Abstract
Aryl CH hydrogen bonds (HBs) are now commonly recognized as important factors in a number of fields, including molecular biology, stereoselective catalysis, and anion supramolecular chemistry. As the utility of CH HBs has grown, so to has the need to understand the structure–activity relationship for tuning both their strength and selectivity. Although there has been significant computational effort in this area, an experimental study of the substituent effects on CH HBs has not been previously undertaken. Herein we disclose a systematic study of a single CH HB by using traditional urea donors as directing groups in a supramolecular binding cavity. Experimentally determined association constants are examined by a combination of computational (electrostatic potential) and empirical (σm and σp) values for substituent effects. The dominance of electrostatic parameters, as observed in a computational DFT study, is consistent with current CH HB theory; however, a novel anion dependence of the substituent effects is revealed in solution.
Graphical abstract
INTRODUCTION
CH hydrogen bonds (HBs) are now understood to be a ubiquitous structural feature in chemistry and biology.1,2 CH donors play important and previously unrecognized roles in the multidisciplinary fields of molecular biology, supramolecular chemistry,3–5 and catalysis.1,6 CH⋯O HBs are common in protein folding and are found in the minor groove of DNA.7,8 Catalysts have also been found to include CH⋯O HBs as an important factor in stereoselectivity.6 Chemists are now widely using CH groups as HB donors in designed systems for anion capture1,9–11 and catalysis.12,13
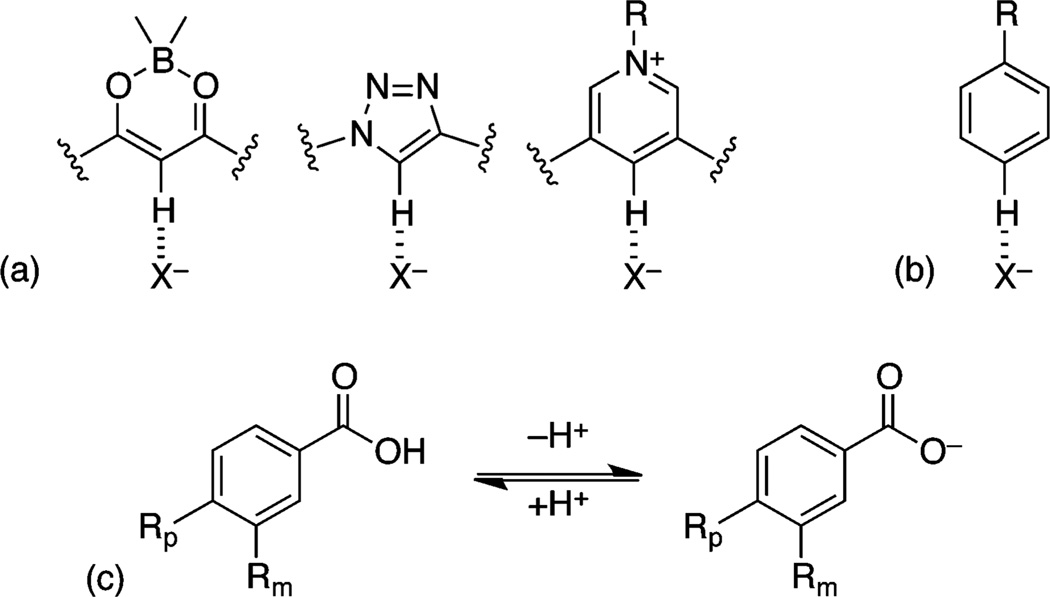
New CH donors have been developed to maximize the strength of a CH HB, including triazoles, bisketo-boronates, and pyridinium ions (Figure 1a).14 These strong, acidic CH donors, when incorporated within heterocycles with electron-poor atoms, are aligned to maximize the C←H dipole. The development of such new CH donors has increased the utility of these nonclassical HBs in structural design.
Figure 1.
(a) Prototypical examples of polarized, strong CH donors. (b) Preferred benzene hydrogen bond geometries. (c) Equilibrium of benzoic acids for derivation of Hammett parameters.
Despite the numerous computational studies on strong CH HB donors, experimental studies that quantify the energetic components of CH HBs, especially for weaker donors like benzene, have proven very difficult to obtain. As such, explanations and descriptions for CH HB strengths have been overwhelmingly dominated by the electrostatic component. Notable computational and experimental studies by Flood et al. have sought to dissect the strength of an arene CH vs an alkyl CH.15 A similar structure was used by Garcia Mancheño and co-workers to examine the influence of structure and electronics on catalysis;12,13 however, neither of these investigations measured the direct effects of substituents or anions on the CH donor energy or the CH component of the total Gibbs free energy of association for these HBs. The lack of studies on these aspects of CH HBs has led to some confusion on the characteristics, i.e., strength and selectivity, compared to traditional HB donors.
Conversely, the related interactions between anions and electron-deficient aromatic rings have been the subject of extensive computational and experimental studies that have resulted in the complete dissection of the energetic components and substituent effects.16,17 It has also been recognized that an anion can interact with the face of an arene via anion–π or weak–σ interactions, leading to differentiation in both substituent and anion effects.18–20 Computational studies indicate this type of dual anion and substituent dependence is important in CH HBs as well, although this has not been reduced to practice experimentally.21 As a standard and classical physical organic tool, the use of linear free energy relationships (LFERs), particularly the Hammett equation, for probing dynamic interactions is increasingly popular.22
Hay et al. performed an initial study to quantify the binding energy of benzene CH HBs, wherein they described both quite strong HBs to anions in the gas phase and a linear dependence of the binding energy on substituent electron-withdrawing ability, as measured by different substituent effects.23,24 The substituent effects in their model (Figure 1b) could be described by a Hammett σm or electrostatic potential (ESP).24 The use of a Hammett σm parameter to describe an interaction at the para position is typically assumed to indicate a mostly electrostatic interaction, due to the lack of conjugation at the meta position in the prototypical Hammett reaction, namely, the ionization of benzoic acid (Figure 1c).25
The assumption of electrostatic dominance is supported by the additional correlation with ESP. A recent analysis by Scheiner et al.21 of HBs to trifluoromethane revealed subtle energetic parameters. As included in the definition of a hydrogen bond, there exists a bond critical point between the H and X− (anion), as well as a shift in the vibrational frequency of the CH stretch.26 These effects are driven by the partial covalency of the HB and can be accentuated by examining the changes across a series of anions.
In the course of designing selective, fluorescent anion receptors, our group recently introduced the benzene CH HB donor into our existing bisarylethynyl urea scaffold to produce 1a (Figure 2a).27 In this report we have sought to better understand the parameters for controlling aryl CH HB acidity with anions by substitution para to the CH donor, 1b–g. The modularity of our scaffold allows us the unique chance to study a single CH⋯X− HB by easily accessible solution techniques: 1H NMR and UV–vis spectroscopic titrations with multiple anions (Cl−, Br−, I−, and NO3−). Association constants, Ka, are reported for seven receptors (1a–g) in water-saturated CHCl3. Combined solution experiments, crystallography, and computations provide new insight into the preferred CH binding geometry and electronic control. Linear free energy relationships using Hammett parameters and ESP reveal ρ dependence on the anion being titrated. Multivariate analysis with Swain–Lupton field (F) and resonance (R) parameters provides a deeper understanding of the percent resonance contribution to aryl CH acidity.28,29 Our combined experimental and computational approach for understanding CH HBs provides renewed support for the role of resonance in CH HBs. In addition, consideration of the anion in a supramolecular structure activity relationship identifies a new avenue for understanding and predicting anion binding selectivity.
Figure 2.
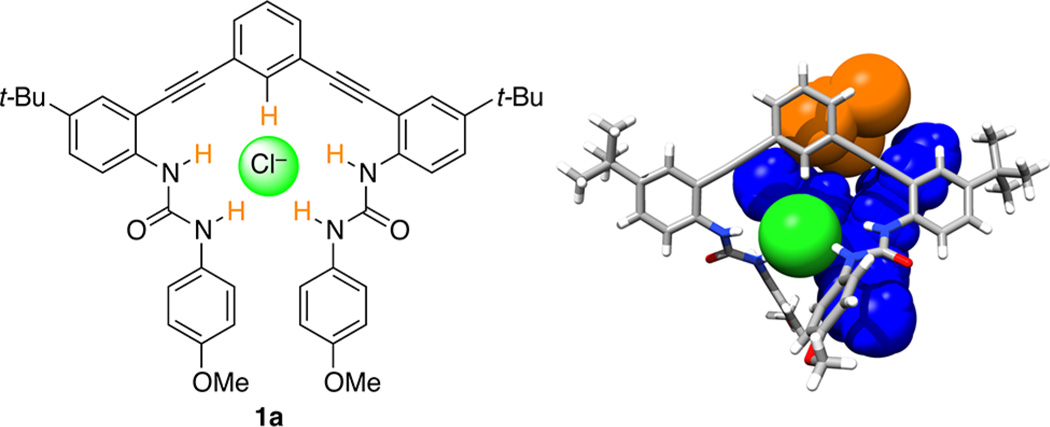
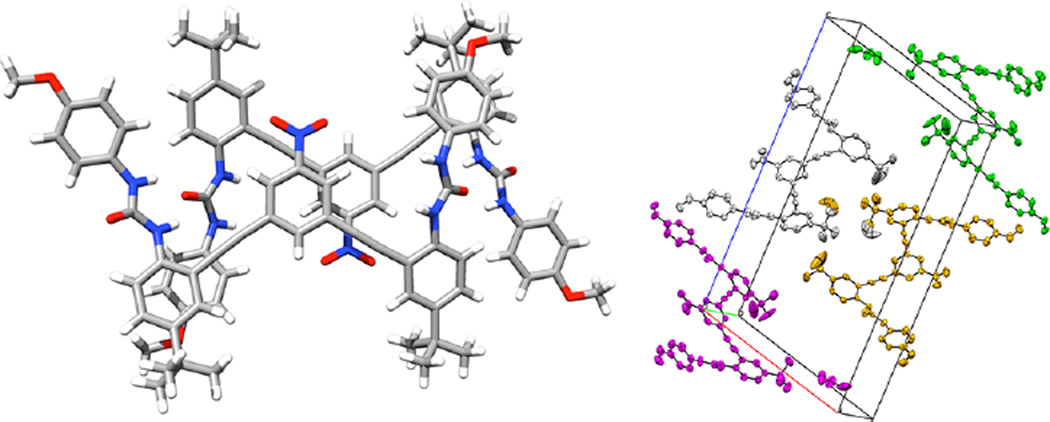
(a, left) Urea anion receptor 1a shown in optimal binding geometry for Cl−. (b, right) X-ray crystal structure of 1a⊂Cl− with solvent (CHCl3, orange) and counterion (TBA+, blue) included as space-filling models. CCDC no. 929532.
RESULTS AND DISCUSSION
Synthesis and Characterization
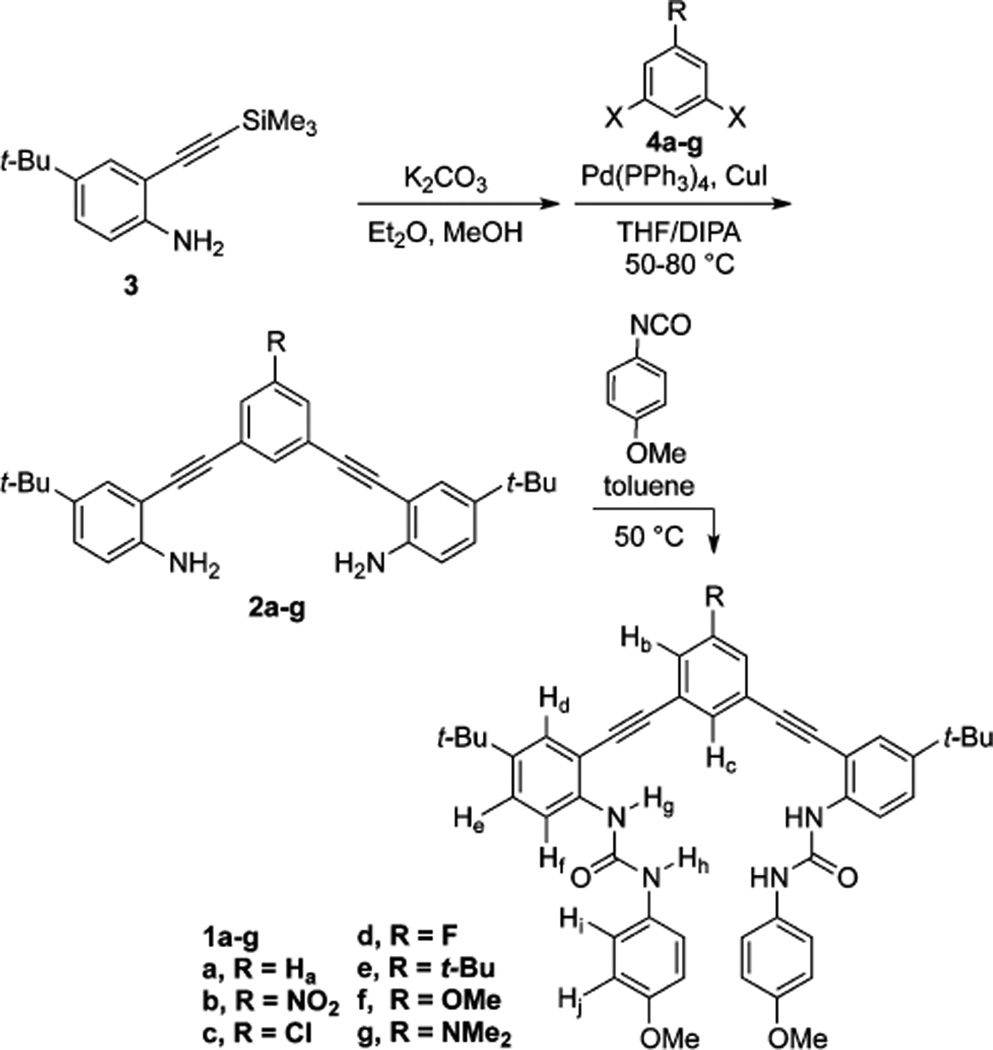
The substituted receptors used for this study are part of the bis(2-anilinoethynyl) arene family of conjugated, fluorescent receptors we have reported previously.30–36 In this case, pendent methoxy substituted phenylureas act as additional HB donors to direct the anion binding into a single site, as illustrated in Figure 2b.19,32,35 The synthesis of 1a has been previously reported and forms the parent scaffold for our study of substituent effects.27 These receptors are highly modular and easily broken into three key units for stepwise synthesis—a core arene, an alkynyl aniline, and an isocyanate.36 For the current investigation, the core arene can be any 3,5-dibromo- or 3,5-diiodobenzene suitable for Sonogashira cross-coupling possessing either an electron-withdrawing or electron-donating substituent in the 1-position. The t-Bu group on the alkynyl aniline was used to provide solubility in noncompetitive organic solvents (i.e., CHCl3). The 4-methoxy unit was chosen for the pendent phenylurea due to its simpler monomeric speciation in solution and to modulate the strength of the competing urea HBs. Such electron-rich ureas provide less competition with the CH⋯X− HB and have proven to be less prone toward self-aggregation.
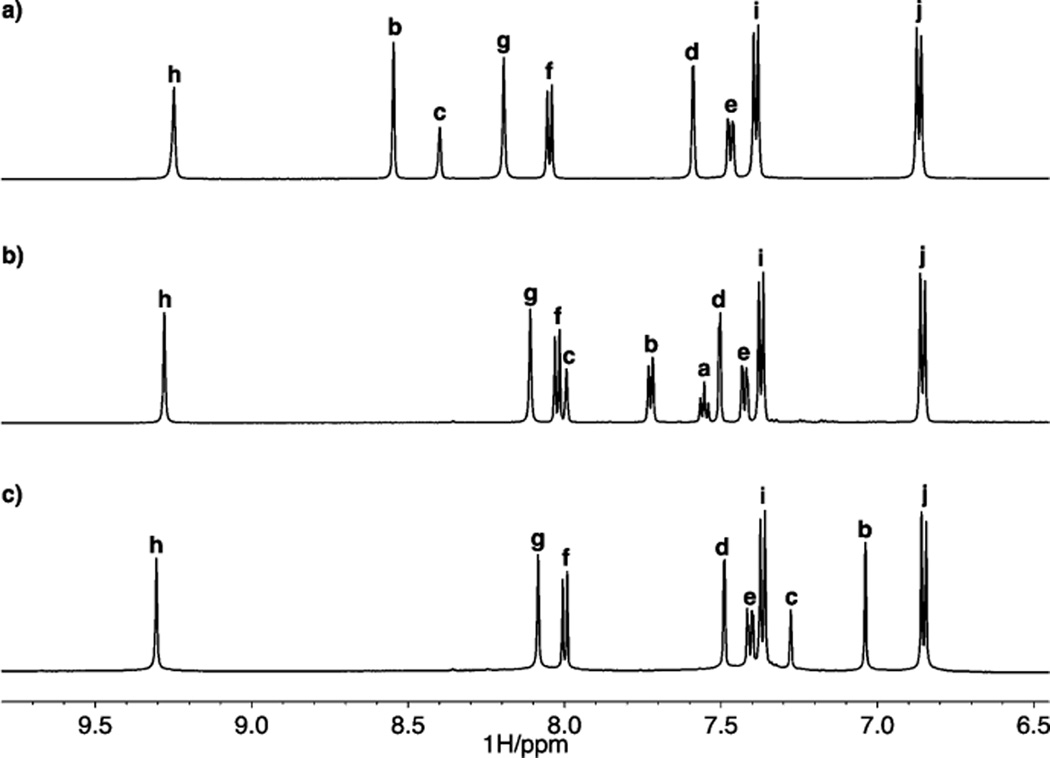
Dianilines 2a–g were synthesized by Sonogashira cross-coupling of 2-ethynyl-4-tert-butylaniline (desilylated 3) with the corresponding dihaloarenes 4a–g (Scheme 1). Reaction of the resulting dianilines with 4-methoxyphenyl isocyanate afforded the bisureas 1a–g. In most cases, the bisurea could be purified by trituration with EtOH to provide analytically pure samples. Receptors 1a–g were characterized by 1H and 13C NMR spectroscopy and high-resolution mass spectrometry. Complete synthetic procedures can be found in the Supporting Information (SI). The pendent ureas on our receptors are necessary to boost the overall binding energy high enough to observe in solution by 1H NMR and UV–vis spectroscopy.37 Previous efforts to study substituent effects in noncovalent interactions have been complicated by substituents altering peripheral HBs.16,38 Gratifyingly, the 1H NMR spectra of these receptors in DMSO-d6 show a small shift in the urea protons (Hh and Hg, Figure 3), suggesting substitution is far enough away to minimize, but not completely mitigate, the substituent effects on the ureas while still modulating the core CH HB donor acidity.39 The closest aromatic proton to the central ring, Hd, shifts <0.01 ppm between the –NO2 (1b) and –NMe2 (1g) substituted receptors. The central ring protons, however, show a strong substitution dependence, with Hb ranging from 7.0 (1f) to 8.5 ppm (1b). The isolation of substituent effects on the Δδ to just the central ring is necessary to measure only the effects on the CH⋯X− interactions without complicating secondary effects. 1H NMR chemical shifts are subject to conformational changes and are insufficient evidence alone; however, calculations of Mulliken charge and ESP also support a small influence on the urea HB donors.
Scheme 1.
Synthesis of Bisarylethynyl Urea Receptors 1a–g
Figure 3.
Stacked NMR spectra of (a) 1b, (b) 1a, and (c) 1g in DMSO-d6. Proton assignments refer to Scheme 1.
The solid-state structure determined by single-crystal X-ray diffraction is consistent with our structural characterization and solution behavior as studied previously by NMR, including 2D 1H–13C HSQC.27 A single crystal of 1a·Cl− was obtained by slow evaporation of CHCl3 containing a 2-fold excess of TBACl. The previously reported structure of 1a·Cl− has a short, linear C(H)⋯Cl contact of 3.579(3) Å and 169° (∠C–H⋯Cl). The asymmetric unit is a 1:1 receptor:anion complex with a cocrystallized tetrabutylammonium (TBA) cation and solvent molecule (Figure 2b). The presence of a 1:1 complex is encouraging for binding in solution and is consistent with other examples of this scaffold.32,34,36,40,41 The packing of this structure is dominated by ion pairing between Cl− and TBA+, with dispersion interactions playing a secondary role. The lack of interhost HBs or π–π stacking interactions also suggested a decreased likelihood of aggregation in solution.
The structure of bisurea 1b was determined from a single crystal grown by vapor diffusion of n-hexane into CH3CN. Host 1b, in the absence of a guest anion, forms long columnar stacks with urea HBs and π–π interactions stitching the layers together (Figure 4a). Columns are held together by dispersion forces between alkyl groups (t-Bu and Me) and arrange into a herringbone pattern (Figure 4b). The propensity for 1b to form hydrogen-bonded aggregates is embodied by poor solubility and aggregation at high concentrations in solution limiting the maximum concentration during 1H NMR titration experiments.
Figure 4.
(a, left) X-ray crystal structure of 1b, showing hydrogen-bonded stacks. (b, right) Packing of 1b.
NMR Titrations
1H NMR titrations were performed to study the substituent effects in solution on the anion binding conformation and CH chemical shift. The magnitude and direction of the change in chemical shift are additional parameters set out in the HB definition for the presence and strength of a HB (instead of an alternative attractive force, such as dispersion).26 Consistent with previous studies on anion–π interactions,17 water-saturated CHCl3 was used as the solvent, and anions were added as their TBA salts. Titrations were performed keeping the host concentrations constant (starting at 0.5–1.5 mM) during an experiment and titrating in a solution of concentrated anion in a solution of the host. Urea proton chemical shift changes between hosts are similar to the small changes observed in DMSO (see SI).
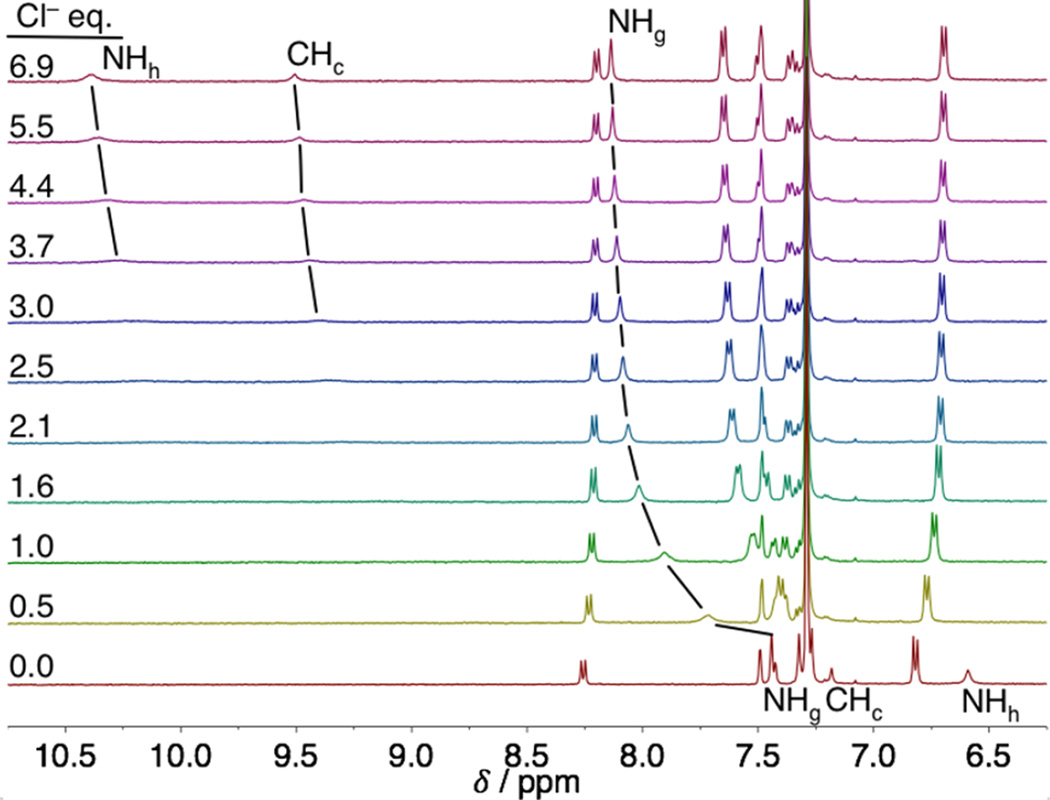
A representative titration is shown in Figure 5. This example of unsubstituted receptor 1a follows the trends for all of the receptors. The alkyl protons (t-Bu and OMe) remain unchanged throughout the course of anion addition (see the SI for complete titration data). The urea proton (Hh) and the CH proton (Hc) are unresolved during most of the titration, which may be due to the large shifts (Δδ = 4.0 and 2.5 ppm) between the free host and the saturated host:Cl− complex. Although the broadening prevents fitting these peaks for an association constant (Ka), the large, downfield shifts indicate strong HBs with Hh and Hc. Fortunately, the other aromatic and urea protons remain well resolved throughout the titration, except for brief periods of overlap for some peaks. Urea Hg shifts downfield with anion binding, while the aromatic protons remain stationary or move upfield slightly. The decreased broadening and smaller Δδ for Hg, observed for the halides with all hosts, is evidence for an overall weaker HB to this urea proton.
Figure 5.
Representative stacked plot for a Cl− titration with host 1a in water-saturated CHCl3 using a TBA salt.
The chemical shift change of urea NHg was fit using nonlinear regression analysis in MatLab to a 1:1 host:guest model.42 This model was selected based on the crystallographic evidence for 1:1 binding and quality of fit compared to higher order models. The trend for association constants follows the general electron-withdrawing ability of the substituents and the expected Hoffmeister bias (Cl− > NO3− > Br− ≫ I−).43,44 The large association constants measured for Cl− (>105 M−1 in some cases) indicates NMR spectroscopy is not the ideal technique for determining high quality association constants. Strong EWG hosts (1b) with Cl− are at the upper limit for measuring Kas by NMR titrations and the fit is based on a single proton shift. We can make a qualitative analysis of the bound geometry and trends, but the further quantitative analysis is based on UV–vis titrations (Table 1).
Table 1.
Complete Association Constants and Binding Energy for Receptors 1a–g at 298 K
| Cl− a | Br− a | I− b | NO3− a | |||||
|---|---|---|---|---|---|---|---|---|
| host (R) | Ka (M−1) | ΔG (kcal mol−1) | Ka (M−1) | ΔG (kcal mol−1) | Ka (M−1) | ΔG (kcal mol−1) | Ka (M−1) | ΔG (kcal mol−1) |
| 1a (H) | 6750 ± 600c | −5.22 ± 0.05 | 1630 ± 120c | −4.38 ± 0.04 | 150 ± 10c | −2.97 ± 0.03 | 3310 ± 170 | −4.80 ± 0.03 |
| 1b (NO2) | 24600 ± 3500 | −5.99 ± 0.09 | 5830 ± 510 | −5.13 ± 0.05 | 580 ± 30 | −3.76 ± 0.03 | 10600 ± 800 | −5.49 ± 0.04 |
| 1c (Cl) | 7900 ± 1300 | −5.32 ± 0.09 | 2680 ± 230 | −4.67 ± 0.05 | 290 ± 10 | −3.36 ± 0.03 | 5520 ± 640 | −5.10 ± 0.07 |
| 1d (F) | 6760 ± 620 | −5.22 ± 0.05 | 2340 ± 180 | −4.59 ± 0.05 | 240 ± 10 | −3.25 ± 0.03 | 4810 ± 320 | −5.02 ± 0.04 |
| 1e (tBu) | 4560 ± 720 | −4.99 ± 0.09 | 1370 ± 100 | −4.28 ± 0.03 | 150 ± 10 | −2.98 ± 0.04 | 2700 ± 420 | −4.68 ± 0.09 |
| 1f (OMe) | 4730 ± 240 | −5.01 ± 0.03 | 1000 ± 70 | −4.09 ± 0.04 | 170 ± 10 | −3.02 ± 0.03 | 3060 ± 360 | −4.75 ± 0.07 |
| 1g (NMe2) | 2660 ± 320 | −4.67 ± 0.07 | 800 ± 40 | −3.96 ± 0.03 | 100 ± 10 | −2.74 ± 0.07 | 1720 ± 160 | −4.41 ± 0.05 |
Determined using UV–vis titrations in H2O-saturated CHCl3; error is the standard deviation (SD) of at least three titrations.
Determined using 1H NMR titrations in H2O-saturated CDCl3; error is the SD of at least two titrations. The minimum error is assumed to be 5% in cases where the SD is <5%.
Previously reported (ref 27).
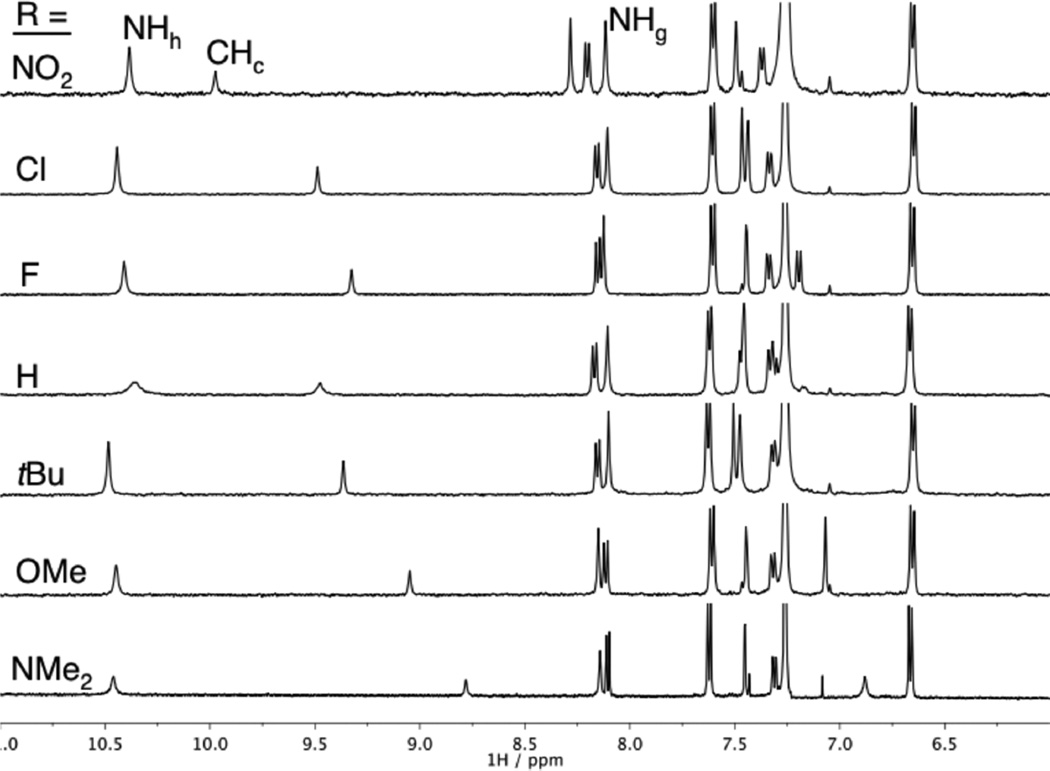
The 1H NMR spectra of the bound receptors are remarkably similar considering the variety of substituents used (Figure 6). As with the free receptors, the largest variation of Δδ is seen for two equivalent phenyl Hb resonances. The final δ for the urea protons, Hg and Hh, changes <0.1 ppm for all of the receptors. The only peak that shows a large change is the aromatic core CHc, where there is a difference of 1.2 ppm between 1b·Cl− and 1g·Cl−. Subtracting the difference before anion binding (Figure 3) leaves Δδ = 0.2 ppm due to a change in the HB strength. Also of note, the final position follows the trend of EWG strength. A similar trend is observed for Br− and I− binding, albeit with smaller Δδ for Hh and Hc. The chemical shift of the internal urea proton Hg changes minimally when bound to Br−, I−, or Cl−, consistent with it being mostly peripheral to halide binding.
Figure 6.
1H NMR spectra (CDCl3) of receptors 1a–g near the saturation point with Cl−. The peak showing the largest shift between 8.8 and 10.0 ppm is that of aryl proton Hc. Peak assignment refers to Scheme 1.
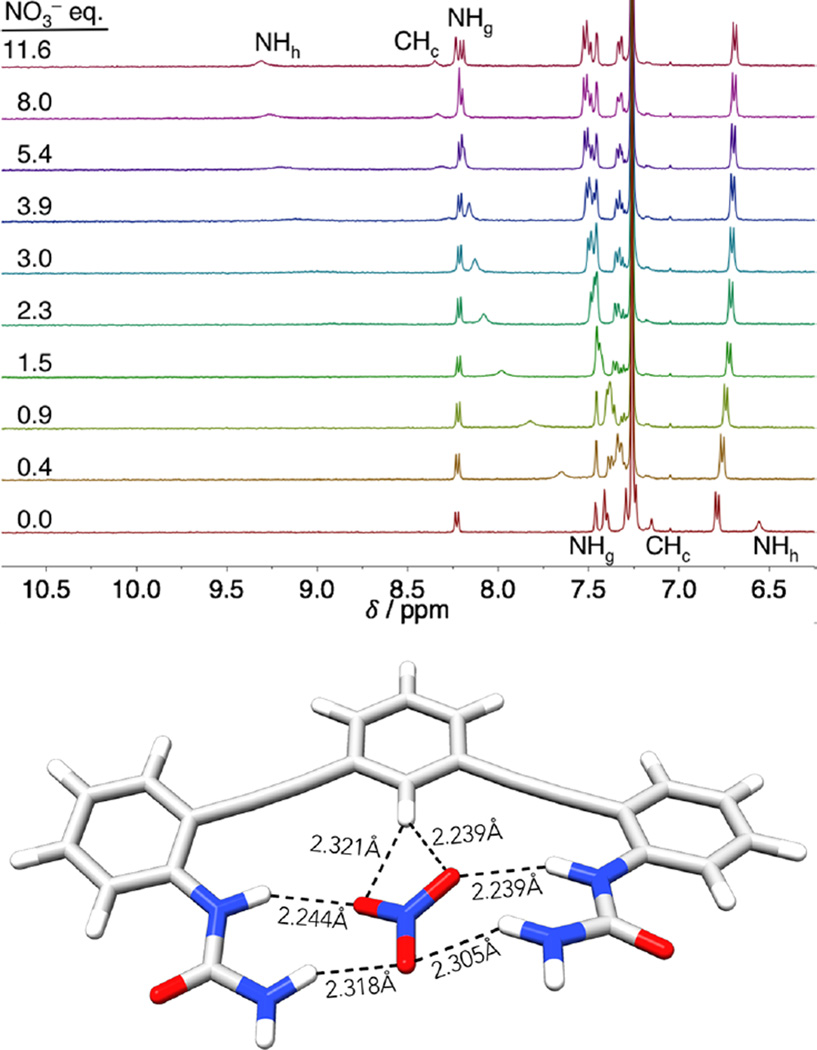
In addition to the halides, titrations were also performed with nitrate to consider shape as a variable in the binding studies (Figure 7, top). While the halides are spherical and have very small preference for HB arrangements, nitrate is trigonal planar and prefers a bifurcated, O⋯(C)H⋯O structure (Figure 7, bottom).21 In this case, the hydrogen-bonding protons NHh and CHc shifted less than observed with the halides, and urea NHg ends up slightly farther downfield. Considering the likely geometries for nitrate binding, the relative chemical shifts point to a geometry where two oxygens are bound by NHg and bifurcated by CHc; the third oxygen only weakly interacts with NHh. Modeling of the nitrate complex in Figure 7 supports this hypothesis, with two local minima, from divergent starting structures, found with nitrate parallel to the CH bond. With confirmation that the anions were bound in a similar manner by all of the receptors in solution, we sought to obtain quantitative association constants by performing UV–vis titrations.
Figure 7.
(Top) Stacked plots showing the NO3− titration of 1a. (Bottom) Local minima of truncated 5a·NO3−, with distances showing the preference of NO3− for maximized urea contacts and a bifurcated CH hydrogen bond. B3LYP/6-31g(d).
UV–Vis Titrations and Association Constants
The rigid, conjugated arylethynyl backbone used in these receptors has the added benefit of providing a convenient absorbance for performing UV–vis titrations.36 Although they do not provide as much structural information as NMR, UV–vis titrations are more accurate in determining Kas for our system because the required receptor concentrations are lowered (limiting aggregation), and problems with disappearing or overlapping peaks present in 1H NMR studies cease. The conditions for UV–vis titrations were chosen to most closely match the 1H NMR binding experiments: water-saturated CHCl3 was used as solvent and anions as their TBA salts were monitored at 298 K. Association constants were determined using the HYPERquad 2006 package to fit the complete spectral window with nonlinear regression.45 Consistent with the 1H NMR experiments, all titrations were fit to a 1:1 binding isotherm. Job’s plot analysis also confirms the best fit model for selected host and anion combinations (see SI).
Table 1 contains the compiled association constants for receptors 1a–g with Cl−, Br−, I−, and NO3−. The selectivity of these receptors follows this preference, with the trend of Cl− > NO3− > Br− ≫ I− holding for all hosts. The chloride association constants are typically 3-fold higher than the bromide Kas. Interestingly, nitrate is not able to outcompete Cl− despite nitrate’s ability to maximize NH HBs. The extremely low association for iodide prevented the accurate determination of binding constant by UV–vis spectrophotometry and necessitated the use of 1H NMR binding data for further analysis.
The range of association constants for Cl− alone spans an order of magnitude, with just altering a single arene substituent. Consistent with the changes in chemical shift, the association constants for a given anion can be ranked according to the relative electron-withdrawing ability of the substituent. Surprisingly, fluorine is an outlier for the trend in eletronegativity of the substituent. Fluorine typically acts as an electron-withdrawing group for electrostatic interactions, except when resonance is a contributor. Fluorine acts as both a strong electron-withdrawing group due to induction and an electron-donating group by resonance with one of its lone pairs. Other groups in this table (OMe, NMe2, Cl) share this dual function and are important for differentiating between induction and resonance effects. The association constants have also been converted to ΔG (kcal mol−1) for comparison to other supramolecular receptors. The total binding energy can be tuned by substituent effects by 1.02–1.32 kcal mol−1 depending on the anion being titrated; i.e., Cl− is bound more strongly than I− by 1.92–2.23 kcal mol−1 throughout this class of receptors.
Computations
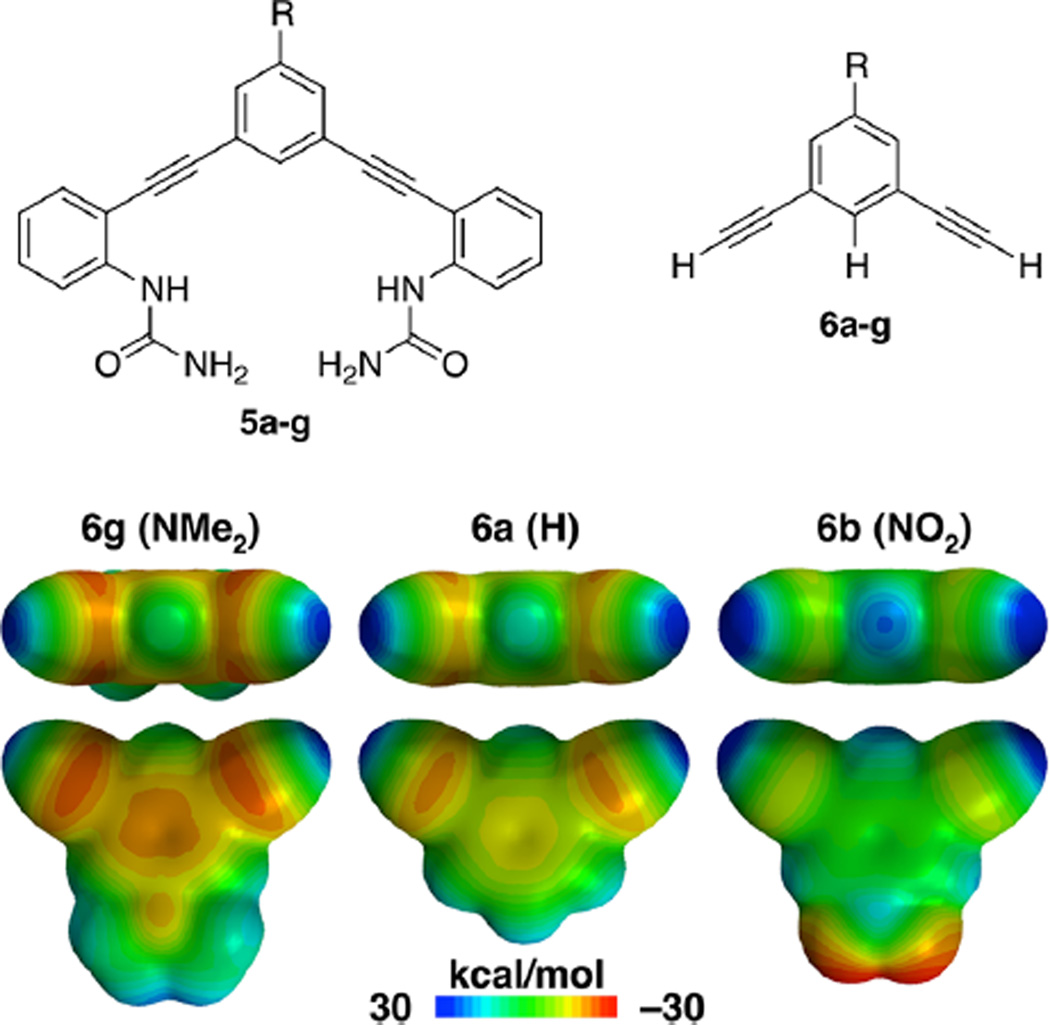
Prior computations on model structures of chloride and nitrate with benzene showed CH HB strength (ΔH) follows linearly with the Hammett σ parameters and ESP.23,24 We have expanded upon these prior computations by calculating the ESP surfaces for the model systems 5 and 6 (Figure 8) to measure electrostatic contributions in the bisurea receptors. The primary metric from these calculations is the ESP of 6a–g (Table 2) at the point where the C–H axis intercepts the 0.002 Å isoelectronic surface. The ESP at this point trends with the electron-withdrawing ability of the substituents. Hammett plots of the ESP (6a–g, Table 2 and Figure S52) and σ parameters favor σp over σm, with R2 = 0.97 and 0.88, respectively. This is a first indication that interactions with the CH are dependent on both field/inductive and resonance contributions of the substituents.
Figure 8.
(Top) Structures of the truncated model compounds 5 and 6 used for computational studies. (Bottom) MESP maps showing effects of substituent on ESP at a 0.002 Å isoelectric surface, calculated using B3LYP/6-31+g(d) in Spartan ’10.
Table 2.
Computational and Empirical Values for LFER Analysis
| host (R) | ESP | σm25 | σp25 | F29 | R29 |
|---|---|---|---|---|---|
| 1b (NO2) | 28.9 | 0.71 | 0.78 | 1.00 | 1.00 |
| 1c (Cl) | 22.1 | 0.37 | 0.23 | 0.72 | −0.24 |
| 1d (F) | 21.4 | 0.34 | 0.06 | 0.74 | −0.39 |
| 1a (H) | 17.9 | 0.00 | 0.00 | 0.00 | 0.00 |
| 1e (tBu) | 16.3 | −0.10 | −0.20 | −0.11 | −0.29 |
| 1f (OMe) | 16.3 | 0.12 | −0.27 | 0.54 | −1.68 |
| 1g (NMe2) | 12.0 | −0.15 | −0.83 | 0.69 | −3.81 |
ESP (kcal mol−1) at the point where the CH bond intersects a 0.002 Å isoelectric surface, calculated using B3LYP/6-31+g(d) in Spartan ’10.
The bisurea model system 5a–g is also useful to measure whether substitution at the central arene affects the urea group. A change in the Mulliken charge on the hydrogen-bond-donating carbon and nitrogens is representative of the effects at each of these positions. The Hammett plot of Mulliken charge at the hydrogen-bond-donating carbon in 5 is linear with σp, R2 = 0.90 and ρ = −0.10 ± 0.02. Mulliken charges on the urea nitrogens produce Hammett plots for σp and σm with very poor fits, R2 ≤ 0.70 and ρ ≈ 0.007 ± 0.002 (Table S43). Substitution on the central arene has very weak through-bond effect on the ureas in this system. The through-space effect is better estimated by the ESP near the urea hydrogens. In this case, Hammett plots reveal the change at the urea is <50% of the change at the CH donor. This model, however, does not account for the electron-donating pendent phenyls in the full receptor, 1a–g, which would further diminish the influence of the ureas.
Linear Free Energy Relationships
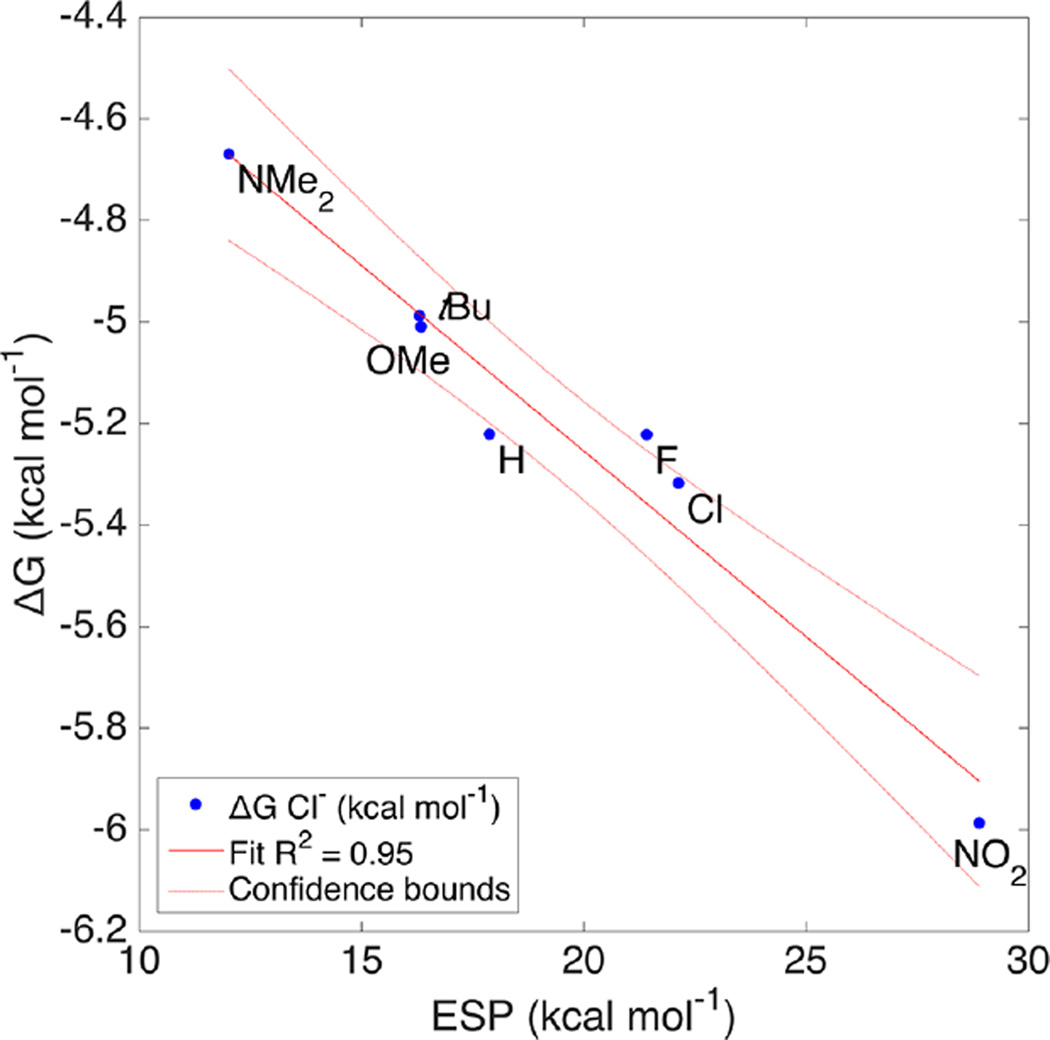
Linear free energy plots of the ESP and association constants are one way of comparing computational and experimental results, bridging the gap between gas phase and solution.46 Non-normalized plots in kcal mol−1 are linear, with R2 > 0.95 for all four anions (Figure 9). A break in the trend of the fitted slopes appears between the harder anions (Cl− and Br−) and soft anions (I− and NO3−). The hardness of anions has been used to explain the selectivity of Cl− transport in micelles, although alternative explanations have not been conclusively ruled out.47
Figure 9.
Linear free energy relationship of the solution Gibbs free energy (ΔG) of binding for Cl− with the ESP at the CH bond. Intercept predicts a hypothetical system where electrostatic potential is zero.
Another interesting implication falls out of the intercept in these plots. When the ESP is reduced to zero at the intercept, the remaining binding energy is due to the non-CH interactions. This is based on an assumption that the electrostatics model completely or mostly describes the CH⋯X interaction. As would be expected the intercept follows the same trend in energies (kcal mol−1), Cl− = −3.79, NO3− = −3.67, Br− = −3.06, I− = −1.98. The remaining “CH hydrogen bond energies” after subtracting the intercept from the solution ΔG for 1b are −2.20, −1.82, −2.07, and −1.78 kcal mol−1 for each anion, respectively. Previous studies with this system compared 1a to a pyridine receptor and estimated the CH HB energy at −1.33 kcal mol−1 to Cl−.27 In this case, the estimated energy for 1a with Cl− is quite close, at −1.43 kcal mol−1. Nitrate is a clear outlier in this series based on its preference for a bifurcated HB. The CH HB is less important for nitrate; however, using this model 47% of the total binding energy for iodide originates from the CH HB solely.
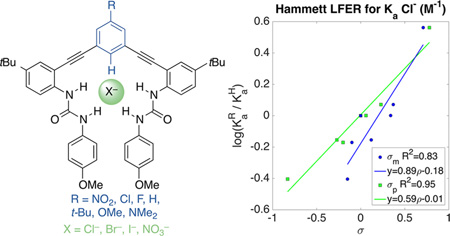
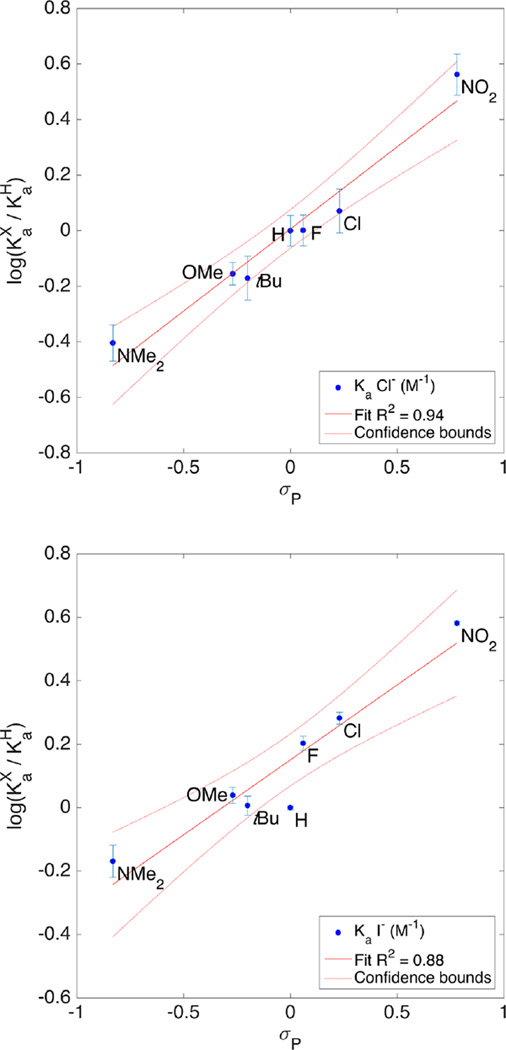
Since the substitution is only on the central arene and appears to only affect the C–Hc proton, we hypothesized that trends in our association constants should, also, be well-described by the σ parameter for substituents. The Hammett plots for Cl−, Br−, I−, and NO3− were prepared for both σm and σp constants. Figure 10 compares the fit for σp with Ka (Cl−) and Ka (I−). Association constants and substituent parameters were fit using the Hammett equation (eqs 1 and 2) in MatLab. The intercept acts as another measure for the quality of fit. In this case, the large intercept for I−, a poor fit for σp, places 1a outside of the confidence bounds (Figure 10 bottom). Table 3 contains the complete results for fitting all four anions to σp and σm.48
| (1) |
| (2) |
Figure 10.
Hammett plots of Ka (Cl−) (top) and Ka (I−) (bottom) with σp (Table 2). Goodness of fit (R2) indicates Cl− is well described by σp, while I− is less well-described.
Table 3.
Coefficients and Fitting Statistics for Hammett Plots for Each Anion Studied
| Ka (X−) | ρ | Ia | Nb | R2 c | Fd |
|---|---|---|---|---|---|
| Cl− (σp) | 0.59(±0.06) | 0.01(±0.03) | 7 | 0.95 | 100 |
| Br− (σp) | 0.57(±0.07) | 0.07(±0.03) | 7 | 0.93 | 68 |
| I− (σp) | 0.47(±0.07) | 0.15(±0.03) | 7 | 0.90 | 46 |
| NO3− (σp) | 0.50(±0.05) | 0.09(±0.02) | 7 | 0.95 | 98 |
| Cl− (σm) | 0.89(±0.18) | −0.18(±0.06) | 7 | 0.83 | 25 |
| Br− (σm) | 0.87(±0.16) | −0.11(±0.06) | 7 | 0.85 | 28 |
| I− (σm) | 0.78(±0.08) | −0.01(±0.03) | 7 | 0.95 | 102 |
| NO3− (σm) | 0.80(±0.09) | −0.08(±0.03) | 7 | 0.94 | 83 |
Intercept obtained from the linear fit.
Number of points used for fitting.
Coefficient of determination for quality of fit determination.
F-value for comparison of models.
The ρ values for all combinations of σ and anion are <1, an average of 0.54 for σp and 0.85 for σm. Reactions with ρ values >1 are considered more sensitive than benzoic acid and ρ values <1 are less sensitive to ionization by substituent effects. If CH⋯X− is a HB and incorporates some covalent character, then it follows that there is a small proton transfer event contributing to the binding energy.26 The method of fitting anion association to a Hammett σp is also applicable to other CH HB anion receptors. For instance, it is interesting to note, at least preliminarily, that a Hammett plot of Cl− association to a CH hydrogen-bond-donating rotaxane host has ρ = 0.53.49 The similarity of this relationship to a very different host in a different solvent is encouraging, and suggests this understanding is extendable to other such systems.
The question remains, has the influence of the urea HB donors been sufficiently accounted for? The ρ for Hammett plots of Mulliken charge and ESP at the ureas is consistently <0.06. The change in association constant due to the ureas is estimated to be <10% based on this and is insufficient to explain the effects on the binding event.
The small ρ value is consistent with the CH bond being much less acidic than benzoic acid. Also consistent with a traditional hydrogen bond definition, the σp parameter has a better fit for Cl− than σm (R2 (σp) = 0.95 vs (σm) = 0.83). σp is often thought to represent a greater resonance contribution; however, two points conflict with this observation: (1) DFT calculations favored σm for Cl−,24 and (2) the results for I− do not match Cl−, where Hammett plots for I− are a better fit using σm. Further confounding the issue, NO3− is well described by both σp and σm.
The unusual results for NO3− can be described by the geometry and altered binding mode in this case. The oxoanion is trigonal planar and can maximize contacts to the NH donors, as discussed above; however, the CH donor is still important to the overall binding energy. The CH proton still shifts downfield by nearly the same magnitude as the NH protons. The observed ρ values are the result of both inductive and resonance contributions. The degree of resonance contribution is a key difference between the σm and σp parameters.
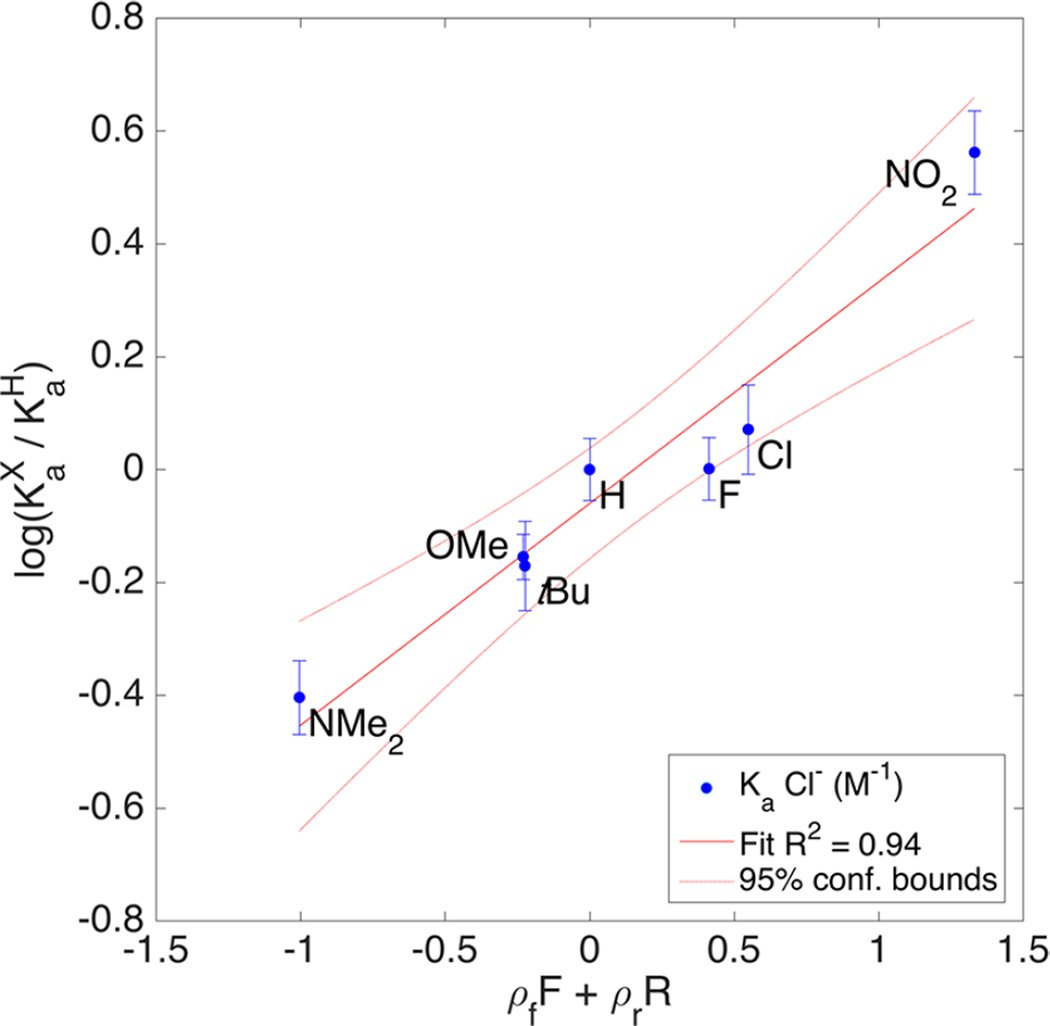
A more accurate method for determining resonance contribution is to perform multivariable fitting with field (F) and resonance (R) parameters, such as those derived by Swain and Lupton.28,29 While many methods for determining field and resonance contribution have been proposed, the F and R parameters (Table 4) developed by Swain–Lupton most closely match Hammett’s σ parameters in their derivation (eq 3). MatLab is capable of handling large, multivariable linear regressions and can easily handle fitting values for F and R from the data presented above. The method was applied to the experimental Ka values and the results for Cl− are plotted in Figure 11. A figure of merit for simultaneous F and R fitting is the percent resonance contribution, %R (eq 4).
| (3) |
| (4) |
Table 4.
Field and Resonance Fitting Parameters
| ρf | ρr | R2 | %Ra | |
|---|---|---|---|---|
| Cl− | 0.36(±0.09) | 0.17(±0.02) | 0.937 | 32 ± 7 |
| Br− | 0.37(±0.03) | 0.15(±0.01) | 0.995 | 30 ± 6 |
| I− | 0.40(±0.05) | 0.14(±0.01) | 0.977 | 25 ± 3 |
| NO3− | 0.37(±0.02) | 0.14(±0.01) | 0.996 | 28 ± 1 |
Calculated from eq 4 for percent resonance.
Figure 11.
LFER plot of Ka (Cl−) and ρfF + ρrR to determine resonance contribution from Swain–Lupton field and resonance parameters.
This reports the resonance contribution observed in the reaction and is the percent of R from the combined ρf and ρr coefficients. Of note, the resonance contribution for Cl− and Br− is higher, but the values drop off for I− and NO3−. The error values in %R exceed 10% in most cases, which is due to the small sample size to variable ratios. As a result of the increasing number of variables, the difference in the %R contribution among the various anions studied is well below the 95% confidence interval by t test. A similar trend, however, in anion effects was observed by Scheiner et al. using computation to study anion binding to trifluoromethane.21 By their computations, the overall binding energy and charge transfer from the anion were correlated, consistent with the effect of resonance contribution we have observed. As in the extreme HB example, increased charge on the carbon can be dissipated by resonance and contribute additional HB energy beyond inductive effects alone.
CONCLUSIONS
The resonance contribution of a substituent clearly plays a role in dictating hydrogen bond strength, even for CH donors. We have observed a weak dependence of σp and σm contribution on the anion being bound. Hammett parameters remain a powerful tool for predicting changes in CH HB strength. It is important to also consider the HB acceptor, not only its charge but also size, shape, and polarizability. The change in resonance contributions as a result of substituent effects as calculated by Swain–Lupton parameters is too small to differentiate between the anions studied, suggesting that emerging hypotheses offering hard/soft acid/base theory as a model for understanding anion binding specificity are overly simplistic. Anion effects on resonance contribution are also supported by calculations of the charge transfer energy and bond stretching in model CH⋯X− systems.
Computationally determined binding energies are a valuable tool for understanding solution binding events, especially in the case of weak interactions. Remarkably, venerable empirical substituent constants such as σ, F, and R can also effectively describe substituent effects in CH HBs, which are increasingly appreciated as rivals to more well-studied, highly polar HB donors (e.g., N–H, O–H). We have found through experimental results that the strength of a single CH HB is tunable across a range of 1.02–1.23 kcal mol−1 by modifying substituents on the receptor and that these interactions vary up to 0.42 kcal mol−1 by changing the anion accepting the CH HBs. Although these values are small, they represent a 10-fold and 3-fold change in anion binding, respectively, and hint at the nature of anion binding selectivity in such receptors. Considering multivalent effects in the largest hydrogen-bond-donating receptors that bring to bear many such interactions in targeting a single anion, the combined effect can be used to dramatically alter the binding event in selectivity and strength. While hard/soft acid/base theory remains a useful tool in understanding coordination chemistry, the present studies add to the evidence that this theory is too simplistic to describe accurately the nature of selectivity in anion binding using hydrogen-bonding receptors; ESPs and other empirical substituent constants appear to provide a more robust understanding.
Aryl CH HBs have seen increasing study in numerous fields, including anion transport, organocatalysis, molecular/ion recognition, and biological ligand/receptor binding. New methods for understanding and controlling the strength and selectivity of these interactions are vital for continued progress in these fields. For instance, ligand and/or drug binding to proteins can be improved by studying and optimizing important CH hydrogen-bonding interactions, and enhancing such interactions in organocatalysis and receptor design may enable improved stereo- and regioselectivity. Although the CH donor cannot be easily categorized as hard or soft, we have made the more important discovery that the possibility exists to influence the preference of this interaction for different anions. A concerted effort to maximize both the resonance withdrawing ability of substituents and the number of CH HB donors should lead to an increased affinity for hard anions. Conversely, the same should be possible by maximizing the inductive substituents to bind soft anions. The results from this study provide important insights to aid chemists and biologists in accomplishing such CH HB optimization.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01-GM087398. The authors acknowledge the Biomolecular Mass Spectrometry Core of the Environmental Health Sciences Core Center at Oregon State University (NIH P30ES000210). We gratefully acknowledge UO Prof. Michael D. Pluth and an anonymous reviewer(s) for helping to improve the detailed physical organic discussion.
The authors declare the following competing financial interest(s): NIH grant R01-GM087398 funded early-stage intellectual property that was licensed by SupraSensor Technologies, a company co-founded by the principal investigators.
Footnotes
ASSOCIATED CONTENT
Supporting Information
REFERENCES
- 1.Cai J, Sessler JL. Chem. Soc. Rev. 2014;43:6198–6213. doi: 10.1039/c4cs00115j. [DOI] [PubMed] [Google Scholar]
- 2.Steiner T, Desiraju GR. The Weak Hydrogen Bond in Structural Chemistry and Biology. Oxford: Oxford University Press; 1999. [Google Scholar]
- 3.Lee S, Flood AH. In: Topics in Heterocyclic Chemistry. Gale PA, Dehaen W, editors. Vol. 28. Berlin: Springer; 2012. pp. 85–107. [Google Scholar]
- 4.Hua Y, Flood AH. Chem. Soc. Rev. 2010;39:1262–1271. doi: 10.1039/b818033b. [DOI] [PubMed] [Google Scholar]
- 5.McDonald KP, Hua Y, Flood AH. In: Topics in Heterocyclic Chemistry. Gale PA, Dehaen W, editors. Vol. 24. Berlin: Springer; 2010. pp. 341–366. [Google Scholar]
- 6.Johnston RC, Cheong PH-Y. Org. Biomol. Chem. 2013;11:5057–5064. doi: 10.1039/c3ob40828k. [DOI] [PubMed] [Google Scholar]
- 7.Desiraju GR. Acc. Chem. Res. 1996;29:441–449. doi: 10.1021/ar950135n. [DOI] [PubMed] [Google Scholar]
- 8.Horowitz S, Trievel RC. J. Biol. Chem. 2012;287:41576–41582. doi: 10.1074/jbc.R112.418574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Flood AH. J. Phys. Org. Chem. 2013;26:79–86. [Google Scholar]
- 10.Hua Y, Liu Y, Chen C-H, Flood AH. J. Am. Chem. Soc. 2013;135:14401–14412. doi: 10.1021/ja4074744. [DOI] [PubMed] [Google Scholar]
- 11.Maeda H, Kitaguchi K, Haketa Y. Chem. Commun. 2011;47:9342–9344. doi: 10.1039/c1cc12822a. [DOI] [PubMed] [Google Scholar]
- 12.Beckendorf S, Asmus S, Mück Lichtenfeld C, García Mancheño O. Chem. - Eur. J. 2013;19:1581–1585. doi: 10.1002/chem.201203360. [DOI] [PubMed] [Google Scholar]
- 13.Asmus S, Beckendorf S, Zurro M, Mück Lichtenfeld C, Fröhlich R, García Mancheño O. Chem. - Asian J. 2014;9:2178–2186. doi: 10.1002/asia.201402237. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, Chen C-H, Flood AH. Nat. Chem. 2013;5:704–710. doi: 10.1038/nchem.1668. [DOI] [PubMed] [Google Scholar]
- 15.Hua Y, Ramabhadran RO, Uduehi EO, Karty JA, Raghavachari K, Flood AH. Chem. - Eur. J. 2011;17:312–321. doi: 10.1002/chem.201002340. [DOI] [PubMed] [Google Scholar]
- 16.Ballester P. Acc. Chem. Res. 2013;46:874–884. doi: 10.1021/ar300080f. [DOI] [PubMed] [Google Scholar]
- 17.Adriaenssens L, Gil-Ramírez G, Frontera A, Quiñonero D, Escudero-Adán EC, Ballester P. J. Am. Chem. Soc. 2014;136:3208–3218. doi: 10.1021/ja412098v. [DOI] [PubMed] [Google Scholar]
- 18.Berryman OB, Hof F, Hynes MJ, Johnson DW. Chem. Commun. 2006:506–508. doi: 10.1039/b511570a. [DOI] [PubMed] [Google Scholar]
- 19.Watt MM, Collins MS, Johnson DW. Acc. Chem. Res. 2013;46:955–966. doi: 10.1021/ar300100g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D-X, Wang M-X. J. Am. Chem. Soc. 2013;135:892–897. doi: 10.1021/ja310834w. [DOI] [PubMed] [Google Scholar]
- 21.Nepal B, Scheiner S. Chem. - Eur. J. 2015;21:1474–1481. doi: 10.1002/chem.201404970. [DOI] [PubMed] [Google Scholar]; Scheiner S, Grabowski SJ, Kar T. J. Phys. Chem. A. 2001;105:10607–10612. [Google Scholar]
- 22.Hwang J, Li P, Carroll WR, Smith MD, Pellechia PJ, Shimizu KD. J. Am. Chem. Soc. 2014;136:14060–14067. doi: 10.1021/ja504378p. [DOI] [PubMed] [Google Scholar]; Wheeler SE, McNeil AJ, Müller P, Swager TM, Houk KN. J. Am. Chem. Soc. 2010;132:3304–3311. doi: 10.1021/ja903653j. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou Y, Yuan Y, You L, Anslyn EV. Chem. - Eur. J. 2015;21:8207–8213. doi: 10.1002/chem.201500105. [DOI] [PubMed] [Google Scholar]; Zhou Y, Ye H, You L. J. Org. Chem. 2015;80:2627–2633. doi: 10.1021/jo502801g. [DOI] [PubMed] [Google Scholar]
- 23.Bryantsev VS, Hay BP. J. Am. Chem. Soc. 2005;127:8282–8283. doi: 10.1021/ja0518272. [DOI] [PubMed] [Google Scholar]
- 24.Bryantsev VS, Hay BP. Org. Lett. 2005;7:5031–5034. doi: 10.1021/ol0520119. [DOI] [PubMed] [Google Scholar]
- 25.Hansch C, Leo A, Taft RW. Chem. Rev. 1991;91:165–195. [Google Scholar]
- 26.Arunan E, Desiraju GR, Klein RA, Sadlej J, Scheiner S, Alkorta I, Clary DC, Crabtree RH, Dannenberg JJ, Hobza P, Kjaergaard HG, Legon AC, Mennucci B, Nesbitt DJ. Pure Appl. Chem. 2011;83:1637–1641. [Google Scholar]
- 27.Tresca BW, Zakharov LN, Carroll CN, Johnson DW, Haley MM. Chem. Commun. 2013;49:7240–7242. doi: 10.1039/c3cc44574g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swain CG, Lupton EC. J. Am. Chem. Soc. 1968;90:4328–4337. [Google Scholar]
- 29.Swain CG, Unger SH, Rosenquist NR, Swain MS. J. Am. Chem. Soc. 1983;105:492–502. [Google Scholar]
- 30.Johnson CA, II, Berryman OB, Sather AC, Zakharov LN, Haley MM, Johnson DW. Cryst. Growth Des. 2009;9:4247–4249. [Google Scholar]
- 31.Gavette JV, Mills NS, Zakharov LN, Johnson CA, Johnson DW, Haley MM. Angew. Chem., Int. Ed. 2013;52:10270–10274. doi: 10.1002/anie.201302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gavette JV, Evoniuk CJ, Zakharov LN, Carnes ME, Haley MM, Johnson DW. Chem. Sci. 2014;5:2899–2905. doi: 10.1039/c4sc00950a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watt MM, Engle JM, Fairley KC, Robitshek TE, Haley MM, Johnson DW. Org. Biomol. Chem. 2015;13:4266–4270. doi: 10.1039/c4ob02409e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watt MM, Zakharov LN, Haley MM, Johnson DW. Angew. Chem., Int. Ed. 2013;52:10275–10280. doi: 10.1002/anie.201303881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engle JM, Carroll CN, Johnson DW, Haley MM. Chem. Sci. 2012;3:1105–1110. doi: 10.1039/C2SC00975G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vonnegut CL, Tresca BW, Johnson DW, Haley MM. Chem. - Asian J. 2015;10:522–535. doi: 10.1002/asia.201403212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon D-W, Gross DE, Lynch VM, Sessler JL, Hay BP, Lee C-H. Angew. Chem., Int. Ed. 2008;47:5038–5042. doi: 10.1002/anie.200801426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGrath JM, Pluth MD. J. Org. Chem. 2014;79:11797–11801. doi: 10.1021/jo502325w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DMSO was chosen in this case because it is a highly competitive solvent and allows for high receptor concentration without fear of aggregation or saturation. The DMSO also acts to bind to the ureas, as indicated by their downfield shifts, decreasing the anion binding strength and making it a poor solvent for titrations. As well, the hydroscopic nature of DMSO hinders attempts to accurately control the water content between experiments.
- 40.Carroll CN, Berryman OB, Johnson CA, Zakharov LN, Haley MM, Johnson DW. Chem. Commun. 2009:2520–2522. doi: 10.1039/b901643k. [DOI] [PubMed] [Google Scholar]
- 41.Carroll CN, Coombs BA, McClintock SP, Johnson CA, II, Berryman OB, Johnson DW, Haley MM. Chem. Commun. 2011;47:5539–5541. doi: 10.1039/c1cc10947b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thordarson P. Chem. Soc. Rev. 2011;40:1305–1323. doi: 10.1039/c0cs00062k. [DOI] [PubMed] [Google Scholar]
- 43.Salis A, Ninham BW. Chem. Soc. Rev. 2014;43:7358–7377. doi: 10.1039/c4cs00144c. [DOI] [PubMed] [Google Scholar]
- 44.Carnegie RS, Gibb CLD, Gibb BC. Angew. Chem., Int. Ed. 2014;53:11498–11500. doi: 10.1002/anie.201405796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gans P, Sabatini A, Vacca A. Talanta. 1996;43:1739–1753. doi: 10.1016/0039-9140(96)01958-3. [DOI] [PubMed] [Google Scholar]
- 46.Busschaert N, Bradberry SJ, Wenzel M, Haynes CJE, Hiscock JR, Kirby IL, Karagiannidis LE, Moore SJ, Wells NJ, Herniman J, Langley GJ, Horton PN, Light ME, Marques I, Costa PJ, Felix V, Frey JG, Gale PA. Chem. Sci. 2013;4:3036–3045. [Google Scholar]
- 47.Lisbjerg M, Valkenier H, Jessen BM, Al-Kerdi H, Davis AP, Pittelkow M. J. Am. Chem. Soc. 2015;137:4948–4951. doi: 10.1021/jacs.5b02306. [DOI] [PubMed] [Google Scholar]
- 48.Hammett analysis was also performed using the alternative parameters σ+ and σ− to better represent the build-up or depletion of charge however, in all cases the fit was worse (Table S44). The build-up of charge is small in this case.
- 49.Hancock LM, Gilday LC, Carvalho S, Costa PJ, Felix V, Serpell CJ, Kilah NL, Beer PD. Chem. - Eur. J. 2010;16:13082–13094. doi: 10.1002/chem.201002076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.