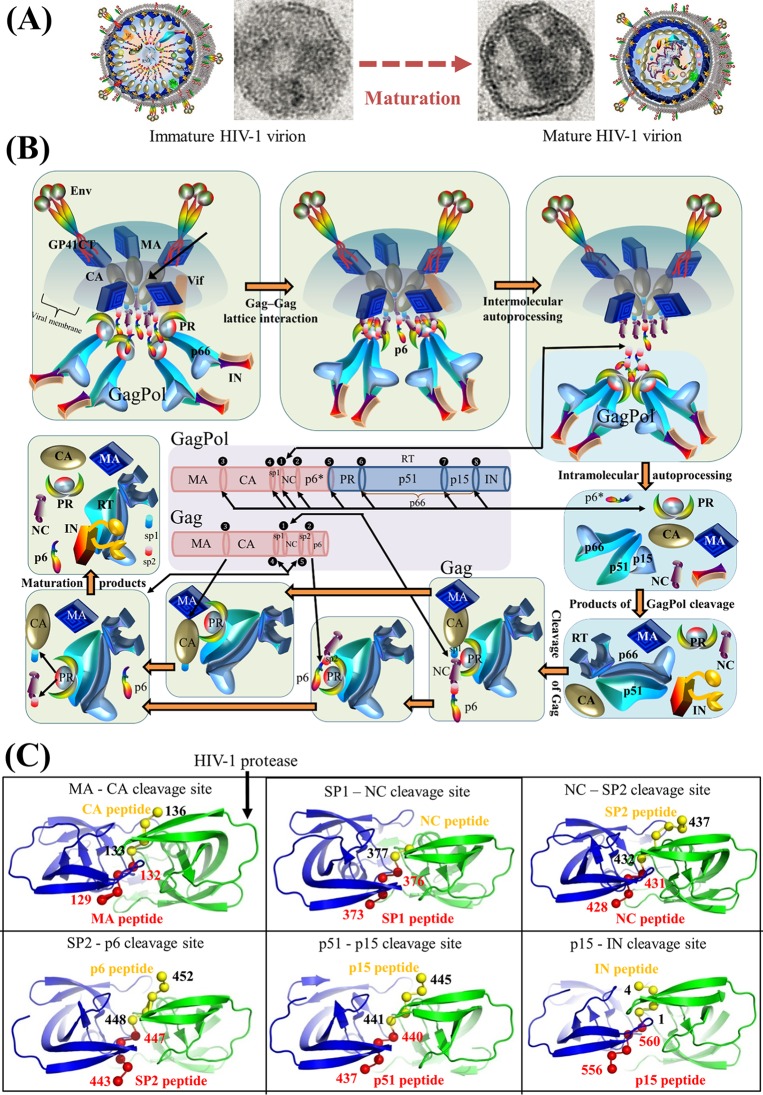
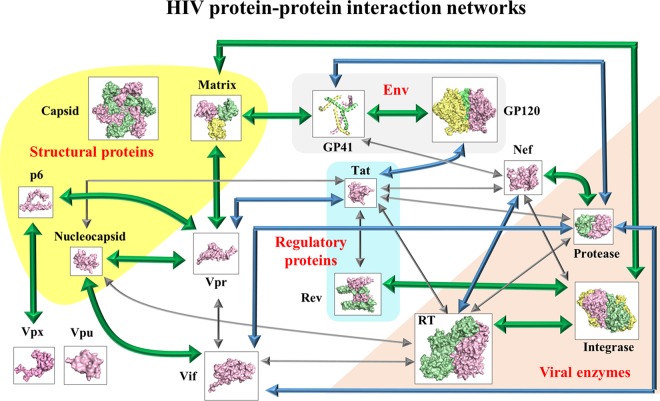
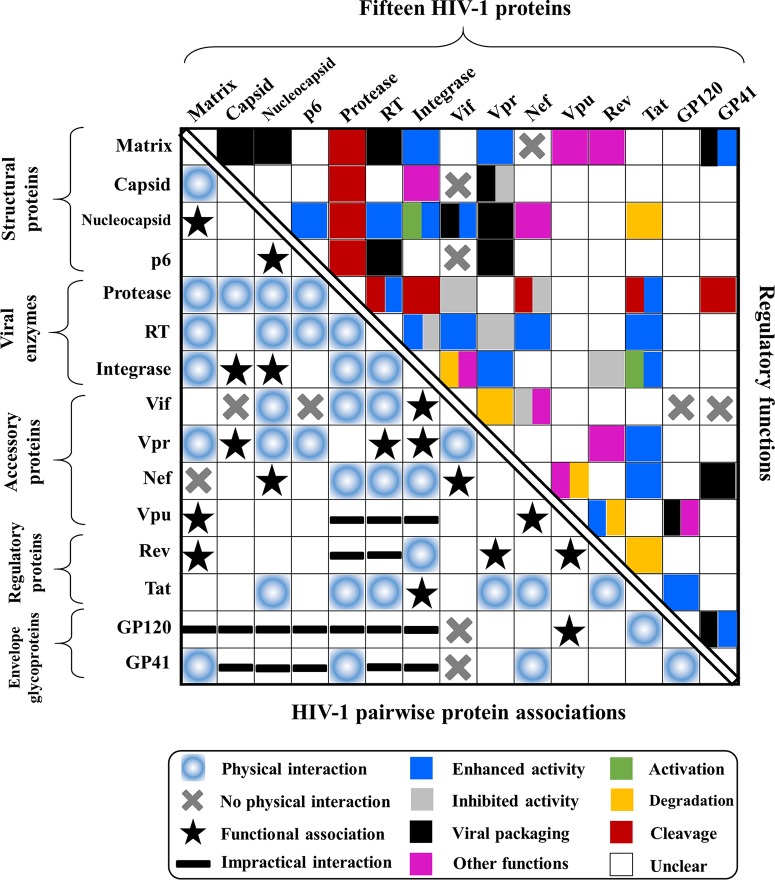
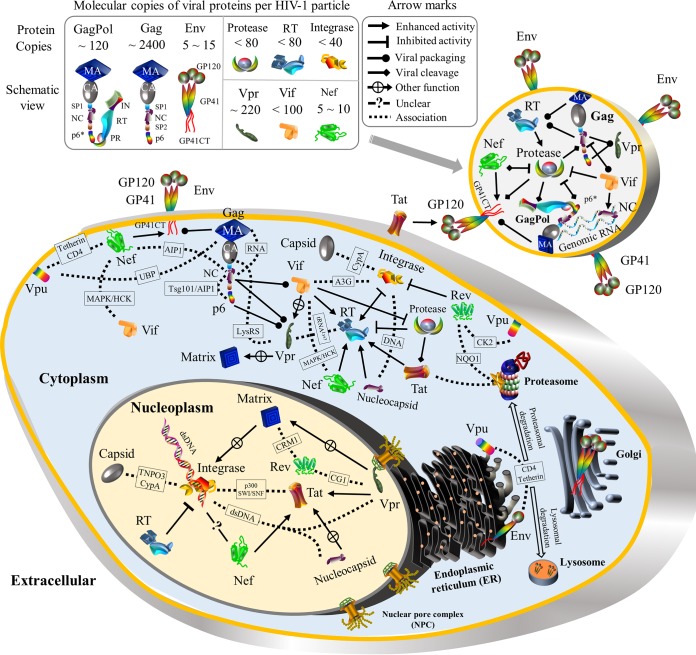
SUMMARY
The HIV genome encodes a small number of viral proteins (i.e., 16), invariably establishing cooperative associations among HIV proteins and between HIV and host proteins, to invade host cells and hijack their internal machineries. As a known example, the HIV envelope glycoprotein GP120 is closely associated with GP41 for viral entry. From a genome-wide perspective, a hypothesis can be worked out to determine whether 16 HIV proteins could develop 120 possible pairwise associations either by physical interactions or by functional associations mediated via HIV or host molecules. Here, we present the first systematic review of experimental evidence on HIV genome-wide protein associations using a large body of publications accumulated over the past 3 decades. Of 120 possible pairwise associations between 16 HIV proteins, at least 34 physical interactions and 17 functional associations have been identified. To achieve efficient viral replication and infection, HIV protein associations play essential roles (e.g., cleavage, inhibition, and activation) during the HIV life cycle. In either a dispensable or an indispensable manner, each HIV protein collaborates with another viral protein to accomplish specific activities that precisely take place at the proper stages of the HIV life cycle. In addition, HIV genome-wide protein associations have an impact on anti-HIV inhibitors due to the extensive cross talk between drug-inhibited proteins and other HIV proteins. Overall, this study presents for the first time a comprehensive overview of HIV genome-wide protein associations, highlighting meticulous collaborations between all viral proteins during the HIV life cycle.
INTRODUCTION
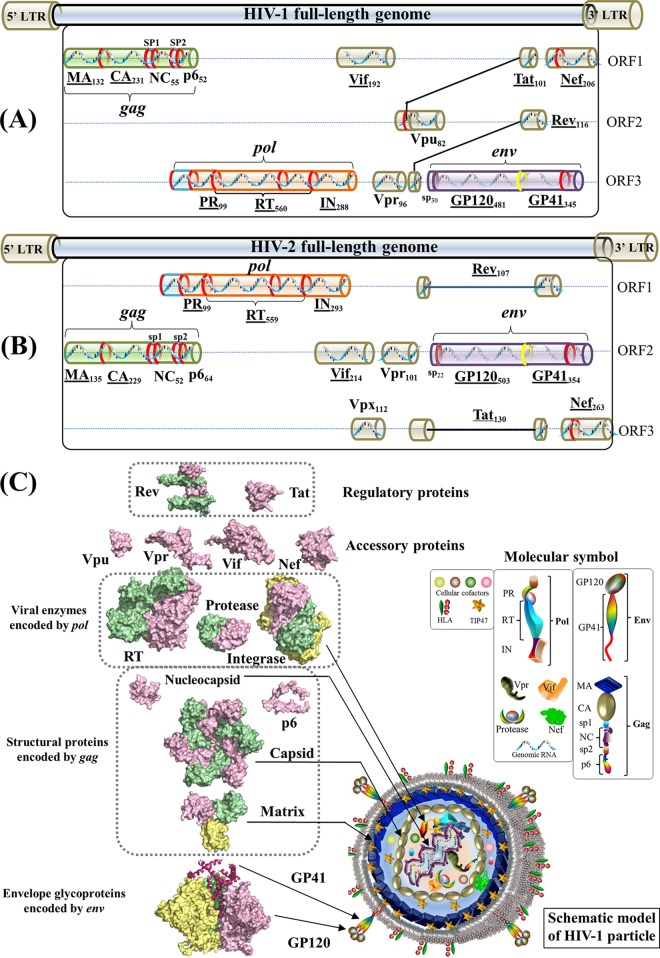
The genome of human immunodeficiency virus (HIV) encodes 16 viral proteins playing essential roles during the HIV life cycle (Fig. 1). Three major genes, gag, pol, and env, code for structural proteins (matrix, capsid, nucleocapsid, and p6), viral enzymes (protease, reverse transcriptase [RT], and integrase), and envelope proteins (GP120 and GP41) (1, 2) (see Text S1 in the supplemental material). The remaining genes code for regulatory proteins (Tat and Rev) and accessory proteins (Vif, Vpu/Vpx, Vpr, and Nef) (3). Vpu is found exclusively in HIV type 1 (HIV-1), whereas Vpx is carried by HIV-2.
FIG 1.
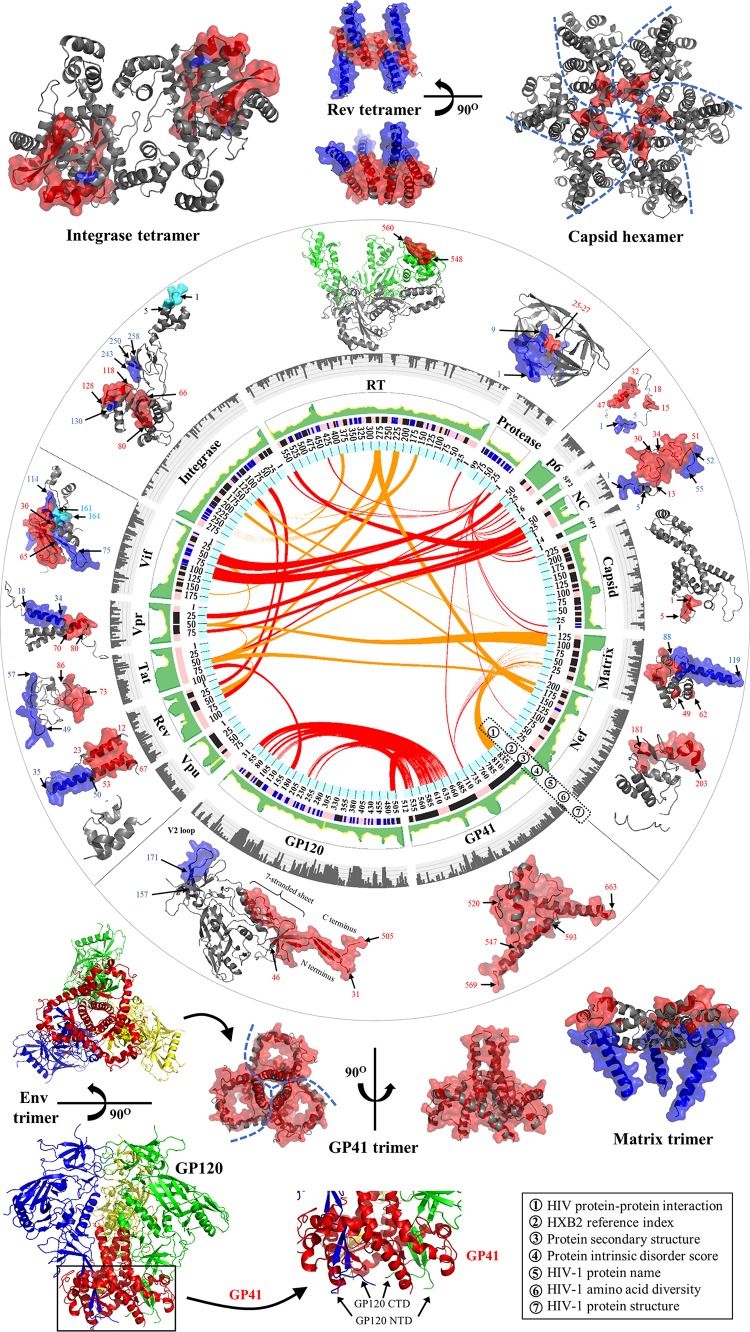
Gene maps and protein structures of HIV-1 and HIV-2. (A) Schematic model of the HIV-1 full-length genome (reference strain HXB2). HIV protein names and amino acid lengths are shown beneath the colored protein regions in three open reading frames (ORFs). Eleven multimeric proteins have underlined names. In the gene map, red rings mark the locations where viral protease cleaves during viral maturation. In the env gene, a yellow ring shows the cleavage position of human proteases (e.g., furin) (594). The 5′ and 3′ long terminal regions (LTRs) are also indicated in the full-length genome. (Adapted from reference 44.) (B) Schematic model of the HIV-2 full-length genome (reference strain SIVmac239). (Adapted from reference 44.) (C) Surface representations of HIV-1 protein structures and schematic view of the HIV-1 particle. Surface representations of 15 HIV-1 protein structures are clustered according to their functional roles. HIV-1 monomeric proteins are shown in pink, and different subunits of multimeric proteins are distinguished with different colors (green, yellow, and red). HIV-1 protein structures are scaled precisely for a direct and intuitive comparison. At the bottom right, a schematic model of a mature viral particle is displayed, and the key shows protein annotations. Proteins in the schematic view are shown for illustration purposes; their structures and sizes here are not necessarily identical to the real protein structures and sizes. Additional information about HIV genomic reference sequences and natural polymorphisms is available online (see http://www.virusface.com/). (Adapted from reference 44.)
Although HIV genomes code for only 16 viral proteins (Fig. 2), a great number of physical interactions between pairs of HIV proteins, so-called HIV pairwise protein interactions, provide essential mechanisms for HIV to achieve efficient viral replication at different stages of the HIV life cycle (4). For instance, the HIV-1 envelope glycoprotein GP120 physically interacts with GP41 during viral entry (5). In addition to HIV pairwise protein interactions, HIV-host protein interactions are known to play essential roles for HIV to hijack human cellular systems (6–11). Because of this, functional associations between HIV proteins can be mediated via host molecules (e.g., CD4). Taking the functional association of Vpu-CD4-Env as an example, the binding of Vpu to CD4 facilitates the proper assembly of Env into HIV-1 particles, because Vpu interacts with CD4 to trigger the rapid degradation of newly synthesized CD4, thereby preventing the aggregation of CD4-Env structural complexes in the endoplasmic reticulum (ER) (12–19). Overall, physical interactions and functional associations between 16 HIV proteins delineate a global perspective of HIV genome-wide associations that play essential roles during the HIV life cycle.
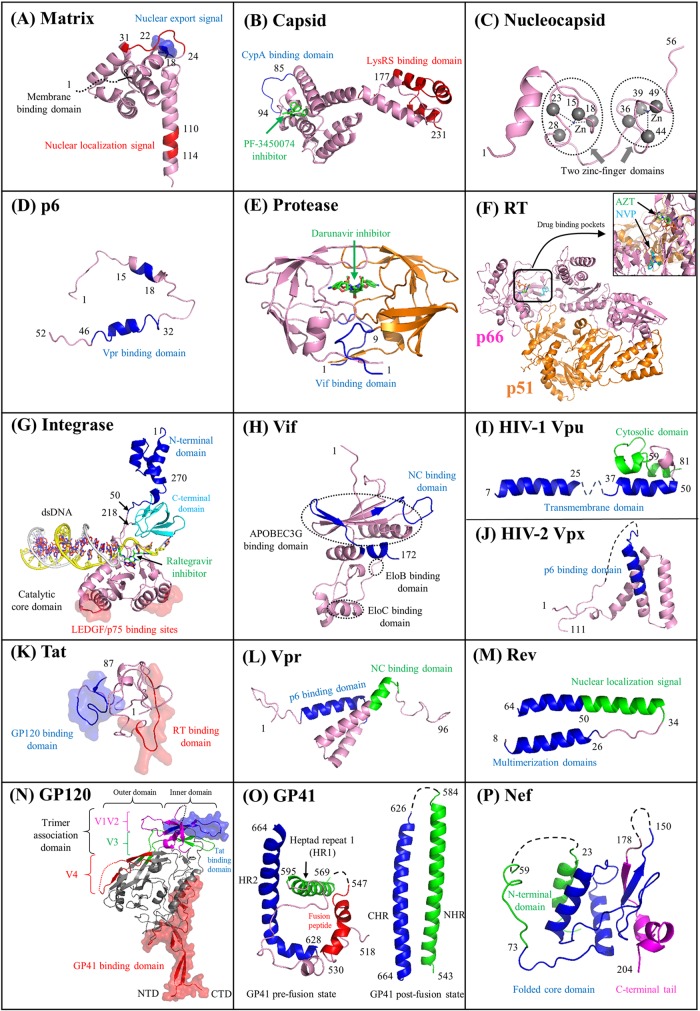
FIG 2.
Functional domains of 16 HIV proteins. Cartoon representations of 16 HIV proteins (matrix, capsid, nucleocapsid, p6, protease, RT, integrase, Vif, Vpu, Vpx, Tat, Vpr, Rev, GP120, GP41, and Nef) are visualized. For each panel, protein domains involved with HIV pairwise protein interactions are marked accordingly. Surface representations indicate protein interaction interfaces. Distinct functional domains are annotated in different colors, such as the N-terminal heptad repeat (NHR) and the C-terminal heptad repeat (CHR) of GP41 in panel O. The V1 to V4 flexible loop regions of GP120 (see details in reference 50) are mapped in panel N. Five small molecules shown in green elucidate protein inhibitors such as the capsid inhibitor PF-3450074 (503) (B), the protease inhibitor darunavir (E), the nucleoside analogue reverse transcriptase inhibitor zidovudine (AZT) (F), the nonnucleoside analogue reverse transcriptase inhibitor nevirapine (NVP) (F), and the integrase inhibitor raltegravir (G). HIV-1 protein domains that interact with cellular proteins are mapped, such as AIP1 (376) (C); LEDGF/p75 (168) (G); and APOBEC3G, EloB, and EloC (595) (H). For some HIV-1 multimeric proteins (matrix, capsid, integrase, Rev, GP120, and GP41), only a subunit is demonstrated, and their multimeric structures are shown in Fig. 1. Text S1 in the supplemental material describes major functions of 16 HIV proteins. Except for HIV-2 Vpx, PDB data for the other 15 proteins were obtained for HIV-1. A list of PDB accession numbers used for our structural visualization is available in Table S1 in the supplemental material. The integrase structure of prototype foamy virus is used for visualization purposes, because the full-length structure of HIV integrase is lacking. HIV structural movies and teaching slides are available at our online platform (see http://www.virusface.com/).
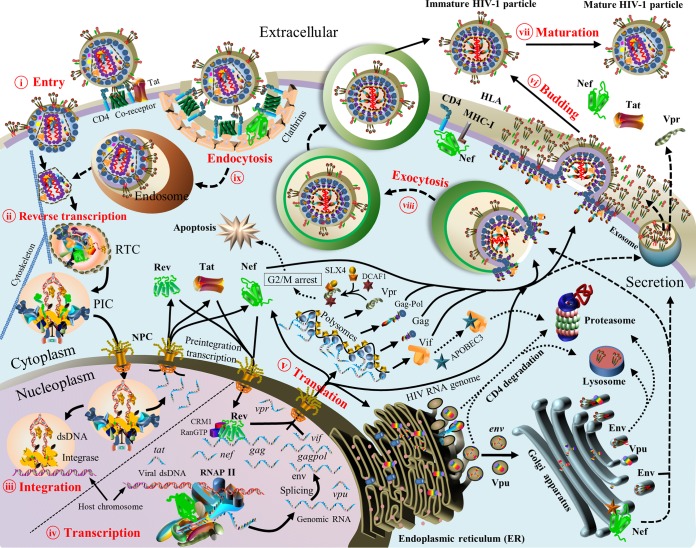
To our knowledge, a systematic review that provides a genome-wide perspective on HIV pairwise protein associations is still lacking in spite of many studies focusing on individual protein associations. In theory, 16 HIV proteins would generate 120 pairwise protein associations, but some associations might be absent during the HIV life cycle. To disclose the mystery of HIV protein associations from a genome-wide perspective, we thus performed the first systematic review to establish experimental evidence for HIV pairwise protein associations and their functional activities at major stages of the HIV life cycle: viral entry (20–22), reverse transcription (23), viral integration (24–26), viral transcription and translation (27–29), viral assembly and budding (2, 30), and viral maturation (2, 30, 31) (Fig. 3).
FIG 3.
Overview of the HIV-1 life cycle. Nine stages are described. (i) Viral entry (viral fusion). Mature HIV virions target host cells through direct binding to the cellular receptor CD4 and chemokine coreceptors (e.g., CCR5 and CXCR4) (20, 21). (ii) Reverse transcription. HIV reverse transcriptase in the reverse transcriptase complex (RTC) produces double-stranded DNA (dsDNA) from single-stranded RNA (23). (iii) Viral integration. The HIV preintegration complex (PIC) transports viral dsDNA into the nucleus by entering the nuclear pore complex (NPC). During preintegration transcription, three viral proteins (Rev, Tat, and Nef) are synthesized from unintegrated dsDNA (25). In the presence of cellular cofactors (e.g., LEDGF/p75), the PIC targets host chromosomal regions with high transcriptional activity, where viral dsDNA is integrated into host chromosomes (24). (iv) Viral transcription. Viral proteins (Tat and Nef) hijack cellular transcription machineries to activate viral mRNA synthesis from integrated viral dsDNA (29). HIV Rev recruits CRM1, RanGTP, and other host proteins to export viral mRNAs into the cytoplasm (213). (v) Viral translation. Viral mRNAs are translated into (precursor) proteins in the cellular compartments. Viral mRNAs of Gag, GagPol, and most accessory proteins are translated in cytosolic polysomes, except for Env and Vpu (30). In the cytoplasm, mature Vpr interacts with host proteins (DCAF1 and SLX4) to induce G2/M cell cycle arrest (236, 596), Vif activates the degradation of APOBEC3 proteins (34, 233), and Nef plays multiple roles in different cellular compartments based on HIV-host protein interactions (597). In the ER, HIV-1 Vpu mRNA is translated, and mature Vpu retains newly synthesized CD4 (19). The dislocated CD4 thereafter undergoes lysosomal and/or proteasomal degradation (19, 451). Tetherin is also targeted by Vpu in the ER, and it is subsequently delivered to the lysosomal and/or proteasomal degradation pathways (459, 461). In the ER, modifications of Env such as signal peptide cleavage, folding, trimerization, and glycosylation occur (598). In the Golgi apparatus, cellular proteases (e.g., furin) cleave Env glycoproteins into GP120 and GP41, which are subsequently assembled into Env spikes via noncovalent interactions (599). Most Env proteins retained in the ER or Golgi apparatus are degraded, and only a small proportion reaches the cell membrane (600). Env, Vpr, Tat, and Nef travel to the cell membrane via secretory pathways (30, 601–603). (vi) Viral assembly and budding. Nascent HIV virions are assembled with two genomic mRNAs, viral proteins (Gag, GagPol, Env, Vif, Vpr, and Nef), and cellular cofactors (e.g., actin, tRNALys3, and TIP47) (2, 30, 277). Nascent HIV particles pinch off from the cellular membrane to infect other host cells (30). HIV-1 Nef induces CD4 degradation to prevent the Env-CD4 interaction on the extracellular membrane (597, 604). (vii) Virus maturation. HIV protease cleaves Gag and GagPol precursors into structural proteins (matrix, capsid, nucleocapsid, and p6) and viral enzymes (protease, RT, and integrase), thus transforming immature virions into mature virions for new infections (2, 30, 31). (viii) Virus exocytosis. As an alternative route of viral budding, nascent HIV virions are released by using exocytosis pathways (605). (ix) Virus endocytosis. As an alternative route of viral entry, mature HIV virions enter host cells through endocytic pathways (46). Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale. MHC-I, major histocompatibility complex class I.
Based on a large body of publications accumulated from 1985 to 2015, our review is focused on the following three aspects. (i) What molecular experiments were used to report HIV protein associations? (ii) Where and when do HIV protein associations achieve their functional activities during the HIV life cycle? (iii) Which viral protein domains are responsible for protein interactions at the molecular level? Clinical relevance and therapeutic implications of HIV genome-wide protein associations are discussed from two aspects: novel mechanisms of HIV drug resistance and HIV-derived peptide inhibitors. The former provides new insights into why HIV-infected patients could fail highly active antiretroviral therapies (HAARTs) in the absence of drug-resistant mutations. The latter may shed light on the development of anti-HIV agents.
Our review begins with the procedure for literature selection. Thereafter, physical interactions and functional associations between HIV proteins are described, depending on their activities at major stages of the HIV life cycle. For each physical interaction or functional association, its biological activities and interaction domains are summarized. For a better understanding of HIV protein associations, we visualize protein interactions during the HIV life cycle, discuss their clinical relevance and therapeutic implications, and establish an online platform to update the information on HIV genome-wide protein associations (http://www.virusface.com/). Newly created structural movies have been shared online to highlight protein interaction domains. Challenges and future perspectives are discussed at the end of this review.
LITERATURE SELECTION
This section describes the procedure for our literature selection (Fig. 4). We performed an electronic literature search by querying English articles from three sources (PubMed, Google Scholar, and Cochrane Library) plus reference lists of retrieved articles published from January 1985 until December 2015. Moreover, we extracted literature from the HIV-1 Human Interaction Database (HHID) to collect information about HIV-host protein interactions (32). This extraction allows the identification of any cellular protein that physically interacts with two HIV proteins. For instance, the Vif-APOBEC3G-integrase association exists because the cellular protein APOBEC3G physically interacts with viral Vif and integrase during the HIV life cycle (33–43). In summary, three major steps were carried out by selecting studies that reported protein associations involving 16 HIV proteins (matrix, capsid, nucleocapsid, p6, protease, RT, integrase, Vif, Vpr, Vpu, Vpx, Tat, Rev, GP120, GP41, and Nef) and/or 2 precursor proteins (Gag and Env).
FIG 4.
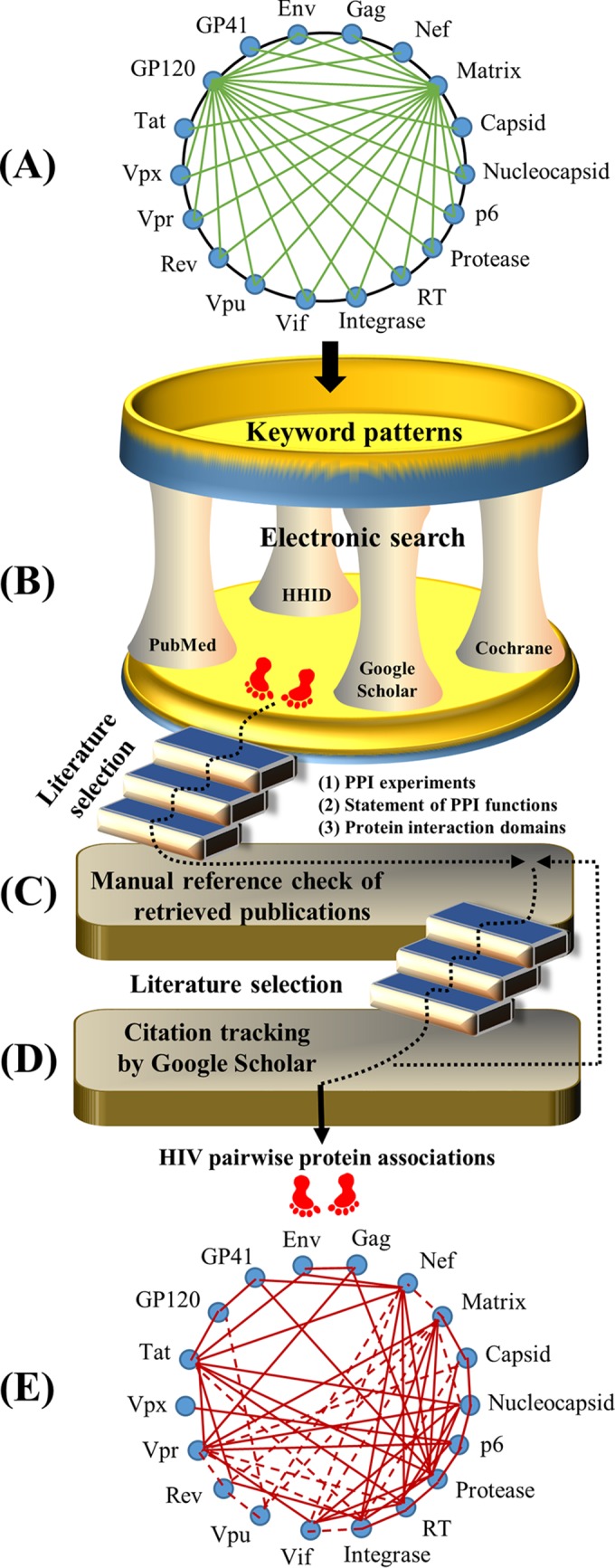
Work flow for our literature selection. (A) HIV keyword patterns. Sixteen HIV proteins and two precursor proteins (Gag and Env) are shown surrounding a circle. Pairwise HIV proteins are annotated to exhibit keyword patterns (e.g., matrix-capsid, matrix-nucleocapsid, and matrix-p6), exemplified by green links bridging one protein to the other proteins. (B) Electronic search. Given the 18 HIV (precursor) proteins, there are 153 pairwise keyword patterns (i.e., combinations of two proteins). Each pairwise keyword pattern was queried by using four sources (PubMed, Google Scholar, the Cochrane Library, and the HIV-1 Human Interaction Database [HHID]). Publications were thereafter retrieved based on three selection criteria: PPI experiments, statement of PPI functions, and protein interaction domains (see Literature Selection). (C) Reference search. We manually checked the reference lists of articles retrieved as described above for panel B. These articles were also examined by the three selection criteria described above. (D) Citation search. Google Scholar was used to retrieve publications that cited the articles retrieved as described above for panel C. If articles supporting HIV pairwise protein associations were newly identified, we returned back to the step described above for panel C to manually check their reference lists. The searching procedure was terminated when new publications could not be identified. (E) Summary of HIV protein associations. Literature information was summarized, for instance, by networks of HIV pairwise protein associations.
Step 1 was an electronic search. We searched English articles through four sources (PubMed, Google Scholar, the Cochrane Library, and the HHID), given the publication period from January 1985 until December 2015. Search terms covered all pairwise associations between 18 HIV (precursor) proteins, resulting in 153 keyword patterns (e.g., “HIV matrix capsid,” “HIV matrix nucleocapsid,” and “HIV matrix p6”). Article titles and abstracts from these databases were scrutinized, except for Google Scholar, by which we examined only the top 100 publications for each keyword pattern due to a great mass of results found. We also queried review articles about the functions and interactions of individual HIV proteins. Thereafter, we gathered publications that met three selection criteria (see below).
Step 2 included manual reference checks of extracted publications. To search the literature on HIV pairwise protein associations, we manually checked the reference list of each publication extracted by using step 1. Publications that met the selection criteria (see below) were selected.
Step 3 included citation tracking in Google Scholar. Using Google Scholar, we manually checked publications that cited those articles retrieved by using step 2. Thereafter, newly identified publications were collected for the next search round through step 2. The search process was terminated if new publications could not be found.
Articles were selected for our review if they met any of the following selection criteria:
Protein-protein interaction (PPI) experiments. We retrieved English articles that demonstrated HIV PPIs or their biological functions using in vitro or in vivo experiments (e.g., coimmunoprecipitation assays, glutathione S-transferase [GST] pulldown assays, two-hybrid assays, enzyme-linked immunosorbent assays [ELISAs], Western blot assays, dot blot assays, electron microscopy analysis, X-ray crystallography, nuclear magnetic resonance [NMR] spectroscopy, and surface plasmon resonance analysis). Articles that reported the absence of a physical interaction between two HIV proteins were also selected. However, we discarded prediction-based studies that only hypothesized PPIs without any experimental proof of physical interactions.
Statement of PPI functions. We retrieved articles that clearly expressed the functional relationship between two HIV proteins with terms such as “interact,” “bind,” “associate,” “packaging,” “incorporate,” “inhibit,” “activate,” “promote,” “cleave,” “enhance,” “degradation,” “upregulate,” and/or “downregulate.”
Protein interaction domains. We retrieved articles that reported interaction domains of HIV pairwise protein interactions. Amino acid positions were indexed by using HIV reference strains (HIV-1 reference strain HXB2 and HIV-2 reference strain BEN) (44).
As for HIV-host protein interactions, we also performed a similar literature search. If different studies suggested incomparable results on HIV protein interactions, they were presented for discussion. Different results for interaction positions are listed separately based on the original publications (Table 1). Biological experiments used for the identification of HIV protein interactions are summarized in Table 2. HIV functional associations mediated by a host protein or a viral factor are summarized in Table 3.
TABLE 1.
Summary of HIV pairwise protein interactionsa
| Protein 1-protein 2 | Life stage(s) | Major function(s) | Positions in protein 1 (reference[s]) | Positions in protein 2 (reference[s]) | Reference(s) |
|---|---|---|---|---|---|
| GP120-GP41 | Entry, budding | Promotes viral entry, promotes viral budding, promotes Env packaging | 31–46, 50–54, 70–75, 84–89, 91, 103, 106, 107, 110, 111, 114, 215, 220–224, 226, 244, 246, 489–505 (50) | 520–528, 530, 533, 534, 536, 537, 540, 541, 543–546, 569–572, 574, 575, 577–579, 581, 582, 585, 586, 588–592, 593, 596–598, 601–610, 614, 617–619, 622, 623, 628, 629, 631–633, 635, 636, 639, 642, 643, 646, 650, 651, 654, 658, 659, 661–663 (50) | 48–58 |
| GP41Env-matrix | Entry, budding | Promotes viral entry, promotes Env packaging | 712, 713–856 (70, 84); 764–856 (591); 814–844 (73) | 12, 30, 34 (76); 18, 20, 22, 32, 33 (83, 592); 6, 29, 31, 62 (74); 49 (84); 62 (72); 84, 99 (75) | 69–76, 83, 84, 591, 592 |
| GP120-Tat | Entry | Promotes viral entry | 166–171 (89); 157–171 (86) | 73–86 (86) | 86, 89 |
| RT-integrase | Reverse transcription, integration | Enhances RT activity, inhibits integrase activity | 1–242, 387–421 (108) | 201–288 (108); 243, 250, 258, 220–270 (102); 213–288 (106) | 102, 106, 108 |
| RT-nucleocapsid | Reverse transcription | Enhances RT activity | 548–560, p51 domain (113) | 13–30, 34–51 (110, 111) | 110, 111, 113 |
| RT-Vif | Reverse transcription | Enhances RT activity | ? | 161–164 | 131 |
| RT-Tat | Reverse transcription | Enhances RT activity | ? | 49–57 | 139 |
| RT-Nef | Reverse transcription | Enhances RT activity | p51 domain | 154–172 | 148 |
| Protease-Tat | Reverse transcription | Protease cleaves Tat to enhance reverse transcription | Protease catalytic site | 49–57 | 152 |
| Integrase-Rev | Integration | Inhibits integrase activity | 66–80, 118–128 (173) | 12–23, 53–67 (173) | 173–175, 180 |
| Integrase-matrix | Integration | Enhances nuclear import of the PIC | 50–212 | 132 | 181 |
| Matrix-Vpr | Integration | Enhances nuclear import of the PIC | 88–132 | ? | 183 |
| Integrase-Nef | Integration | ? | ? | 58–206 | 149 |
| Tat-Vpr | Transcription | Enhances viral transcription | 50–67 | ? | 220 |
| Tat-Rev | Transcription | Proteasomal degradation of Tat | ? | 35–50 | 216 |
| Tat-Nef | Transcription | Enhanced viral transcription | ? | ? | 223 |
| Tat-nucleocapsid | Transcription | NC induces Tat degradation | ? | ? | 228 |
| Vif-Vpr | Transcription | Vif mediates Vpr degradation | ? | ? | 232 |
| NCGag-Vif | Budding | Vif packaging, viral core stability, inhibits PR cleavage | 44–55 (301) | 75–114 (300) | 300, 301, 303 |
| NCGag-Vpr | Budding | Vpr packaging | 13–30, 34–51 | 70–80 | 313 |
| p6Gag-Vpr | Budding | Vpr packaging | 15–18 (329); 32–46 (325); 34–36 (326); 35–47 (330); 41–46 (322) | 18–34 (322); 1–71 (327) | 322, 325–327, 329, 330 |
| p6Gag-Vpx | Budding | Vpx packaging | 15–40 (333) | 73–89 (333) | 314, 332, 333 |
| GP41Env-Nef | Budding | Env packaging | 712–715 | 181–210 | 336 |
| Gag-RT | Budding | RT packaging | 183–305 (347) | ? | 347, 348 |
| Protease-Gag/GagPol | Maturation | Gag and GagPol cleavage | Protease catalytic site | Gag/GagPol cleavage sites | 495–497 |
| Protease-Vif | Maturation | Inhibits protease activity | 1–9 (516, 517) | 30–65 (519); 41–65 (520); 78–98 (516); 81–88, 88–98 (521) | 516, 517, 519–521 |
| Protease-RT | Maturation | Protease cleaves RTGagPol, RT promotes protease activity, protease inhibits RT activity | Protease catalytic site | 440 | 441 | 522, 593 |
| Protease-Nef | Maturation | Protease cleaves Nef, Nef inhibits protease activity | Protease catalytic site | 57 | 58 | 528 |
| Protease-GP41CT | Maturation | Protease cleaves GP41CT | Protease catalytic site | 714 | 715, 716 | 717 | 538 |
A question mark indicates that the corresponding information is not available. A vertical line indicates a protease cleavage site (e.g., 57 | 58 suggests that HIV protease cleaves the substrate protein between amino acid positions 57 and 58). Position indices of GP120 and GP41 are based on the Env protein sequence in the HIV-1 HXB2 reference strain. Except for HIV-2 Vpx, interaction domains are reported for HIV-1.
TABLE 2.
Summary of experiments that confirm HIV pairwise protein interactionsa
| Protein interaction | Confirmation of protein interaction by: |
Reference(s) | |||||
|---|---|---|---|---|---|---|---|
| IP | Pulldown | Two-hybrid assay | Western/dot blotting | X-ray/EM/NMR/SPR | Others | ||
| GP120-GP41 | Yes | Yes | 48–58 | ||||
| GP41Env-matrix | Yes | Yes | Yes | 69–76 | |||
| GP120-Tat | Yes | Yes | Yes | 86–89 | |||
| RT-integrase | Yes | Yes | Yes | Yes | 101–108 | ||
| RT-nucleocapsid | Yes | Yes | Yes | 110–113 | |||
| RT-Vif | Yes | 131 | |||||
| RT-Tat | Yes | Yes | Yes | 139 | |||
| RT-Nef | Yes | Yes | Yes | 148, 149 | |||
| Integrase-Rev | Yes | Yes | 173–175 | ||||
| Integrase-matrix | Yes | 181 | |||||
| Matrix-Vpr | Yes | Yes | 183 | ||||
| Integrase-Nef | Yes | Yes | Yes | 149 | |||
| Tat-Vpr | Yes | Yes | 220 | ||||
| Tat-Rev | Yes | Yes | Yes | 216 | |||
| Tat-Nef | Yes | Yes | Yes | 223 | |||
| Tat-nucleocapsid | Yes | Yes | Yes | Yes | 228 | ||
| Vif-Vpr | Yes | 232 | |||||
| NCGag-Vif | Yes | Yes | Yes | Yes | 300–303 | ||
| NCGag-Vpr | Yes | Yes | Yes | 312–314 | |||
| p6Gag-Vpr | Yes | Yes | Yes | Yes | 314, 315, 320–330 | ||
| p6Gag-Vpx | Yes | Yes | Yes | Yes | 314, 320, 332, 333 | ||
| GP41Env-Nef | Yes | 336 | |||||
| Gag-RT | Yes | Yes | 347, 348 | ||||
| Protease-Gag/GagPol | Yes | Yes | 495–497 | ||||
| Protease-Vif | Yes | Yes | 282, 516, 517 | ||||
| Protease-RT | Yes | Yes | 522 | ||||
| Protease-Tat | Yes | 152 | |||||
| Protease-Nef | Yes | 528–532 | |||||
| Protease-GP41CT | Yes | 538, 539 | |||||
IP, co- or radioimmunoprecipitation assay; pulldown, GST pulldown assay; two-hybrid assay, yeast or mammalian two-hybrid assay; Western/dot blotting, (far-)Western blot or dot blot assay; X-ray/EM/NMR/SPR, X-ray crystallography, electron microscopy analysis, NMR spectroscopy analysis, or surface plasmon resonance analysis; others, other cell culture or cell-free experiments (e.g., mass spectrometry) used for the identification of HIV pairwise protein interactions.
TABLE 3.
Summary of HIV pairwise protein associations
| Protein associationa | Life stage | Major function(s) | References |
|---|---|---|---|
| RT-tRNALys3-Vpr | Reverse transcription | Vpr interacts with tRNALys3 to inhibit initiation of reverse transcription | 155–159 |
| Integrase-dsDNA-Vpr | Integration | Vpr promotes binding of integrase to dsDNA | 24, 187, 189 |
| Integrase-dsDNA-nucleocapsid | Integration | Nucleocapsid stabilizes integrase binding to DNA and promotes the integrase strand transfer reaction | 191–194 |
| Integrase-TNPO3/CypA-Capsid | Integration | Integrase and capsid interact with TNPO3/CypA to facilitate PIC nuclear import and viral integration | 195–197, 203, 204 |
| Rev-CRM1-matrixGag | Translation | Rev and matrixGag recruit CRM1 and cellular cofactors for nuclear export of viral mRNA | 239–242 |
| Rev-CG1-Vpr | Translation | Rev and Vpr bind to CG1 for mRNA nuclear export | 246–248 |
| Tat-p300/SWI/SNF-integrase | Transcription | p300/SWI/SNF promotes Tat-mediated viral transcription and integrase-mediated viral integration | 251–256, 266–272 |
| MatrixGag-RNA-NCGag | Budding | Viral genomic RNA binds to matrixGag and NCGag for viral RNA incorporation | 350–360 |
| CapsidGag-LysRS-Vpr | Budding | LysRS binds to capsidGag for LysRS packaging, but Vpr inhibits the enzymatic activity of LysRS | 159, 367–369, 372, 373 |
| Gag-AIP1-Nef | Budding | Gag and Nef recruit AIP1 to promote viral budding | 375–378 |
| NCGag-Tsg101/AIP1-p6Gag | Budding | NCGag and p6Gag recruit Tsg101 and AIP1 to promote viral budding | 378, 385–390 |
| Vif-A3G-integrase | Budding | A3G binds to integrase for prohibiting proviral DNA formation, but Vif induces A3G degradation | 33–43, 395 |
| Vif-MAPK/HCK-Nef | Budding | Nef activates the HCK pathway to downregulate cell surface receptors, but Vif counteracts HCK-mediated inhibition of viral release; MAPK phosphorylates Vif, but Nef inhibits the kinase activity of MAPK | 407, 408, 415–421 |
| Vpu-CD4-GP120Env | Budding | CD4 prevents GP120 transport for viral budding, but HIV-1 Vpu induces CD4 degradation | 14, 15, 427–432, 435, 436, 450 |
| Vpu-tetherin/CD4-Nef | Budding | HIV-1 Vpu and Nef antagonize tetherin and CD4 to promote viral budding | 456–463, 465–468, 475 |
| Vpu-CK2-Rev | Budding | Rev promotes CK2 activity, which phosphorylates HIV-1 Vpu for CD4 degradation | 453, 482–484 |
| Vpu-UBP-matrixGag | Budding | UBP mediates a functional association between HIV-1 Vpu and matrixGag | 486–490 |
Protein association indicates that two HIV proteins either independently or dependently interact with a third molecule (e.g., CD4 or dsDNA). Note that two HIV proteins in a protein association are not necessary to construct a structural complex or to undertake biological activities at the same time during the viral life cycle. A3G, APOBEC3G.
VIRAL ENTRY
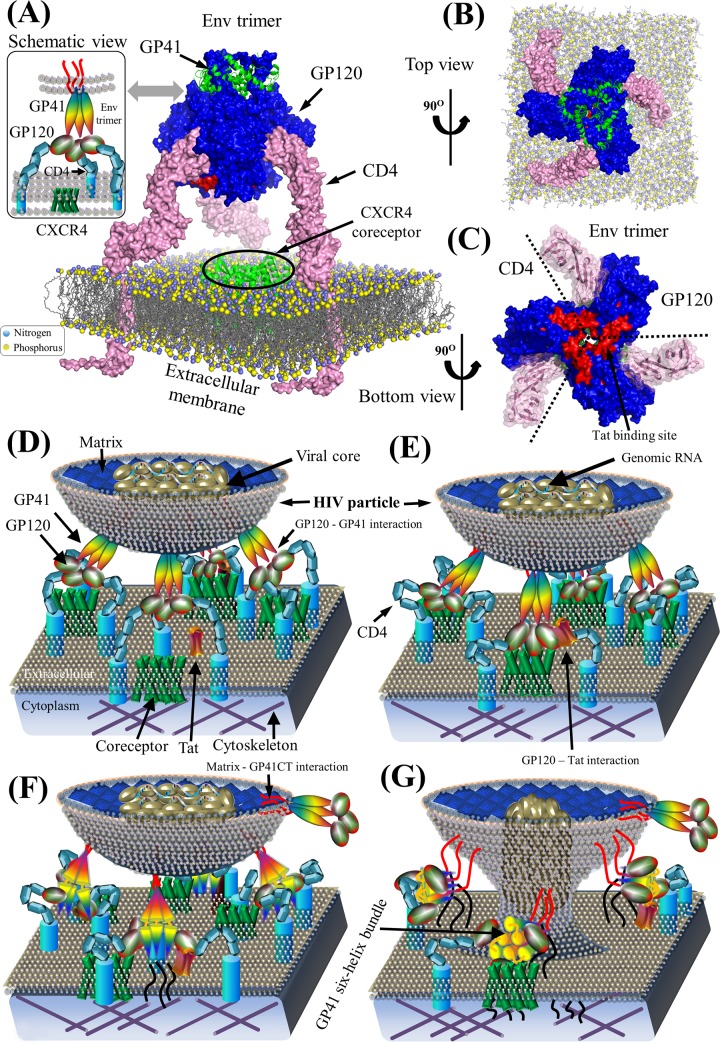
During viral entry, HIV particles penetrate host cells and initiate cell infection (Fig. 3). Host cells (e.g., T-helper cells, monocytes, macrophages, and dendritic cells), which express the CD4 (cluster of differentiation 4) glycoprotein on the cell surface, are the primary targets of HIV Env spikes, structural complexes formed by HIV GP120 and GP41 (Fig. 5). HIV entry pathways, entry inhibitors, and HIV-associated human proteins have been reviewed elsewhere (20, 21, 45–47). Here, we focus on HIV pairwise protein interactions during viral entry.
FIG 5.
Env structure complex and schematic model of HIV-1 pairwise protein interactions during viral entry. (A) Structural model of a prefusion HIV-1 Env spike associated with CD4 on the extracellular membrane. Surface representations of GP120, GP41, and CD4 proteins are shown in blue, green, and pink, respectively. Lipid bilayers of the extracellular membrane (606) are shown at the bottom, where nitrogen and phosphorus are indicated by blue and yellow spheres, respectively. The crystallized structure of the CXCR4 coreceptor in green is placed in the center across the extracellular membrane. Red areas on the GP120 surface illustrate the Tat-binding site (86, 89). Table S1 in the supplemental material provides a list of PDB accession numbers used for our structural visualization. PyMOL V1.7 visualization software was used (see http://www.pymol.org/). (B) Top view of a prefusion HIV-1 Env spike in complex with CD4 on the extracellular membrane. (C) Bottom view of the prefusion HIV-1 Env spike in complex with CD4. GP120 subunits within the trimeric Env spike bind to CD4. Red areas indicate Tat-binding sites. (E) Schematic view of the binding of Env to CD4 and coreceptors for viral attachment to the host membrane. GP120 on the mature virion surface interacts with CD4 to induce the aggregation of CD4 and chemokine coreceptors (e.g., CCR5 and CXCR4) (607, 608). Thereafter, GP120 binds to chemokine coreceptors on the host membrane. (F) Construction of GP41 six-helix bundles. Interactions between GP120 and chemokine coreceptors induce conformation rearrangements in Env spikes, which expose GP41 to construct the six-helix bundles (20). (G) Viral entry. GP41 six-helix bundles pull the extracellular membrane to create a fusion pore, which might consist of 1 to 7 Env spikes depending on divergent HIV strains (61, 62). The viral core in the HIV particle is then injected into the host cytoplasm by entering the newly created fusion pore. Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale.
GP120-GP41 Interaction
On the surface of HIV particles, GP120 physically interacts with GP41 to construct trimeric Env spikes via noncovalent interactions (48–58). During viral entry, HIV Env spikes undergo dynamic structural rearrangements to invade host cells (51, 52, 59) (Fig. 5). When GP120 binds to the cellular receptor CD4 on the host cell, this binding induces an outward domain shift of GP120 subunits to disrupt noncovalent interactions between GP120 and GP41 and to expose coreceptor-binding sites (60). Thereafter, GP41 helices at the core of Env spikes serve as anchors by which the rest of Env can be reorganized into open structural conformations for viral entry (48). Specifically, prefusion GP41 wraps its hydrophobic core around the extended N-terminal domain (NTD) and the C-terminal domain (CTD) of GP120 to construct a GP41-tryptophan clasp (50, 56). In comparisons of the prefusion and postfusion conformations of GP41 (Fig. 2), it has been shown that the spike rearrangements open the GP41-tryptophan clasp to expel GP120 termini, thereby constructing a fusion pore for viral entry (50, 61). Although it remains debated (22), the construction of a fusion pore may require 1 to 7 Env spikes for entry stoichiometry, with most HIV strains depending on 2 to 3 Env spikes (62).
Extensive studies have elaborated interaction domains of the GP120-GP41 interaction (48–58). It is generally agreed that the inner domain and the N- and C-terminal domains of GP120 maintain noncovalent interactions with the heptad repeat 1 and disulfide-bonded domains of GP41 (Fig. 2). Particularly, the GP120 inner domain can modulate the GP120-GP41 interaction and CD4 binding (51), while GP120 terminal regions mainly interact with the disulfide-bonded region of GP41 (55, 63–65). Amino acid substitutions (e.g., W596A and W610A) within the GP41 disulfide-bonded region disrupt the GP120-GP41 interaction (66). In addition, broadly neutralizing antibodies (e.g., 3BC315) have been identified to interrupt the GP120-GP41 interaction, but the dynamic nature of the Env trimers may influence the exposure of antibody epitopes (67, 68). Table 1 summarizes findings from a recent X-ray crystallographic study that unveils the interaction positions of GP120 and GP41 in the atomic structure of HIV-1 Env (50). Last but not least, the GP120-GP41 interaction exerts an impact on drug resistance to HIV entry inhibitors, a novel mechanism of HIV drug resistance which is described in detail below.
GP41Env-MatrixGag Interaction
GP41Env has been detected to physically interact with HIV-1 matrix in Gag precursors (matrixGag) (69–76) (Table 2). The cytoplasmic tail of GP41 (GP41CT) not only enhances Env packaging during viral budding (77, 78) but also drives the rearrangements of Env prebundle structures during viral entry (79). The GP41Env-matrixGag interaction undertakes multiple activities. (i) HIV-1 entry is suppressed by the interaction between the GP41CT and unprocessed Gag in immature HIV-1 particles (80). However, this suppression is dismissed when HIV-1 protease cleaves Gag and GagPol precursors, a maturation process that transforms immature HIV-1 particles into mature HIV-1 particles (80). (ii) Differential localization of Env trimers on the viral surface depends on the GP41Env-matrixGag interaction, because the proteolysis of Gag rearranges the inner protein lattice to alter the clustering of Env for viral entry (81). (iii) MatrixGag prevents access of the GP41CT to biotinylation (82).
Regarding the interaction domains, the basic and C-terminal domains of HIV-1 matrixGag physically interact with the GP41CT (72, 75, 83). Mutagenesis analyses suggest that the matrix substitution L49D destabilizes the GP120-GP41 interaction, but this impairment can be rescued by a Y710S substitution at the GP41CT (84). The last 13 to 43 amino acid positions in the GP41CT are critical for the GP41Env-matrixGag interaction (73). In addition, GP41CT mutations may confer resistance to HIV protease inhibitors (PIs) (85), a mechanism which is described in detail below.
GP120-Tat Interaction
HIV-1 Tat can physically interact with GP120, an interaction detected by isothermal titration calorimetry, pulldown assays, ELISAs, electron cryomicroscopy, and surface plasmon resonance analyses (86–89). Although Tat is dispensable for viral entry, the binding of Tat to GP120 contributes to efficient viral entry (86), and Tat-mediated viral entry promotes the infection of monocyte-derived dendritic cells (88). After HIV-infected cells release Tat to the extracellular space (86, 90), the GP120-Tat interaction undertakes multiple activities. (i) Extracellular Tat binds to Env spikes, a process which blocks the recognition of anti-Env antibodies, allowing HIV to escape from Env neutralization (88). Furthermore, extracellular Tat interacts with chemokine receptors (e.g., CCR2 and CCR3) to recruit chemokine receptor-expressing monocytes and macrophages toward HIV-infected cells (91). (ii) The released Tat physically interacts with GP120 on the extracellular membrane of uninfected cells (86). By doing so, Tat induces the aggregation of Env trimers to adapt conformational changes for viral entry (92). (iii) The GP120-Tat interaction might affect the switch of viral coreceptor tropism, because after its interaction with Tat, GP120 of an X4-tropic virus efficiently interacts with CXCR4 and CCR5 (92). Although the GP120-Tat interaction has an impact on viral entry, it does not affect Tat-mediated transactivation (86).
Regarding the GP120-Tat interaction domains, molecular docking analyses suggest that the CD4-binding site and the V3 loop of GP120 may interact with the cysteine-rich domain of Tat (87, 88). Other studies have also proposed the binding of the V1/V2 loop of GP120 to the second exon of Tat (86, 89) (Fig. 2). Further analyses are still needed to examine whether this disagreement is due to dynamic protein interactions on the extracellular membrane or due to different experimental settings.
REVERSE TRANSCRIPTION
During HIV reverse transcription, RT produces a double-stranded DNA (dsDNA) genome from a single-stranded RNA genome (23) (Fig. 3). After viral entry, a series of events take place in the viral core for the establishment of the reverse transcriptase complex (RTC) (Fig. 6). Although its exact composition is still a topic of debate, the HIV-1 RTC may consist of RT, protease, integrase, matrix, capsid, nucleocapsid, Vif, Tat, Nef, Vpr, and host proteins (93–95). Notably, HIV capsid and a small subset of phosphorylated matrix are weakly associated with viral DNA (95, 96). During reverse transcription, the RTC produces viral dsDNA with a high content of uracil that protects viral dsDNA from viral autointegration (97). HIV autointegration is a suicidal process in which viral dsDNA is integrated within itself by viral integrase (98).
FIG 6.
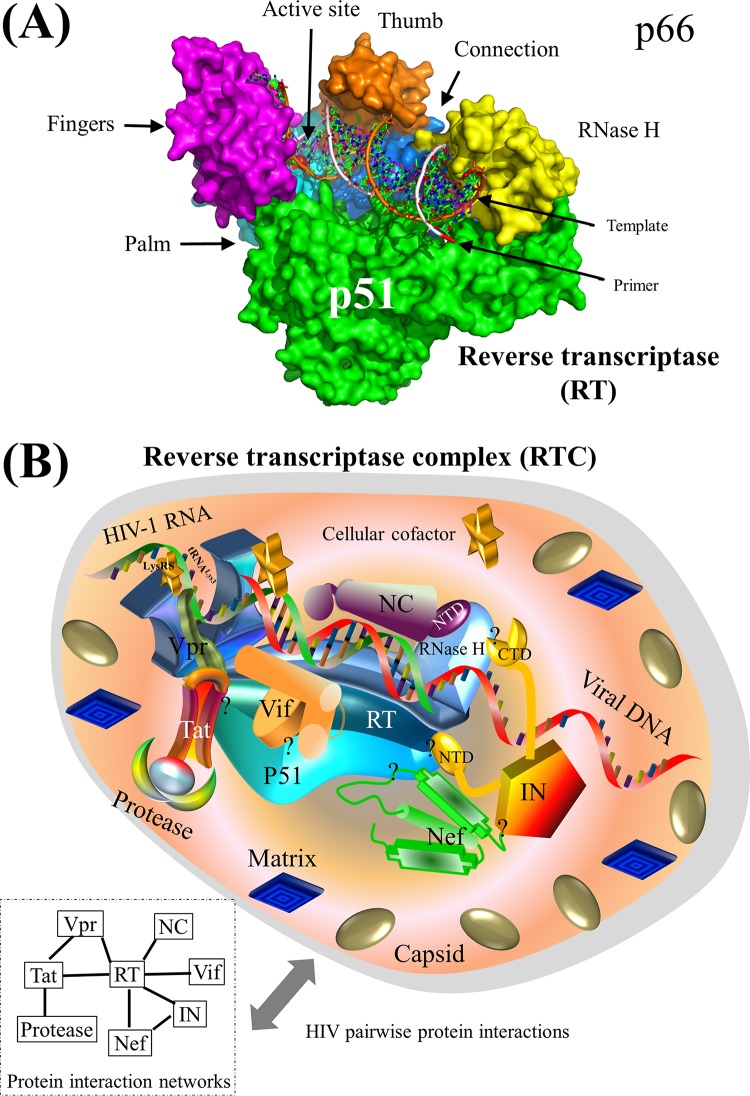
Surface representation of HIV-1 reverse transcriptase and schematic model of the HIV-1 RTC. (A) Surface representation of HIV-1 RT (PDB accession number 3KLG). Two major subunits, p51 and p66, are annotated. The p66 subunit consists of fingers, thumb, palm, connection, and RNase H domains (609). HIV-1 RNA/DNA is shown in the middle, and the active site of RT is mapped to the 3′ end of the DNA located between the fingers and thumb domains of HIV-1 RT. See Movie S1 in the supplemental material for a structural movie of HIV-1 RT. (B) Schematic model of HIV-1 pairwise protein interactions in the RTC. The HIV-1 RTC consists of RT, protease, integrase, matrix, capsid, nucleocapsid, Vif, Tat, Nef, Vpr, and many cellular proteins, although the exact composition of the RTC remains debated (93–95). HIV-1 capsid and a subset of phosphorylated matrix are weakly associated with the RTC (95, 96). Seven HIV-1 pairwise protein associations, including the RT-integrase, RT-nucleocapsid, RT-Vif, RT-Tat, RT-Nef, and protease-Tat interactions as well as the RT-tRNALys3-Vpr association, are mapped (Tables 1 and 3). Cellular cofactors (e.g., eEF1A), marked by yellow stars, may interact with the RTC to facilitate HIV-1 reverse transcription (610). Localization of the RTC in the cytoplasm is mediated by the interaction between HIV-1 matrix in the RTC and the actin cytoskeleton (611), although only a small subset of matrix is present in the RTC (95). A protein-protein interaction network is shown at the bottom left to demonstrate the physical protein interactions. Question marks indicate unclear interaction domains. Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale.
As of today, it remains a topic of debate as to where and when HIV reverse transcription occurs. Recent evidence favors the hypothesis that reverse transcription takes place in the intact capsid core (96) and is triggered by the presence of massive amounts of deoxyribonucleotides in the cytoplasm (99). Thereafter, the intact capsid core moves toward the nuclear pore, during which the RTC is reconstructed into the preintegration complex (PIC) (Fig. 6). Different aspects of HIV reverse transcription have been reviewed elsewhere, for instance, enzymatic activities of HIV-1 RT (23), the maturation of the RTC (93), strand transfer reactions, and recombinant events (100). Here, we focus on HIV-1 pairwise protein interactions and associations that take place during reverse transcription.
RT-Integrase Interaction
HIV-1 RT has been identified to physically interact with viral integrase by using GST pulldown assays, coimmunoprecipitation assays, dot blot assays, NMR spectroscopy analyses, and surface plasmon resonance analyses (101–108). The binding of integrase to RT does not require multimeric integrase or an integrase with complete enzymatic activity (108). Owing to the integrase-RT interaction, HIV-1 integrase plays an important role in the initiation of reverse transcription (104). Although viral integrase exerts no influence on steps at or before template-primer annealing, it acts at the early stages of reverse transcription by stimulating the initiation and elongation of viral DNA synthesis (109). Of interest, the RT-integrase interaction exerts an impact on drug resistance to HIV RT inhibitors (RTIs) and integrase inhibitors (INIs), a novel drug resistance mechanism that is described below.
Regarding the interaction domains, the C-terminal domain of integrase may interact with RT (102, 104, 106, 107). Mutagenesis analyses also suggest that integrase mutations at the catalytic core domain (e.g., C130S) and the C-terminal domain (e.g., W243E, V250E, and K258A) could severely diminish the RT-integrase interaction, thereby impairing reverse transcription (102, 106). Moreover, the finger-palm domain (positions 1 to 242) and the C terminus of the connection subdomain (positions 387 to 421) in RT may interact with integrase (108). However, the exact interaction positions remain unclear.
RT-Nucleocapsid Interaction
HIV-1 RT physically interacts with nucleocapsid according to far-Western blot, chemical cross-linking, and coimmunoprecipitation assays (110–113). The binding of HIV-1 nucleocapsid to RT contributes to the increased production of long proviral DNA transcripts (114, 115). At the early stage of reverse transcription, HIV-1 nucleocapsid interacts with RT to facilitate the annealing of primer tRNALys3 onto viral genomic RNA (110, 116). At the final stage of reverse transcription, a 99-nucleotide DNA flap is established in the center of the proviral DNA genome to mediate the nuclear import of the HIV-1 genome (117, 118). The construction of this central flap requires nucleocapsid chaperone activity, RT-mediated DNA synthesis, and the critical interaction between nucleocapsid and RT (117). Multiple activities of nucleocapsid take place during reverse transcription. (i) The nucleic acid-binding and chaperoning properties of nucleocapsid stabilize the RT-DNA complex to promote reverse transcription (119–121). The chaperoning activity of nucleocapsid also protects HIV-1 RNA from degradation induced by the RNase H domain of RT (122). Moreover, nucleocapsid improves the stability of RT-substrate complexes by reducing dissociation rate constants (120). (ii) Nucleocapsid not only promotes the RT strand transfer reaction (112, 122–124) but also increases RT processivity and primer extension at specific DNA template sites (125). The binding of nucleocapsid to RT counteracts the decreased strand transfer efficiency of RT mutants (113). (iii) At the early stage of HIV-1 reverse transcription, nucleocapsid can destabilize the stem-loop structure of the primer-binding site that governs the initiation-to-elongation transition and causes the major pauses during primer extension (126). Moreover, nucleocapsid inhibits primer extension prior to the formation of the RT-primer/template-deoxynucleoside triphosphate (dNTP) structural complex (125). In line with this evidence, mutations at the zinc finger domains of nucleocapsid cause premature reverse transcription (127). (iv) The excision repair activity of RT, a mechanism by which RT corrects mismatches at the cDNA polymerization site, is stimulated by nucleocapsid (119). The nucleocapsid-mediated annealing of the primer template promotes RT activity by reducing the rate of incorrect nucleotide incorporation (128).
Regarding the interaction domains, two zinc finger domains in nucleocapsid may interact with RT (110), and they are crucial for the efficient unfolding of highly structured RNA and DNA intermediates during the RT strand transfer reactions (124). HIV-1 nucleocapsid improves the RNase activity of the RNase H domain in HIV-1 RT (129), while RT regulates the nucleocapsid architecture to coordinate HIV-1 preintegration processes (130). The exact interaction domains in RT are yet to be discovered by future studies.
RT-Vif Interaction
As a component of the RTC (94), Vif has been detected to interact with RT by using GST pulldown assays (131). During the early stage of reverse transcription, the RT-Vif interaction stimulates primer annealing and increases the polymerization rate (132, 133). Multiple activities of Vif take place during reverse transcription. (i) Vif can modulate nucleic acid components in the viral genomic RNA and tRNALys3 to promote efficient reverse transcription (134), although this process happens mainly as an early event after viral entry (135). (ii) Vif not only stimulates the formation of loose HIV-1 genomic RNA dimers but also collaborates with nucleocapsid to enhance single-stranded DNA (ssDNA) synthesis (133). At an early stage of reverse transcription, Vif inhibits the hybridization of tRNALys3 and prevents the nucleotide-mediated formation of RNA dimers (133). (iii) The stable accumulation of HIV-1 reverse transcripts is mediated by Vif (136). Vif-defective mutants cause impaired DNA synthesis as well as reduced RT activity in nonpermissive cells (137, 138). However, Vif neither exerts an impact on genomic RNA dimerization nor affects the stability of the RNA dimer linkage (135).
Regarding the interaction domains, the C-terminal domain of Vif (positions 161 to 164) physically interacts with RT to stimulate reverse transcription (131). To our knowledge, RT functional domains that interact with Vif remain unclear. Additional studies are also required to verify the reproducibility of the RT-Vif interaction.
RT-Tat Interaction
The direct interaction between RT and Tat has been detected by GST pulldown assays, coimmunoprecipitation assays, and mammalian two-hybrid assays (139). An HIV-1 Tat mutant called nullbasic, whose entire arginine-rich domain is replaced by either glycine or alanine, has also been proven to interact with RT by using coimmunoprecipitation assays, pulldown assays, and biolayer interferometry assays (140). As an antiviral protein, nullbasic reduces viral core stability to prevent HIV-1 reverse transcription (140). Although Tat is dispensable for reverse transcription, Tat in complex with RT stimulates viral DNA synthesis (139). In comparison with its activity in gene expression, Tat uses distinct mechanisms to regulate HIV-1 reverse transcription (141). First, the nucleic acid chaperone activity of Tat not only promotes the placement of tRNALys3 onto viral RNA but also suppresses nonspecific DNA polymerization (142). Second, HIV-1 Tat prevents the synthesis of deleterious DNA products and interrupts DNA polymerization during the late stages of reverse transcription (143). Third, Tat acts cooperatively with nucleocapsid to promote nucleic acid annealing for the RT strand transfer reaction (144). Overall, Tat contributes to efficient reverse transcription, as HIV-1 strains lacking Tat are defective in endogenous assays of reverse transcription (145).
The RT-Tat interaction domains in the RT partner are mapped to the p51 subunit by GST pulldown and immunoprecipitation assays and to the p51 and p66 subunits by mammalian two-hybrid assays (139). The basic region of Tat (positions 44 to 61) may promote the RNA-annealing reaction by HIV-1 RT (144, 146). Two cysteine-rich domains of Tat (positions 21 to 39 and 40 to 47) suppress DNA elongation during reverse transcription (142). By altering the positive-charge distribution, the acetylation of Tat residues K28, K50, and K51 can regulate the activity of Tat in reverse transcription and transcriptional activity (146, 147). Overall, the basic domain of Tat plays a role in efficient reverse transcription, but the exact RT domains that interact with Tat remain unclear. Additional studies are still required to verify the reproducibility of the RT-Tat interaction.
RT-Nef Interaction
HIV-1 Nef can physically interact with RT according to GST pulldown assays, coimmunoprecipitation assays, and in vitro binding assays (148, 149). In fact, Nef can stimulates proviral DNA synthesis during reverse transcription (150). Being independent of its binding to viral RNA, Nef increases the binding affinity of HIV-1 RT for viral RNA (148). In the absence of Nef, RT generates 5- to 10-fold-fewer DNA products (151). Regarding protein interaction domains, data from mutagenesis analyses suggest that the p51 unit of RT may interact with the disorder loop in the C-terminal domain of Nef (positions 154 to 172) (148). Although HIV-1 Nef may play a role during reverse transcription, future studies are still needed to verify the reproducibility of the RT-Nef interaction.
Protease-Tat Interaction
Data from cell-free and cell culture assays suggest that HIV-1 protease cleaves Tat (152). This protease-mediated cleavage requires the basic domain of Tat (positions 49 to 57) (152). The Tat motif R49KKR52 plays a critical role in modulating HIV-1 reverse transcription (152). Moreover, a Tat mutant harboring a single mutation, Y47N, near the protease cleavage site can downregulate Tat-stimulated reverse transcription, suggesting that the protease-mediated cleavage of Tat influences Tat-enhanced reverse transcription (152). Future studies are still required to verify protease-mediated cleavage on HIV-1 Tat, as it has been reported in only a single study.
RT-tRNALys3-Vpr Association
Although both RT and Vpr are colocalized in the RTC, the PIC, and the viral core (153, 154), a direct interaction between RT and Vpr has not been reported to our knowledge. Based on HIV-host protein interactions, cellular primer tRNALys3 physically interacts with RT (155–158) and Vpr (159). During reverse transcription, RT initiates minus-strand DNA synthesis from the 3′ end of primer tRNALys3 (160). To influence the initiation of reverse transcription, Vpr interacts with tRNALys3 and prohibits the LysRS-mediated aminoacylation of tRNALys3 (159). Of interest, tRNALys3 is packaged into HIV-1 virions with ∼20 molecules per virion (161).
For efficient DNA synthesis, the thumb subdomain in the p66 unit of RT may interact with the anticodon loop in tRNALys3 (155). The RT connection domain may take part in tRNALys3 annealing but not in tRNALys3 packaging (156). The V241QPI244 peptide in the cross-link between the thumb and the palm subdomains of RT (Fig. 6A) may interact with primer tRNALys3 (158). Despite the fact that interaction domains in Vpr are yet to be resolved, peptides derived from two Vpr regions (positions 57 to 71 and 61 to 75) can interact with RT to inhibit HIV-1 reverse transcription (162).
VIRAL INTEGRATION
After HIV reverse transcription, the RTC reorganizes into the PIC in the cytoplasm (Fig. 7). Although the exact composition of the PIC remains debated (163), the PIC is likely comprised of cellular cofactors, dsDNA, integrase, RT, matrix, nucleocapsid, Vpr, and a small amount of capsid (96, 164–166). During viral integration, a series of actions take place (24–26, 167, 168). The first action is 3′-end processing. In the cytoplasm, the viral integrase tetramer removes 2 nucleotides at each 3′ end of dsDNA to generate a reactive intermediate that contains a 3′-hydroxyl group (168). The second action is nuclear import. The PIC-containing dsDNA is imported from the cytoplasm to the nucleus through nucleus pore complexes (168). The third step is nuclear localization. The PIC is localized to host chromosomal domains with high transcriptional activity (24). This process is assisted by cellular cofactors such as lens epithelium-derived growth factor (LEDGF)/p75, a cellular transcriptional coactivator serving as a tethering protein between the PIC and host chromosomes (167). The fourth action is the strand transfer reaction. Viral dsDNA is inserted into host chromosomes through the integrase strand transfer reaction (168). The final action is gap repair. Unpaired regions between HIV and host dsDNA are repaired under the assistance of cellular cofactors (24).
FIG 7.
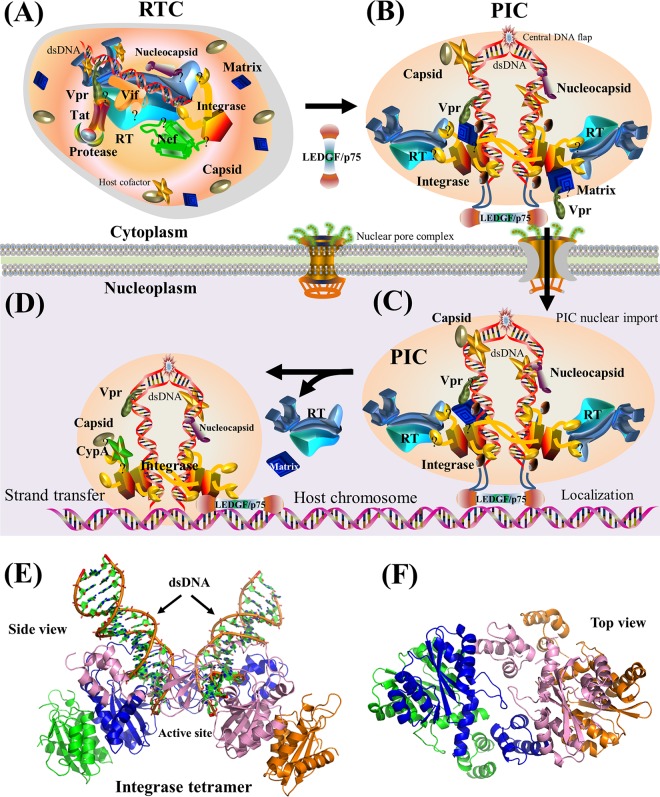
Schematic model of viral integration and cartoon representation of viral integrase. The HIV-1 PIC is comprised of cellular cofactors, viral dsDNA, integrase, RT, matrix, nucleocapsid, Vpr, and a small amount of capsid (96, 164–166), although the exact composition of the PIC remains debated (163). (A) Viral uncoating. The RTC turns into the PIC with the recruitment of host factors (e.g., LEDGF/p75). During this process, Nef, Tat, and most capsid proteins are dissociated from the PIC (169). (B) Nuclear import. The PIC is imported from the cytoplasm to the nucleoplasm. Vpr physically interacts with the nuclear pore complex for PIC nuclear import (612). The formation of the central DNA flap promotes viral uncoating at the nuclear pore (613). Although it remains debated, a small amount of capsid is associated with the PIC to enhance its nuclear import (96). At the late stages of viral integration, Rev may interact with integrase to prevent the nuclear import of the overexpressed PIC (179). (C) Chromosomal localization. Assisted by cellular proteins such as LEDGF/p75, HIV-1 dsDNA in the PIC is tethered to the host chromosome (170). (D) Integration. HIV-1 dsDNA is integrated into host chromosomes by viral integrase (168). (E) Cartoon representation of a prototype foamy virus integrase in complex with dsDNA (PDB accession number 3OY9). The active site of integrase is shown in the middle. See Movie S2 in the supplemental material for a structural movie of viral integrase. (F) Cartoon representation of the HIV-1 integrase tetramer in the absence of dsDNA (PDB accession number 1K6Y). Four subunits of the integrase tetramer are shown in green, blue, pink, and orange, respectively. For the schematic models in panels A to D, the protein shapes do not represent the exact protein structures, nor are the protein sizes to scale. Question marks indicate that interaction domains remain unclear.
The mechanisms of PIC nuclear import, preintegration transcription, and integration-associated host proteins (e.g., LEDGF/p75) have been reviewed elsewhere (24–26, 167–170). Here, we focus on physical interactions and functional associations between these HIV proteins that take place during viral integration.
Integrase-RT Interaction
HIV integrase has been determined to physically interact with RT by using GST pulldown assays, coimmunoprecipitation assays, dot blot assays, NMR spectroscopy, and surface plasmon resonance analyses (101–107). Two functions of the integrase-RT interaction have been proposed. First, RT in the PIC inhibits both the 3′-end endonuclease and the strand transfer activity of integrase (101, 103). Second, RT can inhibit the DNA disintegration activity of integrase before viral integration, although HIV-1 integrase may promote RT activity during reverse transcription (171). Note that DNA disintegration is a reverse reaction of viral integration that releases viral dsDNA and repairs the continuity of host chromosomes (172). Overall, RT can efficiently regulate the activity of integrase through the integrase-RT interaction.
Regarding the interaction domains, the CTD of integrase is necessary and sufficient for the interaction with RT (102, 106). For instance, amino acid substitutions (W243E, V250E, and K258A) at the integrase CTD severely impair the integrase-RT interaction (102). As for the interaction domains in HIV-1 RT, the finger-palm domain and the C-terminal half of the connection domain of the RT heterodimer may interact with the integrase CTD (108). In terms of different interaction domains reported during viral integration and reverse transcription, additional analyses are still needed to verify whether these differences are detected in different cell lines, HIV-1 strains, or conformation rearrangements adapted for different activities of the RTC and the PIC.
Integrase-Rev Interaction
HIV-1 Rev has been found to physically interact with the integrase dimer or tetramer by using GST pulldown assays and coimmunoprecipitation assays (173–175). It is known that integrase interacts with the cellular LEDGF/p75 protein (168, 176, 177). Experimental evidence suggests that Rev may disrupt the interaction between integrase and LEDGF/p75, a mechanism that inhibits premature viral integration before the nuclear localization of viral dsDNA (173, 178). At the postintegration stage, Rev expressed at the pretranscription processing step can prevent the nuclear import of integrase through the Rev-integrase interaction, thereby limiting the massive number of copies of viral DNA integrated into host chromosomes (175, 179). Since increased integration has been postulated to cause excessive cell death, Rev thus protects HIV-1-infected cells from premature cell death (175).
Regarding the interaction domains, two Rev domains (positions 13 to 23 and 53 to 67) may interact with the central regions of integrase (positions 118 to 128 and 66 to 80) (173). Interestingly, Rev-derived peptides (positions 13 to 23 and 53 to 67) inhibit the activity of integrase, whereas integrase-derived peptides (positions 66 to 80 and 118 to 128) rescue the Rev-mediated inhibitory effect (180).
Integrase-Matrix Interaction
HIV-1 integrase has been identified to interact with matrix by using coimmunoprecipitation assays (181). The integrase-matrix interaction promotes the nuclear import of the PIC in nondividing cells such as macrophages (181). Although viral integrase and matrix are components of the PIC, the entire matrix is dispensable for viral nuclear import (182). Regarding the interaction domains, the catalytic core domain of integrase (positions 50 to 212) may bind to matrix, while C-terminal tyrosine phosphorylation of matrix is crucial for the integrase-matrix interaction (181). Replacing tyrosine with phenylalanine at matrix position 132 can block PIC nuclear import (181). Independent analyses are still required to verify the reproducibility of the integrase-matrix interaction.
Matrix-Vpr Interaction
HIV-1 matrix has been found to interact with Vpr by using yeast two-hybrid assays and coimmunoprecipitation assays (183). As nucleophilic proteins, HIV-1 matrix and Vpr collaboratively improve the stoichiometry of nucleophilic components in the PIC and promote PIC nuclear import in nondividing cells (184). Although Vpr-mediated nuclear export is dispensable for viral packaging (185), HIV-1 Vpr promotes the nuclear import of the PIC in macrophages (186). In fact, HIV-1 Vpr in the cytoplasm is transported into the nucleus by hijacking cellular proteins such as importin alpha (186). As described previously, matrix also takes part in nuclear import due to its interaction with HIV-1 integrase (181). Regarding the interaction domains, the C-terminal domain of matrix (positions 88 to 132) may interact with Vpr (183), but the interaction domains in Vpr remain unclear. Future studies are still required to verify the reproducibility of the matrix-Vpr interaction.
Integrase-Nef Interaction
The physical interaction between integrase and Nef has consistently been detected by using yeast two-hybrid assays, coimmunoprecipitation assays, and GST pulldown assays (149). Although biological functions of this interaction remain unclear, it is speculated that Nef may take part in HIV-1 reverse transcription and integration (149). More studies are required to investigate the activities of this interaction as well as the binding domains. Additional analyses are still required to verify the reproducibility of the integrase-Nef interaction, as it has been reported in only a single study.
Integrase-dsDNA-Vpr Association
HIV-1 integrase, Vpr, and dsDNA are key components of the HIV PIC (Fig. 7). During viral integration, it is known that integrase inserts viral dsDNA into host chromosomes (24). As the most abundant viral protein in the PIC (187), HIV-1 Vpr promotes the nuclear localization of viral dsDNA during the nuclear import of the HIV-1 PIC (184, 188). To enhance PIC nuclear import, HIV-1 Vpr acts as a DNA architectural protein to bridge two or more DNA helices into synaptic and stretched nucleofilaments (187). The binding of HIV-1 Vpr to DNA also induces double-strand breaks in chromosomal DNA, which might influence viral integration (189). Moreover, full-length Vpr and its C terminus (positions 52 to 96) not only stimulate the strand transfer reaction but also enhance the binding of integrase to viral dsDNA (190). Interestingly, Vpr-derived peptides (positions 57 to 71 and 61 to 75) can inhibit the activity of HIV-1 integrase (162). Overall, Vpr may promote integrase activity via its interaction with dsDNA, although a direct interaction between integrase and Vpr remains unclear.
Integrase-dsDNA-Nucleocapsid Association
Although a direct interaction between integrase and nucleocapsid has not been reported, the integrase-dsDNA-nucleocapsid structural complex plays multiple roles during viral integration. (i) The chaperone activity of nucleocapsid on viral DNA enhances HIV-1 integration (191). Specifically, the zinc finger domains of nucleocapsid not only stabilize the binding of integrase to viral dsDNA but also complement DNA binding to promote the integrase strand transfer reaction (192). (ii) HIV-1 nucleocapsid can promote coupled integration by >1,000-fold under in vitro conditions (193). During viral DNA integration, viral integrase takes part in the coupled joining that merges two ends of the viral genome into the host genome (193). (iii) In the presence of Mg2+, a high concentration of viral integrase is required for HIV-1 integration (194). For an efficient DNA strand transfer reaction, nucleocapsid counteracts this defect by keeping a low concentration of integrase in the presence of Mg2+ (194). Overall, nucleocapsid interacts with viral dsDNA to promote integrase activities during viral integration.
Integrase-TNPO3/CypA-Capsid Association
Although a direct interaction between capsid and integrase has not been reported, transportin 3 (TNPO3, transportin-SR2, or TRN-SR2) interacts with viral integrase and capsid to facilitate the nuclear transport of the viral PIC (195–198). As a member of the importin-β family, TNPO3 recognizes serine/arginine-rich repeats within precursor mRNA splicing factors and transports these factors from the cytoplasm to the nucleus (199). Regarding the interaction domains, it has been reported that TNPO3 interacts with amino acid positions in the integrase CTD (e.g., R262 to K264, K266, and R269) (198). HIV-1 integrase mutants with R262A and K264A mutations display a weak interaction with TNPO3, showing a 2.3-fold-lower affinity than that of the wild-type integrase (200). Although it is not a major determinant of HIV-1 nuclear import, the integrase-TNPO3 interaction may take place when the PIC enters the nucleus but before viral integration (201, 202). Other studies also suggest that viral capsid, not integrase, dictates the TNPO dependency of PIC nuclear import (195, 201).
Cyclophilin A (CypA) is an important cellular peptidyl-prolyl isomerase that participates in the uncoating of viral core (203, 204). CypA physically interacts with viral capsid (204–206), while viral integrase is required to maintain the physical interaction between capsid and CypA (203). Owing to the capsid-CypA interaction, HIV-1 capsid can be either stabilized or destabilized by CypA (207, 208). On the one hand, viral capsid is stabilized by CypA when it travels to the nuclear pore in the cytoplasm (205). On the other hand, viral capsid escapes from CypA dependence through conformational dynamics (206). Accumulated evidence also implies a direct association between HIV-1 capsid and integrase, because capsid mutants (Q63A and Q67A) exert a deleterious effect on viral integration (164). Moreover, the presence of integrase mutants (e.g., C130S) induces the degradation of capsid in the cytoplasm, thus decreasing viral core stability (203).
Overall, viral capsid and integrase are associated with cellular proteins (e.g., TNPO3 and CypA) in order to facilitate PIC nuclear import and viral integration.
VIRAL TRANSCRIPTION AND TRANSLATION
Two different HIV transcription pathways have been observed before and after viral dsDNA integration. (i) A small amount of regulatory proteins (Rev, Tat, and Nef) can be synthesized from unintegrated viral DNA, a process called preintegration transcription (25, 209) (Fig. 3). These synthesized viral proteins interact with cellular machineries to regulate viral production at subsequent stages of the HIV life cycle (Fig. 8). For instance, Rev transports viral RNAs from the nucleus to the cytoplasm (210). (ii) A large number of viral mRNAs are produced by cellular microRNA (miRNA) machineries, which synthesize mRNA from viral dsDNA integrated into host chromosomes (27). Viral mRNAs are then processed (polyadenylation, methylation, capping, and splicing) for protein maturation (211). Posttranslational modifications of viral proteins (e.g., phosphorylation, methylation, and acetylation) are also essential during this process (147, 212).
FIG 8.
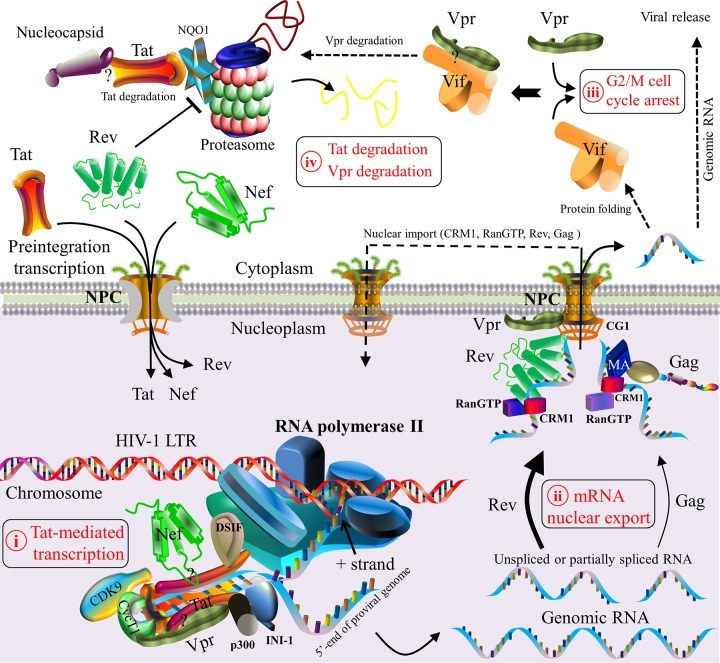
Schematic model of HIV-1 protein interactions during viral transcription and translation. Five steps are described. (i) HIV-1 Tat initiates viral transcription by its interaction with the TAR of viral RNA, which is a regulatory element located downstream of the HIV-1 LTR (212). Tat subsequently recruits the subunits of the positive transcription elongation factor (e.g., cyclin T1 [CycT1] and cyclin-dependent kinase 9 [CDK9]) to construct a transcription complex (212). This complex activates the kinase CDK9 for the hyperphosphorylation of RNA polymerase II, which interacts with Tat and other host factors (e.g., DSIF [5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor] [614]) to produce viral genomic mRNAs (615, 616). Genomic mRNAs are spliced thereafter. Although many host proteins take part in viral transcription (23), only CDK9, cyclin T1, p300, DSIF, INI1, and RNA polymerase II are shown. (ii) Nuclear export of newly synthesized viral RNAs is accomplished by either Rev- or Gag-mediated pathways (239–242). In the former case, Rev recruits cellular factors (e.g., CRM1 and RanGTP) to export viral genomic RNAs as well as incompletely spliced and unspliced mRNAs from the nucleoplasm to the cytoplasm (30, 213). The binding of Vpr to the nuclear pore complex (NPC) is predominantly localized in the nuclear envelope of the nucleus (247, 248). In the latter case, RNA nuclear export is activated by the nuclear export signal (NES) of matrixGag, which interacts with CRM1 (214, 242). The nuclear export of viral RNA allows viral protein maturation in the cytoplasm and cellular compartments. CRM1, RanGTP, Rev, and Gag are then imported back to the nucleus (213, 214, 242). (iii) HIV-1 Vif and Vpr, both of which are expressed in the cytoplasm, can independently trigger G2/M cell cycle arrest (233). Vif may also interact with Vpr to mediate the degradation of Vpr and to reduce Vpr-induced cell cycle arrest (232). (iv) At the late stage of viral transcription, the proteasomal degradation of Tat is induced by viral nucleocapsid (228). Moreover, Rev induces Tat degradation by downregulating the expression level of the cellular protein NQO1 (216). Question marks indicate unclear interaction domains. Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale.
Previous studies have reviewed mechanisms of HIV transcription and translation (27–29), Rev-mediated nuclear export (213), Gag-mediated nuclear localization (214), and interactions between HIV proteins and cellular transcription factors (28, 29, 215). Here, we focus on physical interactions and functional associations between HIV-1 proteins, which play important roles in viral transcription and translation.
Tat-Rev Interaction
The direct interaction between HIV-1 Rev and Tat has been detected by two-hybrid assays, pulldown assays, and coimmunoprecipitation assays (216). The nuclear export signal region of HIV-1 Rev (Fig. 2) takes part in the proteasomal degradation of cytoplasmic Tat at the posttranslational level, leading to a significant decrease of HIV-1 gene expression (216). The Rev-mediated downregulation of Tat might be associated with HIV-1 latency, because the decrease of the intracellular level of Tat below a critical threshold potentially marks the rise of HIV-1 latency (216). Moreover, the regulatory proteins Rev and Tat shuttle between the nucleus and the cytoplasm to interact with various cellular factors (4, 216). HIV-1 Rev and Tat expressed via viral preintegration transcription (Fig. 3) are frequently targeted by human cytotoxic T lymphocytes for the immune control of viral infections (217).
Although the Tat-Rev interaction does not induce Rev-mediated Tat degradation, Rev causes Tat degradation in the cytoplasm by downregulating the level of a host protein called NAD(p)H:quinine oxidoreductase 1 (NQO1) (216). A host protein called DEAD box RNA helicase (DDX1) has also been reported to interact with both HIV-1 Rev and Tat (218). During nuclear export, Rev interacts with DDX1 to promote Rev multimerization on the Rev response element (RRE) of viral mRNA (219). Moreover, an HIV-1 Tat mutant, called nullbasic, interacts with DDX1 to disrupt the subcellular localization of Rev, thereby decreasing the expression of Rev-dependent viral mRNA (218). To counteract this defect, wild-type Tat interacts with DDX1 to restore the Rev-mediated export of viral mRNA (218). Regarding the interaction domains, the nuclear localization signal (positions 35 to 50) of Rev is vital for the Tat-Rev interaction (216), whereas interaction domains in Tat remain unclear. Additional studies are still required to verify the reproducibility of the Tat-Rev interaction.
Tat-Vpr Interaction
Data from GST pulldown and coimmunoprecipitation assays suggest that HIV-1 Vpr physically interacts with Tat and cyclin T1 in the nucleus (220). Tat is a regulatory protein known for its interaction with positive transcription elongation factor b (pTEFb), a structural complex consisting of cyclin-dependent kinase 9 (CDK9) and cyclin T1 (221, 222). During viral transcription, a tertiary complex, Tat-Vpr-pTEFb, is constructed to promote the superactivation of the HIV-1 long terminal region (LTR), leading to increased transcriptional activity (220).
Regarding the interaction domains, Vpr may interact with the Tat domain within amino acid positions 50 to 67 (220). Moreover, a single substitution, R73S, in Vpr severely reduces the Tat-induced transcription of the HIV-1 LTR, suggesting a key role of Vpr R73 in modulating Tat activity (220). The exact interaction domains in Vpr are yet to be clarified. More studies are still needed to verify the Tat-Vpr interaction, because this interaction has been reported in only a single study.
Tat-Nef Interaction
The direct interaction between HIV-1 Tat and Nef has been identified by using coimmunoprecipitation assays, GST pulldown assays, and transient-transfection assays (223). Colocalized in the nucleus, both Tat and Nef can be expressed during preintegration transcription (Fig. 3). First, Nef induces many host factors (e.g., CDK9, Tat-SF1, and IRF2) to promote Tat-mediated transcriptional activity (224). Second, Nef-mediated signaling can enhance Tat-mediated transcriptional activity via an extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase (MAPK)-dependent pathway (225). Third, Nef promotes Tat-mediated transcription via the heterogeneous nuclear ribonucleoprotein K (hnRNP-K)-nucleated signaling complex (226). Note that hnRNP-K plays essential roles in transcriptional processes and molecular interactions (227). Overall, Nef exerts an impact on Tat-mediated transcription either by direct interaction or by signaling pathways mediated via cellular cofactors (223–226). The exact protein domains that mediate the Tat-Nef interaction remain unclear. Future studies are still required to verify the Tat-Nef interaction and its functions.
Tat-Nucleocapsid Interaction
The direct interaction between Tat and nucleocapsid has been detected by using yeast two-hybrid assays, GST pulldown assays, coimmunoprecipitation assays, and subcellular colocalization assays (228). For HIV-1 and HIV-2, both Tat and nucleocapsid are chaperone proteins that mediate the proper folding of viral RNA (229, 230). Interestingly, the proteasomal degradation of Tat is induced by viral nucleocapsid in a ubiquitin-independent manner, subsequently reducing Tat-mediated transcription at the late stage of viral transcription (228). In the absence of Tat, nucleocapsid is localized predominantly in the cytoplasm (228), even though nucleocapsid can shuttle from the cytoplasm to the nucleus (231). The exact interaction domains in Tat and nucleocapsid remain unclear. Additional studies are still required to verify the Tat-nucleocapsid interaction, because it has been reported in only a single study.
Vif-Vpr Interaction
HIV-1 Vif may interact with Vpr according to coimmunoprecipitation assays (232). HIV-1 Vpr and Vif share common activities during the viral life cycle: (i) both HIV-1 proteins independently cause T-cell cytopathicity (233), (ii) they can promote viral infections by the induction of cell cycle arrest at the G2/M phase in dividing cells (234–237), and (iii) HIV-1 Vpr and Vif downregulate the antiviral cellular factor APOBEC3G through the proteasomal degradation pathway (238). On the other hand, Vif may interact with Vpr to mediate the degradation of Vpr via the ubiquitin and proteasome pathways (232). It has been speculated that this interaction may modulate Vpr activity in order to decrease the accumulation of HIV-infected cells at the stage of G2/M cell cycle arrest (232). Experimental evidence also suggests that the elimination of both the vif and vpr genes from the HIV-1 genome, but not each gene individually, prevents cell death and G2/M cell cycle arrest of HIV-infected cells (233). To our knowledge, the exact protein domains that mediate the Vpr-Vif interaction remain unclear. More studies are still required to verify the Vif-Vpr interaction, as it has been reported in only a single study.
Rev-CRM1-MatrixGag Association
HIV-1 Rev and matrixGag physically interact with a cellular protein called chromosome region maintenance 1 (CRM1) to export viral RNA via the Rev-mediated or the Gag-mediated export pathways (239–242). As a major pathway, Rev recruits CRM1, RanGTP, and other host proteins (e.g., DDX3) to export viral mRNA from the nucleus to the cytoplasm (213, 243). The CRM1-Rev interaction has been demonstrated by pulldown assays, mammalian two-hybrid assays, gel mobility shift assays, and protein footprinting assays (239, 240). During the early stage of HIV-1 infection, the nuclear export signal of Rev binds to CRM1 and other cellular factors, leading to the dynamic trafficking of Rev between the nucleus and the cytoplasm (239, 240).
The CRM1-matrixGag interaction, identified by two-hybrid assays, paves the way for the Gag-mediated nuclear export pathway (242). MatrixGag harbors one nuclear export signal (NES) (positions 18 and 22) (242) and two nuclear localization signals (NLSs) (positions 24 to 31 and 110 to 114) (244) (Fig. 2). During the early stage, matrixGag NLSs promote the nuclear localization of the PIC in nondividing cells (244). During the late stages of viral translation, the matrixGag NES is a dominant signal that counteracts the nuclear import activity of the matrixGag NLS to keep Gag in the cytoplasm (242). Although the localization of HIV-1 Gag proteins in the nucleus has been proposed (214, 242), the nuclear trafficking ability of HIV-1 Gag remains debated (245). Overall, the binding of CRM1 to Rev and matrixGag plays a key role in the nuclear export of viral mRNA. Further investigation of Gag-mediated nuclear trafficking is still warranted.
Rev-CG1-Vpr Association
Human nucleoporin-like protein 1 (NLP-1 or CG1), which interacts with Rev and Vpr, is an important component of the nuclear pore complex (NPC) (246, 247). On the one hand, data from mammalian two-hybrid assays suggest a direct interaction between Rev and CG1 (246). The Rev-CG1 interaction, which requires the nuclear export signal of Rev (positions 75 to 83), plays a role in Rev-mediated nuclear export after viral transcription (246). On the other hand, GST pulldown assays, coimmunoprecipitation assays, and yeast two-hybrid assays demonstrate that CG1 physically interacts with Vpr (247). The Vpr-CG1 interaction enhances the docking of Vpr at the nuclear pore complex, leading to the accumulation of Vpr in the nuclear envelope (247). Although it is localized predominantly in the nuclear envelope, Vpr harboring two nuclear localization signals (positions 17 to 34 and 46 to 74) shuttles rapidly between the nuclear and cytoplasmic compartments (247–249). Regarding the interaction domains, alpha-helix regions of Vpr (positions 17 to 46 [247], L23, and K27 [248]) may interact with the N-terminal region of CG1 (positions 94 to 170 [247]). Overall, the binding of the human protein CG1 to HIV-1 Rev and Vpr plays an important role in HIV-1 nuclear export.
Tat-p300/SWI/SNF-Integrase Association
Although a direct interaction between Tat and integrase has not been reported, HIV Tat and integrase are colocalized in the nucleus (Fig. 3), permitting a possible association through nuclear proteins. The transcriptional coactivator and histone acetyltransferase p300 is known to regulate chromatin conformation and DNA transcription (250). Concrete evidence suggests that p300 physically interacts with HIV-1 Tat (251–254) and integrase (255, 256). On the one hand, the acetyltransferase p300 acetylates two lysine residues (K50 and K51) in HIV-1 Tat, resulting in an improvement of the Tat-mediated transcriptional activation of the HIV-1 promoter as well as an increased binding affinity of acetylated Tat for core histones (251, 253). In addition to K50 and K51, p300 also acetylates K28 in the activation domain of Tat to promote HIV-1 transcription (257). On the other hand, HIV-1 integrase is subject to posttranslational modifications by cellular cofactors (e.g., p300) (258, 259). During viral integration, p300 acetylates three lysine residues (K264, K266, and K273) in the C-terminal domain of viral integrase (255, 256). Although it remains debated (258), the p300-mediated acetylation of HIV-1 integrase might increase the binding affinity of integrase for nucleosomal DNA and promote the integrase strand transfer reaction (255). In addition to p300, other cellular cofactors (e.g., CREB and GCN5) are recruited by Tat to promote HIV-1 transactivation (260–263). Overall, p300 acts as an acetyltransferase to alter the activities of HIV-1 Tat and integrase.
The SWI/SNF (switch/sucrose nonfermenting) complexes are a family of ATP-dependent chromatin-remodeling complexes that utilize the energy of ATP hydrolysis to remodel the nucleosome in order to make the DNA accessible during transcription, replication, and DNA repair (264, 265). Human SWI/SNF complexes consist of multiple subunits, such as a single ATPase (either BRM or BRG1), three core subunits (BAF47 [also called INI1 or SNF5], BAF155, and BAF170), and several accessory subunits (e.g., β-actin) (264). Among these subunits, INI1 (266, 267), BRM (268), BRG1 (269), and β-actin (269) can physically interact with HIV-1 Tat, while INI1 binds to HIV-1 integrase (270–272). To activate viral transcription, Tat binds to the transactivation response (TAR) element, which is a 59-nucleotide stem-loop in viral RNA (29, 273). Subsequently, Tat recruits pTEFb (a structural complex with CDK9 and cyclin T1), SWI/SNF chromatin-remodeling complexes, and other cellular cofactors to the HIV-1 promoter (268, 274). In this process, p300 acts synergistically with INI1 and BRG1 to activate the HIV-1 promoter (269). As for the integrase-SWI/SNF interaction, the binding of the INI1 subunit of SWI/SNF to HIV-1 integrase not only promotes the efficient integration of viral DNA into stable nucleosomes (275, 276) but also enhances the packaging of INI1 (270). Overall, the meticulous associations between viral proteins (Tat and integrase) and cellular cofactors (p300 and SWI/SNF) in the nucleus play an important role in HIV-1 integration and transcription.
VIRAL ASSEMBLY AND BUDDING
As illustrated in Fig. 9, HIV genomic RNA, Vif, Nef, Vpr, Env, Gag/GagPol precursors, and host factors (e.g., TIP47 [tail-interacting protein of 47 kDa] and lipid) are assembled into nascent HIV virions, subsequently pinching off from the cell membrane (2, 30, 277). The presence of multiple HIV proteins in nascent virions thus establishes a basis for HIV pairwise protein interactions. The amounts of HIV-1 proteins per virion have been quantified in the literature.
FIG 9.
Schematic model of HIV-1 protein interactions during viral budding. Env trimers are exported to the extracellular membrane through a secretory pathway (617). Gag and GagPol are targeted to glycolipid-enriched membrane lipid rafts, where cholesterol, sphingolipids, and glycosylphosphatidylinositol-linked proteins are abundant (618). Several HIV protein interactions have been observed. The first interaction is the matrixGag-GP41Env interaction. Mediated by the cellular cofactor TIP47, matrixGag interacts with GP41Env for Env packaging into HIV particles (297). Another is the Vpr-Gag interaction. Vpr is incorporated into nascent virions through the binding of Vpr to NCGag and p6Gag (315, 316, 324). A third interaction is the Vpx-p6Gag interaction. HIV-2 Vpx binds to p6Gag for Vpx packaging (333). A fourth interaction is the NCGag-Vif interaction. NCGag interacts with Vif for the packaging of Vif (300–303). A fifth interaction is the Env-Nef interaction. Nef enhances the packaging of Env (339, 341). On the viral membrane, ∼50 to 63 HLA-II complexes are incorporated per virion (619). As a capsid-encoding virus (620), HIV is featured by its fullerene cone harboring ∼250 hexamers and exactly 12 pentamers of HIV-1 capsid (285). Protein shapes do not indicate exact protein structures, nor are the protein sizes to scale.
For Gag and GagPol, an immature HIV-1 particle contains ∼2,400 ± 700 copies of Gag precursors, quantified by electron tomography and scanning transmission electron microscopy (278). Dot blot assays also measured the ratio of Gag to GagPol to be 20:1 (279), corresponding to ∼120 GagPol copies per virion.
For Env, cryo-electron microscopy tomography shows ∼14 Env spikes per HIV-1 virion (280). Typically, each HIV virion contains 5 to 15 Env spikes (22).
For Vif, semiquantitative Western blot analyses estimated an average of 30 to 80 copies of Vif per virion (281). Although controversial results have been reported, it is generally agreed that there are <100 Vif copies per virion (43, 282, 283).
For Vpr, phosphorimage analyses estimated the molar ratio of Vpr to capsid to be ∼1:7 (284). Findings from X-ray crystallographic analyses suggest that the HIV-1 capsid lattice consists of ∼250 hexamers and exactly 12 pentamers of capsid (285), which correspond to ∼1,560 capsid monomers per virion. Therefore, ∼220 copies of Vpr might be encapsulated per virion.
For Nef, autoradiography and bioimager analyses estimated that ∼5 to 10 copies of Nef are incorporated per virion (286).
Previous studies have reviewed the roles of Env trafficking and packaging (5, 287), HIV-1 genome packaging (288), membrane lipids (289), and cellular cofactors (290) in promoting viral budding. Here, we focus on HIV-1 pairwise protein interactions and associations during viral budding.
MatrixGag-GP41Env Interaction
The GP41CT can physically interact with matrixGag (69–76). The matrixGag-GP41CT interaction takes part in multiple activities during viral budding. (i) This interaction promotes Env packaging (76, 291). (ii) Although the entire matrixGag protein is dispensable for Env packaging (182), the matrixGag-GP41Env interaction enhances the proper intracellular transport of Env glycoproteins in HIV-infected CD4+ T lymphocytes (292). (iii) Given that Gag and Env proteins are colocalized in the plasma membrane (293), the matrixGag-GP41CTEnv interaction permits the efficient association of Env glycoproteins anchored with lipid rafts on the extracellular membrane (294). (iv) Gag may determine the inhibition of Env internalization because in the absence of Gag, Env on the extracellular membrane is rapidly internalized through endocytosis (295). The presence of Gag decreases the rate of Env internalization by targeting the internalization motif in the HIV-1 GP41CT (295). (v) Matrix trimerization that builds a lattice capable of accommodating the GP41CT is crucial for Env packaging (296). (vi) Human proteins may exert an impact on the matrixGag-GP41Env interaction. For instance, the cellular cofactor TIP47 interacts with both matrixGag and Env during viral budding (297). Because of this, the overexpression of TIP47 promotes Env packaging, whereas TIP47 depletion can abolish Env packaging (297).
Regarding the interaction domains, data from mutagenesis analyses suggest that amino acid substitutions (e.g., E16K, K29E, K31E, and E99V) in matrixGag can impair Env packaging (74, 75, 292, 298). Substitutions at serine positions (S9, S67, S72, and S77) in HIV-1 matrixGag dramatically reduce the phosphorylation of matrixGag and inhibit the binding of matrixGag to lipid rafts, thereby causing an impairment of Env packaging (299). Moreover, three matrixGag substitutions (L12E, L30E, and V34I) efficiently block Env packaging (71, 76), whereas Y712C/F substitutions in the GP41CT compensate for the impaired infectivity of virions with the three matrixGag substitutions described above (70). Other studies have also confirmed that L12 and V34 in matrixGag play a critical role in Env packaging (69, 76).
NCGag-Vif Interaction
Vif physically interacts with nucleocapsid in the Gag precursor (NCGag), an interaction identified by coimmunoprecipitation assays, GST pulldown assays, phage display assays, and mammalian two-hybrid assays (300–303). This interaction works in three aspects. (i) Vif packaging into nascent HIV-1 particles not only requires the binding of Vif to two zinc finger domains in NCGag but also depends on the interaction between Vif and viral genomic RNA (300). In the absence of Vif, NCGag is less stably packaged into the HIV-1 core (304). Moreover, Vif and NCGag share common binding sites on tRNALys3, a cellular primer which is incorporated into HIV-1 particles (305). (ii) Despite a small amount of Vif being imported into HIV-1 particles (306), the Vif-NCGag interaction promotes the stability of HIV-1 core and prevents the premature degradation or the disassembly of nucleoprotein complexes (304). Vif negatively regulates the NC-assisted maturation of the viral RNA dimer in nucleoprotein complexes, which in turn prevents the premature initiation of reverse transcription (133). (iii) Vif selectively inhibits protease-mediated proteolytic cleavage between NCGag and SP1Gag (307). In order to keep a low level of expression of Vif that inhibits protease activity, most newly synthesized Vif proteins are rapidly degraded by cellular proteases (307). (iv) Vif induces the degradation of the human cytidine deaminase APOBEC3G via the ubiquitin-proteasome pathway, thereby preventing the packaging of APOBEC3G into nascent HIV particles (308, 309). When this Vif-mediated degradation is impaired under certain circumstances, APOBEC3 proteins are efficiently incorporated into nascent HIV-1 particles through their interactions with NCGag or matrixGag (310).
Regarding the interaction domains, two zinc finger domains of nucleocapsid are essential for Vif packaging (300). Early studies suggested that nucleocapsid may interact with the motif at the C terminus of Vif (positions 171 to 192) (303) or Vif domains (positions 68 to 81, 89 to 100, 162 to 173, and 177 to 189) (301). Subsequent studies reanalyzed the role of the Vif C terminus and refuted its interaction with NCGag (302, 311). Instead, the N-terminal domain (positions 1 to 22) and the central domain (positions 70 to 100) of Vif might interact with NCGag (302).
NCGag-Vpr Interaction
HIV-1 Vpr can interact with nucleocapsid (NCGag) in Gag precursors, an interaction detected by two-hybrid assays, far-Western blot assays, Vpr binding assays, and competition experiments using agarose bead-immobilized avidin (312–314). During the early assembly of Gag, Vpr is encapsulated into nascent viral particles via its interaction with Gag (312–316). Vpr multimerization is crucial for the Gag-Vpr interaction (315). On the one hand, NCGag cooperates with p6Gag to promote Vpr packaging (313), although NCGag is dispensable for Vpr packaging (317). On the other hand, Vpr enhances the transcription of unspliced gag transcripts expressed from unintegrated viral DNA (318). In contrast to the Vpr-NCGag interaction in HIV, Vpr and Vpx in SIVsm do not bind to NCGag, suggesting a distinct difference between HIV and SIVsm proteins (314). In addition, the NC-Vpr structural complex can activate phosphatase 2A0, a cellular protein that inhibits the transition of the cell cycle (319).
Regarding the interaction domains, the C-terminal helix domain of Vpr (positions 70 to 80) may interact with NCGag (313). An early study proposed that two zinc finger domains of NCGag may interact with Vpr (313), but this result was refuted by a subsequent study (314). Future studies are needed to verify their interaction domains.
p6Gag-Vpr Interaction
Vpr can be incorporated into nascent viral particles via its interaction with the p6 domain (p6Gag) in Gag precursors (314, 320–330). This interaction has been identified by using yeast two-hybrid assays, immunoprecipitation assays, maltose-binding protein pulldown assays, nuclear magnetic resonance, fluorescence lifetime imaging microscopy, and plasmon waveguide resonance spectroscopy techniques (314, 315, 322, 328). The binding affinity between Vpr and p6Gag is increased under the environment of lipid bilayer membranes (328). Notably, p6Gag cooperates with NCGag to promote Vpr packaging (313).
Regarding the interaction domains, the F15RFG18 motif (329) and the leucine-rich motif (L41XXLF45) of p6Gag (314, 320, 325–327, 330) may interact with the N-terminal domain of Vpr (positions 18 to 34 [322] and 1 to 71 [327]). The phosphorylation of position S487 in p6Gag promotes the Gag-Vpr interaction and enhances Vpr packaging (331).
p6Gag-Vpx Interaction
HIV-2 and simian immunodeficiency virus (SIV) Vpx proteins can interact with p6Gag according to coimmunoprecipitation assays, GST pulldown assays, yeast two-hybrid assays, and in vitro binding assays (314, 320, 332, 333). The Vpx-p6Gag interaction drives Vpx packaging into HIV-2 and SIV particles (314). In contrast to the HIV-1 Vpr-NCGag interaction, SIV Vpx does not interact with NCGag (314). In nondividing cells, the conserved domain of Vpx (positions 60 to 85) in HIV-2 and SIV is essential for PIC nuclear import (334, 335). Regarding the interaction domains, the leucine-containing motif D17XAXXLL23 in p6Gag (320) may bind to SIVmac Vpx (320) and HIV-2 Vpx (positions 73 to 89) (332, 333).
GP41Env-Nef Interaction
HIV-1 binding assays suggest that Nef can physically interact with the GP41CT (336). This interaction potentially offers a distinct feature for Nef to protect HIV-1 virions from potent neutralizing monoclonal antibodies (e.g., 2F5 and 4E10) that target the GP41CT (337). As an accessory protein with multiple activities, Nef not only is involved in CD4 downregulation (338, 339) but also enhances viral entry involving CD4 and chemokine receptors (286, 340). In the former case, Nef downregulates CD4 to prevent the aggregation of Env-CD4 complexes in the endoplasmic reticulum (17, 341, 342), thereby inhibiting CD4 packaging into HIV-1 particles of either CCR5- or CXCR4-tropic strains (343). HIV-1 Nef also collaborates with Env to activate plasmacytoid dendritic cells for the production of interferon alpha (IFN-α) based on CD4-dependent mechanisms (338). Overall, Nef counteracts the inactivation of trimeric Env spikes to promote Env packaging (344), although Nef is dispensable for viral infection (345).
Regarding the interaction domains, data from mutagenesis analyses suggest that the C-terminal domain of Nef (positions 181 to 210) may interact with the C-terminal dileucine motif (positions 712 to 715) of the GP41CT (336). Deletion of the GP41CT abrogates the Nef-induced enhancement of viral infectivity in HIV-infected CD4+ T lymphocytes (336). Moreover, the tyrosine-based sorting motif (Y712XXL715) in the GP41CT is required for the efficient intracellular trafficking of Env glycoproteins (346). Future studies are still needed to verify the reproducibility of the GP41CT-Nef interaction.
Gag-RT Interaction
Findings from coimmunoprecipitation assays and Western blot analyses suggest that HIV-1 RT interacts with Gag for RT packaging (347, 348). HIV-1 matrixGag and p6Gag in Gag might interact with RT, covering the thumb domain of RT (347). However, the occurrence of HIV-1 RT packaging is likely limited, because it is rare for RT encoded by GagPol to be cleaved by viral protease and to become mature before viral budding. Precise interaction domains in RT and Gag remain unclear. For future studies, independent experiments are still needed to verify the reproducibility of the Gag-RT interaction.
MatrixGag-RNA-NCGag Association
To our knowledge, a direct interaction between matrix and nucleocapsid has not been reported, despite the finding that the N-terminal domain of matrixGag in Gag might fold back onto the C-terminal domain of NCGag for the regulation of Gag assembly (349). During viral budding, viral genomic RNA interacts with two Gag domains: matrixGag (350–355) and NCGag (351, 353, 356–360). The basic residues of HIV-1 NCGag and the N-terminal region of matrixGag (K26KQYK30) are required for the packaging of viral genomic RNA (351, 352, 358). Before viral budding, and after viral budding and maturation, NCGag preferentially binds to the psi and Rev response elements in the viral genomic RNA, while NCGag binds to many sites on the HIV-1 genome (353). In addition to viral RNA, many host proteins (e.g., importin-α/β) also interact with matrixGag and NCGag for Gag packaging and intracellular trafficking (361).
The matrixGag-RNA-NCGag association allows multiple activities. (i) This association is critical for genomic RNA packaging (356). From a broad perspective, viral matrixGag generally contributes to RNA binding and genomic RNA packaging in deltaretroviruses (e.g., bovine leukemia virus) (362). (ii) The binding of Gag to genomic RNA contributes to Gag multimerization (351, 357, 363). (iii) Before the delivery of Gag to appropriate budding sites, the binding of viral RNA to matrixGag protects matrixGag from its association with inadequate cellular membranes (350, 355). Thiadiazolanes that target the matrixGag-RNA interaction can inhibit HIV-1 replication (354). (iv) To promote virus budding, viral RNA can downregulate Gag membrane binding (364). In the absence of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], viral RNA interacts with matrixGag to abolish the binding of Gag to liposomes (364). (v) MatrixGag inhibits the annealing of primer tRNALys3 onto viral genomic RNA, whereas NCGag is essential for tRNALys3 annealing (365). MatrixGag exclusively interacts with cellular tRNAs (e.g., tRNALys3) in the cytosol, thereby regulating Gag binding to cell membranes (353). Overall, the matrixGag-RNA-NCGag association plays a critical role in viral budding.
CapsidGag-LysRS-Vpr Association
Recognized as a conserved cellular enzyme, lysyl-tRNA synthetase (LysRS) takes part in protein synthesis and circulates in multiple compartments (the nucleus, mitochondria, and plasma membrane) for transcriptional regulation, cytokine-like signaling, and the transport of proteins to the cell membrane (366). Although a direct interaction between capsid and Vpr has not been reported to our knowledge, LysRS physically interacts with both HIV-1 Vpr (159) and capsidGag (367–369). On the one hand, HIV-1 Vpr interacts with LysRS to inhibit the LysRS-catalyzed aminoacylation of tRNALys3, although the accessory protein Vpr is dispensable for viral production (159). During viral budding, tRNALys3 is incorporated into HIV-1 particles (367, 370, 371). On the other hand, newly synthesized LysRS binds rapidly to capsidGag on the plasma membrane during the course of viral budding, thereby avoiding the localization of LysRS to other cellular compartments (367). This process permits the incorporation of ∼25 LysRS molecules per HIV-1 particle (372).
Regarding the interaction domains, the C-terminal domain of capsid (positions 177 to 231) may interact with amino acid positions 208 to 259 within the dimerization helix of LysRS (369, 373). The homodimerization of Gag and LysRS indeed contributes to the capsidGag-LysRS interaction (368). LysRS interacts with the N-terminal domain of HIV-1 Vpr (positions 1 to 39) (159). Overall, LysRS binds to capsidGag for its packaging and interacts with Vpr to prevent the LysRS-mediated aminoacylation of tRNALys3.
Gag-AIP1-Nef
As an accessory protein known for its multiple activities, Nef not only increases the total amount of Gag proteins localized at the plasma membrane but also enhances cell-to-cell viral infection in primary CD4+ lymphocytes (374). Although a direct interaction between Gag and Nef has not been reported to our knowledge, both NCGag and Nef bind to AIP1 for efficient viral budding (375–378). AIP1 (apoptosis-linked gene 2 [ALG2]-interacting protein 1) (also known as Alix or PDCD6IP) is associated with the endosomal sorting complex required for transport (ESCRT) machinery (377). The ESCRT machinery is known for promoting cargo sorting and multivesicular body biogenesis (379). On the one hand, GST pulldown assays demonstrate the physical interaction between NCGag and the Bro1 domain of AIP1 (378). On the other hand, data from GST pulldown and coimmunoprecipitation assays suggest that Nef physically binds to AIP1 (375, 377). The Nef-AIP1 interaction not only promotes the proliferation of multivesicular bodies (375) but also facilitates CD4 degradation through lysosomal pathways (377). HIV-1 Nef and glycosylated Gag cooperatively downregulate two transmembrane proteins, serine incorporator 3 (SERINC3) and SERINC5, from the cell surface to prevent their packaging, consequently counteracting their antiviral activity (380).
Regarding interaction domains, the Brol1 and V domains of AIP1 interact with Nef (377). The first 202 positions in the Brol1 domain of AIP1 bind to the zinc finger and N-terminal domains of NCGag (376, 378, 381), particularly key positions such as R3, R7, R10, K11, K14, K20, and R26 (382). Data from crystallization analyses also suggest that amino acid position F105 at the unique extended loop of AIP1 is crucial for HIV-1 budding (383). Moreover, the Y135PLT138 motif in Nef may interact with AIP1 (375).
NCGag-Tsg101/AIP1-P6Gag Association
Two cellular proteins, Tsg101 and AIP1 (also termed Alix), are important components of the ESCRT machinery, which initiates protein sorting into late endosomes (379, 384). During the early stage of viral budding, the Gag protein in its ubiquitinated form recruits AIP1 and Tsg101/ESCRT-I to initiate ESCRT-mediated assembly (385). Thereafter, the downstream ESCRT-III and VPS4 factors are recruited to complete viral budding (386). Experimental evidence suggests that HIV-1 NCGag physically interacts with AIP1 (378) and Tsg101 (387). In addition, HIV-1 p6Gag interacts with AIP1 (386, 388, 389) and Tsg101 (390). During viral budding, AIP1 is packaged into viral particles through the interaction between the Bro1 domain of AIP1 and the zinc finger domains of NCGag (378). Moreover, the binding of Gag to the ESCRT machinery is vital for virus scission from the extracellular membrane of HIV-infected cells (382). Notably, the NCGag-AIP1 interaction requires the involvement of RNA (376) and the cellular protein galectin-3 (391). In the former case, viral RNA bridges the interaction between NCGag and AIP1 (376). In the latter case, galectin-3 interacts with AIP1 to promote the AIP1-p6Gag interaction (391).
Regarding the interaction domains, the Bro1 and V domains of AIP1 can bind to NCGag and p6Gag, respectively (376, 378). The N-terminal basic residues and zinc finger domains of NCGag interact with AIP1 (381). Two motifs, P7TAP10 and L35YPXnL, in p6Gag interact with the cellular proteins Tsg101 and AIP1, respectively (381). Moreover, the C-terminal proline-rich domain of AIP1 (positions 391 to 510) may interact with p6Gag (388), while the N-terminal domain of Tsg101 binds to the P7TAPP11 motif in p6Gag (390). Overall, NCGag and p6Gag in Gag cooperatively interact with Tsg101 and AIP1 to recruit the ESCRT machinery for viral budding.
Vif-APOBEC3G-Integrase Association
Although a direct interaction between HIV-1 integrase and Vif has not been reported, integrase and Vif interact with apolipoprotein B mRNA-editing catalytic polypeptide-like 3G (APOBEC3G or A3G) (33–43). APOBEC3 proteins from the human APOBEC3 family of DNA cytosine deaminases are known as anti-HIV cellular proteins that impair viral DNA synthesis and integration by introducing G-to-A hypermutation to the viral genome (43, 392–394). APOBEC3G physically interacts with HIV-1 integrase to prohibit the formation of proviral DNA (33). To counteract this restriction, Vif physically interacts with APOBEC3G for the degradation of APOBEC3G (35, 36, 39–42, 395). HIV-1 Vif in complex with cellular proteins (e.g., Cul5, Rbx1, and elongins B and C) induces the ubiquitination and proteasomal degradation of APOBEC3G in the cytoplasm (38). Moreover, Vif interacts with the transcription cofactor CBF-β to enhance the degradation of APOBEC3G (396). In studies of four subspecies of African green monkey, it has been shown that APOBEC3G is adaptively diversifying within hosts because of the antagonism-driven coevolution between Vif and APOBEC3G (397). In addition to APOBEC3G, Vif binds to other proteins in the human APOBEC3 family (e.g., APOBEC3C [398, 399] and APOBEC3F [43, 400]). Alanine-scanning analyses revealed six Vif residues (D14, R15, M16, W79, D172, and W174) in three conserved motifs that are essential for the degradation of APOBEC3C and APOBEC3F (401).
Interaction domains of integrase-APOBEC3G and APOBEC3G-Vif interactions have been investigated by extensive studies. In the former case, the C-terminal domain of HIV-1 integrase (positions 213 to 288) may interact with the link domain of APOBEC3G (positions 104 to 156) (33). In the latter case, HIV-1 Vif binds to the N-terminal domain of APOBEC3G within the α1-β1, β2-α2, and β4-α4 loop regions (395). The P161PLP164 motif of Vif is essential because mutations at this motif disrupt the Vif-APOBEC3G interaction, triggering the escape of APOBEC3G, which allows APOBEC3G packaging into nascent HIV particles (36). In summary, many Vif motifs have been found to interact with APOBEC3G, such as D14RMR17 (39), W21xSLVK26 (402), Y40RHHY44 (39), V55xIPLx4–5Lxϕx2YWxL72 (403), Y69xxL72 (404), L81GxGxxIxW89 (41), P161PLP164 (36, 405), E171DRW174 (41), and T(Q/D/E)x5ADx2(I/L), located between positions 96 and 107 (42). Additional studies are still needed to investigate whether these Vif motifs vary due to different protein interaction interfaces or due to different experimental settings.
Vif-MAPK/HCK-Nef Association
MAPK is a serine/threonine/tyrosine-selective protein kinase that phosphorylates multiple HIV-1 proteins, such as matrix, Vif, Tat, Rev, and Nef (406). Specifically, MAPK phosphorylates HIV-1 Vif positions T96 and S165 to downregulate viral replication (407). To counteract this defect, the proline-rich repeat region of Nef (positions 69 to 83) physically interacts with MAPK (408). This interaction allows HIV-1 Nef to inhibit the kinase activity of MAPK (408). In fact, Nef not only alters the activity of the MAPK pathway for T-cell receptor stimulation in primary CD4 T cells (409) but also induces many transcription factors (e.g., activator protein 1) (410). In astrocytes, MAPK and hematopoietic cell kinase (HCK) signaling pathways take part in the production of Nef-induced interleukin-6 (IL-6), interleukin-8 (IL-8), and chemokine (C-C motif) ligand 5 (CCL5) (411, 412).
As a member of the Src family of tyrosine kinases, HCK plays a role in the innate immune response and many signaling pathways (413, 414). HCK physically interacts with HIV-1 Vif (415, 416) and Nef (417–422). Specifically, the SH3 domain of HCK binds to the proline-rich motif P72QVP75 in the N-terminal anchor domain of Nef (422, 423) as well as the P161PLP164 motif in HIV-1 Vif (415). The SH2 and SH3 domains of HCK physically interact with the P72xxP75 motif of HIV-1 Nef to stabilize the functional conformation of the Nef dimer (421). Multiple functions of the Vif-HCK-Nef association have been characterized. (i) Vif interacts with HCK to counteract the HCK-mediated inhibition of viral release (416). (ii) HIV-1 Nef can selectively activate HCK (418). This activation can inhibit the functions of macrophage colony-stimulating factor (M-CSF) receptor in monocytes and macrophages (424). The direct interaction between Nef and HCK induces the activation of HCK, which is indispensable for the downregulation of the M-CSF receptor Fms accumulated as an immature protein at the Golgi apparatus (420). (iii) The Nef-HCK interaction promotes viral growth of Nef-positive (Nef+) viruses but does not alter CD4 downregulation (425). (iv) HCK is involved in the Nef-mediated downregulation of CD1 expression in dendritic cells (426). In addition to HCK, other Src family members (e.g., Lyn and c-Src) interact with Nef (418).
Overall, Nef activates the HCK pathway to downregulate cell surface receptors (419, 420), but Vif counteracts the HCK-mediated inhibition of viral release. Moreover, MAPK phosphorylates Vif, whereas Nef inhibits the kinase activity of MAPK.
Vpu-CD4-GP120Env Association
As a key receptor on cell membranes, CD4 is known to physically interact with GP120 (427–432) and Vpu (14, 19, 433). Due to the flexible nature of HIV-1 Env, the closed and open conformations of Env drive the dynamic binding of Env to CD4, coreceptors, and antibodies (56, 434). To promote viral entry, GP120 interacts with CD4 to trigger profound dynamic structural rearrangements and to induce the aggregation of CD4 and coreceptors (e.g., CCR5) (427, 435–439). During viral budding, this aggregation, however, prohibits the packaging of Env into viral particles (12–17). As of today, a large number of antibodies (e.g., VRC01) that target CD4-binding sites in GP120 have been demonstrated to have a broad and potent activity of neutralization (440–446). Many antiviral agents (e.g., NBD556) against the GP120-CD4 interaction are under investigation (447–449).
As an accessory protein with high sequence variability, HIV-1 Vpu not only enhances viral budding but also induces CD4 degradation (14, 15, 450). HIV-1 Vpu is colocalized with Env and Gag in the trans-Golgi network (451). The binding of HIV-1 Vpu to newly synthesized CD4 causes the retention of CD4 in the ER and, subsequently, the delivery of CD4 to the ER-associated degradation pathway (12–19). The rapid downregulation of CD4 reduces the amount of Env-CD4 structural complexes in the ER, thus promoting the transport of Env to the Golgi apparatus, where oligosaccharide modifications take place (12, 13, 19, 452). The Vpu-CD4 interaction requires the phosphorylated Vpu monomer as the active structure, because monomeric but not multimeric Vpu acts on CD4 downregulation (19), and phosphorylated but not nonphosphorylated Vpu triggers CD4 degradation (453). Many cellular proteins, such as βTrCP, also take part in the Vpu-dependent degradation of CD4 (15, 18). Overall, HIV-1 Vpu induces CD4 degradation to promote Env packaging.
Vpu-Tetherin/CD4-Nef Association
Although a direct interaction between HIV-1 Vpu and Nef is yet to be explored, Vpu and Nef share common activities to downregulate various host proteins (e.g., tetherin, CD4, and CD62L) (341, 454, 455). It has been shown that human tetherin (also termed BST2) interacts with Vpu (456–463) and Nef (464). Meanwhile, CD4 physically binds to Vpu (14, 19, 433) and Nef (465). To explore possible associations between HIV-1 Vpu and Nef, we focus on the downregulation of tetherin and CD4 induced by Vpu and Nef.
Human tetherin is known for its antiviral activity that restricts viral budding, whereas this mechanism is antagonized by HIV-1 Vpu and Nef, resulting in the promotion of viral budding (457, 466–468). Although the exact mechanisms of Vpu action remain debated (469), the binding of HIV-1 Vpu to tetherin leads to the capture of tetherin (459) and subsequently to the downregulation of tetherin via lysosomal and/or proteasomal degradation (459, 461). Different aspects of this mechanism have been reported. (i) HIV-1 Vpu hijacks trafficking pathways of the clathrin adaptor protein complex 1 (AP1) and adaptor protein complex 2 (AP2) to induce postendocytic membrane trafficking events that remove tetherin from the cell membrane (456, 458). (ii) Despite the fact that tetherin enhances the susceptibility of HIV-infected cells to antibodies, HIV-1 Vpu and Nef antagonize tetherin to protect HIV-infected cells from antibody-dependent cell-mediated cytotoxicity, a type of human immune response where virus-specific antibodies activate the killing of HIV-infected cells (470–472). (iii) Among four HIV-1 groups (groups M, N, O, and P), only HIV-1 group M encodes Vpu that robustly counteracts human tetherin (473). Interestingly, the Nef-mediated antagonism of human tetherin is conceived to have evolved before the spread of HIV-1 group O (474).
HIV-1 Vpu and Nef act synergistically to counteract CD4 expression on the HIV-infected cell membrane that deleteriously blocks Env packaging into nascent HIV particles (341). On the one hand, HIV-1 Vpu determines CD4 downregulation by capturing newly synthesized CD4 in the ER, and the ER-retained CD4 is subsequently redistributed to the ER-associated degradation pathway (19). On the other hand, HIV-1 Nef in complex with AP2 interacts with the cytosolic tail of cell surface CD4, causing the internalization of cell surface CD4 to endosomes (475, 476). HIV-1 Nef also mediates the postendocytic targeting of internalized CD4 from the endosomes to the multivesicular body pathway, leading to the eventual degradation of CD4 in lysosomes (477). In addition to CD4, HIV-1 Vpu and Nef downregulate a broad spectrum of more than 32 cell surface receptors (454). Significant downregulation has been observed in the tetraspanin protein family harboring various cellular proteins (e.g., CD9, CD53, CD63, CD81, and CD82) that take part in membrane-based processes (454, 478). As another example, HIV-1 Nef and Vpu inhibit the adhesion and the signaling of L-selection (CD62L) to keep HIV-infected cells away from lymph nodes, a mechanism for HIV-1 to escape from host immune surveillance (455). In the presence of HIV-1 Vpu, Nef acts synergistically to downregulate PVR, a ligand that activates the receptor CD226 in natural killer cells and CD8+ T cells against HIV-1 infections (479). Overall, HIV-1 Vpu and Nef can downregulate various cellular proteins to enhance viral budding.
Vpu-CK2-Rev Association
Casein kinase 2 (CK2) is a ubiquitous serine/threonine-selective protein kinase in all eukaryotes (480, 481). CK2 not only phosphorylates serine residues S52 and S56 at the cytoplasmic domain of HIV-1 Vpu (453, 482) but also phosphorylates HIV-1 Rev serine residues S5 and S8 to downregulate viral production (483, 484). In the former case, CK2-catalyzed phosphorylation is critical for Vpu-mediated CD4 degradation (453, 482). In the latter case, the regulatory beta subunit of CK2 binds to the N-terminal domain of Rev via electrostatic and hydrophobic interactions (483, 485). HIV-1 Rev harboring its arginine-rich domain (positions 35 to 50) activates CK2 to promote viral replication (484). Overall, the Vpu-CK2-Rev association makes it possible for Rev to trigger the activity of CK2, which phosphorylates HIV-1 Vpu for CD4 degradation.
Vpu-UBP-MatrixGag Association
Although a direct interaction between HIV-1 Vpu and Gag has not been reported, a Vpu-binding protein (UBP) has been identified to mediate the functional association between HIV-1 Vpu and matrixGag in Gag (486–489). HIV-1 Vpu can redistribute UBP and Gag to the intracellular membrane of HIV-1-infected cells (488). Newly synthesized Gag is initially transported to the extracellular membrane, where the endocytic uptake of Gag is dismissed in the presence of HIV-1 Vpu (490). In contrast, the absence of HIV-1 Vpu causes a significant amount of Gag to be redistributed to internal membranes for endocytosis (490). Interestingly, Vpu start codon mutants can rescue the impaired viral infectivity of a matrix mutant with the amino acid substitution L30E (491). Overall, HIV-1 Vpu collaborates with UBP to inhibit the endosomal accumulation of Gag in late endosomes, an inhibition process that enhances viral budding (492–494).
VIRAL MATURATION
After viral budding, immature HIV virions undergo maturation processes during which Gag and GagPol precursors are cleaved into mature proteins based on protease-mediated proteolytic processing (Fig. 10). Information about HIV maturation, core morphology, and Gag protein structures has been reviewed elsewhere (1, 2, 30, 31). Here, we focus on HIV-1 pairwise protein interactions that take place during viral maturation.
FIG 10.
HIV-1 morphology, schematic model of protease-mediated proteolytic processing, and protease-substrate structural complexes. (A) Morphology of HIV-1 immature and mature particles during viral maturation. Schematic models of HIV-1 immature and mature particles are also illustrated. Protease-mediated proteolytic processing triggers the maturation of viral proteins inside HIV particles, resulting in significant changes of virion morphology. Electron micrograph images of HIV virions were obtained from the Centers for Disease Control and Prevention (CDC/Maureen Metcalfe and Tom Hodge; http://phil.cdc.gov/phil/details_linked.asp?pid=13472). (B) Protease-mediated proteolytic processing on Gag and GagPol precursors during viral maturation. Five steps have been recorded. First, GagPol dimerization within two GagPol precursors induces the activation of protease dimers with low enzymatic activity (507). Gag and GagPol dimerization occurs before their anchoring to the cell membrane, while GagPol is efficiently packaged into nascent viral particles through its interaction with Gag (621). The transframe region p6* in GagPol prevents premature protease-mediated proteolytic processing (508). The second step is protease-mediated intramolecular (cis) autoprocessing. The connection link between SP1 and nucleocapsid is cleaved by protease within the same GagPol dimer (622). The third step is protease-mediated intermolecular (trans) autoprocessing. HIV protease in one GagPol dimer further cleaves multiple cleavage sites (NC-p6*, p6*-protease, MA-CA, CA-SP1, protease-p51, protease-p66, p66-integrase, p51-p15, and p15-integrase) in another GagPol dimer (593, 623). After GagPol cleavage, HIV protease, integrase, RT, capsid, and nucleocapsid are folded properly. The fourth step is protease-mediated proteolytic processing of Gag. Mature viral protease cleaves Gag precursors in a specific order: SP1-NC, SP2-p6, MA-CA, CA-SP1, and NC-SP2 (31). This process releases MA, CA, SP1, NC, SP2, and p6 from Gag precursors. In the fifth step, during the maturation of HIV-1 proteins, a series of conformation rearrangements turns immature particles into mature particles (624). Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale. (C) HIV-1 protease structures crystallized with 6 substrate peptides derived from Gag and GagPol cleavage sites. Upstream and downstream amino acids of Gag cleavage sites are shown in red and yellow, respectively. See Table S1 in the supplemental material for PDB accession numbers.
Protease-Gag/GagPol Interaction
HIV protease cleaves its substrates (Gag and GagPol) at specific cleavage sites (495–497), a mechanism called protease-mediated proteolytic processing (Fig. 10). This important mechanism allows the maturation of HIV structural proteins (matrix, capsid, nucleocapsid, and p6) and enzymatic proteins (protease, RT, and integrase), thereby transforming immature virions into mature virions with the full capacity for new viral infections (498). As of today, it remains a challenge to resolve the protein structures of full-length Gag and protease in order to identify exact interaction positions in Gag that bind to viral protease. Although the full-length structure of Gag in complex with viral protease has not been resolved, the tertiary structures of protease in complex with peptides near the Gag cleavage sites reveal that protease or Gag substitutions contribute to weak binding (499), low interaction energies (500), and reduced van der Waals contacts (501). Recent findings also suggest that amino acid substitutions in Gag induce significant resistance to protease inhibitors (PIs), causing treatment failure in patients receiving PI-based therapies (502–505). In the presence of protease drug-resistant mutations, cleavage site mutations in Gag and GagPol compensate for impaired protease-mediated cleavages (497, 505). This novel mechanism of HIV drug resistance is discussed below.
GagPol precursors harbor the transframe region p6* at the neighboring region of protease (Fig. 10). At the initial stage of viral maturation, the autoprocessing of HIV protease at the GagPol dimer produces mature viral enzymes (506, 507). As demonstrated by kinetic analyses and cross-linking experiments, p6* inhibits HIV-1 protease activity via a direct interaction (508–510). Multiple activities of p6* have also been reported. (i) p6* prevents protease activities until viral assembly is completed, therefore counteracting premature enzymatic processing (510). In line with this evidence, a complete deletion of p6* enhances protease-mediated proteolytic processing (511). (ii) Cleavages on incorrect p6* positions may significantly reduce the maturation of viral particles, highlighting the importance of accurate cleavages at p6* for protease activation (512). (iii) Although GagPol precursors lacking the p6* region could be incorporated into HIV-1 particles, p6* exerts negative regulation on protease dimerization, and it is indispensable for protease-mediated maturation (513).
Although interaction domains are yet to be explored, non-active-site residues and the C-terminal domain of p6* may regulate embedded protease functions by modulating protease conformations (514). The hydrophilic tripeptides (E4DL6 and E4DF6) at the N-terminal domain (509) and S65FNF68 at the C-terminal domain (508) of p6* may block the substrate-binding cleft of HIV-1 protease. Moreover, the central domain of p6* is unlikely to modulate protease activity (515).
Protease-Vif Interaction
Findings from coimmunoprecipitation assays, ELISAs, and HIV-1 binding assays suggest that HIV-1 Vif physically interacts with protease (282, 516, 517). To inhibit the premature activation of viral protease, HIV-1 Vif interacts with protease to interfere with protease dimerization (516–518). The binding of Vif to protease might take place inside HIV-1-infected cells prior to viral budding, potentially interrupting the autoprocessing of Gag and GagPol precursors (518). Accumulated evidence suggests that Vif selectively inhibits protease-mediated proteolytic processing at specific cleavage sites (e.g., matrix | capsid [518] and SP1 | NC [307]). Although it remains debated, the Vif-protease interaction is speculated to inhibit the protease digestion of cellular cofactors (516, 519).
Regarding the interaction domains, the N-terminal domain of protease (positions 1 to 9 [516, 517]) may interact with the central domain of Vif (positions 78 to 98 [516, 519]). Of interest, Vif-derived peptides (positions 21 to 65 [520], 30 to 65 [519], 81 to 88 [521], 78 to 98 [519], and 88 to 98 [521]) may efficiently inhibit HIV-1 protease activity.
Protease-RT Interaction
Data from coimmunoprecipitation analyses and ELISAs suggest a direct interaction between HIV-1 protease and RT (522). On the one hand, HIV-1 heterodimeric RT contributes to efficient protease-mediated proteolytic processing at many cleavage sites (e.g., RT-integrase) (523). HIV-1 RT upregulates protease activity in a concentration-dependent manner, but this upregulation is independent of pH and ionic strength (523). On the other hand, HIV-1 protease may inhibit DNA synthesis by heterodimeric RT (522). However, HIV-1 protease does not affect the activity of the RNase H domain in RT (522). Future studies are still needed to verify the protease-RT interaction, because this interaction has been reported by only a single study.
Regarding the interaction positions, alanine substitutions (T128A, Y146A, W398A, W401A, and W406A) in RT severely impair the efficiency of protease cleavages on Gag and GagPol precursors (524, 525). The decreased stability of RT mutants (e.g., F130W), however, increases the susceptibility of HIV-1 RT to protease-mediated degradation (526). Moreover, HIV-1 RT substitutions (e.g., L264S, I274T, L279S, and L310S) induce the misfolding and misprocessing of GagPol precursors, leading to the impairment of protease-mediated cleavages (527).
Protease-Nef Interaction
HIV-1 protease cleaves most Nef molecules at a location between positions W57 and L58 (AW57 | L58E) (528–531), while the cleavage site in HIV-2 Nef is located between Y39 and S40 (EY39 | S40Q) (532). This protease-mediated cleavage may take place inside HIV particles given the fact that a small amount of Nef is incorporated into HIV virions (286, 531, 533). Being independent of Nef myristoylation (532) and CD4 downregulation (534), this cleavage dissociates Nef into two parts: the N-terminal myristoylated membrane anchor domain and the C-terminal core domain (286, 530, 533). The former domain determines Nef packaging and CD4 downregulation (535). The latter domain might be stably associated with the viral core within mature HIV-1 particles (536). Full-length Nef inhibits protease activity, while the absence of Nef could decrease the production of mature viral particles (537). In comparisons of the activities of HIV-1 and HIV-2 proteases, it was found that Nef is cleaved more selectively by HIV-1 protease than by HIV-2 protease (532).
Regarding the cleavage sites, the fully conserved residue W57 and the relatively conserved residue L58 in HIV-1 Nef have been demonstrated in large-scale sequence data sets (4). Mutagenesis analyses also suggest that the alanine substitution W57A substantially decreases the efficiency of Nef processing, whereas L58A has little to no impact (530).
Protease-GP41CT Interaction
During viral maturation, HIV-1 protease cleaves the GP41CT (538, 539). This cleavage produces large truncations in the GP41CT, causing resistance to an entry inhibitor called amphotericin B methyl ester (AME) (538, 539). Three protease-mediated cleavage sites (P203 | L204, S205 | F206, and R236 | L237) at the GP41CT generate large truncations of ∼140 amino acids, a mechanism that has been speculated to induce conformation changes to the external regions of Env glycoproteins (538, 539). Although the protease-mediated cleavage of the GP41CT allows the virus to develop AME resistance, large truncations of the GP41CT may impair viral entry and reduce viral fitness (539). Apart from the fact that AME has not been approved for HIV treatment, it remains unclear whether the cleavage of the GP41CT takes part in other biological activities.
ABSENCE OF HIV PAIRWISE PROTEIN INTERACTIONS
Although more than 34 HIV pairwise protein interactions have been discovered (Fig. 11 and 12), the absence of several HIV protein interactions has also been investigated.
FIG 11.
HIV protein-protein interaction networks. Surface or cartoon representations of 16 HIV protein structures are shown in squares. Three types of colored arrows represent HIV pairwise protein interactions between 16 mature HIV proteins. Green arrows, well-known interactions that have been cited >300 times or have been reported by at least 3 publications with more than 100 citations in total (see Table S2 in the supplemental material); gray arrows, little-known interactions that have been reported by a single paper with fewer than 100 citations; blue arrows, lesser-known interactions, including the remaining interactions. Four protein groups are shown background areas. Gray, Env glycoproteins; blue, regulatory proteins; pink, viral enzymes; yellow, structural proteins. According to our current knowledge, neither mature capsid nor Vpu physically interacts with any other HIV proteins.
FIG 12.
Summary of possible HIV-1 pairwise protein associations. The names of 15 HIV-1 proteins are shown at the left and top. Five HIV-1 protein classes (envelope glycoproteins, regulatory proteins, accessory proteins, viral enzymes, and structural proteins) are shown on the left. The biological functions of HIV-1 pairwise protein associations are shown at the top right. Protein functions are mapped with different colors. If one protein association bears multiple functions, at most two major functions are annotated, and the mapped squares combine different colors inside. At the bottom is a summary of HIV-1 pairwise protein associations. Black stars indicate protein associations where two HIV-1 proteins are associated via a third molecule (Table 3), and black horizontal lines indicate impractical interactions between HIV-1 proteins, which are unlikely to take place during the HIV-1 life cycle (see the text for details). Empty squares without any mark inside imply that HIV protein associations or interactions remain unclear. Note that interactions involved with Gag and Env precursors are decomposed into individual protein interactions. For instance, the Gag-RT interaction (347) is decomposed into the matrix-RT and p6-RT interactions. The Gag-Vpr interaction leads to the NC-Vpr and p6-Vpr interactions. MatrixGag connecting with capsidGag in Gag results in the matrix-capsid interaction. Due to the limited number of reports on HIV-2, only information on HIV-1 protein associations is presented.
Absence of Vif-Capsid Interaction
A possible association between Vif and capsidGag was initially proposed because short deletions in capsidGag (positions 284 to 304 and 350 to 362) may enhance Vif packaging (306). Independent analyses, however, suggested that Vif could not interact with mature capsid in HIV-1-infected H9 cells (303). In fact, Vif packaging does not require a direct interaction between Vif and capsidGag; instead, it requires the binding of Vif to NCGag and/or to viral genomic RNA (300–303, 311).
Absence of p6Gag-Vif Interaction
It has been shown that p6Gag is dispensable for Vif packaging, because a complete truncation of p6Gag does not prevent Vif packaging (540). As a matter of fact, Vif packaging requires the binding of Vif to NCGag and/or viral genomic RNA (300–303, 311).
Absence of Matrix-Nef Interaction
HIV-1 matrix is dispensable for Nef-enhanced virus infectivity (541), even though the serine phosphorylation of HIV-1 matrix is promoted by the interaction between Nef and the human serine kinase PAK65 (542). More importantly, results from GST pulldown assays suggest that Nef cannot physically interact with HIV-1 Gag (543).
Absence of p6*-Nef Interaction
An early study suggested that Nef might interact with the transframe domain p6* in GagPol precursors (Fig. 10) by using GST pulldown and coimmunoprecipitation assays (543, 544). It was proposed that this interaction could enhance Nef packaging and viral infectivity (543, 544). However, subsequent analyses refuted these results. First, clustered substitutions in p6* did not affect Nef packaging (545). Second, viral infectivity is reduced in HIV-1 mutants with 40 substitutions of the 56 total residues in p6*, but this phenomenon is observed to be in a Nef-independent manner (545). Third, Nef packaging is mediated by the plasma membrane via a bipartite membrane location signal of Nef (533). Importantly, Nef can be packaged in the absence of GagPol (533). Fourth, the presence of Nef inside viral particles does not increase HIV-1 infectivity (546). To unveil how Nef improves viral infectivity, a recent study suggests that Nef inhibits the packaging of the host transmembrane proteins SERINC3 and SERINC5 to counteract their antiviral activity that inhibits HIV-1 production in primary human blood cells (547). Overall, p6* is unlikely to take part in Nef packaging or in Nef-mediated viral infectivity.
Absence of Vif-Env Interaction
An early study suggested that Vif may regulate Env structural conformations via its interaction with the GP41CT (548). Nevertheless, three subsequent studies refuted such an association by demonstrating that (i) the replication capacity of HIV-1 mutants with deletions in the GP41CT was independent of Vif activity (549); (ii) Vif functions are unrelated to Env, and Vif does not influence Env packaging (550); and (iii) GP41 is intact in cells infected with either the wild type or a Vif mutant (551).
Absence of Vpx-NCGag Interaction
Data from yeast two-hybrid assays suggest that SIV Vpx does not interact with NCGag (314), although HIV-1 Vpr interacts with both NCGag and p6Gag (315, 316, 324).
Absence of Other HIV Protein Interactions
Based on protein activities and localizations during the HIV-1 life cycle, the absence of the following HIV-1 protein interactions could be speculated based on our current knowledge:
HIV-1 Vpu circulates mainly in the cytoplasm, and it is rarely observed in the nucleus, the RTC, the PIC, and nascent HIV-1 particles, where protease, RT, and integrase are localized (Fig. 3). It is therefore tempting to speculate that HIV-1 Vpu might be unlikely to interact with these viral enzymes during the HIV-1 life cycle.
GP120 is an envelope glycoprotein anchored mostly on cellular membranes and the membrane surface of HIV-1 particles (Fig. 5). This limits the possible interactions between GP120 and HIV structural/enzymatic proteins (matrix, capsid, nucleocapsid, p6, protease, RT, and integrase).
GP41 in Env spikes is anchored mostly on cellular and viral membranes. GP41 is cleaved by protease and interacts with matrix, whereas it is absent in the nucleus, the viral core, the RTC, and the PIC (Fig. 7). It is thus tempting to speculate that GP41 might be unlikely to interact with either capsid, nucleocapsid, p6, RT, or integrase.
Rev is absent in HIV particles, the RTC, and the PIC. Thus, Rev might be unlikely to physically interact with HIV-1 protease and RT to affect their enzymatic activities.
In addition to the above-mentioned speculations, other HIV pairwise protein interactions are yet to be discovered. Here, we present a few examples. (i) A direct interaction between Vpr and Nef has not been reported, but Vpr is required for Nef expression from the unintegrated HIV-1 DNA during preintegration transcription (318). (ii) Although it remains unclear whether p6Gag physically interacts with capsidGag, amino acid mutations in HIV-1 p6Gag may interrupt the proteolytic cleavage between capsidGag and SP1Gag (552). (iii) Neutralizing responsiveness to Nef and glycosylated Gag is determined by GP120 V1/V2 regions (344), whereas it remains unclear whether GP120 physically interacts with Nef or glycosylated Gag. (iv) A recent study speculates a possible interaction between integrase and capsid (553), but experimental evidence is still lacking. Overall, future investigations of HIV genome-wide interactions are still needed to unveil all possible HIV protein interactions and to characterize their biological mechanisms and interaction domains.
CLINICAL RELEVANCE AND THERAPEUTIC IMPLICATIONS
Despite the many anti-HIV inhibitors approved by the FDA, a curative vaccine or drug against worldwide HIV infections has not been discovered (4, 554, 555). To enlighten the possible applications of HIV genome-wide protein associations, this section focuses on the clinical relevance and therapeutic implications from two aspects. First, novel mechanisms of HIV drug resistance are discussed to shed light on why some HIV-infected patients have failed antiviral treatments without drug-resistant mutations in drug-targeted proteins. Second, we provide an overview of HIV-derived peptide inhibitors that target viral enzymes on the basis of HIV pairwise protein interactions.
Novel Mechanisms of HIV Drug Resistance
Why do some HIV-infected patients fail highly active antiretroviral therapies (HAARTs) without detectable drug-resistant mutations in drug-targeted proteins? HIV genome-wide protein associations may provide insights into this mystery. It is known that drug-resistant mutations in drug-targeted proteins lead to treatment failure in HIV-infected patients (556). However, recent studies also suggest that protein-protein interactions may provide novel drug resistance mechanisms for HIV to escape the inhibition of antiviral drugs (4, 85, 557, 558). In principle, current findings rely on the hypothesis that the drug resistance of anti-HIV inhibitors could be established by sequence changes outside drug-inhibited proteins, owing to the physical interactions between drug-inhibited proteins and other HIV proteins. In fact, HIV protein interactions have a strong impact on viral genomic diversity, potentially affecting antiviral treatments and vaccine outcomes (44). Here, we provide proof-of-concept examples to highlight novel mechanisms of drug resistance driven by HIV genome-wide protein associations.
GP120 mutations may confer resistance to GP41 inhibitors.
As an approved entry inhibitor, enfuvirtide (T20) is a peptide with 36 amino acids derived from the C-terminal heptad repeat region of HIV-1 GP41 (559). Sequence changes in this heptad repeat are known to confer drug resistance to enfuvirtide (556). However, the V3 loop of GP120, which interacts with host coreceptors (e.g., CCR5), also harbors mutations that confer resistance to enfuvirtide (560, 561). In an independent manner, both the GP41 heptad repeat and the GP120-coreceptor interaction contribute to sensitivity to enfuvirtide (560). To explain this finding, a mechanism has been proposed whereby conformation changes in GP41 are driven by the GP120-coreceptor interaction, thereby affecting sensitivity to the entry inhibitor enfuvirtide (561).
GP41 mutations may confer resistance to CCR5 and protease inhibitors.
It is known that CCR5 inhibitors (e.g., maraviroc) interrupt virus entry by inhibiting the CCR5-GP120 interaction; therefore, sequence changes in GP120 may induce drug resistance to CCR5 inhibitors (556, 562). Surprisingly, clinical and experimental evidence suggests that GP41 mutants (e.g., G516V, M518V, and F519I) confer resistance to the CCR5 inhibitor vicriviroc (563–565). To explain this observation, a novel mechanism has been proposed whereby sequence changes in GP41 may shift the GP120-GP41 interaction to compensate for different conformations of CCR5, causing resistance to vicriviroc (566).
HIV-1 protease cleaves the GP41CT (538, 539), while the GP41CT physically interacts with the matrix region of Gag (78). Because of this physical interaction, GP41 mutations can impair protease-mediated proteolytic processing, conferring resistance to protease inhibitors (85). Although clinical evidence seems to support this mechanism, the list of GP41 mutations associated with protease drug resistance has yet to be fully described.
Integrase mutations may confer resistance to RT inhibitors.
HIV integrase physically interacts with viral RT (101–106), driving integrase mutations to confer resistance to RT inhibitors (RTIs) (e.g., efavirenz) (557). For instance, viruses with integrase mutations (G140S and Q148H) and the RT mutation K103N have significantly increased fold changes in 50% inhibitory concentrations (IC50s) of efavirenz compared to those of viruses with the RT drug-resistant mutation K103N alone (557). This finding is in agreement with data from viral fitness analyses, which suggest that integrase mutations (G140S and Q148H) rescue the replicative fitness of HIV-1 harboring RT drug-resistant mutations (K103N and E138K) under the selective pressure of efavirenz (557). The list of RTI-resistant mutations outside the RT coding region has yet to be fully described.
RT mutations may confer resistance to integrase inhibitors.
HIV RT physically interacts with viral integrase (101–106), permitting RT mutations to confer resistance to integrase inhibitors (e.g., raltegravir) (557). HIV-1 strains with RT mutations (E138K) and integrase mutations (G140S and Q148H) have significantly increased IC50s of raltegravir compared to those of viruses with integrase mutations alone (557). This observation is in agreement with viral fitness changes in which RT drug-resistant mutations (K103N and Y181C) rescue the replicative fitness of viruses harboring integrase drug-resistant mutations under the selective pressure of raltegravir (557). In the absence of drug pressure, protease and RT drug-resistant mutations may decrease the replicative fitness of viruses harboring integrase mutations (567). A list of INI-resistant mutations outside the integrase coding region has yet to be fully described.
Gag mutations may confer resistance to protease inhibitors.
HIV protease cleaves Gag precursors during viral maturation (Fig. 10). Amino acid mutations in Gag may cause an impaired enzymatic activity of HIV protease, inducing treatment failure of PI-based therapies in patient populations (502, 504, 505). Clinical and experimental studies have generally agreed that the protease-mediated processing of Gag establishes an alternative mechanism for HIV to select specific amino acid mutations in Gag, thus conferring resistance to protease inhibitors (502, 504, 505). In analyses of Gag amino acid mutations associated with PI resistance, it has been shown that PI-associated Gag mutations are located mainly at the C-terminal domain and cleavage sites of Gag (502, 504, 556, 568, 569). Moreover, Gag mutations such as V128I, Y132F, K415R, Q430R, A431V, L449FV, S451GT, R452S, and P453TL are significantly associated with protease drug resistance (502, 505). As new PI-associated Gag mutations have been consistently reported (558, 570), further studies are required to map all PI-associated Gag mutations and to evaluate their impacts in studies with large patient populations.
Vif mutations may confer resistance to protease inhibitors.
HIV-1 Vif can physically interact with protease during viral maturation (282, 516, 517). In analyses of Vif sequences sampled from PI-treated and PI-naive patients, amino acid substitutions at five positions of Vif (R36, P47, E101, D117, and L124) are associated with protease drug resistance (571). Due to the limited number of copies of Vif molecules incorporated into HIV particles (Fig. 13), a question that remains to be explored is whether Vif-associated drug resistance plays a significant role in large patient populations.
FIG 13.
Overview of HIV-1 pairwise protein associations during the HIV-1 life cycle. The approximate numbers of viral proteins per HIV-1 particle are annotated in the box at the top left (see Viral Assembly and Budding). Direct physical interactions between HIV-1 proteins are shown by different arrow marks, annotated in the key at the top. If an interaction exerts multiple activities, only the major function is annotated by an arrow mark. Dash lines demonstrate functional protein associations in which two HIV-1 proteins interact with a third molecule, whose name is placed within a text box. Tables 1 and 3 summarize physical protein interactions and functional associations at each stage of the HIV-1 life cycle. Detailed signaling pathways and mechanisms of action are described in the text. Annotated protein associations indicate only their presence at a certain stage of the HIV-1 life cycle and do not indicate the simultaneous existence of all possible associations. Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale.
Apart from the novel mechanisms of HIV drug resistance described above, other HIV pairwise protein associations may have provided alternative pathways for HIV to develop resistance against anti-HIV drugs, marking a new era for the investigation of HIV drug resistance. Importantly, our survey indicates that HIV genomic sequencing is needed to detect novel drug-resistant mutations occurring outside drug-targeted regions, whereas this strategy has not been implemented in current commercial genotypic or phenotypic resistance assays. Overall, a comprehensive survey of HIV genome-wide protein associations sheds light on novel mechanisms of HIV drug resistance from a genome-wide perspective, contributing to the advancement of antiviral treatments.
Development of HIV-Derived Peptide Inhibitors
Based on publications accumulated over the past 3 decades, our review provides a comprehensive overview of HIV pairwise protein associations at different stages of the HIV life cycle, potentially contributing to the development of anti-HIV inhibitors. In light of the FDA approval of enfuvirtide (572, 573), this section highlights HIV-derived peptide inhibitors being developed based on HIV pairwise protein associations (Table 4).
TABLE 4.
HIV-derived peptide inhibitors that interfere with HIV protein associations
| HIV protein association | Drug target(s) | HIV protein(s) (positions), sequence of HIV-derived peptide inhibitora | Mechanism(s) of drug action | Reference |
|---|---|---|---|---|
| RT-integrase | Integrase | RT (176–195), PDIVIYQYMDDLYVGSDLEI | RT-derived peptides inhibit 3′-end processing and the strand transfer reaction of integrase | 575 |
| Integrase | RT (366–385), KQLTEAVQKITTESIVIWGK | |||
| Integrase | RT (396–415), ETWETWWTEYWQATWIPEWE | |||
| RT | Integrase (46–65), KGEAMHGQVDCSPGIWQLDC | IN-derived peptide binds to RT and inhibits RT activity | 576 | |
| Vpr-integrase | Integrase, RT | Vpr (57–71), VEAIIRILQQLLFIH | Vpr-derived peptides inhibit activities of integrase and RT | 162 |
| Vpr-RT | Integrase, RT | Vpr (61–75), IRILQQLLFIHFRIG | ||
| Integrase-Rev | Integrase | Rev (13–23), FRKLIYLTKVL | Rev-derived peptides bind to integrase and inhibit enzymatic activities of HIV-1 integrase | 174 |
| Integrase | Rev (53–67), GLYRTSPSGRIWSI | |||
| Rev | Integrase (66–80), WTHLEGKIILVAVHVA | Integrase-derived peptides abrogate the inhibitory effect of Rev upon viral integration | 180 | |
| Rev | Integrase (118–128), WGSNFTSTTVKA | |||
| Protease-Vif | Protease | Vif (1–9), MENRWQVMI | The N-terminal domain of Vif inhibits the enzymatic activity of HIV-1 protease | 516 |
| Protease | Vif (21–65), WKSLVKHHMYVSGKARGWFYRHHYESPHPRISSEVHIPLGDARLV | 520 | ||
| Protease-p6* | Protease | p6* (65–68), SFNF | The C terminus of p6* inhibits HIV-1 protease activity | 508 |
| Protease | PR (1–5), Tat (49–61), p6* (65–68), PR (95–99), PQITLRKKRRQRRRPPQVSFNFATLNF | Inhibits protease dimerization and activities | 580 |
Peptide information begins with the HIV protein name followed by the peptide-derived region in the HIV protein and the peptide sequence. For instance, “RT (176–195), PDIVIYQYMDDLYVGSDLEI” shows a peptide with the sequence “PDIVIYQYMDDLYVGSDLEI,” which is derived from HIV RT between amino acid positions 176 and 195. Only representative peptide inhibitors with potent inhibitory activity were collected from the literature. Note that p6* is a transframe region in GagPol precursor proteins (Fig. 10).
RT-integrase interaction.
On the one hand, peptide inhibitors derived from HIV-1 RT (positions 166 to 185 and 519 to 532) can efficiently inhibit 3′-end processing and the strand transfer reaction of integrase (574, 575). On the other hand, a peptide derived from HIV-1 integrase (positions 46 to 65) can inhibit the polymerase activity of HIV-1 RT (576) (Table 4).
Vpr-integrase association.
Peptide inhibitors (positions 58 to 72, 57 to 71, and 61 to 75) derived from an alpha-helix structure of Vpr can inhibit the activity of HIV-1 integrase (162, 577). By screening a peptide library built from HIV-1 protein sequences, Vpr-derived peptides containing the motif L64QQLLF69 exhibit promising inhibitory activity against HIV-1 integrase (578).
Vpr-RT association.
Although RT-binding domains in Vpr remain unclear, peptides derived from two Vpr regions (positions 57 to 71 and 61 to 75) physically interact with HIV-1 RT to inhibit reverse transcription (162).
Integrase-Rev association.
Rev-derived peptides (positions 13 to 23 and 53 to 67) efficiently inhibit the enzymatic activity of integrase (180). Moreover, integrase-derived peptides (positions 66 to 80 and 118 to 128) can rescue Rev-mediated inhibitory effects (180).
Protease-Vif association.
Peptide inhibitors derived from the N-terminal domain of HIV-1 protease (positions 1 to 9) can mimic the interface of the protease-Vif interaction to block protease-mediated proteolytic processing (517). Moreover, Vif-derived peptides (positions 30 to 65 [519], 41 to 65 [520], 78 to 98 [519], 81 to 88 [521], and 88 to 98 [521]) efficiently inhibit protease activity.
Protease-p6* interaction.
A peptide inhibitor derived from four parts (p6* CTD, Tat cell-permeable domain, and protease NTD and CTD) (Table 4) blocks protease dimerization and interrupts protease-mediated proteolytic processing (579, 580). Note that four amino acids (S65FNF68) at the p6* CTD are essential for inhibiting the activity of viral protease (508).
Overall, anti-HIV peptides derived from protease, RT, integrase, Vif, Rev, Vpr, and p6* have shown potent activities against enzymatic activities of HIV protease, RT, and integrase. For this reason, an in-depth understanding of HIV genome-wide protein interactions may provide insights into anti-HIV drug development.
CONCLUSIONS AND FUTURE PERSPECTIVES
Armed with publications accumulated over the past 3 decades, this review provides for the first time a comprehensive overview of HIV genome-wide protein associations at major stages of the HIV life cycle (Fig. 13). Our genome-wide perspective on HIV pairwise protein associations reveals intrinsic cross talk between HIV proteins, contributing to the investigation of novel drug resistance and the development of novel antiviral agents (581–584). Bear in mind that the HIV genome encodes only 16 proteins; a high level of HIV pairwise protein associations is therefore expected. Given 120 possible HIV pairwise protein associations between 16 viral proteins, our review summarizes experimental evidence on 34 direct physical interactions (Fig. 11) and 17 functional associations (Fig. 12). To provide a global perspective of HIV genome-wide protein interactions, we have also mapped the protein interaction domains to HIV protein structures, along with integrated information on protein secondary structure, protein intrinsic disorder, and protein sequence diversity (Fig. 14). Overall, this in-depth overview of HIV genome-wide protein associations reveals a high level of mutual collaborations between HIV proteins during the HIV life cycle.
FIG 14.
Integrated map of HIV-1 pairwise protein interactions in the full-length genome. Fifteen HIV-1 proteins are plotted in a circle with seven layers. In layer 1, red links in the center indicate interaction domains for HIV-1 pairwise protein interactions (Table 1). Orange links indicate physical interactions between HIV-1 proteins, but their interaction domains are yet to be resolved (Table 1). In layer 2, indices of amino acid positions are annotated by using HIV-1 HXB2 as a reference. Layer 3 shows protein secondary structures (dark blue, helix structures; light blue, beta-strand structures; pink, random-coil structures) (44). In layer 4, intrinsic disorder scores of individual amino acid positions are shown in green. The range of intrinsic disorder scores is between 0 and 1 (the higher the value, the higher the structural variability) (44). In layer 5, 15 HIV-1 proteins have their names annotated accordingly. In layer 6, amino acid genetic diversity of the HIV-1 subtype B genome is exhibited in gray. Diversity values of between 0 and 1 are mapped on five sublayers (44). In layer 7, protein interaction domains are mapped on cartoon representations of crystallized HIV-1 protein structures. HIV-1 multimeric proteins are shown outside the circle. Data sets for HIV-1 intrinsic disorder scores and amino acid diversity were gathered from our recent report (44). Human proteins involved in HIV-1 pairwise protein associations are not illustrated. See Table S1 in the supplemental material for a list of PDB accession numbers used for structural visualization. PyMOL V1.7 (see http://www.pymol.org/) and Circos V0.64 (http://circos.ca/) visualization software were used.
Our survey suggests that every HIV protein is associated with another viral protein. Among 16 viral proteins, only HIV-1 Vpu and structural proteins are unlikely to interact with other viral proteins due to their functional roles (see Text S1 in the supplemental material). HIV-1 Vpu is known to interact with host proteins for CD4 downregulation and tetherin antagonism (585), while structural proteins (e.g., capsid) are expected to maintain stable HIV structures with lesser associations (503). In comparison, HIV-1 regulatory and accessory proteins (Tat, Rev, Vpr, Vif, and Nef) have more opportunities to engage in a dialogue with other viral and host proteins in many cell compartments, because they undertake multiple activities during the HIV life cycle (Fig. 13).
Despite many findings accumulated over the past 3 decades, investigation of HIV pairwise protein associations from a genome-wide perspective is still warranted. Previous studies unveiled the global landscape of HIV-host protein interactions from high-throughput data (6–11). However, most of these studies have underestimated HIV pairwise protein interactions, providing little to no information on the sophisticated associations between HIV proteins. Exploration of HIV genome-wide protein associations requires the accurate detection of protein associations at different stages of the HIV life cycle. Here, we highlight several challenges for future studies.
HIV macromolecular structures such as the RTC and the PIC have not been resolved in spite of countless attempts being made over the past decades.
A single protein interaction (e.g., RT-integrase) may have different activities at several HIV life stages, making the elucidation of such interactions difficult.
HIV proteins harbor intrinsically disordered structures to interact with other proteins (44, 586). Intrinsically disordered structures usually have dynamic forms, which might hinder accurate detections of protein interaction domains (586).
Many HIV pairwise protein interactions and their accurate interaction domains have yet to be fully described (Fig. 14 and Table 1). Of 34 pairwise protein interactions, more than 10 interactions are known, and their biological functions are well characterized in the literature (Fig. 11). Nevertheless, future studies are still required to address the reproducibility of the other HIV protein interactions, especially those interactions reported in only a single article (Table 2).
Many experiments performed in cell-free settings or with nonnatural target cells may have underestimated the nature of HIV protein interactions because of different protein expression levels and/or the lack of host proteins in cell-free and nonnatural settings. Furthermore, cell type-specific factors in different HIV strains might have been underestimated, because most cell culture experiments have been performed by using cell lines infected with HIV-1 subtype B strains (e.g., HXB2). Therefore, the development of cell culture experiments that represent dynamic protein interactions in a real biological context remains a challenge.
Our review focuses mainly on HIV-1 and less on HIV-2/SIV, because HIV-1 causes major infections worldwide, and the number of reports on HIV-2 and SIV is limited. Although both HIV-1 and HIV-2 originated from SIV (587), they have distinct gene maps (Fig. 1). Particularly, Vpu in HIV-1 and Vpx in HIV-2/SIV, which play different roles during the HIV life cycle, mark a distinct difference (588, 589). For instance, the packaging of SIV Vpx is absolutely dependent on the L41XXLF45 motif of SIV p6Gag (320), but HIV-1 Vpu is not packaged into virions (590). Therefore, it warrants further investigations to explore the differences of genome-wide protein associations between HIV-1 and other simian immunodeficiency viruses (e.g., HIV-2 and SIV). On the other hand, our review identified HIV protein interactions based on keyword searches of reports published between 1985 and 2015, but additional HIV protein interactions will be (or might have been) reported in the literature. For this reason, we have established an online platform (http://www.virusface.com/) to update the information on HIV protein interactions.
Overall, our review provides insights into HIV pairwise protein associations from a genome-wide perspective, shedding light on potential therapeutic targets for drug discovery. Importantly, a comprehensive map of HIV genome-wide protein associations has been established to support the hypothesis that all HIV proteins collaborate meticulously to facilitate viral infections during the viral life cycle.
Supplementary Material
ACKNOWLEDGMENTS
We wholeheartedly thank Christiane Callebaut for her proficient editorial assistance. We acknowledge Anne-Mieke Vandamme and Kristof Theys for helpful discussions.
Guangdi Li was supported by the National Basic Research Program of China (grant 2014CB910500), the National Nature Science Foundation of China (grant 31571368), and the Project of Innovation-Driven Plan of Central South University (grant 2016CX031).
Biographies

Guangdi Li, Ph.D., received his doctoral degree in Biomedical Sciences from the Faculty of Medicine, KU Leuven, Belgium, in 2014. During his Ph.D., he received extensive training from the Master of Molecular and Cellular Biophysics, Faculty of Science, KU Leuven. In 2006, he obtained his bachelor's degree in Applied Mathematics at Hunan University. Between 2006 and 2009, he pursued his master's degree in Computer Science at Shandong University and received full scholarships to work as a research assistant at the Technical University of Madrid, Spain. Between 2009 and 2014, he undertook his doctorate training in the Faculty of Medicine, KU Leuven, Belgium. Since 2015, he has become a research fellow at the Second Xiangya Hospital, Central South University, Hunan, China. His research interests are focused on the genome-wide diversity, coevolution, and interaction of HIV, HBV, and HCV (see http://www.virusface.com/).

Erik De Clercq, M.D., Ph.D., is a professor emeritus at KU Leuven, Belgium, and a visiting professor at the University of South Bohemia in the Czech Republic. His research interests are embraced within the broad areas of microbiology, virology, molecular biology, and antiviral research, focusing on HIV, hepatitis B virus, hepatitis C virus, influenza virus, varicella-zoster virus, herpesvirus, and emerging viruses (e.g., Ebola, dengue, and chikungunya viruses). Over a period of five decades, he has developed new antiviral medicines, including nucleotide analogues (e.g., tenofovir), and he has coinvented several approved antiviral drugs, such as BVDU (brivudine), amino acyl esters of acyclovir (e.g., valacyclovir), acyclic nucleoside phosphonates (ANPs) (cidofovir, adefovir, and tenofovir), nonnucleoside reverse transcriptase inhibitors, and HEPT and TIBO derivatives leading to rilpivirine for the treatment of AIDS. Since the start of his academic career in 1967, Prof. De Clercq has published more than 2,700 research articles (see http://www.virusface.com/).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00065-15.
REFERENCES
- 1.Engelman A, Cherepanov P. 2012. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat Rev Microbiol 10:279–290. doi: 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Li G. 2014. HIV genome-wide diversity, interaction and coevolution. PhD thesis. University of Leuven, Leuven, Belgium. [Google Scholar]
- 5.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. 2012. Global landscape of HIV-human protein complexes. Nature 481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 10.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. 2009. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rato S, Maia S, Brito PM, Resende L, Pereira CF, Moita C, Freitas RP, Moniz-Pereira J, Hacohen N, Moita LF, Goncalves J. 2010. Novel HIV-1 knockdown targets identified by an enriched kinases/phosphatases shRNA library using a long-term iterative screen in Jurkat T-cells. PLoS One 5:e9276. doi: 10.1371/journal.pone.0009276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willey RL, Maldarelli F, Martin MA, Strebel K. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol 66:7193–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenburg ME, Landau NR. 1993. Vpu-induced degradation of CD4: requirement for specific amino acid residues in the cytoplasmic domain of CD4. J Virol 67:7238–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margottin F, Benichou S, Durand H, Richard V, Liu LX, Gomas E, Benarous R. 1996. Interaction between the cytoplasmic domains of HIV-1 Vpu and CD4: role of Vpu residues involved in CD4 interaction and in vitro CD4 degradation. Virology 223:381–386. doi: 10.1006/viro.1996.0491. [DOI] [PubMed] [Google Scholar]
- 15.Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell 1:565–574. doi: 10.1016/S1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 16.Bour S, Boulerice F, Wainberg MA. 1991. Inhibition of gp160 and CD4 maturation in U937 cells after both defective and productive infections by human immunodeficiency virus type 1. J Virol 65:6387–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crise B, Buonocore L, Rose JK. 1990. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol 64:5585–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. 2010. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog 6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magadan JG, Bonifacino JS. 2012. Transmembrane domain determinants of CD4 downregulation by HIV-1 Vpu. J Virol 86:757–772. doi: 10.1128/JVI.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilen CB, Tilton JC, Doms RW. 2012. HIV: cell binding and entry. Cold Spring Harb Perspect Med 2:a006866. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melikyan GB. 2014. HIV entry: a game of hide-and-fuse? Curr Opin Virol 4:1–7. doi: 10.1016/j.coviro.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandenberg OF, Magnus C, Regoes RR, Trkola A. 2015. The HIV-1 entry process: a stoichiometric view. Trends Microbiol 23:763–774. doi: 10.1016/j.tim.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Hu WS, Hughes SH. 2012. HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2:a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craigie R, Bushman FD. 2012. HIV DNA integration. Cold Spring Harb Perspect Med 2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloan RD, Wainberg MA. 2011. The role of unintegrated DNA in HIV infection. Retrovirology 8:52. doi: 10.1186/1742-4690-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debyser Z, Christ F, De Rijck J, Gijsbers R. 2015. Host factors for retroviral integration site selection. Trends Biochem Sci 40:108–116. doi: 10.1016/j.tibs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Ott M, Geyer M, Zhou Q. 2011. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu H, Li Z, Xue Y, Zhou Q. 2013. Viral-host interactions that control HIV-1 transcriptional elongation. Chem Rev 113:8567–8582. doi: 10.1021/cr400120z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karn J, Stoltzfus CM. 2012. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell NM, Lever AM. 2013. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol 21:136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Ako-Adjei D, Fu W, Wallin C, Katz KS, Song G, Darji D, Brister JR, Ptak RG, Pruitt KD. 2015. HIV-1, human interaction database: current status and new features. Nucleic Acids Res 43:D566–D570. doi: 10.1093/nar/gku1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol 81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith JL, Pathak VK. 2010. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol 84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donahue JP, Vetter ML, Mukhtar NA, D'Aquila RT. 2008. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology 377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A 101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 39.Russell RA, Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol 81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwatani Y, Chan DS, Liu L, Yoshii H, Shibata J, Yamamoto N, Levin JG, Gronenborn AM, Sugiura W. 2009. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc Natl Acad Sci U S A 106:19539–19544. doi: 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang Y, Davis RW, York IA, Zheng YH. 2010. Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J Virol 84:5741–5750. doi: 10.1128/JVI.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang Y, Wang X, York IA, Zheng YH. 2010. Identification of a critical T(Q/D/E)x5ADx2(I/L) motif from primate lentivirus Vif proteins that regulate APOBEC3G and APOBEC3F neutralizing activity. J Virol 84:8561–8570. doi: 10.1128/JVI.00960-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. 2009. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol Mol Biol Rev 73:211–232. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G, Piampongsant S, Faria NR, Voet A, Pineda-Pena AC, Khouri R, Lemey P, Vandamme A-M, Theys K. 2015. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 12:18. doi: 10.1186/s12977-015-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchil PD, Mothes W. 2009. HIV entry revisited. Cell 137:402–404. doi: 10.1016/j.cell.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Permanyer M, Ballana E, Este JA. 2010. Endocytosis of HIV: anything goes. Trends Microbiol 18:543–551. doi: 10.1016/j.tim.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Vigant F, Santos NC, Lee B. 2015. Broad-spectrum antivirals against viral fusion. Nat Rev Microbiol 13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guttman M, Lee KK. 2013. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J Virol 87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drummer HE, Hill MK, Maerz AL, Wood S, Ramsland PA, Mak J, Poumbourios P. 2013. Allosteric modulation of the HIV-1 gp120-gp41 association site by adjacent gp120 variable region 1 (V1) N-glycans linked to neutralization sensitivity. PLoS Pathog 9:e1003218. doi: 10.1371/journal.ppat.1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Do Kwon Y, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, et al. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alsahafi N, Debbeche O, Sodroski J, Finzi A. 2015. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One 10:e0122111. doi: 10.1371/journal.pone.0122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. 2013. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci U S A 110:12438–12443. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moscoso CG, Sun Y, Poon S, Xing L, Kan E, Martin L, Green D, Lin F, Vahlne AG, Barnett S, Srivastava I, Cheng RH. 2011. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc Natl Acad Sci U S A 108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melikyan GB. 2008. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology 5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandenberg OF, Magnus C, Rusert P, Regoes RR, Trkola A. 2015. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog 11:e1004595. doi: 10.1371/journal.ppat.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen J, Jacobs A, Caffrey M. 2008. Role of the HIV gp120 conserved domain 5 in processing and viral entry. Biochemistry 47:7788–7795. doi: 10.1021/bi800227z. [DOI] [PubMed] [Google Scholar]
- 64.Khasawneh AI, Laumaea A, Harrison DN, Bellamy-McIntyre AK, Drummer HE, Poumbourios P. 2013. Forced virus evolution reveals functional crosstalk between the disulfide bonded region and membrane proximal ectodomain region of HIV-1 gp41. Retrovirology 10:44. doi: 10.1186/1742-4690-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Sen J, Rong L, Caffrey M. 2008. Role of the HIV gp120 conserved domain 1 in processing and viral entry. J Biol Chem 283:32644–32649. doi: 10.1074/jbc.M806099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.York J, Nunberg JH. 2004. Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J Virol 78:4921–4926. doi: 10.1128/JVI.78.9.4921-4926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JH, Leaman DP, Kim AS, Torrents de la Pena A, Sliepen K, Yasmeen A, Derking R, Ramos A, de Taeye SW, Ozorowski G, Klein F, Burton DR, Nussenzweig MC, Poignard P, Moore JP, Klasse PJ, Sanders RW, Zwick MB, Wilson IA, Ward AB. 2015. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat Commun 6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burton DR, Mascola JR. 2015. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murakami T, Freed EO. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol 74:3548–3554. doi: 10.1128/JVI.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.West JT, Weldon SK, Wyss S, Lin X, Yu Q, Thali M, Hunter E. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J Virol 76:3338–3349. doi: 10.1128/JVI.76.7.3338-3349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freed EO, Martin MA. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol 69:1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tedbury PR, Ablan SD, Freed EO. 2013. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog 9:e1003739. doi: 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan WE, Wang YL, Lin HH, Chen SS. 2004. Effect of extension of the cytoplasmic domain of human immunodeficiency type 1 virus transmembrane protein gp41 on virus replication. J Virol 78:5157–5169. doi: 10.1128/JVI.78.10.5157-5169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tedbury PR, Mercredi PY, Gaines CR, Summers MF, Freed EO. 2015. Elucidating the mechanism by which compensatory mutations rescue an HIV-1 matrix mutant defective for Gag membrane targeting and envelope glycoprotein incorporation. J Mol Biol 427:1413–1427. doi: 10.1016/j.jmb.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brandano L, Stevenson M. 2012. A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles. J Virol 86:2347–2359. doi: 10.1128/JVI.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Freed EO, Martin MA. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol 70:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dorfman T, Mammano F, Haseltine WA, Gottlinger HG. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol 68:1689–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murakami T, Freed EO. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci U S A 97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abrahamyan LG, Mkrtchyan SR, Binley J, Lu M, Melikyan GB, Cohen FS. 2005. The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J Virol 79:106–115. doi: 10.1128/JVI.79.1.106-115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murakami T, Ablan S, Freed EO, Tanaka Y. 2004. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J Virol 78:1026–1031. doi: 10.1128/JVI.78.2.1026-1031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 82.Ritchie C, Cylinder I, Platt EJ, Barklis E. 2015. Analysis of HIV-1 Gag protein interactions via biotin ligase tagging. J Virol 89:3988–4001. doi: 10.1128/JVI.03584-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhatia AK, Campbell N, Panganiban A, Ratner L. 2007. Characterization of replication defects induced by mutations in the basic domain and C-terminus of HIV-1 matrix. Virology 369:47–54. doi: 10.1016/j.virol.2007.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis MR, Jiang J, Zhou J, Freed EO, Aiken C. 2006. A mutation in the human immunodeficiency virus type 1 Gag protein destabilizes the interaction of the envelope protein subunits gp120 and gp41. J Virol 80:2405–2417. doi: 10.1128/JVI.80.5.2405-2417.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rabi SA, Laird GM, Durand CM, Laskey S, Shan L, Bailey JR, Chioma S, Moore RD, Siliciano RF. 2013. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest 123:3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marchio S, Alfano M, Primo L, Gramaglia D, Butini L, Gennero L, De Vivo E, Arap W, Giacca M, Pasqualini R, Bussolino F. 2005. Cell surface-associated Tat modulates HIV-1 infection and spreading through a specific interaction with gp120 viral envelope protein. Blood 105:2802–2811. doi: 10.1182/blood-2004-06-2212. [DOI] [PubMed] [Google Scholar]
- 87.Poon S, Moscoso CG, Xing L, Kan E, Sun Y, Kolatkar PR, Vahlne AG, Srivastava IK, Barnett SW, Cheng RH. 2013. Putative role of Tat-Env interaction in HIV infection. AIDS 27:2345–2354. doi: 10.1097/01.aids.0000432453.60733.b2. [DOI] [PubMed] [Google Scholar]
- 88.Monini P, Cafaro A, Srivastava IK, Moretti S, Sharma VA, Andreini C, Chiozzini C, Ferrantelli F, Cossut MR, Tripiciano A, Nappi F, Longo O, Bellino S, Picconi O, Fanales-Belasio E, Borsetti A, Toschi E, Schiavoni I, Bacigalupo I, Kan E, Sernicola L, Maggiorella MT, Montin K, Porcu M, Leone P, Leone P, Collacchi B, Palladino C, Ridolfi B, Falchi M, Macchia I, Ulmer JB, Butto S, Sgadari C, Magnani M, Federico MP, Titti F, Banci L, Dallocchio F, Rappuoli R, Ensoli F, Barnett SW, Garaci E, Ensoli B. 2012. HIV-1 tat promotes integrin-mediated HIV transmission to dendritic cells by binding Env spikes and competes neutralization by anti-HIV antibodies. PLoS One 7:e48781. doi: 10.1371/journal.pone.0048781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cardaci S, Soster M, Bussolino F, Marchio S. 2013. The V1/V2 loop of HIV-1 gp120 is necessary for Tat binding and consequent modulation of virus entry. FEBS Lett 587:2943–2951. doi: 10.1016/j.febslet.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 90.Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC. 1993. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol 67:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Albini A, Ferrini S, Benelli R, Sforzini S, Giunciuglio D, Aluigi MG, Proudfoot AE, Alouani S, Wells TN, Mariani G, Rabin RL, Farber JM, Noonan DM. 1998. HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci U S A 95:13153–13158. doi: 10.1073/pnas.95.22.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poon S, Moscoso CG, Yenigun OM, Kolatkar PR, Cheng RH, Vahlne A. 2013. HIV-1 Tat protein induces viral internalization through Env-mediated interactions in dose-dependent manner. AIDS 27:2355–2364. doi: 10.1097/01.aids.0000432452.83604.59. [DOI] [PubMed] [Google Scholar]
- 93.Warrilow D, Tachedjian G, Harrich D. 2009. Maturation of the HIV reverse transcription complex: putting the jigsaw together. Rev Med Virol 19:324–337. doi: 10.1002/rmv.627. [DOI] [PubMed] [Google Scholar]
- 94.Carr JM, Davis AJ, Coolen C, Cheney K, Burrell CJ, Li P. 2006. Vif-deficient HIV reverse transcription complexes (RTCs) are subject to structural changes and mutation of RTC-associated reverse transcription products. Virology 351:80–91. doi: 10.1016/j.virol.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 95.Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campbell EM, Hope TJ. 2015. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yan N, O'Day E, Wheeler LA, Engelman A, Lieberman J. 2011. HIV DNA is heavily uracilated, which protects it from autointegration. Proc Natl Acad Sci U S A 108:9244–9249. doi: 10.1073/pnas.1102943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. 2009. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog 5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arhel N. 2010. Revisiting HIV-1 uncoating. Retrovirology 7:96. doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. 2008. Strand transfer events during HIV-1 reverse transcription. Virus Res 134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 101.Tasara T, Maga G, Hottiger MO, Hubscher U. 2001. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett 507:39–44. doi: 10.1016/S0014-5793(01)02945-3. [DOI] [PubMed] [Google Scholar]
- 102.Wilkinson TA, Januszyk K, Phillips ML, Tekeste SS, Zhang M, Miller JT, Le Grice SF, Clubb RT, Chow SA. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J Biol Chem 284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Herschhorn A, Oz-Gleenberg I, Hizi A. 2008. Quantitative analysis of the interactions between HIV-1 integrase and retroviral reverse transcriptases. Biochem J 412:163–170. doi: 10.1042/BJ20071279. [DOI] [PubMed] [Google Scholar]
- 104.Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol 73:2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chakraborty A, Sun GQ, Mustavich L, Huang SH, Li BL. 2013. Biochemical interactions between HIV-1 integrase and reverse transcriptase. FEBS Lett 587:425–429. doi: 10.1016/j.febslet.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 106.Zhu K, Dobard C, Chow SA. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol 78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tekeste SS, Wilkinson TA, Weiner EM, Xu X, Miller JT, Le Grice SF, Clubb RT, Chow SA. 2015. Interaction between reverse transcriptase and integrase is required for reverse transcription during HIV-1 replication. J Virol 89:12058–12069. doi: 10.1128/JVI.01471-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hehl EA, Joshi P, Kalpana GV, Prasad VR. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J Virol 78:5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dobard CW, Briones MS, Chow SA. 2007. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J Virol 81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Druillennec S, Caneparo A, de Rocquigny H, Roques BP. 1999. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J Biol Chem 274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- 111.Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. 1998. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem 273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- 112.Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 113.Cameron CE, Ghosh M, Le Grice SF, Benkovic SJ. 1997. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc Natl Acad Sci U S A 94:6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Drummond JE, Mounts P, Gorelick RJ, Casas-Finet JR, Bosche WJ, Henderson LE, Waters DJ, Arthur LO. 1997. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res Hum Retroviruses 13:533–543. doi: 10.1089/aid.1997.13.533. [DOI] [PubMed] [Google Scholar]
- 115.Anthony RM, Destefano JJ. 2007. In vitro synthesis of long DNA products in reactions with HIV-RT and nucleocapsid protein. J Mol Biol 365:310–324. doi: 10.1016/j.jmb.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, Gruninger-Leitch F, Barre-Sinoussi F, LeGrice SF, Darlix JL. 1989. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J 8:3279–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hameau L, Jeusset J, Lafosse S, Coulaud D, Delain E, Unge T, Restle T, Le Cam E, Mirambeau G. 2001. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement DNA synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J Virol 75:3301–3313. doi: 10.1128/JVI.75.7.3301-3313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 119.Bampi C, Bibillo A, Wendeler M, Divita G, Gorelick RJ, Le Grice SF, Darlix JL. 2006. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J Biol Chem 281:11736–11743. doi: 10.1074/jbc.M600290200. [DOI] [PubMed] [Google Scholar]
- 120.Grohmann D, Godet J, Mely Y, Darlix JL, Restle T. 2008. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry 47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- 121.Ramalanjaona N, de Rocquigny H, Millet A, Ficheux D, Darlix JL, Mely Y. 2007. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J Mol Biol 374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 122.Tanchou V, Gabus C, Rogemond V, Darlix JL. 1995. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol 252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 123.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem 270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 124.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol 74:8980–8988. doi: 10.1128/JVI.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ji X, Klarmann GJ, Preston BD. 1996. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry 35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 126.Liu S, Harada BT, Miller JT, Le Grice SF, Zhuang X. 2010. Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat Struct Mol Biol 17:1453–1460. doi: 10.1038/nsmb.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. 2008. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J Virol 82:9318–9328. doi: 10.1128/JVI.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim J, Roberts A, Yuan H, Xiong Y, Anderson KS. 2012. Nucleocapsid protein annealing of a primer-template enhances (+)-strand DNA synthesis and fidelity by HIV-1 reverse transcriptase. J Mol Biol 415:866–880. doi: 10.1016/j.jmb.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roda RH, Balakrishnan M, Hanson MN, Wohrl BM, Le Grice SF, Roques BP, Gorelick RJ, Bambara RA. 2003. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J Biol Chem 278:31536–31546. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- 130.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, Le Cam E. 2007. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS One 2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kataropoulou A, Bovolenta C, Belfiore A, Trabatti S, Garbelli A, Porcellini S, Lupo R, Maga G. 2009. Mutational analysis of the HIV-1 auxiliary protein Vif identifies independent domains important for the physical and functional interaction with HIV-1 reverse transcriptase. Nucleic Acids Res 37:3660–3669. doi: 10.1093/nar/gkp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cancio R, Spadari S, Maga G. 2004. Vif is an auxiliary factor of the HIV-1 reverse transcriptase and facilitates abasic site bypass. Biochem J 383:475–482. doi: 10.1042/BJ20040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Henriet S, Sinck L, Bec G, Gorelick RJ, Marquet R, Paillart JC. 2007. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res 35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dettenhofer M, Cen S, Carlson BA, Kleiman L, Yu XF. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J Virol 74:8938–8945. doi: 10.1128/JVI.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Goncalves J, Korin Y, Zack J, Gabuzda D. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol 70:8701–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Simon JH, Malim MH. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol 70:5297–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dornadula G, Yang S, Pomerantz RJ, Zhang H. 2000. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J Virol 74:2594–2602. doi: 10.1128/JVI.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nascimbeni M, Bouyac M, Rey F, Spire B, Clavel F. 1998. The replicative impairment of Vif− mutants of human immunodeficiency virus type 1 correlates with an overall defect in viral DNA synthesis. J Gen Virol 79(Part 8):1945–1950. [DOI] [PubMed] [Google Scholar]
- 139.Apolloni A, Meredith LW, Suhrbier A, Kiernan R, Harrich D. 2007. The HIV-1 Tat protein stimulates reverse transcription in vitro. Curr HIV Res 5:473–483. [DOI] [PubMed] [Google Scholar]
- 140.Lin MH, Apolloni A, Cutillas V, Sivakumaran H, Martin S, Li D, Wei T, Wang R, Jin H, Spann K, Harrich D. 2015. A mutant Tat protein inhibits HIV-1 reverse transcription by targeting the reverse transcription complex. J Virol 89:4827–4836. doi: 10.1128/JVI.03440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ulich C, Dunne A, Parry E, Hooker CW, Gaynor RB, Harrich D. 1999. Functional domains of Tat required for efficient human immunodeficiency virus type 1 reverse transcription. J Virol 73:2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kameoka M, Morgan M, Binette M, Russell RS, Rong L, Guo X, Mouland A, Kleiman L, Liang C, Wainberg MA. 2002. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J Virol 76:3637–3645. doi: 10.1128/JVI.76.8.3637-3645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kameoka M, Rong L, Gotte M, Liang C, Russell RS, Wainberg MA. 2001. Role for human immunodeficiency virus type 1 Tat protein in suppression of viral reverse transcriptase activity during late stages of viral replication. J Virol 75:2675–2683. doi: 10.1128/JVI.75.6.2675-2683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boudier C, Storchak R, Sharma KK, Didier P, Follenius-Wund A, Muller S, Darlix JL, Mely Y. 2010. The mechanism of HIV-1 Tat-directed nucleic acid annealing supports its role in reverse transcription. J Mol Biol 400:487–501. doi: 10.1016/j.jmb.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 145.Harrich D, Ulich C, Garcia-Martinez LF, Gaynor RB. 1997. Tat is required for efficient HIV-1 reverse transcription. EMBO J 16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Boudier C, Humbert N, Chaminade F, Chen Y, de Rocquigny H, Godet J, Mauffret O, Fosse P, Mely Y. 2014. Dynamic interactions of the HIV-1 Tat with nucleic acids are critical for Tat activity in reverse transcription. Nucleic Acids Res 42:1065–1078. doi: 10.1093/nar/gkt934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fournier C, Cortay JC, Carbonnelle C, Ehresmann C, Marquet R, Boulanger P. 2002. The HIV-1 Nef protein enhances the affinity of reverse transcriptase for RNA in vitro. Virus Genes 25:255–269. doi: 10.1023/A:1020971823562. [DOI] [PubMed] [Google Scholar]
- 149.Ciuffi A, Munoz M, Bleiber G, Favre M, Stutz F, Telenti A, Meylan PR. 2004. Interactions of processed Nef(58-206) with virion proteins of HIV type 1. AIDS Res Hum Retroviruses 20:399–407. doi: 10.1089/088922204323048140. [DOI] [PubMed] [Google Scholar]
- 150.Aiken C, Trono D. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol 69:5048–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schwartz O, Marechal V, Danos O, Heard JM. 1995. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol 69:4053–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Apolloni A, Hooker CW, Mak J, Harrich D. 2003. Human immunodeficiency virus type 1 protease regulation of tat activity is essential for efficient reverse transcription and replication. J Virol 77:9912–9921. doi: 10.1128/JVI.77.18.9912-9921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. 2006. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology 3:4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Le Rouzic E, Benichou S. 2005. The Vpr protein from HIV-1: distinct roles along the viral life cycle. Retrovirology 2:11. doi: 10.1186/1742-4690-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arts EJ, Miller JT, Ehresmann B, Le Grice SF. 1998. Mutating a region of HIV-1 reverse transcriptase implicated in tRNA(Lys-3) binding and the consequences for (−)-strand DNA synthesis. J Biol Chem 273:14523–14532. doi: 10.1074/jbc.273.23.14523. [DOI] [PubMed] [Google Scholar]
- 156.Cen S, Niu M, Kleiman L. 2004. The connection domain in reverse transcriptase facilitates the in vivo annealing of tRNALys3 to HIV-1 genomic RNA. Retrovirology 1:33. doi: 10.1186/1742-4690-1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Khorchid A, Javanbakht H, Wise S, Halwani R, Parniak MA, Wainberg MA, Kleiman L. 2000. Sequences within Pr160gag-pol affecting the selective packaging of primer tRNA(Lys3) into HIV-1. J Mol Biol 299:17–26. doi: 10.1006/jmbi.2000.3709. [DOI] [PubMed] [Google Scholar]
- 158.Dufour E, Reinbolt J, Castroviejo M, Ehresmann B, Litvak S, Tarrago-Litvak L, Andreola ML. 1999. Cross-linking localization of a HIV-1 reverse transcriptase peptide involved in the binding of primer tRNALys3. J Mol Biol 285:1339–1346. doi: 10.1006/jmbi.1998.2430. [DOI] [PubMed] [Google Scholar]
- 159.Stark LA, Hay RT. 1998. Human immunodeficiency virus type 1 (HIV-1) viral protein R (Vpr) interacts with Lys-tRNA synthetase: implications for priming of HIV-1 reverse transcription. J Virol 72:3037–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kleiman L, Jones CP, Musier-Forsyth K. 2010. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett 584:359–365. doi: 10.1016/j.febslet.2009.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang Y, Mak J, Cao Q, Li Z, Wainberg MA, Kleiman L. 1994. Incorporation of excess wild-type and mutant tRNA(3Lys) into human immunodeficiency virus type 1. J Virol 68:7676–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gleenberg IO, Herschhorn A, Hizi A. 2007. Inhibition of the activities of reverse transcriptase and integrase of human immunodeficiency virus type-1 by peptides derived from the homologous viral protein R (Vpr). J Mol Biol 369:1230–1243. doi: 10.1016/j.jmb.2007.03.073. [DOI] [PubMed] [Google Scholar]
- 163.Ambrose Z, Aiken C. 2014. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 454–455:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol 80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Delelis O, Carayon K, Saib A, Deprez E, Mouscadet JF. 2008. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology 5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ciuffi A, Bushman FD. 2006. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet 22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 168.Engelman A, Cherepanov P. 2008. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog 4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Suzuki Y, Craigie R. 2007. The road to chromatin—nuclear entry of retroviruses. Nat Rev Microbiol 5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 170.Poeschla EM. 2008. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci 65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Oz I, Avidan O, Hizi A. 2002. Inhibition of the integrases of human immunodeficiency viruses type 1 and type 2 by reverse transcriptases. Biochem J 361:557–566. doi: 10.1042/0264-6021:3610557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chow SA, Vincent KA, Ellison V, Brown PO. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 173.Levin A, Rosenbluh J, Hayouka Z, Friedler A, Loyter A. 2010. Integration of HIV-1 DNA is regulated by interplay between viral rev and cellular LEDGF/p75 proteins. Mol Med 16:34–44. doi: 10.2119/molmed.2009.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rosenbluh J, Hayouka Z, Loya S, Levin A, Armon-Omer A, Britan E, Hizi A, Kotler M, Friedler A, Loyter A. 2007. Interaction between HIV-1 Rev and integrase proteins: a basis for the development of anti-HIV peptides. J Biol Chem 282:15743–15753. doi: 10.1074/jbc.M609864200. [DOI] [PubMed] [Google Scholar]
- 175.Levin A, Hayouka Z, Brack-Werner R, Volsky DJ, Friedler A, Loyter A. 2009. Novel regulation of HIV-1 replication and pathogenicity: Rev inhibition of integration. Protein Eng Des Sel 22:753–763. doi: 10.1093/protein/gzp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A 102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cherepanov P, Devroe E, Silver PA, Engelman A. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem 279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 178.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 179.Levin A, Hayouka Z, Friedler A, Loyter A. 2010. Nucleocytoplasmic shuttling of HIV-1 integrase is controlled by the viral Rev protein. Nucleus 1:190–201. doi: 10.4161/nucl.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Levin A, Hayouka Z, Helfer M, Brack-Werner R, Friedler A, Loyter A. 2009. Peptides derived from HIV-1 integrase that bind Rev stimulate viral genome integration. PLoS One 4:e4155. doi: 10.1371/journal.pone.0004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gallay P, Swingler S, Song J, Bushman F, Trono D. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 182.Reil H, Bukovsky AA, Gelderblom HR, Gottlinger HG. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J 17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sato A, Yoshimoto J, Isaka Y, Miki S, Suyama A, Adachi A, Hayami M, Fujiwara T, Yoshie O. 1996. Evidence for direct association of Vpr and matrix protein p17 within the HIV-1 virion. Virology 220:208–212. doi: 10.1006/viro.1996.0302. [DOI] [PubMed] [Google Scholar]
- 184.Heinzinger NK, Bukinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci U S A 91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jenkins Y, Sanchez PV, Meyer BE, Malim MH. 2001. Nuclear export of human immunodeficiency virus type 1 Vpr is not required for virion packaging. J Virol 75:8348–8352. doi: 10.1128/JVI.75.17.8348-8352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nitahara-Kasahara Y, Kamata M, Yamamoto T, Zhang X, Miyamoto Y, Muneta K, Iijima S, Yoneda Y, Tsunetsugu-Yokota Y, Aida Y. 2007. Novel nuclear import of Vpr promoted by importin alpha is crucial for human immunodeficiency virus type 1 replication in macrophages. J Virol 81:5284–5293. doi: 10.1128/JVI.01928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lyonnais S, Gorelick RJ, Heniche-Boukhalfa F, Bouaziz S, Parissi V, Mouscadet JF, Restle T, Gatell JM, Le Cam E, Mirambeau G. 2013. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Res 171:287–303. doi: 10.1016/j.virusres.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vodicka MA, Koepp DM, Silver PA, Emerman M. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev 12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tachiwana H, Shimura M, Nakai-Murakami C, Tokunaga K, Takizawa Y, Sata T, Kurumizaka H, Ishizaka Y. 2006. HIV-1 Vpr induces DNA double-strand breaks. Cancer Res 66:627–631. doi: 10.1158/0008-5472.CAN-05-3144. [DOI] [PubMed] [Google Scholar]
- 190.Bischerour J, Tauc P, Leh H, de Rocquigny H, Roques B, Mouscadet JF. 2003. The (52-96) C-terminal domain of Vpr stimulates HIV-1 IN-mediated homologous strand transfer of mini-viral DNA. Nucleic Acids Res 31:2694–2702. doi: 10.1093/nar/gkg364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Buckman JS, Bosche WJ, Gorelick RJ. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol 77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Poljak L, Batson SM, Ficheux D, Roques BP, Darlix JL, Kas E. 2003. Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J Mol Biol 329:411–421. doi: 10.1016/S0022-2836(03)00472-8. [DOI] [PubMed] [Google Scholar]
- 193.Carteau S, Gorelick RJ, Bushman FD. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol 73:6670–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Carteau S, Batson SC, Poljak L, Mouscadet JF, de Rocquigny H, Darlix JL, Roques BP, Kas E, Auclair C. 1997. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol 71:6225–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. 2010. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol 84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, Debyser Z. 2008. Transportin-SR2 imports HIV into the nucleus. Curr Biol 18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 197.Valle-Casuso JC, Di Nunzio F, Yang Y, Reszka N, Lienlaf M, Arhel N, Perez P, Brass AL, Diaz-Griffero F. 2012. TNPO3 is required for HIV-1 replication after nuclear import but prior to integration and binds the HIV-1 core. J Virol 86:5931–5936. doi: 10.1128/JVI.00451-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.De Houwer S, Demeulemeester J, Thys W, Taltynov O, Zmajkovicova K, Christ F, Debyser Z. 2012. Identification of residues in the C-terminal domain of HIV-1 integrase that mediate binding to the transportin-SR2 protein. J Biol Chem 287:34059–34068. doi: 10.1074/jbc.M112.387944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Luban J. 2008. HIV-1 infection: going nuclear with TNPO3/transportin-SR2 and integrase. Curr Biol 18:R710–R713. doi: 10.1016/j.cub.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 200.De Houwer S, Demeulemeester J, Thys W, Rocha S, Dirix L, Gijsbers R, Christ F, Debyser Z. 2014. The HIV-1 integrase mutant R263A/K264A is 2-fold defective for TRN-SR2 binding and viral nuclear import. J Biol Chem 289:25351–25361. doi: 10.1074/jbc.M113.533281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cribier A, Segeral E, Delelis O, Parissi V, Simon A, Ruff M, Benarous R, Emiliani S. 2011. Mutations affecting interaction of integrase with TNPO3 do not prevent HIV-1 cDNA nuclear import. Retrovirology 8:104. doi: 10.1186/1742-4690-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.De Iaco A, Luban J. 2011. Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus. Retrovirology 8:98. doi: 10.1186/1742-4690-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Briones MS, Dobard CW, Chow SA. 2010. Role of human immunodeficiency virus type 1 integrase in uncoating of the viral core. J Virol 84:5181–5190. doi: 10.1128/JVI.02382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu C, Perilla JR, Ning J, Lu M, Hou G, Ramalho R, Himes BA, Zhao G, Bedwell GJ, Byeon IJ, Ahn J, Gronenborn AM, Prevelige PE, Rousso I, Aiken C, Polenova T, Schulten K, Zhang P. 2016. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat Commun 7:10714. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lu M, Hou G, Zhang H, Suiter CL, Ahn J, Byeon IJ, Perilla JR, Langmead CJ, Hung I, Gor'kov PL, Gan Z, Brey W, Aiken C, Zhang P, Schulten K, Gronenborn AM, Polenova T. 2015. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc Natl Acad Sci U S A 112:14617–14622. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. 2013. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J Virol 87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li Y, Kar AK, Sodroski J. 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J Virol 83:10951–10962. doi: 10.1128/JVI.00682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wu Y, Marsh JW. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- 210.Daugherty MD, Liu B, Frankel AD. 2010. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wilusz J. 2013. Putting an ‘end’ to HIV mRNAs: capping and polyadenylation as potential therapeutic targets. AIDS Res Ther 10:31. doi: 10.1186/1742-6405-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dahiya S, Nonnemacher MR, Wigdahl B. 2012. Deployment of the human immunodeficiency virus type 1 protein arsenal: combating the host to enhance viral transcription and providing targets for therapeutic development. J Gen Virol 93:1151–1172. doi: 10.1099/vir.0.041186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rausch JW, Grice SF. 2015. HIV Rev assembly on the rev response element (RRE): a structural perspective. Viruses 7:3053–3075. doi: 10.3390/v7062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Parent LJ. 2011. New insights into the nuclear localization of retroviral Gag proteins. Nucleus 2:92–97. doi: 10.4161/nucl.2.2.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.He N, Zhou Q. 2011. New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC). J Neuroimmune Pharmacol 6:260–268. doi: 10.1007/s11481-011-9267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lata S, Ali A, Sood V, Raja R, Banerjea AC. 2015. HIV-1 Rev downregulates Tat expression and viral replication via modulation of NAD(P)H:quinine oxidoreductase 1 (NQO1). Nat Commun 6:7244. doi: 10.1038/ncomms8244. [DOI] [PubMed] [Google Scholar]
- 217.Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Philips MN, Habeeb K, Khatri A, Brander C, Robbins GK, Mazzara GP, Goulder PJ, Walker BD, HIV Controller Study Collaboration . 2001. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci U S A 98:1781–1786. doi: 10.1073/pnas.98.4.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lin MH, Sivakumaran H, Jones A, Li D, Harper C, Wei T, Jin H, Rustanti L, Meunier FA, Spann K, Harrich D. 2014. A HIV-1 Tat mutant protein disrupts HIV-1 Rev function by targeting the DEAD-box RNA helicase DDX1. Retrovirology 11:121. doi: 10.1186/s12977-014-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Robertson-Anderson RM, Wang J, Edgcomb SP, Carmel AB, Williamson JR, Millar DP. 2011. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J Mol Biol 410:959–971. doi: 10.1016/j.jmb.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sawaya BE, Khalili K, Gordon J, Taube R, Amini S. 2000. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J Biol Chem 275:35209–35214. doi: 10.1074/jbc.M005197200. [DOI] [PubMed] [Google Scholar]
- 221.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462. doi: 10.1016/S0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 222.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. 2010. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Joseph AM, Ladha JS, Mojamdar M, Mitra D. 2003. Human immunodeficiency virus-1 Nef protein interacts with Tat and enhances HIV-1 gene expression. FEBS Lett 548:37–42. doi: 10.1016/S0014-5793(03)00725-7. [DOI] [PubMed] [Google Scholar]
- 224.Simmons A, Aluvihare V, McMichael A. 2001. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 14:763–777. doi: 10.1016/S1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 225.Witte V, Laffert B, Gintschel P, Krautkramer E, Blume K, Fackler OT, Baur AS. 2008. Induction of HIV transcription by Nef involves Lck activation and protein kinase C theta raft recruitment leading to activation of ERK1/2 but not NF kappa B. J Immunol 181:8425–8432. doi: 10.4049/jimmunol.181.12.8425. [DOI] [PubMed] [Google Scholar]
- 226.Wolf D, Witte V, Clark P, Blume K, Lichtenheld MG, Baur AS. 2008. HIV Nef enhances Tat-mediated viral transcription through a hnRNP-K-nucleated signaling complex. Cell Host Microbe 4:398–408. doi: 10.1016/j.chom.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 227.Barboro P, Ferrari N, Balbi C. 2014. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett 352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 228.Hong HW, Lee SW, Myung H. 2013. Induced degradation of Tat by nucleocapsid (NC) via the proteasome pathway and its effect on HIV transcription. Viruses 5:1143–1152. doi: 10.3390/v5041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Godet J, Boudier C, Humbert N, Ivanyi-Nagy R, Darlix JL, Mely Y. 2012. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus Res 169:349–360. doi: 10.1016/j.virusres.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pachulska-Wieczorek K, Stefaniak AK, Purzycka KJ. 2014. Similarities and differences in the nucleic acid chaperone activity of HIV-2 and HIV-1 nucleocapsid proteins in vitro. Retrovirology 11:54. doi: 10.1186/1742-4690-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zhang J, Crumpacker CS. 2002. Human immunodeficiency virus type 1 nucleocapsid protein nuclear localization mediates early viral mRNA expression. J Virol 76:10444–10454. doi: 10.1128/JVI.76.20.10444-10454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang J, Shackelford JM, Selliah N, Shivers DK, O'Neill E, Garcia JV, Muthumani K, Weiner D, Yu XF, Gabuzda D, Finkel TH. 2008. The HIV-1 Vif protein mediates degradation of Vpr and reduces Vpr-induced cell cycle arrest. DNA Cell Biol 27:267–277. doi: 10.1089/dna.2007.0707. [DOI] [PubMed] [Google Scholar]
- 233.Sakai K, Dimas J, Lenardo MJ. 2006. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc Natl Acad Sci U S A 103:3369–3374. doi: 10.1073/pnas.0509417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang J, Shackelford JM, Casella CR, Shivers DK, Rapaport EL, Liu B, Yu XF, Finkel TH. 2007. The Vif accessory protein alters the cell cycle of human immunodeficiency virus type 1 infected cells. Virology 359:243–252. doi: 10.1016/j.virol.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wang J, Reuschel EL, Shackelford JM, Jeang L, Shivers DK, Diehl JA, Yu XF, Finkel TH. 2011. HIV-1 Vif promotes the G(1)- to S-phase cell-cycle transition. Blood 117:1260–1269. doi: 10.1182/blood-2010-06-289215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Laguette N, Bregnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. 2014. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 237.Davy C, Doorbar J. 2007. G2/M cell cycle arrest in the life cycle of viruses. Virology 368:219–226. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhou D, Wang Y, Tokunaga K, Huang F, Sun B, Yang R. 2015. The HIV-1 accessory protein Vpr induces the degradation of the anti-HIV-1 agent APOBEC3G through a VprBP-mediated proteasomal pathway. Virus Res 195:25–34. doi: 10.1016/j.virusres.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 239.Hakata Y, Yamada M, Mabuchi N, Shida H. 2002. The carboxy-terminal region of the human immunodeficiency virus type 1 protein Rev has multiple roles in mediating CRM1-related Rev functions. J Virol 76:8079–8089. doi: 10.1128/JVI.76.16.8079-8089.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem 273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 241.Kohler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 242.Dupont S, Sharova N, DeHoratius C, Virbasius CM, Zhu X, Bukrinskaya AG, Stevenson M, Green MR. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 243.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 244.Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, Nadler SG, Bukrinsky M. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J Mol Biol 299:359–368. doi: 10.1006/jmbi.2000.3768. [DOI] [PubMed] [Google Scholar]
- 245.Baluyot MF, Grosse SA, Lyddon TD, Janaka SK, Johnson MC. 2012. CRM1-dependent trafficking of retroviral Gag proteins revisited. J Virol 86:4696–4700. doi: 10.1128/JVI.07199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Farjot G, Sergeant A, Mikaelian I. 1999. A new nucleoporin-like protein interacts with both HIV-1 Rev nuclear export signal and CRM-1. J Biol Chem 274:17309–17317. doi: 10.1074/jbc.274.24.17309. [DOI] [PubMed] [Google Scholar]
- 247.Le Rouzic E, Mousnier A, Rustum C, Stutz F, Hallberg E, Dargemont C, Benichou S. 2002. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J Biol Chem 277:45091–45098. doi: 10.1074/jbc.M207439200. [DOI] [PubMed] [Google Scholar]
- 248.Jacquot G, Le Rouzic E, David A, Mazzolini J, Bouchet J, Bouaziz S, Niedergang F, Pancino G, Benichou S. 2007. Localization of HIV-1 Vpr to the nuclear envelope: impact on Vpr functions and virus replication in macrophages. Retrovirology 4:84. doi: 10.1186/1742-4690-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kamata M, Nitahara-Kasahara Y, Miyamoto Y, Yoneda Y, Aida Y. 2005. Importin-alpha promotes passage through the nuclear pore complex of human immunodeficiency virus type 1 Vpr. J Virol 79:3557–3564. doi: 10.1128/JVI.79.6.3557-3564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Dancy BM, Cole PA. 2015. Protein lysine acetylation by p300/CBP. Chem Rev 115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Deng L, de la Fuente C, Fu P, Wang L, Donnelly R, Wade JD, Lambert P, Li H, Lee CG, Kashanchi F. 2000. Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology 277:278–295. doi: 10.1006/viro.2000.0593. [DOI] [PubMed] [Google Scholar]
- 252.Hottiger MO, Nabel GJ. 1998. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol 72:8252–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E. 1999. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol 9:1489–1492. doi: 10.1016/S0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 254.Wong K, Sharma A, Awasthi S, Matlock EF, Rogers L, Van Lint C, Skiest DJ, Burns DK, Harrod R. 2005. HIV-1 Tat interactions with p300 and PCAF transcriptional coactivators inhibit histone acetylation and neurotrophin signaling through CREB. J Biol Chem 280:9390–9399. doi: 10.1074/jbc.M408643200. [DOI] [PubMed] [Google Scholar]
- 255.Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. 2005. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J 24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Topper M, Luo Y, Zhadina M, Mohammed K, Smith L, Muesing MA. 2007. Posttranslational acetylation of the human immunodeficiency virus type 1 integrase carboxyl-terminal domain is dispensable for viral replication. J Virol 81:3012–3017. doi: 10.1128/JVI.02257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.D'Orso I, Frankel AD. 2009. Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc Natl Acad Sci U S A 106:3101–3106. doi: 10.1073/pnas.0900012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Al-Mawsawi LQ, Neamati N. 2007. Blocking interactions between HIV-1 integrase and cellular cofactors: an emerging anti-retroviral strategy. Trends Pharmacol Sci 28:526–535. doi: 10.1016/j.tips.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 259.Allouch A, Cereseto A. 2011. Identification of cellular factors binding to acetylated HIV-1 integrase. Amino Acids 41:1137–1145. doi: 10.1007/s00726-009-0444-3. [DOI] [PubMed] [Google Scholar]
- 260.Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. 1998. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem 273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 261.Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F. 2006. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 3:48. doi: 10.1186/1742-4690-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Deng L, Wang D, de la Fuente C, Wang L, Li H, Lee CG, Donnelly R, Wade JD, Lambert P, Kashanchi F. 2001. Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA. Virology 289:312–326. doi: 10.1006/viro.2001.1129. [DOI] [PubMed] [Google Scholar]
- 263.Terreni M, Valentini P, Liverani V, Gutierrez MI, Di Primio C, Di Fenza A, Tozzini V, Allouch A, Albanese A, Giacca M, Cereseto A. 2010. GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology 7:18. doi: 10.1186/1742-4690-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. 2015. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 265.Kadoch C, Crabtree GR. 2015. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yung E, Sorin M, Pal A, Craig E, Morozov A, Delattre O, Kappes J, Ott D, Kalpana GV. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat Med 7:920–926. doi: 10.1038/90959. [DOI] [PubMed] [Google Scholar]
- 267.Ariumi Y, Serhan F, Turelli P, Telenti A, Trono D. 2006. The integrase interactor 1 (INI1) proteins facilitate Tat-mediated human immunodeficiency virus type 1 transcription. Retrovirology 3:47. doi: 10.1186/1742-4690-3-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S. 2006. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M, Verdin E. 2006. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem 281:19960–19968. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- 270.Yung E, Sorin M, Wang EJ, Perumal S, Ott D, Kalpana GV. 2004. Specificity of interaction of INI1/hSNF5 with retroviral integrases and its functional significance. J Virol 78:2222–2231. doi: 10.1128/JVI.78.5.2222-2231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Morozov A, Yung E, Kalpana GV. 1998. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc Natl Acad Sci U S A 95:1120–1125. doi: 10.1073/pnas.95.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 273.Ippolito JA, Steitz TA. 1998. A 1.3-A resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc Natl Acad Sci U S A 95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mousseau G, Mediouni S, Valente ST. 2015. Targeting HIV transcription: the quest for a functional cure. Curr Top Microbiol Immunol 389:121–145. doi: 10.1007/82_2015_435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, Arneodo A, Andreola ML, Lavigne M, Parissi V. 2011. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog 7:e1001280. doi: 10.1371/journal.ppat.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Maillot B, Levy N, Eiler S, Crucifix C, Granger F, Richert L, Didier P, Godet J, Pradeau-Aubreton K, Emiliani S, Nazabal A, Lesbats P, Parissi V, Mely Y, Moras D, Schultz P, Ruff M. 2013. Structural and functional role of INI1 and LEDGF in the HIV-1 preintegration complex. PLoS One 8:e60734. doi: 10.1371/journal.pone.0060734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Pornillos O, Garrus JE, Sundquist WI. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol 12:569–579. doi: 10.1016/S0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 278.Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunewald K, Krausslich HG. 2008. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Shehu-Xhilaga M, Crowe SM, Mak J. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol 75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 281.Fouchier RA, Simon JH, Jaffe AB, Malim MH. 1996. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J Virol 70:8263–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Khan MA, Akari H, Kao S, Aberham C, Davis D, Buckler-White A, Strebel K. 2002. Intravirion processing of the human immunodeficiency virus type 1 Vif protein by the viral protease may be correlated with Vif function. J Virol 76:9112–9123. doi: 10.1128/JVI.76.18.9112-9123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Bernacchi S, Henriet S, Dumas P, Paillart JC, Marquet R. 2007. RNA and DNA binding properties of HIV-1 Vif protein: a fluorescence study. J Biol Chem 282:26361–26368. doi: 10.1074/jbc.M703122200. [DOI] [PubMed] [Google Scholar]
- 284.Müller B, Tessmer U, Schubert U, Krausslich HG. 2000. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J Virol 74:9727–9731. doi: 10.1128/JVI.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Welker R, Kottler H, Kalbitzer HR, Krausslich HG. 1996. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology 219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 287.Tedbury PR, Freed EO. 2014. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol 22:372–378. doi: 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lu K, Heng X, Summers MF. 2011. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Waheed AA, Freed EO. 2009. Lipids and membrane microdomains in HIV-1 replication. Virus Res 143:162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Weiss ER, Gottlinger H. 2011. The role of cellular factors in promoting HIV budding. J Mol Biol 410:525–533. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yu X, Yuan X, Matsuda Z, Lee TH, Essex M. 1992. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol 66:4966–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lee YM, Tang XB, Cimakasky LM, Hildreth JE, Yu XF. 1997. Mutations in the matrix protein of human immunodeficiency virus type 1 inhibit surface expression and virion incorporation of viral envelope glycoproteins in CD4+ T lymphocytes. J Virol 71:1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Hermida-Matsumoto L, Resh MD. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J Virol 74:8670–8679. doi: 10.1128/JVI.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Bhattacharya J, Repik A, Clapham PR. 2006. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J Virol 80:5292–5300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Egan MA, Carruth LM, Rowell JF, Yu X, Siliciano RF. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol 70:6547–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Tedbury PR, Novikova M, Ablan SD, Freed EO. 2016. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A 113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Lopez-Verges S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. 2006. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci U S A 103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ono A, Huang M, Freed EO. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol 71:4409–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bhatia AK, Kaushik R, Campbell NA, Pontow SE, Ratner L. 2009. Mutation of critical serine residues in HIV-1 matrix result in an envelope incorporation defect which can be rescued by truncation of the gp41 cytoplasmic tail. Virology 384:233–241. doi: 10.1016/j.virol.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Khan MA, Aberham C, Kao S, Akari H, Gorelick R, Bour S, Strebel K. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J Virol 75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Huvent I, Hong SS, Fournier C, Gay B, Tournier J, Carriere C, Courcoul M, Vigne R, Spire B, Boulanger P. 1998. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J Gen Virol 79(Part 5):1069–1081. [DOI] [PubMed] [Google Scholar]
- 302.Syed F, McCrae MA. 2009. Interactions in vivo between the Vif protein of HIV-1 and the precursor (Pr55(GAG)) of the virion nucleocapsid proteins. Arch Virol 154:1797–1805. doi: 10.1007/s00705-009-0520-8. [DOI] [PubMed] [Google Scholar]
- 303.Bouyac M, Courcoul M, Bertoia G, Baudat Y, Gabuzda D, Blanc D, Chazal N, Boulanger P, Sire J, Vigne R, Spire B. 1997. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J Virol 71:9358–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ohagen A, Gabuzda D. 2000. Role of Vif in stability of the human immunodeficiency virus type 1 core. J Virol 74:11055–11066. doi: 10.1128/JVI.74.23.11055-11066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sleiman D, Bernacchi S, Xavier Guerrero S, Brachet F, Larue V, Paillart JC, Tisne C. 2014. Characterization of RNA binding and chaperoning activities of HIV-1 Vif protein. Importance of the C-terminal unstructured tail. RNA Biol 11:906–920. doi: 10.4161/rna.29546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sova P, Volsky DJ, Wang L, Chao W. 2001. Vif is largely absent from human immunodeficiency virus type 1 mature virions and associates mainly with viral particles containing unprocessed gag. J Virol 75:5504–5517. doi: 10.1128/JVI.75.12.5504-5517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Akari H, Fujita M, Kao S, Khan MA, Shehu-Xhilaga M, Adachi A, Strebel K. 2004. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J Biol Chem 279:12355–12362. doi: 10.1074/jbc.M312426200. [DOI] [PubMed] [Google Scholar]
- 308.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem 279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 309.Stopak K, de Noronha C, Yonemoto W, Greene WC. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell 12:591–601. doi: 10.1016/S1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 310.Wang T, Zhang W, Tian C, Liu B, Yu Y, Ding L, Spearman P, Yu XF. 2008. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology 377:71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Henzler T, Harmache A, Herrmann H, Spring H, Suzan M, Audoly G, Panek T, Bosch V. 2001. Fully functional, naturally occurring and C-terminally truncated variant human immunodeficiency virus (HIV) Vif does not bind to HIV Gag but influences intermediate filament structure. J Gen Virol 82:561–573. doi: 10.1099/0022-1317-82-3-561. [DOI] [PubMed] [Google Scholar]
- 312.Li MS, Garcia-Asua G, Bhattacharyya U, Mascagni P, Austen BM, Roberts MM. 1996. The Vpr protein of human immunodeficiency virus type 1 binds to nucleocapsid protein p7 in vitro. Biochem Biophys Res Commun 218:352–355. doi: 10.1006/bbrc.1996.0061. [DOI] [PubMed] [Google Scholar]
- 313.de Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix JL, Roques BP. 1997. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem 272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- 314.Selig L, Pages JC, Tanchou V, Preveral S, Berlioz-Torrent C, Liu LX, Erdtmann L, Darlix J, Benarous R, Benichou S. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol 73:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Fritz JV, Dujardin D, Godet J, Didier P, De Mey J, Darlix JL, Mely Y, de Rocquigny H. 2010. HIV-1 Vpr oligomerization but not that of Gag directs the interaction between Vpr and Gag. J Virol 84:1585–1596. doi: 10.1128/JVI.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Lavallée C, Yao XJ, Ladha A, Gottlinger H, Haseltine WA, Cohen EA. 1994. Requirement of the Pr55gag precursor for incorporation of the Vpr product into human immunodeficiency virus type 1 viral particles. J Virol 68:1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Accola MA, Strack B, Gottlinger HG. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol 74:5395–5402. doi: 10.1128/JVI.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Poon B, Chang MA, Chen IS. 2007. Vpr is required for efficient Nef expression from unintegrated human immunodeficiency virus type 1 DNA. J Virol 81:10515–10523. doi: 10.1128/JVI.00947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tung HY, De Rocquigny H, Zhao LJ, Cayla X, Roques BP, Ozon R. 1997. Direct activation of protein phosphatase-2A0 by HIV-1 encoded protein complex NCp7:vpr. FEBS Lett 401:197–201. doi: 10.1016/S0014-5793(96)01470-6. [DOI] [PubMed] [Google Scholar]
- 320.Accola MA, Bukovsky AA, Jones MS, Gottlinger HG. 1999. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J Virol 73:9992–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Accola MA, Ohagen A, Gottlinger HG. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6(gag). J Virol 74:6198–6202. doi: 10.1128/JVI.74.13.6198-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bachand F, Yao XJ, Hrimech M, Rougeau N, Cohen EA. 1999. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J Biol Chem 274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 323.Paxton W, Connor RI, Landau NR. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol 67:7229–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Kondo E, Mammano F, Cohen EA, Gottlinger HG. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol 69:2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Kondo E, Gottlinger HG. 1996. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol 70:159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Salgado GF, Marquant R, Vogel A, Alves ID, Feller SE, Morellet N, Bouaziz S. 2009. Structural studies of HIV-1 Gag p6ct and its interaction with Vpr determined by solution nuclear magnetic resonance. Biochemistry 48:2355–2367. doi: 10.1021/bi801794v. [DOI] [PubMed] [Google Scholar]
- 327.Jenkins Y, Pornillos O, Rich RL, Myszka DG, Sundquist WI, Malim MH. 2001. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6(Gag). J Virol 75:10537–10542. doi: 10.1128/JVI.75.21.10537-10542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Salgado GF, Vogel A, Marquant R, Feller SE, Bouaziz S, Alves ID. 2009. The role of membranes in the organization of HIV-1 Gag p6 and Vpr: p6 shows high affinity for membrane bilayers which substantially increases the interaction between p6 and Vpr. J Med Chem 52:7157–7162. doi: 10.1021/jm901106t. [DOI] [PubMed] [Google Scholar]
- 329.Zhu H, Jian H, Zhao LJ. 2004. Identification of the 15FRFG domain in HIV-1 Gag p6 essential for Vpr packaging into the virion. Retrovirology 1:26. doi: 10.1186/1742-4690-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lu YL, Bennett RP, Wills JW, Gorelick R, Ratner L. 1995. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol 69:6873–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kudoh A, Takahama S, Sawasaki T, Ode H, Yokoyama M, Okayama A, Ishikawa A, Miyakawa K, Matsunaga S, Kimura H, Sugiura W, Sato H, Hirano H, Ohno S, Yamamoto N, Ryo A. 2014. The phosphorylation of HIV-1 Gag by atypical protein kinase C facilitates viral infectivity by promoting Vpr incorporation into virions. Retrovirology 11:9. doi: 10.1186/1742-4690-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Jin L, Zhou Y, Ratner L. 2001. HIV type 2 Vpx interaction with Gag and incorporation into virus-like particles. AIDS Res Hum Retroviruses 17:105–111. doi: 10.1089/08892220150217193. [DOI] [PubMed] [Google Scholar]
- 333.Pancio HA, Ratner L. 1998. Human immunodeficiency virus type 2 Vpx-Gag interaction. J Virol 72:5271–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Pancio HA, Vander Heyden N, Ratner L. 2000. The C-terminal proline-rich tail of human immunodeficiency virus type 2 Vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J Virol 74:6162–6167. doi: 10.1128/JVI.74.13.6162-6167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Mahalingam S, Van Tine B, Santiago ML, Gao F, Shaw GM, Hahn BH. 2001. Functional analysis of the simian immunodeficiency virus Vpx protein: identification of packaging determinants and a novel nuclear targeting domain. J Virol 75:362–374. doi: 10.1128/JVI.75.1.362-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Schiavoni I, Trapp S, Santarcangelo AC, Piacentini V, Pugliese K, Baur A, Federico M. 2004. HIV-1 Nef enhances both membrane expression and virion incorporation of Env products. A model for the Nef-dependent increase of HIV-1 infectivity. J Biol Chem 279:22996–23006. doi: 10.1074/jbc.M312453200. [DOI] [PubMed] [Google Scholar]
- 337.Lai RP, Yan J, Heeney J, McClure MO, Gottlinger H, Luban J, Pizzato M. 2011. Nef decreases HIV-1 sensitivity to neutralizing antibodies that target the membrane-proximal external region of TMgp41. PLoS Pathog 7:e1002442. doi: 10.1371/journal.ppat.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Reszka-Blanco NJ, Sivaraman V, Zhang L, Su L. 2015. HIV-1 Env and Nef cooperatively contribute to pDCs activation via CD4-dependent mechanisms. J Virol 89:7604–7611. doi: 10.1128/JVI.00695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Arganaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. 2003. Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. J Biol Chem 278:33912–33919. doi: 10.1074/jbc.M303679200. [DOI] [PubMed] [Google Scholar]
- 340.Pizzato M, Popova E, Gottlinger HG. 2008. Nef can enhance the infectivity of receptor-pseudotyped human immunodeficiency virus type 1 particles. J Virol 82:10811–10819. doi: 10.1128/JVI.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Lama J, Mangasarian A, Trono D. 1999. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol 9:622–631. doi: 10.1016/S0960-9822(99)80284-X. [DOI] [PubMed] [Google Scholar]
- 342.Lundquist CA, Tobiume M, Zhou J, Unutmaz D, Aiken C. 2002. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J Virol 76:4625–4633. doi: 10.1128/JVI.76.9.4625-4633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Lundquist CA, Zhou J, Aiken C. 2004. Nef stimulates human immunodeficiency virus type 1 replication in primary T cells by enhancing virion-associated gp120 levels: coreceptor-dependent requirement for Nef in viral replication. J Virol 78:6287–6296. doi: 10.1128/JVI.78.12.6287-6296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Usami Y, Gottlinger H. 2013. HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep 5:802–812. doi: 10.1016/j.celrep.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Abraham L, Fackler OT. 2012. HIV-1 Nef: a multifaceted modulator of T cell receptor signaling. Cell Commun Signal 10:39. doi: 10.1186/1478-811X-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Day JR, Munk C, Guatelli JC. 2004. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J Virol 78:1069–1079. doi: 10.1128/JVI.78.3.1069-1079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Liao WH, Huang KJ, Chang YF, Wang SM, Tseng YT, Chiang CC, Wang JJ, Wang CT. 2007. Incorporation of human immunodeficiency virus type 1 reverse transcriptase into virus-like particles. J Virol 81:5155–5165. doi: 10.1128/JVI.01796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Saadatmand J, Guo F, Cen S, Niu M, Kleiman L. 2008. Interactions of reverse transcriptase sequences in Pol with Gag and LysRS in the HIV-1 tRNALys3 packaging/annealing complex. Virology 380:109–117. doi: 10.1016/j.virol.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 349.Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. 2007. Conformation of the HIV-1 Gag protein in solution. J Mol Biol 365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Alfadhli A, Still A, Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol 83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Ott DE, Coren LV, Gagliardi TD. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J Virol 79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Purohit P, Dupont S, Stevenson M, Green MR. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576–584. doi: 10.1017/S1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Alfadhli A, McNett H, Eccles J, Tsagli S, Noviello C, Sloan R, Lopez CS, Peyton DH, Barklis E. 2013. Analysis of small molecule ligands targeting the HIV-1 matrix protein-RNA binding site. J Biol Chem 288:666–676. doi: 10.1074/jbc.M112.399865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Alfadhli A, McNett H, Tsagli S, Bachinger HP, Peyton DH, Barklis E. 2011. HIV-1 matrix protein binding to RNA. J Mol Biol 410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Clever JL, Taplitz RA, Lochrie MA, Polisky B, Parslow TG. 2000. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J Virol 74:541–546. doi: 10.1128/JVI.74.1.541-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.El Meshri SE, Dujardin D, Godet J, Richert L, Boudier C, Darlix JL, Didier P, Mely Y, de Rocquigny H. 2015. Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: a fluorescence lifetime imaging microscopy investigation. J Mol Biol 427:1480–1494. doi: 10.1016/j.jmb.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 358.Cimarelli A, Sandin S, Hoglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol 74:3046–3057. doi: 10.1128/JVI.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Berkowitz RD, Goff SP. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 360.Bell NM, Kenyon JC, Balasubramanian S, Lever AM. 2012. Comparative structural effects of HIV-1 Gag and nucleocapsid proteins in binding to and unwinding of the viral RNA packaging signal. Biochemistry 51:3162–3169. doi: 10.1021/bi2017969. [DOI] [PubMed] [Google Scholar]
- 361.Parent LJ, Gudleski N. 2011. Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation. J Mol Biol 410:553–564. doi: 10.1016/j.jmb.2011.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Sun M, Grigsby IF, Gorelick RJ, Mansky LM, Musier-Forsyth K. 2014. Retrovirus-specific differences in matrix and nucleocapsid protein-nucleic acid interactions: implications for genomic RNA packaging. J Virol 88:1271–1280. doi: 10.1128/JVI.02151-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol 73:8527–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A 107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Jones CP, Datta SA, Rein A, Rouzina I, Musier-Forsyth K. 2011. Matrix domain modulates HIV-1 Gag's nucleic acid chaperone activity via inositol phosphate binding. J Virol 85:1594–1603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Motzik A, Nechushtan H, Foo SY, Razin E. 2013. Non-canonical roles of lysyl-tRNA synthetase in health and disease. Trends Mol Med 19:726–731. doi: 10.1016/j.molmed.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 367.Halwani R, Cen S, Javanbakht H, Saadatmand J, Kim S, Shiba K, Kleiman L. 2004. Cellular distribution of lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J Virol 78:7553–7564. doi: 10.1128/JVI.78.14.7553-7564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Kovaleski BJ, Kennedy R, Hong MK, Datta SA, Kleiman L, Rein A, Musier-Forsyth K. 2006. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J Biol Chem 281:19449–19456. doi: 10.1074/jbc.M601189200. [DOI] [PubMed] [Google Scholar]
- 369.Javanbakht H, Halwani R, Cen S, Saadatmand J, Musier-Forsyth K, Gottlinger H, Kleiman L. 2003. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J Biol Chem 278:27644–27651. doi: 10.1074/jbc.M301840200. [DOI] [PubMed] [Google Scholar]
- 370.Rigourd M, Bec G, Benas P, Le Grice SF, Ehresmann B, Ehresmann C, Marquet R. 2003. Effects of tRNA 3 Lys aminoacylation on the initiation of HIV-1 reverse transcription. Biochimie 85:521–525. doi: 10.1016/S0300-9084(03)00045-2. [DOI] [PubMed] [Google Scholar]
- 371.Cen S, Khorchid A, Javanbakht H, Gabor J, Stello T, Shiba K, Musier-Forsyth K, Kleiman L. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J Virol 75:5043–5048. doi: 10.1128/JVI.75.11.5043-5048.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Cen S, Javanbakht H, Kim S, Shiba K, Craven R, Rein A, Ewalt K, Schimmel P, Musier-Forsyth K, Kleiman L. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J Virol 76:13111–13115. doi: 10.1128/JVI.76.24.13111-13115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Dewan V, Wei M, Kleiman L, Musier-Forsyth K. 2012. Dual role for motif 1 residues of human lysyl-tRNA synthetase in dimerization and packaging into HIV-1. J Biol Chem 287:41955–41962. doi: 10.1074/jbc.M112.421842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Malbec M, Sourisseau M, Guivel-Benhassine F, Porrot F, Blanchet F, Schwartz O, Casartelli N. 2013. HIV-1 Nef promotes the localization of Gag to the cell membrane and facilitates viral cell-to-cell transfer. Retrovirology 10:80. doi: 10.1186/1742-4690-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Costa LJ, Chen N, Lopes A, Aguiar RS, Tanuri A, Plemenitas A, Peterlin BM. 2006. Interactions between Nef and AIP1 proliferate multivesicular bodies and facilitate egress of HIV-1. Retrovirology 3:33. doi: 10.1186/1742-4690-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Sette P, Dussupt V, Bouamr F. 2012. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J Virol 86:11608–11615. doi: 10.1128/JVI.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Amorim NA, da Silva EM, de Castro RO, da Silva-Januario ME, Mendonca LM, Bonifacino JS, da Costa LJ, daSilva LL. 2014. Interaction of HIV-1 Nef protein with the host protein Alix promotes lysosomal targeting of CD4 receptor. J Biol Chem 289:27744–27756. doi: 10.1074/jbc.M114.560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Popov S, Popova E, Inoue M, Gottlinger HG. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol 82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Olmos Y, Carlton JG. 2016. The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol 38:1–11. doi: 10.1016/j.ceb.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog 5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. 2011. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol 85:2304–2315. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. 2011. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PLoS One 6:e27466. doi: 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Hurley JH. 2015. ESCRTs are everywhere. EMBO J 34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Sette P, Nagashima K, Piper RC, Bouamr F. 2013. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10:79. doi: 10.1186/1742-4690-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Ku PI, Bendjennat M, Ballew J, Landesman MB, Saffarian S. 2014. ALIX is recruited temporarily into HIV-1 budding sites at the end of gag assembly. PLoS One 9:e96950. doi: 10.1371/journal.pone.0096950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Chamontin C, Rassam P, Ferrer M, Racine PJ, Neyret A, Laine S, Milhiet PE, Mougel M. 2015. HIV-1 nucleocapsid and ESCRT-component Tsg101 interplay prevents HIV from turning into a DNA-containing virus. Nucleic Acids Res 43:336–347. doi: 10.1093/nar/gku1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Lazert C, Chazal N, Briant L, Gerlier D, Cortay JC. 2008. Refined study of the interaction between HIV-1 p6 late domain and ALIX. Retrovirology 5:39. doi: 10.1186/1742-4690-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699. doi: 10.1016/S0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 390.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc Natl Acad Sci U S A 98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Wang SF, Tsao CH, Lin YT, Hsu DK, Chiang ML, Lo CH, Chien FC, Chen P, Arthur Chen YM, Chen HY, Liu FT. 2014. Galectin-3 promotes HIV-1 budding via association with Alix and Gag p6. Glycobiology 24:1022–1035. doi: 10.1093/glycob/cwu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Henderson S, Fenton T. 2015. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med 21:274–284. doi: 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 393.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Holmes RK, Malim MH, Bishop KN. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci 32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 395.Kouno T, Luengas EM, Shigematsu M, Shandilya SM, Zhang J, Chen L, Hara M, Schiffer CA, Harris RS, Matsuo H. 2015. Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat Struct Mol Biol 22:485–491. doi: 10.1038/nsmb.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2012. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Compton AA, Hirsch VM, Emerman M. 2012. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe 11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. 2012. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol 19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 399.Stauch B, Hofmann H, Perkovic M, Weisel M, Kopietz F, Cichutek K, Munk C, Schneider G. 2009. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proc Natl Acad Sci U S A 106:12079–12084. doi: 10.1073/pnas.0900979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Siu KK, Sultana A, Azimi FC, Lee JE. 2013. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun 4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, Tsuzuki S, Matsuoka K, Nemoto M, Hachiya A, Sugiura W, Yokomaku Y, Watanabe N, Iwatani Y. 2016. Structural insights into HIV-1 Vif-APOBEC3F interaction. J Virol 90:1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Dang Y, Wang X, Zhou T, York IA, Zheng YH. 2009. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol 83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.He Z, Zhang W, Chen G, Xu R, Yu XF. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol 381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 404.Pery E, Rajendran KS, Brazier AJ, Gabuzda D. 2009. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol 83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Walker RC Jr, Khan MA, Kao S, Goila-Gaur R, Miyagi E, Strebel K. 2010. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J Virol 84:5201–5211. doi: 10.1128/JVI.02318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yang X, Gabuzda D. 1999. Regulation of human immunodeficiency virus type 1 infectivity by the ERK mitogen-activated protein kinase signaling pathway. J Virol 73:3460–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Yang X, Gabuzda D. 1998. Mitogen-activated protein kinase phosphorylates and regulates the HIV-1 Vif protein. J Biol Chem 273:29879–29887. doi: 10.1074/jbc.273.45.29879. [DOI] [PubMed] [Google Scholar]
- 408.Greenway A, Azad A, Mills J, McPhee D. 1996. Human immunodeficiency virus type 1 Nef binds directly to Lck and mitogen-activated protein kinase, inhibiting kinase activity. J Virol 70:6701–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Schrager JA, Der Minassian V, Marsh JW. 2002. HIV Nef increases T cell ERK MAP kinase activity. J Biol Chem 277:6137–6142. doi: 10.1074/jbc.M107322200. [DOI] [PubMed] [Google Scholar]
- 410.Biggs TE, Cooke SJ, Barton CH, Harris MP, Saksela K, Mann DA. 1999. Induction of activator protein 1 (AP-1) in macrophages by human immunodeficiency virus type-1 NEF is a cell-type-specific response that requires both hck and MAPK signaling events. J Mol Biol 290:21–35. doi: 10.1006/jmbi.1999.2849. [DOI] [PubMed] [Google Scholar]
- 411.Liu X, Kumar A. 2015. Differential signaling mechanism for HIV-1 Nef-mediated production of IL-6 and IL-8 in human astrocytes. Sci Rep 5:9867. doi: 10.1038/srep09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Liu X, Shah A, Gangwani MR, Silverstein PS, Fu M, Kumar A. 2014. HIV-1 Nef induces CCL5 production in astrocytes through p38-MAPK and PI3K/Akt pathway and utilizes NF-kB, CEBP and AP-1 transcription factors. Sci Rep 4:4450. doi: 10.1038/srep04450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Musumeci F, Schenone S, Brullo C, Desogus A, Botta L, Tintori C. 2015. Hck inhibitors as potential therapeutic agents in cancer and HIV infection. Curr Med Chem 22:1540–1564. doi: 10.2174/0929867322666150209152057. [DOI] [PubMed] [Google Scholar]
- 414.Poh AR, O'Donoghue RJ, Ernst M. 2015. Hematopoietic cell kinase (HCK) as a therapeutic target in immune and cancer cells. Oncotarget 6:15752–15771. doi: 10.18632/oncotarget.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Marcsisin SR, Narute PS, Emert-Sedlak LA, Kloczewiak M, Smithgall TE, Engen JR. 2011. On the solution conformation and dynamics of the HIV-1 viral infectivity factor. J Mol Biol 410:1008–1022. doi: 10.1016/j.jmb.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Hassaine G, Courcoul M, Bessou G, Barthalay Y, Picard C, Olive D, Collette Y, Vigne R, Decroly E. 2001. The tyrosine kinase Hck is an inhibitor of HIV-1 replication counteracted by the viral vif protein. J Biol Chem 276:16885–16893. doi: 10.1074/jbc.M009076200. [DOI] [PubMed] [Google Scholar]
- 417.Lee CH, Leung B, Lemmon MA, Zheng J, Cowburn D, Kuriyan J, Saksela K. 1995. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J 14:5006–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Trible RP, Emert-Sedlak L, Smithgall TE. 2006. HIV-1 Nef selectively activates Src family kinases Hck, Lyn, and c-Src through direct SH3 domain interaction. J Biol Chem 281:27029–27038. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Hung CH, Thomas L, Ruby CE, Atkins KM, Morris NP, Knight ZA, Scholz I, Barklis E, Weinberg AD, Shokat KM, Thomas G. 2007. HIV-1 Nef assembles a Src family kinase-ZAP-70/Syk-PI3K cascade to downregulate cell-surface MHC-I. Cell Host Microbe 1:121–133. doi: 10.1016/j.chom.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 420.Hiyoshi M, Suzu S, Yoshidomi Y, Hassan R, Harada H, Sakashita N, Akari H, Motoyoshi K, Okada S. 2008. Interaction between Hck and HIV-1 Nef negatively regulates cell surface expression of M-CSF receptor. Blood 111:243–250. doi: 10.1182/blood-2007-04-086017. [DOI] [PubMed] [Google Scholar]
- 421.Alvarado JJ, Tarafdar S, Yeh JI, Smithgall TE. 2014. Interaction with the Src homology (SH3-SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment. J Biol Chem 289:28539–28553. doi: 10.1074/jbc.M114.600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Jung J, Byeon IJ, Ahn J, Gronenborn AM. 2011. Structure, dynamics, and Hck interaction of full-length HIV-1 Nef. Proteins 79:1609–1622. doi: 10.1002/prot.22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Kuo LS, Baugh LL, Denial SJ, Watkins RL, Liu M, Garcia JV, Foster JL. 2012. Overlapping effector interfaces define the multiple functions of the HIV-1 Nef polyproline helix. Retrovirology 9:47. doi: 10.1186/1742-4690-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Suzu S, Harada H, Matsumoto T, Okada S. 2005. HIV-1 Nef interferes with M-CSF receptor signaling through Hck activation and inhibits M-CSF bioactivities. Blood 105:3230–3237. doi: 10.1182/blood-2004-06-2084. [DOI] [PubMed] [Google Scholar]
- 425.Saksela K, Cheng G, Baltimore D. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J 14:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Shinya E, Shimizu M, Owaki A, Paoletti S, Mori L, De Libero G, Takahashi H. 2016. Hemopoietic cell kinase (Hck) and p21-activated kinase 2 (PAK2) are involved in the down-regulation of CD1a lipid antigen presentation by HIV-1 Nef in dendritic cells. Virology 487:285–295. doi: 10.1016/j.virol.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 427.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Bowman MR, MacFerrin KD, Schreiber SL, Burakoff SJ. 1990. Identification and structural analysis of residues in the V1 region of CD4 involved in interaction with human immunodeficiency virus envelope glycoprotein gp120 and class II major histocompatibility complex molecules. Proc Natl Acad Sci U S A 87:9052–9056. doi: 10.1073/pnas.87.22.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Bour S, Geleziunas R, Wainberg MA. 1995. The human immunodeficiency virus type 1 (HIV-1) CD4 receptor and its central role in promotion of HIV-1 infection. Microbiol Rev 59:63–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Diskin R, Marcovecchio PM, Bjorkman PJ. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol 17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Singh SK, Mockel L, Thiagarajan-Rosenkranz P, Wittlich M, Willbold D, Koenig BW. 2012. Mapping the interaction between the cytoplasmic domains of HIV-1 viral protein U and human CD4 with NMR spectroscopy. FEBS J 279:3705–3714. doi: 10.1111/j.1742-4658.2012.08732.x. [DOI] [PubMed] [Google Scholar]
- 434.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Yi L, Fang J, Isik N, Chim J, Jin T. 2006. HIV gp120-induced interaction between CD4 and CCR5 requires cholesterol-rich microenvironments revealed by live cell fluorescence resonance energy transfer imaging. J Biol Chem 281:35446–35453. doi: 10.1074/jbc.M607302200. [DOI] [PubMed] [Google Scholar]
- 436.Agrawal-Gamse C, Lee FH, Haggarty B, Jordan AP, Yi Y, Lee B, Collman RG, Hoxie JA, Doms RW, Laakso MM. 2009. Adaptive mutations in a human immunodeficiency virus type 1 envelope protein with a truncated V3 loop restore function by improving interactions with CD4. J Virol 83:11005–11015. doi: 10.1128/JVI.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. 2012. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol 86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Moscoso CG, Xing L, Hui J, Hu J, Kalkhoran MB, Yenigun OM, Sun Y, Paavolainen L, Martin L, Vahlne A, Zambonelli C, Barnett SW, Srivastava IK, Cheng RH. 2014. Trimeric HIV Env provides epitope occlusion mediated by hypervariable loops. Sci Rep 4:7025. doi: 10.1038/srep07025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, Connors M, Burton DR, Wilson IA, Klasse PJ, Ward AB, Moore JP, Sanders RW. 2015. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. 2015. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog 11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Ko SY, Pegu A, Rudicell RS, Yang ZY, Joyce MG, Chen X, Wang K, Bao S, Kraemer TD, Rath T, Zeng M, Schmidt SD, Todd JP, Penzak SR, Saunders KO, Nason MC, Haase AT, Rao SS, Blumberg RS, Mascola JR, Nabel GJ. 2014. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature 514:642–645. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Gardner MR, Kattenhorn LM, Kondur HR, von Schaewen M, Dorfman T, Chiang JJ, Haworth KG, Decker JM, Alpert MD, Bailey CC, Neale ES Jr, Fellinger CH, Joshi VR, Fuchs SP, Martinez-Navio JM, Quinlan BD, Yao AY, Mouquet H, Gorman J, Zhang B, Poignard P, Nussenzweig MC, Burton DR, Kwong PD, Piatak M Jr, Lifson JD, Gao G, Desrosiers RC, Evans DT, Hahn BH, Ploss A, Cannon PM, Seaman MS, Farzan M. 2015. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519:87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, Karita E, Kilembe W, Lakhi S, Inambao M, Kamali A, Sanders EJ, Anzala O, Edward V, Bekker LG, Tang J, Gilmour J, Kosakovsky-Pond SL, Phung P, Wrin T, Crotty S, Godzik A, Poignard P. 2016. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog 12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Matz J, Kessler P, Bouchet J, Combes O, Ramos OH, Barin F, Baty D, Martin L, Benichou S, Chames P. 2013. Straightforward selection of broadly neutralizing single-domain antibodies targeting the conserved CD4 and coreceptor binding sites of HIV-1 gp120. J Virol 87:1137–1149. doi: 10.1128/JVI.00461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG. 2014. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat Chem Biol 10:845–852. doi: 10.1038/nchembio.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Curreli F, Choudhury S, Pyatkin I, Zagorodnikov VP, Bulay AK, Altieri A, Kwon YD, Kwong PD, Debnath AK. 2012. Design, synthesis, and antiviral activity of entry inhibitors that target the CD4-binding site of HIV-1. J Med Chem 55:4764–4775. doi: 10.1021/jm3002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schon A, Freire E, Routy JP, Smith AB III, Park J, Jones DM, Courter JR, Melillo BN, Kaufmann DE, Hahn BH, Permar SR, Haynes BF, Madani N, Sodroski JG, Finzi A. 2015. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci U S A 112:E2687–E2694. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Pickering S, Hue S, Kim EY, Reddy S, Wolinsky SM, Neil SJ. 2014. Preservation of tetherin and CD4 counter-activities in circulating Vpu alleles despite extensive sequence variation within HIV-1 infected individuals. PLoS Pathog 10:e1003895. doi: 10.1371/journal.ppat.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Van Damme N, Guatelli J. 2008. HIV-1 Vpu inhibits accumulation of the envelope glycoprotein within clathrin-coated, Gag-containing endosomes. Cell Microbiol 10:1040–1057. doi: 10.1111/j.1462-5822.2007.01101.x. [DOI] [PubMed] [Google Scholar]
- 452.Pal R, Hoke GM, Sarngadharan MG. 1989. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:3384–3388. doi: 10.1073/pnas.86.9.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Schubert U, Strebel K. 1994. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J Virol 68:2260–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Haller C, Muller B, Fritz JV, Lamas-Murua M, Stolp B, Pujol FM, Keppler OT, Fackler OT. 2014. HIV-1 Nef and Vpu are functionally redundant broad-spectrum modulators of cell surface receptors, including tetraspanins. J Virol 88:14241–14257. doi: 10.1128/JVI.02333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Vassena L, Giuliani E, Koppensteiner H, Bolduan S, Schindler M, Doria M. 2015. HIV-1 Nef and Vpu interfere with l-selectin (CD62L) cell surface expression to inhibit adhesion and signaling in infected CD4+ T lymphocytes. J Virol 89:5687–5700. doi: 10.1128/JVI.00611-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Kueck T, Foster TL, Weinelt J, Sumner JC, Pickering S, Neil SJ. 2015. Serine phosphorylation of HIV-1 Vpu and its binding to tetherin regulates interaction with clathrin adaptors. PLoS Pathog 11:e1005141. doi: 10.1371/journal.ppat.1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.McNatt MW, Zang T, Bieniasz PD. 2013. Vpu binds directly to tetherin and displaces it from nascent virions. PLoS Pathog 9:e1003299. doi: 10.1371/journal.ppat.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, Xiong Y. 2014. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. eLife 3:e02362. doi: 10.7554/eLife.02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. 2009. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog 5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Skasko M, Wang Y, Tian Y, Tokarev A, Munguia J, Ruiz A, Stephens EB, Opella SJ, Guatelli J. 2012. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem 287:58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, Tokunaga K. 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem 284:35060–35072. doi: 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Kueck T, Neil SJ. 2012. A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog 8:e1002609. doi: 10.1371/journal.ppat.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Lim ES, Malik HS, Emerman M. 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J Virol 84:7124–7134. doi: 10.1128/JVI.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Serra-Moreno R, Zimmermann K, Stern LJ, Evans DT. 2013. Tetherin/BST-2 antagonism by Nef depends on a direct physical interaction between Nef and tetherin, and on clathrin-mediated endocytosis. PLoS Pathog 9:e1003487. doi: 10.1371/journal.ppat.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Rossi F, Gallina A, Milanesi G. 1996. Nef-CD4 physical interaction sensed with the yeast two-hybrid system. Virology 217:397–403. doi: 10.1006/viro.1996.0130. [DOI] [PubMed] [Google Scholar]
- 466.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 467.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Gotz N, Sauter D, Usmani SM, Fritz JV, Goffinet C, Heigele A, Geyer M, Bibollet-Ruche F, Learn GH, Fackler OT, Hahn BH, Kirchhoff F. 2012. Reacquisition of Nef-mediated tetherin antagonism in a single in vivo passage of HIV-1 through its original chimpanzee host. Cell Host Microbe 12:373–380. doi: 10.1016/j.chom.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Arias JF, Iwabu Y, Tokunaga K. 2012. Sites of action of HIV-1 Vpu in BST-2/tetherin downregulation. Curr HIV Res 10:283–291. doi: 10.2174/157016212800792423. [DOI] [PubMed] [Google Scholar]
- 470.Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, Evans DT. 2014. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc Natl Acad Sci U S A 111:6425–6430. doi: 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. 2014. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol 88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Pham TN, Lukhele S, Hajjar F, Routy JP, Cohen EA. 2014. HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology 11:15. doi: 10.1186/1742-4690-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Iwami S, Sato K, Morita S, Inaba H, Kobayashi T, Takeuchi JS, Kimura Y, Misawa N, Ren F, Iwasa Y, Aihara K, Koyanagi Y. 2015. Pandemic HIV-1 Vpu overcomes intrinsic herd immunity mediated by tetherin. Sci Rep 5:12256. doi: 10.1038/srep12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, Usmani SM, Sauter D, Joas S, Hotter D, Bibollet-Ruche F, Plenderleith LJ, Peeters M, Geyer M, Sharp PM, Fackler OT, Hahn BH, Kirchhoff F. 2014. Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell Host Microbe 16:639–650. doi: 10.1016/j.chom.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Ren X, Park SY, Bonifacino JS, Hurley JH. 2014. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. eLife 3:e01754. doi: 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Gondim MV, Wiltzer-Bach L, Maurer B, Banning C, Arganaraz E, Schindler M. 2015. AP-2 is the crucial clathrin adaptor protein for CD4 downmodulation by HIV-1 Nef in infected primary CD4+ T cells. J Virol 89:12518–12524. doi: 10.1128/JVI.01838-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.daSilva LL, Sougrat R, Burgos PV, Janvier K, Mattera R, Bonifacino JS. 2009. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J Virol 83:6578–6590. doi: 10.1128/JVI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Lambele M, Koppensteiner H, Symeonides M, Roy NH, Chan J, Schindler M, Thali M. 2015. Vpu is the main determinant for tetraspanin downregulation in HIV-1-infected cells. J Virol 89:3247–3255. doi: 10.1128/JVI.03719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. 2012. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol 86:4496–4504. doi: 10.1128/JVI.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Mulekar JJ, Huq E. 2014. Expanding roles of protein kinase CK2 in regulating plant growth and development. J Exp Bot 65:2883–2893. doi: 10.1093/jxb/ert401. [DOI] [PubMed] [Google Scholar]
- 481.Meggio F, Pinna LA. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J 17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 482.Paul M, Jabbar MA. 1997. Phosphorylation of both phosphoacceptor sites in the HIV-1 Vpu cytoplasmic domain is essential for Vpu-mediated ER degradation of CD4. Virology 232:207–216. doi: 10.1006/viro.1997.8541. [DOI] [PubMed] [Google Scholar]
- 483.Meggio F, Marin O, Boschetti M, Sarno S, Pinna LA. 2001. HIV-1 Rev transactivator: a beta-subunit directed substrate and effector of protein kinase CK2. Mol Cell Biochem 227:145–151. doi: 10.1023/A:1013177326481. [DOI] [PubMed] [Google Scholar]
- 484.Ohtsuki K, Maekawa T, Harada S, Karino A, Morikawa Y, Ito M. 1998. Biochemical characterization of HIV-1 Rev as a potent activator of casein kinase II in vitro. FEBS Lett 428:235–240. doi: 10.1016/S0014-5793(98)00538-9. [DOI] [PubMed] [Google Scholar]
- 485.Marin O, Sarno S, Boschetti M, Pagano MA, Meggio F, Ciminale V, D'Agostino DM, Pinna LA. 2000. Unique features of HIV-1 Rev protein phosphorylation by protein kinase CK2 (‘casein kinase-2′). FEBS Lett 481:63–67. doi: 10.1016/S0014-5793(00)01971-2. [DOI] [PubMed] [Google Scholar]
- 486.Callahan MA, Handley MA, Lee YH, Talbot KJ, Harper JW, Panganiban AT. 1998. Functional interaction of human immunodeficiency virus type 1 Vpu and Gag with a novel member of the tetratricopeptide repeat protein family. J Virol 72:5189–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Deora A, Spearman P, Ratner L. 2000. The N-terminal matrix domain of HIV-1 Gag is sufficient but not necessary for viral protein U-mediated enhancement of particle release through a membrane-targeting mechanism. Virology 269:305–312. doi: 10.1006/viro.1999.0094. [DOI] [PubMed] [Google Scholar]
- 488.Handley MA, Paddock S, Dall A, Panganiban AT. 2001. Association of Vpu-binding protein with microtubules and Vpu-dependent redistribution of HIV-1 Gag protein. Virology 291:198–207. doi: 10.1006/viro.2001.1166. [DOI] [PubMed] [Google Scholar]
- 489.Lee YH, Schwartz MD, Panganiban AT. 1997. The HIV-1 matrix domain of Gag is required for Vpu responsiveness during particle release. Virology 237:46–55. doi: 10.1006/viro.1997.8711. [DOI] [PubMed] [Google Scholar]
- 490.Harila K, Prior I, Sjoberg M, Salminen A, Hinkula J, Suomalainen M. 2006. Vpu and Tsg101 regulate intracellular targeting of the human immunodeficiency virus type 1 core protein precursor Pr55gag. J Virol 80:3765–3772. doi: 10.1128/JVI.80.8.3765-3772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Gautam A, Bhattacharya J. 2013. Evidence that Vpu modulates HIV-1 Gag-envelope interaction towards envelope incorporation and infectivity in a cell type dependent manner. PLoS One 8:e61388. doi: 10.1371/journal.pone.0061388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Gottlinger HG, Dorfman T, Cohen EA, Haseltine WA. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci U S A 90:7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog 2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Harila K, Salminen A, Prior I, Hinkula J, Suomalainen M. 2007. The Vpu-regulated endocytosis of HIV-1 Gag is clathrin-independent. Virology 369:299–308. doi: 10.1016/j.virol.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 495.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369–381. doi: 10.1016/S0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 496.Alvizo O, Mittal S, Mayo SL, Schiffer CA. 2012. Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease. Protein Sci 21:1029–1041. doi: 10.1002/pro.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Ozen A, Lin KH, Kurt Yilmaz N, Schiffer CA. 2014. Structural basis and distal effects of Gag substrate coevolution in drug resistance to HIV-1 protease. Proc Natl Acad Sci U S A 111:15993–15998. doi: 10.1073/pnas.1414063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Hill M, Tachedjian G, Mak J. 2005. The packaging and maturation of the HIV-1 Pol proteins. Curr HIV Res 3:73–85. doi: 10.2174/1570162052772942. [DOI] [PubMed] [Google Scholar]
- 499.Liu Z, Wang Y, Brunzelle J, Kovari IA, Kovari LC. 2011. Nine crystal structures determine the substrate envelope of the MDR HIV-1 protease. Protein J 30:173–183. doi: 10.1007/s10930-011-9316-2. [DOI] [PubMed] [Google Scholar]
- 500.Chaudhury S, Gray JJ. 2009. Identification of structural mechanisms of HIV-1 protease specificity using computational peptide docking: implications for drug resistance. Structure 17:1636–1648. doi: 10.1016/j.str.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Tie Y, Boross PI, Wang YF, Gaddis L, Liu F, Chen X, Tozser J, Harrison RW, Weber IT. 2005. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J 272:5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Li G, Verheyen J, Theys K, Piampongsant S, Van Laethem K, Vandamme A-M. 2014. HIV-1 Gag C-terminal amino acid substitutions emerging under selective pressure of protease inhibitors in patient populations infected with different HIV-1 subtypes. Retrovirology 11:79. doi: 10.1186/s12977-014-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Li G, Verheyen J, Rhee SY, Voet A, Vandamme A-M, Theys K. 2013. Functional conservation of HIV-1 gag: implications for rational drug design. Retrovirology 10:126. doi: 10.1186/1742-4690-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Li G, Theys K, Verheyen J, Pineda-Pena A, Khouri R, Piampongsant S, Eusebio M, Ramon J, Vandamme A-M. 2015. A new ensemble coevolution system for detecting HIV-1 protein coevolution. Biol Direct 10:1. doi: 10.1186/s13062-014-0031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Fun A, Wensing AM, Verheyen J, Nijhuis M. 2012. Human immunodeficiency virus Gag and protease: partners in resistance. Retrovirology 9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. 2004. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J Virol 78:8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. 2008. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature 455:693–696. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Paulus C, Hellebrand S, Tessmer U, Wolf H, Krausslich HG, Wagner R. 1999. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J Biol Chem 274:21539–21543. doi: 10.1074/jbc.274.31.21539. [DOI] [PubMed] [Google Scholar]
- 509.Louis JM, Dyda F, Nashed NT, Kimmel AR, Davies DR. 1998. Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease. Biochemistry 37:2105–2110. doi: 10.1021/bi972059x. [DOI] [PubMed] [Google Scholar]
- 510.Yu FH, Chou TA, Liao WH, Huang KJ, Wang CT. 2015. Gag-Pol transframe domain p6* is essential for HIV-1 protease-mediated virus maturation. PLoS One 10:e0127974. doi: 10.1371/journal.pone.0127974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Partin K, Zybarth G, Ehrlich L, DeCrombrugghe M, Wimmer E, Carter C. 1991. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 88:4776–4780. doi: 10.1073/pnas.88.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Ludwig C, Leiherer A, Wagner R. 2008. Importance of protease cleavage sites within and flanking human immunodeficiency virus type 1 transframe protein p6* for spatiotemporal regulation of protease activation. J Virol 82:4573–4584. doi: 10.1128/JVI.02353-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Chiu HC, Wang FD, Chen YM, Wang CT. 2006. Effects of human immunodeficiency virus type 1 transframe protein p6* mutations on viral protease-mediated Gag processing. J Gen Virol 87:2041–2046. doi: 10.1099/vir.0.81601-0. [DOI] [PubMed] [Google Scholar]
- 514.Huang L, Li Y, Chen C. 2011. Flexible catalytic site conformations implicated in modulation of HIV-1 protease autoprocessing reactions. Retrovirology 8:79. doi: 10.1186/1742-4690-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Paulus C, Ludwig C, Wagner R. 2004. Contribution of the Gag-Pol transframe domain p6* and its coding sequence to morphogenesis and replication of human immunodeficiency virus type 1. Virology 330:271–283. doi: 10.1016/j.virol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 516.Baraz L, Hutoran M, Blumenzweig I, Katzenellenbogen M, Friedler A, Gilon C, Steinitz M, Kotler M. 2002. Human immunodeficiency virus type 1 Vif binds the viral protease by interaction with its N-terminal region. J Gen Virol 83:2225–2230. doi: 10.1099/0022-1317-83-9-2225. [DOI] [PubMed] [Google Scholar]
- 517.Hutoran M, Britan E, Baraz L, Blumenzweig I, Steinitz M, Kotler M. 2004. Abrogation of Vif function by peptide derived from the N-terminal region of the human immunodeficiency virus type 1 (HIV-1) protease. Virology 330:261–270. doi: 10.1016/j.virol.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 518.Kotler M, Simm M, Zhao YS, Sova P, Chao W, Ohnona SF, Roller R, Krachmarov C, Potash MJ, Volsky DJ. 1997. Human immunodeficiency virus type 1 (HIV-1) protein Vif inhibits the activity of HIV-1 protease in bacteria and in vitro. J Virol 71:5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Baraz L, Friedler A, Blumenzweig I, Nussinuv O, Chen N, Steinitz M, Gilon C, Kotler M. 1998. Human immunodeficiency virus type 1 Vif-derived peptides inhibit the viral protease and arrest virus production. FEBS Lett 441:419–426. doi: 10.1016/S0014-5793(98)01602-0. [DOI] [PubMed] [Google Scholar]
- 520.Potash MJ, Bentsman G, Muir T, Krachmarov C, Sova P, Volsky DJ. 1998. Peptide inhibitors of HIV-1 protease and viral infection of peripheral blood lymphocytes based on HIV-1 Vif. Proc Natl Acad Sci U S A 95:13865–13868. doi: 10.1073/pnas.95.23.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Friedler A, Blumenzweig I, Baraz L, Steinitz M, Kotler M, Gilon C. 1999. Peptides derived from HIV-1 Vif: a non-substrate based novel type of HIV-1 protease inhibitors. J Mol Biol 287:93–101. doi: 10.1006/jmbi.1998.2585. [DOI] [PubMed] [Google Scholar]
- 522.Bottcher M, Grosse F. 1997. HIV-1 protease inhibits its homologous reverse transcriptase by protein-protein interaction. Nucleic Acids Res 25:1709–1714. doi: 10.1093/nar/25.9.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Goobar-Larsson L, Luukkonen BG, Unge T, Schwartz S, Utter G, Strandberg B, Oberg B. 1995. Enhancement of HIV-1 proteinase activity by HIV-1 reverse transcriptase. Virology 206:387–394. doi: 10.1016/S0042-6822(95)80054-9. [DOI] [PubMed] [Google Scholar]
- 524.Chiang CC, Tseng YT, Huang KJ, Pan YY, Wang CT. 2012. Mutations in the HIV-1 reverse transcriptase tryptophan repeat motif affect virion maturation and Gag-Pol packaging. Virology 422:278–287. doi: 10.1016/j.virol.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 525.Nishitsuji H, Yokoyama M, Sato H, Yamauchi S, Takaku H. 2011. Identification of amino acid residues in HIV-1 reverse transcriptase that are critical for the proteolytic processing of Gag-Pol precursors. FEBS Lett 585:3372–3377. doi: 10.1016/j.febslet.2011.09.034. [DOI] [PubMed] [Google Scholar]
- 526.Olivares I, Mulky A, Boross PI, Tozser J, Kappes JC, Lopez-Galindez C, Menendez-Arias L. 2007. HIV-1 protease dimer interface mutations that compensate for viral reverse transcriptase instability in infectious virions. J Mol Biol 372:369–381. doi: 10.1016/j.jmb.2007.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Dunn LL, Boyer PL, Clark PK, Hughes SH. 2013. Mutations in HIV-1 reverse transcriptase cause misfolding and miscleavage by the viral protease. Virology 444:241–249. doi: 10.1016/j.virol.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Gaedigk-Nitschko K, Schon A, Wachinger G, Erfle V, Kohleisen B. 1995. Cleavage of recombinant and cell derived human immunodeficiency virus 1 (HIV-1) Nef protein by HIV-1 protease. FEBS Lett 357:275–278. doi: 10.1016/0014-5793(94)01370-G. [DOI] [PubMed] [Google Scholar]
- 529.Miller MD, Warmerdam MT, Ferrell SS, Benitez R, Greene WC. 1997. Intravirion generation of the C-terminal core domain of HIV-1 Nef by the HIV-1 protease is insufficient to enhance viral infectivity. Virology 234:215–225. doi: 10.1006/viro.1997.8641. [DOI] [PubMed] [Google Scholar]
- 530.Chen YL, Trono D, Camaur D. 1998. The proteolytic cleavage of human immunodeficiency virus type 1 Nef does not correlate with its ability to stimulate virion infectivity. J Virol 72:3178–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Bukovsky AA, Dorfman T, Weimann A, Gottlinger HG. 1997. Nef association with human immunodeficiency virus type 1 virions and cleavage by the viral protease. J Virol 71:1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Schorr J, Kellner R, Fackler O, Freund J, Konvalinka J, Kienzle N, Krausslich HG, Mueller-Lantzsch N, Kalbitzer HR. 1996. Specific cleavage sites of Nef proteins from human immunodeficiency virus types 1 and 2 for the viral proteases. J Virol 70:9051–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Welker R, Harris M, Cardel B, Krausslich HG. 1998. Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: analysis of its role in enhancement of viral infectivity. J Virol 72:8833–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Pandori M, Craig H, Moutouh L, Corbeil J, Guatelli J. 1998. Virological importance of the protease-cleavage site in human immunodeficiency virus type 1 Nef is independent of both intravirion processing and CD4 down-regulation. Virology 251:302–316. doi: 10.1006/viro.1998.9407. [DOI] [PubMed] [Google Scholar]
- 535.Akari H, Arold S, Fukumori T, Okazaki T, Strebel K, Adachi A. 2000. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol 74:2907–2912. doi: 10.1128/JVI.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Kotov A, Zhou J, Flicker P, Aiken C. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J Virol 73:8824–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Mendonca LM, Poeys SC, Abreu CM, Tanuri A, Costa LJ. 2014. HIV-1 Nef inhibits protease activity and its absence alters protein content of mature viral particles. PLoS One 9:e95352. doi: 10.1371/journal.pone.0095352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Waheed AA, Ablan SD, Roser JD, Sowder RC, Schaffner CP, Chertova E, Freed EO. 2007. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc Natl Acad Sci U S A 104:8467–8471. doi: 10.1073/pnas.0701443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Waheed AA, Ablan SD, Sowder RC, Roser JD, Schaffner CP, Chertova E, Freed EO. 2010. Effect of mutations in the human immunodeficiency virus type 1 protease on cleavage of the gp41 cytoplasmic tail. J Virol 84:3121–3126. doi: 10.1128/JVI.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Bardy M, Gay B, Pebernard S, Chazal N, Courcoul M, Vigne R, Decroly E, Boulanger P. 2001. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: co-encapsidation and interference with viral protease-mediated Gag processing. J Gen Virol 82:2719–2733. doi: 10.1099/0022-1317-82-11-2719. [DOI] [PubMed] [Google Scholar]
- 541.Dorfman T, Popova E, Pizzato M, Gottlinger HG. 2002. Nef enhances human immunodeficiency virus type 1 infectivity in the absence of matrix. J Virol 76:6857–6862. doi: 10.1128/JVI.76.13.6857-6862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Swingler S, Gallay P, Camaur D, Song J, Abo A, Trono D. 1997. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J Virol 71:4372–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Costa LJ, Zheng YH, Sabotic J, Mak J, Fackler OT, Peterlin BM. 2004. Nef binds p6* in GagPol during replication of human immunodeficiency virus type 1. J Virol 78:5311–5323. doi: 10.1128/JVI.78.10.5311-5323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Ono T, Iwatani Y, Nishimura A, Ishimoto A, Sakai H. 2000. Functional association between the nef gene product and gag-pol region of HIV-1. FEBS Lett 466:233–238. doi: 10.1016/S0014-5793(99)01791-3. [DOI] [PubMed] [Google Scholar]
- 545.Leiherer A, Ludwig C, Wagner R. 2009. Influence of extended mutations of the HIV-1 transframe protein p6 on Nef-dependent viral replication and infectivity in vitro. Virology 387:200–210. doi: 10.1016/j.virol.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 546.Laguette N, Benichou S, Basmaciogullari S. 2009. Human immunodeficiency virus type 1 Nef incorporation into virions does not increase infectivity. J Virol 83:1093–1104. doi: 10.1128/JVI.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Guy B, Geist M, Dott K, Spehner D, Kieny MP, Lecocq JP. 1991. A specific inhibitor of cysteine proteases impairs a Vif-dependent modification of human immunodeficiency virus type 1 Env protein. J Virol 65:1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Akari H, Yoshida A, Fukumori T, Adachi A. 2000. Host cell-dependent replication of HIV-1 mutants with deletions in gp41 cytoplasmic tail region is independent of the function of Vif. Microbes Infect 2:1019–1023. doi: 10.1016/S1286-4579(00)01256-9. [DOI] [PubMed] [Google Scholar]
- 550.Akari H, Uchiyama T, Fukumori T, Iida S, Koyama AH, Adachi A. 1999. Pseudotyping human immunodeficiency virus type 1 by vesicular stomatitis virus G protein does not reduce the cell-dependent requirement of vif for optimal infectivity: functional difference between Vif and Nef. J Gen Virol 80(Part 11):2945–2949. doi: 10.1099/0022-1317-80-11-2945. [DOI] [PubMed] [Google Scholar]
- 551.Ma XY, Sova P, Chao W, Volsky DJ. 1994. Cysteine residues in the Vif protein of human immunodeficiency virus type 1 are essential for viral infectivity. J Virol 68:1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Votteler J, Neumann L, Hahn S, Hahn F, Rauch P, Schmidt K, Studtrucker N, Solbak SM, Fossen T, Henklein P, Ott DE, Holland G, Bannert N, Schubert U. 2011. Highly conserved serine residue 40 in HIV-1 p6 regulates capsid processing and virus core assembly. Retrovirology 8:11. doi: 10.1186/1742-4690-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. 2015. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol 89:9765–9780. doi: 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.De Clercq E. 2007. The design of drugs for HIV and HCV. Nat Rev Drug Discov 6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 555.De Clercq E, Holý A. 2005. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat Rev Drug Discov 4:928–940. doi: 10.1038/nrd1877. [DOI] [PubMed] [Google Scholar]
- 556.Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2015. 2015 update of the drug resistance mutations in HIV-1. Top Antivir Med 23:132–141. [PMC free article] [PubMed] [Google Scholar]
- 557.Hu Z, Kuritzkes DR. 2014. Altered viral fitness and drug susceptibility in HIV-1 carrying mutations that confer resistance to nonnucleoside reverse transcriptase and integrase strand transfer inhibitors. J Virol 88:9268–9276. doi: 10.1128/JVI.00695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Flynn WF, Chang MW, Tan Z, Oliveira G, Yuan J, Okulicz JF, Torbett BE, Levy RM. 2015. Deep sequencing of protease inhibitor resistant HIV patient isolates reveals patterns of correlated mutations in Gag and protease. PLoS Comput Biol 11:e1004249. doi: 10.1371/journal.pcbi.1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.De Clercq E. 2009. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int J Antimicrob Agents 33:307–320. doi: 10.1016/j.ijantimicag.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 560.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Derdeyn CA, Decker JM, Sfakianos JN, Zhang Z, O'Brien WA, Ratner L, Shaw GM, Hunter E. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J Virol 75:8605–8614. doi: 10.1128/JVI.75.18.8605-8614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Roche M, Salimi H, Duncan R, Wilkinson BL, Chikere K, Moore MS, Webb NE, Zappi H, Sterjovski J, Flynn JK, Ellett A, Gray LR, Lee B, Jubb B, Westby M, Ramsland PA, Lewin SR, Payne RJ, Churchill MJ, Gorry PR. 2013. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology 10:43. doi: 10.1186/1742-4690-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Anastassopoulou CG, Ketas TJ, Klasse PJ, Moore JP. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc Natl Acad Sci U S A 106:5318–5323. doi: 10.1073/pnas.0811713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Anastassopoulou CG, Ketas TJ, Sanders RW, Klasse PJ, Moore JP. 2012. Effects of sequence changes in the HIV-1 gp41 fusion peptide on CCR5 inhibitor resistance. Virology 428:86–97. doi: 10.1016/j.virol.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Anastassopoulou CG, Ketas TJ, Depetris RS, Thomas AM, Klasse PJ, Moore JP. 2011. Resistance of a human immunodeficiency virus type 1 isolate to a small molecule CCR5 inhibitor can involve sequence changes in both gp120 and gp41. Virology 413:47–59. doi: 10.1016/j.virol.2010.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.De Feo CJ, Weiss CD. 2012. Escape from human immunodeficiency virus type 1 (HIV-1) entry inhibitors. Viruses 4:3859–3911. doi: 10.3390/v4123859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Weber J, Rose JD, Vazquez AC, Winner D, Margot N, McColl DJ, Miller MD, Quinones-Mateu ME. 2013. Resistance mutations outside the integrase coding region have an effect on human immunodeficiency virus replicative fitness but do not affect its susceptibility to integrase strand transfer inhibitors. PLoS One 8:e65631. doi: 10.1371/journal.pone.0065631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Arts EJ, Hazuda DJ. 2012. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Siliciano JD, Siliciano RF. 2013. Recent trends in HIV-1 drug resistance. Curr Opin Virol 3:487–494. doi: 10.1016/j.coviro.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Stray KM, Callebaut C, Glass B, Tsai L, Xu L, Muller B, Krausslich HG, Cihlar T. 2013. Mutations in multiple domains of Gag drive the emergence of in vitro resistance to the phosphonate-containing HIV-1 protease inhibitor GS-8374. J Virol 87:454–463. doi: 10.1128/JVI.01211-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Adekale MA, Cane PA, McCrae MA. 2005. Changes in the Vif protein of HIV-1 associated with the development of resistance to inhibitors of viral protease. J Med Virol 75:195–201. doi: 10.1002/jmv.20256. [DOI] [PubMed] [Google Scholar]
- 572.Fumakia M, Yang S, Gu J, Ho EA. 26 August 2015. Protein/peptide-based entry/fusion inhibitors as anti-HIV therapies: challenges and future direction. Rev Med Virol doi: 10.1002/rmv.1853. [DOI] [PubMed] [Google Scholar]
- 573.Chong H, Wu X, Su Y, He Y. 15 May 2016. Development of potent and long-acting HIV-1 fusion inhibitors. AIDS doi: 10.1097/QAD.0000000000001073. [DOI] [PubMed] [Google Scholar]
- 574.Oz Gleenberg I, Avidan O, Goldgur Y, Herschhorn A, Hizi A. 2005. Peptides derived from the reverse transcriptase of human immunodeficiency virus type 1 as novel inhibitors of the viral integrase. J Biol Chem 280:21987–21996. doi: 10.1074/jbc.M414679200. [DOI] [PubMed] [Google Scholar]
- 575.Zawahir Z, Neamati N. 2006. Inhibition of HIV-1 integrase activity by synthetic peptides derived from the HIV-1 HXB2 Pol region of the viral genome. Bioorg Med Chem Lett 16:5199–5202. doi: 10.1016/j.bmcl.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 576.Oz Gleenberg I, Herschhorn A, Goldgur Y, Hizi A. 2007. Inhibition of human immunodeficiency virus type-1 reverse transcriptase by a novel peptide derived from the viral integrase. Arch Biochem Biophys 458:202–212. doi: 10.1016/j.abb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 577.Nomura W, Aikawa H, Ohashi N, Urano E, Metifiot M, Fujino M, Maddali K, Ozaki T, Nozue A, Narumi T, Hashimoto C, Tanaka T, Pommier Y, Yamamoto N, Komano JA, Murakami T, Tamamura H. 2013. Cell-permeable stapled peptides based on HIV-1 integrase inhibitors derived from HIV-1 gene products. ACS Chem Biol 8:2235–2244. doi: 10.1021/cb400495h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Suzuki S, Urano E, Hashimoto C, Tsutsumi H, Nakahara T, Tanaka T, Nakanishi Y, Maddali K, Han Y, Hamatake M, Miyauchi K, Pommier Y, Beutler JA, Sugiura W, Fuji H, Hoshino T, Itotani K, Nomura W, Narumi T, Yamamoto N, Komano JA, Tamamura H. 2010. Peptide HIV-1 integrase inhibitors from HIV-1 gene products. J Med Chem 53:5356–5360. doi: 10.1021/jm1003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Davis DA, Tebbs IR, Daniels SI, Stahl SJ, Kaufman JD, Wingfield P, Bowman MJ, Chmielewski J, Yarchoan R. 2009. Analysis and characterization of dimerization inhibition of a multi-drug-resistant human immunodeficiency virus type 1 protease using a novel size-exclusion chromatographic approach. Biochem J 419:497–506. doi: 10.1042/BJ20082068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Davis DA, Brown CA, Singer KE, Wang V, Kaufman J, Stahl SJ, Wingfield P, Maeda K, Harada S, Yoshimura K, Kosalaraksa P, Mitsuya H, Yarchoan R. 2006. Inhibition of HIV-1 replication by a peptide dimerization inhibitor of HIV-1 protease. Antiviral Res 72:89–99. doi: 10.1016/j.antiviral.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 581.Fry DC. 2012. Small-molecule inhibitors of protein-protein interactions: how to mimic a protein partner. Curr Pharm Des 18:4679–4684. doi: 10.2174/138161212802651634. [DOI] [PubMed] [Google Scholar]
- 582.Morelli X, Bourgeas R, Roche P. 2011. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I). Curr Opin Chem Biol 15:475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 583.de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, Andre P, Lotteau V. 2012. New horizons for antiviral drug discovery from virus-host protein interaction networks. Curr Opin Virol 2:606–613. doi: 10.1016/j.coviro.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 584.Koes DR, Camacho CJ. 2012. Small-molecule inhibitor starting points learned from protein-protein interaction inhibitor structure. Bioinformatics 28:784–791. doi: 10.1093/bioinformatics/btr717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Dube M, Bego MG, Paquay C, Cohen EA. 2010. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology 7:114. doi: 10.1186/1742-4690-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Xue B, Mizianty MJ, Kurgan L, Uversky VN. 2012. Protein intrinsic disorder as a flexible armor and a weapon of HIV-1. Cell Mol Life Sci 69:1211–1259. doi: 10.1007/s00018-011-0859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Hemelaar J. 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol Med 18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 588.Wertheim JO, Worobey M. 2009. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol 5:e1000377. doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther 9:57–65. [PubMed] [Google Scholar]
- 590.Varthakavi V, Smith RM, Bour SP, Strebel K, Spearman P. 2003. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc Natl Acad Sci U S A 100:15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Gabuzda DH, Lever A, Terwilliger E, Sodroski J. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 66:3306–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Casella CR, Raffini LJ, Panganiban AT. 1997. Pleiotropic mutations in the HIV-1 matrix protein that affect diverse steps in replication. Virology 228:294–306. doi: 10.1006/viro.1996.8355. [DOI] [PubMed] [Google Scholar]
- 593.Louis JM, Clore GM, Gronenborn AM. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat Struct Biol 6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 594.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.da Costa KS, Leal E, dos Santos AM, Lima e Lima AH, Alves CN, Lameira J. 2014. Structural analysis of viral infectivity factor of HIV type 1 and its interaction with A3G, EloC and EloB. PLoS One 9:e89116. doi: 10.1371/journal.pone.0089116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Rice AP, Kimata JT. 2015. Subversion of cell cycle regulatory mechanisms by HIV. Cell Host Microbe 17:736–740. doi: 10.1016/j.chom.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Foster JL, Garcia JV. 2008. HIV-1 Nef: at the crossroads. Retrovirology 5:84. doi: 10.1186/1742-4690-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. doi: 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 600.Santos da Silva E, Mulinge M, Perez Bercoff D. 2013. The frantic play of the concealed HIV envelope cytoplasmic tail. Retrovirology 10:54. doi: 10.1186/1742-4690-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Ferrucci A, Nonnemacher MR, Wigdahl B. 2011. Human immunodeficiency virus viral protein R as an extracellular protein in neuropathogenesis. Adv Virus Res 81:165–199. doi: 10.1016/B978-0-12-385885-6.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM. 2010. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Loret E. 2015. HIV extracellular Tat: myth or reality? Curr HIV Res 13:90–97. doi: 10.2174/1570162X12666141202125643. [DOI] [PubMed] [Google Scholar]
- 604.Roeth JF, Collins KL. 2006. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev 70:548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Wu L, KewalRamani VN. 2006. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat Rev Immunol 6:859–868. doi: 10.1038/nri1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC. 2014. Lipid14: the amber lipid force field. J Chem Theory Comput 10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Kwong PD, Mascola JR, Nabel GJ. 2011. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harb Perspect Med 1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Ward AB, Wilson IA. 2015. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Zheng X, Perera L, Mueller GA, DeRose EF, London RE. 3 June 2015. Asymmetric conformational maturation of HIV-1 reverse transcriptase. eLife doi: 10.7554/eLife.06359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Li D, Wei T, Rawle DJ, Qin F, Wang R, Soares DC, Jin H, Sivakumaran H, Lin MH, Spann K, Abbott CM, Harrich D. 2015. Specific interaction between eEF1A and HIV RT is critical for HIV-1 reverse transcription and a potential anti-HIV target. PLoS Pathog 11:e1005289. doi: 10.1371/journal.ppat.1005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Bukrinskaya A, Brichacek B, Mann A, Stevenson M. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med 188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Fouchier RA, Meyer BE, Simon JH, Fischer U, Albright AV, Gonzalez-Scarano F, Malim MH. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol 72:6004–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. 2007. HIV-1 DNA flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J 26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Ping YH, Rana TM. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 276:12951–12958. doi: 10.1074/jbc.M006130200. [DOI] [PubMed] [Google Scholar]
- 615.Zhou C, Rana TM. 2002. A bimolecular mechanism of HIV-1 Tat protein interaction with RNA polymerase II transcription elongation complexes. J Mol Biol 320:925–942. doi: 10.1016/S0022-2836(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 616.Isel C, Karn J. 1999. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol 290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- 617.Murakami T. 2012. Retroviral env glycoprotein trafficking and incorporation into virions. Mol Biol Int 2012:682850. doi: 10.1155/2012/682850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Nguyen DH, Hildreth JE. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol 74:3264–3272. doi: 10.1128/JVI.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Trubey CM, Chertova E, Coren LV, Hilburn JM, Hixson CV, Nagashima K, Lifson JD, Ott DE. 2003. Quantitation of HLA class II protein incorporated into human immunodeficiency type 1 virions purified by anti-CD45 immunoaffinity depletion of microvesicles. J Virol 77:12699–12709. doi: 10.1128/JVI.77.23.12699-12709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Koonin EV, Dolja VV. 2014. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev 78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Hill MK, Hooker CW, Harrich D, Crowe SM, Mak J. 2001. Gag-Pol supplied in trans is efficiently packaged and supports viral function in human immunodeficiency virus type 1. J Virol 75:6835–6840. doi: 10.1128/JVI.75.15.6835-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Sadiq SK, Noe F, De Fabritiis G. 2012. Kinetic characterization of the critical step in HIV-1 protease maturation. Proc Natl Acad Sci U S A 109:20449–20454. doi: 10.1073/pnas.1210983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Shehu-Xhilaga M, Kraeusslich HG, Pettit S, Swanstrom R, Lee JY, Marshall JA, Crowe SM, Mak J. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J Virol 75:9156–9164. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Frank GA, Narayan K, Bess JW Jr, Del Prete GQ, Wu X, Moran A, Hartnell LM, Earl LA, Lifson JD, Subramaniam S. 2015. Maturation of the HIV-1 core by a non-diffusional phase transition. Nat Commun 6:5854. doi: 10.1038/ncomms6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.