A cyclic nucleotide-gated channel that regulates plant immunity has multiple C-terminal calmodulin domains and is regulated both positively and negatively by calmodulin, challenging the current model.
Abstract
Ca2+ signaling is critical to plant immunity; however, the channels involved are poorly characterized. Cyclic nucleotide-gated channels (CNGCs) are nonspecific, Ca2+-permeable cation channels. Plant CNGCs are hypothesized to be negatively regulated by the Ca2+ sensor calmodulin (CaM), and previous work has focused on a C-terminal CaM-binding domain (CaMBD) overlapping with the cyclic nucleotide binding domain of plant CNGCs. However, we show that the Arabidopsis thaliana isoform CNGC12 possesses multiple CaMBDs at cytosolic N and C termini, which is reminiscent of animal CNGCs and unlike any plant channel studied to date. Biophysical characterizations of these sites suggest that apoCaM interacts with a conserved isoleucine-glutamine (IQ) motif in the C terminus of the channel, while Ca2+/CaM binds additional N- and C-terminal motifs with different affinities. Expression of CNGC12 with a nonfunctional N-terminal CaMBD constitutively induced programmed cell death, providing in planta evidence of allosteric CNGC regulation by CaM. Furthermore, we determined that CaM binding to the IQ motif was required for channel function, indicating that CaM can both positively and negatively regulate CNGC12. These data indicate a complex mode of plant CNGC regulation by CaM, in contrast to the previously proposed competitive ligand model, and suggest exciting parallels between plant and animal channels.
INTRODUCTION
Ca2+ serves as a universal second messenger in eukaryotic signaling pathways, and transient changes in cytosolic Ca2+ levels are rapidly induced by diverse stimuli in plants (Sanders et al., 2002; Kudla et al., 2010). Despite such a central role for Ca2+ in plant biology, relatively little is known regarding the Ca2+ channels of plants (Spalding and Harper, 2011). Plant genomes examined to date lack homologs to animal voltage-gated cation channels and instead possess expanded families of ligand-gated cation channels, represented primarily by the ionotropic glutamate receptor-like channel and cyclic nucleotide-gated channel (CNGC) families (Mäser et al., 2001). CNGCs are nonselective cation channels that are hypothesized to function as Ca2+ channels in plants (Dietrich et al., 2010; Jammes et al., 2011; Zelman et al., 2012) and are characterized by conserved structural components, including a short cytosolic N terminus, six transmembrane helices (S1-S6) with a pore-forming region between S5 and S6, and a cytosolic C terminus containing a cyclic nucleotide binding domain (CNBD; Kaupp and Seifert, 2002; Matulef and Zagotta, 2003). The CNBD mediates channel gating by cyclic nucleotide monophosphates such as cAMP and/or cGMP (Mäser et al., 2001; Kaupp and Seifert, 2002; Matulef and Zagotta, 2003; Kaplan et al., 2007; Zelman et al., 2012), though studies of this mechanism are largely restricted to animal isoforms.
All plant and animal CNGCs studied to date also possess at least one calmodulin (CaM) binding domain (CaMBD) (Kaplan et al., 2007; Ungerer et al., 2011). CaM is a ubiquitous eukaryotic Ca2+ sensor, which binds four Ca2+ ions via EF-hand motifs arranged in N- and C-terminal globular domains. Upon binding Ca2+, CaM changes conformation from a closed, Ca2+-free state (apoCaM) to an extended Ca2+/CaM conformation with high affinity for a broad range of target proteins (Hoeflich and Ikura, 2002; Bouché et al., 2005; DeFalco et al., 2009; Poovaiah et al., 2013). This structural flexibility, along with the ability of some proteins to interact with CaM independently of Ca2+, allows CaM to regulate numerous protein targets in diverse signaling pathways (Crivici and Ikura, 1995; Yamniuk and Vogel, 2004). Mammalian CNGCs possess diverse sites for CaM binding, with at least nine CaMBDs found across both the N and C termini of the six different CNGC isoforms (Ungerer et al., 2011). However, to date, experiments have provided evidence of a functional role for only the N-terminal CaMBDs (Liu et al., 1994; Weitz et al., 1998; Trudeau and Zagotta, 2002; Zheng et al., 2003; Song et al., 2008), particularly the N-terminal LQ site of the regulatory CNGB1 subunit (Ungerer et al., 2011). Generally, CaM is hypothesized to function in the feedback regulation of CNGCs by binding to one or more CaMBD(s) at elevated cytosolic Ca2+ levels and allosterically inhibiting CNGC conductance (Zheng et al., 2003; Trudeau and Zagotta, 2002, 2004; Bradley et al., 2001, 2004; Ungerer et al., 2011; Liu et al., 1994; Chen and Yau, 1994; Weitz et al., 1998; Song et al., 2008).
Initial studies on individual isoforms from barley (Hordeum vulgare) (Schuurink et al., 1998), tobacco (Nicotiana tabacum) (Arazi et al., 1999), and Arabidopsis thaliana (Köhler and Neuhaus, 2000) confirmed that plant CNGCs are also CaM-binding proteins. However, in contrast to animal isoforms, a single CaMBD was mapped to a site that overlaps the C-terminal α-helix (αC) of the CNBD (Arazi et al., 2000; Köhler and Neuhaus, 2000). This αC CaMBD is characterized by a four-residue, Trp-Arg-Thr-Trp (WRTW) motif required for CaM binding (Arazi et al., 2000), which is widely conserved across the Arabidopsis CNGC family (Chin et al., 2010). Previous studies have hypothesized that the location of the CaMBD within the CNBD allows CaM to compete with cyclic nucleotide monophosphate as a ligand in the allosteric gating of channel conductance (Hua et al., 2003; Kaplan et al., 2007; Swarbreck et al., 2013). It was originally suggested that this site is the only CaMBD in plant CNGCs, while sequence conservation of this region varies considerably between CNGC isoforms (Zelman et al., 2012). Recently, Arabidopsis CNGC20 was found to bind CaM via a distinct isoleucine-glutamine (IQ) motif adjacent to but not overlapping the αC-helix (Fischer et al., 2013), suggesting that plant CNGCs, like mammalian CNGCs, may possess a variety of CaMBDs. A long-standing hypothesis holds that plant CNGCs, like animal isoforms, are negatively regulated by CaM (Hua et al., 2003; Kaplan et al., 2007), but direct in planta evidence to support this is currently lacking.
Though the importance of Ca2+ signaling in immunity is well established, the molecular components involved in generating and regulating Ca2+ signals during defense responses are only beginning to be characterized (Seybold et al., 2014). Furthermore, despite the importance of CNGCs in plant biology, as implied by the expanded size of CNGC gene families and phenotypes of some CNGC mutants, very little is known regarding the structure-function of these channels. Individual CNGC isoforms have been implicated in immune signaling, including the positive regulator of Arabidopsis immunity, CNGC12 (Yoshioka et al., 2006; Moeder et al., 2011). CaM is also involved in immunity, wherein it plays both positive and negative regulatory roles (Cheval et al., 2013; Poovaiah et al., 2013). Thus, CNGCs represent a poorly studied junction between Ca2+, CaM, and immunity. To further understand the regulation and physiological roles of CNGCs, we undertook a thorough characterization of the CaM binding properties and function of CNGC12. In this work, we demonstrate that CNGC12 has multiple CaMBDs at both its cytosolic N and C termini, including both Ca2+-dependent and -independent sites, and which mediate both positive and negative regulation of channel function, revealing complexity in CNGC regulation by Ca2+.
RESULTS
At-CNGC12 Contains CaMBDs at Both Its N and C Termini
To characterize the regulation of CNGCs by CaM during immunity, we investigated the CaM binding properties of CNGC12. The region corresponding to the conventional αC CaMBD within the CNBD of CNGC12 is poorly conserved relative to other members of the CNGC family in Arabidopsis (Zelman et al., 2012). Thus, we empirically investigated the CaM binding properties of this region and searched for novel CaMBDs in CNGC12 (see Supplemental Figure 1 for delineation data). A recombinant 6xHis-tagged fragment corresponding to the predicted cytosolic C terminus of the channel (CNGC12358-649) clearly bound HRP-CaM in our overlay assay. Seven truncated fragments of this region were subsequently expressed and assayed for CaM binding (Supplemental Figure 1). In contrast with studies of other plant CNGCs, our delineations revealed that an alternative CaMBD exists outside of the CNBD, as a region C-terminal to the CNBD (CNGC12561-626) was able to bind CaM. This suggested the presence of a novel CaMBD within the C terminus of CNGC12. Our assays showed that a minimal region of amino acids 595 to 626 contained the novel CaMBD; notably, this region had not been previously characterized in any plant CNGC. Concomitant with the C-terminal delineation, the cytosolic N-terminal region of CNGC12 (amino acids 1 to 43) was expressed as a GST fusion and assayed for CaM binding. No N-terminal CaMBDs have been previously reported among plant CNGC isoforms; however, we observed clear CaM binding in our overlay assay with this N-terminal region. Following these in vitro delineations, we performed in silico analyses to predict putative CaMBD motifs within these empirically derived regions. On the basis of sequence, electrostatics, and helical propensity (Yap et al., 2000), two putative motifs were predicted, which we named NT (CNGC1217-42) and CT (CNGC12600-623). In addition to the sites identified with our binding assays, a putative IQ motif was predicted at CNGC12568-588; this site is highly conserved across the entire Arabidopsis CNGC family and has been validated as a CaMBD in the case of CNGC20 (Fischer et al., 2013). The positions of the NT, IQ, and CT CaMBDs are shown in Figure 1A.
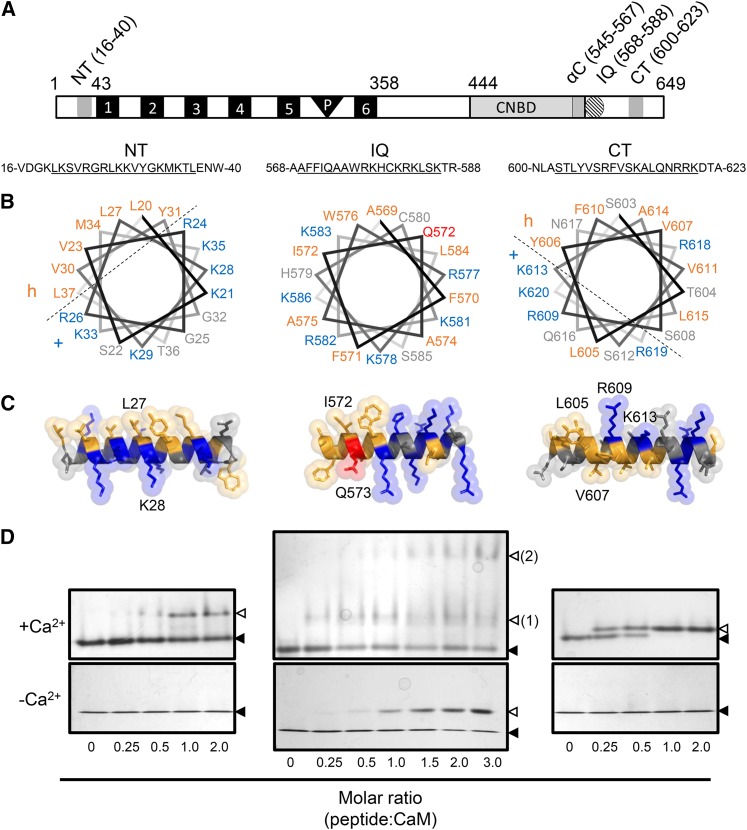
Figure 1.
Structural Modeling and ND-PAGE Mobility Shift Assay of CNGC12 CaMBD Peptides.
(A) Location of the CaMBDs of CNGC12 (see delineation of NT and CT sites in Supplemental Figure 1). Numbers indicate amino acid position. 1 to 6, Transmembrane helices; P, pore region.
(B) Helical wheel projections of motifs for the NT, IQ, and CT peptides (underlined sequences). Dashed lines separate proposed hydrophobic (h) and basic (+) faces of the NT and CT wheels.
(C) 3D model of full-length CaMBD peptides. Specific residues used for mutant analysis of each motif in this work are indicated in each model. In both (B) and (C), hydrophobic and basic residues are colored orange and blue, respectively, while the Q573 residue of the IQ motif is also colored (red) due to its suspected involvement in CaM binding.
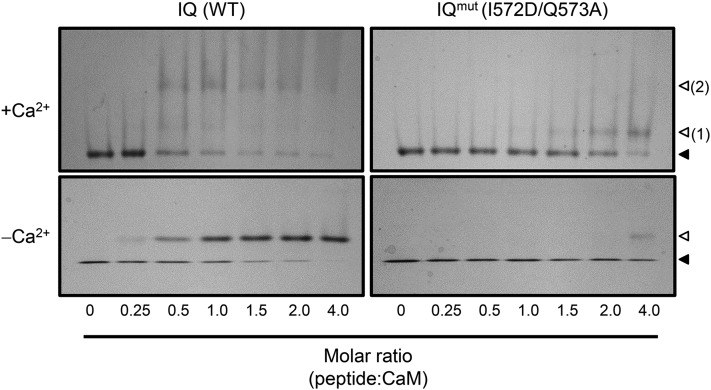
(D) ND-PAGE mobility shift assays in the presence of 0.1 mM CaCl2 (top panels) or 2 mM EGTA (bottom panels). Closed triangles indicate the migration size of CaM alone, while open triangles indicate the migration size of CaM-peptide complex. Two distinct IQ-CaM complexes consistently migrated separately in Ca2+/CaM assays with IQ peptide (top panel, numbered 2); such a pattern was never observed with NT or CT peptides or in any apoCaM assays.
Prediction of CNGC12 secondary structure using the PSIPRED server (Jones, 1999; Buchan et al., 2013) suggested that the IQ and CT motifs form helices, while the cytosolic N terminus of the channel was predicted to exist in a random coil orientation. However, given the ability of CaM binding to induce helical conformation in target sequences (Yamniuk and Vogel, 2004), the NT, IQ, and CT regions were all modeled as helices and visualized as helical wheels (rzlab.ucr.edu/) and three-dimensional helices (PyMOL version 1.3) (Figures 1B and 1C). The NT and CT models produced amphipathic helices, with hydrophobic and basic faces characteristic of Ca2+-dependent CaMBDs (Yamniuk and Vogel, 2004), while the IQ motif lacked such clear amphipathic character (Figures 1B and 1C). The CNGC12 IQ motif did not bind CaM in our overlay assays (Supplemental Figure 1), which were performed under stringent conditions that may not detect relatively weak binding.
To characterize these sites in greater detail, we obtained synthetic peptides corresponding to each site (Supplemental Table 1). Each of these peptides exhibited CaM binding in our nondissociating PAGE (ND-PAGE) assays (Figure 1D). The NT and CT peptides caused a shift in the mobility of CaM only in the presence of Ca2+, while the IQ peptide was able to cause a shift in the mobility of both Ca2+/CaM and apoCaM. However, we consistently observed multiple complexes formed between the IQ peptide and CaM that migrated separately and were observed only in the presence of Ca2+, suggesting that the IQ peptide interactions with Ca2+/CaM and apoCaM have different biophysical profiles, whereby Ca2+ induces the formation of a higher order complex between the IQ peptide and CaM (Figure 1D), though the nature of this complex remains unknown.
The NT, IQ, and CT Motifs Bind CaM with Different Biophysical Profiles
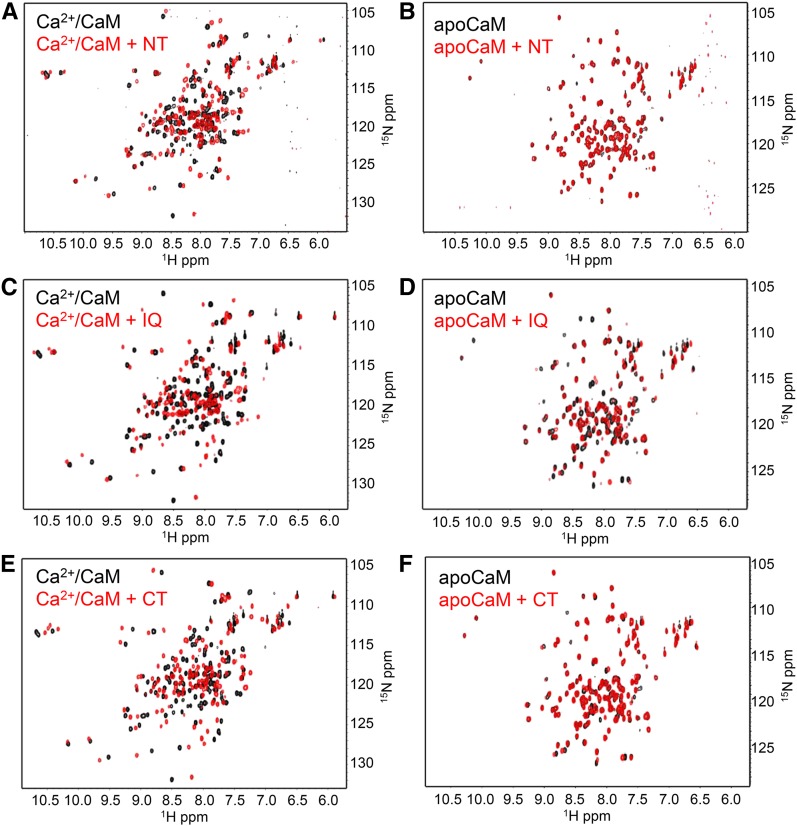
To further examine the interactions of these CaMBDs with CaM, we performed 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectroscopy on uniformly 15N-labeled CaM in the absence and presence of CaMBD peptide. Each of the three peptides induced chemical shift perturbations in the spectra of 15N-Ca2+/CaM, clearly indicative of an interaction with CaM (Figures 2A, 2C, and 2D). These results further corroborated our ND-PAGE findings, as both the NT and CT peptides caused minimal chemical shift perturbations in the spectra of 15N-apoCaM (Figures 2B and 2F), while the IQ peptide induced a clear shift in the apoCaM spectra (Figure 2D). Taken together, these findings suggested that the NT and CT are Ca2+-dependent CaMBDs, while the IQ motif may be a Ca2+-independent CaMBD.
Figure 2.
HSQC-NMR Spectroscopic Analysis of CaM-Peptide Interactions.
Overlaid spectra of uniformly 15N-labeled CaM in the absence (black) or presence (red) of equimolar peptide. In all assays, spectra of 0.2 mM 15N-CaM were collected in either Ca2+ buffer (10 mM HEPES, 100 mM NaCl, and 5 mM CaCl2, pH 7.5) or apo buffer (10 mM HEPES, 100 mM NaCl, and 1 mM EGTA, pH 7.5), as indicated. Spectra of Ca2+/CaM (A) or apoCaM +/− NT peptide (B). Spectra of Ca2+/CaM (C) or apoCaM +/− IQ peptide (D). Spectra of Ca2+/CaM (E) or apoCaM +/− CT peptide (F).
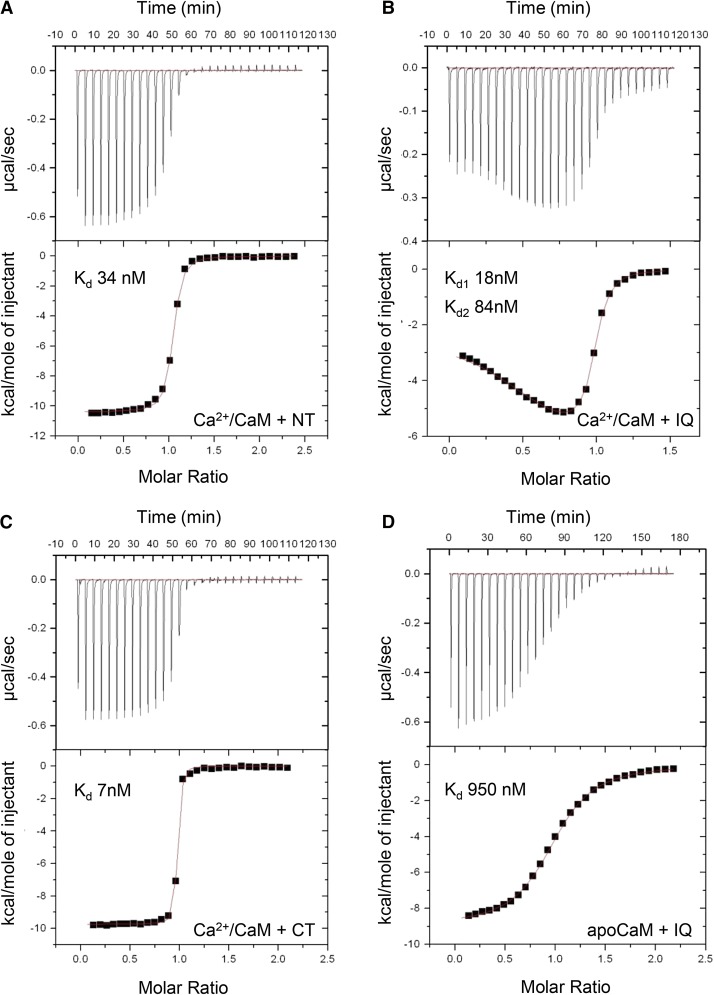
To determine the binding affinities of each interaction, isothermal titration calorimetry (ITC) was performed with each of the CaMBD peptides. In agreement with our previous results, the NT, IQ, and CT peptides each exhibited high-affinity interactions with Ca2+/CaM with 1:1 stoichiometries (Figures 3A, 3B, and 3C). Of these three CaMBDs, the CT had the highest affinity, with a measured Kd of 7 nM (Figure 3C), whereas the measured affinity for the NT peptide was ∼5-fold lower (Kd 34 nM; Figure 3A). In the case of the IQ peptide, we also performed titrations into apoCaM (Figure 3D). Titration of IQ peptide into either Ca2+/CaM or apoCaM each gave clear binding data with overall 1:1 molar stoichiometry; however, the IQ peptide bound Ca2+/CaM with higher affinity and the data were best fitted to a two-site model (Kd1 18 nM and Kd2 84 nM), while the IQ-apoCaM data modeled as a lower affinity interaction (Kd 950 nM). Parameters calculated from these ITC measurements are shown in Supplemental Table 2.
Figure 3.
ITC Analysis of Peptide-CaM Interactions.
For all experiments, calorimetric titrations (top panels) and the least-squares fitted model of binding are shown (bottom panels). Calculated Kd values are shown for each modeled binding site.
(A) Titration of 150 µM NT peptide into 16 µM CaM in the presence of 5 mM CaCl2.
(B) Titration of 205 µM IQ peptide into 20 µM CaM in the presence of 5 mM CaCl2.
(C) Titration of 150 µM CT peptide into 16 µM CaM in the presence of 5 mM CaCl2.
(D) Titration of 138 µM IQ peptide into 20 µM CaM in the presence of 1 mM EGTA.
Mutations Disrupt Site-Specific Binding of CaM to CNGC12
To determine residues required for CaM binding within these sites, residues within the NT, IQ, and CT motifs were selected for mutagenesis based on the helical wheel projections shown in Figure 1B. Hydrophobic and/or basic residues were mutated to acidic residues (Glu or Asp) to disrupt CaMBD interactions with the hydrophobic clefts and acidic residues of CaM.
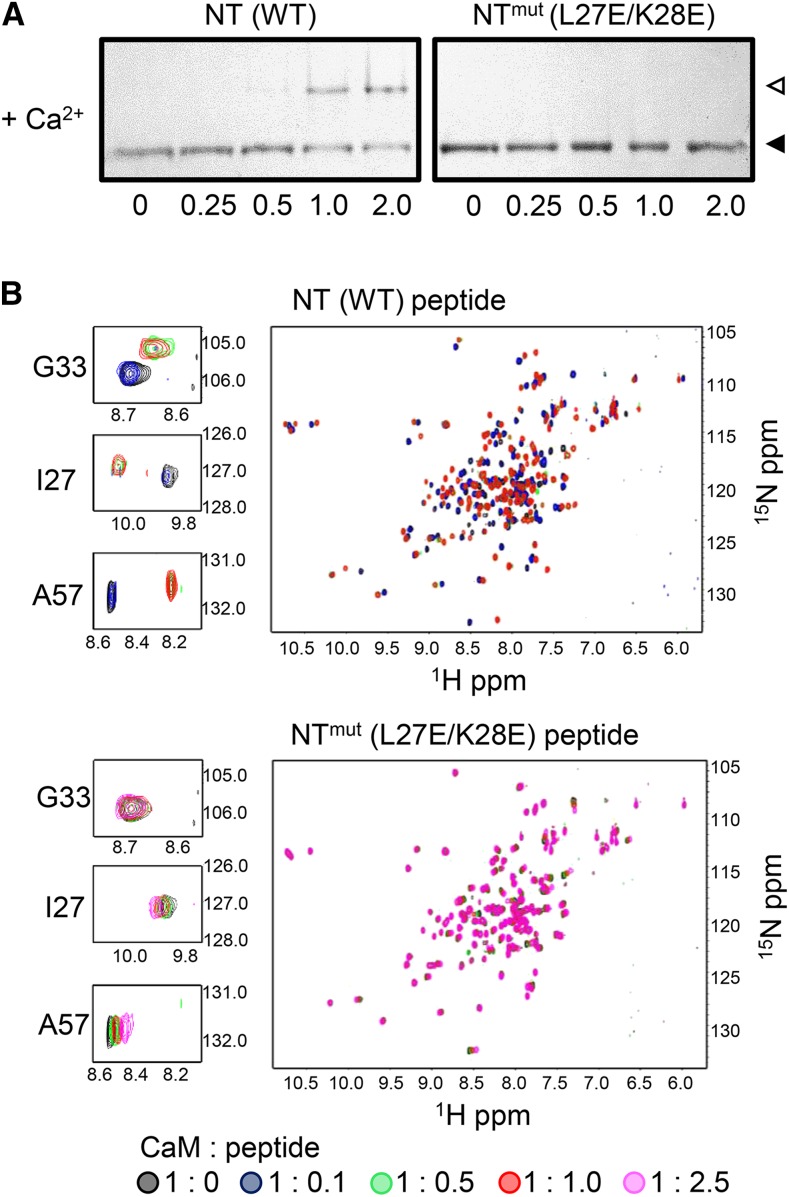
In the case of the NT site, a double substitution mutant (CNGC12L27E/K28E) was designed to disrupt both the hydrophobic and basic faces of the helix, and a peptide corresponding to this mutant CaMBD (named NTmut) was synthesized (Supplemental Table 1). This mutant peptide did not cause a shift in the mobility of CaM in our ND-PAGE assay (Figure 4A). The ability of NTmut peptide and wild-type NT peptide to bind CaM was further compared in detail via an HSQC-NMR titration assay (Figure 4B), in which increasing amounts of each peptide were incubated with CaM in the presence of Ca2+. The wild-type NT peptide caused chemical shift changes, exhibiting slow exchange on the chemical shift time scale in the spectra of 15N-CaM, which appear saturated at a 1:1 molar ratio, indicating high affinity binding. In contrast, addition of increasing amounts of NTmut peptide caused much smaller chemical shift changes, which exhibited fast exchange on the chemical shift time scale and did not saturate even with addition of 2.5-fold molar excess amounts of NTmut peptide, indicating that the L27E/K28E mutation substantially reduced CaM binding. This result was confirmed via ITC measurement with the NTmut peptide (Supplemental Figure 2A).
Figure 4.
The L27E/K28E Mutation Disrupts CaM Binding to the CNGC12 NT Motif.
(A) ND-PAGE of Ca2+/CaM with NT peptide or a peptide bearing the double mutation L27E/K28E (NTmut) at molar ratios indicated. Closed triangles indicate the migration size of CaM alone, while open triangles indicate the migration size of CaM-peptide complex.
(B) Overlaid 1H-15N HSQC-NMR spectra of 0.2 mM uniformly 15N-labeled CaM with increasing molar ratios (as indicated under the figure) of NT (top) or NTmut (bottom) peptide. Spectra of 0.2 mM uniformly 15N-labeled CaM in the presence of increasing molar ratios of NTmut peptide. NMR samples were prepared in 10 mM Tris-Cl, 150 mM NaCl, and 10 mM CaCl2 pH 7.0. The peaks corresponding to three individual CaM residues (Gly-33, Ile-27, and Ala-57) are shown in enlarged panels.
A two-residue substitution was also designed to disrupt CaM binding to the IQ motif, and a synthetic peptide bearing this mutation (I564D/Q565A, referred to as IQmut) was tested for CaM binding. This IQmut peptide showed reduced CaM binding in our ND-PAGE assays, suggesting that the I564D/Q565A mutation substantially reduces both apoCaM and Ca2+/CaM binding (Figure 5). This reduction in binding was quantified via ITC measurement, which showed that the affinity of IQmut peptide was significantly lower for Ca2+/CaM compared with the IQ peptide (Supplemental Figure 2B).
Figure 5.
The I572D/Q573A Mutation Disrupts CaM Binding to the CNGC12 IQ Motif.
ND-PAGE was performed with IQ (wild-type sequence) or IQmut peptide (bearing the I672D/Q573A mutation). Closed and open arrows indicate the migration of free or peptide-bound CaM, respectively, while the two distinct bands formed by the IQ-Ca2+/CaM complex are numbered 1 and 2.
The CT site lacks a canonical CaM target motif; thus, residues Leu-605, Val-607, Arg-609, and Lys-613 were each individually mutagenized to Glu to determine if any individual substitutions could disrupt CaM binding. Fragments containing the CT (CNGC12581-649) were expressed as 6xHis-tagged fusion proteins, each containing a single mutation, and assayed for CaM binding in an overlay assay. Substitution of any of these individual basic or hydrophobic residues to an acidic residue (Glu) reduced CaM binding relative to the wild type, with the V607E mutation exhibiting the most drastic reduction (Supplemental Figure 3). We further analyzed a peptide containing this mutation and observed a loss of CaM binding in our ND-PAGE assay, while ITC measurement showed this CTmut peptide had an ∼16-fold decrease in affinity for CaM relative to the CT peptide (Supplemental Figure 3).
Loss of CaM Binding to the NT Site Triggers CNGC12-Induced Programmed Cell Death in Planta
Previously, a chimeric channel comprising a fusion of the N terminus of CNGC11 and the C terminus of CNGC12 (CNGC11/12) was isolated as responsible for the phenotype of the Arabidopsis lesion-mimic mutant cpr22 (constitutive expressor of PR genes 22) (Yoshioka et al., 2001, 2006). CNGC12 is a positive regulator of immunity (Moeder et al., 2011), and CNGC11/12 was hypothesized to represent a misregulated form of CNGC12 that constitutively induces autoimmune phenotypes including hypersensitive response (HR)-like programmed cell death (PCD) in a constitutive manner (Yoshioka et al., 2006; Urquhart et al., 2007; Baxter et al., 2008). Activation of such autoimmunity can be assessed via Agrobacterium tumefaciens-mediated transient expression of At-CNGC11/12 in Nicotiana benthamiana, where CNGC11/12-GFP, but not CNGC12-GFP, can induce PCD (Yoshioka et al., 2006). As such, we used this system to evaluate whether the novel CaMBDs of CNGC12 may have a role(s) in regulating the induction of PCD.
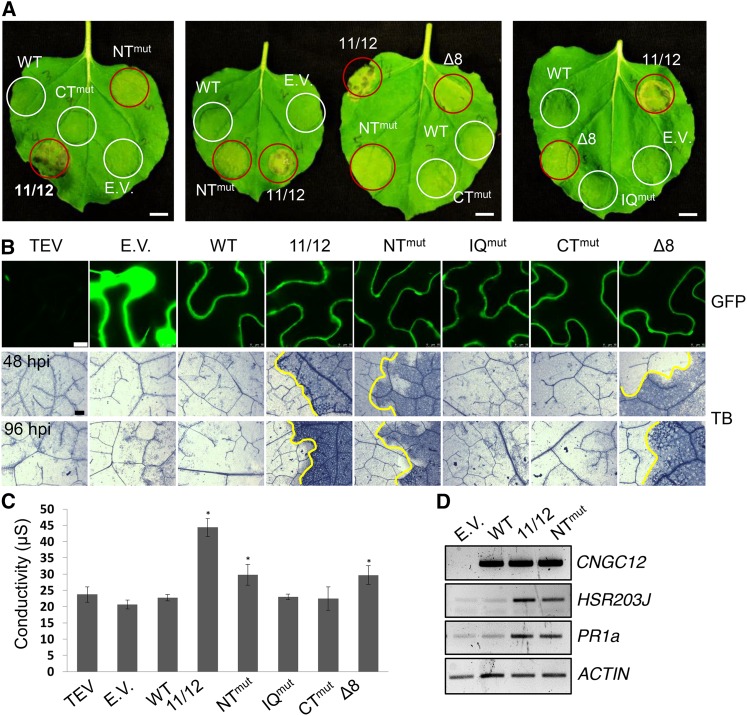
Interestingly, expression of CNGC12-GFP fusion constructs with a mutation disrupting either the IQ motif (CNGC12I572D/Q573A) or CT (CNGC12V607E) did not induce PCD, while PCD was induced by expression of the NT mutant CNGC12L27E/K28E (Figure 6A). Although induction of PCD by the NT mutant was slower relative to that induced by the chimeric CNGC11/12 channel, this cell death was never observed by the expression of WT-CNGC12 (Figure 6) and was consistently visible at the macroscopic level 4 to 5 d postinfiltration (dpi), in comparison to the PCD induced by CNGC11/12, which was consistently visible 2 to 3 dpi. CNGC12L27E/K28E induced cell death was microscopically confirmed by Trypan blue staining and quantitatively measured by ion leakage (Figures 6B and 6C, respectively), and the results of both assays confirmed the induction of PCD specifically by the NT mutant. None of the mutations significantly altered the expression levels or subcellular localization of CNGC12-GFP (Figure 6B), thus implicating loss of the N-terminal CaM binding in the misregulation of CNGC12 and activation of autoimmunity. To further confirm this result, another NT mutant, CNGC12Δ32-39 (deletion of amino acids 32 to 39), was tested. As shown in Figure 6 (as Δ8), this mutant induced PCD in a similar manner to that of the NT mutant CNGC12L27E/K28E. This eight-residue deletion is a naturally occurring polymorphism in the N terminus of CNGC11 from Arabidopsis ecotype Wassilewskija-2 (Ws-2) and, therefore, also the CNGC11/12 chimeric channel. Unlike the N terminus of CNGC12, this Δ8 N-terminal region did not bind CaM in our in vitro assays when we expressed it as a GST fusion (Supplemental Figure 4). Overall, our results with two NT site mutations in CNGC12 support our hypothesis that the NT site is required for the proper regulation of channel function.
Figure 6.
Transient Expression of NT Mutants Induces PCD in N. benthamiana.
(A) Appearance of N. benthamiana leaves at 4 dpi (bar = 1 cm). Areas were infiltrated with Agrobacterium carrying different constructs. Areas showing cell death are circled in red.
(B) GFP confocal microscopy of leaf areas 30 hpi (bar = 10 µm) and Trypan blue staining (TB) for cell death of N. benthamiana leaves expressing wild-type or mutant CNGC12 constructs at 48 and 96 hpi, as indicated (bar = 0.1 mm). Infiltrated and uninfiltrated leaf areas are separated by a yellow line in samples showing cell death.
(C) Ion leakage of infiltrated leaf areas 4 dpi. Values shown are averages of three replicates (error bars = sd; *P < 0.05 compared with empty vector, Student’s t test). E.V., empty vector (GFP); WT, At-CNGC12; 11/12, CNGC11/12; NTmut, CNGC12L27E/K28E; IQmut, CNGC12I572D/Q573A; CTmut, CNGC12V607E; Δ8, CNGC12Δ32-39.
(D) RT-PCR of cDNA from N. benthamiana leaves 24 hpi expressing wild-type or mutant CNGC12 constructs. Nb-ACTIN was used as a loading control (26 cycles).
To further corroborate a role for this CaMBD in the immune response, we analyzed cell death- and immunity-related marker genes. As shown in Figure 6D, the well established immunity-related cell death HR marker Nb-HSR203J (Pontier et al., 1994) was induced by expression of At-CNGC12L27E/K28E and At-CNGC11/12, but not by WT-At-CNGC12. In addition, the activation of immune signaling was further confirmed by the expression of N. benthamiana pathogenesis related gene 1 (Nb-PR1a). Both CNGC11/12 and CNGC12L27E/K28E induced Nb-PR1a, whereas WT-CNGC12 did not (Figure 6D). We were also able to observe more robust PCD induction by our mutants at lower temperature (Supplemental Figure 5), a hallmark of the immune response (Hua, 2013).
Taken together, these data suggest that CaM binding at the NT site negatively regulates CNGC12-mediated immunity activation.
The IQ Motif Is Required for Channel Function
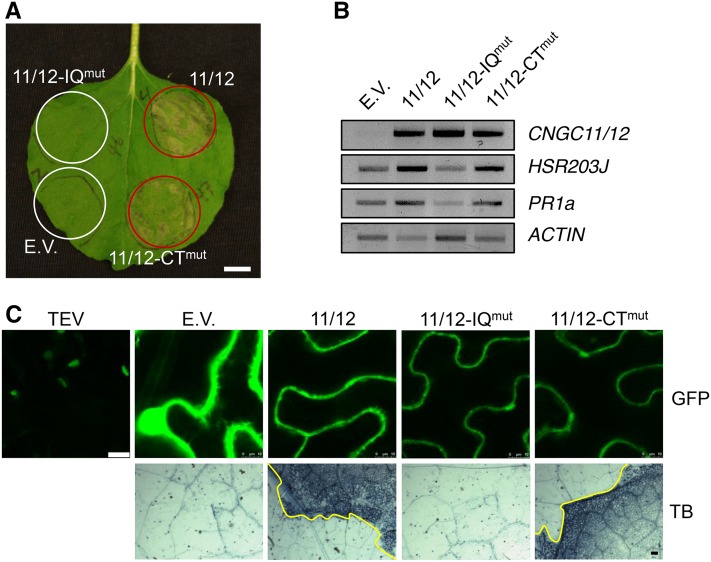
Because the chimeric CNGC11/12 is hypothesized to comprise a misregulated form of CNGC12 and constitutively activates PCD, it has proven to be an excellent tool to study channel structure-function (Baxter et al., 2008; Chin et al., 2010; Abdel-Hamid et al., 2013). CNGC11/12 possesses the C-terminal half of CNGC12 and therefore shares the CNGC12 IQ and CT motifs. We were thus able to use CNGC11/12 to study whether either of these CaMBDs influences channel function, and toward this end the CNGC12 IQmut and CTmut mutations were each introduced into CNGC11/12 [i.e., we generated 11/12-IQmut (CNGC11/12I564D/Q565A) and 11/12-CTmut (CNGC11/12V599E)], and these constructs were expressed as GFP fusions in N. benthamiana. Although no significant change in PCD induction was observed with CNGC11/12-CTmut (CNGC11/12V599E), CNGC11/12-IQmut (CNGC11/12I564D/Q565A) failed to induce PCD, indicating that disruption of the IQ site interferes with its function (Figure 7). The lack of PCD formation with 11/12-IQmut expression relative to CNGC11/12 and 11/12-CTmut was clearly visible in our N. benthamiana assays at both the macroscopic and microscopic levels (Figures 7A and 7C). The lack of PCD was also mirrored in the absence of Nb-HSR203J and Nb-PR1a induction (Figure 7B). These findings were unexpected given that CaM binding has previously been posited to provide only a negative feedback mechanism for CNGCs. To further corroborate our in planta findings, we performed the same PCD experiments using myc-tagged constructs and observed similar results (Supplemental Figure 6).
Figure 7.
The IQ Motif Is Required for CNGC11/12-Induced PCD.
(A) N. benthamiana leaf 4 dpi (bar = 1 cm).
(B) RT-PCR of cDNA from N. benthamiana leaves 24 hpi expressing CNGC11/12 or site-directed mutant constructs. Nb-ACTIN was used as a loading control (26 cycles).
(C) GFP confocal microscopy of N. benthamiana leaves 30 hpi (bar = 10 µm) and Trypan blue (TB) staining for cell death in N. benthamiana leaves 3 dpi (bar = 0.1 mm). E.V., empty vector (GFP); 11/12, CNGC11/12; 11/12-IQmut, CNGC11/12I564D/Q565A; 11/12-CTmut, CNGC11/12V599E.
We also tested a peptide bearing the CNGC11/12 S58 mutation (R577C, which is adjacent to the IQ motif) for CaM binding in our NMR assay and found that this peptide could partially bind CaM under reducing conditions (Supplemental Figure 7). The R577C mutation was previously found to suppress CNGC11/12 PCD induction at normal temperatures, while PCD was restored at low temperature (Chin et al., 2010). While we again observed CNGC11/12R577C-induced PCD at lower temperature, 11/12-IQmut failed to induce PCD even at low temperature (Supplemental Figure 8), further indicating that this IQ site mutation leads to a full loss-of-function phenotype. Taken together, these data revealed the complex regulation of CNGC12 by multiple CaMBDs and demonstrated that CaM and Ca2+ are involved in different aspects of channel function and regulation.
DISCUSSION
The mechanisms of mammalian CNGC allosteric regulation are well characterized, whereby binding of the cyclic nucleotide monophosphates cAMP and/or cGMP to the C-terminal CNBD provide a gating mechanism for channel conductance (Kaupp and Seifert, 2002; Matulef and Zagotta, 2003). Allosteric inhibition of mammalian CNGC function is in turn regulated by CaM binding, which decreases the affinity of the channel for cAMP/cGMP (Liu et al., 1994; Chen and Yau, 1994; Trudeau and Zagotta, 2004; Bradley et al., 2004; Ungerer et al., 2011). Mammalian CNGCs isoforms possess CaMBD(s) in their cytosolic N and/or C termini; however, the functional data appear to be limited to N-terminal CaMBDs (Ungerer et al., 2011).
By contrast, plant CNGC function has been largely inferred from genetic studies, which have implicated various CNGCs in diverse physiological processes, including immunity (reviewed in Moeder et al., 2011; Ma and Berkowitz, 2011), while CNGC structure-function has proven recalcitrant to direct study. Early studies with select isoforms led to the hypotheses that Ca2+/CaM provides allosteric negative regulation of plant CNGC function and that a single CaMBD is conserved within the αC-helix of every plant CNGC (Arazi et al., 2000; Hua et al., 2003; Köhler and Neuhaus, 2000; Li et al., 2005; Kaplan et al., 2007); however, discovery-driven investigations into how CaM interacts with and regulates various plant CNGC isoforms have been lacking. CNGC12 (specifically) had been previously suggested to not bind CaM due to low conservation at its αC-helix region (Zelman et al., 2012); however, our study demonstrates that this isoform possesses novel CaMBDs at both its cytosolic N and C termini, providing evidence that an individual plant CNGC isoform possesses multiple CaMBDs.
The structure of the CNGC12 C-linker and CNBD regions was previously modeled (Chin et al., 2010; Abdel-Hamid et al., 2013) based on homology to the mammalian channels SpIH and HCN2 (PDB ID: 2PTM and PDB ID:IQ50, respectively) (Flynn et al., 2007; Zagotta et al., 2003). Such models have provided important insight into CNGC structure-function; however, they could not include the C-terminal region downstream of the CNBD due to lack of homology to mammalian templates (i.e., amino acids ∼565 to 649 in At-CNGC12). As such, modeling could not incorporate two CaMBDs (IQ and CT motifs). Thus, while sequence-based interrogations of CNGC structure-function are useful, our study emphasizes the value of empirical analyses and suggests that CaMBDs in other CNGCs across taxa may have been overlooked hitherto. In addition to delineating these novel CaMBDs, our biochemical and biophysical assays of each site have demonstrated that the three CaMBDs of CNGC12 interact differently with CaM, as the NT, IQ, and CT motifs each bind CaM with different affinities and Ca2+ dependency. While our experiments have displayed 1:1 molar ratios for the interactions between each of the NT, IQ, and CT peptides with CaM, the overall stoichiometry of CaM to full-length CNGC12 in vivo is unknown, though we speculate that it may vary depending on the presence of Ca2+. It is unclear in our current model whether CaM binds to the various CNGC12 CaMBDs independently or in a competitive manner. Thus, it is likely that apoCaM is preloaded at the IQ motif at basal Ca2+ levels and that elevation of cytosolic Ca2+ induces recruitment of both preloaded and cytosolic Ca2+/CaM to the three CaMBDs. Similar mechanisms are present in numerous vertebrate cation channels (Kovalevskaya et al., 2013), but their presence in plants was previously unclear.
Transient heterologous expression of chimeric At-CNGC11/12 in N. benthamiana triggers Ca2+-dependent spontaneous HR-like PCD, suggesting that CNGC11/12 is a constitutively active and/or misregulated form of CNGC12, which is a positive regulator of immunity (Baxter et al., 2008; Urquhart et al., 2007). Low temperature is known to enhance immune responses in plants (Hua, 2013), and shifting to lower temperature has been previously shown to enhance the PCD phenotype induced by CNGC11/12 mutants (Mosher et al., 2010; Chin et al., 2010). We observed a similar effect in our assays, wherein incubation at 16°C increased the PCD induced by both of our NT mutant CNGC12 constructs (Supplemental Figure 5). Together with our RT-PCR, these data suggest that disregulation of CNGC12 due to loss of CaM binding at the NT site leads to the activation of defense responses.
The discovery that CNGC12 possesses an N-terminal CaMBD was unexpected given long-standing hypotheses regarding the location of a C-terminal CaMBD in plant CNGCs. However, our results did implicate this novel NT site in the inhibition of channel function, providing in planta validation of the hypothesis that CaM provides negative feedback regulation. In contrast, our finding that the IQ motif is required for CNGC11/12-induced PCD suggests that CaM binding is also required for channel function. This discovery stands in contrast to the long-held expectation that CaM functions solely in the negative feedback inhibition of CNGC function. These data provide new context for our previous finding that the loss of the C-terminal region (including the IQ motif) due to a premature stop codon lead to complete loss of CNGC11/12-induced phenotypes and channel function in yeast complementation assays (Chin et al., 2010). Interestingly, the same study also showed a point mutation adjacent to the IQ motif (cpr22 suppressor 58, S58, caused by the point mutation CNGC11/12 R577C) was able to partially suppress CNGC11/12 function (Chin et al., 2010). When we performed NMR assays with an extended peptide bearing this R577C mutation that overlapped with the IQ motif, we observed no interaction with Ca2+/CaM; however, binding was partially restored under reducing conditions (Supplemental Figure 7). We previously reported that S58 was only a partial loss-of-function mutation in CNGC11/12, as PCD induction could be restored at low temperature (Chin et al., 2010). In contrast, 11/12-IQmut was still unable to induce any cell death at low temperature (Supplemental Figure 8), in keeping with our hypothesis that the S58 mutation partially reduced the accessibility of CaM to the IQ motif, while 11/12-IQmut directly disrupted CaM binding. Taken together, our data suggest that the IQ motif is essential for CNGC11/12-induced phenotypes and that indirect or direct disruption of this CaMBD leads to corresponding partial or full loss of channel function.
According to our ND-PAGE assay, the IQmut mutation that suppressed CNGC11/12-induced PCD disrupted apoCaM binding more severely than Ca2+/CaM binding, as we were only able to observe a shift in the migration of apoCaM with the addition of a high concentration of IQmut peptide (Figure 5). We postulate that CaM (via constitutive association with the IQ motif) comprises a subunit of the functional CNGC complex in vivo, though further work will be needed to determine whether this is specific to CNGC12 or more broadly applicable across plant CNGCs. As noted previously, the IQ motif is conserved at the sequence level across the At-CNGC family (Fischer et al., 2013). Such constitutive association between CaM and IQ motifs is a widespread phenomenon among animal cation channels (Kovalevskaya et al., 2013) but had not previously been confirmed in plants. Our data suggest that CaM may be responsible for Ca2+-dependent intra- and/or intersubunit interactions, another feature thought to be specific to animal cation channels (Fallon et al., 2009; Liu and Vogel, 2012; Sarhan et al., 2012). This is an interesting avenue for investigation, given that homo- or heterotetramers are hypothesized to comprise functional CNGCs in vivo (Kaplan et al., 2007; Zelman et al., 2012), though to date we have only minimal data regarding how different plant CNGC subunits associate (Chin et al., 2013). We observed no detectable effect of mutation at the CT motif on the induction of PCD by CNGC12 or 11/12. It is possible that the V607E mutation does not completely abolish CaM binding in planta. Indeed, ITC measurements show that our V607E mutation significantly reduces CaM binding but does not completely ablate it (Supplemental Figure 3); whether mutations that have a more drastic loss of CaM binding at the CT site have effects on channel function is an interesting avenue for further exploration.
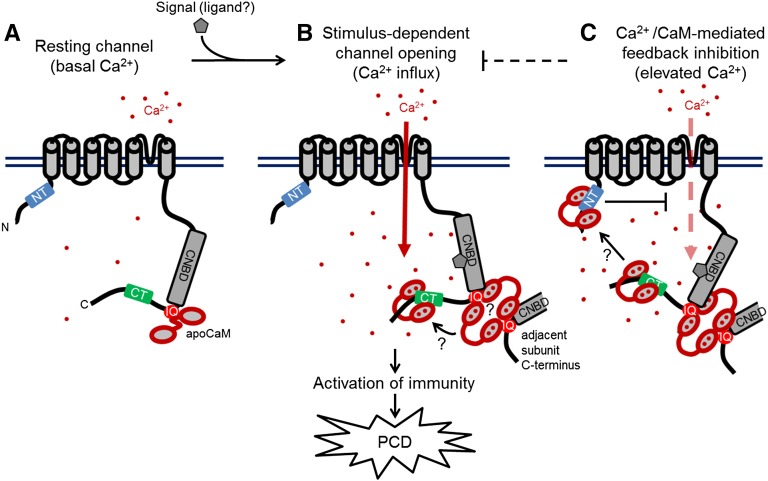
Given our biochemical and physiological dissection of its CaMBDs, we have proposed a model for the regulation of CNGC12 by CaM (Figure 8). This model posits that CNGC12 is constitutively calmodulated via the IQ motif in a Ca2+-independent manner, while Ca2+/CaM may sequentially bind to multiple CaMBDs. Our model shows CNGC12 mediating Ca2+ influx directly; however, we cannot exclude the possibility that CNGC12 signaling triggers Ca2+ current(s) mediated by other channel(s). Our model includes speculative features that are consistent with our results to date but which require further experimental investigation and some of which bear clear similarities to various animal channels, such as CaM-mediated bridging of inter- and intrasubunit interactions and dynamic movement of CaM between multiple CaMBDs (Fallon et al., 2009; Liu and Vogel, 2012; Sarhan et al., 2012; Morales et al., 2013; Ben Johny et al., 2013; Shao et al., 2014). Interestingly, such CaM regulatory features are widely distributed among various animal voltage- or Ca2+-gated cation channels for which there are no clear homologs in plants (Edel and Kudla, 2015). Generally, our combined biochemical and physiological data demonstrate interesting parallels and distinctions between plant CNGCs and animal cation channels. These results open exciting avenues for investigation, particularly the possibility that plant CNGC isoforms comprise both functional and regulatory subunits, as is the case among the animal CNG family (Matulef and Zagotta, 2003), and that the regulatory role of specific subunits may be at least partially determined by the orientation and binding properties of CaMBD(s). As such, it will be informative to determine how different combinations of CaMBD(s) are conserved in other plant CNGC isoforms. Recent work has demonstrated that in planta assays can be used to test pairwise combinations of CNGC isoforms for interaction (Chin et al., 2013) and that some plant CNGC isoforms are amenable to electrophysiological study (Zhou et al., 2014; Gao et al., 2014). The combination of additional structural-functional, physiological, and electrophysiological studies will help to elucidate the complex mode of CaM-mediated CNGC regulation by Ca2+. Given the important roles of CNGCs and CaM in immunity (Moeder et al., 2011; Poovaiah et al., 2013), further characterizing CaM-CNGC interactions will be critical to understanding CNGC-mediated Ca2+ signaling during immune responses.
Figure 8.
Proposed Model of CNGC12 Regulation by CaM.
The model draws from the biophysical and physiological results of this study, previous data cited in the Discussion, and speculative components that currently remain unexplored, but which are compatible with our findings to date.
(A) At resting conditions and basal Ca2+ levels, the channel is calmodulated via interaction between CaM and the IQ motif.
(B) Upon perception of a signal (the molecular nature of which remains to be empirically determined), the channel opens and cytosolic Ca2+ levels increase. Given the necessity of CaM binding to the IQ motif for channel function as well as the higher-order interactions that occur between the IQ motif and Ca2+/CaM, channel activation may require CaM-dependent interactions between adjacent channel subunits as depicted. Furthermore, current data do not resolve whether individual CaM molecules move between different CaMBDs in a Ca2+-dependent manner or multiple CaMs can bind sites independently.
(C) We propose that at higher Ca2+ concentration, Ca2+/CaM provides inhibition of CNGC12 function via the NT CaMBD; whether this inhibition involves the bridging of channel termini remains unclear.
METHODS
Plasmid Construction and Recombinant Protein Expression
All in vitro experiments were performed with a conserved plant CaM isoform (petunia [Petunia hybrida] CaM81, 100% amino acid identity to Arabidopsis thaliana CaM7), with the exception of HSQC-NMR spectroscopy experiments, which were performed with a conserved vertebrate isoform from Xenopus laevis (xCaM, 90%/97% identity/similarity to At-CaM7). Untagged CaM81 was expressed using the pET5a vector in BL21 pLysS Escherichia coli strain (Fromm and Chua, 1992), whereas xCaM was expressed using pAS vector in the AR58 strain (Ikura et al., 1990b) and both purified via phenyl-Sepharose hydrophobic interaction chromatography. CNGC12 fragments were expressed using either the pET28 or pGEX4T3 vectors (for 6xHis- or GST-tagged fusion proteins, respectively, as indicated in the figures) in BL21 (DE3) codon plus E. coli cells. Constructs for in planta expression as GFP fusions were cloned into the pJP (pMBP3) binary plasmid (Baxter et al., 2008) in frame with a C-terminal GFP tag. Site-directed mutagenesis was performed in vitro using overlapping PCR reactions. For myc-tagged constructs, CNGC12 (wild type or mutants) were cloned into the pGreen binary vector (Hellens et al., 2000) in frame with a C-terminal myc epitope. The fidelity of all constructs was confirmed by DNA sequencing. Primers used for cloning and mutagenesis are listed in Supplemental Table 3.
HRP-CaM Overlay Assays
Horseradish peroxidase-conjugated CaM (HRP-CaM) was generated and used as a probe as described previously (DeFalco et al., 2010).
In Silico CaMBD Modeling
The amino acid sequence corresponding to each of the CaMBD peptides (shown in Supplemental Table 1) was individually modeled as a perfect α-helix using the PyMOL Molecular Graphics System, version 1.3 (Schrödinger). The side chains of specific residues in each model were colored as indicated in Figure 1.
ND-PAGE
Peptides and CaM were each diluted into sterile 25 mM Tris-Cl (pH 7.5) to a final concentration of 1 mg/mL and subsequently used in CaM binding ND-PAGE gel shift assays as described previously (DeFalco et al., 2010).
ITC
ITC was performed using a VP-ITC calorimeter (Microcal) at 30°C in 25 mM HEPES, 100 mM NaCl, pH 7.5, with 5 mM CaCl2 or 1 mM EGTA as indicated. In all experiments, peptide was titrated into CaM in 29 injections of 10 μL each with a 360-s equilibration between injections. In the case of NT and CT experiments, 150 μM peptide was titrated into 16 μM CaM, while in the case of IQ experiments, 135 μM peptide was titrated into 20 μM CaM. Data were analyzed and fitted using Origin 7.0 software (OriginLab).
1H-15N HSQC-NMR Spectroscopy
15N-labeled recombinant CaM was expressed in E. coli and purified to homogeneity as previously described (Ikura et al., 1990a). Lyophilized CaM was dissolved in buffer in the presence or absence of Ca2+ as described in the figure captions. Synthetic peptides were dissolved in buffer (corresponding to figure captions) without Ca2+ or EGTA and washed with excess buffer using Amicon Ultracel MWCO 3000 spin columns (Millipore) to equilibrate pH before being added to CaM stocks (at the ratios indicated in the figure captions). Combined samples were washed again and concentrated to a final volume of 35 µL, resulting in a final concentration of 15N-CaM of 0.2 mM. 15N -1H HSQC spectra were collected with 16 scans using a Bruker 600-MHz Avance II NMR spectrometer equipped with a 1.7-mm microcryoprobe.
Plant Growth Conditions and Agrobacterium-Mediated Transient Expression
Nicotiana benthamiana was grown on Sunshine mix soil (Sun Gro Horticulture Canada) in a growth chamber under a 9/15-h light/dark regimen at 22°C (day) and 20°C (night) (60% relative humidity and ∼140 µE m−2 s−1). Low-temperature treatments were conducted under the same conditions at 16°C. Transient expression was performed via infiltration of N. benthamiana with Agrobacterium tumefaciens strain GV2260 as described previously (Urquhart et al., 2007). Agrobacterium carrying CaMV35S:HC-Pro from Tobacco etch virus (labeled TEV in the figures) was infiltrated alone or coinfiltrated with constructs as described in the figure captions.
Trypan Blue Staining
Trypan blue staining for cell death detection was performed as described previously (Abdel-Hamid et al., 2013; Yoshioka et al., 2006).
Ion Leakage Analysis
In accordance with an established protocol (Abdel-Hamid et al., 2013), three leaf discs (1 cm each, from independent leaves) were floated in 4 mL of deionized water for 15 min and the conductivity of each sample was determined using an Oakton Con5 Acorn series conductivity meter (Oakton Instruments). Assays were performed in triplicate and values were averaged.
RNA Extraction and RT-PCR
RNA extraction and cDNA synthesis were performed as described previously (Abdel-Hamid et al., 2013). RT-PCR analysis of At-CNGC12 and N. benthamiana ACTIN, HSR203J, and PR1a transcripts was performed using primers shown in Supplemental Table 3.
Confocal Microscopy
Confocal microscopy as performed described previously with minor changes (Abdel-Hamid et al., 2013). Briefly, 1-cm sections of Agrobacterium-infiltrated N. benthamiana leaves were excised ∼30 h postinfiltration (hpi) for microscopy using the Leica TCS SP8 confocal system (Leica Microsystems). GFP (500 to 600 nm) or chloroplast autofluorescence (650 to 700 nm) was detected under the 63× oil immersion objective lens (numerical aperture 1.40) with 3× zoom using the 488-nm OPSL laser set to 25%.
Experimental Replicates
Experiments were performed a minimum of three times and representative results are displayed in the figures (with the exception of ITC measurements, which are representative of at least two replicates). Where appropriate, measurements have been averaged and subjected to statistical analysis as indicated in the figure captions.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At-CNGC12 (At2g46450, ACD81988), At-CNGC11 (At2g46440, ACD81989), Nb-PR1a (Niben101Scf00107g03008.1), Nb-ACTIN (NbS00000307g0001.1), Nb-HSR203J (Niben101Scf05283g00016.1), CaM81 (M80836), xCaM (NP_001080864.1), At-CaM7 (At3g43810), At-CaM4 (At1g66410), and At-CML9 (At3g51920).
Supplemental Data
Supplemental Figure 1. N- and C-terminal regions of At-CNGC12 possess CaMBDs.
Supplemental Figure 2. ITC analysis of the NTmut (L27E/K28E) and IQmut (I672D/Q573A) peptides.
Supplemental Figure 3. The V607E mutation disrupts CaM binding to the CT motif.
Supplemental Figure 4. The Δ8 mutation (CNGC12 amino acids 32 to 39) disrupts the CaM binding to the N terminus of CNGC12.
Supplemental Figure 5. PCD induction by NT mutants is increased at lower temperature.
Supplemental Figure 6. Phenotypic analysis of myc-tagged At-CNGC12 construct expression in N. benthamiana.
Supplemental Figure 7. The S58 mutant of CNGC11/12 (R577C) partially disrupts CaM binding to the IQ motif.
Supplemental Figure 8. Low temperature does not restore PCD induction by CNGC11/12-IQmut.
Supplemental Table 1. CNGC12 CaMBD peptides.
Supplemental Table 2. Calculated parameters from ITC measurements.
Supplemental Table 3. Primers used for experiments.
Supplementary Material
Acknowledgments
We thank Huda Abdel-Hamid for the initial data collection. This work was supported by doctoral scholarships from the Natural Sciences and Engineering Research Council (NSERC) and the Ontario government to T.A.D. and an NSERC Discovery Grant (no. 2014-04114) to K.Y. This work was partly supported by an NSERC Discovery Grant to W.A.S. and by a CIHR grant (MOP-13552) to M.I. M.I. holds a Canada Research Chair. NMR spectrometers were funded by Canada Foundation for Innovation (CFI). A Multi-disciplinary Internal Research Grant from Texas State University and National Science Foundation Grant (IOS-1553613) supported H.-G.K. The authors thank Purva Karia for assistance with microscopy and Ji Chul Nam for his technical support. pGreen plasmid was a kind gift of Raymond Zielinski (University of Illinois).
AUTHOR CONTRIBUTIONS
T.A.D., W.M., and K.Y. designed the research. T.A.D. performed most of the experiments. C.B.M. and M.I. contributed to the NMR analyses. K.M. performed ITC experiments and W.A.S. contributed to data analysis of ITC. H.-G.K. helped with cloning and transformations. T.A.D., W.M., and K.Y. analyzed the data, and T.A.D., W.M., and K.Y. wrote the article.
Glossary
- CNGC
cyclic nucleotide-gated channel
- CNBD
cyclic nucleotide binding domain
- CaMBD
CaM binding domain
- CaM
calmodulin
- ND-PAGE
nondissociating PAGE
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- HR
hypersensitive response
- PCD
programmed cell death
- dpi
days postinfiltration
- hpi
hours postinfiltration
References
- Abdel-Hamid H., Chin K., Moeder W., Shahinas D., Gupta D., Yoshioka K. (2013). A suppressor screen of the chimeric AtCNGC11/12 reveals residues important for intersubunit interactions of cyclic nucleotide-gated ion channels. Plant Physiol. 162: 1681–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T., Kaplan B., Fromm H. (2000). A high-affinity calmodulin-binding site in a tobacco plasma-membrane channel protein coincides with a characteristic element of cyclic nucleotide-binding domains. Plant Mol. Biol. 42: 591–601. [DOI] [PubMed] [Google Scholar]
- Arazi T., Sunkar R., Kaplan B., Fromm H. (1999). A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 20: 171–182. [DOI] [PubMed] [Google Scholar]
- Baxter J., Moeder W., Urquhart W., Shahinas D., Chin K., Christendat D., Kang H.G., Angelova M., Kato N., Yoshioka K. (2008). Identification of a functionally essential amino acid for Arabidopsis cyclic nucleotide gated ion channels using the chimeric AtCNGC11/12 gene. Plant J. 56: 457–469. [DOI] [PubMed] [Google Scholar]
- Ben Johny M., Yang P.S., Bazzazi H., Yue D.T. (2013). Dynamic switching of calmodulin interactions underlies Ca2+ regulation of CaV1.3 channels. Nat. Commun. 4: 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N., Yellin A., Snedden W.A., Fromm H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56: 435–466. [DOI] [PubMed] [Google Scholar]
- Bradley J., Bönigk W., Yau K.-W., Frings S. (2004). Calmodulin permanently associates with rat olfactory CNG channels under native conditions. Nat. Neurosci. 7: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J., Reuter D., Frings S. (2001). Facilitation of calmodulin-mediated odor adaptation by cAMP-gated channel subunits. Science 294: 2176–2178. [DOI] [PubMed] [Google Scholar]
- Buchan D.W., Minneci F., Nugent T.C., Bryson K., Jones D.T. (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41: W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-Y., Yau K.-W. (1994). Direct modulation by Ca(2+)-calmodulin of cyclic nucleotide-activated channel of rat olfactory receptor neurons. Nature 368: 545–548. [DOI] [PubMed] [Google Scholar]
- Cheval C., Aldon D., Galaud J.-P., Ranty B. (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta 1833: 1766–1771. [DOI] [PubMed] [Google Scholar]
- Chin K., DeFalco T.A., Moeder W., Yoshioka K. (2013). The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol. 163: 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., Moeder W., Abdel-Hamid H., Shahinas D., Gupta D., Yoshioka K. (2010). Importance of the alphaC-helix in the cyclic nucleotide binding domain for the stable channel regulation and function of cyclic nucleotide gated ion channels in Arabidopsis. J. Exp. Bot. 61: 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivici A., Ikura M. (1995). Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24: 85–116. [DOI] [PubMed] [Google Scholar]
- DeFalco T.A., Bender K.W., Snedden W.A. (2009). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425: 27–40. [DOI] [PubMed] [Google Scholar]
- DeFalco T.A., Chiasson D., Munro K., Kaiser B.N., Snedden W.A. (2010). Characterization of GmCaMK1, a member of a soybean calmodulin-binding receptor-like kinase family. FEBS Lett. 584: 4717–4724. [DOI] [PubMed] [Google Scholar]
- Dietrich P., Anschütz U., Kugler A., Becker D. (2010). Physiology and biophysics of plant ligand-gated ion channels. Plant Biol (Stuttg) 12 (Suppl 1): 80–93. [DOI] [PubMed] [Google Scholar]
- Edel K.H., Kudla J. (2015). Increasing complexity and versatility: how the calcium signaling toolkit was shaped during plant land colonization. Cell Calcium 57: 231–246. [DOI] [PubMed] [Google Scholar]
- Fallon J.L., Baker M.R., Xiong L., Loy R.E., Yang G., Dirksen R.T., Hamilton S.L., Quiocho F.A. (2009). Crystal structure of dimeric cardiac L-type calcium channel regulatory domains bridged by Ca2+ calmodulins. Proc. Natl. Acad. Sci. USA 106: 5135–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C., Kugler A., Hoth S., Dietrich P. (2013). An IQ domain mediates the interaction with calmodulin in a plant cyclic nucleotide-gated channel. Plant Cell Physiol. 54: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn G.E., Black K.D., Islas L.D., Sankaran B., Zagotta W.N. (2007). Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure 15: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm H., Chua N. (1992). Cloning of plant cDNAs encoding calmodulin-binding proteins using 35S-labeled recombinant calmodulin as a probe. Plant Mol. Biol. Report. 10: 199–206. [Google Scholar]
- Gao Q.-F., Fei C.-F., Dong J.-Y., Gu L.-L., Wang Y.-F. (2014). Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant 7: 739–743. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hoeflich K.P., Ikura M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108: 739–742. [DOI] [PubMed] [Google Scholar]
- Hua B.G., Mercier R.W., Zielinski R.E., Berkowitz G.a. (2003). Functional interaction of calmodulin with a plant cyclic nucleotide gated cation channel. Plant Physiol. Biochem. 41: 945–954. [Google Scholar]
- Hua J. (2013). Modulation of plant immunity by light, circadian rhythm, and temperature. Curr. Opin. Plant Biol. 16: 406–413. [DOI] [PubMed] [Google Scholar]
- Ikura M., Kay L.E., Bax A. (1990a). A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry 29: 4659–4667. [DOI] [PubMed] [Google Scholar]
- Ikura M., Marion D., Kay L.E., Shih H., Krinks M., Klee C.B., Bax A. (1990b). Heteronuclear 3D NMR and isotopic labeling of calmodulin. Towards the complete assignment of the 1H NMR spectrum. Biochem. Pharmacol. 40: 153–160. [DOI] [PubMed] [Google Scholar]
- Jammes F., Hu H.-C., Villiers F., Bouten R., Kwak J.M. (2011). Calcium-permeable channels in plant cells. FEBS J. 278: 4262–4276. [DOI] [PubMed] [Google Scholar]
- Jones D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202. [DOI] [PubMed] [Google Scholar]
- Kaplan B., Sherman T., Fromm H. (2007). Cyclic nucleotide-gated channels in plants. FEBS Lett. 581: 2237–2246. [DOI] [PubMed] [Google Scholar]
- Kaupp U.B., Seifert R. (2002). Cyclic nucleotide-gated ion channels. Physiol. Rev. 82: 769–824. [DOI] [PubMed] [Google Scholar]
- Köhler C., Neuhaus G. (2000). Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 471: 133–136. [DOI] [PubMed] [Google Scholar]
- Kovalevskaya N.V., van de Waterbeemd M., Bokhovchuk F.M., Bate N., Bindels R.J.M., Hoenderop J.G.J., Vuister G.W. (2013). Structural analysis of calmodulin binding to ion channels demonstrates the role of its plasticity in regulation. Pflugers Arch. 465: 1507–1519. [DOI] [PubMed] [Google Scholar]
- Kudla J., Batistic O., Hashimoto K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Borsics T., Harrington H.M., Christopher D.A. (2005). Arabidopsis AtCNGC10 rescues potassium channel mutants of E. coli, yeast and Arabidopsis and is regulated by calcium/calmodulin and cyclic GMP in E. coli. Funct. Plant Biol. 32: 643–653. [DOI] [PubMed] [Google Scholar]
- Liu M., Chen T.Y., Ahamed B., Li J., Yau K.W. (1994). Calcium-calmodulin modulation of the olfactory cyclic nucleotide-gated cation channel. Science 266: 1348–1354. [DOI] [PubMed] [Google Scholar]
- Liu Z., Vogel H.J. (2012). Structural basis for the regulation of L-type voltage-gated calcium channels: interactions between the N-terminal cytoplasmic domain and Ca(2+)-calmodulin. Front. Mol. Neurosci. 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Berkowitz G.A. (2011). Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 190: 566–572. [DOI] [PubMed] [Google Scholar]
- Mäser P., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126: 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K., Zagotta W.N. (2003). Cyclic nucleotide-gated ion channels. Annu. Rev. Cell Dev. Biol. 19: 23–44. [DOI] [PubMed] [Google Scholar]
- Moeder W., Urquhart W., Ung H., Yoshioka K. (2011). The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant 4: 442–452. [DOI] [PubMed] [Google Scholar]
- Morales P., Garneau L., Klein H., Lavoie M.-F., Parent L., Sauvé R. (2013). Contribution of the KCa3.1 channel-calmodulin interactions to the regulation of the KCa3.1 gating process. J. Gen. Physiol. 142: 37–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S., Moeder W., Nishimura N., Jikumaru Y., Joo S.-H., Urquhart W., Klessig D.F., Kim S.-K., Nambara E., Yoshioka K. (2010). The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol. 152: 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D., Godiard L., Marco Y., Roby D. (1994). hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 5: 507–521. [DOI] [PubMed] [Google Scholar]
- Poovaiah B.W., Du L., Wang H., Yang T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 163: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 (suppl.): S401–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan M.F., Tung C.C., Van Petegem F., Ahern C.A., Van Petegem F. (2012). Crystallographic basis for calcium regulation of sodium channels. Proc. Natl. Acad. Sci. USA 109: 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink R.C., Shartzer S.F., Fath A., Jones R.L. (1998). Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. USA 95: 1944–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold H., Trempel F., Ranf S., Scheel D., Romeis T., Lee J. (2014). Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 204: 782–790. [DOI] [PubMed] [Google Scholar]
- Shao D., et al. (2014). The individual N- and C-lobes of calmodulin tether to the Cav1.2 channel and rescue the channel activity from run-down in ventricular myocytes of guinea-pig heart. FEBS Lett. 588: 3855–3861. [DOI] [PubMed] [Google Scholar]
- Song Y., Cygnar K.D., Sagdullaev B., Valley M., Hirsh S., Stephan A., Reisert J., Zhao H. (2008). Olfactory CNG channel desensitization by Ca2+/CaM via the B1b subunit affects response termination but not sensitivity to recurring stimulation. Neuron 58: 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E.P., Harper J.F. (2011). The ins and outs of cellular Ca(2+) transport. Curr. Opin. Plant Biol. 14: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck S.M., Colaço R., Davies J.M. (2013). Plant calcium-permeable channels. Plant Physiol. 163: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau M.C., Zagotta W.N. (2004). Dynamics of Ca2+-calmodulin-dependent inhibition of rod cyclic nucleotide-gated channels measured by patch-clamp fluorometry. J. Gen. Physiol. 124: 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau M.C., Zagotta W.N. (2002). Mechanism of calcium/calmodulin inhibition of rod cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. USA 99: 8424–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer N., Mücke N., Broecker J., Keller S., Frings S., Möhrlen F. (2011). Distinct binding properties distinguish LQ-type calmodulin-binding domains in cyclic nucleotide-gated channels. Biochemistry 50: 3221–3228. [DOI] [PubMed] [Google Scholar]
- Urquhart W., Gunawardena A.H.L., Moeder W., Ali R., Berkowitz G.A., Yoshioka K. (2007). The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol. Biol. 65: 747–761. [DOI] [PubMed] [Google Scholar]
- Weitz D., Zoche M., Müller F., Beyermann M., Körschen H.G., Kaupp U.B., Koch K.W. (1998). Calmodulin controls the rod photoreceptor CNG channel through an unconventional binding site in the N-terminus of the beta-subunit. EMBO J. 17: 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamniuk A.P., Vogel H.J. (2004). Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27: 33–57. [DOI] [PubMed] [Google Scholar]
- Yap K.L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000). Calmodulin target database. J. Struct. Funct. Genomics 1: 8–14. [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Kachroo P., Tsui F., Sharma S.B., Shah J., Klessig D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26: 447–459. [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H.-G., Kachroo P., Masmoudi K., Berkowitz G., Klessig D.F. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta W.N., Olivier N.B., Black K.D., Young E.C., Olson R., Gouaux E. (2003). Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425: 200–205. [DOI] [PubMed] [Google Scholar]
- Zelman A.K., Dawe A., Gehring C., Berkowitz G.A. (2012). Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 3: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Varnum M.D., Zagotta W.N. (2003). Disruption of an intersubunit interaction underlies Ca2+-calmodulin modulation of cyclic nucleotide-gated channels. J. Neurosci. 23: 8167–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Lan W., Jiang Y., Fang W., Luan S. (2014). A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant 7: 369–376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.