Abstract
Dehydrins are a group of intrinsically disordered proteins that protect plants from damage caused by drought, cold, and high salinity. Like other intrinsically disordered proteins, dehydrins can gain structure when bound to a ligand. Previous studies have shown that dehydrins are able to protect liposomes from cold damage, but the interactions that drive membrane binding and the detailed structure of the bound and unbound forms are not known. We use an ensemble-structure approach to generate models of a dehydrin known as K2 in the presence and absence of sodium dodecyl sulfate micelles, and we docked the bound structure to the micelle. The collection of residual dipolar coupling data, amide protection factors, and paramagnetic relaxation enhancement distances, in combination with chemical shifts and relaxation measurements, allows for determining plausible structures that are not otherwise visible in time-averaged structural data. The results show that in the bound structure, the conserved lysines are important for membrane binding, whereas the flanking hydrophobic residues play a lesser role. The unbound structure shows a high level of disorder and an extended structure. We propose that the structural differences between bound and unbound forms allow dehydrins to act as molecular shields in their unbound state and as membrane protectants in their bound state. Unlike α-synuclein, the significant gain of α-helicity in K2 at low concentrations of sodium dodecyl sulfate is not due to a decrease in the critical micelle concentration. The study provides structural insight into how a disordered protein can interact with a membrane surface.
Introduction
Because plants cannot move, they have developed extensive protective mechanisms against abiotic stresses such as cold, drought, and salinity. One example is a family of abiotic stress proteins, named dehydrins (a shortened version of “dehydration proteins”) (1, 2, 3, 4, 5, 6), which have been shown in several studies to be important in protecting enzymes (7, 8), DNA and RNA (9), and membranes (10, 11) from cold and desiccation damage. These multiple functions are reflected in the multiple intracellular locations where dehydrins can be found. Localization studies have shown that they can be found in the cytosol (12), nucleus (13), plasma membranes (14), and mitochondria (15). Like many other abiotic plant stress proteins (16, 17, 18), dehydrins have highly hydrophilic sequences, contain a paucity of hydrophobic residues, and are especially poor in Trp and Cys amino acids (1). Structurally, they have been shown to be intrinsically disordered proteins (IDPs) (19, 20, 21); that is, they do not have a well defined three-dimensional structure and contain minimal amounts of regular secondary structure.
The dehydrin protein family is defined by the presence of three different conserved motifs. The most important is the K-segment, since by definition a protein must have at least one K-segment to be classified as a dehydrin (1, 2, 6), although a K-segmentless dehydrin has been recently described (22). As the name suggests, the K-segment is rich in Lys residues. The canonical K-segment is often written as the sequence EKKGIMDKIKEKLPG, but a recent comprehensive analysis of >650 dehydrin sequences shows that this segment is better defined by a position-weighted matrix (Malik et al., unpublished data). This motif likely has a role in membrane binding (23). The other two conserved motifs, the Y- and S-segments, are not present in all dehydrins. The Y-segment consists of the sequence DEX1GNPX2, where X1 is Tyr 75% of the time, but can also be Phe or His, and X2 is generally a hydrophobic amino acid ((2); Malik et al., unpublished data). The Y-segment has been proposed to bind nucleotides, but this has not yet been experimentally established. The S-segment has a core conserved tract of four to eight Ser residues (1), although it can also be described as a longer motif that may contain an Snf-1-related kinase consensus sequence ((24); Malik et al., unpublished data). The S-segment can be phosphorylated in vivo and in vitro (25, 26); it has been shown that a phosphorylated S-segment has a role in nuclear localization (13) and in calcium binding (27). Regions between the conserved motifs are designated as ϕ-segments, and contain amino acids that are mostly small and polar. A specific function has not been discovered for the ϕ-segments, but they may have a role in the positioning of the other motifs for optimal ligand binding (6).
Dehydrins have very modular sequences with a variable number of conserved segments. The K-segment, by definition, must be present in at least one copy but can be present as often as 13 times (28) and is usually present near the C-terminus of the protein. The Y- and S-segments, when present, are usually found in one or two copies and are located N-terminal to the K-segment (1). The resulting common architectures are Kn, YnKn, SKn, and YnSKn, with architectures such as KnS occurring more rarely. The functional role of the different architectures has not yet been established, but there are two possibilities: either the different architectures are expressed during different abiotic stresses (6) or they determine which ligands they may interact with inside a cell.
To date there have been only low-resolution structural analyses of dehydrins, generally involving circular dichroism (CD) spectroscopy (29, 30, 31) and NMR approaches that did not result in three-dimensional (3D) structure calculations (32, 33). One study found that there is a modest gain in α-helicity when a dehydrin binds to a membrane (34). This may not be a universal effect; the Arabidopsis dehydrins ERD10 and ERD14 were shown to bind to liposomes, but CD experiments did not detect any change in secondary structure (35), and the study of a dehydrin from Thellungiella salsuginea by Fourier-transformed infrared spectroscopy showed a modest gain in the β-strand character of the protein (36). One study showed that dehydrins preferred to bind to negatively charged lipids such as phosphatidyl serine, phosphatidyl glycerol and phosphatidic acid (30). Gains in α-helicity were also shown for several different dehydrins in the presence of the membrane-mimicking sodium dodecyl sulfate (SDS) micelle (30, 34, 37).
Previously, we showed that the model Vitis riparia dehydrin known as K2 is able to prevent liposome fusion after freeze-thaw treatment and lower the transition temperature to maintain membrane fluidity (11). In this work, we use NMR to examine the ensemble structure of this IDP in the presence and absence of micelles, and use in silico docking to determine how this protein can interact with membrane surfaces.
Materials and Methods
Expression, purification, and modification of the dehydrin
The Vitis riparia K2 dehydrin was expressed and purified as described previously (11). The Thr24Cys mutation was created using the Quikchange mutagenesis protocol (Stratagene, La Jolla, CA) by following the manufacturer’s protocol using the forward primer 5′- AAGCAAACGAGCGCCTGCAGCACGCCGGGGCAGG-3′ and reverse primer 5′- CCTGCCCCGGCGTGCTGCAGGCGCTCGTTTGCTT-3′, where the underlining indicates the bases that are to be mutated. Positive clones of the Thr24Cys mutation were confirmed by sequencing the resulting plasmid construct.
K2 with the Thr24Cys modification (K2-T24C) was purified as described above for the wild-type protein. K2-T24C was treated with 5 mM dithiothreitol and subsequently purified by reverse-phase high-performance liquid chromatography to ensure that the Cys residue was completely reduced. The paramagnetic spin label (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanesulfonate (MTSL; Toronto Research Chemicals, Toronto, Ontario, Canada) was coupled to Cys-24 as described previously (38). Unlabeled protein was separated from labeled protein using reverse-phase high-performance liquid chromatography (8).
Secondary-structure analysis in the presence and absence of micelles
K2 CD data were collected using a Jasco-815 CD spectropolarimeter (Easton, MD). The protein was dissolved in 10 mM sodium phosphate, pH 7.4, at a protein concentration of 0.16 mg/mL (30 μM). A quartz cuvette with a 2 mm pathlength (Hellma, Concord, Ontario, Canada) containing the protein sample with and without SDS (50 mM) was scanned from 250 to 190 nm. For the SDS titration experiment, the concentration of the detergent was varied between 0 and 50 mM. The spectra were averaged over eight accumulations. All CD experiments were performed at 25°C.
Determination of the SDS critical micelle concentration in the presence and absence of K2
The critical micelle concentration (CMC) of SDS was determined by both conductivity measurements and changes in 1-anilonapthalene-8-sulfonate (ANS) fluorescence. For the conductivity measurements, increasing amounts of SDS (0–12 mM) were added to a solution containing 10 μM K2 in 20 mM sodium phosphate buffer, pH 7.4. Conductivity was measured using a CDM210 conductivity meter (Radiometer Analytical, Lyon, France). For the fluorescence measurements, SDS was dissolved in 20 mM phosphate buffer, pH 6.0. After the addition of K2, ANS was added to a final concentration of 20 μM. Samples were allowed to equilibrate for 30 min before the fluorescence was measured (λex at 370 nm and λem at 495 nm) using a PTI fluorometer (Photon Technology International, London, Ontario, Canada). Intensities were recorded using FeLiX software, version 1.41.
NMR experiments
Lyophilized 15N-K2, MTSL-15N-K2-T24C, or 13C/15N-K2 samples were resuspended in 600 μL of NMR buffer (20 mM sodium phosphate, pH 6.0, 10 mM NaCl, 0.01% sodium azide, 0.1 mM dimethyl-4-silapentane-1-sulfonic acid (DSS), and 10% D2O (v/v)). Data were collected on a Bruker Avance DRX600 spectrometer equipped with a cryogenic triple resonance probe with the temperature set to 300 K. 1H, 13C, and 15N referencing was performed relative to DSS as described previously (39). The chemical shifts of K2 alone (32) and in the presence of 50 mM SDS (11) have been previously assigned. All new NMR experiments were processed using NMRPipe (40), and the resulting spectra were visualized and analyzed using the CCPNMR analysis software, version 2.2 (41).
Residual dipolar coupling experiments
15N-K2 (1 mM) was partially aligned with the magnetic field using an alkyl-poly(ethylene glycol) C8E5 (Sigma-Aldrich, St. Louis, MO), 1-octanol, and water mix as described by Rückert and Otting (42). We attempted to use the same system to align 1 mM 15N-K2 in the presence of 50 mM SDS, but the aligning medium is not compatible with the detergent, resulting in sample precipitation. We therefore used a stretched polyacrylamide gel to partially align the protein with the magnetic field, as outlined by (43). The in-phase, anti-phase experiments (44) were collected at 277.15 K (5% (w/w) C8E5 with 1-octanol samples) or 300 K (stretched gel) using 0.5 mM 15N-K2. The spectra contained 1024 (1HN) and 128 (15N) complex data points with a total of 16 transients per t1 increment using spectral widths of 7211.539 Hz (1HN) and 1337.991 Hz (15N). The 1H-15N residual dipolar coupling (RDC) values were calculated as the difference between the J-coupling with RDC (D + J) in the aligned sample and the J-coupling (J) in the unaligned sample.
Paramagnetic relaxation enhancement experiments—structural constraints
For the paramagnetic relaxation enhancement (PRE) experiments, 15N heteronuclear single-quantum coherence (15N-HSQC) experiments were collected with 0.65 mM of MTSL-15N-K2-T24C protein in the presence and absence of 50 mM 2H-SDS at 300 K in the oxidized and reduced states. The MTSL label was reduced by the addition of ascorbic acid, and the intensity measurements were corrected for the volume change. For all PRE experiments, 1024 (1HN) and 256 (15N) complex data points were acquired with 16 t1 increments using spectral widths of 7211.539 Hz (1HN) and 1337.925 (15N) Hz. The ratio of the intensities can be expressed as
| (1) |
where Iox and Ired are the peak intensities of the oxidized and reduced forms, R2 is the intrinsic relaxation rate, R2sp is the paramagnetic relaxation rate, and t is the duration of the INEPT transfer (5.6 ns). R2sp can be used to determine the distance between the electron spin of the nitroxide spin label and the nuclear spin of interest from (45)
| (2) |
where r is the distance between the nuclear and electronic spins, τc is the correlation time for the electron-nuclear dipole interaction (estimated from the rotational correlation time, τm, i.e., the time taken by the protein to tumble one radian), ωH is the Larmor frequency of the proton (600 MHz), and K is 1.23 × 10−23 cm6 s−2 as determined by the equation of physical constants:
| (3) |
where S is the quantum spin of an electron, γ is the gyromagnetic ratio, g is the electronic Landé factor, and β is the Bohr magnetron.
Water-amide solvent-exchange experiments
To determine which residues are protected from solvent exchange, CLEANEX-PM experiments (46) were performed on K2 in the presence and absence of 50 mM 2H-SDS at 300 K. For all CLEANEX-PM experiments, 1024 (1HN), and 128 (15N) complex data points were acquired with 16 t1 increments using spectral widths of 7211.539 Hz (1HN) and 1337.698 (15N) Hz. Mixing times of 10, 20, 30, 75, and 100 ms were used, and the water-amide exchange rates and protection factors were calculated as previously described (46, 47).
ENSEMBLE calculation of K2 structures in the presence and absence of micelles
A series of sample K2 structures were generated using TraDES (48), and plausible, distinct IDP structures in the bound and unbound states (49, 50) were extracted using ENSEMBLE 2.1 (51). In brief, structures were calculated using constraints from the chemical shifts of HN, Hα, Cα, Cβ, and C′ atoms, R2 relaxation rates, 1H-15N RDC data, PRE distance data from MTSL-labeled K2, and accessible surface area. Default parameters were used unless otherwise indicated. An initial soup of 100,000 random conformations of K2 was generated using TraDES (48), and the pool in ENSEMBLE is a selection of 5000 structures from the soup (51). The ENSEMBLE program then selected an ensemble of structures using a Monte Carlo method from the pool, and the expected observables were back-calculated from these structures (52, 53, 54). The selection and analysis process was repeated until the ensemble fitted the experimental data. Fitting of each experimental restraint is based on a semiempirical pseudo-energy function as defined in ENSEMBLE 2.1 (Table 1) and is based on the properties of the entire structure ensemble rather than on any single structure in the ensemble (55). Two final pools of 100 structures were generated for K2 alone and for K2 in the presence of SDS micelles. Previous studies have shown that the generation of a large number of structures results in an ensemble that has similar properties to that of 10–25 independent ensemble calculations (56). Constraints were added incrementally in the following order: chemical shifts, R2 data, 1H-15N RDC data, PRE distance data, and surface accessibility. Since the RDC back calculation uses a 15-residue window, the three terminal residues at both ends of the protein were omitted from the calculation. Converting protection factors into surface accessibility constraints was performed as described previously (55).
Table 1.
Goodness of Fit of ENSEMBLE Structures to Experimental Constraints
| Restraint | Target Energy Term | Target Energy Value | Calculated Energy Value |
|---|---|---|---|
| K2 alone | |||
| RDC | 15.000 | 1.326 | |
| PRE | 1.733 | 0.964 | |
| ACCESS | 6.641 | 5.099 | |
| R2 | r | 5.050 | 3.699 |
| δC′ | 11.640 | 11.611 | |
| δCα | 23.709 | 23.632 | |
| δCβ | 9.944 | 9.490 | |
| δHN | 1.414 | 1.146 | |
| δHα | 1.130 | 0.110 | |
| K2 with SDS | |||
| RDC | 10.500 | 1.659 | |
| PRE | 1.402 | 1.108 | |
| R2 | r | 5.049 | 3.393 |
| δC′ | 8.266 | 6.575 | |
| δCα | 6.269 | 6.197 | |
| δCβ | 7.321 | 6.299 | |
| δHN | 1.458 | 1.310 | |
| δHα | 0.336 | 0.331 | |
Target energy functions are defined by Marsh and Forman-Kay (53). ACCESS, accessible surface area; R2, transverse relaxation rate, δC′, carbonyl chemical shift; δCα, carbon chemical shift; δCβ, β-carbon chemical shift; δHN, amide proton chemical shift; δHα, α-proton chemical shift; N, value of the restraint; σN, average standard deviation of the restraint; r, correlation coefficient for fitting R2. Energy term units are arbitrary.
Several tools were used to analyze the final 100 structures generated by ENSEMBLE. The Ramachandran plots were created using PROCHECK-NMR (57) and the secondary structure was determined using STRIDE (58). Rg histogram plots and heavy-atom contact maps were generated using scripts written by the Forman-Kay group (http://pound.med.utoronto.ca/∼JFKlab/). The predicted Rg value for K2 was calculated using (59)
| (4) |
where Rg is the radius of gyration, R0 is a constant (1.927), N is the number of residues (48 for K2) and is the exponential scaling factor (0.598).
Cluster analysis on the final 100 structures was performed using the NMRCLUST protocol by Kelley et al. (60) as implemented in the program Chimera (61). NMRCLUST is an automated method to cluster protein structures that simultaneously minimizes the number of clusters and spread in each cluster. Clusters with only one structure were rejected on the basis of being too small for further analysis.
Probing protein-micelle interactions
To determine which residues are interacting with the detergent micelle, we measured the changes in resonance intensities from the 15N-HSQC spectra using the PRE compounds MnCl2 and 5-DOXYL-steric acid (5-DSA). The soluble Mn2+ will affect residues exposed to the solvent, whereas 5-DSA will affect residues bound to the micelle (62). The intensity of the affected residues will be attenuated by the signals from residues close to the paramagnetic probe. The MnCl2 was added at concentrations of 0.25 mM and 1 mM to 1 mM of 15N-K2. The 5-DSA was incorporated into the SDS micelles at a concentration such that, assuming an aggregation number of ∼60 for SDS (63), there would be on average one molecule of 5-DSA in every micelle. For these experiments, 1024 (1HN) and 128 (15N) complex data points were acquired, with a total of eight transients per t1 increment using spectral widths of 7211.539 Hz (1HN) and 1337.991 Hz (15N).
Docking of K2 structures to the micelle surface
Docking of K2 to SDS micelles was performed using HADDOCK 2.1 (64, 65) with a method using a constraint from the protein to the center of mass of the micelle, as described by Dancea et al. (66). This includes assigning residues that showed decreased intensity in the 15N-HSQC spectrum in the presence of the 5-DSA micelles (residues 2–11 and 37–46) at a distance of 14.8 Å (i.e., the distance from sulfhydryl S to C4 in the SDS molecule, which is the equivalent distance of the carboxyl C to the 5-doxyl position in 5-DSA) to the geometric center of the micelle. The center of the micelle was defined as an uncharged pseudoatom in the micelle structure. Passive residues were not defined. All 67 SDS bound structures from the most populated ENSEMBLE cluster (Table 2) were used in the docking exercise; only the core K-segments (equivalent to residues 6–15 in the K-segment motif) were used for docking, since the data show that the ϕ-segment is extremely flexible and not involved in the interaction (see Fig. S1 in the Supporting Material). Preequilibrated SDS micelle coordinates were kindly provided by Dr. A. MacKerell (67). The topology and parameter files were generated using PRODRG (68), and the charges were obtained from the molecular dynamics work on SDS micelles by Bruce et al. (69). The numtrees.cns script was modified to increase numtrees by 60 in each iteration because of the number of SDS molecules in the docking calculation, and the SDS residue was defined in the covalions.cns script.
Table 2.
ENSEMBLE Clusters
| Cluster | Fractional Population |
|---|---|
| K2 alone | |
| 1 | 0.52 |
| 2 | 0.46 |
| 3 | 0.02 |
| K2 with SDS | |
| 1 | 0.67 |
| 2 | 0.10 |
| 3 | 0.08 |
| 4 | 0.08 |
| 5 | 0.05 |
Clusters are sorted in order of decreasing fractional population.
In the docking calculation, side-chain flexibility was allowed while the backbone was kept rigid to maintain consistency with the experimental data used to generate the ENSEMBLE structures. Models were selected for further analysis by clustering the results using a 5 Å root mean-square deviation cut-off and a cluster size cut-off of 10 structures based on the lowest HADDOCK score (70). The structures of all 670 docked models were evaluated for electrostatic contacts with the sulfhydryl oxygen atoms using a modification of the HADDOCK “print_hbonds.inp” analysis scripts to look for atoms that are within hydrogen-bonding distance (≤2.5 Å between donor and acceptor) of the oxygen atoms in the SDS headgroup. Hydrophobic contacts (<4 Å) were evaluated using the “print_nb.inp” analysis scripts. All SDS molecules were treated as equivalent for the analysis.
Results
Interaction of K2 with a micellar membrane
Our goal was to determine the structure of K2 when bound to the membrane using several biophysical techniques, including solution-state NMR. NMR studies on membrane-bound proteins can be challenging due to the large molecular weight of the liposome, which causes a dramatic decrease in the tumbling rate of the protein. SDS micelles, with their smaller size, are especially suitable to the study of membrane proteins by NMR (71, 72). This smaller size also causes less light scattering when using spectrophotometric techniques. To that end, we used SDS micelles as a model membrane, since they are regarded as a good substitute to mimic the interactions that occur between proteins and lipids (73).
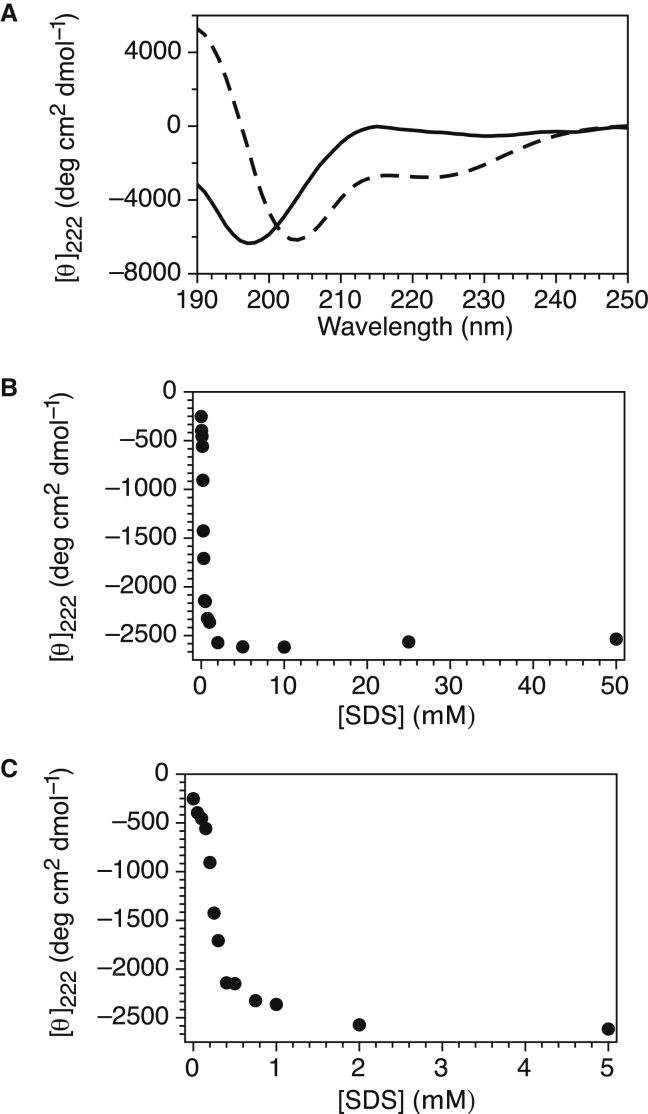
We acquired the CD spectrum in the presence and absence of SDS micelles (Fig. 1 A) to see if the structural changes of K2 upon binding were similar to those previously observed in the presence of liposomes (11). The 50 mM SDS concentration represents the same amount of detergent used in the NMR experiments described below. The K2 signal minimum shifted from 198 to 204 nm, and a significant gain in the CD signal was observed at 222 nm. This shows that K2 has lost ∼15% coil structure and gained ∼10% α-helicity in the presence of micelles (74). To examine this change more closely, a titration was performed by adding increasing amounts of SDS and monitoring the gain in helicity by plotting the CD signal at 222 nm (Fig. 1 B). An expanded view of the graph at low SDS concentrations (Fig. 1 C) shows that initially (between 0 and 0.2 μM SDS), there was a small gain of helicity, whereas between 0.2 and 0.4 μM of SDS there was a large gain. Next, a minimal gain of α-helicity was observed between 0.4 mM and 2 mM SDS, which may represent a shift of the K2 more toward the bound state. After 2 mM, the change in helicity becomes nearly asymptotic.
Figure 1.
The K2 dehydrin gains helicity upon binding to micelles below the CMC of SDS. (A) CD spectrum of K2 in the absence (solid line) and presence (dashed line) of 50 mM SDS. (B) Gain of secondary structure in the presence of SDS. The relative amount of α-helicity was determined by measuring the molar ellipticity of K2 at 222 nm, and the SDS concentration was varied between 0 and 50 mM. (C) Expanded view of (B) between 0 and 5 mM SDS.
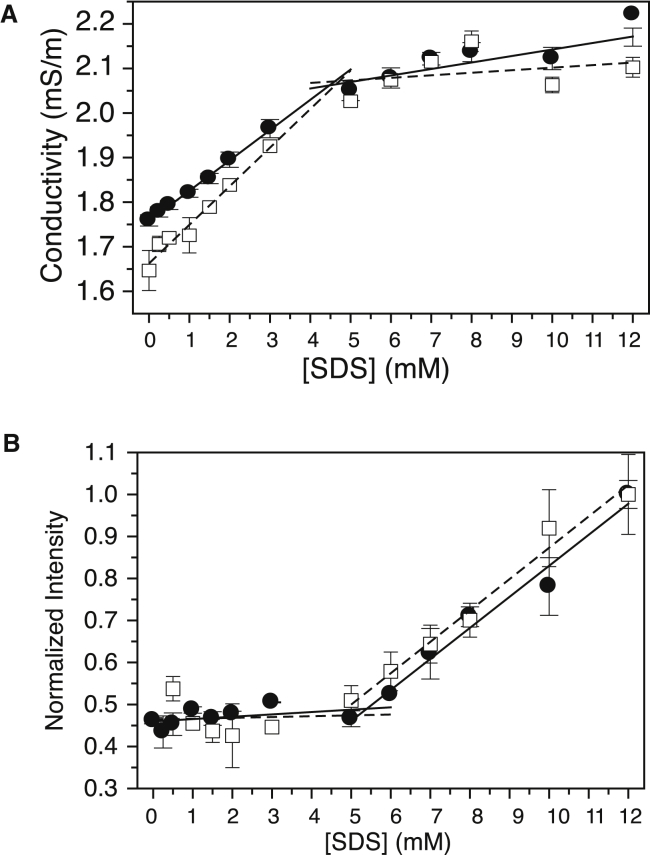
The typically reported critical micelle concentration (CMC) should be on the order of 5 mM in our buffer system (75), yet the results in Fig. 1 C show that the largest amount of structural gain occurred between 0.2 and 0.4 mM before reaching a shallower slope. In the work by Südhof et al. with α-synuclein (76), a very similar pattern by CD was observed, such that they speculated that this IDP was lowering the CMC of the detergent. We tested this proposal by measuring the CMC of SDS in the presence of 10 μM K2 (i.e., a protein concentration similar to that used in the CD experiments) using two separate techniques (conductivity and ANS fluorescence). In both cases, a sudden change in slope represents the change in SDS going from a premicellar to a micellar state, and the intercept of the two slopes gives the CMC. In the absence of K2, the two methods (conductivity and fluorescence (Fig. 2, A and B, respectively, solid circles) gave similar CMC values of ∼5 mM. In the presence of K2 (Fig. 2, A and B, open squares), the measured CMC values from the two techniques are virtually identical (∼5 mM). This proves that K2 has not altered the CMC of SDS, and that the plateauing effect of the helical content after 0.4 mM of SDS is due to another phenomenon.
Figure 2.
K2 does not alter the CMC of SDS. (A) CMC of SDS in the presence of K2 as measured by conductivity. The measurements were made in the presence (open squares and dashed lines) and absence (closed circles and solid lines) of K2. The lines represent linear fits before and after the transition points of the slopes. (B) CMC of SDS in the presence of K2 as measured by the change in ANS fluorescence. Symbols and lines are the same as in (A).
Structure of K2 in the presence and absence of micelles
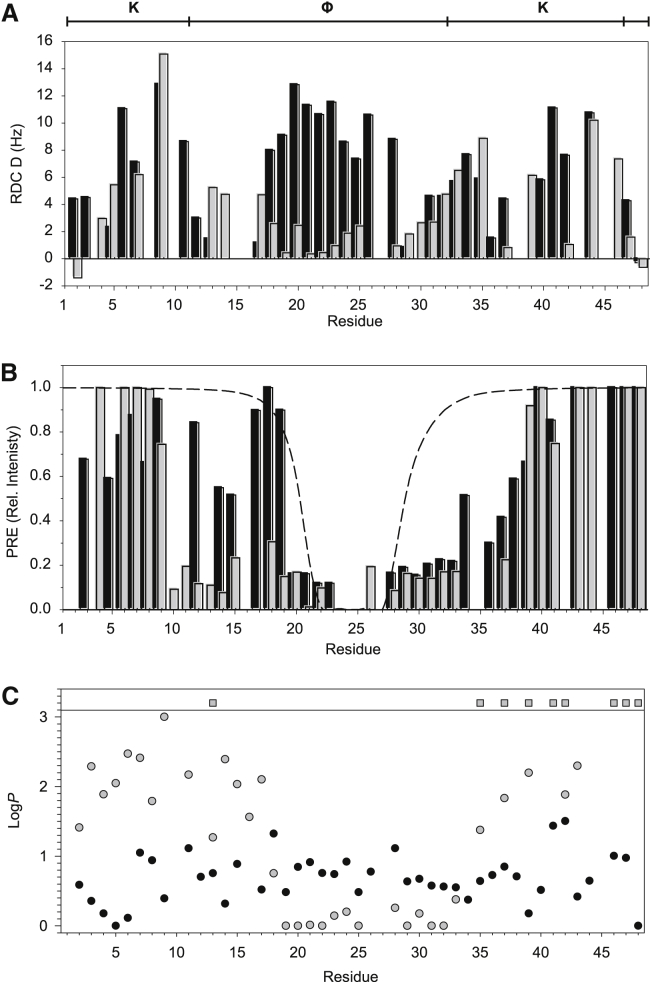
We next sought to determine the structure of K2 in the presence and absence of SDS micelles. Ensemble descriptions (51) based on NMR data can be used to calculate possible structures that an IDP can sample in both the presence and absence of a ligand. The chemical shifts of K2 alone have been previously measured and are available under Biological Magnetic Resonance Data Bank (BMRB) entry 16445 (32), whereas the chemical shifts for K2 in the presence of SDS are available under BMRB entry 26554 (11), as are relaxation data for both states (11). In addition to the chemical shift assignments and relaxation data, we also collected RDC (1H-15N RDC, Fig. 3 A), PRE data (Fig. 3 B), and solvent-exchange factors (Fig. 3 C) to see what structures K2 could acquire. For K2 alone and in the presence of SDS, the PRE data showed very few contacts between the probe (located at Cys24) and residues in the K-segments, and between the probe and the N-terminal end of the ϕ-segment (Fig. 3 B, black bars). Several contacts closer than that predicted by a purely random coil structure are observed between the probe and residues 29–32, with possibly weaker contacts up to residue 39. In the SDS sample (Fig. 3 B, gray bars), several PRE contacts can be seen within the ϕ-segment, including residues 10–20 and 29–37. For the solvent-exchange data (Fig. 3 C), there is little observed protection over any part of the protein in the absence of SDS. In the presence of SDS, there is some protection of residues around the first K-segment, with significant protection observed for residues in the second K-segment (Fig. 3 C). RDCs are important for ensemble calculations, because they provide local and long-range orientation information (77). Note that because the RDC experiments with K2 alone and K2 in the presence of SDS needed to be collected in two different alignment media (see Materials and Methods for details), a direct comparison of the two results is not easily possible. The RDC data can still be used in the structure calculation despite this difference.
Figure 3.
RDC and PRE data of bound and unbound K2. (A) A plot of RDC values. The diagram at the top of the figure shows the location of the various segments in the K2 sequence. (B) A plot of the PRE caused by Cys24-MTSL. The intensity of the signal is normalized to that in the unbound spectrum. The dashed line represents the theoretical intensity of a random-coil structure (38). In (A) and (B), the black bars represent free K2 and the gray bars bound K2. (C) Protection factors of amide residues. The exchange rates of backbone amide groups were converted to protection factors. Black circles represent free K2 and gray symbols bound K2. Circles represent calculated protection factors and squares represent protection factors that could not be accurately quantified.
The PRE, RDC, chemical shift, and relaxation data were used to calculate a series of potential K2 structures with and without SDS present. This was performed using the ENSEMBLE program (51) and its submodules (48, 52, 54). The target energy term, target energy value, and calculated energy value of the ENSEMBLE structures for each experimental parameter are shown in Table 1; optimal target energy values are based on empirical analyses of previously calculated structures (56). For our runs, all restraints were simultaneously satisfied over the whole ensemble structure.
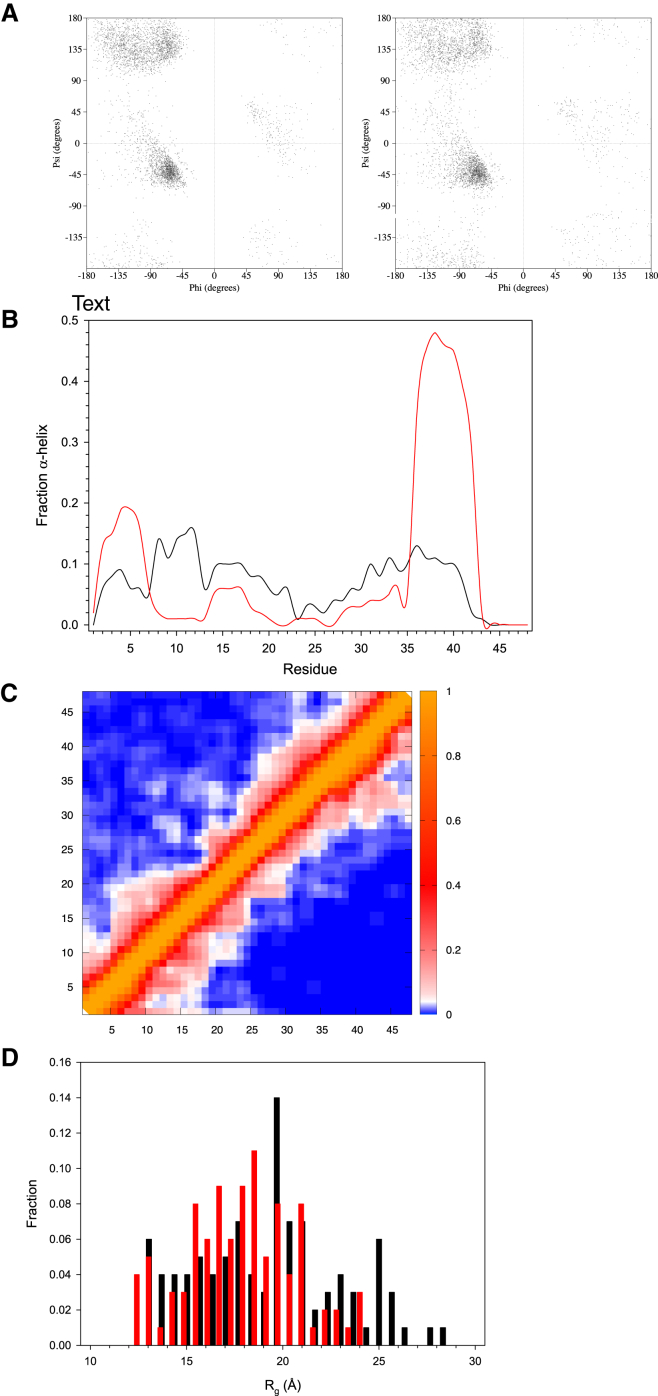
We examined differences in secondary structure between K2-alone and SDS-bound-K2 ENSEMBLE structures. Cluster analysis shows that the K2-alone structure results in several structure clusters, whereas in the presence of SDS, 67 of the 100 structures group into one cluster (Table 2). The Ramachandran plots (Fig. 4 A) show that residues occupy all of the secondary structure space, with the SDS-bound form showing a greater preference for α-helical space compared to unbound K2. This is also reflected in the fraction of SDS-bound K2 residues that are in an α-helical structure (Fig. 4 B). For K2 alone, weak α-helical structure was observed at the N-terminus and nearer the C-terminus, where both regions represent the two K-segments. In the presence of micelles, the N-terminal K-segment gains nearly 20% α-helicity at its peak, whereas the second C-terminal K-segment gains nearly 50% α-helicity. Both are in agreement with previous NMR experiments on K2 (11). Zero β-strand structure was observed for either condition (K2 alone or in the presence of micelles).
Figure 4.
Differences in the structure of K2 in free and SDS-bound states. (A) Ramachandran plots of all residues in the final ensemble of 100 structures for K2 alone (left) and K2 in the presence of SDS (right). (B) Fraction of α-helical structures on a per-residue basis for K2 alone (black line) and K2 in the presence of SDS (red line). (C) Fractional contact plot map of the final 100 structures. Residues are said to be in contact if a heavy atom of one residue is within 6 Å of another heavy atom. The fractional color scale is shown to the right of the map. The map above the diagonal represents the results for K2 alone and that below the diagonal represents the results in the presence of SDS. (D) Radius of gyration of the final 100 structures, shown as a histogram plot, for K2 alone (black bars) and K2 in the presence of SDS (red bars). To see this figure in color, go online.
Subsequently, differences in tertiary structure were inspected using fractional contact maps and plots of the radius of gyration (Rg). Over the whole protein, the K2-alone contact map (Fig. 4 C, upper) shows a few local contacts (<5 residues distant), with a very small number of long-range contacts (>10 residues distant). In the presence of SDS, the contact map has changed subtly, but contains similar local contacts (Fig. 4 C, lower). Differences in the presence of SDS include a lack of long-range contacts and a gain of more consistent medium-range contacts in residues 37–44, likely a result of the formation of α-helix in this region (see Fig. 4 B). Differences in tertiary structure can also be seen in the radius of gyration (Rg, Fig. 4 D). If K2 were to behave as a random coil, one would expect an Rg of 19.5 Å (59). The average observed values in the structure prediction are 19 ± 4 Å for K2 alone and 17 ± 2 Å for the SDS-bound K2. Both conditions have starting Rg values of ∼13 Å. K2 alone shows more structures at the upper range of values, with some structures ranging out to 28 Å, whereas the bound form has no structure with an Rg value of >23 Å.
Only K-segment residues are involved in membrane binding
The docking of K2 to micelles, described in detail below, required us to split the protein structure into K-segment 1 and K-segment 2. To justify this approach, we present several lines of evidence from previously published work (11) that it is only the K-segments that bind SDS. The first piece of evidence is the HN, Hα, Cα, Cβ, C′, and backbone N chemical shifts of K2 in the presence of SDS, which were used as input for the program δ2Δ (78). The result (Fig. S1 A) shows that the largest gain in α-helicity occurs in residues 3–9 and 34–44, which represents residues in the middle of the K-segment. Likewise, R2 relaxation data (Fig. S1 B) demonstrate that residues 2–12 and 36–45, again covering the K-segments, are fairly rigid in the presence of SDS. We also mapped the exchange rates of residues in K2 with SDS (Fig. S1 C). Residues in the K-segment are colored blue, which shows that the exchange rate in those regions is occurring on a slow timescale. Taken together, all three of these results demonstrate that it is the K-segments that are interacting with the SDS micelles, and that residues outside these two regions have no role in binding.
Structure of K2 bound to a micelle
To determine what the structure of K-segments might look like when bound to a micelle, we confirmed which residues in the K-segments were interacting with the SDS molecules and which residues were interacting with the solvent. To do this, we followed the disappearance of 1H-15N resonances in the 15N-HSQC spectra due to PRE through the use of two different paramagnetic probes, Mn2+ and 5-DSA (62). Mn2+ is a soluble metal cation, so signals attenuated in its presence show which HN atoms are interacting with the solvent. 5-DSA is a fatty acid that can be readily incorporated into SDS micelles; this molecule contains a free radical at the 5-carbon position that can be used to probe which residues are interacting with the acyl chain just below the sulfate headgroup of SDS.
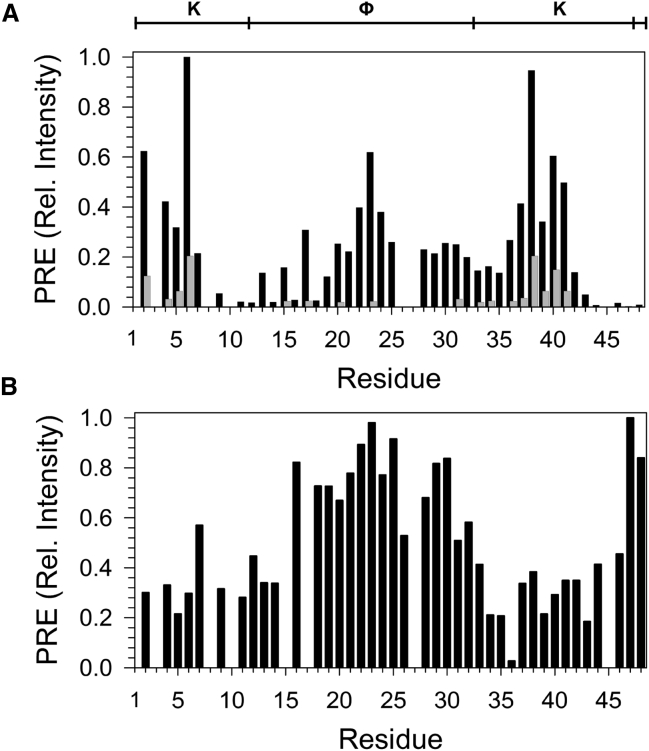
In Fig. 5 A, two different concentrations of MnCl2 were added (0.25 mM (black bars) and 1 mM (gray bars)) to see how resonances disappeared with low and high concentrations of this paramagnetic agent. At the lower concentration, nearly all of the resonance intensities decreased, which is likely a reflection of the binding interaction being not very strong (16 μM (11)) and the K2, therefore, coming on and off the membrane surface and/or changing conformation at an appreciable rate. Nonetheless, Lys6 and Lys39 showed almost no decrease in intensity. The K-segments showed some protection, where several residues (Lys2, Lys4, Lys6, Glu38, Lys39, Ile40, and Lys41) showed >0.4 fractional intensity compared to the control sample. Residues 22–24, located in the ϕ-segment, showed a moderate amount of protection, which is surprising since the NMR chemical shift and relaxation data showed that there was no interaction with the surface. This may be only a weak interaction or some kind of nonspecific sequestration near the surface, since the addition of 1 mM MnCl2 resulted in essentially no signal in this region, whereas residues Lys2, Lys6, Lys39, and Lys41 in the K-segments still showed 0.15–0.20 fractional intensity compared to the control. This interpretation is also supported by the water-amide exchange data (Fig. 3 C), where in the presence of micelles, residues in the K-segment show better protection from exchange than residues in the ϕ-segment. To corroborate the identity of the surface-exposed residues, we also measured the changes in signal intensity of K2 in the presence of micelles containing 5-DSA (Fig. 5 B). Residues in the ϕ-segment (12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30) showed very little loss in intensity, whereas residues in the K-segments (residues 34–43) showed <0.4 intensity of the control. Taken together, this provides further evidence that the K-segments are involved in membrane binding. The results from both paramagnetic probes indicate that the pair of Lys residues in the center of the K-segments have the strongest binding interactions.
Figure 5.
Lysine residues in the K-segment are key to SDS micelle binding. (A) PRE of signals in the 15N-HSQC in the presence of MnCl2. Signal intensities were normalized to those in the absence of the paramagnetic agent. Black bars represent 0.25 mM MnCl2 and gray bars 1 mM MnCl2. (B) PRE of signals in the 15N-HSQC in the presence of 5-doxyl stearate. Signal intensities were normalized to those in the absence of the paramagnetic agent. The diagram at the top of the figure shows the location of the various segments in the K2 sequence.
In silico docking of the dehydrin to the micelles was performed to analyze interactions between the side chains and the SDS molecules. Using the software program HADDOCK (64, 65), the docking problem was approached by dividing the protein into two parts (the core of each K-segment (residues 2–11 and 37–46), which has been numbered Met6–Gly15 in the subsequent analysis to simplify their description). This could be done since the NMR data showed that the ϕ-segment would not be required for docking (see Fig. S1). In addition, HADDOCK runs with the ϕ-segment present often caused ϕ-segment residues to be in contact with the micelle surface despite not being defined in HADDOCK as active residues (data not shown). Structural integrity of the distance between the two K-segment fragments was maintained a posteriori by only selecting models where the distance between the end of the first K-segment and the beginning of the second K-segment is in the same range as those distances found in the representative models of the ENSEMBLE calculated structures. Cluster analysis on the 100 ENSEMBLE generated structures in the presence of SDS was performed as described in Materials and Methods. The results showed that 67 of these structures were similar (Table 2), and all 67 structures were used in the HADDOCK docking calculation.
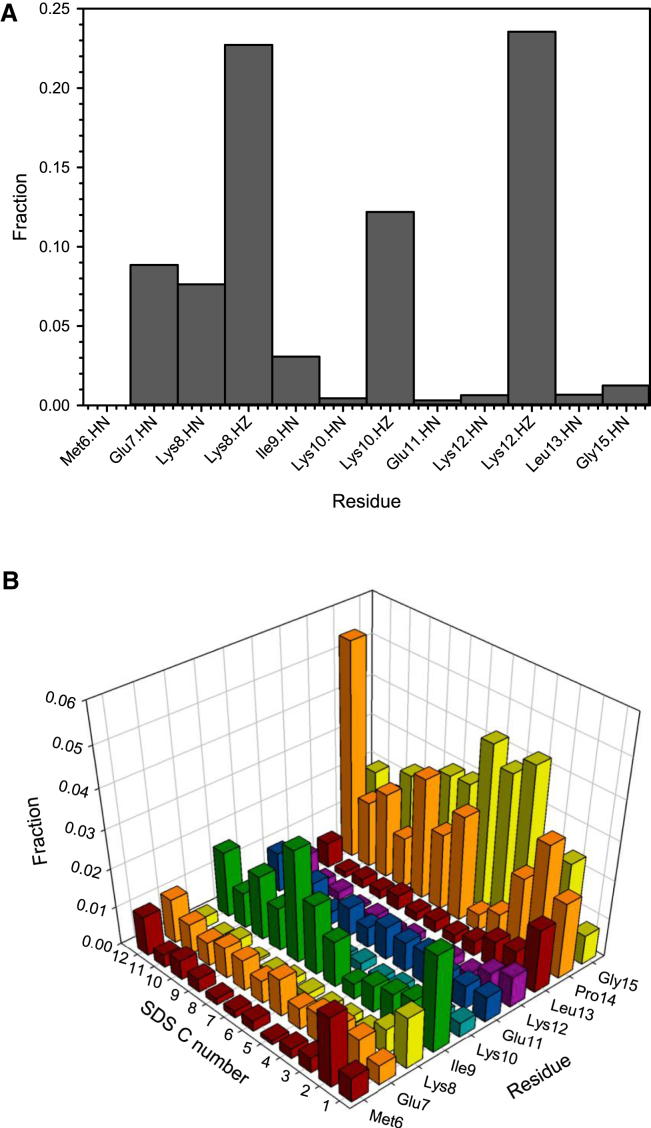
Further analysis was performed to determine which interactions (electrostatic bonds, hydrogen bonds, or hydrophobic interactions) could be involved in the binding interaction. The structures were analyzed for functional groups that are capable of interacting with the sulfate headgroup through hydrogen bonds or electrostatic interactions. Fig. 6 A counts which HN groups could form hydrogen bonds and which HZ groups from Lys could form electrostatic bonds with the sulfate. Electrostatic interactions dominate; both Lys8.HZ and Lys12.HZ interactions are found in nearly 25% of all of the docked models, whereas Lys10.HZ interactions appear half as frequently. For hydrogen bonds, the most common interactions are nearer the N-terminus of the K-segment, with Glu7.HN and Lys8.HN found in 7–8% of the interactions. Other functional groups do not appear to make notable contributions. The ensemble structures were similarly analyzed for possible hydrophobic contacts between functional groups in the K-segment and micelles. Unlike hydrogen and electrostatic bonds, hydrophobic contacts are less frequently seen for residues 37–44 (4–10%), with the exception of Ile9, which makes several contacts at the lower end of the SDS tail. Although less hydrophobic, Pro14 and Gly15, found in 27% and 31%, respectively, of all of the docked models, make many contacts with the SDS tail.
Figure 6.
Micelle-bound K2 shows interactions between the lysine amide and aliphatic groups with SDS. (A) Hydrogen bonds and electrostatic interactions between the K-segment and micelles. Analysis was performed as described in Materials and Methods. (B) Hydrophobic interactions between the K-segment and micelles. Analysis was performed as described in Materials and Methods. The SDS C number axis shows which aliphatic carbon is in a potential contact, with carbon 1 being the carbon attached to the sulfate group. To see this figure in color, go online.
Discussion
The docking of K2 to the micelle surface can explain why at some positions in the K-segment, specific amino acids are conserved and hence functionally important for membrane binding. As suggested in Fig. 6 A, the electrostatic interactions from Lys8 and Lys12 are the ones most consistently seen in the docked models, with interactions from Lys10 occurring at half the frequency. In the K-segment, lysine residues are highly conserved at positions 8, 10, and 12 (Malik et al., unpublished data). The lack of arginine at these positions may relate to the recently observed phenomenon that charged amino groups strengthen hydrophobic interactions, whereas guanidinium groups eliminate them (79). The negatively charged aspartate and glutamate residues at positions 7 and 11 make few contacts with the micelle; their role may be to help orient the lysine and hydrophobic residues toward the membrane surface (see the helical wheel in Fig. 3 F in (11)). The role of Lys and Glu residues is supported by the observation that dehydrins prefer to bind to negatively charged lipids rather than neutral or positively charged ones (30). In contrast, hydrophobic interactions appear to be less significant contributors to the docked interactions. Ile9, a highly conserved hydrophobic residue, does make several contacts with the acyl tails, generally at the seventh carbon or higher (Fig. 6 B). Despite isoleucine's highly hydrophobic nature, more frequent contacts are observed with Pro14 and Gly15 (i.e., the end of the K-segment). These residues are also highly conserved in the K-segment, but their function is not known. We propose that they are important for helping the K-segment to bind a membrane surface, though proof requires further experimental data.
The docking interaction also provides an example of a fuzzy complex that forms between an IDP and its ligand (80); the ϕ-segment remains fully disordered when K2 is interacting with the micelle. Fig. S1 A shows that residues flanking the center of the K-segment are 80–95% helical compared to ∼100% for the middle lysine, whereas Fig. S1 B shows that the same residues have more flexibility compared to the middle lysine. These two observations suggest that the bound K-segments are sampling other possible conformations that still allow for membrane binding.
An analysis of the ENSEMBLE structures of K2 in the unbound and micelle-bound forms (Fig. 4) shows that the largest changes occurred in the secondary structure, without major changes in the tertiary contacts. Nevertheless, the Rg plot suggests that the K2 alone structure can sample larger radii of gyration than the SDS-bound form. We propose that this difference allows for dehydrins to function optimally in their protective roles. The K2 maintains its disorder in the unbound form so that it can act as a molecular shield for enzyme cryoprotection (i.e., maintain its large hydrodynamic radius) (8, 31), yet gain some structure in its membrane-bound form so that it can optimally interact with the membrane surfaces and protect them from cold-stress damage (11). This represents an example of an IDP “moonlighting” (performing more than one function), using its disorder as an advantage in one case and its ability to gain structure in another (81).
Like many other IDPs, K2 gains structure in the presence of a ligand. As shown in Fig. 1 B, K2 gains more α-helicity as more SDS is initially added, followed by a plateau where the subsequent addition of more SDS results in a minor gain in helicity. This is unusual, since the CMC of SDS under these conditions would be expected to be on the order of 5 mM (75), yet the largest change happens before 0.4 mM SDS (Fig. 1 C). Below the CMC, one would expect that free SDS molecules would be unable to support the gain in α-helicity (i.e., a surface would be required to support the secondary structure). A very similar observation was made for α-synuclein, where the authors speculated that this protein may be lowering the CMC (76); in the case of α-synuclein, it was subsequently shown that the protein is able to decrease the CMC approximately 10-fold (82). Fig. 2 demonstrates that this is not the case for the K2 dehydrin. Comparing these two proteins is complicated by the observation that SDS induces α-synuclein fibrillization, which may alter micelle formation in a way that monomeric K2 would not. Support for some IDPs gaining structure below the critical micelle concentration comes from a study with amelogenin (83). This protein shares some broad structural similarity with K2 in that the N- and C termini gain α-helical structure, whereas the centers of the proteins, which are rich in Gly, show no change.
Many studies have suggested that SDS molecules can associate and form a structure at submicellar concentrations (see, for example, Hadgiivanova et al. (84)). The interaction of acyl coenzyme A binding protein at submicellar concentrations was examined using small-angle x-ray scattering (85). At moderate, but still submicellar, concentrations, the detergent could be modeled as metastable aggregates around the protein; these metastable aggregates have also been observed in pure SDS solutions (84). This has been described as the difference between the macroscopic CMC (which we observed with our methods, see Fig. 2) versus microscopic CMC (which we would not observe). In the case of K2, it is possible that these effects are combined. First, K2 may stabilize the aggregates through the numerous positively charged Lys residues, and these microscopic CMC micelles in turn are providing a stable enough surface for the K-segments to fold into α-helices, all the while leaving the macroscopic CMC unaffected.
Author Contributions
J.A. and S.P.G. designed the study and wrote the article. M.W.C. and K.F.B. collected the CD data. J.M.W. and J.A. collected the NMR data and J.A. and S.P.G. calculated the structures. S.P.G. performed the in silico docking. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
The authors thank the University of Guelph NMR Centre for use of the facility and Sameer Al-Abdul-Wahid for running the CLEANEX-PM pulse sequence, the Merrill laboratory for use of the CD spectropolarimeter, the Wood laboratory for the use of the conductivity meter, and the Sharom laboratory for use of the fluorometer. They also thank Julie Forman-Kay, Mickaël Krzeminski, and Joseph A. Marsh for providing and troubleshooting the ENSEMBLE program and analysis tools, and Christopher Hogue for providing and troubleshooting the TraDES scripts and programs.
This work was supported by a Natural Sciences and Engineering Research Council Discovery Grant to S.P.G. Infrastructure support to S.P.G. comes from the Canadian Foundation for Innovation and the Ontario Innovation Trust.
Editor: David Eliezer.
Footnotes
One figure is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30515-X.
Supporting Material
References
- 1.Close T.J. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 1996;97:795–803. [Google Scholar]
- 2.Close T.J. Dehydrins: a commonalty in the response of plants to dehydration and low temperature. Physiol. Plant. 1997;100:291–296. [Google Scholar]
- 3.Allagulova ChR., Gimalov F.R., Vakhitov V.A. The plant dehydrins: structure and putative functions. Biochemistry (Mosc.) 2003;68:945–951. doi: 10.1023/a:1026077825584. [DOI] [PubMed] [Google Scholar]
- 4.Kosová K., Vítámvás P., Prášil I.T. Expression of dehydrins in wheat and barley under different temperatures. Plant Sci. 2011;180:46–52. doi: 10.1016/j.plantsci.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson S.K., Harryson P. Dehydrins: molecular biology, structure and function. In: Lüttge U., Beck E., Bartels D., editors. Plant Desiccation Tolerance. Springer; Berlin, Germany: 2011. pp. 289–305. [Google Scholar]
- 6.Graether S.P., Boddington K.F. Disorder and function: a review of the dehydrin protein family. Front. Plant Sci. 2014;5:576. doi: 10.3389/fpls.2014.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X., Rikkerink E.H.A., Uversky V.N. Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell. 2013;25:38–55. doi: 10.1105/tpc.112.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes S., Graether S.P. Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 2011;20:42–50. doi: 10.1002/pro.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara M., Shinoda Y., Kuboi T. DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ. 2009;32:532–541. doi: 10.1111/j.1365-3040.2009.01947.x. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson S.K., Kutzer M., Harryson P. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell. 2011;23:2391–2404. doi: 10.1105/tpc.111.085183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke M.W., Boddington K.F., Graether S.P. Structural and functional insights into the cryoprotection of membranes by the intrinsically disordered dehydrins. J. Biol. Chem. 2015;290:26900–26913. doi: 10.1074/jbc.M115.678219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisniewski M., Webb R., Griffith M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: a dehydrin from peach (Prunus persica) Physiol. Plant. 1999;105:600–608. [Google Scholar]
- 13.Goday A., Jensen A.B., Pagès M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell. 1994;6:351–360. doi: 10.1105/tpc.6.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danyluk J., Perron A., Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1998;10:623–638. doi: 10.1105/tpc.10.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara M., Terashima S., Kuboi T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta. 2003;217:290–298. doi: 10.1007/s00425-003-0986-7. [DOI] [PubMed] [Google Scholar]
- 16.Tunnacliffe A., Wise M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- 17.Battaglia M., Olvera-Carrillo Y., Covarrubias A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hincha D.K., Thalhammer A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 2012;40:1000–1003. doi: 10.1042/BST20120109. [DOI] [PubMed] [Google Scholar]
- 19.Dunker A.K., Lawson J.D., Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 20.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 21.Uversky V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perdiguero P., Barbero M.C., Collada C. Novel conserved segments are associated with differential expression patterns for Pinaceae dehydrins. Planta. 2012;236:1863–1874. doi: 10.1007/s00425-012-1737-4. [DOI] [PubMed] [Google Scholar]
- 23.Koag M.-C., Wilkens S., Close T.J. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009;150:1503–1514. doi: 10.1104/pp.109.136697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlad F., Turk B.E., Merlot S. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008;55:104–117. doi: 10.1111/j.1365-313X.2008.03488.x. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X., Wang Y. β-Elimination coupled with tandem mass spectrometry for the identification of in vivo and in vitro phosphorylation sites in maize dehydrin DHN1 protein. Biochemistry. 2004;43:15567–15576. doi: 10.1021/bi0483965. [DOI] [PubMed] [Google Scholar]
- 26.Alsheikh M.K., Svensson J.T., Randall S.K. Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant Cell Environ. 2005;28:1114–1122. [Google Scholar]
- 27.Alsheikh M.K., Heyen B.J., Randall S.K. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 2003;278:40882–40889. doi: 10.1074/jbc.M307151200. [DOI] [PubMed] [Google Scholar]
- 28.Liu C.-C., Li C.-M., Yang C.-P. Genome-wide identification and characterization of a dehydrin gene family in poplar (Populus trichocarpa) Plant Mol. Biol. Report. 2012;30:848–859. [Google Scholar]
- 29.Lisse T., Bartels D., Jaenicke R. The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. Biol. Chem. 1996;377:555–561. doi: 10.1515/bchm3.1996.377.9.555. [DOI] [PubMed] [Google Scholar]
- 30.Koag M.-C., Fenton R.D., Close T.J. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003;131:309–316. doi: 10.1104/pp.011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes S.L., Schart V., Graether S.P. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013;163:1376–1386. doi: 10.1104/pp.113.226803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Findlater E.E., Graether S.P. NMR assignments of the intrinsically disordered K2 and YSK2 dehydrins. Biomol. NMR Assign. 2009;3:273–275. doi: 10.1007/s12104-009-9192-2. [DOI] [PubMed] [Google Scholar]
- 33.Szalainé Ágoston B., Kovács D., Perczel A. Full backbone assignment and dynamics of the intrinsically disordered dehydrin ERD14. Biomol. NMR Assign. 2011;5:189–193. doi: 10.1007/s12104-011-9297-2. [DOI] [PubMed] [Google Scholar]
- 34.Ismail A.M., Hall A.E., Close T.J. Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol. 1999;120:237–244. doi: 10.1104/pp.120.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovacs D., Kalmar E., Tompa P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman L.N., McKay F., Dutcher J.R. Interactions of Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 with membranes at cold and ambient temperatures-surface morphology and single-molecule force measurements show phase separation, and reveal tertiary and quaternary associations. Biochim. Biophys. Acta. 2013;1828:967–980. doi: 10.1016/j.bbamem.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 37.Soulages J.L., Kim K., Cushman J.C. Conformation of a group 2 late embryogenesis abundant protein from soybean. Evidence of poly (L-proline)-type II structure. Plant Physiol. 2003;131:963–975. doi: 10.1104/pp.015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teilum K., Kragelund B.B., Poulsen F.M. Transient structure formation in unfolded acyl-coenzyme A-binding protein observed by site-directed spin labelling. J. Mol. Biol. 2002;324:349–357. doi: 10.1016/s0022-2836(02)01039-2. [DOI] [PubMed] [Google Scholar]
- 39.Wishart D.S., Bigam C.G., Sykes B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 40.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 42.Rückert M., Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- 43.Chou J.J., Gaemers S., Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 44.Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 45.Battiste J.L., Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 46.Hwang T.L., van Zijl P.C., Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 47.Ono Y.-I., Miyashita M., Nishimura C. Comparison of residual α- and β -structures between two intrinsically disordered proteins by using NMR. Biochim. Biophys. Acta. 2015;1854:229–238. doi: 10.1016/j.bbapap.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Feldman H.J., Hogue C.W. A fast method to sample real protein conformational space. Proteins. 2000;39:112–131. [PubMed] [Google Scholar]
- 49.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittag T., Marsh J., Forman-Kay J.D. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krzeminski M., Marsh J.A., Forman-Kay J.D. Characterization of disordered proteins with ENSEMBLE. Bioinformatics. 2013;29:398–399. doi: 10.1093/bioinformatics/bts701. [DOI] [PubMed] [Google Scholar]
- 52.Neal S., Nip A.M., Wishart D.S. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 53.García De La Torre J., Huertas M.L., Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marsh J.A., Baker J.M.R., Forman-Kay J.D. Calculation of residual dipolar couplings from disordered state ensembles using local alignment. J. Am. Chem. Soc. 2008;130:7804–7805. doi: 10.1021/ja802220c. [DOI] [PubMed] [Google Scholar]
- 55.Marsh J.A., Forman-Kay J.D. Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J. Mol. Biol. 2009;391:359–374. doi: 10.1016/j.jmb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Marsh J.A., Forman-Kay J.D. Ensemble modeling of protein disordered states: experimental restraint contributions and validation. Proteins. 2012;80:556–572. doi: 10.1002/prot.23220. [DOI] [PubMed] [Google Scholar]
- 57.Laskowski R.A., Macarthur M.W., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 58.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 59.Kohn J.E., Millett I.S., Plaxco K.W. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley L.A., Gardner S.P., Sutcliffe M.J. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng. 1996;9:1063–1065. doi: 10.1093/protein/9.11.1063. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Singarapu K.K., Tonelli M., Markley J.L. Structural characterization of Hsp12, the heat shock protein from Saccharomyces cerevisiae, in aqueous solution where it is intrinsically disordered and in detergent micelles where it is locally α-helical. J. Biol. Chem. 2011;286:43447–43453. doi: 10.1074/jbc.M111.306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quina F.H., Nassar P.M., Bales B.L. Growth of sodium dodecyl sulfate micelles with detergent concentration. J. Phys. Chem. 1995;99:17028–17031. [Google Scholar]
- 64.Dominguez C., Boelens R., Bonvin A.M.J.J. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 65.de Vries S.J., van Dijk A.D.J., Bonvin A.M.J.J. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 66.Dancea F., Kami K., Overduin M. Lipid interaction networks of peripheral membrane proteins revealed by data-driven micelle docking. Biophys. J. 2008;94:515–524. doi: 10.1529/biophysj.107.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacKerell A.D. Molecular dynamics simulation analysis of a sodium dodecyl sulfate micelle in aqueous solution: decreased fluidity of the micelle hydrocarbon interior. J. Phys. Chem. 1995;99:1846–1855. [Google Scholar]
- 68.Schüttelkopf A.W., van Aalten D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 69.Bruce C.D., Berkowitz M.L., Forbes M.D.E. Molecular dynamics Simulation of sodium dodecyl sulfate micelle in water: micellar structural characteristics and counterion distribution. J. Phys. Chem. B. 2002;106:3788–3793. [Google Scholar]
- 70.van Zundert G.C.P., Bonvin A.M.J.J. Modeling protein-protein complexes using the HADDOCK webserver “modeling protein complexes with HADDOCK”. Methods Mol. Biol. 2014;1137:163–179. doi: 10.1007/978-1-4939-0366-5_12. [DOI] [PubMed] [Google Scholar]
- 71.Buchko G.W., Rozek A., Kennedy M.A. The use of sodium dodecyl sulfate to model the apolipoprotein environment. Evidence for peptide-SDS complexes using pulsed-field-gradient NMR spectroscopy. Biochim. Biophys. Acta. 1998;1392:101–108. doi: 10.1016/s0005-2760(98)00028-9. [DOI] [PubMed] [Google Scholar]
- 72.Eliezer D., Kutluay E., Browne G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 73.Tulumello D.V., Deber C.M. SDS micelles as a membrane-mimetic environment for transmembrane segments. Biochemistry. 2009;48:12096–12103. doi: 10.1021/bi9013819. [DOI] [PubMed] [Google Scholar]
- 74.Greenfield N., Fasman G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 75.Fuguet E., Rafols C., Bosch E. Critical micelle concentration of surfactants in aqueous buffered and unbuffered systems. Anal. Chim. Acta. 2005;548:95–100. [Google Scholar]
- 76.Chandra S., Chen X., Südhof T.C. A broken α-helix in folded α-synuclein. J. Biol. Chem. 2003;278:15313–15318. doi: 10.1074/jbc.M213128200. [DOI] [PubMed] [Google Scholar]
- 77.Tjandra N., Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 78.Camilloni C., De Simone A., Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 79.Ma C.D., Wang C., Abbott N.L. Modulation of hydrophobic interactions by proximally immobilized ions. Nature. 2015;517:347–350. doi: 10.1038/nature14018. [DOI] [PubMed] [Google Scholar]
- 80.Fuxreiter M., Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 81.Tompa P., Szász C., Buday L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Rivers R.C., Kumita J.R., Christodoulou J. Molecular determinants of the aggregation behavior of α- and β-synuclein. Protein Sci. 2008;17:887–898. doi: 10.1110/ps.073181508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chandrababu K.B., Dutta K., Moradian-Oldak J. Structural adaptation of tooth enamel protein amelogenin in the presence of SDS micelles. Biopolymers. 2014;101:525–535. doi: 10.1002/bip.22415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hadgiivanova R., Diamant H. Premicellar aggregation of amphiphilic molecules. J. Phys. Chem. B. 2007;111:8854–8859. doi: 10.1021/jp070038z. [DOI] [PubMed] [Google Scholar]
- 85.Andersen K.K., Oliveira C.L., Otzen D. The role of decorated SDS micelles in sub-CMC protein denaturation and association. J. Mol. Biol. 2009;391:207–226. doi: 10.1016/j.jmb.2009.06.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.