Abstract
The vitamin A metabolite all-trans retinoic acid (ATRA) induces a gut-homing phenotype in activated CD4+ conventional T cells (Tconv) by upregulating the integrin α4β7 and the chemokine receptor CCR9. We report that, in contrast to mouse Tconv, only about 50% of regulatory T cells (Treg) upregulate CCR9 when stimulated by physiological levels of ATRA, even though Tconv and Treg express similar levels of the retinoic acid receptor (RAR). The resulting bimodal CCR9 expression is not associated with differences in the extent of their proliferation, level of Foxp3 expression, or affiliation with naturally-occurring Treg (nTreg) or induced Treg (iTreg) in the circulating Treg pool. Furthermore, we find that exposure of Treg to the mechanistic target of rapamycin (mTOR) inhibitor rapamycin suppresses upregulation of both CCR9 and α4β7, an effect that is not evident with Tconv. This suggests that in Treg, ATRA-induced upregulation of CCR9 and α4β7 is dependent on activation of an mTOR signaling pathway. The involvement of mTOR is independent of Akt activity, since specific inhibition of Akt, pyruvate dehydrogenase kinase-1, or its downstream target glycogen synthase kinase-3, did not prevent CCR9 expression. Additionally, Rictor (mTOR complex [C] 2)-deficient Treg showed unaltered ability to express CCR9, whereas Raptor (mTORC1)-deficient Treg were unable to upregulate CCR9, suggesting the selective participation of mTORC1. These findings reveal a novel difference between ATRA signaling and chemokine receptor induction in Treg versus Tconv and provide a framework via which the migratory behavior of Treg versus Tconv might be regulated differentially for therapeutic purposes.
Keywords: rodent, T cells, cytokine receptors, cell trafficking, spleen and lymph nodes, signal transduction, gene regulation
Introduction
CD4+CD25+ forkhead box p3+ (Foxp3+) regulatory T cells (Treg) are a well-studied subset of immunosuppressive T cells. They have shown considerable promise in the treatment of experimental autoimmune disease (1–3), prevention of skin or organ allograft rejection (4–6) and suppression of graft-versus-host disease following bone marrow transplantation (7–9). Challenges to the clinical development of Treg therapy include the generation of sufficient numbers of these cells ex vivo (10, 11), the functional stability and longevity of the Treg in an inflammatory milieu, and their homing to lymphoid tissue versus inflammatory sites (12). In addressing these issues, the mechanistic target of rapamycin (mTOR) antagonist rapamycin and the vitamin A metabolite all-trans retinoic acid (ATRA), have both been shown to improve the selective expansion of Treg and to enhance their stability and resistance to pro-inflammatory cytokines (13–16).
There is also evidence that rapamycin and ATRA can affect chemokine receptor expression by T cells and consequently, their migration. Thus, ATRA induces upregulation of the gut-homing receptors CCR9 and α4β7 integrin (17, 18), whereas rapamycin increases the expression of CCR7 and consequently promotes more of a lymph node (LN)-homing Treg phenotype (19, 20). Given that the migratory ability of Treg directly impacts their therapeutic efficacy in vivo (21–24), a deeper understanding of how these agents may affect the migratory phenotype and behavior of Treg holds the potential to further improve their therapeutic application.
Previously, we reported (20) that rapamycin and ATRA synergize with TGF-β to generate induced Treg (iTreg) from conventional T cells (Tconv), but that they confer distinct migratory behavior on the resulting iTreg. We thus questioned how rapamycin and ATRA might affect the migration of naturally-occurring (n)Treg (a mixture of thymic and peripherally-induced Treg) when administered during their ex-vivo expansion. We show that, in contrast to our findings regarding the induction of Treg, those Treg that are already differentiated (representing the pool of cells accessible for ex vivo expansion and clinical use) respond differently to ATRA and rapamycin. Thus, nTreg show less sensitivity to ATRA-induced upregulation of CCR9. Moreover, this upregulation is dependent on the concomitant activity of mTOR complex 1 (mTORC1) in Treg, but not in Tconv, since rapamycin or the absence of Raptor, an essential component of this complex, blocks upregulation of CCR9 by the former, but not the latter population. Mechanistically, ATRA-induced upregulation of CCR9 in nTreg, rather unexpectedly does not require activation of the conventional upstream mediators pyruvate dehydrogenase kinase (PDK)1 or Akt, suggesting an alternate mechanism of mTORC1 activation. Ultimately, this translates into an altered in vivo distribution of Treg that is a function of their specific conditioning during ex vivo expansion, Our findings reveal a novel role of mTORC1 in determining the migratory behavior of nTreg that is distinct from that of Tconv and that could be exploited for selective manipulation of the in vivo distribution of Treg and Tconv populations for therapeutic purposes.
Materials and Methods
Animals
Eight- to 12-wk-old C57BL/6 (B6; H2Kb) mice and Foxp3-enhanced green fluorescent protein (eGFP) B6 reporter mice were obtained from The Jackson Laboratory and maintained in the specific pathogen-free central animal facility of University of Pittsburgh School of Medicine. Conditional Rictor gene disruption was accomplished by crossing (25) floxed Rictor mice with mice expressing tamoxifen-inducible Cre under the ROSA26 promoter (ROSA26-Cre ERT2). Rictorfl/fl ROSA26-Cre ERT2 mice or ROSA26 wild-type mice bred in our facility were given tamoxifen (82 mg/kg i.p.; Sigma-Aldrich; T5648) before experimentation, as described (26). Mice with conditional Raptor gene disruption selectively in T cells were a gift from Dr. Jonathan Powell (Johns Hopkins University) and were obtained by crossing floxed Raptor mice (The Jackson Laboratory; Stock No: 013188) with CD4-Cre transgenic mice (The Jackson Laboratory; Stock No: 017336). Experiments were conducted under an institutional animal care and use committee-approved protocol and in accordance with National Institutes of Health-approved guidelines.
Reagents and Abs
Complete medium (CM) comprised RPMI-1640 (Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% v/v FCS (Atlanta Biologicals, Lawrenceville, GA), non-essential amino acids, L-glutamine, sodium pyruvate, penicillin-streptomycin and 2-ME (all from Invitrogen, Carlsbad, CA). The mouse CD4-negative isolation kit for T cell isolation and αCD3/αCD28-coated beads (artificial APC, Dynal) for ex-vivo T cell expansion were from Invitrogen. Anti-PE microbeads and magnetic separation columns were from Miltenyi Biotec (Auburn, CA). ATRA, 2-deoxyglucose (2-DG), bexarotene and lithium chloride (LiCl) were from Sigma Aldrich (St. Louis, MO). Mouse rIL-2 and human rTGF-β1 (CHO cell-derived) were from PeproTech (Rocky Hills, NJ), rapamycin was from LC labs (Woburn, MA), the PDK inhibitor BX-912 was from Selleck Chemicals (Houston, TX) and the Akt inhibitor Akt-VIII from Santa Cruz Biotechnology (Dallas, TX). Anti-mouse CD4 (clone L3T4), -Foxp3 (FJK-16s), -CD25 (PC61.5), -CCR9 (eBioCW-1.2), -α4β7/LPAM-1 (DATK32) and -Helios (22F6) Abs were obtained from eBioscience (San Diego, CA). Anti-mouse CD44 (1M7) and -CD62L (MEL-14) were from BD Biosciences (San Diego, CA). Anti-mouse retinoic acid receptor alpha (RARα; EP1823Y) was from Abcam (Cambridge, MA).
Tconv and Treg purification
T cells were purified from mouse spleens and lymph nodes (LN) as described (5, 20). Briefly, single cell suspensions were incubated with purified anti-CD11b, -CD8, -B220, anti-Gr-1, -CD16/32, -Ter119 (BD Bioscience, San Jose, CA) and -I-A/E (eBioscience) mAbs. The cells were then washed and incubated with anti-rat dynabeads (Dynal, Invitrogen) according to the manufacturer’s instructions. Bulk CD4+ T cells were then purified by magnetic negative selection of the unwanted cells. For Treg isolation, these bulk CD4+ T cells were incubated with anti-CD25-PE mAb and CD4+CD25+ T cells isolated by positive selection using anti-PE microbeads and MS separation columns (Miltenyi Biotec). Purity was assessed by cytofluorometric analysis and was consistently 90–95%. The remaining cells were used as CD4+CD25− Tconv.
Cell culture and flow cytometry
Freshly-isolated Tconv and Treg were labeled with violet proliferation dye (VPD, CellTrace™ Violet Cell Proliferation Kit; Invitrogen) following the manufacturer’s protocol and cultured for 3–4 days in 96-well, round-bottom plates with αCD3/αCD28 beads (1.5 to 2 beads per cell), 10 ng/ml IL-2, with or without ATRA, rapamycin and other reagents, as indicated for each experiment. Each well contained 105 cells and 1.5 to 2.0 × 105 αCD3/αCD28 beads in a total of 200 μl final volume. For hypoxia experiments, 96-well plates were placed in a hypoxia chamber (1% O2). At the end of culture, the cells were stained with viability dye (Fixable Viability Dye; eBioscience), washed then stained with the mAbs indicated and analyzed by flow cytometry using a BD LSR II. Data were analyzed with FlowJo and events gated on viable, CD4+ Foxp3+ or CD4+ Foxp3− cells.
In vitro suppression assay
Freshly-isolated, naïve CD4+CD25+ cells were cultured for 3–4 days with IL-2 and αCD3/αCD28 beads, in the presence of either ATRA or a combination of ATRA and rapamycin. The ATRA-cultured Treg were then flow-sorted into eGFP-Foxp3+CCR9+ and eGFP-Foxp3+CCR9− populations and the ATRA/rapamycin-cultured Treg flow-sorted for eGFP-Foxp3+ cells. The three separate populations were then each co-cultured with freshly-isolated, naive CD4+CD25− VPD-stained Tconv and αCD3/αCD28 beads in 96-well plates. Each well contained 105 beads and 105 Tconv. Treg were added to achieve the Treg:Tconv ratios indicated. Co-cultures were maintained for 4 days, followed by staining and flow cytometry. T cell proliferation was quantified by analysis of the resulting VPD dilution profile using FlowJo.
In vivo migration
Freshly-isolated, naïve CD4+ CD25+ cells were cultured and flow-sorted, as described above, to generate ATRA-CCR9+ Treg, ATRA-CCR9− Treg and ATRA-rapamycin Treg. Each cell type was labeled with CFSE alone, VPD alone or a combination of CFSE and VPD, as indicated. After labeling, the cells were mixed and an aliquot analyzed by flow cytometry to determine the percentage breakdown of injected cells. One to 2 million total cells in 200 μl PBS were injected into congenic mice via the lateral tail vein. After 36 hours, spleens and LNs were harvested as described above and single cell suspensions analyzed by flow cytometry to determine the recovered percentages of ATRA-CCR9+, ATRA-CCR9− and ATRA-rapamycin Treg. The relative recovery was calculated as the percentage of recovered cells divided by the percentage of injected cells.
Statistical analyses
Means ± 1SD are shown, unless otherwise indicated. One-way ANOVA, followed by Tukey-Kramer multiple comparisons test was performed on datasets when comparing multiple groups simultaneously. One-way ANOVA, followed by Dunnett’s multiple comparison test was performed on datasets when comparing individual samples with a single control sample. Normalized datasets were compared using the repeated measures ANOVA, followed by the Tukey-Kramer multiple comparisons test.
Results
Treg and Tconv respond differently to ATRA stimulation
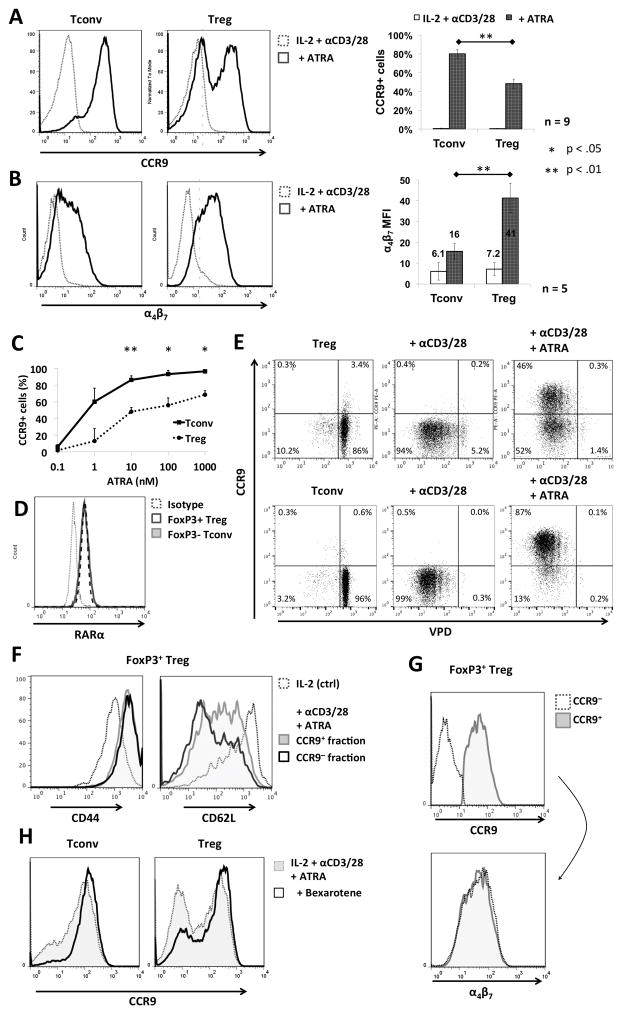
To determine the influence of ATRA on chemokine receptor expression by nTreg during their ex-vivo expansion, we characterized the response of freshly-isolated nTreg stimulated with αCD3/αCD28 beads for 4 days in the presence of ATRA by measuring cell surface expression of α4β7 and CCR9. Whereas both Tconv and nTreg upregulated α4β7 in response to ATRA, with an overall higher mean fluorescence intensity (MFI) induced in nTreg, only ~ 50% of nTreg became CCR9+ by day 4, compared to Tconv that displayed homogenous upregulation of CCR9 by day 3 of culture (Fig. 1 A and B). This bimodal distribution of CCR9+ and CCR9− nTreg was observed both at physiological (1 nM) (27) and supra-physiological (1000 nM) concentrations of ATRA (Fig. 1C), with a dose-response increase in the proportion of CCR9+ cells evident for both Tconv and Treg populations, although to a different extent. Since this observation suggested that nTreg respond differently to ATRA than Tconv, we investigated whether this was due to a heterogeneous expression of the RA receptor (RAR) in the nTreg starting population that would then give rise to the two subpopulations of Treg observed. Intracellular staining revealed that expression of RARα, the principal receptor isoform involved in T cell RA signaling (28), was homogeneous and identical on freshly-isolated nTreg and Tconv (Fig. 1D). Notably, gene array data (29) have also indicated similar levels of mRNA expression for all Rar and Rxr isoforms other than Rarg, which is actually increased in Treg. We can therefore conclude that the bimodal expression of CCR9 in nTreg following stimulation with ATRA was not due to comparatively low RARα expression by a subset of cells.
FIGURE 1. Differential responses of Tconv and Treg to ex-vivo stimulation in the presence of ATRA.
(A and B) Freshly-isolated B6 mouse Tconv or Treg were cultured for 3–4 days with IL-2/αCD3/αCD28 in the absence or presence of ATRA, as described in the Materials and Methods, then analyzed by flow cytometry for (A) CCR9 and (B) α4β7 expression. Plots are gated on CD4+Foxp3− (Tconv) or CD4+Foxp3+ (Treg). Bar graphs on the right depict means ±1SD across 5–9 individual experiments. The α4β7 MFIs are displayed above the bars. (C) Percentages of CCR9+ Tconv and Treg following culture with the indicated concentrations of ATRA. (D) Freshly-isolated Tconv and Treg were examined for intracellular levels of retinoic acid receptor alpha (RARα) by flow cytometry. Plots were gated on CD4+Foxp3− (Tconv) or CD4+Foxp3+ (Treg). (E and F) Treg and Tconv were labeled with violet proliferation dye (VPD) and cultured for 3–4 days in the absence or presence of αCD3/αCD28 beads ± ATRA, then analyzed by flow cytometry to assess (E) CCR9 expression and the extent of cell division, or (F) CD44 and CD62L expression in each Treg fraction. (G) CCR9+ and CCR9− cells were gated and analyzed for α4β7 expression. (H) Tconv and Treg were cultured as above, but with the addition of the RXR agonist bexarotene. CCR9 expression was then analyzed by flow cytometry. *, p< 0.05; **, p< 0.01
Previous studies (28) have implicated nuclear factor of activated T cells (NFAT) signaling in the modulation of CCR9 expression after TCR stimulation in the presence of RA. Since nTreg are considered anergic in vitro, and to proliferate less readily than Tconv in response to TCR ligation (30), we tested whether lack of CCR9 upregulation in a subpopulation of ATRA-treated nTreg was the result of insufficient cell activation. Staining with the proliferation dye VPD showed that both CCR9− and CCR9+ populations of ATRA-treated Treg underwent similar extents of proliferation (Fig. 1E) and comparable to the proliferation observed in Tconv. Moreover, each Treg subset displayed a similar degree of activation based on CD44 upregulation and CD62L downregulation (Fig. 1F) though CCR9+ Treg comprised a significant population with intermediate expression of CD62L rather than low. Furthermore, proliferating CCR9− and CCR9+ cells showed very similar cell surface expression of α4β7, suggesting that RA signaling was functional and activated in both populations (Fig. 1G).
Earlier reports (28, 31) have suggested that retinoid X receptor (RXR) activation can potentiate CCR9 mRNA transcription in the presence of RAR signaling. Since our results indicated that RA signaling was intact, but only approx. 50% of Treg upregulated CCR9, we questioned whether adding an RXR agonist would increase the proportion of CCR9+ nTreg obtained with ATRA stimulation. As shown in Fig. 1H, addition of the RXR agonist bexarotene at the start of cultures increased the proportion of CCR9+ Treg from 50% to about 80%. Thus, in addition to RAR, RXR signaling appears to be important in promoting CCR9 expression by Treg.
ATRA-stimulated CCR9+ and CCR9− nTreg exhibit similar Treg phenotypes and function
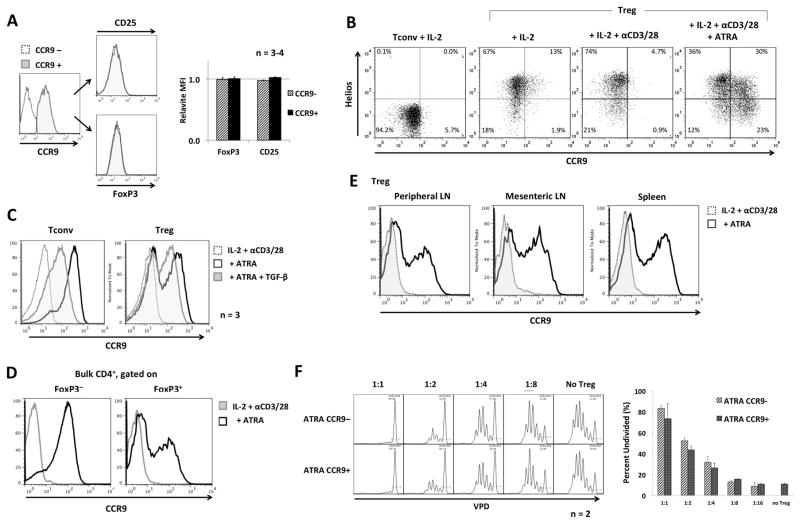
To obtain a better understanding of the bimodal expression of CCR9 by nTreg expanded in the presence of ATRA, we investigated whether the CCR9+ and CCR9− populations exhibited different phenotypes or function. As shown in Fig. 2A, the two populations expressed very similar levels of Foxp3 and CD25. Moreover, analysis of Helios expression, reported to distinguish naturally-occurring thymic-derived (tTreg) from peripherally-induced Treg (pTreg) (32), revealed that, in response to ATRA stimulation, both populations contributed to CCR9+ and CCR9− cells. Helios− pTreg made a greater relative contribution to CCR9+ cells than Helios+ tTreg (Fig. 2B), in agreement with our previous observations on the effect of ATRA during the induction of Treg (20). However, this limited contribution suggests that intrinsic properties of tTreg versus pTreg play only a limited role in regulating the differential expression of CCR9.
FIGURE 2. ATRA-expanded Treg display a suppressive phenotype that is unrelated to the differential expression of CCR9.
(A) ATRA-expanded CCR9− and CCR9+ Treg (generated as indicated in Figure 1) were analyzed by flow cytometry for CD25 and Foxp3 expression. (B) Tconv or Treg were cultured for 3–5 days with IL-2/αCD3/αCD28 in the absence or presence of ATRA and analyzed for CCR9 and Helios expression (n=2). (C) Tconv or Treg were cultured as described above, with or without addition of TGF-β, then analyzed for CCR9 expression (n=3). (D), Bulk CD4+ T cells were cultured in the presence or absence of ATRA, then gated based on FoxP3 expression and analyzed for CCR9 expression (n=2). (E) Treg were isolated from spleens, mesenteric lymph nodes (LN) or axillary/cervical (peripheral) LN, cultured in IL-2/αCD3/αCD28, with or without ATRA for 3–4 days, then analyzed for CCR9 expression by flow cytometry. (F) ATRA-expanded Treg were flow-sorted into CCR9+ and CCR9− populations and tested in a suppression assay, as described in the Materials and Methods, at the indicated Treg to Tconv ratios. Data in the lower panel are means ±1SD. Results were obtained from at least 2 experiments.
We then considered whether TGF-β, the principal cytokine responsible for generation of induced Treg, might play a role in CCR9 expression, since a previous study (33) had shown TGF-β could modulate expression of CCR9 in response to ATRA in Tconv. We asked if TGF-β secreted in culture, possibly by a subset of cells, might be responsible for the differences in CCR9 expression observed between Tconv and nTreg, as only Treg secrete TGF-β (34). Addition of recombinant TGF-β during their in vitro expansion decreased the overall intensity of CCR9 expression on both nTreg and Tconv. However, it did not alter the distribution between CCR9+ and CCR9− subsets in either nTreg or Tconv (Fig. 2C). Additionally, culturing Treg together with Tconv during expansion did not enhance conversion of Treg into CCR9+ cells compared to culturing each sorted population alone (Fig. 2D). This suggests that the heterogeneity in CCR9 expression was not due to a paracrine or autocrine effect of TGF-β secreted by nTreg (or that no factor secreted by activating T cells could support CCR9 up-regulation).
Since a proportion of the Treg that we investigated were derived from mesenteric LN that drain the intestines, we considered whether these cells (that might have been pre-exposed to ATRA in situ) were the dominant source of the resulting CCR9+ fraction. We stimulated, separately, nTreg harvested from mesenteric LN, spleen, or peripheral (cervical/axillary) LN in the absence or presence of ATRA. Bimodal CCR9 expression was exhibited by all 3 populations, ruling out the possibility that the CCR9+ population had originated solely from mesenteric LN (Fig. 2E), or that the ability to upregulate CCR9 was imprinted by the environment in which the nTreg resided before expansion.
We then assessed whether the differential expression of CCR9 was associated with altered Treg suppressive activity in vitro. Employing a conventional CFSE-based dye-dilution T cell proliferation assay (5), we determined whether the addition of CCR9− or CCR9+ Treg sorted following expansion in the presence of ATRA displayed differential inhibitory effects at various Treg to effector cell ratios. As shown in Fig. 2F, the two populations displayed similar suppressive activity, excluding the possibility that the CCR9 expression correlated with a different regulatory function.
ATRA-induced CCR9/α4β7 expression by nTreg is regulated by mTORC1
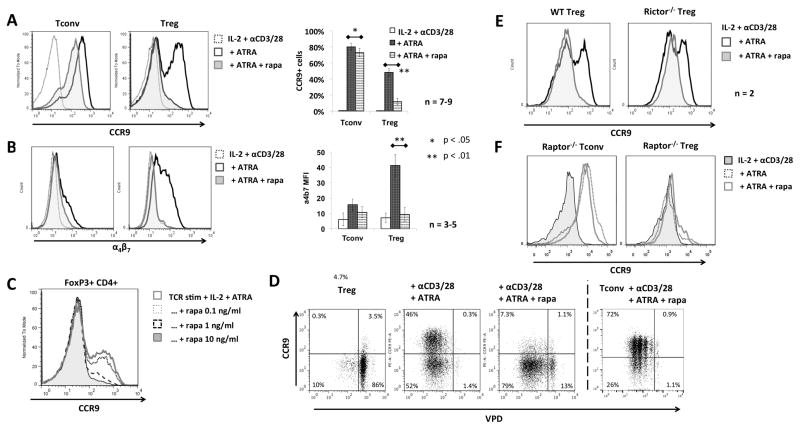
Given the different responses of nTreg and Tconv to ATRA (compared to our previous findings regarding iTreg (20)), we investigated the influence of rapamycin on CCR9 and α4β7 expression by stimulated nTreg when added in conjunction with ATRA. While addition of rapamycin at the start of cultures had only a modest inhibitory effect on CCR9 and α4β7 expression by Tconv, it strikingly blocked the upregulation of CCR9 and α4β7 by stimulated Treg (Fig. 3A–B). Concentrations of rapamycin as low as 0.1 ng/ml rapamcin (the approximate IC50 of rapamycin for the protein kinase mTORC1) (35) exerted an inhibitory effect on Treg (Fig. 3C), whereas 10 ng/ml exerted only a minimal effect on Tconv, suggesting that Treg but not Tconv require mTORC1 activity to upregulate α4β7 and CCR9.
FIGURE 3. ATRA-induced upregulation of CCR9 in Treg is dependent on mTORC1.
(A, B) Freshly-isolated Tconv or Treg were cultured for 3–4 days in IL-2/αCD3/αCD28 in the absence or presence of ATRA or with ATRA and rapamycin, then analyzed by flow cytometry for (A) CCR9 and (B) α4β7 expression. Panels on the right show means +1SD across multiple experiments. (C) Treg were also cultured in various concentrations of rapamycin and analyzed for CCR9 expression. (D) Freshly-isolated Treg and Tconv were stained with VPD and cultured as above, then analyzed by flow cytometry for the extent of cell proliferation. A representative result is shown. (E, F) Freshly-isolated Treg from wild-type (wt) or (E) Rictor-deficient or (F) Raptor-deficient mice were cultured in IL-2/αCD3/αCD28 without or with ATRA or ATRA and rapamycin, then analyzed by flow cytometry. Data were obtained from n=3–9 experiments (A, B) or at least n=2 experiments (C–F). *, p<0.05; **, p<0.01.
The sensitivity of CCR9 upregulation on nTreg to mTORC1 inhibition was rather unexpected, since we are aware of no reports of a T cell subset-specific intersection of the ATRA and mTOR pathways. Moreover, compared to Tconv, nTreg exhibit constitutively higher levels of already-activated mTORC1 (36). Since rapamycin is known to block T cell cycle progression in response to IL-2 signaling (37), we questioned whether the absence of CCR9 upregulation on nTreg was due to a selective blockade of cell proliferation, as most of the CCR9+ ATRA only-exposed Treg had undergone at least one cell division. Although the addition of rapamycin did cause a minor reduction in cell proliferation, almost all of the nTreg were CCR9−, even after 5 divisions (Fig. 3D). In contrast, despite an even more profound impact on proliferation, the majority of rapamycin-treated Tconv were CCR9+. Furthermore, the non-proliferating Treg population was also predominantly CCR9−, suggesting that the blocking effect of rapamycin was not (strictly) related to its influence on cell division.
There is evidence that prolonged exposure to rapamycin can indirectly inhibit mTORC2 by blocking its assembly (38). Since our culture period in the presence of rapamycin was 3–5 days, we investigated whether the block in CCR9 upregulation might be due to inhibition of mTORC1 or mTORC2. Treg from Rictor-deficient mice that lack functional mTORC2 were cultured with ATRA and showed a similar, biphasic profile of CCR9 expression as wild-type nTreg (Fig. 3E). This ATRA-dependent upregulation was also blocked in Rictor-deficient nTreg by the addition of rapamycin, suggesting that the blockade of CCR9 upregulation was not due to inhibition of mTORC2. To confirm that inhibition of mTORC1 was indeed responsible for blockade of CCR9 upregulation, nTreg and Tconv from Raptor-deficient mice (that lack functional mTORC1) were also examined (Fig. 3F). While Raptor-deficient Tconv upregulated CCR9 in the presence of ATRA, Raptor-deficient Treg did not do so. This finding confirms that ATRA-induced upregulation of CCR9 in Treg requires mTORC1, whereas this complex is dispensable for the same response in Tconv, highlighting an important difference between effector and regulatory T cell subsets.
CCR9+ and CCR9− Treg and ATRA-rapamycin Treg display similar suppressive function in vitro, but different lymphoid tissue-homing patterns in vivo
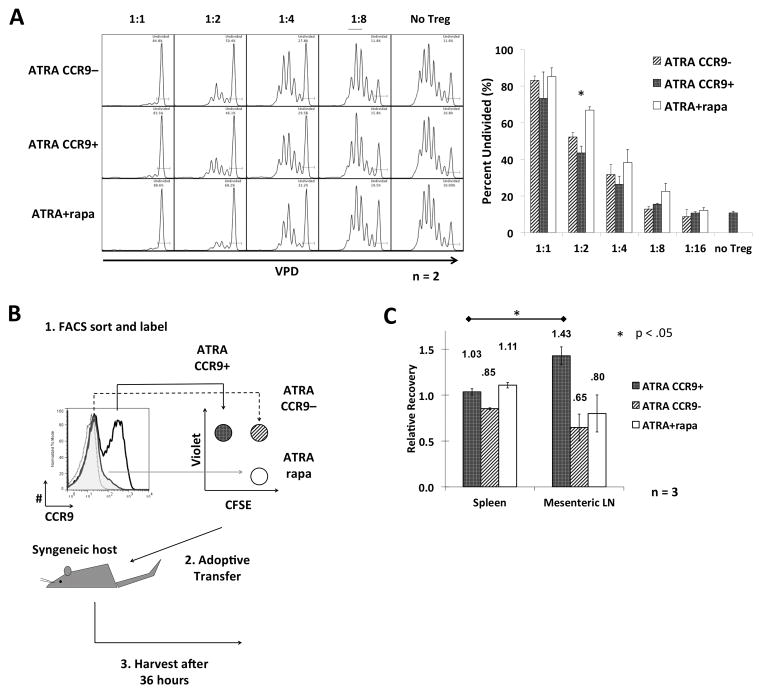
Previous studies (16) suggest that expanding Treg ex vivo in the presence of ATRA and rapamycin enhances their suppressive capacity and confers an epigenetic status more conducive to Treg stability in vivo. However, their migratory capacity impacts significantly on their therapeutic efficacy in vivo (21–23). Based on our finding that mTORC1 is required for upregulation of CCR9 by nTreg, we investigated whether ex vivo-expanded nTreg generated with ATRA + rapamycin would have different suppressive activity and, more importantly, impaired ability to migrate to the gut (due to the different CCR9 expression profile). We used an adoptive transfer model to compare the migration of Treg expanded either with ATRA alone or with the combination of ATRA + rapamycin. Further, since the ATRA-expanded Treg consist of a bimodal population, the CCR9− and CCR9+ fractions were separated via fluorescence-activated cell sorting. This yielded a total of 3 distinct Treg populations being analyzed. First, suppressive activity was assessed in vitro via CFSE-proliferation assay measuring the proportion of Tconv remaining undivided following stimulation with αCD3/αCD28 when co-cultured with titrations of each Treg subset. The degrees of suppression were similar between the 3 distinct Treg groups, with a trend towards the ATRA + rapamycin-expanded cells being slightly more suppressive at some ratios (Fig. 4A). To allow assessment of the migration of each population in the same animal (and to minimize differences due to biological variability between mice), each sorted Treg population was labeled distinctly with combinations of CFSE and VPD and a mixture (1:1:1) of the 3 Treg transferred into syngeneic B6 hosts (Fig. 4B). As predicted by their phenotype, 36 h after their adoptive transfer CCR9+ ATRA-expanded Treg accumulated in mesenteric LN at higher frequency compared to the spleen, whereas both CCR9− ATRA-expanded Treg and ATRA + rapamycin Treg accumulated in the mesenteric LN and the spleen to a similar extent (Fig. 4B).
FIGURE 4. mTOR does not impact Treg function but controls effective gut tropism.
(A) Freshly-isolated Treg were expanded in the presence of ATRA or ATRA and rapamycin. The ATRA-expanded Treg were then flow-sorted into CCR9+ and CCR9− populations. Each of the 3 types of Treg were then assessed for their suppressive function, at the Treg to Tconv ratios indicated, and for their in vivo migratory activity. (B) Schema illustrating how each of the 3 Treg populations was stained with either VPD or CFSE alone, or a combination of both. (C) A total of 2×106 cells was injected i.v. into each normal syngeneic host. Mice were euthanized 36 hours after Treg injection and single cell suspensions from the spleen and mesenteric LNs analyzed by flow cytometry to determine numbers of migrated Treg. The relative recovery ratios are displayed above the bars. Data were obtained from n=2–3 experiments. *, p<0.05.
Treg CCR9 upregulation is independent of PI3K/Akt, PDK-1, and GSK-3β pathways
Having confirmed that inhibition of CCR9 expression by Treg following exposure to rapamycin translated into altered in vivo cell distribution, we undertook preliminary characterization of the mechanism of rapamycin-mediated suppression of CCR9 upregulation. We explored several signaling pathway components upstream of mTORC1. mTORC1 is recognized mainly as a downstream target of PI3K and the serine-threonine kinase Akt and differences in Akt signaling have been reported between nTreg and Tconv (39). We thus investigated whether the PI3K/Akt pathway was responsible for the observed difference in CCR9 upregulation between nTreg and Tconv. nTreg and Tconv were cultured with Akt-VIII, an Akt inhibitor that preferentially inhibits the Akt isoforms Akt-1 and Akt-2, and then stimulated in the presence of ATRA. Unexpectedly, no inhibition of CCR9 upregulation was observed across a concentration range of 16 to 2000 nM, the upper limit being ~40 × the IC50 for Akt-1 and 10 × the IC50 for Akt-2 (Fig. 5A). This finding suggests that the PI3K/Akt axis is dispensable for the activation of mTORC1 necessary for CCR9 upregulation on nTreg.
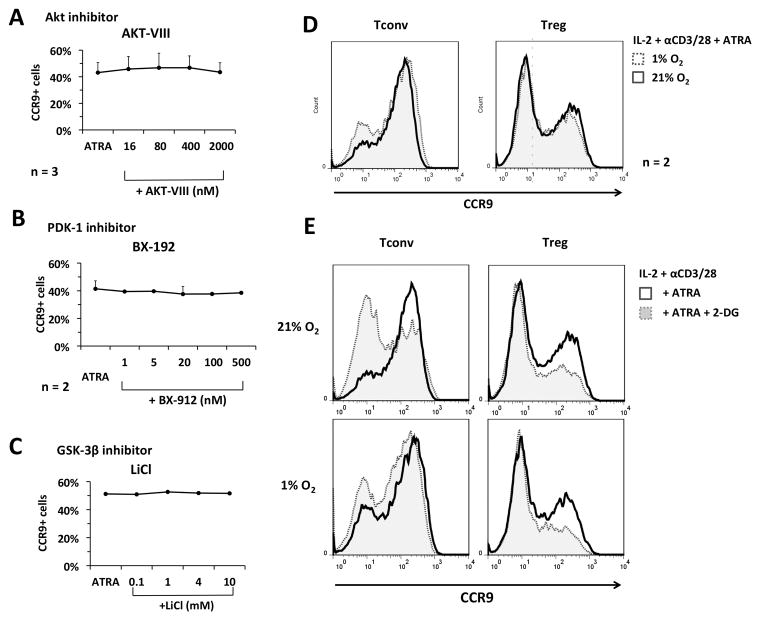
FIGURE 5. mTORC1-dependent CCR9 upregulation in Treg is independent of Akt, PDK-1 or GSK-3β activity and cannot be bypassed by stimulation of HIFα-controlled glycolysis.
(A–C) Freshly-isolated Treg were expanded in IL-2/αCD3/αCD28 for 4 days in the presence of ATRA, together with various concentrations of (A) the Akt inhibitor Akt-VIII, (B) the PDK-1 inhibitor BX-912, or (C) the GSK-3 inhibitor LiCl. (D–E) Freshly-isolated Tconv or Treg were expanded with ATRA under (D) normoxic versus hypoxic (1% O2) conditions or (E) in the absence or presence of the glycolytic inhibitor 2-deoxyglucose (2-DG). Data were obtained from 3 separate experiments (A–C), or are representative of n=2 experiments (D, E).
Apart from Akt, PDK-1 is an intermediary in mTOR signaling. A previous report (40) has related chemokine receptor expression and migration of CD8+ T cells to PDK-1 activity. We thus tested whether PDK-1 might play a similar role in nTreg in relation to ATRA signaling and CCR9 upregulation. However, addition of a PDK-1 inhibitor (BX-192) at the start of nTreg cultures did not affect their level of CCR9 expression when stimulated in the presence of ATRA (Fig. 5B), excluding its involvement in mTORC1 activation.
There is evidence that Treg have enhanced activity of glycogen synthase kinase-3β (GSK-3β) that inhibits mTOR by phosphorylating tuberous sclerosis complex 2 (TSC2) in the Wnt signaling pathway (41). With our new understanding of an important role of mTOR in the ability of Treg to respond to ATRA, we tested whether the decreased upregulation of CCR9 observed in Treg (Fig 1) was due to increased GSK-3β activity and whether augmenting mTOR activity by inhibiting GSK-3β would enhance upregulation of CCR9 by Treg. Addition of the GSK-3β inhibitor lithium chloride (LiCl) did not increase CCR9 expression by Treg (Fig. 5C). This excluded another possible explanation of the limited response of Treg to ATRA in comparison to Tconv.
mTORC1-dependency of Treg CCR9 upregulation is not explained by glycolysis and HIF-1α activity
Tconv and Treg exhibit different metabolic phenotypes, with the former displaying anaerobic glycolysis and Treg exhibiting oxidative phosphorylation (42). Since mTOR is a master regulator of metabolism (43, 44), we considered whether the mTORC1-dependency of CCR9 upregulation by Treg but not Tconv, was due to differences in their metabolism. Moreover, mTOR can modulate the migration of CD8+ T cells to lymphoid tissues by activating hypoxia-induced factor-1α (HIF-1α) that promotes glycolytic activity (40). We therefore questioned whether this effect was paralleled in CD4+ T cells, in that the selective requirement of mTORC1 in Treg CCR9 upregulation might be due to an inherently lower level of glycolytic activity in Treg, which would be more easily blocked by rapamycin. When Tconv or Treg were cultured under hypoxic (1% O2) versus normoxic conditions (Fig. 5D), or in the presence of the HIF-1α-inducer cobalt chloride (CoCl2) (Supplemental Fig. 1), neither condition increased levels of CCR9 expression by Treg, but slightly decreased CCR9 expression by Tconv (Fig. 5D). Moreover, the addition of rapamycin during stimulation under hypoxia or in the presence of CoCl2 resulted in complete inhibition of CCR9 up-regulation. This suggested that the mTORC1-dependency of Treg CCR9 upregulation was not mediated via mTORC1-induced activation of HIF-1α.
To test whether glycolytic activity independent of HIF-1α activation was necessary for CCR9 upregulation, we cultured Tconv or Treg in the absence or presence of the glycolytic inhibitor 2-deoxyglucose (2-DG). Although 2-DG modestly decreased the extent of CCR9 upregulation by Treg (but not to the extent induced by rapamycin), the effect was unexpectedly more pronounced in Tconv (Fig. 5E; upper panels). Moreover, the blocking effect of 2-DG in Tconv was almost completely abrogated under hypoxic conditions that enhance HIF-1α activity (Fig. 5E; lower panels). These results suggest that, while glycolysis and HIF-1α activity may play an important role in upregulation of CCR9 by Tconv, glycolysis and HIF-1α activity are not unique mediators of mTORC1-dependent CCR9 upregulation by Treg in response to ATRA.
Discussion
In this study, we have uncovered differential and novel effects of retinoic acid (ATRA) and rapamycin on expression of the gut-homing receptors CCR9 and α4β7 by mouse Treg versus Tconv. Thus, Treg display bimodality of cell surface CCR9 expression in response to ex-vivo αCD3/αCD28 stimulation in the presence of ATRA and are less sensitive than their Tconv counterparts to ATRA, despite similar levels of RAR isoform expression. Surprisingly, this behavior is restricted to regulation of CCR9, as upregulation of α4β7 in response to ATRA is more pronounced (and homogeneous) in Treg than Tconv. These findings suggest that, compared to Tconv, an intrinsic property of a substantial proportion of Treg attenuates their expression of an effective gut-homing phenotype. This observation cannot be explained simply by the presence of thymic- versus peripherally-derived Treg in the bulk population of Treg from secondary lymphoid tissue that we studied, since both Helios+ and Helios− cells were present in the expanded CCR9+ and CCR9− Treg populations.
Our studies exclude the involvement of multiple additional possible causes: a difference in proliferation from the starting cell population, a difference in the expression of Foxp3 that could compete with RAR for binding to the CCR9 promoter, a heterogeneity in Treg activation, or an impact of limiting amounts of TGFβ that counteract CCR9 upregulation. Overall, our results indicate that an intracellular molecular network regulates the ability of ATRA to induce CCR9 upregulation by Treg. In T cells, CCR9 expression requires cooperation between RAR and NFATc2 (28). Given that Treg have a significantly lower NFAT activity than Tconv in response to TCR engagement (45–47), we can speculate that reduced activation of NFATc2 in Treg may explain their reduced response to ATRA. Moreover, as activation of NFATc2 is digital after graded TCR signaling in Tconv (while NFκB activation is analog) (48), it is reasonable to suggest that such a property also pertains to Treg (and is further strengthened by competition with Foxp3 for binding to consensus sequences) that may explain the clear distinction between CCR9+ and CCR9− populations (with no intermediate expression). The lower activity of NFAT in Treg could also enhance the dependency of CCR9 gene expression on the activity of the heterodimer RAR/RXR (28, 31). The increase in the proportion of CCR9+ Treg we observed with the addition of the RXR agonist bexarotene (Fig. 1H) suggests a strong dependency on the full activation of the RAR/RXR heterodimer for proper expression of CCR9 in Treg. All these possible molecular explanations are the subject of ongoing investigations.
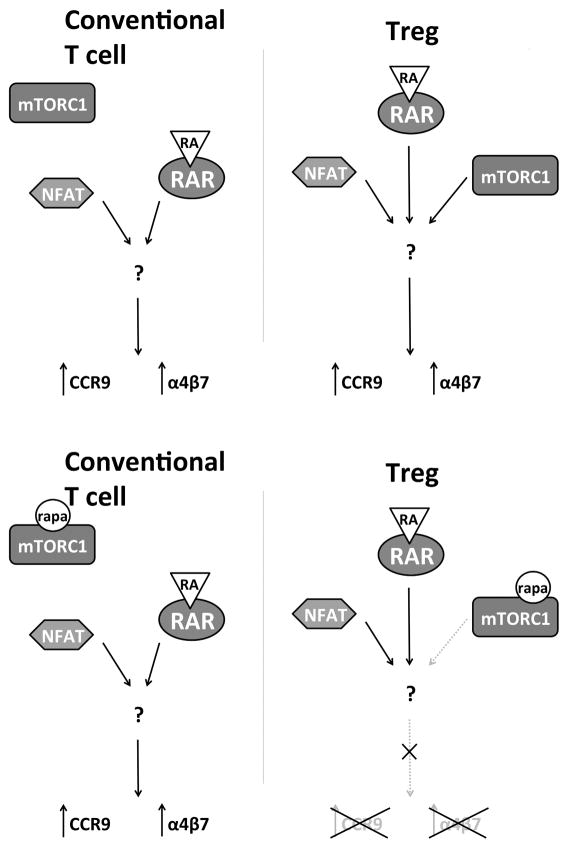
We also report the novel observation that upregulation of both CCR9 and α4β7 on Treg, but not on Tconv, is inhibited by the mTOR inhibitor rapamycin and that mTOR function, in the form of mTORC1 activity in Treg, is required for expression of an effective gut-homing phenotype in response to ATRA. This difference between Treg and Tconv is very striking and a clearer molecular understanding could reveal strategies to regulate immune responses via selective modification of Tconv or Treg migration. In our studies, CCR9 upregulation by Treg was unaffected by selective blockade of either Akt, PDK-1 or GSK-3β signaling. This raises the question that, if inhibiting Akt or PDK-1 does not block CCR9 expression by Treg, what signaling transduction pathway initiates mTORC1 activity? There are multiple potential pathways. It has been reported that, in addition to being a ligand for RAR, ATRA can bind peroxisome proliferator-activated receptor (PPAR) delta and promote expression of PDK-1 that then drives Akt and mTOR activation (49, 50). However, this pathway should have been affected by the inhibitors we investigated and thus we exclude this mechanism. Another possible explanation is the observation that, in cell lines sensitive to ATRA, addition of this molecule causes direct activation of mTOR and downstream induction of p70 S6 kinase activity (51). There is also the finding that Treg display greater constitutive mTOR activity than Tconv (36), in some circumstances induced by signaling of the leptin receptor (52), which could explain the absence of effect we observed with inhibitors of Akt or PDK-1. Finally, it is possible that mTOR activation is completely independent of ATRA and is induced by TCR engagement in a PI3K/AKT-independent fashion. Through activation of the signaling complex formed by caspase recruitment domain, CARD, membrane-associated guanylate kinase, MAGUK, protein 1 (CARMA1), B-cell lymphoma 10 (BCL10) and mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), Tconv activation stimulates glutamine uptake that, in turn, activates mTORC1 (53). Dissecting these possibilities would provide the necessary insight to design the aforementioned approaches to selectively modulate Tconv or Treg migration in vivo.
Our work does not address directly whether the influence of mTORC1 on CCR9 upregulation is at the transcriptional or translational level. It is unlikely that the inhibitory effect of rapamycin on upregulation of CCR9 by Treg is the result of global inhibition of protein translation, as rapamycin prevents downregulation of proteins such as CD62L and CCR7 in T cells (54). However, the suppression of mTORC1 activity by rapamycin has a probable translational component, as we observed a modest downshift in CCR9 expression (MFI) by Tconv exposed to rapamycin in addition to ATRA. This suggests that Tconv can use mTOR in promoting the expression of CCR9 in response to ATRA, but the activity of an alternative pathway (absent in Treg) excludes total dependency (Fig. 6). RAR activity is modulated by its interaction with co-repressors and co-activators. The aforementioned interaction with NFATc2 and NFATc1 is a clear example, but many more interactions have been characterized (18). This suggests that in Treg, activation of mTORC1 is necessary to either enable the activation and nuclear translocation of a transcription factor that acts as co-activator for RAR, or to inactivate a co-repressor. Further studies are needed to clarify whether mTORC1 plays a selective role in transcription or translation of gut-homing molecule expression by nTreg.
FIGURE 6. Suggested model of differential regulation of CCR9 and α4β7 induction in Treg and Tconv.
In Tconv, NFAT and RAR cooperate to upregulate CCR9 and α4β7 independently of mTOR activity. In Treg, mTROC1 activity is required for upregulation of CCR9 and α4β7.
During the past decade, we have learned that mTOR activity is pivotal for the integration of immune signals and metabolic clues necessary to maintain homeostasis, as well as for appropriate shaping of T cell activation and differentiation (43, 55–57). This intersection between metabolism and immunity has been extended further to the relationship between T cell metabolism and migration and examined in recent studies aimed at characterizing the role of glycolysis in CD8+ T cells (40, 58) and CCL5-dependent chemotaxis of effector T cells (59). In our preliminary exploration of this relationship, we observed that, in agreement with the general theme of these previous reports, upregulated CCR9 expression by Tconv may be at least partially dependent on glycolysis and can be rescued, in part, by increased HIF-1α activity. However, upregulation of CCR9 on Treg appears much less dependent on HIF-1α-induced glycolysis, suggesting that glycolytic activity does not play a major role in the unique Treg requirement of mTOR function for CCR9 expression. We believe that understanding this novel role of mTOR in controlling the migratory behavior of Treg will help clarify the controversy regarding the role of mTOR in Treg differentiation and function (43, 56) and better define similarities and differences between Treg and Tconv. For example, a recent study has shown that mTORC1-deficient Treg are less protective in a mouse model of inflammatory bowel disease based on adoptive transfer of T cells (36). In the light of our results, it is also possible that an unappreciated deficiency in gut migration may have contributed to the diminished in vivo therapeutic efficacy of mTORC1-deficient Treg. The extent to which the dependency on mTOR for CCR9 expression is restricted to Treg remains an open question. Our data show that, in Tconv, a redundant alternative pathway is present, suggesting that the main difference is the absence of this alternative pathway in Treg. Moreover, in line with investigations at the intersection between metabolism and immunity, it is noteworthy that Treg share many metabolic functions with memory T cells. Interestingly, it has been reported recently that, despite the ability of low doses of rapamycin to increase memory CD8 T cell numbers during the response to infection in secondary lymphoid tissues, the same treatment prevents accumulation of resident memory CD8+ T cells in intestinal and vaginal mucosa (60). Although the study did not investigate requirements for ATRA-induced upregulation of CCR9, it suggests that dependence on mTOR for CCR9 expression could extend to resident memory CD8 T cells.
Our findings bring to light the novel concept of targetable differences in migratory behavior between Treg and Tconv. As many studies have linked the migration and in vivo distribution of Treg to their in vivo function (21–24, 61–63), our finding that Treg, but not Tconv, are dependent on mTORC1 for their gut-homing tropism may have implications for the situational modulation of immunity by selectively targeting Treg (either in vivo or ex vivo). One such example would be to augment protective immunity in response to gastrointestinal pathogens and toxins by limiting Treg migration to effector sites. A recent report showed that subcutaneous immunization in the presence of exogenous RA improved homing of protective T cells to the gut in mice (64). It is reasonable to conceive that co-administration of RA and rapamycin would promote the migration of Tconv while inhibiting the migration of Treg to the gut, enhancing the immune response to infection and toxin. Based on the recent observation that intranasal administration of antigens promotes the generation of gastrointestinal tract-targeting effector T cells (via lung CD103+ DC producing ATRA) (65), it is possible that this vaccination strategy could be potentiated by co-administration of low doses of rapamycin. In a similar vein, a targetable, selective deficit in Treg migration may have beneficial applications in boosting anti-tumor immunity. Of course, these promising approaches could be affected by the immunosuppressive effect of rapamycin when used at too high a dose. This issue could then be obviated by a better understanding of the molecular synergy between mTOR and RAR and the identification of more selective pharmacological modifications. To our knowledge, our findings are the first to identify a selective requirement for mTORC1 activity in determining the gut-homing phenotype of Treg but not Tconv. Our future studies will determine the extent of differences in migratory behavior between Treg and Tconv and how these differences can be targeted for therapeutic application.
Supplementary Material
Acknowledgments
We thank Dr. Jonathan Powell for providing both the Raptor-conditional KO mice and for valuable discussion. We also thank Drs. David Geller and Allan Tsung for assistance with the hypoxia experiments. We thank Dr. Hongmei Shen of the Starzl Transplantation Institute Flow Core (University of Pittsburgh) and Xiaoling Zhang of the Ross Research Flow Cytometry Core (Johns Hopkins University) for invaluable assistance with all flow cytometry-based experiments.
This work was supported by National Institutes of Health grant R01 AI 67541 (to AWT), by grant number UL1TR000005 in the form of a CTSI-PEIR (to GR), by an American Diabetes Association Junior Faculty Award (1-10-JF-43 to GR) and by a Howard Hughes Medical Institute Predoctoral Fellowship (to LCC).
Non-standard abbreviations used in this article
- Akt
protein kinase B
- ATRA
all-trans retinoic acid
- eGFP
enhanced green fluorescent protein
- Foxp3
forkhead boxp3
- HIF-1α
hypoxia-induced factor-1α
- iTreg
induced regulatory T cell(s)
- MFI
mean fluorescence intensity
- mTOR
mechanistic target of rapamycin
- nTreg
naturally-occurring regulatory T cell(s)
- PDK-1
pyruvate dehydrogenase kinase-1
- RAR
retinoic acid receptor
- RXR
retinoic X receptor
- Treg
regulatory T cell(s)
- Tconv
conventional T cell(s)
- VPD
violet proliferation dye
References
- 1.Mottet C, Uhlig HH, Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. Journal of immunology. 2003;170:3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- 2.Tang Q, Henriksen KJ, Bi M, Finger EB, Szot G, Ye J, Masteller EL, McDevitt H, Bonyhadi M, Bluestone JA. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. The Journal of experimental medicine. 2004;199:1455–1465. doi: 10.1084/jem.20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bluestone JA, Buckner JH, Fitch M, Gitelman SE, Gupta S, Hellerstein MK, Herold KC, Lares A, Lee MR, Li K, Liu W, Long SA, Masiello LM, Nguyen V, Putnam AL, Rieck M, Sayre PH, Tang Q. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci Transl Med. 2015;7:315ra189. doi: 10.1126/scitranslmed.aad4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joffre O, Santolaria T, Calise D, Al Saati T, Hudrisier D, Romagnoli P, van Meerwijk JP. Prevention of acute and chronic allograft rejection with CD4+CD25+Foxp3+ regulatory T lymphocytes. Nature medicine. 2008;14:88–92. doi: 10.1038/nm1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raimondi G, Sumpter TL, Matta BM, Pillai M, Corbitt N, Vodovotz Y, Wang Z, Thomson AW. Mammalian target of rapamycin inhibition and alloantigen-specific regulatory T cells synergize to promote long-term graft survival in immunocompetent recipients. J Immunol. 2010;184:624–636. doi: 10.4049/jimmunol.0900936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore C, Tejon G, Fuentes C, Hidalgo Y, Bono MR, Maldonado P, Fernandez R, Wood KJ, Fierro JA, Rosemblatt M, Sauma D, Bushell A. Alloreactive regulatory T cells generated with retinoic acid prevent skin allograft rejection. European journal of immunology. 2015;45:452–463. doi: 10.1002/eji.201444743. [DOI] [PubMed] [Google Scholar]
- 7.Di Ianni M, Falzetti F, Carotti A, Terenzi A, Castellino F, Bonifacio E, Del Papa B, Zei T, Ostini RI, Cecchini D, Aloisi T, Perruccio K, Ruggeri L, Balucani C, Pierini A, Sportoletti P, Aristei C, Falini B, Reisner Y, Velardi A, Aversa F, Martelli MF. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood. 2011;117:3921–3928. doi: 10.1182/blood-2010-10-311894. [DOI] [PubMed] [Google Scholar]
- 8.Taylor PA, Lees CJ, Blazar BR. The infusion of ex vivo activated and expanded CD4(+)CD25(+) immune regulatory cells inhibits graft-versus-host disease lethality. Blood. 2002;99:3493–3499. doi: 10.1182/blood.v99.10.3493. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann P, Ermann J, Edinger M, Fathman CG, Strober S. Donor-type CD4(+)CD25(+) regulatory T cells suppress lethal acute graft-versus-host disease after allogeneic bone marrow transplantation. The Journal of experimental medicine. 2002;196:389–399. doi: 10.1084/jem.20020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hippen KL, Merkel SC, Schirm DK, Nelson C, Tennis NC, Riley JL, June CH, Miller JS, Wagner JE, Blazar BR. Generation and large-scale expansion of human inducible regulatory T cells that suppress graft-versus-host disease. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:1148–1157. doi: 10.1111/j.1600-6143.2011.03558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Net JB, Bushell A, Wood KJ, Harden PN. Regulatory T cells: first steps of clinical application in solid organ transplantation. Transpl Int. 2016;29:3–11. doi: 10.1111/tri.12608. [DOI] [PubMed] [Google Scholar]
- 12.Tang Q, Bluestone JA. Regulatory T-cell therapy in transplantation: moving to the clinic. Cold Spring Harbor perspectives in medicine. 2013;3:a015552. doi: 10.1101/cshperspect.a015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao S, Jin H, Korn T, Liu SM, Oukka M, Lim B, Kuchroo VK. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. Journal of immunology. 2008;181:2277–2284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Kong N, Wang J, Fan H, Zou H, Horwitz D, Brand D, Liu Z, Zheng SG. Cutting edge: all-trans retinoic acid sustains the stability and function of natural regulatory T cells in an inflammatory milieu. Journal of immunology. 2010;185:2675–2679. doi: 10.4049/jimmunol.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scotta C, Esposito M, Fazekasova H, Fanelli G, Edozie FC, Ali N, Xiao F, Peakman M, Afzali B, Sagoo P, Lechler RI, Lombardi G. Differential effects of rapamycin and retinoic acid on expansion, stability and suppressive qualities of human CD4(+)CD25(+)FOXP3(+) T regulatory cell subpopulations. Haematologica. 2013;98:1291–1299. doi: 10.3324/haematol.2012.074088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golovina TN, Mikheeva T, Brusko TM, Blazar BR, Bluestone JA, Riley JL. Retinoic acid and rapamycin differentially affect and synergistically promote the ex vivo expansion of natural human T regulatory cells. PloS one. 2011;6:e15868. doi: 10.1371/journal.pone.0015868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Brown C, Ortiz C, Noelle RJ. Leukocyte homing, fate, and function are controlled by retinoic acid. Physiological reviews. 2015;95:125–148. doi: 10.1152/physrev.00032.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battaglia M, Stabilini A, Migliavacca B, Horejs-Hoeck J, Kaupper T, Roncarolo MG. Rapamycin promotes expansion of functional CD4+CD25+FOXP3+ regulatory T cells of both healthy subjects and type 1 diabetic patients. Journal of immunology. 2006;177:8338–8347. doi: 10.4049/jimmunol.177.12.8338. [DOI] [PubMed] [Google Scholar]
- 20.Jhunjhunwala S, Chen LC, Nichols EE, Thomson AW, Raimondi G, Little SR. All-trans retinoic acid and rapamycin synergize with transforming growth factor-beta1 to induce regulatory T cells but confer different migratory capacities. J Leukoc Biol. 2013;94:981–989. doi: 10.1189/jlb.0312167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Schroppel B, Lal G, Jakubzick C, Mao X, Chen D, Yin N, Jessberger R, Ochando JC, Ding Y, Bromberg JS. Regulatory T cells sequentially migrate from inflamed tissues to draining lymph nodes to suppress the alloimmune response. Immunity. 2009;30:458–469. doi: 10.1016/j.immuni.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegmund K, Feuerer M, Siewert C, Ghani S, Haubold U, Dankof A, Krenn V, Schon MP, Scheffold A, Lowe JB, Hamann A, Syrbe U, Huehn J. Migration matters: regulatory T-cell compartmentalization determines suppressive activity in vivo. Blood. 2005;106:3097–3104. doi: 10.1182/blood-2005-05-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Issa F, Hester J, Milward K, Wood KJ. Homing of regulatory T cells to human skin is important for the prevention of alloimmune-mediated pathology in an in vivo cellular therapy model. PLoS One. 2012;7:e53331. doi: 10.1371/journal.pone.0053331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gratz IK, Campbell DJ. Organ-specific and memory treg cells: specificity, development, function, and maintenance. Front Immunol. 2014;5:333. doi: 10.3389/fimmu.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosborough BR, Raich-Regue D, Matta BM, Lee K, Gan B, DePinho RA, Hackstein H, Boothby M, Turnquist HR, Thomson AW. Murine dendritic cell rapamycin-resistant and rictor-independent mTOR controls IL-10, B7-H1, and regulatory T-cell induction. Blood. 2013;121:3619–3630. doi: 10.1182/blood-2012-08-448290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raich-Regue D, Rosborough BR, Watson AR, McGeachy MJ, Turnquist HR, Thomson AW. mTORC2 Deficiency in Myeloid Dendritic Cells Enhances Their Allogeneic Th1 and Th17 Stimulatory Ability after TLR4 Ligation In Vitro and In Vivo. Journal of immunology. 2015;194:4767–4776. doi: 10.4049/jimmunol.1402551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi H, Yokota-Nakatsuma A, Ohoka Y, Kagechika H, Kato C, Song SY, Iwata M. Retinoid X receptor agonists modulate Foxp3(+) regulatory T cell and Th17 cell differentiation with differential dependence on retinoic acid receptor activation. Journal of immunology. 2013;191:3725–3733. doi: 10.4049/jimmunol.1300032. [DOI] [PubMed] [Google Scholar]
- 28.Ohoka Y, Yokota A, Takeuchi H, Maeda N, Iwata M. Retinoic acid-induced CCR9 expression requires transient TCR stimulation and cooperativity between NFATc2 and the retinoic acid receptor/retinoid X receptor complex. Journal of immunology. 2011;186:733–744. doi: 10.4049/jimmunol.1000913. [DOI] [PubMed] [Google Scholar]
- 29.Heng TS, Painter MW C Immunological Genome Project. The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi T, Kuniyasu Y, Toda M, Sakaguchi N, Itoh M, Iwata M, Shimizu J, Sakaguchi S. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. International immunology. 1998;10:1969–1980. doi: 10.1093/intimm/10.12.1969. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi H, Yokota A, Ohoka Y, Kagechika H, Kato C, Song SY, Iwata M. Efficient induction of CCR9 on T cells requires coactivation of retinoic acid receptors and retinoid X receptors (RXRs): exaggerated T Cell homing to the intestine by RXR activation with organotins. Journal of immunology. 2010;185:5289–5299. doi: 10.4049/jimmunol.1000101. [DOI] [PubMed] [Google Scholar]
- 32.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. Journal of immunology. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi H, Yokota A, Ohoka Y, Iwata M. Cyp26b1 regulates retinoic acid-dependent signals in T cells and its expression is inhibited by transforming growth factor-beta. PloS one. 2011;6:e16089. doi: 10.1371/journal.pone.0016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. The Journal of experimental medicine. 2001;194:629–644. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards SR, Wandless TJ. The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. The Journal of biological chemistry. 2007;282:13395–13401. doi: 10.1074/jbc.M700498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. Journal of immunology. 1999;162:2775–2784. [PubMed] [Google Scholar]
- 38.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Crellin NK, Garcia RV, Levings MK. Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood. 2007;109:2014–2022. doi: 10.1182/blood-2006-07-035279. [DOI] [PubMed] [Google Scholar]
- 40.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. The Journal of experimental medicine. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham JA, Fray M, de Haseth S, Lee KM, Lian MM, Chase CM, Madsen JC, Markmann J, Benichou G, Colvin RB, Cosimi AB, Deng S, Kim J, Alessandrini A. Suppressive regulatory T cell activity is potentiated by glycogen synthase kinase 3{beta} inhibition. The Journal of biological chemistry. 2010;285:32852–32859. doi: 10.1074/jbc.M110.150904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michalek RD, V, Gerriets A, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. Journal of immunology. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, Chapman NM, Karmaus PW, Zeng H, Chi H. mTOR and metabolic regulation of conventional and regulatory T cells. Journal of leukocyte biology. 2015;97:837–847. doi: 10.1189/jlb.2RI0814-408R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunological reviews. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumpter TL, Payne KK, Wilkes DS. Regulation of the NFAT pathway discriminates CD4+CD25+ regulatory T cells from CD4+CD25− helper T cells. J Leukoc Biol. 2008;83:708–717. doi: 10.1189/jlb.0507321. [DOI] [PubMed] [Google Scholar]
- 46.Torgerson TR, Genin A, Chen C, Zhang M, Zhou B, Anover-Sombke S, Frank MB, Dozmorov I, Ocheltree E, Kulmala P, Centola M, Ochs HD, Wells AD, Cron RQ. FOXP3 inhibits activation-induced NFAT2 expression in T cells thereby limiting effector cytokine expression. J Immunol. 2009;183:907–915. doi: 10.4049/jimmunol.0800216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaeth M, Schliesser U, Muller G, Reissig S, Satoh K, Tuettenberg A, Jonuleit H, Waisman A, Muller MR, Serfling E, Sawitzki BS, Berberich-Siebelt F. Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2012;109:16258–16263. doi: 10.1073/pnas.1203870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Podtschaske M, Benary U, Zwinger S, Hofer T, Radbruch A, Baumgrass R. Digital NFATc2 activation per cell transforms graded T cell receptor activation into an all-or-none IL-2 expression. PLoS One. 2007;2:e935. doi: 10.1371/journal.pone.0000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Molecular and cellular biology. 2009;29:3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lal L, Li Y, Smith J, Sassano A, Uddin S, Parmar S, Tallman MS, Minucci S, Hay N, Platanias LC. Activation of the p70 S6 kinase by all-trans-retinoic acid in acute promyelocytic leukemia cells. Blood. 2005;105:1669–1677. doi: 10.1182/blood-2004-06-2078. [DOI] [PubMed] [Google Scholar]
- 52.Procaccini C, De Rosa V, Galgani M, Abanni L, Cali G, Porcellini A, Carbone F, Fontana S, Horvath TL, La Cava A, Matarese G. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, Blonska M, Lin X, Sun SC. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of experimental medicine. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. The Journal of clinical investigation. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan O, Burke JD, Gao DF, Fish EN. The chemokine CCL5 regulates glucose uptake and AMP kinase signaling in activated T cells to facilitate chemotaxis. The Journal of biological chemistry. 2012;287:29406–29416. doi: 10.1074/jbc.M112.348946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting edge: generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J Immunol. 2014;193:2067–2071. doi: 10.4049/jimmunol.1400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sather BD, Treuting P, Perdue N, Miazgowicz M, Fontenot JD, Rudensky AY, Campbell DJ. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. The Journal of experimental medicine. 2007;204:1335–1347. doi: 10.1084/jem.20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacDonald KG, Orban PC, Levings MK. T regulatory cell therapy in transplantation: stability, localization and functional specialization. Current opinion in organ transplantation. 2012;17:343–348. doi: 10.1097/MOT.0b013e328355aaaf. [DOI] [PubMed] [Google Scholar]
- 63.Menning A, Loddenkemper C, Westendorf AM, Szilagyi B, Buer J, Siewert C, Hamann A, Huehn J. Retinoic acid-induced gut tropism improves the protective capacity of Treg in acute but not in chronic gut inflammation. Eur J Immunol. 2010;40:2539–2548. doi: 10.1002/eji.200939938. [DOI] [PubMed] [Google Scholar]
- 64.Hammerschmidt SI, Friedrichsen M, Boelter J, Lyszkiewicz M, Kremmer E, Pabst O, Forster R. Retinoic acid induces homing of protective T and B cells to the gut after subcutaneous immunization in mice. J Clin Invest. 2011;121:3051–3061. doi: 10.1172/JCI44262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruane D, Brane L, Reis BS, Cheong C, Poles J, Do Y, Zhu H, Velinzon K, Choi JH, Studt N, Mayer L, Lavelle EC, Steinman RM, Mucida D, Mehandru S. Lung dendritic cells induce migration of protective T cells to the gastrointestinal tract. J Exp Med. 2013;210:1871–1888. doi: 10.1084/jem.20122762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.