Abstract
Mechanisms of self-tolerance often result in CD8+ tumor-infiltrating lymphocytes (TIL) with a hypofunctional phenotype incapable of tumor clearance. Using a transplantable colon carcinoma model, we found that CD8+ T cells became tolerized in less than 24 hours in an established tumor environment. To define the collective impact of pathways suppressing TIL function, we compared genome-wide mRNA expression of tumor-specific CD8+ T cells from the tumor and periphery. Notably, gene expression induced during TIL hypofunction more closely resembled self-tolerance than viral-exhaustion. Differential gene expression was refined to identify a core set of genes that defined hypofunctional TIL; these data comprise the first “molecular profile” of tumor-specific TIL that are naturally responding and represent a polyclonal repertoire. The molecular profile of TIL was further dissected to determine the extent of overlap and distinction between pathways that collectively restrict T cell functions. As suggested by the molecular profile of TIL, protein expression of inhibitory receptor LAG-3 was differentially regulated throughout prolonged late-G1/early-S phase of the cell cycle. Our data may accelerate efficient identification of combination therapies to boost anti-tumor function of TIL specifically against tumor cells.
Keywords: tumor-infiltrating lymphocytes, TIL, T cell dysfunction, T cell hypofunction, tolerance, exhaustion, transcriptome, gene expression profile, genome-wide mRNA expression profile, co-therapies, cancer immunotherapy
Introduction
For some cancers, immunological data from tumor samples are better predictors of patient survival than those currently used to stage disease (1, 2). Compelling evidence associates functional CD8+ tumor-infiltrating lymphocytes (TIL2) with survival of cancer patients. For example, patient survival positively correlates with TIL that produce pro-inflammatory cytokines such as IFNγ, kill target cells, and progress through the cell cycle. These data suggest that the immunological status of TIL is paramount for patient response.
To acquire cytotoxic functions, the antigen receptor on naïve T cells must be engaged in a pro-inflammatory environment (3). Cell-intrinsic pathways are then initiated to drive T cell proliferation and differentiation to cytotoxic T cells that kill cells expressing the associated antigen. CD8+ T cell activation and differentiation are substantially altered by slight changes in the environment and antigen encounter. In the context of cancer, tumor environments evolve to suppress and evade CD8+ T cells (4); therefore, TIL typically have a hypofunctional phenotype incapable of mounting an effective attack against an established solid tumor (5).
Exhaustion, tolerance, anergy, senescence, and ignorance are states of CD8+ T cell hypofunction that have been tied to TIL (3). Of these programs, hypofunctional TIL are most commonly referred to as exhausted or tolerant. T cell exhaustion occurs during prolonged inflammation and/or TCR exposure to an antigen, such as during a chronic viral infection. Peripheral T cell tolerance occurs more quickly when antigen is presented in the absence of immunostimulatory signals to avoid self-reactivity and autoimmunity. Unlike naïve or memory CD8+ T cells that, upon TCR and co-stimulation, enter the cell cycle to undergo clonal expansion to functional effector T cells, a functional response by exhausted and self-tolerant CD8+ T cells is constrained.
The promise of promoting functional TIL to treat cancer patients has been developing for decades to produce a recent explosion of clinical successes (6). Such therapies include blockade of receptors, such as PD-1 (7), that inhibit TIL function. However, a therapy that targets just one inhibitory pathway is often ineffective against established solid tumors. Co-therapies targeting multiple pathways inhibiting TIL function have recently translated from mouse models to the clinic (8). For instance, dual blockade of T cell inhibitory receptors PD-1 and TIM-3 was first shown to augment clearance of the CT26 solid tumor model in mice (9) and is now in clinical trials (Trial registration ID: NCT02608268).
The transplantable colon carcinoma CT26 (10) reflects both the infiltrating immune cells and pathways of immune suppression found in many solid human tumors that are immunogenic (11, 12). As with many antigens being pursued in cancer patients (13, 14), the immunodominant CD8+ T cell response against CT26 is directed against a tumor/self-antigen, GP70423-431, also known as AH1 (15, 16). CT26 TIL exhibit an exhausted phenotype similar to CD8+ T cells from melanoma patients that bear TCRs specific for a tumor/self-antigen (9, 17). The majority of CT26 TIL express multiple inhibitory receptors and are hypofunctional (9). Nevertheless, mobilizing a functional CD8+ T cell response is necessary for successful anti-CT26 immunotherapy (18).
Here we show within just 24 h of transfer into a CT26 tumor-bearing host, AH1-specific CD8+ T cells that were protective in a prophylactic setting (19) were markedly hypofunctional in the tumor compared to those in the periphery. To molecularly define the collective impact of pathways that suppress AH1-specific TIL function, we compared genome-wide mRNA expression of AH1-specific CD8+ T cells from the CT26 tumor and periphery. Gene expression of TIL hypofunction more closely resembled self-tolerance than viral-exhaustion. Differential gene expression identified a core set of genes, or “molecular profile,” that defined TIL hypofunction. Further scrutiny of the molecular profile of TIL delineated overlapping and distinct pathways that restrict function of polyclonal TIL to highlight key mechanisms that are known, hypothesized, and potentially novel drivers of immune suppression and evasion. The T cells from the tumor were hypofunctional and expressed more inhibitory receptor protein when compared to those in the periphery. Known restrictors of TIL function were predicted to converge at cell cycle inhibition. Inhibitory receptor LAG-3 and cell cycle progression validated a simplified approach for pathway analysis of small gene clusters within the molecular profile of TIL. TIL progressed more slowly through late-G1 and early S-phase, associated with expression of inhibitory receptor LAG-3. Our molecular dissection of overlapping and distinct pathways that restrict TIL function may accelerate identification of efficient co-therapy combinations to boost naturally-responding TIL specifically against tumors.
Materials and Methods
Mice
Female BALB/c (BALB/cAnNCr) mice were purchased from the National Cancer Institute and Charles River Laboratories. Mice congenic for thymus cell antigen-1 (Thy1), a kind gift of Dr. Charles D. Surh of The Scripps Research Institute, were bred onto the BALB/cAnCr background for 6 generations. For all experiments, mice were 6- to 12-weeks old unless otherwise indicated; all animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee at National Jewish Health or the University of Colorado – School of Medicine (CU-SOM).
Peptides, vaccine, and tumor challenge
Peptides used were βgal (TPHPARIGL) (20), AH1 (SPSYVYHQF) (15), and a TCR agonist of AH1 (amino acid substitution underlined), A5 (SPSYAYHQF) (21). Soluble synthetic peptides were ≥ 95% pure (Chi Scientific) and stocks were diluted to 10 mg/ml in double distilled H2O. CD8+ T cells bearing TCRs specific for the immunodominant CT26 T cell epitope AH1 (15) were expanded in vivo through vaccination with peptide A5, as described (18). Mice were challenged subcutaneously with 5×104 CT26 tumor cells on the hind flank (21). Tumors were measured with calipers, and mice were sacrificed when indicated or the tumor size reached 1 cm in the longest diameter. Spleens from 5 vaccinated mice were combined and enriched for CD8+ T cells with a Mouse CD8 Negative Selection Kit (Invitrogen), then divided equally among 10 recipient mice (approximately 1×107 cells/mouse) for intravenous injection into Thy1 congenic naïve or tumor-bearing hosts. Spleens and tumors were harvested at the indicated time points and assayed for inhibitory receptor expression and IFN γ production as specified below.
Isolation of tumor-specific CD8+ T cells
Tumors were collected from BALB/c mice, minced using a razor blade, and treated for 45 min at 37°C with 0.1 mg/ml Liberase (Research Grade, Dispase Low) in serum-free RPMI medium according to manufacturer’s instructions (Roche Life Science). Large clumps of tumor were broken-up through an 18 gauge needle, filtered through a 100 μm cell strainer, and washed in Complete Medium [RPMI with L-glutamine, 10% FBS, 100 units/ml each of penicillin and streptomycin, 1 mM sodium pyruvate, 10 mM HEPES, 1× MEM nonessential amino acids, 2 mM additional L-glutamine, and 0.1 mM 2-mercaptoethanol (BME, Sigma-Aldrich), as described (21)]. When indicated, Ficoll-Paque PLUS (GE Healthcare) was used according to manufacturers instructions to enrich for live lymphocytes. Splenocytes were harvested from mice, mechanically dissociated, treated for approximately 2 min with ammonium chloride-potassium lysis buffer (ACK) (22), filtered through a 100 μm cell strainer, and washed in Complete Medium. Before FAC-sorts, spleens were processed into single cell suspensions through Liberase digestion, as performed for tumors. Before FAC-sorts and adoptive transfer experiments, CD8+ T cells were enriched from splenocytes using the Mouse CD8 Negative Selection Kit (Invitrogen) according to manufacturer’s instructions. Cells were used directly for staining, in vitro stimulation assays, or adoptive cell transfer.
Antibodies, staining reagents, and flow cytometry
For flow cytometry analyses, 2×106 live cells were incubated in 30 μl stains containing an antibody to block Fc receptors (FcR, 2.4G2), and fluorochrome-conjugated antibodies for at least 45 min at RT while protected from light. All staining was done in flow cytometry wash buffer [FACs buffer, 1× PBS, 2% FBS, 10 mM HEPES (pH 7.5), and 0.1% (w/v) sodium azide, as described (21)] for at least 45 min at RT. Stain time was increased to 1.5 h when peptide-loaded H-2Ld-tetramer (Ld Tet3) (23) was included.
Fluorochrome-conjugated antibodies purchased from Biolegend, unless otherwise noted, included the following: mouse CD8α (53-6.7), CD11a (M17/4), CD4 (RM4-5), B220 (RA3-6B2), major histcompatibility complex class II (I-A/I-E; M5/114.15.2), CD279/PD-1 (RMP1-30), CD366/TIM-3 (B8.2C12), CD223/LAG-3 (C9B7W), CD244.1/2B4 (C9.1, BD Bioscience), and Thy1.1 (53-2.1; eBioscience). Other staining reagents included viability-discriminating agents 7-aminoactinomycin D (7-AAD, 1 mg/ml, Sigma-Aldrich), and Fixable Viability Dye eFluor® 450 (eBioscience) that were used according to manufacturer’s instructions. CD4/B220/I-A/I-E antibodies and viability discriminating agents are collectively referred to as the “dump.”
Cells were stained for sorts at 35×107 cells/ml then diluted to approximately 5×107 cells/ml Sorting Buffer (1× PBS, 1 mM EDTA, 25 mM HEPES pH 7.0, 1% FBS) and FAC-sorted into Complete Medium. Intracellular cytokine were stained as described (24) following peptide stimulations at 10 pg/ml, 10 ng/ml, and 10 μg/ml, but stains contained an antibody directed against Thy1.1 rather than CD11a. Samples were analyzed on a variety of flow cytometers and data were analyzed using Kaluza (Beckman Coulter) or FlowJo software (Tree Star).
Microarray
Single cell suspensions were stained with Fixable Viability Dye, then with AH1-Ld Tet (23), and fluorochrome conjugated antibodies against CD8α, CD11a, CD4, B220, and I-A/I-E. Tumor-specific live CD8+ T cells were FAC-sorted to a purity of 90 – 100% from tumors on the MoFlo XDP100 (Beckman Coulter) and from spleens on the MoFlo XDP70 (Beckman Coulter) at the CU-SOM Cancer Center Flow Cytometry Shared Resource.
RNA was immediately extracted using the RNeasy Mini Kit (QIAGEN) and treated once with approximately 30 Kunitz units of DNase I (QIAGEN) for 15 min. RNA quality was determined by BioAnalyzer (Agilent) and quantified by Qubit® (Life Technologies). Samples were narrowed to those that had an RNA integrity number (RIN) of 9.6-10 and a minimum of 5 ng total RNA (25) before submission to the CU-SOM Genomics and Microarray Core to be amplified by the Ovation® RNA Amplification System V2 (NuGEN) (25) and hybridized to Mouse Gene1.0 ST Arrays (Affymetrix) according to manufacturer’s instructions.
Cell cycle analysis
Tumors or spleens were processed into single cell suspensions and live lymphocytes were enriched from tumors using Ficoll-Paque PLUS (GE Healthcare) according to manufacturer’s instructions. Cells were plated at 2×106 cells/ml Complete Medium in 24-well tissue culture plates and stimulated by plate-bound CD3 and CD28, each at 2 μg/ml, and spun for 2 min at approximately 340 ×g to synchronize cell cycle progression. After 48 h, cells were pulsed for 1 or 2 h with 5-ethynyl-2’-deoxyuridine (EdU, 10 μM, Sigma-Aldrich), washed in Complete Medium, and stained for live CD8+ T cells and EdU using the Click-iT® Plus EdU Flow Cytometry Assay Kit (Life Technologies) according to manufacturer’s instructions. Briefly, cells were fixed, permeabilized, and EdU was detected with 1/5th the recommended Click-iT reaction to decrease background. To stain total DNA, 7-AAD was diluted 1:400 in permeabilization buffer (Life Technologies) and used in combination with RNaseA (0.2 mg/ml, QIAGEN). Cell cycle phase was determined for live CD8+ T cells by flow cytometry.
Statistical Analyses
High-throughput microarray data were processed by Affymetrix Expression Console software (affymetrix.com), performing robust multichip analysis (RMA) background correction, quantile normalization, and median polish summarization. The gene-expression data described in this paper have been deposited in the NCBI Gene Expression Omnibus GEO accession number GSE79858, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79858. Data sets were compared by one-way analysis of variance ANOVA4 to delineate significant differential gene expression when p ≤ 0.01, false discovery rate (FDR) ≤ 0.01 (Benjamini-Hochberg method (26)), and |log2 (ratio)| ≥ (+2-fold). The molecular profile of the Tumor Antigen Specific Polyclonal Repertoire (TASPR5) TIL consists of differentially expressed genes that have been narrowed by multiple statistical analysis as described above for ANOVA and significance analysis of microarray (SAM6) (27, 28), where q-value ≤ 0.05 (Benjamini-Hochberg method, FDR ≤ 0.01 (26)), and |log2 (ratio)| ≥ (+2-fold). SAM was done in MeV version 4.8 (29). Principal component analysis (PCA7), ANOVA, and corresponding Volcano Plots were done using Partek® software. The PCA was done on scaled data. Copyright, Partek Inc. Partek and all other Partek Inc. product or service names are registered trademarks or trademarks of Partek Inc., St. Louis, MO, USA.
All gene set enrichment analyses (GSEAs8) were run simultaneously using the GSEA software from the Broad Institute (30) and employed the following parameters recommended for expression data sets that contain a sample with fewer than 7 replicates: all data were converted into gene symbols, redundant probe sets were collapsed using probe medians, a Signal2Noise metric was used for ranking genes, the weighted enrichment statistic was employed, and 1000 gene set permutations were used. All comparison populations and statistics used to generate gene sets have been published (17, 31-34). For gene sets corresponding to human data, gene symbols were converted to mouse gene symbols by the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 (35, 36), and gene symbols were manually verified to contain the gene symbol identifier(s) corresponding to our microarray Probe Set ID’s. A compiled list of all gene sets used for GSEA has been included (Supplemental Table 1). Leading edge comparisons were done in Pangloss Venn diagram generator.
Figure of Merit (FOM) (37) was used to estimate the appropriate number of clusters to divide these data for the K means clusters (KMC9) algorithm (38, 39). The smallest cluster number was used for which FOM values did not continue to decrease. To verify the appropriate number of clusters to divide differentially expressed genes for pathway analysis, we analyzed these data divided into 5, 7, and 13 clusters by Ingenuity® Pathway Analysis. Thirteen clusters were ultimately used as it was the smallest number of clusters for which the FOM value leveled out (at 6) and had the lowest trend in overall p-values associated with biofunctions that were predicted to be enriched among clusters by pathway analysis. All pathway analyses were done through QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN, Redwood City, CA, USA). MeV was used for FOM, KMC, and to generate the histogram (29). Expression profiles for Clusters 1, 3, and 5 were made in using custom R scripts. Gene type was determined in PANTHER through Gene Ontology protein class (40); unidentified genes in PANTHER were manually assigned a gene type through a Gene Ontology molecular function backed with stringent evidence in mice by inference from a mutant phenotype (IMP) or a direct assay (IDA) in the National Center for Biotechnology Information (NCBI) database. All gene products that did not meet the criteria to merit a specified gene type were listed as “Miscellaneous.” TRANSFAC provided by the GATHER webtool was utilized to analyze promoter regions of differentially expressed genes for enrichment of transcription factor motifs (41).
Unless otherwise indicated, all other statistical significance was analyzed with using an unpaired two-tailed Student’s t test [Prism version 4.0 (GraphPad)]. A p-value of ≤ 0.01 was considered statistically significant and error bars represent SEM. Statistical significance is denoted by a “*”, where ****p ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01, and *p ≤ 0.05.
Results
TIL rapidly lose function in the CT26 tumor environment
We previously showed that vaccine strategies that are protective against CT26 tumor growth do not work as well in a therapeutic setting (19). These results led us to determine how quickly tumor-specific CD8+ T cells become hypofunctional in an established tumor environment. We investigated the loss of the production of the anti-tumor cytokine, IFNγ (4). Although hypofunction of early exhausted CD8+ T cells often cannot be detected without analysis of multiple functions, reduced capacity to produce IFNγ generally occurs more slowly than loss of target cell lysis, proliferative potential, as well as IL-2 and TNFα production (42). Deficient IFNγ production is also a hallmark of CD8+ T cell tolerance (43). We expected functional analysis of IFNγ production to distinguish between exhausted and tolerant TIL, as loss of IFNγ production should occur overtime during exhaustion and immediately in tolerance (3).
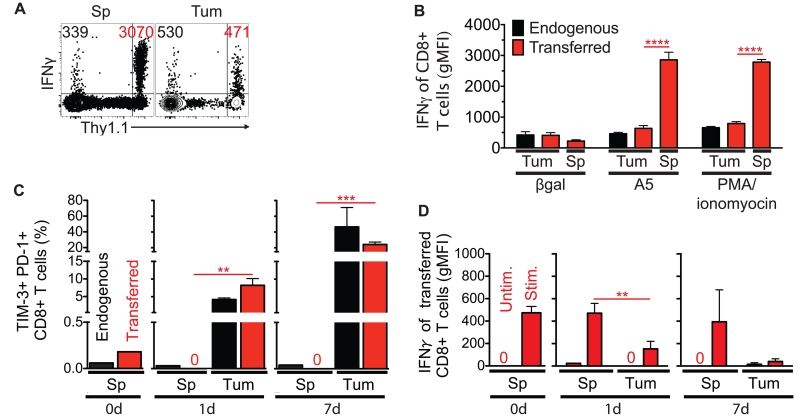
Functional tumor-specific CD8+ T cells were expanded in vivo by a vaccine strategy that is protective against CT26 tumor challenge (18). We transferred these cells into congenic hosts bearing an established CT26 tumor. Within 24 h of transfer, tumor-specific TIL became markedly hypofunctional relative to peripheral counterparts (Figure 1A-B); a phenotype that became more pronounced over 1 wk (Figure 1C-D). Transferred T cells were similarly functional in the spleens of tumor-bearing and non-tumor-bearing mice (unpublished observation). IFNγ protein expression in response to PMA/ionomyocin stimulation (a means to bypass TCR signaling) was also decreased, suggesting that functional defects of TIL were cell-intrinsic. In addition, the immediate loss of function and accumulation of multiple inhibitory receptors suggested that self-tolerance of TIL is established quickly in a solid tumor environment.
FIGURE 1.
Effector CD8+ T cells become hypofunctional within 24 h in a CT26 tumor environment. Transferred live CD8+ T cells, known to protect against tumor challenge, were adoptively transferred into a tumor-bearing host and monitored at the indicated time points from the tumor (Tum) and spleen (Sp). (A) One day after adoptive transfer into a tumor-bearing host, transferred (Thy1.1+) CD8+ T cells from the Tum and Sp were assayed for IFNγ protein in response to A5 peptide (10 μg/ml) stimulation ex vivo. Geometric mean fluorescent intensities (gMFIs) in representative dot plots from IFNγ+ endogenous (upper left quadrant, black) and transferred (upper right quadrant, red) live CD8+ T cells are shown. (B) Expression level of IFNγ in transferred CD8+ T cells from the Tum and Sp was measured in response to A5 peptide (10 μg/ml) and PMA/ionomyocin stimulation ex vivo one day after adoptive transfer into a tumor-bearing host. βgal (10 μg/ml) is an H-2Ld binding irrelevant peptide . (C) Co-expression of inhibitory receptors was monitored over time on transferred CD8+ T cells from the Tum and Sp. “0d” represents immediately before transfer, and a frequency of “0” designates no dual PD-1+/TIM-3+ cells of interest. (D) Transferred CD8+ T cells from the Tum and Sp were monitored over time for IFNγ protein production following ex vivo PMA/ionomyocin stimulation. A gMFI of “0” designates no IFNγ+ among cells of interest. Data represent at least 2 independent experiments, n=2-3 biological replicates per group, and error bars=standard deviation of the mean (SD).
Alternatively, differences among transferred T cells in the spleen and tumor may have been due to altered trafficking of differentially activated polyclonal T cells after vaccination rather than a suppressive tumor environment. However, when the vaccination is given prophylactically, functional CD8+ T cells traffic to eliminate CT26 tumors (18). Therefore, immediate differential expression of proteins, such as IFNγ, in T cells from the spleen and tumor strongly suggested that the tumor and spleen environments differentially affect the functionality of T cells.
Genome-wide mRNA expression of TIL and functional validation
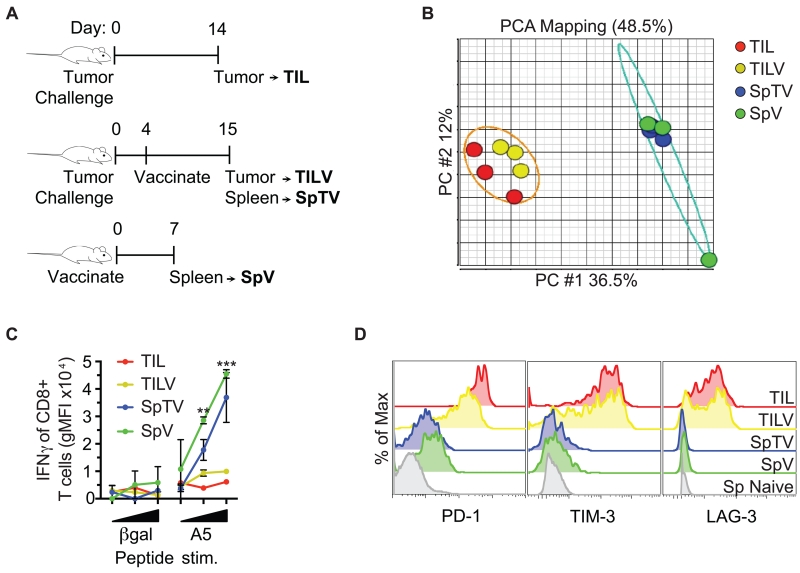
To ultimately define the collective impact of multiple pathways suppressing CT26 TIL function (44), a comparison of genome-wide mRNA expression of tumor-specific CD8+ T cells from the tumor was required (Figure 2A). Tumor bearing mice were vaccinated to expand tumor-specific CD8+ T cells for detection in the periphery. Although this vaccination strategy is protective when given prophylatically (18), under the therapeutic conditions used, vaccination only slightly delayed tumor growth. Therefore, we hypothesized that the TIL from vaccinated tumor-bearing mice (TILV10) have a hypofunctional phenotype similar to naturally-responding TIL in comparison to a peripheral counterpart in vaccinated tumor-bearing mice (SpTV11). Because tumors have systemic effects on the immune system, we also included a cohort of tumor-specific CD8+ T cells from vaccinated non-tumor bearing mice (SpV12) as a positive control for effective anti-tumor function. Rather than transferred T cells, we defined the hypofunction of naturally responding TIL after chronic exposure to a tumor environment. Tumor-specific CD8+ T cells were tetramer-sorted from tumors (TIL and TILV) and spleens (SpTV and SpV) (Supplemental Figure 1) for genome-wide mRNA expression analysis.
FIGURE 2.
Genome-wide mRNA expression of tumor-specific CD8+ T cells segregates into two functionally distinct clusters. (A) Schematic of the origin of tumor antigen-specific T cells is shown. AH1-specific CD8+ T cells were tetramer-sorted from CT26 tumors of mice that had (TILV) or had not (TIL) been vaccinated, and spleens of vaccinated mice that either harbored (SpTV) or did not harbor tumors (SpV). (B) Messenger RNA from the groups described in A was interrogated by microarray. Principal component analysis (PCA) divided genome-wide mRNA expression of the indicated cell populations into two distinct clusters, tumor-specific CD8+ T cells from the spleen or the tumor. Five to ten mice were pooled per group, and each group was repeated 3 times. (C) AH1-specific CD8+ T cells described in A were stimulated ex vivo with increasing concentrations of the indicated peptide and assayed for IFNγ production. (D) AH1-specific CD8+ T cells described in A were analyzed for inhibitory receptor expression. The grey histogram represents naive CD8+ T cells from a spleen as a negative control for inhibitory receptor expression. C and D are representative data of at least 2 independent experiments, n=1-4 biological replicates, and error bars=SD.
To identify genome-wide similarities and differences in the mRNA of samples and to determine the most appropriate grouping of samples, we performed Principal Component Analysis (PCA) (45). PCA is a statistical technique to reduce a large set of variables to a small set of principal components that contain most of the original variation. The two largest principal components, PC #1 and PC #2, accounted for 48.5% of the variance between samples and divided tumor-specific CD8+ T cells into two distinct clusters, those from the spleen and the tumor (Figure 2B). Therefore, genome-wide mRNA expression of TIL and TILV were most similar to each other and diverged from peripheral counterparts SpTV and SpV.
We next determined if tumor-specific CD8+ T cells from the spleen and tumor also represented functionally distinct cohorts and expressed the expected inhibitory receptors. As determined by IFNγ production, TIL from vaccinated mice were comparably hypofunctional to TIL from non-vaccinated mice, while function of tumor-specific CD8+ T cells from the spleens of vaccinated tumor-bearing mice was similar to that of cells from vaccinated non-tumor-bearing mice (Figure 2C). TIL and TILV had higher protein expression of multiple inhibitory receptors than SpTV and SpV (Figure 2D); the expression data were comparable to those obtained in Figure 1 and previously published studies of naturally-responding CD8+ TIL of CT26 tumors that did not consider antigen specificity (9). These results suggested that our genome-wide mRNA expression data reflect differential gene expression responsible for hypofunction of TIL in the tumor environment. For the remaining analyses, the “Tumor Antigen Specific Polyclonal Repertoire” of CD8+ T cells from tumors (TIL and TILV) and spleens (SpTV and SpV) will collectively be referred to as “TASPR” T cells from the CT26 tumor.
Gene expression of heterogeneous TASPR TIL resembles self-tolerance
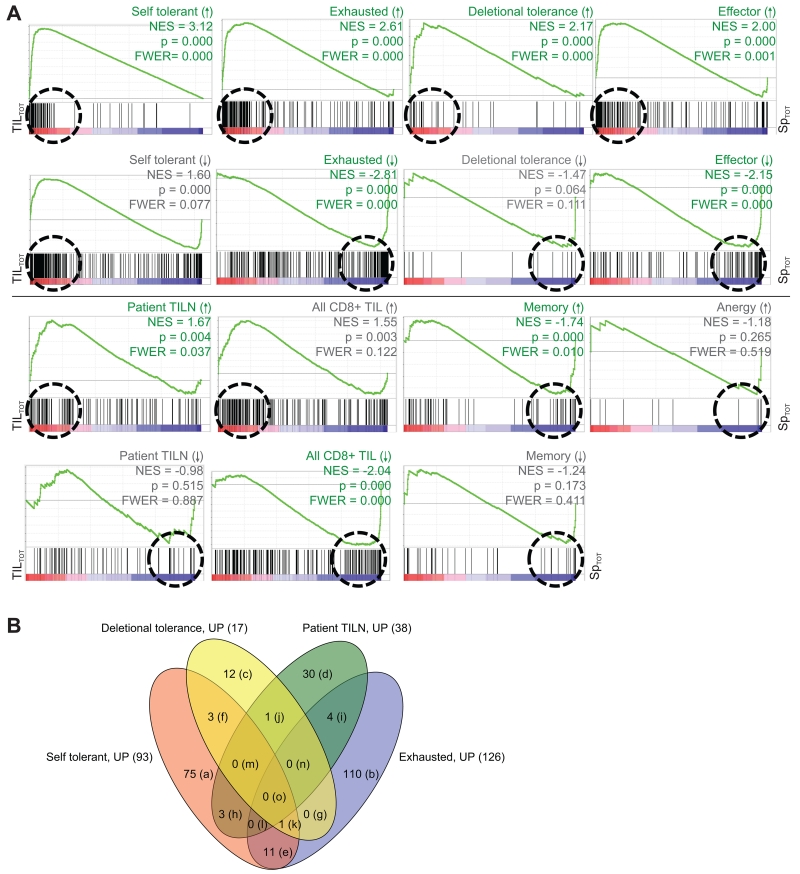
Gene Set Enrichment Analysis (GSEA) is a statistical method commonly used to compare gene expression results between data sets that are derived from independent experiments (17, 30, 46). We utilized GSEA to determine if TASPR TIL had similar gene expression to other published gene expression profiles of hypofunctional T cells that have been associated with TIL. These included exhaustion, tolerance, anergy, and other TIL profiles (Figure 3A and Supplemental Table 1) (17, 31-34). TASPR TIL were also compared by GSEA to other functional T cell subsets that can infiltrate tumors, such as memory and effector T cells (31). Statistical significance, as reflected by the Normalized Enrichment Scores (NES13), p values, and familywise error rates (FWER), was analyzed for enrichment of genes associated with T cell subsets among TASPR T cells.
FIGURE 3.
Genome-wide mRNA expression of tumor-specific TIL from the CT26 tumor is similar to transcription profiles of other hypofunctional CD8+ T cells; similarities are driven by distinct gene expression. Genome-wide mRNA expression of TASPR T cells, acquired as described in Figure 2A, was simultaneously compared to other published CD8+ T cell transcriptional profiles for similarities by gene set enrichment analysis (GSEA) (30). (A) Plots show enrichment of genes associated with the indicated T cell profile, or “gene set.” Vertical black lines show where gene sets match genes expressed by TASPR T cells from the tumor (red, TILTOT15 = TIL and TILV) and spleen (blue, SpTOT16 = SpTV and SpV). The Normalized Enrichment Scores (NES) reflect the degree to which a gene set is overrepresented at the extremes of the entire ranked list of gene expression differences between TASPR T cells from the tumor and spleen; an NES value farther from 0 indicates greater enrichment of the indicated profile genes among TASPR T cells from the tumor (positive NES) or spleen (negative NES). The green line is a visual representation of gene set enrichment along the ranked list. The Leading Edge, roughly shown by a dashed circle, includes the core genes from a gene set that contribute most to the deviation of an NES from 0. Profile gene sets were determined to significantly cluster among genes highly expressed by TASPR T cells from the tumor or spleen when p-value ≤ 0.001 (30) and familywise error rate ≤ 0.05 (FWER, which suggests the probability of type 1 errors), and are delineated by green rather than grey labels. (B) The majority of Leading Edge genes differ between the hypofunctional CD8+ T cell profiles. These core genes drive the NES away from 0 in the Leading Edge (circled in A) and are compared in the Venn diagram and Supplemental Table 2.
TASPR TIL were most similar to a self-tolerant profile (33) followed by the virally exhausted profile (31). Exhaustion occurs over time with chronic TCR antigen or inflammatory cytokine exposure (47, 48), whereas self-tolerance generally occurs quickly in response to initial TCR exposure to self-antigen in the absence of immunostimulatory signals (3). As expected, similarities were also identified between TASPR TIL, deletional tolerance (32), and effector T cells (9, 31). We compared TASPR TIL with tumor-specific CD8+ T cells isolated from lymph node metastases (TILN14) of previously vaccinated melanoma patients (17) and total naturally-responding CD8+ TIL from a mouse model of melanoma for which antigen specificity was not restricted (34). Curiously, while genes highly expressed by vaccinated TILN (17) were enriched among TASPR TIL, genes highly expressed by the total naturally-responding CD8+ TIL (34) were not. A likely explanation for such a large differential impact is the lack of TCR specificity for a tumor antigen in the total CD8+ TIL (34). For example, in the CT26 tumor model, protein expression of inhibitory receptors by total CD8+ TIL is much lower and more variable than by CD8+ TIL that have been narrowed by tumor-specificity (unpublished observation). Genes highly expressed by memory CD8+ T cells were in direct opposition of TASPR TIL and similar to functional TASPR T cells in the spleen. Unique enrichment of individual CD8+ T cell programs among TASPR TIL (Figure 3B and Supplemental Table 2) and rapid loss of T cell function in a tumor that became more pronounced over time (Figure 1) strongly suggested that pathways do not just overlap between exhausted and self-tolerant CD8+ T cells, but that TASPR TIL are a heterogeneous population that is largely self-tolerant and exhausted.
The gene set highly expressed by anergic T cells was approximately 15 genes (32) and was too small (30) to obtain an accurate estimate of enrichment among TASPR T cells (see statistics in anergy enrichment plot, Figure 3A). The gene consistently down-regulated by anergic T cells could not be systematically compared to TASPR T cells by GSEA (30, 32) (Supplemental Table 1). Notably, a gene whose decreased expression has been heavily studied in the context of T cell anergy (32) was also down-regulated by TASPR TIL. Although not available for systematic comparisons by GSEA, we also manually compared the TASPR T cells to signature genes expressed by ignorant and senescent T cells. Genes such as KLRG1 and CD57, that are associated with senescent CD8+ T cells in a non-reversible state of cell cycle arrest (49), were not up-regulated by TASPR TIL. Finally, TASPR TIL were antigen-experienced and did not down-regulate the TCR-CD3 complex, and therefore did not correspond to T cells “ignorant” of the growing tumor (3, 50).
Molecular profile of tumor-specific TIL
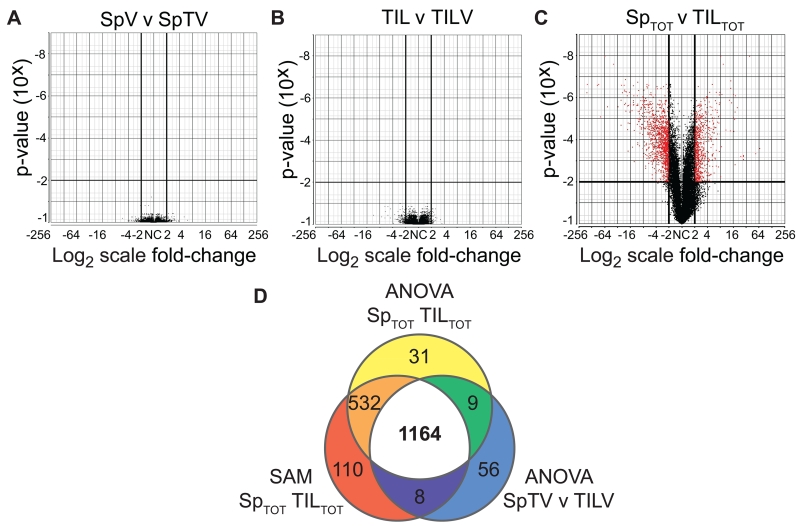
To delineate significant differential gene expression between TASPR T cells, genome-wide mRNA expression was compared by one-way analysis of variance (ANOVA) and narrowed by a twofold-change (Figure 4A-C). There were no significant differences among TASPR TIL, regardless of vaccination, or between TASPR T cells from the periphery, regardless of tumor growth (Figure 4A-B). However, when TASPR T cells from the tumor were compared to those from the periphery, we identified a total of 1973 genes that were differentially expressed (Figure 4C). We further narrowed the list to 1164 differentially expressed genes by significance analysis of microarray (SAM) (Figure 4D and Supplemental Table 3) (27, 28). Unlike ANOVA, which assumes an underlying normal distribution, SAM is a non-parametric technique for determining differential expression (27, 28). Both statistical analyses are commonly used to assess genome-wide mRNA expression data: we reasoned dual criteria would increase confidence that genes are not falsely included (51) in the molecular profile of TASPR TIL hypofunction.
FIGURE 4.
Molecular profile of tumor-specific TIL that are naturally-responding and polyclonal. Genome-wide mRNA expression of TASPR T cells acquired in Figure 2A (TILTOT=TIL and TILV and SpTOT=SpTV and SpV) were compared for differential gene expression. (A-C) Volcano plots comparing the indicated groups illustrate significant 2-fold differential gene-expression by one-way ANOVA in red. (D) Differentially-expressed genes were narrowed by ANOVA, as in C, and by SAM. Genes common to all three groups were designated the “molecular profile” of TASPR TIL.
Molecular profile of TIL highlights cell cycle inhibition
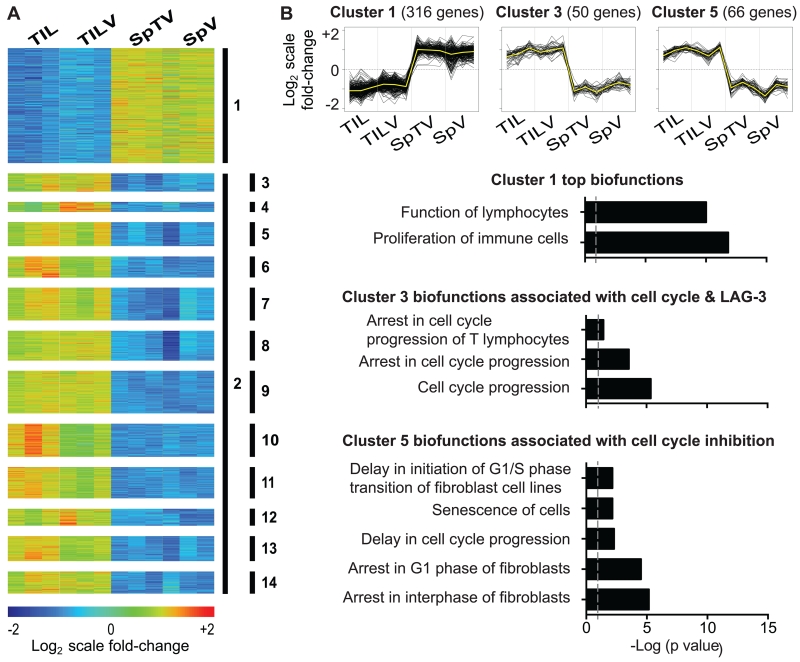
To dissect the downstream impact of collective pathways suppressing CT26 TIL function, the 1164 genes comprising the molecular profile of TASPR TIL were sequestered into smaller gene clusters enriched for biological pathways using K-means clustering (KMC, Figure 5A and Supplemental Table 3) (38, 39). Ingenuity® Pathway Analysis of individual clusters indicated that genes up-regulated by TASPR T cells in the periphery were associated with lymphocyte proliferation (Figure 5B, Cluster 1), while those up-regulated in the tumor were broadly associated with regulation of cell cycle progression (Figure 5A, Cluster 2, p=2.79×10−28). KMC further divided Cluster 2 into 13 smaller clusters by similar patterns of gene expression that were enriched for more specialized biological pathways (Figure 5A, Clusters 3-14). These clusters within the molecular profile of TASPR TIL allow systematic focus of individual pathways associated with TIL immunosuppression. For instance, Cluster 5 was enriched for many gene products associated with arrest in cell cycle (Figure 5B), and the top biofunction related to the cell cycle in Cluster 5 was G1/S-phase transition (p=2.12×10−6). Additionally, genes up-regulated in the periphery were associated with TCR signaling and CD28 co-stimulation (Cluster 1, p=3.16×10−7 and 1.37×10−7, respectively), while genes induced in TASPR TIL included inhibitory receptors, such as PD-1 (PDCD1, Cluster 12, Supplemental Table 3), and reflected an environment of strong TGF-β signaling (Cluster 2, p=4.9×10−36). Suboptimal TCR and co-stimulation, PD-1, and TGF-β, are all known to restrict TIL function and to inhibit cell cycle progression (5, 52-54).
FIGURE 5.
Molecular profile of tumor-specific TIL from the CT26 tumor highlights transcriptional regulation of cell cycle inhibition. The molecular profile of TASPR T cells acquired in Figure 2A was analyzed in clusters for enrichment of biological pathways. (A) K-means clustering (KMC) divided differentially-expressed genes into 13 clusters for focused pathway analyses. Heat map shows log2-transformed expression intensities mean-centered at the probe level. (B) Pathway analysis of gene products from Clusters 1, 3, and 5 revealed corresponding biofunctions enriched in TASPR T cells from the spleen and tumor. Grey dashed lines represent p value = 0.05.
Interestingly, the majority of inhibitory receptors were distributed between different clusters. Cluster 9 contained two surface receptors, TIM-3 (HAVCR2) and 2B4 (CD244), that can broadly inhibit function of CD8+ T cells (55, 56), while another such inhibitory receptor, TIGIT (57), was contained separately in Cluster 13. PD-1 (PDCD1) (7) segregated into Cluster 12 that included other surface receptors not usually expressed on CD8+ T cells such as, CD80, a molecule that is expressed on CD8+ T cells from HIV-infected patients (58). Inhibitory receptor CTLA-4 and the controversial surface receptor IL2RA (CD25), that are associated with negative and positive effects on anti-tumor T cell function (59-61), respectively, were grouped together in Cluster 4 that was broadly associated with cell-to-cell signaling and interactions (p=2.1×10−7).
Clusters that contained inhibitory receptors were also enriched for other gene products that have been independently tied to some of the same biological pathways (Figure 5A and B). For instance, inhibitory receptor LAG-3 (62) was among the gene products grouped into Cluster 3 that was associated with arrest in cell cycle, including CDKN2C, CDKN3, CHN2, LYN, and PLK1. Besides LAG-3, many gene products associated with cell cycle arrest in K-means Clusters 3 and 5 have recently attracted attention for functions that may ultimately regulate DNA replication and licensing cell cycle progression beyond the G1 and S phases (63-66). Since much of the downstream signaling that mediates the effect of these inhibitory receptors on TIL hypofunction is unknown, these clusters may provide a valuable and fluid tool to dissect the extent of distinction and overlap between such pathways.
LAG-3 expression associated with cell cycle progression
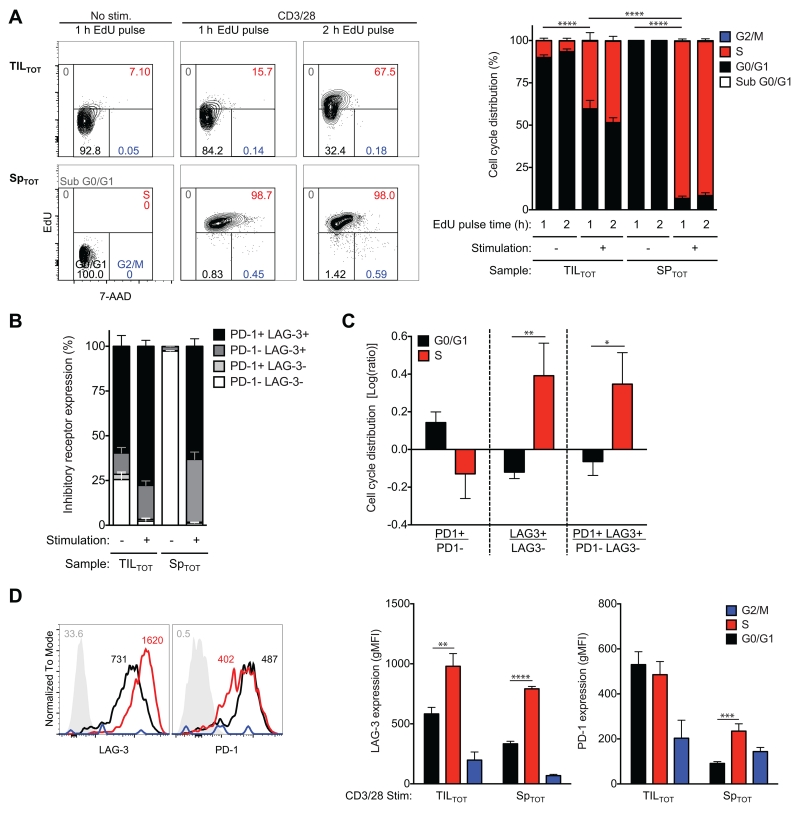
Hypofunctional TIL incapable of tumor clearance have decreased capacity to progress through the cell cycle (9, 48, 67). This foundation was largely laid by studies that distinguish cycling cells by staining for Ki-67 (9, 67), which groups non-cycling cells in the G0/G1 phase from cycling cells in the late-G1, S, and G2/M phases (68), and total DNA staining to set apart the G2/M phase (9, 69). Because many of the same gene products are associated with cell cycle inhibition and progression depending on posttranslational modifications (66, 70), we determined cell cycle progression of CD8+ T cells from the CT26 tumor relative to those from the periphery (Figure 6A). As expected (3, 71), TIL did not progress through the cell cycle as readily as peripheral counterparts in response to TCR- and co-stimulation. Most stimulated and unstimulated TIL were in the late-G1/early-S phase.
FIGURE 6.
Slowed cell cycle progression of CD8+ T cells in the CT26 tumor corresponds to a novel pattern of inhibitory receptor expression predicted by the molecular profile of TIL. Total CD8+ T cells from CT26 tumors (TILTOT) and spleens (SpTOT) from timelines shown in Figure 2A were untreated or treated with antibodies to CD3 and CD28 for 48 h before a 1 h (or indicated time) pulse with the thymidine analogue EdU. Flow cytometric analyses identified cell cycle phase and inhibitory receptor expression of live CD8+ T cells. (A) Cell cycle phase was resolved by staining for EdU and by the DNA dye 7-AAD. Frequencies are shown in sub G0/G1 (grey), G0/G1 (black), S (red), and G2/M (blue) phase. (B) Inhibitory receptor expression (PD-1 and LAG-3) was determined in the presence and absence of stimulation. (C) Inhibitory receptor expression on TILTOT during G0/G1 and S phase was determined. (D) Representative histograms (left) of naive CD8+ T cells from spleen (grey, negative control) and stimulated TILTOT cells [G0/G1 (black), S (red), and G2/M (blue) phase] are shown. Numbers indicate the gMFI. At this time point (48 h), frequency of synchronized T cells in G2/M phase (blue) was too small to confidently determine inhibitory receptor expression. All data (right) represent three independent experiments, n=3-10 biological replicates per group, error bars=SEM.
Dual PD-1 and LAG-3 positive CD8+ T cells were the most abundant among CT26 TIL (Figure 6B). Although previous studies have tied PD-1 more strongly to the cell cycle, specifically arrest in the G0/G1 phase (53), we were interested in analyzing both PD-1 and LAG-3 expression in TIL since TIL were blocked in late-G1/early S-phase and LAG-3 surprisingly clustered with many cell cycle-regulated genes. Stimulated TIL in G0/G1 phase were enriched among those that express PD-1; whereas stimulated TIL in S phase were enriched among those that express LAG-3 and those that express both PD-1 and LAG-3 (Figure 6C). Stimulated TIL also expressed higher levels of PD-1 and LAG-3 in the G0/G1 and S phase, respectively (Figure 6D). Although expected to peak in the S phase (72), LAG-3 protein expression has not previously been investigated on wild-type CD8+ T cells throughout the cell cycle. As suggested by cluster analysis of the molecular profile of TASPR TIL, these data functionally tie control of LAG-3 expression with the cell cycle and corroborate that progression of CT26 TIL through the G1/S-phase boundary is inhibited upon TCR- and co-stimulation.
Discussion
The molecular profile of TASPR TIL from CT26 tumors identifies many known, hypothesized, and potentially novel pathways that underlie TIL hypofunction. As expected, CT26 TIL express high levels of multiple inhibitory receptors and are hypofunctional; however, immediate accumulation of inhibitory receptors and lack of function in the tumor environment revealed a robust similarity between the molecular profiles of TASPR TIL and self-tolerance. Differential gene expression between additional hypofunctional and functional CD8+ T cell subsets, as well as between pathways that restrict T cell function, provide a map to navigate the intricacies of TIL hypofunction.
The molecular profile of TASPR TIL may provide a novel resource for studies to characterize and boost the response of naturally responding TIL against a tumor. The TASPR profile differs from other relevant strategies as follows. Tumor-specific CD8+ T cells isolated from lymph node metastases of previously vaccinated melanoma patients, or “TILN”, likely express T cell genes reflective of an anatomical location of the metastases in a lymph node rather than a primary tumor tissue (73). Another molecular profile corresponding to total CD8+ TIL is not restricted to tumor reactivity (34); both profiles were not as similar to TASPR TIL as exhausted and self-tolerant T cells. Genome-wide mRNA expression of TASPR TIL reflect a molecular profile of T cells that bear TCRs of relatively low to intermediate affinity for tumor antigen (24). Conversely, a molecular profile of tumor-specific TIL that have been genetically modified to all express the same tumor-specific TCR may not model inhibitory pathways, such as suboptimal TCR stimulation, that are known to restrict function of a naturally-responding polyclonal repertoire of TIL.
Although the profile of patient TILN expressed TCRs specific for a tumor/self antigen, GSEA showed that TILN hypofunction most resembles murine viral exhaustion (17, 30, 46). The more recent genome-wide mRNA expression profile of total CD8+ TIL from a mouse model of melanoma also found TIL hypofunction similar to viral exhaustion (34). Neither molecular profile was systematically compared to self-tolerance (17). In addition, others have previously observed that total CD8+ TIL that naturally respond to CT26 tumors, regardless of TCR specificity, are comprised largely of exhausted T cells and a smaller fraction of effector T cells (9). Therefore, we expected the profile of TASPR TIL from CT26 tumors to reflect a hypofunctional T cell program most similar to exhausted CD8+ T cells during chronic viral infection (31).
TASPR TIL have gene expression and functional capacities most similar to a recently published molecular profile of self-tolerance (33) followed by exhaustion (31). Functional tumor-specific CD8+ T cells do not become gradually hypofunctional over weeks, but become tolerized within one day of trafficking into this solid tumor model. Although both self-tolerance and viral-exhaustion can be at least temporarily overridden for a functional T cell response (3), peripheral self-tolerance is a T cell program acquired quickly in response to initial TCR exposure to self-antigen in the absence of immunostimulatory signals (3) whereas viral-exhaustion is a T cell program acquired gradually during chronic non-self TCR or immunostimulatory signals (47, 48). Distinctions between T cell programs of self-tolerance and exhaustion are therefore not restricted semantics, but have wide-spread implications for development of cancer immunotherapies that mobilize patient TIL against a mutated or non-mutated “self” tumor antigen (13-16).
We have since reanalyzed the patient TILN data and found significant overlap between gene expression of TILN (5) and self-tolerant murine T cells (33) that was also associated with control of cell cycle progression (74). The line between self-tolerance and exhaustion gene sets among TILN and TASPR TIL is blurred and pathways may overlap. However, there is little overlap between the exhausted and self-tolerant gene sets that define hypofunction of TILN (4 of 114 genes) and TASPR TIL from CT26 tumors (12 of 195 genes) as similar to viral-exhaustion and self-tolerance. These data suggest that TILN (5) and CT26 TIL are heterogeneously exhausted and self-tolerant, or that TILN (5) and CT26 TIL hypofunction is a T cell program that is distinct yet overlapping with other T cell subsets. Both scenarios are in line with current literature; CD8+ T cell hypofunction varies by patient, assaulting malignancy, and over time (5, 48). A shift in focus towards overlapping mechanisms known to underlie multiple hypofunctional T cell subsets may enhance the functional response in the majority of heterogeneous TILN and TIL (44).
The tumor environment varies in response to different antigens, in different tumor models, and across species (75, 76). Nevertheless, CT26 TIL have a hypofunctional phenotype (9) and molecular profile similar to exhausted patient TILN (9, 17), and exhausted TIL from an inducible model of melanoma (34). CT26 is therefore an immunogenic tumor model with widespread implications as to whether or not TIL tolerance can be overridden against tumor/self antigens or if efforts should be focused on restoring function of exhausted TIL against mutated/foreign tumor antigens (11, 76, 77). We predict as molecular profiles of TIL specific for mutated tumor antigens become available, comparison to the CT26 TIL that are “self”-specific will identify a highly conserved gene expression signature among TIL.
Transcriptional regulators interpret multiple extracellular signals to determine T cell function and differentiation; such intracellular targets represent promising avenues to simultaneously disrupt multiple inhibitory pathways that restrict anti-tumor function of TIL (44). FOXP1, a transcription factor that has recently been reported as highly expressed in TIL and a key regulator of T cell hypofunction in many cancers (78), is down-regulated in TASPR TIL from CT26 tumors. FOXP1 expression in TASPR TIL is similar to the genome-wide mRNA expression profiles of virally exhausted T cells (79), hypofunctional TILN (17), and naturally responding total CD8+ TIL from a mouse melanoma model (34). In the model melanoma system, MAF was the alternatively pursued transcriptional regulator of TIL hypofunction (34), but MAF is not differentially expressed between TASPR T cells from the tumor and periphery. Among gene products corresponding to transcriptional regulators that are up-regulated between TASPR T cells from the tumor versus the periphery are HIF1A, E2F7, IRF8, IKZF2, and STAT3; conversely, among those down-regulated in TASPR TIL are TCF7, TOB1, FOS, and RNX1 (Supplemental Table 3). Differential expression of genes involved in regulating transcription in TASPR TIL suggests an altered pattern of differentiation among T cells from the tumor and spleen.
E2F1, a transcription factor closely associated with late-G1/early-S phase was predicted to be a key mediator between extracellular stimuli and the decreased proliferative capacity of TIL, although increased expression did not meet the strict twofold-change criterion used for true difference among TASPR T cells (q-value=0.00, 1.98 fold-change). Nevertheless, it is not uncommon for transcriptional regulators to be controlled post-translationally, and for subtle changes in expression of transcriptional regulators to have global effects (44, 70). The top five transcription factor motifs enriched in promoter regions (41) of genes up-regulated by TASPR TIL were all variants for E2F1 (p<1×10−10, FDR<1×10−9, Fisher’s exact test). Additionally, E2F1 was among the top transcriptional regulators predicted by Ingenuity® Pathway Analysis to be a key upstream regulator of differential gene expression in TASPR TIL (p=2.6×10−21, Cluster 2). E2F activity is extremely complex and regulates some of the most basic cell functions, such as cell division and apoptosis (80, 81). During late-G1/early-S phase, E2F1 activity determines if cells progress through the cell cycle or arrest and apoptose (66). E2F1 is also a potent regulator of central and peripheral tolerance (80, 82-85). Implications of these data are that E2F1 regulates self-tolerance (44) and is likely to compromise function of CT26 TIL in late-G1/early-S phase.
Additionally, TIL hypofunction is downstream of many known inhibitory pathways targeted individually in patients to restore anti-tumor function of T cells (86), which we analyzed by KMC, an unsupervised sequestering of differentially expressed genes into smaller clusters by similar patterns of gene expression (38). These clusters allow focus on gene products associated with specific biological pathways that have not yet been investigated together or in TIL. The TASPR profile predicts that heterogeneous CT26 TIL receive many inhibitory signals through overlapping mechanisms to restrict cell cycle progression. Cell cycle progression and LAG-3 protein expression confirm pathway analysis of the TASPR TIL profile. We identified novel control of inhibitory receptor LAG-3 expression on cycling CD8+ T cells, which peaks in S phase. Although LAG-3 is highly expressed on NK T cells in S phase (72), variations in LAG-3 expression on cycling wild-type CD8+ T cells has not been investigated prior to this study.
While PD-1 and CTLA-4 specifically restrict cell cycle progression of CD8+ T cells beyond G0/G1 phase (87-89), LAG-3 has been more broadly demonstrated to restrict T cell expansion in many inflammatory contexts in vivo (90). Although rare, CT26 TIL singularly positive for PD-1 were slightly enriched in G0/G1 phase; cycling TIL did not express significantly higher levels of PD-1 in any phase. Although these data do not confirm in vitro experiments regarding PD-1 regulation of cycling CD8+ T cells (53), IPA of the cluster in which PD-1 was grouped predicts less association of PD-1 and similarly expressed gene products with the cell cycle (p=5.10×10−7) than other biofunctions in TASPR TIL, such as cell death and survival (p=6.62×10−8) and immune cell trafficking (p=6.45×10−8) (49). As with co-therapies that target both PD-1 and CTLA-4 (8), deletion of both PD-1 and LAG-3 in tumor-specific CD8+ T cells increases control of tumor burden more than singularly targeting one of these inhibitory receptors (62, 91), and represent attractive therapeutic targets to restore TIL function (92). Notably, deletion of both PD-1 and LAG-3 accounts for less autoimmunity relative to deletion of CTLA-4 (91, 93).
LAG-3 gene expression was among the few to overlap between TASPR TIL hypofunction, self-tolerance, and viral-exhaustion (Supplemental Table 2). Although the entire gene set up-regulated by self-tolerant CD8+ T cells (33) was associated with the cell cycle (p=2.59×10−18, Supplemental Table 1), overlapping gene expression of TASPR TIL and self-tolerant CD8+ T cells (33) was further enriched for cell cycle gene products (p=8.60×10−20, Supplemental Table 2). Genes associated with the cell cycle were similarly noted among vaccinated TILN and virally exhausted T cells when compared to their functional counterparts (17, 31). These data are striking in that alterations in cell cycle progression/proliferation modify TIL susceptibility to apoptosis and memory T cell development (69, 94, 95). Genes highly expressed by memory CD8+ T cells (31) were collectively down-regulated by TASPR TIL. Our analyses dissect the extent of overlap and distinction between the pathways that cooperatively restrict T cell functions and may accelerate identification of effective co-therapy combinations and intracellular targets to efficiently boost anti-tumor function of TIL.
As evident by the lack of published studies, isolating a sufficient number of tumor-specific TIL has been historically limiting for high-throughput analyses. Only differences between the most abundant transcripts are reliably detectable after the necessary amplification methods to detect genome-wide mRNA expression (96). Subtleties that underlie genome-wide mRNA expression profiles of small numbers of patient TIL, such as differential expression of transcription factors, are difficult to biologically replicate due to heterogeneity (17, 96). We avoided these problems by tetramer-sorting many tumor-specific CD8+ T cells (≥40,000 cells, 5 ng RNA) from pooled samples of genetically identical mice. Future mRNA expression profiles from relatively small numbers of patient TIL may be validated by comparison to the TASPR profile, as technologies are rapidly advancing (96). We predict that genes with high expression differences between patient TIL and their functional counterparts will have similar expression to the TASPR profile, but that subtle yet significant differences among TASPR T cells will elucidate of the intricacies of collective pathways that restrict anti-tumor function of patient TIL.
Supplementary Material
Acknowledgements
We would like to thank the exceptional personnel of the CU-SOM Cancer Center Flow Cytometry Shared Resource, especially Lester Acosta and Karen Helm. Additionally, thanks to Todd Woessner, Curtis J. Henry, Cydney Rios, Brian P. O’Connor, James DeGregori, Kathryn Tuttle, Max A. Seibold, John W. Kappler, Keith P. Smith, Daniel Munson, Michelle Hoffmann, and Linda F. van Dyk for scientific advice, help with experimental design, and manuscript corrections.
Footnotes
KAW was supported by the American Cancer Society Grant RSG-08-184-01-L1B, the National Institute of Health Grants 5T32AI007405 and 5R25GM083333, and a generous donation from Richard and Donna Hammel.
TIL Tumor infiltrating lymphocytes
H-2Ld-tetramer
Analysis of variance
Tumor antigen specific polyclonal repertoire
Significance analysis of microarray
Principal component analysis
Gene set enrichment analysis
K-means clustering
AH1-specific TIL from tumors of vaccinated CT26 tumor-bearing mice
AH1-specific CD8+ T cells from spleens CT26 tumor-bearing mice
AH1-specific CD8+ T cells from spleens non-tumor-bearing mice
Normalized Enrichment Scores
Tumor-specific CD8+ T cells isolated from lymph node metastases
AH1-specific TILV and TIL from Figure 2A
AH1-specific SpTV and SpV from Figure 2A
Disclosures
The authors have no financial conflict of interest.
References
- 1.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 3.Schietinger A, Greenberg PD. Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol. 2014;35:51–60. doi: 10.1016/j.it.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 5.Baitsch L, Fuertes-Marraco SA, Legat A, Meyer C, Speiser DE. The three main stumbling blocks for anticancer T cells. Trends Immunol. 2012;33:364–372. doi: 10.1016/j.it.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125:3335–3337. doi: 10.1172/JCI83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano F, Kaneko K, Tamura H, Dong H, Wang S, Ichikawa M, Rietz C, Flies DB, Lau JS, Zhu G, Tamada K, Chen L. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089–1096. [PubMed] [Google Scholar]
- 8.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett TH, Griswold DP, Jr., Roberts BJ, Peckham JC, Schabel FM., Jr. Evaluation of single agents and combinations of chemotherapeutic agents in mouse colon carcinomas. Cancer. 1977;40:2660–2680. doi: 10.1002/1097-0142(197711)40:5+<2660::aid-cncr2820400940>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, Ohlfest JR, Hu P, Epstein AL. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J Immunother. 2013;36:477–489. doi: 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castle JC, Loewer M, Boegel S, de Graaf J, Bender C, Tadmor AD, Boisguerin V, Bukur T, Sorn P, Paret C, Diken M, Kreiter S, Tureci O, Sahin U. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics. 2014;15:190. doi: 10.1186/1471-2164-15-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, Matrisian LM. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melero I, Gaudernack G, Gerritsen W, Huber C, Parmiani G, Scholl S, Thatcher N, Wagstaff J, Zielinski C, Faulkner I, Mellstedt H. Therapeutic vaccines for cancer: an overview of clinical trials. Nat Rev Clin Oncol. 2014;11:509–524. doi: 10.1038/nrclinonc.2014.111. [DOI] [PubMed] [Google Scholar]
- 15.Huang AY, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, Engelhard VH, Pasternack G, Cotter R, Hunt D, Pardoll DM, Jaffee EM. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci U S A. 1996;93:9730–9735. doi: 10.1073/pnas.93.18.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McWilliams JA, Sullivan RT, Jordan KR, McMahan RH, Kemmler CB, McDuffie M, Slansky JE. Age-dependent tolerance to an endogenous tumor-associated antigen. Vaccine. 2008;26:1863–1873. doi: 10.1016/j.vaccine.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baitsch L, Baumgaertner P, Devevre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, Rufer N, Speiser DE. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan KR, McMahan RH, Kemmler CB, Kappler JW, Slansky JE. Peptide vaccines prevent tumor growth by activating T cells that respond to native tumor antigens. Proc Natl Acad Sci U S A. 2010;107:4652–4657. doi: 10.1073/pnas.0914879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan KR, McMahan RH, Oh JZ, Pipeling MR, Pardoll DM, Kedl RM, Kappler JW, Slansky JE. Baculovirus-infected insect cells expressing peptide-MHC complexes elicit protective antitumor immunity. J Immunol. 2008;180:188–197. doi: 10.4049/jimmunol.180.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overwijk WW, Surman DR, Tsung K, Restifo NP. Identification of a Kb-restricted CTL epitope of beta-galactosidase: potential use in development of immunization protocols for “self” antigens. Methods. 1997;12:117–123. doi: 10.1006/meth.1997.0461. [DOI] [PubMed] [Google Scholar]
- 21.Slansky JE, Rattis FM, Boyd LF, Fahmy T, Jaffee EM, Schneck JP, Margulies DH, Pardoll DM. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529–538. doi: 10.1016/s1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 22.Kruisbeek AM. Isolation and fractionation of mononuclear cell populations. In: Coligan AMKJE, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. John Wiley & Sons, Inc.; United States: 1993. pp. 3.1.1–3.1.5. [Google Scholar]
- 23.McMahan RH, McWilliams JA, Jordan KR, Dow SW, Wilson DB, Slansky JE. Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J Clin Invest. 2006;116:2543–2551. doi: 10.1172/JCI26936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buhrman JD, Jordan KR, Munson DJ, Moore BL, Kappler JW, Slansky JE. Improving antigenic peptide vaccines for cancer immunotherapy using a dominant tumor-specific T cell receptor. J Biol Chem. 2013;288:33213–33225. doi: 10.1074/jbc.M113.509554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clement-Ziza M, Gentien D, Lyonnet S, Thiery JP, Besmond C, Decraene C. Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling. BMC Genomics. 2009;10:246. doi: 10.1186/1471-2164-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Y. H. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995:289–300. Series B (Methodological) [Google Scholar]
- 27.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B. Differential gene expression detection using penalized linear regression models: the improved SAM statistics. Bioinformatics. 2005;21:1565–1571. doi: 10.1093/bioinformatics/bti217. [DOI] [PubMed] [Google Scholar]
- 29.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Parish IA, Rao S, Smyth GK, Juelich T, Denyer GS, Davey GM, Strasser A, Heath WR. The molecular signature of CD8+ T cells undergoing deletional tolerance. Blood. 2009;113:4575–4585. doi: 10.1182/blood-2008-10-185223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schietinger A, Delrow JJ, Basom RS, Blattman JN, Greenberg PD. Rescued tolerant CD8 T cells are preprogrammed to reestablish the tolerant state. Science. 2012;335:723–727. doi: 10.1126/science.1214277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, Baitsch L, Vanhille L, Sieweke MH, Speiser DE, Auphan-Anezin N, Schmitt-Verhulst AM, Verdeil G. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 2015;34:2042–2058. doi: 10.15252/embj.201490786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- 38.Hartigan JA, W. MA. Algorithm AS 136: A K-Means Clustering Algorithm. Journal of the Royal Statistical Society. 1979;28:100–108. Series C. [Google Scholar]
- 39.Selvaraj S, Natarajan J. Microarray data analysis and mining tools. Bioinformation. 2011;6:95–99. doi: 10.6026/97320630006095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 42.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez J, Aung S, Redmond WL, Sherman LA. Phenotypic and functional analysis of CD8(+) T cells undergoing peripheral deletion in response to cross-presentation of self-antigen. J Exp Med. 2001;194:707–717. doi: 10.1084/jem.194.6.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waugh KA, Leach SM, Slansky JE. Targeting Transcriptional Regulators of CD8+ T Cell Dysfunction to Boost Anti-Tumor Immunity. Vaccines (Basel) 2015;3:771–802. doi: 10.3390/vaccines3030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 46.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 47.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015;36:265–276. doi: 10.1016/j.it.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reichert TE, Day R, Wagner EM, Whiteside TL. Absent or low expression of the zeta chain in T cells at the tumor site correlates with poor survival in patients with oral carcinoma. Cancer Res. 1998;58:5344–5347. [PubMed] [Google Scholar]
- 51.Macready DH, a. W, G. W. No Free Lunch Theorems for Optimization. IEEE Trans. on Evolutionary Computation. 1997;1:67–82. [Google Scholar]
- 52.Shi M, Lin TH, Appell KC, Berg LJ. Cell cycle progression following naive T cell activation is independent of Jak3/common gamma-chain cytokine signals. J Immunol. 2009;183:4493–4501. doi: 10.4049/jimmunol.0804339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patsoukis N, Sari D, Boussiotis VA. PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle. 2012;11:4305–4309. doi: 10.4161/cc.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKarns SC, Schwartz RH. Distinct effects of TGF-beta 1 on CD4+ and CD8+ T cell survival, division, and IL-2 production: a role for T cell intrinsic Smad3. J Immunol. 2005;174:2071–2083. doi: 10.4049/jimmunol.174.4.2071. [DOI] [PubMed] [Google Scholar]
- 55.Anderson AC. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol Res. 2014;2:393–398. doi: 10.1158/2326-6066.CIR-14-0039. [DOI] [PubMed] [Google Scholar]
- 56.Waggoner SN, Kumar V. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front Immunol. 2012;3:377. doi: 10.3389/fimmu.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, Eaton DL, Grogan JL. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Wolthers KC, Otto SA, Lens SM, Kolbach DN, van Lier RA, Miedema F, Meyaard L. Increased expression of CD80, CD86 and CD70 on T cells from HIV-infected individuals upon activation in vitro: regulation by CD4+ T cells. Eur J Immunol. 1996;26:1700–1706. doi: 10.1002/eji.1830260806. [DOI] [PubMed] [Google Scholar]
- 59.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 60.Ladanyi A, Somlai B, Gilde K, Fejos Z, Gaudi I, Timar J. T-cell activation marker expression on tumor-infiltrating lymphocytes as prognostic factor in cutaneous malignant melanoma. Clin Cancer Res. 2004;10:521–530. doi: 10.1158/1078-0432.ccr-1161-03. [DOI] [PubMed] [Google Scholar]
- 61.Churlaud G, Pitoiset F, Jebbawi F, Lorenzon R, Bellier B, Rosenzwajg M, Klatzmann D. Human and Mouse CD8(+)CD25(+)FOXP3(+) Regulatory T Cells at Steady State and during Interleukin-2 Therapy. Front Immunol. 2015;6:171. doi: 10.3389/fimmu.2015.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grosso JF, Kelleher CC, Harris TJ, Maris CH, Hipkiss EL, De Marzo A, Anders R, Netto G, Getnet D, Bruno TC, Goldberg MV, Pardoll DM, Drake CG. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest. 2007;117:3383–3392. doi: 10.1172/JCI31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mandal R, Strebhardt K. Plk1: unexpected roles in DNA replication. Cell Res. 2013;23:1251–1253. doi: 10.1038/cr.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koulintchenko M, Vengrova S, Eydmann T, Arumugam P, Dalgaard JZ. DNA polymerase alpha (swi7) and the flap endonuclease Fen1 (rad2) act together in the S-phase alkylation damage response in S. pombe. PLoS One. 2012;7:e47091. doi: 10.1371/journal.pone.0047091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schreiber M, Muller WJ, Singh G, Graham FL. Comparison of the effectiveness of adenovirus vectors expressing cyclin kinase inhibitors p16INK4A, p18INK4C, p19INK4D, p21(WAF1/CIP1) and p27KIP1 in inducing cell cycle arrest, apoptosis and inhibition of tumorigenicity. Oncogene. 1999;18:1663–1676. doi: 10.1038/sj.onc.1202466. [DOI] [PubMed] [Google Scholar]
- 66.Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anat. 2008;213:30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H, Ohtani H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 68.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 69.Radoja S, Saio M, Frey AB. CD8+ tumor-infiltrating lymphocytes are primed for Fas-mediated activation-induced cell death but are not apoptotic in situ. J Immunol. 2001;166:6074–6083. doi: 10.4049/jimmunol.166.10.6074. [DOI] [PubMed] [Google Scholar]
- 70.Zheng S, Moehlenbrink J, Lu YC, Zalmas LP, Sagum CA, Carr S, McGouran JF, Alexander L, Fedorov O, Munro S, Kessler B, Bedford MT, Yu Q, La Thangue NB. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol Cell. 2013;52:37–51. doi: 10.1016/j.molcel.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 72.Byun HJ, Jung WW, Lee DS, Kim S, Kim SJ, Park CG, Chung HY, Chun T. Proliferation of activated CD1d-restricted NKT cells is down-modulated by lymphocyte activation gene-3 signaling via cell cycle arrest in S phase. Cell Biol Int. 2007;31:257–262. doi: 10.1016/j.cellbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 73.Nolz JC, Starbeck-Miller GR, Harty JT. Naive, effector and memory CD8 T-cell trafficking: parallels and distinctions. Immunotherapy. 2011;3:1223–1233. doi: 10.2217/imt.11.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waugh KA, Leach SM, Slansky JE. Tolerance of Tumor-Specific T cells in Melanoma Metastases. Journal of Clinical and Cellular Immunology. 2016;7 doi: 10.4172/2155-9899.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dranoff G. Experimental mouse tumour models: what can be learnt about human cancer immunology? Nat Rev Immunol. 2012;12:61–66. doi: 10.1038/nri3129. [DOI] [PubMed] [Google Scholar]
- 76.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 78.Stephen TL, Rutkowski MR, Allegrezza MJ, Perales-Puchalt A, Tesone AJ, Svoronos N, Nguyen JM, Sarmin F, Borowsky ME, Tchou J, Conejo-Garcia JR. Transforming growth factor beta-mediated suppression of antitumor T cells requires FoxP1 transcription factor expression. Immunity. 2014;41:427–439. doi: 10.1016/j.immuni.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 81.Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr., Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 82.Murga M, Fernandez-Capetillo O, Field SJ, Moreno B, Borlado LR, Fujiwara Y, Balomenos D, Vicario A, Carrera AC, Orkin SH, Greenberg ME, Zubiaga AM. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- 83.Li FX, Zhu JW, Tessem JS, Beilke J, Varella-Garcia M, Jensen J, Hogan CJ, DeGregori J. The development of diabetes in E2f1/E2f2 mutant mice reveals important roles for bone marrow-derived cells in preventing islet cell loss. Proc Natl Acad Sci U S A. 2003;100:12935–12940. doi: 10.1073/pnas.2231861100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu JW, DeRyckere D, Li FX, Wan YY, DeGregori J. A role for E2F1 in the induction of ARF, p53, and apoptosis during thymic negative selection. Cell Growth Differ. 1999;10:829–838. [PubMed] [Google Scholar]
- 85.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–645. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- 86.Galluzzi L, Vacchelli E, Bravo-San Pedro JM, Buque A, Senovilla L, Baracco EE, Bloy N, Castoldi F, Abastado JP, Agostinis P, Apte RN, Aranda F, Ayyoub M, Beckhove P, Blay JY, Bracci L, Caignard A, Castelli C, Cavallo F, Celis E, Cerundolo V, Clayton A, Colombo MP, Coussens L, Dhodapkar MV, Eggermont AM, Fearon DT, Fridman WH, Fucikova J, Gabrilovich DI, Galon J, Garg A, Ghiringhelli F, Giaccone G, Gilboa E, Gnjatic S, Hoos A, Hosmalin A, Jager D, Kalinski P, Karre K, Kepp O, Kiessling R, Kirkwood JM, Klein E, Knuth A, Lewis CE, Liblau R, Lotze MT, Lugli E, Mach JP, Mattei F, Mavilio D, Melero I, Melief CJ, Mittendorf EA, Moretta L, Odunsi A, Okada H, Palucka AK, Peter ME, Pienta KJ, Porgador A, Prendergast GC, Rabinovich GA, Restifo NP, Rizvi N, Sautes-Fridman C, Schreiber H, Seliger B, Shiku H, Silva-Santos B, Smyth MJ, Speiser DE, Spisek R, Srivastava PK, Talmadge JE, Tartour E, Van Der Burg SH, Van Den Eynde BJ, Vile R, Wagner H, Weber JS, Whiteside TL, Wolchok JD, Zitvogel L, Zou W, Kroemer G. Classification of current anticancer immunotherapies. Oncotarget. 2014;5:12472–12508. doi: 10.18632/oncotarget.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci Signal. 2012;5:ra46. doi: 10.1126/scisignal.2002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brunner MC, Chambers CA, Chan FK, Hanke J, Winoto A, Allison JP. CTLA-4-Mediated inhibition of early events of T cell proliferation. J Immunol. 1999;162:5813–5820. [PubMed] [Google Scholar]
- 89.Greenwald RJ, Oosterwegel MA, van der Woude D, Kubal A, Mandelbrot DA, Boussiotis VA, Sharpe AH. CTLA-4 regulates cell cycle progression during a primary immune response. Eur J Immunol. 2002;32:366–373. doi: 10.1002/1521-4141(200202)32:2<366::AID-IMMU366>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 90.Workman CJ, Cauley LS, Kim IJ, Blackman MA, Woodland DL, Vignali DA. Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. J Immunol. 2004;172:5450–5455. doi: 10.4049/jimmunol.172.9.5450. [DOI] [PubMed] [Google Scholar]
- 91.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL, Tangsombatvisit S, Grosso JF, Netto G, Smeltzer MP, Chaux A, Utz PJ, Workman CJ, Pardoll DM, Korman AJ, Drake CG, Vignali DA. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72:917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nguyen LT, Ohashi PS. Clinical blockade of PD1 and LAG3--potential mechanisms of action. Nat Rev Immunol. 2015;15:45–56. doi: 10.1038/nri3790. [DOI] [PubMed] [Google Scholar]
- 93.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 94.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kinjyo I, Qin J, Tan SY, Wellard CJ, Mrass P, Ritchie W, Doi A, Cavanagh LL, Tomura M, Sakaue-Sawano A, Kanagawa O, Miyawaki A, Hodgkin PD, Weninger W. Real-time tracking of cell cycle progression during CD8+ effector and memory T-cell differentiation. Nat Commun. 2015;6:6301. doi: 10.1038/ncomms7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marinov GK, Williams BA, McCue K, Schroth GP, Gertz J, Myers RM, Wold BJ. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.