Abstract
Hydrochlorothiazide (HCTZ) is among the most commonly prescribed antihypertensives, yet <50% of HCTZ treated patients achieve blood pressure (BP) control. Herein, we integrated metabolomic and genomic profiles of HCTZ treated patients to identify novel genetic markers associated with HCTZ BP response. The primary analysis included 228 white hypertensives treated with HCTZ from the Pharmacogenomic Evaluation of Antihypertensive Response study. Genome-wide analysis was conducted using Illumina Omni 1M-Quad Chip, and untargeted metabolomics was performed on baseline fasting plasma samples using a gas chromatography-time-of-flight mass spectrometry platform. We found thirteen metabolites significantly associated with HCTZ systolic BP (SBP) and diastolic BP (DBP) responses (FDR<0.05). Additionally, integrating genomics and metabolomics data revealed three polymorphisms (rs2727563 PRKAG2, rs12604940 DCC, and rs13262930 EPHX2) along with arachidonic acid, converging in the netrin signaling pathway (p=1×10−5), as potential markers significantly influencing HCTZ BP response. We successfully replicated the three genetic signals in 212 white hypertensives treated with HCTZ, and created a response score by summing their BP lowering alleles. We found patients carrying one response allele had a significantly lowest response than carriers of six alleles (ΔSBP/ΔDBP: −1.5/1.2 vs −16.3/−10.4 mmHg, respectively, SBP-score p=1×10−8 and DBP-score p=3×10−9). This score explained 11.3% and 11.9% of the variability in HCTZ SBP and DBP responses, respectively, and was further validated in another independent study of 196 whites treated with HCTZ (DBP-score p=0.03, SBP-score p=0.07). This study suggests that PRKAG2, DCC and EPHX2 might be important determinants of HCTZ BP response.
Keywords: Pharmacogenomics, Metabolomics, Hydrochlorothiazide, Hypertension, GWAS
Introduction
Hydrochlorothiazide (HCTZ), a thiazide diuretic, is among the most commonly prescribed antihypertensive medications in the U.S, with over 50 million prescriptions annually.1 It is highly recommended as first line treatment for most patients with uncomplicated essential hypertension (HTN), and for patients requiring more than one antihypertensive therapy for blood pressure (BP) control.2 Despite its importance, patients’ response to HCTZ varies widely, and studies have shown that less than 50% of HCTZ treated patients achieve BP control.3, 4 This wide inter-individual variability in response to HCTZ and other antihypertensive medications reveals that the current approach for therapy selection and BP control is suboptimal. Thus, identifying predictors of BP response to HCTZ and other antihypertensive medications, which could be utilized in therapy selection, would help optimize antihypertensive treatment selection and improve BP control.
In the past decade, HTN pharmacogenetic studies have advanced our understanding of the potential role of genetics in the observed variable response to antihypertensive medications.5, 6 However, most of these studies focused on candidate genes, which revealed few reliable predictors of HCTZ antihypertensive response.6, 7 Recently, we have reported on genome wide association studies (GWAS) using data generated in studies called Pharmacogenomics Evaluation of Antihypertensive Responses (PEAR) and Genetic Epidemiology of Responses to Antihypertensives (GERA), and have identified and replicated genetic signals associated with HCTZ BP response including protein kinase C, alpha (PRKCA) and G-protein alpha subunit-endothelia-3 (GNAS-EDN3) in European Americans (whites).8 However, these genetic signals only explain a small proportion of the variability associated with HCTZ BP response and many more remain to be found. Additionally, in GWAS, the stringent significance threshold (p<5×10−8) limits our success in identifying additional significant single nucleotide polymorphisms (SNPs) influencing HCTZ BP response, particularly with the small sample sizes of the globally available HTN pharmacogenomics studies. This suggests that the standard GWAS approach will not be able to yield all or even the majority of the genetic variance associated with variability in drug response.
In recent years, metabolomics approaches have been successfully employed to identify novel biomarkers associated with different diseases and traits, and bridge the gap between genomics and phenotype.9, 10 Additionally, integrating metabolomics with genomics has been successful in identifying novel key regulators, and pathways for various diseases and traits, including drug response.11 Thus, in the current study we aimed to extend the genetic findings from the PEAR study by incorporating metabolomics data with the genomics data on which we have previously reported8 and sought to: (A) identify metabolites significantly associated with the BP response to HCTZ, (B) use a metabolomics-genomics integrative approach to identify novel genetic variants associated with HCTZ BP response, and then (C) create a response score using replicated genetic signals from this study to evaluate their relative contribution toward the inter-individual variability in BP response to HCTZ.
Methods
Study Participants
The primary analysis of the current study included clinical data and biological samples from white participants recruited as part of the Pharmacogenomic Evaluation of Antihypertensive Response (PEAR) trial (clinicaltrials.gov # NCT00246519).12 In brief, PEAR was a prospective, multicenter study that recruited mild to moderate hypertensive participants, aged 17–65 years, with a primary goal of evaluating the role of genetics on BP response of HCTZ and atenolol monotherapy and combination therapy of both drugs. Further details of the PEAR study design are illustrated in the online-only Data Supplement (Supplementary Figure S1). Of note, the discovery analysis in this study included PEAR whites treated with HCTZ monotherapy (n=228), which will be referred to as HCTZ monotherapy. PEAR whites who started HCTZ after atenolol will be referred to as HCTZ add-on. Data from the latter group of participants (n=214) were used for replication efforts in this study.
A total of 196 white participants, from the Genetic Epidemiology of Responses to Antihypertensives (GERA) study (clinicaltrials.gov # NCT00005520),13 were also used for the replication efforts within this study. In brief, GERA was a prospective, multicenter study that recruited hypertensive participants, aged 30 to 59 years, to examine the role of genetics on the BP response of HCTZ and candesartan monotherapy. More details about the GERA study are presented in the online-only Data Supplement. The PEAR and GERA study protocols were approved by the Institutional Review Board at each study site. All participants provided voluntary written informed consent prior to participation in the study.
Blood Pressure Response Measurement
In PEAR, participants had their BP measured pre-HCTZ (baseline) and after 9 weeks of HCTZ. Home, office and ambulatory BP were measured, as previously described12 (please see online-only Data Supplement). For the analysis of HCTZ monotherapy and HCTZ add-on participants, we used a weighted composite BP of the home, office and ambulatory daytime and night time BP data, which we have shown to be a more accurate measurement of BP response with a better signal-to-noise ratio and more power to identify genetic predictors of BP response.14
In GERA, participants had their BP measured in triplicate by a trained assistant using a random zero sphygmomanometer (Hawksley and Sons, Ltd., England).13 HCTZ BP response was measured by calculating the difference between post- and pre-HCTZ BP readings.
Untargeted Metabolomics Profiling
Baseline fasting plasma samples from 123 PEAR whites treated with HCTZ were used for the metabolomics analysis. Samples were selected based on participants with a large waist circumference (men ≥40 in. and women ≥35 in.), and there was a good representation of HCTZ BP response among participants within the metabolomics dataset (Supplementary Figure S2). Untargeted metabolite profiling was conducted using Gas Chromatography-Time-of-Flight Mass Spectrometry (GC-TOF MS). Plasma samples were prepared for analysis using a two-step methoximation/silylation protocol.15 For each metabolite, peak height was measured, which represents the maximum intensity of all data points forming the mass spectrometry peak. Further details are presented in the online-only Data Supplement.
Genotyping
Details of the genome-wide genotyping, quality control and imputation performed on PEAR and GERA participants are described in the online-only Data Supplement.
Statistical Analyses
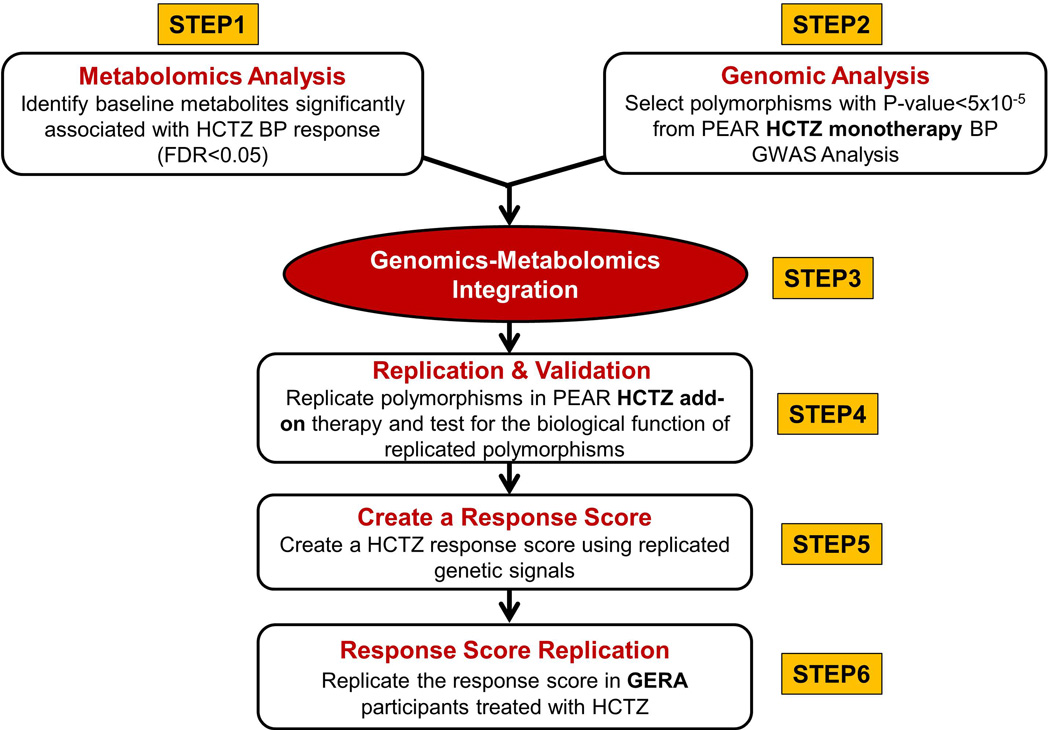
The overall analyses framework used in this study included six steps, as illustrated in Figure 1. In brief, ingenuity pathway analysis software (Ingenuity Systems, www.ingenuity.com) was used to integrate metabolites significantly associated with HCTZ BP response (FDR<0.05) with SNPs at p<5×10−5 from the GWAS analysis. Of note, this p-value threshold was selected based on the quantile-quantile (Q-Q) plots which reveal that SNPs with p<5×10−5 deviated above the diagonal, that is, deviated from the expectation under the null hypothesis of no relationship between SNPs and HCTZ BP response in PEAR white participants treated with HCTZ (Supplementary Figure S3). From the pathway analysis, we focused on significant SNPs/genes and metabolites converging within the significant pathways (FDR<0.05) and further confirmed their association with HCTZ BP response by testing them for replication in PEAR HCTZ add-on white participants treated with HCTZ (n=212). Additionally, to confirm the specificity of the genetic replicated signals to HCTZ, we also tested their association in whites treated with atenolol monotherapy in the PEAR study (n=214) and with candesartan (angiotensin II receptor antagonist) monotherapy in the GERA study (n=198).
Figure 1. The overall analyses framework of the study.
To assess the effect of multiple response alleles on HCTZ BP response, and to examine the relative contribution of our genetic findings toward our phenotype, we constructed a genetic response score based on replicated SNPs. Alleles with BP lowering effect were summed up for inclusion in a regression model, with adjustment for age, sex, baseline BP, and PC1 and 2. To replicate the association of this score with HCTZ BP response, we tested this score in data from HCTZ treated participants (n=196) within the GERA study. In silico analysis was conducted using HaploReg v4.1 http://www.broadinstitute.org/mammals/haploreg/haploreg.php to test whether the replicated SNPs identified in this study affect gene expression levels. A separate analysis was also conducted to test another response score including the replicated genetic signals identified in this study combined with other well-replicated SNPs (NEDD4L rs4149601, PRKCA rs16960228 and GNAS-EDN3 rs2273359) that have previously been reported to be associated with HCTZ BP response.7, 8 More details about the analysis approach are illustrated in the online-only Data Supplement.
Results
Baseline characteristics and HCTZ BP responses of participants included in the genomics and metabolomics analyses are presented in Table 1. Age, sex and body mass index (BMI) baseline characteristics were similar between the PEAR HCTZ monotherapy (genomics and metabolomics datasets), PEAR HCTZ add-on and GERA HCTZ studies. Pre-treatment SBP and DBP were lower within the PEAR HCTZ add-on participants compared to the PEAR HCTZ monotherapy and GERA participants, due to atenolol treatment before starting HCTZ therapy, whereas all other groups were untreated at baseline. Because we have previously shown that baseline BP is the most significant predictor of BP response,13 we adjusted for baseline BP in all the analyses. Of note, HCTZ produced greater BP lowering when used as monotherapy in PEAR HCTZ and GERA HCTZ compared to its use when added to atenolol as HCTZ add-on therapy.
Table 1.
Characteristics of participants included in the genomics and metabolomics analyses
| Characteristics | PEAR HCTZ monotherapy (N=228)# |
PEAR HCTZ monotherapy (N=123)§ |
PEAR HCTZ add- on therapy (N=214)# |
GERA HCTZ monotherapy (N=196)# |
|---|---|---|---|---|
| Age, mean (SD) years |
50 ± 9.5 | 50.7 ± 8.9 | 50.2 ± 9.2 | 48.5 ± 7.3 |
| Women, N (%) | 91 (40) | 57 (46.7) | 98 (42) | 84 (43) |
| BMI, mean (SD) kg*m−2 |
30.30 ± 4.90 | 33 ± 4.90 | 30.23 ± 5.50 | 31.30 ± 5.57 |
| Pre-treatment office SBP, mean (SD) mmHg |
151.80 ± 12.40 | 153.46 ± 12.24 | 136.22 ± 14.15 | 142.70 ± 12.60 |
| Pre-treatment office DBP, mean (SD) mmHg |
98.10 ± 5.80 | 98.12 ± 6.30 | 86.31 ± 8.74 | 95.60 ± 5.70 |
| Office SBP response, mean (SD) mmHg |
−11.00 ± 12.80 | −10.80 ± 12.94 | −7.23 ± 12.82 | −10.90 ± 13.00 |
| Office DBP response, mean (SD) mmHg |
−5.01 ± 7.17 | −4.39 ± 6.97 | −3.47 ± 8.69 | −6.26 ± 8.83 |
| Composite SBP response, mean (SD) mmHg |
−8.50 ± 7.02 | −9.3 ± 6.90 | −6.68 ± 6.54 | NA |
| Composite DBP response, mean (SD) mmHg |
−4.68 ± 4.79 | −5.11 ± 4.87 | −3.79 ± 4.40 | NA |
Continuous variables are presented as mean ± standard deviation (SD); categorical variables are presented as numbers and percentage. PEAR, Pharmacogenomic Evaluation of Antihypertensive Responses; GERA, Genetic Epidemiology of Responses to Antihypertensives; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure;
represents genomics data;
represents metabolomics data.
Metabolomics Analysis
Using a GC-TOF MS platform, we were able to identify 212 structurally known metabolites from PEAR HCTZ monotherapy participants. Thirteen metabolites, out of the 212, were significantly associated with both DBP and SBP responses to HCTZ (FDR<0.05), after adjustment for age, sex and baseline BP (Supplementary Table S1). Those thirteen metabolites were then integrated with PEAR HCTZ monotherapy GWAS top signals (p<5×10−5) using a pathway integrative approach as shown below.
Genomics Metabolomics Integration (Step 3, Figure 1)
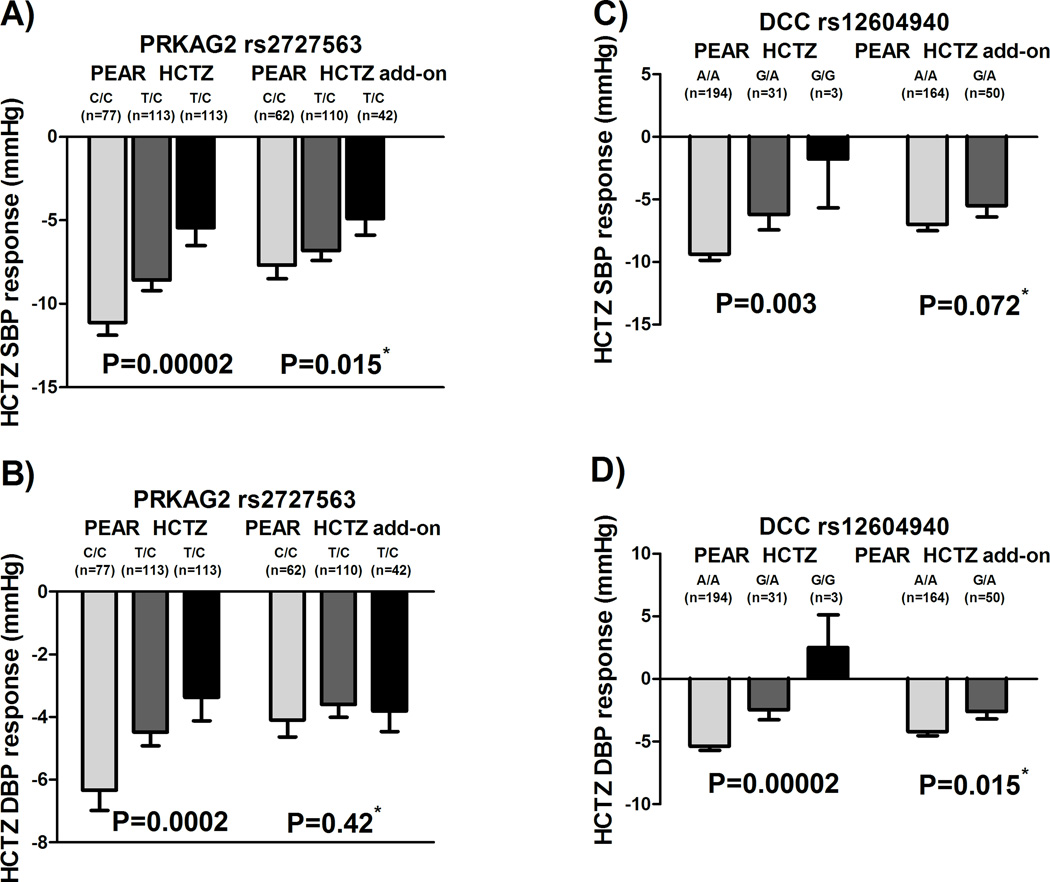
A total of 103 SNPs were selected, from PEAR HCTZ monotherapy SBP and DBP GWAS analyses, based on our suggestive cut off p-value (i.e. p<5×10−5) (Supplementary Table S2). Integrating those 103 SNPs with the thirteen significant metabolites identified the netrin signaling pathway as the only significant pathway (p=1.54×10−5), after adjusting for multiple comparisons (FDR<0.05), with rs2727563 in PRKAG2 and rs12604940 in DCC converging with the arachidonic acid metabolite in the same pathway (Supplementary Figure S4). We found that carriers of the PRKAG2 rs2727563 C allele had better responses to HCTZ in a manner consistent with an additive genetic model (p=2×10−5, Figure 2A). We also found that DCC rs12604940 carriers of the CC genotypes had a better response to HCTZ compared to participants with CG and GG genotypes (p=2×10−5, Figure 2B).
Figure 2. Effects of rs2727563 and rs12604940 polymorphisms on the BP responses of whites treated with HCTZ in the PEAR HCTZ monotherapy and HCTZ add-on.
A) rs2727563 on SBP responses in PEAR monotherapy and PEAR HCTZ add-on. B) rs2727563 on DBP response in the PEAR monotherapy and PEAR HCTZ add-on. C) rs12604940 on SBP response in PEAR monotherapy and PEAR HCTZ add-on. D) rs12604940 on DBP response in the PEAR monotherapy and PEAR HCTZ add-on. BP responses were adjusted for baseline BP, age, sex, and principal components 1 and 2. P-values represented are for contrast of adjusted means between different genotype groups. Error bars represent standard error of the mean. *One sided p-value based on a one-sided hypothesis tested in the replication study.
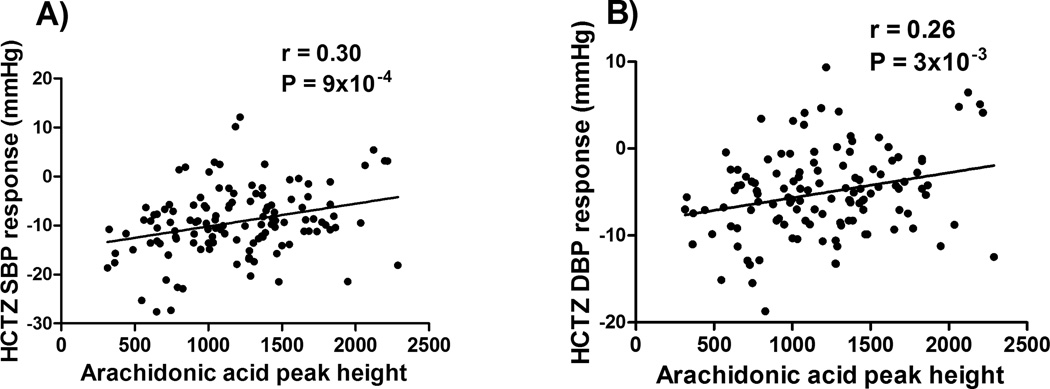
We also showed that arachidonic acid is involved in the netrin signaling pathway and had a significant association with HCTZ BP response (SBP adjusted-p=1×10−4, DBP adjusted-p=7×10−4; Figure 3), after adjustment for age, gender and baseline BP. Since arachidonic acid and its metabolites have been associated with cardiovascular traits and BP regulation,16, 17 we hypothesized that the arachidonic acid association with HCTZ BP response might also be mediated via polymorphisms within genes involved in the arachidonic acid metabolic pathway. Therefore, we tested our hypothesis by investigating SNPs within eleven genes directly involved in the synthesis and degradation of arachidonic acid and have been previously reported to be associated with BP regulation (Supplementary Table S3). From this analysis, we were able to identify rs324425, within the candidate region of the FAAH (fatty acid amide hydrolase) gene, and rs7816586 and rs13262930 in the EPHX2 gene, with statistical significant association with HCTZ BP response (FDR<0.05, Supplementary Table S4). Those three SNPs along with the PRKAG2 rs2727563 and DCC rs12604940 SNPs were then moved for replication in PEAR HCTZ add-on participants as shown below.
Figure 3. Correlation between HCTZ BP response and arachidonic acid peak height.
P-values and correlation coefficient (r-values) were generated using Pearson correlation test. A) systolic blood pressure. B) diastolic blood pressure. Peak height represents the maximum intensity of all data points forming the mass spectrometry peak.
Replication and Validation (Step 4, Figure 1)
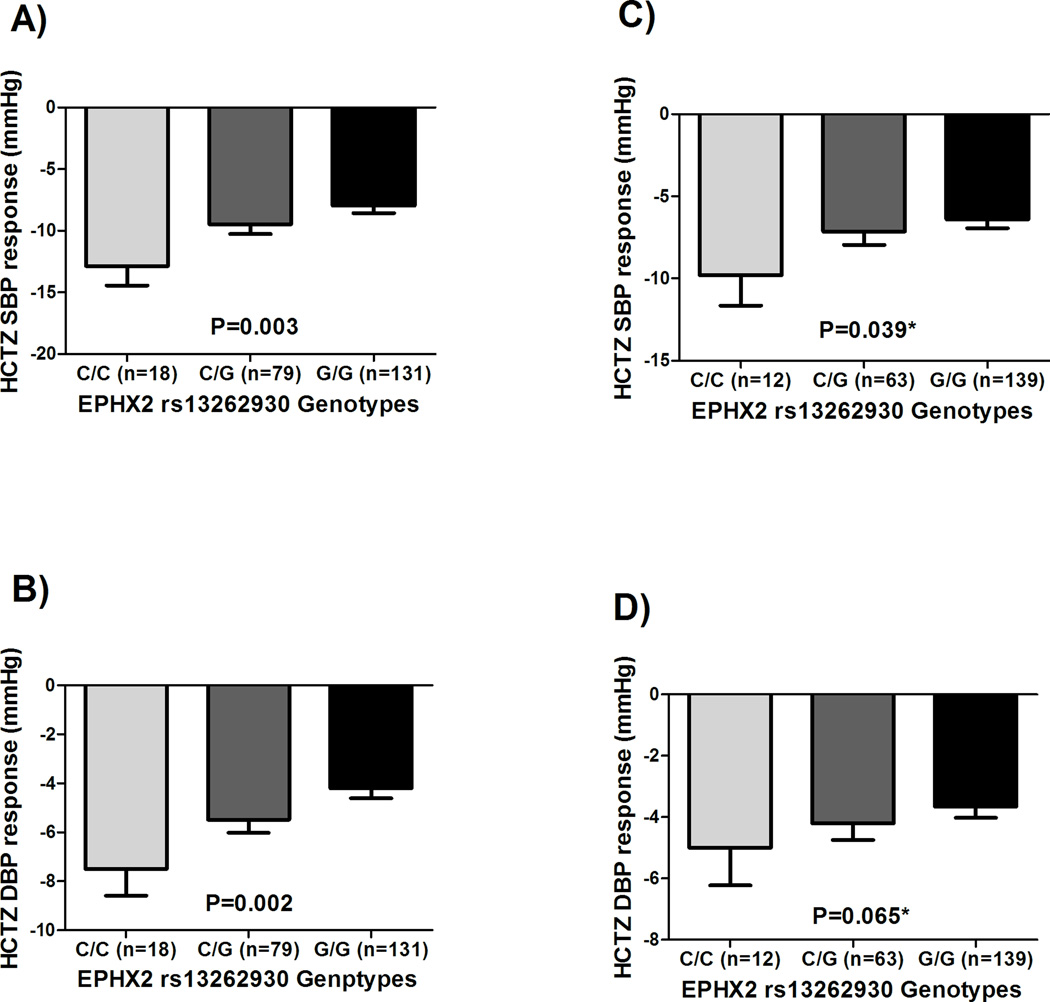
Three SNPs (PRKAG2 rs2727563, DCC rs12604940 and EPHX2 rs13262930), out of the five tested SNPs, were replicated in the same direction as shown in Figures 2A, 2D, and 4, respectively. In silico analyses revealed that rs13262930 and rs2727563 affect the expression levels of EPHX2 and PRKAG2 in blood, respectively (Supplementary Table S5). Additionally, rs13262930 has been shown to affect the expression levels of EPHX2 in several other tissues including the heart (Supplementary Table S5). Interestingly, we also found that PRKAG2 rs2727563 has a significant association on arachidonic acid baseline levels (p=0.03, Supplementary Figure S5). Similarly, EPHX2 rs13262930 has also shown a marginally significant association with arachidonic acid (p=0.055, Supplementary Figure S5). For DCC rs12604940 SNP, we didn’t find any effect on DCC gene expression or on arachidonic acid levels.
Figure 4. Effects of rs13262930 polymorphism on the BP response of whites treated with HCTZ in the PEAR HCTZ monotherapy and PEAR HCTZ add-on.
A) SBP response in the PEAR HCTZ monotherapy. B) DBP response in the PEAR HCTZ monotherapy. C) Replicating the effect on SBP response in the PEAR HCTZ add-on. D) Replicating the effect on DBP response in the PEAR HCTZ add-on. BP responses were adjusted for baseline BP, age, sex and principal components 1 and 2. P-values represented are for contrast of adjusted means between different genotype groups. Error bars represent standard error of the mean. *One sided p-value based on a one-sided hypothesis tested in the replication study.
We confirmed the specificity of these three replicated signals to HCTZ BP response by testing them in whites treated with other antihypertensives like atenolol in PEAR and candesartan in GERA. We found that neither of these SNPs were significantly associated with atenolol BP response (rs2727563 SBP p=0.24, DBP p=0.47; rs12604940 SBP p=0.96, DBP p=0.79; rs13262930 SBP p=0.56, DBP p=0.93) nor with candesartan BP response (rs2727563 SBP p=0.64, DBP p=0.71; rs12604940 SBP p=0.37, DBP p=0.29; rs13262930 SBP p=0.81, DBP p=0.77). These results suggest that these signals might be important determinants of HCTZ BP response in particular.
Create a Response Score (Step 5 and 6, Figure 1)
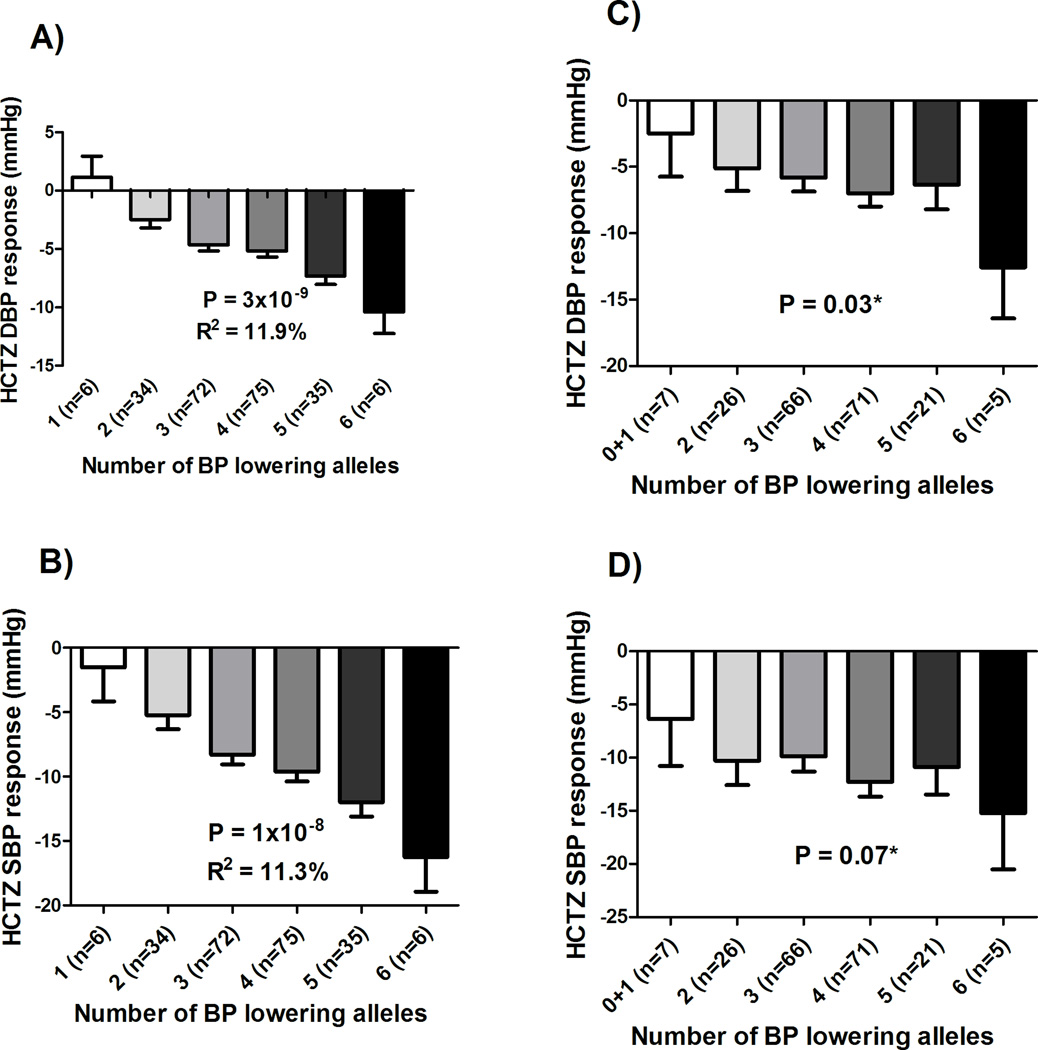
Linear regression analysis adjusting for age, sex, baseline BP, and PCs 1 and 2, revealed that individuals with a higher score had a better HCTZ SBP (p=1×10−8) and DBP (p=3×10−9) responses compared to lower score participants (Figure 5A, and 5B, respectively). We found that this genetic response score, by itself, explained 11.3% and 11.9% of HCTZ SBP and DBP responses, respectively. Additionally, we also found a significant association when we tested this response score with home BP, 24hrs ambulatory BP and office BP responses measured (Supplementary Figure S6). Moreover, we validated this score in whites treated with HCTZ in GERA in which we found a significant association with DBP response (1-sided p=0.03, Figure 5C), and a trend towards significance with SBP response (1-sided p=0.07, Figure 5D).
Figure 5. HCTZ response score in PEAR and GERA studies.
A) Tested against DBP response in the PEAR HCTZ monotherapy. B) Tested against SBP response in the PEAR HCTZ monotherapy. C) Tested against DBP response in the GERA study. D) Tested against SBP response in the GERA study. BP responses were adjusted for baseline BP, age, sex and principal components 1 and 2. P-values represented are for contrast of adjusted means between different genotype groups. Error bars represent standard error of the mean. *One sided p-value based on a one-sided hypothesis tested in the replication study.
Additional Analysis
In a separate analysis, we added another three well-replicated SNPs, that have previously been reported to be associated with HCTZ BP response in whites (NEDD4L rs4149601, PRKCA rs16960228 and GNAS-EDN3 rs2273359),7, 8 to our response score. Interestingly, adding those three SNPs to the response score improved the significance and the predictability of the score in both PEAR (DBP p=5×10−10, r2=16.2% and SBP p=5×10−11, r2=14.9%) and GERA (DBP p=2×10−4, r2=4.5% and SBP p=3×10−3, r2=3.3%), as shown in Supplementary Figure S7.
Discussion
The genomics-metabolomics integrative approach used in this study helped identify three signals, PRKAG2 rs2727563, DCC rs12604940, and EPHX2 rs13262930, which were significantly associated with HCTZ BP response, and their associations were replicated in a second cohort. Using these three replicated signals, we constructed a genetic response score with a stronger association with HCTZ BP response compared to individual SNPs. This is not surprising for a complex phenotype like antihypertensive response, because it is known to be affected by multiple genetic contributors. This response score by itself explained 11.3% and 11.9% of HCTZ SBP and DBP responses, respectively, and was further validated in a third independent study, which emphasizes the importance of this response score and its signals to be considered in future models for guiding the selection of HCTZ therapy.
HCTZ is known to inhibit the Na+/Cl− co-transporter (NCC) in the distal convoluted tubule within the kidney. This inhibition initially contracts plasma volume and decreases cardiac output leading to BP lowering, however, the plasma volume and cardiac output returns to normal after 4–6 weeks of thiazide initiation. This suggests that the long term BP lowering effects of HCTZ might be controlled by other unknown mechanisms.6 The genomics-metabolomics pathway analysis performed in this study highlighted the netrin signaling pathway as a significant pathway, including metabolic and genetic signatures associated with HCTZ BP response. This pathway is activated by netrins, a class of proteins that play a crucial role in neuronal migration and in axon guidance. Netrin-1 is the most studied member of the family and has been shown as a potent endothelial mitogen stimulating the production of nitric oxide via a DCC-ERK1/2 dependent mechanism.18 Additionally, a recent study has shown that netrin-1 and its receptor, DCC, control sympathetic arterial innervation and play an important role in the regulation of the blood flow to peripheral organs.19 Moreover, netrin-1 binding to specific receptors like DCC has been shown to activate multiple pathways including MAPKs, PKC, src, Rac and Rho kinase, and focal adhesion kinase20–22, which all have been previously reported to be associated with HTN and BP regulation.23–25 Furthermore, a recent study has demonstrated that netrin-1 activates PRKC alpha, and FAK/Fyn, which are important for the activation of the ERK, JNK and NF-kB.26 Of note, we recently identified and replicated a signal within the PRKC alpha gene with a clinically significant influence on the BP response to HCTZ.8 Collectively, this highlights the importance of the netrin signaling pathway and suggests that it might be a novel and substantial pathway through which HCTZ produces its long term antihypertensive effects.
The genomics metabolomics integrative analyses have also identified rs2727563 SNP within the PRKAG2 with a significant association to HCTZ BP response. PRKAG2 has been shown as an important regulator of cellular energy metabolism including de novo biosynthesis of fatty acids, and also acts as a regulator of cellular polarity by remodeling the actin cytoskeleton.27 Additionally, PRKAG2 has previously shown to be significantly associated with BP,28 ventricular pre-excitation (Wolff-Parkinson-White syndrome), chronic kidney disease,29 and left ventricular hypertrophy resembling cardiomyopathy.30 Altogether, the literature evidence supporting the association of the PRKAG2 with BP and cardiovascular diseases, and the evidence from our results which included the identification and replication of PRKAG2 rs2727563 association with HCTZ BP response suggest PRKAG2 as a potential determinant of HCTZ BP response. Future work still needed to demonstrate the mechanistic relation between this gene and HCTZ BP response mechanism.
Our results have also revealed arachidonic acid, within the netrin signaling pathway, as a significant metabolomic signature influencing the BP response to HCTZ therapy. Arachidonic acid and its metabolites have been well-known for their role in the regulation of renal vascular tone, BP and sodium transport.17, 31 Testing genetic variants within genes, directly involved in the synthesis and degradation of arachidonic acid, revealed EPHX2 rs13262930 SNP significantly associated with HCTZ BP response. EPHX2 is well-known for encoding the soluble epoxide hydrolase (sEH) enzyme, which converts epoxyeicosatrienoic acid (EET), a strong vasodilator and antiinflammatory compound, to the biologically less active compound, dihydroxyeicosatrienoic acid (DHET).32 Studies have shown the expression of the sEH enzyme is positively correlated with BP and inhibiting this enzyme increases the production of the EETs and ultimately reduces BP.33 Additionally, Ma et. al. recently reported that HCTZ might be mediating its antihypertensive BP response through the inhibition of the sEH.34 Collectively, our results along with Ma et. al. study results emphasize that EPHX2 might be involved in the BP lowering mechanism of thiazide diuretics and suggest that it might be an important predictor of thiazide diuretics’ BP response.
Our study has several strengths. To our knowledge, this is the first study to use a genomics-metabolomics integrative approach to identify novel biomarkers associated with HCTZ BP response. This integrative approach was successful in identifying novel genetic variants that we were not able to identify using GWAS data alone. We believe that using such an approach can help us to take forward the large investment in GWAS and utilize the output of this approach to identify additional genetic variants and biologically relevant pathways associated with drug response. Secondly, replicating our genetic signals and further validating them in another independent study, as a combined alleles response score, emphasize the importance of our findings and their significant influential effect on HCTZ BP response.
Our study also has several limitations. First, our samples size was relatively small which limited our power to identify additional SNPs within the GWAS analysis. However, integrating the metabolomics and the genomics profiles of participants treated with HCTZ added to the breadth and the depth of our analyses and helped us to overcome this limitation and to identify novel genetic signals that we were not able to identify using GWAS data alone. Secondly, the approach that we used to identify EPHX2 rs13262930 might have some flaws like 1) neglecting important genes that haven’t been well studied with HTN yet, or 2) obtaining false positive results by adjusting for multiple comparisons in SNPs within certain genes. However, the EPHX2 rs13262930 SNP that we identified using this approach was confirmed by replicating it in two other independent studies. Additionally, we showed that this SNP has biologically functional properties as it affects the expression levels of the EPHX2 gene in different tissues, including the heart, and has a marginally significant association with arachidonic acid in fasting plasma. These multiple levels of replication and validation highlight the legitimacy of our approach and its results and suggest that the EPHX2 rs13262930 SNP is likely a true and important genetic predictor of HCTZ BP response.
In conclusion, this study illustrates the power of integrating different types of omics data to identify novel genetic variants underlying drug response. Future use of this approach would improve the breadth and depth of studying complex phenotypes, as antihypertensive response, and might provide more knowledge and insight in to the mechanism underlying BP response. This knowledge might facilitate the development of new drugs and therapeutic approaches based on a deeper understanding of the determinants of the BP response.
Perspectives
Thiazide diuretics have been the mainstay anti-HTN therapy for decades and are still ranked among the most commonly prescribed medications worldwide. However, the wide inter-individual variability in response to this class of drugs highlights the need for identifying predictors that can be used for improving the BP response of this therapy. Using both genomics and metabolomics data in this study revealed the netrin signaling pathway as an important pathway associated with HCTZ BP response. Future work on this pathway might provide more insights in the mechanism underlying HCTZ antihypertensive effect, and help in identifying novel drug targets of new antihypertensive medications. The results of this study have also shed light on DCC, PRKAG2 and EPHX2 genes as important determinants of HCTZ BP response. The response score created using SNPs within these genes should be further tested in other independent cohorts to further confirm its utility in guiding the selection of HCTZ therapy.
Supplementary Material
Novelty and Significance.
What is new?
To our knowledge, this is the first study to use a genomics-metabolomics integrative approach to identify novel genetic markers associated with blood pressure response to hydrochlorothiazide.
We identified and replicated three novel genetic signals, PRKAG2 rs2727563, DCC rs12604940 and EPHX2 rs13262930 with clinically relevant effects on the blood pressure response of European Americans treated with hydrochlorothiazide.
A response score to hydrochlorothiazide was created using the three replicated genetic signals, and was further validated in independent participants treated with hydrochlorothiazide.
What is relevant?
The multiple level of replication for the three identified genetic signals, individually and combined in the response score, suggests their importance as predictors of hydrochlorothiazide blood pressure response, which may help in optimizing the selection of antihypertensive therapy. Additionally, these signals may provide insight into the antihypertensive mechanisms underlying thiazide diuretics, which may lead to the identification of novel drug targets.
Summary
Integrating the genomics and metabolomics profiles of European American participants treated with hydrochlorothiazide succeeded in identifying novel genetic signals with clinically relevant effects on the blood pressure response to hydrochlorothiazide. Replicating these signals in an independent study substantiated the importance of these genetic signals as important determinants of hydrochlorothiazide blood pressure response. Combining the three replicated signals in a response score explained 11.3–11.9% of the blood pressure response to hydrochlorothiazide. Validating this response score in another independent study emphasized the importance of considering this score and its signals in future models for guiding the selection of hydrochlorothiazide therapy.
Acknowledgments
We acknowledge the valuable contributions of the PEAR and GERA study participants, support staff and study physicians.
Sources of Funding
PEAR was supported by the National Institute of Health Pharmacogenetics Research Network grant U01-GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University) and UL1 TR000135 (Mayo Clinic). PEAR was also supported by funds from the Mayo Foundation. The metabolomics work was funded by NIGMS (National Institute of General Medical Sciences) grant RC2-GM092729 “Metabolomics Network for Drug Response Phenotype”. Additional support for this work includes: M.H.S. is supported by AHA pre-doctoral fellowship award #14PRE20460115 and O.F. is funded through NIH DK097154.
Footnotes
Conflict of Interest
None
References
- 1.Ims institute. [Accessed June 1, 2016];Medicines use and spending shifts: A review of the use of medicines in the U.S. In 2014. 2014 https://www.Imshealth.Com/en/thought-leadership/ims-institute/reports/medicines-use-in-the-us-2014. [Google Scholar]
- 2.James PA, Oparil S, Carter BL, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: Report from the panel members appointed to the eighth joint national committee (jnc 8) JAMA. 2014;311:507–520. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 3.Materson BJ. Variability in response to antihypertensive drugs. Am J Med. 2007;120:S10–S20. doi: 10.1016/j.amjmed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, Hamburger RJ, Fye C, Lakshman R, Gottdiener J. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The department of veterans affairs cooperative study group on antihypertensive agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JA. Pharmacogenomics of antihypertensive drugs: Past, present and future. Pharmacogenomics. 2010;11:487–491. doi: 10.2217/pgs.10.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahin MH, Johnson JA. Mechanisms and pharmacogenetic signals underlying thiazide diuretics blood pressure response. Curr Opin Pharmacol. 2016;27:31–37. doi: 10.1016/j.coph.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonough CW, Burbage SE, Duarte JD, Gong Y, Langaee TY, Turner ST, Gums JG, Chapman AB, Bailey KR, Beitelshees AL, Boerwinkle E, Pepine CJ, Cooper-DeHoff RM, Johnson JA. Association of variants in nedd4l with blood pressure response and adverse cardiovascular outcomes in hypertensive patients treated with thiazide diuretics. J Hypertens. 2013;31:698–704. doi: 10.1097/HJH.0b013e32835e2a71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner ST, Boerwinkle E, O'Connell JR, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62:391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper-Dehoff RM, Hou W, Weng L, et al. Is diabetes mellitus-linked amino acid signature associated with beta-blocker-induced impaired fasting glucose? Circulation. Cardiovascular Genetics. 2014;7:199–205. doi: 10.1161/CIRCGENETICS.113.000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaddurah-Daouk R, Weinshilboum R. Metabolomic signatures for drug response phenotypes: Pharmacometabolomics enables precision medicine. Clin Pharmacol Ther. 2015;98:71–75. doi: 10.1002/cpt.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, Gums J, Curry RW, Gong Y, Beitelshees AL, Schwartz G, Turner ST. Pharmacogenomics of antihypertensive drugs: Rationale and design of the pharmacogenomic evaluation of antihypertensive responses (pear) study. Am. Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman AB, Schwartz GL, Boerwinkle E, Turner ST. Predictors of antihypertensive response to a standard dose of hydrochlorothiazide for essential hypertension. Kidney Int. 2002;61:1047–1055. doi: 10.1046/j.1523-1755.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- 14.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-Dehoff RM, Boerwinkle E, Johnson JA, Bailey KR. Power to identify a genetic predictor of antihypertensive drug response using different methods to measure blood pressure response. J Transl Med. 2012;10:47. doi: 10.1186/1479-5876-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, Moon S, Nikolau B. Quality control for plant metabolomics: Reporting msi-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 16.Rahman M, Wright JT, Jr, Douglas JG. The role of the cytochrome p450-dependent metabolites of arachidonic acid in blood pressure regulation and renal function: A review. Am J Hypertens. 1997;10:356–365. doi: 10.1016/s0895-7061(96)00381-0. [DOI] [PubMed] [Google Scholar]
- 17.Sarkis A, Roman RJ. Role of cytochrome p450 metabolites of arachidonic acid in hypertension. Curr Drug Metab. 2004;5:245–256. doi: 10.2174/1389200043335603. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Cai H. Netrin-1 prevents ischemia/reperfusion-induced myocardial infarction via a dcc/erk1/2/enos s1177/no/dcc feed-forward mechanism. J Mol. Cell Cardiol. 2010;48:1060–1070. doi: 10.1016/j.yjmcc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunet I, Gordon E, Han J, et al. Netrin-1 controls sympathetic arterial innervation. J Clin Invest. 2014;124:3230–3240. doi: 10.1172/JCI75181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai Wing Sun K, Correia JP, Kennedy TE. Netrins: Versatile extracellular cues with diverse functions. Development. 2011;138:2153–2169. doi: 10.1242/dev.044529. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Beggs H, Jurgensen C, Park HT, Tang H, Gorski J, Jones KR, Reichardt LF, Wu J, Rao Y. Netrin requires focal adhesion kinase and src family kinases for axon outgrowth and attraction. Nat Neurosci. 2004;7:1222–1232. doi: 10.1038/nn1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forcet C, Stein E, Pays L, Corset V, Llambi F, Tessier-Lavigne M, Mehlen P. Netrin-1-mediated axon outgrowth requires deleted in colorectal cancer-dependent mapk activation. Nature. 2002;417:443–447. doi: 10.1038/nature748. [DOI] [PubMed] [Google Scholar]
- 23.Buchholz RA, Dundore RL, Cumiskey WR, Harris AL, Silver PJ. Protein kinase inhibitors and blood pressure control in spontaneously hypertensive rats. Hypertension. 1991;17:91–100. doi: 10.1161/01.hyp.17.1.91. [DOI] [PubMed] [Google Scholar]
- 24.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andre G, Sandoval JE, Retailleau K, Loufrani L, Toumaniantz G, Offermanns S, Rolli-Derkinderen M, Loirand G, Sauzeau V. Smooth muscle specific rac1 deficiency induces hypertension by preventing p116rip3-dependent rhoa inhibition. J Am Heart Assoc. 2014;3:e000852. doi: 10.1161/JAHA.114.000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SJ, Jung YH, Oh SY, Yong MS, Ryu JM, Han HJ. Netrin-1 induces mmp-12-dependent e-cadherin degradation via the distinct activation of pkcalpha and fak/fyn in promoting mesenchymal stem cell motility. Stem Cells Dev. 2014;23:1870–1882. doi: 10.1089/scd.2013.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp BE, Mitchelhill KI, Stapleton D, Michell BJ, Chen ZP, Witters LA. Dealing with energy demand: The amp-activated protein kinase. Trends. Biochem Sci. 1999;24:22–25. doi: 10.1016/s0968-0004(98)01340-1. [DOI] [PubMed] [Google Scholar]
- 28.Tragante V, Barnes MR, Ganesh SK, et al. Gene-centric meta-analysis in 87,736 individuals of european ancestry identifies multiple blood-pressure-related loci. Am J Hum Genet. 2014;94:349–360. doi: 10.1016/j.ajhg.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arad M, Maron BJ, Gorham JM, Johnson WH, Jr, Saul JP, Perez-Atayde AR, Spirito P, Wright GB, Kanter RJ, Seidman CE, Seidman JG. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 31.Lasker JM, Chen WB, Wolf I, Bloswick BP, Wilson PD, Powell PK. Formation of 20-hydroxyeicosatetraenoic acid, a vasoactive and natriuretic eicosanoid, in human kidney. Role of cyp4f2 and cyp4a11. The Journal of Biological Chemistry. 2000;275:4118–4126. doi: 10.1074/jbc.275.6.4118. [DOI] [PubMed] [Google Scholar]
- 32.Spector AA, Fang X, Snyder GD, Weintraub NL. Epoxyeicosatrienoic acids (eets): Metabolism and biochemical function. Prog Lipid Res. 2004;43:55–90. doi: 10.1016/s0163-7827(03)00049-3. [DOI] [PubMed] [Google Scholar]
- 33.Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8:794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma F, Lin F, Chen C, Cheng J, Zeldin DC, Wang Y, Wang DW. Indapamide lowers blood pressure by increasing production of epoxyeicosatrienoic acids in the kidney. Mol Pharmacol. 2013;84:286–295. doi: 10.1124/mol.113.085878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.