Summary
Different modes of bacterial taxis play important roles in environmental adaptation, survival, colonization and dissemination of disease. One mode of taxis is flotation due to the production of gas vesicles. Gas vesicles are proteinaceous intracellular organelles, permeable only to gas, that enable flotation in aquatic niches. Gene clusters for gas vesicle biosynthesis are partially conserved in various archaea, cyanobacteria, and some proteobacteria, such as the enterobacterium, S erratia sp. ATCC 39006 (S39006). Here we present the first systematic analysis of the genes required to produce gas vesicles in S39006, identifying how this differs from the archaeon H alobacterium salinarum. We define 11 proteins essential for gas vesicle production. Mutation of gvpN or gvpV produced small bicone gas vesicles, suggesting that the cognate proteins are involved in the morphogenetic assembly pathway from bicones to mature cylindrical forms. Using volumetric compression, gas vesicles were shown to comprise 17% of S39006 cells, whereas in E scherichia coli heterologously expressing the gas vesicle cluster in a deregulated environment, gas vesicles can occupy around half of cellular volume. Gas vesicle production in S39006 and E . coli was exploited to calculate the instantaneous turgor pressure within cultured bacterial cells; the first time this has been performed in either strain.
Introduction
Bacteria can use multiple modes of taxis, including swimming, twitching, swarming and flotation. Flotation is enabled through the regulated production of intracellular buoyancy chambers: gas vesicles. Gas vesicle‐driven buoyancy facilitates the movement of photosynthetic cyanobacteria into water column positions, allowing exposure to wavelengths of light that can support phototrophy (Pfeifer, 2012). Gas vesicles can also increase bacterial surface area‐to‐volume ratios, and thereby help survival under environmental and nutritional stresses (Houwink, 1956; Walsby, 1972).
Gas vesicles are visible as light‐refracting structures under phase‐contrast microscopy (PCM), and bacterial colonies producing gas vesicles can be opaque on plates. All gas vesicles identified to date appear to be constructed from homologous proteins (Pfeifer, 2012). The primary gas vesicle structural protein, GvpA, assembles into tandem arrays that form ribs of the cylindrical vesicle (Englert and Pfeifer, 1993; Walsby, 1994). A second protein, GvpC, forms an exterior mesh on the gas vesicle surface, providing further structural support and influencing gas vesicle shape (Hayes et al., 1988; 1992; Walsby and Hayes, 1988; Englert and Pfeifer, 1993; Walsby, 1994; Offner et al., 2000). Recently, the structure of GvpF from the cyanobacterium, Microcystis aeruginosa was solved, and through electron microscopy, GvpF was shown to form a part of the gas vesicle structure (Xu et al., 2014). However, the precise biochemical roles of many other proteins found in gas vesicle gene clusters are unknown, although their stoichiometry is thought to be important (Shukla and DasSarma, 2004; Chu et al., 2011; Pfeifer, 2012).
Recently, we discovered gas vesicles in the enterobacterium, Serratia spp. ATCC39006 (S39006), a genetically tractable host (Ramsay et al., 2011). S39006 is a Gram‐negative bacillus that is pathogenic to plant and nematode hosts and produces two secondary metabolites: the tripyrrole red pigment, 2‐methyl‐3‐pentyl‐6‐methoxyprodigiosin (prodigiosin) and the β‐lactam antibiotic, 1‐carbapen‐2‐em‐3‐carboxylic acid (a carbapenem) (Coulthurst et al., 2005; Williamson et al., 2006). The genetic locus for gas vesicle biosynthesis in S39006 was determined, and gas vesicles were shown to be responsible for the opaque colony phenotype and buoyancy. Transcription of the gas vesicle gene cluster is cell density‐dependent [under the control of the SmaIR quorum‐sensing (QS) system], responsive to oxygen status, and controlled by the gas vesicle regulatory protein A, GvrA (a member of the NtrC family of regulators) encoded within the gas vesicle cluster (Ramsay et al., 2011; Ramsay and Salmond, 2012). Although there are obvious similarities between various gas vesicle gene clusters, the S39006 cluster does not encode the two widely studied regulatory proteins, GvpD and GvpE, that are found in Halobacterium salinarum strains and in Haloferax mediterranei (Englert et al., 1992; Ng et al., 2000; Zimmermann and Pfeifer, 2003; Hofacker et al., 2004). However, as in various organisms, the functionality of other proteins encoded by the gas vesicle genetic locus of S39006 is unknown.
Here, we describe a comprehensive analysis of the 16.6 kb gas vesicle cluster in S39006. In frame deletions were created for each of the 19 genes in the cluster and impacts on gas vesicle production investigated. Pressure nephelometry was used to determine the collapse pressure of gas vesicles, and volumetric occupancy of the bacterial cytoplasm by vesicles in S39006, and an engineered E. coli‐carrying gas vesicles, was assessed. We also exploited gas vesicles to calculate the instantaneous turgor pressure of both S39006 cells and the recombinant E. coli strain by examining the collapse pressure of gas vesicles under different growth conditions. This study presents a complete analysis of a single gas vesicle gene cluster and further demonstrates how gas vesicles can be exploited in E. coli, generating cells that are composed of ∼50% gas vesicles.
Results
The core gas vesicle gene composition in proteobacteria
S39006 carries a 16.6 kb cluster of 19 genes responsible for gas vesicle synthesis (Ramsay and Salmond, 2012). This gas vesicle gene composition is similar to that of other proteobacteria, particularly β‐, γ‐ and δ‐proteobacteria, possessing a gas vesicle gene cluster (Fig. 1). S39006 has three genes (gvpA1, gvpA2 and gvpA3), encoding isoforms of the primary gas vesicle structural protein, GvpA; this is consistent with six proteobacteria that possess a gas vesicle gene cluster. A phylogenetic tree of GvpA showed that GvpA2 and GvpA3 are homologous to proteins previously annotated as GvpS and GvpJ, respectively, in other bacteria (Fig. S1 and Table S1). Despite differing annotations across organisms, all three GvpA variants contain a canonical GvpA domain. In addition, S39006 has three genes (gvpF1, gvpF2 and gvpF3) encoding homologues of a reportedly minor structural protein, GvpF, and each is conserved in γ‐ and δ‐proteobacteria that carry a gas vesicle gene cluster (Fig. S1). Other genes encoding the gas vesicle‐associated proteins GvpC, GvpG, GvpH, GvpK and GvpN are also conserved in seven proteobacteria (Fig. 1 and Fig. S1). S39006 has five candidate gas vesicle proteins of unknown function: GvpV, GvpW, GvpX, GvpY and GvpZ, and of these, GvpV and GvpZ are conserved in four other β‐, γ‐ and δ‐proteobacteria gas vesicle gene clusters (Fig. 1).
Figure 1.
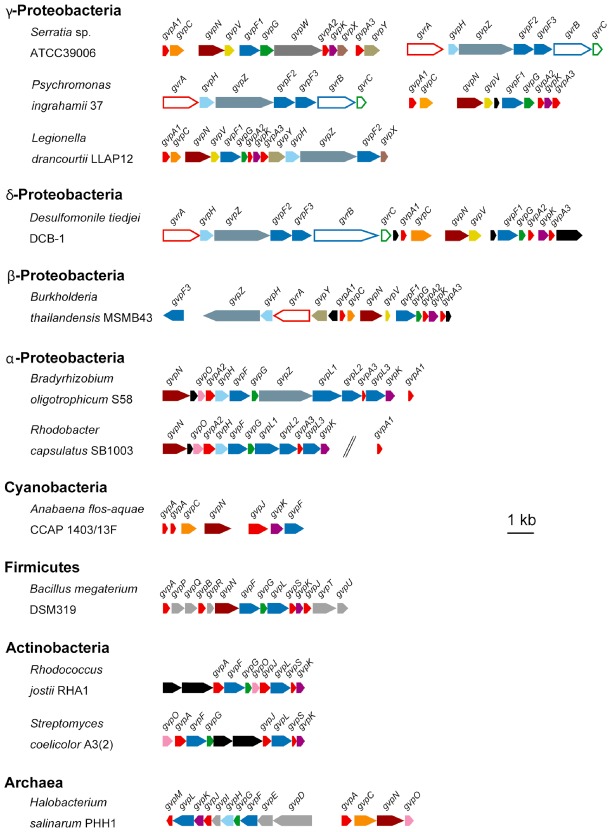
Organization of gas vesicle gene clusters. Gas vesicle gene clusters from the indicated organisms were annotated and compared. Genes of predicted similar function are denoted in the same colour. Genes predicted to encode gas vesicle regulatory proteins (Gvr proteins) are indicated as hollow arrows and gas vesicle proteins (Gvp proteins) are indicated as filled arrows. The scale bar indicates 1 kb.
The gas vesicle locus in S39006 is defined by two distinct operons; one directly regulated by the QS morphogen signal, BHL
The 16.6 kb gas vesicle morphogenesis locus in S39006 is predicted to contain two operons, beginning with gvpA1 and gvrA respectively. Reverse transcription polymerase chain reaction (PCR) and 5′‐RACE analysis showed that the two operons did not overlap and that there was no detectable read through between them (Fig. S2). The corresponding transcriptional start sites upstream of gvpA1 and gvrA were determined (Fig. 2). Using reporter gene fusions, we showed previously that transcription of gvpA1 is controlled by the σ54, NtrC‐like regulator GvrA. This is consistent with the location of a consensus σ54 binding site lying upstream of the gvpA1 transcriptional start site.
Figure 2.
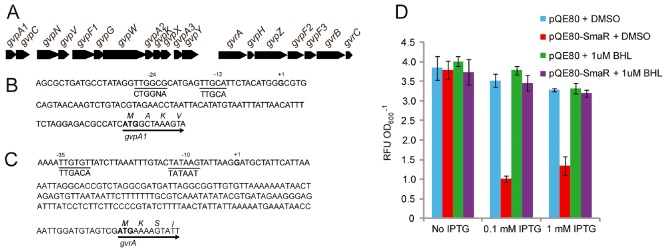
Operonic determination of S39006 gas vesicle cluster.
A. Genetic organization of the S39006 gas vesicle gene cluster locus.
B. The gvpA 1 transcriptional start site determined by 5′RACE is denoted in bold with a +1 above, and potential −12 and −24 sites of its promoter are underlined. Consensus sequences for σ54 are shown below. The translational start site is indicated with an arrow.
C. The gvrA transcriptional start site, determined by 5′RACE, is denoted in bold with a +1 above, and potential −10 and −30 sites of its promoter are underlined. The consensus sequences for σ70 are shown below and the translational start site is indicated with an arrow.
D. SmaR regulates gvpA 1 directly, but not gvrA. E scherichia coli strains carrying pRW50‐gvpA 1pro were grown in the presence of either pQE80 or pQE80‐SmaR with added dimethylsulphoxide (DMSO) or 1 μM BHL dissolved in DMSO, where indicated. After 8 h of growth, samples were taken and β‐gal activity assayed (represented as RFU OD 600 −1). The values are the average of three biological replicates ± standard deviation (SD).
The production of gas vesicles in S39006 is cell density dependent, controlled by the quorum‐sensing locus, SmaIR (Ramsay et al., 2011). SmaI produces the diffusible signalling molecule, N‐butanoyl‐L‐homoserine lactone (BHL) and, above a concentration threshold, BHL binds to SmaR, derepressing target gene expression (Slater et al., 2003). In a smaI mutant (BHL−), gas vesicles are absent but production can be restored by the addition of 1 μM BHL (Ramsay et al., 2011). BHL is therefore defined as a diffusible morphogen. To determine whether SmaR directly regulated expression of gas vesicles through either the gvrA or gvpA1 operons, their respective promoters were first inserted upstream of a β‐galactosidase (β‐gal) reporter fusion. We investigated the expression of these two promoters in E. coli carrying either the gvrA or gvpA1 promoter in the presence of a plasmid encoding SmaR, or with an empty vector control. In the presence or in the absence of SmaR, β‐gal expression from the gvrA promoter was the same (Fig. S2). In contrast, activity of the gvpA1 promoter decreased by over 75% in the presence of SmaR, but this repression could be relieved by the addition of 1 μM BHL (Fig. 2). Together, these results suggest that QS‐dependent regulation of flotation in S39006 acts via direct transcriptional repression of gvpA1, but not gvrA.
Comprehensive deletion analysis of the gas vesicle operons
Previous studies of the proteins required for construction of gas vesicles in archaea and cyanobacteria have focused largely on the main structural protein, GvpA, and the strengthening protein, GvpC (Walsby and Hayes, 1988; Walsby, 1994; Offner et al., 1996). In an earlier study on S39006, transposon insertion mutants and strains with reporter fusions in gvpA1 and gvrA were analysed, but these insertion mutations were polar (Ramsay et al., 2011) so it was not possible to assess the impacts of the 19 individual gvp or gvr genes (mostly with no known function) on gas vesicle morphogenesis. Furthermore, as our bioinformatic analysis suggests, the core gene set found within gas vesicle clusters is likely to be widely conserved (Fig. 1). To elucidate the role of all 19 genes in the S39006 locus and to try to define the ‘minimum gene set’ required for gas vesicle morphogenesis, in frame deletions were constructed for each gene. Each mutant was scored on plates for the opaque colony morphotype (a facile indicator of gas vesicle presence); cultures were examined directly for phase‐bright vacuoles by PCM; and flotation assays were performed to determine if mutants were buoyant (Fig. 3). Transmission electron microscopy (TEM) images were also taken to observe the presence and shapes of gas vesicles (Fig. 4). From these data, we concluded that gvpA1, F1, G, A2, K, A3, F2 and F3 and gvrA and B were required for the formation of phase‐bright structures and for cell buoyancy (Fig. 3); none of the corresponding mutant strains produced gas vesicles detectable by TEM or by PCM under any growth conditions tested. In a gvrC mutant, a few gas vesicles were observed in some cells taken from a colony on a plate, but not in planktonic cells. Ectopic synthesis of the corresponding wild‐type proteins restored gas vesicle production in each deletant strain, confirming that each in‐frame knockout only affected one gene (Fig. S3). From these data, we concluded that the protein products of these 10 genes are each required for gas vesicle formation in S39006.
Figure 3.
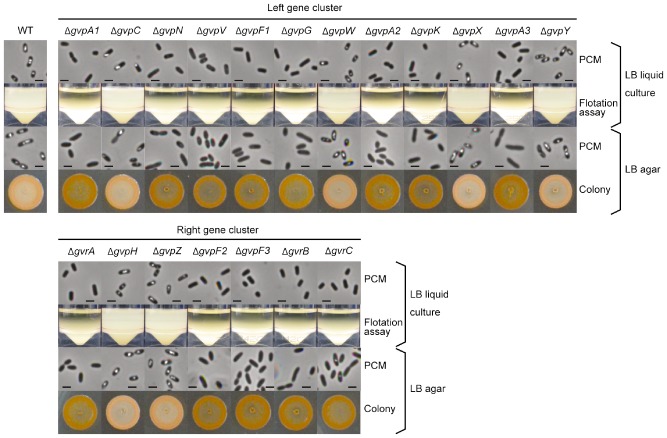
Comprehensive mutational analysis of the gas vesicle production locus. Gas vesicle production in the WT and each gvp or gvr mutant was assessed throughout both the gvpA1 operon (top section) and gvrA operon (bottom section). Cells were grown in LB in sealed universals or on LB agar at 30°C for 24 h. Gas vesicle formation was confirmed by PCM observations from liquid culture (top row), flotation assays from liquid culture (second row), PCM from colonies grown on plates (third row) or colony morphology (fourth row). The scale bars in the PCM images represent 1 μm.
Figure 4.
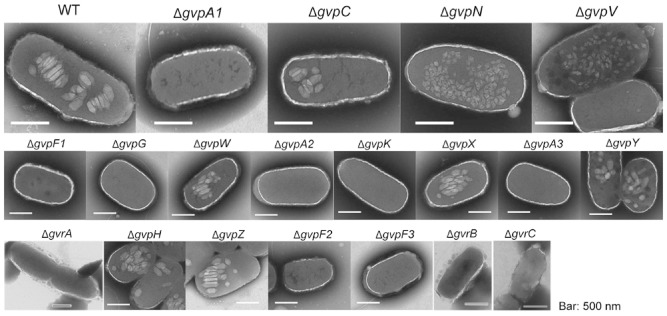
Transmission electron microscopy images of wild type (WT) and several gvp mutants. WT cells and mutants were grown in LB in sealed universals for 24 h and gas vesicles were observed by TEM (scale bars represent 500 nm).
S39006 mutants defective in gvpN and gvpV did not float but contained small bicone‐shaped gas vesicles visible by TEM (Fig. 4). It has been observed in other bacteria that gas vesicles initially assemble in a bicone form before morphogenetic development into the full cylindrical form (for comprehensive reviews, see Walsby, 1994; Pfeifer, 2012). When wild‐type versions of either gvpN or gvpV were expressed in these S39006 mutants, in trans, gas vesicles were observed by PCM, and full‐size mature gas vesicles were seen by TEM (Fig. S4A). However, neither a deletion of gvpN nor gvpV could be complemented by excess of the other, suggesting that the two proteins perform independent functions (data not shown). Previous work demonstrated that applying pressure to wild‐type cells could lead to generation of similar bicone vesicles in S39006 and, in the absence of further stresses, these grew into the larger form (Ramsay et al., 2011). However, when we applied pressure to either a gvpN or gvpV mutant, bicone vesicles were still visible by TEM, suggesting that these bicone structures are robust (Fig. S4). The measurements of gas vesicles within gvpN or gvpV mutants showed that they were the same size in both strains, although significantly different from gas vesicles seen in the wild‐type strain (Fig. S4B). Taken together, these data suggest that GvpN and GvpV are essential for the morphogenetic assembly of gas vesicles from simple bicones into mature, functional gas vesicles in an S39006 developmental program.
There are three GvpA isoforms in S39006. Ramsay et al postulated that each of the GvpA proteins might relate to a different size of vesicle (2011). However, we found that loss of any one of the gvpA protein isoforms prevented gas vesicle formation (Fig. 3). Thus, we believe that all three GvpA isoforms seem to be essential for the production of mature gas vesicles in S39006.
Robustness of gas vesicles under pressure
In other organisms, GvpC is thought to form a meshed structure on the surface of gas vesicles, providing additional strength (Walsby and Hayes, 1988; Hayes et al., 1992; Pfeifer, 2012). Gas vesicles are phase bright within cells, but when collapsed, they no longer refract light. By applying incremental pressure and examining the change in light refraction, the mean critical pressure (p c), when 50% of gas vesicle has collapsed, could be determined. The p c of gas vesicles in S39006 was 0.4334 MPa and, in a gvpC mutant, it decreased to 0.1435 MPa (Fig. 5A). This decrease in p c was counteracted by expression of gvpC in trans (Fig. S5A). It was formally possible that this change in collapse pressure could have been because the gas vesicles in a gvpC mutant were a different shape and size. However, no significant differences in gas vesicle sizes were observed between a gvpC mutant and wild type (Fig. S4) and so these data confirm that GvpC39006 strengthens gas vesicles.
Figure 5.
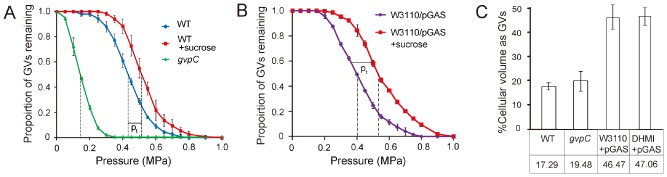
Physical characterization of gas vesicles and volumetric calculations in E . coli and S39006. (A) Collapse pressure experiments on S39006 or (B) E . coli carrying the pGAS cosmid. At 0.05 MPa pressure increments, the relative proportion of gas vesicles present in each sample was ascertained. The values are the average of three biological replicates ± SD. The black dashed lines indicate the mean collapse pressure, p c, for each sample and the continuous black line indicates p t, the turgor pressure. Where indicated, the collapse pressure was measured in cells exposed to LB medium containing 0.35 M sucrose. (C) Volumetric calculation of the percentage of gas vesicles in cells. Values indicated are the mean gas vesicle percentage of three biological replicates ± SD.
Gas vesicles can be produced in E. coli strain W3110 carrying the S39006 gas vesicle cluster expressed from the cosmid, pGAS (Ramsay et al., 2011). We used this strain to compare the behaviour of gas vesicles in E. coli and S39006. The p c of gas vesicles in E. coli strain W3110 carrying pGAS was very similar to that of the gas vesicles in S39006 (0.4012 MPa in E. coli and 0.4334 MPa in S39006), suggesting structural similarity in both natural and engineered hosts (Fig. 5B).
Gas vesicles as tools for measurement of instantaneous turgor pressure of E . coli and S39006
Cells maintain a cytoplasmic solute balance relative to their environment, and the difference between external and internal concentrations results in turgor pressure. Instantaneous measurements of turgor within cells have proved problematic, but a method for measuring turgor pressure within live cells containing gas vesicles exists (Holland and Walsby, 2009). By measuring the relative change in p c when cells are exposed to medium containing 0.35 M sucrose, altering the balance of solute concentration relative to the local environment, turgor pressure (pt) in live cells can be assessed. However, until now, this has only been used in cyanobacteria and never in proteobacteria such as E. coli or S39006 (Holland and Walsby, 2009).
Based on TEM observations and collapse pressure profiles, gas vesicles in E. coli and S39006 were indistinguishable. Thus, we were able to exploit them to calculate corresponding instantaneous turgor pressures by pressure nephelometry. We first examined the change in p c in S39006 between cells in Lysogeny Broth – Lennox (LB) or cells exposed to LB + 0.35 M sucrose. The difference in collapse pressures between these two conditions (p t) was 0.1036 MPa (Fig. 5A), suggesting that the turgor pressure in S39006 was around 0.1 MPa. Similar experiments testing the difference in p c in E. coli indicated that p t was 0.1423 MPa (Fig. 5B). Thus, we have been able to exploit the presence of gas vesicles to determine turgor pressure for the first time in enterobacteria such as E. coli.
Transcription of gvpA 1 is independently regulated by GvrA, GvrB and GvrC
We showed previously that transcription of gvpA1 was dependent on prior expression of the gvrA‐gvrC operon, but it was unknown which specific genes were involved in this regulation (Ramsay et al., 2011) To study this further, we combined in frame mutations in gvrA, gvrB or gvrC in S39006 with a gvpA1::uidA reporter fusion and analysed β‐glucuronidase (β‐gluc) activity in these strains throughout growth. It was reported previously that β‐gluc activity increased exponentially in stationary phase in a strain carrying a gvpA1::uidA fusion alone. However, a significant decrease in β‐gluc activity, reflecting gvpA1 expression, was observed in strains carrying gvrA, gvrB or gvrC mutations (Fig. S6). Taken together, these results suggest that GvrA, GvrB and GvrC are required independently for full expression of the gvpA1 operon and gas vesicle synthesis in S39006.
Discussion
Different taxis systems enable bacterial dissemination between niches. The components that make up the apparatus required for swimming and twitching motility have been widely studied and their assembly pathways are well defined (Macnab, 2003; Busch and Waksman, 2012). Although the importance of gas vesicles in facilitating flotation is understood, many organisms that produce gas vesicles, such as the archaeon H. salinarum, contain gas vesicle genetic clusters that contain genes absent from S39006, for example gvpI and gvpO, and a different regulatory hierarchy, most notably well‐characterized regulators GvpD and GvpE (Kruger et al., 1998; Hofacker et al., 2004; Scheuch et al., 2008; Bleiholder et al., 2012; Marschaus and Pfeifer, 2012). Therefore, comprehensive studies of gas vesicles in H. salinarum or the cyanobacterium Anabaena flos‐aquae are not necessarily readily comparable with those in S39006 (Hayes and Powell, 1995; Offner et al., 1996; 1998; 2000; Li and Cannon, 1998; Hofacker et al., 2004). Furthermore, not all bacteria that possess a putative gas vesicle genetic cluster may actually produce gas vesicles. For example, the actinomycete, Streptomyces coelicolor contains a cryptic gas vesicle gene cluster, but no conditions have yet been identified where gas vesicles are formed, implying that gas vesicles may perform another, unidentified, function in this soil bacterium (van Keulen et al., 2005). An alternative explanation may be that the organism simply lacks one or more proteins that are essential for gas vesicle production. The gas vesicle gene cluster in S. coelicolor lacks gvpN, gvpV, gvpF2, gvrA, gvrB and gvrC, all genes required for full cylindrical gas vesicle formation in S39006. Further, the GvpA protein encoded in the S. coelicolor cluster differs significantly from other gas vesicle‐producing organisms studied so far (Fig. S1 and van Keulen et al., 2005). Although there is variation between the different gene clusters, the required gene set described here may provide a starting point for ‘turning on’ gas vesicles in S. coelicolor and other bacterial species.
Although not all gas vesicle gene clusters contain the same genes in an identical order, they follow a general pattern, at least within the Proteobacteria. The gene cluster is divided into two operons, one starting with gvpA1, as in S39006. The second operon almost always begins with gvrA and includes genes with known regulatory function such as gvrB and gvrC. However, in Psychromonas ingrahamii 37 and Desulfomonile tiedjei DCB‐1 the order of the operons has been inverted, and in Burkholderia thailandensis MSMB43 the two operons are divergently transcribed. Despite these clear differences, the arrangement of genes is largely similar across many bacterial species (Fig. 1).
Gas vesicles are produced by the enterobacterium, S39006, a highly genetically tractable strain (Ramsay et al., 2011). Furthermore, the gas vesicle genetic module from 39006 can be functionally reconstituted in E. coli, and so the roles of each S39006 gene involved in gas vesicle production in both natural and heterologous hosts can also be addressed. Here, we have performed the first full mutational analysis of the two operons that form the gas vesicle cluster in S39006 and determined the minimal gene set required for gas vesicle synthesis. We have ascertained further information about proteins such as GvpN and GvpV, potentially acting as chaperones, that facilitate formation of cylindrical vesicles from small bicones in S39006. In H. salinarum PHH1, the deletion of gvpN resulted in smaller bicone gas vesicles, but this study was unable to define the function of GvpN and the corresponding mutations could not be complemented (Offner et al., 2000). Furthermore, in H. salinarum NRC‐1, deletion of gvpN from the mini‐chromosome pNRC100 also led to the formation of smaller vesicles (DasSarma et al., 1994). However, neither of these gas vesicle gene clusters contains a copy of gvpV, similar to that found in S39006. GvpV homologues have been identified in only five proteobacterial species, suggesting that this protein may perform a function specific to these bacteria (Fig. 1). However, we have shown that both GvpN and GvpV act independently to extend the gas vesicle shape from bicone to the full cylindrical form. Although this study has clarified the function of some gas vesicle genes, there are also differences in functions between organisms. For example, gvpH is found in all six proteobacterial strains we analysed, but it is absent from A. flos‐aquae and Bacillus megaterium (Fig. 1). In the archaeon H. salinarum PHH1, gvpH is present and, as in S39006, its deletion is not detrimental to gas vesicle formation, yet in a gvpH deletion of H. salinarum, gas vesicles were less stable when isolated and could not be visualized by TEM (Offner et al., 2000). However, an earlier study demonstrated that an insertion in gvpH resulted in significantly reduced production of gas vesicles from the gene cluster on the mini‐chromosome pNRC100 in H. salinarum NRC‐1 (DasSarma et al., 1994). In contrast, in S39006, we found no change in gas vesicle shape or stability in a strain carrying a gvpH mutation.
From previous TEM studies, it appeared that differently sized vesicles were made by S39006 (Ramsay et al., 2011), but an extensive microscopy analysis in this study has not shown statistically robust evidence supporting this interpretation. Our results could suggest a finely tuned assembly process with few visually obvious intermediate steps, as almost all of our mutants were either gas vesicle negative or revealed gas vesicles with no discernible phenotypic difference from those of the wild type. These results echo those observed in other studies of gas vesicle genetic clusters from H. salinarum NRC‐1 and PHH1 and in M. aeruginosa, where very few intermediate assembly steps were identified (DasSarma et al., 1994; Offner et al., 2000; Mlouka et al., 2004).
It seems unlikely that the deletion of these ‘non‐essential’ genes has no impact whatsoever in gas vesicle biogenesis. For example, in these experiments, we cannot assess the kinetics of gas vesicle assembly because we only monitor the behaviour of cell populations. Thus, it is formally possible that some of these ‘non essential’ genes might play very subtle roles in modulating gas vesicle formation that cannot be detected in crude assays or seen by TEM. It is also possible that S39006 modulates the structure of gas vesicles under varying physiological conditions that were not investigated in this work.
In addition to defining the minimum gene set required for gas vesicle formation in S39006, we were also able to physically characterize these gas vesicles. The collapse pressure of gas vesicles in S39006 (0.4334 MPa) is similar to that found in the cyanobacterium, Microcystis sp. 8401 (0.468 MPa) (Holland and Walsby, 2009) but is very different than the collapse pressure of gas vesicles in Microcystis sp. BC 84/1 (0.77 MPa; Walsby and Bleything, 1988) or H. salinarum (0.09 MPa; Walsby, 1972). Furthermore, as in Anabaena and other systems, removal of GvpC caused more than a threefold decrease in the critical collapse pressure (Buchholz et al., 1993). Thus, it appears that the structural robustness function of GvpC is conserved in S39006, Anabaena and Microcystis sp. 8401. Similarly, the role of GvpA1 (or GvpA in other systems) appears to be the main structural protein. However, the role of GvpA2/A3 (or the corresponding proteins GvpJ/S in other systems) is still unclear. In S39006, we have annotated these three proteins as variants of GvpA because all three isoforms contain clear GvpA domains, despite the different annotations used in other organisms. Finally, and most importantly, we have demonstrated that all three of these GvpA variants are required for the morphogenesis of functional gas vesicles in S39006.
An ongoing challenge in the study of cellular turgor pressure has been the difficulty of ascertaining instantaneous turgor in a living culture without damaging the cells. For example, atomic force microscopy has been used to measure the deformability of the cellular membrane (Arnoldi et al., 2000; Deng et al., 2011). Although this technique is useful, it can only be done at a single‐cell level, which may not reflect the average turgor pressure across a bacterial culture. However, the biosynthesis of gas vesicles can be exploited to determine the turgor pressure across cells in a heterogeneous culture. As gas vesicles are opaque, their collapse results in a change in culture turbidity. By monitoring the changes in pressure collapse under different conditions (say one where turgor pressure has been removed by placing the cells in an isotonic solution), the turgor pressure across a culture can be calculated. As we can now reproduce this in E. coli, for the first time, the effect of particular mutations in solute transport pathways, on internal cellular pressure can be determined. Potentially, this has widespread applications and may have utility in other bacterial systems, wherever they are capable of producing gas vesicles.
This study significantly enhances our understanding of gas vesicles in S39006, and raises multiple questions. First, although some roles for GvpA and GvpC have been documented in other systems, important questions remain about the functionality of proteins GvpA2 and A3 (or GvpJ and S in other systems). All three isoforms of GvpA (A1, A2 and A3) in S39006 contain a putative ‘GvpA’ domain, and our mutational and complementation analyses have now proved that all three GvpA variants are essential for gas vesicle formation in 39006. More information is clearly required about the structures of all three GvpA variants, how they may perhaps dock together, and their precise morphogenetic roles in gas vesicle development.
This work with in frame mutants has also defined several new proteins as essential for gas vesicle formation in S39006. For example, what are the precise roles of GvrA, B and C in the regulation of gas vesicle synthesis? Each of the three proteins is essential for gas vesicle formation under the growth conditions tested here, and each independently regulates transcription of the gvpA1 operon, although they could be integrating different environmental or physiological cues in the signal transduction pathway to gas vesicle development. The specific role of the three GvpF variants in gas vesicle assembly also remains unclear. GvpF is widely conserved across different gas vesicle gene clusters (Fig. 1), and the protein has been identified in two proteomic gas vesicle analyses (Shukla and DasSarma, 2004; Chu et al., 2010). A recent report described the structure of GvpF from M. aeruginosa and showed that it localized to the gas‐facing surface of gas vesicles, suggesting that this protein does play a structural role (Xu et al., 2014). However, the precise role of GvpF in the structure or assembly of gas vesicles has never been elucidated. It also remains unclear why S39006 contains three GvpF isoforms. An earlier hypothesis that each GvpF variant might pair with one of the three GvpA proteins (Ramsay and Salmond, 2012) now seems unlikely, but further study of the structures and functions of these essential GvpF variants is required to clarify their role(s).
In summary, although the production of gas vesicles was originally described in 1895, the specific role of each protein required for gas vesicle formation is still being dissected (DasSarma et al., 1994; Offner et al., 2000; Mlouka et al., 2004; Pfeifer, 2012; Tavlaridou et al., 2013; 2014; Xu et al., 2014). This work describes the minimal gene set required for gas vesicles in S39006 and demonstrates that gas vesicles formed in E. coli and S39006 are physically indistinguishable. Gas vesicles have potential applications in multiple fields, including in nanoparticles (DasSarma et al., 2014), as a contrast agent in ultrasound imaging (Shapiro et al., 2014a, 2014b), and for delivery of antigens as fusion proteins (DasSarma et al., 2014). In the synthetic biology, chemical factory, biotechnology and bioprocessing arenas, the gas vesicle cluster could be exploited in E. coli, or other organisms, to engineer cells that float or sink in a tightly controlled way – perhaps in response to specific synthetic chemical signals. Furthermore, through the use of pressure nephelometry, the exploitation of the gas vesicle cluster in E. coli can now allow quantifiable assessments of the impact of any mutation or environmental condition on the instantaneous turgor pressure of a whole culture.
Experimental procedures
Bacterial strains, plasmids and culture conditions
Serratia sp. ATCC39006 LacA was the wild‐type parental strain for all mutant strains constructed and used in this work. Bacterial strains and plasmids used in this study are listed in Table S2. Additional details of construction of strains and plasmids are found in the Strain construction and plasmid cloning section below. Oligonucleotides used are listed in Table S3. S39006 and E. coli were grown in LB (10 g tryptone l−1, 5 g NaCl l−1, 5 g yeast extract l−1) or LB agar (1.5%) at 30°C or 37°C, respectively, and supplemented with antibiotics where necessary.
Strain construction and plasmid cloning
In frame mutants carrying deletions of each gene in the S39006 gas vesicle cluster were constructed by homologous recombination using derivatives of the suicide vector pKNG101 listed in Table S2. For construction of gvpA1 mutant, fragments flanking the open reading frame were amplified with oligonucleotide pairs, gvpA1_5F_BamHI/gvpA1_5R_XhoI, and gvpA1_3F_XhoI/gvpA1_3R_SpeI (all oligonucleotides are listed in Table S3). These fragments were used to perform overlap extension PCR, creating a DNA fragment lacking the gvpA1 open reading frame, but containing the flanking DNA. The resulting DNA fragment was digested with BamHI/SpeI and ligated into compatibly digested pKNG101 to yield pKNG101‐ΔgvpA1. The Escherichia coli β 2163 harbouring this plasmid was used as a donor for conjugation, and marker‐exchange mutagenesis was carried out. The confirmation of the gene deletion was by PCR and sequence analysis. The construction of other mutants was performed in the same way with restriction enzymes corresponding to their respective oligonucleotides.
For the construction of pQE80oriT‐gvpA1, the gvpA1 gene was amplified from the S39006 chromosome with oligonucleotides gvpA1F‐EcoRI and gvpA1R‐HindIII (Table S3), digested with EcoRI/HindIII and cloned in pQE80oriT under control of the lac promoter. The plasmid‐producing GvpA1 without a His6 tag was introduced into Serratia by conjugation using E. coli β 2163 as a donor strain. The construction of other plasmids containing gvp or gvr gene was performed similarly but with restriction enzymes corresponding to their respective oligonucleotides.
The promoter fusions of gvrApro and gvpA1pro were created by PCR using oligonucleotides oREM399/400 and oREM397/398 respectively. These fragments were digested with EcoRI/HindIII and ligated into compatibly digested pRW50 and used to transform E. coli strain DH5α, creating pRW50‐gvpA1 pro and pRW50‐gvrA pro. Each of these strains was transformed with either pQE80 or pQE80‐SmaR (Slater et al., 2003).
RNA studies
The cells from cultures grown in LB medium were pelleted by centrifugation, and RNA was extracted from the samples using the RNeasy mini kit (Qiagen) according to the manufacturer's instruction. Residual DNA was removed using the TURBO DNA‐free kit (Ambion) according to the manufacturer's instructions. The transcriptional start sites of gvpA1 and gvrA were determined using a 5′/3′RACE kit (Roche). Reverse transcription PCR was carried out to determine if the left gene cluster is operonic. The synthesis of cDNA was performed with random hexamers using SuperScript II (Life Technologies). PCR was conducted using cDNA as a template with oligonucleotide pairs: RTGVL1/RTGVL2, RTGVL3/RTGVL4, RTGVL5/RTGVL6, RTGVL7/RTGVL8, RTGVL9/RTGVL10, RTGVL11/RTGVL12, RTGVL13/RTGVL14, RTGVL15/RTGVL16 and RTGVL17/RTGVL18.
The transcriptional start site of gvpA1 was determined by 5′RACE using a 5′/3′ RACE kit (Roche). The synthesis of cDNA was carried out with an oligonucleotide RTgvpCR and cDNA was treated with terminal transferase and dATP as per the manufacturer's instruction. PCR for 5′RACE was carried out using specific oligonucleotides complementary for the gvpA1 (gvpA1R‐HindIII) and oligo‐dT. The PCR product was cloned into pBLUEScript SK+, yielding pBluescript + gvpA1‐5′RACE. Both the original and cloned 5′RACE products were sequenced.
5′RACE analysis was also used to identify the transcriptional start site of gvrA and to determine if the right gene cluster was operonic. cDNA synthesis was carried out using an oligonucleotide complementary to the 3′ end of gvrC, the final gene in the predicted operon containing gvrA, gvpH, gvpZ, gvpF2, gvpF3, gvrB and gvrC. cDNA was treated with terminal transferase and dGTP to attach a poly‐G tract to the 3′ of the cDNA. PCR for 5′RACE was carried out using specific oligonucleotides complementary to gvrA (gvrASP1bam) and a poly‐C oligonucleotide (polyDRACE). Nested oligonucleotides were then used to further amplify from this reaction (gvrASP2bam and Anchorprimer, see Roche instructions), which resulted in amplification of a single product that was subsequently cloned into pBLUEScript SK+. Both the original and the cloned 5′RACE products were sequenced.
Confirmation of gas vesicle formation
A total of 5 ml of overnight cultures was grown in 25 ml sealed universals on a roller wheel at 30°C and then used to subculture into a second 5 ml broth, which was grown for 24 h under the same conditions. For flotation assays, cultures were then kept statically at room temperature for 48 h before imaging by PCM, TEM or flotation in the universal tube was assessed. Cells from liquid cultures or agar plates were observed by PCM or TEM as described previously (Ramsay et al., 2011). Cells were imaged for PCM using an Olympus BX‐51 microscope with a 100x oil‐immersion lens, with a QICAM monochrome camera and qcapture pro‐6 software. For TEM, samples were observed in a FEI Tecnai G2 TEM. Images were captured with an AMT XR60B digital camera running deben software at the Cambridge University Advanced Imaging Centre. For colony morphology experiments, 3 µl of normalized culture were spotted onto LB agar plates, incubated at 30°C for 24 h and colony morphologies observed. Cells scraped from plates were suspended in PBS and observed by PCM.
Pressure nephelometry
Pressure collapse experiments were performed largely using the same apparatus, and as previously described, in Holland and Walsby (2009). Cultures were grown overnight on a roller wheel at 30°C and then left to settle for 24 h at room temperature before application of incremental pressure using the apparatus shown in Fig. S5. As intracellular gas vesicles collapsed under increasing pressure, changes in culture turbidity were assessed. The collapse pressures of gas vesicles were determined by pressure nephelometry. The machine was set to zero with 4 ml of LB and 500 μl of the indicated culture was added and mixed. This mixture was carefully placed into the nephelometer, ensuring that 100% of gas vesicles remained intact, and the millivoltmeter set to 100. Pressure was applied as N2 gas in 0.05 MPa increments, up to 1 MPa, and the change in culture turbidity monitored after 20 s of equilibration using an analogue millivoltmeter. At each pressure increment, the proportion of gas vesicle remaining (G p) was calculated using the following formula:
where Gm is the measurement taken at the indicated pressure and G 1MPa and G 0MPa are the measurements of turbity taken at 1 MPa and 0 MPa respectively.The mean critical pressure (p c) was calculated by determining the pressure when G p = 0.5. When cultures were exposed to 0.35 M sucrose solutions, the mean apparent critical pressure (p a) was also determined by identifying the pressure when G p = 0.5. The turgor pressure (p t) was calculated by subtracting p a from p c.
Volume calculation of gas vesicles
Calculation of gas volume in cultures was performed as described in Walsby (1982). Briefly, cultures were grown as described for pressure nephelometry, and the relative change in volume was calculated using the apparatus shown in Fig. S5. The change in culture volume was recorded under pressure that caused the collapse of 100% of vesicles (0.8 MPa). The change in culture volume was normalized to the total cell numbers to determine the percentage of cell volume that was due to gas vesicles. Volumetric calculations were performed using a compression tube (Walsby, 1982; Walsby et al., 1992). Tubes, with a capillary 0.2 mm in internal diameter, were filled to capacity (∼2.5 ml of culture). The volume of gas vesicles was determined by observing the relative change in the meniscus inside the capillary (L) upon application of 0.8 MPa of pressure, via N2 gas, causing the collapse of all gas vesicles within the culture. A change in 1 mm of the meniscus within a capillary 0.2 mm in diameter is equivalent to 31.4 ηl. The mean gas vesicle volume per cell was calculated using the change in volume normalized to the number of cells in each culture, determined by colony counts.
Measurements of gas vesicles
TEM images were analysed using imagej (Schneider et al., 2012). Each image was calibrated against its scale bar, and gas vesicles were measured. Mean heights and widths of gas vesicles were then calculated.
β‐Galactosidase and β‐glucuronidase activity
S39006 Cultures were grown at 30°C with shaking at 250 r.p.m. and the optical densities (OD600) examined every 2 h. After 6 h of incubation, samples were taken and frozen at −80°C until required. Activity of β‐gluc, the protein product of uidA, was determined using the fluorogenic substrate, 4′‐Methylumbelliferyl–d‐glucuronide (Melford Laboratories), as described previously (Ramsay et al., 2011). Briefly, frozen samples were thawed and diluted to appropriate ratios where a linear increase in activity could be observed, and 10 µl of diluted samples was frozen and thawed again. Then, 100 µl of reaction buffer (400 g ml−1 lysozyme, 250 g ml−1 4′‐Methylumbelliferyl–d‐glucuronide in PBS) was added, and fluorescence was immediately monitored (excitation 360 nm, emission 450 nm, cut‐off 435 nm) every 30 s for 30 min using a Gemini XPS plate reader. β‐Gluc expression was measured by the relative fluorescence units per second, produced from cleavage of 4′‐Methylumbelliferyl‐β‐d‐glucuronide, normalized to the optical density of the culture (RFU s−1 OD600 −1). β‐Gal activity was monitored in a similar fashion but using 4′‐Methylumbelliferyl‐β‐d‐galactopyranoside (Melford Laboratories) as a fluorogenic substrate.
Bioinformatic analyses
Sequences were accessed from NCBI, and putative gas vesicle clusters in other organisms were identified using psi‐blast (Altschul et al., 1997). Amino acid sequences were aligned using clustalx (Larkin et al., 2007) and phylogenetic trees were constructed by njplot (Perriere and Gouy, 1996). Phylogenetic distances were determined by neighbour‐joining analysis. The numbers on branching points are percentages of bootstrap values with 1000 replicates.
Supporting information
Fig. S1. Phylogenetic relationships of GvpA, GvpF/L, GvpC, GvpG, GvpH, GvpK, GvpN, GvpV and GvpZ.
Fig. S2. The gas vesicle production locus of S39006 is composed of two operons. (A) The locus of the amplified region in the gvp cluster. (B) RT‐PCR results showing that the left cluster, starting from gvpA1, is operonic. (C) RT‐PCR result showing that there is no read through between the left and right operon. indicates the marker lane, with indicated sizes on the side. P indicates a sample including reverse transcriptase, C indicates a positive control sample containing only genomic DNA, and N indicates a negative control sample with no reverse transcriptase. (D) SmaR does not regulate gvrA directly. Escherichia coli strains carrying pRW50‐gvrApro were grown in the presence of either pQE80 or pQE80‐SmaR with added DMSO or 1 μM BHL dissolved in DMSO, where indicated. After 8 h of growth, samples were taken and β‐gal activity assayed (represented as RFU OD600 −1). The values are the average of three biological replicates ± SD.
Fig. S3. Complementation of GV formation in in frame mutations. Mutations in the GV cluster that failed to produce GVs were grown with the indicated plasmid, with or without 0.1 mM IPTG, and the cells were observed by PCM, flotation assays and colony morphology. (A) Analyses of ΔgvpA1, ΔgvpA2 and ΔgvpA3 mutants. (B) Analyses of ΔgvpF1, ΔgvpF and ΔgvpF3 mutants. (C) Analyses of ΔgvpG and ΔgvpK mutants. (D) Analyses of ΔgvpN, ΔgvpV, ΔgvrA, ΔgvrB and ΔgvrC mutants. Scale bar indicates 1 μm.
Fig. S4. Measurements of GVs in different mutant strains and with application of pressure. (A) gvpV or gvpN mutant strains were imaged by TEM with or without 1.0 MPa pressure or with either gvpV or gvpN added back in trans. Scale bars indicate 500 nm. (B) Measurement of GVs in different strains. TEM images of GVs from the indicated strains, grown in sealed universals, were analysed using imagej and the height and widths of the indicated number (below) of vesicles measured. The values presented are the average heights and widths ± SD.
Fig. S5. (A) Restoration of the collapse pressure phenotype in the gvpC mutant, by an in trans copy of gvpC. (B–C) Apparatus used for pressure nephelometry (B) or volumetric calculations (C).
Fig. S6. Expression from the gvpA1::uidA transcriptional fusion throughout growth in S39006 WT, ΔgvrA, ΔgvrB, and ΔgvrC backgrounds. Dashed lines show growth and solid lines show gene expression levels via reporter enzymes. β‐Gluc activity was assayed and represented as RFU OD600 −1. Values are the mean of three biological replicates ± SD.
Table S1. Comparison of amino acid sequences of GvpA and GvpF proteins from S39006.
Table S2. Bacterial strains and plasmids used in this study.
Table S3. Oligonucleotides used in this study.
Acknowledgements
The authors would like to thank Professor Tony Walsby (Emeritus, Bristol University) for advice, technical help and donation of the pressure nephelometry and volumetric calculation apparatus. We would also like to thank Alison Drew for technical support, and Chin Mei Lee and Andrew Day for critical reading. REM and GPCS were supported through the BBSRC (Grant ID BB/K001833/1). YT was supported by a Scientific Research Fellowship from the Japan Society for the Promotion of Sciences (JSPS) and JPR was supported by a Herschel Smith Post Doctoral Fellowship while at Cambridge University. The authors declare that they have no conflict of interest.
References
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. , and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldi, M. , Fritz, M. , Bauerlein, E. , Radmacher, M. , Sackmann, E. , and Boulbitch, A. (2000) Bacterial turgor pressure can be measured by atomic force microscopy. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 62: 1034–1044. [DOI] [PubMed] [Google Scholar]
- Bleiholder, A. , Frommherz, R. , Teufel, K. , and Pfeifer, F. (2012) Expression of multiple tfb genes in different Halobacterium salinarum strains and interaction of TFB with transcriptional activator GvpE. Arch Microbiol 194: 269–279. [DOI] [PubMed] [Google Scholar]
- Buchholz, B.E. , Hayes, P.K. , and Walsby, A.E. (1993) The distribution of the outer gas vesicle protein, GvpC, on the Anabaena gas vesicle, and its ratio to GvpA. J Gen Microbiol 139: 2353–2363. [DOI] [PubMed] [Google Scholar]
- Busch, A. , and Waksman, G. (2012) Chaperone‐usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci 367: 1112–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, L.J. , Chen, M.C. , Setter, J. , Tsai, Y.S. , Yang, H. , Fang, X. , et al (2010) New structural proteins of Halobacterium salinarum gas vesicle revealed by comparative proteomics analysis. J Proteome Res 10: 1170–1178. [DOI] [PubMed] [Google Scholar]
- Chu, L.J. , Chen, M.C. , Setter, J. , Tsai, Y.S. , Yang, H. , Fang, X. , et al (2011) New structural proteins of Halobacterium salinarum gas vesicle revealed by comparative proteomics analysis. J Proteome Res 10: 1170–1178. [DOI] [PubMed] [Google Scholar]
- Coulthurst, S.J. , Barnard, A.M. , and Salmond, G.P. (2005) Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol 3: 295–306. [DOI] [PubMed] [Google Scholar]
- DasSarma, P. , Negi, V.D. , Balakrishnan, A. , Karan, R. , Barnes, S. , Ekulona, F. , et al (2014) Haloarchaeal gas vesicle nanoparticles displaying Salmonella SopB antigen reduce bacterial burden when administered with live attenuated bacteria. Vaccine 32: 4543–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma, S. , Arora, P. , Lin, F. , Molinari, E. , and Yin, L.R. (1994) Wild‐type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J Bacteriol 176: 7646–7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Sun, M. , and Shaevitz, J.W. (2011) Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys Rev Lett 107: 158101. [DOI] [PubMed] [Google Scholar]
- Englert, C. , and Pfeifer, F. (1993) Analysis of gas vesicle gene expression in Haloferax mediterranei reveals that GvpA and GvpC are both gas vesicle structural proteins. J Biol Chem 268: 9329–9336. [PubMed] [Google Scholar]
- Englert, C. , Kruger, K. , Offner, S. , and Pfeifer, F. (1992) Three different but related gene clusters encoding gas vesicles in halophilic archaea. J Mol Biol 227: 586–592. [DOI] [PubMed] [Google Scholar]
- Hayes, P. , and Powell, R. (1995) The gvpA/C cluster of Anabaena flos‐aquae has multiple copies of a gene encoding GvpA. Arch Microbiol 164: 50–57. [DOI] [PubMed] [Google Scholar]
- Hayes, P.K. , Lazarus, C.M. , Bees, A. , Walker, J.E. , and Walsby, A.E. (1988) The protein encoded by gvpC is a minor component of gas vesicles isolated from the cyanobacteria Anabaena flos‐aquae and Microcystis sp. Mol Microbiol 2: 545–552. [DOI] [PubMed] [Google Scholar]
- Hayes, P.K. , Buchholz, B. , and Walsby, A.E. (1992) Gas vesicles are strengthened by the outer‐surface protein, GvpC. Arch Microbiol 157: 229–234. [DOI] [PubMed] [Google Scholar]
- Hofacker, A. , Schmitz, K.M. , Cichonczyk, A. , Sartorius‐Neef, S. , and Pfeifer, F. (2004) GvpE‐ and GvpD‐mediated transcription regulation of the p‐gvp genes encoding gas vesicles in Halobacterium salinarum . Microbiology 150: 1829–1838. [DOI] [PubMed] [Google Scholar]
- Holland, D.P. , and Walsby, A.E. (2009) Digital recordings of gas‐vesicle collapse used to measure turgor pressure and cell‐water relations of cyanobacterial cells. J Microbiol Methods 77: 214–224. [DOI] [PubMed] [Google Scholar]
- Houwink, A.L. (1956) Flagella, gas vacuoles and cell‐wall structure in Halobacterium halobium; an electron microscope study. J Gen Microbiol 15: 146–150. [DOI] [PubMed] [Google Scholar]
- van Keulen, G. , Hopwood, D.A. , Dijkhuizen, L. , and Sawers, R.G. (2005) Gas vesicles in actinomycetes: old buoys in novel habitats? Trends Microbiol 13: 350–354. [DOI] [PubMed] [Google Scholar]
- Kruger, K. , Hermann, T. , Armbruster, V. , and Pfeifer, F. (1998) The transcriptional activator GvpE for the halobacterial gas vesicle genes resembles a basic region leucine‐zipper regulatory protein. J Mol Biol 279: 761–771. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, N. , and Cannon, M.C. (1998) Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli . J Bacteriol 180: 2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab, R.M. (2003) How bacteria assemble flagella. Annu Rev Microbiol 57: 77–100. [DOI] [PubMed] [Google Scholar]
- Marschaus, L. , and Pfeifer, F. (2012) A dual promoter region with overlapping activator sequences drives the expression of gas vesicle protein genes in haloarchaea. Microbiology 158: 2815–2825. [DOI] [PubMed] [Google Scholar]
- Mlouka, A. , Comte, K. , Castets, A.‐M. , Bouchier, C. , and Tandeau de Marsac, N. (2004) The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to loss of cell buoyancy. J Bacteriol 186: 2355–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.V. , Kennedy, S.P. , Mahairas, G.G. , Berquist, B. , Pan, M. , Shukla, H.D. , et al (2000) Genome sequence of Halobacterium species NRC‐1. Proc Natl Acad Sci USA 97: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offner, S. , Wanner, G. , and Pfeifer, F. (1996) Functional studies of the gvpACNO operon of Halobacterium salinarium reveal that the GvpC protein shapes gas vesicles. J Bacteriol 178: 2071–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offner, S. , Ziese, U. , Wanner, G. , Typke, D. , and Pfeifer, F. (1998) Structural characteristics of halobacterial gas vesicles. Microbiology 144: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Offner, S. , Hofacker, A. , Wanner, G. , and Pfeifer, F. (2000) Eight of fourteen gvp genes are sufficient for formation of gas vesicles in halophilic archaea. J Bacteriol 182: 4328–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriere, G. , and Gouy, M. (1996) WWW‐query: an on‐line retrieval system for biological sequence banks. Biochimie 78: 364–369. [DOI] [PubMed] [Google Scholar]
- Pfeifer, F. (2012) Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol 10: 705–715. [DOI] [PubMed] [Google Scholar]
- Ramsay, J.P. , and Salmond, G.P.C. (2012) Quorum sensing‐controlled buoyancy through gas vesicles: Intracellular bacterial microcompartments for environmental adaptation. Commun Integr Biol 5: 96–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, J.P. , Williamson, N.R. , Spring, D.R. , and Salmond, G.P.C. (2011) A quorum‐sensing molecule acts as a morphogen controlling gas vesicle organelle biogenesis and adaptive flotation in an enterobacterium. Proc Natl Acad Sci USA 108: 14932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuch, S. , Marschaus, L. , Sartorius‐Neef, S. , and Pfeifer, F. (2008) Regulation of gvp genes encoding gas vesicle proteins in halophilic Archaea. Arch Microbiol 190: 333–339. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. , and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, M.G. , Goodwill, P.W. , Neogy, A. , Yin, M. , Foster, F.S. , Schaffer, D.V. , and Conolly, S.M. (2014a) Biogenic gas nanostructures as ultrasonic molecular reporters. Nat Nanotechnol 9: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, M.G. , Ramirez, R.M. , Sperling, L.J. , Sun, G. , Sun, J. , Pines, A. , et al (2014b) Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem 6: 629–634. [DOI] [PubMed] [Google Scholar]
- Shukla, H.D. , and DasSarma, S. (2004) Complexity of gas vesicle biogenesis in Halobacterium sp. strain NRC‐1: identification of five new proteins. J Bacteriol 186: 3182–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, H. , Crow, M. , Everson, L. , and Salmond, G.P. (2003) Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum‐sensing‐dependent and ‐independent pathways. Mol Microbiol 47: 303–320. [DOI] [PubMed] [Google Scholar]
- Tavlaridou, S. , Faist, K. , Weitzel, K. , and Pfeifer, F. (2013) Effect of an overproduction of accessory Gvp proteins on gas vesicle formation in Haloferax volcanii . Extremophiles 17: 277–287. [DOI] [PubMed] [Google Scholar]
- Tavlaridou, S. , Winter, K. , and Pfeifer, F. (2014) The accessory gas vesicle protein GvpM of haloarchaea and its interaction partners during gas vesicle formation. Extremophiles 18: 693–706. [DOI] [PubMed] [Google Scholar]
- Walsby, A.E. (1972) Structure and function of gas vacuoles. Bacteriol Rev 36: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsby, A.E. (1982) The elastic compressibility of gas vesicles. Proceedings of the Royal Society Series B‐Biological Sciences 216: 355–368. [Google Scholar]
- Walsby, A.E. (1994) Gas vesicles. Microbiol Rev 58: 94–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsby, A.E. , and Bleything, A. (1988) The dimensions of cyanobacterial gas vesicles in relation to their efficiency in providing buoyancy and withstanding pressure. Microbiology 134: 2635–2645. [Google Scholar]
- Walsby, A.E. , and Hayes, P.K. (1988) The minor cyanobacterial gas vesicle protein, GvpC, is attached to the outer surface of the gas vesicle. J Gen Microbiol 134: 2647–2657. [Google Scholar]
- Walsby, A.E. , Kinsman, R. , and George, K.I. (1992) The measurement of gas vesicle volume and buoyant density in planktonic bacteria. J Microbiol Methods 15: 293–309. [Google Scholar]
- Williamson, N.R. , Fineran, P.C. , Leeper, F.J. , and Salmond, G.P. (2006) The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4: 887–899. [DOI] [PubMed] [Google Scholar]
- Xu, B.Y. , Dai, Y.N. , Zhou, K. , Liu, Y.T. , Sun, Q. , Ren, Y.M. , et al (2014) Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa . Acta Crystallogr D Biol Crystallogr 70: 3013–3022. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P. , and Pfeifer, F. (2003) Regulation of the expression of gas vesicle genes in Haloferax mediterranei: interaction of the two regulatory proteins GvpD and GvpE. Mol Microbiol 49: 783–794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic relationships of GvpA, GvpF/L, GvpC, GvpG, GvpH, GvpK, GvpN, GvpV and GvpZ.
Fig. S2. The gas vesicle production locus of S39006 is composed of two operons. (A) The locus of the amplified region in the gvp cluster. (B) RT‐PCR results showing that the left cluster, starting from gvpA1, is operonic. (C) RT‐PCR result showing that there is no read through between the left and right operon. indicates the marker lane, with indicated sizes on the side. P indicates a sample including reverse transcriptase, C indicates a positive control sample containing only genomic DNA, and N indicates a negative control sample with no reverse transcriptase. (D) SmaR does not regulate gvrA directly. Escherichia coli strains carrying pRW50‐gvrApro were grown in the presence of either pQE80 or pQE80‐SmaR with added DMSO or 1 μM BHL dissolved in DMSO, where indicated. After 8 h of growth, samples were taken and β‐gal activity assayed (represented as RFU OD600 −1). The values are the average of three biological replicates ± SD.
Fig. S3. Complementation of GV formation in in frame mutations. Mutations in the GV cluster that failed to produce GVs were grown with the indicated plasmid, with or without 0.1 mM IPTG, and the cells were observed by PCM, flotation assays and colony morphology. (A) Analyses of ΔgvpA1, ΔgvpA2 and ΔgvpA3 mutants. (B) Analyses of ΔgvpF1, ΔgvpF and ΔgvpF3 mutants. (C) Analyses of ΔgvpG and ΔgvpK mutants. (D) Analyses of ΔgvpN, ΔgvpV, ΔgvrA, ΔgvrB and ΔgvrC mutants. Scale bar indicates 1 μm.
Fig. S4. Measurements of GVs in different mutant strains and with application of pressure. (A) gvpV or gvpN mutant strains were imaged by TEM with or without 1.0 MPa pressure or with either gvpV or gvpN added back in trans. Scale bars indicate 500 nm. (B) Measurement of GVs in different strains. TEM images of GVs from the indicated strains, grown in sealed universals, were analysed using imagej and the height and widths of the indicated number (below) of vesicles measured. The values presented are the average heights and widths ± SD.
Fig. S5. (A) Restoration of the collapse pressure phenotype in the gvpC mutant, by an in trans copy of gvpC. (B–C) Apparatus used for pressure nephelometry (B) or volumetric calculations (C).
Fig. S6. Expression from the gvpA1::uidA transcriptional fusion throughout growth in S39006 WT, ΔgvrA, ΔgvrB, and ΔgvrC backgrounds. Dashed lines show growth and solid lines show gene expression levels via reporter enzymes. β‐Gluc activity was assayed and represented as RFU OD600 −1. Values are the mean of three biological replicates ± SD.
Table S1. Comparison of amino acid sequences of GvpA and GvpF proteins from S39006.
Table S2. Bacterial strains and plasmids used in this study.
Table S3. Oligonucleotides used in this study.


