Abstract
Bacterial wilt (BW) is a serious disease that affects eggplant (Solanum melongena) production. Although resistance to this disease has been reported, the underlying mechanism is unknown. In this study, we identified a NAC family transcription factor (SmNAC) from eggplant and characterized its expression, its localization at the tissue and subcellular levels, and its role in BW resistance. To this end, transgenic eggplant lines were generated in which the expression of SmNAC was constitutively up regulated or suppressed using RNAi. The results indicated that overexpression of SmNAC decreases resistance to BW. Moreover, SmNAC overexpression resulted in the reduced accumulation of the plant immune signaling molecule salicylic acid (SA) and reduced expression of ICS1 (a gene that encode isochorismate synthase 1, which is involved in SA biosynthesis). We propose that reduced SA content results in increased bacterial wilt susceptibility in the transgenic lines. Our results provide important new insights into the regulatory mechanisms of bacterial wilt resistance in eggplant.
NAC family proteins are plant-specific transcription factors that collectively elicit a range of biological functions, are widely distributed across terrestrial plant genomes, and are considered to be one of the largest families of transcription factors1. NAC family proteins have an N-terminus containing a highly conserved and specific NAC domain2 and were originally characterized and named for their sequence similarity to the petuniahybrida NAM (NO APICAL MERISTEM) and the Arabidopsis thaliana ATAF1, ATAF2, and CUC2 (CUP-SHAPED COTYLEDON) genes2. They have been shown to play a variety of roles in plant growth and development, responses to abiotic and biotic stresses, and fruit maturation and to be components of hormone signal transduction pathways3,4,5,6. For example, Zhao et al.7 reported that a NAC1-type transcription factor (TaNAC-S) from wheat (Triticumaestivum) suppresses leaf senescence and promotes grain yield and protein content7. The silencing of a peach (Prunus persica) NAC gene that is expressed at particularly high levels in blood-fleshed peaches caused a reduction in anthocyanin pigmentation8. You et al.9 characterized BdNAC genes from Brachypodium distachyon and demonstrated that their expression is influenced by abiotic stresses and phytohormones9, and a recent study revealed that two NAC transcription repressors (NAC050 and NAC052) control flowering time by associating with the histone demethylase JMJ1410.
In addition to developmental roles, NAC transcription factors are known to be involved in coordinating responses to attacks by phytopathogens, including fungi, bacteria and viruses. Sun et al.11 found that 63 rice (Oryza sativa) NAC genes exhibited overlapping expression patterns following exposure to a variety of biotic (infection by bacterial, fungal, and viral pathogens, or parasitic plants) and abiotic (cold, salt and drought) stresses11. Moreover, in a study of specific plant-pathogen interactions, the wheat GRAB1 and GRAB2 proteins were found to bind to the wheat dwarf geminivirus (WDV) RepA protein, such that the conserved N-terminal domain of the GRAB proteins activates the RepA protein. The overexpression of the GRAB proteins were found to inhibit WDV DNA replication12. In another report, the two pathogen-responsive rice NAC transcription factors ONAC122 and ONAC131 were described as being localized in the nucleus, to exhibit transcriptional activation activity13, and to be expressed after Magnaporthe grisea infection. In another example, silencing of ONAC122 or ONAC131 expression in transgenic rice lines resulted in their increased susceptibility to M. grisea. These results further support an important role for ONAC122 and ONAC131 in rice disease resistance responses, suggesting their regulation of the expression of defense- and signaling-related genes13. Xia et al.14 showed that the expression of the wheat TaNAC8 gene increased following infection by the stripe rust pathogen Puccinia striiformis f. sp. Tritici14. In another study, the binding of a turnip crinkle virus coat protein to the A. thaliana NAC transcription factor TIP resulted in reduced viral invasion15. Other examples linking NAC transcription factors to disease resistance processes include reports that the A. thaliana NAC083 protein interacts with the mungbean yellow mosaic India virus (MYMIV) Rep protein16 and that suppression of the bread wheat TaNAC1 gene enhances resistance to stripe rust17.
When pathogenic bacteria infect plants, they generally activate the expression of a large number of genes as part of a coordinated defense response that involves the salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) hormone signaling pathways. Bu et al.8 reported that ANAC019 and ANAC055 are involved in JA-mediated defense responses in A. thaliana and that they might regulate the transcription of the JA induced VEGETATIVE STORAGE PROTEIN1 (VSP1) and LIPOXYGENASE2 (LOX2) defense genes. The expression of VSP1 and LOX2 was also enhanced after ANAC019 and ANAC055 were overexpressed, and the double mutant (ANAC019 and ANAC055) exhibited reduced VSP1 and LOX2 expression and resistance to Botrytis cinerea18.
Bacterial Wilt (BW), which is caused by Ralstonia solanacearum, is considered one of the most destructive bacterial plant diseases and is known for extreme aggressiveness, with worldwide geographic distribution and a broad host range (more than 200 plant species). The dominant species of Ralstonia solanacearum affecting eggplant in our country is biovar III and biovar IV, which belong to race 1. Several eggplant studies investing BW include the discussion of host resistance genetic regulation19,20,21,22; pathogen separation23, collection, and identification of disease resistance resources24, and screens of resistant gene-relevant molecular markers21,22,25. The most documented example is the acetyl transferase popP226, recently renamed ripP227, which interacts with the A. thaliana gene RRS1-R28,29 (that belongs to the salicylic acid regulation pathway) and the cysteine protease RD1930, triggering plant immunity, making it the first avirulence gene described in R. solanacearum31. Delaspre et al.32 suggested that the bacterial wilt gene HrpB has an important function in infecting the host32. In addition, Milling et al.33 found that bacterial wilt gene EPS facilitates avirulence and is related to tomato bacterial wilt resistance33. Pensec et al.31 used an original association genetic approach combining resistant eggplant, pepper, and tomato DNA microarray and pathogenicity data to identify type-III effector (T3E) repertoires associated with virulence of the bacterial wilt pathogen Ralstonia solanacearum on Solanaceous crops31.
In this study, we describe the identification of a NAC transcription factor (SmNAC) from eggplant (Solanum melongena) based on RNA-seq data, and present data supporting its role in resistance to bacterial wilt (BW), a complex and devastating soil-borne vascular disease in plants caused by Ralstonia solanacearum. Bacterial wilt occurs in many crop species, including eggplant, tomato, potato, pepper, banana, ginger, cowpea, peanut, papaya, cashew, and olive plant34. Reports on mechanisms of host resistance are limited, and there are no reports of BW resistance regulation by NAC transcription factors. Although transcription factors containing the NAC domain have been identified in many plant species, such as in rice (Oryza.sativa)35,36, A. thaliana37, poplar (Populus trichocarpa)37, soybean (Glycine max), tobacco (Nicotiana tabacum)38, barley (Hordeum vulgare)38, and potato (Solanum tuberosum)39, to date, no NAC domain-containing proteins in eggplant have been described. Here, we present results indicating that SmNAC is involved in the resistance of eggplant to BW and describe its association with hormone signaling.
Results
Identification of SmNAC from S. melongena based on RNA-Seq data
Analysis of the RNA-seq data revealed 1,137 genes that were expressed at higher levels, and 9,048 genes that were expressed at lower levels in A0 than in A1 (Fig. S1a,b). A total of 6,087 genes were expressed at higher levels, and 5,832 were expressed at lower levels in B0 than B1, whereas 738 and 217 were expressed at higher or lower levels, respectively, in B0 compared to A0. Furthermore, 4,712 genes were expressed at higher levels and 12,523 were expressed at lower levels in B1 compared to A1. For a general characterization of the sterm tissue assemblies, an overview of the number of slimmed GO-terms for biological process is showed in Fig. S1c. The complete list of assigned GO terms for each assembly, including different levels of biological process, molecular function and cellular component, is available in it. One significantly differentially expressed gene, SmNAC (Gen accession: KM435267), was expressed at higher levels in BW-susceptible plants after inoculation with the pathogen (B1 plants) but at lower levels in BW-resistant plants after pathogen induction (A1 plants) (Fig. S2a). And the phenotypic symptoms of eggplants after inoculation with Ralstonia solanacearum were exhibited in Fig. S2b.
Sequence analysis showed that SmNAC was 1,708 bp long with an ORF of 1,038 bp. The SmNAC protein is predicted to contain 345 amino acids and to exhibt a molecular weight of 39,035 Da and an isoelectric point of 8.94. The 5′-untranslated region (UTR) 3′-UTR are predicted to be 159 and 511 bp, respectively. The full-length predicted amino acid sequence of SmNAC is annotated as containing a conserved NAM domain within the N-terminal region, similar to that found in NAC proteins from other plant species (Fig. 1, Fig. S3). The SmNAC amino acid sequence exhibits high similarity to ANAC019 (AT1G52890.1), ANAC055 (AT3G15500.1) and ANAC072 (AT4G27410.2) of A. thaliana, and SNAC1 (LOC_Os03g60080), OsNAC3 (LOC_Os07g12340) and OsNAC4 (LOC_Os01g60020) of rice. However, the gene and protein structures of SmNAC are same in the both inbred lines (E-31 and E-32).
Figure 1. Alignment analysis of SmNAC.
Alignment analysis of the SmNAC protein sequence with other plant NAC proteins. SmNAC was aligned with A. thaliana ANAC019 (AT1G52890.1), ANAC032 (AT1G77450.1), ANAC055 (AT3G15500.1), ANAC072 (AT4G27410.2), ANAC081 (AT5G08790.1) and ANAC102 (AT5G63790.1), and rice OsNAC4 (LOC_Os01g60020.1), OsNAC3 (LOC_Os07g12340), and SNAC1 (LOC_Os03g60080). The five highly conserved amino acid motifs (A–E) are indicated by black lines. Alignments were carried out using CLUSTALW v1.83 and the GeneDoc software.
Characterization of SmNAC expression in S. melongena
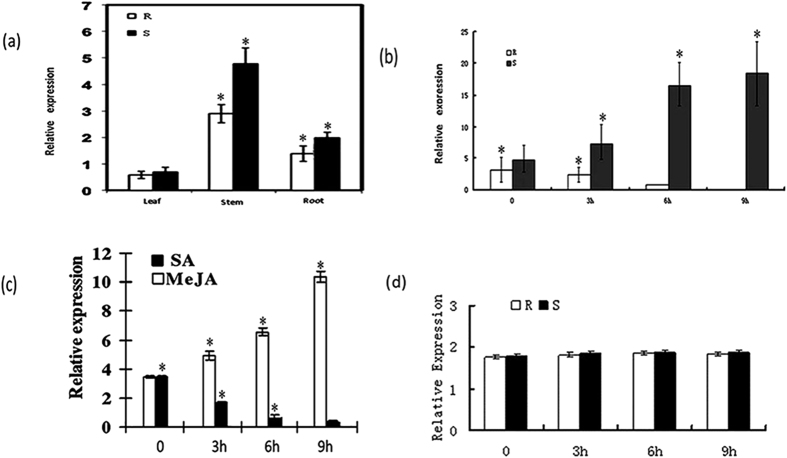
To assess whether SmNAC is involved in the regulation of BW resistance in S. melongena, its expression was analyzed in inoculated resistant (E-31) and susceptible (E-32) plants (Fig. 2). Under uninfected conditions, SmNAC expression was detected in roots, stems and leaves, with the highest expression in stems and the lowest expression in leaves (Fig. 2a). Prior to inoculation, SmNAC expression was higher in susceptible (E-32) than in resistant (E-31) plants. After inoculation with R. solanacearum strains, SmNAC transcript levels increased in susceptible S. melongena (E-32) but decreased in resistant S. melongena (E-31) plants, whereas they were no longer detected at 9 h after inoculation. However, the expression was substantially greater in E-32 plants (Fig. 2b). At the same time, E-32 plants weretreated with MeJA and JA strains, and the results indicate that MeJA, but not SA, can induce the expression of SmNAC. Taken together, these results suggest that SmNAC might play a role in the regulation of BW-resistance in S. melongena.
Figure 2. Evaluation of SmNAC expression in resistant and susceptible plants.
The expression of SmNAC in resistant and susceptible plants under normal conditions (a), in the stems of resistant and susceptible plants after inoculation with Ralstonia solanacearum (b), and in E-32 plants after treatment with MeJA (1.5 mM) and SA (0.2 mM) (c), and Mock (d). R: resistant plants (E-31); S: susceptible plants (E-32).
Immunohistochemical localization of SmNAC
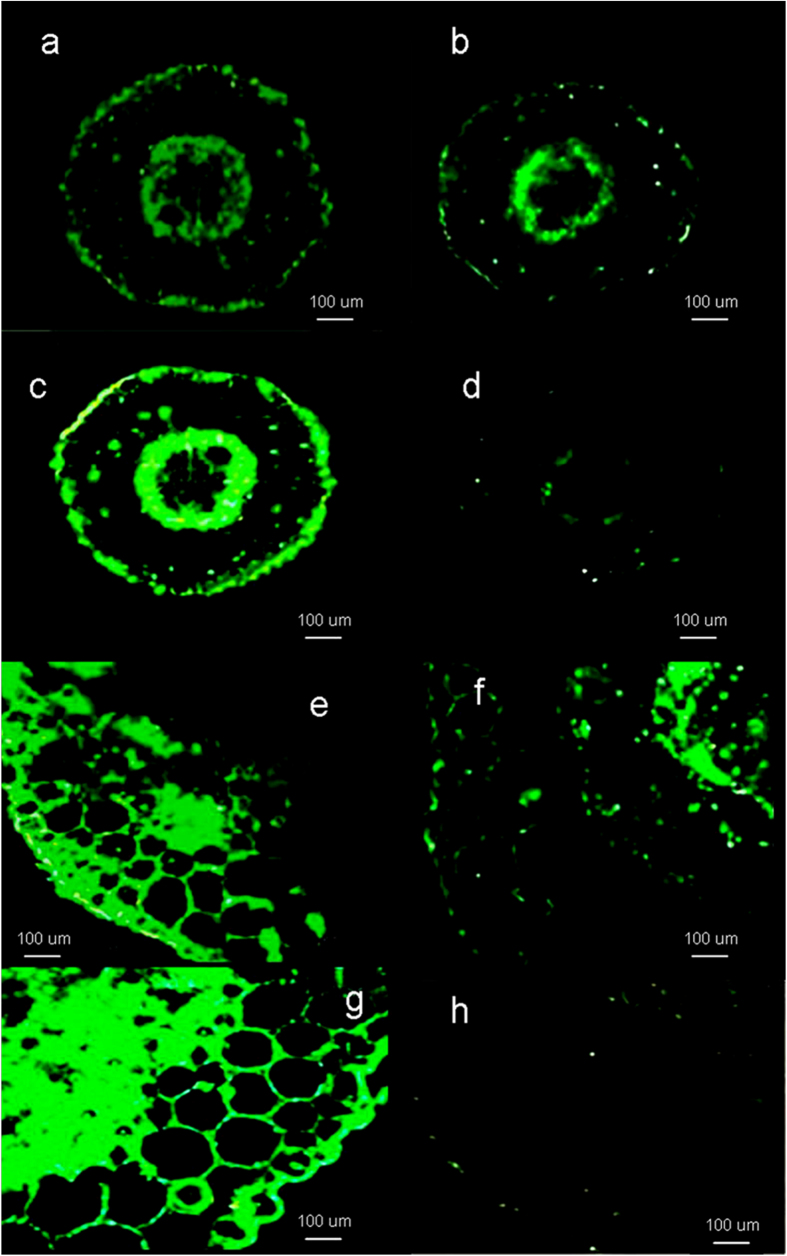
In order to analyze the tissue specific localization of SmNAC in S. melongena, tissue sections of stems and roots were treated with antibodies coupled to a green fluorescent dye. In the root sections, two zones of strong SmNAC antibody staining corresponding to the phloem and xylem were observed (Fig. 3a,b). After inoculation with BW, the staining was stronger in the roots of E-32 plants than in those of E-31 plants (Fig. 3c,d), and the same pattern was observed in stem tissues (Fig. 3e–h). Because the signal intensity increased in E-32 but decreased in E-31 plants, we concluded that R. solanacearum can induce the accumulation of SmNAC in susceptible but not resistant S. melongena plants.
Figure 3. Immunohistochemical localization analysis of SmNAC.
Fresh ‘E-32’ S. melongena stem and root tissues were used, and cross-sections of eggplant tissue labeled with purified SmNAC antibodies and visualized using an anti-rabbit IgG conjugated to green fluorescent protein. Scale bars, 100 μm. Each experiment repeated three times. (a) SmNAC protein distribution in the root of an uninfected susceptible plant (E-32). (b) SmNAC protein distribution in the root of an uninfected resistant plant (E-31). (c) SmNAC protein distribution in the root of a susceptible plant (E-32) after inoculation with Ralstonia solanacearum for 6 h. (d) SmNAC protein distribution in the root of a resistant plant (E-31) after inoculation with R. solanacearum for 6 h. (e) SmNAC protein distribution in the stem of an uninfected susceptible plant (E-32). (f) SmNAC protein distribution in the stem of an uninfected resistant plant (E-31). (g) SmNAC protein distribution in the stem of a susceptible plant (E-32) after inoculation with R. solanacearum for 6 h. (h) SmNAC protein distribution in the stem of a resistant plant (E-31) after inoculation with R. solanacearum for 6 h.
Subcellular localization and transcriptional activation of SmNAC
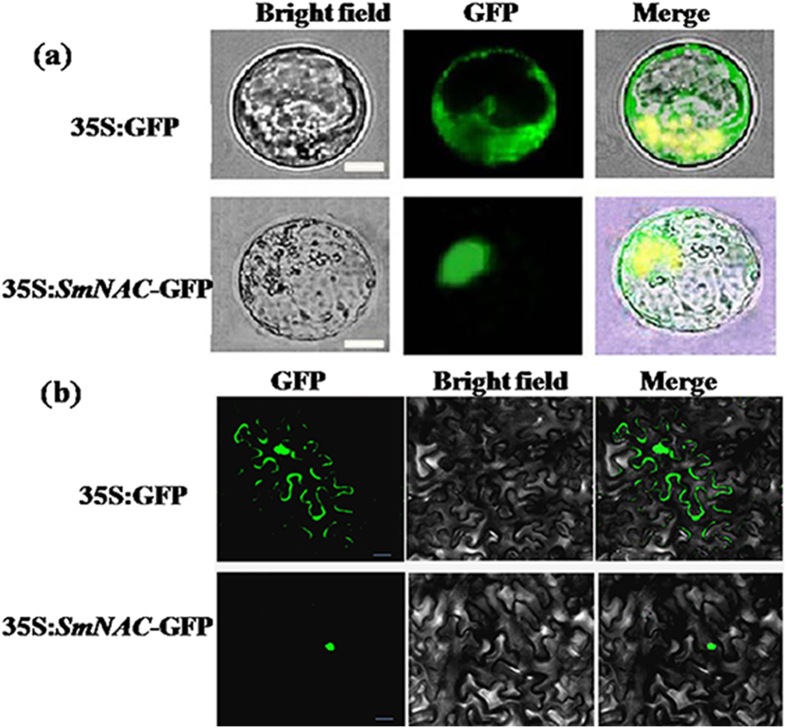
To establish the subcellular localization of SmNAC, a SmNAC and GFP fusion protein under the control of a 35S promoter was transiently expressed in S. melongena (E-32) and Ben’s tobacco protoplasts. Fluorescence signal localized to the nuclei (Fig. 4a,b). In contrast, control cells expressing GFP alone exhibited an even distribution of the GFP signal in both the cytoplasm and the nuclei.
Figure 4. Subcellular localization of SmNAC.
The PCR products were sub-cloned into the pEZS-NL-GFP vector in-frame with the green fluorescent protein (GFP) sequence, resulting in SmNAC-GFP vectors under the control of the CaMV 35S promoter. The fusion construct vector and the control GFP alone vector were introduced into S. melongena (E-32) protoplasts. GFP fluorescence was visualized using a laser scanning confocal microscope. All transient expression assays were repeated at least three times. (a) Eggplant (E-32) protoplasts were transiently transformed with SmNAC-GFP (green fluorescent protein) or GFP expressing constructs and fluorescence was visualized using a fluorescence microscope. Images were acquired in either dark field (green fluorescence) or bright field modes. Scale bars = 25 μm. (b) Ben’s tobacco protoplasts were transiently transformed with SmNAC-GFP (green fluorescent protein) or GFP expressing constructs and fluorescence was visualized using a fluorescence microscope. Images were acquired in either dark field (green fluorescence) or bright field modes. Scale bars = 25 μm.
The transcriptional activation of SmNAC was assessed in a transient expression assay using E-32 protoplasts. Full-length SmNAC fused to the GAL4 DNA-binding domain (GAL4-BD) was used as the effector. A dual luciferase reporter vector containing five copies of the GAL4 DNA-binding element and a minimal TATA region of a 35S promoter was fused to the firefly luciferase (LUC) reporter together witha Renilla luciferase (REN) reporter controlled by a 35S promoter. Activation of these reporters was used as an internal control for successful transformation (Fig. 5a), and an empty GAL4-BD (pBD) vector was used as a negative control. Compared with the negative control (pBD), the presence of SmNAC strongly increased the expression of the LUC reporter gene (Fig. 5b), and the LUC/REN ratio of SmNAC was significantly higher than that of the negative control.
Figure 5. Transcriptional activation of SmNAC.
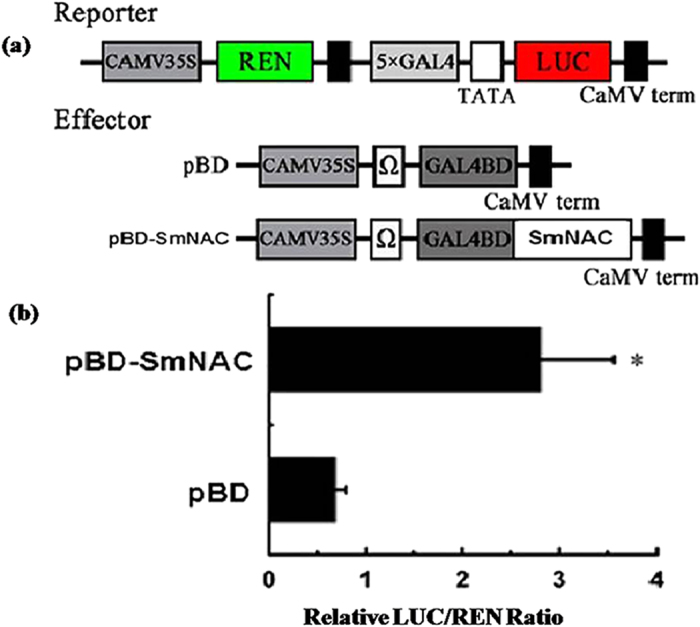
The SmNAC coding sequence without the stop codon was cloned into the reconstructed GAL4-DBD vector. To assay the binding activity of SmNAC to the ICS1 promoter, the ICS1 promoter was cloned into a pGreenII 0800-LUC double reporter vector, whereas SmNAC was inserted into the pGreenII 62-SK vector, generating the effector construct. The effector and reporter plasmids were co-transformed into S. melongena protoplasts as previously described and incubated as described above. LUC and REN luciferase activities were measured using a dual luciferase assay kit (Promega, USA). The analysis was performed using the Luminoskan Ascent Microplate Luminometer (Thermo, USA) with a 5 s delay and 15 s integration time. The binding activity of SmNAC to the ICS1 promoter was measured as a ratio of LUC to REN. At least six transient assays were measured for each assay. (a) The dual luciferase reporter construct contains a LUC reporter gene driven by the 35S (TATA box) promoter with five GAL4-binding elements, whereas each of the effectors contain a GAL4 DNA-binding domain (GAL4-BD); pBD was used as a negative control. SmNAC was linked to the GAL4-BD sequence and expression was driven by a 35S promoter. (b) Transactivation activity of SmNAC. Plasmid combinations of the dual REN/ LUC reporter, and effectors were co-transformed into eggplant protoplasts. After 12 h, the transactivation activity of SmNAC was measured as a ratio of LUC to REN. Each value represents the means of three biological replicates, and vertical bars represent the S.E. The asterisk indicates a significant difference at the 5% level.
Overexpression of SmNAC in S. melongena reduces resistance to BW
In order to analyze the tissue specific localization of SmNAC in SmNAC-overexpressing plants and SmNAC-RNAi transgenic plants, tissue sections of stems were treated with antibodies coupled to a green fluorescent dye. Two zones of strong SmNAC antibody staining corresponding to the phloem and xylem were observed (Fig. S6a,c). After overexpressing SmNAC, the staining was stronger in the stem of a SmNAC-overexpressing plant (EGT1–87) than in those of a non-transgenic plant (E-31) (Fig. S6a,b); After knocking-down SmNAC, the staining was weaken in the stem of the SmNAC-RNAi transgenic plant (RNAi-2-1) than in those of a non-transgenic plant (E-32) (Fig. S6c,d). The potential function of SmNAC in disease resistance was evaluated using transgenic eggplant plant lines overexpressing SmNAC and RNAi-SmNAC in which SmNAC expression was suppressed. We generated a thousand hypocotyl explants of the SmNAC-overexpressing lines, in which transgene expression was driven by a constitutive CaMV 35S promoter; six transgenic S. melongena plants were obtained (EGT0–5, EGT0–23, EGT0–43, EGT0–87, EGT0–145 and EGT0–204). The presence of the transgene was verified by genomic Southern blot, qRT-PCR and Western blot analyses (Fig. S4a–c). SmNAC expression levels were higher in all of the transgenic plants but not in the non-transgenic plants (E-31) (Fig. S4b), the levels of SmNAC protein in all transgenic plants exhibited the same changes (Fig. S4c). Three T1 transgenic lines (EGT1–87, EGT1–145 and EGT1–204) were obtained by selfing the primary transformants (T0) and these were used to assess BW resistance. Resistance was reduced in the T0 and T1 transgenic SmNAC overexpressing plants (Table 1, Fig. 6) and the non-transgenic plants (E-31) did not exhibit any symptoms. We conclude from these results that the overexpression of SmNAC in S. melongena reduces resistance to BW.
Table 1. Evaluation of bacterial wilt resistance in SmNAC overexpressing plants.
| Material | Generation | Evaluation of scale |
||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||
| EGT0–5 | T0 | 0 | 1 | 2 | 22 | 5 |
| EGT0–23 | T0 | 0 | 2 | 3 | 17 | 8 |
| EGT0–43 | T0 | 0 | 2 | 5 | 21 | 2 |
| EGT0–87 | T0 | 0 | 0 | 4 | 20 | 6 |
| EGT0–145 | T0 | 0 | 0 | 5 | 20 | 5 |
| EGT0–204 | T0 | 0 | 1 | 3 | 23 | 3 |
| EGT1–87 | T1 | 0 | 0 | 3 | 25 | 3 |
| EGT1–145 | T1 | 0 | 0 | 1 | 23 | 6 |
| EGT1–204 | T1 | 0 | 0 | 4 | 18 | 8 |
| E-31 | CK | 14 | 12 | 4 | 0 | 0 |
Evaluation of scale: 0 = healthy, 1 = one or two leaves wilted, 2 = three or more leaves wilted, 3 = all leaves wilted, and 4 = dead. Grades 0–2 were considered bacterial wilt resistant, and grades 3–4 susceptible. Bacterial wilt resistance of all plants was assessed after 20 dpi.
Figure 6. Identification of bacterial wilt (BW) resistance in SmNAC overexpressing eggplant lines.
CK represents non-transgenic E-31 plants; EGT0–87, EGT0–145, EGT0–204 are overexpressing SmNAC transgenic lines T0); and EGT1–204 indicates the T1 transgenic plant of EGT0–204. BW resistance was measured 15 d after pathogen inoculation. Whereas the non-transgenic resistant plant E-31 exhibited no wilt symptoms, the T0 and T1 overexpressing SmNAC plants exhibited clear wilt symptoms.
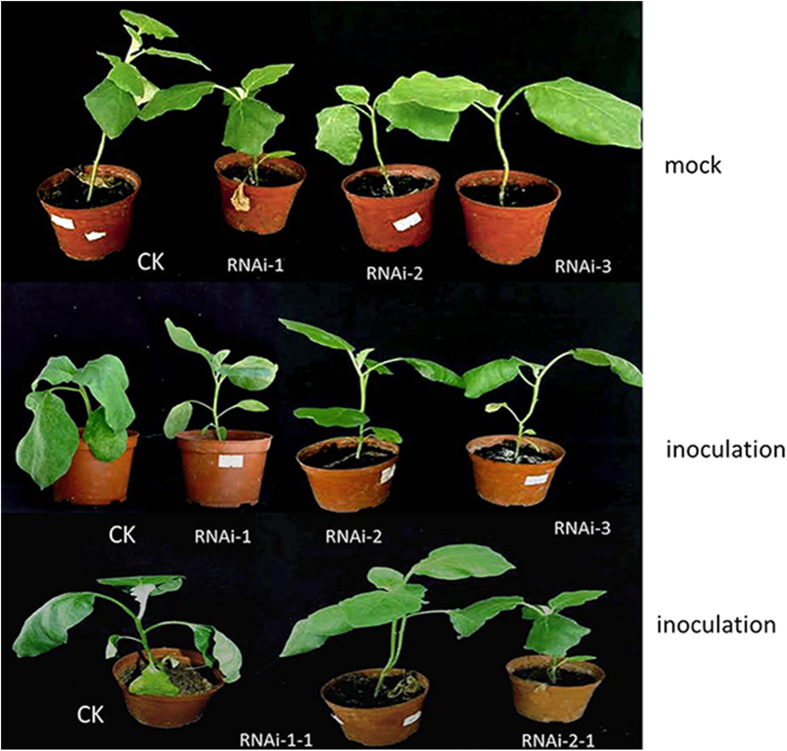
The association that we observed was further supported by our evaluation of the effects of SmNAC silencing in susceptible plants (E-32) using RNAi. We again used 1,000 hypocotyls as explants for transformation, and obtained 5 RNAi-SmNAC transgenic plants, which were screened for the presence of the transgene by Southern blot, qRT-PCR and Western blot analysis (Fig. S5a–c). SmNAC transcript levels were reduced in the SmNAC-RNAi transgenic (T0) plants compared to the non-transgenic plants (E-32), and no hybridization signals of SmNAC protein in any of the RNAi-treated plants were detected (Fig. S5c). The T0 and T1 transgenic plants were BW resistant after silencing of SmNAC (Table 2, Fig. 7). At 7 days after bacterial inoculation, the non-transgenic plants exhibited BW symptoms, whereas the RNAi plants did not; at 13 days after inoculation, more than half of the non-transgenic plants exhibited BW symptoms, and the SmNAC-RNAi plants also began to develop leaf wilt. We conclude that the silencing of SmNAC in susceptible S. melongena delays the presentation of BW symptoms, thereby increasing resistance to BW.
Table 2. Evaluation of bacterial wilt resistance of RNAi-SmNAC transgenic plants.
| Material | Generation | Evaluation of scale |
||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||
| RNAi-1 | T0 | 1 | 4 | 19 | 6 | 0 |
| RNAi-2 | T0 | 1 | 3 | 22 | 5 | 0 |
| RNAi-3 | T0 | 2 | 3 | 20 | 5 | 0 |
| RNAi-4 | T0 | 1 | 4 | 22 | 3 | 0 |
| RNAi-5 | T0 | 2 | 5 | 18 | 5 | 0 |
| RNAi-1-1 | T1 | 2 | 12 | 15 | 10 | 0 |
| RNAi-2-1 | T1 | 3 | 15 | 17 | 12 | 0 |
| RNAi-3-1 | T1 | 3 | 14 | 18 | 10 | 1 |
| E-32 | CK | 0 | 0 | 3 | 15 | 18 |
Evaluation scale: 0 = healthy, 1 = one or two leaves wilted, 2 = three or more leaves wilted, 3 = all leaves wilted, and 4 = dead. Grades 0–2 were considered bacterial wilt resistant, and grades 3–4 susceptible. Bacterial wilt resistance of all plants was assessed after 20 dpi.
Figure 7. Identification of bacterial wilt (BW) resistance in RNAi-SmNAC transgenic eggplants.
CK represents non-transgenic E-32 plants; RNAi-1, -2, and -3 represents RNAi-SmNAC transgenic eggplants (T0); RNAi-1-1 and RNAi-2-1 are T1 progeny. The BW resistance of plants was assessed at 15 days after pathogen inoculation. The non-transgenic susceptible plants from the E-32 line exhibited wilt symptoms, whereas the T0 and T1 transgenic RNAi-SmNAC plants did not.
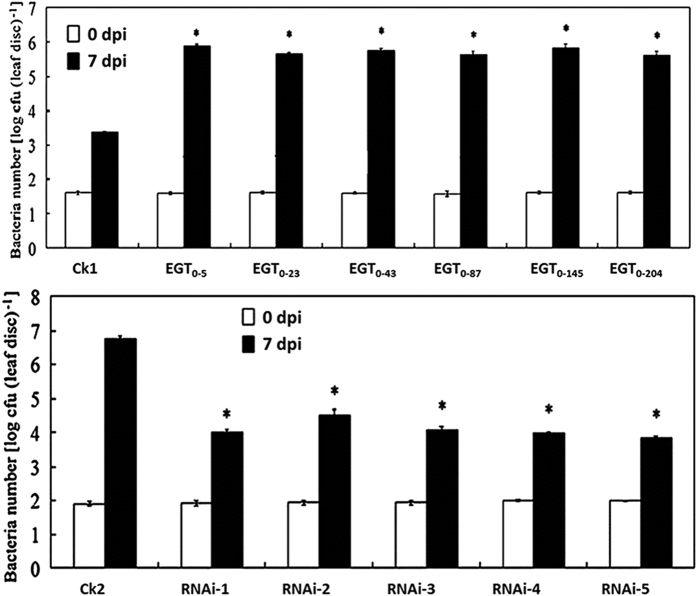
Quantification of bacteria in the pathogen-infected eggplant leaves revealed that the SmNAC-overexpressing plants exhibited a significantly increased bacterial population compared to the non-transgenic plants (E-31) at 7 dpi, whereas the leaves of the SmNAC-RNAi plants exhibited fewer bacteria than those of the non-transgenic plants (E-32) (Fig. 8).
Figure 8. Measurement of bacterial growth in transgenic eggplants.
Briefly, using the syringe inoculation method, bacteria wilt was scraped off a fresh plate, resuspended in sterile water to 105 colony-forming units (c.f.u.) ml−1, and pressure-infiltrated into leaves with a needleless syringe. After 7 days, leaves were harvested and surface sterilized (30 s in 70% ethanol, followed by 30 s in sterile distilled water) for the spray inoculation method. Leaf discs from different leaves were ground in 10 mM MgCl2 using a microfuge tube glass pestle. After homogenization, the samples were thoroughly vortex-mixed and diluted 1:10 serially. Samples were finally plated on TZC solid medium (3 g casein hydrolysate, 5 g peptone, and 10 g glucose (pH 7.0). The plates were incubated at 28 °C for 2 days, after which the colony-forming units were counted. CK1 represents non-transgenic plants from the E-31 line; EGT0–87, EGT0–145, and EGT0–204 are the SmNAC overexpressing transgenic eggplants (T0). CK2 represents non-transgenic plants from the E-32 line; RNAi-1, -2, -3, -4, and -5 are RNAi-SmNAC transgenic plants (T0). Error bars represent standard error, and the experiments were repeated at least three times with similar results. Asterisks indicate a significant difference at P < 0.05 compared with the non-transgenic plants.
The effect of SmNAC on the expression of genes involved in defense signaling pathways
We analyzed the effect of SmNAC on the expression of genes involved in the JA and SA defense signaling pathways. Transcript levels were quantified by qRT-PCR, and it was found that the expression of genes in the JA signaling pathway (JAR1, Pin2, LoxA) was higher in the SmNAC overexpressing lines (Fig. 9a) and lower in the SmNAC-RNAi lines (Fig. 9b) compared with control plants. In contrast, the expression of genes in the SA signaling pathway (EDS1, GluA, NPR1, TGA, SGT1, PAD4, PR-1a) was significantly lower in the SmNAC overexpressing lines (Fig. 9a) and higher in the RNAi-silenced SmNAC lines (Fig. 9b). These results indicate that SmNAC promotes and represses the transcription of genes in the JA and SA pathways, respectively.
Figure 9. qRT-PCR analysis of the expression of defense signaling genes in transgenic eggplants.
Quantitative reverse-transcription PCR (qPCR) was performed using a SYBR Premix Ex Taq kit (TaKaRa, Dalian, China), following the manufacturer’s protocols. Triplicate qPCR reactions were performed for each sample and the relative gene expression data was analyzed using the 2−ΔΔCt method. (a) qRT-PCR analysis of defense signaling genes in SmNAC oveexpressing plants. CK represents non-transgenic plants from the E-31 line, whereas 1–3 show the SmNAC overexpressing transgenic T0 plants EGT0–87, EGT0–145, and EGT0–204, and 4–6 show the SmNAC overexpressing transgenic T1 plants EGT1–87, EGT1–145, and EGT1–204. (b) qRT-PCR analysis of defense signaling genes in RNAi-SmNAC plants. CK represents non-transgenic plants from line E-32. 1–5 show the RNAi-SmNAC transgenic plants (T0) RNAi-1, RNAi-2, RNAi-3, RNAi-4, and RNAi-5.
SmNAC regulates SA biosynthetic genes
We next measured the effects of R. solanacearum infection on SA and JA levels in the SmNAC overexpressing plants, SmNAC-RNAi plants, and non-transgenic plants (E-31, E-32). The SA concentrations were increased in the non-transgenic resistant plants (E-31, CK1) compared to the SmNAC overexpressing plants (Table 3), and they were significantly increased in the SmNAC-RNAi plants compared to the non-transgenic susceptible plants (E-32, CK2). In addition, we observed that the JA content of all the susceptible plants differed significantly from that of the resistant plants after inoculation with the pathogen (Table 4). Based on these findings, we conclude that the SmNAC gene represses SA biosynthesis in S. melongena and might directly mediate genetic control over SA biosynthesis or metabolism.
Table 3. Salicylic acid content of transgenic eggplants after inoculation (μg.g−1 fresh weight).
| Lines | Generation | Time of treatment (d) |
||||
|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | 8 | ||
| EGT0–5 | T0 | 2.83 ± 0.32b | 2.15 ± 0.24B | 1.57 ± 0.11B | 0.74 ± 0.02B | 0.28 ± 0.01B |
| EGT0–23 | T0 | 2.72 ± 0.17b | 2.07 ± 0.12B | 1.61 ± 0.08B | 0.81 ± 0.02B | 0.23 ± 0.01B |
| EGT0–43 | T0 | 2.64 ± 0.25b | 2.13 ± 0.11B | 1.57 ± 0.09B | 0.94 ± 0.03B | 0.31 ± 0.02B |
| EGT0–87 | T0 | 2.55 ± 0.23b | 1.95 ± 0.19B | 1.41 ± 0.03B | 0.73 ± 0.05B | 0.25 ± 0.01B |
| EGT0–145 | T0 | 2.87 ± 0.22b | 2.34 ± 0.15B | 1.54 ± 0.02B | 0.85 ± 0.03B | 0.36 ± 0.03B |
| EGT0–204 | T0 | 2.91 ± 0.19b | 2.04 ± 0.12B | 1.46 ± 0.05B | 0.76 ± 0.01B | 0.18 ± 0.01B |
| EGT1–87 | T1 | 2.87 ± 0.21b | 2.15 ± 0.14B | 1.71 ± 0.01B | 0.81 ± 0.06B | 0.22 ± 0.02B |
| EGT1–145 | T1 | 2.81 ± 0.13b | 2.06 ± 0.11B | 1.63 ± 0.11B | 0.75 ± 0.03B | 0.27 ± 0.01B |
| EGT1–204 | T1 | 2.68 ± 0.24b | 1.93 ± 0.18B | 1.33 ± 0.11B | 0.67 ± 0.01B | 0.21 ± 0.01B |
| E-31 | CK1 | 4.76 ± 0.25a | 6.68 ± 0.44 A | 8.82 ± 0.39A | 13.67 ± 0.87A | 9.58 ± 0.75A |
| RNAi-1 | T0 | 4.15 ± 0.34a | 5.62 ± 0.39A | 7.64 ± 0.63a | 10.35 ± 0.84A | 8.42 ± 0.61A |
| RNAi-2 | T0 | 4.23 ± 0.26a | 5.04 ± 0.19A | 7.21 ± 0.58a | 11.21 ± 0.89A | 8.04 ± 0.52A |
| RNAi-3 | T0 | 4.56 ± 0.41a | 6.13 ± 0.24A | 7.54 ± 0.47a | 10.64 ± 0.93A | 7.95 ± 0.66A |
| RNAi-4 | T0 | 4.31 ± 0.35a | 6.15 ± 0.44A | 7.27 ± 0.62a | 10.83 ± 0.59A | 7.33 ± 0.59A |
| RNAi-5 | T0 | 4.47 ± 0.32a | 5.97 ± 0.43A | 7.18 ± 0.49a | 11.33 ± 0.67A | 7.61 ± 0.45A |
| RNAi-1-1 | T1 | 4.53 ± 0.32a | 6.21 ± 0.29A | 7.37 ± 0.53a | 10.87 ± 0.88A | 6.95 ± 0.55A |
| RNAi-2-1 | T1 | 4.67 ± 0.27a | 6.25 ± 0.35A | 7.55 ± 0.53a | 11.24 ± 0.25A | 7.41 ± 0.51A |
| RNAi-3-1 | T1 | 4.36 ± 0.24a | 6.22 ± 0.33A | 7.46 ± 0.54a | 11.62 ± 0.37A | 7.46 ± 0.61A |
| E-32 | CK2 | 3.87 ± 0.29b | 4.52 ± 0.31B | 6.12 ± 0.54b | 4.33 ± 0.23B | 2.65 ± 0.25B |
Error bars represent standard error. ab, AB, Significantly different from CK: ab, P < 0.05; AB, P < 0.01.
Table 4. Jasmonic acid content in transgenic eggplant after inoculation pathology (μg.g−1 fresh weight).
| Lines | Generation | Time of treatment (d) |
||||
|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | 8 | ||
| EGT0–5 | T0 | 4.23 ± 0.12 | 4.85 ± 0.22a | 5.57 ± 0.32A | 7.14 ± 0.24A | 6.78 ± 0.21A |
| EGT0–23 | T0 | 4.22 ± 0.14 | 4.87 ± 0.13a | 5.61 ± 0.31A | 7.21 ± 0.22A | 6.73 ± 0.33A |
| EGT0–43 | T0 | 4.34 ± 0.22 | 4.83 ± 0.11a | 5.67 ± 0.24A | 7.24 ± 0.25A | 6.81 ± 0.24A |
| EGT0–87 | T0 | 4.15 ± 0.23 | 4.75 ± 0.22a | 5.64 ± 0.23A | 7.23 ± 0.33A | 6.75 ± 0.28A |
| EGT0–145 | T0 | 4.27 ± 0.22 | 4.94 ± 0.15a | 5.71 ± 0.32A | 7.25 ± 0.23A | 6.86 ± 0.24A |
| EGT0–204 | T0 | 4.11 ± 0.19 | 4.91 ± 0.15a | 5.66 ± 0.25A | 7.16 ± 0.19A | 7.06 ± 0.23A |
| EGT1–87 | T1 | 4.12 ± 0.17 | 4.85 ± 0.17a | 5.81 ± 0.21A | 7.21 ± 0.26A | 6.92 ± 0.12A |
| EGT1–145 | T1 | 4.21 ± 0.15 | 4.86 ± 0.28a | 5.93 ± 0.28A | 7.15 ± 0.23A | 6.82 ± 0.27A |
| EGT1–204 | T1 | 4.28 ± 0.23 | 4.93 ± 0.28a | 5.83 ± 0.21A | 7.17 ± 0.24A | 6.88 ± 0.29A |
| E-31 | CK1 | 4.16 ± 0.26 | 3.18 ± 0.31b | 2.72 ± 0.18B | 1.38 ± 0.27B | 0.75 ± 0.05B |
| RNAi-1 | T0 | 5.95 ± 0.14 | 3.92 ± 0.19 | 2.64 ± 0.13B | 1.95 ± 0.14B | 1.32 ± 0.01B |
| RNAi-2 | T0 | 5.98 ± 0.16 | 4.04 ± 0.21 | 2.51 ± 0.24B | 1.97 ± 0.28B | 1.38 ± 0.11B |
| RNAi-3 | T0 | 5.96 ± 0.24 | 3.93 ± 0.14 | 2.54 ± 0.27B | 1.94 ± 0.33B | 1.35 ± 0.11B |
| RNAi-4 | T0 | 5.91 ± 0.25 | 3.95 ± 0.23 | 2.67 ± 0.28B | 1.98 ± 0.18B | 1.33 ± 0.10B |
| RNAi-5 | T0 | 5.93 ± 0.22 | 3.97 ± 0.31 | 2.62 ± 0.19B | 1.95 ± 0.25B | 1.36 ± 0.07B |
| RNAi-1-1 | T1 | 5.97 ± 0.12 | 3.96 ± 0.22 | 2.57 ± 0.23B | 1.97 ± 0.28B | 1.35 ± 0.08B |
| RNAi-2-1 | T1 | 6.07 ± 0.17 | 4.05 ± 0.13 | 2.55 ± 0.15B | 2.02 ± 0.25B | 1.37 ± 0.07B |
| RNAi-3-1 | T1 | 6.02 ± 0.21 | 3.95 ± 0.33 | 2.58 ± 0.14B | 1.96 ± 0.36B | 1.31 ± 0.02B |
| E-32 | CK2 | 5.97 ± 0.17 | 4.02 ± 0.31 | 5.95 ± 0.31A | 7.97 ± 0.27A | 6.41 ± 0.28A |
Error bars represent standard error. ab, AB, Significantly different from CK: ab, P < 0.05; AB, P < 0.01.
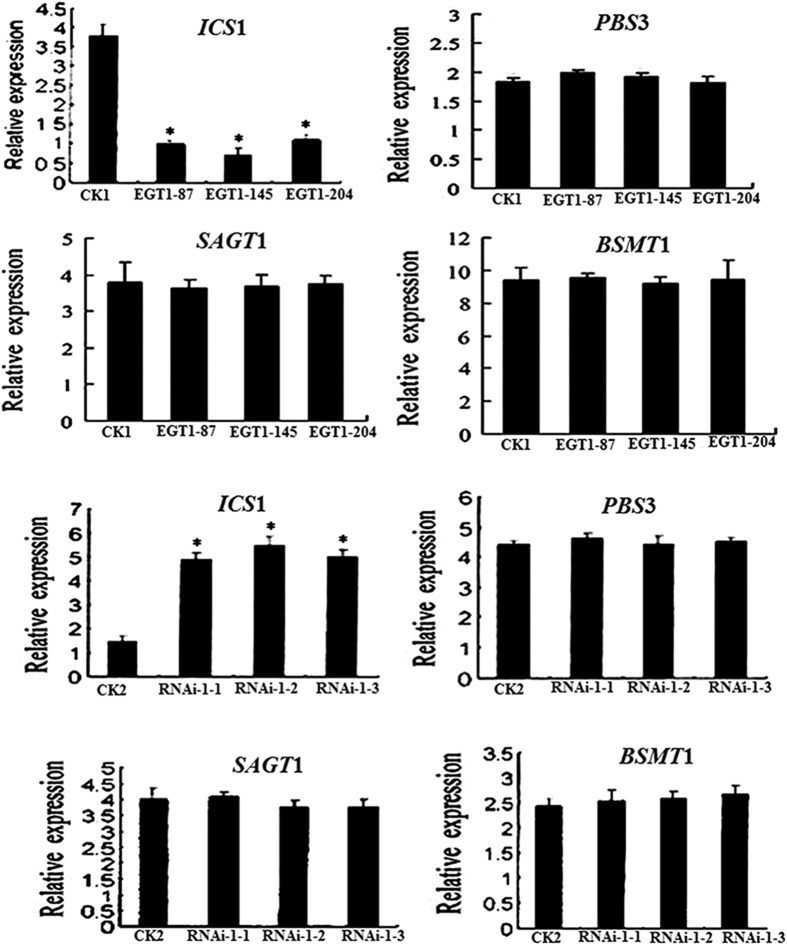
To determine whether SA-biosynthetic or SA-catabolic genes are regulated by SmNAC, we measured the expression levels of four associated genes (ICS1, PBS3, SAGT1, BSMT1). ICS1 expression was increased in the non-transgenic resistant plants (E-31) compared to the SmNAC overexpressing or non-transgenic susceptible plants (E-32), whereas the expression of the other three genes (PBS3, SAGT1, BSMT1) was not significantly different between the non-transgenic resistant plants (E-31) and the SmNAC overexpressing lines. Similarly, we found that ICS1 expression levels were higher in the SmNAC-RNAi lines than in the non-transgenic susceptible plants (E-32), but that there were no significant differences in the expression levels of the other three SA-related genes. This suggests that the three SA catabolic genes are not affected by SmNAC (Fig. 10), but that the SA biosynthetic gene ICS1 is regulated by SmNAC expression.
Figure 10. Expression of salicylic acid biosynthetic and catabolic genes in both SmNAC overexpressing plants and RNAi-SmNAC plants.
EGT1–87, EGT1–145, and EGT1–204 are the T1 generation of SmNAC overexpressing plants from the T0 generation plants, EGT0–87, EGT0–145, and EGT0–204. CK1 shows E-31. RNAi-1-1, RNAi-2-1, and RNAi-3-1 are the T1 generation RNAi-SmNAC plants from the T0 generation plants, RNAi-1, RNAi-2, and RNAi-3. CK2 shows E-32.
SmNAC regulates ICS1 through direct interaction with its promoter
A 1,697 bp region of the predicted ICS1 promoter, which we identified by genome walking PCR, was analyzed using the PLACE and Plant-CARE databases (Table S3). Based on these analyses, we found six NAC core-binding sites (CACG). The promoter sequence also contained the core cis-acting elements TATA and CAAT, several light response cis-acting elements, heat stress response elements, an elicitor-responsive element, and a cis-acting element involved in defense and stress responses.
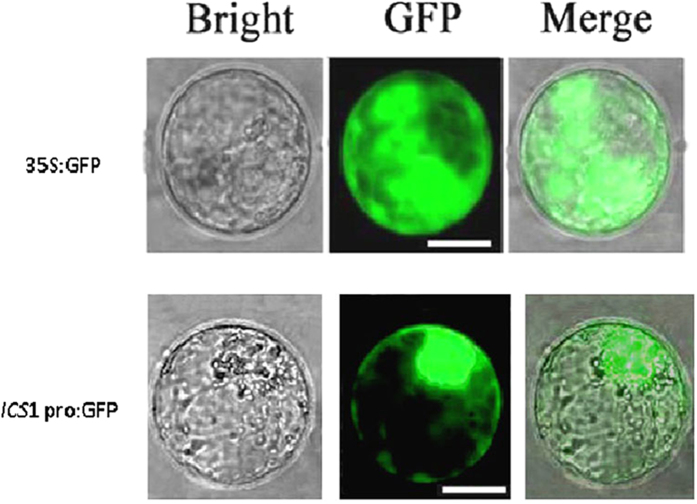
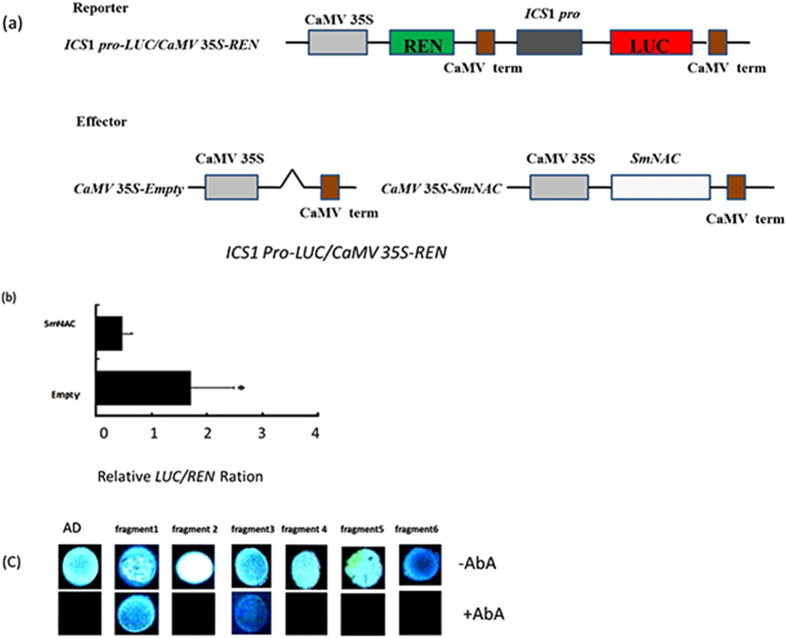
To monitor ICS1 promoter activation, an ICS1 pro-GFP vector and a positive control vector (35 S::GFP) were transformed into S. melongena protoplasts, and due to the fluorescent signal observed in both types of transformed protoplasts (Fig. 11), we conclude that the ICS1 promoter was sufficient to drive GFP expression. To investigate whether SmNAC could directly activate the ICS1 promoter in plant cells, a dual luciferase-based transactivation assay was performed using S. melongena E-32 protoplasts. The dual luciferase reporter plasmid harbored the ICS1 promoter fused to LUC and REN that are driven by the CaMV35S promoter (CaMV35S-REN/ICS1pro-LUC). An effector plasmid harboring SmNAC was expressed under the control of the CaMV 35S promoter (Fig. 12a). We observed that when an ICS1 pro-LUC reporter construct was co-transformed with the effector, CaMV 35S-SmNAC, the LUC/REN ratio was significantly reduced compared to the control ratio (Fig. 12b). Similarly, yeast one hybrid (Y1H) (Fig. 12c) analyses indicated that SmNAC binds to the ICS1 promoter at two binding sites, from −1 to −370 and from −550 to −750. We conclude that SmNAC functions as a transcriptional repressor that regulates ICS1 expression by directly binding to its promoter and repressing the transcription of ICS1.
Figure 11. Detection of ICS1 promoter activity in eggplant (E-32) protoplasts.
GFP reporters driven by the ICS1 promoter (ICS1 pro::GFP) and a CaMV 35S promoter (35S::GFP, positive control) were transiently expressed in eggplant E-32 protoplasts. After incubation for 12 h, GFP fluorescence was visualized by fluorescence microscopy. Scale bar = 25 μm. The experiment was repeated three times with similar results.
Figure 12. SmNAC represses the ICS1 promoter in a dual luciferase assay and yeast-one hybrid assay.
(a) A schematic representation of the double reporter and effector plasmids used. The double-reporter plasmid contained the ICS1 promoter fused to sequences encoding the LUC luciferase and the REN luciferase driven by the CaMV35S promoter. The effector plasmid contained SmNAC driven by a CaMV35S promoter. (b) SmNAC represses the ICS1 promoter. The reporter and effector vectors, as indicated, were co-transformed into eggplant E-32 protoplasts, which were incubated for 12 h. The activation of the ICS1 promoter by SmNAC was based on the ratio of LUC to REN. The asterisk indicates a significant difference at the 5% of the level compared to the empty effector vector. Each value represents the means of three biological replicates. (c) Yeast-1-hybrid (Y1H) assay of SmNAC and ICS1 promoter fragments. Fragment 1: −1 to−370 bp; fragment 2: −380 to −520 bp; fragment 3: −550 to −750 bp; fragment 4: −952 to −1170 bp; fragment 5: −1180 to −1318 bp; fragment 6: −1410 to −1570 bp. +AbA = SD/−Leu/+AbA medium, including the addition of 100 μm AbA. −AbA = SD/−Leu medium, no AbA added.
The effects of exogenous SA on BW resistance in SmNAC overexpressing plants
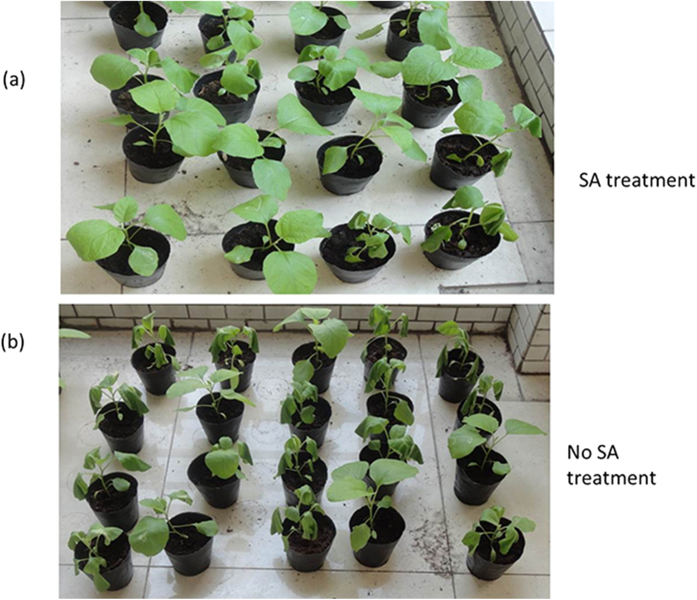
To examine the effects of exogenous SA on BW resistance in S. melongena, 60 SmNAC overexpressing seedlings (EGT1–204) and susceptible S. melongena (E-32) seedlings were sprayed with 0.2 mM SA prior to infection with R. solanacearum. At 7 days after inoculation, 38% of the EGT1–204 and 55% of the E-32 plants that had been treated with SA began to wilt, whereas the SmNAC overexpressing (EGT1–204) and susceptible S. melongena (E-32) plants treated with SA did not exhibit any BW symptoms. At 10 days after infection, 18% of the EGT1–204 plants and 27% of the E-32 plants treated with SA began to wilt, whereas at 15 days after inoculation, all of the non-treated EGT1–204 and E-32 plants presented with significant wilt symptoms. However, all of the plants treated with SA developed wilt at 20 days after inoculation (Table 5). These results suggest that SA affects BW resistance in S. melongena and, further, that reducing SA levels reduces resistance to BW (Fig. 13).
Table 5. Effect of exogenous salicylic acid on bacterial wilt resistance in SmNAC overexpressing plants.
| Material | #of plants | Treatment | % of plants with wilt symptoms after inoculation |
|||
|---|---|---|---|---|---|---|
| 7 dpi | 10 dpi | 15 dpi | 20 dpi | |||
| EGT1–204 | 60 | CK | 38.3 | 81.7 | 100 | |
| 60 | SA treatment | 0 | 18.3 | 61.7 | 100 | |
| E-32 | 60 | CK | 55.0 | 86.7 | 100 | |
| 60 | SA treatment | 0 | 26.7 | 66.7 | 100 | |
CK: plants were sprayed with water, before inoculation with the pathogen. SA (salicylic acid) treatment: plants were treated with 0.2 mM SA before inoculation with the pathogen. dpi, days post-inoculation.
Figure 13. Effect of exogenous salicylic acid on bacterial wilt (BW) resistance in SmNAC overexpressing plants.
Transgenic EGT1–204 plants were sprayed with 0.2 mM salicylic acid and water prior to inoculation with the pathogen. After 10 d, the plants were assessed for bacterial wilt resistance.
Discussion
NAC transcription factors elicit a variety of biological functions in plant development, including the regulation of responses to abiotic or biotic stresses, and they can be broadly classified into two large groups, I and II, based on structural features35. A series of studies have indicated that they are also involved in the regulation of the plant defense networks in response to attacks by a wide range of microbial pathogens and insects. For example, A. thaliana ATAF2 is thought to act as a repressor of pathogenesis-related (PR) gene expression40, whereas ATAF1 suppresses defense responses following pathogen infection41. OsNAC3 was found to be involved in responses to abiotic stress42, rice OsNAC6 and OsNAC19 are thought to promote resistance to the rice blast fungus43, and NAC genes have been found to be induced in oil seed rape after flea beetle colonization and Sclerotinia sclerotiorum infection44.
In this study, SmNAC from eggplant was identified from an analysis of RNA-Seq data and was shown to be most similar in sequence to the NAC proteins described above (OsNAC3, OsNAC4, SNAC1, ANAC081, ANAC102, ANAC32, ANAC019, ANAC055 and ANAC072) (Fig. 1). The expression levels of SmNAC differed between BW-resistant (E-31) and susceptible (E-32) plants (Fig. 2). This might be attributable to different promoter structures of SmNAC between the two plants, although future studies will be needed to confirm this. SmNAC expression was induced by infection with the bacterium R. solanacearum, the causal agent of BW. SmNAC overexpression in resistant S. melongena plants reduced BW resistance (Fig. 6), whereas silencing of SmNAC in susceptible S. melongena resulted in increased BW resistance (Fig. 7). We conclude from these results that SmNAC expression suppresses resistance to BW.
The specific triggers and mechanisms associated with BW resistance in S. melongena are currently unknown. Plant immunity-related signals, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are known to play essential roles in defense against pathogens, and immune responses are coordinated through a complex signaling crosstalk among these hormone-regulated pathways. For example, JA can suppress SA-mediated defenses45,46. Several reports have shown that the ET and SA signaling pathways of tomato plants are activated during the development of BW disease resistance. Furthermore, the JA, ET, and SA defense signaling pathways have been shown to interact synergistically to increase resistance to BW in tomato plants47,48. Milling et al.33 found that the expression of key genes of the JA pathway did not enhance BW resistance in tomato33, and in this study, the expression of three marker genes of the JA signal pathway (JAR1, PIN2, LOXA) increased in SmNAC overexpressing transgenic lines and in susceptible non-transgenic eggplant (E-32) but decreased in the RNAi-silenced SmNAC transgenic plants and in the resistant non-transgenic eggplant (E-31) (Fig. 9). However, the JA content of all the susceptible plants were significantly higher than those of the resistant plants (Table 4) after pathogen inoculation. We therefore deduce that the genes of the JA signaling pathway can reduce BW resistance in eggplant. In contrast, the expression of key genes in the SA signaling pathway (EDS1, GLUA, NPR1, TGA, SGT1, PAD4, and PR-1A) decreased in SmNAC overexpressing transgenic S. melongena lines but increased in RNAi-silenced SmNAC transgenic plants (Fig. 9), and the SA levels of resistant plants were significantly higher than those of the susceptible plants (Table 3). The SA-related defense pathway has been well characterized in A. thaliana, and the NPR1 gene has been shown to be required to promote TGA expression and to activate downstream defense genes47. Our data support this same function for SA in S. melongena. We note that in our analyses, the expression of EIN2 and EIL1 in the ET signaling pathway exhibited no significant differences between the transgenic and non-transgenic plants, suggesting that the ET signaling pathway does not play a role in SmNAC-mediated regulation of BW-resistance in eggplant. The MAPK pathway has also been proposed as a defense mechanism against BW in tomato plants47, although the expressions of genes in this pathway were not evaluated in S. melongena in this or previous studies49. Because SmNAC overexpression in resistant S. melongena reduced BW resistance compared to non-transgenic resistant plants (E-31), SmNAC silencing in susceptible S. melongena might increase BW resistance compared to non-transgenic susceptible S. melongena (E-32). We conclude that the expression of SmNAC reduces BW resistance by suppressing gene expression in the SA signaling pathway. Kloek et al.50 reported that increasing SA levels in A. thaliana enhanced resistance to the bacterium Pseudomonas syringae50. In A. thaliana, coronatine, a toxin produced by P. syringae, activates the expression of ANAC019, ANAC055, and ANAC072 through the action of the MYC2 transcription factor. This results in an inhibition of SA accumulation by repressing the ICS1 gene and activating the BSMT1 gene. The end result is a decrease in resistance to the BW pathogen. In our study, SmNAC suppresses the activity of ICS1 by interacting with the ICS1 promoter in S. melongena (Fig. 12), whereas SmNAC does not regulate the expression of PBS3, SAGT1or BSMT1 (Fig. 10). The SA levels in SmNAC overexpressing transgenic plants were lower than those in non-transgenic plants (E-31), but SA levels in RNAi-silenced SmNAC transgenic plants were higher than those in non-transgenic susceptible S. melongena (E-32). Taken together, these results suggest that SmNAC represses the accumulation of SA by inhibiting ICS1 expression, thereby lowering BW resistance. Our discovery might be important for the development of strategies for reducing loss of eggplant crops to BW infection.
Materials and Methods
Plant material
We used two Solanum melongena inbred lines, ‘E-31’ (R) and ‘E-32’ (S), obtained from South China Agricultural University (Guangzhou, China). ‘E-31’ (R) and ‘E-32’ (S) have been described by Xiao et al.49.
Inoculation with R. solanacearum strains
S. melongena seedlings were grown in a mixture of turf soil and perlite (2:1) in a greenhouse with natural light at 20–25 °C. At the two- or three-leaf stage, seedlings were transferred to a phytotron maintained at (30 ± 2)/(25 ± 2) °C day/night (12 h day length) and 90% relative humidity. A highly virulent R. solanacearum strain (race1) was isolated from the BW susceptible S. melongena strain (E-32). A single colony of the virulent type was grown at 30 °C for 48 h on TZC medium. The inoculum was incubated in liquid TZC medium containing 3 g casein hydrolysate, 5 g peptone, and 10 g glucose (pH 7.0) by shaking in a water bath at 30 °C for 24 h. After incubation, the density of the suspension was determined using a spectrophotometer and adjusted to 1 × 108 colony forming units (cfu)/ml. Seedlings were inoculated at the four- or five-leaf stage by wounding the roots and incubating them in the bacterial suspension for 20 min. The plants were inoculated for 4 weeks. After 7 d of inoculation, susceptible plants began showing symptoms of BW.
Total RNA extraction, construction of RNA-Seq libraries and data analysis
RNA-Seq analysis was performed on resistant (E-31) and susceptible (E-32) eggplant lines. Lines A0 and A1 corresponded to mock-inoculated and pathogen-inoculated E-31 plants at 7 days post-inoculation (dpi). Similarly, B0 and B1 correspond to mock-inoculated and pathogen-inoculated E-32 plants at also at 7 dpi. RNA was extracted from ten plants stems of each of these four lines and used for RNA-Seq library construction, with 2 replicates for each treatment. Total RNA was extracted using TRIZOL reagent (Huayueyang, Beijing, China) and then treated with RNase-free DNaseI (TaKaRa, Dalian, China), according to the manufacturer’s protocols. PolyA mRNAs were purified using oligo-dT-attached magnetic beads. One microgram of purified mRNA was cleaved into 200-500 bp fragments by super-sonication, and the cleaved mRNAs were used as templates for the RNA-Seq libraries and digital gene expression (DGE) profile analyses. First-strand cDNA was synthesized using an M-MLV RTase cDNA synthesis kit (TaKaRa, China) following the manufacturer’s protocols, prior to purification using the QiaQuick PCR extraction kit, end repair, addition of dA-tails, and ligation to Illumina adapters. The ligation products were size resolved by agarose gel electrophoresis, and the fragments were excised for PCR amplification. The amplified fragments were sequenced using an Illumina HiSeq™ 2000 by Gene Denovo Co. (Guangzhou, China).
For sequence assembly, raw reads were filtered to remove adaptor sequences, empty reads, and low-quality sequences. The resulting cleaned reads were de novo assembled51, and unigenes were annotated using the NCBI BLASTx function (http://www.ncbi.nlm.nih.gov/BLAST/) with an E-value threshold of 1E−5, the NCBI nr database (http://www.ncbi.nlm.nih.gov), the Swiss-Prot protein database (http://www. expasy.ch/sprot), the KEGG database (http://www.genome.jp/kegg), and the COG database (http://www.ncbi.nlm.nih.gov/COG). The direction of the unigenes was determined according to the best alignment results. The FPKM (reads per kb per million reads) value was used to estimate the gene expression level and was calculated as previously described52. Blast results of the de novo assemblies were used to retrieve Gene Ontology (GO) terms with Blast2GO53 under different categories: biological processes, molecular function and cellular component, which are hierarchically organized into different levels.
Alignments were performed using CLUSTALW v1.83 and the GeneDoc software54. A phylogenetic tree was constructed using the neighbor-joining method in MEGA6.055 and visualized using TreeView56. The theoretical isoelectric points (pIs) and mass values of the mature peptides were calculated using the PeptideMass program (http://web.expasy.org/peptide_mass/).
Construction of the SmNAC RNAi and SmNAC overexpression vectors and transformation procedures
We generated a 200 bp SmNAC forward fragment that was amplified by PCR (using the primers UP-P1: 5′-CGGATTTAAATGAAAATTAGAAAGATTACAAC-3′; DW-P1: 5′-CCGCCATGG TAAAAGAATATACATGTCCCT-3′), digested with SwaI and NcoI, and ligated into the pFGC5941 vector. The reverse fragment was amplified by PCR (using the primers UP-P2: 5′-CATGGATCCGAAAATTAGAAAGATTACAAC-3′; DW-P2: 5′-CTACCCGGGTAAAAGAA TATACA TGT CCCT-3′), digested with SmaI and BamHI and ligated into the pFGC5941 vector to create the RNAi-SmNAC vector. This binary vector was introduced into Agrobacterium tumefaciens strain EHA105. The PCR protocol used was as follows: 94 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 30 s, and extension at 72 °C for 2 min. The RNAi-SmNAC vector then was transformed into ‘E-32’ eggplant and generated the RNAi-SmNAC transgenic plants.
The open reading frame (ORF) for SmNAC was amplified by PCR (5′-ATGGGTGTTCAAGAAAAAGAT CCTC-3′ and 5′-CTACTGTCTGAACCCGAGATTTAACGT-3′) and cloned into the pMD-T19 vector. The gene fragment and the expression vector pBI121 were then digested with SmaI and SacI, and the SmNAC fragment was ligated into the digested vector using T4 ligase. The resulting overexpression vector, pBI-SmNAC, containing the CaMV35S promoter, Nos terminator, and the NPT-II gene, was then transformed into ‘E-31’ eggplant, using the methods as previously described57. Kanamycin (15 mg/L) was included in the growth medium to select for positive transformants, and resistant buds were obtained after 25 days. The presence of the transgene was verified by Southern blot analysis. T1 transgenic eggplant progeny were obtained following the self-pollination of T0 transgenic plants, and the transgenic T1 lines were tested for BW-resistance. Bacterial wilt growth was measured in transgenic eggplants as described by Zipfel et al.58. Briefly, using the syringe inoculation method, bacteria wilt was scraped off a fresh plate, resuspended in sterile water to 105 colony-forming units (c.f.u.) ml−1, and pressure infiltrated into leaves with a needleless syringe. After 7 days, leaves were harvested and surface sterilized (30 s in 70% ethanol, followed by 30 s in sterile distilled water) for the spray inoculation method. Leaf discs from different leaves were ground in 10 mM MgCl2 using a microfuge tube glass pestle. After homogenization, the samples were thoroughly vortex-mixed and diluted 1:10 serially. Samples were finally plated on TZC solid medium (3 g casein hydrolysate, 5 g peptone, and 10 g glucose (pH 7.0). The plates were incubated at 28 °C for 2 days, after which the colony-forming units were counted.
Immunohistochemical analysis of SmNAC
Fresh ‘E-32’ S. melongena stem and root tissues, and SmNAC-overexpressing and SmNAC-RNAi transgenic plants’ stem were cut into 4 mm pieces and fixed by vacuum infiltration for 2.5 h under ambient pressure in 3% paraformaldehyde, 0.1% Triton X-100 in phosphate buffered saline (PBS) at pH 7.2. The samples were dehydrated by incubation in increasing concentrations of 10%, 30%, 50%, 70% to 95% ethanol for 30 mins at each step, and placed in a polyethylene glycol (PEG) solution containing PEG1500:4000 (2:1) at 55 °C. After cooling at 4 °C, embedded material was sliced into 10 μm sections using a microtome. The sections were transferred to PBS to remove the PEG, washed for 10 min in PBS (pH 7.2), 5 min in 0.1 M NH4Cl, and then 5 min in PBS (pH 7.2). The tissues were next incubated in a blocking buffer consisting of 5% bovine serum albumin (BSA) in PBS (pH 7.2) for 1 h, then labeled with a polyclonal anti-SmNAC being generated by Invitrogen (Shanghai, China) or rabbit pre-immune serum for 12 h at 4 °C. The tissue sections were then washed three times with 0.1% BSA in PBS and immersed in a solution with anti-rabbit IgG conjugated to Alexa-Fluor 488 (Invitrogen, USA) at 37 °C for 2 h. After 4 washes with PBS, tissue sections were visualized using a reflected light microscope (Zeiss, Axioskop, Germany) with a dichroic filter (460–490 nm), and fluorescent images were acquired.
Quantitative reverse-transcription (qRT-PCR) analysis
Quantitative reverse-transcription PCR (qPCR) was performed using gene-specific primers (Table S1) and a SYBR Premix Ex Taq kit (TaKaRa, Dalian, China), following the manufacturer’s protocols. The qRT-PCR cycles were as follows: an initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 56 °C for 20 s, and extension at 72 °C for 35 s. Triplicate qPCR reactions were performed for each sample, and the relative gene expression data were analyzed using the 2−ΔΔCt method59.
Southern blot and Western blot analysis
DNA was extracted from leaves of the putative transgenic eggplant lines using the CTAB method, and Southern blot analysis was performed using a DNA labellling and detection kit (Boehringer Mannheim, Germany). The primers for the NPT-II probe used were F: 5′-TCGGCTATGACTGGGCACA-3′ and R: 5′-GATACCGTAAAGCACGAGGAAG-3′. The Bar primer sequences used were pBar1: 5′-ATGAGCCCAGAACGACGC-3′ and pBar2: 5′-TCT CAAATCTCGGTGACG-3. The PCR cycles were as follows: 5 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 56 °C, 2 min at 72 °C, and one final 10 min step at 72 °C. The products were stored at 4 °C.
For Western-blot analysis, 30 μg of total proteins from plant tissue was separated by 10% SDS/PAGE gels and electro blotted onto a nitrocellulose membrane (GE Healthcare, Munich, Germany), and the membranes were preblocked with Tris-buffered saline (20 mM Tris, pH 7.5, and 150 mM NaCl) containing 5% (w/v) skim milk powder and 0.01% (v/v) Tween20. The membranes were probed with a primary anti-SmNAC, followed by a secondary goat anti-rabbit IgG antibody (Bio-Rad). Signals were detected using the ECL Prime Western Blotting Detection Reagent following the manufacturer’s protocols (GE Healthcare, Munich, Germany).
Salicylic acid (SA) and jasmonic acid (JA) analysis
After inoculation, free SA was detected in R. solanacearum by HPLC at 0, 2, 4, 6 and 8 d, as previously described60. Quantitative determination of JA content was performed following the protocol described by Zhang et al.61. Each treatment repeated 3 times, and statistical analysis was used by the SPSS software.
Subcellular localization of SmNAC
The coding region of Sm-NAC, without the stop codon, was amplified by PCR with the sub-SmNACF (5′-ATGGGTGTTCAAGAAAAAGATCC-3′) and sub-SmNAC R (5′-CTACTGTCTGAACCCGAGATTTAAC-3′) primers. The PCR products were sub-cloned into the pEZS-NL-GFP vector in-frame with the green fluorescent protein (GFP) sequence, resulting in SmNAC-GFP vectors under the control of a CaMV 35S promoter. The fusion construct vector and the control GFP-alone vector were introduced into S. melongena (E-32) protoplasts using 40% PEG solution as previously described62. GFP fluorescence was visualized using a laser scanning confocal microscope (Leica TCS SP2, Leica Microsystems, Wetzlar, Germany). All transient expression assays were repeated at least three times.
Promoter isolation
Genomic DNA was extracted from fresh ‘E-32’ (S) S. melongena leaves using the DNeasy Plant Mini Kit (Qiagen, Germany). The promoter of the eggplant isochorismate synthase 1 (ICS1) gene was isolated using a Genome Walker Kit (Clontech, USA) by nested PCR (primers shown in Table S2) according to the manufacturer’s protocols. After sequencing, conserved cis-element motifs of the promoter were predicted using the PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and Plant-CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) databases.
Protoplast transformation assay
For transactivation analysis of SmNAC, the SmNAC coding sequence without the stop codon was cloned into the reconstructed GAL4-DBD vector (Clontech, USA), producing the effector construct. The double reporter vectors were kindly provided by Professor Jianye Chen (Horticultural College of South Agricultural University) and included a GAL4-LUC and an internal control, REN, driven by the 35S promoter. GAL4-LUC contains five copies of a GAL4-binding element and a minimal TATA region of the 35S promoter, which are all located upstream of the LUC gene. To assay the binding activity of SmNAC to the ICS1 promoter, the ICS1 promoter was cloned into a pGreenII 0800-LUC double reporter vector, whereas SmNAC was inserted into the pGreenII 62-SK vector, generating the effector construct63. The effector and reporter plasmids were co-transformed into S. melongena protoplasts as previously described64 and incubated as described above. LUC and REN luciferase activities were measured using a dual luciferase assay kit (Promega, USA). The analysis was performed using the Luminoskan Ascent Microplate Luminometer (Thermo, USA) with a 5 s delay and 15 s integration time. The binding activity of SmNAC to the ICS1 promoter was measured as a ratio of LUC to REN. At least six transient assays were measured for each assay.
Yeast One-Hybrid assay
The sequence encoding the SmNAC domain (amino acids 1-345) was inserted into pGADT7 (pAD) vector (Clontech, USA) to generate the pAD-SmNAC construct. Six DNA fragments of the eggplant ICS1 promoter (−1 to −370 bp, −380 to −520 bp, −550 to −750 bp, −952 to −1170 bp, −1180 to −1318 bp, and −1410 to −1570 bp) were ligated into the pAbAi vector (Clontech, USA). Transformation of the Y1H yeast strain AH109 with these pAbAi vectors containing the promoter fragments yielded the bait yeast strains, which were further transfected with pAD-SmNAC. Growth of the transformants on SD/-Leu/+AbA medium was considered as an indicator for SmNAC binding to the corresponding DNA fragment.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD). Statistical analysis was used by the SPSS (Statistical Product and Service Solutions) software. A value of p < 0.05, p < 0.01 was considered to be statistically significant. Statistical analysis was performed with Excel 2010.
Additional Information
How to cite this article: Na, C. et al. Overexpression of the Eggplant (Solanum melongena) NAC Family Transcription Factor SmNAC Suppresses Resistance to Bacterial Wilt. Sci. Rep. 6, 31568; doi: 10.1038/srep31568 (2016).
Supplementary Material
Acknowledgments
This work was supported by the State Natural Science Foundation of China, the Natural Science Fund of the Guangdong Province [Grant No.: S2011030001410], Programs for Science and Technology of the Guangdong Province (2015B020202004, 2015A020209154), and Programs of the Science Innovation of Guangdong Higher Education Institutes (2014KTSCX033). We thank PlantScribe (www.plantscribe.com) for editing this manuscript.
Footnotes
Author Contributions C.N. performed experiments and analyzed the data. C.B. designed the study and wrote the manuscript. L.J. and C.C. supervised the study and modified the manuscript. W.S., F.J. and J.J. gave help to the experiments. All authors reviewed the manuscript.
References
- Zhu T., Nevo E., Sun D. & Peng J. Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 66, 1833–1866 (2012). [DOI] [PubMed] [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H. & Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X. Characterization of a novel Medicago sativa NAC transcription factor gene involved in response to drought stress. Mol. Biol. Rep 40, 6451–6458 (2013). [DOI] [PubMed] [Google Scholar]
- Xu Q. F., He Q., Li S. & Tian Z. D. Molecular characterization of StNAC2 in potato and its overexpression confers drought and salt tolerance. Acta Physiol. Plant 36, 1841–1851 (2014). [Google Scholar]
- Yang X., Hu Y. X., Li X. L., Yu X. D. & Li Q. L. Molecular characterization and function analysis of SlNAC2 in Suaeda liaotungensis K. Gene. 543, 190–197 (2014). [DOI] [PubMed] [Google Scholar]
- Yu X. W. et al. CarNAC2, a novel NAC transcription factor in chickpea (Cicer arietinum L.), is associated with drought-response and various developmental processes in transgenic Arabidopsis. J. Plant Biol. 57, 55–66 (2014). [Google Scholar]
- Zhao D., Derkx A. P., Liu D. C., Buchner P. & Hawkesford M. J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 17, 904–913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121 (2015). [DOI] [PubMed] [Google Scholar]
- You J., Zhang L., Song B., Qi X. O. & Chan Z. L. Systematic Analysis and Identification of Stress-Responsive Genes of the NAC Gene Family in Brachypodium distachyon. PloS one 10, e0122027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y. Q. et al. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 43, 1469–1484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. J. et al. Comprehensive Analysis Suggests Overlapping Expression of Rice ONAC Transcription Factors in Abiotic and Biotic Stress Responses. Int. J. Mol. Sci. 16, 4306–4326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A. P., Guo H. S., García J. A. & Gutiérrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a Gemini virus protein. Plant Mol. Biol. 39, 647–656 (1999). [DOI] [PubMed] [Google Scholar]
- Sun L. et al. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol. 81, 41–56 (2013). [DOI] [PubMed] [Google Scholar]
- Xia N. et al. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 37, 3703–3712 (2010). [DOI] [PubMed] [Google Scholar]
- Donze T., Qu F., Twigg P. & Morris T. J. Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology 449, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyal G., Rana V. S., Mukherjee S. K., Wajid S. & Choudhury N. R. Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) Rep protein. Virus genes 48, 486–493 (2014). [DOI] [PubMed] [Google Scholar]
- Wang F. T. et al. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front. plant Sci. 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q. et al. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18, 756–767 (2008). [DOI] [PubMed] [Google Scholar]
- Gao Y. M. Genetic analysis and molecular marking of resistance of bacterial wilt of eggplant. Chinese Academy of Agricultural (2006). [Google Scholar]
- Li M., Wang Y. Q., Tian S. B., Luo Z. Y. & Wang X. J. Genetic Analysis of Resistance to Bacterial Wilt and Identification of an Associated AFLP Marker in Eggplant (Solanum melongena). Acta Horticulturae Sinica 33, 869–872 (2006). [Google Scholar]
- Cao B. H., Lei J. J., Wang Y. & Chen G. J. Inheritance and identification of SCAR marker linked to bacterial wilt-resistance in eggplant. Afr. J. Biotechnol. 8, 5201–5207 (2009). [Google Scholar]
- Lebeau A. et al. Genetic mapping of a major dominant gene for resistance to Ralstonia solanacearum in eggplant. Theor. Appl. Genet. 126, 143–158 (2013). [DOI] [PubMed] [Google Scholar]
- Yao J. Q., Lin B. Q. & Xiao Y. H. Histopathological research of eggplant verticillium wilt resistant material. Acta Phytopathologica Sinica. 26, 159–163 (1996). [Google Scholar]
- Liu F. Z., Lian Y., Feng D. X., Song Y. & Chen Y. H. Identification and Evaluation of Resistance to Bacterial Wilt in Eggplant Germplasm Resources. J. Plant Genetic Resources 6, 381–385 (2005). [Google Scholar]
- Zhu H. W. et al. Studies on RAPD Marker of Bacterial Wilt Resistance Gene in Eggplant (Solanum melongena). Acta Horticultutae Sinica. 32, 321–323 (2005). [Google Scholar]
- Deslandes L. et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. P. Natl. Acad. Sci. 100, 8024–8029 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N. et al. Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC genomics 14, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L. et al. Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soil borne pathogen Ralstonia solanacearum. Molplant Microbe In. 11, 659–667 (1998). [DOI] [PubMed] [Google Scholar]
- Deslandes L. et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. P. Natl. Acad. Sci. 99, 2404–2409 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M. et al. RD19, an Arabidopsis cysteine protease required for RRS1-R–mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20, 2252–2264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensec F. et al. Towards the Identification of Type III Effectors Associated with Ralstonia solanacearum Virulence on Tomato and Eggplant. Phytopathology 105, 1529–1544 (2015). [DOI] [PubMed] [Google Scholar]
- Delaspre F. et al. The Ralstonia solanacearum pathogenicity regulator HrpB induces 3-hydroxy-oxindole synthesis. P. Natl. Acad. Sci. 104, 15870–15875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milling A., Babujee L. & Allen C. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. Plos One 6, e15853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A. C. The hosts of Pseudomonas solanacearum. In Hayward A. C., Harhnan G. L. (eds) Bacterial wilt: the disease and its causative agent, Pseudomonas. CAB International, Wallingford. pp, 9–25 (1994). [Google Scholar]
- Ooka H. et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247 (2003). [DOI] [PubMed] [Google Scholar]
- Fang Y., You J., Xie K., Xie W. & Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563 (2008). [DOI] [PubMed] [Google Scholar]
- Hu R. B. et al. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10, 145–167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Plant TFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 39, 1114–1117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Sharma V., Pal A. K., Acharya V. & Ahuja P. S. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20, 403–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delessert C. et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43, 745–757 (2005). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe In. 22, 1227–1238 (2009). [DOI] [PubMed] [Google Scholar]
- Sun L. et al. Comprehensive analysis suggests overlapping expression of rice ONAC transcription factors in abiotic and biotic stress responses. Int. J. Mol. Sci. 16, 4306–4326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Zhao W., Meng X., Wang M. & Peng Y. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci. 172, 120–130 (2007). [Google Scholar]
- Hegedus D. et al. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397 (2003). [DOI] [PubMed] [Google Scholar]
- Grant M. R. & Jones J. D. Hormone (dis) harmony moulds plant health and disease. Science 324, 750–752 (2009). [DOI] [PubMed] [Google Scholar]
- Zheng X. Y. et al. Coronatine Promotes Pseudomonas syringae Virulence in Plants by Activating a Signaling Cascade that Inhibits Salicylic Acid Accumulation. Cell Host Microbe 11, 587–596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Y. et al. Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol. Plantarum. 136, 324–335 (2009). [DOI] [PubMed] [Google Scholar]
- Ho F. I., Chen Y., Lin Y., Cheng C. & Wang J. F. A tobacco rattle virus-induced gene silencing system for a soil-borne vascular pathogen Ralstonia solanacearum. Bot. Studies 50, 413–424 (2009). [Google Scholar]
- Xiao X. O. et al. Functional Characterization of a Putative Bacterial Wilt Resistance Gene (RE-bw) in Eggplant. Plant Mol. Biol. Rep. 33, 1058–1073 (2015). [Google Scholar]
- Kloek A. P. et al. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522 (2001). [DOI] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: a universal tool for annotation, visualzation and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Nicholas K. B. & Nicholas H. B. J. GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the author 14 (available at http://www.psc.edu/biomed/genedoc/gddl.htm) (1997). [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl.Biosci. 12, 357–358 (1996). [DOI] [PubMed] [Google Scholar]
- Cao B. H., Huang Z. Y., Chen G. J. & Lei J. J. Restoring pollen fertility in transgenic male-sterile eggplant byCre/loxp-mediated site-specific recombination system. Genet Mol Biol. 33, 298–307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 (2004). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using realtime quantitative PCR 2ΔΔ Ct method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Mercedes E. C., Sylvia V. C. & Victor H. A. Relationships between salicylic acid content, phenylalanine ammonia-lyase (PAL) activity, and resistance of barley to aphid infestation. J. Agr. Food Chem. 51, 2227–2231 (2003). [DOI] [PubMed] [Google Scholar]
- Zhang F. J., Jin Y. J., Xu X. Y., Lu R. C. & Chen H. J. Study on the extraction, purification and quantification of jasmonic acid, abscisic acid and indole-3-acetic acid in plants. Phytochem. Analysis 19, 560–567 (2008). [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H. & Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- Hellens R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activityand RNA silencing in plants. Plant Methods 1, 13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S. & Theologis A. Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 5, 421–427 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.