Abstract
Introduction:
Metabolic enzyme variation and other patient and environmental characteristics influence smoking behaviors, treatment success, and risk of related disease. Population-specific variation in metabolic genes contributes to challenges in developing and optimizing pharmacogenetic interventions. We applied a custom genome-wide genotyping array for addiction research (Smokescreen), to three laboratory-based studies of nicotine metabolism with oral or venous administration of labeled nicotine and cotinine, to model nicotine metabolism in multiple populations. The trans-3′-hydroxycotinine/cotinine ratio, the nicotine metabolite ratio (NMR), was the nicotine metabolism measure analyzed.
Methods:
Three hundred twelve individuals of self-identified European, African, and Asian American ancestry were genotyped and included in ancestry-specific genome-wide association scans (GWAS) and a meta-GWAS analysis of the NMR. We modeled natural-log transformed NMR with covariates: principal components of genetic ancestry, age, sex, body mass index, and smoking status.
Results:
African and Asian American NMRs were statistically significantly (P values ≤ 5E-5) lower than European American NMRs. Meta-GWAS analysis identified 36 genome-wide significant variants over a 43 kilobase pair region at CYP2A6 with minimum P = 2.46E-18 at rs12459249, proximal to CYP2A6. Additional minima were located in intron 4 (rs56113850, P = 6.61E-18) and in the CYP2A6-CYP2A7 intergenic region (rs34226463, P = 1.45E-12). Most (34/36) genome-wide significant variants suggested reduced CYP2A6 activity; functional mechanisms were identified and tested in knowledge-bases. Conditional analysis resulted in intergenic variants of possible interest (P values < 5E-5).
Conclusions:
This meta-GWAS of the NMR identifies CYP2A6 variants, replicates the top-ranked single nucleotide polymorphism from a recent Finnish meta-GWAS of the NMR, identifies functional mechanisms, and provides pan-continental population biomarkers for nicotine metabolism.
Implications:
This multiple ancestry meta-GWAS of the laboratory study-based NMR provides novel evidence and replication for genome-wide association of CYP2A6 single nucleotide and insertion–deletion polymorphisms. We identify three regions of genome-wide significance: proximal, intronic, and distal to CYP2A6. We replicate the top-ranking single nucleotide polymorphism from a recent GWAS of the NMR in Finnish smokers, identify a functional mechanism for this intronic variant from in silico analyses of RNA-seq data that is consistent with CYP2A6 expression measured in postmortem lung and liver, and provide additional support for the intergenic region between CYP2A6 and CYP2A7.
Introduction
The overall smoking rate in developed countries has significantly declined over the last 50 years.1 However, tobacco-attributable disease remains the largest potentially modifiable cause of mortality,2 with an annual (2004–2009) United States mortality between 380000 and 480000.1,3 In 2012, more than 42 million adults in the United States were current cigarette smokers.4 Lung cancer, the classic tobacco-attributable disease, continues to be the single largest cause of death due to cancer in the United States, and to cancer among males in the world.5 The greatest incidence and mortality rates in the United States have been observed among African American males.6,7 Overall smoking prevalence among Asian Americans is lower than other racial groups,8 but reflects a complex relationship with native country differences in smoking rates by gender and acculturation.9 Although lung cancer rates are falling in some groups of Asian Americans, they are rising in others.10 Reduction of tobacco product use and associated morbidity and mortality will require enhanced tobacco control strategies, personalization of tobacco dependence treatment, and improved access to lung cancer treatment.
Nicotine metabolism influences cigarette consumption,11 tobacco exposures,12,13 tobacco attributable disease, for example, lung cancer risk,14,15 and response to smoking cessation treatments.16,17 Nicotine metabolism18 is primarily regulated by aldehyde oxidase (AO) and cytochrome oxidase 2A6 (CYP2A6), for the successive oxidation of nicotine to cotinine, by CYP2A6, for the oxidation of cotinine to trans-3′-hydroxycotinine, and by uridine diphosphate glycosyltransferase 2 family, members B10 (UGT2B10) for the N-glucuronidation of nicotine and cotinine, and B17 (UGT2B17),19 for the O-glucuronidation of trans-3′-hydroxycotinine. Variation in nicotine metabolism activity is influenced by multiple patient characteristics,20,21 including genetic variation in nicotine metabolism pathway genes.18,22
Development of biomarkers to assess nicotine pharmacokinetics and pharmacodynamics may help optimize (personalize) smoking cessation treatment.23,24 The ratio of trans-3′-hydroxycotinine/cotinine, termed the nicotine metabolite ratio (NMR), reflecting enzymatic activity of the major pathway for cotinine metabolism, has been shown to be a useful nicotine metabolism biomarker, due to its high correlation with the clearance of nicotine,11 heritability (0.67 [95% CI 0.56–0.76]25 and 0.81 [95%CI 0.70–0.88]26), stability, and measurement reliability.27,28 Recent modeling of CYP2A6 variants in European American individuals and laboratory study derived metabolite ratios has demonstrated that such modeling can account for nearly half of NMR variance,29 and that this metabolic activity metric can predict smoking cessation.16
Motivated by the need for a more comprehensive model of nicotine metabolism, we present the first meta-genome-wide association scan (GWAS) of the laboratory study-based NMR data from multiple ancestries. We use several knowledge-bases to identify potential functional mechanisms for top associations, and compare our results with recent literature. This meta-GWAS provides evidence that specific polymorphisms can predict nicotine metabolism in multiple continental populations and suggests further analyses to characterize these findings in diverse ancestries. This work supports development of biomarkers for use in clinical and population-based studies of nicotine metabolism,30 effects on tobacco product use,31 related disease,1 and cessation.32
Methods
Human Subjects
Institutional Review Board (IRB) approval for each study, and informed written consent from each participant, was obtained by the Principal Investigators of each study. IRB approval for these analyses was obtained from the Committee on Human Research at the University of California, San Francisco and the Human Subjects Committee at SRI International.
Study Design
For our GWAS, we selected data and DNA samples from three existing laboratory studies of nicotine metabolism. These studies were based on clinical administration of deuterium-substituted nicotine and cotinine followed by body fluid collection and analysis of nicotine metabolites via established gas chromatography/tandem mass spectroscopy methods.11 We selected DNA samples from unrelated African-American, Asian-American, and European-American ancestry individuals (N = 326 total) for genotyping on the Smokescreen Genotyping Array.33 For all individuals in this analysis, the NMR is defined as the trans-3′-hydroxycotinine to cotinine ratio obtained from the 6h, postadministration biospecimen (blood or saliva).
Pharmacokinetics in Twins (“PKTWIN”)34: European-American participants were recruited from the Northern California Twin Registry to investigate heritable components of nicotine metabolism. Participants consented to 30 min venous administration of deuterium-labeled nicotine and cotinine, followed by an 8 h hospital stay for blood and urine sample collections.
Pharmacogenetic Study of Nicotine Metabolism (“588”)11: European-, African-, and Asian-American smokers and nonsmokers of both sexes were recruited from the San Francisco Bay Area through multi-media advertisements for a nicotine metabolism study. Participants consented to morning oral administration of labeled nicotine and cotinine. The following biospecimens were collected: saliva up to 60h after dosing; blood up to 480min; and urine up to 8h.
SMOking in FAMilies (“SMOFAM”)35: Individuals from 61 pedigrees with at least three ever-smokers per pedigree were recruited from the Pacific Northwest to assess the relations between genetic factors, environmental factors, and tobacco use. Participants completed a clinical study of nicotine metabolism and consented to oral administration of a fixed dose of deuterium-labeled nicotine and cotinine (non-nicotine users were given unlabeled cotinine) at home monitored by a nurse, followed by salivary sample collection at multiple timepoints as well as a blood sample for DNA analysis.
Sample Genotyping
DNA samples were genotyped on the Smokescreen Genotyping Array at RUCDR Infinite Biologics (Piscataway, NJ). The Smokescreen Genotyping Array is a custom, genome-wide array for research on smoking behavior, addiction, pharmacological treatment, and related disease.33 It contains 646247 single nucleotide polymorphisms (SNPs) and indels for discovery and characterization studies. Genes (N = 1014) relevant to addiction and smoking-related phenotypes were identified by literature search, expert nomination, and biological knowledge-bases. The array contains dense coverage of common variation (MAF ≥ 0.05) for these genes (±20 kilo basepairs [kbp], 255862 markers) in African (specifically Yoruba in Ibadan, Nigeria, 97.5% at r 2 ≥ 0.80, 95.2% at r 2 ≥ 0.9), East Asian (98.1% at r 2 ≥ 0.80, 97.0% at r 2 ≥ 0.9), and European (98.1% at r 2 ≥ 0.80, 97.0% at r 2 ≥ 0.9) populations. About 17632 rare exonic coding variants are also included for these genes. The array contains nearly complete coverage of the chr19q13.2 nicotine-metabolizing enzyme genes, CYP2A6 (612 markers) and CYP2B6 (1628 markers). Additional markers are included for tagging of common SNPs over the entire region (chr19:41285047–41574301), including EGLN2, CYP2A7, CYP2G1P, and CYP2B7P1.
Genotyping Quality Control and Imputation
Genotypes and sample-level quality statistics were generated from raw CEL files using the Affymetrix Power Tools software suite (v. 1.16.0)36 and Smokescreen library and annotation files (v. r2). In brief, samples with a Dish QC value <0.82, stage I genotyping call rate <97%, scan failures, positive controls added by the genotyping lab, samples with discordant genotyped versus reported sex, monozygotic twins, siblings, and replicate samples with the lowest call rate were removed. The Affymetrix SNPolisher software (v. 1.5.0) was used to determine the best probeset for each marker and categorize markers based on quality. Markers with their best probeset categorized as CallRateBelowThreshold (N = 5987), Hemizygous (N = 181), Other (N = 49 999), Off-target variant (N = 1866), or which failed previous validation by the manufacturer and were categorized as MonoHighResolution (N = 7918) were removed.
All markers overlapping the 1000 Genomes Project reference set (Phase 3, version 5 downloaded from the Minimac3 website37), were matched to the forward strand. Study genotypes were pre-phased using MACH (v1.0),38 using 200 states, 20 rounds, and all other default settings. Markers having a concordance <95% (N = 22554) in previously Smokescreen-genotyped 1000 Genomes samples (N = 61) were excluded from the inference set before imputation. Minimac3 (v1.0.7) was used to impute markers in the 1000 Genomes Project reference set in 5 million basepair (Mbp) chunks for chromosomes 1–22, using a 250 kbp buffer for each chunk and all other default settings.
Statistical Analysis
Sample characteristics were summarized by study and ancestry. Chi square tests (for categorical variables) and F-tests (for continuous variables) were used to test for differences in groups. A significance level of 0.05 was used for these tests. Linear regression was used to model the relationship between the natural log transformed NMR and imputation estimated allelic dosages, adjusting for age, ancestry, gender, body mass index, and smoking status. We incorporated covariates available to us across the three studies, based on prior knowledge18 and practice,25 and recently validated in a large (N = 1672) sample of individuals screened for a clinical trial.21 Ancestry was estimated using principal-components analysis of 5545 ancestry informative markers.39 Genome-wide association scans were performed separately for self-reported European, African, and Asian American individuals, then combined in an inverse-variance based meta-analysis.40 All regression analysis was performed in [R].41 We excluded variants from reporting when the estimate of the squared correlation between imputed and true genotypes was less than 0.3 (cutoff recommended in the MACH software documentation) or the variant was rare (observed allele frequency < 0.01) in one or more of the study groups. P-values were evaluated for deviations from the expected distribution using quantile-quantile (QQ) plots and summarized in genome-wide association plots using the [R] qqman package.42 All variant-NMR associations with meta-GWAS P-values <5.0E-5 were reported. A P <5.0E-8 was considered genome-wide significant (GWS). Additional model goodness of fit and conditional analyses were performed for top associations. After initial review, we performed a post hoc smoking status × SNP interaction analysis of the CYP2A6 region. Extraction of epigenetic annotation from the Roadmap Epigenomics Consortium43 was performed for four top-ranked SNPs using HaploReg version 4.144 and the Epigenome Browser.45 Splicing Based Analysis of Variants (SPANR),46 a bioinformatics resource using RNA-seq data from the Human Body Map Project (GSE30611, 16 tissues from 16 individuals), was used to estimate effects of one intronic variant on CYP2A6 exon 5 splicing. The Genotype-Tissue Expression (GTEx) project47 (Version 4, build 200) was queried to investigate the relationship between three top-ranked SNPs and CYP2A6 expression in two relevant tissues; we report effect sizes and P-values obtained from GTEx and apply a Bonferroni correction to GTEx results. Chromosome coordinates refer to the hg19 assembly. Quanto48 was used to estimate sample sizes for future GWAS studies.
Results
Of 326 study participants genotyped, three samples failed genotyping; four samples were unexpectedly related (full siblings or monozygotic twins) or replicates; and four had differences in chromosomal and clinical genders. Three European American ancestry individuals had missing clinical variables and were excluded.
Forty-nine African, 51 Asian, and 212 European American ancestry individuals, comprising 312 individuals total, were included in statistical analyses (Table 1). The 588 study was the source of all African American and Asian American individuals while European American individuals originated from all three laboratory studies. The African American group had the highest mean age. A majority of the PKTWIN and SMOFAM samples were female. SMOFAM participants had the highest mean body mass index. The highest proportion of current smokers were found among European American individuals from the 588 study. We observed significantly lower NMRs in African American and Asian American individuals than in European American individuals (P = 5.0E-5 and P = 2.4E-6, respectively). In this study, we did not detect a statistically significant difference between African American and Asian American NMRs (P = 0.58).
Table 1.
Characteristics of Individuals from Nicotine Metabolism Laboratory Studies (N = 312)
| Study | 588 | PKTWINa | SMOFAMa | P | ||
|---|---|---|---|---|---|---|
| Ancestry | African | Asian | European | European | European | |
| N | 49 | 51 | 67 | 95 | 50 | |
| Age, M (SD) | 38.3 (11.3) | 28.3 (7.41) | 34.3 (12.2) | 36.9 (11.8) | 28.1 (1.38) | <.0001 |
| Sex (% F) | 38.7 | 49.0 | 44.8 | 74.7 | 58.0 | <.0001 |
| BMI, M (SD) | 26.5 (4.20) | 23.8 (2.87) | 24.9 (3.71) | 25.1 (4.13) | 28.6 (7.28) | <.0001 |
| Smoking (%) | 53.1 | 29.4 | 56.7 | 18.9 | 46.0 | <.0001 |
| NMR | −1.79 (0.50) | −1.84 (0.56) | −1.42 (0.47) | −1.47 (0.57) | −1.46 (0.45) | 0.0076 |
588 and SMOFAM = oral administration of labeled nicotine and cotinine and collection and analysis of saliva (SMOFAM, only saliva) and blood (plasma); PKTWIN = venous administration of labeled nicotine and cotinine and analysis of blood (plasma); Natural log transformed nicotine metabolite ratio (NMR) values are reported.
aUnrelated individuals only.
After filtering rare and poorly imputed genetic variants, 5891179 variants remained in the analyses. Quantile-quantile plots of each genome-wide scan and the meta-analysis indicated no inflation of the observed P-values over expected (Supplementary Figure 1); genomic inflation factors were 0.996, 0.993, 0.998, and 1.039 for European, African and Asian American GWAS analyses, and meta-GWAS analysis, respectively.
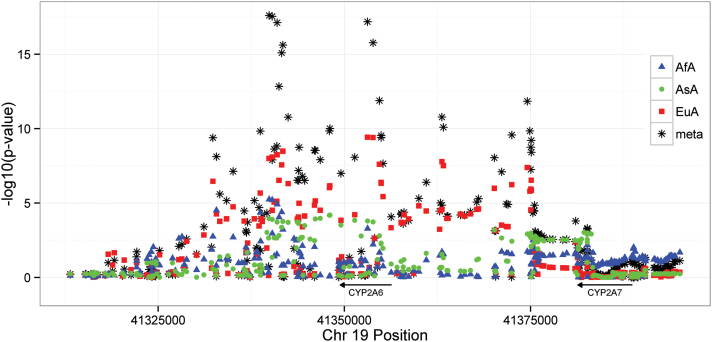
Meta-GWAS analysis of the natural log transformed, laboratory study-based NMR identified a single region of 43 kbp with 36 GWS results (34 are SNPs, and 2 are insertion–deletion polymorphisms) at the chr19q13.2 CYP2A6 locus (Figure 1 and Supplementary Table 1). While most GWS findings in the meta-GWAS were with common variants, 5, 1, and 5 SNPs of 36 GWS findings had minor allele frequencies between 0.01 and 0.05, in African, Asian, and European American samples, respectively. Regional associations in the CYP2A6-CYP2A7 region are presented in Figure 1 with select details presented in Table 2. A region in the 3′ flanking region of the transcript, ~9.5 kbp proximal to CYP2A6, and a region in intervening sequence 4 (IVS4), have the most statistically significant meta-GWAS results. The 3′ flank top SNP (rs12459249, chr19:41339896), and the IVS4 top SNP (rs56113850, chr19:41353107) exhibit P values 2.46E-18 and 6.61E-18. A third region intergenic between CYP2A6 and CYP2A7 (a distance of 25 kbp) contained two local P value minima, ~7 and ~18 kbp distal of CYP2A6 (rs7247098, chr19:41363098 and rs34226463, chr19:41374558), with P values 1.68E-11 and 1.45E-12. There is substantial linkage disequilibrium between the three regions in East Asian (EAS) and European (EUR) 1000 Genomes Project populations (all r 2 > 0.51) with more modest linkage disequilibrium observed in African (AFR) 1000 Genomes Project populations (all r 2 < 0.40) (Supplementary Figures 3–5). Conditional analyses adjusting for rs12459249 (smallest P-value in the meta-GWAS) resulted in no additional GWS findings; rs7247903 (GWS in the original meta-GWAS) in the CYP2A6-CYP2A7 intergenic region had the smallest P-value (P = 1.22E-6) (Table 2 and Supplementary Figure 3). Linkage disequilibrium values between the post-conditional top ranked SNP rs7247903 and the top ranked proximal, intronic, and intergenic meta-GWAS findings are low in 1000 Genomes Project populations (all r 2 < 0.14), suggesting that this variant represents a second intergenic signal. Of the five SNPs cited above, rs7247098 and rs34226463 were imputed, and rs12459249, rs56113850 and rs7247903 were genotyped. No results at the GWS (P < 5E-8) or reporting (P < 5E-5) thresholds were observed for a smoking status × SNP interaction analysis of the CYP2A6 region.
Figure 1.
Region of GWS meta-analysis results plus flanking 20 kbp by ancestry and overall.
Table 2.
Selected chr19q13.2 Ancestry-Specific GWAS and Meta-GWAS Analysis Results
| GWAS | RSID | rs12459249 | rs4001926 | rs56113850 | rs7247098 | rs7247903a | rs7247903b | rs34226463 |
|---|---|---|---|---|---|---|---|---|
| Coor | 41339896 | 41348062 | 41353107 | 41363098 | 41372475 | 41372475 | 41374558 | |
| African American | AltAF | 0.31 | 0.86 | 0.49 | 0.48 | 0.16 | 0.16 | 0.60 |
| B | −0.58 | −0.03 | −0.40 | −0.38 | −0.30 | −0.28 | −0.43 | |
| SE | 0.11 | 0.16 | 0.11 | 0.16 | 0.13 | 0.10 | 0.17 | |
| P | 5.69E-06c | 8.62E-01 | 5.22E-04 | 1.93E-02 | 2.72E-02 | 8.91E-03 | 1.81E-02 | |
| R 2 | 0.51 | 0.17 | 0.39 | 0.28 | 0.27 | 0.59 | 0.28 | |
| Asian American | AltAF | 0.52 | 0.56 | 0.58 | 0.38 | 0.18 | 0.18 | 0.57 |
| B | −0.40 | −0.38 | −0.36 | −0.27 | −0.38 | −0.19 | −0.37 | |
| SE | 0.09 | 0.08 | 0.09 | 0.12 | 0.12 | 0.13 | 0.11 | |
| P | 1.12E-04 | 6.51E-05d | 1.62E-04 | 2.86E-02 | 3.69E-03 | 1.61E-01 | 1.20E-03 | |
| R 2 | 0.51 | 0.52 | 0.50 | 0.38 | 0.43 | 0.53 | 0.46 | |
| European American | AltAF | 0.32 | 0.49 | 0.32 | 0.34 | 0.05 | 0.05 | 0.46 |
| B | −0.31 | −0.25 | −0.33 | −0.32 | −0.49 | −0.37 | −0.31 | |
| SE | 0.05 | 0.05 | 0.05 | 0.05 | 0.10 | 0.09 | 0.05 | |
| P | 1.02E-08 | 6.96E-07 | 3.81E-10 e | 1.67E-08 | 5.87E-07 | 1.30E-04 | 4.14E-08 | |
| R 2 | 0.29 | 0.26 | 0.31 | 0.29 | 0.26 | 0.34 | 0.28 | |
| Meta | B | −0.37 | −0.26 | −0.34 | −0.32 | −0.42 | −0.30 | −0.33 |
| SE | 0.04 | 0.04 | 0.04 | 0.05 | 0.07 | 0.06 | 0.05 | |
| P | 2.46E-18 f | 9.94E-11 | 6.61E-18 | 1.68E-11 | 2.66E-10 | 1.22E-06g | 1.45E-12 |
RSID = SNP identifier; Coor = chr19 basepair location; AltAF = alternative allele frequency; B = beta; SE = standard error. R 2 = proportion of natural log NMR variance. P = GWAS natural log NMR model or meta-analysis inverse variance P value. Bold values were to indicate the SNPs with P values < 5E-8, the GWS threshold.
aGWAS and meta-GWAS analysis results.
bGWAS and meta-GWAS analysis results, adjusting for rs12459249.
cTop ranked African American SNP.
dTop ranked Asian American SNP.
eTop ranked European American SNP.
fTop ranked meta-GWAS analysis result.
gTop ranked meta-GWAS analysis result, adjusting for rs1245249.
Ancestry-specific GWS results were only found for the largest ancestry group, covering 35 kbp, extending from the 3′ to the 5′ flanks of the CYP2A6 transcript. The top SNP in the European American ancestry group GWAS was rs56113850 (B = −0.31, SE = 0.05, P = 3.81E-10), found 151bp 5′ of exon 5; in the African and Asian American samples, this SNP had Β and P values of −0.40 and 5.2E-4 and of −0.36 and 1.6E-4, respectively (Table 2). The top CYP2A6 SNP in the African American GWAS was rs12459249 (Β = −0.58, SE = 0.11, P = 5.69E-6), found ~9.5 kbp proximal to CYP2A6; in the Asian American and European American samples, this SNP had Β and P values of −0.40 and 1.1E-4 and of −0.31 and 1.0E-8, respectively. The top CYP2A6 SNP in the Asian American GWAS was rs4001926 (Β = −0.38, SE = 0.09, P = 6.5E-5), found ~1.3 kbp proximal to CYP2A6; this SNP had no evidence of association in African Americans, but did have evidence of association in European Americans (Table 2).
HaploReg identified epigenetic annotations from lung and liver tissues for the top-ranked SNPs (Supplementary Table 2). Observed epigenetic annotations reported by the EpiGenomics Consortium included H3K4me1 marks at rs12459249 and at rs56113850 in liver, H3K4me1 histone marks at rs56113850 in lung, and H3K27ac and H3K9ac histone marks at rs56113850 in liver. Histone methylation marks identified the edge of an enhancer at rs12459249 in liver, and weak transcription at rs56113850 in lung and liver. Using SPANR, the C (alternate) and T alleles of rs56113850 predict inclusion of exon 5 in 11% and in 46.5% of CYP2A6 transcripts, respectively. GTEx eQTL analyses identified statistically significant reductions of CYP2A6 expression in postmortem lung (N = 278) and liver (N = 97) (Table 3). For rs12459249, effect sizes were similar in both tissues, but not statistically significant. For rs56113850, effects were stronger and statistically significant (P values < 0.0005) in both tissues. Neither intergenic GWS SNP was available in the GTEx eQTL database, however, rs4001921 (203 base pairs distal, with meta-GWAS P = 8.1E-11 and in LD with rs7247098, see Supplementary Table 1) exhibited similar effects in both tissues as rs56113850, but with statistical significance only in lung.
Table 3.
Genome-Wide Significant Single Nucleotide Polymorphisms and Association with CYP2A6 Expressiona
| SNP | rs12459249 | rs56113850 | rs4001921b |
|---|---|---|---|
| GWS locus | proximal | intronic | Intergenic |
| Lung effect | −0.15 | −0.28 | −0.24 |
| Lung P | 0.037 | 0.00048 | 0.0019 |
| Liver effect | −0.14 | −0.23 | −0.22 |
| Liver P | 0.091 | 0.00038 | 0.016 |
aFrom GTEx.
b r 2 = 0.92, 0.98, and 0.91 with rs7247098 in AFR, EAS, and EUR 1000 Genome Project populations.
Discussion
We have identified a region of GWS association with the laboratory-based NMR among individuals from three continental ancestries, that is, with SNPs proximal (rs12459249), intronic (rs56113850), and distal (rs7247098 and rs34226463) to CYP2A6 with P values < 1.0E-17 and < 1.0E-10, respectively. The distal intergenic region shows evidence of independent association after conditioning on rs12459249. This study replicates the top ranked finding in a meta-GWAS of the NMR in a Finnish population,26 that is, rs56113850, which is second ranked in our meta-GWAS and top ranked in our European American sample. Based on databases derived from molecular analyses of postmortem human tissues, we propose that reduced transcription explains the reduction in the NMR associated with the intronic variant rs56113850.
The significance and effect size of the top ranked CYP2A6 SNPs in our meta-GWAS of the laboratory-based NMR demonstrates the power of performing analyses with metabolite ratios.49 This meta-GWAS significantly improved upon the candidate gene-based analyses of the laboratory-based NMR previously performed using samples from two of the three cohorts,50 identifying SNPs of greater effect size and significance in a smaller sample of unrelated individuals from three cohorts. This is due to the much greater density of markers for drug metabolizing enzyme genes on the Smokescreen array than on the earlier DMET Plus array.51 The Smokescreen array was designed to capture rare and common variants found in the 1000 Genomes Project and Exome Sequencing Project for the CYP2A6-CYP2B6 region (chr19:41285047–41574301). Another strength is that we were able to use three independent sources of epigenetic and transcript data to propose functional mechanisms. In particular, GWS replication and mechanistic data unambiguously supports use of rs56113850 in future European ancestry nicotine and tobacco research.
Sample size was a limitation for the African American and Asian American NMR GWAS analyses. Linkage disequilibrium patterns differ among ancestries (Table 2 and Supplementary Figures 3–5), influencing power to detect effects on the NMR in a meta-GWAS. Here, we nominate SNPs that may serve as candidates for NMR research in three ancestries (we indicated in Table 2 the most significant SNP in each ancestry-specific GWAS). Larger studies are required before these candidate SNPs can be considered as defined markers for tobacco research in African American and Asian American populations. A minor limitation of this research is that we lacked information on all possible covariates influencing the NMR18 in all three studies. Thus, we did not include alcohol consumption, information on menopausal status, use of estrogen-based prophylactic hormones, or hormone replacement therapy; with one post hoc exception, we did not incorporate covariate interactions into the model.21 Observing GWS at classical functional alleles will require larger sample sizes than available in this analysis.52
CYP2A6 Region SNPs Previously Identified at GWS With Nicotine Metabolism and Related Phenotypes
The recent meta-GWAS of the NMR in three Finnish cohorts totaling 1518 smokers,26 identified hundreds of GWS SNPs in a 4.2 Mbp region on chr19q13.2. The top ranked SNP in that study, rs56113850 (β = −0.65, P = 5.8E-86), explained 0.14–0.23 of NMR variance. In a drug metabolizing enzyme and transporter candidate gene study, rs4803381, located in the CYP2A6 promoter (c.−1013), and rs1137115, located in exon 1 (c.51) of CYP2A6, were associated with the NMR at GWS in treatment-seeking smokers (β = −0.280, P = 1.25E-21, N = 633 individuals, and β = −0.240, P = 7.30E-14, N = 614 individuals), explaining 0.13 and 0.09 of NMR variance.50 The CYP2A6 locus has been associated in meta-GWAS analyses with reduced cigarette consumption at rs4105144, found ~2.7 kbp distal with P = 2.2E-12 in 83317 European smokers, with an effect size of 0.39 cigarettes/day.53 CYP2A6 SNPs rs8102683 (intergenic, ~7 kbp distal and tagging a large copy number polymorphism), and rs11878603 (proximal to CYP2A6), were identified with P values 4.3E-26 and 9.7E-30, and accounted for 2% of the variance in cigarette consumption in 17158 Japanese smokers.54 A meta-GWAS in 9614 non-Hispanic Whites and African Americans identified association of rs56113850 with moderate centrilobular emphysema (overall P = 1.3E-9, non-Hispanic White P = 1.2E-6, African American P = 2.1E-4), with an allele effect size reduction of 2% in both groups.55
Future Analyses of the NMR and Related Phenotypes
Analytic validity,28 clinical validity,17 and clinical utility56 of the NMR have been recently established. Our analysis and that of Loukola et al. find the same GWS variant in European ancestry studies with modest sample sizes. We consider what sample sizes are needed to identify GWS for future analyses of the NMR or related phenotypes. The only ancestry sample that yielded GWS signals in this study was the European American sample of 212 individuals. Based on univariate power analyses, ancestry-specific NMR distributions (Table 1) and effect sizes (Table 2) of top ranked SNPs in the African American (rs12459249) and Asian American (rs4001926) GWAS analyses, modest sample sizes (<100 participants) will be sufficient to observe GWS association with the NMR at these SNPs. Based on univariate power analyses, prior effect sizes of CYP2A6 SNPs associated with cigarette consumption at GWS in European (rs4105144) and Japanese (rs8102683) smokers, and recent cigarette consumption in the European Union57 and in Japan,54 sample sizes of ~31500 and ~1900 smokers would be required to observe GWS association with cigarette consumption of these CYP2A6 variants in European and Japanese populations.
Implications for Exposure, Attributable Disease, and Cessation Research
Genetics has an overarching influence across components of nicotine dependence.58 The CYP2A6 gene product plays a major role in the nicotine metabolism pathway,59,60 which influences nicotine intake.22,61 Nicotine binds to nicotinic cholinergic receptors (nAChRs), triggering neurotransmitter release, which, over time, leads to nicotine dependence. The development of nicotine dependence is influenced by variants at cholinergic receptor genes62 and CYP2A6,63 among other genes.53,64–67 Given the global nature of the tobacco epidemic and efforts to personalize prevention and treatment, additional ancestries in the United States and in other countries need to be included in tobacco biomarker genomic analyses to discover and characterize variants at CYP2A6 and elsewhere in the genome for use in nicotine and tobacco research. Analyses beyond single SNP models, that is, integrating haplotypic, functional genomic, and clinical data, will improve our understanding of these genes’ influence on nicotine metabolism and smoking behaviors and can help characterize tobacco product exposures and risks for smoking-attributable diseases14,68 and co-morbidities,69 as well as development of personalized therapies in multiple populations.70
Supplementary Material
Supplementary Tables 1 and 2 and Figures 1–6 can be found online at http://www.ntr.oxfordjournals.org
Funding
This work was supported by the National Institute on Drug Abuse, National Institutes of Health, United States Department of Health and Human Services (SMOKESCREEN: GENETIC SCREENING TOOL FOR TOBACCO DEPENDENCE & TREATMENT [HHS 271201300004C-0-0-1 to JWB] and DMET Genes, Nicotine Metabolism and Prospective Abstinence [DA033813 to AWB]). Computing was supported by an Amazon Web Services Research Grant to JWB. Previous funding that supported collection, analysis, and curation of the three datasets included in this analysis was received from the National Institute on Drug Abuse: Pharmacokinetics and Pharmacodynamics of Nicotine (DA002277 to NLB), Young Adult Substance Use—Predictors and Consequences (DA003706 to HH), Pharmacokinetics of Nicotine in Twins (DA011170 to GES), Pharmacogenetics of Nicotine Addiction Treatment Consortium (DA020830 to NLB, Rachel F. Tyndale, and Caryn Lerman); and from the Tobacco-Related Disease Research Program of the University of California: Nicotine Metabolism in Families (7PT-2004 to NLB).
Declaration of Interests
JWB reports ownership of BioRealm LLC and intention to commercialize the Smokescreen Genotyping Array as a potential competing interest. CKE reports employment by BioRealm LLC and a financial interest in the Smokescreen Genotyping Array as potential competing interests. CIP reports employment by BioRealm LLC as a potential competing interest. DVC, RK, HSJ, HH, GES, and NLB report no potential competing interests. HSJ and NLB disclose service as paid expert witnesses in litigation against tobacco companies. NLB discloses consultation with pharmaceutical companies that market smoking cessation medications. AWB reports past employment at SRI International as a potential competing interest.
Supplementary Material
Acknowledgments
We thank the laboratory study participants from the PKTWIN, SMOFAM, and 588 studies for their contributions to research. We thank Denise Nishita (SRI International) for preparing the DNA samples. We also thank the genotyping team at RUCDR Infinite Biologics for helping validate the Smokescreen array and the processing of DNA samples from this study.
References
- 1. U.S. Department of Health and Human Services. Executive Summary. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. Atlanta, GA: United States Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2014:36. [Google Scholar]
- 2. Bauer UE, Briss PA, Goodman RA, Bowman BA. Prevention of chronic disease in the 21st century: elimination of the leading preventable causes of premature death and disability in the USA. Lancet. 2014;384(9937):45–52. [DOI] [PubMed] [Google Scholar]
- 3. Rostron B. Smoking-attributable mortality by cause in the United States: revising the CDC’s data and estimates. Nicotine Tob Res. 2013;15:238–246. [DOI] [PubMed] [Google Scholar]
- 4. Agaku IT, King BA, Dube SR, Centers for Disease Control and Prevention (CDC) Current cigarette smoking among adults—United States, 2005–2012. MMWR Morb Mortal Wkly Rep. 2014;63(2):29–34. www.ncbi.nlm.nih.gov/pubmed/24430098 Accessed October 22, 2015. [PMC free article] [PubMed] [Google Scholar]
- 5. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 6. Underwood JM, Townsend JS, Tai E, et al. Racial and regional disparities in lung cancer incidence. Cancer. 2012;118(7):1910–1918. doi:10.1002/cncr.26479. [DOI] [PubMed] [Google Scholar]
- 7. O’Keefe EB, Meltzer JP, Bethea TN. Health disparities and cancer: racial disparities in cancer mortality in the United States, 2000–2010. Front Public Health. 2015;3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chae DH, Gavin AR, Takeuchi DT. Smoking prevalence among Asian Americans: findings from the National Latino and Asian American Study (NLAAS). Public Health Rep. 2006;121(6):755–763. www.ncbi.nlm.nih.gov/pubmed/17278411 Accessed October 22, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorman BK, Lariscy JT, Kaushik C. Gender, acculturation, and smoking behavior among U.S. Asian and Latino immigrants. Soc Sci Med. 2014;106:110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomez SL, Noone A-M, Lichtensztajn DY, et al. Cancer incidence trends among Asian American populations in the United States, 1990–2008. J Natl Cancer Inst. 2013;105(15):1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dempsey D, Tutka P, Jacob P, III, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. [DOI] [PubMed] [Google Scholar]
- 12. Strasser AA, Benowitz NL, Pinto AG, et al. Nicotine metabolite ratio predicts smoking topography and carcinogen biomarker level. Cancer Epidemiol Biomarkers Prev. 2011;20(2):234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derby KS, Cuthrell K, Caberto C, et al. Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rotunno M, Yu K, Lubin JH, et al. Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS One. 2009;4(5):e5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen LS, Bloom AJ, Baker TB, et al. Pharmacotherapy effects on smoking cessation vary with nicotine metabolism gene (CYP2A6). Addiction. 2014;109:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lerman C, Schnoll RA, Hawk LW, Jr, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med. 2015;3(2):131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hukkanen J, Jacob P, III, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57(1):79–115. [DOI] [PubMed] [Google Scholar]
- 19. Chen G, Giambrone NE, Jr, Dluzen DF, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: importance of functional UGT2B10 and UGT2B17 polymorphisms. Cancer Res. 2010;70(19):7543–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubinstein ML, Shiffman S, Rait MA, Benowitz NL. Race, gender, and nicotine metabolism in adolescent smokers. Nicotine Tob Res. 2013;15(7):1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chenoweth MJ, Novalen M, Hawk LW, Jr, et al. Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol Biomarkers Prev. 2014;23(9):1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tyndale RF, Sellers EM. Variable CYP2A6-mediated nicotine metabolism alters smoking behavior and risk. Drug Metab Dispos. 2001;29(4 Pt 2):548–552. www.ncbi.nlm.nih.gov/pubmed/11259349 Accessed November 1, 2015. [PubMed] [Google Scholar]
- 23. Bough KJ, Lerman C, Rose JE, et al. Biomarkers for smoking cessation. Clin Pharmacol Ther. 2013;93:526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen L-S, Bierut LJ. Genomics and personalized medicine: CHRNA5-CHRNA3-CHRNB4 and smoking cessation treatment. J Food Drug Anal. 2013;21(4):S87–S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3′ hydroxycotinine to cotinine in plasma and urine. Pharmacogenet Genomics. 2009;19(5):388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loukola A, Buchwald J, Gupta R, et al. A genome-wide association study of a biomarker of nicotine metabolism. PLoS Genet. 2015;11(9):e1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamilton DA, Mahoney MC, Novalen M, et al. Test–retest reliability and stability of the nicotine metabolite ratio among treatment-seeking smokers. Nicotine Tob Res. 2015;17(12):1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanner J-A, Novalen M, Jatlow P, et al. Nicotine metabolite ratio (3-hydroxycotinine/cotinine) in plasma and urine by different analytical methods and laboratories: implications for clinical implementation. Cancer Epidemiol Biomarkers Prev. 2015;24(8):1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bloom J, Hinrichs AL, Wang JC, et al. The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans. Pharmacogenet Genomics. 2011;21(7):403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patel YM, Stram DO, Wilkens LR, et al. The contribution of common genetic variation to nicotine and cotinine glucuronidation in multiple ethnic/racial populations. Cancer Epidemiol Biomarkers Prev. 2015;24(1):119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vugrin ED, Rostron BL, Verzi SJ, et al. Modeling the potential effects of new tobacco products and policies: a dynamic population model for multiple product use and harm. PLoS One. 2015;10(3):e0121008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang H, Abramsohn E, Park H-Y, Cowling DW, Al-Delaimy WK. Using a cessation-related outcome index to assess California’s cessation progress at the population level. Tob Control. 2010;19(Suppl 1):i56–i61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baurley JW, Edlund CK, Pardamean CI, Conti DV, Bergen AW. Smokescreen: a targeted genotyping array for addiction research. BMC Genomics. 2016;17(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swan GE, Benowitz NL, Jacob P, III, et al. Pharmacogenetics of nicotine metabolism in twins: methods and procedures. Twin Res. 2004;7(5):435–448. [DOI] [PubMed] [Google Scholar]
- 35. Swan GE, Hudmon KS, Jack LM, et al. Environmental and genetic determinants of tobacco use: methodology for a multidisciplinary, longitudinal family-based investigation. Cancer Epidemiol Biomarkers Prev. 2003;12(10):994–1005. www.ncbi.nlm.nih.gov/pubmed/14578134 Accessed October 22, 2015. [PMC free article] [PubMed] [Google Scholar]
- 36. Affymetrix Power Tools. Affymetrix. www.affymetrix.com/estore/partners_programs/programs/developer/tools/powertools.affx Accessed October 22, 2015. [Google Scholar]
- 37. Minimac3—Genome Analysis Wiki http://genome.sph.umich.edu/wiki/Minimac3 Accessed October 22, 2015.
- 38. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 40. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. www.R-project.org/ Accessed October 22, 2015. [Google Scholar]
- 42. Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. January 2014:005165. [Google Scholar]
- 43. Roadmap Epigenomics Consortium , Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou X, Maricque B, Xie M, et al. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8(12):989–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218):1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gauderman WJ. Sample size requirements for matched case–control studies of gene-environment interaction. Stat Med. 2002;21(1):35–50. [DOI] [PubMed] [Google Scholar]
- 49. Suhre K, Raffler J, Kastenmüller G. Biochemical insights from population studies with genetics and metabolomics. Arch Biochem Biophys. 2016. 589:168–176. [DOI] [PubMed] [Google Scholar]
- 50. Bergen AW, Michel M, Nishita D, et al. Drug metabolizing enzyme and transporter gene variation, nicotine metabolism, prospective abstinence, and cigarette consumption. PLoS One. 2015;10(7):e0126113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deeken J. The Affymetrix DMET platform and pharmacogenetics in drug development. Curr Opin Mol Ther. 2009;11(3):260–268. www.ncbi.nlm.nih.gov/pubmed/19479659 Accessed October 22, 2015. [PubMed] [Google Scholar]
- 52. McDonagh EM, Wassenaar C, David SP, et al. PharmGKB summary: very important pharmacogene information for cytochrome P-450, family 2, subfamily A, polypeptide 6. Pharmacogenet Genomics. 2012;22(9):695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumasaka N, Aoki M, Okada Y, et al. Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS One. 2012;7(9):e44507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Castaldi PJ, Cho MH, San José Estépar R, et al. Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190(4):399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaufmann A, Hitsman B, Goelz PM, et al. Rate of nicotine metabolism and smoking cessation outcomes in a community-based sample of treatment-seeking smokers. Addict Behav. 2015;51:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. European Commission. Attitudes of Europeans towards Tobacco 2012. http://ec.europa.eu/health/tobacco/docs/eurobaro_attitudes_towards_tobacco_2012_en.pdf Accessed November 4, 2015.
- 58. Maes HH, Sullivan PF, Bulik CM, et al. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34:1251–1261. www.ncbi.nlm.nih.gov/pubmed/?term=15697051 Accessed November 1, 2015. [DOI] [PubMed] [Google Scholar]
- 59. Nakajima M, Yamamoto T, Nunoya K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–1217. www.ncbi.nlm.nih.gov/pubmed/?term=8937855 Accessed November 1, 2015. [PubMed] [Google Scholar]
- 60. Nakajima M, Yamamoto T, Nunoya K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277:1010–1015. www.ncbi.nlm.nih.gov/pubmed/?term=8627511 Accessed November 1, 2015. [PubMed] [Google Scholar]
- 61. Jarvik ME, Madsen DC, Olmstead RE, Iwamoto-Schaap PN, Elins JL, Benowitz NL. Nicotine blood levels and subjective craving for cigarettes. Pharmacol Biochem Behav. 2000;66(3):553–558. www.ncbi.nlm.nih.gov/pubmed/10899369 Accessed November 1, 2015. [DOI] [PubMed] [Google Scholar]
- 62. Cannon DS, Mermelstein RJ, Hedeker D, et al. Effect of neuronal nicotinic acetylcholine receptor genes (CHRN) on longitudinal cigarettes per day in adolescents and young adults. Nicotine Tob Res. 2014;16(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cannon DS, Medina TR, Mermelstein RJ, et al. CYP2A6 longitudinal effects in young smokers. Nicotine Tob Res. 2016;18(2):196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gelernter J, Kranzler HR, Sherva R, et al. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77(5):493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hancock DB, Reginsson GW, Gaddis NC, et al. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hong LE, Yang X, Wonodi I, et al. A CHRNA5 allele related to nicotine addiction and schizophrenia. Genes Brain Behav. 2011; 10(5):530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dempsey DA, St Helen G, Jacob P, III, Tyndale RF, Benowitz NL. Genetic and pharmacokinetic determinants of response to transdermal nicotine in white, black, and Asian nonsmokers. Clin Pharmacol Ther. 2013;94(6):687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.