Abstract
High grade prostatic intraepithelial neoplasia (HGPIN) is widely believed to represent a precursor to invasive prostatic adenocarcinoma. However, recent molecular studies have suggested that retrograde spread of invasive adenocarcinoma into pre-existing prostatic ducts can morphologically mimic HGPIN. Thus, previous molecular studies characterizing morphologically-identified HGPIN occurring in radical prostatectomies or needle biopsies with concurrent invasive carcinoma may be partially confounded by intraductal spread of invasive tumor. To assess ERG and PTEN status in HGPIN foci likely to represent true precursor lesions in the prostate, we studied isolated HGPIN occurring without associated invasive adenocarcinoma in cystoprostatectomies performed at Johns Hopkins between 2009 and 2014. Of 344 cystoprostatectomies, 33% (115/344) contained invasive prostatic adenocarcinoma in the partially submitted prostate (10 blocks/case on average) and were excluded from the study. Of the remaining cases without sampled cancer, 32% (73/229) showed 133 separate foci of HGPIN and were immunostained for ERG and PTEN using genetically validated protocols. Of foci of HGPIN with evaluable staining, 7% (8/107) were positive for ERG. PTEN loss was not seen in any HGPIN lesion (0/88). Because these isolated HGPIN foci at cystoprostatectomy are unlikely to represent retrograde spread of invasive tumor, our study suggests that ERG rearrangement, but not PTEN loss, is present in a minority of potential neoplastic precursor lesions in the prostate.
Keywords: Prostatic carcinoma, prostatic intraepithelial neoplasia (PIN), PTEN, ERG, immunohistochemistry, radical cystoprostatectomy
Introduction
High-grade prostatic intraepithelial neoplasia (HGPIN) is widely considered the main precursor lesion to invasive prostatic adenocarcinoma [1-4]. HGPIN is characterized by a proliferation of atypical luminal cells with nuclear and nucleolar enlargement within ducts and acini with an intact basal cell layer [3, 4]. Architecturally, HGPIN may show a tufted, micropapillary or flat growth pattern. Though HGPIN is more commonly seen in prostates with invasive prostatic adenocarcinoma than those without invasive tumors, meta-analyses have found that the short-term risk of subsequent diagnosis of invasive carcinoma after an initial diagnosis of HGPIN on needle biopsy is not markedly elevated over baseline unless HGPIN is multifocal [5]. Historically, the evidence supporting HGPIN as a precursor lesion for invasive adenocarcinoma has largely been based on its cytologic resemblance to, and frequent association with, invasive tumors [1, 2]. In addition, common molecular features have been reported for invasive adenocarcinoma and HGPIN. Of these, the presence of ERG gene rearrangement in HGPIN has been one of the most compelling findings. ERG or other ETS gene family members are rearranged in approximately 50% of invasive prostatic adenocarcinoma cases from patients of European descent, resulting in over-expression of ERG protein [6, 7]. In more than 20 prior studies, ERG gene rearrangement has been reported to occur in HGPIN associated with invasive cancers, and ERG status of the invasive tumor and the adjacent HGPIN are frequently concordant, suggesting a likely clonal relationship between the two lesions [8-28]. Though these data do not prove that HGPIN is a precursor lesion to invasive adenocarcinoma, they are consistent with this hypothesis.
Recently, however, there is increasing recognition that HGPIN may have considerable morphologic overlap with another intraductal lesion in the prostate that has a markedly different natural history. Intraductal carcinoma of the prostate (IDC-P) is widely regarded to commonly result from retrograde intraductal spread of a pre-existing high grade invasive adenocarcinoma [29, 30]. Though the current strict morphologic definition of IDC-P is designed to preclude over-diagnosis of HGPIN as IDC-P, an unavoidable consequence of this specificity is that some true cases of IDC-P are likely underdiagnosed as HGPIN [29, 30]. Two recent studies have highlighted this pitfall. Patients with atypical intraductal lesions that fail to meet morphologic criteria for diagnosis of IDC-P (and thus may currently be diagnosed as HGPIN) have a substantially increased risk of subsequent diagnosis with high grade invasive carcinoma [28]. These data suggest that a wider morphologic spectrum of intraductal proliferations than are currently included in the definition of IDC-P may, in fact, represent retrograde spread of invasive carcinoma rather than true precursor lesions [14, 21, 28]. In addition, a recent study of ERG-positive intraductal lesions (some resembling HGPIN and some more recognizable as intraductal carcinoma) that were associated with nearby invasive carcinoma demonstrated identical ERG rearrangement breakpoints in the HGPIN-like and intraductal lesions and the concurrent invasive adenocarcinomas [31]. Further, the presence of heterogeneous PTEN loss in the invasive tumor with homogeneous PTEN loss in the intraductal and HGPIN-like lesions in these cases strongly suggests that the HGPIN-like and intraductal carcinoma lesions actually represent late stage ductal colonization by the invasive tumor in at least some cases.
If morphologically-identified HGPIN may not always be a precursor lesion in prostate cancer, but may in some cases represent later stage retrograde spread of adjacent invasive carcinoma, then many prior studies of molecular changes in HGPIN are likely confounded [32]. Because the prevalence of ERG gene rearrangement and PTEN loss in HGPIN have only been studied in radical prostatectomies (which invariably harbor concurrent invasive tumor) or in biopsies (where the presence of concurrent invasive tumor is uncertain) may have inadvertently included cases of intraductal spread of invasive carcinoma masquerading as HGPIN [8-28, 32]. Thus, the prevalence of these genetic alterations in true precursor lesions to invasive prostate carcinoma remains unclear. To address this, we studied a series of cystoprostatectomy specimens with isolated HGPIN in the absence of concurrent sampled invasive adenocarcinoma that are likely to represent true precursor lesions. Using genetically validated immunohistochemistry assays, we demonstrate that ERG expression occurs in a minority of isolated HGPIN lesions, while PTEN loss is extremely uncommon at this early stage in tumorigenesis.
Methods
Patient and Tissue Selection
This study, including tissue collection and immunohistochemistry (IHC) staining, was approved by Johns Hopkins Institutional Review Board. A search of the Johns Hopkins Pathology database between 2009 and 2014 for cystoprostatectomy specimens performed for urothelial carcinoma without concurrent reported prostate cancer was made. At Hopkins, gross sectioning of the prostate in cystoprostatectomy cases is geared towards detecting significant prostate carcinomas and urothelial carcinoma involving the prostate and prostatic urethra. In general, 10 blocks of prostate are submitted, focusing on the posterior peripheral zone, with 1-2 separate blocks submitted to examine the prostatic urethra and transition zone. Submitted sections comprise around 30% of the total prostate volume in most cases, unless a gross lesion is detected, in which case the prostate is entirely submitted.
A total of 229 cystoprostatectomy specimens were retrieved from the surgical pathology archives and all of the prostate slides were reviewed by one pathologist (CLM) to select cases with HGPIN. HGPIN was defined as a proliferation of atypical luminal cells with crowding, stratification and/or irregular spacing, involving ducts and acini. These lesions, generally visible at low power, showed one of the following architectural patterns: tuffing, micropapillary cribriform or flat [3, 4]. Enlargement of the nuclei and nucleoli, with nucleoli visible at 20× magnification was required. One to three separate blocks containing HGPIN were selected for each case. Some cases had multiple foci of HGPIN identified within each block (range: 1-3), with a minimal distance of 4 mm between individual foci required to consider the foci separate. Atypical intraductal lesions as described previously were not observed in any cystoprostatectomy specimen [28].
Immunohistochemistry
One hematoxylin and eosin (H&E) stained section to verify presence of HGPIN on deeper sections of the block and two 4 μm sections were prepared for immunostaining. ERG immunostaining was performed by using a mouse monoclonal antibody (clone 9FY, Biocare Medical). In brief, following deparaffinization and rehydration, antigen unmasking was done by EDTA buffer (pH 8.0) for 45 minutes. Endogenous peroxidase activity was blocked by incubation in dual endogenous hydrogen peroxidase and alkaline phosphatase enzyme blocker solution for 5 min at room temperature. After washing with PBS with Tween 20, non-serum protein block was applied for 5 min (ULTRA V Block, Thermo Scientific). Then the primary antibody was allowed to react in dilution of 1:50 for 45min at room temperature. After washing in phosphatase-buffer saline, a horseradish peroxidase-labeled polymer (UltraVision Quanto Detection System HRP DAB, Thermo Scientific) was then applied for 10 min at room temperature. Peroxidase was visualized by DAB (3,3’-diaminobenzidine tetrahydrochloride) chromogen. Slides were counterstanined with hematoxylin, dehydrated, and mounted.
PTEN IHC was performed as previously described on the Ventana Discovery Ultra automated staining platform utilizing CC1 antigen retrieval buffer (Roche-Ventana Medical Systems, Tucson, AZ) for 32 minutes at 100°C, followed by incubation with a rabbit anti-human PTEN antibody (Clone D4.3 XP; Cell Signaling, Danvers, MA; 1:75 dilution) at 36°C for 32 minutes, followed by the Optiview HRP multimer secondary detection system [33, 34].
PIN4 immunostaining in a subset of cases was performed using a prediluted antibody cocktail for p63, cytokeratin 903 and AMACR (Zeta Corporation, Sierra Madre, CA) on the Ventana Benchmark Ultra (Ventana-Roche) automated immunostainer.
Immunohistochemistry Scoring
Nuclear ERG protein was visually scored using a previously validated dichotomous scoring system by two pathologists (CLM and TLL) [35]. All glands on the standard slide were scored once they met morphologic criteria for HGPIN, based on side-by-side comparisons with a hematoxylin- and eosin-stained section. Staining for nuclear ERG was assessed in comparison with stromal endothelial cell staining, which provided an internal positive control for ERG in each section. Staining for ERG was considered positive if any lesional cells showed nuclear positivity, even those with somewhat weaker staining when compared with endothelial cells, and negative if no lesional cells were positive.
PTEN immunohistochemistry was blindly scored using a previously genetically validated dichotomous scoring system [33, 34, 36] by two pathologists (LG and TLL). A tissue core was considered to have PTEN protein loss if the intensity of cytoplasmic and nuclear staining was markedly decreased or entirely negative across >10% of tumor cells compared to surrounding benign glands and/or stroma, which provide internal positive controls for PTEN protein expression. This simple dichotomous scoring system has been shown to be highly correlated with underlying homozygous genetic deletion of PTEN [36].
Results
A search of the Johns Hopkins Pathology database revealed that a total of 344 cystoprostatectomies were performed at the Johns Hopkins Hospital from January of 2009 to December of 2014. Of these, 114 specimens had an incidental diagnosis of prostate carcinoma and one cystoprostatectomy was performed for a prostatic stromal sarcoma and all of these were excluded from the study. Among the 229 cystoprostatectomy specimens where the prostate was reported as free of cancer, 32% (73/229) had identifiable high grade prostatic intraepithelial neoplasia (HGPIN) and comprised the study cohort. The age of the 73 patients ranged from 46 to 81 years with a median of 66 years. Between 1 and 3 tissue blocks containing HGPIN were sampled for each case, resulting in the inclusion of 133 separate HGPIN foci, varying from 1 to 3 foci for each case.
From 133 HGPIN foci in 73 cystoprostatectomy specimens, 110 HGPIN foci (110/133; 83%) in 61 cystoprostatectomy specimens (84%) met inclusion criteria for the study after performing deeper levels for IHC. The 23 excluded HGPIN foci included 22 foci where the HGPIN focus was no longer present on deeper recuts performed for IHC and 1 case where a small focus of atypical glands, suspicious for carcinoma, appeared on deeper levels. Of the 61 cystoprostatectomy specimens included in the study, 24 (39%) had multifocal HGPIN defined as presence of HGPIN on multiple slides.
Overall, ERG IHC was positive in 7% (8/107) of individual HGPIN foci (Figures 1, 2, 3). Three HGPIN foci (3% or 3/110) were uninterpretable due to weak staining of internal control endothelial nuclei. Interestingly, no specimen had more than one focus of ERG-positive HGPIN, thus a total of 13% (8/61) of individual cystoprostatectomy specimens contained an ERG-positive HGPIN focus (Figure 3). The rate of ERG positivity was not significantly different among cases with HGPIN present on only one slide (5/37 or 13.5%) versus cases with multifocal HGPIN present on multiple slides (3/24 or 12.5%). In order to confirm that the ERG-positive foci did not represent occult infiltrating cancer with a HGPIN-like morphology, 4 of the positive foci with smaller HGPIN glands were assessed for presence of basal cells using PIN4 immunostaining cocktail. All evaluated foci (4/4; 100%) were positive for basal cell markers, p63 and high molecular weight cytokeratin (Figures 2, 3).
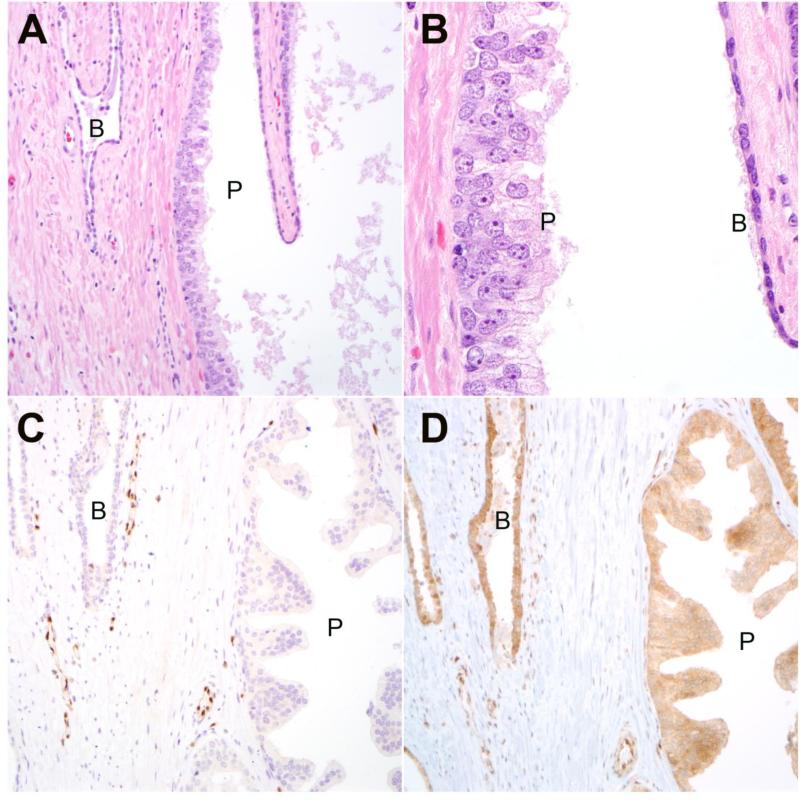
Figure 1.
A Representative HGPIN lesion (P) from radical cystoprostatectomy with adjacent benign glandular epithelium (B) (200× magnification). B: prominent nucleoli are apparent in HGPIN cells (P) compared to adjacent benign luminal cells (B) (630× magnification). C: Immunostaining for ERG is negative in HGPIN (P) and benign (B) glands, with positive staining in adjacent endothelial nuclei as an internal positive control (200× magnification). D: PTEN immunostaining is intact in HGPIN (P) lesion and adjacent benign (B) glands (200× magnification).
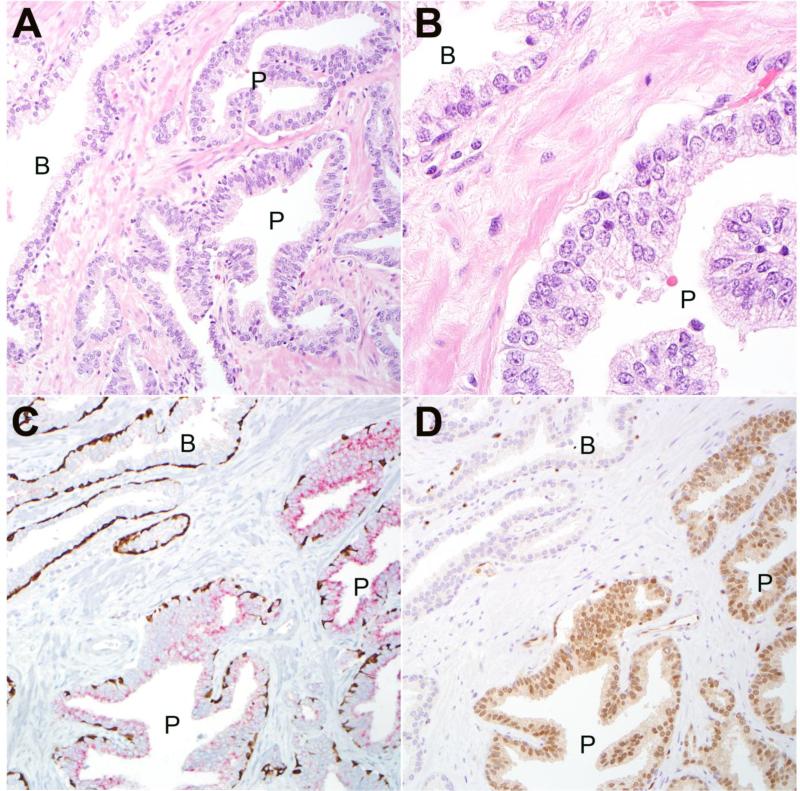
Figure 2.
A: Representative HGPIN lesion (P) from radical cystoprostatectomy with adjacent benign glandular epithelium (B) (200× magnification). B: prominent nucleoli are apparent in HGPIN cells (P) compared to adjacent benign luminal cells (B) (630× magnification). C: Immunostaining with PIN4 cocktail for high molecular weight keratin and p63 demonstrates positively staining basal cells (brown) in both benign (B) and HGPIN (P) glands (200× magnification). Racemase positivity (red) is seen in HGPIN lesion. D: ERG is expressed in nuclei of HGPIN (P) lesion but is negative in adjacent benign (B) glands (200× magnification).
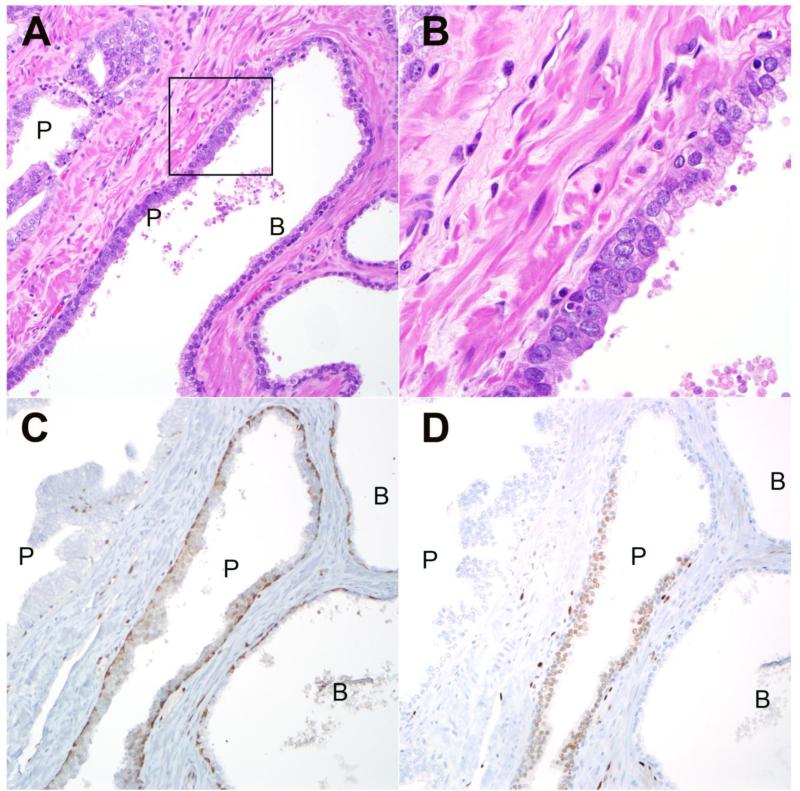
Figure 3.
A: Representative HGPIN lesions (P) from radical cystoprostatectomy with adjacent benign glandular epithelium (B) within the same gland. B: High power image of boxed region from upper left demonstrates prominent nucleoli in HGPIN cells compared to adjacent benign luminal cells (200× magnification) (630× magnification). C: Immunostaining with PIN4 cocktail for high molecular weight keratin and p63 demonstrates positively staining basal cells (brown) in both benign (B) and HGPIN (P) glands. Racemase positivity (red) is absent in this HGPIN lesion (200× magnification). D: Immunostaining for ERG is positive in luminal cells from one of two HGPIN (P) glands and negative and negative in adjacent benign (B) luminal cells within the same gland. An adjacent HGPIN lesion (P) is negative for ERG in the same field (200× magnification).
PTEN immunohistochemistry was interpretable on 80% (88/110) of the HGPIN foci remaining on unstained deeper levels on 60 individual cystoprostatectomy specimens. None of the HGPIN foci (0/88) showed PTEN loss (Figure 1).
Discussion
While a number of histological lesions referred to by various names have been described previously as potential prostate cancer precursor lesions [1], intraductal dysplasia (later referred to as high grade prostatic intraepithelial neoplasia) was first described with detailed morphological criteria as a potential pre-malignant lesion by McNeal and Bostwick in 1986 [2]. Over the years, a combination of morphological, epidemiological and molecular evidence has been used to support the hypothesis that HGPIN is a precursor to invasive carcinoma [4]. The cytologic similarity between the atypical luminal epithelial cells in HGPIN and invasive carcinoma and the frequent presence of HGPIN adjacent to micro-invasive foci of carcinoma (“PINATYP”) have long been used to link the two lesions [37, 38]. In prior cystoprostatectomy series, men with incidental invasive carcinomas are more likely to have HGPIN compared to those without carcinoma (100% vs 63%) [39, 40] and the vast majority of tumors occur in a background of multi-focal HGPIN (91%) [41]. Further, in autopsy series, the prevalence of HGPIN increases dramatically with age just as prostate cancer does, and a higher age-related prevalence of HGPIN is seen in ethnic groups with a higher prevalence of prostate cancer [42]. Finally, as an isolated finding in needle biopsy specimens, multifocal HGPIN is associated with an increased risk of cancer in subsequent biopsies [5].
The molecular evidence that HGPIN is a precursor lesion to prostate cancer has consisted mainly of molecular alterations in common between invasive adenocarcinoma and HGPIN. Discrete molecular lesions, such as GSTP1 methylation [43, 44] and telomere shortening [45, 46] are seen in HGPIN as well as in the majority of adenocarcinomas. Similarly, chromosomal copy number alterations involving chromosome 8p [47, 48] and 8q24 [49-51] have been reported in both HGPIN as well as invasive adenocarcinomas. However, perhaps the most convincing alteration reported in both HGPIN and invasive adenocarcinoma involves TMPRSS2-ERG gene fusions. Present in nearly half of prostate carcinomas and extremely specific to this tumor type, the prevalence of ERG fusions has been repeatedly documented in HGPIN over the past decade. Altogether, our literature review retrieved at least 21 independent studies of ERG fusion rates in HGPIN, with over 1100 separate HGPIN lesions queried for ERG fusion by FISH, RT-PCR or IHC (Table 1) [8-28]. Examining either radical prostatectomy specimens or needle biopsies with HGPIN sampled with or without concurrent invasive adenocarcinoma, these studies have found evidence of ERG rearrangement in a median of 17% of HGPIN lesions (range: 0-36%). Because 20/21 of these prior studies utilized radical prostatectomy specimens or needle biopsies with HGPIN and concurrent or subsequent invasive carcinoma, many examined the concordance between the two lesions and this concordance has generally been high, especially whether the HGPIN and invasive carcinoma are located in close proximity to one another (Table 1). These data have been used to support the argument that HGPIN is a precursor to invasive prostate cancers.
Table 1.
Summary of studies on ERG status in HGPIN.
| Reference | % ERG+ HGPIN | Method | Tissue | % with concurrent invasive cancer | Geographic relationship between HGPIN and invasive cancer | Concordance between HGPIN and invasive cancer for ERG status |
|---|---|---|---|---|---|---|
| Cerveira, 2006 (8) | 21% (4/19) | RT-PCR | RP | 100% (19/19) | NS | NS |
| Perner, 2007 (9) | 19% (5/26) | FISH | RP (TMA) | 100% (26/26) | 100% of ERG+ HGPIN in “tight proximity” to cancer | 80% (4/5) for ERG+ HGPIN |
| Furusato, 2008 (10) | 14% (2/14) | RT-PCR | RP | 100% (14/14) | 36% (5/14) adjacent; 64% (9/14) distant | NS |
| Mosquera, 2008 (11) | 16% (23/143) | FISH | RP, Bx | 87% (124/143) | NS | 73% (91/124) |
| Carver et al, 2009 (12) | 10% (4/10) | FISH | RP (TMA) | 100% (10/10) | adjacent | 100% (10/10) |
| Han, 2009 (13) | 15% (5/33) | FISH | RP (TMA) | 100% (33/33) | Adjacent = <3 mm; away = >3 mm | 75% (15/20) for HGPIN adjacent to ERG+ cancer; 0% (0/10) for HGPIN away from ERG+ cancer |
| Han, 2010 (14) | 0% (0/16) | FISH | RP | 100% (16/16) | >3 mm | NS |
| van Leenders, 2011 (15) | 52% (11/21) | IHC | Bx | 90% (19/21) | Same slide | 95% (18/19) |
| Yaskiv, 2011 (16) | 29% (5/17) | IHC | Bx | 100% (17/17) | 71% (12/17) immediately adjacent to invasive carcinoma | 100% (5/5) for ERG+ HGPIN |
| Gao, 2012 (17) | 36% (59/162) | FISH | Bx | 37% (61/162) with subsequent invasive carcinoma | NS | NS |
| He, 2012 (18) | 5% (5/94) | IHC | Bx | 38% (36/94) with subsequent invasive carcinoma | NS | NS |
| Tomlins, 2012 (19) | 18% (12/68) | IHC | Bx | 22% (15/68) | Separate core | 40% (6/15) |
| Liu, 2013 (20) | 22% (4/18) | IHC | Bx | 72% (13/18) | Separate core | NS |
| Lotan, 2013 (21) | 13% (5/39) | IHC | RP | 100% (39/39) | 23 PIN <3mm from PCa and 16 PIN >3mm from Pca | 57% (17/30) |
| Teng, 2013 (22) | 7% (2/29) | IHC | RP (TMA) | 100% (29/29) | NS | 100% (2/2) for ERG+ HGPIN |
| Teng, 2013 (23) | 6% (4/69) | IHC | RP (TMA) | 100% (69/69) | NS | NS |
| Verdu, 2013 (24) | 0% (0/10) | IHC | RP (TMA) | 100% (10/10) | “Distant” | 80% (8/10) |
| Park, 2014 (25) | 11% (51/461) | IHC | Bx | 37% (170/461) with subsequent invasive carcinoma | NS | NS |
| Taris, 2014 (26) | 18% (10/57) | IHC | RP (TMA) | 100% (57/57) | NS | NS |
| Lee, 2015 (27) | 27% (12/45) | IHC | Bx | 20% (9/45) | Same core | 100% (9/9) |
| Morais, 2015 (28) | 0% (0/19) | IHC | Bx | 0% (0/19) | NA | NA |
Although the morphologic, epidemiologic and molecular data presented above support a close or even clonal relationship between HGPIN and invasive carcinoma, these data do not help us to discern the temporal or evolutionary relationship between these two lesions. HGPIN could evolve into invasive adenocarcinoma in some cases, however the possibility that HGPIN could represent late stage retrograde spread of invasive adenocarcinoma into pre-existing benign ducts would be equally consistent with all of the data summarized above [32]. Ultimately, multiple molecular alterations must be simultaneously examined in HGPIN and adjacent carcinoma to determine whether there is a clonal relationship between the two, and if so, whether one lesion likely gave rise to the other. In a recent study that used ERG breakpoint analysis and PTEN gene deletion to begin to address this question, it appears that at least a subset of lesions meeting morphologic criteria for HGPIN may have evolved from (rather than into) adjacent invasive carcinoma [31].
Thus, it is likely the case that morphologically identified HGPIN may actually represent a spectrum of intraductal lesions in the natural history of prostate cancer. While some HGPIN lesions may precede carcinoma and have the capacity to evolve into invasive tumors, others may represent intraductal spread of previously invasive tumor. Because of this, many previous molecular studies of HGPIN are likely confounded since nearly all studies of HGPIN have been performed using radical prostatectomy specimens (which all contain invasive tumor) or needle biopsies (where the existence of concurrent invasive tumor is unknown) [32]. Indeed, because many of the morphologically-identified HGPIN lesions in these previous studies are adjacent to invasive tumors, it is likely that at least some of the lesions studied represent retrograde spread of invasive tumors. Thus, inclusion of these cases may make the prevalence of cancer-related molecular alterations in HGPIN look artificially high.
This confounding issue could also potentially explain the results of the few studies that found that the presence of cancer-associated molecular alterations in HGPIN on needle biopsy, such as presence of ERG gene rearrangement, is associated with a higher risk of developing subsequent invasive cancer [17, 25, 32]. Indeed, if the rate of ERG positivity in true precursor HGPIN is low, it is possible that the group of ERG-expressing HGPIN lesions was relatively enriched for retrograde intraductal spread of concurrent unsampled invasive tumors compared to the ERG-negative HGPIN group. This scenario would also explain a higher rate of subsequent invasive cancer in these ERG-positive HGPIN cases when the prostates were resampled by needle biopsy.
To begin to understand the true prevalence of molecular alterations in HGPIN, it is necessary to study HGPIN occurring in the absence of invasive carcinoma, as these are likely to represent true precursor lesions. Though autopsy specimens would be ideal for these studies, poor tissue preservation in autopsies makes identification of the characteristic cytologic features of HGPIN difficult. Here, we studied prostate tissue from cystoprostatectomies performed for urothelial carcinoma where the submitted prostate tissue did not contain prostatic adenocarcinoma. Though one important weakness of our study is that the prostate in cystoprostatectomy specimens is not submitted in totality for histologic examination at our institution, our prostate sampling procedure in these specimens is likely adequate to detect most significant prostate tumors (with at least 30% of total prostate volume submitted in most cases). In addition, we could clearly exclude the presence of invasive tumor within 3 millimeters of the HGPIN lesion examined. Ultimately, this study will need to be confirmed in a series of cystoprostatectomy specimens containing HGPIN where the prostate is entirely submitted for histologic examination to exclude the possibility of occult invasive cancer in some cases. However, overall, it is highly likely that the HGPIN lesions identified in our cystoprostatectomy study did not result from retrograde spread of invasive tumor (which typically happen in the context of clinically significant, high stage tumors [29]), but are instead representative of a spectrum of true precursor lesions in the prostate.
In the current study, we found that 7% of these precursor HGPIN lesions express ERG protein by immunohistochemistry, a proven surrogate for the presence of ERG gene rearrangement. This prevalence of ERG expression in HGPIN is at the low end of the range reported in previous large studies of radical prostatectomy and needle biopsy specimens. In one of the largest studies utilizing 143 radical prostatectomies and biopsies, ERG rearrangement (measured by FISH) was found in 16% of lesions [11]. In the largest study of biopsies alone (from a prostate cancer prevention trial, GTx Protocol G300104), ERG expression (by IHC) was seen in 11% (51/461) cases overall [25]. Interestingly, most of the tumors found subsequent to HGPIN in this study were low grade (Gleason score 6) tumors that may not have had the capacity for retrograde intraductal spread. Thus the rate of ERG positivity in this study is not significantly different from what we observed in the current study using cystoprostatectomies. However, another large study of prostate biopsies using FISH found that 36% (59/162) of HGPIN lesions showed ERG rearrangement [17]. Thus it is possible that the inadvertent inclusion of some cases of intraductal retrograde spread of tumor may have artificially inflated the prevalence of ERG expression reported in some prior HGPIN studies.
Given that ERG rearrangement can occur in a minority of isolated HGPIN lesions, it is unlikely that the presence of ERG-positive PIN will be useful to distinguish true HGPIN from retrograde intraductal spread of cancer. However, PTEN may be useful in this context. We have previously reported that PTEN loss occurs at a high frequency in IDC-P, and can be seen in a majority of IDC-P lesions occurring with concurrent invasive adenocarcinoma [21, 28]. In contrast, we found that PTEN loss rarely occurs in HGPIN lesions, either occurring in radical prostatectomies (adjacent or distant from invasive adenocarcinoma) or in needle biopsies where HGPIN was an isolated finding. Here, we add to these data by showing that PTEN loss does not occur at a detectable frequency in isolated HGPIN occurring in cystoprostatectomy specimens. These data add additional support to the concept that PTEN loss in HGPIN or other atypical intraductal lesions may have a high positive predictive value for the presence of concurrent invasive adenocarcinoma, suggesting that these cases should get additional and very close follow-up to exclude this possibility.
Acknowledgments
Grant Support: Funding for this research was provided in part by a CDMRP award supporting the Precision Medicine Validating Center, a Prostate Cancer Foundation Young Investigator Award (TLL), and a generous gift from Mr. David H. Koch (TLL), and the NIH Cancer Center Support Grant 5P30CA006973-52
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure/Conflicts of Interest: TLL has received research funding from Ventana Medical Systems and JAS has consulted for CymoGen Dx, LLC.
References
- 1.McNeal J. Origin and development of carcinoma in the prostate. Cancer. 1969;23:24–34. doi: 10.1002/1097-0142(196901)23:1<24::aid-cncr2820230103>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 2.McNeal J, Bostwick D. Intraductal dysplasia: A premalignant lesion of the prostate. Hum Pathol. 1986;17:64–71. doi: 10.1016/s0046-8177(86)80156-3. [DOI] [PubMed] [Google Scholar]
- 3.Epstein J. Precursor lesions to prostatic adenocarcinoma. Virchows Arch. 2009;454:1–16. doi: 10.1007/s00428-008-0707-5. [DOI] [PubMed] [Google Scholar]
- 4.Bostwick D, Cheng L. Precursors of prostate cancer. Histopathology. 2012;60:4–27. doi: 10.1111/j.1365-2559.2011.04007.x. [DOI] [PubMed] [Google Scholar]
- 5.Epstein J, Herawi M. Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: Implications for patient care. J Urol. 2006;175:820–34. doi: 10.1016/S0022-5347(05)00337-X. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins S, Rhodes D, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Pettersson A, Graff R, Bauer S, Pitt MJ, Lis RT, Stack EC, et al. The TMPRSS2: ERG rearrangement, ERG expression, and prostate cancer outcomes: A cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerveira N, Ribeiro F, Peixoto A, Costa V, Henrique R, Jerónimo C. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–32. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perner S, Mosquera J, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: An early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–8. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 10.Furusato B, Gao C, Ravindranath L, Chen Y, Cullen J, McLeod DG, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2007;21:67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 11.Mosquera J, Perner S, Genega E, Sanda M, Hofer MD, Mertz KD, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–5. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carver B, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han B, Mehra R, Lonigro R, Wang L, Suleman K, Menon S, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han B, Suleman K, Wang L, Siddiqui J, Sercia L, Magi-Galluzzi C, et al. ETS gene aberrations in atypical cribriform lesions of the prostate: Implications for the distinction between intraductal carcinoma of the prostate and cribriform high-grade prostatic intraepithelial neoplasia. Am J Surg Pathol. 2010;34:478–85. doi: 10.1097/PAS.0b013e3181d6827b. [DOI] [PubMed] [Google Scholar]
- 15.Van Leenders G, Boormans J, Vissers C, Hoogland AM, Bressers AA, Furusato B, et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: Implications for pathological practice. Mod Pathol. 2011;24:1128–38. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 16.Yaskiv O, Zhang X, Simmerman K, Daly T, He H, Falzarano S, et al. The utility of ERG/P63 double immunohistochemical staining in the diagnosis of limited cancer in prostate needle biopsies. Am J Surg Pathol. 2011;35:1062–8. doi: 10.1097/PAS.0b013e318215cc03. [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Li L, Zhou F, Kie KJ, Shao CK, Su ZL, et al. ERG rearrangement for predicting subsequent cancer diagnosis in high-grade prostatic intraepithelial neoplasia and lymph node metastasis. Clin Cancer Res. 2012;18:4163–72. doi: 10.1158/1078-0432.CCR-11-2449. [DOI] [PubMed] [Google Scholar]
- 18.He H, Osunkoya A, Carver P, Falzarano S, Klein E, Magi-Galluzzi C, et al. Expression of ERG protein, a prostate cancer specific marker, in high grade prostatic intraepithelial neoplasia (HGPIN): Lack of utility to stratify cancer risks associated with HGPIN. BJU Int. 2012;110:E751–5. doi: 10.1111/j.1464-410X.2012.11557.x. [DOI] [PubMed] [Google Scholar]
- 19.Tomlins S, Palanisamy N, Siddiqui J, Chinnaiyan A, Kunju L. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136:935–46. doi: 10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Shi J, Wilkerson M, Yang XJ. Immunohistochemical evaluation of ERG expression in various benign and malignant tissues. Ann Clin Lab Sci. 2013;43:3–9. [PubMed] [Google Scholar]
- 21.Lotan T, Gumuskaya B, Rahimi H, Hicks JL, Iwata T, Robinson BD, et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol. 2012;26:587–603. doi: 10.1038/modpathol.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng L, Wang C, Bégin L, Dolph M, Yilmaz A, Trpkov K, et al. ERG protein expression and gene rearrangements are present at lower rates in metastatic and locally advanced castration-resistant prostate cancer compared to localized disease. Urology. 2013;82:394–9. doi: 10.1016/j.urology.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Teng L, Hong L, Wang C, Dolph M, Donnelly B, Bismar TA. ERG protein expression is of limited Prognostic value in men with localized prostate cancer. ISRN urology. 2013;2013:786545. doi: 10.1155/2013/786545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verdu M, Trias I, Roman R, Rodon N, Garcia-Pelaez B, Calvo M, et al. ERG expression and prostatic adenocarcinoma. Virchows Arch. 2013;462:639–44. doi: 10.1007/s00428-013-1415-3. [DOI] [PubMed] [Google Scholar]
- 25.Park K, Bostwick DG, Dalton JT, Hancock ML, Narayanan R, Barbieri CE, et al. TMPRSS2: ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade Prostatic intraepithelial neoplasia. J Clin Oncol. 2013;32:206–11. doi: 10.1200/JCO.2013.49.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taris M, Irani J, Blanchet P, Multigner L, Cathelineau X, Fromont G. ERG expression in prostate cancer: The prognostic paradox. J Clin Oncol. 2014;74:1481–7. doi: 10.1002/pros.22863. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Yu D, Wang C, Saba R, Liu S, Trpkov K, et al. ERG expression in prostate needle biopsy: Potential diagnostic and Prognostic implications. Appl Immunohistochem Mol Morphol. 2014;23:499–505. doi: 10.1097/PAI.0000000000000119. [DOI] [PubMed] [Google Scholar]
- 28.Morais C, Han J, Gordetsky J, Nagar MS, Anderson AE, Lee S, et al. Utility of PTEN and ERG immunostaining for distinguishing high-grade PIN from intraductal carcinoma of the prostate on needle biopsy. Am J Surg Pathol. 2014;39:169–78. doi: 10.1097/PAS.0000000000000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C, Epstein J. Intraductal carcinoma of the prostate on needle biopsy: Histologic features and clinical significance. Mod Pathol. 2006;19:1528–35. doi: 10.1038/modpathol.3800702. [DOI] [PubMed] [Google Scholar]
- 30.Cohen R, Wheeler T, Bonkhoff H, Rubin M. A proposal on the identification, histologic reporting, and implications of intraductal prostatic carcinoma. Arch Pathol Lab Med. 2007;131:1103–9. doi: 10.5858/2007-131-1103-APOTIH. [DOI] [PubMed] [Google Scholar]
- 31.Haffner M, Weier C, Xu M, Vaghasia A, Gürel B, Gümüşkayaet B, et al. Molecular evidence that invasive adenocarcinoma can mimic prostatic intraepithelial neoplasia (PIN) and intraductal carcinoma through retrograde glandular colonization. J Pathol. 2015;238:31–41. doi: 10.1002/path.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Marzo AM, Haffner MC, Lotan TL, Yegnasubramanian S, Nelson WG, Hopkins J. Premalignancy in prostate cancer: Rethinking what we know. Can Prev Res. 2016 doi: 10.1158/1940-6207.CAPR-15-0431. in press. [DOI] [PubMed] [Google Scholar]
- 33.Lotan T, Wei W, Morais CL, Hawley ST, Fazli L, Hurtado-Coli A, et al. PTEN loss as determined by clinical-grade Immunohistochemistry assay is associated with worse recurrence-free survival in prostate cancer. Eur Urol Focus. 2015 doi: 10.1016/j.euf.2015.07.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahearn TU, Pettersson A, Ebot EM, Gerke T, Graff RE, Morais CL, et al. A Prospective Investigation of PTEN Loss and ERG Expression in Lethal Prostate Cancer. J Natl Cancer Inst. 2016;108:djv346. doi: 10.1093/jnci/djv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lotan T, Gupta N, Wang W, Toubaji A, Haffner MC, Chaux A, et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24:820–8. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotan TL, Morais CL, Wei W, Jamaspishvili T, McKenney J, Feng Z, et al. PTEN status determination in prostate cancer: Comparison of IHC and FISH in a large multi-center cohort. Mod Pathol. 2015;28:241A. [Google Scholar]
- 37.Bostwick D, Brawer M. Prostatic intra-epithelial neoplasia and early invasion in prostate cancer. Cancer. 1987;59:788–94. doi: 10.1002/1097-0142(19870215)59:4<788::aid-cncr2820590421>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 38.Kronz J, Shaikh A, Epstein J. High-grade prostatic intraepithelial neoplasia with adjacent small atypical glands on prostate biopsy. Hum Pathol. 2001;32:389–95. doi: 10.1053/hupa.2001.23522. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Yang X. Prevalence of high-grade prostatic intraepithelial neoplasia and its relationship to serum prostate specific antigen. Int Braz J Urol. 2002;28:413–6. [PubMed] [Google Scholar]
- 40.Troncoso P, Babaian R, Ro J, Grignon D, Eschenbach von, Ayala A. Prostatic intraepithelial neoplasia and invasive prostatic adenocarcinoma in cystoprostatectomy specimens. Urology. 1989;34:52–6. [PubMed] [Google Scholar]
- 41.Wiley E, Davidson P, McIntire D, Sagalowsky A. Risk of concurrent prostate cancer in cystoprostatectomy specimens is related to volume of high-grade prostatic intraepithelial neoplasia. Urology. 1997;49:692–6. doi: 10.1016/S0090-4295(96)00627-9. [DOI] [PubMed] [Google Scholar]
- 42.Sakr W, Grignon D, Haas G, Heilbrun L, Pontes J, Crissman J. Age and racial distribution of prostatic intraepithelial neoplasia. Eur Urol. 1996;30:138–44. doi: 10.1159/000474163. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama M, Bennett C, Hicks J, Epstein JI, Platz EA, Nelson WG, et al. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: A detailed study using laser- capture microdissection. Am J Pathol. 2003;163:923–33. doi: 10.1016/s0002-9440(10)63452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks J, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Cancer Epidemiol Biomarkers Prev. 1998;7:531–6. [PubMed] [Google Scholar]
- 45.Meeker A, Hicks J, Platz E, March GE, Bennett CJ, Delannoy MJ, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–9. [PubMed] [Google Scholar]
- 46.Vukovic B, Park P, Al-Maghrabi J, Beheshti B, Sweet J, Evans A, et al. Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma. Oncogene. 2003;22:1978–87. doi: 10.1038/sj.onc.1206227. [DOI] [PubMed] [Google Scholar]
- 47.Emmert-Buck, Vocke C, Pozzatti R, Duray PH, Jennings SB, Florence CD, et al. Allelic loss on chromosome 8p12-21 in microdissected prostatic intraepithelial neoplasia. Cancer Res. 1995;55:2959–62. [PubMed] [Google Scholar]
- 48.Häggman M, Wojno K, Pearsall C, Macoska J. Allelic loss of 8p sequences in prostatic intraepithelial neoplasia and carcinoma. Urology. 1997;50:643–7. doi: 10.1016/S0090-4295(97)00304-X. [DOI] [PubMed] [Google Scholar]
- 49.Qian J, Bostwick D, Takahashi S, Borrell TJ, Herath JF, Lieber MM, et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Res. 1995;55:5408–14. [PubMed] [Google Scholar]
- 50.Qian J, Jenkins R, Bostwick D. Detection of chromosomal anomalies and c-myc gene amplification in the cribriform pattern of prostatic intraepithelial neoplasia and carcinoma by fluorescence in situ hybridization. Mod Pathol. 1997;10:1113–9. [PubMed] [Google Scholar]
- 51.Bethel C, Faith D, Li X, Guan B, Hicks JL, Lan F, Jenkins RB, et al. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: Association with gleason score and chromosome 8p deletion. Cancer Res. 2006;66:10683–90. doi: 10.1158/0008-5472.CAN-06-0963. [DOI] [PubMed] [Google Scholar]