The colugo genome reveals hidden biodiversity and the sister group to primates.
Keywords: Colugo, population dynamics, phylogenomics, biogeography, Southeast Asia, genetics, genomic analysis
Abstract
Colugos are among the most poorly studied mammals despite their centrality to resolving supraordinal primate relationships. Two described species of these gliding mammals are the sole living members of the order Dermoptera, distributed throughout Southeast Asia. We generated a draft genome sequence for a Sunda colugo and a Philippine colugo reference alignment, and used these to identify colugo-specific genetic changes that were enriched in sensory and musculoskeletal-related genes that likely underlie their nocturnal and gliding adaptations. Phylogenomic analysis and catalogs of rare genomic changes overwhelmingly support the contested hypothesis that colugos are the sister group to primates (Primatomorpha), to the exclusion of treeshrews. We captured ~140 kb of orthologous sequence data from colugo museum specimens sampled across their range and identified large genetic differences between many geographically isolated populations that may result in a >300% increase in the number of recognized colugo species. Our results identify conservation units to mitigate future losses of this enigmatic mammalian order.
INTRODUCTION
As members of a strictly arboreal lineage of Southeast Asian gliding mammals, colugos (Order Dermoptera) have been known to science for centuries. However, the absence of captive individuals and a cryptic, nocturnal lifestyle have left basic questions surrounding their ecology and evolutionary history unanswered (1–3). At various times within the past century, colugos have been allied to mammals as divergent as insectivores and bats, and have played a central role in discussions of primate ancestry (4, 5). The phylogenetic position of colugos relative to other euarchontan orders remains highly controversial, with competing studies favoring an association of colugos with either primates or treeshrews (6–12). Current taxonomy describes the order Dermoptera as one of the least speciose within all of Mammalia, consisting of just two species in monotypic genera: the Sunda colugo, Galeopterus variegatus, and the Philippine colugo, Cynocephalus volans (13). This low species richness is surprising because colugos are widely distributed throughout the Southeast Asian mainland and archipelago, a region of otherwise remarkable biodiversity (14). Colugos also have the most elaborate gliding membrane among living vertebrates, which inhibits terrestrial movement and dispersal outside of forests (1, 2). Population differentiation has been supported by morphology (3) and high mitochondrial divergence between some Sunda colugo populations (15, 16). These distinctions may reflect valid species isolated in allopatry but remain unsubstantiated in the absence of broader geographic and genomic sampling and comparison. Thus, evolutionary questions surrounding dermopteran origins and taxonomic diversity remain unresolved, despite their importance to the interpretation of early primate origins and evolution, and to developing effective conservation strategies, respectively (8, 17, 18).
RESULTS AND DISCUSSION
Assembling a colugo reference genome
To produce the first detailed genetic insights into the poorly known history of this enigmatic mammalian order, we produced a draft genome assembly from a male Sunda colugo from West Java. We generated ~55× depth of coverage using Illumina sequence reads and produced an assembly (G_variegatus-3.0.2) that is 3.2 giga–base pairs (Gbp) in length (see Materials and Methods). This assembly is longer than most eutherian genomes, with a scaffold N50 of 245.2 kbp and contig N50 of 20.7 kbp. The assembly was annotated with the National Center for Biotechnology Information (NCBI) annotation pipeline and colugo RNA sequencing (RNAseq) libraries (see Materials and Methods), which identified 23,081 protein-coding genes. To test competing hypotheses concerning the relationship of colugos to other mammals, we performed comparative genomic analyses with a 2.5-Mbp one-to-one orthologous coding DNA sequence alignment between colugo and 17 other sequenced mammalian genomes (see Materials and Methods and table S1). These alignments were augmented with reference assemblies from a male Philippine colugo on the basis of 14× Illumina sequencing coverage, and from a pentailed treeshrew (Ptilocercus lowii) on the basis of ~5× coverage, to mitigate long-branch attraction effects (see Materials and Methods).
The closest living relative of primates
The relationship of colugos to other mammalian orders remains a topic of considerable debate. Craniodental characters primarily support colugos as sister to treeshrews (Sundatheria) (10), whereas retrotransposon insertions and some interpretations of postcranial skeleton characters suggest that colugos are sister to primates (Primatomorpha) (4, 6). Previous studies have demonstrated that evolutionary relationships based solely on phenomic data sets generate numerous polyphyletic relationships of convergently evolved mammals positioned within different superorders of placental mammals (10, 19). To address these conflicting hypotheses, we constructed maximum likelihood and coalescent-based phylogenies with the genome-wide supermatrix of 21 species from 8 eutherian orders (fig. S1). Our results consistently supported the Primatomorpha hypothesis (4), confirming colugos as the sister group of primates (Fig. 1 and fig. S1) (7, 9, 18). Because tree-building methods applied to deep, star-like radiations with shallow terminal lineages may be confounded by long-branch attraction artifacts (12), we also searched the whole-genome alignments for two independent types of phylogenetic character support that are not influenced in this way: (i) in-frame protein-coding indels (insertion/deletions) and (ii) near homoplasy-free retrotransposon insertions (see Materials and Methods). We identified 20, 5, and 5 coding indels (P = 4.5 × 10−5, χ2 test) and 16, 1, and 0 retrotransposon insertions [P = 2.7 × 10−7, Kuritzin-Kischka-Schmitz-Churakov test (20)] supporting Primatomorpha, Sundatheria, and primates + treeshrews, respectively (Fig. 1, figs. S2 to S4, and tables S2 to S4). These statistically robust reconstructions of Primatomorpha stand in contrast to the phenomic data set of O’Leary et al. (10), who identified 69 morphological characters uniting colugos with treeshrews (Sundatheria). Our results imply that any morphological similarities shared by colugos and treeshrews (10, 21) are due to convergent evolution or represent primitive characters lost in the primate ancestor.
Fig. 1. Phylogenetic placement of Dermoptera.
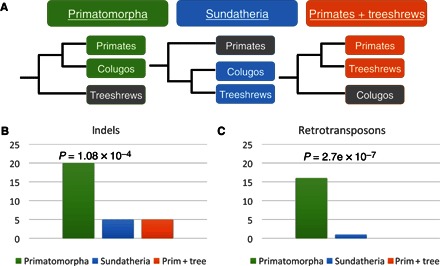
(A) Phylogenies depicting alternative hypotheses for dermopteran relationships relative to primates and treeshrews. (B) Number of indels supporting each evolutionary relationship. (C) Number of transposed elements supporting each evolutionary relationship.
Colugo adaptive evolution
We used a comparative genomic approach to explore the annotated gene sets of multiple euarchontans to investigate two major events in their early evolution: (i) lineage-specific genic changes that plausibly underpin colugo adaptations and provide insight into their distinctive biology, and (ii) lineage-specific genic changes that underpin ancestral primate innovations. We annotated the colugo olfactory receptor (OR) and vomeronasal (V1R) gene superfamilies (see Materials and Methods), which encode odorant and pheromone receptors, and found that they were intermediate in size between treeshrews and primates (Fig. 2A and table S5). This finding supports a progressive loss of OR gene repertoires that began in the ancestral lineage of Primatomorpha. We also found evidence for an increased importance of vision and hearing in the colugo lineage based on significant enrichment for positively selected genes involved in these sensory modalities (Padj = 0.0097 and Padj = 0.004, respectively) including genes that, when mutated, cause sensorial hearing loss and a variety of visual pathologies, notably macular degeneration (Fig. 2C and databases S1 and S2). The increased number of loss-of-function OR gene mutations in colugos is consistent with the view that selection for enhanced visual processing in a nocturnal, arboreal milieu corresponds with a relaxation of selection on olfaction (22). The magnitude of this hypothesized trade-off is greatest among mammals that experienced adaptive shifts from nocturnality to diurnality (23), but here, we show prevalence in a decidedly nocturnal lineage.
Fig. 2. Functional gene evolution and positive selection in colugos and ancestral primates.
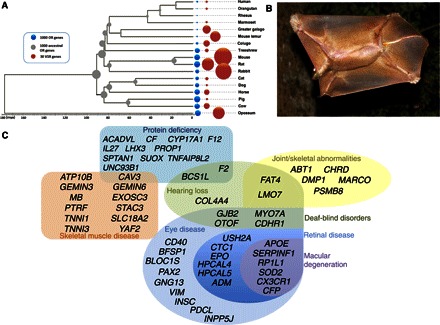
(A) Relative abundance of functional V1R (orange) and OR (blue) genes across sequenced mammals. The size of the circles is proportional to the number of functional genes. (B) Colugo gliding with patagium fully extended. (C). Venn diagram showing relationship between categories of enriched disease gene categories of colugo positively selected genes.
Positive selection was detected on similar vision-related genes on the ancestral primate branch, notably those in which mutations are implicated in night blindness and retinal degeneration (for example, NXNL1 and C8orf37). The ancestral primate branch also showed significant enrichment for positively selected genes that underlie brain function (Padj = 0.0001), including neurotransmitter genes implicated in behavioral disorders, such as schizophrenia and bipolar disorder, and neurodegenerative disease (Padj = 0.0004) (databases S3 and S4). The latter category includes ATXN10 and SACS, two genes in which mutations are associated with autosomal recessive spastic ataxia of Charlevoix-Saguenay, a human genetic disorder characterized by early-onset spastic ataxia, nystagmus, distal muscle wasting, finger and foot deformities, and retinal hypermyelination (24). It is highly plausible that positive selection on this suite of genes underpins the early morphological and behavioral evolution from ground-dwelling ancestors to early arboreal primates adept at grasping and climbing (25). The colugo patagium is the most extensive gliding membrane of any living vertebrate and stretches to the extremes of the digits and the tail when fully extended, resembling a living kite (Fig. 2B). Enriched disease gene categories within the dermopteran positively selected gene set include muscular atrophy (Padj = 0.0086) and protein deficiency (Padj = 0.0002), including genes involved in muscle contraction (for example, SLC18A2, TNNI1, and TNNI3) (database S2). Eight positively selected genes are also associated with joint/digital deformities in a variety of disorders (database S2). We speculate that adaptive changes in this suite of genes contribute to the gross anatomical transformations of the musculature and skeleton that evolved in the arboreal ancestors of these skilled gliders (4).
Colugo museomics, species diversity, and Southeast Asian biogeography
Colugos are widely distributed throughout Sundaland, a region well known for species richness and complex biogeographic patterns because of fluctuations in temperature, sea level, and vegetation throughout the Neogene (14, 26). Although current Sundaic forest distributions are in a refugial state with high sea stands, sea levels have been more than 40 m below current levels for ~92% of the past 1 million years (14, 26). The exposure of the Sunda shelf connected many islands with the mainland and with each other. It has been difficult to decipher the geographical extent of forested connections during low sea stands because geological, biotic, and climatic evidence remains inconclusive. Sundaic phylogeography is potentially informative in this regard; however, most widely distributed species that have been studied are either volant (for example, birds or bats) or highly vagile (for example, carnivorans), and many have diversified very recently within the region (26, 27). We hypothesized that colugos should track ancient Sundaic forest distributions because of their probable origin and widespread diversification within Sundaland; preference for closed-canopy forests; and reported isolation by rivers, disturbed forests, and savannahs (1, 2).
Given the scarcity of modern colugo samples with which to test this hypothesis, we exploited capture-based next-generation sequencing technologies (16) to retrieve orthologous DNA sequences from a broad sampling of museum specimens (see Materials and Methods, tables S6 to S8, and database S5) distributed across Sundaland and the southern Philippine islands of Greater Mindanao (Fig. 3A). We targeted ~140 kbp of biparental and Y chromosome loci from 66 colugo museum specimens that were between 28 and 121 years old and yielded adequate DNA (see Materials and Methods and database S6). We also obtained mitogenomic sequences from both off-target nuclear capture reads and direct low-coverage genome sequencing (see Materials and Methods and table S6). The colugo molecular timetrees were calibrated with the 95% confidence interval of our molecular estimate (average, 11.3 Ma) of divergence time between Galeopterus and Cynocephalus (see Materials and Methods, figs. S5 and S6, and table S9). Maximum likelihood–based maternal, paternal, and biparental phylogenies for both genera sort strongly by geography, showing major colugo lineages diversified in the Miocene or Pliocene (Fig. 3 and figs. S7 to S12). In addition, principal component analyses (PCAs) of X chromosome single-nucleotide polymorphism (SNP) variation (fig. S15) sort Sunda colugos largely by geographic location. PCA of 19 craniodental measurements (figs. S13 and S14, tables S15 and S16, and database S7) also sorts colugos roughly by geography to subsets of a continuous distribution but is insufficient to definitively separate colugos from different geographic origins or evolutionary lineages (see Materials and Methods). Notably, the large island of Borneo harbors multiple, deeply divergent colugo lineages, with eastern and western populations spanning the oldest bifurcation within Galeopterus (Fig. 3, figs. S7 to S12, and tables S12 and S13). This finding supports our prediction that ecological or topographic features such as mountains or major river systems presented substantial dispersal barriers to colugos despite simulations that predict forested connections throughout Borneo up to the last glacial maximum (26).
Fig. 3. Colugo phylogeography based on museomics.
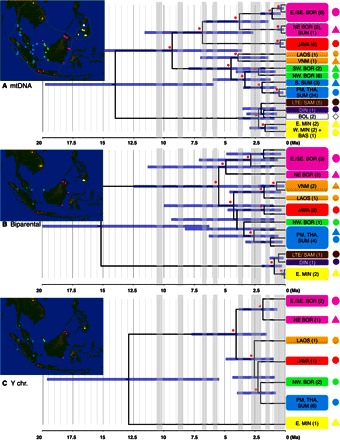
(A to C) Timetrees based on major lineages within phylogenies representing maternal (mtDNA; 16.6 kb), biparental (autosome + chrX; 115.6 kb), and paternal (chrY; 24.3 kb) evolutionary histories (figs. S7 to S12). Nodes with 100% maximum likelihood bootstrap support are denoted with red asterisks. Maps depict sample collection locations for each tree with corresponding colored symbol. Boxes indicate highly supported monophyletic clusters or divergent lineages representing putative species. Gray vertical bars denote times of low sea stands. VNM, Vietnam; THA, Thailand; PM, Peninsular Malaysia; S. SUM, south Sumatra; E/SE. BOR, east/southeast Borneo; NE BOR, northeast Borneo; SW BOR, southwest Borneo; NW. BOR, northwest Borneo; BUN, Bunguran; BAS, Basilan; E. MIN, eastern Mindanao; W. MIN, western Mindanao; BOL, Bohol; LTE, Leyte; SAM, Samar; DIN, Dinagat.
Ongoing debates argue for the presence/absence of a Pleistocene savannah corridor separating Borneo from Peninsular Malaysia and Sumatra, which may have prevented dispersal of forest-dependent species while allowing dispersal of larger terrestrial mammals between Indochina and Java (27, 28). We observed complete sorting of colugo mitochondrial and nuclear haplotypes from Peninsular Malaysia/Sumatra and western Borneo (Fig. 3), suggesting an absence of Pleistocene genetic exchange despite evidence for a forested connection at the last glacial maximum (26, 29). We infer that forested dispersal corridors during late Pleistocene glacial maxima were fragmentary or rare, or that strong reproductive isolating barriers to gene flow had accumulated in allopatry throughout the Pliocene, limiting introgression and retaining geographic structure. In contrast, colugos from Thailand, Peninsular Malaysia, and Sumatra show less than 1.0% mitochondrial divergence from most of their satellite islands (table S14), supporting recent, geographically limited dispersal and colonization following repeated insular submergence during the late Pleistocene.
Similar to the separation of colugos from Borneo versus Java and Peninsular Malaysia/Sumatra, Philippine colugo mitochondrial lineages are private to different islands and coalesce to the early Pleistocene (>1.5 Ma) (Fig. 3A and figs. S7 to S9). These dates mirror similar genetically structured patterns in several Philippine mammals, including tarsiers (30) and Apomys (forest mice) (31), suggesting that current deepwater channels formed effective barriers to interisland dispersal of arboreal lineages for much of the early Pleistocene. Nuclear gene loci also support monophyly of sampled islands (Fig. 3B and fig. S10), but divergences coalesce instead to the late Pleistocene, suggesting more recent nuclear gene flow between islands at low sea stands via forested connections.
Despite the overall similarity in phylogeographic patterns between mitochondrial and nuclear genomes, some discordance in phylogenetic topologies suggests that inheritance patterns reflected by male and female colugos differed (Fig. 3). Nuclear timetrees indicate Javan colugos diverged ~4.0 Ma from Indochinese, Peninsular Malaysian/Sumatran, and west Bornean colugos. However, in the mitochondrial DNA (mtDNA) phylogeny, Javan colugos are sister to east Bornean colugos and diverged much earlier at ~9.3 Ma (Fig. 3). This earlier Miocene mitochondrial divergence time and alternative topological placement of Javan colugos suggests that colugos most likely colonized Java from the ancestral east Borneo population. During Pliocene glacial maxima, eastwardly migrating colugo males could have dispersed into Java from western Sundaic source populations and captured the local mtDNA genome that is more similar to East Bornean colugos. This finding is consistent with the observation that male-biased dispersal is common in mammals and usually results in introgression of a local population’s mitochondria into the genome of the dispersing population, often with little nuclear introgression (32). Similar scenarios of male-mediated nuclear gene flow would explain the much older mtDNA versus nuclear divergence times of Philippine colugos (Fig. 3).
To underscore the magnitude of population genetic differentiation within both colugo genera, we calculated between-group maximum likelihood genetic distances for populations represented in the biparental and mitogenomic phylogenies (Fig. 3) and compared them to genetic distances between well-established mammal species (Fig. 4). The average between-group Sunda colugo mtDNA genetic distance was 11.7% (SD, 3.57%; minimum, 5.8%), and the neutral X chromosome distance was 0.58% (SD, 0.16%; minimum, 0.27%) between groups (tables S10 and S11), exceeding divergences between numerous well-accepted primate and Sundaic species (Fig. 4) (14). Partitioning of mtDNA genetic variation was similarly high among seven Sundaic populations [fixation indices (FST) = 0.89, P < 1× 10−5] and little within populations (table S15). Philippine colugo mtDNA population differentiation between and within islands was also very high (FST = 0.96, P < 1× 10−5; 4.42% mean divergence), whereas the average X chromosome divergence is 0.12% (SD, 0.02%; minimum, 0.1% between three sampled populations; tables S10 and S11), with five mitochondrial and three nuclear lineages displaying equivalent or greater genetic divergence than is observed between many primate species (Fig. 4) (30). Bayesian phylogenetics and phylogeography (BPP) analyses show the most significant support (PP ≥ 0.95) for six Sundaic and two Philippine species (tables S18 and S19). Considering these results, we argue that Sundaic and Philippine lineages each comprise multiple distinct species based on the application of modern species concepts (for example, genetic, general lineage) that recognize separately evolving lineages (33).
Fig. 4. Comparison of genetic distance between well-established species and proposed species groups for Galeopterus and Cynocephalus.
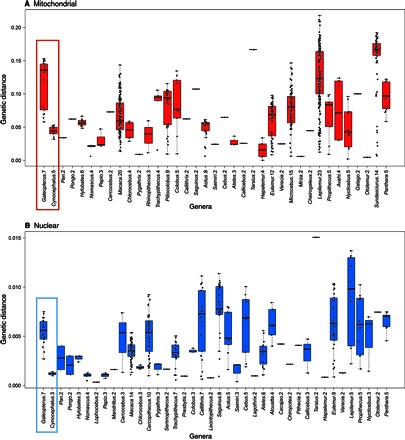
(A and B) mtDNA and nuclear DNA. The x axis lists the name of each genus followed by the number of species in that genus that were compared. For colugo genera, this number represents a conservative number of proposed groups/species. Colugo genera are highlighted in red and blue boxes.
Conservation
Our findings have far-reaching implications for the conservation of Sunda and Philippine colugos, which are presently listed as “least concern” on the International Union for Conservation of Nature (IUCN) Red List when considered as just two species. Here, we present concordant mitochondrial and nuclear genetic evidence for seven to eight colugo lineages that should be recognized as evolutionary significant units (ESUs) (34), or even distinct species, deserving of a conservation management strategy. Inclusion of additional, deeply divergent (that is, >4 to 5%) mitochondrial lineages from populations that currently lack nuclear DNA data may increase the number of ESUs to 14 (fig. S8 and table S16). The current and future conservation status of many of these smaller, isolated species-level taxa (for example, West Java) is uncertain, given their present low conservation status in the IUCN Red List and a general lack of population monitoring throughout their distribution. By 2010, ~70% of the primary lowland forests within Sundaland had been removed (35). Much of this land has been converted into oil palm and rubber plantations, and deforestation continues apace. Logging in the Philippines has also led to >90% reduction in forest cover over the past century (36). Population and ecological assessments within Singapore report that while colugos can persist quite well within secondary tropical forests with >95% canopy cover, these folivorous generalists are rarely found within boundaries of monoculture plantations (1, 2). Therefore, preserving minimally disturbed forests with high-density canopies within the range of these newly defined species will be critical for their future persistence and may facilitate the survival of many other endangered species in this region.
MATERIALS AND METHODS
G_variegatus-3.0.2 genome assembly and annotation
Sample and DNA extraction
The DNA used for sequencing Galeopterus variegatus was derived from a single male animal collected in West Java by M. Baba (Kitakyushu Museum of Natural History and Human History, Japan) under Indonesian Institute of Sciences Research Permits 6541/I/KS/1999, 3452/SU/KS/2002, and 3380/SU/KS/2003 (15). Ethanol-preserved tissue was used to extract genomic DNA with a Qiagen DNeasy Blood and Tissue Kit.
Genome sequencing and assembly
Total input sequence coverage of Illumina reads was 60× (45× 230- to 330-bp short inserts, 15× 3-kb mate pairs, and 5× 8-kb mate pairs) using a genome size estimate of 3.0 Gb. The assembled sequence coverage was 55×. The combined sequence reads were assembled using the SOAPdenovo2 software (37). The assembly was improved using an unpublished program designed to close gaps, and SSPACE (38). This draft assembly was referred to as Galeopterus_variegatus-3.0.2. This version had been gap-filled, error-corrected with approximately 12× Illumina reads, and cleaned of contaminating contigs. The assembly was made up of a total of 179,513 scaffolds with an N50 scaffold length of 249 kb (N50 contig length was 20.8 kb). The assembly spanned over 3.2 Gb.
RNA sequencing
Annotation for the G_variegatus-3.0.2 genome assembly was performed by NCBI and used RNAseq data from sequence read archive (SRA) samples SAMN02736899 and SAMN02736900.
Genome data set construction and analyses
Constructing coding sequences and determining orthology
One-to-one orthologous amino acid and nucleotide raw alignments for nine taxa (Homo sapiens, Pan troglodytes, Macaca mulatta, Callithrix jacchus, Otolemur garnettii, Ochotona princeps, Oryctolagus cuniculus, Canis familiaris, and Felis catus) (Ensembl v.79) were downloaded from OrthoMaMv9 (39). Coding DNA sequences were extracted from genome scaffolds for Galeopterus and Tupaia belangeri chinensis using their respective .gff annotation files. Coding DNA sequences were constructed for Cynocephalus using the Galeopterus.gff annotation file and consensus sequences derived from aligning Cynocephalus reads to the Galeopterus reference genome with Burrows-Wheeler Alignment (BWA) (settings: -n 0.001 -o 1 -l 24 -k 2). All coding DNA sequences were translated toamino acid sequences and only the longest isoform was kept for Galeopterus, Tupaia, and Cynocephalus. Orthologous sequences were determined by a three-way protein Basic Local Alignment Search Tool (BLAST) of human, mouse, and dog amino acid sequences to the longest amino acid isoforms from Galeopterus, Cynocephalus, and Tupaia followed by BLAST filtration. Filtration required that each query taxon (human, mouse, and dog) must have only one best BLAST hit per database species (Galeopterus, Tupaia, or Cynocephalus), the query sequence must have had >50% of database sequence bases covered, and the best BLAST hit bit score must be 2% greater than its second bit score.
Aligning orthologous amino acid isoforms to existing OrthoMaMv9 amino acid alignment
Unaligned orthologous amino acid longest isoforms from Galeopterus, Tupaia, and Cynocephalus were aligned to the existing raw amino acid alignment using Prank (40). Any stop codon characters (“*” or “_”) were masked with “X” before alignment. This ensured proper reverse translation of amino acid alignments to amino acid–guided nucleotide alignments. The stop codons were reintroduced to the amino acid–guided nucleotide alignments. Stop codon positions were recorded, and any nucleotide gene alignment with an internal stop codon not at the terminus was removed. Nucleotides corresponding to terminal stop codons were removed before analysis.
Genomic alignment filtration
Amino acid genome-wide coding DNA sequence alignments were filtered by Gblocks v0.9.1 (41) and subsequently filtered with two custom python scripts. The first removes the entire gene alignment if one or more individuals include frameshift mutations, which result in poor alignments for the length of the gene. The second removes highly divergent windows, which is useful for masking poorly aligned isoforms, and highly divergent regions of dubious orthology. Filtrations were applied to amino acid alignments with a window size of 10 amino acids, using a pairwise divergence calculation between each taxon and human. Pairwise deletion was applied to divergence calculations within windows such that gaps and missing data were skipped and matched nucleotide pairs = the numerator and matched + mismatched nucleotide pairs = the denominator. Windows were excluded (masked with X’s) if the amino acid divergence for one taxon was greater than 2 SDs away from the mean amino acid divergence across windows from identical alignment coordinates assuming a normal distribution, and the amino acid window had to be more than 20% diverged from human. In addition, all postfiltration amino acid sequences required >50% amino acid coverage and ≥20 amino acids in total length. Filtered amino acid sequences were back-translated to nucleotide sequences using custom python scripts. Nucleotide sequences only contained bases corresponding to unambiguous (that is, not X) amino acid bases, and skip blocks that were removed by Gblocks (41).
We also applied a filter to identify and remove poor alignments and putative paralogous sequences. We constructed maximum likelihood phylogenies for each gene alignment. Genes were excluded if the phylogenetic distance between any two pairs of taxa was more than 2.5× the observed genetic distance between human and mouse sequences, or over 3.75 SDs away from the mean branch length between terminal nodes for the 21-taxon data set. The same filter was applied to the 12-taxon data set to estimate the divergence time between colugo genera; however, we used a branch length cutoff of 2.5× the distance between human and dog as the criterion for gene exclusion. We chose human, mouse, and dog for these calculations because these species have the highest quality mammalian genome assemblies, with relatively large phylogenetic distances that we could use to compare to other taxa in the tree.
Phylogenetic analyses
RAxML v8.1.17 was used for all phylogenetic analyses with rapid bootstrap algorithm “-f a,” GTR + Γ “-m GTRGAMMA,” and 1000 bootstrap replicates for all nucleotide phylogenies (42). Amino acid phylogenies were constructed with similar settings and JTT amino acid substitution matrix + Γ “-m PROTGAMMAJTT.” SVDquartets analyses (43) were conducted with PAUP* 4.0a147 (D. Swofford). SVDquartets was originally designed for SNP data, but this method also performed well with simulated, multilocus data (43). All possible quartets were evaluated. Tree inference was based on the QFM quartet assembly method with the multispecies coalescent tree model. We performed a search for the optimal tree and a bootstrap analysis with 500 pseudoreplicate data sets.
Coding gene indels
A pool of potentially phylogenetically informative indels was identified by a custom python script that searches amino acid gene alignments for indels supporting a specific phylogenetic hypothesis. Input gene alignments were unfiltered whole-genome amino acid–coding sequence alignments (OrthoMaMv9) (39). Galeopterus, Cynocephalus, and Tupaia amino acid–coding sequences were then added to these alignments, as described in the Aligning orthologous amino acid isoforms to existing OrthoMaMv9 amino acid alignment section. Deletions were identified by shared gaps in taxa specified for a given hypothesis, whereas insertions were identified as gaps shared by all taxa exclusive of a specific hypothesis. For the Primatomorpha hypothesis (Homo, Pan, Gorilla, Pongo, Nomascus, Papio, Macaca, Callithrix, Tarsius, Otolemur, Microcebus, and Galeopterus), we specified that all 12 taxa had a shared gap of equal length in a sequence alignment. In the first stage of screening, we relaxed indel identification to only require a subset of these taxa (Homo, Macaca, and Galeopterus) to have the deletion, whereas the rest of the taxa (Pan, Gorilla, Pongo, Nomascus, Papio, Callithrix, Tarsius, Otolemur, and Microcebus) could, but were not required to, have the deletion. This was done because of errors in the lower quality genome assemblies and gene annotation for the remaining nine primates, which would prevent identification of the indel if all taxa were required to have the indel. Similarly, Galeopterus and Tupaia alignments were required to support Sundatheria indels, and Homo, Macaca, and Tupaia alignments were required to support primate + treeshrew indels. Candidate indels were filtered from the final list if any of the nonspecified taxa had a deletion (because of homoplasy or alignment error) with the same coordinates as those specified in the hypothesis. Candidate gene alignments were manually curated to identify the final set of phylogenetically informative indels (fig. S4). We required good indels to be flanked by conserved amino acid residues. We tested for statistical significance of each of the three specified hypotheses (table S4) following previously described methods (44).
Retrotransposons
Given the newly sequenced colugo genome, we further explored the question of how closely colugos were related to primates by focusing on another more complex but reliable set of rare genomic changes by screening for and analyzing the integration patterns of retroposed elements, virtually non-homoplastic phylogenetic markers. In mammals, retrotransposons integrated continuously over time and were accompanied by duplications of randomly selected 4 to 30 nucleotides [4 to 12 for long terminal repeats (LTRs) and 8 to 30 for long interspersed element 1 (LINE1s)] of the coincidental genomic target sites. Target site duplications enabled verification of orthology of diagnostic retroposon insertions in different taxa. Identical retroposon insertions in two species and an orthologous empty site in a third species supported the monophyly of the two and provided no support for relatedness of the third. We systematically tested all possible evolutionary hypotheses relating colugos (Dermoptera) to treeshrews (Scandentia) and human (Primates), by statistically considering the following three possible evolutionary scenarios: a phylogenetic group composed of (i) colugo + human, (ii) colugo + treeshrew, or (iii) treeshrew + human.
On the basis of a previous successful screening in Euarchontoglires, we searched the newly sequenced colugo genome (fig. S3) for LTRs (MLT1A/MSTD) and LINE (L1MA5/6) retrotransposon subfamilies, which were both active during the euarchontan speciation (11, 45). We then compared the 29,222 LTR-MLT1A and 12,983 LINE-L1MA5/6 hits we received along with their flanking target site duplications to other euarchontan genomes, which yielded 221 pairwise-aligned regions. After comparing these to the genomes of additional outgroup species (Ochotona princeps, Oryctolagus cuniculus, Mus musculus, Dipodomys ordii, Cavia porcellus, Ictidomys tridecemlineatus, Canis familiaris, Felis catus, Pteropus vampyrus, Equus caballus, Loxodonta africana, Procavia capensis, Choloepus hoffmanni, and Dasypus novemcinctus), we generated a retroposon presence/absence pattern for these species (table S2). Seventeen of these retroposons were phylogenetically informative; 12 LTR-MLT1A/MSTD elements and 4 LINE-L1MA5/6 elements were present in both colugo and human but were absent in treeshrew and other mammals. One additional LTR-MSTD element was present in both colugo and treeshrew but absent in human and outgroups. To specifically test the third hypothesis (treeshrews + primates), we also screened 66,860 loci of the two-way (University of California, Santa Cruz) human-treeshrew alignment (34,703 MLT1A/MSTD and 32,157 LiMA5/6), yielding 198 orthologous elements in human and treeshrew, but none were found that were also absent in colugo and the outgroup species (figs. S2 and S3). Sixteen retroposon elements shared between Primates and Dermoptera support the sister group relatedness of these two eutherian orders (table S2). The KKSC statistical test for genomic insertion data (http://retrogenomics.uni-muenster.de:3838/KKSC_significance_test/) (20) was significant (P = 2.7 × 10−7).
Sensory gene family expansions and positive selection
Published V1R and OR gene sequences from human, mouse, rat, cow, dog, and opossum were used as the query sequences for BLAST searches against the domestic cat genome. We enforced an E value threshold of 10−5 for filtering BLAST results. All identified sequences were extended 1 kb on either side for open reading frame identification and assessment of functionality. If multiple start codons were found, then the alignment results of known intact mammalian V1R and OR amino acid sequences were used as guidance. Any putative genes containing early stop codons, frameshift mutations, and/or incomplete gene structure (that is, three extracellular regions, seven transmembrane regions, and three intracellular regions) were designated as pseudogenes. To confirm orthology, we aligned all members of the V1R and OR gene families and constructed maximum likelihood trees. We compared the V1R and OR gene trees generated above to a mammalian species tree (9) to estimate gene gain and loss using the software Notung (46). Notung uses a given species tree and gene family numbers for each terminal taxon, and then uses an automated event-inference parsimony approach to estimate ancestral gene family sizes and gene duplication histories on each branch of the phylogeny.
Two data sets were constructed to test for positive selection: (i) a seven-taxon data set (H. sapiens, C. jacchus, O. garnettii, G. variegatus, Tupaia belangeri chinensis, M. musculus, and C. familiaris) and (ii) an eight-taxon data set (H. sapiens, C. jacchus, O. garnettii, G. variegatus, C. volans, T. belangeri chinensis, M. musculus, and C. familiaris). Amino acid sequences were downloaded from OrthoMaMv9. The initial seven-taxon data set contained 8514 gene orthologs, and the eight-taxon data set contained 4899 gene orthologs. Individual genes were removed if at least one taxon had a frameshift mutation or premature stop codon. A Perl script pipeline was applied, which removed poorly aligned or incorrectly annotated amino acid residues caused by obvious gene annotation errors within the genome assemblies. Aligned amino acid sequences were used to guide nucleotide coding sequences by adding insertion gaps and removing poorly aligned regions. Sequences were then back-translated to nucleotide sequences, and then genes still containing sequences for all taxa were aligned with Prank (40) and filtered as described in the Genomic alignment filtration section. We estimated nonsynonymous and synonymous substitution rates using the software PAML 4.0 (47). We used both branch-site and branch models as described in the study by Montague et al. (48) to identify accelerated rates of genes on specific branches of each evolutionary tree and specific amino acid residues that were potentially under positive selection. Paired models representing different hypotheses consisted of branch tests and branch-site tests (fixed ω = 1 versus variable ω). For the branch tests, free-ratio tests versus one-ratio tests were used to identify putatively positively selected genes. These genes were subsequently tested by two- and one-ratio models to identify genes with significant positive selection of one branch versus all other branches (two-branch test) (databases S1 and S3). Significance of likelihood ratio test results used a threshold of P < 0.05. We assessed enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and disease gene association tests using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/), and gene symbols as input (organism of interest: H. sapiens) (databases S2 and S4). Only significant KEGG Pathways and Disease Association categories were reported using a hypergeometric test, and the significance level was set at 0.05, implementing the Benjamini and Hochberg multiple test adjustment to control for false discovery. The most significant enrichment for genes in the colugo lineage was related to various categories of cardiovascular disease and lipid metabolism in humans, notably those encoding apolipoproteins (that is, APOE and APOH) that function in phospholipid and lipoprotein metabolism (Fig. 2C). However, we argue that this enrichment category was likely driven by a large number of genes with pleiotropic effects in both sensory systems and skeletal muscular function (shown in red in Fig. 2C; databases S1 to S4).
Colugo population data set construction and phylogenetic analyses
Sampling and DNA extraction
Ethanol-preserved tissues for CVO were obtained from the Field Museum of Natural History (Chicago, IL). DNA was extracted using Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s specifications.
Dried museum tissues were sampled from three different institutions: the Smithsonian Institution, National Museum of Natural History, Washington, DC; the American Museum of Natural History, New York, NY; and the Raffles Museum of Biodiversity Research, National University of Singapore (database S5). For most of the specimens, we removed ~5 mg of adherent tissue from inside the cranial cavity or nasal turbinate system. For some specimens, we collected multiple sample types, including hair, skin, cartilage, and bone. DNA was extracted from all tissues through proteinase K digestion, protein precipitation and removal, and ethanol DNA precipitation. Digestion was performed with 520 μl of Cell Lysis Solution (Puregene D-5002; Gentra, Qiagen), 600 μg of proteinase K, and 50 μg of linear acrylamide (Ambion) and incubated with a rotating heat block at 60°C for 48 hours. Undigested samples were disrupted with a pestle after 24 hours. Protein precipitation and ethanol DNA precipitation followed the standard guidelines of the Gentra/Puregene DNA isolation protocol (Qiagen).
Illumina library preparation, low-coverage sequencing, and nuclear capture
Illumina libraries for low-coverage sequencing were prepared with the Illumina TruSeq HT Dual Indexing kit following the manufacturer’s specifications, except that Microcon-30 centrifugal filters (Millipore) were used following the blunt-ending step, before adapter ligation, to retain degraded, low-molecular-weight DNA fragments (≥50 bp). Illumina libraries for nuclear capture were prepared with the NEXTflex Rapid DNA Sequencing Kit following the manufacturer’s specifications. Libraries were sequenced on the Illumina HiSeq 2000.
Nuclear capture was performed following the study by Mason et al. (16) with slight modifications, including a 72-hour hybridization reaction with Illumina adapter blocking oligos (49, 50). Primers for probe amplification were designed from the G_variegatus-3.0.2 genome assembly (database S6). Capture probes were generated through polymerase chain reaction (PCR) amplification of ~1 kb DNA fragments from modern Galeopterus (GVA), and Cynocephalus (CVO) DNA extracts (15). Four separate probe pools were generated, including individuals from different geographical locations. These were applied to different target samples, on the basis of locality, to minimize sequence divergence between the probe and the sample during capture experiments. Probes were amplified from high–molecular weight DNA extracted from the following frozen tissue samples: GVA_03 (Singapore, Peninsular Malaysia), GVA_04 (West Java), and CVO_02 (Leyte, Philippines). Three probe pools [(i) Peninsular Malaysia, (ii) West Java, and (iii) Peninsular Malaysia + West Java] were used to perform hybrid capture from Galeopterus samples, and the CVO_02 probes were used to perform hybrid capture from Cynocephalus samples. Amplifications were performed with Platinum Taq DNA polymerase (Invitrogen), 1.5 mM MgCl2, 0.8 mM deoxynucleotide triphosphates, and 2 μM primers under the following cycling conditions: 2-min hot start at 94°C, denaturation for 30 s at 94°C, touchdown annealing at 2 cycles each of 60°, 58°, 56°, 54°, and 52°C, followed by 30 cycles at 50°C, extension for 1 min at 72°C, and final extension for 5 min at 72°C. Successful amplicons were pooled (equal volume) and labeled with biotin using Biotin-High Prime (Roche).
Probe design and generation
X chromosome probes: We queried the draft colugo genome assembly using a set of human and mouse 1:1 orthologous X chromosome coding sequences (Ensembl v67) using BLAST to identify candidate colugo X chromosome contigs. We then performed reverse BLAST of the top scoring colugo contigs back to the human (GRCh37.p7), mouse (NCBI m37), and dog (CanFam 2.0) genome assemblies. Colugo contigs that had top-scoring BLAST hits to the X chromosome sequence from all three species were considered orthologous. We generated nearly neutral X chromosome capture probes by designing PCR-based amplicons ~1 kb in length from within each retrieved scaffold (database S6). We specifically targeted nonrepetitive sequence that was the greatest possible distance from any annotated coding sequence within the scaffold.
Y chromosome probes: Human and mouse single-copy Y chromosome genes (51, 52) were queried against Galeopterus_variegatus-3.0.2 using BLAST. We then performed reverse BLAST of the top-scoring colugo contigs back to the human genome (GRCh37.p7). We selected those contigs with a best BLAST hit to the same single-copy Y chromosome sequences and/or to the X chromosome with >15% sequence divergence. We also constructed maximum likelihood trees with each candidate contig and annotated X and Y orthologs to validate reciprocal monophyly of X and Y gene sequences. We generated nearly neutral Y chromosome capture probes in the same manner as described above for the X chromosome probes (database S6). Primers were validated as Y-specific by simultaneous PCR screening of male and female DNA samples.
Autosomal gene probes: We designed capture probes to target a subset of selected protein-coding genes that influence vision, coat color, and body size in mammals (database S6). Candidate human protein-coding genes were queried against Galeopterus_variegatus-3.0.2 using BLAST. Identified exons were aligned to human genes and trimmed to human exon boundaries. Orthology versus paralogy was determined by maximum likelihood phylogenetic tree construction using a sequence matrix of known mammalian orthologs, as well as closely related paralogs. We selected all exon-containing contigs that formed a monophyletic group with orthologous mammalian exons for the genes of interest. Primers were designed for all targeted sequences with BatchPrimer3 (53).
Modern DNA sequence trimming and filtration
Illumina sequences were filtered with TrimGalore! v0.3.3 to remove Illumina adapter sequences and trim low-quality bases (parameters: --paired --retain_unpaired -q 20 --length 30 --stringency 1 --length_1 31 length_2 31) (www.bioinformatics.babraham.ac.uk/projects/trim_galore/).
Modern genome reference assemblies
We generated ~14× paired-end Illumina read coverage for Cynocephalus from a 300-bp average insert size Illumina library created from DNA of CVO_02. A reference assembly was constructed by aligning quality-filtered Illumina sequences to the G_variegatus-3.0.2 genome assembly with BWA v0.7.5a-r405, bwa-mem (http://bio-bwa.sourceforge.net/bwa.shtml). Approximately 5× coverage of pen-tailed treeshrew (Ptilocercus lowii) Illumina reads (SRA accession no. SRP064536) was reference-aligned to the Tupaia genome scaffolds (54) using bwa-mem.
Museum DNA sequence trimming and filtration
Raw Illumina sequences were filtered with SeqPrep to remove Illumina adapter sequences, trim low-quality bases, and merge overlapping sequence pairs (parameters: -A AGATCGGAAGAGCACACGTC -B AGATCGGAAGAGCGTCGTGT -q 13 -o 15 -L 30 -g) (https://github.com/jstjohn/SeqPrep). We removed the first and last three bases of each sequence read, because these bases are highly susceptible to chemical damage (55). However, a previous analysis of a subset of these DNA samples failed to indicate significant levels of DNA damage (16).
mtDNA, biparental, and Y chromosome sequence assembly
Mitochondrial genomes were assembled using both de novo (SOAP and CAP3, http://seq.cs.iastate.edu/cap3.html) (37, 56) and reference-based (BWA aln v0.7.5a-r405) (57) assembly strategies. Multiple assemblies based on a range of k-mer values were performed on sequences from each individual. De novo assemblies that produced complete mitogenomes were used as reference sequences for other individuals, and selected on the basis of geographic proximity, to reduce sequence divergence between the reference sequence and the assembled reads. The reference sequences were three complete Galeopterus mtDNA genomes from GenBank (AJ428849.1, JN800721.1, and AF460846.1) and five de novo mitogenome assemblies from samples GVA_22 (Palembang, Sumatra), GVA_45 (Sabah, Borneo), GVA_49 (Pulau Sebuko, Borneo), CVO_06 (Samar Island, Philippines), and CVO_08 (Tupi, Mindanao). Sequence reads from each sample were aligned to reference mitogenomes from several of the geographically closest candidates, with BWA aln parameters -n 0.0001 -o 1 -l 24 -k 3. The assembly with the highest percentage of reference bases covered and the highest average depth of sequence was chosen for final consensus sequence generation.
Biparentally inherited target loci were extracted from the G_variegatus-3.0.2 genome assembly and used as the reference sequence for reference-based assemblies for all Galeopterus samples. Sample CVO_08 was chosen as the reference sequence for all Cynocephalus assemblies because it mapped to the G_variegatus-3.0.2 probe sequences with the highest reference base coverage and depth. Reference assemblies were mapped using BWA with parameters -n 0.001 -o 1 -l 24 -k 2.
Y chromosome sequence alignments for male Sunda colugos were constructed by aligning to Y chromosome scaffolds identified within the G_variegatus-3.0.2 genome assembly. Philippine colugos were aligned to a Cynocephalus reference-based Y chromosome consensus sequence generated from aligning the reads of the highest quality male DNA specimen, CVO_10, to G_variegatus-3.0.2 Y chromosome scaffolds.
Consensus sequences
Consensus sequences were called using SAMtools (58) [v1.1-26-g29b0367 (htslib 1.1-90-g9a88137)] mpileup, vcftools [v1.1-88-g4c0d79d (htslib 1.1-90-g9a88137)], and vcfutils.pl. All biparental sequences were called as diploid requiring a minimum read depth of 3. Y chromosome sequences were called as haploid and required a minimum depth of 2.
Sequence alignments and phylogenetic analyses
Mafft v7.127 (59) was used to align all consensus sequences. Alignments were manually curated to remove poorly aligned regions. RAxML v8.1.17 was used for all phylogenetic analyses with rapid bootstrap algorithm -f a, GTR + Γ -m GTRGAMMA, and 1000 bootstrap replicates for all nucleotide phylogenies (42).
Genetic distance estimates
MEGA6 v6.06 (60) was used to calculate between-group mean genetic distances. Groups were defined as strongly supported (100% bootstrap support) monophyletic groups or lineages that had at least ~3 to 5% between-group mtDNA divergence and, in some cases, corroborative nuclear phylogenetic structure. MEGA-CC v7.0.7 (61) was used to calculate between-species divergence levels within primate genera sampled from the study by Perelman et al. (62). Species comparisons were required to have >20% total sequence coverage for mitogenome data and >50% coverage for nuclear data. We used the maximum composite likelihood + Γ distance and pairwise deletion (removing all ambiguous bases for each sequence pair) for all comparisons. We required a minimum of two species per genus for comparisons to other primate and Sundaic mitochondrial and nuclear gene sequences. Panthera interspecific distance calculations were derived from whole-genome alignments between several species (63). Boxplots were constructed in R v3.1.2 (R Core Team, 2014).
Analysis of molecular variance
To estimate the degree of differentiation among populations, we estimated FST using an analysis of molecular variance (AMOVA) approach calculated in Arlequin v3.5.2.2 (64). Mitochondrial haplotype data were compiled in DnaSP v5 using a complete deletion-option alignment (7176-bp final sites) for 45 Sunda colugos to make the input Arlequin file (65). We defined seven populations for the AMOVA, with 16,000 permutations.
PCA of genetic variation
We analyzed 1340 SNPs present in our X chromosome capture data from 12 individuals that represent the 7 major clades of Sunda colugos present in the biparental phylogenies. Specimen read group (@RG) and sample (SM:) information were introduced to each .bam alignment file during the alignment. SNPs were called through SAMtools mpileup, bcftools, and vcfutils.pl. SNPs were required to have a minimum root mean squared mapping quality of 30, a minimum depth of 3, and a maximum depth of 100.
We used the R package SNPRelate to perform the PCA from the .vcf file. The first five principal components explained 19.4, 13.7, 12.2, 10.9, and 9.1% of the total variation. Therefore, only 65.3% of the total X chromosome SNP variation was explained by the first five principal components. Although only a small subset of the variation can be explained in two-dimensional space, we still see clear separation of five of the seven proposed Sundaic species from the first two principal components (fig. S13). Biplots for principal components 1 to 6 are shown in figs. S15 to S19. The biplot for PC1 versus PC2 illustrated almost no variation between peninsular Malaysian/Sumatran individuals and the west Bornean individual (GVA_16). However, the first two principal components are inappropriate for comparisons to GVA_16, because GVA_16 is only minimally correlated with PC1 (r2 = 0.11) and PC2 (r2 = −0.05) and therefore does not vary along dimensions described by PC1 and PC2. On the other hand, GVA_16 is highly correlated with PC5 (r2 = −0.35) and PC6 (r2 = −0.73) (table S19), meaning GVA_16 varies along these axes and the total variance explained by PC5 and PC6 is still substantial: 9.1 and 7.5%, respectively. Because the total variance explained by each principal component is known and the sum of squares of one principal component’s loadings is equal to 1, we can calculate the proportion of variance explained by each variable (individual in this case) for each principal component. vi = (l)^2*(v), where vi is the percentage of total variance explained by one variable (individual) for one principal component, l is the variable component loading, and v is the percentage of total variation explained by this one principal component. The calculated percentages of total variation explained by GVA_16 for PC1 and PC2 were 0.23 and 0.03%, whereas those for PC5 and PC6 were 1.15 and 4.06%, respectively.
Divergence dating
Calibrations
The poor dermopteran fossil record precluded the application of internal fossil calibrations. Therefore, we calibrated these phylogenies with the 95% confidence intervals of the Galeopterus-Cynocephalus divergence date estimated from 2729 genome-wide orthologous coding gene alignments (final matrix length, 3,515,409 bp) extracted from 12 mammalian genomes (H. sapiens, P. troglodytes, M. mulatta, C. jacchus, O. garnettii, O. cuniculus, O. princeps, C. familiaris, F. catus, G. variegatus, C. volans, and T. belangeri chinensis) (OrthoMaMv9) (39) and 7 external fossil calibrations (table S9) as minimum and maximum constraints.
MCMCTree
Divergence time estimation was performed under several different analysis conditions that varied both rates (independent and autocorrelated) and calibration (hard and soft) following the study by Meredith et al. (9). We chose an approximate-likelihood method under a GTR + Γ model of sequence evolution in the MCMCTree package (47). We used v4.8a, which implements a revised dirichlet prior, enabling proper estimation of rgene_gamma (substitution rate through unit time) and proper retention of uncertainty in confidence intervals from fossil calibrations (66). All MCMCTree calculations were performed twice to ensure convergence. Timetree branch lengths were averaged from multiple runs, and the maximal range of 95% confidence intervals was kept to represent maximal uncertainty for each node.
The rgene_gamma prior shape and scale (α and β) values were estimated by first calculating the clock-like substitution rate per unit time in baseml for the whole phylogeny given a nucleotide alignment and rooted phylogeny with branch lengths, and point estimates of divergence time at available calibrated nodes. Prior values α = (m/s)^2 and β = m/s^2, where m is the mean and s is the SD. Mean is the SD of the Γ distribution when the shape parameter α = 1. If α = 1, then we solve for β with m = s.
Molecular divergence time estimate between colugo genera
The mean divergence time between Galeopterus and Cynocephalus was estimated as 11.3 Ma (with a 95% credibility interval of 5.1 to 24.3 Ma). The point estimate coincided approximately with the lowest sea stand of any prior to that during the Tertiary (67) and represented the first glacial period that lowered sea levels below present-day levels during the Miocene (68, 69). We note, however, that conflicting long- and short-term eustatic sea level curves have been reported throughout the Neogene (70). We hypothesized that a forested corridor between Borneo and the Philippines must have been present to facilitate colonization of the Philippines, likely via a route formed along the current Sulu archipelago.
Molecular divergence time estimates within colugo genera
We estimated divergence times within each colugo genus using MCMCTree, assuming an autocorrelated rates model with a soft calibration for the basal split between Cynocephalus and Galeopterus derived from the 95% confidence intervals of our molecular supermatrix-based estimate (Molecular divergence time estimate between colugo genera section). Calculations were performed with exact likelihood and an HKY-85 + Γ model of sequence evolution. Data sets were reduced to one taxon per divergent lineage, selecting individuals with the greatest capture probe coverage.
Craniodental morphometric analyses
Morphometric data included 19 linear craniodental measurements taken from 82 Sunda colugo skulls after sampling tissue from museum specimens (database S7). Data were log-normalized before PCA. PCA was performed with the R package “prcomp” by singular value decomposition. We conducted PCA with and without normalizing for body size (fig. S16). Condylobasal length (CBL) is a measurement of skull length and is correlated with body size; therefore, we divided measurements by CBL to normalize measurements for body size (3).
We observed the most geographic sorting in PCAs without normalizing for body size and when recently diverged dwarf individuals were removed (fig. S18). After males, females, and dwarfs were normalized for body size, we saw little geographic structuring, which indicated that most of the variation in craniodental measurements was due to body size variation, confirming observations by Stafford and Szalay (3) (figs. S18 and S19). This is expected because all measurements are highly correlated with CBL (mean = 0.68 and SD = 0.18). However, the measurement of “min.w.temps” (the minimum distance between the temporal lines on the roof of the skull) is least correlated with body size (r = 0.26). The min.w.temps (minimum width between temporal lines) vector is of significant magnitude and tends to sort Bornean colugos to a subset of the distribution after body size normalization (fig. S17 and table S19). No PCAs based on morphology were capable of sorting colugo populations to mutually exclusive clusters; however, they did generally sort on the basis of regional geographic distribution.
Dwarf colugos were defined as individuals with a 10% reduction in CBL compared to the average CBL of neighboring populations from large islands or the mainland. Dwarf individuals residing on satellite islands represent recent deviations in phenotype when compared to the morphology of larger islands and therefore are not representative of the deeper evolutionary history of Sunda colugo species. This again agrees with Stafford and Szalay (3), who concluded that dwarf populations did not warrant species-level classifications based on body size reduction alone.
Species classifications, BPP, and conservation units
The genetic species concept by Baker and Bradly (33) argues that species can be classified on the basis of genetic isolation rather than reproductive isolation. We present evidence for multiple, genetically divergent lineages within the Sunda and Philippine colugo that, in most of the cases, is consistent across two or more genetic transmission components (that is, mtDNA, Y chromosome, and biparentally inherited loci). The genetic divergence levels between six and seven Sunda colugo populations (tables S10 to S13) exceeded those observed between numerous well-established species within other mammalian orders. Nuclear DNA and mtDNA genetic divergence levels conservatively supported a classification scheme that recognizes a minimum of six species within Galeopterus. Three additional ESUs, and potentially valid species, may be recognized within the genus [southwest Borneo (GVA_58 and GVA_61), southeast Borneo (GVA_49), and south Sumatra (GVA_21, GVA_22, and GVA_28)] on the basis of divergent (>4%) mitochondrial haplotypes.
The nuclear sequence divergence estimated for all Cynocephalus pairwise comparisons exceeded that of at least seven pairs of described primate species (Fig. 4), potentially supporting up to three species-level taxa within the Philippines: Leyte, Dinagat, and eastern Mindanao. Only mtDNA was obtained from specimens of colugos sampled from western Mindanao (Zamboanga Peninsula), Basilan, and Bohol. Mitochondrial divergence between Basilan and western Mindanao was less than 1%. However, the mtDNA sequence divergence between Bohol and western Mindanao and between these two populations and all other Philippine populations was between 3.2 and 4.1% (estimated divergence time, ≥1.5 Ma) (table S12 and Fig. 3A). Given the general concordance between divergent nuclear and mitochondrial lineages, we consider each of the following five Philippine populations as ESUs worthy of formal recognition and separate conservation strategies: (i) eastern Mindanao, (ii) western Mindanao + Basilan, (iii) Dinagat, (iv) Leyte, and (v) Bohol.
We used BPP, which uses multispecies coalescent models (MSCs) (71), to test our proposed colugo species groups against alternative species models. We provided a fixed guide tree based on the structure recovered from phylogenies constructed in RAxML to serve as our species delimitation model. We used transmodel inference, where two MSC models [rjMCMC (reversible-jump Markov chain Monte Carlo) algorithm 0 and rjMCMC algorithm 1] were used to calculate the posterior probabilities for the splitting of each node within the phylogeny from multilocus data (A10: species delimitation = 1 and species tree = 0). BPP implements the rjMCMC algorithm (72) and was used to test species delimitation with two parameter settings. The species delimitation parameters were set as species delimitation = 102 and as species delimitation = 1121 in the BPP control file. The first model, species delimitation = 102, can be translated as 1 (which means that species delimitation is not fixed), 0 (which means that rjMCMC algorithm 0 is used), and 2 [which means that ε (a fine-tuning parameter) is equal to 2] (reasonable values for ε are 1, 2, 5, etc.) [(72), bppDOC.pdf]. The second model, species delimitation = 112 1, can be translated as 1 (which means that species delimitation is not fixed), the second 1 (which means that rjMCMC algorithm 1 is used), 2 [which means that α (shape) is equal to 2], and 1 [which means that m (mean) is equal to 1], where α and m are fine-tuning parameters. Equal prior probabilities for rooted trees were specified by speciesmodelprior = 1. The provided fixed phylogeny represented the seven proposed species groups of the Sunda colugo “((east Borneo and northeast Borneo), ((Vietnam, Laos), (Java, (west Borneo, Peninsular Malaysia + Sumatra)))),” with 3121214 individuals representing each species group, respectively. For the Philippine colugo, we proposed three species for which we successfully captured adequate nuclear DNA: “(Mindanao, (Leyte, Dinagat))” using 2 1 1 individuals per species group. The rjMCMC model imitates the biological species concept for speciation because it assumes that no recent gene flow (migration) is experienced between populations (72). This model includes two main parameters θs and τs.
We estimated the Γ priors theta (θs), the parameter for modern and ancestral species population sizes, and tau (τs), the divergence time parameter for the root in the species tree [(71), bppDOC.pdf]. A Γ prior’s distribution is defined by two parameters: shape parameter (α) and rate parameter (β). The shape parameter should be changed depending on how accurately the prior represents the data (71). High-confidence prior values might have a high α value, whereas low-confidence priors should have a low α value. Increasing α restricts the Γ distribution, reducing how much parameters can vary in the posterior, whereas low α results in a diffuse Γ distribution, where estimated values can vary more freely in the Bayesian posterior (71). To lessen restrictions on parameter estimates in the posterior, we chose a diffuse shape parameter α = 2. We estimated the rate parameter β for Γ priors theta (θs) and tau (τs) with α = 2, the mean (m) of the Γ distribution, and the SD (s) of the Γ distribution. The Γ prior theta is based on the population size, and the mean of the Γ distribution is calculated as the average proportion of different sites between any two random sequences. The average between-group genetic divergence for Sunda colugos is ~0.5%; therefore, the average proportion of differing sites is ~0.005, which is equal to the mean (m) of the Γ distribution. For Cynocephalus, m = ~0.001. The relationship between the mean and SD of a Γ distribution changes as α changes. The SD of the Γ distribution is s = m/√α. For Galeopterus, s = 0.005/√2 = ~0.0035. The mean and SD are used to calculate β = m/s2 = 0.005/0.00352 = 408. Calculated α and β for theta prior were 2408. The mean for tau was calculated as the years of divergence of the root of the tree divided by the mutation rate (1 × 10−9). The mean of the Γ distribution for tau for Galeopterus was 5.5 × 106/1 × 10−9 = 0.0055. Divergence time for the root was derived from the biparental timetree, which is ~5.5 Ma. The same procedure was followed for calculating priors for tau and Cynocephalus. Estimating priors followed the study by Yang (71) and the BPP documentation. All BPP runs were executed twice to confirm convergence. The sensitivity of BPP analyses was also assessed by varying the rate parameter (β) for theta and tau priors (71). The Galeopterus biparental data set was sensitive to variations of β for theta when varied from 10 to 1000; however, it was not sensitive to variation in β for tau (table S11). The sensitivity of the biparental data set for Galeopterus only changed the support values between the six- and seven-species models and the probability for the presence of a node separating Laos from Vietnam; however, only six species were strongly supported, with posterior probability >0.95 (table S17). The Cynocephalus data set was neither sensitive to variation in β for theta, when varied from 1000 to 2040, nor to variation in β from 10 to 1000 for tau (table S18).
Biogeography notes
The presence of a north-south savannah corridor running through the South China Sea and the Javan Sea during Pleistocene glacial maxima would likely have prevented dispersal of forest-dependent taxa, like colugos, between Borneo and Sumatra + Peninsular Malaysia, and between Borneo and Java. However, the extent to which this savannah corridor was present, and the continuity of the corridor across its proposed distribution, is debated (26, 27, 29). In general, glacial maxima are characterized as dry periods with less precipitation, accompanied by drastically lowered sea levels (−120 m), which expose sandy seabed soils (73). Simulations have predicted that the last glacial maximum was very dry and cold, suggesting the possibility of a continuous savannah corridor. This finding is supported by genetic evidence from many forest-dependent vertebrate species distributed between Borneo, Peninsular Malaysia, and Sumatra, which have estimated divergence times predating the last glacial maximum (28). However, even if there was a savannah corridor present at the last glacial maximum, there were many interglacial periods during prior millenia when forested corridors likely would have existed to connect these present-day landmasses (14, 26, 74).
Supplementary Material
Acknowledgments
We thank the following individuals for providing samples and/or technical assistance during the course of this project: M. Baba, B. Covert, H. Minh Duc, L. Gordon, L. Heaney, J. Janečka, D. Lunde, P. Ng, T. Raudsepp, and M. Roberts. Funding: This work was supported in part by the NSF (grant EF0629849 to W.J.M.), Texas A&M Institute for Genome Sciences and Society (to W.J.M. and V.C.M.), Texas A&M College of Veterinary Medicine and Biomedical Sciences (to V.C.M.), the German Research Foundation (grant SCHM1469/3-2 to J.S.), and the NIH–National Human Genome Research Institute (grant 5U54HG00307907 to R.K.W.). Author contributions: W.J.M., V.C.M., and G.L. conceived and designed the phylogenomic and population genomic experiments. K.M.H., V.C.M., and W.J.M. performed museum specimen sampling. N.T-L.L. provided specimens. W.C.W., P.M., and R.K.W. generated and assembled the Galeopterus draft genome sequence. V.C.M. generated the sequence data and reference assembly for Cynocephalus, performed mammalian phylogenomic analysis, and wrote code for and performed the coding indel analysis. A.D.M. and N.J.D. provided Ptilocercus sequence data. J.S. and G.C. conceived the retroposon presence/absence analysis. G.C. extracted potential informative retroposons. G.C., J.S., and L.D. analyzed phylogenetically diagnostic retroposons. G.C. compiled the genomic transposon landscape. V.C.M. and G.L. performed DNA ortholog curation and developed code for filtration pipelines. G.L. performed olfactory and vomeronasal gene annotation and gene family analysis. G.L. performed positive selection analyses. G.L. and W.J.M. performed positive selected gene curation and enrichment analyses. W.J.M., G.L., A.D.M., and N.J.D. analyzed sensory gene results. V.C.M. and M.S.S. performed relaxed molecular clock analyses. M.S.S. performed SNP coalescent analyses. V.C.M. isolated museum DNA; designed and created capture probes; performed museum DNA capture; analyzed sequence data; and performed population genetic analysis, molecular dating, and BPP analyses. K.M.H. scored morphometric data. V.C.M. analyzed morphometric data. W.J.M., V.C.M., J.S., A.D.M., M.J.D., M.S.S., and K.M.H. wrote the article with input from the remaining authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Data reported in this article are available under NCBI Genome accession no. GCA_000696425.1 G_variegatus-3.0.2, short read archive accession numbers SRX1854847-1854914, and Dryad accession number doi:10.5061/dryad.6q738. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/8/e1600633/DC1
fig. S1. Maximum likelihood phylogeny of Euarchontoglires based on an alignment of 631 orthologous protein-coding sequences.
fig. S2. Insertion-deletion evidence for colugo phylogenetic relationships based on transposons.
fig. S3. Description of the transposon screening strategy.
fig. S4. Phylogenetically informative protein-coding gene indels supporting Primatomorpha (P1 to P15), Sundatheria (S1 to S5), and Primates + Scandentia (PS1 to PS5).
fig. S5. Maximum likelihood timetree showing seven nodes with external fossil calibrations.
fig. S6. Maximum likelihood timetree showing seven nodes with external fossil calibrations, with euarchontan monophyly enforced.
fig. S7. Maximum likelihood mtDNA tree based on a matrix of 53 colugos with ≥90% mitogenome coverage.
fig. S8. Maximum likelihood mtDNA tree based on a matrix of 65 colugos with ≥30% mitogenome coverage.
fig. S9. Maximum likelihood tree based on a combined nuclear (biparental + Y) matrix of 17 taxa.
fig. S10. Maximum likelihood tree based on biparental data set (18 taxa).
fig. S11. Maximum likelihood Y chromosome tree (depth 2) (15 taxa).
fig. S12. Maximum likelihood Y chromosome tree (depth 3) (14 taxa).
fig. S13. Biplot of PC1 and PC2 from PCA of colugo X chromosome variants.
fig. S14. Biplot of PC2 and PC3 from PCA of colugo X chromosome variants.
fig. S15. Biplot of PC3 and PC4 from PCA of colugo X chromosome variants.
fig. S16. Biplot of PC4 and PC5 from PCA of colugo X chromosome variants.
fig. S17. Biplot of PC5 and PC6 from PCA of colugo X chromosome variants.
fig. S18. Biplot of morphometric PCA with males and females, and no dwarf specimens removed.
fig. S19. Biplot of colugo morphometric PCA, with male, female, and dwarf specimens, where all measurements are normalized by body size.
table S1. Mammals used in phylogenetic analyses.
table S2. Presence/absence of phylogenetically informative retroposon markers and species distribution.
table S3. Location of diagnostic retroposon markers in the human genome.
table S4. χ2 calculation for phylogenetically informative indels.
table S5. Colugo and select mammal V1R and OR gene annotation summaries.
table S6. Nuclear capture efficiency.
table S7. Adapter blocking oligos.
table S8. Off-target mtDNA assembly statistics.
table S9. External fossil calibrations used to calculate divergence time between the two colugo genera.
table S10. Principal component loadings for morphometric PCA using males and females with no dwarf individuals.
table S11. Principal component loadings for morphometric PCA using males, females, and dwarf individuals, and all measurements were normalized for body size.
table S12. mtDNA maximum composite likelihood + Γ distance matrix calculated between seven Sunda colugo groups and five Philippine groups.
table S13. X chromosome maximum composite likelihood + Γ distance matrix calculated between seven Sunda colugo groups and three Philippine groups.
table S14. Between-group mtDNA composite likelihood + Γ distance matrix for Sunda colugos from Thailand, Peninsular Malaysia, and Sumatra, and their satellite islands.
table S15. Within-group mtDNA maximum composite likelihood + Γ distance matrix.
table S16. Between-group mtDNA composite likelihood + Γ distance matrix for 10 Sunda colugo groups and 5 Philippine groups.
table S17. BPP species estimation results for Sunda colugos.
table S18. BPP species estimation results for Philippine colugos.
table S19. Principal component loadings for X chromosome genetic variants.
database S1. Dermopteran positively selected genes.
database S2. Dermopteran WebGestalt results.
database S3. Ancestral primate positively selected genes.
database S4. Ancestral primate WebGestalt results.
database S5. Museum specimen information.
database S6. Primers used to amplify nuclear DNA capture probes.
database S7. Craniodental morphometric measurements for 125 Sunda colugos.
REFERENCES AND NOTES
- 1.N. T-L. Lim, Colugo: The Flying Lemur of South-East Asia (Draco Publishing, Singapore, 2007), 80 pp. [Google Scholar]
- 2.Lim N. T-L., Giam X., Byrnes G., Clements G. R., Occurrence of the Sunda colugo (Galeopterus variegatus) in the tropical forests of Singapore: A Bayesian approach. Mamm. Biol. 78, 63–67 (2013). [Google Scholar]
- 3.Stafford B. J., Szalay F. S., Craniodental functional morphology and taxonomy of dermopterans. J. Mammal. 81, 360–385 (2000). [Google Scholar]
- 4.K. C. Beard, in Primates and Their Relatives in Phylogenetic Perspective, R. D. E. MacPhee, Ed. (Springer, New York, 1993), pp. 63–90. [Google Scholar]
- 5.Jackson S. M., Thorington R. W. Jr, Gliding mammals: Taxonomy of living and extinct species. Smithson Contrib. Zool. 10.5479/si.00810282.638.1 (2012). [Google Scholar]
- 6.Schmitz J., Ohme M., Suryobroto B., Zischler H., The colugo (Cynocephalus variegatus, Dermoptera): The Primates’ gliding sister? Mol. Biol. Evol. 19, 2308–2312 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Janečka J. E., Miller W., Pringle T. H., Wiens F., Zitzmann A., Helgen K. M., Springer M. S., Murphy W. J., Molecular and genomic data identify the closest living relative of primates. Science 318, 792–794 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Martin R. D., Colugos: Obscure mammals glide into the evolutionary limelight. J. Biol. 7, 13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meredith R. W., Janečka J. E., Gatesy J., Ryder O. A., Fisher C. A., Teeling E. C., Goodbla A., Eizirik E., Simão T. L. L., Stadler T., Rabosky D. L., Honeycutt R. L., Flynn J. J., Ingram C. M., Steiner C., Williams T. L., Robinson T. J., Burk-Herrick A., Westerman M., Ayoub N. A., Springer M. S., Murphy W. J., Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (2011). [DOI] [PubMed] [Google Scholar]
- 10.O’Leary M. A., Bloch J. I., Flynn J. J., Gaudin T. J., Giallombardo A., Giannini N. P., Goldberg S. L., Kraatz B. P., Luo Z.-X., Meng J., Ni X., Novacek M. J., Perini F. A., Randall Z. S., Rougier G. W., Sargis E. J., Silcox M. T., Simmons N. B., Spaulding M., Velazco P. M., Weksler M., Wible J. R., Cirranello A. L., The placental mammal ancestor and the post–K-Pg radiation of placentals. Science 339, 662–667 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Kriegs J. O., Churakov G., Jurka J., Brosius J., Schmitz J., Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 23, 158–161 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lin J., Chen G., Gu L., Shen Y., Zheng M., Zheng W., Hu X., Zhang X., Qiu Y., Liu X., Jiang C., Phylogenetic affinity of tree shrews to Glires is attributed to fast evolution rate. Mol. Phylogenet. Evol. 71, 193–200 (2014). [DOI] [PubMed] [Google Scholar]
- 13.D. E. Wilson, D. M. Reeder, Mammal Species of the World: A Taxonomic and Geographic Reference (Johns Hopkins Univ. Press, Baltimore, MD, 2005). [Google Scholar]
- 14.de Bruyn M., Stelbrink B., Morley R. J., Hall R., Carvalho G. R., Cannon C. H., van den Bergh G., Meijaard E., Metcalfe I., Boitani L., Maiorano L., Shoup R., von Rintelen T., Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Syst. Biol. 63, 879–901 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Janečka J. E., Helgen K. M., Lim N. T-L., Baba M., Izawa M.,Boeadi , Murphy W. J., Evidence for multiple species of Sunda colugo. Curr. Biol. 18, R1001–R1002 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Mason V. C., Li G., Helgen K. M., Murphy W. J., Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Res. 21, 1695–1704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moritz G. L., Lim N. T-L., Neitz M., Peichl L., Dominy N. J., Expression and evolution of short wavelength sensitive opsins in colugos: A nocturnal lineage that informs debate on primate origins. Evol. Biol. 40, 542–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melin A. D., Wells K., Moritz G. L., Kistler L., Orkin J. D., Timm R. M., Bernard H., Lakim M. B., Perry G. H., Kawamura S., Dominy N. J., Euarchontan opsin variation brings new focus to primate origins. Mol. Biol. Evol. 33, 1029–1041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Springer M. S., Meredith R. W., Teeling E. C., Murphy W. J., Technical comment on “The placental mammal ancestor and the post–K-Pg radiation of placentals”. Science 341, 613 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Kuritzin A., Kischka T., Schmitz J., Churakov G., Incomplete lineage sorting and hybridization statistics for retroposon insertion data. PLOS Comput. Biol. 12, e1004812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloch J. I., Silcox M. T., Boyer D. M., Sargis E. J., New Paleocene skeletons and the relationship of plesiadapiforms to crown-clade primates. Proc. Natl. Acad. Sci. U.S.A. 104, 1159–1164 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G., Shi P., Zhu Z., Zhang Y.-P., More functional V1R genes occur in nest-living and nocturnal terricolous mammals. Genome Biol. Evol. 2, 277–283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton R. A., Purvis A., Harvey P. H., Evolutionary radiation of visual and olfactory brain systems in primates, bats and insectivores. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348, 381–392 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Storey E., Genetic cerebellar ataxias. Semin. Neurol. 34, 280–292 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Sussman R. W., Tab Rasmussen D., Raven P. H., Rethinking primate origins again. Am. J. Primatol. 75, 95–106 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Cannon C. H., Morley R. J., Bush A. B. G., The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc. Natl. Acad. Sci. U.S.A. 106, 11188–11193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bird M. I., Taylor D., Hunt C., Palaeoenvironments of insular Southeast Asia during the Last Glacial Period: A savanna corridor in Sundaland? Quat. Sci. Rev. 24, 2228–2242 (2005). [Google Scholar]
- 28.Leonard J. A., den Tex R. J., Hawkins M. T. R., Muñoz-Fuentes V., Thorington R., Maldonado J. E., Phylogeography of vertebrates on the Sunda Shelf: A multi-species comparison. J. Biogeogr. 42, 871–879 (2015). [Google Scholar]
- 29.Sheldon F. H., Lim H. C., Moyle R. G., Return to the Malay Archipelago: The biogeography of Sundaic rainforest birds. J. Ornithol. 156, 99–113 (2015). [Google Scholar]
- 30.Brown R. M., Weghorst J. A., Olson K. V., Duya M. R. M., Barley A. J., Duya M. V., Duya M. V., Shekelle M., Neri-Arboleda I., Esselstyn J. A., Dominy N. J., Ong P. S., Moritz G. L., Luczon A., Diesmos M. L. L., Diesmos A. C., Siler C. D., Conservation genetics of the Philippine tarsier: Cryptic genetic variation restructures conservation priorities for an island archipelago primate. PLOS One 9, e104340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steppan S. J., Zawadzki C., Heaney L. R., Molecular phylogeny of the endemic Philippine rodent Apomys (Muridae) and the dynamics of diversification in an oceanic archipelago. Biol. J. Linn. Soc. 80, 699–715 (2003). [Google Scholar]
- 32.Petit R. J., Excoffier L., Gene flow and species delimitation. Trends Ecol. Evol. 24, 386–393 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Baker R. J., Bradley R. D., Speciation in mammals and the genetic species concept. J. Mammal. 87, 643–662 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moritz C., Defining ‘evolutionary significant units’ for conservation. Trends Ecol. Evol. 9, 373–375 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Wilcove D. S., Giam X., Edwards D. P., Fisher B., Koh L. P., Navjot’s nightmare revisited: Logging, agriculture, and biodiversity in Southeast Asia. Trends Ecol. Evol. 28, 531–540 (2013). [DOI] [PubMed] [Google Scholar]
- 36.R. M. Brown, A. C. Diesmos, in Encyclopedia of Islands, R. Gillespie, D. Clague, Eds. (University of California Press, Berkeley, 2009), pp. 723–732. [Google Scholar]
- 37.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., Tang J., Wu G., Zhang H., Shi Y., Liu Y., Yu C., Wang B., Lu Y., Han C., Cheung D. W., Yiu S.-M., Peng S., Xiaoqian Z., Liu G., Liao X., Li Y., Yang H., Wang J., Lam T.-W., Wang J., SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W., Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Douzery E. J. P., Scornavacca C., Romiguier J., Belkhir K., Galtier N., Delsuc F., Ranwez V., OrthoMaM v8: A database of orthologous exons and coding sequences for comparative genomics in mammals. Mol. Biol. Evol. 31, 1923–1928 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Löytynoja A., Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Talavera G., Castresana J., Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chifman J., Kubatko L., Quartet inference from SNP data under the coalescent model. Bioinformatics 30, 3317–3324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waddell P. J., Kishino H., Ota R., A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 12, 141–154 (2001). [PubMed] [Google Scholar]
- 45.Kriegs J. O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Retroposed elements as archives for the evolutionary history of placental mammals. PLOS Biol. 4, e91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen K., Durand D., Farach-Colton M., NOTUNG: A program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7, 429–447 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Montague M. J., Li G., Gandolfi B., Khan R., Aken B. L., Searle S. M. J., Minx P., Hillier L. W., Koboldt D. C., Davis B. W., Driscoll C. A., Barr C. S., Blackistone K., Quilez J., Lorente-Galdos B., Marques-Bonet T., Alkan C., Thomas G. W. C., Hahn M. W., Menotti-Raymond M., O’Brien S. J., Wilson R. K., Lyons L. A., Murphy W. J., Warren W. C., Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc. Natl. Acad. Sci. U.S.A. 111, 17230–17235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maricic T., Whitten M., Pääbo S., Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLOS One 16, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. G. Del Mastro, M. Lovett, in Gene Isolation and Mapping Protocols (Springer, New York, 1997), pp. 183–199. [Google Scholar]
- 51.Skaletsky H., Kuroda-Kawaguchi T., Minx P. J., Cordum H. S., Hillier L. D., Brown L. G., Repping S., Pyntikova T., Ali J., Bieri T., Chinwalla A., Delehaunty A., Delehaunty K., Du H., Fewell G., Fulton L., Fulton R., Graves T., Hou S.-F., Latrielle P., Leonard S., Mardis E., Maupin R., McPherson J., Miner T., Nash W., Nguyen C., Ozersky P., Pepin K., Rock S., Rohlfing T., Scott K., Schultz B., Strong C., Tin-Wollam A., Yang S.-P., Waterston R. H., Wilson R. K., Rozen S., Page D. C., The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Soh Y. Q. S., Alföldi J., Pyntikova T., Brown L. G., Graves T., Minx P. J., Fulton R. S., Kremitzki C., Koutseva N., Mueller J. L., Rozen S., Hughes J. F., Owens E., Womack J. E., Murphy W. J., Cao Q., de Jong P., Warren W. C., Wilson R. K., Skaletsky H., Page D. C., Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You F. M., Huo N., Gu Y. Q., Luo M.-c., Ma Y., Hane D., Lazo G. R., Dvorak J., Anderson O. D., BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9, 253 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan Y., Huang Z.-Y., Cao C.-C., Chen C.-S., Chen Y.-X., Fan D.-D., He J., Hou H.-L., Hu L., Hu X.-T., Jiang X.-T., Lai R., Lang Y.-S., Liang B., Liao S.-G., Mu D., Ma Y.-Y., Niu Y.-Y., Sun X.-Q., Xia J.-Q., Xiao J., Xiong Z.-Q., Xu L., Yang L., Zhang Y., Zhao W., Zhao X.-D., Zheng Y.-T., Zhou J.-M., Zhu Y.-B., Zhang G.-J., Wang J., Yao Y.-G., Genome of the Chinese tree shrew. Nat. Commun. 4, 1426 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Dabney J., Knapp M., Glocke I., Gansauge M.-T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.-L., Meyer M., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X., Madan A., CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S., Stecher G., Peterson D., Tamura K., MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28, 2685–2686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perelman P., Johnson W. E., Roos C., Seuánez H. N., Horvath J. E., Moreira M. A. M., Kessing B., Pontius J., Roelke M., Rumpler Y., Schneider M. P. C., Silva A., O’Brien S. J., Pecon-Slattery J., A molecular phylogeny of living primates. PLOS Genet. 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li G., Davis B. W., Eizirik E., Murphy W. J., Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 26, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Excoffier L., Lischer H. E. L., Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Librado P., Rozas J., DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Dos Reis M., Zhu T., Yang Z., The impact of the rate prior on Bayesian estimation of divergence times with multiple loci. Syst. Biol. 63, 555–565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.E. Meijaard, Solving mammalian riddles: A reconstruction of the Tertiary and Quaternary distribution of mammals and their palaeoenvironments in island South-East Asia, unpublished thesis, Australian National University, Canberra, Australia (2004). [Google Scholar]
- 68.Haq B. U., Hardenbol J., Vail P. R., Chronology of fluctuating sea levels since the Triassic. Science 235, 1156–1167 (1987). [DOI] [PubMed] [Google Scholar]
- 69.Uba C. E., Strecker M. R., Schmitt A. K., Increased sediment accumulation rates and climatic forcing in the central Andes during the late Miocene. Geology 35, 979–982 (2007). [Google Scholar]
- 70.Kominz M. A., Browning J. V., Miller K. G., Sugarman P. J., Mizintseva S., Scotese C. R., Late Cretaceous to Miocene sea-level estimates from the New Jersey and Delaware coastal plain coreholes: An error analysis. Basin Res. 20, 211–226 (2008). [Google Scholar]
- 71.Yang Z., The BPP program for species tree estimation and species delimitation. Curr. Zool. 61, 854–865 (2015). [Google Scholar]
- 72.Yang Z., Rannala B., Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. U.S.A. 107, 9264–9269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raes N., Cannon C. H., Hijmans R. J., Piessens T., Saw L. G., van Welzen P. C., Slik J. W. F., Historical distribution of Sundaland’s Dipterocarp rainforests at Quaternary glacial maxima. Proc. Natl. Acad. Sci. U.S.A. 111, 16790–16795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lohmann D. J., de Bruyn M., Page T., von Rintelen K., Hall R., Ng P. K. L., Shih H.-T., Carvalho G. R., von Rintelen T., Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 42, 205–226 (2011). [Google Scholar]
- 75.Emerling C. A., Huynh H. T., Nguyen M. A., Meredith R. W., Springer M. S., Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proc. Biol. Sci. 282, 20151817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rose K. D., DeLeon V. B., Missiaen P., Rana R. S., Sahni A., Singh L., Smith T., Early Eocene lagomorph (Mammalia) from Western India and the early diversification of Lagomorpha. Proc. R. Soc. B 275, 1203–1208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kapur V. V., Bajpai S., Oldest South Asian tapiromorph (Perissodactyla, Mammalia) from the Cambay Shale Formation, western India, with comments on its phylogenetic position and biogeographic implications. The Palaeobotanist 64, 95–103 (2015). [Google Scholar]
- 78.Benton M. J., Donoghue P. C. J., Asher R. J., Friedman M., Near T. J., Vinther J., Constraints on the timescale of animal evolutionary history. Palaeontol. Electron. 18, 1–106 (2015). [Google Scholar]
- 79.Chester S. G. B., Bloch J. I., Boyer D. M., Clemens W. A., Oldest known euarchontan tarsals and affinities of Paleocene Purgatorius to Primates. Proc. Natl. Acad. Sci. U.S.A. 112, 1487–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. J. Benton, P. C. J. Donoghue, R. J. Asher, in The Timetree of Life, S. B. Hedges, S. Kumar, Eds. (Springer, New York, 2009), pp. 35–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/8/e1600633/DC1
fig. S1. Maximum likelihood phylogeny of Euarchontoglires based on an alignment of 631 orthologous protein-coding sequences.
fig. S2. Insertion-deletion evidence for colugo phylogenetic relationships based on transposons.
fig. S3. Description of the transposon screening strategy.
fig. S4. Phylogenetically informative protein-coding gene indels supporting Primatomorpha (P1 to P15), Sundatheria (S1 to S5), and Primates + Scandentia (PS1 to PS5).
fig. S5. Maximum likelihood timetree showing seven nodes with external fossil calibrations.
fig. S6. Maximum likelihood timetree showing seven nodes with external fossil calibrations, with euarchontan monophyly enforced.
fig. S7. Maximum likelihood mtDNA tree based on a matrix of 53 colugos with ≥90% mitogenome coverage.
fig. S8. Maximum likelihood mtDNA tree based on a matrix of 65 colugos with ≥30% mitogenome coverage.
fig. S9. Maximum likelihood tree based on a combined nuclear (biparental + Y) matrix of 17 taxa.
fig. S10. Maximum likelihood tree based on biparental data set (18 taxa).
fig. S11. Maximum likelihood Y chromosome tree (depth 2) (15 taxa).
fig. S12. Maximum likelihood Y chromosome tree (depth 3) (14 taxa).
fig. S13. Biplot of PC1 and PC2 from PCA of colugo X chromosome variants.
fig. S14. Biplot of PC2 and PC3 from PCA of colugo X chromosome variants.
fig. S15. Biplot of PC3 and PC4 from PCA of colugo X chromosome variants.
fig. S16. Biplot of PC4 and PC5 from PCA of colugo X chromosome variants.
fig. S17. Biplot of PC5 and PC6 from PCA of colugo X chromosome variants.
fig. S18. Biplot of morphometric PCA with males and females, and no dwarf specimens removed.
fig. S19. Biplot of colugo morphometric PCA, with male, female, and dwarf specimens, where all measurements are normalized by body size.
table S1. Mammals used in phylogenetic analyses.
table S2. Presence/absence of phylogenetically informative retroposon markers and species distribution.
table S3. Location of diagnostic retroposon markers in the human genome.
table S4. χ2 calculation for phylogenetically informative indels.
table S5. Colugo and select mammal V1R and OR gene annotation summaries.
table S6. Nuclear capture efficiency.
table S7. Adapter blocking oligos.
table S8. Off-target mtDNA assembly statistics.
table S9. External fossil calibrations used to calculate divergence time between the two colugo genera.
table S10. Principal component loadings for morphometric PCA using males and females with no dwarf individuals.
table S11. Principal component loadings for morphometric PCA using males, females, and dwarf individuals, and all measurements were normalized for body size.
table S12. mtDNA maximum composite likelihood + Γ distance matrix calculated between seven Sunda colugo groups and five Philippine groups.
table S13. X chromosome maximum composite likelihood + Γ distance matrix calculated between seven Sunda colugo groups and three Philippine groups.
table S14. Between-group mtDNA composite likelihood + Γ distance matrix for Sunda colugos from Thailand, Peninsular Malaysia, and Sumatra, and their satellite islands.
table S15. Within-group mtDNA maximum composite likelihood + Γ distance matrix.
table S16. Between-group mtDNA composite likelihood + Γ distance matrix for 10 Sunda colugo groups and 5 Philippine groups.
table S17. BPP species estimation results for Sunda colugos.
table S18. BPP species estimation results for Philippine colugos.
table S19. Principal component loadings for X chromosome genetic variants.
database S1. Dermopteran positively selected genes.
database S2. Dermopteran WebGestalt results.
database S3. Ancestral primate positively selected genes.
database S4. Ancestral primate WebGestalt results.
database S5. Museum specimen information.
database S6. Primers used to amplify nuclear DNA capture probes.
database S7. Craniodental morphometric measurements for 125 Sunda colugos.


