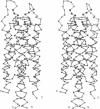Abstract
We used mixed, mutagenic oligonucleotides to create single amino acid substitutions in the bacterial chemoreceptor Trg. Mutagenesis was directed at a 20-residue segment of the periplasmic domain implicated in ligand recognition. Transmembrane signaling by the mutant receptors was assayed in vivo by monitoring adaptational covalent modification. Among 20 functionally altered but stable receptors there were two distinct signaling phenotypes. Insensitive receptors did not signal upon stimulation and thus appeared defective in productive ligand interaction. Mimicked-occupancy receptors exhibited transmembrane signaling without ligand. Many mimicked-occupancy receptors produced additional signaling upon ligand binding and in appropriate conditions mediated effective chemotaxis; most insensitive receptors did not. Like normal receptors with one binding site occupied, mimicked-occupancy proteins adapted to persistent transmembrane signaling by increased methylation and thus could respond to other stimuli. Signaling phenotypes were strikingly segregated by residue position. Substitutions mimicking ligand occupancy occurred in half the segment, and those creating insensitive phenotypes occurred in the other half. These observations could be related to the three-dimensional structure of the periplasmic domain of the Tar(s) chemoreceptor. Insensitive substitutions occurred near the distal end of helix 1, where bulky protein ligands could interact; occupancy-mimicking substitutions were on the same helix at positions buried in the subunit interface between helices 1 and 1'. Thus perturbation of the interface induced transmembrane signaling, implicating changes at that interface in signal transduction, a conclusion consistent with differences in crystal structures of unoccupied and ligand-occupied Tar(s).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames P., Chen J., Wolff C., Parkinson J. S. Structure-function studies of bacterial chemosensors. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):59–65. doi: 10.1101/sqb.1988.053.01.010. [DOI] [PubMed] [Google Scholar]
- Ames P., Parkinson J. S. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell. 1988 Dec 2;55(5):817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci U S A. 1980 May;77(5):2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci U S A. 1980 May;77(5):2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Salvo J. J., Grindley N. D. A simple and efficient procedure for saturation mutagenesis using mixed oligodeoxynucleotides. Gene. 1986;46(2-3):145–152. doi: 10.1016/0378-1119(86)90398-7. [DOI] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Gardina P., Conway C., Kossman M., Manson M. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J Bacteriol. 1992 Mar;174(5):1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Park C., Nowlin D. M. Adaptational "crosstalk" and the crucial role of methylation in chemotactic migration by Escherichia coli. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1448–1452. doi: 10.1073/pnas.86.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L. The bacterial chemosensory system. Can J Microbiol. 1988 Apr;34(4):466–474. doi: 10.1139/m88-080. [DOI] [PubMed] [Google Scholar]
- Kossmann M., Wolff C., Manson M. D. Maltose chemoreceptor of Escherichia coli: interaction of maltose-binding protein and the tar signal transducer. J Bacteriol. 1988 Oct;170(10):4516–4521. doi: 10.1128/jb.170.10.4516-4521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee L., Imae Y. Role of threonine residue 154 in ligand recognition of the tar chemoreceptor in Escherichia coli. J Bacteriol. 1990 Jan;172(1):377–382. doi: 10.1128/jb.172.1.377-382.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Mizuno T., Imae Y. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J Bacteriol. 1988 Oct;170(10):4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Intrasubunit signal transduction by the aspartate chemoreceptor. Science. 1991 Dec 13;254(5038):1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Mowbray S. L., Koshland D. E., Jr Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell. 1987 Jul 17;50(2):171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- Mutoh N., Oosawa K., Simon M. I. Characterization of Escherichia coli chemotaxis receptor mutants with null phenotypes. J Bacteriol. 1986 Sep;167(3):992–998. doi: 10.1128/jb.167.3.992-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner S. S., Goodin D. B., Smith M. A simple and efficient procedure for generating random point mutations and for codon replacements using mixed oligodeoxynucleotides. DNA. 1988 Mar;7(2):127–134. doi: 10.1089/dna.1988.7.127. [DOI] [PubMed] [Google Scholar]
- Nowlin D. M., Bollinger J., Hazelbauer G. L. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins. 1988;3(2):102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- Nowlin D. M., Bollinger J., Hazelbauer G. L. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1987 May 5;262(13):6039–6045. [PubMed] [Google Scholar]
- Ordal G. W., Adler J. Isolation and complementation of mutants in galactose taxis and transport. J Bacteriol. 1974 Feb;117(2):509–516. doi: 10.1128/jb.117.2.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutation plus amplification of a transducer gene disrupts general chemotactic behavior in Escherichia coli. J Bacteriol. 1986 Dec;168(3):1378–1383. doi: 10.1128/jb.168.3.1378-1383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Wolff C., Parkinson J. S. Aspartate taxis mutants of the Escherichia coli tar chemoreceptor. J Bacteriol. 1988 Oct;170(10):4509–4515. doi: 10.1128/jb.170.10.4509-4515.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]