Structured Abstract
INTRODUCTION
The ubiquitin-proteasome system comprises hundreds of distinct pathways of degradation, which converge at the step of ubiquitin recognition by the proteasome. Five proteasomal ubiquitin receptors have been identified, two that are intrinsic to the proteasome (Rpn10 and Rpn13) and three reversibly associated proteasomal ubiquitin receptors (Rad23, Dsk2, and Ddi1).
RATIONALE
We found that the five known proteasomal ubiquitin receptors of yeast are collectively nonessential for ubiquitin recognition by the proteasome. We therefore screened for additional ubiquitin receptors in the proteasome and identified subunit Rpn1 as a candidate. We used nuclear magnetic resonance (NMR) spectroscopy to characterize the structure of the binding site within Rpn1, which we term the T1 site. Mutational analysis of this site showed its functional importance within the context of intact proteasomes. T1 binds both ubiquitin and ubiquitin-like (UBL) proteins, in particular the substrate-delivering shuttle factor Rad23. A second site within the Rpn1 toroid, T2, recognizes the UBL domain of deubiquitinating enzyme Ubp6, as determined by hydrogen-deuterium exchange mass spectrometry analysis and validated by amino acid substitution and functional assays. The Rpn1 toroid thus serves a critical scaffolding role within the proteasome, helping to assemble multiple proteasome cofactors as well as substrates.
RESULTS
Our results indicate that proteasome subunit Rpn1 can recognize both ubiquitin and UBL domains of substrate shuttling factors that themselves bind ubiquitin and function as reversibly-associated proteasomal ubiquitin receptors. Recognition is mediated by the T1 site within the Rpn1 toroid, which supports proteasome function in vivo. We found that the capacity of T1 to recognize both ubiquitin and UBL proteins was shared with Rpn10 and Rpn13. The surprising multiplicity of ubiquitin-recognition domains within the proteasome may promote enhanced, multipoint binding of ubiquitin chains. The structures of the T1 site in its free state and complexed with monoubiquitin or K48-linked diubiquitin were solved, revealing that three neighboring outer helices from the T1 toroid engage two ubiquitins. This binding mode leads to a preference for certain ubiquitin chain types, especially K6- and K48-linked chains, in a distinct configuration that can position substrates close to the entry port of the proteasome. The fate of proteasome-docked ubiquitin conjugates is determined by a competition between deubiquitination and substrate degradation. We find that proximal to the T1 site within the Rpn1 toroid is a second UBL-binding site, T2, that does not assist in ubiquitin chain recognition, but rather in chain disassembly, by binding to the UBL domain of deubiquitinating enzyme Ubp6. Importantly, the UBL interactors at T1 and T2 are distinct, assigning substrate localization to T1 and substrate deubiquitination to T2.
CONCLUSION
A ligand-binding hotspot was identified in the Rpn1 toroid, consisting of two adjacent receptor sites, T1 and T2. The Rpn1 toroid represents a novel class of binding domains for ubiquitin and UBL proteins. This study thus defines a novel two-site recognition domain intrinsic to the proteasome that uses homologous ubiquitin/UBL-class ligands to assemble substrates, substrate shuttling factors, and a deubiquitinating enzyme in close proximity.
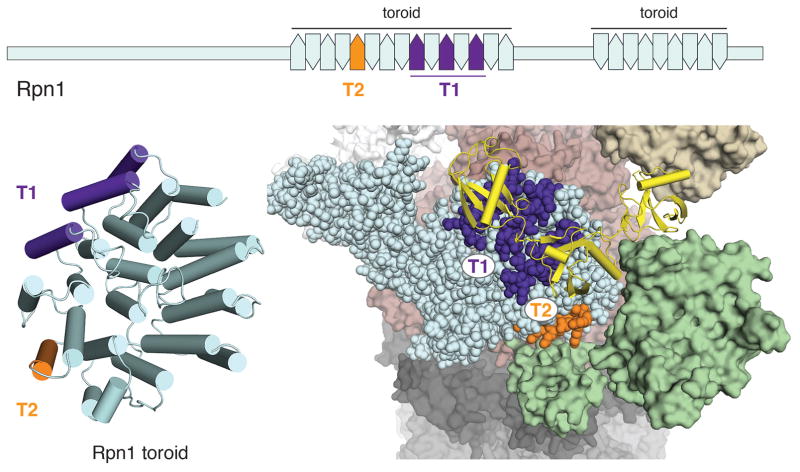
A ligand-binding hotspot in the proteasome for assembling substrates and cofactors.
Schematic (top) and model structure (bottom, left) mapping the UBL-binding Rpn1 T1 (indigo) and T2 (orange) sites. (Bottom, right) Enlarged region of the proteasome to illustrate the Rpn1 T1 and T2 sites bound to a ubiquitin chain (yellow) and deubiquitinating enzyme Ubp6 (green), respectively. PDB 4CR2 and 2B9R were used for this figure.
Hundreds of pathways for degradation converge at ubiquitin recognition by proteasome. Here we found that the five known proteasomal ubiquitin receptors are collectively nonessential for ubiquitin recognition, and identified a sixth receptor, Rpn1. A site (T1) in the Rpn1 toroid recognized ubiquitin and ubiquitin-like (UBL) domains of substrate shuttling factors. T1 structures with monoubiquitin or K48 diubiquitin show three neighboring outer helices engaging two ubiquitins. T1 contributes a distinct substrate-binding pathway with preference for K48-linked chains. Proximal to T1 within the Rpn1 toroid is a second UBL-binding site (T2) that assists in ubiquitin chain disassembly, by binding the UBL of deubiquitinating enzyme Ubp6. Thus a two-site recognition domain intrinsic to the proteasome uses homologous ubiquitin/UBL-class ligands to assemble substrates, shuttling factors, and a deubiquitinating enzyme.
Protein fates are regulated on a global scale by the covalent conjugation of ubiquitin, which can direct target proteins to the proteasome for degradation. Misregulation of the ubiquitin-proteasome system (UPS) is associated with a broad range of human diseases, such as cancer, developmental disorders, neurodegenerative diseases, immune disorders, and microbial infections. In the vast network of the UPS, the recognition of ubiquitin by the proteasome constitutes a central node. Defects in proteasomal ubiquitin receptors have been implicated in both ALS and Alzheimer’s disease (1, 2). Although the mechanisms by which the proteasome achieves its substrate specificity are largely unresolved, it is clear that myriad substrates are first committed to degradation through an initial receptor-mediated recognition step.
There are two distinct modes of ubiquitin recognition by proteasome. The intrinsic recognition pathway utilizes specific proteasome subunits (Rpn10 and Rpn13) for ubiquitin binding (3–7), while the extrinsic recognition pathway uses shuttling factors (Rad23, Dsk2, and Ddi1 in yeast) to escort ubiquitinated substrates to the proteasome (8, 9). The shuttling factors are known as UBL-UBA proteins, as each binds the proteasome via a ubiquitin-like (UBL) domain while binding ubiquitin chains with a ubiquitin-associated domain (UBA). The intrinsic and extrinsic pathways are functionally redundant, at least in part, and cooperate in mediating substrate degradation by the proteasome (6, 8, 9). These receptors exhibit some substrate specificity, but with a mechanistic basis that is poorly understood (9–13).
Existence of an Unidentified Ubiquitin Receptor in the Proteasome
To investigate ubiquitin binding by the proteasome, we first obtained an RPN13 allele that abrogates ubiquitin binding while having no discernable effect on proteasome assembly. (Existing alleles of Rpn13 decrease affinity to ubiquitin by 8-fold; (6)). Guided by NMR data, we introduced two substitutions into the loop segments of the PRU domain, which, when combined with three previously characterized substitutions, completely abolished Rpn13 binding to ubiquitin, as judged by the absence of spectral changes in NMR titration experiments performed at high concentrations of ubiquitin and Rpn13 (fig. S1). The resulting rpn13 E41K E42K L43A F45A S93D mutant is referred to as rpn13-pru below.
To test whether the five known proteasomal ubiquitin receptors are together required for ubiquitin recognition by the proteasome, we introduced the rpn13-pru allele into a strain carrying mutations in the other four receptors, all of which appear to be nulls for ubiquitin recognition. Perhaps surprisingly, this strain, hereafter known as the quintuple mutant, proved to be viable and only moderately defective in growth under standard conditions (Fig. 1A). This strain does display a range of physiological defects, including heat sensitivity, canavanine sensitivity, and high salt sensitivity (Fig. 1A and data not shown); evidently its UPS is significantly attenuated. In summary, the viability of the quintuple mutant led us to hypothesize the existence of still unidentified ubiquitin receptors on the proteasome.
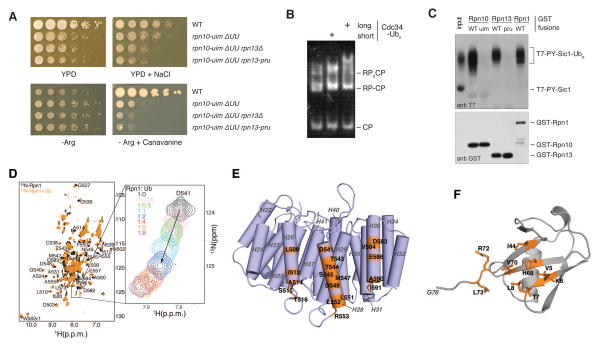
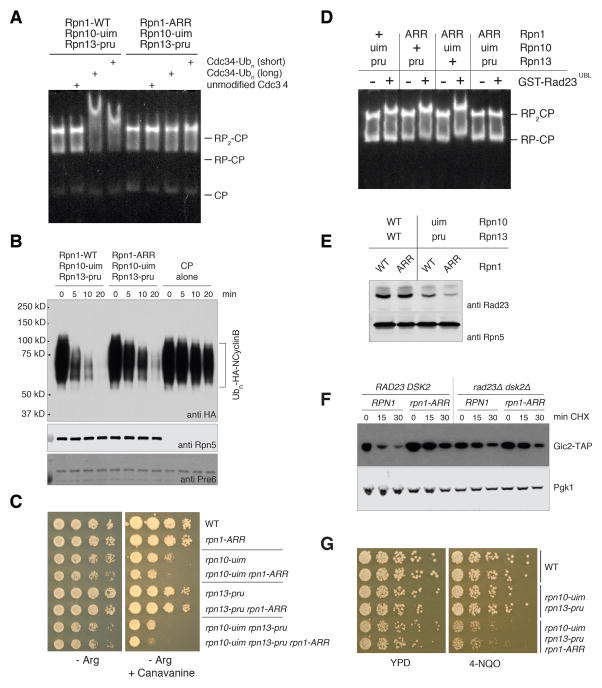
Figure 1. Evidence for an unidentified ubiquitin receptor in yeast.
(A) Yeast strains with the indicated genotypes were serially diluted, transferred to agar plates, and incubated at 30°C. Media were YPD, YPD with 1M NaCl (top row), synthetic medium lacking arginine (Arg−), or Arg− with 6 mg/ml canavanine (bottom row). ΔUU designates the rad23 dsk2 ddi1 background. (B) Proteasome association with ubiquitin conjugates evaluated by a mobility shift assay. RP complexes were purified from an rpn10-uim rpn13-pru strain and incubated with core particle. Reconstituted proteasomes were incubated with autoubiquitinated Cdc34, resolved via native PAGE, and visualized with a fluorogenic activity stain. (C) Known and candidate ubiquitin receptor proteins were expressed in bacteria as GST fusion proteins and immobilized on glutathione resin. Ubiquitinated T7-PY-Sic1 conjugates were used as ligand. Resin-bound proteins were resolved by SDS-PAGE, blotted, and probed with antibodies as indicated. Direct loading of ubiquitinated T7-PY-Sic1 is also included (input). (D) 1H, 15N HSQC spectra of 15N Rpn1412–625 (black) and with equimolar ubiquitin (orange). A titration series is displayed to the right for D541 with increasing ubiquitin, as indicated. (E) Model ribbon diagram of the Rpn1 toroidal domain (PDB 4CR2 (47)) showing residues that shift following ubiquitin addition (D) in orange. (F) Ubiquitin amino acids with amide signals that shift by one standard deviation above average following Rpn1 T1 addition, as shown in fig S8B and S8D, are labeled and highlighted in orange on a ribbon diagram (PDB 2K39 (48)).
Proteasome subunit Dss1/Sem1 was recently proposed to be a novel ubiquitin receptor for this complex (14). In this study, recombinant Sem1 was shown to bind ubiquitin, but whether proteasomes lacking Sem1 are defective in ubiquitin recognition was not examined. To evaluate Sem1, we tested the ubiquitin binding activity of sem1Δ proteasomes. We could not resolve any defect, even in genetic backgrounds in which other ubiquitin receptors had been eliminated (fig. S2), and thus further studies may be required to show that Sem1 is a proteasomal ubiquitin receptor. Because the ubiquitin-binding surface of Sem1 overlaps with its proteasome-binding surface (14), simultaneous binding of the proteasome and ubiquitin by Sem1 might not be possible. Though mutations in the ubiquitin-binding region of Sem1 lead to hypomorphic proteasome phenotypes, these mutations are also associated with proteasome assembly defects.
To test further for novel proteasomal ubiquitin receptors, we characterized proteasomes in which all five known ubiquitin receptors were either functionally inactivated by making amino acid substitutions in their ubiquitin-binding sites or removed during purification by salt wash. When such proteasomes were electrophoresed on a native gel in the presence of ubiquitin conjugates, their migration, visualized by an in-gel activity assay, was retarded, indicating productive binding (Fig. 1B). This binding activity was further localized to the 9-subunit base subcomplex using native gel-shift assays (fig. S3). With the number of candidate receptors narrowed down in this way, we tested proteins individually for ubiquitin binding. Rpn1 in particular proved to form a stable complex (Fig. 1C) with a model proteasome substrate, ubiquitinated PY-Sic1 (15). Rpn1 binding to this substrate was specific to the ubiquitin moiety because non-ubiquitinated Sic1, an internal control, failed to bind (Fig. 1C). Rpn1, the largest subunit of the base, had been previously suggested to serve in the recognition of ubiquitin-like proteins such as Rad23 by the proteasome (16–18).
Rpn1 is paralogous to Rpn2, the structure of which has been solved by X-ray crystallography (19). Both proteins have a centrally located domain composed of 11 repeats of 30–40 residues each that form helix-turn-helix hairpins, known as PC repeats (20). Collectively, these repeats form a closed toroid (19). Based on these data, we generated a series of deletions in Rpn1. Using ubiquitinated PY-Sic1 as a ligand, the binding activity of Rpn1 was mapped to residues S376–A635 (fig. S4). This fragment was trimmed further to obtain a recombinant fragment suitable for NMR, G412-T625 (fig. S5A). NMR titration experiments in which unlabeled ubiquitin was added to 15N-labeled Rpn1 (412–625) showed clear binding (Fig. 1D). The signals in the NMR spectrum were assigned to Rpn1 amino acids (fig. S5A–B), as described below and in supplementary materials. In an Rpn1 model structure, those residues that contact ubiquitin according to the titration experiment (fig. S6) map to three outer helices of the toroid (Fig. 1E). We thus name this ubiquitin-binding region T1 (Toroid 1). Plots of the spectral changes observed for each amino acid with ubiquitin addition revealed two distinct binding affinities, with amino acids from helix 26 (H26) presenting Kd values of 350 ± 70 μM and those from H28 and H30 with values of 120 ± 10 μM (fig. S7). Thus, two weak binding sites for monoubiquitin are suggested on T1.
When unlabeled Rpn1 (412–625) was added to 15N- or 13C-labeled ubiquitin, significant shifting was observed for amino acids located in the ubiquitin β-strand surface (Fig. 1F and fig. S8) that is recognized as well by Rpn10 and Rpn13 (7, 21). Included in this surface are L8, R42, I44, H68, V70, and R74, which shift significantly upon Rpn1 addition.
The Rpn1 T1 Site Binds Two Ubiquitins with Three Outer Toroid Helices
Since only a model structure was available for Rpn1, we used NMR to solve the structure of the T1 site. The N- and C-terminal portions of Rpn1 (412–625) were poorly behaved, with Y608–T625 absent from the recorded spectra and G412–K484 disordered (fig. S5B and S5C). In addition, we observed a concentration-dependent dimerization as determined by dynamic light scattering (fig. S9A), which NMR data suggested to be mediated by the inner helices (H27, H29, and H31) of the toroid (Fig. 1E, fig. S9B and S9C). These effects are no doubt the result of truncation artifacts, though larger deletions only destabilized the protein fold further (fig. S10). Nonetheless, by using standard NMR techniques, as well as stereo-selective methyl labeling and 4D NOESY experiments to record interactions between methyl groups (fig. S11), we were able to solve the structure of the Rpn1 T1 site. This region folds into 3.5 PC repeats (Materials and Methods, Table S1), with H25 forming an incomplete hairpin structure (Fig. 2A). H26–H31, which forms three complete hairpins, converge to a backbone root mean square deviation (RMSD) of 0.8Å. The 4D NOESY spectrum (fig. S11) and an evaluation based on chemical shift values (fig. S5C) revealed H25 to be helical, but loss of its hairpin partner (presumed to range from A470–T479) apparently destabilized it and resulted in this helix adopting a dynamic orientation relative to H26–H31.
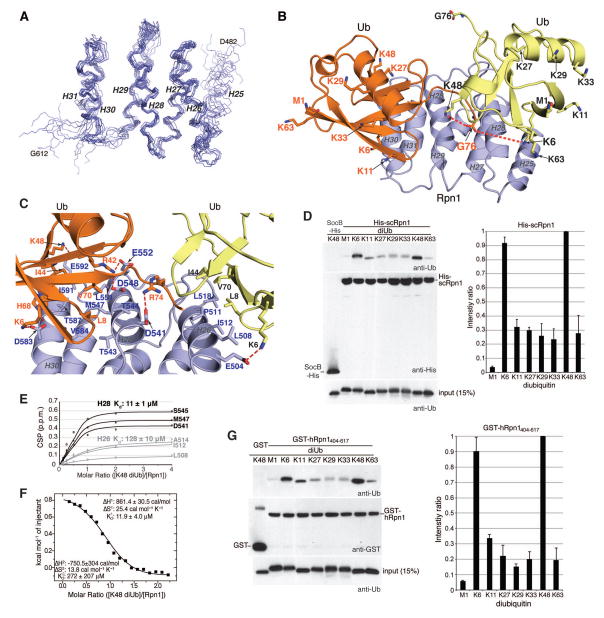
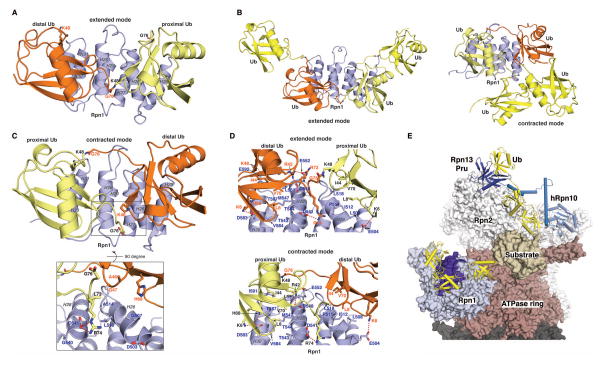
Figure 2. The Rpn1 T1 site engages two ubiquitin molecules in a mode suited for K48-linked chains.
(A) Backbone heavy atoms for the ten lowest energy structures of the Rpn1 T1 site with H26–H31 superimposed. The N- and C-terminal residues and the individual helices are labeled. (B) Ribbon diagram of the lowest energy structure of the Rpn1 T1 site (blue) bound to two monoubiquitin molecules (orange and yellow). Heavy atoms for G76, the M1 backbone, and lysine sidechains are displayed with their oxygen and nitrogen atoms in red and blue respectively. Displayed sidechains for the ubiquitin at H28/H30 are labeled in orange whereas those from the ubiquitin centered at H26 are labeled in black. Dashed orange lines are drawn between G76 from the ubiquitin at H28/H30 and the closest lysine sidechains (K6 and K48) from the ubiquitin at H26; the distance between these two pairs of amino acids is <10Å and the flexibility of the lysine sidechain and C-terminal tail of monoubiquitin allows this distance to be readily shortened. (C) Zoomed-in view of (B) highlighting the Rpn1:ubiquitin contact surface with key amino acids displayed and labeled. Electrostatic interactions are indicated with a red dashed line. (D) Pull-down assay with His-scRpn1 full length protein and M1-, K6-, K11-, K27-, K29-, K33-, K48-, and K63-diubiquitin, as indicated (top and middle panels, left). Immunoblotting was done with anti-ubiquitin (top, left) or anti-His (middle, left) antibodies. Intrinsically disordered protein SocB-His (49) was used as a negative control with K48 diubiquitin, as indicated. Direct loading for 15% of the diubiquitin input for each chain type with immunoblotting by anti-ubiquitin antibody is included (bottom, left). The pull-down assay was repeated four times and the diubiquitin signal intensities were separately normalized to the strongest signal by using ImageJ. The average value and standard deviation is plotted (right). (E) Shifting for indicated Rpn1 T1 site residues plotted with increasing K48 diubiquitin and fitting to the listed Kd values. (F) ITC analysis of Rpn1412–625 binding to K48 diubiquitin. 1.91mM K48 diubiquitin was injected into a calorimeter cell containing 0.18mM Rpn1412–625 and the data were fit to a 2-site sequential binding mode to yield the indicated thermodynamic values. (G) As in (D) but with GST-hRpn1404–617 or GST (as a control). In this case, quantification on the right was done with two independent pull-down assays.
We next solved the structure of the Rpn1 T1 site complexed with ubiquitin, as described in Materials and Methods and (22). Spectra collected on this protein complex showed higher signal-to-noise ratio (fig. S12) compared to those recorded on free Rpn1 and indicated that the structure of the Rpn1 T1 site does not change upon binding to ubiquitin (fig. S13A, S13B, and S13D). Moreover, NOESY spectra recorded on the complex were used to generate new intramolecular constraints for Rpn1 to be used in the calculation of the Rpn1 T1:ubiquitin complex (Table S2). Half-filtered NOESY experiments specialized to record interactions between Rpn1 and ubiquitin revealed many contacts involving ubiquitin L8, T9, I44, V70, L71, and L73 (fig. S14A and S14B), as expected from the titration experiment (Fig. 1F). Intermolecular contacts involving these amino acids validated the presence of two binding sites. For example, L8 contacted L508 and T544 (fig. S14A), spanning a 16Å distance across the toroid (fig. S14C). Ultimately, 106 unambiguous intermolecular distance constraints were identified between Rpn1 and two ubiquitin molecules (Table S2). Twenty-four intermolecular paramagnetic relaxation enhancement (PRE)-derived constraints were also collected with a spin label substituted for ubiquitin G75 (fig. S15A). The resulting structures converged to a backbone RMSD of 0.93Å (fig. S16) with one ubiquitin at H28/H30 (Fig. 2B, orange) and the other at H26 (Fig. 2B, yellow). Individually, the H28/H30:ubiquitin region converged better than the H26:ubiquitin region, with backbone RMSD values of 0.59 and 0.74, respectively (Table S2), owing to fewer detected interactions for the lower-affinity H26 site (figs. S14A, S14B, and S7). Both sites use hydrophobic amino acids to engage ubiquitin L8-I44-V70 (Fig. 2C).
One of seven lysines or the N-terminal methionine in ubiquitin is used to generate ubiquitin chains, which differ in their signaling properties (23). To evaluate Rpn1 ubiquitin chain preference experimentally, resin-bound His-scRpn1 was incubated separately with diubiquitin of each linkage type, washed, and complex formation assayed by immunoblotting with anti-ubiquitin antibody. This experiment validated the structure-based hypothesis to find Rpn1 T1 demonstrating greatest affinity for K6 and K48 diubiquitin (Fig. 2D), the latter of which is an established linkage type for signaling substrate proteolysis by the proteasome (23). Consistent with this result, G76 from ubiquitin at H28/H30 is spatially close to K48 and K6 of the ubiquitin at H26 (Fig. 2B and fig. S14D). An NMR titration experiment revealed Rpn1 to prefer K48-linked ubiquitin chains to monoubiquitin with 11- and 2.7-fold increased affinity at H28/H30 and H26, respectively (Fig. 2E), as supported by isothermal titration calorimetry (ITC, Fig. 2F). The 11 μM affinity of Rpn1 for K48 diubiquitin is similar to the 13 μM affinity reported for scRpn10 binding to K48 diubiquitin (24).
Human Rpn1 (404–617) is 43.8% identical to scRpn1 (412–625, fig. S17). GST-hRpn1 (404–617) or GST (as a negative control) was bound to glutathione S-sepharose resin and incubated with diubiquitin of each linkage type. Following removal of unbound proteins by washing, binding between hRpn1 T1 and ubiquitin was assessed by immunoblotting. This experiment confirmed that human Rpn1 also harbors the ubiquitin-binding T1 site, which prefers K6 and K48 linkages (Fig. 2G).
The T1 Plays a Dual Role in Binding Ubiquitin Chains and Shuttle Factor Rad23
Rpn1 H28 contacts both ubiquitins with electrostatic interactions with D541, D548, and E552 (Fig. 2C). We substituted these residues in the Rpn1 T1 site to generate a D541A D548R E552R triple mutant. The structural integrity of this ARR mutant was maintained, as indicated by 2D NMR analysis (fig. S18). These substitutions eliminated ubiquitin interaction at 5-fold molar excess (Fig. 3A) as well as Rpn1 T1 binding to K6 and K48 diubiquitin in GST-pull down (Fig. 3B) and ITC (fig. S19) experiments. Using the D541A D548R E552R mutant (hereafter rpn1-ARR), we tested in vitro ubiquitin binding activity in the context of full-length Rpn1. The Rpn1-ARR protein showed significantly decreased binding activity to ubiquitinated PY-Sic1 (Fig. 3C).
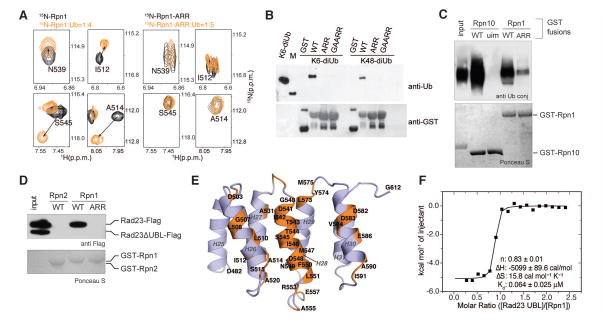
Figure 3. H28 of the Rpn1 T1 site plays a dual role in binding ubiquitin chains and shuttle factor Rad23.
(A) Selected regions from 1H, 15N HSQC spectra of 15N wild-type Rpn1412–625 (left) and 15N Rpn1412–625-ARR (D541A/D548R/E552R, right) alone (black) and with ubiquitin (orange) at 4- and 5-fold molar excess, respectively. (B) GST pull-down assay with GST, GST-Rpn1412–625, GST-Rpn1412–625-ARR, or GST-Rpn1412–625 A514G/D517A/D541A/D548R/E552R (GAARR) and the indicated ubiquitin species. Immunoblotting was done with anti-ubiquitin (top) or anti-GST (bottom) antibodies. (C) Defective ubiquitin binding by the Rpn1-ARR mutant protein. Full length GST-Rpn1 fusion protein was expressed, purified, and tested for binding to ubiquitinated T7-PY-Sic1. GST-Rpn10 and GST-Rpn10-uim were included as positive and negative controls, respectively. (D) Rad23 variants were tested for binding to Rpn1-WT and Rpn1-ARR. A mixture of recombinant Rad23-Flag and Rad23 UBL-Flag (serving as an internal negative control) were used as ligands. UBA domains were absent from both constructs. (E) Rpn1 amino acids at the Rad23 UBL domain contact surface as determined by the data in fig S20 are highlighted in orange on the ribbon diagram of the Rpn1 T1 site. (F) ITC analysis of the Rpn1 T1 site binding to Rad23 UBL. 0.407mM Rad23 UBL was injected into a calorimeter cell containing 0.036mM Rpn1412–625 and the data were fit to a 1-site binding mode with the indicated thermodynamic parameters.
We also tested the binding of Rad23 to the Rpn1-ARR protein. Rpn1-ARR proved to be almost completely defective in Rad23 binding as well (Fig. 3D). This binding was mediated by the UBL domain of Rad23, in agreement with previous studies of the Rad23-Rpn1 interaction (16, 18). Thus the Rpn1-ARR mutation appears to define a binding site for both ubiquitin and UBL-UBA proteins. To test this model further, we added unlabeled Rad23 UBL domain to 15N labeled Rpn1 T1 and used 2D NMR to visualize the affected amino acids. This experiment validated the positioning of H28 at the UBL contact surface (Fig. 3E and fig. S20). ITC demonstrated that Rpn1 (412–625) binds Rad23 UBL with an affinity of 64 ± 25 nM (Fig. 3F), and binding was not detectable with the triple mutant (fig. S21). Thus, H28 is a hotspot within the Rpn1 T1 site for directly and indirectly receiving ubiquitinated substrates.
Phenotypic Effects of the rpn1-ARR Mutation
To assess the ubiquitin-binding activity of Rpn1 in vivo, we integrated the ARR mutation into the genomic RPN1 locus. Rpn1-ARR proteasomes were found to resemble those of wild-type in their assembly and stability, based on several assays: proteasomes were purified from rpn1-ARR cells without apparent difference in composition or yield (fig. S22A); the level of Rpn1-ARR protein was comparable to that of wild type (fig. S22); the relative levels among different subcomplexes of the proteasome were unaffected by the rpn1-ARR mutation (fig. S22C); the SDS-PAGE and native gel profiles of wild-type and Rpn1-ARR proteasomes were indistinguishable (fig. S22A–C); using an anti-Rpn1 antibody to probe endogenous Rpn1 levels in total cell extracts of yeast, no mutant-specific fragmentation of the protein was observed (fig. S22D).
To assess the affinity of the proteasome for ubiquitin conjugates and UBL proteins in a defined biochemical system, we used proteasomes reconstituted from their two major subassemblies, the 19-subunit regulatory particle (RP), which contains Rpn1, and the 28-subunit proteolytic core particle (CP). The RP was purified from strains bearing RPN1-WT or rpn1-ARR alleles in the rpn13-pru rpn10-uim background (fig. S23A), and reconstituted into holoenzyme by adding back purified wild-type CP. Reconstituted proteasomes have a strong ubiquitin-binding activity that is lost in the rpn1-ARR mutant (Fig. 4A). This activity cannot be attributed to contaminating UBL-UBA proteins, because the molar ratio of Rad23 to RP is very low in these preparations (between 1:340 to 1:680; fig. S23B–C). Moreover, the dependence of ubiquitin binding on Rpn1 was also seen using RPs purified from the ubl-ubaΔ background (fig. S23D). These results show that the Rpn1 T1 site can function as a ubiquitin receptor on the proteasome, and that this ubiquitin-binding surface in whole proteasomes is the same as that characterized by NMR.
Figure 4. Rpn1, Rpn10, and Rpn13 play dual roles in recruiting ubiquitin conjugates and shuttling receptors to the proteasome.
(A) Proteasome association with ubiquitin conjugates evaluated by mobility shift assay. Regulatory particle (RP) complexes were purified from the indicated yeast strains, incubated with purified core particle (CP) complex to form holoenzyme, and tested for association with ubiquitin conjugates as described for Figure 1B. (B) Proteasomes were prepared as described for (A), and assayed for degradation of a ubiquitinated fragment of cyclin B1 (Ubn-HA-NCyclinB). Rpn5 was probed as a loading control. (C) Synthetic canavanine sensitivity of rpn1-ARR with other intrinsic ubiquitin receptor mutants. All strains were prepared in the rad23 dsk2 ddi1 background, and carry additional mutations as indicated. Yeast cultures were serially diluted, transferred to synthetic media agar plates lacking arginine (Arg−), or Arg− with 2 mg/ml canavanine, followed by incubation at 30°C. (D) Proteasome association with Rad23 evaluated by mobility shift assay. Proteasomes were purified from rad23 dsk2 ddi1 yeast strains bearing the indicated intrinsic ubiquitin receptor mutations, and incubated with ligand at 100-fold molar excess. Complexes including GST-Rad23-UBL were resolved by native PAGE, and assayed as described. (E) Proteasomes were purified in the absence of salt from yeast strains bearing mutations in intrinsic ubiquitin receptors, as indicated. Proteasomes and their associated proteins were resolved by SDS-PAGE, blotted, and probed with antibodies to proteasome-associated UBL protein Rad23, and to proteasome subunit Rpn5 as a loading control. (F) In vivo degradation of proteasome substrate Gic2. rpn10-uim rpn13-pru yeast strains bearing TAP-tagged Gic2 integrated at the native genomic locus, and other alleles as indicated, were treated with cycloheximide for the indicated times. Lysates were prepared and resolved by SDS-PAGE. Proteins were blotted and probed for Gic2-TAP, with Pgk1 as a loading control. (G) The rpn1-ARR allele confers sensitivity to 4-NQO. Cultures of strains carrying the indicated mutations were serially diluted, transferred to YPD plates either lacking or containing 0.1 mg/ml 4-NQO, and incubated at 30°C for two days.
To test whether conjugate binding to the T1 site could be productive of protein degradation, we used an established in vitro degradation assay based on ubiquitinated cyclin B1 (25–27), with all proteasomes tested being from the rpn13-pru rpn10-uim ubl-ubaΔ background. The rpn1-ARR mutation led to stabilization of cyclin B1 in this assay (Fig. 4B). Stabilization was incomplete however, pointing to the possible existence of a fourth intrinsic ubiquitin receptor in the proteasome. Similar results were obtained using ubiquitinated PY-Sic1 (fig. S24).
We next examined the rpn1-ARR phenotype for signs of proteasome dysfunction in vivo. Proteasome function in yeast is typically assayed in the presence of the arginine analog canavanine, whose presence in proteins induces misfolding and consequent proteasome-mediated degradation. rpn1-ARR exhibited a canavanine-sensitive phenotype when in the presence of the rpn10-uim allele (Fig. 4C). Its synthetic interaction with rpn10-uim was similar to that of rpn13-pru. In these strains, all genes encoding UBL-UBA proteins had been deleted, so that proteasome output should be driven only by direct ubiquitin recognition by intrinsic ubiquitin receptors. Thus, the canavanine-sensitive phenotype of the rpn1-ARR allele suggests that Rpn1 can support ubiquitin-dependent substrate recognition in cells. Its ability to do so is at some level redundant with other proteasomal ubiquitin receptors, a common characteristic of these proteins.
As discussed above, the isolated toroid domain of Rpn1 can bind both ubiquitin and the ubiquitin-like protein Rad23. To assess which of the intrinsic proteasomal ubiquitin receptors can also bind UBL proteins within the context of intact proteasomes, we purified proteasomes from yeast strains that express mutant forms of two of the three intrinsic receptors, leaving only a single receptor functional. As a control, one strain carried mutant forms of all three receptors. Using a gel-shift assay, we found that all three receptors could bind Rad23 (Fig. 4D). Therefore, the dual ubiquitin/UBL binding mode seen for Rpn1 is, interestingly, a general feature of proteasomal ubiquitin receptors. These data were confirmed using endogenous material; co-purification of Rad23 with rpn10-uim rpn13-pru proteasomes was reduced by the rpn1-ARR mutation (Fig. 4E). This effect is not due to a reduced level of Rad23 in the mutant strain (fig. S25). The T1 site was also found to be a Dsk2 binding site, using recombinant material (fig. S26).
Notably, control proteasomes lacking ubiquitin/UBL-binding sites in all three receptors showed no detectable interaction with Rad23 (Fig. 4D), suggesting that all major proteasomal receptors for this UBL-UBA protein have now been identified. The contribution of Rpn10 to Rad23 binding was unexpected because several previous studies had found that Rpn10 did not appear to contribute to UBL protein recognition by intact proteasomes (16, 28, 29). The difficulty in resolving this binding interaction in the past was likely the result of high background binding due to Rpn1 and Rpn13. One possible caveat to these data is that some fraction of copurified Rad23 may be indirectly pulled down by ubiquitin conjugates via the ubiquitin-binding UBA domains that are present in Rad23. This is addressed however by the reconstitution experiments of Fig. 4D. In summary, our results reveal a deep interconnectivity between the intrinsic and extrinsic ubiquitin receptors of the proteasome, where each intrinsic receptor can dock ubiquitin conjugates either by direct ubiquitin recognition or indirectly via an extrinsic receptor.
In phenotypic studies, we found that the T1 site of Rpn1 may promote protein degradation as well as DNA repair through UBL binding. In particular, the proteasome substrate Gic2, in its TAP-tagged form (30), is strongly stabilized by the T1 site mutation, and this effect depends on the expression of UBL-UBA proteins (Fig. 4F). Similar effects were observed for Gcn4 (fig. S27). Mutation of the T1 site also leads to a phenotype of sensitivity to 4-nitroquinoline 1-oxide (4-NQO), suggesting a defect in DNA repair (Fig. 4G). No difference was seen between mutant and wild-type forms of T1 in the UBL-UBA null genetic background (data not shown), consistent with the 4-NQO-sensitive phenotype being mediated by an interaction between the proteasome and Rad23, a known DNA repair factor (31). In summary, phenotypic data suggest that the T1 site functions via docking of ubiquitin conjugates at the proteasome, but can do so both directly and through docking of ubiquitin receptors.
Structure of Rpn1 T1:K48 Diubiquitin Suggests a New Pathway at the Proteasome for Ubiquitin Chains
Because of the importance of K48 ubiquitin chains in targeting substrates for proteolysis, we characterized Rpn1 binding to this chain type more extensively. As observed with monoubiquitin (fig. S13A and S13B), binding to K48 diubiquitin did not change the Rpn1 T1 fold, as demonstrated by contacts between methyl groups (fig. S13A and S13C). NMR experiments designed to record intermolecular contacts between Rpn1 and each ubiquitin of the K48 dimer revealed the presence of two binding modes. In particular, each ubiquitin moiety can bind to either the H28/H30 or the H26 binding surface (fig. S28). We thus calculated two sets of Rpn1 T1:K48 diubiquitin structures (Materials and Methods, Table S2). In addition to NOE-derived distance constraints, we obtained intermolecular distances between the Rpn1 T1 site and each ubiquitin of the K48 dimer by using PRE data obtained by placing a spin label in the loop connecting H26 and H27 at L518 (fig. S15B). In one set of calculations, 104 NOE- and nineteen PRE-derived intermolecular distance constraints placed distal ubiquitin at Rpn1 H28/H30 and proximal ubiquitin was centered at Rpn1 H26 (Fig. 5A, fig. S29A); proximal ubiquitin is so-designated for its free G76, which can in principle be conjugated to a substrate. We refer to this binding mode as “extended” because addition of ubiquitin moieties at either end of the chain yields an opened configuration across the Rpn1 T1 site, as illustrated in a model structure of Rpn1482–612 bound to K48 tetraubiquitin (Fig. 5B, left).
Figure 5. Structures of the Rpn1 T1:K48 diubiquitin complex suggest a new pathway for ubiquitin chain recognition at the proteasome.
(A, C) Lowest energy structures of Rpn1482–612:K48 diubiquitin in the extended (A) or contracted (C) binding mode. These structures were solved experimentally by using a suite of NMR experiments, as described in Materials and Methods by using the data listed in Table S2. An enlarged view is included in (C) to illustrate restricted accessibility of proximal ubiquitin G76. Displayed amino acid sidechains from distal ubiquitin (orange) are labeled in orange whereas those from proximal ubiquitin (yellow) are labeled in black. Blue coloring is used for Rpn1 with helices labeled in grey. (B) Model of Rpn1 T1:K48 tetraubiquitin by adding a ubiquitin (yellow) to each end of the K48 diubiquitin chain for the extended (left) and contracted (right) experimentally determined Rpn1 T1:K48 diubiquitin structure. The Rpn1 T1:K48 tetraubiquitin structures were energy minimized by using Schrödinger (www.schrodinger.com). (D) Expanded view of the extended (top) or contracted (bottom) binding mode for Rpn1 T1:K48 diubiquitin to illustrate hydrophobic and electrostatic interactions at the contact surface. (E) Model of proteasome engaging a ubiquitinated substrate, generated with Rpn1 T1:K48 diubiquitin in the extended binding mode, Rpn13 Pru:ubiquitin (PDB 2Z59), hRpn10:K48 diubiquitin (PDB 2KDF), and human cyclin B1 (PDB 2B9R) placed into a proteasome cryoEM-based model (PDB 4CR2). Rpn1, blue and indigo; ubiquitin, yellow; Rpn13 Pru, navy; Rpn10, light blue; substrate, beige; ATPase ring, burgundy; CP, grey; remaining RP, white.
The second set of structural calculations uses 100 NOE- and eight PRE-derived intermolecular distance constraints that place proximal ubiquitin at H28/H30 and distal ubiquitin at H26 (Fig. 5C, fig. S29B). In this binding mode, G76 from ubiquitin at H28/H30 is spatially close to ubiquitin at H26 (Fig. 5C, expanded region), a configuration caused by a H-bond between proximal ubiquitin R74 and Rpn1 D541 and by van der Waals interactions between proximal ubiquitin L73 and Rpn1 L510 and A514 (Fig. 5C, expanded region). This binding orientation results in a “contracted” ubiquitin chain, best illustrated in a model structure with K48-linked tetraubiquitin (Fig. 5B, right).
Many electrostatic and hydrophobic interactions used to bind monoubiquitin are preserved when the Rpn1 T1 site binds K48 diubiquitin (Fig. 2C and 5D) The backbone RMSD between the contracted and extended binding mode is 0.49 and 0.36 Å for ubiquitin binding at Rpn1 H26 and H28/H30, respectively. This similarity reflects that contacts between ubiquitin L8, T9, I44, V70, L71, L73 and Rpn1 H26, H28, and H30 are retained in all cases, as supported by the experimental data (figs. S14 and S28).
We computationally generated a model substrate (cyclin B1) with an attached K48 triubiquitin chain and docked it in the extended binding mode into the Rpn1 T1 site of a proteasome cryoEM-based model structure (PDB 4CR2). The resulting model illustrated the substrate to be directed toward the ATPase ring and its substrate entry channel (fig. S30). We next adapted this model to include our Rpn13 Pru:ubiquitin (7) and hRpn10:K48 diubiquitin (32) structures (Fig. 5E). The full model represents an additional pathway for ubiquitin chain binding contributed by the Rpn1 T1 site that exists on the same proteasome face as the ubiquitin-binding pathway generated by hRpn13 and hRpn10. Additional experiments will be needed to test this model further.
A Second UBL-Recognizing Site on the Rpn1 Toroid Binds a Deubiquitinating Enzyme
In transit from Rpn1 and the other proteasome ubiquitin receptors to the catalytic center, substrates are deubiquitinated before passing through a narrow gating mechanism, as reviewed in (33). We have previously suggested that the principal binding site for Ubp6 on the proteasome may be in Rpn1 (18, 34), and more recently Rpt1 has also been implicated as an interaction site (35, 36). Using purified Rpn1 and purified Ubp6, we were unable to map the putative Ubp6 binding site of Rpn1 by deletion analysis. We turned to hydrogen exchange mass spectrometry (HX MS), which has proven useful for structural characterization of protein systems intractable by other techniques. HX MS reports on backbone amide hydrogens of proteins, which undergo isotopic exchange in deuterated solvent (37–39). Exchange rates can vary by many orders of magnitude (40) and are influenced by both hydrogen bonding of the backbone and solvent accessibility.
During HX MS analysis of free Rpn1, it became apparent that extensive regions of Rpn1 underwent localized cooperative unfolding, visualized as multiple peaks/populations that incorporate deuterium at different, nonsynchronized rates. Such exchange kinetics, termed EX1 (41–43), are a hallmark of a heterogeneous protein population, often reporting on long-lived conformational fluctuations (44) that result from changes in the conformational ensemble during the deuterium labeling. We hypothesized that Rpn1 incorporated into the base complex might be stabilized by intermolecular interactions within this complex, and therefore attempted to map the Rpn1-Ubp6 interaction in the context of the base.
A base-Ubp6 complex was subjected to deuterium labeling, followed by proteoytic digestion, and the resulting peptide mixture was analyzed by UPLC, ion mobility spectrometry, and mass spectrometry (fig. S31). We identified and measured the temporal deuterium uptake for 40 Rpn1 peptides covering ~60% of the sequence (Fig. 6A and figs. S32 and S33). As hypothesized, when incorporated into the base complex, Rpn1 was dramatically stabilized in conformation relative to when it was free in solution (Fig. 6A). In particular, the EX1 uptake kinetics indicative of conformational heterogeneity was no longer observable. The addition of Ubp6 to the base provided further protection from exchange exclusively for a highly localized region, suggestive of a well-defined binding site rather than further stabilization of the Rpn1 conformation (Fig. 6B). HX was suppressed most strongly for peptic peptide 419–436, corresponding to inner helix 21 and outer helix 22 of the toroid (Fig. 6B; figs. S4 and S33). These experiments represent to our knowledge the largest unique-sequence multi-protein complex studied to date by HX MS.
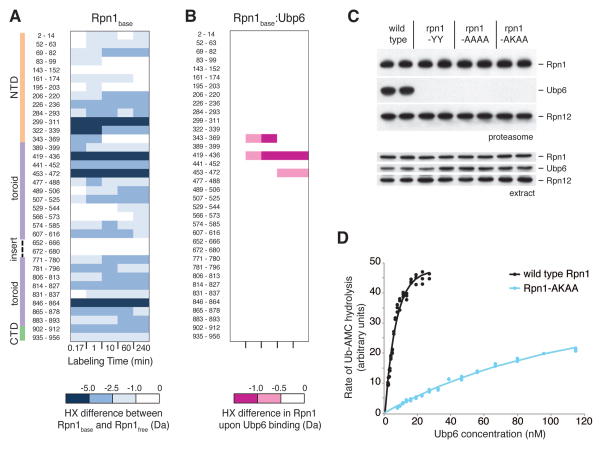
Figure 6. Evidence for a second UBL-specific receptor site on the Rpn1 toroid, which recognizes Ubp6.
(A) Deuteration of recombinant Rpn1 (Rpn1free) compared with deuteration of Rpn1 in the context of the base (Rpn1base). Peptide residue numbers are shown at left, as well as the Rpn1 domain organization as described in the Supplement. Differences at each time point were calculated using Eq. 6 (see Supplement) and color-coded according to the scale at the bottom. (B) Deuteration differences between Rpn1base when bound to Ubp6 minus Rpn1base without Ubp6. Peptide residue numbers are identical to those in (A), and deuteration differences are indicated by the scale at the bottom. (C) Proteasomes were purified from strains bearing wild type or mutant Rpn1 alleles as indicated, employing mild washes. The YY mutant is D431Y Q434Y; AAAA is L430A D431A Q434A Q435A; and AKAA is L430A D431K Q434A Q435A. Aliquots of extracts and purified proteasomes were resolved by 10% SDS-PAGE, blotted, and probed with antibodies against Rpn1, Ubp6, and Rpn12. (D) Activation of Ubp6 by wild type and mutant regulatory particle. Ubp6 was incubated with RP purified from wild type and rpn1-AKAA strains and assayed for Ub-AMC hydrolysis activity. Concentration of RP was constant at 1 nM, and the concentration of Ubp6 was graded. Curve-fitting as shown yields a Kd value of 4.7 nM for wild type. For the AKAA mutant, a 29-fold higher concentration of Ubp6 would be required to achieve a hydrolytic rate corresponding to half-maximal for wild-type.
Having apparently localized the Ubp6 binding site to residues 419–436 of Rpn1, we introduced three sets of mutations into this region of the toroid and in all cases Ubp6 failed to co-purify with proteasomes upon affinity purification (Fig. 6C). We refer to this site, a second and distinct UBL binding site within the Rpn1 toroid, as T2. We purified RP complexes from the T2 mutant rpn1-L430A D431K Q434A Q435A (hereafter rpn1-AKAA), and found, using the fluorogenic substrate ubiquitin-7-Amino-4-Methylcoumarin (ubiquitin-AMC) (34), that these mutations result in a dramatic loss of affinity for recombinant Ubp6 (Fig. 6D).
We next tested whether the T1 and T2 sites can be loaded independently and whether their specificities are truly distinct. Native gel binding assays carried out with T1 site and T2 site mutants showed that the two sites have nonoverlapping specificities (Fig. 7A). In particular Rpn1-AKAA demonstrated a specific defect in Ubp6 and not Rad23 binding, whereas Rpn1-ARR bound to Ubp6 but was defective for Rad23 binding (Fig. 7A). This experiment also mapped the T2 site interacting element of Ubp6 to its UBL domain (Fig. 7A), consistent with cryoelectron microscopy analysis of the proteasome (35, 36). The lack of effect of T1 site mutations on Ubp6 binding was confirmed with endogenous material (fig. S34). We tested whether the T1 site influences the activity of Ubp6 by using the ubiquitin-AMC assay. Neither the T1 site, nor the ubiquitin-binding sites of Rpn13 and Rpn10, influenced Ubp6-mediated deubiquitinating activity (Fig. 7B), thus demonstrating high specificity among these sites despite their common usage of UBL class domains as ligands. In summary, the Rpn1 toroid contains two proximal binding sites for ubiquitin or UBL domains, as shown in Fig. 7C, and thus Rpn1 serves as a key receptor and spatial organizer of both substrates and cofactors of the proteasome.
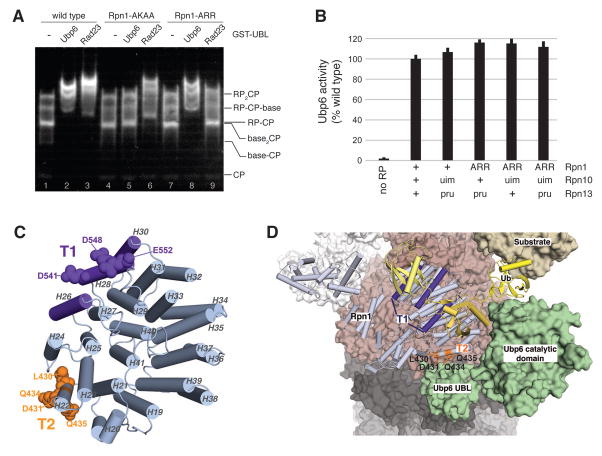
Figure 7. The Rpn1 toroid spatially registers ubiquitin chains and Ubp6.
(A) Reconstituted proteasomes, prepared with regulatory particles isolated from wild type, rpn1-AKAA, or rpn1-ARR strains, were incubated with GST-Ubp6UBL or GST-Rad23UBL in molar excess, resolved by native PAGE, and assayed as described. (B) Ubp6 in RP complexes as indicated was evaluated for activation by the proteasome. Proteins were incubated with 1 μM Ub-AMC, and hydrolytic activity was monitored by the fluorescence of released AMC. (C) Toroidal domain of Rpn1 highlighting the T1 and T2 sites. Residues required for interaction with ubiquitin chains and Rad23 are shown in purple, and exposed residues implicated in Ubp6 binding are highlighted in orange. This image is generated by using PDB 4CR2. (D) Rpn1 T1 and T2 sites in the context of the proteasome, with sites colored as indicated as in (C). This image was generated by docking Ubp6 onto PDB 4CR2 based on the experimental data of Fig. 6B and 6C. The coloring scheme follows that of Fig. 5E and with Ubp6 in green. T2 amino acids L430, D431, Q434, and Q435 are highlighted in orange.
We used a molecular modeling program (HADDOCK 2.1) to dock the Ubp6 UBL domain against Rpn1 amino acids L430, D431, Q434, and Q435. Although the orientation of the UBL is not defined experimentally, the resulting model demonstrates the relative positioning of the T1 and T2 sites and was integrated within the overall architecture of the proteasome as determined by cryoelectron microscopy (Fig. 7D).
Discussion
We report here a hotspot in the proteasome for assembling and localizing substrates and cofactors. This region is contributed by the toroid domain of Rpn1 and spans at least four of its eleven hairpin repeats. Interestingly, we were able to identify three distinct binding positions for the ubiquitin/UBL structure, two of which combine to form a bifurcated binding site for a diubiquitin element, especially when linked by K48 or K6. This novel ubiquitin-binding motif in the Rpn1 toroid also binds to the UBL domain of substrate shuttling factor Rad23 and we name this substrate receptor site T1. The third UBL-binding site, which is adjacent to the T1 site, provides a tether point for deubiquitinating enzyme Ubp6. The location of the T1 and T2 sites within the proteasome places substrate ubiquitin chains and Ubp6 in the neighborhood of the ATPase hexamer, where the Ubp6 catalytic domain is known to form contacts with Rpt1 (35, 36). It is furthermore anticipated that the positioning of these sites on the same proteasome face as the ubiquitin-binding pathway generated by hRpn10 and hRpn13 allows for avidity effects that could increase proteasome affinity for ubiquitinated substrates, especially those that carry multiple ubiquitin chains. A recent single molecule study indeed demonstrated greater degradation efficiency for model substrates in which ubiquitin groups are dispersed into multiple chains (45). Like Rpn1, Rpn13 functions not only as a ubiquitin receptor but also as a receptor for Uch37, a second proteasomal deubiquitinating enzyme. Thus, both Ubp6 and Uch37 are positioned in apposition to ubiquitin receptors, and it will be interesting to explore the functional implications of this localization in future work.
The ability of the T1 site of Rpn1 to bind both ubiquitin and UBL proteins that shuttle ubiquitinated substrates to the proteasome applies to all intrinsic substrate receptor sites of the proteasome. The multiplicity of intrinsic substrate receptors on the proteasome, taken together with the combinatorial array of states associated with loading of multiple extrinsic receptors on any intrinsic receptor, should allow for tens and possibly hundreds of distinct receptor states for binding of ubiquitin conjugates. The utilization of multiple sites for substrate recognition, not all showing high affinity, is consistent with a multipoint, avid mode of ubiquitin chain recognition, which may be associated with highly dynamic interactions between ubiquitin receptors and the proteasome-bound substrate (45). Substrate binding is thought to be productive for proteolysis only if it results in the presentation of an unstructured initiation site into the substrate translocation channel of the ATPase ring (46). A dynamic mode of ubiquitin chain binding may allow for the body of the substrate to present alternative orientations to the proteasome so as to achieve productive positioning of the initiation site. In summary, this work defines a new pathway in the proteasome that engages substrates by using avid low affinity binding sites, and places them in the neighborhood of deubiquitinating enzyme Ubp6 and the ATPase ring.
Materials and Methods
Strain construction
Yeast strains built for this study were either prepared by crossing of haploids bearing existing alleles, or by transformation of diploids with gene disruption constructs followed by sporulation and dissection to retrieve the target haploid. The tandem protein A sequence fused to the C-terminus of RPN11 has been described previously (34), though the strains designated as ProA2 in Table S3 bear the ADH1 terminator following the tandem ProA tag. The tandem protein A sequence fused to RPT1 is also as previously described (34), except the modification was integrated into the endogenous RPT1 locus. Modifications in the RPN10 and RPN13 genes are as described previously (6, 8). Modifications in the RPN1 gene were made using the integrative construct YSp97, which bears klTRP1 as a selective marker (50) flanked by the TEF promoter and terminator (51). The rad23::hphMX allele corresponds to that found in SY722 (6). The dsk2::CaURA3MX allele was prepared by integrating a SalI-SacII fragment from YSp244, the marker having been derived from the C. albicans homolog (52). The ddi1::LEU2MX allele was prepared by integrating a SalI-SacII fragment from YSp249. The sem1::hphMX allele was prepared as a precise deletion of the ORF, using a cassette previously described (53). The ubp6::URA3 locus is as previously described (34). The GIC2-TAP::HIS3 and GCN4-TAP::HIS3 alleles were generated using DNA amplified from suitable strains from the yeast TAP-tagged library (30) All strains used in this study are listed in Table S3.
Plate Assays
Yeast cells were inoculated in YPD media and grown overnight at 30°C. Cell density was measured and the cultures diluted with fresh YPD media to OD600 = 0.1. Cultures were allowed to grow at 30°C for another 3–5 hours, until the OD600 reached 0.4. Cultures were then serially diluted four-fold in arginine dropout media, spotted onto plates with a pin array, and incubated at 30°C. All plates were prepared using media supplemented with 2% agar. The canavanine plates in Fig. 1A were prepared as synthetic media without arginine (2% dextrose, 0.67% yeast nitrogen base without amino acids, 50 mg/L adenine hemisulfate, 500 mg/L uridine, 200 mg/L phenylalanine, 200 mg/L isoleucine, 300 mg/L valine, 40 mg/L tyrosine, 600 mg/L proline, 100 ml/L glycine, 300 mg/L alanine, 300 mg/L serine, 200 mg/L threonine, 400 mg/L glutamate, 200 mg/L aspartate, 800 mg/L glutamine, 200 mg/L asparagine, 150 mg/L histidine, 450 mg/L lysine, 150 mg/L methionine, 400 mg/L leucine and 400 mg/L tryptophan), and canavanine was added to a final concentration of 6 μg/ml. The canavanine plates used in Fig. 4C were prepared similarly, except that all amino acids were present at 25% of the concentrations given above, and canavanine was added to a final concentration of 2 μg/ml. For the 4-NQO assay shown in Fig. 4G, plates were prepared with YPD, and serial dilutions prior to plating were prepared in YPD.
Antibodies
The antibodies used in this study and their sources are detailed in Table S4.
Protein Turnover Assays
Yeast cells were grown in YPD until an OD600 of 0.8 was reached, and cycloheximide was then added to the media to a final concentration of 100 μg/ml. At each time point, yeast cells were withdrawn and mixed with an equal volume of cold stop buffer (150 mM NaCl, 50 mM NaF, 1 mM NaN3, 10 mM EDTA, 60 mM 2-deoxy-D-glucose), and placed on ice. Cells were then recovered by centrifugation and, for each OD-ml of cells, 10 μl of water supplemented with Protease Inhibitor Cocktail (PIC, 1 μg/ml leupeptin, 2 μg/ml antipain, 10 μg/ml benzamidine hydrochloride, 5–10 KIU/ml aprotinin, 1 μg/ml chymostatin, 1 μg/ml pepstatin, and 2 mM AEBSF) was added to resuspend the pellet, and the sample was placed on ice. An equal amount of 2X Laemmli loading buffer was mixed with the resuspended cells and the sample heated immediately at 70°C for 10 min. Cell lysates were clarified by centrifugation and supernatants were resolved by SDS-PAGE.
Expression and purification of recombinant proteins
Plasmids used for the expression of all recombinant proteins are listed in Table S5. Rosetta (DE3) cells (EMD Millipore) transformed with expression plasmids were grown in selective LB media to OD600 0.6 ~ 0.8. IPTG was then added to a final concentration of 200 μM, and cells were transferred to a 16°C shaker for overnight protein induction. Cells were collected by centrifugation at 3300 x g, and resuspended in lysis buffer as described below.
For the purification of GST-fusion recombinant proteins, cells were collected by centrifugation, resuspended in lysis buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA, 1 mM DTT) supplemented with 1X protease inhibitor cocktail (PIC), then lysed via French press. Lysates were clarified by centrifugation at 17,000 x g for 30 min at 4°C. Clarified cell lysate was incubated with Glutathione Sepharose 4B resin (GE Healthcare) at a ratio of 1 L initial culture to 500 μl resin. After one hour incubation, resin was washed with 100 bed volumes of lysis buffer, and step-eluted with lysis buffer supplemented with 20 mM reduced glutathione (pH 7.5). Elution fractions that contained significant amounts of protein were pooled and dialyzed against lysis buffer containing 10% glycerol.
For intein fusion proteins, cells were lysed similarly with 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, and cell lysate was incubated with chitin resin (New England Biolabs) at a ratio of 1 L culture to 20 ml resin. After a one-hour incubation, resin was washed with 100 bed volumes of lysis buffer containing 1 M NaCl, and intein-induced cleavage was performed in lysis buffer containing 50 mM DTT at 30°C for 2 hours followed by 4°C overnight. Cleaved fractions were pooled and dialyzed against lysis buffer containing 10% glycerol.
For protein fused with an N-terminal His6 tag, 2ml Ni-NTA (Qiagen) beads was used for cell lysate from 1 L E. coli culture and incubated for 1 hour at 4°C, followed by washing with 30 bed-volumes of lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% glycerol, 20 mM imidazole). Elution was achieved by flushing the beads with 10 ml elution buffer containing 250 mM imidazole.
For the HXMS experiments, free proteasomal base was prepared as previously described (54). Rpn1 was prepared using the intein-based construct, and Ubp6 was prepared using the HIS-tagged construct. These proteins were further purified by anion-exchange FPLC with Hitrap Q HP column (GE Healthecare), followed by size-exclusion FPLC on Superdex 200 column (GE Healthcare). The purified proteins were stored in 20 mM Tris-HCl (pH 7.5) and 100 mM NaCl.
Ubiquitin Conjugate Preparation
Cdc34 ubiquitin conjugates (Figs. 1B, 4A, S3C and S23D) were prepared as previously described (55). Long conjugates of autoubiquitinated Cdc34 were prepared by incubating 1.5 μM E1 (HIS-Uba1), 1 μM HIS-Cdc34, and 50 μM bovine ubiquitin (Sigma) in the presence of 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.1 mM DTT and 2 mM ATP, at 30°C for 16 hours. Short conjugates of autoubiquitinated Cdc34 were prepared similarly, except that 2 μM HIS-Cdc34 was used in the reaction. Conjugates were prepared as aliquots and stored at −80°C.
PY-Sic1 ubiquitin conjugates (Figs. 1C and 3C) were prepared as previously described (56), and as follows: 4 μM PY-Sic1 was incubated with by 0.1 μM E1 (Uba1), 0.64 μM Ubc4, 0.72 μM Rsp5, and 20 μM ubiquitin in the presence of conjugation buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 5 mM ATP) at room temperature for 90 min. After synthesis, substrate was diluted five-fold with recovery buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl) to decrease the DTT concentration, absorbed to Ni-NTA (Qiagen) resin, and eluted in recovery buffer containing 40% glycerol and 200 mM imidazole. Eluent was dialyzed against storage buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 10% glycerol) and stored at −80°C.
Ubiquitin conjugates of an N-terminal fragment of cyclin B (Figs. 4B) were prepared as previously described (27).
GST Binding Assays
Equal molar amounts (~260 pmol) of GST fusion proteins were immobilized on 50 μl glutathione resin. For the binding reactions carried out in Figs. 1C and S4C, a 300 μl cocktail containing 13.3 nM ubiquitinated PY-Sic1, 13.3 nM unmodified PY-Sic1, binding buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA, 1 mM DTT), and 0.4 mg/ml BSA carrier protein was prepared. Binding cocktails were added to resin, and the binding was carried out at room temperature with tumbling for 20 min. Resins were then washed with 300 μl of binding buffer three times, and proteins were eluted from resin with 50 mM reduced glutathione (pH 7.5). In Fig. 3D, the 300 μl binding cocktail contained 3 pmol Rad23 UBA-flag protein, 4 pmol Rad23 UBA UBL protein, binding buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.1% NP-40), and 0.4 mg/ml BSA as carrier protein. Similarly, the binding reaction was carried out at room temperature with tumbling for 20 min. Resin was then washed with 300 μl binding buffer each time, for a total of four times, and proteins finally eluted with 50 mM reduced glutathione (pH 7.5).
Proteasome Purification
Proteasomes were purified via affinity tags previously described (57). Cells were grown to an OD600 of ~7, then harvested. The cell pellet was washed in PBS once, and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 1 mM EDTA, 1 mM ATP) supplemented with protease inhibitor cocktail (PIC). Cells were then lysed by passing the cell suspension through a French press at a pressure of 20,000 psi. Clarified yeast lysate was incubated with IgG resin (MP Biomedicals; at a ratio of 1 L culture to 1 ml resin) for one hour at 4°C. After incubation, the resin was washed with 100 bed volumes of wash buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA, 1 mM ATP), followed by an equilibration with 20 bed volumes of elution buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, 10% glycerol). TEV cleavage was carried out by incubating resin with 2 bed volumes of elution buffer supplemented with 3 units of AcTEV protease (Life Technologies), at 30°C for 1 hour. The cleaved fraction was then collected and the resin was further washed with 5 bed volumes of elution buffer. All eluted fractions were pooled and passed through equilibrated Ni-NTA resin to remove the AcTEV protease. Proteasomes were then concentrated by ultrafiltration using devices with a 30-kDa molecular weight cut-off (EMD Millipore), and protein concentration was measured using Coomassie Plus reagent (Thermo Scientific). Regulatory particle and core particle were purified similarly, except 500 mM NaCl was used during the washes to disrupt the RP-CP interface. Base was likewise similarly purified except that 1M NaCl was used to disrupt the base-CP and base-lid interface. For the holoenzyme and RP preps in Figs. 6, 7 and S2, the PBS wash of the cell pellets, protease inhibitors during lysis, and the glycerol during elution were omitted. The holoenzymes in Fig. 6C and S2 were prepared using a wash buffer with 50 mM NaCl, rather than 100 mM NaCl. The holoenzymes in Fig. 4E were prepared using wash buffer containing no NaCl.
Reconstitution of Proteasome and Subcomplexes
To reconstitute the proteasome, RP and CP were incubated at a 3:1 molar ratio (which corresponds to a mass ratio of 3.6) in the presence of 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 1 mM ATP, at 30°C for 30 min. After incubation, reconstituted RP-CP complexes were directly used for binding or degradation assays. To reconstitute base-CP, base and CP were incubated at a 3:1 molar ratio in the presence of 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1mM ATP, and 10% glycerol at 30°C for 20 min.
Native Gel Electrophoresis
Native PAGE of samples containing proteasome and in-gel activity assay were carried out as previously described (55). Native gel solution (90 mM Tris Base, 90 mM boric acid, 5 mM MgCl2, 0.5 mM EDTA, 3.5% acryl:bis [37.5:1]) was prepared and degassed. After degassing, 1 mM ATP, 0.1% APS, and 0.1% TEMED were added, and the solution was poured to form a 1.5-mm gel. Polymerized gels were used directly or stored at 4°C for no more than one night. Samples containing proteasome in no more than 10 μl of volume were prepared with loading buffer (10 mM Tris-HCl [pH 7.5], 10% glycerol and 100 ng/ml xylene cyanol) and applied to the native gel. Electrophoresis was carried out for 3 hours with a constant voltage of 110 V at 4°C, with the buffer containing 90 mM Tris-Base, 90 mM boric acid, 5 mM MgCl2, 0.5 mM EDTA, and 1 mM ATP. After electrophoresis, the gel was gently dislodged from the plates and incubated with developing buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM ATP) supplemented with 100 μM suc-LLVY-AMC (Bachem) for 12–15 min at 30°C. SDS was added to a final concentration of 0.02% to activate CP in Figs. 1B and 4A. The gels were then visualized on a documentation system with UV light filtered through a 365 nm filter.
Native Gel Mobility Shift Assays
Binding reactions were carried out in a buffer containing 20 mM Tris-HCl [pH7.5], 50 mM NaCl, 5 mM MgCl2, and 1 mM ATP, in a total volume of 14–16 ml, and at 30°C for 15 min, using 1–5 pmol proteasome per sample. Reactions with autoubiquitinated Cdc34 were carried out using a 5–10 fold molar excess of conjugates relative to proteasome (Figs. 1B, 4A, S2, and S23D). Reactions with GST-Rad23UBL, GST-Dsk2UBL, and GST-Ubp6UBL were carried out using either a 50-fold molar excess of ligand (Fig. 7A) or 200-fold molar excess of ligand (Fig. 4D and S26A) over proteasome.
In vitro Protein Degradation Assays
20 nM CP alone or CP reconstituted with 60 nM RP was incubated with ~100 nM substrate, 1 mM ATP, 0.5 mM DTT, and 0.4 mg/ml BSA. At each time point, 10 μl of the reaction mixture was withdrawn, and the reaction was terminated by adding Laemmli loading buffer. Samples were then analyzed by SDS-PAGE and immunoblotting.
Ub-AMC Hydrolysis Assays
To measure the enzymatic activation of Ubp6 by RP, 4 nM Ubp6 was incubated with 1 nM RP in Ub-AMC assay buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM ATP, 5 mM MgCl2, 1 mM DTT, and 1 mg/ml ovalbumin) at room temperature for 30 min. The reaction was then initiated by adding 1 μM Ub-AMC, whose cleavage was monitored by measuring fluorescence in real time for 50 min at Ex365/Em460 with an EnVision Plate reader (Perkin Elmer).
Kd measurement for Ubp6
To measure the affinity of Ubp6 for the regulatory particle, activation of Ubp6 for Ub-AMC hydrolysis was used as a proxy for association. Assays were carried out as above, but using Ubp6 concentrations ranging from 1 nM to 115 nM. Reaction rates were determined by sampling every twenty seconds for 4 min. Hydrolysis rates of Ubp6 alone and RP alone were subtracted from the rates observed for Ubp6 in the presence of RP in order to isolate Ubp6 activity stimulated by the proteasome. The concentration and rate data shown in Fig. 6D were fit with an exponential function:
| (Eq. 1) |
by minimizing the variance along the rate axis. “Activity” is the observed rate of Ub-AMC hydrolysis, “conc” is the reaction concentration of Ubp6, “Amax” is the theoretical maximal reaction rate, and M is a constant. The uncertainties in the fitted parameters were estimated using bootstrap resampling of the data. The constants for the wild type RP curve are Amax = 47.6 ± 0.73 and M = 0.148 ± 0.0061. The constants for the Rpn1-AKAA RP curve are Amax = 32.9 ± 2.5 and M = 0.00953 ± 0.000867.
NMR Sample Preparation
All Rpn1 constructs were expressed in Escherichia coli as fusion proteins with glutathione S-transferase (GST) and a thrombin cleavage site. Cells were grown at 37 °C to OD600 of 0.5–0.6 and protein expression induced by 0.4 mM IPTG at 16 °C overnight. The cells were frozen in liquid nitrogen and stored at −80 °C for ~4 hours, followed by resuspension in lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA and 2 mM DTT at pH 7.2) supplemented with protease inhibitor cocktail tablets (Roche Diagnostics). Cells were lysed by sonication and spun down at 27,000 g for 30 mins. The supernatant was incubated with pre-washed glutathione S-sepharose resin for 3 hours. After extensive washing in lysis buffer, Rpn1 was separated from GST and the resin by cleaving with thrombin in FPLC buffer (50 mM HEPES, 50 mM NaCl, 1 mM EDTA, and 2 mM DTT at pH 6.7). Further purification was achieved by size exclusion chromatography on an FPLC system ÄKTA pure (GE Healthcare) using a HiLoad 16/600 Superdex 200 prep grade column in FPLC buffer. His-tagged full length scRpn1 and Rad23 UBL domain (amino acids 1–78) were purified in an identical manner, but with Ni-NTA agarose resin (Qiagen) and 250 mM imidazole for elution. Rpn13 Pru domain (amino acids 1–150) with no substitutions or with E41K/E42K/S93D and E41K/E42K/L43A/F45A/S93D (6) as well as isotope-labeled ubiquitin (58) were prepared as described. 15N ammonium chloride, 13C glucose, and 2H2O were used for isotope labeling. 2H, 15N, 13C1H3 -methyl (Ileδ1, Leu, Val) labeled Rpn1412–625 (sample A) or 2H, 15N, 13C1H3 -methyl (Ileδ1, Ala, Met), Leu/Val 13C1H3proS labeled Rpn1412–625 (sample B) was produced as described (59) with the precursors 2-keto-(3-methyl-13C)-butyric-4-13C, 3-d acid sodium salt (Sigma-Aldrich) and 2-ketobutyric acid-4-13C, 3, 3-d2 sodium salt (Cambridge Isotope Laboratories, Inc.) (sample A) or precursor kit QLAM-Aβ/Iδ1/LVproS (NMR-Bio) and L-Methionine (2,3,3,4,4-D5, methyl-13CH3) (Cambridge Isotope Laboratories, Inc.) (sample B). Amino acid substitutions were introduced by the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and validated by standard DNA sequencing (Macrogen). M1-, K6-, K11-, K48- and K63-linked diubiquitin were produced from plasmids supplied by Keith Wilkinson, David Komander, or Cecile Pickart. All NMR and ITC samples were validated by LC/mass spectrometry. Unlabeled monoubiquitin was purchased (Sigma-Aldrich). K27-, K29-, and K33-linked diubiquitin were purchased (UBPBio).
NMR Experiments for Rpn1 Assignments and Structure
15N- or 13C-labeled samples used for titration experiments were at 0.3–0.4 mM, whereas experiments acquired for structure determination were performed with 0.6–0.7 mM 15N-, 13C-, or 15N-,13C-labeled samples. All experiments were conducted in 50 mM HEPES, 50 mM NaCl, 1 mM EDTA, 2 mM DTT, 0.1% NaN3, and 5% 2H2O / 95% 1H2O at pH 6.7 except as noted. NMR experiments were acquired on spectrometers equipped with cryogenically cooled probes at 25°C. For 1H, 15N, and 13C assignments, 2D HSQC and 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, and HNCACB, and were acquired on 15N, 13C, and 70% 2H labeled Rpn1412–625 and HCCHTOCSY and (H)CCHTOCSY spectra were acquired on 13C labeled Rpn1412–625. Distance constraints for structure calculations were obtained by using a 15N-dispersed NOESY spectrum on 15N, 50% 2H labeled Rpn1412–625 with a 200 ms mixing time and a 13C-dispersed NOESY on 13C-labeled Rpn1412–625 dissolved in D2O (80 ms mixing time). 4D methyl–methyl HMQC-NOESY-HMQC experiments were also used for unambiguous assignments of inter-residue NOEs between methyl groups of Ileδ1, Ala, Met, and Leu/Val. These experiments were acquired on sample A and B (defined above) in D2O (200 ms mixing time) by using an NUS sampling scheme and the data were processed with NESTA-NMR, as described in (60).
NMR Experiments for Rpn1 complexes with monoubiquitin and diubiquitin
To solve the structures of the protein complexes, 15N/13C NOESY experiments were acquired on 15N/13C-labeled Rpn1412–625 mixed with monoubiquitin or K48-linked diubiquitin at equimolar ratio to assign Rpn1412–625 in its bound states and to generate corresponding intramolecular NOE distance constraints for use in the structure calculations. Intermolecular distance constraints were determined by 13C-half-filtered 3D NOESY experiments (100 ms) performed on samples with equimolar ratio of 13C-labeled monoubiquitin:unlabeled Rpn1412–625, 13C-labeled Rpn1412–625:unlabeled monoubiquitin, and 13C-labeled Rpn1412–625:K48-linked diubiquitin. Selectivity was achieved for diubiquitin by 13C-labeling proximal or distal ubiquitin. 13C-half-filtered 3D NOESY experiments (100 ms) were acquired as controls on free 13C-labeled Rpn1412–625 or ubiquitin samples under identical conditions to distinguish break-through intramolecular NOEs from intermolecular NOE signals.
NMR Titration Experiments
Chemical shift perturbation (CSP) analysis was used to determine protein contact surfaces as described (22). 2D (1H, 15N) HSQC or (1H, 13C) HSQC experiments were recorded on 15N- or 13C- labeled protein with increasing molar ratios of unlabeled binding partner. The concentration of 15N-labeled Rpn13 wild-type and E41K/E42K/L43A/F45A/S93D ranged from 0.17 to 0.40 mM and experiments were in 20 mM NaPO4, 50 mM NaCl, and 4 mM DTT at pH 6.5. The concentration of 15N-labeled Rpn1 wild-type and D541A/D548R/E552R mutant as well as of 15N- or 13C-labeled ubiquitin ranged from 0.20 to 0.40 mM and experiments were in FPLC buffer. Resulting CSP values were derived from Equation 2.
| (Eq. 2) |
ΔδH, change in proton value (in parts per million); Δδs, change in nitrogen or carbon value (in parts per million); x = 0.2 for nitrogen and 0.3 for carbon (22, 61).
Kd values for Rpn1412–625 binding to monoubiquitin or K48-linked diubiquitin were fit by Equation 3 in Microsoft Excel as described in (22) by using NMR titration data acquired with 0.35 mM 15N-labeled Rpn1412–625 and addition of unlabeled monoubiquitin or K48-linked diubiquitin.
| (Eq. 3) |
[U] is the concentration of monoubiquitin or K48-linked diubiquitin; [R] is the concentration of Rpn1412–625. CSPobs and CSPmax are CSP values of selected Rpn1412–625 residue signals at a corresponding [U]/[R] molar ratio and at saturation, respectively.
Spin labeling experiments
Rpn1412–625 with L518C, C538A, and C606A substitutions and ubiquitin G75C were generated, expressed, and purified as described above for the wild-type proteins except with low concentrations of DTT. Spin labeling was performed as described (62) by using S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) or N-(1-oxyl-2,2,6,6-tetramethyl-4-piperidinyl) maleimide (TEMPO-maleimide) (Toronto Research Chemicals Inc.). 1H, 15N HSQC experiments were collected on a Bruker 850 MHz spectrometer at 25°C on samples of 0.3–0.4 mM concentration. For the Rpn1:monoubiquitin complex, data were collected for 15N-labeled Rpn1412–625 mixed with equimolar ubiquitin G76C spin labeled with TEMPO in the paramagnetic and diamagnetic states. The diamagnetic state was generated by adding 10-fold molar excess ascorbic acid for 1 hour at 25°C.
Two sets of paramagnetic relaxation enhancement (PRE) experiments were collected for the Rpn1:K48-linked diubiquitin complex. The two Rpn1 endogenous cysteines (C538 and C606) were replaced with alanine while L518 from the loop between H26 and H27 was substituted with cysteine. The resulting Rpn1412–625 (L518C, C538A, C606A) was then spin labeled with MTSL and mixed with K48-linked diubiquitin that was 15N-labeled at either the proximal or distal ubiquitin.
PRE distances were calculated from the intensity ratio of recorded HSQC spectra acquired on the samples in their paramagnetic and diamagnetic states according Equations 4 and 5 (63–65).
| (Eq. 4) |
| (Eq. 5) |
Ipara and Idia are the integrated peak values of signals in the HSQC spectra for the paramagnetic or diamagnetic state, respectively; R2,dia is the transverse relaxation rate of the amide 1H in its diamagnetic state; Γ2 is the amide 1H PRE rate; t is the time used in the experiment for the PRE to occur; r is the distance between the unpaired electron center and amide 1H atom; K= 1.23 x 10−44 m6 s−2; τc is the protein rotational correlation time; ωH is the proton Larmor frequency. The PRE distance constraints were defined as previously described (62, 63). Protons with Ipara/Idia < 15%, including protons whose resonances were no longer detectable in the paramagnetic spectra, were assigned distance constraints ranging from 1.8 to 15 Å. Constraints were assigned between the ubiquitin G75 Cα atom and attenuated Rpn1 amide protons for the Rpn1 T1:monoubiquitin complex, or between the Rpn1412–625 L518 Cγ atom and attenuated ubiquitin amide protons for the Rpn1 T1:K48 diubiquitin complex.
Structure determination
TALOS+ (http://spin.niddk.nih.gov/bax/software/TALOS) was used to derive backbone ϕ and ψ torsion angle constraints, which were combined with H-bond constraints as well as NOE- and, for the complexes, PRE-derived distance constraints (Table S1–2) to determine the structure of Rpn1482–612, Rpn1482–612:ubiquitin and Rpn1482–612:K48 diubiquitin with XPLOR-NIH 2.33 (http://nmr.cit.nih.gov/xplor-nih/) on a Linux operating system. H-bonds were generated from secondary structure assignments and NOE connectivities with defined distance from the acceptor oxygen to the donor hydrogen and nitrogen of 1.8–2.1 Å and 2.5–2.9 Å, respectively. When calculating the structures of Rpn1482–612, a total of 50 linear starting structures were subjected to 100,000 simulated annealing and cooling steps of 0.005ps. All resulting structures had no distance or dihedral angle violation greater than 0.3 Å or 5°, respectively. The ten lowest energy structures were chosen for visualization and statistical analysis.
When calculating the structures of the Rpn1 complexes with monoubiquitin or K48-linked diubiquitin, intermolecular distance constraints determined from the 13C-half-filtered 3D NOESY and PRE experiments were used, in addition to intramolecular constraints for Rpn1482–612 that were generated from 15N/13C NOESY spectra acquired on the complexes (Table S2). The structure of ubiquitin did not change in the complexes, as determined by 2D HSQC spectra acquired on 15N/13C-labeled ubiquitin or K48-linked diubiquitin titrated with unlabeled Rpn1412–625. We therefore used published constraints for free ubiquitin (PDB: 1D3Z).
All complexed structures were calculated from 50 linear starting structures and subjected to 19,400 simulated annealing and cooling steps of 0.005ps. Initial structure calculations for the Rpn1 T1:monoubiquitin complex revealed that all constraints could only be satisfied with two ubiquitins bound, one at the H28/H30 site and the other centered at H26 (fig. S14). Intermolecular NOEs were manually assigned to a ubiquitin at H28/H30 or H26. Initial calculations were done without inclusion of the PRE data, which was sorted manually and added at the refinement stage. In the final round of structure calculations, all of the data was able to be included and the resulting structures had no distance or dihedral angle violation greater than 0.3 Å or 5°, respectively. This process was also applied to the calculations of the Rpn1 T1:K48 diubiquitin structures. The ten lowest energy structures were chosen for visualization and statistical analysis. Structures were visualized with and figures generated by PYMOL (The PyMOL Molecular Graphics System, http://www.pymol.org/).
The model Rpn1:K48 tetraubiquitin structures were created by adding a ubiquitin to each end of K48 diubiquitin for the extended or contracted binding mode of the experimentally determined Rpn1:K48 diubiquitin structure. The model structures were then energy minimized in Schrodinger (www.schrodinger.com) by using 1000 steps and the OPLS_2005 force field.
Modeling the Rpn1:Ubp6 UBL domain complex
HADDOCK 2.1 (High Ambiguity Driven protein-protein DOCKing) (66) was used to generate a model structure of the Ubp6 UBL:Rpn1 complex. The atomic coordinates for mouse Ubp6 UBL domain and Rpn1 were obtained from PDB entries 1WGG and 4CR2, respectively. Based on the results of the mutagenesis experiments in Figure 6C, L430, D431, Q434, and Q435 of Rpn1 were defined as “active” residues and their neighboring amino acids, with solvent accessibility > 40%, as “passive” residues. The “active” and “passive” residues of Ubp6 UBL domain were defined by using CPORT, a bioinformatics interface prediction software within HADDOCK (67). This analysis focused on amino acids with exposed side chains, identifying 26 and 23 “active” and “passive” residues, respectively, that distribute uniformly around the Ubp6 UBL domain. Ambiguous interaction restraints (AIRs) restricted the “active” residues to be within 2.0 Å of any atom from “active” or “passive” residues of the binding partner (66).
1000 structures were generated in the first step of rigid body docking, followed by energy minimization. The top scoring 200 structures were used for semi-flexible refinement in torsion angle space, followed by water refinement, scoring and clustering of all 200 structures. During semi-flexible simulated annealing, “active” and “passive” amino acids were allowed to move, but constrained by the AIR restraints. After water refinement, Ubp6 UBL:Rpn1 complexes were clustered according to a 7.5 Å cut-off criterion, resulting in 12 clusters. The most populated cluster included 30% (60) of the structures, whereas the next populated cluster had 15% (29). The average backbone RMSD value for this cluster compared to its lowest energy structure is 1.5 Å. This lowest energy structure was selected to represent the Ubp6 UBL:Rpn1 model, which was integrated into the cryoEM-based pdb file (PDB 4CR2).
Dynamic Light Scattering (DLS) experiments
DLS experiments were recorded on a DynaPro Titan Dynamic Light Scattering System (Wyatt Technologies) at 25°C on Rpn1412–625 samples with concentrations of 25, 50, 100, 150, 200, 300, and 390 μM.
Isothermal titration calorimetry (ITC) experiments
ITC was performed at 25 °C on a MicroCal iTC200 system. Rpn1412–625, Rpn1412–625 D541A/D548R/E552R, Rad23 UBL domain, and K48-linked diubiquitin samples were dialyzed extensively against 20 mM NaPO4, 50 mM NaCl, and 1 mM tris(2-carboxyl) phosphine (TCEP) at pH 6.7. Eighteen aliquots of 2.1 μL 1.91 mM K48-linked diubiquitin, 0.407 mM Rad23 UBL, 1.96 mM K48-linked diubiquitin, and 0.442 mM Rad23 UBL were injected at 1000 rpm into a calorimeter cell (volume 200.7 μL), which contained 0.18 mM Rpn1412–625, 0.036 mM Rpn1412–625, 0.17 mM Rpn1412–625 D541A/D548R/E552R, and 0.04 mM Rpn1412–625 D541A/D548R/E552R, respectively. Blank experiments were performed by replacing protein samples with buffer and this blank data was subtracted from the experimental data during analysis. The integrated interaction heat values were normalized as a function of the molar ratio of Rad23 UBL or K48-linked diubiquitin to Rpn1, and the data were fit with MicroCal Origin 7.0 software. Binding was assumed to be at one site to yield the binding affinity Ka (1/Kd), stoichiometry, and other thermodynamic parameters. A model invoking a 2-site sequential binding mode provided a better fit for the binding of K48-linked diubiquitin to Rpn1412–625.
GST and His pull-down analyses
800 pmol of purified His-tagged scRpn1 full-length was added to 20 μL of pre-washed Ni-NTA agarose resin for one hour and washed once with 50 mM HEPES, 50 mM NaCl, 5% (v/v) glycerol, and 15 mM 2-mercaptoethanol at pH 6.7. The resin was next incubated with 800 pmol of M1-, K6-, K11-, K27-, K29-, K33-, K48-, and K63-linked diubiquitin for one hour. Intrinsically disordered protein SocB with a C-terminal His-tag (49) was used as a negative control with K48-linked diubiquitin. Unbound protein was removed by extensive washing in the above buffer. Proteins that were retained on the resin were fractionated by electrophoresis and transferred to a PVDF membrane. The membrane was treated with denaturing buffer 20 mM Tris-HCl, 6 M guanidine hydrochloride, 1 mM PMSF, and 5 μM 2-mercaptoethanol at pH 7.4 for 30 mins at 4°C, extensively washed with Tris-buffer saline +0.1% Tween (TBS-T) buffer and analyzed by immunoblotting. Experiments for GST-tagged hRpn1404–617 or GST protein (Thermo Scientific) (as a negative control) were performed similarly to that described above, but with pre-washed glutathione S-sepharose resin and a 3-hour incubation period. 50 mM HEPES, 50 mM NaCl, 5% (v/v) glycerol, 1 mM EDTA, and 2 mM DTT at pH 6.7 was used for washing. Diubiquitin was detected with mouse anti-Ub antibody (1:1000) (EMD Millipore) followed by HRP-conjugated anti-mouse IgG (1:5,000) (Sigma-Aldrich). His-tagged scRpn1 or SocB-His was detected with mouse anti-His antibody (1:1,000) (Thermo Scientific) followed by HRP-conjugated anti-mouse IgG (1:5,000) (Sigma-Aldrich). GST-tagged hRpn1 or GST protein was detected with rabbit anti-GST antibody (1:5,000) (Cell Signaling Technology) followed by HRP-conjugated anti-rabbit IgG (1:5,000) (Thermo Scientific).
For the experiments to check Rpn1 variant binding to diubiquitin, 800 pmol GST-tagged scRpn1412–625 wild type, D541A/D548R/E552R, A514G/D517A/D541A/D548R/E552R, or GST protein (as a negative control) was added to 20 μL of pre-washed glutathione S-sepharose resin for three hours, washed once with washed with 50 mM HEPES, 50 mM NaCl, 5% (v/v) glycerol, 1 mM EDTA, and 2 mM DTT at pH 6.7, and then incubated with 1 nmol K6- or K48-linked diubiquitin for one hour. Unbound protein was removed by extensive washing in the above buffer. Proteins retained on the resin were fractionated by electrophoresis, transferred to a PVDF membrane, and analyzed by immunoblotting. K6- and K48-linked diubiquitin was detected with mouse anti-Ub antibody (1:1000) (EMD Millipore) followed by HRP-conjugated anti-mouse IgG (1:5,000) (Sigma-Aldrich). GST-tagged scRpn1 or GST was detected with rabbit anti-GST antibody (1:5,000) (Cell Signaling Technology) followed by HRP-conjugated anti-rabbit IgG (1:5,000) (Thermo Scientific).
Hydrogen/Deuterium Exchange Mass Spectrometry
Materials
Deuterium oxide (99.96% D) was obtained from Cambridge Isotopes Laboratories (Andover, MA). Porcine pepsin was purchased from Sigma-Aldrich (St. Louis, MO) and immobilized in-house (68) via aldehyde coupling to AL-20 POROS beads from Life Technologies/Applied Biosystems (Carlsbad, CA). Tris hydrochloride and sodium chloride were obtained from RPI (Mount Prospect, IL).
Deuterium Exchange
Proteasome base complex samples were labeled with D2O in the presence and absence of Ubp6. Labeling of the base complex was initiated via 14-fold dilution into exchange buffer (20 mM Tris [pD 7.5], 100 mM NaCl,) made with D2O. Ubp6-base complex samples were prepared at 10:1 Ubp6:base, followed by dilution into deuterated buffer (resulting in >90% bound Rpn1) (18). Aliquots were taken and quenched after times ranging from 10 seconds to 4 hours by mixing with quench solution (0.8 M GdnHCl + 0.8% formic acid) in a 1.5:1 ratio. The resulting 50 μL solution (~60 pmol 19S base complex) had a pH of 2.5 and was flash frozen on dry ice followed by storage at −80°C. All samples were prepared and analyzed in duplicate. Undeuterated samples were made in the identical manner as the labeled samples except that H2O buffer was substituted for D2O buffer.
Mass Spectrometry
Quenched and frozen samples were rapidly thawed and mixed with 7.5 μL immobilized pepsin slurry. Digestion proceeded for 5 min on ice. Resultant peptides were separated from protease beads using a 0.45 μm Spin-X centrifuge tube filter (Sigma-Aldrich, St. Louis, MO) and a microcentrifuge operating at 7800 g and 4°C for 30 s. The flow-through was injected into a Waters nanoACQUITY UPLC HDX Manager (69) (Waters, Milford, MA) and peptides were trapped and desalted for 3 min at 100 μL min−1. Chromatographic separation was performed on a 1 x 100-mm Waters ACQUITY UPLC C18 (BEH) reversed-phase column using a 9 min linear 5–40% acetonitrile:water gradient.
Mass spectra were acquired using a Waters Synapt G2Si, operated in data-independent ion mobility mode, at a capillary voltage of 3 kV and a desolvation temperature of 200°C. The IMS and transfer travelling wave ion guide (TWIG) wave velocities were 800 and 380 m s−1, respectively. To maximize peptide separation, the IMS wave height was set to 40 V while the transfer wave height was maintained at 4 V. The trap and transfer TWIGs were purged with Ar to 2.36 x 10−2 mbar, the He cell was pressurized to 4.73 mbar, and the IMS TWIG was filled with 2.92 mbar N2. The trap CE was maintained at 6 V for both precursor and fragmentation scans, while the transfer CE was toggled between 0 V and 32 V. Peptides in all experiments were identified using ProteinLynx Global Server 2.5 and deuterium incorporation was calculated using DynamX software. As all measurements were relative and performed using highly similar instrumental conditions, no corrections were made for deuterium loss due to back exchange during analysis (39, 70). HX differences were calculated using:
| (Eq. 6) |
where Dfree(t) corresponds to the centroid mass of the isotopic envelope for a peptic peptide resulting from free protein (eg. Rpn1base) at a given labeling time, and Dbound(t) refers to the centroid mass of the identical peptide arising from the corresponding protein complex (eg. Rpn1base:Ubp6). The mean intensity-weighted standard deviation of the relative deuterium uptake was 0.15 Da and was calculated for each peptide and time point using multiple charge states in each replicate. A 95% confidence interval for the mean relative deuterium uptake of ±0.5 Da was calculated as described (71) and used as a threshold for significance, i.e. changes larger than 0.5 Da were considered significant at 95% confidence.
Supplementary Material
Acknowledgments
For advice, assistance, or comments on the manuscript, we thank D. Chandler-Militello, M. Gill, J. Hanna, A. Kajava, R. King, Y. Lu, Y. Li, M. Pahre, and J. Roelofs. For gifts of reagents we thank D. Clarke, M. Glickman, F. He, Y. Ye, D. Komander, C. Larsen, J. Li, U. Nowicka, S. Sadis, and W. Tansey. This research was funded by grants from the National Institutes of Health (R37-GM043601 to D.F., CA136472 to K.J.W., and R01-GM101135 to J.R.E.), by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research to K.J.W., and by a research collaboration with the Waters Corporation (J.R.E). Atomic coordinates for the Rpn1 T1 site, Rpn1 T1:ubiquitin, and Rpn1 T1:K48 diubiquitin are available through the Protein Data Bank with accession codes 2n3t, 2n3u, and 2n3v/2n3w, respectively.
Footnotes
REFERENCES AND NOTES
- 1.Zhang T, Jia Y. Meta-analysis of Ubiquilin1 gene polymorphism and Alzheimer’s disease risk. Med Sci Monit. 2014;20:2250–2255. doi: 10.12659/MSM.891030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 4.van Nocker S, et al. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu H, et al. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 6.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiner P, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 9.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci U S A. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplun L, et al. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol. 2005;25:5355–5362. doi: 10.1128/MCB.25.13.5355-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voloshin O, Bakhrat A, Herrmann S, Raveh D. Transfer of Ho Endonuclease and Ufo1 to the Proteasome by the UbL-UbA Shuttle Protein, Ddi1, Analysed by Complex Formation In Vitro. PLoS One. 2012;7:e39210. doi: 10.1371/journal.pone.0039210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Paraskevopoulos K, et al. Dss1 is a 26S proteasome ubiquitin receptor. Mol Cell. 2014;56:453–461. doi: 10.1016/j.molcel.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeki Y, Isono E, Toh-E A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 16.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 17.Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9:33. doi: 10.1186/1741-7007-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287:14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, et al. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Kajava AV. What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J Biol Chem. 2002;277:49791–49798. doi: 10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Walters KJ. Identifying and studying ubiquitin receptors by NMR. Methods Mol Biol. 2012;832:279–303. doi: 10.1007/978-1-61779-474-2_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, Walters KJ. Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci. 2010;35:352–360. doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, et al. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 26.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 27.Dimova NV, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matiuhin Y, et al. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Gillette TG, et al. The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 2001;15:1528–1539. doi: 10.1101/gad.869601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang N, et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finley D, Chen X, Walters KJ. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci. 2015 doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 35.Aufderheide A, et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1510449112. 201510449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bashore C, et al. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 38.Percy AJ, Rey M, Burns KM, Schriemer DC. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-A review. Anal Chim Acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 39.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 40.Skinner JJ, Lim WK, Bédard S, Black BE, Englander SW. Protein hydrogen exchange: Testing current models. Protein Sci. 2012;21:987–995. doi: 10.1002/pro.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konermann L, Tong X, Pan Y. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 42.Weis DD, Wales TE, Engen JR, Hotchko Ma. Identification and Characterization of EX1 Kinetics in H/D Exchange Mass Spectrometry by Peak Width Analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Xiao H, et al. Mapping protein energy landscapes with amide hydrogen exchange and mass spectrometry: I. A generalized model for a two-state protein and comparison with experiment. Protein Sci. 2005;14:543–557. doi: 10.1110/ps.041001705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006;357:1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y, Lee B-h, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 47.Unverdorben P, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A. 2014;111:5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange OF, et al. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 49.Nowicka U, et al. Mycobacterium tuberculosis copper-regulated protein SocB is an intrinsically disordered protein that folds upon interaction with a synthetic phospholipid bilayer. Proteins. 2015 doi: 10.1002/prot.24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 51.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 52.Goldstein AL, Pan X, McCusker JH. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 54.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elsasser S, Shi Y, Finley D. Binding of ubiquitin conjugates to proteasomes as visualized with native gels. Methods Mol Biol. 2012;832:403–422. doi: 10.1007/978-1-61779-474-2_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leggett DS, Glickman MH, Finley D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol Biol. 2005;301:57–70. doi: 10.1385/1-59259-895-1:057. [DOI] [PubMed] [Google Scholar]
- 58.Raasi S, Pickart CM. Ubiquitin chain synthesis. Methods Mol Biol. 2005;301:47–55. doi: 10.1385/1-59259-895-1:047. [DOI] [PubMed] [Google Scholar]
- 59.Plevin MJ, Boisbouvier J. In: Recent Developments in Biomolecular NMR. Clore GM, Potts J, editors. chap 1. The Royal Society of Chemistry; 2012. pp. 1–24. [Google Scholar]
- 60.Sun S, Gill M, Li Y, Huang M, Byrd RA. Efficient and generalized processing of multidimensional NUS NMR data: the NESTA algorithm and comparison of regularization terms. J Biomol NMR. 2015;62:105–117. doi: 10.1007/s10858-015-9923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 64.Iwahara J, Tang C, Marius Clore G. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solomon I, Bloembergen N. Nuclear magnetic interactions in the HF molecule. J Chem Phys. 1956;25:261–266. [Google Scholar]
- 66.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 67.de Vries SJ, Bonvin AM. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 69.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Engen JR, Wales TE. Analytical Aspects of Hydrogen Exchange Mass Spectrometry. Annu Rev Anal Chem. 2015;8:150605180052009. doi: 10.1146/annurev-anchem-062011-143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diaz-Martinez LA, Kang Y, Walters KJ, Clarke DJ. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006;1:28. doi: 10.1186/1747-1028-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geng F, Tansey WP. Similar temporal and spatial recruitment of native 19S and 20S proteasome subunits to transcriptionally active chromatin. Proc Natl Acad Sci U S A. 2012;109:6060–6065. doi: 10.1073/pnas.1200854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.