Abstract
In many viruses molecular motors forcibly pack single DNA molecules to near-crystalline density into ~50–100 nm prohead shells1, 2. Unexpectedly, we found that packaging frequently stalls in conditions that induce net attractive DNA-DNA interactions3. Here, we present findings suggesting that this stalling occurs because the DNA undergoes a nonequilibrium jamming transition analogous to that observed in many soft-matter systems, such as colloidal and granular systems4–8. Experiments in which conditions are changed during packaging to switch DNA-DNA interactions between purely repulsive and net attractive reveal strongly history-dependent dynamics. An abrupt deceleration is usually observed before stalling, indicating that a transition in DNA conformation causes an abrupt increase in resistance. Our findings suggest that the concept of jamming can be extended to a single polymer molecule. However, compared with macroscopic samples of colloidal particles5 we find that single DNA molecules jam over a much larger range of densities. We attribute this difference to the nanoscale system size, consistent with theoretical predictions for jamming of attractive athermal particles.9, 10
ATP-powered motors package DNA into viral proheads via a portal nanochannel, overcoming large forces resisting DNA confinement arising from DNA bending rigidity, electrostatic self-repulsion, and entropy loss1, 11–14. In addition to being of biological interest, viral packaging is an experimentally accessible model for investigating effects of spatial confinement on polymer dynamics, a topic of fundamental interest in polymer physics15–19. Although the physics of DNA packaging has been modeled theoretically using a wide variety of analytic and simulation methods11–14, a full understanding has remained elusive.
In aqueous solutions containing monovalent and divalent salt ions DNA-DNA self-interactions are purely repulsive20, 21. We obtain this condition here with a standard packaging buffer containing 25 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, and 0.5 mM ATP. Addition of +3 or +4 ions induces attractive DNA-DNA interactions and above a critical ion concentration, the interaction becomes net attractive, which causes condensation of DNA in solution into a densely-packed form20, 21. We obtain this condition here by adding 5 mM spermidine or 20 mM spermine to the standard packaging buffer. With 100% of the wildtype genome length packaged, X-ray scattering measurements on phage lambda virus, which packages to similar density as the phage phi29 we study here, show that +3 or +4 ions do not induce a net attractive DNA-DNA interaction because the strands are confined to such a small spacing that they are in a repulsive portion of the distance-dependent interaction potential22. However at lower packing density, e.g. with 78% of the wildtype genome length packaged, the X-ray measurements show that a net attractive interaction is induced22. A net attractive interaction is induced in our studies when we add polyamine ions because we are in this regime of lower packing density.
Addition of a low concentration of +3 ions, below the threshold for DNA condensation, speeds up packaging in phage phi29 viruses23 and increases the yield of lambda phages in an in vitro assembly assay24, consistent with packaging being facilitated by a reduction in strength of the net repulsive DNA-DNA interaction due to increased ionic screening. However with a larger concentration of +3 ions, when the DNA-DNA interaction becomes net attractive, we found that frequent stalling of DNA packaging occurs3. This finding was unexpected because theoretical models predicted that forces resisting DNA confinement would be greatly reduced with a net attractive interaction11–13, 25, 26. We present evidence here suggesting that this stalling occurs because the DNA undergoes a nonequilibrium jamming transition.
Many soft matter systems, including colloids, granular materials, suspensions, clays, pastes, and foams exhibit nonequilibrium transitions from a fluid-like to solid-like state4–8. The concept of a jamming transition was proposed to be a universal mechanism for such transitions based on the formation of a disordered stress-bearing network of geometrically-constrained particles4, 8, 27. Characteristic features of this transition are that it occurs above a critical packing density and/or below a critical load force, and is promoted by attractive interactions5. Unjamming can be induced by application of a force that exceeds the “yield force” of the jammed material. Another characteristic feature is that these systems exhibit history-dependent dynamics or “memory effects”4, 6, 28.
The concept of jamming has been shown to be applicable to the packing of macroscopic "granular polymer chains", governed by jamming of semi-rigid loops29. Here, we provide evidence that jamming can also occur in a submicroscopic polymer chain with increasing density under confinement and that net attractive self- interactions promote this. We also present data suggesting that an unjamming transition can be induced by force or by changing the DNA-DNA interaction from net attractive to purely repulsive.
We measure packaging of single DNA molecules into single bacteriophage phi29 proheads using optical tweezers (Fig. 1a)2, 30–32. In the purely repulsive DNA-DNA interaction condition the 6.6 µm genome length of DNA is efficiently packaged into the ~50 nm diameter prohead in ~5 minutes, reaching a final density of ~0.5 g/ml. However, in the net attractive interaction condition, 75% ± 2.5% (mean ± standard error; n = 293) of complexes stopped abruptly before packaging 80% of the genome length, an effect we refer to as "stalling"3. In the purely repulsive condition stalling was only observed in 8% ± 1.5% (n = 343) of complexes. Our interpretation is that stalling occurs because the packaged DNA undergoes a jamming transition and translocation halts because the yield force of the jammed DNA exceeds the maximum force the motor can exert. This finding is consistent with theoretical predictions and experiments on colloid systems showing that attractive interactions promote jamming5, 10.
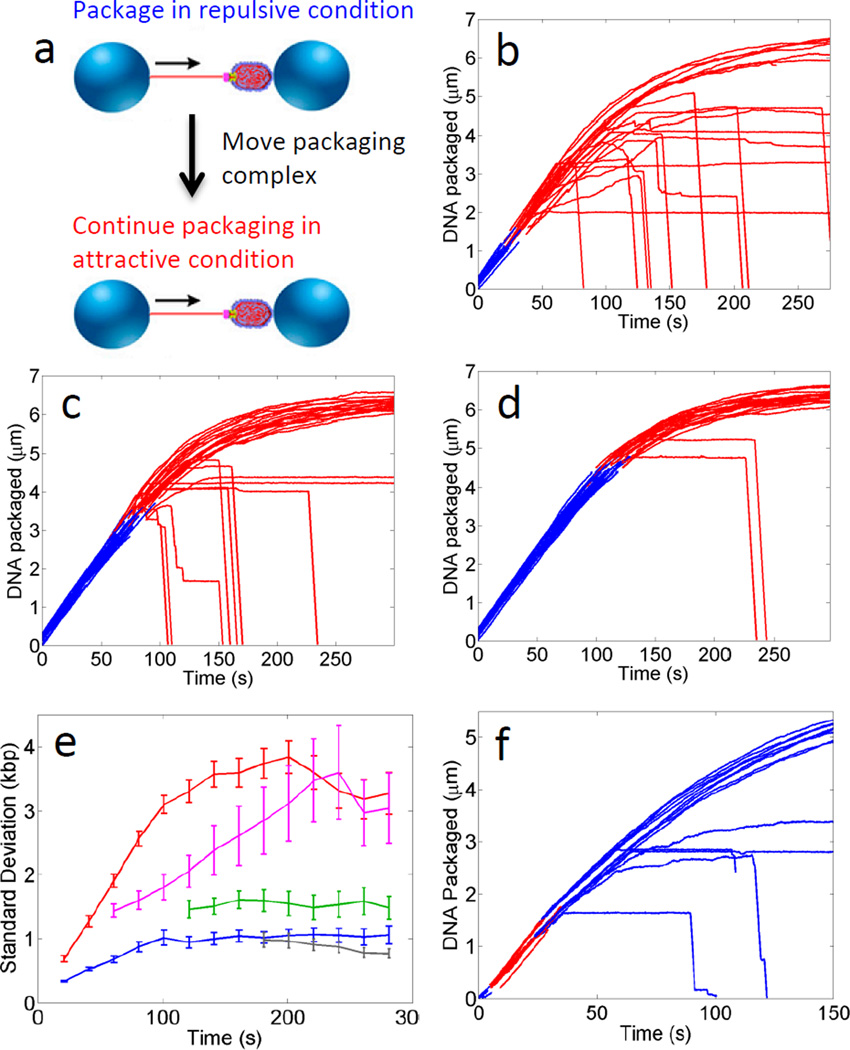
Fig. 1. Viral DNA packaging exhibits history-dependent dynamics.
(A) Prohead-motor complexes are attached to one trapped microsphere and DNA is attached to a second trapped microsphere. Packaging is initiated by bringing the two microspheres into proximity, whereupon the motor translocates the DNA. (B-D) Typical measurements of complexes that were initiated in the repulsive DNA-DNA interaction condition (blue lines) and allowed to proceed to ~20% (B), ~48% (C), or ~66% filling (D) prior to moving the complexes to the net attractive condition (red lines). (E) Standard deviation in length of DNA packaged vs. time for complexes measured in the repulsive condition (blue), net attractive condition (red), and after packaging ~20% (magenta), ~48% (green), or ~66% (grey) in the repulsive condition prior to moving them to the net attractive condition. Error bars indicate standard errors in the means calculated by applying the bootstrap method to the ensemble of datasets recorded on different complexes. (F) Typical measurements of complexes that were exposed to the net attractive condition (red) at low filling and subsequently moved to the repulsive condition (blue).
To investigate whether viral packaging exhibits history-dependent dynamics, a signature observed in many other systems that undergo jamming4, 6, 28, we conducted experiments in which the solution was changed during packaging to switch the DNA-DNA interactions between purely repulsive and net attractive. First, we initiated packaging in the purely repulsive condition and proceeded to various filling levels (fraction of genome length packaged). The complex was then moved rapidly, within ~1 s, into the net attractive condition. We find that the fraction of complexes that stall per unit time after switching the condition decreases with increasing DNA length pre-packaged in the repulsive condition (Fig. 1b–d). Specifically, 41% ± 3% (n=210) of complexes stalled within one minute after switching to the net attractive condition at 20% (standard deviation, 6%) filling, versus 28% ± 5% (n=89) of complexes after switching at 49% (standard deviation, 9%), versus 8 ± 3% (n=113) of complexes after switching at 66% (standard deviation, 6%) filling. This effect results in a decrease in the heterogeneity in length DNA packaged vs. time with increasing length pre-packaged under repulsive interactions (Fig. 1e). Thus, the dynamics are indeed history-dependent. Our interpretation is that the repulsive condition promotes formation of more favorable (lower energy) packed DNA conformations that persist and influence the dynamics of rearrangement of the packaged DNA after switching to the net attractive condition. History dependence was also observed in the motor velocity at some filling levels—compared with that measured after switching at 20% filling (standard deviation, 6%), the average velocity was ~6% higher over the range from 40–50% filling after switching in this range, and ~16% higher over the range from 60–70% filling after switching in this range (Supplementary Fig. 1).
We also conducted experiments in which complexes were briefly exposed to net attractive conditions at low filling (for ~20 s at filling levels up to 26%; standard deviation, 8%) then moved to the repulsive condition (Fig. 1f). Whereas no stalling was induced during the brief exposure to net attractive conditions, 39% ± 7.6% (n = 41) of complexes stalled after a return to repulsive conditions compared with only 8% ± 1.5% (n = 343) packaged with continuously repulsive interactions. This again shows that the DNA translocation dynamics are strongly history dependent and suggests that unfavorable DNA conformations formed during early stages of packaging can influence the subsequent dynamics and promote stalling.
We next analyzed in further detail the dynamics of DNA translocation in the continuously net attractive condition. Strikingly, we find that stalling is usually preceded by an abrupt and large deceleration event (Fig. 2a&b). Examples of these events are shown in Supplementary Fig. 2. 77% ± 2.5% (n = 293) of complexes exhibited maximum decelerations greater than three standard deviations below the average at the same filling level in the repulsive condition (Fig. 2c). The sudden decreases in motor velocity indicate that the DNA has undergone an abrupt transition in conformation that causes a large increase in resistance. One possible interpretation is that, at this point, the DNA undergoes a jamming transition but that the motor can generate a force that exceeds the yield force of the DNA and rapidly induces an unjamming transition, allowing translocation to continue. Given the time resolution of our measurement (~1 s) we would not expect to observe arrest because the high rate of translocation (~50 nm/s) would rapidly increase the applied force and induce unjamming. Within this interpretation, our results would imply that the yield force of individual jammed complexes is highly variable—in some it is high enough to cause irreversible stalling while in others it is lower and the motor can induce unjamming.
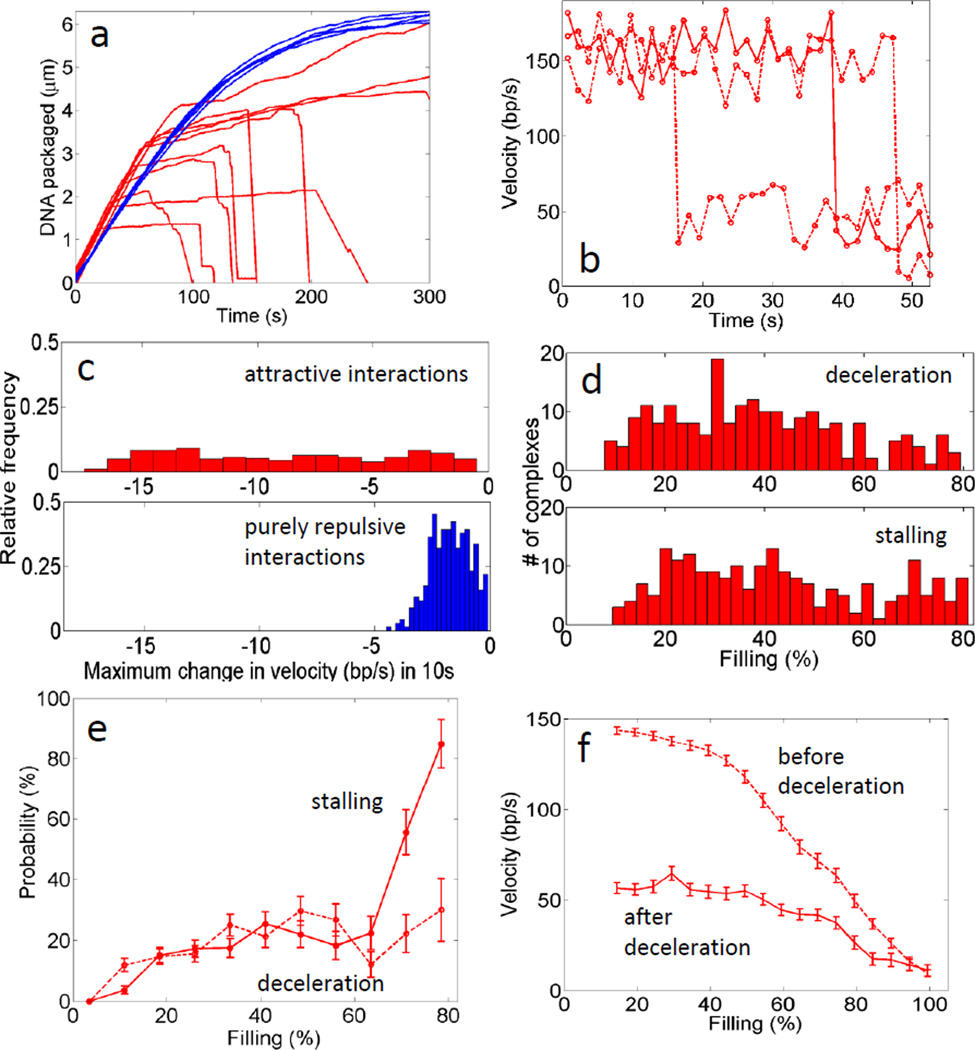
Fig. 2. Deceleration and stalling events.
(a) Typical measurements showing abrupt decelerations and stalling of DNA translocation measured in the continuously net attractive condition (red), not observed in the continuously repulsive condition (blue). (b) Examples of velocity changes calculated in a 1 s window during deceleration events. (c) Maximum decelerations before 80% filling calculated in a 10 s window for the repulsive (bottom/blue) and net attractive condition (top/red). (d) Histogram of filling levels at which complexes exhibited deceleration events (top) and stalling events (bottom) when packaging in the continuously net attractive condition. (e) Probability, p, of a deceleration event vs. filling (dashed line) and probability of a stalling event vs. filling (solid line), calculated as the number of complexes that exhibit an event in each filling range divided by the number, N, that package to or through that range before stalling. Error bars indicate standard errors in the means, calculated as the standard deviation of the binomial distribution . (f) Mean motor velocity vs. filling for all sections of packaging before a deceleration event (including complexes that did not exhibit a deceleration event) (dashed line). Mean velocity vs. filling for all sections of packaging after a deceleration event (solid line). Error bars indicate standard errors in the means, computed as standard deviation divided by square root of the number of complexes.
Notably, both deceleration and stalling events during packaging in the net attractive condition occur over a wide range of filling levels (Fig. 2d), suggesting that the density at which jamming occurs for individual complexes is also highly variable. However, for the ensemble of complexes the probability of both deceleration and stalling events increases with prohead filling (Fig. 2e), as expected for jamming. 88% ± 2.9% (n = 226) of the complexes exhibiting deceleration events subsequently stalled. Our interpretation is that although the DNA can be un-jammed by motor force its conformation is such that it is predisposed to jam again, ultimately stalling the motor irreversibly. This finding demonstrates another form of history-dependent behavior in which the translocation dynamics of a complex depends on its past translocation dynamics. After a deceleration event the packaging rate also remains low and does not increase back to the original value. The average motor velocity exhibited by complexes following a deceleration event is lower at every filling level compared with that of complexes that did not undergo or had not yet undergone a deceleration event (Fig. 2f). It is therefore possible that the deceleration does not represent a jamming (fluid-solid) transition followed by unjamming, but rather a transition from a fluid-like state to another more viscous fluid-like state which presents higher resistance and slows the motor.
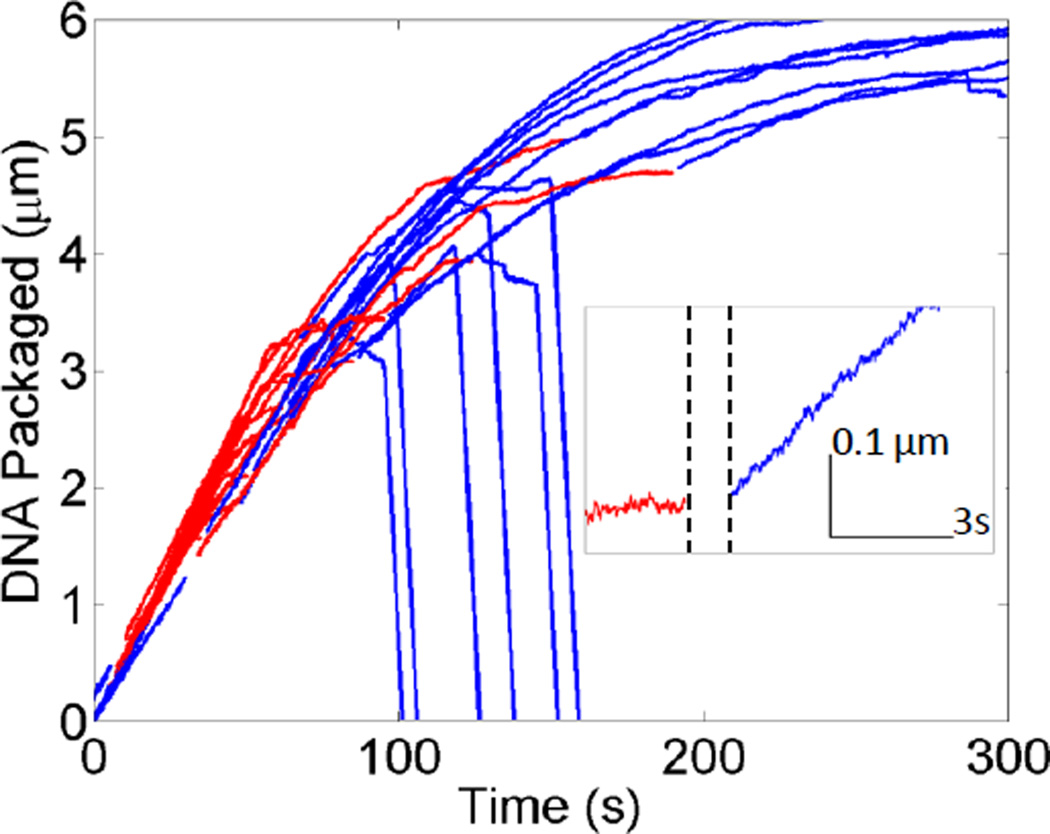
Behavior more clearly like an unjamming (solid-to-fluid) transition was observed in a different experiment in which complexes that stalled in the net attractive condition were switched into the purely repulsive condition (Fig. 3). Restarting of DNA translocation was observed in 49% ± 5.1% (n = 96) of trials, consistent with the expectation that unjamming can be induced at constant density and constant force by reducing the strength of the attractive interactions4, 5. On the other hand, roughly half of complexes did not restart, suggesting that the DNA may be trapped in an unfavorable conformation, formed in the net attractive condition, which cannot quickly relax to allow packaging to resume on the timescale of the measurement. A long relaxation time is plausible given that our previous measurements revealed DNA relaxation times as long as 10 minutes even in the purely repulsive condition33.
Fig. 3. Rescue of stalled complexes.
Typical measurements in which complexes that stalled in the net attractive condition (red) were rapidly moved back to the repulsive condition (blue) causing translocation to restart. Inset: Zoomed-in plot showing one example where a stalled complex suddenly restarted (dashed lines indicate period when the complex was moved).
Almost all of the complexes which stall then later exhibit slipping of the DNA back out of the prohead. In only 2% of packaging events in the attractive condition did data recording end before a slip was observed. The average time between stalling and slipping was similar in the continuously net attractive condition (20 ± 2 sec) and in the experiments where the condition was switched after stalling from net attractive to repulsive (17 ± 4 sec). It has been shown that proheads can be perforated by freezing and thawing such that the DNA leaks out of the prohead as it is translocated and there is no buildup of force resisting DNA confinement34. In the net attractive condition we find that only 2% ± 2% of complexes with perforated proheads slip after 1 minute compared with 21% ± 2% for regular proheads. We therefore attribute the frequent slipping after stalling observed with regular proheads not to weakened DNA grip, but to high resistance force presented by jammed DNA that causes failure of the motor-DNA grip.
The single DNA molecule jamming we provide evidence for exhibits a striking difference from jamming measured in macroscopic samples of colloidal particles in that the transition in the latter case occurs over a much smaller range of packing densities. In Ref. 5 a sharp fluid-to-solid transition was measured for carbon black particles as the volume fraction was increased from ~0.05 to 0.053, whereas the individual complexes in our studies stalled at DNA volume fractions ranging from ~0.05 to 0.4. This difference is likely attributable to the much smaller size of our system. Theoretical studies of disordered systems of athermal particles predict that the range of densities over which jamming occurs increases with decreasing system size (number of particles) and that attractive interactions cause jamming at lower densities 9, 10. The colloidal samples studied in Ref. 5 contained >109 particles (volumes large enough to be measured in a rheometer), whereas the viral DNA can be modeled as a polymer with only ~100 persistence lengths (statistically uncorrelated segments in terms of tangent vector directions in free solution)35.
Interestingly, evidence has been presented that DNA packaged in mature lambda phages (100% wildtype genome length) undergoes a solid-to-fluid like transition at increased temperature36. The transition was reported to occur in a narrow temperature range of a few °C in conditions with net repulsive DNA-DNA interactions. Notably, in our measurements we observe much less heterogeneity in the packaging dynamics in the repulsive condition than in the net attractive condition (Fig. 1e). Some of the evidence for the temperature-dependent transition in Ref. 36 was from bulk X-ray scattering and calorimetry measurements, providing statistical averages, which may not reveal heterogeneity in individual complexes. However, spring constants of individual viruses were also measured by atomic force microscopy and there appears to be higher variability in these at a temperature slightly below the transition point36. In another potentially related study, attractive DNA-DNA interactions were found to partly suppress ejection of DNA from phage lambda and cause larger variation in lengths of DNA remaining unejected than when ejection was suppressed by osmotic pressure in purely repulsive conditions37. The conclusion of this study that attractive interactions promotes the formation of nonequilibrium DNA conformations that effect DNA ejection is consistent with our conclusion that attractive interactions promotes formation of nonequilibrium conformations that affect the packaging dynamics.
Our findings suggest that at a particular filling level some viral proheads contain jammed DNA arrangements while others do not. At the ensemble level, the probability of jamming increases only gradually with packing density, not sharply as in macroscopic samples. Such behavior is generally consistent with the theoretical prediction that nanoscale systems should exhibit less-sharp phase transitions than macroscopic systems38.
Supplementary Material
Acknowledgments
We thank Damian delToro for technical assistance. This work was supported by NSF Grants PHY-0848905 and MCB-1158328 and NIH Grant R01-GM088186.
Footnotes
Contributions
D.E.S. and N.K. conceived the research. P.J.J. and S.G. prepared the phi29 proheads and motor protein. N.K. conducted the measurements. N.K. and D.E.S. analyzed the data. D.E.S. and N.K. wrote the manuscript, which was discussed by and edited by all authors.
References
- 1.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nature Reviews Microbiology. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 2.Smith DE. Single-molecule studies of viral DNA packaging. Curr. Opin. Virol. 2011;1:134–141. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller N, Grimes S, Jardine PJ, Smith DE. Repulsive DNA-DNA interactions accelerate viral DNA packaging in phage phi29. Phys. Rev. Lett. 2014;112:248101. doi: 10.1103/PhysRevLett.112.248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu AJ, Nagel SR, editors. Jamming and Rheology: Constrained Dynamics on Microscopic and Macroscopic Scales. London, England: Taylor & Francis; 2001. [Google Scholar]
- 5.Trappe V, Prasad V, Cipelletti L, Segre P, Weitz D. Jamming phase diagram for attractive particles. Nature. 2001;411:772–775. doi: 10.1038/35081021. [DOI] [PubMed] [Google Scholar]
- 6.Cipelletti L, Weeks ER. Glassy dynamics and dynamical heterogeneity in colloids. In: Berthier L, Biroli G, Bouchaud JP, Cipelletti L, van Saarloos W, editors. Dynamical Heterogeneities in Glasses, Colloids, and Granular Media. Oxford, England: Oxford University Press; 2011. pp. 110–143. Ch. 4. [Google Scholar]
- 7.Dauchot O, Durian DJ, van Hecke M. Dynamical heterogeneities in grains and foams. In: Berthier L, Biroli G, Bouchaud JP, Cipelletti L, van Saarloos W, editors. Dynamical Heterogeneities in Glasses, Colloids, and Granular Media. Oxford, England: Oxford University Press; 2011. pp. 203–224. Ch. 6. [Google Scholar]
- 8.Jaeger HM. Celebrating Soft Matter’s 10th Anniversary: Toward jamming by design. Soft matter. 2015;11:12–27. doi: 10.1039/c4sm01923g. [DOI] [PubMed] [Google Scholar]
- 9.O'Hern CS, Langer SA, Liu AJ, Nagel SR. Random packings of frictionless particles. Phys. Rev. Lett. 2002;88:075507. doi: 10.1103/PhysRevLett.88.075507. [DOI] [PubMed] [Google Scholar]
- 10.Head D. Well defined transition to gel–like aggregates of attractive athermal particles. The European Physical Journal E. 2007;22:151–155. doi: 10.1140/epje/e2007-00022-1. [DOI] [PubMed] [Google Scholar]
- 11.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purohit PK, et al. Forces during bacteriophage DNA packaging and ejection. Biophys. J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys. J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey SC, Petrov AS, Devkota B, Boz MB. Viral assembly: a molecular modeling perspective. Phys Chem Chem Phys. 2009;11:10553–10564. doi: 10.1039/b912884k. [DOI] [PubMed] [Google Scholar]
- 15.De Gennes PG. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell University Press; 1979. [Google Scholar]
- 16.Ali I, Marenduzzo D, Yeomans JM. Polymer packaging and ejection in viral capsids: Shape matters. Phys. Rev. Lett. 2006;96:208102. doi: 10.1103/PhysRevLett.96.208102. [DOI] [PubMed] [Google Scholar]
- 17.Sakaue T. Semiflexible polymer confined in closed spaces. Macromolecules. 2007;40:5206–5211. [Google Scholar]
- 18.Reisner W, Pedersen JN, Austin RH. DNA confinement in nanochannels: physics and biological applications. Rep Prog Phys. 2012;75:106601. doi: 10.1088/0034-4885/75/10/106601. [DOI] [PubMed] [Google Scholar]
- 19.Balducci A, Hsieh C, Doyle P. Relaxation of stretched DNA in slitlike confinement. Phys. Rev. Lett. 2007;99:238102. doi: 10.1103/PhysRevLett.99.238102. [DOI] [PubMed] [Google Scholar]
- 20.Rau DC, Parsegian VA. Direct Measurement of the Intermolecular Forces between Counterion-Condensed DNA Double Helices - Evidence for Long-Range Attractive Hydration Forces. Biophys. J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 22.Qiu X, et al. Salt-dependent DNA-DNA spacings in intact bacteriophage λ reflect relative importance of DNA self-repulsion and bending energies. Phys. Rev. Lett. 2011;106:028102. doi: 10.1103/PhysRevLett.106.028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller DN, et al. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nurmemmedov E, Castelnovo M, Medina E, Catalano CE, Evilevitch A. Challenging packaging limits and infectivity of phage λ. J. Mol. Biol. 2012;415:263–273. doi: 10.1016/j.jmb.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Comolli LR, et al. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 26.Petrov AS, Harvey SC. Role of DNA–DNA interactions on the structure and thermodynamics of bacteriophages Lambda and P4. J. Struct. Biol. 2011;174:137–146. doi: 10.1016/j.jsb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majmudar TS, Behringer RP. Contact force measurements and stress-induced anisotropy in granular materials. Nature. 2005;435:1079–1082. doi: 10.1038/nature03805. [DOI] [PubMed] [Google Scholar]
- 28.Josserand C, Tkachenko AV, Mueth DM, Jaeger HM. Memory effects in granular materials. Phys. Rev. Lett. 2000;85:3632–3635. doi: 10.1103/PhysRevLett.85.3632. [DOI] [PubMed] [Google Scholar]
- 29.Zou LN, Cheng X, Rivers ML, Jaeger HM, Nagel SR. The packing of granular polymer chains. Science. 2009;326:408–410. doi: 10.1126/science.1177114. [DOI] [PubMed] [Google Scholar]
- 30.Smith DE, et al. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 31.Chemla YR, Smith DE. Single Molecule Studies of Viral DNA Packaging. In: Rao V, Rossmann MG, editors. Viral Molecular Machines. New York, NY USA: Springer; 2012. pp. 549–584. [Google Scholar]
- 32.Hetherington CL, Moffitt JR, Jardine PJ, Bustamante C. Viral DNA Packaging Motors. In: Goldman YE, Ostap EM, editors. Comprehensive Biophysics. Vol. 4. Amsterdam, Netherlands: Elsevier B.V.; 2012. pp. 420–446. [Google Scholar]
- 33.Berndsen ZT, Keller N, Grimes S, Jardine PJ, Smith DE. Nonequilibrium dynamics and ultraslow relaxation of confined DNA during viral packaging. Proc Natl Acad Sci USA. 2014;111:8345–8350. doi: 10.1073/pnas.1405109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, et al. A Viral Packaging Motor Varies Its DNA Rotation and Step Size to Preserve Subunit Coordination as the Capsid Fills. Cell. 2014;157:702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bustamante C, Smith SB, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu T, et al. Solid-to-fluid-like DNA transition in viruses facilitates infection. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1321637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin Y, Knobler CM, Gelbart WM. Controlling the extent of viral genome release by a combination of osmotic stress and polyvalent cations. Physical Review E. 2015;92:022708. doi: 10.1103/PhysRevE.92.022708. [DOI] [PubMed] [Google Scholar]
- 38.Hill TL. Thermodynamics of small systems. Mineola, NY, USA: Dover Publications; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.