Abstract
Computational modeling is essential for structural characterization of biomolecular mechanisms across the broad spectrum of scales. Adequate understanding of biomolecular mechanisms inherently involves our ability to model them. Structural modeling of individual biomolecules and their interactions has been rapidly progressing. However, in terms of the broader picture, the focus is shifting toward larger systems, up to the level of a cell. Such modeling involves a more dynamic and realistic representation of the interactomes in vivo, in a crowded cellular environment, as well as membranes and membrane proteins, and other cellular components. Structural modeling of a cell complements computational approaches to cellular mechanisms based on differential equations, graph models, and other techniques to model biological networks, imaging data, etc. Structural modeling along with other computational and experimental approaches will provide a fundamental understanding of life at the molecular level and lead to important applications to biology and medicine. A cross section of diverse approaches presented in this review illustrates the developing shift from the structural modeling of individual molecules to that of cell biology. Studies in several related areas are covered: biological networks; automated construction of three-dimensional cell models using experimental data; modeling of protein complexes; prediction of non-specific and transient protein interactions; thermodynamic and kinetic effects of crowding; cellular membrane modeling; and modeling of chromosomes. The review presents an expert opinion on the current state-of-the-art in these various aspects of structural modeling in cellular biology, and the prospects of future developments in this emerging field.
Keywords: modeling of biological mesoscale, protein interactions, macromolecular crowding, cellular membranes, chromosome modeling
Graphical Abstract
Introduction
Structural characterization of biomolecular mechanisms across a broad spectrum of scales is key to our understanding of life at the molecular level. Along with experimental techniques, computational modeling is an essential part of this characterization, as a source of structural information and the means of predicting new, experimentally unobserved/unobservable phenomena. An adequate understanding of biomolecular mechanisms inherently involves our ability to model them.
Structural modeling of individual biomolecules and their interactions has been rapidly progressing [1, 2], with many challenges to be addressed in the coming years. However, in large part due to this progress, in terms of a broader picture, the focus is inevitably shifting toward larger systems, up to the level of a cell. Such modeling should involve a more dynamic and realistic representation of the interactomes in vivo, in a crowded cellular environment, as well as of membranes, membrane proteins, and other cellular components. The atomistic modeling methodology requires vigorous development. The efforts in structural modeling of a cell do not negate the need for this development. To the contrary - they will spur it, and expand its scope to larger and more heterogeneous systems.
Whole cell modeling is “the grand challenge of the 21st century” [3]. Specifically it is important for a variety of reasons, including integration of heterogeneous datasets into a unified representation of knowledge about a given organism, prediction of complex multi-network phenotypes, identification of gaps in our knowledge of cellular processes, and development of our ability to modulate them [4].
Emerging experimental techniques, such as femtosecond crystallography with X-ray free-electron lasers, small angle x-ray scattering, and advances in widely adopted methods such as high-resolution cryoelectron microscopy [5-10] provide new data and experimental validation for the modeling. Great examples of joint experimental and computational techniques are rapidly developing approaches to identification of biological assemblies and construction of large protein complexes with hybrid methods, such as integrative modeling [11].
At this point, molecular and cellular modelers use substantially different approaches and, in fact, speak largely different “scientific languages.” Modeling of structures in molecular biology usually means predicting the structure or simulating the folding of a protein, or modeling the interactions between two isolated molecules. It is usually assumed that folding or binding occurs in dilute solutions, so the only environmental concerns are modeling the effect of water and possibly ionic strength. While these calculations are far from easy, and require substantive understanding of biophysics, sophisticated simulation software and significant computational efforts, modeling in cellular biology generally deals with much more complex systems. Accordingly, simulations at the cellular level usually require substantial coarse-graining and simplifications, and existing models of “virtual cell” are largely based on differential equations, imaging data, and other integrative approaches [12, 13]. It is clear that closing the gap between such different levels of approximation will require significant effort, and a number of groups with backgrounds in structural modeling at the molecular level have made progress toward the development of multi-scale approaches to introduce a higher degree of structural information into the modeling of cells. Some of these approaches are coarse-grained and some are atomic resolution, but if put together would potentially provide an integral and self-consistent model of the whole system.
A cross section of diverse approaches presented in this review illustrates the developing shift from the structural modeling of individual molecules to that of cell biology. Studies in several related areas are covered: biological networks; automated construction of three-dimensional (3D) molecular cell models using experimental data; structural modeling of protein complexes; prediction of the non-specific and transient protein-protein interactions that occur in the crowded environment of the cell; atomistic modeling of the thermodynamic and kinetic effects of crowding on folding and binding of macromolecules; all-atom cellular membrane modeling and simulation; and modeling of chromosomes.
This review originated from a discussion at the 2014 meeting on Modeling of Protein Interactions (http://conferences.compbio.ku.edu), and presents an expert opinion on the current state-of-the-art in various aspects of structural modeling in cellular biology, and the prospects of future developments in this emerging field.
From biophysics of molecules to cellular phenotypes
Structures of proteins, nucleic acids, and their complexes have provided foundational knowledge to gain insight into the molecular mechanisms of biological processes [14]. An overall understanding of complex cellular phenotypes, however, requires considering multiple molecular species and their interactions. There are growing interests in understanding the nature of protein-protein and protein-nucleic acid interactions, as well as in computational docking studies [15]. Since many different species of molecules participate in determining the outcome of cellular processes, the discovery of relevant molecular players, their post-translational modifications, and the formation of networks for gene regulation and signal transduction have been the focus of many experimental investigations.
There are a large number of resources where experimental knowledge and computational tools of networks are organized and curated. The Systems Biology Markup Language (SBML) provides an open interchange format for modeling metabolic networks, cell signaling networks, and other biological processes [16]. The Biological Pathway Exchange BioPAX provides a standard language to facilitate integration, exchange, visualization and analysis of biological pathway data [17]. The KEGG (Kyoto Encyclopedia of Genes and Genomes) database contains rich information on genomes, biological pathways, diseases, drugs, and chemical substances [18]. The BioModels is a repository of computational models of biological processes curated from literature and enriched with cross-references, providing valuable resources for studying behavior of metabolic and signal transduction networks [19].
Developing quantitative models to account for experimental facts and to predict emergent biological behaviors are key to gaining mechanistic understanding of cellular processes. As an example, much has been learned from quantitative models of the lysogeny-lysis decision network based on classic studies of phage lambda, including understanding of system stability against perturbations, robustness against genetic mutations, regulation of cellular fate and rare-event transitions, and as network architectural determinants for heritable epigenetic state [20-22]. Studies based on quantitative models of network systems biology integrating many cellular components will continue to contribute to our understanding of broad biological questions such as stem cell differentiations [23-27] and cancer development [28, 29].
There exists a hierarchy of modeling frameworks for studying biological networks. These include graph models [30], Boolean networks [31], ordinary differential equations (ODE) [32], stochastic differential equations (SDE) [33], and chemical master equations (CME) [34-38], in increasing accuracy but also complexity. There are several important considerations in modeling complex biological phenomena. First, we need to gather sufficient and unambiguous biological facts to construct an insightful and appropriate biological network model. This often requires in-depth biological knowledge and can benefit from large amount of data from the convergence of modern high-throughput measuring techniques. Second, we need to obtain a model at the appropriate level of details. Different choice of ODE models, stochastic differential equation model, or the chemical master equation model may lead to different conclusion [38, 39]. At high concentrations such as those found in a metabolic network, an ODE model is preferred, since detailed stochastic models limit the scale of problems that can be examined, with no additional advantages. However, at low concentrations as in gene regulatory networks and signal transduction networks, the copy numbers of involved molecules may be very small (e.g., nM concentration), and stochasticity often plays an important role [40]. At this level, the choice of SDE (Langevin or Fokker-Planck) or CME formulation may be required. As these different model choices may yield different results, a challenging issue is to develop hybrid models and to determine when a particular modeling formalism is appropriate and when an alternative approach is necessary, e.g. when will an ODE model break down and an SDE model be used, and when will an SDE model break down and a CME model be used. Third, describing the complex geometry of the cellular environment may also be necessary for the modeling of transport and communication among its various spatial regions and compartments [13], requiring the construction of a more realistic “virtual cell” and the use of partial differential equations (PDE). Fourth, we need to ensure that computational methods and algorithms can yield the correct solutions, or at least we should be aware of the limitation of the algorithms and recognize possible errors in the computational answers. Correct computational results may not be found even though the problem is formulated correctly. This is especially relevant for problems where stochasticity is significant and when sampling methods are employed. For example, the widely used method of Stochastic Simulation Algorithm does not work well in studying rare events important in many biological phenomena [41-43]. Recent progress at the most detailed level in finding an exact solution of the probability landscape governed by the chemical master equation, in developing optimal method for state enumeration, and in formulating a theoretical framework for a priori estimation of truncation error of finite state space, and in biased Monte Carlo sampling of reaction trajectories for rare events have shown promise in resolving these issues [37, 38, 41-47].
Developing quantitative models requires knowledge of reaction rates and binding constants as parameters. Whether it is a large comprehensive network or a minimalistic network most germane to the question at hand, the availability and validity of model parameters are a challenging issue, as it is unrealistic to expect to have in vivo measurement of every model parameter. The study of the epigenetic circuit of phage lambda showed that introduction of modest protein-protein cooperative interaction of CI-dimers can lead to desired probabilistic landscape of deep-threshold and efficient switch [38], illustrating the importance of protein-protein binding in regulating network phenotypes. Studies of computational biophysics on binding affinities and reaction rates therefore are of growing importance for developing effective models of systems biology [48-51]. However, there is a gap between single valued rate and binding affinity parameters and the rich information contained in ensembles of interacting protein-protein or protein-nucleic acid complexes examined in studies such as protein docking. Identification of potential interactions and binding partners from experimental and computational structural biology studies of protein-protein complexes will provide valuable information for improving network models of cellular processes. How biophysical studies of protein stability and binding interactions can inform developing systems biology models to go beyond single parameter and homogeneous systems remains open and progress will likely be fruitful [48-51].
Building complex cellular environments at molecular detail
The cell is a hierarchy of structures that span from atoms to organelles, all of which interact in an intricate choreography with tempos that range from femtoseconds to hours. The biological mesoscale range includes biological structures from 10 to 100 nanometers. Structures of this size include viruses, cellular organelles, large molecular complexes, and any other internal cellular environments within that range. The mesoscale is important because it represents the scale of cellular systems that is not fully accessible to a single experimental technique.
Structural data is now available at a wide range of length scales – from atomic resolution structures of cellular protein and nucleic acid components to organelle and larger cellular structures. Biophysical techniques range from atomic resolution X-ray crystallography and NMR spectroscopy, to electron and light microscopy. In addition, spatial distributions and dynamics are accessible by a variety of fluorescence microscopy methods, and expression and concentration levels are obtainable via technologies ranging from chip arrays and other mRNA technologies to mass spectrometry and other proteomic analyses.
Over the past several years there have been a number of efforts to build complete structural models of cellular environments at molecular detail. This type of work has typically focused on a particular portion of a cell, for example E. coli cytoplasm [52], M. genitalium cytoplasm [53], bacterial division machinery [54], synaptic vesicles [55], and an entire synaptic bouton [56]. Because of the size and complexity of cellular structure, there are numerous challenges that must be faced before building a structural model of a complete cell becomes a reality. Among these challenges are: 1) development of a model building framework that can unify the various cellular components at multiple scales; 2) the implementation of accelerated computation through parallelization and custom hardware solutions; 3) the data analysis and visualization software capable of handling large complex models; 4) the development of metrics to quantify and validate the models; and 5) the development of communities and collaborations to be able to approach such large and complex modeling tasks, and to continually improve and curate the models.
Here we focus on cellPACK [57, 58] which has been developed as a computational framework that attempts to address some of these challenges. The cellPACK software uses structural and distribution data for a given mesoscale environment gathered from different experimental methods and automatically synthesizes one or many 3D models that are statistically consistent with all of this available information. For a given cellular or subcellular structure, the geometry of the large components such as organelles or intact virions seen with electron microscopy can define specific volumes and surfaces to fill with the smaller molecular entities. Since the locations of the contents of these larger components are constantly changing, cellPACK uses statistical measures to place these molecular components into the compartmental volumes and membrane surfaces. Thus, a filled model is one snapshot of many possible fills.
cellPACK uses distance field grid to discretize and describe a volume, enabling multiple modular packing algorithms to interoperate on the same model and can combine several complex packing algorithms to integrate three different major localization modes – volumetric, surface, and procedural – into unified models. It has numerous modules for cell/molecule-specific packing. In the resultant model, each molecular object retains a connection to various other forms of data to enable deeper analysis, in preparation for systems integration or large-scale simulations, or for modifications of the molecule’s representations.
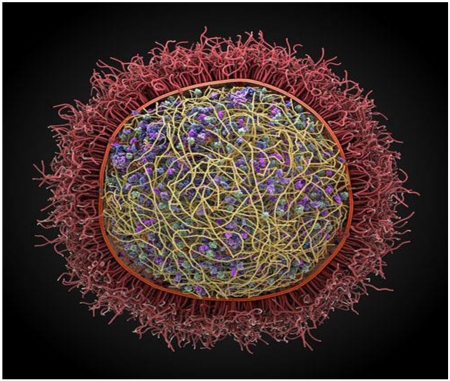
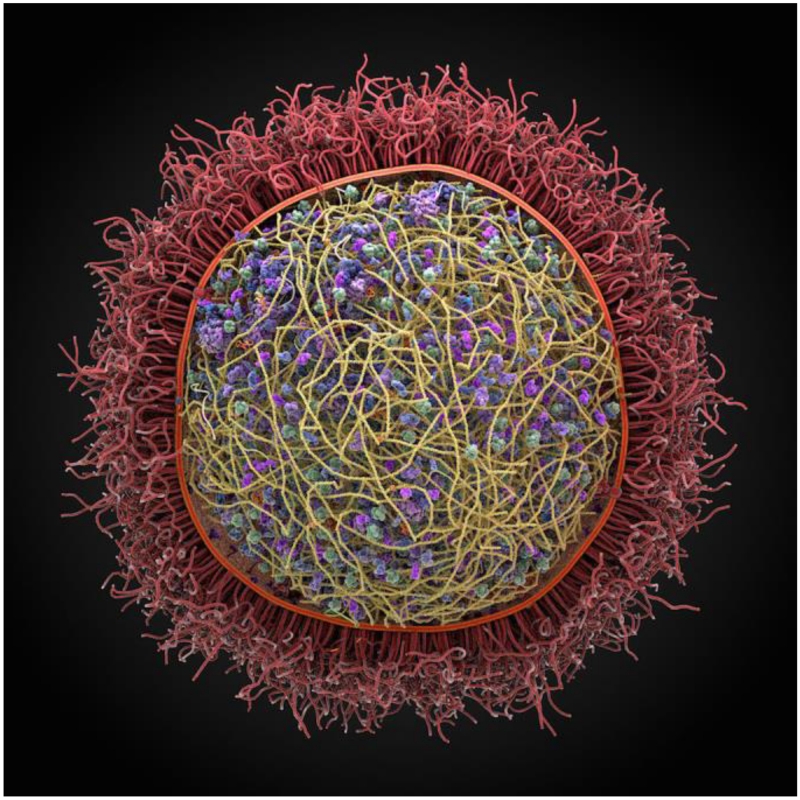
To date, cellPACK has been used to generate models of blood plasma, the immature and mature HIV virion, the packing of synaptic vesicles and a preliminary model of a mycoplasma (Fig. 1). These models contain thousands to 10s of thousands of individual biomolecules in the context of cellular environments. Prior to cellPACK, such models had to be built by hand taking weeks or months, and presented a serious bottleneck in preparing the starting conditions for input to large-scale simulations such as Brownian dynamics [52, 59, 60]. With the automated procedures in cellPACK, such models can be produced in minutes, making possible the construction of a large ensemble of models each different in detail, but each consistent with the input experimental data. This enables the possibility to run many parallel simulations, each with a different initial model. Additionally, cellPACK enables the exploration of different structural hypotheses, creating models that can be compared with experimental observation.
Figure 1. A preliminary computer assembled and generated 3D model of Mycoplasma genitalium, a parasitic bacterium found in human urogenital and respiratory tracts.
This pathogen has one of the smallest genomes of any free-living organism (525 genes). It was produced using the autoPACK/cellPACK software.
Frameworks such as cellPACK will enable the structural modeling community to create and share models of complex molecular environments and make possible new analyses and simulations of these environments (see http://cellpack.org).
Structural modeling of protein complexes
Protein-protein interactions are central for cellular processes. Experimental approaches to determining the interaction networks have limited reliability [61, 62]. Thus computational prediction of interactors is important [63]. For proper training and validation of such approaches one needs representative databases of interacting proteins [64], as well as those that do not interact [65]. Structural characterization of proteins is essential for understanding molecular processes in the cell. However, only a fraction of known proteins have experimentally determined structures. That fraction is even smaller for protein-protein complexes. Thus, modeling is key to their structural determination [66-68].
An important insight into the basic rules of protein recognition is provided by the studies of large-scale structural recognition factors in macromolecular assemblies [69], and binding-related anisotropy of protein shape [70, 71]. Such factors in protein association have to do with the funnel-like intermolecular energy landscape [72]. It has been shown that simple energy functions, including coarse-grained (low-resolution) models, reveal major landscape characteristics, such as the number and distribution of the funnel-like energy basins, transition between low and high resolution, and funnel size [73]. The intermolecular energy landscapes are further characterized by conformational properties of interacting proteins [74-76].
The docking degrees of freedom involve six external degrees of the rigid body movement (3 translation coordinates and 3 angles of rotation), as well as internal degrees of freedom, which determine the conformation of the proteins. To make the number of the internal degrees of freedom manageable, approximations are essential. The rigid-body approximation leaves only the external degrees of freedom and approximates internal degrees of freedom by making the proteins soft and thus tolerant to local structural mismatches. The rigid-body approximation is adequate for bound docking (separated proteins from co-crystallized complexes), low-resolution unbound docking (structures determined outside of the complex), as well as in some cases of high-resolution unbound docking. However, in general, for the atomic resolution unbound docking, some form of conformational sampling is required. For most crystallographically determined complexes, the unbound to bound conformational change is largely restricted to the surface side chains [77], thus drastically limiting the combinatorics of the conformational search. Protein docking approaches are extensively evaluated in the community-wide experiment on Critical Assessment of Predicted Interactions (CAPRI) [2], and in numerous studies based on benchmarking sets (e.g. [77, 78]). Protein docking procedures were also shown to be successful in packing protein structural motifs [79] and predicting complexes of membrane proteins [80].
The coarse-graining of protein structures allows exploration of structural dynamics of large-scale (microseconds or longer) processes [81, 82]. It also allows comparison with low-resolution experimental data, which often is the only available structural information on the system [83]. Coarse-grained elastic networks modeling of structure fluctuations showed that, on average, the interface is more rigid than the rest of the protein surface [84, 85], and the interface mobility is correlated with the interface type, size and obligate nature of the complex [85]. In structural modeling of protein-protein complexes, the coarse-graining approaches are used to model structural flexibility in protein assembly [75, 81, 86, 87]. Low-resolution allows implicit accounting for local conformational flexibility without sampling the internal degrees of freedom, and thus is useful in docking [88, 89].
The number of experimentally determined protein structures accounts only for a fraction of known proteins. Thus docking often has to rely on the modeled structures of the interactors, especially in the case of large protein-protein interaction (PPI) networks. Structures of modeled proteins are typically less accurate than the ones determined by X-ray crystallography or NMR. The goal of the modeling should determine the accuracy of the models. The accuracy of the output complex cannot be higher than the accuracy of the input structures. Thus the necessary level of structural accuracy of the complex determines the required accuracy of the modeling of the individual proteins. The question then is: what is that necessary level of structural accuracy for protein complexes? In protein-protein interactions, many experimental (and theoretical) studies require simple knowledge of the residues at the interfaces (e.g. for further experimental analysis) and have no use for atomic resolution structural details of the complex (specific atom-atom, or even residue-residue contacts across the interface). The same is true for small ligand – protein docking, when the goal is identification of the binding/functional site on the protein. For the interface (binding, functional site) prediction, the high-resolution protein structures, generally, are not needed [90-92]. That has been extensively demonstrated by systematic studies over a number of years [66, 89]. However, when a high-resolution structure of the complex is required, in protein-protein interactions (e.g. for estimation of the binding affinity) or in small ligand – protein docking (e.g. for identification of specific ligands), higher accuracy protein models are needed.
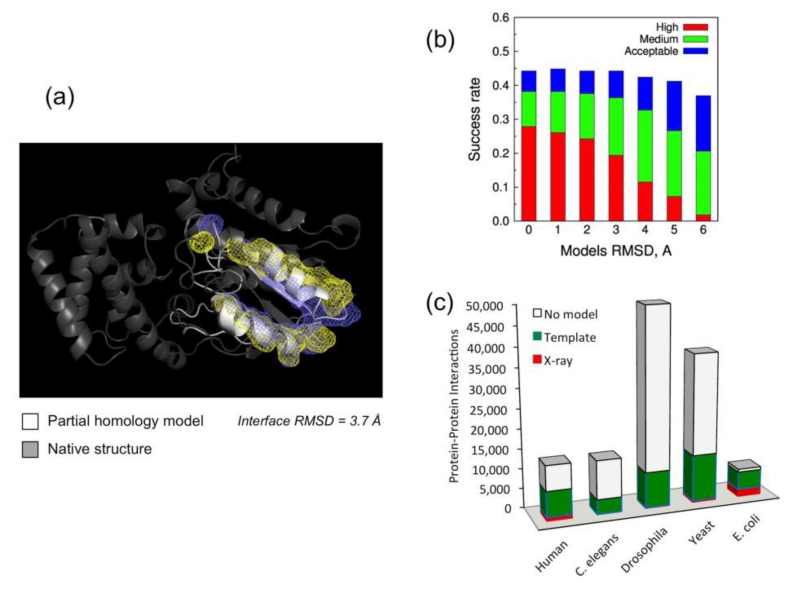
High-throughput modeling for entire genomes requires a computationally tractable methodology. A statistical analysis of target-template sequence alignments for systematic evaluation of potential accuracy in high-throughput modeling of binding sites was performed on a representative set of protein complexes [93]. The modeling was performed in a high-throughput fashion based on standard sequence alignment and comparative modeling, as opposed to more detailed and sophisticated (but also more computationally expensive) multi-template procedures. Overall, ~50% of protein pairs with the interfaces modeled by high-throughput techniques had accuracy suitable for structural modeling of their complexes (Fig. 2a).
Figure 2. Structural modeling of protein interactome.
(a) ~50% of protein complexes with interfaces modeled by high-throughput techniques have accuracy suitable for docking. (b) Models by template-based docking classified according to CAPRI-like criteria (clashes and contacts not considered). High accuracy: ligand (the smaller protein in the complex) RMSD < 1.0 Å or interface RMSD (measured on the interface residues Cα) < 1.0 Å. Medium accuracy: ligand RMSD < 5.0 Å or interface RMSD < 2.0 Å. Acceptable accuracy: ligand RMSD < 10.0 Å or interface RMSD < 4.0 Å. The data indicates that even inaccurate models typically dock with good success rate. (c) Availability of protein-protein structures in genomes with the largest number of known protein interactions (according to BIND [227] and DIP [228] databases). “No model” indicates no template for the interactors, not the complex (almost all structurally characterized interactors have templates for their complexes).
Although structural modeling of protein complexes primarily has to rely on modeled structures of the individual proteins, such “double” modeling remains so far largely untested in a systematic way, largely due to the absence of an adequate benchmark set that would contain protein structures with accuracy levels according to a full array of pre-defined root-mean-square-deviation (RMSD) values. Such sets were generated based on crystallographically determined complexes from the Dockground resource [78, 94, 95]. A comprehensive benchmarking of template-based docking by structure alignment [96] and free docking [97, 98] techniques was performed on a set of 165×6 model protein structures with accuracy levels at 1 - 6 Å Cα RMSD. The results (Anishchenko et al. submitted) show that many docking models fall into acceptable quality category, according to the CAPRI Challenge [2] criteria, even for highly distorted models (Fig. 2b). The template-based methodology is less sensitive to the inaccuracies of protein models compared to the free docking. However, both can be applied to the structural modeling of the protein interactome.
Proteome-scale modeling of PPI networks [99-102] is essential for modeling of a cell. Templates are available for a significant part of soluble proteins in genomes [103], including those in known PPIs [104]. The approaches to genome-wide structural modeling of PPIs are either “traditional” template-free docking [105, 106] or the template-based docking [63, 104, 107-111]. The latter, while potentially providing much greater success rate [96], critically depends on the availability of the templates [63, 104, 108, 109]. The X-ray structures of the proteins were complemented by homology models and the templates for their complexes were detected in PDB [104]. Figure 2c shows the results for five genomes with the largest number of known PPIs. Structural alignments yielded a dramatic increase in the structural coverage of complexes, from the coverage provided by the sequence alignment. The structural templates were found for nearly all (33,537 out of 33,840, or 99%) complexes in which both components could be built. Thus, the limiting factor in interactome modeling is actually the availability of the templates for the individual proteins (more protein-protein templates are still needed for greater accuracy of modeling). Still the free docking is necessary, and its importance growing, for many protein encounters in the crowded cell environment, which are not likely to correspond to energetically stable co-crystallized templates.
The challenge in the development of protein docking methodology is to adequately incorporate internal degrees of freedom into the docking protocols. This includes structural flexibility of the interacting proteins, especially in case of significant conformational changes upon binding, as well as structural inaccuracies of the proteins, especially models. Another grand challenge is to understand and simulate the environment in which proteins interact in vivo. This environment is densely populated, which strongly affects protein diffusion, binding and conformational transitions. For large-scale structural modeling of PPI networks, such approaches have to be high-throughput, taking advantage of new algorithms and hardware resources.
Atomistic modeling of thermodynamic and kinetic effects of crowding in cellular environment
There is growing recognition that the cellular context and the cellular environment have fundamental influences on biochemical processes [112, 113]. Missing in typical in vitro biophysical studies done in dilute solution are the many “bystander” macromolecules, which have considerable consequences on the biomolecules of direct interest.
The many bystander macromolecules together occupy a high fraction of the volume of the given cellular compartment. An early focus was on how this condition, known as macromolecular crowding, impacts thermodynamic and kinetic properties of protein folding, binding, and aggregation. Of particular note are in vitro experiments in which crowding agents are added to mimic bystander macromolecules in cellular compartments [114-116]. Experimental and computational studies have now converged on the conclusion that effects of macromolecular crowding are relatively modest, on the order of 0.5 kcal/mol in energetic terms, for the folding and binding of single-domain proteins, and become progressively greater as the sizes of the reactant species increase, and reach striking magnitudes for protein aggregation [52, 112, 117].
Protein folding under macromolecular crowding has been modeled in two complementary approaches. In the direct simulation approach, one mixes the protein of interest with crowders (i.e., bystander macromolecules), similar to the in vitro experiments with crowding agents. To adequately sample the folding and unfolding transitions while also simulating the movements of the crowders, it was necessary to use coarse-grained representations for the protein and crowders [118], although some aspects of folding have been studied using an all-atom representation [119]. In the alternative approach [48, 52], now known as postprocessing [120], one runs simulations of the crowders by themselves and runs separate simulations of the protein at end states, e.g., the folded and unfolded states. In this way, one avoids the expensive simulations of the rare transitions between the end states. One then computes the transfer free energies of the protein in the end states from a dilute solution into the crowder solution.
The basis for the transfer free energy calculations was provided by Widom’s particle insertion method [121]. A brute-force implementation turned out to incur “very significant computational expense” [52]. Recognizing that this problem has much similarity to the docking of a ligand to a protein and the use of the fast Fourier transform (FFT) technique in the latter problem [97, 122], FMAP (FFT-based method for Modeling Atomistic Proteins-crowder interactions) was developed for computing the transfer free energies [123, 124].
To model subcellular problems, a reasonable (perhaps necessary) choice is to represent water (and other small molecules of the solvent) implicitly. A number of groups have carried out simulations of subcellular compartments, modeled with implicit solvent [52, 59, 60]. A focus of these simulation studies is the diffusion coefficient of a tracer protein in these crowded environments. Then one faces the problem of parameterizing the effective protein-crowder interaction energies. These interactions contain hard-core strong repulsion and longer-distance weak attraction or repulsion. The soft attraction can lead to protein-crowder weak association. Parameterizing energy functions is of course not a new problem, but data from in vitro experiments with crowding agents can be very useful. For example, the second virial coefficient in the expansion of the osmotic pressure in terms of macromolecular concentration contains rich information on intermolecular interactions and can be easily measured by techniques such as static light scattering [125].
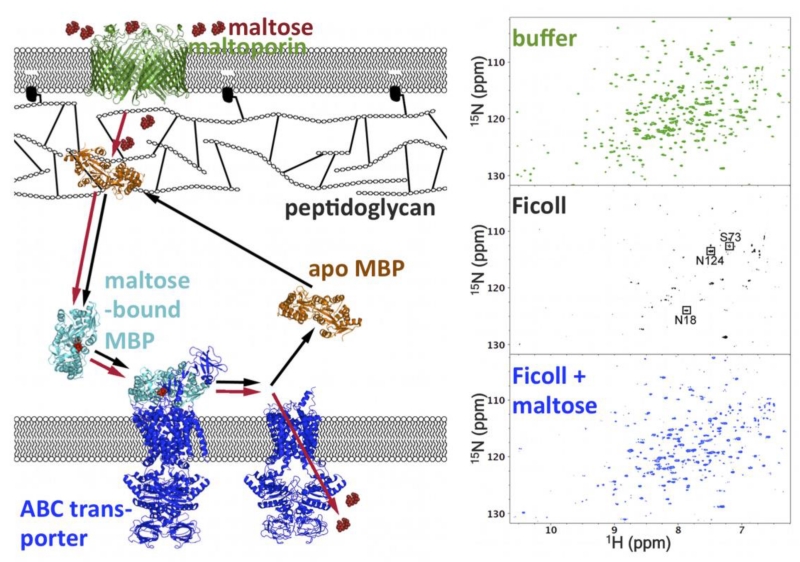
Injecting new interest into modeling cellular context and the cellular environment are experimental studies demonstrating emergent behaviors of proteins and nucleic acids under crowded conditions. The first is the nonrandom nature of protein-crowder weak association. In particular, some proteins were found to associate with specific cellular targets. For example, the neural protein tau when injected into X. laevis oocytes binds to microtubules [126]. In E. coli, the MetJ repressor forms extensive nonspecific interactions with genomic DNA [127]. In other cases, there is evidence implicating a specific site of a protein in the nonspecific interactions. Pin1 uses the substrate recognition site for nonspecific interactions. Nonspecific interactions are apparently abrogated when either the substrate recognition site is phosphorylated or a substrate is bound [128]. Similarly, the maltose binding protein (MBP) forms nonspecific interactions with proteins and synthetic polymers, but this ability is weakened or lost when maltose is bound [129] (Fig. 3). FMAP-enabled calculations are capturing the nonrandom nature of the weak association (Qin and Zhou, to be published).
Figure 3. Ligand binding of MBP in vivo and in vitro.
Left: possible shuttling of MBP in the E. coli periplasm for transport of maltose into the cytoplasm. Right: competition of Ficoll and maltose for interaction with MBP, shown by NMR spectroscopy. In buffer, apo MBP shows well-resolved 1H-15N TROSY spectra. With 200 g/l Ficoll, most of the TROSY peaks are broadened beyond detection, indicating MBP-Ficoll association. Upon further addition of 1 mM maltose, the peaks are recovered, indicating that the ligand has competed out the weakly associated Ficoll. Adapted from Miklos et al. [129].
The weak nonspecific association with bystander macromolecules often can be inferred to impart biological function. For example, the binding of tau to microtubules is thought to be important for the latter’s stability. Nonspecific binding of the MetJ repressor to genomic DNA may facilitate the search for a specific site. Nonspecific association with endogenous proteins via the substrate recognition site may be the mechanism for subcellular localization. For MBP, it has been proposed that nonspecific association with the outer membrane-attached peptidoglycan primes the protein for receiving maltose; binding of maltose releases the protein, allowing it to diffuse to the inner membrane-bound ABC transporter and hand over the maltose for translocation into the cytoplasm [129] (Fig. 3).
It is remarkable that nonspecific association can be tuned out by phosphorylation or substrate binding [128], or by ligand binding [129]. Calmodulin gains nonspecific interactions upon binding Ca2+ but loses this ability again upon further binding a substrate peptide [130]. Apparently, nonspecific association can be regulated by some of the same mechanisms, e.g., phosphorylation or ligand or substrate binding, as for specific association.
Another emergent behavior is the formation of mesoscale cellular structures. The cytoskeleton offers a prime example, but other subcellular organizations are being recognized as well. In particular, it is now well known that enzymes in the same metabolic pathway are co-localized [131], possibly to facilitate substrate channeling between successive enzymes.
Perhaps the most exciting emergent behavior is liquid-liquid phase separation, between the protein-poor cytoplasm and the protein-rich cellular bodies. These bodies, commonly referred to as droplets, are membrane-less organelles and are implicated in many cellular functions, such as for protein or RNA storage [132-136]. Interestingly, phase separation has been achieved in vitro using reconstituted or even designed components [137]. While this protein-rich phase is liquid-like, other proteins can form crystalline assemblies; and macromolecular crowding can drive their formation [138]. Modeling such as that enabled by FMAP (Qin and Zhou, submitted) and other techniques and in vitro experiments mimicking cellular conditions will allow us to reach quantitative understanding of all these emergent behaviors in the cellular context.
Modeling nonspecific interactions and aggregation of proteins
To recognize a specific partner, a protein must align its binding interface, usually a small fraction of the total surface, with a similarly small binding interface on the other protein. The goal of docking methods is to identify this specific association as the global minimum of a free energy landscape. However, nonspecific interactions among macromolecules are also important, particularly in a crowded environment of a cell, since the high frequency of such encounters can substantially affect the stability of the equilibrium state [48, 52, 112, 113, 139-141]. Indeed, it was shown in crowding experiments that the energetics of interactions with crowders impacts the formation of specific complexes and of non-native aggregates beyond simple excluded volume effects [140, 141]. Since global docking methods systematically sample the entire conformational space of protein-protein complexes, in principle such methods can be used to study both specific and non-specific associations.
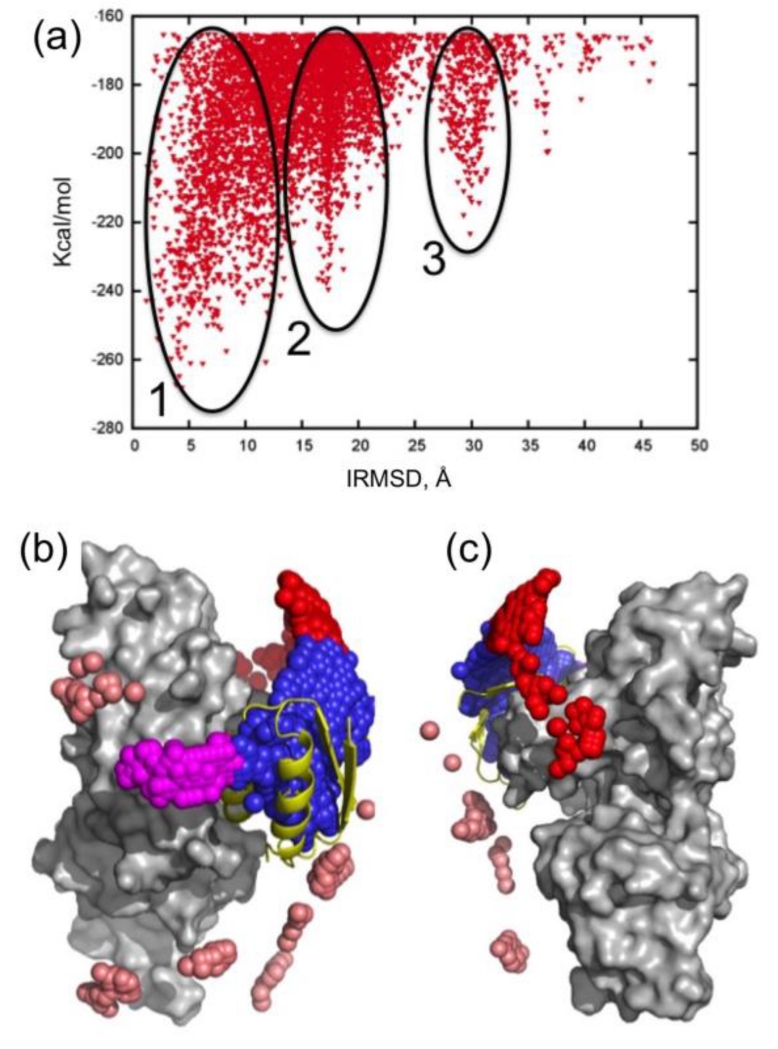
The first step toward modeling nonspecific association is the analysis of encounter complexes [142]. An encounter complex can be thought of as an ensemble of transition states in which the two molecules can rotationally diffuse along each other, or participate in a series of “microcollisions.” A particular type of encounter complex, a late near-native intermediate, referred to as the transient complex, is a key concept in modeling the kinetics of specific association and predicting the association rate constant [49, 143]. A well-studied example of non-specific association is the N-terminal domain of Enzyme I (EIN) and the histidine-containing phosphocarrier protein (HPr) [142, 144]. For a computational study of this interaction we systematically sampled the relative orientations of the two molecules. Fig. 4a shows the interface root mean square deviation (IRMSD) from the native EIN/HPr complex versus the interaction energy score of the docked structures, and reveals 3 large clusters. We note that the interaction energy is given in kcal/mol units, but it does not account either for any entropy loss or for the desolvation of the component proteins, and hence has no absolute thermodynamic meaning. In fact, it was shown that the probability of each cluster of low energy docked structures is proportional to the relative population of the cluster [145, 146], and hence one can use cluster size rather than energy values for selecting putative complex models.
Figure 4. Energy surface and encounter complexes.
(a) IRMSD from the native complex vs. the energy function obtained by docking histidine-containing phosphocarrier protein (HPr) to the N-terminal domain of Enzyme I (EIN). Clusters around the three lowest energy minima are indicated. (b) Native complex formed by EIN (gray surface) and HPr (shown as yellow cartoon). Centers of HPr structures in the encounter complex ensemble are shown as small spheres. Colors indicate classification as follows: Cluster 1, overlapping with the final complex, blue; Cluster 2, encounter complex within 20Å RMSD from the final complex, but not overlapping with it; Cluster 3, encounter complex around 30Å RMSD from the final complex. A number of low energy structures that do not belong to any of these clusters are shown in pink. (c) Same as b, but after rotating 180° around the vertical axis (the bound HPr is now on the left side, almost completely hidden by EIN.
The structures in the largest cluster (Cluster 1, shown in blue in Fig. 4b) overlap with the native state. The structures in this cluster, akin to the aforementioned transient complex, are the results of rigid body rotations and small translations around the native binding mode. The two other clusters (red and magenta in Figures 4b and 4c) consist of structures that can coexist with the native complex. The existence of the three clusters was experimentally verified using NMR paramagnetic relaxation enhancement (PRE), a technique that is exquisitely sensitive to the presence of lowly populated states in the fast exchange regime [144, 147, 148], indicating that these docked structure have physical meaning and represent encounter complexes.
The specific association between EIN and HPr has an equilibrium dissociation constant of 7 μM [149], whereas the KD value of encounter complexes may be as high as 10 mM [144]. In spite of the large difference in binding affinity, the existence of transitional nonspecific association has biological implications. It is estimated that under cellular conditions at least 1% of HPr exists in form of a tertiary complex HPrnonspecific ∕EIN∕HPr, in which the native EIN∕HPr complex nonspecifically binds an additional HPr molecule [144]. The formation of transient HPrnonspecific∕EIN∕HPr ternary complexes may help EIN compete for the cellular pool of HPr. Intracellular overcrowding and compartmentalization may favor the ternary complex further, possibly making these nonspecific interactions even more important for enhancing enzymatic turnover in vivo [144].
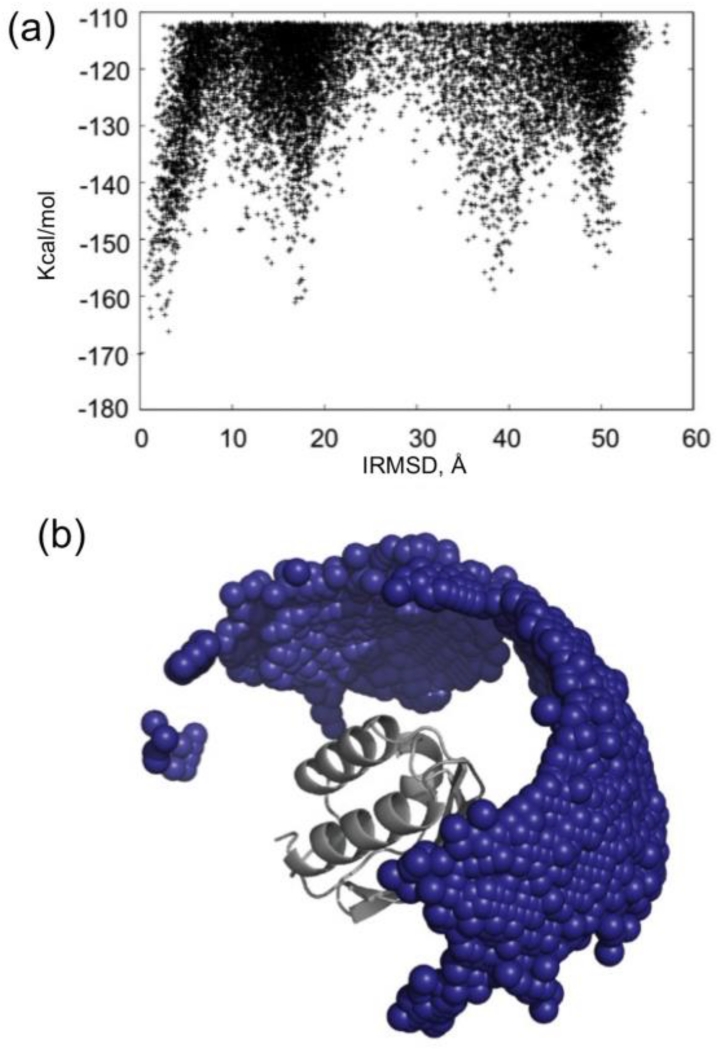
A large fraction of protein pairs that are present in the cell do not form specific complexes with any measurable affinity, as demonstrated by the Negatome database [65]. Nevertheless, some level of nonspecific association always occurs at high protein concentrations [150]. As an example, Fig. 5a shows the energy landscape of the interaction between two HPr monomers that are known not to form a specific stable homodimer. Since there is no native structure in this case, the IRMSD is calculated from an arbitrary structure in the lowest energy region. In contrast to the specific association between EIN and HPr, resulting in a deep and broad minimum (Fig. 4a), the nonspecific binding results in a higher number of minima with comparable energies, some of them > 50 Å from each other. The small blue spheres in Fig. 5b represent the centers of low energy docked structures of the second HPr molecule, and show that the distribution covers a large fraction of the surface. However, the energy-IRMSD plot still shows some large low energy clusters, indicating that some docked structures are more likely than some others, supporting the nonrandom nature of weak association noted above. Indeed, protein aggregation generally does not lead to the formation of entirely amorphous globules, but to the occurrence of some highly preferred interactions between monomeric proteins, or interactions that differ from those seen in the native ones if the protein tends to form a complex.
Figure 5. Energy surface and encounter complexes in non-specific association.
(a) IRMSD vs. the energy function obtained by docking phosphocarrier protein (HPr) to another copy of HPr. The IRMSD is calculated from an arbitrary low energy structure. (b) Centers of docked structures of the second HPr molecule are shown as small spheres.
The biomedical importance of nonspecific interactions is due to the fact that neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis and prion diseases appear to have common cellular and molecular mechanisms including protein aggregation [151]. The aggregates usually consist of fibers containing misfolded protein with a β-sheet conformation, termed amyloid. The likelihood of aggregation is generally increased by increasing protein concentration, which can be caused by genetic dosage alterations. In the case of protein-coding mutations, the altered primary structure can also make the protein more prone to aggregate. Another important factor modulating aggregation is covalent modification, particularly phosphorylation. For example, α-synuclein purified from Lewy bodies in Parkinson’s disease patients is extensively phosphorylated [152]. Some of the factors modulating the interactions have already been discussed in the context of crowding.
In spite of its well-recognized importance, modeling aggregation is challenging, and substantial methodology development is needed. While hydrophobic patches signal aggregation prone regions of proteins [153], no reliable and computationally feasible methods can predict the stability of aggregates and the rate of aggregation. These problems occur in many areas of molecular interactions, as scoring functions do not provide adequate estimates of the binding free energy, whereas more sophisticated tools such as free energy perturbation require detailed structural information and high computational efforts. It is clear that exploring aggregation in the crowded and inhomogeneous cellular environment is even more difficult. Another difficulty is caused by the limited availability of experimental data. Some data are available on peptides that can form amyloid-fibrils or amorphous β-aggregates and on potential aggregation prone regions in proteins, but aggregation rates upon mutations have been experimentally determined only for a dozen of amyloidogenic proteins [154].
Modeling cell membranes and membrane proteins
Cell membranes made up of a wide variety of lipids act as a matrix to host integral membrane proteins, to recruit peripheral membrane proteins, and thus to actively participate in cellular membrane functions together with these proteins. Complexity of biological membrane systems arises from a considerable heterogeneity in the spatial distribution of lipids and proteins on the cell membrane and between the bilayer leaflets. The outer membrane of gram-negative bacteria provides an extreme example, where the lipid component of the outer leaflet is predominantly lipopolysaccharides and those of the inner leaflet are typical phospholipids (Fig. 6) [155]. To a lesser extent, the outer leaflet of the plasma membrane contains more lipids with the phosphatidylcholine head group and sphingolipids (e.g., sphingomyelin) than the inner leaflet, and glycosphingolipids (e.g., gangliosides) exist only in the outer leaflet [156].
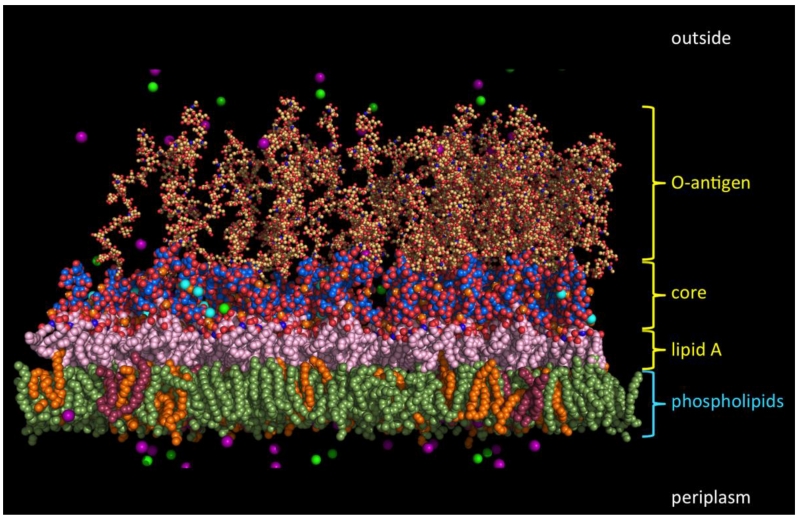
Figure 6. Gram-negative bacterial outer membrane molecular complexity.
The image illustrates a typical E. coli outer membrane and the molecular system used to represent the complexity in molecular dynamics simulations. Molecules represent the bilayer composed of (from the top, external leaflet) glycosylated amphipathic molecules known as lipopolysaccharide consisting of an O-antigen polysaccharide, a core oligosaccharide, and lipid A and (the bottom, periplasmic leaflet) consisting of various phospholipid molecules such as phosphatidylethanolamine (PE; green), phosphatidylglycerol (PG; orange), and cardiolipin (CL; magenta) in a ratio of PE : PG : CL = 15 : 4 : 1. The cyan atoms interspersed with the core oligosaccharides are Ca2+ ions, which immobilize the membrane by mediating the cross-linking electrostatic interaction network with phosphate and carboxyl groups attached to the lipid A and core sugars. Magenta and yellow spheres represent K+ and Cl− ions, respectively.
Membrane proteins play important roles in many cellular processes, such as transmembrane signaling [157, 158], transport of ions and small molecules [159-163], energy transduction [164, 165], and cell-cell recognition [166]. They are quantitatively significant as well: 20-30% of the protein-encoding regions of known genomes encode membrane proteins [167]. Furthermore, about 50% of these membrane proteins are considered putative drug targets [168]. Hydrophobic match between the hydrophobic length of the protein transmembrane domain and that of the lipid bilayer has been thought to play an important role in membrane protein function and organization [169, 170]. Responses to an energetically unfavorable hydrophobic mismatch include lipid-induced changes in conformation and association of the transmembrane domains, as well as protein-induced changes in the lipid chain order, and bilayer thickness and curvature. Therefore, membrane proteins require optimal lipid compositions (lipid types and cholesterol concentration) in a bilayer for their optimal function, and membrane protein organization is also largely dependent on lipid compositions [171-178].
Given the aforementioned complexity of cell membranes, membrane proteins, and their distribution and organization, as well as the importance of delicate protein-lipid (or protein-bilayer) interactions in the structural integrity of membrane proteins [179] and in cellular membrane functions, carrying out molecular dynamics simulations of these complex systems based on realistic all-atom models presents a difficult challenge. Even the construction of initial simulation systems requires knowledge of structural models of individual proteins and lipids, as well as their ratios and locations. Furthermore, considerable computational resources are required to simulate such large systems for a sufficiently long time to obtain meaningful information. Such difficulties are the reason why most all-atom simulation studies of membrane proteins have been limited to either one or a few proteins, and mostly one or two lipid types [180-182]. Simpler low-resolution coarse-grained models have, however, been used to simulate systems with a large number of membrane proteins [183-186]. In addition, the long timescales required present unique challenges in studying both the folding and insertion of transmembrane and peripheral membrane proteins using traditional molecular dynamics simulations. Recently, Tajkhorshid and coworkers developed the highly mobile membrane-mimetic (HMMM) model with accelerated lipid motion by replacing the lipid tails with small organic molecules [187]. The HMMM model provides accelerated lipid diffusion by 1-2 orders of magnitude, and is particularly useful in studying membrane-protein associations [188, 189].
There are various approaches to construct bilayers around membrane proteins [190-197]. Membrane Builder [190-192] (http://www.charmm-gui.org/input/membrane) in CHARMM-GUI [198] uses lipid-like pseudo atoms that are first distributed and packed around a protein and then replaced by lipid molecules one at a time. This so-called “replacement” method [199, 200] (which essentially corresponds to a reverse coarse-graining operation) allows easy control of the lipid types and the number of each lipid type based on the area per each lipid type (estimated from pure bilayer simulations) in a complex membrane system. Similar packing algorithm is adopted in packmol [197], MembraneEditor [194], MemBuilder [201], and insane [202], but detailed protocol may have varying degrees of complexity. For example, insane program can only build coarse-grained membrane and the lipid molecules with straight conformations are used, and packmol provides a sophisticated packing algorithm and any lipid molecule can be adopted if provided by a user. InflateGRO2 [193] adopts an approach that removes the lipids that are overlapping to the membrane protein upon insertion. Deleting overlapping lipid molecules can be tricky because membrane proteins may have cavities in the transmembrane region. Therefore, InflateGRO2 adopts a grid-based search that detects protein cavities and assigns scores to the lipids based on the degree of overlap with protein, so the lipids that are ranked high can be deleted. GRIFFIN [203] and g_membed [195] adopt similar algorithms, where proteins are inserted into the membrane and overlapping lipids are either pushed away slowly or simply deleted.
Over the years, the complexity of the membrane systems that one has been able to build and simulate has increased, in line with the developmental stages of CHARMM-GUI Membrane Builder. Membrane Builder was first developed in 2007 as a publicly available web resource [190]. The first implementation allowed users to generate an initial configuration of a protein in homogeneous lipid bilayers; three lipid types were available. In 2009, Membrane Builder was further developed to allow generation of membrane-only and a protein-membrane system in heterogeneous bilayers of multiple lipid types [191]; 35 lipid types were available. In 2014, Membrane Builder was expanded to handle > 180 lipid types including phosphoinositides, cardiolipin, sphingolipids, bacterial lipids, and ergosterol, which make it possible to build biologically realistic membrane systems for many single-celled organisms and models for membranes in the human body [192]. Importantly, Membrane Builder also provides well-validated equilibration and production inputs for many molecular dynamics packages (CHARMM [204], NAMD [205], GROMACS [206], AMBER [207], OpenMM [208], and CHARMM/OpenMM) [209].
In the context of any biomolecular simulation, simulation timescale and molecular force field accuracy have always been challenging issues, and they will be so, as we are all interested in more challenging biological problems that require lager system size and longer simulation time ever. Simulation time and force field accuracy are also coupled as we often see inaccuracies in force fields as simulation timescales of complex systems are extended. In membrane simulations, one might ask the following challenging questions: Can all-atom molecular simulations predict lateral lipid organization and domain formation? Are the current force fields good for liquid ordered (rafts), ripple, or gel phases? Can simulations reveal specific protein-lipid interactions that activate protein functions? Can all-atom modeling and simulation handle interactions of peripheral membrane proteins with specific lipid types on the membrane surface?
With these questions in mind, we would like to turn into challenges and progress of cellular membrane modeling that requires various lipids and general assembly procedures. Having most (phospho- and sphingo-) lipid types covered, the next challenges are in building biological membranes containing glycolipids such as gangliosides, glycophosphatidylinositol (GPI) linkages, and lipopolysaccharide (LPS in Gram-negative bacterial outer membranes) [210], as the CHARMM force fields already cover a variety of carbohydrates [211-213]. These various types of lipids containing carbohydrates are necessary to model realistic extracellular membrane surface, but it is challenging to model and assemble them together, as glycans come in a diversity of sequences and structures by linking individual sugar units in a multitude of ways. Together Figs. 6 and 7 show the current progress of all-atom modeling and simulation of complex membrane systems [214-216] including LPS and GPI-anchor. It is also now possible to build various glycolipid models in Glycolipid Modeler in CHARMM-GUI (http://www.charmm-gui.org/input/glycolipid), and it will be possible to incorporate Glycolipid Modeler into Membrane Builder in the future to model realistic cellular membranes.
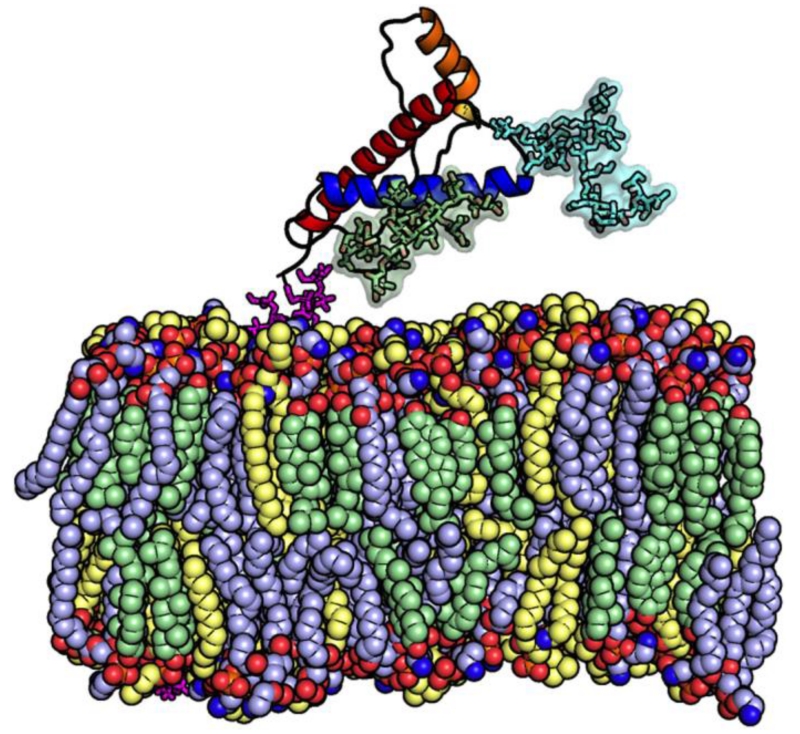
Figure 7. GPI-anchored glycosylated prion protein in raft-like membranes.
Prion protein, N-glycan 1, N-glycan 2, GPI-anchor, cholesterol, POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), and PSM (N-palmitoyl sphingomyelin) are represented in cartoon (HA: orange, HB: blue, HC: red), green surface, blue surface, magenta sticks, light green spheres, light blue spheres, and light yellow spheres, respectively. The bilayer composition is about 1 : 1 : 1 of cholesterol : POPC : PSM. Water molecules and KCl ions are omitted for clarity.
Spatial architecture of chromosome in cell nucleus
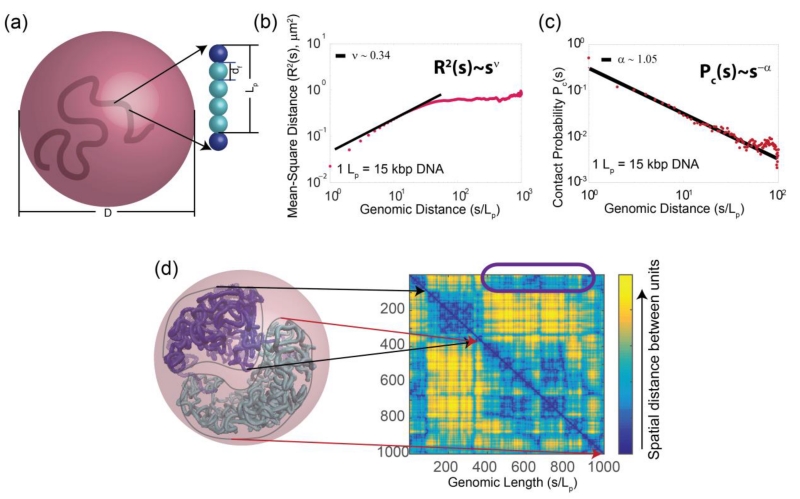
Understanding the spatial organization of chromatin in the cell nucleus is key to gaining insights into the mechanism of gene activities, nuclear functions and maintenance of cellular epigenetic states [217, 218]. Chromosome conformation capture (3C) and related techniques as well as single cell imaging studies have provided a wealth of information on the spatial architecture of the cell nucleus [217, 218]. Ensemble models of 3D structures of chromatins can help decipher physical mechanisms of long-range gene interactions and control of gene expression [219-221]. A challenging task is to infer 3D structures of folded chromosomes from frequency maps measured in 3C-based studies. This bears some resemblance to the problem of inferring protein structures from contact maps obtained from NMR measurements, despite the obvious difference in size and in scale. While chromatin chains are fundamentally different from protein chains and unlikely to fold into a unique native structure, both possess basic physical properties such as constraints on excluded volume, chain connectivity, and spatial confinement. It is likely that techniques developed in studying protein structures will have some relevance in modeling 3D structures of chromosomes. For example, built upon methods developed in protein folding studies [222, 223], the chromosome self-avoiding chain (C-SAC) model and the geometric sequential importance sampling technique were developed (Fig. 8). As a result, the equilibrium ensemble of randomly folded chromosomes in the confined nuclear volume was successfully generated, a challenging task as effective sampling under small volume constraint is extremely difficult. The results explain various experimentally observed scaling properties of spatial distance and looping probability [224]. These results suggest that spatial confinement has dominant effects and offers an alternative interpretation of 3C studies to the earlier fractal globule model and the Strings and Binders Switch (SBS) model [217, 225]. It further suggests that the formation of topological domains can arise spontaneously from basic chain connectivity in severe spatial confinement. It is expected that experimental development such as high-resolution and single-cell Hi-C measurement will provide detailed information to understand how chromosome folding and its dynamic changes related to the control of cellular phenotype during development of individual cells [226]. Further development in modeling 3D chromosome structures will help understand the overall architecture of chromosomes in the nuclear space, identify novel specific spatial interactions among gene elements, and gain mechanistic understanding of changes in the folding landscape of chromosomes which undergoes significant tissue- and developmental stage-specific size and shape changes.
Figure 8. Chromatin chain models and scaling properties.
(a) In the C-SAC model, a chromatin fiber is a self-avoiding polymer chain with a persistence length Lp, consisting of beads with a diameter df, with blue spheres at the boundaries of Lp, cyan spheres interpolated in-between. Chromatin chains are generated by a chain growth algorithm inside a confined space of a diameter D. (b) The scaling of mean-square spatial distance R2(s) in log10 scale derived from 10,000 chains of length 1000Lp in a confinement of diameter D proportional to ~11 8#x00B5;m diameter of an average human cell. R2(s) follows a power law of ~s2ν, with ν ~ 0.34, similar to measured ν of ~ 0.33. (c) The scaling of contact probability Pc(s) follows a power law of ~1/sα, with α ~1.05, similar to the measured α of 1.08. (d) A random C-SAC chromatin chain with two interacting sub-structures that can give rise to topologically associated domains. The two domain-like substructures and their corresponding spatial distance matrices can be seen, where distances between loci are color-coded. Inter-substructure interactions are highlighted in the purple box. Details can be found in Gursoy et al. [224].
Conclusions
Biological science is on the cusp of a new and transformational way to view living systems – the creation of physical molecular models of the fundamental unit of life, the cell. Developing 3D models to account for experimental observations and to predict emergent biological behaviors is key to gaining mechanistic understanding of cellular processes. We described emerging approaches to the structural modeling on a broad scale, from individual molecules to cell biology. The cross section of these diverse approaches covers 3D molecular cell models based on experimental data, genome-wide structural modeling of protein interactions, atomistic modeling of protein-crowder interactions, nonspecific protein interactions, cellular membrane modeling and simulation, and modeling of chromosomes. The list is far from a complete roster of methodologies needed for structural modeling of a cell, and simply represents one sample of approaches and techniques for such modeling.
Structural modeling of a cell complements computational approaches to cellular mechanisms based on differential equations, graph models, and other techniques to model biological networks, imaging data, etc. The structural modeling along with other computational and experimental approaches will provide a fundamental understanding of life at the molecular level and lead to important applications to biology and medicine.
Highlights.
Structural characterization is key to our understanding of biomolecular mechanisms
Modeling of biomolecules and their interactions has been rapidly progressing
The current focus is shifting toward larger systems, up to the level of a cell
The review describes structural approaches to cell modeling and future development
Acknowledgments
WI acknowledges grant support from NIH R01 GM092950, NIH U54 GM087519, NSF MCB1516154, NSF MCB1157677, NSF DBI1145987, and NSF IIA1359530; JL thanks Youfang Cao, Gamze Gursoy, Yun Xu, and Jieling Zhao for their work on chromatin folding, and acknowledges grant support from NIH R01 GM079804, NSF MCB1415589, and the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust; AJO thanks Graham Johnson, Ludovic Autin, Michel Sanner and David Goodsell for their work on cellPACK, and acknowledges grant support from P41 GM103426 an NIH Research Resource (R. Amaro, Director); HXZ acknowledges grant support from NIH R01 GM088187; SV thanks Dima Kozakov for contributing to research on encounter complexes and nonspecific protein-protein interactions, and acknowledges grant support from NIH R01 GM093147, NIH R01 GM061867, and NSF DBI1147082; IAV thanks Petras Kundrotas and Ivan Anishchenko for their work on modeling of protein interactome, and acknowledges grant support from NIH R01GM074255, NSF DBI1262621 and NSF CNS1337899.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP) — round X. Proteins. 2014;82(Suppl 2):1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lensink MF, Wodak SJ. Docking, scoring, and affinity prediction in CAPRI. Proteins. 2013;81:2082–95. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- [3].Tomita M. Whole-cell simulation: A grand challenge of the 21st century. Trends Biotech. 2001;19:205–10. doi: 10.1016/s0167-7799(01)01636-5. [DOI] [PubMed] [Google Scholar]
- [4].Carrera J, Covert MW. Why build whole-cell models? Trends Cell Biol. 2015;25:719–22. doi: 10.1016/j.tcb.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kuhlbrandt W. Cryo-EM enters a new era. eLife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Glaeser RM. How good can cryo-EM become? Nature Methods. 2016;13:28–32. doi: 10.1038/nmeth.3695. [DOI] [PubMed] [Google Scholar]
- [7].Graewert MA, Svergun DI. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS) Curr. Opin. Struct. Biol. 2013;23:748–54. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- [8].Rambo RP, Tainer JA. Super-resolution in solution X-Ray scattering and its applications to structural systems biology. Ann. Rev. Bioph. 2013;42:415–41. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- [9].Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–6. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Barends TR, Foucar L, Ardevol A, Nass K, Aquila A, Botha S, et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015;350:445–50. doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- [11].Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, et al. Putting the pieces together: Integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Takahashi K, Kaizu K, Hu B, Tomita M. A multi-algorithm, multi-timescale method for cell simulation. Bioinformatics. 2004;20:538–46. doi: 10.1093/bioinformatics/btg442. [DOI] [PubMed] [Google Scholar]
- [13].Cowan AE, Moraru II, Schaff JC, Slepchenko BM, Loew LM. Spatial modeling of cell signaling networks. Methods Cell Biol. 2012;110:195–221. doi: 10.1016/B978-0-12-388403-9.00008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- [15].Janin J, Rodier F, Chakrabarti P, Bahadur RP. Macromolecular recognition in the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2007;63:1–8. doi: 10.1107/S090744490603575X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- [17].Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–42. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Juty N, Ali R, Glont M, Keating S, Rodriguez N, Swat MJ, et al. BioModels: Content, features, functionality, and use. CPT Pharmacometrics Syst. Pharmacol. 2015;4:e3. doi: 10.1002/psp4.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–48. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Aurell E, Brown S, Johanson J, Sneppen K. Stability puzzles in phage lambda. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2002;65:051914. doi: 10.1103/PhysRevE.65.051914. [DOI] [PubMed] [Google Scholar]
- [22].Zhu XM, Yin L, Hood L, Ao P. Robustness, stability and efficiency of phage lambda genetic switch: dynamical structure analysis. J. Bioinform. Comput. Biol. 2004;2:785–817. doi: 10.1142/s0219720004000946. [DOI] [PubMed] [Google Scholar]
- [23].Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- [24].Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144:910–25. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang J, Xu L, Wang E, Huang S. The potential landscape of genetic circuits imposes the arrow of time in stem cell differentiation. Biophys. J. 2010;99:29–39. doi: 10.1016/j.bpj.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang J, Zhang K, Xu L, Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:8257–62. doi: 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang B, Wolynes PG. Stem cell differentiation as a many-body problem. Proc. Natl. Acad. Sci. USA. 2014;111:10185–90. doi: 10.1073/pnas.1408561111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ao P, Galas D, Hood L, Zhu X. Cancer as robust intrinsic state of endogenous molecular-cellular network shaped by evolution. Med. Hypotheses. 2008;70:678–84. doi: 10.1016/j.mehy.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 2009;10:336–42. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- [30].Barabasi AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- [31].Wang RS, Saadatpour A, Albert R. Boolean modeling in systems biology: an overview of methodology and applications. Phys. Biol. 2012;9:055001. doi: 10.1088/1478-3975/9/5/055001. [DOI] [PubMed] [Google Scholar]
- [32].Tyson JJ, Chen K, Novak B. Network dynamics and cell physiology. Nat. Rev. Mol. Cell Biol. 2001;2:908–16. doi: 10.1038/35103078. [DOI] [PubMed] [Google Scholar]
- [33].Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an autoregulatory genetic module. Proc. Natl. Acad. Sci. USA. 2003;100:7714–9. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gillespie DT. Stochastic simulation of chemical kinetics. Ann. Rev. Phys. Chem. 2007;58:35–55. doi: 10.1146/annurev.physchem.58.032806.104637. [DOI] [PubMed] [Google Scholar]
- [35].Qian H, Bishop LM. The chemical master equation approach to nonequilibrium steady-state of open biochemical systems: linear single-molecule enzyme kinetics and nonlinear biochemical reaction networks. Int. J. Mol. Sci. 2010;11:3472–500. doi: 10.3390/ijms11093472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liang J, Qian H. Computational cellular dynamics based on the chemical master equation: A challenge for understanding complexity. J. Comput. Sci. Technol. 2010;25:154–68. doi: 10.1007/s11390-010-9312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cao Y, Lu HM, Liang J. Stochastic probability landscape model for switching efficiency, robustness, and differential threshold for induction of genetic circuit in phage lambda. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2008;2008:611–4. doi: 10.1109/IEMBS.2008.4649227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cao Y, Lu HM, Liang J. Probability landscape of heritable and robust epigenetic state of lysogeny in phage lambda. Proc. Natl. Acad. Sci. USA. 2010;107:18445–50. doi: 10.1073/pnas.1001455107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Paulsson J, Berg OG, Ehrenberg M. Stochastic focusing: Fluctuation-enhanced sensitivity of intracellular regulation. Proc. Natl. Acad. Sci. USA. 2000;97:7148–53. doi: 10.1073/pnas.110057697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].McAdams HH, Arkin A. It’s a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 1999;15:65–9. doi: 10.1016/s0168-9525(98)01659-x. [DOI] [PubMed] [Google Scholar]
- [41].Kuwahara H, Mura I. An efficient and exact stochastic simulation method to analyze rare events in biochemical systems. J. Chem. Phys. 2008;129:165101. doi: 10.1063/1.2987701. [DOI] [PubMed] [Google Scholar]
- [42].Roh MK, Daigle BJ, Gillespie DT, Petzold LR. State-dependent doubly weighted stochastic simulation algorithm for automatic characterization of stochastic biochemical rare events. J. Chem. Phys. 2011;135:234108. doi: 10.1063/1.3668100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cao Y, Liang J. Adaptively biased sequential importance sampling for rare events in reaction networks with comparison to exact solutions from finite buffer dCME method. J. Chem. Phys. 2013;139:025101. doi: 10.1063/1.4811286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Munsky B, Khammash M. The finite state projection algorithm for the solution of the chemical master equation. J. Chem. Phys. 2006;124:044104. doi: 10.1063/1.2145882. [DOI] [PubMed] [Google Scholar]
- [45].Peles S, Munsky B, Khammash M. Reduction and solution of the chemical master equation using time scale separation and finite state projection. J. Chem. Phys. 2006;125:204104. doi: 10.1063/1.2397685. [DOI] [PubMed] [Google Scholar]
- [46].Cao Y, Terebus A, Liang J. State space truncation with quantified errors for accurate solutions to discrete chemical master equation. Bull. Math. Biol. 2016 doi: 10.1007/s11538-016-0149-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cao Y, Terebus A, Liang J. Accurate chemical master equation solution method with multi-finite buffers for time-evolving and steady state probability landscapes and first passage times. SIAM Multiscale Modeling and Simulation. 2016 doi: 10.1137/15M1034180. in pess. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Qin S, Zhou HX. Atomistic modeling of macromolecular crowding predicts modest increases in protein folding and binding stability. Biophys. J. 2009;97:12–9. doi: 10.1016/j.bpj.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Qin S, Pang X, Zhou HX. Automated prediction of protein association rate constants. Structure. 2011;19:1744–51. doi: 10.1016/j.str.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Qin S, Cai L, Zhou HX. A method for computing association rate constants of atomistically represented proteins under macromolecular crowding. Phys. Biol. 2012;9:066008. doi: 10.1088/1478-3975/9/6/066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhou HX, Bates PA. Modeling protein association mechanisms and kinetics. Curr. Opin. Struct. Biol. 2013;23:887–93. doi: 10.1016/j.sbi.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comp. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Feig M, Harada R, Mori T, Yue I, Takahashi K, Sugita Y. Complete atomistic model of a bacterial cytoplasm for integrating physics, biochemistry, and systems biology. J. Mol. Graph. Mod. 2015;58:1–9. doi: 10.1016/j.jmgm.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vendeville A, Lariviere D, Fourmentin E. An inventory of the bacterial macromolecular components and their spatial organization. FEMS Microbiol. Rev. 2011;35:395–414. doi: 10.1111/j.1574-6976.2010.00254.x. [DOI] [PubMed] [Google Scholar]
- [55].Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–46. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- [56].Wilhelm BG, Mandad S, Truckenbrodt S, Krohnert K, Schafer C, Rammner B, et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–8. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- [57].Johnson GT, Goodsell DS, Autin L, Forli S, Sanner MF, Olson AJ. 3D molecular models of whole HIV-1 virions generated with cellPACK. Faraday Discuss. 2014;169:23–44. doi: 10.1039/c4fd00017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Johnson GT, Autin L, Al-Alusi M, Goodsell DS, Sanner MF, Olson AJ. cellPACK: A virtual mesoscope to model and visualize structural systems biology. Nature Methods. 2015;12:85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ando T, Skolnick J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc. Natl. Acad. Sci. USA. 2010;107:18457–62. doi: 10.1073/pnas.1011354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mereghetti P, Gabdoulline RR, Wade RC. Brownian dynamics simulation of protein solutions: structural and dynamical properties. Biophys. J. 2010;99:3782–91. doi: 10.1016/j.bpj.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comp. Biol. 2007;3:337–44. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Piehler J. New methodologies for measuring protein interactions in vivo and in vitro. Curr. Opin. Struct. Biol. 2005;15:4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- [63].Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–60. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Douguet D, Chen HC, Tovchigrechko A, Vakser IA. DOCKGROUND resource for studying protein-protein interfaces. Bioinformatics. 2006;22:2612–8. doi: 10.1093/bioinformatics/btl447. [DOI] [PubMed] [Google Scholar]
- [65].Blohm P, Frishman G, Smialowski P, Goebels F, Wachinger B, Ruepp A, et al. Negatome 2.0: A database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis. Nucl. Acids Res. 2014;42:D396–D400. doi: 10.1093/nar/gkt1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Vakser IA. Low-resolution structural modeling of protein interactome. Curr. Opin. Struct. Biol. 2013;23:198–205. doi: 10.1016/j.sbi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lua RC, Marciano DC, Katsonis P, Adikesavan AK, Wilkins AD, Lichtarge O. Prediction and redesign of protein-protein interactions. Prog. Bioph. Mol. Biol. 2014;116:194–202. doi: 10.1016/j.pbiomolbio.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Schwede T. Protein modeling: What happened to the “protein structure gap”? Structure. 2013;21:1531–40. doi: 10.1016/j.str.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lasker K, Sali A, Wolfson HJ. Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins. 2010;78:3205–11. doi: 10.1002/prot.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Vacha R, Frenkel D. Relation between molecular shape and the morphology of self-assembling aggregates: A simulation study. Biophys. J. 2011;100:1432–9. doi: 10.1016/j.bpj.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kundrotas PJ, Vakser IA. Protein-protein alternative binding modes do not overlap. Protein Sci. 2013;22:1141–5. doi: 10.1002/pro.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tovchigrechko A, Vakser IA. How common is the funnel-like energy landscape in protein-protein interactions? Protein Sci. 2001;10:1572–83. doi: 10.1110/ps.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Vakser IA. Low-resolution recognition factors determine major characteristics of the energy landscape in protein-protein interaction. In: Schreiber G, Nussinov R, editors. Computational Protein-Protein Interactions. CRC press; Taylor and Francis: 2009. pp. 21–42. [Google Scholar]
- [74].Trizac E, Levy Y, Wolynes PG. Capillarity theory for the fly-casting mechanism. Proc. Natl. Acad. Sci. USA. 2010;107:2746–50. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ravikumar KM, Huang W, Yang S. Coarse-grained simulations of protein-protein association: An energy landscape perspective. Biophys. J. 2012;103:837–45. doi: 10.1016/j.bpj.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc. Natl. Acad. Sci. USA. 2009;106:19819–23. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vreven T, Moal IH, Vangone A, Pierce BG, Kastritis PL, Torchala M, et al. Updates to the integrated protein-protein interaction benchmarks: Docking Benchmark Version 5 and Affinity Benchmark Version 2. J. Mol. Biol. 2015;427:3031–41. doi: 10.1016/j.jmb.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gao Y, Douguet D, Tovchigrechko A, Vakser IA. DOCKGROUND system of databases for protein recognition studies: Unbound structures for docking. Proteins. 2007;69:845–51. doi: 10.1002/prot.21714. [DOI] [PubMed] [Google Scholar]
- [79].Jiang S, Tovchigrechko A, Vakser IA. The role of geometric complementarity in secondary structure packing: A systematic docking study. Protein Sci. 2003;12:1646–51. doi: 10.1110/ps.0304503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kaczor AA, Selent J, Sanz F, Pastor M. Modeling complexes of transmembrane proteins: Systematic analysis of protein-protein docking tools. Mol. Inf. 2013;32:717–33. doi: 10.1002/minf.201200150. [DOI] [PubMed] [Google Scholar]
- [81].Saunders MG, Voth GA. Coarse-graining of multiprotein assemblies. Curr. Opin. Struct. Biol. 2012;22:144–50. doi: 10.1016/j.sbi.2012.01.003. [DOI] [PubMed] [Google Scholar]
- [82].Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: Bridging between structure and function. Ann. Rev. Bioph. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhang Z, Voth GA. Coarse-grained representations of large biomolecular complexes from low-resolution structural data. J. Chem. Theory Comput. 2010;6:2990–3002. doi: 10.1021/ct100374a. [DOI] [PubMed] [Google Scholar]
- [84].Ruvinsky AM, Vakser IA. Sequence composition and environment effects on residue fluctuations in protein structures. J. Chem. Phys. 2010;133:155101. doi: 10.1063/1.3498743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zen A, Micheletti C, Keskin O, Nussinov R. Comparing interfacial dynamics in protein-protein complexes: An elastic network approach. BMC Struct. Biol. 2010;10:26. doi: 10.1186/1472-6807-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Karaca E, Bonvin AMJJ. Multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes. Structure. 2011;19:555–65. doi: 10.1016/j.str.2011.01.014. [DOI] [PubMed] [Google Scholar]
- [87].Burton B, Zimmermann MT, Jernigan RL, Wang Y. A computational investigation on the connection between dynamics properties of ribosomal proteins and ribosome assembly. PLoS Comp. Biol. 2012;8:e1002530. doi: 10.1371/journal.pcbi.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–99. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- [89].Vakser IA, Matar OG, Lam CF. A systematic study of low-resolution recognition in protein-protein complexes. Proc. Natl. Acad. Sci. USA. 1999;96:8477–82. doi: 10.1073/pnas.96.15.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhou HX, Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins. 2001;44:336–43. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- [91].Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: Test against NMR data. Proteins. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- [92].Zhou HX, Qin S. Interaction-site prediction for protein complexes: A critical assessment. Bioinformatics. 2007;23:2203–9. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- [93].Kundrotas PJ, Vakser IA. Accuracy of protein-protein binding sites in high-throughput template-based modeling. PLoS Comp. Biol. 2010;6:e1000727. doi: 10.1371/journal.pcbi.1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Anishchenko I, Kundrotas PJ, Tuzikov AV, Vakser IA. Protein models: The Grand Challenge of protein docking. Proteins. 2014;82:278–87. doi: 10.1002/prot.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Anishchenko I, Kundrotas PJ, Tuzikov AV, Vakser IA. Protein models docking benchmark 2. Proteins. 2015;83:891–7. doi: 10.1002/prot.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein-protein interfaces. Proteins. 2010;78:3235–41. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. USA. 1992;89:2195–9. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Vakser IA. Protein docking for low-resolution structures. Protein Eng. 1995;8:371–7. doi: 10.1093/protein/8.4.371. [DOI] [PubMed] [Google Scholar]
- [99].Kuzu G, Keskin O, Gursoy A, Nussinov R. Constructing structural networks of signaling pathways on the proteome scale. Curr. Opin. Struct. Biol. 2012;22:367–77. doi: 10.1016/j.sbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- [100].Stein A, Mosca R, Aloy P. Three-dimensional modeling of protein interactions and complexes is going ‘omics. Curr. Opin. Struct. Biol. 2011;21:200–8. doi: 10.1016/j.sbi.2011.01.005. [DOI] [PubMed] [Google Scholar]
- [101].Kar G, Keskin O, Nussinov R, Gursoy A. Human proteome-scale structural modeling of E2-E3 interactions exploiting interface motifs. J. Proteome Res. 2012;11:1196–207. doi: 10.1021/pr2009143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wass MN, David A, Sternberg MJE. Challenges for the prediction of macromolecular interactions. Curr. Opin. Struct. Biol. 2011;21:382–90. doi: 10.1016/j.sbi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- [103].Levitt M. Nature of the protein universe. Proc. Natl. Acad. Sci. USA. 2009;106:11079–84. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc. Natl. Acad. Sci. USA. 2012;109:9438–41. doi: 10.1073/pnas.1200678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Mosca R, Pons C, Fernandez-Recio J, Aloy P. Pushing structural information into the yeast interactome by high-throughput protein docking experiments. PLoS Comp. Biol. 2009;5:e1000490. doi: 10.1371/journal.pcbi.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhu Z, Tovchigrechko A, Baronova T, Gao Y, Douguet D, O’Toole N, et al. Large-scale structural modeling of protein complexes at low resolution. J. Bioinformatics Comp. Biol. 2008;6:789–810. doi: 10.1142/s0219720008003679. [DOI] [PubMed] [Google Scholar]
- [107].Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–9. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- [108].Gao M, Skolnick J. Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc. Natl. Acad. Sci. USA. 2010;107:22517–22. doi: 10.1073/pnas.1012820107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc. Natl. Acad. Sci. USA. 2010;107:10896–901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kundrotas PJ, Zhu Z, Vakser IA. GWIDD: A comprehensive resource for genome-wide structural modeling of protein-protein interactions. Human Genomics. 2012;6:7. doi: 10.1186/1479-7364-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kundrotas PJ, Zhu Z, Vakser IA. GWIDD: Genome-Wide Protein Docking Database. Nucl. Acid Res. 2010;38:D513–D7. doi: 10.1093/nar/gkp944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhou HX. Influence of crowded cellular environments on protein folding, binding, and oligomerization: biological consequences and potentials of atomistic modeling. FEBS Lett. 2013;587:1053–61. doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Ann. Rev. Biophys. 2008;37:375–97. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Miklos AC, Sarkar M, Wang Y, Pielak GJ. Protein crowding tunes protein stability. J. Am. Chem. Soc. 2011;133:7116–20. doi: 10.1021/ja200067p. [DOI] [PubMed] [Google Scholar]
- [115].Phillip Y, Harel M, Khait R, Qin S, Zhou HX, Schreiber G. Contrasting factors on the kinetic path to protein complex formation diminish the effects of crowding agents. Biophys. J. 2012;103:1011–9. doi: 10.1016/j.bpj.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Batra J, Xu K, Qin S, Zhou HX. Effect of macromolecular crowding on protein binding stability: Modest stabilization and significant biological consequences. Biophys. J. 2009;97:906–11. doi: 10.1016/j.bpj.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hatters DM, Minton AP, Howlett GJ. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J. Biol. Chem. 2002;277:7824–30. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- [118].Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. USA. 2005;102:4753–8. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Feig M, Sugita Y. Variable interactions between protein crowders and biomolecular solutes are important in understanding cellular crowding. J. Phys. Chem. B. 2012;116:599–605. doi: 10.1021/jp209302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Qin S, Minh DD, McCammon JA, Zhou HX. Method to predict crowding effects by postprocessing molecular dynamics trajectories: Application to the flap dynamics of HIV-1 protease. J. Phys. Chem. Lett. 2010;1:107–10. doi: 10.1021/jz900023w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Widom B. Some topics in theory of fluids. J. Chem. Phys. 1963;39:2808–12. [Google Scholar]
- [122].Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- [123].Qin S, Zhou HX. An FFT-based method for modeling protein folding and binding under crowding: benchmarking on ellipsoidal and all-atom crowders. J. Chem. Theory Comput. 2013;9:4633–43. doi: 10.1021/ct4005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Qin S, Zhou HX. Further development of the FFT-based method for atomistic modeling of protein folding and binding under crowding: Optimization of accuracy and speed. J. Chem. Theory Comput. 2014;10:2824–35. doi: 10.1021/ct5001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Wu D, Minton AP. Quantitative characterization of nonspecific self- and hetero-interactions of proteins in nonideal solutions via static light scattering. J. Phys. Chem. B. 2015;119:1891–8. doi: 10.1021/jp510918d. [DOI] [PubMed] [Google Scholar]
- [126].Bodart JF, Wieruszeski JM, Amniai L, Leroy A, Landrieu I, Rousseau-Lescuyer A, et al. NMR observation of Tau in Xenopus oocytes. J. Magn. Reson. 2008;192:252–7. doi: 10.1016/j.jmr.2008.03.006. [DOI] [PubMed] [Google Scholar]
- [127].Augustus AM, Reardon PN, Spicer LD. MetJ repressor interactions with DNA probed by in-cell NMR. Proc. Natl. Acad. Sci. USA. 2009;106:5065–9. doi: 10.1073/pnas.0811130106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Luh LM, Hansel R, Lohr F, Kirchner DK, Krauskopf K, Pitzius S, et al. Molecular crowding drives active pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition. J. Am. Chem. Soc. 2013;135:13796–803. doi: 10.1021/ja405244v. [DOI] [PubMed] [Google Scholar]
- [129].Miklos AC, Sumpter M, Zhou HX. Competitive interactions of ligands and macromolecular crowders with maltose binding protein. PLoS One. 2013;8:e74969. doi: 10.1371/journal.pone.0074969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Latham MP, Kay LE. Is buffer a good proxy for a crowded cell-like environment? A comparative NMR study of calmodulin side-chain dynamics in buffer and E. coli lysate. PLoS One. 2012;7:e48226. doi: 10.1371/journal.pone.0048226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].O’Connell JD, Zhao A, Ellington AD, Marcotte EM. Dynamic reorganization of metabolic enzymes into intracellular bodies. Ann. Rev. Cell. Dev. Biol. 2012;28:89–111. doi: 10.1146/annurev-cellbio-101011-155841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–32. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- [133].Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4334–9. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Ann. Rev. Cell. Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- [135].Garber K. CELL BIOLOGY. Protein ‘drops’ may seed brain disease. Science. 2015;350:366–7. doi: 10.1126/science.350.6259.366. [DOI] [PubMed] [Google Scholar]
- [136].Strzyz P. Molecular networks: Protein droplets in the spotlight. Nat. Rev. Mol. Cell Biol. 2015;16:639. doi: 10.1038/nrm4070. [DOI] [PubMed] [Google Scholar]
- [137].Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–40. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Petrovska I, Nuske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife. 2014 doi: 10.7554/eLife.02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Feig M, Sugita Y. Reaching new levels of realism in modeling biological macromolecules in cellular environments. J. Mol. Graph. Model. 2013;45:144–56. doi: 10.1016/j.jmgm.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Kuznetsova IM, Turoverov KK, Uversky VN. What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 2014;15:23090–140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Breydo L, Reddy KD, Piai A, Felli IC, Pierattelli R, Uversky VN. The crowd you’re in with: Effects of different types of crowding agents on protein aggregation. Biochim. Biophys. Acta. 2014;1844:346–57. doi: 10.1016/j.bbapap.2013.11.004. [DOI] [PubMed] [Google Scholar]
- [142].Kozakov D, Li K, Hall DR, Beglov D, Zheng J, Vakili P, et al. Encounter complexes and dimensionality reduction in protein-protein association. eLife. 2014;3:e01370. doi: 10.7554/eLife.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Alsallaq R, Zhou HX. Electrostatic rate enhancement and transient complex of protein-protein association. Proteins. 2008;71:320–35. doi: 10.1002/prot.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Fawzi NL, Doucleff M, Suh JY, Clore GM. Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. USA. 2010;107:1379–84. doi: 10.1073/pnas.0909370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins. 2013;81:2159–66. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Vajda S, Hall DR, Kozakov D. Sampling and scoring: A marriage made in heaven. Proteins. 2013;81:1874–84. doi: 10.1002/prot.24343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Clore GM. Visualizing lowly-populated regions of the free energy landscape of macromolecular complexes by paramagnetic relaxation enhancement. Mol. Biosyst. 2008;4:1058–69. doi: 10.1039/b810232e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–39. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Garrett DS, Seok YJ, Peterkofsky A, Gronenborn AM, Clore GM. Solution structure of the 40,000 M-r phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nature Struct. Biol. 1999;6:166–73. doi: 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- [150].Camacho CJ, Kimura SR, DeLisi C, Vajda S. Kinetics of desolvation-mediated protein-protein binding. Biophys. J. 2000;78:1094–105. doi: 10.1016/S0006-3495(00)76668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- [152].Iwatsubo T, Yamaguchi H, Fujimuro M, Yokosawa H, Ihara Y, Trojanowski JQ, et al. Purification and characterization of Lewy bodies from the brains of patients with diffuse Lewy body disease. Am. J. Pathol. 1996;148:1517–29. [PMC free article] [PubMed] [Google Scholar]
- [153].Agrawal NJ, Kumar S, Wang X, Helk B, Singh SK, Trout BL. Aggregation in protein-based biotherapeutics: Computational studies and tools to identify aggregation-prone regions. J. Pharm. Sci. 2011;100:5081–95. doi: 10.1002/jps.22705. [DOI] [PubMed] [Google Scholar]
- [154].Thangakani AM, Nagarajan R, Kumar S, Sakthivel R, Velmurugan D, Gromiha MM. CPAD, Curated Protein Aggregation Database: A repository of manually curated experimental data on protein and peptide aggregation. PLoS One. 2016;11:e0152949. doi: 10.1371/journal.pone.0152949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Wang X, Quinn PJ. Endotoxins: Lipopolysaccharides of gram-negative bacteria. In: Wang X, Quinn PJ, editors. Endotoxins: Structure, Function and Recognition, Subcellular Biochemistry. 2010/07/02 ed. Springer Science+Business Media B.V.; Dordrecht: 2010. pp. 3–25. [DOI] [PubMed] [Google Scholar]
- [156].van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nature Rev. Mol. Cell. Biol. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Jordan JD, Landau EM, Iyengar R. Signaling networks: The origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Hunter T. Signaling--2000 and beyond. Cell. 2000;100:113–27. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- [159].Khademi S, O’Connell JD, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science. 2004;305:1587–94. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- [160].Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, et al. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- [161].Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–22. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- [162].Yellen G. The voltage-gated potassium channels and their relatives. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- [163].Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–6. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- [164].Dong J, Yang G, McHaourab HS. Structural basis of energy transduction in the transport cycle of MsbA. Science. 2005;308:1023–8. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- [165].Elston T, Wang H, Oster G. Energy transduction in ATP synthase. Nature. 1998;391:510–3. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- [166].Alberts B. Molecular biology of the cell. 4th ed. Garland Science; New York: 2002. [Google Scholar]
- [167].Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–38. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Terstappen GC, Reggiani A. In silico research in drug discovery. Trends Pharmacol. Sci. 2001;22:23–6. doi: 10.1016/s0165-6147(00)01584-4. [DOI] [PubMed] [Google Scholar]
- [169].Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: an energetic perspective. Ann. Rev. Biophys. Biomol. Struct. 2007;36:107–30. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- [170].Kim T, Im W. Revisiting hydrophobic mismatch with free energy simulation studies of transmembrane helix tilt and rotation. Biophys. J. 2010;99:175–83. doi: 10.1016/j.bpj.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Sandermann H. Regulation of membrane enzymes by lipids. Biochim. Biophys. Acta. 1978;515:209–37. doi: 10.1016/0304-4157(78)90015-1. [DOI] [PubMed] [Google Scholar]
- [172].McElhaney RN. The influence of membrane lipid composition and physical properties of membrane structure and function in Acholeplasma Laidlawii. Crit. Rev. Microbiol. 1989;17:1–32. doi: 10.3109/10408418909105720. [DOI] [PubMed] [Google Scholar]
- [173].Bienvenue A, Marie JS. Modulation of protein function by lipids. In: Dick H, editor. Current Topics in Membranes. Academic Press; 1994. pp. 319–54. [Google Scholar]
- [174].Dowhan W. Molecular basis for membrane phospholipid diversity: Why are there so many lipids? Ann. Rev. Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- [175].Lee AG. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- [176].Kucerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J, Sachs JN. The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering. Biophys. J. 2008;95:2792–805. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lin M, Gessmann D, Naveed H, Liang J. Outer membrane protein folding and topology from a computational transfer free energy scale. J. Am. Chem. Soc. 2016 doi: 10.1021/jacs.5b10307. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Naveed H, Xu Y, Jackups R, Liang J. Predicting three-dimensional structures of transmembrane domains of beta-barrel membrane proteins. J. Am. Chem. Soc. 2012;134:1775–81. doi: 10.1021/ja209895m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Ann. Rev. Bioph. 2013;42:361–92. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Khalili-Araghi F, Gumbart J, Wen PC, Sotomayor M, Tajkhorshid E, Schulten K. Molecular dynamics simulations of membrane channels and transporters. Curr. Opin. Struct. Biol. 2009;19:128–37. doi: 10.1016/j.sbi.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Stansfeld PJ, Sansom MS. Molecular simulation approaches to membrane proteins. Structure. 2011;19:1562–72. doi: 10.1016/j.str.2011.10.002. [DOI] [PubMed] [Google Scholar]
- [182].Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–42. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Wan CK, Han W, Wu YD. Parameterization of PACE force field for membrane environment and simulation of helical peptides and helix-helix association. J. Chem. Theory Comput. 2012;8:300–13. doi: 10.1021/ct2004275. [DOI] [PubMed] [Google Scholar]
- [184].Qi Y, Cheng X, Han W, Jo S, Schulten K, Im W. CHARMM-GUI PACE CG builder for solution, micelle, and bilayer coarse-grained simulations. J. Chem. Inf. Model. 2014;54:1003–9. doi: 10.1021/ci500007n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Ingolfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, et al. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014;136:14554–1459. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- [186].Qi Y, Ingolfsson HI, Cheng X, Lee J, Marrink SJ, Im W. CHARMM-GUI martini maker for coarse-grained simulations with the martini force field. J. Chem. Theory Comput. 2015;11:4486–94. doi: 10.1021/acs.jctc.5b00513. [DOI] [PubMed] [Google Scholar]
- [187].Ohkubo YZ, Pogorelov TV, Arcario MJ, Christensen GA, Tajkhorshid E. Accelerating membrane insertion of peripheral proteins with a novel membrane mimetic model. Biophys. J. 2012;102:2130–9. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Qi Y, Cheng X, Lee J, Vermaas JV, Pogorelov TV, Tajkhorshid E, et al. CHARMM-GUI HMMM builder for membrane simulations with the highly mobile membrane-mimetic model. Biophys. J. 2015;109:2012–22. doi: 10.1016/j.bpj.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Vermaas JV, Baylon JL, Arcario MJ, Muller MP, Wu Z, Pogorelov TV, et al. Efficient exploration of membrane-associated phenomena at atomic resolution. J. Membr. Biol. 2015;248:563–82. doi: 10.1007/s00232-015-9806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–8. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Schmidt TH, Kandt C. LAMBADA and InflateGRO2: Efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J. Chem. Inf. Model. 2012;52:2657–69. doi: 10.1021/ci3000453. [DOI] [PubMed] [Google Scholar]
- [194].Sommer B, Dingersen T, Gamroth C, Schneider SE, Rubert S, Kruger J, et al. CELLmicrocosmos 2.2 MembraneEditor: A modular interactive shape-based software approach to solve heterogeneous membrane packing problems. J. Chem. Inf. Model. 2011;51:1165–82. doi: 10.1021/ci1003619. [DOI] [PubMed] [Google Scholar]
- [195].Wolf MG, Hoefling M, Aponte-Santamaria C, Grubmuller H, Groenhof G. g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 2010;31:2169–74. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- [196].Kutzner C, Van der Spoel D, Fechner M, Lindahl E, Schmitt UW, De Groot BL, et al. Software news and update - Speeding up parallel GROMACS on high-latency networks. J. Comput. Chem. 2007;28:2075–84. doi: 10.1002/jcc.20703. [DOI] [PubMed] [Google Scholar]
- [197].Martinez L, Andrade R, Birgin EG, Martinez JM. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009;30:2157–64. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- [198].Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–65. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- [199].Woolf TB, Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipd bilayer. Proc. Natl. Acad. Sci. USA. 1994;91:11631–5. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Im W, Roux B. Ions and counterions in a biological channel: A molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 2002;319:1177–97. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- [201].Ghahremanpour MM, Arab SS, Aghazadeh SB, Zhang J, van der Spoel D. MemBuilder: A web-based graphical interface to build heterogeneously mixed membrane bilayers for the GROMACS biomolecular simulation program. Bioinformatics. 2014;30:439–41. doi: 10.1093/bioinformatics/btt680. [DOI] [PubMed] [Google Scholar]
- [202].Wassenaar TA, Ingolfsson HI, Bockmann RA, Tieleman DP, Marrink SJ. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015;11:2144–55. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- [203].Staritzbichler R, Anselmi C, Forrest LR, Faraldo-Gomez JD. GRIFFIN: A versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J. Chem. Theory Comput. 2011;7:1167–76. doi: 10.1021/ct100576m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Brooks BR, Brooks CL, III, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- [207].Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Eastman P, Pande VS. Efficient nonbonded interactions for molecular dynamics on a graphics processing unit. J. Comput. Chem. 2010;31:1268–72. doi: 10.1002/jcc.21413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory. Comput. 2016;12:405–13. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Wu EL, Engstrom O, Jo S, Stuhlsatz D, Yeom MS, Klauda JB, et al. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–55. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, et al. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008;29:2543–64. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Guvench O, Hatcher E, Venable RM, Pastor RW, MacKerell AD. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–70. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Hatcher E, Guvench O, MacKerell AD. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–76. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Wu EL, Fleming PJ, Yeom MS, Widmalm G, Klauda JB, Fleming KG, et al. E. coli outer membrane and interactions with OmpLA. Biophys. J. 2014;106:2493–502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Wu EL, Qi Y, Park S, Mallajosyula SS, MacKerell AD, Klauda JB, et al. Insight into early-stage unfolding of GPI-anchored human prion protein. Biophys. J. 2015;109:2090–100. doi: 10.1016/j.bpj.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Patel DS, Re S, Wu EL, Qi Y, Klebba PE, Widmalm G, et al. Dynamics and interactions of OmpF and LPS: Influence on pore accessibility and ion permeability. Biophys. J. 2016 doi: 10.1016/j.bpj.2016.01.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Hu M, Deng K, Qin Z, Dixon J, Selvaraj S, Fang J, et al. Bayesian inference of spatial organizations of chromosomes. PLoS Comp. Biol. 2013;9:e1002893. doi: 10.1371/journal.pcbi.1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert JP, et al. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–88. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Varoquaux N, Ay F, Noble WS, Vert JP. A statistical approach for inferring the 3D structure of the genome. Bioinformatics. 2014;30:i26–33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Beglov D, Hall D, Brenke R, Shapovalov MV, Dunbrack RL, Kozakov D, et al. Minimal ensembles of side chain conformers for modeling protein-protein interactions. Proteins. 2011;80:591–601. doi: 10.1002/prot.23222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chemistry & Biology. 2014;21:1102–14. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Gursoy G, Xu Y, Kenter AL, Liang J. Spatial confinement is a major determinant of the folding landscape of human chromosomes. Nucl. Acids Res. 2014;42:8223–30. doi: 10.1093/nar/gku462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, Pombo A, et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA. 2012;109:16173–8. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucl. Acids Res. 2005;33:D418–D24. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucl. Acids Res. 2004;32:D449–D51. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]