Abstract
Down syndrome (DS) is caused by a triplication of chromosome 21 (HSA21). Increased oxidative stress, decreased neurogenesis and synaptic dysfunction from HSA21 gene overexpression are thought to cause mental retardation, dementia and seizure in this disorder. Recent epigenetic studies have raised the possibility that DNA methylation has significant effects on DS neurodevelopment. Here, we performed methylome profiling in normal and DS fetal cortices and observed a significant hypermethylation in ∼4% of probes in the DS samples compared with age-matched normals. The probes with differential methylation were distributed across all chromosomes, with no enrichment on HSA21. Functional annotation and pathway analyses showed that genes in the ubiquitination pathway were significantly altered, including: BRCA1, TSPYL5 and PEX10. HSA21 located DNMT3L was overexpressed in DS neuroprogenitors, and this overexpression increased the promoter methylation of TSPYL5 potentially through DNMT3B, and decreased its mRNA expression. DNMT3L overexpression also increased mRNA levels for TP53 and APP, effectors of TSPYL5. Furthermore, DNMT3L overexpression increased APP and PSD95 expression in differentiating neurons, whereas DNMT3LshRNA could partially rescue the APP and PSD95 up-regulation in DS cells. These results provide some of the first mechanistic insights into causes for epigenetic changes in DS, leading to modification of genes relevant for the DS neural endophenotype.
Introduction
Down syndrome (DS) is a genetic disorder caused by an extra copy of chromosome 21 (HSA21). The most profound neurological features of DS include mental retardation, seizures and early onset Alzheimer disease (AD) and are thought to arise predominantly for aberrant gene expression from chromosomal triplication. We have previously used a reverse genetic approach by generating DS human neuroprogenitors (HNPs) (1), and performing high-throughput microarray screening and gene network analyses to identify candidate HSA21 genes that contribute to the DS neurological phenotype during neurogenesis. These initial studies have led to the identification of three HSA21 genes S100B, APP and OLIG2 that share an interactive role in promoting a gliocentric shift in the progenitor pool, leading to oxidative stress, mitochondria impairment, cell death and inhibition of neural proliferation (2–5). These studies suggest the neurological phenotypes in DS may be attributed not only to the genomic imbalance from triplication of HSA21 genes, but also to additive influences on associated genes within a given network.
DNA methylation is a biochemical process that involves the addition of a methyl group from a methyl donor S-adenosylmethionine to the 5′ carbon of the cytosine pyrimidine ring that typically occurs in a CpG (cytosine phosphate guanine) dinucleotide site (6). This reaction is carried out by DNA methyltransferases (DNMTs), including DNMT1, DNMT3A, DNMT3B and DNMT3L. Alternatively, this reaction can be reversed by DNA demethylation enzymes, such as AID, APOBEC and TETs (7). DNA methylation itself may physically impede the binding of transcriptional proteins to the gene, and methylated DNA may be bound by proteins known as methyl-CpG-binding domain (MBD) proteins. MBD proteins then recruit additional proteins to the locus, such as histone deacetylases and other chromatin remodeling proteins that can modify histones. DNA methylation can occur at any CpG site in the genome, but methylation in promoter regions usually cause reduced gene expression (8). Substantial alterations in DNA methylation occur during stem-cell differentiation (9–11), suggesting that these epigenetic effects might also be important not only during cortical development, but also in DS HNPs pathological processes.
Several reports have shown global DNA methylation changes in DS. DS methylation profiling in adult leukocytes and T lymphocytes identified a subset of genes involved in lymphocyte development (12). Studies in DS adult buccal epithelial cells led to the selection of differentially methylated genes, correlated to cognitive impairment (13). Global DNA hypermethylation was also observed in DS placenta villi samples (14). Finally, the quantification of DNA methylation status in varied DS somatic tissues (the brain, kidney, lung, muscle and skin) suggest that DNA methylation is a dynamic, temporally and spatially changing process that is tissue and age-specific (15–17). The extent to and manner by which DNA methylation contributes to the DS neurological phenotypes in HNPs remain unclear.
In the current studies, we performed methylation profiling on age-matched DS and control HNPs. We observed a global hypermethylation in DS samples with particular involvement of genes in ubiquitination pathway. We also observed the up-regulation of the HSA21 localized DNMT3L and showed that overexpression of this methylation gene can enhance methylation and impair mRNA expression of the ubiquitin associated TSPYL5, leading to changes in APP and PSD95, presumably via TP53, and thereby altering neural proliferation, survival and synaptic connectivity.
Results
Global hypermethylation in T21 fetal cortical DNA samples
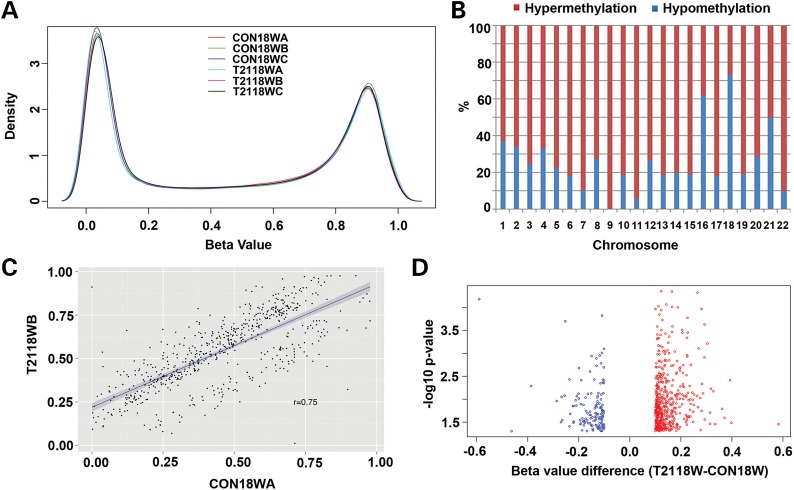
Several reports have shown global DNA methylation changes in DS lymphocytes, buccal epithelial cells and placenta villi cells, but not in neural cells. DNA methylation could potentially affect DS neurodevelopment by modulating gene expression in neurons (18–21). However, the specific methylation changes, cause of methylation changes and their contribution to the DS neurological phenotypes are unknown. We, therefore, compared the methylation profiles of control (CON) and DS (trisomy 21, T21) frontal cortex from 18 gestational weeks (18W GA) fetal brain using Infinium HumanMethylation450 BeadChip. The platform interrogates methylation state of 482 421 CpG sites and covers promoters of about 20 602 genes. We first excluded the confounding probes (see details in the ‘Materials and Methods’ section), and established the β-value distribution (a measurement of the DNA methylation level between 0 and 1, where 0 reflects no methylation and 1 corresponds to 100% methylation). Within the refined 410 876 probes, we observed a bimodal distribution with methylation levels either exhibiting high-methylation (≥75%) or low-methylation (≤25%) status (Fig. 1A). Next, we compared the methylation difference of these 410 876 probes between CON18W (three cases) and T2118W (three cases) by Student's t-test with cut-off P-value of 0.05, which resulted in 17 166 probes (4%) showing significant differences between CON and T21. We then calculated the absolute difference between the β-value means for T2118W and CON18W. Of the 17 166 significant probes, 744 had a β-value difference of >10%, referred to as Group I. Using the β-values in Group I, we first looked at the distribution of methylation changes in 22 chromosomes, showing more hypermethylation (β-value difference: T2118W > CON18W) than hypomethylation (β-value difference: T2118W < CON18W) in most chromosomes except chromosome 16, 18 and 21 (Fig. 1B). The correlation analysis of β-values between CON18W and T2118W showed a significant difference of these probes in Group I (Fig. 1C), with a correlation coefficient of 0.75, much lower than that of 0.94 or 0.95 for the correlations between different CON18W samples (Supplementary Material, Fig. S1A) or different T2118W samples (Supplementary Material, Fig. S1B). This difference was further enlightened by a volcano plot of the probes in Group I, plotting the log of P-value with β-value difference, which also showed more hypermethylation (red circles) than hypomethylation (blue circles) (Fig. 1D), though most differences were small between 10 and 20%. Next, by comparing the methylation change from the original methylation state of CON samples (methylated with β-value ≥ 0.75 or unmethylated with β- value ≤ 0.25) in Group I probes, we found there were more methylated probes that were hypomethylated than hypermethylated (84.4 versus 15.6%), whereas more unmethylated probes were hypermethylated than hypomethylated (97.44 versus 2.56%) (Supplementary Material, Fig. S1C). Though limited, these comparisons suggest a possible global hypermethylation change in DNA methylation in DS fetal cortex, with large differences noted only in a small, specific subgroup of the promoter sites.
Figure 1.
Global hypermethylation in T21 fetal cortical DNA samples. (A) A density plot demonstrates the distribution of the DNA methylation level (β-value) for 410 876 probes in three CON (CON18WA, CON18WB and CON18WC) and three T21 (T2118WA, T2118WB and T2118WC) cases, which shows bimodally distributed methylation levels enriching highly methylated (≥0.75) and unmethylated (≤0.25) probes. (B) A percentage chart demonstrates the distribution of hypermethylation and hypomethylation in differential methylated probes of T2118W and CON18W comparison on 22 auto chromosomes (P-value ≤ 0.05, β-value difference ≥ 0.1), which shows more hypermethylation than hypomethylation in all chromosomes except chromosome 16, 18 and 21. (C) A correlation plot of β-value between CON18W and T2118W cases, with a correlation coefficient of 0.75, showing the significant difference of the two groups (P-value ≤ 0.05, β-value difference ≥ 0.1). (D) A volcano plot demonstrates the distribution of differential methylated probes of T2118W and CON18W with P-value levels, showing more hypermethylation (red circles) than hypomethylation (blue circles).
Disturbed DNA methylation in ubiquitin proteolysis pathway indicated by DAVID analysis
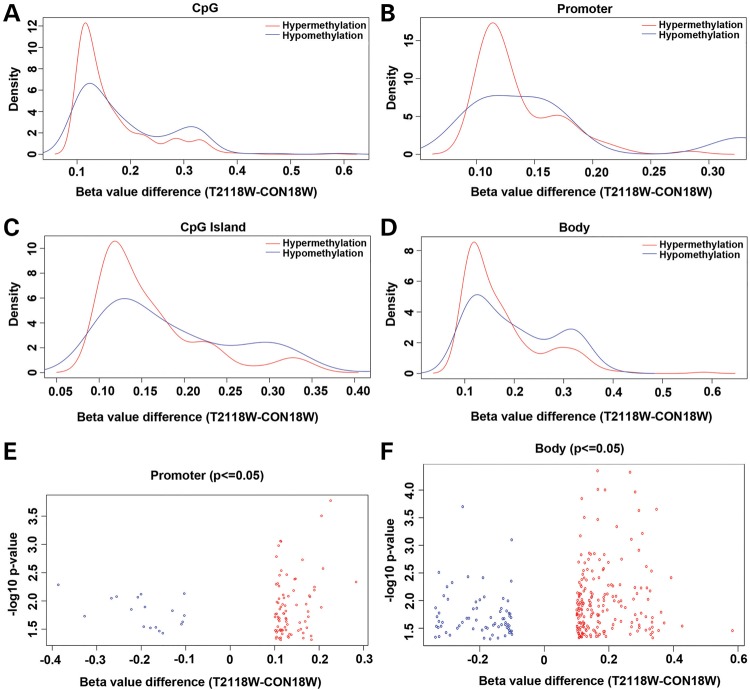
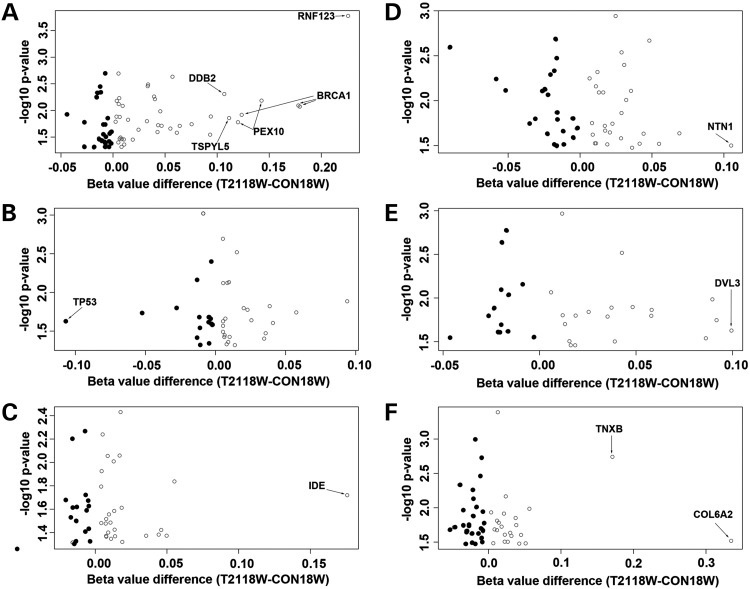
The illumina probes cover the whole genome, corresponding to 99% of RefSeq genes. With 17 CpGs per gene region on average, the probes distribute across the promoter region, 5′untranslated region (UTR), first exon, gene body and 3′UTR, and cover 96% of the CpG islands. To further detect how and where our differential Group I probes are located on the genome, we made density plots of the β-value differences in different genomic regions. The results showed similar patterns for all genomic regions including the CpG sites, promoter, CpG islands and gene body (Fig. 2). Most differential probes had β-value difference of between 10 and 20%, with greater degrees of hypermethylation than hypomethylation. These differential probes were located more on the non-promoter-associated gene bodies (0.199%, 263/132387) than they were on the promoters (0.111%, 100/89769) or islands (0.195%, 261/133765). The differential distribution difference between that on the gene body and that on the promoter was further demonstrated by the volcano plots (Fig. 2E and F). Under less stringent conditions (17166 probes showing significant difference between CON and T21 with P ≤ 0.05), the differences among regions decreased: 4.41% (5839/132387) on the non-promoter-associated gene bodies, 3.61% (3242/89769) on the promoters and 3.77% (5050/133765) on the islands. Although methylation in gene body showed more changes than that in the promoter region, DNA methylation in promoter regions is more strongly correlated with the absence of gene expression than that in gene-body regions (22). Therefore, we used 3242 probes with P ≤ 0.05 and associated with promoter regions (referred to as Group II) to perform the functional annotation analysis with DAVID. We identified several genes involved in the ubiquitin proteolysis as the most significant kyoto encyclopedia of genes and genomes (KEGG) pathway (Supplementary Material, Table S3 and Fig. S2), with a P-value of 5.69E-06, covering 40 genes. In the alternative functional annotation clusters assay, this group of genes had an enrichment score of 2.06, which was higher than the significant level of 1.3, though the top one cluster pulled out were genes involved in organelle lumen. Since we were more interested in the signal pathways that were disturbed with DNA methylation changes, we looked into the genes involved in the top 2 KEGG pathways (40 genes in ubiquitin mediated proteolysis and 36 genes in cell cycle) and 35 genes in Alzheimer's disease (AD) pathway. Nine extra ubiquitination genes involved in methylation change that were not in the KEGG were also included for the analysis. The data on these significant gene probes were listed in Supplementary Material, Table S4. Using a volcano plot, we can see the distribution of β-value difference for these probes for ubiquitin mediated proteolysis (Fig. 3A), cell cycle (Fig. 3B) and AD (Fig. 3C) pathways. The β-values differences of most probes were within 0.1, with greater hypermethylations (open circles) than hypomethylations. The gene probes with β-value difference of >0.1 were labeled, which showed more affected genes in ubiquitin mediated proteolysis pathways than the other two. Specifically, three probes on BRCA1, two probes on PEX10 and one probe on TSPYL5, RNF123 and DDB2, respectively. To increase stringency, we established a data set with 100 probes that have a P-value of ≤0.05, a β-value difference (between CON18W and T2118W) of >10%, and were located in the promoter region (referred to as Group III). The same analysis as in Group II was done in Groups III and I, which gave similar albeit less significant results due to smaller data size (Supplementary Material, Tables S3 and S4). To look into the methylation changes in gene-body regions, as they were more robust than those in promoter regions (Fig. 2E and F), we used 4095 probes with P≤0.05 and localized within non-promoter-associated gene-body regions (referred to as Group IV) to perform the functional annotation analysis with DAVID. We demonstrated the distribution of β-value difference for the probes from top three KEGG pathways (Supplementary Material, Table S3) with volcano plots, including axon guidance (Fig. 3D), Notch signaling (Fig. 3E) and extracellular matrix (ECM)-receptor interaction pathways (Fig. 3F) (Supplementary Material, Table S4). Most values were within 0.1, and very few showed >10% difference (labeled with arrows). Then we did the same KEGG pathway search with increased stringency using 263 probes that have a P-value of ≤0.05, a β-value difference (between CON18W and T2118W) of >10%, and were localized within non-promoter-associated gene-body regions (referred to as Group V). With the only two pathways pulled out (TGF-β-signaling pathway and cytokine–cytokine receptor interaction pathway), the significance was limited by the small-data size (Supplementary Material, Table S3). Next, we analyzed the data from Groups IV and V with the alternative functional annotation clusters assay, where the top candidates included the protocadherin alpha gene clusters and morphogenesis molecules involved in neural adhesion (Supplementary Material, Fig. S3F) and differentiation (Supplementary Material, Fig. S3G) for Group IV, and genes associated with ECM (Supplementary Material, Fig. S3H), ion channel (Supplementary Material, Fig. S3I) and voltage gated channel activity (Supplementary Material, Fig. S3J) for Group V (Supplementary Material, Tables S3 and S5). These volcano plots were compared with those of the analyses with the data from Groups II and III (Supplementary Material, Tables S3 and S5), which were promoter-associated, including genes associated with organelle lumen (Supplementary Material, Fig. S3A and C), cell cycle (Supplementary Material, Fig. S3B), peroxisome (Supplementary Material, Fig. S3D) and DNA damage (Supplementary Material, Fig. S3E). In summary, the functional annotation clusters assay of probes localized on the non-promoter-associated gene body suggests some of the genes on cell adhesion and differentiation are significantly methylated at the gene-body region, including CELSR3, GDF7, ADAMTS10, ADAMTS16, SMOC2 and TNXB, as well as some ion channel genes including KCNH2 and SCNN1A. The significantly methylated or demethylated genes at the promoter region by functional annotation cluster assay include MDC1, DDB2, BRCA1, TGOLN2, PEX5 and PEX10.
Figure 2.
Distribution of differential DNA methylation in different regions of genome. Density plots demonstrate similar pattern on the distribution of differential DNA methylation (β-value difference) between T2118W and CON18W cases (P-value ≤ 0.05, β-value difference ≥ 0.1) in CpG sites (A), promoter (B), CpG island (C) and body (D). Most methylation changes occur between 10 and 20% for all groups. The distribution of differential DNA methylation was also shown with two volcano plots for promoter (E) and body (F).
Figure 3.
The distribution of differential DNA methylation in different KEGG pathways Volcano plots demonstrate the distribution of differential methylated probes between T2118W and CON18W (P-value ≤ 0.05, promoter-associated) with P-value levels in ubiquitin proteolysis pathway (A), cell cycle pathway (B) and AD pathway (C). The same analyses with the data set that have a P-value of ≤0.05, and are gene body-associated show the distribution of differential methylated probes between T2118W and CON18W in axon guidance pathway (D), Notch signaling pathway (E) and ECM-receptor interaction pathway (F). Probes with >10% change are labeled with gene names. Most changes are <10%, and there are more hypermethylation (open circles) than hypomethylation (filled circles).
Disturbed DNA methylation in ubiquitination genes indicated by cluster analysis
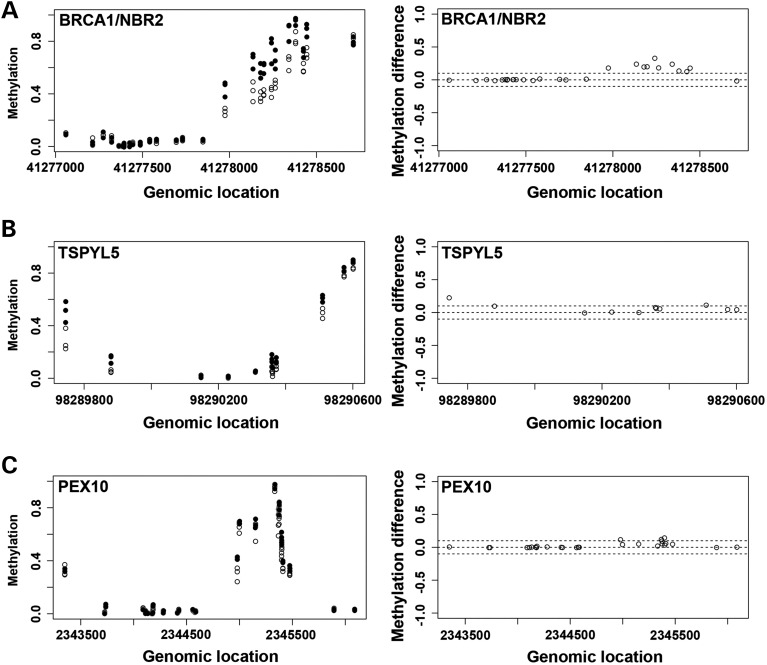
There are multiple CpG sites in a gene, including the promoter regions that control gene expression. Single-CpG methylation changes, however, may not have a significant effect on the gene regulation. To look for differential methylation regions containing multiple differential methylated probes, we ran a Bumphunter package in R (23) to identify differentially methylated regions or significantly different clusters. Combining with P ≤ 0.05, and β-value difference ≥ 0.1, we found 37 clusters in 37 genes (Supplementary Material, Tables S6 and S7), which include the ubiquitination genes we found significant in the previous annotation assays. Specifically they were BRCA1, TSPYL5 and PEX10. The others were CYP2E1, CPT1B, LOC100130522, ZSCAN12P1, PON1, STK19, DOM3Z, POU6F2, PIWIL1, IZUMO2, CYTH2, C2orf61, SDHAP3, FAM83H, LOC387646, CD151, RIN1, LMF1, SPRED3, TNFRSF6B, VIPR2, LRRC24, HN1L, LMTK3, ZNF837, ADAMTS10, ZNF264, GYPC, CELSR3, DOK7, SMOC2, B3GALT4, GLI4 and PLEC1. The methylation differences of the probes on three genes (BRCA1, TSPYL5 and PEX10) were illustrated along the genomic locations of each gene (Fig. 4), showing both methylation levels and methylation changes between CON (open circle) and T21 (filled circle) in clusters. These clusters were located around promoter region of these genes, and all showed hypermethylation change. Interestingly, CELSR3, ADAMTS10 and SMOC2, also showed hypermethylation changes, and were among the other genes that were pulled out in previous annotation assays.
Figure 4.
Probes clusters showing hypermethylation in three genes on ubiquitin proteolysis pathway. Plots demonstrate the distribution of DNA methylation levels and methylation difference (β-value difference) between T2118W (filled circle) and CON18W (open circle) along the genomic locations for the probes clusters on BRCA1/NBR2 (A), TSPYL5 (B) and PEX10 (C) in ubiquitin proteolysis pathway. These cluster changes are pulled out by bumphunter. The dash line indicates the levels of methylation difference at 0.1 and −0.1.
Disturbed DNA methylation in selected ubiquitination genes confirmed by bisulfite sequencing
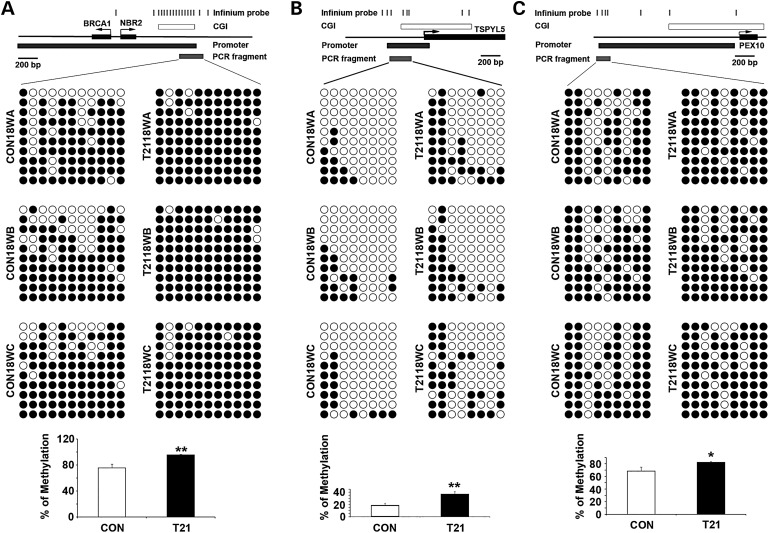
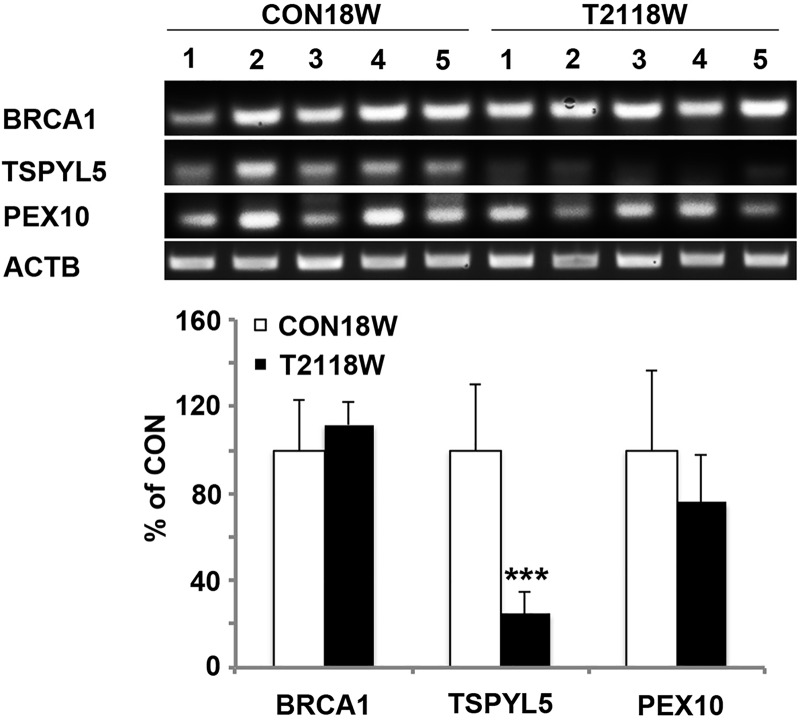
To confirm the DNA methylation changes in the three candidate genes (BRCA1, TSPYL5 and PEX10) that were involved in ubiquitination, we performed bisulfite sequencing using primers around the promoter region of each gene overlapped with the infinium probe clusters that showed significant differences. The percentage of methylation for each gene was calculated with all the CpG sites tested, which showed significant increase of methylation in all three genes, especially for TSPYL5 with 50% difference (Fig. 5). To test if the increased methylation correlated with decreased expression in these three genes, we did semi-quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) using the same case samples. As expected, TSPYL5 RNA showed significant reduction in T21 samples compared with CON samples; PEX10 RNA showed the trend of reduction and BRCA1 RNA did not show any change (Fig. 6). These differential results may reflect the different extent of methylation change in these three genes, with greater DNA methylation modification resulting in stronger gene expression regulation.
Figure 5.
Bisulfite sequencing confirming hypermethylation in three genes on ubiquitin proteolysis pathway. Bisulfite sequencing results of BRCA1/NBR2 (A), TSPYL5 (B) and PEX10 (C) demonstrate increased DNA methylation in T2118W compared with CON18W cases. The aligned circles stand for CpG sites tested (white: unmethylated and black: methylated). The above maps show the locations of significant Infinium probes, CpG island (CGI), promoter and PCR fragment along the genome for each gene. The graph below show the statistic difference of the methylation changes (*P ≤ 0.05 and **P ≤ 0.01).
Figure 6.
mRNA changes in three genes on ubiquitin proteolysis pathway. Semi-quantitative RT-PCR demonstrates the mRNA levels of BRCA1 (A), TSPYL5 (B) and PEX10 (C) in five CON18W and five T2118W cases, with significant down-regulation in TSPYL5. The quantification is shown below (***P ≤ 0.001).
DNMT3L overexpression causes TSPYL5 hypermethylation and reduced mRNA
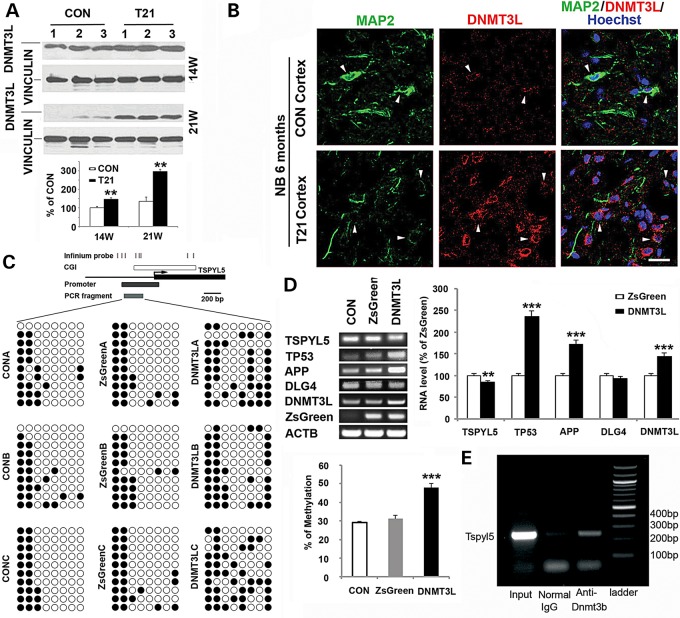
Mechanisms underlying the methylation changes in T21 are unknown in DS. As DNMT3L is on HAS21, we first asked whether the expression of DNMT3L was up-regulated at the protein level in DS. We found that the methylation-associated protein was increased in several ages of frontal cortex by western blot and immunostaining (Fig. 7A and B). Next, we overexpressed DNMT3L in normal HNPs by infecting with lentivirus carrying the DNMT3L gene at ∼1.5-fold increase through viral titration, with ZsGreen serving as an internal control, and tested the most significantly changed ubiquitination gene TSPYL5 from the previous study for DNA methylation by bisulfite sequencing and mRNA changes by semi-quantitative RT-PCR. DNMT3L overexpression increased the TSPYL5 DNA methylation ∼1.5-fold compared with controls, and the mRNA level of TSPYL5 was down-regulated ∼15% (Fig. 7C). TSPYL5 have been shown to suppress TP53 expression by promoting its ubiquitination through reducing the activity of USP7, a deubiquitylase for TP53 (24). Therefore, we tested the mRNA levels of TP53 after DNMT3L overexpression, which was significantly increased >2-fold (Fig. 7D), consistent with the TSPYL5 down-regulation.
Figure 7.
DNMT3L overexpression causes hypermethylation of TSPYL5 gene, and down-regulation of its mRNA expression DNMT3L is significantly up-regulated in 14W and 21W of T21 frontal cortex shown by western blot (A) and in 6-month-old newborn T21 patient shown by immunofluorescence, with DNMT3L (Cy3) and MAP2 (Cy2) double staining (B). Bisulfite sequencing results of TSPYL5 demonstrate increased DNA methylation in DNMT3L overexpressed HNPs 3d after the lentiviral infection (C). The aligned circles stand for CpG sites tested (white: unmethylated and black: methylated). The above map shows the locations of significant Infinium probes, CpG island (CGI), promoter and PCR fragment along the genome for TSPYL5. The graph on the right shows the statistic difference of the methylation changes (***P ≤ 0.001). Semi-quantitative RT-PCR demonstrates the mRNA changes in TSPYL5, TP53, APP and DLG4 after DNMT3L overexpression in CON HNPs 3d after the lentiviral infection (D). The quantification graph is below (***P ≤ 0.001). (E) Chromatin immunoprecipitation assay demonstrates that DNMT3B interacts with the promoter site of TSPYL5. Antibodies to Dnmt3b are able to pull-down the Tspyl5 promoter site, suggesting that DNMT3L could modify Tspyl5 methylation through its effects on Dnmt3b.
DNMT3L is not thought to function as a DNMT as it does not contain the amino acid residues necessary for methyltransferase activity. Rather, DMNT3L stimulates DNMT3A/3B activity. Given that DNMT3B is expressed during brain development and the initial screening was performed on neural progenitors, we addressed whether DNMT3B could bind the promoter site of TSPYL5 (Fig. 7E). The ChIP assay shows that antibodies to Dnmt3b can effectively immunoprecipitate the promoter region of Tspyl5 in NIH 3T3 cells, providing a mechanism whereby DNMT3L overexpression from trisomy HSA21 could alter DNMT3B function and TSPYL5 methylation.
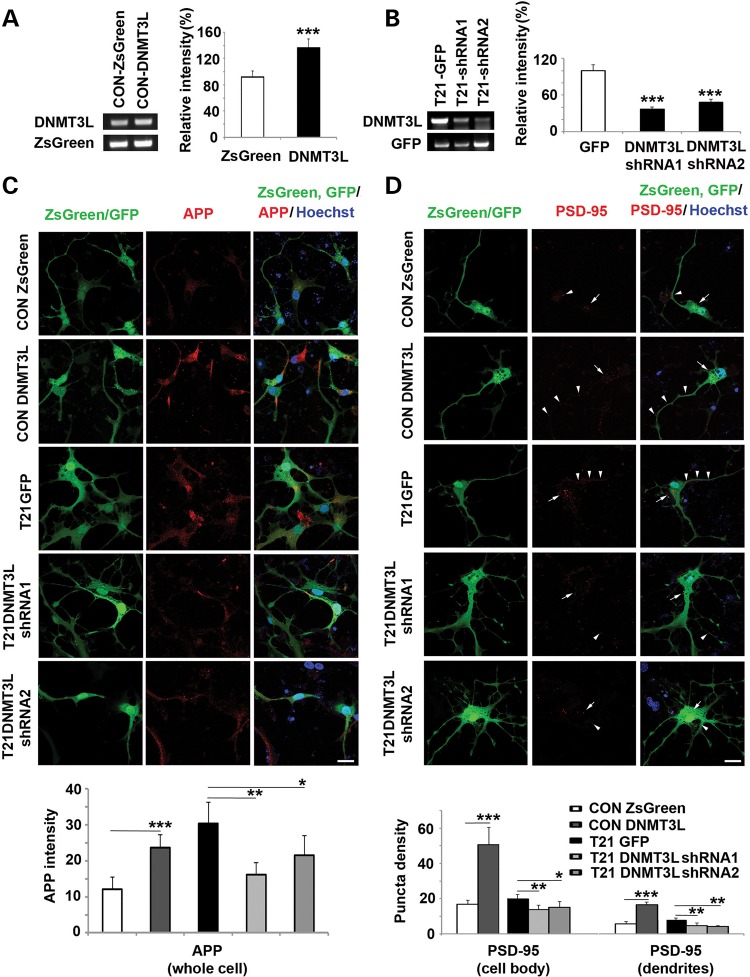
DNMT3L overexpression increases APP/PSD95 expression in differentiated neurons
The cognitive impairment seen in DS has been linked to synaptic dysfunction through PSD95 and APP-dependent AD like dementia. HSA21 localized APP has previously been shown to drive apoptosis through TP53-dependent mechanisms, whereas TP53 has also been linked to increased PSD95 interactions (25,26). Given the current studies showing DNMT3L overexpression leads to TSPYL5 methylation/mRNA and TP53 mRNA changes, we asked if DNMT3L overexpression could also affect these genes, implicated in DS cortical development. APP showed more than a 1.5-fold increase after DNMT3L overexpression in neural progenitors. Moreover, DNMT3L overexpression in differentiated neurons through lentiviral infection of constructs carrying DNMT3L led to increased APP expression. Conversely, normalization of DNMT3L mRNA levels through shRNA lentiviral infection led to reduction in the up-regulated APP levels in T21 differentiated neurons. The mRNA levels were confirmed by RT-PCR after lentiviral infection (Fig. 8A–C). While we did not observe a significant change in DLG4 mRNA (DLG4 encodes for PSD95) in neural progenitors, we did find an analogous increase in PSD95 expression in both cell bodies and dendrites of neurons overexpressing DNMT3L and reduction of PSD95 expression in T21 neurons following DNMT3LshRNA lentiviral infection (Fig. 8D). Given the previously reported TP53 association with APP and PSD95, these studies would suggest that epigenetic modification through DNMT3L could direct DS endophenotypes through this ubiquitin-related pathway.
Figure 8.
DNMT3L overexpression increases APP and PSD95 expression in young differentiated neurons. The overexpression of DNMT3L in CON human neurons is confirmed by RT-PCR after the HNPs are infected with lentivirus carrying DNMT3L for 3 days and differentiated into neurons for 7 days (A); and the normalized DNMT3L in T21 human neurons is confirmed by RT-PCR after the T21 HNPs are infected with lentivirus carrying DNMT3LshRNA and differentiated into neurons for 7 days (B). With the same culture and infection protocol as those in (A and B), DNMT3L overexpression can increase APP expression in CON neurons shown by immunofluorescence staining of (Cy3), whereas the overexpression of APP in T21 neurons is down-regulated by DNMT3LshRNA (C). DNMT3L overexpression can increase PSD95 puncta density in cell body (white arrow, Cy3) and dendrites (white arrow head, Cy3), whereas the increased PSD95 puncta density in T21 neurons is down-regulated by DNMT3LshRNA (D). The quantification graphs are below (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001). Bar = 25 μm.
Discussion
In the current studies, we performed DNA methylation profiling on human DS neural progenitors and found global epigenetic changes due to trisomy 21 with global hypermethylation. Confirmation through bisulfate sequencing, followed by cross correlation with mRNA microarray profiling and pathway analyses yielded identification of altered signaling in several genes involved in ubiquitination, including TSPYL5. TSPYL5 has been implicated in the TP53 pathway. To understand potential mechanisms that would drive the epigenetic changes seen in DS, we focused on DNMT3L, a methylation-associated gene found on HSA21 that we found to be overexpressed in DS neural progenitors. DNMT3L activates DNMT3A/B, two proteins which alter neurogenesis and synaptic plasticity through methylation, and DNMT3B can bind the TSPYL5 by ChIP assay. Overexpression of DNMT3L led to hypermethylation of TSPYL5, down-regulation of TSPYL5 mRNA and consequent increase in TP53. TP53 has been implicated in APP associated cell death/proliferation and changes in PSD95—two proteins thought to be involved in the development of DS associated-dementia and mental retardation. DNMT3L overexpression also led to increased levels of APP and PSD95 expression. Conversely, inhibition of DNMT3L in DS progenitors rescued these effects. Collectively, these studies provide some of the first mechanistic insight into how epigenetic changes arise in DS and are associated with DS endophenotypes.
Prior methylation studies in DS tissue samples
Several studies have compared the DNA methylation profiles between normal and DS patients, and our data shared some common characteristics with these findings, which included global hypermethylation, no enrichment on chromosome 21 and tissue and developmental-stage-specific nature of DNA methylation. We compared our data with two other reports that also used the Infinium chip to detect DNA methylation changes between normal and DS patients. Kerkel et al. (12) profiled the methylation in adult human leukocytes and T lymphocytes, whereas Jones et al. (13) performed analogous studies in adult human buccal epithelial cells. Kerkel et al. identified 118 differentially methylated CpG sites whereas Johns et al. found differences between DS and control in 3300 CpGs, including 495 CpGs, which overlapped with clusters of differentially methylated probes. We identified 88 differentially methylated CpG sites, which were in common with the methylation changes found in the buccal epithelial cells, but only four sites overlapped with the Kerkel findings (Supplementary Material, Table S8). These differences likely reflect the differences in cell origin and age between the three studies. Among the 88 probes shared between the adult buccal epithelium and fetal cortex, several genes with multiple probes (≥3) showed significant difference (P ≤ 0.05, β-value difference ≥ 0.1) with same direction (hypermethylation or hypomethylation). These genes are CELSR3, ZSCAN12L1, STK19, GLI4, FAM83H, LRRC24, BRCA1/NBR2, LOC100130522 and CPT1B. These genes have diverse functions and no significant functional annotation clusters or KEGG pathways were pulled out from these genes. However, the methylation of CPT1B, the rate-controlling enzyme of the long-chain fatty acid β-oxidation pathway in muscle mitochondria, seems to be affected in all the studies.
Methylation changes in the promoter region and gene-body region
The data analyses in the current study indicate that the most significant methylation changes involve hypermethylation, but not hypomethylation. Among the most significant 37 genes from the cluster searching, only 5 genes have probes showing hypomethylation (C2orf61, LMF1, HN1L and DOK7 in the gene body; and LOC100130522 in the promoter). On the other hand, hypermethylation occurs more often in the gene-body region than the promoter region (17 genes versus 13 genes, Supplementary Material, Table S7), and they are mutually exclusive. Promoter methylation usually correlates with decreased gene expression, whereas gene body methylation may correlate with increased gene expression in dividing cells, but not non-dividing cells (27). A study in Dnmt3a knockout mice suggests that Dnmt3a can promote non-promoter DNA methylation of neurogenesis genes such as Dlx2, Gbx2 and Sp8 in neural progenitor cells by functionally antagonizing Polycomb repression, resulting in increased expression of these genes (21). In this regard, among the four genes (ADAMTS10, PLEC1, CPT1B and B3GALT4) that show mRNA changes in our previous microarray data, only B3GALT4 and ADAMTS10 show the methylation-gene expression correlation in promoter and gene body, respectively (Supplementary Material, Table S7). However, the functional analyses suggest some of the genes involved in ubiquitination pathway (BRCA1, TSPYL5 and PEX10) or neural adhesion and differentiation (CELSR3, ADAMTS10 and SMOC2) are worth further investigation.
Ubiquitination pathway genes in DS neurodevelopment
The ubiquitin proteasome/lysome system (UPLS) is responsible for the transport, processing and degradation of proteins from multiple cellular compartments (especially mitochondria and synapses) in order to maintain normal cellular function (28,29). Progression in DS cognitive impairment has been associated with accumulation of NF plaques and tangles containing ubiquitin (30). Dystrophic neurites in DS also contain ubiquitin and the UPLS-associated molecules PSMA5 and USP5 are up-regulated in the DS fetal brain (31). Beta amyloid could regulate synaptic protein degradation and function through the ubiquitin pathway (32,33). Moreover, several E3 ubiquitin ligases have been shown to promote APP degradation (34,35). Additionally, HSA21 located genes AIRE and UBE2G2 are directly involved in the ubiquitin pathway and could contribute to the phenotype. Thus, DNA methylation may directly impair synaptic function and structure (through beta amyloid or synaptic proteins) by impairing UPLS, but would also possibly enhance oxidative stress and mitochondrial/peroxisomal dysfunction.
In the current study, DNA methylation profiling between normal and DS fetal cortex revealed several genes from the ubiquitination pathway, which suggests that this pathway may play a significant role in the development of DS neuropathology. BRCA1 is an E3 ubiquitin-protein ligase that specifically mediates the formation of ‘Lys-6’-linked poly-ubiquitin chains and helps DNA repair by facilitating cellular responses to DNA damage (36,37). However, the methylation level of BRCA1 in normal cells already approaches 80%, so the increased methylation in DS did not show any change in its mRNA expression, and the biological function of this methylation change is unknown. PEX10 encodes an ubiquitin-protein ligase anchored in the peroxisomal membrane, involved in the import of peroxisomal matrix proteins (38). Its down-regulation may damage the function of peroxisome in reducing reactive oxygen species, specifically hydrogen peroxide, causing more apoptosis and gliosis. TSPYL5 can suppress TP53 expression by promoting its ubiquitination through reducing the activity of USP7, a deubiquitylase for TP53 (24). Thus down-regulation of TSPYL5 by its increasing methylation increase TP53, which may account for the increasing neuronal death during early cortical development.
Methylation changes in other trisomic states
Several observations suggest that changes in DNA methylation from T21 derive from both alterations in expression of genes involved in methylation (i.e. DNMT3L) as well as increased oxidative stress. Our current studies clearly show that overexpression of DNMT3L can lead to changes in methylation within the promoter region of genes such as TSPYL5. Overexpression of this methylation-associated protein also leads to increased levels of APP, which have been associated with increased oxidative stress, and linked to changes in methylation pattern. While limited in sample number, we did compare DNA methylation patterns among individual samples of T13, T18 and T21 at 14W and 18W. Lacking statistical power, we chose a β-value of >0.1 as a cut-off for methylation difference with T2118W (β-value difference ≥ 0.1) as reference. Using this standard, we found the global hypermethylation in T21 was not observed in T13 or T18 samples, which showed essentially a 50–50 ratio (Supplementary Material, Fig. S4A–D). However, the pattern found in T21 whereby methylated genes became hypomethylated, and unmethylated genes became hypermethylated was also found in T13 and T18 samples, though the ratios were lower than that in T21 (Supplementary Material, Fig. S4E–H). Another common pattern shared by all trisomies was that most methylation changes were between 10 and 20% (Supplementary Material, Fig. S5). In addition, the correlation plots suggested that age causes more methylation changes than extra chromosomes in T13, T18 and T21 between GA14W and GA18W (Supplementary Material, Fig. S6). Our correlation analysis among these samples showed that DNA methylation in normal 14W fetal tissues correlated more strongly with the DNA methylation in the trisomic 14W fetal tissues than the comparable 18W tissues. This is consistent with other studies that compared the DNA methylation of T21 and T18 fetal tissue, and revealed DNA methylation changes caused more by development than by trisomies (17). In finding more hypermethylated probes being hypomethylated and more hypomethylated probes being hypermethylated among all trisomies, it is suggested that this methylation fluctuation may be caused by the oxidative stress or proteotoxic stress from an extra copy of chromosomes (39). Nevertheless, more hypermethylation was seen in T21 than that in T13 and T18, indicating a possible effect from specific genes on HSA21, such as DNMT3L. Examination of additional cases would provide greater insight into the contributions from specific HSA21 genes and/or oxidative changes.
Methylation change caused by DNMT3L and its consequential effects
HSA21 localized DNMT3L can physically binds and stimulates the methylation activity of DNMT3A/3B (40–42), which has been implicated in regulating gliogenesis, neuronogenesis and synaptic plasticity by regulating-related genes expression, such as Bdnf, Reln, Dlx2, Gbx2, Sp8 and Stat1 (18–21). Therefore, its overexpression in DS may cause abnormal neurodevelopment we observed in previous studies. Indeed, as DNMT3L overexpression increases TSPYL5 gene methylation in the promoter region, it also down-regulates its mRNA expression, and up-regulates mRNA expressions of TP53 and APP. The current studies also show that DNMT3B, a direct effector of DNMT3L, can bind the TSPYL5 promoter site, thereby providing a means to regulate TSPYL5 methylation and mRNA expression. Since TSPYL5 suppresses TP53 expression by promoting its ubiquitination through reducing the activity of USP7, a deubiquitylase for TP53 (24), down-regulation of TSPYL5 by DNMT3L overexpression will increase TP53 expression, thus increase apoptosis, the most significant endophenotype during DS cortical development (4). Though the mechanism of increasing APP by DNMT3L overexpression is not known, either modified directly by DNMT3L or indirectly through ubiquitination/deubiquitination genes, it will for certain enhance the toxic effects of this gene, inducing more apoptosis and gliosis. The increased APP staining during neuronal differentiation after DNMT3L overexpression supports the mRNA changes.
Histologically, DS cortex has spine dysgenesis (sparse, small, short stalks intermingled with unusually long spines (43) in early development), suggesting that both increased and decreased expression of PSD95 could cause cognitive impairment. Prior studies using Ts65Dn mice show a decrease in PSD95 (44) and these mice exhibit cognitive impairment. However, we actually see increased PSD95 expression in both the cell bodies and synapses of DS neurons. This discrepancy might extend from the fact that the Ts65Dn mouse model contains extra copies of chromosome 16 and 17, but does not have the extra copy of Dnmt3 l, which is found on mouse chromosome 10. Additionally, increased PSD95 and spine density, but LTP inhibition are seen in the DS-associated Dyrk1a transgenic mice, which also exhibit cognitive deficits (45). Finally, overexpression of Dnmt3a, an effector of Dnmt3l, can increase spine density, consistent with our observed increase in PSD95 (19).
It is likely that other factors mediate epigenetic changes in DS. The current studies suggest that DNMT3L clearly provides one mechanism for regulation of aberrant gene expression in DS. To understand the extent of contribution of other factors such as oxidative stress in regulation of DNA methylation, further studies will be required.
Materials and Methods
Human tissue, ethical and licensing considerations
The Institutional Review Board (IRB) at the Beth Israel Deaconess Medical Center and Brigham and Women's Hospital has approved this study. The identified human discarded tissues were obtained from pathological samples obtained during planned, induced abortions with post-mortem intervals under 4 h. The tissue from T13, T18, T21 and age-matched control brains (GA 14W, 18W and 21W) as well as newborn tissue from the NICHD Brain and Tissue Bank for Developmental Disorders were used in this study for each of the experiments described. Studies were performed in accordance to guidelines from the IRB review committee at BIDMC (protocol number 2004-P-000299/5). Detailed information of samples is listed in Supplementary Material, Table S1.
Genomic DNA extraction and methylation array
The genomic DNA of each sample was extracted from the thawed frozen cells, which were dissociated from the frontal cortex of the discarded brain tissue as described previously (1). The DNA was isolated and cleaned following the manufacturer's protocol using QIAamp DNA Mini kit (Qiagen 51304). Then the DNA samples were processed for array analysis using Infinium HumanMethylation450 BeadChip Kit as per manufacturer's protocol (Illumina, CA, USA). Following bisulfite treatment of 500 ng genomic DNA using the EZ DNA Methylation kit (Zymo Research, CA, USA), the bisulfite-converted DNA samples were chemically denatured and neutralized using 0.1 N NaOH and amplified at 37°C for 20–24 h. The amplified products were then enzymatically fragmented at 37°C for 1 h. The fragmented DNA was precipitated using 100% 2-propanol at 4°C for 30 min and centrifuged (3000g) at 4°C for 20 min to collect a tight pellet. The supernatant was decanted by quick inversion and pellets were allowed to dry at room temperature (22°C) for 1 h. The pellets were resuspended in Illumina's custom hybridization buffer and incubated at 48°C for 1 h followed by 95°C for 20 min. The samples were loaded onto the HumanMethylation450 BeadChips and hybridized at 48°C for 16–24 h. The chips were washed in Illumina's wash solutions and placed into TecanTe-Flow chambers in preparation for staining. Unhybridized and non-specific hybridized DNA was washed off the BeadChips. Labeled nucleotides were added to extend the primers hybridized to the DNA. The BeadChips were stained with Cy3 and Cy5 fluorescent dye, followed by washing, drying and a coating to protect against ozone and other elements that may be detrimental to the dyes.
Infinium methylation data normalization and filter
The BeadChips were scanned using the IlluminaiScan Reader. The image data were then transferred to IlluminaGenomeStudio for data processing, validation of assay controls, controls normalization with background subtraction and report generation using the methylation module. The level of methylation for the interrogated locus (β-value) was determined by calculating the ratio of the fluorescent signals from the methylated versus unmethylated sites (method from https://support.illumina.com/content/dam/illumina-support/documents/myillumina/90666eaa-c66-48b4-8199-3be99b2b3ef9/genomestudio_methylation_v1.8_user_guide_11319130_b.pdf, p48-50, Illumina). The normalized data contained 485577 probes. Due to the sequence overlap between autosomal and sex-linked probes on the IlluminaInfinium Human Methylation450 BeadChip assay (46), first, 11 648 probes on the X or Y chromosome, together with 3091 probes for CpH sites and 65 probes for single-nucleotide polymorphisms (SNPs) were removed from further analysis. Secondly, SNP probes with polymorphisms at the target C or G can confound the measurement of DNA methylation, and non-specific probes targeting multiple genomic sites can also confound the results. Thus, 20 869 probes that are known to be polymorphic at the CpG sites, and 41 937 probes which have non-specific binding to the X, Y chromosome or autosome were removed (47). Finally, the probes with missing data for any sample were also removed. Together, we eliminated 74 701 probes, leaving a total of 410 876 probes for further analysis.
Differential methylation analysis by R
The filtered data were analyzed using the R software (version 3.1.1). The comparison of methylation difference on 410 876 probes between CON18W (three cases) and T2118W (three cases) was first examined by Student's t-test with a cut-off of P ≤ 0.05, which gave 17166 probes that were significantly different. These probes were further filtered to have a β-value difference (between CON18W and T2118W) of >10%, which gave a subsection of 744 probes. Then the distribution of differential methylation in these 744 probes was displayed using density plot, correlation plot, volcano plot and column chart. In addition to t-test, we also tried lmfit with the data on 410 876 probes using limma package, a moderated t-statistic with the empirical Bayesian variance method and Benjamini–Hochberg correction to control the false-discovery rate (48). To look for differential methylation regions containing multiple differential methylated probes, we ran a Bumphunter package (23) to get significantly different clusters.
Functional annotation by DAVID
We explored the function annotation clustering and KEGG pathways using the online tool (DAVID Bioinformatics Resources 6.7, NIAID/NIH) with high classification stringency on three groups of genes following the published standard where clusters with enrichment scores >1.3 were considered significant (49). The first group of genes (387) were extracted from the 744 probes that have a P-value of ≤0.05, and a β-value difference (between CON18W and T2118W) of >10%; the second group of genes (2574) were extracted from the 3242 probes that have a P–value of ≤0.05, and are promoter-associated; the third group of genes (72) were extracted from the 100 probes that have a P-value of≤0.05, a β-value difference (between CON18W and T2118W) of >10%, and are promoter-associated. The fourth group of genes (2988) were extracted from the 4095 probes that have a P-value of ≤0.05, and are gene body-associated; the fifth group of genes (210) were extracted from the 263 probes that have a P-value of ≤0.05, a β-value difference (between CON18W and T2118W) of >10%, and are gene body-associated. The five groups of genes are within DAVID's gene database.
Bisulfite sequencing
500 ng of genome DNA was taken for bisulfite conversion using EpiTect Bisulfite Kit (Qiagen, 59104, USA). The candidate gene region for bisulfite sequencing was selected based on the location of Illumina array probes: the sequence around significant changed CpG sites and promoter region was used for primer designs. PCR primers for bisulfite sequencing was designed using the online software (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) (50). PCR was performed with EmeraldAmp® MAX PCR Master Mix (Clontech RR320B, USA) for 35 cycles (1 min denatures at 94°C, 2 min annealing at 55°C and 1 min extension at 72°C). The primer sequences and corresponding unconverted genomic sequences were listed in Supplementary Material, Table S2. The PCR fragments were cloned into pL253 plasmid (NCI-Frederick, Frederick, MD, USA) at KpnI and SalI restriction sites for the subsequent sequencing. At least 10 clones were randomly selected from each sample for sequencing, and the results were aligned for quantification.
Reverse transcriptase–polymerase chain reaction
The mRNA level was determined using the semi-quantitative RT-PCR method. The total mRNA was extracted from cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) as directed by the manufacturer. One microgram of RNA was used for each reaction using SuperScript® First-Strand Synthesis System for RT-PCR (Life Technologies) following the described protocol. The PCR primers were listed in Supplementary Material, Table S2.
Constructs, lentivirus production and infection
pHAGE-CMV-MCS-IZsGreenW-DNMT3L was constructed by inserting DNMT3L CDNA into NotI/NheI restriction sites of pHAGE-CMV-MCS-IZsGreenW. pSicoR-GFP-DNMT3LshRNA constructs were constructed by inserting DNMT3LshRNA sequence into XhoI/BamHI restriction sites of psicoR-GFP. The DNMT3LshRNA sequences are 5′-CGACGATGACGGGTACCAAGAGATCTCTTGGTACCCGTCATCGTCGTTTTT-3′(DNMT3LshRNA1) and 5′-CGGCCAAGTGGCCCACCAAGAGATCTCTTGGTGGGCCACTTGGCCGTTTTT-3′(DNMT3LshRNA2) (51). Production of lentiviruses was done in 293T cells as described by Richard Mulligan's lab (52). The multiplicity of infection of viruses was 1. Dissociated neural progenitor cells were infected with lentivirus carrying the target genes, and kept in the neural stem-cell medium (StemPro NSC SFM, Life Technologies) for 3 days, then the cells were processed for analyses for DNA methylation and mRNA expression, or differentiation for 7 days in the Neurobasal-A + 2% B27 medium (Life Technologies) without virus before the immunostaining.
Human neural progenitor cell and neuron cultures
Samples were obtained along the periventricular zone within the frontal cortex, minced and washed in cold Hank's-buffered saline solution and mechanically dissociated with pipettes. The sample was then strained through a 40 µm cell strainer (Falcon, San Jose, CA, USA). The dissociated cells were spun down, the media aspirated and cells were placed at low dilution (1 × 106 per 5 ml) in neurosphere medium (StemPro NSC SFM, Life Technologies,) for expansion. The neuronal culture was maintained in neuronal differentiation medium (NeuralbasalA + 2%B27, Invitrogen, Carlsbad, CA, USA) for 7 days. The differentiation of the neurons after 7 days were confirmed by immunostaining with neuron-specific antibodies (mouse anti-MAP2, 1:100, Sigma; mouse anti-PSD95, 1:100, Millipore; rabbit anti-Synapsin I, 1:100, Millipore; mouse anti-Synaptophysin, 1:100, Millipore).
Immunostaining
Frozen cortices from NICHD brain bank were cut into 14 µm thick sections. The sections or cells were fixed in 4% paraformaldehyde for 15 min, and then placed in blocking solution with phosphate-buffered saline (PBS) containing 3% goat serum, incubated overnight in the appropriate primary antibody (mouse anti-MAP2, 1: 100, Sigma; rabbit anti-DNMT3L, 1:100, Sigma; mouse anti-APP, 1:100, Millipore; mouse anti-PSD95, 1:100, Millipore) and processed through standard fluorescent secondary antibodies (Cy2, Cy3, Jackson Immunoresearch Laboratories, Westgrove, PA, USA). Specimens were examined using confocal fluorescence microscopy after mounting in appropriate media. Cells were mounted on slides and scanned by fluorescence microscope. The intensity of APP staining was measured by ImageJ and the PSD95 positive puncta were counted in 10 randomly chosen ZsGreen/GFP-expressed cells from each treatment, and puncta on cell body or 100 µm long-proximal processes were counted separately.
Western blot
Proteins were extracted from the cortical tissue according to the previously described methods (2). Briefly, cells were solubilized in lysis buffer, separated on a 7.5% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis gel and transferred onto polyvinylidene fluoride membrane. The membrane was probed with the appropriate antibody and detected by enhanced chemiluminescence. The exposed blots were then analyzed with Image J using integrated density (the product of Area and Mean Gray Value) as the measurement for immuno-stained bands. Primary antibodies used for western blot included mouse anti-VINCULIN (1:5000, Abcam, USA; rabbit anti-DNMT3L, 1:500, Sigma, USA).
Chromatin immunoprecipitation assay
X-ChIP assay followed the protocol according to the cross-linking chromatin immunoprecipitation X-ChIP protocol (Abcam). Briefly, 3–5 × 107subconfluent NIH 3T3 cells were cultured in 10% fetal bovine serum/Dulbecco's modified Eagle's medium and treated with 0.75% formaldehyde for 15 min. Cultures were washed with PBS in triplicate, harvested in CHIP buffer [50 mm HEPES (pH7.5) 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail] and sonicated for 15 cycles (30″ON/30″ OFF) with Bioruptor 300 sonicator. The sonicated cell lysates were centrifuged at 8000 g for 10 min, and the supernatants were diluted for 10 times with RIPA buffer and incubated at 4°C with normal IgG or anti-Dnmt3b antibodies and protein A/G beads overnight. The beads were washed once with low-salt buffer, high-salt buffer and LiCl wash buffer, respectively. The DNAs were eluted and PCR-amplified for 35 cycles with Tspyl5 primers (forward: 5-ATTATGGTTCTGGCTTTCAC and reverse: 5-ATTGATAGCAAGCGTGTC). The PCR products were examined on 2% agarose gel.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
Some of the human tissue (frontal cortex sections of newborn, UMB 390, UMBM1974M) was obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, USA. This work was supported by grants to V.S. from National Institute of Neurological Disorders and Stroke (1R01NS092062-01). Part of this work was carried out by the Genomics Shared Resource supported by Roswell Park Cancer Institute and National Cancer Institute (NCI) grant no. P30 CA016056.
Supplementary Material
References
- 1.Lu J., Delli-Bovi L.C., Hecht J., Folkerth R., Sheen V.L. (2011) Generation of neural stem cells from discarded human fetal cortical tissue. J. Vis. Exp., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esposito G., Imitola J., Lu J., De Filippis D., Scuderi C., Ganesh V.S., Folkerth R., Hecht J., Shin S., Iuvone T. et al. (2008) Genomic and functional profiling of human Down syndrome neural progenitors implicates S100B and aquaporin 4 in cell injury. Hum. Mol. Genet., 17, 440–457. [DOI] [PubMed] [Google Scholar]
- 3.Esposito G., Scuderi C., Lu J., Savani C., De Filippis D., Iuvone T., Steardo L. Jr, Sheen V., Steardo L. (2008) S100B induces tau protein hyperphosphorylation via Dickopff-1 up-regulation and disrupts the Wnt pathway in human neural stem cells. J. Cell. Mol. Med., 12, 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J., Esposito G., Scuderi C., Steardo L., Delli-Bovi L.C., Hecht J.L., Dickinson B.C., Chang C.J., Mori T., Sheen V. (2011) S100B and APP promote a gliocentric shift and impaired neurogenesis in down syndrome neural progenitors. PLoS One, 6, e22126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J., Lian G., Zhou H., Esposito G., Steardo L., Delli-Bovi L.C., Hecht J.L., Lu Q.R., Sheen V. (2012) OLIG2 over-expression impairs proliferation of human Down syndrome neural progenitors. Hum. Mol. Gen., 21, 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Gen., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 7.Carey N., Marques C.J., Reik W. (2011) DNA demethylases: a new epigenetic frontier in drug discovery. Drug Discov. Today, 16, 683–690. [DOI] [PubMed] [Google Scholar]
- 8.Gopalakrishnan S., Van Emburgh B.O., Robertson K.D. (2008) DNA methylation in development and human disease. Mutat. Res., 647, 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunner A.L., Johnson D.S., Kim S.W., Valouev A., Reddy T.E., Neff N.F., Anton E., Medina C., Nguyen L., Chiao E. et al. (2009) Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Gen. Res., 19, 1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen N.M., Dighe V., Landan G., Reynisdottir S., Palsson A., Mitalipov S., Tanay A. (2009) DNA methylation programming and reprogramming in primate embryonic stem cells. Gen. Res., 19, 2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straussman R., Nejman D., Roberts D., Steinfeld I., Blum B., Benvenisty N., Simon I., Yakhini Z., Cedar H. (2009) Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol., 16, 564–571. [DOI] [PubMed] [Google Scholar]
- 12.Kerkel K., Schupf N., Hatta K., Pang D., Salas M., Kratz A., Minden M., Murty V., Zigman W.B., Mayeux R.P. et al. (2010) Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet., 6, e1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones M.J., Farre P., McEwen L.M., Macisaac J.L., Watt K., Neumann S.M., Emberly E., Cynader M.S., Virji-Babul N., Kobor M.S. (2013) Distinct DNA methylation patterns of cognitive impairment and trisomy 21 in Down syndrome. BMC Med. Gen., 6, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin S., Lee Y.K., Lim Y.C., Zheng Z., Lin X.M., Ng D.P., Holbrook J.D., Law H.Y., Kwek K.Y., Yeo G.S. et al. (2013) Global DNA hypermethylation in down syndrome placenta. PLoS Genet., 9, e1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kangaspeska S., Stride B., Metivier R., Polycarpou-Schwarz M., Ibberson D., Carmouche R.P., Benes V., Gannon F., Reid G. (2008) Transient cyclical methylation of promoter DNA. Nature, 452, 112–115. [DOI] [PubMed] [Google Scholar]
- 16.Metivier R., Gallais R., Tiffoche C., Le Peron C., Jurkowska R.Z., Carmouche R.P., Ibberson D., Barath P., Demay F., Reid G. et al. (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature, 452, 45–50. [DOI] [PubMed] [Google Scholar]
- 17.Yuen R.K., Neumann S.M., Fok A.K., Penaherrera M.S., McFadden D.E., Robinson W.P., Kobor M.S. (2011) Extensive epigenetic reprogramming in human somatic tissues between fetus and adult. Epigenetics Chromatin, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt J.D., Silva A.J., Fan G. (2010) Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci., 13, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaPlant Q., Vialou V., Covington H.E. III, Dumitriu D., Feng J., Warren B.L., Maze I., Dietz D.M., Watts E.L., Iniguez S.D. et al. (2010) Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci., 13, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levenson J.M., Roth T.L., Lubin F.D., Miller C.A., Huang I.C., Desai P., Malone L.M., Sweatt J.D. (2006) Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem., 281, 15763–15773. [DOI] [PubMed] [Google Scholar]
- 21.Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E. (2010) Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science, 329, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Rohde C., Tierling S., Jurkowski T.P., Bock C., Santacruz D., Ragozin S., Reinhardt R., Groth M., Walter J. et al. (2009) DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet., 5, e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe A.E., Murakami P., Lee H., Leek J.T., Fallin M.D., Feinberg A.P., Irizarry R.A. (2012) Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidem., 41, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epping M.T., Meijer L.A., Krijgsman O., Bos J.L., Pandolfi P.P., Bernards R. (2011) TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat. Cell Biol., 13, 102–108. [DOI] [PubMed] [Google Scholar]
- 25.Bianchetta M.J., Lam T.T., Jones S.N., Morabito M.A. (2011) Cyclin-dependent kinase 5 regulates PSD-95 ubiquitination in neurons. J. Neurosci., 31, 12029–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozaki T., Li Y., Kikuchi H., Tomita T., Iwatsubo T., Nakagawara A. (2006) The intracellular domain of the amyloid precursor protein (AICD) enhances the p53-mediated apoptosis. Biochem. Biophys. Res. Commun., 351, 57–63. [DOI] [PubMed] [Google Scholar]
- 27.Moore L.D., Le T., Fan G. (2013) DNA methylation and its basic function. Neuropsychopharmacology, 38, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livnat-Levanon N., Glickman M.H. (2011) Ubiquitin-proteasome system and mitochondria - reciprocity. Biochim. Biophys. Acta, 1809, 80–87. [DOI] [PubMed] [Google Scholar]
- 29.Mabb A.M., Ehlers M.D. (2010) Ubiquitination in postsynaptic function and plasticity. Annu. Rev. Cell Dev. Biol., 26, 179–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattiace L.A., Kress Y., Davies P., Ksiezak-Reding H., Yen S.H., Dickson D.W. (1991) Ubiquitin-immunoreactive dystrophic neurites in Down's syndrome brains. J. Neuropathol. Exp. Neurol., 50, 547–559. [DOI] [PubMed] [Google Scholar]
- 31.Engidawork E., Juranville J.F., Fountoulakis M., Dierssen M., Lubec G. (2001) Selective upregulation of the ubiquitin-proteasome proteolytic pathway proteins, proteasome zeta chain and isopeptidase T in fetal Down syndrome. J. Neural. Transm. Suppl., 61, 117–130. [DOI] [PubMed] [Google Scholar]
- 32.Roselli F., Livrea P., Almeida O.F. (2011) CDK5 is essential for soluble amyloid beta-induced degradation of GKAP and remodeling of the synaptic actin cytoskeleton. PloS One, 6, e23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roselli F., Tirard M., Lu J., Hutzler P., Lamberti P., Livrea P., Morabito M., Almeida O.F. (2005) Soluble beta-amyloid1-40 induces NMDA-dependent degradation of postsynaptic density-95 at glutamatergic synapses. J. Neurosci., 25, 11061–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko M., Saito R., Okuma Y., Nomura Y. (2012) Possible involvement of ubiquitin ligase HRD1 insolubilization in amyloid beta generation. Biol. Pharm. Bull., 35, 269–272. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe T., Hikichi Y., Willuweit A., Shintani Y., Horiguchi T. (2012) FBL2 Regulates amyloid precursor protein (APP) metabolism by promoting ubiquitination-dependent APP degradation and inhibition of APP endocytosis. J. Neurosci., 32, 3352–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans T.A., Raina A.K., Delacourte A., Aprelikova O., Lee H.G., Zhu X., Perry G., Smith M.A. (2007) BRCA1 may modulate neuronal cell cycle re-entry in Alzheimer disease. Int. J. Med. Sci., 4, 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X., Fu S., Lai M., Baer R., Chen J. (2006) BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev., 20, 1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkhart S.E., Kao Y.T., Bartel B. (2014) Peroxisomal ubiquitin-protein ligases peroxin2 and peroxin10 have distinct but synergistic roles in matrix protein import and peroxin5 retrotranslocation in Arabidopsis. Plant Physiol., 166, 1329–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y.C., Williams B.R., Siegel J.J., Amon A. (2011) Identification of aneuploidy-selective antiproliferation compounds. Cell, 144, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chedin F., Lieber M.R., Hsieh C.L. (2002) The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA, 99, 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurkowska R.Z., Rajavelu A., Anspach N., Urbanke C., Jankevicius G., Ragozin S., Nellen W., Jeltsch A. (2011) Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J. Biol. Chem., 286, 24200–24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. (2004) DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem., 279, 27816–27823. [DOI] [PubMed] [Google Scholar]
- 43.Marin-Padilla M. (1976) Pyramidal cell abnormalities in the motor cortex of a child with Down's syndrome. A Golgi study . J. Comp. Neurol., 167, 63–81. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez F., Trinidad J.C., Blank M., Feng D.D., Burlingame A.L., Garner C.C. (2009) Normal protein composition of synapses in Ts65Dn mice: a mouse model of Down syndrome. J. Neurochem., 110, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomazeau A., Lassalle O., Iafrati J., Souchet B., Guedj F., Janel N., Chavis P., Delabar J., Manzoni O.J. (2014) Prefrontal deficits in a murine model overexpressing the Down syndrome candidate gene dyrk1a. J. Neurosci., 34, 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y.A., Choufani S., Ferreira J.C., Grafodatskaya D., Butcher D.T., Weksberg R. (2011) Sequence overlap between autosomal and sex-linked probes on the Illumina HumanMethylation27 microarray. Genomics, 97, 214–222. [DOI] [PubMed] [Google Scholar]
- 47.Price M.E., Cotton A.M., Lam L.L., Farre P., Emberly E., Brown C.J., Robinson W.P., Kobor M.S. (2013) Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin, 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth G.K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl.. Gen. Mol. Biol., 3, Article3. [DOI] [PubMed] [Google Scholar]
- 49.Huangda W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 50.Li L.C., Dahiya R. (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics, 18, 1427–1431. [DOI] [PubMed] [Google Scholar]
- 51.Minami K., Chano T., Kawakami T., Ushida H., Kushima R., Okabe H., Okada Y., Okamoto K. (2010) DNMT3L is a novel marker and is essential for the growth of human embryonal carcinoma. Clin. Cancer Res, 16, 2751–2759. [DOI] [PubMed] [Google Scholar]
- 52.Mostoslavsky G., Fabian A.J., Rooney S., Alt F.W., Mulligan R.C. (2006) Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl Acad. Sci. USA, 103, 16406–16411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.