisome proliferator-activated receptor alpha
- PXR
pregnane X receptor
- RXR
retinoid X receptor
- SFN
Sulforaphane
- SN-38
7-ethyl-10-hydroxycamptothecin
- SRC
steroid receptor co-activator
- SREBP-1
sterol regulatory element-binding protein 1
- SULT
sulfotransferase
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- TNBS
2,4,6-trinitrobenzene sulfonic acid
- TNC
trans-nonachlor
- UGT
UDP-glucoronyltransferase
- Wild-type
WT
- XREM
xenobiotic responsive enhancer module
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 2.Wang YM, Ong SS, Chai SC, Chen TS. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Met. 2012;8:803–817. doi: 10.1517/17425255.2012.685237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic Metabolomics: Major Impact on the Metabolome. Annu Rev Pharmacol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tolson AH, Wang HB. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv Drug Deliver Rev. 2010;62:1238–1249. doi: 10.1016/j.addr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YM, Lin WW, Chai SC, Wu J, Ong SS, Schuetz EG, Chen TS. Piperine activates human pregnane X receptor to induce the expression of cytochrome P450 3A4 and multidrug resistance protein 1. Toxicol Appl Pharm. 2013;272:96–107. doi: 10.1016/j.taap.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin WW, Wu J, Dong HQ, Bouck D, Zeng FY, Chen TS. Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem. 2008;283:30650–30657. doi: 10.1074/jbc.M806132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichti-Kaiser K, Brobst D, Xu CS, Staudinger JL. A Systematic Analysis of Predicted Phosphorylation Sites within the Human Pregnane X Receptor Protein. J Pharmacol Exp Ther. 2009;331:65–76. doi: 10.1124/jpet.109.157180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pondugula SR, Brimer-Cline C, Wu J, Schuetz EG, Tyagi RK, Chen TS. A Phosphomimetic Mutation at Threonine-57 Abolishes Transactivation Activity and Alters Nuclear Localization Pattern of Human Pregnane X Receptor. Drug Metab Dispos. 2009;37:719–730. doi: 10.1124/dmd.108.024695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doricakova A, Novotna A, Vrzal R, Pavek P, Dvorak Z. The role of residues T248, Y249 and T422 in the function of human pregnane X receptor. Arch Toxicol. 2013;87:291–301. doi: 10.1007/s00204-012-0937-9. [DOI] [PubMed] [Google Scholar]
- 10.Elias A, High AA, Mishra A, Ong SS, Wu J, Peng JM, Chen TS. Identification and characterization of phosphorylation sites within the pregnane X receptor protein. Biochem Pharmacol. 2014;87:360–370. doi: 10.1016/j.bcp.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen TS. Overcoming drug resistance by regulating nuclear receptors. Adv Drug Deliver Rev. 2010;62:1257–1264. doi: 10.1016/j.addr.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YM, Chai SC, Lin WW, Chai XJ, Elias A, Wu J, Ong SS, Pondugula SR, Beard JA, Schuetz EG, Zeng S, Xie W, Chen TS. Serine 350 of human pregnane X receptor is crucial for its hetrodimerization with retinoid X receptor alpha and transactivation of target genes in vitro and in vivo. Biochem Pharmacol. 2015;96:357–368. doi: 10.1016/j.bcp.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinz M, Kim S, Zhu ZR, Chen TS, Anthony M, Dickinson K, Rodrigues AD. Evaluation of 170 xenobiotics as transactivators of human pregnane X receptor (hPXR) and correlation to known CYP3A4 drug interactions. Curr Drug Metab. 2006;7:375–388. doi: 10.2174/138920006776873535. [DOI] [PubMed] [Google Scholar]
- 14.Sinz MW. Evaluation of pregnane X receptor (PXR)-mediated CYP3A4 drug-drug interactions in drug development. Drug Metab Rev. 2013;45:3–14. doi: 10.3109/03602532.2012.743560. [DOI] [PubMed] [Google Scholar]
- 15.Wang YM, Chai SC, Brewer CT, Chen TS. Pregnane X receptor and drug-induced liver injury. Expert Opin Drug Met. 2014;10:1521–1532. doi: 10.1517/17425255.2014.963555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekins S, Mirny L, Schuetz EG. A ligand-based approach to understanding selectivity of nuclear hormone receptors PXR, CAR, FXR, LXR alpha, and LXR beta. Pharm Res-Dordr. 2002;19:1788–1800. doi: 10.1023/a:1021429105173. [DOI] [PubMed] [Google Scholar]
- 17.Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 18.Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Bruntk EM, Guzelian PS, Evans RM. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 19.Gong HB, Singh SV, Singh SP, Mu Y, Lee JH, Saini SPS, Toma D, Ren SR, Kagan VE, Day BW, Zimniak P, Xie W. Orphan nuclear receptor pregnane X receptor sensitizes oxidative stress responses in transgenic mice and cancerous cells. Mol Endocrinol. 2006;20:279–290. doi: 10.1210/me.2005-0205. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Shah Y, Cheung C, Guo GL, Feigenbaum L, Krausz KW, Idle JR, Gonzalez FJ. The pregnane X receptor gene-humanized mouse: A model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab Dispos. 2007;35:194–200. doi: 10.1124/dmd.106.012831. [DOI] [PubMed] [Google Scholar]
- 21.Ma X, Cheung C, Krausz KW, Shah YM, Wang T, Idle JR, Gonzalez FJ. A Double Transgenic Mouse Model Expressing Human Pregnane X Receptor and Cytochrome P450 3A4. Drug Metab Dispos. 2008;36:2506–2512. doi: 10.1124/dmd.108.022723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheer N, Ross J, Rode A, Zevnik B, Niehaves S, Faust N, Wolf CR. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest. 2008;118:3228–3239. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi K, Kitajima S, Aisaki K, Tanemura K, Taquahashi Y, Moriyama N, Ikeno E, Matsuda N, Saga Y, Blumberg B, Kanno J. Development of humanized steroid and xenobiotic receptor mouse by homologous knock-in of the human steroid and xenobiotic receptor ligand binding domain sequence. J Toxicol Sci. 2012;37:373–380. doi: 10.2131/jts.37.373. [DOI] [PubMed] [Google Scholar]
- 24.Cheng J, Shah YM, Gonzalez FJ. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci. 2012;33:323–330. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmann T, Moehle C, Mauerer R, Scharl M, Liebisch G, Zahn A, Stremmel W, Schmitz G. Loss of detoxification in inflammatory bowel disease: Dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Dring MM, Goulding CA, Trimble VI, Keegan D, Ryan AW, Brophy KM, Smyth CM, Keeling PWN, O'Donoghue D, O'Sullivan M, O'Morain C, Mahmud N, Wikstrom AC, Kelleher D, McManus R. The pregnane X receptor locus is associated with susceptibility to inflammatory bowel disease. Gastroenterology. 2006;130:341–348. doi: 10.1053/j.gastro.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Shah YM, Ma XC, Pang XY, Tanaka T, Kodama T, Krausz KW, Gonzalez FJ. Therapeutic Role of Rifaximin in Inflammatory Bowel Disease: Clinical Implication of Human Pregnane X Receptor Activation. J Pharmacol Exp Ther. 2010;335:32–41. doi: 10.1124/jpet.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK, Talley NJ, Moayyedi P. Antibiotic Therapy in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Am J Gastroenterol. 2011;106:661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 29.Zhou C, Tabb MM, Nelson EL, Grun F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, Blumberg B. Mutual repression between steroid and xenobiotic receptor and NF-kappa B signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest. 2006;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sepe V, Ummarino R, D'Auria MV, Mencarelli A, D'Amore C, Renga B, Zampella A, Fiorucci S. Total Synthesis and Pharmacological Characterization of Solomonsterol A, a Potent Marine Pregnane-X-Receptor Agonist Endowed with Anti-Inflammatory Activity. J Med Chem. 2011;54:4590–4599. doi: 10.1021/jm200241s. [DOI] [PubMed] [Google Scholar]
- 31.Burk O, Arnold KA, Nussler AK, Schaeffeler E, Efimova E, Avery BA, Avery MA, Fromm MF, Eichelbaum M. Antimalarial artemisinin drugs induce cytochrome p450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2005;67:1954–1965. doi: 10.1124/mol.104.009019. [DOI] [PubMed] [Google Scholar]
- 32.Hu DH, Wang YG, Chen ZW, Ma ZC, You Q, Zhang XX, Zhou T, Xiao Y, Liang QD, Tan HL, Xiao CR, Tang XL, Zhang BL, Gao Y. Artemisinin protects against dextran sulfate-sodium-induced inflammatory bowel disease, which is associated with activation of the pregnane X receptor. Eur J Pharmacol. 2014;738:273–284. doi: 10.1016/j.ejphar.2014.04.050. [DOI] [PubMed] [Google Scholar]
- 33.Dou W, Zhang JJ, Zhang EY, Sun AN, Ding LL, Chou GX, Wang ZT, Mani S. Chrysin Ameliorates Chemically Induced Colitis in the Mouse through Modulation of a PXR/NF-kappa B Signaling Pathway. J Pharmacol Exp Ther. 2013;345:473–482. doi: 10.1124/jpet.112.201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou W, Zhang JJ, Li H, Kortagere S, Sun K, Ding LL, Ren GY, Wang ZT, Mani S. Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR-dependent pathway. J Nutr Biochem. 2014;25:923–933. doi: 10.1016/j.jnutbio.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau AJ, Chang TKH. 3-Hydroxyflavone and structural analogues differentially activate pregnane X receptor: Implication for inflammatory bowel disease. Pharmacol Res. 2015;100:64–72. doi: 10.1016/j.phrs.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 36.Ma XC, Shah YM, Guo GL, Wang T, Krausz KW, Idle JR, Gonzalez FJ. Rifaximin is a gut-specific human pregnane X receptor activator. J Pharmacol Exp Ther. 2007;322:391–398. doi: 10.1124/jpet.107.121913. [DOI] [PubMed] [Google Scholar]
- 37.Gotoh S, Negishi M. Statin-activated nuclear receptor PXR promotes SGK2 dephosphorylation by scaffolding PP2C to induce hepatic gluconeogenesis. Sci Rep–Uk. 2015;5:14076. doi: 10.1038/srep14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 39.Aune GJ, Furuta T, Pommier Y. Ecteinascidin 743: a novel anticancer drug with a unique mechanism of action. Anti-Cancer Drug. 2002;13:545–555. doi: 10.1097/00001813-200207000-00001. [DOI] [PubMed] [Google Scholar]
- 40.van Kesteren C, de Vooght MMM, Lopez-Lazaro L, Mathot RAA, Schellens JHM, Jimeno JM, Beijnen JH. Yondelis (R) (trabectedin, ET-743): the development of an anticancer agent of marine origin. Anti-Cancer Drug. 2003;14:487–502. doi: 10.1097/00001813-200308000-00001. [DOI] [PubMed] [Google Scholar]
- 41.Zhou CC, Poulton EJ, Grun F, Bammler TK, Blumberg B, Thummel KE, Eaton DL. The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol Pharmacol. 2007;71:220–229. doi: 10.1124/mol.106.029264. [DOI] [PubMed] [Google Scholar]
- 42.Ekins S, Kholodovych V, Ai N, Sinz M, Gal J, Gera L, Welsh WJ, Bachmann K, Mani S. Computational discovery of novel low micromolar human pregnane X receptor antagonists. Mol Pharmacol. 2008;74:662–672. doi: 10.1124/mol.108.049437. [DOI] [PubMed] [Google Scholar]
- 43.Poulton EJ, Levy L, Lampe JW, Shen DD, Tracy J, Shuhart MC, Thummel KE, Eaton DL. Sulforaphane is not an effective antagonist of the human pregnane X-receptor in vivo. Toxicol Appl Pharm. 2013;266:122–131. doi: 10.1016/j.taap.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atwell LL, Zhang Z, Mori M, Farris PE, Vetto JT, Naik AM, Oh KY, Thuillier P, Ho E, Shannon J. Sulforaphane bioavailability and chemopreventive activity in women scheduled for breast biopsy. Cancer Prev Res (Phila) 2015;8:1184–1191. doi: 10.1158/1940-6207.CAPR-15-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang HW, Li H, Moore LB, Johnson MDL, Maglich JM, Goodwin B, Ittoop ORR, Wisely B, Creech K, Parks DJ, Collins JL, Willson TM, Kalpana GV, Venkatesh M, Xie W, Cho SY, Roboz J, Redinbo M, Moore JT, Mani S. The phytoestrogen Coumestrol is a naturally occurring antagonist of the human pregnane x receptor. Mol Endocrinol. 2008;22:838–857. doi: 10.1210/me.2007-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim YP, Ma CY, Liu CL, Lin YH, Hu ML, Chen JJ, Hung DZ, Hsieh WT, Huang JD. Sesamin: A Naturally Occurring Lignan Inhibits CYP3A4 by Antagonizing the Pregnane X Receptor Activation. Evid Based Complement Alternat Med. 2012:242810. doi: 10.1155/2012/242810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YK, Tang Y, Robbins GT, Nie DT. Camptothecin Attenuates Cytochrome P450 3A4 Induction by Blocking the Activation of Human Pregnane X Receptor. J Pharmacol Exp Ther. 2010;334:999–1008. doi: 10.1124/jpet.110.168294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mani S, Dou W, Redinbo MR. PXR antagonists and implication in drug metabolism. Drug Metab Rev. 2013;45:60–72. doi: 10.3109/03602532.2012.746363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeshita A, Taguchi M, Koibuchi N, Ozawa Y. Putative role of the orphan nuclear receptor SXR (steroid and xenobiotic receptor) in the mechanism of CYP3A4 inhibition by xenobiotics. J Biol Chem. 2002;277:32453–32458. doi: 10.1074/jbc.M111245200. [DOI] [PubMed] [Google Scholar]
- 50.Ekins S, Chang C, Mani S, Krasowski MD, Reschly EJ, Iyer M, Kholodovych V, Ai N, Welsh WJ, Sinz M, Swaan PW, Patel R, Bachmann K. Human pregnane X receptor antagonists and Agonists define molecular requirements for different binding sites. Mol Pharmacol. 2007;72:592–603. doi: 10.1124/mol.107.038398. [DOI] [PubMed] [Google Scholar]
- 51.Wang HW, Huang HY, Li H, Teotico DG, Sinz M, Baker SD, Staudinger J, Kalpana G, Redinbo MR, Mani S. Activated pregnenolone X-receptor is a target for ketoconazole and its analogs. Clin Cancer Res. 2007;13:2488–2495. doi: 10.1158/1078-0432.CCR-06-1592. [DOI] [PubMed] [Google Scholar]
- 52.Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26:258–268. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 53.Li H, Redinbo MR, Venkatesh M, Ekins S, Chaudhry A, Bloch N, Negassa A, Mukherjee P, Kalpana G, Mani S. Novel Yeast-based Strategy Unveils Antagonist Binding Regions on the Nuclear Xenobiotic Receptor PXR. J Biol Chem. 2013;288:13655–13668. doi: 10.1074/jbc.M113.455485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das BC, Madhukumar AV, Anguiano J, Kim S, Sinz M, Zvyaga TA, Power EC, Ganellin CR, Mani S. Synthesis of novel ketoconazole derivatives as inhibitors of the human Pregnane X Receptor (PXR; NR1I2; also termed SXR, PAR) Bioorg Med Chem Lett. 2008;18:3974–3977. doi: 10.1016/j.bmcl.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 55.Venkatesh M, Wang HW, Cayer J, Leroux M, Salvail D, Das B, Wrobel JE, Mani S. In Vivo and In Vitro Characterization of a First-in-Class Novel Azole Analog That Targets Pregnane X Receptor Activation. Mol Pharmacol. 2011;80:124–135. doi: 10.1124/mol.111.071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, Moore DD. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- 58.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- 60.Masson D, Qatanani M, Sberna AL, Xiao R, Pais de Barros JP, Grober J, Deckert V, Athias A, Gambert P, Lagrost L, Moore DD, Assem M. Activation of the constitutive androstane receptor decreases HDL in wild-type and human apoA-I transgenic mice. J Lipid Res. 2008;49:1682–1691. doi: 10.1194/jlr.M700374-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Wei P, Zhang J, Dowhan DH, Han Y, Moore DD. Specific and overlapping functions of the nuclear hormone receptors CAR and PXR in xenobiotic response. Pharmacogenomics J. 2002;2:117–126. doi: 10.1038/sj.tpj.6500087. [DOI] [PubMed] [Google Scholar]
- 62.Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell. 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 63.Luo G, Cunningham M, Kim S, Burn T, Lin J, Sinz M, Hamilton G, Rizzo C, Jolley S, Gilbert D, Downey A, Mudra D, Graham R, Carroll K, Xie J, Madan A, Parkinson A, Christ D, Selling B, LeCluyse E, Gan LS. CYP3A4 induction by drugs: correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab Dispos. 2002;30:795–804. doi: 10.1124/dmd.30.7.795. [DOI] [PubMed] [Google Scholar]
- 64.Tamasi V, Juvan P, Beer M, Rozman D, Meyer UA. Transcriptional activation of PPARalpha by phenobarbital in the absence of CAR and PXR. Molecular pharmaceutics. 2009;6:1573–1581. doi: 10.1021/mp9001552. [DOI] [PubMed] [Google Scholar]
- 65.Roth A, Looser R, Kaufmann M, Blattler SM, Rencurel F, Huang W, Moore DD, Meyer UA. Regulatory cross-talk between drug metabolism and lipid homeostasis: constitutive androstane receptor and pregnane X receptor increase Insig-1 expression. Mol Pharmacol. 2008;73:1282–1289. doi: 10.1124/mol.107.041012. [DOI] [PubMed] [Google Scholar]
- 66.Li L, Sinz MW, Zimmermann K, Wang H. An Insulin-Like Growth Factor 1 Receptor Inhibitor Induces CYP3A4 Expression through a Pregnane X Receptor-Independent, Noncanonical Constitutive Androstane Receptor-Related Mechanism. The Journal of Pharmacology and Experimental Therapeutics. 2012;340:688–697. doi: 10.1124/jpet.111.188854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodwin B, Hodgson E, D'Costa DJ, Robertson GR, Liddle C. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol. 2002;62:359–365. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- 68.Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol. 2008;74:443–453. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci U S A. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–2915. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- 71.Sugatani J, Kojima H, Ueda A, Kakizaki S, Yoshinari K, Gong QH, Owens IS, Negishi M, Sueyoshi T. The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology. 2001;33:1232–1238. doi: 10.1053/jhep.2001.24172. [DOI] [PubMed] [Google Scholar]
- 72.Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- 73.Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol. 2005;67:1257–1267. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- 74.Sugatani J, Sueyoshi T, Negishi M, Miwa M. Regulation of the Human UGT1A1 Gene by Nuclear Receptors Constitutive Active/Androstane Receptor, Pregnane X Receptor, and Glucocorticoid Receptor. In: Helmut S, Lester P, editors. Methods in Enzymology. Vol. 400. Academic Press; Waltham, Massachusetts: 2005. pp. 92–104. [DOI] [PubMed] [Google Scholar]
- 75.Qatanani M, Zhang J, Moore DD. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology. 2005;146:995–1002. doi: 10.1210/en.2004-1350. [DOI] [PubMed] [Google Scholar]
- 76.Mathas M, Burk O, Qiu H, Nusshag C, Godtel-Armbrust U, Baranyai D, Deng S, Romer K, Nem D, Windshugel B, Wojnowski L. Evolutionary history and functional characterization of the amphibian xenosensor CAR. Mol Endocrinol. 2012;26:14–26. doi: 10.1210/me.2011-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanno Y, Aoki S, Mochizuki M, Mori E, Nakahama T, Inouye Y. Expression of Constitutive Androstane Receptor Splice Variants in Rat Liver and Lung and Their Functional Properties. Biol Pharm Bull. 2005;28:2058–2062. doi: 10.1248/bpb.28.2058. [DOI] [PubMed] [Google Scholar]
- 78.Gray MA, Peacock JN, Squires EJ. Characterization of the porcine constitutive androstane receptor (CAR) and its splice variants. Xenobiotica. 2009;39:915–930. doi: 10.3109/00498250903330348. [DOI] [PubMed] [Google Scholar]
- 79.DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol Pharmacol. 2009;75:1005–1013. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- 81.Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lynch C, Pan Y, Li L, Heyward S, Moeller T, Swaan PW, Wang H. Activation of the constitutive androstane receptor inhibits gluconeogenesis without affecting lipogenesis or fatty acid synthesis in human hepatocytes. Toxicol Appl Pharm. 2014;279:33–42. doi: 10.1016/j.taap.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lake BG. Species differences in the hepatic effects of inducers of CYP2B and CYP4A subfamily forms: relationship to rodent liver tumour formation. Xenobiotica. 2009;39:582–596. doi: 10.1080/00498250903098184. [DOI] [PubMed] [Google Scholar]
- 84.Yamada T, Okuda Y, Kushida M, Sumida K, Takeuchi H, Nagahori H, Fukuda T, Lake BG, Cohen SM, Kawamura S. Human hepatocytes support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogen sodium phenobarbital in an in vivo study using a chimeric mouse with humanized liver. Toxicol Sci. 2014;142:137–157. doi: 10.1093/toxsci/kfu173. [DOI] [PubMed] [Google Scholar]
- 85.Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz E. Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther. 2004;311:811–821. doi: 10.1124/jpet.104.069310. [DOI] [PubMed] [Google Scholar]
- 86.Shmueli O, Horn-Saban S, Chalifa-Caspi V, Shmoish M, Ophir R, Benjamin-Rodrig H, Safran M, Domany E, Lancet D. GeneNote: whole genome expression profiles in normal human tissues. C R Biol. 2003;326:1067–1072. doi: 10.1016/j.crvi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 87.Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003;31:3194–3207. doi: 10.1093/nar/gkg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kanno Y, Aoki S, Nakahama T, Inouye Y. Role of the Defective Splicing of mRNA in the Lack of Pulmonary Expression of Constitutively Active Receptor in Rat. J Health Sci. 2003;49:541–546. [Google Scholar]
- 89.Arnold KA, Eichelbaum M, Burk O. Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nuclear receptor. 2004;2:1. doi: 10.1186/1478-1336-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jinno H, Tanaka-Kagawa T, Hanioka N, Ishida S, Saeki M, Soyama A, Itoda M, Nishimura T, Saito Y, Ozawa S, Ando M, Sawada J. Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol Pharmacol. 2004;65:496–502. doi: 10.1124/mol.65.3.496. [DOI] [PubMed] [Google Scholar]
- 91.Ross J, Plummer SM, Rode A, Scheer N, Bower CC, Vogel O, Henderson CJ, Wolf CR, Elcombe CR. Human Constitutive Androstane Receptor (CAR) and Pregnane X Receptor (PXR) Support the Hypertrophic but not the Hyperplastic Response to the Murine Nongenotoxic Hepatocarcinogens Phenobarbital and Chlordane In Vivo. Toxicol Sci. 2010;116:452–466. doi: 10.1093/toxsci/kfq118. [DOI] [PubMed] [Google Scholar]
- 92.Savkur RS, Wu Y, Bramlett KS, Wang M, Yao S, Perkins D, Totten M, Searfoss G, 3rd, Ryan TP, Su EW, Burris TP. Alternative splicing within the ligand binding domain of the human constitutive androstane receptor. Mol Genet Metab. 2003;80:216–226. doi: 10.1016/j.ymgme.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 93.Auerbach SS, Dekeyser JG, Stoner MA, Omiecinski CJ. CAR2 displays unique ligand binding and RXRalpha heterodimerization characteristics. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2007;35:428–439. doi: 10.1124/dmd.106.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ikeda S, Kurose K, Jinno H, Sai K, Ozawa S, Hasegawa R, Komamura K, Kotake T, Morishita H, Kamakura S, Kitakaze M, Tomoike H, Tamura T, Yamamoto N, Kunitoh H, Yamada Y, Ohe Y, Shimada Y, Shirao K, Kubota K, Minami H, Ohtsu A, Yoshida T, Saijo N, Saito Y, Sawada J. Functional analysis of four naturally occurring variants of human constitutive androstane receptor. Mol Genet Metab. 2005;86:314–319. doi: 10.1016/j.ymgme.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 95.Lau AJ, Chang TK. Fetal bovine serum and human constitutive androstane receptor: evidence for activation of the SV23 splice variant by artemisinin, artemether, and arteether in a serum-free cell culture system. Toxicol Appl Pharm. 2014;277:221–230. doi: 10.1016/j.taap.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 96.Omiecinski CJ, Coslo DM, Chen T, Laurenzana EM, Peffer RC. Multi-species analyses of direct activators of the constitutive androstane receptor. Toxicol Sci. 2011;123:550–562. doi: 10.1093/toxsci/kfr191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma D, Lau AJ, Sherman MA, Chang TK. Differential activation of human constitutive androstane receptor and its SV23 and SV24 splice variants by rilpivirine and etravirine. Br J Pharmacol. 2014;172:1263–1276. doi: 10.1111/bph.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lau AJ, Chang TK. Indirect activation of the SV23 and SV24 splice variants of human constitutive androstane receptor: analysis with 3-hydroxyflavone and its analogues. Br J Pharmacol. 2013;170:403–414. doi: 10.1111/bph.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DeKeyser JG, Laurenzana EM, Peterson EC, Chen T, Omiecinski CJ. Selective phthalate activation of naturally occurring human constitutive androstane receptor splice variants and the pregnane X receptor. Toxicol Sci. 2011;120:381–391. doi: 10.1093/toxsci/kfq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paul KB, Thompson JT, Simmons SO, Vanden Heuvel JP, Crofton KM. Evidence for triclosan-induced activation of human and rodent xenobiotic nuclear receptors. Toxicol In Vitro. 2013;27:2049–2060. doi: 10.1016/j.tiv.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 101.Thompson EE, Kuttab-Boulos H, Krasowski MD, Di Rienzo A. Functional constraints on the constitutive androstane receptor inferred from human sequence variation and cross-species comparisons. Hum Genomics. 2005;2:168–178. doi: 10.1186/1479-7364-2-3-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cortes CP, Siccardi M, Chaikan A, Owen A, Zhang G, la Porte CJ. Correlates of efavirenz exposure in Chilean patients affected with human immunodeficiency virus reveals a novel association with a polymorphism in the constitutive androstane receptor. Ther Drug Monit. 2013;35:78–83. doi: 10.1097/FTD.0b013e318274197e. [DOI] [PubMed] [Google Scholar]
- 103.Urano T, Usui T, Shiraki M, Ouchi Y, Inoue S. Association of a single nucleotide polymorphism in the constitutive androstane receptor gene with bone mineral density. Geriatr Gerontol Int. 2009;9:235–241. doi: 10.1111/j.1447-0594.2009.00527.x. [DOI] [PubMed] [Google Scholar]
- 104.Choi EJ, Jang YJ, Cha EY, Shin JG, Lee SS. Identification and characterization of novel alternative splice variants of human constitutive androstane receptor in liver samples of Koreans and Caucasians. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2013;41:888–896. doi: 10.1124/dmd.112.049791. [DOI] [PubMed] [Google Scholar]
- 105.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The Repressed Nuclear Receptor CAR Responds to Phenobarbital in Activating the Human CYP2B6 Gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 106.Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E. Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem. 2004;279:29295–29301. doi: 10.1074/jbc.M400580200. [DOI] [PubMed] [Google Scholar]
- 107.Carazo Fernandez A, Smutny T, Hyrsova L, Berka K, Pavek P. Chrysin baicalein and galangin are indirect activators of the human constitutive androstane receptor (CAR) Toxicol Lett. 2015;233:68–77. doi: 10.1016/j.toxlet.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 108.Yueh M-F, Taniguchi K, Chen S, Evans RM, Hammock BD, Karin M, Tukey RH. The commonly used antimicrobial additive triclosan is a liver tumor promoter. Proceedings of the National Academy of Sciences. 2014;111:17200–17205. doi: 10.1073/pnas.1419119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of Acetaminophen-Induced Hepatotoxicity by the Xenobiotic Receptor CAR. Science. 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
- 110.Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett. 2003;548:17–20. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- 111.Mutoh S, Osabe M, Inoue K, Moore R, Pedersen L, Perera L, Rebolloso Y, Sueyoshi T, Negishi M. Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3) J Biol Chem. 2009;284:34785–34792. doi: 10.1074/jbc.M109.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mutoh S, Sobhany M, Moore R, Perera L, Pedersen L, Sueyoshi T, Negishi M. Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci Signal. 2013;6:ra31. doi: 10.1126/scisignal.2003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-Responsive Nuclear Translocation of the Receptor CAR in Induction of the CYP2B Gene. Mol Cell Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joannard F, Rissel M, Gilot D, Anderson A, Orfila-Lefeuvre L, Guillouzo A, Atfi A, Lagadic-Gossmann D. Role for mitogen-activated protein kinases in phenobarbital-induced expression of cytochrome P450 2B in primary cultures of rat hepatocytes. Toxicol Lett. 2006;161:61–72. doi: 10.1016/j.toxlet.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 115.Koike C, Moore R, Negishi M. Extracellular Signal-Regulated Kinase Is an Endogenous Signal Retaining the Nuclear Constitutive Active/Androstane Receptor (CAR) in the Cytoplasm of Mouse Primary Hepatocytes. Mol Pharmacol. 2007;71:1217–1221. doi: 10.1124/mol.107.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bauer D, Wolfram N, Kahl GF, Hirsch-Ernst KI. Transcriptional regulation of CYP2B1 induction in primary rat hepatocyte cultures: repression by epidermal growth factor is mediated via a distal enhancer region. Mol Pharmacol. 2004;65:172–180. doi: 10.1124/mol.65.1.172. [DOI] [PubMed] [Google Scholar]
- 117.Li H, Chen T, Cottrell J, Wang H. Nuclear Translocation of Adenoviral-Enhanced Yellow Fluorescent Protein-Tagged-Human Constitutive Androstane Receptor (hCAR): A Novel Tool for Screening hCAR Activators in Human Primary Hepatocytes. Drug Metab Dispos. 2009;37:1098–1106. doi: 10.1124/dmd.108.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zelko I, Sueyoshi T, Kawamoto T, Moore R, Negishi M. The Peptide Near the C Terminus Regulates Receptor CAR Nuclear Translocation Induced by Xenochemicals in Mouse Liver. Mol Cell Biol. 2001;21:2838–2846. doi: 10.1128/MCB.21.8.2838-2846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lynch C, Zhao J, Huang R, Xiao J, Li L, Heyward S, Xia M, Wang H. Quantitative high-throughput identification of drugs as modulators of human constitutive androstane receptor. Sci Rep. 2015;5:10405. doi: 10.1038/srep10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang H, Wang H. Signaling control of the constitutive androstane receptor (CAR) Protein & Cell. 2014;5:113–123. doi: 10.1007/s13238-013-0013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dong B, Qatanani M, Moore DD. Constitutive androstane receptor mediates the induction of drug metabolism in mouse models of type 1 diabetes. Hepatology. 2009;50:622–629. doi: 10.1002/hep.23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem. 2009;284:25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tamashiro KL, Terrillion CE, Hyun J, Koenig JI, Moran TH. Prenatal stress or high-fat diet increases susceptibility to diet-induced obesity in rat offspring. Diabetes. 2009;58:1116–1125. doi: 10.2337/db08-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lahtela JT, Arranto AJ, Sotaniemi EA. Enzyme inducers improve insulin sensitivity in non-insulin-dependent diabetic subjects. Diabetes. 1985;34:911–916. doi: 10.2337/diab.34.9.911. [DOI] [PubMed] [Google Scholar]
- 125.Goerdt C, Keith M, Rubins HB. Effects of Phenytoin on Plasma High-Density Lipoprotein Cholesterol Levels in Men with Low Levels of High-Density Lipoprotein Cholesterol. J Clin Pharmacol. 1995;35:767–775. doi: 10.1002/j.1552-4604.1995.tb04118.x. [DOI] [PubMed] [Google Scholar]
- 126.Pelkonen R, Fogelholm R, Nikkila EA. Increase in serum cholesterol during phenytoin treatment. Br Med J. 1975;4:85. doi: 10.1136/bmj.4.5988.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Masuyama H, Hiramatsu Y. Treatment with a constitutive androstane receptor ligand ameliorates the signs of preeclampsia in high-fat diet-induced obese pregnant mice. Mol Cell Endocrinol. 2012;348:120–127. doi: 10.1016/j.mce.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 128.Wagner M, Halilbasic E, Marschall HU, Zollner G, Fickert P, Langner C, Zatloukal K, Denk H, Trauner M. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology. 2005;42:420–430. doi: 10.1002/hep.20784. [DOI] [PubMed] [Google Scholar]
- 129.Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]
- 130.Yamamoto Y, Moore R, Flavell RA, Lu B, Negishi M. Nuclear Receptor CAR Represses TNFα-Induced Cell Death by Interacting with the Anti-Apoptotic GADD45B. PloS One. 2010;5:e10121. doi: 10.1371/journal.pone.0010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Elcombe CR, Peffer RC, Wolf DC, Bailey J, Bars R, Bell D, Cattley RC, Ferguson SS, Geter D, Goetz A, Goodman JI, Hester S, Jacobs A, Omiecinski CJ, Schoeny R, Xie W, Lake BG. Mode of action and human relevance analysis for nuclear receptor-mediated liver toxicity: A case study with phenobarbital as a model constitutive androstane receptor (CAR) activator. Crit Rev Toxicol. 2014;44:64–82. doi: 10.3109/10408444.2013.835786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang D, Li L, Yang H, Ferguson SS, Baer MR, Gartenhaus RB, Wang H. The constitutive androstane receptor is a novel therapeutic target facilitating cyclophosphamide-based treatment of hematopoietic malignancies. Blood. 2013;121:329–338. doi: 10.1182/blood-2012-06-436691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fukumasu H, Rochetti AL, Pires PR, Silva ER, Mesquita LG, Strefezzi RF, De Carvalho DD, Dagli ML. Constitutive androstane receptor ligands modulate the anti-tumor efficacy of paclitaxel in non-small cell lung cancer cells. PloS One. 2014;9:e99484. doi: 10.1371/journal.pone.0099484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chakraborty S, Kanakasabai S, Bright JJ. Constitutive androstane receptor agonist CITCO inhibits growth and expansion of brain tumour stem cells. Br J Cancer. 2011;104:448–459. doi: 10.1038/sj.bjc.6606064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang Y, Masuyama H, Nobumoto E, Zhang G, Hiramatsu Y. The inhibition of constitutive androstane receptor-mediated pathway enhances the effects of anticancer agents in ovarian cancer cells. Biochem Pharmacol. 2014;90:356–366. doi: 10.1016/j.bcp.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 136.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 137.Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 138.Shan L, Vincent J, Brunzelle JS, Dussault I, Lin M, Ianculescu I, Sherman MA, Forman BM, Fernandez EJ. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16:907–917. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Noble SM, Carnahan VE, Moore LB, Luntz T, Wang H, Ittoop OR, Stimmel JB, Davis-Searles PR, Watkins RE, Wisely GB, LeCluyse E, Tripathy A, McDonnell DP, Redinbo MR. Human PXR forms a tryptophan zipper-mediated homodimer. Biochemistry. 2006;45:8579–8589. doi: 10.1021/bi0602821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.di Masi A, De Marinis E, Ascenzi P, Marino M. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol Aspects Med. 2009;30:297–343. doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 141.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol. 2003;331:815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 142.Khan JA, Camac DM, Low S, Tebben AJ, WenseI DL, Wright MC, Su JL, Jenny V, Das Gupta R, Ruzanov M, Russo KA, Bell A, An YM, Bryson JW, Gao MA, Gambhire P, Baldwin ET, Gardner D, Cavallaro CL, Duncia JV, Hynes J. Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR. J Mol Biol. 2015;427:924–942. doi: 10.1016/j.jmb.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 143.Xue Y, Chao E, Zuercher WJ, Willson TM, Collins JL, Redinbo MR. Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorgan Med Chem. 2007;15:2156–2166. doi: 10.1016/j.bmc.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cheng Y, Redinbo MR. Activation of the human nuclear xenobiotic receptor PXR by the reverse transcriptase-targeted anti-HIV drug PNU-142721. Protein Sci. 2011;20:1713–1719. doi: 10.1002/pro.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, Redinbo MR. Crystal structure of the pregnane X receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol. 2007;21:1028–1038. doi: 10.1210/me.2006-0323. [DOI] [PubMed] [Google Scholar]
- 146.Teotico DG, Bischof JJ, Peng L, Kliewer SA, Redinbo MR. Structural Basis of Human Pregnane X Receptor Activation by the Hops Constituent Colupulone. Mol Pharmacol. 2008;74:1512–1520. doi: 10.1124/mol.108.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, Lambert MH, Kliewer SA, Redinbo MR. 2.1 angstrom crystal structure of human PXR in complex with the St. John's wort compound hyperforin. Biochemistry. 2003;42:1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 148.Windshugel B, Jyrkkarinne J, Vanamo J, Poso A, Honkakoski P, Sippl W. Comparison of homology models and X-ray structures of the nuclear receptor CAR: assessing the structural basis of constitutive activity. J Mol Graph Model. 2007;25:644–657. doi: 10.1016/j.jmgm.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 149.Moore DD. CAR: three new models for a problem child. Cell Metab. 2005;1:6–8. doi: 10.1016/j.cmet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 150.Wallace BD, Betts L, Talmage G, Pollet RM, Holman NS, Redinbo MR. Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXR alpha. J Mol Biol. 2013;425:2561–2577. doi: 10.1016/j.jmb.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19:1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 152.Delfosse V, Dendele B, Huet T, Grimaldi M, Boulahtouf A, Gerbal-Chaloin S, Beucher B, Roecklin D, Muller C, Rahmani R, Cavailles V, Daujat-Chavanieu M, Vivat V, Pascussi JM, Balaguer P, Bourguet W. Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds. Nat Commun. 2015;6:8089. doi: 10.1038/ncomms9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tabb MM, Kholodovych V, Grun F, Zhou CC, Welsh WJ, Blumberg B. Highly chlorinated PCBs inhibit the human xenobiotic response mediated by the steroid and xenobiotic receptor (SXR) Environ Health Persp. 2004;112:163–169. doi: 10.1289/ehp.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kublbeck J, Jyrkkarinne J, Molnar F, Kuningas T, Patel J, Windshugel B, Nevalainen T, Laitinen T, Sippl W, Poso A, Honkakoski P. New in Vitro Tools to Study Human Constitutive Androstane Receptor (CAR) Biology: Discovery and Comparison of Human CAR Inverse Agonists. Mol Pharmaceut. 2011;8:2424–2433. doi: 10.1021/mp2003658. [DOI] [PubMed] [Google Scholar]
- 155.Krausova L, Stejskaova L, Wang HW, Vrzal R, Dvorak Z, Mani S, Pavek P. Metformin suppresses pregnane X receptor (PXR)-regulated transactivation of CYP3A4 gene. Biochem Pharmacol. 2011;82:1771–1780. doi: 10.1016/j.bcp.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Healan-Greenberg C, Waring JF, Kempf DJ, Blomme EAG, Tirona RG, Kim RB. A human immunodeficiency virus protease inhibitor is a novel functional inhibitor of human pregnane x receptor. Drug Metab Dispos. 2008;36:500–507. doi: 10.1124/dmd.107.019547. [DOI] [PubMed] [Google Scholar]
- 157.Cherian MT, Lin WW, Wu J, Chen TS. CINPA1 Is an Inhibitor of Constitutive Androstane Receptor That Does Not Activate Pregnane X Receptor. Mol Pharmacol. 2015;87:878–889. doi: 10.1124/mol.115.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lin W, Yang L, Chai SC, Lu Y, Chen T. Development of CINPA1 analogs as novel and potent inverse agonists of constitutive androstane receptor. Eur J Med Chem. 2016;108:505–528. doi: 10.1016/j.ejmech.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]