Abstract
Background
Color patterns in angiosperm flowers are produced by spatially and temporally restricted deposition of pigments. Identifying the mechanisms responsible for restricted pigment deposition is a topic of broad interest. Some dicots species develop bicolor petals, which are often caused by the post-transcriptional gene silencing (PTGS) of chalcone synthase (CHS) genes. An Asiatic hybrid lily (Lilium spp.) cultivar Lollypop develops bicolor tepals with pigmented tips and white bases. Here, we analyzed the global transcription of pigmented and non-pigmented tepal parts from Lollypop, to determine the main transcriptomic differences.
Results
De novo assembly of RNA-seq data yielded 49,239 contigs (39,426 unigenes), which included a variety of novel transcripts, such as those involved in flavonoid-glycosylation and sequestration and in regulation of anthocyanin biosynthesis. Additionally, 1258 of the unigenes exhibited significantly differential expression between the tepal parts (false discovery rates <0.05). The pigmented tepal parts accumulated more anthocyanins, and unigenes annotated as anthocyanin biosynthesis genes (e.g., CHS, dihydroflavonol 4-reductase, and anthocyanidin synthase) were expressed 7–30-fold higher than those in non-pigmented parts. These results indicate that the transcriptional regulation of biosynthesis genes is more likely involved in the development of bicolor lily tepals rather than the PTGS of CHS genes. In addition, the expression level of a unigene homologous to LhMYB12, which often regulates full-tepal anthocyanin pigmentation in lilies, was >2-fold higher in the pigmented parts. Thus, LhMYB12 should be involved in the transcriptional regulation of the biosynthesis genes in bicolor tepals. Other factors that potentially suppress or enhance the expression of anthocyanin biosynthesis genes, including a WD40 gene, were identified, and their involvement in bicolor development is discussed.
Conclusions
Our results indicate that the bicolor trait of Lollypop tepals is caused by the transcriptional regulation of anthocyanin biosynthesis genes and that the transcription profile of LhMYB12 provides a clue for elucidating the mechanisms of the trait. The tepal transcriptome constructed in this study will accelerate investigations of the genetic controls of anthocyanin color patterns, including the bicolor patterns, of Lilium spp.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2995-5) contains supplementary material, which is available to authorized users.
Keywords: Anthocyanin, Flower color pattern, De novo assembly, LhMYB12, R2R3-MYB, Transcriptional regulation, WD40
Background
Lilies are among the most important floricultural crops, owing to their large flowers with unique and diverse colors. The genus Lilium consists of >90 species, which are classified into several sections [1, 2], and since species belonging to the same section have relatively high interspecific crossing abilities, interspecific hybridization is the principal method of lily breeding. Among the resulting hybrids, the Asiatic hybrid lilies (Lilium spp.) are one of the main groups and are derived from interspecific crosses among the species of section Sinomartagon, which are mainly distributed in East Asia [3].
In lilies, flower color is a commercially important characteristic and much interest has been placed in cultivars that bear flowers with unique colors. Asiatic hybrid lilies, specifically, exhibit large variations in color hue that result from the accumulation of anthocyanins and carotenoids, which result in pink and yellow/orange coloration, respectively, or red coloration with the combination of anthocyanins and orange carotenoids [4–6]. Flavonols, flavones, and cinnamic acid derivatives (CADs) in higher plants are colorless flavonoid or phenylpropanoid compounds having ultraviolet-absorbing characteristics and, in floral organs, these show co-pigmentation effects with anthocyanins [7–9]. In the lily tepals, high amounts of CADs accumulate, whereas flavonols and flavones are often scarcely present [10]. In addition to the wide variation in color hue, the Asiatic hybrid lilies also exhibit variation in color patterns, including the occurrence of several types of spots [11] and bicolor phenotypes, in which two distinct colors occur on individual tepals [4]. For example, the Asiatic hybrid lily cultivar Lollypop has bicolor (pink and white) tepals, in which anthocyanin pigments are heavily accumulated in the upper tepals but less so in the bases.
Anthocyanins are among the most studied and best understood compounds in plant science, and their metabolic pathways have been extensively described (Additional file 1: Figure S1) [12]. The activity of anthocyanin biosynthesis enzymes is primarily controlled at the transcriptional level and is regulated by complexes that consist of the R2R3-MYB and basic helix-loop-helix (bHLH) transcription factors and WD40 proteins (hereafter, MBW complexes) [13, 14]. In angiosperm flowers, large variations in color patterns are observed, owing to the spatially and temporally restricted deposition of anthocyanin pigments, and color pattern variations include bicolor phenotypes, vein-associated anthocyanin pigmentation (venation), stripes and spots, and light-induced pigment accumulation on exposed petal surfaces (bud-blush). Clarifying these mechanisms is a topic of broad interest [15, 16]. The mechanisms of restricted pigment deposition have been characterized in some model plants, and studies have shown that individual species possess multiple R2R3-MYB genes that are responsible for regulating the biosynthesis of anthocyanins in flower petals. In petunias, for example, AN2 generates fully pigmented petals, whereas PURPLE HAZE (PHZ) determines bud-blush, and DEEP PURPLE (DPL) causes venation in the flower tubes [17]. In snapdragon, Rosea1 and Rosea2 determine whether the petals are fully pink but have different activities, and Venosa regulates venation [18, 19]. Furthermore, monkeyflowers (Mimulus spp.) have two R2R3-MYB genes, PELAN and NEGAN, which control anthocyanin production in the petal lobes and in the spots of the nectar guide, respectively [20]. In non-model lilies, several R2R3-MYBs have been shown to control anthocyanin pigmentation, together with LhbHLH2, in flowers [21–24]. For example, LhMYB12 controls anthocyanin pigmentation in whole tepals [22], Latvia allele of LhMYB12 determines splatter-type tepal spots [23], and LrMYB15 regulates a bud-blush phenotype in Lilium regale [24]. These observations in both model plants and lilies indicate that R2R3-MYB transcription factors are principally involved in color patterning.
Mechanisms other than the regulation by R2R3-MYB transcription factors cause bicolor patterns. The bicolor phenotypes of star and marginal picotee (white margin, pigmented center) petunias are caused by the post-transcriptional gene silencing (PTGS) of the chalcone synthase (CHS) A gene, which is one of the anthocyanin biosynthesis genes, in white areas [25, 26]. Another type of marginal picotee petunias that exhibit a pigmented margin and white center is not caused by the down-regulation of anthocyanin biosynthesis genes. At the white centers, increased accumulations of flavonols and flavonol synthase (FLS) transcripts have been detected [27], which indicates that there is competition between the enzymes FLS and dihydroflavonol-4-reductase (DFR) for the common substrate, dihydroflavonol, and that flavonol biosynthesis is more dominant than anthocyanin biosynthesis at the white centers. These findings indicate that several mechanisms are involved in bicolor generation. However, the mechanisms of bicolor tepal development in lilies have yet to be analyzed.
Lilium spp. have a huge genome (33–36 Gb) [28], often exhibit gametophytic self-incompatibility, and have long life cycles (3–7 years from sowing to anthesis). Although a few molecular linkage maps have been developed, using PCR-based markers [29, 30], high-density genetic maps are lacking. Mutant and tagging lines are not developed yet, and both stable and transient transformation are still challenging. Thus, the genetic analysis of agricultural traits in lilies is difficult. To date, several biosynthesis and regulatory genes that are involved in anthocyanin biosynthesis in lily flowers have been identified [21, 22, 31–34]; however, the overall molecular mechanisms that underlie tepal pigmentation remains limited; e.g., no sequence information of WD40 proteins or anthocyanin-glycosylating enzymes. In addition, although several negatively regulating transcription factors have been reported in other species [35–37], such information is not available in lilies.
Enrichment of genetic resources, including transcriptome sets, is essential in order to provide effective tools for further extensive and intensive research on more complicated flower traits in lilies. The whole transcriptome sequencing based on next-generation sequencing (NGS) and de novo assembly enable transcriptome studies of non-model organisms without reference sequences [38, 39]. The strategy has been widely applied to non-model plants, e.g., to floricultural crops, in order to obtain mass sequence data for the molecular mechanisms responsible for color diversity [40–42]. Because of the huge genome size, whole genome sequencing is currently unavailable in lilies. Thus, de novo transcriptome sequencing has been applied to Lilium spp., and several NGS studies have already been published. More than two thousand simple sequence repeats (SSR) were detected in expressed sequences used to develop SSR markers [43, 44], the global transcription of lily bulb meristems after cold-treatment was profiled to understand the molecular regulation of vernalization [45, 46], and genome-wide transcription was analyzed to clarify the genetic response to cold-stress [47]. However, there have been no NGS studies concerning color diversification in lily tepals.
In the present study, whole transcriptome sequencing was conducted using the bicolor Asiatic hybrid lily cultivar Lollypop. RNA samples from pink (pigmented) and white (non-pigmented) tepal parts were sequenced using Illumina NGS, and the gene expression levels of the two tepal regions were compared. The objectives of the present study were to sequence the whole transcriptome of lily tepals and to define the main transcriptomic differences between the two tepal parts, in order to characterize the mechanisms involved in complicated bicolor traits.
Results and discussion
Qualitative and quantitative analyses of anthocyanins and CADs
The Lollypop cultivar exhibited bicolor tepals, with upper tepals that were pink and tepal bases that were white or pale yellow with red raised spots, and the color of the tepals during floral development were as follows: stage (St) 1, tepals were not pigmented yet; St 2, red spots appeared on bases; St 3, pigmentation began in upper tepals; St 4 (one day before anthesis), the content of tepal anthocyanin was highest; and St 5, flowering (Fig. 1). First, we measured the amount of pigments accumulated in the pink upper tepals and the white bases using high-performance liquid chromatography (HPLC). The segments of upper tepals and tepal bases were cut out from the same inner tepals at St 4, and pigments were evaluated with three replicates (each replicate derived from a different flower). The Lollypop tepals included a single anthocyanin pigment, and its retention time and absorbance spectrum were identical to those of cyanidin 3-O-β-rutinoside (Additional file 1: Figure S2). However, flavones and flavonols were not detected (data not shown). Furthermore, at least two major peaks were detected by absorbance at 340 nm (Additional file 1: Figure S2), and a wavelength scan of each of the peaks suggested that they included a caffeic acid moiety. Because such products are derived from cinnamic acid, we hereafter refer to these compounds as CADs [10, 27]. These products were further separated by silica gel column chromatography and thin-layer chromatography (TLC), and one of the major compounds was identified using 1H- and 13C-NMR as 1-O-β-D-caffeoylglucose [48].
Fig. 1.

Tepal development of the Asiatic hybrid lily Lollypop. Inner tepal (right) and outer tepal (left) at each stage are shown. Flower bud development stages (St) are as follows: St 1, no anthocyanin pigmentation; St 2, red spots appeared on tepal bases; St 3, beginning of pigmentation in upper tepals; St 4 (1 d before anthesis), maximum pigmentation; St 5, blossoming. Scale bar = 1 cm. Colored boxes indicate tepal parts used for the experiments (black boxes–upper tepals, red boxes–tepal bases)
When the amounts of anthocyanins and CADs of the pink upper tepals and white bases were compared, high amounts of anthocyanins were detected in the upper tepals, whereas much less was found in the bases (Fig. 2a). In contrast, the amount of CADs was higher in the bases than in the upper tepals (Fig. 2b), confirming that the accumulation profiles of the two tepal parts were different.
Fig. 2.
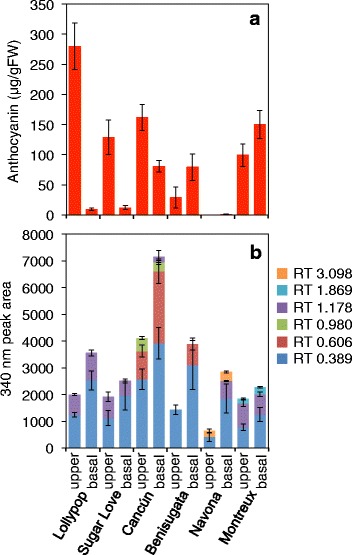
Anthocyanins and cinnamic acid derivatives in the tepals of six Asiatic hybrid lily (Lilium) cultivars. a Anthocyanin content. b The six cinnamic acid derivatives were distinguished by HPLC retention time (RT, min). Values and error bars indicate the means ± standard error (n = 3). FW: fresh weight
In the marginal picotee cultivars of petunia, which exhibit pigmented margins and white centers, a dramatic increase in flavonol accumulation is detected in the white centers [27]. To verify whether the higher accumulation of CADs in the bases specifically occurred in bicolor lily cultivars, the anthocyanin and CAD contents of five additional cultivars, which included two other bicolor cultivars, were examined (Fig. 2). In the bicolor cultivars Sugar Love (pink upper tepals and white bases) and Cancún (red upper tepals and yellow bases), more anthocyanins were found in upper tepals, in accordance with their color. In the Cancún cultivar, because of pink abaxial surface of inner tepals, a relatively small difference was observed in the anthocyanin content of the upper tepals and tepal bases. Meanwhile, in the non-bicolor cultivars Benisugata (red tepals) and Montreux (pink tepals), more anthocyanins were found in the bases, and no or very little anthocyanins were detected in the white tepal cultivar Navona.
Among the six Asiatic hybrid cultivars, six types of CADs were detected and were distinguished by retention time (RT). The total amounts of CADs were higher in the tepal bases than in upper tepals in all of the tested cultivars. The most abundant compound (RT = 0.389 min) was commonly detected among the six cultivars, and its amounts were also higher in tepal bases (Fig. 2b). These results indicate that the higher accumulation of CADs in the bases of lily tepals is not specific to bicolor cultivars.
Segregation of the bicolor trait in F1 progeny
To determine the genetic background of the bicolor trait, segregation of the bicolor trait was analyzed using F1 plants that were derived from crosses between Lollypop and another Asiatic hybrid lily cultivar (Blackout) that has tepals fully pigmented by anthocyanins. Wild species and cultivars of Lilium are heterozygous, and thus, traits often segregate in F1 progenies. Of the 240 F1 plants, 226 exhibited bicolor tepals, and 14 exhibited fully pigmented tepals. This segregation was skewed to the Lollypop parent (bicolor trait) and did not fit the segregation ratios of 1:1 or 3:1, which indicates that the bicolor trait is not inherited in a Mendelian fashion and is controlled by at least several genes.
Sequencing, de novo assembly, and detection of differentially expressed genes (DEGs)
To elucidate the genetic controls of complicated traits, we performed RNA-seq for the pink upper part and white basal part of St 3 tepals with two replicates. Two replicates were derived from distinct plants and, in each replicate, segments of upper tepals and tepal bases were collected from three inner tepals of a single flower. These samples were analyzed in a single run on the Illumina MiSeq to obtain relatively highly expressed sequences. Approximately 50 million raw reads (~24 million fragments) were obtained as a result and de novo assembly yielded 39,426 unigenes (Additional file 1: Table S1).
We compared the expression of the 39,426 unigene in the upper tepals and tepal bases and 1258 exhibited significantly differential expression (false discovery rate [FDR] <0.05, Additional file 2: Table S2). The positive and negative log2 Fold Change (LogFC) values respectively indicated the up- and down-regulation of the unigenes in upper tepals, relative to the bases, and indicated that 665 unigenes (53 %) were up-regulated in upper tepals.
Unigenes related to anthocyanin and CAD accumulation
One of the aims of the present study was to obtain the sequences of all the genes involved in anthocyanin and CAD biosyntheses in Lilium. These biosynthesis pathways are branched from the general phenylpropanoid pathway, and a number of enzymes are involved (Additional file 1: Figure S1). Phenylalanine ammonia-lyase (PAL), CHS, chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), DFR, and anthocyanidin synthase (ANS) genes have been identified in Lilium but the other genes have not. Thus, the unigene sequences of putatively encoded proteins were BLASTed against the Swiss-Prot database and 15, 31, and three unigenes in Lollypop sequence database were predicted to encode phenylpropanoid, anthocyanin, and cinnamic acid biosynthesis enzymes, respectively. In addition, 27 candidates were predicted to encode proteins that are required for flavonoid sequestration in vacuoles, including members of the multidrug and toxic compound extrusion (MATE) gene family and glutathione S-transferase (GST) genes (Table 1) [49, 50]. All of the main genes necessary for anthocyanin and CAD biosyntheses were included in the Lollypop transcriptome. Among the candidates for CHI, F3H, F3′H, DFR, and ANS, unigenes with relatively high FPKM (Fragments Per Kb per Million mapped reads) values (hereafter, major unigenes) had high sequence similarity to the corresponding genes isolated from the other Asiatic hybrid lily cultivar or L. speciosum [10, 31, 34]. In some cases, unigenes did not correspond well to previously isolated genes in lilies: Unigene c29955_g1 (annotated as PAL) consisted of three PAL gene sequences (LhPAL1, LhPAL2, and LhPAL3) and unigene c30110_g1 (annotated as CHS) contained LhCHSa and LhCHSb sequences (Additional file 1: Figure S3). It was expected that these gene sequences were not distinguished during de novo assembly.
Table 1.
Annotation and expression of unigenes involved in anthocyanin and cinnamic acid derivative biosyntheses and transport
| Putative gene (according to Swiss-Prot) | Trinity transcript id | Homologous gene in Lilium | Blastp top hit | Length | Identity | E-value | FPKM (expression) in | logFC | logCPM | FDR | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Upper tepals | Tepal bases | ||||||||||
| Phenylpropanoid pathway genes | |||||||||||
| PAL | c29955_g1 | LhPAL1/2/3 | phenylalanine ammonia lyase [Lilium hybrid division I] | 729 | 99 % | 0 | 631.93 | 472.92 | 0.41 | 10.09 | 0.7580164 |
| PAL | c36922_g1 | LhPAL3 | phenylalanine ammonia lyase [Lilium hybrid division I] | 114 | 95 % | 2E-64 | 44.13 | 5.04 | 3.14 | 3.09 | 2.07E-05 |
| PAL | c30134_g17 | phenylalanine ammonia lyase, partial [Musa acuminata AAA Group] | 436 | 81 % | 0 | 12.30 | 19.98 | −0.65 | 4.32 | 0.6531477 | |
| PAL | c30134_g11 | phenylalanine ammonia-lyase [Betula pendula] | 115 | 64 % | 6E-44 | 10.26 | 14.29 | −0.37 | 1.37 | 0.9748477 | |
| PAL | c30134_g1 | phenylalanine ammonia-lyase, partial [Rhizophora apiculata] | 191 | 84 % | 4E-113 | 8.62 | 14.20 | −0.62 | 2.34 | 0.7807557 | |
| C4H | c26579_g1 | cinnamate-4-hydroxylase [Canarium album] | 507 | 88 % | 0 | 1030.58 | 803.78 | 0.37 | 10.53 | 0.6668013 | |
| C4H | c25090_g1 | cinnamate 4-hydroxylase, partial [Tulipa fosteriana] | 330 | 90 % | 0 | 3.54 | 10.24 | −1.43 | 2.61 | 0.1219183 | |
| 4CL | c29891_g1 | 4-hydroxycinnamoyl-CoA ligase 2 [Coffea arabica] | 518 | 70 % | 0 | 344.61 | 485.55 | −0.39 | 9.02 | 0.6668013 | |
| 4CL | c27769_g1 | 4-coumarate-CoA ligase-like protein [Arabidopsis thaliana] | 561 | 66 % | 0 | 112.74 | 114.24 | −0.01 | 7.48 | 1 | |
| 4CL | c27493_g1 | coumaroyl-CoA ligase [Vitis vinifera] | 522 | 72 % | 0 | 91.97 | 63.02 | 0.57 | 6.96 | 0.6023584 | |
| 4CL | c26975_g1 | 4-coumarate--CoA ligase-like 7 [Vitis vinifera] | 572 | 62 % | 0 | 34.27 | 32.43 | 0.09 | 5.87 | 0.9889067 | |
| 4CL | c26242_g1 | 4-coumarate--CoA ligase-like 9 [Vitis vinifera] | 482 | 61 % | 0 | 14.85 | 26.40 | −0.79 | 4.82 | 0.4774231 | |
| 4CL | c29687_g1 | 4-coumarate--CoA ligase-like 6 [Glycine soja] | 582 | 62 % | 0 | 12.42 | 13.44 | −0.07 | 4.41 | 1 | |
| 4CL | c20611_g1 | 4cl [Lilium hybrid cultivar] | 549 | 97 % | 0 | 5.39 | 16.11 | −1.53 | 4.22 | 0.04040995 | |
| 4CL | c25512_g2 | 4-coumaroyl-coenzyme A ligase [Allium cepa] | 225 | 65 % | 3E-99 | 6.45 | 6.72 | 0.03 | 2.02 | 1 | |
| Cinnamic acid derivative biosynthesis genes | |||||||||||
| C3H | c28376_g1 | 4-coumarate 3-hydroxylase [Eucalyptus globulus] | 508 | 77 % | 0 | 6.19 | 7.07 | −0.17 | 2.89 | 0.9748477 | |
| HCT | c27695_g1 | Hydroxycinnamoyl-CoA:quinate hydroxycinnamoyltransferase [Zostera marina] | 431 | 49 % | 2E-125 | 34.28 | 37.59 | −0.12 | 5.65 | 0.9748477 | |
| HCT | c30288_g1 | hydroxycinnamoyl transferase [Nicotiana benthamiana] | 324 | 63 % | 6E-147 | 5.02 | 46.05 | −3.14 | 4.45 | 0.0001381333 | |
| Anthocyanin biosynthesis genes | |||||||||||
| CHS | c30110_g1 | LhCHSa/b | chalcone synthase [Lilium hybrid division I] | 267 | 99 % | 0 | 1209.02 | 421.80 | 1.47 | 9.77 | 0.01588364 |
| CHS | c29657_g1 | LhCHSc | chalcone synthase C [Lilium hybrid division I] | 249 | 98 % | 1E-173 | 267.20 | 63.11 | 2.10 | 7.05 | 0.001083655 |
| CHS | c29657_g2 | LhCHSc | chalcone synthase C [Lilium hybrid division I] | 249 | 97 % | 1E-174 | 48.68 | 104.39 | −1.12 | 6.01 | 0.1175434 |
| CHS | c30110_g2 | chalcone synthase, partial [Tulipa fosteriana] | 242 | 72 % | 4E-120 | 52.81 | 72.32 | −0.47 | 5.73 | 0.6531477 | |
| CHS | c29920_g1 | chalcone synthase, partial [Tulipa fosteriana] | 389 | 78 % | 0 | 34.11 | 19.43 | 0.65 | 4.42 | 0.4774231 | |
| CHI | c28538_g1 | LsCHIb | chalcone isomerase, partial [Lilium speciosum] | 210 | 99 % | 4E-135 | 1232.45 | 816.96 | 0.60 | 9.77 | 0.4774231 |
| CHI | c28136_g1 | LsCHIa | chalcone isomerase [Lilium speciosum] | 234 | 97 % | 2E-156 | 276.09 | 178.28 | 0.63 | 7.91 | 0.4774231 |
| CHI | c27139_g1 | Chalcone-flavanone isomerase family protein isoform 1 [Theobroma cacao] | 420 | 50 % | 2E-133 | 34.06 | 22.96 | 0.56 | 5.63 | 0.5573998 | |
| CHI | c23754_g1 | Chalcone-flavanone isomerase family protein isoform 3, partial [Theobroma cacao] | 299 | 64 % | 6E-85 | 12.87 | 16.08 | −0.32 | 3.55 | 0.7828791 | |
| F3H | c28413_g1 | LhF3H | flavanone 3-hydroxylase [Lilium hybrid division I] | 370 | 97 % | 0 | 666.13 | 330.01 | 1.03 | 9.21 | 0.1196772 |
| F3'H | c27194_g1 | LhF3'H | flavonoid 3'-hydroxylase [Lilium hybrid division I] | 515 | 99 % | 0 | 195.25 | 33.85 | 2.54 | 7.58 | 1.38E-06 |
| DFR | c30307_g2 | LhDFR | dihydroflavonol 4-reductase [Lilium hybrid division I] | 376 | 98 % | 0 | 3143.14 | 1550.13 | 0.98 | 11.27 | 0.1175434 |
| DFR | c24884_g1 | dihydroflavonol-4-reductase-like [Elaeis guineensis] | 348 | 68 % | 3E-163 | 87.02 | 71.25 | 0.28 | 6.49 | 0.7807557 | |
| DFR | c29645_g2 | cinnamoyl-CoA reductase 1-like [Phoenix dactylifera] | 303 | 70 % | 2E-138 | 35.55 | 34.81 | −0.02 | 4.50 | 1 | |
| DFR | c27848_g1 | dihydroflavonol-4-reductase [Vitis vinifera] | 334 | 73 % | 0 | 15.05 | 15.08 | 0.01 | 4.00 | 1 | |
| DFR | c12034_g1 | Dihydroflavonol-4-reductase [Aegilops tauschii] | 92 | 57 % | 3E-27 | 11.78 | 4.96 | 1.20 | 0.42 | 0.6318245 | |
| DFR | c27774_g2 | Dihydroflavonol-4-reductase [Aegilops tauschii] | 104 | 57 % | 2E-34 | 8.23 | 5.01 | 0.70 | 1.25 | 0.6668013 | |
| DFR | c9045_g1 | Bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [Morus notabilis] | 86 | 65 % | 9E-32 | 0.00 | 10.40 | −5.20 | −0.17 | 0.1055062 | |
| ANS | c26135_g1 | LhANS | anthocyanidin synthase [Lilium hybrid division I] | 362 | 98 % | 0 | 1767.61 | 876.34 | 1.02 | 10.75 | 0.1055062 |
| ANS | c28679_g2 | Gibberellin 3-beta-dioxygenase 4 [Morus notabilis] | 316 | 42 % | 2E-76 | 62.99 | 243.34 | −1.97 | 7.33 | 0.0006236071 | |
| ANS | c27368_g1 | Leucoanthocyanidin dioxygenase [Morus notabilis] | 377 | 62 % | 1E-160 | 68.14 | 50.96 | 0.42 | 6.02 | 0.6531477 | |
| ANS | c19298_g1 | Leucoanthocyanidin dioxygenase [Gossypium arboreum] | 362 | 67 % | 7E-175 | 8.51 | 29.34 | −1.79 | 4.38 | 0.002704819 | |
| 3GT | c35034_g1 | UDP-glucose: anthocyanidin 3-O-glucosyltransferase [Tulipa fosteriana] | 484 | 78 % | 0 | 1650.78 | 1337.40 | 0.31 | 11.18 | 0.7580164 | |
| 3GT | c16413_g1 | anthocyanidin 3-O-glucosyltransferase [Tulipa fosteriana] | 450 | 71 % | 0 | 120.55 | 90.36 | 0.41 | 7.09 | 0.6531477 | |
| 3GT | c29247_g1 | anthocyanidin 3-O-glucosyltransferase [Zea mays] | 489 | 50 % | 8E-137 | 19.03 | 20.65 | −0.10 | 4.92 | 0.9889067 | |
| 3GT | c26782_g1 | UDP glucose:flavonoid 3-O-glucosyltransferase [Pyrus x bretschneideri] | 109 | 54 % | 3E-32 | 21.19 | 16.46 | 0.45 | 2.17 | 0.9367663 | |
| 3GT | c26782_g5 | anthocyanidin 3-O-glucosyltransferase [Tulipa fosteriana] | 346 | 57 % | 6E-115 | 11.65 | 13.85 | −0.18 | 3.53 | 0.9889067 | |
| 3RT | c25725_g1 | anthocyanidin-3-glucoside rhamnosyltransferase [Petunia x hybrida] | 465 | 55 % | 0 | 2144.38 | 1507.37 | 0.51 | 11.53 | 0.6023584 | |
| 7GT | c29610_g1 | scopoletin glucosyltransferase-like [Phoenix dactylifera] | 489 | 60 % | 0 | 53.84 | 80.60 | −0.64 | 5.55 | 0.4774231 | |
| 7GT | c28503_g1 | UDP-glycosyltransferase 1 [Linum usitatissimum] | 418 | 44 % | 6E-107 | 16.58 | 25.74 | −0.57 | 4.50 | 0.6318245 | |
| 7GT | c26782_g3 | phenylpropanoid:glucosyltransferase 2, partial [Nicotiana tabacum] | 377 | 49 % | 1E-117 | 9.78 | 10.58 | −0.09 | 3.36 | 0.9889067 | |
| Transporters of anthocyanins and/or cinnamic acid derivatives, to vacuole | |||||||||||
| MATE | c27999_g1 | MATE efflux family protein [Arabidopsis thaliana] | 535 | 62 % | 0 | 483.91 | 412.47 | 0.25 | 9.61 | 0.8141854 | |
| MATE | c29991_g1 | MATE efflux family protein [Arabidopsis thaliana] | 507 | 72 % | 0 | 106.41 | 103.68 | 0.03 | 7.40 | 1 | |
| MATE | c30035_g1 | transparent testa 12 protein [Zea mays] | 489 | 69 % | 0 | 50.45 | 116.90 | −1.18 | 7.11 | 0.06782638 | |
| MATE | c25329_g1 | transparent testa 12 protein [Zea mays] | 527 | 78 % | 0 | 66.84 | 94.90 | −0.50 | 7.10 | 0.6318245 | |
| MATE | c23411_g1 | MATE efflux family protein LAL5-like isoform X1 [Phoenix dactylifera] | 517 | 73 % | 0 | 37.53 | 41.30 | −0.16 | 5.82 | 0.9243257 | |
| MATE | c1112_g1 | MATE efflux family protein 5 [Phoenix dactylifera] | 510 | 69 % | 0 | 14.64 | 21.87 | −0.52 | 4.84 | 0.6668013 | |
| MATE | c19647_g1 | MATE efflux family protein 5 [Vitis vinifera] | 481 | 55 % | 0 | 13.65 | 13.72 | −0.05 | 3.77 | 1 | |
| MATE | c23623_g1 | transparent testa 12 protein [Zea mays] | 480 | 78 % | 0 | 9.71 | 8.01 | 0.32 | 3.81 | 0.8141854 | |
| MATE | c27478_g1 | MATE efflux family protein [Theobroma cacao] | 258 | 65 % | 1E-108 | 6.83 | 10.52 | −0.50 | 3.06 | 0.6668013 | |
| MATE | c12223_g2 | MATE efflux family protein 5-like [Musa acuminata subsp. malaccensis] | 212 | 78 % | 2E-112 | 3.74 | 8.87 | −1.26 | 1.79 | 0.3152999 | |
| MATE | c17372_g1 | MATE efflux family protein 5-like [Phoenix dactylifera] | 148 | 74 % | 3E-63 | 7.98 | 3.35 | 1.24 | 0.91 | 0.4774231 | |
| MATE | c29524_g1 | MATE efflux family protein 5-like [Elaeis guineensis] | 196 | 77 % | 3E-103 | 5.02 | 5.95 | −0.40 | 0.93 | 0.9243257 | |
| GST | c24611_g1 | glutathione S-transferase [Vitis amurensis] | 228 | 57 % | 9E-88 | 1251.26 | 959.39 | 0.40 | 9.90 | 0.7580164 | |
| GST | c27339_g1 | glutathione S-transferase [Allium cepa] | 215 | 56 % | 7E-82 | 554.35 | 774.28 | −0.49 | 8.90 | 0.6318245 | |
| GST | c28555_g1 | elongation factor 1, partial [Lilium regale] | 418 | 89 % | 0 | 162.71 | 190.06 | −0.22 | 8.00 | 0.8141854 | |
| GST | c27591_g1 | glutathione S-transferase T1-like [Musa acuminata subsp. malaccensis] | 244 | 73 % | 1E-122 | 170.25 | 170.91 | 0.00 | 7.01 | 1 | |
| GST | c25698_g2 | glutathione S-transferase [Tulipa fosteriana] | 229 | 84 % | 2E-133 | 87.22 | 128.32 | −0.83 | 5.88 | 0.3152999 | |
| GST | c24296_g1 | elongation factor 1-gamma 2 [Zea mays] | 417 | 78 % | 0 | 90.46 | 123.33 | −0.44 | 7.27 | 0.6318245 | |
| GST | c25698_g1 | glutathione S-transferase [Arachis hypogaea] | 207 | 71 % | 8E-94 | 90.79 | 120.45 | −0.40 | 5.57 | 0.6668013 | |
| GST | c22668_g1 | glutathione S-transferase 2 [Oryza sativa Japonica Group] | 221 | 69 % | 8E-103 | 93.72 | 80.32 | 0.20 | 6.25 | 0.9131831 | |
| GST | c27057_g1 | glutathione s-transferase [Elaeis guineensis] | 221 | 67 % | 4E-104 | 20.61 | 21.78 | −0.09 | 4.04 | 0.9889067 | |
| GST | c27057_g2 | glutathione s-transferase [Elaeis guineensis] | 258 | 65 % | 2E-98 | 28.04 | 5.32 | 2.46 | 3.63 | 0.100767 | |
| GST | c21745_g1 | Glutathione S-transferase Phi class [Zostera marina] | 221 | 65 % | 1E-96 | 3.32 | 19.01 | −2.49 | 2.80 | 0.003065782 | |
| GST | c20342_g1 | glutathione S-transferase family protein [Populus trichocarpa] | 149 | 64 % | 1E-53 | 8.57 | 12.71 | −0.54 | 1.72 | 0.7580164 | |
| GST | c27063_g1 | Tetrachloro-P-hydroquinone reductive dehalogenase [Zea mays] | 266 | 71 % | 3E-146 | 9.96 | 10.03 | 0.10 | 3.65 | 0.9889067 | |
| GST | c19621_g1 | glutathione S-transferase [Musa acuminata AAA Group] | 111 | 66 % | 6E-43 | 13.16 | 2.27 | 2.40 | 1.02 | 0.2599001 | |
| GST | c33779_g1 | Glutathione S-transferase Phi class [Zostera marina] | 99 | 60 % | 1E-35 | 0.00 | 10.44 | −5.65 | 0.05 | 0.1057929 | |
Most wild species and cultivars of Lilium, including Lollypop, mainly accumulate cyanidin 3-O-β-rutinoside, although some also accumulate cyanidin 3-O-β-rutinoside-7-O-β-glucoside [5]. In the present study, several unigenes were annotated as anthocyanidin 3-O-glucosyltransferase (3GT), anthocyanidin-3-glucoside rhamnosyltransferase (3RT), and anthocyanidin-3-rutinoside 7-glucosyltransferase (7GT), among which unigenes c35034_g1 (3GT) and c25725_g1 (3RT) exhibited relatively high FPKM values.
Among the unigenes involved in phenylpropanoid and anthocyanin biosyntheses, the major unigenes annotated as LhPAL3 (c36922_g1), LhCHSa/b (c30110_g1), LhCHSc (c29657_g1), and LhF3′H (c27194_g1) were highly expressed in the upper tepals (FDR <0.05). These expression profiles were coincident with higher accumulation of anthocyanin pigments in upper tepals. One ANS-annotated unigene (c28679_g2), which showed relatively high FPKM values, was significantly high in its expression in tepal bases (FDR <0.05). However, according to NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), c28679_g2 showed no hits for ANS and was annotated as gibberellin 3-ß-dioxygenase. Thus, it is still unknown whether the protein encoded by this unigene has anthocyanidin synthase activity.
Unigenes related to the regulation of anthocyanin accumulation
In higher plants, the expression of anthocyanin biosynthesis genes is commonly regulated by MBW complexes [13, 14]. Although the functions of LhMYB12 and LhbHLH2 in regulating anthocyanin biosynthesis in the tepals of Lilium spp. are well evaluated, WD40 orthologues have not been isolated. To detect WD40 orthologues in Lilium, we BLASTed the amino acid sequence of AtTTG1 (WD40 in Arabidopsis) against the Lollypop sequence database. Then, unigene c2514_g1 was obtained, which exhibited 78 % amino acid identity to PhAN11 (WD40 in Petunia, data not shown).
R2R3-MYB genes constitute the largest MYB gene family in plants, with 125 R2R3-MYB genes in Arabidopsis and 109 in rice, and regulate plant-specific processes, including the biosynthesis of secondary metabolites [51, 52]. In order to conduct a comprehensive search for R2R3-MYB genes in Lilium, we BLASTed the R2R3 repeat motif sequence [51] against the Lollypop tepal transcriptome. As a result, 51 MYB unigenes were detected, including two MYB3R, 44 R2R3-MYB, three R3-MYB, and two unusual MYB genes that had two or more repeats (Table 2). We have isolated 14 R2R3-MYB cDNA sequences (LhMYB1–12, LhMYB15, and LhMYB16) from wild species and cultivars of Lilium. Of the 14 R2R3-MYB sequences, 10 were highly similar to 11 Lollypop unigenes (Table 2), although four (LhMYB4, 6, 9, and 11) were not represented in the Lollypop sequence database.
Table 2.
Annotation and expression of unigenes showing the homology with R2R3-MYB, R3-MYB, and MYB3R genes
| Type and sub-group (SG)a) | Trinity transcript id | Homologous gene in Lilium | Homologous gene in Arabidopsis | Homologous gene in other species | Blastp top hit | Length | Identity | E-value | FPKM (expression) in | logFC | logCPM | FDR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upper tepals | Tepal bases | ||||||||||||
| MYB3R type | |||||||||||||
| c19823_g1 | AtMYB3R-1 | myb-related protein 3R-1-like isoform X2 [Phoenix dactylifera] | 242 | 74 % | 2E-115 | 4.24 | 7.13 | −0.61 | 1.90 | 1 | |||
| c27498_g5 | AtMYB3R-3,3R-5 | myb-related protein 3R-1-like [Musa acuminata subsp. malaccensis] | 502 | 50 % | 7E-154 | 10.74 | 11.67 | −0.11 | 4.65 | 1 | |||
| R2R3-MYB type | |||||||||||||
| SG 1 | c10428_g1 | AtMYB30,94,96 | Am306 | R2R3-MYB transcriptional factor, partial [Lilium hybrid division I] | 124 | 75 % | 2E-42 | 2.96 | 2.18 | 0.30 | −0.14 | 1 | |
| SG 1 | c19652_g1 | LhMYB7 | AtMYB30,94,96 | Am306 | R2R3-MYB transcriptional factor, partial [Lilium hybrid division I] | 193 | 99 % | 2E-112 | 6.94 | 3.00 | 1.20 | 1.56 | 0.5056056 |
| SG 1 | c19652_g2 | LhMYB7 | AtMYB30,94,96 | Am306 | R2R3-MYB transcriptional factor, partial [Lilium hybrid division I] | 109 | 100 % | 1E-63 | 1.72 | 5.00 | −1.19 | 0.26 | 0.9933287 |
| SG 1 | c23337_g2 | LhMYB5 | AtMYB30,94,96 | Am306 | R2R3-MYB transcriptional factor, partial [Lilium hybrid division I] | 309 | 98 % | 6E-141 | 7.72 | 14.20 | −0.85 | 3.12 | 0.5775627 |
| SG 2 | c2281_g1 | AtMYB13,14,15 | putative MYB transcription factor [Rosa rugosa] | 89 | 83 % | 3E-48 | 10.39 | 7.12 | 0.65 | 1.35 | 1 | ||
| SG 2 | c8990_g1 | AtMYB13,14,15 | myb-like protein Myb15 [Citrus maxima] | 83 | 83 % | 8E-47 | 1.73 | 8.46 | −2.10 | −0.42 | 0.9933287 | ||
| SG 2 | c12438_g1 | AtMYB13,14,15 | myb-related protein Zm1-like [Elaeis guineensis] | 259 | 51 % | 1E-64 | 14.05 | 12.72 | 3.19 | 2.36 | 0.004292067 | ||
| SG 2 | c18397_g1 | AtMYB13,14,15 | Myb-related protein Zm1 [Aegilops tauschii] | 206 | 51 % | 1E-43 | 4.60 | 4.21 | 0.21 | 1.28 | 1 | ||
| SG 2 | c18397_g2 | AtMYB13,14,15 | myb-related protein Myb4-like [Brachypodium distachyon] | 43 | 72 % | 2E-14 | 5.15 | 1.96 | 1.99 | 0.35 | 0.4220982 | ||
| SG 2 | c35333_g1 | AtMYB13,14,15 | MYB transcription factor [Zea mays] | 121 | 87 % | 3E-62 | 1.58 | 4.09 | −1.24 | −0.07 | 1 | ||
| SG 2 | c36958_g1 | AtMYB13,14,15 | myb-related protein Myb4 [Zea mays] | 98 | 69 % | 7E-35 | 1.81 | 1.79 | −0.05 | −0.66 | 1 | ||
| SG 4 | c10735_g1 | LhMYB3 | AtMYB3,4,7,32 | FaMYB1, Am308, Am330 | R2R3-MYB transcriptional factor [Lilium hybrid division I] | 119 | 97 % | 5E-81 | 5.83 | 3.37 | 0.91 | 1.16 | 1 |
| SG 4 | c25442_g1 | LhMYB8 | AtMYB3,4,7,32 | FaMYB1, Am308, Am330 | R2R3-MYB transcriptional factor [Lilium hybrid division I] | 214 | 99 % | 2E-156 | 273.93 | 218.16 | 0.31 | 7.76 | 1 |
| SG 4 | c25442_g2 | AtMYB3,4,7,32 | FaMYB1, Am308, Am330 | myb-related protein 308-like [Elaeis guineensis] | 252 | 69 % | 1E-119 | 72.64 | 61.13 | 0.25 | 6.28 | 1 | |
| SG 6 | c22900_g1 | LhMYB12 | PAP1,PAP2 | PhAN2 | transcription factor R2R3-MYB [Lilium hybrid division I] | 162 | 95 % | 6E-97 | 276.37 | 216.83 | 0.30 | 7.89 | 1 |
| SG 6 | c24386_g1 | LrMYB15 | PAP1,PAP2 | PhAN2 | transcription factor R2R3-MYB [Lilium regale] | 242 | 88 % | 4E-155 | 7.23 | 15.11 | −1.05 | 3.20 | 0.3424106 |
| SG 6 | c24386_g2 | LhMYB16 | PAP1,PAP2 | PhAN2 | R2R3-MYB transcriptional factor, partial [Lilium hybrid division I] | 240 | 84 % | 5E-75 | 5.47 | 8.71 | −0.69 | 2.32 | 0.9933287 |
| SG 7 | c25442_g3 | AtMYB11,12,111 | ZmP | MYB12 [Arabidopsis thaliana] | 287 | 70 % | 1E-72 | 30.92 | 11.36 | 1.49 | 3.84 | 0.3424106 | |
| SG 9 | c10215_g1 | AtMYB17 | AmMIXTA | Myb domain protein 17 isoform 2 [Theobroma cacao] | 72 | 66 % | 1E-19 | 0.00 | 4.63 | −3.28 | −0.92 | 1 | |
| SG 9 | c10215_g2 | AtMYB17 | AmMIXTA | myb domain protein 17 [Zostera marina] | 80 | 91 % | 2E-46 | n.d. | n.d. | n.d. | n.d. | n.d. | |
| SG 9 | c22634_g1 | LhMYB10 | AtMYB17 | AmMIXTA | R2R3-MYB transcriptional factor, partial [Lilium hybrid division I] | 245 | 98 % | 1E-143 | 7.57 | 11.78 | −0.62 | 2.96 | 0.9933287 |
| SG 9 | c22634_g2 | AtMYB17 | AmMIXTA | Transcription factor [Morus notabilis] | 107 | 99 % | 5E-72 | 13.11 | 16.98 | −0.29 | 2.07 | 1 | |
| SG 13 | c4482_g2 | AtMYB55,86 | putative transcription factor MYB55 [Arabidopsis thaliana] | 105 | 96 % | 3E-62 | 2.67 | 1.34 | 0.69 | −0.51 | 1 | ||
| SG 13 | c16280_g2 | AtMYB55,86 | MYB transcription factor MYB89 [Glycine max] | 107 | 48 % | 6E-25 | 2.58 | 4.98 | −0.64 | 0.04 | 1 | ||
| SG 13 | c35026_g1 | AtMYB55,86 | transcription factor MYB34-like [Nicotiana tomentosiformis] | 86 | 90 % | 2E-50 | 2.17 | 1.79 | 0.42 | −0.55 | 1 | ||
| SG 18 | c19459_g1 | AtMYB65,101 | HvGAMYB | transcription factor GAMYB [Vitis vinifera] | 480 | 43 % | 7E-119 | 5.55 | 4.82 | 0.16 | 2.80 | 1 | |
| SG 18 | c32967_g1 | AtMYB65,101 | HvGAMYB | transcription factor GAMYB-like [Musa acuminata subsp. malaccensis] | 115 | 67 % | 2E-49 | 2.44 | 2.07 | 0.41 | 0.41 | 1 | |
| SG 19 | c7757_g1 | LhMYB2 | AtMYB21,24 | Am305, Am340, PhEOBII | R2R3-MYB transcriptional factor [Lilium hybrid division I] | 191 | 100 % | 4E-142 | 2609.05 | 2336.57 | 0.16 | 11.12 | 1 |
| SG 19 | c23501_g1 | LhMYB1 | AtMYB21,24 | Am305, Am340, PhEOBII | LhMyb [Lilium hybrid division I] | 192 | 99 % | 5E-131 | 2929.08 | 2745.59 | 0.09 | 11.22 | 1 |
| SG 21 | c7270_g1 | AtMYB105,117 | MlRCP1 | myb-like protein Q [Musa acuminata subsp. malaccensis] | 156 | 52 % | 3E-18 | 3.36 | 0.32 | 3.04 | 0.02 | 0.3424106 | |
| SG 21 | c16635_g1 | AtMYB105,117 | MlRCP1 | MYB143 protein [Gossypium hirsutum] | 73 | 70 % | 0.0000004 | 20.94 | 0.95 | 4.16 | 2.15 | 0.00079 | |
| SG 22 | c12801_g2 | AtMYB44, 70, 73, 77 | transcription factor MYB44-like [Vitis vinifera] | 410 | 51 % | 2E-100 | 23.84 | 29.80 | −0.29 | 5.21 | 1 | ||
| SG 22 | c13168_g1 | AtMYB44, 70, 73, 77 | MYB transcription factor MYB178 [Glycine max] | 75 | 78 % | 3E-24 | 0.00 | 3.69 | −3.20 | −0.93 | 1 | ||
| SG 22 | c21029_g1 | AtMYB44, 70, 73, 77 | sucrose responsive element-binding protein [Elaeis guineensis] | 205 | 61 % | 7E-72 | 5.17 | 13.50 | −1.35 | 2.23 | 0.3424106 | ||
| SG 22 | c21511_g2 | AtMYB44, 70, 73, 77 | Myb domain protein 73 [Theobroma cacao] | 222 | 59 % | 4E-55 | 62.90 | 87.74 | −0.45 | 5.24 | 0.9953253 | ||
| SG 22 | c37985_g1 | AtMYB44, 70, 73, 77 | transcription factor MYB44-like [Musa acuminata subsp. malaccensis] | 92 | 53 % | 5E-24 | 0.73 | 1.16 | −0.13 | −0.93 | 1 | ||
| SG 23 | c1544_g1 | AtMYB1 | myb-related protein B-like [Gossypium raimondii] | 135 | 65 % | 3E-50 | 2.32 | 3.46 | −0.30 | 0.18 | 1 | ||
| SG 23 | c26297_g1 | AtMYB1 | transcription factor MYB86 [Cucumis sativus] | 331 | 57 % | 5E-116 | 14.64 | 13.84 | 0.06 | 3.86 | 1 | ||
| SG 25 | c14426_g1 | AtMYB98 | transcription factor MYB98-like [Prunus mume] | 77 | 61 % | 1E-23 | 9.25 | 1.90 | 2.24 | −0.35 | 0.7101734 | ||
| c14549_g1 | AtMYB20, 85 | PhODO1 | protein ODORANT1 [Sesamum indicum] | 125 | 54 % | 4E-36 | 4.46 | 4.21 | −0.01 | 0.35 | 1 | ||
| c14549_g2 | AtMYB20, 85 | PhODO1 | myb-related protein 315-like [Nicotiana tomentosiformis] | 67 | 88 % | 4E-40 | 0.00 | 7.78 | −4.67 | −0.44 | 0.3424106 | ||
| c16655_g1 | AtMYB20, 85 | PhODO1 | protein ODORANT1-like [Elaeis guineensis] | 263 | 64 % | 6E-118 | 3.83 | 5.67 | −0.47 | 1.75 | 1 | ||
| c28204_g2 | AtMYB88,124 | Myb124, putative isoform 1 [Theobroma cacao] | 509 | 45 % | 3E-119 | 6.92 | 9.76 | −0.19 | 3.60 | 1 | |||
| c24798_g1 | AtMYB91/AS1 | AmMYBPHAN | transcription factor AS1 [Populus euphratica] | 345 | 69 % | 3E-165 | 10.58 | 7.27 | 0.52 | 3.44 | 0.9933287 | ||
| R3-MYB type | |||||||||||||
| c24227_g1 | AtMYBL2 | transcription repressor MYB4-like [Nelumbo nucifera] | 157 | 42 % | 3E-37 | 4.82 | 3.86 | 0.41 | 0.79 | 1 | |||
| c24227_g2 | AtMYBL2 | myb-related protein 308-like [Phoenix dactylifera] | 157 | 43 % | 3E-34 | 28.20 | 3.00 | 3.29 | 2.42 | 0.00079 | |||
| c18278_g2 | AtCAPRICE | PhMYBx | transcription factor CPC-like [Nelumbo nucifera] | 90 | 62 % | 6E-23 | 5.87 | 4.64 | 0.28 | −0.13 | 1 | ||
| 'Unusual' MYB genes with two or more repeats | |||||||||||||
| c28856_g1 | AtCDC5 | cell division cycle 5-like protein [Arabidopsis thaliana] | 1049 | 73 % | 0 | 12.74 | 13.64 | −0.06 | 5.40 | 1 | |||
| c23757_g4 | AtMYB4R1 | myb-like protein L [Phoenix dactylifera] | 197 | 74 % | 3E-98 | 6.93 | 5.83 | −0.20 | 1.21 | 1 | |||
a)The grouping follows [51]
R2R3-MYBs are further classified into subgroups, and the genes belonging to the same subgroup often share similar functions [51, 53]. Of the 44 R2R3-MYBs expressed in Lollypop tepals, 39 were clustered into 13 subgroups. The Lollypop unigenes c22900_g1 (LhMYB12), c24386_g1 (LrMYB15), and c24386_g2 (LhMYB16) were assigned to subgroup 6, the members of which are often involved in the regulation of anthocyanin biosynthesis. LhMYB12 usually regulates full-tepal pigmentation in lilies [22]. In contrast, LrMYB15 has been shown to regulate the anthocyanin pigmentation responsible for bud-blush in L. regale [24], although its function in Asiatic hybrid lilies is unknown, and the functions of LhMYB16 have yet to be investigated.
In addition, the three unigenes c10735_g1 (LhMYB3), c25442_g1 (LhMYB8), and c25442_g2 were designated as subgroup 4 R2R3-MYBs, which are suppressors and often inhibit anthocyanin or lignin biosyntheses [54, 55]; however, the functions of LhMYB3 and LhMYB8 are not yet understood. R3-MYBs are short MYBs that only include the R3 repeat, and AtMYBL2 and AtCAPRICE in Arabidopsis and PhMYBx in petunias are R3-MYBs that suppress anthocyanin biosynthesis [17, 35, 36]. In the present study, the three unigenes c24227_g1, c24227_g2, and c18278_g2 were identified as R3-MYBs (Table 2).
In lilies, the two bHLH genes LhbHLH1 (petunia JAF13 type) and LhbHLH2 (petunia AN1 type) have been shown to regulate anthocyanin biosynthesis, especially LhbHLH2 [21]. We BLASTed these sequences against the Lollypop transcriptome. As a result, four short unigenes (c16322_g1–4) exhibited similarities of >90 % to the amino acid sequence of LhbHLH1 in Montreux. Therefore, these unigenes should be considered a single gene, even though they were not assembled during de novo assembly. Similarly, the two unigenes c7085_g1 and c27198_g1 exhibited high similarities to the N- and C-terminal halves of the LhbHLH2 sequence in Montreux, respectively. Thus, the LhbHLH2 cDNA sequence should be split into two unigenes. These results indicate that the orthologous genes of both LhbHLH1 and LhbHLH2 are expressed in Lollypop tepals, although the FPKM values were higher in c7085_g1 and c27198_g1 (LhbHLH2) than in c16322_g1–4 (LhbHLH1; data not shown).
Transcription in upper tepals and tepal bases during floral development
To determine whether the expression of MBW complex genes corresponded to the development of tepal color, we performed quantitative reverse transcription-PCR (qRT-PCR) using the total RNA of the upper tepals and tepal bases collected at various stages of floral development (St 1–5). The upper and basal tepal segments were collected from a single inner tepal and each stage included three replicates, each of which was derived from a different flower. The expression of c22900_g1 (LhMYB12) increased with the progression of floral development showing the highest at St 4, and was >2-fold higher in the upper tepals than in the tepal bases at St 3 and 4 (Fig. 3). This expression pattern was coincident with the accumulation of anthocyanins, which also increased at St 3 and 4 and was higher in the upper tepals. The expression of c2514_g1 (WD40) increased gently during tepal development, although it was also higher in upper tepals than in tepal bases at St 3. In contrast, c27198_g1 (LhbHLH2) was consistently expressed, with no significant differences in expression between development states or tepal location.
Fig. 3.
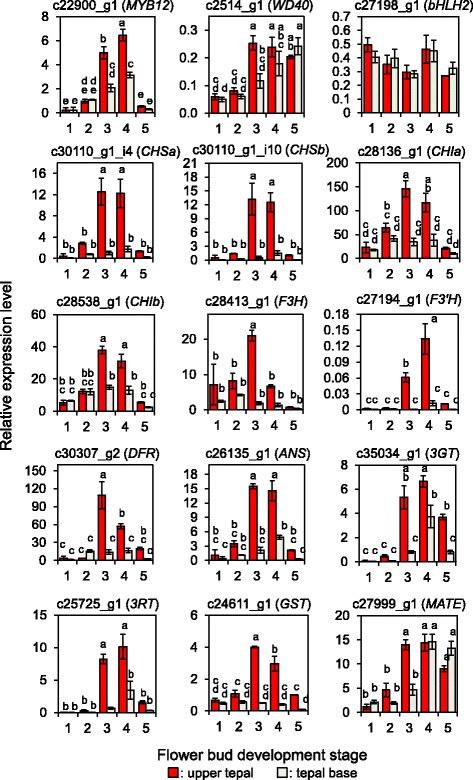
Relative expression of 15 unigenes in Asiatic hybrid lily Lollypop tepals during floral development. The 15 target genes included c22900_g1 (LhMYB12), c2514_g1 (WD40), c27198_g1 (LhbHLH2), c30110_g1_i4 (LhCHSa), c30110_g1_i10 (LhCHSb), c28136_g1 (CHIa), c28538_g1 (CHIb), c28413_g1 (LhF3H), c27194_g1 (LhF3′H), c30307_g2 (LhDFR), c26135_g1 (LhANS), c35034_g1 (3GT), c25725_g1 (3RT), c24611_g1 (GST), and c27999_g1 (MATE), and their expression was normalized using ACTIN expression. Floral development stages (1–5) are shown in Fig. 1. Values and error bars indicate the means ± standard error (n = 3). The same letters above the columns indicate that the values are not statistically significant (p <0.05) by Tukey’s HSD
In addition, the expression of major unigenes involved in anthocyanin biosynthesis was also evaluated by qRT-PCR. We found that the expression of unigenes annotated as LhCHSa, LhCHSb, LhF3H, LhF3′H, LhDFR, and LhANS were higher at St 3 and/or 4 during tepal development and were 7–30-fold higher in the upper tepals than in tepal bases at St 3 when anthocyanin pigments started to accumulate in upper tepals (Fig. 3). The expression of c28136_g1 (CHIa), c28538_g1 (CHIb), c35034_g1 (3GT), c25725_g1 (3RT), and c24611_g1 (GST) was higher at St 3 and/or 4 during floral development and was 2–12-fold higher in the tepal bases than in upper tepals (Fig. 3). c27999_g1 (MATE) exhibited high expression at St 3, 4, and 5 but no clear difference of the expression between the two tepal parts at St 4 and 5. These results indicate that most of the unigenes involved in anthocyanin biosynthesis and sequestration, other than MATE, show higher expression in the pigmented upper part at St 3 and St 4.
St 3 tepals were used for RNA-seq in this study. All of DEGs (e.g., c30110_g1 and c27194_g1, Table 1) showed clear difference in expression at St 3 in qRT-PCR analysis but not all of unigenes showing significant difference at St 3 by qRT-PCR were detected as DEGs. This may be due to the low number of fragments (~24 million fragments) in our RNA-seq analysis, which should lower the power to detect DEGs.
Factors involved in bicolor development
Some of the mechanisms involved in the development of bicolor petals have been identified in model plants. For example, the star and marginal picotee (white margin, pigmented center) patterns in petunias are caused by PTGS of CHS-A in the white petal regions, rather than changes in the expression of anthocyanin biosynthesis genes [25, 26]. In the present study, unigenes annotated as LhCHSa, LhCHSb, LhF3H, LhF3′H, LhDFR and LhANS exhibited >7-fold higher expression in upper tepals than in the tepal bases (Fig. 3), which indicated a strong correlation between the expression of anthocyanin biosynthesis genes and anthocyanin pigmentation, and the coordinated down-regulation of these genes in the tepal bases indicates that the bicolor trait is not caused by PTGS.
Another mechanism of bicolor development has been reported in the marginal picotee pattern of petunias with pigmented margins and white centers; at the white center, the increase of flavonol production should prevent anthocyanin production [27]. Although no flavonols and flavones were detected in lily tepals, during the present study, high amounts of CADs accumulated and reached greater levels in the tepal bases than in the upper tepals (Fig. 2). This observation suggests that competition for p-coumaroyl CoA, which occurs at a branching point of the anthocyanin and CAD biosynthesis pathways and is a common substrate of CHS and shikimate O-hydroxycinnamoyltransferase (HCT, Additional file 1: Figure S1), occurs between the biosynthesis of anthocyanins and CADs in lily tepals. This idea is supported by previous studies suggesting that the suppression of HCT expression results in an increase in flavonoid biosynthesis and an accumulation of anthocyanins [56]. However, in Lollypop, although a unigene annotated as HCT (c30288_g1) exhibited a higher FPKM value in tepal bases at St 3 (Table 1), its transcription at St 3 and 4 were relatively low during floral development (Additional file 1: Figure S4). In addition, higher amounts of CADs in tepal bases were not a specific feature to bicolor cultivars and were also observed in the full-pink and white tepal cultivars (Fig. 2). Furthermore, in petunias, elevated FLS expression failed to affect the expression of either DFR or ANS [27], whereas in lilies, the expression of major unigenes annotated as anthocyanin biosynthesis were all suppressed in non-pigmented regions. Therefore, the active biosynthesis of CADs is not likely responsible for the bicolor trait, and novel mechanisms are likely involved.
Because the expressions of the LhCHSa, LhCHSb, LhF3H, LhF3′H, LhDFR, and LhANS annotated unigenes were coordinately up-regulated in the upper tepals of the Lollypop cultivar, the transcriptional regulation of these biosynthesis genes is most likely to influence the bicolor phenotype. In higher plants, MBW complexes predominantly regulate the transcription of anthocyanin biosynthesis genes, and R2R3-MYB transcription factors have a key role in the spatial and temporal restriction of anthocyanin biosynthesis [13, 14]. Anthocyanin pigmentation is regulated by subgroup 6 R2R3-MYBs in many plant species and by subgroup 5 R2R3-MYBs in Gramineae and Orchidaceae [57, 58]. In the Lollypop tepal transcriptome, three unigenes were assigned to subgroup 6, although none was assigned to subgroup 5. Among the three unigenes in subgroup 6, c22900_g1 (LhMYB12) exhibited the highest FPKM (Table 2) and its expression was correlated with pigment deposition, with higher expression at St 3 and 4 of floral development (Fig. 3). Meanwhile, unigenes c24386_g1 (LhMYB15) and c24386_g2 (LhMYB16) exhibited relatively low FPKM values (Table 2). Thus, c22900_g1 (LhMYB12) is more likely to regulate the biosynthesis of anthocyanins in Lollypop tepals. Since LhMYB12 regulates both early (e.g., CHS) and late (e.g., DFR and ANS) biosynthesis genes in Asiatic hybrid lilies [10], c22900_g1 (LhMYB12) transcripts might be responsible for the coordinated expression of LhCHSa, LhCHSb, LhF3H, LhF3′H, LhDFR, and LhANS unigenes. In addition, the expression of c22900_g1 (LhMYB12) was >2-fold higher in upper tepals than in tepal bases (Fig. 3). In another bicolor cultivar Sugar Love, the expression of LhMYB12 was >4-fold higher in upper tepals (Additional file 1: Figure S5), whereas a previous study has demonstrated that LhMYB12 expression was higher in the tepal bases of full-pink tepal cultivar Montreux [22]. Therefore, the expression profiles of c22900_g1 (LhMYB12) are correlated with bicolor creation.
Factors that suppress the transcription of anthocyanin biosynthesis genes
Although the expression of c22900_g1 (LhMYB12) was >2-fold different in the upper tepals and tepal bases, the expression of the anthocyanin biosynthesis unigenes exhibited a >7-fold difference (Fig. 3). This pattern suggests that the expression of c22900_g1 is partly responsible for the low expression of biosynthesis genes in tepal bases, although it does not explain it completely, and that other factors contribute to the complete suppression of biosynthesis genes. WD40 proteins stabilize the MBW complexes and increase the activity of the complexes as a result [59–61]. In Lollypop tepals, the expression of c2514 (WD40) was higher in upper tepals at St 3 (Fig. 3). The similar expression profiles of WD40 were also observed in the other bicolor cultivar Sugar Love but were not detected in the full-pink cultivar Montreux (Additional file 1: Figure S5). These observations suggest that the lower expression of c2514 (WD40) in non-pigmented tepal regions is specific to bicolor cultivars and synergistically reduces the expression of the biosynthesis genes in tepal bases.
Putative unigenes that could suppress the expression of anthocyanin biosynthesis genes were identified in the Lollypop transcriptome, e.g., subgroup 4 R2R3-MYBs, R3-MYBs, and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes [62] (Table 3). If suppressor genes are involved in bicolor development, their expressions should be higher in tepal bases at St 3 and/or 4. However, qRT-PCR analysis during the floral development indicated that c18278_g2 (R3-MYB) expression was higher in tepal bases at St 5 and the expressions of other unigenes were higher in upper tepals or similar between the two tepal parts (Table 3, Additional file 1: Figure S4). Thus, the functions of these unigenes remain to be determined.
Table 3.
Expression profiles of unigenes that potentially suppress the expression of anthocyanin biosynthesis genes
| Unigene | Homologous gene in lilies | Homologous genes in other species | Putative function | Reference | Expression profilea |
|---|---|---|---|---|---|
| c10735_g1 | LhMYB3 | AtMYB4, FaMYB1, Am308, Am330 | Subgroup 4 R2R3-MYB that inhibits anthocyanin or lignin biosyntheses | [54, 55] | Higher in upper tepals at St 3 and 4 |
| c25442_g1 | LhMYB8 | AtMYB4, FaMYB1, Am308, Am330 | Subgroup 4 R2R3-MYB that inhibits anthocyanin or lignin biosyntheses | [54, 55] | Similar between the two tepal parts |
| c25442_g2 | AtMYB4, FaMYB1, Am308, Am330 | Subgroup 4 R2R3-MYB that inhibits anthocyanin or lignin biosyntheses | [54, 55] | Similar between the two tepal parts | |
| c24227_g1 | AtMYBL2 | R3-MYB that suppresses anthocyanin biosynthesis | [35] | Higher in upper tepals at St 4 | |
| c24227_g2 | AtMYBL2 | R3-MYB that suppresses anthocyanin biosynthesis | [35] | Higher in upper tepals at St 3 and 4 | |
| c18278_g2 | AtCAPRICE, PhMYBx | R3-MYB that suppresses anthocyanin biosynthesis | [17, 36] | Higher in tepal bases at St 5 | |
| c36339_g1 | AtSPL9, AtSPL15 | SPL9 and 15 directly prevent the expression of anthocyanin biosynthetic genes by destabilizing MBW complexes | [62] | Similar between the two tepal parts |
aExpression during floral development is shown in Additional file 1: Figure S4
Factors that influence the expression of MYB12
In the Lollypop cultivar, the expression of c22900_g1 (LhMYB12) was higher in the upper tepals, whereas a previous study has demonstrated that the expression of LhMYB12 is higher in the bases of fully pink tepals [22]. In LhMYB12, six allelic sequences, Montreux [22], Renoir [63], Landini [63], Vermeer [33], Latvia [23], and Sugar Love [unpublished result] alleles, have been detected in Asiatic hybrid lilies. The sequence of c22900_g1 was identical to that of the Landini allele of LhMYB12, which suggests that the Landini allele shows differential expression profiles in the Lollypop and the fully pink cultivar Landini. In addition, Cancún also possesses the Landini allele, while Sugar Love possesses the Sugar Love allele, which demonstrate that not all bicolor cultivars possess the same LhMYB12 allele. These observations indicate that other factors that function upstream of LhMYB12 are responsible for the unique expression profiles of LhMYB12 observed in bicolor cultivars.
The upstream transcription factors of R2R3-MYB are well characterized in the fruits, seeds, and vegetative organs of other species [64–68] but not in flowers. However, Reduced Carotenoid Pigmentation 1 (RCP1) has recently been shown to down-regulate the expression of an anthocyanin biosynthesis activator in Mimulus flowers [69]. RCP1 is an R2R3-MYB that belongs to subgroup 21, to which the two Lollypop unigenes c7270_g1 and c16635_g1 were also assigned (Table 2). Since c16635_g1 showed higher expression at St 4 when anthocyanin contents were highest (Additional file 1: Figure S4), their role should be further examined.
In addition to transcriptional regulation by transcription factors, post-transcriptional regulation by microRNA should also be considered. Transcripts of R2R3-MYB that regulate anthocyanin biosynthesis are often targeted by microRNA828 (miR828) [70]. Sequences of miR828 target sites are well conserved in the R2R3-MYB genes of dicots and monocots [71], and in the present study, one such sequence was found in c22900_g1 (LhMYB12, Additional file 1: Figure S6). Thus, primary-miR828 (pri-miR828) in Vitis (VvPri-miR828a) was used in a BLAST search of the Lollypop transcriptome, and a unigene c13793_g1, encoding putative pri-miR828, was obtained. Although the sequences and lengths of pri-miR828 sequences are highly divergent, the 22-bp sequence of the putative miR828 was completely conserved in pri-miR828 sequences (Additional file 1: Figure S6). Differences in the FPKM values of c13793_g1 were small in upper tepals and tepal bases. However, because multiple regulation systems affect microRNA processing efficiency [72], further analyses of c13793_g1 will be necessary.
Conclusions
In the present study, we analyzed the global transcription in the bicolor lily cultivar Lollypop to determine the main transcriptomic differences in pigmented and non-pigmented tepal parts. We identified and annotated a large number of unigenes, including those for both anthocyanin modification and sequestration, thus providing an excellent platform for future genetic and functional genomic research. In addition, the expression profiles of flower color-related genes in both upper tepals and tepal bases provide new insight into the molecular mechanisms underlying the development of bicolor tepals in Asiatic hybrid lilies. Our results also indicate that the development of bicolor tepals is the result of the transcriptional regulation of anthocyanin biosynthesis genes, rather than PTGS or substrate competition, and moreover, the unique transcription profile of the transcription factor LhMYB12 indicates that it plays a key role. Therefore, the up- and down-stream factors associated with LhMYB12 function should be further investigated. Fortunately, the tepal transcriptome generated by the present study will accelerate such investigations.
Methods
Plant material
The Asiatic hybrid lily (Lilium sp.) Lollypop was grown in a greenhouse (unheated and natural photoperiod) at the experimental farm of Hokkaido University, Sapporo, Japan. Flower tepals for transcriptome sequencing were collected at St 3 (Fig. 1, the flower stages followed [31]). To analyze the segregation of the bicolor trait, Lollypop was crossed with the Asiatic hybrid lily Blackout, and their F1 progeny were grown under the same conditions.
Pigment measurement
Anthocyanins and related compounds were extracted with a solvent mixture of methanol, acetic acid, and water (4:1:5, v:v:v), and the extracts were filtered through a ZORBAX Eclipse Plus C18 column (Agilent Technologies, Tokyo, Japan). The extract solution of each sample was then analyzed using HPLC with an Agilent 1290 infinity system (Agilent Technologies). A linear gradient of 10-60 % of solvent B (1.5 % H3PO4, 20 % acetic acid, and 25 % acetonitrile) in solvent A (1.5 % H3PO4) was run over a 9-min period. The anthocyanins and CADs were identified on the basis of absorption spectra obtained using a photodiode array detector, and pigment quantity was determined on the basis of absorption values at wavelengths of 340 nm (CADs) and 530 nm (anthocyanins).
Structural determination of CADs
Tepals of Lollypop (15 g) were soaked in 100 % ethanol, and after filtering the ethanol solution with filter paper (No. 1; Toyo, Tokyo, Japan), the filtrate was concentrated in vacuo. The resulting material was dissolved in 1 L 80 % ethanol to obtain the precipitate, and after filtering the solution with filter paper, the supernatant was again concentrated in vacuo and was subsequently purified using silica gel column chromatography (methanol/chloroform, 1:15, v/v) and preparative TLC (methanol/chloroform, 1:15, v/v) to obtain caffeoylglucose. The silica gel column chromatography was performed using 50 g of Kanto silica gel 60 N (60–210 mesh; Kanto Kagaku, Tokyo, Japan) and the preparative TLC was performed using Merck silica gel 60 F254 (Art 5554; Merck, Darmstadt, Germany). Nuclear magnetic resonance (NMR) spectra were recorded using a Bruker AM-500 (1H at 500 MHz; Bruker, Billerica, MA, USA) and a JEOL JNM-EX 270 FT-NMR system (1H at 270 MHz; 13C at 67.5 MHz; JEOL, Tokyo, Japan).
Transcriptome sequencing
Four cDNA samples were prepared from total RNA isolated from the pink upper tepals and white basal tepals of Lollypop at St 3. The upper tepal and tepal base samples included two replicates, which were collected from two different plants. Total RNA was extracted from the tepal parts using TriPure Isolation Reagent (Roche, Basel, Switzerland) and Fruit-mate for RNA Purification (TaKaRa, Shiga, Japan), according to the manufacturer’s protocols. To prevent contamination by genomic DNA, the RNA was treated with deoxyribonuclease I (Thermo Fisher Scientific, Yokohama, Japan) and purified using an RNeasy Mini Kit (Qiagen, Tokyo, Japan). The RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies), and the samples were prepared for transcriptome analyses using a SureSelect Strand Specific RNA Library Prep Kit (Agilent Technologies), following the manufacturer’s instructions. First, poly(A) RNA was purified from 4 μg total RNA using oligo (dT) magnetic beads, and then the poly(A) RNA was chemically fragmented at 94 °C for 4 min. Next, first- and second-strand cDNA was synthesized using fragmented RNA as templates, and the resulting double-stranded cDNA was end repaired, adenylated, and ligated using SureSelect Oligo Adaptor (Agilent Technologies). These products were selectively amplified using PCR and were sequenced (75 bp, paired-end) using a MiSeq System (Illumina Inc., San Diego, California, USA). Then, we obtained approximately 24 million fragments (paired-end reads), which consisted of 7,589,796 (upper tepals, replicate 1), 4,653,736 (upper tepals, replicate 2), 5,002,031 (tepal bases, replicate 1), and 6,909,815 (tepal bases, replicate 2) fragments.
Raw data processing and de novo assembly
Raw data processing, base calling, and quality control were performed using RTA, OLB, and CASAVA (Illumina), according to the manufacturer’s pipeline. The output sequence quality was inspected using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the reads were cleaned using cutadapt (version 1.8.1) [73] to trim low-quality ends (<QV30), the 76th nucleotides, and adapter sequences, and to discard reads shorter than 50 bp. Then, 48,897,598 high quality reads (98 % of the raw data) were selected. The clean reads were then assembled de novo using the Trinity program version r20140413p1 [74], yielding 49,239 contigs that clustered into 39,426 subcomponents (i.e., unigenes). The unigene sizes were 201 to 10,772 bp, with a mean length of 427 bp, N50 length of 1228 bp, and total combined length of 29,260,585 bp (Additional file 1: Table S1).
Differential expression analysis
We identified DEGs using scripts bundled with the Trinity package according to “Trinity Transcript Quantification” (https://github.com/trinityrnaseq/trinityrnaseq/wiki/Trinity-Transcript-Quantification) and “Differential Expression Analysis Using a Trinity Assembly” (https://github.com/trinityrnaseq/trinityrnaseq/wiki/Trinity-Differential-Expression) with a default setting to process the strand-specific RNA-seq data generated from the upper tepals and tepal bases, with two biological replicates from each tepal part. The RNA-seq reads were aligned to full-transcript sequences using bowtie 1.0.0 [75]. Transcript abundance was then estimated by RSEM 1.2.11 [76], and DEGs were identified using edgeR 3.6.8 [77], with trimmed mean of M-values (TMM) normalization. The FDR values were recalculated from the edgeR p-values by considering only the unigenes listed in Tables 1, 2, and Additional file 2: Table S2.
Functional annotation and identification of orthologous genes
For functional annotation, the sequences of putatively encoded proteins were BLASTed against the public protein databases (Swiss-Prot [78], Pfam [79], eggNOG [80], and Gene Ontology [81]) using the BLASTX algorithm and a typical cut off value of E <0.00001. Of the 39,426 unigenes, 24,835 (63 %) were annotated, and the number of unigenes annotated by the Swiss-Prot, Pfam, eggNOG, and GO databases were 21,649; 24,835; 8575; and 16,084, respectively. The Swiss-Prot and Pfam databases accounted for a large proportion of the annotations, and the unigenes that were annotated with gene descriptions from eggNOG and GO databases were all included in those annotated to the Swiss-Prot and/or Pfam databases. Therefore, the unigenes annotated as being related to the phenylpropanoid, anthocyanin, and monolignol metabolic pathways were selected according to the Swiss-Prot annotations (Table 2).
In order to detect WD40, R2R3-MYB, bHLH, and Pri-miR828 related sequences in Lollypop, we BLASTed the amino acid sequence of AtTTG1 (WD40 in Arabidopsis [60]), the R2R3 repeat motif sequence [51], two lily bHLH (LhbHLH1 and LhbHLH2) [21], and the nucleotide sequence of VvPri-miR828a, against the Lollypop sequence database. We used a typical cut off value of E <0.00001 in most case but a cut off value of E <0.001 for Pri-miR828, because of the large variation between non-coding RNA sequences.
qRT-PCR analysis
The total RNA used for qRT-PCR analysis was extracted from upper tepals and tepal bases that were collected at five floral development stages (St 1–5), using a NucleoSpin RNA II Kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany). Total RNA was converted into first strand cDNA using the ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Tokyo, Japan). SYBR Premix Ex Taq (TaKaRa) was used to intercalate SYBR Green I into the amplified products, and signals were monitored using the CFX Connect Real-Time system (Bio-Rad, Tokyo, Japan) with the following reaction conditions: preheating at 95 °C for 30 s; 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 20 s; and a final extension step at 72 °C for 5 min. Primer specificity was confirmed by melting curve analysis, and the amount of ACTIN mRNA in each sample was used to normalize the level of each target mRNA. Relative fold-change values of three biological replicates (three different flowers) were used to calculate mean values and standard errors (SE). A post hoc analysis using Tukey’s HSD was performed using R version 3.2.1 (http://cran.r-project.org/doc/manuals/r-release/R-intro.pdf). All primers used for analysis are listed in Additional file 1: Table S3.
Abbreviations
3GT, anthocyanidin 3-O-glucosyltransferase; 3RT, anthocyanidin-3-glucoside rhamnosyltransferase; 4CL, 4-coumaroyl: CoA-ligase; 7GT, anthocyanidin-3-rutinoside 7-glucosyltransferase; ANS, anthocyanidin synthase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CAD, cinnamic acid derivative; CHI, chalcone isomerase; CHS, chalcone synthase; DEGs, differentially expressed genes; DFR, dihydroflavonol 4-reductase; F3′H, flavonoid 3′-hydroxylase; F3H, flavanone 3-hydroxylase; FDR, false discovery rates; FLS, flavonol synthase; FPKM, fragments per kb per million mapped reads; GO, gene ontology; GST, glutathione S-transferase; HCT, shikimate O-hydroxycinnamoyl transferase; HPLC, high-performance liquid chromatography; MATE, multidrug and toxic compound extrusion transporter; MBW complex, R2R3-MYB–bHLH–WDR protein complex; NGS, next-generation sequencing; PAL, phenylalanine ammonia-lyase; pri-miR, primary-microRNA; PTGS, post-transcriptional gene silencing; qRT-PCR, quantitative reverse transcription-PCR; RT, retention time; SE, standard error; SSR, simple sequence repeat; St, floral development stage; TLC, thin layer chromatography
Acknowledgments
Funding
This work was supported by a Grant-In-Aid for Scientific Research (No. 15H04447) from the Japan Society for the Promotion of Science.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article (and its Additional files 1 and 2).
Authors’ contributions
KS and MY designed the research and drafted the manuscript. KS mainly performed the experiments. TS, TN, and HD contributed to the NGS analysis and data processing. TN and HM conducted the pigment analyses. KM participated in the real-time PCR analysis. MY maintained the lilies and performed the segregation analysis. All authors carefully read and approved the final manuscript.
Competing interests
The authors declare that the present study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Data deposition
The sequence dataset was submitted to the DNA Databank of Japan Sequence Read Archive (DRA) under accession number DRA004630 with the following BioProject accession number PRJDB4732 and BioSample accession numbers SAMD00050882–SAMD00050883.
Additional files
Summary of assembled sequences from the tepal parts of the Asiatic hybrid lily Lollypop. Table S3. Primers used for quantitative RT-PCR (qRT-PCR) analysis. Figure S1. Phenylpropanoid, anthocyanin, and cinnamic acid derivative biosynthesis pathways in lily tepals. Enzymes whose genes are up-regulated in upper tepals (estimated by qRT-PCR) are shown in blue. 3GT, anthocyanidin 3-O-glucosyltransferase; 3RT, anthocyanidin-3-glucoside rhamnosyltransferase; 4CL, 4-coumaroyl: CoA-ligase; 7GT, anthocyanidin-3-rutinoside 7-glucosyltransferase; ANS, anthocyanidin synthase; CHI, chalcone isomerase; CHS, chalcone synthase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; GST, glutathione S-transferase; HCT, shikimate O-hydroxycinnamoyl transferase; MATE, multidrug and toxic compound extrusion transporter; PAL, phenylalanine ammonia-lyase. Figure S2. HPLC analysis of anthocyanins and CADs in upper tepals (upper) and tepal bases (basal) of lily cultivars. A: Absorbance at 525 nm (anthocyanins) of the tepal extracts in Lollypop, and cyanidin 3-O-glycoside (Cy3G) and cyanidin 3-O-rutinoside (Cy3R) standards. B: Absorbance at 340 nm (CADs) of the tepal extracts in six cultivars. Figure S3. Alignment of predicted amino acid sequences of isoforms annotated as LhPAL1, LhPAL2, and LhPAL3 (A), and LhCHSa and LhCHSb (B). Letters on black and grey backgrounds indicate identical and similar amino acids, respectively. Asterisks indicate stop codons. Figure S4. Relative expression levels of c30288_g1 (HCT), c10735_g1 (MYB3), c25442_g1 (MYB8), c25442_g2, c24227_g1 (R3-MYB), c24227_g2 (R3-MYB), c18278_g2 (R3-MYB), c36339_g1 (SPL9), and c16635-g1 (RCP1) in upper tepals and tepal bases of Lollypop during floral development (St 1–5). ACTIN was used to normalize the expression of target genes. Values and vertical bars indicate the mean ± standard error (n = 3). The same letters above the columns indicate that the values are not statistically significant (p <0.05) by Tukey’s HSD. Figure S5. Relative expression levels of LhMYB12 in Sugar Love and WD40 in Sugar Love and Montreux in upper tepals and tepal bases during floral development (St 1–5, A) and flowers of the cultivars Montreux and Sugar Love (B). ACTIN was used to normalize the expression of target genes. Values and vertical bars indicate the means ± standard error (n = 3). The same letters above the columns indicate that the values are not statistically significant (p <0.05) by Tukey’s HSD. Figure S6. Putative miR828 and pri-miR828 sequences in Lollypop. A: Putative miR828 and its target site appeared in c22900_g1 (MYB12). B: Sequence alignment of c13793_g1 and pri-miR828 in Glycine max [GmPri-miR828a (NR_126648) and GmPri-miR828b (NR_126651)], Vitis vinifera [VvPri-miR828a (NR_127861) and VvPri-miR828b (LM611741)], and Malus domestica [MdPri-miR828b (NR_120979) and MdPri-miR828a (NR_120978)]. (PDF 1261 kb)
Annotation and expression summary of differentially expressed genes. (XLSX 216 kb)
Contributor Information
Kazuma Suzuki, Email: kazuma114515@gmail.com.
Tomohiro Suzuki, Email: suzukit@cc.utsunomiya-u.ac.jp.
Takashi Nakatsuka, Email: nakatsuka.takashi@shizuoka.ac.jp.
Hideo Dohra, Email: dora.hideo@shizuoka.ac.jp.
Masumi Yamagishi, Email: yamagisi@res.agr.hokudai.ac.jp.
Kohei Matsuyama, Email: ko-matsu@res.agr.hokudai.ac.jp.
Hideyuki Matsuura, Email: matsuura@chem.agr.hokudai.ac.jp.
References
- 1.Comber HF. A new classification of the genus Lilium. In: Lily yearbook, vol. 13. London: The Royal Horticultural Society; 1949. p. 85–105.
- 2.Smyth DR, Kongsuwan K, Wisudharomn S. A survey of C-band patterns in chromosomes of Lilium (Liliaceae) Plant Syst Evol. 1989;163:53–69. doi: 10.1007/BF00936153. [DOI] [Google Scholar]
- 3.Leslie AC. The international lily register. 3. London: The Royal Horticultural Society; 1982. [Google Scholar]
- 4.Yamagishi M. How genes paint lily flowers: regulation of colouration and pigmentation patterning. Sci Hortic. 2013;163:27–36. doi: 10.1016/j.scienta.2013.07.024. [DOI] [Google Scholar]
- 5.Nørbæk R, Kondo T. Anthocyanin from flowers of Lilium (Liliaceae) Phytochem. 1999;50:1181–4. doi: 10.1016/S0031-9422(98)00661-X. [DOI] [Google Scholar]
- 6.Yamagishi M, Kishimoto S, Nakayama M. Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 2010;129:100–7. doi: 10.1111/j.1439-0523.2009.01656.x. [DOI] [Google Scholar]
- 7.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, et al. Transcriptional repression by AtMYB4 controls production of UV–protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–61. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T, Toyama-Kato Y, Yoshida K. Essential structure of co-pigment for blue sepal-color development of hydrangea. Tetrahedron Lett. 2005;46:6645–9. doi: 10.1016/j.tetlet.2005.07.146. [DOI] [Google Scholar]
- 9.Nishihara M, Nakatsuka T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol Lett. 2011;33:433–41. doi: 10.1007/s10529-010-0461-z. [DOI] [PubMed] [Google Scholar]
- 10.Lai YS, Shimoyamada Y, Nakayama M, Yamagishi M. Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.) Plant Sci. 2012;193:136–47. doi: 10.1016/j.plantsci.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Yamagishi M, Akagi K. Morphology and heredity of tepal spots in Asiatic and Oriental hybrid lilies (Lilium spp.) Euphytica. 2013;194:325–34. doi: 10.1007/s10681-013-0937-8. [DOI] [Google Scholar]
- 12.Tanaka Y, Ohmiya A. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotech. 2008;19:190–7. doi: 10.1016/j.copbio.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–42. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–85. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Davies KM, Albert NW, Schwinn KE. From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct Plant Biol. 2012;39:619–38. doi: 10.1071/FP12195. [DOI] [PubMed] [Google Scholar]
- 16.Glover BJ, Walker RH, Moyroud E, Brockington SF. How to spot a flower. New Phytol. 2013;197:687–9. doi: 10.1111/nph.12112. [DOI] [PubMed] [Google Scholar]
- 17.Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–84. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 18.Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, et al. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–51. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, et al. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011;189:602–15. doi: 10.1111/j.1469-8137.2010.03498.x. [DOI] [PubMed] [Google Scholar]
- 20.Yuan YW, Sagawa JM, Frost L, Vela JP, Bradshaw HD., Jr Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus) New Phytol. 2014;204:1013–27. doi: 10.1111/nph.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N. Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci Hortic. 2009;121:84–91. doi: 10.1016/j.scienta.2009.01.008. [DOI] [Google Scholar]
- 22.Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K. Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 2010;51:463–74. doi: 10.1093/pcp/pcq011. [DOI] [PubMed] [Google Scholar]
- 23.Yamagishi M, Toda S, Tasaki K. The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.) New Phytol. 2014;201:1009–20. doi: 10.1111/nph.12572. [DOI] [PubMed] [Google Scholar]
- 24.Yamagishi M. A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale. Mol Breed. 2016;36:3. doi: 10.1007/s11032-015-0426-y. [DOI] [Google Scholar]
- 25.Koseki M, Goto K, Masuta C, Kanazawa A. The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 2005;46:1879–83. doi: 10.1093/pcp/pci192. [DOI] [PubMed] [Google Scholar]
- 26.Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, et al. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J. 2012;70:739–49. doi: 10.1111/j.1365-313X.2012.04908.x. [DOI] [PubMed] [Google Scholar]
- 27.Saito R, Fukuta N, Ohmiya A, Itoh Y, Ozeki Y, Kuchitsu K, et al. Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in Petunia. Plant Sci. 2006;170:828–34. doi: 10.1016/j.plantsci.2005.12.003. [DOI] [Google Scholar]
- 28.Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot. 2011;107:467–590. doi: 10.1093/aob/mcq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe H, Nakano M, Nakatsuka A, Nakayama M, Koshioka M, Yamagishi M. Genetic analysis of floral anthocyanin pigmentation traits in Asiatic hybrid lily using molecular linkage maps. Theor Appl Genet. 2002;105:1175–82. doi: 10.1007/s00122-002-1053-7. [DOI] [PubMed] [Google Scholar]
- 30.Shahin A, Arens P, van Heusden AW, van der Linden G, van Kaauwen M, Khan N, et al. Genetic mapping in Lilium: mapping of major genes and quantitative trait loci for several ornamental traits and disease resistances. Plant Breed. 2011;130:372–82. doi: 10.1111/j.1439-0523.2010.01812.x. [DOI] [Google Scholar]
- 31.Nakatsuka A, Izumi Y, Yamagishi M. Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci. 2003;165:759–67. doi: 10.1016/S0168-9452(03)00254-1. [DOI] [Google Scholar]
- 32.Yamagishi M. Oriental hybrid lily ‘Sorbonne’ homologue of LhMYB12 regulates anthocyanin biosyntheses in flower tepals and tepal spots. Mol Breed. 2011;28:381–9. doi: 10.1007/s11032-010-9490-5. [DOI] [Google Scholar]
- 33.Yamagishi M, Ihara H, Arakawa K, Toda S, Suzuki K. The origin of the LhMYB12 gene, which regulates anthocyanin pigmentation of tepals, in Oriental and Asiatic hybrid lilies (Lilium spp.) Sci Hortic. 2014;174:119–25. doi: 10.1016/j.scienta.2014.05.017. [DOI] [Google Scholar]
- 34.Suzuki K, Tasaki K, Yamagishi M. Two distinct spontaneous mutations involved in white flower development in Lilium speciosum. Mol Breed. 2015;35:193. doi: 10.1007/s11032-015-0389-z. [DOI] [Google Scholar]
- 35.Matsui K, Umehara Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–67. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant. 2009;2:790–802. doi: 10.1093/mp/ssp030. [DOI] [PubMed] [Google Scholar]
- 37.Nakatsuka T, Yamada E, Saito M, Fujita K, Nishihara M. Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers. Plant Cell Rep. 2013;32:1925–37. doi: 10.1007/s00299-013-1504-4. [DOI] [PubMed] [Google Scholar]
- 38.Bräutigam A, Mullick T, Schliesky S, Weber APM. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C3 and C4 species. J Exp Bot. 2011;62:3093–102. doi: 10.1093/jxb/err029. [DOI] [PubMed] [Google Scholar]
- 39.Shahin A, van Gurp T, Peters SA, Visser RGF, van Tuyl JM, Arens P. SNP markers retrieval for a non-model species: a practical approach. BMC Res Notes. 2012;5:79. doi: 10.1186/1756-0500-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Jiang C, Wen Q, Wang N, Tao Y, Xu L. Deep sequencing of the Camellia chekiangoleosa transcriptome revealed candidate genes for anthocyanin biosynthesis. Gene. 2014;538:1–7. doi: 10.1016/j.gene.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Lou Q, Liu Y, Qi Y, Jiao S, Tian F, Jiang L, et al. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J Exp Bot. 2014;65:3157–64. doi: 10.1093/jxb/eru168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Mao Y, Liu H, Yu F, Li S, Yin T. Transcriptome analysis of differentially expressed genes relevant to variegation in peach flowers. PLoS One. 2014;9:e90842. doi: 10.1371/journal.pone.0090842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shahin A, van Kaauwen M, Esselink D, Bargsten JW, van Tuyl JM, Visser RGF, et al. Generation and analysis of expressed sequence tags in the extreme large genomes Lilium and Tulipa. BMC Genomics. 2012;13:640. doi: 10.1186/1471-2164-13-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M, Jiang L, Zhang D, Jia G. De novo transcriptome characterization of Lilium ‘Sorbonne’ and key enzymes related to the flavonoid biosynthesis. Mol Genet Genomics. 2015;290:399–412. doi: 10.1007/s00438-014-0919-0. [DOI] [PubMed] [Google Scholar]
- 45.Huang J, Liu X, Wang J, Lü Y. Transcriptomic analysis of Asiatic lily in the process of vernalization via RNA-seq. Mol Biol Rep. 2014;41:3839–52. doi: 10.1007/s11033-014-3250-2. [DOI] [PubMed] [Google Scholar]
- 46.Villacorta-Martin C, González FFNC, de Haan J, Huijben K, Passarinho P, Hamo MLB, et al. Whole transcriptome profiling of the vernalization process in Lilium longiflorum (cultivar White Heaven) bulbs. BMC Genomics. 2015;16:550. doi: 10.1186/s12864-015-1675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Yang Y, Liu X, Huang J, Wang Q, Gu J, et al. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genomics. 2014;15:203. doi: 10.1186/1471-2164-15-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galland S, Mora N, Abert-Vian M, Rakotomanomana N, Dangles O. Chemical synthesis of hydroxycinnamic acid glucosides and evaluation of their ability to stabilize natural colors via anthocyanin copigmentation. J Agric Food Chem. 2007;55:7573–9. doi: 10.1021/jf071205v. [DOI] [PubMed] [Google Scholar]
- 49.Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell. 2007;19:2023–38. doi: 10.1105/tpc.106.046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller LA, Goodman CD, Silady RA, Walbot V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000;123:1561–70. doi: 10.1104/pp.123.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–56. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 52.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–24. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 53.Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008;8:83. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, et al. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006;140:499–511. doi: 10.1104/pp.105.067231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Q, Dixon RA. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 2011;16:227–33. doi: 10.1016/j.tplants.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.) New Phytol. 2011;190:627–39. doi: 10.1111/j.1469-8137.2010.03621.x. [DOI] [PubMed] [Google Scholar]
- 57.Allan AC, Hellens RP, Laing WA. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008;13(3):99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Hsu CC, Chen YY, Tsai WC, Chen WH, Chen HH. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 2015;168:175–91. doi: 10.1104/pp.114.254599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Vetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–34. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- 60.Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–50. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–80. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 62.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–22. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamagishi M, Yoshida Y, Nakayama M. The transcription factor LhMYB12 determines anthocyanin pigmentation in the tepals of Asiatic hybrid lilies (Lilium spp.) and regulates pigment quantity. Mol Breed. 2012;30:913–25. doi: 10.1007/s11032-011-9675-6. [DOI] [Google Scholar]
- 64.Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, et al. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–79. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sagasser M, Lu GH, Hahlbrock K, Weisshaar B. A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Gene Dev. 2002;16:138–49. doi: 10.1101/gad.212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009;50(12):2210–22. doi: 10.1093/pcp/pcp159. [DOI] [PubMed] [Google Scholar]
- 67.Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koshimäki JJ, Hohtola A, et al. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010;153:1619–29. doi: 10.1104/pp.110.158279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubin G, Tohge T, Matsuda F, Saito K, Sheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–84. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sagawa JM, Stanley LE, LaFountain AM, Frank HA, Liu C, Yuan YW. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016;209:1049–57. doi: 10.1111/nph.13647. [DOI] [PubMed] [Google Scholar]
- 70.Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–32. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo QJ, Mittal A, Jia F, Rock DC. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol Biol. 2012;80:117–29. doi: 10.1007/s11103-011-9778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 73.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10–2. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 74.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–70. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Hueruta-Cepas J, et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:D231–9. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article (and its Additional files 1 and 2).


