Abstract
Retrotransposons have generated about 40 % of the human genome. This review examines the strategies the cell has evolved to coexist with these genomic “parasites”, focussing on the non-long terminal repeat retrotransposons of humans and mice. Some of the restriction factors for retrotransposition, including the APOBECs, MOV10, RNASEL, SAMHD1, TREX1, and ZAP, also limit replication of retroviruses, including HIV, and are part of the intrinsic immune system of the cell. Many of these proteins act in the cytoplasm to degrade retroelement RNA or inhibit its translation. Some factors act in the nucleus and involve DNA repair enzymes or epigenetic processes of DNA methylation and histone modification. RISC and piRNA pathway proteins protect the germline. Retrotransposon control is relaxed in some cell types, such as neurons in the brain, stem cells, and in certain types of disease and cancer, with implications for human health and disease. This review also considers potential pitfalls in interpreting retrotransposon-related data, as well as issues to consider for future research.
Keywords: Alu, Autoimmunity, Epigenetics, LINE-1, Methylation, Restriction, Retrovirus, RNAi, SINE, SVA
Background
Sixty-five years on from Barbara McClintock’s seminal discovery of mobile DNA [1] we now understand that genomes are dynamic and changeable, with transposable elements (TEs) being major contributors to their fluidity. We recognize that TEs, sometimes called “junk DNA”, are major players in genome evolution and have helped shape the form and function of many genes [2]. Nevertheless, TEs are foremost parasitic DNA, and parasites must be controlled or they will destroy a host. There is far more junk than treasure in mobilomes.
DNA transposons comprise about 3 % of the human genome and most move by a “cut and paste” mechanism involving excising an element and reinserting it elsewhere (Fig. 1 [3]). With the exception of at least one family of piggyBac elements in little brown bats [4], no active DNA transposons are known in mammals. There are two classes of retrotransposon. Both move by a “copy and paste” mechanism, involving reverse transcription of an RNA intermediate and insertion of its cDNA copy at a new site in the genome. LTR retrotransposons are named for the long terminal repeats that flank their sequences (reviewed in [5–7]). Endogenous retroviruses (ERVs) are relics of past germline viral infections and for the most part are highly mutated. However, some intracisternal A-particle (IAP) and Etn/MusD family LTR elements remain insertionally active in mice [8], and formation of infective virions by recombination or phenotypic mixing of intact proteins from different ERV proviruses has been reported [9–12]. Among the 31 human endogenous retrovirus subfamilies extant in the human genome, no replication-competent HERVs are known, although their existence has not been ruled out [13, 14], and recently an unfixed fully intact HERV-K (HML2) provirus was identified in some individuals [15]. Many HERVs, and their lone LTRs that populate the genome as a consequence of non-homologous recombination, remain capable of expression and may act as transcriptional regulatory elements for genes (reviewed in [16]).
Fig. 1.
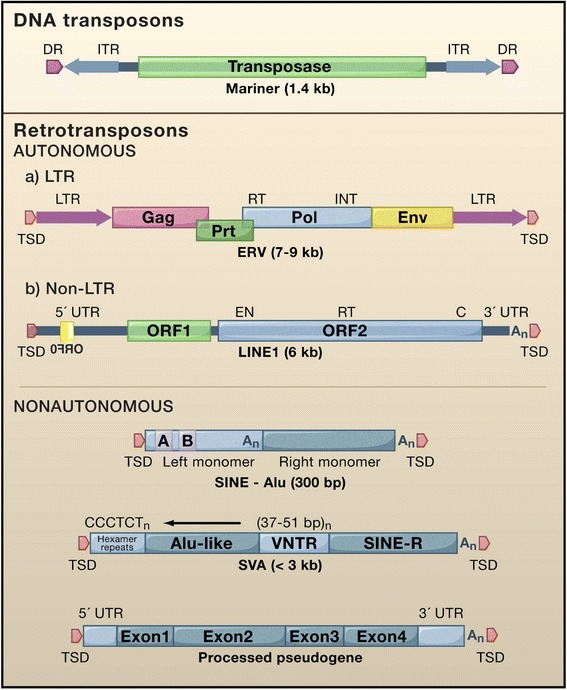
Types of transposable elements in mammals. Abbreviations: DR, direct repeat; ITR, inverted terminal repeat; Gag, group-specific antigen; Prt, protease; Pol, polymerase; Env, envelope; RT, reverse transcriptase domain; INT, integrase domain; TSD, target site duplication; LTR, long terminal repeat; EN, endonuclease domain; C, zinc knuckle domain; An, poly (A); A/B, A- and B-box Pol III promoter; SVA, SINE-R, VNTR, Alu element; VNTR, variable number tandem repeats (reproduced from [3]; Elsevier license number 3803340576977)
Non-LTR retrotransposons are as old as the earliest multi-cellular organisms and their 28 clades have origins in the Precambrian Era of 600 million years ago [17, 18]. Long Interspersed Elements (LINEs) and Short Interspersed Elements (SINEs) comprise most of this group in mammals. LINE-1s (L1s), the only currently active autonomous mobile DNA in humans, have been evolving during at least 150 million years of mammalian radiation. Multiple active L1 lineages coexisted in ancestral primates, but for the past 40 Myr there has been a single unbroken lineage of subfamilies [19, 20]. Expansion of L1s was massive, and roughly 500,000 copies now occupy about 17 % of the human genome. Remnant copies of extinct L2 and L3 family elements comprise an additional 4 % [21]. L1s have also been responsible for genomic insertion of 8000 processed pseudogenes and over a million non-autonomous SINEs [22]. B1s and Alus, the predominant SINEs of mice and men, respectively, originate from the 7SL RNA component of the signal recognition particle. Alus are about 300 base pairs in length with a dimeric structure; B1s are monomeric (reviewed in [23]). SVAs are hominid-specific SINEs, and the youngest family of active human retrotransposons. Their name is an acronym reflecting their composite nature: a HERV-K(HML-2)-derived SINE-R, variable-number-of-tandem-repeats (VNTR), and an Alu-like region. There are roughly 2700 SVA copies per human genome, most of which are full-length and about 50 of which may be active [24–31]. SVA-like variants have been described, including a human-specific subfamily generated by fusion of the first intron of the MAST2 gene with an SVA [29, 32, 33], and the LAVA, PVA and FVA elements of non-human primates [34–36].
From 12 Myr ago, the primate LINE-1 expansion slowed, and most insertions are molecular fossils, truncated, rearranged, or mutated [20]. However, although most L1s no longer “jump”, at least 100 remain potentially mobile in any individual diploid human genome [37, 38]. Many more L1s are transcribed. Interestingly, only a small number of the active L1s are ”hot” for retrotransposition and these have accounted for most de novo insertions. However, when several of these “hot” Ta-1 L1s were examined across diverse human populations, considerable individual allelic variation affected their ability to retrotranspose [39]. Up to 5 % of newborn children have a new retrotransposon insertion, and to date there are 124 known human disease-causing germline insertions of L1s, Alus, and SVAs [40–42]. The current residual activity of human retrotransposons is the background that escapes a variety of mechanisms that have evolved to limit replication of mobile DNA. This review focuses on mammalian non-LTR retrotransposons and how the cell controls them.
Non-LTR retrotransposons are mobilized by a mechanism very different from that used by retroviruses and LTR retrotransposons. Extensive biochemical analyses of insect R1 and R2 elements, together with genomic sequence analyses, indicate that L1s likely retrotranspose by a process known as target-primed reverse transcription (TPRT) that occurs at the site of DNA insertion. According to this model, L1-encoded endonuclease nicks the bottom strand of target DNA exposing a 3'-hydoxyl that primes reverse transcription of bound L1 RNA. Second-strand DNA synthesis follows and the integrant is resolved in a manner still poorly understood [43]. Short target site duplications (TSDs) of variable length, and occasionally deletions, are generated at new L1 insertion sites.
The 6 kilobase bicistronic L1 has a 5' untranslated region (UTR) that functions as an internal promoter, a 3' UTR that ends in a poly (A) signal and tail, and two open reading frames (ORF1 and ORF2) on the sense strand. A weak promoter on the antisense strand of the human 5' UTR [44] lies upstream of a recently identified 216-nt translation-competent ORF0 [45]. Unlike human L1s, mouse L1s have a 5′ UTR consisting of tandemly repeated ∼ 200 bp sequences called monomers [46]. ORF2 encodes a 150 kD protein with endonuclease and reverse transcriptase (RT) activities. While the 40 kD ORF1p RNA-binding protein is essential for LINE-1 retrotransposition, its precise function remains unclear, although it possesses chaperone activity in vitro [47, 48]. Early L1 investigations showed ORF1p to be predominantly cytoplasmic where it forms large aggregates, subsequently identified as stress granules (SGs) and processing bodies (PBs) [49–51]. Endogenous L1 RNA has also been detected in PBs [52]. SGs are discrete cytoplasmic aggregates which can be induced by a range of stress conditions, including heat shock, osmotic shock, oxidative stress, viral infection, and overexpression of some proteins. PBs are dynamic cytoplasmic compartments containing molecules involved in mRNA decay and translation inhibition (reviewed in [53, 54]). ORF1p can also concentrate at the perinucleus, is detected faintly in the nucleus, and is seen in nucleoli of a small fraction of cells [55–57] (Fig. 2). Expressed from a full-length L1 construct, ORF1p is present in SGs as a ribonucleoprotein (RNP) complex together with L1 RNA, ORF2p, and many other RNA-binding proteins [58, 59]. Recently, endogenous ORF1p and ORF2p have been reported to also colocalize in nuclear foci of cancer cells [60].
Fig. 2.
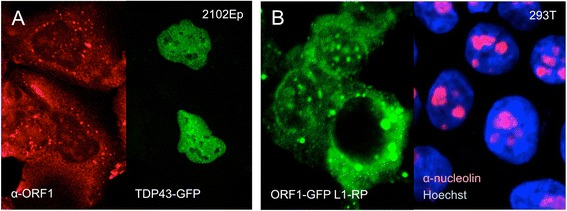
Subcellular distribution of LINE-1 ORF1 protein. a. Endogenous ORF1p detected in human embryonal carcinoma 2102Ep cells by a monoclonal antibody [57]. ORF1p is mostly cytoplasmic where it concentrates in SGs and PBs and occasionally at the nuclear membrane. It is faintly detectable in some nuclei and concentrates in nucleoli of a small percentage of cells. Expression of GFP-tagged TDP43 in nuclei but not in nucleoli is shown as a marker. b. Exogenously expressed GFP-tagged ORF1p strongly concentrates at the nuclear membrane and in perinucleolar foci of 5 % or fewer human embryonic kidney (HEK) 293T cells, with attendant reduction in size and number of cytoplasmic granules (left panel). Construct ORF1-EGFP L1-RP contains a CMV promoter, ORF1 C-terminally tagged with EGFP, followed by intact downstream L1 sequence. Nucleoli are marked by α-C23 (nucleolin) antibody (Santa Cruz) and nuclei are stained with Hoechst (right panel)
How retrotransposons impact the mammalian cell and genome has been the subject of many other reviews [3, 41, 42, 61–67]. These effects extend beyond simple mutation by genomic insertion. L1 RNA and protein overexpression has been linked with apoptosis, DNA damage and repair, tumor progression, cellular plasticity, and stress response [68–72]. Consequently, the cell has evolved a battery of defenses to protect against the dangers of unfettered retrotransposition. It is not surprising that many of the known anti-retrotransposon restriction factors are also anti-retroviral. Phylogenetic analyses suggest that eukaryote non-LTR retrotransposons predate LTR retrotransposons, which in turn gave rise to retroviruses through the acquisition of an envelope (env) gene [73–76]. Indeed, some restriction factors may have first evolved to control ancient endogenous retroelements and were later recruited to the fight against exogenous invaders. It is reasonable to presume that from the study of factors controlling endogenous retrotransposition new insights into the control of viral infections will emerge.
Until recently, our knowledge of the cellular factors that interact with mammalian retrotransposons to facilitate or frustrate their activity lagged behind our understanding of such factors in yeast and flies [77–80]. Nevertheless, in recent years, with the aid of mouse transgenic models, improved antibodies, efficient strategies for immunoprecipitating retrotransposon RNP complexes from cells, new high-throughput (HT) DNA resequencing strategies, and cell culture retrotransposition assays, we have significantly increased our understanding of how the mammalian cell attempts to coexist with a molecular parasite whose unchecked activity could be bad news indeed.
Of all the tools in the toolbox of mammalian retrotransposon research, after twenty years the cell culture assay for retrotransposition remains the most important (Fig. 3; reviewed in [81, 82]). It built upon earlier assays that tracked Ty1 LTR retrotransposition in budding yeast [83]. A reporter gene cassette, interrupted by a backwards intron and inserted in opposite transcriptional orientation into the 3' UTR of a retrotransposition-competent L1, is expressed only when the L1 transcript is spliced, reverse-transcribed, its cDNA inserted in the genome, and the reporter gene expressed from its own promoter. The original neomycin phosphotransferase gene reporter [84–86] was later joined by enhanced green fluorescent protein, blasticidin S-resistance, firefly luciferase, and secreting gaussia luciferase gene constructs [51, 87–89] (Fig. 3). Alu, SVA, and mouse SINE non-LTR, and IAP and HERV LTR retrotransposition assays have also been established [10, 90–96]. While immensely effective in revealing cis- and trans-acting factors of retrotransposition, the degree to which these plasmid-based assays truly reflect endogenous levels of retrotransposition is often uncertain. Fortunately, cell culture results can now be confirmed by HT genome sequencing [97].
Fig. 3.
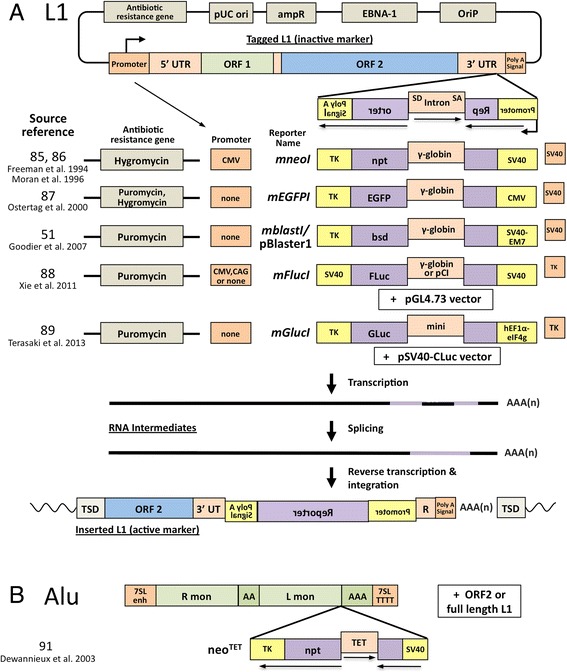
Cell culture retrotransposition assay reporter gene cassettes come in a variety of flavors. a. LINE-1 assays. A retrotransposition-competent L1 and reporter cassette is cloned in pCEP4 (Invitrogen)-based vectors, which encode EBNA-1 and OriP and so replicate in primate cells. Variants of the vectors also contain or lack an exogenous promoter upstream of the L1, and encode resistance to hygromycin or puromycin permitting antibiotic selection of transfected cells. mneoI and mblastI reporter cassettes confer drug resistance to cells having a retrotransposition event. These cells are expanded in culture to form colonies, fixed, stained, and the number of colonies scored. The mEGFPI cassette fluorescently marks cells with retrotransposon insertions and allows their numbers to be counted by flow cytomentry. Firefly luciferase gene mFlucI reporter vectors may be cotranfected with pGL4.73 (Promega) or other vector which constitutively expresses renilla luciferase from transfected cells. Following cell lysis, retrotransposition levels, indicated by firefly luciferase, are adjusted to renilla expression to control for differences in transfection efficiency. The mGlucI cassette expresses Gaussia luciferase which when secreted into the media serves as an effective read-out of accumulated retrotransposition events. Levels of Gluc may be normalized to those of Cypridina luciferase (which is also secreted and does not cross-react with Gluc) constiitutively expressed from the cotransfected pSV40-CLuc vector (NEB). Simply by sampling small aliquots of cell culture media, retrotransposition may be assessed in a single well at multiple time points without cell lysis. Luciferase-based reporter cassettes are amenable to HT retrotransposition screening. b. The Alu assay. An active Ya5 Alu and neoTET cassette interrupted by a Tetrahymena thermophila self-splicing 23S rRNA Group I intron is cloned between the 7SL pol III enhancer and terminator. When this construct is co-expressed with L1 ORF2 alone or a full-length retrotransposition-competent L1, Alu RNAs are reverse transcribed along with the spliced npt gene and integrated into the genome to confer neomycin resistance. Abbreviations: 7SL enh, 7SL enhancer; 7SL TTTT, 7SL transcription terminator; ampR, ampicillin resistance gene; bsd, blasticidin S deaminase gene; CMV, cytomegalovirus promoter; EBNA-1, Epstein-Barr nuclear antigen 1; EGFP, enhanced green fluorescent protein; L mon, left monomer; mini, chimeric mini-intron of the plasmid psiCHECK-2 (Promega); npt, neomycin phosphotransferase gene; oriP, latent origin of replication; pCI: synthetic intron from pCI (Promega); R mon, right monomer; SA, splice acceptor; SD, splice donor; SV40, simian virus 40 early enhancer/promoter; TET, T. thermophila self-splicing intron; TK, herpes simplex virus thymidine kinase poly(A) signal
Lines of defense
To a significant degree, non-LTR retrotransposon sequence itself and the nature of TPRT mitigate genomic insertions. Most L1s die at the time of TPRT, undergoing 5' truncations or inversions, or internal deletions. Most of the insertions that remain intact ultimately lose their ability to remobilize due to DNA recombination or mutation [98]. It has also been suggested that the adenosine richness of the L1 template retards processivity of transcription and limits retrotransposition [99]. Mutations to binding sites for transcription/enhancer factors, including E2F1/RB1, ETS, p53, RUNX3, SOX2, SP1, TCF-LEF, and YY1 for L1s and AHR, CTCF, RAR and SLUG for mouse SINEs, modulate TE expression and in some cases retrotransposition [100–116] (All factors and their full names are listed in Table 1). Cryptic splice sites in L1 RNA transcripts induce a complex pattern of splicing that may remove portions of the ORFs or the 5' UTR [117, 118]. Alus also contain cryptic splice sites, and when resident in genes are frequently exonized into mRNA transcripts and are occasionally translated [119–122]. Interestingly, heterogeneous nuclear ribonucleoprotein C (HNRNPC) protects the cell from Alu-mediated aberrant exonization by competing with splicing factor U2AF2 for binding at Alu splice sites [123]. Alu lacks its own Pol III transcription termination signal, requiring termination at signals in downstream flanking DNA with possible loss of retrotransposition efficiency. The L1 possesses a poly (A) termination signal that is inherently weak and permits occasional read-through of L1 transcripts, necessitating their termination at signals downstream. Interestingly, in silico studies show that approximately 15 % of L1s have transduced 3’ flanking DNA to a new genomic location, in the process generating between 19 and 30.5 Mb of new DNA or as much as 1 % of the human genome [124–127]. Cryptic polyadenylation signals are also scattered along the A-rich length of the L1, and consequently a majority of L1 RNAs are prematurely truncated and incapable of forming functional RNPs [99, 128]. Post-translational protein modifications, including phosphorylation of ORF1p [129, 130], may also modulate retrotransposition.
Table 1.
Cellular factors associated with mammalian non-LTR retrotransposon activity
| Protein Symbol (Hs) (1) | Alternative Name (Hs or Mm) (1) | Protein Name | Source species of factor (1) | Comment | Altered retrotransposition reported | References |
|---|---|---|---|---|---|---|
| Post-transcriptional | ||||||
| AICDA | AID | Activation-induced cytidine deaminase | various | RNA-editing cytidine deaminase | Y | [142, 144] |
| APOBEC1 | Apolipoprotein B mRNA editing enzyme catalytic subunit 1 | various | RNA-editing cytidine deaminase | Y | [143, 145] | |
| APOBEC3 | Apolipoprotein B mRNA editing enzyme catalytic subunit 3 | Hs | RNA-editing cytidine deaminase | Y | [136, 145, 149, 152, 153, 160, 515–521] | |
| ATG5 | Autophagy related 5 | Hs | Autophagy E1-like activating enzyme | Y | [52] | |
| BECN1 | ATG6 | Beclin 1 | Mm | Autophagy factor | Y | [52] |
| CALCOCO2 | NDP52 | Calcium binding and coiled-coil domain 2 | Hs | Macroautophagy receptor | Y | [52] |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein L | Hs | Y | [56, 236, 240, 261] | ||
| KIAA0430 | MARF1, LKAP | Meiosis arrest female protein 1 | Hs | Regulator of oogenesis | [266, 267] | |
| MOV10 | Mov10 RISC complex RNA helicase | Hs | Putative ATP-dependent RNA helicase | Y | [232, 234–237] | |
| MTNR1A | MT1 | Melatonin receptor 1A | Hs | Y | [258] | |
| PABPC1 | PABP1 | Poly(A) binding protein cytoplasmic 1 | Hs | Y | [259] | |
| RNASEL | Ribonuclease L | Hs | Endoribonuclease/component of 2-5A system | Y | [254] | |
| SAMHD1 | SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 | Hs,Mm | Y | [168, 169, 171, 172] | ||
| SQSTM1 | P62 | Sequestosome 1 | Hs | Macroautophagy receptor | Y | [52] |
| TEX19 | TEX19.1 | Testis expressed 19 | Mm | [264, 265] | ||
| TREX1 | AGS1 | Three prime repair exonuclease 1 | Mm | 3' exonuclease | Y | [257] |
| ZC3HAV1 | PARP13, ZAP | Zinc finger CCCH-type containing, antiviral 1 | Hs | Antiviral protein | Y | [236, 240, 250] |
| piRNA/RNAi-Pathways | ||||||
| ASZ1 | GASZ | Ankyrin repeat, SAM and basic leucine zipper domain containing 1 | Mm | [206, 217] | ||
| DDX4 | MVH, VASA | DEAD-box helicase 4 | Mm | ATP-dependent RNA helicase | [210, 217] | |
| DGCR8 | pasha | DGCR8 microprocessor complex subunit | Hs | Subunit of microprocessor | Y | [196, 197] |
| DICER1 | DCR1 | Dicer 1, ribonuclease type III | Hs | dsRNA endoribonuclease | Y | [191, 194] |
| DROSHA | RNASEN | Drosha ribonuclease type III | Hs | dsRNA-specific endoribonuclease/subunit of microprocessor | Y | [197] |
| EXD1 | Exonuclease 3'-5' domain containing 1 | Mm | 3'-5' exonuclease activity | [223] | ||
| FKBP6 | FK506 binding protein 6 | Mm | Cis-trans peptidyl-prolyl isomerase | [215] | ||
| GTSF1 | CUE110 | Gametocyte specific factor 1 | Mm | UPF0224 (FAM112) family member | [209] | |
| HENMT1 | HEN1 methyltransferase homolog 1 | Mm | 2'-O-methylation pf piRNAs | [221] | ||
| HSP90AA1 | HSP90N | Heat shock protein 90 alpha family class A member 1 | Mm | Molecular chaperone | [220] | |
| MAEL | Maelstrom spermatogenic transposon silencer | Mm | [204] | |||
| MIR128-1 | microRNA 128-1 | Hs | Y | [192] | ||
| MOV10L1 | Mov10 RISC complex RNA helicase like 1 | Mm | Putative ATP-dependent RNA helicase | [244, 245] | ||
| Nct1/2 | Non-coding in testis 1/2 | Mm | piRNA encoding non-coding RNAs | [205] | ||
| PIWIL1 | MIWI | Piwi like RNA-mediated gene silencing 1 | Mm | Argonaute family member | [212] | |
| PIWIL2 | MILI | Piwi like RNA-mediated gene silencing 2 | Mm | Argonaute family member | [200, 211, 216, 217] | |
| PIWIL4 | MIWI2 | Piwi like RNA-mediated gene silencing 4 | Mm | Argonaute family member | [202, 203, 211, 212, 222] | |
| PLD6 | MITOPLD | Phospholipase D family member 6 | Mm | [213] | ||
| TDRD1 | Tudor domain containing 1 | Mm | [207] | |||
| TDRD5 | Tudor domain containing 5 | Mm | [214] | |||
| TDRD9 | Tudor domain containing 9 | Mm | Putative ATP-dependent RNA helicase | [208, 217] | ||
| TDRD12 | Tudor domain containing 12 | Mm | Putative ATP-dependent RNA helicase | [218] | ||
| TDRKH | TDRD2 | Tudor and KH domain containing | Mm | [219] | ||
| Epigenetic/Nuclear Factors | ||||||
| ALKBH1 | alkB homolog 1, histone H2A dioxygenase | Mm | 3-methylcytosine demethylase | [366] | ||
| ATM | ATM serine/threonine kinase | Hs,Mm | PI3/PI4-kinase family member | Y | [369–371] | |
| CHAF1 | CAF1 | Chromatin assembly factor 1 | Mm | Assembles histone octamer | [297] | |
| DCLRE1C | ARTEMIS | DNA cross-link repair 1C | Gg | Roles in NHEJ DNA repair and V(D)J recombination | Y | [367] |
| DNMT1 | DNA methyltransferase 1 | Hs,Mm | [313, 340] | |||
| DNMT3A | DNA methyltransferase 3 alpha | Hs,Mm | [313, 340, 342, 344] | |||
| DNMT3B | DNA methyltransferase 3 beta | Hs,Mm | [313, 340, 342, 344] | |||
| DNMT3L | DNA methyltransferase 3 like | Mm | DNA methyltransferase cofactor | [222, 342, 345] | ||
| EHMT2 | G9A | Euchromatic histone lysine methyltransferase 2 | Mm | Histone H3K9me1 and H3K9me2 methyltransferase | [289] | |
| ERCC1 | RAD10 | ERCC excision repair 1, endonuclease non-catalytic subunit | Cg | Nucleotide excision repair | Y | [368] |
| ERCC4 | XPF | ERCC excision repair 4, endonuclease catalytic subunit | Hs,Cg | Nucleotide excision repair (heterodimer with ERCC1) | Y | [368] |
| KDM1A | LSD1 | Lysine demethylase 1A | Mm | Histone H3K4me and H3K9me demethylase | [363] | |
| LIG4 | DNA ligase 4 | Gg | Roles in NHEJ DNA repair and V(D)J recombination | Y | [367] | |
| MECP2 | Methyl CpG binding protein 2 | Hs,Mm | Binds methylated DNA | Y | [355–357] | |
| MORC1 | MORC family CW-type zinc finger 1 | Mm | Role in early spermatogenesis | [352] | ||
| PRKDC | XRCC7, DNA-PKcs | Protein kinase, DNA-activated, catalytic polypeptide | Cg | NHEJ DNA double-strand break repair | Y | [68, 72] |
| SIRT6 | Sirtuin 6 | Mm | NAD-dependent protein deacetylase | Y | [473] | |
| SUV39H | Suppressor of variegation 3–9 homolog 1 | Hs,Mm | Histone H3-K9 methyltransferase 1 | [286, 288, 291] | ||
| TRIM28 | KAP1 | Tripartite motif containing 28 | Hs,Mm | Nuclear corepressor for KRAB-ZFPs | [313] | |
| UHRF1 | NP95, ICBP90 | Ubiquitin-like with PHD and ring finger domains 1 | Mm | RING-finger type E3 ubiquitin ligases | [351] | |
| XRCC4 | X-ray repair cross complementing 4 | Cg | DNA single-strand break repair | Y | [68, 72] | |
| XRCC6 | KU70 | X-ray repair cross complementing 6 | Gg | ssDNA-dependent ATP-dependent helicase | Y | [367] |
| Krüppel-associated box domain-containing zinc finger proteins (KRAB-ZFPs) | ||||||
| GM6871 | Predicted gene 6871 | Mm | [313] | |||
| ZBTB16 | PLZF | Zinc finger and BTB domain containing 16 | Hs,Mm | Y | [317] | |
| ZFP819 | Zinc finger protein 819 | Mm | [314] | |||
| ZNF91 | Zinc finger protein 91 | Hominidae | [315] | |||
| ZNF93 | Zinc finger protein 93 | Hominidae | Y | [315] | ||
| Other transcription factors | ||||||
| AHR | Aryl hydrocarbon receptor | Mm,Dr | Ligand-activated transcription factor | [109, 114] | ||
| CTCF | CCCTC-binding factor | various | BORIS + CTCF gene family member | [108, 116] | ||
| ETS1 | ETS proto-oncogene 1, transcription factor | Hs | ETS transcription family member | [102] | ||
| RAR | Retinoic acid receptor | Hs | Thyroid-steroid hormone receptor superfamily member | [107, 115] | ||
| RB/E2F1 | RB transcriptional corepressor proteins/E2F transcription factor 1 | Hs,Mm | Transcription repressor complex | [112, 298] | ||
| RUNX3 | Runt-related transcription factor 3 | Hs | Runt domain-containing transcription family member | Y | [104] | |
| SNAI2 | SLUG | Snail family transcriptional repressor 2 | Mm,Dr | Snail C2H2-type zinc finger transcription family member | [109, 114] | |
| SOX2 | SRY-box 2 | Hs,Mm | SRY-related HMG-box (SOX) transcription family member | [103, 106, 111] | ||
| SP1 | Sp1 transcription factor | Hs | zinc finger transcription factor | [102] | ||
| TCF-LEF | T-cell factor/lymphoid enhancer factor | Rn | Wnt transcription factors | [111] | ||
| TP53 | p53 | tumor protein p53 | Dr,Hs,Mm | tumor suppressor protein | Y | [110, 299, 300] |
| YY1 | YY1 transcription factor | Hs,Mm | GLI-Kruppel zinc finger transcription family member | [101, 105, 113] | ||
(1) Cg, Cricetulus griseus; Cl, Canis lupus; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus, Rn, Rattus norvegicus
The cell has also evolved a phalanx of trans-acting restriction factors that function as an early defense against both viral infection and endogenous retroelements. Many of these proteins are involved in nucleic acid metabolism and may be constitutively expressed or induced, often by type I interferons. Typically they form a rapid response to infection, and act in the cytoplasm. Early examples were found by comparing cell lines that were permissive or restrictive for viral infection.
Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC)/ Activation-induced cytidine deaminase (AID) proteins
The first anti-retrotransposon restriction factors identified were AID/APOBEC proteins, an evolutionarily conserved, vertebrate-specific family of cytidine deaminases. While rodents have a single APOBEC3 family member, humans have seven APOBEC3s (A3A-D, A3F, A3G and A3H). It was discovered that A3G is packaged into virions of Vif-deficient HIV-1, where during reverse transcription it deaminates cytosines to uracils in the nascent first-strand HIV cDNA. Uracils in the cDNA cause dG > dA hypermutations during second strand synthesis, limiting viability of the viral progeny [131–133]. In the past 10 years, A3B and A3F have also been shown to have antiretroviral activity, and some APOBEC3 proteins are effective against other classes of virus (reviewed in [134]).
The discoveries that AID/APOBEC proteins restrict not only infecting viruses but also LTR and non-LTR retrotransposons have been summarized in previous reviews [135–141]. All APOBEC3 proteins inhibit LINE-1 retrotransposition to varying degrees, with A3A and A3B being most effective. AID and APOBEC1 proteins both inhibit cell culture L1 and LTR element retrotransposition [142–145]. AID may also promote methylation of TEs in the nuclei of primordial germ cells (PGCs) [146]. Interestingly, Khatua et al. [147] revealed a way in which restriction may be transferred from one cell to another, showing that extruded exosome vesicles can encapsulate A3F and A3G mRNAs and be taken up by other cells to inhibit their ability to support Alu and L1 retrotransposition. Tumor-derived microvesicles are also enriched in LINE-1, Alu, and especially HERV RNAs [148]. Apart from being a potential conduit for moving TEs between cells, these tumor-derived microvesicles may make useful cancer biomarkers if they can be confidently detected in human blood or sera.
Unexpectedly, catalytically inactive APOBEC3s still inhibit non-LTR retrotransposons, and several investigations found scant genomic evidence for L1 editing by cytidine deamination [149–151]. Deamination-independent mechanisms of APOBEC action were therefore proposed, including sequestration of retrotransposon RNPs in high molecular weight cytoplasmic complexes and their targeting to SGs and PBs for possible degradation by RNAi silencing [152–158]. However, in silico analyses by Carmi et al. [159] confirmed extensive editing of LTR retrotransposons and found strong evidence for editing of SVAs (20 %) and mouse L1 elements (0.74 %), but minimal editing of human L1s (which occurred mostly within older subfamilies). Richardson et al. [160] then proposed that annealed L1 RNA normally protects first-strand cDNA from deamination, but that transiently exposed single-stranded (ss) cDNA occurring during TPRT becomes accessible to deamination by A3A. Normally the cell repairs U mutations, but by inhibiting uracil DNA glycosylase in cell culture, these authors detected A3A-induced L1 mutations. Moreover, overexpression of both A3A and RNase H, which degrades RNA:DNA hybrids, increased L1 cDNA mutation in an in vitro RT assay [161]. HIV, unlike L1, encodes RNase H activity, which may make its cDNA more susceptible to APOBEC3-mediated deamination.
SAM domain and HD domain 1 (SAMHD1)
Another important member of the anti-retroviral arsenal is SAMHD1, a dGTP-activated deoxynucleoside triphosphate triphosphohydrolase. It has been proposed that SAMHD1 degrades the dNTP pool in non-dividing cells to levels below that necessary for reverse transcription of retroviruses and replication of some DNA viruses [162–166]. Loss of SAMHD1 has been linked to Aicardi-Goutières syndrome (AGS), an early-onset inflammatory disorder affecting particularly the brain [167].
Overexpression of SAMHD1 inhibits, while coexpression of SIV-encoded accessory protein viral protein X (Vpx) or depletion of endogenous SAMHD1 increases non-LTR retrotransposition in cell culture. Seven of eight AGS-related mutations in SAMHD1 reduced inhibition of cell culture LINE-1 retrotransposition by 40 % or more [168]. On the other hand, nine naturally occurring polymorphisms failed to alter SAMHD1 inhibition of retrotransposition [169]. One might expect patients with mutant SAMHD1 alleles to show increased retrotransposition; however, sequencing of bulk tissue and single neurons from the brain of one AGS patient revealed no increase of L1 insertions compared with controls [170].
Although SAMHD1 restricts HIV and SIV in non-dividing cells only, non-LTR retrotransposition is reduced in dividing cells where dNTPs are constantly replenished. Furthermore, SAMHD1 proteins with mutations in the NTPase catalytic domain or at a residue whose phosphorylation is important for retroviral restriction still inhibit cell culture retrotransposition [168] (although Hu et al. [171] reported an NTPase mutant that failed to inhibit retrotransposition). Tetramer formation by SAMHD1 is required for both dNTPase activity and regulation of HIV-1 and LINE-1s [172].
These data predict a mechanism other than dNTPase activity for restricting L1s. SAMHD1 also possesses ribonuclease activity, which even in the absence of functional dNTPase inhibits HIV-1 replication [173]: its effect on retrotransposons remains to be tested. Zhao et al. [168] reported that SAMHD1 reduced L1 reverse transcription by inhibiting ORF2p but not ORF1p. Hu et al. [171] proposed a novel mechanism whereby SAMHD1 enhances assembly of cytoplasmic stress granules that then sequester L1 RNPs and prevent their retrotransposition. Depletion of SG proteins G3BP1 (which binds the L1 RNP) or TIA1 prevented SG formation and reduced SAMHD1 inhibition of LINE-1s. While LINE-1 proteins and RNA concentrate in SGs and PBs along with factors linked with their restriction, a direct role for cytoplasmic granules in modulating retrotransposition remains unclear. Previous experiments investigated PBs and LTR retrotransposons only, and results were conflicted. PBs were required for yeast Ty1 and Ty3 virus-like particle (VLP) assembly and retrotransposition [174–176], but PBs inhibited mouse IAPs [157]. It remains to be determined if cytoplasmic aggregates are a retrotransposition dead-end or an integral part of the L1 life cycle.
RNA-induced Silencing Complex (RISC) and Piwi-interacting RNA (piRNA) pathway proteins
Small interfering RNA (siRNA)-mediated post-transcriptional gene silencing is an ancient strategy for limiting the spread of mobile genetic elements. RNA interference (RNAi) can act at the post-transcriptional level by causing RNA degradation and loss of translation, or at the transcriptional level by inducing epigenetic modifications. Several lines of evidence suggest a direct role for small RNAs in mammalian retrotransposon silencing (reviewed in [177–183]. A large number of endogenous retrotransposon-related small RNAs of a size consistent with siRNAs, miRNAs and piRNAs have been detected in cells [179, 184–188] (reviewed in [189]). Treating cells with in vitro diced L1 siRNAs hindered cell culture retrotransposition [190], and L1-related endo-siRNAs decreased retrotransposon activity, apparently by promoter hypermethylation [191]. Recently, a specific microRNA, mir-128, was found to bind L1 RNA and repress its integration in HeLa and induced pluripotent stem cells (iPSCs) [192]. Indeed, it has been proposed that miRNAs originally evolved from TEs [189]. The question remains, however, as to whether RNAi pathways evolved to silence TEs themselves or gene transcripts that happened to contain target TE sequences [193].
In the nucleus, DGCR8 binds DROSHA, an RNase III-type enzyme, to form the Microprocessor complex. Microprocessor cleaves primary miRNAs (pri-miRNAs), which are then further processed in the cytoplasm to mature miRNAs by DICER and loaded into Argonaute (AGO)-containing RISCs. Knockdown of DICER1 (which also processes siRNAs from dsRNAs) or AGO2 causes an increase in the rate of retrotransposition of tagged L1s in cell culture [191, 194]. Elevated transcription of murine L1 and IAP elements has been observed in embryonic stem cells (ESCs) of Dicer-null mice [195]. Interestingly, DGCR8 also directly binds L1- and SINE-derived RNAs, presumably at hairpin structures, which are apparently cleaved by Microprocessor in a manner independent of DICER and miRNAs. Both DROSHA and DGCR8 affect cell culture retrotransposition [196–198]. Non-LTR retrotransposon RNAs that escape Microprocessor surveillance in the nucleus may be captured in the cytoplasm for further processing by DICER and RISC loading.
piRNAs are small RNAs slightly longer than siRNAs (24–30 nt) that are processed independently of DICER and silence TEs specifically in the germline. They mediate both PIWI protein endonuclease-slicer activity [199] and de novo methylation of TE sequences (discussed below). A large proportion of mouse prepachytene piRNAs derives from retrotransposon sequences [200–202], and the importance of piRNA pathway proteins in repressing retrotransposons in prenatal gonad development and spermatogenesis has repeatedly been demonstrated in mutant mouse lines. Loss of EXD1, FKBP6, GASZ/ASZ1, GTSF1, HENMT1, HSP90α, MAEL, MILI/PIWIL2, MIWI/PIWIL1, MIWI2/PIWIL4, MVH/DDX4, PLD6/MITOPLD, TDRD1, TDRD5, TDRD9, TDRD12, or TDRKH/TDRD2 protein, or the piRNA-encoding non-coding RNAs Nct1/2 is accompanied by derepression of LINE-1 and IAP retrotransposons [201–223]. These studies generated much discussion in the RNAi and retrotransposon fields. However, they failed to provide a crucial piece of information: do the observed accumulation of retrotransposon RNAs and proteins mean increased numbers of endogenous insertion events? Also, it remains to be determined if increased retrotransposition contibutes to the male germline defects and sterility observed in many of these knockout (KO) mice. With the advance of HT genome sequencing, this information can now be obtained.
Moloney leukemia virus 10, homolog (mouse) (MOV10)/ Moloney leukemia virus 10-like 1, homolog (mouse) (MOV10L1)
MOV10 is a member of the UPF1-like superfamily1 of ATP-dependent RNA helicases and was first identified as a protein that prevents infection of mice by Moloney leukemia virus [224, 225]. It is a homolog of SDE3, a helicase for RNAi in Arabidopsis, and Armitage, a protein involved in RISC assembly and piRNA control of RNA viruses and endogenous retroelements in Drosophila [226, 227]. In humans, MOV10 associates with APOBEC3 proteins and components of RISC in SGs and PBs [156, 228]. Several groups examined the role of MOV10 in limiting HIV-1 replication but results were conflicted [229–233]. However, MOV10 strongly inhibits all human non-LTR retrotransposons in cell culture, consistent with its subcellular colocalization with L1 ORF1p in cytoplasmic granules, co-immunoprecipitation (co-IP) with the L1 RNP, and binding of L1 transcripts [232, 234–236]. Li et al. [237] showed that overexpression of MOV10 strongly reduced levels of exogenously expressed IAP and L1 RNAs at a post-transcriptional step, while inhibition of endogenous MOV10 increased RNA levels of transfected L1s. On the other hand, Lu et al. [238] found that MOV10 decreased IAP RT products but not IAP RNA or protein. The exact mode of MOV10 restriction remains uncertain. MOV10 binds mRNA surveillance protein UPF1 and promotes UPF1-induced nonsense-mediated decay, possibly by unwinding mRNA secondary structure and displacing proteins from 5' UTRs [235]. UPF1 itself binds both L1 ORF1p and ORF2p RNPs, and conceivably could recruit MOV10 to the L1 RNP. Paradoxically, however, while depletion of endogenous UPF1 increases L1 expression, cell culture retrotransposition is reduced [239]. Overexpression of UPF1 has no effect on retrotransposition [240].
MOV10L1, a MOV10 paralog, is expressed specifically in the mouse male germline and is required for both fertility and meiosis. Its RNA helicase activity is necessary for the proper biogenesis of pre-pachytene and pachytene piRNAs [241, 242] (reviewed in [243]). Loss of MOV10L in mice leads to depletion of MILI- and MIWI2-associated piRNAs, DNA demethylation in the testes, severe DNA damage in spermatids, and elevated expression of LINE-1 and IAP retrotransposons [244, 245].
Zinc finger CCCH-type, antiviral 1 (ZC3HAV1/ZAP/PARP13)
ZAP is a member of the poly (ADP-ribose) polymerase (PARP) family of proteins. Human ZAP is a predominantly cytoplasmic protein that exists in two alternatively spliced isoforms, the shorter form being inducible by interferon (IFN) [246]. The longer isoform possesses a defective C-terminal PARP-like domain incapable of poly-ADP ribosylation. An N-terminal CCCH-type zinc finger domain binds and induces the degradation of transcripts from several positive and negative-strand RNA viruses, possibly by recruiting the RNA processing exosome and targeting viral RNA to cytoplasmic granules [247–249].
Both ZAP isoforms potently restrict cell culture insertion of non-LTR and mouse IAP retrotransposons through loss of retroelement RNA. ZAP closely colocalizes with L1 ORF1p and RNA in SGs, and binds the L1 RNP [236, 250]. While it is likely that ZAP recruits RNA degradation proteins to retrotransposon transcripts, inhibition of translation by ZAP has been reported for some viruses and cannot be excluded for L1s [251, 252]. Any roles for the exosome and SGs in ZAP-mediated retrotransposon restriction remain to be determined. Observations that both ZAP and MOV10 co-IP, overlap in cytoplasmic granules together with the L1 RNP, and promote loss of L1 RNA and proteins suggest the two proteins may act in the same pathway [250].
Ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent ribonuclease) (RNASEL)
RNaseL is an IFN-inducible endoribonuclease that binds and cleaves single-stranded regions of viral and cellular RNAs, and upon prolonged activation induces autophagy and apoptosis and the death of virus-containing cells. Viral double-strand (ds) RNAs activate oligoadenylate synthetase (OAS), which uses ATP to synthesize 2',5'-linked oligoadenylates (2-5As). 2-5A molecules bind latent RNASEL inducing its active dimer form (reviewed in [253]). RNASEL restricts retrotransposition of both IAP and L1 elements in cultured human cells, and causes loss of L1 RNA. Zhang et al. [254] hypothesized that RNASEL is activated by double-stranded regions existing within L1 RNA or that are formed by annealing of complementary transcripts generated by the sense and antisense promoters of the L1 5' UTR,
Three prime repair exonuclease 1 (TREX1)
TREX1, the most abundant 3′–5′ DNA exonuclease in mammalian cells, targets reverse-transcribed retroviral cDNAs to prevent their accumulation in the cytosol. Paradoxically, TREX1 has also been identified as a cofactor for HIV-1 replication, and it has been proposed that HIV in part evades host innate immunity by exploiting TREX1 to clear its non-pre-integration complex cDNAs to levels unable to trigger cytosolic DNA receptors [255, 256]. Stetson et al. [257], showed that overexpression of TREX1 dramatically reduced retrotransposition of L1 and IAP elements in cell culture, and that ssDNA fragments from endogenous retroelements, including LINE-1s, SINEs and ERVs, accumulate in heart cells of Trex1 KO mice, demonstrating that TREX1 metabolizes reverse transcribed cDNA.
Others
Other cellular proteins strongly inhibit retrotransposition, mostly by unknown mechanisms. For example, melatonin, the hormonal regulator of circadian rhythms and sleep, and its MT1 receptor suppress L1 expression in an in vivo cancer model and dramatically decrease retrotransposition in cultured cells [258]. Poly-A binding protein C1 (PABPC1) is important for L1 RNP formation, and perturbing its levels alters cell culture retrotransposition and subcellular localization of ORF1p [259]. An affinity capture screen of factors that bind the internal ribosome entry site (IRES) of mouse L1 RNA revealed HNRNPL and nucleolin, whose depletion, respectively, increased and decreased mouse L1 cell culture retrotransposition 10-fold [260, 261]. Evidence suggested that while nucleolin functions as an IRES-dependent trans-acting factor for mouse ORF2 translation, HNRNPL behaves like a host restriction factor by decreasing levels of L1 RNA and protein. In separate studies, human HNRNPL bound the L1 RNP and strongly reduced cell culture retrotransposition [56, 236, 240].
TEX19.1 is a mammalian-specific protein of unknown function whose expression is limited to germ and pluripotent stem cells, and the placenta [262]. Mouse TEX19.1 is important for normal placenta development and spermatogenesis. It also represses expression of transposable elements, including MMERVK10C LTR elements in the male germline and LINE-1 in embryonic stem cells (ESCs) and hypomethylated trophectoderm-derived cells of the placenta [263–265]. Although the mouse Tex19.1 KO phenotype resembles those of Miwi2 and Mili mutants, there are indications that TEX19.1 protein may inhibit retroelements at a post-transcriptional step, distinct from the piRNA pathway [263].
MARF1 is an essential regulator of mouse oogenesis, and loss of function causes infertility in females only. LINE-1 and IAP retrotransposon expression is upregulated in mutant Marf1 oocytes coincident with an increase in dsDNA breaks. While its mechanism of inhibition is unknown, structural similarities have been noted between MARF sequence and RNase-like and RNA binding-motifs of PIWI and TDRD5/7 proteins, respectively [266, 267]. Limkain B, the human orthologue of MARF1, is a component of P-bodies [268].
Macroautophagy traps cellular components in double-walled vesicles called autophagosomes and delivers them to lysosomes for degradation. Autophagy also plays a role in the metabolism of Alu and L1 transcripts, which colocalize and copurify with autophagosomes. Knockdown of autophagy receptor proteins increased Alu and L1 cell culture retrotransposition, and qPCR analyses showed LINE-1 insertions to increase in mice lacking the autophagy regulatory protein Beclin1 (BECN1/ATG6) [52]. Autophagic control of retrotransposition is a strategy also conserved in Saccharomyces cerevisiae, which targets Ty1 VLPs to autophagosomes via interaction with Atg19p [269].
Adenosine deaminase acting on RNA (ADAR) proteins bind dsRNAs and convert adenosines to inosines. Both antiviral and proviral roles have been reported for ADARs (reviewed in [270]). In humans A-I RNA editing occurs primarily in Alus present in the non-coding regions of pre-mRNA transcripts [271–274]. Alus with inverted orientations and proximal to each other, and there are many in the human genome, form dsRNA stem loop structures that are preferred templates for ADAR editing. Editing of Alus has been linked with alternative splicing, gene silencing, and altered RNA transport (reviewed in [275–277]). While roles for ADAR editing in the evolution of Alu subfamilies and the suppression of their retrotransposition by mutation has not yet been determined, it is logical to assume they exist.
Yeast Two Hybrid assays and recent affinity-capture and co-IP experiments have identified many other predominantly RNA-binding proteins that bind and colocalize with L1 RNP complexes. Some of these proteins strongly repress cell culture retrotransposition when overexpressed and are obvious candidates for future investigation [51, 56, 236, 239]. CSDA, DDX39, HNRNPA1, HNRNPU, MX2, PURA, SRSF1, and YB1 form a partial list. Roles for many of these proteins in viral replication are known.
The nuclear option
Most of the restriction factors described so far function largely in the cytoplasm, limiting retrotransposition by post-transcriptional mechanisms. Other factors function in the nucleus, suppressing transcription at the first step of retrotransposition, or interfering with DNA integration at the last (reviewed in [278, 279]).
In plants much crosstalk exists between DNA methylation, histone modification, and RNA interference, each of which has been implicated in transcriptional silencing of retrotransposons [280, 281]. Our understanding of their united effects in mammals is less developed and derives mainly from studies in mouse ESCs and embryos and extensive work on the regulation of ERVs. Repression of IAP retrotransposons in early mouse embryogenesis is maintained primarily by histone methylation, but in post-mitotic germ and other differentiated cells DNA methylation assumes importance [222, 282–284]. SINEs, LINEs and SVAs typically bear histone H3 methylated at Lys9 (H3K9me2/me3) repressed chromatin marks, and H3K9 methyltransferases EHMT2/G9A and SUV39H have been implicated in their repression [285–289] (although Dong et al. [290] failed to detect increased expression of LINE-1s in a G9A−/− cell line despite their hypomethylation). Inhibition of SUV39H In human cells reduces H3K9 histone trimethylation and stimulates recruitment of polymerase III together with increased expression of some subfamilies of Alu [291]. Loss of ESET/SETDB1 methyltransferase in mouse PGCs is marked by a decrease of H3K9me3 and H3K27me3 marks on LTRs and LINE-1s, with widespread transcriptional derepression of ERVs but not L1s [284, 292]. Other repressive histone marks may be enriched on non-LTR retrotransposons, although the predominant mark may vary with cell type and species, and discrepancies between study results exist [285, 286, 293–295]. Fadloun et al. [296], for example, found that repression of L1s during preimplantation follows loss of active chromatin marks such as H3K4me3 rather than gain of repressed H3K9me3 marks.
Additional chromatin-associated proteins have been implicated in repression of non-LTR retrotransposons. Loss of histone chaperone chromatin assembly factor 1 (CAF-1) leads to significant up-regulation of L1s, B2 SINES, and IAPs in morula-stage mouse embryos, together with increased histone H2AX phosphorylation and developmental arrest. Treatment with RT inhibitors rescues some of these embryos and so implicates retrotransposon activation in their arrest [297]. Mouse embryonic fibroblasts (MEFs) deficient for all retinoblastoma susceptibility protein family members show upregulation of L1 expression and diminished HDAC1, HDAC2 and NuRD (nucleosomal and remodeling deacetylase) corepressor complex recruitment with consequent epigenetic changes at the L1 promoter [112, 298]. Retrotransposition has salted the human genome with p53 transcription factor binding sites present in the L1 5' UTR, with potentially significant effects on the expression of neighboring genes [110]. Functional p53 represses DNA damage-induced SINE transcription [299]. Loss of p53 increases activity of Drosophila non-LTR retrotransposons, and a human L1 introduced into tp53-mutant zebrafish showed increased retrotransposition and loss of H3K9me3 marks on the 5'UTR. Elevated LINE-1 expression is also a feature of p53 mutant cancer cell lines [300].
KRAB-associated protein 1 (KAP1/TRIM28) is a transcriptional corepressor essential for normal development and cell differentiation. KAP1 mediates the recruitment of chromatin remodeling complexes to DNA by binding Krüppel-associated box domain-containing zinc finger proteins (KRAB-ZFPs) and other DNA-binding proteins (reviewed in [301, 302]). Roles for KAP1 and its KRAB-ZFPs in the control of both exogenous and endogenous retroviruses of mice are well established [303–307] (reviewed in [31, 308]). In mouse ESCs, silencing of many ERV elements is maintained by SETDB1-mediated H3K9me3 methylation, and KAP1 is required for their repression [309–312]. In human ESCs, KAP1 is recruited to older L1PA6 to L1PA3 subfamilies, but is largely absent from young human-specific L1Hs elements. Its binding is associated with H3K9me3 enrichment and its depletion with expression of these older elements [313]. Some non-LTR retrotransposons are bound by species-specific KRAB-ZFPs, including mouse ZFP819, which inactivates LINES and SINES [314], mouse GM6871, which weakly suppresses two relatively young but retrotransposition-incompetent mouse L1 subfamilies (L1MdF2 and L1MdF3) [313], and ZNF91 and ZNF93, for which there is evolutionary evidence for suppression of now inactive human SVA and L1 subfamilies [315] (reviewed in [316]). ZBTB16/PLZF, a regulator of cell growth and differentiation, also binds L1 DNA, altering local chromatin acetylation and methylation, and repressing L1 expression in germ and progenitor cells and retrotransposition in the cell culture assay [317].
Methylation regulation of retrotransposons is complex and controlled by interacting factors whose activity has been linked to mammalian germline development. Half the CpGs in the human genome reside in repeats, 25 % of them in Alus and 12 % in LINEs [318, 319]. While most CpG islands in gene promoters are undermethylated if the genes are expressed, an island in the L1 5' UTR is typically heavily methylated in somatic cells and L1 expression is suppressed [320–323]. Indeed, it has been proposed that DNA methylation of CpGs evolved primarily as a host defense mechanism against TEs [324]. Garcia-Perez et al. [325] made the interesting observation that in human embryonic carcinoma-derived cell lines an L1 reporter was strongly silenced by methylation during or shortly after retrotransposition and could be desilenced with histone deacetylase inhibitors; a control reporter gene inserted alone into the genome was not. This suggested that epigenetic silencing specifically targets the TPRT event. Significantly, methylation and other repressive chromatin marks may spread beyond LTR and non-LTR insertions into flanking DNA, and effects on the expression of nearby genes are possible [326–330].
In adult mice, IAP elements are heavily methylated in mature eggs and sperm, but L1s are undermethylated in eggs compared with sperm and somatic cells [331, 332]. Successive waves of demethylation occur in the developing mouse embryo. The first wave is shortly after fertilization until the morula stage and involves LINES, SINES and LTRs. Demethylation occurs again around E8.5 in post-implantation primordial germ cells that are entering the hindgut endoderm, and continues through to E12.5 to E13.5 when PGCs have colonized the genital ridges. LINE-1 methylation is largely erased in PGCs, but IAP CpGs remain more resistant (summarized in [181, 333–337]). Some retrotransposons evade the remethylation that occurs post-E13.5 and are subject to piRNA pathway-mediated methylation from E16.5 [202, 338]. However, substantial demethylation of L1s is not always mirrored by transcriptional activation [337].
DNMT1, the most abundant DNA methyltransferase (MTase) in mammals, preferentially methylates hemimethylated DNA (maintenance methylation); DNMT3 MTases are more involved in de novo methylation of unmethylated CpGs. However, the three MTases function cooperatively. Both IAPs and L1s are demethylated in Dnmt1−/− mouse ES cells [339–344]. B1 SINEs are methylated by DNMT3A, and IAP and LINE-1 elements are methylated by both DNMT3A and DNMT3B. DNMT3L regulatory factor lacks catalytic activity, but recruits DNMT3A and DNMT3B to their targets. In nondividing prospermatogonia, DNMT3L functions mainly in establishing methylation of retrotransposons, including L1s and IAPs. Deletion of Dnmt3a or Dnmt3l results in uncontrolled transposon expression in the mouse male germline with spermatogenesis failure and sterility [342, 345]. Zamudio et al. [222] found that DNA methylation is dispensable for TE silencing prior to male germ cell meiosis. With the onset of meiosis and programmed loss of chromatin repression, L1s were activated in Dnmt3l−/− mice, accompanied by precocious loss of H3K9me2 marks and gain of H3K4me3 marks and SPO11-induced dsDNA breaks in TEs. However, no attendant increase in TE genomic copy number was detected by qPCR.
In addition to MTases, several cofactors are involved in DNA methylation and suppression of TEs. Lymphoid-specific helicase (LSH/HELLS) belongs to the SNF2 helicase family of chromatin remodeling proteins and may recruit DNMT3B to chromatin [346]. LSH is essential for normal embryonic development, and its loss in mouse embryos or the female germline is accompanied by DNA demethylation, altered histone acetylation, abnormal heterochromatinization, and hypomethyation of pericentromeric satellite repeats and IAP elements [347, 348]. In the absence of LSH, IAPs are upregulated, but LINE-1 sequences remain repressed despite being hypomethylated [349]. UHRF1 (also known as NP95 in mice and ICBP90 in humans) is another cofactor that recruits DNMT1 to hemimethylated CpGs. Its loss leads to hypomethylation and upregulation of IAPs, L1s, and SINEs in mouse embryos [350, 351]. In the mouse male germ line, MORC1, a member of the Microrchidia (Morc) family of GHKL ATPases, regulates repression of various families of retrotransposons, including L1s. Although KO mice suffer germ cell loss and infertility similar to animals defective for piRNA proteins, MORC1 appears to act independently of the piRNA pathway and may facilitate DNA methylation of TEs [352]. Morc family homologs in Arabidopsis also repress transposons [353].
DNMT3A and DNMT3B also mediate methylation-independent gene repression through their association with heterochromatin protein 1 (HP1), methyl-CpG-binding proteins (MeCP), and histone MTase activity [354]. MECP2 binds methylated CpG dinucleotides and forms complexes with DNA MTases or histone deacetylases. In cell culture, MECP2 binds the L1 5' UTR to limit expression and retrotransposition [355]. Retrotransposon expression is increased, and, as shown by qPCR, L1 copy numbers are elevated in brains of Mecp2 KO mice as well as in patients with Rett syndrome, a rare neurodevelopmental disorder caused by mutations in MECP2 [356, 357].
The piRNA pathway not only degrades RNAs post-transcriptionally but may induce gene silencing by promoting DNA and histone methylation (reviewed in [358]). PIWI proteins play important roles in de novo DNA methylation. The fact that de novo CpG DNA methylation and transcriptional silencing of transposable elements is reduced during fetal spermatogenesis of Mili and Miwi2 KO mice suggests that piRNA RISC recruits methylation proteins to TE loci, including the L1 5' UTR [200–203, 359]. The piRNA pathway is also important in mouse germ cells for deposition of H3K9me3 marks on young active L1s [329], as well as on TEs in flies [360, 361].
The enzymes that catalyze DNA and histone methylation are well characterized; those that remove methyl-groups much less so. Deficiency of maternal LSD1/KDM1A histone H3 mono- and dimethyl K4 demethylase during early mouse development leads to desilencing of muERV-L/MERVL elements and LINE-1s, and an increase in L1 ORF1p expression that is most obvious in cell nuclei [362, 363]. TET1 (ten-eleven translocation) enzyme oxidizes 5mC to 5-hydroxymethylcytosine (5hmC), an intermediate for the removal of 5mC [364]. 5hmC is enriched at the promoters of L1s in murine ESCs [365]. Recently the notion that DNA methylation in mammals occurs only as 5-methylcytosines was challenged with the discovery of N6-methyladenine modification in mouse ES cells. Knockout of the demethylase gene Alkbh1 caused increased N6-mA deposition on the 5' ends of young but not old L1s that correlated with an increase in other repressive marks on the L1s and transcriptional silencing of nearby genes and enhancers [366]. Discoveries such as these add new layers of complexity to the regulation of retrotransposons by DNA methylation
DNA repair proteins have been implicated in modulation of retrotransposition, although their roles in TPRT are unclear. Endonuclease-independent retrotransposition is strongly elevated in Chinese hamster ovary cells lacking non-homologous end-joining (NHEJ) repair proteins [68, 72]. Chicken DT40 cell lines defective for NHEJ genes, including DCLRE1C (artemis), LIG4, and XRCC6/KU70, show restricted retrotransposition of transfected human L1 and zebrafish ZfL2-2 LINE2 elements [367]. The ERCC1/XPF heterodimer is involved in nucleotide excision, recombination, and inter-strand crosslink repair, and limits non-LTR retrotransposition in cell culture [368]. Ataxia telangiectasia mutated (ATM), a serine/threonine protein kinase activated by dsDNA breaks, has been linked with retrotransposition, although results are contradictory. Cell lines mutated for ATM, or with ATM protein levels reduced by expression of human papillomavirus E6 oncoprotein, had attenuated L1 activity, implying a supportive role for ATM in retrotransposition [369, 370]. On the other hand, ATM-deficient neuronal precursor cells and the brains of Atm KO mice showed elevated activity of an L1-mEGFPI reporter transgene (Fig. 3), and ataxia telangiectasia patients had increased L1 copy numbers as detected by PCR [371]. Thomas et al. [372] suggested that use of G418 antibiotic to select the L1-mneoI reporter construct used by Gasior et al. [369] may have caused cell toxicity and affected results (although, GFP-induced cytotoxicity has also been reported [373]).
Despite the numerous cellular proteins implicated in the control of mammalian non-LTR retrotransposon integration, beyond first strand synthesis, no comprehensive model of integrant resolution and repair exists. This failure has been in part due to the lack of an effective in vitro assay that recapitulates later steps of the TPRT reaction. Such assays have been instrumental in detailing the mechanisms of genome insertion by bacterial and yeast Group II introns and insect R1 and R2 retrotransposons [374, 375]. Although Cost et al. [376] reconstituted the initial stages of L1 element transposition in vitro, the field has failed to take up the challenge to refine this assay and apply it to mechanistic investigations.
An arms race
It has been proposed that cells are engaged in a genetic “arms race” with infecting retroviruses and endogenous retrotransposons, and must constantly evolve new strategies to fight infection or transposition. This places selective pressure on the parasitic element, which contrives to evolve measures to evade repression, which in turn may be countered by new changes within a host restriction factor [377]. One signature of the struggle between host and pathogen is positive (diversifying) selection for alleles that confer fitness benefit. For examples, the C-terminal PARP-like domain of ZAP-L displays recurrent positive selection and enhanced ZAP-mediated anti-viral and anti-retrotransposon activites [250, 378]. APOBEC3A has undergone diversifying selection in response to a changing repertoire of viral pathogens, while maintaining the ability to inhibit L1s through 40 million years of primate evolution [379]. A recent study by Jacobs et al. [315] provided two examples of dynamic coevolution between KRAB-ZFPs and their retrotransposon targets. Eight to 12 million years ago, a series of structural modifications enabled ZNF91 to bind and repress SVA elements. In turn, until about 12.5 million years ago, ZNF93 suppressed early primate L1s until there arose the younger L1PA3 subfamily that lacked the ZNF93 target sequence. It has been proposed that KRAB-ZFP genes reflect a classic arms race between retroelements and their hosts, with ZFP repressors evolving novel DNA binding specificities that target retrotransposon subfamilies as they became newly active in the genome [380]. Indeed, of 18 KRAB-ZFPs tested, 16 bound specific classes of endogenous retroelements [381]. Moreover, the predicted ages of the ZFPs and the retroelements they bound were correlated with only two exceptions, ZNF33A, which primarily bound SVAs, and ZNF382, which associated with younger L1Hs elements.
LINE-1 type Transposase Domain-containing 1 (L1TD1/ECAT11), the sole known example of functional domestication of LINE-1-derived protein sequence, contains two ORF1-like domains. L1TD1 is associated with self-renewal and the maintenance of pluripotency in embryonic stem cell culture (although not in KO mice) [382, 383]. Its loss or pseudogenization in multiple mammalian lineages, together with evidence for diversifying selection, prompted McLaughlin et al. [384] to propose that L1TD1 originally evolved as a host restriction factor against retrotransposons that was later coopted as a pluripotency factor. Like L1 ORF1p, endogenous L1TD1 is detected in PBs [382], although any effect on L1 activity is so far unknown. Conceivably, expression of L1TD1 protein might exert a dominant-negative effect on ORF1p function.
There is some evidence for positive selection within and in the near vicinity of L1s. Using genome-wide analyses, Kuhn et al. [385] detected extended haplotype homozygosity around some L1 insertions with evidence for recent positive selection; this predicts potential phenotypic effects of the L1s, although, no supporting functional studies were attempted. Since the mammalian radiation, a single lineage of L1s has been active in both mice and humans, each subfamily losing activity due to mutations, to be then supplanted by the next, until today there remains one active subfamily in humans (L1PA1) and three in mice (A, TF, and GF) [386, 387]. Positive selection is evident in the coiled-coil domain of human L1 ORF1p; coiled-coils mediate protein-protein interactions [19, 20, 388]. Although the coiled-coil domain of mouse ORF1 fails to show positive selection, there has been considerable structural instability in this region. It has been suggested that the diversity of 5' UTRs and novel ORF1 sequence variants that distinguish mouse L1 subfamilies arose from recombination and may reflect an evolutionary drive for the L1 to adapt to cellular host factors [46, 47, 389].
Some lentiviruses have evolved small accessory proteins that both modify cellular functions and mute the cell’s antiviral response. Vif and Vpx, for example, target APOBEC3G and SAMHD1 for ubiquitination and degradation, and BST-2 is neutralized by HIV-1 Vpu, SIV Nef, and HIV-2 Env (reviewed in [390–392]). It is therefore reasonable to consider that by disrupting host restriction factors, HIV infection might stimulate retrotransposition, Indeed, this effect was observed [393], and expression of Vif or Vpr was necessary for maximal induction of HIV-infected Jurkat cell culture retrotransposition. An increase in L1 and Alu DNA copy numbers was also detected by qPCR of DNA from infected CD4+ T cells, but could not be confirmed to represent new insertions [393]. Vpr is a multifunctional accessory protein that regulates nuclear import of the HIV-1 preinitiation complex; it is not known if it targets a host restriction factor [394]. Recombinant Vpr protein added to cell culture increased tagged L1 retrotransposition, and when injected into transgenic mice caused an increase in genomic L1 copy number as determined by qPCR [395, 396].
At this point, a note of caution may be in order. Commencing with investigations of retrotransposition in brain tissue samples [356, 371, 397, 398], the use of sensitive qPCR strategies to assess variation in the copy number of L1 genomic insertions is becoming de rigeur in the field. This trend is likely to increase with the development of more sensitive digital droplet PCR protocols [399]. Apparent changes in retrotransposon copy number are never confirmed by downstream genome sequencing to detect new insertions. Previously, I proposed a possible source of bias for such PCR-based studies [400]. Cellular conditions that stimulate expression of L1s or HERVs, and therefore their encoded reverse transcriptases, might also induce promiscuous reverse transcription of retrotransposon RNAs not engaged in TPRT at the site of chromatin integration. The cDNAs so generated would be amenable to qPCR amplification, biasing upwards estimates of genomic L1 copy numbers. Although an unverified concern, recent studies suggest that it is not an unreasonable one. For example, elevated levels of Alu- and LINE-1-containing hybrid RNA/DNA molecules have been detected in cancer cell lines and are lost upon treatment with RT inhibitor [401]. cDNA complementary to infecting viruses and cellular mRNA is generated independent of genomic integration in the presence of LINE-1 ORF2p [402, 403]. Protocols that isolate only high molecular weight DNA (such as gel purification) and that apply RNase H (to degrade RNA/DNA substrates) and ssDNA nucleases prior to qPCR could remove contaminating molecules that might confound data interpretation.
Since the 1980s, extrachomosomal small polydispersed circular (spc) DNAs containing retrotransposon sequences, including SINEs and L1s, have been reported in cells. Recombination, replicon misfiring, modified TPRT, and reverse transcription models have been proposed to explain these extrachromosomal DNAs [404–407]. Their copy numbers are elevated in cancer cells and associated with genome instability [408, 409]. Still a poorly studied class of mammalian copy number variants, spcDNAs could conceivably be an additional off-target source of amplicons for some PCR-based analyses of genomic retrotransposon insertions. That said, standardized PCR protocols that reliably detect retrotransposon insertion copy numbers would be a boon to the field.
RT-PCR assessment of retrotransposon expression is also prone to misinterpretation. One must be confident that only L1 RNAs transcribed from the 5' UTR promoter are amplified. Amplification of unrelated mRNAs that by chance contain retrotransposon PCR target sequence will bias results. When designing primer pairs, in silico analyses of potential off-target binding sites in mRNAs should be performed. Studies published to date do not report such analyses, and frequently qPCR results are not validated by other techniques measuring L1 expression. As well as being part of longer mRNA transcripts, L1s generate full length, spliced and prematurely polyadenylated products from their sense promoters. Northern blotting protocols that provide information about the 5' ends and length of L1 transcripts should be the gold standard of analysis (these issues are discussed in [410]).
When the defenses fail
If we think of retrotransposons as genetic parasites, it makes sense they should have evolved to be active in the germline and transmit to future generations, but remain inactive in somatic cells and not risk harming the host. This notion was dispelled by F. Gage and colleagues at the Salk Institute who showed that L1 retrotransposition occurs in neuronal precursor cells, especially in the hippocampus [106], and by the Boeke and Kazazian labs who showed retrotransposition in early mouse and human development, implying that each of us is a mosaic of somatic genomes [411–413]. Other papers using HT genome sequencing have since concurred that there is endogenous somatic L1 retrotransposition in neural precursors and the adult brain, although estimates of insertion frequency differ by more than an order of magnitude [170, 371, 414–416] (reviewed in [372, 417–419]. The Faulkner group [170], using retrotransposon-capture sequencing (RC-seq) of single cells, estimated high rates of L1 retrotransposition in the hippocampus (averaging 13.7 insertions per neuron) and cerebral cortex (16.3 per neuron). The Walsh and Park groups [415, 416] reported a much lower average of <0.6 of an insertion per neuron using L1Hs insertion profiling (L1-IP). Following somatic transfer and expansion in oocytes of six post-mitotic nuclei of mouse MT (middle temporal visual area) neurons, Hazen et al. [420] found an average of 1.3 new insertions per neuron. Recently, Evrony et al. [421] reanalyzed the Faulkner group data, criticized aspects of its bioinformatic and validation approaches, and concluded a revised estimate of 0.2 of an event per neuron. It has been suggested, however, that this reanalysis made both inappropriate use of a post-filtered dataset and erroneous assumptions in concluding chimeric artifacts in the Upton et al. [170] PCR-validations (G. Faulkner, pers. comm.). Nevertheless, even the low estimate of 0.2 of an insertion per neuron predicts 20 billion unique insertion events in a human brain. There is evidence based on tagged engineered L1 assays that retrotransposition is not limited to neuronal precursor cells but can take place in non-dividing mature neuronal cells as well (J. Garcia-Perez, pers. comm.). It has been proposed that retrotransposition contributes to neuronal plasticity (reviewed in [422]), although brain tumors seem as likely a consequence. However, to date no de novo L1 insertions have been detected in glioblastoma or medulloblastoma brain cancers [423–425].
Why suppression of non-LTR retrotransposons is perturbed in some but not other cell types is unclear, but has implications for development and disease. L1 promoter hypomethylaton, elevated L1 expression, and cell culture retrotransposition have been demonstrated in human iPSC and ESC lines [426–433] (reviewed in [434, 435]). Interestingly, iPSCs from non-human primates support greater cell culture retrotransposition than human iPSCs, correlating with lower levels of APOBEC3B and PIWIL2 proteins in the former, and the significantly larger pool of chimpanzee-specific L1 elements [432]. Recently, retrotransposition of endogenous L1, Alu and SVA elements has been shown to occur during reprogramming of human iPSCs and in pluripotent stem cell culture [436, 437].
There is limited data on endogenous retrotransposition in normal somatic adult tissues other than the brain, except for the finding of a single potential somatic insertion in hepatocytes [438] and small numbers of insertions detected in DNA of esophagus, stomach and colon [439–441]: at least some of these insertions may have occurred during early embryogenesis. On the other hand, many de novo insertions have been detected during HT sequencing analyses of bulk cancer tissues. In 2010, Iskow et al. [423] first reported tumor-only L1 insertions in lung cancer, and subsequent studies have made it clear that somatic retroelement insertions are detectable at varying frequencies in a subset of tumors, especially those of epithelial origin [424, 425, 438–444]. These insertions may have sequence characteristics that differ from typical germline insertions, such as a higher degree of 5’ truncation and more frequent insertions independent of L1-encoded endonuclease cleavage [445].
A role for retrotransposition in the etiology of cancer remains an open question, however (reviewed in [446–448]). To date there have been a few “smoking gun” examples of tumor-specific L1 insertions presumed to have led to cancer. In 1992 Miki et al. [449] showed that an L1 had inserted into an exon of the APC tumor suppressor gene in a colon cancer, but that the insertion was undetectable in normal colon of the affected individual. Over 20 years later, tumor-specific L1 insertions were found in the Suppression of Tumorigenicity18 (ST18) gene of a hepatocarcinoma [438], in an exon of the PTEN gene in endometrial cancer [425], and in the APC gene of a colorectal cancer [450]. Recent work indicates that many insertion events detected in tumors were already present in precancerous lesions, and were perhaps present in the somatic cells that gave rise to the tumors. Some insertions were detected only in metastases and not in the primary tumors, suggesting late cancer-specific events [440, 444]. We do not, however, know the background levels of retrotransposition in normal somatic cells and if these levels are sufficiently frequent to be of significance for cancer progression or somatic disease. We do not know if retrotransposition may drive cancer, or cancer accelerates retrotransposition. Interestingly, there is evidence for the misregulation of AID and APOBEC3 in some cancers leading to increased mutation and perhaps contributing to clonal evolution and tumor progression [451, 452] (summarized in [453, 454]). Increased cytosine deaminase activity could tamp-down retrotransposition in tumors; it might also be an induced response to increased retroelement expression.
Expression of L1s and their ORF1 or ORF2 proteins is altered in various tumor types compared with their normal tissues, phenomena that may prove useful as diagnostic markers of cancer progression [57, 60, 439, 455–458]. Hypomethylation of L1 DNA has been observed in many cancers and is associated with increased L1 expression [459, 460]. One might therefore expect increased expression to mean increased retrotransposition. However, no study to date can conclude that endogenous retrotransposition frequency is specifically elevated in cancer since only tissues in bulk have been sequenced. In non-tumor tissue, an individual de novo insertion will be present in only a small subset of cells among the large total population of cells sampled, and may exist in too low a copy number to be detected by standard amplification methods. An insertion initially present within a normal cell is more easily detected once that cell clonally expands as a tumor, which, upon sampling, sequencing and PCR validation, would falsely appear to possess a tumor-only event (discussed in [400]). Single-cell sequencing protocols should provide true estimates of the rates of retrotransposition in tumor versus normal somatic cells.
Misregulated expression of retrotransposons can damage the genome. The endonuclease activity of the L1 ORF2 protein generates a dsDNA break that recruits repair proteins to the retrotransposon insertion site. This is a normal part of TPRT. However, transient transfection of L1s in cell culture induces DNA breaks far in excess of what would be expected for TPRT alone [369]. DNA damage caused by overexpression of ORF2p may induce genotoxic stress and cellular apoptosis [69, 369, 461, 462]. Elevated ORF2 endonuclease and RT activities in mice have also been linked with increased meiotic prophase I defects and fetal oocyte attrition, a mysterious process that involves loss of a majority of oocytes prior to birth [463]. The fact that treating mice with the nucleoside analog AZT blocks oocyte attrition, suggests that RT inhibitors might be applied to suppress retrotransposons and perhaps extend the female reproductive lifespan [464]. One might also wonder if epigenetic misregulation or loss of a restriction factor causing elevated retrotransposon activity could trigger diminished fertility or even spontaneous abortions in humans, topics worthy of further investigation.
Several reviews have linked the aging process with progressive changes in chromatin architecture and increased expression of retrotransposons [465–469]. Increased mobilization of gypsy and non-LTR R1 and R2 retrotransposons in the aging fly brain is accompanied by neural and cognitive decline [470]. Senescence of fibroblasts and aging mouse tissues are marked by progressive epigenetic reorganization, depression of retrotransposons, and increased insertions at late-stage senescence as determined by qPCR [471, 472]. Longevity-linked protein Sirtuin-6 (SIRT6) represses L1s by binding their 5' UTRs, and promotes heterochromatinization through mono-ADP ribosylation of KAP1. SIRT6 vacates L1 loci in senescent cells and brain tissues of aging mice, with an accompanying increase in L1 transcription and PCR-detected insertions [473]. However, while senescence may foster retrotransposition, the notion that retrotransposition hastens aging, as with the notion that it significantly promotes cancer, remains speculative. Elevated ORF2 endonuclease expression and TPRT cause DNA damage and genomic lesions, and certainly DNA damage increases with age. However, a direct connection between these phenomena is unclear.
Most links between retrotransposons and disease involve endogenous retroviruses. Altered HERV expression occurs in SLE, Sjogren’s syndrome, multiple sclerosis, schizophrenia, psoriasis, Creutzfeldt-Jakob disease, amyotrophic lateral sclerosis, and various cancer conditions, although the specific HERV loci that contribute to the transcriptional misregulation observed have been only partially documented and a causative role for these sequences in disease is largely speculative [474–478] (reviewed in [479–484]). Among the more convincing studies, elevated expression of HERV-K transcripts in cortical and spinal neurons of ALS patients is supported by evidence of neurotoxic effects of the HERV-K env protein in a mouse model [485]. Syncytin1 protein, which derives from the envelope gene of the HERV-W ERVWE-1 locus, has essential functions during placental development and is upregulated in multiple sclerosis. Its expression causes cytotoxicity of astrocytes in vitro and oligodendrocyte loss and demyelination in transgenic mice (reviewed in [486]).
Links between misregulated non-LTR retrotransposon expression and disease are fewer. Expression of Alus is altered in certain neurodegenerative conditions, including Creutzfeldt-Jakob and Alzheimer diseases [487]. DICER deficiency in geographic atrophy, a form of age-related macular degeneration, induces accumulation of Alu RNA, which in turn activates the NLRP3 inflammasome complex and downstream caspases leading to retinal pigment epithelial cell death [488, 489]. In a study that proved Sjögren’s syndrome autoantigen RO60/SS-A binds Alu RNAs, transfection of the bound Alu motif into peripheral blood cells stimulated proinflammatory cytokine secretion, while IFN-α treatment of RO60-null lymphocyte cells activated Alu transcription [490]. Yu et al. [491] further showed that increased L1 expression in human fibrosarcoma cells and in testes and MEFs of Mov10l KO mice is marked by induction of IFN-β and IFN-stimulated genes (although, not unexpectedly, elevated IFN levels also inhibit L1 cell culture retrotransposition, perhaps by inducing restriction factors [250, 491]). Such results link retrotransposon RNA metabolism with the immune response.
Aicardi–Goutières syndrome is a rare inflammatory disorder with no known cure. Within the first year of life, patients usually experience severe brain dysfunction and neurological damage that clinically mimics in-utero viral infection. AGS is characterized by increased IFN and IFN-stimulated gene expression. The condition has been associated with mutations in seven genes, including SAMHD1, TREX1, RNASEH2A, RNASEH2B, RNASEH2C, ADAR1, and IFIH1 [492, 493]. TREX1 mutations are also implicated in other autoimmune conditions, including systemic lupus erythematous, chilblain lupus, and retinal vasculopathy with cerebral leukodystrophy [494, 495]. Significantly, AGS-associated genes are involved in DNA or RNA metabolism and are components of the intrinsic immune response against exogenous retroviruses or endogenous retroelements. It has been proposed that a defect in an AGS-related gene prevents the cell from efficiently “clearing” endogenously-produced nucleic acids, causing their accumulation in the cytoplasm where they bind pattern recognition receptors and trigger an innate type I interferon response. One possibility is that these self-nucleic acids derive from the RNA or reverse transcribed cDNA of retroelements, although this remains untested [257, 482, 496]. Significantly, Stetson et al. [257] found that ssDNA fragments from endogenous retroelements accumulate in heart cells of Trex1-null mice, possibly contributing to their characteristic inflammatory myocarditis and death [497] (although a more recent study failed to detect increased ERV expression in Trex1−/− mouse dendritic and macrophage cells [498]). Significantly, a combination of nucleoside RT inhibitors expected to inhibit both retroviruses and L1 retrotransposons attenuated the autoimmune myocarditis [499]. Encouraged by such insights, an early stage RT inhibitor clinical trial for children with AGS has begun in Paris (https://clinicaltrials.gov/ct2/show/NCT02363452).
Aftermath
Retrotransposons pose an ongoing threat to the human genome. In the past six million years, 1174 fixed LINE-1s, 5530 Alus, and 864 SVAs have accumulated in hominins [500]. In addition, each of us possesses rare and potentially active insertions. Extrapolating, the world’s population of 7 billion may therefore harbor millions of active unique or low allele frequency human-specific non-LTR retrotransposons [66]. While low levels of retrotransposition and other mutations in the germline maintain genetic variation, high levels may jeopardize viability of the cell. Moreover, if, as our field is coming to believe, retrotransposition in somatic cells is much greater than in the germline, it could be a significant source of cell-to-cell variation. Retrotransposon activity may play key roles not only in Mendelian disorders, but in multifactorial disorders and cancers as well. It is therefore important to better understand how the cell limits retrotransposons and why its defenses occasionally falter.
Retotransposition is elevated in early embryogenesis, stem cells, neuronal progenitor cells, and perhaps some cancers. Misregulation of RT expression is detected in some disease conditions, and RT inhibition has been reported to inhibit cell proliferation and tumor growth, alter embryogenesis, and promote cell differentiation [401, 501–503] (see reviews [504, 505]). These observations deserve greater investigation. While not reviewed here, a rich literature also demonstrates that environmental stress, including carcinogens, heat shock, heavy metals, addictive stimulant drugs, ionizing radiation and steroids, induce mammalian non-LTR retrotransposons, with consequences for humans that remain to be determined (reviewed in [506–512]).
The repression of retrotransposons occurs on two main fronts: post-transcriptionally and mostly in the cytoplasm, and in the nucleus where histone modification and DNA methylation limit transcription (Table 1). The epigenetic derepression that occurs during early embryogenesis and germ cell development in mammals leaves the cell vulnerable to retrotransposition. Thus, piRNA pathway proteins have evolved to be important guardians of germline genomes. Some cytoplasmic restriction factors are part of the intrinsic immune system of the cell, although their battle strategies may sometimes be unclear. APOBEC3 proteins, for example, may sequester non-LTR retrotransposon RNPs in cytoplasmic aggregates or may deaminate their cDNAs during TPRT. While SAMHD1 may help target L1 RNPs to SGs, its ability to degrade dNTPs, and so perhaps limit reverse transcription of non-LTR element RNAs, bears further investigation. The study of restriction factors that control retrotransposition has been dominated by cell culture retrotransposition assays: these may not always reflect in vivo reality. There are numerous mouse models with mutations in putative restriction factors that can now be screened by HT sequencing for altered endogenous retrotransposition. Many of these mice have mutations in RNAi pathway proteins; others are models for cancer and disease. For example, KO mice exist for most of the genes implicated in Aicardi Goutières Syndrome [513].
Some anti-viral defense strategies that are part of the intrinsic immune system of the cell have been coopted to inhibit non-LTR retrotransposition; the converse is likely also true. The coevolution of strategies by the cell to enforce and in turn by a retroelement to evade restriction perpetuates a molecular arms race. It has been proposed, for example, that the diversity of KRAB-ZFPs arose from their functions in silencing a diverse range of TEs [380]. The ability of retroelements to counter host restriction systems will determine which new subfamilies assume active dominance in a genome.
So far, the study of retrotransposon suppression has yielded surprising insights into gene regulation, epigenetics, DNA repair, and RNA interference. Intriguingly, some factors restrict both retroviruses and retrotransposons but by mechanisms that differ. It is logical, therefore, to study retrotransposon restriction factors as a means of gleaning new insights into viral control. For example, insights down the road may foster a better understanding of retroviral latency and perhaps contribute to its treatment. The continued presence of a reservoir of silenced HIV-1 proviral DNA integrated in the genomes of CD4+ T cells is a major obstacle to eradication of the virus in AIDs patients. Strategies under investigation attempt to overcome innate proviral repression, and then use standard antiretroviral therapy to kill cells with reactivated virus. Trials to date have had limited success and new insights are needed [514].
In cancers, widespread demethylation promotes retroelement transcription, and one would presume increased mobilization as well. Interestingly, to date bulk tissue sequencing efforts have detected new insertion events almost exclusively in tumors of epithelial cell types. To accurately assess the extent and tissue-specificity of retrotransposition, concerted efforts are needed to sequence many single cells from a large number of cell types (normal and cancerous) from many individuals. These efforts will help us to determine the extent to which ongoing insertions are drivers or passengers of cancer and disease. They will tell us if retrotransposition significantly influences brain diversity. To expedite these efforts, the field needs consensus concerning the best current protocols for capturing and identifying rare retrotransposon integrations in bulk tissues or single cells. We need effective algorithms that map retrotransposon transcript sequences in HT RNA-Seq data to their genomic source loci, a difficult task at present. With these algorithms, we will better determine how retrotransposon transcription changes under different cellular conditions and how it may modulate expression of cellular genes and impinge upon cell health. It is likely the physiological consequences of altered retrotransposon expression will be found to far exceed those of increased genome insertions.
While the great mass of the mobilome may be junk, the majority is benign, some is toxic waste, and occasionally bits of treasure may be found.
Abbreviations
AGS. Aicardi–Goutières syndrome; ds. double-stranded; ESC. embryonic stem cell; HERV. human endogenous retrovirus; HNRNP. heterogeneous nuclear ribonucleoprotein; HT. high-throughput; IAP. intracisternal A-particle; IFN. interferon; iPSC. induced pluripotent stem cell; IRES. internal ribosome entry site; kD. kilodalton; KO. knock-out; KRAB-ZFP. Krüppel-associated box domain-containing zinc finger protein; LINE. long interspersed element; LTR. long-terminal repeat; MEF. mouse embryonic fibroblast; miRNA. microRNA; MTase. methyltransferase; ORF. open reading frame; PB. processing (P-) body; PGC. primordial germ cell; piRNA. Piwi-interacting RNA; qPCR. quantitative polymerase chain reaction; RISC. RNA-induced silencing complex; RNP. ribonucleoprotein; RT, reverse transcriptase; SG. stress granule; SINE. short interspersed element; siRNA. small interfering RNA; spcDNA. small polydispersed circular DNA; ss. single-stranded; SVA. SINE-R, VNTR, and Alu; TPRT. target-primed reverse transcription; TSD. target site duplication; UTR. untranslated region.
Acknowledgments
The author is supported by grants from the NIH National Institute of Neurological Disorders and Stroke (1R03NS087290-01) and the ALS Therapy Alliance. Able assistance was provided by G. Pereira. I would like to thank Drs. J. Mayer (U. of Saarland), J. Garcia-Pérez (U. of Edinburgh), and I. Adams (U. of Edinburgh) for ongoing discussions and critical reading of the manuscript, Dr. C. Feschotte (U. of Utah) and two anonymous reviewers for comments, and Dr. K. Burns (Johns Hopkins) for sharing the monoclonal ORF1p antibody.
Competing interests
The author declares that he has no competing interests.
References
- 1.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci. 1950;36(6):344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazazian HH. Mobile DNA: Finding Treasure in Junk. New Jersey: FT Press, Upper Saddle River; 2011.
- 3.Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135(1):23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Mitra R, Li X, Kapusta A, Mayhew D, Mitra RD, Feschotte C, et al. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc Natl Acad Sci. 2013;110(1):234–239. doi: 10.1073/pnas.1217548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stocking C, Kozak CA. Endogenous retroviruses. Cell Mol Life Sci. 2008;65(21):3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012. [DOI] [PubMed]
- 7.Mager DL, Stoye JP. Mammalian Endogenous Retroviruses. Mobile DNA III: Washington, DC: American Society for Microbiology; 2015. p. 1079–100.
- 8.Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. Genome-Wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4(2):e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins NA, Copeland NG. High frequency germline acquisition of ecotropic MuLV proviruses in SWR/J-RF/J hybrid mice. Cell. 1985;43(3):811–819. doi: 10.1016/0092-8674(85)90254-5. [DOI] [PubMed] [Google Scholar]
- 10.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16(12):1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491(7426):774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contreras-Galindo R, Kaplan MH, Dube D, Gonzalez-Hernandez MJ, Chan S, Meng F, et al. Human Endogenous Retrovirus Type K (HERV-K) Particles Package and Transmit HERV-K–Related Sequences. J Virol. 2015;89(14):7187–7201. doi: 10.1128/JVI.00544-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchi E, Kanapin A, Magiorkinis G, Belshaw R. Unfixed endogenous retroviral insertions in the human population. J Virol. 2014;88(17):9529–9537. doi: 10.1128/JVI.00919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naveira H, Bello X, Abal-Fabeiro JL, Maside X. Evidence for the persistence of an active endogenous retrovirus (ERVE) in humans. Genetica. 2014;142(5):451–460. doi: 10.1007/s10709-014-9789-y. [DOI] [PubMed] [Google Scholar]
- 15.Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci. 2016;113(16):E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson PJ, Macfarlan TS, Lorincz MC. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol Cell. 2016;62(5):766–776. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16(6):793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 18.Kapitonov VV, Tempel S, Jurka J. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene. 2009;448(2):207–213. doi: 10.1016/j.gene.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18(12):2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- 20.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16(1):78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13(12):2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ade C, Roy-Engel AM, Deininger PL. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol. 2013;3(6):639–645. doi: 10.1016/j.coviro.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono M, Kawakami M, Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15(21):8725–8737. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, et al. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269(11):8466–8476. [PubMed] [Google Scholar]
- 26.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73(6):1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168(2):933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, et al. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354(4):994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 29.Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, et al. 5'-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19(11):1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancks DC, Kazazian HH., Jr SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol. 2010;20(4):234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol. 2015;31(1):429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- 32.Bantysh O, Buzdin A. Novel family of human transposable elements formed due to fusion of the first exon of gene MAST2 with retrotransposon SVA. Biochem Mosc. 2009;74(12):1393–1399. doi: 10.1134/S0006297909120153. [DOI] [PubMed] [Google Scholar]
- 33.Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH. Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19(11):1983–1991. doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbone L, Harris RA, Mootnick AR, Milosavljevic A, Martin DIK, Rocchi M, et al. Centromere remodeling in hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol Evol. 2012;4(7):648–658. doi: 10.1093/gbe/evs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hara T, Hirai Y, Baicharoen S, Hayakawa T, Hirai H, Koga A. A novel composite retrotransposon derived from or generated independently of the SVA (SINE/VNTR/Alu) transposon has undergone proliferation in gibbon genomes. Genes Genet Syst. 2012;87(3):181–190. doi: 10.1266/ggs.87.181. [DOI] [PubMed] [Google Scholar]
- 36.Ianc B, Ochis C, Persch R, Popescu O, Damert A. Hominoid composite non-LTR retrotransposons--variety, assembly, evolution, and structural determinants of mobilization. Mol Biol Evol. 2014;31(11):2847–2864. doi: 10.1093/molbev/mst256. [DOI] [PubMed] [Google Scholar]
- 37.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci. 2003;100(9):5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141(7):1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.del Carmen SM, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci. 2006;103(17):6611–6616. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 41.Hancks DC, Kazazian HH. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7(1):1. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 44.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21(6):1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denli Ahmet M, Narvaiza I, Kerman Bilal E, Pena M, Benner C, Marchetto Maria CN, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163(3):583–593. doi: 10.1016/j.cell.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 46.Adey NB, Schichman SA, Graham DK, Peterson SN, Edgell MH, Hutchison C. Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol Biol Evol. 1994;11(5):778–789. doi: 10.1093/oxfordjournals.molbev.a040158. [DOI] [PubMed] [Google Scholar]
- 47.Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J Biomed Biotechnol. 2006;2006:1–6. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7(6):706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin SL, Branciforte D. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol Cell Biol. 1993;13(9):5383–5392. doi: 10.1128/MCB.13.9.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hohjoh H, Singer MF. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J Mol Biol. 1997;271(1):7–12. doi: 10.1006/jmbi.1997.1159. [DOI] [PubMed] [Google Scholar]
- 51.Goodier JL, Zhang L, Vetter MR, Kazazian HH. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27(18):6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun. 2014;5:5276. doi: 10.1038/ncomms6276. [DOI] [PubMed] [Google Scholar]
- 53.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172(6):803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Cold Spring Harb Perspect Biol. 2011;3(12):a002774-a. doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodier JL, Ostertag EM, Engleka K, Seleme M, Kazazian HH. A potential role for the nucleolus in L1 retrotransposition. Hum Mol Genet. 2004;13(10):1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 56.Goodier JL, Cheung LE, Kazazian HH. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013;41(15):7401–7419. doi: 10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodić N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184(5):1280–1286. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6(10):e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodier JL, Mandal PK, Zhang L, Kazazian HH. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet. 2010;19(9):1712–1725. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Luca C, Guadagni F, Sinibaldi-Vallebona P, Sentinelli S, Gallucci M, Hoffmann A, et al. Enhanced expression of LINE-1-encoded ORF2 protein in early stages of colon and prostate transformation. Oncotarget. 2015;7(4):4048–4061. doi: 10.18632/oncotarget.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18(3):343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 62.Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105(1):92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 63.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12(1):187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149(4):740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardson SR, Moran JV, Kopera HC, Doucet AJ, Moldovan JB, Garcia-Perez JL. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Mobile DNA III: Washington, DC: American Society for Microbiology; 2015. p. 1165–208. [DOI] [PMC free article] [PubMed]
- 67.Mita P, Boeke JD. How retrotransposons shape genome regulation. Curr Opin Genet Dev. 2016;37:90–100. doi: 10.1016/j.gde.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 69.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6(1):1. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinibaldi‐Vallebona P, Lavia P, Garaci E, Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosom Cancer. 2006;45(1):1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- 71.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616(1–2):46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446(7132):208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 73.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9(10):3353. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doolittle RF, Feng DF. Tracing the origin of retroviruses. Curr Top Microbiol Immunol. 1992;176:195–211. doi: 10.1007/978-3-642-77011-1_13. [DOI] [PubMed] [Google Scholar]
- 75.Malik HS. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10(9):1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 76.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11(7):1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 77.Maxwell PH, Curcio MJ. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryotic Cell. 2007;6(7):1069–1080. doi: 10.1128/EC.00092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42(1):587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siomi MC, Saito K, Siomi H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 2008;582(17):2473–2478. doi: 10.1016/j.febslet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 80.Eickbush TH, Eickbush DG. Integration, Regulation, and Long-Term Stability of R2 Retrotransposons. Mobile DNA III: American Society for Microbiology; 2015. p. 1127–46. [DOI] [PMC free article] [PubMed]
- 81.Rangwala SH, Kazazian HH. The L1 retrotransposition assay: a retrospective and toolkit. Methods. 2009;49(3):219–226. doi: 10.1016/j.ymeth.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kopera HC, Larson PA, Moldovan JB, Richardson SR, Liu Y, Moran JV. LINE-1 Cultured Cell Retrotransposition Assay. Methods Mol Biol. 2016;1400:139–156. doi: 10.1007/978-1-4939-3372-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 84.Heidmann T, Heidmann O, Nicolas JF. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc Natl Acad Sci. 1988;85(7):2219–2223. doi: 10.1073/pnas.85.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Freeman J, Goodchild N, Mager D. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques. 1994;17(1):46–52. [PubMed] [Google Scholar]
- 86.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/S0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 87.Ostertag EM, Prak E, DeBerardinis R, Moran JV, Kazazian HH. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28(6):1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie Y, Rosser JM, Thompson TL, Boeke JD, An W. Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res. 2011;39(3):e16-e. doi: 10.1093/nar/gkq1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Terasaki N, Goodier JL, Cheung LE, Wang YJ, Kajikawa M, Kazazian HH, et al. In vitro screening for compounds that enhance human L1 mobilization. PLoS ONE. 2013;8(9):e74629. doi: 10.1371/journal.pone.0074629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Esnault C, Casella J-F, Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30(11):e49-e. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35(1):41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 92.Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet. 2004;36(5):534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- 93.Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol. 2005;349(2):241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 94.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20(17):3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012;40(4):1666–1683. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bock A, Schumann GG. The Engineered SVA Trans-mobilization Assay. Methods Mol Biol. 2016;1400:203–222. doi: 10.1007/978-1-4939-3372-3_14. [DOI] [PubMed] [Google Scholar]
- 97.Ewing AD. Transposable element detection from whole genome sequence data. Mobile DNA. 2015;6(1). [DOI] [PMC free article] [PubMed]
- 98.Ostertag EM, Kazazian HH. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11(12):2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429(6989):268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 100.Minakami R, Kurose K, Etoh K, Furuhata Y, Hattori M, Sakaki Y. Identification of an internal cis-element essential for the human Li transcription and a nuclear factor (s) binding to the element. Nucleic Acids Res. 1992;20(12):3139–3145. doi: 10.1093/nar/20.12.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Becker KG, Swergold G, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2(10):1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 102.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem. 1998;273(2):891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]
- 103.Tchénio T, Casella J, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28(2):411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang N, Kazazian HH. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31(16):4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32(13):3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 107.Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18(11):1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Román AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc Natl Acad Sci. 2008;105(5):1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harris C, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28(44):3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12(9):1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, et al. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res/Fundam Mol Mech Mutagen. 2009;665(1–2):20–28. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S-H, Cho S-Y, Shannon MF, Fan J, Rangasamy D. The impact of CpG island on defining transcriptional activation of the mouse L1 retrotransposable elements. PLoS ONE. 2010;5(6):e11353. doi: 10.1371/journal.pone.0011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Román AC, González-Rico FJ, Moltó E, Hernando H, Neto A, Vicente-Garcia C, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21(3):422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu Q, Tanasa B, Trabucchi M, Li W, Zhang J, Ohgi KA, et al. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat Struct Mol Biol. 2012;19(11):1168–1175. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves Â, Kutter C, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148(1):335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34(5):1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411(1–2):38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12(7):1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons. Science. 2003;300(5623):1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 121.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8(6):R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci. 2011;108(7):2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7(2):143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 125.Moran JV, DeBerardinis R, Kazazian HH. Exon shuffling by L1 retrotransposition. Science. 1999;283(5407):1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 126.Pickeral OK, Makalowski W, Boguski M, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10(4):411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Goodier JL, Ostertag EM, Kazazian HH. Transduction of 3'-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9(4):653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 128.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35(4):363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 129.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15(3):630. [PMC free article] [PubMed] [Google Scholar]
- 130.Cook PR, Jones CE, Furano AV. Phosphorylation of ORF1p is required for L1 retrotransposition. Proc Natl Acad Sci. 2015;112(14):4298–4303. doi: 10.1073/pnas.1416869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 132.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4(11):868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 133.Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. BioMed Res Int. 2013;2013:1–18. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Willems L, Gillet NA. APOBEC3 Interference during Replication of Viral Genomes. Viruses. 2015;7(6):2999–3018. doi: 10.3390/v7062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci. 2007;32(3):118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 136.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35(9):2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schumann G. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35(3):637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- 138.Chiu Y-L, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26(1):317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 139.Koito A, Ikeda T. Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements. Mobile Genetic Elements. 2011;1(3):197–202. doi: 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arias JF, Koyama T, Kinomoto M, Tokunaga K. Retroelements versus APOBEC3 family members: No great escape from the magnificent seven. Front Microbio. 2012;3:275. doi: 10.3389/fmicb.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbio. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37(6):1854–1867. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ikeda T, Abd El Galil KH, Tokunaga K, Maeda K, Sata T, Sakaguchi N, et al. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res. 2011;39(13):5538–5554. doi: 10.1093/nar/gkr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Metzner M, Jäck HM, Wabl M. LINE-1 retroelements complexed and inhibited by activation induced cytidine deaminase. PLoS ONE. 2012;7(11):e49358. doi: 10.1371/journal.pone.0049358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lindič N, Budič M, Petan T, Knisbacher BA, Levanon EY, Lovšin N. Differential inhibition of LINE1 and LINE2 retrotransposition by vertebrate AID/APOBEC proteins. Retrovirology. 2013;10(1):156. doi: 10.1186/1742-4690-10-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Khatua AK, Taylor HE, Hildreth JEK, Popik W. Inhibition of LINE-1 and Alu retrotransposition by exosomes encapsidating APOBEC3G and APOBEC3F. Virology. 2010;400(1):68–75. doi: 10.1016/j.virol.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F Inhibit L1 Retrotransposition by a DNA Deamination-independent Mechanism. J Biol Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 150.Esnault C, Priet S, Ribet D, Heidmann O, Heidmann T. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology. 2008;5(1):75. doi: 10.1186/1742-4690-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Anwar F, Davenport MP, Ebrahimi D. Footprint of APOBEC3 on the genome of human retroelements. J Virol. 2013;87(14):8195–8204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci. 2006;103(23):8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The Anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281(39):29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 155.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2(5):e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81(5):2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lu C, Contreras X, Peterlin BM. P bodies inhibit retrotransposition of endogenous intracisternal A particles. J Virol. 2011;85(13):6244–6251. doi: 10.1128/JVI.02517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Horn AV, Klawitter S, Held U, Berger A, Vasudevan AAJ, Bock A, et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2014;42(1):396–416. doi: 10.1093/nar/gkt898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carmi S, Church GM, Levanon EY. Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution. Nat Commun. 2011;2:519. doi: 10.1038/ncomms1525. [DOI] [PubMed] [Google Scholar]
- 160.Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife. 2014;3:e02008. [DOI] [PMC free article] [PubMed]
- 161.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13(7):655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 162.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HCT, Rice GI, Christodoulou E, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 163.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13(3):223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Baldauf H-M, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat Med. 2012;18(11):1682–1689. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gramberg T, Kahle T, Bloch N, Wittmann S, Müllers E, Daddacha W, et al. Restriction of diverse retroviruses by SAMHD1. Retrovirology. 2013;10(1):26. doi: 10.1186/1742-4690-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kim ET, White TE, Brandariz-Núñez A, Diaz-Griffero F, Weitzman MD. SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J Virol. 2013;87(23):12949–12956. doi: 10.1128/JVI.02291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao K, Du J, Han X, Goodier John L, Li P, Zhou X, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Rep. 2013;4(6):1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.White TE, Brandariz-Nuñez A, Valle-Casuso JC, Knowlton C, Kim B, Sawyer SL, et al. Effects of human SAMHD1 polymorphisms on HIV-1 susceptibility. Virology. 2014;460–461:34–44. doi: 10.1016/j.virol.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Upton K, Gerhardt D, Jesuadian J, Richardson S, Sánchez-Luque F, Bodea G, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161(2):228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 2015;11(7):e1005367. doi: 10.1371/journal.pgen.1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu C, Gao W, Zhao K, Qin X, Zhang Y, Peng X et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nature Communications. 2013;4. [DOI] [PubMed]
- 173.Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim S-Y, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20(8):936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beliakova-Bethell N, Terry LJ, Bilanchone V, DaSilva R, Nagashima K, Wente SR, et al. Ty3 nuclear entry is initiated by viruslike particle docking on GLFG nucleoporins. J Virol. 2009;83(22):11914–11925. doi: 10.1128/JVI.01192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. P-Body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol. 2010;30(2):382–398. doi: 10.1128/MCB.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bilanchone V, Clemens K, Kaake R, Dawson AR, Matheos D, Nagashima K, et al. Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes. PLoS Genet. 2015;11(9):e1005528. doi: 10.1371/journal.pgen.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129(1):37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364(1513):99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 180.Castañeda J, Genzor P, Bortvin A. piRNAs, transposon silencing, and germline genome integrity. Mutat Res/Fundam Mol Mech Mutagen. 2011;714(1–2):95–104. doi: 10.1016/j.mrfmmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 181.Bao J, Yan W. Male germline control of transposable elements. Biol Reprod. 2012;86(5):162. doi: 10.1095/biolreprod.111.095463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guo M, Wu Y. Fighting an old war with a new weapon--silencing transposons by Piwi-interacting RNA. IUBMB Life. 2013;65(9):739–747. doi: 10.1002/iub.1192. [DOI] [PubMed] [Google Scholar]
- 183.Yang F, Wang PJ. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Seminars in Cell & Developmental Biology. 2016;pii: S1084-9521(16)30066-0. [DOI] [PMC free article] [PubMed]
- 184.Smalheiser N, Torvik V. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21(6):322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 185.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453(7194):534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 187.Oey HM, Youngson NA, Whitelaw E. The characterisation of piRNA-related 19mers in the mouse. BMC Genomics. 2011;12(1):315. doi: 10.1186/1471-2164-12-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lukic S, Chen K. Human piRNAs are under selection in Africans and repress transposable elements. Mol Biol Evol. 2011;28(11):3061–3067. doi: 10.1093/molbev/msr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Roberts JT, Cardin SE, Borchert GM. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mobile Genetic Elements. 2014;4(3):e29255. doi: 10.4161/mge.29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Soifer HS, Zaragoza A, Peyvan M, Behlke M, Rossi J. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 2005;33(3):846–856. doi: 10.1093/nar/gki223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen L, Dahlstrom JE, Lee S-H, Rangasamy D. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics. 2012;7(7):758–771. doi: 10.4161/epi.20706. [DOI] [PubMed] [Google Scholar]
- 192.Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, et al. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol. 2015;22(10):824–831. doi: 10.1038/nsmb.3090. [DOI] [PubMed] [Google Scholar]
- 193.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang N, Kazazian HH. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13(9):763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 195.Kanellopoulou C, Muljo S, Kung A, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Cáceres JF. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19(8):760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, et al. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20(10):1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Heras SR, Macias S, Cáceres JF, Garcia-Perez JL. Control of mammalian retrotransposons by cellular RNA processing activities. Mobile Genetic Elements. 2014;4(2):e28439. doi: 10.4161/mge.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5'end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 200.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 201.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carmell MA, Girard A, van de Kant HJG, Bourc’his D, Bestor TH, de Rooij DG, et al. MIWI2 Is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 204.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15(2):285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, et al. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod. 2008;79(1):51–57. doi: 10.1095/biolreprod.108.068072. [DOI] [PubMed] [Google Scholar]
- 206.Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5(9):e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain–containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16(6):639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 208.Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 209.Yoshimura T, Toyoda S, Kuramochi-Miyagawa S, Miyazaki T, Miyazaki S, Tashiro F, et al. Gtsf1/Cue110, a gene encoding a protein with two copies of a CHHC Zn-finger motif, is involved in spermatogenesis and retrotransposon suppression in murine testes. Dev Biol. 2009;335(1):216–227. doi: 10.1016/j.ydbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 210.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24(9):887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480(7376):259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 212.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480(7376):264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 213.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20(3):364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192(5):781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47(6):970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 216.Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50(4):601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 217.Lim AK, Lorthongpanich C, Chew TG, Tan CWG, Shue YT, Balu S, et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development. 2013;140(18):3819–3825. doi: 10.1242/dev.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pandey RR, Tokuzawa Y, Yang Z, Hayashi E, Ichisaka T, Kajita S, et al. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc Natl Acad Sci. 2013;110(41):16492–16497. doi: 10.1073/pnas.1316316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32(13):1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ichiyanagi T, Ichiyanagi K, Ogawa A, Kuramochi-Miyagawa S, Nakano T, Chuma S, et al. HSP90 plays an important role in piRNA biogenesis and retrotransposon repression in mouse. Nucleic Acids Res. 2014;42(19):11903–11911. doi: 10.1093/nar/gku881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lim SL, Qu ZP, Kortschak RD, Lawrence DM, Geoghegan J, Hempfling A-L, et al. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 2015;11(10):e1005620. doi: 10.1371/journal.pgen.1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zamudio N, Barau J, Teissandier A, Walter M, Borsos M, Servant N, et al. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 2015;29(12):1256–1270. doi: 10.1101/gad.257840.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yang Z, Chen K-M, Pandey Radha R, Homolka D, Reuter M, Janeiro BK, et al. PIWI Slicing and EXD1 Drive Biogenesis of Nuclear piRNAs from Cytosolic Targets of the Mouse piRNA Pathway. Mol Cell. 2016;61(1):138–152. doi: 10.1016/j.molcel.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jaenisch R, Jähner D, Nobis P, Simon I, Löhler J, Harbers K, et al. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24(2):519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- 225.Mooslehner K, Müller U, Karls U, Hamann L, Harbers K. Structure and expression of a gene encoding a putative GTP-binding protein identified by provirus integration in a transgenic mouse strain. Mol Cell Biol. 1991;11(2):886–893. doi: 10.1128/MCB.11.2.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post‐transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20(8):2069–2077. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116(6):817–829. doi: 10.1016/S0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 228.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15(23):2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 229.Furtak V, Mulky A, Rawlings SA, Kozhaya L, Lee K, KewalRamani VN, et al. Perturbation of the P-Body component Mov10 inhibits HIV-1 infectivity. PLoS ONE. 2010;5(2):e9081. doi: 10.1371/journal.pone.0009081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, et al. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J Biol Chem. 2010;285(19):14346–14355. doi: 10.1074/jbc.M110.109314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, et al. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J Virol. 2010;84(19):10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9(1):53. doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Huang F, Zhang J, Zhang Y, Geng G, Liang J, Li Y, et al. RNA helicase MOV10 functions as a co-factor of HIV-1 Rev to facilitate Rev/RRE-dependent nuclear export of viral mRNAs. Virology. 2015;486:15–26. doi: 10.1016/j.virol.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 234.Goodier JL, Cheung LE, Kazazian HH. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8(10):e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gregersen L, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, et al. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell. 2014;54(4):573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 236.Moldovan JB, Moran JV. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015;11(5):e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, et al. The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem. 2013;288(29):21148–21160. doi: 10.1074/jbc.M113.465856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lu C, Luo Z, Jager S, Krogan NJ, Peterlin BM. Moloney leukemia virus type 10 inhibits reverse transcription and retrotransposition of intracisternal A particles. J Virol. 2012;86(19):10517–10523. doi: 10.1128/JVI.00868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Taylor MS, LaCava J, Mita P, Molloy K, Huang CR, Li D, et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell. 2013;155(5):1034–1048. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Moldovan JB. Identification of cellular host factors that associate with LINE-1 ORF1p and the effect of the Zinc Finger Antiviral Protein ZAP on LINE-1 retrotransposition [Ph.D. Thesis]: University of Michigan; 2015.
- 241.Zheng K, Wang PJ. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012;8(11):e1003038. doi: 10.1371/journal.pgen.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, et al. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29(6):617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhu X, Zhi E, Li Z. MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis. Reproduction. 2015;149(5):R229–R235. doi: 10.1530/REP-14-0569. [DOI] [PubMed] [Google Scholar]
- 244.Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci. 2010;107(26):11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci. 2010;107(26):11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol. 2011;12(1):37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 247.Guo X, Carroll JWN, MacDonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol. 2004;78(23):12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci. 2007;104(1):151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lee H, Komano J, Saitoh Y, Yamaoka S, Kozaki T, Misawa T, et al. Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc Natl Acad Sci. 2013;110(30):12379–12384. doi: 10.1073/pnas.1310604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet. 2015;11(5):e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77(21):11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci. 2011;108(38):15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Silverman RH. Viral Encounters with 2',5'-Oligoadenylate Synthetase and RNase L during the Interferon Antiviral Response. J Virol. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhang A, Dong B, Doucet AJ, Moldovan JB, Moran JV, Silverman RH. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 2014;42(6):3803–3820. doi: 10.1093/nar/gkt1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11(11):1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hasan M, Yan N. Safeguard against DNA sensing: the role of TREX1 in HIV-1 infection and autoimmune diseases. Front Microbiol. 2014;5:193. doi: 10.3389/fmicb.2014.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.deHaro D, Kines KJ, Sokolowski M, Dauchy RT, Streva VA, Hill SM, et al. Regulation of L1 expression and retrotransposition by melatonin and its receptor: implications for cancer risk associated with light exposure at night. Nucleic Acids Res. 2014;42(12):7694–7707. doi: 10.1093/nar/gku503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Dai L, Taylor MS, O’Donnell KA, Boeke JD. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol Cell Biol. 2012;32(21):4323–4336. doi: 10.1128/MCB.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Li P, Li J, Timmerman S, Krushel L, Martin S. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition. Nucleic Acids Res. 2006;34(3):853–864. doi: 10.1093/nar/gkj490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Peddigari S, Li PW-L, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013;41(1):575–585. doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kuntz S, Kieffer E, Bianchetti L, Lamoureux N, Fuhrmann G, Viville S. Tex19, a Mammalian-Specific Protein with a Restricted Expression in Pluripotent Stem Cells and Germ Line. Stem Cells. 2008;26(3):734–744. doi: 10.1634/stemcells.2007-0772. [DOI] [PubMed] [Google Scholar]
- 263.Ollinger R, Childs AJ, Burgess HM, Speed RM, Lundegaard PR, Reynolds N, et al. Deletion of the pluripotency-associated Tex19.1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS Genet. 2008;4(9):e1000199. doi: 10.1371/journal.pgen.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Reichmann J, Reddington JP, Best D, Read D, Ollinger R, Meehan RR, et al. The genome-defence gene Tex19.1 suppresses LINE-1 retrotransposons in the placenta and prevents intra-uterine growth retardation in mice. Hum Mol Genet. 2013;22(9):1791–1806. doi: 10.1093/hmg/ddt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tarabay Y, Kieffer E, Teletin M, Celebi C, Van Montfoort A, Zamudio N, et al. The mammalian-specific Tex19.1 gene plays an essential role in spermatogenesis and placenta-supported development. Hum Reprod. 2013;28(8):2201–2214. doi: 10.1093/humrep/det129. [DOI] [PubMed] [Google Scholar]
- 266.Su Y-Q, Sugiura K, Sun F, Pendola JK, Cox GA, Handel MA, et al. MARF1 regulates essential oogenic processes in mice. Science. 2012;335(6075):1496–1499. doi: 10.1126/science.1214680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Su Y-Q, Sun F, Handel MA, Schimenti JC, Eppig JJ. Meiosis arrest female 1 (MARF1) has nuage-like function in mammalian oocytes. Proc Natl Acad Sci. 2012;109(46):18653–18660. doi: 10.1073/pnas.1216904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Bloch DB, Li P, Bloch EG, Berenson DF, Galdos RL, Arora P, et al. LMKB/MARF1 localizes to mRNA processing bodies, interacts with Ge-1, and regulates IFI44L gene expression. PLoS ONE. 2014;9(4):e94784. doi: 10.1371/journal.pone.0094784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Suzuki K, Morimoto M, Kondo C, Ohsumi Y. Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Dev Cell. 2011;21(2):358–365. doi: 10.1016/j.devcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 270.Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411(2):180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Blow M, Futreal P, Wooster R, Stratton M. A survey of RNA editing in human brain. Genome Res. 2004;14(12):2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kim D, Kim T, Walsh T, Kobayashi Y, Matise T, Buyske S, et al. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14(9):1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22(8):1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 275.Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21(2):77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 276.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7(12):919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. NatRev Mol Cell Biol. 2016;17(2):83-96. [DOI] [PMC free article] [PubMed]
- 278.Crichton JH, Dunican DS, MacLennan M, Meehan RR, Adams IR. Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell Mol Life Sci. 2014;71(9):1581–1605. doi: 10.1007/s00018-013-1468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: war and peace. Mol Cell Biol. 2015;35(5):770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Gebert D, Rosenkranz D. RNA‐based regulation of transposon expression. Wiley Interdiscip Rev: RNA. 2015;6(6):687–708. doi: 10.1002/wrna.1310. [DOI] [PubMed] [Google Scholar]
- 282.Hutnick LK, Huang X, Loo T-C, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285(27):21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8(6):e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci. 2012;37(4):127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 285.Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J Biol Chem. 2003;278(30):27658–27662. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 286.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24(4):800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker ME, Datson NA, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci. 2012;109(43):17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014;55(2):277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 289.Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O’Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7(1):24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27(20):2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun. 2015;6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Liu S, Brind’Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28(18):2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Huda A, Mariño-Ramírez L, Jordan IK. Epigenetic histone modifications of human transposable elements: genome defense versus exaptation. Mob DNA. 2010;1(1):2. doi: 10.1186/1759-8753-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Rangasamy D. Distinctive patterns of epigenetic marks are associated with promoter regions of mouse LINE-1 and LTR retrotransposons. Mob DNA. 2013;4(1):27. doi: 10.1186/1759-8753-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Su M, Han D, Boyd-Kirkup J, Yu X, Han J-DJ. Evolution of Alu elements toward enhancers. Cell Rep. 2014;7(2):376–385. doi: 10.1016/j.celrep.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 296.Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, et al. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20(3):332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- 297.Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, et al. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci. 2015;112(47):14641–14646. doi: 10.1073/pnas.1512775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Montoya-Durango DE, Ramos KA, Bojang P, Ruiz L, Ramos IN, Ramos KS. LINE-1 silencing by retinoblastoma proteins is effected through the nucleosomal and remodeling deacetylase multiprotein complex. BMC Cancer. 2016;16(1):1. doi: 10.1186/s12885-016-2068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Hagan CR, Rudin CM. DNA cleavage and Trp53 differentially affect SINE transcription. Genes Chromosomes Cancer. 2007;46(3):248–260. doi: 10.1002/gcc.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wylie A, Jones AE, D’Brot A, Lu W-J, Kurtz P, Moran JV, et al. p53 genes function to restrain mobile elements. Genes Dev. 2015;30(1):64–77. doi: 10.1101/gad.266098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286(30):26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Cheng C-T. KAPtain in charge of multiple missions: Emerging roles of KAP1. World J Biol Chem. 2014;5(3):308. doi: 10.4331/wjbc.v5.i3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131(1):46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 304.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458(7242):1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 306.Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140(3):519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, et al. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014;24(8):1260–1270. doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mobile DNA. 2015;6(1). [DOI] [PMC free article] [PubMed]
- 309.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464(7290):927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 310.Karimi M, Goyal P, Maksakova Irina A, Bilenky M, Leung D, Tang Jie X, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8(6):676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Maksakova IA, Thompson PJ, Goyal P, Jones SJM, Singh PB, Karimi MM, et al. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin. 2013;6(1):15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc Natl Acad Sci. 2015;112(27):8367–8372. doi: 10.1073/pnas.1422187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, et al. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28(13):1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, et al. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013;11(3):1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 315.Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Castro-Diaz N, Friedli M, Trono D. Drawing a fine line on endogenous retroelement activity. Mobile Genetic Elements. 2015;5(1):1–6. doi: 10.1080/2159256X.2015.1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Puszyk W, Down T, Grimwade D, Chomienne C, Oakey RJ, Solomon E, et al. The epigenetic regulator PLZF represses L1 retrotransposition in germ and progenitor cells. EMBO J. 2013;32(13):1941–1952. doi: 10.1038/emboj.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16(2):157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Xie H, Wang M, Bonaldo MF, Smith C, Rajaram V, Goldman S, et al. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 2009;37(13):4331–4340. doi: 10.1093/nar/gkp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Crowther P, Doherty J, Linsenmeyer M, Williamson M, Woodcock D. Revised genomic consensus for the hypermethylated CpG island region of the human L1 transposon and integration sites of full length L1 elements from recombinant clones made using methylation-tolerant host strains. Nucleic Acids Res. 1991;19(9):2395–2401. doi: 10.1093/nar/19.9.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Thayer RE, Singer MF, Fanning TG. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1 -encoded protein. Gene. 1993;133(2):273–277. doi: 10.1016/0378-1119(93)90651-I. [DOI] [PubMed] [Google Scholar]
- 322.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189(2):227–234. doi: 10.1016/S0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 323.Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272(12):7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 324.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–340. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 325.Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466(7307):769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de Novo methylation. J Biol Chem. 1997;272(35):22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 327.Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7(9):e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zhang Y, Shu J, Si J, Shen L, Estecio MRH, Issa JPJ. Repetitive elements and enforced transcriptional repression co-operate to enhance DNA methylation spreading into a promoter CpG-island. Nucleic Acids Res. 2012;40(15):7257–7268. doi: 10.1093/nar/gks429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Pezic D, Manakov SA, Sachidanandam R, Aravin AA. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 2014;28(13):1410–1428. doi: 10.1101/gad.240895.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Grandi FC, Rosser JM, Newkirk SJ, Yin J, Jiang X, Xing Z, et al. Retrotransposition creates sloping shores: a graded influence of hypomethylated CpG islands on flanking CpG sites. Genome Res. 2015;25(8):1135–1146. doi: 10.1101/gr.185132.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Sanford JP, Clark HJ, Chapman VM, Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987;1(10):1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- 332.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113(1):119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 333.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117(1–2):15–23. doi: 10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 334.Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35(2):88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 335.Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134(14):2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 336.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139(1):15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 337.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48(6):849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, et al. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 2014;28(14):1544–1549. doi: 10.1101/gad.244350.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20(2):116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 340.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22(2):480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Gaudet F, Rideout WM, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24(4):1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16(19):2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 343.Takebayashi S, Tamura T, Matsuoka C, Okano M. Major and essential role for the DNA Methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions. Mol Cell Biol. 2007;27(23):8243–8258. doi: 10.1128/MCB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T et al. Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol. 2015;16(1):115. [DOI] [PMC free article] [PubMed]
- 345.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 346.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28(1):215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq H, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32(17):5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8(12):1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 349.Dunican DS, Cruickshanks HA, Suzuki M, Semple CA, Davey T, Arceci RJ, et al. Lsh regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol. 2013;14:R146. doi: 10.1186/gb-2013-14-12-r146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 351.Sharif J, Muto M, Takebayashi S-i, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 352.Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S et al. MORC1 represses transposable elements in the mouse male germline. Nature Communications. 2014;5:5795. [DOI] [PMC free article] [PubMed]
- 353.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336(6087):1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Fuks F, Hurd P, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31(9):2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Yu F, Zingler N, Schumann GG, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29(21):4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37(4):457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Sampey GC, Guendel I, Das R, Jaworski E, Klase Z, Narayanan A, et al. Transcriptional Gene Silencing (TGS) via the RNAi Machinery in HIV-1 Infections. Biology. 2012;1(3):339–369. doi: 10.3390/biology1020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Aravin AA, Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22(8):970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Sienski G, Batki J, Senti K-A, Dönertas D, Tirian L, Meixner K, et al. Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015;29(21):2258–2271. doi: 10.1101/gad.271908.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25(6):594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ancelin K, Syx L, Borensztein M, Ranisavljevic N, Vassilev I, Briseño-Roa L et al. Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife. 2016:e08851. [DOI] [PMC free article] [PubMed]
- 364.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 365.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 366.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532(7599):329-33. [DOI] [PMC free article] [PubMed]
- 367.Suzuki J, Yamaguchi K, Kajikawa M, Ichiyanagi K, Adachi N, Koyama H, et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5(4):e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair. 2008;7(6):983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357(5):1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Wallace NA, Gasior SL, Faber ZJ, Howie HL, Deininger PL, Galloway DA. HPV 5 and 8 E6 expression reduces ATM protein levels and attenuates LINE-1 retrotransposition. Virology. 2013;443(1):69–79. doi: 10.1016/j.virol.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MCN, Muotri AR, Mu Y, et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci. 2011;108(51):20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Thomas CA, Paquola AC, Muotri AR. LINE-1 retrotransposition in the nervous system. Annu Rev Cell Dev Biol. 2012;28:555–573. doi: 10.1146/annurev-cellbio-101011-155822. [DOI] [PubMed] [Google Scholar]
- 373.Liu H-S, Jan M-S, Chou C-K, Chen P-H, Ke N-J. Is green fluorescent protein toxic to the living cells? Biochem Biophys Res Commun. 1999;260(3):712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 374.Zimmerly S, Guo H, Perlman PS, Lambowitz AM. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82(4):545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 375.Eickbush DG, Luan DD, Eickbush TH. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol Cell Biol. 2000;20(1):213–223. doi: 10.1128/MCB.20.1.213-223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target‐primed reverse transcription in vitro. EMBO J. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc B Biol Sci. 1979;205(1161):489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 378.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4(1):e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.McLaughlin RN, Gable JT, Wittkopp C, Emerman M, Malik HS. Conservation and innovation of APOBEC3A restriction functions during primate evolution. Mol Biol Evol. 2016;33(8):1889-901. [DOI] [PMC free article] [PubMed]
- 380.Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411(2):273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 381.Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 2015;33(5):555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 382.Närvä E, Rahkonen N, Emani MR, Lund R, Pursiheimo J-P, Nästi J, et al. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30(3):452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Iwabuchi KA, Yamakawa T, Sato Y, Ichisaka T, Takahashi K, Okita K, et al. ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSCGeneration, but it is dispensable for the maintenance and induction of pluripotency. PLoS ONE. 2011;6(5):e20461. doi: 10.1371/journal.pone.0020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.McLaughlin RN, Young JM, Yang L, Neme R, Wichman HA, Malik HS. Positive selection and multiple losses of the LINE-1-derived L1TD1 gene in mammals suggest a dual role in genome defense and pluripotency. PLoS Genet. 2014;10(9):e1004531. doi: 10.1371/journal.pgen.1004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Kuhn A, Ong YM, Cheng C-Y, Wong TY, Quake SR, Burkholder WF. Linkage disequilibrium and signatures of positive selection around LINE-1 retrotransposons in the human genome. Proc Natl Acad Sci. 2014;111(22):8131–8136. doi: 10.1073/pnas.1401532111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol. 2000;64:255–294. doi: 10.1016/S0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- 387.Goodier JL, Ostertag EM, Du K, Kazazian HH. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11(10):1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Furano A, Duvernell D, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20(1):9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 389.Sookdeo A, Hepp CM, McClure MA, Boissinot S. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA. 2013;4(1):1. doi: 10.1186/1759-8753-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Strebel K. HIV accessory proteins versus host restriction factors. Current Opinion in Virology. 2013;3(6):692–699. doi: 10.1016/j.coviro.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Collins DR, Collins KL. HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathog. 2014;10(1):e1003851. doi: 10.1371/journal.ppat.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16(6):546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Jones RB, Song H, Xu Y, Garrison KE, Buzdin AA, Anwar N, et al. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J Virol. 2013;87(24):13307–13320. doi: 10.1128/JVI.02257-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Romani B, Cohen ÉA. Lentivirus Vpr and Vpx accessory proteins usurp the cullin4–DDB1 (DCAF1) E3 ubiquitin ligase. Current Opinion in Virology. 2012;2(6):755–763. doi: 10.1016/j.coviro.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Iijima K, Okudaira N, Tamura M, Doi A, Saito Y, Shimura M, et al. Viral protein R of human immunodeficiency virus type-1 induces retrotransposition of long interspersed element-1. Retrovirology. 2013;10(1):83. doi: 10.1186/1742-4690-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Doi A, Iijima K, Kano S, Ishizaka Y. Viral protein R of HIV type-1 induces retrotransposition and upregulates glutamate synthesis by the signal transducer and activator of transcription 1 signaling pathway. Microbiol Immunol. 2015;59(7):398–409. doi: 10.1111/1348-0421.12266. [DOI] [PubMed] [Google Scholar]
- 397.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460(7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, et al. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81(2):306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 399.White TB, McCoy AM, Streva VA, Fenrich J, Deininger PL. A droplet digital PCR detection method for rare L1 insertions in tumors. Mob DNA. 2014;5(1):30. doi: 10.1186/s13100-014-0030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Goodier JL. Retrotransposition in tumors and brains. Mob DNA. 2014;5(1):11. doi: 10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Sciamanna I, Gualtieri A, Cossetti C, Osimo EF, Ferracin M, Macchia G, et al. A tumor-promoting mechanism mediated by retrotransposon-encoded reverse transcriptase is active in human transformed cell lines. Oncotarget. 2013;4(12):2271–2287. doi: 10.18632/oncotarget.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Dhellin O, Maestre J, Heidmann T. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for invivo mRNA reverse transcription. EMBO J. 1997;16(21):6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Shimizu A, Nakatani Y, Nakamura T, Jinno-Oue A, Ishikawa O, Boeke JD, et al. Characterisation of cytoplasmic DNA complementary to non-retroviral RNA viruses in human cells. Sci Rep. 2014;4:5074. doi: 10.1038/srep05074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Fujimoto S, Tsuda T, Toda M, Yamagishi H. Transposon-like sequences in extrachromosomal circular DNA from mouse thymocytes. Proc Natl Acad Sci. 1985;82(7):2072–2076. doi: 10.1073/pnas.82.7.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Jones RS, Potter SS. L1 sequences in HeLa extrachromosomal circular DNA: evidence for circularization by homologous recombination. Proc Natl Acad Sci. 1985;82(7):1989–1993. doi: 10.1073/pnas.82.7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yamagishi H. Role of mammalian circular DNA in cellular differentiation. Bioessays. 1986;4(5):218–221. doi: 10.1002/bies.950040508. [DOI] [PubMed] [Google Scholar]
- 407.Han JS, Shao S. Circular retrotransposition products generated by a LINE retrotransposon. Nucleic Acids Res. 2012;40(21):10866–10877. doi: 10.1093/nar/gks859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14(8):977–985. doi: 10.1038/sj.onc.1200917. [DOI] [PubMed] [Google Scholar]
- 409.Schmidt H, Taubert H, Lange H, Kriese K, Schmitt WD, Hoffmann S, et al. Small polydispersed circular DNA contains strains of mobile genetic elements and occurs more frequently in permanent cell lines of malignant tumors than in normal lymphocytes. Oncol Rep. 2009;22(2):393–400. [PubMed] [Google Scholar]
- 410.Deininger P, Belancio VP. Detection of LINE-1 RNAs by Northern Blot. Methods Mol Biol. 2016;1400:223–236. doi: 10.1007/978-1-4939-3372-3_15. [DOI] [PubMed] [Google Scholar]
- 411.An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, et al. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci. 2006;103(49):18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.van den Hurk JA, Meij IC, del Carmen SM, Kano H, Nikopoulos K, Hoefsloot LH, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16(13):1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 413.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23(11):1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Evrony G, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann Hillel S, et al. Single-Neuron Sequencing Analysis of L1 Retrotransposition and Somatic Mutation in the Human Brain. Cell. 2012;151(3):483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Evrony G, Lee E, Mehta Bhaven K, Benjamini Y, Johnson Robert M, Cai X, et al. Cell Lineage Analysis in Human Brain Using Endogenous Retroelements. Neuron. 2015;85(1):49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J Neurosci. 2013;33(45):17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15(8):497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet. 2014;48:1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- 420.Hazen JL, Faust GG, Rodriguez AR, Ferguson WC, Shumilina S, Clark RA, et al. The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning. Neuron. 2016;89(6):1223–1236. doi: 10.1016/j.neuron.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. eLife. 2016;5:e12966. doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33(8):345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural Mutagenesis of Human Genomes by Endogenous Retrotransposons. Cell. 2010;141(7):1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, et al. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24(7):1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15(17):2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- 427.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O’Shea KS, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16(13):1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 428.Macia A, Muñoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, et al. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol. 2011;31(2):300–316. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 2011;286(42):36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Lee S-H, Rangasamy D. LINE-1 based insertional mutagenesis screens to identify genes involved in embryonic stem cell differentiation. J Stem Cell Res Ther. 2012;2:117.
- 431.Wissing S, Muñoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, et al. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet. 2012;21(1):208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503(7477):525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Díaz N, Rowe HM, et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24(8):1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Macia A, Blanco-Jimenez E, García-Pérez JL. Retrotransposons in pluripotent cells: Impact and new roles in cellular plasticity. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2015;1849(4):417–426. doi: 10.1016/j.bbagrm.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 435.Gerdes P, Richardson SR, Mager DL, Faulkner GJ. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 2016;17(1):1. doi: 10.1186/s13059-016-0965-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Arokium H, Kamata M, Kim S, Kim N, Liang M, Presson AP, et al. Deep sequencing reveals low incidence of endogenous LINE-1 retrotransposition in human induced pluripotent stem cells. PLoS ONE. 2014;9(10):e108682. doi: 10.1371/journal.pone.0108682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Klawitter S, Fuchs NV, Upton KR, Muñoz-Lopez M, Shukla R, Wang J, et al. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun. 2016;7:10286. doi: 10.1038/ncomms10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Shukla R, Upton Kyle R, Muñoz-Lopez M, Gerhardt Daniel J, Fisher Malcolm E, Nguyen T, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153(1):101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Doucet-O’Hare TT, Rodić N, Sharma R, Darbari I, Abril G, Choi JA, et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc Natl Acad Sci. 2015;112(35):E4894–E4900. doi: 10.1073/pnas.1502474112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim M-S, et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015;25(10):1536–1545. doi: 10.1101/gr.196238.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Doucet-O’Hare TT, Sharma R, Rodić N, Anders RA, Burns KH, Kazazian HH. Somatically acquired LINE-1 insertions in normal esophagus undergo clonal expansion in esophageal squamous cell carcinoma. Hum Mutat. 2016. doi:10.1002/humu.23027. [DOI] [PMC free article] [PubMed]
- 442.Ewing AD, Ballinger TJ, Earl D, Harris CC, Ding L, Wilson RK, et al. Retrotransposition of gene transcripts leads to structural variation in mammalian genomes. Genome Biol. 2013;14(3):R22. doi: 10.1186/gb-2013-14-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345(6196):1251343. [DOI] [PMC free article] [PubMed]
- 444.Rodić N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med. 2015;21(9):1060–1064. doi: 10.1038/nm.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22(12):2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Seminars in Cancer Biology. 2010;20(4):200–10. [DOI] [PMC free article] [PubMed]
- 447.Carreira PE, Richardson SR, Faulkner GJ. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 2014;281(1):63–73. doi: 10.1111/febs.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Kemp JR, Longworth MS. Crossing the LINE toward genomic instability: LINE-1 retrotransposition in cancer. Front Chem. 2015;3:68. doi: 10.3389/fchem.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–645. [PubMed] [Google Scholar]
- 450.Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016;26(6):745–755. doi: 10.1101/gr.201814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45(9):977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45(9):970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med. 2015;21(5):274–284. doi: 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 454.Rebhandl S, Huemer M, Greil R, Geisberger R. AID/APOBEC deaminases and cancer. Oncoscience. 2015;2:320. doi: 10.18632/oncoscience.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Asch H, Eliacin E, Fanning T, Connolly J, Bratthauer G, Asch B. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res. 1996;8(6):239. [PubMed] [Google Scholar]
- 456.Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, et al. Association of nuclear localization of a Long Interspersed Nuclear Element-1 Protein in breast tumors with poor prognostic outcomes. Genes Cancer. 2010;1(2):115–124. doi: 10.1177/1947601909360812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Chen L, Dahlstrom JE, Chandra A, Board P, Rangasamy D. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res Treat. 2012;136(1):129–142. doi: 10.1007/s10549-012-2246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Gualtieri A, Andreola F, Sciamanna I, Sinibaldi Vallebona P, Serafino A, Spadafora C. Increased expression and copy number amplification of LINE-1 and SINE B1 retrotransposable elements in murine mammary carcinoma progression. Oncotarget. 2013;4((11):1882–1893. doi: 10.18632/oncotarget.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176(1–2):39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- 460.Kitkumthorn N, Mutirangura A. Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clinical Epigenetics. 2011;2(2):315–330. doi: 10.1007/s13148-011-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotransposition-Competent Human LINE-1 Induces Apoptosis in Cancer Cells With Intact p53. J Biomed Biotechnol. 2004;2004(4):185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Farkash EA, Prak ETL. DNA damage and L1 retrotransposition. J Biomed Biotechnol. 2006;2006:1–8. doi: 10.1155/JBB/2006/37285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Malki S, van der Heijden GW, O’Donnell Kathryn A, Martin Sandra L, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell. 2014;29(5):521–533. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Chuma S. LINE-1 of evidence for fetal oocyte attrition by retrotransposon. Dev Cell. 2014;29(5):501–502. doi: 10.1016/j.devcel.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 465.St. Laurent G, Hammell N, McCaffrey TA. A LINE-1 component to human aging: Do LINE elements exact a longevity cost for evolutionary advantage? Mech Ageing Dev. 2010;131(5):299–305. doi: 10.1016/j.mad.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Front Genet. 2013;4. [DOI] [PMC free article] [PubMed]
- 467.Sedivy JM, Kreiling JA, Neretti N, Cecco MD, Criscione SW, Hofmann JW, et al. Death by transposition - the enemy within? BioEssays. 2013;35(12):1035–1043. doi: 10.1002/bies.201300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Belancio VP, Blask DE, Deininger P, Hill SM, Jazwinski SM. The aging clock and circadian control of metabolism and genome stability. Front Genet. 2015;5. [DOI] [PMC free article] [PubMed]
- 469.Maxwell PH. What might retrotransposons teach us about aging? Curr Genet. 2015. [DOI] [PMC free article] [PubMed]
- 470.Li W, Prazak L, Chatterjee N, Grüninger S, Krug L, Theodorou D, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16(5):529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12(2):247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 2013;5(12):867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K (HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9(1):354. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Perron H, Hamdani N, Faucard R, Lajnef M, Jamain S, Daban-Huard C, et al. Molecular characteristics of Human Endogenous Retrovirus type-W in schizophrenia and bipolar disorder. Transl Psychiatry. 2012;2(12):e201. doi: 10.1038/tp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K (HML-2) loci in melanoma. Genome Biol Evol. 2013;5(2):307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Schmitt K, Richter C, Backes C, Meese E, Ruprecht K, Mayer J. Comprehensive analysis of human endogenous retrovirus group HERV-W locus transcription in multiple sclerosis brain lesions by high-throughput amplicon sequencing. J Virol. 2013;87(24):13837–13852. doi: 10.1128/JVI.02388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Douville RN, Nath A. Human endogenous retroviruses and the nervous system. Handb Clin Neurol. 2014;123:465–485. doi: 10.1016/B978-0-444-53488-0.00022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Perl A, Fernandez D, Telarico T, Phillips PE. Endogenous retroviral pathogenesis in lupus. Curr Opin Rheumatol. 2010;22(5):483–492. doi: 10.1097/BOR.0b013e32833c6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Lee Y-J, Jeong B-H, Choi E-K, Kim Y-S. Involvement of endogenous retroviruses in prion diseases. Pathogens. 2013;2(3):533–543. doi: 10.3390/pathogens2030533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Katoh I, Kurata S. Association of endogenous retroviruses and long terminal repeats with human disorders. Front Oncol. 2013;3:234. [DOI] [PMC free article] [PubMed]
- 482.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol. 2014;15(5):415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Kassiotis G. Endogenous retroviruses and the development of cancer. J Immunol. 2014;192(4):1343–1349. doi: 10.4049/jimmunol.1302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Suntsova M, Garazha A, Ivanova A, Kaminsky D, Zhavoronkov A, Buzdin A. Molecular functions of human endogenous retroviruses in health and disease. Cell Mol Life Sci. 2015;72(19):3653–3675. doi: 10.1007/s00018-015-1947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Li W, Lee MH, Henderson L, Tyagi R, Bachani M, Steiner J, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7(307):307ra153–307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Antony JM, DesLauriers AM, Bhat RK, Ellestad KK, Power C. Human endogenous retroviruses and multiple sclerosis: Innocent bystanders or disease determinants? Biochim Biophys Acta. 2011;1812(2):162–176. doi: 10.1016/j.bbadis.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Kiesel P, Gibson TJ, Ciesielczyk B, Bodemer M, Kaup F-J, Bodemer W, et al. Transcription of Alu DNA elements in blood cells of sporadic Creutzfeldt-Jakob disease (sCJD) Prion. 2010;4(2):87–93. doi: 10.4161/pri.4.2.11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Tarallo V, Hirano Y, Gelfand Bradley D, Dridi S, Kerur N, Kim Y, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149(4):847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Hung T, Pratt GA, Sundararaman B, Townsend MJ, Chaivorapol C, Bhangale T, et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350(6259):455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Yu Q, Carbone CJ, Katlinskaya YV, Zheng H, Zheng K, Luo M, et al. Type I Interferon controls propagation of Long Interspersed Element-1. J Biol Chem. 2015;290(16):10191–10199. doi: 10.1074/jbc.M114.612374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet. 2015;167(2):296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Crow YJ, Manel N. Aicardi–Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15(7):429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 494.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee Y-A, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39(9):1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 495.Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutières syndrome. Am J Hum Genet. 2007;80(4):811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. 2009;18(R2):R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3'- > 5' DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24(15):6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Ahn J, Ruiz P, Barber GN. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol. 2014;193(9):4634–4642. doi: 10.4049/jimmunol.1401337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Beck-Engeser GB, Eilat D, Wabl M. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology. 2011;8(1):91. doi: 10.1186/1742-4690-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78(4):671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Sciamanna I, Landriscina M, Pittoggi C, Quirino M, Mearelli C, Beraldi R, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24(24):3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 502.Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69(1):141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Patnala R, Lee S-H, Dahlstrom JE, Ohms S, Chen L, Dheen ST, et al. Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast Cancer Res Treat. 2014;143(2):239–253. doi: 10.1007/s10549-013-2812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Sciamanna I, Gualtieri A, Piazza PV, Spadafora C. Regulatory roles of LINE-1-encoded reverse transcriptase in cancer onset and progression. Oncotarget. 2014;5(18):8039–8051. doi: 10.18632/oncotarget.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Rangasamy D, Lenka N, Ohms S, Dahlstrom J, Blackburn A, Board P. Activation of LINE-1 Retrotransposon Increases the Risk of Epithelial-Mesenchymal Transition and Metastasis in Epithelial Cancer. Curr Mol Med. 2015;15(7):588–597. doi: 10.2174/1566524015666150831130827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Ishizaka Y, Okudaira N, Tamura M, Iijima K, Shimura M, Goto M, et al. Modes of retrotransposition of long interspersed element-1 by environmental factors. Front Microbiol. 2012;3:191. doi: 10.3389/fmicb.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Mourier T, Nielsen LP, Hansen AJ, Willerslev E. Transposable elements in cancer as a by-product of stress-induced evolvability. Front Genet. 2014;5:156. eCollection 2014. [DOI] [PMC free article] [PubMed]
- 508.Okudaira N, Ishizaka Y, Nishio H. Retrotransposition of long interspersed element 1 induced by methamphetamine or cocaine. J Biol Chem. 2014;289(37):25476–25485. doi: 10.1074/jbc.M114.559419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Hunter RG, Gagnidze K, McEwen BS, Pfaff DW. Stress and the dynamic genome: Steroids, epigenetics, and the transposome. Proc Natl Acad Sci. 2015;112(22):6828–6833. doi: 10.1073/pnas.1411260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Miousse IR, Chalbot M-CG, Lumen A, Ferguson A, Kavouras IG, Koturbash I. Response of transposable elements to environmental stressors. Mutat Res/Rev Mutat Res. 2015;765:19–39. doi: 10.1016/j.mrrev.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Morales ME, Servant G, Ade C, Roy-Engel AM. Altering genomic integrity: heavy metal exposure promotes transposable element-mediated damage. Biol Trace Elem Res. 2015;166(1):24–33. doi: 10.1007/s12011-015-0298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Moszczynska A, Flack A, Qiu P, Muotri AR, Killinger BA. Neurotoxic Methamphetamine Doses Increase LINE-1 Expression in the Neurogenic Zones of the Adult Rat Brain. Sci Rep. 2015;5:14356. [DOI] [PMC free article] [PubMed]
- 513.Behrendt R, Roers A. Mouse models for Aicardi–Goutières syndrome provide clues to the molecular pathogenesis of systemic autoimmunity. Clin Experiment Immunol. 2014;175(1):9–16. doi: 10.1111/cei.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Blankson JN, Siliciano JD, Siliciano RF. Finding a cure for human immunodeficiency virus-1 infection. Infect Dis Clin N Am. 2014;28(4):633–650. doi: 10.1016/j.idc.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J Biol Chem. 2004;279(42):43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 516.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, et al. APOBEC3A is a potent Inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 518.Muckenfuss H, Hamdorf M, Held U, Perković M, Lower J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 519.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390(1):199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Tan L, Sarkis PT, Wang T, Tian C, Yu X-F. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23(1):279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Liang W, Xu J, Yuan W, Song X, Zhang J, Wei W, Yu XF, Yang Y. APOBEC3DE inhibits LINE-1 retrotransposition by interacting with ORF1p and influencing LINE reverse transcriptase activity. PLoS ONE. 2016;11(7):e0157220. [DOI] [PMC free article] [PubMed]


