Abstract
Chromatin insulators orchestrate gene transcription during embryo development and cell differentiation by stabilizing interactions between distant genomic sites. Mutations in genes encoding insulator proteins are generally lethal, making in vivo functional analyses of insulator proteins difficult. In Drosophila, however, mutations in the gene encoding the Suppressor of Hairy wing insulator protein [Su(Hw)] are viable and female sterile, providing an opportunity to study insulator function during oocyte development. Whereas previous reports suggest that the function of Su(Hw) in oogenesis is independent of its insulator activity, many aspects of the role of Su(Hw) in Drosophila oogenesis remain unexplored. Here we show that mutations in su(Hw) result in smaller ring canal lumens and smaller outer ring diameters, which likely obstruct molecular and vesicle passage from nurse cells to the oocyte. Fluorescence microscopy reveals that lack of Su(Hw) leads to excess accumulation of Kelch (Kel) and Filament-actin (F-actin) proteins in the ring canal structures of developing egg chambers. Furthermore, we found that misexpression of the Src oncogene at 64B (Src64B) may cause ring canal development defects as microarray analysis and real-time RT-PCR revealed there is a three fold decrease in Src64B expression in su(Hw) mutant ovaries. Restoration of Src64B expression in su(Hw) mutant female germ cells rescued the ring phenotype but did not restore fertility. We conclude that loss of su(Hw) affects expression of many oogenesis related genes and down-regulates Src64B, resulting in ring canal defects potentially contributing to obstruction of molecular flow and an eventual failure of egg chamber organization.
Keywords: chromatin insulators, suppressor of Hairy wing, Su(Hw), Drosophila, oogenesis, ring canals, Src64B
Introduction
While DNA provides the blueprint for eukaryotic cell structure and function, chromatin structure is critical for regulating gene expression, (AGALIOTI et al. 2000; GUCCIONE et al. 2006; KOUZARIDES 2007; LI et al. 2007). In addition, regulatory sequences such as enhancers may act over tens of kilobases of DNA in conjunction with cognate promoters in order to activate the expression of a target gene (MARSMAN and HORSFIELD 2012; ONG and CORCES 2011). Chromatin insulators are one class of genomic elements that were initially characterized because of their ability to block communication between enhancers and promoters and to protect genes from heterochromatin spread (BRASSET and VAURY 2005; GASZNER and FELSENFELD 2006; YANG and CORCES 2012). However, recent progress in high throughput technologies has revealed that not all insulators sites in the genome seem to block enhancers (NEGRE et al. 2010), and evidence for the heterochromatin barrier function of insulators has been questioned based on the lack of barrier activity at sites in the genome that flank Polycomb domains (VAN BORTLE et al. 2012). Because insulators facilitate long-range interactions between distant genomic sites, and because recent developments in chromosome conformation capture techniques have allowed to determine precise genome-wide long-range interactions, a new paradigm is emerging suggesting that the major function of insulators is to help organize the tridimensional organization of the genome to ensure proper temporal and spatial gene expression (LABRADOR and CORCES 2002; ONG and CORCES 2014; PHILLIPS-CREMINS and CORCES 2013; PHILLIPS-CREMINS et al. 2013; RAO et al. 2014; SCHOBORG and LABRADOR 2014; WALLACE and FELSENFELD 2007). Albeit these advances in our understanding of the role of insulators in genome organization, the precise mechanism by which insulators regulate gene expression is not known.
Chromatin insulators have been discovered in a variety of organisms ranging from yeast to humans (SCHOBORG et al. 2013). One of the best-characterized insulators is the Drosophila gypsy insulator, which requires the function of three major proteins: Su(Hw), which directly binds insulator DNA, Modifier of mdg4 protein [Mod(mdg4)67.2], and Centrosomal protein 190 (CP190), which bind Su(Hw), allowing chromatin insulator function (GERASIMOVA et al. 1995; GHOSH et al. 2001; PAI et al. 2004). Although the two binding partners of Su(Hw), Mod(mdg4)67.2 and CP190 proteins are required for chromatin insulator activity, only Su(Hw) is essential for oogenesis (BAXLEY et al. 2011).
In Drosophila, oogenesis begins at the first asymmetric division of a germline stem cell located at the far anterior-tip of the germarium. This asymmetric cell division gives rise to a daughter stem cell and a cystoblast, which will later form an egg chamber by generating sixteen cells following four incomplete mitotic divisions. In each developing egg chamber only one cell adopts the oocyte cell fate while the remaining fifteen cells become nurse cells, which will produce essential nutrients to provide support for the oocyte and later embryo development. In the germarium, each mitotic division ends with an incomplete cytokinesis generating cytoplasmic bridge structures known as ring canals. These ring canals will eventually interconnect all germline cells in the egg chamber. In the germarium, a germline-specific organelle called fusome grows within the cystocytes as a continuous branched structure that winds through and plugs all ring canals. Each cell division produces another branch of the fusome, connecting the new cell to the cluster of previously formed cells. This process continues until all sixteen cells form, and the plugs eventually break down once the cystocytes leave the germarium. During the following stages of oogenesis, the ring canals remain open, functioning as channels that allow transport of cytoplasmic constituents such as mRNA, proteins, organelles and vesicles, which ultimately travel to the developing oocyte (DE CUEVAS and SPRADLING 1998; LIN et al. 1994).
This molecular flow towards the developing oocyte occurs in two phases: the early and slow phase and the late and fast phase. The first is a process releasing specific and selected molecules for transport to the oocyte, and the second is a rapid process beginning at stage 10B when nurse cells dump the entirety of their cytoplasmic contents into the oocyte (BATE and MARTINEZ ARIAS 1993; BUSZCZAK and COOLEY 2000; HAGLUND et al. 2011). Phenotypes described as “dumpless” (i.e. defective yolk deposit phenotype) commonly arise from mutations in genes encoding components of protein complexes involved in cytoskeleton organization pathways or ring canal formation, such as Hu-li tai shao (hts), kelch, and Src64B (DODSON et al. 1998; XUE and COOLEY 1993; YUE and SPRADLING 1992).
Su(Hw) is detected in the nucleus of both somatic follicle cells and germ cells in ovaries, and loss of Su(Hw) results in female sterility. It was first noticed that su(Hw) mutations suppressed yolk deposition and sequentially arrested ovary development at mid-oogenesis, thereby causing sterility (BAXLEY et al. 2011; HARRISON et al. 1993; KLUG et al. 1968; KLUG et al. 1970). The su(Hw) allele su(Hw)f encodes a protein with a defective Zinc-finger 10, which causes only a partial loss of insulator activity even though germline development remains normal (HARRISON et al. 1993). Analyzing the global role of Su(Hw) during oogenesis, Baxley et al. (2011) concluded that the defects of Su(Hw) in female germline development are independent of its insulator function, and suggested that the functions of Su(Hw) in regulating insulator activity and female germline development are separable (BAXLEY et al. 2011). Additionally, a recent report suggests that Su(Hw) may function as a classic transcriptional regulator of oogenesis and that a major effect of the absence of Su(Hw) during oogenesis is failure to repress expression of RNA-binding protein 9 (Rbp9). In fact, reducing Rbp9 expression by half within ovaries largely rescues oogenesis defects in su(Hw) mutants, although fertility was not completely restored given that eggs produced by rescued females contained patterning defects and did not produce viable offspring (SOSHNEV et al. 2013; SOSHNEV et al. 2012).
In this study, we used a cell type and stage specific Gal4-UAS binary system to examine spatial and temporal expression of Su(Hw) and determine its precise role in different stages of oogenesis and ovary development. We show that germline specific expression of Su(Hw) driven by Gal4 in su(Hw) mutant ovaries is necessary for yolk-deposition and normal oocyte development. At the same time, Gal4 driven expression of Su(Hw) in somatic follicle cells is not sufficient to rescue oogenesis. Interestingly, we found that intracellular transport within egg chambers is blocked prior to stage 8 in su(Hw) mutants, suggesting that this blockage may result from defective ring canal development during oogenesis. Our results show that ring canals in su(Hw) mutant ovaries have an abnormal morphology with excess accumulation of F-actin and Kelch proteins, yielding a small lumen phenotype similar to that of unrelated ring canal mutations that prevent molecular passage. Furthermore, microarray data shows that eighty-two misregulated genes in su(Hw) mutants participate in oogenesis and among these genes Src64B is significantly down-regulated. Overexpression of Src64B in su(Hw) mutants rescues the ring canal phenotype but cannot completely restore intracellular transport within egg chambers, suggesting that Su(Hw) is required for other aspects of egg chamber organization necessary for proper transport and development in addition to ring canal formation.
Materials and methods
Fly stocks and culture conditions. All fly stocks were cultured using cornmeal-agar food and yeast in a 25°C incubator. Fly stocks used in this study included su(Hw) mutant lines: w1118;PBac(RB)su(Hw) e04061/TM6B, and y2 w ct6; su(Hw) v/TM6B (HARRISON et al. 1992). Expression of Su(Hw)::eGFP [y w; P{su(Hw)::eGFP,w+}] (SCHOBORG et al. 2013) was driven by various Gal4 drivers including w*; P{en2.4-Gal4}e22C; w*;P{GAL4-nos.NGT40}; w*; P{nos-Gal4::VP16} (VAN DOREN et al. 1998), w*; P{matalpha4-GAL-VP16}V37, and y w; P{Tj-Gal4}. For ectopic expression of Src64B we used w*; P{UAS-Src64B.C}2.
Egg chamber staining and image processing. Three to five-day-old female flies were collected and their ovaries were dissected for whole mount ovary immunostaining following standard protocols (PAGE and HAWLEY 2001). Tissues were fixed with heptane in 4% paraformaldehyde and washed with PBST. Fixed tissues were incubated with blocking solution. Multiple primary antibodies were utilized for staining using the following dilutions: 1:100 rabbit anti-GFP antibody (Invitrogen), 1:200 mouse anti-Orb antibody (4H8), 1:200 mouse anti-Kel antibody (Kel-1B), 1:200 mouse anti-Hts F antibody (1B1), 1:200 anti-Hts RC and 1:200 Lamin Dm0 antibody (Hybridoma bank). Secondary antibodies were used with a 1:200 dilution and are as follow: FITC-conjugated anti-rabbit IgG, TexRed-conjugated anti-rabbit IgG and FITC-conjugated anti-mouse IgG (The Jackson Laboratory). F-actin staining was performed using TexRed-phallodin (Life Technologies). DNA was stained with 4?, 6-diamidino-2-phenylindole (DAPI, 0.5 μg/ml) and all samples were mounted in Vectashield mounting medium (Vector Laboratories).
Slides were analyzed using a Leica DM6000B wide-field epifluorescence microscope equipped with a Hamamatsu ORCA-ER CCD camera and a HC PL FLUOTAR 20x /0.50NA objective. Image acquisition was performed using Simple PCI v6.6 (Hamamatsu Photonics). Images were processed using the AutoQuant's 3D Deconvolution Algorithm utilizing an adaptive (blind) PSF implemented into Leica Deblur (v2.3.2) software. All wildtype and mutant samples were processed and imaged under identical conditions of immunostaining, microscope, camera and software settings. Egg chambers were measured using Image J software and specific stages were determined based on size (SULLIVAN et al. 2000).
Microarray and data analysis. 20 three-day-old wildtype (Oregon R) and homologous mutant (su(Hw)2) female flies were collected for ovary dissection. Total RNA was extracted from ovaries using TRIzol reagent (Invitrogen) and used for microarray hybridization of Affymetrix Drosophila 2.0 arrays (Cat. #900532). Microarray hybridization was performed by the Affymetrix microarray facility at the University of Tennessee, Knoxville. Three biological repeats of each genotype were analyzed. Microarray analysis was performed using R version 3.0.2. Raw expression data were normalized using the gcrma package (WU et al. 2004). The mas5calls function from the affy package (GAUTIER et al. 2004) was used to call each expression value present, absent, or marginal. Genes that were present in all replicates of at least one treatment group were kept for further analysis, resulting in 7324 genes. The limma package was used to compare gene expression between su(Hw) mutants and wildtype flies (SMYTH et al. 2005), and the p-values were adjusted by the FDR method (BENJAMINI and HOCHBERG 1995) to control the false discovery rate. Affymetrix probe IDs were matched with gene symbols and FlyBase IDs using the “drosophila2.db” annotation package from Bioconductor version 2.14. Oogenesis-related genes were identified using the QuickSearch tool at FlyBase (flybase.org) version FB2014_03 to search for “oogenesis”.
Real-time PCR. Three to five-day-old female flies were collected for ovary dissection, and late stage egg chambers after stage 9 were manually removed under the dissecting scope. Ovarian total RNA was purified using TRIzol reagent (Invitrogen) and was then converted to cDNA using the SuperScript First-strand cDNA synthesis kit (Invitrogen). For each genotype sample, three independent biological RNA samples were prepared. Real-time PCR was performed using specific primers of targeted genes and iQ SYBR Green Supermix (Biorad) while the reactions were set up on a BioRad iQ5 Multicolor Real-Time PCR Detection System. For each gene amplification three independent technical repeats were set up. Each amplification condition was optimized, and primer specificity was determined using the melting curve method. The transcriptional level of Src64B was normalized to the internal control Rp49 (ΔCt value), and the relative abundance of target gene transcripts among each genotype was determined using the relative quantitative method (ΔΔCt value). Primers used in this study: Src64B F- CATTCTGCTGATGGAGCTGT; Src64B R- CCGGGAAGTAGT GATTCGTT and Rp49 primers (WALLACE et al. 2010).
Fertility assay. Rescue of the su(Hw) sterility phenotype was determined by counting the number of eggs laid by two to three-day-old su(Hw) mutant females carrying either the su(Hw)::eGFP or Src64B transgenes driven by different Gal4 drivers. Eggs were collected for three days using grape juice agar plates containing wet yeast paste (SULLIVAN et al. 2000). The fertility rescue rate was calculated using the total number of eggs laid by rescued females divided by the total number laid by the same numbers of wildtype females.
Results
Oocyte development is defective in su(Hw) v/e04061 mutants. su(Hw) mutant females are sterile resulting from incomplete oogenesis, as mutant egg chambers ultimately undergo apoptosis following arrested development at mid-oogenesis (BAXLEY et al. 2011; HARRISON et al. 1993; HARRISON et al. 1992; KLUG et al. 1968; KLUG et al. 1970). In order to further characterize the role of Su(Hw) in oogenesis, we used the su(Hw)e04061 mutant, created by an insertion of a piggyBac transposon at the 5’ end of the second exon, as well as the su(Hw)v mutant, which carries a deletion of the su(Hw) promoter (HARRISON et al. 1992). Both homozygous su(Hw)e04061 and trans-heterozygous su(Hw) v/e04061 mutant flies show a loss of insulator activity and fertility (Supplementary Figure 1 ) (BAXLEY et al. 2011; SCHOBORG et al. 2013). The oogenesis phenotype of both mutant genotypes is practically indistinguishable, and to avoid genetic interference from second site mutations, we used trans-heterozygous su(Hw) v/e04061 mutant flies for phenotypic characterization.
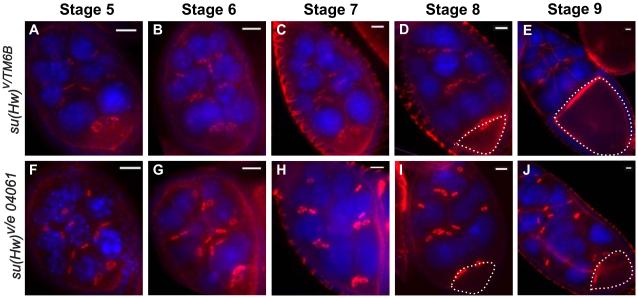
First, we analyzed the structures of egg chambers throughout oogenesis using TexRed-phalloidin staining as a filamentous actin probe and verified that su(Hw) mutant oocytes cease growing after stage 8 of oogenesis (Figure 1). At stage 9, the volume of wildtype oocytes reaches more than one-third of the overall egg chamber size; however, the mutant oocyte does not expand dramatically from stage 8 to 9 as it does in wildtype ovaries (Figure 1 D-E and I-J). Some mutant egg chambers continued growing beyond stage 9, yet the size of these oocytes never expanded and instead showed a shrunken nuclear lamina in the oocyte and nurse cells, an early indication these cells were undergoing apoptosis (PRITCHETT et al. 2009). This consequently leads to degeneration of the entire egg chamber (Supplementary Figure 2). Since oocyte development depends on transport of essential factors from nurse cells to oocyte through the ring canals, the observed increase in egg-chamber volume and lack of oocyte growth suggest that this process is impaired in su(Hw) mutant ovaries.
Figure 1.
su(Hw) mutant ovaries show oocyte and ring canal developmental defects. F-actin staining using Phalloidin on wildtype and su(Hw) mutant ovaries appears red and DAPI staining of DNA appears blue. The oocyte was observed at different stages of each genotype and the scale bar is 50 μm in each image. The dash lines highlight oocytes at stage 8 and 9.
Continuous spatial and temporal expression of Su(Hw) is critical for normal ovary development. The previous observation suggested that loss of su(Hw) leads to oocyte developmental defects that may be derived from failed communications between nurse cells and developing oocytes. Su(Hw) is detected in somatic follicle cells and post-mitotic nurse cells in egg chambers (BAXLEY et al. 2011) and introducing ectopic su(Hw) expression in both types of cells rescues the su(Hw) mutant phenotype (HARRISON et al. 1993). To determine which cell type and stage of Su(Hw) expression is necessary for oogenesis, we took advantage of the GAL4-UAS binary system to express a su(Hw)::eGFP transgene in su(Hw)v/e04061 mutant flies (SCHOBORG et al. 2013). We used traffic jam (tj-Gal4) to drive gene expression in all somatic follicle cells throughout oogenesis and en2.4-Gal4 to drive expression of Su(Hw)::eGFP in follicle stem cells specifically (LEATHERMAN and DINARDO 2010; SOKOL and COOLEY 2003). We used three different Gal4 drivers to control Su(Hw)::eGFP expression in germline cells at different stages of oogenesis: meta-Gal4 expresses Gal4 under the alphaTub67C promoter starting at stage 4 of oogenesis, whereas both nanos-Gal4 (w*; P{nos-Gal4::VP16}) and nos-Gal4 (w*;P{GAL4-nos.NGT40} express Gal4 throughout oogenesis, although nanos-gal4 gives specific expression peaks during the germarium stage and later in egg chambers during stage 9 (RORTH 1998; VAN DOREN et al. 1998). Expression of Su(Hw)::eGFP for each driver was confirmed by immunofluorescence staining using anti-GFP specific antibodies (Supplementary Figure 3).
Virgin females with Su(Hw)::eGFP expression driven under all 5 Gal4 drivers were collected and crossed with y w wildtype male flies. We quantified the number of eggs laid by each rescued female within three days after eclosion to determine the fertility rescue rate. Mutant flies expressing Su(Hw)::eGFP driven by tj-Gal4 and en2.4-Gal4 in follicle cells were infertile and manifested the same incomplete oogenesis as mutant flies not overexpressing Su(Hw)::eGFP (Table 1). On the other hand, introducing Su(Hw)::eGFP expression in germline cells restored mutant fertility to different degrees depending upon the specific driver. All three overexpression lines showed less than a 50% fertility rescue rate (Table 1), and only mutant females with nanos-Gal4 driven Su(Hw)::eGFP expression were able to lay a small number of wildtype eggs (24%) that hatched successfully. Furthermore, mutant flies rescued with Su(Hw)::eGFP expression driven by meta-Gal4 and nos-Gal4 laid significantly fewer eggs, indicating that fewer egg chambers were able to complete oogenesis, likely as a result of insufficient Su(Hw) expression. In addition, all embryos produced by meta-Gal4 and nos-Gal4 females displayed axis defects, revealing the possibility that Su(Hw) may affect axis determination. In summary, the expression of Su(Hw) in follicle cells alone is not sufficient for completing oogenesis, corroborating that the Su(Hw) expression in germline cells is necessary. These results indicate that normal oocyte differentiation requires precise temporal and spatial expression of Su(Hw).
Table 1.
Spatial and temporal expression of Su(Hw) is critical for ovary development.
| Driver | Cell Type | Stage | Fertility Rate (%) |
|---|---|---|---|
| nanos-Gal4 | Germline | G to S13 | 41.4 (235/567) |
| nos-Gal4 | Germline | G to S13 | 6.1 (33/539) |
| met -Gal4 | Germline | S4 to S13 | 1.6 (2/128) |
| en2.4-Gal4 | Somatic and germline |
G-S1 | 0 |
| tj-Gal4 | Somatic | G to S13 | 0 |
Different Gal4 drivers were used in su(Hw)v/e04061 mutants to control su(Hw)::eGFP expression within specific cell types and ovary developmental stages (G: germarium, S: oogenesis stage as listed in the table.) The fertility rescue rate for each line was determined as number of eggs from rescued females divided by eggs from wildtype females.
Intercellular transport between nurse cells and the oocyte is partially blocked in su(Hw) mutant ovaries. The transport of cytoplasm from nurse cells to the oocyte is divided into two phases: the slow phase, which is longer and takes place from early stages to stage 10 of oogenesis, and the fast dumping phase, which takes place while the oocyte doubles in volume from stage 10B to 11. After observing a delay in oocyte development from stage 8 to 9, which is marked by a lack of oocyte volume expansion, we speculated that loss of Su(Hw) may affect nurse cell dumping. An indicator of nurse cell preparation for the fast dumping phase is the formation of actin filament cables, called actin bundles, that is derived from the cortex extending toward the nucleus at stage 10 (GUILD et al. 1997; GUTZEIT 1986). To determine whether Su(Hw) is required for fast dumping, we used TexRed-phallodin staining to observe actin bundle formation in wildtype flies, su(Hw)v/e04061 mutant flies and nos-Gal4 >Su(Hw)::GFP rescued flies. Whereas no Actin bundle was found in su(Hw)v/e04061 mutant flies, we observed that the oocyte enlargement at stage 10B as well as the actin bundles normally appeared in egg chambers with overexpression of Su(Hw) using the nos-Gal4 driver (Supplementary Figure 4). These data suggest that lack of expression of Su(Hw) is at least indirectly responsible for the failed process of fast dumping in female mutant germ cells.
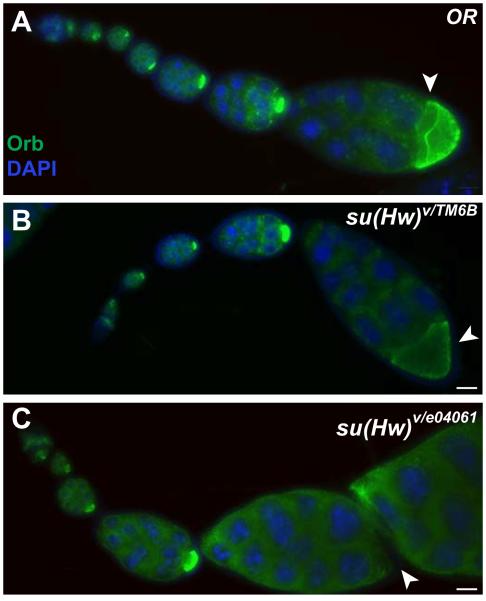
In addition to nutrients released during the fast dumping phase, the slow phase also releases maternal morphogens, which will be later required for proper determination of the embryo dorsal-ventral patterning during development (BATE and MARTINEZ ARIAS 1993). To determine whether these slow phase molecules can also travel from nurse cells to the oocyte in the earlier stages of oogenesis in su(Hw) mutant ovaries, we used the oo18 RNA binding protein Orb (POKRYWKA and STEPHENSON 1995), as a marker to evaluate molecular flow efficiency in wildtype and mutant ovary egg chambers. As expected, Orb translocated from nurse cells to the oocyte and specifically accumulated at the posterior of wildtype oocytes (Figure 2-A). In mutant egg chambers, Orb localization appears normal in most early stage chambers (Figure 2-C), indicating that lack of Su(Hw) does not cause major problems with oocyte determination or molecular transport during early stages of oogenesis. However, in both heterozygous and trans-heterozygous mutants, we detected an abnormally high accumulation of Orb in the cytoplasm of nurse cells and a striking reduction of Orb in stage 7 and 8 oocytes (Figure 2-B and C). These data suggest that an inefficient translocation of essential maternal morphogens from nurse cells to the oocyte in su(Hw) mutant egg chambers may be one of the causes of severe developmental defects.
Figure 2.
Orb mislocalizes to the cytoplasm of nurse cells in stage 8 egg chambers from su(Hw) mutant ovaries. Egg chambers were stained with Orb antibody (green) and DAPI (blue). High concentration of Orb is observed in oocytes through oogenesis in wildtype (A). Oocyte localization of Orb is significantly reduced at stage 8 in su(Hw)v/TM6B heterozygotes (B), and is totally absent and limited to nurse cell cytoplasm in su(Hw)v/e04061 mutant (C). Arrowheads point to stage 8 oocytes. Scale bar is 50 μm.
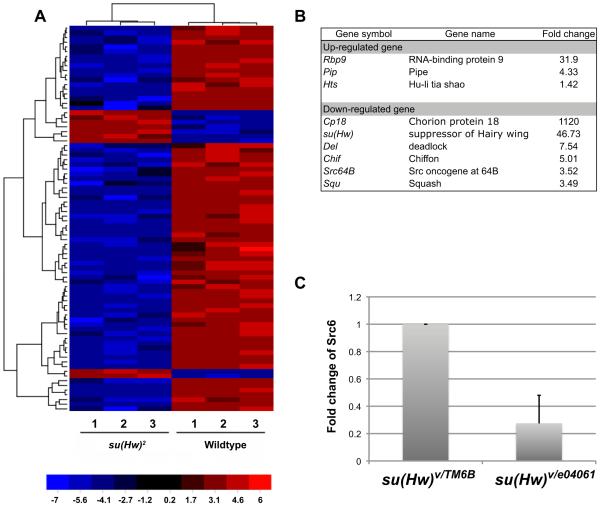
Genes involved in nurse cell-oocyte transport are misexpressed in su(Hw) mutant ovaries. To understand whether the phenotype of defective transport in su(Hw) mutants results from misregulation of genes involved in molecular transport, microarray analysis was performed using the total RNA from wildtype (OR) and su(Hw)2 homozygous mutant ovaries. su(Hw)2 is a hypomorphic allele resulting from a jockey element insertion into su(Hw) (HARRISON et al. 1993; PARKHURST et al. 1988). We used ovaries from su(Hw)2 mature mutant females to extract information related to genes involved in ring canal development or nurse cytoplasm-oocyte transportation, and we also compared the transcriptional changes in response to loss of su(Hw) function using array data generated from ovaries of su(Hw)2/e04061, su(Hw)2/v and su(Hw)f young virgin female mutants published elsewhere (SOSHNEV et al. 2013). Given that ovaries from three to five days mated females in our microarray samples should present significant developmental differences with ovaries from young virgin females in Soshnev et al. (2013), we reasoned that gene sets that have a similar transcriptional response to su(Hw) mutations in both samples are likely to be strongly influenced by the lack of Su(Hw) protein. Hierarchical clustering based on oogenesis-related genes and most other gene sets showed that our flies clustered separately from Soshnev et al. flies (2013), likely indicating that the large differences are due to differences in developmental stages between samples. However, when data was clustered based on genes in “eggshell chorion assembly” (GO:0007306), “structural constituent of chorion” (GO:0005213) and “multicellular organismal development” (GO:0007275), which contain mostly chorion-related genes, su(Hw) mutants from both data sets clustered tightly together. These results suggest that certain chorion-related genes may be specifically regulated by Su(Hw) (Supplementary Figure 6). Microarray data also show significant changes in the expression of eighty-two genes (p < 0.01 and absolute mean fold-change > 3) known to have a role in oogenesis (Figure 3-A). The relative amount of change for a select group of these genes is shown in Figure 3-B.
Figure 3.
Su(Hw) regulates expression of many genes involved in oogenesis. Heatmap of eighty-two genes related to ovary development with a significant change in expression greater than three-fold with up-regulation showed in red and down-regulation showed in blue. Corresponding fold changes of gene symbols are shown in supplementary table 1. Color key is shown at bottom with numbers indicating fold (A). Table in B shows a group of selected genes with their corresponding change in expression. qRT-PCR shows that expression of Src64B is reduced 30% in ovaries from su(Hw) mutants (C).
Specifically, nine oogenesis-related genes were highly up-regulated (greater than 3-fold, p < 0.01) in su(Hw)2 mutants, including rbp9 (32-fold, p = 0.00006). Other important genes were up-regulated, but to a lesser degree, including hts (1.76-fold, p = 0.003). On the other hand, seventy-three oogenesis-related genes were highly down-regulated (greater than 3-fold, p < 0.01), including Src64B (3.5-fold, p = 0.002). In particular, Src64B (down-regulated) and hts (up-regulated) have a role directly related with the structure and function of ring canals, and misexpression of these genes could have effects in the structure and function of rings and in the overall transportation of substances from the nurse cells to the oocyte.
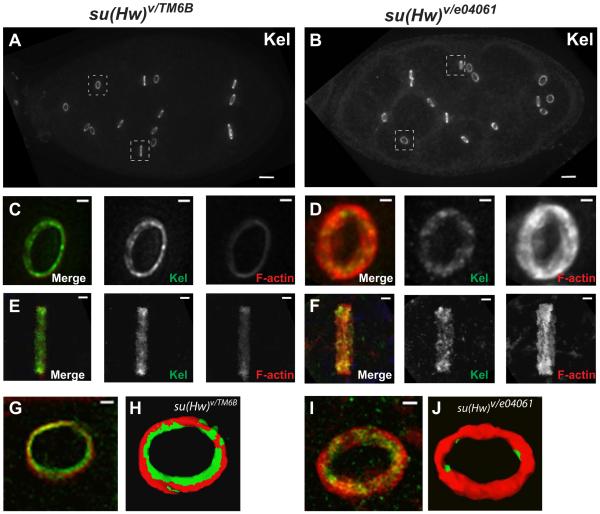
Loss of Su(Hw) causes structural defects in ring canals during oogenesis. Given the finding of misexpression of hts and Src64B in ovaries, we performed immunofluorescence experiments to determine whether su(Hw) mutant ring canals show defects that could be related to inefficient molecular transport. Interestingly, comparison of su(Hw) mutant and wildtype ring canal sizes using F-actin fluorescence staining revealed a remarkable difference in thickness, brightness and average size of the rings (Figure 1). We further confirmed this observation through immunostaining experiments in mutant and wildtype ovaries using specific antibodies anti-Kelch, a structural component of ring canals that functions in cross-linking F-actin within the ring (KELSO et al. 2002; ROBINSON and COOLEY 1997). Results showed that amounts of cytoplasmic Kelch in the nurse cells cytoplasm of mutant egg chambers were above normal and that ring canals appeared thicker and longer as a consequence of excessive accumulation of F-actin in the ring structure (Figure 4).
Figure 4.
Rings from su(Hw) mutant egg chambers show significant morphological differences compared to wildtype. Ring canals in wildtype and mutant egg chambers are stained with antibody anti-Kelch in green and Phalloidin in red. Stage 8 egg chambers stained with Kelch are shown in wildtype (A) and su(Hw) mutant (B). Zoom in images of wildtype individual rings from dashed squares in A are shown in C and E. Zoom in images of su(Hw) mutant individual rings from dashed squares in B are shown in D and F. Isosurface images of individual rings in wildtype (G and H) and su(Hw) mutant (I and J) rings were generated using Leica Deblur software, and show the accumulation of actin in rings. The scale bars in egg chamber images represent 10 µm, and in individual ring images represent 1µm.
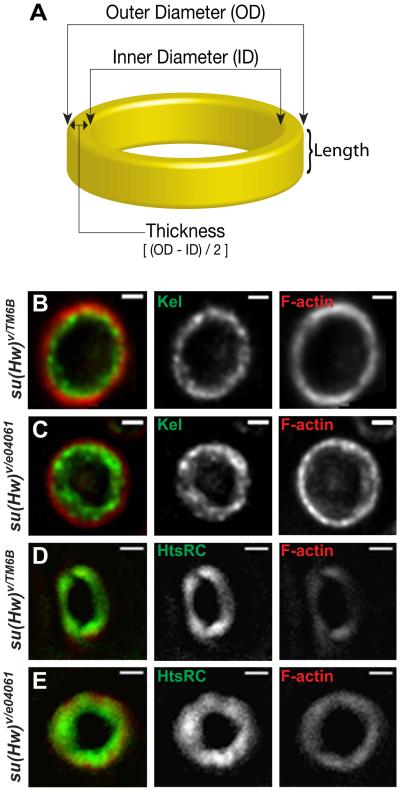
The hts gene is essential for fertility in Drosophila, and loss of hts causes female sterility (DING et al. 1993; YUE and SPRADLING 1992). It encodes Ovhts, a polyprotein that is cleaved to yield two proteins. One, Ovhts-Fus, localizes to the fusome in mitotic cells within the early germarium, while the other, Ovhts-RC, serves as a ring canal structure protein in late oogenesis (PETRELLA et al. 2007). We used an antibody against Ovhts-RC to visualize detailed structures of ring canals in immunostaining experiments. Interestingly, su(Hw) mutant rings at stage 6 are not only thicker but also have smaller inner diameters due to accumulation of structural proteins, Kelch and Ovhts-RC (Figure 5), which is consistent with microarray results showing up-regulation of hts (1.76-fold, p = 0.003). These thicker rings create smaller lumens that may cause the potential obstruction responsible for slowing down molecular flow through ring canals.
Figure 5.
Ring canals in su(Hw) mutant egg chambers show excess accumulation of structural proteins. A cartoon ring image illustrating the structure and organization of a ring canal (A). Staining of rings at stage 6 using antibodies against ring structural proteins Kelch (B and C), Ovhts-RC (D and E) and F-actin (B-E). Kelch and Ovhts-RC show accumulation at the inner rim in su(Hw) mutants (C and E) but not in wildtype (B and D). Scale bars represent 1µm.
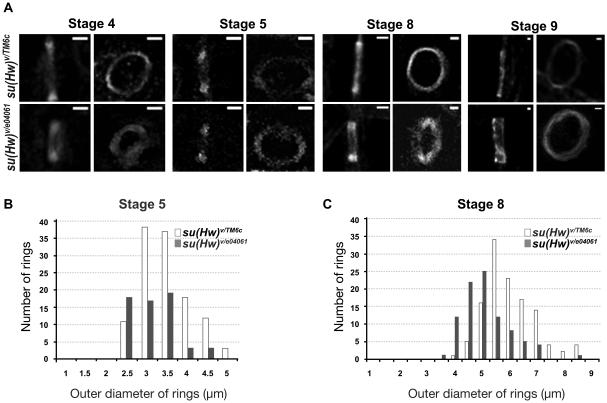
Ring canals show developmental defects in su(Hw) mutant ovaries. To closely monitoring growth differences between wildtype and mutant egg chambers during development, we measured ring canal outer diameters from stages four to eight (Figure 6-A). Given that ring sizes vary within each egg chamber depending upon ring age, such that older rings (formed earlier during mitosis within the germarium) appear larger, we used fluorescence microscopy to measure all fifteen rings in each egg chamber, recording data only from images clearly displaying all fifteen rings. The ring size distributions at stages five and eight in mutant and wild-type egg chambers are shown in Figures 6-B and C. The average ring size at stage 5 is 3.2 µm in wildtype and 2.9 µm in mutants, revealing no significant difference. Ring sizes at stages six and seven do not show a significant difference either. However, wildtype rings at stage 8 expanded to 5.7 µm (N 120, SD 0.9), whereas mutant rings expanded only to 4.8 µm (N 90, SD 0.8), confirming that mutant ring canals are significantly smaller (Student’s t-Test, p = 0.0001) (Figure 6-B and C). Although outer ring diameter expansion is significantly delayed at stage 8 in su(Hw) mutants, smaller ring lumens can be already observed at earlier stages (Figure 5), suggesting that the smaller rings at stage 8 may be an accumulative effect of an abnormal ring development initiated in earlier stages.
Figure 6.
Ring canal development is defective in su(Hw) mutants.
From stage 4 to 9, ring canals were detected using F-actin staining (A), and the sizes of rings at each stage were quantified using the measurement tool in ImageJ. The measurements of ring outer diameter in each genotype at stage 5 and 8 are shown in histograms (B and C). Scale bars represent 1µm.
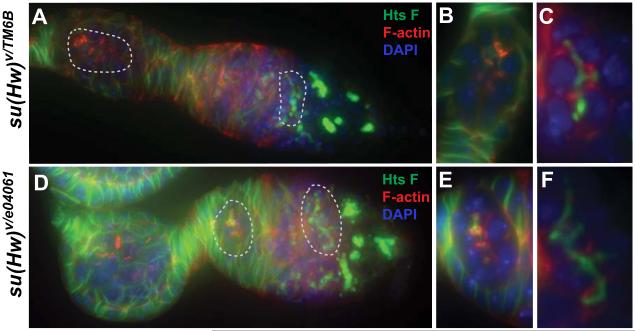
The observation of abnormal rings at different stages in su(Hw) mutants (Figure 6) and the abnormal accumulation of Ovhts-RC (Figure 5) led us to ask whether this phenotype correlates to fusome development defects in the germarium, and whether this phenotype is due to hts gene misexpression. We used an anti-Hts Fus monoclonal antibody, specifically against Ovhts-Fus to perform immunostaining in wildtype and mutant germarium. These experiments revealed seemingly normal fusomes in su(Hw) mutants that plugged ring canals during initial mitotic divisions and formed branched structures that disappear at stage 1, consistent with observations in wildtype ovaries (Figure 7). We concluded that using fluorescence microscopy we could not detect significant fusome organization defects in su(Hw) mutant ovaries. Consequently, these results suggest that defects in ring canals do not originate from hts overexpression in the germarium stage, and that hts overexpression does not cause major defects in the formation and structure of the fusome.
Figure 7.
Fusome development in su(Hw) mutants show no differences with wildtype. Ovary staining was performed using 1B1 antibody (green) to detect fusomes. F-actin is shown in red and DAPI in blue. Early germarium stage egg chambers in wildtype (A) and su(Hw) mutants (D) are shown. Dashed areas are shown in detail for wildtype (B and C) and mutant (E and F), showing no major differences in fusome organization between su(Hw) mutants and wildtype.
Misexpression of Src64B in su(Hw) mutant ovaries causes structural defects in ring canals. In addition to hts, Src64 was found to be down-regulated in su(Hw) mutant ovaries (Figure 3). Src64B mutant females produce abnormally small eggs due to unsuccessful nurse cell dumping, caused in part by defects in fusome development, ring canal growth and morphogenesis (COOLEY 1998; DJAGAEVA et al. 2005; DODSON et al. 1998). Orb retention in nurse cells such as that observed in su(Hw) mutant egg chambers in this work, is also a phenotype identified in female flies carrying mutations of the Src64B oncogene (DJAGAEVA et al. 2005). Src64B is a protein tyrosine kinase that plays an important role in regulation of ring canal growth and morphogenesis during Drosophila oogenesis. Src64B kinase activity regulates Kel function through phosphorylation, and both a mutation of tyrosine 627 in kelch and a null mutation of Src64B, cause a dramatic reduction of actin monomer turnover resulting in thicker rings with small lumens (DODSON et al. 1998; KELSO et al. 2002; ROBINSON and COOLEY 1997; XUE and COOLEY 1993), a phenotype similar to the ring phenotype described here in su(Hw) mutant egg chambers (Figure 4 and 5). The actin binding protein Kelch functions in cross-linking actin monomers during ring canal formation, consequently stabilizing F-actin by protecting it from depolymerization (ROBINSON et al. 1994). F-actin polymerization and depolymerization are dynamic processes during ring canal development. At stage 6, for example, the ring canal size expands rapidly in preparation for nurse cell dumping during the subsequent stages. When the outer ring canal diameter rapidly expands to increase the lumen, F-actin must depolymerize in the inner ring rim to prepare for ring size expansion. kel null mutants show disorganized actin filaments starting at stage 4 and present a completely disrupted organization at stage 6 when ring expansion is necessary for nurse cell dumping (ROBINSON and COOLEY 1997; XUE and COOLEY 1993).
Taken together, these observations suggest not only that the abnormal ring canal structure in su(Hw) mutants may impact molecular transport within the egg chamber, thereby causing oogenesis failure and sterility, but also that this phenotype might be at least partially due to misexpression of Src64B. To exclude the possibility that decreased expression of Src64B observed in microarray experiments stems from a developmental factor, we performed real-time RT-PCR to compare Src64B expression in wildtype and su(Hw) mutants by manually removing egg chambers older than stage 9. Results showed that Src64B expression is down-regulated more than three-fold in su(Hw) mutant ovaries compared to wildtype, a result consistent with the microarray data (Figure 3-C). Since we detected an abnormal accumulation of F-actin in ring canals and found that Src64B is under-expressed in su(Hw) mutants, we hypothesize that the thick ring phenotype is caused by a reduction in the levels of Src64B expression.
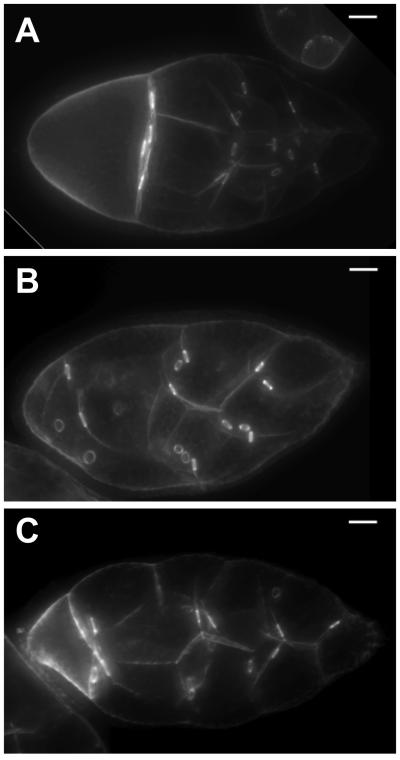
To test this hypothesis, we used nos-Gal4 to drive Src64B expression in su(Hw) mutants and then observed ring canal morphology in Src64B rescued females. Ovary immunostaining in Src64 rescued females showed rings became thinner and shorter, similar to wildtype rings (Figure 8). These data suggest that the abnormal ring canal morphology in su(Hw) mutants is caused by Src64B misregulation. In addition, the abnormal position of rings and egg chamber interior organization observed in mutants are improved in rescued flies. Nevertheless, fertility of these Src64B rescued females is not recovered, indicating that other factors remain critical to cause oogenesis failure in addition to Src64B misregulation.
Figure 8.
Restoration of Src64B expression rescues ring phenotype in su(Hw) mutants. Ring morphology was imaged using F-actin staining in wildtype (A), su(Hw)v/e04061 mutant (B), and nosGal4>> Src64B rescued su(Hw)v/e04061 mutant egg chambers (C). Scale bars represent 20 µm.
Discussion
Loss of Su(Hw) chromatin insulator protein in ovaries causes a complex female sterility phenotype resulting from incomplete oocyte development and egg chamber degeneration beginning at mid-oogenesis (BAXLEY et al. 2011; HARRISON et al. 1993; KLUG et al. 1968; KLUG et al. 1970). To further understand the causes of this phenotype we investigated the structure of egg chambers and their molecular flow dynamics in ovaries from mutant females. Ultimately, we found that these mutants lack normal flow of molecules and vesicles from nurse cells to oocyte. Further structural and microarray analyses revealed that this flow is disrupted by defective ring canals in the egg chamber, which undergo abnormal development resulting from down-regulation of Src64B, which in turn disrupts Kel function and actin organization in the rings.
Oocyte development depends upon Su(Hw) expression in germline cells. Our initial observation was that mutant egg chambers continue increasing in size through oogenesis, whereas oocytes cease their enlargement at stage 9 (Figure 1), indicating absence of fast dumping from nurse cells. We also determined that Orb remained in the cytoplasm of nurse cells, revealing a similar impact of su(Hw) mutations on molecular transport between nurse cells and the oocyte before stage 9 (Figure 2). Specific morphogens traveling into the oocyte are important for oocyte maturation and embryo development, and loss or mislocation of these morphogens causes oogenesis failure and abnormal embryo production in a variety of mutants, including actin-binding proteins, actin-dependent motor protein and transcription factors Lark RNA-binding proteins (MCNEIL et al. 2004; MYSTER et al. 2000; WHEATLEY et al. 1995; XUE and COOLEY 1993). Moreover, we show that restoration of Su(Hw) expression using germline specific Gal4 drivers rescues nurse cell dumping, oocyte development, and female fertility (Table 1 and Supplementary Figure 4), ruling out the possibility that lack of dumping or structural defects of ring canals are due to secondary mutations in the chromosomes of mutant stocks. Using several Gal4 drivers we concluded that the fertility rescue rate is also dependent on the appropriate spatial and temporal expression of Su(Hw) during oogenesis. The observation that the fertility rescue rate was increased from 1.6% using meta-Gal4 to 6.1% using nos-Gal4, suggests that Su(Hw) expression in germline cells is already necessary during early oogenesis, before stage 4. On the other hand, flies expressing Su(Hw) under nanos-Gal4 showed the highest rescue rate (41.4%), compared to flies with expression under a stronger driver such as nos-Gal4 (6.1%), which, unlike nanos-Gal4, restricts the higher expression of Gal4 in earlier germarium and later during stage 9, and has mild expression during intermediate stages of oogenesis. In addition, although Su(Hw) expression in germline cells is necessary for normal oogenesis, production of abnormal embryos in partially rescued females suggests that adequate spatial-temporal Su(Hw) expression in ovaries is also necessary for proper embryo development after fertilization.
Su(Hw) affects the expression of genes involved in ring canal function. We performed microarray analysis using mature ovaries from su(Hw)2 mutant females and compared the results with transcriptional changes in response to loss of su(Hw) function using published microarray data generated from virgin female ovaries from su(Hw)2/e04061, su(Hw)2/v and su(Hw)f (SOSHNEV et al. 2013). We were interested in identifying genes with a robust transcriptional response to su(Hw) mutations all through oogenesis, which we reasoned would be manifested in both types of samples. Accordingly, hierarchical clustering analysis of chorion-related genes clustered together su(Hw) mutants from both data sets (su(Hw)2 from mature ovaries, and su(Hw)2/e04061, su(Hw)2/v and su(Hw)f, from young virgin females), suggesting that expression of chorion genes is not only developmentally regulated, but may also be specifically regulated by Su(Hw) (Supplementary Figure 6). Consistently with this result, Rbp9 and hts are also up-regulated in all samples, whereas Src64B, is down-regulated, both in our microarray analysis using su(Hw)2 mutant as well as in su(Hw)e04061/2 and su(Hw)v/2 virgin females. Transcription levels were confirmed for Src64B using qRT-PCR, which revealed a three-fold down-regulation of Src64B in su(Hw)v/e04061 mutants (Figure 3).
These results suggest that ring canal genes hts and Src64B are misregulated in su(Hw) mutants. Su(Hw) can regulate gene expression throughout the genome as a consequence of its genome-wide chromatin insulator function. Some reports suggest that it can also regulate gene transcription directly through binding to gene regulatory sequences at sites nearby specific genes, rather functioning as a transcription factor (BAXLEY et al. 2011; SOSHNEV et al. 2013; SOSHNEV et al. 2012). However, it is difficult to establish a clear distinction between these two roles given our limited understanding of the mechanisms of insulator function. In addition, insulator proteins such as Mod(mdg4), CTCF, CP190, BEAF and GAF are frequently found associated to promoters of genes, making it even more difficult to distinguish between insulator and transcriptional regulatory function. A direct role of Su(Hw) in transcription regulation has been previously suggested. In su(Hw) mutant ovaries, for example, Rbp9 is de-repressed, and a experimentally induced reduction of Rbp9 expression rescues female fertility (SOSHNEV et al. 2013). A decrease in Su(Hw) binding at the Rbp9 promoter region was experimentally shown in su(Hw) mutant ovaries, suggesting that Su(Hw) may have a direct role in Rbp9 repression during oogenesis (SOSHNEV et al. 2013). We analyzed the location of Su(Hw) binding sites using published ChIP on chip data (BUSHEY et al. 2009; NEGRE et al. 2011; SOSHNEV et al. 2012), and found that for Src64, Mod(mdg4), CP190 and GAF are present in the promoter region of the gene, whereas Su(Hw) is located 20 kb downstream in the 3rd intron. This site is not directly bound by Mod(mdg4) or Cp190, suggesting the site may not have the properties of other insulator sites such those mediated by the gypsy insulator. However, there is no available data to explain how all these binding sites contribute to regulate Src64 expression during oogenesis.
Accumulation of actin and thickening of ring canals in su(Hw) mutants results from Src64B down-regulation. The position and orientation of ring canals within egg chambers hinges on the specific arrangement and positioning of neighboring nurse cells. We have observed that the position and layout of rings in mutant ovaries is atypical, indicating that the arrangement of nurse cells in mutant egg chambers is different from that found in wildtype females (Figure 1). This unusual organization may contribute to inefficient molecular transport, but it is not clear whether mutations in su(Hw) directly cause defects in the layout of nurse cells in egg chambers, or are deficiencies in ring canal development which cause an abnormal layout of nurse cells. The abnormal thickening of the ring structure observed in su(Hw) mutant egg chambers is evident from stage 4 to stage 9 (Figures 4 and 5). To rule out the possibility that this observation is the response to a delayed ring expansion caused by an early egg chamber degeneration in mutants, we examined the structure of rings in egg chambers older than stage 8, noting that the outer diameter of rings continuously increased as opposed to shrinking, as it normally occurs in wildtype females (Supplementary Figure 5.). The excessive accumulation F-actin found in rings from these mutants suggests that actin organization is misregulated upon loss of su(Hw) expression.
Our results suggest that misexpression of Src64B may cause actin disorganization in ring canals due to dysfunctional Kel, which normally maintains a rapid turnover of the actin cytoskeleton in the rings. Src64B kinase activity regulates Kel function through Kel phosphorylation, and Kel is a structural component of ring canals that helps cross-linking F-actin within the ring (KELSO et al. 2002; ROBINSON and COOLEY 1997). We show that Kel is disproportionately enriched in the cytoplasm of nurse cells from mutant egg chambers at the same time that ring canals appeared thicker and longer because of an excessive accumulation of F-actin in the ring structure. More importantly, restoration of Src64B expression in su(Hw) mutants rescues the morphology of ring canals (Figure 8).
Altogether, we have shown a novel su(Hw) mutant ring canal phenotype resulting from significant Src64B down-regulation during oogenesis. Src64B down-regulation is not the only factor leading to infertility, given that loss of su(Hw) function induces a pleiotropic effect in oogenesis, generating a complex phenotype that culminates with sterility in su(Hw) mutant females. Our results show that structural defects in the formation of ring canals during development may block molecular flow and contribute to oogenesis arrest, but they cannot explain the full extent of the sterility phenotype. We have shown that overexpression of Src64B rescues the ring canal wildtype phenotype but does not restore fertility. On the other hand, a reduction in the expression of Rbp9 in su(Hw) mutants seems to restore fertility (SOSHNEV et al. 2012), and subsequently ring and dumping phenotypes, prompting the question of whether expression of Src64B may be regulated by Rbp9 . This gene encodes a RNA-binding protein that belongs to the ELAV/ Hu gene family, which participates in regulating gene expression by influencing mRNA splicing and translation in animals (HILGERS et al. 2012; SOLLER et al. 2010). In Drosophila, Rbp9 interacts with U-rich mRNA and regulates the turnover of its target mRNAs (KIM and BAKER 1993; PARK et al. 1998), which is in agreement with a role of Rbp9 in repressing expression of Src64B.
However, early reports show that while overexpression of Rbp9 driven by nos-Gal4 causes incomplete oocyte development and apoptosis at stage 10, a normal enlargement of oocytes at stage 10 is still observed (JEONG and KIM-HA 2003). This observation indicates that oocytes reach a significantly advanced development and normal dumping in Rbp9 overexpression egg chambers, compared to oocytes in su(Hw) mutants. This comparison also suggests that factors other than Rbp9 contribute to failed oocyte development and egg chamber degeneration before stage 10 in su(Hw) mutants. If defects in the development of ring canals are independent of Rbp9 overexpression, how the reduction of expression of Rbp9 is capable of restoring normal oogenesis remains an open question, but it underscores the complex nature of phenotypes induced by mutations in chromatin insulator proteins, and emphasizes the need of further studies to characterize critical factors regulating oogenesis failure in su(Hw) mutants and the role of chromatin insulators in the regulation of such factors.
Supplementary Material
Highlights.
Drosophila su(Hw) female mutants lack molecular flow from nurse cells to oocyte
Ring canals in su(Hw) mutant egg chambers are smaller and have narrow lumens
Ring canals in su(Hw) mutants have excess accumulation of Kelch, Hts and F-actin
Src64B expression is reduced three-fold in su(Hw) mutant ovaries
Restoration of Src64B expression alone rescues the ring phenotype but not fertility
Acknowledgements
We thank Dr. Steven DiNardo at the University of Pennsylvania for providing y w; P{Tj-Gal4} stock, Dr. Laura Lee at the University of Vanderbilt for P{GAL4-nos.NGT40} stock , Dr. Victor Corces at Emory University for y2 w ct6; su(Hw) v/TM6B stock and Dr. Bruce McKee at the University of Tennessee for P{nos-Gal4::VP16} , w* and P{matalpha4-GAL-VP16}V37 stocks. We appreciate very much the discussions provided by Dr. Albrecht von Arnim at the University of Tennessee and the former Lab member Dr. Todd Schoborg for his support, discussions and several reagents used in this work. Also, we would like to thank Tim Wesley for editing and making figures. The anti-Orb (4H8), anti-Hts F (1B1), Lamin Dm0, anti-Kel (Kel-1B) and anti-Hts RC monoclonal antibodies, were developed by Drs. Paul Schedl, Howard D. Lipshitz, Paul A. Fisher and Lynn Cooley, respectively and were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by grants NIH GM78132-2 and NSF MCB-0616081, plus additional support from the College of Arts & Sciences, the Department of Biochemistry & Cellular & Molecular Biology and the Office of Research at the University of Tennessee, Knoxville.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AGALIOTI T, LOMVARDAS S, PAREKH B, YIE J, MANIATIS T, et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- BATE M, MARTINEZ ARIAS A. Cold Spring Harbor Laboratory. Cold Spring Harbor; N.Y.: 1993. The development of Drosophila melanogaster. [Google Scholar]
- BAXLEY RM, SOSHNEV AA, KORYAKOV DE, ZHIMULEV IF, GEYER PK. The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev Biol. 2011;356:398–410. doi: 10.1016/j.ydbio.2011.05.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENJAMINI Y, HOCHBERG Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- BRASSET E, VAURY C. Insulators are fundamental components of the eukaryotic genomes. Heredity. 2005;94:571–576. doi: 10.1038/sj.hdy.6800669. [DOI] [PubMed] [Google Scholar]
- BUSHEY AM, RAMOS E, CORCES VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes & Development. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSZCZAK M, COOLEY L. Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ. 2000;7:1071–1074. doi: 10.1038/sj.cdd.4400755. [DOI] [PubMed] [Google Scholar]
- COOLEY L. Drosophila ring canal growth requires Src and Tec kinases. Cell. 1998;93:913–915. doi: 10.1016/s0092-8674(00)81196-4. [DOI] [PubMed] [Google Scholar]
- DE CUEVAS M, SPRADLING AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- DING D, PARKHURST SM, LIPSHITZ HD. Different genetic requirements for anterior RNA localization revealed by the distribution of Adducin-like transcripts during Drosophila oogenesis. Proc Natl Acad Sci U S A. 1993;90:2512–2516. doi: 10.1073/pnas.90.6.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DJAGAEVA I, DORONKIN S, BECKENDORF SK. Src64 is involved in fusome development and karyosome formation during Drosophila oogenesis. Dev Biol. 2005;284:143–156. doi: 10.1016/j.ydbio.2005.05.012. [DOI] [PubMed] [Google Scholar]
- DODSON GS, GUARNIERI DJ, SIMON MA. Src64 is required for ovarian ring canal morphogenesis during Drosophila oogenesis. Development. 1998;125:2883–2892. doi: 10.1242/dev.125.15.2883. [DOI] [PubMed] [Google Scholar]
- GASZNER M, FELSENFELD G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- GAUSE M, MORCILLO P, DORSETT D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21:4807–4817. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAUTIER L, COPE L, BOLSTAD BM, IRIZARRY RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- GERASIMOVA TI, GDULA DA, GERASIMOV DV, SIMONOVA O, CORCES VG. A Drosophila Protein That Imparts Directionality on a Chromatin Insulator Is an Enhancer of Position-Effect Variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- GHOSH D, GERASIMOVA TI, CORCES VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUCCIONE E, MARTINATO F, FINOCCHIARO G, LUZI L, TIZZONI L, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- GUILD GM, CONNELLY PS, SHAW MK, TILNEY LG. Actin filament cables in Drosophila nurse cells are composed of modules that slide passively past one another during dumping. J Cell Biol. 1997;138:783–797. doi: 10.1083/jcb.138.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUTZEIT HO. The role of microfilaments in cytoplasmic streaming in Drosophila follicles. Journal of Cell Science. 1986;80:159–169. doi: 10.1242/jcs.80.1.159. [DOI] [PubMed] [Google Scholar]
- HAGLUND K, NEZIS IP, STENMARK H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun Integr Biol. 2011;4:1–9. doi: 10.4161/cib.4.1.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRISON DA, GDULA DA, COYNE RS, CORCES VG. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 1993;7:1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- HARRISON DA, MORTIN MA, CORCES VG. The RNA polymerase II 15-kilodalton subunit is essential for viability in Drosophila melanogaster. Mol Cell Biol. 1992;12:928–935. doi: 10.1128/mcb.12.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILGERS V, LEMKE SB, LEVINE M. ELAV mediates 3' UTR extension in the Drosophila nervous system. Genes Dev. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JEONG K, KIM-HA J. Expression of Rbp9 during mid-oogenesis induces apoptosis in egg chambers. Mol Cells. 2003;16:392–396. [PubMed] [Google Scholar]
- KELSO RJ, HUDSON AM, COOLEY L. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J Cell Biol. 2002;156:703–713. doi: 10.1083/jcb.200110063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM YJ, BAKER BS. The Drosophila gene rbp9 encodes a protein that is a member of a conserved group of putative RNA binding proteins that are nervous system-specific in both flies and humans. J Neurosci. 1993;13:1045–1056. doi: 10.1523/JNEUROSCI.13-03-01045.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLUG WS, BODENSTEIN D, KING RC. Oogenesis in the suppressor of hairy-wing mutant of Drosophila melanogaster. I. Phenotypic characterization and transplantation experiments. J Exp Zool. 1968;167:151–156. doi: 10.1002/jez.1401670203. [DOI] [PubMed] [Google Scholar]
- KLUG WS, KING RC, WATTIAUX JM. Oogenesis in the suppressor of hairy-wing mutant of Drosophila melanogaster. II. Nucleolar morphology and in vitro studies of RNA protein synthesis. J Exp Zool. 1970;174:125–140. doi: 10.1002/jez.1401740203. [DOI] [PubMed] [Google Scholar]
- KOUZARIDES T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- LABRADOR M, CORCES VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- LEATHERMAN JL, DINARDO S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI B, CAREY M, WORKMAN JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- LIN H, YUE L, SPRADLING AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- MARSMAN J, HORSFIELD JA. Long distance relationships: enhancer-promoter communication and dynamic gene transcription. Biochim Biophys Acta. 2012;1819:1217–1227. doi: 10.1016/j.bbagrm.2012.10.008. [DOI] [PubMed] [Google Scholar]
- MCNEIL GP, SMITH F, GALIOTO R. The Drosophila RNA-binding protein Lark is required for the organization of the actin cytoskeleton and Hu-li tai shao localization during oogenesis. Genesis. 2004;40:90–100. doi: 10.1002/gene.20069. [DOI] [PubMed] [Google Scholar]
- MYSTER DL, BONNETTE PC, DURONIO RJ. A role for the DP subunit of the E2F transcription factor in axis determination during Drosophila oogenesis. Development. 2000;127:3249–3261. doi: 10.1242/dev.127.15.3249. [DOI] [PubMed] [Google Scholar]
- NEGRE N, BROWN CD, MA L, BRISTOW CA, MILLER SW, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEGRE N, BROWN CD, SHAH PK, KHERADPOUR P, MORRISON CA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONG CT, CORCES VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONG CT, CORCES VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAGE SL, HAWLEY RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAI CY, LEI EP, GHOSH D, CORCES VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- PARK SJ, YANG ES, KIM-HA J, KIM YJ. Down regulation of extramacrochaetae mRNA by a Drosophila neural RNA binding protein Rbp9 which is homologous to human Hu proteins. Nucleic Acids Res. 1998;26:2989–2994. doi: 10.1093/nar/26.12.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARKHURST SM, HARRISON DA, REMINGTON MP, SPANA C, KELLEY RL, et al. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988;2:1205–1215. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- PETRELLA LN, SMITH-LEIKER T, COOLEY L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- PHILLIPS-CREMINS JE, CORCES VG. Chromatin Insulators: Linking Genome Organization to Cellular Function. Molecular Cell. 2013;50:461–474. doi: 10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILLIPS-CREMINS JE, SAURIA ME, SANYAL A, GERASIMOVA TI, LAJOIE BR, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POKRYWKA NJ, STEPHENSON EC. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–370. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- PRITCHETT TL, TANNER EA, MCCALL K. Cracking open cell death in the Drosophila ovary. Apoptosis. 2009;14:969–979. doi: 10.1007/s10495-009-0369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAO SS, HUNTLEY MH, DURAND NC, STAMENOVA EK, BOCHKOV ID, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON DN, CANT K, COOLEY L. Morphogenesis of Drosophila ovarian ring canals. Development. 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- ROBINSON DN, COOLEY L. Drosophila kelch is an oligomeric ring canal actin organizer. Journal of Cell Biology. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RORTH P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- SCHOBORG T, LABRADOR M. Expanding the roles of chromatin insulators in nuclear architecture, chromatin organization and genome function. Cellular and molecular life sciences : CMLS. 2014 doi: 10.1007/s00018-014-1672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHOBORG T, RICKELS R, BARRIOS J, LABRADOR M. Chromatin insulator bodies are nuclear structures that form in response to osmotic stress and cell death. J Cell Biol. 2013;202:261–276. doi: 10.1083/jcb.201304181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMYTH GK, MICHAUD J, SCOTT HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- SOKOL NS, COOLEY L. Drosophila filamin is required for follicle cell motility during oogenesis. Dev Biol. 2003;260:260–272. doi: 10.1016/s0012-1606(03)00248-3. [DOI] [PubMed] [Google Scholar]
- SOLLER M, LI M, HAUSSMANN IU. Determinants of ELAV gene-specific regulation. Biochem Soc Trans. 2010;38:1122–1124. doi: 10.1042/BST0381122. [DOI] [PubMed] [Google Scholar]
- SOSHNEV AA, BAXLEY RM, MANAK JR, TAN K, GEYER PK. The insulator protein Suppressor of Hairy wing is an essential transcriptional repressor in the Drosophila ovary. Development. 2013 doi: 10.1242/dev.094953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOSHNEV AA, HE B, BAXLEY RM, JIANG N, HART CM, et al. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 2012;40:5415–5431. doi: 10.1093/nar/gks225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUBRAMANIAN A, TAMAYO P, MOOTHA VK, MUKHERJEE S, EBERT BL, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SULLIVAN W, ASHBURNER M, HAWLEY RS. Drosophila protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- VAN BORTLE K, RAMOS E, TAKENAKA N, YANG J, WAHI JE, et al. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Research. 2012;22:2176–2187. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DOREN M, WILLIAMSON AL, LEHMANN R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Current Biology. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- WALLACE HA, PLATA MP, KANG HJ, ROSS M, LABRADOR M. Chromatin insulators specifically associate with different levels of higher-order chromatin organization in Drosophila. Chromosoma. 2010;119:177–194. doi: 10.1007/s00412-009-0246-0. [DOI] [PubMed] [Google Scholar]
- WALLACE JA, FELSENFELD G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHEATLEY S, KULKARNI S, KARESS R. Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development. 1995;121:1937–1946. doi: 10.1242/dev.121.6.1937. [DOI] [PubMed] [Google Scholar]
- WU ZJ, IRIZARRY RA, GENTLEMAN R, MARTINEZ-MURILLO F, SPENCER F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- XUE F, COOLEY L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- YANG J, CORCES VG. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev. 2012;22:86–92. doi: 10.1016/j.gde.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUE L, SPRADLING AC. hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev. 1992;6:2443–2454. doi: 10.1101/gad.6.12b.2443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.