Abstract
The evolution of functional magnetic resonance imaging to resting state (R-fMRI) allows measurement of changes in brain networks attributed to state changes, such as in neuropsychiatric diseases versus healthy controls. Since these networks are observed by comparing normalized R-fMRI signals, it is difficult to determine the metabolic basis of such group differences. To investigate the metabolic basis of R-fMRI network differences within a normal range, eyes open versus eyes closed in healthy human subjects was used. R-fMRI was recorded simultaneously with fluoro-deoxyglucose positron emission tomography (FDG-PET). Higher baseline FDG was observed in the eyes open state. Variance-based metrics calculated from R-fMRI did not match the baseline shift in FDG. Functional connectivity density (FCD)-based metrics showed a shift similar to the baseline shift of FDG, however, this was lost if R-fMRI “nuisance signals” were regressed before FCD calculation. Average correlation with the mean R-fMRI signal across the whole brain, generally regarded as a “nuisance signal,” also showed a shift similar to the baseline of FDG. Thus, despite lacking a baseline itself, changes in whole-brain correlation may reflect changes in baseline brain metabolism. Conversely, variance-based metrics may remain similar between states due to inherent region-to-region differences overwhelming the differences between normal physiological states. As most previous studies have excluded the spatial means of R-fMRI metrics from their analysis, this work presents the first evidence of a potential R-fMRI biomarker for baseline shifts in quantifiable metabolism between brain states.
Key words: : baseline state, default mode, energy metabolism, functional connectivity, neuronal activity, resting state
Introduction
Resting state functional magnetic resonance imaging (R-fMRI) has emerged as a popular mode of imaging networks in the human brain (Biswal et al., 1995; van den Heuvel and Hulshoff Pol, 2010). Changes in these networks occur under various diseases, and can closely match changes in brain metabolism due to that disease, for example, in Alzheimer's (Perrotin et al., 2015). This has created much interest in better understanding the metabolic basis of R-fMRI networks (Duncan et al., 2013; Hyder et al., 2013; Nugent et al., 2015; Tomasi et al., 2013; Vaishnavi et al., 2010). R-fMRI is noninvasive and, compared to other methods (e.g., positron emission tomography, PET), easy to acquire. Therefore, if R-fMRI can be used to generate metabolic biomarkers of certain neurological diseases, it would be useful for monitoring the progress of such diseases and potentially for diagnosis as well.
There are several R-fMRI-derived candidates for measuring state changes in metabolic activity in the brain at rest. First, metrics based on per-voxel signal variance/power, including frequency-band-limited measures such as the amplitude of low-frequency fluctuations (ALFF) and fractional ALFF (FALFF), hypothetically measure inherent levels of the spontaneous local neural activity that is generating the networks (Zang et al., 2007; Zou et al., 2008). FALFF and ALFF show a state change between the eyes open and eyes closed states (Jao et al., 2013; Yan et al., 2009) and correlate well with simultaneously recorded maps of glucose metabolism (Aiello et al., 2015). Second, to hypothetically measure hubs of communication across the brain, connections between voxels can be quantified to produce maps of the density of connections. Methods for this include regional homogeneity, degree of centrality and functional connectivity density (FCD) (Buckner et al., 2009; Tomasi and Volkow, 2010; Zang et al., 2004). Like FALFF, regional homogeneity and FCD correlate well with glucose metabolism (Aiello et al., 2015; Tomasi et al., 2013). Third is the impact of BOLD time-series signal fluctuations common to all voxels of the brain, also called the whole-brain signal or global signal. This is poorly understood, but appears to reflect global neural electrical activity (Scholvinck et al., 2010) and, like FALFF, shows a state change between the eyes open and eyes closed states (Wong et al., 2013).
Previous studies of these potential biomarkers and metabolism have examined the spatial similarity between the derived R-fMRI metrics and glucose metabolism by using spatial correlation within a single brain state (Aiello et al., 2015; Tomasi et al., 2013) and comparing specific networks (Di et al., 2012; Wehrl et al., 2013). This study innovates on their work in two ways. First [as in Riedl et al. (2014)] eyes closed versus eyes open was used herein as a state change in the brain. (Different subjects were used in each group.) Second, and more importantly, previous studies have discarded the baseline glucose activity either directly or through using correlation coefficients that remove the mean glucose activity level from each subject. While these methods are highly effective at finding topological similarity, emerging evidence suggests that the state changes in brain metabolism may be global rather than local (Wong et al., 2013) and thus such normalization may obscure the very state change we wish to search for within the R-fMRI signal (Hyder et al., 2013).
Previously reported data of subjects with either eyes closed or eyes open (Riedl et al., 2014) were used herein, where R-fMRI was recorded simultaneously with fluoro-deoxyglucose PET (FDG-PET). Average nuclear decay counts (Becquerel/mL [Bq/mL]) of FDG were calculated in 16 brain networks derived from the R-fMRI data. Averages of variance-based, FCD-based, and whole-brain signal-based R-fMRI metrics were also calculated within these 16 networks. The change in baseline Bq/mL could be compared to the change in the R-fMRI signals (which are inherently relative and thus lack their own baselines). Thus, a link can be made between the inherently relative R-fMRI metrics and nonrelative baseline brain metabolism from PET.
The results presented herein suggest that FCD (when calculated from short-range connections with zeros removed), and whole-brain correlation are potential biomarkers of a metabolic state change in the brain, with important caveats and considerations. First, if “nuisance signal” regression was done before FCD calculation this result was lost. Second, part of the network to network difference in these potential biomarkers was observed to be based on R-fMRI signal variance not correlated with the measured glucose metabolism (but well described by FALFF).
Materials and Methods
Data collection
This study used simultaneously recorded FDG-PET, anatomical MRI, and R-fMRI from 22 subjects, 11 with eyes open (7 male, 4 female, mean ± SD: 56.7 ± 9.6 years old, 75.4 ± 16.4 kg, 172.7 ± 7.8 cm tall) and 11 with eyes closed (8 male, 3 female, mean ± SD: 52.2 ± 10.4 years old, 77.0 ± 13.3 kg, 177.7 ± 9.1 cm tall). These data have been previously published; see Riedl et al. (2014) for detailed methods. To summarize, subjects were instructed to keep their eyes either closed or open, relax, and not to fall asleep. R-fMRI (echo-planar imaging [EPI], TR 2 sec, TE 30 msec, 35 slices with 0.6 mm gap, 192 × 192 mm FOV, 64 × 64 matrix size, 300 volumes, 10 min, 8 sec) data acquisition began simultaneously with injection of the FDG-PET tracer. This allowed R-fMRI acquisition to coincide with the most sensitive period for the FDG-PET imaging: the initial minutes after injection. Following completion of R-fMRI, all subjects closed their eyes and anatomical MRI (MP-RAGE, TR 2.3 sec, TE 2.98 msec, 160 slices with 0.5 mm gap, 256 × 256 mm FOV, 256 × 256 matrix size, 5 min 3 sec) images were then acquired. Thirty minutes postinjection, the FDG-PET images were acquired until 40 min postinjection (saturated list mode, 128 slices with 0.5 mm gap, 192 × 192 mm matrix size, 3.7 × 2.3 × 2.7 mm3 voxel sizes, 10 min).
Before data recording, light from the window of the console room was used to perform the FDG tracer injection. Following this, during all data recording, all lights were switched off during the experiment, including lights within the scanner bore.
FDG-PET data are used in the originally recorded units of Bq/mL corresponding to the number of nuclear decay events of FDG per milliliter of brain tissue.
Data registration and network creation
These methods are described in detail in the Supplementary Data Sections S1 to S11 (Supplementary Data are available online at www.liebertpub.com/brain). To summarize, fMRI data were slice-timing corrected and motion corrected. Segmentation produced gray matter, white matter, cerebrospinal fluid (CSF), and whole-brain masks. Each subject's R-fMRI and FDG-PET data were registered to the same subject's anatomical MRI gray matter maps. Each subject's data in anatomical space were then registered to Montreal Neurological Institute (MNI) space with 2 mm isotropic voxels.
Twenty brain networks were generated using MNI space, blurred (full-width, half-maximum [FWHM] = 8 mm, size =6 mm) R-fMRI data, and the Group ICA of fMRI Toolbox (GIFT, mialab.mrn.org/software/gift/) (Correa et al., 2007). Networks were visually identified and classified solely for the purpose of excluding white matter, CSF, or noise networks; four such networks were excluded (IC#1, #3, #6, and #20). Classifications of networks are listed in Supplementary Table S1.
Comparison of FDG-PET counts to quantitative units
Bq/mL units correspond to the amount of radioactive decay of FDG in each brain location, and thus correlate with baseline glucose metabolism. However, there was concern that differential uptake by different cell types could cause Bq/mL to differ spatially from the actual cerebral metabolic rate of glucose consumption (CMRglc).
To test this, CMRglc data in 41 Brodmann regions (left and right sides combined, Supplementary Table S2) from a previous study, recorded with arterial blood sampling to provide absolute CMRglc units, were used (Hyder et al., 2016). From this study, mean Bq/mL (from FDG-PET) and mean T1 MPRAGE contrast (from anatomical MRI) in the same 41 Brodmann regions were calculated for each subject. The mean for each Brodmann region across all subjects with eyes closed (as eyes were closed in Hyder et al.) was then taken and mean Bq/mL was correlated with both mean CMRglc from Hyder et al. and mean T1 MPRAGE contrast.
Calculation of R-fMRI metrics
From the R-fMRI data we calculated three signal variance based metrics, three FCD based metrics, and a final metric based on whole-brain correlation. Before calculation of metrics, all R-fMRI data were masked to the brain, blurred (FWHM = 8, size = 6 mm) and were analyzed both (i) without any nuisance signal regression, and (ii) with regression of nine signals. The nine signals were as follows: the mean whole-brain signal, mean white matter signal, mean CSF signal, and time courses for six motion parameters. See Supplementary Data Section S3 for full details. Additional subsets of regression were also performed, however, each proved similar to either i or ii, and therefore these will be the focus of the main text (Supplementary Data Section S7 for full details).
Note that the use of high-pass filters, Pearson correlation coefficients, and variance in calculating R-fMRI metrics removed the temporal means of the BOLD signals. However, these metrics are based on correlation and variance rather than intensity, and thus the results may have very different spatial means between subjects and between brain regions.
FCD-based R-fMRI metrics: short-range, long-range, and total FCD
Before FCD calculation, R-fMRI images were spatially downsampled by 4.5 (to reduce computational time of FCD; Supplementary Data Section S4), and filtered to 0.01–0.08 Hz (For FCD preprocessing in detail see Supplementary Data Section S5). FCD is defined as the number of voxels that correlate with a given voxel above a certain threshold; here, we used 0.6 as was used in the original FCD work (Tomasi and Volkow, 2010).
Three FCD metrics were calculated. (1) Total FCD resulting from the connections between a voxel and all other voxels in the brain. (2) Short-range FCD resulting from the connections between a voxel and other voxels within six voxels (12 mm) distance in any direction (thus a sphere with 24 mm diameter). While small, this is comparable and slightly larger than has been used for the similar, distance-limited measure of regional homogeneity in similar recent work (Aiello et al., 2015). (3) The short-range FCD map was subtracted from the total FCD map to provide long-range FCD.
For averaging purposes, the three FCD metrics were both calculated including zero values (FCD) and not including zero values (nzFCD) for a total of six FCD metrics in each network. (Zero values occur when no connections are above the correlation threshold.)
Nuisance signal-based metric: whole-brain correlation
A final metric was calculated, referred to as “whole-brain correlation.” The mean over all voxels in the whole-brain mask was calculated at each time point for BOLD. This “whole-brain signal” was then correlated (Pearson correlation) with each voxel's own time signal. The resulting correlation value (r) was then assigned to that voxel's spatial location to provide a map of “whole-brain correlation.” A quadratic detrend was done on the whole-brain signal before correlation to remove global drift (subtract quadratic fit; see Supplementary Data Section S11 and Supplementary Fig. S1 for frequencies affected). Other than that, no filtering, regression, or normalization was done to the BOLD data or whole-brain signal before this calculation. Finally, the r values were converted to Z values, which represented the number of standard deviations from a null hypothesis of no correlation using a normalizing fisher transformation [equation (1) in Thompson et al. (2013)]. Note that, despite averaging first, this method is mathematically equivalent to calculating the correlation between each voxel and every other voxel in the brain (Saad et al., 2013). Also note that whole-brain correlation can also be considered a form of weighted FCD (Cole et al., 2010).
Variance-based R-fMRI metrics: full-band variance, ALFF, FALFF
Three signal variance-based metrics were calculated from the R-fMRI signal. (1) Standard variance (σ2) per-voxel (full-band variance). (2) The ALFF. (3) FALFF. Calculation was as in Zang et al. (2007) and Zou et al. (2008) including pass-bands of 0.01–0.08 Hz for both and a 0–0.25 Hz normalizing band for FALFF.
Significance testing on metrics
Significance testing
Two-way analysis of variance (ANOVA2) with eyes open versus eyes closed as the first variable, and network versus network as the second variable, was conducted on each metric. The metrics upon which significance testing was conducted were FDG, three variance-based metrics (full-band variance, ALFF, and FALFF), six FCD-based metrics (total, short-range, and long-range, with and without zeros included in averaging) and whole-brain correlation calculated from R-fMRI data. The variance and FCD-based metrics were tested both with and without nuisance signal regression. This produces 20 tests, which are listed in Table 1.
Table 1.
Two-Dimensional ANOVA Results
| Eyes open vs. eyes closed | Network vs. network | ||||
|---|---|---|---|---|---|
| p | % Change | p | Mean% change | Interaction p | |
| FDG (Bq/mL) | 1.1 × 10−6 | 11 | 0.51 | 5.0 | 1.0 |
| Full-band variance | |||||
| No regression | 0.27 | 7.9 | 0.98 | 11 | 0.97 |
| Regression | 0.59 | 3.2 | 0.99 | 8.0 | 0.98 |
| ALFF | |||||
| No regression | 8.9 × 10−4 | 79 | 1.6 × 10−25 | 19 | 1.0 |
| Regression | 0.0029 | 470 | 3.1 × 10−27 | −35 | 1.0 |
| FALFF | |||||
| No regression | 0.93 | 0.62 | 6.5 × 10−41 | 1400 | 1.0 |
| Regression | 0.065 | 20 | 2.1 × 10−32 | 240 | 1.0 |
| Total FCD (voxels) | |||||
| No regression | 0.23 | 26 | 0.98 | 33 | 1.0 |
| Regression | 0.22 | 13 | 0.011 | 36 | 0.85 |
| Short-range FCD (voxels) | |||||
| No regression | 0.0024 | 19 | 0.0016 | 24 | 1.0 |
| Regression | 0.56 | 1.9 | 3.4 × 10−14 | 22 | 1.0 |
| Long-range FCD (voxels) | |||||
| No regression | 0.23 | 26 | 0.98 | 33 | 1.0 |
| Regression | 0.18 | 18 | 0.036 | 43 | 0.83 |
| Total nzFCD (voxels) | |||||
| No regression | 0.17 | 27 | 0.99 | 29 | 1.0 |
| Regression | 0.41 | 7.5 | 0.073 | 29 | 0.95 |
| Short-range nzFCD (voxels) | |||||
| No regression | 8.0 × 10−4 | 17 | 0.0037 | 17 | 1.0 |
| Regression | 0.20 | 2.9 | 2.6 × 10−12 | 14 | 1.0 |
| Long-range nzFCD (voxels) | |||||
| No regression | 0.12 | 28 | 1.0 | 23 | 1.0 |
| Regression | 0.88 | 1.2 | 0.82 | 17 | 1.0 |
| Whole-brain correlation (Z) | 4.4 × 10−5 | 20 | 0.32 | 12 | 1.0 |
Results from 2D ANOVA testing with between-condition (eyes open vs. eyes closed) as one dimension and between-network as the other dimension. Each row is a different metric, with FDG as the first metric, R-fMRI-derived potential biomarkers following, and whole-brain correlation last. Where applicable, results with and without regression of nuisance signals are shown. Bolded values are significant at p ≤ 0.05 corrected with SGoF for multiple comparisons across all p values. The first two columns show the p value and percentage change between the eyes open and eyes closed conditions. This is significant (p ≤ 0.00089 from SGoF) for ALFF (without regression), FDG, short-range nzFCD (without regression), and whole-brain correlation. The middle two columns show the p value and (mean) percentage change between networks. This is significant for ALFF and FALFF (with or without regression) and short-range FCD and nzFCD (with regression). The final column shows the p value for interaction between network and condition. This was not significant (p > 0.50) in every case.
ALFF, amplitude of low-frequency fluctuation; ANOVA, analysis of variance; FALFF, fractional ALFF; FCD, functional connectivity density; FDG, fluoro-deoxyglucose; nzFCD, FCD with zeros removed for averaging purposes only; SGoF, sequential goodness of fit.
The mean percentage change was calculated by taking each pair of values in a set, finding the percentage change from the lower of the two values to the higher, then taking the mean of all of these percentage changes. (Mean percentage change can be negative if calculated from negative values.)
Multiple comparisons correction
All p values resulting from all ANOVA2 tests in the main study (60 p values total, Table 1) were concatenated and tested with sequential goodness of fit (SGoF) a binomial method of controlling Type I (false positive) errors (Carvajal-Rodriguez et al., 2009) at a threshold of p ≤ 0.05 (Supplementary Data Section S6 for Type II errors).
Comparison of network to network mean differences
For reasons stated in the Results Section, the FDG, FALFF, short-range nzFCD, and whole-brain correlation metrics were chosen to be compared in terms of their mean value in-network versus the 16 different networks. The mean values in-network had the mean taken across all subjects in either the eyes closed or eyes open state, and these 32 values (16 networks, two states) were concatenated end to end to produce a “signal” of mean value versus network/state. The four “signals” from FDG, FALFF, short-range nzFCD, and whole-brain correlation were each set to zero mean, unit variance and the shared variance in terms of metric mean per network/state were calculated between each pair of “signals” by taking Pearson correlation squared.
Significance testing on motion
Separate from metric significance testing (see Significance testing on metrics Section) the maximum deviations in motion for each R-fMRI run were also tested with ANOVA2 with eyes open versus eyes closed as the first variable and which motion direction as the second variable. Two separate ANOVA2 tests were conducted for the three translation parameters and the three rotation parameters. Results were also corrected with SGoF. Variance was also tested in Supplementary Data Section S10.
Results
Statistical significance threshold
The 60 p values resulting from ANOVA2 were used to generate a new threshold corrected for Type I errors with a threshold of p ≤ 0.05. The threshold produced was p ≤ 8.9 × 10−4, resulting in 10 of 60 tests being significant. These 10 tests are shown in bold on Table 1.
Change in FDG
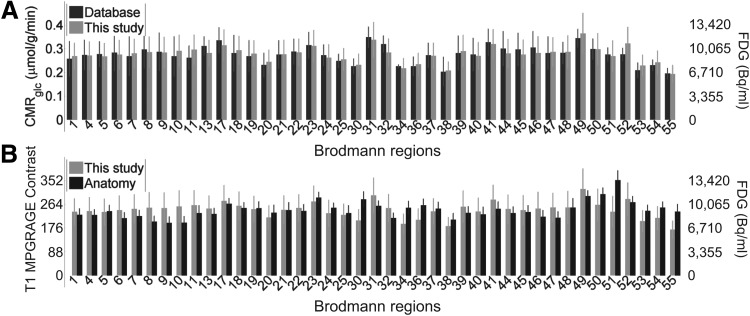
Mean FDG in the eyes closed state in 41 Brodmann regions (left and right sides combined) from this study was compared to mean CMRglc in the eyes closed state from Hyder et al. (2016) (Fig. 1). The FDG data and the anatomical T1 MPRAGE contrast (structural MRI) were also compared. Correlation between this study's FDG (in Bq/mL) and CMRglc from Hyder et al. was very high, r = 0.895/80%. Correlation between this study's FDG and its matching structural MRI was lower, r = 0.20/4.0%. This indicates that the region-to-region variation in FDG (measured in Bq/mL) closely follows CMRglc rather than anatomical peculiarities of this study.
FIG. 1.
Mean values across 41 Brodmann regions (listed in Supplementary Table S2). Mean is taken across the region first, then across all subjects with eyes closed. (A) Mean FDG (Bq/mL, N = 11) from this study (light gray bars) compared to mean CMRglc (N = 13) from Hyder et al. (2016) (dark gray bars). Error bars are one standard deviation. (B) Mean FDG (Bq/mL, N = 11) from this study (light gray bars) compared to mean anatomical contrast (T1 MPRAGE, N = 11) also from this study (black bars). Note that Bq/mL correlates very highly with quantitative CMRglc, even more than the mean anatomical contrast from the same subjects. CMRglc, cerebral metabolic rate of glucose consumption; FDG, fluorodeoxyglucose.
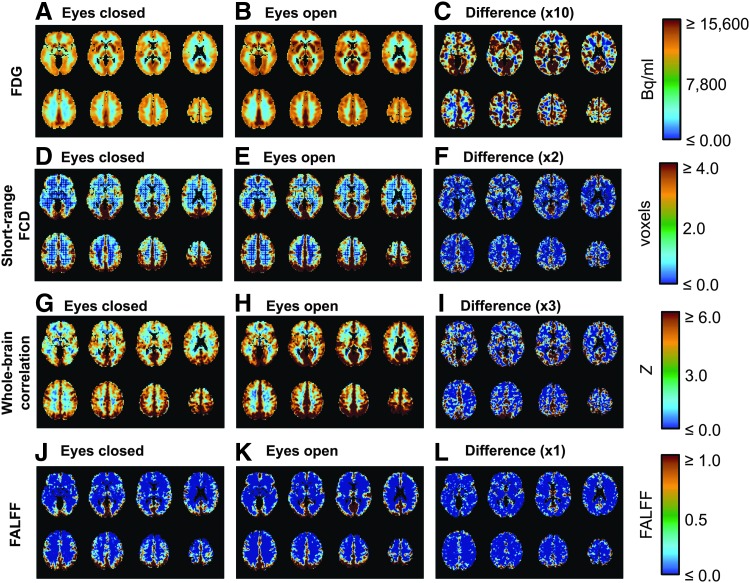
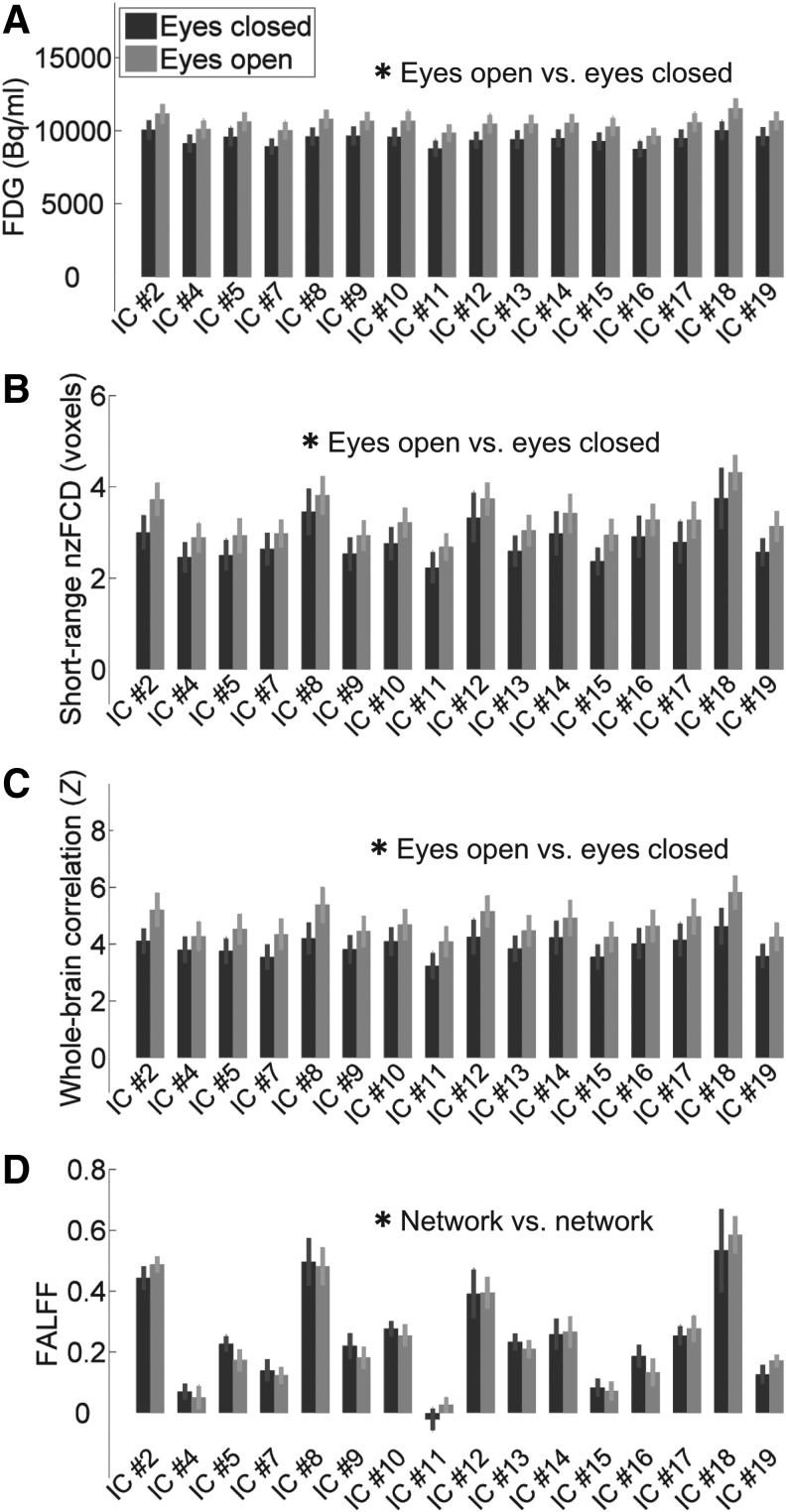
Globally higher FDG was observed in the eyes open condition as compared to the eyes closed condition (Figs. 2A–C and Fig. 3A), this difference was significant (Table 1). While some region to region differences are visible, such as IC #18 (the posterior cingulate cortex) being higher than IC #16 (the dorsal somatomotor cortex), these differences are comparatively small compared to inter-subject variance and were not significant (Table 1). Overall, there is a very large baseline FDG in each state, this baseline changes between states, and the network to network differences atop this baseline are comparatively small.
FIG. 2.
Mean (N = 11) brain images showing the global increase in FDG and some potential R-fMRI biomarkers. (A) FDG, lower in eyes closed condition. (B) FDG, higher in eyes open condition. (C) Difference between (A) and (B), times 10 to show on same scale. (D) Short-range FCD, lower in the eyes closed condition. (E) Short-range FCD, higher in the eyes open condition. (F) Difference between (D) and (E), times two to show on the same scale. (G) Whole-brain correlation, lower in the eyes closed condition. (H) Whole-brain correlation, greater in the eyes open condition. (I) Difference between (G) and (H), times three to show on the same scale. (J) FALFF in the eyes closed condition. (K) FALFF in the eyes open condition. (L) Difference between (J) and (K) on the same scale. From (C), (F) and (I) it can be noted that these metrics are consistently higher across gray matter in the eyes open condition. In (L) as there are changes in both directions (Fig. 3). ALFF, amplitude of low-frequency fluctuation; FALFF, fractional ALFF; FCD, functional connectivity density; R-fMRI, resting state functional magnetic resonance imaging.
FIG. 3.
Mean (N = 11) of metrics is plotted as bars for 16 numbered networks along the X-axis. Error bars are one standard error. Dark gray is N = 11 in the eyes closed condition, light gray is N = 11 in the eyes open condition. All significance from ANOVA2 (Table 1) is shown with asterisks (*), nonsignificant not shown. (A) FDG. (B) Short-range nzFCD. (C) Whole-brain correlation. (D) FALFF. Note that FDG, short-range nzFCD and whole-brain condition are relatively constant within a given state, but globally greater in the eyes open state. FALFF has strong network to network variation, but does not show the global effect. nzFCD, FCD with zeros removed for averaging purposes only.
FCD-based metrics
Short-range (<6 voxels or 12 mm sphere) nzFCD with no nuisance signal regression was observed to be globally higher in the eyes open condition as compared to the eyes closed condition (Figs. 2D–F and 3B), this difference was significant. There were greater network versus network differences in short-range nzFCD than in FDG, but it was nonsignificant (Table 1; see also Supplementary Data Section S6 and Supplementary Fig. S2 for consideration of Type II errors).
When nuisance signal regression was not done, results for other distances of FCD and including zeros in the averaging followed the same trend as short-range nzFCD, but were nonsignificant (Table 1 and Supplementary Fig. S3). However, if nuisance signal regression was done, no eyes open versus eyes closed differences remain significant, and some network versus network differences become significant (Table 1 and Supplementary Fig. S3). For additional subsets of regression, see Supplementary Data Section S7.
These results suggest that short-range FCD can potentially be a biomarker of the globally higher values in the eyes open condition observed in FDG. However, FCD varies between networks more than FDG. Regression of nuisance signals amplifies this variation and removes the eyes open versus eyes closed difference.
Whole-brain correlation as a metric
The mean correlation between in-network voxels and the whole-brain signal, between the eyes open versus eyes closed states, is shown in Figures 2G–I and 3C. There is significant higher whole-brain correlation in all networks in the eyes open condition than in the eyes closed condition, and network versus network differences are small (Table 1). This is similar to FDG, though network versus network has a higher percentage change for whole-brain correlation (20% vs. 11%).
Variance-based metrics
FALFF (without nuisance signal regression) had large and statistically significant network versus network differences, but was not significantly different eyes open versus eyes closed (Figs. 2J–L and 3D). Results for ALFF, and FALFF/ALFF without nuisance signal regression were generally similar, though ALFF without nuisance signal regression also showed a significant increase in the eyes open state (Table 1 and Supplementary Fig. S4). There were no significant results for full-band variance (Table 1 and see also Supplementary Data Section S6 and Supplementary Fig. S5 for consideration of Type II errors).
Thus, variance-based metrics were not similar to FDG, but FALFF well-characterized large network to network differences.
Network to network variation
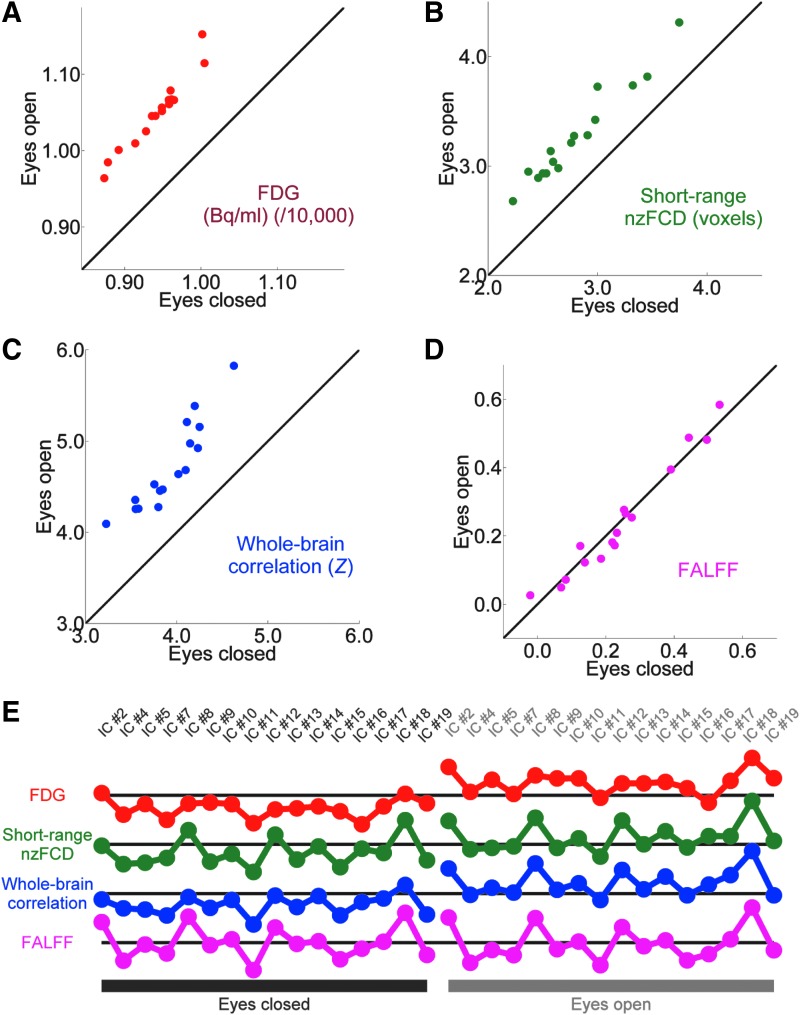
To better understand the R-fMRI signal, the two best potential biomarkers (short-range nzFCD without nuisance signal regression and whole-brain correlation) were chosen. FALFF without regression was also chosen as it had the greatest mean network to network difference (1400%), is considered less susceptible to physiological noise than ALFF (Zou et al., 2008), and was compared to the nzFCD calculated without regression. Network to network values in FDG (Fig. 4A), short-range nzFCD (no regression, Fig. 4B), whole-brain correlation (Fig. 4C), and FALFF (no regression, Fig. 4D) were converted to signals (Fig. 4E) and the network to network variation correlated between modalities.
FIG. 4.
(A) Scatter plot with each network as a point, mean FDG (N = 11) in the eyes closed condition as the X-axis, mean FDG (/10,000, N = 11) in the eyes open condition as the Y-axis. Line is Y = X. All values are in Bq/mL divided by 10,000. (B) As (A), but short-range nzFCD. (C) As (A), but whole-brain correlation. (D) As (A), but FALFF. FDG, short-range nzFCD and whole-brain correlation show a shift from the Y = X line, while FALFF instead occupies a larger dynamic range along it. Voxel-wise scatter plots for (A–D) are shown in Supplementary Figure S7. (E) The Y-axis plots the relative values of each metric in different networks, with the X-axis corresponding to 16 networks in the eyes closed condition (left) and eyes open condition (right). For each metric the black horizontal line represents the average value across all networks and both conditions. Peaks represent a network having a higher value than other networks in that condition (mean of N = 11). Note that, as shown in Figure 3, FDG, short-range nzFCD, and whole-brain correlation all are consistently greater in the eyes open condition, while FALFF has approximately the same mean value across conditions. Also note that some peaks in short-range FCD and whole-brain correlation occur in FALFF, but do not occur in FDG. This suggests that short-range FCD and whole-brain correlation relate to both FDG and FALFF. Statistics are shown in Table 2. Color images available online at www.liebertpub.com/brain
The shared variance between the network to network mean metrics is shown in Table 2. All shared variance values are high (22% or greater) indicating substantial topological similarity between FALFF, short-range FCD and FDG, as has been previously reported (Aiello et al., 2015).
Table 2.
Network to Network Shared Variance Between Different Metrics
| Short-range nzFCD (voxels) (%) | Whole-brain correlation (Z) (%) | FALFF (%) | |
|---|---|---|---|
| FDG (Bq/mL) | 51 | 71 | 22 |
| Short-range nzFCD (voxels) | 85 | 65 | |
| Whole-brain correlation (Z) | 43 |
Correlation across different metrics, between the mean values each metric produces in each network per condition (i.e., correlation of the traces in Fig. 1). FDG, short-range nzFCD (without regression), whole-brain correlation, and FALFF (without regression) are shown. The least shared variance (22%) is between FDG and FALFF. Short-range nzFCD and whole-brain correlation have 43–71% shared variance with both FALFF and FDG. The greatest shared variance is between short-range nzFCD and whole-brain correlation (85%).
It can be noted that, while FDG, short-range nzFCD, and whole-brain correlation all show the trend of being higher in the eyes open condition, FALFF does not (Fig. 3). While some peaks are present in all four, some peaks present in whole-brain correlation and short-range nzFCD are present in FALFF, but are not present in FDG (Fig. 4E). Short-range nzFCD and whole-brain correlation have comparatively high shared variance with FDG and FALFF, compared with FDG shared variance with FALFF (Table 2). Thus, components of both FALFF and FDG network to network variation correlate with the network to network variation of these potential biomarkers.
Short-range nzFCD and whole-brain correlation also show 85% shared variance in network to network differences (Table 2). As whole-brain correlation is much quicker to compute than FCD (Supplementary Data Section S4), this suggests it might be possible to use whole-brain correlation to approximate FCD.
Effect of motion
SGoF correction for Type I errors on the six p values for motion parameters (eyes open vs. eyes closed, the three directions vs. each other, and interaction between direction and state, for both rotation and translation) produced a threshold of p ≤ 5.8 × 10−4 for significance, with two significant tests.
There were significant differences in the amount of motion depending on the direction (p = 2.1 × 10−7, means: 0.43 mm right, 0.34 mm forward, and 1.0 mm up) and the angle (p = 5.9 × 10−4, means: 0.019 rad for pitch, 0.0093 rad for roll, and 0.011 rad for yaw) but there was no significant difference for either depending on whether a subject was in the eyes open or eyes closed group (p = 0.093 for translation, p = 0.93 for rotation, 0.68 mm, 0.013 rad for eyes closed, 0.52 mm, 0.013 rad for eyes open) and there was also no significant interaction between these terms (p = 0.49 for translation, p = 0.87 for rotation).
This suggests motion did not influence observed differences between the eyes open and eyes closed states. In addition, a motion effect would be expected to have a differential effect on long-range connections versus short-range connections in the FCD calculations (Power et al., 2012). The relationship between FDG and the FCD metrics (and whole-brain correlation) is largely constant network to network (Fig. 3), whereas the effect of motion has been shown to vary network to network (Van Dijk et al., 2012).
Discussion
Our study uses a simple yet common state difference that is physiological (eyes open vs. eyes closed) to model a state change in the brain. Within a given state the FDG was constant across networks (not individual brain regions), however, globally higher FDG was observed across all networks in the eyes open state compared with the eyes closed state (Table 1 and Fig. 2). The ICA-derived networks spanned the entire cerebrum, from the frontal cortex through the posterior cingulate cortex to the visual cortex (Supplementary Table S1). Unlike FDG, broadband variance and FALFF showed an increase in some networks but a decrease in others, and ALFF and FALFF showed significant variation between networks within a given state. Thus, these variance-based metrics were not observed to reflect the trends in FDG (Fig. 3). Conversely, all measures of FCD were, like FDG, globally higher in the eyes open state. However, FCD was only statistically significant at r = 0.60 for short-range nzFCD (Table 1 and Fig. 3) and at r = 0.25 for short-range and long-range nzFCD (Supplementary Fig. S2). Whole-brain correlation had a statistically identical result to short-range nzFCD (with r = 0.60) and FDG (Table 1 and Fig. 3C). In terms of network to network variation, whole-brain correlation was strongly correlated with short-range nzFCD, both of these were strongly correlated to both FALFF and FDG, but FDG and FALFF themselves were less correlated (Table 2 and Fig. 4E).
Limitations
FDG-PET data were acquired without arterial blood sampling and, while the original radiation counts numbers (Bq/mL) were used, these counts could vary from the actual CMRglc. However, there is a strong correlation in the eyes closed condition between this study's Bq/mL counts and our previous work where arterial blood sampling was used to obtain absolute CMRglc (Fig. 1), indicating the FDG numbers in this study do reflect the underlying metabolism.
Image registration was generally very consistent for each modality (BOLD, FDG, anatomical MRI). However, there was a small (<0.5%) but significant difference between eyes open and eyes closed for the registrations of the anatomical MRI images versus the registrations of the FDG-PET images (see Supplementary Data Section S8 and Supplementary Fig. S6). This did not affect our results as we compared BOLD metrics to FDG-PET metrics, which did not significantly differ in their registrations. However, future studies that compare anatomy to FDG-PET data should account for this variation.
ICA has “inherent global signal regression built into it” [p. 16, Murphy et al. (2009)]. Therefore, the networks generated by ICA could potentially be biased. However, a similar positive shift can be seen without network selection (Fig. 2 and Supplementary Fig. S7A). Additionally, when different methods of generating networks were used (including seed-based networks with no whole-brain signal regression), a statistically identical result is produced in terms of FDG: a significant increase in the eyes open condition and no significant network to network difference [Conference abstract: Thompson et al. (2015)].
Non-neuronal and nonmetabolic effects impact the measured BOLD signal including changes in field homogeneity, coil receiver gain, analog-to-digital conversion, and subject motion (as motion correction is not perfect). Time-variant and spatially variant effects may average together to impact the potential biomarkers, in particular the whole-brain signal, for example, through bulk motion. Future work including other simultaneously measured and time-variant modalities (e.g., electrophysiology or in vivo microdialysis in animal models) is needed to confirm R-fMRI-derived biomarkers are actually due to metabolism or brain activity.
Neurometabolic basis of potential R-fMRI biomarkers
The state change in glucose metabolism seen between conditions was reflected with the potential R-fMRI biomarkers of FCD and whole-brain correlation, but was only somewhat reflected by ALFF and was not reflected by FALFF or broadband variance. However, a previous study has shown that the topological variation in FALFF does match the topological variation in CMRglc (Aiello et al., 2015). This similarity in spatial correlation was replicated in this study (Fig. 4E and Table 2; Supplementary Fig. S8D and Supplementary Table S3). This raises an important question: How can CMRglc/R-fMRI similarity seen with spatial correlation not directly translate to the state difference? One possibility is that different potential R-fMRI biomarkers represent varying levels of regional tissue differences versus sensitivity to baseline metabolism, and these two elements are not completely correlated.
Tomasi et al. (2013) hypothesize that voxels with greater degree of connectivity (here, higher FCD score) have greater energy demand due to the property of Hopfield neural networks (Hopfield, 1982) where energy demand scales with number of direct functional connections. Cole et al. (2010) indicated that whole-brain connectivity also measures the amount of connections, but in a weighted manner, and thus it would also reflect energy demand. While Aiello et al. (2015) also use connections as a surrogate for measuring energy demand, they also include FALFF, which is not a direct measure of connections. Their justification for FALFF as a correlate of glucose comes from the assumption that low-frequency fluctuations are an inherent part of functional connectivity. Thus, since low-frequency fluctuations create the functional connections we would otherwise measure with FCD, FALFF should also correlate with metabolism.
While this assumption is valid and holds well for differences in FALFF across different regions of the brain, it does not consider that there are both region to region and state to state changes in metabolism. Different brain regions vary in cell type, cell density, vascular density, large vein proximity, and nonphysiological parameters such as the signal-to-noise ratio (Logothetis, 2008; Logothetis et al., 2009), all of which affect either metabolism or the ability to accurately measure it. It is possible that, due to these regional differences in metabolic, hemodynamic and physiological factors, the level of R-fMRI fluctuations measured in a given region may have a high level of variance owed to being in that particular region (e.g., Fig. 3D).
Thus, in the context of a global change in brain metabolism, FALFF will continue to reflect the regional variance differences (and ALFF will still be dominated by them, though it may be more sensitive to baseline metabolism as it is not divided by the wider frequency range), even if the change to baseline FDG does have an effect. FCD and whole-brain correlation are, conversely, less sensitive to the regional variance differences as Pearson correlation normalizes every voxel's temporal variance to one.
Comparison with previous studies
Comparison with previous studies of eyes closed versus eyes open
Even a condition as simple as opening one's eyes creates a large, global metabolic change in the brain. This has been defined as an “interoceptive” state when the eyes are closed, versus an “exteroceptive” state when the eyes are open, that is, either internally or externally focused (Marx et al., 2003).
Many previous studies have examined the difference between the eyes open and eyes closed states using fMRI. However, unlike this study, those studies were interested in local differences so justifiably discarded the mean of each R-fMRI metric with each conditions through performing statistical parametric mapping using the general linear model (Bianciardi et al., 2009; Brandt, 2006; Gonzalez-Hernandez et al., 2005; Marx et al., 2004; Qin et al., 2012; Riedl et al., 2014; Wicker et al., 2003), performing global signal regression to find local significance (Xu et al., 2014; Yuan et al., 2014; Zou et al., 2009), or using paired T-tests on a per voxel basis (which can hypothetically detect a global change if region-to-region variation is small, but will not necessarily as it only compares per-voxel changes and global means are never calculated) (Patriat et al., 2013). All of these methods will amplify local region to region, voxel to voxel variation, but obscure the relations to the global change observed in FDG (Fig. 2 and see also Supplementary Data Section S9 and Supplementary Fig. S9). Some studies have had conflicting results, for example, whether the visual system is more “active” in the eyes open or eyes closed state (Marx et al., 2003; Yuan et al., 2014) or whether functional connectivity is higher or lower (Jao et al., 2013; Yan et al., 2009). The global change in ALFF, short-range nzFCD and whole-brain correlation observed here followed changes in baseline metabolism as measured by absolute Bq/mL values. Thus, various data processing methods that retain different amounts of the global metabolic change may be behind some of the inconsistencies in the literature.
Previous studies have been able to find all networks in both states (Xu et al., 2014). Patriat et al. (2013) reported differences between states, but comparison to this study is difficult as they used correlation between regions of interest rather than per-voxel metrics in defined networks.
Reliability has been shown to be improved in certain networks in the eyes open condition versus eyes closed (Zou et al., 2015). This may be related to the greater baseline metabolic activity observed in this study. Overall, the results of this study support this general consensus that R-fMRI studies can be conducted with either eyes open or eyes closed, with the caveat that baseline metabolic activity (and thus potential reliability of metabolic biomarkers) will be higher with eyes open.
Comparison with previous studies of R-fMRI metrics as metabolic biomarkers
Short- and long-range FCD have both been previously shown to correlate with FDG when recorded separately (Tomasi et al., 2013). Studies of cerebral perfusion also strongly suggest a metabolic link between both short and long range connections (Liang et al., 2013). The regional homogeneity (a measure of short-distance similarity, likely comparable to short-range FCD) has also been shown to correlate with FDG when recorded simultaneously as data were herein (Aiello et al., 2015). In our study, long-range FCD was not significant but demonstrated an identical trend to short-range FCD and FDG (Table 1 and Supplementary Fig. S3). This may be due to high spatial similarity between long-range and short-range FCD and higher subject to subject variance in long-range FCD (Tomasi and Volkow, 2011), corroborated by long-range FCD becoming significant if the r threshold was lowered (Supplementary Fig. S2). Aiello et al. also did not observe as strong a relationship between FDG and the measure of “degree of centrality,” which was calculated in a similar manner to long-range FCD. For comparison to Aiello et al., voxel-wise scatter plots between R-fMRI metrics and CMRglc are shown in Supplementary Figure S8.
Yan et al. showed increased FALFF in the default mode network in the eyes open condition (Yan et al., 2009). Our study replicates that result (IC #19, Fig. 3D). Jao et al. (2013) more recently showed decreased variance and FALFF in the default mode network in the eyes open condition. Results of Jao et al. use much smaller nodes than the networks used in our study, and also used correlation matrices rather than per-voxel FCD, thus many of their results are difficult to compare to our study. Results of Jao et al. regarding FALFF differ from that of Yan et al., and thus there is no consensus with which to compare the results of this study on whether FALFF should increase or decrease with increased CMRglc.
Our result supports the hypothesis put forth by Tomasi et al. (2013) that the metabolism is reflected by the number of connections (from a region) rather than the amplitude of individual fluctuations (within that region). A recent study by Di et al. (2012) showed more spatial correlation between homotopic R-fMRI networks and metabolism, as compared to widely separated networks. Results from the same data as used here, using the general linear model, also agreed that areas of greater local PET activity were spatially localized to areas where functional connectivity increased (Riedl et al., 2014). Similarly, high glucose uptake has been linked to the most strongly connected default mode network nodes (Passow et al., 2015).
While the source of the global signal in the brain is not well understood, there is increasing evidence that part of it has a neural origin (even if other parts are influenced by non-neural or even nonmetabolic hardware-based effects). Schölvinck et al. (2010) showed that an implanted electrode correlated with fMRI voxels from all over the brain. Conversely, for local correlations between two sites in short time windows, regression of the global signal increased correlation between changes in neuroelectric and BOLD signals (Thompson et al., 2013). Wong et al. (2013) demonstrated a relationship between the variance of the global signal itself and vigilance. They observed greater variance in the eyes closed state, which may correspond to the lower mean correlation observed here. Schizophrenia shows increased cortical power and variance across the whole brain (Yang et al., 2014) and lower global brain connectivity overall (Hahamy et al., 2014). Obsessive–compulsive disorder shows changes in local areas in terms of how closely they relate to the global signal (Anticevic et al., 2014). Global brain connectivity also increases across the entire brain under acute ketamine (Driesen et al., 2013).
Variations in cerebrovascular reactivity between different areas of the brain, states and subjects also likely influence the observed results. In particular, the close mapping between resting state physiological fluctuation amplitude and cerebrovascular reactivity observed previously (Kannurpatti and Biswal, 2008; Liu et al., 2013) and the large network to network differences in variance-based metrics observed here (Table 1) suggest that network to network variation in R-fMRI metrics may be influenced by cerebrovascular reactivity. This may apply even to the metrics where network versus network comparisons were not significant (Fig. 4E and Table 2).
Finally, it must be considered that non-neuronal and nonmetabolic factors influence BOLD, and in particular the whole-brain signal (see Limitations Section).
Comparison with previous studies comparing different R-fMRI metrics
Cole et al. referred to a metric similar to FCD as “unweighted global brain connectivity” and an equivalent metric to whole-brain correlation “weighted global brain connectivity” (Cole et al., 2010). They demonstrated that weighted and unweighted global brain connectivities were very similar. Our study mirrors this as high, shared variance was observed between short-range nzFCD and whole-brain correlation (Table 2 and Supplementary Fig. S8B).
Nugent et al. (2015) compared ALFF, whole-brain correlation, and glucose consumption in individuals with temporal lobe epilepsy and healthy controls using spatial correlation. They found ALFF more similar to glucose consumption in healthy controls, but whole-brain correlation more similar in patients (Nugent et al., 2015). It is difficult to compare these results to our study, however, as spatial correlation is independent of spatial mean, which were shifted between states (Fig. 3C).
Aiello et al. (2015) compared multiple R-fMRI metrics to simultaneously recorded brain glucose metabolism in the state of eyes open. They observed strong similarity between FALFF, regional homogeneity, and CMRglc. However, to facilitate topological comparison of these metrics, they set all R-fMRI and PET metrics to zero mean and unit variance before analysis. While this step was necessary for correlation, it meant that they could not have observed a change in global means of potential R-fMRI biomarkers. This makes comparisons with this study difficult as herein results were dominated by the change in the global means of ALFF, short-range nzFCD, and whole-brain correlation. However, corroborating Aiello et al., substantial shared variance of 22% (r = 0.47) or more was observed between all pairings of short-range FCD, FALFF, and CMRglc in terms of topological comparison between networks (Fig. 4E and Table 2). For comparison to Aiello et al., voxel-wise scatter plots between different R-fMRI metrics are shown in Supplementary Figure S8. Compare parts B, E, and F to Figures 2 and 3 of Aiello et al.
Future work
In Figure 2A and B it can be seen that an increase in white matter is observed as well, though to a smaller extent than gray matter. Supplementary Figure S7A illustrates that this is not a partial volume effect as the majority of white matter voxels show an increase similar to gray matter. While this has remained unstudied to a large extent, some evidence suggests that functional connectivity, measurable with R-fMRI, exists in white matter (Ding et al., 2013). In addition, regression of the white matter signal alone produced effects similar to regression of the whole-brain signal, which is unusual, considering that signal variance and temporal features have normally been reported to differ between gray matter and white matter (Birn et al., 2008; Chang and Glover, 2009; Chang et al., 2009) (see Supplementary Data Section S7, Supplementary Table S4). As neurons exist in white matter it is possible these results reflect an actual change in neural activity, but much work beyond the scope of this study would be required to demonstrate this with certainty.
Interaction between state to state differences and network to network differences always had a high probability of accepting the null hypothesis (p > 0.5, often p ≈ 1, Table 2). As this study only had two states, but many networks, it is hard to say definitively if this means that such interactions do not exist. Future work with multiple states (such as different stimuli in healthy subjects or different disease states) could better illuminate how global state changes interact with network to network differences in these potential biomarkers.
The two significant potential biomarkers of FDG in the main study, short-range nzFCD and whole-brain correlation, vary network to network in a manner similar to FALFF (Fig. 4E and Table 2). At a lower threshold for connections, short-range nzFCD also shows the same significance pattern as ALFF (Supplementary Fig. S2A). These results suggest that FCD (and maybe ALFF) reflect two different types of information: both state-dependent metabolic information and network-dependent information containing both metabolic and hemodynamic components. Future studies using calibrated fMRI (combining blood flow and BOLD measurements) could more accurately establish the potential of both short-range FCD and whole-brain correlation as alternatives for metabolic imaging.
A limitation of the dataset of this study is that subjects either had eyes closed or eyes open, but not both. While this does highlight a potential strength of our analytical method (it can used for disease states where a subject cannot be in both groups), intra-subject comparisons would improve interpretation and the authors are currently collecting such data for future work.
Conclusion
We have demonstrated that the globally higher glucose metabolism in the eyes open state is reflected by a similar state change within short-range nzFCD along with an even more similar state change within whole-brain correlation. Thus, these two R-fMRI metrics are promising as potential biomarkers of state changes in glucose metabolism in the brain. Variance-based metrics, conversely, vary network to network more strongly and may elucidate network to network variation better than the other potential biomarkers. Future combined PET/R-fMRI studies with a larger extent of changes in glucose metabolism (e.g., anesthetized or sleep states) would be useful in establishing these R-fMRI metrics as biomarkers for brain state changes.
Supplementary Material
Acknowledgments
Funding for this research included NIH P30 NS-052519 and NIH R01 MH-067528 to F.H., Kommission für klinische Forschung Grant 8762754 at the Klinikum Rechts der Isar der Technischen Universität München to V.R., and a fellowship supporting G.J.T. from NIH 2T32DA022975-06A1. Thanks to Reinder Vos de Wael for proofreading. The acknowledged funding sources were not involved in the design of this study. Informed consent was given by all subjects and all procedures were approved by the Ethics Review Board of the Klinikum Rechts der Isar, Technische Universität München (TUM). Parts of this work were originally presented on June 1, 2015 at the 23rd Annual Meeting of the International Society for Magnetic Resonance in Medicine.
Author Disclosure Statement
No competing financial interests exist.
References
- Aiello M, Salvatore E, Cachia A, Pappata S, Cavaliere C, Prinster A, et al. 2015. Relationship between simultaneously acquired resting-state regional cerebral glucose metabolism and functional MRI: a PET/MR hybrid scanner study. Neuroimage 113:111–121 [DOI] [PubMed] [Google Scholar]
- Anticevic A, Hu S, Zhang S, Savic A, Billingslea E, Wasylink S, et al. 2014. Global resting-state functional magnetic resonance imaging analysis identifies frontal cortex, striatal, and cerebellar dysconnectivity in obsessive-compulsive disorder. Biol Psychiatry 75:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciardi M, Fukunaga M, van Gelderen P, Horovitz SG, de Zwart JA, Duyn JH. 2009. Modulation of spontaneous fMRI activity in human visual cortex by behavioral state. Neuroimage 45:160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. 2008. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage 40:644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34:537–541 [DOI] [PubMed] [Google Scholar]
- Brandt T. 2006. How to see what you are looking for in fMRI and PET—or the crucial baseline condition. J Neurol 253:551–555 [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, et al. 2009. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 29:1860–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Rodriguez A, de Una-Alvarez J, Rolan-Alvarez E. 2009. A new multitest correction (SGoF) that increases its statistical power when increasing the number of tests. BMC Bioinformatics 10:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. 2009. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage 44:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. 2009. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage 47:1381–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Pathak S, Schneider W. 2010. Identifying the brain's most globally connected regions. Neuroimage 49:3132–3148 [DOI] [PubMed] [Google Scholar]
- Correa N, Adali T, Calhoun VD. 2007. Performance of blind source separation algorithms for fMRI analysis using a group ICA method. Magn Reson Imaging 25:684–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X, Biswal BB, Alzheimer's Disease Neuroimaging Initiative. 2012. Metabolic brain covariant networks as revealed by FDG-PET with reference to resting-state fMRI networks. Brain Connect 2:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Newton AT, Xu R, Anderson AW, Morgan VL, Gore JC. 2013. Spatio-temporal correlation tensors reveal functional structure in human brain. PLoS One 8:e82107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driesen NR, McCarthy G, Bhagwagar Z, Bloch M, Calhoun V, D'Souza DC, et al. 2013. Relationship of resting brain hyperconnectivity and schizophrenia-like symptoms produced by the NMDA receptor antagonist ketamine in humans. Mol Psychiatry 18:1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan NW, Wiebking C, Tiret B, Marjanska M, Hayes DJ, Lyttleton O, et al. 2013. Glutamate concentration in the medial prefrontal cortex predicts resting-state cortical-subcortical functional connectivity in humans. PLoS One 8:e60312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hernandez JA, Cespedes-Garcia Y, Campbell K, Scherbaum WA, Bosch-Bayard J, Figueredo-Rodriguez P. 2005. A pre-task resting condition neither ‘baseline’ nor ‘zero’. Neurosci Lett 391:43–47 [DOI] [PubMed] [Google Scholar]
- Hahamy A, Calhoun V, Pearlson G, Harel M, Stern N, Attar F, et al. 2014. Save the global: global signal connectivity as a tool for studying clinical populations with functional magnetic resonance imaging. Brain Connect 4:395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ. 1982. Neural networks and physical systems with emergent collective computational abilities. Proc Natl Acad Sci U S A 79:2554–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyder F, Fulbright RK, Shulman RG, Rothman DL. 2013. Glutamatergic function in the resting awake human brain is supported by uniformly high oxidative energy. J Cereb Blood Flow Metab 33:339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyder F, Herman P, Bailey CJ, Moller A, Globinsky R, Fulbright RK, et al. 2016. Uniform distributions of glucose oxidation and oxygen extraction in gray matter of normal human brain: no evidence of regional differences of aerobic glycolysis. J Cereb Blood Flow Metab [Epub ahead of print]; DOI: 10.1177/0271678X15625349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao T, Vertes PE, Alexander-Bloch AF, Tang IN, Yu YC, Chen JH, et al. 2013. Volitional eyes opening perturbs brain dynamics and functional connectivity regardless of light input. Neuroimage 69:21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannurpatti SS, Biswal BB. 2008. Detection and scaling of task-induced fMRI-BOLD response using resting state fluctuations. Neuroimage 40:1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zou Q, He Y, Yang Y. 2013. Coupling of functional connectivity and regional cerebral blood flow reveals a physiological basis for network hubs of the human brain. Proc Natl Acad Sci U S A 110:1929–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Hebrank AC, Rodrigue KM, Kennedy KM, Park DC, Lu H. 2013. A comparison of physiologic modulators of fMRI signals. Hum Brain Mapp 34:2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK. 2008. What we can do and what we cannot do with fMRI. Nature 453:869–878 [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Murayama Y, Augath M, Steffen T, Werner J, Oeltermann A. 2009. How not to study spontaneous activity. Neuroimage 45:1080–1089 [DOI] [PubMed] [Google Scholar]
- Marx E, Deutschlander A, Stephan T, Dieterich M, Wiesmann M, Brandt T. 2004. Eyes open and eyes closed as rest conditions: impact on brain activation patterns. Neuroimage 21:1818–1824 [DOI] [PubMed] [Google Scholar]
- Marx E, Stephan T, Nolte A, Deutschlander A, Seelos KC, Dieterich M, et al. 2003. Eye closure in darkness animates sensory systems. Neuroimage 19:924–934 [DOI] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. 2009. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage 44:893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent AC, Martinez A, D'Alfonso A, Zarate CA, Theodore WH. 2015. The relationship between glucose metabolism, resting-state fMRI BOLD signal, and GABA-binding potential: a preliminary study in healthy subjects and those with temporal lobe epilepsy. J Cereb Blood Flow Metab 35:583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passow S, Specht K, Adamsen TC, Biermann M, Brekke N, Craven AR, et al. 2015. Default-mode network functional connectivity is closely related to metabolic activity. Hum Brain Mapp 36:2027–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriat R, Molloy EK, Meier TB, Kirk GR, Nair VA, Meyerand ME, et al. 2013. The effect of resting condition on resting-state fMRI reliability and consistency: a comparison between resting with eyes open, closed, and fixated. Neuroimage 78:463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotin A, Desgranges B, Landeau B, Mezenge F, La Joie R, Egret S, et al. 2015. Anosognosia in Alzheimer disease: disconnection between memory and self-related brain networks. Ann Neurol 78:477–486 [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59:2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Duncan NW, Wiebking C, Gravel P, Lyttelton O, Hayes DJ, et al. 2012. GABA(A) receptors in visual and auditory cortex and neural activity changes during basic visual stimulation. Front Hum Neurosci 6:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl V, Bienkowska K, Strobel C, Tahmasian M, Grimmer T, Forster S, et al. 2014. Local activity determines functional connectivity in the resting human brain: a simultaneous FDG-PET/fMRI study. J Neurosci 34:6260–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Jo HJ, Gotts SJ, Chen G, Martin A, et al. 2013. Correcting brain-wide correlation differences in resting-state FMRI. Brain Connect 3:339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. 2010. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci U S A 107:10238–10243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Merritt MD, Pan WJ, Magnuson ME, Grooms JK, Jaeger D, et al. 2013. Neural correlates of time-varying functional connectivity in the rat. Neuroimage 83C:826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Riedl V, Grimmer T, Drzezga A, Herman P, Hyder F. Metabolic basis for the “rest” condition in FMRI: comparison of eyes open vs. closed states reveals constancy of glucose metabolism across networks. In Proceedings of the International Society for Magnetic Resonance in Medicine, Toronto, Canada, 2015 [Google Scholar]
- Tomasi D, Volkow ND. 2010. Functional connectivity density mapping. Proc Natl Acad Sci U S A 107:9885–9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. 2011. Functional connectivity hubs in the human brain. Neuroimage 57:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Wang GJ, Volkow ND. 2013. Energetic cost of brain functional connectivity. Proc Natl Acad Sci U S A 110:13642–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. 2010. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci U S A 107:17757–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Hulshoff Pol HE. 2010. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol 20:519–534 [DOI] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. 2012. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage 59:431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrl HF, Hossain M, Lankes K, Liu CC, Bezrukov I, Martirosian P, et al. 2013. Simultaneous PET-MRI reveals brain function in activated and resting state on metabolic, hemodynamic and multiple temporal scales. Nat Med 19:1184–1189 [DOI] [PubMed] [Google Scholar]
- Wicker B, Ruby P, Royet JP, Fonlupt P. 2003. A relation between rest and the self in the brain? Brain research. Brain Res Rev 43:224–230 [DOI] [PubMed] [Google Scholar]
- Wong CW, Olafsson V, Tal O, Liu TT. 2013. The amplitude of the resting-state fMRI global signal is related to EEG vigilance measures. Neuroimage 83:983–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Huang R, Wang J, Van Dam NT, Xie T, Dong Z, et al. 2014. Different topological organization of human brain functional networks with eyes open versus eyes closed. Neuroimage 90:246–255 [DOI] [PubMed] [Google Scholar]
- Yan C, Liu D, He Y, Zou Q, Zhu C, Zuo X, et al. 2009. Spontaneous brain activity in the default mode network is sensitive to different resting-state conditions with limited cognitive load. PLoS One 4:e5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GJ, Murray JD, Repovs G, Cole MW, Savic A, Glasser MF, et al. 2014. Altered global brain signal in schizophrenia. Proc Natl Acad Sci U S A 111:7438–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan BK, Wang J, Zang YF, Liu DQ. 2014. Amplitude differences in high-frequency fMRI signals between eyes open and eyes closed resting states. Front Hum Neurosci 8:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Jiang T, Lu Y, He Y, Tian L. 2004. Regional homogeneity approach to fMRI data analysis. Neuroimage 22:394–400 [DOI] [PubMed] [Google Scholar]
- Zang YF, He Y, Zhu CZ, Cao QJ, Sui MQ, Liang M, et al. 2007. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev 29:83–91 [DOI] [PubMed] [Google Scholar]
- Zou Q, Long X, Zuo X, Yan C, Zhu C, Yang Y, et al. 2009. Functional connectivity between the thalamus and visual cortex under eyes closed and eyes open conditions: a resting-state fMRI study. Hum Brain Mapp 30:3066–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Miao X, Liu D, Wang DJJ, Zhuo Y, Gao JH. 2015. Reliability comparison of spontaneous brain activities between BOLD and CBF contrasts in eyes-open and eyes-closed resting states. Neuroimage 121:91–105 [DOI] [PubMed] [Google Scholar]
- Zou QH, Zhu CZ, Yang Y, Zuo XN, Long XY, Cao QJ, et al. 2008. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods 172:137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.