Abstract
The data presented here are associated with the article “The blister fluid proteome of paediatric burns” (Zang et al., 2016) [1]. Burn injury is a highly traumatic event for children. The degree of burn severity (superficial-, deep-, or full-thickness injury) often dictates the extent of later scar formation which may require long term surgical operation or skin grafting. The data were obtained by fractionating paediatric burn blister fluid samples, which were pooled according to burn depth and then analysed using data dependent acquisition LC–MS/MS. The data includes a table of all proteins identified, in which burn depth category they were found, the percentage sequence coverage for each protein and the number of high confidence peptide identifications for each protein. Further Gene Ontology enrichment analysis shows the significantly over-represented biological processes, molecular functions, and cellular components of the burn blister fluid proteome. In addition, tables include the proteins associated with the biological processes of “wound healing” and “response to stress” as examples of highly relevant processes that occur in burn wounds.
Specifications Table
| Subject area | Biochemistry |
| More specific subject area | Proteomics |
| Type of data | Table, Figures, and Cytoscape file |
| How data was acquired | LC-MS/MS, Eksigent ekspert 400 nanoLC system tandem TripleTOF 5600+ mass spectrometer (SCIEX) |
| Data format | Raw, filtered and analysed |
| Experimental factors | The blister fluid samples were pooled based on the depth classification, fractionated using 4 different methods, digested by trypsin and de-salted and enriched using Stage-Tips. |
| Experimental features | Data dependent acquisition LC- MS/MSGene ontology analysis |
| Data source location | Institute of Health and Biomedical Innovation (IHBI), Queensland University of Technology (QUT), Kelvin Grove, Queensland, Australia |
| Data accessibility | Data is provided with this article |
Value of the data
-
•
First and most comprehensive proteome of burn blister fluid that can be used to compare against other disease states/patient populations.
-
•
These data provide a reference list of known, observed proteins within paediatric burn blister fluid, which will be of interest for the burn wound research community and clinicians.
-
•
Qualitative evaluation of the biochemical differences between burns of different depths will enable future targeted quantitative analyses of protein abundance based on burn depth.
-
•
The dataset allows for extensive Gene Ontology (GO) term analysis of the burn blister fluid proteome and the interaction/interrogation of this proteome through the Cytoscape data file provided herein.
1. Data
Presented in this publication is an inventory of proteins identified in paediatric burn blister fluid (1% FDR corrected), and the depths at which they were detected (Supplementary Table S1). For each protein, the following elements are provided: the UniProt accession number; description; detected presence in three different burn depths (superficial, deep partial and full thickness); ProteinPilot confidence score; percent sequence coverage; and the number of ≥95% confident peptides identified per protein. An example of the quality of the mass spectrometry data acquired is shown in Fig. 1. The Gene Ontology (GO) enrichment analysis of the whole protein library categorised by biological processes, molecular functions and cellular components in response to burn injury is shown in Fig. 2 and provided online as a Cytoscape file. The proteins specifically involved in the GO term biological process annotations for ‘wound healing’ and ‘response to stress’ are shown in Tables A1 and B1.
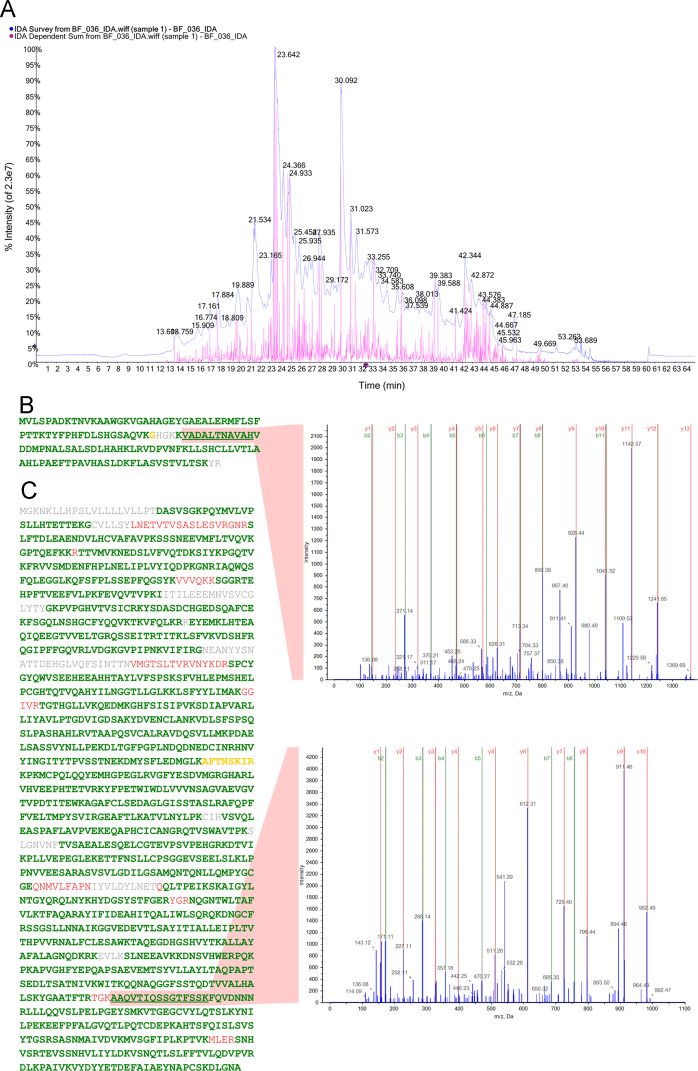
Fig. 1.
The mass spectra of an example sample. (A) Total ion chromatogram (blue) shows the complexity of ion information acquired from a single blister fluid sample. The dependent scan chromatogram (pink) shows the number of MS/MS ions detected. The total protein sequence coverage obtained and the corresponding MS2 spectra, with good y-ion series, of the underlined peptide are exemplified for burn relevant proteins, haemoglobin subunit alpha (B) and alpha-2 macroglobulin (C), respectively. Sequence coverage indicated by ≥95% confident peptides (green), ≥75% confident peptides (yellow), and ≥50% confident peptides (red). Grey amino acids were not detected.
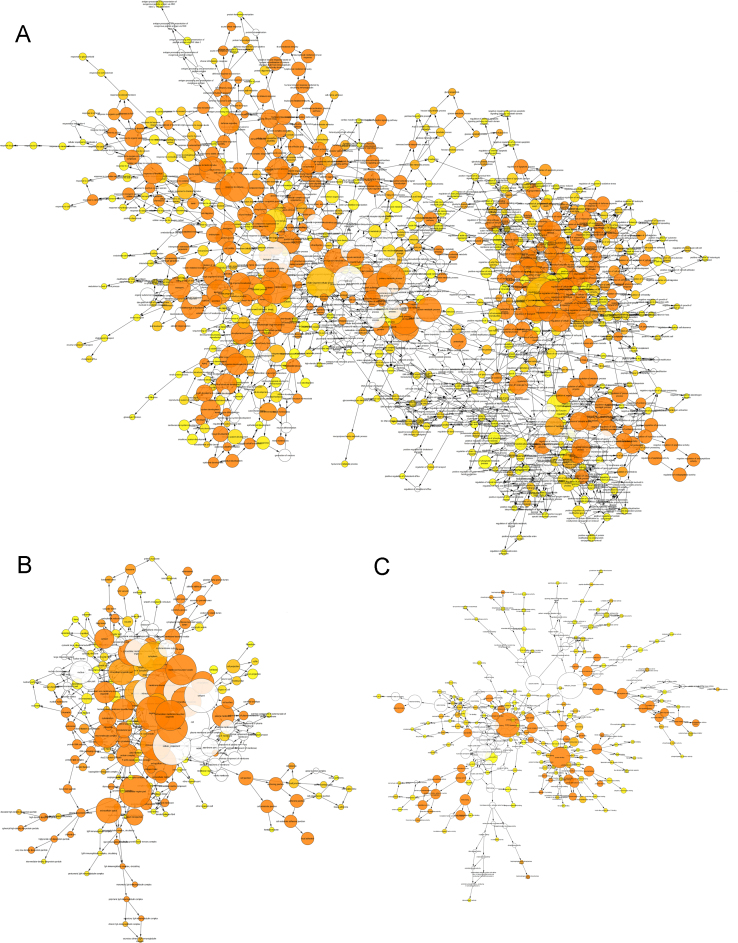
Fig. 2.
Gene ontology enrichment analysis for the entire protein inventory of burn blister fluid. The number of proteins involved in the annotation is in proportion to the size of nodes. The colour of the node represents the (corrected) p-value with a darker colour indicative of greater significance of over-representation for that GO term. Uncoloured nodes are the parents of over-represented downstream categories without over-representation themselves. (A) The network of biological process. (B) The network of cellular component. (C) The network of molecular function.
2. Experimental design, materials and methods
Methodology for blister fluid sample collection, sample preparation, liquid chromatography tandem mass spectrometry analysis, protein identification and GO analysis are described elsewhere [1].
2.1. Liquid chromatography tandem mass spectrometry (LC–MS/MS)
The quality of mass spectrometry data acquired and subsequently analysed was of a high standard (Fig. 1). The generation of complex and information rich ion chromatograms ensure that robust identifications of proteins are made (Fig. 1A). Furthermore, data acquired for proteins of relevance to burn injury were also of a high standard, with excellent sequence coverage and y-ion and b-ion series in MS2 spectra (Fig. 1B and C).
2.2. GO analysis
The over-represented biological processes (BP), molecular functions (MF), and cellular components (CC) were determined through Gene Ontology (GO) enrichment analysis of the whole blister fluid proteome using the BiNGO app within Cytoscape (Version 3.2.1, National Resource for Network Biology) (Fig. 2 and the Cytoscape file available online). Detected proteins within the two over-represented GO terms, ‘wound healing’ and ‘response to stress’ were compared across three burn depths (Tables A1 and B1, respectively). Subsets of these data with additional interpretation relevant to burn injury can be found elsewhere [1].
Acknowledgements
The authors acknowledge the patients and their families for the kind donation of their blister fluids and the assistance of the clinical staff at the Royal Children׳s Hospital and Lady Cilento Children׳s Hospital with sample collection. The authors also acknowledge Dr. Pawel Sadowski and Dr. Rajesh Gupta of the Central Analytical Research Facility (CARF) at the Queensland University of Technology, for their assistance with the mass spectrometry. Research support for this project was provided by the Wound Management Innovation Cooperative Research Centre (WMICRC) (project number 1-19) in addition to TZ’s and CT’s scholarship support and salary support for CT, DAB and JAB. LC was supported by a National Health and Medical Research Council Fellowship (#APP1035907). The authors have no other relevant affiliations or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.dib.2016.07.033.
Supplementary data associated with this article can be found in the online version at 10.1016/j.dib.2016.07.033.
Appendix
See
Table A1.
Distribution of significance level between burn severities for the biological process GO term, wound healing, and the burn depth that proteins associated with this GO term were detected. S=superficial thickness, D=deep partial thickness, F=full-thickness.
|
Biological process ofwound healing | ||||
|---|---|---|---|---|
| UniProt accession number | Cohort | S | D | F |
| heatmap | ||||
| corrected p-value | 1.55E−30 | 2.09E−35 | 3.11E−48 | |
| P11021 | 78 kDa glucose-regulated protein | X | X | ✓ |
| P60709 | Actin, cytoplasmic 1 | ✓ | ✓ | ✓ |
| P63261 | Actin, cytoplasmic 2 | ✓ | ✓ | ✓ |
| Q01518 | Adenylyl cyclase-associated protein 1 | X | X | ✓ |
| P01009 | Alpha-1-antitrypsin | ✓ | ✓ | ✓ |
| P08697 | Alpha-2-antiplasmin | ✓ | ✓ | ✓ |
| P01023 | Alpha-2-macroglobulin | ✓ | ✓ | ✓ |
| P12814 | Alpha-actinin-1 | X | X | ✓ |
| O43707 | Alpha-actinin-4 | ✓ | ✓ | ✓ |
| P01008 | Antithrombin-III | ✓ | ✓ | ✓ |
| P08519 | Apolipoprotein | ✓ | ✓ | ✓ |
| P02647 | Apolipoprotein A-I | ✓ | ✓ | ✓ |
| P04114 | Apolipoprotein B-100 | ✓ | ✓ | ✓ |
| P05090 | Apolipoprotein D | ✓ | X | ✓ |
| P02749 | Beta-2-glycoprotein 1 | ✓ | ✓ | ✓ |
| P20851 | C4b-binding protein beta chain | ✓ | X | X |
| Q96IY4 | Carboxypeptidase B2 | ✓ | ✓ | ✓ |
| P16070 | CD44 antigen | ✓ | X | ✓ |
| O00299 | Chloride intracellular channel protein 1 | X | X | ✓ |
| P10909 | Clusterin | ✓ | ✓ | ✓ |
| P00740 | Coagulation factor IX | ✓ | ✓ | ✓ |
| A0A0A0MRJ7 | Coagulation factor V | X | X | ✓ |
| P00742 | Coagulation factor X | ✓ | ✓ | ✓ |
| P03951 | Coagulation factor XI | ✓ | X | ✓ |
| P00748 | Coagulation factor XII | ✓ | ✓ | ✓ |
| P00488 | Coagulation factor XIII A chain | X | X | ✓ |
| P05160 | Coagulation factor XIII B chain | ✓ | ✓ | ✓ |
| P02452 | Collagen alpha-1 | X | ✓ | ✓ |
| P02461 | Collagen alpha-1 | X | X | ✓ |
| P02741 | C-reactive protein | ✓ | ✓ | ✓ |
| P07585 | Decorin | X | X | ✓ |
| P15924 | Desmoplakin | ✓ | ✓ | X |
| P02671 | Fibrinogen alpha chain | ✓ | ✓ | ✓ |
| P02675 | Fibrinogen beta chain | ✓ | ✓ | ✓ |
| P02679 | Fibrinogen gamma chain | ✓ | ✓ | ✓ |
| P02751 | Fibronectin | X | ✓ | ✓ |
| P23142 | Fibulin-1 | ✓ | ✓ | ✓ |
| P04075 | Fructose-bisphosphate aldolase A | ✓ | X | ✓ |
| P06396 | Gelsolin | ✓ | ✓ | ✓ |
| P04792 | Heat shock protein beta-1 | ✓ | ✓ | ✓ |
| P68871 | Haemoglobin subunit beta | ✓ | ✓ | ✓ |
| P02042 | Haemoglobin subunit delta | X | ✓ | ✓ |
| P69891 | Haemoglobin subunit gamma-1 | X | X | ✓ |
| P05546 | Heparin cofactor 2 | ✓ | ✓ | ✓ |
| P04196 | Histidine-rich glycoprotein | ✓ | ✓ | ✓ |
| P68431 | Histone H3.1 | ✓ | X | ✓ |
| Q71DI3 | Histone H3.2 | X | X | ✓ |
| P03956 | Interstitial collagenase | ✓ | ✓ | ✓ |
| Q92876 | Kallikrein-6 | X | ✓ | ✓ |
| P02538 | Keratin, type II cytoskeletal 6A | X | ✓ | ✓ |
| P01042 | Kininogen-1 | ✓ | X | X |
| P14151 | L-selectin | X | ✓ | ✓ |
| P01033 | Metalloproteinase inhibitor 1 | ✓ | ✓ | ✓ |
| P35579 | Myosin-9 | X | X | ✓ |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | ✓ | ✓ | ✓ |
| P03952 | Plasma kallikrein | ✓ | ✓ | ✓ |
| P05155 | Plasma protease C1 inhibitor | ✓ | ✓ | ✓ |
| P05154 | Plasma serine protease inhibitor | ✓ | ✓ | X |
| P00747 | Plasminogen | ✓ | ✓ | ✓ |
| P02775 | Platelet basic protein | X | ✓ | ✓ |
| P07737 | Profilin-1 | ✓ | ✓ | ✓ |
| P05109 | Protein S100-A8 | ✓ | ✓ | ✓ |
| P00734 | Prothrombin | ✓ | ✓ | ✓ |
| P02787 | Serotransferrin | ✓ | ✓ | ✓ |
| P02768 | Serum albumin | ✓ | ✓ | ✓ |
| P0DJI8 | Serum amyloid A-1 protein | ✓ | ✓ | ✓ |
| P00441 | Superoxide dismutase [Cu-Zn] | ✓ | ✓ | ✓ |
| P18827 | Syndecan-1 | X | X | ✓ |
| Q9Y490 | Talin-1 | X | X | ✓ |
| P07996 | Thrombospondin-1 | X | ✓ | ✓ |
| P18206 | Vinculin | X | X | ✓ |
| P07225 | Vitamin K-dependent protein S | ✓ | ✓ | ✓ |
| P04275 | von Willebrand factor | X | ✓ | ✓ |
Table B1.
Distribution of significance level between burn severities for the biological process GO term, response to stress, and the burn depth that proteins associated with this GO term were detected. S=superficial thickness, D=deep partial thickness, F=full-thickness.
|
Biological process of response to stress | ||||
|---|---|---|---|---|
| UniProt accession number | Cohort | S | D | F |
| heatmap | ||||
| corrected p-value | 1.10E−61 | 1.01E−72 | 1.37E−76 | |
| P31946 | 14-3-3 protein beta/alpha | X | X | ✓ |
| P62258 | 14-3-3 protein epsilon | ✓ | X | X |
| P31947 | 14-3-3 protein sigma | ✓ | ✓ | ✓ |
| P11021 | 78 kDa glucose-regulated protein | X | X | ✓ |
| P60709 | Actin, cytoplasmic 1 | ✓ | ✓ | ✓ |
| P63261 | Actin, cytoplasmic 2 | ✓ | ✓ | ✓ |
| P61160 | Actin-related protein 2 | ✓ | X | X |
| O15145 | Actin-related protein 2/3 complex subunit 3 | ✓ | X | X |
| O15511 | Actin-related protein 2/3 complex subunit 5 | ✓ | ✓ | X |
| P61158 | Actin-related protein 3 | X | X | ✓ |
| Q01518 | Adenylyl cyclase-associated protein 1 | X | X | ✓ |
| Q15848 | Adiponectin | ✓ | X | X |
| P02763 | Alpha-1-acid glycoprotein 1 | ✓ | ✓ | ✓ |
| P19652 | Alpha-1-acid glycoprotein 2 | ✓ | ✓ | ✓ |
| P01011 | Alpha-1-antichymotrypsin | ✓ | ✓ | ✓ |
| P01009 | Alpha-1-antitrypsin | ✓ | ✓ | ✓ |
| P08697 | Alpha-2-antiplasmin | ✓ | ✓ | ✓ |
| P02765 | Alpha-2-HS-glycoprotein | ✓ | ✓ | ✓ |
| P01023 | Alpha-2-macroglobulin | ✓ | ✓ | ✓ |
| P12814 | Alpha-actinin-1 | X | X | ✓ |
| O43707 | Alpha-actinin-4 | ✓ | ✓ | ✓ |
| P01019 | Angiotensinogen | ✓ | ✓ | ✓ |
| P04083 | Annexin A1 | ✓ | X | ✓ |
| P01008 | Antithrombin-III | ✓ | ✓ | ✓ |
| P08519 | Apolipoprotein | ✓ | ✓ | ✓ |
| P02647 | Apolipoprotein A-I | ✓ | ✓ | ✓ |
| P06727 | Apolipoprotein A-IV | ✓ | ✓ | ✓ |
| P04114 | Apolipoprotein B-100 | ✓ | ✓ | ✓ |
| P05090 | Apolipoprotein D | ✓ | X | ✓ |
| P02649 | Apolipoprotein E | ✓ | ✓ | ✓ |
| O14791 | Apolipoprotein L1 | ✓ | ✓ | ✓ |
| O75882 | Attractin | ✓ | X | X |
| P02749 | Beta-2-glycoprotein 1 | ✓ | ✓ | ✓ |
| P04003 | C4b-binding protein alpha chain | ✓ | ✓ | ✓ |
| P20851 | C4b-binding protein beta chain | ✓ | X | X |
| P27797 | Calreticulin | X | X | ✓ |
| P00918 | Carbonic anhydrase 2 | X | X | ✓ |
| Q96IY4 | Carboxypeptidase B2 | ✓ | ✓ | ✓ |
| P31944 | Caspase-14 | ✓ | X | ✓ |
| P04040 | Catalase | X | X | ✓ |
| J3KNB4 | Cathelicidin antimicrobial peptide | X | ✓ | X |
| P07858 | Cathepsin B | X | X | ✓ |
| P16070 | CD44 antigen | ✓ | X | ✓ |
| O43866 | CD5 antigen-like | ✓ | ✓ | ✓ |
| O00299 | Chloride intracellular channel protein 1 | X | X | ✓ |
| P10909 | Clusterin | ✓ | ✓ | ✓ |
| P00740 | Coagulation factor IX | ✓ | ✓ | ✓ |
| A0A0A0MRJ7 | Coagulation factor V | X | X | ✓ |
| P00742 | Coagulation factor X | ✓ | ✓ | ✓ |
| P03951 | Coagulation factor XI | ✓ | X | ✓ |
| P00748 | Coagulation factor XII | ✓ | ✓ | ✓ |
| P00488 | Coagulation factor XIII A chain | X | X | ✓ |
| P05160 | Coagulation factor XIII B chain | ✓ | ✓ | ✓ |
| P02452 | Collagen alpha-1 | X | ✓ | ✓ |
| P02461 | Collagen alpha-1 | X | X | ✓ |
| P02745 | Complement C1q subcomponent subunit A | ✓ | ✓ | ✓ |
| P02747 | Complement C1q subcomponent subunit C | ✓ | ✓ | ✓ |
| Q9NZP8 | Complement C1r subcomponent-like protein | ✓ | ✓ | ✓ |
| P09871 | Complement C1s subcomponent | ✓ | ✓ | ✓ |
| P06681 | Complement C2 | ✓ | ✓ | ✓ |
| P01024 | Complement C3 | ✓ | ✓ | ✓ |
| P0C0L4 | Complement C4-A | ✓ | ✓ | ✓ |
| P0C0L5 | Complement C4-B | ✓ | ✓ | ✓ |
| P01031 | Complement C5 | ✓ | ✓ | ✓ |
| P13671 | Complement component C6 | ✓ | ✓ | ✓ |
| P10643 | Complement component C7 | ✓ | ✓ | ✓ |
| P07357 | Complement component C8 alpha chain | ✓ | ✓ | ✓ |
| P07358 | Complement component C8 beta chain | ✓ | ✓ | ✓ |
| P07360 | Complement component C8 gamma chain | ✓ | ✓ | ✓ |
| P02748 | Complement component C9 | ✓ | ✓ | ✓ |
| P08603 | Complement factor H | ✓ | ✓ | ✓ |
| P31146 | Coronin-1A | ✓ | X | ✓ |
| P02741 | C-reactive protein | ✓ | ✓ | ✓ |
| P01034 | Cystatin-C | ✓ | ✓ | ✓ |
| P99999 | Cytochrome c | ✓ | X | X |
| P07585 | Decorin | X | X | ✓ |
| P81605 | Dermcidin | ✓ | X | ✓ |
| P15924 | Desmoplakin | ✓ | ✓ | X |
| P78527 | DNA-dependent protein kinase catalytic subunit | ✓ | ✓ | X |
| Q16610 | Extracellular matrix protein 1 | ✓ | ✓ | ✓ |
| P08294 | Extracellular superoxide dismutase [Cu–Zn] | ✓ | ✓ | ✓ |
| Q01469 | Fatty acid-binding protein, epidermal | ✓ | ✓ | ✓ |
| P02671 | Fibrinogen alpha chain | ✓ | ✓ | ✓ |
| P02675 | Fibrinogen beta chain | ✓ | ✓ | ✓ |
| P02679 | Fibrinogen gamma chain | ✓ | ✓ | ✓ |
| P02751 | Fibronectin | X | ✓ | ✓ |
| P23142 | Fibulin-1 | ✓ | ✓ | ✓ |
| Q15485 | Ficolin-2 | X | X | ✓ |
| O75636 | Ficolin-3 | ✓ | ✓ | ✓ |
| P04075 | Fructose-bisphosphate aldolase A | ✓ | X | ✓ |
| P17931 | Galectin-3 | ✓ | X | ✓ |
| Q08380 | Galectin-3-binding protein | X | X | ✓ |
| P06396 | Gelsolin | ✓ | ✓ | ✓ |
| A0A087X1J7 | Glutathione peroxidase | ✓ | ✓ | ✓ |
| H0YBE4 | Glutathione peroxidase 3 | ✓ | X | X |
| P09211 | Glutathione S-transferase P | ✓ | ✓ | ✓ |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | ✓ | ✓ | ✓ |
| P00738 | Haptoglobin | ✓ | ✓ | ✓ |
| P00739 | Haptoglobin-related protein | ✓ | ✓ | ✓ |
| P34931 | Heat shock 70 kDa protein 1-like | X | ✓ | X |
| P11142 | Heat shock cognate 71 kDa protein | ✓ | X | ✓ |
| P04792 | Heat shock protein beta-1 | ✓ | ✓ | ✓ |
| P08238 | Heat shock protein HSP 90-beta | ✓ | ✓ | X |
| P69905 | Haemoglobin subunit alpha | ✓ | ✓ | ✓ |
| P68871 | Haemoglobin subunit beta | ✓ | ✓ | ✓ |
| P02042 | Haemoglobin subunit delta | X | ✓ | ✓ |
| P69891 | Haemoglobin subunit gamma-1 | X | X | ✓ |
| P05546 | Heparin cofactor 2 | ✓ | ✓ | ✓ |
| P04196 | Histidine-rich glycoprotein | ✓ | ✓ | ✓ |
| O60814 | Histone H2B type 1-K | X | ✓ | X |
| P68431 | Histone H3.1 | ✓ | X | ✓ |
| Q71DI3 | Histone H3.2 | X | X | ✓ |
| Q9BYW2 | Histone-lysine N-methyltransferase SETD2 | X | ✓ | X |
| P01876 | Ig alpha-1 chain C region | ✓ | ✓ | ✓ |
| P01877 | Ig alpha-2 chain C region | X | X | ✓ |
| P01859 | Ig gamma-2 chain C region | ✓ | ✓ | ✓ |
| P01861 | Ig gamma-4 chain C region | ✓ | ✓ | ✓ |
| P01602 | Ig heavy chain V-I region 5 | ✓ | ✓ | ✓ |
| P01814 | Ig heavy chain V-II region OU | ✓ | X | ✓ |
| P01764 | Ig heavy chain V-III region 23 | ✓ | X | ✓ |
| P01767 | Ig heavy chain V-III region BUT | ✓ | X | X |
| P01781 | Ig heavy chain V-III region GAL | ✓ | X | X |
| P01780 | Ig heavy chain V-III region JON | X | ✓ | ✓ |
| P01762 | Ig heavy chain V-III region TRO | ✓ | X | X |
| P01779 | Ig heavy chain V-III region TUR | X | ✓ | ✓ |
| P01776 | Ig heavy chain V-III region WAS | ✓ | X | X |
| P01593 | Ig kappa chain V-I region AG | X | X | ✓ |
| P01613 | Ig kappa chain V-I region Ni | X | ✓ | X |
| P01608 | Ig kappa chain V-I region Roy | ✓ | X | X |
| P01609 | Ig kappa chain V-I region Scw | X | ✓ | X |
| P06310 | Ig kappa chain V-II region RPMI 6410 | ✓ | X | X |
| P01617 | Ig kappa chain V-II region TEW | ✓ | ✓ | ✓ |
| P18136 | Ig kappa chain V-III region HIC | ✓ | ✓ | ✓ |
| P01621 | Ig kappa chain V-III region NG9 | X | X | ✓ |
| P06314 | Ig kappa chain V-IV region B17 | X | X | ✓ |
| P06313 | Ig kappa chain V-IV region JI | ✓ | ✓ | ✓ |
| P06316 | Ig lambda chain V-I region BL2 | ✓ | X | X |
| P01701 | Ig lambda chain V-I region NEW | X | X | ✓ |
| P04208 | Ig lambda chain V-I region WAH | ✓ | X | ✓ |
| P80748 | Ig lambda chain V-III region LOI | X | X | ✓ |
| P01714 | Ig lambda chain V-III region SH | X | ✓ | ✓ |
| P0CG06 | Ig lambda-3 chain C regions | ✓ | X | X |
| Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | ✓ | ✓ | ✓ |
| P03956 | Interstitial collagenase | ✓ | ✓ | ✓ |
| Q92876 | Kallikrein-6 | X | ✓ | ✓ |
| P08779 | Keratin, type I cytoskeletal 16 | ✓ | ✓ | ✓ |
| P04264 | Keratin, type II cytoskeletal 1 | ✓ | ✓ | ✓ |
| P02538 | Keratin, type II cytoskeletal 6A | X | ✓ | ✓ |
| P01042 | Kininogen-1 | ✓ | X | X |
| E7ER44 | Lactotransferrin | ✓ | ✓ | ✓ |
| E7EQB2 | Lactotransferrin | X | ✓ | X |
| P09960 | Leukotriene A-4 hydrolase | X | X | ✓ |
| P18428 | Lipopolysaccharide-binding protein | ✓ | ✓ | ✓ |
| P14151 | L-selectin | X | ✓ | ✓ |
| F8VV32 | Lysozyme | X | X | ✓ |
| P61626 | Lysozyme C | ✓ | ✓ | ✓ |
| P14174 | Macrophage migration inhibitory factor | ✓ | ✓ | ✓ |
| P40925 | Malate dehydrogenase, cytoplasmic | ✓ | ✓ | ✓ |
| P48740 | Mannan-binding lectin serine protease 1 | ✓ | ✓ | X |
| O00187 | Mannan-binding lectin serine protease 2 | X | X | ✓ |
| P11226 | Mannose-binding protein C | ✓ | X | X |
| P01033 | Metalloproteinase inhibitor 1 | ✓ | ✓ | ✓ |
| P08571 | Monocyte differentiation antigen CD14 | ✓ | ✓ | ✓ |
| P05164 | Myeloperoxidase | ✓ | ✓ | ✓ |
| P35579 | Myosin-9 | X | X | ✓ |
| Q96PD5 | N-acetylmuramoyl-L-alanine amidase | ✓ | ✓ | ✓ |
| P59666 | Neutrophil defensin 3 | ✓ | ✓ | ✓ |
| P08246 | Neutrophil elastase | ✓ | X | ✓ |
| P06748 | Nucleophosmin | ✓ | ✓ | ✓ |
| O95497 | Pantetheinase | X | ✓ | ✓ |
| P26022 | Pentraxin-related protein PTX3 | ✓ | X | X |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | ✓ | ✓ | ✓ |
| Q06830 | Peroxiredoxin-1 | ✓ | X | X |
| P32119 | Peroxiredoxin-2 | ✓ | ✓ | ✓ |
| P03952 | Plasma kallikrein | ✓ | ✓ | ✓ |
| P05155 | Plasma protease C1 inhibitor | ✓ | ✓ | ✓ |
| P05154 | Plasma serine protease inhibitor | ✓ | ✓ | X |
| P00747 | Plasminogen | ✓ | ✓ | ✓ |
| P02775 | Platelet basic protein | X | ✓ | ✓ |
| P02545 | Prelamin-A/C [Cleaved into: Lamin-A/C | ✓ | ✓ | ✓ |
| P07737 | Profilin-1 | ✓ | ✓ | ✓ |
| P25788 | Proteasome subunit alpha type-3 | ✓ | X | X |
| P28066 | Proteasome subunit alpha type-5 | ✓ | X | X |
| O14818 | Proteasome subunit alpha type-7 | X | X | ✓ |
| P28072 | Proteasome subunit beta type-6 | ✓ | ✓ | ✓ |
| P07237 | Protein disulphide-isomerase | X | X | ✓ |
| P30101 | Protein disulphide-isomerase A3 | X | X | ✓ |
| P13667 | Protein disulphide-isomerase A4 | X | X | ✓ |
| Q92597 | Protein NDRG1 | X | X | ✓ |
| Q9HCY8 | Protein S100-A14 | ✓ | X | X |
| P31151 | Protein S100-A7 | X | X | ✓ |
| P05109 | Protein S100-A8 | ✓ | ✓ | ✓ |
| P06702 | Protein S100-A9 | ✓ | ✓ | ✓ |
| A0A096LPE2 | Protein SAA2-SAA4 | ✓ | ✓ | ✓ |
| P00734 | Prothrombin | ✓ | ✓ | ✓ |
| P55786 | Puromycin-sensitive aminopeptidase | X | X | ✓ |
| P02787 | Serotransferrin | ✓ | ✓ | ✓ |
| P02768 | Serum albumin | ✓ | ✓ | ✓ |
| E9PQD6 | Serum amyloid A protein | X | ✓ | X |
| P0DJI8 | Serum amyloid A-1 protein | ✓ | ✓ | ✓ |
| P0DJI9 | Serum amyloid A-2 protein | X | ✓ | ✓ |
| P02743 | Serum amyloid P-component | ✓ | ✓ | ✓ |
| P08254 | Stromelysin-1 | X | X | ✓ |
| P00441 | Superoxide dismutase [Cu-Zn] | ✓ | ✓ | ✓ |
| P18827 | Syndecan-1 | X | X | ✓ |
| Q9Y490 | Talin-1 | X | X | ✓ |
| P10599 | Thioredoxin | ✓ | ✓ | ✓ |
| P07996 | Thrombospondin-1 | X | ✓ | ✓ |
| P55072 | Transitional endoplasmic reticulum ATPase | ✓ | ✓ | ✓ |
| P35030 | Trypsin-3 | X | X | ✓ |
| P07437 | Tubulin beta chain | ✓ | ✓ | X |
| P68371 | Tubulin beta-4B chain | ✓ | X | X |
| P62987 | Ubiquitin-60S ribosomal protein L40 | ✓ | ✓ | ✓ |
| P18206 | Vinculin | X | X | ✓ |
| P07225 | Vitamin K-dependent protein S | ✓ | ✓ | ✓ |
| P04004 | Vitronectin | ✓ | ✓ | ✓ |
| P04275 | von Willebrand factor | X | ✓ | ✓ |
Transparency document. Supplementary material
Supplementary material
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Reference
- 1.Zang T., Broszczak D.A., Cuttle L., Broadbent J.A., Tanzer C., Parker T.J. The blister fluid proteome of paediatric burns. J. Proteom. 2016 doi: 10.1016/j.jprot.2016.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material