Abstract
Mutualism is ubiquitous in nature and plays an integral role in most communities. To predict the eco-evolutionary dynamics of mutualism it is critical to extend classic pairwise analysis to include additional species. We investigated the effect of adding a third species to a pair-wise mutualism in a spatially structured environment. We tested the hypotheses that selection for costly excretions in a focal population i) decreases when an exploiter is added ii) increases when a third mutualist is added relative to the pair-wise scenario. We assayed the selection acting on Salmonella enterica when it exchanges methionine for carbon in an obligate mutualism with an auxotrophic Escherichia coli. A third bacterium, Methylobacterium extorquens, was then added and acted either as an exploiter of the carbon or third obligate mutualist depending on the nitrogen source. In the tri-partite mutualism M. extorquens provided nitrogen to the other species. Contrary to our expectations, adding an exploiter increased selection for methionine excretion in S. enterica. Conversely, selection for cooperation was lower in the tri-partite mutualism relative to the pair-wise system. Genome-scale metabolic models helped identify the mechanisms underlying these changes in selection. Our results highlight the utility of connecting metabolic mechanisms and eco-evolutionary dynamics.
Keywords: Cooperation, exploitation, mutualism, metabolic modeling, E. coli, S. enterica, M. extorquens
Introduction
Mutualisms play an important role in many fundamental biological processes from energy generation to reproduction, and involve organisms spread across the tree of life. These interactions are generally studied as pair-wise associations, although it is broadly appreciated that additional species interactions can have significant ecological and evolutionary impacts on the strength and stability of mutualisms (Stanton 2003, Jones et al 2012, Chamberlain et al 2014, Afkhami 2014). As we attempt to build a general framework for predicting the eco-evolutionary dynamics of mutualism it is important to know how and why selection in a pair-wise system is influenced by third-party interactions that range from exploitation to mutualism.
Ultimately, the evolution of mutualism is driven by the costs and benefits associated with aiding another species. If the benefits of mutualism are shared equally by all members of a population, then cheaters that do not pay the costs of mutualisms will be favored (Williams 1966). To maintain mutualism, genotypes that express cooperative traits must receive a greater share of the benefits (Trivers 1971, Axelrod and Hamilton 1981, Queller 1985). The costs and benefits of mutualism are known to be context-dependent, however, it is often difficult to predict how they will respond to specific changes in the environment. Thrall et al (2007) suggest that increasing diversity will typically correlate with less beneficial interactions, though this may lead to selection for specialization ultimately increasing investment in pair-wise mutualisms. Similarly, it is often asserted that reduced nutrient availability will increase the benefit of mutualism (Bronstein 1994, Thrall et al 2007, Pringle et al 2013), however a variety of outcomes have been observed (Chamberlain et al 2014). The variance in these outcomes is influenced by the metabolic mechanisms that relate nutrient utilization to species interactions. Across habitats nutrient availability can vary both temporally and spatially, giving rise to a mosaic of selection strengths (Thompson 2005). A metabolically explicit framework that incorporates spatial structure will aid attempts to understand and predict the eco-evolutionary dynamics of species interactions.
A third species is likely to affect selection for mutualism differently based on how the additional player interacts with the species in a pair-wise association. For clarity, it is useful to focus on one species in a pair-wise mutualism, deemed the focal species. The second member in the initial pair-wise association is referred to as the partner. The third species can then be classified as an exploiter or a mutualist based upon how it interacts with the focal species. Exploiters that acquire benefits from a mutualism but provide nothing in return are generally expected to destabilize mutualisms (Afkhami 2014). Indeed, one of the fundamental questions about the evolution of mutualism is how mutualisms can persist in the presence of exploiting competitors. Models suggest that adding an exploiter will generally select for a focal species to reduce cooperation with its partner (Ferriere et al 2007, Jones 2009). Conversely, adding a third mutualistic species has been suggested to increase selection for mutualism, as it makes partner choice more efficient (Jones et al 2012, Foster and Kokko 2006). Alternatively, if a third species provides services that do not overlap with pre-existing partners, addition of the third species should increase the fitness of a focal mutualist (Afkhami et al 2014). Selection for mutualism would then increase if cooperation by the focal species generated both direct and indirect benefits. This previous research leads to the hypotheses that i) adding an exploiter to a pair-wise mutualism should reduce selection for cooperation, while ii) adding a third mutualist should increase selection for cooperation.
The effect of additional species on pair-wise interactions is likely to be influenced by spatial structure (Thompson 2005). Growth in a spatially structured environment can favor mutualism by concentrating the benefits of cooperation in the vicinity of cooperators (Maynard Smith 1964). The ability of spatial structure to select for cooperation has been demonstrated both with models (Wilson 1975, Nowak and May 1992, Frank 1994, Doebli and Knowlton 1998, Yamamura et al 2004, Bull and Harcombe 2009, Mitri et al 2011, Ezoe and Ikegawa 2013, Momeni et al 2013), and empirically (Griffin et al 2004, Maclean and Gudelj 2006, Chuang et al 2009, Harcombe et al 2010, Momeni et al 2013). However, less is known about how additional species influence the selection for mutualism in spatially structured environments. Mitri et al. (2011) assert that adding species to a pair-wise interaction can segregate mutualists from their partners, thereby reducing selection for mutualism in structured environments. Their simulations suggests that spatial structure may decrease the selection for mutualism as systems become more diverse.
Microbial systems are ideal for investigating the evolution of mutualism. Cooperation is common in microbial communities (Zelezniak et al 2015), and hence plays a role in processes from human health to global nutrient cycling. Additionally, experiments with bacteria can be tightly controlled to test the mechanistic basis of cooperation (Shou et al 2007, Hillesland and Stahl 2010, Celliker and Gore 2012, Lawrence et al 2012, Muller et al 2013, Pande et al 2014). For example, Harcombe (2010) showed that spatial structure and byproduct utilization were essential for the evolution of a novel mutualism between Escherichia coli and Salmonella enterica. On agar surfaces S. enterica evolved that excreted a costly amino acid needed by E. coli, however, in the absence of spatial structure or byproduct utilization S. enterica that did not excrete the amino acid outcompeted the cooperative genotype. Microbial systems allow rigorous control to investigate fundamental processes, as well as being relevant for understanding the dynamics in microbial communities.
Novel computational approaches based upon flux balance analysis (FBA) have been developed for microbes that make it possible to explicitly investigate how community dynamics emerge from organismal metabolism (Harcombe et al 2014). In essence, FBA uses evolutionary optimality to predict how an organism will mediate the tradeoff inherent at every branch point in its metabolic network. A matrix of stoichiometrically balanced equations can be generated to describe all of the metabolic reactions in a cell, thereby defining feasible patterns of steady state metabolite flow through a population of microbes (Orth et al 2010). Constraints can be imposed for known directionality and upper limits of flux through individual reactions. Optimization calculations can then identify the flow through the network that maximizes growth or other optimality criteria (Orth et al 2010). FBA predicts metabolite uptake, pathway activity, excretion profiles and has had noted success at predicting microbial growth in different environments (Varma & Palsson 1994, Lewis et al 2010, Harcombe et al 2013). Further, a method has been developed to use FBA to predict interactions between species in spatially structured environments (Harcombe et al 2014). Models of different genotypes can be distributed in a lattice and then FBA and diffusion calculations can be integrated over short time intervals to simulate how intra-cellular metabolic mechanisms generate nutrient gradients that ultimately drive species interactions. Saturation constants (Km) determine how local metabolite concentrations influence the rate of each species’ nutrient uptake reactions. Generating hypotheses for experiments with complex interactions is inherently difficult, and these novel computational approaches enable not only consideration of underlying mechanisms a posteriori, but also experimental predictions a priori.
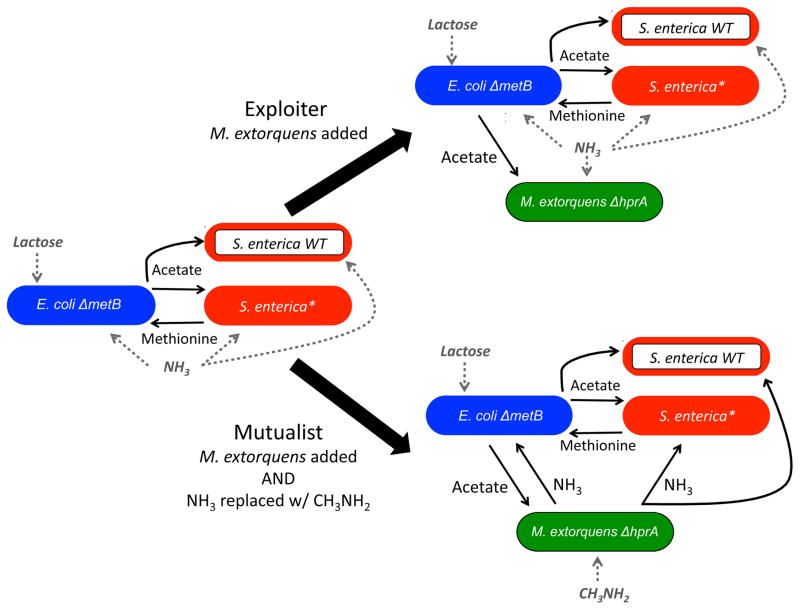
Here, we investigated how selection for mutualism changes as one moves from a 2- to a 3-species system in a spatially structured environment. The starting pair-wise mutualism was the system introduced above in which S. enterica exchanges costly methionine for carbon byproducts from E. coli (Fig 1). S. enterica is the focal species, and the selection for mutualism was assayed by tracking changes in the frequency of methionine-excreting S. enterica relative to a non-excreting genotype. Three species communities were created by adding a mutant Methylobacterium extorquens strain which requires carbon from E. coli. When this M. extorquens was added to the pair-wise system with no change in media, the strain acted as an exploiter competing with S. enterica for the carbon byproducts excreted by E. coli. In contrast, if the nitrogen source was changed from ammonia to methylamine, the M. extorquens acted as a third obligate mutualist, providing essential nitrogen to the other two species in return for carbon. We tracked the frequency of genotypes in the S. enterica population in different biotic contexts in the laboratory to test the hypotheses that adding an exploiter reduces selection for cooperation while adding a third mutualist increases selection for cooperation. We subsequently used genome-scale metabolic models to investigate the mechanisms underlying the patterns observed. Finally, we returned to the lab to test computationally-derived explanations. We demonstrate that adding biotic complexity can have counterintuitive impacts on the selection for mutualism, but these outcomes can be predicted from genome-scale metabolic mechanisms.
Figure 1.
Interactions in the 2 and 3-species systems. S. enterica is the focal strain and the selection for mutualism in each system is determined by tracking the relative frequency of methionine-excreting cooperators (solid red) to non-excreting genotypes (red outline). All S. enterica genotypes require acetate and ammonia. E. coli (blue) consumes lactose, excretes acetate and requires methionine and ammonia. M. extorquens (green) requires acetate. If ammonia is provided in the media then M. extorquens acts as an exploiter with S. enterica for acetate. In contrast, if methylamine is the only nitrogen source then M. extorquens acts as a mutualist providing the other two species with ammonia.
Material and Methods
Bacterial strains and culture conditions
Our experimental communities consisted of strains of E. coli K-12, S. enterica LT2 and M. extorquens AM1. The E. coli was ΔmetB from the Keio collection (CGSC# 10824, (Baba et al., 2006)) with the lac operon replaced via conjugation with E. coli HfrH PO1 relA1 thi-1 spoT supQ80 nad57::Tn10. Two genotypes of S. enterica were used. The first was a wildtype S. enterica LT2. The second was a methionine-excreting mutant that was created through a combination of engineering and selection as described in Harcombe (2010) and Chubiz et al (2014). For the liquid culture assays YFP and CFP were inserted into the S. enterica chromosome for cooperating and non-excreting genotypes, respectively. In repeated tests, swapping fluorescent markers led to no change in growth. The ΔhprA M. extorquens lacking hydroxypyruvate reductase was previously described (Marx 2008). This mutation blocks carbon assimilation from single-carbon compounds (like methylamine) and thus forces M. extorquens to rely on multi-carbon compounds from E. coli.
The community was grown on two types of media. The two-species community was grown on plates of lactose Hypho minimal media (2.92 mM lactose, 7.26 mM K2HPO4, 9.38 mM NaH2PO4, 1.89 mM (NH4)2SO4, 0.41 mM MgSO4, 0.6 μM ZnSO4, 9.98 μM CaCl2, 0.5 μM MnCl2, 1 μM (NH4)6Mo7, 0.5 μM CUSO4, 1 μM CoCl2, 0.169 μM Na2WO4, 8.88 μM FeSO4 – Based on Delaney et al 2013a). This media was also used for experiments in which M. extorquens acted as an exploiter. To force M. extorquens to act as a third mutualist we used methylamine-lactose minimal medium plates ((NH4)2SO4 replaced with 1.89 mM Na2SO4, and 1.16 mM methylamine•HCl added). The same media compositions were also used for liquid culture assays.
Distinguishing S. enterica genotypes
A lawn of the auxotrophic E. coli was spread on lactose minimal media plates with an indicator that turned blue when cells grew (X-Gal; 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside). Colonies of S. enterica were then stabbed onto the lawn in a grid and plates were incubated at 37 °C. Growth was only observed in grid boxes with cooperative methionine excreting S. enterica, as neither the E. coli nor the S. enterica could grow in the absence of mutualistic interactions. In liquid assays S. enterica genotypes were distinguished by fluorescence using a blue light transilluminator (Maestrogen, Las Vegas).
Investigating the spread of S. enterica cooperators
S. enterica populations were started with a ratio of 30:70 by CFU of methionine-excreting cooperators to non-excreting genotypes. 5 μL of the S. enterica mixture was then combined with 5 μL of each other species in an experimental treatment and plated on the corresponding minimal media plate. Three replicates were set up for each treatment. After 96 hrs incubation at 30 °C, 720 μL of liquid minimal media was used to scrub the biomass from the surface of the plate. 5 μL of the resultant suspension was then transferred to a fresh plate, spread and incubated for 96 hrs. A total of four transfers were completed and at each transfer CFU were counted and the ratio of S. enterica genotypes in each replicate was determined.
To test if the results of adding an exploiter were affected by spatial structure, experiments were carried out in liquid cultures. Strains were mixed in 5 mL volumes of the appropriate minimal media at a ratio of 30:70 as before. Three replicates were carried out for both pair-wise communities and 3-species communities in which the M. extorquens acted as an exploiter. Cultures were grown for 96 hrs at 30 °C with vigorous shaking. At the end of growth, each culture was plated on LB plates with X-Gal to count colonies. S. enterica genotypes were distinguished by fluorescence using a blue light transilluminator (Maestrogen, Las Vegas).
The effect of nitrogen concentration on cost of cooperation was assayed by measuring the growth rate of S. enterica isolates. Media was made that contained NH4 at concentrations of 0.56, 1.12, 1.68, 2.24 and 2.80 mM. Four replicates for each genotype were assayed at each nitrogen concentration. Assays were carried out in 48-well plates that were shaking at 30 °C. Optical densities were obtained every 30 min to 1 h on a Wallac Victor 2 plate reader (Perkin-Elmer, Waltham) until cultures reached saturation, using a previously described automated measurement system (Delaney et al 2013b). Growth rates were determined by fitting an exponential growth model using custom analysis software, Curve Fitter (Delaney et al., 2013a; http://www.evolvedmicrobe.com/Software.html).
Genome-scale metabolic modeling
To determine the metabolic mechanism underlying observed evolutionary patterns we used constraint-based metabolic modeling. Genome-scale metabolic networks were obtained for E. coli (iJO_1366) (Orth et al., 2011), S. enterica (iRR_1083) (Raghunathan et al., 2009) and M. extorquens (Klitgord and Segrè, 2010). The models were modified to incorporate known genetic constraints in each species as described in Harcombe et al (2014). Briefly the gene knockouts in E. coli and M. extorquens were modeled by constraining flux through the associated enzymatic steps to zero. Methionine excretion in the mutualist S. enterica was modeled by connecting excretion of the amino acid to the biomass equation that serves as the objective function. FBA models were converted to COMETS format with the script provided on the COMETS website (http://www.bu.edu/segrelab/comets/). Michaelis-Menten parameters are used to determine how local nutrient concentrations influence the maximum rate of uptake (Vmax) of metabolites by each unit of biomass. The maximum nutrient uptake (Vmax) was set to 10 mmol/g/hr for every transport reaction in each species. Similarly, the saturation constant (Km) was initially set to 10 μM for each reaction as done previously (Harcombe et al 2014, Chubiz et al 2015). The Km of ammonia in the cooperative S. enterica model was subsequently doubled to investigate the selective affect of reducing the ability of cooperators to compete for low concentrations of ammonia.
COMETS v. 2.2.11 was used to simulate the metabolic interactions and growth of each species in a spatially structured community. Sixty units of biomass of each relevant strain were randomly distributed on a 2-dimensional lattice that was 100×100 boxes in size. Each unit consisted of 2.5×10−7 g of biomass and biomass overlap was allowed. The lattice was also seeded with the same nutrients as in experimental media, and oxygen was set to 1×10−5 mM. The relative abundance of strains was compared after the system reached carrying capacity. To control for effects of orientation four different random distributions of E. coli and S. enterica (a population containing both cooperating and non-excreting genotypes) were generated, and the arrangement of these bacteria was maintained in 3-species simulations. The results in a 3-species simulation could thereby be compared to those from a 2-species simulation with the same orientation of E. coli and S. enterica. The randomized placement of biomass in each of the four replicates generated variance between replicates; within a replicate the randomized order of biomass update led to a variance in outcomes of <0.1% so simulations were each run once. Each box in the lattice was 0.5 mm in diameter and step size was set to 0.1 hours. Metabolite diffusion was set to 5×10−6 cm2/s and biomass diffusion was set to 3×10−10 cm2/s. Biomass was set as the objective function for all models, and GNU Linear Programming Kit (GLPK) was used as the optimizer. In addition to tracking the biomass, the concentration of metabolites was monitored across the lattice. Total concentration of relevant nutrients was obtained by adding the concentrations in each box on the lattice, while patchiness of resource distribution was determined by taking the coefficient of variation of concentrations across the lattice. Metabolite calculations were done after 40 simulated hours of growth when communities were in mid-growth phase.
Results
Adding biotic complexity alters the selection on cooperation
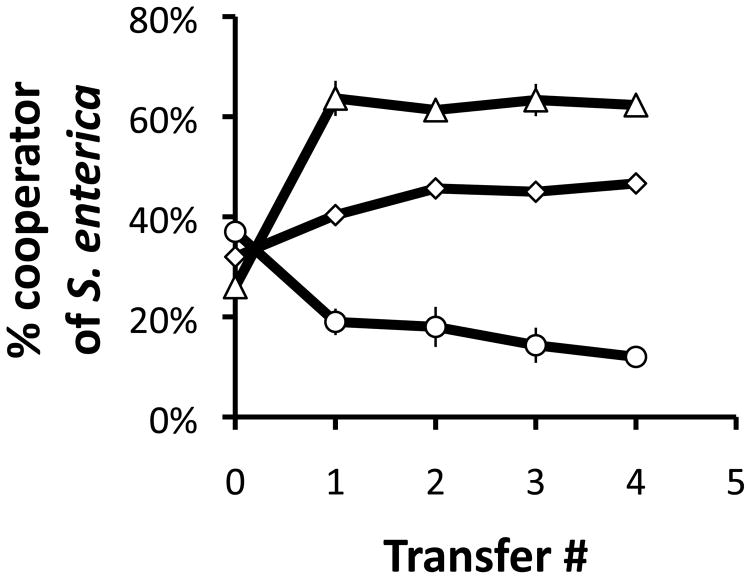
In the 2-species community there was frequency-dependent selection on cooperation. In laboratory experiments on plates, cooperators reached an equilibrium frequency of 46±1 % of the S. enterica population (mean ± standard deviation - Fig 2). This intermediate frequency of cooperators was reached by transfer 2 and then remained constant through transfer 4. The S. enterica population had a density of 2.63×0.27×108 cfu/ml at the end of the experiment.
Figure 2.
Change in percentage of cooperators in the S. enterica population over time in the laboratory. In the 2-species system (diamonds) cooperators in the S. enterica population stabilized at an intermediate frequency. In the 3-species system with M. extorquens as an exploiter (triangles) cooperators reached a higher equilibrium frequency. When M. extorquens acts as mutualist (circles) cooperators reached a lower equilibrium frequency. Three replicates for each treatment were transferred to new plates every 96 hours. Error bars represent standard errors; some are smaller than the symbols.
Adding a third player that functioned as an exploiter increased the equilibrium frequency of cooperators in the S. enterica population. In the same media as used in the 2-species scenario, M. extorquens interacts with S. enterica through competition for the acetate that E. coli excretes. Under these conditions the equilibrium frequency of cooperators increased to 62±2 % of the S. enterica population (Fig 2, 2-tailed T-test P=2×10−4). As before the frequency of cooperators was stable after transfer three. The final S. enterica population size (3.97±0.60×108 cfu/ml) was not significantly different than in the 2-species community (2-tailed T-test, P>0.05).
When the same third player functioned as a mutualist, its addition reduced the equilibrium frequency of cooperators in the S. enterica population. In media with methylamine (rather than ammonia) as the sole nitrogen source, M. extorquens acted as a mutualist, excreting ammonia upon which E. coli and S. enterica relied. In this context the mean frequency of cooperators stabilized at 12±2 % of the S. enterica population (Fig 2) with no significant changes in frequency after transfer three. The mean frequency of cooperators was significantly lower (2-tailed T-test, P=8×10−6) than in the 2-species case. Again adding a third species had no significant effect on the S. enterica population size (1.59±0.28×108 cfu/ml, 2-tailed T-test, P>0.05).
Adding an exploiter changes the net benefit of cooperation
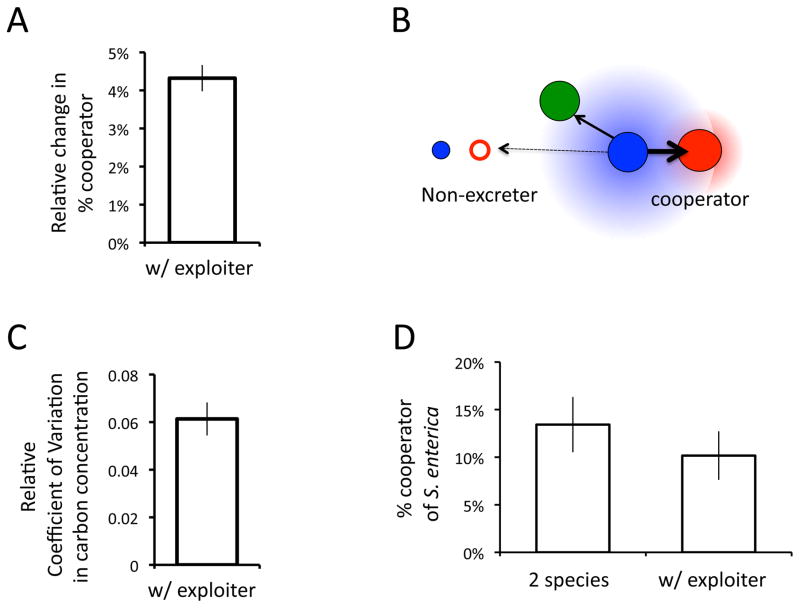
We used constraint-based metabolic modeling to understand the mechanisms that caused cooperator frequency to change when an exploiter was added to the pair-wise mutualism. A genome-scale model was made that incorporated known mutations for each of the four strains (cooperative S. enterica, non-excreting S. enterica, E. coli, and M. extorquens). COMETS was then used to simulate the metabolic interactions and growth of each species in a spatially structured community. The relative frequency of S. enterica genotypes in each 3-species simulation was standardized to the 2-species simulation with matched arrangements of E. coli and S. enterica biomass. In all cases, adding M. extorquens as an exploiter increased the frequency of cooperators in the S. enterica population. The average increase of 4.3±0.3% was significant (Fig 3A, 2-tailed T-test with unequal variance P=0.0002). While these results do not quantitatively match those observed in laboratory experiments, they do suggest that metabolic mechanisms are sufficient to increase selection for cooperation when an exploiter is added to a pair-wise mutualism.
Figure 3.
Effects of M. extorquens acting as an exploiter. A) The average change in percentage of cooperators in 3-species simulations relative to paired 2-species simulations after growing to the carrying capacity. B) Hypothesized mechanism by which M. extorquens increases the benefit of cooperation. Cooperative S. enterica colonies (solid red) secrete methionine (diffuse red) in the environment. Access to methionine allows E. coli (solid blue) to grow and excrete carbon byproducts (diffuse blue). M. extorquens (green) consume carbon excretions reducing the relative amount of byproducts (arrows) reaching non-excreting S. enterica (open red). C) The average change in the coefficient of variation for concentration of carbon byproducts in the presence of the exploiter relative to the pair-wise scenario. Coefficient of variation was calculated between boxes on a lattice after 40 simulated hours of growth. D) Laboratory data on the percentage of cooperators in the S. enterica population in 2 and 3-species communities after growth in shaking flasks for 96 hours. All error bars represent standard error.
We hypothesized that M. extorquens increased the frequency of cooperative S. enterica by increasing the patchiness of rewards for cooperating. If carbon byproducts from E. coli become more localized around cooperators, the cooperators should have a competitive advantage over carbon-limited non-excreters (Fig 3B). We calculated the coefficient of variation in carbon byproduct concentration in 2- and 3-species simulations at mid-growth phase (i.e. 40 hours). Consistent with our hypothesis, we found that the presence of an exploiter increased the coefficient of variation in carbon byproduct concentration across the environment by an average of 6.13±.07% (Fig 3C, 2-tailed T-test with unequal variance, P=0.004).
Finally, we experimentally tested the mechanism underlying the change in selection for mutualism in the presence of an exploiter. By growing the bacteria in liquid cultures rather than plates we created an environment in which chemicals rapidly diffuse and patches of resources cannot build up. In this environment we found that after one growth phase adding an exploiter had no significant effect on cooperator frequency (Fig 3D, 2-tailed T-test, P=0.45), though cooperator frequency was lower than on plates, consistent with previous results (Harcombe 2010). Growth in liquid may change more than the diffusion of metabolites however, laboratory results further support that spatial structure is critical for the observed increase in cooperation in the face of exploitation.
Adding a third mutualist changes the cost of cooperation
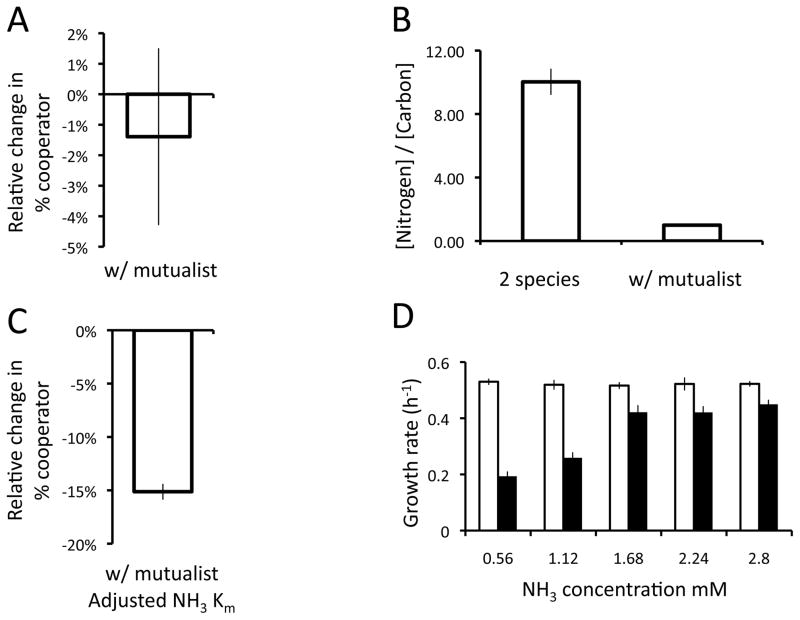
We also used constraint-based metabolic modeling to understand the mechanisms that caused cooperator frequency to decrease when M. extorquens acted as a mutualist. COMETS simulations found no change in selection for S. enterica cooperators as the system was changed from a pair-wise to a tri-partite mutualism (Fig 4A, 2-tailed T-test with unequal variance, P=0.70). However, we found that the ratio of available nitrogen to carbon in the environment was dramatically different in the two scenarios (Fig 4B, 2-tailed T-test, P=6.62×10−5). Ammonia was more limiting relative to carbon in the tripartite mutualism, leading us to suspect that nitrogen limitation might alter the competitive ability of cooperators. Specifically, we hypothesized that methionine secretion might alter the saturation constant (Km) of nitrogen uptake, as this would reduce the fitness of cooperators at low nitrogen concentrations. Such an effect could not have been predicted a priori by our model, as saturation constants are set by the user as a defined parameter. S. enterica contains multiple nitrogen assimilation pathways that vary by orders of magnitude in Km as discussed further below. To test the effect of nitrogen uptake on selection for cooperation, we conservatively doubled the Km for ammonia uptake in the cooperator model. This increased the amount of nitrogen necessary to reach the maximum uptake rate in cooperators (i.e. reduced the uptake rates at low concentrations). Consistent with our hypothesis, we found that if the Km was increased in the cooperator, then simulations predicted a significant decrease in cooperators when M. extorquens was added as a mutualist (Fig 4C – 2-tailed, T-test with unequal variance P=3.5×10−4).
Figure 4.
Effects of M. extorquens acting as a mutualist. A) The average change in percentage of cooperators in the S. enterica population in 3-species simulations relative to paired 2-species simulations after growing to the carrying capacity. B) The ratio of nitrogen to carbon byproducts in 2 and 3-species simulations after 80 simulated hours of growth. C) The average change in percentage of cooperators with adjusted nitrogen uptake in the S. enterica population in 3-species simulations relative to paired 2-species simulations after growing to the carrying capacity. D) Laboratory data on the growth rate of cooperator S. enterica (black bars) and non-excreting S. enterica (open bars) in monocultures in 48-well plates with glucose as the carbon source and variable concentrations of ammonia. All error bars represent standard error.
Based on simulation results, we hypothesized that nitrogen limitation was altering the cost of methionine excretion in the tripartite mutualism. As a test, we grew cooperative and non-excreting S. enterica alone on glucose with different nitrogen concentrations (0.56–2.8 mM NH4). Isolate growth rates were measured for five replicates of each genotype at each concentration. Consistent with model predictions, the growth rate of cooperators relative to non-excreters decreased substantially as nitrogen was reduced (Fig 4D). In this scenario, differences in growth are driven by the cost of methionine excretion, as the benefits of cooperation (both carbon and nitrogen) are the same for excreting and non-excreting genotypes. These results further support the assertion that adding M. extorquens as a third mutualist increased the cost of cooperation in S. enterica.
Discussion
It has long been appreciated that mutualisms happen not in isolation but rather in a “tangled bank” of species interactions. Here we investigated how adding a third species to a two species mutualism influenced the selection for cooperation in a spatially structured environment. We initially hypothesized that adding an exploiter to the system would decrease the frequency of methionine-excreting genotypes in the S. enterica population, while adding a third mutualist would increase the relative frequency of excreters. Contrary to these expectations, selection for mutualism increased when an exploiter was added, and decreased when a third mutualist was added. Genome-scale metabolic modeling suggested that the reason for these changes in selection had to do with how the third player influenced the costs and benefits associated with mutualism.
It is well established that mutualisms are context-dependent, but it has been challenging to predict exactly how costs and benefits will be influenced by the abiotic and biotic environment (Bronstein 1994, Chamberlain et al 2014). For example, some have suggested that low nutrient environments will select for greater mutualism (Thrall et al 2007). This is based on the assumption that as nutrients become scarce it should increase the relative advantage of obtaining nutrients from other species. However, low quality environments are also likely to change the cost of cooperation, and hence it is not clear what should be expected (Denison 2014). Many mutualisms involve nutrient provisioning (Sachs et al 2004), and in these cases costs arise from underlying metabolic tradeoffs. Greater incorporation of metabolic mechanisms into studies of mutualisms will improve our ability to predict the costs and benefits of cooperation in diverse environments. Below, we first discuss how metabolism influenced the effect of adding a third mutualist and then transition to discussing the effect of adding an exploiter.
Adding a third mutualist decreased selection for mutualism in our focal species by altering the cost of cooperation. We hypothesized that the opposite would be observed as increasing the size of the mutualistic network was expected to increase the benefits of cooperating. Specifically, it was thought that adding M. extorquens to the network would cause methionine excretion in S. enterica to generate indirect benefits of nitrogen in addition to the direct carbon benefits. Instead we found that cooperation became more costly. Importantly, this outcome was tied to the changes in nitrogen source that we used to get M. extorquens to act as a mutualist. This alteration of the environment changed the limiting nutrient in our system. Our results should not be extrapolated to all cases of adding a third mutualist to a pairwise system, but rather highlight the extent to which selection for mutualism is context-dependent. The results also provide another clear example that decreasing environmental quality (in this case reducing available nitrogen) does not universally select for more mutualism, but rather can increase the cost of mutualistic behavior.
Metabolic models provided insight into the mechanisms underlying the effect of adding a third mutualist. First, the metabolic modeling helped identify that the limiting nutrient had changed from carbon to nitrogen. This outcome was not obvious a priori as it depended on the relative metabolic rates of E. coli and M. extorquens. The realization that the limiting nutrient had changed led to the testable hypothesis that costs of methionine excretion would be higher as nitrogen became limiting. Further, the metabolic modeling helped highlight a likely molecular basis for the changing cost of methionine excretion in S. enterica. Simulations demonstrated that altering the saturation constant (Km) of nitrogen uptake caused methionine excreting S. enterica to increase in frequency in the tri-partite mutualism. S. enterica has two pathways for the assimilation of nitrogen (Gottschalk 1986). At high nitrogen concentrations, glutamate dehydrogenase catalyzes ammonia assimilation. However, this enzyme has a Km of ~10−1 M, and hence is ineffective when nitrogen is rare in the environment. At low concentrations of nitrogen, S. enterica switches to using a combination of glutamine synthetase and glutamine:2-oxoglutarate amidotransferase (i.e. the GS-GOGAT system). This pathway requires ATP but has a Km of approximately 10−4 M. If the mutations that alter the regulation of methionine biosynthesis also influence the ability of cells to switch between nitrogen assimilation pathways, then cooperators would be less effective than non-excreting genotypes at competing for low levels of ammonia. Our computational and empirical results are consistent with this supposition, but future work will be necessary to test the mechanism that drove observed change in nitrogen assimilation in our methionine-excreting S. enterica.
We were also surprised that adding an exploiter increased selection for cooperation in S. enterica. We hypothesized that adding an exploiter would have the opposite effect as exploiters are typically expected to destabilize mutualisms. Indeed, Ferriere et al (2007) mathematically demonstrated that exploiters reduced selection for investment in cooperation. In their model, the exploiters change the abundance of resources flowing to the focal mutualist population but do not change the relative payoff received by cooperative genotypes. In contrast, in our system adding exploiters increased the patchiness of the environment and hence the correlation between costs and benefits increased. The exploiter consumed acetate, a public good that diffuses away from patches of growing cooperators. Reducing the amount of acetate between patches increased the variance in concentration of carbon byproducts across the landscape. By disproportionately decreasing the resources available to non-excreters, the benefits of cheating were reduced, and higher levels of cooperation were favored. This result is consistent with the observation by Momeni et al (2013) that selection for cooperation is enhanced if mutualists excrete less of a public good.
Exploiters increasing selection for cooperation has also been observed in other systems. Celiker and Gore (2012) showed that adding bacterial exploiters increased selection for public good production in yeast. They nicely demonstrate that the increased intra-specific cooperation is driven by density-dependent dynamics. Several groups found that exploiters increased selection for mutualism by enhancing the efficacy of partner choice (Foster and Kokko 2006, Jones et al 2012). These results are each different than those presented here as the density of the S. enterica population did not change and there is no mechanism of partner choice in our system. The mechanisms underlying our observed effect of adding exploiters are most similar to those shown by Mitri et al (2011). They modeled the growth of bacteria in biofilms and demonstrated that heterospecific exploiters can insulate patches of cooperators, blocking the diffusion of public goods to cheating genotypes. Interestingly, Mitri et al (2011) argue that social insulation is unlikely to function in mutualisms, because a species that “insulates” cooperators from cheaters would also “insulate” cooperators from their mutualistic partner. However, their system had weaker selection and less capacity for between-patch competition. Strong selection in spatially structured habitats is expected to often lead to negative-frequency dependent selection for cooperation in microbial systems (Levin 1988, Ross-Gillespie et al 2007).
Genome-scale metabolic models are useful for understanding how different abiotic and biotic environments influence selection for mutualism. By incorporating genome-scale models of metabolic tradeoffs and demands, our models were able to identify that the effect of adding a mutualist was driven by changes in the limiting nutrient. Specifically, modeling suggested that nitrogen byproducts from M. extorquens were more limiting than carbon byproducts from E. coli, and the computation identified a likely molecular mechanism mediating this cost in S. enterica. Further, by incorporating diffusion calculations, COMETS was able to identify how a third species influenced the nutrient gradients in a spatially structured environment. COMETS provided quantitative predictions about ecological and evolutionary dynamics based on systems-level, intracellular metabolic mechanisms. It should be noted that COMETS under-predicted the size of effects observed in the lab, in part because of computational limitations on the scale of simulations. The evolution of mutualism in this system depends on a heterogeneous distribution of cells into patches, and differential growth of patches based on the frequency of cooperators in them. With 105 cells of each species spread across a plate, many patches will form, and selection on these patches will be efficient. In contrast in a 100×100 lattice the number of distinct patches is inherently limited, thereby diminishing the strength of selection. Indeed, we found that the predicted strength of selection fell as lattice size was further reduced to 40×40 boxes (data not shown). Additionally, there are likely aspects of biology such as gene regulation that influence the selection for mutualism in the laboratory but are not currently incorporated in COMETS simulations. Despite these effects, COMETS was able to provide useful insight into why adding biotic complexity changed the selection acting on a mutualist population. Broadly applicable approaches for identifying the selection acting in communities will be critical as we attempt to predict the dynamics of vital microbial systems.
While it is known that adding species to a pair-wise system often changes the selection pressures, it is typically hard to predict how the selection will change (Chamberlain et al 2014). Conceptual frameworks that explicitly incorporate resources have been suggested to improve our ability to predict the effect of increasing biotic complexity (Holland et al 2005, Afkhami et al 2014). Our results suggest that going further and incorporating intracellular metabolic mechanisms may provide even more insight. Metabolic mechanisms explicitly link resources to fitness, and constrain the shape of nutrient-based tradeoffs. Incorporating metabolism into modeling frameworks can enable prediction of the nutrient gradients that arise from resource utilization, and ultimately how species interactions emerge from intracellular mechanisms. Such metabolically explicit modeling approaches are most readily available for microbial systems, but are also being developed for eukaryotes (Mo et al 2007, Libourel and Shachar-Hill 2008). More broadly, as we attempt to understand the diversity of mutualistic interactions observed in the natural world, it is critical to focus on how biotic context influences the costs, benefits and ultimately selection acting in systems.
Acknowledgments
The authors thank Emily Jones, Sara Mitri, Ford Denison, Jeremy Yoder and three reviewers for useful comments that improved the manuscript. WRH was supported by NIH NRSA 1F32GM090760 and a DOE award to CJM (DE-SC0006731).
Resources cited
- Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- Afkhami ME, Rudgers JA, Stachowicz JJ. Multiple mutualist effects: conflict and synergy in multispecies mutualisms. Ecology. 2014;95:833–844. doi: 10.1890/13-1010.1. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takaei Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein JL. Conditional outcomes in mutualistic interactions. Trends Ecol Evol. 1994;9:214–217. doi: 10.1016/0169-5347(94)90246-1. [DOI] [PubMed] [Google Scholar]
- Bull J, Harcombe WR. Population dynamics constrain the cooperative evolution of cross-feeding. PLoS One. 2009;4:e4115. doi: 10.1371/journal.pone.0004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiker H, Gore J. Competition between species can stabilize public-goods cooperation within a species. Mol Syst Biol. 2012;8:621. doi: 10.1038/msb.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SA, Bronstein JL, Rudgers JA. How context dependent are species interactions? Ecol Letters. 2014;17:881–890. doi: 10.1111/ele.12279. [DOI] [PubMed] [Google Scholar]
- Chuang JS, Rivoire O, Leibler S. Simpson’s paradox in a synthetic microbial system. Science. 2009;323:272–275. doi: 10.1126/science.1166739. [DOI] [PubMed] [Google Scholar]
- Chubiz LM, Douglas SM, Harcombe WR. Combining engineering and evolution to create novel metabolic mutualisms between species. Methods Mol Biol. 2014;1151:39–47. doi: 10.1007/978-1-4939-0554-6_3. [DOI] [PubMed] [Google Scholar]
- Chubiz LM, Granger BR, Segrè D, Harcombe WR. Species interactions differ in their genetic robustness. Front Microbiol. 2015;6:271. doi: 10.3389/fmicb.2015.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney NF, Kaczmarek ME, Ward LM, Swanson PK, Lee MS, Marx CJ. Development of an optimized medium, strain, and high-throughput culturing methods for Methylobacterium extorquens. PLoS One. 2013;8:e62957. doi: 10.1371/journal.pone.0062957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney NF, Rojas Echenique JI, Marx CJ. Clarity: an open-source manager for laboratory automation. J Lab Autom. 2013;18:171–177. doi: 10.1177/2211068212460237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison RF. Drowning out the protection racket: partner manipulation or drought can strengthen ant-plant mutualism. Trends Plant Sci. 2014;19:411–413. doi: 10.1016/j.tplants.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Doebli M, Knowlton N. The evolution of interspecific mutualisms. Proc Natl Acad Sci. 1998;95:8676–8680. doi: 10.1073/pnas.95.15.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezoe H, Ikegawa Y. Coexistence of mutualists and non-mutualists in a dual-lattice model. J Theor Biol. 2013;332:1–8. doi: 10.1016/j.jtbi.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Ferriere R, Gauduchon M, Bronstein JL. Evolution and persistence of obligate mutualists and exploiters: competition for partners and evolutionary immunization. Ecol Lett. 2007;10:115–126. doi: 10.1111/j.1461-0248.2006.01008.x. [DOI] [PubMed] [Google Scholar]
- Foster KR, Kokko H. Cheating can stabilize cooperation in mutualisms. Proc Biol Sci. 2006;273:2233–2239. doi: 10.1098/rspb.2006.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. Genetics of mutualism: The evolution of altruism between species. J Theor Biol. 1994;170:393–400. doi: 10.1006/jtbi.1994.1200. [DOI] [PubMed] [Google Scholar]
- Gottschalk G. Bacterial Metabolism. 2. Springer-Verlag; New York: 1986. pp. 40–42. [Google Scholar]
- Griffin AS, West SA, Buckling A. Cooperation and competition in a pathogenic bacterium. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- Harcombe WR, Delaney NF, Leiby N, Klitgord N, Marx CJ. The ability of flux balance analysis to predict evolution of central metabolism scales with the initial distance to the optimum. PLoS Comput Biol. 2013;9:e1003091. doi: 10.1371/journal.pcbi.1003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, Bonilla G, Kar A, Leiby N, Mehta P, Marx CJ, Segrè D. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JN, Ness JH, Boyle AL, Bronstein JL. Ecology of Predator Prey Interactions. Oxford University Press; New York: 2005. Mutualisms as consumer-resource interactions; pp. 17–33. [Google Scholar]
- Jones EI, Ferriere R, Bronstein JL. Eco-evolutionary dynamics of mutualists and exploiters. Am Nat. 2009;174:780–794. doi: 10.1086/647971. [DOI] [PubMed] [Google Scholar]
- Jones EI, Bronstein JL, Ferriere R. The fundamental role of competition in the ecology and evolution of mutualisms. Ann N Y Acad Sci. 2012;1256:66–88. doi: 10.1111/j.1749-6632.2012.06552.x. [DOI] [PubMed] [Google Scholar]
- Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems. PLoS Comput Biol. 2010;6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D, Fiegna F, Behrends V, Bundy JG, Philimore AB, Bell T, Barraclough TG. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR. Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond B Biol Sci. 1988;319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D, Weitz KK, Elis R, Konig R, Smith RD, Palsson BØ. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libourel IG, Shachar-Hill Y. Metabolic flux analysis in plants: from intelligent design to rational engineering. Annu Rev Plant Biol. 2008;59:625–650. doi: 10.1146/annurev.arplant.58.032806.103822. [DOI] [PubMed] [Google Scholar]
- Maclean RC, Gudelj I. Resource competition and social conflict in experimental populations of yeast. Nature. 2006;441:498–501. doi: 10.1038/nature04624. [DOI] [PubMed] [Google Scholar]
- Marx CJ. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res Notes. 2008;1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. Group selection and kin selection. Nature. 1964;201:1145–1147. [Google Scholar]
- Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA. 2011;108:10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo ML, Jamshidi N, Palsson BØ. A genome-scale, constraint-based approach to systems biology of human metabolism. Mol Biosyst. 2007;3:598–603. doi: 10.1039/b705597h. [DOI] [PubMed] [Google Scholar]
- Momeni B, Brileya KA, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLife. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MJI, Neugeboren BI, Nelson DR, Murray AW. Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci. 2013;111:1037–1042. doi: 10.1073/pnas.1313285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, May RM. Evolutionary games and spatial chaos. Nature. 1992;359:826–829. [Google Scholar]
- Orth JD, Thiele I, Palsson BØ. What is flux balance analysis. Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BØ. A comprehensive genome-scale reconstruction of Escherichia coli metabolism. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 2014;8:953–962. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle EG, Akcay E, Raab TK, Dirzo R, Gordon DM. Water stress strengthens mutualism among ants, trees, and scale insects. PLoS Biology. 2013;11:e1001705. doi: 10.1371/journal.pbio.1001705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC. Kinship, reciprocity and synergism in the evolution of social behavior. Nature. 1985;318:366–367. [Google Scholar]
- Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol. 2009;3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Gillespie A, Gardner A, West SA, Griffin AS. Frequency-dependence and cooperation: theory and a test with bacteria. Am Nat. 2007;170:331–342. doi: 10.1086/519860. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolution of cooperation Q. Rev Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- Shou W, Ram S, Vilar J. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton ML. Interacting guilds: moving beyond the pairwise perspective on mutualisms. Am Nat. 2003;162:S10–23. doi: 10.1086/378646. [DOI] [PubMed] [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution (Interspecific Interactions) 1. University of Chicago Press; 2005. [Google Scholar]
- Thrall PH, Hochberg ME, Burdon JJ, Bever JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol Evol. 2007;22:120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Trivers R. The evolution of reciprocal altruism. Q Rev Biol. 1971;46:35–57. [Google Scholar]
- Varma A, Palsson BØ. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol. 1994;60:3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC. Adaptation and Natural Selection. Princeton NJ: Princeton University Press; 1966. [Google Scholar]
- Wilson DS. A theory of group selection. Proc Natl Acad Sci. 1975;72:143–146. doi: 10.1073/pnas.72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura N, Higashi M, Behera N, Wakano JY. Evolution of mutualism through spatial effects. J Theor Biol. 2004;226:421–428. doi: 10.1016/j.jtbi.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112:6649–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]