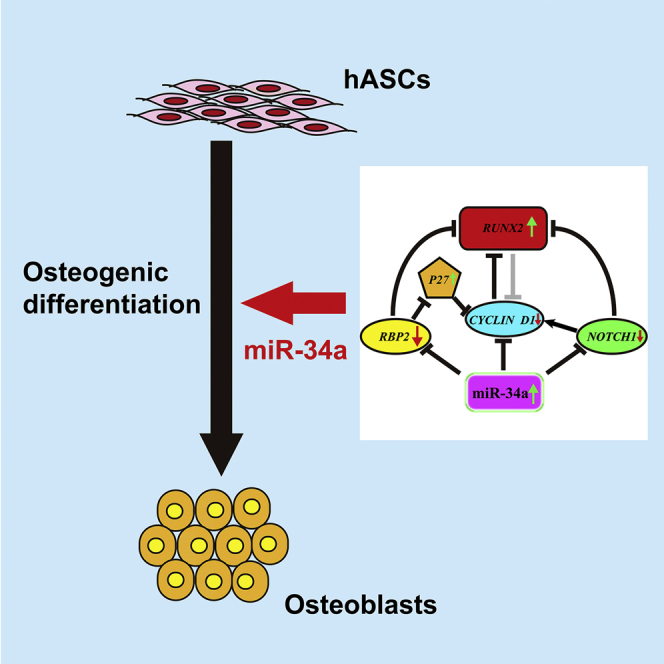
Summary
MiR-34a was demonstrated to be upregulated during the osteogenic differentiation of human adipose-derived stem cells (hASCs). Overexpression of miR-34a significantly increased alkaline phosphatase activity, mineralization capacity, and the expression of osteogenesis-associated genes in hASCs in vitro. Enhanced heterotopic bone formation in vivo was also observed upon overexpression of miR-34a in hASCs. Mechanistic investigations revealed that miR-34a inhibited the expression of retinoblastoma binding protein 2 (RBP2) and reduced the luciferase activity of reporter gene construct comprising putative miR-34a binding sites in the 3′ UTR of RBP2. Moreover, miR-34a downregulated the expression of NOTCH1 and CYCLIN D1 and upregulated the expression of RUNX2 by targeting RBP2, NOTCH1, and CYCLIN D1. Taken together, our results suggested that miR-34a promotes the osteogenic differentiation of hASCs via the RBP2/NOTCH1/CYCLIN D1 coregulatory network, indicating that miR-34a-targeted therapy could be a valuable approach to promote bone regeneration.
Graphical Abstract
Highlights
-
•
MiR-34a promotes osteogenesis of hASCs in vitro and in vivo
-
•
MiR-34a directly binds to the 3′ UTR of RBP2 mRNA in hASCs
-
•
MiR-34a promotes osteogenesis of hASCs via the RBP2/NOTCH1/CYCLIN D1 network
In this article, Zhou and colleagues show that miR-34a promotes the osteogenic differentiation of hASCs in vitro and in vivo. Dual-luciferase reporter assay demonstrated that miR-34a directly targets RBP2 mRNA and promotes osteogenesis of hASCs via the RBP2/NOTCH1/CYCLIN D1 network, indicating that miR-34a-targeted therapy could be a valuable approach to promote bone regeneration.
Introduction
Tissue engineering technology has become one of the most promising therapeutic approaches for bone regeneration in bone defects (Zou et al., 2011, Ye et al., 2011, Xiao et al., 2011). As a source of mesenchymal stem cells (MSCs), human adipose-derived stem cells (hASCs) are receiving more attention in bone tissue engineering (Bosnakovski et al., 2005, Zuk et al., 2002, Wang et al., 2011). However, the paucity of available information about the molecular pathways that govern the osteogenic differentiation of hASCs has hampered further development of hASC-based cell therapies.
MicroRNAs (miRNAs) are a class of endogenously expressed, small non-coding RNA molecules that negatively regulate gene expression at the post-transcriptional level by base pairing with the 3′ UTR of their target mRNAs (Thomas et al., 2010). They play vital roles in various biological processes, including the cell fate of embryonic stem cells, cell proliferation, apoptosis, differentiation, morphogenesis, carcinogenesis, and angiogenesis (Ambros, 2004, Hua et al., 2006, Xu et al., 2004). A single miRNA is often involved in several gene regulatory networks (Bartel, 2004, Krek et al., 2005), and overexpression or inhibition of miRNAs can regulate the endogenous expression of multiple growth factors simultaneously (Yau et al., 2012). Therefore, we hypothesized that the delivery of a desired miRNA may result in optimization of bone regeneration. Recent studies have reported that several miRNAs, such as miR-22, -100, -106a, -146a, and -148b, are involved in the osteogenic differentiation of stem cells (Cho et al., 2010, Huang et al., 2012, Li et al., 2013a, Liao et al., 2014, Qureshi et al., 2013, Zeng et al., 2012). However, further regulatory mechanisms of miRNAs in the osteogenesis of MSCs still await investigation.
Our previous study showed that the inhibition of retinoblastoma binding protein 2 (RBP2) significantly improved the in vitro and in vivo osteogenic capacity of hASCs (Ge et al., 2011). Based on these data, we aimed to screen and select miRNAs that positively regulate the osteogenic differentiation of hASCs by targeting RBP2. Microarray analyses revealed that after osteogenic induction, 21 miRNAs were upregulated in hASCs (Zhang et al., 2012) and 51 miRNAs were upregulated in bone marrow-derived MSCs (BMSCs) (Gao et al., 2011), suggesting that 72 upregulated miRNAs had potential effects on the osteogenic differentiation of MSCs. Moreover, RNA22 prediction software indicated that 122 miRNAs might bind to the 3′ UTR of RBP2 mRNA. These two categories of miRNAs were combined and an intersection of five miRNAs was produced: miR-663, -34a, -26a, -17, and -155. The RNA22 prediction software predicted their corresponding folding energy (ΔG) was −14.00 kcal/mol, −16.8 kcal/mol, −12.50 kcal/mol, −13.20 kcal/mol, and −13.30 kcal/mol. According to the results predicted by RNA22 prediction software, miR-34a possessed the maximum likelihood for binding to the 3′ UTR of RBP2 mRNA (ΔG = −16.8 kcal/mol); therefore, we selected miR-34a for further investigation (Figure S1).
NOTCH1 and CYCLIN D1 are direct target genes of miR-34a in tumor cells (Hermeking, 2010, Pang et al., 2010), and have effects on the proliferation and osteogenic differentiation of MSCs by regulating runt-related transcription factor 2 (RUNX2) (Engin et al., 2008), a key osteogenesis-associated transcription factor. Thus, NOTCH1 and CYCLIN D1 pathways were integrated into our hypothetical regulatory network of miR-34a.
In this study, we investigated the functional roles of miR-34a in the osteogenic differentiation of hASCs both in vitro and in vivo, and explored whether miR-34a regulated this biological process through the RBP2/NOTCH1/CYCLIN D1 coregulatory network. Our study provided a better understanding of the role and mechanism of miR-34a in hASCs' osteogenic differentiation and suggested that miR-34a could be a therapeutic target in future bone regeneration therapy, which will lead to advances in clinical bone tissue engineering.
Results
Expression Levels of miR-34a during the Osteogenic Differentiation of hASCs
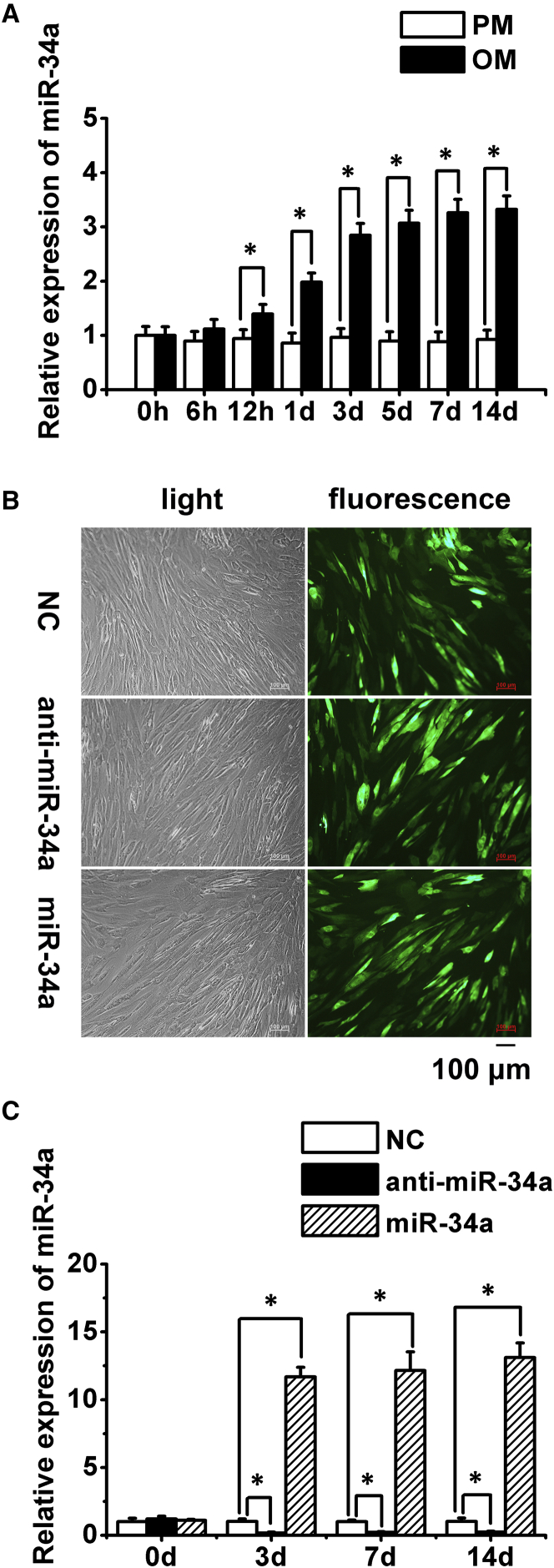
After culturing hASCs in osteogenic medium (OM) for 12 hr, miR-34a expression increased significantly, and further increased with prolonged osteogenic induction. However, no significant change was detected in hASCs cultured in proliferation medium (PM) when compared with the 0-hr time point (Figure 1A). These data suggested that miR-34a might play a role in the regulation of hASCs' osteogenic differentiation.
Figure 1.
Expression of Endogenous miR-34a during hASCs' Osteogenic Induction, and Determination of Lentiviral Transduction Efficiency and Effect
(A) Quantitative real-time PCR analysis of miR-34a expression in hASCs cultured in PM and OM.
(B) Microscopic images of GFP-positive hASCs under ordinary and fluorescent light. Scale bar, 100 μm.
(C) Quantitative real-time PCR analysis of miR-34a in transduced hASCs cultured in PM.
PM, proliferation medium; OM, osteogenic medium; NC, lentivirus negative control; anti-miR-34a, lentivirus anti-sense miR-34a; miR-34a, lentivirus miR-34a mimics. Data represent the means ± SD of three independent experiments. ∗p < 0.05 versus the NC group.
Promotion Effects of miR-34a on the Osteogenic Differentiation of hASCs In Vitro
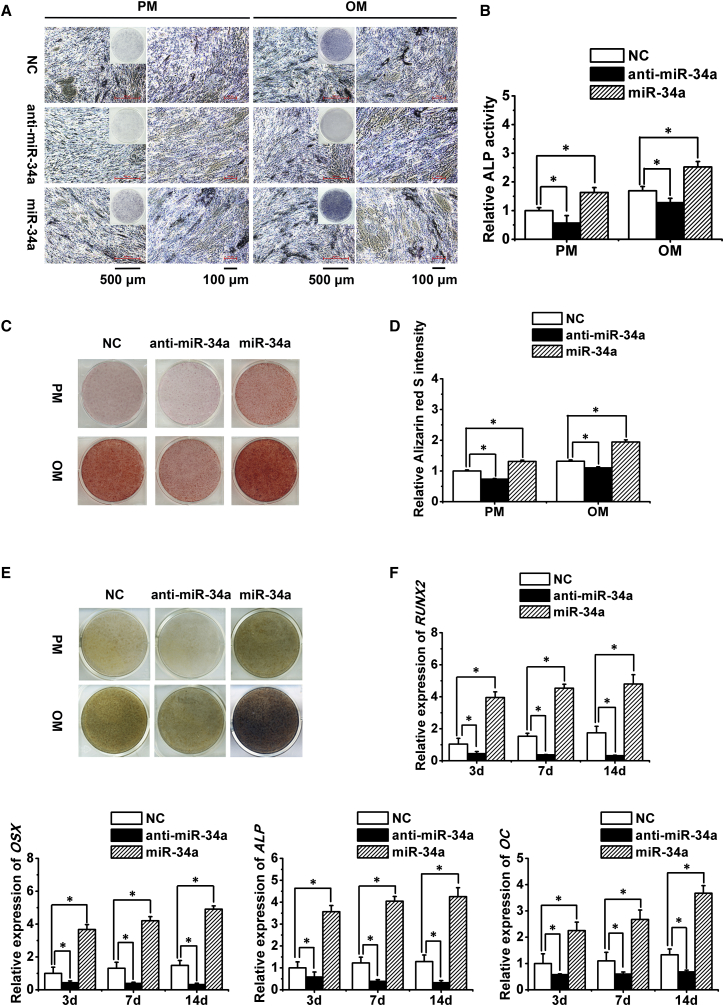
The transduction efficiency of lentivirus was estimated to be 80%–90%, as evaluated by the percentage of GFP-positive cells under an inverted fluorescence microscope 72 hr after transduction (Figure 1B). Quantitative real-time PCR analysis of miR-34a expression in transduced hASCs cultured in PM at 0, 3, 7, and 14 days showed a >10-fold increase in the miR-34a overexpression group and >75% reduction in the miR-34a knockdown group when compared with the negative control (NC) group (Figure 1C). Alkaline phosphatase staining and quantification showed that overexpression of miR-34a enhanced the osteogenic differentiation of hASCs cultured in PM or OM at 7 days, while miR-34a knockdown inhibited alkaline phosphatase activity of hASCs (Figures 2A and 2B). The extracellular mineralization of hASCs, as tested by Alizarin red S (ARS) and von Kossa staining in PM or OM at 14 days, displayed similar results to the alkaline phosphatase tests (Figures 2C–2E). Moreover, overexpression of miR-34a significantly increased the expression of osteogenesis-associated genes, including RUNX2, osterix (OSX), alkaline phosphatase (ALP), and osteocalcin (OC), while miR-34a knockdown led to the opposite tendency (Figures 2F and S2).
Figure 2.
Promotion of hASCs' Osteogenic Differentiation by miR-34a In Vitro
(A and B) ALP staining (A) and quantification (B) of transduced hASCs. Scale bar of the left panel in PM or OM group, 500 μm; scale bar of the right panel in PM or OM group, 100 μm.
(C and D) ARS staining (C) and quantification (D) of transduced hASCs.
(E) von Kossa staining of transduced hASCs.
(F) Quantitative real-time PCR analysis of RUNX2, OSX, ALP, and OC expression in transduced hASCs.
ALP, alkaline phosphatase; ARS, Alizarin red S; RUNX2, runt-related transcription factor 2; OSX, osterix; OC, osteocalcin. Data represent the means ± SD of three independent experiments. ∗p < 0.05 versus the NC group.
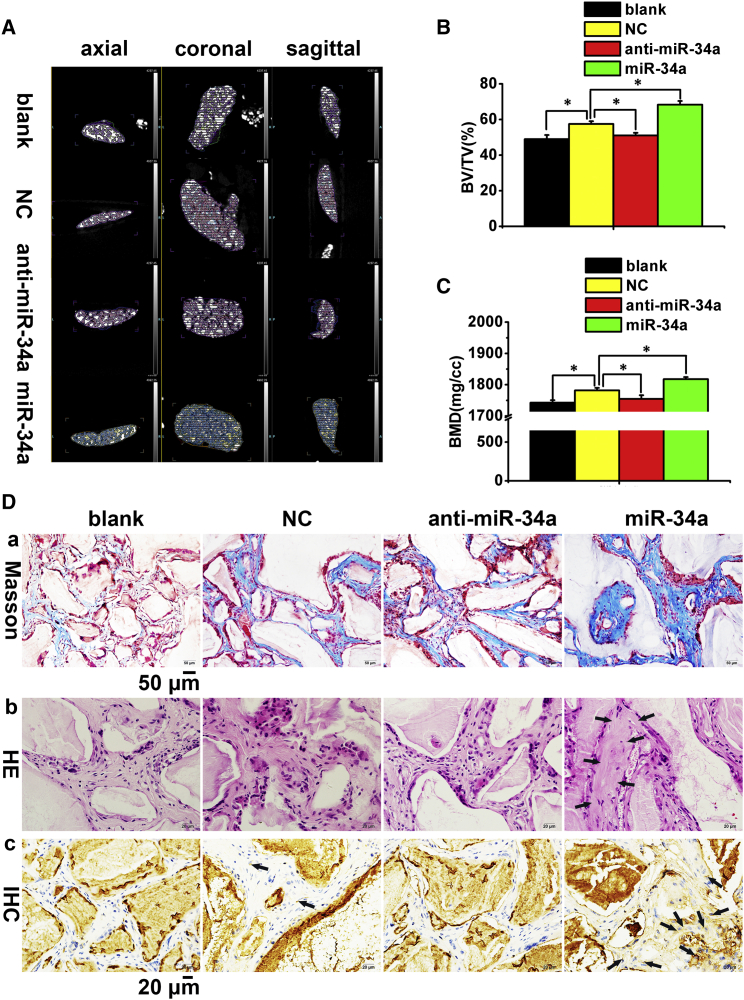
Promotion Effects of miR-34a on the Osteogenic Differentiation of hASCs In Vivo
Given the existence of other biological factors and the uncontrollability of the microenvironment, sometimes the in vivo results may be different and even opposite to those in vitro. Therefore, the investigation of miR-34a's in vivo effect was necessary. The microstructure of the newly formed bone was evaluated by micro-computed tomography (CT) imaging. The representative images in the blank and miR-34a knockdown groups showed less newly formed bone and more scaffold remnants. In contrast, the miR-34a overexpression group exhibited the most newly formed bone with the fewest scaffold remnants when compared with other groups (Figure 3A). By quantifying the amount of new bone, the percentages of new bone volume to tissue volume (BV/TV) in the miR-34a overexpression group showed a greater than 2-fold increase, whereas the blank and miR-34a knockdown groups showed a decrease when compared with the NC group (Figure 3B). Similarly, the bone mineral density (BMD) of the miR-34a overexpression group was the highest among these four groups, while the blank and miR-34a knockdown groups were lower than the NC group (Figure 3C).
Figure 3.
Promotion of hASCs' Osteogenic Differentiation by miR-34a In Vivo
(A) Newly formed bone in Bio-Oss collagen scaffolds are indicated in different colors; scaffold remnants appear as white irregular lumps.
(B and C) Quantitative analysis of BV/TV and BMD. Data represent the means ± SD of three independent experiments. ∗p < 0.05 versus the NC group.
(D) Histological assessment of ectopic bone formation. (a) Masson trichrome staining. The collagen in the bone matrix was stained blue-green. (b) H&E staining. New bone structures are indicated by black arrows. (c) IHC staining for OC. Dark-brown granules indicating positive staining are marked by black arrows. Scale bar, 50 μm in (a) and 20 μm in (b) and (c).
BV/TV, percentage of new bone volume to tissue volume; BMD, bone mineral density; IHC, immunohistochemistry; blank, scaffolds without hASCs; NC/anti-miR-34a/miR-34a, scaffolds seeded with hASCs transfected by lentivirus negative control/anti-sense miR-34a/miR-34a mimics.
Collagen deposition, as assessed by Masson trichrome staining, demonstrated that the most bone matrix was found in the miR-34a overexpression group (Figure 3Da). H&E staining of each group showed that no new bone was found in the blank, NC, or miR-34a knockdown group, but osteoid was formed in the miR-34a overexpression group (Figures 3Db and S3). Immunohistochemical (IHC) staining for OC indicated that both the range and intensity of the stained granules in osteoblasts were generally increased in the miR-34a overexpression group (Figure 3Dc), suggesting that miR-34a induced the expression of OC.
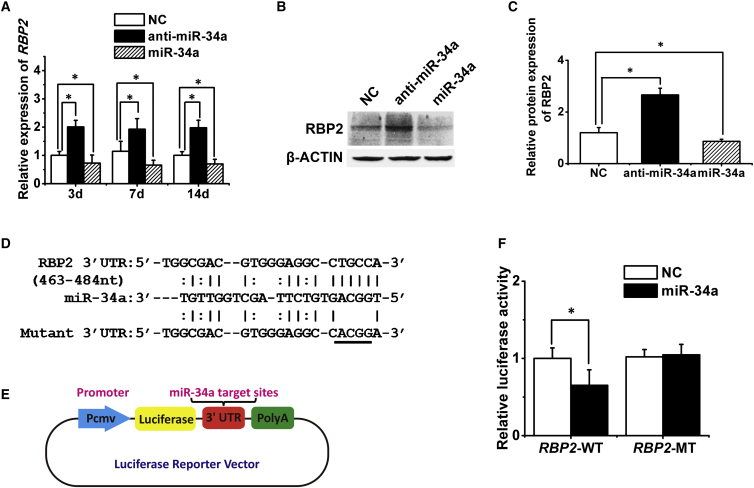
Direct Targeting of RBP2 by miR-34a
RNA22 prediction software identified RBP2 as a potential target gene of miR-34a, and RBP2 inhibition improves the osteogenic capacity of hASCs (Ge et al., 2011); therefore, we investigated its effect on RBP2 expression to clarify the molecular mechanism underlying the osteogenic regulation of hASCs by miR-34a. As predicted, overexpression of miR-34a resulted in downregulation of RBP2, whereas miR-34a knockdown increased the expression of RBP2 at both the mRNA and protein levels (Figures 4A–4C). The putative binding sites of miR-34a in the 3′ UTR of RBP2 was predicted by RNA22 prediction software (Figure 4D). Thus, we constructed a luciferase reporter vector (Figure 4E) to determine whether miR-34a could directly target these sites. Luciferase activity analysis showed that miR-34a repressed the luciferase expression of vectors containing the 3′ UTR of wild-type RBP2 (RBP2-WT), but had no effect on the mutant-type RBP2 (RBP2-MT) group compared with the NC group (Figure 4F). These results indicated that miR-34a negatively regulated RBP2 by directly binding to the 3′ UTR of its mRNA.
Figure 4.
Validation of RBP2 as a Direct Target Gene of miR-34a
(A–C) Quantitative real-time PCR (A) and western blotting (B and C) analysis of the effects of miR-34a on RBP2 expression.
(D) Predicted binding sites of miR-34a in the 3′ UTR of RBP2-WT mRNA (underlined part indicates mutated base sequences in the 3′ UTR of RBP2-MT).
(E) Schematic showing the constructed luciferase reporter system containing the binding sites of miR-34a.
(F) Luciferase activity of cells with miR-34a overexpression in the RBP2-WT or RBP2-MT group.
RBP2, retinoblastoma binding protein 2; RBP2-WT, wild-type RBP2 mRNA; RBP2-MT, mutant-type RBP2 mRNA. Data represent the means ± SD of three independent experiments. ∗p < 0.05 versus the NC group.
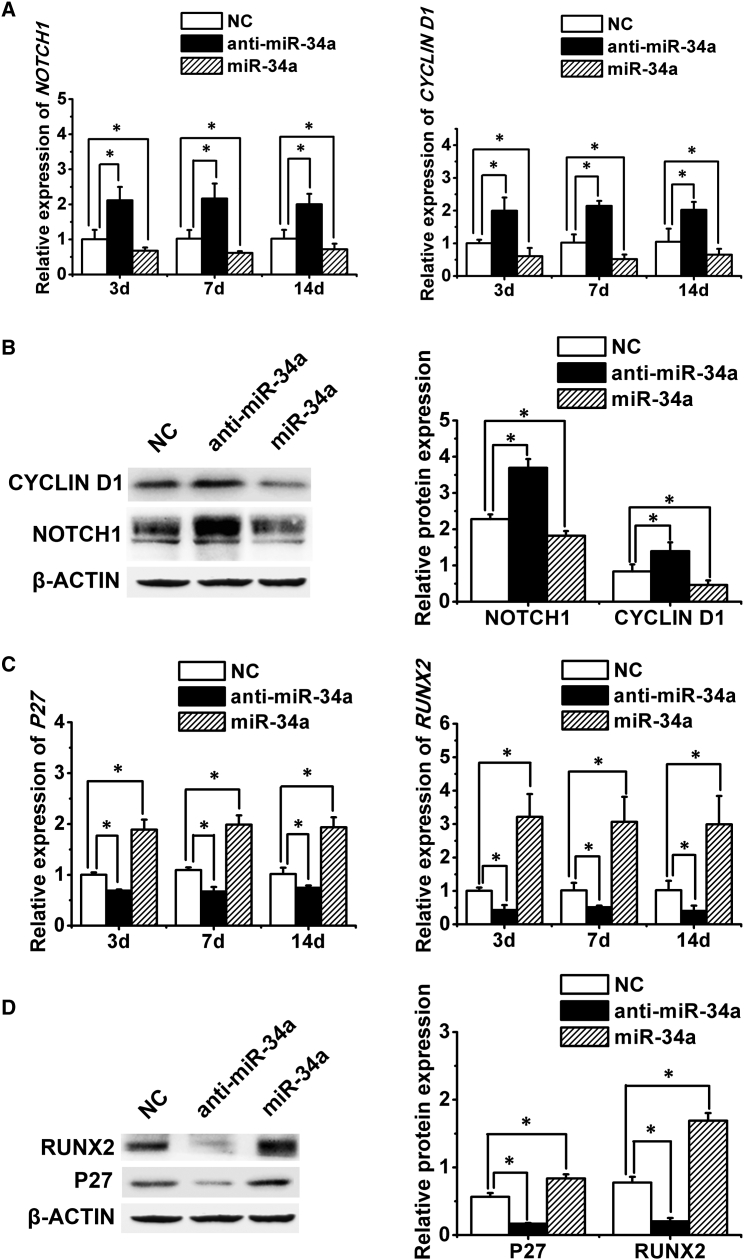
Direct Repression of NOTCH1 and CYCLIN D1 by miR-34a
NOTCH1 and CYCLIN D1 were previously identified as direct target genes of miR-34a in tumor cells (Hermeking, 2010, Pang et al., 2010). Moreover, the miR-34a potential target sites in NOTCH1 and CYCLIN D1 transcripts were predicted by RNA22 software (Figure S4). Our results confirmed that in hASCs, miR-34a overexpression suppressed the expression of NOTCH1 and CYCLIN D1 at both the mRNA and protein levels, while miR-34a knockdown resulted in the upregulation of these two genes (Figures 5A and 5B).
Figure 5.
miR-34a Repressed NOTCH1 and CYCLIN D1 Expression and Upregulated P27 and RUNX2 Expression
(A and B) Quantitative real-time PCR (A) and western blotting (B) analysis of NOTCH1 and CYCLIN D1 with miR-34a knockdown or overexpression.
(C and D) Quantitative real-time PCR (C) and western blotting (D) analysis of P27 and RUNX2 with miR-34a knockdown or overexpression.
Data represent the means ± SD of three independent experiments. ∗p < 0.05 versus the NC group.
Upregulated Expression of RUNX2 by miR-34a
Previous studies reported that RBP2 could directly downregulate P27, leading to the upregulation of CYCLIN D1 expression (Liang et al., 2013, Teng et al., 2013, Zeng et al., 2010). Thus, we explored the effect of miR-34a on P27 expression and demonstrated that miR-34a increased the expression of P27 at both the mRNA and protein levels (Figures 5C and 5D). Importantly, miR-34a overexpression significantly upregulated the expression of RUNX2, a major osteogenesis-associated transcription factor, and RUNX2 expression was downregulated in response to miR-34a knockdown (Figures 5C and 5D).
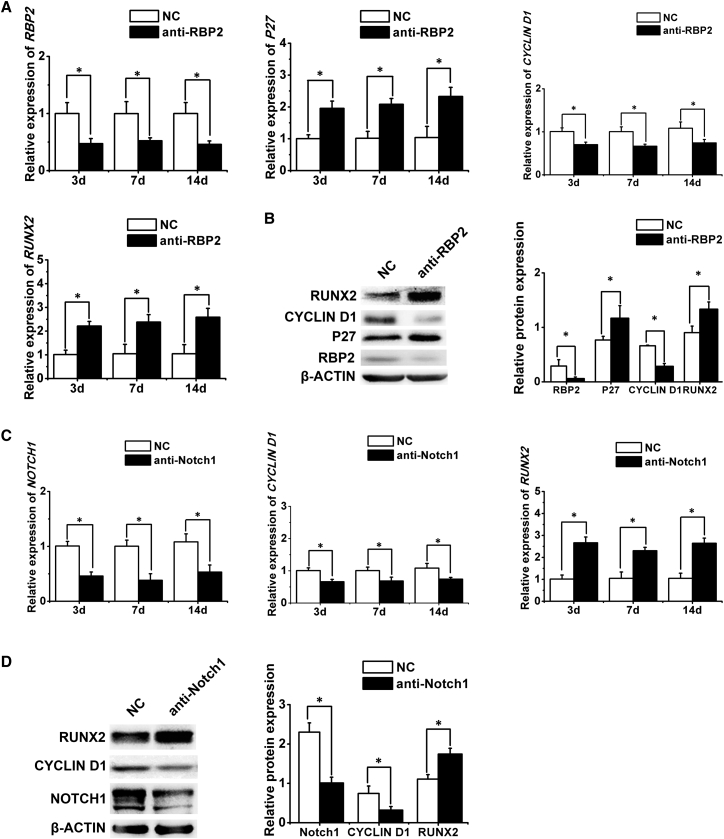
Coregulation of the RBP2/NOTCH1/CYCLIN D1 Network by miR-34a in the Osteogenic Differentiation of hASCs
To further confirm the interactions among RBP2, NOTCH1, CYCLIN D1, and RUNX2, we transfected hASCs with anti-RBP2 and anti-NOTCH1 lentivirus and then evaluated the expression of possible downstream target genes through quantitative real-time PCR and western blotting. The transduction effects of anti-RBP2 and anti-NOTCH1 lentivirus showed ∼50% reductions in the expression of respective genes (Figures 6A–6D). We found that RBP2 knockdown upregulated the expression of P27 and RUNX2, and downregulated CYCLIN D1 expression (Figures 6A and 6B). Similarly, NOTCH1 knockdown resulted in the downregulation of CYCLIN D1 and upregulation of RUNX2 (Figures 6C and 6D). Taken together, these results indicated that miR-34a indirectly increased the expression of P27 and RUNX2 by directly suppressing RBP2, NOTCH1, and CYCLIN D1, suggesting that miR-34a modulated the osteogenic differentiation of hASCs through the RBP2/NOTCH1/CYCLIN D1 coregulatory network.
Figure 6.
Confirmation of the Relationships among RBP2, NOTCH1, P27, CYCLIN D1, and RUNX2
(A and B) Quantitative real-time PCR (A) and western blotting (B) analysis of RBP2, P27, CYCLIN D1, and RUNX2 expression after RBP2 knockdown.
(C and D) Quantitative real-time PCR (C) and western blotting (D) analysis of NOTCH1, CYCLIN D1, and RUNX2 expression after NOTCH1 knockdown.
Data represent the means ± SD of three independent experiments. ∗p < 0.05 versus the NC group.
Discussion
The development of miRNA-based therapy approaches has become one of the most attractive areas of tissue engineering (Lian et al., 2012). Recently, many studies reported that miRNAs are important regulators for the therapy or differentiation of stem cells (Cho et al., 2010, Huang et al., 2012, Li et al., 2013a, Li et al., 2013b, Liao et al., 2014, Qureshi et al., 2013, Zeng et al., 2012, Deng et al., 2013). In this study, we observed a significant increase in miR-34a expression during the osteogenic differentiation of hASCs, which was consistent with previous studies (Ambros, 2004, Hua et al., 2006). This upregulation during osteogenic induction indicated that miR-34a might have an effect on the osteogenic differentiation of hASCs.
A combination of in vitro and in vivo experiments indicated that miR-34a overexpression promoted the osteogenic differentiation and ectopic osteogenesis of hASCs, while miR-34a knockdown inhibited osteogenic capacity when compared with the NC group. We found that miR-34a promoted the alkaline phosphatase activity and mineralization capacity and also increased the expression of osteogenesis-associated genes including RUNX2, OSX, ALP, and OC in both PM and OM. This indicated that miR-34a overexpression promoted osteogenic differentiation in vitro. Moreover, heterotopic bone formation after in vivo implantation of hASCs and scaffold hybrids was evaluated by micro-CT and histological staining. As stated above, the osteogenic effect of miR-34a shown in scanning images and quantitative analysis by BV/TV and BMD were in accordance with those in vitro, and histological assessment including Masson, H&E, and IHC staining verified this conclusion as well.
Although miR-34a was once reported to inhibit proliferation and osteogenic differentiation in BMSCs by targeting a number of known cell-cycle proteins and JAGGED1 (Chen et al., 2014), two other studies report that it enhances osteogenesis by inhibiting osteoclastogenesis of osteoclasts from bone marrow (Krzeszinski et al., 2014) or promoting osteogenic differentiation of apical papilla stem cells (Sun et al., 2014). In our study, both the mRNA and protein expression of JAGGED1 did not show significant difference by overexpressing or knocking down miR-34a in hASCs (Figure S5). The contradictory results might partly be attributed to the varied characteristics of the different cell lines and distinct post-transcriptional regulation of the osteogenic differentiation in tissue-specific MSCs. In addition, Park et al. (2015) found that overexpression of miR-34a decreased the cell proliferation and downregulated the expression of various cell-cycle regulators such as CDKs (-2, -4, -6) and CYCLINs (-E, -D). These results were consistent with ours, but found too that the potential of adipogenesis and osteogenesis of hASCs was also diminished by miR-34a overexpression. As we know, a mutually exclusive relationship usually exists between osteoblastogenesis and adipogenesis, with factors stimulating one of these processes while at the same time inhibiting the other (Huang et al., 2012, Wang et al., 2013). Thus we need to further investigate the effects of miR-34a on the osteogenesis and adipogenesis of hASCs. Moreover, there are some reports concerning the contradictory effects of other miRNAs on osteogenic differentiation. For example, miR-26a can either promote (Wang et al., 2015a, Wang et al., 2015b) or suppress (Luzi et al., 2008, Luzi et al., 2012) the osteogenic differentiation of hASCs. Therefore, further research is needed to elucidate the osteogenic regulation of miRNAs. Taken together, our study indicated that other signal molecules and pathways might be involved in the regulation of osteogenic differentiation by miR-34a.
Furthermore, we demonstrated that RBP2 was a target gene of miR-34a in hASCs. MiR-34a repressed the luciferase activity of reporter vectors containing putative binding sites in the 3′ UTR of RBP2, indicating that miR-34a directly binds to the 3′ UTR of RBP2 mRNA. In addition, RBP2 directly binds to the promoter of P27 (Liang et al., 2013, Teng et al., 2013, Zeng et al., 2010), which is a member of cyclin-dependent kinase inhibitors (CDKIs) and plays a critical role in inhibiting the transition of the cell cycle from the G1 to the S phase by binding and inhibiting CYCLIN/CDKs, including CYCLIN D1 (Perisanidis et al., 2012, Pestell, 2013, Wang et al., 2012, Zhang et al., 2014b, Chen et al., 2013). In the present study, both miR-34a overexpression and RBP2 knockdown caused upregulation of P27 and downregulation of CYCLIN D1, suggesting the suppressed proliferation of hASCs. Moreover, we previously demonstrated that RBP2 physically and genetically repressed the transcriptional activity of RUNX2 (Ge et al., 2011). The upregulation of RUNX2 after knockdown of RBP2 confirmed this conclusion. These results suggested that miR-34a could upregulate the expression of RUNX2 by directly binding to the RBP2 mRNA.
Our study also confirmed that miR-34a overexpression induced the downregulation of NOTCH1 and CYCLIN D1, which have been identified as target genes of miR-34a in tumor cells (Wei et al., 2012, Bae et al., 2012, Li et al., 2012, Sun et al., 2008, Zhang et al., 2014a). In addition, CYCLIN D1 is a direct target gene of NOTCH1 (Perisanidis et al., 2012, Cohen et al., 2010). We found that CYCLIN D1 expression was reduced significantly after knockdown of NOTCH1. Moreover, gain of NOTCH inhibits osteoblast maturation by directly repressing RUNX2, as well as by repressing the anti-proliferative effects of RUNX2 via CYCLIN D1 upregulation (Engin et al., 2008). This study showed upregulated expression of RUNX2 in hASCs with NOTCH1 knockdown. Our current findings and those from previous studies suggested that miR-34a could also upregulate the expression of RUNX2 by directly depressing NOTCH1 and CYCLIN D1 expression.
RUNX2 is an important osteoblast lineage-determining transcription factor that induces the expression of bone sialoprotein (BSP), OC, OSX, and osteopontin (OPN), which are required to finalize terminal osteogenic differentiation (Komori, 2002, Komori, 2003, Komori, 2008). The key effect of RUNX2 in osteogenesis is suppressing osteoblast proliferation and promoting osteoblast maturation by supporting exit from the cell cycle (Galindo et al., 2005, Pratap et al., 2003). Our study proved that overexpression of miR-34a and knockdown of RBP2 or NOTCH1 eventually induced the upregulation of RUNX2, which probably accounted for the positive regulation of hASCs' osteogenic differentiation by miR-34a.
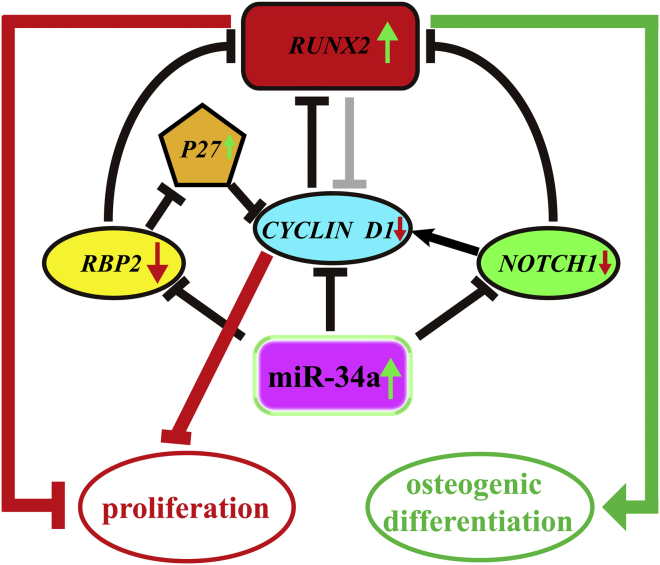
Our sorting data displayed the interrelationship between miR-34a, RBP2, NOTCH1, CYCLIN D1, and RUNX2. The positive effect of miR-34a on hASCs' osteogenic differentiation could be attributed to the ultimate downregulation of CYCLIN D1 and upregulation of RUNX2 via the RBP2/NOTCH1/CYCLIN D1 coregulatory network, which might result in the inhibition of proliferation and promotion of osteogenic differentiation in hASCs (Figure 7). These findings supported miR-34a as a potential target for bone tissue engineering and provided valuable information on the management of bone-related diseases via epigenetic intervention.
Figure 7.
Schematic Representation of the RBP2/NOTCH1/CYCLIN D1 Coregulatory Network Involved in the Osteogenic Differentiation of hASCs by miR-34a
MiR-34a directly targeted the RBP2, NOTCH1, and CYCLIN D1 transcripts, leading to the downregulation of these target genes and subsequent upregulation of P27 and RUNX2. The final repression of CYCLIN D1 and upregulation of RUNX2 mediated a switch from proliferation to osteogenic differentiation in hASCs.
Experimental Procedures
Cell Culture and Osteogenic Differentiation
The hASCs and 293T cells were obtained separately from ScienCell Research Laboratories and the American Type Culture Collection. Stem cells from three donors of the third passage were used for the in vitro and in vivo experiments. All cell-based experiments were repeated three times using hASCs from the three donors, respectively. Cells were cultured in PM containing DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Osteogenic differentiation of hASCs was induced after the cells reached 70%–80% confluence using OM (standard PM supplemented with 100 nM dexamethasone, 0.2 mM ascorbic acid, and 10 mM β-glycerophosphate).
Lentivirus Transduction and Establishment of Stably Expressing Transductants
Lentivirus containing GFP-labeled plasmid vectors of the negative control (NC), anti-sense miR-34a (anti-miR-34a), miR-34a mimics (miR-34a), RBP2 short hairpin RNA (shRNA) (anti-RBP2), and NOTCH1 shRNA (anti-NOTCH1) were synthesized and packaged by GenePharma. The hASCs were stably transfected with these lentiviruses at an MOI of 50 in the presence of 8 μg/ml polybrene (Sigma). After 24 hr, the lentivirus-containing medium was removed and replaced with fresh medium. Transduction efficiency was evaluated by the percentage of GFP-positive cells observed under an inverted fluorescence microscope (Nikon, TE2000-U). Puromycin at 1 μg/ml was used to select infected cells.
Alkaline Phosphatase Staining and Quantification
The hASCs were seeded at a density of 2 × 105 cells per well in 24-well plates and transfected with the lentiviruses mentioned above. Cells were cultured in PM or OM for 7 days and then evaluated for alkaline phosphatase activity. Alkaline phosphatase staining was performed using an NBT/BCIP staining kit (CoWin Biotech) with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP). For quantification of alkaline phosphatase activity, cells were rinsed three times with PBS followed by 1% Triton X-100, scraped in distilled water, and subjected to three freeze-thaw cycles. The alkaline phosphatase activity was determined at 405 nm using p-nitrophenyl phosphate as the substrate. The total protein content was determined using the BCA method with the Pierce BCA protein assay kit (Thermo Scientific). Aliquots of the same samples were read at 562 nm and calculated against a series of BSA standards. Relative alkaline phosphatase activity was compared with that in the controls and calculated after normalization to the total protein content.
Mineralization Assays and Von Kossa Staining
The infected hASCs were cultured in 24-well plates with PM or OM for 14 days, and matrix mineralization was determined by ARS (Sigma) and von Kossa staining. ARS staining and quantification were performed as follows. Plates were washed three times with PBS and then stained with 0.1% ARS in distilled water (pH 4.2) for 1 hr at room temperature. After staining, the cultures were washed three times with distilled water. For quantification of matrix mineralization, ARS-stained cultures were incubated in 100 mM cetylpyridinium chloride (Sigma) for 1 hr to solubilize and release calcium-bound ARS into solution. The absorbance of the released ARS was measured at 562 nm. Relative ARS intensity was compared with that in the control treatment and calculated after normalization to the total protein content. For von Kossa staining, culture plates were fixed in 4% paraformaldehyde for 30 min and washed three times with PBS, then 1 ml of 5% silver nitrate was added and the cells were exposed to a 100 W UV lamp for 60 min. After being washed with distilled water three times, 1 ml of 5% sodium thiosulfate solution was added for 2 min and rinsed with distilled water. Finally, the mineralized extracellular matrix was observed under the microscope.
RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
Total cellular RNA from infected hASCs cultured in PM or OM (both for 3, 7, and 14 days) were isolated using TRIzol reagent (Invitrogen) and used for first-strand cDNA synthesis with a Reverse Transcription System (Takara). Quantification of all gene transcripts was performed by quantitative real-time PCR using a Power SYBR Green PCR Master Mix (Roche) and a 7500 Real-Time PCR Detection System (Applied Biosystems). The following thermal settings were used: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The internal control for mRNAs and miR-34a were GAPDH and U6, respectively (Ge et al., 2014). The products of quantitative real-time PCR were sequenced as previously described (Jia et al., 2014). The primers used are listed in Table S1. The data were analyzed using the 2−ΔΔCt method.
Western Blotting
Western blotting was performed as described previously (Jia et al., 2014). Primary antibodies against RBP2, NOTCH1, P27, CYCLIN D1, RUNX2, and β-ACTIN (all from Cell Signaling Technology; catalog numbers 3876, 3608, 3686, 2978, 8486, and 4970, respectively) were diluted 1:1,000 and incubated with the blots at 4°C overnight. Horseradish peroxidase-conjugated anti-rabbit secondary antibodies (Cell Signaling Technology) were diluted 1:10,000 and incubated at room temperature for 1 hr.
Reporter Vectors Construction and Dual-Luciferase Reporter Assay
The functional alignment of the target region of RBP2 was predicted by RNA22 prediction software. The 3′ UTR of RBP2, containing the predicted miR-34a binding sites, were synthesized and cloned into a modified version of pcDNA3.1(+) that contained a firefly luciferase reporter gene (a gift from Brigid L.M. Hogan, Duke University), at a position downstream of the luciferase reporter gene, thus forming a wild-type (WT)-RBP2 luciferase reporter plasmid. Site-directed mutagenesis of the miR-34a binding site in the RBP2 3′ UTR was performed using a Site-Directed Mutagenesis Kit (SBS Genetech) and named as mutant-type (MT)-RBP2 luciferase reporter plasmid. All constructs were confirmed by DNA sequencing. Luciferase assays were performed as described previously (Jia et al., 2014). In brief, 293T cells at a density of 5 × 104 per well were grown in a 48-well plate and expanded to 105 per well before being transfected with 400 ng of either pcDNA3.0 or pcDNA3.0-miR-34a, 40 ng of the firefly luciferase reporter plasmid (RBP2-WT or -MT), and 4 ng of pRL-TK, a plasmid expressing Renilla luciferase (Promega). Luciferase activity was measured 24 hr after transfection using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well. Each experiment was repeated in triplicate.
In Vivo Implantation of hASCs and Bio-Oss Collagen Scaffold Hybrids
The hASCs of the third passage transfected with lentivirus (NC, anti-miR-34a, and miR-34a) were cultured in PM before the in vivo study. After being trypsinized and resuspended directly in DMEM, the cells were incubated with 6 × 4 × 2 mm3 Bio-Oss collagen scaffolds (Geistlich; GEWO) for 1 hr at 37°C, followed by centrifugation at 150 g for 5 min, and implanted into four symmetrical sites, together with blank scaffolds (without cells), on the dorsal subcutaneous space of 5-week-old, BALB/c homozygous nude (nu/nu) mice (n = 6 per group). This study was approved by the Institutional Animal Care and Use Committee of the Peking University Health Science Center (LA2014233), and all animal experiments were performed in accordance with the Institutional Animal Guidelines.
Analyses of Bone Formation In Vivo
Specimens were harvested 6 weeks after implantation, and the animals were euthanized by CO2 asphyxiation. After fixation in 4% paraformaldehyde, the specimens were analyzed using a high-resolution Inveon micro-CT (Siemens). In brief, an X-ray voltage of 80 kV, a node current of 500 μA, and an exposure time of 500 ms for each of the 360 rotational steps were used. For quantitative analysis of the images, percentages of BV/TV and BMD were calculated (Inveon Research Workplace). The specimens were then decalcified in 10% EDTA (pH 7.4) for 14 days, followed by embedding in paraffin. Sections (5 μm thickness) were cut and stained with Masson trichrome and H&E. Meanwhile, IHC staining was also performed with primary antibodies against OC (Santa Cruz Biotechnology, catalog no. sc-240750) to evaluate osteogenesis. Finally, the tissue slices were observed under a light microscope (Olympus).
Statistical Analysis
Statistical analysis was performed using SPSS Statistics 20.0 software (IBM). Differences between two groups were analyzed by Student's t test. In cases of multiple-group testing, one-way ANOVA in conjugation with Tukey's test was conducted. A two-tailed value of p < 0.05 was considered statistically significant. All data are presented as the means ± SD of three independent experiments.
Author Contributions
C.F., L.J., and Y.Z. designed and conducted experiments, collected and analyzed data, and wrote the manuscript. C.J., H.L., and Y.L. assisted in conducting experiments and analyzed data. Y.Z. designed the concept, and wrote and approved the manuscript.
Acknowledgments
This study was financially supported by grants from the National Natural Science Foundation of China (No. 81371118), the Program for New Century Excellent Talents in University from Ministry of Education of China (NCET-11-0026), and the Ph.D. Programs Foundation of Ministry of Education of China (No. 20130001110101).
Published: July 21, 2016
Footnotes
Supplemental Information includes five figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.06.010.
Supplemental Information
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bae Y., Yang T., Zeng H.C., Campeau P.M., Chen Y., Bertin T., Dawson B.C., Munivez E., Tao J., Lee B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D., Mizuno M., Kim G., Takagi S., Okumura M., Fujinaga T. Isolation and multilineage differentiation of bovine bone marrow mesenchymal stem cells. Cell Tissue Res. 2005;319:243–253. doi: 10.1007/s00441-004-1012-5. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang T., Shi J., Xu P., Gu Z., Sandham A., Yang L., Ye Q. Notch1 signaling regulates the proliferation and self-renewal of human dental follicle cells by modulating the G1/S phase transition and telomerase activity. PLoS One. 2013;8:e69967. doi: 10.1371/journal.pone.0069967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Holmstrøm K., Qiu W., Ditzel N., Shi K., Hokland L., Kassem M. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. 2014;32:902–912. doi: 10.1002/stem.1615. [DOI] [PubMed] [Google Scholar]
- Cho H.H., Shin K.K., Kim Y.J., Song J.S., Kim J.M., Bae Y.C., Kim C.D., Jung J.S. NF-κB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J. Cell Physiol. 2010;223:168–177. doi: 10.1002/jcp.22024. [DOI] [PubMed] [Google Scholar]
- Cohen B., Shimizu M., Izrailit J., Ng N.F., Buchman Y., Pan J.G., Dering J., Reedijk M. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Res. Treat. 2010;123:113–124. doi: 10.1007/s10549-009-0621-9. [DOI] [PubMed] [Google Scholar]
- Deng Y., Zhou H., Zou D., Xie Q., Bi X., Gu P., Fan X. The role of miR-31-modified adipose tissue-derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials. 2013;34:6717–6728. doi: 10.1016/j.biomaterials.2013.05.042. [DOI] [PubMed] [Google Scholar]
- Engin F., Yao Z., Yang T., Zhou G., Bertin T., Jiang M.M., Chen Y., Wang L., Zheng H., Sutton R.E. Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M., Pratap J., Young D.W., Hovhannisyan H., Im H.J., Choi J.Y., Lian J.B., Stein J.L., Stein G.S., van Wijnen A.J. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Yang T., Han J., Yan K., Qiu X., Zhou Y., Fan Q., Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell Biochem. 2011;112:1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- Ge W., Shi L., Zhou Y., Liu Y., Ma G.E., Jiang Y., Xu Y., Zhang X., Feng H. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells. 2011;29:1112–1125. doi: 10.1002/stem.663. [DOI] [PubMed] [Google Scholar]
- Ge W., Liu Y., Chen T., Zhang X., Lv L., Jin C., Jiang Y., Shi L., Zhou Y. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials. 2014;35:6015–6025. doi: 10.1016/j.biomaterials.2014.04.055. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Hua Z., Lv Q., Ye W., Wong C.K., Cai G., Gu D., Ji Y., Zhao C., Wang J., Yang B.B. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Wang S., Bian C., Yang Z., Zhou H., Zeng Y., Li H., Han Q., Zhao R.C. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L.F., Wei S.B., Mitchelson K., Gao Y., Zheng Y.F., Meng Z., Gan Y.H., Yu G.Y. MiR-34a inhibits migration and invasion of tongue squamous cell carcinoma via targeting MMP9 and MMP14. PLoS One. 2014;9:e108435. doi: 10.1371/journal.pone.0108435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. Runx2, a multifunctional transcription factor in skeletal development. J. Cell Biochem. 2002;87:1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- Komori T. Requisite roles of Runx2 and Cbfb in skeletal development. J. Bone Miner. Metab. 2003;21:193–197. doi: 10.1007/s00774-002-0408-0. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and maintenance by Runx2. Front Biosci. 2008;13:898–903. doi: 10.2741/2730. [DOI] [PubMed] [Google Scholar]
- Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krzeszinski J.Y., Wei W., Huynh H., Jin Z., Wang X., Chang T.C., Xie X.J., He L., Mangala L.S., Lopez-Berestein G. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431–435. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li X.J., Ji M.H., Zhong S.L., Zha Q.B., Xu J.J., Zhao J.H., Tang J.H. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Arch. Med. Res. 2012;43:514–521. doi: 10.1016/j.arcmed.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Li H., Li T., Wang S., Wei J., Fan J., Li J., Han Q., Liao L., Shao C., Zhao R.C. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res. 2013;10:313–324. doi: 10.1016/j.scr.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Li Y., Fan L., Liu S., Liu W., Zhang H., Zhou T., Wu D., Yang P., Shen L., Chen J. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34:5048–5058. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- Lian J.B., Stein G.S., van Wijnen A.J., Stein J.L., Hassan M.Q., Gaur T., Zhang Y. MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zeng J., Wang L., Fang M., Wang Q., Zhao M., Xu X., Liu Z., Li W., Liu S. Histone demethylase retinoblastoma binding protein 2 is overexpressed in hepatocellular carcinoma and negatively regulated by hsa-miR-212. PLoS One. 2013;8:e69784. doi: 10.1371/journal.pone.0069784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.H., Chang Y.H., Sung L.Y., Li K.C., Yeh C.L., Yen T.C., Hwang S.M., Lin K.J., Hu Y.C. Osteogenic differentiation of adipose-derived stem cells and calvarial defect repair using baculovirus-mediated co-expression of BMP-2 and miR-148b. Biomaterials. 2014;35:4901–4910. doi: 10.1016/j.biomaterials.2014.02.055. [DOI] [PubMed] [Google Scholar]
- Luzi E., Marini F., Sala S.C., Tognarini I., Galli G., Brandi M.L. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J. Bone Miner. Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- Luzi E., Marini F., Tognarini I., Galli G., Falchetti A., Brandi M.L. The regulatory network menin-microRNA 26a as a possible target for RNA-based therapy of bone diseases. Nucleic Acid Ther. 2012;22:103–108. doi: 10.1089/nat.2012.0344. [DOI] [PubMed] [Google Scholar]
- Pang R.T., Leung C.O., Ye T.M., Liu W., Chiu P.C., Lam K.K., Lee K.F., Yeung W.S. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–1044. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- Park H., Park H., Pak H.J., Yang D.Y., Kim Y.H., Choi W.J., Park S.J., Cho J.A., Lee K.W. MiR-34a inhibits differentiation of human adipose tissue-derived stem cells by regulating cell cycle and senescence induction. Differentiation. 2015;90:91–100. doi: 10.1016/j.diff.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Perisanidis C., Perisanidis B., Wrba F., Brandstetter A., El Gazzar S., Papadogeorgakis N., Seemann R., Ewers R., Kyzas P.A., Filipits M. Evaluation of immunohistochemical expression of p53, p21, p27, cyclin D1, and Ki67 in oral and oropharyngeal squamous cell carcinoma. J. Oral Pathol. Med. 2012;41:40–46. doi: 10.1111/j.1600-0714.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- Pestell R.G. New roles of cyclin D1. Am. J. Pathol. 2013;183:3–9. doi: 10.1016/j.ajpath.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J., Galindo M., Zaidi S.K., Vradii D., Bhat B.M., Robinson J.A., Choi J.Y., Komori T., Stein J.L., Lian J.B. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- Qureshi A.T., Monroe W.T., Dasa V., Gimble J.M., Hayes D.J. miR-148b-nanoparticle conjugates for light mediated osteogenesis of human adipose stromal/stem cells. Biomaterials. 2013;34:7799–7810. doi: 10.1016/j.biomaterials.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Sun F., Fu H., Liu Q., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- Sun F., Wan M., Xu X., Gao B., Zhou Y., Sun J., Cheng L., Klein O.D., Zhou X., Zheng L. Crosstalk between miR-34a and Notch signaling promotes differentiation in apical papilla stem cells (SCAPs) J. Dent. Res. 2014;93:589–595. doi: 10.1177/0022034514531146. [DOI] [PubMed] [Google Scholar]
- Teng Y.C., Lee C.F., Li Y.S., Chen Y.R., Hsiao P.W., Chan M.Y., Lin F.M., Huang H.D., Chen Y.T., Jeng Y.M. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013;73:4711–4721. doi: 10.1158/0008-5472.CAN-12-3165. [DOI] [PubMed] [Google Scholar]
- Thomas M., Lieberman J., Lal A. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- Wang S., Qu X., Zhao R.C. Mesenchymal stem cells hold promise for regenerative medicine. Front Med. 2011;5:372–378. doi: 10.1007/s11684-011-0164-4. [DOI] [PubMed] [Google Scholar]
- Wang M.T., Chen G., An S.J., Chen Z.H., Huang Z.M., Xiao P., Ben X.S., Xie Z., Chen S.L., Luo D.L. Prognostic significance of cyclinD1 amplification and the co-alteration of cyclinD1/pRb/ppRb in patients with esophageal squamous cell carcinoma. Dis. Esophagus. 2012;25:664–670. doi: 10.1111/j.1442-2050.2011.01291.x. [DOI] [PubMed] [Google Scholar]
- Wang J., Guan X., Guo F., Zhou J., Chang A., Sun B., Cai Y., Ma Z., Dai C., Li X. miR-30e reciprocally regulates the differentiation of adipocytes and osteoblasts by directly targeting low-density lipoprotein receptor-related protein 6. Cell Death Dis. 2013;4:e845. doi: 10.1038/cddis.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xie Q., Yu Z., Zhou H., Huang Y., Bi X., Wang Y., Shi W., Sun H., Gu P. A regulatory loop containing miR-26a, GSK3β and C/EBPα regulates the osteogenesis of human adipose-derived mesenchymal stem cells. Sci. Rep. 2015;5:15280. doi: 10.1038/srep15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang D., Hu Z., Cheng J., Zhuo C., Fang X., Xing Y. MicroRNA-26a-modified adipose-derived stem cells incorporated with a porous hydroxyapatite scaffold improve the repair of bone defects. Mol. Med. Rep. 2015;12:3345–3350. doi: 10.3892/mmr.2015.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Shi Y., Zheng L., Zhou B., Inose H., Wang J., Guo X.E., Grosschedl R., Karsenty G. MiR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Zhou H., Liu G., Zhang P., Fu Y., Gu P., Hou H., Tang T., Fan X. Bone marrow stromal cells with a combined expression of BMP-2 and VEGF-165 enhanced bone regeneration. Biomed. Mater. 2011;6:015013. doi: 10.1088/1748-6041/6/1/015013. [DOI] [PubMed] [Google Scholar]
- Xu P.Z., Guo M., Hay B.A. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Yau W.W., Rujitanaroj P.O., Lam L., Chew S.Y. Directing stem cell fate by controlled RNA interference. Biomaterials. 2012;33:2608–2628. doi: 10.1016/j.biomaterials.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Ye J.H., Xu Y.J., Gao J., Yan S.G., Zhao J., Tu Q., Zhang J., Duan X.J., Sommer C.A., Mostoslavsky G. Critical-size calvarial bone defects healing in a mouse model with silk scaffolds and SATB2-modified iPSCs. Biomaterials. 2011;32:5065–5076. doi: 10.1016/j.biomaterials.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Ge Z., Wang L., Li Q., Wang N., Björkholm M., Jia J., Xu D. The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138:981–992. doi: 10.1053/j.gastro.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Qu X., Li H., Huang S., Wang S., Xu Q., Lin R., Han Q., Li J., Zhao R.C. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 2012;586:2375–2381. doi: 10.1016/j.febslet.2012.05.049. [DOI] [PubMed] [Google Scholar]
- Zhang Z.J., Zhang H., Kang Y., Sheng P.Y., Ma Y.C., Yang Z.B., Zhang Z.Q., Fu M., He A.S., Liao W.M. MiRNA expression profile during osteogenic differentiation of human adipose-derived stem cells. J. Cell Biochem. 2012;113:888–898. doi: 10.1002/jcb.23418. [DOI] [PubMed] [Google Scholar]
- Zhang C., Mo R., Yin B., Zhou L., Liu Y., Fan J. Tumor suppressor microRNA-34a inhibits cell proliferation by targeting Notch1 in renal cell carcinoma. Oncol. Lett. 2014;7:1689–1694. doi: 10.3892/ol.2014.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Sakamoto K., Wagner K.U. D-type cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol. Cell Endocrinol. 2014;382:583–592. doi: 10.1016/j.mce.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Zhang Z., Ye D., Tang A., Deng L., Han W., Zhao J., Wang S., Zhang W., Zhu C. Repair of critical-sized rat calvarial defects using genetically engineered bone marrow-derived mesenchymal stem cells overexpressing hypoxia-inducible factor-1α. Stem Cells. 2011;29:1380–1390. doi: 10.1002/stem.693. [DOI] [PubMed] [Google Scholar]
- Zuk P.A., Zhu M., Ashjian P., De Ugarte D.A., Huang J.I., Mizuno H., Alfonso Z.C., Fraser J.K., Benhaim P., Hedrick M.H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.