Abstract
The need for a rapid and accurate method for the detection of fungal pathogens has become imperative as the incidence of fungal infections has increased dramatically. Herein, we tested the Luminex 100, a novel flow cytometer, for the detection of the medically important genus Trichosporon. This genus was selected as our proof-of-concept model due to the close phylogenetic relationship between the species. The method, which is based on a nucleotide hybridization assay, consists of a combination of different sets of fluorescent beads covalently bound to species-specific capture probes. Upon hybridization, the beads bearing the target amplicons are classified by their spectral addresses with a 635-nm laser. Quantitation of the hybridized biotinylated amplicon is based on fluorescence detection with a 532-nm laser. We tested in various multiplex formats 48 species-specific and group-specific capture probes designed in the D1/D2 region of ribosomal DNA, internal transcribed spacer regions, and intergenic spacer region. Species-specific biotinylated amplicons were generated with three sets of primers to yield fragments from the three regions. The assay was specific and fast, as it discriminated species differing by 1 nucleotide and required less than 50 min following amplification to process a 96-well plate. The sensitivity of the assay allowed the detection of 102 genome molecules in PCRs and 107 to 108 molecules of biotinylated amplification product. This technology provided a rapid means of detection of Trichosporon species with the flexibility to identify species in a multiplex format by combining different sets of beads.
The advances of medical technologies and treatments, e.g., chemotherapy, organ transplantation, and antimicrobial therapies, have contributed to the dissemination of fungal infections. For example, the incidence of invasive fungal infestation among organ transplant recipients has been reported as high as 59% (15). Among fungal diseases, deep-seated trichosporonosis has been reported to cause mortality in immunocompromised patients (31, 35). The disease is associated with severe conditions that cause morbidity, such as respiratory and renal failure, intravascular coagulation syndrome, and neutropenic state in immunocompromised patients (23). The causative agents of the disease include Trichosporon species T. inkin, T. ovoides, T. cutaneum, T. asahii, T. asteroides, and T. mucoides. The clinical cases caused by opportunistic fungal infection are constantly rising, and new species within the genus are emerging as new opportunistic pathogens (14, 24, 33). For example, one recent clinical case has confirmed the emergence of T. loubieri as a new human pathogen that can cause death if the disease is left unattended (24). To make matters worse, the prognosis for patients is relatively poor. In view of the severity of the situation, a rapid and correct identification method is important for efficient and prompt therapy. However, most clinical laboratories rely on methods that employ phenotypic characteristics that can be time-consuming and not very accurate. With rapid advances in molecular biology, combined with our in-house fungal database and the public accessibility of microbial sequence data, we developed a rapid and simple assay with a high-throughput capability to identify all the species within the genus Trichosporon. This method uses a novel flow cytometer, the Luminex 100. Luminex xMAP technology uses multiple color fluorescent microspheres by varying the proportion of red and infrared fluorescent dyes within microspheres to create an array of up to 100 separate bead classifications, each of which can represent a single species. Each dye, which is applied in different proportions, generates a 10 by 10 colored palette with a total of 100 sets of unique colors that are visible to a laser. The polystyrene microspheres are coated with carboxyl groups, which bind covalently to species-specific nucleic acid probes by carbodiimide coupling using 1-ethyl-3-(dimethylaminopropyl)carbodiimidede-hydrochloride (EDC) (10). The capture probe microsphere-based hybridization assay uses PCR-biotinylated amplicon target DNA, which is inoculated into the microsphere bead mixture containing species-specific probes of interest. By adding a reporter molecule (streptavidin R-phycoerythrin), all hybridized species-specific amplicons captured by their complementary nucleotide sequence on the microsphere beads are recognized by the fluorescence of the reporter molecule. The median fluorescence intensity (MFI) of the reporter molecule is then used to quantify the amount of DNA bound to the beads.
This technology has been adapted to a wide variety of applications involving human single-nucleotide polymorphisms (38), and bacterial identification (5, 27, 38).
We present a sensitive molecular method which is rapid and simple to perform. The described assay represents the validation of the technology with fungal cultures and details the methods for probe design and testing. Future studies will address the potential application of the assay with clinical specimens. This technology, which was adapted to identify species within the genus Trichosporon, can be expanded to include other pathogenic fungal species. To our knowledge, this is the first application of Luminex technology for the detection of fungal pathogens.
MATERIALS AND METHODS
Strains and DNA isolation.
The examined strains (Table 1) were obtained from Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands, the Portuguese Yeast Culture Collection, and the American Type Culture Collection.
TABLE 1.
List of strains studied
| Genus and species | Strain |
|---|---|
| Trichosporon aquatile | CBS 5973 T |
| Trichosporon asahii | CBS 2479 T |
| CBS 8640 | |
| Trichosporon asteroides | CBS 2481 T |
| Trichosporon brassicae | CBS 6382 T |
| Trichosporon caseorum | CBS 9052 T |
| Trichosporon cutaneum | CBS 2466 T |
| Trichosporon coremiiforme | CBS 2482 T |
| CBS 2478 | |
| Trichosporon dehoogii | CBS 8686 T |
| Trichosporon debeurmannianum | CBS 1896 T |
| Trichosporon dermatis | CBS 2043 T |
| CBS 8381 | |
| Trichosporon domesticum | CBS 8280 T |
| CBS 8111 | |
| Trichosporon dulcitum | CBS 8257 T |
| Trichosporon faecale | CBS 4828 T |
| Trichosporon gamsii | CBS 8245 T |
| Trichosporon gracile | CBS 8189 T |
| CBS 8518 | |
| CBS 8519 | |
| Trichosporon guehoae | CBS 8521 T |
| Trichosporon inkin | CBS 5585 T |
| Trichosporon japonicum | CBS 8641 T |
| Trichosporon jirovecii | CBS 6864 T |
| Trichosporon laibachii | CBS 5790 T |
| Trichosporon lignicola | CBS 22234 T |
| Trichosporon loubierii | CBS 7065 T |
| Trichosporon moniliiforme | ATCC 46490a |
| Trichosporon montevideense | CBS 6721 T |
| Trichosporon mucoides | CBS 7625 T |
| Trichosporon multisporum | CBS 2495 T |
| Trichosporon ovoides | CBS 7556 T |
| Trichosporon porosum | CBS 2040 T |
| Trichosporon scarabeorum | CBS 5601 T |
| Trichosporon smithiae | CBS 8370 T |
| Trichosporon sporotrichoides | CBS 8246 T |
| Trichosporon vadense | CBS 8901 T |
| Trichosporon veenhuisii | CBS 7136 T |
| Cryptococcus curvatus | CBS 570 T |
| Cryptococcus magnus | CBS 140 T |
| Cryptococcus sp. | CBS 7743 |
| Tsuchiyea wingfieldii | CBS 7118 T |
ATCC, American Type Culture Collection.
DNA isolation was obtained from cell cultures grown overnight and employed the QIAmp tissue kit (QIAGEN Inc.) and a lysing enzyme derived from Trichoderma harzianum (Sigma). The extraction method was described by Fell et al. (7).
Phylogenetic analysis.
Phylogenetic analyses, which were based on sequence analyses of the D1/D2 and internal transcribed spacer (ITS) regions, employed PAUP*4.0b10 using maximum likelihood and random step-wise addition.
PCR conditions.
DNA amplification was carried out with DNA extracted from pure-cure cultures using three sets of primers targeting the ribosomal DNA (rDNA) regions: (i) the large-subunit D1/D2 (LrDNA) region; (ii) ITS regions; and (iii) intergenic spacer (IGS) region. The D1/D2 amplicons, which yielded amplicon sizes of ∼630 bp, were generated using the universal forward primer F63 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and the universal reverse primer R635 (5′-GGTCCGTGTTTCAAGACG-3′). The ITS regions (530 bp) were amplified using the forward primer ITS1 (5′-TCCGTAGGTGAACCTGCG-3′) and the reverse primer ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). For IGS amplifications, three different sets of amplicons were generated using the reverse primer 5Srs (5′-AGCTTGACTTCGCAGATCGG-3′) and the forward primers Lr12 (5′-CTGAACGCCTCTAAGTCAGAA-3′; 650 to 875 bp), Lr11 (5′TTACCACAGGGATAACTGGC-3′; 950 to 1,200 bp), and IGS1 (5′CAGACGACTTGAATGGGAACG-3′; 490 to 600 bp). All PCR reverse primers were biotinylated at the 5′ end.
The reactions were carried out in microtubes containing QIAGEN HotStar Taq master mix (QIAGEN Inc.) in a final volume of 50 μl. The master mix contained 100 ng to 1 pg of genomic DNA, 1× PCR buffer, and 0.4 μM concentrations of forward and reverse primer pairs. The PCR was performed for 35 cycles in an MJ Research PTC 100 thermocycler. The PCR program involved 15 min of initial activation at 95°C, 30 s of denaturing at 95°C, 30 s of annealing at 50°C, and 30 s of extension at 72°C, followed by a 7-min final extension at 72°C. Samples were kept at 4°C until further analysis. An agarose gel electrophoresis was performed to confirm the synthesis of amplicons.
Probe development and probe coupling.
Probe design at the species level was based on sequence data from D1/D2, ITS1 and ITS2, and IGS regions (26, 32). Probe selection was facilitated by using visual sequence alignment employing the MegAlign program (DNAStar). Areas displaying sequence divergence among the species were analyzed for probe selection. All probes were designed to be uniform in length (21-mer); however, to avoid potential secondary structures or an unstable ΔG, some probe lengths were modified, resulting in probe sequences of 20 to 24 bp. The quality of the probe was assessed using the software program Oligo (Molecular Biology Insights, Inc.).
The specificity of the prospective sequence was analyzed with a yeast database developed in our laboratory using the Mac Vector program and GenBank BLAST. The database sequences are accessible in GenBank. Further probe validation was achieved by testing the performance of the probe on a capture probe hybridization format. Typically, the probes were tested in a multiplex format of 5. The capture probes, which were complementary in sequence to the biotinylated strand of the target amplicon, were synthesized with a 5′-end Amino C12 modification (IDT, Coralville, Iowa). Each probe was covalently coupled to a different set of 5.6-μm polystyrene carboxylated microspheres using a carbodiimide method (10) with slight modifications. Each microsphere set (Miraibio) contained unique spectral addresses by combining different ratios of red and infrared fluorochromes. A typical reaction involved the coupling of 5 × 106 microspheres resuspended in 25 μl of 0.1 M MES (2[N-morpholino]ethanesulfonic acid), pH 4.5, with a determined amount of probe (0.1 to 0.4 nmol). After successive vortexing and sonication steps, the beads were incubated twice with a final concentration of 0.5 μg of EDC/μl in the dark for 30 min at room temperature. The microspheres were washed with 1 ml of 0.02% Tween 20, followed by 1 ml of 0.1% sodium dodecyl sulfate. The beads were resuspended in 100 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8) and kept in the dark at 4°C.
Capture probe hybridization assay.
This assay is based upon detection of 5′-biotin-labeled PCR amplicons hybridized to specific capture probes covalently bound to the carboxylate surface of the microspheres. The 50-μl-volume reaction, which was carried out in 96-well plates in the presence of a 3 M TMAC solution (tetramethyl ammonium chloride-50 mM Tris [pH 8.0]-1 mM EDTA [pH 8.0]-0.1% sodium dodecyl sulfate), consisted of 5 μl of biotinylated amplicon diluted in 12 μl of 1× TE buffer (pH 8) and 33 μl of 1.5× TMAC solution containing a bead mixture of approximately 5,000 microspheres of each set of probes. Prior to hybridization, the reaction mixture was incubated for 5 min at 95°C in a PTC 100 thermocycler (MJ Research). This step was followed by 15 min of incubation at 55°C. After hybridization, the microspheres were pelleted by centrifugation at 2,250 rpm for 3 min with an Eppendorf 5804 centrifuge. Once the supernatant was carefully removed, the plate was further incubated for 5 min at 55°C and the hybridized amplicons were labeled for 5 min at 55°C with 300 ng of the fluorescent reporter molecule, streptavidin R-phycoerythrin. Reactions were then analyzed on the Luminex 100. One hundred microspheres of each set were analyzed, which represented 100 replicate measurements. MFI values were calculated with a digital signal processor and the Luminex proprietary software. Each assay was run twice, and the samples were run in duplicates. A blank and a set of positive and negative controls were included in the assay. The signal-to-background ratio represents the MFI signals of positive controls versus the background fluorescence of samples containing all components except the amplicon target.
To test the detection limits of the Luminex technology, several assays were conducted with various quantities (100 to 5 fmol) of biotinylated synthetic oligonucleotide targets bearing the reverse and complement of the probe sequence. In addition, the sensitivity of the assay was evaluated using serial dilutions of genomic DNA (10 to 10−3 ng) and amplicons (500 to 10−3 ng). DNA quantification was determined with a NanoDrop ND-1000 spectrophotometer using the absorbance at 260 nm. Prior to quantification, PCR products were purified with QIAGEN Quick-spin apparatus. Reactions were performed in duplicate, and the experiment was run twice.
To test the multiplex capability of the assay, each individual set of D1/D2, ITS, and IGS probes was pooled together into a bead mix and tested in various multiplex formats. The multiplex array of D1/D2, ITS, and IGS probes consisted of 16-, 14-, and 18-plex assays, respectively. In addition, probes were tested in 1- 5-, and 15-plex formats. Each plex assay was tested with amplicons derived from single species.
RESULTS
Trichosporon as a test model.
The genus Trichosporon was selected as our proof-of-concept model to test the Luminex 100 technology, as this group comprises a large number of closely related species, some of which are virulent pathogens. A list of all the tested strains is presented in Table 1. This list includes all 34 species of the basidiomycetous genus Trichosporon (22). One strain belonging to the genus Cryptococcus (Cryptococcus curvatus CBS 570) was included based on the phylogenetic position occupied within the Cutaneum clade (see Fig. 1).
FIG. 1.
Phylogenetic tree (maximum likelihood [PAUP 4.0b10]) of the D1/D2 LSU rDNA, depicting probe numbers developed for Trichosporon species. ⊄, species not included in cluster probes.
The phylogenetic delineation obtained from the D1/D2 analysis segregated the species into four distinct clades: Gracile, Cutaneum, Porosum, and Ovoides (see Fig. 1). Of the four clades, Cutaneum and Ovoides contain most of the medically relevant species. For example T. mucoides T. cutaneum, T. asahii, T. asteroides, T. ovoides, and T. inkin are the most commonly encountered pathogens (17). The other species are prevalent soil fungi, some of which (e.g., T. loubieri) have the potential to become opportunistic pathogens (17, 24).
Probe development.
Species-specific probes and cluster-specific probes were developed from sequence analysis of D1/D2, ITS, and IGS regions. Initial experiments were designed to validate and determine the probe specificity and the stringency conditions required to discriminate among closely related species. The species-specific and cluster probe sequences and the rRNA region chosen for probe design are illustrated in Table 2. The probes were designed to have a GC content higher than 30%, Tms higher than 50°C, and a length of 21 oligonucleotides. Some probes did not follow the described requirements. For example, T. debeurmannianum (P24) and Porosum cluster (P30) probes displayed GC contents of 29 and 24%, respectively, whereas probes targeting T. gracile (P8), T. debeurmannianum (P24), Porosum cluster (P30), T. caseorum (P19), and C. curvatus (P34b) exhibited Tm values ranging from 45 to 49°C. In order to increase the hybridization efficiency, some probes underwent length modification by adding or subtracting 1 to 3 bp at the 5′ and/or 3′ end, i.e., T. porosum (P21b). Probes that seemed to form hairpins or strong secondary structures and a positive ΔG (free energy of reaction) were avoided. Those displaying runs of more than three Gs or Cs at the 5′ or 3′ end were not chosen. An exemption was T. vadense (P44b), which displayed five Gs in a row.
TABLE 2.
List of probe sequences used for Trichosporon species-specific and group-specific identification
| Probe | rDNA region | Specificity | Probe Sequence (5′-3′) | Concn (nM) |
|---|---|---|---|---|
| P1 | D1/D2 | T. brassicae | ATAGCCTAGTATCACATACAC | 0.1 |
| P2 | D1/D2 | T. montevideense/T. domesticum | ATAGCCTAGGTTCACATACAC | 0.1 |
| P3c | IGS | T. scarabeorum | ATTGGCCATATTCCTACTTGC | 0.4 |
| P4 | D1/D2 | T. monoliiforme | TATTATTGCATGCACTGGGTG | 0.1 |
| P5 | D1/D2 | T. jirovecil/T. cutaneum | CAGTCGTGTTCTTCAGATTCA | 0.1 |
| P6 | ITS | T. laibachii/T. multisporum | TGGCTCCTCTCAAAAGAGTTA | 0.1 |
| P7b | IGS | T. multisporum | AGTTCGTACAAGTTCGTGGAT | 0.2 |
| P8 | D1/D2 | T. gracile | GGATAAAGATGCTAGGAATGT | 0.1 |
| P9 | D1/D2 | T. veenhuisii | TTGTCCGGTAGATAAAGGCAG | 0.1 |
| P10 | D1/D2 | T. loubieri | TCAGTTTTGCCCGGTGGATAA | 0.2 |
| P11 | ITS | T. mucoides | ACTTCGGTCGATTACTTTTAC | 0.2 |
| P11b | IGS | T. mucoides | AACGTGCTGCGCTACTAGGTG | 0.4 |
| P12 | ITS | T. cutaneum | TCGCTGGTGTTGACTTGAGAA | 0.1 |
| P12b | IGS | T. cutaneum | CAGTGACATGTGGGCGTTATA | 0.2 |
| P13 | ITS | T. ovoides | GTTTCACTGGTTCCATTGTGT | 0.1 |
| P14 | ITS | T. inkin | CTGGGTCCATGGTGTTGAAGC | 0.1 |
| P15 | D1/D2 | T. asteroides/T. japonicum/T. asahii | TACTTCCTTGGAACGGGTCAA | 0.1 |
| P16b | IGS | T. japonicum | GAGCAGCGAGCGACTTGGCAG | 0.2 |
| P17 | D1/D2 | T. faecale | ACTGCAGCTCACCTTTATGGC | 0.2 |
| P18b | IGS | T. aquatile | CTTAACACGATAACCGGTGCT | 0.2 |
| P19 | D1/D2 | T. caseorum | TTATAGCCTGTTATCACATAC | 0.4 |
| P20 | D1/D2 | T. guehoe | AGGTAGTTTCAATGTAGCTTC | 0.2 |
| P21b | D1/D2 | T. porosum | CATGAATCATGTTTATTGGACTC | 0.1 |
| P22 | ITS | Ovoides cluster | CTGATCGCTCGCCTTAAAAGA | 0.1 |
| P23b | ITS | T. smithiae | TGGATTTGAGTGATGGCAGTT | 0.2 |
| P24 | ITS | T. debeurmannianum | TTTTTACTTCGATCTCAAATC | 0.1 |
| P25b | ITS | T. sporotrichoides | CACTCTGTGTCGATTTTACAA | 0.2 |
| P25c | IGS | T. sporotrichoides | AGTTCATGCTCTAAGTCGGTTC | 0.2 |
| P26 | D1/D2 | T. dehoogii | CCTATTGTCGTATACACTGGA | 0.1 |
| P27b | ITS | Cutaneum cluster | CGGACAATTCTTGAACTCTGGT | 0.2 |
| P28b | IGS | T. jirovecii | TGTGAGTCTATCGGGCGCTTG | 0.2 |
| P29 | ITS | T. lignicola | TGTCCTCTGGAGTAATAAGTT | 0.1 |
| P30 | ITS | Porosum cluster | TGAACGTCTAGTTATTATAACAA | 0.2 |
| P31 | D1/D2 | T. gamsii | ATAAAGGTAATAGGAAGGTGG | 0.2 |
| P32 | D1/D2 | T. laibachii/T. multisporum/T. dulcitum/T. gracile | CAGTCGTGTTTATTGGATTCA | 0.2 |
| P33 | ITS | Gracile cluster | TGAACGTCATTAGATCATAA | 0.1 |
| P34b | IGS | Cryptococcus curvatus | GGTTTAAGATTGTATTGACTG | 0.2 |
| P35 | IGS | T. dulcitum | AACGTACAAGTCCGGACATGA | 0.2 |
| P36 | IGS | T. dermatis | ACGGTAGTTTGGAGTGAAGTGC | 0.2 |
| P37 | IGS | T. asteroides/T. japonicum | GAGCAGCGAGCGACTTGGCAG | 0.2 |
| P38 | IGS | T. asahii | TTCTCGACTGATGGCCTTGGT | 0.2 |
| P39 | IGS | T. montevideense | GGGTCCTAATAGATGCCATGT | 0.2 |
| P41b | IGS | T. coremiiforme | CACAGAGGTGCAAGAGGTGGG | 0.2 |
| P42 | IGS | T. lactis | GCTTCGGAGACTTGGGTTTGC | 0.4 |
| P43 | ITS | T. inkin/T. ovoides | ACTGTTCTACCACTTGACGCA | 0.4 |
| P44b | D1/D2 | T. vadense | TTATCACATGCACTGGGGGAG | 0.4 |
| P46 | IGS | T. domesticum | GCATGAGTGACGGCAGAGGTG | 0.4 |
| P47 | IGS | T. laibachii | GTGCACAAGAAACATACTAAC | 0.2 |
Figure 1 depicts a phylogenetic tree comprising the species within the genus Trichosporon and 48 probes that were designed and tested in the present study. Group-specific probes to identify the four major lineages within the Trichosporon genus were designed in the ITS region, since this region exhibited less variability than the D1/D2 region (26). All other species-specific probes, or mini-cluster probes, which target various species, were designed in one of the three rRNA regions (Table 2). The design of probes to include all members within the individual Cutaneum, Gracile, and Porosum clades represented a challenge. For example, T. porosum and T. debeurmannianum are species not targeted by Porosum (P30) and Cutaneum cluster (P27b) probes, respectively. Similarly, the Gracile cluster probe (P33) excludes the species T. montevideense and T. domesticum. In view of this limitation, specific probes were designed to target all species excluded from cluster probes. The exclusion of the species within their clades was due to inherent sequence divergence within the probe-designing region or because the species diverged significantly from the cluster group. The latter type of divergence applies to T. montevideense and T. domesticum, which showed sequence dissimilarity ranging from 2.5 to 5.7% from members of the Gracile clade. The ITS tree topology depicted both species as a sister clade of the Gracile group (26).
Special attention was given to the medically important yeasts, for which duplicate probes were designed for selected species. For the detection of T. mucoides and T. cutaneum, which are commonly encountered pathogens, two species-specific probes were designed in different regions of the rDNA as follows: T. mucoides, P11(ITS) and P11b (IGS); T. cutaneum, P12 (ITS) and P12b (IGS). Other species, such as T. inkin, T. ovoides, T. cutaneum, and T. asahii, were targeted by species-specific and cluster probes (Table 2). Probes for T. loubieri (P10) and T. dermatis (P36), which are new opportunistic pathogens within the genus (24), were also identified by species-specific probes. Identification of T. asteroides, an agent implicated in superficial infections, relied on a process of elimination, due to the lack of an adequate probe sequence. Thus, two probes with broader specificities were designed to target T. asteroides. These were probe 15, which includes the species T. asteroides, T. japonicum, and T. asahii, and P37, comprising T. asteroides and T. japonicum. With the inclusion of an additional probe, P16b (T. japonicum), we were able to resolve the species T. japonicum from T. asteroides.
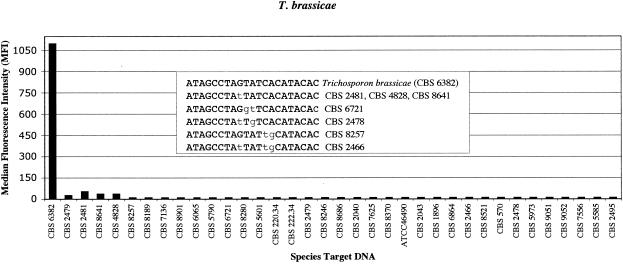
Each species-specific probe was tested against the complementary target amplicon, positive controls (perfect match), negative controls (more than three mismatches), and cross-reactive groups (one to three mismatches). The Luminex assay format, which was employed to test the specificities of the probes, included members of Trichosporon and other fungal genera that can potentially cross-react with the probe sequence. Results on 21-mer-length probes demonstrated that the selected hybridization assay conditions discriminated probe sequences differing by 1 or 2 bp, depending on the position of the mismatch, which influenced the extent of the hybrid destabilization. An example of probe specificity is illustrated in Fig. 2. The D1/D2 probe, T. brassicae (P1), was tested against a battery of DNA (Trichosporon spp.) that displayed several mismatches from the selected probe sequence. The results illustrated that mismatches located in the center or at positions 9 to 10/11 from the 5′ or 3′ end were discriminated under the assay conditions (Fig. 2). However, when the mismatches were located near the 3 or 5′ end, the assay lost specificity (Fig. 3). To illustrate the extent of potential cross-reactivity, an experimental T. scarabeorum probe sequence was designed to differ by 2 bp at the 5′ end from T. dulcitum. The results demonstrated that the location of the mismatches is critical and can lead to false-positive results if mismatches are not centered (Fig. 3).
FIG. 2.
Results of T. brassicae probe (P1) tested with other species of Trichosporon. Nucleotide variations between the most closely related species are depicted in the inset box.
FIG. 3.
Results of T. scarabeorum probe tested with other species of Trichosporon and Cryptococcus sp. Sequences with off-center nucleotide variations, i.e., T. dulcitum (CBS 8257), can yield false-positive results. Samples were run in duplicates, and the background fluorescence was subtracted.
Hybridization assay optimization conditions.
The assay conditions involved the use of TMAC, which is a quaternary ammonium salt agent that increases the stringency conditions, allowing discrimination among oligonucleotides differing by 1 bp. In 3 M TMAC, the hybridization conditions were dependent on the oligonucleotide length and not upon the base composition. The hybridization conditions were optimized by adjusting hybridization temperatures to provide adequate sensitivity and stringency conditions necessary for detection of the target species. Figure 4 shows the performance of T. ovoides (P13) at different hybridization temperatures ranging from 45 to 56°C. Hybridization at 45°C did not provide the discrimination between perfectly matched and mismatched sequences, since other species, e.g., T. aquatile (CBS 5973), T. asahii (CBS 2479), T. faecale (CBS 4828), and T. asteroides (CBS 2481), cross-reacted with the probe. A point to note is that the fluorescent intensity of these species, which differed by only 1 bp, was lower than that of the perfect-matched species, T. ovoides (CBS 7556). In contrast, hybridization at 55°C provided a good signal and the stringent conditions needed to discriminate between these closely related species (Fig. 4).
FIG. 4.
Effect of temperature on the specificity and signal intensity of the T. ovoides probe (P13). T. ovoides is represented by CBS 7556. Nucleotide variations between the most closely related species are depicted in the inset box. Other strains differed by 8 to 9 bp.
Further optimization of assay conditions was achieved by comparing the hybridization efficiency against various amounts of capture probes conjugated to the microspheres. These coupling amounts were optimized for each probe. For example, T. guehoe (P20) and T. faecale (P17) probes showed a 75 and 61% increase in signal performance at 0.2 nmol compared to 0.1 nmol of probe (Fig. 5). Others, like the group-specific ITS T. inkin/T. ovoides (P43) probe and the D1/D2 T. caseorum (P19) probe improved their signals by 51.8 and 75%, respectively, when the capture probe coupling amount was increased from 0.2 to 0.4 nmol (Fig. 5). An increase in the amount of capture probe did not always correlate with a higher signal. For instance, probes like T. sporotrichoides (P25b) showed a 43.5% decrease in signal when the coupling probe amount was increased from 0.2 to 0.4 nmol (Fig. 5). This “hook effect” occurs when the microspheres are overconjugated (Luminex, personal communication). In this scenario, the hybridization efficiency decreases due to steric hindrance on overconjugated microspheres.
FIG. 5.
Effect of varying the amount of capture probe on the signal performance of T. faecale (CBS 4828), T. guehoe (CBS 8521), T. inkin (CBS 5585)/T. ovoides (CBS7556), T. mucoides (CBS 7625), T. caseorum (CBS 9052), and T. sporotrichoides (CBS 8246). Samples were run in duplicates, and the background fluorescence of each set of beads was subtracted.
Probes which did not perform satisfactorily after testing for optimal probe coupling amount underwent sequence or length modification by adding bases at the 3′or 5′ end. For instance, a 74.5 and 45% increase in signal was observed when a total of 2 bp, located at the 3′ end and 5′ end, were added to the probe sequences of T. smithiae (P23b) and T. porosum (P21b), respectively (Fig. 6). Addition of bases can enhance the hybridization efficiency by increasing the amount of hybridized material. However, in some instances, probe lengthening led to a decrease in hybridization efficiency by decreasing probe specificity. For example, when the Porosum cluster probe (Table 2) was modified by adding 2 bp, ATGAACGTCTAGTTATTATAACAA (underlined characters denote addition of base pair), the probe lost specificity and cross-reacted with T. smithiae (P23b).
FIG. 6.
Effect of capture probe modification on the fluorescence signal of T. porosum (CBS 2040) and T. smithiae (CBS 8370). Probe 21 represents the 21-mer oligo ATGAATCATGTTTATTGGACT, whereas probe 21b represents the modified version, represented by CATGAATCATGTTTATTGGACTC. The sequence of probe 23, TGGATTTGAGTGATGGCAGTT, was modified to TGGATTTGAGTGATGGCAGTT (P23b) by adding 1 bp at the 5′ and 3′ ends.
Different probes exhibited different signal intensities, ranging from ∼200 to 2,000 above background levels. The signal-to-background ratio (S/B) for all tested probes fluctuated between ∼3.3 and ∼61.8. The highest S/B was observed for T. jirovecii (P28b), with an S/B of ∼61.8, followed by T. montevideense/T. domesticum (P2) with an S:B of ∼59. In contrast, the ITS probe designed to target the species T. sporotrichoides (P25b) exhibited an S/B of ∼3.30. In view of the poor S/B of P25b, another probe sequence (P25c) was chosen to avoid ambiguous identifications. This new probe (P25c), which was designed in the IGS, exhibited an S/B of ∼28. Overall, positive results corresponded to normalized MFI values, which were twice the background levels.
Amplicon size.
Using three different sets of primers (IGS1/5sR, Lr12/5sR, and Lr11/5sR), we examined the effect of amplicon size on the hybridization signal of the species T. mucoides (P11b), T. aquatile (P18b), T. jirovecii (P28b), T. japonicum (P16b), T. dermatis (P36), T. sporotrichoides (P25c), and T. asahii (P38) (Fig. 7). The primer set IGS1/5sR was used to generate the shortest segments, ranging in length from 490 to 600 bp, whereas Lr12/5sR yielded 650- to 875-bp amplicon fragments. For longer target amplicons (950 to 1,200 bp), the primer combination Lr11/5sR was employed. The wide range of species length polymorphisms, with each set of primers, is attributed to indel and repeat areas, which are common characteristics of the IGS region (4, 32). Surprisingly, lower hybridization signals were documented with the shorter target amplicons (490 to 600 bp) generated with IGS1/5sR (Fig. 7). In contrast, significantly higher signals were observed with amplicon fragments over 600 bp, which were obtained with the primer pairs Lr11/5sR and Lr12/5sR (Fig. 7). A similar trend was observed for the DI/D2 probes, T. gracile (P8) and T. veenhuisii (P9). Both probes exhibited a nearly 38% increase in signal with target amplicons of approximately ∼1,200 bp (ITS1/R635), as opposed to that with the ∼600-bp fragments obtained with the primer set F63-R635 (data not shown). Not all probes performed better with longer fragments. For instance, an ITS probe designed to target the species T. laibachii/T. multisporum (P6) failed to produce hybridization signal when tested against a target sequence of 1,200 bp. However, a positive signal was obtained when a shorter amplicon of ∼600 bp was used (data not shown).
FIG. 7.
Effects of amplicon size on the fluorescence intensities of the following probes: P11b, T. mucoides (CBS 7625); P18b, T. aquatile (CBS 5973); P16b, T. japonicum (CBS 8641); P25c, T. sporotrichoides (CBS 8246); P28b, T. jirovecii (CBS 6864); P36, T. dermatis (CBS 2043); P38, T. asahii (CBS 2749).
Multiplex reactions.
To test the multiplex capability of the Luminex technology, different sets of probes were pooled together and tested using a single target PCR per well. Fluorescence signal intensities for D1/D2, ITS, and IGS probes tested in multiplex formats were found to be similar to those observed in the uniplex (nonmultiplexed) or quintuplex format. For example, Fig. 8 illustrates the performance of P11 and P43 when tested in different multiplex formats consisting of 1-plex (1 probe), 5-plex (5 probes), and 15-plex (15 probes) formats. The signal intensities of probes tested in various plex formats were not significantly different, demonstrating the capability to simultaneously test different sets of probes without compromising the fluorescent signal.
FIG. 8.
Comparison of hybridization signals of P11 and P43 using 1-plex, 5-plex, and 15-plex assay formats. Each set of probes was tested individually and in a bead mix consisting of 1, 5, and 15 probes.
Genomic and amplicon detection limits.
Results on genomic detection limits employing clinically relevant fungal species are shown in Fig. 9. P43 (T. inkin/T. ovoides), P13 (T. ovoides), and P11b (T. mucoides) gave robust signals when the amount of genomic DNA in the PCR mixture ranged from 10 to 1 ng, but lower signals were recorded when genomic DNA ranged between 500 and 100 pg (Fig. 9). Below 10 pg the probe signals were barely detectable, except for T. mucoides (P11b), which displayed an MFI of ∼80. The results demonstrate that the present method is able to detect as low as 10 to 100 pg of genomic DNA with MFI signals ranging from 67 to 150 above background levels. An exception was P12 (T. cutaneum), which exhibited a detection limit of 500 pg. However, better detection limits were obtained when larger amounts of PCR product were used in the assay format. For instance, detection limits as low as 10 pg were obtained for P43 (T. inkin/T. ovoides), P11b (T. mucoides), P38 (T. asahii), and P11 (T. mucoides) when the amount of PCR product was increased to 15 μl (data not shown).
FIG. 9.
Detection limits of genomic DNA using various quantities of genomic DNA. The DNA template in the PCR ranged from 1 pg to 10 ng. After amplification, 5 μl of the PCR product was tested with its complementary probe sequence. The hybridization assay was carried out at 55°C.
To determine the detection limits of the amplification products, amplicons were serially diluted from 500 to 10−3 ng. As shown in Fig. 10, there was a steady increase in signal as the amount of amplification product was increased from 1 to 500 ng. Below 1 ng, no increase in signal was documented and the MFI values were close to background levels. Based on the presented data, this study shows that PCR sensitivity for the tested probes ranges between 1 and 5 ng, with signal intensities over 50 MFI, except for P12, which showed MFI values of <50 at 1 ng of amplicon target.
FIG. 10.
Detection of amplified targets. Amplicons targeting the complementary probe sequence were serially diluted and tested using the described hybridization assay format. Strains and probes tested were the following: P12, T. cutaneum (CBS 2466); P11, T. mucoides (CBS 7625); P13, T. ovoides (CBS 7556); P36, T. dermatis (CBS 2043).
DISCUSSION
There is a need for prompt, accurate, and reliable identification of yeast pathogens. To date, many common fungal species are unnoticed by common serological and microscopic tests (8, 28). Most of the conventional fungal diagnostic kits, such as the API kit and ID 32C, allow identification based on physiological and biochemical characteristics and sometimes can be laborious, inconclusive, and not provide accurate resolution at species level (9). During the last decade, several molecular techniques have been employed for the detection of fungal pathogens by using gene sequence analyses combined with species-specific primers or hybridization probes designed for 18S rDNA (19), 26S rDNA (6, 13), mitochondrial DNA (11), and the ITS region (1, 9, 31). Some of the PCR-based methods have been employed for the detection of clinically relevant Trichosporon species (9, 23, 30). This important genus is normally found in soil and fresh water and has been known to cause white piedra, hypersensitive pneumonia, and deep-seated infections (36). A nested PCR was developed for two of the most common species, T. asahii and T. mucoides, both of which cause deep-seated infections (23). Similarly, Sugita et al. (30) described a PCR analysis that employed one set of genus-specific primers to detect all Trichosporon species. Most recently, a multiplex PCR-based method in conjunction with microchip electrophoresis was developed for the identification of several species of Trichosporon and Candida (9). However, the PCR-microchip electrophoresis technology, which is based on length variability of PCR products, can be of little value for species exhibiting similar-length PCR products, as is the case for Candida albicans and Candida dubliniensis, with both species displaying similar amplicon fragment sizes in the ITS region (9). Also, as observed with any gel electrophoresis identification method, nonspecific bands can translate into ambiguous results. Even though some of the PCR approach methods are fairly fast, these analyses focus on a limited number of species and do not provide the resolution necessary to differentiate among closely related species.
Herein, we have described and tested a reliable molecular technique which combines PCR, oligonucleotide hybridization, and flow cytometry to target group-specific and species-specific isolates of the medically important genus Trichosporon. A total of 48 probes were designed and tested using a hybridization assay format combined with Luminex 100 technology. This technology provided a rapid means of species detection with the flexibility to allow the detection of species in a multiplex format. The present hybridization assay format combined with Luminex technology provided sufficient specificity and discrimination to differentiate closely related species. A probe hierarchical approach was followed to target species-specific probes and group-specific probes encompassing closely related species within a clade. The combined use of several species-specific probes and general probes can alleviate ambiguities and provides further information related to the phylogenetic placement of the species. This approach can be of extreme value in clinical settings, where redundancy in results is needed to ascertain an accurate diagnosis.
As in any hybridization assay with capture probes, optimization of assay parameters was needed to facilitate stable duplex formations with high specificity. The use of 3 M TMAC, in combination with a 55°C hybridization temperature, provided the conditions necessary to achieve the high-stringency conditions for discriminating between sequences differing by only 1 bp. TMAC, which is known to equalize AT and CG by base pair stability, is incorporated in hybridization assay formats because it allows different sets of probes with different characteristics to be used under identical hybridization conditions (16, 20). The equalization of the melting points of different probes with a 3 or 4 M solution of TMAC enhances the duplex yields (21).
To achieve probe specificity, it was of paramount importance to locate any mismatch in the center of the probe sequence; otherwise, the assay led to false-positive results. Mismatches in the center are known to have a more profound effect on the equilibrium state than mismatches near the 5′ or 3′ end (12). A study based on the kinetic effects of mismatches located at the first, fifth, and seventh base pair in a 13-mer oligonucleotide showed that the variation in Ka (association rate constant) was the highest when the location was at midpoint from the 5′ or 3′ end (12).
Other factors, such as probe length and attachment efficiency, can have significant effects on the specificity and performance of some probes. For instance, the addition of 2 bp to the probe sequences of T. smithiae (P23b) and T. porosum (P21b) improved their hybridization performance. Probe lengthening has been reported to enhance hybridization efficiency by increasing the amount of hybridized material (29), and it also can have significant impacts on probe equilibrium states by increasing the stability of the probe-target duplex reaction. The impact on the equilibrium state is dependent upon the base pair composition addition and the sequence context (nearest-neighbor effect) (25, 34). However, adding a few base pairs to some probe sequences does not always improve probe performance, as was the case for the Porosum clade-specific probe (P30b) (results not shown). Similar effects have been reported by others, for which a substantial decrease in resolution and specificity was found when probes underwent length modifications of a few nucleotides (3). Reports in the literature indicate that when a length of a probe is increased, a mismatched base pair in the probe-target duplex will have a marginal effect on the stability of the duplex. In this scenario, the effect of free energy penalty associated with mismatched base pairs becomes a fraction of the total free energy of binding (2). This would explain why mismatches associated in shorter sequences promote higher levels of destabilization in a duplex (3, 18).
Amplicon sequences under 300 bp are usually recommended in multiple-hybridization assay formats, as they allow probe sequences to overcome steric hindrance and successfully compete with the complementary strand of the amplicon (Luminex, personal communication). In contrast, our studies demonstrated that efficient hybridization reactions, as defined by signal intensity, can occur with amplicons longer than 600 bp. Similar results using a bead-based system have been reported with bacterial amplicons ranging from 628 to 728 bp (27).
Overall, factors related to probe design and sequence content were found to be of uttermost importance for the success of this methodology. For instance, a sequence displaying a string of six repeats, as portrayed in one of the T. vadense experimental probes (AGATCATAACATAAAAAAACTT), was found to be nonspecific. Therefore, another probe sequence was selected to target the species (Table 2). Similarly, the location of the probe, represented by the binding site of the amplicon, appeared to have an effect on probe performance. For example, sequences selected near 100 bp from the 5′ end performed poorly or did not yield any signal. On the contrary, sequences selected from the middle or close to the 3′ end of the alignment tended to perform better. Apparently, binding site areas closer to the 3′ end allow better interaction between the capture probe and the target by minimizing potential formation of secondary structures near the duplex formation site.
The observed wide range of fluorescence signals upon hybridization of different probes might be associated with several factors: (i) base-stacking interactions associated with probe sequences. For example, unpaired bases stacking on the end of a duplex, as is the case when the target overlaps the capture probe, may affect the duplex yield (37). (ii) Another factor is sequence complexity and base composition. Several studies have demonstrated that sequences with the same base composition but different sequences give different yields (20, 21). (iii) A third factor is the presence of an internal hairpin structure. Although we avoided probes with hairpin structures, a few internal complementary bases within the sequence of the capture probe might lead to minor secondary structures affecting the formation and duplex yield. (iv) A fourth factor is the presence of secondary structure conformations near the probe-target binding area. The DNA target can easily fold back upon itself to form helices and even more-complicated structures as a result of the Watson-Crick base pairing. These structural conformations, if close to the binding area, might prevent or partially interfere with duplex formation (37). For instance, when comparing the performance of P6 with two amplicon fragments differing by ∼600 bp, a positive signal was only generated with the smaller fragment (∼600 bp). Secondary structures obtained with version 3 of the MFOLD software (data not shown) showed that even though both amplicons displayed similar secondary structures around the target area (positions 360 to 380), the longer fragment exhibited a more complex structural conformation consisting of multiple hairpin loops, stem structures, and a main multibranch with 17 hairpin loops. Most probably, all these secondary structures can induce bending and distortion, which can limit and affect probe accessibility and binding efficiency.
The sensitivity of the assay, as determined with P43 (T. inkin/T. ovoides), P38 (T. asahii), P11, and P11b (T. mucoides), demonstrated that this method enables the detection of 10 pg of genomic DNA template in the PCR, except in the cases of P13 (T. ovoides) and P14 (T. inkin), which required 100 pg of genomic DNA (Fig. 9). Assuming that the genome size of the Trichosporon spp. is similar to that of Cryptococcus neoformans (24 Mb) and that the average molecular mass of a double-stranded DNA base pair is 660 Da, 10 pg of genomic DNA corresponds to a detection limit of ∼380 genome molecules. Detection limits ranging from 1,659 to 189,753 genome molecules have been reported by others using the Luminex technology for identification of bacterial pathogens (5).
After correcting for PCR product length and assuming there are 200 rRNA gene copies in Trichosporon spp., the PCR product limit of detection for P36 (T. dermatis), P13 (T. ovoides), and P11 (T. mucoides) ranged from 20.2 to 25.2 fmol. This represents a detection limit of 6.08 × 107 copies for T. dermatis and 7.55 × 107 copies for T. mucoides and T. ovoides. In contrast, P14 (T. inkin) and P43 (T. inkin/T. ovoides) required 50.4 fmol (1.51 × 108 copies). Other probes, particularly P38 (T. asahii) and P12 (T. cutaneum), displayed detection limits of 189 fmol (5.68 × 108 copies) and 252 fmol (7.58 × 108 copies), respectively. These detection limits represent cutoff values above background signals where the signal is ∼2 times above background level, once the background has been subtracted. Our calculated limit of detection could be more sensitive than the above values; however, at those low levels, species identification can be ambiguous and questionable due to the poor S/B of some of the probes. Relatively higher PCR detection limits, ranging from 0.25 to 0.1 fmol or 106 to 107 amplicon copies, were reported by Dunbar et al. (5), who used 20-mer synthetic oligonucleotide targets to determine the sensitivity of the PCR product in a hybridization assay. In contrast, we employed >600-bp PCR fragments. We speculate that the difference in detection limits was attributable to different hybridization kinetics and efficiencies when longer amplicons were employed. For instance, when we tested synthetic oligonucleotide targets, a much higher sensitivity was observed with signals of ∼1,000 MFI at 5-fmol levels (data not shown).
In summary, we developed a fast and reliable method that can be executed in clinical settings for the identification of Trichosporon species from culture-based material. This medically important fungal pathogen was used as our proof-of-concept model for the development of a comprehensive assay aimed at the identification of all the medically relevant fungal species. This assay uses Luminex technology, which has the potential capability to provide multiplex analysis combined with a high-throughput system. This nonwashed captured probe hybridization assay involves few and simple steps that can be performed in less than 50 min after amplification products are generated. The specificity and sensitivity of the assay allowed discrimination of 1 bp among the species, allowing the detection of 102 to 104 genome copies in the PCR. Limits of detection in the hybridization reaction ranged from 107 to 108 amplicon target copies. In addition to the multiplexing capability, where as many as 100 different species can be analyzed in a single well, the ease of use, accuracy, and low cost of operation are a few of the conveniences of this technology. In addition, this bead-based assay allows the creation of different clinical testing platforms by combining different sets of microspheres. Any modification to the modules will simply involve the mixing of the proper sets of microspheres. In contrast, density microarray methods are less flexible, since they require the printing of new plates with specialized equipment.
Further studies are under way to validate these probes by using isolates derived from clinical cultures and specimens. The results of the second phase of the study will be published and will cover the clinical applicability of the assay in routine laboratory work. In addition, PCR multiplex experiments will be developed to generate IGS, D1/D2, and ITS fragments from a single PCR. This will simplify the assay and will reduce the cost of operation and amount of material to be analyzed.
Acknowledgments
We are grateful to Adele Statzell-Tallman for her valuable assistance with fungal cultures and Michael LaGier for reviewing the manuscript. James Jacobson, Sherry Dunbar, and Allen Ward (Luminex Corp.) are acknowledged for their valuable scientific and technical input on Luminex technology.
This research was supported by National Institutes of Health grant 1-UO1 AI53879-01.
REFERENCES
- 1.Abd-Elsalam, K. A., N. Ibrahim, M. A. Abdel-Satar, M. S. Khalil, and J. A. Verreett. 2003. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr. J. Biotechnol. 2:82-85. [Google Scholar]
- 2.Abou-ela, F., D. Koh, I. Tinoco, Jr., and F. J. Martin. 1985. Base-base mistmatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y= A,C,G,T). Nucleic Acids Res. 13:3944-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, B., M. Stewart, and A. Mazumder. 2000. Suspension arrays for high throughput multiplexed single nucleotide polymorphism genotyping. Cytometry 40:102-108. [PubMed] [Google Scholar]
- 4.Diaz, M. R., J. W. Fell, T. Boekhout, and B. Theelen. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Syst. Appl. Microbiol. 23:535-545. [DOI] [PubMed] [Google Scholar]
- 5.Dunbar, S. A., C. A. Vander Zee, K. G. Oliver, K. L. Karem, and J. W. Jacobson. 2003. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMap system. J. Microbiol. Methods 53:245-252. [DOI] [PubMed] [Google Scholar]
- 6.Fell, J. W. 1995. rDNA targeted oligonucleotide primers for the identification of pathogenic yeasts in a polymerase chain reaction. J. Ind. Microbiol. 14:475-477. [DOI] [PubMed] [Google Scholar]
- 7.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Bacteriol. 50:1351-1371. [DOI] [PubMed] [Google Scholar]
- 8.Fleming, R. V., T. J. Walsh, and E. J. Anaissie. 2002. Emerging and less common fungal pathogens. Infect. Dis. Clin. North Am. 16:915-933. [DOI] [PubMed] [Google Scholar]
- 9.Fujita, S. I., Y. Senda, S. Nakaguchi, and T. Hashimoto. 2001. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 39:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulton, R., R. McDade, P. Smith, L. Kienker, and J. Kettman. 1997. Advanced multiplexed analysis with the FlowMetrix system. Clin. Chem. 43:1749-1756. [PubMed] [Google Scholar]
- 11.Gardes, M., T. J. White, J. A. Fortin, T. D. Bruns, and J. W. Taylor. 1991. Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae by amplification of nuclear and mitochondrial ribosomal DNA. Can. J. Bot. 69:180-190. [Google Scholar]
- 12.Gotoh, M., Y. Hasegawa, Y. Shinohara, M. Schimizu, and M. Tosu. 1995. A new approach to determine the effect of mismatches on kinetic parameters in DNA hybridization using an optical biosensor. DNA Res. 2:285-293. [DOI] [PubMed] [Google Scholar]
- 13.Haynes, K. A., T. J. Westerneng, J. W. Fell, and W. Moens. 1995. Rapid detection of pathogenic fungi by polymerase chain reaction amplification of large subunit ribosomal DNA. J. Med. Vet. Mycol. 33:319-325. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, T., U. Hosokawa, N. Kondera, N. Toyazaki, and Y. Asada. 1996. Disseminated infection with Trichosporon asahii. Mycoses 39:195-199. [DOI] [PubMed] [Google Scholar]
- 15.Kanj, S. S., K. Welty-Wolf, J. Madden, V. Tapson, M. A. Baz, and D. Davis. 1996. Fungal infections in lung and heart-lung transplant recipients, report of 9 cases and review of the literature. Medicine 75:142-156. [DOI] [PubMed] [Google Scholar]
- 16.Kiesling, T., M. R. Diaz, A. Statzell-Tallman, and J. W. Fell. 2002. Field identification of marine yeasts using DNA hybridization macroarrays, p. 69-80. In K. D. Hyde, S. T. Moss, and L. L. P. Vrijmoed (ed.), Fungi in marine environments. Fungal Diversity Press, Hong Kong.
- 17.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology, p. 866. Lea & Febiger, Philadelphia, Pa.
- 18.Livshits, M. A., and A. D. Mirzabekov. 1996. Theoretical analysis of the kinetics of DNA hybridization with gel immobilized oligonucleotides. Biophys. J. 71:2795-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makimura, K., S. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 20.Maskos, U., and E. M. Southern. 1993. A study of oligonucleotide reassociation using large arrays of oligonucleotides synthesized on a large support. Nucleic Acids Res. 21:4663-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maskos, U., and E. M. Southern. 1992. Parallel analysis of oligodeoxyribonucleotide (oligonucleotide) interactions. I. Analysis of factors influencing duplex formation. Nucleic Acids Res. 20:1675-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Middelhoven, W. J., G. Scorzetti, and J. W. Fell. 2004. Systematics of the anamorphic basidiomycetous yeast genus Trichosporon Behrend with the description of five novel species: Trichosporon vadense, T. smithiae, T. dehoogii, T. scarabeorum, and T. gamsii. Int. J. Sys. Evol. Microbiol. 54:975-986. [DOI] [PubMed] [Google Scholar]
- 23.Nagai, H., Y. Yamakami, A. Hashimoto, I. Tokimatsu, and M. Nasu. 1999. PCR detection of DNA specific for Trichosporon species in serum patients with disseminated trichosporonosis. J. Clin. Microbiol. 37:694-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padhye, A. A., S. Verghese, P. Ravichandran, G. Balamurugan, L. Hall, P. Padma, and M C. Fernandez. 2003. Trichosporon loubieri infection in a patient with adult polycystic kidney disease. J. Clin. Microbiol. 41:479-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santa Lucia, J., Jr. 1998. A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scorzetti, G., J. W. Fell, A. Fonseca, and A. Statzell-Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 27.Spiro, A., M. Lowe, and D. Brown. 2000. A bead-based method for the multiplexed quantitation of DNA sequences using flow cytometry. Appl. Environ. Microbiol. 66:4258-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staib, F. 1987. Cryptococcosis in AIDS—mycological, diagnostic and epidemiological observations. AIDS Forsch. 2:363-382. [Google Scholar]
- 29.Steel, A. B., T. M. Herne, and M. J. Tarlov. 1998. Electrochemical quantitation of DNA immobilized on gold. Anal. Chem. 70:4670-4677. [DOI] [PubMed] [Google Scholar]
- 30.Sugita, T. A., M. Nikishikawa, and T. Shinoda. 1998. Rapid detection of species opportunistic yeast Trichosporon by PCR. J. Clin. Microbiol. 36:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugita, T. A., M. Nikishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichoporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugita, T. A., M. Nakajima, R. Ikeda, T. Matsuhima, and T. Shinoda. 2002. Sequence analysis of ribosomal DNA intergenic spacer 1 regions of Trichoporon species. J. Clin. Microbiol. 40:1826-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton, D. A. 2002. Laboratory evaluation of new antifungal agents against rare and refractory mycoses. Curr. Opin. Infect. Dis. 15:576-582. [DOI] [PubMed] [Google Scholar]
- 34.Tsourkas, A., M. A. Behlke, S. D. Rose, and G. Bao. 2003. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 31:1319-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walling, D. M., D. J. McGraw, W. G. Merz, J. E. Karp, and G. M. Hutchins. 1987. Disseminated infection with Trichosporon beigelii. Rev. Infect. Dis. 9:1013-1019. [DOI] [PubMed] [Google Scholar]
- 36.Walsh, T. J. 1989. Trichosporonosis. Infect. Dis. Clin. North Am. 3:43-52. [PubMed] [Google Scholar]
- 37.Williams, J. C., S. C. Case-Green, S. C. Mir., and E. M. Southern. 1994. Studies of oligonucleotide interactions by hybridization to arrays: the influence of dangling ends on duplex yield. Nucleic Acids Res. 22:1365-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye, F., M. S. Li, J. D. Taylor, Q. Nguyen, H. M. Colton, W. M. Casey, M. Wagner, M. P. Weiner, and J. Chen. 2001. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum. Mutat. 17:305-316. [DOI] [PubMed] [Google Scholar]