Abstract
Poly(D,L-lactic-co-glycolic) acid (PLGA)-based submicron particles are uniquely posed to overcome limitations of conventional drug delivery systems. However, tailoring cargo/payload release profiles from PLGA micro/nanoparticles typically requires optimization of the multi-parameter formulation, where small changes may cause drastic shifts in the resulting release profiles. In this study, we aimed to establish whether refining the average diameter of submicron particle populations after formulation alters protein release profiles. PLGA particles were first produced via double emulsion-solvent evaporation method to encapsulate bovine serum albumin. Particles were then subjected to centrifugal fractioning protocols varying in both spin time and force to determine encapsulation efficiency and release profile of differently sized populations that originated from a single batch. We found the average particle diameter was related to marked alterations in encapsulation efficiencies (range: 36.4–49.4%), burst release (range: 15.8–49.1%), and time for total cargo release (range: 38–78 days). Our data corroborate previous reports relating PLGA particle size with such release characteristics, however, this is the first study, to our knowledge, to directly compare particle population size while holding all formulation parameters constant. In summary, centrifugal fractioning to selectively control the population distribution of sub-micron PLGA particles represents a feasible tool to tailor release characteristics.
Keywords: PLGA particles, release profile, centrifugal fractioning, protein encapsulation
INTRODUCTION
The popularity of the FDA-approved biodegradable polymer poly(D,L-lactic-co-glycolic acid) (PLGA) for drug delivery applications is not surprising due to the versatility of PLGA. Specifically, PLGA can encapsulate both water-soluble and insoluble molecules, facilitates tunable cargo release profiles, holds the potential for direct injection into target tissues, and consists of metabolizable degradation products.1–5 Moreover, PLGA matrices maintain prolonged, localized bioavailability and may aid in protecting the encapsulated cargo from degradation, a critical parameter for protein-based cargo.1,6,7 Sub-micron PLGA particles are of particular interest due to the potential engineering opportunities for deeper tissue infiltration, improved cellular internalization and ability to circulate and accumulate in target tissues. 1,3,8–10 However, utility of PLGA particle systems can be compromised by undesirable release profiles, such as the characteristic large burst phase, which is followed by the desired steady state release profile. Therefore, mechanisms to tailor and refine specific aspects of the release profile are highly desirable for drug delivery applications.
One of the most common fabrication methods for PLGA particles is emulsion with solvent evaporation. Here, the fabrication parameters largely dictate the resulting particle morphological characteristics and thus play a critical role in determining cargo loading capacity and the resulting release profile (see reviews 11–13). Previous studies have evaluated the influence of the fabrication parameters on the cargo loading and release from PLGA particles to provide general formulation trends on obtaining broad release profile characteristics. 2,14,15 However, seemingly simple changes in the PLGA particle formulation may ultimately lead to a loss of desirable attribute(s).13,16 For example, altering the polymer concentration in the organic phase reportedly affects both particle size and porosity, which in turn influences the encapsulation efficiency and initial burst release.17,18 In another example, varying the emulsifier concentration significantly affects particle size, zeta potential and encapsulation efficiency.18,19 Each particle formulation must therefore be thoroughly characterized to verify the final release properties. As a result, fine-tuning release profiles based solely on altering formulation parameters may prove substantially challenging and highly laborious.
Here we describe modulation of protein release properties from sub-micron PLGA particles via a centrifugal fractioning technique that refines the particle diameter distribution of an initially polydisperse population. The significance of this approach is that we have applied it to study encapsulation and release of protein from particles of varying diameter that were otherwise prepared identically. We hypothesized that the average particle diameter would directly affect protein loading and subsequent release characteristics. This post-fabrication approach is the first, to our knowledge, to directly evaluate the effect of PLGA particle size on critical release parameters while holding all fabrication parameters constant. Bovine serum albumin (BSA)-loaded sub-micron particles were fabricated with identical formulation conditions and then subjected to centrifugal fractioning. We observed particle size-dependent effects on the encapsulation efficiency, burst release, subsequent protein release rate and total release period. The results from this report significantly impact future PLGA micro/nanoparticle studies that may employ this technique as an additional tool to tune and achieve a desired release profile without altering baseline formulation parameters.
METHODS
PLGA particle formulation
Sub-micron PLGA particles were synthesized via a W/O/W emulsion technique adopted from a method further described by McCall et al.20 In short, the organic phase comprised of 100 mg/mL PLGA (PLGA; 50:50 ester-terminated; inherent viscosity=0.55–0.75dL/g; Lactel, Birmingham, AL) in ethyl acetate (Alfa Aesar; Ward Hill, MA). The first emulsion was generated by vortexing the organic phase with a phosphate-buffered saline (PBS) solution containing 20 mg/ mL BSA (total protein content of 2.0% w/w of PLGA; Sigma-Aldrich, St. Louis, MO). The above mixture was added dropwise to a 3.6× volume excess of an aqueous solution containing 2% (w/v) d-α tocopheryl polyethylene glycol 1000 succinate (TPGS; Sigma-Aldrich) under heavy vortex. The second emulsion was produced by ultrasonicating on ice for three consecutive 15s periods (Omni Ruptor 4000; Omni International; Kennesaw, GA). The emulsion was then quickly transferred to a stirring aqueous bath containing 0.2% TPGS (10× volume excess; 300 rpm) and left undisturbed for 3 hrs to undergo solvent evaporation. The particles were washed three times by replacing the supernatant with deionized water after being centrifuged (Beckman Counter; Allegra 25R; Pasadena, CA) at 15,000 g for 15mins. The particles were frozen with 25% (w/w) D-(+)-trehalose dihydrate; Sigma-Aldrich) and recovered via lyophilization.
Centrifugal fractioning
Following solvent evaporation, PLGA particles were subjected to centrifugal fractioning to obtain separate pellet and supernatant sub-groups (Fig. 1). A total of six groups were used for the study. Fractioning (with varying parameters; Table I) was performed on five groups, whereas the remaining was left unfractioned. Freshly fabricated PLGA particles (35.0 mg) were resuspended in 0.5 mL of deionized water and carefully added atop of 5.5% (w/v) glucose solution with a density of 1.055 g/cm3 (5 mL; Acros Organics, Geel, Belgium). The particles were then size fractioned (Centra CL3R; Thermo Fisher Scientific; Waltham, MA) at specified centrifugal forces and spin times (Table I). The supernatant and pellet fractions were separated and washed three more times with deionized water prior to lyophilization. Subsequent particle size analyses, loading and release assays were conducted for all pellet and supernatant subgroups and compared to the unfractioned group.
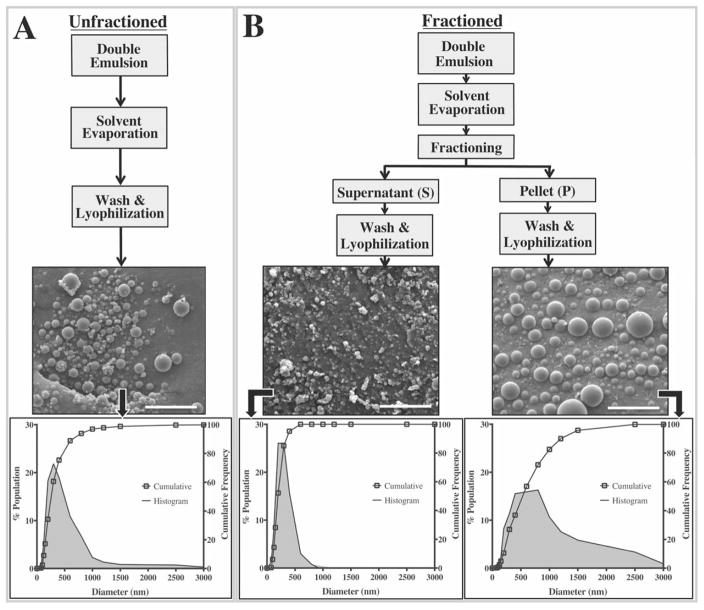
FIGURE 1.
Schematic outlining particle preparation protocols and their resulting size distributions. A) Unfractioned sub-micron PLGA particles were generated using a standard double emulsion technique. B) For the fractioned groups, particles were subjected to a centrifugal size fractioning prior to the washing steps where relatively small particles comprised the supernatant sub-groups, and larger particles formed the pellet subgroups. Cumulative frequency plots along with their respective histograms illustrate significant differences in particle size distributions were achieved without necessitating changes to any formulation parameters. All scale bars represent 5 μm.
TABLE I.
Average Particle Diameters and Protein Encapsulation Efficiencies (EE) Are Significantly Altered as a Function of Centrifugation Force and Time
| Average Diameter (nm) | PDI | Yield (%) | EE (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Unfractioned | 341 | 0.74 | 65.1 | 41.8 ± 1.6% | ||||||
|
| ||||||||||
| Supernatant | Pellet | |||||||||
|
|
||||||||||
| Centrifugal Force | Spin Time (min:s) | Average Diameter (nm) | PDI | Yield (%) | EE (%) | Average Diameter (nm) | PDI | Yield (%) | EE (%) | Total Yield (%) |
| 500 (Low) | 2:15 | 300 | 0.47 | 52.7 | 43.2 ± 0.4% | |||||
| 3:45 | 274a | 0.37 | 48.0 | 41.1 ± 1.1% | 607a | 0.52 | 14.0 | 44.6 ± 0.5% | 62.0 | |
| 7:00 | 276a | 0.45 | 38.4 | 40.4 ± 1.5% | 707a | 0.37 | 20.3 | 48.0 ± 1.1%# | 58.7 | |
| 10:00 | 224a | 0.33 | 41.3 | 38.3 ± 1.0% | 617a | 0.47 | 24.4 | 48.6 ± 0.2%# | 65.7 | |
| 1600 (High) | 10:00 | 211a | 0.18 | 17.3 | 36.4 ± 0.5%# | 541a | 0.47 | 41.2 | 49.4 ± 0.1%# | 58.5 |
Total particle yields (pellet+supernatant) was not significantly affected due to the fractioning process.
represent p<0.05 in comparison to unfractioned.
Particle size analysis
Particle size analysis was performed via scanning electron microscopy (SEM). Briefly, particle samples were extracted from each group after the final deionized water rinse and mounted on to carbon tape after lyophilization. They were then coated with a gold/palladium sputter coater (108-Auto, Cressington Scientific; Watford, UK) to achieve a 5–10 nm thick layer of Au/Pd. Samples were subsequently imaged with a 3–5 kV electron beam (Phillips XL-30; San Francisco, CA). A minimum of seven regions was imaged per particle group. ImageJ (National Institutes of Health, Bethesda, MD) was employed to measure the diameter of at least 85 particles for each SEM image; thus at minimum 595 particles were measured per sample group that make up the size distribution plots. The particle polydispersity index (PDI) was approximated as the square of the standard deviation divided by the mean diameter of each group.
Encapsulation efficiency
Total protein loading was determined by complete dissolution of the particles in dimethyl sulfoxide (DMSO; American Bioanalytical, Natick, MA). The DMSO-polymer solution was then diluted 1:15 in 2.5% (w/v) sodium dodecyl sulfate (Sigma Aldrich)+0.1N sodium hydroxide. The mixture was thoroughly vortexed prior to completing a micro bicinchoninic assay (BCA; G Biosciences; St. Louis, MO) to quantify protein using manufacturer’s protocols. Standards were prepared with known amounts of soluble BSA supplemented with blank PLGA particles (that is, no protein encapsulated, but produced using identical formulation/fractioning protocols). Encapsulation efficiency (EE) was calculated based on the ratio of total protein measured versus the total protein added during fabrication.
Protein release assays
Lyophilized particles were resuspended in PBS supplemented with 0.01% Tween 80 and 0.01% NaN3 (8 mg/mL) and incubated at 37°C under constant agitation. At specified time points, the particle suspensions were centrifuged at 14,000 g for 15mins and supernatant was removed then replaced with fresh PBS. Release media samples were collected at the following time points: 1 hr, 6 hrs, 11 hrs, 24 hrs, 3 days, and subsequently at every five days until day 83. The release media from all groups and time points was then quantified for protein content using the BCA assay following manufacturer’s protocols.
Statistics
All results are depicted as the mean±one standard deviation, unless otherwise noted. Statistical analyses were performed in PRISM (GraphPad Prism, La Jolla, CA) to evaluate differences between groups using analysis of variance (ANOVA), and multiplicity adjusted p-values are reported for Tukey post-hoc comparisons for significance value of α=0.05.
RESULTS
Centrifugal fractioning does not affect particle morphology and yield
The SEM micrographs indicate that the formulation protocol yields spherical particles with a smooth surface morphology and minimal batch-to-batch variability (Fig. 1).20,21 The fractioning protocol did not affect the structural integrity of the particles; qualitative differences in particle shape or surface morphology were not observed across any of the groups. Total yields for all fractioned sub-groups (pellet+supernatant) ranged between 58.5% and 65.7% relative to 65.1% for the unfractioned group indicating that the fractioning process also did not lead to a substantial loss of yield. Collectively, average diameters for all groups evaluated in the study ranged between 211–707 nm with a PDI between 0.18–0.74 (Table I).
Centrifugal fractioning significantly modulates particle size distribution
The initial, unfractioned particles exhibited an average diameter of 341 nm with a PDI of 0.74. The particle diameters ranged between 60 nm and 2600 nm with approximately 93% of the population <750 nm in diameter [Fig. 1(A)]. Upon centrifugal fractioning, we observed a marked change in particle diameter distribution [Fig. 1(B)]. For example, exposing the initial particle population to 550 g for 10mins resulted in a significantly smaller population in the supernatant with an average diameter of 224 nm (PDI=0.33) where 90% of the particles were <400 nm in diameter [Fig. 1(B)]. In contrast, the pellet population for this same fractioning protocol exhibited an average particle diameter of 617 nm (PDI =0.47) with 90% of the particles <1200 nm in diameter [Fig. 1(B)]. Across the board, significant differences in size distributions were observed between the unfractioned population and all fractioned sub-groups (pellets and supernatants) with the exception of supernatant collected from the lowest centrifugal force with the shortest spin time (550 g and 2 min 15s; Table I). The combination of low spin force (550 g) and time (2 min 15s) likely resulted in an insufficient pellet mass to significantly affect the size distribution in the supernatant. The low pellet yield also meant further analysis of size distribution, EE and release profiles were not possible for the 550 g, 2:15 pellet group [Fig. 2(A)].
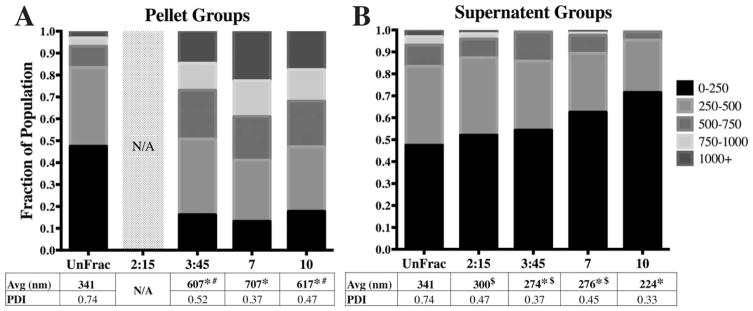
FIGURE 2.
Spin time alters average diameter and diameter distributions. A) All fractioned pellet sub-groups (constant spin force of 550 g; Low G) had significantly larger diameter distributions relative to unfractioned (UnFrac). Size analysis for the 2:15 min group was not conducted due to low yield. B) All Low G supernatant sub-groups were significantly different relative to Unfrac, except for the 2 min 15s group. The 10 min supernatant group was significantly different compared to the rest of the fractioned groups. (* − p<0.01 compared to unfractioned; # − p<0.05 compared to 7 min pellet; $ − p<0.05 compared to 10 min supernatant).
Importance of centrifugation parameters on particle size distribution
Five different fractioning protocols were evaluated in this study, resulting in 10 different particle sub-groups. To compare the size distributions within each sub-group, we generated stacked frequency distribution bar graphs to directly visualize specific population ranges [Figs. 2 and 3]. Nearly half of the population in the unfractioned group exhibited diameters below 250 nm. Fractioning at a centrifugal force of 550 g for times ranging from 2:15 to 10:00mins resulted in a significant decrease in the 0–250 nm particle subpopulation in the pellet groups [15–20%; Fig. 2(A)]. Not surprisingly, we observed that the pelleted sub-populations were dominated by particles >500 nm [50–60%; Fig. 2(A)]. The shift in particle sub-populations resulted in approximately doubling of average pellet diameters (ranging between 607–707 nm) relative to the unfractioned group (341 nm). Conversely, we observed a steady increase in the smallest 0–250 nm sub-population within the supernatant with respect to spin time [Fig. 2(B)]. Specifically, the 10 min supernatant fraction had a population distribution that was significantly different compared to both unfractioned and shorter spin time sup-population groups [Fig. 2(B)]. Here, we observe the 0–250 nm sub-population increase by 30%, while <5% of the particles were >500 nm in diameter. Collectively, this population shift resulted in an average diameter of 224 nm compared to 341 nm for the unfractioned group.
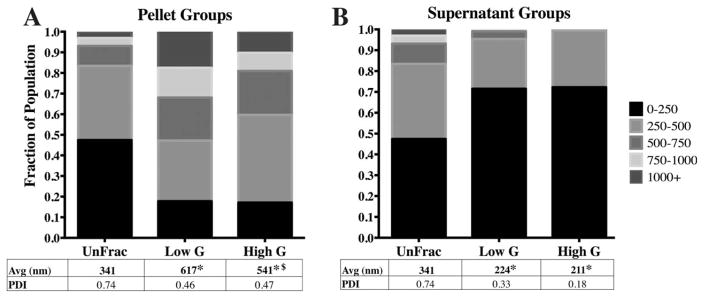
FIGURE 3.
Spin force affects average diameter and diameter distributions. A) Fractioning at 550 g (Low G) and 1600 g (High G) leads to significant changes between Low G and High G pellet sub-groups. B) The supernatant size distributions for both the Low and High G groups were significantly different from UnFrac. Although the differences between the Low and High G supernatant sub-groups were not statistically significant, the PDI is reduced from 0.33 to 0.18. (* - p<0.01 compared to unfractioned; $- p<0.05 compared to 10 min supernatant).
Altering centrifugal force between 550 g (“Low G”) and 1600 g (“High G”) while maintaining a constant spin time (10:00 mins) yielded significant difference in particle size distributions between the pelleted fractions due to nuanced changes in particle sub-populations [Fig. 3(A)]. Specifically, we observed a concurrent shift in two sub-populations: 250–500 nm and >750 nm particles. The High G pellet sub-group shifted to contain a larger portion of 250–500 nm particles (13%) compared to the Low G pellet sub-group [Fig. 3(A)]. At the same time, the Low G pellet sub-group contained a greater number of particles larger than 750 nm. No difference in average diameter was observed between the supernatant groups [Fig. 3(B)]. Yet, the PDI was noticeably lower for the High G supernatant (0.18) compared to the Low G supernatant fraction [0.33; Fig. 3(B)] due to the lack of particles >1000 nm within the High G sub-populations.
Average particle diameter affects protein loading and release characteristics
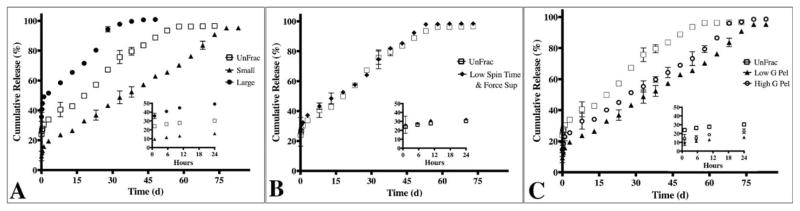
Upon demonstrating that centrifugal fractioning yields significant differences in particle populations, we next investigated the functional effects of average diameter on protein loading (encapsulation efficiency) and cargo release profile of our model protein, BSA. We observed marked differences in the encapsulation efficiencies among the particle groups, ranging from 36.4% and 49.4% (Table I) with a direct relationship between average diameter and the encapsulation efficiency. Additionally, we observed a profound impact of particle size distribution on the resulting protein release profile. Release profiles for all particle sub-groups described in Figures 2 and 3 were collected and compared; for simplicity, we will conduct three main comparisons to highlight the impact of particle size (Supporting Information Fig. 1; S1) on release profile (Fig. 4). First, we compared fractioned sub-groups with the largest difference in average diameter as illustrated by the cumulative frequency plots (Fig. S1). The unfractioned group exhibited a high burst release of 30.3% of total protein in the first 24 hrs followed by sustained release for 57 days (Fig. 4A). Particle populations with the smallest average diameter resulted in higher burst release (49.1%) and shorter total release period (43 days; Fig. 4A). Conversely, the largest average diameter sub-group resulted in a lower burst release (15.8%) and longer protein release period (78 days; Fig. 4A). Secondly, the fractioned group with average diameter and size distributions (Fig. S1B) most similar to the unfractioned group exhibited comparable release profiles (Fig. 4B). Thirdly, fine-tuning of the release profile based on modest yet statistically significant particle diameter distributions (Fig. S1C) was also achieved (Fig. 4C). The High versus Low G pellet sub-groups exhibited altered burst release (15.8% vs. 22.9%, respectively) and significant difference in protein release period (63 days vs. 78 days, respectively; Fig. 4C).
FIGURE 4.
The protein release profile is dependent on the particle size distribution. A) Compares two sub-groups “Small” (1600 g supernatant) and “Large” (Large; 550 g, 7 min pellet) with the most significant differences average diameters relative to unfractioned (UnFrac). The corresponding release profile from Small exhibited a 50% burst release and 40 day release period. Large exhibited an 18% burst release followed by a 75 day release period. In contrast, UnFrac exhibited a burst release of 30.3% with sustained release for 57 days. B) Groups with similar particle size distributions also exhibit comparable burst release, protein release rate and release period. “Low Spin Time & Force Sup” represents the 550 g, 2:15 min supernatant sub-group. C) Modest, yet statistically significant differences in particle size distribution also affects the release profile. Significant differences in cumulative protein release were observed at all time-points except for hours 1, 6 & 11 and days 3, 33 & 48 (p>0.05).
Dependence of loading and release characteristics on particle size
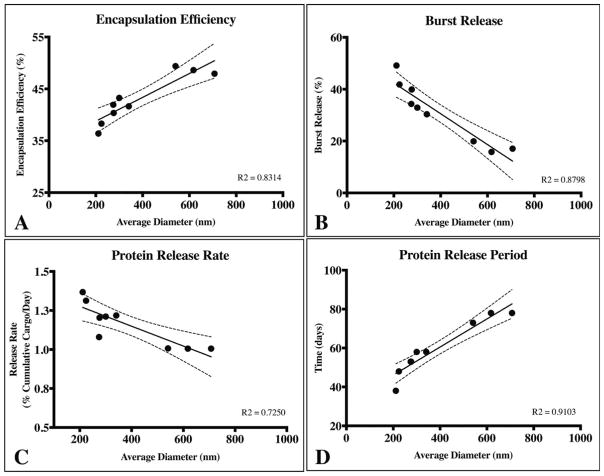
To highlight the key findings from this study, we probed for overarching trends in the encapsulation efficiency, burst release, release rate after burst and protein release period as a function of average particle diameter (Fig. 5). Here, encapsulation efficiency and protein release period were directly related to average particle diameter, whereas, burst release and subsequent protein release rate was inversely proportional to average diameter. These findings provide tangible data regarding the direct impact particle size alone has on specific release characteristics. Thus, similar fractioning techniques may be employed to fine-tune release characteristics without altering sub-micron PLGA particle formulation parameters.
FIGURE 5.
Average particle diameter affects (A) encapsulation efficiency, (B) burst release, (C) protein release rate after the burst phase, and (D) and total protein release period. Encapsulation efficiency and release period was directly proportional to the average particle diameter (A and D). Conversely, the magnitude of the burst release and the rate of protein release subsequently were inversely related to average particle diameter (B and C). The dotted lines represent 95% confidence interval for all cases and a linear regression was used to empirically model the trends for each case.
DISCUSSION
PLGA is one of the most commonly investigated biodegradable polymers for applications in drug and protein delivery, where tailoring release profiles for different cargo/application settings is critical.1,22 In surveying methods to selectively tailor release characteristics, one is typically limited to adjusting particle formulation parameters to tune specific particle properties (for example, size, porosity, surface charge/coatings).4,11–13 Optimization of the release characteristics is thus timely and laborious as a minor change in a formulation parameters may result in a drastic shift in the release profile. Therefore, methods that modulate PLGA particle-based release properties without changing formulation parameters, such as the protocol described here, will reduce time and energy to obtain the desired controlled release system.
Previous studies have reported similar correlations in the encapsulation efficiency and release profile relative to PLGA particle average diameter.12,16,23–25 However, these studies acquired different particle sizes by modifying particle formulation parameters that could be expected to confound encapsulation and release characteristics. Here, the PLGA particle characteristics were based solely on differences in particle size by employing centrifugal fractioning to formulation conditions previously established to yield consistent particle size distributions with minimal batch-to-batch variability.20,21 Our data revealed a direct relationship between average particle diameter, encapsulation efficiency and burst release; we also observed an inverse relationship between average diameter, protein release rate after burst and total release period (Fig. 5). An example of these trends is illustrated in a comparison of the two fractioned subgroups with a large difference in average diameter (High G supernatant and pellet sub-groups). Here, the smaller subgroup (average diameter: 211 nm) exhibited the lowest observed encapsulation efficiency of 36.4% as opposed to 49.4% for the the larger particle sub-group (average diameter: 541 nm; Table I). These results corroborate previous studies that systemically modulated formulation parameters to alter average particle diameter where they also report a positive correlation between encapsulation efficiency and average diameter.12,23,24
Encapsulation efficiency is heavily dependent on the interaction of encapsulated agent with the polymer and water phase during the solvent evaporation stage. Here, the cargo is mobile within the dispersed oil (polymer+organic solvent) phase and is thus free to diffuse into the continuous aqueous phase.16 Due to the higher surface area to volume ratio of smaller oil droplets, diffusion of hydrophilic proteins across the aqueous/organic interface is expected to lower encapsulation efficiency relative to larger droplets. As such, the amount of protein encapsulated is expected to relate directly to particle diameter, as we observed (Fig. 5A). Our maximum encapsulation efficiency of near 50% is lower than previously reported PLGA micro/nanoparticles that obtained 80%, however, these studies altered the emulsifier concentration to increase the amount of protein loaded, whereas we held this parameter constant throughout the study.12,25
The release profiles reported here exhibit an initial burst release followed by a roughly zero-order release rate (Fig. 4). Reported in-vitro release profiles from PLGA micro/nano-particles vary greatly, ranging from zero-order to monophasic, biphasic and triphasic shapes.2 Cargo release profiles depend on interactions within the particle (that is, cargo/polymer, cargo/cargo, and so forth) and release mechanisms (i.e., diffusion, bulk/surface erosion, and so forth) that are unique to each particle formulation and release conditions.2,14 Formulation parameters, such as the type of PLGA polymer (that is, MW, end-group, lactide:glycolide ratio, and so forth) and the resulting initial particle morphology (that is, size, porosity and density), determine which mechanisms dominate control release rate.13 Previous studies indicate that reversible interactions between BSA and PLGA, particularly for carboxyl end-capped polymers, dominate protein encapsulation and release rate properties.2 Irreversible aggregation or adsorption to PLGA polymers/oligomers result in BSA instability and incomplete release.6 Evaluating protein stability/degradation of released BSA was beyond the scope of this study, yet the release assays reliably accounted for at least 90% of the encapsulated protein over the course of the release assays (Fig. 4). In addition to particle morphology, population distribution (PDI) is also an important factor in determining the overall release profile. For example, Berkland et al. combined particles of various sizes to shift the population PDI, resulting in a switch in the release profile of a small, water-soluble molecule from Fickian to zero-order release.26 Here, we observed similar linear release rates after burst for our particle groups, potentially due to the high PDI (PDI <0.1 is considered monodisperse; Table I). In addition, a number of other studies report similar release profiles (semi-linear release profile after a burst phase) from micro- and nanoparticles.27,28
During the early release phase when PLGA particles undergo hydration and wetting of the polymer matrix, diffusion dominates the protein release profile. Surface adsorbed and loosely immobilized cargo diffuse out rapidly, resulting in the burst release.2 This phase of the release profile is largely correlated with the initial particle porosity, cargo properties (size, effective diffusivity, charge, hydrophilicity, and so forth) and hydration rate of the particle.14 In addition, particle size also plays a significant role in affecting the initial burst release.16 Increased surface area to volume ratio of smaller particles results in a higher burst release, since a greater percentage of the cargo is likely to be loosely surface adsorbed and/or pore immobilized in close proximity to the surface. Our data support this model whereby we observed a marked increase in burst release from the smallest and largest average particle diameter groups (Fig. 5B). Conversely, comparing two particle groups with an average diameter and population distribution most similar to each other demonstrated no significant difference in release profile (Fig. 4B). These results suggest that average diameter and diameter distributions of particles formulated using identical parameters plays a significant role in modulating the burst release. The inverse relationship between burst release and the average particle diameter corroborates with general trends reported in literature where different micro/nanoparticle sizes were achieved by altering formulation parameters.16,24,29
Probing further aspects of the release profile, we revealed relationships between the average diameter, the total release period and the release rate of BSA after the burst phenomenon [Fig. 5(C,D)]. Release of encapsulated cargo from PLGA particles occurs by three mechanisms: 1) transport through the polymer, 2) transport through water-filled pores and 3) transport-independent dissolution of encapsulated cargo.2 The rate of water penetration into PLGA matrices is fast relative to the rate of polymer hydrolysis, and thus micro/nanoparticles are primarily degraded via bulk instead of surface processes.30 Cargo diffusion through the PLGA matrix is assumed to be negligible for all but small, hydrophobic molecules.31 Thus, after the burst phase and particle hydration, release of hydrophilic proteins such as BSA (MW=66 kDa) is mediated by diffusion through water-filled pores and is thought to be limited by the rate of PLGA degradation/erosion that produces these pores.2 Specifically, release is attributed to the formation, dilation and coalescence of nanopores (forming mesopores) inside the PLGA matrix. Effective protein diffusivity is directly correlated to the size, interconnectivity and tortuosity of the pore network, as well as protein/polymer and protein/protein interactions.14,32 Subsequent diffusion of proteins through these pores driven mainly by concentration gradients determines the release profile following the initial burst. We observed an overall decrease in the rate of protein release rate as the average particle diameter increased [Fig. 5(C)] potentially due to: (1) additional time required to form an interconnected pore network (assuming identical particle porosity & density) and (2) significantly longer diffusion lengths to reach the release medium in relatively large particles. Blanco et al. generated sub-micron PLGA particles with diameters ranging between 320–523 nm and found that protein release rate range between 0.49–1.45% cumulative release per day after burst.24 Although their protein release rates are similar to the ranges reported here (1.01–1.36% cumulative release/day), they did not report any discernable trends; likely due to the confounding effects of altering formulation parameters.
Increasing the average diameter of a particle population also led to a longer protein release period, most likely due to principles outlined above [Fig. 5(D)]. In our study, the detectable protein release period ranged from 38 days to 75 days relative to the unfractioned group that released protein for 57 days [Fig. 5(D)]. It is important to note that the longer diffusion paths of large particles may also result in accumulation of acidic PLGA degradation products near the core of the particle, leading to pH-driven autocatalytic polymer degradation.14 The heterogeneous rate of polymer degradation and erosion is a proposed mechanism to describe relatively fast cargo release rates for some large microparticles and, in some cases, shorter release periods than smaller particles.33 The effects of autocatalytic polymer degradation is much less pronounced in smaller particles and does not seem to play an appreciable role for the sub-micron particles used in this study.14
CONCLUSION
Due to the complexity and the number of interactions involved in determining PLGA particle properties, changing formulation parameters to achieve desired loading and release characteristics may be time consuming. The methods outlined here demonstrate a direct relationship between release properties and the particle population size characteristics (distribution and average diameter). Encapsulation efficiency and several parameters of the release curve (burst release, protein release rate and protein release period) correlated to the average diameter of the particle population. Thus centrifugal fractioning represents a potential tool for tuning sub-micron PLGA particle properties without modifying formulation parameters. Subsequently, centrifugal fractioning is useful tool in achieving a desired release profile (such as reducing burst release) without potentially compromising other particle attributes due to changes in formulation parameters.
Acknowledgments
Contract grant sponsor: NIH; contract grant number: 1DP2HD084067 (to S.E.S.)
Contract grant sponsor: NSF; contract grant number: 1454282 (to S.E.S.)
Contract grant sponsor: the Ben and Catherine Ivy Foundation (to R.W.S.)
Contract grant sponsor: DoD; contract grant number: W81XWH-14–1-0311 (to R.W.S.)
Contract grant sponsor: ASU Start-up Funds (to S.E.S.)
The authors acknowledge Dr. Vikram Kodibagkar of ASU for his assistance with the ultrasonicator, and the John M. Cowley Center for High Resolution Electron Microscopy (CHREM) for assistance with the SEM.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: An overview of biomedical applications. J Control Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 2.Fredenberg S, Wahlgren M, Reslow M, Axelsson A. The mechanisms of drug release in poly(lactic-co-glycolic acid)-based drug delivery systems—A review. Int J Pharm. 2011;415:34–52. doi: 10.1016/j.ijpharm.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 3.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 4.Azizi M, Farahmandghavi F, Joghataei M, Zandi M, Imani M, Bakhtiary M, Dorkoosh FA, Ghazizadeh F. Fabrication of protein-loaded PLGA nanoparticles: Effect of selected formulation variables on particle size and release profile. J Polym Res. 2013;20:110. [Google Scholar]
- 5.Overstreet DJ, Dutta D, Stabenfeldt SE, Vernon BL. Injectable hydrogels. J Polym Sci Part B Polym Phys. 2012;50:881–903. [Google Scholar]
- 6.Zhu G, Mallery SR, Schwendeman SP. Stabilization of proteins encapsulated in injectable poly (lactide-co-glycolide) Nat Biotechnol. 2000;18:52–57. doi: 10.1038/71916. [DOI] [PubMed] [Google Scholar]
- 7.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: Theory to practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 8.Cartiera MS, Johnson KM, Rajendran V, Caplan MJ, Saltzman WM. The uptake and intracellular fate of PLGA nanoparticles in epithelial cells. Biomaterials. 2009;30:2790–2798. doi: 10.1016/j.biomaterials.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Householder KT, DiPerna DM, Chung EP, Wohlleb GM, Dhruv HD, Berens ME, Sirianni RW. Intravenous delivery of camptothecin-loaded PLGA nanoparticles for the treatment of intracranial glioma. Int J Pharm. 2015;479:374–380. doi: 10.1016/j.ijpharm.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Patel TR, Sirianni RW, Strohbehn G, Zheng M-Q, Duong N, Schafbauer T, Huttner AJ, Huang Y, Carson RE, Zhang Y, Sullivan DJ, Piepmeier JM, Saltzman WM. Highly penetrative, drug-loaded nanocarriers improve treatment of glioblastoma. Proc Natl Acad Sci U S A. 2013;110:11751–11756. doi: 10.1073/pnas.1304504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers. 2011;3:1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feczkó T, Tóth J, Dósa G, Gyenis J. Optimization of protein encapsulation in PLGA nanoparticles. Chem Eng Process Process Intensif. 2011;50:757–765. [Google Scholar]
- 13.Astete CE, Sabliov CM. Synthesis and characterization of PLGA nanoparticles. J Biomater Sci Polym Ed. 2006;17:247–289. doi: 10.1163/156856206775997322. [DOI] [PubMed] [Google Scholar]
- 14.Ford Versypt AN, Pack DW, Braatz RD. Mathematical modeling of drug delivery from autocatalytically degradable PLGA microspheres— A review. J Control Release. 2013;165:29–37. doi: 10.1016/j.jconrel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Kao WJ. Drug release kinetics and transport mechanisms of non-degradable and degradable polymeric delivery systems. Exp Opin Drug Deliv. 2010;7:429–444. doi: 10.1517/17425241003602259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeo Y, Park K. Control of encapsulation efficiency and initial burst in polymeric microparticle systems. Arch Pharm Res. 2004;27:1–12. doi: 10.1007/BF02980037. [DOI] [PubMed] [Google Scholar]
- 17.Mehta RC, Thanoo BC, Deluca PP. Peptide containing microspheres from low molecular weight and hydrophilic poly(d,l-lactide-co-glycolide) J Controlled Release. 1996;41:249–257. [Google Scholar]
- 18.Kwon H-Y, Lee J-Y, Choi S-W, Jang Y, Kim J-H. Preparation of PLGA nanoparticles containing estrogen by emulsification–diffusion method. Colloid Surf Physicochem Eng Asp. 2001;182:123–130. [Google Scholar]
- 19.Yang Y-Y, Chung T-S, Ping Ng N. Morphology, drug distribution, and in vitro release profiles of biodegradable polymeric microspheres containing protein fabricated by double-emulsion solvent extraction/evaporation method. Biomaterials. 2001;22:231–241. doi: 10.1016/s0142-9612(00)00178-2. [DOI] [PubMed] [Google Scholar]
- 20.McCall RL, Sirianni RW. PLGA Nanoparticles Formed by Single-or Double-emulsion with Vitamin E-TPGS. J Vis Exp. :82. doi: 10.3791/51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta D, Fauer C, Mullenaux HL, Stabenfeldt SE. Tunable controlled release of bioactive SDF-1α via protein specific interactions within fibrin/nanoparticle composites. J Mater Chem B. 2015;3:7963–7973. doi: 10.1039/C5TB00935A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumari A, Yadav SK, Yadav SC. Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf B Biointerfaces. 2010;75:1–18. doi: 10.1016/j.colsurfb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Lamprecht A, Ubrich N, Hombreiro Pérez M, Lehr C-M, Hoffman M, Maincent P. Biodegradable monodispersed nanoparticles prepared by pressure homogenization-emulsification. Int J Pharm. 1999;184:97–105. doi: 10.1016/s0378-5173(99)00107-6. [DOI] [PubMed] [Google Scholar]
- 24.Blanco M, Alonso M. Development and characterization of protein-loaded poly(lactide-co-glycolide) nanospheres. Eur J Pharm Biopharm. 1997;43:287–294. [Google Scholar]
- 25.Feczkó T, Tóth J, Gyenis J. Comparison of the preparation of PLGA–BSA nano- and microparticles by PVA, poloxamer and PVP. Colloids Surf Physicochem Eng Asp. 2008;319:188–195. [Google Scholar]
- 26.Berkland C, King M, Cox A, Kim K (Kevin), Pack DW. Precise control of PLG microsphere size provides enhanced control of drug release rate. J Control Release. 2002;82:137–147. doi: 10.1016/s0168-3659(02)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee B, Santra K, Pattnaik G, Ghosh S. Preparation, characterization and in-vitro evaluation of sustained release protein-loaded nanoparticles based on biodegradable polymers. Int J Nanomedicine. 2008;3:487–496. doi: 10.2147/ijn.s3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirby GTS, White LJ, Rahman CV, Cox HC, Qutachi O, Rose FRAJ, Hutmacher DW, Shakesheff KM, Woodruff MA. PLGA-Based Microparticles for the Sustained Release of BMP-2. Polymers. 2011;3:571–586. [Google Scholar]
- 29.Sah H, Toddywala R, Chien YW. Continuous release of proteins from biodegradable microcapsules and in vivo evaluation of their potential as a vaccine adjuvant. J Control Release. 1995;35:137–144. [Google Scholar]
- 30.Batycky RP, Hanes J, Langer R, Edwards DA. A theoretical model of erosion and macromolecular drug release from biodegrading microspheres. J Pharm Sci. 1997;86:1464–1477. doi: 10.1021/js9604117. [DOI] [PubMed] [Google Scholar]
- 31.Wischke C, Schwendeman SP. Principles of encapsulating hydrophobic drugs in PLA/PLGA microparticles. Int J Pharm. 2008;364:298–327. doi: 10.1016/j.ijpharm.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 32.Cohen S, Yoshioka T, Lucarelli M, Hwang LH, Langer R. Controlled delivery systems for proteins based on poly(lactic/glycolic acid) microspheres. Pharm Res. 1991;8:713–720. doi: 10.1023/a:1015841715384. [DOI] [PubMed] [Google Scholar]
- 33.Siepmann J, Elkharraz K, Siepmann F, Klose D. How autocatalysis accelerates drug release from PLGA-based microparticles: A quantitative treatment. Biomacromolecules. 2005;6:2312–2319. doi: 10.1021/bm050228k. [DOI] [PubMed] [Google Scholar]