The regulation of Rho-GAP localization is not well understood. A novel coiled-coil protein Rng10 is characterized that localizes the Rho-GAP Rga7 in fission yeast. Rng10 and Rga7 physically interact and work together to regulate the accumulation and dynamics of glucan synthases for successful septum formation during cytokinesis.
Abstract
Rho GAPs are important regulators of Rho GTPases, which are involved in various steps of cytokinesis and other processes. However, regulation of Rho-GAP cellular localization and function is not fully understood. Here we report the characterization of a novel coiled-coil protein Rng10 and its relationship with the Rho-GAP Rga7 in fission yeast. Both rng10Δ and rga7Δ result in defective septum and cell lysis during cytokinesis. Rng10 and Rga7 colocalize on the plasma membrane at the cell tips during interphase and at the division site during cell division. Rng10 physically interacts with Rga7 in affinity purification and coimmunoprecipitation. Of interest, Rga7 localization is nearly abolished without Rng10. Moreover, Rng10 and Rga7 work together to regulate the accumulation and dynamics of glucan synthases for successful septum formation in cytokinesis. Our results show that cellular localization and function of the Rho-GAP Rga7 are regulated by a novel protein, Rng10, during cytokinesis in fission yeast.
INTRODUCTION
Cytokinesis segregates chromosomes, cytoplasm, and organelles into the two daughter cells during the cell-division cycle. The underlying mechanisms and most players for cytokinesis are conserved from yeast to humans (Balasubramanian et al., 2004; Laporte et al., 2010; Pollard and Wu, 2010). Cytokinesis requires six coordinated events: division-site selection, actomyosin contractile-ring assembly, ring constriction and the associated disassembly, plasma membrane deposition, septum formation or extracellular matrix remodeling, and daughter-cell separation or midbody abscission (Balasubramanian et al., 2004; Barr and Gruneberg, 2007; Wloka and Bi, 2012). The later stages of cytokinesis are poorly understood.
Proper septum formation and cell separation are critical to maintain cell integrity and viability during fungal cytokinesis due to their high cellular turgor pressure (Roncero and Sanchez, 2010; Proctor et al., 2012; Wloka and Bi, 2012; Basu et al., 2014). The septum is a trilaminar structure with the primary septum, which is digested away during daughter-cell separation, sandwiched by the secondary septa. Three conserved glucan synthases are known to be essential for proper septum formation in the fission yeast Schizosaccharomyces pombe. β-Glucan synthase, Bgs1/Cps1, is responsible for primary septum synthesis (Liu et al., 1999; Cortes et al., 2002, 2007). β-Glucan synthase, Bgs4/Cwg1/Orb11, is essential for cell integrity and secondary septum formation (Cortes et al., 2005; Munoz et al., 2013). α-Glucan synthase, Ags1/Mok1, is mainly involved in secondary septum formation (Vos et al., 2007; Cortes et al., 2012). These conserved synthases are also essential for cell wall and septum formation in fungal pathogens and therefore are ideal targets for antifungal drugs (Yamaguchi et al., 1985; Ishiguro et al., 1997; Perlin, 2011; Lackner et al., 2014).
Rho GTPases are important regulators in all stages of cytokinesis (Santos et al., 2003; Jordan and Canman, 2012; Onishi et al., 2013; Chircop, 2014). They are known as “molecular switches” due to their ability to switch between the active, GTP-bound and the inactive, GDP-bound forms (Cherfils and Zeghouf, 2013; Braun and Olayioye, 2015). The activity of Rho GTPases is strictly regulated by Rho guanine nucleotide exchange factors (GEFs) and Rho GTPase-activating proteins (GAPs; Cherfils and Zeghouf, 2013; Zuo et al., 2014; Braun and Olayioye, 2015). Rho-GAPs hydrolyze GTP to GDP to inactivate Rho GTPases (Tcherkezian and Lamarche-Vane, 2007; Cherfils and Zeghouf, 2013). However, cells have more Rho-GAPs than Rho GTPases (Tcherkezian and Lamarche-Vane, 2007; Cherfils and Zeghouf, 2013; Schaefer et al., 2014). For most Rho GTPases, several Rho-GAPs are involved in their regulation (Schmidt et al., 2002; Moon and Zheng, 2003; Soto et al., 2010; Braun and Olayioye, 2015). Therefore it is crucial to tightly regulate Rho-GAP activities to specific sites in cells.
S. pombe has nine known or putative Rho GAPs (Yang et al., 2003; Villar-Tajadura et al., 2008; Arasada and Pollard, 2011, 2015; Martin-Garcia et al., 2014). Most Rho GAPs (Rga1, Rga2, Rga5, Rga7, and Rga8) localize to the cell tips during interphase and the division site during cell division (Nakano et al., 2001; Calonge et al., 2003; Yang et al., 2003; Villar-Tajadura et al., 2008; Martin-Garcia et al., 2014; Arasada and Pollard, 2015). The exceptions are Rga4, which localizes to the cell sides during interphase and the division site during cytokinesis, and Rga6, which localizes throughout the plasma membrane (Matsuyama et al., 2006; Tatebe et al., 2008; Villar-Tajadura et al., 2008; Soto et al., 2010). Rga1 functions as a GAP for Rho1 and is involved in actin-patch localization, cell morphogenesis, septation, and cell wall synthesis (Nakano et al., 2001). Rga2 is a GAP for Rho2 and regulates cell morphology and cell wall integrity (Villar-Tajadura et al., 2008). Rga2 localization depends on polarity markers and actin polymerization (Villar-Tajadura et al., 2008). Rga4 is a GAP for both Cdc42 and Rho2 and has roles in the control of cell diameter and symmetry and cell integrity pathway (Das et al., 2007; Soto et al., 2010). The DYRK protein kinase Pom1 regulates the localization and phosphorylation of Rga4 (Tatebe et al., 2008; Soto et al., 2010). Rga5 is a GAP for Rho1 and regulates cell integrity (Calonge et al., 2003). Rga6 is a less-studied GAP for Rho2 (Villar-Tajadura et al., 2008; Soto et al., 2010). Rga8 is a GAP for Rho1 and a downstream target of p21-activated kinase Shk1 (Yang et al., 2003). Rga3 and Rga9 have not been studied. Rga7 is a Rho2 GAP (Martin-Garcia et al., 2014; Arasada and Pollard, 2015). It cooperates with Fes/CIP4 homology-Bin/amphiphysin/Rvs (F-BAR) proteins Cdc15 and Imp2 in actomyosin ring stability, proper ring disassembly, and successful septum formation and separation to ensure cell integrity (Martin-Garcia et al., 2014). Rga7 GAP activity is important for regulating α-glucan synthase Ags1 through the Rho2 pathway in controlling cell wall and septum formation (Cansado et al., 2010; Soto et al., 2010), although a recent study suggests that the GAP activity is dispensable for Rga7 function in cytokinesis (Martin-Garcia et al., 2014). Rga7 also participates in β-glucan synthase Bgs4 trafficking from the Golgi to the plasma membrane (Arasada and Pollard, 2015). Despite the functional studies on these Rho GAPs, knowledge of the regulation of their localizations and functions is limited.
In this study, we show how the Rho-GAP Rga7 localizes to the division site to aid septum formation in fission yeast. We identify a novel coiled-coil protein, Rng10, which physically interacts with Rga7 and is required for Rga7 localization. Rng10 and Rga7 collaborate to regulate the localization and dynamics of glucan synthases for successful septum formation during cytokinesis.
RESULTS
Rng10 is a novel protein important for cell integrity during cell division
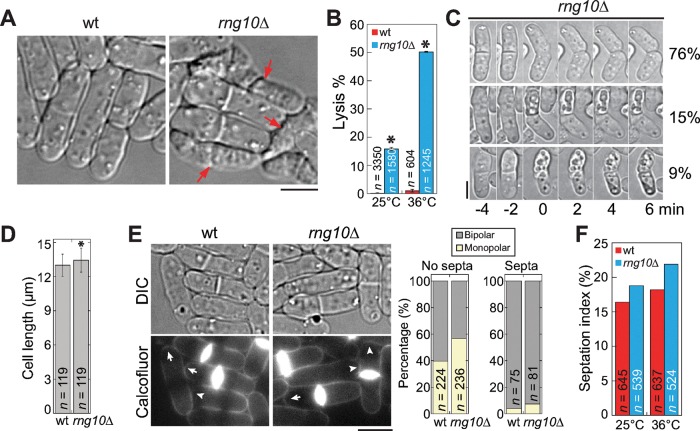
A novel protein, SPAC688.07c, was found to localize to cell tips and the site of cell division in a global protein localization analysis in S. pombe (Matsuyama et al., 2006). It is a 114-kDa protein with unknown functions and will be referred to as Rng10 in this study because of its localization. To investigate its role in S. pombe, we deleted rng10 in a haploid strain, since it is not an essential gene (Hayles et al., 2013). The major phenotype of rng10Δ was cell lysis (Figure 1A). Significantly more rng10Δ cells lysed (16% at 25°C, 50% at 36°C) than wild type (wt; 0.15% at 25°C, 1.0% at 36°C; Figure 1, A and B). Time-lapse movies showed that either one or both daughter cells lysed during cell separation in ∼25% of rng10Δ cells (n = 74 cells) at 25°C (Figure 1C and Supplemental Video S1). rng10Δ cells were slightly longer than wt cells at cell division (Figure 1D). Approximately 1% of rng10Δ cells (n = 1056) partially lost polarity and became rounder. We also examined polarized growth in wt and rng10Δ cells using Calcofluor, which stains cell wall but not birth scars, marking the beginning position in the new end (Figure 1E; Bohnert and Gould, 2012). The growing end has a decent arc stained beyond the birth scar, whereas the nongrowing end has a birth scar very close to the cell tip (Figure 1E, left; Bohnert and Gould, 2012). Quantification of growth patterns showed higher percentages of cells with monopolar growth in rng10Δ cells either with or without septa than in wt cells (Figure 1E, right). Thus Rng10 is crucial for cell integrity during cell division and plays a minor role in cell growth and cell size control.
FIGURE 1:
The novel protein Rng10 is important for cell integrity. (A, B) rng10Δ leads to cell lysis. (A) DIC images of cells grown at 36°C for 4 h. Arrows mark examples of lysed cells, which are evident by the shrunken cytoplasm. (B) Quantification of cell lysis in wt and rng10Δ cells at 25°C and 4 h at 36°C. Data are mean ± 1 SD in this and other figures except where noted. The number n is the total from three experiments. *p < 0.0001 compared with wt in t test. (C) Time-lapse images of cell-lysis scenarios (left) and percentage (right) during cell separation at 25°C. No (top), one (middle), or both (bottom) daughter cells lysed. (D) Quantification of cell length of wt and rng10Δ cells with forming or complete septa at 25°C. *p < 0.01 compared with wt. (E) Micrographs (left) and quantifications (right) of polarized growth in wt and rng10Δ cells stained with Calcofluor. Arrows mark growing ends, and arrowheads mark nongrowing ends that lack Calcofluor staining. (F) Septation index of wt and rng10Δ cells at 25°C or 4 h at 36°C. Bars, 5 μm.
To determine the causes of cell lysis in rng10Δ cells and roles of Rng10, we first examined the actomyosin contractile ring, which is essential for cytokinesis (Laporte et al., 2010; Lee et al., 2012). Using myosin-II regulatory light chain (Rlc1) tagged with tandem dimer Tomato (tdTomato) as a contractile‑ring marker, we found that rng10Δ did not affect ring assembly or constriction at 25°C (Supplemental Figure S1, A, B, and D). The time from the end of ring constriction to cell separation also appeared normal (Supplemental Figure S1D). However, the timings of these events were significantly affected at 36°C (Supplemental Figure S1E). The septation index of rng10Δ cells was slightly higher than that of wild type at both 25 and 36°C (Figure 1F). Next we tested whether cell lysis was due to defects in plasma membrane closure during cytokinesis, using fluorescence loss in photobleaching (FLIP). The exchange of diffusible green fluorescent protein (GFP) between the bleached and unbleached daughter cells stopped at the same time after ring constriction in rng10Δ and wt cells (Supplemental Figure S2, A, B, and D). Thus Rng10 plays a role in the contractile ring at high temperature but has no obvious (or redundant) role in contractile-ring or plasma membrane closure at 25°C during cytokinesis. Because cell lysis also occurs at 25°C, contractile-ring defects may not be the main reason for lysis in rng10Δ cells.
Rng10 localizes to the division site and the cell tips
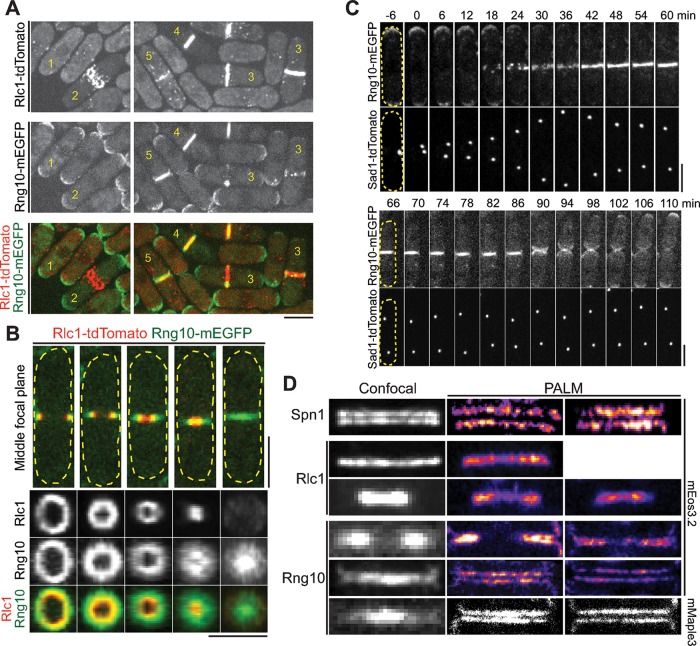
To further investigate Rng10 function during cytokinesis, we tagged rng10 at its native chromosomal locus with monomeric enhanced GFP (mEGFP) and other fluorescent tags to examine Rng10 localization throughout cell cycle (Figure 2). We found that Rng10 localized to cell tips (cells 1 and 2) during interphase and early mitosis and was concentrated at the division site (cells 3–5) during cell division (Figure 2A). After Rlc1 nodes condensed into a compact ring, Rng10 first appeared near the ring as puncta and then became a ring (Figure 2, A, cell 3, and C). Using the spindle pole body (SPB) protein Sad1 as a cell-cycle marker, we found that Rng10 arrived at the division site 17.0 ± 1.9 min (n = 27 cells) after SPB separation (Figure 2C, top, and Supplemental Video S2). Rng10 constricted with the contractile ring but also spread across the division site to form a disk, with a higher concentration at the leading edge of the cleavage furrow (Figure 2, A, cells 4 and 5, and B). After cell separation, Rng10 stayed at the new cell tip and then concentrated at both cell tips (Figure 2C, bottom, and Supplemental Video S2).
FIGURE 2:
Localization of Rng10 to the cells tips and the division site. (A) Rng10-mEGFP (green) localization in the cells with Rlc1-tdTomato (red)–labeled contractile ring at different stages. (B) Side (top) and vertical (bottom) views of Rng10 at the division plane in cells with constricting Rlc1 ring. (C) Time course (in minutes) of Rng10 localization with SPB protein Sad1 as a cell-cycle marker. SPB separation is defined as time 0. The broken lines mark the cell boundary in B and C. (D) Confocal and PALM images of Spn1, Rlc1, and Rng10 localization at the division site in cells tagged with mEGFP (confocal) or mEos3.2 and mMaple3 (PALM). Bars, 5 μm.
Because Rng10 only partially colocalizes with the contractile ring (Figure 2, A and B) and its localization is also distinct from the septin double rings on the confocal microscope (Figure 2D), we used superresolution photoactivated localization microscopy (PALM) to further examine its localization. Surprisingly, Rng10 formed a double-layer invagination that supposedly extended into two disks at the division site (Figure 2D). Rng10 was outside of the contractile ring (Rlc1) but inside of the septin rings (Spn1) along the cell long axis. This localization pattern suggests that Rng10 likely localizes to the plasma membrane at the division plane.
To test whether Rng10 localization depends on F-actin, microtubules, or vesicle trafficking, we treated cells with different drugs, including latrunculin A, CK-666, methyl benzimidazole-2-yl carbamate (MBC), and brefeldin A (BFA; Supplemental Figure S2, E and F). Rng10 tagged with a monomeric enhanced citrine (mECitrine, a yellow fluorescent protein variant; Griesbeck et al., 2001; Lee and Wu, 2012) still localized to the division site under these treatments. Together these data show that Rng10 localizes to the cell tips and the division site, and its localization or maintenance at the plasma membrane at the division site does not require F-actin, microtubules, or vesicle trafficking.
Rng10 localization depends on both its N- and C-termini
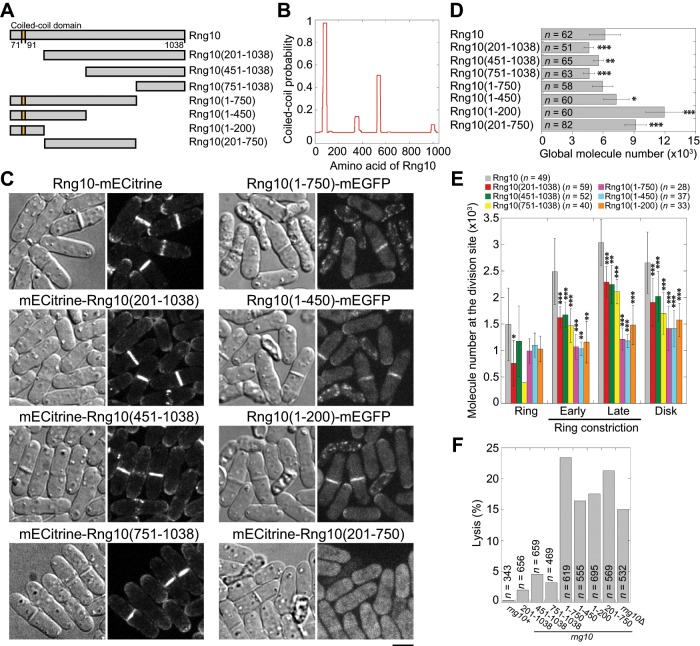
To determine the region of Rng10 required for its localization, we conducted a domain analysis of Rng10. Besides a short coiled-coil domain at the N-terminus, Rng10 contained no other recognizable domains (Figure 3, A and B). We constructed Rng10 N- and C-terminal truncations (Figure 3A) with either mECitrine or mEGFP tag under rng10 promoter based on the predicted domain boundary (Eickholt et al., 2011) and compared their localization and intensity to those of full-length (FL) Rng10 (Figure 3C). Both the N- and C-terminal truncations still localized to the division site (Figure 3C). We measured absolute protein numbers using quantitative fluorescence microscopy with known proteins as standards (Supplemental Figure S3 and Table 1). The molecule numbers of Rng10 truncations at the division site were significantly reduced compared with FL Rng10 in cells with a septum (Figure 3, C and E), although the global protein levels were mostly comparable or higher (Figure 3D, Supplemental Figure S3, and Table 1). Rng10(201-750) with both N- and C-termini truncated did not localize to the division site (Figure 3C). Thus both N (1-200) and C (751-1038) terminal regions of Rng10 are important for its localization. Of interest, cells with Rng10 C-terminal truncations resembled rng10Δ and lysed more frequently than N-terminal truncations (Figure 3F), suggesting that the C-terminus of Rng10 is more important for its function.
FIGURE 3:
Domain analyses for Rng10. (A) Schematics of Rng10 and its truncations constructed. (B) Probability of coiled-coil formation for Rng10 calculated using the program from Lupas et al. (1991) with a window size of 28. (C) Localization of FL and truncated Rng10. DIC images show cell morphology and lysis. Global (D) and division-site (E) molecule numbers of FL and truncated Rng10. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the FL protein. (E) Cells are grouped into four stages: ring (before constriction), ring constriction early (Rng10 occupies <50% of the division plane in the middle focal plane) and late (>50%), and disk (Rng10 fills the whole division site). For most stages, n = 4–31 cells, except for Rng10(1-450) in ring and early ring constriction stages with n = 2. The total cells analyzed for each constructs are listed on the top. (F) Quantification of cell lysis in rng10 mutants. Bars, 5 μm.
TABLE 1:
Global protein levels for Rng10 and its truncations along with known proteins for the standard curve measured by quantitative fluorescence microscopy.
| Protein | Number of cells analyzed | Intensity (mean ± SD) | Molecules per cell (mean ± SD) | Global concentration (μM)a |
|---|---|---|---|---|
| Rng10 | 58 | 1230 ± 300 | 6180 ± 1520 | 0.38 ± 0.09 |
| 62 | 2600 ± 300b | |||
| Rng10(1-200) | 60 | 1780 ± 270 | 11,940 ± 1780 | 0.74 ± 0.11 |
| Rng10(1-450) | 60 | 1340 ± 230 | 7280 ± 1240 | 0.45 ± 0.08 |
| Rng10(1-750) | 58 | 1210 ± 210 | 5960 ± 1020 | 0.37 ± 0.06 |
| Rng10(201-1038) | 51 | 2285 ± 240b | 4620 ± 480 | 0.29 ± 0.03 |
| Rng10(451-1038) | 65 | 2470 ± 220b | 5570 ± 490 | 0.35 ± 0.03 |
| Rng10(751-1038) | 63 | 2290 ± 260b | 4650 ± 530 | 0.29 ± 0.03 |
| Rng10(201-750) | 82 | 3191 ± 360b | 9150 ± 1030 | 0.57 ± 0.06 |
| Myo2 | 64 | 1060 ± 500 | 7300 ± 3450c | 0.45 ± 0.21 |
| Rlc1 | 64 | 1720 ± 230 | 9600 ± 1280c | 0.60 ± 0.08 |
| Spn1 | 61 | 1760 ± 280 | 10,300 ± 1620c | 0.63 ± 0.10 |
| Cdc15 | 64 | 4010 ± 600 | 35,600 ± 5370c | 2.13 ± 0.33 |
aThe global concentration was calculated with a mean cytoplasm volume of 27 μm3, which was derived from a mean cell volume of 92 μm3 and the cytoplasm occupying ∼29% of the cell volume (Wu and Pollard, 2005).
bIntensity for proteins tagged with mECitrine. Other intensities are from the mEGFP tag.
cThe mean numbers are from published data (Wu and Pollard, 2005).
Rng10 physically interacts with the Rho-GAP Rga7 and is crucial for its localization
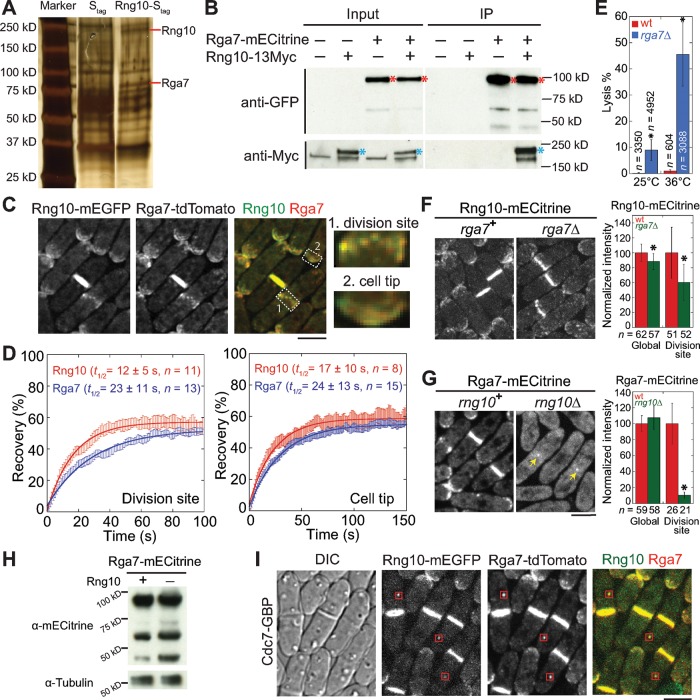
To elucidate the role of Rng10 in cytokinesis and why rng10Δ cells lyse during cell division, we searched for its binding partner. Affinity purification of Rng10-Stag from S. pombe extract and mass spectrometry identified Rga7, a Rho GAP with an F-BAR domain, as a potential Rng10-interacting protein (Figure 4A). Coimmunoprecipitation (coIP) of Rng10-13Myc by Rga7-mECitrine confirmed the interaction between Rng10 and Rga7 (Figure 4B). In addition, Rng10 and Rga7 perfectly colocalized at the cell tips and division site and displayed short-range diffusion motions together (Figure 4C and Supplemental Video S3). Furthermore, fluorescence recovery after photobleaching (FRAP) analyses using mECitrine-tagged strains revealed that Rng10 and Rga7 had similar dynamics at the division site and the cell tips (Figure 4D and Supplemental Figure S4A). The half-times for recovery at the division site were also similar for mEGFP-tagged Rng10 (13 ± 10 s, n = 22 cells) and Rga7 (15 ± 9 s, n = 18 cells). Similar to rng10Δ cells, rga7Δ cells also had significantly higher percentages of cell lysis (9% at 25°C, 46% at 36°C) than did wt (Figure 4E). Together these results suggest that Rng10 and Rga7 function together during cell division.
FIGURE 4:
Rng10 physically interacts with the Rho-GAP Rga7 and is important for Rga7 localization. (A) Silver staining showing S. pombe proteins pulled down by Stag and Rng10-Stag. Red lines mark Rng10 and Rga7 bands. (B) Rng10 and Rga7 coimmunoprecipitate from cell extracts. Asterisks mark Rga7 (red) and Rng10 (blue). (C) Deconvoluted images showing colocalization of Rng10 and Rga7. Right, magnified regions at the division site (1) and the cell tip (2). (D) FRAP analyses of Rng10-mECitrine and Rga7-mECitrine at the division site (left) and the cell tip (right). Cells were bleached at time 0. Mean ± 1 SEM. (E) Quantification of cell lysis in wt and rga7Δ cells at 25°C and 4 h at 36°C. *p < 0.05 compared with wt. (F, G) Localization (left) and protein level (right) of Rng10 (F) and Rga7 (G). Arrows in G mark the Rga7 dot remaining at the division site. *p < 0.0001 compared with wt. (H) Western blotting showing Rga7 protein levels in cells extracts from wt (+) and rng10Δ (–) cells. Tubulin was used as a loading control. (I) Rng10 mislocalizes Rga7 to SPBs (boxes) using Cdc7-GBP. Bars, 5 μm.
To further test the functional interactions between Rga7 and Rng10, we first examined their localization interdependence. rga7Δ resulted in an ∼40% decrease in Rng10 level at the division site (Figure 4F). Surprisingly, Rga7 localization was severely affected by rng10Δ (Figure 4G). Rga7 localization at the cell tips was abolished, and Rga7 no longer localized to the division site as a complete ring or disk in rng10Δ cells. Instead, a dot formed in the center of the division plane in ∼10% (n = 200) of cells. Although Rga7 global level was not affected by rng10Δ, the division-site level was only 11 ± 4% of the rng10+ cells (Figure 4, G and H). Because Rga7 is a GAP for Rho2 GTPase (Villar-Tajadura et al., 2008; Soto et al., 2010), we examined whether Rho2 plays a role in Rga7 localization. Rga7 localization was normal in rho2Δ (Supplemental Figure S4B). In rho2Δ rng10Δ cells, the Rga7 dot at the center of the division site was still observed (Supplemental Figure S4B). This indicated that Rga7 localization did not depend on Rho2.
To test whether Rng10 specifically affects Rga7 localization, we observed the localizations of Rho-GAPs Rga2 and Rga8 in wt and rng10Δ cells. Rga2 has a similar localization pattern as Rga7 and is also a Rho2 GAP (Villar-Tajadura et al., 2008). Rga8 has an F-BAR domain (Yang et al., 2003), and rga8Δ was synthetic lethal with rng10Δ (Table 2). However, Rga2 and Rga8 localizations were not dramatically affected in rng10Δ (Supplemental Figure S4, C and D). These data suggest that the regulation on Rga7 localization by Rng10 may be specific.
TABLE 2:
Genetic interactions of rng10Δ and rga7Δ with other cytokinesis mutations.
| Parent 1a | Parent 2b | Percentage viable double mutants at 25°C | Total number of tetrads | Genetic interaction at 25°Cc |
|---|---|---|---|---|
| rga7Δ | rng10Δ | 0 | 46 | - |
| rho1-596 | rng10Δ | 0 | 86 | - |
| rho1-596 | rga7Δ | 0 | 9 | - |
| rga8Δ | rng10Δ | 0 | 17 | - |
| rga8Δ | rga7Δ | 0 | 46 | - |
| imp2 | rng10Δ | 0 | 13 | - |
| art1∆ | rng10Δ | 0 | 32 | - |
| art1Δ | rga7Δ | 11 | 22 | ++ |
| myo2-E1 | rng10Δ | 33 | 12 | +d |
| myo2-E1 | rga7Δ | 0 | 9 | - |
| pxl1Δ | rng10Δ | 80 | 19 | ++ |
| pxl1Δ | rga7Δ | 0 | 14 | - |
| bgs1-191 | rng10Δ | 100 | 15 | + |
| spn1-Δ2 | rng10Δ | 86 | 16 | + |
| cdc7-24 | rng10Δ | 86 | 19 | ++ |
| cdc7-24 | rga7Δ | 100e | 17 | + |
| myp2-Δ2 | rng10Δ | 100 | 13 | +++ |
| myp2-Δ2 | rga7Δ | 64 | 16 | ++d |
| rho2Δ | rng10Δ | 95 | 20 | +++ |
| rho2Δ | rga7Δ | 57 | 12 | +++ |
| exo70∆ | rng10Δ | 100 | 14 | +++f |
| exo70∆ | rga7Δ | 85 | 25 | +++ |
| mid1-6 | rng10Δ | 75 | 8 | +++ |
| mid1-6 | rga7Δ | 100 | 9 | +++ |
aSingle-parent mutants were 70–100% viable in the crosses on YE5S plates at 25°C.
bSingle-parent mutants were 60–90% viable in the crosses on YE5S plates at 25°C. The majority of nonviable cells are due to failed spore germination.
cCells were freshly grown on YE5S and YE5S plus Phloxin B (PB, which accumulates in dead cells) plates before checking the morphology under DIC. The severity of cytokinesis defects compared with the parents was classified as follows: -, synthetic lethal interaction; +, strong synthetic interaction and prominent cytokinesis defects; ++, mild synthetic interaction and moderate cytokinesis defects; or +++, no additive cytokinesis defects.
dThe interaction was more obvious at 32°C.
eThe double mutants were scored from nonparental ditype tetrads.
fCell-lysis phenotype was slightly rescued.
To test whether Rng10 is sufficient for Rga7 localization, we used GFP-binding protein (GBP) to mislocalize Rng10 (Figure 4I). We confirmed that GBP bound to GFP but not tdTomato (Supplemental Figure S4E; Rothbauer et al., 2008). Mislocalized Rng10-mEGFP by GBP-tagged Cdc7, a kinase in the SIN pathway (Fankhauser and Simanis, 1994; Mehta and Gould, 2006), successfully recruited Rga7-tdTomato to SPBs (Figure 4I) in 98% of cells with Rng10 at SPBs (n = 54 cells). Thus Rng10 is sufficient to recruit Rga7.
Collectively these data suggest that Rng10 works together with Rga7 as a protein complex and is important for Rga7 localization. However, rng10Δ rga7Δ double mutant was synthetically lethal and died from cell lysis (Supplemental Figure S4F and Table 2), indicating that Rng10 and Rga7 also have independent functions. Indeed, genetic interactions of rng10Δ and rga7Δ with other cytokinesis mutations shared many similarities and some differences (Table 2). For example, both were synthetic lethal with rho1-596, rga8Δ, and imp2Δ (Martin-Garcia et al., 2014), and synthetic sick with cdc7-24 (Table 2). rng10Δ and rga7Δ had no genetic interaction with rho2Δ, exo70∆, or mid1-6 (Table 2). However, rng10Δ and rga7Δ had different genetic interactions with art1Δ, myo2-E1, pxl1Δ, and myp2-Δ2 (Table 2). In addition, although rga7Δ cells resembled rng10Δ cells in contractile-ring assembly, cell separation, and plasma membrane closure, ring constriction was significantly slower in rga7Δ cells at 25°C (Supplemental Figures S1 and S2, A–D). Taking the results together, we find that Rng10 interacts with Rga7 in vivo and dictates its localization, although each protein also has some distinct functions during cytokinesis.
Rng10 and Rga7 regulate the localization and dynamics of glucan synthases during septum formation
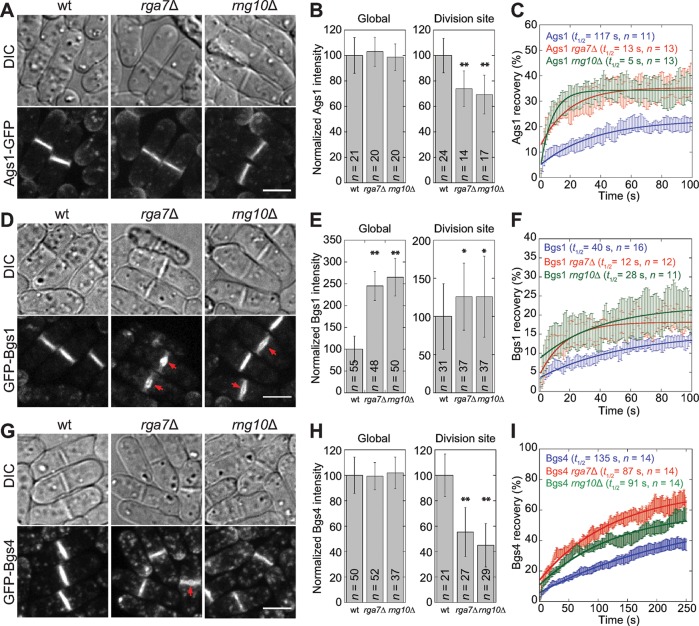
Our data showed that Rng10 is important for Rga7 localization at the division site. We next asked what roles Rng10 and Rga7 play during septum formation. Rga7 is a GAP for Rho2 GTPase, which regulates α-glucan synthase, Ags1, a transmembrane protein essential for septum formation (Calonge et al., 2000; Villar-Tajadura et al., 2008). Thus we first examined Ags1 localization and dynamics at the division site in rga7Δ and rng10Δ cells. Ags1 still localized to the division site in the mutants (Figure 5A). Although the global protein levels were not affected, Ags1 levels at the division site measured by fluorescence intensity were significantly reduced in rga7Δ and rng10Δ cells (Figure 5B). FRAP analysis showed that Ags1 was more dynamic at the division site, with shorter recovery half-times in rga7Δ and rng10Δ cells (Figure 5C). Thus Rng10 and Rga7 are important for Ags1 accumulation and dynamics at the division site.
FIGURE 5:
rng10Δ and rga7Δ affect accumulation and dynamics of glucan synthases at the division site. Localization (A, D, G), protein levels (B, E, H), and dynamics (C, F, I) of Ags1 (A–C), Bgs1 (D–F), and Bgs4 (G–I) in wt, rga7Δ, and rng10Δ cells at 25°C. Arrows in D and G mark the bulges at the center of septa. (B, E, H) Global and division-site protein levels. *p < 0.05 and **p < 0.0001 compared with wt. (C, F, I) FRAP analyses of glucan synthases at the division site. Half of the division site was bleached at time 0. Mean ± 1 SEM. Bars, 5 μm.
Both rga7Δ and rng10Δ are synthetically lethal or sick with mutations in GTPase rho1 and arrestin art1 (Table 2), which regulate septum formation through β-glucan synthases (Arellano et al., 1996; Viana et al., 2013; Davidson et al., 2015). We examined whether rga7Δ and rng10Δ affect the β-glucan synthases Bgs1 and Bgs4, which are also essential for septum formation (Liu et al., 1999; Cortes et al., 2005, 2007; Munoz et al., 2013). Bgs1 still localized to the division site in rga7Δ and rng10Δ (Figure 5D). However, Bgs1 frequently appeared as a bulge (indicated with the arrows; 48% rga7Δ cells, n = 142; 35% rng10Δ cells, n = 188) in the center of the division plane in cells with completed or nearly completed primary septa (Figure 5D). Bgs1 global and division-site levels increased significantly (Figure 5E). Bgs1 was also more dynamic at the division site in rga7Δ and rng10Δ cells, as shown by FRAP analyses (Figure 5F). Because Bgs1 catalytic activity is regulated mostly by Rho1 GTPase (Arellano et al., 1996; Mazur and Baginsky, 1996), we examined whether overexpressing Rho1 could rescue the cell lysis phenotype in rga7Δ and rng10Δ cells. Cells transformed with high-copy Rho1 plasmids under the Rho1 promoter had a significant reduction in cell lysis (Supplemental Figure S5A). These data suggest that in rga7Δ and rng10Δ cells, Bgs1 and Rho1 may be up-regulated through the cell-wall integrity pathway to attenuate cell lysis (Wu et al., 2010).
Rga7 is involved in trafficking of β-glucan synthase, Bgs4, from late Golgi compartments to the plasma membrane (Arasada and Pollard, 2015). We predicted that the Bgs4 level at the division site is affected in rng10Δ cells. Indeed, although Bgs4 global levels were not significantly different in wt, rga7Δ, and rng10Δ cells, its intensities at the division site were significantly lower in the mutants (Figure 5, G and H). In 15% of rga7Δ (n = 54) but not in rng10Δ cells with completed or nearly completed primary septa, Bgs4 appeared as a bulge in the center of the septum (arrow). Moreover, Bgs4 was more dynamic, with a faster signal recovery, in rga7Δ and rng10Δ cells in FRAP assays (Figure 5I).
Taken together, our data imply that Rng10 and Rga7 may contribute to septum formation by regulating the localization and dynamics of glucan synthases at the division site.
rga7Δ and rng10Δ cause defects in septum formation
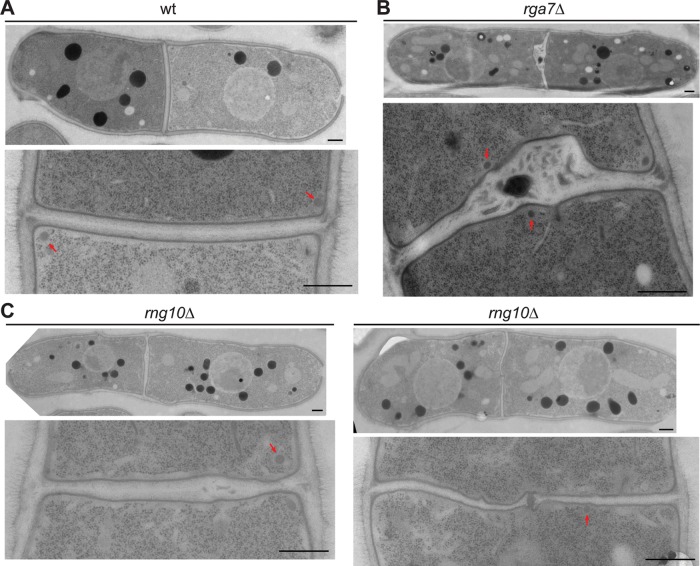
We used electron microscopy (EM) to observe whether the septum is defective in rga7Δ and rng10Δ cells. Compared to the smooth and linear septum with uniform thickness in wt (n = 16 of 17 cells; Figure 6A), 75% of rga7Δ (n = 8) and 94% of rng10Δ cells (n = 16) had abnormal septa (Figure 6, B and C). In rga7Δ cells, the complete septum contained a big bulge in the middle with dark materials trapped inside (Figure 6B), similar to a previous study (Martin-Garcia et al., 2014). In rng10Δ cells, the septum had a heterogeneous thickness (Figure 6C), and some also had a bulge in the center with dark materials trapped inside (Figure 6C, right). Thus the septum is defective in both rga7Δ and rng10Δ cells.
FIGURE 6:
rng10Δ and rga7Δ lead to abnormal septum formation but not secretory vesicle accumulation. (A–C) EM images of wt (A), rga7Δ (B), and rng10Δ cells (C) grown at 36°C for 4 h. Top: whole cell; bottom, the division site. Arrows mark secretory vesicles. Bars, 0.5 μm.
If the abnormal septum was caused by defects in vesicle trafficking, which delivers cell-wall synthases to the plasma membrane, we would expect to see vesicle accumulation near the division site (Wang et al., 2002). However, localization of secretory vesicles labeled with the vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptor v-SNARE Syb1 was not obviously affected in rga7Δ or rng10Δ cells (Supplemental Figure S5B). Compared to the massive accumulation of secretory vesicles near the septum in the exocyst mutant sec8-1 under EM (Supplemental Figure S5C; Wang et al., 2002, 2016), the number of secretory vesicles in rga7Δ and rng10Δ cells was comparable to that in wt (Figure 6, A–C). This suggests that the septum defects and cell lysis in rga7Δ and rng10Δ cells are not due to defective vesicle trafficking. Collectively our data indicate that Rng10 and Rga7 contribute to septum formation by regulating the localization and dynamics of glucan synthases on the plasma membrane.
DISCUSSION
In this study, we characterized a novel coiled-coil protein, Rng10, that localizes the Rho-GAP Rga7 to the cell tips and the division site. Rng10 and Rga7 work together to regulate the localization and dynamics of glucan synthases for successful septum formation in fission yeast cytokinesis.
Rng10 is important for the Rho-GAP Rga7 localization
Previous studies found that Rga7 localization depends on its F-BAR domain, which is a typical and highly conserved domain for binding to the plasma membrane (Martin-Garcia et al., 2014; Arasada and Pollard, 2015). Here we identified a novel protein, Rng10, that is responsible for localizing Rga7 to the plasma membrane at the cell tips and division site. Our data show that Rng10 physically interacts with Rga7 (Figure 4, A and B). Rga7 accumulation to cell tips is abolished, and ∼90% of Rga7 is lost from the division site during cell division without Rng10, although the global Rga7 level is not affected (Figure 4, G and H). Thus Rga7 localization to the plasma membrane requires the assistance of Rng10 besides its F-BAR domain. It is unknown whether Rng10 directly binds to Rga7. In vitro binding assays using purified proteins/domains will be of great interest in future studies.
Rng10 may help Rga7 localize to the plasma membrane through three mechanisms. First, Rng10 might alter lipid composition upon binding to the plasma membrane, and then Rga7 would bind to locally enriched phospholipids through its F-BAR domain. Second, Rng10 might induce membrane curvature itself or with Rga7 through a crowding effect (Stachowiak et al., 2012; Ramesh et al., 2013) that facilitates Rga7 binding. Third, Rng10 functions as an adaptor/stabilizer between the plasma membrane and Rga7, although Rga7 can still bind to the membrane directly. Future experiments are needed to distinguish these possibilities.
Although rng10Δ resulted in the loss of 90% Rga7 from the division site, a small percentage of cells still had a concentrated Rga7 dot in the center, indicating that a minor localization pathway for Rga7 exists in addition to Rng10 (Figure 4G). F-BAR protein Imp2 interacts with C2-domain protein Fic1, paxillin Pxl1, and Rga7 (Martin-Garcia et al., 2014). Because Rga7 is known to interact with Fic1 and Pxl1 as well, they are suggested to form part of a complex to regulate proper ring anchoring, integrity, contraction, and disassembly (Martin-Garcia et al., 2014). Thus proteins in this complex may play a role in regulating Rga7 localization independent of Rng10. Further investigation is needed to shed light on the alternative localization pathway for Rga7.
Regulation of Rga7 localization by Rng10 provides insights into Rho-GAP cellular localization
The Rng10 and Rga7 interaction provides insights into the localization of other Rho GAPs. We have limited understanding of Rho-GAP localization at the plasma membrane. Several mechanisms have been proposed to explain the regulation of their localization, including phosphorylation, secretory trafficking, lipid binding, and interaction with other proteins (Lefèbvre et al., 2012; Zebda et al., 2013; Morishita et al., 2015; Post et al., 2015). In both fission and budding yeast, it has been shown that Rho GEFs interact with other adaptor proteins for their localization (Endo et al., 2003; Krause et al., 2012; Zhu et al., 2013; Davidson et al., 2015). In S. pombe, Rho1 GEF Rgf3 and the arrestin Art1 interact and are interdependent for localization to the division site (Davidson et al., 2015). Another putative Rho-GEF, Gef2, interacts with the adaptor protein Nod1 through their C-termini, and they are interdependent for localization to cortical nodes (Zhu et al., 2013). In Saccharomyces cerevisiae, the GEF Cdc24 is regulated by the adaptor protein Bem1 for cell polarization (Smith et al., 2013). These and other data on Rho GEFs and our findings on Rng10 and Rga7 suggest that Rho GAPs may also depend on their adaptors or binding partners for localization. Consistently, mammalian ArhGAP29 localization to the plasma membrane depends on interaction with the PDZ domain of Radil, and together they regulate the endothelial barrier (Post et al., 2015). Mammalian p190RhoGAP recruitment to the cell periphery requires interaction with the C-terminal domain of p120-catenin, which helps to stabilize cell–cell transmembrane cadherin complexes and regulate actin dynamics (Zebda et al., 2013). Thus adaptor proteins could provide another layer of regulation to the specificity of Rho signaling.
Rga7 and Rng10 function in late stages of cytokinesis
Similar to previous studies (Martin-Garcia et al., 2014; Arasada and Pollard, 2015), we also observed roles of Rga7 in contractile-ring constriction, septum formation, and maintenance of cell integrity (Supplemental Figure S1 and Figure 6). In rga7Δ cells, some ring signals even persist until daughter cell separation (Figure S1C). Completed septa in rga7Δ cells have uneven thickness, with a bulged structure near the center of the division plane in our and a previous study (Figure 6; Martin-Garcia et al., 2014). Because glucan synthases are required for septum formation, previous studies also examined glucan synthases in rga7+ and rga7Δ cells (Martin-Garcia et al., 2014; Arasada and Pollard, 2015). We confirmed that Bgs1 forms a bulge in rga7Δ cells (Figure 5D; Martin-Garcia et al., 2014). However, our data suggest an increase in Bgs1 protein level at the division site (Figure 5E), in contrast to a slightly lower peak value in rga7Δ cells (Arasada and Pollard, 2015). This difference may be due to the imaging conditions (still images vs. time-lapse movies). However, both studies find that Bgs4 level at the division site is lower than in wt. We are the first to examine Ags1 level and dynamics in rga7Δ cells, even though previous studies looked at the function of Rga7 in Rho2 regulation (Villar-Tajadura et al., 2008; Soto et al., 2010; Martin-Garcia et al., 2014). Consistent with its critical role in Rga7 localization, we find that Rng10 has very similar functions to Rga7 in contractile-ring constriction and septum formation.
Future studies are needed to reveal exactly how Rga7 and Rng10 regulate the dynamics and localization of glucan synthases. Rga7 is a Rho2 GAP, which regulates Ags1 activity (Villar-Tajadura et al., 2008; Calonge et al., 2000; Soto et al., 2010), although it is unknown why increased Rho2 activation in rga7Δ cells leads to low Ags1 level at the division site. One possible explanation could be due to a decrease in total Rho2 level in rga7Δ cells (Villar-Tajadura et al., 2008), which leads to reduced active Rho2 and lower Ags1 levels. It is also unknown how Rga7 and Rng10 regulate Bgs1 and Bgs4 levels and dynamics. Because Rho1 overexpression can rescue both rga7Δ and rng10Δ cells and both deletions have synthetic interactions with mutations in rho1 and rho1 regulators (Table 2; Chen et al., 2016), Rga7 and Rng10 may regulate Bgs1 and Bgs4 through Rho1 GTPase and its GEFs and GAPs. Moreover, Rga7 is in a protein complex with F-BAR proteins Cdc15 and Imp2, C2 protein Fic1, and paxillin Pxl1 (Martin-Garcia et al., 2014), and they may work together to regulate the dynamics and localization of glucan synthases. Taken together, our data are consistent with previous studies on Rga7 function in regulating glucan synthases during septum formation and coordinating the late stages of cytokinesis.
Rng10 and Rga7 have independent functions
We find that Rng10 and Rga7 function together in regulating localization and dynamics of glucan synthases and thus septum formation and cell integrity during cytokinesis. However, several lines of evidence suggest that Rng10 and Rga7 have functions independent of each other. First, rng10Δ and rga7Δ are synthetic lethal (Supplemental Figure S4F and Table 2), indicating that they are not in a single linear pathway. Second, the completed septa in rng10Δ cells more frequently have uneven thickness compared with the big bulge in the majority of rga7Δ cells, suggesting that the defects are not caused by the exact same mechanisms or that the residual Rga7 in rng10Δ cells makes the difference (Figure 6, B and C). Third, rng10Δ causes more cell lysis than rga7Δ, and the two deletions are rescued to different degrees by Rho1 overexpression (Figures 1B and 4E and Supplemental Figure S5A), suggesting that Rng10 may affect another pathway besides the Rga7 and Rho2 pathway in septum regulation. We found that rng10Δ is synthetic lethal with rga8Δ (Table 2). Even though Rga8 still localizes to the division site, Rga8 is more diffused and less concentrated in rng10Δ cells (Supplemental Figure S4D). Thus Rng10 may play a role in the Rga8 and Rho1 pathway. Fourth, rng10Δ and rga7Δ have some different genetic interactions with other mutations (Table 2). Fifth, Rng10 has two domains for localization at its N- and C-termini (Figure 3C). It is likely that Rng10 binds to different partners through these domains and the partner other than Rga7 also functions at the division site. Sixth, rga7Δ causes a significant delay in contractile‑ring constriction and disassembly, whereas rng10Δ does not affect the contractile ring at 25°C (Supplemental Figure S1D). This defect is similar to fic1Δ, which causes the persistence of proteins at the division site after cytokinesis and leads to polarity defects in the next cell cycle (Bohnert and Gould, 2012). Rga7 also interacts with Fic1 in coIP (Martin-Garcia et al., 2014). Thus Rga7 may function in a similar way as Fic1 to regulate polarized growth after cytokinesis. Taking the results together, we conclude that Rng10 and Rga7 also have independent functions during cell division and polarized growth, which will be further investigated in the future.
In summary, we characterized a novel coiled-coil protein, Rng10, and identified its binding partner, the Rho-GAP Rga7. Rga7 localization depends on Rng10, and they work together to regulate conserved glucan synthases for successful septum formation and cell integrity during cell division. It will be interesting to elucidate how Rng10 interacts with Rga7. Our findings also provide insight into the regulation of localization and function of other Rho GAPs.
MATERIALS AND METHODS
Strains and genetic, molecular, and cellular methods
Strains used in this study are shown in Supplemental Table S1. PCR-based gene targeting and standard genetics were used to construct the strains (Moreno et al., 1991; Bähler et al., 1998). All tagged and truncated genes are expressed under their endogenous promoters and integrated at their native chromosomal loci. The exceptions are tagged glucan synthases Bgs1, Bgs4, and Ags1, which are regulated by the endogenous promoters but integrated at the leu1 loci with the endogenous copies deleted (Cortes et al., 2005, 2012). For the rescue of the lysis phenotype of rng10Δ or rga7Δ by Rho1 overexpression, the cells were transformed with pUR19-Rho1 or the control pUR19 empty vector plasmid. Transformed cells were grown in Edinburgh minimal medium plus five supplements (EMM5S)-uracil at 25°C for 36 h and then transferred to yeast extract with five supplements (YE5S) medium at 36°C for 4 h before observation and imaging.
The N-terminal tagging and truncations of rng10 were made by first cloning the native promoter of rng10 (−510 to +6) into pFA6a-kanMX6-P3nmt1-mECitrine vector to replace the 3nmt1 promoter with BglII and PacI. The resulting plasmid, pFA6a-kanMX6-Prng10-mECitrine (JQW908), was then used as the template for amplifying Rng10 fragments flanked with homologous sequences. These fragments were transformed into wt strains as described previously (Bähler et al., 1998). Positive transformants were confirmed by PCR and sequencing. Then the kanMX6 marker at the 5′ end of rng10 or its truncations was looped out by crossing h+ and h− strains.
The functionalities of the tagged FL proteins (Rng10, Rga7, Rga2, Rga8, Rlc1, and Spn1) were tested by growing the strains at 25–36°C on YE5S medium or crossing to mutants. mECitrine-, mEGFP-, and tdTomato-tagged rng10 and rga7 strains were crossed to rga7Δ or rng10Δ, respectively. mECitrine-tagged Rga2 and Rga8 strains were crossed to rng10Δ. The growth and morphology of the strains expressing the fusion proteins under different temperatures were similar to those of wt, and the double mutants from crosses were not different from single-deletion mutants, indicating that the tagged proteins were functional.
Cells were grown in an exponential phase in YE5S liquid medium at 25°C for 36–48 h before fluorescence microscopy. To quantify rng10Δ cell lysis at 36°C or the synthetic interactions between mutations, cells were first grown at 25°C for ∼44 h and then transferred to 36°C for 4 h. Drug treatments were performed at room temperature and imaging done on bare slides to maintain the drug concentration as before (Wang et al., 2014a; Davidson et al., 2015). Briefly, to test whether Rng10 localization depended on F-actin or microtubule, cells were treated with latrunculin A at a final concentration of 100 μM in dimethyl sulfoxide (DMSO) for 30 min or MBC at a final concentration of 25 μg/ml in DMSO for 10 min. The Arp2/3 complex inhibitor CK-666 (ChemDiv, San Diego, CA; Nolen et al., 2009) was used at a final concentration of 100 μM in DMSO for 10 min. BFA was used at a final concentration of 50 μg/ml from a stock solution of 5 mg/ml in ethanol for 10 min. For observing and quantifying polarized growth, cells were stained with a final concentration of 10 μl/ml Calcofluor in the dark for 2 min before imaging.
Confocal and superresolution microscopy
Confocal microscopy was done as previously described (Davidson et al., 2015; Wang et al., 2015). Briefly, cells were collected from liquid culture by centrifuging at 3000 rpm for 30 s and then washed with EMM5S twice to reduce autofluorescence. A final concentration of 50 nM n-propyl-gallate (n-PG) from a 10× stock in EMM5S was added during the second wash to protect cells from free radicals (Giloh and Sedat, 1982; Laporte et al., 2011). For counting molecules, 100 μl of n-PG 10× stock in YE5S was added to ∼900 μl of cells in YE5S and centrifuged at 3000 rpm for 30 s before imaging without washing. Live cells were imaged on a thin layer of EMM5S with 20% gelatin (Sigma-Aldrich, St. Loius, MO) and 50 nM n-PG at 23–24°C except where noted. Imaging was done using two microscopy systems with 100×/1.4 numerical aperture (NA) Plan-Apo objective lenses (Nikon, Melville, NY). For fluorescence images and time-lapse movies, cells were imaged using a spinning-disk confocal system (UltraVIEW Vox CSUX1 system; PerkinElmer, Waltham, MA) with 440‑, 488-, 515-, and 561-nm solid-state lasers and a back-thinned electron-multiplying charge-coupled device (EMCCD) camera (C9100-13; Hamamatsu Photonics, Bridgewater, NJ) on a Nikon Ti-E microscope without binning. For the differential interference contrast (DIC) images used for quantification of cell lysis, septation index, and polarized growth using Calcofluor staining, cells were imaged using a Nikon Eclipse Ti inverted microscopy equipped with a Nikon DS-QI1 cooled digital camera.
For superresolution PALM, cells tagged with photoactivatable mEOS3.2 (Zhang et al., 2012) or mMaple3 (Wang et al., 2014b) were washed in the same way as for the confocal microscopy before being loaded into an imaging chamber made as follows. Two pieces of double-sided tape were placed ∼0.5 cm apart in the center of a cleaned slide, covering the whole width of the slide. A cleaned coverslip (22 × 22 mm) was then placed onto the center with two sides sealed by the tape. One unsealed end of the coverslip extended slightly out of the slide. The slide was inverted with the coverslip facing down. A small volume of 1 mg/ml polylysine was added to the extended side of the coverslip and run through the chamber by sucking the liquid through using Kimwipes on the opposite side. The polylysine slide was set for >1 h. Live cells were run through the chamber in the same way and washed with 100 μl of EMM5S plus n-PG twice before imaging. The PALM system was based on inverted microscopy (IX71; Olympus, Tokyo, Japan) with a 100×/1.49 NA oil immersion total internal reflection fluorescence objective and an EMCCD camera (iXon Ultra 897; Andor Technologies, Belfast, United Kingdom) with EM gain of 255 and exposure time of 0.02 s. We used 405 and 488-nm diode lasers (Vortran Laser Technology, Sacramento, CA) and a 561-nm diode-pumped solid-state laser (CrystaLaser, Reno, NV). The 488-nm laser with the filter 525/30 nm was used to check fluorescence signal before photoactivation. The 561-nm laser with the filter 593/40 nm was used for imaging. One DIC image was taken every 1000 fluorescence images. These DIC images were used for lateral drift correction. Proteins were photoactivated by a 405-nm laser from low to high power, depending on the efficiency of fluorophore activation. Usually 4500–8000 images were taken for each field. Superresolution images were reconstructed using MempSTORM (Huang et al., 2015).
Image analysis and counting of protein molecules
Images were analyzed using ImageJ (National Institutes of Health, Bethesda, MD) and Volocity (PerkinElmer). Fluorescence images in figures are maximum-intensity projections with z-sections spaced at 0.4 and 0.5 μm or at 0.05 μm for deconvoluted images, except where noted. Three-dimensional projections were generated from cells with 11 z-sections spaced at 0.6 μm. We used confocal microscopy to count protein molecules by quantifying fluorescence intensity (Coffman et al., 2011; Deng et al., 2014). Cells within the central ∼75% of the imaging field were quantified to ensure almost even illumination. We summed the intensity from all 13 z-sections at 0.5-μm spacing imaged with exposure time of 300-ms per section. For global intensity quantification, the polygon region of interest (ROI) tool in ImageJ was used to draw the outline of the cell to include all signals. The wt cells without any fluorescent tag were used to subtract background. For quantification of division-site intensity, a rectangular ROI1 was drawn to include >90% of ring or septum signal to measure the intensity. Another rectangular ROI approximately twice the size of ROI1 was measured to subtract cytoplasmic background (Davidson et al., 2015; Wang et al., 2015). To obtain molecule numbers, the measured global and local (division site) intensities were normalized using the protein standard curve generated from mEGFP-tagged proteins with known molecule numbers (Wu and Pollard, 2005) and imaged under the same conditions. The ratios of mEGFP- and mECitrine-tagged FL Rng10 and Rga7 were used to convert intensities from mECitrine into molecule numbers.
Data in figures are mean ± 1 SD except where noted. The p values in statistical analyses were calculated using two-tailed Student‘s t tests.
FRAP and FLIP analysis
Both FRAP and FLIP were done using the photokinesis unit on the UltraVIEW Vox confocal system at 23–24°C. In the FRAP assay, the middle focal planes of cells were bleached to <50% of the original fluorescence intensity. For the division site, we photobleached half of the division site in cells with either forming septa or complete septa. For cell tip, >90% of a cell tip of an interphase cell was photobleached. Four prebleach images and 100 postbleach images were collected every second. For image analysis, we corrected for background and photobleaching during image acquisition using empty space among cells and unbleached cells in the same image. The prebleach intensity for the ROI was normalized to 100%, and the first postbleach intensity was normalized to 0%. Intensities of three consecutive postbleach time points were averaged to reduce noise. The intensity data were plotted and fitted using the exponential decay equation y = m1 + m2 exp(−m3x), where m3 is the off-rate (KaleidaGraph; Synergy Software, Reading, PA). The half-time for recovery was calculated from t1/2 = ln2/m3.
The FLIP assay was also imaged at the middle focal plane. Cells with constricting rings (Rlc1-tdTomato) at the final stage of constriction were chosen to be photobleached. A small ROI (0.19 μm2) was drawn in the daughter cell and bleached repeatedly. Two prebleach images and two postbleach images were collected for each bleach-imaging-diffusion cycle for 40 cycles at 30-s interval. To analyze the data, the mean intensities for the unbleached half-cell were measured using the ROI tool and the plugin Z-profiler in ImageJ. Images were corrected for background and photobleaching during image acquisition using cells from the same image that were not bleached. The end of the first photobleaching was defined as time 0. The end of ring constriction was defined when the pixel intensity of Rlc1 at the division site reached its peak. The end of membrane closure was defined as when the mEGFP exchange stopped between the two daughter cells. The time from the end of ring constriction to membrane closure was quantified and compared between wt and mutants.
Affinity purification, mass spectrometry, IP, and Western blotting
We performed Stag protein purification to identify the Rng10 binding partner (Liu et al., 2010). We expressed Stag control under the 41nmt1 promoter and expressed Rng10-Stag under the rng10 native promoter. Cells were grown in YE5S medium at 25°C for 48 h. Lyophilized cells were ground with a mortar and pestle to a homogeneous fine powder at room temperature. Then we mixed them with cold HK extraction buffer (25 mM Tris, pH 7.5, 1% NP-40, 300 mM NaCl, 5 mM EDTA, 15 mM ethylene glycol tetraacetic acid, 60 mM β-glycerophosphate, 500 μM Na3VO4, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and protease inhibitors [Roche, Mannheim, Germany]). Cell extract was then centrifuged twice at 4°C (21,000 rpm for 30 min and 21,000 rpm for 10 min). Protein concentration was measured using the Bradford assay. Then cell extract was incubated with S protein–conjugated agarose beads at 4°C for 2 h. The beads were collected by centrifugation (4000 rpm) and washed with an equal volume of HK extraction buffer, four times with an equal volume of washing buffer (5 mM Tris, pH 7.5, 300 mM NaCl, 5 mM EDTA, 500 μM Na3VO4, 10 mM NaF, 1 mM PMSF, and 1 mM DTT), and twice with 1 ml of washing buffer. The proteins were then eluted with sample buffer by boiling the beads. We then separated the protein samples in SDS–PAGE gel and detected them by silver staining using Pierce Silver Stain Kit (24612; Thermo Fisher Scientific, Waltham, MA). For mass spectrometry analysis, the protein sample was just run through the stacking SDS–PAGE gel (Liu et al., 2010). All protein bands were excised as one sample and send for mass spectrometry (Mass Spectrometry and Proteomics Facility, The Ohio State University, Columbus, OH).
IP assay and Western blotting were carried out as previously described (Laporte et al., 2011; Lee and Wu, 2012; Wang et al., 2014a). Briefly, lyophilized cells (∼30 mg) were broken into a homogeneous fine powder using toothpick in a 2-ml tube and mixed with IP buffer (1% NP-40 buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [Hepes], pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% NP-40, 50 mM NaF, 20 mM glycerophosphate, and 0.1 mM Na3VO4], 1 mM PMSF, and protease inhibitor [Roche]). Then mECitrine-tagged Rga7 was pulled down from S. pombe cell extract using protein G covalently coupled magnetic Dynabeads (100.04D; Invitrogen, Carlsbad, CA) with polyclonal anti-GFP antibodies (NB600-308; Novus Biologicals, Littleton, CO). Then the beads were washed five times with 1% NP-40 buffer and boiled in sample buffer to elute the proteins. The protein samples were then separated in SDS–PAGE gel and detected by Western blotting with monoclonal anti-GFP antibody (1:2000 dilution; 11814460001; Roche, Mannheim, Germany) or monoclonal anti-Myc antibody (1:5000 dilution; 9E10, Santa Cruz Biotechnology, Dallas, TX). Secondary antibody anti-mouse immunoglobulin G (A4416, Sigma-Aldrich) was then used at 1:5000 dilution. The blot was reacted with SuperSignal Maximum Sensitivity Substrate (Thermo Fisher Scientific) and exposed to film (BioMax MR; Kodak, Rochester, NY).
Electron microscopy
EM was done at the Boulder Electron Microscopy Services at the University of Colorado, Boulder (Boulder, CO) as described previously (Lee et al., 2014). Briefly, cells were grown at 25°C for ∼48 h and then shifted to 36°C for 4 h before being harvested using Millipore filters. Samples were prepared using high-pressure freezing with a Wohlwend Compact 02 Freezer and freeze substitution in the presence of 2% osmium tetroxide and 0.1% uranyl acetate in acetone. Serial sections with a thickness of 70 nm were prepared from cells embedded in Epon-Araldite epoxy resin. Then the sections were poststained with uranyl acetate and lead citrate for observation under a Philips CM100 transmission electron microscope (FEI, Hillsboro, OR).
Supplementary Material
Acknowledgments
We thank Mohan Balasubramanian, Takayoshi Kuno, Dannel McCollum, Stephen Osmani, Pilar Pérez, Thomas Pollard, Juan Carlos Ribas, Shelley Sazer, and Ning Wang for strains and plasmids; the Anita Hopper, James Hopper, and Stephen Osmani laboratories, the Mass Spectrometry and Proteomics Facility at The Ohio State University, and Mary Morphew and Thomas Giddings, Jr., at the University of Colorado at Boulder for technical support; and members of the Wu laboratory for helpful discussions and suggestions. This work was supported by National Institute of General Medical Sciences of the National Institutes of Health Grant GM118746 and American Cancer Society Grant RSG-13-005-01-CCG to J.-Q.W.
Abbreviations used:
- coIP
coimmunoprecipitation
- DIC
differential interference contrast
- F-BAR
Fes/CIP4 homology-Bin/amphiphysin/Rvs
- FL
full length
- FLIP
fluorescence loss in photobleaching
- GAP
GTPase-activating protein
- GBP
GFP-binding protein
- GEF
guanine nucleotide exchange factor
- mECitrine
monomeric enhanced citrine
- mEGFP
monomeric enhanced GFP
- n-PG
n-propyl-gallate
- PALM
photoactivated localization microscopy
- ROI
region of interest
- SPB
spindle pole body
- tdTomato
tandem dimer Tomato
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0156) on July 6, 2016.
REFERENCES
- Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. A role for F-BAR protein Rga7p during cytokinesis in S. pombe. J Cell Sci. 2015;128:2259–2268. doi: 10.1242/jcs.162974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Duran A, Perez P. Rho1 GTPase activates the (1-3)β-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15:4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Basu R, Munteanu EL, Chang F. Role of turgor pressure in endocytosis in fission yeast. Mol Biol Cell. 2014;25:679–687. doi: 10.1091/mbc.E13-10-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet. 2012;8:e1003004. doi: 10.1371/journal.pgen.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun AC, Olayioye MA. Rho regulation: DLC proteins in space and time. Cell Signal. 2015;27:1643–1651. doi: 10.1016/j.cellsig.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Calonge TM, Arellano M, Coll PM, Perez P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol Microbiol. 2003;47:507–518. doi: 10.1046/j.1365-2958.2003.03312.x. [DOI] [PubMed] [Google Scholar]
- Calonge TM, Nakano K, Arellano M, Arai R, Katayama S, Toda T, Mabuchi I, Perez P. Schizosaccharomyces pombe Rho2p GTPase regulates cell wall α-glucan biosynthesis through the protein kinase Pck2p. Mol Biol Cell. 2000;11:4393–4401. doi: 10.1091/mbc.11.12.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansado J, Soto T, Gacto M, Perez P. Rga4, a Rho-GAP from fission yeast: finding specificity within promiscuity. Commun Integr Biol. 2010;3:436–439. doi: 10.4161/cib.3.5.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-S, Beckley JR, Ren L, Feoktistova A, Jensen MA, Rhind N, Gould KL. Discovery of genes involved in mitosis, cell division, cell wall integrity and chromosome segregation through construction of Schizosaccharomyces pombe deletion strains. Yeast 2016. 2016 doi: 10.1002/yea.3172. doi: 10.1002/yea.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Chircop M. Rho GTPases as regulators of mitosis and cytokinesis in mammalian cells. Small GTPases. 2014;5:e29770. doi: 10.4161/sgtp.29770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes JC, Carnero E, Ishiguro J, Sanchez Y, Duran A, Ribas JC. The novel fission yeast (1,3)β-D-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J Cell Sci. 2005;118:157–174. doi: 10.1242/jcs.01585. [DOI] [PubMed] [Google Scholar]
- Cortes JC, Ishiguro J, Duran A, Ribas JC. Localization of the (1,3)β-D-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J Cell Sci. 2002;115:4081–4096. doi: 10.1242/jcs.00085. [DOI] [PubMed] [Google Scholar]
- Cortes JC, Konomi M, Martins IM, Munoz J, Moreno MB, Osumi M, Duran A, Ribas JC. The (1,3)β-D-glucan synthase subunit Bgs1p is responsible for the fission yeast primary septum formation. Mol Microbiol. 2007;65:201–217. doi: 10.1111/j.1365-2958.2007.05784.x. [DOI] [PubMed] [Google Scholar]
- Cortes JC, Sato M, Munoz J, Moreno MB, Clemente-Ramos JA, Ramos M, Okada H, Osumi M, Duran A, Ribas JC. Fission yeast Ags1 confers the essential septum strength needed for safe gradual cell abscission. J Cell Biol. 2012;198:637–656. doi: 10.1083/jcb.201202015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Wiley DJ, Medina S, Vincent HA, Larrea M, Oriolo A, Verde F. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol Biol Cell. 2007;18:2090–2101. doi: 10.1091/mbc.E06-09-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R, Laporte D, Wu J-Q. Regulation of Rho-GEF Rgf3 by the arrestin Art1 in fission yeast cytokinesis. Mol Biol Cell. 2015;26:453–466. doi: 10.1091/mbc.E14-07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kabeche R, Wang N, Wu J-Q, Moseley JB. Megadalton node assembly by binding of Skb1 to the membrane anchor Slf1. Mol Biol Cell. 2014;25:2660–2668. doi: 10.1091/mbc.E14-04-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickholt J, Deng X, Cheng J. DoBo: protein domain boundary prediction by integrating evolutionary signals and machine learning. BMC Bioinformatics. 2011;12:43. doi: 10.1186/1471-2105-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shirouzu M, Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J Biol Chem. 2003;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 1994;13:3011–3019. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloh H, Sedat JW. Fluorescence microscopy: reduced photobleaching of rhodamine and fluorescein protein conjugates by n-propyl gallate. Science. 1982;217:1251–1255. doi: 10.1126/science.7112126. [DOI] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3:130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gumpper K, Chi Y, Sun M, Ma J. Fast two-dimensional super-resolution image reconstruction algorithm for ultra-high emitter density. Opt Lett. 2015;40:2989–2992. doi: 10.1364/OL.40.002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro J, Saitou A, Duran A, Ribas JC. cps1+, a Schizosaccharomyces pombe gene homolog of Saccharomyces cerevisiae FKS genes whose mutation confers hypersensitivity to cyclosporin A and papulacandin B. J Bacteriol. 1997;179:7653–7662. doi: 10.1128/jb.179.24.7653-7662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SN, Canman JC. Rho GTPases in animal cell cytokinesis: an occupation by the one percent. Cytoskeleton (Hoboken) 2012;69:919–930. doi: 10.1002/cm.21071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause SA, Cundell MJ, Poon PP, McGhie J, Johnston GC, Price C, Gray JV. Functional specialisation of the yeast Rho1 GTP exchange factors. J Cell Sci. 2012;125:2721–2731. doi: 10.1242/jcs.100685. [DOI] [PubMed] [Google Scholar]
- Lackner M, Tscherner M, Schaller M, Kuchler K, Mair C, Sartori B, Istel F, Arendrup MC, Lass-Florl C. Positions and numbers of FKS mutations in Candida albicans selectively influence in vitro and in vivo susceptibilities to echinocandin treatment. Antimicrob Agents Chemother. 2014;58:3626–3635. doi: 10.1128/AAC.00123-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee I-J, Wu J-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Zhao R, Wu J-Q. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol. 2010;21:892–898. doi: 10.1016/j.semcdb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Coffman VC, Wu J-Q. Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives. Cytoskeleton (Hoboken) 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wang N, Hu W, Schott K, Bähler J, Giddings TH, Jr, Pringle JR, Du L-L, Wu J-Q. Regulation of spindle pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol Biol Cell. 2014;25:2735–2749. doi: 10.1091/mbc.E13-11-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wu J-Q. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J Cell Sci. 2012;125:2973–2985. doi: 10.1242/jcs.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèbvre F, Prouzet-Mauléon V, Hugues M, Crouzet M, Vieillemard A, McCusker D, Thoraval D, Doignon F. Secretory pathway-dependent localization of the Saccharomyces cerevisiae Rho GTPase-activating protein Rgd1p at growth sites. Eukaryot Cell. 2012;11:590–600. doi: 10.1128/EC.00042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-L, Osmani AH, Ukil L, Son S, Markossian S, Shen KF, Govindaraghavan M, Varadaraj A, Hashmi SB, De Souza CP, et al. Single-step affinity purification for fungal proteomics. Eukaryot Cell. 2010;9:831–833. doi: 10.1128/EC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, McCollum D, Balasubramanian MK. Drc1p/Cps1p, a 1,3-β-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 1999;153:1193–1203. doi: 10.1093/genetics/153.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Martin-Garcia R, Coll PM, Perez P. F-BAR domain protein Rga7 collaborates with Cdc15 and Imp2 to ensure proper cytokinesis in fission yeast. J Cell Sci. 2014;127:4146–4158. doi: 10.1242/jcs.146233. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Mazur P, Baginsky W. In vitro activity of 1,3-β-D-glucan synthase requires the GTP-binding protein Rho1. J Biol Chem. 1996;271:14604–14609. doi: 10.1074/jbc.271.24.14604. [DOI] [PubMed] [Google Scholar]
- Mehta S, Gould KL. Identification of functional domains within the septation initiation network kinase, Cdc7. J Biol Chem. 2006;281:9935–9941. doi: 10.1074/jbc.M600160200. [DOI] [PubMed] [Google Scholar]
- Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morishita Y, Tsutsumi K, Ohta Y. Phosphorylation of serine 402 regulates RacGAP protein activity of FilGAP protein. J Biol Chem. 2015;290:26328–26338. doi: 10.1074/jbc.M115.666875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J, Cortes JC, Sipiczki M, Ramos M, Clemente-Ramos JA, Moreno MB, Martins IM, Perez P, Ribas JC. Extracellular cell wall β(1,3)glucan is required to couple septation to actomyosin ring contraction. J Cell Biol. 2013;203:265–282. doi: 10.1083/jcb.201304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Mutoh T, Mabuchi I. Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2001;6:1031–1042. doi: 10.1046/j.1365-2443.2001.00485.x. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol. 2013;202:311–329. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin DS. Current perspectives on echinocandin class drugs. Future Microbiol. 2011;6:441–457. doi: 10.2217/fmb.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post A, Pannekoek WJ, Ponsioen B, Vliem MJ, Bos JL. Rap1 spatially controls ArhGAP29 to inhibit Rho signaling during endothelial barrier regulation. Mol Cell Biol. 2015;35:2495–2502. doi: 10.1128/MCB.01453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor SA, Minc N, Boudaoud A, Chang F. Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr Biol. 2012;22:1601–1608. doi: 10.1016/j.cub.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh P, Baroji YF, Reihani SNS, Stamou D, Oddershede LB, Bendix PM. FBAR syndapin 1 recognizes and stabilizes highly curved tubular membranes in a concentration dependent manner. Sci Rep. 2013;3:1565. doi: 10.1038/srep01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C, Sanchez Y. Cell separation and the maintenance of cell integrity during cytokinesis in yeast: the assembly of a septum. Yeast. 2010;27:521–530. doi: 10.1002/yea.1779. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Santos B, Gutierrez J, Calonge TM, Perez P. Novel Rho GTPase involved in cytokinesis and cell wall integrity in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2003;2:521–533. doi: 10.1128/EC.2.3.521-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Reinhard NR, Hord PL. Toward understanding RhoGTPase specificity: structure, function and local activation. Small GTPases. 2014;5:6. doi: 10.4161/21541248.2014.968004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schmelzle T, Hall MN. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol Microbiol. 2002;45:1433–1441. doi: 10.1046/j.1365-2958.2002.03110.x. [DOI] [PubMed] [Google Scholar]
- Smith SE, Rubinstein B, Mendes Pinto I, Slaughter BD, Unruh JR, Li R. Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J Cell Biol. 2013;202:1091–1106. doi: 10.1083/jcb.201304180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto T, Villar-Tajadura MA, Madrid M, Vicente J, Gacto M, Perez P, Cansado J. Rga4 modulates the activity of the fission yeast cell integrity MAPK pathway by acting as a Rho2 GTPase-activating protein. J Biol Chem. 2010;285:11516–11525. doi: 10.1074/jbc.M109.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nat Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Tatebe H, Nakano K, Maximo R, Shiozaki K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr Biol. 2008;18:322–330. doi: 10.1016/j.cub.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Viana RA, Pinar M, Soto T, Coll PM, Cansado J, Pérez P. Negative functional interaction between cell integrity MAPK pathway and Rho1 GTPase in fission yeast. Genetics. 2013;195:421–432. doi: 10.1534/genetics.113.154807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Tajadura MA, Coll PM, Madrid M, Cansado J, Santos B, Perez P. Rga2 is a Rho2 GAP that regulates morphogenesis and cell integrity in S. pombe. Mol Microbiol. 2008;70:867–881. doi: 10.1111/j.1365-2958.2008.06447.x. [DOI] [PubMed] [Google Scholar]
- Vos A, Dekker N, Distel B, Leunissen JAM, Hochstenbach F. Role of the synthase domain of Ags1p in cell wall α-glucan biosynthesis in fission yeast. J Biol Chem. 2007;282:18969–18979. doi: 10.1074/jbc.M605147200. [DOI] [PubMed] [Google Scholar]
- Wang H, Tang X, Liu J, Trautmann S, Balasundaram D, McCollum D, Balasubramanian MK. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol Biol Cell. 2002;13:515–529. doi: 10.1091/mbc.01-11-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lee I-J, Rask G, Wu J-Q. Roles of the TRAPP-II complex and the exocyst in membrane deposition during fission yeast cytokinesis. PLoS Biol. 2016;14:e1002437. doi: 10.1371/journal.pbio.1002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lo Presti L, Zhu Y-H, Kang M, Wu Z, Martin SG, Wu J-Q. The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J Cell Biol. 2014a;205:357–375. doi: 10.1083/jcb.201308146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang M, Zhu Y-H, Grosel TW, Sun D, Kudryashov DS, Wu J-Q. The Rho-GEF Gef3 interacts with the septin complex and activates the GTPase Rho4 during fission yeast cytokinesis. Mol Biol Cell. 2015;26:238–255. doi: 10.1091/mbc.E14-07-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci USA. 2014b;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Ye Y, Wang N, Pollard TD, Pringle JR. Cooperation between the septins and the actomyosin ring and role of a cell-integrity pathway during cell division in fission yeast. Genetics. 2010;186:897–915. doi: 10.1534/genetics.110.119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Hiratani T, Baba M, Osumi M. Effect of aculeacin A, a wall-active antibiotic, on synthesis of the yeast cell wall. Microbiol Immunol. 1985;29:609–623. doi: 10.1111/j.1348-0421.1985.tb00865.x. [DOI] [PubMed] [Google Scholar]
- Yang P, Qyang Y, Bartholomeusz G, Zhou X, Marcus S. The novel Rho GTPase-activating protein family protein, Rga8, provides a potential link between Cdc42/p21-activated kinase and Rho signaling pathways in the fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2003;278:48821–48830. doi: 10.1074/jbc.M306819200. [DOI] [PubMed] [Google Scholar]
- Zebda N, Tian Y, Tian X, Gawlak G, Higginbotham K, Reynolds AB, Birukova AA, Birukov KG. Interaction of p190RhoGAP with C-terminal domain of p120-catenin modulates endothelial cytoskeleton and permeability. J Biol Chem. 2013;288:18290–18299. doi: 10.1074/jbc.M112.432757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- Zhu Y-H, Ye Y, Wu Z, Wu J-Q. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol Biol Cell. 2013;24:3187–3204. doi: 10.1091/mbc.E13-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Oh W, Frost JA. Controlling the switches: Rho GTPase regulation during animal cell mitosis. Cell Signal. 2014;26:2998–3006. doi: 10.1016/j.cellsig.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.