Arabidopsis histone methyl transferases SET DOMAIN GROUP8 and 25 regulate pep1-, flg22-, and effector-triggered immunity as well as systemic acquired resistance through regulation of the basal and induced transcriptome.
Abstract
Posttranslational modification of histones modulates gene expression affecting diverse biological functions. We showed that the Arabidopsis thaliana histone methyl transferases SET DOMAIN GROUP8 (SDG8) and SDG25 regulate pep1-, flg22-, and effector-triggered immunity as well as systemic acquired resistance. Genome-wide basal and induced transcriptome changes regulated by SDG8 and/or SDG25 showed that two genes of the SDG-dependent transcriptome, CAROTENOID ISOMERASE2 (CCR2) and ECERIFERUM3 (CER3), were also required for plant immunity, establishing mechanisms in defense functions for SDG8 and SDG25. CCR2 catalyzes the biosynthesis of carotenoids, whereas CER3 is involved in the biosynthesis of cuticular wax. SDG8 and SDG25 affected distinct and overlapping global and locus-specific histone H3 lysine 4 (H3K4) and histone H3 lysine 36 (H3K36) methylations. Loss of immunity in sdg mutants was attributed to altered global and CCR2- and CER3-specific histone lysine methylation (HLM). Loss of immunity in sdg, ccr2, and cer3 mutants was also associated with diminished accumulation of lipids and loss of cuticle integrity. In addition, sdg8 and sdg25 mutants were impaired in H2B ubiquitination (H2Bubn) at CCR2, CER3, and H2Bubn regulated R gene, SNC1, revealing crosstalk between the two types of histone modifications. In summary, SDG8 and SDG25 contribute to plant immunity directly through HLM or indirectly through H2Bubn and by regulating expression of plant immunity genes, accumulation of lipids, biosynthesis of carotenoids, and maintenance of cuticle integrity.
INTRODUCTION
Plant immunity to microbial infection depends on a variety of mechanisms that either exist prior to infection or are induced in response to attempted infection. Effector-triggered immunity (ETI) is based on recognition of pathogen virulence effectors by highly polymorphic plant resistance proteins, conferring a highly effective form of plant resistance (Jones and Dangl, 2006). Recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (Boller and Felix, 2009) results in PAMP-triggered immunity (PTI), which confers quantitative resistance. Also, pathogen recognition results in a primed state of gene expression and heightened systemic pathogen resistance to secondary infection (Conrath et al., 2002; Jaskiewicz et al., 2011). ETI and PTI pathways regulate activation of overlapping sets of immune responses (such as accumulation of pathogenesis-related proteins, reactive oxygen species, callose, and many secondary metabolites) with differing speed and intensity (Tsuda and Katagiri, 2010; Thomma et al., 2011). Transcriptional reprogramming of immune response genes and their spatial and temporal regulation are important for effective immunity. The details of how transcription reprogramming, coregulation, and the primed state are achieved are not well known.
Posttranslational modifications of histones and DNA methylation are epigenetic marks that turn genes on or off without altering the underlying DNA sequences. Recent studies underscore the role of epigenetic mechanisms in plant and animal immunity to pathogens (Walley et al., 2008; Dhawan et al., 2009; Berr et al., 2010; Palma et al., 2010; Bierne et al., 2012), but the details of how such mechanisms impact plant immunity are not completely understood. Environmental stress and pathogen infection cause DNA methylation, histone methylation, and increased accumulation of microRNAs (Ruiz-Ferrer and Voinnet, 2009; Bierne et al., 2012; De-La-Peña et al., 2012; Dowen et al., 2012). It is through such biochemical modifications that environmental factors like diet, stress, and prenatal nutrition can affect genes in humans that are passed from one generation to the next (Whitelaw and Whitelaw, 2008). Similarly, plants appear to remember exposure to pathogens long after the initial infection as is revealed by the occurrence of systemic acquired resistance (Ryals et al., 1995). For example, the exposure of seeds, trees, or seedlings to low temperature (vernalization) breaks dormancy, hastens plant development, and determines the onset of flowering (Kim et al., 2009). Plants remember the vernalization period long after the cold spell through an epigenetic memory involving histone/DNA methylation (Jaskiewicz et al., 2011; Song et al., 2012). Transgenerational inheritance of acquired disease and insect resistance has also been linked to epigenetic mechanisms (Luna et al., 2012; Rasmann et al., 2012; Slaughter et al., 2012). Although the robustness of transgenerational inheritance of acquired stress and disease resistance is still debatable (Paszkowski and Grossniklaus, 2011; Gutzat and Mittelsten Scheid, 2012; Pecinka and Mittelsten Scheid, 2012), it is likely that many agronomic and horticultural traits are modulated through epigenetic mechanisms although they may not involve transgenerational inheritance.
In eukaryotes, DNA is wrapped in a nucleoprotein complex known as chromatin. The basic subunit of chromatin is the nucleosome, which contains ∼147 DNA base pairs packed and wrapped around a core of eight histone molecules consisting of two copies each of histones H2A, H2B, H3, and H4 (Schneider and Grosschedl, 2007). Histones are targets of various posttranslational modifications that modulate their interaction with DNA (Heyse et al., 2009). Histone lysine methylation (HLM) and histone H2B monoubiquitination (H2Bubn) activate or repress transcription depending on the specific residue that is modified (Shilatifard, 2006). Trimethylations of H3 lysine 4 (H3K4me3), H3 lysine 36 (H3K36me3), and H2Bubn are enriched at actively expressed genes, whereas H3K27me3 correlates with a repressive state of chromatin that attenuates gene expression. H3K9me2 and H4K20me1 are enriched at heterochromatin and silenced transposon regions (Liu et al., 2007; Quan and Hartzog, 2010; Berke et al., 2012). These and other histone modifications play an important role in maintaining and reprogramming gene expression underlying different plant processes, such as flowering time, cell wall and seed development, and pathogen responses (Liu et al., 2007; Dhawan et al., 2009; Berr et al., 2010; Palma et al., 2010; Xia et al., 2013).
In addition, plant chromatin or chromatin-modifying enzymes are targets of plant pathogens that attenuate plant immunity (Brosch et al., 1995). Pathogen infection or chemical inducers of resistance promote changes in histone acetylation and methylation by affecting the promoter of defense-related genes (Jaskiewicz et al., 2011; De-La-Peña et al., 2012). Mutant plants defective in histone-modifying enzymes show altered plant immunity to different pathogens (Zhou et al., 2005; Berr et al., 2010; Xia et al., 2013). Loss of H2Bubn due to mutations in the histone ubiquitination enzymes HUB1 and HUB2 abrogates fungal resistance (Dhawan et al., 2009; Laluk et al., 2011). Histone modification regulates plant autoimmunity, an ectopic cell death due to deregulation of plant resistance gene expression (Palma et al., 2010; Xia et al., 2013). In addition, the R genes SNC1, RPP4, and LAZ5 are all regulated by HLM (Palma et al., 2010; Xia et al., 2013). Interestingly, systemic acquired resistance (SAR) and defense priming due to prior exposure to pathogens or other chemical elicitors have been correlated with histone modifications at chromatin of defense/stress response genes (Alvarez et al., 2010; Jaskiewicz et al., 2011; De-La-Peña et al., 2012; Luna et al., 2012; Slaughter et al., 2012). Overall, the accumulating genetic data reveal the direct and indirect contributions of histone modifications in the control of plant immunity.
The biological function of histone methyl transferases is attributed to their role in regulating gene expression. Although correlations between certain histone modifications and gene expression have been documented, the mechanisms by which HMTs function in plant immunity and the interaction between different HLMs and with H2Bubn are poorly understood. We showed that SDG8 and SDG25, encoding HMTs belonging to two different subfamilies of methyl transferases, regulated multiple aspects of plant immunity to bacterial and fungal pathogens, including PTI, ETI, and SAR. The sdg8 and sdg25 single and double mutants accumulated reduced levels of lipids and displayed enhanced cuticle permeability accompanied by reduced expression of genes that function in plant immunity and lipid-related functions. In addition, SDG8 and SDG25 target genes CAROTENOID ISOMERASE2 (CCR2) and ECERIFERUM3 (CER3), which encode enzymes in the biosynthesis of carotenoids and cuticular wax, respectively, were also required for plant immunity, accumulation of lipids, and cuticle integrity. Loss of SDG8 and SDG25 singly or in combination altered HLM profiles globally and at specific target loci. Interestingly, H2Bubn at CCR2 and CER3 and the HUB1-regulated resistance gene SNC1 were diminished in sdg single and double mutants, revealing the crosstalk between the two histone modifications. Altogether, our data suggest that SDG8 and SDG25 contribute to plant immunity through the regulation of HLM, immune response transcriptome, carotenoid and lipid accumulation, and cuticle integrity.
RESULTS
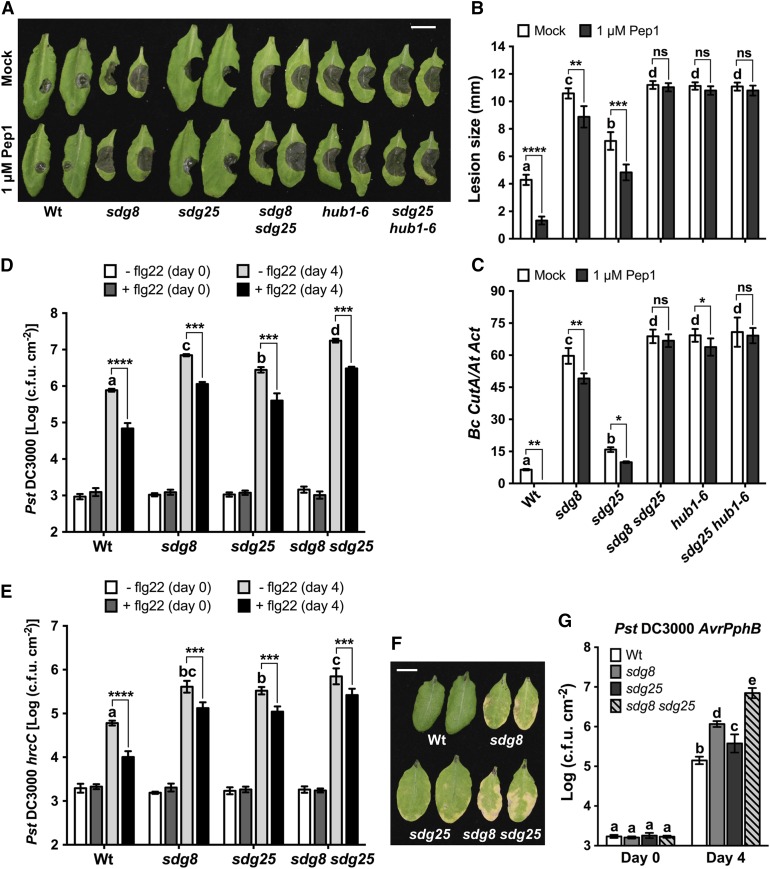
Histone Lysine Methyl Transferases SDG8 and SDG25 Are Required for Peptide-Triggered Immunity
We conducted a reverse genetic screen aimed at identifying histone methyl transferases (HMTs) that regulate PTI by focusing on pathogen-responsive HMT genes. Arabidopsis thaliana HMTs encode Su(var)3-9 and Enhancer of zeste (SET) domain-containing proteins and are thus named SET Domain Group (SDG) genes. T-DNA insertion mutants in 10 Botrytis cinerea-responsive SDGs were tested for fungal resistance (Supplemental Figures 1A to 1C and Supplemental Table 1) and/or PTI. Because SDG2 is a major H3K4 methyl transferase in Arabidopsis (Ng et al., 2007), the sdg2 mutant was also screened for altered responses to fungal infection (Supplemental Figure 1D). SDG25 and SDG8 were identified as regulators of pep1-triggered immunity to fungal infection, suggesting a significant role of SDG25 and SDG8 in PTI in Arabidopsis (Figures 1A to 1C). To confirm the role of SDG25 in fungal resistance, we tested an additional T-DNA insertion allele, sdg25-2 (Col-3 background), for resistance to B. cinerea. The sdg25-2 mutant was also more susceptible to B. cinerea than the wild type (Supplemental Figure 2), confirming the disease susceptibility in the mutants is due specifically to loss of SDG25.
Figure 1.
Histone H3 Methyltransferase Participates in Plant Immunity at Multiple Levels.
(A) to (C) Pep1-triggered immunity to B. cinerea is compromised in H3 lysine methylation and H2B ubiquitination mutants. Plants were inoculated with B. cinerea 1 d after infiltration with double deionized water (Mock) or 1 μM Pep1. Fungal growth was determined by qPCR amplification of the B. cinerea Cutinase DNA (Bc CutA). Relative DNA levels were calculated by the comparative cycle threshold method with Arabidopsis Actin2 as the reference gene.
(A) B. cinerea disease symptoms at 3 d after inoculation. Bar = 1 cm.
(B) Size of B. cinerea-disease lesions. Data represent means ± se (n ≥ 36 disease lesions from three independent experiments).
(C) Quantification of fungal growth in mock- or pep1-treated plants at 3 d after inoculation. Data represent means ± sd (n = 9).
(D) and (E) Flagellin-triggered immunity to bacterial infection in H3 lysine methylation mutants. Arabidopsis plants were inoculated with bacterial suspensions (Pst DC3000, OD600 = 0.0002; Pst DC3000 hrcC, OD600 = 0.001), and bacterial growth was determined at 0 and 4 d after inoculation. Data represent mean log (colony-forming units [cfu]/cm2) ± sd (n = 6).
(D) Growth of Pst DC3000 in mock (white and light-gray bars) or 1 μM flg22 (dark gray and black bars) pretreated plants.
(E) Growth of Pst DC3000 hrcC in mock (white and light-gray bars) or 1 μM flg22 (dark-gray and black bars) pretreated plants.
(F) and (G) SDG25 and SDG8 are required for ETI. Plants were inoculated with the Pst DC3000 (AvrPphB) at OD600 = 0.002. Data represent mean log (cfu/cm2) ± sd from three replicates.
(F) Disease symptoms of Pst DC3000 (AvrPphB) at 4 d after inoculation. Bar = 1 cm.
(G) Growth of Pst DC3000 (AvrPphB) at 0 and 4 d after inoculation.
In (B) to (G), different letters indicate significant difference among genotypes (Student’s t test, P < 0.05). Asterisks indicate significant difference between mock and pep1 or flg22 treatments (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; two-way ANOVA). The experiments were repeated at least three times with similar results. Mock, plants pretreated with double deionized water; flg22, flagellin peptide; Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1; ns, not significant.
Arabidopsis endogenous peptide 1 (pep1) is a typical damage-associated molecular pattern whose recognition by PEPR1 and PEPR2 receptors activates plant immunity (Huffaker and Ryan, 2007; Yamaguchi et al., 2010), including resistance to necrotrophic fungi (Liu et al., 2013). The sdg8 and sdg25 mutant plants showed enhanced susceptibility to B. cinerea as revealed by larger disease lesions and increased fungal growth both prior to and after pretreatment with pep1 (Figures 1A to 1C). The sdg8 and sdg25 mutants displayed significantly attenuated pep1-triggered immunity to B. cinerea compared with wild-type plants. The sdg8 mutant was more susceptible than sdg25 both prior to and after treatment with pep1. The sdg8 sdg25 double mutant abolished pep1-triggered immunity more than either of the single mutants. The data suggest additive contributions of SDG8 and SDG25 to pep1-triggered immunity to fungal infection. Analyses of the expression of SDG25 and SDG8 using qRT-PCR confirmed that both genes were significantly upregulated by B. cinerea inoculation and pep1 treatment of plants (Supplemental Figure 3A). Interestingly, the Arabidopsis hub1-6 mutant, which is impaired in an E3 ligase for histone H2Bubn, also completely lacked pep1-triggered immunity to B. cinerea (Figures 1A to 1C). We further explored the impact of the interaction between histone methylation and ubiquitination in pep1-triggered immunity. The sdg25 hub1-6 double mutant completely eliminated pep1-triggered immunity. Thus, HLM and H2Bubn are required for pep1-triggered immunity. The sdg8 and sdg25 mutant alleles and their growth features are shown in Supplemental Figures 3B to 3E.
The sdg8 and sdg25 mutants were also susceptible to Alternaria brassicicola, suggesting that their contributions to basal resistance extend beyond B. cinerea (Supplemental Figure 4). The severity of disease, as measured by the size of disease lesions and fungal growth, was significantly greater in the sdg8 mutant than in the sdg25 mutant, and the phenotype of the double mutant was more extreme than either single mutant. Thus, SDG8 and SDG25 acted synergistically for A. brassicicola resistance as revealed by the significantly increased fungal growth on the sdg8 sdg25 double mutant.
SDG8 and SDG25 Mediate Flagellin-Triggered Immunity
To evaluate the specificity of the SDGs and determine the impact on plant immunity to distinct pathogen groups, the sdg8 and sdg25 single and double mutants were tested for responses to bacterial pathogens. These mutants supported increased bacterial growth at 4 d postinoculation (dpi) with the virulent strains of Pseudomonas syringae pv tomato DC3000 (Pst DC3000), causing enhanced disease symptom and bacterial growth (Figure 1D). The double mutant was significantly more susceptible than either of the single mutants. Similarly, the mutant plants supported enhanced growth of the hrcC strain of Pst DC3000 (Pst DC3000 hrcC; Figure 1E), which is normally nonpathogenic on wild-type Arabidopsis plants due to its defects in the type III secretion system but retains a collection of bacterial PAMPs that trigger plant immune responses. At 4 dpi, the growth of the Pst DC3000 hrcC strain was significantly higher in the single and double mutants than in the wild-type controls, and the double mutant was significantly more susceptible than either of the single mutants, indicating impaired PTI (Figure 1E). To confirm the observed defects in PTI, we further analyzed single and double mutants for flagellin-triggered immunity (flg22-PTI). Plants were pretreated with a synthetic peptide derived from the conserved part of the flagellin protein of the bacterial flagella. The wild-type plants pretreated with flg22 had significantly improved greater resistance to the virulent strain of Pst DC3000 as well as the nonpathogenic Pst DC3000 hrcC strain (Figures 1D and 1E). By contrast, the sdg25 and sdg8 single and double mutants showed a loss of flg22-PTI to both of these strains. The sdg8 sdg25 double mutant had a slightly greater loss of flg22-PTI than the sdg8 mutant (Figures 1D and 1E). Thus, full flg22-PTI requires both SDG8 and SDG25. The loss of flagellin-triggered immunity in the single and double mutants suggests that the role of these HMTs is not limited to pep1-triggered immunity.
SDG25 Is Required Specifically for Resistance to AvrPphB, but SDG8 Has Broader Functions in ETI
ETI results from direct or indirect recognition of pathogen effectors by R proteins (Jones and Dangl, 2006). The sdg8 and sdg25 mutants were analyzed for their impact on ETI after inoculation with bacterial strains expressing various effectors. The sdg25 mutant showed no altered responses (Supplemental Figure 5), demonstrating that SDG25 is dispensable for ETI to the Pst DC3000 strains expressing AvrRpm1, AvrB, and AvrRpt2 effectors. However, both sdg8 and sdg25 supported enhanced growth of Pst DC3000 (AvrPphB) strain (Figures 1F and 1G). The ETI function of SDG25 appears to be specific to AvrPphB-triggered immunity, whereas sdg8 is affected in many of these responses consistent with published reports (Palma et al., 2010). Therefore, although SDG8 affects ETI responses more broadly, the function of SDG25 in ETI is specific to AvrPphB strain.
SDG8 and SDG25 Contribute to SAR
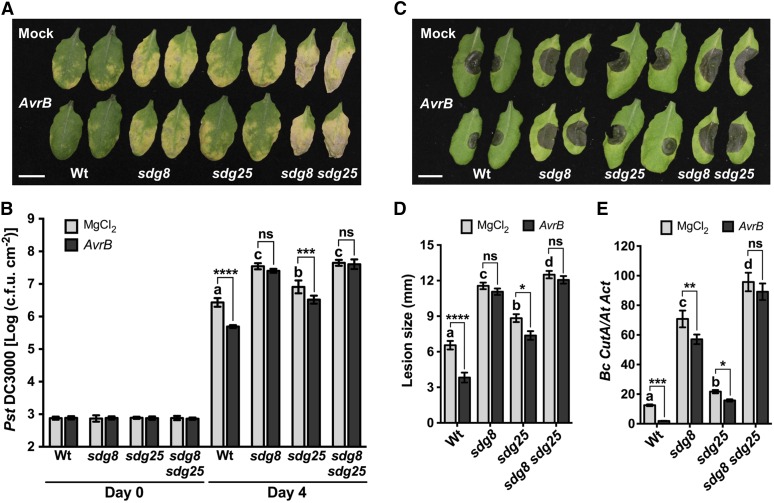
SAR assays were conducted to determine the possible role of SDGs in systemic immunity. To establish SAR, the lower leaves of the wild type and sdg mutants were infiltrated with either MgCl2 or a strain of Pst DC3000 expressing AvrB. After 2 dpi, secondary leaves of all plants were inoculated with the virulent strain Pst DC3000. The wild-type plants that were preinoculated with an avirulent strain showed a 7-fold reduction in growth of Pst DC3000 compared with plants that were preinfiltrated with MgCl2, suggesting induction of SAR (Figures 2A and 2B). By contrast, SAR was clearly attenuated in both single and double mutants as clearly observed by the increased bacterial growth in secondary leaves challenged with Pst DC3000 (Figures 2A and 2B). SAR does not increase resistance to B. cinerea as shown by at least one previous study (Govrin and Levine, 2002). To test whether SDGs contribute to SAR to necrotrophic fungal pathogens, secondary leaves of the wild type and sdg mutants were inoculated with B. cinerea 2 d after lower leaves were infiltrated with MgCl2 or a strain of Pst DC3000 expressing AvrB to activate SAR. Unexpectedly, activation of SAR induced by Pst DC3000 (AvrB) increased resistance to B. cinerea in systemic tissues of wild-type plants (Figures 2C and 2D). However, the sdg8 mutant exhibited a complete loss of SAR, but sdg25 exhibited a significantly reduced SAR and the double mutant exhibited a loss of SAR comparable to sdg8 (Figures 2C and 2D). Therefore, these observations suggest that SDGs are important for SAR to bacterial and fungal pathogens.
Figure 2.
SDG8 and SDG25 Are Required for SAR to Bacterial and Fungal Pathogens.
(A) and (B) Impaired systemic resistance of sdg mutants to bacterial infection. Disease symptoms (A) at 4 d after bacterial inoculation and bacterial growth (B) in secondary leaves at 0 and 4 d after inoculation with the virulent bacterial strain Pst DC3000 with 10 mM MgCl2. Data represent mean log (cfu/cm2), and error bars indicate ± sd from six replicates. Bar = 1 cm.
(C) to (E) Impaired systemic resistance of sdg mutants to fungal infection. Disease symptoms (C), disease lesion size (D), and fungal growth (E) in secondary leaves at 3 d after drop inoculation with B. cinerea. Fungal growth was determined by qPCR amplification of the B. cinerea Cutinase DNA (Bc CutA). The relative DNA levels were calculated by the comparative cycle threshold method with Arabidopsis Actin2 as the reference gene. Bar = 1 cm.
Data in (D) represent means ± se (n ≥ 39 disease lesions) and in (E) represent mean ± sd (n = 9). In all cases, Pst DC3000 (OD600 = 0.001) or B. cinerea (2.5 × 105 spores/mL) were inoculated on secondary leaves 2 d after lower leaves were inoculated with 10 mM MgCl2 or avirulent strain Pst DC3000 carrying AvrB (OD600 = 0.02) to activate SAR. Asterisks indicate significant difference between mock and AvrB treatments (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; two-way ANOVA). Different letters indicate significant differences among genotypes (P < 0.05, Student’s t test). The experiments were repeated at least three times with similar results. Mock, plants pretreated with 10 mM MgCl2; Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1; ns, no significance.
SDG8 and SDG25 Have a Global Impact on Basal and Pathogen-Induced Gene Expression
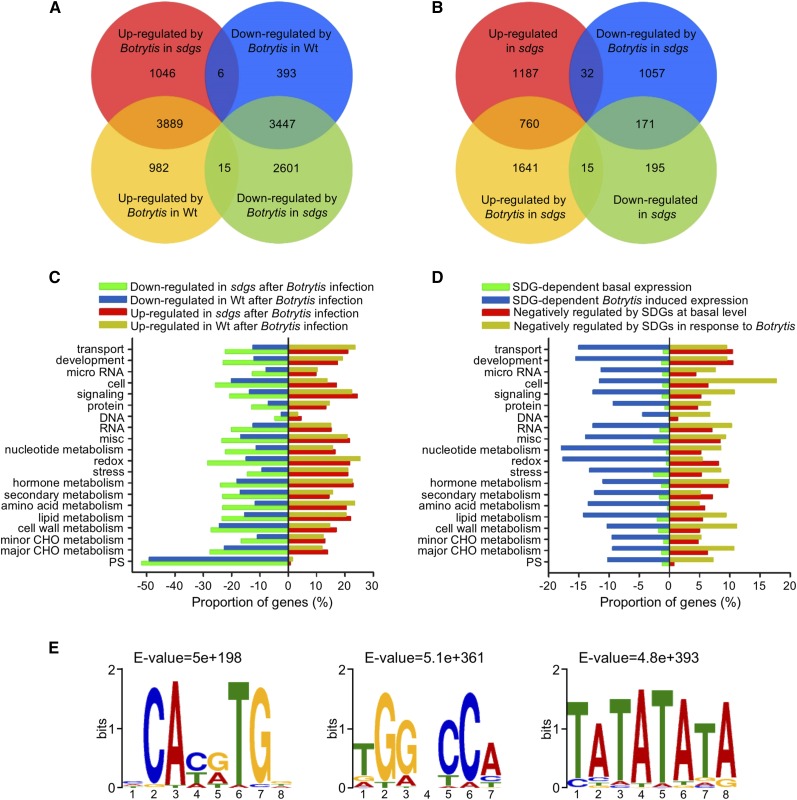
To elucidate the genome-wide regulatory impact of HLM-mediated by SDG8 and SDG25, we generated transcriptome profiles of mock- or pathogen-inoculated wild-type and sdg8 sdg25 double mutant plants using RNA-seq. RNA samples were extracted at 36 h after mock or B. cinerea inoculation. This time point represents a critical infection stage prior to activation of secondary stress responses associated with the aggressive virulence of B. cinerea. The data were analyzed to determine gene expression patterns underlying the impaired plant immune responses in the double mutant and to identify genes regulated by SDG8 and/or SDG25. For each sample with count data, reads per kilobase of exon model per million mapped reads values were calculated. Based on at least a 2-fold change in gene expression and the false discovery rate value (P ≤ 0.05), 4886 B. cinerea-induced and 3846 B. cinerea-suppressed genes were identified from infected wild-type plants (Figure 3A). By comparison, in the double mutant, 4941 genes were induced by B. cinerea and 6063 genes were suppressed, of which 3889 genes are shared between the wild type and the double mutant (Figure 3A). About 15 genes were induced in the wild-type but suppressed in sdg8 sdg25, and six genes were induced in sdg8 sdg25 but downregulated by B. cinerea in the wild type (Figure 3A). Overall, B. cinerea triggered significant transcriptional changes in both the wild type and sdg8 sdg25. In particular, in the sdg8 sdg25 double mutant, 6063 genes failed to be induced by B. cinerea. Thus, the loss of SDGs significantly impacts the Arabidopsis transcriptome with roughly 25% of the Arabidopsis genome not responsive to infection. By comparison, in the sdg8 sdg25 double mutant, 4941 genes were upregulated to a greater extent than in the corresponding wild-type plants inoculated with B. cinerea, suggesting that these genes are likely targets of negative regulation by SDGs (Figure 3A). In the mock-inoculated sdg8 sdg25 double mutant, 1979 genes were upregulated, whereas only 381 genes were expressed at a lower level than in the wild type, indicating the influence of the sdg8 sdg25 genotypic impact on the basal transcriptome (Figure 3B).
Figure 3.
RNA-Seq Analysis of Arabidopsis Wild-Type and sdg Mutant Plants after Fungal Infection.
(A) Venn diagram showing the number of differentially expressed genes in response to B. cinerea infection in sdg8 sdg25 (sdgs) and wild-type (Wt) plants. The numbers in the Venn diagram were obtained by comparing expression in B. cinerea-inoculated sdgs versus mock-inoculated sdgs; B. cinerea-inoculated wild type versus mock-inoculated wild type.
(B) Venn diagram showing differentially expressed genes in the sdg8 sdg25 double mutant compared with the wild type with or without B. cinerea infection. The numbers in the Venn diagram were obtained by comparing expression of genes in mock-inoculated sdgs versus mock-inoculated wild type and B. cinerea-inoculated sdgs versus B. cinerea-inoculated wild type.
(C) Functional categories of genes showing altered gene expression in response to B. cinerea in the wild type and sdg8 sdg25 double mutant.
(D) Functional categorization of SDG-dependent up- or downregulated genes in the mock- or B. cinerea-inoculated sdg8 sdg25 double mutant. In (C) and (D), MapMan functional categories were used for data from genes with altered expression before and after inoculation with B. cinerea. Horizontal bars represent percentage of up- or downregulated genes.
(E) Identification of cis-regulatory elements in SDG-dependent B. cinerea-induced genes. Genes that are statistically significantly overrepresented were analyzed for cis-regulatory binding motifs in promoter regions of the downregulated genes in sdg8 sdg25 after B. cinerea infection.
Genes in the SDG-dependent basal and induced transcriptome were categorized into functional groups based on Gene Ontology (GO). GO terms overrepresented in differentially expressed genes after mock or B. cinerea inoculation in an SDG-dependent manner are summarized in Figures 3C and 3D. Genes encoding nucleotide metabolism, carbohydrate and amino acid metabolism, redox, plant development, transport, lipid metabolism, and stress response were among the most overrepresented categories. Interestingly, more genes were downregulated in the sdg double mutant than upregulated for all the functional groups closely examined. A small proportion of genes showed reduced gene expression prior to inoculation but a significantly higher number were upregulated in sdg8 sdg25 prior to inoculation, suggesting that SDG8 and SDG25 play a significant negative regulatory role in gene expression at the basal level. Many genes in different functional groups were expressed at a reduced level in the sdg8 sdg25 mutant in response to inoculation, suggesting a genetic requirement for induced gene expression consistent with impaired induced resistance in the mutants. The network of cellular processes that are significantly overrepresented, based on genes that are up- or downregulated in the sdg8 sdg25 double mutant after B. cinerea infection, is summarized in Supplemental Figures 6 and 7. Overall, the proportions of genes that required SDG for their B. cinerea-induced expression were higher than those with differential gene expression at a basal level, suggesting a critical role of SDGs for induced gene expression.
In addition to the GO term classification, we examined the composition of the SDG8 and/or SDG25 regulated transcriptome. In particular, 2757 genes that showed 2.5-fold lower expression induced by B. cinerea in the double mutant relative to the mock-inoculated plants were closely analyzed. Interestingly, we found a clear overrepresentation of genes involved in cell wall metabolism, pathogen perception and signaling, pathogenesis-related proteins, defensins, cytochrome P450s, and fatty acid and lipid metabolism (Supplemental Data Set 1). Thirty genes were expressed at least 10-fold lower in sdg8 sdg25 double mutant inoculated with B. cinerea than the mock-inoculated controls. This includes genes involved in lipid biosynthesis or metabolism and cell wall biosynthesis, especially cellulose synthase-like genes (CSLA01), glycosyl transferases, and LRR-RLKs. A total of 111 genes implicated in lipid and fatty acid-related functions, including acyl-CoA synthases that catalyze the synthesis of omega-hydroxy fatty acyl-CoA intermediates in the pathway to cutin synthesis, sterol biosynthesis, and acyl transferases, were identified. Finally, 26 lipid transfer protein genes, 20 GDSL-like lipase/acylhydrolase, 28 cytochrome P450, and 67 LRR-RLKs exhibited lower gene expression in the sdg8 sdg25 mutant (Supplemental Data Set 1). Interestingly, 21 genes encoding putative S-adenosyl-l-methionine-dependent methyltransferases were also expressed at a lower level in the inoculated double mutant.
Promoters of SDG-Regulated Genes Contain Conserved cis-Regulatory Elements
Genes that showed 2.5-fold lower gene expression in the sdg8 sdg25 double mutant, despite a clear upregulation by B. cinerea in wild-type plants, were selected and their promoter regions analyzed for conserved cis-regulatory motifs. MEME motif analysis was conducted on the 1000-bp upstream sequences of 2757 genes of interest obtained from the Arabidopsis database (TAIR10 BLAST database). Three cis-regulatory motifs, CAc/tg/aTG (E-value = 5E+198), TGGcCA (E-value = 5.1E+361), and TATATATA (E-value = 4.8+393), were overrepresented in the promoter of the 2757 genes selected as SDG8 and/or SDG25 target genes (Figure 3E). CACGTG is a known bZIP family binding motif, and the FORCA motif is present in Arabidopsis promoters known to integrate light- and defense-related signals (Evrard et al., 2009). The third cis-motif (TATATATA) is known to bind to AT-hook motif DNA binding (AHL) proteins. The AT-hook motif is a small DNA binding protein motif that has been found in the high mobility group of nonhistone chromosomal proteins (Kim et al., 2011). Examples of SDG-regulated genes that contain the cis-motifs identified above include CER3, CCR2, and PDF1.2. We compared the 2757 genes to the published list of 728 genes that were reported to directly bind SDG8 (Li et al., 2015b). Of the 2757 genes, ∼30 B. cinerea-induced genes that showed reduced expression in the double mutant were also bound by SDG8 (Supplemental Table 2). Interestingly, these genes are annotated to function in different biological roles and contain the conserved cis-elements described above.
SDG8 and SDG25 Regulate Gene Expression during the Immune Response
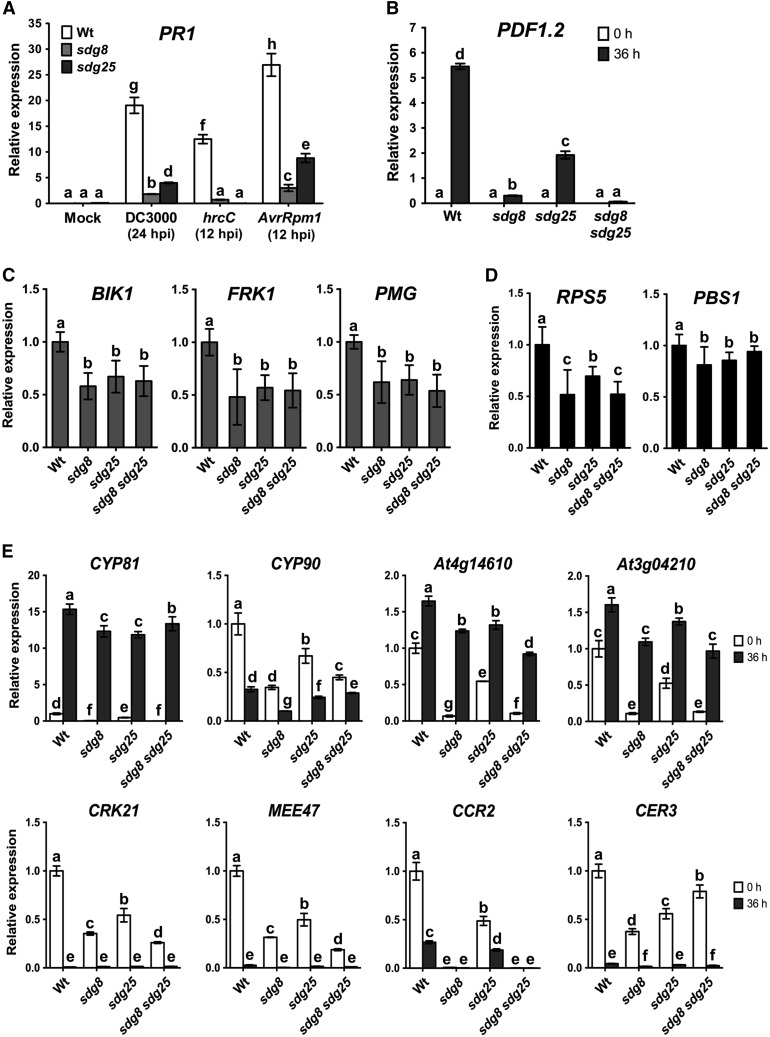
The RNA-seq data revealed that SDGs regulate diverse immune response genes implicated in bacterial and fungal resistance (Supplemental Data Set 1; Figures 3A to 3D). This includes a variety of genes with direct and indirect roles in plant immunity, including immune response marker genes as well as potential pattern recognition receptors. Pathogenesis-related proteins (chitinases, glucanases, and peroxidases) and plant defensins exhibited significantly reduced B. cinerea-induced expression in the inoculated sdg mutants. Some of these genes were further analyzed through qRT-PCR. Induced expression of PR1 was reduced in sdg8 and sdg25 mutant plants inoculated with Pst DC3000, Pst DC3000 hrcC, and Pst DC3000 (AvrRpm1) (Figure 4A). The plant defensin gene PDF1.2 was induced in response to B. cinerea infection in wild-type plants, but this induced expression was nearly abolished in the sdg8 mutant (Figure 4B). PDF1.2 expression was also severely reduced in sdg25, but a significant level of PDF1.2 expression occurred relative to sdg8 and despite the susceptibility of sdg25 to fungal infection (Figure 4B). The sdg8 sdg25 double mutant was comparable to the sdg8 single mutant in the impact on PDF1.2 expression (Figure 4B). The expression of these immune response markers was consistent with their known roles in defense response pathways. In addition, the expression of BIK1 (BOTRYTIS-INDUCED KINASE1), a critical regulator of PTI to fungal and bacterial pathogens, was significantly reduced in all of the single and double mutants tested (Figure 4C). The FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE1) expression was strongly reduced in all of the sdg mutants relative to the wild type (Figure 4C). Expression of PMG (At2g17740) gene, also a defense response marker, was lower in the single and double mutants. These reduced expression patterns correlated with a defective PTI. Although we identified many LRR-RLKs that are regulated by SDG8 and SDG25, the expression of PEPR1 and FLS2 was not affected by these genes, suggesting that SDGs display an effect downstream of pathogen recognition. The expression of RPS5, which determines the activation of race-specific resistance to the AvrPphB strain, was significantly reduced in the single and double mutants (Figure 4D). By contrast, PBS1, which is required for RPS5-mediated resistance was only slightly reduced in the sdg mutants (Figure 4D). Thus, the altered ETI response was correlated with transcriptional downregulation of the R gene RPS5 but not PBS1.
Figure 4.
SDG8 and SDG25 Are Required for Expression of Defense Genes.
(A) Expression of PR1 in response to Pst DC3000 strains.
(B) Expression of PDF1.2 in response to B. cinerea.
(C) Basal expression levels of PAMP receptor and PTI marker genes.
(D) Expression of RPS5 and PBS1, which are required for resistance to AvrPphB.
(E) Expression of SDG8- and SDG25-regulated genes.
In (A) and (B), expression levels were analyzed by qRT-PCR after pathogen inoculation in the wild type and sdg mutants. Data are normalized by the comparative cycle threshold method with Actin2 as the internal control and presented as relative expression. Data represent means ± sd (n = 3). In (C) and (D), qRT-PCR data were normalized to Actin2 and presented as relative expression compared with the wild type, which was set to 1. Values represent mean ± sd from three technical replicates of six independent biological repeats (n = 18). In (E), qRT-PCR was conducted on samples collected from B. cinerea-inoculated plants. Expression of the wild type at 0 h was set to 1.0. Error bars indicate ± sd and n = 3. Different letters indicate significant differences (P < 0.05, Student’s t test). Similar results were obtained in at least two independent biological experiments. Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1.
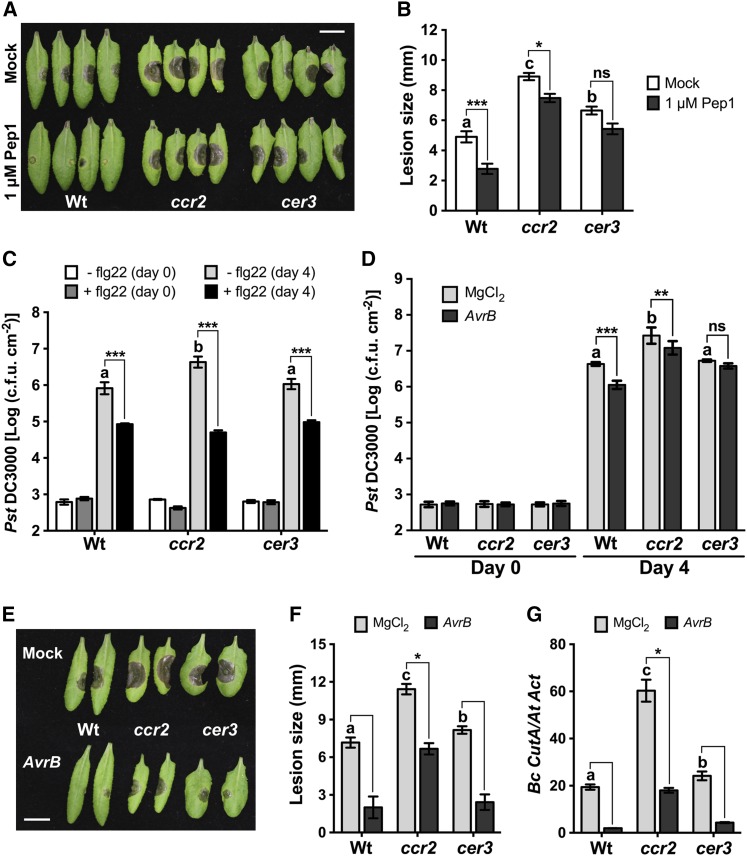
The SDG8- and SDG25-Regulated Genes CCR2 and CER3 Are Required for Plant Immunity
We screened for downstream target genes that may account for the defense functions of SDG8 and SDG25. Ten genes with differing patterns of gene expression in the single and double mutants were selected for further functional analysis (Figure 4E). The basal and/or induced expressions of these genes were significantly reduced in the single and/or double mutants (Figure 4E). These genes encode two cytochrome P450s (CYP81, At5g57220; CYP90, At5g05690) implicated in glucosinolate synthesis, fungal resistance, and the brassinosteroid biosynthesis process; RLKs (CRK21, At4g23290); a pseudo-gene with a CC-NBS-LRR domain (At4g14610); and putative TIR-NBS-LRR disease resistance genes (At3g04210); CER3 (WAX2, At5g57800), maternal effect embryo arrest 47 (MEE47, At4g00950), and CCR2 (CAROTENOID ISOMERASE, At1g06820) gene, which was previously described as SDG8 target gene (Cazzonelli et al., 2009). Expression of CCR2 was eliminated in sdg8 and sdg8 sdg25 and significantly reduced in sdg25 (Figure 4E). CER3 also showed reduced gene expression in the sdg single and the double mutant (Figure 4E).
T-DNA insertion mutants in 10 selected SDG8-and/or SDG25-regulated genes were tested for their pathogen responses to determine the link between the biological functions of SDG and their potential target genes. The ccr2 and cer3 mutants showed enhanced susceptibility to B. cinerea and A. brassicicola similar to sdg8 and sdg25 single and double mutants but showed no major growth defects (Supplemental Figures 8A to 8G). In addition, pep1-triggered immunity to B. cinerea was significantly reduced in both ccr2 and cer3 mutants, although the severity of the disease symptoms and the level of fungal growth varied between the two mutants (Figures 5A and 5B). By contrast, the ccr2 mutant displayed wild-type flg22-PTI and responses to the hrcC strain of Pst DC3000 but was susceptible to Pst DC3000 with significantly increased bacterial growth (Figure 5C; Supplemental Figure 8H). AvrB-induced SAR to bacterial infection was also impaired in the ccr2 and cer3 mutants (Figure 5D). By contrast, AvrB-induced SAR to fungal infection was slightly impaired in the ccr2 mutant relative to the wild type but not in the cer3 mutant (Figures 5E to 5G). The cer3 mutant was comparable to the wild type in all aspects of bacterial resistance (Figures 5C and 5D; Supplemental Figure 8H). Our data suggest that SDGs function in pathogen responses, at least partially, through CCR2- and CER3-regulated downstream processes, including carotenoid and cuticular wax accumulation. Carotenoids are essential photoprotective and antioxidant pigments synthesized by all photosynthetic organisms. CER3 encodes a transmembrane protein with similarity to the sterol desaturase implicated in cuticle membrane and wax biosynthesis (Chen et al., 2003).
Figure 5.
Arabidopsis CCR2 and CER3 Are Required for PTI and SAR.
(A) and (B) Pep1-triggered immunity to B. cinerea is compromised in ccr2 and cer3 mutants. Disease symptoms at 3 d after fungal inoculation (A) and comparison of B. cinerea disease lesion size (B) in mock- or pep1-treated plants. Plants were inoculated with B. cinerea 1 d after mock or 1 μM Pep1 treatment. Asterisks indicate a significant difference between mock and pep1 treatments (mean ± se; n ≥ 40; *P < 0.05 and ***P < 0.001; two-way ANOVA). ns, not significant. Different letters indicate significant differences among mock-treated genotypes (P < 0.05, Student’s t test). Bar = 1 cm.
(C) Flagellin-triggered immunity is independent of CCR2 and CER3. Plants were inoculated with Pst DC3000 (OD600 = 0.0002) 1 d after mock or 1 μM flg22 pretreatment. Bacterial growth was determined at 4 dpi. Data represent mean log (cfu/cm2), and error bars indicate ± sd of six replicates. Mock-treated plants were infiltrated with double deionized water.
(D) CCR2 and CER3 are required for SAR to bacterial infection. Data show bacterial growth in secondary leaves of SAR-induced plants and represent mean log (cfu/cm2), and error bars indicate ±sd from six replicates. Secondary leaves were tested for systemic resistance to bacterial infection 2 d after lower leaves were inoculated with Pst DC3000 (AvrB) to activate SAR. In (C) and (D), asterisks indicate significant difference between mock and flg22 or AvrB treatments (**P < 0.01 and ***P < 0.001; two-way ANOVA). Different letters indicate significant differences among mock-treated genotypes (P < 0.05, Student’s t test).
(E) to (G) CCR2 and CER3 are required for SAR to fungal infection. B. cinerea disease symptoms (E), comparison of disease lesion size (F), and fungal growth (G) in systemic leaves of ccr2 and cer3 mutants showing loss of SAR. Data in (F) represent means ± se (n ≥ 24) and in (G) represent means ± sd (n = 9). Asterisks indicate significant difference between mock and AvrB treatments from the wild type (one-way ANOVA followed by Tukey’s HSD, P < 0.05). Different letters indicate significant differences among mock-treated genotypes (P < 0.05, Student’s t test). Secondary leaves were tested for systemic resistance to bacterial or fungal infection 2 d after lower leaves were inoculated with Pst DC3000 (AvrB) to activate SAR. Fungal growth was determined by qPCR amplification of the B. cinerea Cutinase DNA (Bc CutA). The relative DNA levels were calculated by the comparative cycle threshold method with Arabidopsis Actin2 as the reference gene. The experiments were repeated at least two times with similar results. Bar = 1 cm. Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1.
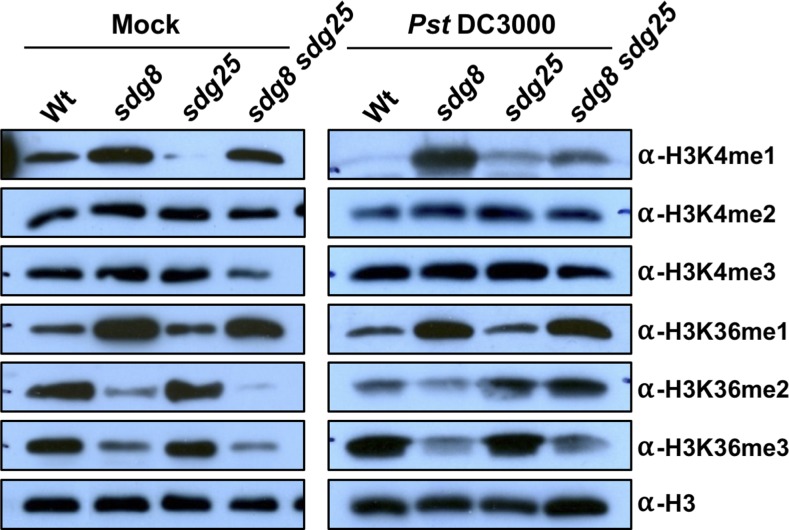
Global Histone Lysine Methylation Is Mediated by SDG8 and SDG25
Global changes in HLM were studied in mock and bacterial inoculated plants to determine the role of SDGs on basal and induced HLMs. Pst DC3000 was used instead of B. cinerea to avoid interference from fungal HLM. The levels of the various H3Kme were determined on an immunoblot with antibodies specific to the different HLMs (Figure 6). In wild-type plants, inoculation with Pst DC3000 increased accumulation of both H3K4me3 and H3K36me3, two modifications that positively correlate with gene expression. Unexpectedly, H3K4me1, H3K4me2, and H3K36me2 were reduced after inoculation with Pst DC3000, although these HLMs are also associated with active gene expression (Figure 6).
Figure 6.
Global Histone Methylation in Arabidopsis Wild Type and sdg Single and Double Mutants.
Analysis of global levels of H3K4me1∼3 and H3K36me1∼3 in the wild type and sdg8, sdg25, and sdg8 sdg25 mutants by immunoblot analysis. Histone-enriched protein extracts obtained at 2 d after mock or Pst DC3000 (OD600 = 0.001) inoculation were immunoblotted and probed with antibodies that recognize specific histone methylations. Total amounts of histone H3 are shown as loading controls. Similar results were obtained in two independent experiments. Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1.
In the sdg8 mutant, the global levels of H3K4me2 and H3K4me3 were unchanged, but H3K36me2 and H3K36me3 were significantly reduced, suggesting positive regulation of these modifications by SDG8. By contrast, H3K4me1 and H3K36me1 levels increased significantly in the sdg8 mutant, indicating negative control by SDG8. Pst DC3000 infection increased H3K4me1 and H3K4me3 levels in the sdg8 mutants but decreased H3K4me2 and H3K36me levels.
The sdg25 mutant had significantly reduced global levels of H3K4me1, which accumulated to a higher level in the sdg8 mutant, suggesting distinct methods of regulation of these modifications by the two HMTs. Intriguingly, some of the H3K4 modifications, including H3K4 di- and trimethylations, do not appear to be affected or were only slightly higher in the sdg25 mutant but were increased in response to Pst DC3000 inoculation. In the sdg8 sdg25 double mutant, H3K4me3, H3K36me2, and H3K36me3 were reduced significantly, but H3K4me1 and H3K36me1 were intermediate between the wild type and the sdg8 mutant. After Pst DC3000 inoculation, H3K4me3, H3K36me2, and H3K36me3 increased but H3K4me1 decreased compared with mock inoculated plants.
In general, these analyses revealed complex interactions among the various HLMs. Overall, global HLM analyses demonstrated that SDG25 primarily affected H3K4me1, but SDG8 affected H3K36me2 and H3K36me3 positively and H3K4me1 and H3K36me1 negatively. The SDG8 effect was often epistatic to the SDG25-mediated modifications.
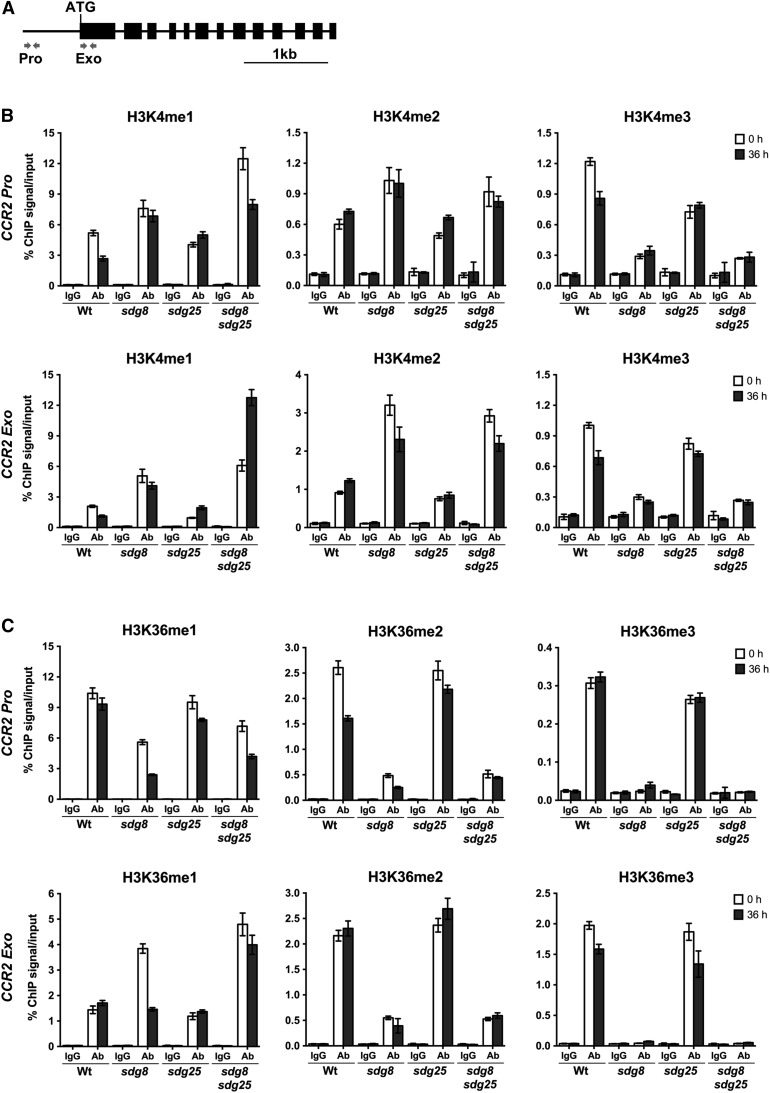
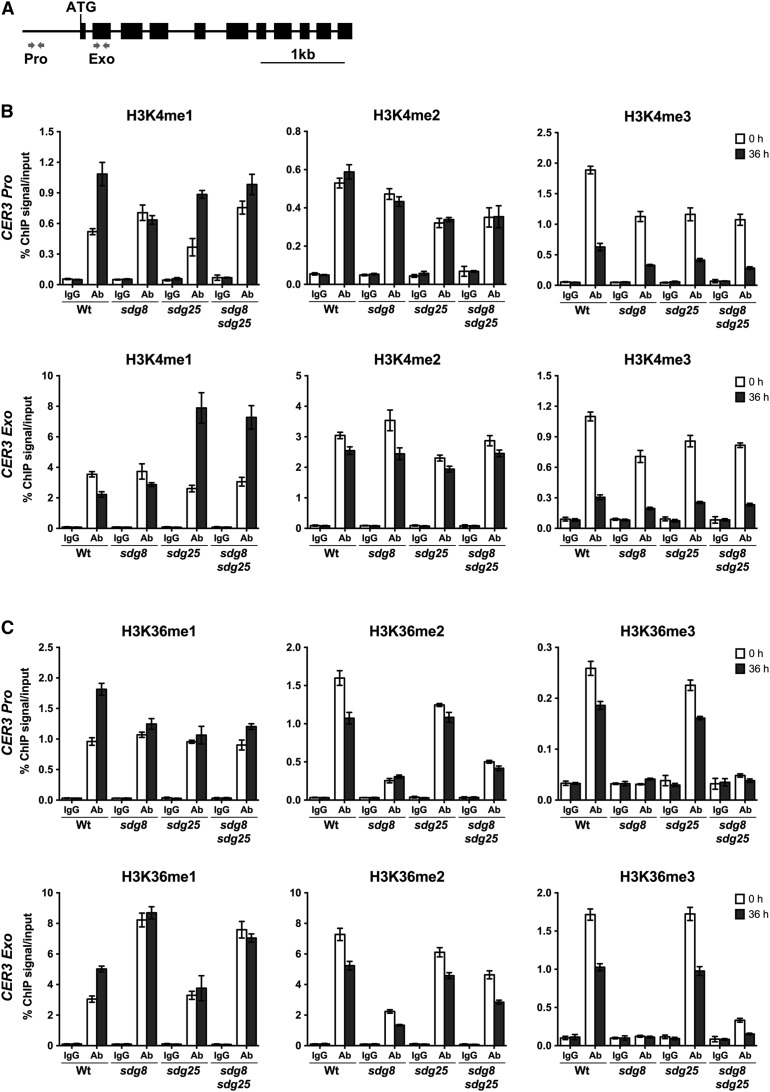
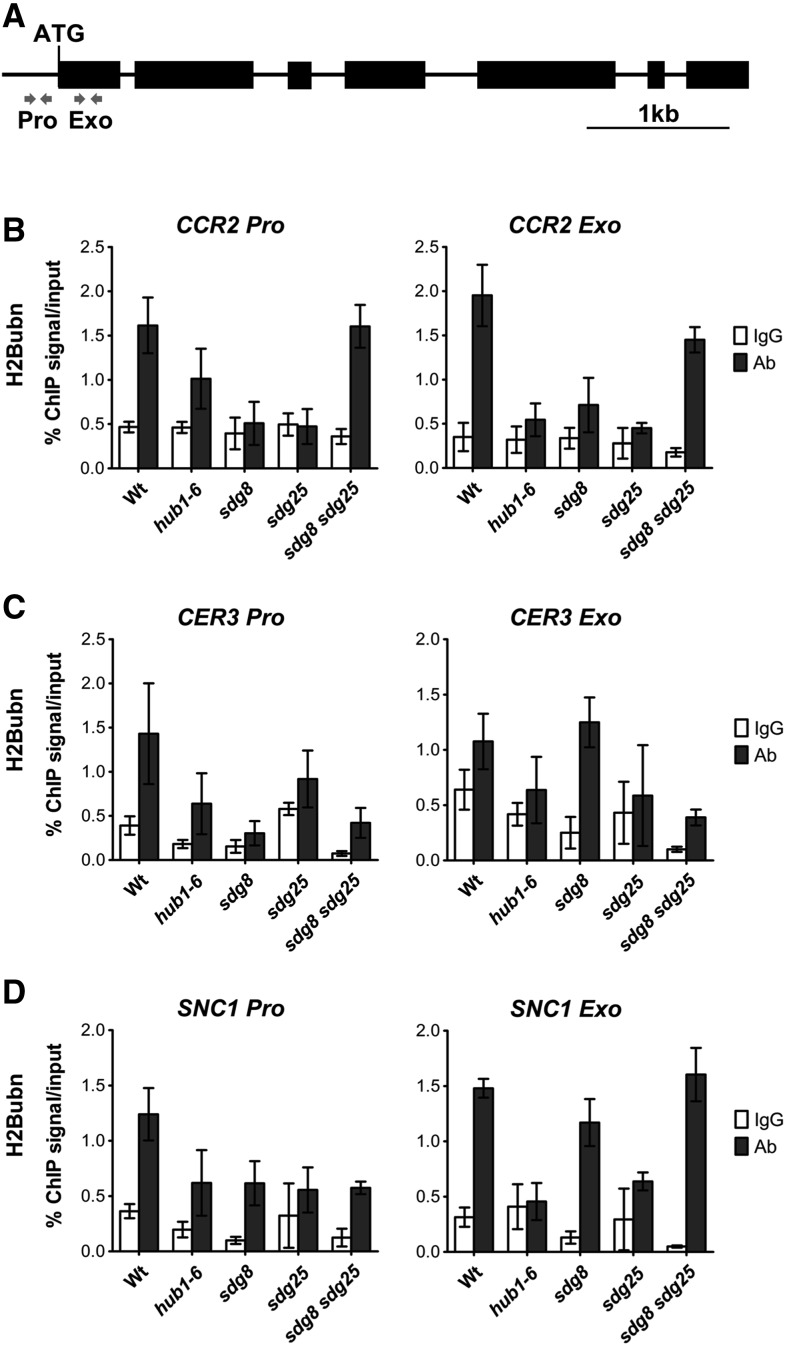
The Chromatin of CCR2 and CER3 Loci Contain Altered Histone H3 Methylations in sdg Mutants
To determine the link between HLM at CCR2 and CER3 chromatin and gene expression and their immune response functions, we analyzed HLM at CCR2 and CER3 loci in the sdg8 and sdg25 single and double mutants. The basal expression of CCR2 and CER3 genes is dependent on SDGs, and the two genes share similar defense functions with SDGs. Chromatin immunoprecipitation (ChIP) assays were conducted with H3K4- and H3K36-specific antibodies and quantified by real-time qPCR with primers at the promoter and exon regions of CCR2 and CER3 genes (Figures 7A and 8A).
Figure 7.
Regulation of H3K4 and H3K36 Methylations at CCR2 Chromatin by SDG8 and SDG25 Methyl Transferases.
(A) Schematics showing CCR2 genomic region. The location of the primers at the promoter (Pro) and coding regions (Exo) used to analyze the level of H3K4 and H3K36 methylations by ChIP assays are indicated by arrows. The solid lines indicate promoter and intron regions and black boxes indicate exons.
(B) Relative enrichment levels of H3K4me1, H3K4me2, and H3K4me3 at chromatin of CCR2 gene before and after inoculation with B. cinerea.
(C) Relative enrichment levels of H3K36me1, H3K36me2, and H3K36me3 at chromatin of CCR2 gene before and after inoculation with B. cinerea. ChIP was conducted on chromatin extracts with antibodies that recognize different histone methylations and IgG was used as a background control. Data from each experiment were normalized to SAM and are presented as percentage of IP/input. The data represent one biological experiment with three technical replicates. Error bars show ±sd (n = 3). Similar results were obtained in two independent experiments. Ab, antibody; Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1.
Figure 8.
Regulation of SDG25 and SDG8 on H3K4 and H3K36 Methylations on Chromatin Surrounding the CER3 Locus.
(A) Schematic showing the genomic region of CER3. The location of primers at the promoter (Pro) and coding regions (Exo) that were used to analyze the level of H3K4 and H3K36 methylations by ChIP assays are shown as arrows. The solid lines indicate promoter and intron regions and black boxes indicate exons.
(B) Relative enrichment levels of H3K4me1, H3K4me2, and H3K4me3 on CER3 before and after inoculation with B. cinerea.
(C) Relative enrichment levels of H3K36me1, H3K36me2, and H3K36me3 on CER3 before and after inoculation with B. cinerea. ChIP was performed on chromatin extracts using antibodies that recognize different histone methylations as indicated, and IgG serves as a background level. Data from each experiment were normalized to SAM and are presented as percentage of IP/input. Data are representative of one biological experiment with three technical replicates. Error bars show ±sd (n = 3). Similar results were obtained in two independent biological experiments. Ab, antibody; Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1.
Clearly, loss of SDG25 significantly impaired H3K4me1, H3K4me2, and H3K4me3, whereas SDG8 affected only H3K4me3 in all tested regions of the CCR2 and CER3 loci (Figures 7B and 8B). Unexpectedly, in the sdg8 mutant, H3K4me1 and H3K4me2 at the CCR2 locus were enriched significantly more than in the wild type similar to the pattern observed in the sdg8 sdg25 double mutant, which hyperaccumulated these lysine modifications (Figure 7B). However, their levels were unchanged at the CER3 locus except in the promoter region, which was enriched for H3K4me1 (Figure 8B), consistent with the global methylation patterns presented in Figure 6. Interestingly, the H3K4me profiles of the sdg8 sdg25 double mutant mostly mirror those of the single sdg8 mutant, suggesting an epistatic nature of sdg8 mutation over sdg25 (Figures 7B and 8B).
The sdg8 mutation drastically reduced H3K36me2 and H3K36me3 at the CCR2 and CER3 loci, whereas H3K36me1 was significantly increased in all gene-specific regions studied except the CCR2 promoter region, which showed strongly reduced H3K36me1 (Figures 7C and 8C). In the sdg25 mutant, H3K36me2 and H3K36me3 were slightly decreased at CER3 and CCR2 loci, respectively, whereas the level of H3K36me1 was unchanged at both loci (Figures 7C and 8C). These data indicate that SDG25 had little to moderate impact on H3K36me2 and H3K36me3. The H3K36me profile of the double mutant was similar to that of sdg8 single mutant (Figures 7C and 8C).
We also analyzed changes in HLMs at CCR2 and CER3 loci in B. cinerea-inoculated plants. Generally, the pattern of H3K4 and H3K36 methylations after B. cinerea inoculation was similar to the pattern in mock-inoculated plants, suggesting the critical role of SDGs in basal HLMs. There were either slight decreases or no significant changes in H3K4me1 and H3K36me1 levels at the CCR2 locus after B. cinerea inoculation in the wild-type plants (Figures 7B and 7C). However, there was a significant increase or decrease, particularly in the sdg8 sdg25 double mutant, in response to B. cinerea. H3K4me1 levels in the exons of the CCR2 locus increased in sdg25 single and sdg8 sdg25 double mutants, and H3K4me2 increased at the CCR2 locus in the wild type and sdg25 mutant (Figure 7B). The level of H3K36me3 and H3K36me2 at the promoter and exon regions of the CCR2 locus were comparable in mock- and B. cinerea-inoculated plants but H3K36me2 at the promoter of CCR2 decreased (Figure 7C).
The wild-type plants showed increased H3K4me1 and H3K36me1 levels in most regions of the CER3 locus after B. cinerea inoculation (Figures 8B and 8C). The sdg25 single and double mutants displayed increased H3K4me1 levels in response to B. cinerea at the CER3 locus. However, H3K36me1 levels were unchanged at the CER3 locus in the sdg single and double mutants (Figures 8B and 8C). Unexpectedly, in response to B. cinerea but consistent with reduced gene expression, the wild-type and sdg mutants exhibited reduced levels of di- and trimethylation at H3K4 and H3K36 in all examined regions of the CER3 gene, except the promoter region, which contained comparable H3K4me2 levels in mock and B. cinerea-inoculated plants (Figures 8B and 8C). Our data suggest complex interactions between the different H3Kme patterns mediated by SDG8 and SDG25 as well as complex patterns following B. cinerea inoculation. Occasionally, the single mutations acted antagonistically for H3Kme despite sharing similar disease response profiles. Overall, the changes due to SDG mutations on the H3Kme at the CER3 locus were not as pronounced as in the CCR2 locus and also correlated with the level of CCR2 and CER3 gene expression in sdg mutants.
Crosstalk between Histone Lysine Methylation and H2B Ubiquitination
Studies in metazoan cells suggest crosstalk between various histone modifications (Suganuma and Workman, 2011). In Arabidopsis, HLM at chromatin of flowering genes is reduced in the hub1 mutant with impaired H2Bubn (Cao et al., 2008). The interactions between H3Kme and H2Bubn were examined in the Arabidopsis mutant hub1-6 (defective in H2Bubn), sdg8, and sdg25, which are altered in HLM. SDG8 and SDG25 target genes, CER3 and CCR2, as well as SNC1 were used to determine the impact of H3Kme on H2Bubn. SNC1 is a TIR-NB-LRR-type protein whose gene expression is regulated by H2Bubn (Zou et al., 2014). As expected, the hub1 mutation significantly reduced H2Bubn in SNC1, CCR2, and CER3 promoters and exons (Figures 9B to 9D). Interestingly, the sdg single mutants carrying a functional HUB1 showed a significant reduction in H2Bubn at these loci, establishing the significant regulatory impact of HLM on H2Bubn (Figures 9B to 9D). The sdg8 sdg25 double mutant had wild type or only slightly reduced levels of H2Bubn, even when the single mutants had severely attenuated H2Bubn, suggesting a complex interaction between the various histone modifications (Figures 9B to 9D). Although the SDG8 and SDG25 mutations impacted the H2Bubn at the SNC1, CER3, and CCR2 loci, the magnitude of the impact differed depending on the locus. Conversely, analysis of the H3K4me and H3K36me at the SNC1 locus showed that SDG8 is required for H3K4me3 and H3K36me3 at promoter and exon regions of SNC1, whereas loss of SDG25 had only a minor effect on H3K4me2 (Supplemental Figure 9). The sdg8 sdg25 double mutant showed significantly reduced H3K4me2/3 and H3K36me3 at the SNC1 promoter and exons similar to the CCR2 and CER3 loci (Supplemental Figure 9). The sdg mutants showed reduced H3Kme at SNC1 locus, demonstrating the broader effect of SDGs on HLM at other loci. In turn, the hub1 mutation impacted H3K4me2 and H3K4me3 at the SNC1 promoter and exons but had no obvious effect on H3K36me2 and H3K36me3 (Supplemental Figure 9). Thus, our study established that HLMs impact immune responses and gene expression directly through HLM and indirectly through H2Bubn and that significant crosstalk occurs between the two modifications.
Figure 9.
Modulation of Histone 2B Monoubiquitination by Histone Lysine Methyl Transferases.
(A) Schematic of the genomic region of SNC1 and the location of the primers (gray arrows) at the promoter (Pro) and coding regions (Exo) are used to analyze the level of H2B monoubiquitination by ChIP assays. The solid lines indicate promoter and intron regions and black boxes indicate exons.
(B) to (D) Histone 2B ubiquitination is cross-regulated by histone lysine modification. Relative H2B monoubiquitination levels at chromatin of CCR2 (B), CER3 (C), and SNC1 (D). ChIP was conducted on chromatin extracts from the indicated plant genotypes with H2B monoubiquitination antibody and IgG was used as a background control. Data from each experiment were normalized to SAM and are presented as percentage of IP/input. Representative data from one experiment with three technical repeats are presented. Error bars show ±sd (n = 3). Similar results were obtained in at least two independent experiments. Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1.
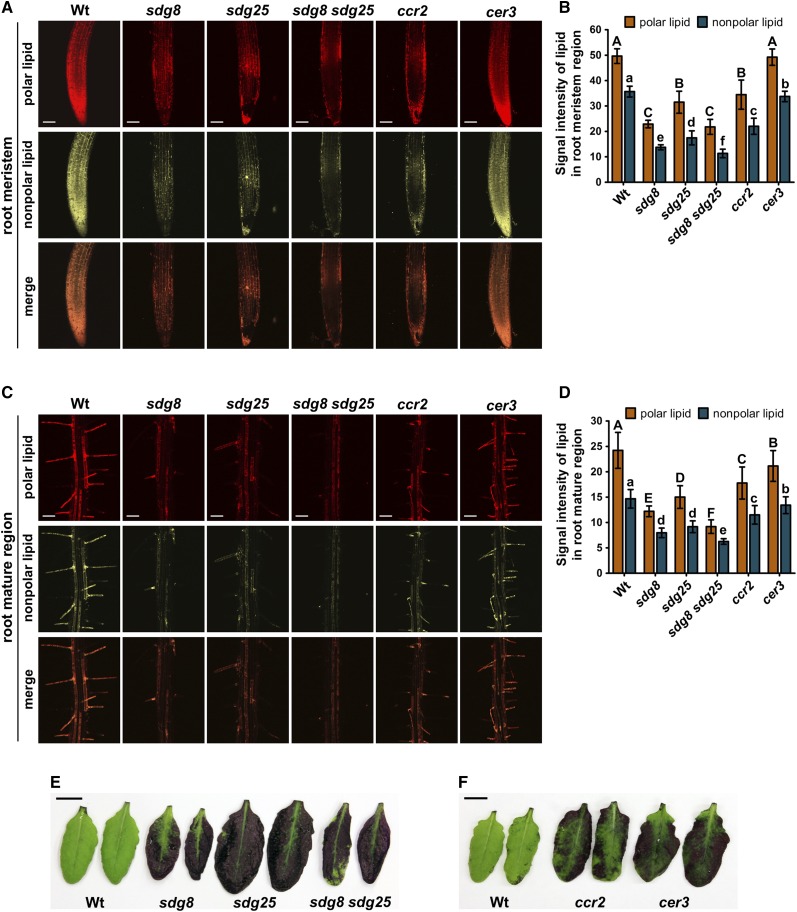
SDG8, SDG25, CCR2, and CER3 Regulate Lipid Accumulation and Cuticle Integrity
The changes in expression of a large number of lipid biosynthesis genes observed in the RNA-seq data of the sdg8 sdg25 double mutant and changes in cuticular wax reported for cer3 prompted us to examine changes in lipid accumulation and cuticle permeability. All of the mutant plants were analyzed for the accumulation of lipids in the root meristem and mature regions by staining with Nile red, a commonly used lipid stain, as described recently (Li et al., 2015a). Interestingly, the sdg8 and sdg25 single and double mutants accumulated significantly reduced levels of polar and nonpolar lipids based on their excitation and emission wavelengths after staining with Nile red (Figures 10A to 10D). The ccr2 mutant accumulated significantly reduced amounts of polar and nonpolar lipids similar to the sdg mutants. In addition, the cer3 mutation, which is known to affect the levels of cuticular wax, reduced the accumulation of the nonpolar lipids in meristem and mature regions of the root when compared with the wild type (Figures 10A to 10D). The single and double mutants contained significantly lower amounts of lipids relative to the wild-type plants with 2- to 3-fold lower lipids levels in the sdg8 sdg25 double mutant. In conjunction with the loss of B. cinerea-induced expression of diverse lipid biosynthesis genes (Supplemental Data Set 1), the loss of lipid accumulation in the sdg, ccr2, and cer3 mutants suggested that SDGs regulate lipid accumulation, and the reduced lipid levels correlate with the altered pathogen responses of the mutants.
Figure 10.
SDG8 and SDG25 Regulate Lipid Accumulation and Cuticle Permeability.
(A) and (B) Lipid accumulation in root meristem regions of sdg, ccr2, and cer3 mutants.
(A) Nile red staining of polar and nonpolar lipids.
(B) Quantification of the fluorescence intensities of polar and nonpolar lipids in the root meristem regions.
(C) and (D) Lipid accumulation in root mature regions of sdg, ccr2, and cer3 mutants. Nile red staining of polar and nonpolar lipids (C) and quantification of the fluorescence intensities of polar and nonpolar lipids in the root mature regions (D). The mean values followed by different letters indicate significant differences (P < 0.05, Student’s t test). Top panels, polar lipids; middle panels, nonpolar lipids; bottom panels, merged image from the other two panels. Bars = 100 μm. The lipid levels were visualized by Nile red staining of root tissues as described in Methods. In (B) and (D), the fluorescence intensities were quantified with the ImageJ software (http://rsb.info.nih.gov/ij/download.html). Values represent mean of 10 measurements ±sd.
(E) and (F) Increased cuticle permeability in sdg, ccr2, and cer3 mutants. Leaves from 4- to 5-week-old plants were incubated in 0.05% toluidine blue solution for 30 min. The experiments were performed three times with similar results. Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1. Bars = 1 cm.
We next tested cuticle permeability, which has been linked to changes in cuticular wax and plant responses to pathogens (Kurdyukov et al., 2006; Chassot et al., 2007; Tang et al., 2007). When leaves were stained with toluidine blue to reveal permeability of the cuticle, the wild-type leaves failed to take up the stain, suggesting an intact cuticular barrier, but the sdg8 and sdg25 single and double mutants stained intensely indicating increased permeability of the cuticle (Figure 10E). Similarly, the ccr2 and cer3 mutants showed increased cuticle permeability as reveled by the densely stained leaves after incubation with toluidine blue (Figure 10F). Thus, sdg, ccr2, and cer3 mutations alter cuticle functions, and the cuticular defects that cause cuticle permeability are associated with enhanced susceptibility to pathogens. In summary, SDGs mediate cuticle functions through HLM of genes, including CER3 and CCR2.
DISCUSSION
In eukaryotic organisms, transcription and other DNA-dependent processes occur within the context of chromatin. Cellular processes that modulate the structure of chromatin are one of several epigenetic mechanisms that modulate transcription, including histone modifications and microRNA and DNA methylation. In this report, we examined the impacts of altered histone lysine methylation profiles caused by single or double mutations in two histone methyl transferase genes SDG8 and SDG25. We show that (1) sdg8 and sdg25 single and double mutants broadly compromise plant immunity pathways, including peptide- and flg22-triggered immunity, ETI, and SAR. (2) The SDG8- and SDG25-regulated plant immunity is mediated, at least partially, through CCR2 and CER3, which are essential regulators of carotenoid and cuticular wax biosynthesis, respectively. Thus, our data establish a strong link between immune response functions of the two HMTs to lipid and carotenoid accumulation. The reduced lipid accumulation and increased permeability of the leaf cuticle in the sdg, ccr2, and cer3 mutants establish a definite contribution to plant immunity. (3) SDG8 affects both H3K36 and H3K4 methylation, whereas SDG25 affects primarily H3K4 methylation. The double mutant was severely attenuated for global H3K4me3, H3K36me2, and H3K36me3, consistent with the observed greater deficiencies in defense responses. In addition, H3K4me3 and H3K36me3 at the CCR2 and CER3 loci were severely reduced in the sdg8 mutant. (4) Interactions between the different HLMs and between HLM and H2Bubn were observed, with both modifications impacting each other and establishing unexpected crosstalk between the different histone modifications. Interestingly, sdg25 attenuated H3K4me1 was unexpectedly increased in the sdg8 mutant. (5) Transcriptome analysis revealed the genome-wide impact of altered H3K4 and H3K36 methylations on basal and induced expression of genes in lipid and fatty acid metabolism and on those acting at different levels of the plant immune signaling hierarchy. (6) SDG8 is broadly epistatic to SDG25 for some HLMs but additive for the disease resistance. This is consistent with SDG25 and SDG8 having overlapping and distinct functions in plant immunity, HLMs, lipid accumulation, and maintaining the integrity of the plant cuticle.
The multilayered plant immune mechanism is composed of constitutive and inducible defenses regulated by a complex web of cellular processes. When pathogens breach constitutive defenses, inducible defenses become pivotal to limit further pathogen ingress into the host tissue. Transcriptional activation of genes encoding various inducible defenses underlies the major differences between resistant and susceptible plants. The state of chromatin in the vicinity of genes brings about either a permissive or a repressive state of gene expression. Certain HLMs confer a permissive state for transcription, whereas others correlate with repression (Martin and Zhang, 2005). H3K4me3 in promoter and coding regions is often considered permissive, but the functions of H3K4me2 and H3K4me1 are more complex (Liu et al., 2010). RNA-seq data revealed that SDGs affect gene expression positively and negatively at basal as well as in response to infection. Basal expression of CCR2 and CER3 genes is SDG dependent. The reduced expression of CCR2 and CER3 after infection is consistent with reduced HLMs but is intriguing in light of the positive role of the two genes in immune responses.
Thus, it is likely that plant immunity mediated by CCR2 and CER3 is primarily dependent on basal HLM. SDG8 and SDG25 affect HLMs that are basally present at CER3 and CCR2 loci and may be required to establish a permissive chromatin state by either limiting the spread of repressive chromatin marks and/or potentiating a rapid transcriptional activation in response to infection as recently suggested (Berr et al., 2012). It is also possible that HLMs on SDG target genes that are not analyzed here are inducible which then activate the transcription of defense-related genes.
Beyond functions in PTI, both SDG25 and SDG8 were recently implicated in the regulation of resistance gene expression (Palma et al., 2010; Xia et al., 2013). SDG8 is required for autoimmunity responses in the Arabidopsis acd11 mutant whereby the acd11 sdg8 double mutant recovers from the accelerated cell death, and this has been attributed to the regulation of an RPS4-like R protein by SDG8 (Palma et al., 2010). SDG8 has been implicated in fungal resistance, and our observations extend its role in PTI (Berr et al., 2010). Interestingly, the expression of plant defense marker genes and known signaling components MKK3 and MKK5 are dependent on SDG8-mediated H3K36 methylation for full expression. Thus, SDG8 has a significant regulatory role consistent with its strong phenotypes in disease resistance. SDG25 also regulates the expression of the NBS-LRR genes SNC1 and RPP4 via H3K4me3 (Xia et al., 2013). Interestingly, the expression of BIK1, PBS1, and RPS5 is dependent on functional SDG25 and SDG8, consistent with the susceptibility of sdg mutants to the strain of Pst DC3000 expressing AvrPphB, although the mechanism involved in this particular case needs further study. BIK1 and PBS1 are both cleaved by the AvrPphB cysteine protease and RPS5 is the resistance protein that mediated ETI to AvrPphB (Shao et al., 2003; Zhang et al., 2010). Overall, the outcome in the form of plant immunity and other phenotypes is a result of complex interactions between the various histone modifications on plant gene expression.
RNA-seq analysis revealed the genome-wide regulatory impact of H3K4 and H3K36 methylations. The basal and induced expression of a wide variety of genes is dependent on SDG8 and SDG25. Direct association of SDG8 and/or SDG25 with three cis-regulatory motifs identified in this report or in previous studies (Li et al., 2015b) is likely to mediate basal and B. cinerea-induced gene expression. The functions of SDG8 in light, carbon response, and energy metabolism (Li et al., 2015b) are linked to lipid and carotenoid biosynthesis and ultimately to plant immunity. The bZIP (G-box), FORCA cis-element, and the AT-hook protein binding elements suggest the role of transcriptional regulators that may function with SDGs to mediate responses to pathogens, light, and carbon (Evrard et al., 2009). The AT-hook motif is a DNA binding protein motif that has been found in the high mobility group of nonhistone chromosomal proteins (Kim et al., 2011). The function of most AHL genes has not been studied but their nuclear localization and their annotation as nonhistone protein components of chromatin is consistent with the functions of SDG in HLM. The first two elements in SDG8- and/or SDG25-regulated genes were previously identified in carbon and light-regulated SDG-dependent genes. Among the list of SDG8 bound genes identified previously, 30 B. cinerea-induced genes were identified in this report, suggesting that their expression requires direct association with the SDG8 and/or SDG25 genes, potentially altering HLM on target genes which in turn recruits factors that modulate transcription.
This report establishes the impact of global and specific HLMs mediated by SDG8 and/or SDG25 on interactions between HLM and H2Bubn, another histone modification known to regulate gene expression. Interestingly, the sdg8 sdg25 double mutant has severe defects in plant immunity due to extensive changes in H3K4 and H3K36 methylations and in H2Bubn, both globally and at SDG target loci. The influence of HLM-regulated genes on plant lipid accumulation and cuticle integrity has been clearly established. H2Bubn has been suggested to act upstream of H3K4me3 (Soares and Buratowski, 2013), facilitating rapid modulation of gene expression during photomorphogenesis in Arabidopsis (Bourbousse et al., 2012). In the yeast Saccharomyces cerevisiae, the ubiquitin conjugating enzyme Rad6 is required for H3K4 methylation. H2Bubn on lysine 123 serves as the signal for methylation of histone H3, which leads to silencing of genes located near the telomeres (Dover et al., 2002). In Arabidopsis, distribution of genome-wide H2Bubn together with transcription profiling revealed that H2Bubn levels increase in concert with increased gene expression. Loss of H2Bubn results both in weaker responses of genes to light stimuli and in impaired defense and flowering (Cao et al., 2008; Dhawan et al., 2009). Interestingly, the contrasting impacts of SDG8 and SDG25 on certain HLMs were also unexpected considering the largely overlapping phenotypes of the two mutants. The sdg8 mutant had the most significant and broader impact on plant immunity and on HLM, which was often epistatic to sdg25.
Our data demonstrate that SDGs are critical regulators of plant immunity pathways, including PTI, ETI, and SAR, through various downstream target processes. CCR2 and CER3 genes, which have a clear role in plant defense, RLKs, and pathogenesis-related proteins, and defensins display SDG25- and/or SDG8-dependent expression, which explains the unusually broader impact of SDGs on plant immunity. The reduced expression of genes in fatty acid and lipid metabolism and lipid-transfer proteins coupled with the reduced accumulation of lipids in sdg and ccr2 mutants establish that mechanisms involved in plant immune responses mediated by SDGs and their target genes are dependent on lipid accumulation. These data establish the regulatory and functional links between HLM, lipid accumulation, and plant immunity pathways.
Some functions of fatty acids and lipids in plant immune pathways have been described recently (Kachroo and Kachroo, 2009). Lipid-transfer proteins can enhance the in vitro transfer of phospholipids between membranes, bind acyl chains, and participate in cutin formation and systemic resistance (Maldonado et al., 2002; Cecchini et al., 2015). In addition, SDGs regulate Arabidopsis CER3, previously implicated in cuticle membrane and cuticular wax production (Chen et al., 2003; Rowland et al., 2007). The exact biochemical function of CER3 is not known, but it encodes a transmembrane protein with similarity to the sterol desaturase family at the N terminus and to the short-chain dehydrogenase/reductase family at the C terminus (Chen et al., 2003). Reconstitution of plant alkane biosynthesis in yeast revealed that CER3 and CER1 form core components of a very-long-chain alkane synthesis complex (Bernard et al., 2012). Interestingly, cer3 is known to affect wax biosynthesis in the cuticle, but our data also demonstrated that the mutant is reduced in nonpolar lipids in both mature and meristem regions of the root as revealed by Nile red staining experiments.
Similarly, CCR2 was previously identified as a SDG8 target gene that is involved in carotenoid biosynthesis (Cazzonelli et al., 2009). The level of lutein, the most abundant carotenoid in higher plants, is reduced in the ccr2 mutant (Cazzonelli et al., 2009). SDG25-mediated HLM and H2Bubn are also required for full expression of CCR2 (Cazzonelli et al., 2009), which in turn is required for plant immunity. In plants, carotenoids are essential components required for photosynthesis, photoprotection and the production of carotenoid-derived phytohormones, including abscisic acid and strigolactone (Cazzonelli, 2011). The protection against active oxygen species is due to the quenching of excited states of photosensitizing molecules, quenching of singlet oxygen, and scavenging of free radicals (Gruszecki and Strzałka, 2005). Carotenoids affect the structural and dynamic properties of lipid membranes (Gruszecki and Strzałka, 2005); thus, protection of biomembranes against oxidative damage can be realized also through the modification of physical properties of the lipid phase of the membranes by carotenoids. Overall, we demonstrate that SDG8- and SDG25-regulated genes CCR2 and CER3 contribute to plant immunity functions through carotenoids, lipids, and cuticular waxes.
SDG8 and SDG25 are likely to have broader biological functions in plants that are consistent with a clear role in regulating a significant part of the plant transcriptome. It is likely that additional SDG target genes mediate the immune response and other functions of SDG8 and/or SDG25. Interestingly, both loss of H2Bubn and HLMs abrogate plant immunity pathways. It can be argued that loss of HLM leads to other modifications resulting in a broader and significant impact on plant physiological functions. HLM has the potential to affect all components in the plant immune response signaling pathway from pathogen perception to signaling and components that act as antimicrobial agents. Finally, how SDGs function with other transcriptional regulators and how they are recruited to specific genomic loci are still unclear. This report provides the foundation for future studies to identify factors that bind to the cis-elements on SDG-regulated genes as well as those that interact with SDG proteins.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes Columbia-0 (Col-0) and Col-3 were used as wild-type controls. The sdg8-2 (SALK_026442), sdg25-1 (SALK_149692C), sdg2 (SALK_021008), sdg4 (SALK_128444), sdg5 (SAIL_55_C04), sdg13 (SALK_021874), sdg18 (SAIL_119_B08), sdg26 (SALK_013895), sdg27 (SALK_149002C), sdg30 (SALK_117262), sdg39 (SALK_024470C), ccr2 (SAIL_791_A10), At4g14610 (SALK_080562C), At3g04210 (SALK_044806), mee47 (SALK_029478), gly61 (SALK_108484), cyp81 (SALK_005861), cyp90 (SALK_090732), cer3 (SALK_206591), crk21 (SALK_035263C), and hub1-6 (WiscDsLox433B10) mutants are in the Col-0 background, whereas sdg25-2 (SAIL_446_F12) is in the Col-3 background. The T-DNA insertion mutants were obtained from the ABRC. The sdg8-2 sdg25-1 and sdg25-1 hub1-6 double mutants were constructed by crossing with homozygous single mutants followed by PCR screens to identify double mutant progenies. Gene-specific primer pairs and T-DNA specific primers were used for genotyping (Supplemental Table 3). Plants were grown in soil in a growth chamber at 23°C under a 12-h-light/12-h-dark cycle, with 70% relative humidity and 140 to 150 μE m−2 s−1 light.
Disease Assays
Disease assays were conducted as described previously (Zheng et al., 2006). Botrytis cinerea and Alternaria brassicicola strain MUCL20297 were cultured on V8 medium agar, and spores were suspended in 1% Sabouraud maltose broth buffer (BD Difco) for plant inoculations. Disease assays for B. cinerea were conducted by drop inoculation (5-μL droplets, 2.5 × 105 spores/mL) on the surface leaves of 4- to 5-week-old plants. The A. brassicicola disease assay was performed on detached leaves by drop inoculation with 5 μL of 2.5 × 105 spores/mL. In both cases, inoculated plants were kept at high humidity for 3 to 5 d before disease lesions were measured.
The bacterial pathogens Pst DC3000, Pst DC3000 hrcC, and Pst DC3000 (AvrRpm1, AvrB, AvrRpt2, and AvrPphB) were cultured on a rotary shaker at 200 rpm at 28°C, in King’s B medium (20 g peptone, 10 g glycerol, 1.5 g K2HPO4, and 6 mL of 1 M MgSO4/L, pH 7.2) with corresponding antibiotics for 16 to 18 h and then resuspended in 10 mM MgCl2. To monitor bacterial growth in plants, leaves of 4- to 5-week-old plants were hand-infiltrated with the virulent Pst DC3000 (OD600 = 0.0002), the nonpathogenic Pst DC3000 hrcC (OD600 = 0.001), or the avirulent Pst DC3000 (OD600 = 0.002) using a needleless syringe. Inoculated plants were covered with a clear plastic lid to maintain high humidity after the liquid was completely absorbed. Leaf discs from three different leaves were sampled with a cork borer and ground in 100 μL of 10 mM MgCl2 in a 1.5-mL tube. The samples were thoroughly mixed with 900 μL of 10 mM MgCl2 and diluted 1:10 serially. The serial dilutions were plated on King’s B medium agar containing corresponding antibiotics. Plates were placed at 28°C and colonies were counted after 2 to 3 d.
PAMP Treatments and Systemic Acquired Resistance Assays
For PTI to pathogens, pep1 (Abbiotec) and flg22 (Alpha Diagnostic International) were used to activate PTI to B. cinerea and Pst DC3000 strains, respectively. Twenty-two hours prior to pathogen inoculation, 4- to 5-week-old plants were infiltrated with 1 μM Pep1 or 1 μM flg22 and each leaf was drop inoculated with 5-μL droplets of B. cinerea (2.5 × 105 spores/mL) or infiltrated with Pst DC3000 (OD600 = 0.0002) or Pst DC3000 hrcC (OD600 = 0.001). Double-deionized water was infiltrated for both pep1 and flg22 as a mock inoculation control. To assay SAR, the lower leaves were inoculated with 10 mM MgCl2 or avirulent bacteria (OD600 = 0.02), and 48 h later, secondary (systemic) leaves were inoculated with the virulent bacterial pathogen Pst DC3000 (OD600 = 0.001) or the fungal pathogen B. cinerea (2.5 × 105 spores/mL).
RNA Extraction and Expression Analysis
Total RNA was extracted from using the Trizol reagent according to the manufacturer’s instructions (Invitrogen). After DNase treatment (Promega), the first-strand cDNA was synthesized from 2 μg of total RNA with the AMV reverse transcriptase (NEB). Quantitative PCR analyses were performed on the Mx3000P real-time PCR detection system (Stratagene) using a SYBR Green Supermix (Bio-Rad) with gene-specific primers. Arabidopsis Actin2 was used as an endogenous control for normalization. At least three technical replicates of the qRT-PCR analysis were used for each sample with a minimum of two biological replicates. Expression levels were calculated by the comparative cycle threshold (Ct) method. Primers used for qRT-PCR are listed in Supplemental Table 3.
Histone Extraction and Immunoblot Analysis
For detection of global levels of H3 lysine methylation, histone proteins were isolated from 4- to 5-week-old plants as described (Li et al., 2012). The proteins were separated on 15% SDS-PAGE gels, transferred to PVDF membranes, and immunoblotted with specific antibodies: anti-H3K4me1 (07-436; EMD Millipore), anti-H3K4me2 (07-030; EMD Millipore), anti-H3K4me3 (07-473; EMD Millipore), anti-H3K36me1 (ab9048; Abcam), anti-H3K36me2 (07-369-I; EMD Millipore), anti-H3K36me3 (ab9050; Abcam), and anti-H3 (ab1791; Abcam) as a loading control. The bound primary antibodies were observed with the enhanced chemiluminescence detection system according to the manufacturer's protocol (Thermo Scientific).
Chromatin Immunoprecipitation
ChIP assays were performed following a published protocol (Saleh et al., 2008) with minor modifications. Chromatin samples were prepared from 4- to 5-week-old plants. Leaf tissues (1.5 to 2 g) were cross-linked by vacuum infiltration with fixation buffer containing 1% (v/v) formaldehyde for 7 min, and the chromatin was sheared to 200- to 1000-bp DNA fragments by sonication. After preclearing with salmon sperm DNA/protein A agarose beads (EMD Millipore), the chromatin samples were immunoprecipitated with specific antibodies to H3 lysine methylation (see previous section) or with a specific antibody to H2B monoubiquitination (MM-0029-P; MediMabs) or Immunoglobulin G (sc‐2027; Santa Cruz) as a negative control. After reverse cross-linking at 65°C for overnight, samples were incubated with 10 μL of 0.5 M EDTA, 20 μL of 1 M Tris-HCl, pH 6.5, and 1 μL of proteinase K (20 mg mL−1; Thermo Scientific) at 45°C for 1.5 h to digest the proteins. DNA samples were purified using a silica membrane column (Macherey-Nagel) and eluted in 50 μL double-deionized water. Two microliters of immunoprecipitated and input DNA was used for qPCR with specific primers as listed in Supplemental Table 3. The enriched regions of ChIP DNA were normalized to the S-Adenosyl Methionine Synthase (SAM) gene, which was used as an internal control. The enrichment level was calculated relative to the input DNA as a percentage of input DNA. Each sample was quantified at least in triplicate.
RNA-Seq and Data Analysis
Plants were grown in soil for 4 weeks and inoculated with B. cinerea (2.5 × 105 spores/mL). At 36 h after mock or B. cinerea inoculation, rosette leaves were collected from three biological replicates (∼12 plants each). Total RNA was extracted, treated with DNase, and purified with the RNA Clean and Concentration-25 (Zymo Research). The quality of the total RNA was determined by a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer.
Total RNA was isolated as described in the protocol of Spectrum Plant Total RNA Kit with on-column DNase digestion (Sigma-Aldrich). For each sample, 3-µg of total RNA was used to prepare the mRNA-seq library according to the TrueSeq RNA Sample Prep Kit protocol (Illumina). Library quality control and quantification were performed with an Experion DNA 1K Chip (Bio-Rad) and a Qubit fluorometer (Invitrogen), respectively. For each library, ∼40 million 100-bp paired-end sequences were generated using an Illumina HiSeq 2500 sequencer. After removing low-quality sequences containing uncalled bases, TopHat 2 software (Kim et al., 2013) was used to align the RNA-seq reads against the reference genome (TAIR 10). TopHat2 alignment parameters were set to allow a maximum of two mismatches and to exclude reads mapping to more than one position on the reference. Moreover, only reads for which both pairs successfully aligned were considered. The gene counts were extracted using the HTSeq python tool (Anders et al., 2015). Differential expression analyses were performed with the edgeR package (Robinson et al., 2010) using empirical Bayesian methods. To filter out weakly expressed genes, only those genes with a minimum expression level of 1 count per million in at least three replicates of one condition were included in the analysis. Genes with a log fold change above 1 (2-fold change), false discovery rate of below 0.05, and P value below 0.01 were considered differentially expressed between conditions. To assess the variability among samples, we performed hierarchical clustering and principal component analysis. Hierarchical clustering was performed based on Euclidean distances. PCA was conducted using the prcomp command with default parameters in the R software package.
AgriGO (http://bioinfo.cau.edu.cn/agriGO/) and ReviGO (http://revigo.irb.hr/) (Du et al., 2010; Supek et al., 2011) were used to identify putative biological functions and biochemical pathways for differentially expressed genes and to find statistically overrepresented GO terms. To expand our functional analysis of differentially expressed genes, we used MapMan software (http://mapman.gabipd.org) to visualize and biochemical pathway overlays as previously described (Thimm et al., 2004).
Nile Red and Toluidine Blue Staining
For Nile red staining, seeds were harvested and surface sterilized by incubation in 10% hypochlorite and 70% ethanol for 5 min each and then rinsed several times with sterile distilled water. The seeds were germinated and grown on vertically on half-strength Murashige and Skoog basal salt mixture (PhytoTechnology Laboratories) containing 1% (w/v) sucrose and 0.7% (w/v) agar, pH adjusted to 5.8, for 7 d. Nile red staining was performed as described in order to determine the lipid content in seedlings (Li et al., 2015a). After Nile red staining, nonpolar lipid were detected (at excitation 514 nm, emission at 520 to 560 nm), and polar lipids were detected (excitation at 534 nm, emission at 600 to 700 nm). The Nikon confocal microscopic images system A1R was used for microscopy images. Confocal images were processed with Photoshop CS6 and fluorescence intensities of images were measured with ImageJ (http://imagej.nih.gov/ij/). Statistical significance was assessed using Student's t test. For Toluidine blue staining, leaf samples were taken from 4- to 5-week-old plants grown in soil. Toluidine blue staining was performed to determine the permeability of the cuticle following a published protocol (Tanaka et al., 2004).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACTIN2 (At3g18780), BIK1 (At2g39660), CCR2 (At1g06820), CER3 (At5g57800), CRK21 (At4g23290), CYP81 (At5g57220), CYP90 (At5g05690), FLS2 (At5g46330), FRK1 (At2g19190), GLY61 (At2g41640), HUB1 (At2g44950), MEET47 (At4g00950), PBS1 (At5g13160), PDF1.2 (At5g44420), PMG (At2g17740), PR1 (At2g14610), RPS5 (At1g12220), SAM (At4g01850), SDG2 (At4g15180), SDG4 (At4g30860), SDG5 (At1g02580), SDG8 (At1g77300), SDG13 (At1g04050), SDG18 (At5g43990), SDG25 (At5g42400), SDG26 (At1g76710), SDG27 (At2g31650), SDG30 (At1g05830), SDG39 (At2g19640), and SNC1 (At4g16890).
Supplemental Data
Supplemental Figure 1. Reverse Genetic Screens on Selected B. cinerea-Responsive SDG Genes for Altered Responses to Fungal Infection.
Supplemental Figure 2. SDG25 Is Involved in Resistance to Botrytis cinerea.
Supplemental Figure 3. Characterization of sdg8-2, sdg25-1, and sdg8-2 sdg25-1 Mutant Plants.
Supplemental Figure 4. SDG25 and SDG8 Are Required for Resistance to Alternaria brassicicola.
Supplemental Figure 5. The Roles of SDG25 and SDG8 in Effector-Triggered Immunity.
Supplemental Figure 6. Network of Processes Affected Based on Genes Upregulated in the sdg8 sdg25 Double Mutant after B. cinerea Infection.
Supplemental Figure 7. Network of Processes Affected Based on Genes Downregulated in the sdg8 sdg25 Double Mutant after B. cinerea Infection.
Supplemental Figure 8. Loss-of-Function Mutation in SDG Target Genes CER3 and CCR2 Causes Enhanced Susceptibility to B. cinerea and A. brassicicola.
Supplemental Figure 9. Regulation of H3K4 and H3K36 Methylations at SNC1 Chromatin by Histone Methyltransferases and Histone 2B Monoubiquitination.
Supplemental Table 1. Microarray-Based Gene Expression Data for Arabidopsis SET Domain Group Genes in Response to Botrytis cinerea Inoculation.
Supplemental Table 2. Examples of SDG8 and/or SDG25 Target Genes that Contain cis-Regulatory Sites within a 1-kb region of their Promoter.
Supplemental Table 3. Sequences of Primers Used in This Study.
Supplemental Data Set 1. Functional Categorization of the Downregulated Genes 36 h after Botrytis cinerea Inoculation in sdg8 sdg25 Plants.
Supplementary Material
Acknowledgments
This research was funded by grants from the National Science Foundation (IOS-1456594) and the Next-Generation BioGreen 21 Program (SSAC, Project PJ01137902), Rural Development Administration, Republic of Korea.
AUTHOR CONTRIBUTIONS
S.L. conducted most of the experiments. S.L and T.M. designed the research, analyzed the data, wrote the article, and directed the project. F.F. performed the analyses of RNA-seq data. S.X. conducted some of the initial reverse genetic screens. S.Y.L. and D.-J.Y. helped in the design and analysis of the data and wrote the article.
Glossary
- ETI
effector-triggered immunity
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- HLM
histone lysine methylation
- SAR
systemic acquired resistance
- HMT
histone methyl transferase
- dpi
days postinoculation
- GO
Gene Ontology
- ChIP
chromatin immunoprecipitation
References
- Alvarez M.E., Nota F., Cambiagno D.A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke L., Sanchez-Perez G.F., Snel B. (2012). Contribution of the epigenetic mark H3K27me3 to functional divergence after whole genome duplication in Arabidopsis. Genome Biol. 13: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Domergue F., Pascal S., Jetter R., Renne C., Faure J.D., Haslam R.P., Napier J.A., Lessire R., Joubès J. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Ménard R., Heitz T., Shen W.H. (2012). Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell. Microbiol. 14: 829–839. [DOI] [PubMed] [Google Scholar]
- Berr A., McCallum E.J., Alioua A., Heintz D., Heitz T., Shen W.H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Hamon M., Cossart P. (2012). Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2: a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bourbousse C., Ahmed I., Roudier F., Zabulon G., Blondet E., Balzergue S., Colot V., Bowler C., Barneche F. (2012). Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 8: e1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch G., Ransom R., Lechner T., Walton J.D., Loidl P. (1995). Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7: 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Dai Y., Cui S., Ma L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C.I. (2011). Carotenoids in nature: insights from plants and beyond. Funct. Plant Biol. 38: 833–847. [DOI] [PubMed] [Google Scholar]
- Cazzonelli C.I., Cuttriss A.J., Cossetto S.B., Pye W., Crisp P., Whelan J., Finnegan E.J., Turnbull C., Pogson B.J. (2009). Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini N.M., Steffes K., Schläppi M.R., Gifford A.N., Greenberg J.T. (2015). Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nat. Commun. 6: 7658. [DOI] [PubMed] [Google Scholar]
- Chassot C., Nawrath C., Métraux J.P. (2007). Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 49: 972–980. [DOI] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U., Pieterse C.M., Mauch-Mani B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7: 210–216. [DOI] [PubMed] [Google Scholar]
- De-La-Peña C., Rangel-Cano A., Alvarez-Venegas R. (2012). Regulation of disease-responsive genes mediated by epigenetic factors: interaction of Arabidopsis-Pseudomonas. Mol. Plant Pathol. 13: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten Scheid O., Mengiste T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A. (2002). Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368–28371. [DOI] [PubMed] [Google Scholar]
- Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard A., Ndatimana T., Eulgem T. (2009). FORCA, a promoter element that responds to crosstalk between defense and light signaling. BMC Plant Biol. 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin E.M., Levine A. (2002). Infection of Arabidopsis with a necrotrophic pathogen, Botrytis cinerea, elicits various defense responses but does not induce systemic acquired resistance (SAR). Plant Mol. Biol. 48: 267–276. [DOI] [PubMed] [Google Scholar]
- Gruszecki W.I., Strzałka K. (2005). Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740: 108–115. [DOI] [PubMed] [Google Scholar]
- Gutzat R., Mittelsten Scheid O. (2012). Epigenetic responses to stress: triple defense? Curr. Opin. Plant Biol. 15: 568–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyse K.S., Weber S.E., Lipps H.J. (2009). Histone modifications are specifically relocated during gene activation and nuclear differentiation. BMC Genomics 10: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Ryan C.A. (2007). Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 104: 10732–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhänsel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kachroo A., Kachroo P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47: 153–176. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. (2009). Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299. [DOI] [PubMed] [Google Scholar]
- Kim H.B., Oh C.J., Park Y.C., Lee Y., Choe S., An C.S., Choi S.B. (2011). Comprehensive analysis of AHL homologous genes encoding AT-hook motif nuclear localized protein in rice. BMB Rep. 44: 680–685. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S., Faust A., Nawrath C., Bär S., Voisin D., Efremova N., Franke R., Schreiber L., Saedler H., Métraux J.P., Yephremov A. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K., Luo H., Chai M., Dhawan R., Lai Z., Mengiste T. (2011). Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 23: 2831–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.X., Sun R., Hicks G.R., Raikhel N.V. (2015a). Arabidopsis ribosomal proteins control vacuole trafficking and developmental programs through the regulation of lipid metabolism. Proc. Natl. Acad. Sci. USA 112: E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qian W., Zhao Y., Wang C., Shen J., Zhu J.K., Gong Z. (2012). Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 11425–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Mukherjee I., Thum K.E., Tanurdzic M., Katari M.S., Obertello M., Edwards M.B., McCombie W.R., Martienssen R.A., Coruzzi G.M. (2015b). The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol. 16: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420. [DOI] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X., Zhou J.M. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110: 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Bruce T.J., Roberts M.R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado A.M., Doerner P., Dixon R.A., Lamb C.J., Cameron R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403. [DOI] [PubMed] [Google Scholar]
- Martin C., Zhang Y. (2005). The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6: 838–849. [DOI] [PubMed] [Google Scholar]
- Ng D.W., Wang T., Chandrasekharan M.B., Aramayo R., Kertbundit S., Hall T.C. (2007). Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta 1769: 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6: e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Grossniklaus U. (2011). Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 14: 195–203. [DOI] [PubMed] [Google Scholar]
- Pecinka A., Mittelsten Scheid O. (2012). Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 53: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan T.K., Hartzog G.A. (2010). Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics 184: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S., De Vos M., Casteel C.L., Tian D., Halitschke R., Sun J.Y., Agrawal A.A., Felton G.W., Jander G. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158: 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ferrer V., Voinnet O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60: 485–510. [DOI] [PubMed] [Google Scholar]
- Ryals J., Lawton K.A., Delaney T.P., Friedrich L., Kessmann H., Neuenschwander U., Uknes S., Vernooij B., Weymann K. (1995). Signal transduction in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 92: 4202–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schneider R., Grosschedl R. (2007). Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 21: 3027–3043. [DOI] [PubMed] [Google Scholar]
- Shao F., Golstein C., Ade J., Stoutemyer M., Dixon J.E., Innes R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75: 243–269. [DOI] [PubMed] [Google Scholar]
- Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares L.M., Buratowski S. (2013). Histone Crosstalk: H2Bub and H3K4 methylation. Mol. Cell 49: 1019–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Angel A., Howard M., Dean C. (2012). Vernalization - a cold-induced epigenetic switch. J. Cell Sci. 125: 3723–3731. [DOI] [PubMed] [Google Scholar]
- Suganuma T., Workman J.L. (2011). Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80: 473–499. [DOI] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Tanaka H., Machida C., Watanabe M., Machida Y. (2004). A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 37: 139–146. [DOI] [PubMed] [Google Scholar]
- Tang D., Simonich M.T., Innes R.W. (2007). Mutations in LACS2, a long-chain acyl-coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol. 144: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Thomma B.P., Nürnberger T., Joosten M.H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13: 459–465. [DOI] [PubMed] [Google Scholar]
- Walley J.W., Rowe H.C., Xiao Y., Chehab E.W., Kliebenstein D.J., Wagner D., Dehesh K. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4: e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw N.C., Whitelaw E. (2008). Transgenerational epigenetic inheritance in health and disease. Curr. Opin. Genet. Dev. 18: 273–279. [DOI] [PubMed] [Google Scholar]
- Xia S., Cheng Y.T., Huang S., Win J., Soards A., Jinn T.L., Jones J.D., Kamoun S., Chen S., Zhang Y., Li X. (2013). Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol. 162: 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605. [DOI] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B., Yang D.L., Shi Z., Dong H., Hua J. (2014). Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol. 165: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.