Abstract
Thrips palmi Karny is a globally distributed polyphagous agricultural pest. It causes huge economic loss by its biological behaviors like feeding, reproduction and transmission of tospoviruses. Since T. palmi shows close morphological similarities with other thrips species, we employed mitochondrial cytochrome oxidase 1 (mtCO1) gene as a molecular marker. BLAST analysis of this sequence helped us to identify the collected specimen as T. palmi. We observed the female to male ratio of about 3:1 from collected samples and suspected the presence of Wolbachia. The presence of Wolbachia was detected by PCR using genus specific primers of 16S rRNA gene. Further confirmation of Wolbachia strain was achieved by conducting PCR amplification of three ubiquitous genes ftsZ, gatB and groEL. A phylogenetic tree was constructed with concatenated sequences of ftsZ and gatB gene to assign supergroup to Wolbachia. Finally, we localized Wolbachia in abdominal region of the insect using fluorescent in situ hybridization with the help of confocal microscope. Our result confirmed the presence of Wolbachia supergroup B strain for the first time in T. palmi.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-016-0567-7) contains supplementary material, which is available to authorized users.
Keywords: mtCO1 gene, Ubiquitous genes, 16S rRNA gene, PCR, FISH
Introduction
Many species of thrips cause huge economic loss in vegetable crops and ornamental plants worldwide [1]. They feed on plant sap by their piercing and sucking mouth parts resulting in curling and silvering of the leaves [2]. The reproductive behavior of thrips also results in fruit injury due to their oviposition [2]. Fourteen species of thrips belonging to the family Thripidae (order: Thysanoptera) are known to transmit 20 different tospoviruses (Tospovirus: Bunyaviridae) affecting around 1100 plant species worldwide [2–5]. T. palmi Karny is known to affect 50 different host plant species and has a global distribution [6]. T. palmi is considered as the one of most economically important thrips species in India [7]. So far, T. palmi is known to vector six tospoviruses; Calla lily chlorotic spot virus, Groundnut bud necrosis virus, Melon yellow necrotic virus, Tomato spotted wilt virus, Watermelon bud necrosis virus and Watermelon silver mottle virus [4, 5].
For an effective pest management strategy of thrips, accurate identification in the field and proper understanding of its biology are an essential pre-requisite. Adult thrips are very small in size (1–2 mm) and show many morphological similarities with other thrips species which make their identification difficult and require a trained manpower [2, 8]. Molecular markers assist in species level identification of such insects. Mitochondrial cytochrome oxidase 1 (mtCO1) gene has been used for identification of a number of organisms [9, 10] and is found to be a more reliable marker for distinguishing inter-specific variation [11]. mtCO1 gene has been effectively used as molecular marker for identification of T. palmi and Thrips tabaci [12].
Wolbachia is a facultative bacterium which comes under the alpha-subdivision of proteobacteria [13]. It is known to infect large number of insects and is reported to be transmitted maternally [13]. Wolbachia induces parthenogenesis, selective male killing and feminization of genetic males or cytoplasmic incompatibility by manipulating host reproduction [14]. Wolbachia has been reported in both thelytokous and arrhenotokous populations of thrips species such as Hercinothrips femoralis, Franklinothrips vespiformis, Echinothrips americanus, and Suocerathrips linguis [15, 16] and shown to induce thelytokous reproduction in Franklinothrips vespiformis [15].
In this study, we have focused on identification of T. palmi Karny in our collected sample. We achieved the same with conventional morphological study followed by molecular identification using mtCO1 gene. Later, we looked for the presence of Wolbachia in three different populations of T. palmi using PCR with its genus specific 16S rRNA gene primers. Three ubiquitous genes namely ftsZ, gatB [17] and groEL [18] were also analyzed to reconfirm presence of Wolbachia.ftsZ and gatB [17] genes were used for genotyping of Wolbachia into particular supergroup strain. To localize Wolbachia as a bacterial endosymbiont in T. palmi, fluorescent in situ hybridization (FISH) was performed using confocal microscopy. This study will provide a base for understanding the role of Wolbachia in reproductive behavior of T. palmi.
Materials and Methods
Sample Collection and DNA Isolation
Three different populations of thrips were collected from agricultural fields of Indian Agricultural Research Institute (IARI), Pusa, New Delhi. Later these specimens were identified by their morphological features with the help of compound microscope as T. palmi (Online Resource 1; Table S1). The identified specimens were stored at −20 °C in 70 % ethanol and absolute acetone for DNA isolation and FISH analysis respectively. For PCR amplification reactions, DNA isolation was done as previously performed by Rana et al. [19].
Molecular Identification of T. palmi Based on mtCO1 Gene
PCR amplification of mtCO1 gene was carried out with two sets of primers (Table 1). PCR reaction conditions and product visualization were same as previously described by Asokan et al. [12]. PCR products were eluted using HiPurA™ Quick Gel Purification Kit and were subsequently sequenced commercially (Macrogen Inc., Korea). The sequences were subjected to BLAST analysis in NCBI server. Sequence alignment was done with Clustal X2.1 and maximum likelihood analysis with Jukes-Cantor correction was performed for phylogenetic tree construction using MEGA6 [20]. Resultant tree topology was evaluated by bootstrap analysis based on 1000 repeats.
Table 1.
Primers used in this study
| Gene | Product | Primer name | Primer sequence (5′–3′) | References |
|---|---|---|---|---|
| mtCO1 | Mitochondrial cytochrome oxidase 1 | mtD7.2F | ATTAGGAGCHCCHGAYATAGCATT | [12] |
| mtD9.2R | GAGGCAAGATTAAAATATAAACTTCTTG | |||
| 1 RA f | TTGACTTCTTCCACCCTCTTTAACTCTT | |||
| 5 RA r | TAGATGTTGATAAAGTACAGGATCT | |||
| 16S rRNA | Ribosomal small subunit 16S | WF | CGGGGGAAAATTTATTGCT | [22] |
| WR | AGCTGTAATACAGAAAGGAAA | |||
| ftsZ | Cell division protein | ftsZ_F1 | ATYATGGARCATATAAARGATAG | [17] |
| ftsZ_R1 | TCRAGYAATGGATTRGATAT | |||
| gatB | Glutamyl-tRNA (Gln) amidotransferase, subunit B | gatB_F1 | GAKTTAAAYCGYCCAGGBGTT | [17] |
| gatB_R1 | TGGYAAYTCRGGYAAAGATGA | |||
| groEL | Chaperon protein | WgroEL_F | GGATCCATGGCTAACATAGT | This study |
| WgroEL_R | GTCGACTTAGAATCCACCCA |
Wolbachia Screening Using its Genus Specific Primer Based on 16S rRNA Gene
Presence of Wolbachia was detected using its genus specific primers of 16S rRNA gene (Table 1). PCR reaction of 25 μL consisted of 100 ng of DNA template isolated from a pool of 20 T. palmi individuals, 2.5 mM dNTP mixture, 10× Taq buffer, 10 pmol of each primer, 1 U Taq polymerase and volume was made up to 25 μL with autoclaved MQ water. PCR parameters were: 94 °C for 1 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 1 min and final extension of 10 min at 72 °C. For positive control, 16S rRNA gene of Wolabachia cloned in vector pGEM-T® Easy (Promega) was taken as template and negative control was performed without DNA template. PCR product was eluted using HiPurA™ Quick Gel Purification Kit and cloned directly into TA cloning vector pGEM-T® Easy (Promega, Madison, WI). E. coli DH5α strain was used for transformation of ligated product. Blue-white screening was performed for selection of positive colonies as per manufacturer’s instructions (Promega, Madison, WI). Three white colonies were selected for plasmid isolation using mdi fastlyse pDNA miniprep kit. Such purified plasmids were sequenced commercially (Macrogen Inc., Korea) followed by BLAST analysis of the sequence on the NCBI for similarity searches.
PCR Amplification of ftsZ, gatB and groEL Genes of Wolbachia
ftsZ, gatB and groEL genes of Wolbachia were amplified using Wolbachia specific primers (Table 1). Chemicals in PCR reaction were same as previously performed experiment. PCR parameters were: 94 °C for 2 min, followed by 42 cycles of 94 °C for 30 s, optimal annealing temperature for 45 s (see below), 72 °C for 1 min and final extension of 10 min at 72 °C. The optimal annealing temperature was 52 °C for ftsZ and gatB and 53 °C for groEL. As a negative control, mock reaction was set up without DNA template. Elution, cloning, sequencing and BLAST analysis of PCR products were performed as described earlier. Clustal X2.1 was used for sequence alignment of a concatenated data set of ftsZ and gatB genes and its phylogenetic analysis was performed as discussed earlier.
FISH Analysis
FISH technique was used as previously performed by Gottlieb et al. [21] with modifications. Insect specimens were incubated overnight at room temperature in Carnoy’s fixative (chloroform: ethanol: glacial acetic acid, 6:3:1) and then decolorized in 6 % H2O2 (Merck) in absolute ethanol for 4 h. Later, the specimens were hybridized overnight at 30 °C in hybridization buffer (20 mM Tris–Cl pH 8.0, 0.9 M NaCl, 0.01 % Sodium dodecyl sulfate, 50 % Formamide) with 0.6 pmol/μL fluorescent labeled probe. For detection of Wolbachia, LNA probe from Exiqon (TEX615-5′-CTTCTGTGAGTACCGTCATTATC-3′) was used. Specimens were then washed with washing buffer (0.3 M NaCl, 0.03 M sodium citrate, 0.01 % SDS) twice for 10 min and then mounted on glass slide using Fluoroshield (Sigma). To check the specificity of FISH, RNase digested control and no probe control were performed. Mounted specimens were observed under Nikon A1 confocal microscope and images were analyzed using NIS-Element (V 3.21.02) software (Nikon).
Results and Discussion
Sex Ratio of T. palmi Populations
Three different populations of thrips were identified by analyzing their morphological features with the help of compound microscope (Online Resource 1; Table S1). We observed the female to male ratio, on an average as 3:1 in the three different populations of T. palmi.
Molecular Identification of T. palmi
T. palmi shows similar morphological appearance to T. flavus Schrank and T. alatus Bhatti. T. flavus can be distinguished by the position of interocellar setae, which arise from inside the ocellar triangle posterior to the anterior ocellus, in contrast, it arise from outside the ocellar triangle in T. palmi [6]. While in T. alatus the metanotal sculpture is striate but not converged on the posterior margin as is usual in T. palmi [6]. To avoid any confusion, morphologically identified specimens were subjected to molecular identification based on mtCO1 gene PCR amplification to make sure that they were T. palmi and not any other thrips species [2, 8]. Sequencing of PCR amplification products of mtCO1 gene resulted in a sequence of 461 and 366 bp with the primer set 1 and set 2 respectively. These resultant sizes were close to the expected sizes of mtCO1 amplified product of T. palmi. Further, the nucleotide sequences from four different T. palmi specimens showed no variation. Similarity search result using BLAST algorithm in NCBI showed that these sequences were 99 % identical to previously deposited sequences of T. palmi in the GenBank. The partial mtCO1 gene sequence of T. palmi obtained in this study was submitted to NCBI GenBank under accession number KM393212. Phylogenetic tree analysis of our specimen and T. palmi mtCO1 gene sequences (retrieved from NCBI database) showed that our specimen was closest to T. palmi of India and formed a separate cluster (Online Resource 1; Fig. S1).
Detection and Reconfirmation of Wolbachia in T. palmi
During our attempts to collect the three different populations of T. palmi, it was observed that the female to male ratio, on an average was 3:1 indicating a strong female bias population. Since, Wolbachia was already reported in a few species of thrips and in Franklinothrips vespiformis, it was reported to induce thelytoky reproduction [15, 16]. The occurrence of female biased population in T. palmi prompted us to investigate for the presence of Wolbachia. Genomic DNA samples of three populations of well identified T. palmi were subjected to PCR for detection of Wolbachia with genus specific 16S rRNA gene primers. Conventionally, a 16S rRNA gene library is made to study bacterial endosymbiont population, but this technique does not detect bacterial endosymbionts which are rare [22]. Genus specific primers have been reported to be more sensitive for the detection of less populated bacterium [21].
We observed an amplification product of 632 bp which was in accordance with the expected size of 16S rRNA gene with genus specific primers of Wolbachia [22] in all three populations. Similarity search result using BLAST algorithm (search was performed against nucleotide sequence nr/nt, scoring parameter; match/mismatch scores were 1/−2 and gap cost set as linear) in NCBI showed that sequence of this product was 99 % identical to previously deposited sequences of Wolbachia from other insects in the GenBank. Thus sequence and BLAST analysis of this product confirmed the presence of Wolbachia in T. palmi. Partial 16S rRNA gene sequence of Wolbachia was submitted to NCBI GenBank under accession number KP889218.
Furthermore, all three populations of T. palmi were subjected to PCR amplification of three ubiquitous gene ftsZ, gatB [17] and groEL [18] for reconfirmation of presence of Wolbachia in T. palmi. We got amplification of ftsZ, gatB and groEL genes in all the three populations of T. palmi. Sequencing and BLAST analysis of these products reconfirmed the presence of Wolbachia in T. palmi. Partial sequences of ftsZ, gatB and groEL genes were submitted under accession numbers KT250944, KT250945 and KP889217 respectively. Phylogenetic tree analysis of concatenated sequences of ftsZ and gatB genes of Wolbachia from T. palmi and other species helped us to identify strain of Wolbachia in T. palmi as belonging to supergroup B (Online Resource 1; Fig. S2).
Localization of Wolbachia in T. palmi by FISH
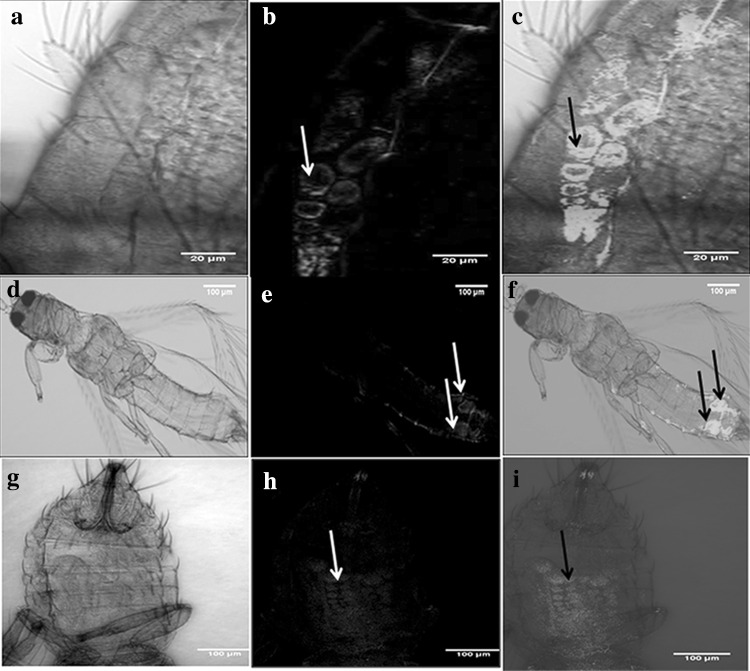
We performed FISH using confocal microscopy for localization of Wolbachia in T. palmi. A strong signal was detected which indicated the presence of Wolbachia in the abdominal region of T. palmi (Fig. 1). The presence of such precise Wolbachia signals were seen on an average in 20 % of specimens screened from all the three populations (100 individuals from each population). Besides this, we could observe some auto fluorescence in the legs and chitinous parts of the body which was also observed in no probe and RNase digested controls. Such structures have earlier been observed to show auto fluorescence in RNase digested controls and in absence of probe during Rickettsia sp. detection in mealybug [22].
Fig. 1.
FISH staining of Wolbachia in the whole mount of T. palmi (Female). a, d and g show bright field images. b, e and h show bacterial endosymbiont Wolbachia localized in abdomen region. c, f and i show merged images
Thus, through this study, we were able to detect and localize Wolbachia supergroup B strain in T. palmi, a condition which has not been reported yet. T. palmi are economically important to agriculture because of their ability to transmit viruses and monitoring their population and understanding their biology are important aspects. Ascertaining the role of bacterial endosymbionts like Wolbachia in biology and population dynamics of T. palmi is a critical part which may give us better insight for understanding the life history parameters of this insect. Further study is required to understand the functional role of Wolbachia in T. palmi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
ORP-SP and NFBSFARA, of the Indian Council for Agricultural Research (ICAR), Govt. of India and Faculty Research Grants of University of Delhi funded this work. SP was provided research fellowship by UGC/DSKPDF. GD was provided research fellowship by UGC. We also thank Mr. Ashok Pal for laboratory assistance.
References
- 1.Kirk WD, Terry IL. The spread of the western flower thrips Frankliniella occidentalis (Pergnade) Agric For Entomol. 2003;5:301–310. doi: 10.1046/j.1461-9563.2003.00192.x. [DOI] [Google Scholar]
- 2.Riley D, Shimat VJ, Srinivasan R, Diffie S. Thrips vectors of tospoviruses. J. Integr Pest Manag. 2011;1:1–10. doi: 10.1603/IPM10020. [DOI] [Google Scholar]
- 3.Parrella G, Gognalons P, Gebre-Selassie K, Vovlas C, Marchoux G. An update of the host range of tomato spotted wilt virus. J Plant Pathol. 2003;85:227–264. [Google Scholar]
- 4.Jones DR. Plant viruses transmitted by thrips. Eur J Plant Pathol. 2005;113:119–157. doi: 10.1007/s10658-005-2334-1. [DOI] [Google Scholar]
- 5.Pappu HR, Jones RAC, Jain RK. Global status of Tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009;141:219–236. doi: 10.1016/j.virusres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Mound LA, Azidah AA. Species of the genus Thrips (Thysanoptera) from Peninsular Malaysia, with a checklist of recorded Thripidae. Zootaxa. 2009;2023:55–68. [Google Scholar]
- 7.Mandal B, Jain RK, Krishnareddy M, Krishna Kumar NK, Ravi KS, Pappu HR. Emerging problem of tospoviruses (Bunyaviridae) and their management in the Indian subcontinent. Plant Dis. 2012;96:468–479. doi: 10.1094/PDIS-06-11-0520. [DOI] [PubMed] [Google Scholar]
- 8.Brunner PC, Flemming C, Frey JE. A molecular identification key for economically important thrips species (Thysanoptera: Thripidae) using direct sequencing and a PCR-RFLP based approach. Agric For Entomol. 2002;4:127–136. doi: 10.1046/j.1461-9563.2002.00132.x. [DOI] [Google Scholar]
- 9.Brunner PC, Chatzivassiliou EK, Katis NI, Frey JE. Host-associated genetic differentiation in Thrips tabaci (Insecta: Thysanoptera), as determined from mtDNA sequence data. Heredity. 2004;93:364–370. doi: 10.1038/sj.hdy.6800512. [DOI] [PubMed] [Google Scholar]
- 10.Frey JE, Frey B. Origin of intra-individual variation in PCR-amplified mitochondrial cytochrome oxidase I of T. tabaci (Thysanoptera: Thripidae): mitochodrial heteroplasmy or nuclear integration? Heriditas. 2004;140:92–98. doi: 10.1111/j.1601-5223.2004.01748.x. [DOI] [PubMed] [Google Scholar]
- 11.Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philos Trans R Soc Ser B. 2005;360:1805–1811. doi: 10.1098/rstb.2005.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asokan R, Krishna Kumar NK, Kumar V, Ranganath HR. Molecular differences in the mitochondrial cytochrome oxidase I (mtCOI) gene and development of a species-specific marker for onion thrips, Thrips tabaci Lindeman, and melon thrips, T. palmi Karny (Thysanoptera: Thripidae), vectors of tospoviruses (Bunyaviridae) Bull Entomol Res. 2007;97:461–470. doi: 10.1017/S0007485307005147. [DOI] [PubMed] [Google Scholar]
- 13.Jiggins FM, Bentley JK, Majerus MEN, Hurst GDD. How many species are infected with Wolbachia? Cryptic sex ratio distorters revealed to be common by intensive sampling. Proc R Soc Lond Ser B. 2001;268:1123–1126. doi: 10.1098/rspb.2001.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Arakaki N, Miyoshi T, Noda H. Wolbachia-mediated parthenogenesis in the predatory thrips Franklinothrips vespiformis (Thysanoptera: Insect) Proc R Soc Lond B. 2001;268:1011–1016. doi: 10.1098/rspb.2001.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumm S, Moritz G. First detection of Wolbachia in Arrhenotokous populations of Thrips species (Thysanoptera: Thripidae and Phelaeothripidae) and its role in reproduction. Environ Entomol. 2008;37:1422–1428. doi: 10.1603/0046-225X-37.6.1422. [DOI] [PubMed] [Google Scholar]
- 17.Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MC, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casiraghi M, Bordenstein SR, Baldo L, Lo N, Beninati T, Wernegreen JJ, Werren JH, Bandi C. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology. 2005;151:4015–4022. doi: 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- 19.Rana VS, Singh ST, Priya NG, Kumar J, Rajagopal R. Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci. PLoS One. 2012;7:421–468. doi: 10.1371/journal.pone.0042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, et al. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae) Appl Environ Microbiol. 2006;72:3646–3652. doi: 10.1128/AEM.72.5.3646-3652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh ST, Kumar J, Thomas A, Ramamurthy VV, Rajapoal R. Detection and localization of Rickettsia sp in Mealybug. Environ Entomol. 2013;42:711–716. doi: 10.1603/EN13032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.