Abstract
Genetic variation is critical to the persistence of populations and their capacity to adapt to environmental change. The distribution of genetic variation across a species' range can reveal critical information that is not necessarily represented in species occurrence or abundance patterns. We identified environmental factors associated with the amount of intraspecific, individual-based genetic variation across the range of a widespread freshwater fish species, the Murray cod Maccullochella peelii. We used two different approaches to statistically quantify the relative importance of predictor variables, allowing for nonlinear relationships: a random forest model and a Bayesian approach. The latter also accounted for population history. Both approaches identified associations between homozygosity by locus and both disturbance to the natural flow regime and mean annual flow. Homozygosity by locus was negatively associated with disturbance to the natural flow regime, suggesting that river reaches with more disturbed flow regimes may support larger, more genetically diverse populations. Our findings are consistent with the hypothesis that artificially induced perennial flows in regulated channels may provide greater and more consistent habitat and reduce the frequency of population bottlenecks that can occur frequently under the highly variable and unpredictable natural flow regime of the system. Although extensive river regulation across eastern Australia has not had an overall positive effect on Murray cod numbers over the past century, regulation may not represent the primary threat to Murray cod survival. Instead, pressures other than flow regulation may be more critical to the persistence of Murray cod (for example, reduced frequency of large floods, overfishing and chemical pollution).
Introduction
Genetic variation is fundamentally linked to effective population size and evolutionary potential (that is, capacity to adapt to environmental change) (Frankham, 1996; Reed and Frankham, 2003; Scoble and Lowe, 2010; Harrisson et al., 2014). By integrating information about evolutionary and demographic processes, the distribution of genetic variation across a species' range can reveal critical insights about the viability of populations that is not necessarily represented in species occurrence or abundance patterns (Thomassen et al., 2010; Kovach et al., 2015). Understanding the distribution of intraspecific genetic variation and how it relates to environmental factors can assist better allocation of conservation resources and more robust predictions about species responses to environmental change (Frankel, 1974; Moritz, 2002; Vandergast et al., 2008; Scoble and Lowe, 2010; Thomassen et al., 2010; Sgrò et al., 2011; Gotelli and Stanton-Geddes, 2015). Although many studies have explored links between environmental factors and species' occurrence or abundance patterns (Elith and Leathwick, 2009), few studies have explicitly and rigorously quantified associations between the environment and levels of intraspecific genetic variation across species' ranges (that is, with individual- or site-based measures of the amount of genetic variation as the response variable) (but see Thomassen et al., 2010; Kovach et al., 2015). Methods based on individual- or site-based measures of genetic variation (that is, amount of genetic diversity) are distinguished here from related methods based on population structure and/or patterns of genetic differentiation (for example, Foll and Gaggiotti, 2006; Jay et al., 2012; Manel and Holderegger, 2013; Wang and Bradburd, 2014; Fitzpatrick and Keller, 2015).
Often characterised by strong environmental gradients, rivers are useful systems for exploring links between environmental factors and species' distributions. Flow-related variables are commonly identified as important predictors of species occurrence patterns in rivers, with flow considered a ‘master variable' strongly associated with habitat complexity, geomorphology and many key physicochemical properties of rivers including temperature, pH and oxygen concentration (Poff et al., 1997). Disruption to a river's natural flow regime as a result of human activities (for example, flow regulation, construction of dams and weirs and water extraction) can have negative consequences for biodiversity, but may benefit some species (Bond et al., 2010; Koehn et al., 2014). For example, in environments with highly variable and unpredictable natural flow regimes, flow-dependent species may benefit from artificially induced perennial flows (Bond et al., 2010). In addition to flows, a wide range of other factors are commonly implicated as drivers of species' occurrences in rivers, including annual climatic extremes, riparian vegetation, presence of introduced species and levels of basin connectivity (Rathert et al., 1999).
The Murray–Darling Basin (MDB) covers more than 1 million km2 of south eastern Australia and is characterised by a flow regime that, in its natural state, is among the most variable in the world (Finlayson and McMahon, 1988). The MDB has experienced considerable change in land use and river condition over the past two centuries following European settlement. As a result, native fish populations in the MDB have declined to ∼10% of pre-European levels because of the combined influences of multiple pressures including habitat loss and fragmentation, altered flow regimes, introduced fishes and overfishing—pressures consistent with reductions in freshwater fish numbers in many river systems globally (Barrett, 2004).
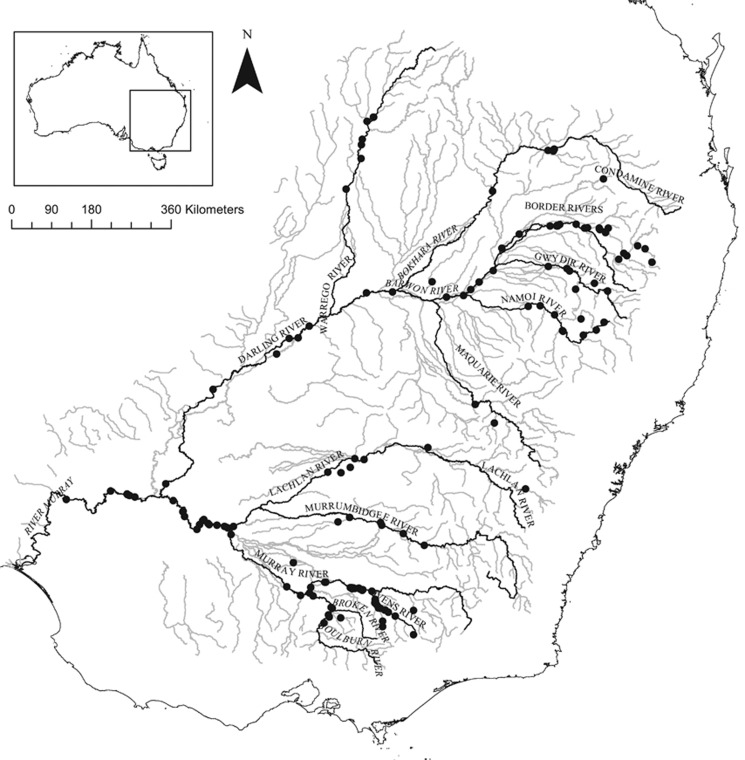
The Murray cod Maccullochella peelii (Mitchell, 1838) is a large, long-lived, endemic freshwater fish species that occurs through much of the MDB. The Murray cod is estimated to live for up to ~50 years (possibly >100 years) and can grow to nearly 2 m and >100 kg, although individuals >50 kg are now rare (Rowland, 1989). Murray cod naturally occur in both faster-flowing, cooler, rocky, upstream river reaches and larger, turbid, slower-flowing lowland rivers, with an apparent preference for deep water and habitat with overhanging vegetation and in-stream woody habitat, the latter being important for shelter and spawning (Lintermans et al., 2005). The Murray cod is capable of large migratory and dispersal distances of up to 1500 km, but movements are typically limited to distances <200 km (Koehn et al., 2009). These movements are typically associated with upstream spawning migrations, with subsequent downstream larval drift, and Murray cod tend to exhibit strong site-fidelity outside of the spawning season (Koehn et al., 2009). Spawning is temperature-cued, occurring in spring once temperatures reach 15 °C (Koehn and Harrington, 2006). Subsequent larval survival and recruitment success is thought to be linked to the occurrence of overbank flooding, although there remains uncertainty surrounding the influence of flows on the distribution and abundance of Murray cod (Rowland, 1989; Koehn and Harrington, 2006; King et al., 2009). Although thermal impacts (cold-water pollution) are known to influence egg and larval survival (Todd et al., 2005), the species has been demonstrated to spawn annually in both regulated and unregulated systems (Humphries, 2005; Koehn and Harrington, 2006; King et al., 2009). The larval duration of the Murray cod is typically <1 month, and sexual maturity is reach at ∼4–6 years of age (Rowland, 2004). Terminal wetlands partially isolate the Lachlan, Macquarie and Gwydir catchments, and these catchments are modestly genetically differentiated (mean pairwise FST=0.06; range: 0.03–0.13) (Figure 1; FST values from Appendix VI in Rourke et al., 2011). The remaining major river catchments in the Murray Darling Basin are connected by moderate to high levels of gene flow (mean pairwise FST=0.01; range: 0–0.04)(Figure 1; FST values from Appendix V1 in Rourke et al., 2011).
Figure 1.
Sampling locations of Murray cod (N=616) across the MDB are indicated by black circles. Major streams in the MDB are shown in grey and the major rivers sampled are labelled and highlighted in black. Genotype data for samples used in this study were previously published in Rourke et al. (2011).
Despite being relatively widespread, the Murray cod is listed nationally as vulnerable under the Environment Protection and Biodiversity Conservation (EPBC) Act 1999 and as a threatened species in the State of Victoria under the Victorian Flora and Fauna Guarantee (FFG) Act 1988. Overharvesting as a result of commercial and recreational fishing contributed to substantial declines of Murray cod through the first half of the twentieth century, particularly in the southern tributaries (Rowland, 1989; Lintermans et al., 2005). Additional pressures that likely exacerbated declines across the MDB included chemical pollution, removal of woody in-stream habitat (‘de-snagging'), introduction of alien fish species, establishment of dams and weirs that pose barriers to fish movement and changes to flow, flood and temperature regimes associated with river flow regulation (Rowland, 1989; Lintermans et al., 2005). A contraction in the distribution of Murray cod through the 1900s was particularly evident in waters impounded by major dams, the upper reaches of major rivers and smaller tributaries (Lintermans et al., 2005).
After a population crash in the 1960s, commercial fisheries were no longer viable, and Murray cod stocks have remained at relatively low levels since (Lintermans et al., 2005). Although wild Murray cod are no longer fished commercially (the last fishery closed in 2003), recreational fishing is still permitted (Lintermans et al., 2005). Supplementation of wild populations using hatchery-reared stock has been used since the 1970s to support the continuation of recreational fishing, with tens of millions of fish released across the MDB (mostly as fingerlings into impoundments) (Lintermans et al., 2005; Rourke et al., 2010; Rourke et al., 2011). Limited monitoring of recruitment success of stocked individuals means that the effects of stocking on wild populations are not well understood. Evidence of localised effects of stocking exists in some catchments. For example, a proportion of genetically nonadmixed individuals in the Warrego, Namoi and Border rivers may indicate the presence of stocked individuals (Rourke et al., 2011). However, across large areas of the MDB (thousands of river km across the southern parts of the basin and the more northern Darling and Condamine catchments), stocking has not had detectable effects on patterns of genetic diversity or population structure (Rourke et al., 2010, 2011).
Given the cultural and socioeconomic importance of the Murray cod, there is strong interest in ensuring its persistence. Understanding the environmental conditions associated with large, healthy populations will be key to effective conservation management. In this study we explored associations between patterns of individual-based genetic variation across the Murray cod's range and a set of environmental variables considered relevant to the biology and life history of Murray cod (including variables related to flow regime, habitat, connectivity, temperature and invasive species).
Materials and methods
Sampling
We used existing genotype data for 16 microsatellite loci that were previously published in Rourke et al. (2011), removing individuals with data missing for >20% of loci. Murray cod samples used here (N=616) were sourced from 15 major river catchments across the MDB (Figure 1 and Table 1) and collected between 1994 and 2006 (87% were sampled between 2000 and 2006). Given Murray cod reach sexual maturity at ∼4–6 years of age, tend to be more fecund at older ages and can live up to ~50 years, the genetic composition of populations is expected to change over decadal—rather than annual—timescales (Lintermans et al., 2005). Because the Murray cod is a large-bodied fish, the species (and therefore sampling) is typically associated with more permanent water bodies (larger streams and river channels). Fish were predominantly sampled from rivers (cf., dams or reservoirs) to avoid sampling large numbers of closely related stocked fish (Rourke et al., 2011) (Table 1). Previous analysis of age class distributions and genetic information suggested that populations did not consist of batches of closely related fish from hatchery or wild spawnings (Rourke et al., 2011).
Table 1. Number of individuals (N individuals), number of sites (N sites), mean number of individuals per site (mean N individuals per site), number of individuals stocked between 1978 and 2006 (N individuals stocked) and the percentage stocked into impoundments (as opposed to rivers) (% stocked into impoundments) and mean homozygosity by locus (mean HL) for each catchment analysed in this study.
| Catchment | N individuals | N sites | Mean N individuals per site | N individuals stocked | % Stocked into impoundments | Mean HL |
|---|---|---|---|---|---|---|
| Benanee | 31 | 10 | 3 | 84 000 | 0 | 0.27 |
| Border | 90 | 22 | 4 | 732 000 | 66 | 0.29 |
| Condamine | 31 | 11 | 3 | 181 000 | 95 | 0.23 |
| Darling | 22 | 7 | 3 | 60 000 | 32 | 0.26 |
| Goulburn-Broken | 19 | 7 | 3 | 1 117 000 | 47 | 0.25 |
| Gwydir | 30 | 6 | 5 | 508 000 | 90 | 0.29 |
| Kiewa | 12 | 1 | — | 80 000 | 0 | 0.28 |
| Lachlan | 40 | 7 | 6 | 707 000 | 93 | 0.26 |
| Lower Murray | 72 | 7 | 10 | 152 000 | 17 | 0.35 |
| Macquarie | 16 | 2 | 8 | 1 095 000 | 89 | 0.30 |
| Murray-Riverina | 123 | 27 | 5 | 656 000 | 1 | 0.33 |
| Murrumbidgee | 41 | 7 | 6 | 2 007 000 | 90 | 0.32 |
| Namoi | 44 | 9 | 5 | 853 000 | 88 | 0.28 |
| Ovens | 38 | 14 | 3 | 106 000 | 2 | 0.27 |
| Warrego | 7 | 7 | 1 | 8000 | 82 | 0.18 |
Environmental data
We selected 11 variables that represent the environmental attributes considered most relevant to the biology and life history of Murray cod populations (such as flow, connectivity, invasive species, habitat and temperature; details in Table 2). Because of the large geographic scale of our sampling, data availability placed some limitations on the environmental variables we were able to include. For example, detailed physicochemical data (for example, pH, oxygen concentration, water temperature) and site-specific habitat information were unavailable, and hence we relied on proxies (Table 2). Similarly, in the absence of measured flow data, we relied on partially modelled accumulated soil water surplus (run-off) to characterise flow attributes (Table 2). Use of accumulated soil water surplus as a proxy for flow is an established practice and the two are usually highly correlated (Stein et al., 2002). Most of the environmental data were sourced from the National Environmental Stream Attributes Database v1.1.5 (available at: http://www.ga.gov.au/) that is linked to the Geofabric digital elevation model (DEM)-derived stream network (Australian Hydrological Geospatial Fabric Version 2; available at: https://data.gov.au/). Each segment in the stream network was linked with its natural and anthropogenic characteristics from the National Environmental Stream Attributes Database based on its stream segment number. Mean stream segment length in our study was 6.9 km (range 0.2–43.5 km). Additional environmental variables were sourced from the Sustainable Rivers Audit (MDBA, 2012) and the Bureau of Meteorology (available at: http://www.bom.gov.au) (Table 2). The selected variables were chosen based on expert advice and limited to those that showed substantial variation across sampled sites. All pairs of variables had Pearson's correlation coefficients (r) <0.7, and hence our inferences were unlikely to be affected by collinearity of variables (Dormann et al., 2013) (Supplementary Appendix S1). Environmental data were extracted for each sampling location using ArcMap 10.2 (ESRI).
Table 2. List of environmental variables included in the models, with their category, unit of measurement, time period over which environmental data were calculated, justification for inclusion, references supporting inclusion and source of environmental data.
| Variable | Category | Units | Calculation period | Justification | Supporting references | Source |
|---|---|---|---|---|---|---|
| Annual mean accumulated soil water surplus | Flow | ML | 1970–2008 | Proxy for flow; considered a ‘master variable' likely to be correlated with habitat size, habitat complexity and physicochemical properties of rivers | (Poff et al., 1997; Stein et al., 2002; Bond et al., 2011) | NESAD |
| Coefficient of variation of annual totals of accumulated soil water surplus | Flow | NA | 1970–2008 | Proxy for variability in flows across years; should reflect long-term stability of water availability | (Bond et al., 2011) | NESAD |
| Mean accumulated soil water surplus for summer | Flow | ML | 1970–2008 | Proxy for summer flows; should reflect annual stability of water availability | (Bond et al., 2010; Bond et al., 2011) | NESAD |
| Flow regime disturbance index | Flow | Index scored between zero and one | 1989–1997 | Indicative of disruption to the natural flow regime; should reflect extent of flow regulation in rivers | (Bond et al., 2010; Koehn et al., 2014) | NESAD |
| Barrier free flow path length | Connectivity | % | 1990 | Measure of connectedness of streams; reflects number of dams and weirs that may pose barriers to fish movement | (Rathert et al., 1999; Lintermans et al., 2005) | NESAD |
| Baseline annual mean net primary productivity | Energy | tC ha−1 | 1788 | Proxy for long-term energy availability; should reflect long-term productivity of rivers | (Guégan et al., 1998) | NESAD |
| Stream and valley percentage extant forest cover | Habitat | % | 2001–2004 | Type and extent of vegetation surrounding stream; may influence the amount and type of overhanging vegetation and in-stream woody habitat | (Lintermans et al., 2005) | NESAD |
| Stream and valley percentage extant woodland cover | Habitat | % | 2001–2004 | Type and extent of vegetation surrounding stream; may influence the amount and type of overhanging vegetation and in-stream woody habitat | (Lintermans et al., 2005) | NESAD |
| Fish condition index | Habitat | Index scored between zero and one | 2008–2010 | Index that should predict healthy recruitment of native fish species | (MDBA, 2012) | SRA |
| Mean spring temperature | Temperature | °C | 1961–1990 | Proxy for water temperature | (Rowland, 1989; Lintermans et al., 2005; Koehn and Harrington, 2006) | BOM |
| Fish nativeness score | Invasive species | Index scored between zero and one | 2008–2010 | Score reflects abundance, biomass and species present that are native relative to alien | (Lintermans et al., 2005) | SRA |
Abbreviations: BOM, Bureau of Meteorology; ML, mega litre; NA, not applicable; NESAD, Sustainable Rivers Audit; SRA, Sustainable Rivers Audit; tC ha−1, tons of carbon per hectare.
Data were sourced from the NESAD v1.1.5 (available at: http://www.ga.gov.au/), SRA (MDBA, 2012) and the BOM (available at: http://www.bom.gov.au).
Individual-based measure of genetic variation
We used an individual-based measure of genetic variation: homozygosity by locus (HL), calculated in GENHET v3 in R (Coulon, 2010; R Core Team, 2014). Using an individual-based metric allowed us to resolve finer-scale associations with environmental variables, as compared with population-based metrics (for example, allelic richness). Higher HL indicates that an individual has proportionally more homozygote genotypes, reflecting greater genetic similarity of its parents. Lower HL indicates that an individual has proportionally more heterozygote genotypes, reflecting greater genetic dissimilarity of its parents, expected for populations with high genetic diversity (Frankham, 1996). HL accounts for locus variability by attributing more weight to homozygosity at more variable loci.
The amount of genetic variation present in a population is expected to be positively correlated with the general capacity of that population to adapt to environmental change and thus to represent a generalised measure of evolutionary potential that accounts for many of the uncertainties surrounding both the genetic basis of adaptation and predictions about environmental change (Harrisson et al., 2014). Levels of variation at a relatively small panel of microsatellite markers may not always be correlated strongly with fitness, trait-based measures of evolutionary potential (that is, narrow-sense heritability) or genome-wide variation (Reed and Frankham, 2001; Sgrò et al., 2011). However, correlations between heterozygosity at a small panel of microsatellite markers and fitness measures (heterozygosity–fitness correlations) are weakest in populations with very large current effective population size (Reed et al., 2003; Miller et al., 2014). Murray cod populations in our study are not characterised by large effective population sizes, having undergone strong population contractions over the last two centuries (Ne estimates across sampled catchments ranged from 22 to 407; Supplementary Appendix S2). For a standard panel of 10–15 loci, in open populations (for example, populations with migration and/or admixture, as for Murray cod), HL should also be more correlated with genome-wide homozygosity and inbreeding coefficients than are other common individual-based measures: internal relatedness and the uncorrected proportion of homozygous loci (Aparicio et al., 2006). Our panel of loci is comparable to other studies that have found associations between intraspecific genetic variation and environment (for example, Faulks et al., 2011; Kovach et al., 2015).
Identifying environmental correlates of individual-based intraspecific genetic variation
To identify environmental variables correlated with HL, we applied two different methods. First, we fitted a random forest model that uses binary recursive partitioning to quantify the amount of variation in the response variable that is explained by each predictor variable (Breiman, 2001). The random forest model makes no a priori assumptions about relationships between the predictors and response variables, allowing nonlinear responses and complex interactions (Breiman, 2001). Second, we fitted a hierarchical Bayesian regression model that quantifies the relative importance of each predictor variable, accounting for linear and nonlinear relationships. Compared with the random forest model, the Bayesian model also accounted for population history (see below).
Random forest model
We fitted the random forest model using the randomForest package in R (Breiman, 2001; Liaw and Wiener, 2002). We allowed a maximum of 1000 trees in the model, sampled one variable randomly at each proposed split and set a minimum terminal node size of 5 individuals. We used a permutation approach to quantify variable importance, based on 10 permutations per tree (Breiman, 2001). We defined variable importance as the reduction in mean-squared errors when including a given predictor variable in the model (higher values indicate that a given variable is more important).
Bayesian model
We used a hierarchical Bayesian regression model to estimate linear and nonlinear relationships between HL and the selected environmental covariates (Table 2; Thomson et al., 2010). The approach is a Bayesian variant of multivariate adaptive regression splines (Friedman, 1991). Model selection (that is, the identification of which variables to include) was performed using reversible-jump Markov chain Monte Carlo that estimates the posterior probability that a given predictor variable has an association with the response variable, accounting for linear, quadratic and cubic associations (Lunn et al., 2006, 2009). River catchment and sampling site were included as clustering variables (given exchangeable priors; equivalent to random effects in a standard mixed model) to control for population history (for example, founder effects and drift) and unmeasured environmental variables by accounting for variation in HL among sites and catchments that was not explained by the remaining predictor variables. The response variable (HL) was assumed to be normally distributed and all predictor variables were standardised to zero mean and unit variance. All parameters were assigned uninformative prior distributions. Models were fitted with the reversible-jump add-in for WinBUGS 1.4 (Lunn et al., 2000) and model outputs were managed in R (R Core Team, 2014). Models were run using three chains of 50 000 iterations following a 50 000 iteration burn-in period. Full model details and WinBUGS code are provided in Supplementary Information (see Supplementary Appendices S3 and S4, respectively).
Inferences were based on the Bayesian model averaged parameter estimates. Bayesian model averaging estimates the average parameter value over all possible models (that is, each possible combination of variables), where the contribution of each possible model is weighted by its posterior probability (Raftery et al., 1997). Model posterior probabilities emerge naturally from a reversible-jump Markov chain Monte Carlo sampling scheme because the sampler visits each possible model in proportion to its posterior probability; the Bayesian model averaged parameter estimate is simply the average of the parameter over all Markov chain Monte Carlo iterations. The relative importance of each predictor variable was given by the posterior probability of variable inclusion for each variable. The prior probability of variable inclusion was 0.5, and hence posterior probabilities of variable inclusion >0.5 indicate evidence in favour of variable inclusion, with values >0.75 providing strong evidence for variable inclusion (odds ratio >3).
Assessing model fit
We used root-mean-square errors (RMSEs) and coefficients of determination (r2) to measure model fit. RMSEs measure the average difference between observed and fitted values. The r2 values were based on Pearson's r and measure the proportion of linear variation explained by a given model. RMSE and r2 values were based on out-of-bag samples for random forest models because these models have a tendency to overfit (Breiman, 2001). For the Bayesian model, RMSE and r2 values were based on the fitted model because marginalisation over all possible models avoids overfitting (Gelman and Hill, 2006).
We used posterior predictive diagnostics to assess Bayesian model adequacy (Gelman et al., 1996). We calculated a summed discrepancy measure (Sobs=Σ(HLobs−HLexp)2, where HLobs and HLexp are observed and modelled values of HL, respectively) and generated a reference distribution for this discrepancy measure (Ssim) based on simulated data drawn from the posterior distribution of the model. If the posterior distributions of Sobs and Ssim overlapped (0.1<Pr(Sobs>Ssim)<0.9) then the fitted model was capable of generating the observed data, indicating an appropriate model structure.
Visualising patterns of individual-based intraspecific genetic variation
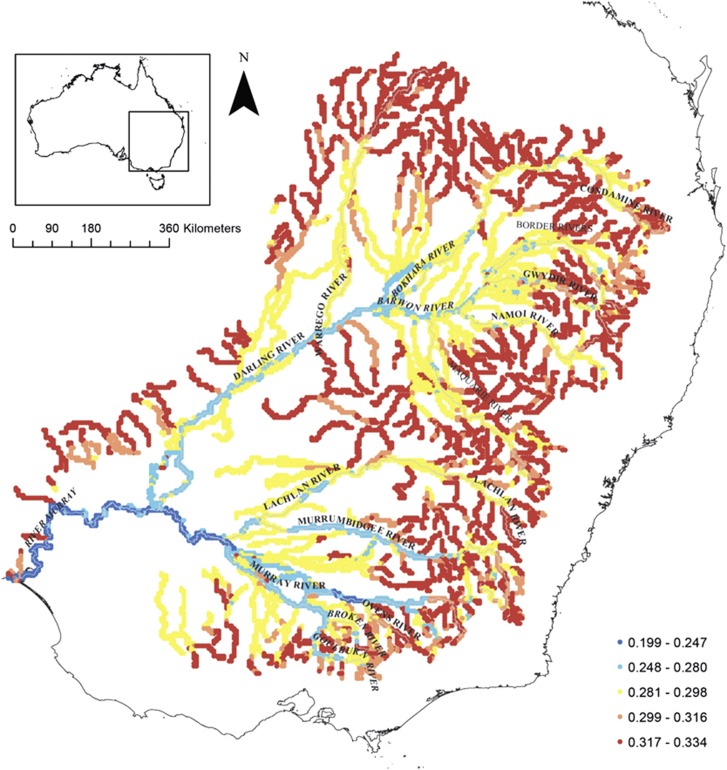
For conservation management it is useful to be able to predict and visualise relationships between intraspecific genetic diversity and environment (Thomassen et al., 2010). To demonstrate how our approach could be used to generate spatially explicit predictions of intraspecific genetic diversity based on environment, we first extracted data for all environmental predictor variables for each stream segment across all major streams in the MDB (‘major' was defined based on the stream segment classification under stream hierarchy in the Geofabric stream network database). These environmental predictors were then combined with the Bayesian model averaged parameter estimates to predict HL values for each stream segment along the major stream network. Predicted HL values across the MDB were visualised in ArcMap.
Testing for effects of stocking and degree of admixture on HL
Stocking and/or admixture could potentially have an influence on HL. We used a standard linear regression to test for an association between the intensity of stocking and mean HL in each catchment (Table 1). To quantify the degree of admixture, we ran STRUCTURE analysis for K-values 1–10 using the admixture model with correlated allele frequencies (Pritchard et al., 2000; Falush et al., 2003). Twenty replicate runs of 3 × 106 Markov chain Monte Carlo, after an initial burn-in period of 106 repetitions, were performed for each value of K. Results were summarised using the standard pipeline on the Clumpak web server (Kopelman et al., 2015). The most likely number of clusters (K) was selected using the Evanno et al. (2005) ΔK method that finds the point of greatest change in the distribution of LnP(D). Each individual was proportionally assigned to each of the three genetic clusters identified by the STRUCTURE analysis. Individuals with a Q-value of >0.2 were considered assigned to a cluster. Thus, each individual was assigned an admixture coefficient of 1, 2 or 3 to reflect how many clusters it was assigned to (that is, degree of admixture). To test for an association between degree of admixture and HL, we performed a standard analysis of variance of HL against admixture coefficient.
Results
Environmental correlates of individual-based intraspecific genetic variation
Random forest model
For the random forest model the overall model fit was poor (Pearson's r=0.20), suggesting that environmental variables explained ∼4% of the variation in HL. The RMSE was 0.12, indicating that fitted HL values differed from observed values by 0.12 on average. Annual mean accumulated soil water surplus and flow regime disturbance index had the strongest associations with HL and had importance values that were twice as large as other included variables.
Bayesian model
For the Bayesian method the overall model fit was moderate (Pearson's r=0.40), suggesting that environmental and clustering variables explained ∼16% of the variation in HL. The RMSE was 0.11, indicating that fitted HL values difference from observed HL values by 0.11 on average. The posterior predictive probability was 0.74, suggesting that the fitted model structure was adequate. There was strong evidence for flow regime disturbance index to be included in the regression model (inclusion probability=0.85, equivalent to an odds ratio=5.7; Table 3), with weak evidence for inclusion of annual mean accumulated soil water surplus (a proxy for mean annual flow; inclusion probability=0.56; Table 3) and little evidence for other variables (inclusion probabilities <0.5; Table 3). There was some evidence that flow regime disturbance had a nonlinear relationship with HL, whereas all other variables had linear (if any) relationships (Table 3).
Table 3. Relative variable importance estimated with a random forest model and a hierarchical Bayesian model.
| Environmental predictor variable | Random forest model | Bayesian model | ||
|---|---|---|---|---|
| Importance Values | Probability of variable inclusion | Direction of effect (slope±1 s.d.) | Degree | |
| Flow regime Disturbance index | 0.002 | 0.85 | Negative (–0.015±0.010) | 1.5 |
| Mean accumulated soil water surplus for summer | 0.001 | 0.39 | Negative (–0.001±0.004) | 0.6 |
| Stream and valley percentage extant woodland cover | 0.001 | 0.34 | Negative (–0.001±0.003) | 0.5 |
| Fish condition index | 0.001 | 0.34 | Positive (0.001±0.004) | 0.5 |
| Annual mean net primary productivity | 0.001 | 0.42 | Negative (–0.002±0.006) | 0.7 |
| Coefficient of variation of annual totals of accumulated soil water surplus | 0.001 | 0.39 | Positive (0.001±0.005) | 0.6 |
| Annual mean accumulated soil water surplus | 0.002 | 0.56 | Negative (–0.006±0.009) | 1.0 |
| Barrier free flow path length | 0.001 | 0.38 | Negative (–0.001±0.004) | 0.6 |
| Average spring temperature | 0.001 | 0.38 | Negative (–0.002±0.006) | 0.6 |
| Stream and valley percentage extant forest cover | 0.001 | 0.40 | Negative (–0.002±0.004) | 0.6 |
| Fish nativeness score | 0.001 | 0.40 | Negative (–0.003±0.006) | 0.7 |
Importance values for the random forest model are proportional to the reduction in root-mean-square errors when a given predictor variable is included in the model; higher values indicate a more important variable. Importance values for the Bayesian model are posterior probabilities of inclusion for each environmental predictor variable. For the Bayesian model, we also present the model averaged parameter estimates for the slope and degree. The prior probability of variable inclusion was 0.5, and hence values >0.75 correspond with odds ratios >3. The slope of a given effect indicates the direction of a variable's effect, and the degree is the number of knots included in the spline term for a given variable, indicating the nonlinearity of a variable's effect (0 is no effect, 1 is a linear effect, 2 is a quadratic effect and 3 is a cubic effect).
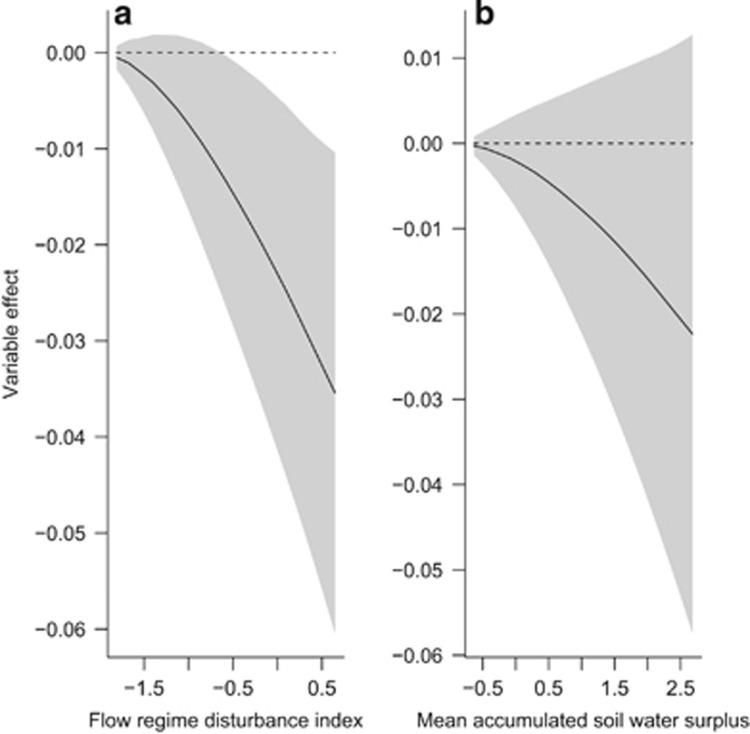
There was strong evidence of a negative, nonlinear effect of flow regime disturbance on HL: individual-based homozygosity decreased with disturbance to the natural flow regime (odds ratio >3; Table 3 and Figure 2a). The effect was modest, corresponding to a ∼0.015 change in HL with each 1 s.d. change in the predictor variable that is equal to ∼2% of the observed range of HL. There was also weak evidence of a negative, approximately linear effect of annual mean accumulated soil water surplus (a proxy for mean annual flow) on HL (odds ratio=1.4; Figure 2b). The effect was about half as strong as that estimated for flow regime disturbance, corresponding to a ∼0.006 change in HL with each 1 s.d. change in the predictor, equal to ∼1% of the observed range of HL.
Figure 2.
Bayesian model averaged parameter estimates for variables with probability of inclusion >0.5: (a) flow regime disturbance index and (b) annual mean accumulated soil water surplus. The y axis represents the deviation from the mean HL value (in units of HL) for a given standardised value of the predictor variable. The grey shaded region denotes 1 s.d. either side of the mean fitted effect.
Visualising patterns of individual-based intraspecific genetic variation
Spatially explicit Bayesian model predictions of HL were visualised for all major rivers in the MDB (Figure 3). Levels of HL predicted by the model were lowest in the most heavily regulated, downstream major river channels (Murray, Darling, Murrumbidgee and Goulburn Rivers) and highest in the upper reaches of major rivers and smaller tributaries (Figure 3).
Figure 3.
Visualisation of patterns of HL predicted by the Bayesian model for Murray cod across all major streams in the MDB, Australia. The largest rivers are shown in grey.
Testing for effects of stocking and degree of admixture on HL
There was no evidence of an association between stocking intensity and mean HL (slope=0.014, R2=0.13, P>0.05). STRUCTURE detected three major genetic clusters across the study region, consistent with the patterns previously reported in Rourke et al. (2011) (see Supplementary Appendix S5). There was no evidence that degree of admixture had an effect on HL (F=3.4, P>0.05; mean HL values for admixture coefficients 1, 2 and 3 were 0.27, 0.31 and 0.29, respectively).
Discussion
We identified potential ecological factors associated with patterns of genetic variation across the range of a widespread freshwater fish species. In contrast to most comparable studies, the approaches used in this study focus on an individual-based measure of genetic diversity (cf., population-based measures of diversity and/or measures based on genetic differentiation/distance) and statistically quantified the relative importance of a given predictor variable, accounting for linear and nonlinear relationships. Our Bayesian method also accounted for population history (for example, differences in levels of genetic variation among catchments resulting from processes such as founder effect and drift). Both the random forest and Bayesian model supported effects of disturbance to the natural flow regime and mean annual flow on HL. The Bayesian model performed best, explaining ∼16% variation in HL as compared with ∼4% explained by the random forest model. Better performance of the Bayesian model may reflect the fact that it also accounts for variation in HL that is due to differences among sites and catchments. We were also able to make spatially explicit predictions about the distribution of genetic diversity across all major streams in the MDB, demonstrating a useful visualisation and management approach for conservation planning. However, given our model explained only ∼16% of the variation in HL, we stress that our predictions of HL across the MDB should be interpreted with caution.
The Bayesian model indicated strong support for a negative association between HL and disturbance to the natural flow regime. Populations that are associated with lower HL values are likely to be healthier, have larger effective population size and higher genetic diversity than populations of individuals with higher HL values (Frankham, 1996; Reed and Frankham, 2003; Aparicio et al., 2006). From an evolutionary perspective, higher genetic diversity is also likely to be positively correlated with a population's evolutionary potential, at least in a general sense (Harrisson et al., 2014). Thus, our findings suggest that river reaches across the MDB that have more disturbed flow regimes are likely to support larger, more genetically diverse populations of Murray cod. Although this result seems counterintuitive because many native fish species are adversely affected by river regulation (Koehn et al., 2014), more consistent, perennial flows observed in regulated channels may reduce the frequency of population bottlenecks that can occur during the dry phases that typify the highly variable and unpredictable natural flow regime in the MDB (Bond et al., 2010). In the MDB the greatest disturbance to the natural flow regime typically occurs in lowland river reaches where river regulation and water extraction for irrigation support agricultural production. Thus, relative to rivers with less disturbed flow regimes, heavily regulated lowland rivers may have greater habitat availability and reduced exposure to low flow conditions and associated harsh water conditions (for example, low oxygen and high temperature) in the summer months or drought periods (Reich et al., 2010). Our study is correlative rather than mechanistic and we do not claim a direct causal link between flow regulation and HL. However, our findings are consistent with other studies that report positive associations between intensity of flow regulation and Murray cod occurrences (Bond et al., 2010; Reich et al., 2010) and population trajectories (Yen et al., 2013).
In addition to strong support for a negative association between flow regulation and HL, we also detected weak evidence for a negative association between mean annual soil water surplus (a proxy for mean annual flow and river size) and HL. As the main lowland river channels that typically experience the highest levels of disturbance to the natural flow regime are also typically larger rivers (with larger flow volumes), the relative effects of flow regulation and mean annual soil water surplus are partially (and unavoidably) confounded. However, the flow regime disturbance index and mean annual soil water surplus were not highly correlated in our study (r=0.48), and support for a stronger association between flow regime disturbance and HL (cf., mean annual soil water surplus and HL) suggests that associations between genetic variation and flow regulation are not solely the result of large (more regulated) rivers supporting larger populations of Murray cod. Differences in HL among catchments resulting from differences in river size were accounted for by inclusion of river catchment as a clustering variable in the Bayesian model.
All large rivers in the MDB experience disturbance to the natural flow regime, at least to a moderate extent. As large, completely unregulated rivers no longer exist in the MDB, and thus are not included in our sampling, we have no information on HL levels in large unregulated rivers. Similarly, despite negative associations between flow-related variables and HL, our study focuses only on relative levels of genetic variation across the MDB for the period of sampling (1994–2006), and does not reflect historical patterns or assess whether levels of genetic diversity are sufficient for long-term population persistence (that is, ‘high' genetic diversity is relative only to very recent levels of genetic diversity in Murray cod that might still be lower than historical genetic diversity). Given that Murray cod populations have experienced severe declines over the past two centuries and population numbers remain very low relative to historical levels, our results should not be taken as evidence that extensive river regulation across the MDB has had an overall positive effect on Murray cod populations (Barret, 2004; Lintermans et al., 2005). Instead, our study suggests that within the contemporary system, the greater amount and stability of habitat afforded by flow regulation may have some benefits for the large-bodied Murray cod, and that other pressures (either independent of environment or not included in our model) may be more critical to the persistence of Murray cod, for example, habitat loss, overfishing and chemical pollution (Rowland, 1989; Lintermans et al., 2005).
The environmental and clustering variables included in our Bayesian model were able to explain ∼16% of the variation in HL, comparable to other studies of associations between genetic variation and environmental variables (see, for example, Faulks et al., 2011). Additional factors that likely explain the remaining variation in HL include: environmental and demographic stochasticity, a relatively large migration range, the necessarily relatively low resolution (spatially and temporally) of the environmental data used in analysis and the modest power of our (16-locus) genetic assay. A complex history of stocking and illegal translocations of adult fish across all catchments analysed here may also contribute to variation in HL that was unexplained by our model. However, because there was no significant relationship between the numbers of fish stocked and mean HL at the catchment level, we argue that stocking effects are more likely to add noise than drive the observed pattern.
Here we demonstrate two approaches that can identify environmental variables associated with range-wide patterns of individual-based intraspecific genetic variation. Our genetic modelling approach is likely to provide information about the viability of populations (notably effective population size, evolutionary potential) not captured by more commonly studied abundance or occurrence patterns, and could be readily applied to other wide-ranging taxa. There is scope for our approach to be extended and incorporated into a species distribution modelling framework. Combining genetic data with presence and abundance data from across species' ranges in a single framework could improve understanding of links between species distributions and underlying environmental drivers, and allow for better predictions of the capacity of species to respond to future environmental change through both range shifts and in situ adaptation (Scoble and Lowe, 2010; Fitzpatrick and Keller, 2015; Gotelli and Stanton-Geddes, 2015).
Data archiving
Genetic data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.4bg42 and PANGAEA. Model code is supplied in Supplementary Material.
Acknowledgments
This work was supported by ARC Grant LP110200017 to Monash University, Flinders University of South Australia and the University of Canberra, with Partner Organization University of Montana. Funding and other support was contributed by industry Partner Organizations ACTEW Corporation, Department of Sustainability and Environment (Victoria) (now within Department of Environment, Land, Water & Planning), Fisheries Victoria (now within Department of Economic Development, Jobs, Transport and Resources), Melbourne Water and Fisheries New South Wales. KAH was supported by the Holsworth Wildlife Research Endowment and an Australian Postgraduate Award through Monash University. We thank Jim Thomson for statistical support and the associate editor and three anonymous reviewers for comments on earlier drafts.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Aparicio JM, Ortego J, Cordero PJ. (2006). What should we weigh to estimate heterozygosity, alleles or loci? Mol Ecol 15: 4659–4665.. [DOI] [PubMed] [Google Scholar]
- Barrett J. (2004). Introducing the Murray-Darling Basin Native Fish Strategy and initial steps toward demonstration reaches. Ecol Manage Restor 5: 15–23.. [Google Scholar]
- Bond N, McMaster D, Reich P, Thomson JR, Lake P. (2010). Modelling the impacts of flow regulation on fish distributions in naturally intermittent lowland streams: an approach for predicting restoration responses. Freshw Biol 55: 1997–2010.. [Google Scholar]
- Bond N, Thomson J, Reich P, Stein J. (2011). Using species distribution models to infer potential climate change-induced range shifts of freshwater fish in south-eastern Australia. Mar Freshwater Res 62: 1043–1061.. [Google Scholar]
- Breiman L. (2001). Random forests. Mach Learn 45: 5–32.. [Google Scholar]
- Coulon A. (2010). GENHET: an easy-to-use R function to estimate individual heterozygosity. Mol Ecol Res 10: 167–169.. [DOI] [PubMed] [Google Scholar]
- Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G et al. (2013). Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36: 27–46. [Google Scholar]
- Elith J, Leathwick JR. (2009). Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40: 677–697.. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164: 1567–1587.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulks LK, Gilligan DM, Beheregaray LB. (2011). The role of anthropogenic vs. natural in-stream structures in determining connectivity and genetic diversity in an endangered freshwater fish, Macquarie perch (Macquaria australasica). Evol Appl 4: 589–601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson B, McMahon T. (1988). Australia vs the world: a comparative analysis of streamflow characteristics. In: Werner RF ed Fluvial Geomorphology of Australia. Academic Press: Sydney, Australia, pp 17–40.. [Google Scholar]
- Fitzpatrick MC, Keller SR. (2015). Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol Lett 18: 1–16. [DOI] [PubMed] [Google Scholar]
- Frankel OH. (1974). Genetic conservation: our evolutionary responsibility. Genetics 78: 53–65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. (1996). Relationship of genetic variation to population size in wildlife. Conserv Biol 10: 1500–1508.. [Google Scholar]
- Friedman JH. (1991). Multivariate adaptive regression splines. Ann Stat 19: 1–141.. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. (2006). Identifying the environmental factors that determine the genetic structure of populations. Genetics 174: 875–891.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Meng X-L, Stern H. (1996). Posterior predictive assessment of model fitness via realized discrepancies. Stat Sin 6: 733–787.. [Google Scholar]
- Gelman A, Hill J. (2006) Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge University Press: New York, NY, USA. [Google Scholar]
- Gotelli NJ, Stanton-Geddes J. (2015). Climate change, genetic markers and species distribution modelling. J Biogeogr 42: 1577–1585.. [Google Scholar]
- Guégan J-F, Lek S, Oberdorff T. (1998). Energy availability and habitat heterogeneity predict global riverine fish diversity. Nature 391: 382–384.. [Google Scholar]
- Harrisson KA, Pavlova A, Telonis-Scott M, Sunnucks P. (2014). Using genomics to characterize evolutionary potential for conservation of wild populations. Evol Appl 7: 1008–1025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries P. (2005). Spawning time and early life history of Murray cod, Maccullochella peelii peelii (Mitchell) in an Australian river. Environ Biol Fishes 72: 393–407.. [Google Scholar]
- Jay F, Manel S, Alvarez N, Durand EY, Thuiller W, Holderegger R et al. (2012). Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol Ecol 21: 2354–2368.. [DOI] [PubMed] [Google Scholar]
- King AJ, Tonkin Z, Mahoney J. (2009). Environmental flow enhances native fish spawning and recruitment in the Murray River, Australia. River Res Appl 25: 1205–1218. [Google Scholar]
- Koehn J, McKenzie J, O'mahony D, Nicol S, O'Connor J, O'Connor W. (2009). Movements of Murray cod (Maccullochella peelii peelii) in a large Australian lowland river. Ecol Freshw Fish 18: 594–602.. [Google Scholar]
- Koehn JD, Harrington D. (2006). Environmental conditions and timing for the spawning of Murray cod (Maccullochella peelii peelii) and the endangered trout cod (M. macquariensis) in southeastern Australian rivers. River Res Appl 22: 327–342.. [Google Scholar]
- Koehn JD, King AJ, Beesley L, Copeland C, Zampatti BP, Mallen-Cooper M. (2014). Flows for native fish in the Murray-Darling Basin: lessons and considerations for future management. Ecol Manage Restor 15: 40–50.. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15: 1179–1191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach RP, Muhlfeld CC, Wade AA, Hand BK, Whited DC, DeHaan PW et al. (2015). Genetic diversity is related to climatic variation and vulnerability in threatened bull trout. Global Change Biol 21: 2510–2524.. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. (2002). Classification and regression by randomForest. R News 2: 18–22.. [Google Scholar]
- Lintermans M, Rowland S, Koehn J, Butler G, Simpson B, Wooden I. (2005) Management of Murray Cod in the Murray-Darling Basin: Statement, Recommendations and Supporting Papers. Workshop, 3–4 June 2004. Murray-Darling Basin Commission Canberra: Australia, pp 15–29.. [Google Scholar]
- Lunn DJ, Best N, Whittaker JC. (2009). Generic reversible jump MCMC using graphical models. Stat Comput 19: 395–408.. [Google Scholar]
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. (2000). WinBUGS-a Bayesian modelling framework: concepts, structure, and extensibility. Statist Comput 10: 325–337.. [Google Scholar]
- Lunn DJ, Whittaker JC, Best N. (2006). A Bayesian toolkit for genetic association studies. Genet Epidemiol 30: 231–247.. [DOI] [PubMed] [Google Scholar]
- Manel S, Holderegger R. (2013). Ten years of landscape genetics. Trends Ecol Evol 28: 1–8.. [DOI] [PubMed] [Google Scholar]
- MDBA. (2012) Sustainable Rivers Audit 2: The Ecological Health of Rivers in the Murray-Darling Basin at the End of the Millenium Drought [2008-2010). Murray-Darling Basin Authority: Canberra, Australia. [Google Scholar]
- Miller J, Malenfant R, David P, Davis C, Poissant J, Hogg J et al. (2014). Estimating genome-wide heterozygosity: effects of demographic history and marker type. Heredity 112: 240–247.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C. (2002). Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst Biol 51: 238–254.. [DOI] [PubMed] [Google Scholar]
- Poff NL, Allan JD, Bain MB, Karr JR, Prestegaard KL, Richter BD et al. (1997). The natural flow regime. Bioscience 47: 769–784.. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery AE, Madigan D, Hoeting JA. (1997). Bayesian model averaging for linear regression models. J Am Stat Assoc 92: 179–191.. [Google Scholar]
- Rathert D, White D, Sifneos J, Hughes R. (1999). Environmental correlates of species richness for native freshwater fish in Oregon, USA. J Biogeogr 26: 257–273.. [Google Scholar]
- Reed DH, Frankham R. (2001). How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55: 1095–1103.. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. (2003). Correlation between fitness and genetic diversity. Conserv Biol 17: 230–237.. [Google Scholar]
- Reed DH, Lowe EH, Briscoe DA, Frankham R. (2003). Fitness and adaptation in a novel environment: effect of inbreeding, prior environment, and lineage. Evolution 57: 1822–1828.. [DOI] [PubMed] [Google Scholar]
- Reich P, McMaster D, Bond N, Metzeling L, Lake PS. (2010). Examining the ecological consequences of restoring flow intermittency to artificially perennial lowland streams: Patterns and predictions from the Broken–Boosey creek system in northern Victoria, Australia. River Res Appl 26: 529–545.. [Google Scholar]
- R Core Team. (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Rourke ML, McPartlan HC, Ingram BA, Taylor AC. (2010). Biogeography and life history ameliorate the potentially negative genetic effects of stocking on Murray cod (Maccullochella peelii peelii). Mar Freshwater Res 61: 918–927.. [Google Scholar]
- Rourke ML, McPartlan HC, Ingram BA, Taylor AC. (2011). Variable stocking effect and endemic population genetic structure in Murray cod Maccullochella peelii. J Fish Biol 79: 155–177.. [DOI] [PubMed] [Google Scholar]
- Rowland S. (1989). Aspects of the history and fishery of the Murray cod, Maccullochella peeli (Mitchell)(Percichthyidae). Proc Linn Soc NSW 111: 201–213.. [Google Scholar]
- Rowland S. (2004). Overview of the history, fishery, biology and aquaculture of Murray cod (Maccullochella peelii peelii). In: Management of Murray Cod in the Murray-Darling Basin: Statement, recommendations and supporting papers. Proceedings of a workshop held in Canberra: Australia, pp 38–61.. [Google Scholar]
- Scoble J, Lowe AJ. (2010). A case for incorporating phylogeography and landscape genetics into species distribution modelling approaches to improve climate adaptation and conservation planning. Divers Distrib 16: 343–353.. [Google Scholar]
- Sgrò CM, Lowe AJ, Hoffmann AA. (2011). Building evolutionary resilience for conserving biodiversity under climate change. Evol Appl 4: 326–337.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J, Stein J, Nix H. (2002). Spatial analysis of anthropogenic river disturbance at regional and continental scales: identifying the wild rivers of Australia. Landscape Urban Plan 60: 1–25.. [Google Scholar]
- Thomassen HA, Cheviron ZA, Freedman AH, Harrigan RJ, Wayne RK, Smith TB. (2010). Spatial modelling and landscape-level approaches for visualizing intra-specific variation. Mol Ecol 19: 3532–3548.. [DOI] [PubMed] [Google Scholar]
- Thomson JR, Kimmerer WJ, Brown LR, Newman KB, Nally RM, Bennett WA et al. (2010). Bayesian change point analysis of abundance trends for pelagic fishes in the upper San Francisco Estuary. Ecol Appl 20: 1431–1448.. [DOI] [PubMed] [Google Scholar]
- Todd CR, Ryan T, Nicol SJ, Bearlin AR. (2005). The impact of cold water releases on the critical period of post-spawning survival and its implications for Murray cod (Maccullochella peelii peelii): a case study of the Mitta Mitta River, southeastern Australia. River Res Appl 21: 1035–1052.. [Google Scholar]
- Vandergast AG, Bohonak AJ, Hathaway SA, Boys J, Fisher RN. (2008). Are hotspots of evolutionary potential adequately protected in southern California? Biol Conserv 141: 1648–1664.. [Google Scholar]
- Wang IJ, Bradburd GS. (2014). Isolation by environment. Mol Ecol 23: 5649–5662.. [DOI] [PubMed] [Google Scholar]
- Yen JDL, Bond NR, Shenton W, Spring DA, Mac Nally R. (2013). Identifying effective water-management strategies in variable climates using population dynamics models. J Appl Ecol 50: 691–701.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.