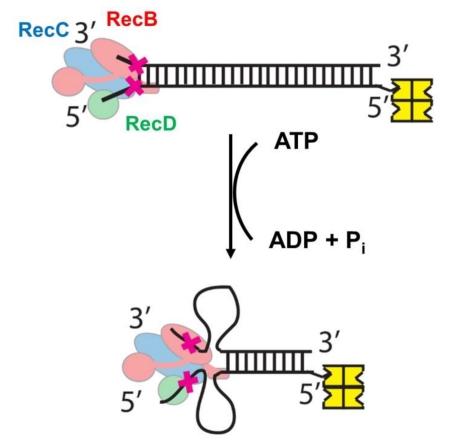
Abstract
Escherichia coli RecBCD is a DNA helicase/nuclease that functions in double-stranded DNA break repair. RecBCD possesses two motors (RecB, a 3′ to 5′ translocase, and RecD, a 5′ to 3′ translocase). Current DNA unwinding models propose that motor translocation is tightly coupled to base pair (bp) melting. However, some biochemical evidence suggests that DNA melting of multiple bp may occur separately from single stranded DNA translocation. To test this hypothesis, we designed DNA substrates containing reverse backbone polarity (RP) linkages that prevent ssDNA translocation of the canonical RecB and RecD motors. Surprisingly, we find that RecBCD can processively unwind DNA for at least 80 bp beyond the RP linkages. This ability requires an ATPase active RecB motor, the RecB “arm” domain and also the RecB nuclease domain, but not its nuclease activity. These results indicate that RecBCD can unwind duplex DNA processively in the absence of ssDNA translocation by the canonical motors and that the nuclease domain regulates the helicase activity of RecBCD.
Keywords: allostery, fluorescence, recombination, SF1 helicase
Graphical abstract
Introduction
E. coli RecBCD is a rapid and highly processive helicase/nuclease that functions to degrade foreign DNA and to repair double stranded DNA breaks[1, 2]. The RecBCD hetero-trimer[3] is a bipolar helicase composed of two superfamily 1 (SF1) ssDNA translocating motors, RecB (134 kDa), a 3′ to 5′ directed ATPase motor, and RecD (67 kDa), a 5′ to 3′ directed ATPase motor[4-6] (see Fig. 1A). Although the RecB and RecD canonical motors translocate with opposite directionalities along single stranded (ss) DNA, they unwind DNA in the same net direction within RecBCD by translocating along the complementary DNA strands of the duplex. RecB also contains a ~30 kDa nuclease domain attached to the motor domain via a ~60 amino acid linker[7-9]. RecC (129 kDa) is a processivity factor that interacts with both RecB and RecD and is structurally homologous to RecB but with non-functional helicase and nuclease domains[3, 10].
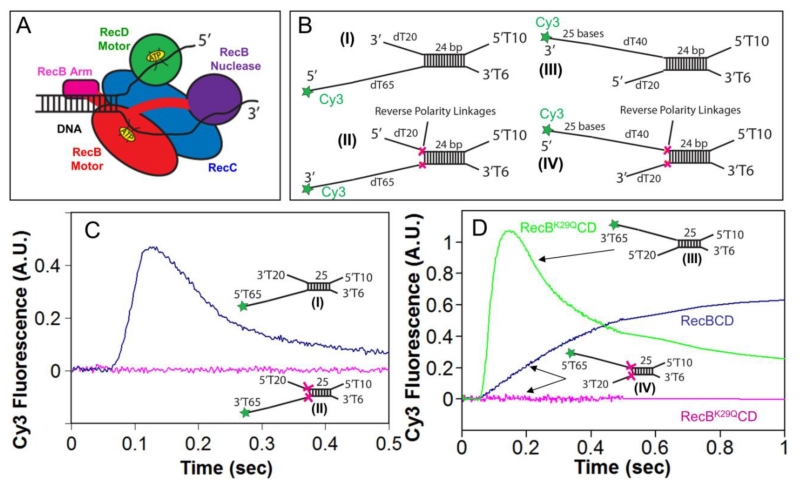
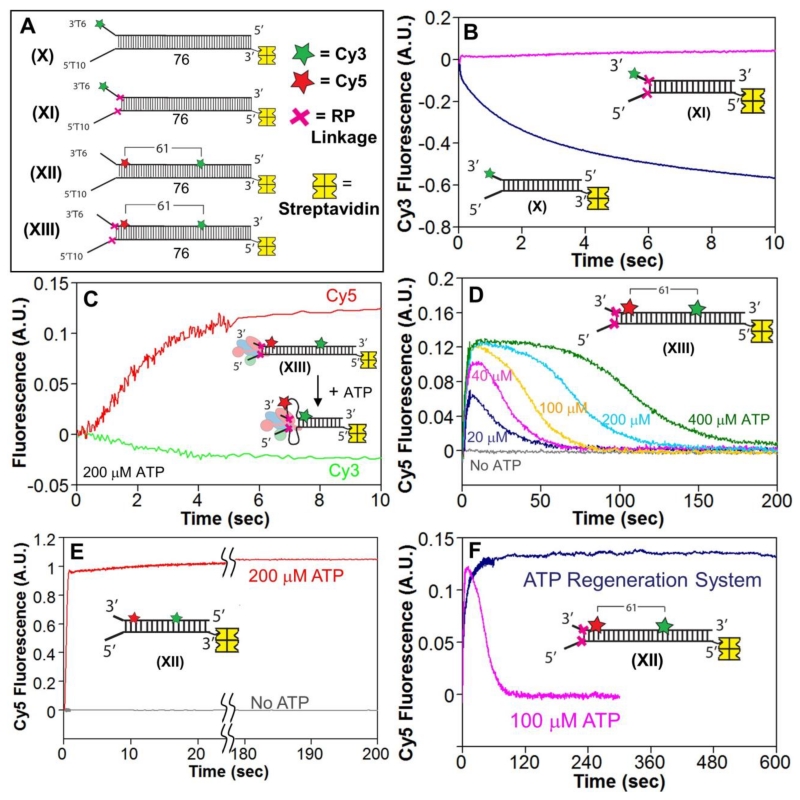
Figure 1. Reversal of the sugar-phosphate backbone polarity prevents ssDNA translocation of the RecB and RecD canonical motors.
A. Cartoon of RecBCD bound to a dsDNA end based on a crystal structure[3]. RecB, RecC, and RecD are shown in red, blue, and green, respectively, with the RecB arm and nuclease domains shown in pink and purple.
B. DNA substrates used to test the effects of reverse polarity (RP) linkages on ssDNA translocation. The red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages. A green star indicates the position of a Cy3 fluorophore.
C. Stopped-flow time courses monitoring Cy3 fluorescence were obtained by pre-incubating RecBCD (18.75 nM) with DNA I (blue trace) or DNA II (red trace) (25 nM) in Buffer M (250 mM NaCl) with ATP (1 mM) and heparin (8 mg/mL) in buffer M (8 mM NaCl) at 1:10 volumetric ratio yielding a final NaCl concentration of 30 mM (concentrations of RecBCD and DNA listed are after mixing) at 25° C.
D. Stopped-flow time courses monitoring Cy3 fluorescence performed as described for panel C. RecBK29QCD pre-incubated with DNA III (green trace) or DNA IV (red trace); RecBCD pre-incubated with DNA IV (blue trace).
To initiate dsDNA break repair, RecBCD first binds to and unwinds from the DNA end using its helicase activity[1, 2]. A current model is that the RecD motor initially moves faster and acts as the lead helicase[5, 11] while the RecB nuclease activity degrades both DNA strands, with a preference for the 3′-ssDNA end. Once the enzyme recognizes a “chi” (crossover hotspot instigator) sequence (5′-GCTGGTGG) within the unwound 3′-ssDNA, RecBCD pauses then continues to unwind DNA at a ~ two-fold reduced rate, with the RecB motor now acting as the lead helicase[11]. Also, the nuclease selectivity of RecB switches to act exclusively on the 5′-ssDNA end, producing a 3′-ssDNA end onto which RecA enzyme is loaded. The resulting RecA-ssDNA filament then initiates homologous recombination to repair the dsDNA break[1, 2]. In the absence of RecD, RecBC forms a stable hetero-dimer that retains highly processive and rapid helicase activity, but is no longer influenced by a chi site and loads RecA constitutively[5, 12-16].
Although it had been proposed that the motor activities of RecD and RecB are independent and uncoupled with RecD being the faster motor before chi recognition and RecB being the faster motor after chi recognition[11], our recent findings[15, 17, 18] suggest a more complex scenario. We found that RecBC (without RecD) possesses two distinct translocase activities that are controlled by the single RecB motor[15, 17]. The primary translocase enables RecBC to move along ssDNA in the 3’ to 5’ direction, consistent with the directionality of the canonical RecB motor, while a secondary translocase facilitates translocation along the other DNA strand, although this secondary translocase is not sensitive to the polarity of the ssDNA backbone[15]. As such, RecBC can move along two unpaired strands of ssDNA at the same rate in a concerted mechanism in which both translocases are tightly coupled to ATP hydrolysis by the RecB motor[15, 17]. This secondary translocase activity, that might represent a double stranded DNA translocase activity[15], also functions within RecBCD, hence the RecB and RecD motors are functionally, but asymmetrically, coupled due to the action of the secondary RecBC translocase[18]. That is, before chi, RecB regulates both 3’ to 5’ and 5’ to 3’ translocation, whereas RecD regulates only 5’ to 3’ translocation[18]. The region of RecBC responsible for the secondary translocase activity has not been identified, although the RecB arm that interacts with the duplex DNA in a crystal structure is a prime candidate[15]. Furthermore, the secondary RecBC translocase activity might actually represent a double stranded DNA translocase activity that functions during DNA unwinding[15].
RecBCD and RecBC are active helicases[19-22] that participate directly in base pair (bp) melting[17, 23]. Most structural models for DNA unwinding by helicases[24] propose that bp melting occurs as the ssDNA translocase motor pulls the duplex DNA across a protein wedge or pin positioned at the ss/dsDNA junction. In the case of RecBCD, the proposed pin is located in RecC with RecB pulling on the 3′ strand and RecD pulling on the 5′ strand[3]. In these models, bp disruption is tightly coupled to ssDNA translocation. However, there is evidence that bp melting and ssDNA translocation might occur as separate processes[17, 25, 26]. Upon binding of a blunt-ended DNA, both RecBCD[27] and RecBC[28] can use their binding free energy to melt 6 base pairs in an ATP-independent reaction. In addition, a kinetic step size of 4±2 bp has been measured for RecBCD[25, 26, 29] and RecBC[14] during processive DNA unwinding. Furthermore, a recent single molecule study has shown that RecBCD facilitates a dynamic opening and closing of ~ 4 bp of duplex DNA in an ATP-independent reaction[23]. Finally, the amount of ATP hydrolyzed by RecBC during processive ssDNA translocation is the same as during processive DNA unwinding[17], suggesting that most of the ATP is used for translocation rather than DNA unwinding.
In this report we find that RecBCD is able to unwind duplex DNA in the absence of translocation by the RecB and RecD motors. For this purpose we used DNA substrates that contain a block in each strand that prevents ssDNA translocation of the canonical RecB and RecD motors. Surprisingly, processive unwinding occurs for at least 80 bp beyond the blocks indicating that DNA unwinding does not occur by the RecB and RecD motors pulling the duplex DNA across a pin or wedge.
Results
ssDNA translocation of the canonical RecB and RecD motors within RecBCD is blocked by reversing the phosphodiester backbone polarity of the DNA
The canonical SF1 ATPase motors translocate with strict directionality along ssDNA[30-34], either 3′ to 5′ as for RecB[12, 15] or 5′ to 3′ as for RecD[5, 6]. We synthesized ssDNA in which the polarity of the phosphodiester backbone is reversed at a unique point in the chain by inserting either a 5′-5′ linkage or a 3′-3′ linkage[35] (Supp. Fig. S1), resulting in ssDNA possessing either two 3′ ends or two 5′ ends[35]. We refer to these as reversed polarity (RP) linkages. Insertion of a 5′-5′ RP linkage within ssDNA blocks ssDNA translocation of the canonical RecB motor within RecBC[15] and RecBCD[18]. Similarly, insertion of a 3′-3′ RP linkage within ssDNA blocks ssDNA translocation of the canonical RecD motor within RecBCD[18]. A demonstration of this is shown in Fig. 1C for RecB and in Fig. 1D for RecD.
We have previously used DNA substrates of the type shown in Figure 1B (I and III) to estimate the ensemble rates of RecBC and RecBCD translocation along ssDNA[15, 18]. These substrates consist of a short 24 bp duplex containing a high affinity RecBCD binding site on one end (unpaired 3′-(dT)6 and 5′-(dT)10 ssDNA tails)[14, 15, 18, 36]. The other end of the duplex possesses oligodeoxythymidylate ssDNA of length L (dTL) extending from either one or both ends of the duplex. A Cy3 fluorophore at the end of the (dT)L, which displays a fluorescence intensity enhancement upon interacting with RecBC or RecBCD, serves to monitor arrival of the ssDNA translocase at the DNA end[15, 18, 31, 37-40]. In such experiments, pre-incubation of an excess of DNA (25 nM final concentration) over RecBCD (18.75 nM final concentration) in Buffer M with 250 mM NaCl ensures that RecBCD binds preferentially to the 5’T10/3’T6 site and not the ssDNA extensions[18]. This is then mixed in a 1:10 volumetric ratio in a stopped-flow apparatus, with a second syringe containing ATP (Buffer M, 8 mM NaCl, for a final NaCl concentration 30 mM) to start the reaction and an excess of heparin that serves to trap any free enzyme or enzyme that dissociates from the DNA during the reaction. The helicase first unwinds the 24 bp duplex and then continues to translocate along the ssDNA (dTL) extensions until the Cy3 is reached. The time course for such a single round reaction displays a lag phase before the onset of Cy3 enhancement. The time of the lag phase for a translocase that initiates at a unique site on the DNA, as in this case, is proportional to the ssDNA length, L, that is labeled with Cy3[15, 18, 37, 38].
A representative time course monitoring ssDNA translocation of RecBCD in the 3′ to 5′ direction is shown in Fig. 1C (blue curve). It shows a lag phase followed by enhancement of Cy3 fluorescence demonstrating that the RecB motor reaches the Cy3 fluorophore at the 5′ end of DNA I. The peak in Cy3 fluorescence is followed by a decrease in fluorescence reflecting RecBCD dissociation from the Cy3 end. However, when DNA II is used that possesses a 5′-5′ RP linkage in the strand along which the canonical RecB motor translocates, no Cy3 enhancement is observed indicating that the RP linkage block translocation of the RecB motor.
To determine whether the 5′ to 3′ directional RecD motor is blocked by a 3′-3′ RP linkage, we examined the RecBK29QCD enzyme on DNA III and IV (Fig. 1B). The K29Q mutation in RecB eliminates its ATPase activity[41] and thus its primary 3′ to 5′ translocation activity as well as its secondary translocation activity[18]. With this mutant we monitor only ssDNA translocation by the RecD motor. The time course for RecBK29QCD on normal DNA (III) shows the characteristics of 5’ to 3’translocation by the RecD motor (Fig. 1D). However, when RecBK29QCD initiates on DNA IV containing a 3′-3′ RP linkage in the strand along which the RecD motor initiates, translocation of the RecD motor is blocked.
Experiments performed with wild type RecBCD and DNA IV show a Cy3 fluorescence enhancement reflecting ssDNA translocation activity (Fig. 1D). This is due to the action of the secondary translocase driven by the RecB motor acting within RecBCD as shown previously[15, 18]. This secondary translocase activity is able to move along ssDNA regardless of the backbone polarity[15, 18]. We note that no Cy3 fluorescence decrease is observed after the Cy3 fluorescence enhancement indicating no dissociation of wt RecBCD from DNA IV (Figure 1D). This is due to the fact that the RecBCD motors become physically stuck behind the two RP linkages. Thus, an RP linkage prevents ssDNA translocation by the canonical RecB and RecD motors, but not the secondary translocase that is driven by the RecB motor within the context of RecBC or RecBCD. Further demonstrations of this are presented below.
RecBCD can melt duplex DNA beyond the reversed polarity linkages
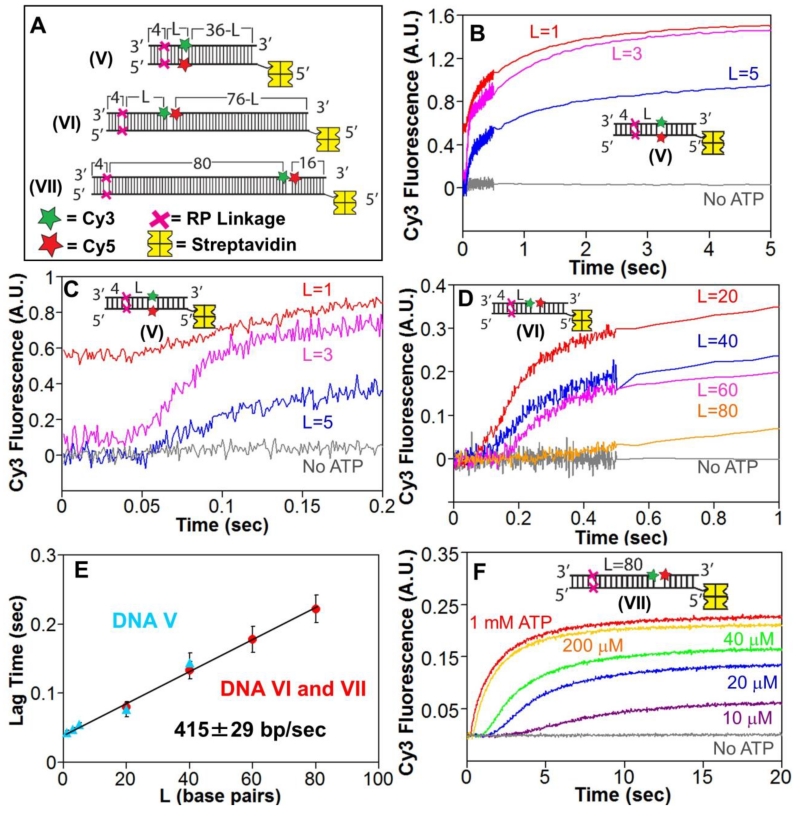
To examine whether RecBCD is able to melt duplex DNA when the canonical RecB and RecD motors are blocked by RP linkages, we examined DNA substrates (Fig. 2A) containing an RP linkage in each strand four bp in from the blunt end. These DNA also contain Cy3-Cy5 fluorophores, positioned “L” bp after the RP linkages, either within the backbone of the complementary DNA strands (DNA V) of across a nick in one strand (DNA VI and VII). In all depictions of DNA substrates in the Figures, a red X indicates the positions of the RP linkages, a green star represents Cy3, and a red star represents Cy5. The Cy3-Cy5 fluorophores undergo Fö rster Resonance Energy Transfer (FRET) when in close proximity, thus any DNA melting that increases the distance between Cy3 and Cy5 should result in an anti-correlated increase in Cy3 fluorescence and decrease in Cy5 fluorescence (loss of FRET). A biotin molecule bound with streptavidin (yellow) is attached to the opposite end of the dsDNA substrate, thus blocking RecBCD from initiating unwinding from that end (Supp. Fig. S2A).
Figure 2. RecBCD can unwind DNA when canonical motor translocation is blocked by RP linkages.
A. DNA used to test if RecBCD can unwind DNA when ssDNA translocation is blocked by RP linkages. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages, separated by L bp from a Cy3 (green star) and Cy5 (red star) fluorophores. One end of each DNA is biotinylated so that streptavidin (yellow) can bind and block binding of RecBCD.
B. Stopped-flow time courses were obtained by mixing pre-incubated RecBCD (15 nM) and DNA V (20 nM) with ATP (5 mM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C. Cy3 fluorescence was excited at 505 nm and Cy3 and Cy5 fluorescence were monitored simultaneously. Cy3 fluorescence time courses are shown for DNA V with L=1 bp (red), L=3 bp (pink) and L=5 bp (blue).
C. The first 0.2 seconds of the time courses shown in panel B showing increase in lag time with increasing L.
D. Stopped-flow time courses showing RecBCD unwinding of DNA VI and DNA VII. Experiments were performed as described in panel B. Cy3 time courses for DNA series VI with L=20 bp (red), L=40 bp (blue), L=60 bp (pink) and DNA VII (L=80 bp).
E. Lag times for RecBCD unwinding of RP DNA (5 mM ATP) with Cy3/Cy5 positioned in the DNA backbone (DNA series V) (blue triangles) or across a nick (DNA VI and VII) (red).
F. ATP-dependence of RecBCD unwinding of DNA VII. Experiments were performed as described in panel B, at the indicated [ATP].
Stopped-flow experiments were performed by pre-incubating RecBCD with DNA V (L= 1, 3 or 5 bp) in one syringe (Buffer M, 30 mM NaCl) and mixing with ATP and heparin to start the reaction. The time courses show a lag phase followed by an increase in Cy3 fluorescence for all three DNA V substrates (Fig. 2B and C). There was also a concomitant decrease in Cy5 fluorescence indicating a loss of FRET efficiency (Supp. Fig. S2B), consistent with DNA melting. No FRET change was observed in the absence of ATP. The Cy5 fluorescence signal also showed a transient increase in fluorescence intensity before its decrease (Supp. Fig. S2B). As observed previously[29], the initial increase in Cy5 fluorescence is due to a RecBCD-induced enhancement of Cy3 fluorescence that occurs before DNA melting and is transferred to Cy5. The lag times increased with increasing number of bp (1, 3 and 5 bp) separating the RP linkages and the Cy3/Cy5 fluorophores (Fig. 2C) indicating that DNA melting occurs progressively from the DNA end at which RecBCD initiates to beyond the RP linkages.
When RecBCD is bound to a DNA end it covers 20-22 bp[3, 42]. Furthermore, upon binding to a blunt DNA end, RecBCD is able to melt 5-6 bp with ATP binding or hydrolysis[3, 27, 36], hence it was not surprising that RecBCD could melt 5 bp of DNA beyond the RP linkages. As a control, we examined DNA V in which the Cy3/Cy5 FRET pair was positioned 20 or 40 bp beyond the RP linkages. For the L=40 bp DNA, the FRET pair is well outside the 20-22 bp footprint of the RecBCD. Unexpectedly, upon addition of ATP, we also observed anti-correlated Cy3/Cy5 FRET changes with these DNA substrates consistent with DNA melting of 20 and 40 bp beyond the RP linkages (Supp. Fig. S2C, S2D). The lag times also increase linearly with duplex length consistent with DNA unwinding initiating from the DNA end containing the RP linkages (Fig. 2E). These results indicate that RecBCD is able to unwind DNA processively for at least 40 bp beyond the RP linkages that prevent ssDNA translocation by the canonical RecB and RecD motors.
We next examined DNA VI (L=20, 40, and 60 bp) and VII (L=80 bp) (Fig. 2A). These substrates are similar to DNA V, but the Cy3/Cy5 fluorophores are positioned across a nick in the DNA strand that initially starts with a 3′-end. This is the strand along which the canonical RecB motor is expected to translocate. If RecBCD is able to unwind the DNA and reach the nick, a loss of FRET should occur. The time courses displayed a lag in the Cy3 fluorescence increase (Fig. 2D) with an anti-correlated decrease in Cy5 fluorescence, consistent with DNA unwinding. Surprisingly, DNA unwinding was observed even for DNA VII in which L= 80 bp (Fig. 2D, 2E, Supp. Fig. S2E). Hence, RecBCD is able to unwind at least 80 bp processively in the absence of ssDNA translocation of the canonical RecB and RecD motors.
The RP linkages in DNA substrates V, VI and VII are positioned only 4 bp from the DNA end to which RecBCD binds and initiates unwinding. Since RecBCD melts 6 bp upon binding a blunt DNA duplex end [3, 27, 36], we were concerned that the presence of the RP linkages within the binding site RecBCD could influence the ability of RecBCD to unwind DNA beyond the RP linkages. We therefore examined a DNA substrate in which the RP linkages were placed further away (15 bp) from the single strand/double strand junction at the high affinity (3-dT6/5′-dT10) RecBCD binding site and thus 21 bp (15 + 6) from the 3′-DNA end. This places the RP linkages outside of the 20 bp RecBCD-DNA binding footprint[3, 42] so that they should not influence RecBCD binding. The Cy3/Cy5 FRET probes are positioned 32 bp from the RP linkages. Stopped-flow experiments (Supp. Fig. S2F) with this substrate show Cy3 time courses with the expected lag phase that is [ATP]-dependent indicating that RecBCD is still able to unwind the DNA beyond the RP linkages.
RecBCD unwinds reversed polarity DNA with only a 2-fold slower rate
The lag times for all RP DNA (V, VI and VII) increase linearly with the number of bp, L, separating the RP linkages from the FRET reporters (Fig. 2E), indicating an unwinding rate of 415±29 bp/sec at 5 mM ATP. The lag time also increases with decreasing [ATP] indicating a slower unwinding rate at lower [ATP] (Fig. 2F).
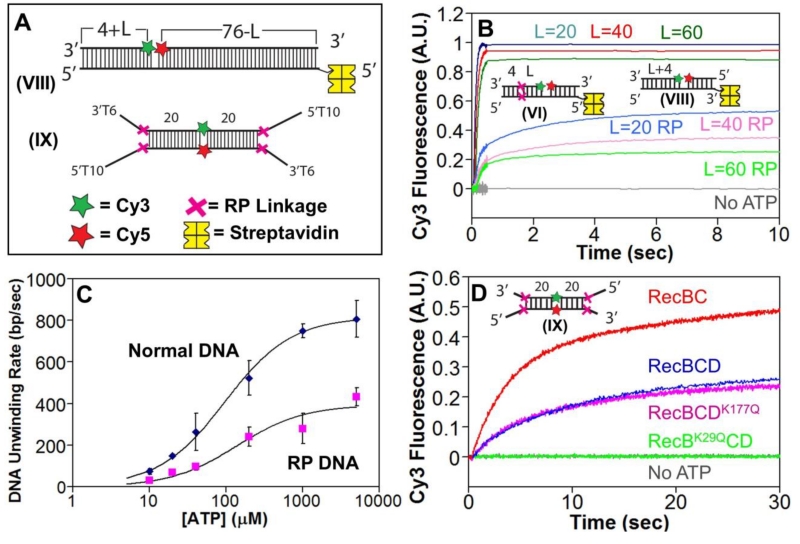
For comparison, we examined RecBCD unwinding of DNA substrates that are identical in sequence to the nicked DNA VI (Fig. 2A), but lack the RP linkages (DNA VIII, L=20, 40, and 60 bp, Fig. 3A). Stopped-flow experiments showed the expected DNA length dependent lag times (Fig. 3B, Supp. Fig. S3A), yielding an unwinding rate of 808±88 bp/sec (Supp. Fig. S3B), the same, within error, as previously determined[25, 29]. Hence RecBCD unwinds RP containing DNA only ~2-fold slower than normal DNA. The ATP-dependence of the unwinding rates for normal and RP DNA shows that RecBCD unwinds the RP DNÃ2-fold slower at all [ATP]. Best fits of the data in Figure 3C yield Vmax = 815±16 bp/s and 392±38 bp/s for normal vs. RP DNA, respectively.
Figure 3. RecBCD unwinds RP DNA with a two-fold slower rate and requires an active RecB motor.
A. DNA VIII used to monitor unwinding of normal DNA. Each end of DNA IX contains a high affinity site for RecBCD binding. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages, separated by L bp from a Cy3 (green star) and Cy5 (red star) fluorophores. DNA VIII is biotinylated at one end so that streptavidin (yellow) can bind and block binding of RecBCD.
B. Comparison of RecBCD unwinding time courses with normal DNA (series VIII) vs. RP DNA (series VI) performed by mixing pre-incubated RecBCD (15 nM) and DNA (20 nM) with ATP (5 mM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C. Cy3 fluorescence was excited at 505 nm, and Cy3 and Cy5 fluorescence were monitored simultaneously. Cy3 fluorescence time courses for DNA with L=20 bp (blue), L=40 bp (red), and L=60 bp (green) are shown for normal DNA Series VIII and RP DNA Series VI.
C. . ATP dependence of DNA unwinding rates for normal DNA (blue) and RP DNA (pink). Smooth curves are the best fits to the Michaelis-Menten equation yielding Km=96±8 μM, Vmax=815±16 bp/sec for normal DNA and Km=138±57, Vmax=392±38 for RP DNA.
D. Stopped flow time courses, monitoring Cy3 fluorescence, for RecBCD (blue), RecBC (red), RecBCDK177Q (pink), and RecBK29QCD (green) with DNA IX.
Although the DNA unwinding rates, as calculated from the length dependence of the lag times, differ only by a factor of ~2, unwinding of the regular DNA by RecBCD was complete within less than one second at saturating ATP concentrations (Fig. 3B; Supp. Fig. S3A). Conversely, unwinding of the RP DNA by RecBCD took much longer to reach completion (Fig. 3B). We also note that the unwinding amplitudes for the RP DNA is much lower than for normal DNA (Fig. 3B). This reduced amplitude suggests either a lower processivity for unwinding RP DNA or that only a fraction of the RecBCD initially bound can unwind past the RP linkages or that RP DNA unwinding is in a steady state between unwinding and rewinding and thus complete unwinding is never reached.
RecB is required to unwind RP DNA
We next examined the role of the two RecBCD motors in unwinding the RP DNA using DNA IX (Fig. 3A) that has identical ends containing high affinity binding sites for RecBCD and RecBC. RP linkages are positioned at the ss/ds DNA junctions and Cy3 and Cy5 are incorporated into the backbone at the center of the DNA, 20 bp removed from the RP linkages. RecBC binds with highest affinity to a duplex DNA end possessing a 3’-T6 tail and a 5’-dT6 tail[36]. RecBCD binds with highest affinity to a duplex DNA end possessing 5’-dT10 and 3’-T6 tails[36]. The longer 5’-dT10 tail is needed to make contact with the RecD motor[36, 43]. This DNA does not require a biotin-streptavidin block since both ends are identical and contain RP linkages.
Stopped-flow experiments performed with DNA in excess over enzyme ensured initiation only at one end of the DNA. We examined RecBCD, RecBC, RecBK29QCD, and RecBCDK117Q, the latter two variants containing a mutation eliminating ATPase activity in the RecB or RecD motor, respectively, so that those motors cannot translocate [41, 44-46]. The time courses indicate that wt RecBCD, RecBC and RecBCDK117Q are able to unwind RP DNA (Fig. 3D). However, RecBK29QCD was unable to unwind RP DNA (Fig. 3D). Therefore an active RecB motor is needed to unwind RP DNA, but not an active RecD motor.
ssDNA loops form during RecBCD unwinding of RP DNA
Recall that the kinetic time course for RecBCD translocation on ssDNA using DNA IV containing two RP linkages does not show a dissociation phase that is clearly evident with normal DNA (Fig. 1D). This suggests that RecBCD remains bound to the DNA, but stuck behind the RP linkages, as it unwinds the DNA. As a test of this, we compared RecBCD unwinding of DNA X (normal DNA) vs. DNA XI (RP DNA) (Fig. 4A). These DNA substrates have a high affinity RecBCD binding site (3’-dT6/5’-dT10), with a Cy3 label at the 3′ end of the 3′-dT6 tail. When RecBCD binds to a DNA end labeled with Cy3, the Cy3 fluorescence is enhanced[36]. Hence, we can use Cy3 fluorescence to monitor whether RecBCD leaves the high affinity binding site as it unwinds the DNA. If it can move past the RP linkages and leave the binding site we should observe a decrease in Cy3 fluorescence as it moves away from the Cy3 at the 3′-end. If RecBCD is stuck behind the RP linkages and thus stays at the binding site, Cy3 fluorescence should remain unchanged.
Figure 4. The RecBCD motors remain stuck behind the RP linkages and induce ssDNA loops during DNA unwinding.
A. DNA substrates used to test for ssDNA loop formation during RecBCD unwinding of RP DNA. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages. Cy3 (green star) and Cy5 (red star) fluorophores are separated by 61 base pairs. One end of each DNA is biotinylated so that streptavidin (yellow) can bind and block binding of RecBCD.
B. Stopped-flow time courses, monitoring Cy3 fluorescence, for RecBCD unwinding of normal DNA X (blue) and RP DNA XI (pink). RecBCD (18.75 nM) and DNA (25 nM) were pre-incubated and then mixed with ATP (5 mM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C.
C. Stopped-flow time courses showing an increase in FRET signal accompanying RecBD1080ACD unwinding of RP DNA XIII. Experiments were performed by mixing pre-incubated RecBCD (37.5 nM) and DNA XIII (50 nM) with ATP (200 μM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C. Cy3 fluorescence was excited at 505 nm, and Cy3 (green) and Cy5 (red) fluorescence were monitored.
D. Cy3 fluorescence time courses for experiments performed with RecBD1080ACD and RP DNA XIII as described in panel C, at 400 μM ATP (green), 200 μM ATP (light blue), 100 μM ATP (gold), 40 μM ATP (pink), 20 μM ATP (dark blue), and no ATP (gray). Inset-identical stopped-flow experiment performed with RecBD1080ACD and DNA XII at 200 μM ATP.
Stopped-flow experiments pre-incubating excess DNA with RecBCD were initiated with ATP and heparin (Buffer M, 30 mm NaCl). The resulting time courses indicate no change in Cy3 fluorescence with RP DNA XI indicating that RecBCD remains at the high affinity binding site (Fig. 4B). In contrast the Cy3 fluorescence decreased for DNA XI that lacks the RP linkages, indicating that RecBCD moves away from the initial binding site during unwinding of normal DNA (Fig. 4B). Hence, the RecB and RecD motors remain bound, but are prevented from moving past the RP linkages when unwinding DNA XI.
As a further test, we found that RecBCD, pre-bound at a high affinity DNA end that contains RP linkages, is unable to reinitiate DNA unwinding on a FRET DNA substrate that is not covalently attached to the RP DNA (Supp. Fig. S4A). This was also true in the absence of the heparin trap (Supp. Fig. S4B). This further confirms that RecBCD is not only prevented from moving beyond the RP linkages, but remains bound to the RP DNA and cannot dissociate in order initiate unwinding of the FRET DNA.
Since the RecB and RecD motors are stuck behind the RP linkages, another region of RecBCD must interact with the duplex DNA in order to carry out DNA unwinding. In addition, the DNA ahead of the RP linkages must be brought closer to the RecBCD that is stuck at the RP linkages during DNA unwinding. This predicts that the intervening ssDNA should form looped intermediates. To address this, we examined DNA XII (normal DNA) and XIII (RP DNA) (Fig. 4A), which are identical in sequence, and contain a Cy5 fluorophore in the DNA backbone of the 3′-dT6 ended strand, one bp ahead of the ss/dsDNA junction, and a Cy3 fluorophore in the backbone of the same DNA strand, but 61 bp away from Cy5. Before any unwinding by RecBCD, the Cy3 and Cy5 fluorophores are separated by 61 bp (>200 Å), yielding no initial FRET signal. Upon unwinding RP DNA XIII, the RecB and RecD motors should remain stuck behind the RP linkages and close to Cy5, resulting in formation of a loop in the ssDNA that would bring the Cy3 closer to the Cy5, resulting in an increased FRET signal.
In fact, stopped-flow experiments with RP DNA XIII show a FRET increase indicating that the Cy3 and Cy5 fluorophores are brought closer together as the DNA is unwound (Fig. 4C), consistent with formation of a loop in the DNA between the RP linkages and the Cy3 fluorophore. At high [ATP] (>100 μM), the Cy3 and Cy5 fluorescence signals reach a plateau after ~5 sec (Fig. 4C). However, at longer times, the Cy5 and Cy3 fluorescence signals return to their initial values before the start of the reaction (Fig. 4D), indicating dissipation of the DNA loops. Furthermore, the time required for loss of the FRET signal decreased with decreasing ATP concentration (Fig. 4D), suggesting that DNA loop formation reverses after all of the ATP is hydrolyzed. As a further test of this, we performed this experiment in the presence of an ATP regenerating system (Materials and Methods). Under these conditions, the high FRET signal remained for > 10 minutes (Fig. 4E). These results indicate that the RecBCD motors remain stuck behind the RP linkages during DNA unwinding, preventing complete strand separation, and that the DNA duplex reforms after all ATP is hydrolyzed.
Upon performing the same experiment with normal DNA (XII) that is identical to DNA XIII, but lacks the RP linkages, a much larger FRET increase was observed upon DNA unwinding by RecBCD, but this FRET signal did not return to baseline even at long times (Fig. 4F). This is consistent with full unwinding and strand separation of the DNA, with no duplex DNA reformation. The FRET increase observed with DNA XII (normal DNA) does not necessarily reflect formation of a DNA loop during unwinding. Rather, after RecBCD unwinds the DNA, the newly formed ssDNA is flexible allowing the Cy3 and Cy5 fluorophores to come in close proximity resulting in an increase in FRET signal.
RecB Arm domain is needed for RecBCD to unwind RP DNA
We next asked what regions of RecB are required for this activity. One possibility is the arm domain since in the crystal structure[3] it contacts the DNA duplex ~22 bp from the blunt DNA end (Figure 1A).
We constructed a RecB variant, RecBΔArm, in which the arm (residues 218-323) was replaced with 4 glycines. We purified a stable RecBΔArmCD hetero-trimer (see Materials and Methods), however, RecBΔArmCD did not display any detectable DNA unwinding activity on a blunt-ended DNA substrate (Supp. Fig. S5A). We considered that this could be due to an inability of RecBΔArmCD to melt out a blunt-ended DNA. We then examined RecBΔArmCD activity on DNA XIV (Fig. 5A) possessing a high affinity 3’-dT6/5’-dT10 DNA end. We found that RecBΔArmCD binds with high affinity to a DNA end possessing 3’-dT6/5’-dT6 tails (Supp. Fig. S5B), hence RecBΔArmCD should bind tightly to DNA XIV. The longer 5’-dT10 tail in DNA XIV also ensures DNA binding to the RecD motor[36, 43]. Stopped-flow experiments showed some unwinding of DNA XIV by RecBΔArmCD, although the amplitude was only ~15% of that observed with wt RecBCD (Fig. 5B).
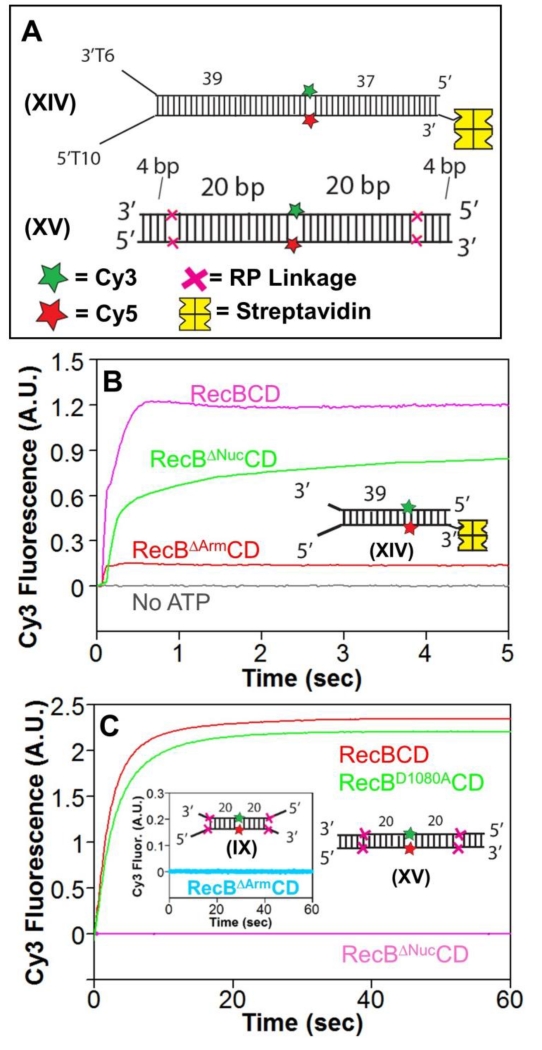
Figure 5. The RecB arm and nuclease domains are needed for RecBCD to unwind RP DNA.
A. DNA substrates used to test for unwinding by RecBΔArmCD and RecBΔNucCD. DNA XIV contains a high affinity binding site on one end and is biotinylated on the opposite end to bind streptavidin (yellow) to block RecBCD binding. Cy3 (green star) and Cy5 (red star) are on opposite strands 39 bp from the binding site. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages in DNA XV separated by L bp from a Cy3 (green star) and Cy5 (red star) fluorophores.
B. Stopped-flow Cy3 fluorescence time courses showing unwinding of DNA XIV (20 nM) by RecBCD, RecBΔNucCD, and RecBΔArmCD (15 nM) in Buffer M (30 mM NaCl) (5 mM ATP and 8 mM heparin) at 25° C.
C. Stopped-flow Cy3 fluorescence time courses with RP DNA XV and RecBCD (15 nM) (red), RecBD1080ACD (15 nM) (green), and RecBΔNucCD (15 nM) (pink) in Buffer M30 (5 mM ATP and 8 mg/mL heparin) at 25° C. Inset-RecBΔArmCD is unable to unwind RP DNA IX containing a high affinity binding site under the same conditions.
RecBΔarmCD displays a low level of 5’ to 3’ ssDNA translocation activity (Supp. Fig. S5C) with a rate of 2058±42 bp/sec (Supp. Fig. S5D) similar to wt RecBCD[18], but shows little secondary translocase activity, at least on a DNA with a (dT65) tail (Supp. Fig. S5E). Furthermore, RecBΔArmCD showed no ability to unwind RP DNA IX, indicating a requirement of the RecB arm for this activity (Fig 5C, inset).
RecB nuclease domain, but not nuclease activity, is needed to unwind RP DNA
We next examined whether the nuclease activity of RecB might be involved in the ability of RecBCD to unwind past RP linkages. We examined two nuclease deficient variants of RecBCD. In the first, RecBD1080ACD, the D1080 mutation in the catalytic site eliminates its nuclease activity[8, 47]. In the second, RecBΔNucCD, the entire RecB nuclease domain (residues 930-1180) was deleted. Both RecBD1080ACD and RecBΔNucCD retain processive DNA helicase activity[7].
Using DNA XIV (Fig. 5A), we found that RecBΔNucCD unwinds normal DNA (Fig. 5B) as shown previously[7]; however, the unwinding rate is ~65% slower than for RecBCD (Vmax = 531±51 bp/s vs. 815±16 bp/s at 5 mM ATP), (Supp. Fig. S5F). This clearly indicates that the nuclease domain plays a role in regulating the rate of DNA unwinding by RecBCD. This is especially interesting in light of the fact that the rate of DNA unwinding by RecBCD is reduced by ~two-fold after recognition of a chi sequence, consistent with the suggestion that RecBΔNucCD might mimic RecBCD after chi recognition[7]. Interestingly, RecBΔNucCD also displayed a higher apparent KM for ATP (KM = 246±87 μM) than wt RecBCD (96±8 μM) (Supp. Fig. S5F).
We next used DNA XV (Fig. 5A) to determine if RecBΔNucCD can unwind RP DNA. Surprisingly, we found that RecBΔNucCD cannot unwind DNA XV (Fig. 5C). However, the RecBD1080ACD mutant, which also lacks nuclease activity but retains the nuclease domain[47], is able to unwind DNA XIV as well as wt RecBCD (Fig. 5C). These unexpected findings indicate that the RecB nuclease domain, but not its nuclease activity, is required for RecBCD to unwind past the RP linkages.
As a further test of these results, DNA unwinding experiments were performed using a standard gel electrophoresis assay. In these experiments, fluorescently labeled DNA was run on a native polyacrylamide gel after denaturing the enzyme in order to detect unwound and strand separated DNA (Materials and Methods). Using this approach, RecBCD, RecBD1080ACD and RecBC showed ATP-dependent unwinding and strand separation of RP DNA, whereas RecBΔNucCD and RecBΔArmCD showed no unwinding (Supp. Fig. S5G). Recall that our stopped-flow experiments indicate that the unwound RP DNA remains bound to the enzyme and can reanneal to reform the original DNA substrate once all of the ATP is hydrolyzed (Fig. 4D). DNA strand separation is only observed when enzyme is denatured and thus cannot remain bound to the DNA.
RecBCD unwinding of RP DNA requires its secondary translocase activity
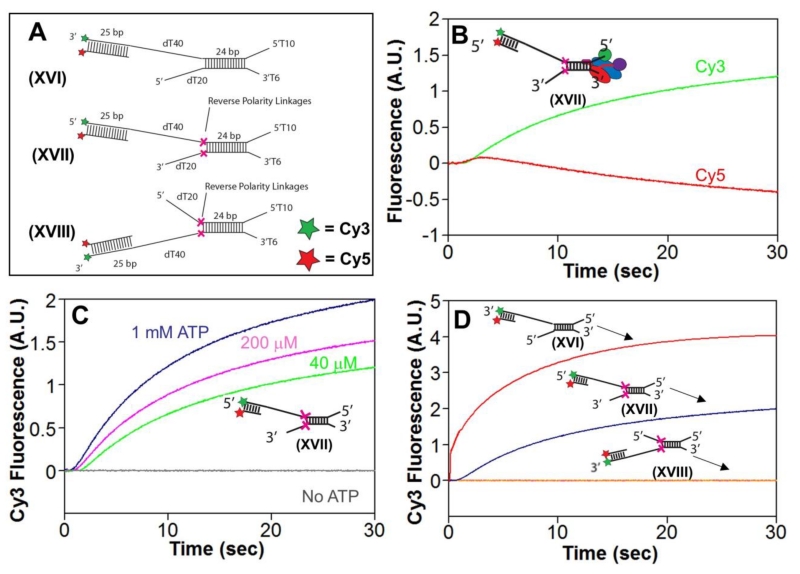
Using a DNA substrate similar to DNA XVI (Figure 6A), we previously found that RecBC is able to unwind a proximal 24 bp duplex, then translocate 5′ to 3′ along a stretch of ssDNA (dT40) and re-initiate unwinding of a 25 bp distal duplex[15]. We therefore examined whether RecBCD could re-initiate DNA unwinding beyond two RP linkages using DNA XVII, and XVIII (Fig. 6A). These DNA substrates contained a 5’-dT10/3’-dT6 DNA end providing a high affinity binding site for RecBCD on the proximal 24 bp duplex. RP linkages were placed in each DNA strand directly after the proximal duplex, followed by a stretch of dT40 on one strand and a distal 25 bp duplex that has a Cy3/Cy5 FRET pair to monitor DNA unwinding of the distal duplex DNA. The other DNA strand possesses a dT20 ssDNA after the RP linkage. DNA XVII has the distal FRET duplex attached to the 5′ end of the DNA, which engages the RecD canonical motor, and thus can only be reached due to action by the RecBC secondary translocase activity. DNA XVIII has the distal FRET duplex attached to the 3′ end of the DNA, which engages the RecB canonical motor, and thus can only be reached by the primary RecB translocase activity.
Figure 6. RecBCD unwinding of RP DNA requires its secondary translocase activity.
A. Substrates used to test re-initiation of unwinding by RecBCD.
B. Stopped-flow experiments were performed by mixing pre-incubated RecBCD and DNA Substrate XVII (18.75 and 25 nM, respectively) in Buffer M (250 mM NaCl) in a 1:10 volumetric ratio with ATP (1 mM) and heparin (8 mg/mL) in buffer M (8 mM NaCl) yielding the final concentrations listed above and a final NaCl concentration of 30 mM at 25°C. Cy3 fluorescence was excited at 505 nm, and Cy3 fluorescence (green) was monitored at 570 nm and Cy5 fluorescence (red) was monitored at > 665 nm.
C. Cy3 fluorescence traces for experiments as performed in panel B, at 1 mM ATP (dark blue), 200 μM ATP (pink), 40 μM ATP (light blue), and no ATP (gray).
D. Cy3 fluorescence traces for experiments as performed in panel B at 1 mM ATP using Substrate XVI (red), XVII (blue), or XVIII (orange).
Stopped-flow experiments were performed by pre-incubation of excess DNA XVII with RecBCD at 250 mM NaCl to ensure that RecBCD binds preferentially to the 5’-dT10/3’-dT6 DNA end and not the longer ssDNA regions or double-stranded DNA[14, 15, 36]. Upon initiation of the reaction by mixing with ATP and heparin in the same buffer, the Cy3 fluorescence signal exhibited a lag phase followed by an increase, while the Cy5 fluorescence signal exhibited a lag phase followed by a short enhancement and then a decrease below the initial baseline (Fig. 6B) indicating unwinding of the distal 25 bp FRET DNA[29]. These DNA unwinding time courses are ATP dependent, showing an increasing lag time at lower ATP concentrations (Fig. 6C). Since the RecBC secondary translocase is the only motor activity that can bypass the two RP linkages in DNA XVII[15], these results indicate that the RecBC secondary translocase is the motor activity that is required to re-initiate DNA unwinding beyond the RP linkages.
As discussed above, RecBCD that is pre-bound at a high affinity DNA end that contains RP linkages is unable to reinitiate DNA unwinding on a FRET DNA substrate that is not covalently attached to the RP DNA (Supp. Fig. S4A), even in the absence of a heparin trap (Supp. Fig. S4B).
Discussion
A current mechanism for how RecBCD unwinds DNA proposes that the two translocating motors, RecB and RecD, act on the complementary strands of ssDNA to pull the duplex DNA across a wedge or pin[3, 48]. In fact, similar wedge models have been proposed for other helicases based solely on structural considerations[24]. All of these models propose that DNA unwinding is tightly coupled to and requires motor translocation along ssDNA. The experiments reported here indicate that both RecBCD and RecBC helicases are able to unwind DNA in the absence of ssDNA translocation by the canonical RecB and RecD motors, ruling out these simple mechanical models. An alternative mechanism is that the free energy of RecBCD binding to DNA is used to melt multiple DNA bp that provide the ssDNA tracks along which the RecB and RecD motors translocate. Translocation proceeds, hydrolyzing 1 ATP per nucleotide moved per motor, until the ss/ds DNA junction is reached. ATP hydrolysis then resets the enzyme so that it can again use its binding free energy to melt out another stretch of duplex DNA to continue the unwinding process.
This mechanism is supported by the finding that both RecBCD and RecBC can melt out 5-6 bp upon binding to a blunt DNA end in a reaction that does not require ATP[3, 23, 27, 28, 49]. Furthermore, the extent of ATP hydrolysis by RecBC is the same during processive DNA unwinding (DNA melting plus ssDNA translocation) as during only ssDNA translocation[17]. It is also consistent with the finding that processive DNA unwinding by RecBCD and RecBC occurs with a kinetic step size of 4±2 bp, suggesting that a slow kinetic step is repeated during the unwinding process and that 4±2 bp are unwound between successive slow steps[14, 18, 25, 26, 29].
In this study we used our finding that reversing the phosphodiester polarity within the DNA backbone will block ssDNA translocation of the canonical SF1 RecB and RecD motors[15, 18]. Yet, the secondary translocation activity that functions within the context of both RecBC and RecBCD is able to function past a 3′-3′ RP linkage as shown previously[15, 18] and in this study. This activity enables RecBCD to unwind DNA past the RP linkages. Our results suggest that the secondary translocase activity resides, at least in part, within the RecB arm domain and is controlled by the RecB ATPase motor.
We present experiments with many different DNA substrates showing that RecBCD and RecBC are able to unwind DNA processively even when its canonical motors are prevented from undergoing ssDNA translocation past the RP linkages. FRET signals indicative of DNA melting are observed when a Cy3/Cy5 pair are placed either in the DNA backbone or across a nick. Stopped-flow experiments show the expected lag phase before the Cy3/Cy5 FRET pair become separated due to DNA unwinding and the lag time increases linearly with DNA length from which we estimate the rate of unwinding[25, 26, 29, 50]. We find that ssDNA loops form between the RP linkages and the FRET pairs due to the fact that the RecBCD motors remain stuck at the RP linkages. Interestingly, these ssDNA loops dissipate and the duplex DNA reforms after all ATP is hydrolyzed. This provides further evidence that the unwound DNA strands remain bound to the RecBCD during the unwinding process.
We observe lower amplitudes for unwinding of the RP DNA vs. normal DNA in the stopped-flow experiments. The lower amplitudes may reflect a lower processivity for unwinding past the RP linkages. However, it also might reflect establishment of a steady state of DNA unwinding due to a repetition of unwinding/rewinding events while RecBCD is stuck at the RP linkages, hence complete unwinding of all RP DNA is never achieved.
We found that the ability of RecBCD to unwind RP DNA processively requires ATP hydrolysis by the RecB motor, but not the RecD motor. It also requires the RecB arm domain. Since deletion of the RecB arm prevents initiation of DNA unwinding on a blunt ended DNA, RecBΔArmCD can only initiate unwinding at a DNA end possessing a high affinity site consisting of pre-melted 3′-dT6/5′-dT10 tails. This result also implicates the RecB arm in the DNA melting process.
Our results indicate that ATP hydrolysis by the RecB motor not only drives ssDNA translocation of that motor, but also transmits conformational changes allosterically to the RecB arm to generate a force that enables the arm region to move along the DNA ahead of the RP linkages. As previously proposed, the RecB arm appears to be responsible for the secondary translocase activity of RecBC [15]. Although this secondary translocase activity was identified as a ssDNA translocase, it is possible that it might actually function as a double stranded DNA translocase during DNA unwinding[15]. Since the RecB and RecD motors are stuck behind the RP linkages, the coupled ATP-dependent activity of such a dsDNA translocase might result in the “reeling in” of the dsDNA resulting in ssDNA loops as illustrated in Fig. 7, consistent with our observation of simultaneous DNA unwinding and ssDNA loop formation by RecBCD when operating on DNA containing RP linkages.
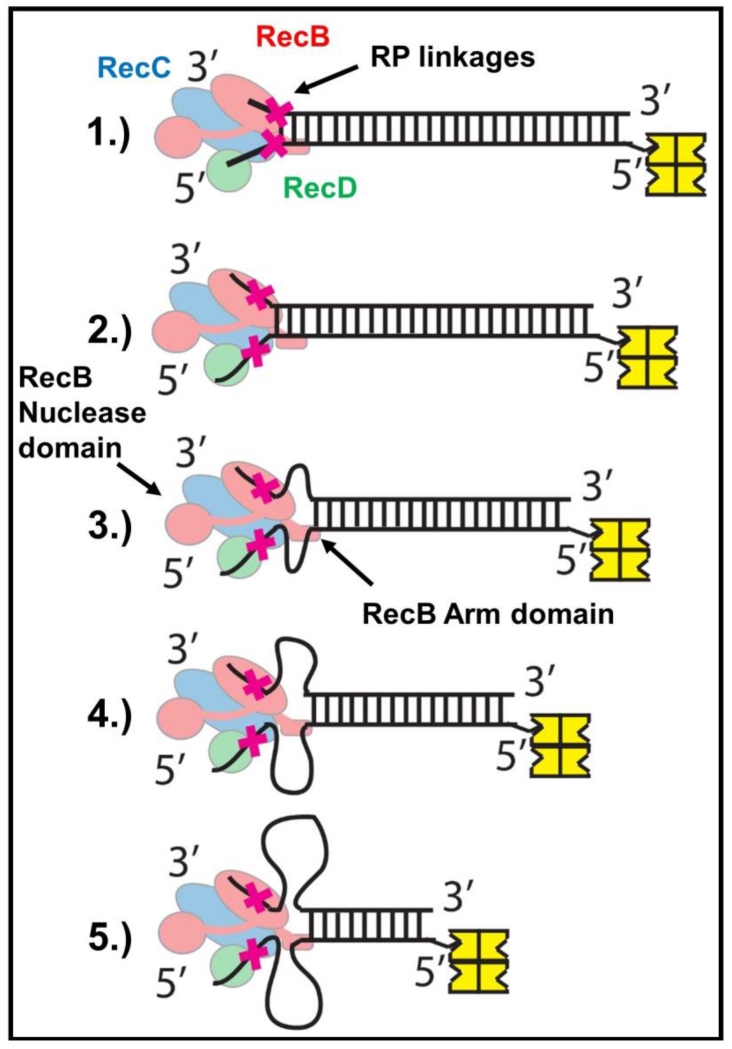
Figure 7. Proposed mechanism for unwinding of reversed polarity DNA by RecBCD.
(Step 1) RecBCD binds to and melts 6 base pairs at a normal DNA end. (Step 2) Canonical ssDNA motors (RecB (red) and RecD (green)) translocate along the ssDNA tracks until the RP linkages (red X) are encountered. (Step 3-5) RP linkages block ssDNA translocation by the canonical RecB and RecD motors, but the secondary translocase, fueled by ATP hydrolysis in the RecB motor, simultaneously pulls dsDNA toward RecBCD, resulting in the formation of single stranded DNA loops. The RecB Arm and RecB Nuclease domains (highlighted) are necessary for this activity.
Interestingly, recent structural studies of the B. subtilis AddAB helicase/nuclease, that is structurally similar to E. coli RecBC, have proposed a model for DNA unwinding that involves a role for the AddA “arm” domain, that is similar to the RecB “arm” domain[51]. In a crystal structure[51], the AddA arm and the C-terminal nuclease domain of AddB contact the duplex DNA ahead of the fork as does the RecB arm in the RecBCD-DNA structure[3]. Krajewski et al.[51] propose that in an ATP-coupled reaction, the AddA arm pulls on the duplex DNA in the opposite direction, but in concert with the AddA canonical motor that pulls on the 3′-ended ssDNA. This is proposed to create stress in the duplex DNA that results in base pair opening. In fact, Krajewski et al.[51] further suggest that this proposed role for the arm domain may provide a basis for the RecBC secondary translocase activity that we have now demonstrated is involved in DNA unwinding by RecBCD. This model is consistent with our suggestion that the arm domain may actually operate as a double stranded DNA translocase driven by the ATPase activity of the AddA, or RecB motor.
Most surprisingly, we find that the ability of RecBCD to unwind RP DNA requires the RecB nuclease domain, although not its nuclease activity. Furthermore, although RecBΔNucCD can unwind normal DNA, its rate of unwinding is reduced ~two-fold compared with RecBCD. This finding indicates that the nuclease domain regulates the helicase activity of RecBCD. This is noteworthy since after chi recognition, the nuclease, helicase and RecA loading activities of RecBCD are altered[2, 11, 52-57]. In particular, the rate of DNA unwinding after chi recognition is reduced ~two-fold, similar to the rates we observe here for RecBΔNucCD unwinding of normal DNA as well as for wt RecBCD unwinding of RP DNA. Hence, both of these situations may mimic aspects of RecBCD after chi recognition. There have been suggestions that the RecB nuclease domain becomes repositioned after RecBCD recognizes a chi site[7, 58, 59]. In particular, the nuclease domain is involved in loading of RecA onto unwound ssDNA by directly binding to RecA[58]. Yet, the interface of the nuclease domain that interacts with RecA is found buried within the RecBCD crystal structure[58]. Hence, the nuclease domain would need to move from its position in the crystal structure in order to interact with RecA[58]. Recently, Taylor et al.[59] also presented evidence for a conformational change upon chi recognition that involves movement of the nuclease domain.
Two possibilities exist for why the nuclease domain is required for RecBCD to unwind past RP linkages. First, the nuclease domain could indirectly promote this activity through an allosteric effect. Second, the nuclease domain might participate directly in DNA unwinding. The nuclease domain is tethered to the RecB motor by a peptide linker of ~60 amino acids. This linker is long enough to potentially enable the nuclease domain to move from its proposed position behind the DNA duplex as in the crystal structure[3] (Fig. 1a) to a position ahead of the fork near the arm domain and interact directly with the duplex to facilitate DNA unwinding.
Although DNA containing RP linkages is not a natural substrate for RecBCD, the ability of RecBCD to unwind DNA past these RP linkages indicates that its mechanism of DNA unwinding is more complex than the two motors pulling the duplex DNA across a pin or wedge as previously suggested. Such behavior may occur if RecBCD encounters physical blocks to its motors, such as some forms of DNA damage. Furthermore, our results indicate that previously unsuspected regions of the enzyme, in particular the RecB arm and nuclease domains, play key roles in the unwinding mechanism.
Materials and Methods
Buffers
Buffers were made from reagent-grade chemicals using double distilled water that was further deionized with a Milli-Q purification system (Millipore Corp., Bedford, MA). Buffer C is 20 mM potassium phosphate (pH 6.8), 0.1 mM 2-mercaptoethanol, 0.1 mM EDTA, 10% (v/v) glycerol. Buffer M is 20 mM MOPS-KOH pH (pH 7.0), NaCl as listed in text, 10 mM MgCl2, 1 mM 2-mercaptoethanol, 5% (v/v) glycerol. Stock concentrations of MgCl2 was determined by refractometry.
Proteins
RecBCD, RecBD1080ACD, RecBK29QCD, and RecBCDK177Q were overexpressed in E. coli strain V2831[41]. RecB and RecC were overexpressed in E. coli strain V186[60]. RecBΔNucCD, and RecBΔArmCD were expressed in E. coli strain V2601, a variant of V186 expressing the lacIq gene. V186, V2601, and V2831 contain deletions of the chromosomal recB, recC, and recD genes. E. coli RecB and RecC proteins were purified and reconstituted to form RecBC as described[15]. RecBC concentrations were determined spectrophotometrically in Buffer C using an extinction coefficient of ε280 = 3.9×105 M−1 cm−1 for the RecBC heterodimer. E. coli RecBCD protein, the mutants RecBK29QCD, RecBCDK177Q, and RecBD1080ACD, and RecBΔNucCD, and RecBΔArmCD were purified as described [25, 29]. However, the RecBΔArmCD construct did not bind to the ssDNA column as in the RecBCD purification. RecBCD purified as a mixture of heterotrimer and hexamer[61]. A Hi-Trap heparin column was used to isolate the heterotrimer, which was used for experiments described in this paper. Proteins were dialyzed into Buffer C, aliquoted, and flash-frozen in liquid nitrogen for storage at −80° C. For stopped-flow experiments, protein was dialyzed into Buffer M and stored at 4 °C for up to a week, since ~5% loss of activity was observed after this time. RecBCD, RecBK29QCD, RecBCDK177Q, and RecBD1080ACD concentrations were determined spectrophotometrically in Buffer C using ε280 = 4.5×105 M−1 cm−1. RecBΔNucCD and RecBΔArmCD concentrations were determined using ε280 = 4.11×105 M−1 cm−1 and 4.32×105 M−1 cm−1, respectively. Bovine serum albumin (BSA) was from Sigma (St. Louis, MO) and its concentration determined using ε280 = 4.3×104 M−1 cm−1 in buffer C[36].
Fluorescence Stopped-Flow Kinetics
Stopped-flow experiments were performed in Buffer M at 25°C using an Applied Photophysics SX.18MV instrument (Applied Photophysics Ltd, Leatherhead, UK). For Cy3 and Cy5 FRET experiments, the Cy3 fluorophore was excited using a 505 nm LED (Applied Photophysics Ltd, Leatherhead, UK) and the fluorescence emission was monitored at 570 nm using an interference filter (Oriel Corp. model 53905). The sensitized Cy5 fluorescence emission was monitored simultaneously at wavelengths >665 nm using a long pass filter (Oriel Corp. model 51330) using the dual-wavelength detection mode. Since the excitation path lengths and filters differ for the two observation channels, the resulting fluorescence intensities are not directly comparable. Cy3 excitation by the 505nm LED showed no photo bleaching over the time-scale of the experiments.
Stopped-flow experiments were performed by incubating the DNA substrate (at twice the final concentrations noted in the figure legends for equal volume mixing experiments and 11 times the final concentrations in the figure legends for 1:10 volumetric mixing) with three times its molar concentration of streptavidin for 5 minutes in the presence of 6 μM BSA. Subsequently, RecBCD or its variants (at twice or 11 times listed concentration as noted above) was mixed with the DNA on ice for 5 minutes before loading into the stopped-flow syringe where it was allowed to equilibrate for another 5 minutes at 25°C. The RecBCD mixture was then rapidly mixed in a 1:1 ratio with a solution containing ATP (various concentrations), and heparin trap (16 mg/mL) in buffer M. This results in final reaction conditions of RecBCD, DNA, and ATP as noted in figure legends, 8 mg/mL heparin trap, 20 mM Mops-KOH (pH 7.0), 10 mM MgCl2, 30 mM NaCl, 1 mM 2-ME, and 5% (v/v) glycerol at 25 °C. For experiments with a 1:10 volumetric mixing ratio, 200 μL of RecBCD mixture in buffer M (250 mM NaCl) was rapidly mixed with a 2 mL solution containing ATP (various concentrations) and heparin trap (8.8 mg/mL) in buffer M (8 mM NaCl), for identical final reaction conditions as 1:1 experiments.
Analysis of DNA Unwinding Kinetics
DNA unwinding rates were estimated from the inverse of the slope of plots of the lag time of the Cy3 kinetic time course vs. L, the number of base pairs between the reverse-polarity linkages and the Cy3-Cy5 fluorophores. The lag times were estimated from the time corresponding to the intersection of two lines corresponding to the baseline in the lag phase and the initial increase in the Cy3 fluorescence signal as described[15, 18]. The ATP dependence of the unwinding rate was fit to a Michaelis-Menten model as described[15].
Gel electrophoresis helicase assay
Reactions (9 μL) contained 20 nM RecBCD and 25 nM DNA in buffer M (30 mM NaCl) were incubated at 25°C for 5 minutes, followed by addition of ATP (final [ATP] = 5 mM), and reaction for 3 minutes before being stopped by addition of EDTA and SDS (20 mM and 1.5% final concentrations, respectively). The samples were then placed on ice until loading to prevent re-annealing of DNA. Negative controls omitted RecBCD or added buffer M instead of 50 mM ATP. Positive controls included only 10 nM DNA, heated to 95 °C for 5 minutes then rapidly cooled to prevent re-annealing. 3.25 μL of 5× loading buffer (.25% bromophenol blue, 0.25% xylene cyanol, 50% glycerol) was added to each sample before electrophoresis on a 10% polyacrylamide gel (19:1 acrylamide to bis-acrylamide). Fluorescence of the Cy3 labeled strand was observed on a Molecular Dynamics Typhoon imaging system (Molecular Dynamics, Sunnyvale, CA).
Fluorescence Titrations
Fluorescence titrations were performed as described [36], using a PTI QM-4 fluorimeter (Photon Technology International, Lawrenceville, NJ). All slit-widths were set at 0.5 mM. The temperature of the sample was controlled using a Lauda RM6 recirculating water bath (Brinkmann, Westbury, NY). Stirring was maintained using a P-73 cylindrical cell stirrer (NSG Precision Cells, Inc., Farmingdale, NY).
DNA Substrate Sequences and Annealing Procedures
All oligodeoxynucleotides were synthesized on a MerMade 4 synthesizer (Bioautomation, Plano, TX) using phosphoramidate reagents (Glen Research, Sterling, VA) and purified as described [62]. The concentration of each DNA strand was determined by digesting the strand with phosphodiesterase I (Worthington, Lakewood, NJ) in 100 mM Tris-HCl (pH 9.2), 3 mM MgCl2, at 25 °C and measuring the absorbance of the resulting mixture of mononucleotides at 260 nm. The extinction coefficients at 260 nm used in this analysis are: 15,340 M−1 cm−1 for AMP, 7,600 M−1 cm−1 for CMP, 1,2160 M−1 cm−1 for GMP, 8,700 M−1 cm−1 for TMP, 5,000 M−1 cm−1 for Cy3, and 10,000 M−1 for Cy5 (Glenn Research).
Double Stranded DNA
Sequences of each oligonucleotide used are given in Supplementary Table S1. Each oligonucleotide is labeled with a Roman numeral and a strand number (1, 2, or 3). Each DNA in a series (e.g., DNA series V), differs in length, designated by L. For example, DNA Substrate Series V (L=5) Strand-1 and DNA Substrate Series V (L=5) Strand-2 were annealed to make DNA V (L=5) (Fig. 1b). Each double-stranded DNA is formed by annealing two oligodeoxynucleotides, with Strand 2 (unlabeled or containing Cy5) in 25% excess over Strand 1. These strands were mixed, heated to 95°C for 5 minutes, and cooled slowly to 25 °C. For DNA with biotin, a 3× molar concentration of streptavidin was added to the mixture and equilibrated for 5 minutes before RecBCD was added. The unlabeled RP DNA substrate was made by annealing Strand II-2 and Strand IV-2 from Substrate II and IV in equi-molar quantities.
Nicked Substrates
Each substrate is formed by annealing three DNA strands, with the template strand in 25% excess and the Cy5 labeled strand (Strand 2) in 50% molar excess over the Cy3 labeled Strand 1 to ensure no un-annealed Strand 1 was present in the experiment. These were mixed, heated to 95°C for 5 minutes, and cooled slowly to 25 °C. For DNA with biotin, a 3× molar excess of streptavidin was added to the reaction mixture and allowed to equilibrate for 5 minutes before RecBCD was added.
Supplementary Material
Highlights.
E. coli RecBCD can unwind duplex DNA processively in the absence of ssDNA translocation by the canonical motors
DNA melting by RecBCD can occur separately from DNA translocation
This activity requires the RecB ATPase motor, the RecB arm and nuclease domains, but not its nuclease activity
The RecBC secondary translocase activity is dependent on the RecB arm domain
The RecB nuclease domain regulates the helicase activity of RecBCD
DNA unwinding by RecBCD does not occur solely by the RecB and RecD motors pulling the duplex DNA across a pin or wedge
Acknowledgements
We thank Thang Ho for synthesis and purification of the DNA, Dr. Chris Ho for constructing views of potential movements of the nuclease domain, Dr. Roberto Galletto for many fruitful discussions, suggestions and comments on the ms. and Drs. Aaron Lucius and Gerald R. Smith for comments on the ms. This work was supported, in part, by NIH (GM045948 to TML) and American Cancer Society fellowship to J.S. (PF-15-040-01-DMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Dillingham MS, Kowalczykowski SC. RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiology and molecular biology reviews : MMBR. 2008;72:642–71. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Smith GR. How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist’s view. Microbiology and molecular biology reviews : MMBR. 2012;76:217–28. doi: 10.1128/MMBR.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–93. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- [4].Chen HW, Ruan B, Yu M, Wang J, Julin DA. The RecD subunit of the RecBCD enzyme from Escherichia coli is a single-stranded DNA-dependent ATPase. J Biol Chem. 1997;272:10072–9. doi: 10.1074/jbc.272.15.10072. [DOI] [PubMed] [Google Scholar]
- [5].Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–93. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- [6].Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–7. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- [7].Yu M, Souaya J, Julin DA. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:981–6. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yu M, Souaya J, Julin DA. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J Mol Biol. 1998;283:797–808. doi: 10.1006/jmbi.1998.2127. [DOI] [PubMed] [Google Scholar]
- [9].Sun JZ, Julin DA, Hu JS. The nuclease domain of the Escherichia coli RecBCD enzyme catalyzes degradation of linear and circular single-stranded and double-stranded DNA. Biochemistry. 2006;45:131–40. doi: 10.1021/bi051150v. [DOI] [PubMed] [Google Scholar]
- [10].Rigden DJ. An inactivated nuclease-like domain in RecC with novel function: implications for evolution. BMC Struct Biol. 2005;5:9. doi: 10.1186/1472-6807-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD Enzyme Switches Lead Motor Subunits in Response to chi Recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bianco PR, Kowalczykowski SC. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–72. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- [13].Korangy F, Julin DA. Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry. 1994;33:9552–60. doi: 10.1021/bi00198a022. [DOI] [PubMed] [Google Scholar]
- [14].Wu CG, Lohman TM. Influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. J Mol Biol. 2008;382:312–26. doi: 10.1016/j.jmb.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wu CG, Bradford C, Lohman TM. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat Struct Mol Biol. 2010;17:1210–7. doi: 10.1038/nsmb.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: the gene for an essential third subunit of exonuclease V. ProcNatlAcadSciUSA. 1986;83:5558–62. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu CG, Xie F, Lohman TM. The primary and secondary translocase activities within E. coli RecBC helicase are tightly coupled to ATP hydrolysis by the RecB motor. J Mol Biol. 2012;423:303–14. doi: 10.1016/j.jmb.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xie F, Wu CG, Weiland E, Lohman TM. Asymmetric regulation of bipolar single-stranded DNA translocation by the two motors within Escherichia coli RecBCD helicase. J Biol Chem. 2013;288:1055–64. doi: 10.1074/jbc.M112.423384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lohman TM. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- [20].Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- [21].Betterton MD, Julicher F. Opening of nucleic-acid double strands by helicases: active versus passive opening. Physical review E, Statistical, nonlinear, and soft matter physics. 2005;71:011904. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- [22].Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–26. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Carter AR, Seaberg MH, Fan HF, Sun G, Wilds CJ, Li HW, et al. Sequence-dependent nanometer-scale conformational dynamics of individual RecBCD-DNA complexes. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bhattacharyya B, Keck JL. Grip it and rip it: structural mechanisms of DNA helicase substrate binding and unwinding. Protein science : a publication of the Protein Society. 2014;23:1498–507. doi: 10.1002/pro.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lucius AL, Vindigni A, Gregorian R, Ali JA, Taylor AF, Smith GR, et al. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J Mol Biol. 2002;324:409–28. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- [26].Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:751–71. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- [27].Farah JA, Smith GR. The RecBCD Enzyme Initiation Complex for DNA Unwinding: Enzyme Positioning and DNA Opening. JMolBiol. 1997;272:699–715. doi: 10.1006/jmbi.1997.1259. [DOI] [PubMed] [Google Scholar]
- [28].Wong CJ, Lohman TM. Kinetic control of Mg2+-dependent melting of duplex DNA ends by Escherichia coli RecBC. J Mol Biol. 2008;378:759–75. doi: 10.1016/j.jmb.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lucius AL, Jason Wong C, Lohman TM. Fluorescence stopped-flow studies of single turnover kinetics of E.coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:731–50. doi: 10.1016/j.jmb.2004.04.009. [DOI] [PubMed] [Google Scholar]
- [30].Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–51. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- [31].Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- [32].Tomko EJ, Fischer CJ, Lohman TM. Single-Stranded DNA Translocation of E. coli UvrD Monomer Is Tightly Coupled to ATP Hydrolysis. J Mol Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–81. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA Monomer Is a Single-stranded DNA Translocase but Not a Processive Helicase in Vitro. J Biol Chem. 2007;282:27076–85. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- [35].Amaratunga M, Lohman TM. Escherichia coli Rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–20. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- [36].Wong CJ, Lucius AL, Lohman TM. Energetics of DNA end binding by E.coli RecBC and RecBCD helicases indicate loop formation in the 3′-single-stranded DNA tail. J Mol Biol. 2005;352:765–82. doi: 10.1016/j.jmb.2005.07.056. [DOI] [PubMed] [Google Scholar]
- [37].Fischer CJ, Tomko EJ, Wu CG, Lohman TM. Fluorescence methods to study DNA translocation and unwinding kinetics by nucleic acid motors. Methods Mol Biol. 2012;875:85–104. doi: 10.1007/978-1-61779-806-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fischer CJ, Wooten L, Tomko EJ, Lohman TM. Kinetics of motor protein translocation on single-stranded DNA. Methods Mol Biol. 2009;587:45–56. doi: 10.1007/978-1-60327-355-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tomko EJ, Fischer CJ, Lohman TM. Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods. 2010;51:269–76. doi: 10.1016/j.ymeth.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A Nonuniform Stepping Mechanism for E. coli UvrD Monomer Translocation along Single-Stranded DNA. Molecular Cell. 2007;26:335–47. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hsieh S, Julin DA. Alteration by site-directed mutagenesis of the conserved lysine residue in the consensus ATP-binding sequence of the RecB protein of Escherichia coli. Nucleic Acids Res. 1992;20:5647–53. doi: 10.1093/nar/20.21.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ganesan S, Smith GR. Strand-specific binding to duplex DNA ends by the subunits of the Escherichia coli RecBCD enzyme. JMolBiol. 1993;229:67–78. doi: 10.1006/jmbi.1993.1008. [DOI] [PubMed] [Google Scholar]
- [43].Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J Biol Chem. 2005;280:37069–77. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- [44].Korangy F, Julin DA. A mutation in the consensus ATP-binding sequence of the RecD subunit reduces the processivity of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:3088–95. [PubMed] [Google Scholar]
- [45].Korangy F, Julin DA. Alteration by site-directed mutagenesis of the conserved lysine residue in the ATP-binding consensus sequence of the RecD subunit of the Escherichia coli RecBCD enzyme. J Biol Chem. 1992;267:1727–32. [PubMed] [Google Scholar]
- [46].Korangy F, Julin DA. Enzymatic effects of a lysine-to-glutamine mutation in the ATP-binding consensus sequence in the RecD subunit of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:1733–40. [PubMed] [Google Scholar]
- [47].Zhang XJ, Julin DA. Isolation and characterization of the C-terminal nuclease domain from the RecB protein of Escherichia coli. Nucleic Acids Res. 1999;27:4200–7. doi: 10.1093/nar/27.21.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- [49].Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. Embo J. 2008;27:2222–9. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J. 2003;85:2224–39. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Krajewski WW, Fu X, Wilkinson M, Cronin NB, Dillingham MS, Wigley DB. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature. 2014;508:416–9. doi: 10.1038/nature13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Taylor AF, Smith GR. RecBCD enzyme is altered upon cutting DNA at a chi recombination hotspot. Proc Natl Acad Sci U S A. 1992;89:5226–30. doi: 10.1073/pnas.89.12.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Taylor AF, Schultz DW, Ponticelli AS, Smith GR. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell. 1985;41:153–63. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- [54].Dixon DA, Kowalczykowski SC. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- [55].Taylor AF, Smith GR. Strand Specificity of Nicking of DNA at Chi Sites by RecBCD Enzyme. JBiolChem. 1995;270:24459–67. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- [56].Anderson DG, Kowalczykowski SC. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–81. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- [57].Anderson DG, Kowalczykowski SC. The Translocating RecBCD Enzyme Stimulates Recombination by Directing RecA Protein onto ssDNA in a c-regulated Manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- [58].Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–80. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- [59].Taylor AF, Amundsen SK, Guttman M, Lee KK, Luo J, Ranish J, et al. Control of RecBCD enzyme activity by DNA binding- and Chi hotspot-dependent conformational changes. J Mol Biol. 2014;426:3479–99. doi: 10.1016/j.jmb.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Boehmer PE, Emmerson PT. The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J Biol Chem. 1992;267:4981–7. [PubMed] [Google Scholar]
- [61].Taylor AF, Smith GR. Monomeric RecBCD enzyme binds and unwinds DNA. J Biol Chem. 1995;270:24451–8. doi: 10.1074/jbc.270.41.24451. [DOI] [PubMed] [Google Scholar]
- [62].Nguyen B, Sokoloski J, Galletto R, Elson EL, Wold MS, Lohman TM. Diffusion of human replication protein A along single-stranded DNA. J Mol Biol. 2014;426:3246–61. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.