Abstract
Background and aims
ABO blood type is associated with cardiovascular disease, although the underlying mechanisms are presumed to be complex. While the relationship between non-O blood types and von Willebrand Factor (vWF) is well-established, associations with cellular adhesion molecules (CAMs) across diverse populations are understudied.
Methods
We genetically inferred ABO alleles for N=6202 participants from the Multi-Ethnic Study of Atherosclerosis. Linear regression was used to evaluate associations between major ABO allele dosages and log-transformed measurements of vWF (N=924), soluble E-selectin (sE-selectin, N=925), soluble P-selectin (sP-selectin, N=2392), and soluble ICAM-1 (sICAM-1, N=2236) by race/ethnicity.
Results
For the selectins, the A1 allele was associated with significantly lower levels for all races/ethnicities, with each additional allele resulting in a 28-39% decrease in sE-selectin and 10-18% decrease in sP-selectin relative to Type O subjects. However, the A2 allele demonstrated effect heterogeneity across race/ethnicity for sE-selectin, with lower levels for non-Hispanic whites (p=0.0011) but higher levels for Hispanics (p=0.0021). We also identified elevated sP-selectin levels for B-allele carriers solely in Hispanic participants (p=1.0E-04). ABO-by-race/ethnicity interactions were significant for both selectins (p <0.0125). More modest associations were observed between A1 allele dosage and levels of sICAM-1, with ABO alleles explaining 0.8-1.1% of the total phenotypic variation within race/ethnicity. ABO associations with vWF activity were consistent across race/ethnicity, with B allele carriers corresponding to the highest vWF activity levels.
Conclusions
ABO blood type demonstrates complex associations with endothelial markers that are largely generalizable across diverse populations.
Keywords: cellular adhesion, von willebrand factor, multi-ethnic, ABO
INTRODUCTION
The ABO histo-blood group is one of the major human blood antigen systems, classified by the presence of A and B antigens on the surface of red blood cells. These antigens are produced by glycosyltransferases encoded by the ABO gene, which modify terminal oligosaccharides of the H precursor antigen. In contrast to the A and B alleles of ABO, the O allele is enzymatically inactive due to the presence of functionally deleterious mutations, and homozygous O allele carriers (i.e., Type O) lack modified H antigens. ABO glycosyltransferases are also known to modify surface glycoproteins of platelets, vascular endothelium and epithelium, and other cell types1.
ABO blood type has been associated with phenotypic variation of cardiovascular disease (CVD) risk factors as well as susceptibility to CVD across multiple studies2. Increased risk of venous thromboembolism and stroke has consistently been observed for non-O blood types3-6. Associations of genetic variation at ABO have also been reported for myocardial infarction7, atherosclerosis8, and coronary heart disease9, with variants tagging non-O alleles associated with increased disease risk. The association between ABO and stroke is likely mediated by the relationship between glycosyltransferase activity and plasma levels of the procoagulant von Willebrand Factor (vWF)10, 11. However, a postulated mechanism relating ABO to atherosclerotic CVD is that ABO glycosyltransferases additionally impact circulation of cellular adhesion molecules (CAMs). CAMs are comprised of multiple protein families that are expressed on the vascular endothelium and recruit leukocytes in response to inflammatory stimuli, and their soluble forms present in blood are the product of shedding or proteolytic cleavage of the ectodomain. Soluble forms of CAMs E-selectin (sE-Selectin), P-selectin (sP-selectin), and ICAM-1 (sICAM-1) are biomarkers of inflammation, and increased circulation with one or more of these markers has been associated with coronary artery disease12, myocardial infarction13 and atherosclerosis14-16.
Recent studies have indicated substantial racial/ethnic differences in endothelial markers among healthy individuals17. Hwang et al.14 identified lower circulation of sICAM-1 in African Americans compared to subjects of European ancestry, while significant racial/ethnic differences in sICAM-1 concentrations were similarly observed between Asian, Hispanic, black, and non-Hispanic white subjects in the Women’s Health Study18. Racial/ethnic differences have also been observed for sP-selectin19, sE-selectin17, 20 and vWF21, 22. Although circulating levels of CAMs are highly heritable23, 24, and genetic studies have consistently identified associations between soluble levels and the ABO locus25-29, associations between ABO blood type and endothelial markers in diverse populations have not been extensively studied. Herein, we examine associations of circulating protein measurements of sE-selectin, sP-selectin, sICAM-1, as well as vWF with genetically inferred ABO blood group alleles in a large, multi-ethnic cohort: the Multi-Ethnic Study of Atherosclerosis (MESA).
MATERIALS AND METHODS
Study participants
The Multi-Ethnic Study of Atherosclerosis (MESA), described in greater detail elsewhere30, enrolled N=6814 participants aged 45-84 and without existing clinical CVD from 2000-2002. This study population included 38% non-Hispanic White American (EUR), 28% African American (AFA), 22% Hispanic American (HIS), and 12% Chinese American (CHN) participants. MESA participants were examined at six field centers located in Baltimore, MD; Chicago, IL; Forsyth County, NC; Los Angeles County, CA; northern Manhattan, NY; and Saint Paul, MN. At each visit, information on demographics, cardiovascular risk factors, past medical history and co-morbidities, social history, family history, and medications was collected through a combination of self-administered questionnaires and interview-administered questionnaires. Height was measured while participants were standing without shoes, heels together against a vertical mounted ruler. Body mass index (BMI) was calculated as weight (kg)/height2 (m2).
For this study, stored DNA was available for additional genotyping of key ABO polymorphisms on N=6276 MESA participants who consented to genetic studies. MESA and its ancillary studies were approved by the Institutional Review Boards at participating centers and all participants gave written informed consent.
Protein measurements
Circulating levels of sE-selectin and vWF were measured at the baseline exam (2000-2002) in a random subset of 1000 participants, of which 997 have available genetic information. sE-selectin was measured in serum using a high sensitivity quantitative sandwich enzyme-linked immunosorbent assay (ELISA) (Parameter Human sE-Selectin Immunoassay; R&D Systems, Minneapolis, MN). The inter-assay CV of the assay ranged from 5.7 – 8.8% with a minimum detectable level of 0.1 ng/mL. vWF activity was measured in citrated plasma by an immunoturbidimetric assay on the Sta-R analyzer (Diagnostica Stago, Parsippany, NJ) with an inter-assay CV of 4.5%. For circulating concentrations of sP-selectin and sICAM-1, a stratified random sample including 720 individuals for each of the four races/ethnicities represented in MESA at Exam 2 (2002-2004) was used (N=2880), of which 2574 participants and 2441 participants had available plasma and serum, respectively. sP-selectin was measured in EDTA plasma on 2529 MESA participants by ELISA using the Human soluble P-selectin/CD62P Immunoassay kit (R&D Systems), with an inter-assay CV was 6.7%, and a minimum detectable level of 0.5 ng/mL. For sICAM-1, measurements were collected in serum on 2374 participants by ELISA using the Human sICAM-1 Instant ELISA (Bender MedSystems GmbH) with an inter-assay CV of 9.1% and a minimum detectable level of 2.17 ng/mL.
Genotyping and annotation
The genotype data consisted of three individual genotyping panels: the Illumina Exome BeadChip31, the Illumina Cardio-MetaboChip32, and the Illumina iSelect ITMAT/Broad/CARe (IBC) Chip33. Each of the three panels had quality control measures individually performed on the genotype data prior to merging them together using Plink v1.0734 under NCBI genome build 37. Population stratification was assessed using EIGENSTRAT35 and the first three ancestry-informative principal components (PCs) were considered for covariate adjustment. Additional genotyping of key functional ABO genetic variants necessary for ABO blood type prediction was conducted using a custom Sequenom (San Diego, CA) panel that included 10 ABO variants as part of a 27 variant panel design, three of which (rs8176746, rs1053878, rs7853989) were previously assayed by the Illumina arrays. Quality control for the Sequenom genotype data was based upon sample call rate (>90%), genotype call rate (>85%), and Hardy-Weinberg equilibrium (p >0.05/40=0.00125; evaluated separately by race/ethnicity and adjusted for multiple testing). Each plate included a CEPH trio for quality control purposes, along with a random selection of 73 samples with duplicates. Discordant duplicate samples (N=2) and samples with discordant genotypes for the three previously genotyped SNPs (N=7) were excluded from further analyses.
ABO blood type allele prediction
Genetic evaluation of ABO blood type was determined on the basis of all ABO coding variants in the union of the merged chip genotype data and the ABO variants interrogated on the Sequenom panel. To fully characterize all identifiable ABO alleles, we first applied de novo haplotyping methods using the R statistical software package haplo.stats, which employs an expectation-maximization type algorithm to estimate subject haplotype pairs (i.e., diplotypes) while accommodating missing data. This analysis produces a posterior probability for each unique diplotype for a given individual’s genotypes. Haplotype analyses were applied to each race/ethnicity separately to accommodate differences in population allele frequencies.
Mapping of estimated ABO haplotypes to previously observed ABO alleles was conducted using genotables from the Blood Group Antigen Gene Mutation Database (BGMUT), an online curated genetic ABO allele database36, 37. All haplotypes with no direct match in the BGMUT database were assigned novel O1, O2, A2, or B allele IDs in a hierarchical fashion (see Supplementary Material for details). For purposes of association analysis with protein measurements, all detected alleles were assigned to either one of the two major A subtype alleles (A1 or A2), B, or O by summing over all posterior probabilities supporting the same major ABO diplotype. ABO blood type was also assigned following the well-established co-dominance relationship between A, B, and O alleles. For example, a given subject with respective posterior probabilities of 0.8 and 0.2 for diplotypes (A101/A103) and (A101/A102) would have a posterior probability of (0.8 + 0.2)=1.0 of diplotype (A1/A1), corresponding to Type A.
Statistical analyses
Outlier protein measurements were defined per race/ethnicity as being greater than four standard deviations away from the mean on the log scale, and excluded from further analysis. Race/ethnicity-stratified association analyses of ABO blood type with log-transformed protein values was conducted using linear regression with the statistical software R 3.0.1. Adjusting covariates included age (continuous), sex, BMI (continuous), and ancestry informative PCs, with values corresponding to the exam the protein data were collected where appropriate. ABO was defined under an additive genetic model by dosage of each major allele type (i.e., number of A1, A2, and B alleles, with O as the reference allele). All analyses were restricted to the subset of individuals that met the ABO major allele diplotype posterior probability threshold of >0.90 (two participants excluded).
ABO association analyses for ABO additive allele dosage effects across race/ethnicity and evaluation of potential race/ethnicity-specific effects based on participants from all four racial/ethnic strata were conducted in two manners: pooled sample regression and random effects meta-analysis. Pooled regression estimates were adjusted for the same set of covariates as the stratified analyses, with the exception of ancestry by self-reported race/ethnicity instead of PCs. Testing for race/ethnicity-specific ABO effects was conducted based upon likelihood ratio tests for race/ethnicity-by-ABO interaction terms. These terms were coded as race/ethnicity dummy variables multiplied by the ABO allele dosage, and testing was conducted per ABO allele (3 degree-of-freedom test) as well as for all alleles (9 degree-of-freedom test) in aggregate as an omnibus test. Random effects meta-analysis using the Sidik-Jonkman38 estimator with inverse-variance weights was applied to the race/ethnicity-stratified analysis results. Along with effect estimates and standard errors, Cochran’s Q statistics and I2 heterogeneity measurements39 were reported for each non-reference allele. All reported p-values are unadjusted; to account for multiple testing, we declared a given ABO-protein association (e.g., A1 allele dosage and vWF) or Q test for heterogeneity to be significant adjusting for the number of evaluated ABO alleles (three) and number of proteins (four), for a Bonferroni adjusted p-value threshold of 0.05/12=0.00417. Significance of omnibus tests for race/ethnicity-ABO interaction was declared based upon adjustment for the total number of proteins (0.05/4=0.0125).
RESULTS
ABO blood types
After quality control exclusions, 6255 MESA participants (AFA=1613, CHN=750, EUR=2469, HIS=1416) were evaluated for genetic prediction of ABO blood type, summarized in Table 1. Overall, 99.2% (6202/6255) of participants corresponded to specific major allele (A1,A2,B,O) ABO diplotypes with a posterior probability>0.90 based upon 28 ABO variants (Tables S1 and S2). Predicted ABO blood type distributions differed significantly across race/ethnicity (χ2 test p <1E-08), with EUR participants corresponding to the highest proportion of Type A (~43%) while the majority of HIS participants (57%) were Type O. Novel A2, B, and O alleles with no genetic match to those present in the BGMUT database accounted for 0.5-15.9% of all alleles observed, dependent upon race/ethnicity, with AFA corresponding to the highest and CHN the lowest proportions (Supplemental Table S3).
Table 1. Baseline characteristics, predicted ABO types and diplotypes, and protein measurement summaries of MESA participants.
Reported as N (%) or mean ± SD.
| Characteristics | African American |
Chinese American |
Non-Hispanic White |
Hispanic American |
p-valuea |
|---|---|---|---|---|---|
| N (All participants with ABO) | 1581 | 747 | 2453 | 1411 | |
| Age (years) | 62.1 ± 10.1 | 62.2 ± 10.4 | 62.6 ± 10.2 | 61.4 ± 10.3 | 0.005 |
| Sex (female) | 832 (53.8) | 383 (51.2) | 1287 (52.1) | 733 (51.9) | 0.59 |
| Body mass index (kg/m2) | 30.2 ± 5.8 | 24.0 ± 3.3 | 27.7 ± 5.0 | 29.5 ± 5.1 | <1e-08 |
| ABO | <1e-08 | ||||
| Type O | 795 (50.3) | 289 (38.7) | 1057 (42.9) | 806 (57.1) | |
| Type A | 420 (26.6) | 194 (26.0) | 1053 (42.9) | 434 (30.8) | |
| A1/A1 | 23 (1.5) | 29 (3.9) | 111 (4.5) | 24 (1.7) | |
| A2/A2 | 4 (0.3) | 0 (0) | 5 (0.2) | 1 (0.1) | |
| A1/A2 | 22 (1.4) | 0 (0) | 74 (3.0) | 14 (1.0) | |
| A1/O | 238 (15.1) | 165 (22.1) | 614 (24.9) | 307 (21.8) | |
| A2/O | 133 (8.4) | 0 (0) | 249 (10.1) | 88 (6.2) | |
| Type B | 286 (18.1) | 199 (26.6) | 255 (10.4) | 138 (9.8) | |
| B/B | 28 (1.8) | 22 (2.9) | 19 (0.8) | 7 (0.5) | |
| B/O | 258 (16.3) | 177 (23.7) | 236 (9.6) | 131 (9.3) | |
| Type AB | 80 (5.0) | 65 (8.7) | 98 (4.0) | 33 (2.3) | |
| A1/B | 53 (3.4) | 65 (8.7) | 71 (2.9) | 25 (1.8) | |
| A2/B | 27 (1.7) | 0 (0) | 27 (1.1) | 8 (0.6) | |
| vWF | |||||
| N | 175 | 95 | 440 | 218 | |
| Mean ± SD | 155.7 ± 62.1 | 134.4 ± 53.40 | 135.3 ± 54.4 | 134.8 ± 52.1 | 0.0005 |
| sE-selectin (ng/mL) | |||||
| N | 176 | 95 | 439 | 219 | |
| Mean ± SD | 59.0 ± 27.3 | 53.7 ± 28.6 | 49.0 ± 21.1 | 62.7 ± 27.1 | <1e-08 |
| sP-selectin (ng/mL) | |||||
| N | 541 | 620 | 637 | 608 | |
| Mean ± SD | 28.4 ± 9.1 | 27.9 ± 8.6 | 28.5 ± 9.4 | 31.8 ± 9.8 | <1e-08 |
| sICAM-1 (ng/mL) | |||||
| N | 489 | 581 | 599 | 573 | |
| Mean ± SD | 248.4 ± 83.0 | 228.1 ± 76.3 | 251.8 ± 77.3` | 259.8 ± 96.4 | <1e-08 |
p-values comparing results across race/ethnicity from Kruskal-Wallis tests for continuous variables or χ2 tests for categorical variables.
Protein measurements
The mean value of vWF activity was 15% higher in AFA (155.7) compared to other races/ethnicities, which were comparable in value (range: 134.4-135.3). Mean concentrations (ng/mL) of sE-selectin ranged from 49.0 (EUR) to 62.7 (HIS), while sP-selectin concentrations (ng/mL) were highest amongst HIS participants (31.8) and lowest in CHN (27.9). HIS participants also corresponded to the highest concentrations (ng/mL) of sICAM-1 (259.8), 13.9% higher than the lowest observed mean (CHN: 228.1). Overall, all proteins measurements demonstrated significant mean differences across race/ethnicity.
ABO association analyses
Race/ethnicity-stratified ABO association results for all protein measurements are presented in Table 2, with evaluations of race/ethnicity-specific effects in Table 3. In accordance with previous findings, vWF activity levels were significantly higher for all non-O blood types regardless of race/ethnicity. We observed higher vWF for A1 and B allele carriers, with comparable effect estimates across racial/ethnic groups and the B allele corresponding to higher effect estimates than the A1 allele, relative to the reference O allele. The A2 allele was significantly associated only in AFA (p=0.0037), with comparable effect estimates to A1, although formal testing for race/ethnicity-specific effects of A2 failed to reject the null (p >0.05).
Table 2. Race/ethnicity-stratified ABO association analysis results.
Analyses with log-transformed protein values of vWF, sE-selectin, sP-selectin, and sICAM-1, adjusted for age, sex, BMI, and genetic ancestry. Betas correspond to coefficient estimates from the regression analyses.
| vWF | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Allele (O ref.) |
African American n=175 |
Chinese Americana n=95 |
Non-Hispanic White n=440 |
Hispanic American n=218 |
|||||
|
| |||||||||
| Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | ||
| A1 | 0.29 (0.06) | 1.9E-05 | 0.24 (0.06) | 2.5E-04 | 0.25 (0.03) | <1E-08 | 0.24 (0.04) | 1.1E-07 | |
| A2 | 0.22 (0.08) | 3.7E-03 | - | - | 0.04 (0.05) | 0.40 | 0.10 (0.08) | 0.24 | |
| B | 0.34 (0.05) | <1E-08 | 0.26 (0.06) | 8.1E-05 | 0.36 (0.04) | <1E-08 | 0.31 (0.07) | 2.4E-05 | |
|
| |||||||||
| sE-selectin | |||||||||
|
| |||||||||
| n=176 | n=95 | n=439 | n=219 | ||||||
|
| |||||||||
| Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | ||
|
| |||||||||
| A1 | −0.35 (0.08) | 3.2E-05 | −0.49 (0.08) | 2.7E-08 | −0.51 (0.03) | <1E-08 | −0.41 (0.04) | <1E-08 | |
| A2 | −0.05 (0.09) | 0.63 | - | - | −0.15 (0.04) | 0.0011 | 0.26 (0.08) | 0.0021 | |
| B | −0.04 (0.07) | 0.51 | −0.01 (0.08) | 0.86 | −0.02 (0.04) | 0.70 | −0.06 (0.07) | 0.47 | |
|
| |||||||||
| sP-selectin | |||||||||
|
| |||||||||
| n=541 | n=620 | n=637 | n=608 | ||||||
|
| |||||||||
| Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | ||
|
| |||||||||
| A1 | −0.11 (0.03) | 1.7E-04 | −0.20 (0.02) | <1E-08 | −0.18 (0.02) | <1E-08 | −0.17 (0.02) | <1E-08 | |
| A2 | 0.02 (0.04) | 0.62 | - | - | −0.03 (0.03) | 0.45 | −0.01 (0.04) | 0.90 | |
| B | 0.07 (0.03) | 0.011 | −0.01 (0.02) | 0.51 | −0.01 (0.03) | 0.88 | 0.13 (0.03) | 1.0E-04 | |
|
| |||||||||
| sICAM-1 | |||||||||
|
| |||||||||
| n=489 | n=581 | n=599 | n=573 | ||||||
|
| |||||||||
| Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | ||
|
| |||||||||
| A1 | −0.05 (0.03) | 0.085 | −0.07 (0.02) | 0.0021 | −0.06 (0.02) | 0.0026 | −0.01 (0.03) | 0.59 | |
| A2 | 0.04 (0.05) | 0.44 | - | - | −0.03 (0.03) | 0.40 | −0.09 (0.05) | 0.085 | |
| B | 0.01 (0.03) | 0.78 | 0.01 (0.02) | 0.55 | −0.03 (0.03) | 0.37 | 0.06 (0.04) | 0.19 | |
SE: standard error.
A2 allele not present in Chinese Americans.
Table 3. Evaluation of potential race/ethnicity effect heterogeneity of ABO alleles on protein measurements.
Results based on race/ethnicity-pooled association analysis and random effects meta-analysis.
| vWF | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Pooled Analysis | Meta-Analysis | ||||||||
|
| |||||||||
| Allele | Beta (SE) | p | Interaction Test p |
Beta (SE) | p | Heterogeneity |
|||
| Allele | Omnibus | Q | p | I2 | |||||
| A1 | 0.25 (0.02) | <1E-08 | 0.99 | 0.83 | 0.25 (0.02) | <1E-08 | 0.40 | 0.94 | 2.0% |
| A2 | 0.08 (0.04) | 0.020 | 0.27 | - | 0.10 (0.06) | 0.051 | 4.23 | 0.12 | 52.3% |
| B | 0.33 (0.03) | <1E-08 | 0.64 | - | 0.32 (0.03) | <1E-08 | 1.60 | 0.66 | 13.3% |
|
| |||||||||
| sE-selectin | |||||||||
|
| |||||||||
| Pooled Analysis | Meta-Analysis | ||||||||
|
| |||||||||
| Allele | Beta (SE) | p | Interaction Test p |
Beta (SE) | p | Heterogeneity |
|||
| Allele | Omnibus | Q | p | I2 | |||||
|
| |||||||||
| A1 | −0.47 (0.02) | <1E-08 | 0.20 | 0.0085 | −0.45 (0.04) | <1E-08 | 5.80 | 0.12 | 51.0 % |
| A2 | −0.06 (0.04) | 0.13 | 7.5E-04 | - | 0.02 (0.12) | 0.88 | 18.33 | 1.0E-04 | 88.0% |
| B | −0.04 (0.03) | 0.13 | 0.73 | - | −0.03 (0.03) | 0.33 | 0.35 | 0.95 | 0.9% |
|
| |||||||||
| sP-selectin | |||||||||
|
| |||||||||
| Pooled Analysis | Meta-Analysis | ||||||||
|
| |||||||||
| Allele | Beta (SE) | p | Interaction Test p |
Beta (SE) | p | Heterogeneity |
|||
| Allele | Omnibus | Q | p | I2 | |||||
|
| |||||||||
| A1 | −0.17 (0.01) | <1E-08 | 0.16 | 0.012 | −0.17 (0.02) | <1E-08 | 6.53 | 0.088 | 62.4% |
| A2 | −0.01 (0.02) | 0.67 | 0.58 | - | −0.00 (0.02) | 0.87 | 0.81 | 0.67 | 7.1% |
| B | 0.03 (0.01) | 0.025 | 0.011 | - | 0.05 (0.03) | 0.19 | 11.22 | 7.0E-04 | 82.5% |
|
| |||||||||
| sICAM-1 | |||||||||
|
| |||||||||
| Pooled Analysis | Meta-Analysis | ||||||||
|
| |||||||||
| Allele | Beta (SE) | p | Interaction Test p |
Beta (SE) | p | Heterogeneity |
|||
| Allele | Omnibus | Q | p | I2 | |||||
|
| |||||||||
| A1 | −0.05 (0.01) | 2.4E-05 | 0.40 | 0.41 | −0.05 (0.01) | 2.8E-04 | 2.61 | 0.46 | 27.0% |
| A2 | −0.02 (0.02) | 0.29 | 0.21 | - | −0.02 (0.04) | 0.48 | 3.20 | 0.20 | 51.6% |
| B | 0.01 (0.01) | 0.49 | 0.53 | - | 0.01 (0.02) | 0.63 | 2.68 | 0.44 | 32.7% |
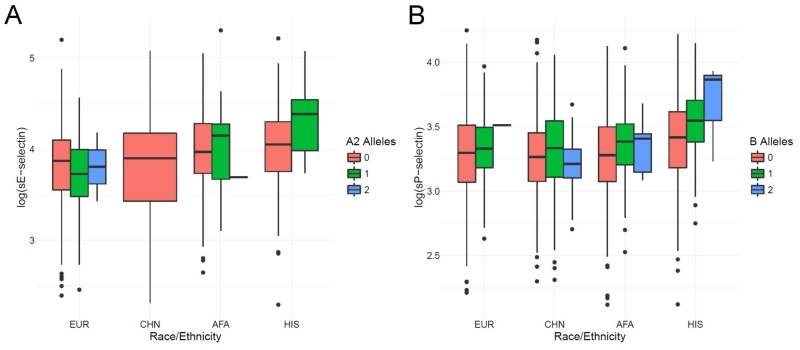
There were significant associations of A1 allele dosage with lower circulating levels of sE-selectin across all racial/ethnic strata, with each additional allele resulting in approximately 30-40% lower sE-selectin levels compared to the Type O participants. The B allele was not associated with protein variation in any analysis. The A2 allele demonstrated heterogeneous effects across race/ethnicity (Figure 1A), associated with lower sE-selectin levels in EUR (β=−0.15; p=0.0011) but increased levels in HIS (β=0.26; p=0.0021). The likelihood ratio test for race/ethnicity-specific A2 allelic effects was statistically significant (p=7.5E-04), as was the Q test for effect heterogeneity from the trans-ethnic meta-analysis (p=1.0E-04).
Fig.1. Distributions of log-transformed circulating selectin levels represented as boxplots, separated by racial/ethnic strata for ABO-protein associations indicating potential race/ethnicity-specific effects.
(A) log-transformed sE-selectin levels by dosage of estimated ABO A2 alleles and (B) log-transformed sP-selectin levels by estimated dosage of ABO B alleles.
A1 allele dosage was associated with the lower sP-selectin levels for all racial/ethnic strata, although effect estimates were lower in magnitude for AFA participants (β=−0.11, p=1.7E-04) than the other racial/ethnic groups (e.g., EUR: β=−0.18). The B allele was also significantly associated with increased plasma sP-selectin levels solely in HIS participants (β=0.13; p=1.0E-04), as indicated in Figure 1B. Although the allele-specific testing for race/ethnicity interaction in the pooled regression analysis was not significant after multiple testing adjustment, the meta-analysis Q heterogeneity statistic was significant (p=7.0E-04; I2=82.5%). Previous genome-wide association studies have demonstrated significant associations between circulation of sP-selectin and missense variants rs6136 and rs6133 within the P-selectin coding gene SELP25, 40, 41, which are known to vary in allele frequency by population. To evaluate the potential modifying role of SELP polymorphisms on the relationship between ABO and sP-selectin, we conducted sensitivity analyses additionally adjusting for minor allele dosages of rs6133 and rs6136 (Supplemental Table S4). Although these variants demonstrated significant independent associations with sP-selectin, there was no evidence indicating SELP polymorphisms modify the association between ABO and soluble protein levels.
Analysis results for MESA participants with serum sICAM-1 measurements indicated modest ABO effects compared to the selectins. The A1 allele was significantly associated with lower sICAM-1 levels in only the CHN (β=−0.07; p=0.0021) and EUR (β=−0.06; p=0.0026) strata, although the trend of association was consistent for AFA participants (β=−0.05; p=0.085). No significant ABO association results were observed for HIS.
For vWF, sE-selectin, and sP-selectin, ABO alleles explained a substantial proportion of the total trait variation (Supplemental Table S5) beyond the effects of age, sex, BMI, and genetic ancestry. Notably, African Americans had much lower phenotypic variation explained by ABO for the selectins than the other racial/ethnic groups.
DISCUSSION
We evaluated associations of ABO blood group alleles with protein measurements of circulating vWF activity, sE-selectin, sP-selectin, and sICAM-1 in a large, multi-ethnic cohort. Our results were consistent with previous findings observed in studies of predominantly non-Hispanic white subjects and demonstrated that the relationships between ABO and endothelial markers are largely generalizable across diverse populations, while also indicating potential race/ethnicity-specific effects for selectins.
The procoagulant vWF is a glycoprotein that mediates thrombosis and adhesion of molecules in the blood, and has been shown to be related with coronary heart disease, venous thromboembolism, and atherosclerosis.42-44 We observed considerably higher vWF in non-O type participants, regardless of race/ethnicity, and B allele carriers exhibited the largest effects. Effect sizes were comparable across race/ethnicity, with ABO alleles accounting for 17-24% of the total trait variation. However, the A2 allele was significantly associated with vWF solely in African Americans, demonstrating similar magnitude of effect to the A1 allele. Previous results from Song et al.45 investigating vWF associations with ABO in the Atherosclerosis Risk in Communities cohort found approximately 8% higher mean vWF for Type A2 African Americans relative to Type O, although this effect is smaller in magnitude relative to that observed in our analysis.
Findings for sICAM-1 were also largely consistent with previous analyses, which characterized the effects of ABO allele A1 to be relatively modest compared to other CAMs25, 26 in subjects of European and African descent. Although race/ethnicity-stratified associations were only significant in Chinese and non-Hispanic white MESA participants, heterogeneity statistics from our meta-analysis across race/ethnicity did not indicate substantial effect differences of the A1 allele associations and null findings in the other racial/ethnic strata are likely due to lack of power to detect the modest effects.
Circulating sE-selectin has been previously shown to be associated with the ABO blood group locus ABO, with the minor allele of rs651007 (tagging the A1 allele) corresponding to lower protein levels27, 28. Similar results were found when evaluating the effect of rs579459 (r2 with rs651007=1.0) in the Italian Bruneck cohort26. While our analyses corroborated the association of the A1 allele with lower sE-selectin levels, we discovered potential racial/ethnic divergence in the effects of the A2 allele. Particularly, HIS participants carrying the A2 allele demonstrated higher mean protein levels than corresponding Type O participants, while analysis results for the EUR participants indicated A2 dosage was associated with modestly lower concentrations. The latter finding is consistent with molecular biology of the A alleles, as the A1 allele has 30-50 fold higher A glycosyltransferase activity than the A2 allele and it has been hypothesized that this mechanism influences the clearance of adhesion molecules25. However, explanations for the positive correlation of A2 allele dosage with higher sE-selectin observed in HIS participants relative to Type O are not immediately evident. Given the relatively low number of HIS A2 carriers with sE-selectin measurements (N=17), chance confounding with relevant lifestyle factors may be possible; however, sensitivity analyses additionally adjusting for alcohol use, smoking status, or diabetes did not attenuate the association and further studies are warranted. Contrary to the findings of Qi et al.28, we did not identify any association with the B-allele in any of the racial/ethnic groups or combined across race/ethnicity via pooled regression or meta-analysis.
Barbalic et al.25 previously identified lower levels of serum P-selectin in subjects of European descent carrying the A1 allele. Similarly, Chen et al.46 also identified lower serum (but not platelet) P-selectin levels in Han Chinese, a population in which the A2 allele is not present. In MESA, HIS, EUR, and CHN participants all demonstrated a mean decrease of approximately 19% per A1 allele, in contrast to AFA, which only corresponded to a mean decrease of 9.5%. The relationship between ABO and sP-selectin was comparable to that of sE-selectin, with the presence of A1 alleles corresponding to lower plasma levels. However, the race/ethnicity-specific significance of the B allele in HIS participants is novel, indicating not only that ABO may be associated with different effects across populations for specific proteins, but across different selectins within populations. Contributing factors to these race/ethnicity-specific effects are not immediately clear, as often effect heterogeneity in genome-wide association analyses across diverse populations is postulated to be due to differential patterns of linkage disequilibrium, impacting polymorphisms tagging of true underlying functional variation. However, the direct genotyping of well-characterized functional variants in ABO applied in the current study largely mitigates that possibility. Similarly, investigation of previously identified missense SELP variants associated with sP-selectin variation did not indicate any effect modification in the relationship between ABO and circulating protein levels.
While a large body of research47, 48 has demonstrated vWF to express ABH antigens via glycan modification by ABO glycosyltransferases, resulting in reduced clearance and proteolysis, the biological mechanisms for ABO modulating soluble CAMs are less clear. The glycosyltransferase activity of A alleles results in the addition of an N-acetyl galactosamine to the H-antigen, in contrast to the B allele, which transfers a galactose, and it has been suggested that decreased cleavage of adhesion molecules from endothelium with A antigen present may result in reduced protein circulation25. Alternatively, CAMs may also be directly modified by ABO glycosyltransferases, altering their function, structure, secretion, and rate of clearance. As noted by others25, 26, this relationship between increased A1 allele dosage and reduced CAM concentrations seemingly contradicts the increased risk of CVD associated with non-O allele carriers in conjunction with the positive relationship between CVD risk and soluble CAM levels. Although our findings to not provide additional mechanistic insight into the seemingly paradoxical relationship connecting ABO, CAMs, and CVD, they do support its preservation across diverse populations.
Our study benefits from a well-characterized and multi-ethnic cohort as well as the quality and high density of directly genotyped functional ABO variation. However, due to sample size limitations within racial/ethnic strata, some ABO diplotypes were quite rare due to low allele frequencies (e.g., A1/A2) and our analyses were likely underpowered to detect modest effects or deviations from additivity. Additionally, we did not examine the effects of rarer allele subtypes, including O alleles that may have weak A antigen synthetic activity49. Finally, race/ethnicity-specific associations identified in our analyses will need to be validated in independent cohorts.
In summary, these results provide new insights into the relationships between ABO blood type, race/ethnicity, and endothelial markers as well as further evidence that ABO glycosyltransferases play a complex role in multiple CVD-related molecular pathways. Additional research is necessary to evaluate potential genetic and/or environmental modifying factors and further our mechanistic understanding of ABO associations with endothelial markers.
Supplementary Material
HIGHLIGHTS.
Cardiovascular disease (CVD) is the leading cause of death in every major racial/ethnic group the United States. Markers of endothelial dysfunction are promising predictors of CVD, and large proportions of natural variation in these biomarkers have been attributed to ABO blood type. However, the relationships between ABO and endothelial markers have not been extensively studied in diverse populations. In this study, we evaluated associations between genetically predicted ABO blood type and circulating measurements of vWF, E-selectin, P-selectin, and ICAM-1 in four racial/ethnic groups. While we observed generally consistent relationships across race/ethnicity, soluble levels of P-selectin and E-selectin demonstrated racial/ethnic heterogeneity, indicating potential differences in modifying factors by population.
ACKNOWLEDGEMENTS
The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
FINANCIAL SUPPORT
Cardiometabochip genotyping data was supported in part by grants and contracts R01HL98077, N02-HL-64278, HL071205, UL1TR000124, DK063491, RD831697, and P50 ES015915. Although the research described in this presentation has been funded in part by the United States Environmental Protection Agency through RD831697 to the University of Washington, it has not been subjected to the Agency’s required peer and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred. Funding for adhesion protein levels was provided by NHLBI by grant R01HL98077. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declared that they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
REFERENCES
- 1.Oriol R, Mollicone R, Coullin P, Dalix AM, Candelier JJ. Genetic regulation of the expression of abh and lewis antigens in tissues. APMIS Suppl. 1992;27:28–38. [PubMed] [Google Scholar]
- 2.Zhang H, Mooney CJ, Reilly MP. Abo blood groups and cardiovascular diseases. Int J Vasc Med. 2012;2012:641917. doi: 10.1155/2012/641917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohira T, Cushman M, Tsai MY, Zhang Y, Heckbert SR, Zakai NA, Rosamond WD, Folsom AR. Abo blood group, other risk factors and incidence of venous thromboembolism: The longitudinal investigation of thromboembolism etiology (lite) J Thromb Haemost. 2007;5:1455–1461. doi: 10.1111/j.1538-7836.2007.02579.x. [DOI] [PubMed] [Google Scholar]
- 4.Sode BF, Allin KH, Dahl M, Gyntelberg F, Nordestgaard BG. Risk of venous thromboembolism and myocardial infarction associated with factor v leiden and prothrombin mutations and blood type. Cmaj. 2013;185:E229–237. doi: 10.1503/cmaj.121636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiggins KL, Smith NL, Glazer NL, Rosendaal FR, Heckbert SR, Psaty BM, Rice KM, Lumley T. Abo genotype and risk of thrombotic events and hemorrhagic stroke. J Thromb Haemost. 2009;7:263–269. doi: 10.1111/j.1538-7836.2008.03243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakai NA, Judd SE, Alexander K, McClure LA, Kissela BM, Howard G, Cushman M. Abo blood type and stroke risk: The reasons for geographic and racial differences in stroke study. J Thromb Haemost. 2014;12:564–570. doi: 10.1111/jth.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Beckerath N, Koch W, Mehilli J, Gorchakova O, Braun S, Schomig A, Kastrati A. Abo locus o1 allele and risk of myocardial infarction. Blood Coagul Fibrinolysis. 2004;15:61–67. doi: 10.1097/00001721-200401000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Gong P, Luo SH, Li XL, Guo YL, Zhu CG, Xu RX, Li S, Dong Q, Liu G, Chen J, Zeng RX, Li JJ. Relation of abo blood groups to the severity of coronary atherosclerosis: An gensini score assessment. Atherosclerosis. 2014;237:748–753. doi: 10.1016/j.atherosclerosis.2014.10.107. [DOI] [PubMed] [Google Scholar]
- 9.He M, Wolpin B, Rexrode K, Manson JE, Rimm E, Hu FB, Qi L. Abo blood group and risk of coronary heart disease in two prospective cohort studies. Arterioscler Thromb Vasc Biol. 2012;32:2314–2320. doi: 10.1161/ATVBAHA.112.248757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirado I, Mateo J, Soria JM, Oliver A, Martinez-Sanchez E, Vallve C, Borrell M, Urrutia T, Fontcuberta J. The abo blood group genotype and factor viii levels as independent risk factors for venous thromboembolism. Thromb Haemost. 2005;93:468–474. doi: 10.1160/TH04-04-0251. [DOI] [PubMed] [Google Scholar]
- 11.Souto JC, Almasy L, Muniz-Diaz E, Soria JP, Borrell M, Bayen L, Mateo J, Madoz P, Stone W, Blangero J, Fontcuberta J. Functional effects of the abo locus polymorphism on plasma levels of von willebrand factor, factor viii, and activated partial thromboplastin time. Arterioscl Throm Vas. 2000;20:2024–2028. doi: 10.1161/01.atv.20.8.2024. [DOI] [PubMed] [Google Scholar]
- 12.Barbaux SC, Blankenberg S, Rupprecht HJ, Francomme C, Bickel C, Hafner G, Nicaud V, Meyer J, Cambien F, Tiret L. Association between p-selectin gene polymorphisms and soluble p-selectin levels and their relation to coronary artery disease. Arterioscler Thromb Vasc Biol. 2001;21:1668–1673. doi: 10.1161/hq1001.097022. [DOI] [PubMed] [Google Scholar]
- 13.Ridker PM, Hennekens CH, Roitman-Johnson B, Stampfer MJ, Allen J. Plasma concentration of soluble intercellular adhesion molecule 1 and risks of future myocardial infarction in apparently healthy men. Lancet. 1998;351:88–92. doi: 10.1016/S0140-6736(97)09032-6. [DOI] [PubMed] [Google Scholar]
- 14.Hwang SJ, Ballantyne CM, Sharrett AR, Smith LC, Davis CE, Gotto AM, Jr., Boerwinkle E. Circulating adhesion molecules vcam-1, icam-1, and e-selectin in carotid atherosclerosis and incident coronary heart disease cases: The atherosclerosis risk in communities (aric) study. Circulation. 1997;96:4219–4225. doi: 10.1161/01.cir.96.12.4219. [DOI] [PubMed] [Google Scholar]
- 15.Ridker PM. Intercellular adhesion molecule (icam-1) and the risks of developing atherosclerotic disease. Eur Heart J. 1998;19:1119–1121. doi: 10.1053/euhj.1998.1101. [DOI] [PubMed] [Google Scholar]
- 16.Polgar J, Matuskova J, Wagner DD. The p-selectin, tissue factor, coagulation triad. J Thromb Haemost. 2005;3:1590–1596. doi: 10.1111/j.1538-7836.2005.01373.x. [DOI] [PubMed] [Google Scholar]
- 17.Lutsey PL, Cushman M, Steffen LM, Green D, Barr RG, Herrington D, Ouyang P, Folsom AR. Plasma hemostatic factors and endothelial markers in four racial/ethnic groups: The mesa study. J Thromb Haemost. 2006;4:2629–2635. doi: 10.1111/j.1538-7836.2006.02237.x. [DOI] [PubMed] [Google Scholar]
- 18.Albert MA, Glynn RJ, Buring JE, Ridker PM. Relation between soluble intercellular adhesion molecule-1, homocysteine, and fibrinogen levels and race/ethnicity in women without cardiovascular disease. Am J Cardiol. 2007;99:1246–1251. doi: 10.1016/j.amjcard.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bielinski SJ, Berardi C, Decker PA, Kirsch PS, Larson NB, Pankow JS, Sale M, de Andrade M, Sicotte H, Tang W, Hanson NQ, Wassel CL, Polak JF, Tsai MY. P-selectin and subclinical and clinical atherosclerosis: The multi-ethnic study of atherosclerosis (mesa) Atherosclerosis. 2015;240:3–9. doi: 10.1016/j.atherosclerosis.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MA, Sagnella GA, Kerry SM, Strazzullo P, Cook DG, Cappuccio FP. Ethnic differences in circulating soluble adhesion molecules: The wandsworth heart and stroke study. Clin Sci (Lond) 2003;104:591–598. doi: 10.1042/CS20020333. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Z, Yu F, Buchanan A, Fu Y, Campos M, Wu KK, Chambless LE, Folsom AR, Boerwinkle E, Dong JF. Possible race and gender divergence in association of genetic variations with plasma von willebrand factor: A study of aric and 1000 genome cohorts. PLoS One. 2014;9:e84810. doi: 10.1371/journal.pone.0084810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folsom AR, Wu KK, Conlan MG, Finch A, Davis CE, Marcucci G, Sorlie PD, Szklo M. Distributions of hemostatic variables in blacks and whites: Population reference values from the atherosclerosis risk in communities (aric) study. Ethn Dis. 1992;2:35–46. [PubMed] [Google Scholar]
- 23.Lee M, Czerwinski SA, Choh AC, Demerath EW, Sun SS, Chumlea WC, Towne B, Siervogel RM. Quantitative genetic analysis of cellular adhesion molecules: The fels longitudinal study. Atherosclerosis. 2006;185:150–158. doi: 10.1016/j.atherosclerosis.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 24.Schnabel RB, Lunetta KL, Larson MG, Dupuis J, Lipinska I, Rong J, Chen MH, Zhao Z, Yamamoto JF, Meigs JB, Nicaud V, Perret C, Zeller T, Blankenberg S, Tiret L, Keaney JF, Jr., Vasan RS, Benjamin EJ. The relation of genetic and environmental factors to systemic inflammatory biomarker concentrations. Circ Cardiovasc Genet. 2009;2:229–237. doi: 10.1161/CIRCGENETICS.108.804245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbalic M, Dupuis J, Dehghan A, Bis JC, Hoogeveen RC, Schnabel RB, Nambi V, Bretler M, Smith NL, Peters A, Lu C, Tracy RP, Aleksic N, Heeriga J, Keaney JF, Jr., Rice K, Lip GY, Vasan RS, Glazer NL, Larson MG, Uitterlinden AG, Yamamoto J, Durda P, Haritunians T, Psaty BM, Boerwinkle E, Hofman A, Koenig W, Jenny NS, Witteman JC, Ballantyne C, Benjamin EJ. Large-scale genomic studies reveal central role of abo in sp-selectin and sicam-1 levels. Hum Mol Genet. 2010;19:1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiechl S, Pare G, Barbalic M, Qi L, Dupuis J, Dehghan A, Bis JC, Laxton RC, Xiao Q, Bonora E, Willeit J, Xu Q, Witteman JC, Chasman D, Tracy RP, Ballantyne CM, Ridker PM, Benjamin EJ, Ye S. Association of variation at the abo locus with circulating levels of soluble intercellular adhesion molecule-1, soluble p-selectin, and soluble e-selectin: A meta-analysis. Circ Cardiovasc Genet. 2011;4:681–686. doi: 10.1161/CIRCGENETICS.111.960682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakas M, Baumert J, Kleber ME, Thorand B, Dallmeier D, Silbernagel G, Grammer TB, Rottbauer W, Meisinger C, Illig T, Marz W, Koenig W. A variant in the abo gene explains the variation in soluble e-selectin levels-results from dense genotyping in two independent populations. PLoS One. 2012;7:e51441. doi: 10.1371/journal.pone.0051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi L, Cornelis MC, Kraft P, Jensen M, van Dam RM, Sun Q, Girman CJ, Laurie CC, Mirel DB, Hunter DJ, Rimm E, Hu FB. Genetic variants in abo blood group region, plasma soluble e-selectin levels and risk of type 2 diabetes. Hum Mol Genet. 2010;19:1856–1862. doi: 10.1093/hmg/ddq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paterson AD, Lopes-Virella MF, Waggott D, Boright AP, Hosseini SM, Carter RE, Shen E, Mirea L, Bharaj B, Sun L, Bull SB. Genome-wide association identifies the abo blood group as a major locus associated with serum levels of soluble e-selectin. Arterioscler Thromb Vasc Biol. 2009;29:1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieva J, Wendel M, Luttgen MS, Marrinucci D, Bazhenova L, Kolatkar A, Santala R, Whittenberger B, Burke J, Torrey M, Bethel K, Kuhn P. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: A longitudinal analysis. Phys Biol. 2012;9:016004. doi: 10.1088/1478-3975/9/1/016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, Chines PS, Teslovich TM, Romm JM, Ling H, McMullen I, Ingersoll R, Pugh EW, Doheny KF, Neale BM, Daly MJ, Kuusisto J, Scott LJ, Kang HM, Collins FS, Abecasis GR, Watanabe RM, Boehnke M, Laakso M, Mohlke KL. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, Frayling TM, Heid IM, Jackson AU, Johnson T, Kilpelainen TO, Lindgren CM, Morris AP, Prokopenko I, Randall JC, Saxena R, Soranzo N, Speliotes EK, Teslovich TM, Wheeler E, Maguire J, Parkin M, Potter S, Rayner NW, Robertson N, Stirrups K, Winckler W, Sanna S, Mulas A, Nagaraja R, Cucca F, Barroso I, Deloukas P, Loos RJ, Kathiresan S, Munroe PB, Newton-Cheh C, Pfeufer A, Samani NJ, Schunkert H, Hirschhorn JN, Altshuler D, McCarthy MI, Abecasis GR, Boehnke M. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. Plos Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SFA, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo YR, Li MY, DerOhannessian S, de Bakker PIW, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai XW, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CWK, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, FitzGerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k snp array for large-scale genomic association studies. PLoS One. 2008:3. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Patnaik SK, Helmberg W, Blumenfeld OO. Bgmut database of allelic variants of genes encoding human blood group antigens. Transfus Med Hemother. 2014;41:346–351. doi: 10.1159/000366108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patnaik SK, Helmberg W, Blumenfeld OO. Bgmut: Ncbi dbrbc database of allelic variations of genes encoding antigens of blood group systems. Nucleic Acids Res. 2012;40:D1023–1029. doi: 10.1093/nar/gkr958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidik K, Jonkman JN. Simple heterogeneity variance estimation for meta-analysis. J Roy Stat Soc C-App. 2005;54:367–384. [Google Scholar]
- 39.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiner AP, Carlson CS, Thyagarajan B, Rieder MJ, Polak JF, Siscovick DS, Nickerson DA, Jacobs DR, Jr., Gross MD. Soluble p-selectin, selp polymorphisms, and atherosclerotic risk in european-american and african-african young adults: The coronary artery risk development in young adults (cardia) study. Arterioscler Thromb Vasc Biol. 2008;28:1549–1555. doi: 10.1161/ATVBAHA.108.169532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee DS, Larson MG, Lunetta KL, Dupuis J, Rong J, Keaney JF, Jr., Lipinska I, Baldwin CT, Vasan RS, Benjamin EJ. Clinical and genetic correlates of soluble p-selectin in the community. J Thromb Haemost. 2008;6:20–31. doi: 10.1111/j.1538-7836.2007.02805.x. [DOI] [PubMed] [Google Scholar]
- 42.Medalie JH, Levene C, Papier C, Goldbourt U, Dreyfuss F, Oron D, Neufeld H, Riss E. Blood groups, myocardial infarction and angina pectoris among 10,000 adult males. N Engl J Med. 1971;285:1348–1353. doi: 10.1056/NEJM197112092852404. [DOI] [PubMed] [Google Scholar]
- 43.Kingsbury KJ. Relation of abo blood-groups to atherosclerosis. Lancet. 1971;1:199–203. doi: 10.1016/s0140-6736(71)90945-7. [DOI] [PubMed] [Google Scholar]
- 44.Koster T, Blann AD, Briet E, Vandenbroucke JP, Rosendaal FR. Role of clotting factor viii in effect of von willebrand factor on occurrence of deep-vein thrombosis. Lancet. 1995;345:152–155. doi: 10.1016/s0140-6736(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 45.Song J, Chen F, Campos M, Bolgiano D, Houck K, Chambless LE, Wu KK, Folsom AR, Couper D, Boerwinkle E, Dong JF. Quantitative influence of abo blood groups on factor viii and its ratio to von willebrand factor, novel observations from an aric study of 11,673 subjects. PLoS One. 2015;10:e0132626. doi: 10.1371/journal.pone.0132626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Zhuo X, Lin Y, Huang W, Xiao J, Zeng J, Jiang L, Chen C, Lin H, Dettke M. Association of abo blood group with p-selectin levels in chinese han healthy volunteers. Transfusion. 2015 doi: 10.1111/trf.13212. [DOI] [PubMed] [Google Scholar]
- 47.O’Donnell J, Boulton FE, Manning RA, Laffan MA. Amount of h antigen expressed on circulating von willebrand factor is modified by abo blood group genotype and is a major determinant of plasma von willebrand factor antigen levels. Arterioscler Thromb Vasc Biol. 2002;22:335–341. doi: 10.1161/hq0202.103997. [DOI] [PubMed] [Google Scholar]
- 48.Gallinaro L, Cattini MG, Sztukowska M, Padrini R, Sartorello F, Pontara E, Bertomoro A, Daidone V, Pagnan A, Casonato A. A shorter von willebrand factor survival in o blood group subjects explains how abo determinants influence plasma von willebrand factor. Blood. 2008;111:3540–3545. doi: 10.1182/blood-2007-11-122945. [DOI] [PubMed] [Google Scholar]
- 49.Yazer MH, Hosseini-Maaf B, Olsson ML. Blood grouping discrepancies between abo genotype and phenotype caused by o alleles. Curr Opin Hematol. 2008;15:618–624. doi: 10.1097/MOH.0b013e3283127062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.